- College of Horticulture, Sichuan Agricultural University, Chengdu, Sichuan, China
Sugars and organic acids are the main factors determining the flavor of citrus fruit. The WRKY transcription factor family plays a vital role in plant growth and development. However, there are still few studies about the regulation of citrus WRKY transcription factors (CsWRKYs) on sugars and organic acids in citrus fruit. In this work, a genome-wide analysis of CsWRKYs was carried out in the citrus genome, and a total of 81 CsWRKYs were identified, which contained conserved WRKY motifs. Cis-regulatory element analysis revealed that most of the CsWRKY promoters contained several kinds of hormone-responsive and abiotic-responsive cis-elements. Furthermore, gene expression analysis and fruit quality determination showed that multiple CsWRKYs were closely linked to fruit sugars and organic acids with the development of citrus fruit. Notably, transcriptome co-expression network analysis further indicated that three CsWRKYs, namely, CsWRKY3, CsWRKY47, and CsWRKY46, co-expressed with multiple genes involved in various pathways, such as Pyruvate metabolism and Citrate cycle. These CsWRKYs may participate in the metabolism of fruit sugars and organic acids by regulating carbohydrate metabolism genes in citrus fruit. These findings provide comprehensive knowledge of the CsWRKY family on the regulation of fruit quality.
Introduction
Citrus fruit is one of the largest fruit crops in the world and is rich in various nutrients, such as sugars, organic acids, amino acids, carotenoids, and flavonoids (Guo et al., 2016; Sheng et al., 2017). Citrus fruit flavor is mainly determined by the content of sugars and organic acids and is one of the important factors affecting consumer purchases (Lin et al., 2015). The main components of soluble sugars in citrus fruit are sucrose, fructose, and glucose (Wu et al., 2021), and citric acid is the decisive organic acid, which accounts for 70% to 90% of organic acids in citrus fruit (Sheng et al., 2017). Studies on fruit quality reveal that the metabolism of sugars and organic acids is regulated by different genes, such as sucrose phosphate synthase (SPS), citrate synthetase (CS), and isocitrate dehydrogenase (IDH) (Etienne et al., 2013; Ruan, 2014; Guo et al., 2016; Hussain et al., 2017; Wan et al., 2018; Zhang et al., 2021). Notably, the metabolism of sugars and organic acids is reported to be regulated by multiple transcription factors (TFs), such as WRKYs, MYBs, and ERFs. Previously, CitERF16 was found to promote sucrose accumulation by activating the expression of CitSWEET11d, and CitZAT5 regulated the accumulation of hexose by mediating the expression of CitSUS5 and CitSWEET6 (Hu et al., 2021; Fang et al., 2023). Moreover, R3-MYB transcription factor TRIPTYCHON-LIKE acted as a repressor of the accumulation of citric acid and anthocyanin, and CitWRKY1 and CitERF6 participated in citric acid degradation of citrus fruit (Li et al., 2017; Li et al., 2020; He et al., 2022).
WRKY TFs are known as one of the largest families of transcriptional regulators in plants and are identified to bind to W-box [(C/T)TGAC(C/T)] cis-elements in the promoter of their target genes (Ciolkowski et al., 2008; Rushton et al., 2010). So far, a large number of WRKY genes have been identified in plants, and genome-wide analyses of WRKYs are also conducted in multiple species. The WRKY proteins always contain one or two conserved WRKYGQK motifs at the N-terminus and a typical zinc-finger motif (C2H2 and C2HC) at the C-terminus (Rushton et al., 1995; Eulgem et al., 2000). Meanwhile, WRKYs are further divided into three groups (I, II, and III) based on their WRKY domains and zinc-finger motifs (Mangelsen et al., 2008). Plant WRKY proteins are well-known to be involved in various stress responses and plant metabolite biosynthesis (Rushton et al., 2010; Han et al., 2021b; Wang et al., 2023b).
Recently, multiple studies have revealed that members of WRKY TFs are widely involved in different processes throughout the growth and development of plants. For example, Arabidopsis WRKYs (such as AtWRKY18 and AtWRKY63) were also found to regulate seed germination and root growth involved in hormone signaling (Chen et al., 2010; Ren et al., 2010). The mutations of AtWRKY12, OsWRKY36, and OsWRKY102 led to the accumulation of secondary cell wall and stem biomass, thereby changing the stem morphology (Yu et al., 2013; Miyamoto et al., 2020). Meanwhile, heterologous overexpression of CpWRKY71, GmWRKY58, and GmWRKY76 induced the expression of flowering-promoting genes (including FT, LFY, and FUL) and promoted flowering in Arabidopsis (Yang et al., 2016; Huang et al., 2019). Notably, several WRKY members were determined to participate in the formation of fruit quality. MaWRKY49 positively regulated pectate lyase genes to modulate fruit ripening (Liu et al., 2023). VvWRKY22 and CdWRKY2 directly regulated the expression of sugar-related genes to mediate sugar biosynthesis (Huang et al., 2021; Huang et al., 2022). Meanwhile, AtWRKY46 and SlWRKY42 were proven to bind the upstream W-box of ALMT9 and alter malate accumulation (Ding et al., 2013; Ye et al., 2017).
In citrus, several WRKY TFs were reported to regulate the biotic and abiotic stresses, but only a few members were found to take part in the metabolism of fruit quality. Hence, additional studies are required to identify more WRKY TFs linked to fruit quality building. In this study, a comprehensive analysis was performed to identify and characterize WRKY TFs in citrus genomes. Additionally, RNA-Seq data and gene expression were further analyzed to study the potential function of WRKY TFs in the metabolism of sugars and organic acids during citrus fruit development. Our results will provide new insights into the regulation of citrus sugar and organic acid metabolism pathways.
Materials and methods
Identification and characteristic analysis of the WRKY gene family
Identification of citrus WRKY genes was performed according to previous studies (Hu et al., 2015; He et al., 2016). The genomic sequences of different citrus species (Citrus clementina v1.0, Citrus grandis ‘Wanbaiyou’ v1.0, Poncirus trifoliata v1.0, and Citrus sinensis v2.0) were obtained from the Citrus Pan-genome to Breeding Database (CPBD: http://citrus.hzau.edu.cn/index.php). The Arabidopsis WRKY gene members and their protein sequences were obtained from the Arabidopsis Information Resource (TAIR: https://www.arabidopsis.org/browse/genefamily/index.jsp). C. sinensis WRKY (CsWRKY) transcription factors were identified using HMMER software version 3.0 and the PFAM protein family database using the WRKY domain (PF03106) as a query (Eddy, 2011; Mistry et al., 2021). Further, BLASTP was constructed to validate all non-overlapping WRKY genes.
The chromosome location image of CsWRKYs was generated using MapGene2Chrom software (MG2C_v2.1). The exon/intron structure of all CsWRKYs was displayed using the TBtools software. The relative molecular weight (RMW) and isoelectric point (pI) of CsWRKY proteins were analyzed using ProtParam (http://web.expasy.org/protparam/).
Analysis of gene structure and conserved motifs of CsWRKYs
The conserved motifs of WRKY proteins were detected with the multiple EM for motif elicitation (MEME software) (Brown et al., 2013). The optimum width of each motif ranged from 6 to 20, the maximum number of motifs to search was 8, and other parameter settings were default values (Bailey et al., 2006). All candidate WRKYs were further validated by using the National Center for Biotechnology Information (NCBI) Conserved Domain Database (CCD) to ensure that they contained the WRKY domains. The WRKY domains and zinc-finger motifs were predicted based on multiple sequence alignment with BioEdit.
Phylogenetic relationship analysis of CsWRKY gene family
Multiple sequence alignments of C. sinensis and Arabidopsis thaliana WRKY protein sequences were performed using molecular evolutionary genetics analysis (MEGA) version 6.0 with 1,000 bootstrap replications, pairwise deletion, and Poisson model. Subsequently, neighbor-joining phylogenetic trees were constructed.
RNA-Seq data and qRT-PCR analysis
RNA-Seq data were obtained from published data (Wang et al., 2017; Feng et al., 2021). The data that support the findings of this study have been deposited in the NCBI BioProject database under accession numbers PRJNA517400 and PRJNA387319. The method used for RNA-Seq data analysis was performed as described in a previous study (Pertea et al., 2016). In brief, raw reads of RNA-Seq were preprocessed for quality using the FastQC software, and low-quality reads (q < 20) and adapters were trimmed using Trimmomatic. Next, the obtained reads were aligned to the C. sinensis genome using HISAT2 with default parameters, and the mapped reads were further assembled by StringTie. Fragments per kilobase per million mapped fragments (FPKM) was used to represent the gene expression levels. The expression profiles of CsWRKYs were extracted from the analyzed RNA-Seq data for further study.
Isolation of total RNA from different tissues was carried out according to the procedure reported by Liu et al. (Liu et al., 2007). Specific primer pairs for amplification of CsWRKYs were designed by using the Primer Express software (Applied Biosystems, Foster City, CA, USA). The specificity and amplification efficiency of the primers were validated by BLASTN in the sweet orange genome. With the citrus β-actin gene as the internal reference gene, relative gene expression values were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). The sequences of RT-PCR primers are displayed in Table S7. All expression data were processed by the Z-score standardization method.
Co-expression network analysis
Weighted gene co-expression network analysis (WGCNA) (v1.71) package in R was used to construct the co-expression networks (Langfelder and Horvath, 2008). From the total of 29,138 genes, 16,961 genes with a sum FPKM of all samples lower than 1 were removed, and the reserved genes were used for the WGCNA co-expression network analysis. The one-step network construction and module detection function were conducted using an unsigned type of topological overlap matrix (TOM) with a soft-thresholding power β of 14 (R2 > 0.9), a minimal module size of 30, and a branch merge cut height of 0.25. The co-expression network of candidate CsWRKYs was visualized by Cytoscape (version 3.6.1) (Shannon et al., 2003).
Plant materials
‘Newhall’ navel orange (C. sinensis Osbeck) fruits were harvested from a commercial orchard in Liangshan Yi autonomous prefecture, Sichuan province, China. Fruits with uniform size and free of visible injury were harvested in August (Aug), October (Oct), November (Nov), and December (Dec). After the harvest, the fruits without obvious damage were transported to the laboratory for further experiments. The pulp was sampled, frozen, homogenized in liquid nitrogen, and kept at −80°C for later analyses. Three replications each containing six fruits were analyzed, and measurements were performed.
Fruit quality determination
‘Newhall’ navel orange fruits at each stage were chosen for determination of the total soluble solid (TSS) and titratable acid (TA). At each development stage, more than 18 fruits were used for quality determination with three replications. TSS (%) and TA (%) were measured by a digital acidity meter (Pocket PAL-BXIACID1, ATAGO, Tokyo, Japan) following the manufacturer’s instructions.
Statistical analysis
All data are shown as means ( ± SD) of one representative experiment. Significant differences between different samples were determined using ANOVA followed by Tukey’s test. The heatmaps were performed using R studio software by the pheatmap package. The correlation analysis was performed using R studio software. Figures were drawn using GraphPad Prism (GraphPad Software, CA, USA).
Results
Identification and phylogenetic analysis of CsWRKYs
Genome-wide identification of WRKYs was performed in the C. sinensis genome. After the removal of redundant sequences and those sequences lacking WRKY motifs, 81 unique CsWRKY transcripts belonging to 47 genes were finally obtained. Consequently, phylogenetic analysis with protein sequences was constructed together with the orthologue WRKYs from the genomes of A. thaliana (Figure 1). All CsWRKYs were clustered into three distinct groups (group I, group II, and group III), and members of group II were further clustered into five subgroups (group IIa, group IIb, group IIc, group IId, and group IIe). Among these WRKYs, 19 proteins were grouped into group I, 55 proteins were grouped into group II, and seven proteins were grouped into group III (Figure 2 and Table S1). Furthermore, multiple sequence alignments indicated that all CsWRKYs contained the conserved WRKYGQK domains and the C2H2 or C2HC zinc finger-like motifs (Figure S1). Most of group I WRKYs were found to have two WRKYGQK domains, and only group III had C2HC zinc finger-like motifs (Figures S1A, G). Meanwhile, the corresponding WRKYs were also identified in C. clementina (47), C. grandis (53), P. trifoliata (50), Atalantia buxifolia (52), Fortunella hindsii (53), and Citrus ichangensis (50) (Table S1).
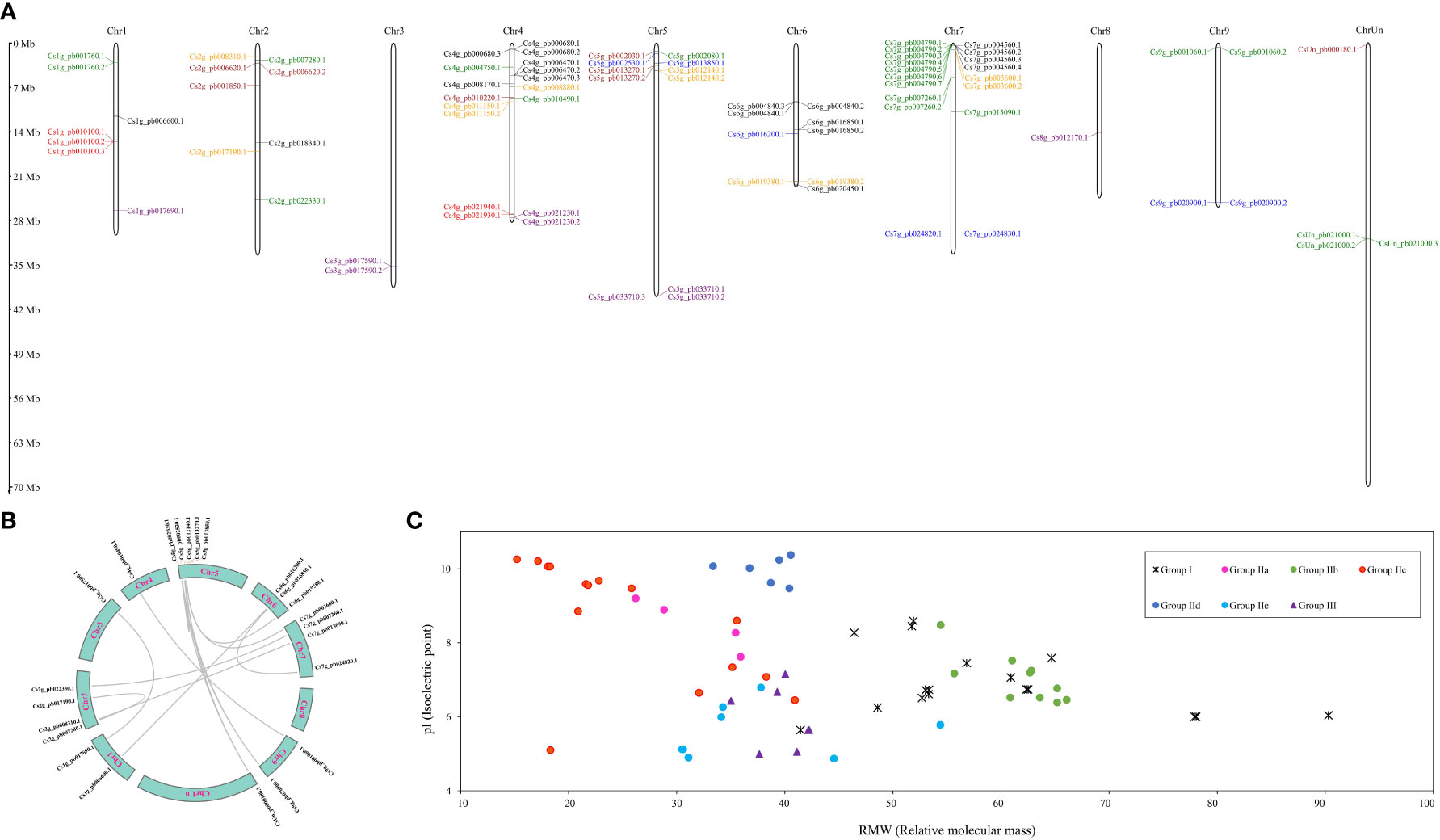
Figure 1 Characterizations of WRKY gene family in Citrus sinensis. (A) The chromosome distribution of WRKY gene family. (B) Repeated events of CsWRKY in C. sinensis genome. (C) Protein properties of CsWRKYs.
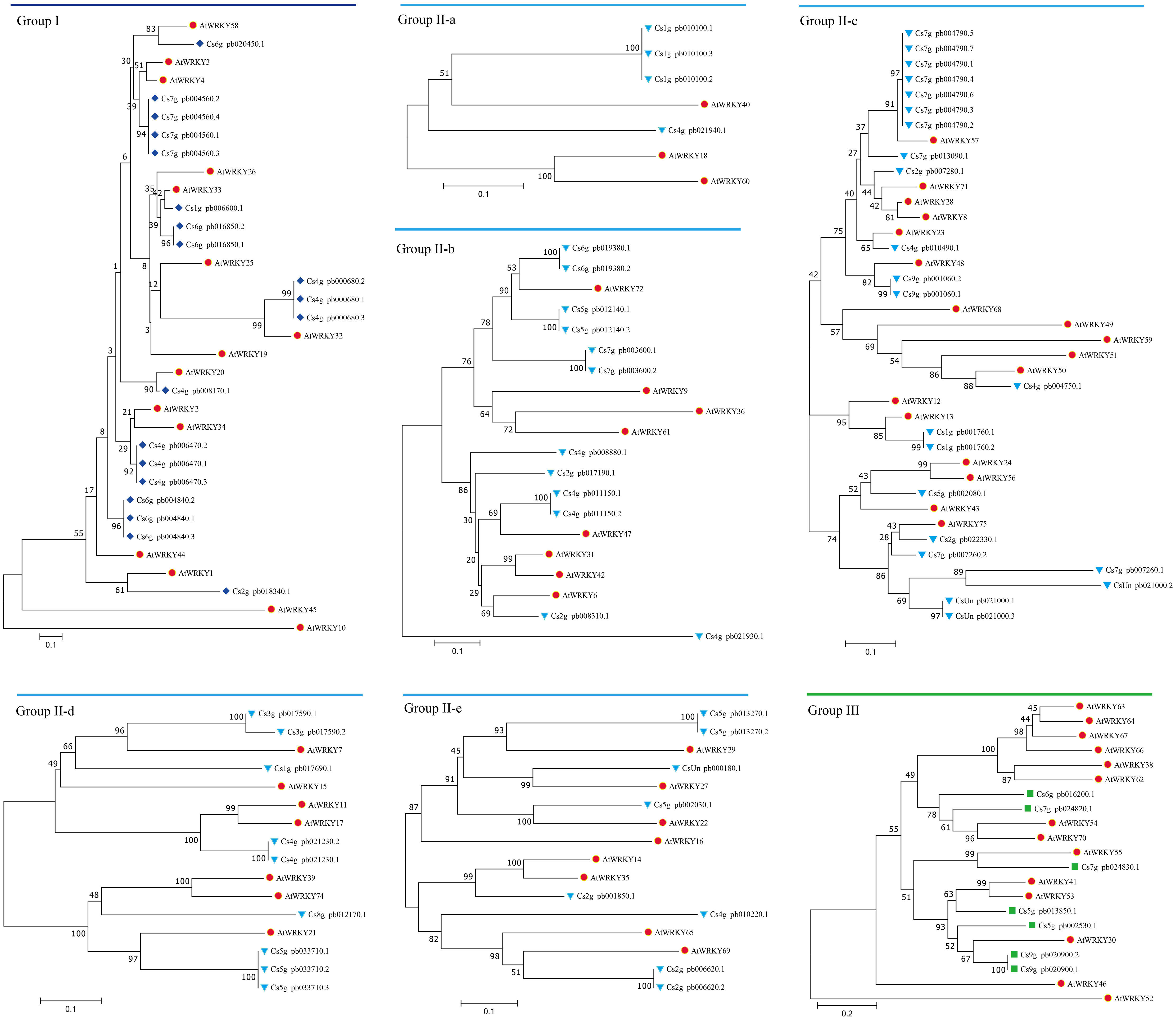
Figure 2 Phylogenetic relationship of WRKY proteins among Citrus sinensis and Arabidopsis thaliana. Phylogenetic analysis was performed using the neighbor-joining method in MEGA v5.1, and phylogenetic trees were constructed from the conserved WRKY domains of citrus and Arabidopsis WRKY proteins. WRKY proteins in A. thaliana were represented by red circles. Citrus WRKY proteins were divided into three groups (I, II, and III, which were represented by blue diamonds, blue triangles, and green squares, respectively). Group II included five subgroups (IIa, IIb, IIc, IId, and IIe).
Chromosomal location, putative pI, and RMW of CsWRKYs
To determine CsWRKYs’ physical locations, all 81 CsWRKYs were mapped onto the chromosomes in the C. sinensis genome sequence (Figure 1A). Their distribution on chromosomes greatly varied from one in Chr8 to 18 in Chr7. All chromosomes showed the presence of at least one group of CsWRKYs, and only group IId WRKYs were localized in Chr3 and Chr8. Meanwhile, four chromosomes (Chr1, Chr2, Chr4, Chr5, and Chr7) were found to contain not less than four WRKY groups. Fourteen pairs of repetitive fragments in the CsWRKYs were identified, and three of them had two repeats (Figure 1B).
The RMW of the predicted 81 CsWRKY proteins ranged from 15.22 to 90.29 kDa, and the pI ranged from 4.87 to 10.37 (Figure 1C; Table S1). Notably, the RMW of CsWRKY proteins in groups I (41.25 to 90.29 kDa) and IIb (54.42 to 66.08 kDa) was much bigger than that of other members, while most of CsWRKY proteins (10/18) in group IIc showed smaller RMW, which was less than 26 kDa. The pI of all CsWRKY proteins in group IId ranged from 9.47 to 10.37, while the pI of CsWRKY proteins in groups IIe and III was less than 8.
Protein motif, gene sequence, and promoter analysis of CsWRKYs
Furthermore, motif analysis was carried out with the MEME web server, resulting in the discovery of a total of eight conserved motifs in CsWRKY proteins (Figure 3A; Table S2). The results indicated that all CsWRKY proteins shared similar motifs, especially motifs 1 and 2, which contained the WRKYGQK domain and part of C2H2 or C2HC. Motif 5 was a unique motif of group I proteins, and motifs 6 and 8 were mainly distributed in group IIb and group IIc proteins. Notably, motif 4 was missing in group III proteins. The distribution of conserved motifs is consistent with the phylogenetic tree result.

Figure 3 The conserved motifs, gene structure, and predicted cis-elements of CsWRKYs. (A) The conserved motifs, numbers 1–8, are shown in different colored boxes. The details of each motif are provided in Table S3. (B) The exon–intron structure of CsWRKYs. Green boxes represent 5′ UTR and 3′ UTR, yellow boxes represent exons, and gray lines present introns. (C) Predicted cis-elements in the CsWRKY prompters. Different cis-elements are represented by different colors.
The exon–intron structures of CsWRKYs were further analyzed. Generally, most of CsWRKYs (78/81) contained two to five exons (Figure 3B). Most of the CsWRKYs in the same group have similar intron–exon structures. For example, CsWRKYs (17/19) in group I contained four to five exons, and only two genes had 10 or six exons. Moreover, seven CsWRKYs in group III showed three exons and two introns. However, one member of the group I CsWRKYs had 11 exons, while group IIc CsWRKYs had only one exon.
To analyze the potential cis-elements, the 2-kb sequences upstream of the translation start sites in each CsWRKY were submitted to the online tool PlantCARE. A total of 14 types of cis-elements associated with responses to different hormones, external environmental signals, cell cycle (MSA-like), circadian control, and zein metabolism were finally identified (Figure 3C; Table S3). Our results indicated that the light-responsive element existed in the promoters of all CsWRKYs. Meanwhile, the promoters of most CsWRKYs were found to have the abscisic acid-responsive element (64/81), anaerobic regulatory elements (70/81), and MeJA-responsive element (70/81). The drought-responsive elements, gibberellin-responsive elements, low-temperature-responsive elements, and zein metabolism regulatory elements exist in nearly one-third of CsWRKYs.
Expression profiles of CsWRKYs in the pulp of citrus fruit
To further explore the expression pattern of CsWRKY genes in citrus fruit, the expression of 47 CsWRKY genes at different fruit development stages based on publicly available transcriptomic data was analyzed. The results showed that all CsWRKYs had different expression patterns among different varieties (Figure 4). In the pulp of ‘Fengjie 72-1’ fruit (Figure 4A), most CsWRKYs (27/47) were highly expressed at 50 days after flowering (DAF) and showed an overall downregulated expression pattern with fruit ripening. Several genes showed high transcription levels in young fruit (50–120 DAF). Moreover, the transcript level of some CsWRKYs (11/47) was generally upregulated with the development of fruit, but two of them showed the lowest transcript level during the expansion period (155 DAF). In the pulp of ‘Zaohong’ and ‘Twenty-first century’ orange fruits, the expression patterns of all CsWRKYs are divided into two groups. As shown in Figure 4B, the transcript abundance of most CsWRKYs (29/47) was much higher at 302 DAF in both ‘Zaohong’ and ‘Twenty-first century’ orange fruits and dramatically decreased with fruit development. Notably, many of these genes (25/29) had higher expression levels in the pulp of ‘Zaohong’ orange fruit. Meanwhile, some CsWRKYs showed high expression at 131 DAF, while a significant reduction was observed with fruit development.
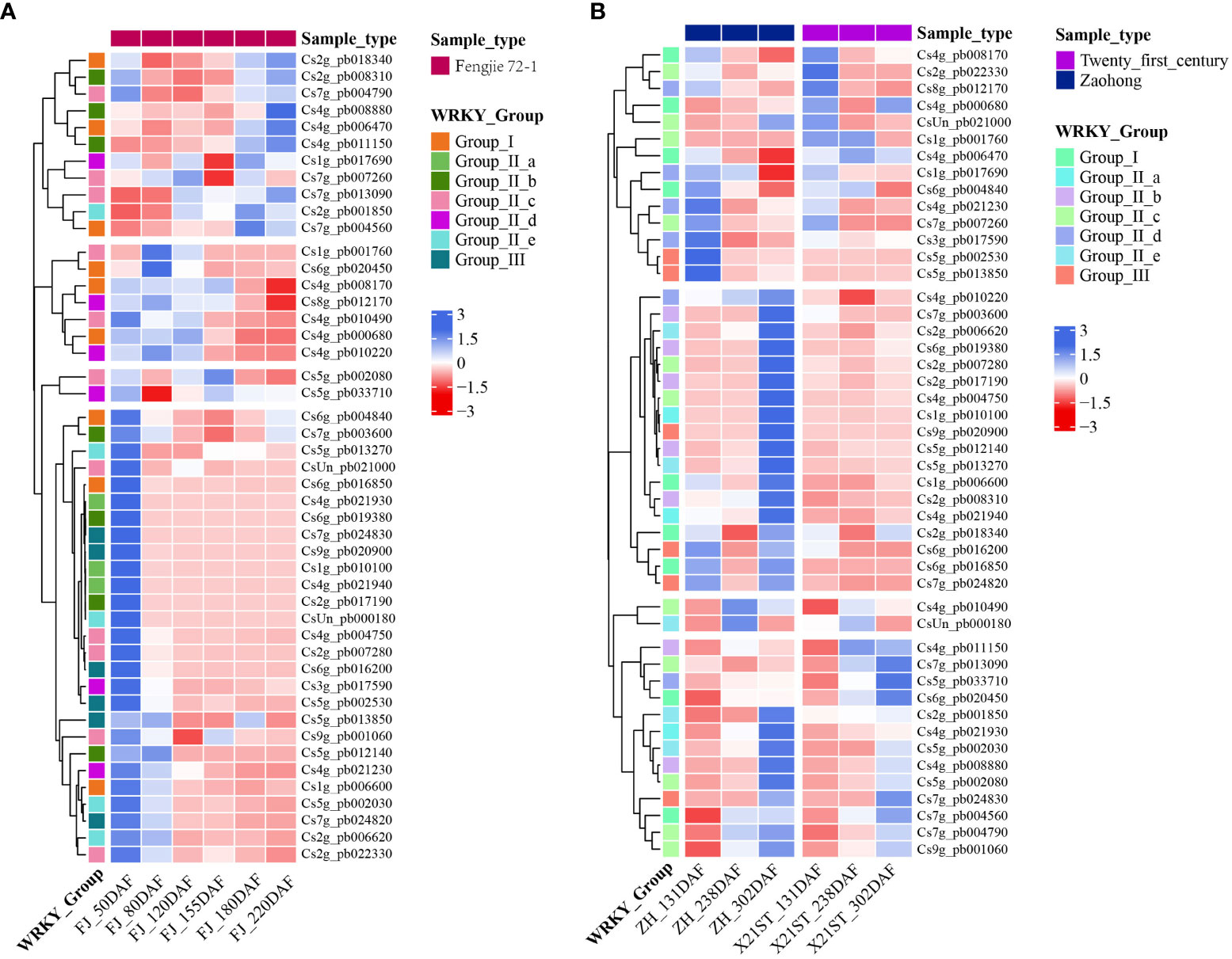
Figure 4 Expression of CsWRKYs in different development stages of citrus fruit. The transcription levels of the CsWRKYs in the pulp of ‘Fengjie 72-1’ (A), ‘Zaohong’, and ‘Twenty-first century’ (B) orange fruits at different development stages. Different CsWRKY groups are represented by different colors. The color scale shows increasing expression levels from red to blue.
Correlation analysis between CsWRKYs and sugars and organic acids in citrus fruit
To further identify the potential functional CsWRKYs involved in the formation of fruit quality, Pearson’s correlation coefficients between CsWRKY expression and sugar and organic acid contents were analyzed. In different citrus varieties, the number of CsWRKYs related to sugars and organic acids was different (Figures 5A–C). Many CsWRKYs were negatively correlated with sugar content in ‘Fengjie 72-1’ fruit, but a number of them showed a positive relationship with sugars in ‘Zaohong’ and ‘Twenty-first century’ fruits. Meanwhile, few genes had a relatively strong relationship (correlation coefficient >0.5 or <−0.5) with the content of organic acids in ‘Fengjie 72-1’ fruit, but more genes were strongly correlated in ‘Zaohong’ and ‘Twenty-first century’ fruits. CsWRKYs with a strong relationship (correlation coefficient >0.6 or <−0.6) with sugars and organic acids were selected for Venn diagram analysis. Four CsWRKYs (Cs5g_pb002530, Cs6g_pb004840, Cs8g_pb012170, and Cs4g_pb021230) were negatively correlated with sugars, while two (Cs7g_pb004560 and Cs2g_pb001850) were positively correlated (Figure 5D). Moreover, three CsWRKYs (Cs4g_pb011150, Cs7g_pb004790, and Cs5g_pb033710) were found to be negatively correlated with organic acids, while two (Cs4g_pb008170 and Cs8g_pb012170) were positively correlated (Figure 5E). Cs8g_pb012170 was found to be positively correlated with sugars and negatively correlated with organic acids. The expression profiles indicated that these CsWRKYs had tissue-specific expression patterns (Figures 5F, G). Two CsWRKYs (Cs2g_pb001850 and Cs8g_pb012170) are rarely expressed in fruit but highly expressed in root. Notably, the transcript abundance of two CsWRKYs (Cs5g_pb002530 and Cs4g_pb021230) in young fruit was higher than that in mature fruit, while three CsWRKYs (Cs6g_pb004840, Cs7g_pb004560, and Cs4g_pb011150) showed higher expression levels in mature fruit. These 10 CsWRKYs may be candidate genes involved in sugar and organic acid metabolism.
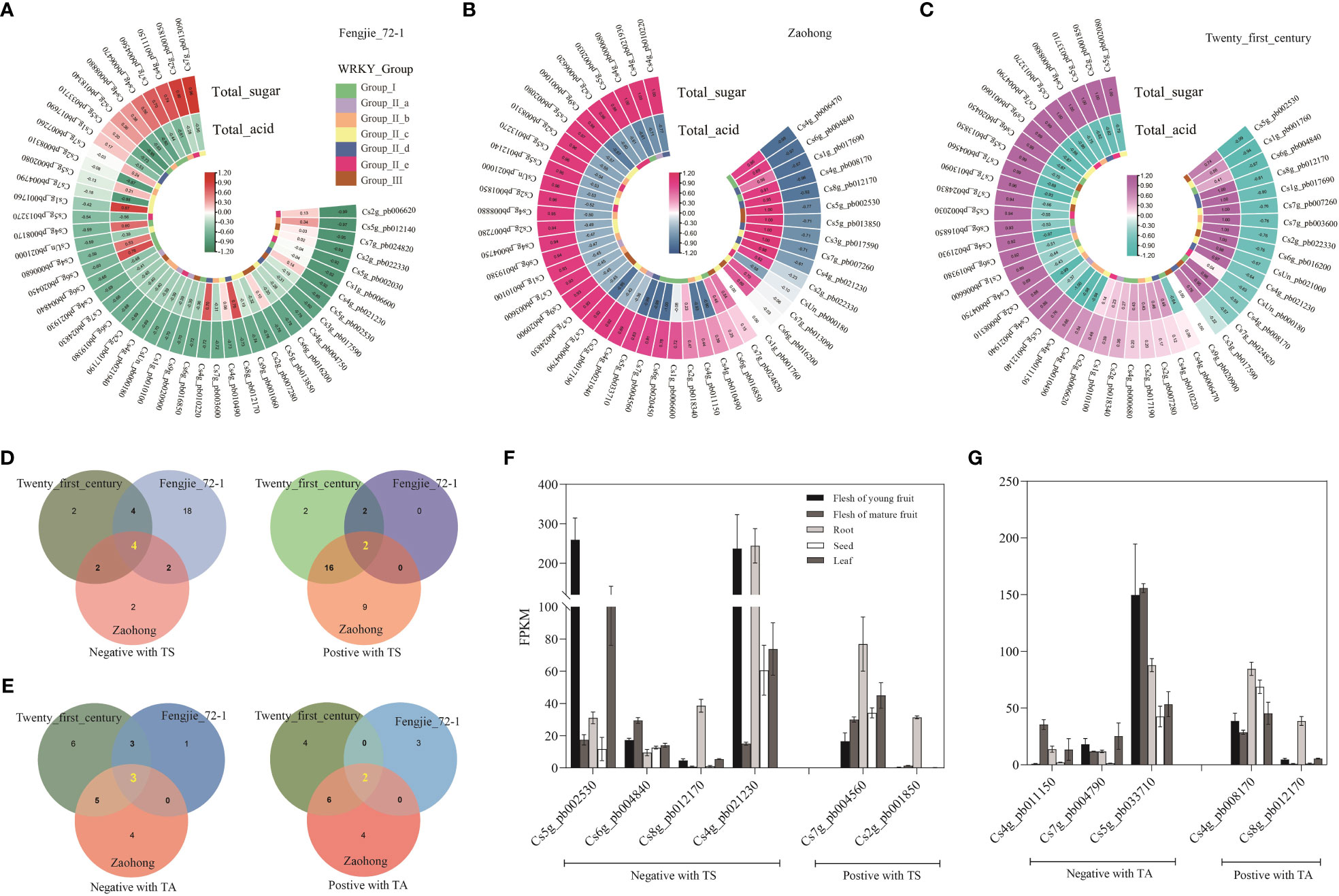
Figure 5 Correlation analysis of CsWRKYs with sugars and organic acids in citrus fruit. Heatmap of Pearson’s correlation coefficients between CsWRKYs and sugar and acid contents in the pulp of ‘Fengjie 72-1’ (A), ‘Zaohong’ (B), and ‘Twenty-first century’ (C) orange fruits. Venn diagrams of CsWRKYs co-expressing with the content of total sugar (D) and total organic acid (E) (correlation coefficient >0.6 or <−0.6). The expression profile of co-expressing CsWRKYs in different tissues of orange fruit (F, G).
Expression analysis of CsWRKYs and determination of TSS and TA in orange fruit
To further explore the potential function of CsWRKYs in the metabolism of sugars and organic acids, qRT-PCR analysis was performed on candidate CsWRKYs. The expression profiles indicated that two CsWRKYs (Cs5g_pb002530 and Cs4g_pb021230) showed the highest transcription levels in August, while their expression dramatically decreased with fruit development. The expression of Cs6g_pb004840 showed a fluctuating trend; meanwhile, the expression of Cs8g_pb012170 increased with fruit development followed by a significant decrease in December (Figure 6A). Moreover, two candidate CsWRKYs positively correlated with sugars had different expression trends in the pulp of orange fruit. Cs7g_pb004560 had an overall upregulated expression trend, while the expression of Cs2g_pb001850 showed slight changes (Figure 6B). As shown in Figure 6C, the transcriptional level of Cs4g_pb011150 increased with fruit development, Cs7g_pb004790 showed the highest level in December, and Cs5g_pb033710 showed the lowest level in October. CsWRKYs (Cs2g_pb001850 and Cs8g_pb012170) positively correlated with organic acid and showed different expression patterns (Figures 6A, D). Furthermore, the main fruit quality, namely, TSS and TA, were determined at different stages of fruit development (Figure 6E). The TSS in fruit showed a consistently decreasing trend throughout the fruit development and reached approximately 12.5% in December (Figure 6F). Meanwhile, the TA content in fruit showed a significant decrease after August, followed by a slight decrease with fruit development (Figure 6G). Combining expression profile and fruit quality, three CsWRKYs (Cs5g_pb002530, Cs7g_pb004560, and Cs4g_pb011150) with high transcriptional abundance in fruit had strong correlations with fruit sugars or organic acids.
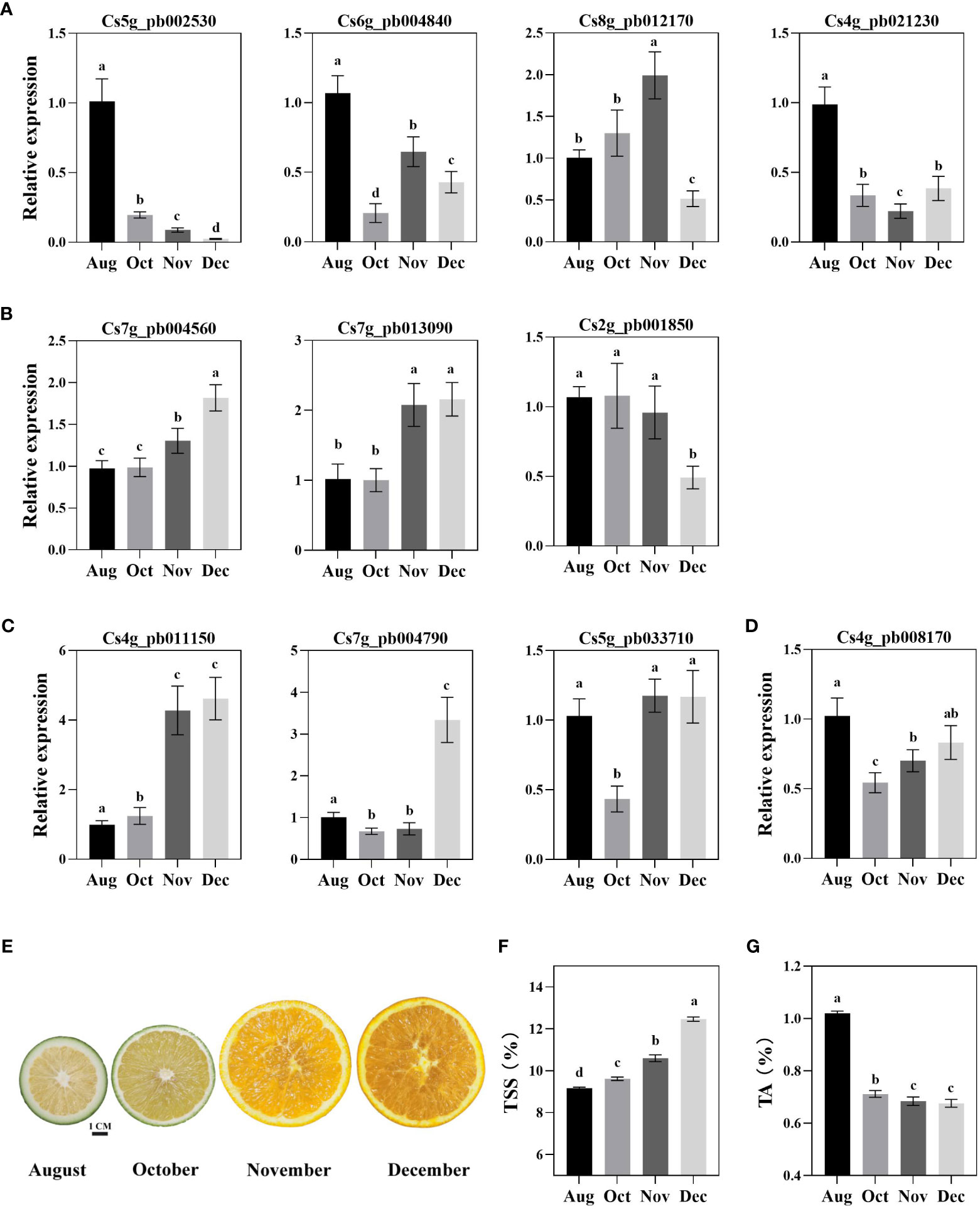
Figure 6 Expression of CsWRKYs and content of total soluble solid (TSS) and titratable acidity (TA) in the pulp of Newhall navel orange fruits at different development stages. Gene expression of CsWRKYs correlated with sugar (A, B) organic acid (C, D) in fruit pulp. The phenotype of orange fruit at development stages (E). TSS (F) and TA (G) in the pulp of orange fruit. Lowercase letters indicate significant differences (p < 0.05) analyzed using Duncan’s test.
Co-expression network analysis of candidate CsWRKYs during fruit development
To further investigate the regulatory mechanism of candidate CsWRKYs, WGCNA was performed using the published transcriptome data of ‘Fengjie 72-1’ with five development stages. A total of 20 merged co-expression modules were finally identified (Figure 7A). Interestingly, the module in turquoise color (6,466 genes) was negatively associated with total sugar (correlation coefficient = −0.9) but positively associated with organic acid (correlation coefficient = 0.9). The module in blue color (4,871 genes) was positively associated with total sugar (correlation coefficient = 0.69) but negatively associated with organic acid (correlation coefficient = −0.82) (Figure 7B; Table S4). In addition, the candidate CsWRKYs (Cs7g_pb004560 named CsWRKY3 and Cs4g_pb011150 named CsWRKY47) with an upregulated trend were found in blue modules, and Cs5g_pb002530 named CsWRKY46 with a downregulated trend was found in the turquoise module. Furthermore, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis showed that most genes in the turquoise module were significantly enriched in various pathways (p < 0.05), such as Pyruvate metabolism (ko00620) and Citrate cycle (ko00020) (Figure 7C). Meanwhile, genes in the turquoise module were also significantly enriched in carbohydrate metabolism pathways, including Pyruvate metabolism (ko00620), Ascorbate and aldarate metabolism (ko00053), and Citrate cycle (ko00020) (Figure 7D; Table S5).
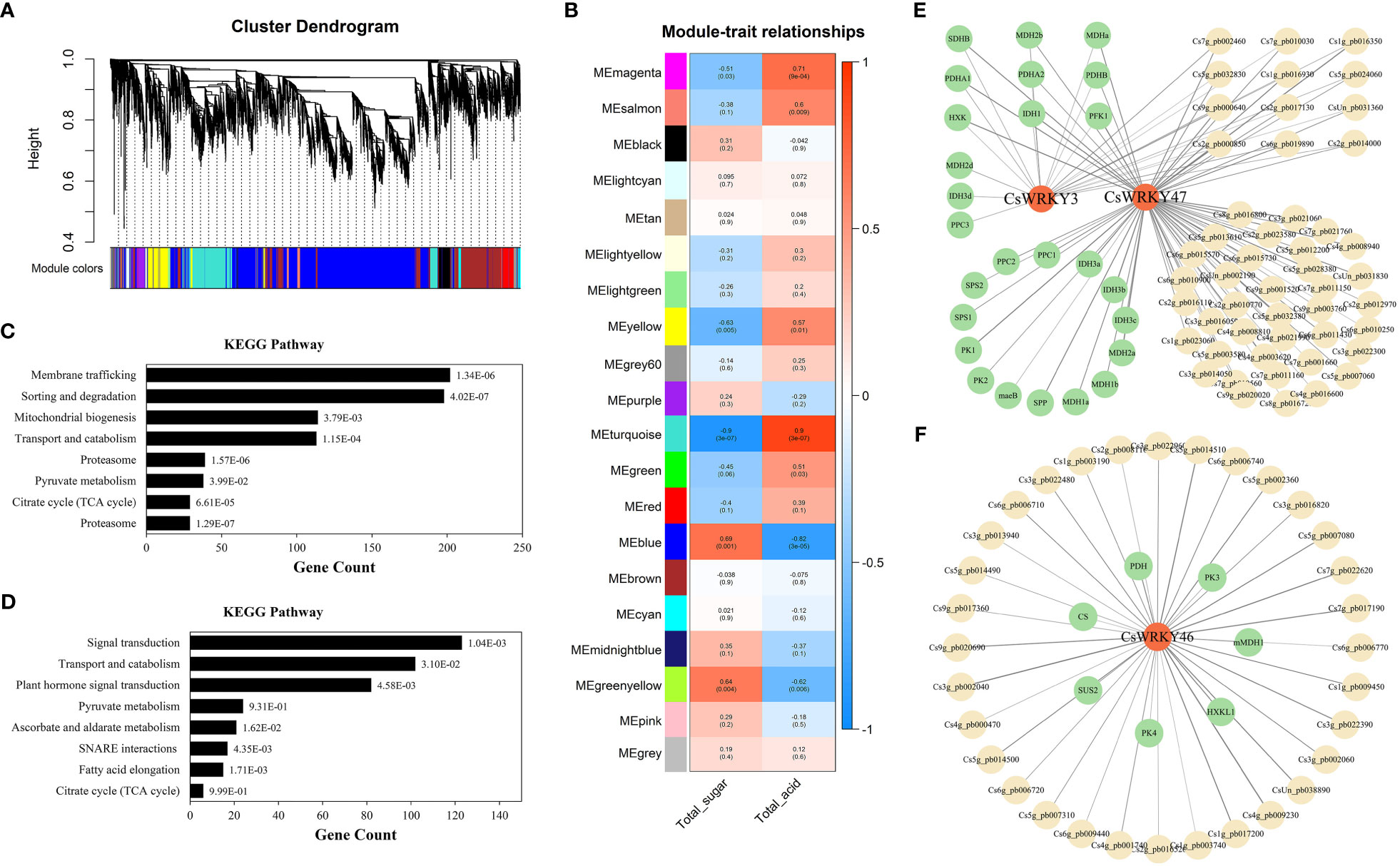
Figure 7 (A) Hierarchical clustering tree showing co-expression modules identified by WGCNA. Different modules are marked with different colors. Each leaf of the cluster tree represents a gene. (B) Module–trait correlations and corresponding p-values. TA and TSS. (C, D) KEGG pathway enrichment analysis of modules. (E, F) Co-expression network analysis of CsWRKYs. WGCNA, weighted gene co-expression network analysis; TA, titratable acid; TSS, total soluble solid; KEGG, Kyoto Encyclopedia of Genes and Genomes.
The co-expression networks further revealed that multiple genes involved in carbohydrate metabolism were closely linked to the candidate CsWRKYs (weight > 0.15). As shown in Figure 7E and Table S6, CsWRKY3 was observed to co-express with succinate dehydrogenase B (SDHB), three malate dehydrogenase genes (MDH2b, MDH2d, and MDHa), three pyruvate dehydrogenase genes (PDHA1, PDHB, and PDHA2), hexokinase (HXK), and two isocitrate dehydrogenase genes (IDH1 and IDH3d) 6-phosphofructokinase 1 (PFK1) and phosphoenolpyruvate carboxylase 3 (PPC3). Meanwhile, there were three PPCs (PPC1, PPC2, and PPC3), two sucrose-phosphate synthase genes (SPS1 and SPS2), two pyruvate kinase genes (PK1 and PK2), four IDHs (IDH1, IDH3a, IDH3b, and IDH3c), five MDHs (MDHa, MDH2b, MDH2a, MDH1a, and MDH1b), malate dehydrogenase B (maeB), and sucrose-6-phosphatase (SPP). Among these carbohydrate metabolism genes, three PDHs, HXK, IDH1, SDHB, and two MDHs were shown to be linked to both CsWRKY3 and CsWRKY47. Moreover, the downregulated CsWRKY46 was found to co-express with two PKs (PK3 and PK4), PDH, HXKL1, mMDH1, sucrose synthase 2 (SUS2), and citrate synthase (CS) (Figure 7F). These results indicated that CsWRKYs participated in the modulation of fruit sugar and organic acid metabolism by regulating different genes during fruit development.
Discussion
Citrus fruit is an economically important fresh fruit worldwide. The contents of sugars and organic acids in fruit are considered the crucial factor that determines fruit flavor and commodity value (Lin et al., 2015). To date, more and more studies have been conducted to investigate the regulatory mechanisms of sugar and organic acid metabolism at the transcriptional level. As one of the largest transcription factor families in higher plants, the WRKY gene family has been found to play an essential role in plant growth, development, and biotic and abiotic stresses (Eulgem et al., 2000; Rushton et al., 2010; Han et al., 2018b; Han et al., 2021a). However, there is still limited information on the functional CsWRKYs in the formation of citrus fruit quality.
Characteristics of CsWRKY in citrus genome
Recently, genome-wide analysis of WRKY gene family has been reported in multiple plants, such as A. thaliana, tomato, tobacco, and rice (Kim and Zhang, 2004; Wu et al., 2005; Ross et al., 2007; Chen et al., 2015; Liu et al., 2022a). In the present study, a total of 81 CsWRKYs were identified in the C. sinensis genome, and the number was greater than that in other citrus genomes, which may account for more transcripts in the genome (Ayadi et al., 2016; Mosharaf et al., 2020). The sequence alignment and phylogenetic analysis showed that all CsWRKYs were divided into three groups and had the conserved N-terminal WRKYGQK domains and zinc finger-like motifs that were consistent with WRKY structures in other species (Figures 2, 3; Table S1). As is known, these prominent features of the WRKY family directly establish their roles in gene regulation (Wei et al., 2016; Chen et al., 2021). In this study, all of the CsWRKY proteins contained at least one WRKY domain, whereas several of them lacked zinc finger-like motifs (Figures 3A, S1). Moreover, two CsWRKYs (Cs7g_pb004560.2 and Cs7g_pb004560.4) in group I lost the second WRKY domains and zinc finger-like motifs, which may result in functional changes.
Cis-elements are the regulatory sites located at the 5′ upstream sequence of genes, and their variety always determines the diversity of regulatory functions of genes. In our results, CsWRKYs were found to be enriched in hormone-responsive and light-responsive cis-elements, as well as various abiotic-responsive cis-elements (Figure 3C; Table S3). The enrichment of different cis-elements in different CsWRKYs indicated their diverse functions involved in plant development and stress response. Notably, the ABA-responsive element was observed to exist in promoters of more than 60 CsWRKYs. As reported previously, ABA is an important phytohormone regulating plant growth and development (Yoshida et al., 2019). Multiple studies have demonstrated that ABA signaling is involved in various metabolic pathways that contribute to fruit growth and ripening (Liao et al., 2018; Feng et al., 2021; Zhu et al., 2023). Meanwhile, several WRKYs were also found to participate in ABA-regulated fruit development, such as VvWRKY22 (Huang et al., 2021). Herein, the enrichment of ABA-responsive elements in CsWRKYs indicated their functional roles in the formation of fruit quality and development of fruit. The mechanism of their responsiveness to ABA signals, as well as transcriptional regulation, deserves more study.
CsWRKYs are involved in the formation of fruit quality in citrus fruit
With the development of citrus fruit, the content of total sugar showed an obvious upward trend, while the content of organic acid significantly reduced (Figures 6F, G). The trend of sugar and organic acid changes is a universal pattern in most citrus fruits (Li et al., 2017; Shi et al., 2019; Bi et al., 2022). As an important regulatory factor in plants, WRKY TFs have been investigated to play critical roles in fruit development, as well as the stress response (Han et al., 2020; Liu et al., 2022b; Guan et al., 2023; Shao et al., 2023). Meanwhile, transcriptome analysis of different tangor mandarins also revealed that multiple WRKYs had potential functions in fruit quality formation throughout fruit development (Bi et al., 2022; Liao et al., 2022). In this study, the expression profiles of WRKY TFs and fruit quality (sugars and organic acids) of orange fruit were conjointly analyzed and revealed that several of these genes may be involved in the regulation of fruit quality formation (Figures 4, 5). Among them, three CsWRKYs with significant correlation with fruit sugars and organic acids, namely, CsWRKY3, CsWRKY47, and CsWRKY46, were regarded as candidate genes (Figures 6, 7). Notably, ABA-responsive elements were also identified in the promoters of these CsWRKYs (eight in CsWRKY3 and CsWRKY47 and four in CsWRKY46) (Table S3). As the key components in ABA signaling, WRKYs participate in regulating ABA-induced responses to plant growth and stresses (Han et al., 2021a; Wang et al., 2023a). Therefore, our findings further indicated that the WRKY-regulated sugar/organic acid metabolism may be modulated by ABA.
Subsequent co-expression network analysis revealed that the genes co-expressed with fruit sugars and organic acids were enriched in different metabolism pathways (Figures 7A, B; Table S5). Many of these genes were further clustered into several carbohydrate metabolism pathways, including Glycolysis metabolism, Pyruvate metabolism, and Citrate cycle, which directly regulated the metabolism of sugars and organic acids (Figures 7C, D). Previous studies have revealed that the accumulation and degradation of sugar and organic acid components are influenced by the changes in transcriptional levels of carbohydrate metabolism genes (for example, SUSs, PFKs, HXKs, CSs, and IDHs) and are regulated by different TFs (for example, ZATs, HLHs, MYBs, and WRKYs) (Chen et al., 2017; Wei et al., 2019; Yu et al., 2021; Liu et al., 2022c; Yu et al., 2022; Fang et al., 2023). Herein, multiple genes (such as IDHs, PDHs, HXKs, PKs, SPSs, and MDHs) involved in carbohydrate metabolism were identified to be closely linked to CsWRKYs with the formation of fruit quality. Notably, the upregulated CsWRKYs (CsWRKY3 and CsWRKY47) were found to co-express with most carbohydrate metabolism genes (Figures 7E, F). Similarly, WRKY3 was reported to co-express with sugar-related genes and further promote the sucrose metabolism in Arachis hypogaea L. and Hylocereus (Han et al., 2018a; Kiranmai et al., 2018; Wei et al., 2019). However, most homologous genes of WRKY47 in different plants were identified to participate in abiotic stress responses, such as selenium tolerance (Wu et al., 2020), boron tolerance (Feng et al., 2020), and drought response (Raineri et al., 2015). There is little information on the roles of WRKY47 in the formation of fruit quality. Hence, the candidate CsWRKYs identified in this study still need more studies, and the results will provide more information in analyzing the regulatory network of fruit quality.
Conclusion
In the present study, a total of 81 CsWRKYs were identified in the C. sinensis genome. All CsWRKYs were classified into three major groups: I, II, and III. CsWRKYs of group II were further divided into five distinct subgroups: IIa, IIb, IIc, IId, and IIe. All identified CsWRKY proteins had the conserved WRKY domains and zinc-finger motifs. Furthermore, expression analysis revealed that multiple CsWRKYs co-expressed with sugars and organic acids during citrus fruit growth. Notably, three CsWRKYs, named CsWRKY3, CsWRKY47, and CsWRKY46, were identified as the key genes involved in the metabolism of fruit sugars and organic acids. The expression network further indicated that these candidate CsWRKYs may modulate the fruit quality by regulating carbohydrate metabolism genes. Our results provided new insights into the regulatory mechanism of CsWRKYs in the formation of fruit sugars and organic acids.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
MZ: Conceptualization, Funding acquisition, Investigation, Resources, Validation, Writing – review & editing. WL: Formal Analysis, Investigation, Writing – original draft. XY: Investigation, Resources, Validation, Writing – original draft. QL: Investigation, Resources, Writing – original draft. XL: Investigation, Writing – original draft. KL: Investigation, Writing – original draft. CY: Investigation, Writing – original draft. BX: Resources, Supervision, Writing – review & editing. LL: Resources, Supervision, Writing – review & editing. GS: Resources, Supervision, Writing – review & editing. SH: Resources, Supervision, Writing – review & editing. JH: Resources, Supervision, Writing – review & editing. XW: Conceptualization, Funding acquisition, Writing – review & editing. ZW: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Sichuan Province Science and Technology Department Project (2023NSFSC1248) and the National Key Research and Development Program of China (2021YFD1600800).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1264283/full#supplementary-material
References
Ayadi, M., Hanana, M., Kharrat, N., Merchaoui, H., Marzoug, R. B., Lauvergeat, V., et al. (2016). The WRKY transcription factor family in citrus: Valuable and useful candidate genes for citrus breeding. Appl. Biochem. Biotechnol. 180, 516–543. doi: 10.1007/s12010-016-2114-8
Bailey, T. L., Williams, N., Misleh, C., Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Bi, X., Liao, L., Deng, L., Jin, Z., Huang, Z., Sun, G., et al. (2022). Combined transcriptome and metabolome analyses reveal candidate genes involved in tangor (Citrus reticulata x Citrus sinensis) fruit development and quality formation. Int. J. Mol. Sci. 23, 5457. doi: 10.3390/ijms23105457
Brown, P., Baxter, L., Hickman, R., Beynon, J., Moore, J. D., Ott, S. (2013). MEME-LaB: motif analysis in clusters. Bioinformatics 29, 1696–1697. doi: 10.1093/bioinformatics/btt248
Chen, Y. S., Chao, Y. C., Tseng, T. W., Huang, C. K., Lo, P. C., Lu, C. A. (2017). Two MYB-related transcription factors play opposite roles in sugar signaling in Arabidopsis. Plant Mol. Biol. 93, 299–311. doi: 10.1007/s11103-016-0562-8
Chen, Y., Jing, X., Wang, S., Wang, J., Zhang, S., Shi, Q. (2021). Genome-wide analysis of WRKY transcription factor family in melon (Cucumis Melo L.) and Their Response to Powdery Mildew. Plant Mol. Biol. Rep. 39, 686–699. doi: 10.1007/s11105-020-01271-6
Chen, H., Lai, Z., Shi, J., Xiao, Y., Chen, Z., Xu, X. (2010). Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 10, 281. doi: 10.1186/1471-2229-10-281
Chen, L., Yang, Y., Liu, C., Zheng, Y., Xu, M., Wu, N., et al. (2015). Characterization of WRKY transcription factors in Solanum lycopersicum reveals collinearity and their expression patterns under cold treatment. Biochem. Biophys. Res. Commun. 464, 962–968. doi: 10.1016/j.bbrc.2015.07.085
Ciolkowski, I., Wanke, D., Birkenbihl, R. P., Somssich, I. E. (2008). Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function. Plant Mol. Biol. 68, 81–92. doi: 10.1007/s11103-008-9353-1
Ding, Z. J., Yan, J. Y., Xu, X. Y., Li, G. X., Zheng, S. J. (2013). WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum-induced malate secretion in Arabidopsis. Plant J. 76, 825–835. doi: 10.1111/tpj.12337
Eddy, S. R. (2011). Accelerated profile HMM searches. PloS Comput. Biol. 7, e1002195. doi: 10.1371/journal.pcbi.1002195
Etienne, A., Génard, M., Lobit, P., Mbeguié, A. M. D., Bugaud, C. (2013). What controls fleshy fruit acidity? A review of malate and citrate accumulation in fruit cells. J. Exp. Bot. 64, 1451–1469. doi: 10.1093/jxb/ert035
Eulgem, T., Rushton, P. J., Robatzek, S., Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/s1360-1385(00)01600-9
Fang, H., Shi, Y., Liu, S., Jin, R., Sun, J., Grierson, D., et al. (2023). The transcription factor CitZAT5 modifies sugar accumulation and hexose proportion in citrus fruit. Plant Physiol. 192 (3), 1858–1876. doi: 10.1093/plphys/kiad156
Feng, Y., Cui, R., Wang, S., He, M., Hua, Y., Shi, L., et al. (2020). Transcription factor BnaA9.WRKY47 contributes to the adaptation of Brassica napus to low boron stress by up-regulating the boric acid channel gene BnaA3.NIP5;1. Plant Biotechnol. J. 18, 1241–1254. doi: 10.1111/pbi.13288
Feng, G., Wu, J., Xu, Y., Lu, L., Yi, H. (2021). High-spatiotemporal-resolution transcriptomes provide insights into fruit development and ripening in Citrus sinensis. Plant Biotechnol. J. 19, 1337–1353. doi: 10.1111/pbi.13549
Guan, K., Yang, Z., Zhan, M., Zheng, M., You, J., Meng, X., et al. (2023). Two Sweet Sorghum (Sorghum bicolor L.) WRKY transcription factors promote aluminum tolerance via the reduction in callose deposition. Int. J. Mol. Sci. 24, 10288. doi: 10.3390/ijms241210288
Guo, L. X., Shi, C. Y., Liu, X., Ning, D. Y., Jing, L. F., Yang, H., et al. (2016). Citrate accumulation-related gene expression and/or enzyme activity analysis combined with metabolomics provide a novel insight for an orange mutant. Sci. Rep. 6, 29343. doi: 10.1038/srep29343
Han, D., Han, J., Xu, T., Li, T., Yao, C., Wang, Y., et al. (2021a). Isolation and preliminary functional characterization of MxWRKY64, a new WRKY transcription factor gene from Malus xiaojinensis Cheng et Jiang. In. Vitro Cell. Dev. Biol. Plant 57, 202–213. doi: 10.1007/s11627-021-10171-7
Han, D. G., Hou, Y. J., Ding, H. B., Zhou, Z. Y., Li, H. X., Yang, G. H. (2018b). Isolation and preliminary functional analysis of MbWRKY4 gene involved in salt tolerance in transgenic tobacco. Int. J. Agric. Biol. 20, 2045–2052. doi: 10.17957/ijab/15.0728
Han, D., Xu, T., Han, J., Liu, W., Wang, Y., Li, X., et al. (2021b). Overexpression of MxWRKY53 increased iron and high salinity stress tolerance in Arabidopsis thaliana. In. Vitro Cell. Dev. Biol. Plant 58, 266–278. doi: 10.1007/s11627-021-10241-w
Han, D., Zhang, Z., Ding, H., Wang, Y., Liu, W., Li, H., et al. (2018a). Molecular cloning and functional analysis of MbWRKY3 involved in improved drought tolerance in transformed tobacco. J. Plant Interact. 13, 329–337. doi: 10.1080/17429145.2018.1478994
Han, D., Zhou, Z., Du, M., Li, T., Wu, X., Yu, J., et al. (2020). Overexpression of aMalus xiaojinensisWRKY transcription factor gene (MxWRKY55) increased iron and high salinity stress tolerance in Arabidopsis thaliana. In. Vitro Cell. Dev. Biology-Plant. 56, 600–609. doi: 10.1007/s11627-020-10129-1
He, Y., Mao, S., Gao, Y., Zhu, L., Wu, D., Cui, Y., et al. (2016). Genome-Wide identification and expression analysis of WRKY transcription factors under multiple stresses in Brassica napus. PloS One 11, e0157558. doi: 10.1371/journal.pone.0157558
He, J., Xu, Y., Huang, D., Fu, J., Liu, Z., Wang, L., et al. (2022). TRIPTYCHON-LIKE regulates aspects of both fruit flavor and color in citrus. J. Exp. Bot. 73, 3610–3624. doi: 10.1093/jxb/erac069
Hu, W., Hou, X., Huang, C., Yan, Y., Tie, W., Ding, Z., et al. (2015). Genome-wide identification and expression analyses of aquaporin gene family during development and abiotic stress in banana. Int. J. Mol. Sci. 16, 19728–19751. doi: 10.3390/ijms160819728
Hu, X., Li, S., Lin, X., Fang, H., Shi, Y., Grierson, D., et al. (2021). Transcription factor CitERF16 is involved in citrus fruit sucrose accumulation by activating CitSWEET11d. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.809619
Huang, X., Cao, L., Fan, J., Ma, G., Chen, L. (2022). CdWRKY2-mediated sucrose biosynthesis and CBF-signalling pathways coordinately contribute to cold tolerance in Bermudagrass. Plant Biotechnol. J. 20, 660–675. doi: 10.1111/pbi.13745
Huang, R., Liu, D., Huang, M., Ma, J., Li, Z., Li, M., et al. (2019). CpWRKY71, a WRKY transcription factor gene of wintersweet (Chimonanthus praecox), promotes flowering and leaf senescence in Arabidopsis. Int. J. Mol. Sci. 20, 5325. doi: 10.3390/ijms20215325
Huang, T., Yu, D., Wang, X. (2021). VvWRKY22 transcription factor interacts with VvSnRK1.1/VvSnRK1.2 and regulates sugar accumulation in grape. Biochem. Biophys. Res. Commun. 554, 193–198. doi: 10.1016/j.bbrc.2021.03.092
Hussain, S. B., Shi, C.-Y., Guo, L.-X., Kamran, H. M., Sadka, A., Liu, Y.-Z. (2017). Recent advances in the regulation of citric acid metabolism in citrus fruit. Crit. Rev. Plant Sci. 36, 241–256. doi: 10.1080/07352689.2017.1402850
Kim, C. Y., Zhang, S. (2004). Activation of a mitogen-activated protein kinase cascade induces WRKY family of transcription factors and defense genes in tobacco. Plant J. 38, 142–151. doi: 10.1111/j.1365-313X.2004.02033.x
Kiranmai, K., Lokanadha Rao, G., Pandurangaiah, M., Nareshkumar, A., Amaranatha Reddy, V., Lokesh, U., et al. (2018). A novel WRKY transcription factor, MuWRKY3 (Macrotyloma uniflorum Lam. Verdc.) enhances drought stress tolerance in transgenic groundnut (Arachis hypogaea L.) plants. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00346
Langfelder, P., Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 9, 559. doi: 10.1186/1471-2105-9-559
Li, S. J., Wang, W. L., Ma, Y. C., Liu, S. C., Grierson, D., Yin, X. R., et al. (2020). Citrus CitERF6 contributes to citric acid degradation via upregulation of CitAclα1, encoding ATP-citrate lyase subunit α. J. Agric. Food Chem. 68, 10081–10087. doi: 10.1021/acs.jafc.0c03669
Li, S. J., Yin, X. R., Wang, W. L., Liu, X. F., Zhang, B., Chen, K. S. (2017). Citrus CitNAC62 cooperates with CitWRKY1 to participate in citric acid degradation via up-regulation of CitAco3. J. Exp. Bot. 68, 3419–3426. doi: 10.1093/jxb/erx187
Liao, L., Li, Y., Bi, X., Xiong, B., Wang, X., Deng, H., et al. (2022). Transcriptome analysis of Harumi tangor fruits: Insights into interstock-mediated fruit quality. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.995913
Liao, X., Li, M., Liu, B., Yan, M., Yu, X., Zi, H., et al. (2018). Interlinked regulatory loops of ABA catabolism and biosynthesis coordinate fruit growth and ripening in woodland strawberry. Proc. Natl. Acad. Sci. U.S.A. 115, E11542–E11550. doi: 10.1073/pnas.1812575115
Lin, Q., Wang, C., Dong, W., Jiang, Q., Wang, D., Li, S., et al. (2015). Transcriptome and metabolome analyses of sugar and organic acid metabolism in Ponkan (Citrus reticulata) fruit during fruit maturation. Gene 554, 64–74. doi: 10.1016/j.gene.2014.10.025
Liu, F., Dou, T., Hu, C., Zhong, Q., Sheng, O., Yang, Q., et al. (2023). WRKY transcription factor MaWRKY49 positively regulates pectate lyase genes during fruit ripening of Musa acuminata. Plant Physiol. Biochem. 194, 643–650. doi: 10.1016/j.plaphy.2022.12.015
Liu, W., Liang, X., Cai, W., Wang, H., Liu, X., Cheng, L., et al. (2022b). Isolation and functional analysis of VvWRKY28, a Vitis vinifera WRKY transcription factor gene, with Functions in tolerance to cold and salt stress in transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 23, 13418. doi: 10.3390/ijms232113418
Liu, Q., Xu, J., Liu, Y., Zhao, X., Deng, X., Guo, L., et al. (2007). A novel bud mutation that confers abnormal patterns of lycopene accumulation in sweet orange fruit (Citrus sinensis L. Osbeck). J. Exp. Bot. 58, 4161–4171. doi: 10.1093/jxb/erm273
Liu, G., Zhang, D., Zhao, T., Yang, H., Jiang, J., Li, J., et al. (2022a). Genome-wide analysis of the WRKY gene family unveil evolutionary history and expression characteristics in tomato and its wild relatives. Front. Genet. 13. doi: 10.3389/fgene.2022.962975
Liu, Y., Zhu, L., Yang, M., Xie, X., Sun, P., Fang, C., et al. (2022c). R2R3-MYB transcription factor FaMYB5 is involved in citric acid metabolism in strawberry fruits. J. Plant Physiol. 277, 153789. doi: 10.1016/j.jplph.2022.153789
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mangelsen, E., Kilian, J., Berendzen, K. W., Kolukisaoglu, U. H., Harter, K., Jansson, C., et al. (2008). Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genomics 9, 194. doi: 10.1186/1471-2164-9-194
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–d419. doi: 10.1093/nar/gkaa913
Miyamoto, T., Takada, R., Tobimatsu, Y., Suzuki, S., Yamamura, M., Osakabe, K., et al. (2020). Double knockout of OsWRKY36 and OsWRKY102 boosts lignification with altering culm morphology of rice. Plant Sci. 296, 110466. doi: 10.1016/j.plantsci.2020.110466
Mosharaf, M. P., Rahman, H., Ahsan, M. A., Akond, Z., Ahmed, F. F., Islam, M. M., et al. (2020). In silico identification and characterization of AGO, DCL and RDR gene families and their associated regulatory elements in sweet orange (Citrus sinensis L.). PloS One 15, e0228233. doi: 10.1371/journal.pone.0228233
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Raineri, J., Wang, S., Peleg, Z., Blumwald, E., Chan, R. L. (2015). The rice transcription factor OsWRKY47 is a positive regulator of the response to water deficit stress. Plant Mol. Biol. 88, 401–413. doi: 10.1007/s11103-015-0329-7
Ren, X., Chen, Z., Liu, Y., Zhang, H., Zhang, M., Liu, Q., et al. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63, 417–429. doi: 10.1111/j.1365-313X.2010.04248.x
Ross, C. A., Liu, Y., Shen, Q. J. (2007). The WRKY gene family in rice (Oryza sativa). J. Integr. Plant Biol. 49, 827–842. doi: 10.1111/j.1744-7909.2007.00504.x
Ruan, Y. L. (2014). Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 65, 33–67. doi: 10.1146/annurev-arplant-050213-040251
Rushton, P. J., Macdonald, H., Huttly, A. K., Lazarus, C. M., Hooley, R. (1995). Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of alpha-Amy2 genes. Plant Mol. Biol. 29, 691–702. doi: 10.1007/bf00041160
Rushton, P. J., Somssich, I. E., Ringler, P., Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shao, A., Xu, X., Amombo, E., Wang, W., Fan, S., Yin, Y., et al. (2023). CdWRKY2 transcription factor modulates salt oversensitivity in Bermudagrass [Cynodon dactylon (L.) Pers.]. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1164534
Sheng, L., Shen, D., Luo, Y., Sun, X., Wang, J., Luo, T., et al. (2017). Exogenous γ-aminobutyric acid treatment affects citrate and amino acid accumulation to improve fruit quality and storage performance of postharvest citrus fruit. Food Chem. 216, 138–145. doi: 10.1016/j.foodchem.2016.08.024
Shi, C. Y., Hussain, S. B., Yang, H., Bai, Y. X., Khan, M. A., Liu, Y. Z. (2019). CsPH8, a P-type proton pump gene, plays a key role in the diversity of citric acid accumulation in citrus fruits. Plant Sci. 289, 110288. doi: 10.1016/j.plantsci.2019.110288
Wan, H., Wu, L., Yang, Y., Zhou, G., Ruan, Y. L. (2018). Evolution of sucrose metabolism: the dichotomy of invertases and beyond. Trends Plant Sci. 23, 163–177. doi: 10.1016/j.tplants.2017.11.001
Wang, H., Chen, W., Xu, Z., Chen, M., Yu, D. (2023a). Functions of WRKYs in plant growth and development. Trends Plant Sci. 28, 630–645. doi: 10.1016/j.tplants.2022.12.012
Wang, H., Cheng, X., Yin, D., Chen, D., Luo, C., Liu, H., et al. (2023b). Advances in the research on plant WRKY transcription factors responsive to external stresses. Curr. Issues Mol. Biol. 45, 2861–2880. doi: 10.3390/cimb45040187
Wang, J. H., Liu, J. J., Chen, K. L., Li, H. W., He, J., Guan, B., et al. (2017). Comparative transcriptome and proteome profiling of two Citrus sinensis cultivars during fruit development and ripening. BMC Genomics 18, 984. doi: 10.1186/s12864-017-4366-2
Wei, W., Cheng, M. N., Ba, L. J., Zeng, R. X., Luo, D. L., Qin, Y. H., et al. (2019). Pitaya HpWRKY3 is associated with fruit sugar accumulation by transcriptionally modulating sucrose metabolic genes HpINV2 and HpSuSy1. Int. J. Mol. Sci. 20, 1890. doi: 10.3390/ijms20081890
Wei, Y., Shi, H., Xia, Z., Tie, W., Ding, Z., Yan, Y., et al. (2016). Genome-wide identification and expression analysis of the WRKY gene family in cassava. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00025
Wu, K. L., Guo, Z. J., Wang, H. H., Li, J. (2005). The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 12, 9–26. doi: 10.1093/dnares/12.1.9
Wu, S., Li, M., Zhang, C., Tan, Q., Yang, X., Sun, X., et al. (2021). Effects of phosphorus on fruit soluble sugar and citric acid accumulations in citrus. Plant Physiol. Biochem. 160, 73–81. doi: 10.1016/j.plaphy.2021.01.015
Wu, X., Tao, M., Meng, Y., Zhu, X., Qian, L., Shah, A., et al. (2020). The role of WRKY47 gene in regulating selenium tolerance in Arabidopsis thaliana. Plant Biotechnol. Rep. 14, 121–129. doi: 10.1007/s11816-019-00560-1
Yang, Y., Chi, Y., Wang, Z., Zhou, Y., Fan, B., Chen, Z. (2016). Functional analysis of structurally related soybean GmWRKY58 and GmWRKY76 in plant growth and development. J. Exp. Bot. 67, 4727–4742. doi: 10.1093/jxb/erw252
Ye, J., Wang, X., Hu, T., Zhang, F., Wang, B., Li, C., et al. (2017). An InDel in the promoter of Al-ACTIVATED MALATE TRANSPORTER9 selected during tomato domestication determines fruit malate contents and aluminum tolerance. Plant Cell 29, 2249–2268. doi: 10.1105/tpc.17.00211
Yoshida, T., Christmann, A., Yamaguchi-Shinozaki, K., Grill, E., Fernie, A. R. (2019). Revisiting the basal role of ABA - roles outside of stress. Trends Plant Sci. 24, 625–635. doi: 10.1016/j.tplants.2019.04.008
Yu, J. Q., Gu, K. D., Sun, C. H., Zhang, Q. Y., Wang, J. H., Ma, F. F., et al. (2021). The apple bHLH transcription factor MdbHLH3 functions in determining the fruit carbohydrates and malate. Plant Biotechnol. J. 19, 285–299. doi: 10.1111/pbi.13461
Yu, J. Q., Gu, K. D., Zhang, L. L., Sun, C. H., Zhang, Q. Y., Wang, J. H., et al. (2022). MdbHLH3 modulates apple soluble sugar content by activating phosphofructokinase gene expression. J. Integr. Plant Biol. 64, 884–900. doi: 10.1111/jipb.13236
Yu, Y., Hu, R., Wang, H., Cao, Y., He, G., Fu, C., et al. (2013). MlWRKY12, a novel Miscanthus transcription factor, participates in pith secondary cell wall formation and promotes flowering. Plant Sci. 212, 1–9. doi: 10.1016/j.plantsci.2013.07.010
Zhang, X., Feng, C., Wang, M., Li, T., Liu, X., Jiang, J. (2021). Plasma membrane-localized SlSWEET7a and SlSWEET14 regulate sugar transport and storage in tomato fruits. Hortic. Res. 8, 186. doi: 10.1038/s41438-021-00624-w
Keywords: WRKY gene family, citrus, fruit quality, sugar, organic acid, expression network
Citation: Zhang M, Lu W, Yang X, Li Q, Lin X, Liu K, Yin C, Xiong B, Liao L, Sun G, He S, He J, Wang X and Wang Z (2023) Comprehensive analyses of the citrus WRKY gene family involved in the metabolism of fruit sugars and organic acids. Front. Plant Sci. 14:1264283. doi: 10.3389/fpls.2023.1264283
Received: 20 July 2023; Accepted: 24 August 2023;
Published: 15 September 2023.
Edited by:
Agnieszka Ludwików, Adam Mickiewicz University in Poznań, PolandReviewed by:
Mingjun Li, Northwest A&F University, ChinaDeguo Han, Northeast Agricultural University, China
Copyright © 2023 Zhang, Lu, Yang, Li, Lin, Liu, Yin, Xiong, Liao, Sun, He, He, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xun Wang, d3gwMTA0QHNpY2F1LmVkdS5jbg==; Zhihui Wang, d2FuZ3poaWh1aTMxOEAxMjYuY29t
†These authors have contributed equally to this work
 Mingfei Zhang
Mingfei Zhang Wen Lu†
Wen Lu† Qin Li
Qin Li Xingyu Lin
Xingyu Lin Bo Xiong
Bo Xiong Zhihui Wang
Zhihui Wang