- 1Crop Improvement division, ICAR-Indian Institute of Wheat and Barley Researh, Karnal, India
- 2Division of AgriBioinformatics, ICAR-Indian Agricultural Statistics Research Institute, New Delhi, India
Salt stress affects plant growth and development, resulting in the loss of crop yield across the world, and sodium-proton antiporters (NHXs) are one of the genes known to promote salt tolerance in transgenic plants. In this study, we conducted a comprehensive genome-wide analysis and expression profile of NHX genes in wheat under salinity stress. We identified 30 TaNHX genes in wheat based on the Na+/H+ exchanger domain, with all genes containing an amiloride motif except one, a known for inhibiting Na+ ions in plants. Phylogenetic analysis classified these genes into three classes with subfamilies: 12 were localized in vacuoles, while 18 were in the endoplasmic reticulum and plasma membrane. Promoter analysis revealed stress-related cis-acting elements, indicating their potential role in abiotic stress tolerance. The non-synonymous (Ka)/synonymous (Ks) ratios highlighted that the majority of TaNHX genes experienced robust purifying selection throughout their evolutionary history. Transcriptomis data analysis and qRT-PCR demonstrated distinct expression patterns for TaNHX genes across various tissues when subjected to salt stress. Additionally, we predicted 20 different miRNA candidates targeting the identified TaNHX genes. Protein-protein interaction prediction revealed NHX6’s involvement in the SOS1 pathway, while NHX1 gene exhibit proton antiporter activity. Molecular dynamics (MD) simulations were also conducted to examine the interactions of TaNHX1, TaNHX2, and TaNHX3. These results represent a significant advancement in our understanding of the molecular mechanisms governing Na+ transporters. This may also offer promising avenues for future studies aimed at unraveling the intricate details of their biological roles and applications.
1 Introduction
Soil salinity stands as a formidable challenge with far-reaching consequences for agriculture on a global scale, impacting a staggering 45 million hectares of irrigated land (Wu et al., 2019). This issue carries immense significance, as it directly influences the capacity to sustain agricultural productivity. This becomes particularly critical when we consider that irrigated lands, renowned for their ability to yield twice the food production compared to rain-fed areas (Aharon et al., 2003), are pivotal in addressing the pressing concerns of food security and resource sustainability. Furthermore, projections indicate that around 50% of cultivable land could be affected by excessive salinization by 2050 (Gaxiola et al., 1999; Pehlivan et al., 2016). The increasing global population further underscores the need to resolve this issue to secure food security and the sustainability of agricultural practices.
In response to the adverse effects of salt stress, plants activate a range of sophisticated mechanisms to ensure their survival. These include complex hormonal regulation of plant growth and metabolism, precise osmotic regulation, and ion homeostasis maintenance (Wu et al., 2019a). The role of sodium/hydrogen antiporter (NHX) proteins is pivotal in regulating ion homeostasis. These proteins intricately modulate the equilibrium of ions, as underscored by their interplay with the essential proton pumps, the H+-PPase and H+-ATPase enzymes. Together, they meticulously orchestrate the translocation of sodium ions (Na+) from the dynamic cytoplasmic milieu to designated cellular destinations, including vacuolar compartments and extracellular regions. This ion flux operation serves as a vigilant guardian, staunchly preventing the potential deleterious accrual of Na+ ions within the intricate confines of cellular compartments (Aharon et al., 2003; Dragwidge et al., 2019).
The TaNHX gene family encodes widely distributed transmembrane proteins classified under the monovalent cation/proton antiporter 1 (CPA1) category (Fukuda et al., 2011). Prior studies (Wu et al., 2019a) have revealed that NHX proteins typically encompass 10–12 transmembrane helices (TMs) and predominantly inhabit vacuolar, endosomal, and plasma membrane locales (Aharon et al., 2003; Pehlivan et al., 2016). In A. thaliana, eight distinct NHX genes were identified (Gaxiola et al., 1999). These genes exhibit diverse subcellular distributions, with two NHX genes (AtNHX7 and AtNHX8) residing within the plasma membrane (PM-class), four inhabiting vacuoles (Vac-class), and two localizing to endosomes (Endo-class) (Shi et al., 2000; Aharon et al., 2003; Brett et al., 2005; Bassil et al., 2011). NHX proteins are distributed in a way that they reflects the specific roles they play in maintaining ion homeostasis in different environmental conditions.
Understanding the different mechanisms of NHX gene function is critical for decoding the plants complex response to salt stress. We can gain a valuable understanding of the critical roles that TaNHX genes play in regulating salt stress in various plant species by examining their regulatory functions, and evolutionary relationships. Such bioinformatics studies and experimental investigations pave the way for future research and manipulation of NHX genes, contributing to the production of salt-tolerant crop varieties and enhancing agricultural productivity in salt-affected regions.
NHX genes are intricately involved in a multitude of biological processes, including response to salt stress, regulation of cell growth, modulation of membrane vesicle trafficking, and maintenance of pH homeostasis (Dragwidge et al., 2019). After inactivating the mutation in AtNHX5 and AtNHX6, it was observed that there were development-related disorders and anomalies in cell division in tissues of the embryo and roots (Dragwidge et al., 2019). OsNHX1, OsNHX2, OsNHX3, and OsNHX5 in rice become activated when they are exposed to salt stress, hyperosmotic stress, and ABA stress conditions. Determining the salt tolerance mechanisms within the rice plant may be delineated by the nuanced expression patterns of NHX genes across diverse tissue types (Fukuda et al., 2011). In other experiments, rye grass engineered with the rice vacuolar membrane OsNHX1 gene endured high salt conditions (350 mM) for 10 weeks, while transgenic B. napus plants thrived even in the presence of 200 mM NaCl. Introducing the Vac-class membrane gene AgNHX1 from A. gmelinii into rice plants conferred the ability to withstand 300 mM NaCl for three days, surpassing the salt sensitivity of wild-type rice. Overexpression of AtNHX1 from A. thaliana into tomato and SsNHX1 from Salsola collina into M. sativa significantly improves salt tolerance in plants. These findings underscore the potential of NHX genes to enhance salt tolerance in various plant species (Manik et al., 2015; Wilkins et al., 2016; Kumari et al., 2018; Wu et al., 2019a). The most widely grown crop in the world is wheat. Its growth is significantly impeded by salt stress since it is not a halophyte. When exposed to salt, wheat cells quickly increase their Na+ concentration. Due to the relatively low net uptake compared to unidirectional inflow (Khare et al., 2015), it was expected that wheat roots would experience high rates of Na+ efflux. The ability of a wheat Na+/H+ antiporter (TaSOS1) to extrude Na+ across the plasma membrane has been experimentally studied (Parveen et al., 2021). Overexpression of the vacuolar NHX antiporter AtNHX1 from Arabidopsis led to an increase in wheat salt tolerance (Parveen et al., 2021). The putative wheat Na+/H+ antiporter TaNHX1 provided salt tolerance to transgenic Arabidopsis plants (Ramesh et al., 2019). In a previous study, TaNHX2 was cloned from bread wheat, and it was examined for its expression and function of the protein and yeast. The proposition that TaNHX2 mediates the compartmentalization of Na+ into vacuoles holds promise for elucidating its pivotal role in enhancing a plant’s salt tolerance. Nevertheless, the precise mechanisms underlying TaNHX2’s participation in Na+/H+ exchange and its subcellular localization within plant cells remain unverified (Shuai et al., 2013). In this study, we performed a genome-wide investigation of all TaNHX gene family members in wheat. The TaNHX genes were identified using gene structure, chromosomal locations, phylogenetic relationships, cis-element, and expression profiling, as well as molecular dynamic simulations. The expression patterns obtained from in-silico analysis were then confirmed using qRT-PCR. TaNHX can be used to breed salt tolerance using genome-editing methods. The results taken here are necessary to systematically explore the gene function of the wheat TaNHX gene family.
2 Materials and methods
2.1 Identification, characterization, phylogenetic analysis and gene ontology of sodium proton antiporters
To identify TaNHX genes in wheat, we employed a bioinformatics approach. Firstly, we obtained protein sequences of NHX from Arabidopsis, cotton, sorghum, and barley, which served as query sequences. These query sequences were then searched against the wheat genome available at the Ensembl Genomes database (fp://fp.ensemblgenomes.org/pub/plants/release-51/fasta/triticumaestivum/pep/) using the BLASTP algorithm. We selected all homologous protein sequences of NHX candidates that met the specified criteria, including an e-value threshold of 1e-10 and a bit score value higher than 100 percent. To further validate the presence of the Na+_H+_Exchanger domain (PF00999) characteristic of NHX transporters, the obtained sequences were scanned against this domain using the HMMER 3.1b2 online software (https://www.ebi.ac.uk/Tools/hmmer/) (Potter et al., 2018).
For the subcellular localization, Wolf Psort (https://wolfpsort.hgc.jp/) was used. The physio-chemical properties were predicted using the online programme ProtParam (http://web.expasy.org/protparam/).To study the evolutionary linkage, Clustal W software was used to analyze NHX protein sequences from Arabidopsis, barley, sorghum, cotton, and wheat (Larkin et al., 2007). Following that, an unrooted phylogenetic tree was built in MEGA-X with 1000 bootstrap repetitions using Maximum likelihood (ML) with default parameters the JTT model with uniform rates and 4 number of threads (Kumar et al., 2018). NHX members in wheat were classified into subfamilies based on their Arabidopsis, barley, sorghum, and cotton homologs.
Functional annotation of the identified genes was performed using Blast2GO (https://www.blast2go.com/). This analysis enabled the elucidation of molecular functions, biological processes, and cellular components intricately linked to the genes under investigation. Additionally, we harnessed the KEGG (Kyoto Encyclopaedia of Genes and Genomes) database (http://www.genome.jp/kegg/) to annotate the metabolic pathways, thus furnishing invaluable insights into the precise pathways where these genes actively participate. The combination of Blast2GO and KEGG analysis allowed for a comprehensive understanding of the functional properties and metabolic roles of the identified genes.
2.2 Conserved motifs, gene structure, promoter analysis and physical mapping of NHX genes
To unravel the presence of conserved motifs within the identified genes, we used the MEME webserver (Multiple Em for Motif Elicitation). Employing default parameters, which encompassed the exploration of 2-20 motif sites, the extraction of up to 10 motifs, and a motif width range extending from 6 to 50 (Bailey et al., 2009), this analysis provided an intricate glimpse into the genetic signatures. Furthermore, we delved into the gene structure using the Gene Structure Display Server (GSDS) tool (Hu et al., 2015), unravelling the intricacies of these genes’ architectural features. This comprehensive approach not only unveiled conserved motifs but also shed light on the structural nuances inherent to the genes under study. The promoter sequences, of up to 2 kb in size, were used for cis-acting element analysis and submitted to the PLANTCARE webserver. The cis-acting elements that were produced were categorized according to their functional class.
Based on the position sites, the chromosomal locations of the wheat TaNHX genes were examined. The Ensembl database is used to retrieve information about the TaNHX gene’s locations. Physical mapping of the transporters was constructed using Map-Chart software to construct the location on the wheat genome (Voorrips, 2002).
2.3 Protein-protein interaction, miRNA targeting, and gene duplication events
For the identification of functional protein-protein interactions, the STRING v1054 databases were employed (Szklarczyk et al., 2015). However, Arabidopsis was used as the reference species to search the interactive network in the database. Blast the sequences with set parameters to an e-value of 1e-10, and the A. thaliana genome was searched against all known interaction partners. The best-hit gene for each gene was chosen using Cytoscape 58 to create a PPI network. Using Cytoscape’s cyto Hubba plugin, the top hub gene from the interaction network was finally determined. Furthermore, we conducted an analysis to pinpoint significant Gene Ontology (GO) terms (FDR ≤ 0.01) related to the molecular functions and biological processes of the interaction network nodes using the iDEP webtool (Ge et al., 2018). The transcript sequences of the TaNHX gene were obtained using the Ensembl plant database. Now, the transcript sequences of TaNHX and the mature miRNA sequences from miRbase (Kozomara and Griffiths-Jones, 2014) were analyzed by using the psRNATarget service’s default settings (Dai and Zhao, 2011) to find the targets of miRNAs.
We accessed the coding sequences (CDS) of T. urartu (A genome), A. tauschii (D genome), and T. dicoccoides (AB genome) from Ensembl plants (fp://fp.ensemblgenomes.org/- pub/plants/release-42/fasta/triticum_urartu,fp://fp.ensemblgenomes.org/pub/release-42/plants/fasta/aegilops_tauschii/cds,fp://fp.ensemblgenomes.org/pub/release-42/plants/fasta/triticum_dicoccoides/cds/). These genomes served as references for the A, D, and AB genomes, respectively. We conducted BLASTn searches using these sequences against the TaNHX CDS sequences in T. aestivum to identify orthologous genes. Top hits were designated as orthologs within each species, adhering to stringent criteria, including an e-value cut-off of 1e-10 and a 150-bit-score cut-off. This approach was similarly applied to search for orthologous genes in other monocots and dicots, including O. sativa, H. vulgare, Z. mays, and A. thaliana. To examine the synteny relationships of the NHX gene family within and across various species, we employed TBtools for a comprehensive analysis.
2.4 Gene expression profiling using RNA-seq data
To determine in-silico gene expression in different tissues under salinity stress, the log2 value of the FPKM value was calculated using the SRA data (SRP304900) from NCBI. Heatmaps were created with Clustvis (https://biit.cs.ut.ee/clustvis/).
2.5 Sample collection and treatments
To investigate the expression patterns of TaNHX genes in response to salinity stress, we sourced wheat genotypes, specifically KRL213 and HD2009, from the Germplasm Unit of the ICAR-Indian Institute of Wheat and Barley Research, located in Karnal, India. In a controlled environment, we initiated the germination process by placing the seeds in Petri dishes at a temperature of 22°C. Prior to germination, the seeds were sterilization using a 1% sodium hypochlorite solution for duration of 10 minutes, followed by thorough rinsing with distilled water, ensuring the removal of any residual sterilization agents. After five days of germination, seedlings were transferred to full-strength Hoagland’s solution phytojars and incubated for 14 days in a BOD incubator with two sets of three biological replicates of each genotype. For salt stress, two contrasting wheat genotypes, HD2009 (salt sensitive) and KRL213 (salt tolerant), were used. Both genotypes at the two-leaf seedling stage were stressed with 150 mM NaCl. After the treatment, the leaf samples were taken at 0, 3, 24, and 48 hours. For total RNA isolation, all acquired samples were immediately wrapped in foil and frozen in liquid nitrogen at -80°C.
2.6 RNA extraction and qRT-PCR analysis
To isolate RNA, we employed TRIzol reagent following the manufacturer’s guidelines. The extracted RNA was further processed to eliminate any residual DNA through DNase I treatment (NEB, USA). Utilizing Superscript-III reverse transcriptase (Invitrogen, USA), 1 µg of total RNA was converted into the first strand of cDNA. This resulting cDNA was subsequently diluted at 1:2 ratio, and 1 µl of the diluted cDNA was employed as a template within a 10 µl reaction volume for real-time qRT-PCR analysis. We conducted SYBR Green-based real-time quantitative RT-PCR analysis using the BIO-RAD CFX96 system (Bio-Rad). For normalization purposes, we employed wheat actin as an endogenous control (Muthusamy et al., 2016). The expression levels were quantified as relative fold changes through the application of the 2△△-Ct method (Livak and Schmittgen, 2001). All the samples were set up in 3 biological replicates. Experimental data was statistically analyzed by a one-way Analysis of Variance (ANOVA) and Tukey’s multiple range test was used for mean value separation by PAST software (Hammer et al., 2001). P-value of <0.05 was considered statistically significant.
2.7 Molecular modelling and dynamics simulations
The three-dimensional structure for TaNHX was generated using AlphaFold2 (Skolnick et al., 2021), as it employs deep learning techniques, specifically deep neural networks, to predict protein structures with remarkable accuracy. It leverages vast amounts of protein sequence and structural data to generate predictions of protein structures, even for proteins with no known structures.
The molecular dynamics simulations (MDS) were performed using GROMCAS 5.0 (Abraham et al., 2015). The simulations aimed to investigate the conformational changes of the TaNHX protein structures in the presence of a solvent system. To set up the simulations, the OPLS_2005 force field (Dubay et al., 2012) was employed to describe the interactions within the proteins. The system was solvated in a cubic water box using the SPC water model. The protein atoms were kept at a minimum distance of 10 Å from the edges of the box. To achieve a neutral charge for the systems, counter ions were added, and a 0.15 M ionic concentration was maintained by including Na+ and Cl ions. First, each system underwent 50,000 steps of steepest descent energy minimization, which helped eliminate steric overlap and stabilize the initial configurations. Following the energy minimization, a two-step equilibration phase was performed. The first step was NVT equilibration, where the number of particles, volume, and temperature were kept constant. This phase ran for 100 picoseconds (ps) to stabilize the system’s temperature. The V-rescale temperature-coupling method was used for NVT, with a constant coupling time of 1 ps and a target temperature of 303.15 K. The second step was NPT equilibration, where the number of particles, pressure, and temperature were maintained constant. This phase also ran for 100 ps, and it involved using the Nose-Hoover pressure coupling method with a constant coupling time of 1 ps and a target temperature of 303.15 K. During this phase, the system was relaxed, and the protein was restrained using position restraints (h-bonds). To account for electrostatic interactions, the Particle Mesh Ewald method was employed for both NVT and NPT simulations. After the equilibration phases, each system underwent a full production run of 30 nanoseconds (ns) without any restraints. The integration time step used was 0.002 ps, and coordinates were recorded every 10 ps using an xtc collection interval of 5,000 steps.
3 Results
3.1 Characterization, phylogenetic analysis, and gene ontology of TaNHX genes in wheat
In order to identify members of the TaNHX gene family within the wheat genome, we employed a two-step approach. Firstly, utilized known NHX sequences from other species as queries and performed a blast search against the wheat proteome database (Supplementary Table S1). This enabled us to retrieve putative TaNHX genes in wheat. Subsequently, we eliminated redundant sequences and confirmed the gene identifications based on the presence of the Na+_H+_Exchanger domain (PF00999). This stringent filtering process resulted in the identification of 30 TaNHX genes in T. aestivum.
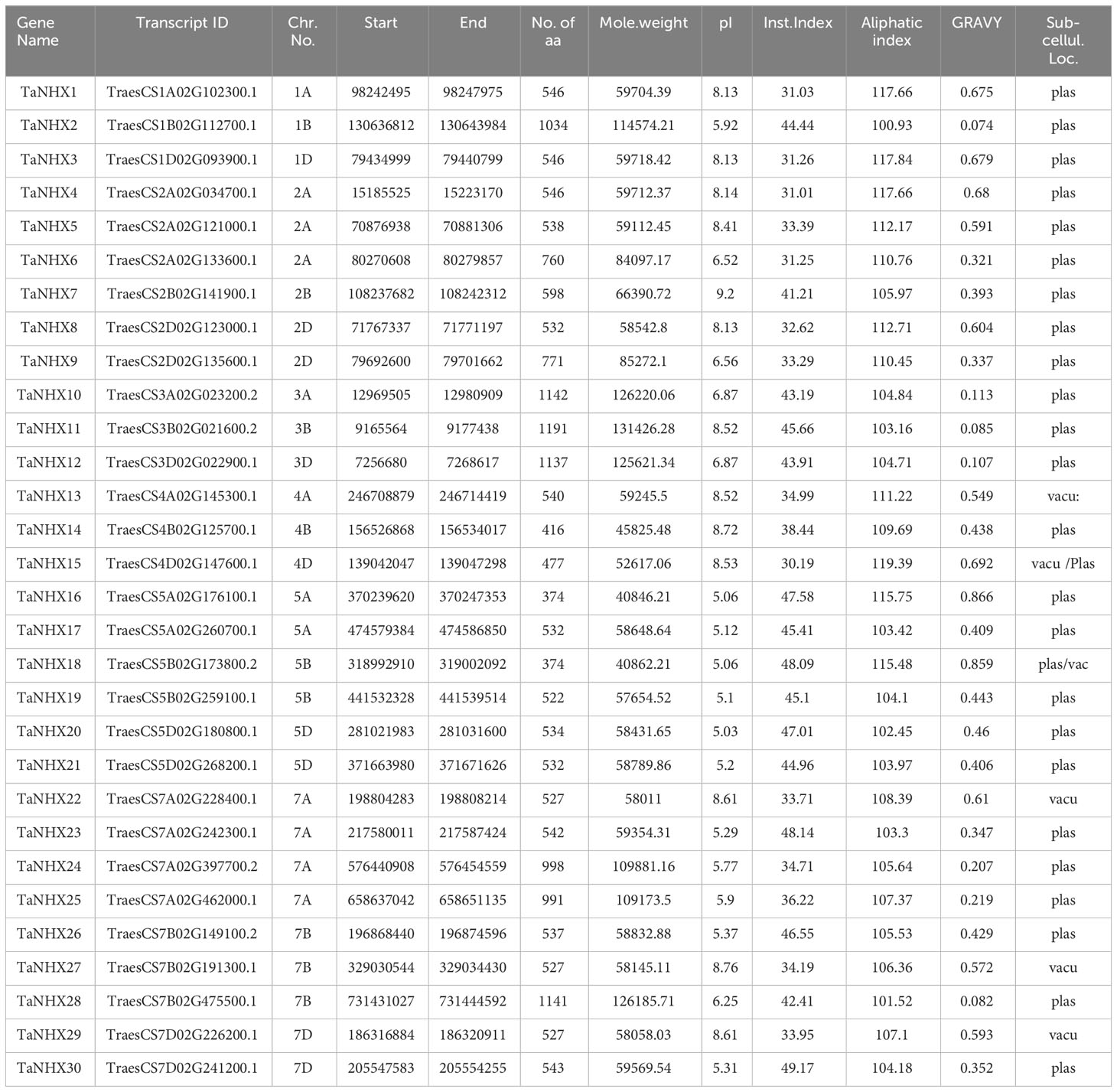
To understand the physio-chemical properties of the wheat TaNHX genes, we conducted a comprehensive analysis, the results of which are summarized in Table 1. Hence, to determine the subcellular localization of NHX proteins, prediction tools have been employed, which were found to predominantly reside on the plasma membrane, endosomes, and vacuoles. Furthermore, we assessed several parameters, including the number of amino acids, which ranged from 374 to 1191, reflecting the diversity in NHX protein lengths. Additionally, the molecular weight of the NHX proteins revealed a wide range of values. For instance, TaNHX16 exhibited a molecular weight of 40.84 kDa, whereas TaNHX11 displayed the highest molecular weight of 131.42 kDa. The findings presented in Table 1 shed light on the distinct physiochemical characteristics of the identified wheat NHX genes.
To analyze the evolutionary lineages of the NHX gene family across different species, a dataset comprising 63 NHX protein sequences was utilized. The sequences were obtained from five species, namely A. thaliana, H. vulgare, G. hirsutum, S. bicolor, and T. aestivum (Supplementary Table S2). These sequences were employed to construct a phylogenetic tree, as depicted in Figure 1. The analysis involved the utilization of previously reported genes, resulting in the classification of the 63 genes into three distinct categories: class I (endo-class), class II (PM-class), and class III (vac-class). Among the five species, the largest group was class III (vac-class), which encompassed 39 proteins. Specifically, this group comprised 4 NHX proteins in A. thaliana, 2 in H. vulgare, 15 in G. hirsutum, 6 in S. bicolor, and 12 in T. aestivum. It is worth noting that 9 TaNHX proteins were distributed evenly between classes I (endo-class) and II (PM-class), as shown in Figure 1.
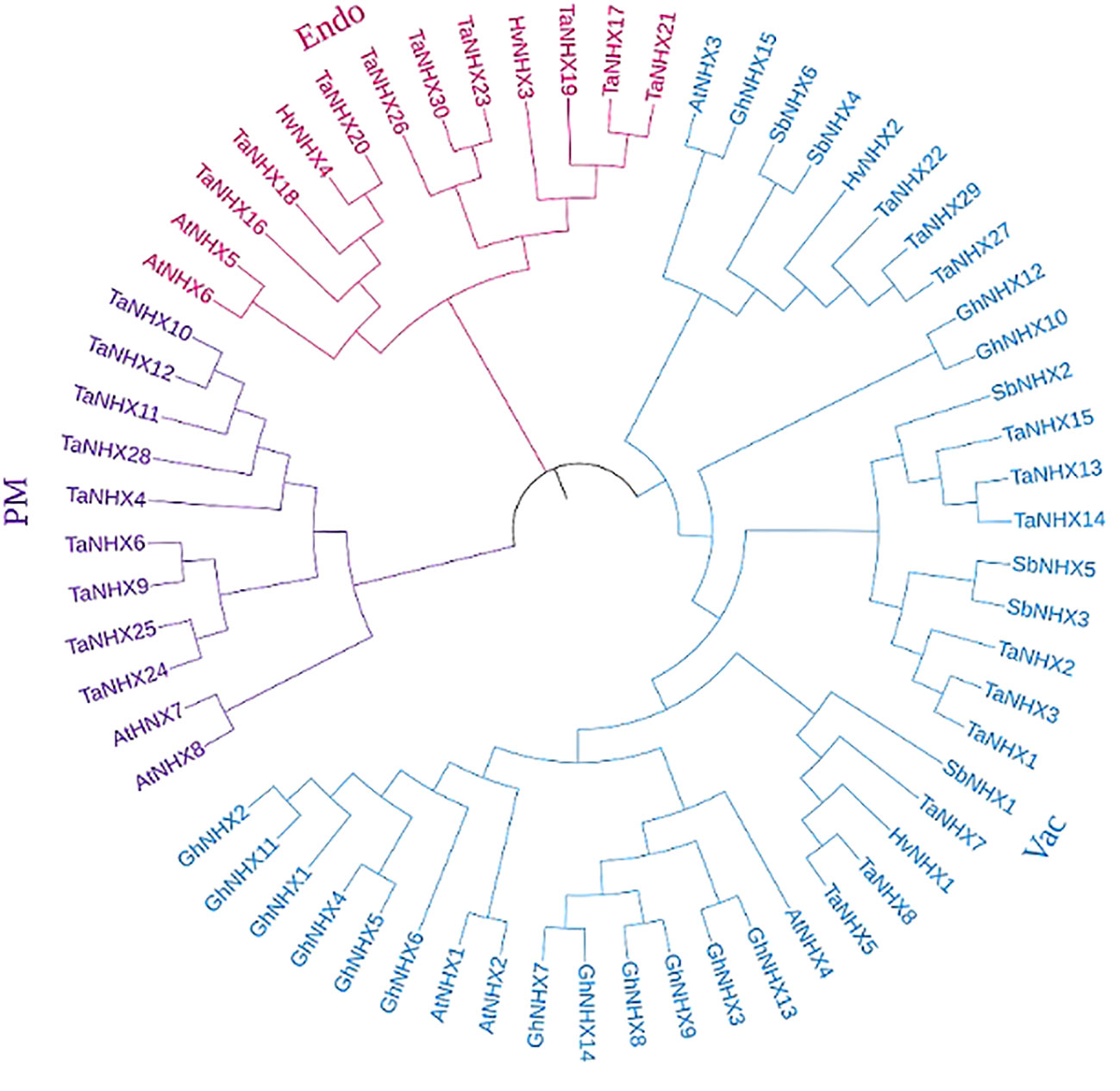
Figure 1 Phylogenetic trees showing relationships of NHX genes family of T. aestivum, A. thaliana, S. bicolor, H. vulgare, and G. hirusitum. The trees were constructed using the maximum likelihood method and a bootstrap replicate of 1000. The trees with the highest bootstrap support for each gene class have been shown here. The three major classes (Vac-, Endo-, and PM-) are marked with different colors.
GO enrichment analysis was conducted to uncover the potential functions of the NHX genes. Among the twenty TaNHX genes, significant enrichment was observed in biological processes (BP), molecular functions (MF), and cellular components (CC). In BP, the enriched terms included “sodium ion transmembrane transport,” “response to salt stress,” “potassium ion homeostasis,” and “regulation of pH,” among others (Supplementary Table S3). The enriched MF terms encompassed “binding,” “antiporter activity,” and “potassium: proton antiporter activity.” Furthermore, the enriched CC terms were associated with “plasma membrane” and “vacuolar membrane.” Notably, individual TaNHX genes exhibited enrichment in specific GO terms, indicating their involvement in distinct processes. For comprehensive information on the enriched GO terms and their functions see Supplementary Table S3.
3.2 Analysis of conserved motifs, gene structure, cis-regulatory elements, and physical mapping
The MEME web-server with default settings was employed to predict conserved motifs, shedding light on the evolutionary preservation of specific functional amino acids within the NHX protein family. Based on the protein sequences of all NHXs, we discovered a total of 10 potential motifs (Figure 2A). The length of the TaNHXs’ anticipated motifs ranged from 6 to 50 amino acids. Members of the same subfamily shared a similar motif arrangement. Logos of the motifs were presented in Supplementary Figure S1. The descriptive details of domains were given in Supplementary Table S4.
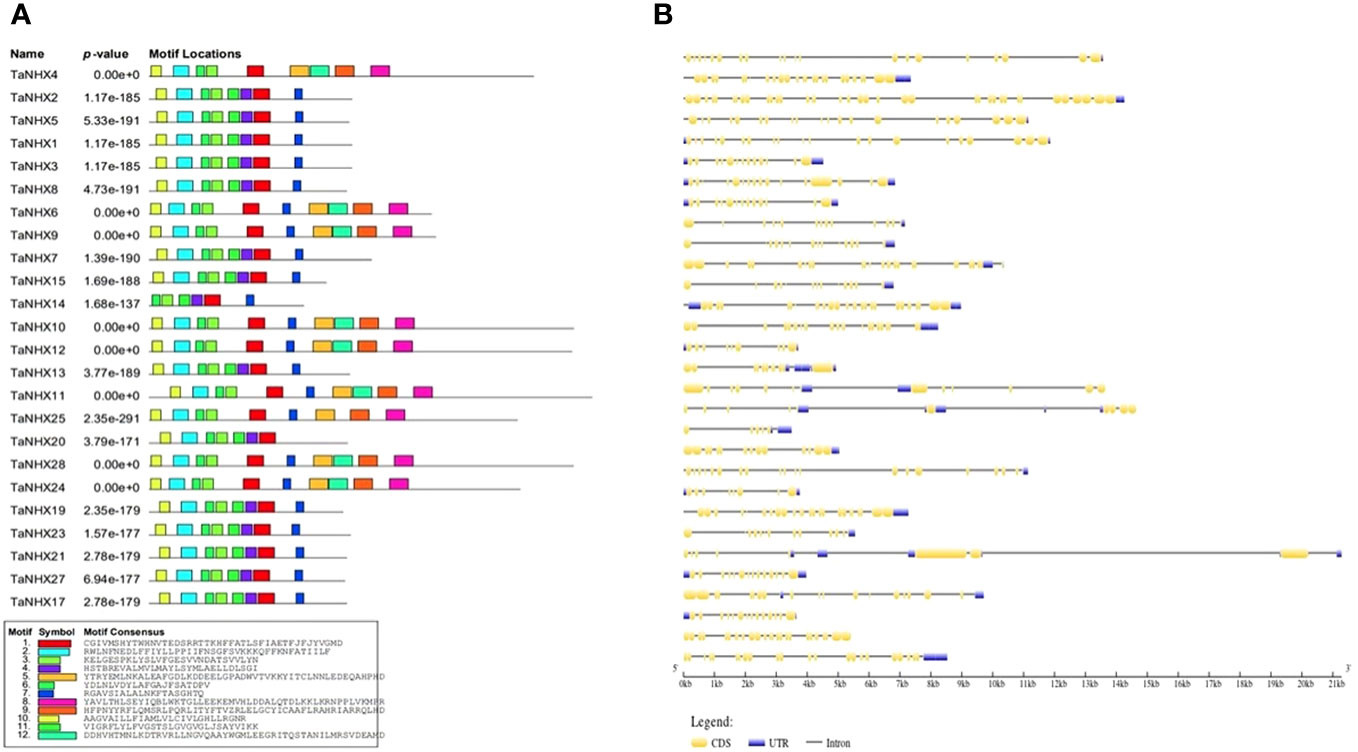
Figure 2 (A) Distribution of conserved motifs identified in proteins encoded by TaNHX. (B) Gene structures showing the organization of exons and introns, TaNHX genes.
Using Gene Structure Display Server v.2.0, the intron/exon architecture of the genes was examined to determine the structural properties of the wheat TaNHXs. The number of introns and exons differed significantly depending on the intron/exon patterns studied, which further explains the diversity in gene length. Non-coding sequences are frequently found in the genome, which is thought to be a sign of genomic complexity. Thus, examining these intron configurations reveals important details about the development, regulation and functionality of the NHXs. The examination of the wheat TaNHX gene architectures revealed significant variations across the three classes in terms of the number of introns and exons. The bulk of the Vac-class family among the 30 wheat TaNHXs included UTR (untranslated region) sequences at both the 5’ and 3’ ends. However, 6 out of the 12 Vac-class TaNHXs had 14 exons, 3 had more than 20 exons, and the rest were less than 10. The endo-class TaNHX had more than 10 exons except TaNHX26 (Figure 2B). However, the PM-class TaNHX (TaNHX9 and TaNHX10) had the highest share of intron-exons, with 27, 37 exons and 14, 20 introns, respectively. It was found that the distribution of introns and exons in the genes belonging to the same clade was very comparable. The exon lengths and intron regions of genes in the same class were mostly conserved. The analysis of amino acid sequence identity further confirmed the wheat TaNHXs’ sequence conservation (Figure 2B).
In order to better understand transcriptional control and gene expression, the promoter sequences of wheat TaNHX were identified by PlantCARE software (Figure 3). The major focus of the analysis was on cis-acting components associated with stress and hormones. Twenty two of the cis-acting components that were discovered had a hormonal connection, including ABA, salicylic acid (SA), gibberellin (GA), and jasmonate. The TaNHX comprised of phytohormones were TaNHX1, TaNHX3, TaNHX5, TaNHX7, TaNHX12, TaNHX13, TaNHX19, TaNHX22, TaNHX23, and TaNHX29 (Figure 3). Eleven cis-acting components, including anaerobic environments and low temperatures, were responsible for stress. The number of light-responsive elements was higher among these components. These cis-acting elements’ findings imply that TaNHXs may be crucial for the control of hormones and the response to stress in wheat.
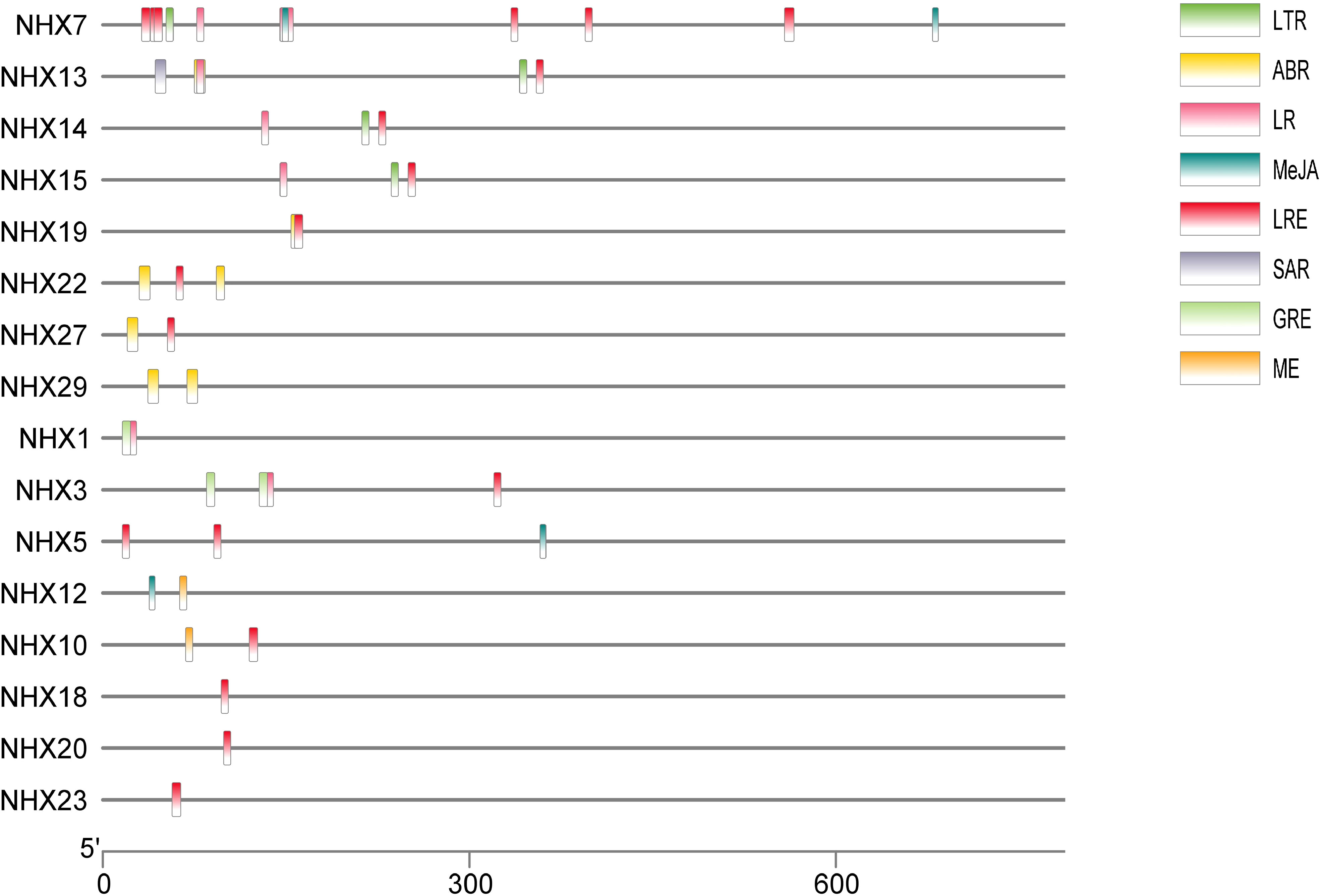
Figure 3 Predicted cis-elements in TaNHX promoters. The upstream length to the translation starting site can be inferred according to the scale at the bottom. The green, yellow, pink, red, orange and violet-colored boxes stand for DSR, SA, MeJA, Me, ABRE and MYB cis-elements, respectively.
To better comprehend how the TaNHX gene family evolved, we further examined the gene duplication occurrences. Thirty TaNHX genes were unevenly mapped onto six (A, B, D) chromosomes of the 21 T. aestivum chromosomes (Figure 4). Two chromosomes (Chr2 and Chr5) contained six TaNHX genes and three chromosomes (Chr1, Chr3, and Chr4) contained only one TaNHX gene on each A, B, and D sub-genome (Figure 4). No genes were present on chromosome 6. Gene duplication events, which are a primary mechanism of gene family growth, gave chances for the synthesis of additional genes and their functional divergence.
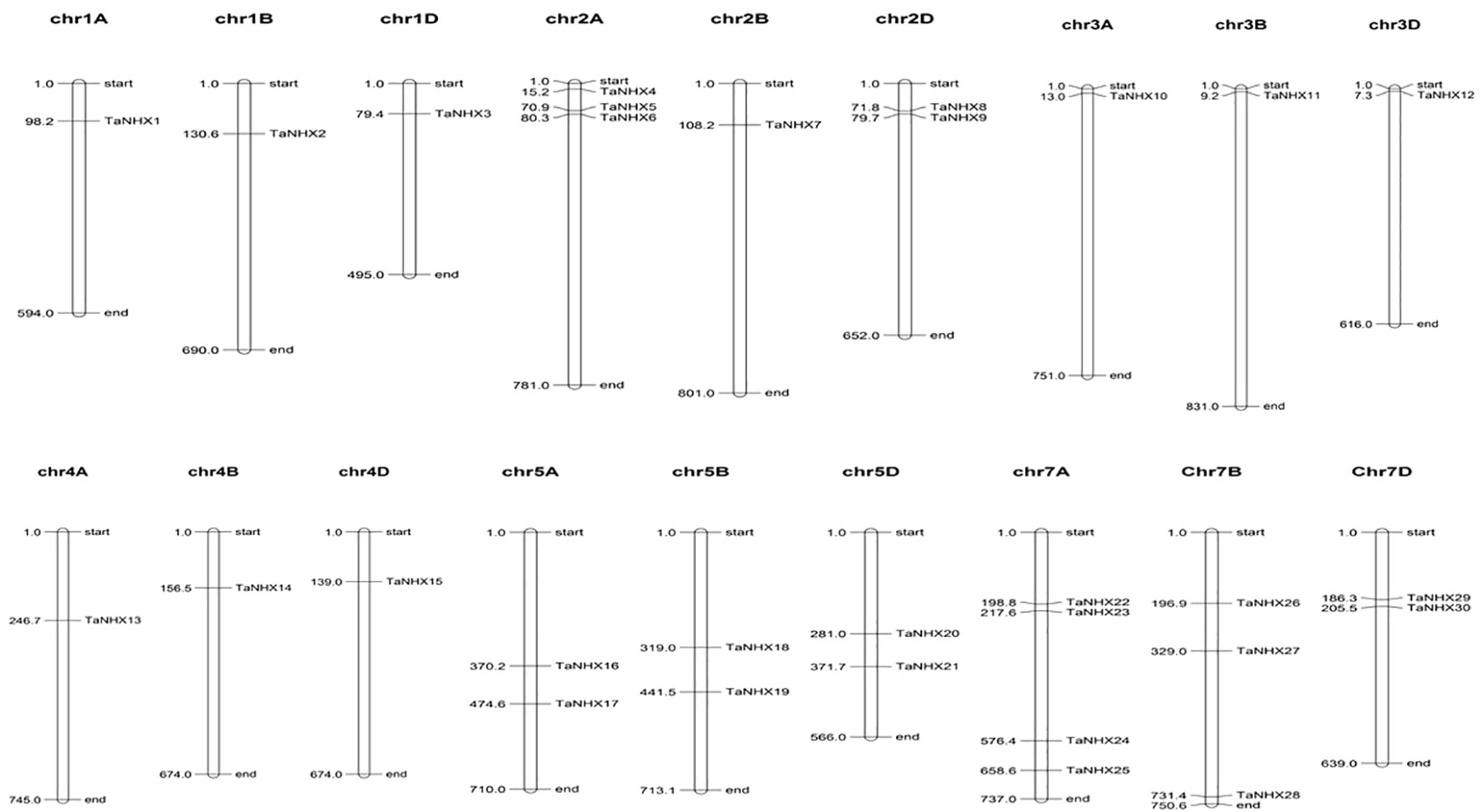
Figure 4 The distribution of NHX genes on chromosomes of T. aestivum. Chromosome numbers have been indicated on the top of each chromosome. The position of each gene on the respective chromosome has been depicted in terms of mega base-pairs by numbers beside each gene.
3.3 Protein-protein interaction, miRNA targeting, and duplication events in NHX genes
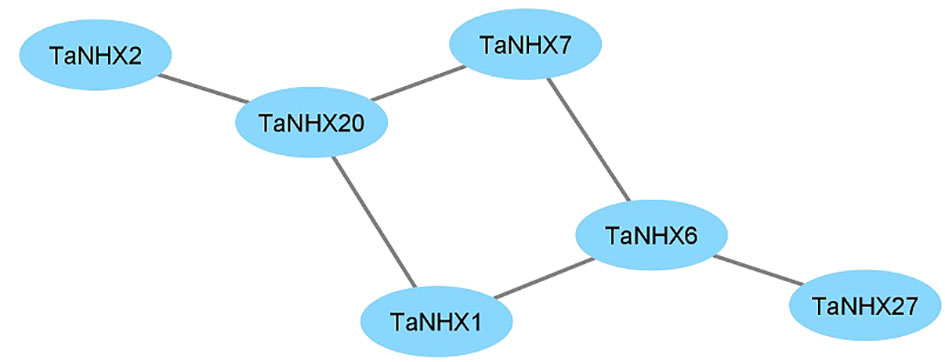
The response to salt stress is one of the major biological processes in which NHXs play a significant role. The PPI network was created by the STRING database to further examine the potential role of TaNHXs during potential interactions with other proteins (Figure 5). TaNHX proteins are not expected to have any direct interactions with one another. Out of thirty TaNHX proteins, six were participating in the protein-protein interaction network. They shared a similar kind of putative interactive protein. TaNHX1, TaNHX2, TaNHX7, and TaNHX20 were involved in proton exchange and belong to monovalent and proton antiporters (Figure 5). Numerous physiological processes, including vesicle trafficking, pH control, K+ homeostasis, protein transport, and growth and development, are regulated by the antiporters. In contrast, TaNHX6 interacted with SOS1, which is crucial for the expulsion of Na+ ions from cells.
This study employed network analysis, using TaNHX gene expression data, to uncover key insights into sodium ion transport mechanisms in plants. Significant Gene Ontology (GO) terms (FDR < 0.001) were identified, revealing TaNHX genes’ involvement in cellular processes, particularly transmembrane transport. Intriguingly, our analysis revealed intricate interactions between sodium ion transport and the regulation of biological quality, chemical homeostasis, and metal ion homeostasis (Supplementary Figure S2). Subcellular localization indicated that TaNHX proteins primarily reside in plasma membranes, endosomes, and vacuoles, emphasizing their role in Na+/-H antiporter activity and ion homeostasis. The findings also imply potential gene interactions affecting activity levels, paving the way for further investigations into these regulatory networks in plant sodium ion transport (Supplementary Figure S2). This research contributes valuable insights to plant biology and ion homeostasis regulation.
The psRNATarget service has been used to look at how miRNAs regulate the expression of TaNHX genes. For twenty distinct miRNAs, we identified 32 TaNHX genes as potential targets (Table 2). Tae-miR395 is implicated in the regulation of eight TaNHX genes (TaNHX6, TaNHX9, TaNHX10, TaNHX11, TaNHX12, TaNHX24, TaNHX25, and TaNHX28). Tae-miR9666 accounted for regulating the expression of four TaNHX genes (TaNHX6, TaNHX9, TaNHX24, and TaNHX25). The expression of TaNHX genes (TaNHX1, TaNHX3, and TaNHX29) may be influenced by Tae-miR9656 (Table 2). Tae-miR9772, Tae-miR9773, and Tae-miR9774 were predicted to regulate the expression of TaNHX22, TaNHX13, and TaNHX9, respectively.
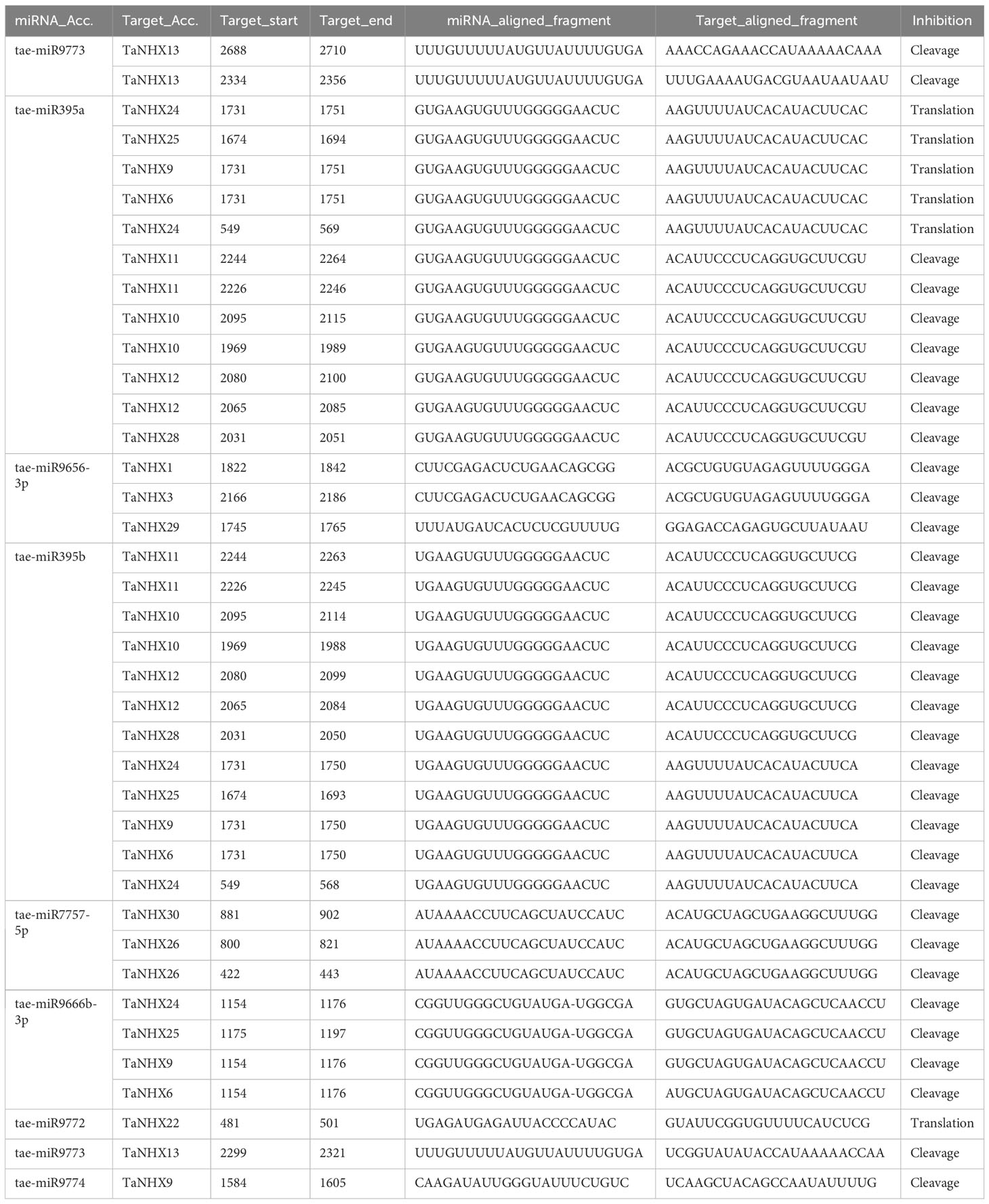
Table 2 Prediction of Tae-MIR genes and their targets by using the psRNATarget server with default parameters.
Comparative analysis was utilized to evaluate the orthologous of TaNHX gene duplication in wheat and A. tauschii. We discovered 28 orthologous gene pairs among all the NHX genes from wheat and A. tauschii. Contrarily, there were 27 NHX orthologous gene pairs discovered between wheat and rice, 29 between wheat and T. dicoccoides, and 30 between wheat and Arabidopsis (Supplementary Table S4). Orthologous associations between several species can be found through the analysis of collinearity correlations. NHX gene pairings between the genomes of T. aestivum and A. thaliana were syntenized. The findings revealed that 12 T. aestivum NHX genes shared syntenic relationships with AtNHX genes (Figure 6), indicating that these genes may have aided in the development of the TaNHX gene family.
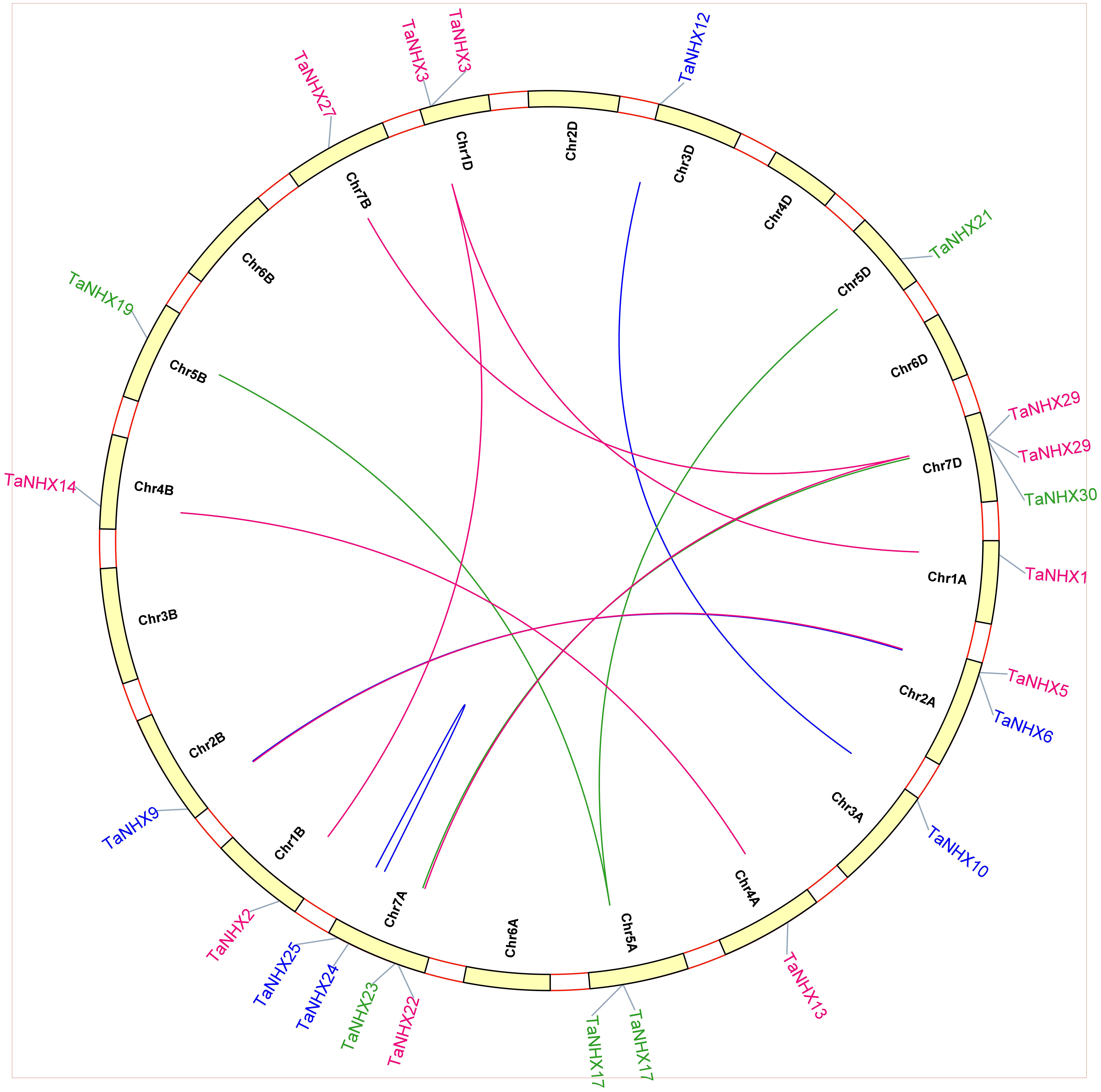
Figure 6 Interspecies synteny of T. aestivum, A. thaliana, O. indica, and H. vulgare on the basis of orthologous genes. Gray lines in the background indicate the collinear blocks within T. aestivum, A. thaliana, O. sativa, and H. vulgare while the red lines highlight the syntenic NHX gene pairs.
We conducted an analysis of synonymous substitutions (Ks) and non-synonymous substitutions (Ka) values to investigate the selective pressures underlying the duplication events of TaNHX genes, considering all nucleotide sequences. Our findings unveiled intriguing patterns, notably identifying six gene pairs (TaNHX7/TaNHX5, TaNHX15/TaNHX14, TaNHX22/TaNHX27, TaNHX1/TaNHX3, TaNHX20/TaNHX18, and TaNHX26/TaNHX23) characterized by Ka/Ks ratios lower than 1. These results suggest that these gene pairs experienced purifying selection, indicative of their evolutionary conservation (Supplementary Table 5). Our analysis detected nine distinct segmental duplication events occurring across various chromosomes, along with one notable tandem duplication event within the same chromosome. These findings underscore the pivotal role of segmental duplications in driving the expansion of TaNHX genes within the wheat genome. Furthermore, our observations suggest that certain TaNHX genes may have originated as a consequence of gene duplication events, shedding light on the evolutionary mechanisms responsible for the diversification of this gene family. But it may be due to selection pressure that the number of TaNHX genes was not expended in comparison to other transcription factors in wheat. Genes from three subfamilies (PM, endo and vac.) participated in the tandem and segmental duplication. We also investigated how frequently tandem duplications occur. This region contained 10 TaNHX gene pairs, all of which were closely linked. Notably, the sequence identities among these genes exceeded 80%, strongly suggesting their involvement in tandem duplication events. Given the profound impact of gene duplication on the emergence of novel functionalities and gene families, we conducted a comprehensive exploration of TaNHX gene duplication events within the wheat genome. The paralogous gene pairs were employed to construct a circos plot (Figure 7), enabling us to visualize and elucidate the intricate patterns of duplication events and their implications.
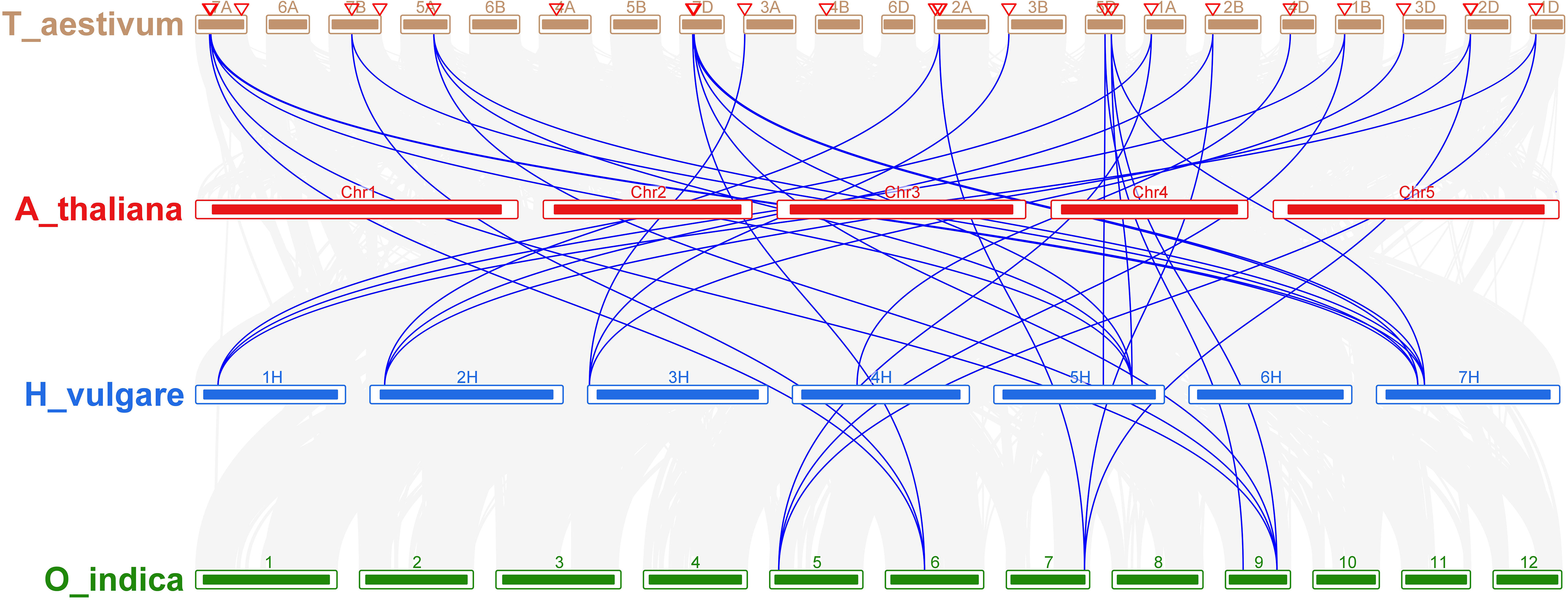
Figure 7 The synteny analysis of TaNHX family in T. aestivum. Different colors represent NHX family on A, B and D sub-genome red lines indicate duplicated TaNHX gene pairs on A sub-genome, green lines indicated on B sub-genome and blue represented D sub-genome. The chromosome number is indicated at the bottom of each chromosome.
3.4 Tissue-specific expression profiles of NHX genes
TaNHX expression levels were analyzed in wheat roots and leaf tissues exposed to salt stress. The expression analysis was done using RNA-seq data (SRP304900) from NCBI. Under normal conditions, TaNHX1, TaNHX2, TaNHX3, TaNHX5, TaNHX10, TaNHX11, and TaNHX20 were found to be upregulated in all tissues except in grain (Figure 8). However, TaNHXs displayed differential gene expression levels among the tissue types in different genes. It was observed that 23 out of 30 TaNHX genes were expressed very highly in both tissues. Whereas, the genes e.g., TaNHX4, TaNHX17, TaNHX21, TaNHX22, TaNHX27, TaNHX28, and TaNHX29 expression were downregulated in all tissues under salt stress conditions (Figure 8). However, it was found that TaNHX3, TaNHX5, TaNHX7, TaNHX11, TaNHX16, TaNHX18, and TaNHX20 expressed upregulation in all tissues under heat, drought, and cold (Supplementary Figure S3) stresses.
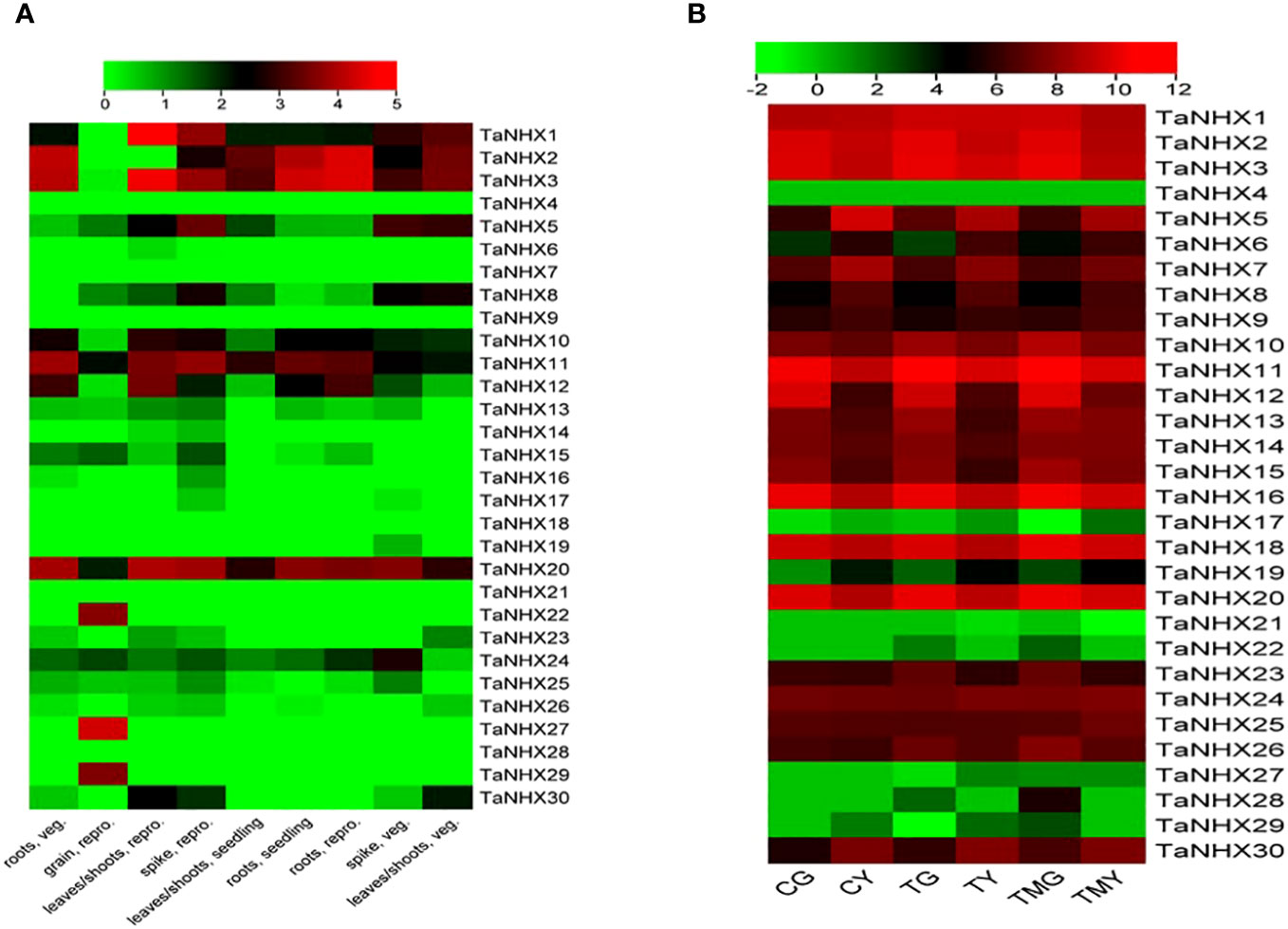
Figure 8 Normalized expression profiles of NHX genes of T. aestivum in various plant tissues under; (A) control and (B) salt stress conditions. CG represented the control wheat root samples, TG: 150mM NaCl treated wheat root, TMG: 5mM 3-Methylamide + 150mM NaCl treated wheat root, CY: control wheat leaves, TY: 150mM NaCl treated wheat leaves, TMY: 5mM 3-MA + 150mM NaCl treated leaves.
3.5 Validation of NHX genes using qRT- PCR under salinity stress
Using qRT-PCR analysis, we quantitatively evaluated the expression patterns of ten TaNHXs genes under control and salt-stress conditions to observe the potential functions that TaNHXs may play in wheat in responses to salinity stress. The findings demonstrated that salt tolerance (KRL213) and salt-sensitive (HD2009) wheat cultivars both stimulated all TaNHX genes in leaf and root tissues in a concentration-dependent manner (Figure 9). The tissues with the highest induction were the roots, followed by the leaves. TaNHX gene overexpression caused by salinity was greater in KRL213 than HD2009. Out of ten TaNHX genes, five genes [TaNHX2 (~14 folds), TaNHX12 (~9 folds), TaNHX16 (~7 folds), TaNHX20 (~10 folds), TaNHX23 (~3 folds)] showed up-regulation, in the roots of KRL213 at 48 h under 150 mM NaCl stress condition (Figure 9A). On the other hand, expression levels of TaNHX2, TaNHX16, and TaNHX20 were expressed more than twofold in roots treated with 150 mM NaCl at different time intervals. It was observed that TaNHX2, TaNHX12, and TaNHX20 expression levels were higher in the leaves of KRL213 (~10, ~5, and ~16 times, respectively) than HD2009 were down-regulated at 48 h of 150 mM NaCl treatment (Figure 9B). Salinity stress clearly shows up-regulation of TaNHXs in the leaves of KRL213, whereas in HD2009 two genes TaNHX2 and TaNHX20, were expressed higher in leaf tissue at 0 h and 3 h of stress. In KRL213 leaf tissue, all genes used for the validation were up-regulated at 3 h of salt stress (Figure 9B). The gene expression profiles of individual TaNHX members were evaluated in both leaf and root tissues under salinity stress conditions, yielding results that consistently mirrored the salt tolerance characteristics of the two wheat genotypes under study.
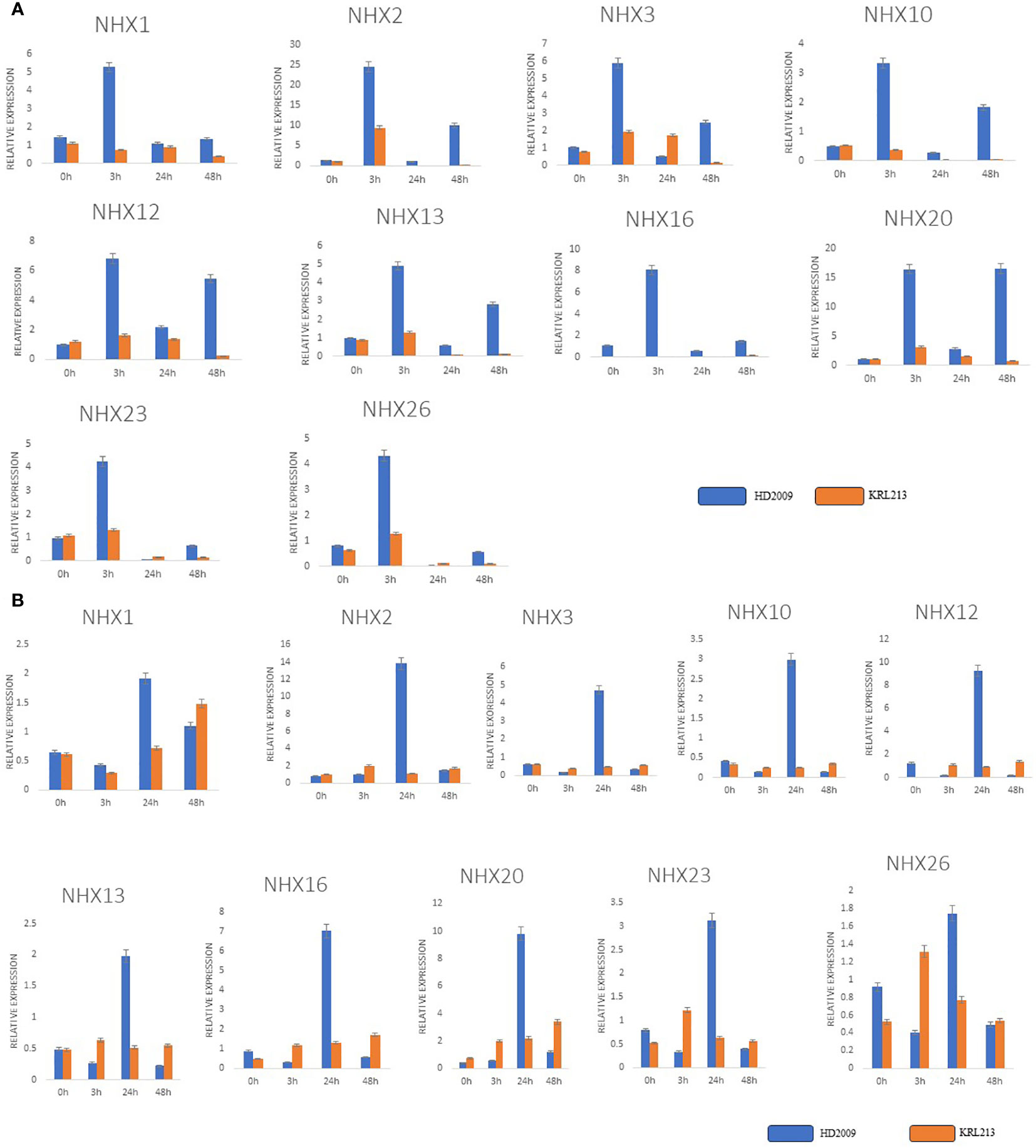
Figure 9 qRT-PCR expression analysis of selected genes in contrasting wheat varieties KRL213 and HD2009 for salt tolerance. (A) leaf tissues and, (B) root tissues under salt stress (150mM NaCl) at different time intervals. The names of the genes are shown in the x-axis, and y-axis represents the fold changes of expression of the genes. The data represents the mean ± standard deviation with n = 3. * Above the bars indicate significant correlation at the 0.05, as analyzed by Student’s T-test.
3.6 MD simulation analysis
The Root Mean Square Deviation (RMSD) serves a crucial statistic for assessing the average changes of a set of atoms concerning a reference frame. Analyzing the RMSD of the protein backbone atoms relative to their initial positions can yield valuable insights into the protein conformational dynamics. In our investigation, the RMSD values for all three proteins were below 2.5 nm. Figure 10A illustrates a stabilizing trend in the RMSD values around 10 ns mark, suggesting the attainment of equilibrium within the simulation. This stabilization milestone serves as an optimal foundation for subsequent analysis.
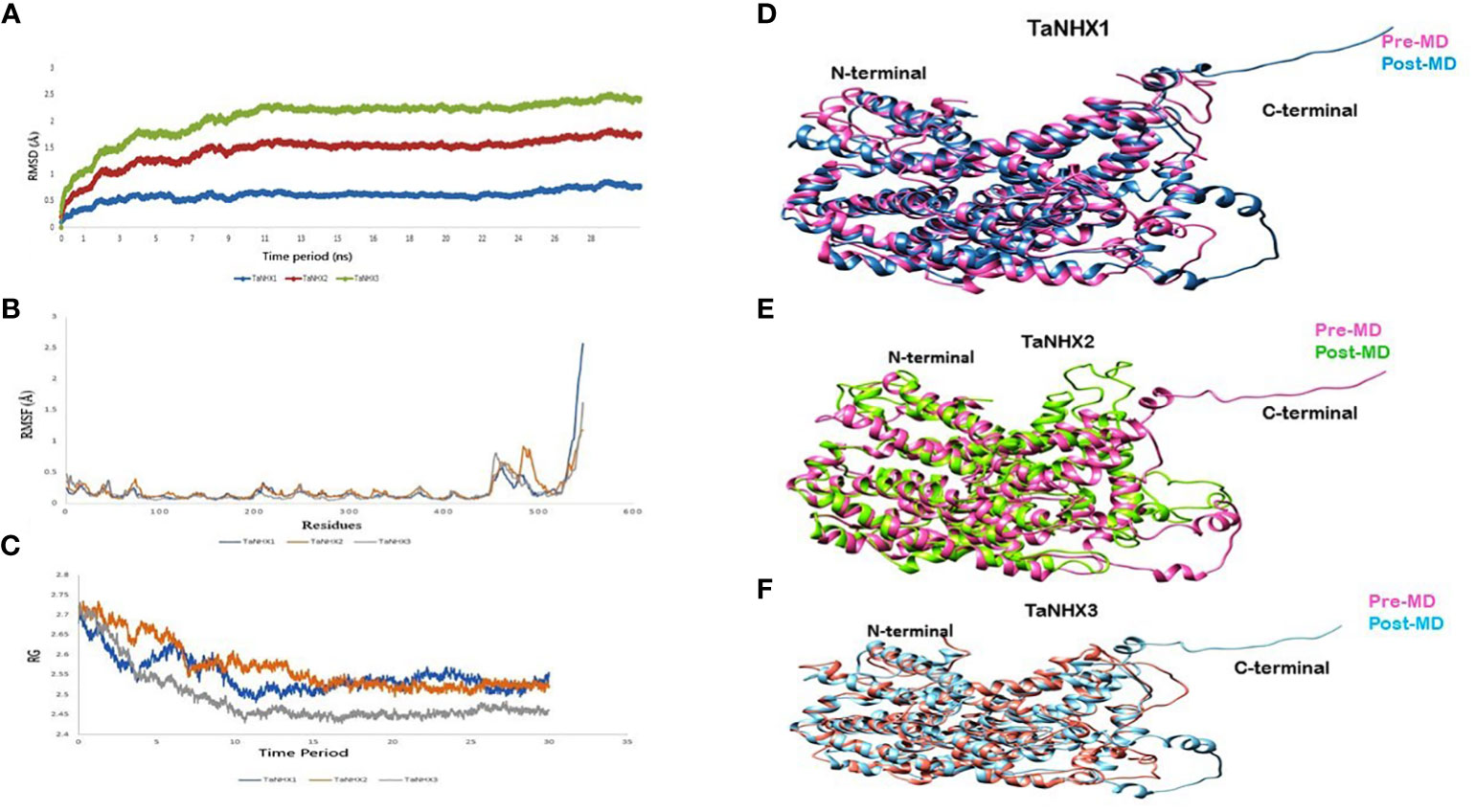
Figure 10 MD simulations analysis of NHX Proteins. (A) Root mean square deviation, (B) Root mean square fluctuations (C) Radius of gyration during 30 ns simulations. Superimposition between pre- and post-MD structure shown with different colours for; (D) TaNHX1, (E) TaNHX2 and, (F) TaNHX3 proteins.
Moreover, the Root Mean Square Fluctuation (RMSF) analysis delves into the localized fluctuations occurring along the protein chain during the molecular dynamics (MD) simulations RMSF graph uncovered fluctuations primarily at the C-terminals of the proteins, while the core of the TaNHX proteins displayed no substantial variations (Figure 10B). Additionally, the radius of gyration (Rg) value for the TaNHX structure exhibited a decreasing trend as the simulation time progressed, indicating a progressive compaction of the modeled structure. This observation aligns with the results from the initial run of the simulations. The radius of gyration is a measure of the overall size and compactness of a protein structure. A lower Rg value suggests a more tightly folded and compact conformation, while a higher Rg value indicates a more extended or flexible structure. Therefore, the lower Rg values obtained for the TaNHX protein structure suggest that they adopt a more compact and stable conformation (Figure 10C).
4 Discussion
Sodium-proton (Na+/H+) antiporters, which are encoded by genes belonging to the NHX family, play a pivotal role in upholding ion equilibrium and aiding in membrane trafficking. Their significance is particularly pronounced in plant cells exposed to the challenges of salinity stress, as elucidated by Pehlivan et al. (2016). The NHX gene families have undergone comprehensive identification and functional characterization in various plant species such as Arabidopsis, rice, wheat, sugar beet, and cotton (Manik et al., 2015; Wilkins et al., 2016; Kumari et al., 2018; and Wu et al., 2019). A comprehensive genome-wide investigation of TaNHX genes in T. aestivum has not been conducted yet. Our study aimed to fill this gap by identifying TaNHX genes in the wheat genome and conducting a thorough characterization, including phylogenetic relationships, their characterization, and expression profiling under varying salinity stress conditions, along with molecular dynamic simulation.
Our analysis revealed that TaNHX members in wheat could be classified into three groups based on multiple sequence alignments and sub-cellular localization. For instance, TaNHX7 and TaNHX8 were found to be localized in the plasma membrane, TaNHX5 and TaNHX6 in the endo-membrane, while the remaining members were localized in the vacuole. This localization pattern was comparable with the prior studies in Arabidopsis (Ramesh et al., 2019; Parveen et al., 2021). Notably, the similarity observed in NHX families across lower to higher plant taxa suggests the retention of functional importance throughout evolutionary processes (Shuai et al., 2013). Sub-cellular localization of NHX transporters significantly influences their functionality. NHX family members situated on both the plasma membrane and tonoplast play an integral role in maintaining ionic equilibrium by sequestering and eliminating excessive sodium ions (Na+).
In our analysis, we identified ten conserved motifs in TaNHXs. Notably, a key element of profound importance is the remarkably conserved membrane-spanning pore and cation-binding domain, exemplified by the nonapeptide sequence “FFIYLLPPI,” renowned as the amiloride-binding site. This distinctive characteristic defines the membrane-bound NHX transporters in plants (Wu et al., 2011). This site is well-known for its role in inhibiting cation/H+ exchange when amiloride is present. Interestingly, we found that this region is conserved in the second motif of all TaNHX peptides, indicating that TaNHXs, as a whole, exhibit functional and sequence-based similarities, with the exception of TaNHX14 (Figure 2A). Similar sequence characteristics have been observed in Arabidopsis (Aharon et al., 2003), soybean (Chen et al., 2015), and tea (Paul et al., 2021). These findings contribute to our understanding of the conserved motifs and functional characteristics of TaNHX proteins in wheat and highlight the evolutionary relationships among TaNHXs based on their gene structures.
cis-acting elements, serving as genetic switches for gene transcription, exert significant control over biological processes, encompassing responses to hormonal fluctuations, environmental stress, and developmental stresses (Ren et al., 2017; Zhang et al., 2020). Phytohormones such as gibberellins, methyl jasmonic acid, and abscisic acid wield immense influence over growth, development, and stress responses (Zhao et al., 2021). It is conceivable that TaNHXs actively participate in hormone signaling throughout wheat’s developmental stages and stress responses. This conjecture finds support in the presence of ABREs (abscisic acid responsive elements) and MeJA (methyl jasmonate) responsive elements within the promoter regions of these genes (Table 2). Our findings are in tune with those in P. trichocarpa (Shuai et al., 2013) and S. bicolor (Kumari et al., 2018), which unveiled stress-responsive elements in NHX gene promoters. Furthermore, the promoter regions of TaNHXs harbor elements associated with light responsiveness and low-temperature sensitivity, suggesting their potential involvement in pivotal regulatory processes encompassing phytohormonal dynamics, stress responses, cellular development, and metabolism. By identifying these cis-acting elements and their association with TaNHXs in wheat, our study sheds light on their potential role in the regulation of TaNHX gene expression and their involvement in crucial physiological processes. These findings deepen our understanding of the intricate signaling networks that coordinate hormone responses, stress adaptation, and overall plant development in wheat.
The association of TaNHXs with other proteins, such as SOS2, contributes to the production of a protein kinase that plays a vital role in alleviating salt stress in Arabidopsis (Wilkins et al., 2016). Another protein, SOS3, acts as a calcium ion sensor and facilitates the transportation of SOS2 and SOS1, leading to the efflux of sodium ions from the cells (Shi et al., 2000). The analysis of protein-protein interactions (PPI) revealed a predicted interaction between PM-bounded TaNHX6 and SOS1. TaNHX6, known to possess several hormone- and stress-related cis-acting elements, plays a pivotal role in facilitating the exclusion of Na+ ions from the cell. Similar results have been observed by Manik et al. (2015) in honeysuckle. However, it is important to note that the regulation of salt tolerance involves a complex and interconnected network of interactions (Figure 5). Among the key players in this network are the membrane-bound pyrophosphatases, namely H+-PPase and H+-ATPase related proteins. These proton transporters, dependent on potassium, are crucial for plant responses to stress as well as growth and development. A balanced Na+/K+ ratio is essential for various vital functions, including proper stomatal function, protein synthesis, cell osmoregulation, photosynthesis, and turgor maintenance (Parveen et al., 2021). Given the importance of TaNHXs and potassium transport-related proteins in maintaining this balance, it is justified to include them in protein interaction networks in current research endeavors.
miRNAs have been previously reported as having abiotic stress-responsiveness (Ramesh et al., 2019), indicating their potential involvement in regulating stress-related processes. Specific miRNA families have known roles in different aspects of plant development and response to abiotic stresses (Shuai et al., 2013; Gupta et al., 2014; Luan et al., 2014; Zhang and Chen, 2021; Liu et al., 2022). We identified a total of 20 different miRNAs targeting 26 distinct TaNHX genes (Table 2). Differential expression patterns of TaNHX genes observed in our study can be linked to the stress-responsive miRNAs that target these genes, further emphasizing their role in stress adaptation and plant development.
The analysis of 30 salt-responsive genes in T. aestivum revealed interesting insights. Collinearity analysis indicated that 18 TaNHX genes resulted from whole-genome duplication or segmental duplication, a pattern observed in various gene families, including cotton MADS-Box, GT47, and soybean WRKY, highlighting the role of gene duplication in their evolution (Cannon et al., 2004; Yin et al., 2013; Ren et al., 2017; Wu et al., 2019). Additionally, the collinearity analysis of NHXs revealed shared ancestry among the A, B, and D genomes of NHXs, signifying their common origin. These findings shed light on the NHX family’s evolution in wheat, primarily driven by WGD or segmental duplication events. This deepens our understanding of gene duplication mechanisms, genetic information exchange between species, and the processes shaping species evolution. Furthermore, the evaluation of non-synonymous (Ka) and synonymous (Ks) substitution rates among duplicated gene pairs stands as a pivotal tool for assessing selection pressure and approximating the timing of duplications. In the context of TaNHX genes in wheat, these findings robustly suggest that purifying selection mechanisms played a prominent role in generating genes with conserved functions and, in some cases, undergoing pseudogenization (Supplementary Table S4). Regarding the anticipated motifs within NHX proteins, it has become evident that genes within the duplicated gene cluster showcase functional conservation, while a few motifs lack discernible functional details, potentially indicating pseudogenization. This intriguing observation underscores the possibility of pseudogenization events, as indicated in (Supplementary Table S4). This phenomenon can be attributed to one or more ancestral polyploidy events that transpired in numerous angiosperm plant lineages. Consequently, these gene duplications within the wheat genome have been instrumental in the emergence of evolutionary innovations. Furthermore, a thorough examination of syntenic blocks among NHX genes across wheat and several other plant species has unveiled that the closest orthologs of the wheat channels trace their origins to barley. These extensive synteny relationships at the gene level serve as compelling evidence affirming the close evolutionary affiliations between these species (Ullah et al., 2022). Variations in these evolutionary relationships can provide valuable insights into the substantial rearrangement events that have left their imprint on the genomes of wheat and its related species throughout the course of evolution.
Considering the association of TaNHX genes with stress-responsive cis-acting elements and miRNAs, we hypothesized that their expression patterns would exhibit variations. In our study, we observed up-regulation of TaNHX genes in both leaf and root tissues of the cultivars under salinity stress, irrespective of the severity of salt exposure and the cultivars’ salt tolerance capacity (Figure 8A). Remarkably, the roots, being the first organs exposed to salt, displayed significantly higher induction of TaNHX genes. Although the induction of TaNHXs in leaves was relatively lower compared to roots, it still contributed to the exclusion and improved compartmentalization of Na+ ions within the cells. This heightened expression of sodium transporters directly facilitated the maintenance of Na+ ion homeostasis. Additionally, we observed a gradual increase in gene expression within 3h of salt treatment in leaf tissue. Similar expression patterns were observed in cotton roots, where gene expression reached its peak at 3h, gradually declined, and then increased again at 48h (Figure 8). These findings align with previous studies on BvNHX genes in sugar beet (Wu et al., 2019). Similarly, the wheat gene TaNHX3 exhibited increased expression within 24h in both roots and leaves, followed by a gradual decrease within 48h. Consistent with previous studies on wheat transgene TaNHX, our findings highlighted the responsiveness of TaNHX2, TaNHX12, and TaNHX20 to salinity stress. Yue et al. (2021) found that the addition of 3-methyladenine (3-MA) inhibits autophagy, increases ROS accumulation and impairs the tolerance of wheat seedlings to NaCl stress. Differential expression patterns of NHXs under salinity stress have also been reported in other plant species, such as Beta vulgaris with five NHXs (Wu et al., 2019), Populus trichocarpa with eight NHXs (Tian et al., 2017), and Sorghum bicolor with six NHXs (Kumari et al., 2018). Hierarchical clustering analysis (Figure 8B) revealed two distinct clusters based on the activities of TaNHXs in all tissues under saline conditions in both cultivars. This clustering suggests different regulatory mechanisms or functional roles of TaNHXs in response to salinity stress.
To gain further insights into the TaNHX proteins, we employed molecular modeling and MD simulations to analyze their three-dimensional structure, stability, and conformational changes. The MD simulations clearly confirmed the stability of the three predicted TaNHX proteins over the entire simulation period. These findings collectively offer valuable insights into the expression patterns of TaNHX genes under salinity stress and their potential role in maintaining Na+ ion homeostasis. By elucidating the distinctive expression patterns and robust stability of TaNHX proteins, our study contributes significantly to the comprehension of the molecular mechanisms that underlie salt tolerance in wheat.
5 Conclusions
A comprehensive analysis of the TaNHX gene family in wheat was carried out in the present study. A total of 30 TaNHXs had been identified and phylogenetically divided into three subfamilies, as supported through highly conserved gene structure and motifs in addition to regulatory elements, revealing their significant role in wheat tolerance under salt stress. These thirty TaNHX genes were unevenly distributed among 18 chromosomes in wheat, except chromosomes 6A, 6B, and 6D, wherein no genes have been present. Collinearity analysis results confirmed that segmental duplication events had been essential for the expansion of TaNHX genes in the wheat genome and that certain TaNHX genes might also had been formed through gene duplication. Through qRT-PCR analysis, we observed that certain TaNHX genes in wheat showed increased expression under salt stress, with varying expression patterns at different time intervals in salt-tolerant and salt-sensitive cultivars of wheat. These findings emphasize the significance of the NHX family in wheat tolerance for salt stress response and offer a strong foundation for in addition exploration of wheat NHX genes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
PS: Conceptualization, Formal analysis, Investigation, Supervision, Writing – original draft, Writing – review & editing, Project administration. SM: Data curation, Formal analysis, Methodology, Software, Validation, Writing – original draft. BP: Data curation, Software, Validation, Writing – review & editing. GS: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The project work was supported by the grants from Indian Council of Agricultural Research, New Delhi. Funding body has no role in the study, data collection, analysis and manuscript writing.
Acknowledgments
The authors acknowledge to Germplasm Resource Unit of ICAR-IIWBR, Karnal for supply of seed material.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1266699/full#supplementary-material
Supplementary Figure 1 | Amino acid sequences of motifs in logos format.
Supplementary Figure 2 | Interaction network of NHX family in wheat list of significant GO term (FDR <0.01) of nodes presented in network.
Supplementary Figure 3 | In-silico expression analysis of NHX genes in various tissues under drought and heat stress.
References
Abraham, M. J., Murtola, T., Schulz, R., Páll, S., Smith, J. C., Hess, B., et al. (2015). Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25. doi: 10.1016/j.softx.2015.06.001
Aharon, G. S., Apse, M. P., Duan, S., Hua, X., Blumwald, E. (2003). Characterization of a family of vacuolar Na + /H + antiporters in. Analysis 253(1):245–256. 10.1023/A:1024577205697
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 37, 1–7. doi: 10.1093/nar/gkp335
Bassil, E., Tajima, H., Liang, Y. C., Ohto, M.a., Ushijima, K., Nakano, R., et al. (2011). The arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23, 3482–3497. doi: 10.1105/tpc.111.089581
Brett, C. L., Donowitz, M., Rao, R. (2005). Evolutionary origins of eukaryotic sodium/proton exchangers. Am. J. Physiol. Cell Physiol. 288 (2), (223–239). doi: 10.1152/ajpcell.00360.2004
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 1–21. doi: 10.1186/1471-2229-4-10
Chen, H. T., Chen, X., Wu, B. Y., Yuan, X. X., Zhang, H. M., Cui, X. Y., et al. (2015). Whole-genome identification and expression analysis of K+ efflux antiporter (KEA) and Na+/H+ antiporter (NHX) families under abiotic stress in soybean. J. Integr. Agric. 14, 1171–1183. doi: 10.1016/S2095-3119(14)60918-7
Dai, X., Zhao, P. X. (2011). PsRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 39, W155–W159. doi: 10.1093/nar/gkr319
Dragwidge, J. M., Scholl, S., Schumacher, K., Gendall, A. R. (2019). NHX-type Na+(K+)/H+ antiporters are required for TGN/EE trafficking and endosomal ion homeostasis in Arabidopsis thaliana. J. Cell Sci. 132, 2–11. doi: 10.1242/jcs.226472
Dubay, K. H., Hall, M. L., Hughes, T. F., Wu, C., Reichman, D. R., Friesner, R. A. (2012). Accurate force field development for modelling conjugated polymers. J. Chem. Theory Comput. 8, 4556–4569. doi: 10.1021/ct300175w
Fukuda, A., Nakamura, A., Hara, N., Toki, S., Tanaka, Y. (2011). Molecular and functional analyses of rice NHX-type Na+/H+ antiporter genes. Planta 233, 175–188. doi: 10.1007/s00425-010-1289-4
Gaxiola, R. A., Rao, R., Sherman, A., Grisafi, P., Alper, S. L., Fink, G. R. (1999). The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc. Natl. Acad. Sci. U. S. A. 96, 1480–1485. doi: 10.1073/pnas.96.4.1480
Ge, S. X., Son, E. W., Yao, R. (2018). iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinf. 19, 1–24. doi: 10.1186/s12859-018-2486-6
Gupta, O. P., Meena, N. L., Sharma, I., Sharma, P. (2014). Differential regulation of microRNAs in response to osmotic, salt and cold stresses in wheat. Mol. Biol. Rep. 41 (7), 4623–4629. doi: 10.1007/s11033-014-3333-0
Hammer, Ø., Harper, A. T., Ryan, P. D. (2001). Past: Paleontological Statistics software package for education and data analysis. Palaeontologia Electronica 4, 9. Available at: http://palaeo-electronica.org/2001_1/past/issue1_01.htm.
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Khare, T., Kumar, V., Kishor, P. B. K. (2015). Na+ and Cl- ions show additive effects under NaCl stress on induction of oxidative stress and the responsive antioxidative defense in rice. Protoplasma 252, 1149–1165. doi: 10.1007/s00709-014-0749-2
Kozomara, A., Griffiths-Jones, S. (2014). MiRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–73. doi: 10.1093/nar/gkt1181
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kumari, P. H., Kumar, S. A., Ramesh, K., Reddy, P. S., Nagaraju, M., Prakash, A. B., et al. (2018). Genome-wide identification and analysis of arabidopsis sodium proton antiporter (NHX) and human sodium proton exchanger (NHE) homologs in Sorghum bicolor. Genes. 9 (5), 236–254. doi: 10.3390/genes9050236
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Liu, C., Ma, D., Wang, Z., Chen, N., Ma, X., He, X. Q. (2022). MiR395c Regulates secondary xylem development through sulfate metabolism in poplar. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.897376
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Luan, M., Xu, M., Lu, Y., Zhang, Q., Zhang, L., Zhang, C., et al. (2014). Family-wide survey of miR169s and NF-YAs and their expression profiles response to abiotic stress in maize roots. PloS One 9, 1–11. doi: 10.1371/journal.pone.0091369
Manik, S. M. N., Shi, S., Mao, J., Dong, L., Su, Y., Wang, Q., et al. (2015). The calcium sensor CBL-CIPK is involved in plant’s response to abiotic stresses. Int. J. Genomics 2015, 10–12. doi: 10.1155/2015/493191
Muthusamy, S. K., Dalal, M., Chinnusamy, V., Bansal, K. C. (2016). Differential regulation of genes coding for organelle and cytosolic ClpATPases under biotic and abiotic stresses in Wheat. Front. Plant Sci. 7, 929. doi: 10.3389/fpls.2016.00929
Parveen, Anwar-Ul-Haq, M., Aziz, T., Aziz, O., Maqsood, L. (2021). Potassium induces carbohydrates accumulation by enhancing morpho-physiological and biochemical attributes in soybean under salinity. Arch. Agron. Soil Sci. 67, 946–959. doi: 10.1080/03650340.2020.1769075
Paul, A., Chatterjee, A., Subrahmanya, S., Shen, G., Mishra, N. (2021). NHX gene family in Camellia sinensis: In-silico genome-wide identification, expression profiles, and regulatory network analysis. Front. Plant Sci. 20. doi: 10.3389/fpls.2021.777884
Pehlivan, N., Sun, L., Jarrett, P., Yang, X., Mishra, N., Chen, L., et al. (2016). Co-overexpressing a plasma membrane and a vacuolar membrane sodium/proton antiporter significantly improves salt tolerance in transgenic arabidopsis plants. Plant Cell Physiol. 57, 1069–1084. doi: 10.1093/pcp/pcw055
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Ramesh, S. V., Govindasamy, V., Rajesh, M. K., Sabana, A. A., Praveen, S. (2019). Stress-responsive miRNAome of Glycine max (L.) Merrill: molecular insights and way forward. Planta 249, 1267–1284. doi: 10.1007/s00425-019-03114-5
Ren, Z., Yu, D., Yang, Z., Li, C., Qanmber, G., Li, Y., et al. (2017). Genome-wide identification of the MIKC-type MADS-Box gene family in Gossypium hirsutum L. unravels their roles in flowering. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00384
Shi, H., Ishitani, M., Kim, C., Zhu, J. K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. 97, 6896–6901. doi: 10.1073/pnas.120170197
Shuai, P., Liang, D., Zhang, Z., Yin, W., Xia, X. (2013). Identification of drought-responsive and novel Populus trichocarpa microRNAs by high-throughput sequencing and their targets using degradome analysis. BMC Genomics 14, 233–246. doi: 10.1186/1471-2164-14-233
Skolnick, J., Gao, M., Zhou, H., Singh, S. (2021). The Alphafold2 predicted structures are not considered accurate enough for virtual screening and further research is required to determine the best way to process and prepare these structures for virtual screening. J. Chem. Inf. Model. 61, 4827–4831. doi: 10.1021/acs.jcim.1c01114.AlphaFold
Szklarczyk, D., Franceschini, A., Wyder, S., Forslund, K., Heller, D., Huerta-Cepas, J., et al. (2015). STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452. doi: 10.1093/nar/gku1003
Tian, F., Chang, E., Li, Y., Sun, P., Hu, J., Zhang, J. (2017). Expression and integrated network analyses revealed functional divergence of NHX-type Na+/H+ exchanger genes in poplar. Sci. Rep. 7, 1–17. doi: 10.1038/s41598-017-02894-8
Ullah, U., Buttar, Z. A., Shalmani, A., Muhammad, I., Ud-Din, A., Ali, H. (2022). Genome-wide identification and expression analysis of CPP-like gene family in Triticum aestivum L. under different hormone and stress conditions. Open Life Sci. 17, 544–562. doi: 10.1515/biol-2022-0051
Voorrips, R. E. (2002). Mapchart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wilkins, K. A., Matthus, E., Swarbreck, S. M., Davies, J. M. (2016). Calcium-mediated abiotic stress signaling in roots. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01296
Wu, A., Hao, P., Wei, H., Sun, H., Cheng, S., Chen, P., et al. (2019a). Genome-wide identification and characterization of Glycosyltransferase family in cotton. Front. Genet. 10. doi: 10.3389/fgene.2019.00824
Wu, G. Q., Wang, Q., Bao, A. K., Wang, S. M. (2011). Amiloride reduces sodiumtransport and accumulation in the succulent xerophyte Zygophyllum xanthoxylum under salt conditions. Biol. Trace Elem. Res. 139, 356–367. doi: 10.1007/s12011-010-8662-9
Wu, G. Q., Wang, J. L., Li, S. J. (2019). Genome-wide identification of Na+/H+ antiporter (NHX) genes in sugar beet (Beta vulgaris L.) and their regulated expression under salt stress. Genes. 10 (5), 401–420. doi: 10.3390/genes10050401
Yin, G., Xu, H., Xiao, S., Qin, Y., Li, Y., Yan, Y., et al. (2013). The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups. BMC Plant Biol. 13, 148–167. doi: 10.1186/1471-2229-13-148
Yue, J., Wang, Y., Jio, J., Wang, H. (2021). Comparative transcriptomic and metabolic profiling provides insight into the mechanism by which the autophagy inhibitor 3-MA enhances salt stress sensitivity in wheat seedlings. BMC Plant Biol. 21, 577. doi: 10.1186/s12870-021-03351-5
Zhang, B., Chen, X. (2021). Secrets of the MIR172 family in plant development and flowering unveiled. PloS Biol. 19, 8–11. doi: 10.1371/journal.pbio.3001099
Zhang, Y., Feng, X., Wang, L., Su, Y., Chu, Z., Sun, Y. (2020). The structure, functional evolution, and evolutionary trajectories of the H+-PPase gene family in plants. BMC Genomics 21, 1–17. doi: 10.1186/s12864-020-6604-2
Keywords: Na +/H + antiporter, amiloride-binding site, molecular dynamics, gene expression, salt stress, wheat
Citation: Sharma P, Mishra S, Pandey B and Singh G (2023) Genome-wide identification and expression analysis of the NHX gene family under salt stress in wheat (Triticum aestivum L). Front. Plant Sci. 14:1266699. doi: 10.3389/fpls.2023.1266699
Received: 25 July 2023; Accepted: 06 November 2023;
Published: 04 December 2023.
Edited by:
Sushil Satish Chhapekar, University of Missouri, United StatesReviewed by:
Shumayla, Texas Tech University, United StatesParviz Heidari, Shahrood University of Technology, Iran
Copyright © 2023 Sharma, Mishra, Pandey and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pradeep Sharma, UHJhZGVlcC5TaGFybWFAaWNhci5nb3YuaW4=
†Present address: Bharti Pandey, ICAR-National Dairy Research Institute, Karnal, India
 Pradeep Sharma
Pradeep Sharma Shefali Mishra1
Shefali Mishra1 Bharati Pandey
Bharati Pandey