
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 20 September 2023
Sec. Plant Biotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1267107
 Quangang Liu1,2†
Quangang Liu1,2† Jiaxing Wen1,2†
Jiaxing Wen1,2† Shipeng Wang1,2
Shipeng Wang1,2 Jianhua Chen1,2
Jianhua Chen1,2 Yongqiang Sun1,2
Yongqiang Sun1,2 Qingbai Liu1,2
Qingbai Liu1,2 Xi Li1,2
Xi Li1,2 Shengjun Dong1,2*
Shengjun Dong1,2*The basic helix-loop-helix (bHLH) family is one of the most well-known transcription factor families in plants, and it regulates growth, development, and abiotic stress responses. However, systematic analyses of the bHLH gene family in Prunus sibirica have not been reported to date. In this study, 104 PsbHLHs were identified and classified into 23 subfamilies that were unevenly distributed on eight chromosomes. Nineteen pairs of segmental replication genes and ten pairs of tandem replication genes were identified, and all duplicated gene pairs were under purifying selection. PsbHLHs of the same subfamily usually share similar motif compositions and exon-intron structures. PsbHLHs contain multiple stress-responsive elements. PsbHLHs exhibit functional diversity by interacting and coordinating with other members. Twenty PsbHLHs showed varying degrees of expression. Eleven genes up-regulated and nine genes down-regulated in −4°C. The majority of PsbHLHs were highly expressed in the roots and pistils. Transient transfection experiments demonstrated that transgenic plants with overexpressed PsbHLH42 have better cold tolerance. In conclusion, the results of this study have significant implications for future research on the involvement of bHLH genes in the development and stress responses of Prunus sibirica.
The bHLH transcription factor (TF) is one of the largest gene families shared by all three eukaryotic kingdoms (Li et al., 2020) and is widely distributed in plants, animals, and fungi (An et al., 2022). bHLH transcription factors include a highly conserved bHLH domain in their sequence that includes the basic, loop, and two helix regions (Zhuang et al., 2019). The basic region is composed of 10–20 amino acids and is located at the N-terminus of the bHLH domain, which recognizes and binds to DNA (Atchley and Fitch, 1997). The His-Glu-Arg sequence that binds to the cis-element E-box (CANNTG) in the plant bHLH domain’s basic region controls the capacity of bHLH transcription factors to bind DNA (Massari and Murre, 2000; Toledo-Ortiz et al., 2003). Two amphipathic α-helices are connected by a loop to form the HLH structure. Approximately 40–50 amino acids make up the HLH domain, which is found at the C-terminus of the bHLH domain. (Wu et al., 2021). Additionally, the HLH loop has a variable length and can form homo- or hetero-dimers, depending on its interaction with the hydrophobic amino acid region of another helix (Matsushima et al., 2004). bHLH TF was first discovered in plants in 1989 (Ludwig et al., 1989). Currently, bHLH superfamily genes have been found and studied in a variety of plant species, including Arabidopsis thaliana (Bailey et al., 2003), Setaria italica (Fan et al., 2021), Prunus avium (Sabir et al., 2022), and Malus domestica (Li et al., 2022). Based on conserved domains, phylogenetic relationships, and sequence homology, plant bHLH TFs are usually classified into 15–26 subfamilies (Buck and Atchley, 2003; Pires and Dolan, 2010), or up to 35 subfamilies if they contain atypical bHLH proteins (Carretero-Paulet et al., 2010).
The bHLH family has a large number of members and performs diverse and complex regulatory functions. It participates in a wide range of biological processes, including light signal regulation (Wang et al., 2020) and hormone signal transduction (Huang et al., 2022). bHLH TFs play important roles in different stages of plant growth and development, especially floral organ growth, anther development, and floral transition. Peach MYB10.1 is overexpression in tobacco plants causes changes in anther filament and pistil length, and developmental defects in male and female gametophytes by repressing NtMYB305 during flower development (Rahim et al., 2019). The SlbHLH22 gene can promote the early flowering of tomatoes (Waseem et al., 2019), whereas SmbHLH13 delays flowering by downregulating key flowering genes (Chen et al., 2023). FcbHLHs are specifically expressed in female flower tissues (Song et al., 2021). This indicates that bHLH is largely involved in floral organ development in plants. Some bHLH TFs may be activated in response to various stresses and bind to the promoters of downstream genes involved in various signaling cascades in order to control plant stress tolerance (Mao et al., 2017), such as responding to high salt, drought, and oxidative stress (Sun et al., 2018). An increasing number of bHLH TFs families have been identified with respect to their functions under low-temperature stress in plants. For example, PubHLH1 in Pyrus ussuriensis activates the transcription and expression of stress response genes to significantly enhance the resistance of transgenic plants to low temperatures and freezing stress (Jin et al., 2016). PtrbHLH improves Poncirus trifoliata’s resistance to low temperatures (Geng et al., 2019). The most well-known low-temperature response route in plants is ICE-CBF-COR (Dong et al., 2022). MdCIbHLH1 enhances the cold tolerance of Malus pumila through the C-repeat-binding factor (CBF) pathway (Gao et al., 2019). The MYC-type bHLH transcription factor ICE is recognized as a core positive regulator of cold stress response. The expression level of CsICE1 in Camellia sinensis significantly increased after low-temperature induction (Wang et al., 2012), ICE1 and its homologous gene ICE2 significantly enhanced the low-temperature resistance of A. thaliana (Xu et al., 2014).
P. sibirica, a deciduous shrub of Rosaceae, is native to Mongolia, eastern Siberia, and northern and northeastern China (Ma et al., 2020a). P. sibirica has excellent characteristics and significant ecological value that prevent wind damage, promote sand fixation, and soil and water conservation (Yin et al., 2019). In addition, P. sibirica has uses in food, medicine, and timber, and is an important ecological and economic tree species unique to Asia. Frost damage during early spring in northern China can easily disrupt the flowering, pollination, and fruiting of P. sibirica. This causes serious loss of flowers and fruits, adversely affecting the ornamental value, fruit yield, and quality of P. sibirica (Ma et al., 2020a). To prevent the reduction in yield and quality of P. sibirica due to cold damage during the flowering period, detecting cold-resistance genes is considered an important strategy for improving its resistance in P. sibirica breeding.
Over the past few decades, bHLH genes have undergone extensive functional analysis and genome-wide identification. However, there hasn’t been any reported functional study on the role of bHLH TFs in P. sibirica’s ability to withstand low temperatures. In the present study, we conducted a comprehensive genome-wide analysis of bHLH TFs in P. sibirica. The expression patterns under low-temperature stress and the tissue-specific expression of PsbHLHs were estimated using Quantitative Real-time PCR (qRT-PCR). Additionally, the gene function of PsbHLH42 in cold tolerance was verified via transient transformation. The results of this study will help predict the function of PsbHLH members and provide a theoretical basis and technical support for the realization of P. sibirica’s stable yield, high-quality cultivation, and provide gene resources for future improvement of the germplasm through genetic engineering.
In total, 104 sequences with conserved bHLH domains were identified as members of the P. sibirica bHLH family. The physical and chemical properties of the PsbHLHs are shown in Table 1. The lengths of the PsbHLHs range from 91–1159 amino acids (aa), and most of these proteins (74%) had lengths of 200–500 aa. The total number of atoms ranged from 1444 (PsbHLH16) to 17840 (PsbHLH18). PsbHLH18 had the largest molecular weight (Mw) of 128.17 kDa, while in other PsbHLHs it ranged from 10.27 kDa (PsbHLH16) to 85.93 kDa (PsbHLH101). The isoelectric point (pI) ranged from 4.6 (PsbHLH22) to 10.31 (PsbHLH32), with the average pI being 6.77. Of these, 66 PsbHLHs had pI <7, indicating acidity, while 38 PsbHLHs had pI >7, indicating alkalinity. The aliphatic index (AI) ranged from 52.34 (PsbHLH50) to 98.96 (PsbHLH80). The instability index (II) ranged from 21.81in PsbHLH16 to 82.05 in PsbHLH5. A total of 101 PsbHLHs greater than 40 were stable proteins, while three were less than 40 and belonged to unstable proteins. The predicted grand average of hydropathicity (GRAVY) values were all negative and ranged between −0.972 (PsbHLH88) and −0.271 (PsbHLH83), representing that all proteins have hydrophilic properties. Subcellular localization assays revealed that 102 PsbHLHs (98.08%) were located in the nucleus; however, PsbHLH47 was located in the chloroplast, and PsbHLH71 was found in the mitochondria.
Multiple sequence alignments were performed to analyze the sequence features and the presence of conserved amino acid residues in PsbHLHs. The bHLH structural domain of P. sibirica consisted of 66 amino acids. Visualizing the results of multiple sequence alignment allowed the identification of four conserved regions, one basic region (ERRR), two helical regions (IRLLLVP or ASLEAIYKLQQL), and a loop region (KD) (Figure 1). The size of the amino acid letters corresponding to each point represents the conservation of amino acids at each point in the bHLH transcription factor gene domain. The conservation of amino acid residues at 19 sites is greater than 50%. The conservation of Arg-11, Arg-12, Pro-27, Leu-22 and Leu-59 residues was greater than or equal to 90% implying the site was highly conserved. In addition, Helix1 is more conserved than Helix2.
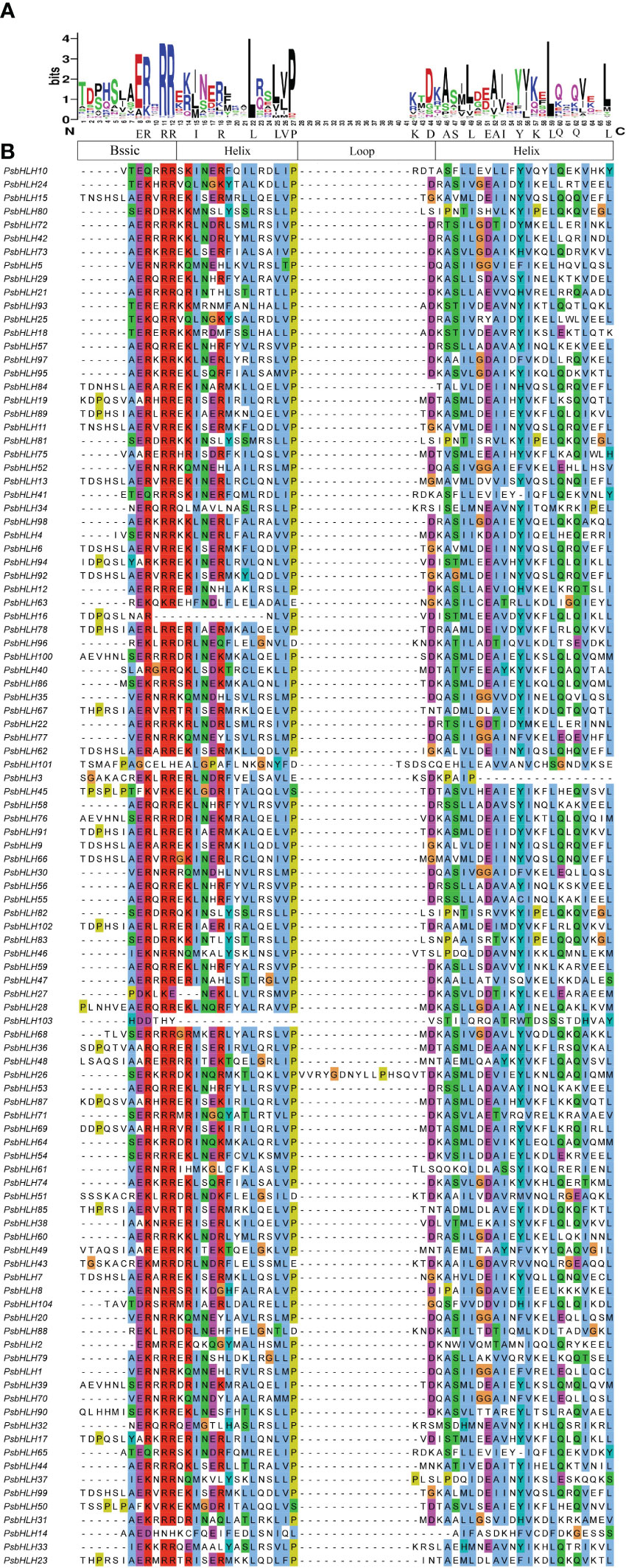
Figure 1 Characterization of PsbHLHs domains. (A) Visualization of conserved amino acids of bHLH domains. (B) Multiple sequence alignments of the bHLH domains using the Clustal color scheme.
Using the protein sequences of the 104 PsbHLHs and 135 A. thaliana bHLHs, a neighbor-joining phylogenetic tree was created to examine the evolutionary relationships and classification of PsbHLHs members. All of the bHLH proteins were divided into 26 subfamilies based on how they were categorized in A. thaliana’s bHLH proteins. The nomenclature of the PsbHLH subfamilies was consistent with that of the A. thaliana bHLH group. Among them, there were multiple members of AtbHLHs, in the Orphans, XIV, and XIII groups, but there were none of the PsbHLHs. As shown in Figure 2, the Ib subfamily had the largest number of family members, containing 12 PsbHLHs; and the smallest were subfamilies VI and XV, each containing only one PsbHLH. According to the statistics of the number of AtbHLH and PsbHLH members in each subfamily, the number of bHLHs in Va, XV, VIIIa, VIIIb, VIIIc,VI, IVb, IVd, III(d+e), IIIb, and III(a+c) were the same.
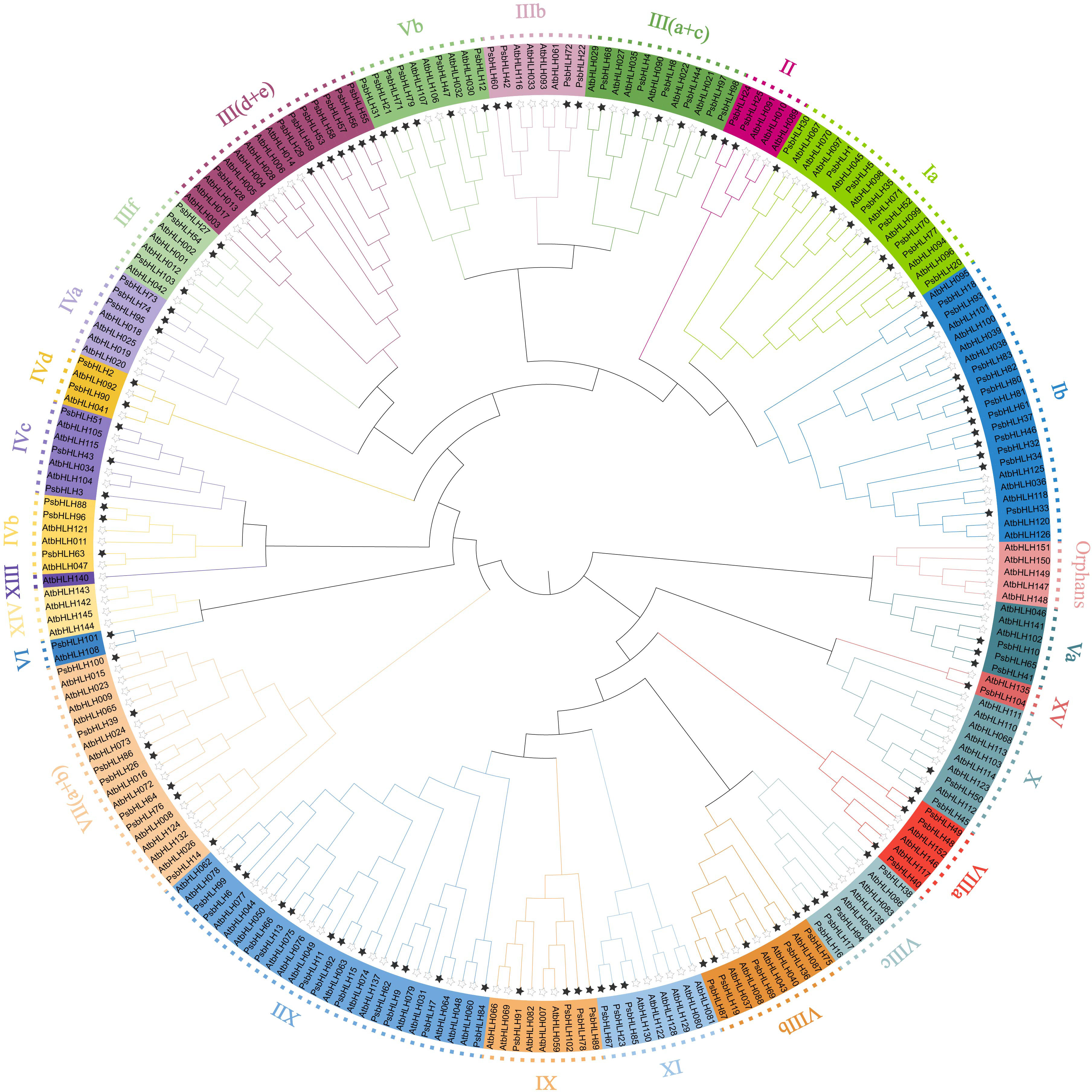
Figure 2 Phylogenetic analysis of the bHLH proteins of P. sibirica and A. tatiana. Different colors represent different groups of P. sibirica bHLH subfamilies. The black stars at the ends of branches represent P. sibirica, and the empty stars represent A. thaliana.
According to the genome annotation information, the 104 PsbHLHs were randomly distributed on eight chromosomes and numbered PsbHLH1–PsbHLH104 based on their chromosomal locations, as shown in Figure 3. The number of PsbHLHs on the chromosomes ranged from seven to 22. Chromosome 1 contained the highest number of PsbHLHs (22). The lowest number of PsbHLHs was present on chromosome 3, which had only seven PsbHLHs. Chromosomes 2, 7, and 8 contained 13 PsbHLHs each, whereas chromosomes 4, 5, and 6 contained 9, 15, and 12 PsbHLHs, respectively (Figure 3). In addition, except for chromosome 3, all the chromosomes exhibited member aggregation.
Gene duplication is considered the primary driving force in the evolution of genomes. Analysis of the segmental and tandem replication of the gene family plays a significant role in explaining the process of gene family expansion. In this study, we investigated the duplication events in the PsbHLH gene family. Gene duplication events were observed on all the chromosomes. Genes with segmental duplications were most frequently found on chromosome 1 (11 genes), followed by chromosome 5 (8 genes). In total, 48 gene duplication events representing approximately 46% (48 of 104) of the total PsbHLHs were found. A total of 34 genes underwent segmental duplication, which accounted for 65% of all syntenic relationships and played an essential role in the evolution of the PsbHLHs. These segmentally duplicated PsbHLHs formed 19 gene clusters that were linked to each other by colored lines, as presented in Figure 4. Among them, PsbHLH10, PsbHLH41, and PsbHLH65 are duplicate gene pairs with each other. PsbHLH18 forms two duplicate genes with PsbHLH86 and PsbHLH93. Moreover, 16 (35%) PsbHLHs, including PsbHLH16/17, PsbHLH32/33, PsbHLH33/34, PsbHLH55/56, PsbHLH56/57, PsbHLH57/58, PsbHLH58/59, PsbHLH73/74, PsbHLH80/81, and PsbHLH97/98, were demonstrated to be tandem duplicated genes. Gene family members formed by tandem replication are closely arranged on the same chromosome and form gene clusters with related sequences and functions. Each gene pair with duplication events belonged to the same subfamily, except for PsbHLH18 and PsbHLH86 segmental duplication gene pairs belonging to subfamilies Ib and XIII(a+b), respectively.
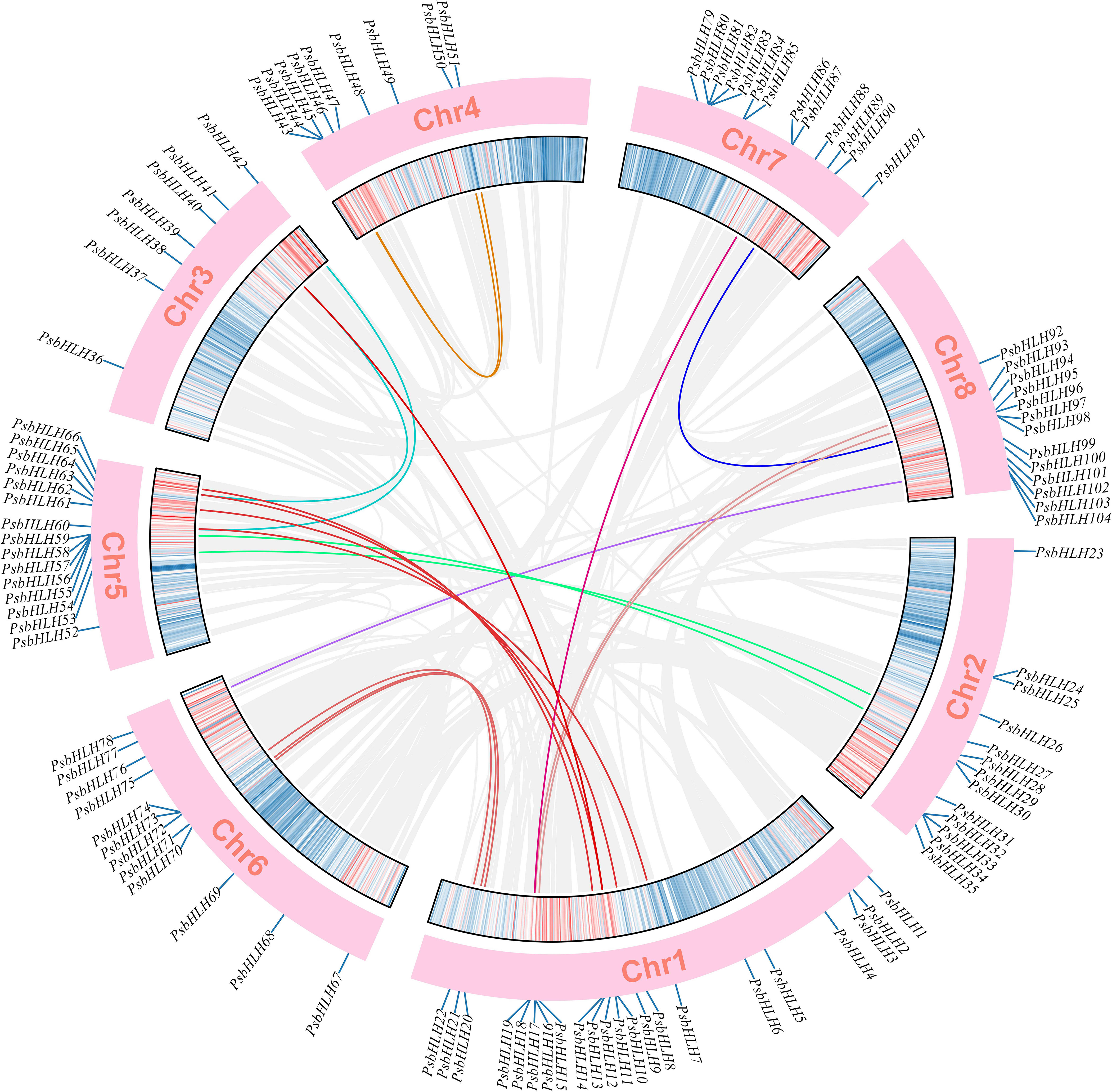
Figure 4 Analysis of PsbHLHs chromosomal distribution and duplication event. The 8 chromosomes of P. sibirica are represented by chr1-chr8. Duplication segments identified were connected by gray ribbons. Different colors are used to connect segmental duplicate PsbHLH gene pairs according to different clades. Positions are in Mb.
The selection pressure on a gene throughout evolution can be determined by Ka/Ks ratio. The Ka/Ks values for the PsbHLH gene family were calculated (Table 2). Ka/Ks >1 indicates positive selection, Ka/Ks<1 indicates purified selection, and Ka/Ks=1 indicates neutral selection. All duplicated PsbHLH pairs with Ka/Ks < 1 might have undergone purifying selection, suggesting that the majority PsbHLHs evolved slowly. Therefore, the Ks value was used to calculate the divergence time of duplicated gene pairs (Table 2). The divergence time of these gene pairs ranged from 12.22 to 78.54 Mya. The earliest gene duplication event occurred at 78.54 Mya.
To learn more about the evolutionary process of the PsbHLH gene family, a syntenic map of the bHLH genes of four Rosaceae species (Prunus salicina, P. mume, P. persica, and P. avium) and the model plant A. thalian was constructed. Red lines represent homologous gene pairs in P. sibirica and other plant genomes. Between P. sibirica and P. avium, 93 orthologous gene pairs were identified, followed by P. persica, P. mume, P. salicina, and A. thalian, with 89, 88, 86, and 66 homologous gene pairs, respectively. P. sibirica had more homologous genes with the four plants in the Rosaceae family, which is indicative of the taxonomic relationships between the species (Figure 5).
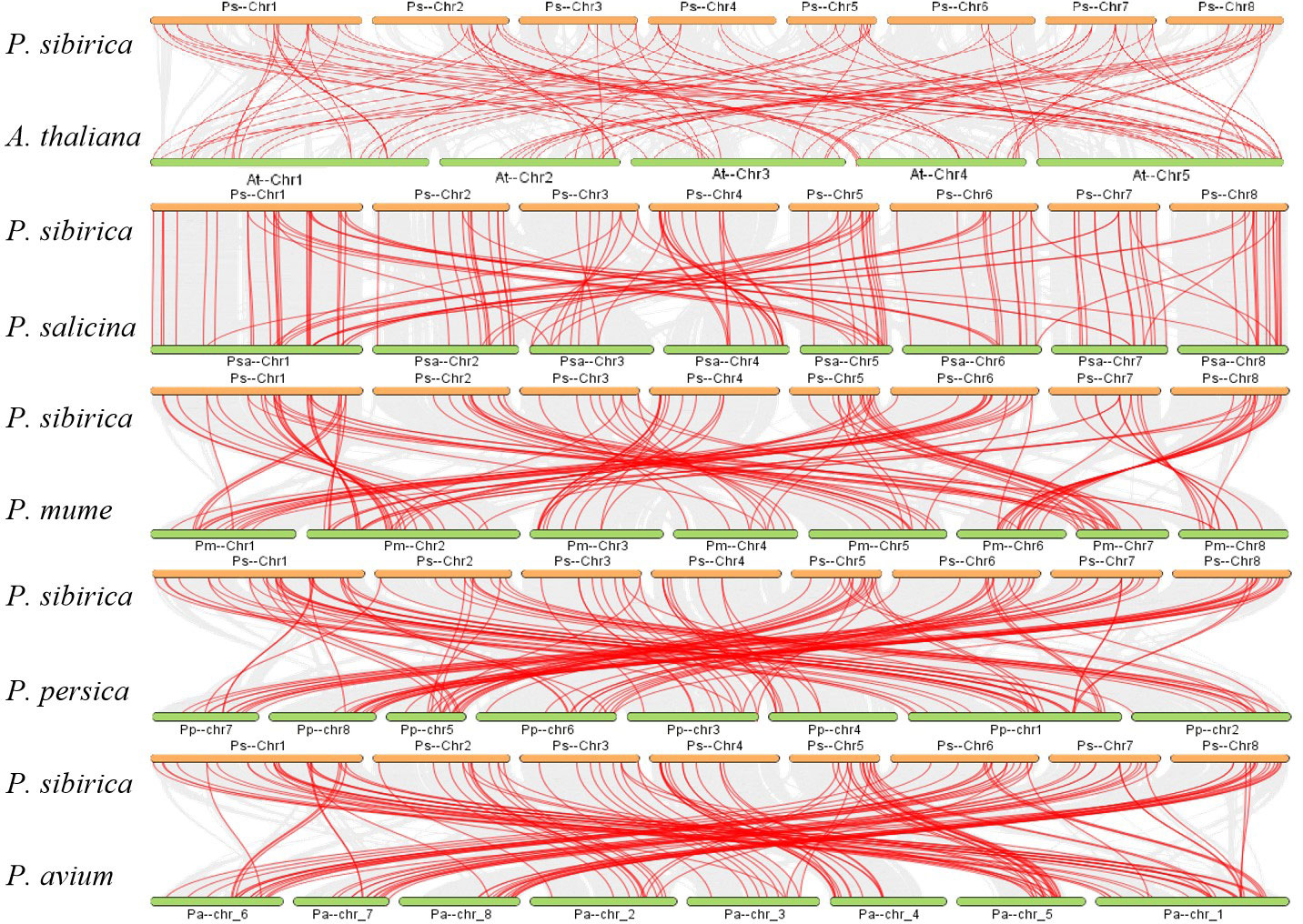
Figure 5 Synteny analysis of the bHLH genes between P. sibirica and five representative plant species. Gray lines are indicative of the collinear blocks within the genomes of P. sibirica and other plants, while red lines are indicative of the syntenic bHLH gene pairs. The plants named with the different prefixes “At,” “Psa,” “Pm,” “Pp,” and “Pa” represent A. thaliana, P. salicina, P. mume, P. persica, and P. avium, respectively.
The prediction of gene structure plays an important role in understanding the evolution of gene family members. The exon-intron structures within PsbHLHs were investigated to determine their genetic features and structural characteristics. Different subfamilies had different intron/exon patterns, but the PsbHLHs sequences within the same subfamily had similar numbers of exons and introns, especially for PsbHLHs on homologous branches (Figure 6). The number of introns ranged from 0 to 17. No introns were present in PsbHLH19, PsbHLH40, PsbHLH69, PsbHLH75, or PsbHLH89. Exon analysis revealed that the number of exons ranged from 1 to 16. In addition, 34 PsbHLHs had no untranslated region (UTR). Gene structure analysis revealed differences in the structural composition of PsbHLHs.

Figure 6 Phylogenetic relationships, conserved motifs, and gene structural analyses of the PsbHLH gene family. (A) Neighbor-joining phylogenetic tree of the PsbHLHs. (B) Gene structure of the PsbHLHs. The green box indicates the exons, the yellow box the untranslational regions (UTRs), and the gray line the introns. (C) PsbHLH proteins conserved motif distribution. Different motifs are represented by different colored boxes.
Twenty motifs were identified in the PsbHLHs using Motif Elicitation (MEME). As shown in Figure 6, different PsbHLHs featured various amounts of conserved motifs, ranging from 1 to 12. Motif 1 and motif 2 constitute the conserved domain of bHLH, motif 1 contains one basic and one helical region, whereas motif2 contains one loop and another helical region (Table S1). Except for PsbHLH2, PsbHLH3, PsbHLH16, PsbHLH27, and PsbHLH103, almost all of the PsbHLHs contained motif1 and motif 2, and both of these conserved domains were close to one another. Interestingly, PsbHLH18 has two conserved domains (motif 1 and motif 2). PsbHLHs grouped in the same subfamily frequently shared consistent motif patterns. In addition, motifs 19, 12, 13, and 17 occurred only in subfamilies XII, VIIIb, III (d+e), and Ib, respectively.
Cis-regulatory elements play an important role in regulating plant growth, development, and stress responses. To predict the regulatory pattern of PsbHLHs, the −2000 bp sequence upstream of ATG were analyzed in the promoter region of 104 PsbHLHs using PlantCARE (Figure 7). The discovery of numerous hormone- and stress-responsive elements suggests that PsbHLHs may possibly be crucial in hormone signal transduction and stress response. Cis-elements we screened were roughly divided into two categories: stress-responsive elements (TC-rich repeats and LTR) and hormone-responsive elements (ABRE, TGA-element, TCA-element, TGACG-motif, CGTCA-motif, GARE-motif, TATC-box, and P-box). Most cis-elements in the promoter regions of PsbHLHs were ABRE (289), followed by the TGACG motif (133) and the CGTCA motif (133). Additionally, 39 members had auxin-responsive element (TGA-element), 47 members had cis-acting element involved in defense and stress responsiveness (TC-rich repeats), 69 members had cis-acting element involved in gibberellin-responsiveness and gibberellin-responsive element (GARE-motif, TATC-boxes and P-box), 52 members had cis-acting element involved in low-temperature responsiveness (LTR), 55 members had cis-acting element that was responsive to salicylic acid (TCA-element), 89 members had cis-acting element that was responsive to the abscisic acid (ABRE), and 67 members had cis-acting regulatory element that was responsive to the MeJA (TGACG-motif and CGTCA-motif). In addition, PsbHLH9 had the most cis-elements (22), followed by PsbHLH86 (20).
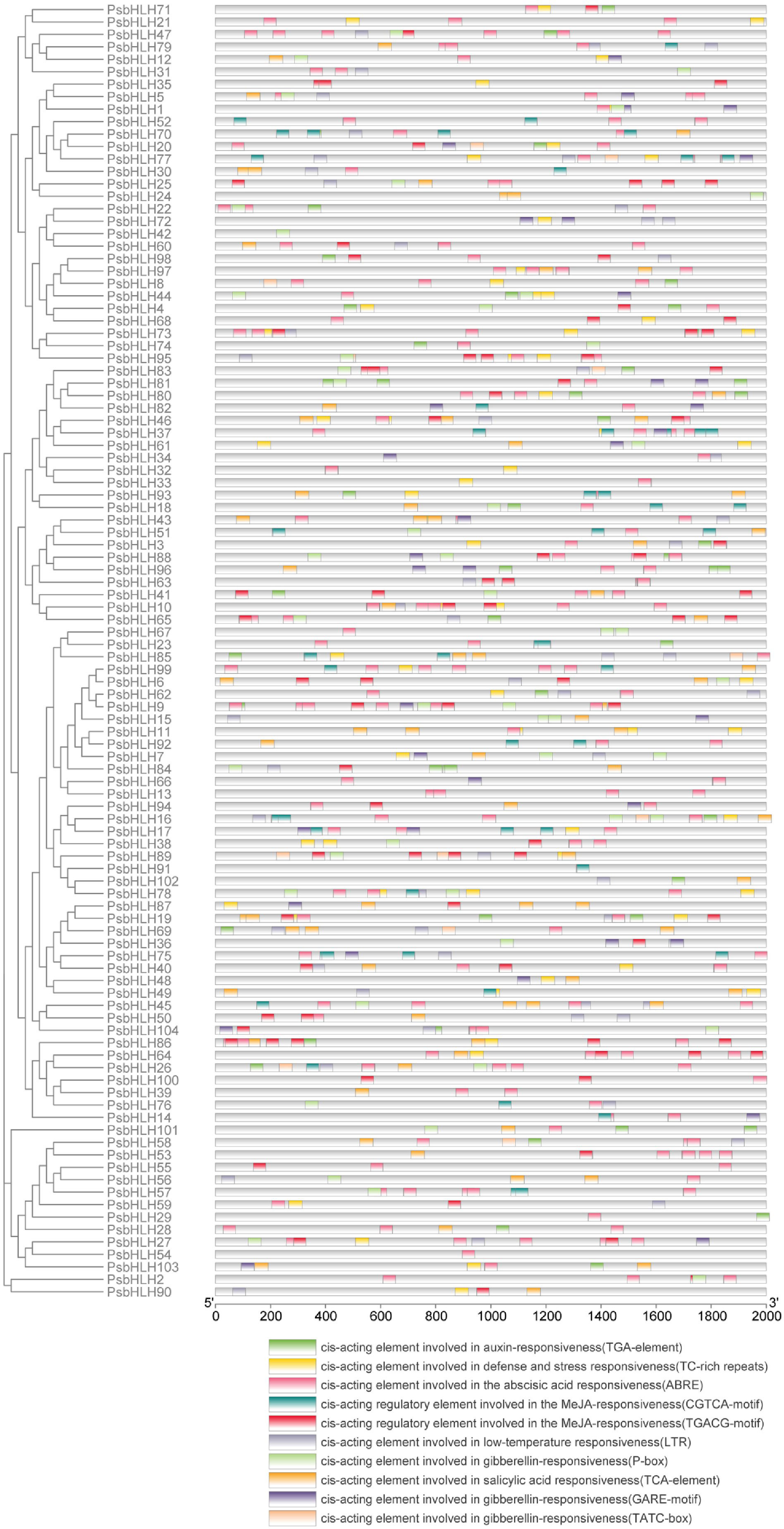
Figure 7 The −2000 bp sequence upstream of ATG was used as the promoter regions to identify the cis-elements. Ten cis-elements were represented in different colored blocks.
To better understand the function of PsbHLHs as TFs, the EggNog website was used for gene ontology (GO) annotation analysis. Among the 104 PsbHLHs, 72 were annotated using GO (Figure 8). The results showed that 72 PsbHLHs were annotated to eight cellular components, 70.83% of the genes were located in cells and cell parts, 69.44% in organelles and less than 20% in other cellular components. Molecular functions include transcriptional regulator activity, binding, molecular transducer activity, and catalytic activity. PsbHLHs were annotated in 17 biological processes. All PsbHLHs were involved in the regulation of biological processes, and 71 genes (98.6%) were involved in cellular and metabolic processes. Thirty genes were involved in response to stimuli, 22 genes in the developmental processes, and 15 genes in reproductive processes. In addition, PsbHLH16 had the most annotations (16), whereas PsbHLH91 was annotated for 11 biological processes.
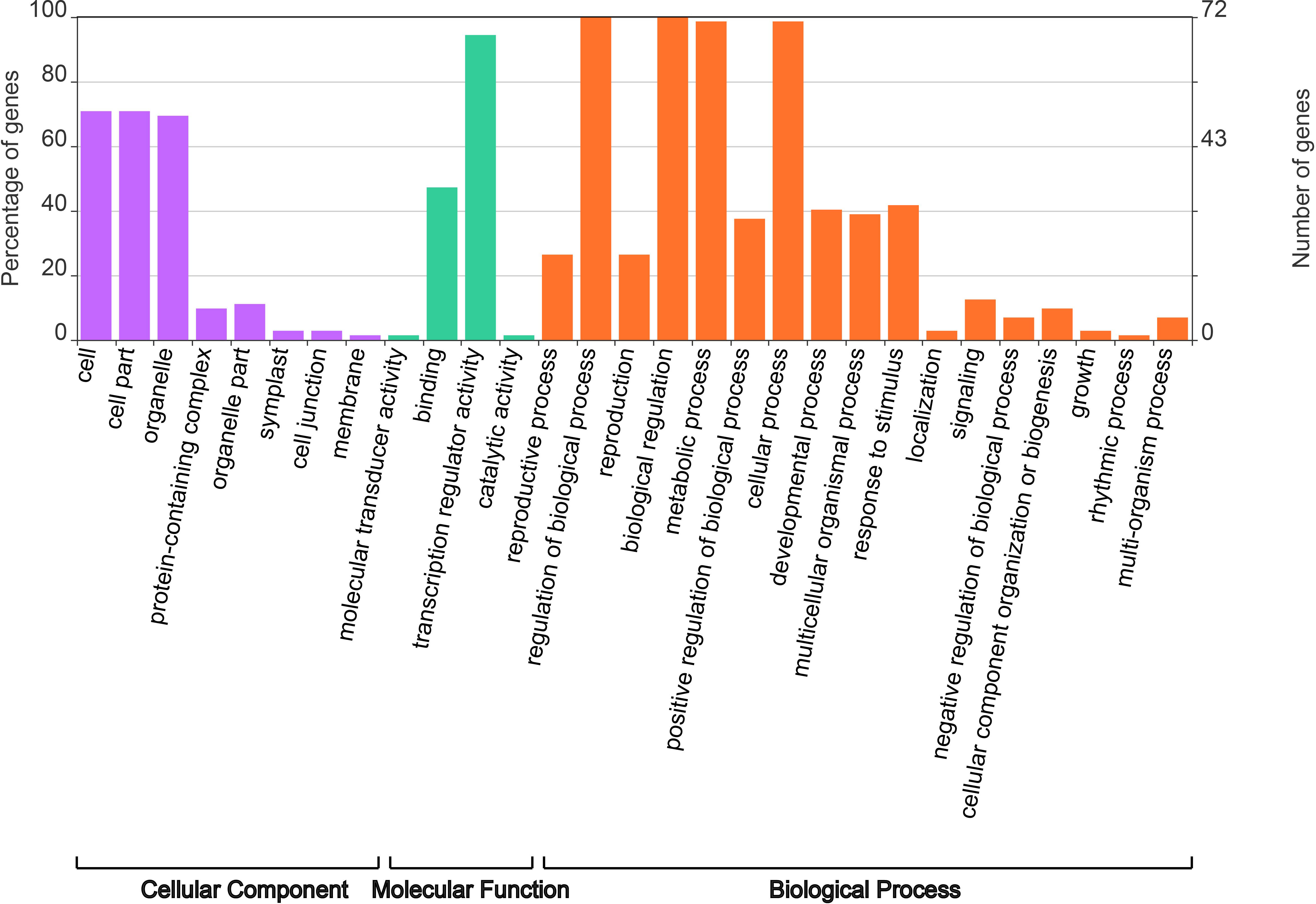
Figure 8 Gene ontology (GO) annotation analysis of PsbHLHs. GO annotation of PsbHLHs was performed by three categories including cellular component, molecular function and biological process.
Twenty candidate genes were identified by integration and overall consideration of the results from the phylogenetic analysis, GO annotation, and cis-element analysis of PsbHLHs to further study the expression patterns and regulation processes in P. sibirica. These 20 genes expression levels were quantified indifferent tissues (roots, stems, leaves, pistils, and stamens) using the expression levels in petals as reference. Nine genes in pistils and 11 genes in the roots were highly expressed. Four genes were highly expressed in the stems and stamens. While, only two genes (PsbHLH54 and PsbHLH88) were highly expressed in leaves. PsbHLH39 was highly expressed in all five tissues. Five PsbHLHs (PsbHLHs19/29/38/44/66) were expressed in one tissue (Figure 9). PsbHLH39 was highly expressed (>60 times) in all tissues except the stamens. The expression levels of seven PsbHLHs were upregulated by more than 60 times in the roots, PsbHLH5/29/60 exceeded by 80 times; PsbHLH11 was upregulated by 80 times in the pistil; PsbHLH35 was upregulated by more than 60 times in the pistil and stem; and PsbHLH73 was upregulated by 90 times in the stamen.
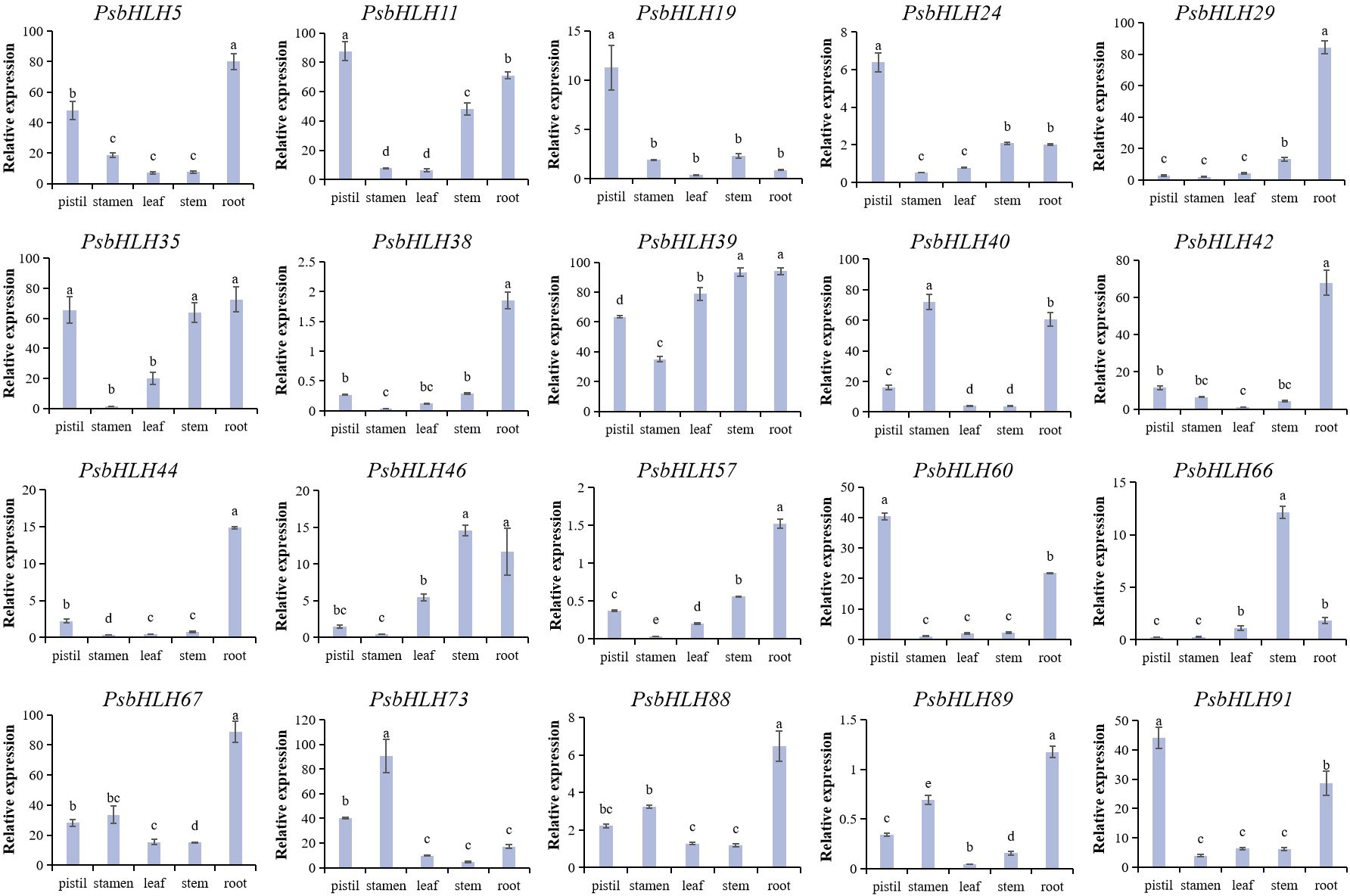
Figure 9 Expression patterns of the 20 PsbHLHs in different tissues (pistil, stamen, root, stem, and leaf) were examined by qRT-PCR. The expression level of the 18S rRNA gene was used as the internal control. Error bars were obtained from three biological replicates and represent standard error. Significant differences achieved by Duncan’s test (p < 0.05) using SPSS are represented by different letters.
In order to further explore the involvement of PsbHLHs in low-temperature tolerance, the expression levels of 20 genes at low temperature (−4°C; 0 h, 15 min, 30 min, 1 h, and 2 h) were detected by qRT-PCR. The relative expression level (0 h) in the control pistils without treatment was normalized to 1. The expression patterns of 11 PsbHLHs (PsbHLH29/35/38/39/40/42/44/46/57/60/89) showed upregulation, whereas nine PsbHLHs (PsbHLH5, 11, 19, 24, 66, 67, 73, 88, and 91) displayed a downregulation. In the upregulated group, all 11 genes were highly expressed, particularly PsbHLH44, which displayed the highest expression level under the low-temperature treatment. Seven PsbHLHs (PsbHLH29/35/38/39/40/42/89) genes were first upregulated at 15 min, followed by slight downregulation at 30 min, and then continued to be upregulated at 1 or 2 h of treatment. In addition, the expression levels of PsbHLH44/46/57/60 reached a maximum at 15 min or 1 h and then decreased with prolonged treatment time (Figure 10).
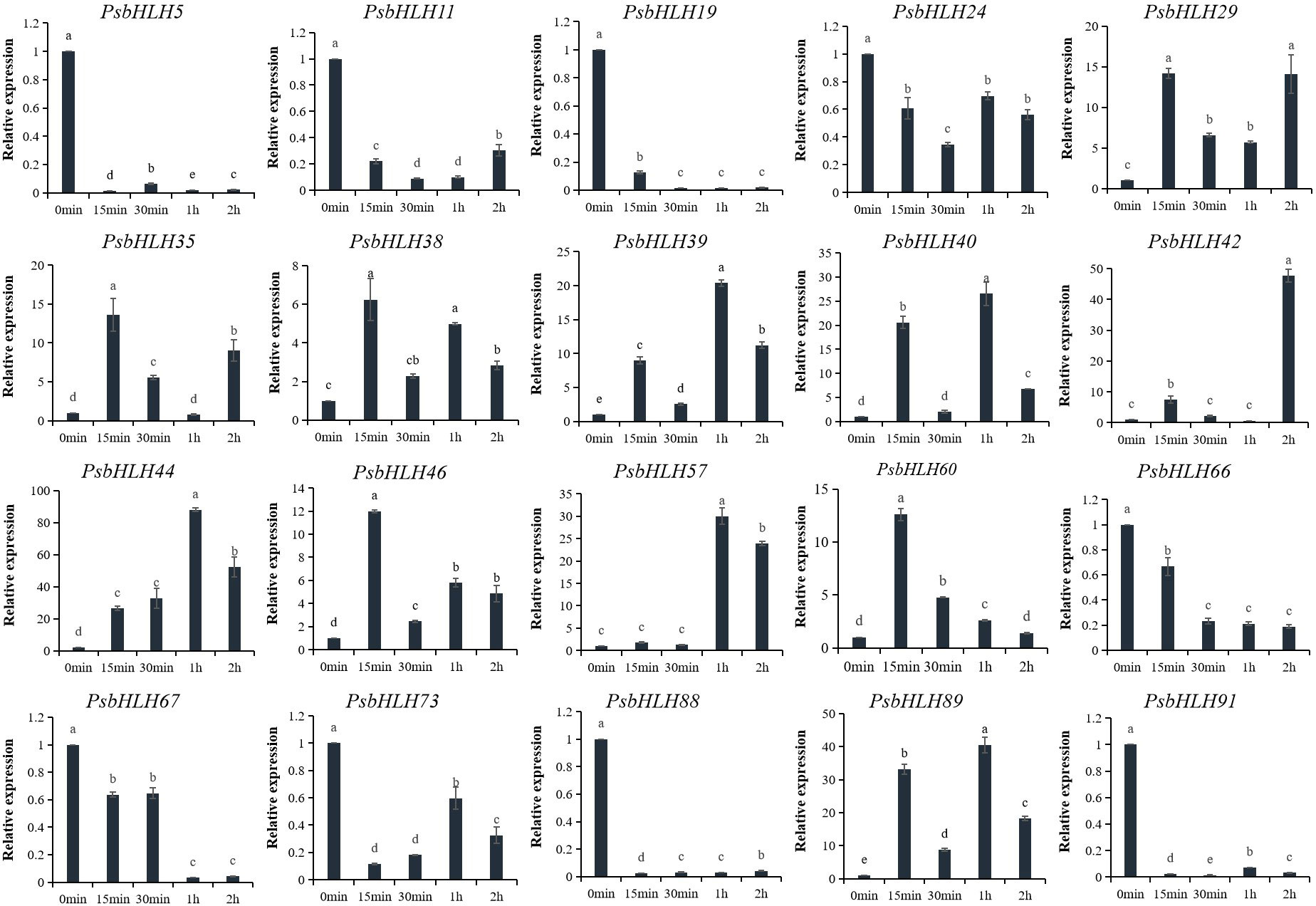
Figure 10 Expression patterns of 20 PsbHLHs in response to low-temperature stress treatment were examined by qRT-PCR. The expression level of the 18S rRNA gene was used as the internal control. Error bars were obtained from three biological replicates and represent standard error. Significant differences achieved by Duncan’s test (p < 0.05) using SPSS are represented by different letters.
Based on these syntenic relationships, orthologous AtbHLHs from 104 PsbHLHs were selected in A. thaliana to predict interactions. Most of the PsbHLHs were predicted to exhibit interactions (Figure 11). Sixty bHLHs interacted with more than one bHLH protein, with PYE (PsbHLH63) having the largest number of interacting partners (24). Detailed information on the interactions is summarized in Table 3. In addition, the gene function of the homolog PsbHLHs was detected based on its verified gene function in A. thaliana. Since PsbHLH42 is homologous to the cold-inducible gene ICE1, it is very likely to take part in P. sibirica’s reaction to low-temperature stress.
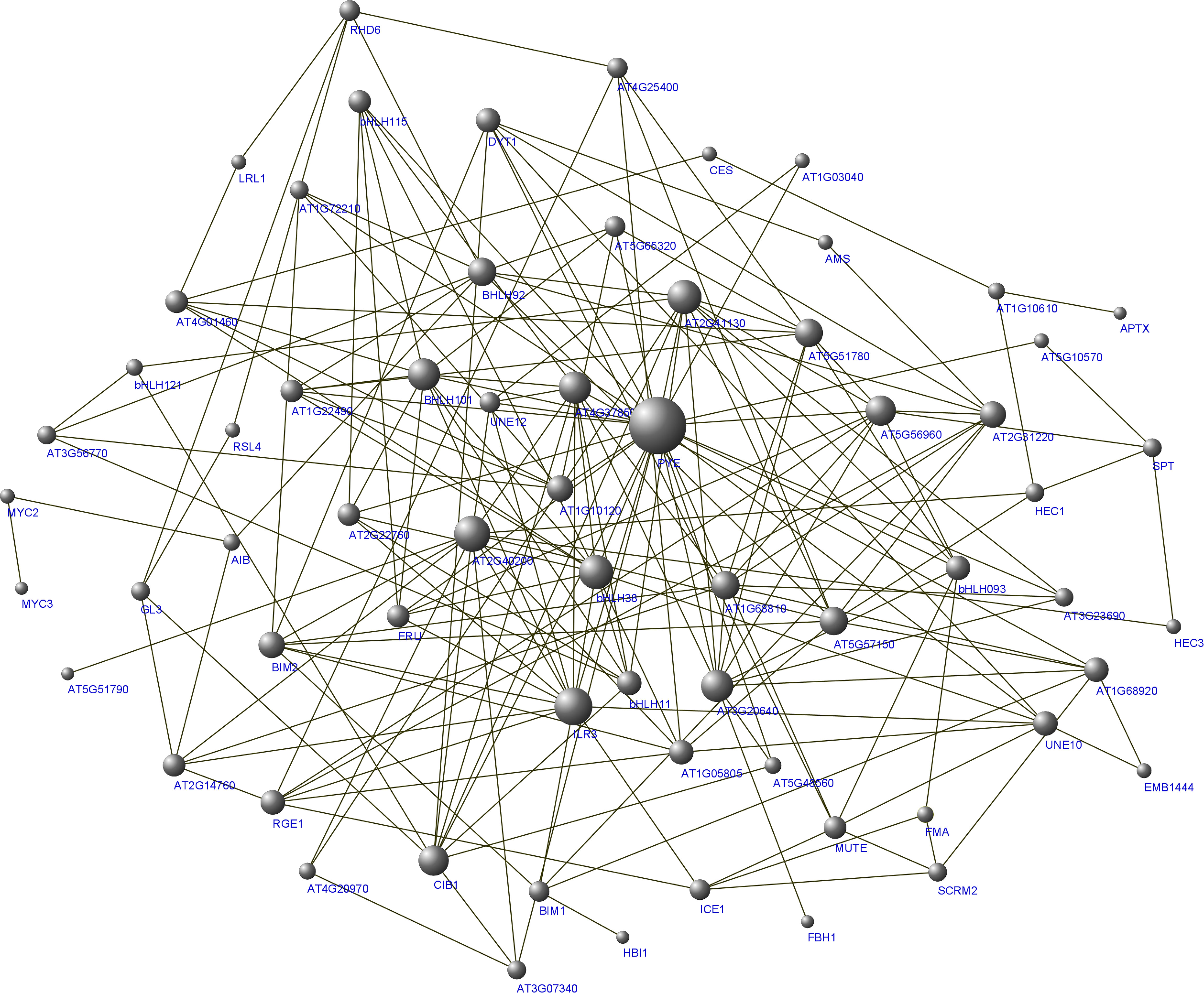
Figure 11 Protein interaction network for PsbHLHs according to bHLH orthologs in A. thaliana. The online tool STRING was used to predict the network.
Based on bioinformatics studies, expression pattern analysis, and homology with ICE1, the involvement of PsbHLH42 in the cold response was hypothesized. To investigate the function of PsbHLH42 in low-temperature stress, the pRI101-PsbHLH42-GFP vector was constructed and transiently transformed tissue culture seedlings of P. ussuriensis by the Agrobacterium-mediated method. An empty vector was transiently transformed into P. ussuriensis as the control. The qRT-PCR results demonstrated that the expression level of PsbHLH42 in the transgenic plants was significantly higher than that in the control plants after transient transformation, indicating that PsbHLH42 was successfully transferred into P. ussuriensis and further analysis could be carried out. The seedlings of the control and transgenic plants were subjected to 4°C stress for 6 h to evaluate the cold resistance of PsbHLH42. Before low-temperature treatment, the physiological signs did not alter significantly between the transgenic and control strains (Figure 12). After low-temperature treatment, compared with the control plants, the transgenic plants that overexpressed PsbHLH42 had higher soluble sugar and soluble protein contents, higher Superoxide dismutase (SOD) and peroxidase (POD) activities, and lower electrical conductivity and Malondialdehyde (MDA) content.
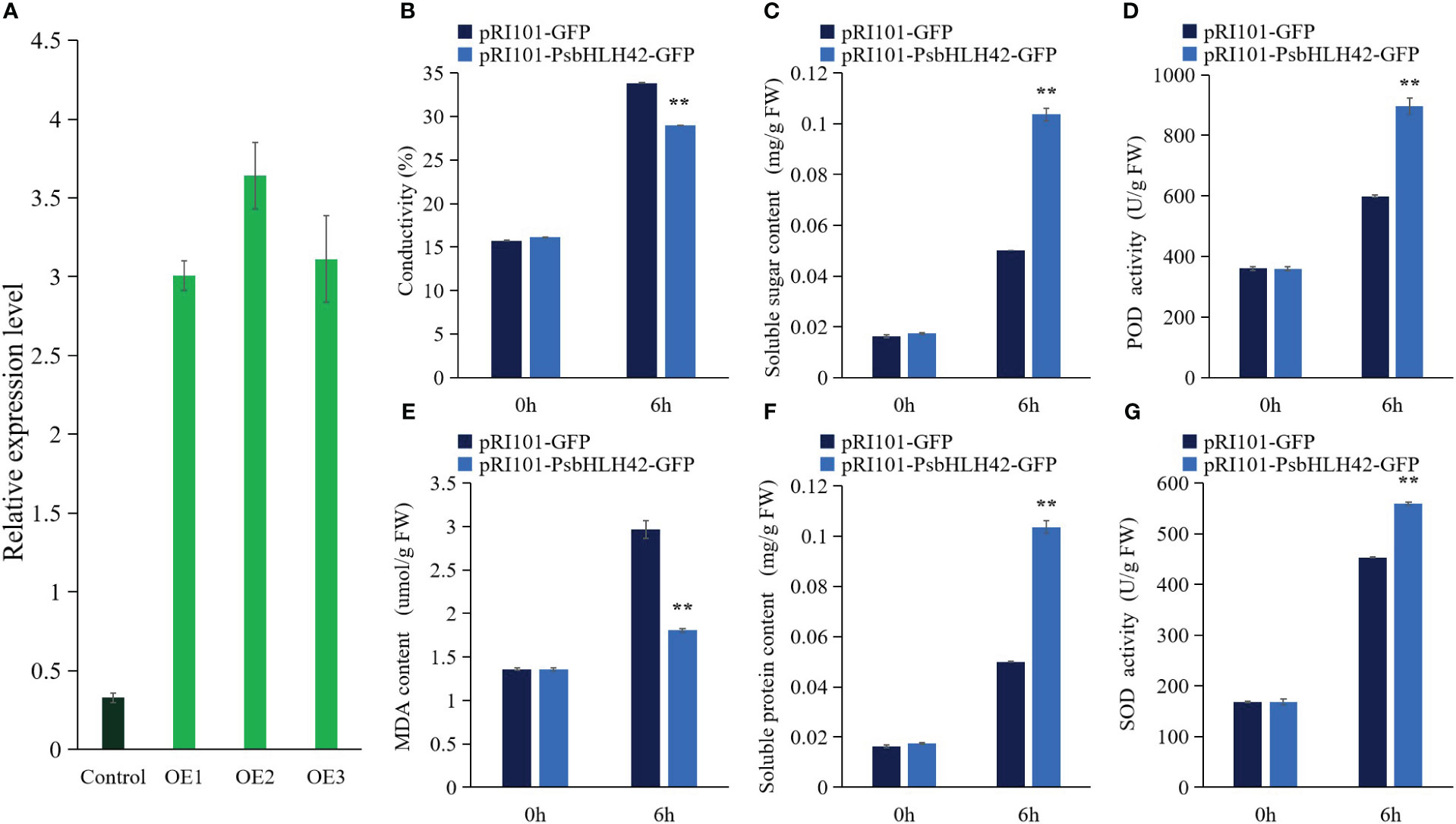
Figure 12 PsbHLH42 can improve plant resistance to low temperature. Physiological indicators were determined in empty vector control and PsbHLH42 overexpressed plants after 0 h and 6 h at 4°C low-temperature treatment. pRI101-PsbHLH42-GFP and pRI101-GFP indicate transgenic plants and empty vector transgenic plants, respectively. (A) Control, empty vector control plants; OE1, OE2 and OE3, three PsbHLH42 transgenic plants. Relative expression levels of PsbHLH42 in the transgenic and control P. ussuriensis plants. 18S rRNA was used as an internal control. (B) Relative conductivity. (C) Soluble sugar content. (D) POD activity. (E) MDA content. (F) Soluble protein content. (G) SOD activity. Error bars were obtained from three biological replicates. The statistical analyses were performed using Student’s t tests (**p < 0.01).
Basic helix-loop-helix (bHLH) TFs play a crucial roles in a variety of processes of plant growth, metabolism, and abiotic stress response (Zuo et al., 2023). However, the characterization of bHLH genes in P. sibirica has not been reported. In the present study, we identified 104 PsbHLHs. The number of bHLH TFs varied greatly among the different plants. The number of members in the P. sibirica bHLH gene family is smaller than that of many other plants, such as Oryza sativa (183) (Wei and Chen, 2018), Triticum aestivum (225) (Guo and Wang, 2017), Gossypium hirsutum (437) (Liu et al., 2019), and Zea mays (208) (Zhang T. et al., 2018). However, it is similar to members of the Rosaceae family, such as P. salicina (95) (Zhang C. et al., 2018), P. mume (100) (Wu et al., 2022), and Rosa chinensis (100) (Zhou et al., 2020). This indicates that the evolution of the bHLH gene family is highly conserved from herbaceous plants to woody plants. The fact that 98.08% of PsbHLHs (102) were located in the nucleus, indicates that PsbHLHs mainly function as TFs in the nucleus. Meanwhile, PsbHLH47 is located in the chloroplasts, and PsbHLH71 is located in the mitochondria. It has been speculated that these genes are mainly involved in the transcriptional regulation of photosynthesis or functional mitochondrial transcription, such as respiration, in P. sibirica.
Structural variation plays a crucial role in the process of gene evolution. Introns evolve with the evolution of the Genome evolution mainly represented in the gain, loss, insertion, or deletion of introns and exons (Zhao et al., 2022). The functions of genes in the identical subfamily can also be comparable or there may additionally be functional differentiation, according to the results of gene structure analysis, which revealed similarities and differences among members of the same subfamily (Zhang et al., 2022). The five genes identified without introns were all distributed in the VIIIb subfamilies, which proved that the subfamily classification was accurate. Except for PsbHLH2/3/16/27/103, all other PsbHLHs contained complete conserved bHLH domain (Motif 1 and Motif 2), suggesting that these five proteins have bHLH domains but are incomplete and belong to special bHLH family members (Carretero-Paulet et al., 2010). In addition, PsbHLH18 has two conserved domains (double motif 1 and motif 2), suggesting that it has undergone certain mutations or replication of the conserved domain during evolution, which may have improved its binding (interaction) ability with downstream genes. In addition to the bHLH domain, other conserved motifs also perform other key functions that are essential for each subfamily. Proteins in the same subfamily have similar conserved motif patterns, indicating that they may have similar functions (Eulgem et al., 2000). Some motifs are unique, with motifs 12, 19, 13, and 17 present in only one subfamily. The uniqueness and conservation of these motifs are also evidence of the evolutionary classification of the PsbHLH gene family. Interestingly, PsbHLH28 and PsbHLH98 belong to the III (d+e) and III (a+c) subfamilies but are closer to the IIIf subfamily in motif patterns. These results indicate both functional conservation and differentiation in PsbHLHs.
Gene duplication plays a crucial role in the acquisition of new genes and supplies the building blocks for the development of genetic diversity (Ma et al., 2021). Duplication analysis indicated that the PsbHLH gene family expanded through segmental (65%) and tandem (35%) duplication events, with segmental duplication being the main mechanism of amplification. The duplicated genes take new functions, whereas the original function of the other gene copy is maintained in the homologous genes (Baldini et al., 2022). Except for PsbHLH18 (subfamily Ib) and PsbHLH86 (subfamily VII (a+b)), all duplication events occurred within the same subfamily, suggesting that after segmental replication, new genes (PsbHLH18 or PsbHLH86) may undergo functional differentiation. Ka/Ks values can be used as indicators of the selection pressure on a gene during evolution (Chen et al., 2022). Due to the degeneracy of the code, sites within codons were defined as synonymous substitutions (Ks) when they did not result in an amino acid change, and as a non-synonymous substitutions (Ka) when they did (Harouaka et al., 2016). Generally, Ka/Ks >1 indicates positive selection, Ka/Ks =1 indicates neutral selection, and Ka/Ks <1 indicates purified selection (Lin et al., 2021). Based on the Ka/Ks ratio, all PsbHLH pairs were subjected to purifying selection.
Due to segmental or tandem duplication and gene loss events, various species have variable numbers of subgroups and genes per subfamily (Sheng et al., 2022). Phylogenetic tree analysis showed that the 104 PsbHLHs could be divided into 23 subfamilies, consistent with the results for Carthamus tinctorius (23) (Hong et al., 2019). The lack of PsbHLHs in the three subgroups (Orphans, XIV, and XIII), when compared to A. thaliana, may be due to gene loss during the evolution of P. sibirica or natural selection leading to elimination; this reflects that the bHLH gene is highly conserved during the evolution from herbs to woody plants. There was only one member each in subclasses XV (PsbHLH104) and VI (PsbHLH101), suggesting that the members of these subclasses evolved relatively slowly, were relatively functionally conserved, and may play an important role in P. sibirica.
Cis-regulatory elements (CREs) in promoter regions exert a significant influence on gene functions (Hou et al., 2021). The promoter prediction results indicated that PsbHLHs contained a large number of hormone and stress response elements. PsbHLH16 contains eight types of elements, PsbHLH37 and PsbHLH86 contains 10 MeJA elements, and PsbHLH9 contains 10 ABA elements, indicating that these genes are most likely to be induced by different hormones to affect development or stress in P. sibirica. GO annotation revealed that the regulatory functions of PsbHLHs were diverse and complex. According to annotations, 69% of PsbHLHs were participating in the regulation of biological processes. Anthocyanins are naturally occurring plant pigments with antioxidant properties (Taghavi et al., 2022). Previous research has demonstrated that TT8 regulates anthocyanin biosynthesis (Ahn et al., 2022), and it has been speculated that the homologous gene PsbHLH103 participates in anthocyanin synthesis in P. sibirica. In addition, PsbHLH44 and PsbHLH97 interact and are both annotated to be involved in reproductive and developmental processes; their homologs, DYT1 (PsbHLH44) and AMS (PsbHLH44), are both involved in stamen development in Arabidopsis, suggesting that PsbHLH44 and PsbHLH97 are most likely to interact and regulate stamen development in P. sibirica. Similarly, PsbHLH36 (HEC3) and PsbHLH100 (SPT) may interact to regulate P. sibirica pistil development.
To a certain extent, gene expression patterns reflect their function. bHLH family members are expressed to varying degrees in different organs of P. sibirica. Many PsbHLHs are highly expressed in the roots, which is in line with the conclusions in P. mume (Wu et al., 2022). PsbHLH38 is expressed in roots, and the homologous gene RHD6 guides root hair development in Arabidopsis through processes involving auxin and ethylene, suggesting that PsbHLH38 is also involved in root development. According to previous reports, MYC participates in the Jasmonic acid (JA) signaling pathway and directly or indirectly controls stamen development through the ABA-JA pathway (Li et al., 2023). PsbHLH40/73 contains five ABAs and 4/6 MeJA, respectively, suggesting that they may be involved in the development of P. sibirica stamens. AtHEC1 is involved in female development and fertility (Gremski et al., 2007), and its homologous gene, PsbHLH19, is highly expressed, and is speculated to regulate pistil development. Therefore, PsbHLHs play an important role in the growth and development of different organs in P. sibirica.
Low-temperature stress (late frost damage) severely affects the yield and quality of P. sibirica. To further investigate the function of PsbHLHs in low temperature regulation, the expression profiles of PsbHLHs under low temperature stress were analyzed. Twenty PsbHLHs exhibited different expression patterns under low-temperature treatment. For example, MYC2 (homolog of PsbHLH29) is a core transcription factor of the JA signaling pathway that reduces damage to plants during abiotic stress by participating in the regulation of JA-inducible genes (Oña Chuquimarca et al., 2020; Xu et al., 2022). Studies have shown that MeJA can improve plants’ freezing tolerance by reducing membrane lipids and maintaining their stability (Ma et al., 2020b). In this study, PsbHLH29 was highly expressed after 15 min and 2 h of cold stress. This may be the result of plasma membrane damage in P. sibirica during the early stages of cold induction. Initially, JA was induced to regulate membrane permeability after reducing plant damage, after that decrease expression; again, JA was expressed as the stress time increased and the degree of cell membrane damage increased. This suggests that PsbHLH29 may regulate cold resistance in plants by regulating JA. AtAIB positively regulates the ABA response in Arabidopsis (Li et al., 2007). PsbHLH53(MYC3)/95(AIB) contained ABA response cis-elements, suggesting that PsbHLH53/95 may also participate in P. sibirica cold response by regulating ABA. PPI analysis revealed that PsbHLH29 (MYC2) interacts with PsbHLH53/95, suggesting that ABA and JA operate together to regulate the freezing tolerance of flowering organs of P. sibirica. The number and kind of cis-acting elements on the promoter may also have a bearing on how well a gene functions, with PsbHLH44 containing three gibberellins-responsive elements that were upregulated 80-fold in response to low-temperature stress. Meanwhile, 89 contained eight MeJA-responsive elements that were also up-regulated 30-fold early (within the first 1 h), suggesting that these two genes are induced by gibberellins and MeJA to regulate the freezing resistance of P. sibirica flowering organs. PsbHLH35, 38, 46, and 60 expression reached its highest level after 15 min of cold treatment. This phenomena could have several causes, one of which is that during the initial phases of cold treatment, active regulatory factors are quickly engaged and operate on the bHLH protein to adapt to the stressful environment, which causes rapid upregulation of bHLH. To avoid an excessive amplification of stress signals, plants must maintain a balance between normal development and adaptation to cold settings when the cold treatment time is extended. At this moment, negative regulators become active and encourage the breakdown of the bHLH protein, delaying the expression of the bHLH gene (Tan et al., 2021).
Studies have demonstrated that ICE1 and ICE2 improve the freezing tolerance of transgenic plants (Liu et al., 2018; Ma et al., 2023). Here, PsbHLH42 (ICE1) was significantly upregulated after −4°C treatment, suggesting that it may play an important role in the cold stress response of P. sibirica. PsbHLH60 interacts with PsbHLH42 (ICE1) and contains a low-temperature response element, whose homologous gene SCRM2 encodes ICE2, suggesting that PsbHLH42 and PsbHLH60 may co-regulate the P. sibirica cold response. In addition, nine genes were downregulated after low-temperature stress, which may have been caused by the brief stress period (2 h) that was insufficient to induce gene expression. In conclusion, the above results suggest that the PsbHLH gene family may play a significant part in the cold stress response pathway.
The homolog of ICE1 gene, PsbHLH42, was used to verify its function in resisting low temperatures. Transient genetic transformation assays can rapidly identify gene function (Han et al., 2023). Because the lack of mature genetic transformation and regeneration systems in P. sibirica, we used the ideal model tree plant P. ussuriensis as the genetic transformation material (Jansson and Douglas, 2007; Movahedi et al., 2014). In this experiment, the PsbHLH42 transgenic strain showed reduced membrane damage, increased osmoregulatory substance content, and increased SOD and POD activities after low-temperature treatment compared to the control. Overexpression of the ICE1 homologous gene PsbHLH42 improved the low-temperature tolerance of P. ussuriensis. In addition, PsbHLH42 interacts with PsbHLH5, and MUTE (PsbHLH5) participates in plant stomatal development (Lampard and Bergmann, 2007); therefore, it is speculated that they may be involved in the low-temperature response by regulating stomata. To further verify the function of this gene, stable genetic transformation experiments will be conducted in the future.
Sequences of P. sibirica (Prunus sibirica F106 Genome v1.0) were obtained from the Genome Database for Rosaceae (GDR, https://www.rosaceae.org/) (Jung et al., 2019). The hidden markov model (HMM) file of the bHLH domain (PF00010) was downloaded from the Pfam database (http://pfam.xfam.org/) (Mistry et al., 2021) and used for protein screening using HMMER software (version 3.3.2). The online CD-search tool (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) (Lu et al., 2020) and Pfam were used to validate the bHLH domains of these protein sequences, and the physical and chemical parameters were calculated using ExPASy (https://www.expasy.org/) (Duvaud et al., 2021). The CELLO program (http://cello.life.nctu.edu.tw/) (Yu et al., 2006) was used to predict the subcellular localization.
MEGA software (version 11.0) and Jalview software (version 2.11.2.0) were used to visualize the sequences of the conserved domains in PsbHLHs. The sequence logo for the bHLH domain was created by submitting multiple alignment sequences to WebLogo (http://weblogo.berkeley.edu/logo.cgi) (Crooks et al., 2004).
The full-length bHLHs protein sequences from P. sibirica and A. thaliana were aligned using ClustalX (version 2.1). Thereafter, a phylogenetic tree was constructed based on the neighbor-joining (NJ) method using MEGA with the following parameter settings: Poisson’s model, pairwise deletion option, and 1000 bootstrap replicates provided for statistical reliability. A phylogenetic tree was constructed using iTOL (https://itol.embl.de/) (Letunic and Bork, 2021). The A. thaliana bHLH protein sequences were downloaded from TAIR (https://www.arabidopsis.org/) (Berardini et al., 2015). Subfamily grouping of PsbHLHs was performed according to the AtbHLH classification scheme.
Chromosomal locations were identified and plotted using TBtools and the P. sibirica genomic database. The nonsynonymous (Ka) and synonymous (Ks) substitution ratios were calculated using the Simple Ka/Ks Calculator in TBtools software to acquire the natural purification selection between target gene pairs. The divergence time was calculated using the formula T = Ks/2r, with Ks being the synonymous substitutions per site, and the r of dicotyledonous plants being 1.5×10−8 synonymous substitutions per site per year (Gaut and Doebley, 1997). Finally, sequences with complete bHLH domains were preserved and numbered PsbHLH1 through PsbHLH104 to correspond with their locations extending from top to bottom on the chromosome.
The GFF and genome data of A. thaliana, P. persica, P. mume, P. persica, and P. avium were derived from the GDR. Syntenic analysis maps of P. sibirica and other representative plants were constructed using TBtools.
The structure of PsbHLHs was analyzed using TBtools. The conserved motifs of PsbHLHs were identified using the online tool MEME (http://meme-suite.org) (Bailey et al., 2015). The sequence of each PsbHLHs was retrieved by TBtools according to the genomic full-length DNA sequences of PsbHLHs.
Cis-elements in the promoter (−2000 bp upstream of ATG) were predicted using PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). Graphs of the structures were drawn using TBtools software.
Eggnog (http://eggnog5.embl.de/) (Huerta-Cepas et al., 2019) was used for GO annotation, and the results were visualized using WeGo (https://wego.genomics.cn/) (Ye et al., 2018).
All PsbHLH sequences were submitted to STRING (version 11.0, http://string-db.org) (Szklarczyk et al., 2019), and A. thaliana was chosen as the reference organism. After the BLAST analysis, the orthologous genes of A. thaliana with the highest scores were used to construct a network.
P. sibirica clone No. 453 (Appraisal of Improved Cultivar of Forest Trees, S-SV-PS-002-2021, Liaoning Provincial Department of Forest and Grassland) from the National Forest Germplasm Resource Preservation Repository for Prunus species at Shenyang Agricultural University (Kazuo, Liaoning, China) was used as the experimental material. The clone No. 453 sprays were placed in an artificial climate box during their peak flowering period and subjected to stress. Samples were collected at 0 h (Control), 15 min, 30 min, 1 h, and 2 h for −4°C low temperature stress. Six different tissue samples (roots, stems, leaves, stamens, pistils, and petals) in a natural environment were collected. All samples were set up with three biological replicates, immediately frozen in liquid nitrogen, and stored at −80°C until required for further analysis.
Total RNA was extracted from the samples using RNA Extraction Kit (Takara, Beijing, China) following the manufacturer’s instructions. The extracted RNA was then reverse transcribed to cDNA using the PrimeScript™ RT Master Mix Perfect Real Time (Takara, Beijing, China), and stored at −80°C. Primer software (version 5.0) was used to design specific primers (Table S2), which were synthesized by SYNBIO Technologies (Suzhou, China). The 18S rRNA gene of P. sibirica was used as the internal reference (Kuchipudi et al., 2012). The qRT-PCR was performed using PerfectStart® Green qPCR SuperMix (TransGen Biotech, Beijing, China) in StepOnePlus™ Real-Time PCR System (Applied Biosystems, Foster City, CA, United States) following the manufacturer’s instructions. The qRT-PCR conditions were as follows: 94°C for 30 s, followed by 45 cycles of 94°C for 5 s and 60°C for 15 s, and 72°C for 10 s. The 2−ΔΔCT method was employed to calculate the relative expression levels of PsbHLHs (Livak and Schmittgen, 2001).
The full-length coding sequence of PsbHLH42 was cloned into a pRI101-GFP vector. The construct was transformed into Agrobacterium tumefaciens GV3101, which was used to transform the tissue culture seedlings of P. ussuriensis (Liu et al., 2021).
The physiological characteristics of pRI101-GFP empty vector and pRI101-PsbHLH42-GFP seedlings (leaves) were determined after 6 h of 4°C stress treatment. SOD and POD activities were quantified using nitrogen blue tetrazolium (NBT), and guaiacol, respectively. MDA levels were determined using the thiobarbituric acid-reactive substances method. The soluble sugar content (SS) was determined using anthrone colorimetry. Soluble protein content (SP) was determined using the Coomassie Brilliant Blue G-250 staining method. Cell membrane permeability (CMP) was determined using a conductivity meter. The CMP was first determined using a conductivity meter after mixing fresh leaf tissue (0.3 g) with distilled water (15 mL) had been mixed for 4 h. The test tubes were then placed in a boiling water bath for 30 min. After standing for 4 h, measurements were performed again using the same conductivity meter (Wang et al., 2021).
In our experience, this is the first genome-wide survey of the bHLH gene family in P. sibirica. A total of 104 PsbHLHs were screened in the P. sibirica genome and grouped into 23 subfamilies. PsbHLHs were randomly distributed across eight chromosomes. Conserved motifs and exon-intron structures are likely to be the same or similar within the identical subfamily. The duplication of segments is a crucial factor in the expansion of the PsbHLH gene family, and the evolution of PsbHLHs is influenced by selective pressure for purification. PsbHLHs are primarily involved in biological regulation processes and contain various low-temperature stress- and hormone-related elements. Twenty PsbHLHs exhibited diverse expression patterns in response to different low-temperature stresses and different tissues. Overexpression of PsbHLH42 improved the low-temperature tolerance of P. ussuriensis. In summary, these results improve our knowledge of the functional characterization of the PsbHLH gene family and provide genetic resources for cultivating high-quality P. sibirica germplasm resources with cold resistance, high yield, and quality.
The original contributions presented in the study are included in the article/Supplementary Files, further inquiries can be directed to the corresponding author.
QL: Data curation, Formal Analysis, Funding acquisition, Writing – original draft, Writing – review and editing, Conceptualization. JW: Data curation, Writing – original draft. SW: Data curation, Writing – original draft. JC: Data curation, Writing – original draft. YS: Data curation, Writing – original draft. QL: Investigation, Writing – original draft. XL: Data curation, Writing – original draft. SD: Data curation, Supervision, Writing – review and editing.
The authors declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Scientific Research Fund of Liaoning Provincial Education Department (LJKMZ20221056) and China Postdoctoral Science Foundation (2022MD723804).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1267107/full#supplementary-material
Ahn, J. Y., Kim, J., Yang, J. Y., Lee, H. J., Kim, S., Cho, K. S., et al. (2022). Comparative transcriptome analysis between two potato cultivars in tuber induction to reveal associated genes with anthocyanin accumulation. Int. J. Mol. Sci. 23, 3681. doi: 10.3390/ijms23073681
An, F., Xiao, X., Chen, T., Xue, J., Luo, X., Ou, W., et al. (2022). Systematic analysis of bHLH transcription factors in cassava uncovers their roles in postharvest physiological deterioration and cyanogenic glycosides biosynthesis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.901128
Atchley, W. R., Fitch, W. M. (1997). A natural classification of the basic helix-loop-helix class of transcription factors. Proc. Natl. Acad. Sci. U S A. 94, 5172–5176. doi: 10.1073/pnas.94.10.5172
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Bailey, P. C., Martin, C., Toledo-Ortiz, G., Quail, P. H., Huq, E., Heim, M. A., et al. (2003). Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell. 15, 2497–2502. doi: 10.1105/tpc.151140
Baldini, L., Robert, A., Charpentier, B., Labialle, S. (2022). Phylogenetic and molecular analyses identify SNORD116 targets involved in the prader-willi syndrome. Mol. Biol. Evol. 39, msab348. doi: 10.1093/molbev/msab348
Berardini, T. Z., Reiser, L., Li, D., Mezheritsky, Y., Muller, R., Strait, E., et al. (2015). The Arabidopsis information resource: Making and mining the "gold standard" annotated reference plant genome. Genesis. 53, 474–485. doi: 10.1002/dvg.22877
Buck, M. J., Atchley, W. R. (2003). Phylogenetic analysis of plant basic helix-loop-helix proteins. J. Mol. Evol. 56, 742–750. doi: 10.1007/s00239-002-2449-3
Carretero-Paulet, L., Galstyan, A., Roig-Villanova, I., Martínez-García, J. F., Bilbao-Castro, J. R., Robertson, D. L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153, 1398–1412. doi: 10.1104/pp.110.153593
Chen, Q., Li, J., Yang, F. (2023). Genome-wide analysis of the mads-box transcription factor family in solanum melongena. Int. J. Mol. Sci. 24, 826. doi: 10.3390/ijms24010826
Chen, Q., Zhang, X., Fang, Y., Wang, B., Xu, S., Zhao, K., et al. (2022). Genome-wide identification and expression analysis of the R2R3-MYB transcription factor family revealed their potential roles in the flowering process in longan (Dimocarpus longan). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.820439
Crooks, G. E., Hon, G., Chandonia, J. M., Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Dong, B., Zheng, Z., Zhong, S., Ye, Y., Wang, Y., Yang, L., et al. (2022). Integrated Transcriptome and Metabolome Analysis of Color Change and Low-Temperature Response during Flowering of Prunus mume. Int. J. Mol. Sci. 23, 12831. doi: 10.3390/ijms232112831
Duvaud, S., Gabella, C., Lisacek, F., Stockinger, H., Ioannidis, V., Durinx, C. (2021). Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 49, W216–W227. doi: 10.1093/nar/gkab225
Eulgem, T., Rushton, P. J., Robatzek, S., Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. . 5, 199–206. doi: 10.1016/s1360-1385(00)01600-9
Fan, Y., Lai, D., Yang, H., Xue, G., He, A., Chen, L., et al. (2021). Genome-wide identification and expression analysis of the bHLH transcription factor family and its response to abiotic stress in foxtail millet (Setaria italica L.). BMC Genomics 22, 778. doi: 10.1186/s12864-021-08095-y
Gao, M., Zhu, Y., Yang, J., Zhang, H. J., Cheng, C. X., Zhang, Y. X., et al. (2019). Identification of the grape basic helix–loop–helix transcription factor family and characterization of expression patterns in response to different stresses. Plant Growth Regul. 88, 19–39. doi: 10.1007/s10725-019-00485-3
Gaut, B. S., Doebley, J. F. (1997). DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. U S A. 94, 6809–6814. doi: 10.1073/pnas.94.13
Geng, J., Wei, T., Wang, Y., Huang, X., Liu, J. H. (2019). Overexpression of PtrbHLH, a basic helix-loop-helix transcription factor from Poncirus trifoliata, confers enhanced cold tolerance in pummelo (Citrus grandis) by modulation of H2O2 level via regulating a CAT gene. Tree Physiol. 39, 2045–2054. doi: 10.1093/treephys/tpz081
Gremski, K., Ditta, G., Yanofsky, M. F. (2007). The HECATE genes regulate female reproductive tract development in Arabidopsis thaliana. Development 134, 3593–3601. doi: 10.1242/dev.011510
Guo, X. J., Wang, J. R. (2017). Global identification, structural analysis and expression characterization of bHLH transcription factors in wheat. BMC Plant Biol. 17, 90. doi: 10.1186/s12870-017-1038-y
Han, X., Rong, H., Feng, Y., Xin, Y., Luan, X., Zhou, Q., et al. (2023). Protoplast isolation and transient transformation system for Ginkgo biloba L. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1145754
Harouaka, D., Engle, R. E., Wollenberg, K., Diaz, G., Tice, A. B., Zamboni, F., et al. (2016). Diminished viral replication and compartmentalization of hepatitis C virus in hepatocellular carcinoma tissue. Proc. Natl. Acad. Sci. U S A. 113, 1375–1380. doi: 10.1073/pnas.1516879113
Hong, Y. Q., Naveed, A., Tian, Y. Y., Liu, J. Y., Wang, L. Y., Wang, G., et al. (2019). Genome-Wide Identification, Expression Analysis, and Subcellular Localization of Carthamus tinctorius bHLH Transcription Factors. Int. J. Mol. Sci. 20, 3044. doi: 10.3390/ijms20123044
Hou, L., Yuan, X., Li, S., Li, Y., Li, Z., Li, J. (2021). Genome-wide identification of CYP72A gene family and expression patterns related to jasmonic acid treatment and steroidal saponin accumulation in dioscorea zingiberensis. Int. J. Mol. Sci. 22, 10953. doi: 10.3390/ijms222010953
Huang, Z., Jin, S. H., Yang, L., Song, L., Wang, Y. H., Jian, L. L., et al. (2022). Heterologous expression of dehydration-inducible mfbHLH145 of myrothamnus flabellifoli enhanced drought and salt tolerance in arabidopsis. Int. J. Mol. Sci. 23, 5546. doi: 10.3390/ijms23105546
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernández-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. doi: 10.1093/nar/gky1085
Jansson, S., Douglas, C. J. (2007). Populus: a model system for plant biology. Annu. Rev. Plant Biol. 58, 435–458. doi: 10.1146/annurev.arplant.58.032806.103956
Jin, C., Huang, X. S., Li, K. Q., Yin, H., Li, L. T., Yao, Z. H., et al. (2016). Overexpression of a bHLH1 Transcription Factor of Pyrus ussuriensis Confers Enhanced Cold Tolerance and Increases Expression of Stress-Responsive Genes. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00441
Jung, S., Lee, T., Cheng, C. H., Buble, K., Zheng, P., Yu, J., et al. (2019). 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 47, D1137–D1145. doi: 10.1093/nar/gky1000
Kuchipudi, S. V., Tellabati, M., Nelli, R. K., White, G. A., Perez, B. B., Sebastian, S., et al. (2012). 18S rRNA is a reliable norMalisation gene for real time PCR based on influenza virus infected cells. Virol. J. 9, 230. doi: 10.1186/1743-422X-9-230
Lampard, G. R., Bergmann, D. C. (2007). A shout-out to stomatal development: How the bHLH proteins SPEECHLESS, MUTE and FAMA regulate cell division and cell fate. Plant Signal Behav. 2, 290–292. doi: 10.4161/psb.2.4.4038
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, S., Hu, Y., Yang, H., Tian, S., Wei, D., Tang, Q., et al. (2023). The regulatory roles of MYC TFs in plant stamen development. Plant Sci. 333, 111734. doi: 10.1016/j.plantsci.2023.11173
Li, T., Shi, Y., Zhu, B., Zhang, T., Feng, Z., Wang, X., et al. (2022). Genome-wide identification of apple atypical bHLH subfamily PRE members and functional characterization of mdPRE4.3 in response to abiotic stress. Front. Genet. 13. doi: 10.3389/fgene.2022.846559
Li, H., Sun, J., Xu, Y., Jiang, H., Wu, X., Li, C. (2007). The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Mol. Biol. 65, 655–665. doi: 10.1007/s11103-007-9230-3
Li, J., Wang, T., Han, J., Ren, Z. (2020). Genome-wide identification and characterization of cucumber bHLH family genes and the functional characterization of CsbHLH041 in NaCl and ABA tolerance in Arabidopsis and cucumber. BMC Plant Biol. 20, 272. doi: 10.1186/s12870-020-02440-1
Lin, R., Zheng, J., Pu, L., Wang, Z., Mei, Q., Zhang, M., et al. (2021). Genome-wide identification and expression analysis of aquaporin family in Canavalia rosea and their roles in the adaptation to saline-alkaline soils and drought stress. BMC Plant Biol. 21, 333. doi: 10.1186/s12870-021-03034-1
Liu, J., Shi, Y., Yang, S. (2018). Insights into the regulation of C-repeat binding factors in plant cold signaling. J. Integr. Plant Biol. 60, 780–795. doi: 10.1111/jipb.12657
Liu, Q., Wang, Z., Yu, S., Li, W., Zhang, M., Yang, J., et al. (2021). Pu-miR172d regulates stomatal density and water-use efficiency via targeting PuGTL1 in poplar. J. Exp. Bot. 72, 1370–1383. doi: 10.1093/jxb/eraa493
Liu, W., Zhang, Z., Zhu, W., Ren, Z., Jia, L., Li, W., et al. (2019). Evolutionary conservation and divergence of genes encoding 3-hydroxy-3-methylglutaryl coenzyme A synthase in the allotetraploid cotton species gossypium hirsutum. Cells. 8, 412. doi: 10.3390/cells8050412
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2020). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Ludwig, S. R., Habera, L. F., Dellaporta, S. L., Wessler, S. R. (1989). Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc. Natl. Acad. Sci. U S A. 86, 7092–7096. doi: 10.1073/pnas.86.18.7092
Ma, Y. X., Wang, S. X., Liu, X. J., Yu, H. Y., Yu, D., Li, G. T., et al. (2020b). Oil content, fatty acid composition and biodiesel properties among natural provenances of Siberian apricot (Prunus sibirica L.) from China. GCB Bioenergy. 13, 112–132. doi: 10.1111/gcbb.12759
Ma, R., Liu, W., Li, S., Zhu, X., Yang, J., Zhang, N., et al. (2021). Genome-wide identification, characterization and expression analysis of the CIPK gene family in Potato (Solanum tuberosum L.) and the role of StCIPK10 in response to drought and osmotic stress. Int J Mol Sci. 22, 13535, 1–24. doi: 10.3390/ijms222413535
Ma, T., Wang, S., Sun, C., Tian, J., Guo, H., Cui, S., et al. (2023). Arabidopsis LFR, a SWI/SNF complex component, interacts with ICE1 and activates ICE1 and CBF3 expression in cold acclimation. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1097158
Ma, M. J., Zhu, Z. Q., Cheng, S. C., Zhou, Q., Zhou, X., Kong, X. M., et al. (2020a). Methyl jasmonate alleviates chilling injury by regulating membrane lipid composition in green bell pepper. Scientia horticulturae. 266, 109308. doi: 10.1016/j.scienta.2020.109308
Mao, K., Dong, Q., Li, C., Liu, C., Ma, F. (2017). Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00480
Massari, M. E., Murre, C. (2000). Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell Biol. 20, 429–440. doi: 10.1128/MCB.20.2.429-440.2000
Matsushima, R., Fukao, Y., Nishimura, M., Hara-Nishimura, I. (2004). NAI1 gene encodes a basic-helix-loop-helix-type putative transcription factor that regulates the formation of an endoplasmic reticulum-derived structure, the ER body. Plant Cell. 16, 1536–1549. doi: 10.1105/tpc.021154
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Movahedi, A., Zhang, J., Amirian, R., Zhuge, Q. (2014). An efficient Agrobacterium-mediated transformation system for poplar. Int. J. Mol. Sci. 15, 10780–10793. doi: 10.3390/ijms150610780
Oña Chuquimarca, S., Ayala-Ruano, S., Goossens, J., Pauwels, L., Goossens, A., Leon-Reyes, A., et al. (2020). The molecular basis of JAZ-MYC coupling, a protein-protein interface essential for plant response to stressors. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01139
Pires, N., Dolan, L. (2010). Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 27, 862–874. doi: 10.1093/molbev/msp288
Rahim, M. A., Resentini, F., Dalla Vecchia, F., Trainotti, L. (2019). Effects on plant growth and reproduction of a peach R2R3-MYB transcription factor overexpressed in tobacco. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01143
Sabir, I. A., Manzoor, M. A., Shah, I. H., Liu, X., Zahid, M. S., Jiu, S., et al. (2022). MYB transcription factor family in sweet cherry (Prunus avium L.): genome-wide investigation, evolution, structure, characterization and expression patterns. BMC Plant Biol. 22, 2. doi: 10.1186/s12870-021-03374-y
Sheng, Y., Yu, H., Pan, H., Qiu, K., Xie, Q., Chen, H., et al. (2022). Genome-wide analysis of the gene structure, expression and protein interactions of the peach (Prunus persica) TIFY gene family. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.792802
Song, M., Wang, H., Wang, Z., Huang, H., Chen, S., Ma, H. (2021). Genome-Wide Characterization and Analysis of bHLH Transcription Factors Related to Anthocyanin Biosynthesis in Fig (Ficus carica L.). Front. Plant Sci. 12. doi: 10.3389/fpls.2021.730692
Sun, X., Wang, Y., Sui, N. (2018). Transcriptional regulation of bHLH during plant response to stress. Biochem. Biophys. Res. Commun. 503, 397–401. doi: 10.1016/j.bbrc.2018.07.123
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
Taghavi, T., Patel, H., Rafie, R. (2022). Anthocyanin extraction method and sample preparation affect anthocyanin yield of strawberries. Natural Product Commun. 17, 1–7. doi: 10.1177/1934578X221099970
Tan, J., Zhou, Z., Feng, H., Xing, J., Niu, Y., Deng, Z. (2021). Data-independent acquisition-based proteome and phosphoproteome profiling reveals early protein phosphorylation and dephosphorylation events in arabidopsis seedlings upon cold exposure. Int. J. Mol. Sci. 22, 12856. doi: 10.3390/ijms222312856
Toledo-Ortiz, G., Huq, E., Quail, P. H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 15, 1749–1770. doi: 10.1105/tpc.013839
Wang, Y., Jiang, C. J., Li, Y. Y., Wei, C. L., Deng, W. W. (2012). CsICE1 and CsCBF1: two transcription factors involved in cold responses in Camellia sinensis. Plant Cell Rep. 31, 27–34. doi: 10.1007/s00299-011-1136-5
Wang, X., Zhang, R., Wang, J., Di, L., Wang, H., Sikdar, A. (2021). The effects of leaf extracts of four tree species on amygdalus pedunculata seedlings growth. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.587579
Wang, J., An, C., Guo, H., Yang, X., Chen, J., Zong, J., et al. (2020). Physiological and transcriptomic analyses reveal the mechanisms underlying the salt tolerance of Zoysia japonica Steud. BMC Plant Biol. 20, 114, 1–16. doi: 10.1186/s12870-020-02330-6
Waseem, M., Li, N., Su, D., Chen, J., Li, Z. (2019). Overexpression of a basic helix-loop-helix transcription factor gene, SlbHLH22, promotes early flowering and accelerates fruit ripening in tomato (Solanum lycopersicum L.). Planta. 250, 173–185. doi: 10.1007/s00425-019-03157-8
Wei, K., Chen, H. (2018). Comparative functional genomics analysis of bHLH gene family in rice, maize and wheat. BMC Plant Biol. 18, 309. doi: 10.1186/s12870-018-1529-5
Wu, C. L., Lin, L. F., Hsu, H. C., Huang, L. F., Hsiao, C. D., Chou, M. L. (2021). Saussurea involucrata (Snow lotus) ICE1 and ICE2 orthologues involved in regulating cold stress tolerance in transgenic arabidopsis. Int. J. Mol. Sci. 22, 10850. doi: 10.3390/ijms221910850
Wu, Y. Y., Wu, S. H., Wang, X. Q., Mao, T. Y., Bao, M. Z., Zhang, J. W., et al. (2022). Genome-wide identification and characterization of the bHLH gene family in an ornamental woody plant Prunus mume. Hortic. Plant J. 8, 531–544. doi: 10.1016/j.hpj.2022.01.004
Xu, W., Jiao, Y., Li, R., Zhang, N., Xiao, D., Ding, X., et al. (2014). Chinese wild-growing Vitis amurensis ICE1 and ICE2 encode MYC-type bHLH transcription activators that regulate cold tolerance in Arabidopsis. PLoS One 9, e102303. doi: 10.1371/journal.pone.0102303
Xu, B. Q., Wang, J. J., Peng, Y., Huang, H., Sun, L. L., Yang, R., et al. (2022). SlMYC2 mediates stomatal movement in response to drought stress by repressing SlCHS1 expression. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.952758
Ye, J., Zhang, Y., Cui, H., Liu, J., Wu, Y., Cheng, Y., et al. (2018). WEGO 2.0: a web tool for analyzing and plotting GO annotations 2018 update. Nucleic Acids Res. 46, W71–W75. doi: 10.1093/nar/gky400
Yin, M. Y., Wuyun, T., Jiang, Z. M., Zeng, J. (2019). Amino acid profiles and protein quality of Siberian apricot (Prunus sibirica L.) kernels from Inner Mongolia. J. Forestry Res. 31, 1391–1397. doi: 10.1007/s11676-019-00882-4
Yu, C. S., Chen, Y. C., Lu, C. H., Hwang, J. K. (2006). Prediction of protein subcellular localization. Proteins. 64, 643–651. doi: 10.1002/prot.21018
Zhang, C., Feng, R., Ma, R., Shen, Z., Cai, Z., Song, Z., et al. (2018). Genome-wide analysis of basic helix-loop-helix superfamily members in peach. PLoS One 13, e0195974. doi: 10.1371/journal.pone.0195974
Zhang, T., Lv, W., Zhang, H., Ma, L., Li, P., Ge, L., et al. (2018). Genome-wide analysis of the basic Helix-Loop-Helix (bHLH) transcription factor family in maize. BMC Plant Biol. 18, 235. doi: 10.1186/s12870-018-1441-z
Zhang, Y., Zhang, J., Yang, D., Jin, Y., Liu, X., Zhang, Z., et al. (2022). Genome-wide identification of NAP1 and function analysis in moso bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 23, 6491. doi: 10.3390/ijms23126491
Zhao, X., Yu, F., Guo, Q., Wang, Y., Zhang, Z., Liu, Y. (2022). Genome-wide identification, characterization, and expression profile analysis of CONSTANS-like genes in woodland strawberry (Fragaria vesca). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.931721
Zhou, X., Liao, Y., Kim, S. U., Chen, Z., Nie, G., Cheng, S., et al. (2020). Genome-wide identification and characterization of bHLH family genes from Ginkgo biloba. Sci. Rep. 10, 13723. doi: 10.1038/s41598-020-69305-3
Zhuang, M., Zhang, Z. M., Jin, L., Wang, B. T., Koyama, Y., Jin, F. J. (2019). The basic-region helix-loop-helix transcription factor devR significantly affects polysaccharide metabolism in aspergillus oryzae. Appl. Environ. Microbiol. 85, e00089–e00019. doi: 10.1128/AEM.00089-19
Keywords: Prunus sibirica, basic helix-loop-helix, genome-wide analysis, low temperature stress, expression pattern, transient transformation
Citation: Liu Q, Wen J, Wang S, Chen J, Sun Y, Liu Q, Li X and Dong S (2023) Genome-wide identification, expression analysis, and potential roles under low-temperature stress of bHLH gene family in Prunus sibirica. Front. Plant Sci. 14:1267107. doi: 10.3389/fpls.2023.1267107
Received: 26 July 2023; Accepted: 30 August 2023;
Published: 20 September 2023.
Edited by:
Ahmad M. Alqudah, Qatar University, QatarReviewed by:
Mohammed Ali Abd Elhammed Abd Allah, Desert Research Center, EgyptCopyright © 2023 Liu, Wen, Wang, Chen, Sun, Liu, Li and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shengjun Dong, ZHNqOTI4QDE2My5jb20=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.