
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 23 October 2023
Sec. Plant Biotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1271757
This article is part of the Research TopicPlant Molecular Farming for the Production of Next-Generation Vaccines and Biologics – Prospects and ChallengesView all 11 articles
 Nataliia Shcherbak1*†
Nataliia Shcherbak1*† Heike Prochaska2†
Heike Prochaska2† Kateryna Lystvan1
Kateryna Lystvan1 Yelizaveta Prokhorova1
Yelizaveta Prokhorova1 Anatoli Giritch2
Anatoli Giritch2 Mykola Kuchuk1
Mykola Kuchuk1Food-borne illnesses caused by pathogenic Escherichia coli strains, especially enterohaemorrhagic E. coli (EHEC), are a serious public health problem, as debilitating disease and even death from such food poisonings have been repeatedly reported. Colicin M (ColM), a non-antibiotic antimicrobial protein produced by some strains of E. coli, has shown promising activity in controlling multiple enteropathogenic strains of E. coli and related pathogens. As contaminated green leafy vegetables are a frequent source of pathogenic E. coli infections, we genetically modified (GM) two edible crops, lettuce (Lactuca sativa L.) and mizuna (Brassica rapa subsp. nipposinica var. laciniata), to stably express the ColM gene and assessed the antibacterial activity of tissue extracts from these plants against selected E. coli strains in vitro. Transgenic plants of these species were developed using Agrobacterium-mediated transformation with a vector containing the ColM-coding gene (cma) under the control of the 35S promoter. Western blot analysis of recombinant ColM protein was performed in selected transgenic plants to confirm cma gene expression and quantify ColM accumulation. Extracts of transgenic plants expressing ColM showed significant activity against two major strains of EHEC (O157:H7 and O104:H4) as well as E. coli strains resistant to beta-lactam- and carbapenem-class antibiotics. Importantly, the antibacterial activity persisted in several subsequent generations of transgenic lettuce and mizuna plants that stably expressed the ColM gene. In addition, our results also show that the antibacterial activity of dried (up to 40°C) biomass of transgenic plants remained stable without a decrease for at least three months.
Food contamination by Shiga-toxin producing Escherichia coli (STEC) strains, especially their subset which can cause hemorrhagic colitis (enterohemorrhagic E. coli, EHEC), has repeatedly resulted in outbreaks of debilitating intestinal infections affecting the health of millions of people worldwide. E. coli of serotype O104:H4, which combines enteroaggregative and Shiga-toxin producing properties (EAEC/EHEC), was responsible for a large outbreak of gastrointestinal disease in multiple countries in Europe in 2011. The high virulence and multidrug resistance (MDR) of this uncommon E. coli strain led to the infection of almost 4,000 people and resulted in 54 deaths (Navarro-Garcia, 2014; Tietze et al., 2015). E. coli O157:H7 is currently the predominant STEC serotype causing severe illnesses with a low infectious dose (50–100 colony-forming units (CFU/g or mL) (Puligundla and Lim, 2022). Frequently the spread of these pathogenic bacteria is associated with the consumption of animal products because they are commonly found in the intestinal tract of cattle (Low et al., 2005; Munns et al., 2015). However, green leafy vegetables, which are usually eaten raw without heat treatment from cooking, can be contaminated in the field through improperly composted manure, or contaminated irrigation water or soil (Alegbeleye et al., 2018; Waltenburg et al., 2021; Xu et al., 2021). A multistate outbreak of STEC O157:H7 associated with the consumption of Romaine lettuce was reported in the USA in 2011 (Slayton et al., 2013). Subsequently, the U.S. Centers for Disease Control and Prevention (USCDC) reported that in 2018 and 2019, several large outbreaks of E. coli O157:H7 linked to Romaine lettuce resulted in 439 illnesses and 5 deaths (USCDC, 2020). In spite of these experiences and the implementation of mitigation measures that followed, contaminated fresh leafy vegetables have continued to cause foodborne disease outbreaks (Irvin et al., 2021; Waltenburg et al., 2021).
The food industry employs various sanitizing methods both for disinfection of food processing equipment and for reduction of common foodborne pathogens associated with fresh-cut vegetables and fruit products. Currently, thermal inactivation is one of the most effective methods for the destruction of pathogenic bacteria in food, but the method is not generally amenable to decontaminating green leafy vegetables or some types of animal feed. For such products, sodium hypochlorite (NaClO) solutions can be used, but their residues are potentially harmful, can impart an undesirable taste, and the process is not environmentally benign (Tomás-Callejas et al., 2011). Antibiotics are often used prophylactically to prevent the spread of pathogenic bacteria on farms and in livestock. In response to the significant increase of MDR bacteria, legislative measures have widely been taken to limit or eliminate the use of antibiotics, including in the form of feed additives for livestock (Urban-Chmiel et al., 2022). According to the U.S. Food and Drug Administration (FDA), medically important antimicrobial drugs approved for use in food-producing animals decreased by 38% since 2015, which was the peak year of sales (FDA, 2021). However, this is only the beginning of the implementation of measures to prevent the increase of MDR bacteria and the problem of the constantly growing number of bacteria resistant to widely used antibiotics, including drugs of last resort, remains relevant. Antibiotics used in food-producing animals are closely related to those used in human medicine and antibiotic-resistant pathogenic bacteria foreshadow a growing global health crisis. Numerous studies have confirmed that bacteria use both phenotypic and genetic strategies that ensure the emergence of antibiotic resistance (Urban-Chmiel et al., 2022). There is already evidence of bacterial resistance to highly effective antibiotics, including transmission among E. coli strains of the resistance gene for the antibiotic colistin, considered the antibacterial of last resort in the management of severe and life-threatening Gram-negative infections (Liu et al., 2016; Paveenkittiporn et al., 2017). Also of concern is the rapidly increasing prevalence of Enterobacteriaceae harbouring carbapenemases and becoming resistant to carbapenem-class antibiotics. The spread of carbapenem resistance has led the World Health Organization (WHO) to designate carbapenem-resistant enterobacteria (CRE) as «priority pathogens» that pose the greatest global threat to human health (Willyard, 2017; Sheu et al., 2019). Alternative management strategies are therefore urgently needed, especially to control Gram-negative bacterial pathogens.
The spread of bacterial infections caused by food contamination with pathogenic microorganisms could be effectively prevented by bacteriocins, which are non-antibiotic antimicrobial proteins produced by bacteria to kill closely related strains, presumably by affording producing strains a competitive advantage for resources in their ecological niche (Gabrielsen et al., 2014; Kumariya et al., 2019; Benitez-Chao et al., 2021). Due to their high species and strain specificity (i.e., narrow spectrum of activity), the use of bacteriocins as antimicrobial agents offers important advantages over current chemistries, including sparing beneficial bacteria and thus avoiding microbial imbalances in the gastrointestinal (GI) tract (Cleveland et al., 2001; Cavera et al., 2015). Further, bacteriocins are natively produced by bacteria resident in the GI tract of humans and other animals, underscoring their evolutionary importance as well as their safety.
Therefore, bacteriocins offer unique advantages as potential alternatives to antibiotics, especially for prophylactic use in both food and feed. Within the bacteriocin family, colicins are antibacterial proteins produced by certain strains of E. coli that are effective against pathogenic strains of the same or closely related species (Callaway et al., 2004; Chérier et al., 2021). Colicins can be recombinantly produced using several platforms, including various plant hosts. Several plant-produced colicins have been approved as GRAS (Generally Recognized as Safe) antimicrobial food processing aids by FDA (GRAS Notice (GRN) No. 593, 2015). Colicin M (ColM) is the only one colicin interfering with peptidoglycan biosynthesis. Targeting such an essential bacterial component potentially makes the ColM-like proteins “universal” antibacterial agents (Chérier et al., 2021). ColM was first produced using transient gene expression methods in Nicotiana benthamiana plants, a well-accepted standard species for recombinant protein manufacturing, as well as in edible species of plants such as spinach and leafy beet (Schulz et al., 2015; Stephan et al., 2017; Hahn-Löbmann et al., 2019). Among the various colicins evaluated, ColM had the broadest antimicrobial activity against the seven major foodborne enterohemorrhagic E. coli (EHEC) strains, including the O157:H7 serotype (Schulz et al., 2015; Hahn-Löbmann et al., 2019) and Klebsiella pneumoniae (Łojewska et al., 2020).
The goal of our work was to create genetically modified edible plants that could stably express recombinant ColM. Previously, ColM had been recombinantly expressed only in non-food species of plants, including transiently and transgenically in N. benthamiana (Hahn-Löbmann et al., 2019) and transgenically in N. tabacum (Łojewska et al., 2020). The ColM isolated from these plants retained its conformation and was fully active. In the present study, we report for the first time the successful development of stable transgenic green leafy vegetables, lettuce and mizuna, expressing ColM and confirm the antibacterial activity of plant extracts for several generations of transgenic plants. We also demonstrate that heat dried (up to 40°C) transgenic plant biomass retained antimicrobial activity without significant reduction for at least 3 months. Our results suggest that bacteriocin-expressing leafy vegetables that are often the vectors for environmentally acquired Gram-negative bacterial infections could be used instead as a functional food to help control foodborne illnesses.
The vector pNMD46772 containing the ColM-coding cma gene under the control of a 35S promoter and selective bar gene under the control of a nos promoter was transformed to Agrobacterium tumefaciens strain GV3101. The transformed bacteria were grown on selective LB (Bertani, 1951) agar medium containing rifampicin 50 mg/L, gentamicin 25 mg/L and kanamycin 100 mg/L. Colonies were confirmed to be transformed with vector pNMD46772 using PCR analysis with primers specific for the cma gene.
Agrobacterium-mediated transformation of lettuce ‘Odeskyj curling’; with the vector pNMD46772 containing the cma gene was carried out as described in Shcherbak et al. (2013). After 48 h of co-cultivation with diluted Agrobacterium suspension supplemented with 100 mM acetosyringone, explants were placed on modified B5 (Gamborg et al., 1968) regeneration medium (B5 medium salts with 2.5% sucrose, 3 mg/L kinetin, 0.5 mg/L NAA) supplemented with 5 mg/L phosfinothricin (PPT) for selection. As shoot appeared, each one was removed and placed on the MS (Murashige and Skoog, 1962) medium supplemented with 0.5 mg/L of indole-3-butyric acid for rooting. Well established plants were transferred into the soil and grown in a greenhouse.
For genetic transformation of mizuna, hypocotyls of 5-7-day-old seedlings were used as explants. After co-cultivation with Agrobacterium, the explants were cultivated on callus induction MS medium containing 2,4-D (2 mg/L) and BAP (0.5 mg/L) for one week. Further regeneration of shoots took place on a nutrient medium containing zeatin (2 mg/L), BAP (1 mg/L) and NAA (0.5 mg/L). PPT at concentrations of 5 mg/L was used for the first step of mizuna transformants selection and after two weeks of cultivation on the regeneration medium the PPT concentration was increased to 10 mg/L. Regenerated plants were also rooted, transferred to the soil and grown in a greenhouse.
Genomic DNA was isolated from plant leaves using the CTAB protocol for isolating DNA from plant tissues. PCR was carried out in 20 μl reaction volume containing 50 ng DNA, 200 μM each of forward and reverse primers, 200 μM dNTPs and 1 U Taq DNA polymerase (Thermo Scientific, USA). Amplification was performed on a Mastercycler® personal (Eppendorf, USA). Sequences of the primers used were as follows:
for bar gene amplification.
bar_fwd 5’-ACATCGAGACAAGCACGGTC-3’.
bar_rev 5’-GCCAGAAACCCACGTCATGC-3’.
for cma gene amplification.
colM_fwd 5’-ATGGAAACCCTTACTGTGCACGCTC-3’.
colM_rev 5’-CCTCTTACCAGACTCTTTGATGTG-3’.
The products of the amplification (411 bp when using primers for bar gene and 1003 bp for cma gene) were separated on 1% (w/v) agarose gels.
RT-PCR was performed for previously selected transgenic plants to detect cma gene transcripts. Total RNA was prepared from 100 mg of transgenic plant leaf tissue using an GeneJET RNA Purification Kit (Thermo Fischer Scientific, Waltham, USA) and reverse-transcribed using a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, USA) with oligo dT primers. Genomic DNA from RNA preparation was removed by DNase I (RNase-free) treatment for 30 min at 37°C. A negative control for each sample consisted of the same reaction mixture without reverse transcriptase. The obtained cDNA was used as a template for PCR with gene-specific primers.
Leaf material of transgenic and non-transgenic plants was ground in liquid nitrogen and total soluble protein (TSP) extracts were prepared with 5 volumes of extraction buffer (50 mM HEPES pH7.0, 10 mM K acetate, 5 mM Mg acetate, 10% glycerol, 0.05% Tween-20, 300 mM NaCl). After incubation for 20 min on ice and centrifugation for 10 min at 13000 rpm at 4°C, supernatant was used for immunoblot analysis and determination of antimicrobial activity. Protein concentration in the extracts was determined by the Bradford method using Bio-Rad Protein Assay (Bio-Rad Laboratories, Hercules, USA) and bovine serum albumin (BSA) (Sigma-Aldrich, Saint Louis, USA) as a standard.
Plant leaf material was harvested, frozen in liquid nitrogen and stored at -80°C. After 262 days, part of the material was thawed and incubated for 1 day at 40°C in an oven for drying. For final drying, material was incubated at room temperature for an additional 10 days. Dried material was further stored at room temperature. Fresh and frozen plant material was extracted with 5 volumes of extraction buffer, while dried material was extracted with 20 volumes of extraction buffer.
To allow direct comparison of activities for fresh and dried plant material, the weight factor for normalization of different extraction volumes and the weight loss during drying process were calculated as follows: Weight factor = fresh weight/dry weight * 5 (vol fresh extraction)/20 (vol dry extraction).
ColM was transiently expressed in N. benthamiana and purified according to Stephan et al. (2017) with minor modifications. For purification plant material was extracted with the buffer containing 20 mM citric acid (pH4.0), 20mM NaH2PO4 and 30 mM NaCl in a 5:1 (v/w) buffer:biomass ratio. Ground leaf material admixed with pre-chilled extraction buffer was incubated on ice for 10 min followed by centrifugation at 10,000 x g for 15 min at 22°C. The supernatant was filtered using Miracloth (Merck KGaA, Darmstadt, Germany) and mixed with 10 mg/ml diatomaceous earth (Sigma-Aldrich, Saint Louis, USA) followed by incubation of the filtrate for 20 min at room temperature and centrifugation for 15 min at 10,000 x g at 22°C. The resulting supernatant was filtrated using filter discs with a pore size of 8–12 µm and the final filtrate was used for further purification by column chromatography. The filtrate was loaded on a HiTrap™Capto™MMC (GE Healthcare, Munich, Germany) column equilibrated with extraction buffer. The column was washed in two steps. The first wash step was performed with extraction buffer. The second wash step was done using diluted (30%) elution buffer consisting of 50 mM Na2HPO4 (pH 7.84), 10 mM citric acid and 1 M NaCl. The elution of ColM was carried out in a linear gradient from 30 to 100% of elution buffer concentration. The eluted fractions were analyzed by SDS-PAGE and Coomassie staining using Instant Blue™ staining solution (Expedeon, San Diego, CA, USA). The colicin containing fractions were pooled and dialyzed overnight at 4°C against 20 mM Na2HPO4 (pH 7.5), 10 mM citric acid and 50 mM NaCl. After dialysis, purified protein was frozen in liquid nitrogen and finally freeze-dried by lyophilization using freeze dryer Alpha 1–2 LDplus (Martin Christ Gefriertrocknungsanlagen, Osterode am Harz, Germany). Purified ColM batch 12 was used as a reference standard for antimicrobial activity and in immunoblot analysis.
Protein-containing plant extracts were mixed 1:1 with 2xLaemmli buffer and denatured at 95°C for 10 min. The total proteins were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using a 15% polyacrylamide gel under reducing conditions and then transferred to a PVDF membrane. The membrane was incubated with a rabbit polyclonal ColM antibody 30365 (diluted 1:5,000) or monoclonal ColM antibody 3F9 (diluted 1:2,000) overnight. Polyclonal ColM antibody was generated against purified full length ColM by BioGenes GmbH (Berlin, Germany). Monoclonal ColM antibody 3F9 was generated against purified full-length ColM by Fraunhofer Institute for Cell Therapy and Immunology (IZI, Halle (Saale), Germany). Secondary antibodies were IgG (whole molecule) peroxidase affinity isolated antibody (Sigma-Aldrich, Saint Louis, USA), anti-mouse (diluted 1:5,000) or anti-rabbit (diluted 1:10,000). Determination of ColM concentration in extracts was done semi-quantitatively by comparison of different amounts of extracts with known amounts of purified ColM batch 12 reference protein.
Microbiological studies were carried out on two laboratory strains of E. coli (DH10B and XL1-Blue) and a set of reference strains (Table 1). The bacteria were incubated at 37°C in LB medium (Bertani, 1951).
Semi-quantitative determinations of antimicrobial bacteriocin activity by a spot-on-lawn soft-agar overlay assay were performed as described in Schulz et al. (2015). Specific activity was calculated by normalization of arbitrary colicin activity units to TSP concentrations determined via Bradford assay to exclude effects of different extraction efficiencies.
Each respective bacterial test culture was grown in LB medium until OD600 of 0.2 and diluted to approximately 2x103 CFU/µl. 5 µl of diluted culture were spotted on LB medium supplemented with 0.75% (w/v) agar, 0.1 mg/ml BSA (AppliChem, Darmstadt, Germany) and a dilution series of purified ColM (10 µg/ml to 1 ng/ml). Plates were incubated for 16-20 h at 37°C before MIC readout. The MIC was defined as the lowest concentration of ColM that inhibited visible growth of the test culture.
We generated transgenic plants of lettuce (Lactuca sativa L.) ‘Odeskyj curling’ and mizuna (Brassica rapa subsp. nipposinica var. laciniata) using Agrobacterium-mediated transformation with a vector carrying the ColM-coding gene (cma). The vector pNMD46772 contains two expression cassettes: a cma gene under the control of a 35S promoter and the selective bar gene under the control of a nos promoter. Expression of the bar gene in transgenic plants provides for their resistance to the herbicide phosphinothricin (PPT). After five- or six-weeks incubation of lettuce cotyledons on the regeneration medium supplemented with the selective agent PPT (5 mg/L), green shoots were formed on the explants (Figure 1A). Regenerated plants developed roots after transfer to the medium supplemented with 0.5 mg/L of indole-3-butyric acid (Figure 1B). The resulting rooted lettuce plants were transferred to soil and grown in a greenhouse (Figure 1C). Mizuna plants were regenerated on the zeatin-containing media after 3-5 weeks of cultivation (Figure 1D). The selected plant lines resistant to 5 mg/L of PPT were cultivated under aseptic conditions (Figure 1E). Two lines were transferred to soil and grown in a greenhouse (Figures 1F, G). Greenhouse-maintained lettuce and mizuna plants bloomed and formed seeds in 3-4 months.
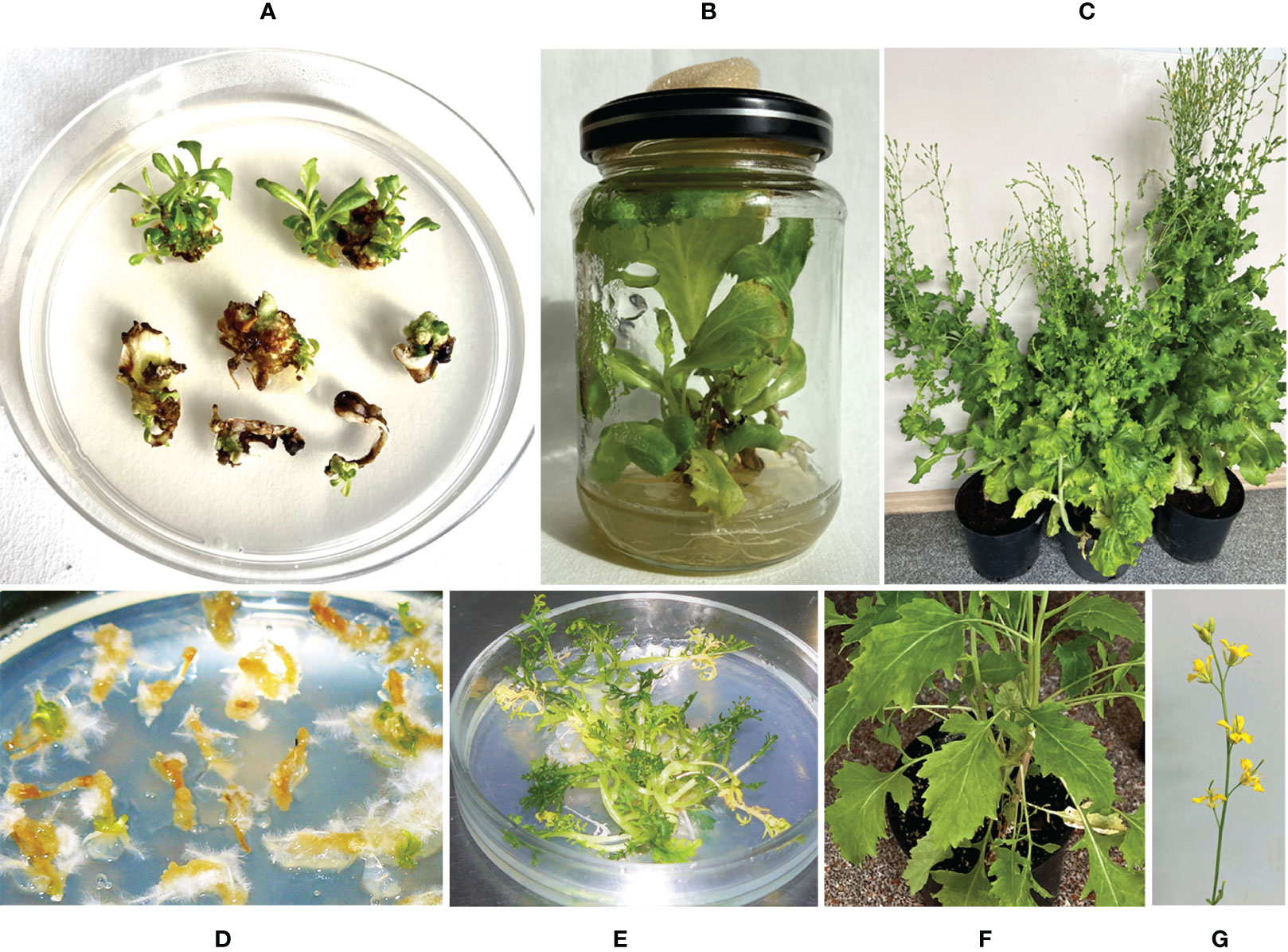
Figure 1 Generation of lettuce and mizuna transgenic plants expressing the gene of ColM. (A) In vitro lettuce plant regeneration from cotyledon explants after six weeks cultivation on selective medium; (B) rooting of regenerated lettuce shoots; (C) transgenic lettuce plants in a greenhouse; (D, E) in vitro regeneration of mizuna plants after four weeks (D) and 3 months (E) of cultivation on selective medium; (F, G) transgenic mizuna plants in a greenhouse.
All putative transgenic plants were initially screened for the integration of bar and cma genes using primers which should produce fragments of 411 bp and 1003 bp respectively. Amplification of the DNA of all plants revealed the expected fragments whose position is indicated by the arrows (Figures 2A, B). This result confirms the presence of cma and bar genes in the genome of the plants generated on the selective medium. A total of 5 independent transgenic lettuce plants and 2 mizuna plants were confirmed as PCR-positive for the transgenes integration. The purified RNA of transgenic plants along with wild type plants were used for cDNAs synthesis by reverse transcription. PCR product (1003 bp) obtained from the cDNAs by PCR with primers complementary to the cma gene confirmed ColM expression at the transcription level for transgenic plants of lettuce and mizuna (Figure 2C). A negative control for each sample consisted of the same reaction mixture without reverse transcriptase and no PCR products have been detected (lanes (1- to 6-), Figure 2C).
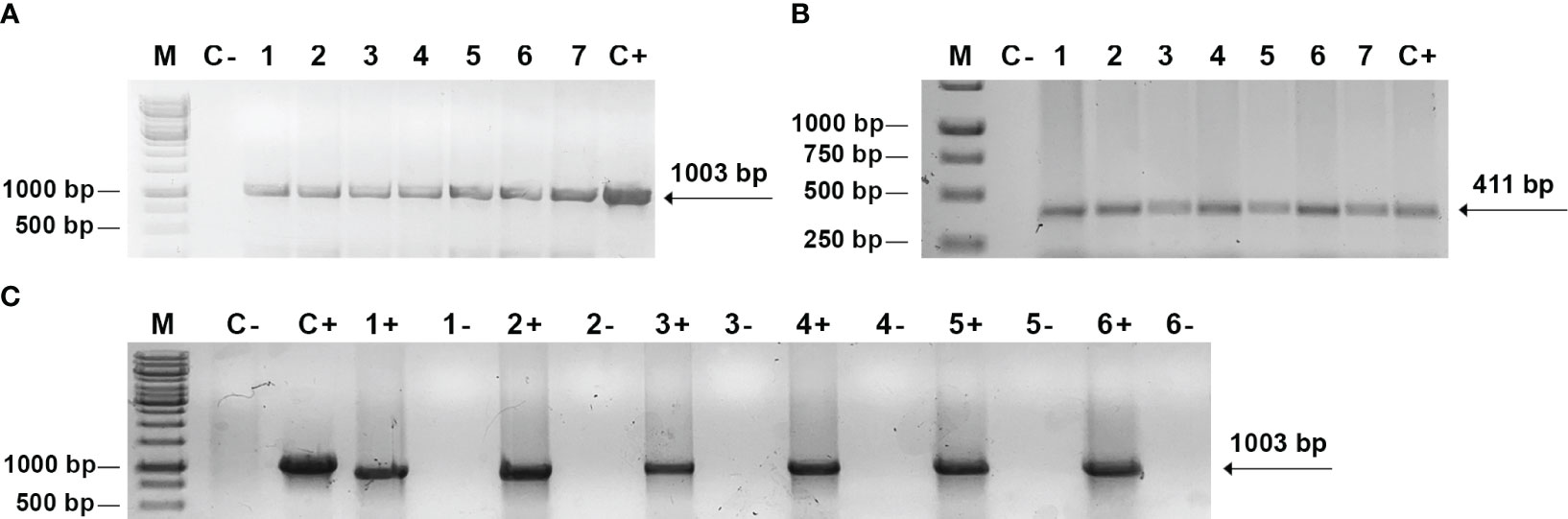
Figure 2 PCR and RT-PCR analysis of transgenic plants. (A, B) PCR analysis of genomic DNA. М - DNA mass ladder O`GeneRuler™ 1 kb DNA Ladder (Thermo Fischer Scientific); C- negative control with DNA from non-transformed plant; C+ - positive control, pNMD46772 plasmid DNA; 1-7 - independent transgenic lettuce (1-5) and mizuna (6-7) lines with primers to cma (A) and bar (B) genes; (C) RT-PCR analysis: C- negative control with RNA from non-transformed plant; C+ - positive control, pNMD46772 plasmid DNA; 1+ - transgenic mizuna RNA, 2+, 3+, 4+, 5+, 6+ - transgenic lettuce RNA; lanes 1-, 2-, 3-, 4-, 5-,6- - negative control for each corresponding sample, consisting of the same reaction mixture without reverse transcriptase.
In an initial screen for antibacterial activity, the transgenic plants were tested by placing explants on agar medium covered with diluted bacterial suspension. Clear bacterial growth inhibition zones were observed around the explants of transgenic lettuce plants grown in a greenhouse (Figure 3А) and two transgenic mizuna plant lines maintained in vitro (Figure 3B).
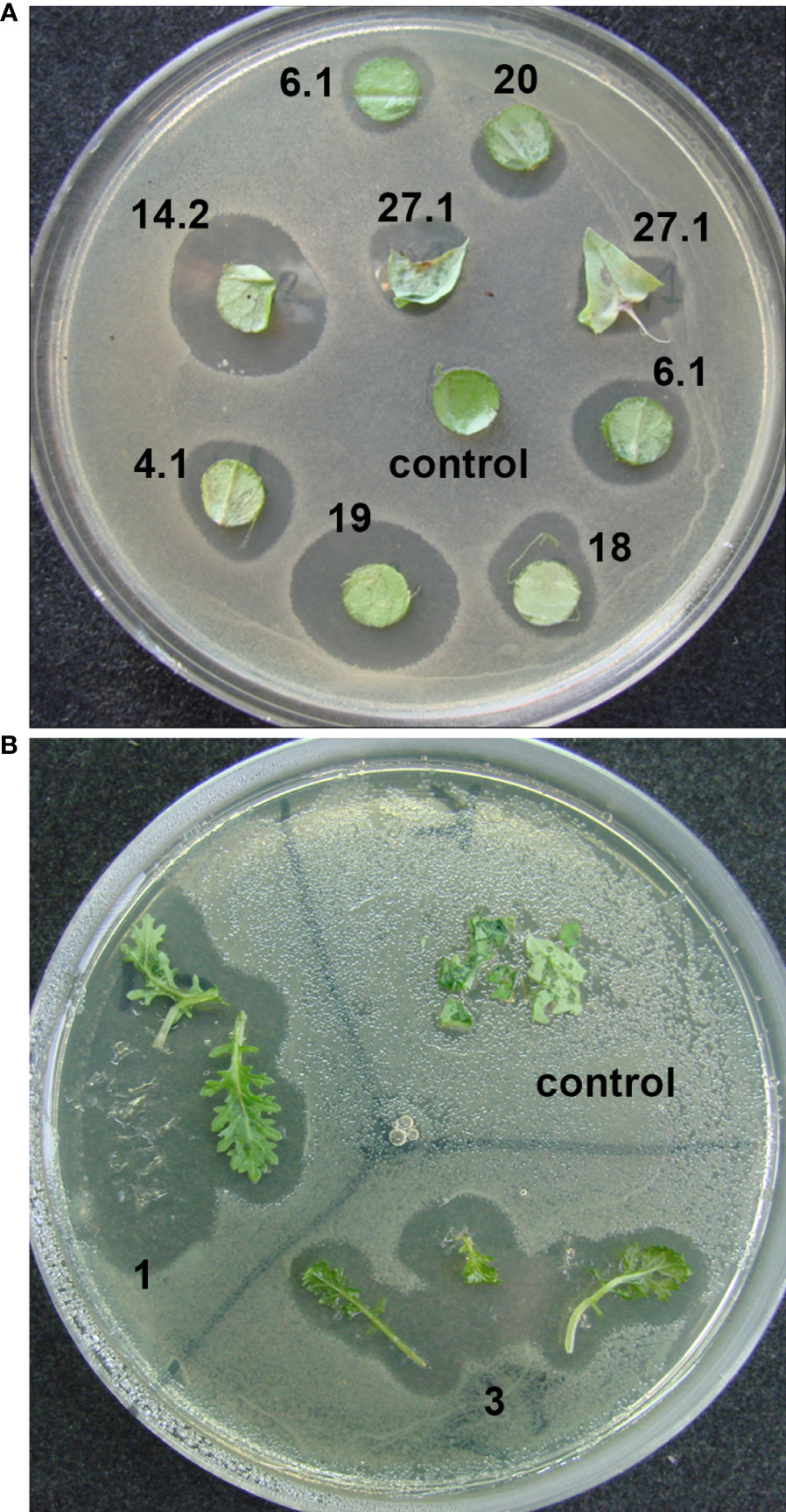
Figure 3 Determination of the antibacterial activity of ColM in transgenic lettuce and mizuna explants using a laboratory strain of E. coli XL1-Blue. (A) explants of different transgenic lines of lettuce; (B) leaves of two different transgenic lines of mizuna (BrT0#1, BrT0#3).
Antimicrobial activity was also evaluated semi-quantitatively using a soft-agar overlay assay with plant extracts based on the highest dilution of each that caused a clearing effect. The results of these experiments are presented in Table 2. The data showed very comparable levels of antibacterial activity in different transgenic lines and even between the two analyzed species. However, a decrease or complete absence of antibacterial activity in some plants of T1 generation was also noted. The antibacterial activity of two T1 generation plants of the transgenic lettuce line Ls#21 (Ls#21/T1#3 and Ls#21/T1#5) differed by almost 10 times (Table 2). This result was also confirmed by analysis of recombinant ColM expression detected using immunoblot: the lower antibacterial activity values correlated with less pronounced immunoblot signals (Figure 4A, these lines are marked in blue).
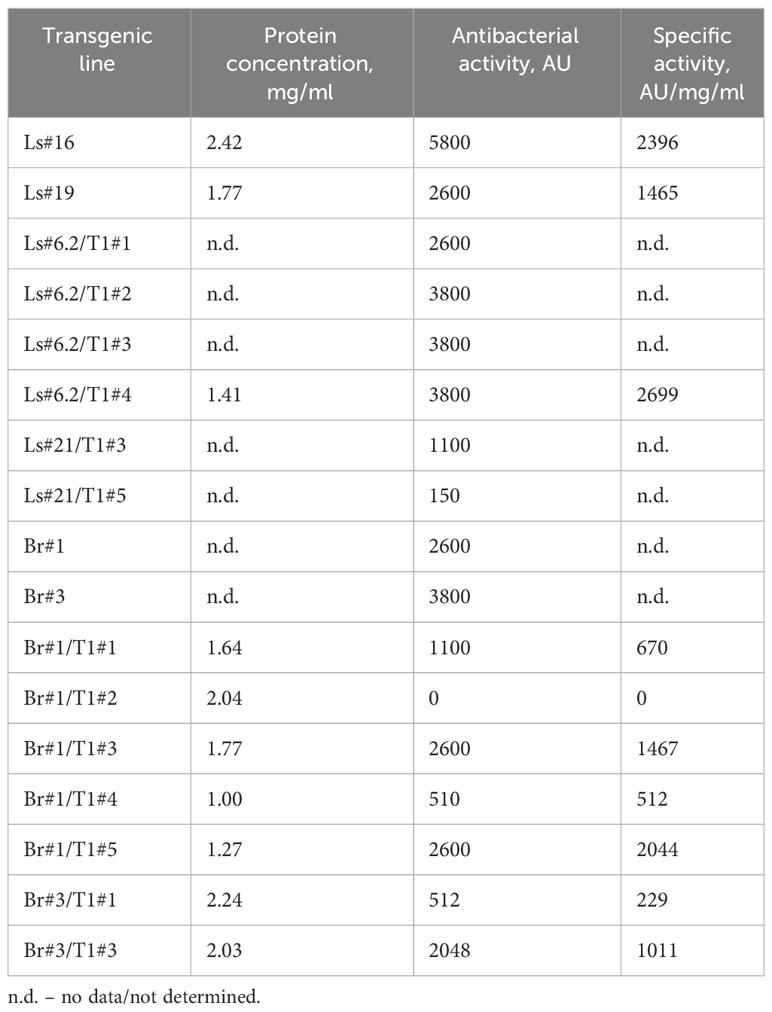
Table 2 Antibacterial activity of transgenic lettuce and mizuna plant extracts against E. coli strain DH10B.
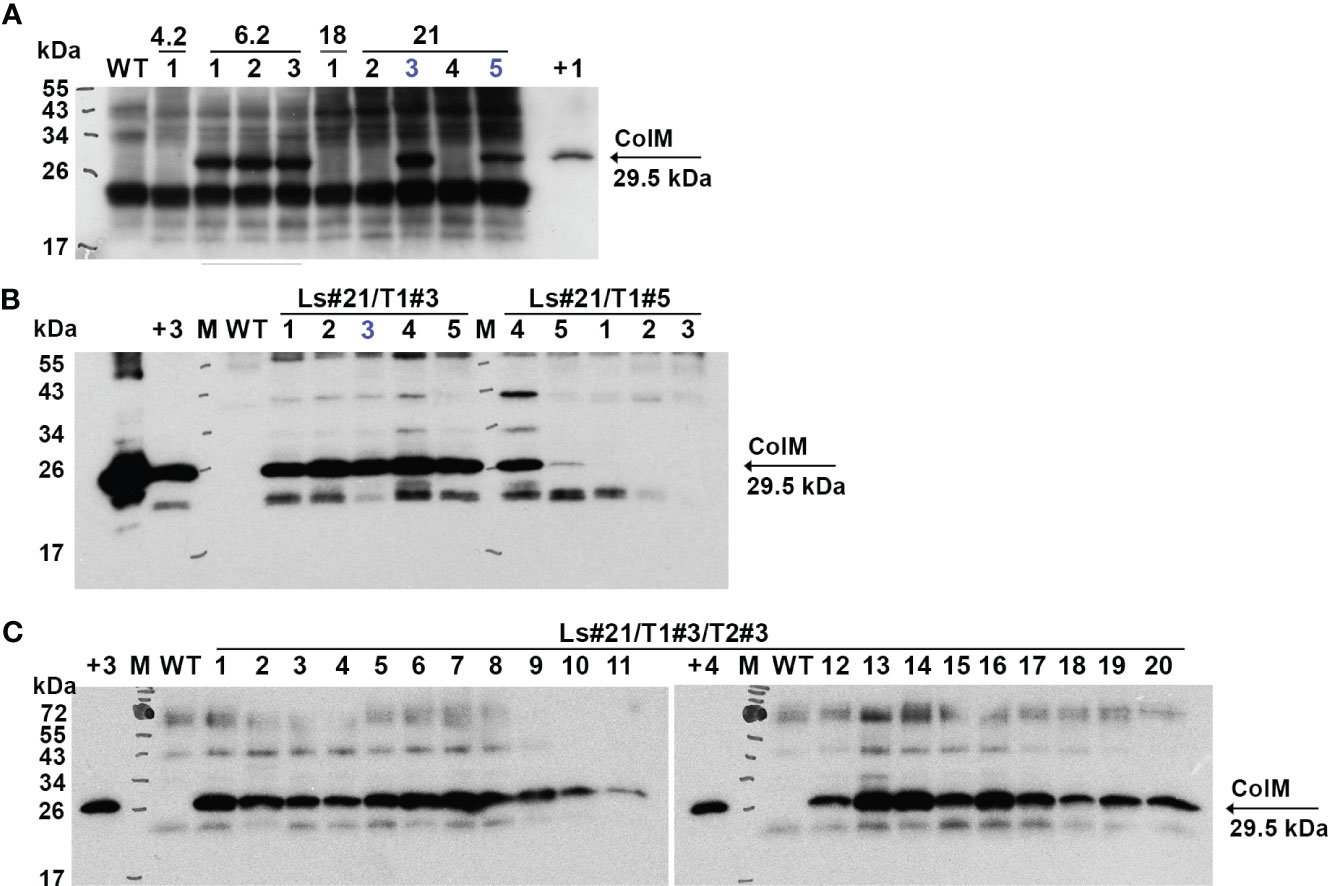
Figure 4 Expression of recombinant ColM detected by іmmunoblot in T1 (A), T2 (B) and T3 (C) generations of transgenic lettuce (Ls46722). Transgenic or wild-type (WT) leaf material was extracted with 5 volumes of buffer. 7.5 µl of TSP were resolved using SDS-PAGE (15% gel) and expression was analyzed by immunoblot with polyclonal ColM antibody. (А) M - molecular weight marker; WT - wild-type non-transgenic lettuce plant; track numbers (1-5) correspond to different lettuce plants of the T1 generation of transgenic lines 4.2; 6.2; 18; 21 (indicated in the top row); +1 - positive control: 80 ng N. benthamiana TSP extract after Agrobacterium-mediated transient expression of ColM; blue color indicates plants which progeny analysis shown in (B). (B) Track numbers correspond to different lettuce plants of the T2 generation of transgenic line 21; +2 - positive control: 1 µg purified ColM; +3 - positive control: TSP extract of Ls#6.2/2; blue color indicates plant which progeny analysis is shown in (C). (C) Track numbers (1-20) correspond to different lettuce plants of the T3 generation of transgenic line 21 (Ls#21/3/3); +4 - positive control: 38 ng purified ColM.
The same correlation between the antibacterial activity of the extracts, determined by soft-agar overlay assay, and the intensity of the immunoblot signals was observed when analyzing plants of the T2 generation of transgenic lettuce. Table 3 summarizes results of analysis of the transgenic trait associated with ColM gene expression in the T2 generation of lettuce plants.
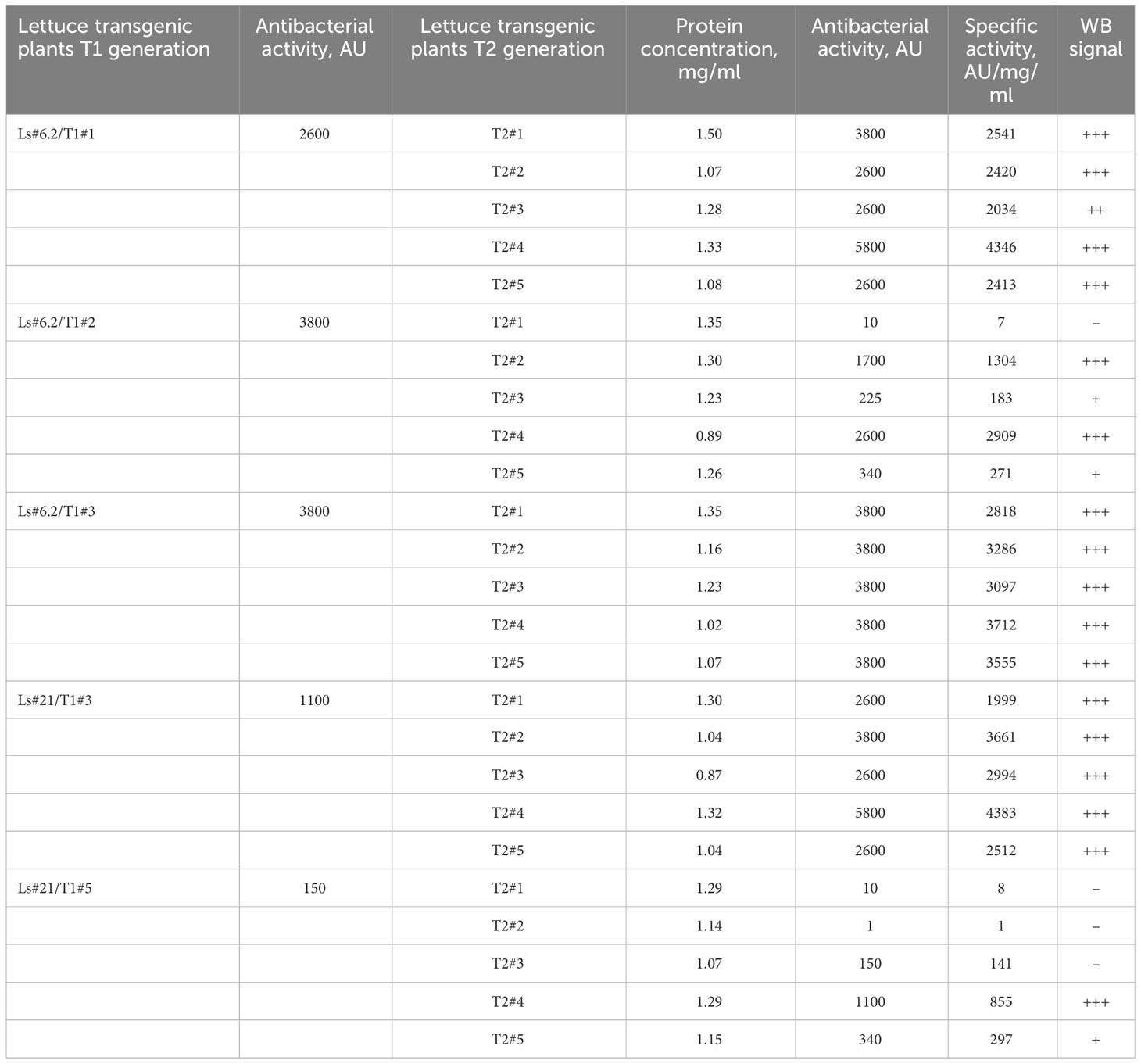
Table 3 The antibacterial activity and expression of the ColM detected by immunoblot in the T1 and T2 generations of transgenic lettuce.
These results show that lettuce transgenic line Ls#6.2 retained significant antibacterial activity in almost all plants of the next generations T1 and T2. Only three plants of T2 generation of Ls#6.2/T1#2 line showed very low or no antibacterial activity (Table 3). One of the plants of T1 generation of lettuce line Ls#21 (namely Ls#21/T1#3) had high antibacterial activity as did all of its T2 descendants (Table 3; Figure 4B). Another T1 plant of line Ls#21 (Ls#21/T1#5) had low antibacterial activity and so did most of its offspring, except line Ls#21/T1#5/T2#4 (Table 3; Figure 4B). This line showed high antibacterial activity despite its parental line showing low activity. This observation could be the result of silencing of the ColM gene in transgenic line Ls#21/T1#5 (T1 generation) and the reversion of silencing in one of the descendants. In addition, the offspring of Ls#21/T1#3/T2#3 was analyzed for ColM expression by immunoblot. All 20 analyzed plants of the T3 generation, which were selected on PPT-containing medium, showed ColM expression (Figure 4C).
For lettuce and mizuna transgenic plants, we selected lines that had high antibacterial activity and calculated the concentration of recombinant ColM in total soluble protein (TSP) extracts generated by addition of 5 volumes of extraction buffer. The content of ColM in transgenic plant extracts (mg/ml) and fresh weight (FW) of plant biomass (mg/g) was calculated via antibacterial activity (Table 4) and by semiquantitative immunoblot analysis (Figure 5).
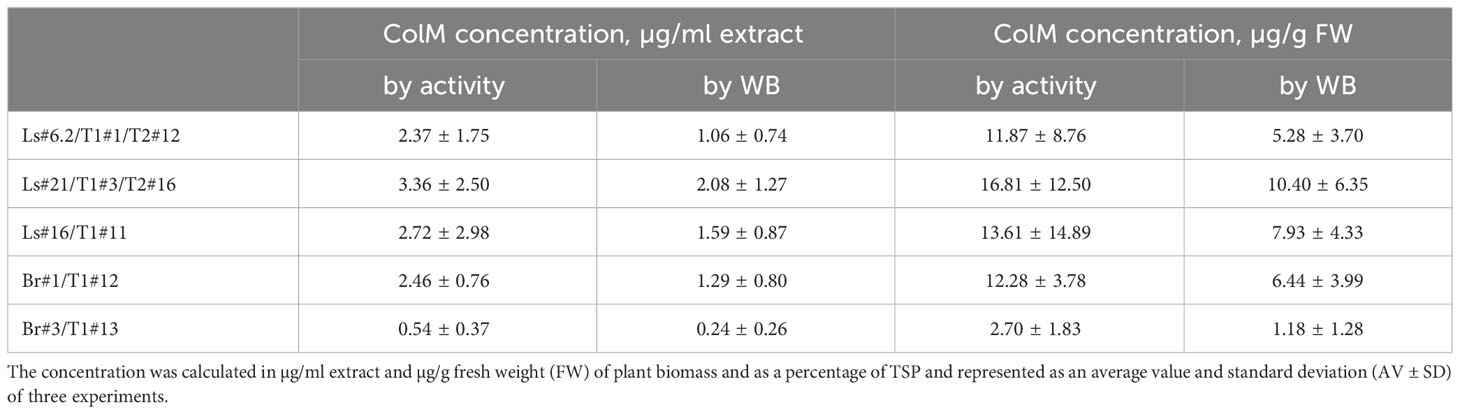
Table 4 Determination of ColM content in transgenic plant extracts by antibacterial activity and semiquantitative immunoblot analysis.
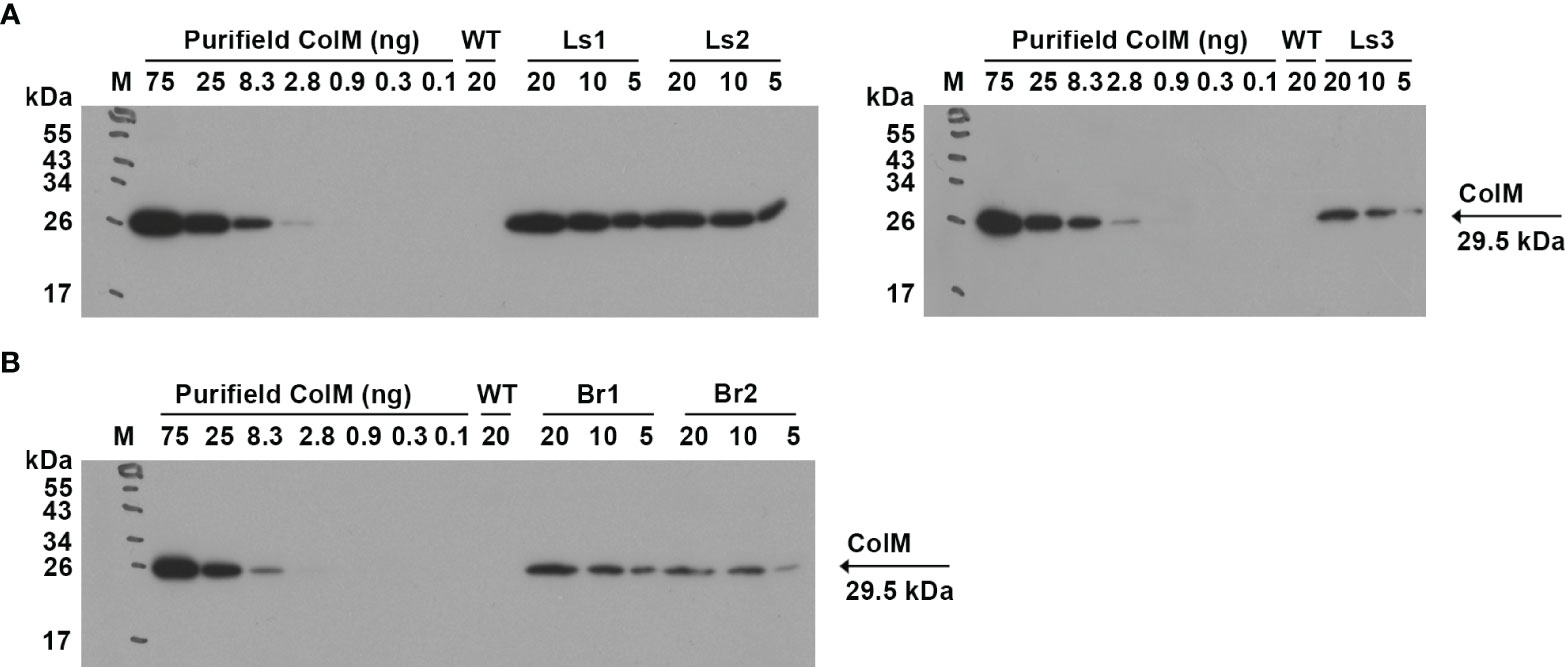
Figure 5 (A) Determination of ColM content in transgenic plant extracts by semiquantitative immunoblot analysis. Leaf material of ColM-expressing transgenic lettuce (A) and mizuna (B) plants was extracted with 5 volumes of buffer. The numbers above the tracks indicate the amount of purified ColM in ng or TSP extracts of transgenic plants in µl. Samples were resolved using SDS-PAGE (15% gel) and expression was analyzed by immunoblot with monoclonal ColM antibody. (A) M - molecular weight marker; WT - wild-type non-transgenic lettuce plant; Ls1 – transgenic lettuce plant of T2 generation Ls#6.2/T1#1/T2#12; Ls2 - transgenic lettuce plant of T2 generation Ls#21/T1#3/T2#16; Ls3 – transgenic lettuce plant of T1 generation Ls#16/T1#11. (B) M - molecular weight marker; WT - wild-type non-transgenic mizuna plant; Br1 – transgenic mizuna plant of T1 generation Br#1/T1#12; Br2 - transgenic mizuna plant of T1 generation Br#3/T1#13.
After finding high antibacterial activity in ColM-containing transgenic plant extracts against E. coli DH10B, we subsequently evaluated the activity of the extracts against a set of reference strains of E. coli, including two EHEC pathogenic serotypes (Figure 6). Аntibacterial activity against the emerging pathogenic serotype O104:H4 was higher than against the other EHEC serotype, O157:H7. In addition, ColM-containing extracts of edible plants, as well as purified ColM, also inhibited the growth of MDR E. coli strains. All three strains, including NCTC13352 that produces beta-lactamase and is therefore resistant to beta-lactam antibiotics, NCTC13919 that produces carbapenemases conferring resistance to carbapenems, and ATCC BAA-2452 that is resistant to both classes of these antibiotics, were sensitive to ColM-containing plant extracts.
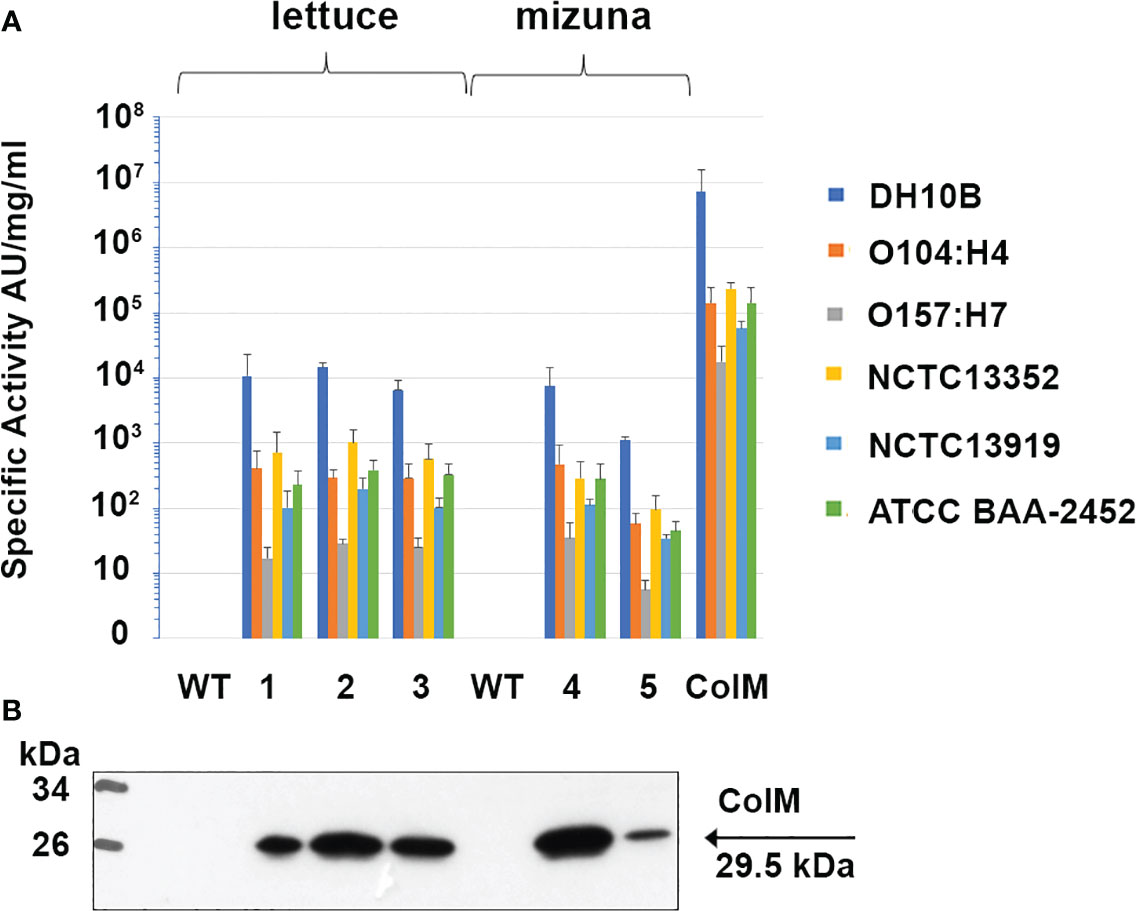
Figure 6 Antibacterial activity of ColM and ColM-containing transgenic plant extracts against E. coli strains O104:H4, O157:H7, O111:108, NCTC13352, NCTC13919 and ATCC BAA-2452. (A) Antimicrobial activity was determined semi-quantitatively on serial dilutions of TSP extracts by soft-agar overlay assay. Error bars indicate standard deviations of biological replicates (n = 3). AU- arbitrary colicin activity units. Transgenic lines: (1) Ls#6.2/T1#1/T2#12; (2) Ls#21/T1#3/T2#16; (3) Ls#16/T1#11; (4) Br#1/T1#12; (5) Br#3/T1#13). (B) Expression of recombinant ColM detected by іmmunoblot with monoclonal ColM antibody in the same transgenic lines. 7.5 µl of TSP extracts were resolved by SDS-PAGE (15% gel).
Plant-derived purified recombinant ColM was used for determined Minimal Inhibitory Concentrations (MIC) for analyzed E. coli strains. The results showed that ColM was active against most of the E. coli strains used in the study with MIC values ranging from 4.0 to 78.0 ng/ml (Table 5). Only for E. coli strain ATCC 35150 O157:H7 the MIC of recombinant ColM was significantly higher (5 µg/ml). The MIC value serves as the basis for assessing the sensitivity or resistance of the pathogen to an antimicrobial agent. The data presented in Table 4 show that the amount of ColM in transgenic lettuce plants extracts and mizuna line Br#1/T1#12 ranged from 1.06 to 3.36 µg/ml and these values are significantly higher than the MIC of purified recombinant ColM for the studied pathogenic bacterial strains (except for ATCC 35150 O157:H7). These results suggest the feasibility of using the obtained edible transgenic plants expressing ColM to help control foodborne disease.
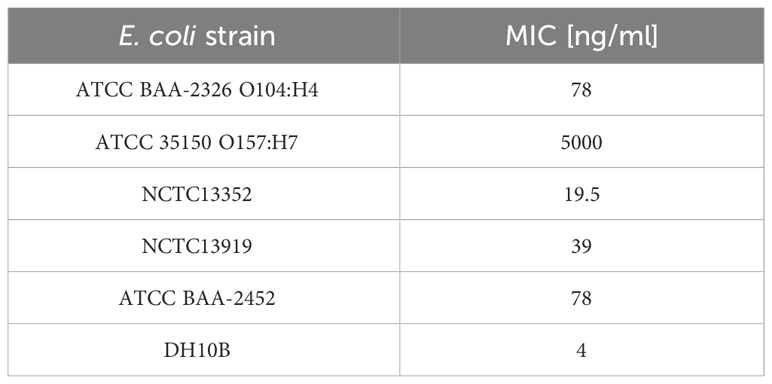
Table 5 Antibacterial activity of colicin M against E. coli strains expressed as a minimal inhibitory concentration (MIC).
For edible plants producing pharmaceuticals, the issue of long-term storage conditions without loss of recombinant protein activity is highly important. Therefore, in our studies we also evaluated the antibacterial activity of dried samples of transgenic lettuce and mizuna plants expressing ColM. In these studies, leaf material stored at -80°C for 9 months was subsequently dried at 40° C and antibacterial activity was evaluated after 1 week, 4 weeks and 3 months of storage at room temperature. The results are shown in Figure 7.
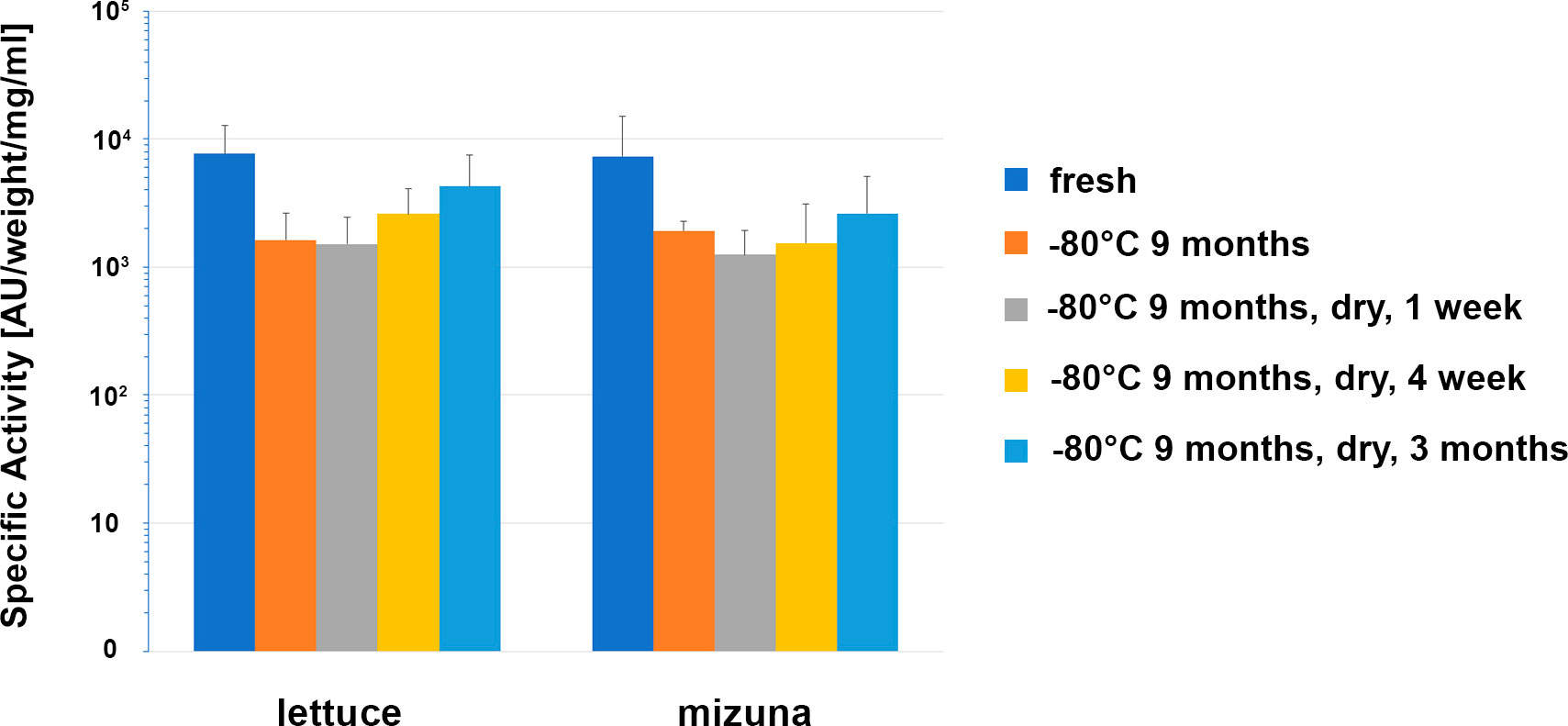
Figure 7 Comparison of the antibacterial activity of lettuce and mizuna samples under different storage conditions. Transgenic leaf material was extracted directly after harvest (fresh), after storage for 274 d at -80°C, and after storage for 262 d at -80°C followed by drying for 1 d at 40°C and subsequent storage for 1 week, 4 weeks or 3 months at room temperature. Activity of extracts was determined using soft-agar overlay assay against E. coli DH10B. In the graph, mean values with standard deviation of 6 different lettuce plants (Ls#6.2/T1#1/T2#11, Ls#6.2/T1#1/T2#12, Ls#21/T1#3/T2#15, Ls#21/T1#3/T2#16, Ls#16/T1#11, Ls#16/T1#12) or four different mizuna plants (Br#1/T1#11, Br#1/T1#12, Br#3/T1#11, Br#3T1#13) are shown. The concentration was calculated in μg/ml extract and μg/g fresh weight (FW) of plant biomass and as a percentage of TSP and represented as an average value and standard deviation (AV ± SD) of three experiments.
Except for an initial drop in ColM antibacterial activity after 9 months of storage at -80°C, no additional activity decrease was observed that could be correlated to either the drying procedure or the length of time of additional storage. These results underscore the advantages of edible transgenic plants over other methods of recombinant ColM production. The levels of specific antimicrobial activity observed in plant extracts (Figure 6), and the stability of ColM under various storage regimes (Figure 7), suggest a new and inexpensive method for producing recombinant pharmaceutical products that does not require costly protein purification steps and is suitable for transportation and long-term storage without loss of activity.
Plants have emerged as a reliable platform for production of recombinant proteins providing a safe and low-cost alternative to bacterial and mammalian cells systems. Transient expression in Nicotiana benthamiana has shown great potential for the manufacturing of recombinant biopharmaceuticals. Consequently, a wide range of medicinal proteins including recombinant vaccine antigens, monoclonal antibodies and other biotherapeutics have been synthesized in plants, some of which have advanced to clinical development (Daniell et al., 2009; Bendandi et al., 2010; Chichester et al., 2012; Gleba et al., 2014; Tusé et al., 2015; Ward et al., 2020; Leroux-Roels et al., 2022). The practice of using plants to produce valuable recombinant proteins has advanced significantly in recent decades (Ghequire and De Mot, 2017; Stander et al., 2022). With regard to antimicrobial proteins specifically, the cost of goods sold (COGS) for such proteins made by transient expression in plants at commercial scale was estimated to be comparable to those of products made by traditional food processing methods (McNulty et al., 2020). Despite the undoubted effectiveness of transient expression systems, the stable expression of antimicrobial proteins in transgenic edible plants makes it possible to reduce the costs of the most expensive part of production, namely, the extraction and purification of the recombinant protein (Łojewska et al., 2016). Such vegetables can be used as an antibacterial food or feed supplement without, or with minimal, downstream purification of the product.
The original idea of obtaining biopharmaceuticals in transgenic plants was implemented primarily for production of edible vaccines, and lettuce (Lactuca sativa L.) has often been used as a host plant for such studies (Matsui et al., 2009; Hamabata et al., 2019; Matsui et al., 2021). The main advantage of lettuce is that it is a fast-growing plant whose leaves are eaten raw and therefore can be used as an oral preparation. Lyophilized lettuce tissue has been used as a vehicle of oral vaccines against tuberculosis (Lakshmi et al., 2013), fasciolosis of cattle and sheep (Wesołowska et al., 2018) as well as to induce tolerance in hemophilia therapy (Su et al., 2015). The acceptance of lettuce plants for such studies is also explained by the fact that in vitro cultivation and genetic transformation techniques have been developed for lettuce, and it is also easy to grow indoors in the greenhouse, in hydroponic systems, and in the field. In addition, lettuce is a very popular crop, which is low in fat, calories and sodium, rich in fiber, iron, folate, and vitamin C and other health-beneficial bioactive compounds (Kim et al., 2016). Another plant in our study, mizuna, is also a fast-growing green leafy vegetable that is eaten raw, is high in phenolic compounds and flavonoids, especially hydroxycinnamic acids, and has high antioxidant activity (Khanam et al., 2012). Mizuna is a traditional and very popular crop in Asia and is increasingly attracting the attention of European and US consumers. Therefore, we developed mizuna plants expressing the ColM gene to impart antibacterial protective functions on a crop that is becoming well-accepted by consumers. In addition, and on a fundamental level, mizuna, in contrast to lettuce, has not been studied as a target for genetic transformation. To our knowledge, we are the first to report not only the successful genetic transformation of mizuna with the cma gene, but also the first genetic transformation of this plant species.
According to published data, the transient expression of ColM has been described in the standard host Nicotiana benthamiana, as well as in edible plant hosts such as spinach and leafy beets. Purified ColM obtained in plants by the transient expression method had the broadest antimicrobial activity against pathogenic strains of E. coli and, in mixtures, supplemented the effectiveness of other colicins (Schulz et al., 2015; Stephan et al., 2017). In addition, transgenic N. benthamiana (Hahn-Löbmann et al., 2019) and N. tabacum L. plants expressing ColM were developed (Łojewska et al., 2020) and the antibacterial activity of purified ColM obtained from these plants was confirmed against an array of pathogenic strains, including clinical isolates of E. coli and Klebsiella pneumonia. In the present study, we describe for the first time the antibacterial activity of edible transgenic plants stably expressing recombinant ColM. Our data demonstrate that ColM can be stably expressed in edible plants at a level that provides the desired antibacterial functionality. We also confirmed both the antibacterial activity of transgenic plant extracts and the inheritance of this trait in subsequent generations. Our studies once again confirm that ColM expressed in plants retains full functional activity against E. coli pathogenic serotype O104:H4 and O157:H7. Furthermore, extracts of edible plants containing ColM were found to inhibit the growth of multidrug-resistant strains of E. coli that produce beta-lactamases and carbapenemases. These results are of practical importance as carbapenems are considered to be antibiotics of last resort and antimicrobial options for the treatment of carbapenem-resistant Enterobacteriaceae (CRE) are limited (Sheu et al., 2019). Our results also show that ColM antimicrobial activity is retained for at least three months without loss of activity in heat-dried (up to 40°C) transgenic lettuce and mizuna biomass. According to published data, ColM is a fairly stable protein. It was demonstrated that pure bacteria-derived СolM remain stable for more than 1 year in solution at concentrations above 1 mg/ml (Schaller et al., 1981). Although the inactivation of ColM occurred when the solution was diluted, this process can be effectively slowed down or stopped by various additives to the medium. High stability was also demonstrated for plant-derived purified recombinant СolM. Recombinant plant-derived СolM remained stable in lyophilized form and in solution stored at 4°C. The solution stored at room temperature was less stable, but ColM was not inactivated for 8 weeks and showed the best stability of all 4 colicins tested (Hahn-Löbmann et al., 2019). However, proteolytic degradation of this bacteriocin is possible in the plant cell. We believe that dried plant biomass retained antimicrobial activity of ColM mostly because of drying procedure which has been applied. Slow drying of entire leaves prevented the decompartmentalization in plant cells thus avoiding massive attack of plant proteases against ColM which could happen, e.g., in case of grinding. In agriculture, dry plant biomass (hay) is known for a good stability of nutritional crude protein. Using a drying procedure for preservation and storage of ColM-containing leaf material was partially inspired by that knowledge. Indeed, this approach appeared to be productive and provided both stable and functional bacteriocine. To our knowledge, this is the first study that demonstrates the antibacterial activity of extracts of edible transgenic plants expressing ColM against various E. coli strains, including pathogenic and MDR pathotypes. The use of edible plants expressing ColM as a food or feed additive for preventive purposes is especially poignant due to the magnitude and recurrence of foodborne E. coli outbreaks worldwide, the role of leafy vegetables in their transmission, and the continued threat posed by MDR bacteria due to the overuse of antibiotics in agriculture.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
NS: Investigation, Writing – original draft. HP: Data curation, Investigation, Writing – original draft. KL: Investigation, Writing – review & editing. YP: Investigation, Writing – review & editing. AG: Conceptualization, Writing – review & editing. MK: Conceptualization, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by National Academy of Sciences of Ukraine: the project 0123U101081 “Synthesis of recombinant pharmaceutical proteins and increasing of content of biologically active natural compounds in plants” and by the National Research Foundation of Ukraine: the project 2020.01/0301 “Plant-based synthesis of recombinant pharmaceutical proteins capable to prevent some infectious diseases of bacterial and viral origin”.
The authors are grateful to Prof. Dr. Yuri Gleba for the support, inspiration and understanding that have made this paper possible. We thank Doreen Bartels for excellent technical assistance and Dr. Anett Stephan and Dr. Simone Hahn-Löbmann (all Nomad Bioscience GmbH, Halle (Saale), Germany) for fruitful discussions. We thank Dr. Daniel Tusé (DT/Consulting Group, Sacramento, CA, USA) for critical reading of manuscript and valuable comments.
Author HP and AG were employed by the company Nomad Bioscience GmbH.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alegbeleye, O. O., Singleton, I., Sant'Ana, A. S. (2018). Sources and contamination routes of microbial pathogens to fresh produce during field cultivation: A review. Food Microbiol. 73, 177–208. doi: 10.1016/j.fm.2018.01.003
Bendandi, M., Marillonnet, S., Kandzia, R., Thieme, F., Nickstadt, A., Herz, S., et al. (2010). Rapid, high-yield production in plants of individualized idiotype vaccines for non-Hodgkin’s lymphoma. Ann. Oncol. 21 (12), 2420–2427. doi: 10.1093/annonc/mdq256
Benitez-Chao, D. F., Leуn-Buitimea, A., Lerma-Escalera, J. A., Morones-Ramirez, J. R. (2021). Bacteriocins: an overview of antimicrobial, toxicity, and biosafety assessment by in vivo models. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.630695
Bertani, L. (1951). Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62, 293–300. doi: 10.1128/jb.62.3.293-300.1951
Callaway, T. R., Stahl, C. H., Edrington, T. S., Genovese, K. J., Lincoln, L. M., Anderson, R. C., et al. (2004). Colicin concentrations inhibit growth of O157:H7 in vitro. J Food Prot 67, 2603–2607. doi: 10.4315/0362-028X-67.11.2603
Cavera, V. L., Arthur, T. D., Kashtanov, D., Chikindas, M. L. (2015). Bacteriocins and their position in the next wave of conventional antibiotics. Int. J. Antimicrob. Agents. 46, 494–501. doi: 10.1016/j.ijantimicag.2015.07.011
Chérier, D., Patin, D., Blanot, D., Touzé, T., Barreteau., H. (2021). The biology of colicin M and its orthologs. Antibiotics 10, 1109. doi: 10.3390/antibiotics.10091109
Chichester, J. A., Jones, R. M., Green, B. J., Stow, M., Miao, F., Moonsammy, G., et al. (2012). Safety and immunogenicity of a plant-produced recombinant hemagglutinin-based influenza vaccine (HAI-05) derived from A/Indonesia/05/2005 (H5N1) influenza virus: A phase 1 randomized, double-blind, placebo-controlled, dose-escalation study in healthy adults. Viruses 4 (11), 3227–3244. doi: 10.3390/v4113227
Cleveland, J., Montville, T. J., Nes, I. F., Chikindas, M. L. (2001). Bacteriocins: safe, natural antimicrobials for food preservation. Int. J. Food Microbiol. 71, 1–20. doi: 10.1016/s0168-1605(01)00560-8
Daniell, H., Singh, N. D., Mason, H., Streatfield, S. J. (2009). Plant-made vaccine antigens and biopharmaceuticals. Trends Plant Sci. 14, 669–679. doi: 10.1016/j.tplants.2009.09.009
Food and Drug Administration. (2021). FDA Releases Annual Summary Report on Antimicrobials Sold or Distributed in 2020 for Use in Food-Producing Animals. Available at: https://www.fda.gov/animal-veterinary/cvm-updates/fda-releases-annual-summary-report-antimicrobials-sold-or-distributed-2020-use-food-producing.
Gabrielsen, C., Brede, D. A., Nes, I. F., Diep, D. B. (2014). Circular bacteriocins: biosynthesis and mode of action. Appl. Environ. Microbiol. 80, 6854–6862. doi: 10.1128/AEM.02284-14
Gamborg, O. L., Miller, R. A., Ojima, K. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50, 151–158. doi: 10.1016/0014-4827(68)90403-5
Ghequire, M. G. K., De Mot, R. (2017). Turning over a new leaf: bacteriocins going green. Trends Microbiol. 26 (1), 1–2. doi: 10.1016/j.tim.2017.11.001
Gleba, Y. Y., Tusé, D., Giritch, A. (2014). Plant viral vectors for delivery by Agrobacterium. Curr. Top. Microbiol. Immunol. 375, 155–192. doi: 10.1007/82_2013_352
GRAS Notice (GRN). Available at: https://fda.report/media/93655/GRAS-Notice-000593 (Accessed 29 July 2021).
Hahn-Löbmann, S., Stephan, A., Schulz, S., Schneider, T., Shaverskyi, A., Tusé, D., et al. (2019). Colicins and salmocins – new classes of plant-made non-antibiotic food antibacterials. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00437
Hamabata, T., Sato, T., Takita, E., Matsui, T., Imaoka, T., Nakanishi, N., et al. (2019). Shiga toxin 2eB-transgenic lettuce vaccine is effective in protecting weaned piglets from edema disease caused by Shiga toxin-producing Escherichia coli infection. Anim. Sci. J. 90 (11), 1460–1467. doi: 10.1111/asj.13292
Irvin, K., Viazis, S., Fields, A., Seelman, S., Blickenstaff, K., Gee, E., et al. (2021). An overview of traceback investigations and three case studies of recent outbreaks of escherichia coli O157:H7 infections linked to romaine lettuce. J. Food Prot. 84, 1340–1356. doi: 10.4315/JFP-21-112
Khanam, U. K. S., Oba, S., Yanase, E., Murakami, Y. (2012). Phenolic acids, flavonoids and total antioxidant capacity of selected leafy vegetables. J. Funct. Foods 4 (4), 979–987. doi: 10.1016/j.jff.2012.07.006
Kim, M., Moon, Y., Tou, J., Mou, B., Waterland, N. (2016). Nutritional value, bioactive compounds and health benefits of lettuce (Lactuca sativa L.). J. Food Composition Anal. 49, 19–34. doi: 10.1016/j.jfca.2016.03.004
Kumariya, R., Garsa, A. K., Rajput, Y. S., Sood, S. K., Akhtar, N., Patel, S. (2019). Bacteriocins: Classification, synthesis, mechanism of action and resistance development in food spoilage causing bacteria. Microb. Pathog. 128, 171–177. doi: 10.1016/j.micpath.2019.01.002
Lakshmi, P. S., Verma, D., Yang, X., Lloyd, B., Daniell, H. (2013). Low cost tuberculosis vaccine antigens in capsules: expression in chloroplasts, bio-encapsulation, stability and functional evaluation in vitro. PloS One 8, e54708. doi: 10.1371/journal.pone.0054708
Leroux-Roels, I., Maes, C., Joye, J., Jacobs, B., Jarczowski, F., Diessner, A., et al. (2022). A randomized, double-blind, placebo-controlled, dose-escalating phase I trial to evaluate safety and immunogenicity of a plant-produced, bivalent, recombinant norovirus-like particle vaccine. Front. Immunol. 13. doi: 10.3389/fimmu.2022.1021500
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Łojewska, E., Kowalczyk, T., Olejniczak, S., Sakowicz, T. (2016). Extraction and purifcation methods in downstream processing of plant-based recombinant proteins. Protein Expr. Purif. 120, 110–117. doi: 10.1016/j.pep.2015.12.018
Łojewska, E., Sakowicz, T., Kowalczyk, A., Konieczka, M., Grzegorczyk, J., Sitarek, P., et al. (2020). Production of recombinant colicin M in Nicotiana tabacum plants and its antimicrobial activity. Plant Biotechnol. Rep. 14, 33–43. doi: 10.1007/s11816-019-00571-y
Low, J. C., McKendrick, I. J., McKechnie, C., Fenlon, D., Naylor, S. W., Currie, C., et al. (2005). Rectal carriage of enterohemorrhagic Escherichia coli O157 in slaughtered cattle. Appl. Environ. Microbiol. 71, 93–97. doi: 10.1128/AEM.71.1.93-97.2005
Matsui, T., Asao, H., Ki, M., Sawada, K., Kato, K. (2009). Transgenic lettuce producing a candidate protein for vaccine against edema disease. Biosci. Biotechnol. Biochem. 73 (7), 1628–1634. doi: 10.1271/bbb.90129
Matsui, T., Takita, E., Oiwa, S., Yokoyama, A., Kato, K., Sawada, K. (2021). Lettuce-based production of an oral vaccine against porcine edema disease for the seed lot system. Plant Biotechnol. (Tokyo) 38 (2), 239–246. doi: 10.5511/plantbiotechnology.21.0414
McNulty, M. J., Gleba, Y., Tusé, D., Hahn-Löbmann, S., Giritch, A., Nandi, S., et al. (2020). Techno-economic analysis of a plant-based platform for manufacturing antimicrobial proteins for food safety. Biotechnol. Prog. 36 (1), e2896. doi: 10.1002/btpr.2896
Munns, K. D., Selinger, L. B., Stanford, K., Guan, L., Callaway, T. R., McAllister, T. A. (2015). Perspectives on super-shedding of Escherichia coli O157:H7 by cattle. Foodborne Pathog. Dis. 12, 89–103. doi: 10.1089/fpd.2014.1829
Murashige, T., Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052
Navarro-Garcia, F. (2014). Escherichia coli O104:H4 Pathogenesis: an Enteroaggregative E. coli/Shiga Toxin-Producing E. coli Explosive Cocktail of High Virulence. Microbiol. Spectr. 2, 6. doi: 10.1128/microbiolspec.EHEC-0008-2013
Paveenkittiporn, W., Kerdsin, A., Chokngam, S., Bunthi, C., Sangkitporn, S., Gregory, C. J. (2017). Emergence of plasmid-mediated colistin resistance and New Delhi metallo-β-lactamase genes in extensively drug-resistant Escherichia coli isolated from a patient in Thailand. Diagn. Microbiol. Infect. Dis. 87, 157–159. doi: 10.1016/j.diagmicrobio.2016.11.005
Puligundla, P., Lim, S. (2022). Biocontrol Approaches against Escherichia coli O157:H7 in Foods. Foods 11, 5. doi: 10.3390/foods11050756
Schaller, K., Dreher, R., Braun, V. (1981). Structural and functional properties of colicin M. J. Bacteriol. 146 (1), 54–63. doi: 10.1128/jb.146.1.54-63.1981
Schulz, S., Stephan, A., Hahn, S., Bortesi, L., Jarczowski, F., Bettmann, U., et al. (2015). Broad and efficient control of major foodborne pathogenic strains of Escherichia coli by mixtures of plant-produced colicins. Proc. Natl. Acad. Sci. U.S.A. 112, E5454–E5460. doi: 10.1073/pnas.1513311112
Shcherbak, N., Kishchenko, O., Sakhno, L., Komarnytsky, I., Kuchuk, M. (2013). Lox-dependent gene expression in transgenic plants obtained via Agrobacterium-mediated transformation. Tsitol. Genet. 47, 21–32. doi: 10.3103/S0095452713030079
Sheu, C. C., Chang, Y. T., Lin, S. Y., Chen, Y. H., Hsueh, P. R. (2019). Infections caused by carbapenem-resistant Enterobacteriaceae: An update on therapeutic options. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00080
Slayton, R. B., Turabelidze, G., Bennett, S. D., Schwensohn, C. A., Yaffee, A. Q., Khan, F., et al. (2013). Outbreak of Shiga toxin-producing Escherichia coli (STEC) O157:H7 associated with romaine lettuce consumption. PloS One 8, 2. doi: 10.1371/journal.pone.0055300
Stander, J., Mbewana, S., Meyers, A. E. (2022). Plant-derived human vaccines: recent developments. BioDrugs 36 (5), 573–589. doi: 10.1007/s40259-022-00544-8
Stephan, A., Hahn-Löbmann, S., Rosche, F., Buchholz, M., Giritch, A., Gleba, Y. (2017). Simple purification of Nicotiana benthamiana-Produced recombinant colicins: high-yield recovery of purified proteins with minimum alkaloid content supports the suitability of the host for manufacturing food additives. Int. J. Mol. Sci. 19, 95. doi: 10.3390/ijms19010095
Su, J., Zhu, L., Sherman, A., Wang, X., Lin, S., Kamesh, A., et al. (2015). Low cost industrial production of coagulation factor IX bioencapsulated in lettuce cells for oral tolerance induction in hemophilia B. Biomaterials 70, 84–93. doi: 10.1016/j.biomaterials
Tietze, E., Dabrowski, P. W., Prager, R., Radonic, A., Fruth, A., Auraß, P., et al. (2015). Comparative genomic analysis of two novel sporadic Shiga toxin-producing Escherichia coli O104:H4 strains isolated 2011 in Germany. PloS One 10, 4. doi: 10.1371/journal.pone.0122074
Tomás-Callejas, A., Martínez-Hernández, G. B., Artés, F., Artés-Hernández, F. (2011). Neutral and acidic electrolyzed water as emergent sanitizers for fresh-cut mizuna baby leaves. Postharvest Biol. Technol. 59, 298–306. doi: 10.1016/j.postharvbio.2010.09.013
Tusé, D., Ku, N., Bendandi, M., Becerra, C., Collins, R., Jr., Langford, N., et al. (2015). Clinical safety and immunogenicity of tumor-targeted, plant-made id-KLH conjugate vaccines for follicular lymphoma. Biomed. Res. Int. 2015, 648143. doi: 10.1155/2015/648143
Urban-Chmiel, R., Marek, A., Stepien-Pysniak, D., Wieczorek, K., Dec, M., Nowaczek, A., et al. (2022). Antibiotic resistance in bacteria - A review. Antibiotics 11, 1079. doi: 10.3390/antibiotics11081079
US Center for Disease Control and Prevention. (2020). Reports of selected E. coli outbreak investigations. Available at: https://www.cdc.gov/ecoli/outbreaks.html.
Waltenburg, M. A., Schwensohn, C., Madad, A., Seelman, S. L., Peralta, V., Koskeet, S. E., et al. (2021). Two multistate outbreaks of a reoccurring Shiga toxin-producing Escherichia coli strain associated with romaine lettuce: USA 2018-2019. Epidemiol. Infect. 150, 16. doi: 10.1017/S0950268821002703
Ward, B. J., Makarkov, A., Séguin, A., Pillet, S., Trépanier, S., Dhaliwall, J., et al. (2020). Efficacy, immunogenicity, and safety of a plant-derived, quadrivalent, virus-like particle influenza vaccine in adults (18–64 years) and older adults (≥65 years): two multicentre, randomised phase 3 trials. Lancet 396, 1491–1503. doi: 10.1016/S0140-6736(20)32014-6
Wesołowska, A., Ljunggren, M. K., Jedlina, L., Basałaj, K., Legocki, A., Wedrychowicz, H., et al. (2018). A preliminary study of a lettuce-based edible vaccine expressing the cysteine proteinase of Fasciola hepatica for fasciolosis control in livestock. Front. Immunol. 9. doi: 10.3389/fimmu.2018.02592
Willyard, C. (2017). The drug-resistant bacteria that pose the greatest health threats. Nature 543, 15. doi: 10.1038/nature.2017.21550
Keywords: bacteriocins, colicin M, transgenic lettuce, transgenic mizuna, pathogenic Escherichia coli, STEC, EHEC, multi-drug resistance
Citation: Shcherbak N, Prochaska H, Lystvan K, Prokhorova Y, Giritch A and Kuchuk M (2023) Accumulation of colicin M protein and its biological activity in transgenic lettuce and mizuna plants. Front. Plant Sci. 14:1271757. doi: 10.3389/fpls.2023.1271757
Received: 02 August 2023; Accepted: 02 October 2023;
Published: 23 October 2023.
Edited by:
Balamurugan Shanmugaraj, Chulalongkorn University, ThailandReviewed by:
Rahul Singh, University of Pennsylvania, United StatesCopyright © 2023 Shcherbak, Prochaska, Lystvan, Prokhorova, Giritch and Kuchuk. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nataliia Shcherbak, bmF0YWxpYUBpY2JnZS5vcmcudWE=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.