
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 24 October 2023
Sec. Plant Pathogen Interactions
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1279502
This article is part of the Research Topic Wheat Disease Resistance: Diagnosis, Germplasm Mining, and Molecular Breeding View all 15 articles
Plant defensins are widely distributed in the leaves, fruits, roots, stems, seeds, and tubers. Research shows that defensin in plants play a significant role in physiological metabolism, growth and development. Plant defensins can kill and suppress a variety of pathogenic bacteria. In this study, we understand the phylogenetic relationships, protein characterization, chromosomal localization, promoter and gene structural features of the TaPDFs family through sequence alignment and conserved protein structural domain analysis. A total of 73 PDF gene members in wheat, 15 PDF genes in maize, and 11 PDF genes in rice were identified. A total of 35, 65, and 34 PDF gene members were identified in the genomes of Ae. tauschii, T. urartu, and T. dicoccoides, respectively. TaPDF4.9 and TaPDF2.15 were constructed into pART27 vector with YFP by homologous recombination for subcellular localization analysis. Subcellular localization results showed that TaPDF4.9 and TaPDF2.15 were basically located in the cell membrane and cytoplasm, and TaPDF4.9 was also located in the nucleus. TaPDF4.9 and TaPDF2.15 could inhibit the infection of Phytophthora infestans strain ‘88069’. The results suggest that TaPDFs may be able to improve disease resistance. The study of wheat defensins will be beneficial for improving wheat yield and provides a theoretical basis for research on resistance to wheat diseases.
Plants have their own defense systems, and when they are infected with external pathogens such as fungi and oomycetes, they will produce a series of antagonistic substances themselves (Sathoff et al., 2019). Antagonistic substances are mainly categorized into some low molecular weight secondary metabolites or some low molecular weight short peptides less than 100 amino acids to prevent the propagation and expansion of the pathogen and further invasion of the host plant (Terras et al., 1992). Plant defenses are among the short, low molecular weight peptides that are widely distributed in plants. (De Coninck et al., 2013).
There are 45-54 amino acid residues in plant defensins. The encoded proteins were approximately 5 kDa, which contain eight conserved cysteine residues forming four pairs of disulfide bonds (Cys4-Cys7, Cys3-Cys6, Cys2-Cys5 and Cys1-Cys8), among which, the most conserved are the two pairs, Cys2-Cys5 and Cys1-Cys8 (Thomma et al., 2002). It was shown that the solution structure of radish defensin (Rs-AFP1) by nuclear magnetic resonance (NMR) consists of three reverse parallel β-folded lamellae and one α-helix, forming a stable βαβββ structure with four pairs of disulfide bonds acting as support, and the Csαβ modal sequence (Cysteine stabilized αβ motif) was found, fixing a pair of disulfide bonds on the lamellae in the α-helix and β-fold (Vriens et al., 2016). The β-folded lamellar signature sequence (Cys-Xaa-Cys) and the α-helical signature sequence (Cys-Xaa-Xaa-Xaa-Cys) are common to most plant defensins (Parisi et al., 2019). The Cysteine stabilized αβ motif indicates that plant defensins are more structurally stable and can be stably expressed in many plants. Defensins have a special resistance mechanism. Defensins act mainly on the cell membranes of pathogenic microorganisms, binding to sphingolipids on the outer side of phospholipids. Defensins are able to disrupt the cell membranes of certain fungi, causing changes in the anions and cations on the cell membrane. Thus defensins disrupt the cell membrane of pathogenic microorganisms, altering their permeability and creating phenomena such as Ca2+ inward flow, K+ outward, and medium liquefaction (Shahmiri et al., 2023).
Most phytodefensins are resistant to fungal pathogens and ward off diseases caused by fungi. It has been found that most defensins are induced to be expressed by fungal infection. GXCX3-9C has antifungal activity and is a core motif in plant defensins. (Sagaram et al., 2011). The core motif peptides of defensins in Medicago truncatula are MtDef4 and MtDef5. And these defensins show high activity against both plant and human bacterial pathogens (Andrew Edward Sathoff et al., 2019). The carrot defensin Rs-AFP2 binds to the nerve sphingolipid glucosylceramide (GlcCer) and acts on the cell membrane, with the binding site being the sphingolipid on the outside of the phospholipid. This in turn disrupts the cell membrane and alters its permeability, resulting in Ca2+ in-flow, K+ out-flow, and liquefaction of the culture medium (Sadhu et al., 2023). In addition, it has been shown that the tolerance of certain plants to the heavy metal zinc is related to phyto-defensin. Reduced Expression of Plant Defensin 1 in Arabidopsis Leads to Increased Resistance to Pathogenic Bacteria and Zinc Toxicity (Nguyen et al., 2023). Phytodefensins have a very broad development prospect as a new antifungal drug. Research has shown that plant-derived AMPs can be used as alternative molecules to overcome pathogen resistance (Lima et al., 2022). This property can therefore be utilized to research new drugs to fight cancer. Candida otitis is a pan-resistant pathogenic yeast that can treat immunocompromised patients. Phytodefensins can influence the virulence properties of clinical strains of C. auris (Kamli et al., 2022). Plant defensins PaDef and γ-thionin are potential angiogenicmodulators of the VEGF activity on endothelial cells (Falcón-Ruiz et al., 2023). Therefore, plant defensins, with their broad-spectrum, antimicrobial and high efficiency, provide new ideas for the research and development of new antifungal and antitumor drugs. With the in-depth exploration of its mechanism of action, it is expected to be put more into the industrial, medical and other fields to play a role.
Wheat is one of the major grain crops in China and has an important position in food production, with more than 1/3 of the world’s population taking it as their staple food. Since the growth of wheat is attacked by diseases, it is necessary to tap the relevant genes to breed disease-resistant varieties. A review of the literature shows that the PDF gene family is more fully reported in Arabidopsis. PDF gene family analysis is not currently reported in wheat, maize and rice. In study, using Arabidopsis PDF proteins (Araport11) as query, PDF genes in wheat were identified. Moreover, PDF protein characters and the phylogenetic relationships were analyzed. The function of PDF was explored through a series of analyses including chromosomal localization, conserved structure and gene structural domains of the wheat PDF gene. This study provides the first systematic certification and classification of the PDF genes family in wheat and its subgenomic donors. Genome-wide identification of TaPDFs will enrich the study of plant disease resistance genes and provide a basis for wheat varieties breeding.
The 15 protein sequences from Arabidopsis PDF family were collected from TAIR database (http://www.Arabidopsis.org/index.jsp) (Mäser et al., 2001), and served as query sequences to research PDF genes in wheat genome (IWGSC v1.1), Maize genome (maizeGDB, B73 RefGen_v5, https://www.maizegdb.org/), and rice genome (RGAP, RGAP7, http://rice.plantbiology.msu.edu/index.shtml) using BLASTp with e-value < 10−10 (Clavijo et al., 2017). Subsequently, the PDF proteins containing the major intrinsic protein structural domain (gamma – thiionin, PF00304) were retained after searching in the Pfam database (http://pfam.xfam.org/) (Finn et al., 2016).
The length of CDS region, molecular weight (MW), isoelectric point (pI), stability, and hydrophilicity characteristics (GRAVY) of PDF proteins in wheat, maize, and rice were analyzed using ExPASy Server 10 (https://prosite.expasy.org/PS50011) (Li et al., 2018). Signal peptide of wheat PDFs was predicted by SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/). The subcellular localization of wheat PDFs was predicted by WoLF PSORT (https://wolfpsort.hgc.jp/) and Plant - mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/).
Comparison of all protein sequences from wheat, rice, maize and Arabidopsis using ClustalW2 (Thompson et al., 1994). Methods Phylogenetic trees were constructed with MEGA 7.0 and calculated according to the maximum likelihood (ML) method (Kumar et al., 2016). Drawing and landscaping the phylogenetic tree was accomplished through the Interactive Tree of Life website (IToL, version 3.2.317, http://itol.embl.de). TaPDF gene annotation file containing information on the chromosome position were extracted from the genome annotation information GFF3 file. Afterwards the physical map was then plotted using MapInspect software (Hu and Liu, 2011). TaPDF genome annotation information was used to map the gene (exon-intron) structure using GSDS2.0 (http://gsds.cbi.pku.edu.cn/index.php) (D.-M. Huang et al., 2022). The conserved motifs of TaPDF proteins were identified by MEME (v4.9.1, http://meme-suite.org/index.html) (Zheng et al., 2017). The conserved motifs are identified by the following criteria: (1) All sequences can include multiple non overlapping motifs; (2) Up to 20 different motifs; (3) The length of the motif is 6-50 aa. The protein sequences were uploaded to SWISS-MODEL website (https://www.swissmodel.expasy.org/) in order to identify protein structure with default parameters (Arnold et al., 2005).
The 1500 bp sequence upstream of the start codon of TaPDFs were submitted to the PlantCARE database to predict the cis-acting elements (Lescot et al., 2002). The enrichment analysis of cis-acting elements was performed using the pattern enrichment analysis (AME) function in the MEME program to identify the regulatory elements (İlhan et al., 2018).
Genomic data for T. dicoccoides (v1.0.43), Ae tauschii (v4.0.43), and T. urartu (v1.43) was obtained from the EnsemblPlants database (http://plants.ensembl.org/index.html) (Alaux et al., 2018). The PDF homologous of urartu wheat (T. urartu), wild dicoccoides (T. dicoccoides), and rough goat grass (Ae. tauschii) were identified by BLASTp (threshold E<10-10, match >80%). The protein which contains the structural domain of PF00304 (Gamma-thionin) was identified as PDF gene family member. The sequences of the identified PDFs were determined by whether the fragments were duplicated or tandem duplicated on the same chromosome, and homology mapping was performed using the R package “circlize”.
In order to further study the expression patterns of TaPDF genes under different stresses, original RNA-seq data under various conditions was downloaded from the SRA database from NCBI (Appendix S1), and was mapped to the wheat reference genome using hisat2. The gene expression level was calculated by Cufflinks, which was normalized through the fragment by the exon base number per kilogram (FPKM) model per million bases (Trapnell et al., 2012). Finally, pheatmap package was used to generate the heatmap of TaPDF genes.
RNA was extracted from wheat leaves using TRIzol™ Reagent (Invitrogen). The cDNA obtained by reverse transcription was used as a template to amplify the target genes. The six highly expressed TaPDF genes TaPDF4.9, TaPDF5.4, TaPDF2.12, TaPDF2.15, TaPDF2.23, and TaPDF2.20, which were highly expressed in the transcriptome analysis, were selected for qRT-PCR analysis during Fusarium graminearum infestation. The wheat varieties used in this experiment were Yangmai 158 and Xinong 98710. The primers used in this study were listed in Appendix S2. The screened genes were analyzed by qRT-PCR on a CFX 96 Real-Time PCR System (Bio-Rad). (Mamidala et al., 2011). The fluorescence quantification reaction system consisted of 10 μL of 2× SYBR Green (Vazyme), 0.4 μL of each of the upstream and downstream primers, 2 μL of 50 ng/μL cDNA, and the addition of 7.2 μL of ddH2O, for a total volume of 20 μL. The reaction conditions were 95°C for 3 min, 95°C for 5 s, primer annealing/extension, and 58°C for 30 s. A total of 45 cycles were started from step 2. Each gene requires the use of three technical repeats. The final resultant data were used to determine relative expression levels using the 2-ΔΔCt method (Livak and Schmittgen, 2001). Use of the ADP-ribosylation factor Ta2291, stably expressed under various conditions, as an internal reference gene for qRT-PCR analysis (Huang et al., 2021).
The target gene were ligated into the pART27 and pART27 vector with YFP using a homologous recombination kit (Vazyme). The connected TaPDF2.15 and TaPDF4.9 were selected for transient expression in Nicotiana benthamiana leaves. After two to three days, the leaves were cut off, and inoculated with Phytophthora infestans strain ‘88069’, then they were placed in a humidor infecting box, stored with thetemperature at 20 °C. After four to six days, measure the lesion diameter of the inoculated leaves were measured (measure the longest and shortest). GFP signals from TaPDF4.9, TaPDF5.4, TaPDF2.12, and TaPDF2.15 trans-overexpressed leaves were observed two days after injection using laser scanning confocal microscope (Leica TCS SP8).
The sequences containing the conserved domain of PF00304 (Gamma-thionin) were retained using Pfam online tool. Finally, 73 wheat PDF members, 15 members of maize, and 11 members of rice were identified (Table 1). The PDF genes of the four species were divided into five groups according to the rootless phylogenetic trees (Figure 1). The genes are named according to the location of the components, e.g. the first gene located in Group I is named as TaPDF1.1. Most of these TaPDFs are concentrated in Group II, IV and V. Group III is only ZmPDF.
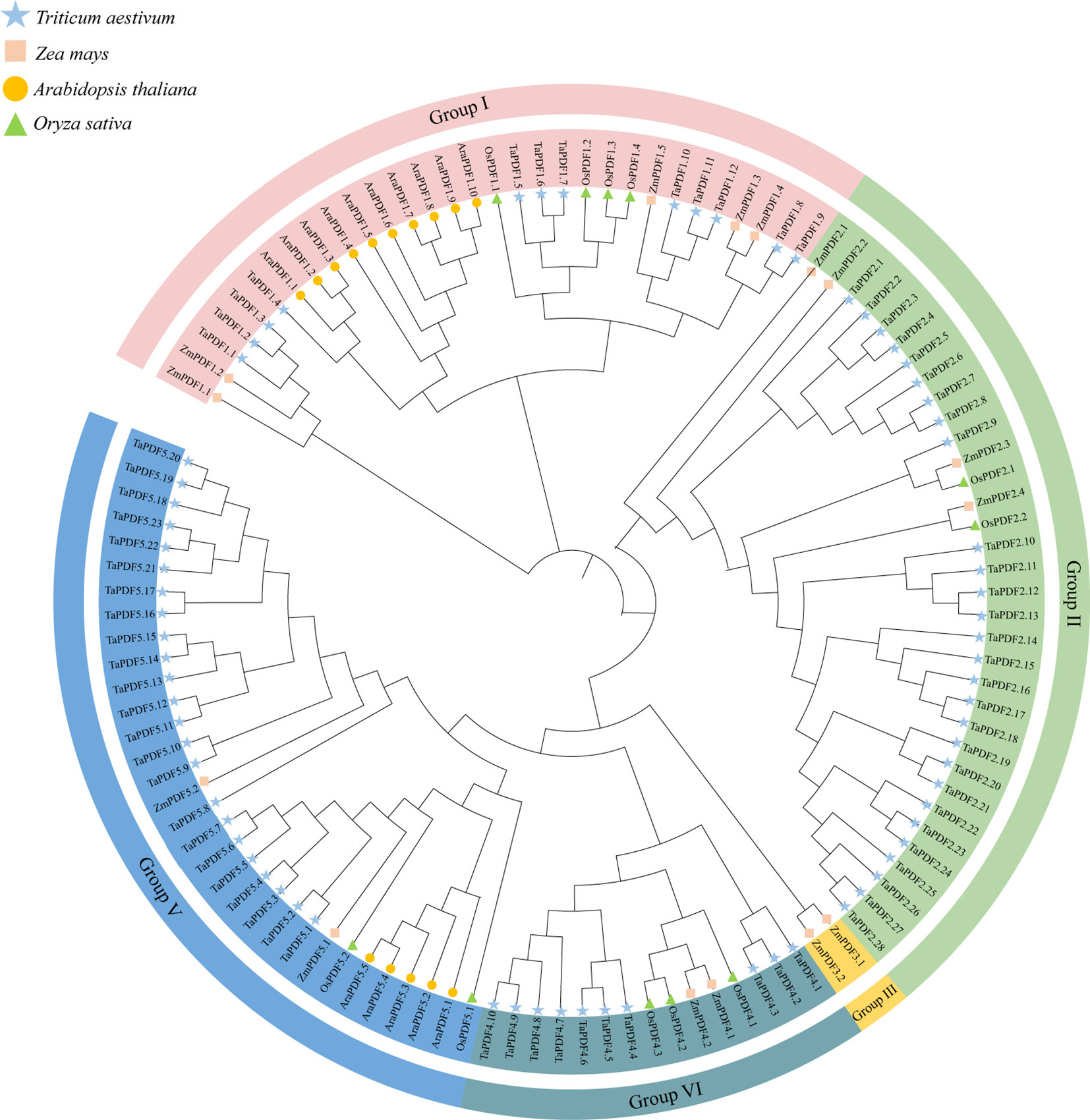
Figure 1 Phylogenetic tree of the PDF family of Triticum aestivum, Zea mays, Oryza sativa, and Arabidopsis thaliana.
As shown in Table 1, the average length of TaPDF is 80.57 aa. The average molecular weight of TaPDFs is 8.8 KD, and the average isoelectric point is 8.27, which indicates that TaPDFs are alkaline proteins. The average instability index is 47.12, which reveals the TaPDFs are unstable proteins. The average value of the aliphatic index is 73.67. The average value of hydrophilicity is 0.06. Multi website prediction of subcellular localization results shows that most TaPDF proteins are located in vacuoles, with a small portion located in the nucleus.
According to the chromosome distribution map (Figure 2), PDF genes were found to be widely distributed on each chromosome, with each chromosome containing multiple genes. As shown in Figure 2, wheat PDF contains multiple genes on its chromosomes during evolution. And each chromosome has duplicate genes. This shows that segmental duplication is the main reason for the expansion of the PDF family during the evolution of wheat.
Based on the TaPDF genomic information, the gene structure was mapped using GSDS2.0 (Figure 3). The vast majority of TaPDF genes contain non-coding regions at the 5’ and 3’ ends, and only a few TaPDFs do not contain non-coding regions. Most TaPDF genes contain two extrons.
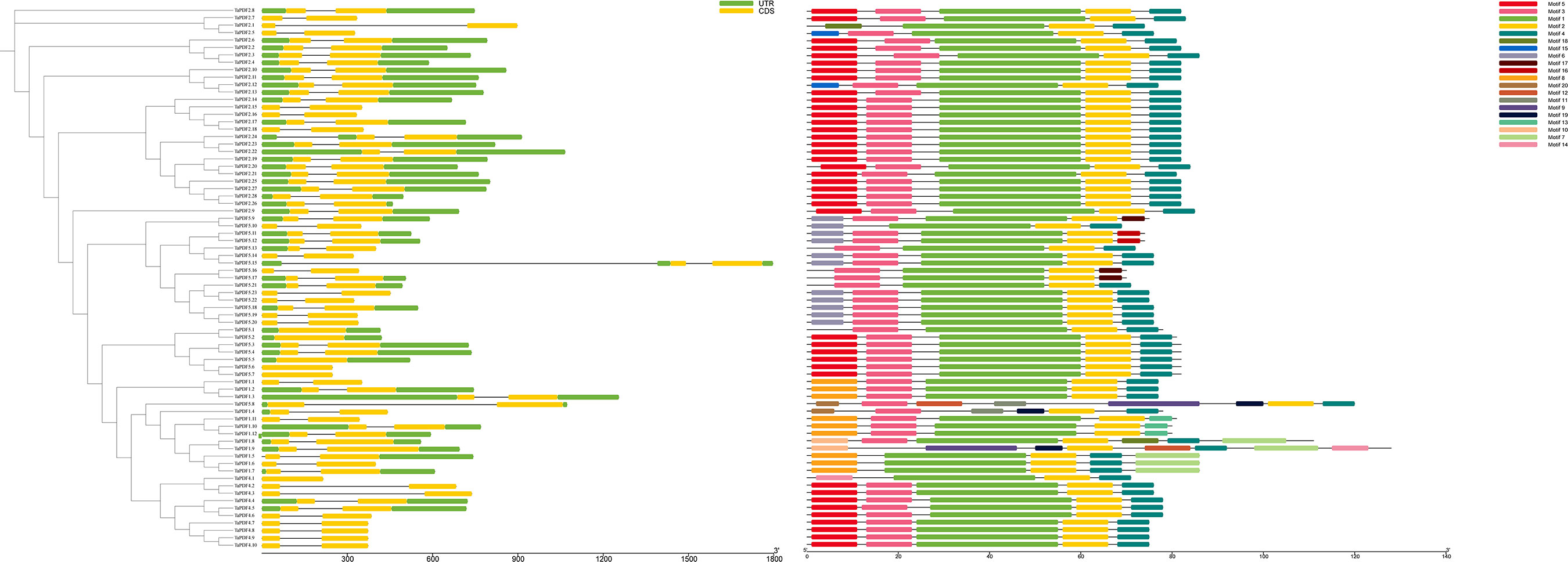
Figure 3 The evolutionary tree, motif analysis and gene structure analysis of 74 wheat PDF families.
TaPDFs all contain motifs 1, 2, 14 and 18. The sequence, position and identification of the conserved motifs in the TaPDF protein are shown in Figure 3. All identified wheat genes contain conserved structural domains of the PDF family. The wheat PDF gene family contains characterized amino acids, including a series of highly conserved active site residues consistent with previous studies on different plant species.
A phylogenetic tree was constructed using MEGA 7.0 maximum likelihood (ML) method (Figure 4A). PDF proteins can be divided into five groups, of which groups I and V contain more PDF protein members. By gene duplication events, 2940 duplicate pairs were obtained. The common wheat positional information was put into the R package for circles plot analysis (Figure 4B). The results suggest that PDFs from each genomic donor may be ancestrally similar to each other. Alternatively, initially different PDFs may be stabilized after a long domestication process and lead to alterations in protein structure and function. Homology analysis of TaPDF suggests that wheat may be repeated multiple times in polyploidy, and we hypothesize that this may have made wheat more adapted to its environment and increased the number of wheat PDF genes during evolution.
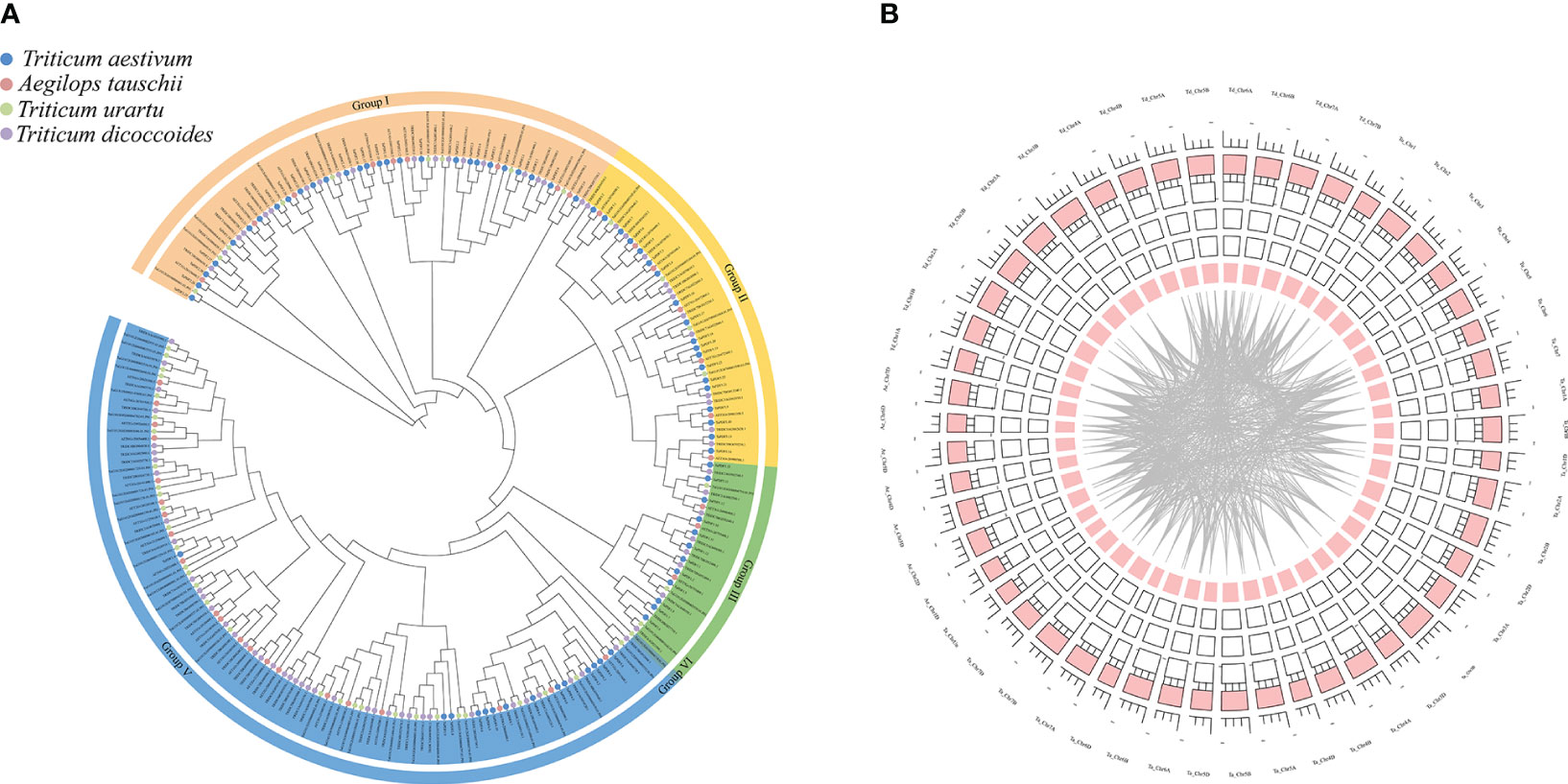
Figure 4 Phylogenetic tree and joint analysis circle diagram of the PDF family in T. aestivum, T. urartu, T. dicoccoides and Ae. tauschii. (A) is the T. aestivum, T. urartu, T. dicoccoides and Ae. tauschii evolutionary tree. (B) is the wheat homology analysis.
A total of 60 cis-acting elements were screened by intercepting a 1500 bp region upstream of the TaPDF gene. These components are divided into three categories: phytohormones, biotic stress-abiotic stress, and growth and development (Figure 5). The results indicate that the promoter sequence of TaPDF gene has multiple cis-regulatory elements, including cis-acting elements involved in defense and stress response, light-responsive elements, low-temperature-inducible elements, anaerobic-inducible elements, and drought-inducible elements biotic-abiotic taxa. The most abundant original in TaPDF is CAAT-box as promoter-associated original. The most widely distributed cis-element is the abscisic acid-responsive cis-acting element ABRE. The two core promoter elements of the same growth and development-related element are TATA-box and CAAT-box. CGTCA motifs and TGACG motifs are the major motifs of jasmonate methyl ester response elements. The most common cis-element among all genes was the jasmonate methyl ester response element. And the light-responsive elements (g-boxes) are also more widely distributed. The next most common cis-elements are the methyl jasmonate response elements (CGTCA motif and TGACG motif), which are distributed in almost all genes. It suggests that PDF is associated with plant growth and development in wheat, and may also be associated with plant disease resistance.
The transcriptome data downloaded and collated from the NCBI website were divided into three categories, including growth and development, abiotic stresses, and biotic stresses. As shown in the heat map in Figure 6, the overall expression of wheat PDF genes was low in the early stages of growth and development. TaPDF5.4 was highly expressed at the apical meristem tiller stage, apical meristem trilobal stage, radicle seedling stage, root seedling stage, root tiller stage, root leaf stage, root trilobal stage and shoot apex meristem seedling stage. Therefore, the function of TaPDF5.4 may be inseparably related to root growth and development. The overall expression of wheat PDF genes was also lower during abiotic stress. However, during the biotic stress treatment, TaPDF4.9, TaPDF5.4, TaPDF2.12, TaPDF2.15, TaPDF2.23, and TaPDF2.20 were more highly expressed in spikelets inoculated by Fusarium graminearum at 3, 6, 12, and 24 h. The results suggest that the TaPDF genes may be associated with plant disease resistance, which was consistent with the prediction of PDF genes function.
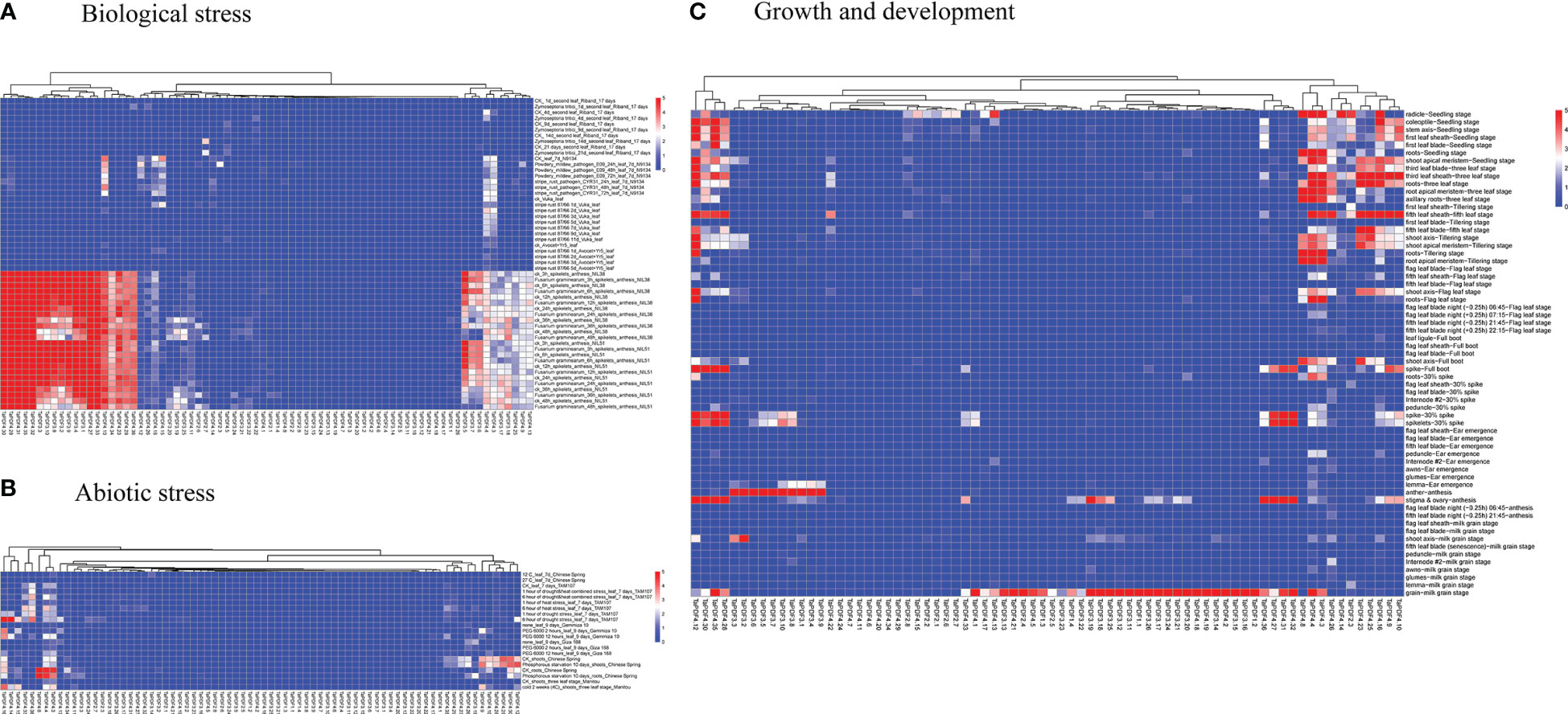
Figure 6 Heat map of growth and development of wheat PDF gene family. A, B and C represents transcriptomic data of TaPDFs in terms of biotic stress, abiotic stress and growth and development.
To further understand the role of TaPDF genes in reaction to biotic stresses, six highly expressed genes (TaPDF2.12, TaPDF2.15, TaPDF2.20, TaPDF2.23, TaPDF4.9, and TaPDF5.4) in the transcriptome (Figure 6) were selected. Expression of PDF genes in Yangmai 158 (disease-resistant) and Xinong 98710 (disease-susceptible) after inoculation with F. graminearum was analyzed by qRT-PCR (Figure 7). The overall expression level of TaPDF genes were up-regulated after inoculation of F. graminearum on the coleoptiles of two wheat species. However, TaPDF4.9 gene expression level was down-regulated in Xinong 98710 and TaPDF2.21 gene expression level was in a down-regulated state in Yangmai 158. According to the quantitative results, PDF gene may play different roles in wheat at 24h and 48h of F. graminearum infestation.
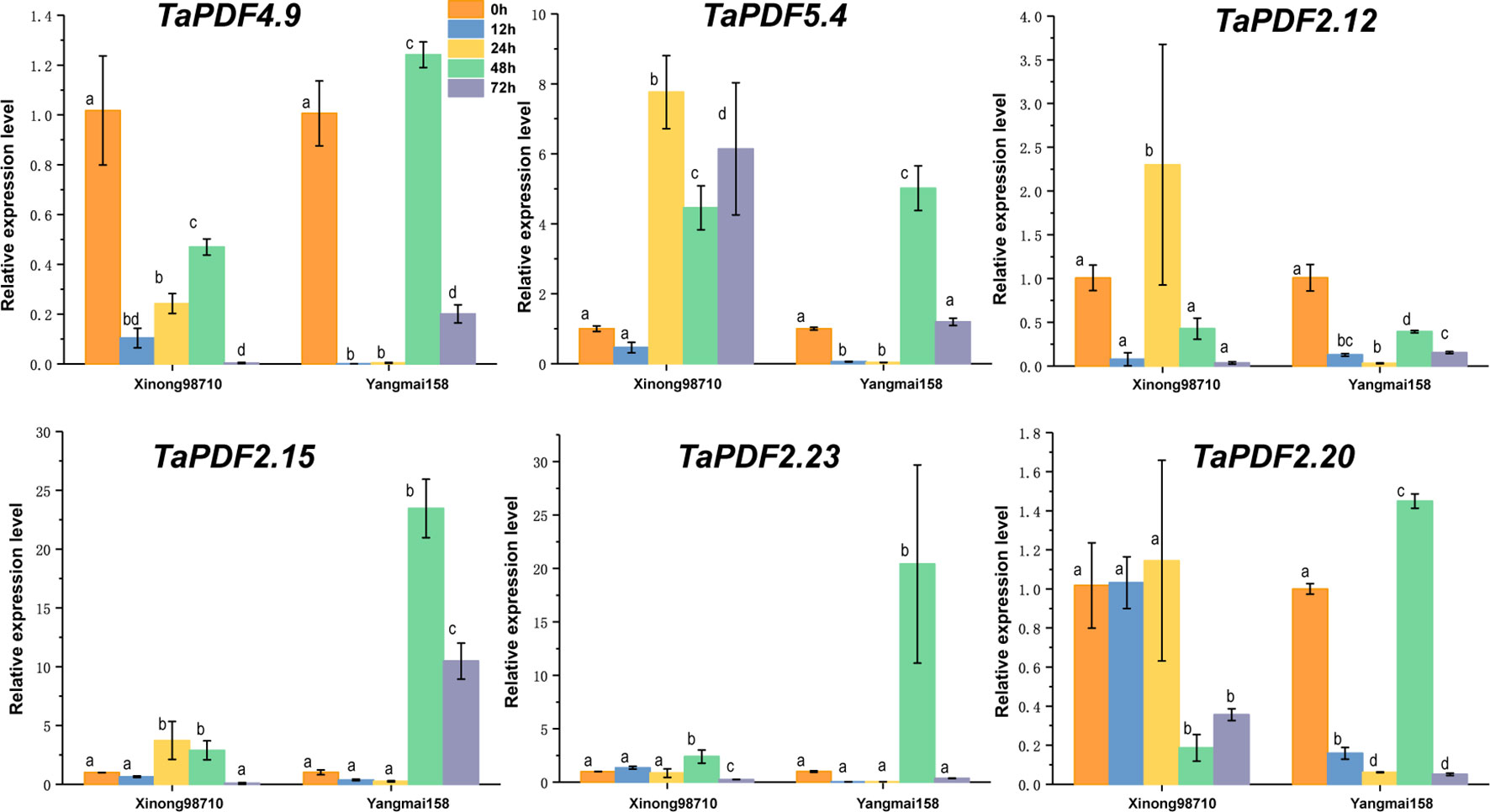
Figure 7 Expression patterns of six wheat (Triticum aestivum) TaPDF genes under F. graminearum treatments.
Most PDF genes were predicted to be located in vesicles, whereas only four genes were predicted to be on vesicles and nucleus. TaPDF4.9, TaPDF5.4, TaPDF2.12, and TaPDF2.15 genes were selected as target genes for homologous recombination with the digested vector pART27. The results of Figure 8 showed that the four target genes were basically localized to the cytoplasm and cell membrane, and TaPDF4.9 was also localized in the nucleus.
After four to six days of inoculation with Phytophthora infestans, the lesion diameter was measured (Figures 9A, C). As shown in Figures 9B, D, the lesion area on the leaves, where TaPDF2.15 and TaPDF4.9 were transient overexpressed, was smaller (P value < 0.01) than the control. This result suggests that TaPDF may play positive roles in resistance to Phytophthora infestans strain ‘88069’.
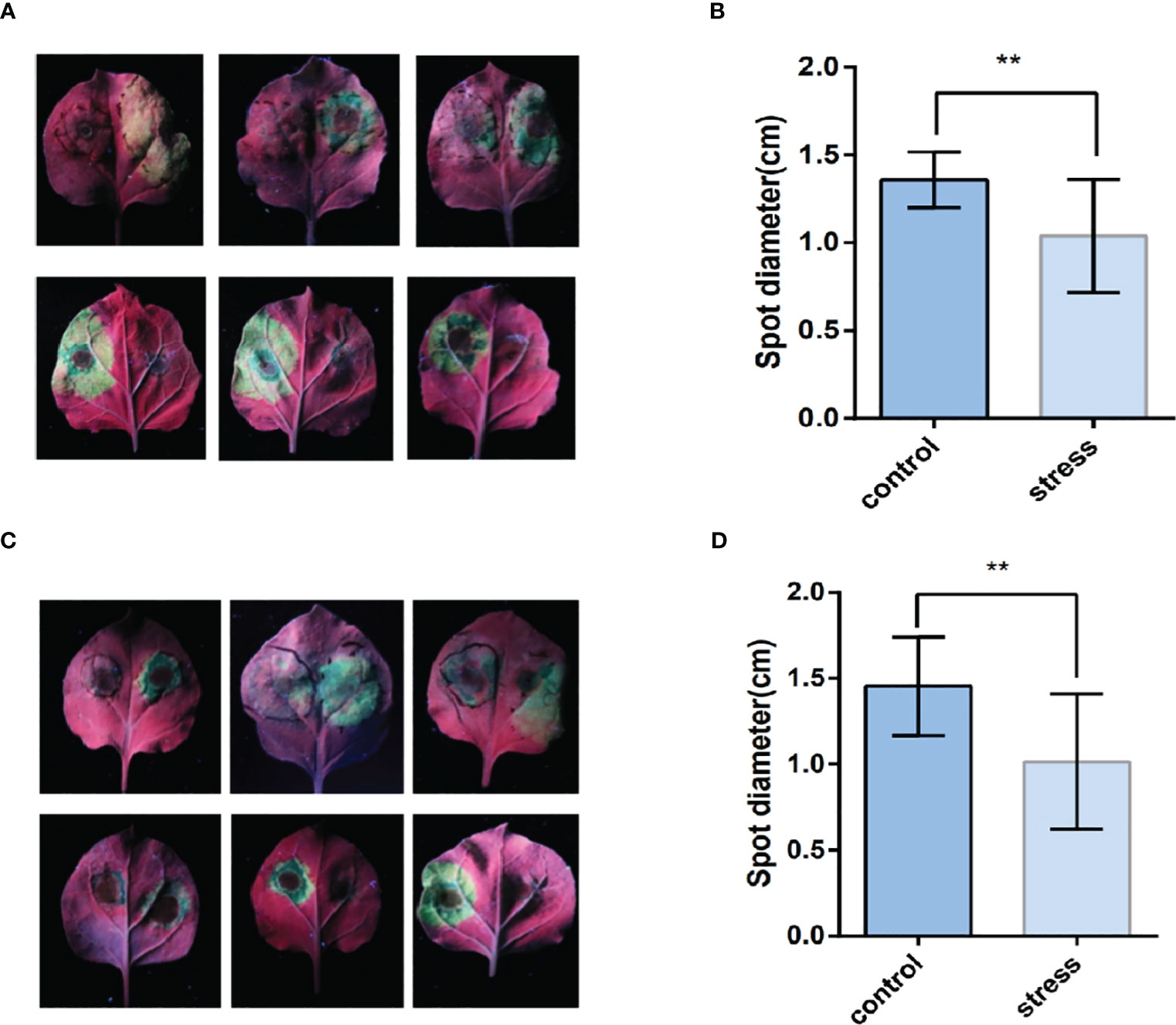
Figure 9 The leaf surface mediated by TaPDF4.9 (AB) and TaPDF2.15 (CD) inoculation with Phytophthora infestans. (A) is TaPDF4.9 infestation on tobacco, (B) is TaPDF4.9 infestation length. (C) is TaPDF2.15 infestation on tobacco and (D) is TaPDF2.15 infestation length. ** represents highly significant differences.
Plant defensins are widely distributed in the leaves, fruits, roots, stems, seeds, and tubers. Based on the genome-wide characterization of PDFs already performed in the literature in Arabidopsis thaliana (Mäser et al., 2001). However, there is a lack of systematic studies on the PDF family in wheat. In order to gain a deeper understanding of the functioning of wheat PDFs, this paper examines them. For the identification of family members, BLASTp searches were performed using known Arabidopsis PDF protein sequences (Kushmerick et al., 1998) and conserved structural domain sequences as query sequences (Clavijo et al., 2017). Sequences were queried in the wheat genome (IWGSC v1.1) and candidate hits were further checked by Pfam and InterProScan to exclude sequences without the Gamma - thiionin structural domain. Seventy-three wheat PDF proteins were finally identified. This study also identified PDF genes in rice and maize, which contained 15 maize PDF genes and 11 rice PDFs. The number of PDF genes varies in different plants. Differences in the number of genes from other species indicate functional redundancy in TaPDFs. However, genes do not correspond to changes in genome size.
Higher numbers of PDFs were identified in wheat than in Arabidopsis, maize and rice. The results illustrate the complexity of genetic traits in wheat. Homology analyses of TaPDF indicate that wheat may repeat multiple times in polyploidy, which we hypothesize may have made wheat more environmentally adapted and increased the number of wheat PDF genes during evolution. Based on gene structure and protein physicochemical property analyses, the wheat PDF gene has a highly conserved gene structure containing eight conserved cysteine residues forming four pairs of disulfide bonds (Cys4-Cys7, Cys3-Cys6, Cys2-Cys5, and Cys1-Cys8) (Thomma et al., 2002). The conclusions in the article are the same as those previously reported for the PDF family having the same structure. And the molecular weight size just shows that wheat PDF belongs to the shorter peptides with smaller molecular weight. The abscisic acid-responsive cis-acting element ABRE, the core promoter elements TATA box and CAAT box in the promoter of the wheat PDF gene are all growth and development-related elements. This suggests that TaPDF is associated with plant growth and development. Whereas methyl jasmonate response elements are distributed in almost all genes, we can infer that TaPDF is associated with plant disease resistance. It has been shown that plant defensins are commonly considered to display antimicrobial activity against only fungi (Sathoff et al., 2019). To further reveal the function of TaPDF, six genes TaPDF2.12, TaPDF2.15, TaPDF2.20, TaPDF2.23, TaPDF4.9 and TaPDF5.4 were analyzed by qRT-PCR. The expression levels of TaPDFs were differentially up-regulated by Fusarium graminearum. The above results indicate that TaPDFs are not only involved in wheat growth and development, but also play a positive regulatory role in the stress response process. The effect of TaPDF on disease resistance was analyzed through the resistance of different wheat varieties to F. graminearu. The results showed that TaPDF has an effect on plant disease resistance. Relevant studies have shown that PsDef1 is significantly increased in Scots pine seedlings during germination and in their response to pathogenic infection with Heterobasidion annosum (Kovaleva et al., 2011).
And after inoculation with Phytophthora infestans, it can be seen that the plaque becomes smaller, indicating that both selected TaPDF2.15 and TaPDF4.9 can inhibit Phytophthora infestans. It shows that AhPDF1s and AtPDF1s were able to confer Zn tolerance and AhPDF1s also displayed antifungal activity (Shahzad et al., 2013). It can be seen that the TaPDF should be functionally similar to that in Arabidopsis thaliana, both of which have the function of inhibiting the infestation of pathogenic bacteria. Although the mechanism of amplification of such gene families is well understood in this paper, the genes are not well understood in different species, which will have to be studied in further depth. The gene family aspect could be further systematically analyzed for defensin gene families by combining more species. The results of subcellular localization showed that the selected genes were basically localized to cytoplasm and cell membrane, but the multisite predictions of the proteins were all on the vesicles and a few on the nucleus. The results indicate that by Agrobacterium alone mediated Nicotiana benthamiana leaves could not be accurately localized to the organelles, but also require organelle-corresponding stains, which need to be investigated in subsequent work.
In this study, we identified 73 wheat PDF family members using bioinformatics methods, 15 members of corn PDF genes and 11 members of rice PDF genes. With the hexaploid wheat as the main research object and the amino acid sequence of PDF identified in Arabidopsis (Kushmerick et al., 1998), maize, and rice as reference, 35, 65, and 34 PDF gene family members were identified in the genomes of T. dicoccoides, Ae. tauschii, and T. urartu, respectively. As can be seen from the basic physicochemical properties, most of the TaPDFs are basic proteins and most are unstable. The genes are relatively evenly distributed across almost every chromosome. The high number of gene duplicate pairs indicates possible redundancy of function. The subcellular localization results of TaPDF4.9, TaPDF5.4, TaPDF2.12, and TaPDF2.15 are basically located in the cytoplasm and cell membrane, and TaPDF 4.9 is also located on the nucleus. Transient overexpression of TaPDF4.9 and TaPDF2.15 could inhibit the infection of Phytophthora infestans strain ‘88069’, which has the function of inhibiting the infection of Phytophthora infestans. According to the expression pattern, TaPDF is resistant to F. graminearum, and most of the selected genes showed an increase in transcript levels after being infested with F. graminearum. In this experiment we performed bioinformatic analysis of the wheat PDF family with subcellular localization, quantitative analysis and pathogenicity analysis. Wheat PDF was found to be effective in suppressing late blight in the Phytophthora infestans. As the genomes of more and more species are sequenced, it is believed that more members of the plant defensin family will be identified. With the development of functional genomics and proteomics, the role of such genes in wheat will become clearer and their mechanism of action will become clearer. Meanwhile, plant defensins have a very vast development prospect as a new antifungal drug, and researchers have also noticed the potential of plant defensins in the development of new anticancer drugs. Therefore, plant defensins, with their broad-spectrum, antimicrobial and high efficiency characteristics, provide new ideas for the research and development of new antifungal and antitumor drugs.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YD: Writing – original draft, Writing – review & editing. YW: Writing – original draft, Writing – review & editing. MT: Writing – original draft, Writing – review & editing. WC: Data curation, Investigation, Validation, Writing – review & editing. YC: Formal Analysis, Methodology, Writing – original draft. WW: Formal Analysis, Project administration, Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Basic and long term scientific and technological work in agriculture (NAES083PP13) and Sichuan Provincial Science and Technology Plan Project (2022YFS0585).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1279502/full#supplementary-material
Alaux, M., Rogers, J., Letellier, T., Flores, R., Alfama, F., Pommier, C., et al. (2018). Linking the International Wheat Genome Sequencing Consortium bread wheat reference genome sequence to wheat genetic and phenomic data. Genome Biol. 19 (1), 111. doi: 10.1186/s13059-018-1491-4
Arnold, K., Bordoli, L., Kopp, J., Schwede, T. (2005). The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22 (2), 195–201. doi: 10.1093/bioinformatics/bti770
Clavijo, B. J., Venturini, L., Schudoma, C., Accinelli, G. G., Kaithakottil, G., Wright, J., Clark, M. D., et al. (2017). An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res. 27 (5), 885–896. doi: 10.1101/gr.217117.116
De Coninck, B., Cammue, B. P. A., Thevissen, K. (2013). Modes of antifungal action and in planta functions of plant defensins and defensin-like peptides. Fungal Biol. Rev. 26 (4), 109–120. doi: 10.1016/j.fbr.2012.10.002
Falcón-Ruiz, E. A., López-Meza, J. E., Ochoa-Zarzosa, A. (2023). The plant defensins PaDef and γ-thionin inhibit the endothelial cell response to VEGF. Peptides 165, 171008. doi: 10.1016/j.peptides.2023.171008
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44 (D1), D279–D285. doi: 10.1093/nar/gkv1344
Hu, L., Liu, S. (2011). Genome-wide identification and phylogenetic analysis of the ERF gene family in cucumbers. Genet. Mol. Biol. 34 (4), 624–633. doi: 10.1590/s1415-47572011005000054
Huang, D.-M., Chen, Y., Liu, X., Ni, D.-A., Bai, L., Qin, Q.-P. (2022). Genome-wide identification and expression analysis of the SWEET gene family in daylily (Hemerocallis fulva) and functional analysis of HfSWEET17 in response to cold stress. BMC Plant Biol. 22 (1), 211. doi: 10.1186/s12870-022-03609-6
Huang, W., He, Y., Yang, L., Lu, C., Zhu, Y., Sun, C., et al. (2021). Genome-wide analysis of growth-regulating factors (GRFs) in Triticum aestivum. PeerJ 9, e10701. doi: 10.7717/peerj.10701
İlhan, E., Büyük, İ., İnal, B. (2018). Transcriptome - Scale characterization of salt responsive bean TCP transcription factors. Gene 642, 64–73. doi: 10.1016/j.gene.2017.11.021
Kamli, M. R., Sabir, J. S. M., Malik, M. A., Ahmad, A. (2022). Characterization of Defensin-like Protein 1 for Its Anti-Biofilm and Anti-Virulence Properties for the Development of Novel Antifungal Drug against Candida auris</i>. J. fungi (Basel Switzerland) 8 (12), 1298. doi: 10.3390/jof8121298
Kovaleva, V., Krynytskyy, H., Gout, I., Gout, R. (2011). Recombinant expression, affinity purification and functional characterization of Scots pine defensin 1. Appl. Microbiol. Biotechnol. 89 (4), 1093–1101. doi: 10.1007/s00253-010-2935-2
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054
Kushmerick, C., de Souza Castro, M., Santos Cruz, J., Bloch, C., Beirão, P. S. L. (1998). Functional and structural features of γ-zeathionins, a new class of sodium channel blockers. FEBS Lett. 440(3), 302–306. doi: 10.1016/S0014-5793(98)01480-X
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/nar/30.1.325
Li, R., An, J.-p., You, C.-x., Shu, J., Wang, X.-f., Hao, Y.-j. (2018). Identification and expression of the CEP gene family in apple (Malus×domestica). J. Integr. Agric. 17 (2), 348–358. doi: 10.1016/s2095-3119(17)61653-8
Lima, A. M., Azevedo, M. I. G., Sousa, L. M., Oliveira, N. S., Andrade, C. R., Freitas, C. D. T., et al. (2022). Plant antimicrobial peptides: An overview about classification, toxicity and clinical applications. Int. J. Biol. Macromol 214, 10–21. doi: 10.1016/j.ijbiomac.2022.06.043
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Mäser, P., Thomine, S., Schroeder, J. I., Ward, J. M., Hirschi, K., Sze, H., et al. (2001). Phylogenetic relationships within cation transporter families of arabidopsis. Plant Physiol. 126 (4), 1646–1667. doi: 10.1104/pp.126.4.1646
Mamidala, P., Rajarapu, S. P., Jones, S. C., Mittapalli, O. (2011). Identification and validation of reference genes for quantitative real-time polymerase chain reaction in Cimex lectularius. J. Med. Entomol 48 (4), 947–951. doi: 10.1603/me10262
Nguyen, N. N. T., Lamotte, O., Alsulaiman, M., Ruffel, S., Krouk, G., Berger, N., et al. (2023). Reduction in PLANT DEFENSIN 1 expression in Arabidopsis thaliana results in increased resistance to pathogens and zinc toxicity. J. Exp. Bot. 74(17): 5374–5393. doi: 10.1093/jxb/erad228
Parisi, K., Shafee, T. M. A., Quimbar, P., van der Weerden, N. L., Bleackley, M. R., Anderson, M. A. (2019). The evolution, function and mechanisms of action for plant defensins. Semin. Cell Dev. Biol. 88, 107–118. doi: 10.1016/j.semcdb.2018.02.004
Sadhu, S., Jogam, P., Gande, K., Marapaka, V., Penna, S., Peddaboina, V. (2023). Expression of radish defensin (RsAFP2) gene in chickpea (Cicer arietinum L.) confers resistance to Fusarium wilt disease. Mol. Biol. Rep. 50 (1), 11–18. doi: 10.1007/s11033-022-08021-9
Sagaram, U. S., Pandurangi, R., Kaur, J., Smith, T. J., Shah, D. M. (2011). Structure-activity determinants in antifungal plant defensins MsDef1 and MtDef4 with different modes of action against Fusarium graminearum. PloS One 6 (4), e18550. doi: 10.1371/journal.pone.0018550
Sathoff, A. E., Velivelli, S. L. S., Shah, D. M., Samac, D. A. (2019). Plant defensin peptides have antifungal and antibacterial activity against human and plant pathogens. Phytopathology 109 3, 402–408. doi: 10.1094/PHYTO-09-18-0331-R
Shahmiri, M., Bleackley, M. R., Dawson, C. S., van der Weerden, N. L., Anderson, M. A., Mechler, A. (2023). Membrane binding properties of plant defensins. Phytochemistry 209, 113618. doi: 10.1016/j.phytochem.2023.113618
Shahzad, Z., Ranwez, V., Fizames, C., Marquès, L., Le Martret, B., Alassimone, J., et al. (2013). Plant Defensin type 1 (PDF1): protein promiscuity and expression variation within the Arabidopsis genus shed light on zinc tolerance acquisition in Arabidopsis halleri. New Phytol. 200 (3), 820–833. doi: 10.1111/nph.12396
Terras, F. R., Schoofs, H. M., De Bolle, M. F., Van Leuven, F., Rees, S. B., Vanderleyden, J., et al. (1992). Analysis of two novel classes of plant antifungal proteins from radish (Raphanus sativus L.) seeds. J. Biol. Chem. 267 (22), 15301–15309. doi: 10.1016/s0021-9258(19)49534-3
Thomma, B. P., Cammue, B. P., Thevissen, K. (2002). Plant defensins. Planta 216 (2), 193–202. doi: 10.1007/s00425-002-0902-6
Thompson, J. D., Higgins, D. G., Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 (22), 4673–4680. doi: 10.1093/nar/22.22.4673
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7 (3), 562–578. doi: 10.1038/nprot.2012.016
Vriens, K., Cools, T. L., Harvey, P. J., Craik, D. J., Braem, A., Vleugels, J., et al. (2016). The radish defensins RsAFP1 and RsAFP2 act synergistically with caspofungin against Candida albicans biofilms. Peptides 75, 71–79. doi: 10.1016/j.peptides.2015.11.001
Keywords: defensins, gene structure, subcellular localization, gene expression, Phytophthora infestans infection
Citation: Dong Y, Wang Y, Tang M, Chen W, Chai Y and Wang W (2023) Bioinformatic analysis of wheat defensin gene family and function verification of candidate genes. Front. Plant Sci. 14:1279502. doi: 10.3389/fpls.2023.1279502
Received: 18 August 2023; Accepted: 11 October 2023;
Published: 24 October 2023.
Edited by:
Xinli Zhou, Southwest University of Science and Technology, ChinaReviewed by:
Manoj Kumar Solanki, University of Silesia in Katowice, PolandCopyright © 2023 Dong, Wang, Tang, Chen, Chai and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Chai, Y2hhaXlpMTIzNDU2QDEyNi5jb20=; Wenli Wang, d2xpd0AxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.