- Coastal Salinity Tolerant Grass Engineering and Technology Research Center, Ludong University, Yantai, Shandong, China
The YUCCAs (YUC) are functionally identified flavin-containing monooxidases (FMOs) in plants that act as an important rate-limiting enzyme functioning in the auxin synthesis IPA (indole-3-pyruvic acid) pathway. In this study, 12 MsYUCs and 15 MtYUCs containing characteristic conserved motifs were identified in M. sativa (Medicago sativa L.) and M. truncatula (Medicago truncatula Gaertn.), respectively. Phylogenetic analysis revealed that YUC proteins underwent an evolutionary divergence. Both tandem and segmental duplication events were presented in MsYUC and MtYUC genes. Comparative syntenic maps of M. sativa with M. truncatula, Arabidopsis (Arabidopsis thaliana), or rice (Oryza sativa L.) were constructed to illustrate the evolution relationship of the YUC gene family. A large number of cis-acting elements related to stress response and hormone regulation were revealed in the promoter sequences of MsYUCs. Expression analysis showed that MsYUCs had a tissue-specific, genotype-differential expression and a differential abiotic stress response pattern based on transcriptome data analysis of M. sativa online. In addition, RT-qPCR confirmed that salt stress significantly induced the expression of MsYUC1/MsYUC10 but significantly inhibited MsYUC2/MsYUC3 expression and the expression of MsYUC10/MsYUC11/MsYUC12 was significantly induced by cold treatment. These results could provide valuable information for functional analysis of YUC genes via gene engineering of the auxin synthetic IPA pathway in Medicago.
1 Introduction
Auxin is a critical plant hormone, involved in diverse developmental events such as cell division, cell differentiation, and flower development. Indole-3-acetic acid (IAA) is the best-studied naturally occurring active auxin, which are synthesized by two pathways: tryptophan-dependent pathway and tryptophan-independent pathway (Zhao, 2010). For tryptophan-dependent IAA synthesis, there are four proposed branches: (1) indole-3-pyruvic acid (IPA); (2) tyramine pathway; (3) index-3-acetamide pathway; and (4) index-3-acetoxime pathway (Stepanova et al., 2011). Among the four branches, the IPA branch is the major route of IAA biosynthesis inferred by the pleiotropic abnormal phenotype of Arabidopsis mutants (Mashiguchi et al., 2011; Won et al., 2011). In the initial step, IPA is catalyzed by Trp aminotransferase 1 (TAA1) and its related proteins TAR1 and TAR2 with Trp as the precursor. Subsequently, the YUCCA (YUC)-encoded enzyme catalyzes the generation of IAA by IPA (Fraaije et al., 2002; Won et al., 2011). The YUC enzyme is the first functionally identified flavin-containing monooxidase (FMOs) in plants. The conserved domain of FMOs contains two conserved motifs, the flavin purine dinucleotide (FAD) binding site and the reduced coenzyme binding site (NADPH-binding site), which have the same GXGxxG characteristic structure in their amino acid sequences (Zhao, 2012).
The YUC gene was originally identified from Arabidopsis mutants with reduced IAA content (Zhao et al., 2001). Genetic and physiological analyses of the loss-of-function mutants of the YUC gene have further demonstrated its important role and rate-limiting enzyme function in the auxin synthesis IPA pathway. Overexpression of transgenic Arabidopsis lines of the YUC gene showed slightly increased auxin levels, accompanied by phenotypic including hypocotyl elongation, cotyledon bias, and enhanced apical dominance (Zhao et al., 2001). Subsequent studies showed that overexpression of the YUC gene in plants such as rice, potato, and strawberry could also produce similar phenotypes of auxin overproduction (Kim et al., 2012; Liu et al., 2014). In addition, inactivation of a single YUC gene in Arabidopsis presented not obvious developmental defects, whereas multiple mutants plants have more severe phenotypes (Cheng et al., 2006), suggesting functional redundancy among YUC members. Moreover, gene and protein expression data in Arabidopsis indicated that YUC1, 2, 4, and 6 were mainly expressed in the stems, whereas YUC 3, 5, 7, 8, and 9 were mainly functional in the roots (Chen et al., 2014). The yuc1yuc2yuc4yuc6 quadruple mutants had severe defects in vascular patterning and failed to produce a normal inflorescence but had no root defects, consistent with their stem-localized expression pattern. YUC3, 5, 7, 8, and 9 are expressed during root development, and the multiple mutants of the five YUC genes developed short and agrotropic roots (Chen et al., 2014). In addition, YUC genes expressed in the shoots (YUC 1, 2, 4.1, and 6) are localized to the cytoplasm, whereas root YUC genes are the ER (endoplasmic reticulum) membrane-binding proteins. In addition, the phenotypes of different sets of individual YUC knockout mutants cannot be complemented by the expression of YUC genes expressed in other tissues (Chen et al., 2014; Zhao, 2018). These studies suggested that different sets of YUC genes exhibited tissue expression specificity, organ-specific subcellular localization patterns, and differential of gene function for auxin biosynthesis.
Plants often respond to environmental stress by regulating hormonal pathways. Several studies have shown that the auxin biosynthetic pathway is upregulated in response to certain abiotic stresses including regulating the expression of YUC genes (Blakeslee et al., 2019). For example, several root-specific YUC genes have been reported to mediate aluminum stress-induced inhibition of root growth in Arabidopsis (Liu et al., 2016). Heat and low-temperature stress can induce ER sheet formation by inducing a specific YUC gene (Pain et al., 2019). In Arabidopsis, heat stress led to an indirect increased expression of YUC8 (Sun et al., 2012), which is similar to the upregulation of CsYUC8/9 in cucumber. Cold stress also led to the upregulation of CsYUC10b but downregulation of other CsYUC proteins in cucumber (Yan et al., 2016). RNA-seq analysis of Arabidopsis under heat and drought stress also revealed a tissue-specific difference in the up- or downregulation of TAA/YUC auxin biosynthesis genes, such as the upregulation of YUC9 expression in leaf tissues after heat stress (Blakeslee et al., 2019). Overexpression of YUC7 in Arabidopsis (Lee et al., 2012), and YUC6 in potato was able to increase drought tolerance with reduced water loss in transgenic plants by reducing the decomposition of IAA (Kim et al., 2012; Cha et al., 2015). An increased free IAA level and improved drought stress tolerance connected with reduced levels of reactive oxygen species and delayed leaf senescence have been observed for plants such as tomato, maize, rice, and petunia (Ke et al., 2015). In contrast to most results in Arabidopsis, increased drought tolerance associated with decreased root IAA levels in rice was found, accompanied by the downregulation of various YUC genes (Du et al., 2013; Naser and Shani, 2016). The different expression patterns of YUC genes in response to different stresses or in different species suggested a possible functional differentiation of YUC genes during stress response.
Medicago sativa L. is a perennial herbaceous legume forages with high yield, nutrient value, and palatability. As a basic component in rations for animals and an important cash crop for biofuel ethanol production, it is widely cultivated (Li et al., 2011). However, the growth and yield of M. sativa could be severely inhibited by external stresses such as salt, cold, and drought stress. Recently, large-scale potential genes involved in M. sativa responsive to adverse stimuli have been investigated by transcriptional profiling and detected several stress-responsive genes and categories (Postnikova et al., 2013; An et al., 2016; Luo et al., 2019; Ma et al., 2021). Root and leaf transcriptomes under salt stress revealed a hormone interaction involved in salinity adaptation (Lei et al., 2018). Overexpressing IAA within root nodules of M. sativa was associated with the improved drought tolerance of plants (Defez et al., 2017). Although YUCs have been identified in several species of plants, such as 11 AtYUCs in Arabidopsis (Mashiguchi et al., 2011), 7 OsYUCs in rice (Yamamoto et al., 2007), 22 TaYUCs in wheat (Yang et al., 2012), 22 GmYUCs in soybean (Wang et al., 2017) and 14 ZmYUCs in maize (Li et al., 2015), the IAA biosynthesis-related YUC genes in M. sativa or its model legume species M. truncatula (Medicago truncatula Gaertn.) has not yet been identified at the genome-wide level and the tissue-specific and abiotic stress expression patterns have not been analyzed (Li and Brummer, 2012), greatly limiting the improvement of stress adaptability of M. sativa by modifying the auxin pathway through genetic engineering.
In this study, a total of 12 MsYUCs in M. sativa and 15 MtYUCs in M. truncatula were identified. The gene structure, motif composition, chromosome location, and gene replication events were analyzed, and the evolutionary relationship of other species associated with M. sativa was constructed. An overall comparative expression analysis in M. sativa was performed to examine the YUC gene expression patterns in different tissues, different varieties, and their responses to cold, drought, and salt stress. These results could provide valuable information for identifying candidate MsYUCs involved in different biological processes and various abiotic stress responses in M. sativa for further gene functional study and for genetic modification.
2 Materials and methods
2.1 Identification of Medicago YUC genes and basic characteristic analysis
Genome sequence and genome annotation information of M. sativa variety Zhongmu No. 1 and M. truncatula used in this study were downloaded from Ensembl Plants (https://plants.ensembl.org). The amino acid sequences of the Arabidopsis YUC family members were downloaded from the TAIR website (https://www.arabidopsis.org/) and used as Query to search the Medicago protein sequences by Local BLAST, and the sequences with e-value less than −20 were reserved. The latest version of all schema database files “Pfam-a.hm.gz” from the Pfam database (https://pfam.xfam.org/) were downloaded, and candidate YUC members containing the FMO-like domain (PFam00743) were identified using TBtools’ (Chen et al., 2020) simple HMM Search plug-in. Results obtained from BLAST and Pfam search were further merged to remove duplicates. Finally, the Batch CD search function in the NCBI website (https://www.ncbi.nlm.nih.gov/) and the SMART database were used to detect and retain the correct and complete sequences of YUC characteristic conserved motifs (http://smart.embl-heidelberg.de/). The basic features such as molecular weight were determined, and isoelectric point analysis was performed using the ExPASy Proteomic Server (https://web.expasy.org/protparam/).
2.2 Chromosome localization and conserved motif and gene structure analyses
According to the chromosomal location data contained in the downloaded Medicago genome annotation information, TBtools was used to map the chromosomal location of YUC members. The YUC members detected in Medicago were named according to their position from top to bottom on chromosomes 1–8. The conserved motif of YUC genes was identified using online motif detection software (http://meme.nbcr.net/meme/), and the length of the motif was set from 2 to 200 bp to detect a maximum of 12 motifs. Visualization was performed with the TBtools software. For gene structure analysis, TBtools’ “gene structure view” function was used to visualize the gene structure (exon and intron number and location) of MsYUC family genes. The “One step build ML tree” plug-in of TBtools was used to get a Newick tree and displayed in the front of the conversed motif and gene structure exhibition.
2.3 Phylogenetic and gene duplication and synteny analysis
A phylogenetic tree was constructed using the YUC amino acid sequences of Arabidopsis, rice, M. truncatula, and M. sativa to analyze the homology relationships. All YUC sequences were aligned to multiple sequences using Clustal W, and the alignment resulted in phylogenetic tree construction using MEGA6.0 software (Larkin et al., 2007; Tamura et al., 2013). The establishment method used the adjacency method (neighbor-joining method) and the P-distance model with the bootstrap test for 1,000 times. Replication events of Medicago YUC genes and collinear blocks of YUC genes within M. sativa, Arabidopsis, M. truncatula, and rice were analyzed using the “One Step MCScanX Wrapper” function of TBtools with the e-value of 1e−3 and number of blast hits of 10. Tandem and segmental duplicates in the YUC gene family were identified using TBtools by searching the final “tandem” and “gene Linked Region” files after running. Phylogenetic analysis of species was performed using “phyliptree.phy” derived from the “NCBI Taxonomy” function. The Ka (nonsynonymous) and Ks (synonymous) substitution rates of gene duplication pairs were calculated using the “Simple Ka/Ks Calculator” function of TBtools. Ka/Ks <1, = 1, and >1 represent purification selection, neutral selection, and positive selection, respectively (Zhang et al., 2006). The divergence time (million years ago/MYA) was calculated through formula T = Ks/2λ * 10−6 (λ = 6.5 × 10−9).
2.4 Protein structure and subcellular localization prediction
Secondary structure prediction of MsYUCs was performed by Phyre2 (http://www.sbg.bio.ic.ac.uk/servers/phyre2/html/page.cgi?id=index). A tertiary structure model of the MsYUC proteins was predicted by SWISS-MODEL (https://swissmodel.expasy.org//). Global model quality estimation (GMQE) was used to obtain the high score-predicted model. Trans-membrane domain (TMD) prediction was constructed using TMHMM based on the hidden Markov model (https://services.healthtech.dtu.dk/services/TMHMM-2.0/). Using the online website CELLOv.2.5, subcellular localization was predicted (http://cello.life.nctu.edu.tw/ ) (Yu et al., 2010).
2.5 Analysis of the promoter-based cis-acting elements
Promoter sequences of the MsYUC genes (2,000 bp upstream of the ATG) were extracted by the “GTF/Gff3 Sequence Extract” function of TBtools using “genome annotation file” and “genome fasta file.” The promoter sequences were submitted to the PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) website for cis-acting element analysis, and the elements represented by different-colored symbols were visualized using TBtools’ “Basic Biosequence viewer” function.
2.6 Analysis of MsYUC gene expression patterns in the RNA-seq data
RNA-seq data of different tissue were downloaded from the online LegumeIP V3 website (https://www.zhaolab.org/LegumeIP/gdp). The expression data of different genotypes and under various abiotic stresses were obtained from previous studies (Zhou et al., 2018; Luo et al., 2019). The different expression profiles were exhibited through a heat map constructed by “Amazing Heatmap” function of TBtools.
2.7 RT-qPCR analysis
Eight-week-old seedlings of Zhongmu No. 1 were exposed to untreated control (CK), cold (4°C), and salt (200 mM NaCl) stresses for 6 h. After treatment, RNeasy Kit (Qiagen) was used to extract the total RNA from three biological replicates under control, salt, and cold stresses, respectively. First-strand cDNA of each sample was synthesized using the TaqMan reverse transcription kit (Applied Biosystems). qPCR was conducted on an ABI real-time PCR system with a total volume of 20 μl containing 10 μl of SYBR Green real-time PCR master mix (Toyobo, Japan), 2 μl of cDNA template, 0.2 μM of upstream and downstream primers. The qPCR program was conducted with denaturation at 95°C or 10 min, followed by 40 cycles of amplification (95°C for 30 s, 60°C for 30 s, and 68°C for 1 min) using the ABI real-time PCR system (Applied Biosystems, Foster City, CA). Transcript levels of each sample were determined and normalized to the untreated control sample (CK) as a calibrator with respect to the internal control gene using the 2−ΔΔCt method (Schmittgen and Livak, 2008). Values represent mean ± SD of three biological replications. One-way ANOVA test was used, and significant differences from CK and treated plants at P < 0.05 are shown by asterisks. All the technical aspects of qPCR experiments fitted the MIQE Guidelines (Bustin et al., 2009). The primers used are listed in Table S1.
2.8 Protein–protein interaction and miRNA target prediction
All MsYUC protein sequences were submitted to the STRING website (http://string-db.org) to build a protein–protein interaction network with their Arabidopsis orthologs as a reference. Using M. truncatula miRNAs as reference, target miRNAs were predicted through the psRNATarget website (https://www.zhaolab.org/psRNATarget/) with default parameters while selecting target accessibility, as previously described (Dai et al., 2018).
3 Results
3.1 Identification and basic characterization of the MsYUC and MtYUC gene families
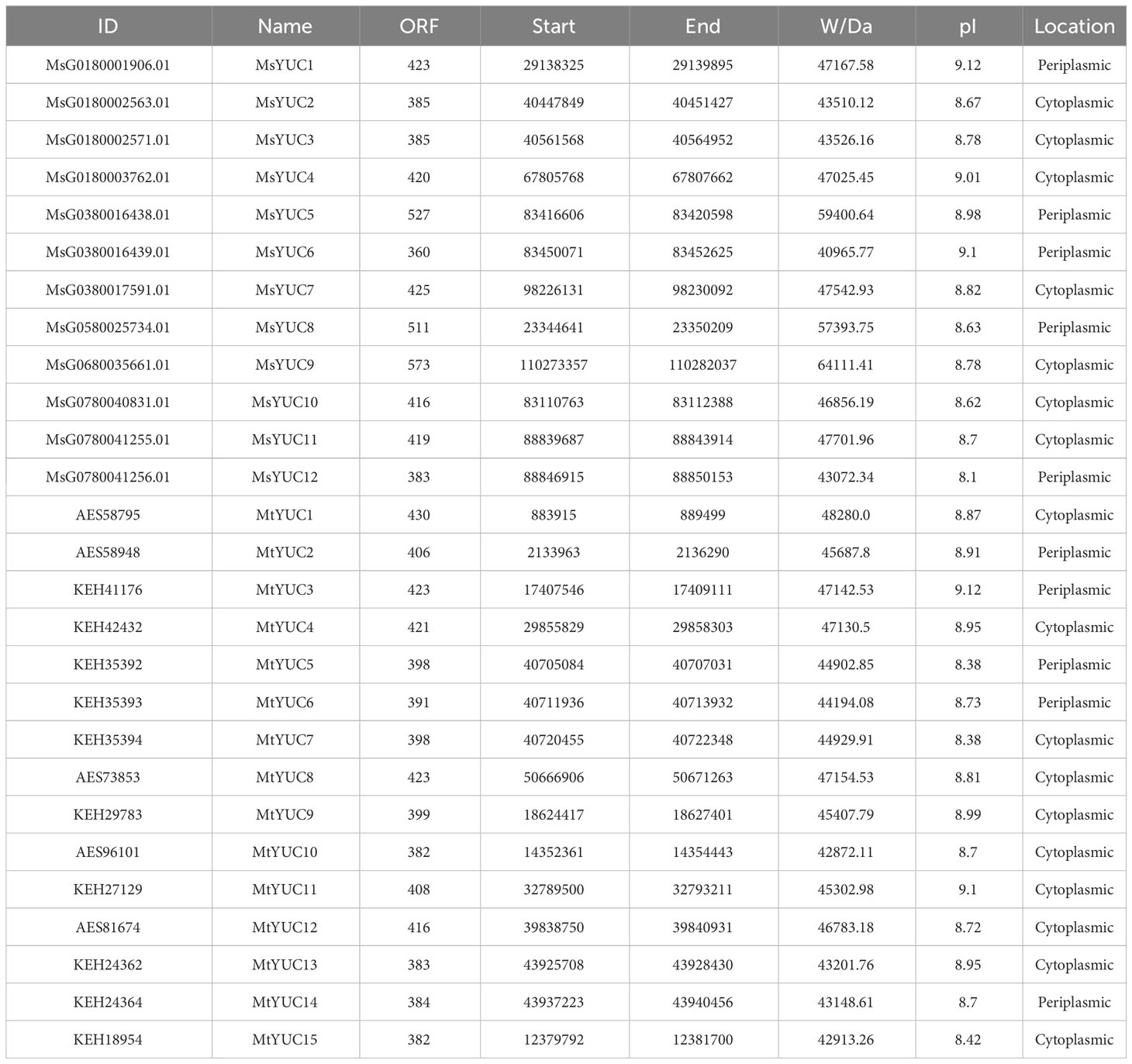
Comparative homology analysis was performed using the downloaded Arabidopsis YUC protein sequences as Query to search the protein sequences and the genome sequence of Medicago, and a total of 12 MsYUCs and 15 MtYUCs were identified from M. sativa and M. truncatula, respectively. All members were designated MsYUC1-MsYUC12 and MtYUC1-MtYUC15 according to their distribution and location information on the chromosome (Table 1; Figure S1). The MsYUC genes showed a significant uneven distribution on eight chromosomes, with the most four MsYUC genes on chromosome 1, three MsYUC genes on chromosomes 3 and 7, and only one MsYUC gene on chromosomes 5 and 6, but no distribution of MsYUC genes on chromosomes 2, 4, and 8 (Figure S1A). However, MtYUC genes were distributed in all chromosomes except for chromosome 2 (Table 1; Figure S1B). Chromosomal localization also showed that all YUC genes could be localized to the Medicago chromosomal genome. As shown in Table 1, the length of the coding region (ORF) of MsYUC genes varied from 360 to 573 amino acids with a molecular weight (MW) from 40.97 to 64.11 kD. The ORF and MW of MtYUC genes varied in relatively small ranges, with The ORF from 382 to 430 amino acids and MW from 42.87 to 48.28 kD (Table 1). All YUC proteins were basic proteins with isoelectric points (pI) greater than 8 (ranging from 8.1 to 9.12). Subcellular location prediction showed that both MsYUC and MtYUC proteins had cytoplasmic and periplasmic locations (Table 1).
3.2 Phylogenetic analysis of Medicago YUC proteins
To further analyze the kinship of YUC genes, YUC protein sequences from M. sativa (12 MsYUC), Arabidopsis (11 AtYUC), M. truncatula (15 MtYUC), and rice (14 OsYUC) were selected, and an evolutionary tree was constructed (Figure 1A). The results showed that 52 YUC proteins in four species can be clustered into two large clusters (clade I and clade II). Clade I can be further subdivided into five small clades, with MsYUC1, 4, 10 and MtYUC3, 4, 12 in clade I-1, MsYUC7, 9 and MtYUC1, 8, 11 in clade I-2. Clade II can be subdivided into four small clades, with MsYUC2, 3, 11, 12 in Clade II-2, MsYUC8, 5 and 6 in clade II-4. MtYUC2 showed a close relationship with AtYUC1 and AtYUC4, which were clustered into clade I-4. MsYUC7/9 and MtYUC1/8/11, belonging to clade I-2, were relatively closely related to AtYUC6. MtYUC10, MtYUC15, and MsYUC8 were closely related to AtYUC10, which belongs to clade II-4 (Figure 1A). A phylogenetic tree of four species was constructed, and the number and distribution of YUC proteins in various subfamilies in four species were counted (Figure 1B). Notably, clade I-3 and cladeII-3 only contained YUC proteins from rice and clade I-4 only contained YUC proteins from the other three species except M. sativa. Clade II-2 only contained YUC members from Medicago with no homologous gene from Arabidopsis. Clade II-4 contained YUC proteins from the other three species except rice. Only two clades, clade I-1 and clade I-2, both contained YUC proteins from the four species (Figure 1C).
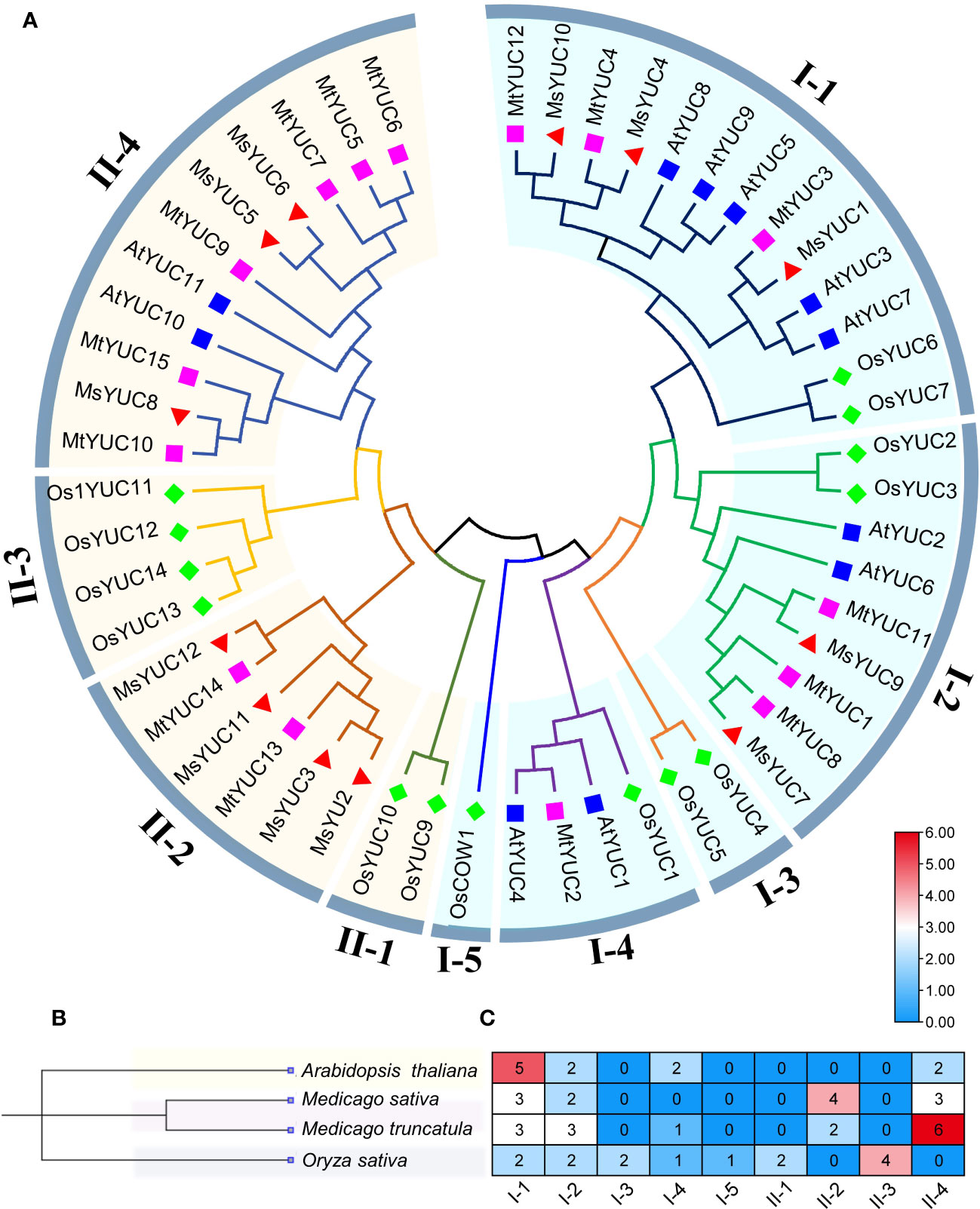
Figure 1 The phylogenetic analysis and subfamily clusters of YUC proteins in plants. (A) Phylogenetic analysis using YUC proteins in Medicago (MsYUC and MtYUC), Arabidopsis (AtYUC), and rice (OsYUC). The phylogenetic tree was constructed using the ClustalX program and the neighbor-joining method. (B) Evolutionary relationships among four species. Phylogenetic analyses of four species were performed using “phyliptree.phy” from the NCBI Taxonomy function. (C) Number of YUC proteins in four species and their distribution in various subfamilies.
3.3 Gene duplication, synteny, and evolution analysis of the YUCs
Tandem and segmental duplication events were analyzed to further investigate the evolutionary pattern of the YUC gene family in Medicago. Results revealed that MsYUC5/MsYUC6 on chromosome 3 and MsYUC11/MsYUC12 on chromosome 7 were obvious tandem duplication genes (Figure S1A). Only one MsYUC gene pair (MsYUC4/MsYUC10) could be identified as segmental duplication events (Figure 2A). The MtYUC gene family has an additional tandem repeat gene pair on chromosome 3 (MtYUC5/6, MtYUC6/7) (Figure S1B; Table S1). Only one MtYUC gene pair (MtYUC1/MtYUC8) of M. truncatula was identified as segmental duplication genes (Figure 2B). Comparative syntenic maps of M. sativa with Arabidopsis, rice, and M. truncatula were constructed to illustrate the evolution relationship of the YUC gene family (Figure 2C). Notably, 10, 10, and 1 orthologous pairs were found between M. sativa and Arabidopsis, M. sativa and M. truncatula, and M. sativa and O. sativa, respectively (Figure 2C). The collinear blocks in which MsYUC4, 5, 7, 9, 10 were located is present in M. truncatula and Arabidopsis except rice. MsYUC1-related collinear blocks were found only in Medicago but not in Arabidopsis or rice (Table S2). Interestingly, one MsYUC family member, MsYUC9, had collinear relationships with gene(s) in all species analyzed (Table S2). The Ka/Ks ratio of homologous MsYUC gene pairs ranged from 0.19 (MsYUC4/10) to 0.49 (MsYUC5/6), whereas the Ka/Ks ratio of MtYUC homologous ranged from 0.09 (MtYUC5/6) to 0.24 (MtYUC6/7), indicating that the YUC genes of Medicago had undergone a great purification selection pressure (Table S3). The evolutionary divergence time (MYA) calculated showed that two homologous gene pairs MsYUC4/10 (47.83 MYA) and MtYUC1/8 (57.28 MYA) were derived from the formation period of genus Medicago. Three gene pairs of MsYUC5/6, MtYUC5/6, and MtYUC6/7 homologous gene pairs were derived around 5 million years ago, and one homologous gene pair (MsYUC11/12) was derived around 75.77 MYA (Table S3).
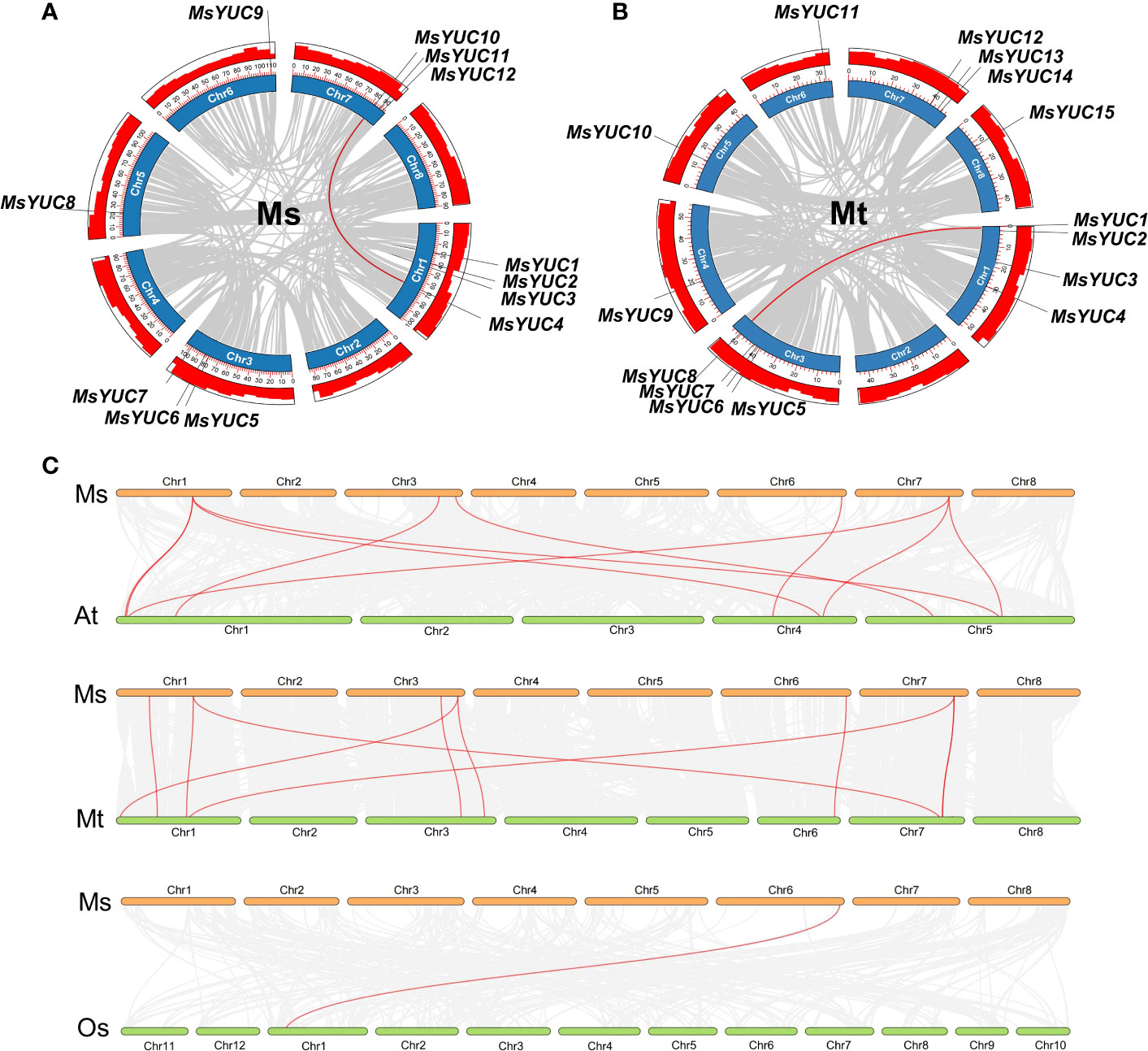
Figure 2 Duplication event analysis for the YUC gene family in the M. sativa (Ms) and M. truncatula (Mt) genome and synteny analysis between M. sativa and the other three species. The duplication events in the M. sativa genome (A) and M. truncatula genome (B). Red-colored lines indicate duplication events of MsYUC family members (MsYUC10/4) and MtYUC family members (MsYUC1/8). (C) Collinearity analysis of M. sativa (Ms) with M. truncatula (Mt) or Arabidopsis (At) or rice (Os). Red-colored lines indicate the YUC family members in different species.
3.4 Motif and gene structure analysis of MsYUC members
Motif analysis showed that 12 MsYUC proteins all contained the conserved FAD-binding motif and NADPH-binding motif (Figure 3A), suggesting a conserved function. Nevertheless, some differences were observed in the FMO-identifying motif of MsYUC5/6/10 proteins, ATG-containing motif1 of MsYUC2/3/9/11/12 proteins, and ATG-containing motif2 in MsYUC2/3/11/12 proteins, which might contribute to the functional divergences (Figure 3A). In the prediction analysis of the conserved motif of MsYUC proteins, 12 relatively conserved motifs (motifs 1~12) were further identified, including motif1 as the FAD binding site and motif2 as the reduced NADPH binding site (Table S4; Figure 3B). Furthermore, eight conserved motifs (motif1, 2, 4, 5, 6, 8, 9, 10) were present in all MsYUCs examined. Each MsYUC protein contained a minimum of 8 to a maximum of 12 of these motifs. and MsYUC6 protein had the least motif. MsYUC1 and MsYUC4 protein had all 12 conserved motifs and MsYUC7 had 11 conserved motifs except motif12. Seven MsYUC proteins (MsYUC9/12/11/2/3/8/5) contained the same 10 conserved motifs (motif1~10). Only MsYUC10 protein lacked motif8 compared with other members (Figure 3B). Gene structure analysis revealed that the number of exons of MsYUCs varied from 3 to 7 whereas MsYUC9 contained the most numerous introns. Five MsYUC genes (MsYUC2/3/5/8/11) had five exons. Three MsYUC genes (MsYUC7/12/6) had four exons, and three MsYUC genes (1/4/10) had three exons. Seven MsYUC genes (MsYUC12/11/2/3/8/5/6) containing the same 10 conserved motifs had four exons, whereas three members had only two exons (MsYUC1/4/10) (Figure 3C).
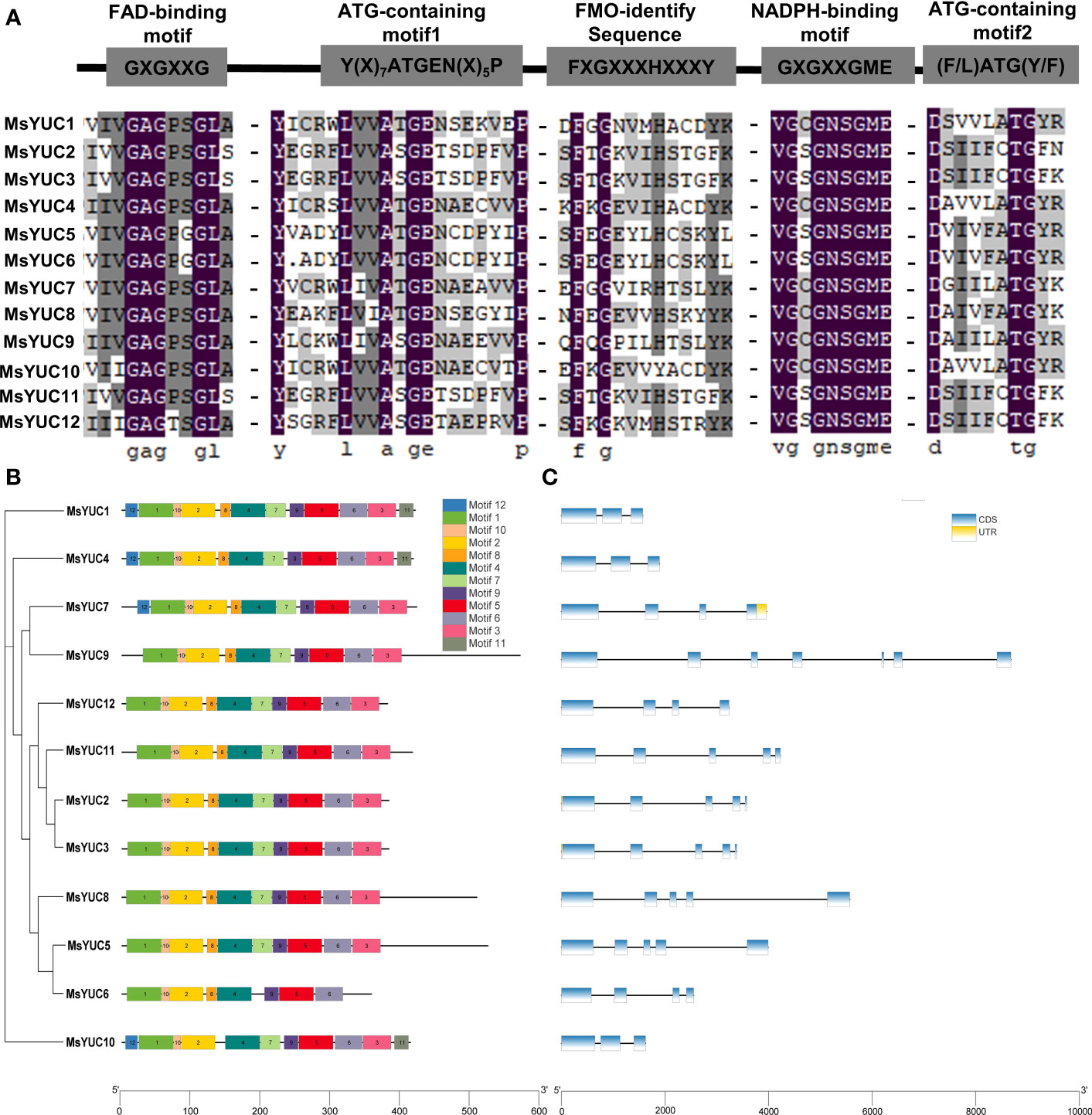
Figure 3 Structure and conversed motifs of MsYUC members. (A) Alignment of conserved domains in MsYUC proteins. (B) Conserved motifs of MsYUC genes predicted by MEME. (C) Gene structure of MsYUC genes. The exons are represented by blue boxes, and black lines are represented by black lines.
3.5 Expression analysis of MsYUCs in different tissues and different genotypes
The expression patterns of 12 MsYUC genes in different tissues were examined using online transcriptome data. Results indicated a tissue expression specificity of different MsYUC genes. For example, MsYUC10, MsYUC12, and MsYUC2 had relatively higher expression levels in specific tissues examined, whereas some members (MsYUC5/6/8) had very low expression levels and were barely detectable. In addition, MsYUC2 had a higher expression level in leaves than in other tissues and MsYUC12 was more highly expressed in both leaves and roots than in other tissues (Figure 4A). In addition, we further analyzed the expression correlation between every two MsYUC genes in five tissues. MsYUC2 showed to be significantly positively correlated with MsYUC3, consistent with their close relationship in the phylogenic tree. MsYUC2 and MsYUC3 showed to be significantly positively correlated with MsYUC4 and MsYUC7, respectively. MsYUC3 showed to be significantly positively correlated with MsYUC4 and MsYUC7, respectively. MsYUC4 and MsYUC7, as well as MsYUC1 and MsYUC9, were significantly positively correlated (Figure 4B). There was also a differential expression pattern of MsYUCs among different genotypes. For example, MsYUC10 and MsYUC12 showed a higher expression level in 95-608 compared with other genotypes. MsYUC11 had a relatively higher expression level in PI251830-K but had the lowest expression detected in 95-608 (Figure 4C).
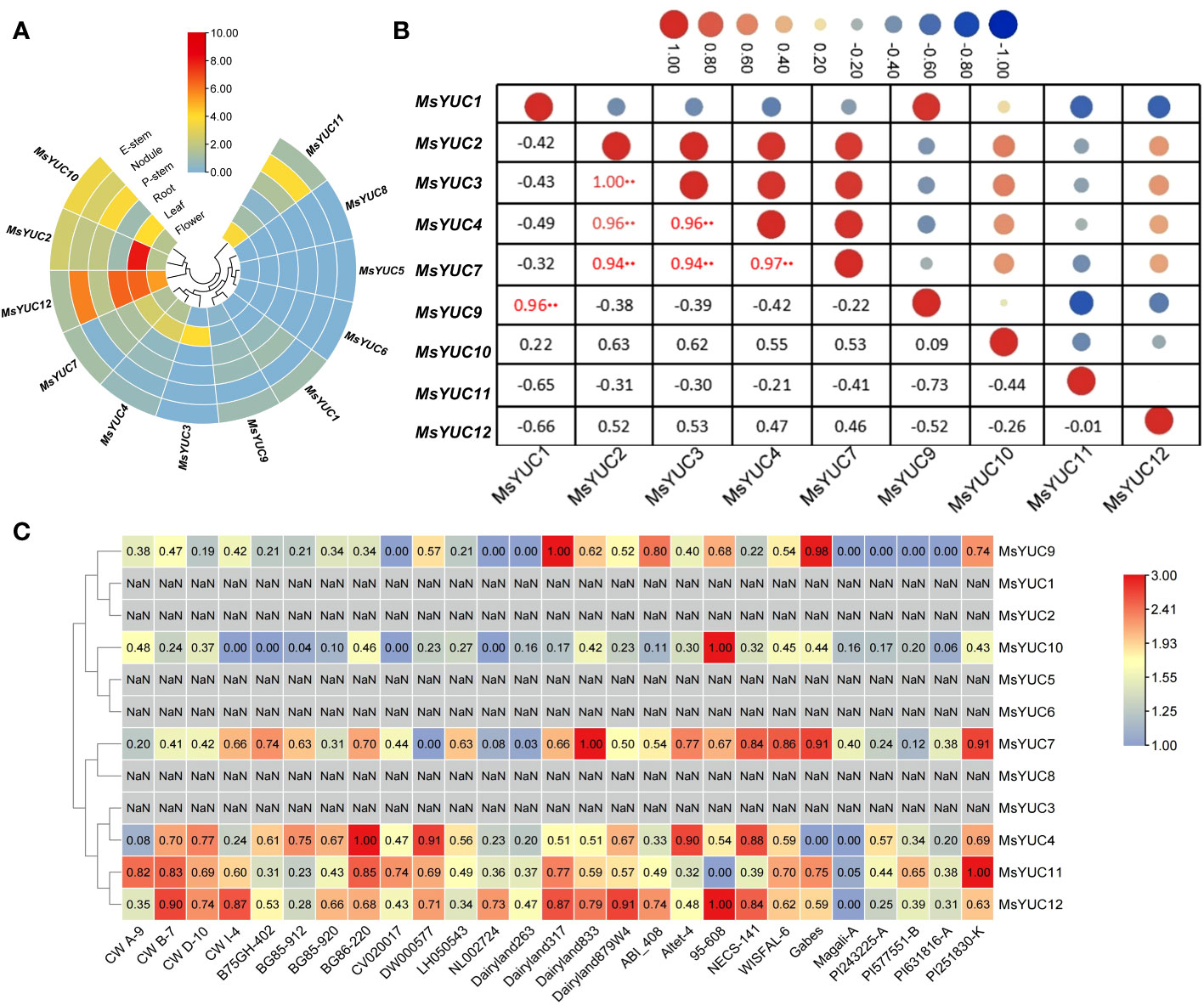
Figure 4 Expression pattern of MsYUC genes in different tissues and in different genotype. (A) Tissue-specific expression analysis of MsYUC genes. (B) The correlation of gene expression patterns between every two MsYUC genes. Red and blue circles represent positive and negative correlations, respectively. (C) MsYUC expression in different genotypes. The color scale of the heatmap refers to the relative expression level.
3.6 Promoter analysis and stress response expression of MsYUCs
To predict the possible regulation of MsYUCs expression, the cis-acting elements included in the promoter sequence of the MsYUC genes were analyzed. Results revealed a variety of stress response elements related to hormone and stress response (Figure 5A). Auxin-responsive elements were found in the promoter region of MsYUC1, 4, 5, 8, 12, and three of their promoters contained AuxRR-core elements. The promoter of MsYUC8 had the most cis-acting elements (6) involved in the abscisic acid responsiveness (ABRE) (Figure 5B). The MsYUC7 promoter region contained five CGTCA motifs, which functions in Me-JA responsive. There were also some GA-responsive elements such as GARE-motif, TATC-box, P-Box, and some SA-responsive elements (TCA-element) in the promoters of certain MsYUCs (Figure 5A). The promoter of MsYUC6, 8, and 10 contained one cis-acting element involved in low-temperature responsiveness LTR (CCGAAA), respectively. Except for MsYUC6, 7, and 8, other members all had one or two MYB-binding site (MBS) involved in drought inducibility. Some anaerobic induction, osmotic pressure-responsive, and defense and stress-responsive elements (TC-rich repeats) were also present on certain MsYUC promoters (Figure 5A). In addition, all the MsYUC promoters had light-response elements. MsYUC1, 2, 8 had the most light-response elements (6) whereas MsYUC12 had the least light-response element (1) (Figure 5B). Based on the abiotic transcriptome data analysis, the expressions of MsYUC1 and MsYUC10 were significantly increased under salt stress (Figure 5C) and the expressions of MsYUC10 and MsYUC12 were induced by cold (Figure 5D). Mannitol treatment significantly induced the expression of MsYUC10 (Figure 5E). RT-qPCR analysis further confirmed that MsYUC1 and MsYUC10 expression could be induced by NaCl (100 mM) and MsYUC10 and MsYUC12 could be elevated by cold (4°C) for 3 h treatments (Figures 5F,G). Expression analysis showed that MsYUC genes might have a tissue-specific expression and differential abiotic stress response pattern.
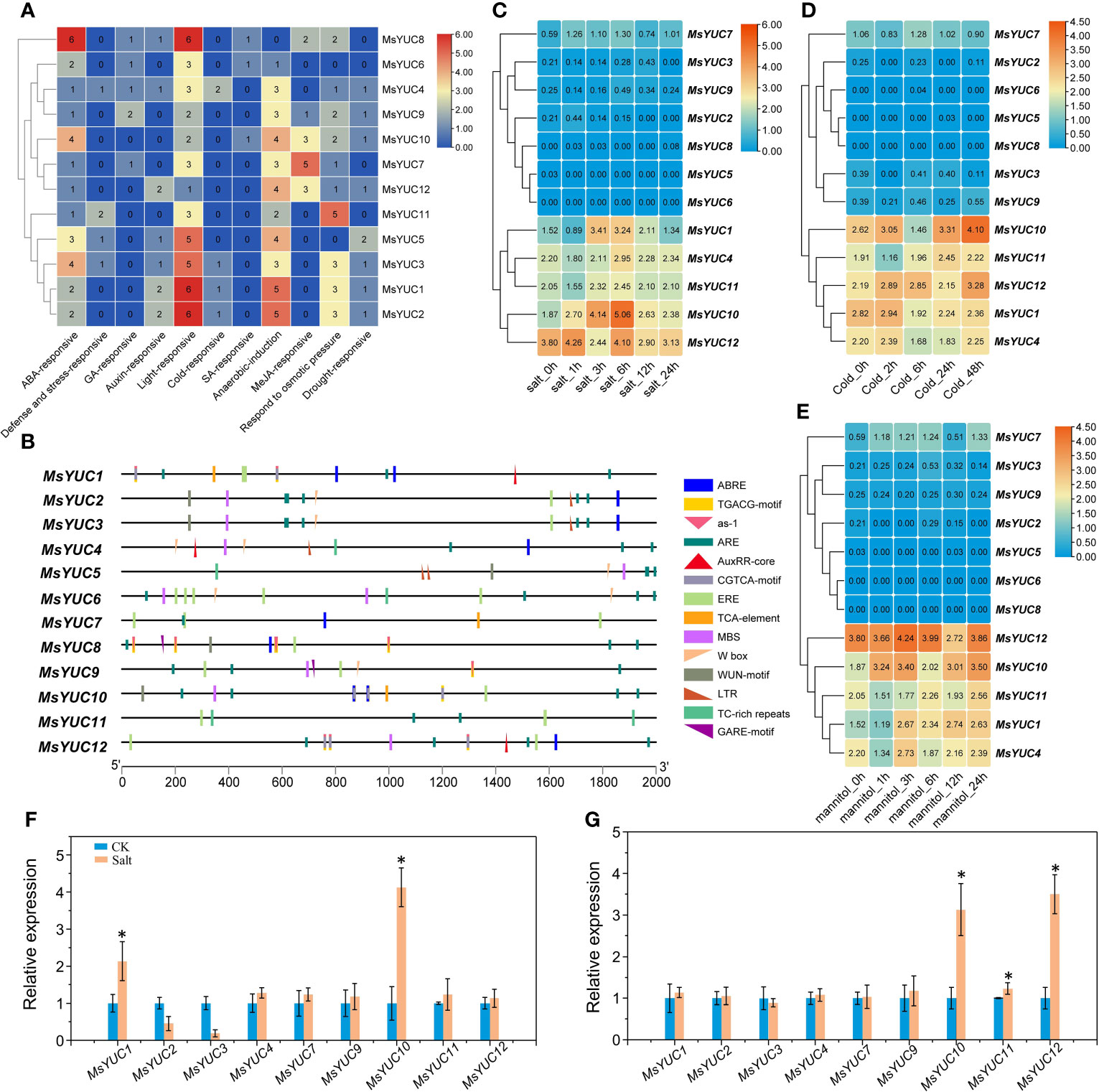
Figure 5 Cis elements of MsYUC promoter prediction and expression analysis of MsYUCs under stress conditions. (A) Number of hormone and stress response-related elements of MsYUCs. (B) Main elements distributed in the promoter region of MsYUC genes. Expression of MsYUC genes under salt stress (C), mannitol treatment (D), and cold stress (E). The color scale of the heatmap refers to the relative expression level. Relative expression of MsYUCs treated by NaCl (F) and cold (G) determined by RT-qPCR. Three replicates were designed for each sample, and M. sativa actin gene expression was used for data normalization. Value represents mean ± SD of three replicates. * indicated significant different from untreated control (CK) plants (p < 0.05, one-way ANOVA).
3.7 Prediction of the protein interaction network and targeted miRNA of MsYUC members
Protein structure prediction showed that MsYUC proteins shared a unique topology containing several α-helices and β-structures (Figures 6A, S2), indicating structural conservation. The trans-membrane prediction showed that MsYUC4, 9, 10, and 11 proteins possessed one TMD, respectively. The TMD regions of MsYUC9 and 11 proteins were localized in the N-terminal, whereas the TMD regions of MsYUC4 and 10 were localized in the middle of the protein (Figure S3). A predicted protein interaction network indicated that MsYUC proteins had multiple interaction partners (Figure 6B). MsYUC10 protein was predicted to interact with transcription factor NAC089 and NAC-like NTL9, and auxin upregulated F-box protein 1 (AUF1), which is a component of E3 ubiquitin ligase complexes. Both MsYUC9 and MsYUC10 proteins were predicted to interact with phytochrome interacting factor 4 (PIF4). MsYUC9 protein could also interact with TAA1 and amidase 1 (AMI1), which functions in auxin biosynthesis. MsYUC1, 7, 9, 10, 12 were predicted to interact with TAA1, TAR1, and TAR2, which function in the first step of the IPA pathway. We next performed miRNA target site prediction for the MsYUC genes. As shown in Figure 6C, MsYUC2, 3, and 11 were predicted to be targeted by a similar miRNA5272f. MsYUC5, 6, and 8 were predicted to be targeted by a similar miRNA5742. All the coding sequences of MsYUCs contained at least three predicted targets for miRNA.
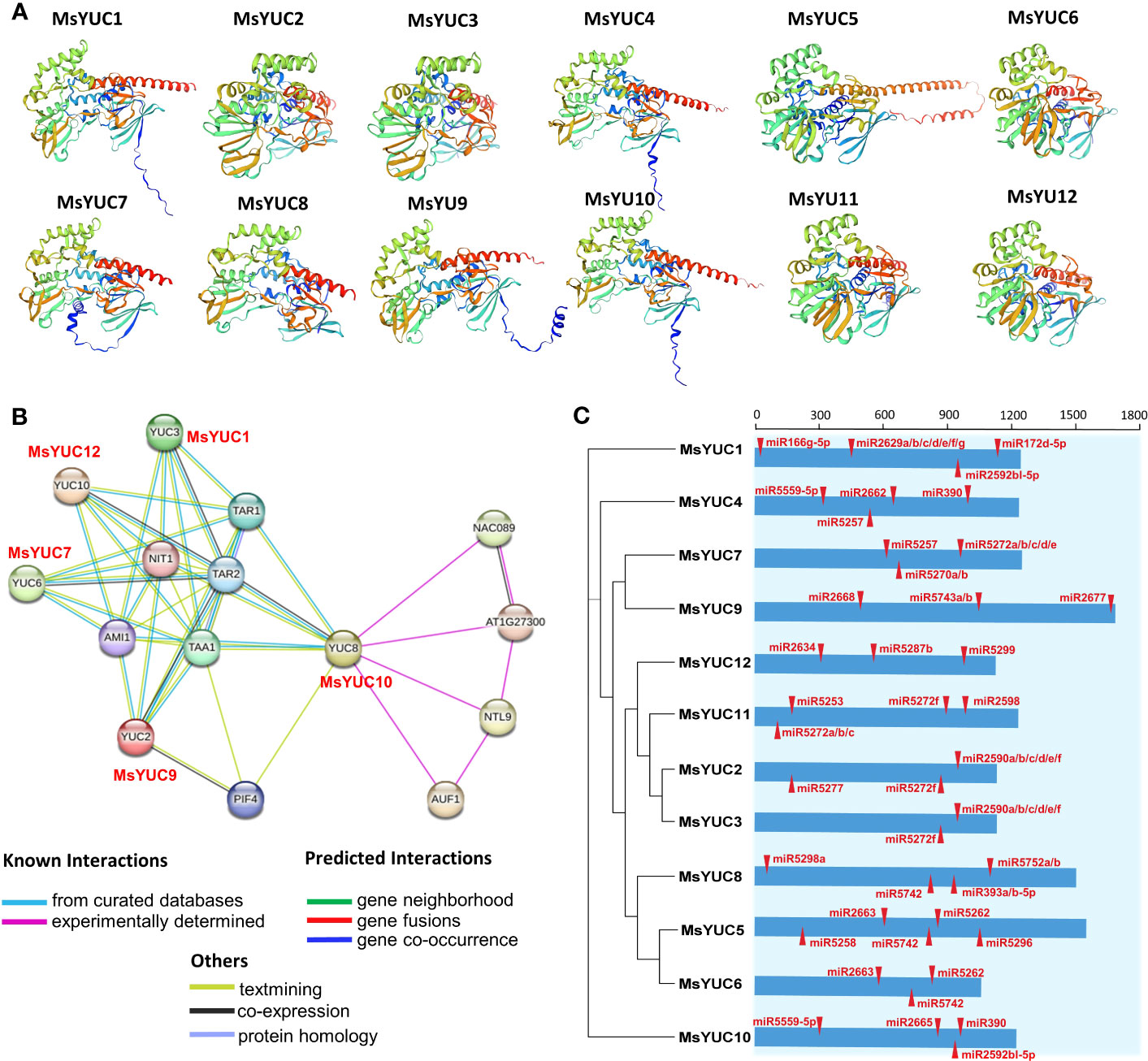
Figure 6 Predicted protein interaction network of MsYUC proteins and miRNA target sites in MsYUC genes. (A) Protein structure prediction of MsYUCs. (B) Protein interaction network predicted using MsYUC orthologs from Arabidopsis. (C) Predicted miRNA targets in the MsYUC coding sequence. The red tangles represent the miRNA-targeted MsYUC sites.
4 Discussion
The YUC gene family proteins involved in auxin biosynthesis are the first identified FMO class family in plants that regulate growth, development, and tolerance in plants (Zhao et al., 2001). In Medicago, the YUC number (12 MsYUCs and 15 MtYUCs) was close to 11 AtYUC in Arabidopsis and 14 OsYUC in rice (Yamamoto et al., 2007; Zhao, 2012). Gene duplication is thought to be the main driver of species evolution and a direct cause of gene family expansion (Lynch and Conery, 2000; Moore and Purugganan, 2003; Maere et al., 2005), and two forms of gene duplication (tandem and segmental) events were identified in Medicago YUC gene families. In the MtYUC gene family, there was a gene cluster containing three MtYUC members (MtYUC5/6/7). Moreover, there was no distribution of MsYUC genes on chromosomes 2, 4, and 8 (Table 1, Figure S1A) whereas MtYUC genes were distributed in all chromosomes except for chromosome 2 (Table 1; Figure S1B). These reasons may together contribute to the more members of MtYUC than that of MsYUC. Notably, most of the YUC proteins in rice (Monocots) and Medicago or Arabidopsis (Dicots) could not gather under the same branch, as clade II-4 had no rice YUC protein and clade I-3 and clade II-3 only contained rice YUC proteins, indicating that YUC proteins underwent an evolutionary divergence, as that there are missing or duplication of YUCs during evolution. (Figure 1). In addition, Ka/Ks was used to evaluate their specific positions under positive selection pressure after duplication (Lynch and Conery, 2000; Mayrose et al., 2007). In this study, the Ka/Ks value of each duplication gene pair of YUCs of Medicago for all gene pairs was less than 1 (Table S3), which suggested that these genes had evolved under strong purifying selection. Since the divergence time of the Papilionoideae subfamily, which includes the genus Medicago, was approximately 34–63.7 millions of years (MYA) (Wang et al., 2023), the evolutionary divergence time of homologous gene pairs MsYUC4/10 and MtYUC1/8 was derived from the formation period of Papilionoideae subfamily. Because of the importance of M. sativa with high yield, nutrient value, and palatability, the mechanisms regulating its growth are of significant interest (Yamamoto et al., 2007). Functional orthologs of YUC genes in model species can provide insight into the functions in Medicago (Wei and Gai, 2008). MtYUC2 showed a close relationship with AtYUC1 and AtYUC4 (Figure 2), which have been reported to play vital roles in the formation of floral organs and vascular tissues in Arabidopsis (Cheng et al., 2006). However, the Arabidopsis AtYUC1 and AtYUC4 had no corresponding homologs in the M. sativa genome. Moreover, some YUCs of M. sativa had no homologs in Arabidopsis or rice, indicating that the gene loss event may have occurred after species divergence (Figure 1; Table S2).
In Arabidopsis, the roots and shoots appear to use two separate sets of YUC genes for auxin biosynthesis: ER-located YUCs functioning in roots or cytoplasmic-located YUCs functioning in shoots (Kriechbaumer et al., 2015). Phylogenetic tree analysis showed that AtYUC3, 5, 7, 8, and 9, which were reported to function in roots with ER location, clustered in clade I-1 (Kriechbaumer et al., 2015). MsYUC4 and MsYUC10, closely related to AtYUC5, 8, 9, also showed a predicted cytoplasmic location (Table 1). In M. sativa, MsYUCs also showed different expression patterns in different tissues. For example, MsYUC10, MsYUC12, and MsYUC2 had relatively higher expression levels in specific tissues examined and MsYUC2 had a higher expression level in leaves than in other tissues (Figure 4A). MsYUC9, closely related to AtYUC2, which was reported to function in shoots, was expressed not only in the shoots but also in roots of M. sativa. MsYUC12, with no homologous genes in Arabidopsis, showed higher expression levels in all tissues and was inferred to have universal roles during plant growth and development (Figure 5A). Therefore, in contrast to AtYUC expression, MsYUC expression does not seem to be clearly divided into shoot or root independent expression, suggesting a specificity in M. sativa compared with Arabidopsis.
The IPA-dependent pathway also plays an important role in integrating environmental stress and hormone signaling, and YUCs were reported to be involved in environmental stress response (Blakeslee et al., 2019). Cis-acting elements on the MsYUCs’ promoter revealed a variety of stress response elements related to hormone such as Auxin-, ABA-, JA-, GA-, and SA-responsive elements in the promoters of certain MsYUC genes (Figure 5A). In Arabidopsis, ABA can inhibit the transcription of YUC2/8 via ABI4, thereby inhibiting primary root elongation (Yu et al., 2014). JA has been reported to promote lateral root growth through a direct regulation of YUC2 by transcription factor ERF109 (Cai et al., 2014). JA also directly activates YUC8/9-dependent auxin biosynthesis to function in mechanical wounding response (Perez-Alonso et al., 2021). In M. sativa, six ABRE elements were found in the promoter of MsYUC8 and five JA response elements were found in the promoter of MsYUC7, respectively. In addition, MsYUC7 showed a closer relationship with AtYUC2, implying a similar function in JA response. In Arabidopsis, expression levels of YUC7, 9, 10, and 11 were upregulated under dehydration conditions (Shi et al., 2014). Activation of YUC7 enhances drought resistance in Arabidopsis (Lee et al., 2012). Overexpressed YUC6 of Arabidopsis in potato and poplar plants or overexpressed BnaYUC6a in Arabidopsis and oilseed rape showed typical auxin overproduction alternation and conferred high drought resistance (Kim et al., 2012; Ke et al., 2015; Hao et al., 2022). Since MsYUC11, which is closely related to AtYUC6, had a five-element response to osmotic stress but no drought response elements, suggesting a function differentiation among species (Figure 5A).
YUC expression was also reported to be affected by cold stress. For example, cucumber CsYUC10b was upregulated by cold stress whereas other CsYUCs were downregulated (Yan et al., 2016). In the hypocotyl, the PIF4-YUC8 regulatory module plays an important role in response to stress signals, including light stress. The accumulation and transcriptional activity of PIF4 are regulated by different proteins, with competition for and interference at the AtYUC8 promoter by other transcription factors affecting the positive regulation of AtYUC8 by PIF4 and consequently affecting biosynthesis of auxin (Ma et al., 2016). In this study, all the MsYUC promoters had light-response elements. MsYUC1, 2, and 8 promoters had the most light-response elements, whereas the MsYUC12 promoter had the least light-response element (Figure 5A). MsYUC10 was further predicted to interact with PIF4, indicating a similar function with AtYUC8 in light response (Figure 6B). Transcription factor AGL21 positively regulates AtYUC5/8 which could be induced by IAA/ABA/JA and a variety of stresses, including salt stress (Yu et al., 2014). MsYUC1 and MsYUC10, which were clustered in the same sub-clade with AtYUC3/5/7/8/9, showed a significantly salt-induced expression (I-1), indicating a salt-response function in M. sativa (Figure 5F). The promoter of MsYUC1, 2, and 3 contained one cis-acting element (LTR) involved in low-temperature responsiveness, respectively (Figure 5A). Cold stress significantly elevated the expression of MsYUC10 and MsYUC12, indicating an LTR-independent cold stress response function (Figure 6B). Moreover, MsYUC10 and MsYUC12 showed a higher expression level in 95-608 compared with other genotypes. Therefore, the stress tolerance of 95-608 should be further compared with other varieties. Studies indicate that miRNA-directed regulation of transcription factors may also play key roles in the precise regulation of IPA-dependent auxin biosynthesis in plants (Luo and Di, 2023). In this study, all MsYUCs contained at least three predicted targets for miRNA, suggesting a miRNA-directed regulation of YUC in M. sativa (Figure 6C).
5 Conclusion
In this study, the YUCs of M. sativa and M. truncatula were identified on a genome-wide scale. The phylogenetic analysis and comparative syntenic maps of M. sativa with other species illustrated their evolution relationship. The tissue and genotype-specific expression and abiotic stress response profiles have also been analyzed to reveal potential functional YUC genes. Moreover, RT-qPCR verified that certain MsYUC members represented salt or cold stress-affected expression patterns. Results in this study could provide valuable information for functional analysis and for the underlying regulation mechanism study of a specific MsYUC gene of M. sativa, especially under different tissues and various abiotic stresses through modification of the auxin synthetic IPA pathway in Medicago.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
AS: Conceptualization, Data curation, Methodology, Writing – original draft. SF: Methodology, Software, Writing – original draft. XX: Validation, Investigation, Writing – review & editing. WW: Funding acquisition, Resources, Writing – review & editing. JF: Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Natural Science Foundation of China (No. 32001389) and the Natural Science Foundation of Shandong Province, China (No. ZR2020QC186).
Acknowledgments
We thank the National Natural Science Foundation of China and the Natural Science Foundation of Shandong Province for the financial support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1268027/full#supplementary-material
References
An, Y., Song, L., Liu, Y., Shu, Y., Guo, C. (2016). De novo transcriptional analysis of alfalfa in response to saline-alkaline stress. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00931
Blakeslee, J. J., Tatiana, S. R., Verena, K. (2019). Auxin biosynthesis: spatial regulation and adaptation to stress. J. Exp. Bot. 70, 5041–5049. doi: 10.1093/jxb/erz283
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE Guidelines: Minimum information for publication of Quantitative Real-Time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Cai, X. T., Xu, P., Zhao, P. X., Liu, R., Yu, L. H., Xiang, C. B. (2014). Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 5, 5833. doi: 10.1038/ncomms6833
Cha, J. Y., Kim, W. Y., Kang, S. B., Kim, J. I., Baek, D., Jung, I. J., et al. (2015). A novel thiol-reductase activity of Arabidopsis YUC6 confers drought tolerance independently of auxin bio-synthesis. Nat. Commun. 6, 8041. doi: 10.1038/ncomms9041
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Q., Dai, X., De-Paoli, H., Cheng, Y., Takebayashi, Y., Kasahara, H., et al. (2014). Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 55, 1072–1079. doi: 10.1093/pcp/pcu039
Cheng, Y., Dai, X., Zhao, Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799. doi: 10.1101/gad.1415106
Dai, X., Zhuang, Z., Zhao, P. X. (2018). psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 46, 49–54. doi: 10.1093/nar/gkr319
Defez, R., Andreozzi, A., Dickinson, M., Charlton, A., Tadini, L., Pesaresi, P., et al. (2017). Improved drought stress response in alfalfa plants modulated by an IAA over-producing rhizobium strain. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02466
Du, H., Wu, N., Chang, Y., Li, X., Xiao, J., Xiong, L. (2013). Carotenoid deficiency impairs ABA and IAA biosynthesis and deferentially affects drought and cold tolerance in rice. Plant Mol. Biol. 83, 475–488. doi: 10.1007/s11103-013-0103-7
Fraaije, M. W., Kamerbeek, N. M., van, Berkel, W. J., Janssen, D. B. (2002). Identification of a Baeyer-Villiger monooxygenase sequence motif. FEBS Lett. 518, 43–47. doi: 10.1016/080394898422544
Hao, M., Wang, W., Liu, J., Wang, H., Zhou, R., Mei, D., et al. (2022). Auxin biosynthesis genes in allotetraploid oilseed rape are essential for plant development and response to drought stress. Int. J. Mol. Sci. 23, 15600. doi: 10.3390/ijms232415600
Ke, Q., Baek, D., Park, H. C., Chun, H. J., Oh, D., Lee, M. K., et al. (2015). Transgenic poplar expressing Arabidopsis YUCCA6 exhibits auxin-overproduction phenotypes and increased tolerance to abiotic stress. Plant Physiol. Biochem. 94, 19–27. doi: 10.1016/j.plaphy.2015.05.003
Kim, J. I., Baek, D., Park, H. C., Chun, H. J., Oh, D. H., Lee, M. K., et al. (2012). Overexpression of Arabidopsis YUCCA6 in potato results in high-auxin developmental phenotypes and enhanced resistance to water deficit. Mol. Plant 6, 337–349. doi: 10.1093/mp/sss100
Kriechbaumer, V., Seo, H., Park, W. J., Hawes, C. (2015). Endoplasmic reticulum localization and activity of maize auxin biosynthetic enzymes. J. Exp. Bot. 66, 6009–6020. doi: 10.1093/jxb/erv314
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lee, M., Jung, J. H., Han, D. Y., Seo, P. J., Park, W. J., Park, C. M. (2012). Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in. Arabidopsis. Planta 235, 923–938. doi: 10.1007/s00425-011-1552-3
Lei, Y., Xu, Y., Hettenhausen, C., Lu, C., Shen, G., Zhang, C., et al. (2018). Comparative analysis of alfalfa (Medicago sativa L.) leaf transcriptomes reveals genotype specific salt tolerance mechanisms. BMC Plant Biol. 18, 35. doi: 10.1186/s12870-018-1250-4
Li, X. H., Brummer, E. C. (2012). Applied genetics and genomics in alfalfa breeding. Agronomy 2, 40–61. doi: 10.3390/agronomy2010040
Li, X., Wei, Y., Moore, K. J., Michaud, R., Viands, D. R., Hansen, J. L., et al. (2011). Association mapping of biomass yield and stem composition in a tetraploid alfalfa breeding population. Plant Genome 4, 24–35. doi: 10.3835/plantgenome2010.09.0022
Li, W., Zhao, X., Zhang, X. (2015). Genome-wide analysis and expression patterns of the YUCCA genes in maize. J. Genet. Genomics 42, 707–710. doi: 10.1016/j.jgg.2015.06.010
Liu, G., Gao, S., Tian, H., Wu, W., Robert, H. S., Ding, Z. (2016). Local transcriptional control of YUCCA regulates auxin promoted root-growth inhibition in response to aluminium stress in Arabidopsis. PloS Genet. 12, e1006360. doi: 10.1371/journal.pgen.1006360
Liu, H., Xie, W. F., Zhang, L., Ye, Z. W., Gao, Q. H., Duan, K. (2014). Auxin biosynthesis by the YUCCA6 flavin monooxygenase gene in woodland strawberry. J. Integr. Plant Biol. 56, 350–363. doi: 10.1111/jipb.12150
Luo, P., Di, D. W. (2023). Precise regulation of the TAA1/TAR-YUCCA auxin biosynthesis pathway in plants. Int. J. Mol. Sci. 24, 8514 doi: 10.3390/ijms24108514
Luo, D., Zhou, Q., Wu, Y., Chai, X., Liu, W., Wang, Y., et al. (2019). Full-length transcript sequencing and comparative transcriptomic analysis to evaluate the contribution of osmotic and ionic stress components towards salinity tolerance in the roots of cultivated alfalfa (Medicago Sativa L.). BMC Plant Biolo. 19, 32. doi: 10.1186/s12870-019-1630-4
Lynch, M., Conery, J. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Ma, D., Li, X., Guo, Y., Chu, J., Fang, S., Yan, C., et al. (2016). Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. U.S.A. 113, 224–229. doi: 10.1073/pnas.1511437113
Ma, Q., Xu, X., Wang, W., Zhao, L., Ma, D., Xie, Y. (2021). Comparative analysis of alfalfa (Medicago sativa L.) seedling transcriptomes reveals genotype-specific drought tolerance mechanisms. Plant Physiol. Biochem. 166, 203–214. doi: 10.21203/rs.3.rs-75482/v1
Maere, S., De Bodt, S., Raes, J., Casneuf, T., Van Montagu, M., Kuiper, M., et al. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 5454–5459. doi: 10.1073/pnas.0501102102
Mashiguchi, K., Tanaka, K., Sakai, T., Sugawara, S., Kawaide, H., Natsume, M., et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18512–18517. doi: 10.1073/pnas.1108434108
Mayrose, I., Doron-Faigenboim, A., Bacharach, E., Pupko, T. (2007). Towards realistic codon models: among site variability and dependency of synonymous and nonsynonymous rates. Bioinformatics 23, 319–327. doi: 10.1093/bioinformatics/btm176
Moore, R. C., Purugganan, M. D. (2003). The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. U.S.A. 100, 15682–15687. doi: 10.1073/pnas.2535513100
Naser, V., Shani, E. (2016). Auxin response under osmotic stress. Plant Mol. Biol. 91, 661–672. doi: 10.1007/s11103-016-0476-5
Pain, C., Kriechbaumer, V., Kittelmann, M., Hawes, C., Fricker, M. (2019). Quantitative analysis of plant ER architecture and dynamics. Nat. Commun. 10, 984. doi: 10.1038/s41467-019-08893-9
Perez-Alonso, M. M., Sanchez-Parra, B., Ortiz-Garcia, P., Santamaria, M. E., Diaz, I., Pollmann, S. (2021). Jasmonic acid-dependent MYC transcription factors bind to a tandem G-box motif in the YUCCA8 and YUCCA9 promoters to regulate biotic stress responses. Int. J. Mol. Sci. 22, 9768. doi: 10.20944/preprints202107.0142.v1
Postnikova, O. A., Shao, J., Nemchinov, L. G. (2013). Analysis of the alfalfa root transcriptome in response to salinity stress. Plant Cell Physiol. 54, 1041–1055. doi: 10.1093/pcp/pct056
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shi, H., Chen, L., Ye, T., Liu, X., Ding, K., Chan, Z. (2014). Modulation of auxin content in Arabidopsis confers improved drought stress resistance. Plant Physiol. Biochem. 82, 209–217. doi: 10.1016/j.plaphy.2014.06.008
Stepanova, A. N., Yun, J., Robles, L. M., Novak, O., He, W., Guo, H., et al. (2011). The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23, 3961–3973. doi: 10.1105/tpc.111.088047
Sun, J., Qi, L., Li, Y., Chu, J., Li, C. (2012). PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PloS Genet. 8, e1002594. doi: 10.1371/journal.pgen.1002594
Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Wang, Y. G., Liu, H. H., Wang, S. P., Li, H. G. (2017). Genome-wide identification and expression analysis of the YUCCA gene family in soybean (Glycine max L.). Plant Growth Regul. 81, 265–275. doi: 10.1007/s10725-016-0203-x
Wang, Y., Ruan, Q., Zhu, X., Wang, B., Wei, B., Wei, X. (2023). Identification of Alfalfa SPL gene family and expression analysis under biotic and abiotic stresses. Sci. Rep. 13, 84. doi: 10.1038/s41598-022-26911-7
Wei, Z. W., Gai, J. Y. (2008). Model legume: Medicago truncatula. Acta Prataculturae Sin. 17, 114–120. doi: 10.1007/s10499-007-9164-4
Won, C., Shen, X., Mashiguchi, K., Zheng, Z., Dai, X., Cheng, Y., et al. (2011). Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18518–18523. doi: 10.1073/pnas.1108436108
Yamamoto, Y., Kamiya, N., Morinaka, Y., Matsuoka, M., Sazuka, T. (2007). Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol. 143, 1362–1371. doi: 10.1104/PP.106.091561
Yan, S., Che, G., Ding, L., Chen, Z., Zhang, X. (2016). Different cucumber CsYUC genes regulate response to abiotic stresses and flower development. Sci. Rep. 6, 20760. doi: 10.1038/srep20760
Yang, Y., Xu, T., Wang, H., Feng, D. (2012). Genome-wide identification and expression analysis of the TaYUCCA gene family in wheat. Mol. Biol. Rep. 48, 1–11. doi: 10.1007/s11033-021-06197-0
Yu, C. S., Lin, C. J., Hwang, J. K. (2010). Predicting subcellular localization of proteins for Gramnegative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 13, 1402–1406. doi: 10.1110/ps.03479604
Yu, L. H., Miao, Z. Q., Qi, G. F., Wu, J., Cai, X. T., Mao, J. L., et al. (2014). MADS-box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals. Mol. Plant 7, 1653–1669. doi: 10.1093/mp/ssu088
Zhang, Z., Li, J., Zhao, X. Q., Wang, J., Wong, K. S., Yu, J. (2006). KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinf. 4, 259–263. doi: 10.1016/S1672-0229(07)60007-2
Zhao, Y. (2010). Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61, 49–64. doi: 10.1146/annurev-arplant-042809-112308
Zhao, Y. (2012). Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 5, 334–338. doi: 10.1093/mp/ssr104
Zhao, Y. (2018). Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu. Rev. Plant Biol. 69, 417–435. doi: 10.1146/annurev-arplant-042817-040226
Zhao, Y., Christensen, S. K., Fankhauser, C., Cashman, J. R., Cohen, J. D., Weigel, D., et al. (2001). A role for flavin monooxygenase- like enzymes in auxin biosynthesis. Science 291, 306–309. doi: 10.1126/science.291.5502.306
Keywords: Medicago, YUC, evolution analysis, expression profile, abiotic stress response
Citation: Shao A, Fan S, Xu X, Wang W and Fu J (2023) Identification and evolution analysis of YUCCA genes of Medicago sativa and Medicago truncatula and their expression profiles under abiotic stress. Front. Plant Sci. 14:1268027. doi: 10.3389/fpls.2023.1268027
Received: 27 July 2023; Accepted: 11 August 2023;
Published: 28 August 2023.
Edited by:
Hui Song, Qingdao Agricultural University, ChinaCopyright © 2023 Shao, Fan, Xu, Wang and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Wang, d2Vpd2FuZ0BsZHUuZWR1LmNu; Jinmin Fu, dHVyZmNuQHFxLmNvbQ==
 An Shao
An Shao Shugao Fan
Shugao Fan Xiao Xu
Xiao Xu Jinmin Fu
Jinmin Fu