- School of Life Sciences, Jiangsu University, Zhenjiang, China
Sclerotinia sclerotiorum (Lib.) de Bary is a highly destructive fungal pathogen that seriously damages the yield and quality of Brassica napus worldwide. The complex interaction between the B. napus and S. sclerotiorum system has presented significant challenges in researching rapeseed defense strategies. Here, we focus on the infection process of S. sclerotiorum, the defense mechanisms of rapeseed, and recent research progress in this system. The response of rapeseed to S. sclerotiorum is multifaceted; this review aims to provide a theoretical basis for rapeseed defense strategies.
Introduction
Rapeseed (Brassica napus) is an allopolyploidy (AACC) resulting from the natural hybridization of Brassica rapa (AA) and Brassica oleracea (CC) (Chalhoub et al., 2014). It is an important and widely grown oil crop rich in oil and protein content (Chen et al., 2015; Chew, 2020). Rapeseed can be classified into three types based on its growth requirements: annual spring type, biennial winter type, and semi-winter type (Wang et al., 2011). Winter types necessitate an extended period of low-temperature vernalization before flowering, while spring types do not require this phase. Semi-winter types exhibit limited cold tolerance and are suitable for regions with moderately cold winter temperatures (>0°C) (Wang et al., 2011; Leijten et al., 2018; Matar et al., 2021). Rapeseed, oil palm, and soybean are the three primary sources of edible plant oil. “Double-low” rapeseed oil, also known as canola oil quality (glucosinolates <30 μmol/g, erucic acid <2%), is rich in unsaturated fatty acids (oleic acid, linoleic acid, linolenic acid, etc.) and various nutrients (phenols, phytosterols, vitamins, etc.). Consequently, the market share of rapeseed oil is consistently on the rise (Beyzi et al., 2019; Ye and Liu, 2023). Rapeseed is also used in animal feed and biofuel production, with high economic value and market potential (Jiang et al., 2019).
Sclerotinia sclerotiorum (Lib.) de Bary is the ascomycetes’ highly destructive fungal pathogen. It is widespread and infects over 700 plant species, including many economically important crops like rapeseed, sunflower (Helianthus annuus), peanut (Arachis hypogaea), soybean (Glycine max), garden lettuce (Lactuca sativa), and sugar beet (Beta vulgaris) (Boland and Hall, 1994; Uloth et al., 2013; Xia et al., 2019; Zhang et al., 2021). Sclerotinia stem rot (SSR) is the primary disease caused by S. sclerotiorum, resulting in the necrosis of stems and leaves in rapeseed, which can lead to yield losses of up to 80% in severe cases (Li et al., 2006a). S. sclerotiorum exists in the form of resting bodies known as sclerotia, which can withstand harsh environmental conditions. When the environment becomes suitable for growth, these sclerotia germinate, adopting either a myceliogenic or carpogenic form, producing hyphae or apothecia. These apothecia release ascospores that infect plant tissue (Bolton et al., 2006). Ascospores land on plant tissues, germinate as mycelium, and secrete oxalic acid (OA), cell wall degrading enzymes (CWDEs), and other substances to facilitate colonization (Kabbage et al., 2015; Xia et al., 2019). Previous studies have shown the critical role of OA synthesis and secretion in the pathogenesis of S. sclerotiorum, with fungal mutants deficient in OA production being non-pathogenic. The pathogen elevates the level of reactive oxygen species (ROS), and it induces a hypersensitive response (HR) in plant cells by secreting OA into plant tissues, ultimately leading to programmed cell death (PCD) (Godoy et al., 1990; Kim et al., 2008). No varieties with complete resistance to SSR have been reported in rapeseed. Disease control primarily relies on field management and fungicide application. However, this requires growers to predict the timing of S. sclerotiorum infection, which may also result in environmental pollution and presents several limitations due to the lack of effective prediction methods (Bolton et al., 2006).
In recent years, the increasing global demand for rapeseed has led to intensified cultivation practices, increasing the urgency for effective strategies against S. sclerotiorum. Developments in various tools, techniques, and expanded research on S. sclerotiorum have generated valuable insights into defense strategies for rapeseed. Here, we introduce the infection process of S. sclerotiorum, the defense strategies of rapeseed, recent research on the identification of defense-related genes in rapeseed, and the resistance strategies of transgenic rapeseed. The B. napus–S. sclerotiorum system has a complex regulatory network, and this review aims to provide a reference for future studies on S. sclerotiorum resistance strategies in rapeseed.
The infection process of S. sclerotiorum
S. sclerotiorum is a typical necrotrophic pathogen, and recent studies have revealed that it undergoes a transient biotrophic phase after initially colonizing plant tissues before transitioning into a necrotrophic pathogen (Kabbage et al., 2015; Liang and Rollins, 2018). The sclerotia present in the soil exhibit two states in relatively high-humidity conditions: myceliogenic germination, which produces hyphae to infect plant roots, and carpogenic germination, leading to the production of apothecia from which ascospores are released into the air (Clarkson et al., 2003; Hegedus and Rimmer, 2005). Typically, ascospores colonize senescent tissues, primarily petals, rather than healthy tissues. Under suitable conditions, they will generate hyphae (Turkington et al., 1991; Jamaux et al., 1995). Subsequently, infection cushions are formed to breach the host cuticle, entering a brief biotrophic phase in the apoplast. During this phase, OA and some hydrolytic enzymes are synthesized and secreted to inhibit the oxidative burst of host cells and host defense, degrade the cell wall, and the composite effect of infection cushions enhances penetration and expedites pathogen colonization. After penetrating the plant’s cuticle, S. sclerotiorum generates subcuticular vesicles and subcuticular infection hyphae that spread below the cuticle, indicating the beginning of the biotrophic phase, accumulate nutrients needed for hyphal growth, and serve as the foundation for colonization by S. sclerotiorum. The branches of subcuticular infection hyphae will produce ramifying hyphae, and they will penetrate anticlinal cell wall junctions, which will reduce the stability of the epidermal cell wall (Jamaux et al., 1995; Huang et al., 2008). Finally, the pathogen enters the necrotrophic phase (Garg et al., 2010; Kabbage et al., 2015; Derbyshire and Denton-Giles, 2016). As the petals fall, the hyphae spread to other healthy plant parts, such as leaves or stems, and may also extend to adjacent plants, causing stem rot and severe yield losses (Figure 1).
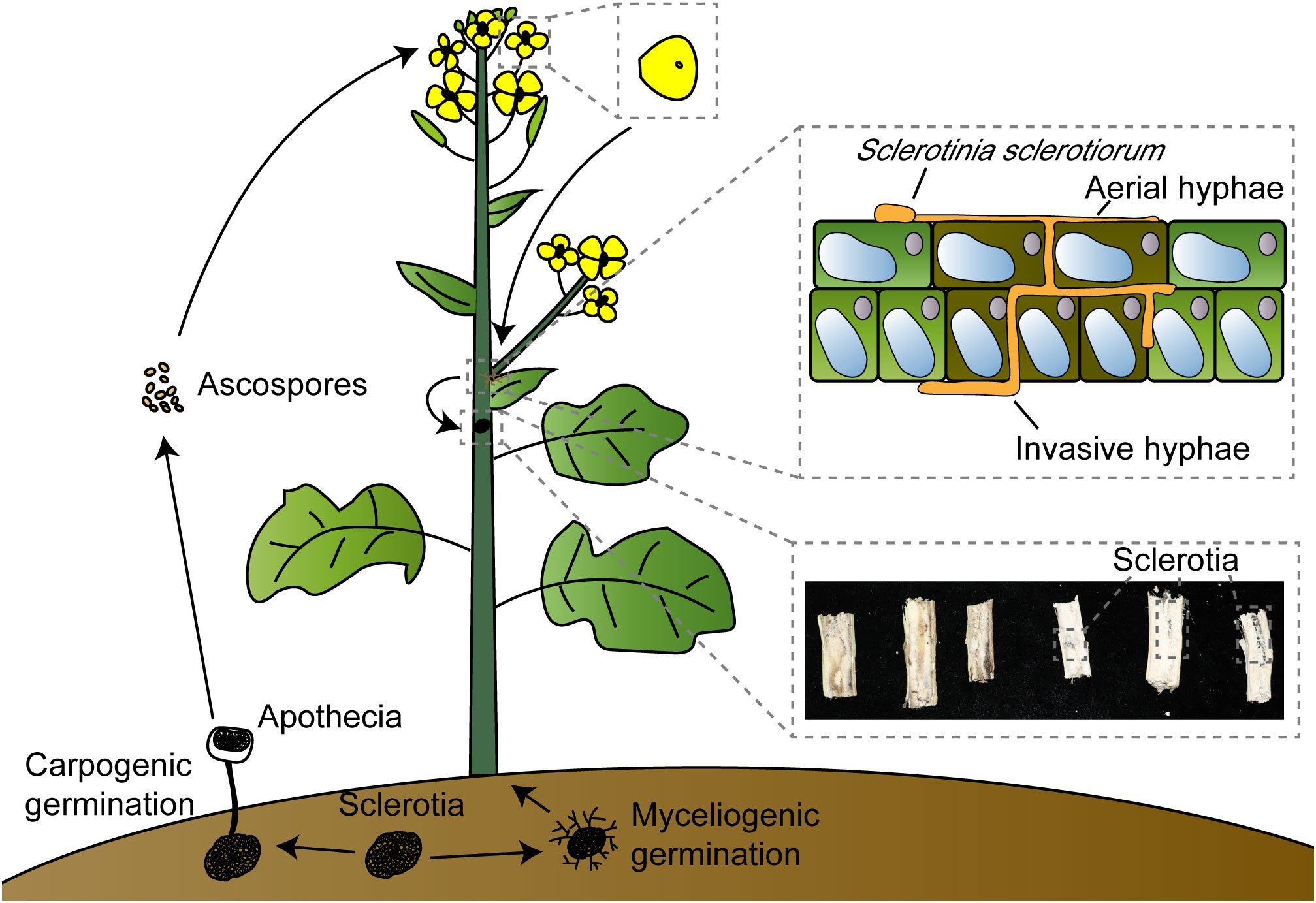
Figure 1 Sclerotinia sclerotiorum invasion model. Sclerotinia sclerotiorum can germinate under suitable conditions, myceliogenic germination produces hyphae to infect plant roots; carpogenic germination produces apothecia, and ascospores are released into the air by apothecia. Ascospores land on petals, which tend to gather in the leaf axils when they fall off. Sclerotinia sclerotiorum will colonize by aerial hyphae and invasive hyphae; invasive hyphae thicken after entering the apoplast and associate with early biotrophic growth, and finally produce sclerotia in the stem leading to hollow and rot.
OA plays a multifaceted role in the pathogenesis of S. sclerotiorum. In the early stages of infection, OA accumulation in infected tissues significantly impacts the host’s redox environment. This effect leads to the suppression of the oxidative burst, the inhibition of callose deposition, and a reduction in ROS production. Consequently, it hampers the plant defense response. OA also contributes to the chelation of Ca2+, influencing Ca2+ signal transduction and pectin structure, and it leads to a decrease in environmental pH, enhancing the activity of CWDEs (Favaron et al., 2004; Williams et al., 2011; Fagundes-Nacarath et al., 2018). In the later stages of infection, the induction of ROS production leads to HR or PCD in host cells (Liang et al., 2009). OA-deficient mutants of S. sclerotiorum are nonpathogenic because they cannot alter the host’s redox environment; the strain cannot colonize but can induce strong HR-like plant defense (Williams et al., 2011). Most hosts of S. sclerotiorum are dicotyledons, which may be associated with OA preference (Derbyshire et al., 2022). Germin-like proteins (GLPs) were found in cereals with oxalate oxidase (OXO) activity and can break down OA into CO2 and H2O2 (Lane et al., 1993; Woo et al., 2000; Davidson et al., 2009). Rapeseed overexpressing the OXO gene showed higher tolerance to OA and stronger resistance to S. sclerotiorum (Zou et al., 2007; Dong et al., 2008; Liu et al., 2015). This may explain why S. sclerotiorum is unable to infect many monocotyledons.
Pathogens secrete effectors that play an essential role in the pathogenesis process, and these small secreted proteins can either promote or inhibit host cell death according to the pathogens (Hogenhout et al., 2009; Lyu et al., 2016). A previous study based on whole genome sequencing estimated the presence of approximately 70 putative effector genes in S. sclerotiorum (Derbyshire et al., 2017). Some effectors have been identified in S. sclerotiorum. PGIP-INactivating Effector 1 (SsPINE1) can inactivate plant polygalacturonase-inhibiting proteins (PGIPs), promoting the dissociation of polygalacturonases (PG)-PGIP and enhancing necrotrophic virulence (Wei et al., 2022). Necrosis and ethylene‐inducing peptides 1 and 2 (SsNep1, SsNep2) caused necrosis by transient expression in tobacco leaves (Dallal Bashi et al., 2010). Integrin alpha N-terminal domain superfamily member SsITL inhibits the expression of PLANT DEFENSIN1.2 (PDF1.2) in A. thaliana, affecting the plant defense response (Zhu et al., 2013). A cutinase, SsCut, causes cell death in some host plants, including B. napus, G. max, and Oryza sativa (Zhang et al., 2014a). SsSSVP1 affects the plants’ energy metabolism and promotes infection (Lyu et al., 2016). Intracellular necrosis‐inducing effector 1 (SsINE1) can enter host cells by the RxLR‐like motif, and SsINE5 causes necrosis by nucleotide‐binding leucine‐rich repeat (NLR) proteins (Newman et al., 2023). Cerato-platanin protein 1 (SsCP1) can target plant PR1 and cause host cell death (Yang et al., 2018). YML079-like cupin protein (SsYCP1) can promote pathogen infection and play an essential role in the pathogenesis of S. sclerotiorum (Fan et al., 2021a). These effectors are crucial elements in the study of interactions between plants and S. sclerotiorum, providing insights into the mechanisms underlying the pathogenic process.
Rapeseed defense response to S. sclerotiorum infection
Plant immunity against pathogens comprises two main branches: pattern-triggered immunity (PTI) and effector-triggered immunity (ETI). PTI relies on transmembrane pattern recognition receptors (PRRs) that recognize pathogen-associated molecular patterns (PAMPs), while ETI involves the use of proteins encoded by intracellular resistance (R) genes. These mechanisms contribute to plant defense together (Jones and Dangl, 2006). The involvement of the PTI has been demonstrated in plants–S. sclerotiorum system (Zhang et al., 2013). Several receptor-like protein (RLP) genes, including candidates such as BnaA02g16770D and BnaC02g22760D, have been identified in B. napus in response to S. sclerotiorum (Li et al., 2022). Various endogenous hormones regulate the downstream immune responses in plants against pathogens through a series of signal transduction pathways. Signal pathways mediated by salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) play significant roles in regulating plant defense responses (Thomma et al., 2001; Pieterse et al., 2012). Studies in Arabidopsis thaliana have revealed that SA can induce PCD and HR reactions at the infection site, providing resistance to biotrophic pathogens. The JA/ET signaling pathway, independent of the SA pathway, confers resistance to necrotic pathogens (Glazebrook, 2005; Li et al., 2019). These signaling pathways have essential roles in the defense of rapeseed against S. sclerotiorum, and in particular, the SA pathway was shown to be involved in the defense response against necrotrophic pathogens (Nováková et al., 2014). SA and JA pathways often exhibit antagonistic interactions. Positive regulatory genes in the SA pathway, such as WRKY70, negatively regulate the JA pathway, while MPK4 plays an opposite role. During S. sclerotiorum infection, SA and JA levels peak at 12 hours post-infection (hpi) and 24 hpi, respectively. The expression changes of SA–JA crosstalk genes such as BnWRKY70, BnNPR1, BnMPK4, and BnEDS1 suggest the importance of the orderly expression of SA and JA pathways (Wang et al., 2012). Additionally, a study in Brassica carinata showed a higher tolerance to S. sclerotiorum infection compared to B. napus, suggesting that differences in the timing of SA and JA pathways may be the contributing factor while indicating the potential importance of the ET pathway in plant defense (Yang et al., 2010). The hormone-mediated complex defense network is essential to the plant immune system. While synergistic and antagonistic effects of SA, JA, and ET pathways are relatively well-understood, research on the ET pathway in the context of the B. napus–S. sclerotiorum system remains somewhat limited, warranting further exploration in future studies.
B. napus–S. sclerotiorum system analysis
Transcriptome analysis of the B. napus–S. sclerotiorum interaction provides a comprehensive understanding of the molecular events during S. sclerotiorum infection and the defense strategies in rapeseed. Illumina sequencing was conducted at various time points following S. sclerotiorum infection, leading to the categorization of differentially expressed genes (DEGs) into several major groups, including hydrolytic enzymes, secondary metabolites, detoxification processes, signaling pathways, developmental processes, secreted effectors, OA regulation, and the production of ROS. The results revealed a noteworthy increase in the expression of genes related to nucleic acid binding during the brief biotrophic phase (12–24 hpi), coinciding with the upregulation of genes such as the pathogen-induced cutinase A gene SsCuta and lipid degradation-related genes SS1G_09557, SS1G_01953, and SS1G_11930; these lipolytic enzymes also seem to be involved in cuticle penetration facilitated by the mechanical pressure applied by infection cushions during the same period (Bashi et al., 2012). Additionally, genes associated with biotrophic interactions exhibited heightened expression during this phase (Lumsden and Wergin, 1980). Plant tissue necrosis typically occurs at 24 hpi, indicating the beginning of the necrotic phase; during this stage, the expression of multiple genes encoding hydrolases and related to secondary metabolite syntheses and toxin increases, such as nonaspartyl acid protease (SsACP1), polyketide synthase (SsPKS), and non-ribosomal peptide synthase (SsNRPS), and genes encoding subtilisin-like serine proteases (SS1G_07655, SS1G_02423, and SS1G_032820) can degrade cell wall glycoproteins and may play an essential role in fungal customization (Olivieri et al., 2002). In late infection, the expression of genes encoding PGs, cellulases, β-1,4-glucanases, arabinogalactan-degrading enzymes, mannosidases, laccases, and necrosis and ET-inducing peptides increased (Dallal Bashi et al., 2010). Genes such as SsOAH encoding oxaloacetate acetylhydrolase showed consistent expression during infection. Still, they exhibited heightened expression in the middle and late stages, paralleling similar trends in the gene encoding oxalate decarboxylase. The dynamic accumulation of OA seems to play an important role in pathogen infection. Detoxification is essential for pathogen resistance against host defenses, involving the modification or degradation of host-produced antitoxins or their removal from host cells. Genes encoding cytochrome P450 enzymes (SS1G_02340) and eburicol 14 alpha-demethylase (CYP51 and SS1G_04805) were upregulated in the early stages of infection (Seifbarghi et al., 2017). The expression of multiple genes encoding membrane transporters, such as significant facilitator superfamily (MFS) and ATP-binding cassette (ABC) transporters, increased in the late stage of infection. Some genes encoding cyanide hydratases/cyanate hydrolases and 2-nitropropane dioxygenases exhibited varying levels of upregulation at different stages of infection (Seifbarghi et al., 2017; Xu et al., 2021). Furthermore, Brassicales plants can produce chemicals for defense, such as isothiocyanates (ITCs), nitriles, and flavonols (Wittstock and Gershenzon, 2002; Cartea et al., 2010). ITCs and nitrile are produced from glucosinolates (GLs) via β-thioglucoside glucohydrolase enzymes (myrosinases) (Rask et al., 2000). Flavonols, including quercetin, kaempferol, isorhamnetin, and other polyphenolic compounds, are also part of this defense system (Cartea et al., 2010). Chen et al. (2019); Chen et al. (2020) identified two detoxification-related genes in S. sclerotiorum, SsQDO (quercetin dioxygenase gene), and Ss12040 (ITC hydrolases gene). SsQDO deletion lines showed decreased pathogenicity and increased sensitivity to flavonols. Ss12040, a homolog of SaxA from Pseudomonas syringae, was essential for pathogenicity on ITC-defended plants. Together, these genes constitute the defense network of S. sclerotiorum against the antitoxins produced by rapeseed. Furthermore, salicylate hydroxylase and specific extracellular effectors, such as SsLysM and SsPINE1, are very important in inhibiting plant defense pathways (Ambrose et al., 2015; Peng et al., 2017; Seifbarghi et al., 2017; Xu et al., 2021). Table 1 contains a list of pathogenicity-related genes in S. sclerotiorum.
Transcriptome analysis revealed the activation of the mitogen-activated protein kinase (MAPK) signaling pathway during the early stages of S. sclerotiorum infection. Multiple MAPKKK, MKK, and MPK genes were induced by S. sclerotiorum. The response of transcription factors (TFs) was intricate, with most TFs in families such as WRKY, MYB, NAC, and others being downregulated. The critical gene ICS1 in SA biosynthesis showed an initial increase in expression followed by a decrease during the infection period (Zheng et al., 2015). Conversely, critical genes in JA biosynthesis, AOS, and LOX2 exhibited increased expression over time (Wasternack, 2007). However, the accumulation of JA could not impede the infection’s progression due to the downregulation of COI1. This downregulation led to an increased abundance of the jasmonate zinc finger inflorescence meristem (ZIM) domain protein (JAZ) (Chini et al., 2007; Ruan et al., 2019). JAZ inhibited the JA signaling pathway and caused the downregulation of MYC2, preventing the activation of downstream genes of JA-related genes (Kazan and Manners, 2013). The ET pathway regulates plant growth development and participates in plant stress response. ETHYLENE RESPONSE FACTOR 1 (ERF1) and OCTADECANOID-RESPONSIVE ARABIDOPSIS ETHYLENE/ERF 59 (ORA59) are considered to be key signaling molecules in the JA-ET pathway (van Loon et al., 2006; Pré et al., 2008). Following S. sclerotiorum infection, the expression of ETHYLENE RESPONSE SENSOR (ERS), ERF, and ORA59 increased, and the activation of the JA/ET pathway resulted in elevated expression of genes responsible for chitinase and pathogenesis-related (PR) genes such as PDF1.2, PR2, PR3, and PR4. This underlines the significance of the JA/ET pathway in defense against S. sclerotiorum. Additionally, the upregulation of PGIPs serves to inhibit PG in the cell wall, preventing cell wall degradation (De Lorenzo and Ferrari, 2002; Joshi et al., 2016b; Wei et al., 2016; Wu et al., 2016; Girard et al., 2017; Jian et al., 2018; Xu et al., 2021). Long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) are essential in regulating the expression of plant genes in response to stress. While the expression of various lncRNAs and miRNAs changes following S. sclerotiorum infection, further research is needed to elucidate their precise roles in the defense response of rapeseed (Cao et al., 2016; Joshi et al., 2016a; Jian et al., 2018).
Defense-related genes in rapeseed
Identifying essential defense genes in rapeseed is a necessary strategy for managing SSR, and it holds significant importance for molecular breeding. The MAPK signaling pathway is the foundation for plant responses to pathogens (Meng and Zhang, 2013; Zhang and Zhang, 2022). Activation of the MEKK1-MKK4/MKK5-MPK3/MPK6 pathway has been established as conferring resistance to fungal pathogens in A. thaliana (Asai et al., 2002). In rapeseed, BnMPK3 and BnMPK6 exhibit heightened responsiveness to S. sclerotiorum infection. BnMPK3-OE and BnMPK6-OE plants showed enhanced resistance to S. sclerotiorum, while their RNA interference (RNAi) plants were more susceptible. Importantly, BnMPK3 and BnMPK6 confer resistance to rapeseed by positively regulating critical genes of the ET pathway, BnACS and BnEIN3 (Alonso et al., 2003; Lin et al., 2009; Wang et al., 2019a; Wang et al., 2020c). Another MAPK pathway, MEKK1–MKK1/MKK2–MPK4/MPK11, is implicated in defense responses in A. thaliana (Zhang and Zhang, 2022). Rapeseeds that overexpress BnMPK4 have been shown to inhibit S. sclerotiorum infection. This overexpression leads to the sustained activation of the expression PDF1.2 but concurrent inhibition of PR1 expression (Wang et al., 2009).
WRKY transcription factors constitute one of the largest TF families in higher plants, participating extensively in regulating plant growth, development, plant hormone signal transduction, and various stress responses. They are characterized by a highly conserved WRKY domain, which recognizes and binds to the TTGAC (C/T) W box region in promoters to inhibit or activate downstream gene expression (Rushton et al., 2010; Jiang et al., 2017; Wani et al., 2021). WRKY33 can be induced by pathogens and regulated by MPK3/MPK6, and WRKY33 can bind to its promoter to regulate expression and induce the synthesis of A. thaliana phytoalexin camalexin (Mao et al., 2011). In rapeseed, BnWRKY33 displays heightened sensitivity to S. sclerotiorum, and the enhanced resistance of BnWRKY33-OE plants to pathogens is likely linked to the activation of both the SA and JA pathways, as well as redox control (Wang et al., 2014b; Liu et al., 2018a). Conversely, BnWRKY15-OE and BnWRKY28-OE plants showed increased susceptibility to S. sclerotiorum, and the negative regulation of BnWRKY15 and BnWRKY28 against pathogen resistance was closely related to the expression of BnWRKY33 and downstream genes (Liu et al., 2018a; Zhang et al., 2022b). WRKY70 is an activator of the SA pathway and an inhibitor of the JA pathway, BnWRKY70-OE plants showed increased susceptibility to S. sclerotiorum, while the Bnwrky70 mutants edited by CRISPR/Cas9 showed stronger resistance to S. sclerotiorum (Li et al., 2006b; Sun et al., 2018). Plant mediators (MEDs) interact with RNA polymerase II (RNAP II) and TFs to regulate gene transcription; AtMED16 regulates the JA/ET pathway and is critical for the recruitment of RNAP II by WRKY33 target genes PDF1.2 and ORA59 (Wang et al., 2015). In rapeseed, the expression of BnMED16 increased following S. sclerotiorum infection, BnMED16 interacted with BnMED25 and BnWRKY33, and BnMED25 was closely related to the JA/ET pathway; BNMED16-OE plants showed increased resistance to S. sclerotiorum (Hu et al., 2021). Heat shock proteins (HSPs) are a class of evolutionarily conserved stress proteins plants produce in response to stress. HSP90, in particular, acts as a molecular chaperone and is essential for the defense response mediated by R proteins (Schulze-Lefert, 2004; Wegele et al., 2004; Sangster and Queitsch, 2005). In rapeseed, 35 HSP90 genes have been identified, with 6 BnHSP90 genes exhibiting altered expression induced by S. sclerotiorum (Wang et al., 2022a). Since HSP90s interact with many proteins, they may be the key to rapeseed defense response (Tichá et al., 2020).
The SA pathway receptor, NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1), is important for plant systemic acquired resistance (SAR). NPR1 is responsible for promoting the expression of downstream defense genes (Silva et al., 2018; Chen et al., 2021). In rapeseed, 19 NPR1-like genes have been identified, and the expression of BnNPR1 significantly decreased following S. sclerotiorum infection. BnNPR1-RNAi plants showed increased sensitivity to S. sclerotiorum, resulting in the accumulation of ROS and the suppression of the SA pathway. However, the expression of JA/ET pathway-related genes increased, and this increased sensitivity may be attributed to the early stage role of the SA pathway (Wang et al., 2020b).
GLPs were initially discovered in Triticum aestivum and are typically characterized by a cupin domain. Various GLP isoforms exhibit diverse enzymatic activities, including OXO (true germins), superoxide dismutase (SOD), polyphenol oxidase (PPO), and ADP glucose pyrophosphatase/phosphodiesterase (AGPPase) (Lane et al., 1993; Breen and Bellgard, 2010; Barman and Banerjee, 2015). In rapeseed, 14 GLP genes have been identified, with BnGLP3 and BnGLP12 playing key roles in plant defense and showed SOD activity in the early stages of S. sclerotiorum infection (Rietz et al., 2012).
Ca2+ is very important in HR and PCD, and they are closely associated with ROS in various plant immune responses (Moeder et al., 2019). Calmodulin-binding activators (CAMTAs) interact with calmodulins (CaMs) to respond to calcium signals, and CAMTA3 has been identified as a participant in plant defense against pathogens in plants (Koo et al., 2009; Zhang et al., 2014b). Eighteen CAMTA genes were identified in the rapeseed genome, and promoter analysis showed that the promoter region of BnCAMTA contained many cis-acting elements, and the expression of BnCAMTA3 was induced by S. sclerotiorum. Atcamta3 mutants of A. thaliana showed increased expression of BAK1 and JIN1 genes, which are important for plant PTI in response to S. sclerotiorum. It appears that CAMTA plays a role in regulating plant immunity to pathogens by negatively regulating PTI and inhibiting the JA pathway (Rahman et al., 2016). Cyclic nucleotide-gated ion channel (CNGC) proteins are known to participate in calcium signal transduction induced by PTI in plants (Tian et al., 2019). In rapeseed, 61 putative BnCNGC genes have been classified into five groups (I, II, III, IV-A, and IV-B). Six of these BnCNGC genes (four in group I and two in group IV-A) were strongly induced by SA and S. sclerotiorum. Susceptible B. oleracea displayed increased calcium signaling during S. sclerotiorum infection, which might be linked to cell death (Mei et al., 2016; Liu et al., 2021b). However, further studies are required to elucidate the precise function of calcium signal transduction in the B. napus–S. sclerotiorum system.
Resistance to S. sclerotiorum in rapeseed is a complex trait controlled by multiple genes’ quantitative (additive) effects. Recent research has identified several quantitative trait loci (QTLs) associated with S. sclerotiorum resistance in rapeseed, with some of these QTLs showing links to flowering time (FT) and yield (Zhao and Meng, 2003; Wei et al., 2014; Li et al., 2015). Doubled-haploid (DH) lines are widely used in QTL mapping studies because they can be consistently identified across different years and experiments (Ding et al., 2021a). Zhao and Meng (2003) used 107 molecular markers to identify six QTLs across various developmental phases. Three QTLs were related to the resistance during the seedling stage. In contrast, the other three were related to resistance during the mature plant stage, and they suggested that different resistance sites may work in different stages or plant organs. The study also observed various epistatic interactions, including dominance × dominance digenic epistasis, in the inheritance of cotyledon resistance (Khan et al., 2020a; Khan et al., 2020b). Wu et al. (2013) mapped 10 QTLs related to stem resistance (SR) during the mature plant stage and 3 QTLs related to leaf resistance (LR) during the seedling stage, SRC6 on the C6 link explained 29.01% to 32.61% of the phenotypic variation, and the study indicated that BnaC.IGMT5.a could be the main candidate gene for this QTL. Yin et al. (2010) used 252 molecular markers and identified a total of 10, 1, and 10 QTLs through mycelial toothpick inoculation (MTI), mycelial plug inoculation (MPI), and infected petal inoculation (IPI), respectively. QTLs on the linkage groups N3, N12, and N17 were detected over 2 years. Zhao et al. (2006) used the petiole inoculation technique to detect 8 QTLs in the HUA population by evaluating days to wilt (DW) and stem lesion length (SLL); four of these QTLs were contributed by resistant parents, and four were contributed by susceptible parents; these results indicated that resistance alleles could be present not only in resistant lines but also in susceptible lines, potentially benefiting breeding efforts. Genome-wide association studies (GWAS) based on linkage disequilibrium (LD) have become a primary tool for identifying gene loci (Wang et al., 2019b). Wei et al. (2016) identified 17 single-nucleotide polymorphisms (SNPs) on A8 and C6 chromosomes by GWAS in different rapeseed accessions with SR to S. sclerotiorum. These C6 SNPs were consistent with findings from previous QTL mapping studies. The integration of QTL information is crucial for the molecular breeding of highly resistant rapeseed. Most known R genes typically contain nucleotide-binding site (NBS) and leucine-rich repeat (LRR) domain (Jia et al., 2015). Li et al. (2015) integrated 35 QTLs on 10 chromosomes and identified two conserved SR QTL regions on chromosomes A9 and C6 with several putative NBS-LRR genes clustering at 22.8 and 33.6 Mb, respectively. Furthermore, Wei et al. (2014) found a weak association between rapeseed resistance to S. sclerotiorum and FT. They detected common QTL regions, suggesting that early FT and high resistance to S. sclerotiorum in rapeseed could benefit future breeding efforts. These findings provide valuable insights into rapeseed resistance to S. sclerotiorum.
Rapeseed transgenic strategy
Rapeseed lacks cultivars with complete resistance to S. sclerotiorum; genetic engineering is an effective way to improve rapeseed resistance. Since the degradation of rapeseed cell walls is significant for S. sclerotiorum colonization, a promising strategy is to impede pathogen colonization as the first line of plant defense. Overexpressing PGIPs typically inhibits pathogen-produced PG and affects pathogen colonization (Federici et al., 2006; Kalunke et al., 2015). Overexpressing O. sativa OsPGIP2 and OsPGIP6 in rapeseed has enhanced resistance to S. sclerotiorum (Wang et al., 2018; Yin et al., 2022). Chitinase can catalyze the hydrolysis of chitin in fungal cell walls, and co-expressing the chitinase gene Chit42 from Trichoderma atroviride with PGIP2 from Phaseolus vulgaris significantly restricts pathogen growth and delays the disease progression (Cletus et al., 2013; Ziaei et al., 2016). Thaumatin-like proteins (TLPs) belong to the PR5 protein family and play an important role in plant defense (Liu et al., 2010; de Jesús-Pires et al., 2020). Co-expressing chimeric chitinase and OsTLP genes have also enhanced rapeseed resistance to S. sclerotiorum (Aghazadeh et al., 2016).
Owing to the significant role of OA in S. sclerotiorum infection, the overexpression of wheat OXO in rapeseed resulted in a remarkable disease reduction rate of up to 90.2% in the sixth-generation lines compared to the parent lines. This breakthrough holds great promise for the development of resistant rapeseed varieties in the future (Dong et al., 2008). However, it is worth noting that while the OXO gene shows substantial potential for enhancing resistance to S. sclerotiorum, it is essential to consider that germin and GLPs have been identified as a class of plant allergens. Further research should be conducted to evaluate any potential allergenic effects carefully (Jensen-Jarolim et al., 2002).
Lignin is a polymer composed of p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units. It serves to reinforce the mechanical strength of the cell wall and plays a positive role in supporting plant growth and development, and has various functions in mitigating biotic and abiotic stresses (Cesarino, 2019; Dong and Lin, 2021). Ferulate-5-hydroxylase (F5H) is responsible for catalyzing the conversion of G-type units into S-type units within lignin. Knocking out BnF5H using CRISPR/Cas9 has been shown to increase the ratio of G-type/S-type units, resulting in enhanced stem strength and improved resistance of stems and leaves to S. sclerotiorum (Cao et al., 2022). Strengthening plant tissue mechanically appears to be a viable auxiliary strategy to reduce the impact of SSR. Furthermore, the plant’s surface is covered by a cuticle layer composed of cutin and wax. Research has indicated that changes in cuticular wax composition can influence resistance to S. sclerotiorum, in addition to the well-documented role of cutinase in cutin degradation (Wang et al., 2020a; Liu et al., 2021a).
GDSL esterase/lipase has been shown to positively affect pathogen defense responses in A. thaliana (Lee et al., 2009). The rapeseed lines overexpressing AtGDSL1 exhibited reduced JA levels, elevated ROS and SA levels, and increased resistance following S. sclerotiorum infection. However, it is worth mentioning that BnGDSL1-OE line did not show any changes in resistance; based on the analysis of candidate genes associated with AtGDSL1 homologs, it was revealed that BnGLIP1 might be a key gene contributing to rapeseed resistance (Tan et al., 2014; Ding et al., 2020).
Petals play a significant role in S. sclerotiorum colonization, and the peptide inflorescence deficient in abscission (IDA) is a key regulator of floral organ abscission (Stø et al., 2015; Shi et al., 2019). An interesting study showed that IDA mutants failed to shed their floral organs properly. When S. sclerotiorum was inoculated into the petals of these mutants, although the petals themselves withered and died, the infection did not spread to the leaves as they could not shed normally. This prevented the pathogen from moving between the petals and leaves, thus restricting its transmission (Geng et al., 2022). While most current research focuses on enhancing resistance to S. sclerotiorum, typically resulting in disease reduction or delay, it is important to note that ascospores of S. sclerotiorum seldom colonize healthy leaves. Therefore, a promising strategy might involve preventing the pathogen’s movement from the heavily colonized petals to healthy leaves or stems, offering an effective means of disease prevention.
Antimicrobial peptides (AMPs) are a class of small cationic peptides known for their wide-ranging antibacterial effects, achieved through mechanisms such as the inhibition of protein transport, cell membrane penetration, and binding to DNA/RNA (Nawrot et al., 2014; Sinha and Shukla, 2019; Bakare et al., 2022). In rapeseed, the identification and validation of the first plant proline-rich antimicrobial peptide (PR-AMP) suggested its potential role in the defense responses of rapeseed (Cao et al., 2015). LjAMP2, a heat-stable antibacterial protein derived from Leonurus japonicus, has exhibited the ability to inhibit a range of plant pathogens, and rapeseed that expresses LjAMP2 showed increased resistance to S. sclerotiorum (Yang et al., 2006; Jiang et al., 2013). Similarly, overexpression of PmAMP1 from Pinus monticola, LTP from O. sativa, and recombinant pathogen-specific antibodies (scFv) in rapeseed also showed increased tolerance to S. sclerotiorum (Yajima et al., 2010; Verma et al., 2012; Fan et al., 2013).
Gene editing technology based on the CRISPR/Cas system has emerged as a powerful tool for investigating plant gene functionality, enhancing crop traits, facilitating breeding efforts, and bolstering plant disease resistance (Cardi et al., 2023; Erdoğan et al., 2023; Li et al., 2023). The knockout of susceptibility (S) genes responsible for susceptibility to powdery mildew and stripe rust in wheat has led to enhanced resistance against these two diseases (Wang et al., 2014a; Wang et al., 2022b). SsSSVP1, a small secreted protein from S. sclerotiorum, has been found to exert an influence on the subcellular localization of QCR8 (the subunit of the cytochrome b-c1 complex) in Nicotiana benthamiana, consequently impairing its biological functions (Lyu et al., 2016). Zhang et al. (2021) identified a homolog of SsSSVP1 in Botrytis cinerea and used CRISPR/Cas9 to reduce the copy number of BnQCR8. The mutants showed strong resistance against not only S. sclerotiorum but also B. cinerea, all without compromising vital agronomic traits in rapeseed. This study provided a novel, effective, and very important strategy for conferring strong resistance in crops against multiple pathogens by editing one gene that encodes a common target of pathogen effectors.
The balance between disease resistance, crop yield, and quality is crucial in agriculture. The discovery of S. sclerotiorum-induced promoters is particularly important, and overexpression of resistance genes can enhance plant resistance and cause energy loss (Ding et al., 2021a). The promoter of glycosyl hydrolase 17 gene (pBnGH17) in rapeseed was shown to be induced by S. sclerotiorum; 5′-deletions and promoter activity analysis showed that a 189-bp region was essential for S. sclerotiorum to induce responses, and the promoter pBnGH17D7, which connects this region to the core promoter region, was induced after S. sclerotiorum infection, but it was less active under normal growth conditions (Lin et al., 2022). This provides an important reference for future work. Because of the particularity of transgenic plants, it is necessary to carefully evaluate the biological safety and other aspects in subsequent work.
Conclusion
In recent years, significant progress in studies of the B. napus–S. sclerotiorum system has been made, and the transcriptomic and bioinformatics analyses have contributed to the search for potential defense-related genes in rapeseed; genetic engineering and CRISPR/Cas9 also provide new approaches for future research. However, there are still some problems in the current studies. First, although transgenic rapeseed (overexpression/knockout) improved the resistance to S. sclerotiorum and delayed disease, no strategy of complete immunity to S. sclerotiorum has been reported. Second, the current studies mainly stay in the laboratory stage and lack field experiments, which will be limited to molecular breeding and application. The relationship between yield, quality, and disease resistance should also be balanced. Third, in addition to the search for R genes, the search and development of S genes are equally important; inactivation of the S genes usually induces long and broad-spectrum resistance with little adverse effect on crop growth and yield (Pavan et al., 2010; Zhang et al., 2017; Wang et al., 2022b). In addition, small RNA (sRNA) and host-mediated pathogen gene silencing (cross-kingdom RNAi) have shown great effects in plant defense against pathogens (Wang et al., 2017; Cai et al., 2018), and may be important directions for future studies.
Author contributions
R-SC: Writing – original draft. J-YW: Writing – review & editing. RS: Writing – review & editing. X-LT: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Jiangsu Agriculture Science and Technology Innovation Fund (CX (21) 2009) and the Key R&D in the Jiangsu Province (BE2022340).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aghazadeh, R., Zamani, M., Motallebi, M., Moradyar, M., Moghadassi Jahromi, Z. (2016). Co-transformation of canola by chimeric chitinase and tlp genes towards improving resistance to Sclerotinia sclerotiorum. World J. Microbiol. Biotechnol. 32, 144. doi: 10.1007/s11274-016-2104-6
Alonso, J. M., Stepanova, A. N., Solano, R., Wisman, E., Ferrari, S., Ausubel, F. M., et al. (2003). Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 100, 2992–2997. doi: 10.1073/pnas.0438070100
Ambrose, K. V., Tian, Z., Wang, Y., Smith, J., Zylstra, G., Huang, B., et al. (2015). Functional characterization of salicylate hydroxylase from the fungal endophyte Epichloë festucae. Sci. Rep. 5, 10939. doi: 10.1038/srep10939
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W.-L., Gomez-Gomez, L., et al. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. doi: 10.1038/415977a
Bakare, O. O., Gokul, A., Fadaka, A. O., Wu, R., Niekerk, L. A., Barker, A. M., et al. (2022). Plant antimicrobial peptides (PAMPs): features, applications, production, expression, and challenges. Molecules 27, 3703. doi: 10.3390/molecules27123703
Barman, A. R., Banerjee, J. (2015). Versatility of germin-like proteins in their sequences, expressions, and functions. Funct. Integr. Genomics 15, 533–548. doi: 10.1007/s10142-015-0454-z
Bashi, Z. D., Rimmer, S. R., Khachatourians, G. G., Hegedus, D. D. (2012). Factors governing the regulation of Sclerotinia sclerotiorum cutinase A and polygalacturonase 1 during different stages of infection. Can. J. Microbiol. 58, 605–616. doi: 10.1139/w2012-031
Beyzi, E., Gunes, A., Buyukkilic Beyzi, S., Konca, Y. (2019). Changes in fatty acid and mineral composition of rapeseed (Brassica napus ssp. oleifera L.) oil with seed sizes. Ind. Crops Products 129, 10–14. doi: 10.1016/j.indcrop.2018.11.064
Boland, G. J., Hall, R. (1994). Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 16, 93–108. doi: 10.1080/07060669409500766
Bolton, M. D., Thomma, B. P., Nelson, B. D. (2006). Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 7, 1–16. doi: 10.1111/j.1364-3703.2005.00316.x
Breen, J., Bellgard, M. (2010). Germin-like proteins (GLPs) in cereal genomes: gene clustering and dynamic roles in plant defence. Funct. Integr. Genomics 10, 463–476. doi: 10.1007/s10142-010-0184-1
Cai, Q., Qiao, L., Wang, M., He, B., Lin, F. M., Palmquist, J., et al. (2018). Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360, 1126–1129. doi: 10.1126/science.aar4142
Cao, H., Ke, T., Liu, R., Yu, J., Dong, C., Cheng, M., et al. (2015). Identification of a novel proline-rich antimicrobial peptide from brassica napus. PloS One 10, e0137414. doi: 10.1371/journal.pone.0137414
Cao, J.-Y., Xu, Y.-P., Zhao, L., Li, S.-S., Cai, X.-Z. (2016). Tight regulation of the interaction between Brassica napus and Sclerotinia sclerotiorum at the microRNA level. Plant Mol. Biol. 92, 39–55. doi: 10.1007/s11103-016-0494-3
Cao, Y., Yan, X., Ran, S., Ralph, J., Smith, R. A., Chen, X., et al. (2022). Knockout of the lignin pathway gene BnF5H decreases the S/G lignin compositional ratio and improves Sclerotinia sclerotiorum resistance in Brassica napus. Plant Cell Environ. 45, 248–261. doi: 10.1111/pce.14208
Cardi, T., Murovec, J., Bakhsh, A., Boniecka, J., Bruegmann, T., Bull, S. E., et al. (2023). CRISPR/Cas-mediated plant genome editing: outstanding challenges a decade after implementation. Trends Plant Sci. 28, 1144–1165. doi: 10.1016/j.tplants.2023.05.012
Cartea, M. E., Francisco, M., Soengas, P., Velasco, P. (2010). Phenolic compounds in Brassica vegetables. Molecules 16, 251–280. doi: 10.3390/molecules16010251
Cesarino, I. (2019). Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr. Opin. Biotechnol. 56, 209–214. doi: 10.1016/j.copbio.2018.12.012
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chen, J., Tan, R. K., Guo, X. J., Fu, Z. L., Wang, Z., Zhang, Z. Y., et al. (2015). Transcriptome analysis comparison of lipid biosynthesis in the leaves and developing seeds of brassica napus. PloS One 10, e0126250. doi: 10.1371/journal.pone.0126250
Chen, J., Ullah, C., Reichelt, M., Beran, F., Yang, Z. L., Gershenzon, J., et al. (2020). The phytopathogenic fungus Sclerotinia sclerotiorum detoxifies plant glucosinolate hydrolysis products via an isothiocyanate hydrolase. Nat. Commun. 11, 3090. doi: 10.1038/s41467-020-16921-2
Chen, J., Ullah, C., Reichelt, M., Gershenzon, J., Hammerbacher, A. (2019). Sclerotinia sclerotiorum circumvents flavonoid defenses by catabolizing flavonol glycosides and aglycones. Plant Physiol. 180, 1975–1987. doi: 10.1104/pp.19.00461
Chen, J., Zhang, J., Kong, M., Freeman, A., Chen, H., Liu, F. (2021). More stories to tell: NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1, a salicylic acid receptor. Plant Cell Environ. 44, 1716–1727. doi: 10.1111/pce.14003
Chew, S. C. (2020). Cold-pressed rapeseed (Brassica napus) oil: Chemistry and functionality. Food Res. Int. 131, 108997. doi: 10.1016/j.foodres.2020.108997
Chini, A., Fonseca, S., Fernández, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Clarkson, J. P., Staveley, J., Phelps, K., Young, C. S., Whipps, J. M. (2003). Ascospore release and survival in Sclerotinia sclerotiorum. Mycol Res. 107, 213–222. doi: 10.1017/S0953756203007159
Cletus, J., Balasubramanian, V., Vashisht, D., Sakthivel, N. (2013). Transgenic expression of plant chitinases to enhance disease resistance. Biotechnol. Lett. 35, 1719–1732. doi: 10.1007/s10529-013-1269-4
Cong, J., Xiao, K., Jiao, W., Zhang, C., Zhang, X., Liu, J., et al. (2022). The coupling between cell wall integrity mediated by MAPK kinases and ssFkh1 is involved in sclerotia formation and pathogenicity of sclerotinia sclerotiorum. Front. Microbiol. 13, 816091. doi: 10.3389/fmicb.2022.816091
Dallal Bashi, Z., Hegedus, D. D., Buchwaldt, L., Rimmer, S. R., Borhan, M. H. (2010). Expression and regulation of Sclerotinia sclerotiorum necrosis and ethylene-inducing peptides (NEPs). Mol. Plant Pathol. 11, 43–53. doi: 10.1111/j.1364-3703.2009.00571.x
Davidson, R. M., Reeves, P. A., Manosalva, P. M., Leach, J. E. (2009). Germins: A diverse protein family important for crop improvement. Plant Sci. 177, 499–510. doi: 10.1016/j.plantsci.2009.08.012
de Jesús-Pires, C., Ferreira-Neto, J. R. C., Pacifico Bezerra-Neto, J., Kido, E. A., De Oliveira Silva, R. L., Pandolfi, V., et al. (2020). Plant thaumatin-like proteins: function, evolution and biotechnological applications. Curr. Protein Pept. Sci. 21, 36–51. doi: 10.2174/1389203720666190318164905
De Lorenzo, G., Ferrari, S. (2002). Polygalacturonase-inhibiting proteins in defense against phytopathogenic fungi. Curr. Opin. Plant Biol. 5, 295–299. doi: 10.1016/S1369-5266(02)00271-6
Derbyshire, M. C., Denton-Giles, M. (2016). The control of sclerotinia stem rot on oilseed rape (Brassica napus): current practices and future opportunities. Plant Pathol. 65, 859–877. doi: 10.1111/ppa.12517
Derbyshire, M., Denton-Giles, M., Hegedus, D., Seifbarghy, S., Rollins, J., Van Kan, J., et al. (2017). The complete genome sequence of the phytopathogenic fungus Sclerotinia sclerotiorum reveals insights into the genome architecture of broad host range pathogens. Genome Biol. Evol. 9, 593–618. doi: 10.1093/gbe/evx030
Derbyshire, M. C., Newman, T. E., Khentry, Y., Owolabi Taiwo, A. (2022). The evolutionary and molecular features of the broad-host-range plant pathogen Sclerotinia sclerotiorum. Mol. Plant Pathol. 23, 1075–1090. doi: 10.1111/mpp.13221
Ding, Y., Chen, Y., Wu, Z., Yang, N., Rana, K., Meng, X., et al. (2022). SsCox17, a copper chaperone, is required for pathogenic process and oxidative stress tolerance of Sclerotinia sclerotiorum. Plant Sci. 322, 111345. doi: 10.1016/j.plantsci.2022.111345
Ding, Y., Chen, Y., Yan, B., Liao, H., Dong, M., Meng, X., et al. (2021b). Host-induced gene silencing of a multifunction gene sscnd1 enhances plant resistance against sclerotinia sclerotiorum. Front. Microbiol. 12, 693334. doi: 10.3389/fmicb.2021.693334
Ding, L.-N., Li, T., Guo, X.-J., Li, M., Liu, X.-Y., Cao, J., et al. (2021a). Sclerotinia stem rot resistance in rapeseed: recent progress and future prospects. J. Agric. Food Chem. 69, 2965–2978. doi: 10.1021/acs.jafc.0c07351
Ding, L.-N., Li, M., Guo, X.-J., Tang, M.-Q., Cao, J., Wang, Z., et al. (2020). Arabidopsis GDSL1 overexpression enhances rapeseed Sclerotinia sclerotiorum resistance and the functional identification of its homolog in Brassica napus. Plant Biotechnol. J. 18, 1255–1270. doi: 10.1111/pbi.13289
Dong, X., Ji, R., Guo, X., Foster, S. J., Chen, H., Dong, C., et al. (2008). Expressing a gene encoding wheat oxalate oxidase enhances resistance to Sclerotinia sclerotiorum in oilseed rape (Brassica napus). Planta 228, 331–340. doi: 10.1007/s00425-008-0740-2
Dong, N.-Q., Lin, H.-X. (2021). Contribution of phenylpropanoid metabolism to plant development and plant–environment interactions. J. Integr. Plant Biol. 63, 180–209. doi: 10.1111/jipb.13054
Duan, Y., Ge, C., Liu, S., Wang, J., Zhou, M. (2013). A two-component histidine kinase Shk1 controls stress response, sclerotial formation and fungicide resistance in Sclerotinia sclerotiorum. Mol. Plant Pathol. 14, 708–718. doi: 10.1111/mpp.12041
Erdoğan, İ., Cevher-Keskin, B., Bilir, Ö., Hong, Y., Tör, M. (2023). Recent developments in CRISPR/cas9 genome-editing technology related to plant disease resistance and abiotic stress tolerance. Biol. (Basel) 12, 1037. doi: 10.3390/biology12071037
Fagundes-Nacarath, I. R. F., Debona, D., Rodrigues, F. A. (2018). Oxalic acid-mediated biochemical and physiological changes in the common bean-Sclerotinia sclerotiorum interaction. Plant Physiol. Biochem. 129, 109–121. doi: 10.1016/j.plaphy.2018.05.028
Fan, Y., Du, K., Gao, Y., Kong, Y., Chu, C., Sokolov, V., et al. (2013). Transformation of LTP gene into Brassica napus to enhance its resistance to Sclerotinia sclerotiorum. Genetika 49, 439–447. doi: 10.1134/S1022795413040042
Fan, H., Yang, W., Nie, J., Lin, C., Wu, J., Wu, D., et al. (2021a). Characterization of a secretory YML079-like cupin protein that contributes to sclerotinia sclerotiorum pathogenicity. Microorganisms 9, 2519. doi: 10.3390/microorganisms9122519
Fan, H., Yang, W., Nie, J., Zhang, W., Wu, J., Wu, D., et al. (2021b). A novel effector protein ssERP1 inhibits plant ethylene signaling to promote sclerotinia sclerotiorum infection. J. Fungi (Basel) 7. doi: 10.3390/jof7100825
Favaron, F., Sella, L., D’ovidio, R. (2004). Relationships among endo-polygalacturonase, oxalate, pH, and plant polygalacturonase-inhibiting protein (PGIP) in the interaction between Sclerotinia sclerotiorum and soybean. Mol. Plant Microbe Interact. 17, 1402–1409. doi: 10.1094/MPMI.2004.17.12.1402
Federici, L., Di Matteo, A., Fernandez-Recio, J., Tsernoglou, D., Cervone, F. (2006). Polygalacturonase inhibiting proteins: players in plant innate immunity? Trends Plant Sci. 11, 65–70. doi: 10.1016/j.tplants.2005.12.005
Garg, H., Li, H., Sivasithamparam, K., Kuo, J., Barbetti, M. J. (2010). The infection processes of Sclerotinia sclerotiorum in cotyledon tissue of a resistant and a susceptible genotype of Brassica napus. Ann. Bot. 106, 897–908. doi: 10.1093/aob/mcq196
Geng, R., Shan, Y., Li, L., Shi, C. L., Zhang, W., Wang, J., et al. (2022). CRISPR-mediated BnaIDA editing prevents silique shattering, floral organ abscission, and spreading of Sclerotinia sclerotiorum in Brassica napus. Plant Commun. 3, 100452. doi: 10.1016/j.xplc.2022.100452
Girard, I. J., Tong, C., Becker, M. G., Mao, X., Huang, J., De Kievit, T., et al. (2017). RNA sequencing of Brassica napus reveals cellular redox control of Sclerotinia infection. J. Exp. Bot. 68, 5079–5091. doi: 10.1093/jxb/erx338
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Godoy, G., Steadman, J. R., Dickman, M. B., Dam, R. (1990). Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris. Physiol. Mol. Plant Pathol. 37, 179–191. doi: 10.1016/0885-5765(90)90010-U
Gong, Y., Fu, Y., Xie, J., Li, B., Chen, T., Lin, Y., et al. (2022). Sclerotinia sclerotiorum ssCut1 modulates virulence and cutinase activity. J. Fungi (Basel) 8, 526. doi: 10.3390/jof8050526
Hegedus, D. D., Rimmer, S. R. (2005). Sclerotinia sclerotiorum: When “to be or not to be” a pathogen? FEMS Microbiol. Lett. 251, 177–184. doi: 10.1016/j.femsle.2005.07.040
Hogenhout, S. A., van der Hoorn, R., Terauchi, R., Kamoun, S. (2009). Emerging concepts in effector biology of plant-associated organisms. Mol. Plant-Microbe Interactions® 22, 115–122. doi: 10.1094/MPMI-22-2-0115
Hu, H., Tang, Y., Wu, J., Chen, F., Yang, Y., Pan, X., et al. (2021). Brassica napus Mediator Subunit16 Induces BnMED25- and BnWRKY33-Activated Defense Signaling to Confer Sclerotinia sclerotiorum Resistance. Front. Plant Sci. 12, 663536. doi: 10.3389/fpls.2021.663536
Huang, L., Buchenauer, H., Han, Q., Zhang, X., Kang, Z. (2008). Ultrastructural and cytochemical studies on the infection process of Sclerotinia sclerotiorum in oilseed rape. J. Plant Dis. Prot. 115, 9–16. doi: 10.1007/BF03356233
Huang, Z., Lu, J., Liu, R., Wang, P., Hu, Y., Fang, A., et al. (2021). SsCat2 encodes a catalase that is critical for the antioxidant response, QoI fungicide sensitivity, and pathogenicity of Sclerotinia sclerotiorum. Fungal Genet. Biol. 149, 103530. doi: 10.1016/j.fgb.2021.103530
Jamaux, I., Gelie, B., Lamarque, C. (1995). Early stages of infection of rapeseed petals and leaves by Sclerotinia sclerotiorum revealed by scanning electron microscopy. Plant Pathol. 44, 22–30. doi: 10.1111/j.1365-3059.1995.tb02712.x
Jensen-Jarolim, E., Schmid, B., Bernier, F., Berna, A., Kinaciyan, T., Focke, M., et al. (2002). Allergologic exploration of germins and germin-like proteins, a new class of plant allergens. Allergy 57, 805–810. doi: 10.1034/j.1398-9995.2002.23686.x
Jia, Y., Yuan, Y., Zhang, Y., Yang, S., Zhang, X. (2015). Extreme expansion of NBS-encoding genes in Rosaceae. BMC Genet. 16, 48. doi: 10.1186/s12863-015-0208-x
Jian, H., Ma, J., Wei, L., Liu, P., Zhang, A., Yang, B., et al. (2018). Integrated mRNA, sRNA, and degradome sequencing reveal oilseed rape complex responses to Sclerotinia sclerotiorum (Lib.) infection. Sci. Rep. 8, 10987. doi: 10.1038/s41598-018-29365-y
Jiang, Y., Fu, X., Wen, M., Wang, F., Tang, Q., Tian, Q., et al. (2013). Overexpression of an nsLTPs-like antimicrobial protein gene (LJAMP2) from motherwort (Leonurus japonicus) enhances resistance to Sclerotinia sclerotiorum in oilseed rape (Brassica napus). Physiol. Mol. Plant Pathol. 82, 81–87. doi: 10.1016/j.pmpp.2012.11.001
Jiang, J., Ma, S., Ye, N., Jiang, M., Cao, J., Zhang, J. (2017). WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 59, 86–101. doi: 10.1111/jipb.12513
Jiang, J., Zhu, S., Yuan, Y., Wang, Y., Zeng, L., Batley, J., et al. (2019). Transcriptomic comparison between developing seeds of yellow- and black-seeded Brassica napus reveals that genes influence seed quality. BMC Plant Biol. 19, 203. doi: 10.1186/s12870-019-1821-z
Jiao, W., Ding, W., Rollins, J. A., Liu, J., Zhang, Y., Zhang, X., et al. (2023). Cross-talk and multiple control of target of rapamycin (TOR) in sclerotinia sclerotiorum. Microbiol. Spectr. 11, e0001323. doi: 10.1128/spectrum.00013-23
Jiao, W., Yu, H., Cong, J., Xiao, K., Zhang, X., Liu, J., et al. (2022). Transcription factor SsFoxE3 activating SsAtg8 is critical for sclerotia, compound appressoria formation, and pathogenicity in Sclerotinia sclerotiorum. Mol. Plant Pathol. 23, 204–217. doi: 10.1111/mpp.13154
Jones, J. D. G., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Joshi, R. K., Megha, S., Basu, U., Rahman, M. H., Kav, N. N. (2016a). Genome Wide Identification and Functional Prediction of Long Non-Coding RNAs Responsive to Sclerotinia sclerotiorum Infection in Brassica napus. PloS One 11, e0158784. doi: 10.1371/journal.pone.0158784
Joshi, R. K., Megha, S., Rahman, M. H., Basu, U., Kav, N. N. V. (2016b). A global study of transcriptome dynamics in canola (Brassica napus L.) responsive to Sclerotinia sclerotiorum infection using RNA-Seq. Gene 590, 57–67. doi: 10.1016/j.gene.2016.06.003
Kabbage, M., Yarden, O., Dickman, M. B. (2015). Pathogenic attributes of Sclerotinia sclerotiorum: Switching from a biotrophic to necrotrophic lifestyle. Plant Sci. 233, 53–60. doi: 10.1016/j.plantsci.2014.12.018
Kalunke, R. M., Tundo, S., Benedetti, M., Cervone, F., De Lorenzo, G., D’ovidio, R. (2015). An update on polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein that protects crop plants against pathogens. Front. Plant Sci. 6, 146. doi: 10.3389/fpls.2015.00146
Kazan, K., Manners, J. M. (2013). MYC2: the master in action. Mol. Plant 6, 686–703. doi: 10.1093/mp/sss128
Khan, M. A., Cowling, W., Banga, S. S., You, M. P., Tyagi, V., Bharti, B., et al. (2020a). Inheritance of leaf resistance to Sclerotinia sclerotiorum in Brassica napus and its genetic correlation with cotyledon resistance. Euphytica 216, 188. doi: 10.1007/s10681-020-02717-4
Khan, M. A., Cowling, W., Banga, S. S., You, M. P., Tyagi, V., Bharti, B., et al. (2020b). Patterns of inheritance for cotyledon resistance against Sclerotinia sclerotiorum in Brassica napus. Euphytica 216, 79. doi: 10.1007/s10681-020-02612-y
Kim, K. S., Min, J. Y., Dickman, M. B. (2008). Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant Microbe Interact. 21, 605–612. doi: 10.1094/MPMI-21-5-0605
Koo, S. C., Choi, M. S., Chun, H. J., Shin, D. B., Park, B. S., Kim, Y. H., et al. (2009). The calmodulin-binding transcription factor OsCBT suppresses defense responses to pathogens in rice. Mol. Cells 27, 563–570. doi: 10.1007/s10059-009-0081-4
Lane, B. G., Dunwell, J. M., Ray, J. A., Schmitt, M. R., Cuming, A. C. (1993). Germin, a protein marker of early plant development, is an oxalate oxidase. J. Biol. Chem. 268, 12239–12242. doi: 10.1016/S0021-9258(18)31377-2
Lee, D. S., Kim, B. K., Kwon, S. J., Jin, H. C., Park, O. K. (2009). Arabidopsis GDSL lipase 2 plays a role in pathogen defense via negative regulation of auxin signaling. Biochem. Biophys. Res. Commun. 379, 1038–1042. doi: 10.1016/j.bbrc.2009.01.006
Leijten, W., Koes, R., Roobeek, I., Frugis, G. (2018). Translating flowering time from arabidopsis thaliana to brassicaceae and asteraceae crop species. Plants 7, 111. doi: 10.3390/plants7040111
Li, J., Brader, G., Kariola, T., Tapio Palva, E. (2006b). WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46, 477–491. doi: 10.1111/j.1365-313X.2006.02712.x
Li, N., Han, X., Feng, D., Yuan, D., Huang, L. J. (2019). Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: do we understand what they are whispering? Int. J. Mol. Sci. 20, 671. doi: 10.3390/ijms20030671
Li, Y., Huang, C., Liu, Y., Zeng, J., Yu, H., Tong, Z., et al. (2023). CRISPR/Cas9-mediated seamless gene replacement in protoplasts expands the resistance spectrum to TMV-U1 strain in regenerated Nicotiana tabacum. Plant Biotechnol. J. 21, 2641–2653. doi: 10.1111/pbi.14159
Li, G. Q., Huang, H. C., Miao, H. J., Erickson, R. S., Jiang, D. H., Xiao, Y. N. (2006a). Biological control of sclerotinia diseases of rapeseed by aerial applications of the mycoparasite coniothyrium minitans. Eur. J. Plant Pathol. 114, 345–355. doi: 10.1007/s10658-005-2232-6
Li, W., Lu, J., Yang, C., Xia, S. (2022). Identification of receptor-like proteins induced by Sclerotinia sclerotiorum in Brassica napus. Front. Plant Sci. 13, 944763. doi: 10.3389/fpls.2022.944763
Li, J., Zhao, Z., Hayward, A., Cheng, H., Fu, D. (2015). Integration analysis of quantitative trait loci for resistance to Sclerotinia sclerotiorum in Brassica napus. Euphytica 205, 483–489. doi: 10.1007/s10681-015-1417-0
Liang, X., Liberti, D., Li, M., Kim, Y. T., Hutchens, A., Wilson, R., et al. (2015a). Oxaloacetate acetylhydrolase gene mutants of Sclerotinia sclerotiorum do not accumulate oxalic acid, but do produce limited lesions on host plants. Mol. Plant Pathol. 16, 559–571. doi: 10.1111/mpp.12211
Liang, X., Moomaw, E. W., Rollins, J. A. (2015b). Fungal oxalate decarboxylase activity contributes to Sclerotinia sclerotiorum early infection by affecting both compound appressoria development and function. Mol. Plant Pathol. 16, 825–836. doi: 10.1111/mpp.12239
Liang, X., Rollins, J. A. (2018). Mechanisms of broad host range necrotrophic pathogenesis in sclerotinia sclerotiorum. Phytopathology® 108, 1128–1140. doi: 10.1094/PHYTO-06-18-0197-RVW
Liang, Y., Strelkov, S. E., Kav, N. N. V. (2009). Oxalic acid-mediated stress responses in Brassica napus L. PROTEOMICS 9, 3156–3173. doi: 10.1002/pmic.200800966
Lin, L., Fan, J., Li, P., Liu, D., Ren, S., Lin, K., et al. (2022). The Sclerotinia sclerotiorum-inducible promoter pBnGH17D7 in Brassica napus: isolation, characterization, and application in host-induced gene silencing. J. Exp. Bot. 73, 6663–6677. doi: 10.1093/jxb/erac328
Lin, Z., Zhong, S., Grierson, D. (2009). Recent advances in ethylene research. J. Exp. Bot. 60, 3311–3336. doi: 10.1093/jxb/erp204
Liu, W., Kang, Y., Ren, R., Liu, Y., Li, W., Xie, P., et al. (2021b). Identification and expression analysis of BnaCNGC family gene in the response to phytohormones, abiotic and biotic stresses in Brassica napus. J. Plant Interact. 16, 575–586. doi: 10.1080/17429145.2021.2007303
Liu, F., Li, X., Wang, M., Wen, J., Yi, B., Shen, J., et al. (2018a). Interactions of WRKY15 and WRKY33 transcription factors and their roles in the resistance of oilseed rape to Sclerotinia infection. Plant Biotechnol. J. 16, 911–925. doi: 10.1111/pbi.12838
Liu, J.-J., Sturrock, R., Ekramoddoullah, A. K. M. (2010). The superfamily of thaumatin-like proteins: its origin, evolution, and expression towards biological function. Plant Cell Rep. 29, 419–436. doi: 10.1007/s00299-010-0826-8
Liu, F., Wang, M., Wen, J., Yi, B., Shen, J., Ma, C., et al. (2015). Overexpression of barley oxalate oxidase gene induces partial leaf resistance to Sclerotinia sclerotiorum in transgenic oilseed rape. Plant Pathol. 64, 1407–1416. doi: 10.1111/ppa.12374
Liu, L., Wang, Q., Zhang, X., Liu, J., Zhang, Y., Pan, H. (2018b). Ssams2, a gene encoding GATA transcription factor, is required for appressoria formation and chromosome segregation in sclerotinia sclerotiorum. Front. Microbiol. 9, 3031. doi: 10.3389/fmicb.2018.03031
Liu, J., Zhu, L., Wang, B., Wang, H., Khan, I., Zhang, S., et al. (2021a). BnA1.CER4 and BnC1.CER4 are redundantly involved in branched primary alcohols in the cuticle wax of Brassica napus. Theor. Appl. Genet. 134, 3051–3067. doi: 10.1007/s00122-021-03879-y
Lumsden, R. D., Wergin, W. P. (1980). Scanning-electron microscopy of infection of bean by species of sclerotinia. Mycologia 72, 1200–1209. doi: 10.1080/00275514.1980.12021302
Lyu, X., Shen, C., Fu, Y., Xie, J., Jiang, D., Li, G., et al. (2016). A small secreted virulence-related protein is essential for the necrotrophic interactions of sclerotinia sclerotiorum with its host plants. PloS Pathog. 12, e1005435. doi: 10.1371/journal.ppat.1005435
Mao, G., Meng, X., Liu, Y., Zheng, Z., Chen, Z., Zhang, S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23, 1639–1653. doi: 10.1105/tpc.111.084996
Matar, S., Kumar, A., Holtgräwe, D., Weisshaar, B., Melzer, S. (2021). The transition to flowering in winter rapeseed during vernalization. Plant Cell Environ. 44, 506–518. doi: 10.1111/pce.13946
Mei, J., Ding, Y., Li, Y., Tong, C., Du, H., Yu, Y., et al. (2016). Transcriptomic comparison between Brassica oleracea and rice (Oryza sativa) reveals diverse modulations on cell death in response to Sclerotinia sclerotiorum. Sci. Rep. 6, 33706. doi: 10.1038/srep33706
Meng, X., Zhang, S. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245–266. doi: 10.1146/annurev-phyto-082712-102314
Moeder, W., Phan, V., Yoshioka, K. (2019). Ca2+ to the rescue – Ca2+channels and signaling in plant immunity. Plant Sci. 279, 19–26. doi: 10.1016/j.plantsci.2018.04.012
Nawrot, R., Barylski, J., Nowicki, G., Broniarczyk, J., Buchwald, W., Goździcka-Józefiak, A. (2014). Plant antimicrobial peptides. Folia Microbiol. (Praha) 59, 181–196. doi: 10.1007/s12223-013-0280-4
Newman, T. E., Kim, H., Khentry, Y., Sohn, K. H., Derbyshire, M. C., Kamphuis, L. G. (2023). The broad host range pathogen Sclerotinia sclerotiorum produces multiple effector proteins that induce host cell death intracellularly. Mol. Plant Pathol. 24, 866–881. doi: 10.1111/mpp.13333
Nováková, M., Šašek, V., Dobrev, P. I., Valentová, O., Burketová, L. (2014). Plant hormones in defense response of Brassica napus to Sclerotinia sclerotiorum – Reassessing the role of salicylic acid in the interaction with a necrotroph. Plant Physiol. Biochem. 80, 308–317. doi: 10.1016/j.plaphy.2014.04.019
Olivieri, F., Eugenia Zanetti, M., Oliva, C. R., Covarrubias, A. A., Casalongué, C. A. (2002). Characterization of an Extracellular Serine Protease of Fusarium eumartii and its Action on Pathogenesis Related Proteins. Eur. J. Plant Pathol. 108, 63–72. doi: 10.1023/A:1013920929965
Pan, Y., Xu, Y., Li, X., Yao, C., Gao, Z. (2015). SsPemG1 encodes an elicitor-homologous protein and regulates pathogenicity in Sclerotinia sclerotiorum. Physiol. Mol. Plant Pathol. 92, 70–78. doi: 10.1016/j.pmpp.2015.08.010
Pavan, S., Jacobsen, E., Visser, R. G. F., Bai, Y. (2010). Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol. Breed. 25, 1–12. doi: 10.1007/s11032-009-9323-6
Peng, Q., Xie, Q., Chen, F., Zhou, X., Zhang, W., Zhang, J., et al. (2017). Transcriptome Analysis of Sclerotinia sclerotiorum at Different Infection Stages on Brassica napus. Curr. Microbiol. 74, 1237–1245. doi: 10.1007/s00284-017-1309-8
Pieterse, C. M. J., van der Does, D., Zamioudis, C., Leon-Reyes, A., Van Wees, S. C. M. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Pré, M., Atallah, M., Champion, A., De Vos, M., Pieterse, C. M., Memelink, J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147, 1347–1357. doi: 10.1104/pp.108.117523
Qin, L., Nong, J., Cui, K., Tang, X., Gong, X., Xia, Y., et al. (2023). SsCak1 regulates growth and pathogenicity in sclerotinia sclerotiorum. Int. J. Mol. Sci. 24, 12610. doi: 10.3390/ijms241612610
Qu, X., Yu, B., Liu, J., Zhang, X., Li, G., Zhang, D., et al. (2014). MADS-box transcription factor SsMADS is involved in regulating growth and virulence in Sclerotinia sclerotiorum. Int. J. Mol. Sci. 15, 8049–8062. doi: 10.3390/ijms15058049
Rahman, H., Xu, Y. P., Zhang, X. R., Cai, X. Z. (2016). Brassica napus Genome Possesses Extraordinary High Number of CAMTA Genes and CAMTA3 Contributes to PAMP Triggered Immunity and Resistance to Sclerotinia sclerotiorum. Front. Plant Sci. 7, 581. doi: 10.3389/fpls.2016.00581
Rana, K., Ding, Y., Banga, S. S., Liao, H., Zhao, S., Yu, Y., et al. (2021). Sclerotinia sclerotiorum Thioredoxin1 (SsTrx1) is required for pathogenicity and oxidative stress tolerance. Mol. Plant Pathol. 22, 1413–1426. doi: 10.1111/mpp.13127
Rask, L., Andréasson, E., Ekbom, B., Eriksson, S., Pontoppidan, B., Meijer, J. (2000). Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol. Biol. 42, 93–113. doi: 10.1023/A:1006380021658
Rietz, S., Bernsdorff, F. E., Cai, D. (2012). Members of the germin-like protein family in Brassica napus are candidates for the initiation of an oxidative burst that impedes pathogenesis of Sclerotinia sclerotiorum. J. Exp. Bot. 63, 5507–5519. doi: 10.1093/jxb/ers203
Rollins, J. A. (2003). The Sclerotinia sclerotiorum pac1 Gene Is Required for Sclerotial Development and Virulence. Mol. Plant-Microbe Interactions® 16, 785–795. doi: 10.1094/MPMI.2003.16.9.785
Ruan, J., Zhou, Y., Zhou, M., Yan, J., Khurshid, M., Weng, W., et al. (2019). Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 20, 2479. doi: 10.3390/ijms20102479
Rushton, P. J., Somssich, I. E., Ringler, P., Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Sangster, T. A., Queitsch, C. (2005). The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Curr. Opin. Plant Biol. 8, 86–92. doi: 10.1016/j.pbi.2004.11.012
Schulze-Lefert, P. (2004). Plant immunity: the origami of receptor activation. Curr. Biol. 14, R22–R24. doi: 10.1016/j.cub.2003.12.017
Seifbarghi, S., Borhan, M. H., Wei, Y., Coutu, C., Robinson, S. J., Hegedus, D. D. (2017). Changes in the Sclerotinia sclerotiorum transcriptome during infection of Brassica napus. BMC Genomics 18, 266. doi: 10.1186/s12864-017-3642-5
Shi, C. L., Alling, R. M., Hammerstad, M., Aalen, R. B. (2019). Control of organ abscission and other cell separation processes by evolutionary conserved peptide signaling. Plants (Basel) 8, 225. doi: 10.3390/plants8070225
Silva, K. J. P., Mahna, N., Mou, Z., Folta, K. M. (2018). NPR1 as a transgenic crop protection strategy in horticultural species. Hortic. Res. 5, 15. doi: 10.1038/s41438-018-0026-1
Sinha, R., Shukla, P. (2019). Antimicrobial peptides: recent insights on biotechnological interventions and future perspectives. Protein Pept. Lett. 26, 79–87. doi: 10.2174/0929866525666181026160852
Stø, I. M., Orr, R. J., Fooyontphanich, K., Jin, X., Knutsen, J. M., Fischer, U., et al. (2015). Conservation of the abscission signaling peptide IDA during Angiosperm evolution: withstanding genome duplications and gain and loss of the receptors HAE/HSL2. Front. Plant Sci. 6, 931. doi: 10.3389/fpls.2015.00931
Sun, Q., Lin, L., Liu, D., Wu, D., Fang, Y., Wu, J., et al. (2018). CRISPR/cas9-mediated multiplex genome editing of the bnWRKY11 and bnWRKY70 genes in brassica napus L. Int. J. Mol. Sci. 19, 2716. doi: 10.3390/ijms19092716
Tan, X., Yan, S., Tan, R., Zhang, Z., Wang, Z., Chen, J. (2014). Characterization and Expression of a GDSL-Like Lipase Gene from Brassica napus in Nicotiana benthamiana. Protein J. 33, 18–23. doi: 10.1007/s10930-013-9532-z
Thomma, B. P. H. J., Penninckx, I., Cammue, B. P. A., Broekaert, W. F. (2001). The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 13, 63–68. doi: 10.1016/S0952-7915(00)00183-7
Tian, W., Hou, C., Ren, Z., Wang, C., Zhao, F., Dahlbeck, D., et al. (2019). A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 572, 131–135. doi: 10.1038/s41586-019-1413-y
Tichá, T., Samakovli, D., Kuchařová, A., Vavrdová, T., Šamaj, J. (2020). Multifaceted roles of HEAT SHOCK PROTEIN 90 molecular chaperones in plant development. J. Exp. Bot. 71, 3966–3985. doi: 10.1093/jxb/eraa177
Turkington, T. K., Morrall, R., Rude, S. V. (1991). Use of petal infestation to forecast sclerotinia stem rot of canola: the impact of diurnal and weather-related inoculum fluctuations. Can. J. Plant Pathol. 13, 347–355. doi: 10.1080/07060669109500920
Uloth, M. B., You, M. P., Finnegan, P. M., Banga, S. S., Banga, S. K., Sandhu, P. S., et al. (2013). New sources of resistance to Sclerotinia sclerotiorum for crucifer crops. Field Crops Res. 154, 40–52. doi: 10.1016/j.fcr.2013.07.013
van Loon, L. C., Rep, M., Pieterse, C. M. J. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
Verma, S. S., Yajima, W. R., Rahman, M. H., Shah, S., Liu, J. J., Ekramoddoullah, A. K., et al. (2012). A cysteine-rich antimicrobial peptide from Pinus monticola (PmAMP1) confers resistance to multiple fungal pathogens in canola (Brassica napus). Plant Mol. Biol. 79, 61–74. doi: 10.1007/s11103-012-9895-0
Wang, Z., Bao, L. L., Zhao, F. Y., Tang, M. Q., Chen, T., Li, Y., et al. (2019a). BnaMPK3 is a key regulator of defense responses to the devastating plant pathogen sclerotinia sclerotiorum in oilseed rape. Front. Plant Sci. 10, 91. doi: 10.3389/fpls.2019.00091
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014a). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Wang, Z., Fang, H., Chen, Y., Chen, K., Li, G., Gu, S., et al. (2014b). Overexpression of BnWRKY33 in oilseed rape enhances resistance to Sclerotinia sclerotiorum. Mol. Plant Pathol. 15, 677–689. doi: 10.1111/mpp.12123
Wang, L., Liu, F., Ju, L., Xue, B., Wang, Y., Wang, D., et al. (2022a). Genome Structures and Evolution Analysis of Hsp90 Gene Family in Brassica napus Reveal the Possible Roles of Members in Response to Salt Stress and the Infection of Sclerotinia sclerotiorum. Front. Plant Sci. 13, 854034. doi: 10.3389/fpls.2022.854034
Wang, Z., Ma, L. Y., Cao, J., Li, Y. L., Ding, L. N., Zhu, K. M., et al. (2019b). Recent advances in mechanisms of plant defense to sclerotinia sclerotiorum. Front. Plant Sci. 10, 1314. doi: 10.3389/fpls.2019.01314
Wang, Z., Ma, L.-Y., Li, X., Zhao, F.-Y., Sarwar, R., Cao, J., et al. (2020b). Genome-wide identification of the NPR1-like gene family in Brassica napus and functional characterization of BnaNPR1 in resistance to Sclerotinia sclerotiorum. Plant Cell Rep. 39, 709–722. doi: 10.1007/s00299-020-02525-z
Wang, Z., Mao, H., Dong, C., Ji, R., Cai, L., Fu, H., et al. (2009). Overexpression of Brassica napus MPK4 Enhances Resistance to Sclerotinia sclerotiorum in Oilseed Rape. Mol. Plant-Microbe Interactions® 22, 235–244. doi: 10.1094/MPMI-22-3-0235
Wang, N., Qian, W., Suppanz, I., Wei, L., Mao, B., Long, Y., et al. (2011). Flowering time variation in oilseed rape (Brassica napus L.) is associated with allelic variation in the FRIGIDA homologue BnaA.FRI.a. J. Exp. Bot. 62, 5641–5658. doi: 10.1093/jxb/err249
Wang, J., Singh, S. K., Geng, S., Zhang, S., Yuan, L. (2020a). Genome-wide analysis of glycerol-3-phosphate O-acyltransferase gene family and functional characterization of two cutin group GPATs in Brassica napus. Planta 251, 93. doi: 10.1007/s00425-020-03384-4
Wang, Z., Tan, X., Zhang, Z., Gu, S., Li, G., Shi, H. (2012). Defense to Sclerotinia sclerotiorum in oilseed rape is associated with the sequential activations of salicylic acid signaling and jasmonic acid signaling. Plant Sci. 184, 75–82. doi: 10.1016/j.plantsci.2011.12.013
Wang, N., Tang, C., Fan, X., He, M., Gan, P., Zhang, S., et al. (2022b). Inactivation of a wheat protein kinase gene confers broad-spectrum resistance to rust fungi. Cell 185, 2961–2974.e2919. doi: 10.1016/j.cell.2022.06.027
Wang, M., Thomas, N., Jin, H. (2017). Cross-kingdom RNA trafficking and environmental RNAi for powerful innovative pre- and post-harvest plant protection. Curr. Opin. Plant Biol. 38, 133–141. doi: 10.1016/j.pbi.2017.05.003
Wang, Z., Wan, L., Xin, Q., Chen, Y., Zhang, X., Dong, F., et al. (2018). Overexpression of OsPGIP2 confers Sclerotinia sclerotiorum resistance in Brassica napus through increased activation of defense mechanisms. J. Exp. Bot. 69, 3141–3155. doi: 10.1093/jxb/ery138
Wang, C., Yao, J., Du, X., Zhang, Y., Sun, Y., Rollins, J. A., et al. (2015). The Arabidopsis Mediator Complex Subunit16 Is a Key Component of Basal Resistance against the Necrotrophic Fungal Pathogen Sclerotinia sclerotiorum. Plant Physiol. 169, 856–872. doi: 10.1104/pp.15.00351
Wang, Z., Zhao, F.-Y., Tang, M.-Q., Chen, T., Bao, L.-L., Cao, J., et al. (2020c). BnaMPK6 is a determinant of quantitative disease resistance against Sclerotinia sclerotiorum in oilseed rape. Plant Sci. 291, 110362. doi: 10.1016/j.plantsci.2019.110362
Wani, S. H., Anand, S., Singh, B., Bohra, A., Joshi, R. (2021). WRKY transcription factors and plant defense responses: latest discoveries and future prospects. Plant Cell Rep. 40, 1071–1085. doi: 10.1007/s00299-021-02691-8
Wasternack, C. (2007). Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100, 681–697. doi: 10.1093/aob/mcm079
Wegele, H., Müller, L., Buchner, J. (2004). Hsp70 and Hsp90–a relay team for protein folding. Rev. Physiol. Biochem. Pharmacol. 151, 1–44. doi: 10.1007/s10254-003-0021-1
Wei, L., Jian, H., Lu, K., Filardo, F., Yin, N., Liu, L., et al. (2016). Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnol. J. 14, 1368–1380. doi: 10.1111/pbi.12501
Wei, D., Mei, J., Fu, Y., Disi, J. O., Li, J., Qian, W. (2014). Quantitative trait loci analyses for resistance to Sclerotinia sclerotiorum and flowering time in Brassica napus. Mol. Breed. 34, 1797–1804. doi: 10.1007/s11032-014-0139-7
Wei, W., Xu, L., Peng, H., Zhu, W., Tanaka, K., Cheng, J., et al. (2022). A fungal extracellular effector inactivates plant polygalacturonase-inhibiting protein. Nat. Commun. 13, 2213. doi: 10.1038/s41467-022-29788-2
Williams, B., Kabbage, M., Kim, H. J., Britt, R., Dickman, M. B. (2011). Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PloS Pathog. 7, e1002107. doi: 10.1371/journal.ppat.1002107
Wittstock, U., Gershenzon, J. (2002). Constitutive plant toxins and their role in defense against herbivores and pathogens. Curr. Opin. Plant Biol. 5, 300–307. doi: 10.1016/S1369-5266(02)00264-9
Woo, E.-J., Dunwell, J. M., Goodenough, P. W., Marvier, A. C., Pickersgill, R. W. (2000). Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat. Struct. Biol. 7, 1036–1040. doi: 10.1038/80954
Wu, J., Cai, G., Tu, J., Li, L., Liu, S., Luo, X., et al. (2013). Identification of QTLs for resistance to sclerotinia stem rot and BnaC.IGMT5.a as a candidate gene of the major resistant QTL SRC6 in Brassica napus. PloS One 8, e67740. doi: 10.1371/journal.pone.0067740
Wu, J., Zhao, Q., Yang, Q., Liu, H., Li, Q., Yi, X., et al. (2016). Comparative transcriptomic analysis uncovers the complex genetic network for resistance to Sclerotinia sclerotiorum in Brassica napus. Sci. Rep. 6, 19007. doi: 10.1038/srep19007
Xia, S., Xu, Y., Hoy, R., Zhang, J., Qin, L., Li, X. (2019). The notorious soilborne pathogenic fungus sclerotinia sclerotiorum: an update on genes studied with mutant analysis. Pathogens 9, 27. doi: 10.3390/pathogens9010027
Xie, C., Shang, Q., Mo, C., Xiao, Y., Wang, G., Xie, J., et al. (2021). Early secretory pathway-associated proteins ssEmp24 and ssErv25 are involved in morphogenesis and pathogenicity in a filamentous phytopathogenic fungus. mBio 12, e0317321. doi: 10.1128/mBio.03173-21
Xu, B., Gong, X., Chen, S., Hu, M., Zhang, J., Peng, Q. (2021). Transcriptome Analysis Reveals the Complex Molecular Mechanisms of Brassica napus-Sclerotinia sclerotiorum Interactions. Front. Plant Sci. 12, 716935. doi: 10.3389/fpls.2021.716935
Xu, T., Li, J., Yu, B., Liu, L., Zhang, X., Liu, J., et al. (2018). Transcription factor ssSte12 was involved in mycelium growth and development in sclerotinia sclerotiorum. Front. Microbiol. 9, 2476. doi: 10.3389/fmicb.2018.02476
Yajima, W., Verma, S. S., Shah, S., Rahman, M. H., Liang, Y., Kav, N. N. V. (2010). Expression of anti-sclerotinia scFv in transgenic Brassica napus enhances tolerance against stem rot. New Biotechnol. 27, 816–821. doi: 10.1016/j.nbt.2010.09.010
Yang, C., Li, W., Huang, X., Tang, X., Qin, L., Liu, Y., et al. (2022). SsNEP2 contributes to the virulence of sclerotinia sclerotiorum. Pathogens 11, 446. doi: 10.3390/pathogens11040446
Yang, X., Li, J., Li, X., She, R., Pei, Y. (2006). Isolation and characterization of a novel thermostable non-specific lipid transfer protein-like antimicrobial protein from motherwort (Leonurus japonicus Houtt) seeds. Peptides 27, 3122–3128. doi: 10.1016/j.peptides.2006.07.019
Yang, B., Rahman, M. H., Liang, Y., Shah, S., Kav, N. N. V. (2010). Characterization of Defense Signaling Pathways of Brassica napus and Brassica carinata in Response to Sclerotinia sclerotiorum Challenge. Plant Mol. Biol. Rep. 28, 253–263. doi: 10.1007/s11105-009-0149-5
Yang, G., Tang, L., Gong, Y., Xie, J., Fu, Y., Jiang, D., et al. (2018). A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 217, 739–755. doi: 10.1111/nph.14842
Ye, Z., Liu, Y. (2023). Polyphenolic compounds from rapeseeds (Brassica napus L.): The major types, biofunctional roles, bioavailability, and the influences of rapeseed oil processing technologies on the content. Food Res. Int. 163, 112282. doi: 10.1016/j.foodres.2022.112282
Yin, M., Wang, R., Li, S., Luo, M., Wei, W., Wang, M., et al. (2022). High Sclerotinia sclerotiorum resistance in rapeseed plant has been achieved by OsPGIP6. Front. Plant Sci. 13, 970716. doi: 10.3389/fpls.2022.970716
Yin, X., Yi, B., Chen, W., Zhang, W., Tu, J., Fernando, W. G. D., et al. (2010). Mapping of QTLs detected in a Brassica napus DH population for resistance to Sclerotinia sclerotiorum in multiple environments. Euphytica 173, 25–35. doi: 10.1007/s10681-009-0095-1
Yu, Y., Du, J., Wang, Y., Zhang, M., Huang, Z., Cai, J., et al. (2019). Survival factor 1 contributes to the oxidative stress response and is required for full virulence of Sclerotinia sclerotiorum. Mol. Plant Pathol. 20, 895–906. doi: 10.1111/mpp.12801
Yu, Y., Xiao, J., Du, J., Yang, Y., Bi, C., Qing, L. (2016). Disruption of the gene encoding endo-β-1, 4-xylanase affects the growth and virulence of sclerotinia sclerotiorum. Front. Microbiol. 7, 1787. doi: 10.3389/fmicb.2016.01787
Yu, Y., Xiao, J., Zhu, W., Yang, Y., Mei, J., Bi, C., et al. (2017). Ss-Rhs1, a secretory Rhs repeat-containing protein, is required for the virulence of Sclerotinia sclerotiorum. Mol. Plant Pathol. 18, 1052–1061. doi: 10.1111/mpp.12459
Zhang, Y., Bai, Y., Wu, G., Zou, S., Chen, Y., Gao, C., et al. (2017). Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 91, 714–724. doi: 10.1111/tpj.13599
Zhang, X., Cheng, J., Lin, Y., Fu, Y., Xie, J., Li, B., et al. (2021). Editing homologous copies of an essential gene affords crop resistance against two cosmopolitan necrotrophic pathogens. Plant Biotechnol. J. 19, 2349–2361. doi: 10.1111/pbi.13667
Zhang, L., Du, L., Shen, C., Yang, Y., Poovaiah, B. W. (2014b). Regulation of plant immunity through ubiquitin-mediated modulation of Ca(2+) -calmodulin-AtSR1/CAMTA3 signaling. Plant J. 78, 269–281. doi: 10.1111/tpj.12473
Zhang, W., Fraiture, M., Kolb, D., Löffelhardt, B., Desaki, Y., Boutrot, F. F., et al. (2013). Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 25, 4227–4241. doi: 10.1105/tpc.113.117010
Zhang, K., Liu, F., Wang, Z., Zhuo, C., Hu, K., Li, X., et al. (2022b). Transcription factor WRKY28 curbs WRKY33-mediated resistance to Sclerotinia sclerotiorum in Brassica napus. Plant Physiol. 190, 2757–2774. doi: 10.1093/plphys/kiac439
Zhang, H., Wu, Q., Cao, S., Zhao, T., Chen, L., Zhuang, P., et al. (2014a). A novel protein elicitor (SsCut) from Sclerotinia sclerotiorum induces multiple defense responses in plants. Plant Mol. Biol. 86, 495–511. doi: 10.1007/s11103-014-0244-3
Zhang, J., Xiao, K., Li, M., Hu, H., Zhang, X., Liu, J., et al. (2022a). SsAGM1-mediated uridine diphosphate-N-acetylglucosamine synthesis is essential for development, stress response, and pathogenicity of sclerotinia sclerotiorum. Front. Microbiol. 13, 938784. doi: 10.3389/fmicb.2022.938784
Zhang, M., Zhang, S. (2022). Mitogen-activated protein kinase cascades in plant signaling. J. Integr. Plant Biol. 64, 301–341. doi: 10.1111/jipb.13215
Zhao, J., Meng, J. (2003). Genetic analysis of loci associated with partial resistance to Sclerotinia sclerotiorum in rapeseed (Brassica napus L.). Theor. Appl. Genet. 106, 759–764. doi: 10.1007/s00122-002-1171-2
Zhao, J., Udall, J. A., Quijada, P. A., Grau, C. R., Meng, J., Osborn, T. C. (2006). Quantitative trait loci for resistance to Sclerotinia sclerotiorum and its association with a homeologous non-reciprocal transposition in Brassica napus L. Theor. Appl. Genet. 112, 509–516. doi: 10.1007/s00122-005-0154-5
Zheng, X. Y., Zhou, M., Yoo, H., Pruneda-Paz, J. L., Spivey, N. W., Kay, S. A., et al. (2015). Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. Proc. Natl. Acad. Sci. U.S.A. 112, 9166–9173. doi: 10.1073/pnas.1511182112
Zhu, W., Wei, W., Fu, Y., Cheng, J., Xie, J., Li, G., et al. (2013). A secretory protein of necrotrophic fungus Sclerotinia sclerotiorum that suppresses host resistance. PloS One 8, e53901. doi: 10.1371/journal.pone.0053901
Ziaei, M., Motallebi, M., Zamani, M. R., Panjeh, N. Z. (2016). Co-expression of chimeric chitinase and a polygalacturonase-inhibiting protein in transgenic canola (Brassica napus) confers enhanced resistance to Sclerotinia sclerotiorum. Biotechnol. Lett. 38, 1021–1032. doi: 10.1007/s10529-016-2058-7
Keywords: plant defense, transcriptome, resistance gene, pathogen effector, oxalic acid, gene editing
Citation: Chen R-S, Wang J-Y, Sarwar R and Tan X-L (2023) Genetic breakthroughs in the Brassica napus–Sclerotinia sclerotiorum interactions. Front. Plant Sci. 14:1276055. doi: 10.3389/fpls.2023.1276055
Received: 11 August 2023; Accepted: 07 November 2023;
Published: 23 November 2023.
Edited by:
Ahmad Fakhoury, Southern Illinois University Carbondale, United StatesReviewed by:
Yang Yu, Southwest University, ChinaNazanin Zamani-Noor, Julius Kühn-Institute, Germany
Begoña Pérez Vich, Spanish National Research Council (CSIC), Spain
Copyright © 2023 Chen, Wang, Sarwar and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiao-Li Tan, eGx0YW5AdWpzLmVkdS5jbg==
 Rong-Shi Chen
Rong-Shi Chen Ji-Yi Wang
Ji-Yi Wang Rehman Sarwar
Rehman Sarwar Xiao-Li Tan
Xiao-Li Tan