
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 07 November 2023
Sec. Crop and Product Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1279823
 Lauren E. Hibbert1,2
Lauren E. Hibbert1,2 Yufei Qian1
Yufei Qian1 Hazel K. Smith3
Hazel K. Smith3 Suzanne Milner3
Suzanne Milner3 Ella Katz1
Ella Katz1 Daniel J. Kliebenstein1
Daniel J. Kliebenstein1 Gail Taylor1*
Gail Taylor1*Watercress (Nasturtium officinale) is a nutrient-dense salad crop with high antioxidant capacity and glucosinolate concentration and with the potential to contribute to nutrient security as a locally grown outdoor aquatic crop in northern temperate climates. However, phosphate-based fertilizers used to support plant growth contribute to the eutrophication of aquatic habitats, often pristine chalk streams, downstream of farms, increasing pressure to minimize fertilizer use and develop a more phosphorus-use efficient (PUE) crop. Here, we grew genetically distinct watercress lines selected from a bi-parental mapping population on a commercial watercress farm either without additional phosphorus (P−) or under a commercial phosphate-based fertilizer regime (P+), to decipher effects on morphology, nutritional profile, and the transcriptome. Watercress plants sustained shoot yield in P− conditions, through enhanced root biomass, but with shorter stems and smaller leaves. Glucosinolate concentration was not affected by P− conditions, but both antioxidant capacity and the concentration of sugars and starch in shoot tissue were enhanced. We identified two watercress breeding lines, with contrasting strategies for enhanced PUE: line 60, with highly plastic root systems and increased root growth in P−, and line 102, maintaining high yield irrespective of P supply, but less plastic. RNA-seq analysis revealed a suite of genes involved in cell membrane remodeling, root development, suberization, and phosphate transport as potential future breeding targets for enhanced PUE. We identified watercress gene targets for enhanced PUE for future biotechnological and breeding approaches enabling less fertilizer inputs and reduced environmental damage from watercress cultivation.
Watercress (Nasturtium officinale R. Br.) is a perennial leafy green crop adapted to growth in aquatic environments. Found in nature in free-flowing alkaline streams and as a crop, it is best grown in hydroponic systems, including open water ponds, hydroponic greenhouses, and vertical farms (Cox, 2009; Schuchardt et al., 2019; Qian et al., 2022). Watercress is a member of the Brassicaceae family, alongside other important food crops such as broccoli (Brassica oleracea var. italica) and oilseed rape (B. napus) (Kiefer et al., 2019). A distinctive characteristic of watercress is its peppery flavor, derived from the hydrolysis of gluconasturtiin, the primary glucosinolate (GSL) in watercress, to phenethyl isothiocyanate (PEITC) (Boyd et al., 2006; Payne, 2011; Jeon et al., 2017). Although the primary function of glucosinolates is for defense against herbivory, it is the isothiocyanates that are responsible for the anti-cancer, antibiotic, and cardioprotective properties of watercress (Newman et al., 1992; Cheung and Kong, 2009; Panahi Kokhdan et al., 2021). Watercress also has high antioxidant (AO) capacity and qualifies as the most nutrient-dense fruit/vegetable, based on the content of 17 nutrients of public health importance (Di Noia, 2014).
Watercress is grown worldwide, including the UK, USA, Spain, Portugal, New Zealand, and China (Li et al., 2007b; Searle, 2019; USDA, 2019). In the UK, 58 hectares of watercress were grown in 2018, representing a total value of £15 million (DEFRA, 2020). It is also a high-value horticultural crop, with a UK market value of £8.90 per kg compared to £4.97 per kg for mixed baby leaf salad bags (DEFRA, 2019). However, there is concern that watercress production is causing environmental pollution, through the direct addition of phosphate-based fertilizers into aquatic systems, including chalk streams in the UK, which are of high conservation value (Cox, 2009; Hibbert and Taylor, 2022). Chalk streams are internationally rare and highly biodiverse environments often referred to as “England’s rainforests”, providing a habitat for species such as the winterbourne stonefly (Nemoura lacustris) and brown trout (Salmo trutta), which depend on its clean low nutrient waters—but they are under threat (White, 2020; CaBA CSRG, 2021; Environment Agency and Natural England, 2021). In these and other aquatic systems, phosphate pollution results in eutrophication of water systems, where excessive flora growth limits light penetration, leading to death of organisms below, deoxygenation of the habitat via microbial decomposition, and ultimately the disruption of community dynamics (Schindler et al., 2008). Naturally, phosphate concentrations in chalk streams are approximately 0.02 mg L−1; however, inputs of phosphorus (P) rapidly increase these concentrations above P targets downstream of watercress farms (Casey and Smith, 1994). Additional P inputs into freshwater systems is predominantly through release from sewage treatment works (STWs), leaking septic tanks, and from excess fertilizer application (Withers et al., 2013; Richards et al., 2016; CaBA CSRG, 2021). However, watercress farms were also shown to contribute 5.4% of the P load in chalk streams and values of up to 62% were reported for some streams, suggesting significant room for improvement (Cox, 2009). Approximately 90% of watercress farms in the UK are on, or upstream of, a Site of Special Scientific Interest (SSSI), increasing the pressure to minimize Prelease. Of the 249 chalk streams in the UK, 39% fail the standards for good ecological P status set out in the Water Framework Directive (European Commission, 2019). Jarvie et al. (2018) also surveyed the nutrient status of British headwater streams and declared that reducing P concentrations in lowland-high-alkalinity rivers (chalk streams) is one of the most important areas to target for improved UK water quality.
P is vital for plant growth, and is used to power cells through the release of phosphate from ATP, for the structure of protein and carbohydrate polymers, cell membranes, and the formation of the phosphodiester bonds that link nucleotides (Westheimer, 1987; Schachtman et al., 1998). Nevertheless, the environmental damage associated with phosphate fertilizer application and the finite nature of P reserves is driving the development of crops with improved P-use efficiency (PUE). For watercress, with commercial production linked to chalk streams, improved PUE is a key target for crop improvement (Hibbert and Taylor, 2022). Our recent review identified the key traits to breed for a PUE ideotype in watercress as (i) increased root surface area through prolific root branching, adventitious root (free floating roots deriving from the stem) formation, and root hair growth, and (ii) increased root aerenchyma formation. Functional genomic traits for improved PUE are (iii) efficacious premobilization and scavenging strategies and (iv) the use of alternative metabolic pathways (Hibbert and Taylor, 2022). Identifying gene-based targets central to the PUE response in watercress is vital to increase the speed of breeding for PUE. Key genomic targets in other species (predominantly based on studies in the soil-grown model plant, Arabidopsis thaliana) have previously been identified as PHT phosphate transporter genes, global transcriptional regulators such as those of the SPX family, and genes involved in galactolipid and sulfolipid biosynthesis such as MGD2/3, PECP1, PSR2, PLDζ1/2, and SQD2 (Hibbert and Taylor, 2022). Although matches for these genes have been found in watercress transcriptome data, the functional significance of these gene targets, particularly in P-deficient growing environments, have not yet been investigated. Other breeding targets for consumers include further improved nutritional quality (AO capacity and GSL concentration) and enhanced sweetness while maintaining yield, and it is therefore important to understand the trade-offs between fertilizer management and nutritional profile, yield, and crop flavor.
There is limited understanding of the effects of nutrient availability on watercress. One study utilized an experimental stream to assess differences in watercress growth rate under varying N:P application, with a focus on N accumulation (Fernandez-Going et al., 2013). As expected, growth rates increased with increasing nutrient availability. Previously, microarray and RNA-sequencing (RNA-seq) approaches have been used to explore differences in AO capacity and GSL concentration, and to study molecular mechanisms underlying contrasting growth responses to submergence in watercress (Payne, 2011; Voutsina et al., 2016; Jeon et al., 2017; Müller et al., 2021). Authors used orthology to A. thaliana to identify key candidate genes involved in GSL/AO biosynthesis pathways and hormone signaling mediating growth responses.
There is an urgent need, therefore, to identify gene targets to enable future breeding for PUE in this nutrient dense leafy green crop. There is currently no relevant breeding for PUE and the literature surrounding the effects of fertilizer on watercress growth is limited and outdated (Austin, 1966; Howard-Williams et al., 1982; Bennett, 1986; Fernandez-Going et al., 2013). This study aims to investigate the genomic basis of PUE in watercress, through a study of the effects of contrasting applications of phosphate-based fertilizer on the growth, biochemistry, and gene expression of selected watercress lines from a bi-parental mapping population using RNA-seq approaches.
An F2 watercress mapping population was previously developed by crossing two lines contrasting for size and nutritional content: WX033 and WX038 (also referred to as Parent A and Parent B, respectively, in Voutsina, 2017). WX033 is the commercialized dwarf leafy “Boldrewood” cultivar with high AO and GSL concentration. WX038 is an accession with a longer stem and lower AO and GSL than WX033 in both field and controlled conditions (Payne, 2011). F2 offspring were self-fertilized to obtain the F3 (F2:3) generation, then multiple plants were grown and seed harvested in bulk (F2:4) (Qian, 2021). This enabled greater seed production for use in this field trial, with a similar homozygosity. Nine watercress lines (referred to as 120, 102, 39, 82, 60, 225, 5, 16, and 173) from this watercress mapping population, the two parent lines (WX033 and WX038), and two commercial control lines (WX001 and WXVITA) were selected for this field study. These lines were selected based on high GSL concentration, AO capacity, vigor, and desirable morphological traits for commercial cultivation observed in previous trials (Qian, 2021; Qian et al., 2023).
F2:4 seeds were sown in peat-filled trays. Tray positioning was randomized in the greenhouse (Vitacress Herbs; Chichester, UK) and irrigated with potable water from an overhead sprinkler four times a day. After 3 weeks, plants were thinned to equal density and transplanted into prepared gravel lined beds utilizing a complete randomized block design at a commercial watercress farm (51°11’42.9”N, 1°32’12.9”W; Hampshire, UK). Blocks were composed of 16 0.25 m2 plots comprising 35 plants per plot. Trial areas were located at the heads of neighboring gravel-lined beds, with a shared flowing spring water supply at an almost constant temperature of 12°C (Figure 1). Beds were covered by an open-ended polytunnel to reduce bird damage and buffer from environmental variation. One bed was given no additional fertilizer (P−) during the trial period, and the other was supplied with a standard commercial fertilizer regime (P+) as follows: one dose of base dressing (Humber Palmer Eco-Cress Base; details of fertilizer in Supplementary 1) on day 16, then one dose of top dressing (Humber Palmer Eco-Cress Plus) on day 23 post-transplanting. The application rates were 200 kg ha−1 of base dressing (30.4 kg ha−1 P2O5) and 100 kg ha−1 of top dressing (12 kg ha−1 P2O5).
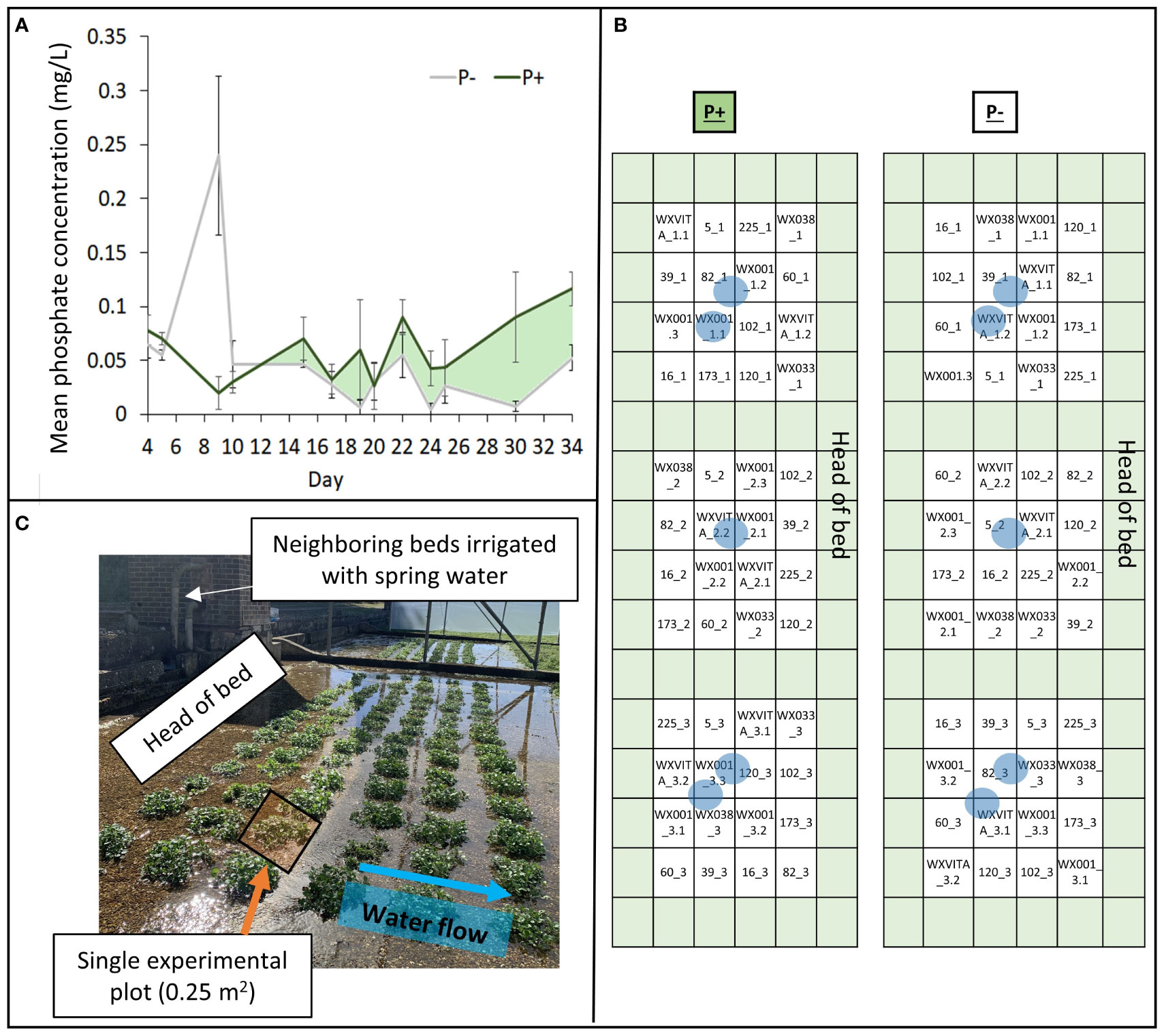
Figure 1 Elements of the field design. (A) Phosphate concentrations within treatments (P+/P−). P+ received doses of fertilizer on days 16 and 23, P− was untreated. Areas shaded light green represent periods where concentration was higher in the P+ treatment. (B) Field plan showing 13 different lines within a randomized complete block design with 3 blocks. Green represents plots of guard plants and blue circles indicate P determination sampling sites (both within and between plots). (C) Images of the field to illustrate plants grouped within 0.25 m2 plots. Neighboring beds were irrigated using the same spring water pump and the direction of water flow across the bed is indicated by the blue arrow.
To monitor P bioavailability during the trial, phosphate in the irrigation supply was quantified using a low range handheld phosphate photometer using an adaption of the ascorbic acid/molybdenum blue method (HI-96713; HANNA instruments). Samples were taken at least every 5 days within the bed (Figure 1A), prior to harvesting plants, 24 h before, and 24 h after each fertilizer application to increase data granularity. Eight additional water samples were collected at the final harvest point and were assessed for ammoniacal nitrogen, nitrate (NO3), P (total unfiltered), and orthophosphate (PO4) concentration by ALS (alsglobal.com) to support handheld phosphate measurements. Though P concentrations fluctuated throughout the trial period in this commercial watercress bed (Figure 1A), overall P concentration was higher in the fertilized bed for the majority of measurements. Additional analyses at the final harvest point showed that nitrogen concentration in both beds did not significantly differ (P+ 34.48 mg/L ± 0.15; P− 34.03 mg/L ± 0.54), increasing evidence that P is the limiting macronutrient at the point of harvest.
After 35 days following transplanting, four plants were sampled per plot for morphological analyses. The following traits were recorded: shoot/root fresh and dry weight, stem length (identified as length of the main stem), and number of leaves. Root:shoot (R:S) ratio was calculated from fresh weight values. Images of dissected plants were used to quantify leaf area parameters (mean and total leaf area) on ImageJ software (Schneider et al., 2012). One plant was also selected from each plot every 5 days to assess morphological changes over time. Watercress forms a densely matted root structure under commercial growing conditions; thus, quantifying aspects of root architecture was not possible in this study.
After 35 days, one plant was selected from each plot, roots were cleaned and cut from the shoot, and both tissue portions were frozen separately in liquid nitrogen. Frozen tissue was ground to a fine homogeneous powder and stored at −80°C prior to further analyses.
The AO capacity of each sample was assessed using the Ferric Reducing Ability of Plasma (FRAP) protocol as described previously (Benzie and Strain, 1996; Payne et al., 2013; Qian et al., 2023). Ground frozen tissue was transferred to QIAshredder homogenizer tubes (Qiagen), weighed, and spun at 20,000 rpm for 5 min. Extracted sap was transferred to 96-well plates alongside a serial dilution of iron sulfate heptahydrate. FRAP reagent solution, containing acetate buffer, TPTZ (2,4,6-Tri(2-pyridyl)-s-triazine), and iron chloride hexahydrate, was added to the plate and immediately read on a spectrophotometer (CLARIOstar Plus; BMG Labtech) at 620 nm. Plates were run in duplicate.
GSLs were quantified by HPLC-DAD as described by Kliebenstein et al. (2001) and used previously for watercress (Qian et al., 2023). Briefly, 20–40 mg of ground frozen tissue was weighed and homogenized in a paint shaker with 90% methanol for 3 min and centrifuged. Ninety-six-well filter plates were loaded with DEAE Sephadex A-25 and the plant supernatants, then washed with water, 90% methanol, and water again. Following an overnight incubation with sulfatase, the Sephadex-bound GSLs were eluted. Desulfoglucosinolates were separated and detected by HPLC-DAD and quantified by comparison to standard curves of purified compounds and results were normalized to fresh weight.
Soluble (sugar) and insoluble (starch) carbohydrates were determined using a modified anthrone method (Leyva et al., 2008; Becerra-Sanchez and Taylor, 2021). One milliliter of buffer (sodium acetate 0.2 mol/L, pH 5.5) was added to 5–50 mg of pre-weighed lyophilized tissue and incubated at 70°C for 15 min. Samples were centrifuged for 10 min at 15,000 rpm, then 50 µL of supernatant was transferred to fresh tubes containing 1 mL of ultrapure water for sugar quantification. The remaining pellets were vortexed and incubated at 100°C for 10 min. Enzymatic digestion was conducted by adding 100 µL of 70 units/mL amyglucosidase and 100 µL of 7 units/mL alpha amylase and incubating pellets for 2 h at 37°C. Tubes were centrifuged at 15,000 rpm for 10 min and 50 µL of supernatant diluted in 1 mL of ultrapure water for starch determination. Samples were plated into 96-well plates in duplicate, alongside glucose calibration curves. A super-standard of pooled watercress shoot samples was also run alongside each batch of samples to standardize runs. Anthrone in sulfuric acid (150 µL; 0.1% w/v) was added per well and incubated for 20 min at 100°C. Absorbance at 620 nm was analyzed on a Multiskan™ FC Microplate Photometer (Thermo Scientific).
Lyophilized root and shoot tissue was also used for quantification of P and potassium (K) by ICP-MS. Sample digestion and ICP-MS analysis was conducted by the Interdisciplinary Center for Plasma Mass Spectrometry at the University of California Davis using an Agilent 8900 ICP-MS (Agilent Technologies, Palo Alto, CA). Samples, duplicate method blacks (50 µL of 18.2 MΩ/cm water), duplicate digestion quality control standards (50 µL), and duplicate standards from tomato (NIST1573a) and spinach (NIST1570a) were digested. Acid digestion involved adding 0.75 mL of 50% HNO3 to samples in two increments, each time allowing gas to evolve and then heating for 35 min and 1 h at 95°C, respectively. After cooling, 50 µL of H2O2 was added incrementally up to 500 µL as samples were heated, then heated for 1 h after the final addition. Finally, samples were allowed to cool and brought to a final volume of 1 mL with 18.2 MΩ/cm water, ready for analysis. ICP-MS analysis was conducted by the Interdisciplinary Center for Plasma Mass Spectrometry at the University of California Davis using an Agilent 8900 ICP-MS (Agilent Technologies, Palo Alto, CA).
RNA was extracted from frozen ground root and shoot tissue taken at the final harvest point (35 days post-transplanting) using a modified cetyltrimethylammonium bromide (CTAB) protocol used previously for watercress (Doyle and Doyle, 1987; Payne et al., 2015; Voutsina, 2017). Tissue (200–300 mg) was weighed and incubated with 900 µL of pre-warmed CTAB (+50 µL of 2-mercaptoethanol) at 65°C for 5 min. CHISAM (Chloroform : Isoamyl alcohol 24:1; 800 µL) was added and tubes were spun at 12,000 rpm for 10 min at room temperature. The aqueous phase was transferred to a fresh tube and the CHISAM step was repeated. A total of 180 µL of 10 M LiCl was then added to the aqueous phase before precipitation at 4°C overnight. Samples were spun at 4°C, then the supernatant was discarded, and the pellet was dissolved in 700 µL of pre-warmed SSTE. Tubes were incubated at 60°C for 5 min followed by a repeat of the CHISAM step. The aqueous phase was transferred to a fresh tube and 700 µL of 100% cold ethanol was added. This was left to precipitate at −20°C for 10 min before centrifugation and removal of supernatant. The remaining pellet was washed with cold 70% ethanol, left to air dry (>45 min) and redissolved in 50 µL of RNase-free H2O.
RNA-seq data for both roots and shoots were processed using a pipeline adapted from that used previously for watercress (Qian, 2021). Novogene Corporation Inc. (Sacramento) provided the eukaryotic RNA-seq service including cDNA library preparation (250–300 bp insert) and sequencing using the Illumina HiSeq (paired-end 150 bp) platform. Raw data processing was conducted using the UC Davis Farm cluster (https://www.hpc.ucdavis.edu/farm-cluster) followed by differential gene expression analysis in R Studio. To check FASTQ file quality, FastQC and MultiQC were conducted. The Trimmomatic preprocessing tool designed to handle paired-end Illumina sequence data was used to trim and remove poor-quality reads (Bolger et al., 2014). The first whole genome sequencing and assembly of the watercress genome has been completed by IGATech (https://igatechnology.com) and provided to the laboratory of Prof. Gail Taylor. Functional genome annotation was done using Interproscan 5.0. and by searching the scanned protein sequence against the UniProt database (https://www.uniprot.org). The genome file was indexed to improve the efficiency of searching the genome using the Hierarchical Graph FM index (HGFM) with the alignment program HISAT2. HISAT2 was also used to align reads to the watercress genome. Then, StringTie was used to assemble read alignments into potential transcripts, and the output was used to generate a count table with featureCounts (Liao et al., 2014; Pertea et al., 2016).
Differential gene expression analysis was conducted in R using the edgeR package using a GLM approach (Robinson et al., 2010). Raw counts were filtered to include only genes with 1 count per million (cpm) in at least two samples and library sizes were normalized. Quasi-likelihood F-tests were performed to provide more robust and reliable error rate control for smaller replicate numbers. Significance testing was conducted using the Benjamini–Hochberg method and differentially expressed genes (DEGs) were selected with a cutoff at FDR < 0.05 (Benjamini and Hochberg, 1995). Finally, gene ontology (GO) enrichment analysis was conducted on DEG lists using ShinyGO with A. thaliana as a reference and Venn diagrams were generated using the online tool VENNY v2.1 (Oliveros, 2015; Ge et al., 2020).
SPSS (version 27, IBM Corp, 2020) and R software (R Core Team, 2021) were used for statistical analyses. Results were averaged per plot to get the genotypic mean per block (n = 3). Normality of residuals was assessed using Q–Q plots and Shapiro–Wilk tests, and homogeneity of variances was checked using Levene’s test of equal variances. Linear mixed effect models were generated with block as a random factor and line and treatment as fixed main effects. Analysis of variance (ANOVA) tests were run on these models to identify differences between treatment and line with a significance threshold of p < 0.05.
Growing watercress in P− conditions impacted several morphological parameters at final harvest (day 35; Table 1; Figure 2; Supplementary 2). The removal of fertilizer application resulted in a 20% increase in root fresh weight (F1,49 = 7.942, p = 0.007), a 46% increase in root dry weight (F1,49 = 12.040, p = 0.001), and a 24% increase in shoot dry weight (F1,49 = 8.819, p = 0.005). However, shoot fresh weight was not affected by treatment (shoot: F1,49 = 0.055, p = 0.815; total: F1,49 = 0.260, p = 0.612). Stem length decreased by approximately 22% in P− (F1,49 = 36.803, p < 0.001). Together, this is reflected in a ~24% increase in root:shoot ratio in P− (F1,48 = 28.340, p < 0.001). Although, the number of leaves (F1,49 < 0.001, p = 0.981) did not change between treatments, mean leaf area decreased by 12% (F1,49 = 5.864, p = 0.019).
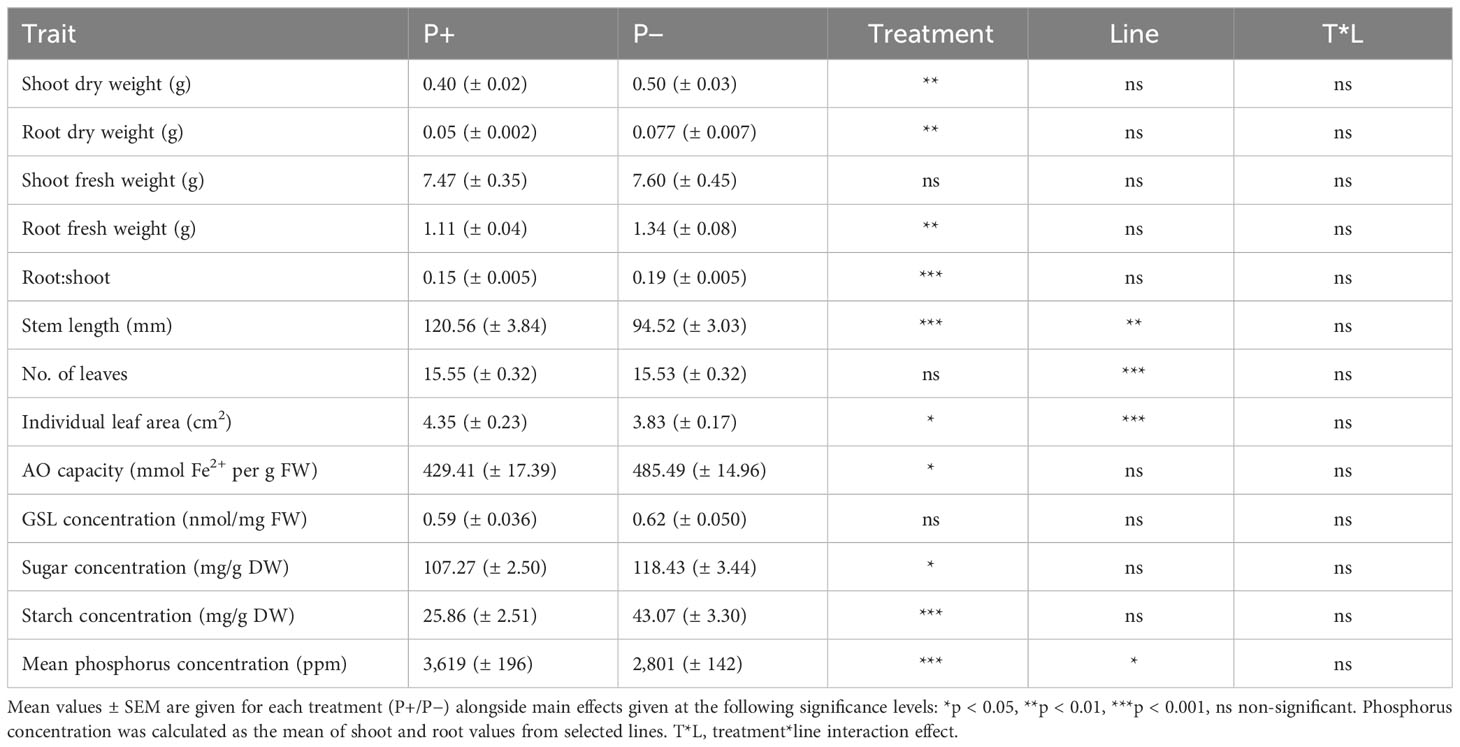
Table 1 Effect of phosphate-based fertilizer application on morphology and biochemistry of watercress at the final harvest point.
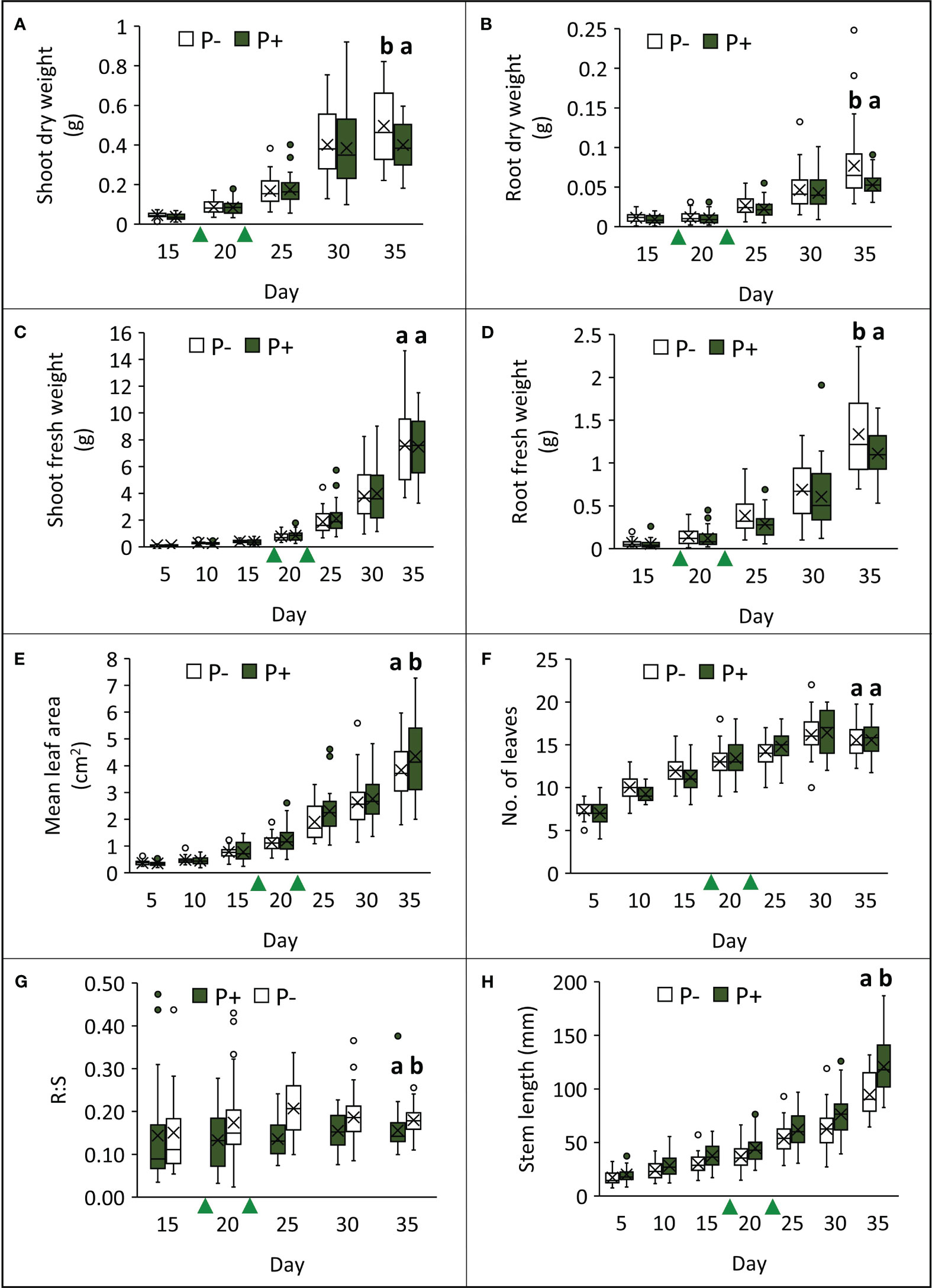
Figure 2 Effects of phosphate-based fertilizer application (P+) on different aspects of watercress morphology (A–H), compared to plants grown without additional phosphate-based fertilizer (P−) over the course of the field trial. Green triangles indicate when P treatments were applied. Crosses within bars denote mean values (n = 39) and letters above bars represent different groups according to Tukey’s LSD tests conducted on data from the final harvest point (p < 0.05).
Plants grown without additional phosphate-based fertilizer exhibited changes to their biochemical profile at harvest point (Table 1; Figure 3). Shoot AO capacity increased by 13% (F1,49 = 6.090, p = 0.017). When considering the concentration of the primary GSL in watercress, PE-GSL (phenylethyl glucosinolate), there was no effect of fertilizer treatment (F1,49 = 0.363, p = 0.550). The concentration of soluble sugars increased by 10% under P− (F1,49 = 6.957, p = 0.011) and starch increased by 67% (F1,49 = 23.835, p < 0.001). The concentration of P and K was assessed in selected lines (WXVITA, 102, 60; line selection is described in the subsequent section). As expected, P concentration of both roots and shoots increased in the P+ treatment (F1,17 = 17.586, p < 0.001): root P concentration increased from 3,274 ppm ( ± 352) to 4,333 ppm ( ± 128), and shoot P concentration increased from 2,575 ppm ( ± 160) to 3,586 ppm ( ± 366), whereas K concentration was unaffected by treatment (F1,17 = 1.19, p = 0.34), providing further evidence that P is the limiting macronutrient in this trial.
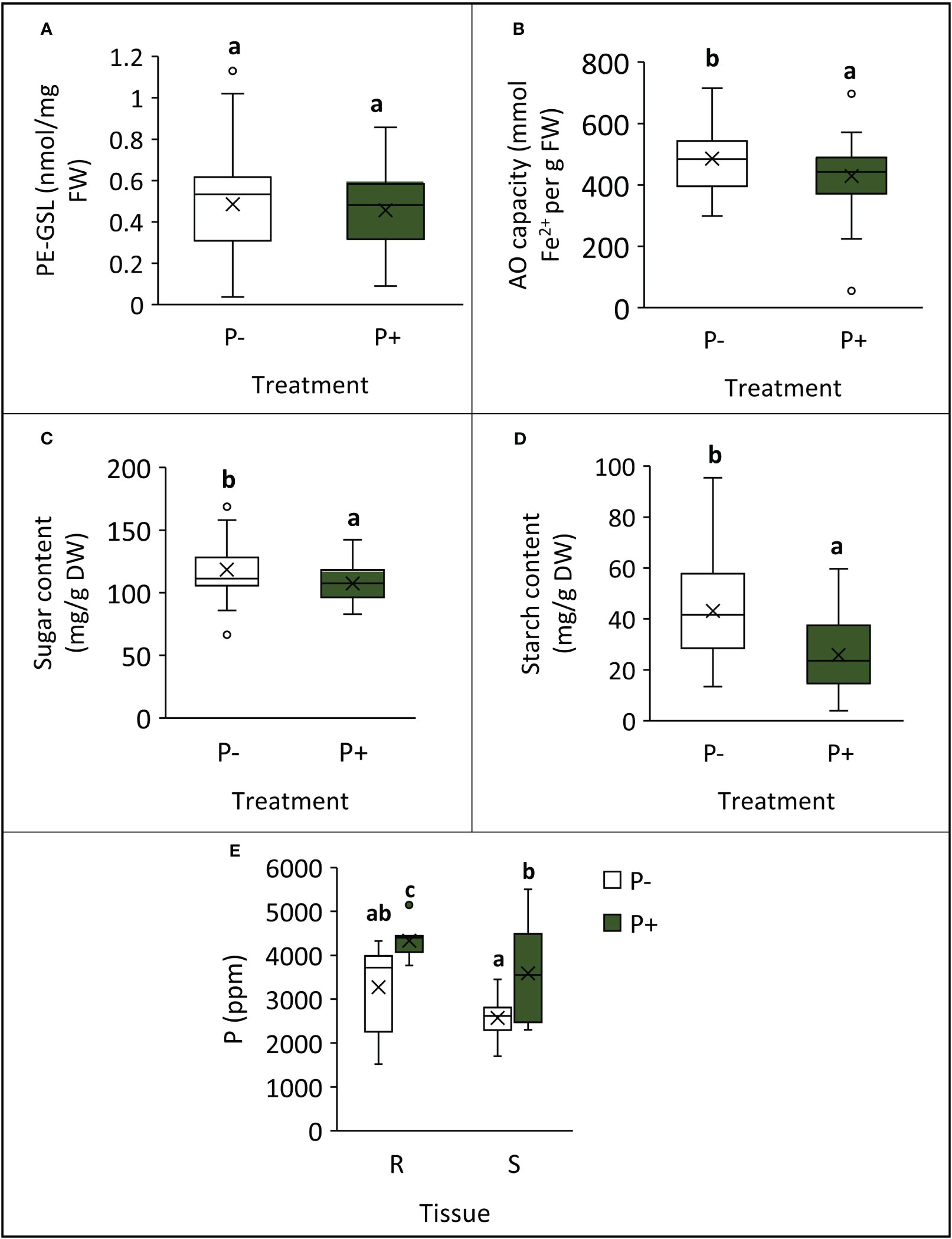
Figure 3 Differences in biochemical traits at final harvest point, following cultivation with (P+) or without (P−) phosphate fertilizer. (A) PE-glucosinolate (PE-GSL) concentration of shoots; (B) antioxidant capacity of shoots; (C) shoot sugar concentration; (D) shoot starch concentration; (E) phosphorus concentration in roots (R) and shoots (S) from selected lines. Means are denoted by crosses within boxes (A–D: n = 39; E: n = 9) and letters represent significantly different groups by Tukey’s LSD tests (p < 0.05).
For each of the 13 lines studied, the following morphological and biochemical traits were quantified: shoot dry weight, root dry weight, shoot fresh weight, R:S, stem length, no. of leaves, individual leaf area, AO capacity, GSL concentration, sugar concentration, and starch concentration. Of these traits, lines varied in stem length (F12,49 = 3.100, p = 0.003), no. of leaves (F12,49 = 4.101, p < 0.001), and mean leaf area (F12,49 = 3.522, p < 0.001; Figure 4). Statistics and figures showing line variation for all other traits are provided in Supplementary 3. To quantify responsiveness to low P conditions, percentage changes under P− conditions were calculated for traits. Line 60 showed a 35% change across all morphology traits under P−, including a 114% increase in mean root fresh weight, suggesting that it is highly responsive to low nutrient conditions. This line is also of note as it ranked highest for root dry weight and had the third highest AO capacity. For other traits such as leaf area and shoot fresh weight, it consistently ranked in the top half. Line 102 is also of interest as it ranked highest for several commercially relevant yield traits such as shoot fresh weight, second highest for no. of leaves, and third highest for PE-GSL concentration. By contrast, this line was far less responsive to P− conditions: there was a 15.5% change across all morphology traits and only a 0.9% increase and 6.6% decrease in shoot dry and fresh weight, respectively. These lines were taken forward for further analysis of P concentration and for RNA-seq analysis. P concentration also differed between these lines (F2,17 = 4.446, p = 0.028; Figure 5). Comparing across treatments, shoot P concentration was 42% higher in 102 compared to the commercial control line WXVITA and line 60 had 58% increase relative to WXVITA (t6 = 3.878, p = 0.019).
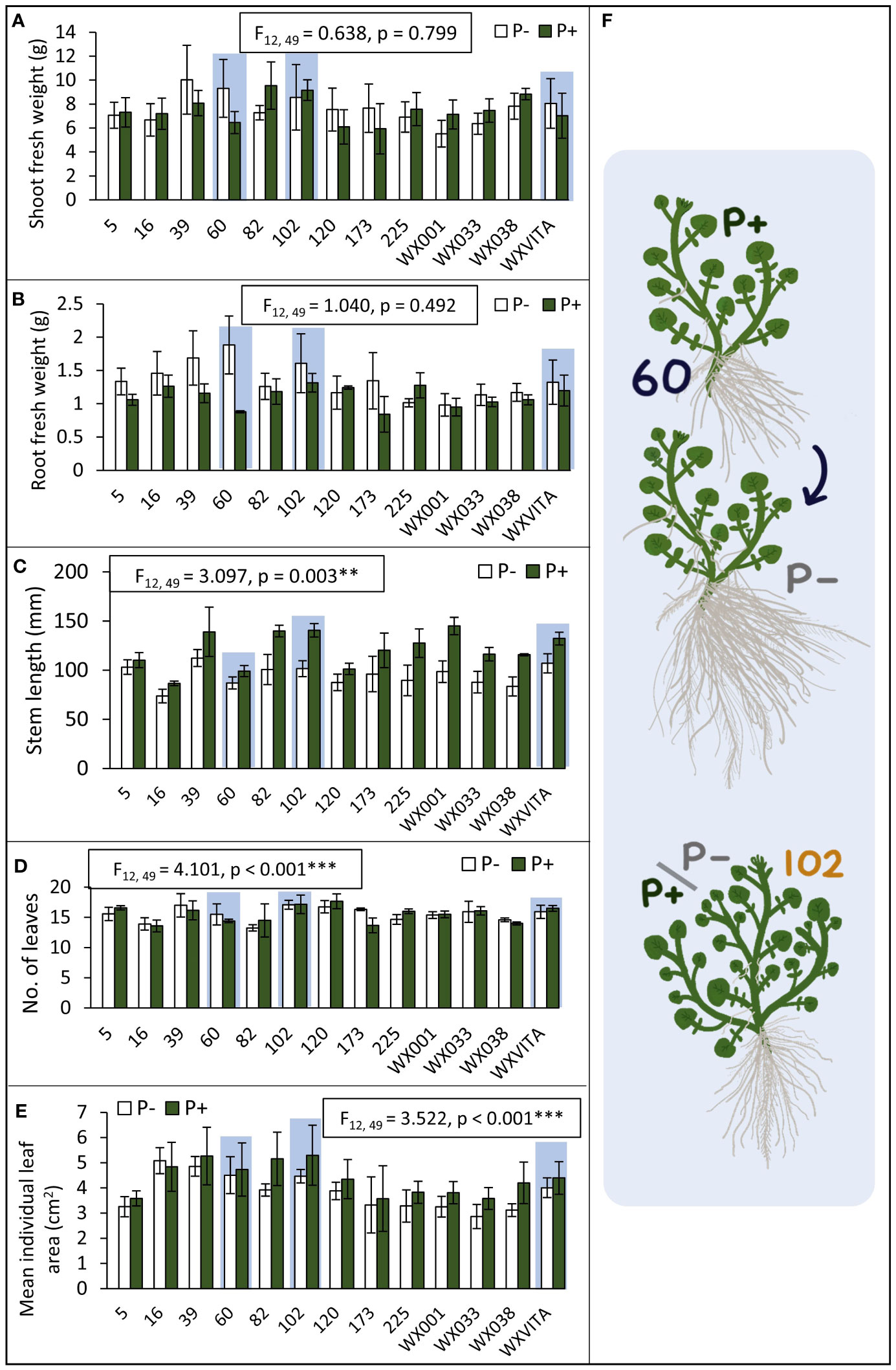
Figure 4 Genetic variation between lines to inform line selection (A–E). Lines that were selected for RNA-seq (60, 102, and WXVITA) are highlighted in light blue. Error bars represent SEM. Statistics for the effect of line on each trait (from ANOVA tests) are given in text boxes. Line effects are significant at the following levels: **p < 0.01, ***p < 0.001. (F) Illustrations of representative plants from selected lines. Line 60 is P-responsive with a large increase in root biomass in P−, whereas 102 maintains high yield under both P conditions.
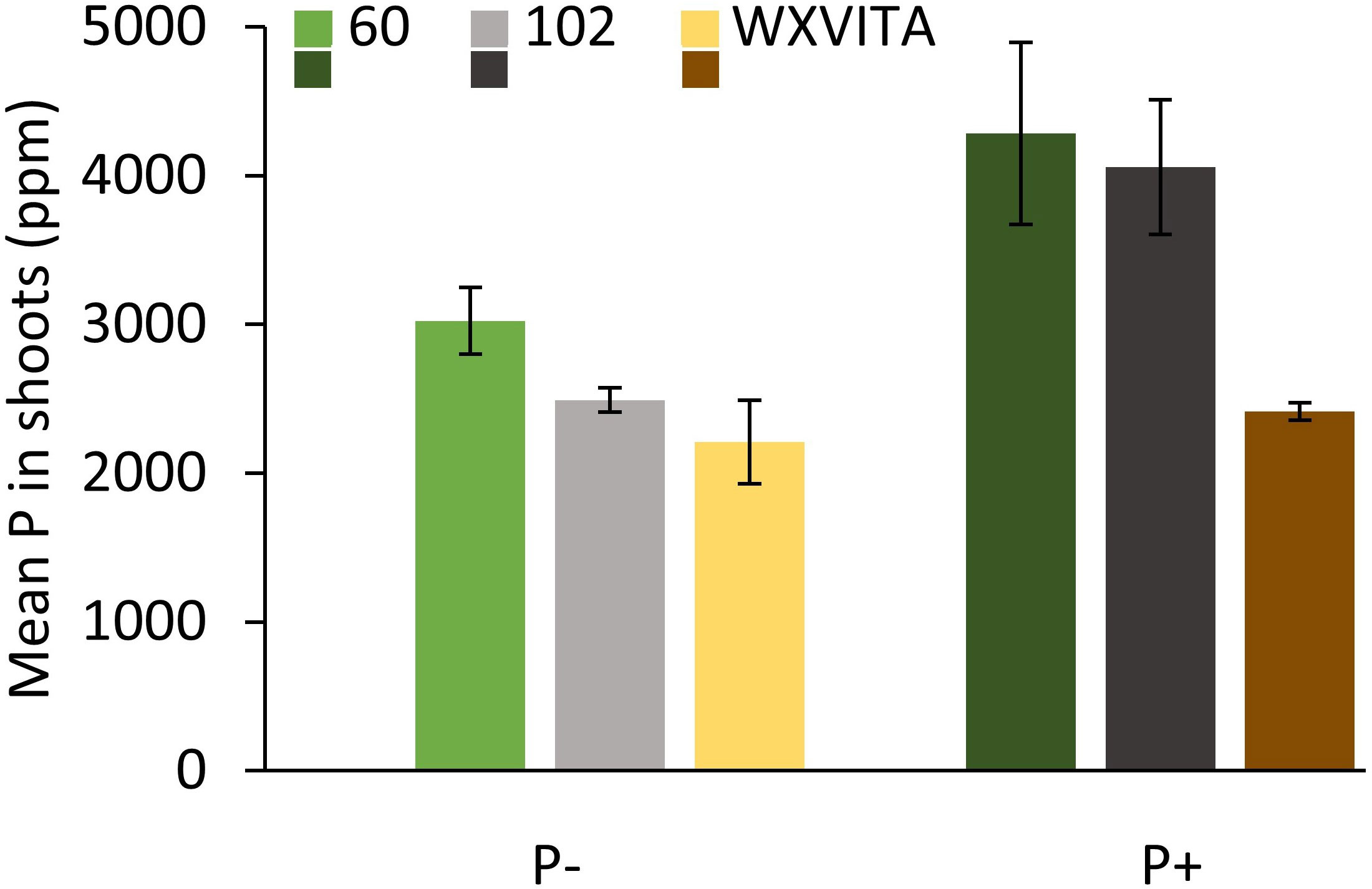
Figure 5 Total phosphorus (P) concentration in dried shoot tissue of selected watercress lines (60, 102, and WXVITA) at final harvest. Error bars are representative of SEM (n = 3).
Between 29 and 43 million reads per library were produced from RNA-seq with all libraries having 70%–91% of reads mapping uniquely to the watercress genome. A total of 44,024 transcripts were identified, and of these, 27,149 were represented at least 1 cpm in at least two shoot samples, and 27,531 were represented at least 1 cpm in at least two root samples. Eighty-eight percent of shoot and root transcripts corresponded to annotated watercress genes. No DEGs were detected in P− shoots compared to P+ shoots (FDR < 0.05). However, 16 genes had an FDR < 0.15 with corresponding p-values < 1e-5 (Supplementary 4). This list included a 0.55- and 1.7-fold upregulation in GDPD5 (AT1G74210) and GDPD1 (AT3G02040), involved in lipid remodeling during P deficiency (Cheng et al., 2011). Genes involved in carbohydrate metabolism, such as CTIMC (AT3G55440) and the phosphoglucan phosphatase SEX4 (AT3G52180), were upregulated in P− conditions (Kötting et al., 2009). Upregulation of VTC4 (AT3G02870) is notable as it encodes a bi-functional enzyme involved in myoinositol and ascorbate synthesis (Torabinejad et al., 2009). Myoinositol has roles for P signaling, storage, and stress tolerance, and ascorbate regulates several abiotic stress signaling pathways and provides AO functions (Xiao et al., 2021; Wu et al., 2023). PLAT1 (AT4G39730), involved in a broad range of abiotic stress response pathways, was also upregulated in P− (Hyun et al., 2014).
In roots, 33 genes were significantly downregulated and 227 genes were significantly upregulated in P− watercress roots with respect to the P+ treatment. Of these, 173 were annotated and almost all annotated genes corresponded to those in A. thaliana. The top 50 annotated DEGs are summarized in Table 2, and the full list of DEGs is shown in Supplementary 5. GO analysis (Figure 6) revealed significant enrichment of pathways involved in known responses to P deficiency. The highest enrichment was observed for genes involved in sulfolipid metabolic and biosynthetic processes (>158-fold enrichment) through upregulation of SQD1 (AT4G33030), SQD2 (AT5G01220), and UGP3 (AT3G56040), followed by a 63-fold enrichment of genes involved in galactolipid biosynthesis including DGD1 (AT3G11670), DGD2 (AT4G00550), MGD3 (AT2G11810), PAH1 (AT3G09560), and PLPZETA2 (AT3G05630) (Yu et al., 2002; Cruz-Ramirez et al., 2006; Kobayashi et al., 2009). Two upregulated genes, PHT1;8 and PHT1;4, encode high-affinity phosphate transporters in A. thaliana (Nussaume et al., 2011). Other genes associated with a cellular response to P starvation included RNS1 (AT2G02990) for P remobilization, and SPX1 (AT5G20150) and SPX3 (AT2G45130), which are essential transcriptional regulators of the P-starvation response (Puga et al., 2014; Zhou et al., 2015).
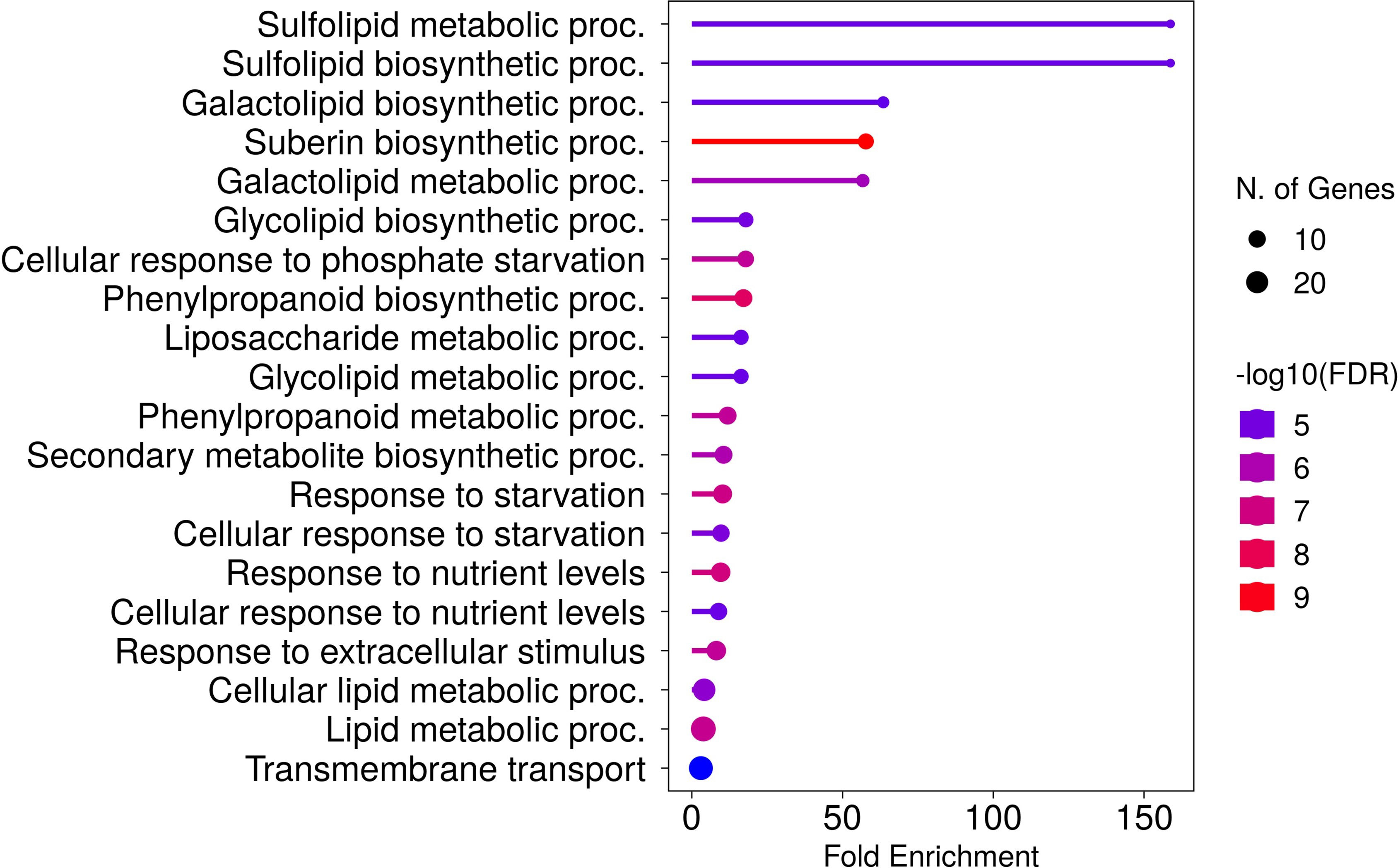
Figure 6 Top 20 enriched biological processes of DEGs in roots of watercress grown without additional phosphate-based fertilizer. Fold enrichment is defined as the percentage of expressed sequences of known annotation belonging to a pathway, divided by the corresponding percentage in the background. Developed using the ShinyGO platform with A. thaliana as a reference.
Multi-dimensional scaling illustrated large transcriptomic differences between line 102, 60, and WXVITA, as shown by differential grouping in Figure 7. To assess responses to P-deficiency unique to the selected lines (60 and 102), DEGs in these lines compared to the commercial control line (WXVITA) were identified in P− conditions (Figure 8; Supplementary 6, 7). A total of 123 DEGs were uniquely upregulated and 130 DEGs were uniquely downregulated in line 60 shoots in P−, relative to WXVITA. Upregulated genes in line 60 included many genes involved in shoot development, such as UGE4 (AT1G64440), MYO6 (AT5G43900), and PSP (AT1G18640). By contrast, 17 were uniquely upregulated and 19 were uniquely downregulated in 102 in P− relative to WXVITA. A total of 21 upregulated and 8 downregulated genes were common to both line 60 and 102 relative to expression in WXVITA. GO enrichment analysis of DEGs common to both selected lines identified that the pathway with highest fold enrichment was phosphate ion homeostasis (Supplementary 8; 162 fold enrichment), due to upregulation of CAX1 (AT2G38170) and downregulation of UBC24 (aka PHO2, AT2G33770) (Liu et al., 2011; Liu et al., 2012).
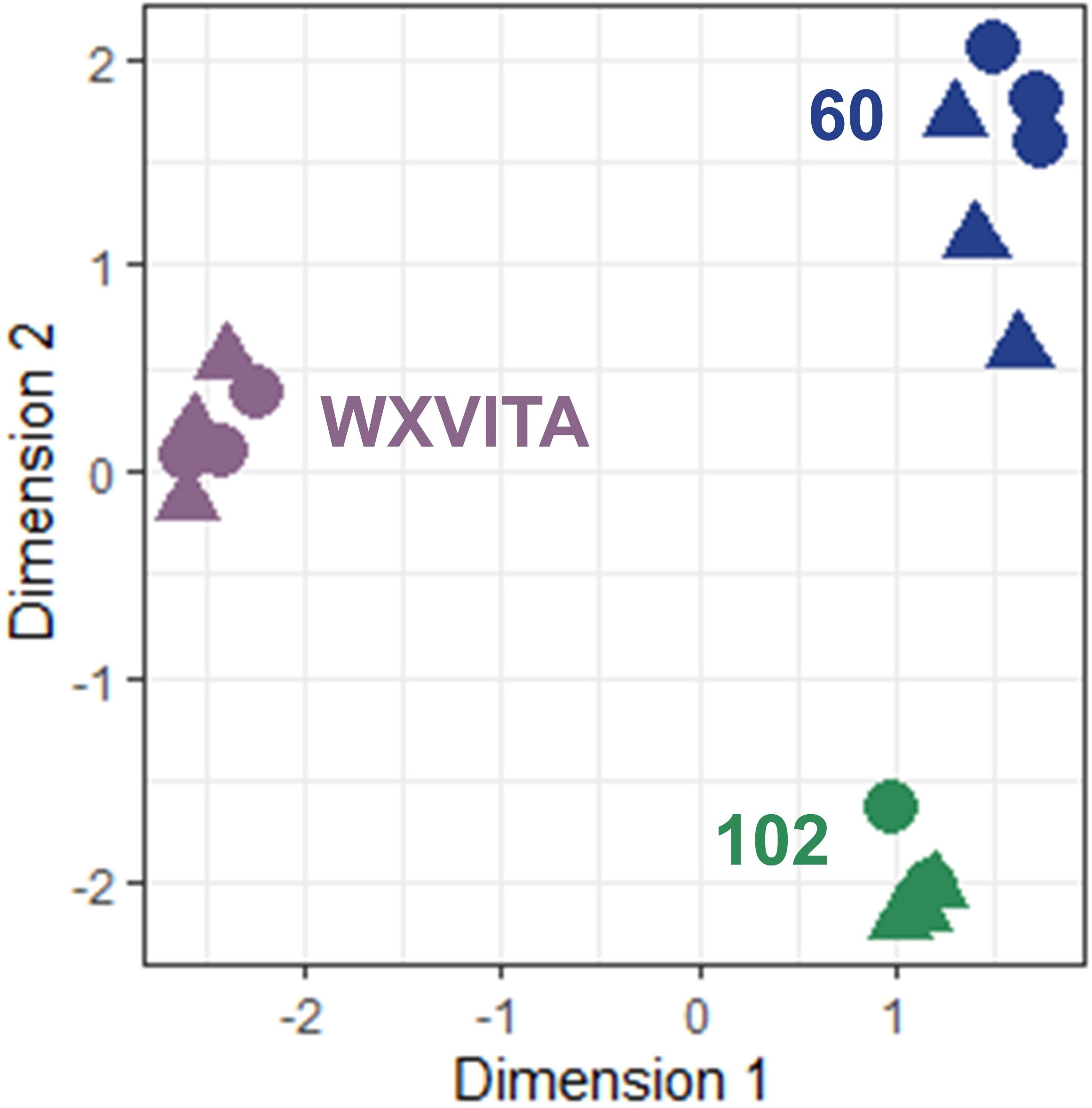
Figure 7 Multidimensional scaling (MDS) of RNA-seq shoot samples. Distances correspond to leading log-fold changes between samples. Circles (◍) and triangles (▲) denote P− and P+ samples, respectively.
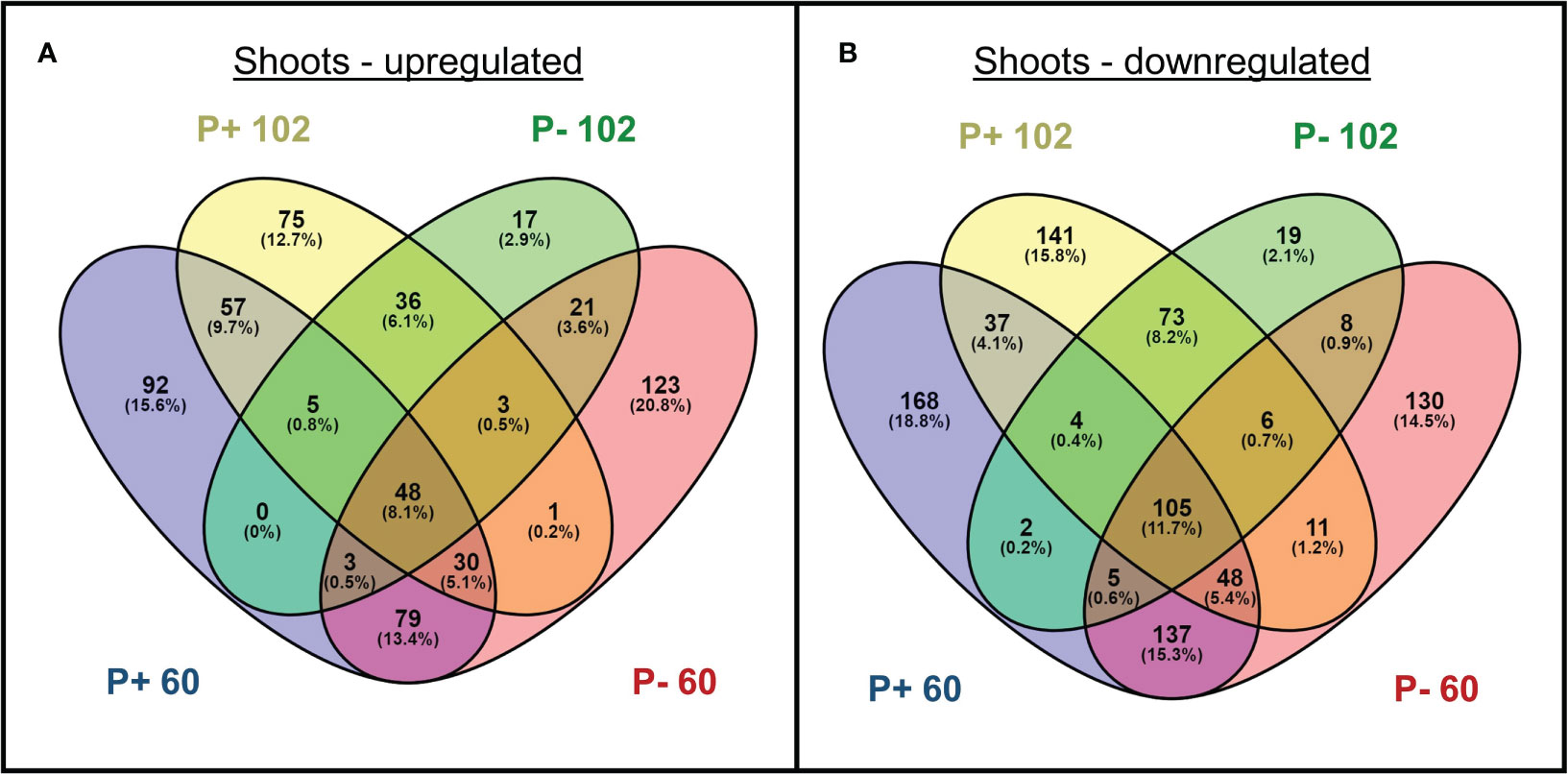
Figure 8 Number of genes (A) upregulated and (B) downregulated in shoots of selected lines (102 and 60) grown with phosphate-based fertilizer (P+) or without additional fertilizer (P−). Differentially expressed genes are respective to expression in WXVITA. Numbers in parentheses indicate the percentage of DEGs from each group falling into each category.
Despite clear environmental issues associated with the leakage of phosphate-based fertilizers into aquatic systems, research targeting breeding for PUE in aquatic crops such as watercress is limited, especially in commercial systems (Austin, 1966; Howard-Williams et al., 1982; Bennett, 1986; Fernandez-Going et al., 2013; Hibbert and Taylor, 2022). In this study, we provide the first comprehensive analysis on the effects of reduced phosphate-based fertilizer supply on watercress morphology, biochemistry, and the transcriptome, and identified novel candidate genes for targeting in future breeding and also new lines with improved tolerance to a low fertilizer cultivation system. These new lines have the potential to be considered as future commercial releases, once these traits are confirmed as fixed.
Watercress root morphology has previously been shown to be made up of adventitious roots and basal roots (defined as a finer root system that anchors the plant and originates from the primary root meristem) (Cumbus and Robinson, 1977). Of these, the adventitious root system absorbs a greater proportion of P than the basal system and here we found that the watercress root systems in this commercial growth environment form a densely matted structure composed almost completely of roots deriving from the stem (adventitious roots; Supplementary 9) at harvest point, suggesting that adventitious roots are most important for P uptake in watercress commercial cultivation and for identifying targets for future watercress selection and breeding.
Growing watercress without additional phosphate-based fertilizer had a significant impact on morphology: mean root biomass was increased, while shoot biomass was either unchanged (fresh biomass) or somewhat increased (dry biomass). Plant stems were shorter with smaller leaves, although the number of leaves was maintained, suggesting that shoot biomass is concentrated in a denser plant. P deprivation results in decreased cell size and is associated with regulation of cell-wall-related genes that cause cellulose synthesis and lignin deposition (Fu et al., 2013; Hoehenwarter et al., 2016; Ogden et al., 2018). The resulting thick-walled small-celled phenotype may explain the dense dwarf shoot phenotype and higher dry mass observed in P−. Most of the evidence for low P-induced cell wall thickening has been investigated only in roots; however, studies in rice have found that plants impaired in the OsGLU3 gene (associated with cellulose content) exhibit reduced shoot cell growth that is P-dependent (Zhou et al., 2006; Zhang et al., 2012). Three endoglucanases involved in hydrolysis of cellulose (AT1G64390, AT1G75680, and AT2G32990) were downregulated in line 60, P− shoots (compared to WXVITA) in our study, suggesting that this cell-wall thickening response is stronger in this line (Libertini et al., 2004). Although it is surprising that watercress was able to maintain shoot fresh biomass, given the lower availability of P, the increase in root biomass suggests that watercress compensates for reduced nutrient availability by investing more in root growth, particularly for line 60, reflected in the increased R:S ratio in P−, which is a common phenomenon observed in low P conditions (Cogliatti and Clarkson, 1983; Yang et al., 2011; Shen et al., 2018; de Souza Campos et al., 2019; Duan et al., 2020; Irfan et al., 2020).
To ensure our study remained commercially relevant, contrasting applications of commercial watercress fertilizer were made in a commercial farm. Despite these treatments also altering N and K application, P remained the limiting macronutrient in P− treatment, in line with what has been reported previously (Crisp, 1970; CaBA CSRG, 2021). This was shown through (a) N concentration remaining constant within the water supply of both treatment beds at harvest point; (b) K concentration of plant tissue being unaffected by treatment but P concentration of plant tissue was lower in P− treatments; and (c) changes to biochemistry typical of P-deficiency were observed in plants grown in P− alongside gene expression changes observed in other species grown in P-deficient conditions.
Mobilizing the resources to ensure continued growth with lower P inputs appears to result from remobilization and scavenging strategies, such as by substituting phospholipids in the cell membrane with other lipids (Yu et al., 2002). This strategy is evident in the DEGs of the P− roots and subsequent GO enrichment analysis, which showed pathways with the highest representation were for sulfolipid and galactolipid biosynthesis (e.g., SQD1/2, DGDG1/2, MGDG3, PLPZETA2, and UGP3). Previous RNA-seq and microarray studies in A. thaliana identified genes related to galactolipid biosynthesis as the largest group within a core subset of P-starvation response (PSR) genes, and here we have shown that these genes were most strongly induced in P− watercress (Lan et al., 2015). These genes function to free P from phospholipids, which accounts for 20% of plant P content (Veneklaas et al., 2012). SQD1/2, DGDG1/2, MGDG3, PLPZETA2, and PECP1 were confirmed here to be primary functional genomic targets for developing more P-use efficient watercress (Hibbert and Taylor, 2022).
Other gene homologs important for watercress PUE include those for P transport, such as high-affinity P transporters PHT1;4 and PHT1;8, which were both upregulated in P− roots. The high affinity carnitine uptake transporter OCT1 was not previously predicted to be important for PUE in watercress; however, it was found as inducible here. This P-inducibility has been shown in other studies and OCT1 is known to regulate lateral root development (Lelandais-Brière et al., 2007; Lan et al., 2015). One study also found its expression to be affected by PHO2, which regulates translocation of P from shoots to roots (Bari et al., 2006). SPX1 and SPX3, transcription factors with multi-functional roles in the PSR, were both upregulated in P-starved roots. SPX-domain-containing genes are part of a core subset of PSR genes in A. thaliana and were predicted to be candidates for PUE in watercress, which is confirmed in this study (Lan et al., 2015; Hibbert and Taylor, 2022). Both are upregulated in A. thaliana under P starvation and play positive roles in adaptation to P−, with SPX3 exerting negative feedback regulation of SPX1 (Duan et al., 2008).
Suberization of roots may also have contributed to the increase in root biomass. Suberization is the deposition of suberin polymers within the root exodermis and endodermis to form hydrophobic barriers (Baxter et al., 2009). GO analysis found that DEGs involved in suberin biosynthesis contributed to a 57-fold enrichment of this pathway in watercress. Suberin has been shown to play a pivotal role in drought, waterlogging, and nutrient homeostasis by providing an apoplastic barrier within the root (Barberon et al., 2016). Studies on effects of P-starvation have found no increase in suberization in P-deficient barley (Hordeum vulgare) and lower suberization in A. thaliana, in contrast to the suggestion of enhanced suberin biosynthesis in this study, suggesting possible fundamental differences in mechanisms between terrestrial and aquatic plants (Andersen et al., 2018; Grünhofer et al., 2021). Müller et al. (2021) showed varying levels of upregulation of numerous PSR genes following whole plant submergence, which may suggest a link between P starvation and watercress submergence responses. Purple acid phosphatases (PAPs) that are secreted outside roots to mobilize P from bound sources are not relevant to watercress cultivation systems where this free P would rapidly wash away. However, intracellular phosphatases are of interest for PUE in aquatic crops as these remobilize P within the plant. Our study identified upregulation of PAP17 (AT3G17790) with dual roles in P mobilization and ROS metabolism (del Pozo et al., 1999). Additionally, phosphatases not previously regarded as central for PUE were also upregulated in P− watercress. This includes DSP2 (AT2G32960), which encodes a tyrosine-specific phosphatase and whose expression was increased 3.2-fold in one study with P-starved A. thaliana (Müller et al., 2007). The gene for the inorganic pyrophosphatase PPA4 (AT3G53620), which releases P from pyrophosphate, was also upregulated in watercress here (Navarro-De la Sancha et al., 2007). These results suggest an increased importance for investigating alternative phosphatases that have previously been overlooked in PUE studies. Key to all these changes is differential gene expression of numerous transcription factors associated with the PSR such as SPX1, SPX3, WRKY75, and HHO2 in watercress roots (Puga et al., 2014; Zhou et al., 2015; Wang et al., 2020).
AO capacity and GSL concentration are of critical importance as nutritional traits, linked to beneficial impacts on human health and central to the watercress nutritional profile, of relevance to the consumer. AO capacity was increased in P− plants, and there were no changes to the total concentration of GSLs. Increased AO capacity could be linked to the impairment of photosynthetic rate (via impairing electron transfer) in P deprivation, leading to ROS generation, which is countered through increased synthesis of AOs and activity of AO enzymes, such as peroxidase (Meng et al., 2021). This is a widely reported response to P−, with evidence in bean (Phaseolus vulgaris), tomato (Solanum lycopersicum), rice (Oryza sativa), and maize (Zea mays) (Juszczuk et al., 2001; Zhang et al., 2014; Muneer and Jeong, 2015; Veronica et al., 2017). The studies in bean and maize also found P− decreased leaf area, as in watercress. Leaf biomass was also higher in phosphate-treated garden sage (Salvia officinalis) in agreement with our results, but this was associated with increased leaf phenolic (AOs) concentration (Nell et al., 2009). However, the P+ concentration used by Nell et al. (136 mg L−1 PO4) far exceeded those used in our field conditions (P+ ~0.06 mg L−1). The increase in AO capacity of P− watercress is supported by transcriptomic changes in our study: VTC4, involved in ascorbate biosynthesis, was upregulated in P− shoot tissue (Conklin et al., 2006). Interestingly, catalytic activity by VTC4 releases phosphate, so this protein may serve a dual function for PUE by freeing bound P and providing AO scavenging (Torabinejad et al., 2009).
No previous studies exist on the effects of fertilizer application on watercress GSL concentration; however, there is literature for other species. Contrary to our results, GSL concentration of Arabidopsis increased in P− conditions; however, the P− conditions used in that trial (3 µM = 0.285 mg L−1 PO4) exceed mean P concentration observed in the irrigation supply here, even in P+ conditions in our trial, so these effects may not be relevant at very low P concentrations (Pant et al., 2015). In rocket (Eruca sativa), increasing P elevated total GSL concentration (Chun et al., 2017). However, once again, even the lowest P concentration (0.5 mM = 45.49 mg L−1 PO4) far exceeded those used in this trial. Additionally, GSLs quantified in this study were methionine-derived, whereas gluconasturtiin (comprising >90% of GSL in watercress) is phenylalanine-derived (Underbill, 1965; Voutsina et al., 2016). Considering methionine is a sulfur-containing amino acid and phenylalanine is not, it is possible that GSL concentration not affected in watercress because there is no trade-off with sulfur allocation between GSL biosynthesis and low P-induced cell membrane remodeling (as sulfolipids). These GSL studies also raise the question of how biologically relevant P-deficient conditions are to field conditions. Although the chalk streams supplying watercress farms naturally contain unusually low concentration of phosphate (<0.04 mg L−1), P available in solution within soils has been reported to be approximately 0.05 mg L−1, but this varies considerably with soil type (Havlin et al., 2005). In several Brassica species (B. campestris and B. juncea) given either 0.1 mM or 0.5 mM P, a similar result was found to watercress in our study: Trejo-Téllez et al. (2019) found that there were no differences in the concentration of alkenyl-GSLs (the primary GSL group in the plants) and several indole-GSLs between different P supplies in either species tested. However, responses of GSLs to treatment varied between species. This demonstrates the variability of GSL responses to P.
Growers are also interested in developing a sweeter watercress crop to appeal to consumers who find its peppery taste polarizing (Dr. H. Smith, Vitacress, 2020, personal communication). Therefore, it is interesting that sugar and starch concentration were also increased in P− shoots. Increases in starch and sugars is a well-reported response to low P (Rychter and Randall, 1994; Ciereszko and Barbachowska, 2000; Müller et al., 2007; Hammond and White, 2011; Li et al., 2021). Sugar is critical for signaling cascades involved in the P-starvation response and many P-responsive genes are sugar-inducible such as PHT1;1 (Karthikeyan et al., 2007; Rouached et al., 2010). Starch accumulation is attributed to an increase in Calvin cycle intermediates (due to decreased exchange of triose-phosphate with cytosolic P) from which P can be liberated (MacNeill et al., 2017). It may also be the case in this study that sugar and starch concentrations were higher as they were more concentrated in dwarfed shoots. RNA-seq results also support the role of sugars in P starvation responses. CTIMC (AT3G55440), involved in gluconeogenesis, had a log fold-change of 0.45 (p < 0.0001) in P− shoots compared to P+ shoots in our study (Dumont et al., 2016). An increase in the CTIMC protein triosephosphate isomerase has been reported to increase in P− maize roots (Li et al., 2007a). SEX4 dephosphorylates starch granules to regulate starch accumulation, and its gene was also upregulated here (Kötting et al., 2009). In P− watercress roots, GWD1 (AT1G10760), which regulates the addition of P to starch, was downregulated (Yu et al., 2001).
It is promising that there was no compromise to the nutritional quality, but the peppery flavor of glucosinolates could be softened by increased sugars. Taste trials are needed to quantify the effects of enhanced sugars on consumer taste preferences. Importantly, increased AO capacity and the maintenance of GSL concentration for commercial crops grown with less P input provide a win–win situation for watercress, with maintained or enhanced nutritional characteristics and an improved environmental footprint. Additional trials are also required to see if these effects are stable across different environments and growing seasons.
To further explore the effects of P deprivation on watercress morphology and biochemistry, an additional aim of this study was to use variation in these responses to P to inform selections for future breeding. Stem length, number of leaves, and individual leaf area all varied significantly between lines and ranking of lines by trait means, and assessing percentage change in response to low P is valuable for making initial line selections for future breeding. This showed that lines 60 and 102 were valuable for quantification of P and K concentration and differential gene expression analysis by RNA-seq. Line 60 was P-responsive (e.g., 114% increase in mean root fresh weight in P−) and had a relatively high AO capacity, whereas line 102 ranked the highest for several yield traits while exhibiting low responsiveness to P treatment. Overall, PUE was the highest in line 60 followed by 102, as reflected by the highest P concentration in dried shoot tissue under both conditions compared to the commercial control. This provides further evidence to progress these lines in breeding for PUE in watercress.
The large increase in root biomass for line 60 in P− suggests that the PUE strategy for this line focuses on altered root morphology, or this could be a by-product of effective P remobilization strategies, whereas the relatively P-unresponsive yield seen in 102 suggests enhanced uptake (e.g., higher activity of P transporters) and more effective utilization internally. The development genes (UGE4, MYO6, and PSP) upregulated in P− in line 60 indicate morphology changes. However, of these, only MYO6 upregulation was unique to line 60 (PSP and UGE4 were also upregulated in 102, with respect to WXVITA). Line 60 has also been shown to have consistently high GSL concentration and leaf area in previous trials in the UK and US (Qian et al., 2023). In addition, DEGs identified in both lines were involved in phosphate ion homeostasis (CAX1 and UBC24, aka PHO2). The downregulation of PHO2 in lines 60 and 102 (with respect to WXVITA) in P− is especially interesting as PHO2 downregulation results in reduced degradation of PHO1, leading to increased P loading to the shoots (Liu et al., 2012). This could partly explain the higher P concentration and yield of these selected lines compared to WXVITA. These genes provide further targets for breeding watercress with improved PUE. The identification of PHO2 in these selected lines with improved PUE also supports our previous review that listed PHO2 as a candidate gene for PUE breeding in watercress (Hibbert and Taylor, 2022).
We provide the first report on the effects of contrasting phosphate-based fertilizer treatments on the morphology, biochemistry, and transcriptome of watercress, alongside the identification of a suite of genes important for PUE in this aquatic species. Taken together, this information will underpin future watercress breeding (Figure 9). Watercress plants sustained shoot yield without additional fertilizer treatment, partially through enhanced root biomass, and had shorter stems and more densely packed leaf area. Strategies (and associated genes) for improved low P tolerance in watercress focused on cell membrane remodeling (e.g., SQD1/2, DGDG1/2, MGDG3, PLPZETA2, UGP3, and PECP1), also regarded as a core gene set for the PSR in A. thaliana (Lan et al., 2015). Homologs of other known PUE genes such as P transporters (e.g., PHT1 and PHO2), transcription factors (e.g., SPX1/3, WRK75, and HHO2), and phosphatases (e.g., PAP17) were also identified as important for watercress PUE. Additional genes not previously regarded as important for their PUE function included numerous genes for root suberization, suggesting a higher emphasis on this strategy for PUE in aquatic crops like watercress. Overall, this study identified new germplasm and genetic targets to assist with advancing PUE in watercress breeding.
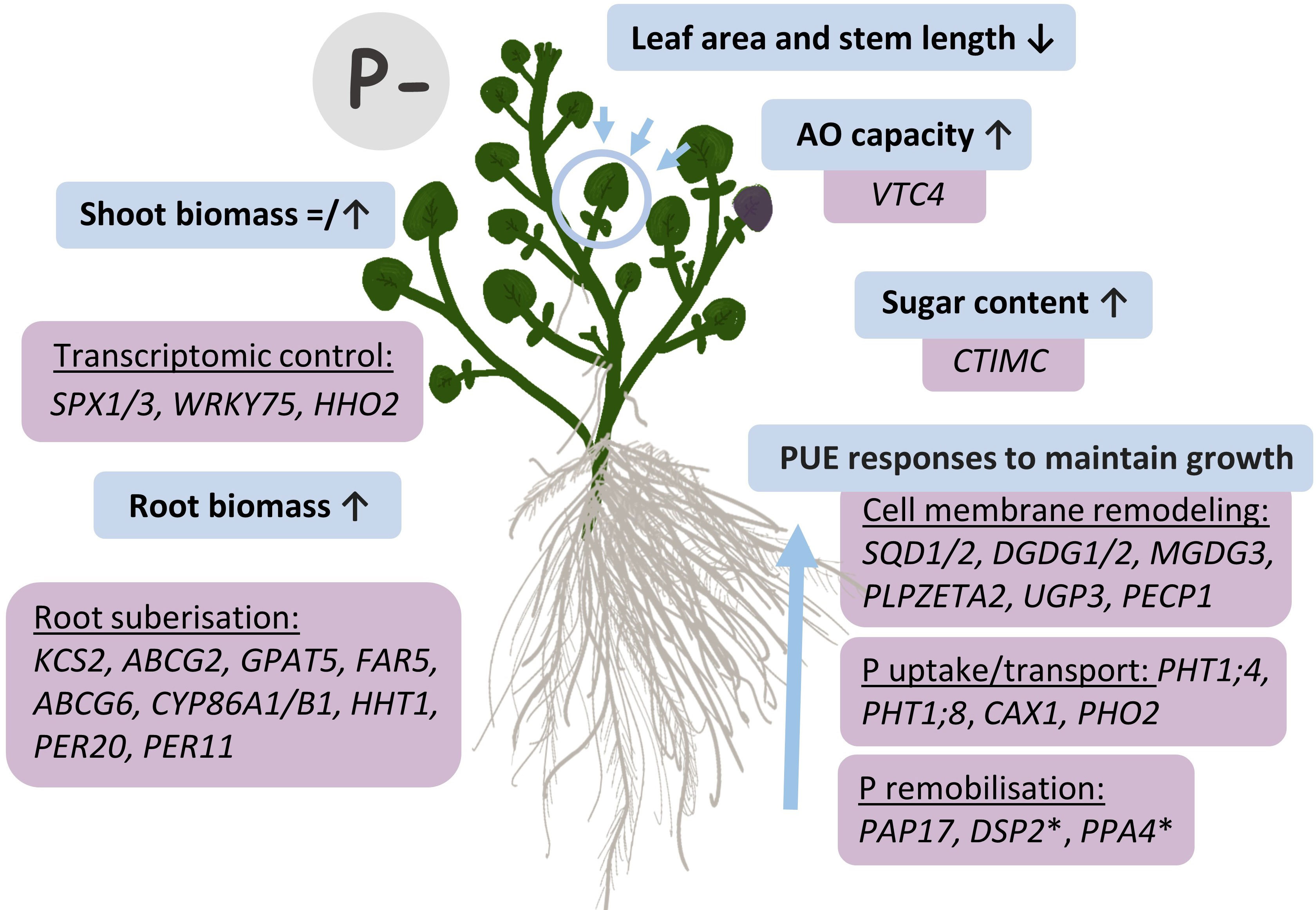
Figure 9 Responses to P deficiency (P−) in watercress and genetic targets identified by RNA-seq. Blue boxes illustrate morphological and biochemical changes observed in P− conditions, with respective genes associated with these changes and identified through RNA-seq results in purple boxes. Up (↑) arrows denote an increase, whereas ↓ arrows denote a decrease. The = icon illustrates the absence of an effect of P− on shoot fresh biomass whereas shoot dry biomass increased. *Other unexplored phosphatases identified here but not previously recognized as PUE genes in A. thaliana that could provide PUE roles.
The data presented in this study are deposited in NCBI's Sequence Read Archive (SRA), BioProject number PRJNA941001.
LH: Data curation, Formal Analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. YQ: Formal Analysis, Methodology, Software, Writing – review & editing. HS: Investigation, Resources, Writing – review & editing. SM: Investigation, Resources, Writing – review & editing. EK: Formal Analysis, Investigation, Methodology, Writing – review & editing. DK: Funding acquisition, Methodology, Resources, Writing – review & editing. GT: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by a PhD studentship funded by Vitacress Salads Ltd. to LH and by the John B Orr endowed chair in Environmental Plant Sciences held by GT at the University of California, Davis.
The authors would like to thank Dr. Felipe Becerra Sanchez for advice on conducting biochemical assays. The authors also thank Lesley Hibbert, Olivia Hibbert, Bethany Richmond, Connor Butler, and Harry Shepherd for their significant contribution to sample processing through a challenging 2020 field trial.
Authors HS and SM were employed by Vitacress Salads Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1279823/full#supplementary-material
Akoh, C. C., Lee, G.-C., Liaw, Y.-C., Huang, T.-H., Shaw, J.-F. (2004). GDSL family of serine esterases/lipases. Prog. Lipid Res. 43, 534–552. doi: 10.1016/j.plipres.2004.09.002
Andersen, T. G., Naseer, S., Ursache, R., Wybouw, B., Smet, W., De Rybel, B., et al. (2018). Diffusible repression of cytokinin signalling produces endodermal symmetry and passage cells. Nature 555, 529–533. doi: 10.1038/nature25976
Austin, R. B. (1966). The growth of watercress (Rorippa nasturtium aquaticum (L) Hayek) from seed as affected by the phosphorus nutrition of the parent plant. Plant Soil 24, 113–120. doi: 10.1007/BF01373077
Barberon, M., Vermeer, J. E. M., De Bellis, D., Wang, P., Naseer, S., Andersen, T. G., et al. (2016). Adaptation of root function by nutrient-induced plasticity of endodermal differentiation. Cell 164, 447–459. doi: 10.1016/j.cell.2015.12.021
Bari, R., Pant, B. D., Stitt, M., Scheible, W.-R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141, 988–999. doi: 10.1104/pp.106.079707
Baxter, I., Hosmani, P. S., Rus, A., Lahner, B., Borevitz, J. O., Muthukumar, B., et al. (2009). Root suberin forms an extracellular barrier that affects water relations and mineral nutrition in arabidopsis. PloS Genet. 5, e1000492. doi: 10.1371/journal.pgen.1000492
Becerra-Sanchez, F., Taylor, G. (2021). Characterising the sweet corn postharvest supply chain: travel from Senegal to the UK. Int. J. Postharvest Technol. Innovation 8, 1–18. doi: 10.1504/IJPTI.2021.10034827
Beisson, F., Li, Y., Bonaventure, G., Pollard, M., Ohlrogge, J. B. (2007). The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell 19, 351–368. doi: 10.1105/tpc.106.048033
Benjamini, Y., Hochberg, Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Society: Ser. B (Methodological) 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bennett, S. M. (1986) Studies on the Growth and Nutrition of Watercress, Nasturtium Officinale R. Br. Available at: https://researchportal.bath.ac.uk/en/studentTheses/studies-on-the-growth-and-nutrition-of-watercress-nasturtium-offi (Accessed June 23, 2022).
Benveniste, I., Tijet, N., Adas, F., Philipps, G., Salaün, J. P., Durst, F. (1998). CYP86A1 from Arabidopsis thaliana encodes a cytochrome P450-dependent fatty acid omega-hydroxylase. Biochem. Biophys. Res. Commun. 243, 688–693. doi: 10.1006/bbrc.1998.8156
Benzie, I. F., Strain, J. J. (1996). The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: the FRAP assay. Anal. Biochem. 239, 70–76. doi: 10.1006/abio.1996.0292
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Borner, G. H. H., Lilley, K. S., Stevens, T. J., Dupree, P. (2003). Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol. 132, 568–577. doi: 10.1104/pp.103.021170
Boyd, L., McCann, M., Hashim, Y., Bennett, R., Gill, C., Rowland, I. (2006). Assessment of the anti-genotoxic, anti-proliferative, and anti-metastatic potential of crude watercress extract in human colon cancer cells. Nutr. Cancer 55, 232–241. doi: 10.1207/s15327914nc5502_15
CaBA CSRG (2021). Catchment Based Approach Chalk Stream Restoration Strategy 2021. (CaBA CSRG). Available at: https://catchmentbasedapproach.org/wp-content/uploads/2021/10/CaBA-CSRG-Strategy-MAIN-REPORT-FINAL-12.10.21-Low-Res.pdf.
Casey, H., Smith, S. M. (1994). The effects of watercress growing on chalk headwater streams in Dorset and Hampshire. Environ. pollut. 85, 217–228. doi: 10.1016/0269-7491(94)90088-4
Chen, X.-Y., Liu, L., Lee, E., Han, X., Rim, Y., Chu, H., et al. (2009). The arabidopsis callose synthase gene GSL8 is required for cytokinesis and cell patterning. Plant Physiol. 150, 105–113. doi: 10.1104/pp.108.133918
Cheng, Y., Zhou, W., El Sheery, N. I., Peters, C., Li, M., Wang, X., et al. (2011). Characterization of the Arabidopsis glycerophosphodiester phosphodiesterase (GDPD) family reveals a role of the plastid-localized AtGDPD1 in maintaining cellular phosphate homeostasis under phosphate starvation. Plant J. 66, 781–795. doi: 10.1111/j.1365-313X.2011.04538.x
Cheung, K. L., Kong, A.-N. (2009). Molecular targets of dietary phenethyl isothiocyanate and sulforaphane for cancer chemoprevention. AAPS J. 12, 87–97. doi: 10.1208/s12248-009-9162-8
Chun, J.-H., Kim, S., Arasu, M. V., Al-Dhabi, N. A., Chung, D. Y., Kim, S.-J. (2017). Combined effect of Nitrogen, Phosphorus and Potassium fertilizers on the contents of glucosinolates in rocket salad (Eruca sativa Mill.). Saudi J. Biol. Sci. 24, 436–443. doi: 10.1016/j.sjbs.2015.08.012
Ciereszko, I., Barbachowska, A. (2000). Sucrose Metabolism in Leaves and Roots of Bean (Phaseolus vulgaris L.) during Phosphate Deficiency. J. Plant Physiol. 156, 640–644. doi: 10.1016/S0176-1617(00)80225-4
Coaker, G., Falick, A., Staskawicz, B. (2005). Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science 308, 548–550. doi: 10.1126/science.1108633
Cogliatti, D. H., Clarkson, D. T. (1983). Physiological changes in, and phosphate uptake by potato plants during development of, and recovery from phosphate deficiency. Physiologia Plantarum 58, 287–294. doi: 10.1111/j.1399-3054.1983.tb04183.x
Compagnon, V., Diehl, P., Benveniste, I., Meyer, D., Schaller, H., Schreiber, L., et al. (2009). CYP86B1 is required for very long chain omega-hydroxyacid and alpha, omega -dicarboxylic acid synthesis in root and seed suberin polyester. Plant Physiol. 150, 1831–1843. doi: 10.1104/pp.109.141408
Conklin, P. L., Gatzek, S., Wheeler, G. L., Dowdle, J., Raymond, M. J., Rolinski, S., et al. (2006). Arabidopsis thaliana VTC4 encodes L-galactose-1-P phosphatase, a plant ascorbic acid biosynthetic enzyme *. J. Biol. Chem. 281, 15662–15670. doi: 10.1074/jbc.M601409200
Cox, J. (2009) Watercress Growing and its Environmental Impacts on Chalk Rivers in England. Available at: http://publications.naturalengland.org.uk/publication/40010 (Accessed November 19, 2019).
Crisp, D. T. (1970). Input and output of minerals for a small watercress bed fed by chalk water. J. Appl. Ecol. 7, 117–140. doi: 10.2307/2401614
Cruz-Ramirez, A., Oropeza-Aburto, A., Razo-Hernandez, F., Ramirez-Chavez, E., Herrera-Estrella, L. (2006). Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc. Natl. Acad. Sci. 103, 6765–6770. doi: 10.1073/pnas.0600863103
Cumbus, I. P., Robinson, L. W. (1977). The function of root systems in mineral nutrition of watercress (Rorippa nasturtium-aquaticum (L) Hayek). Plant Soil 47, 395–406. doi: 10.1007/BF00011498
DEFRA (2019) Wholesale Fruit and Vegetable Prices (GOV.UK). Available at: https://www.gov.uk/government/statistical-data-sets/wholesale-fruit-and-vegetable-prices-weekly-average (Accessed December 8, 2020).
DEFRA (2020) Horticulture Statistics 2019. Available at: https://www.gov.uk/government/statistics/latest-horticulture-statistics (Accessed May 25, 2021).
del Pozo, J. C., Allona, I., Rubio, V., Leyva, A., de la Peña, A., Aragoncillo, C., et al. (1999). A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J. 19, 579–589. doi: 10.1046/j.1365-313x.1999.00562.x
de Souza Campos, P. M., Cornejo, P., Rial, C., Borie, F., Varela, R. M., Seguel, A., et al. (2019). Phosphate acquisition efficiency in wheat is related to root:shoot ratio, strigolactone levels, and PHO2 regulation. J. Exp. Bot. 70, 5631–5642. doi: 10.1093/jxb/erz349
Di Noia, J. (2014). Defining powerhouse fruits and vegetables: a nutrient density approach. Prev. Chronic Dis. 11, e95. doi: 10.5888/pcd11.130390
Dörmann, P., Balbo, I., Benning, C. (1999). Arabidopsis galactolipid biosynthesis and lipid trafficking mediated by DGD1. Science 284, 2181–2184. doi: 10.1126/science.284.5423.2181
Doyle, J. J., Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bull. 19, 11–15.
Duan, X., Jin, K., Ding, G., Wang, C., Cai, H., Wang, S., et al. (2020). The impact of different morphological and biochemical root traits on phosphorus acquisition and seed yield of Brassica napus. Field Crops Res. 258, 107960. doi: 10.1016/j.fcr.2020.107960
Duan, H., Schuler, M. A. (2005). Differential expression and evolution of the Arabidopsis CYP86A subfamily. Plant Physiol. 137, 1067–1081. doi: 10.1104/pp.104.055715
Duan, K., Yi, K., Dang, L., Huang, H., Wu, W., Wu, P. (2008). Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 54, 965–975. doi: 10.1111/j.1365-313X.2008.03460.x
Dumont, S., Bykova, N. V., Pelletier, G., Dorion, S., Rivoal, J. (2016). Cytosolic triosephosphate isomerase from arabidopsis thaliana is reversibly modified by glutathione on cysteines 127 and 218. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01942
Edstam, M. M., Blomqvist, K., Eklöf, A., Wennergren, U., Edqvist, J. (2013). Coexpression patterns indicate that GPI-anchored non-specific lipid transfer proteins are involved in accumulation of cuticular wax, suberin and sporopollenin. Plant Mol. Biol. 83, 625–649. doi: 10.1007/s11103-013-0113-5
Environment Agency and Natural England (2021) Chalk stream strategy launched to protect ‘England’s rainforests.’. Available at: https://www.gov.uk/government/news/chalk-stream-strategy-launched-to-protect-englands-rainforests (Accessed January 2, 2023).
Essigmann, B., Güler, S., Narang, R. A., Linke, D., Benning, C. (1998). Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 95, 1950–1955. doi: 10.1073/pnas.95.4.1950
European Commission (2019) Fitness check of the Water Framework Directive and the Floods Directive. Available at: https://ec.europa.eu/environment/water/fitness_check_of_the_eu_water_legislation/index_en.htm (Accessed January 25, 2021).
Faure, J. D., Vittorioso, P., Santoni, V., Fraisier, V., Prinsen, E., Barlier, I., et al. (1998). The PASTICCINO genes of Arabidopsis thaliana are involved in the control of cell division and differentiation. Development 125, 909–918. doi: 10.1242/dev.125.5.909
Fernandez-Going, B., Even, T., Simpson, J. (2013). The effect of different nutrient concentrations on the growth rate and nitrogen storage of watercress (Nasturtium officinale R. Br.). Hydrobiologia 705, 63–74. doi: 10.1007/s10750-012-1380-x
Fu, Y., Yang, X., Shen, H. (2013). The physiological mechanism of enhanced oxidizing capacity of rice (Oryza sativa L.) roots induced by phosphorus deficiency. Acta Physiologiae Plantarum 36, 179–190. doi: 10.1007/s11738-013-1398-3
Ge, S. X., Jung, D., Yao, R. (2020). ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629. doi: 10.1093/bioinformatics/btz931
Grünhofer, P., Schreiber, L., Kreszies, T. (2021). “Suberin in Monocotyledonous Crop Plants: Structure and Function in Response to Abiotic Stresses,” in Rhizobiology: Molecular Physiology of Plant Roots Signaling and Communication in Plants. Eds. Mukherjee, S., Baluška, F. (Cham, Switzerland: Springer), 333–378. doi: 10.1007/978-3-030-84985-6_19
Hammond, J. P., White, P. J. (2011). Sugar signaling in root responses to low phosphorus availability. Plant Physiol. 156, 1033–1040. doi: 10.1104/pp.111.175380
Hanchi, M., Thibaud, M.-C., Légeret, B., Kuwata, K., Pochon, N., Beisson, F., et al. (2018). The phosphate fast-responsive genes PECP1 and PPsPase1 affect phosphocholine and phosphoethanolamine content. Plant Physiol. 176, 2943–2962. doi: 10.1104/pp.17.01246
Harrar, Y., Bellec, Y., Bellini, C., Faure, J.-D. (2003). Hormonal control of cell proliferation requires PASTICCINO genes. Plant Physiol. 132, 1217–1227. doi: 10.1104/pp.102.019026
Havlin, J., Beaton, J., Tisdale, S., Nelson, W. (2005). “Soil fertility and fertilizers,” in Soil Fertility and Fertilizers (New Jersey, USA: Pearson), 165.
Hibbert, L., Taylor, G. (2022). Improving phosphate use efficiency in the aquatic crop watercress (Nasturtium officinale). Horticulture Res. 9, uhac011. doi: 10.1093/hr/uhac011
Hoehenwarter, W., Mönchgesang, S., Neumann, S., Majovsky, P., Abel, S., Müller, J. (2016). Comparative expression profiling reveals a role of the root apoplast in local phosphate response. BMC Plant Biol. 16, 106. doi: 10.1186/s12870-016-0790-8
Howard-Williams, C., Davies, J., Pickmere, S. (1982). The Dynamics of growth, the effects of changing area and nitrate uptake by watercress nasturtium officinale R. Br. in a New Zealand stream. J. Appl. Ecol. 19, 589–601. doi: 10.2307/2403491
Hsieh, M.-H., Goodman, H. M. (2002). Molecular characterization of a novel gene family encoding ACT domain repeat proteins in Arabidopsis. Plant Physiol. 130, 1797–1806. doi: 10.1104/pp.007484
Hyun, T. K., Graaff, E., van der, Albacete, A., Eom, S. H., Großkinsky, D. K., Böhm, H., et al. (2014). The arabidopsis PLAT domain protein1 is critically involved in abiotic stress tolerance. PloS One 9, e112946. doi: 10.1371/journal.pone.0112946
Irfan, M., Aziz, T., Maqsood, M. A., Bilal, H. M., Siddique, K. H. M., Xu, M. (2020). Phosphorus (P) use efficiency in rice is linked to tissue-specific biomass and P allocation patterns. Sci. Rep. 10, 4278. doi: 10.1038/s41598-020-61147-3
Ishiyama, K., Inoue, E., Watanabe-Takahashi, A., Obara, M., Yamaya, T., Takahashi, H. (2004). Kinetic properties and ammonium-dependent regulation of cytosolic isoenzymes of glutamine synthetase in Arabidopsis. J. Biol. Chem. 279, 16598–16605. doi: 10.1074/jbc.M313710200
Ismond, K. P., Dolferus, R., De Pauw, M., Dennis, E. S., Good, A. G. (2003). Enhanced low oxygen survival in arabidopsis through increased metabolic flux in the fermentative pathway. Plant Physiol. 132, 1292–1302. doi: 10.1104/pp.103.022244
Jamsheer K, M., Laxmi, A. (2015). Expression of Arabidopsis FCS-Like Zinc finger genes is differentially regulated by sugars, cellular energy level, and abiotic stress. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00746
Jarvie, H. P., Smith, D. R., Norton, L. R., Edwards, F. K., Bowes, M. J., King, S. M., et al. (2018). Phosphorus and nitrogen limitation and impairment of headwater streams relative to rivers in Great Britain: A national perspective on eutrophication. Sci. Total Environ. 621, 849–862. doi: 10.1016/j.scitotenv.2017.11.128
Jeon, J., Bong, S. J., Park, J. S., Park, Y.-K., Arasu, M. V., Al-Dhabi, N. A., et al. (2017). De novo transcriptome analysis and glucosinolate profiling in watercress (Nasturtium officinale R. Br.). BMC Genomics 18, 1–14. doi: 10.1186/s12864-017-3792-5
Jiménez-López, D., Muñóz-Belman, F., González-Prieto, J. M., Aguilar-Hernández, V., Guzmán, P. (2018). Repertoire of plant RING E3 ubiquitin ligases revisited: New groups counting gene families and single genes. PloS One 13, e0203442. doi: 10.1371/journal.pone.0203442
Joubès, J., Raffaele, S., Bourdenx, B., Garcia, C., Laroche-Traineau, J., Moreau, P., et al. (2008). The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Mol. Biol. 67, 547–566. doi: 10.1007/s11103-008-9339-z
Juszczuk, I., Malusà, E., Rychter, A. M. (2001). Oxidative stress during phosphate deficiency in roots of bean plants (Phaseolus vulgaris L.). J. Plant Physiol. 158, 1299–1305. doi: 10.1078/0176-1617-00541
Karthikeyan, A. S., Varadarajan, D. K., Jain, A., Held, M. A., Carpita, N. C., Raghothama, K. G. (2007). Phosphate starvation responses are mediated by sugar signaling in Arabidopsis. Planta 225, 907–918. doi: 10.1007/s00425-006-0408-8
Kiefer, C., Willing, E.-M., Jiao, W.-B., Sun, H., Piednoël, M., Hümann, U., et al. (2019). Interspecies association mapping links reduced CG to TG substitution rates to the loss of gene-body methylation. Nat. Plants 5, 846–855. doi: 10.1038/s41477-019-0486-9
Kliebenstein, D. J., Lambrix, V. M., Reichelt, M., Gershenzon, J., Mitchell-Olds, T. (2001). Gene duplication in the diversification of secondary metabolism. Plant Cell 13, 681–694. doi: 10.1105/tpc.13.3.681
Kobayashi, K., Awai, K., Nakamura, M., Nagatani, A., Masuda, T., Ohta, H. (2009). Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J. 57, 322–331. doi: 10.1111/j.1365-313X.2008.03692.x
Kötting, O., Santelia, D., Edner, C., Eicke, S., Marthaler, T., Gentry, M. S., et al. (2009). STARCH-EXCESS4 is a laforin-like Phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana. Plant Cell 21, 334–346. doi: 10.1105/tpc.108.064360
Kurdyukov, S., Faust, A., Nawrath, C., Bär, S., Voisin, D., Efremova, N., et al. (2006). The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in arabidopsis. Plant Cell 18, 321–339. doi: 10.1105/tpc.105.036079
Lan, V. P., Li, W., Schmidt, W. (2015). ‘Omics’ Approaches towards understanding plant phosphorus acquisition and use. Annu. Plant Rev. 8, 65–97. doi: 10.1002/9781118958841.ch3
Lapis-Gaza, H. R., Jost, R., Finnegan, P. M. (2014). Arabidopsis PHOSPHATE TRANSPORTER1 genes PHT1;8 and PHT1;9 are involved in root-to-shoot translocation of orthophosphate. BMC Plant Biol. 14, 1–19. doi: 10.1186/s12870-014-0334-z
Lee, S.-B., Jung, S.-J., Go, Y.-S., Kim, H.-U., Kim, J.-K., Cho, H.-J., et al. (2009). Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J. 60, 462–475. doi: 10.1111/j.1365-313X.2009.03973.x
Lelandais-Brière, C., Jovanovic, M., Torres, G. A. M., Perrin, Y., Lemoine, R., Corre-Menguy, F., et al. (2007). Disruption of AtOCT1, an organic cation transporter gene, affects root development and carnitine-related responses in Arabidopsis. Plant J. 51, 154–164. doi: 10.1111/j.1365-313X.2007.03131.x
Leyva, A., Quintana, A., Sánchez, M., Rodríguez, E. N., Cremata, J., Sánchez, J. C. (2008). Rapid and sensitive anthrone-sulfuric acid assay in microplate format to quantify carbohydrate in biopharmaceutical products: method development and validation. Biologicals 36, 134–141. doi: 10.1016/j.biologicals.2007.09.001
Li, J., Du, A., Liu, P., Tian, X., Jin, Y., Yi, Z., et al. (2021). High starch accumulation mechanism and phosphorus utilization efficiency of duckweed (Landoltia punctata) under phosphate starvation. Ind. Crops Products 167, 113529. doi: 10.1016/j.indcrop.2021.113529
Li, L., Teixeira da Silva, J., Cao, B. (2007b). Aquatic vegetable production and research in China. Asian Australas. J. Plant Sci. Biotechnol. 1, 37–42.
Li, K., Xu, C., Zhang, K., Yang, A., Zhang, J. (2007a). Proteomic analysis of roots growth and metabolic changes under phosphorus deficit in maize (Zea mays L.) plants. PROTEOMICS 7, 1501–1512. doi: 10.1002/pmic.200600960
Liao, Y., Smyth, G. K., Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Libertini, E., Li, Y., McQueen-Mason, S. J. (2004). Phylogenetic analysis of the plant endo-beta-1,4-glucanase gene family. J. Mol. Evol. 58, 506–515. doi: 10.1007/s00239-003-2571-x
Liu, T.-Y., Aung, K., Tseng, C.-Y., Chang, T.-Y., Chen, Y.-S., Chiou, T.-J. (2011). Vacuolar ca2+/H+ Transport activity is required for systemic phosphate homeostasis involving shoot-to-root signaling in arabidopsis. Plant Physiol. 156, 1176–1189. doi: 10.1104/pp.111.175257
Liu, T., Du, L., Li, Q., Kang, J., Guo, Q., Wang, S. (2021). AtCRY2 negatively regulates the functions of atANN2 and atANN3 in drought tolerance by affecting their subcellular localization and transmembrane ca2+ Flow. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.754567
Liu, T.-Y., Huang, T.-K., Tseng, C.-Y., Lai, Y.-S., Lin, S.-I., Lin, W.-Y., et al. (2012). PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in arabidopsis. Plant Cell 24, 2168–2183. doi: 10.1105/tpc.112.096636
Lutfiyya, L. L., Xu, N., D’Ordine, R. L., Morrell, J. A., Miller, P. W., Duff, S. M. G. (2007). Phylogenetic and expression analysis of sucrose phosphate synthase isozymes in plants. J. Plant Physiol. 164, 923–933. doi: 10.1016/j.jplph.2006.04.014
MacGregor, D. R., Deak, K. I., Ingram, P. A., Malamy, J. E. (2008). Root system architecture in arabidopsis grown in culture is regulated by sucrose uptake in the aerial tissues. Plant Cell 20, 2643–2660. doi: 10.1105/tpc.107.055475
MacNeill, G. J., Mehrpouyan, S., Minow, M. A. A., Patterson, J. A., Tetlow, I. J., Emes, M. J. (2017). Starch as a source, starch as a sink: the bifunctional role of starch in carbon allocation. J. Exp. Bot. 68, 4433–4453. doi: 10.1093/jxb/erx291
Martin, T., Sharma, R., Sippel, C., Waegemann, K., Soll, J., Vothknecht, U. C. (2006). A protein kinase family in arabidopsis phosphorylates chloroplast precursor proteins *. J. Biol. Chem. 281, 40216–40223. doi: 10.1074/jbc.M606580200
Maurino, V. G., Grube, E., Zielinski, J., Schild, A., Fischer, K., Flügge, U.-I. (2006). Identification and expression analysis of twelve members of the nucleobase–ascorbate transporter (NAT) gene family in arabidopsis thaliana. Plant Cell Physiol. 47, 1381–1393. doi: 10.1093/pcp/pcl011
May, A., Spinka, M., Köck, M. (2012). Arabidopsis thaliana PECP1: enzymatic characterization and structural organization of the first plant phosphoethanolamine/phosphocholine phosphatase. Biochim. Biophys. Acta 1824, 319–325. doi: 10.1016/j.bbapap.2011.10.003
Meng, X., Chen, W.-W., Wang, Y.-Y., Huang, Z.-R., Ye, X., Chen, L.-S., et al. (2021). Effects of phosphorus deficiency on the absorption of mineral nutrients, photosynthetic system performance and antioxidant metabolism in Citrus grandis. PloS One 16, e0246944. doi: 10.1371/journal.pone.0246944
Müller, R., Morant, M., Jarmer, H., Nilsson, L., Nielsen, T. H. (2007). Genome-Wide Analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol. 143, 156–171. doi: 10.1104/pp.106.090167
Müller, J. T., van Veen, H., Bartylla, M. M., Akman, M., Pedersen, O., Sun, P., et al. (2021). Keeping the shoot above water – submergence triggers antithetical growth responses in stems and petioles of watercress (Nasturtium officinale). New Phytol. 229, 140–155. doi: 10.1111/nph.16350
Muneer, S., Jeong, B. R. (2015). Proteomic analysis provides new insights in phosphorus homeostasis subjected to pi (Inorganic phosphate) starvation in tomato plants (Solanum lycopersicum L.). PloS One 10, e0134103. doi: 10.1371/journal.pone.0134103
Nagarajan, V. K., Satheesh, V., Poling, M. D., Raghothama, K. G., Jain, A. (2016). Arabidopsis MYB-related HHO2 exerts a regulatory influence on a subset of root traits and genes governing phosphate homeostasis. Plant Cell Physiol. 57, 1142–1152. doi: 10.1093/pcp/pcw063
Navarro-De la Sancha, E., Coello-Coutiño, M. P., Valencia-Turcotte, L. G., Hernández-Domínguez, E. E., Trejo-Yepes, G., Rodríguez-Sotres, R. (2007). Characterization of two soluble inorganic pyrophosphatases from Arabidopsis thaliana. Plant Sci. 172, 796–807. doi: 10.1016/j.plantsci.2006.12.011
Nell, M., Vötsch, M., Vierheilig, H., Steinkellner, S., Zitterl-Eglseer, K., Franz, C., et al. (2009). Effect of phosphorus uptake on growth and secondary metabolites of garden sage (Salvia officinalis L.). J. Sci. Food Agric. 89, 1090–1096. doi: 10.1002/jsfa.3561
Newman, R. M., Hanscom, Z., Kerfoot, W. C. (1992). The watercress glucosinolate-myrosinase system: a feeding deterrent to caddisflies, snails and amphipods. Oecologia 92, 1–7. doi: 10.1007/BF00317255
Nussaume, L., Kanno, S., Javot, H., Marin, E., Pochon, N., Ayadi, A., et al. (2011). Phosphate import in plants: focus on the PHT1 transporters. Front. Plant Sci. 2. doi: 10.3389/fpls.2011.00083
Ogden, M., Hoefgen, R., Roessner, U., Persson, S., Khan, G. A. (2018). Feeding the walls: how does nutrient availability regulate cell wall composition? Int. J. Mol. Sci. 19, 2691. doi: 10.3390/ijms19092691
Ohta, D., Fujimori, K., Mizutani, M., Nakayama, Y., Kunpaisal-Hashimoto, R., Münzer, S., et al. (2000). Molecular cloning and characterization of ATP-phosphoribosyl transferase from Arabidopsis, a key enzyme in the histidine biosynthetic pathway. Plant Physiol. 122, 907–914. doi: 10.1104/pp.122.3.907
Oliveros, J. C. (2015) Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available at: https://bioinfogp.cnb.csic.es/tools/venny/index.html.
Panahi Kokhdan, E., Khodabandehloo, H., Ghahremani, H., Doustimotlagh, A. H. (2021). A narrative review on therapeutic potentials of watercress in human disorders. Evid Based Complement Alternat Med. 2021, 5516450. doi: 10.1155/2021/5516450
Pant, B.-D., Pant, P., Erban, A., Huhman, D., Kopka, J., Scheible, W.-R. (2015). Identification of primary and secondary metabolites with phosphorus status-dependent abundance in Arabidopsis, and of the transcription factor PHR1 as a major regulator of metabolic changes during phosphorus limitation. Plant Cell Environ. 38, 172–187. doi: 10.1111/pce.12378
Payne, A. C. (2011). Harnessing the genetic diversity of watercess (Rorippa nasturtium-aquaticum) for improved morphology and anticancer benefits: underpinning data for molecular breeding. Southampton: University of Southampton.
Payne, A. C., Clarkson, G. J. J., Rothwell, S., Taylor, G. (2015). Diversity in global gene expression and morphology across a watercress ( Nasturtium officinale R. Br.) germplasm collection: first steps to breeding. Horticulture Res. 2, 1–8. doi: 10.1038/hortres.2015.29
Payne, A. C., Mazzer, A., Clarkson, G. J. J., Taylor, G. (2013). Antioxidant assays – consistent findings from FRAP and ORAC reveal a negative impact of organic cultivation on antioxidant potential in spinach but not watercress or rocket leaves. Food Sci. Nutr. 1, 439–444. doi: 10.1002/fsn3.71
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Puga, M. I., Mateos, I., Charukesi, R., Wang, Z., Franco-Zorrilla, J. M., de Lorenzo, L., et al. (2014). SPX1 is a phosphate-dependent inhibitor of PHOSPHATE STARVATION RESPONSE 1 in Arabidopsis. Proc. Natl. Acad. Sci. 111, 14947–14952. doi: 10.1073/pnas.1404654111
Qian, Y. (2021). Developing Genomic Resources in Watercress: Plant Morphology and Phytochemical Properties. Davis, California: University of California Davis.
Qian, Y., Hibbert, L. E., Katz, E., Smith, H. K., Kliebenstein, D. J., Taylor, G. (2023). Watercress yield and quality vary depending on both genotype and environment: Results from highly contrasting growing systems of California and UK. Scientia Hortic. 319, 112154. doi: 10.1016/j.scienta.2023.112154
Qian, Y., Hibbert, L. E., Milner, S., Katz, E., Kliebenstein, D. J., Taylor, G. (2022). Improved yield and health benefits of watercress grown in an indoor vertical farm. Scientia Hortic. 300, 111068. doi: 10.1016/j.scienta.2022.111068
Qin, C., Wang, X. (2002). The Arabidopsis phospholipase D family. Characterization of a calcium-independent and phosphatidylcholine-selective PLDζ1 with distinct regulatory domains. Plant Physiol. 128, 1057–1068. doi: 10.1104/pp.010928
R Core Team. (2021). R: A language and Environment for Statistical Computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Remy, E., Cabrito, T. R., Batista, R. A., Teixeira, M. C., Sá-Correia, I., Duque, P. (2012). The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol. 195, 356–371. doi: 10.1111/j.1469-8137.2012.04167.x
Richards, S., Paterson, E., Withers, P. J. A., Stutter, M. (2016). Septic tank discharges as multi-pollutant hotspots in catchments. Sci. Total Environ. 542, 854–863. doi: 10.1016/j.scitotenv.2015.10.160
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C.-Z., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Romano, P. G. N., Horton, P., Gray, J. E. (2004). The Arabidopsis cyclophilin gene family. Plant Physiol. 134, 1268–1282. doi: 10.1104/pp.103.022160
Roppolo, D., Boeckmann, B., Pfister, A., Boutet, E., Rubio, M. C., Dénervaud-Tendon, V., et al. (2014). Functional and evolutionary analysis of the CASPARIAN STRIP MEMBRANE DOMAIN PROTEIN family. Plant Physiol. 165, 1709–1722. doi: 10.1104/pp.114.239137
Rouached, H., Arpat, A. B., Poirier, Y. (2010). Regulation of phosphate starvation responses in plants: signaling players and cross-talks. Mol. Plant 3, 288–299. doi: 10.1093/mp/ssp120
Rychter, A. M., Randall, D. D. (1994). The effect of phosphate deficiency on carbohydrate metabolism in bean roots. Physiologia Plantarum 91, 383–388. doi: 10.1111/j.1399-3054.1994.tb02964.x
Saatian, B., Austin, R. S., Tian, G., Chen, C., Nguyen, V., Kohalmi, S. E., et al. (2018). Analysis of a novel mutant allele of GSL8 reveals its key roles in cytokinesis and symplastic trafficking in Arabidopsis. BMC Plant Biol. 18, 295. doi: 10.1186/s12870-018-1515-y
Sanda, S., Leustek, T., Theisen, M. J., Garavito, R. M., Benning, C. (2001). Recombinant Arabidopsis SQD1 converts udp-glucose and sulfite to the sulfolipid head group precursor UDP-sulfoquinovose in vitro. J. Biol. Chem. 276, 3941–3946. doi: 10.1074/jbc.M008200200
Sandoval, F. J., Roje, S. (2005). An FMN hydrolase is fused to a riboflavin kinase homolog in plants. J. Biol. Chem. 280, 38337–38345. doi: 10.1074/jbc.M500350200
Schachtman, D. P., Reid, R. J., Ayling, S. M. (1998). Phosphorus uptake by plants: from soil to cell. Plant Physiol. 116, 447–453. doi: 10.1104/pp.116.2.447
Schindler, D. W., Hecky, R. E., Findlay, D. L., Stainton, M. P., Parker, B. R., Paterson, M. J., et al. (2008). Eutrophication of lakes cannot be controlled by reducing nitrogen input: Results of a 37-year whole-ecosystem experiment. Proc. Natl. Acad. Sci. U.S.A. 105, 11254–11258. doi: 10.1073/pnas.0805108105
Schluepmann, H., van Dijken, A., Aghdasi, M., Wobbes, B., Paul, M., Smeekens, S. (2004). Trehalose mediated growth inhibition of arabidopsis seedlings is due to trehalose-6-phosphate accumulation. Plant Physiol. 135, 879–890. doi: 10.1104/pp.104.039503
Schneider, C. A., Rasband, W. S., Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Schnurr, J., Shockey, J., Browse, J. (2004). The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 16, 629–642. doi: 10.1105/tpc.017608
Schuchardt, J. P., Hahn, A., Greupner, T., Wasserfurth, P., Rosales-López, M., Hornbacher, J., et al. (2019). Watercress – cultivation methods and health effects. J. Appl. Bot. Food Qual. 92, 232–239. doi: 10.5073/JABFQ.2019.092.032
Searle, F. (2019)The Watercress Company targets Spain. In: Fruitnet. Available at: https://www.fruitnet.com/fresh-produce-journal/the-watercress-company-targets-Spain/179533.article (Accessed January 2, 2023).
Serrano, I., Gu, Y., Qi, D., Dubiella, U., Innes, R. W. (2014). The arabidopsis EDR1 protein kinase negatively regulates the ATL1 E3 ubiquitin ligase to suppress cell death. Plant Cell 26, 4532–4546. doi: 10.1105/tpc.114.131540
Shanmugabalaji, V., Grimm, B., Kessler, F. (2020). Characterization of a plastoglobule-localized SOUL4 heme-binding protein in arabidopsis thaliana. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00002
Shen, Q., Wen, Z., Dong, Y., Li, H., Miao, Y., Shen, J. (2018). The responses of root morphology and phosphorus-mobilizing exudations in wheat to increasing shoot phosphorus concentration. AoB Plants 10, ply054. doi: 10.1093/aobpla/ply054
Smyczynski, C., Roudier, F., Gissot, L., Vaillant, E., Grandjean, O., Morin, H., et al. (2006). The C terminus of the immunophilin PASTICCINO1 is required for plant development and for interaction with a NAC-like transcription factor. J. Biol. Chem. 281, 25475–25484. doi: 10.1074/jbc.M601815200
Strabala, T. J., O’donnell, P. J., Smit, A.-M., Ampomah-Dwamena, C., Martin, E. J., Netzler, N., et al. (2006). Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol. 140, 1331–1344. doi: 10.1104/pp.105.075515
Tannert, M., May, A., Ditfe, D., Berger, S., Balcke, G. U., Tissier, A., et al. (2018). Pi starvation-dependent regulation of ethanolamine metabolism by phosphoethanolamine phosphatase PECP1 in Arabidopsis roots. J. Exp. Bot. 69, 467–481. doi: 10.1093/jxb/erx408
Thieme, C. J., Rojas-Triana, M., Stecyk, E., Schudoma, C., Zhang, W., Yang, L., et al. (2015). Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 1, 15025. doi: 10.1038/nplants.2015.25
Todd, J., Post-Beittenmiller, D., Jaworski, J. G. (1999). KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J. 17, 119–130. doi: 10.1046/j.1365-313x.1999.00352.x
Tognolli, M., Penel, C., Greppin, H., Simon, P. (2002). Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene 288, 129–138. doi: 10.1016/s0378-1119(02)00465-1
Töller, A., Brownfield, L., Neu, C., Twell, D., Schulze-Lefert, P. (2008). Dual function of Arabidopsis glucan synthase-like genes GSL8 and GSL10 in male gametophyte development and plant growth. Plant J. 54, 911–923. doi: 10.1111/j.1365-313X.2008.03462.x
Torabinejad, J., Donahue, J. L., Gunesekera, B. N., Allen-Daniels, M. J., Gillaspy, ,. G. E. (2009). VTC4 is a bifunctional enzyme that affects myoinositol and ascorbate biosynthesis in plants. Plant Physiol. 150, 951–961. doi: 10.1104/pp.108.135129
Trejo-Téllez, L. I., Estrada-Ortiz, E., Gómez-Merino, F. C., Becker, C., Krumbein, A., Schwarz, D. (2019). Flavonoid, nitrate and glucosinolate concentrations in brassica species are differentially affected by photosynthetically active radiation, phosphate and phosphite. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00371
Ulmasov, T., Hagen, G., Guilfoyle, T. J. (1997). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868. doi: 10.1126/science.276.5320.1865
Underbill, E. W. (1965). Biosynthesis of mustard oil glucosides: V. Formation of gluconasturtiin from L-γ-phenylbutyrine-C14-N15 in watercress. Can. J. Biochem. 43, 179–187. doi: 10.1139/o65-025
USDA (2019) Market News - Fruit and Vegetable - Search by Reports. Available at: https://www.marketnews.usda.gov/mnp/fv-report-top-filters?type=termPrice&commAbr=BASIL&locName=&commName=BASIL&startIndex=1&rowDisplayMax=25&portal=fv&navType=byComm&navClass=HERBS&termNavClass=&shipNavClass=&movNavClass=&stateID=&volume=&repType=termPriceDaily&locAbr=&environment=&varName=&organic=&repDate=03%2F01%2F2019&Go=Go (Accessed April 15, 2020).
Veneklaas, E. J., Lambers, H., Bragg, J., Finnegan, P. M., Lovelock, C. E., Plaxton, W. C., et al. (2012). Opportunities for improving phosphorus-use efficiency in crop plants. New Phytol. 195, 306–320. doi: 10.1111/j.1469-8137.2012.04190.x
Veronica, N., Subrahmanyam, D., Kiran, T., Yugandhar, P., Bhadana, V. P., Padma, V., et al. (2017). Influence of low phosphorus concentration on leaf photosynthetic characteristics and antioxidant response of rice genotypes. Photosynthetica 55, 285–293. doi: 10.1007/s11099-016-0640-4
Verrier, P. J., Bird, D., Burla, B., Dassa, E., Forestier, C., Geisler, M., et al. (2008). Plant ABC proteins–a unified nomenclature and updated inventory. Trends Plant Sci. 13, 151–159. doi: 10.1016/j.tplants.2008.02.001
Volkert, K., Debast, S., Voll, L. M., Voll, H., Schießl, I., Hofmann, J., et al. (2014). Loss of the two major leaf isoforms of sucrose-phosphate synthase in Arabidopsis thaliana limits sucrose synthesis and nocturnal starch degradation but does not alter carbon partitioning during photosynthesis. J. Exp. Bot. 65, 5217–5229. doi: 10.1093/jxb/eru282
Voutsina, N. (2017). Elucidating the genomics of nutritional and morphological traits in watercress (Nasturtium officinale R. Br.): The first genomic resources. Southampton: University of Southampton.
Voutsina, N., Payne, A. C., Hancock, R. D., Clarkson, G. J. J., Rothwell, S. D., Chapman, M. A., et al. (2016). Characterization of the watercress (Nasturtium officinale R. Br.; Brassicaceae) transcriptome using RNASeq and identification of candidate genes for important phytonutrient traits linked to human health. BMC Genomics 17, 1–15. doi: 10.1186/s12864-016-2704-4
Wan, J., Wang, R., Zhang, P., Sun, L., Ju, Q., Huang, H., et al. (2021). MYB70 modulates seed germination and root system development in Arabidopsis. iScience 24, 103228. doi: 10.1016/j.isci.2021.103228
Wang, J., Song, J., Clark, G., Roux, S. J. (2018). ANN1 and ANN2 function in post-phloem sugar transport in root tips to affect primary root growth. Plant Physiol. 178, 390–401. doi: 10.1104/pp.18.00713
Wang, X., Wang, H.-F., Chen, Y., Sun, M.-M., Wang, Y., Chen, Y.-F. (2020). The transcription factor NIGT1.2 modulates both phosphate uptake and nitrate influx during phosphate starvation in arabidopsis and maize. Plant Cell 32, 3519–3534. doi: 10.1105/tpc.20.00361
Welinder, K. G., Justesen, A. F., Kjærsgård, I. V. H., Jensen, R. B., Rasmussen, S. K., Jespersen, H. M., et al. (2002). Structural diversity and transcription of class III peroxidases from Arabidopsis thaliana. Eur. J. Biochem. 269, 6063–6081. doi: 10.1046/j.1432-1033.2002.03311.x
Westheimer, F. H. (1987). Why nature chose phosphates. Science 235, 1173–1178. doi: 10.1126/science.2434996
White, B. (2020) Effects of watercress farming on fish populations. Available at: https://research.brighton.ac.uk/en/studentTheses/effects-of-watercress-farming-on-fish-populations (Accessed May 26, 2021).
Withers, P. J., Jordan, P., May, L., Jarvie, H. P., Deal, N. E. (2013). Do septic tank systems pose a hidden threat to water quality? Front. Ecol. Environ. 12, 123–130. doi: 10.1890/130131
Wu, H., Li, L., Du, J., Yuan, Y., Cheng, X., Ling, H.-Q. (2005). Molecular and biochemical characterization of the Fe(III) chelate reductase gene family in Arabidopsis thaliana. Plant Cell Physiol. 46, 1505–1514. doi: 10.1093/pcp/pci163
Wu, Q., Wu, J., Hu, P., Zhang, W., Ma, Y., Yu, K., et al. (2023). Quantification of the three-dimensional root system architecture using an automated rotating imaging system. Plant Methods 19, 11. doi: 10.1186/s13007-023-00988-1
Xiao, M., Li, Z., Zhu, L., Wang, J., Zhang, B., Zheng, F., et al. (2021). The multiple roles of ascorbate in the abiotic stress response of plants: antioxidant, cofactor, and regulator. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.598173
Yadav, V., Molina, I., Ranathunge, K., Castillo, I. Q., Rothstein, S. J., Reed, J. W. (2014). ABCG transporters are required for suberin and pollen wall extracellular barriers in Arabidopsis. Plant Cell 26, 3569–3588. doi: 10.1105/tpc.114.129049
Yang, M., Ding, G., Shi, L., Xu, F., Meng, J. (2011). Detection of QTL for phosphorus efficiency at vegetative stage in Brassica napus. Plant Soil 339, 97–111. doi: 10.1007/s11104-010-0516-x
Yu, T.-S., Kofler, H., Häusler, R. E., Hille, D., Flügge, U.-I., Zeeman, S. C., et al. (2001). The arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell 13, 1907–1918. doi: 10.1105/TPC.010091
Yu, Q., Tian, H., Yue, K., Liu, J., Zhang, B., Li, X., et al. (2016). A P-loop NTPase regulates quiescent center cell division and distal stem cell identity through the regulation of ROS homeostasis in arabidopsis root. PloS Genet. 12, e1006175. doi: 10.1371/journal.pgen.1006175
Yu, B., Xu, C., Benning, C. (2002). Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc. Natl. Acad. Sci. U.S.A. 99, 5732–5737. doi: 10.1073/pnas.082696499
Zhang, K., Liu, H., Tao, P., Chen, H. (2014). Comparative proteomic analyses provide new insights into low phosphorus stress responses in maize leaves. PloS One 9, e98215. doi: 10.1371/journal.pone.0098215
Zhang, J.-W., Xu, L., Wu, Y.-R., Chen, X.-A., Liu, Y., Zhu, S.-H., et al. (2012). OsGLU3, a putative membrane-bound endo-1,4-beta-glucanase, is required for root cell elongation and division in rice (Oryza sativa L.). Mol. Plant 5, 176–186. doi: 10.1093/mp/ssr084
Zhou, H.-L., He, S.-J., Cao, Y.-R., Chen, T., Du, B.-X., Chu, C.-C., et al. (2006). OsGLU1, A putative membrane-bound endo-1,4-ß-D-glucanase from rice, affects plant internode elongation. Plant Mol. Biol. 60, 137–151. doi: 10.1007/s11103-005-2972-x
Keywords: abiotic stress, Brassica, fertilizer, nutrition, Nasturtium officinale, phosphorus, transcriptome
Citation: Hibbert LE, Qian Y, Smith HK, Milner S, Katz E, Kliebenstein DJ and Taylor G (2023) Making watercress (Nasturtium officinale) cropping sustainable: genomic insights into enhanced phosphorus use efficiency in an aquatic crop. Front. Plant Sci. 14:1279823. doi: 10.3389/fpls.2023.1279823
Received: 18 August 2023; Accepted: 10 October 2023;
Published: 07 November 2023.
Edited by:
Antonio Pannico, University of Naples Federico II, ItalyReviewed by:
Maharajan Theivanayagam, Rajagiri College of Social Sciences, IndiaCopyright © 2023 Hibbert, Qian, Smith, Milner, Katz, Kliebenstein and Taylor. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gail Taylor, Z3RheWxvckB1Y2RhdmlzLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.