- 1Beijing Key Laboratory of Ornamental Plants Germplasm Innovation & Molecular Breeding, National Engineering Research Center for Floriculture, Beijing Laboratory of Urban and Rural Ecological Environment, School of Landscape Architecture, Beijing Forestry University, Beijing, China
- 2Genomics and Genetic Engineering Laboratory of Ornamental Plants, Department of Horticulture, College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, China
TCP proteins, part of the transcription factors specific to plants, are recognized for their involvement in various aspects of plant growth and development. Nevertheless, a thorough investigation of TCPs in Chrysanthemum lavandulifolium, a prominent ancestral species of cultivated chrysanthemum and an excellent model material for investigating ray floret (RF) and disc floret (DF) development in Chrysanthemum, remains unexplored yet. Herein, a comprehensive study was performed to analyze the genome-wide distribution of TCPs in C. lavandulifolium. In total, 39 TCPs in C. lavandulifolium were identified, showing uneven distribution on 8 chromosomes. Phylogenetic and gene structural analyses revealed that ClTCPs were grouped into classes I and II. The class II genes were subdivided into two subclades, the CIN and CYC/TB1 subclades, with members of each clade having similar conserved motifs and gene structures. Four CIN subclade genes (ClTCP24, ClTCP25, ClTCP26, and ClTCP27) contained the potential miR319 target sites. Promoter analysis revealed that ClTCPs had numerous cis-regulatory elements associated with phytohormone responses, stress responses, and plant growth/development. The expression patterns of ClTCPs during capitulum development and in two different florets were determined using RNA-seq and qRT-PCR. The expression levels of TCPs varied in six development stages of capitula; 25 out of the 36 TCPs genes were specifically expressed in flowers. Additionally, we identified six key ClCYC2 genes, which belong to the class II TCP subclade, with markedly upregulated expression in RFs compared with DFs, and these genes exhibited similar expression patterns in the two florets of Chrysanthemum species. It is speculated that they may be responsible for RFs and DFs development. Subcellular localization and transactivation activity analyses of six candidate genes demonstrated that all of them were localized in the nucleus, while three exhibited self-activation activities. This research provided a better understanding of TCPs in C. lavandulifolium and laid a foundation for unraveling the mechanism by which important TCPs involved in the capitulum development.
1 Introduction
Chrysanthemum ×morifolium Ramat., a member of the Asteraceae family (Anthemideae), has been under cultivation in China for more than 3,000 years because of its high economic and ornamental values. These species have attractive ornamental traits with colorful and various types of flowers. C. ×morifolium exhibits a prominent ornamental trait in the form of a typical radiate capitulum, commonly referred to as a pseudanthium. This structure comprises central disc florets (DFs) and peripheral ray florets (RFs), making it one of the most notable features of the plant (Bello et al., 2013; Ibanez et al., 2017). Moreover, DFs and RFs display distinct variations concerning gender differentiation, tissue structure, organ fusion, and pigment distribution (Stevens et al., 2001; Li et al., 2010; Bello et al., 2013; Wen et al., 2019b). Investigations on transcriptomics and gene functions have provided many candidate genes potentially responsible for the differences between the two floret types (Broholm et al., 2008; Chapman et al., 2012; Tahtiharju et al., 2012; Juntheikki-Palovaara et al., 2014; Huang et al., 2016; Liu et al., 2016; Zhao et al., 2016; Bello et al., 2017; Zhang et al., 2017; Chen et al., 2018; Wen et al., 2019a; Shen et al., 2021). However, the mechanisms underlying the development of the two floret types are still uncertain.
Transcription factors (TFs) play vital roles in flower development and their temporal and spatial expression differences are usually closely related to flower morphology (Doebley, 1993; Doebley and Lukens, 1998). TCPs are responsible for a variety of plant growth processes, including shoot branching (Aguilar-Martinez et al., 2007; Martin-Trillo et al., 2011), plant height regulation (Daviere et al., 2014; Shi et al., 2016), leaf development (Palatnik et al., 2003; Crawford et al., 2004), flower development (Luo et al., 1996; Feng et al., 2006; Tahtiharju et al., 2012; Garces et al., 2016; Huang et al., 2016; Bello et al., 2017; Chen et al., 2018; Wen et al., 2019a; Shen et al., 2021), fruit (Leng et al., 2019) and seed development (Takeda et al., 2006; Nag et al., 2009), and other biological and abiotic stress response (Viola et al., 2016; Guan et al., 2017). Conducting a genome-wide analysis of TCPs in C. lavandulifolium holds significant importance in gaining deeper insights into the molecular mechanisms regulating flower development.
TCP proteins are plant-specific TFs responsible for several processes of plant growth/development by modulating cell proliferation and differentiation (Cubas et al., 1999; Lopez et al., 2015). These proteins are characterized by a TCP domain, containing a conserved 59-residue-long basic helix-loop-helix (bHLH) structure at the N-terminus. This domain is known for its roles in DNA binding, protein-protein interaction, and facilitating nuclear localization of the protein (Danisman et al., 2013). TCPs were classified as two subfamilies, distinguishing them based on dissimilarities in their TCP domains: class I (known as PCF or TCP-P) and class II (or referred to as TCP-C). The most obvious distinction between the two subfamilies is that class I members lack 4 amino acids in the TCP domain (Martin-Trillo and Cubas, 2010). The class II TCPs are subdivided into 2 subclades (CYC/TB1 and CIN) in accordance with the differences between the TCP domains. The former subclade has eight members in Arabidopsis, out of which five are modulated by miR319 (Palatnik et al., 2003). Some class II members comprise an arginine-rich motif (R domain) that is responsible for the facilitation of protein-protein interaction (Cubas et al., 1999; Martin-Trillo and Cubas, 2010). Phylogenetic analysis indicates that the CYC/TB1 clade is subdivided into 3 types of genes that expanded through duplication within the group: CYC1, 2, and 3. Taking into account that two classes of TCP proteins possess distinct yet partially overlapping DNA binding sites with the consensus sequence GGNCCC (Kosugi and Ohashi, 2002), previous research has indicated that class I and II TCP proteins are responsible for synergistic and antagonistic biological interactions (Li, 2015), which usually occurs between the same class of TCP transcription factors.
Class I TCP family members enhance cell proliferation and growth, however class II members suppress these processes (Herve et al., 2009; Li et al., 2019). Studies have shown that some class I TCP TFs have been found to participate in various hormone-signaling pathways and play crucial roles in regulating inflorescence stem elongation (Daviere et al., 2014), gynoecium development (Lucero et al., 2015), seed germination (Resentini et al., 2015), filament elongation of stamen (Gastaldi et al., 2020) and flowering (Lucero et al., 2017) in Arabidopsis. Compared to class I members, the functions of the majority of class II TCPs have been extensively studied and understood. For instance, Arabidopsis CIN-TCPs are associated with the regulation of flowering time and floral organ development (Kubota et al., 2017; Li et al., 2019), and recent advances in the function and regulation of CIN-like TCPs are well described (Lan and Qin, 2020). It is well known that TB1 participates in the determination of maize axillary meristem fate (Doebley et al., 1997) and CYC controls the asymmetry of petals in Antirrhinum flower (Luo et al., 1996). In Arabidopsis, AtTCP12 and AtTCP18 are two homologs of TB1 that are responsible for inhibiting bud outgrowth (Aguilar-Martinez et al., 2007). Moreover, AtTCP18 is responsible for the suppression of the early floral transition in axillary meristems by binding with florigen proteins (Niwa et al., 2013). AtTCP1 is the homologous gene of CYC, which affects plant growth/development by regulating the expression levels of DWARF4, a brassinosteroid (BR) biosynthesis gene (Guo et al., 2010), and also affects leaf elongation growth (Koyama et al., 2010). Additionally, CYC-like genes have been shown to regulate floral symmetry in many species, such as Primulina heterotricha (Gao et al., 2008; Yang et al., 2012), Torenia fournieri (Su et al., 2017), Saintpaulia ionantha (Hsu et al., 2018), Gerbera hybrida (Elomaa et al., 2018; Zhao et al., 2020), Chrysanthemum (Huang et al., 2016; Chen et al., 2018; Shen et al., 2021) and so on.
Recently, TCPs have been discovered in various plant species, and the number of TCPs varies among different plants. For example, Arabidopsis, Solanum lycopersicum, Oryza sativa, Populus euphratica, P. trichocarpa, Malus domestica and Prunus mume have 24, 30, 28, 33, 36, 52 and 19 TCPs, respectively (Martin-Trillo and Cubas, 2010; Parapunova et al., 2014; Xu et al., 2014; Ma et al., 2016; Zhou et al., 2016). However, the TCP gene family in C. lavandulifolium remains relatively unexplored, despite its significance as an origin species of cultivated chrysanthemum (Dai et al., 2002). This plant also serves as an excellent model material for investigating the development of RFs and DFs in Chrysanthemum. Recognizing the crucial roles of TCPs in plant growth/development, we undertook an extensive examination of the TCP gene family in C. lavandulifolium. Herein, we identified 39 TCPs from the C. lavandulifolium genome and executed a comprehensive analysis, including chromosomal position, phylogenetic relationship, conserved motifs, gene structures, and miRNA target sites. To clarify their functions in flower development, we further analyzed the expression patterns of ClTCPs during capitulum development and in two different florets, as well as the key ClTCPs were tested by qPCR in Chrysanthemum species. Additionally, the subcellular localization characteristics and the self-activation activity in yeast of key TCP proteins were also examined. This study not only provided a basis for a comprehensive understanding of TCPs in Chrysanthemum, but also laid a foundation for revealing the mechanism of TCPs regulating the capitulum development.
2 Materials and methods
2.1 Plant materials and growth conditions
The C. lavandulifolium G1 line, a diploid relative wild species for C. ×morifolium Ramat., which has achieved genomic sequence (Wen et al., 2022), was the main material for gene family analysis and screening key ClTCPs. Other Chrysanthemum samples, including C. indicum (Ci), C. vesticum (CVW), C. ×morifolium ‘C27’, ‘D91’, ‘G70’, ‘T3’, and C. aromaticum (SN) were chosen to detect the expression profiles of target genes in DFs and RFs. All plant materials were planted and managed under conventional field conditions in the chrysanthemum germplasm nursery at Beijing Forestry University (Beijing, China).
2.2 Identification of TCPs in C. lavandulifolium
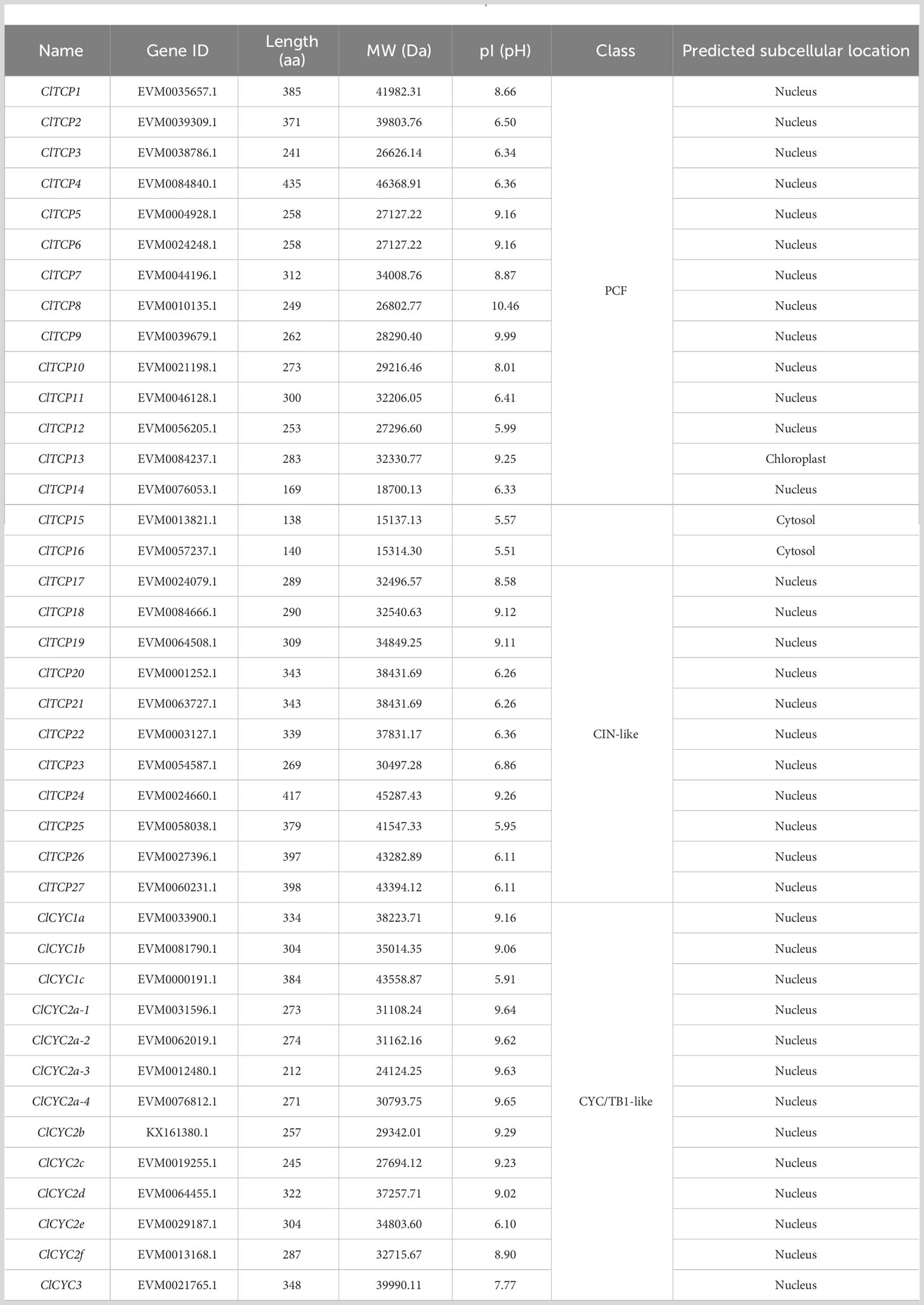
Genome sequences were obtained from the C. lavandulifolium database (Wen et al., 2022). To determine the TCPs in C. lavandulifolium, we used the Arabidopsis Information Resource (TAIR) database (http://www.arabidopsis.org) to download all AtTCP protein sequences as reference sequences and screened the C. lavandulifolium genome database using local BLAST comparisons. All ClTCP protein sequences were verified by the Swissprot database (https://www.sib.swiss/swiss-prot). We removed repeated sequences using Excel and further analyzed the obtained non-redundant ClTCP sequences. Subsequently, we calculated the molecular weights (MW) and isoelectric points (pI) of ClTCP proteins using the ExPasy website (https://web.expasy.org/protparam/) and predicted the subcellular localization of the ClTCPs through WoLF PSORT (http://www.genscript.com/psort/wolf_psort.html).
2.3 Phylogenetic analysis, conserved motif and gene structure identification
To explore the phylogenetic relationships of TCPs between C. lavandulifolium and Arabidopsis, we used MAFFT v7 (https://mafft.cbrc.jp/alignment/server/index.html) to perform multiple sequence alignments and subsequently used MEGA X software to develop a phylogenetic tree according to the multiple alignments data using the Maximum Likelihood (ML) approach with 1000 replicates.
Conserved motifs of TCP proteins were discovered and evaluated by MEME (https://meme-suite.org/) (parameter settings: max. number of motifs 10; max. width of motifs 50; min. width of motifs 6), and the SMART (http://smart.embl-heidelberg.de/) was employed to annotate the identified motifs. The TBtools toolkit (Chen et al., 2020) was used to assess the gene structures with the corresponding DNA sequences and CDSs at default parameter settings.
2.4 Chromosomal position and miR319 target site prediction
Based on positional information from the C. lavandulifolium genome, locations of TCPs on the C. lavandulifolium chromosomes were determined using the TBtools toolkit. To predict target sites for miR319, we used the psRNATarget online application (https://www.zhaolab.org/psRNATarget/analysis?function=3) to analyze ClTCPs nucleotide sequences.
2.5 In silico promoter analysis
The 2,000 bp upstream of the ClTCPs coding regions were extracted from C. lavandulifolium genome data, which was regarded as the promoter sequences. We used the PlantCARE tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) for the identification of the putative cis-acting elements of ClTCP promoters.
2.6 Expression patterns of ClTCPs in various development stages and two different florets
We obtained the expression patterns of ClTCPs in six development stages of capitula (S1, S2, S5, S6, S9, and S10) and two different florets (R1, D1, R2, D2) from the previously reported RNA-seq data (Wen et al., 2019a; Wen et al., 2022). Data normalization was conducted according to the average expression value of each gene across all studied samples. Genes and their expression patterns were systematically grouped according to the average Person’s metric. Finally, the heatmap of ClTCPs was constructed with TBtools (Chen et al., 2020).
2.7 Expression analysis of ClCYC2 genes by quantitative real-time PCR
The qRT-PCR was conducted to analyze the expression patterns of key differential ClTCPs in various Chrysanthemum species. Total RNA was isolated from the DFs and RFs at two development stages (R1, D1, R2, D2) using the RNAprep-Pure-Plant-Kit (Tiangen, China) and used for cDNA synthesis with the FastKing-RT-Kit (Tiangen, China), qRT-PCR analysis was conducted on a CFX96-real-time-system (Bio-Rad, USA) using the SYBR-Premix-Ex-Taq-kit (Takara, Japan). The primer pairs are presented in Supplementary Table 1. We used ClSAND as a reference gene (Qi et al., 2016; Pu et al., 2020; Lu, 2022).
2.8 Subcellular localization of ClCYC2 proteins
The cDNA of the ClCYC2a, ClCYC2b, ClCYC2c, ClCYC2d, ClCYC2e, and ClCYC2f without stop codons were amplified with the primer pairs (Supplementary Table 2) and recombined into pBI121-eGFP to generate six fusion constructs. Agrobacterium tumefaciens strain GV3101 was employed to introduce the recombinants and pBI121-eGFP (used as a control) into the leaves of Nicotiana benthamiana through infiltration. After 48 hours, the expression of GFP was examined under a confocal laser scanning microscope (Leica-TCS-SP8, Wetzlar, Germany).
2.9 Analysis of transactivation activity in yeast
To assess the transactivational activity of six ClCYC2 proteins in yeast, we employed the pGBKT7 vector (BD, Clontech, United States) for the experiments. PCR amplification of the entire ClCYC2 open reading frames was performed with the gene-specific primers pairs (Supplementary Table 3). These amplicons were then independently recombined into the pGBKT7 vector. Following this, we co-transformed the pGBKT7-ClCYC2 recombinant vectors, pGBKT7-53, and pGBKT7-lam, along with pGADT7 (AD, Clontech, United States) empty vectors, into the Y2HGold strain. The transformed strains were then grown in SD/-Leu-Trp and SD/-Ade-His-Leu-Trp media, and then incubated at 30°C for 3-5 days.
3 Results
3.1 Identification and chromosomal location of TCPs in C. lavandulifolium
The C. lavandulifolium genome database offers the references for the genome-wide detection of its candidate genes (Wen et al., 2022). In order to identify the TCPs in C. lavandulifolium, we performed a BLAST search against the C. lavandulifolium genome database using all the Arabidopsis TCP protein sequences as the query. A total of 39 sequences were obtained. Including one sequence isolated from the transcriptome (Wen et al., 2022), there were a total of 40 sequences obtained, which confirmed the occurrence of the TCP domain using the Swissprot database, indicating there were at least 40 TCPs in C. lavandulifolium. Among these genes, 14 TCPs with R domain were identified (Figure 1). In addition, The TCP proteins displayed diversity in their characteristics, including variations in length, MW, and pI. The protein lengths varied between 138 and 435 amino acids, the MW ranged from 15.14 to 46.37 kDa, and the range of pI was 5.51-10.46 pH. According to WoLF PSORT predictions, the majority of ClTCP proteins showed nuclear localization, while a few were found in the cytoplasm and chloroplast (Table 1).
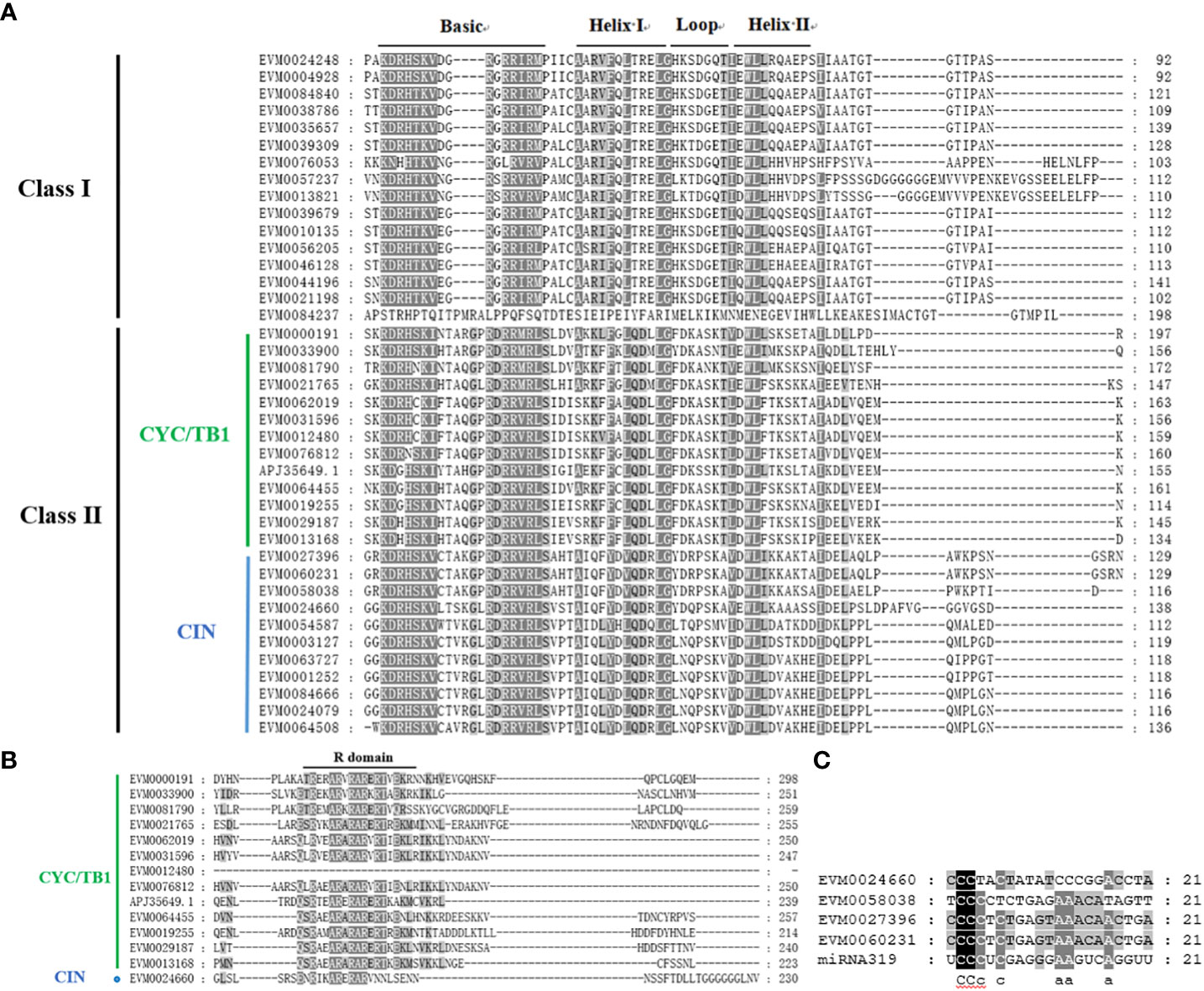
Figure 1 Multiple sequence alignments of TCP proteins in C. lavandulifolium and analysis of the potential miR319-targeted sites. (A) TCP domain alignment for the ClTCP proteins. The conserved amino acids were shaded in grey, and the bHLH regions were marked. (B) R domain alignment of the class II subfamily ClTCP members. Multiple sequence alignments were performed with Genedoc. (C) Alignment of potential target sites for miR319 (aligned in reverse). Target sites were taken from the coding sequences of ClTCPs, and the mature miR319 sequence was obtained from miRBase (http://www.mirbase.org/).
We located a total of 39 ClTCPs on chromosomes (Figure 2), but they were unevenly distributed across the C. lavandulifolium genome. There were no genes identified on Chr 3. Among them, members of the CYC/TB1 subfamily were mapped to Chr 2, 4, 6, 7, and 8, respectively. Especially, four ClTCPs (ClCYC2c, ClCYC2d, ClCYC2e, and ClCYC2f) which belong to the CYC2 subclade formed a gene cluster on Chr 7, and subfunctionalization might have occurred in these genes throughout the evolutionary process of the C. lavandulifolium genome (Wen et al., 2022). While the multi-copy genes of ClCYC2a-1, ClCYC2a-2, and ClCYC2a-3 formed a gene cluster on Chr 6. Two segmentally duplicated gene pairs (ClTCP5 and ClTCP6, ClTCP20 and ClTCP21) were located on Chr 9.
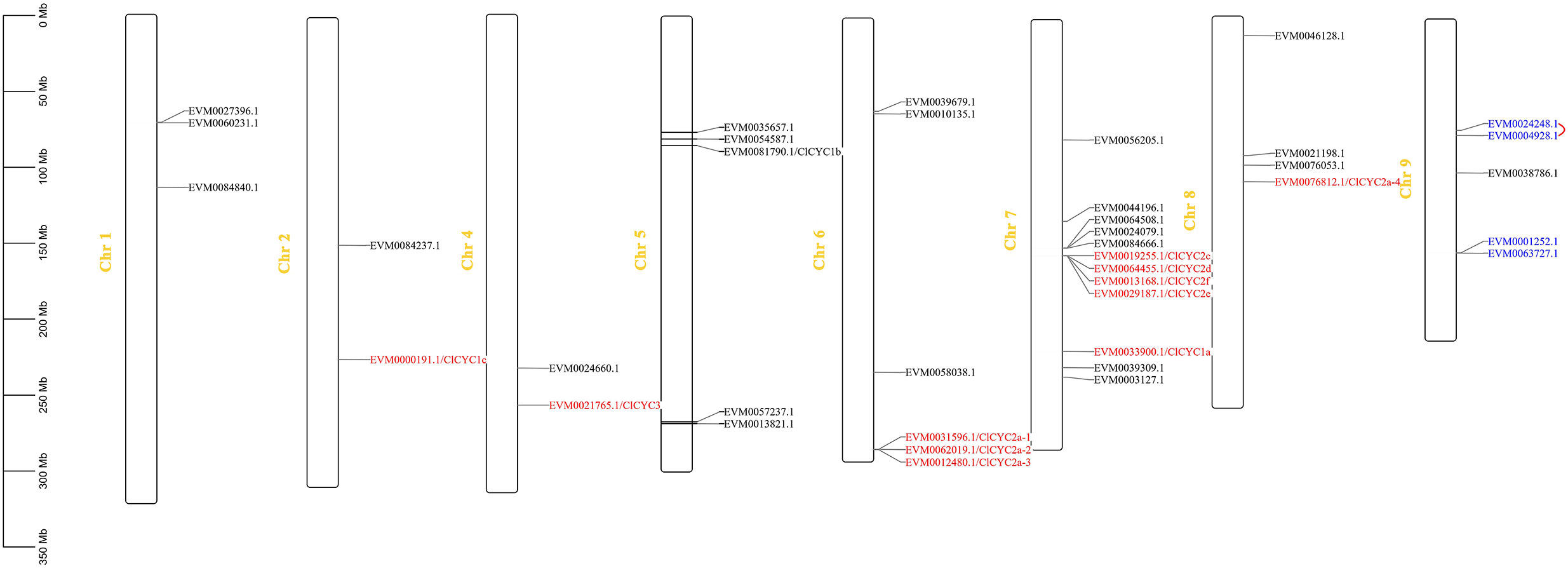
Figure 2 Chromosomal positions of TCPs on C. lavandulifolium chromosomes. The scale shows a chromosomal distance of 50 Mb. All 39 ClTCPs were localized to the chromosomes. Segmentally duplicated genes (EVM0004928.1_ClTCP5 and EVM0024248.1_ClTCP6, EVM0001252.1_ClTCP20 and EVM063727.1_ClTCP21) were marked with blue letters. Eleven ClTCPs belong to CYC/TB1 subclade, which are highlighted in red and located on the chromosomes.
3.2 Phylogenetic analyses and classification of ClTCPs
To investigate the phylogenetic and evolutionary relationships between the C. lavandulifolium TCPs and Arabidopsis TCPs, a phylogenetic tree was established using the complete sequences of 40 TCP proteins from C. lavandulifolium and 24 TCP proteins from Arabidopsis. According to the previous classification of TCPs in Arabidopsis, phylogenetic analysis and multiple sequence alignments all showed that ClTCP proteins were divided into two categories: class I and II, which contain 16 and 24 genes, respectively (Figures 1A, 3). It was clear that class II proteins possess an additional four amino acids in the basic domain compared to class I members (Figure 3A). The R domains were identified at the C-terminus of all ClTCP proteins belonging to class II CYC/TB1, as well as one member from the CIN class (Figure 3B). The phylogenetic analysis indicated that class II was subdivided into two subclades, CIN (11) and CYC/TB1 (13). Then CYC/TB1 is further divided into three branches: CYC1, 2 and 3. In C. lavandulifolium, the numbers of CYC1, CYC2 and CYC3 protein members were 3, 9 and 1, respectively. Compared with Arabidopsis, the Class II TCPs in C. lavandulifolium were significantly expanded, especially the CYC/TB1 subclade with 13 members, and 9 TCPs belonging to the CYC2 branch. Studies on the CYC2-like genes shown that they regulate the floret morphology in G. hybrida, Helianthus annuus, Senecio vulgaris and C. ×morifolium (Tahtiharju et al., 2012; Garces et al., 2016; Huang et al., 2016). The phylogenetic tree suggested that particular genes could have experienced evolution specific to each species.
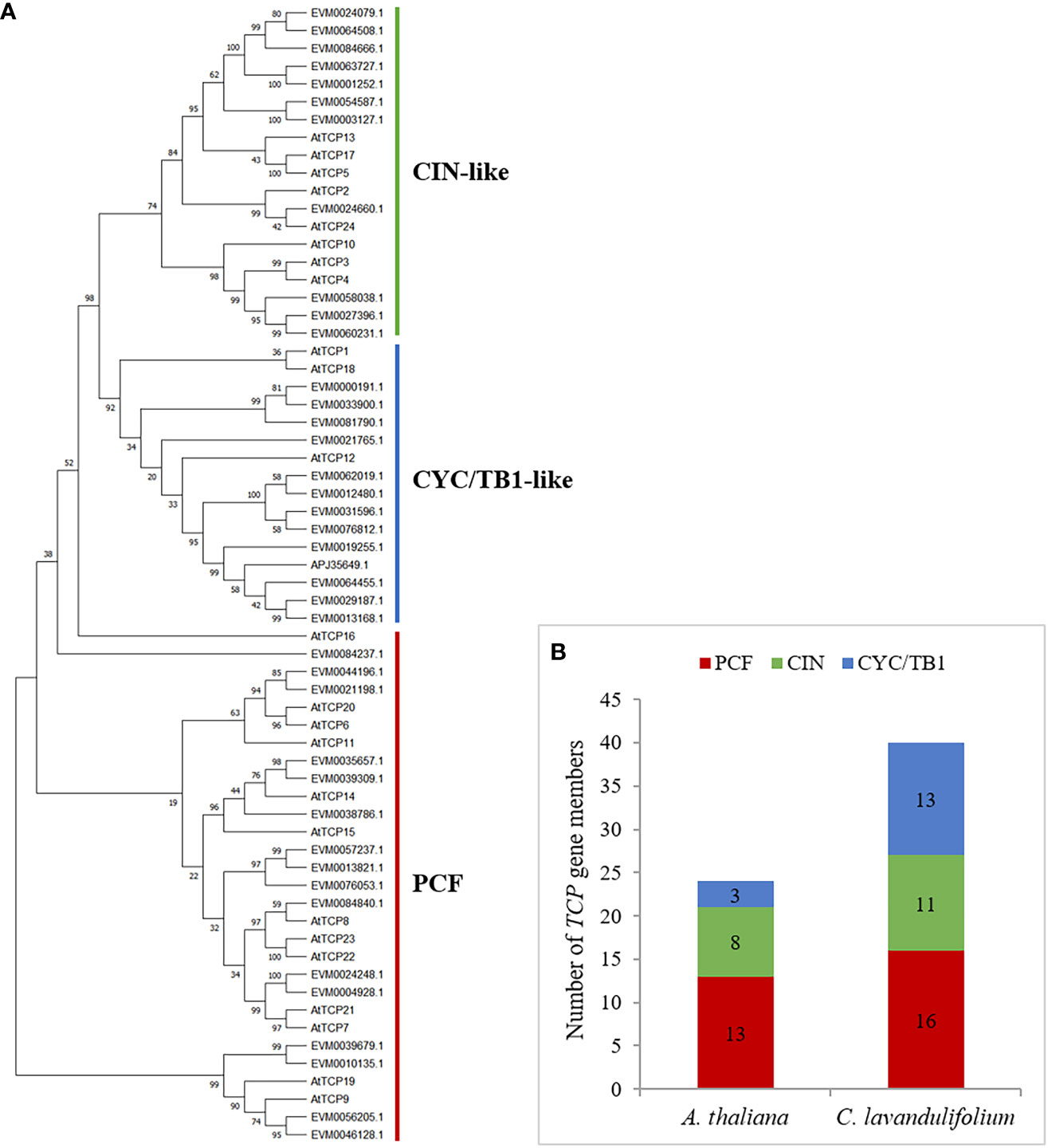
Figure 3 Phylogenetic and statistical analyses of TCP members from C. lavandulifolium and Arabidopsis. (A) The tree was built using the Maximum Likelihood (ML) method with 1000 replicates by MEGA X. Multiple sequence alignments of ClTCPs was performed using MAFFT v7. The bootstrap value was indicated by the number above the branches and values less than 50 would not be displayed. Red, green and blue lines indicate the PCF, CIN and CYC/TB1 subclades, respectively. (B) Statistical analysis of TCP members from C. lavandulifolium and A. thaliana.
Additionally, four CIN subclade genes (ClTCP24, ClTCP25, ClTCP26, and ClTCP27) contained miR319 target sites and showed a high degree of similarity with miR319-targeted TCPs in Arabidopsis (Figure 1C).
3.3 Conserved motif and gene structure identification
The conserved motif and gene structures of ClTCPs were examined to reveal the structural and evolutionary characteristics of the TCP proteins in C. lavandulifolium. By utilizing the complete sequences of the ClTCP proteins, a phylogenetic tree was constructed, effectively categorizing the ClTCP proteins into three distinct subgroups (Figure 4A). Ten conserved motifs were identified by the MEME online tool to gain a further understanding of the variety of motif compositions among ClTCPs. The results indicated that all ClTCP proteins had highly conserved TCP domains (motif 1). Almost all of the CYC/TB1 subclade proteins and one CIN protein hit the conserved R domains (motif 3) (Figure 4B). In contrast, most CIN proteins contained the N-terminal TCP domains of motif 7. Furthermore, motif 2 was only found in PCF, supporting previous research (Wei et al., 2016; Lei et al., 2017) that certain motifs present in a specific subgroup can play particular roles in those genes.
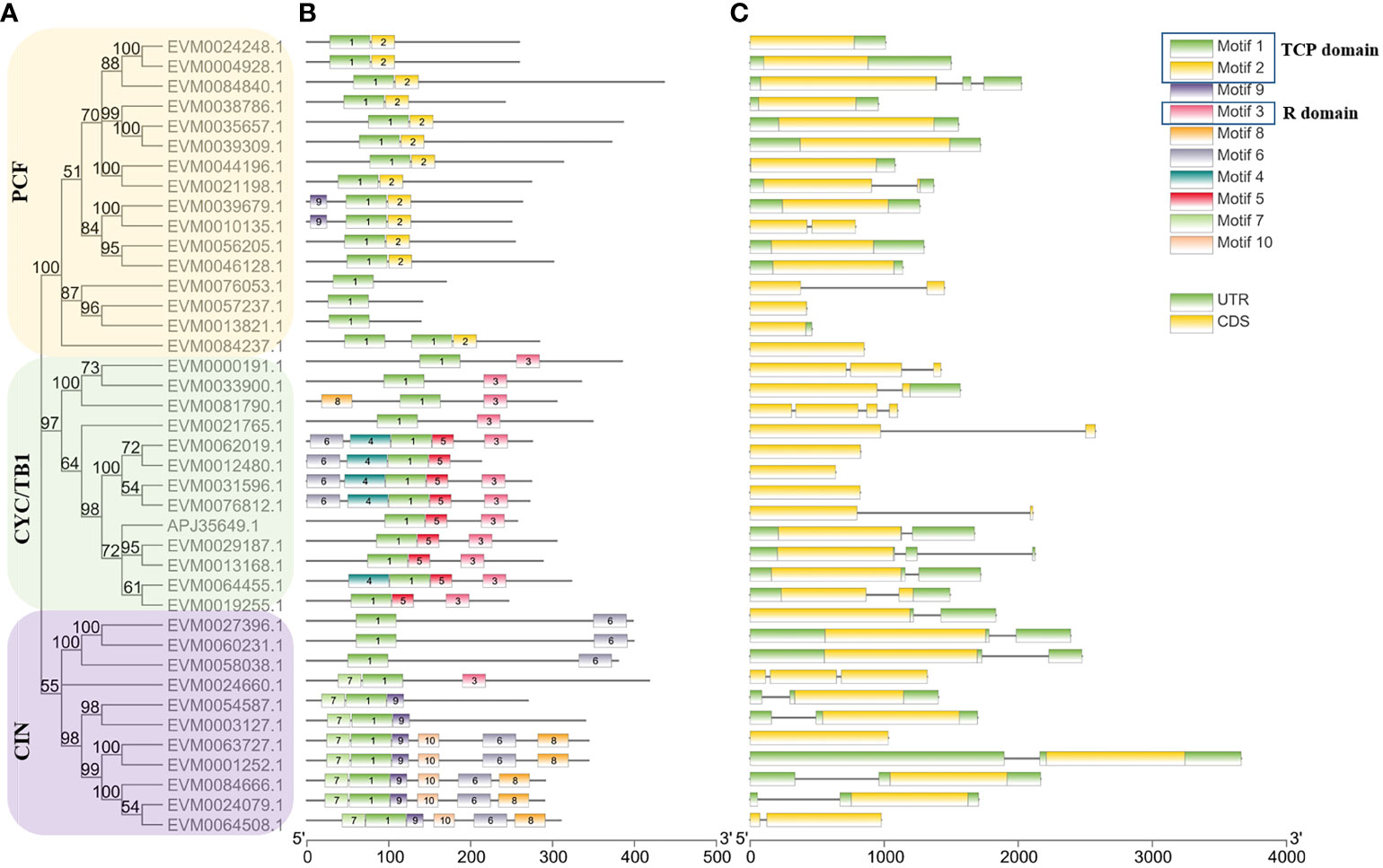
Figure 4 Conserved motif distributions and gene structure of ClTCPs. (A) The tree was generated based on the sequences of ClTCP proteins. The yellow, purple and green rectangles indicated the clustering of genes into the PCF, CIN and CYC/TB1 subclades, respectively. (B) Conserved motifs in C. lavandulifolium TCP members were identified by MEME tools. Each motif is indicated by a rectangular box of a different color and numbered from 1 to 10. (C) Gene structure analysis of ClTCPs. The coding sequence (CDS), untranslated region (UTRs) and introns are indicated by yellow boxes, green boxes and blank lines, respectively.
The intron-exon organization of nearly all ClTCPs was relatively conserved, 28 of the 39 ClTCPs had no introns, while the remaining 11 genes had one to three introns. Except for ClTCP8, ClTCP10, and ClTCP14, we discovered that no TCP-P (PCF) type genes included introns. Sequences of TCP-C genes in C. lavandulifolium had more introns compared to TCP-P genes. Within the TCP-C genes, five of them contained an intron, two contained two introns, and one contained three introns. In summary, the intron-exon distribution patterns of most ClTCPs within the same subfamily are similar, which reinforces the subclade classification and supports the understanding of their evolutionary relationships (Figure 4C).
3.4 Analysis of promoter cis-regulatory elements in ClTCPs
The cis-regulatory elements in the promoter sequences were examined to enhance the comprehension of gene functions and regulatory mechanisms of ClTCPs. The ClTCP gene promoter regions, which are a 2-kb segment of genomic DNA located upstream of the translation start site, were submitted to the PlantCARE database. Numerous cis-acting elements associated with phytohormone responses, stress responses, and plant growth/development were identified apart from the basic CAAT and TATA boxes (Figure 5; Supplementary Table 4). Among them, the largest number of elements were the light-responsive, MeJA-responsive and abscisic acid-responsive elements. Additionally, the elements of auxin-responsive, stress-responsive, cell cycle regulation and meristem regulation were also found in their promoter regions of ClTCPs (Figure 5).
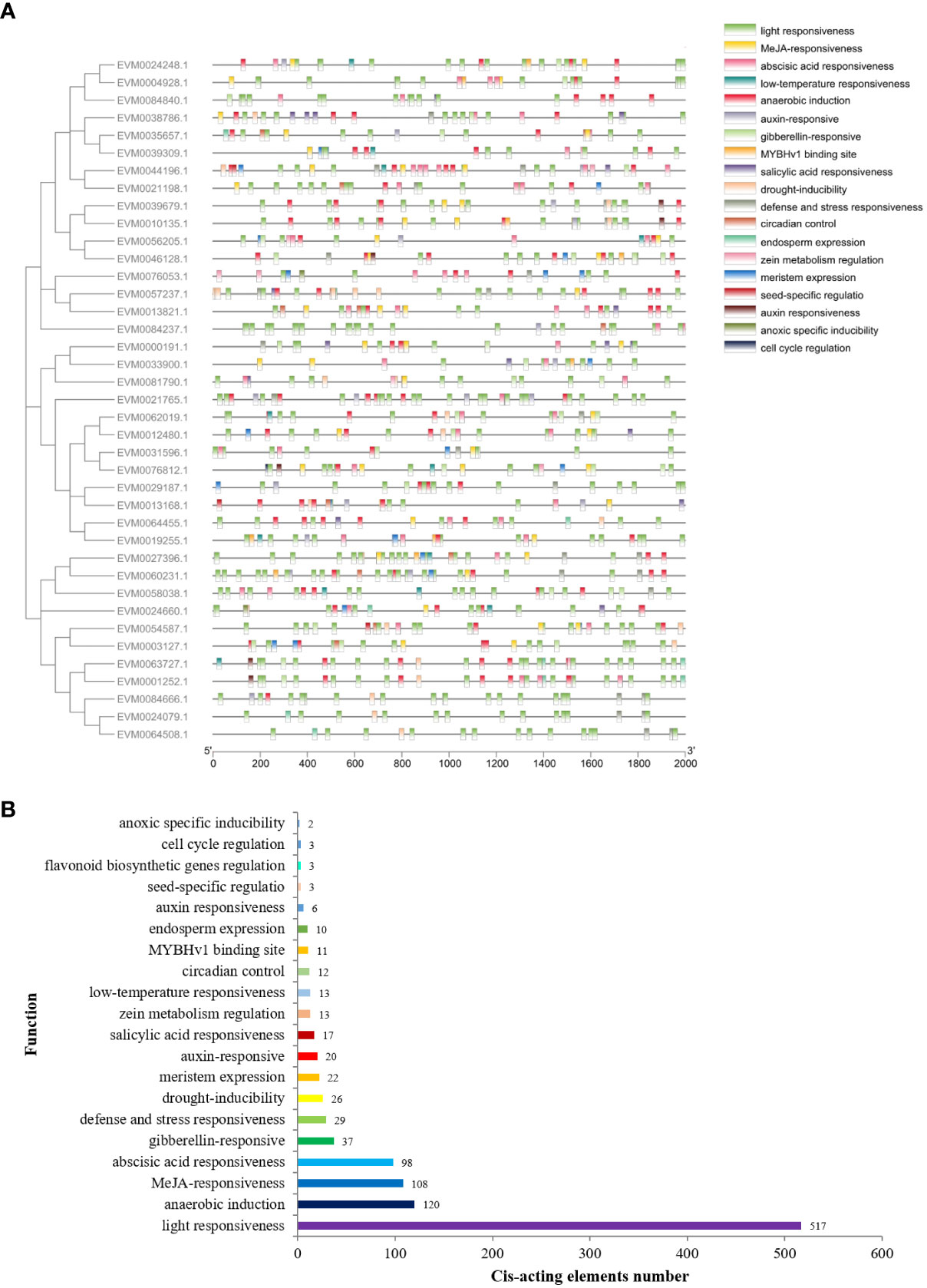
Figure 5 Cis-regulator elements analysis of ClTCP promoters. (A) Distribution of cis-regulator elements in the promoter region (2 kb upstream of the transcription start site) of ClTCPs. (B) Statistical analysis on the number and functional type of cis-regulatory elements in all ClTCP promoters in C. lavandulifolium.
A kind of cis-regulatory elements associated with endosperm expression (GCN4_motif) was determined in the promoter region of 8 ClTCPs, and a kind of elements associated with meristem expression (CAT-box) was determined in 18 ClTCPs (Supplementary Table 4). The circadian control element (circadian) and cell cycle regulation element (MSA-like) were observed in 10 and 3 ClTCPs, respectively. Furthermore, the flavonoid biosynthetic genes regulation (MBSI), and seed-specific regulation element (RY-element) were also detected in the ClTCPs promoter regions (Figure 5; Supplementary Table 4).
In hormone-associated cis-acting elements, the MeJA-responsive element (CGTCA and TGACG motifs), the ABA-responsive element (ABRE) and the gibberellin-responsive element (TATC-box, GARE-motif and P-box) were found in the promoter regions of 28, 37 and 21 ClTCPs, respectively. The salicylic acid (SARE and TCA-element) and auxin-responsive element (TGA-element, AuxRE and AuxRR-core) were discovered in 12 and 18 ClTCPs, respectively. The promoter region of ClTCPs had an abundance of hormone-responsive elements, suggesting that hormones may be crucial regulators of plant growth/development. Moreover, cis-elements associated with stress, such as low-temperature (LTR) responsiveness, drought-inducibility (MBS) and anaerobic induction (ARE) elements were also determined in the promoter regions of 10, 18 and 34 ClTCPs, respectively (Figure 5; Supplementary Table 4).
3.5 Expression patterns of ClTCPs during capitulum development and in two different florets
The TCPs play vital roles in multiple progress during plant development. The previously reported RNA-seq data (Wen et al., 2019a; Wen et al., 2022) were applied to investigate the expression patterns of ClTCPs in six developmental stages of capitula (S1, S2, S5, S6, S9, and S10) and two different florets (R1, D1, R2, D2), to explore their functions in flower development. Through searching and blast alignment, a total of 40 TCP sequences were obtained, and the genes whose expression level FPKM value was 0 in all samples were removed. Finally, 36 and 35 ClTCPs corresponding to the two transcriptomes were used for drawing the expression heatmap (Supplementary Tables 5, 6).
The expression levels of TCPs differed in six developmental stages of capitula, 25 out of the 36 TCP genes were specifically expressed in flowers (Figure 6A). The expression levels of EVM0029420 (ClCYC2d), EVM0028754 (ClCYC2a), EVM0014549 (ClCYC2f), and EVM0021074 (ClCYC2e) in CYC/TB1 subclade in S5 stage were significantly lower than those in S6 stage, and they expressed steadily and continuously in the subsequent S9 and S10 stages. This suggested that CYC2-like genes in C. lavandulifolium were expressed at the early stage of the capitula, and ultimately played a role in the ontogenetic differences between DFs and RFs. The expression level of EVM0034432 (ClCYC1c) in S5 stage was significantly upregulated compared to S6 stage and decreased continuously in S9 and S10 stages.
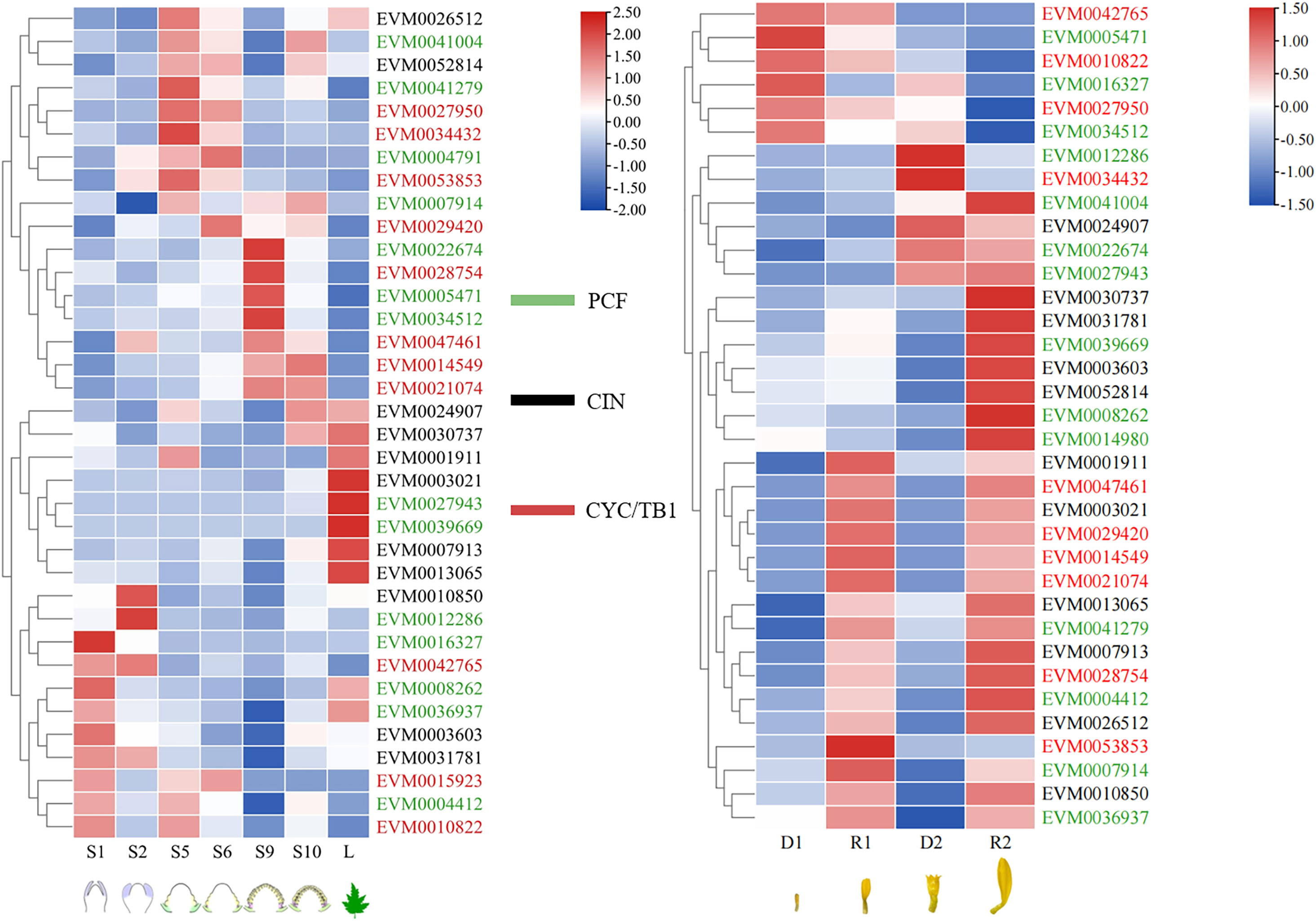
Figure 6 Expression patterns of ClTCPs during capitulum development and in two different florets of C. lavandulifolium. Red and blue boxes show high and low expression levels, respectively. Green, black and red color of gene names belong to PCF, CIN, and CYC/TB1, respectively. S1, S2, S5, S6, S9, and S10 indicated the different development stages of capitula (Wen et al., 2022), L indicated reproductive-stage leaf; D1, D2, R1, R2 indicated disc and ray florets at two stages.
Figure 6B showed the results of hierarchical clustering to further analyze the potential involvement of TCPs in the development of DFs and RFs. According to their expression patterns, the TCPs in C. lavandulifolium are categorized into three clusters. The TCPs in C. lavandulifolium were grouped into three clusters based on their expression profiles. Cluster 1 consisted of the genes whose expression levels in RFs were remarkably lower than those in DFs. Conversely, the genes in Cluster 2 whose expression levels in RFs were markedly elevated compared to DFs, and Cluster 2 is the largest group. In Cluster 3, the expression levels of ClTCPs, such as EVM0024907 (ClTCP22), EVM0022674 (ClTCP15), and EVM0027943 (ClTCP10) showed an increase at the late stage of two florets development. In CYC/TB1 subclade, EVM0047461 (ClCYC2c), EVM0029420 (ClCYC2d), EVM0014549 (ClCYC2f), EVM0021074 (ClCYC2e), EVM0028754 (ClCYC2a) and EVM0053853 (ClCYC2a) were detected to be highly expressed in RFs at two stages, but EVM0027950 (ClCYC1a) was detected to be highly expressed in DFs at two stages.
3.6 Expression analysis of ClCYC2 genes in different Chrysanthemum species
To verify the RNA-seq data and determine ClCYC2 genes, which belong to the class II TCP subclade, and might be responsible for the development of RFs and DFs, qRT-PCR was executed to assess their expression profiles in RFs and DFs during capitulum development. Notably, six ClCYC2 genes were differentially expressed between RFs and DFs at two developmental stages. Almost all of these genes were highly expressed in the RFs of C. lavandulifolium, whereas they showed almost no or very low expression levels in the DFs, which was similar to previous research (Huang et al., 2016; Chen et al., 2018; Wen et al., 2019a). Among them, the expression difference between ClCYC2c and ClCYC2d in the two florets was the most significant (Figure 7).
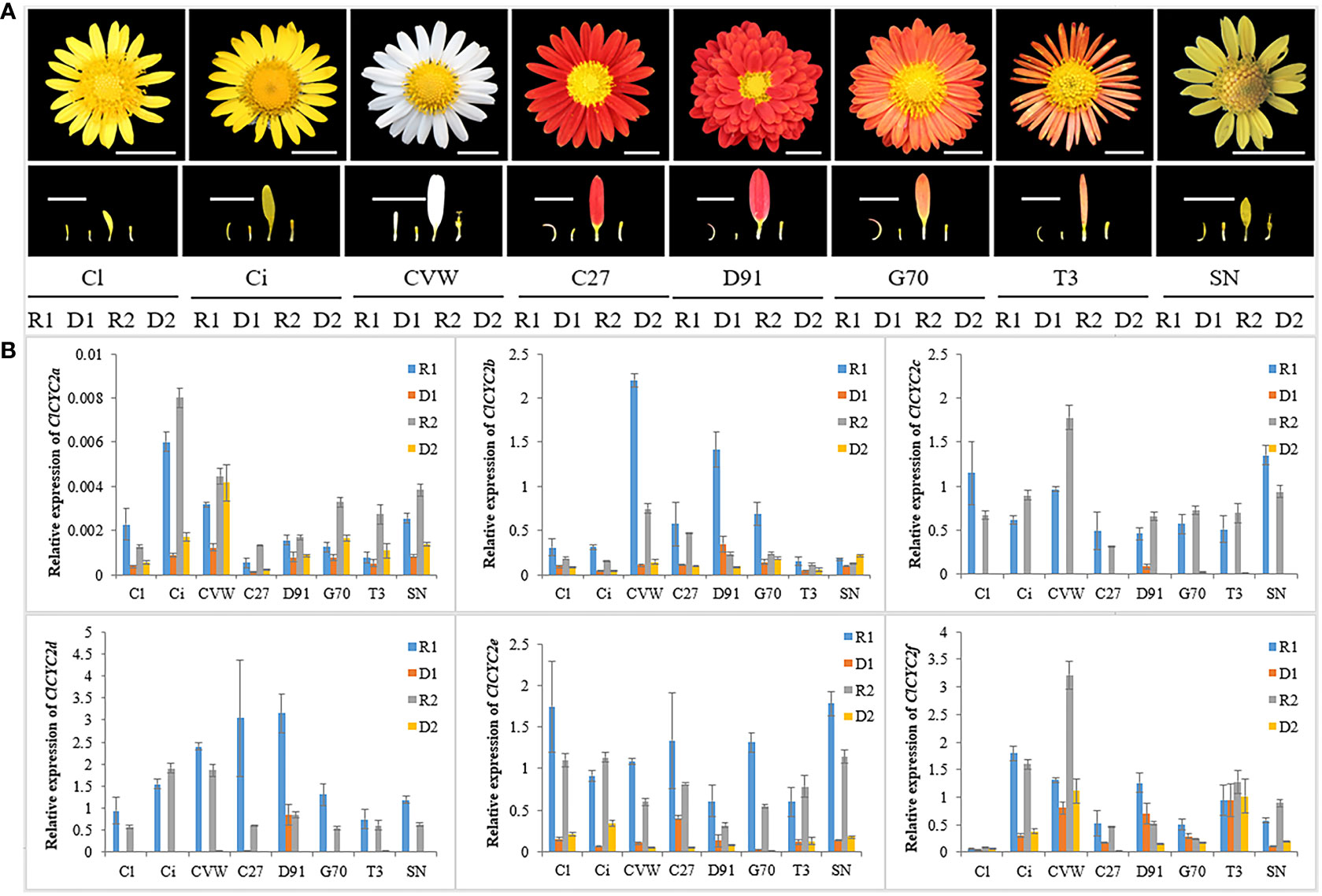
Figure 7 Relative expression levels of ClCYC genes in eight different Chrysanthemum samples using qRT-PCR. (A) Capitulum and DFs and RFs of two development stages of C. lavandulifolium (Cl), C. indicum (Ci), C. vesticum (CVW), C. ×morifolium ‘C27’, ‘D91’, ‘G70’, ‘T3’ and C. aromaticum (SN). Bars=1 cm. (B) Expression levels of six ClCYC2 genes in DFs and RFs of the above samples. Data were normalized to the expression data of ClSAND and expressed as the means ± standard error of three biological replicates.
Next, other seven Chrysanthemum samples from the chrysanthemum germplasm nursery (Figure 7) were used to determine whether these CYC2-like exhibited similar expression patterns in the RFs and DFs of Chrysanthemum species. Notably, the expression patterns of CYC2 genes between two florets of different Chrysanthemum samples were almost similar, which suggested that the functions of the CYC2 gene may be conserved in the development of two florets of Chrysanthemum. In conclusion, we regarded ClCYC2c and ClCYC2d as the key genes responsible for DFs and RFs development in C. lavandulifolium.
3.7 Subcellular localization and transactivation activity of ClCYC2 proteins
Generally, transcription factors (TFs) can regulate the candidate genes by interacting with specific cis-regulatory elements located in their promoter regions, which occur in the nucleus. In this study, according to WoLF PSORT predictions, most ClTCP proteins were anticipated to be located in the nucleus (Table 1). To examine this feature, the CDSs of the ClCYC2a, ClCYC2b, ClCYC2c, ClCYC2d, ClCYC2e, and ClCYC2f without stop codons were amplified with the primer pairs (Supplemental Table 2) and recombined into pBI121-eGFP to generate six fusion constructs, then the fusion proteins and pBI121-eGFP were expressed transiently in N. benthamiana leaves via Agrobacterium-regulated transformation. The results showed that the control vector pBI121-eGFP had a strong fluorescence signal in the whole cell, and the distribution of the fluorescence signal in the cell was not specific. The fluorescence signals of the six ClCYC2 vectors were localized in the nucleus (Figure 8), which was similar to the results in Moso Bamboo (Liu et al., 2018), Vitis vinifera (Jiu et al., 2019) and Ipomoea batatas (Ren et al., 2021). The positive controls for each test to ensure the reliability of the results. These results indicated that six ClCYC2 proteins of C. lavandulifolium were nuclear proteins, and consistent with the general characteristic of nuclear localization of TFs.
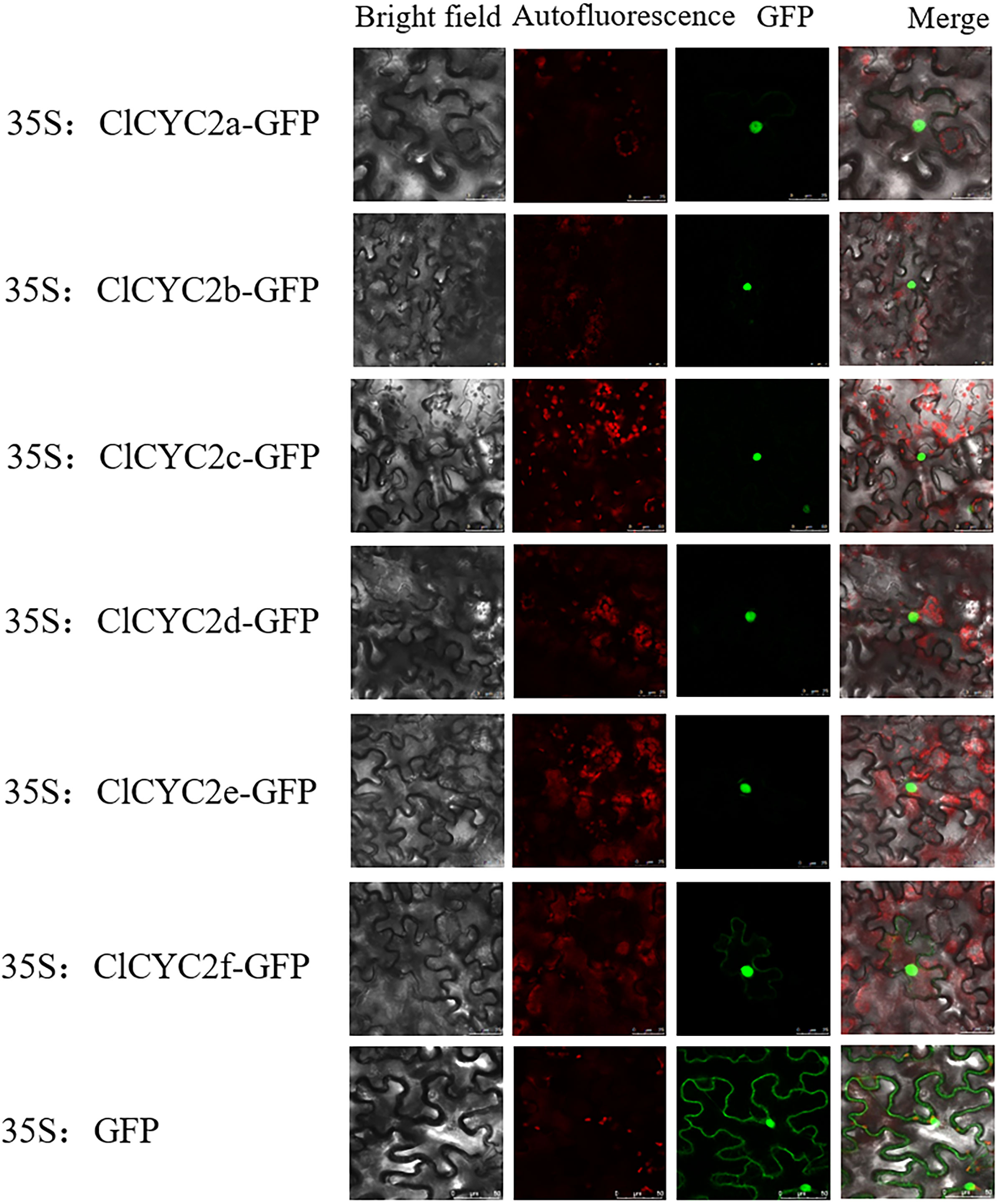
Figure 8 Subcellular localization of six ClCYC2 proteins. Six ClCYC2-GFP fusion recombinants (ClCYC2a-GFP, ClCYC2b-GFP, ClCYC2c-GFP, ClCYC2d-GFP, ClCYC2e-GFP, and ClCYC2f-GFP) and pBI121-GFP as control were transiently expressed in N. benthamiana leaves and observed by a laser scanning confocal microscope 48 hours after injection. Bar=25 µm.
To explore the transactivational activity levels of six ClCYC2 proteins, the pGBKT7-ClCYC2 recombinant vectors (gene-specific primer pairs in Supplementary Table 3), the pGBKT7-53 (positive control) and pGBKT7-lam with pGADT7 empty vectors (negative control) were co-transformed into theY2HGold strain, respectively. All of these transformants grew well and exhibited obvious white colonies on the SD/-Leu-Trp medium (Figure 9). Only the yeast cells carrying ClCYC2c, ClCYC2e, ClCYC2f, and the positive group could grow well on the SD/-Ade-His-Leu-Trp medium. Whereas, the yeast cells harboring ClCYC2a, ClCYC2b, ClCYC2d, and the negative group did not proliferate (Figure 9). Hence, three out of six candidate Class II ClTCP constructs activated the levels of the His3 and Ade2 reporter genes, demonstrating their transactivational activities in the yeast strain. To screen the interacting proteins in the subsequent yeast two-hybridization system, the bait expression vectors with self-activating activities were inhibited by adding 3-AT. The results showed that the bait vectors of ClCYC2c, ClCYC2e, and ClCYC2f could inhibit their self-activating activities by adding 80 mM, 20 mM, and 10 mM 3-AT, respectively.
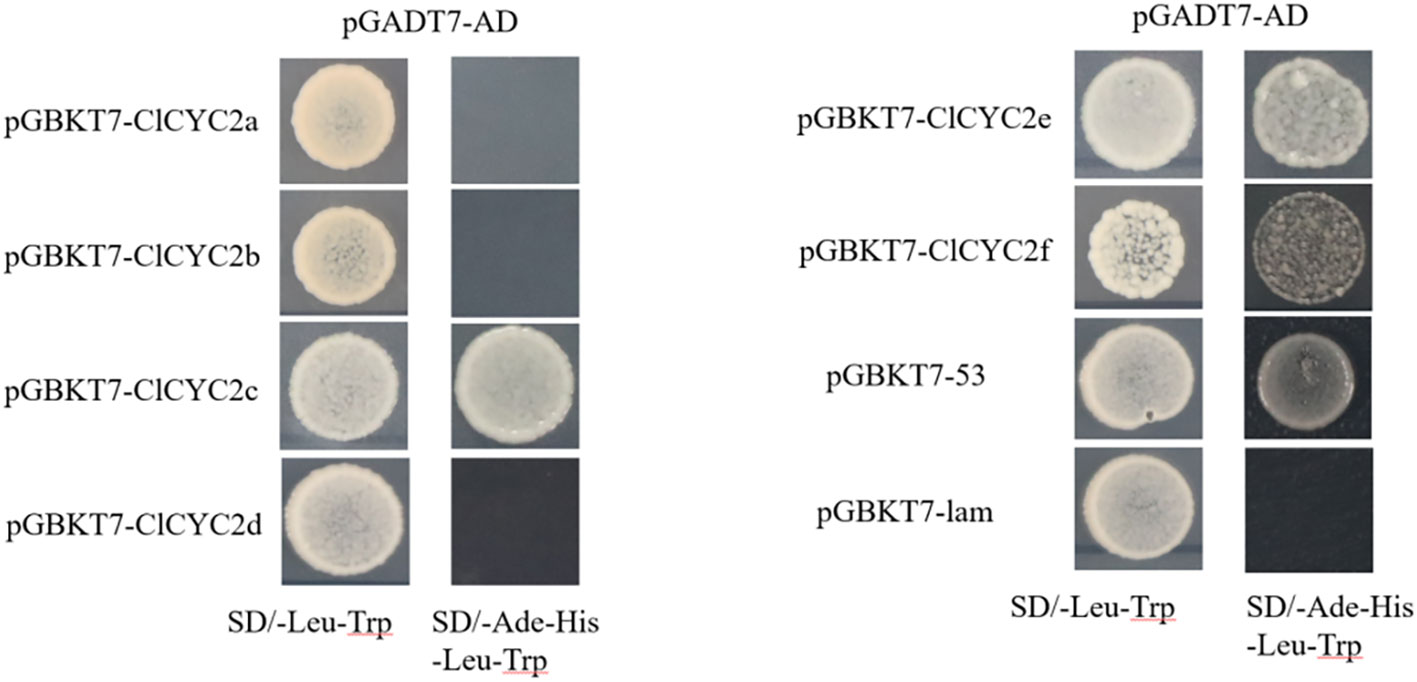
Figure 9 Transactivational analysis of ClCYC2 proteins in Y2HGold strain. The fusion constructs with pGADT7 AD empty, positive constructs (pGBKT7-53) and negative constructs (pGBKT7-lam) were co-transformed into Y2HGold strain and incubated on the SD/-Leu-Trp and SD/-Ade-His-Leu-Trp media.
4 Discussion
TCP protein is a TF specific to plants, which is involved in different aspects of biological and physiological processes throughout plant growth/development (Manassero et al., 2013). TCPs have yet been discovered and characterized in various plants, including Arabidopsis (Riechmann et al., 2000; Li, 2015), M. domestica (Xu et al., 2014), Legume (Ling et al., 2020), Vitis vinifera (Jiu et al., 2019), P. mume (Zhou et al., 2016), Moso Bamboo (Liu et al., 2018) and C. nankingense (Tian et al., 2022). Nonetheless, no comprehensive analysis of TCPs in C. lavandulifolium, a prominent ancestral species of cultivated chrysanthemum has been undertaken. In the study, 40 ClTCPs were determined from C. lavandulifolium genome and transcriptome. Moreover, we performed a comprehensive analysis of the ClTCPs by assessing their phylogenetic relationships, chromosomal position, conserved motifs, gene structures, promoter analysis, expression patterns in different developmental stages and different flower types, and subcellular localization and transactivation activity of key ClTCP proteins. These findings offer a basis for further investigations on the functions of ClTCPs during flowering.
4.1 Evolutionary conservation and divergence of TCPs in C. lavandulifolium
In C. lavandulifolium, 40 TCPs were discovered in the genome and transcriptome, and their distributions on the chromosomes were not uniform. The number of ClTCPs in C. lavandulifolium is about double that of C. nankingense, V. vinifera, and Arabidopsis, which possess 23, 17, and 24 TCP members, respectively (Li, 2015; Jiu et al., 2019; Tian et al., 2022). The collinearity analysis of the V. vinifera and C. lavandulifolium genomes revealed a 1 vs. 3 syntenic association between them, providing compelling evidence for whole-genome triplication (WGT-1) in C. lavandulifolium (Wen et al., 2022). The WGT event was shared by Asteraceae species and may be responsible for the emergence of complex traits in the capitulum. Earlier research has established that CYC2 clade genes play a crucial role in regulating DFs and RFs development in Asteraceae species, including G. hybrida, H. annuus, S. vulgaris, and C. ×morifolium (Tahtiharju et al., 2012; Garces et al., 2016; Huang et al., 2016; Chen et al., 2018). We identified nine CYC2-like genes in C. lavandulifolium (Figure 3). It possesses a larger number of TCPs, amounting to nine members, which surpasses the 7 and 8 members identified in C. nankingense and H. annuus, respectively, but fewer than 25 members found in C. ×morifolium (Song et al., 2023). Besides, tandem duplication events occurred in the evolutionary process in C. lavandulifolium CYC2 genes, which led to CYC2a multicopy genes forming a gene cluster on chromosome 6. The duplication event likely facilitated the proliferation of CYC2c/2d/2e/2f, and subsequent evolutionary processes could have led to subfunctionalization of these genes within the C. lavandulifolium genome (Figure 2). The notable difference in the number of TCPs between C. lavandulifolium and C. nankingense might be due to the divergency time, that C. lavandulifolium and C. nankingense diverged from each other around 7.2 Mya (Wen et al., 2022). The previous result shows the class II TCPs in C. lavandulifolium were significantly expanded (Figure 3).
According to multiple sequence alignments and phylogenetic analyses, all 40 ClTCPs were grouped into 3 subgroups (Figures 1 and 3), which was in accordance with those in Arabidopsis (Li, 2015). Generally, TCPs within a clade have similar gene structures, chromosomal locations and motif distributions, suggesting that TCPs are evolutionarily conserved. For example, we found the highly-conserved TCP domains (motif 1) occurred in all ClTCP proteins (Figure 4), while motif 2 was only present in the class I PCF clade and motif 7 was detected in most of class II CIN proteins (Figure 4B). The close evolutionary relationships are further supported by the fact that the motif compositions and gene structures of ClTCPs exhibit a high degree of consistency. Additionally, our analysis revealed the presence of an arginine-rich R domain at the C-terminus of all ClTCP proteins classified under class II CYC/TB1, and one member (EVM0024660) from CIN. R domain was estimated to increase protein-protein interactions, as do four members (AtTCP2, 12, 18, and 24) in Arabidopsis.
Subcellular localization indicated that six ClCYC2 proteins of C. lavandulifolium were nuclear proteins, but these six CYC2 members have different transcriptional activation activities, which may be affected by several factors, including the binding ability of the TCP transcription factors to the target genes, differences in gene structures and sequences, differences in expression regulatory mechanisms, and the involvement of other cofactors (Martin-Trillo et al., 2011; Tahtiharju et al., 2012; Valsecchi et al., 2013; Parapunova et al., 2014). Further exploration and experimental verification will help to fully understand the regulatory mechanisms of these key ClCYC2 genes.
4.2 The potential roles of ClTCPs in plant growth and development
The function of a gene is closely correlated with its expression patterns (Xu et al., 2015). Herein, we determined the expression patterns of all ClTCPs during flower development by RNA-seq data reported previously (Wen et al., 2019a; Wen et al., 2022) and detailed expression patterns of key class II ClTCPs (ClCYC2) in different Chrysanthemum samples via real-time RT-PCR. The result indicated that these genes exhibited unique expression patterns during capitulum development and in different flower types. In detail, most ClTCPs were specifically expressed in flowers during capitulum development, and TCPs were categorized into three clusters in accordance with their expression patterns in DFs and RFs. The expression levels of CYC2 genes in RFs were upregulated compared to DFs, but the expression profiles of CYC1 and CYC3 group genes were different from those of CYC2 genes in C. lavandulifolium, which presumed that they had different functions during flower development. These suggested that TCPs in C. lavandulifolium were associated with the growth of capitula or two florets.
In earlier research (Sarvepalli and Nath, 2018), the CIN subclade TCPs were found to primarily regulate leaf development, floral organ development, and flowering time. In Arabidopsis, 5 CIN-like genes have been reported to undergo post-transcriptional regulation by microRNA319 (miR-319). Notably, the regulation of TCP4 by miR-319a is crucial for the development of stamens and petals (Nag et al., 2009). In C. lavandulifolium, four homologs of CIN subclade genes (ClTCP24, ClTCP25, ClTCP26 and ClTCP27) included the potential binding sites for miR319 (Figure 1C). ClTCP24, ClTCP25 and ClTCP26 are upregulated in leaves, suggesting their potential roles in leaf development. The conservation of miR319 targeting may extend to other TCPs as well (Palatnik et al., 2003; Seki et al., 2020).
Moreover, promoter analysis shows that the promoter region of ClTCPs was found to contain a substantial number of hormone-responsive elements (Figure 5), implying these genes might exert significant influences on hormone-responsive plant developmental processes. Previous reports have shown that TFs and phytohormones play important roles in determining flower size in Arabidopsis (Krizek and Anderson, 2013). Cytokinin has been demonstrated to influence the duration of cell division in developing floral organs (Bartrina et al., 2011). Considering the abundant hormone-responsive elements distributed in ClTCP gene promoters (Figure 5), we speculate that ClTCPs may serve as important regulators of hormone-induced alterations in plant development. Of course, this hypothesis needs to be further tested.
4.3 Contribution of CYC2 genes to the morphological differences of two florets in Chrysanthemum
Flower development in the angiosperm is a complex physiological and morphological process, which was determined by a set of TFs regulatory networks (Soltis et al., 2002). Much evidence has already suggested that TCPs, especially the CYC2 clade genes affect the morphology of capitula by regulating the reproductive organ development, inflorescence architecture and floral symmetry (Juntheikki-Palovaara et al., 2014; Huang et al., 2016; Chen et al., 2018; Elomaa et al., 2018).
Our investigation revealed the presence of 13 genes in C. lavandulifolium that belong to the CYC/TB1 subclade, CYC2 group has nine members with the largest number. Among them, six TCPs from CYC2 group, EVM0047461 (ClCYC2c), EVM0029420 (ClCYC2d), EVM0014549 (ClCYC2f), EVM0021074 (ClCYC2e), EVM0028754 (ClCYC2a) and KX161380.1 (ClCYC2b) were all highly expressed in the RFs, whereas they shown almost no or very low expression levels in DFs (Figure 7). And the difference in expression was universal in different Chrysanthemum species, which suggested the functions of the CYC2 gene may be conserved in the two florets development of Chrysanthemum, whereas certain members of the CYC/TB1 subclade experienced functional divergence subsequent to gene duplication events. Based on previous studies, it was hypothesized that CYC2c and CYC2d are mainly involved in RF identity and thus affect the development of capitulum, CYC2b and CYC2e tend to regulate petal length in RFs (Shen et al., 2021), and CYC2c can interact with the CYC2f promoter to modulate floral symmetry development (Yuan et al., 2020). Moreover, six CYC2 genes were expressed in C. lavandulifolium at the early capitulum developmental stages, which was consistent with the results of nucleic acid in situ hybridization of Gerbera and S. vulgaris that detected CYC2-like gene expression in the meristem and floret primordium at the early capitula development (Garces et al., 2016; Elomaa et al., 2018). This implied that CYC2 genes in C. lavandulifolium were expressed at the early stages of the capitula, and ultimately played roles in the ontogenetic differences between ray and disc florets.
5 Conclusion
In summary, the first genome-wide analyses of TCPs in C. lavandulifolium were conducted. 40 ClTCPs were discovered and distributed on 8 chromosomes. These ClTCPs were classified into two main groups in accordance with the phylogenetic analysis and structural characteristics. The ClTCP promoter sequences had many different kinds of cis-acting elements, suggesting that ClTCPs were controlled by a complex regulatory network. ClTCPs, especially those from class II CYC/TB1 clade, may be crucial for the two florets development in Chrysanthemum as indicated by their expression patterns and previous studies. Notably, six candidate ClCYC2 genes were higher expressed in RFs than those in DFs, which speculated that the CYC2 genes may be functionally conserved in two florets development in Chrysanthemum. The subcellular localization and transactivation activity analysis of six candidate TCP proteins were also examined in C. lavandulifolium. In short, our study increased the understanding of ClTCP functions during flower development in Chrysanthemum species. Based on this study, in the future we can validate the functions of key TCPs in native plants, and deeply study the interactions between the key ClTCPs and their upstream and downstream regulatory mechanisms, to resolve the molecular network of TCPs involved in the capitulum development of Chrysanthemum.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
XWu: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. JL: Resources, Software, Writing – review & editing. XWe: Resources, Writing – review & editing. QZ: Writing – review & editing. SD: Conceptualization, Funding acquisition, Project administration, Supervision, Validation, Writing – review & editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. The research was supported by the Natural Science Foundation of China (grant no. 32371948 & 31530064) and the National Key Research and Development Program (No. 2018YFD1000405).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1276123/full#supplementary-material
Abbreviations
ABA, Abscisic acid; cDNA, complementary DNA; CIN, CINCINNATA; CYC, CYCLOIDEA; FPKM, Fragments per kilobase of transcript per million mapped reads; DF, Disc floret; GFP, Green fluorescent protein; MeJA, Methyl Jasmonate; PCF, Proliferating cell factor; qRT-PCR, Real-time quantitative polymerase chain reaction; RF, Ray floret; TB1, Teosinte branched 1; TCP, TEOSINTE BRANCHED/CYCLOIDEA/PCF; TF, Transcription factor; 3-AT, 3-Amino-1,2,4-triazole.
References
Aguilar-Martinez, J. A., Poza-Carrion, C., Cubas, P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19 (2), 458–472. doi: 10.1105/tpc.106.048934
Bartrina, I., Otto, E., Strnad, M., Werner, T., Schmuelling, T. (2011). Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23 (1), 69–80. doi: 10.1105/tpc.110.079079
Bello, M. A., Alvarez, I., Torices, R., Fuertes-Aguilar, J. (2013). Floral development and evolution of capitulum structure in Anacyclus (Anthemideae, Asteraceae). Ann. Bot. 112 (8), 1597–1612. doi: 10.1093/aob/mcs301
Bello, M. A., Cubas, P., Alvarez, I., Sanjuanbenito, G., Fuertes-Aguilar, J. (2017). Evolution and expression patterns of CYC/TB1 genes in Anacyclus: phylogenetic insights for floral symmetry genes in Asteraceae. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00589
Broholm, S. K., Taehtiharju, S., Laitinen, R., Albert, V. A., Teeri, T. H., Elomaa, P. (2008). A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl. Acad. Sci. U. S. A. 105 (26), 9117–9122. doi: 10.1073/pnas.0801359105
Chapman, M. A., Tang, S., Draeger, D., Nambeesan, S., Shaffer, H., Barb, J. G., et al. (2012). Genetic analysis of floral symmetry in Van Gogh’s sunflowers reveals independent recruitment of CYCLOIDEA genes in the Asteraceae. PloS Genet. 8 (3), e1002628. doi: 10.1371/journal.pgen.1002628
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, J., Shen, C., Guo, Y., Rao, G. (2018). Patterning the Asteraceae capitulum: duplications and differential expression of the flower symmetry CYC2-like Genes. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00551
Crawford, B. C. W., Nath, U., Carpenter, R., Coen, E. S. (2004). CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol. 135 (1), 244–253. doi: 10.1104/pp.103.036368
Cubas, P., Lauter, N., Doebley, J., Coen, E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18 (2), 215–222. doi: 10.1046/j.1365-313X.1999.00444.x
Dai, S., Wang, W., Huang, J. (2002). Advances of researches on phylogeny of Dendranthema and origin of chrysanthemum. J. Beijing Forest. Univ. 24, 230–234. doi: 10.3321/j.issn:1000-1522.2002.05.045
Danisman, S., van Dijk, A. D. J., Bimbo, A., van der Wal, F., Hennig, L., de Folter, S., et al. (2013). Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J. Exp. Bot. 64 (18), 5673–5685. doi: 10.1093/jxb/ert337
Daviere, J.-M., Wild, M., Regnault, T., Baumberger, N., Eisler, H., Genschik, P., et al. (2014). Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr. Biol. 24 (16), 1923–1928. doi: 10.1016/j.cub.2014.07.012
Doebley, J. (1993). Genetics, development and plant evolution. Curr. Opin. Genet. Dev. 3 (6), 865–872. doi: 10.1016/0959-437x(93)90006-b
Doebley, J., Lukens, L. (1998). Transcriptional regulators and the evolution of plant form. Plant Cell 10 (7), 1075–1082. doi: 10.1105/tpc.10.7.1075
Doebley, J., Stec, A., Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386 (6624), 485–488. doi: 10.1038/386485a0
Elomaa, P., Zhao, Y., Zhang, T. (2018). Flower heads in Asteraceae-recruitment of conserved developmental regulators to control the flower-like inflorescence architecture. Horticult. Res. 5. doi: 10.1038/s41438-018-0056-8
Feng, X., Zhao, Z., Tian, Z., Xu, S., Luo, Y., Cai, Z., et al. (2006). Control of petal shape and floral zygomorphy in Lotus japonicus. Proc. Natl. Acad. Sci. U. S. A. 103 (13), 4970–4975. doi: 10.1073/pnas.0600681103
Gao, Q., Tao, J., Yan, D., Wang, Y., Li, Z. (2008). Expression differentiation of CYC-like floral symmetry genes correlated with their protein sequence divergence in Chirita heterotricha (Gesneriaceae). Dev. Genes Evol. 218 (7), 341–351. doi: 10.1007/s00427-008-0227-y
Garces, H. M. P., Spencer, V. M. R., Kim, M. (2016). Control of floret symmetry by RAY3, SvDIV1B, and SvRAD in the capitulum of Senecio vulgaris. Plant Physiol. 171 (3), 2055–2068. doi: 10.1104/pp.16.00395
Gastaldi, V., Lucero, L. E., Ferrero, L. V., Ariel, F. D., Gonzalez, D. H. (2020). Class-I TCP transcription factors activate the SAUR63 gene subfamily in gibberellin-dependent stamen filament elongation. Plant Physiol. 182 (4), 2096–2110. doi: 10.1104/pp.19.01501
Guan, P., Ripoll, J. J., Wang, R., Vuong, L., Bailey-Steinitz, L. J., Ye, D., et al. (2017). Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. U. S. A. 114 (9), 2419–2424. doi: 10.1073/pnas.1615676114
Guo, Z., Fujioka, S., Blancaflor, E. B., Miao, S., Gou, X., Li, J. (2010). TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 22 (4), 1161–1173. doi: 10.1105/tpc.109.069203
Herve, C., Dabos, P., Bardet, C., Jauneau, A., Auriac, M. C., Ramboer, A., et al. (2009). In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol. 149 (3), 1462–1477. doi: 10.1104/pp.108.126136
Hsu, H., He, C., Kuo, W., Hsin, K., Lu, J., Pan, Z., et al. (2018). Genetic analysis of floral symmetry transition in African Violet suggests the involvement of trans-acting factor for CYCLOIDEA expression shifts. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01008
Huang, D., Li, X., Sun, M., Zhang, T., Pan, H., Cheng, T., et al. (2016). Identification and characterization of CYC-like genes in regulation of ray floret development in Chrysanthemum morifolium. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01633
Ibanez, M. S., Mercado, M. I., Coll Araoz, M. V., Zannier, M. L., Grau, A., Ponessa, G. I. (2017). Flower structure and developmental stages of the capitulum of Smallanthus sonchifolius (Asteraceae): reproductive implications. J. Plant Res. 130 (2), 327–337. doi: 10.1007/s10265-017-0904-x
Jiu, S., Xu, Y., Wang, J., Wang, L., Wang, S., Ma, C., et al. (2019). Genome-wide identification, characterization, and transcript analysis of the TCP transcription factors in Vitis vinifera. Front. Genet. 10. doi: 10.3389/fgene.2019.01276
Juntheikki-Palovaara, I., Tahtiharju, S., Lan, T. Y., Broholm, S. K., Rijpkema, A. S., Ruonala, R., et al. (2014). Functional diversification of duplicated CYC2 clade genes in regulation of inflorescence development in Gerbera hybrida (Asteraceae). Plant J. 79 (5), 783–796. doi: 10.1111/tpj.12583
Kosugi, S., Ohashi, Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30 (3), 337–348. doi: 10.1046/j.1365-313X.2002.01294.x
Koyama, T., Mitsuda, N., Seki, M., Shinozaki, K., Ohme-Takagi, M. (2010). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22 (11), 3574–3588. doi: 10.1105/tpc.110.075598
Krizek, B. A., Anderson, J. T. (2013). Control of flower size. J. Exp. Bot. 6), 1427–1437. doi: 10.1093/jxb/ert025
Kubota, A., Ito, S., Shim, J. S., Johnson, R. S., Song, Y., Breton, G., et al. (2017). TCP4-dependent induction of CONSTANS transcription requires GIGANTEA in photoperiodic flowering in Arabidopsis. PloS Genet. 13 (6). doi: 10.1371/journal.pgen.1006856
Lan, J., Qin, G. (2020). The regulation of CIN-like TCP transcription factors. Int. J. Mol. Sci. 21 (12). doi: 10.3390/ijms21124498
Lei, N., Yu, X., Li, S., Zeng, C., Zou, L., Liao, W., et al. (2017). Phylogeny and expression pattern analysis of TCP transcription factors in cassava seedlings exposed to cold and/or drought stress. Sci. Rep. 7. doi: 10.1038/s41598-017-09398-5
Leng, X., Wei, H., Xu, X., Ghuge, S. A., Jia, D., Liu, G., et al. (2019). Genome-wide identification and transcript analysis of TCP transcription factors in grapevine. BMC Genomics 20 (1). doi: 10.1186/s12864-019-6159-2
Li, S. (2015). The Arabidopsis thaliana TCP transcription factors: A broadening horizon beyond development. Plant Signaling Behav. 10 (7), e1044192–e1044192. doi: 10.1080/15592324.2015.1044192
Li, F., Chen, S., Fang, W., Liu, Z., Zhang, F. (2010). Histological structure observation on the floral development of anemone type chrysanthemum. Acta Hortic. Sin. 37 (12), 1961–1968. doi: 10.16420/j.issn.0513-353x.2010.12.024
Li, D., Zhang, H., Mou, M., Chen, Y., Xiang, S., Chen, L., et al. (2019). Arabidopsis Class II TCP transcription factors integrate with the FT-FD module to control flowering. Plant Physiol. 181 (1), 97–111. doi: 10.1104/pp.19.00252
Ling, L., Zhang, W., An, Y., Du, B., Wang, D., Guo, C. (2020). Genome-wide analysis of the TCP transcription factor genes in five legume genomes and their response to salt and drought stresses. Funct. Integr. Genomics 20 (4), 537–550. doi: 10.1007/s10142-020-00733-0
Liu, H., Li, F., Gao, Y., Chen, F., Xiang, Y. (2018). TCP transcription factors in Moso Bamboo (Phyllostachys edulis): genome-wide identification and expression analysis. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01263
Liu, H., Sun, M., Du, D., Pan, H., Cheng, T., Wang, J., et al. (2016). Whole-transcriptome analysis of differentially expressed genes in the ray florets and disc florets of Chrysanthemum morifolium. BMC Genomics 17. doi: 10.1186/s12864-016-2733-z
Lopez, J. A., Sun, Y., Blair, P. B., Mukhtar, M. S. (2015). TCP three-way handshake: linking developmental processes with plant immunity. Trends Plant Sci. 20 (4), 238–245. doi: 10.1016/j.tplants.2015.01.005
Lu, C. (2022). Transcriptional regulatory mechanism of differential accumulation of carotenoids in the flower of Chrysanthemum ×morifolium. [doctor’s thesis] (Beijing: Beijing Forestry University).
Lucero, L. E., Manavella, P. A., Gras, D. E., Ariel, F. D., Gonzalez, D. H. (2017). Class I and Class II TCP transcription factors modulate SOC1-dependent flowering at multiple levels. Mol. Plant 10 (12), 1571–1574. doi: 10.1016/j.molp.2017.09.001
Lucero, L. E., Uberti-Manassero, N. G., Arce, A. L., Colombatti, F., Alemano, S. G., Gonzalez, D. H. (2015). TCP15 modulates cytokinin and auxin responses during gynoecium development in Arabidopsis. Plant J. 84 (2), 267–282. doi: 10.1111/tpj.12992
Luo, D., Carpenter, R., Vincent, C., Copsey, L., Coen, E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383 (6603), 794–799. doi: 10.1038/383794a0
Ma, X., Ma, J., Fan, D., Li, C., Jiang, Y., Luo, K. (2016). Genome-wide identification of TCP family transcription factors from Populus euphratica and their involvement in leaf shape regulation. Sci. Rep. 6. doi: 10.1038/srep32795
Manassero, N. G. U., Viola, I. L., Welchen, E., Gonzalez, D. H. (2013). TCP transcription factors: architectures of plant form. Biomol. Concepts 4 (2), 111–127. doi: 10.1515/bmc-2012-0051
Martin-Trillo, M., Cubas, P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15 (1), 31–39. doi: 10.1016/j.tplants.2009.11.003
Martin-Trillo, M., Gonzalez Grandio, E., Serra, F., Marcel, F., Luisa Rodriguez-Buey, M., Schmitz, G., et al. (2011). Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J. 67 (4), 701–714. doi: 10.1111/j.1365-313X.2011.04629.x
Nag, A., King, S., Jack, T. (2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 106 (52), 22534–22539. doi: 10.1073/pnas.0908718106
Niwa, M., Daimon, Y., Kurotani, K.-I., Higo, A., Pruneda-Paz, J. L., Breton, G., et al. (2013). BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. Plant Cell 25 (4), 1228–1242. doi: 10.1105/tpc.112.109090
Palatnik, J. F., Allen, E., Wu, X. L., Schommer, C., Schwab, R., Carrington, J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425 (6955), 257–263. doi: 10.1038/nature01958
Parapunova, V., Busscher, M., Busscher-Lange, J., Lammers, M., Karlova, R., Bovy, A. G., et al. (2014). Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 14. doi: 10.1186/1471-2229-14-157
Pu, Y., Huang, H., Wen, X. H., Lu, C. F., Zhang, B. H., Gu, X. Q., et al. (2020). Comprehensive transcriptomic analysis provides new insights into the mechanism of ray floret morphogenesis in chrysanthemum. BMC Genomics 21 (1). doi: 10.1186/s12864-020-07110-y
Qi, S., Yang, L., Wen, X., Hong, Y., Song, X., Zhang, M., et al. (2016). Reference gene selection for RT-qPCR analysis of flower development in Chrysanthemum morifolium and Chrysanthemum lavandulifolium. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00287
Ren, L., Wu, H. X., Zhang, T. T., Ge, X. Y., Wang, T. L., Zhou, W. Y., et al. (2021). Genome-wide identification of TCP transcription factors family in sweet potato reveals significant roles of miR319-targeted TCPs in leaf anatomical morphology. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.686698
Resentini, F., Felipo-Benavent, A., Colombo, L., Blazquez, M. A., Alabadi, D., Masiero, S. (2015). TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol. Plant 8 (3), 482–485. doi: 10.1016/j.molp.2014.11.018
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290 (5499), 2105–2110. doi: 10.1126/science.290.5499.2105
Sarvepalli, K., Nath, U. (2018). CIN-TCP transcription factors: Transiting cell proliferation in plants. IUBMB Life 70 (8), 718–731. doi: 10.1002/iub.1874
Seki, K., Komatsu, K., Tanaka, K., Hiraga, M., Kajiya-Kanegae, H., Matsumura, H., et al. (2020). A CIN-like TCP transcription factor (LsTCP4) having retrotransposon insertion associates with a shift from Salinas type to Empire type in crisphead lettuce (Lactuca sativa L.). Horticult. Res. 7, 15–15. doi: 10.1038/s41438-020-0241-4
Shen, C., Chen, J., Zhang, C., Rao, G., Guo, Y. (2021). Dysfunction of CYC2g is responsible for the evolutionary shift from radiate to disciform flowerheads in the Chrysanthemum group (Asteraceae: Anthemideae). Plant J. 106 (4), 1024–1038. doi: 10.1111/tpj.15216
Shi, P., Guy, K. M., Wu, W., Fang, B., Yang, J., Zhang, M., et al. (2016). Genome-wide identification and expression analysis of the ClTCP transcription factors in Citrullus lanatus. BMC Plant Biol. 16. doi: 10.1186/s12870-016-0765-9
Soltis, D. E., Soltis, P. S., Albert, V. A., Oppenheimer, D. G., dePamphilis, C. W., Ma, H., et al. (2002). Missing links: the genetic architecture of flowers correction of flower and floral diversification. Trends Plant Sci. 7 (1), 22–31; dicussion 31-24. doi: 10.1016/s1360-1385(01)02098-2
Song, A. P., Su, J. S., Wang, H. B., Zhang, Z. R., Zhang, X. T., van de Peer, Y., et al. (2023). Analyses of a chromosome-scale genome assembly reveal the origin and evolution of cultivated chrysanthemum. Nat. Commun. 14 (1). doi: 10.1038/s41467-023-37730-3
Stevens, M., Darris, D. C., Lambert, S. M. (2001). Ethnobotany, culture, management, and use of common camas. Native Plants J. 2 (1), 47–53. doi: 10.3368/npj.2.1.47
Su, S., Xiao, W., Guo, W., Yao, X., Xiao, J., Ye, Z., et al. (2017). The CYCLOIDEA-RADIALIS module regulates petal shape and pigmentation, leading to bilateral corolla symmetry in Torenia fournieri (Linderniaceae). New Phytol. 215 (4), 1582–1593. doi: 10.1111/nph.14673
Tahtiharju, S., Rijpkema, A. S., Vetterli, A., Albert, V. A., Teeri, T. H., Elomaa, P. (2012). Evolution and diversification of the CYC/TB1 gene family in Asteraceae-a comparative study in Gerbera (Mutisieae) and Sunflower (Heliantheae). Mol. Biol. Evol. 29 (4), 1155–1166. doi: 10.1093/molbev/msr283
Takeda, T., Amano, K., Ohto, M.-A., Nakamura, K., Sato, S., Kato, T., et al. (2006). RNA interference of the Arabidopsis putative transcription factor TCP16 gene results in abortion of early pollen development. Plant Mol. Biol. 61 (1-2), 165–177. doi: 10.1007/s11103-006-6265-9
Tian, C., Zhai, L., Zhu, W., Qi, X., Yu, Z., Wang, H., et al. (2022). Characterization of the TCP gene family in Chrysanthemum nankingense and the role of CnTCP4 in cold tolerance. Plants-Basel 11 (7). doi: 10.3390/plants11070936
Valsecchi, I., Guittard-Crilat, E., Maldiney, R., Habricot, Y., Lignon, S., Lebrun, R., et al. (2013). The intrinsically disordered C-terminal region of Arabidopsis thaliana TCP8 transcription factor acts both as a transactivation and self-assembly domain. Mol. Biosyst. 9 (9), 2282–2295. doi: 10.1039/c3mb70128j
Viola, I. L., Camoirano, A., Gonzalez, D. H. (2016). Redox-dependent modulation of anthocyanin biosynthesis by the TCP transcription factor TCP15 during exposure to high light intensity conditions in Arabidopsis. Plant Physiol. 170 (1), 74–85. doi: 10.1104/pp.15.01016
Wei, W., Hu, Y., Cui, M., Han, Y., Gao, K., Feng, J. (2016). Identification and transcript analysis of the TCP transcription factors in the diploid woodland strawberry Fragaria vesca. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01937
Wen, X., Li, J., Wang, L., Lu, C., Gao, Q., Xu, P., et al. (2022). The Chrysanthemum lavandulifolium genome and the molecular mechanism underlying diverse capitulum types. Horticult. Res. 9. doi: 10.1093/hr/uhab022
Wen, X., Qi, S., Huang, H., Wu, X., Zhang, B., Fan, G., et al. (2019a). The expression and interactions of ABCE-class and CYC2-like genes in the capitulum development of Chrysanthemum lavandulifolium and C. ×morifolium. Plant Growth Regul. 88 (3), 205–214. doi: 10.1007/s10725-019-00491-5
Wen, X., Qi, S., Yang, L., Hong, Y., Dai, S. (2019b). Expression pattern of candidate genes in early capitulum morphogenesis of Chrysanthemum lavandulifolium. Scientia Hortic. 252, 332–341. doi: 10.1016/j.scienta.2019.03.064
Xu, R., Sun, P., Jia, F., Lu, L., Li, Y., Zhang, S., et al. (2014). Genomewide analysis of TCP transcription factor gene family in Malus domestica. J. Genet. 93 (3), 733–746. doi: 10.1007/s12041-014-0446-0
Xu, Z., Sun, L., Zhou, Y., Yang, W., Cheng, T., Wang, J., et al. (2015). Identification and expression analysis of the SQUAMOSA promoter-binding protein (SBP)-box gene family in Prunus mume. Mol. Genet. Genomics 290 (5), 1701–1715. doi: 10.1007/s00438-015-1029-3
Yang, X., Pang, H., Liu, B., Qiu, Z., Gao, Q., Wei, L., et al. (2012). Evolution of double positive autoregulatory feedback loops in CYCLOIDEA2 clade genes is associated with the origin of floral zygomorphy. Plant Cell 24 (5), 1834–1847. doi: 10.1105/tpc.112.099457
Yuan, C., Huang, D., Yang, Y., Sun, M., Cheng, T., Wang, J., et al. (2020). CmCYC2-like transcription factors may interact with each other or bind to the promoter to regulate floral symmetry development in Chrysanthemum morifolium. Plant Mol. Biol. 103 (1-2), 159–171. doi: 10.1007/s11103-020-00981-5
Zhang, T., Zhao, Y., Juntheikki, I., Mouhu, K., Broholm, S. K., Rijpkema, A. S., et al. (2017). Dissecting functions of SEPALLATA-like MADS box genes in patterning of the pseudanthial inflorescence of Gerbera hybrida. New Phytol. 216 (3), 939–954. doi: 10.1111/nph.14707
Zhao, Y., Broholm, S. K., Wang, F., Rijpkema, A. S., Lan, T., Albert, V. A., et al. (2020). TCP and MADS-Box transcription factor networks regulate heteromorphic flower type identity in Gerbera hybrida. Plant Physiol. 184 (3), 1455–1468. doi: 10.1104/pp.20.00702
Zhao, Y., Zhang, T., Broholm, S. K., Tähtiharju, S., Mouhu, K., Albert, V. A., et al. (2016). Evolutionary co-option of floral meristem identity genes for patterning of the flower-like Asteraceae inflorescence. Plant Physiol. 172, 284–296. doi: 10.1104/pp.16.01711
Keywords: TCP gene family, expression analysis, capitulum development, ray and disc floret, Chrysanthemum
Citation: Wu X, Li J, Wen X, Zhang Q and Dai S (2023) Genome-wide identification of the TCP gene family in Chrysanthemum lavandulifolium and its homologs expression patterns during flower development in different Chrysanthemum species. Front. Plant Sci. 14:1276123. doi: 10.3389/fpls.2023.1276123
Received: 11 August 2023; Accepted: 13 September 2023;
Published: 29 September 2023.
Edited by:
Dayong Li, Beijing Vegetable Research Center, ChinaReviewed by:
Junming Zhao, Sichuan Agricultural University, ChinaChao Ma, China Agricultural University, China
Copyright © 2023 Wu, Li, Wen, Zhang and Dai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Silan Dai, c2lsYW5kYWlAc2luYS5jb20=
 Xiaoyun Wu
Xiaoyun Wu Junzhuo Li
Junzhuo Li Xiaohui Wen
Xiaohui Wen Qiuling Zhang1
Qiuling Zhang1 Silan Dai
Silan Dai