- 1Ecology Institute of Qilu University of Technology (Shandong Academy of Sciences), Jinan, China
- 2School of Agriculture, Food and Wine, The University of Adelaide, Urrbrae, SA, Australia
As one of the major abiotic stresses, salinity can affect crop growth and plant productivity worldwide. The inoculation of rhizosphere or endophytic microorganisms can enhance plant tolerance to salt stresses, but the potential mechanism is not clear. In this study, Trichoderma harzianum ST02 was applied on sweet sorghum [Sorghum bicolor (L.) Moench] in a field trial to investigate the effects on microbiome community and physiochemical properties in the rhizosphere soil. Compared with the non-inoculated control, Trichoderma inoculation significantly increased the stem yield, plant height, stem diameter, and total sugar content in stem by 35.52%, 32.68%, 32.09%, and 36.82%, respectively. In addition, Trichoderma inoculation improved the nutrient availability (e.g., N, P, and K) and organic matter in the rhizosphere soil and changed the bacterial community structure and function in both bulk and rhizosphere soil by particularly increasing the relative abundance of Actinobacter and N-cycling genes (nifH, archaeal and bacterial amoA). We proposed that T. harzianum ST02 could promote sweet sorghum growth under saline conditions by regulating available nutrients and the bacterial community in the rhizosphere soil.
1 Introduction
The issue of soil salinization is a pressing environmental concern, with an estimated 20% of irrigated land worldwide affected by salinity (Yuan et al., 2016). Although plants have developed various strategies to withstand salt stress (Rojas-Tapias et al., 2012), these mechanisms are insufficient to support growth under severe salt stress conditions. Moreover, the intricate interplay between many microorganisms and plants (Hassani et al., 2018) has a positive impact on the viability and overall health of plants (Rosier et al., 2016; Etesami and Maheshwari, 2018). It has been found that the rhizosphere microorganisms can interact with their hosts to form a root–soil–microbial interaction network, which can detect and react to signal molecules released by plant roots, leading to enhanced tolerance to biotic and abiotic stress factors such as salinity (Qin et al., 2016; Zhang et al., 2017a). Myriad rhizosphere or endophytic microorganisms can effectively improve the salt tolerance of plants by modulating the ion balance (Zhu et al., 2016) and/or increasing osmotic protective substances in plants (Bhattacharyya et al., 2015). Microorganisms can also produce 1-aminocyclopropane-1-carboxylate (ACC) deaminase, siderophores, or indole acetic acid (IAA) to trigger plant defense and metabolic pathways to tolerate salt stress (Xiong et al., 2019).
Trichoderma spp. have been extensively studied as antagonistic fungal agents against plant pathogens as well as plant growth promoters (Woo et al., 2014). In addition, there have been numerous reports of applying Trichoderma spp. to exert beneficial effects on enhancing plant tolerance under saline conditions (Contreras-Cornejo et al., 2016). Some Trichoderma strains can stably colonize plant roots, cause substantial changes in plant metabolism by producing secondary metabolites, and induce plant resistance to abiotic stress (Mastouri et al., 2010). For instance, T. virens (Tv29.8) and T. atroviride (IMI 206040) enhanced root development of Arabidopsis thaliana through auxin signaling and produced osmotic substances to promote plant growth in saline conditions (Contreras-Cornejo et al., 2014). T. echinospora Q1 alleviated the salt stress on cucumber by changing plant hormone levels and phosphate solubilization capacity (Zhao and Zhang, 2015). T. longibrachiatum TL-6 disturbed the intracellular ionic homeostasis and modulated the transcriptional levels of IAA and ethylene synthesis genes in wheat seedlings under salt stress through promoted ACC deaminase activity and increased IAA production (Zhang et al., 2019). Trichoderma spp. were capable of inhibiting the growth of plant pathogens. For example, the antagonistic activity test of a T. cyanodichotomus strain showed that its inhibition ability was high to suppress the growth of Botryosphaeria dothidea, Pythium aphanidermatum, Rhizoctonia solani, and Verticillium dahlia, but poor for Botrytis cinerea and Helminthosporium sativum (Li et al., 2018). Therefore, antagonistic activity of Trichoderma may change the community structure, consequently altering the microbial diversity of soil (Pang et al., 2017; Velmourougane et al., 2017). However, the mechanism of Trichoderma spp. modulating rhizosphere available nutrients and the microbiome community under saline conditions remains poorly understood.
Sweet sorghum [Sorghum bicolor (L.) Moench] is a widely adapted sugar crop with high biomass production and high carbohydrate content (Guo et al., 2018). The sugar-rich stalk of sweet sorghum makes the crop particularly amenable to direct fermentable sugar extraction, and appropriate saline stress can increase sugar accumulation (Sui et al., 2015). The low input costs of sweet sorghum are based on its highly drought tolerance and C4 photosynthesis (Calviño and Messing, 2012). Previously, we isolated T. harzianum strain ST02 from the saline soil of the Yellow River Delta in Dongying City and demonstrated that ST02 was able to enhance the tolerance of tomato to NaCl stress in a greenhouse experiment through improving the antioxidant defense reaction of plants (Zhao et al., 2021). Hence, in the present study, we aimed to investigate the impact of ST02 inoculation on physiological growth, biomass, and sugar content of sweet sorghum, along with the changes of nutrient availability and microbiome community in the rhizosphere soil.
2 Materials and methods
2.1 Plant and microorganism
The seeds of sweet sorghum, variety Aertuo326, were purchased from Hunan Longping Seeds Industry Co. (Changsha, China). Trichoderma harzianum ST02 was isolated from saline soil collected at the Yellow River Delta in Dongying City, as described previously (Zhao et al., 2021). Conidial suspension of ST02 was scraped from a potato dextrose agar (PDA) plate cultured for 72 h, and the concentration of conidia was adjusted to 108 CFU mL−1 with sterile water. The inoculum for field experiment was prepared as wettable powders composed of 10% fresh conidial suspension (2 × 108 CFU g−1) and 90% sterilized diatomite.
2.2 Field experiment
The site of field experiment was located in Guangrao town, Dongying City, Shandong Province (118°37′5ʺE, 37°15′18ʺN). The soil was classified as sandy loam, and the physicochemical characteristics were pH (soil/water ratio of 1:2.5, w/v) of 7.15, electric conductivity of 585 μS cm−1, organic matter of 3.98 mg g−1, total N of 1.39 mg g−1, total P of 0.46 mg g−1, and total K of 11.81 mg g−1. The field experiment was conducted in a completely randomized block design with +/− Trichoderma inoculation as two factors. For the + Trichoderma treatment, 3 kg of ST02 wettable powder was mixed with 20 kg of sweet sorghum seeds before sowing. For the − Trichoderma control, the same amount of sterilized water was mixed with sterilized diatomite to prepare the wettable powder for the application. The field was divided into eight plots (5 m × 30 m) by 0.5-m gaps; four plots were chosen at random as + Trichoderma treatment and the other four plots as − Trichoderma control. Each treatment was sown with 12 rows by a seeding machine. Conventional agricultural management measures were adopted after sowing in all plots.
To determine the inoculum amount of T. harzianum ST02 on each sweet sorghum seed, 50 inoculated seeds were rinsed with 20 mL of 0.9% sterilized sodium chloride solution in a 50-mL triangular bottle (Bazhanov et al., 2017). After being vortexed for 5 min, the resulting suspensions and their serial dilutions were smeared on PDA plates. Trichoderma colonies were counted after 3 days at 26°C.
After sweet sorghum maturity and harvesting, 20 plants were randomly selected from each treatment to measure the plant height, stalk diameter, stalk fresh weight, and total sugar content in the stem. Whole sorghum stems were crushed in a cane-juice squeezer, and the collected juice was used to determine the total sugar content by a handheld refractometer (Lichen LC-DR-328, LC-BX Inc., Shanghai, China), which has been widely used to measure sugar content in the liquid solution. Sugar content was indicated by the light reflectance when it passed through the sample and calibrated by the samples with known sugar concentration. The total yield of sweet sorghum at different plots was recorded at the time of harvesting.
2.3 Soil sampling
Bulk soil and rhizosphere soil samples were collected at five different sites as a biological replicate, with a total of seven replicates for each treatment (28 samples in total). Bulk soil was designated as the soil of 20 to 30 mm away from the roots, and rhizosphere soil was designated as within 1 mm on the root surface. All soil samples were divided into two parts: one part of 2 g was stored at −80°C for DNA extraction, and the other part of 20 g was air dried and stored at room temperature for physicochemical properties analysis.
2.4 Soil physicochemical property analysis
The air-dried soil samples were passed through a 2-mm sieve before the physicochemical property analysis. Soil pH and electric conductivity were tested with a pH meter (Mettler Toledo, Germany) in a 1:2.5 (soil/water ratio of 1:2.5, w/v soil: deionized water W/V) suspension (Qiu et al., 2022). Soil total nitrogen (TN) content was determined using the modified Kjeldahl method (HJ717-2014). The total potassium (TK) content was quantitatively determined with an ultraviolet spectrophotometer (GB/T9836-1988). Soil organic matter (SOM) was measured by redox titration with K2Cr2O7 (HJ615-2011). The alkaline hydrolysis diffusion method was used to determine the available nitrogen (AN) content in the soil (Fu et al., 2019). The available phosphorus (AP) concentration was measured by the molybdenum blue method (Olsen and Sommers, 1982), and the available potassium (AK) was measured by flame photometry in 1 M ammonium acetate soil extracts (NY/T 889-2004).
2.5 Soil DNA extraction and Miseq sequencing
PowerSoil DNA Isolation Kit (Qiagen, Germany) was used to extract total genomic DNA from 0.25-g soil samples following the manufacturer’s instructions. The extracted DNA was electrophoresed with 1% agarose gel, and the concentration and purification of the genomic DNA were measured by a NanoDrop 2000 spectrophotometer (Thermo Scientific, United States). The specific primers 341F (5′-CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′) (Klindworth et al., 2012) of the bacterial 16S rRNA gene were used for amplification. The purified products were quantified by a Qubit 3.0 DNA detection kit, and then samples were mixed and homogenized in equimolar ratio and sequenced on an Illumina MiSeq PE300 platform at Shanghai Sangon Bioengineering Co. The bioinformatic analysis of the DNA sequence is referred to Li et al. (2022). Alpha diversity indices, such as Shannon diversity, Chao1 richness, and Faith’s phylogenetic diversity, were calculated to estimate the bacterial diversity within an individual sample.
2.6 Quantitative PCR analysis of N-cycling genes
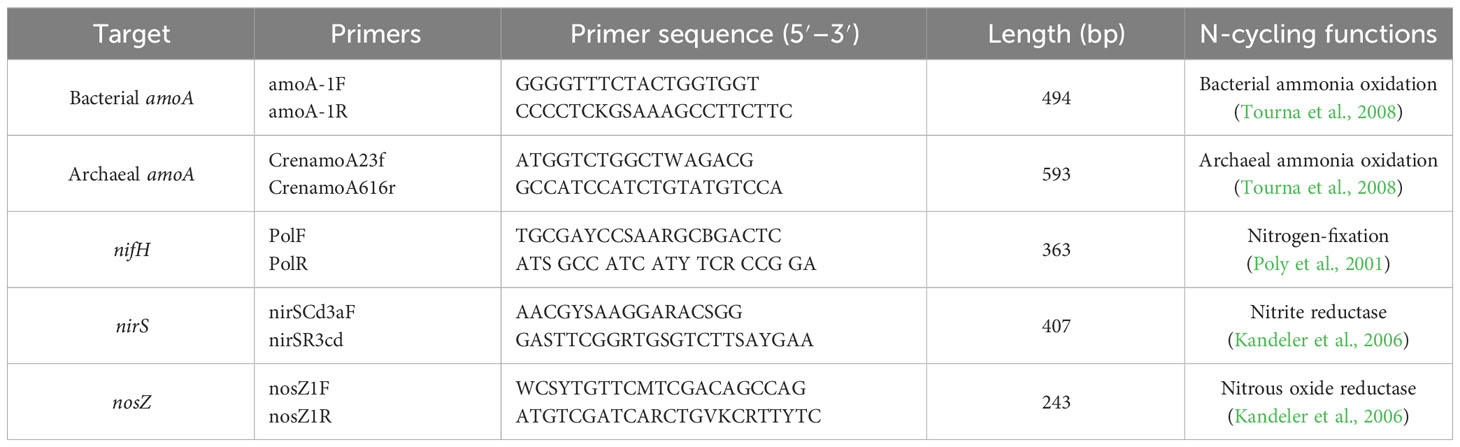
The abundances of five representative N-cycling genes (nifH, bacterial amoA, archaeal amoA, nirS, and nosZ) were determined by quantitative polymerase chain reaction (qPCR) with the CFX96 (Bio-Rad, Hercules, CA, USA) for the rhizosphere and bulk soil DNA samples. The primer sequences are shown in Table 1. The 25-µL qPCR reactions contained 2× Power SYBR® Green PCR Master Mix (TaKaRa, Dalian, China) 12.5 µL, each 10 µM forward and reverse primers 0.5 µL, soil DNA samples 1.0 µL, and sterile DNA-free water 10.5 µL. A standard thermal profile was used: 3 min at 95°C for pre-denaturation, followed by 40 cycles of 10 s at 95°C for denaturation and 30 s at optimal temperature for annealing, then extension at 72°C for 20 s. The copy number of target DNA was calculated according to the standard curves, which were generated from 10-fold serial dilution of a plasmid containing the targeted DNA fragments. Melting curve analysis was conducted after the amplification cycles by setting up the temperature at 95°C, 1 min; 65°C, 1 min; from 65°C for every 0.5°C for 10 s; and then continuous increase to 95°C. Three technical replicates for each sample were used to detect the possible error.
2.7 Statistical analysis
All the results were expressed as the mean ± standard deviation. Duncan’s multiple range test was used to compare the significant difference at the 0.05 level using SPSS 25.0 (IBM Corporation, Armonk, NY, USA). Correlation analyses were carried out using the Spearman correlation method. Bray–Curtis distance was used to investigate the bacterial communities structural variation and then visualized via principal coordinate analysis (PCoA). Redundancy analysis (RDA) was performed to reveal the association of bacterial communities in relation to environmental factors based on relative abundances of bacterial species at different taxa levels using the “vegan” package in R (Lozupone et al., 2010).
3 Results
3.1 Sweet sorghum growth influenced by +/− T. harzianum ST02 in saline soil field test
Field experiments were conducted to study the effects of T. harzianum ST02 on the growth of sweet sorghum. The results showed that the application of Trichoderma significantly promoted the growth and sugar accumulation of sweet sorghum. It significantly increased in plant height (Figure 1A, p < 0.01), stalk diameter (Figure 1B, p < 0.01), and sugar content (Figure 1C, p < 0.01) by 32.68%, 32.09% and 36.82%, respectively, compared with non-inoculated control. The yield of sweet sorghum stems inoculated with Trichoderma ST02 significantly increased by 35.52% to control (Figure 1D, p < 0.01).
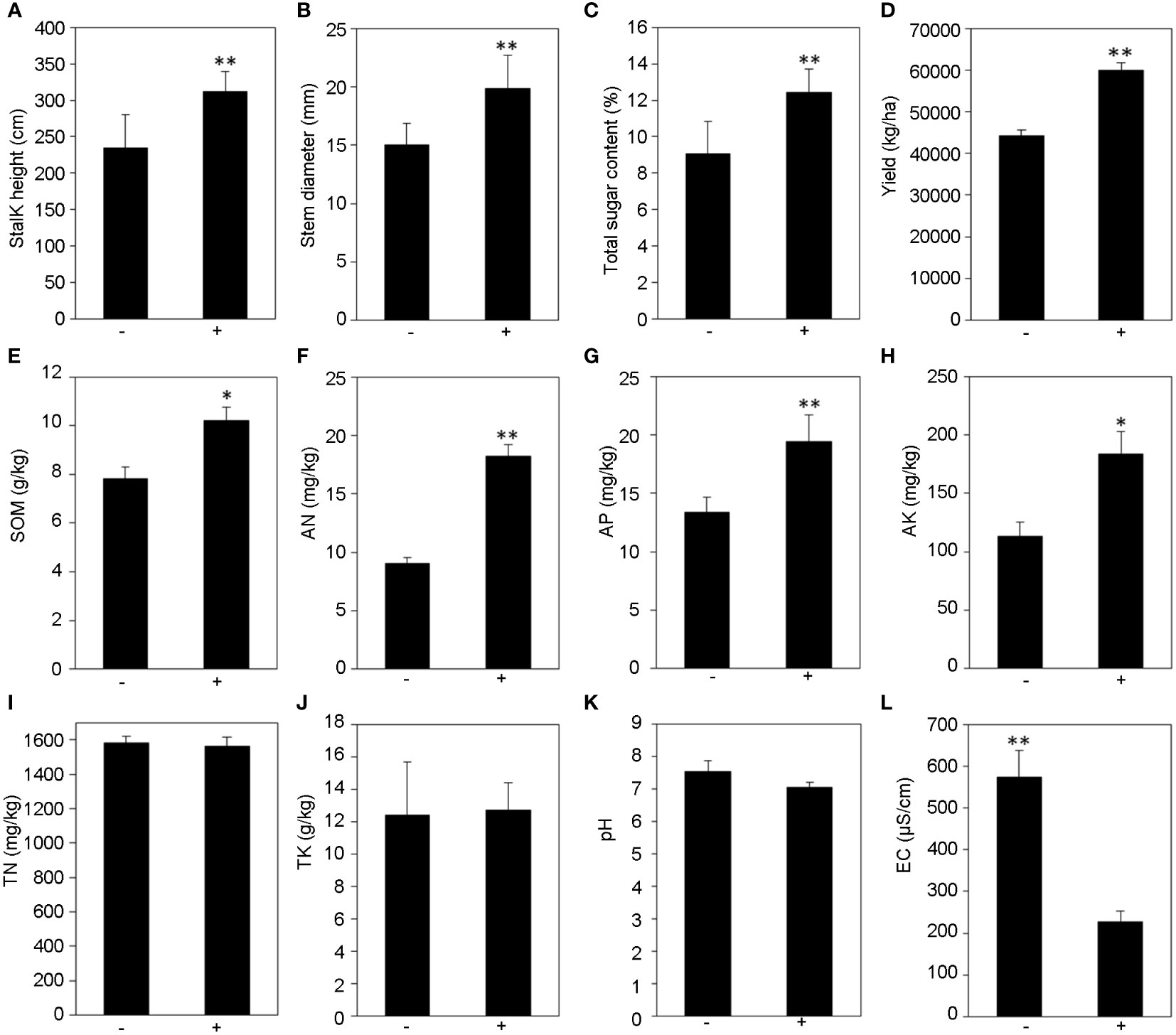
Figure 1 The effect of T. harzianum ST02 inoculation on the growth (A–D) and rhizosphere soil physicochemical properties (E–L) of sweet sorghum. SOM, soil organic matter; TN, total nitrogen; AN, available nitrogen; AP, available phosphorus; TK, total potassium; AK, available potassium; EC, electrical conductivity. The statistically significant values are presented as asterisks (*p < 0.05; **p < 0.01) based on one-way ANOVA with Duncan’s multiple range test. +/− stands for with and without T. harzianum ST02 inoculation, respectively.
3.2 Analysis of soil physical and chemical properties
The physical and chemical properties of rhizosphere soil in the field experiment were analyzed. Our results indicated that sweet sorghum inoculation with Trichoderma had greater contents of SOM, AN, AP, and AK but lower electric conductivity (Figure 1) in the rhizosphere soil than the non-inoculated control. Compared with the untreated control, the contents of SOM, AN, AP, and AK in rhizosphere soil increased by 30.6%, 101.55%, 44.77%, and 61.95%, respectively (Figures 1E–H). Additionally, the electric conductivity was decreased from 574 μS/cm (− ST02) to 226 μS/cm (+ST02) (Figure 1L). For the contents of TN, TK, and pH, there was no significant difference between the soils treated with and without Trichoderma (Figures 1I–K).
3.3 The effects of ST02 inoculation on the soil bacterial community
There were 1,728,736 clean sequences obtained from all samples after filtering with an average length of 416 bp. Shannon indexes and Chao1 indexes of Trichoderma-inoculated treatment were significantly lower (Figures 2A, B, p < 0.05) than the non-inoculated control in the rhizosphere and bulk soil.
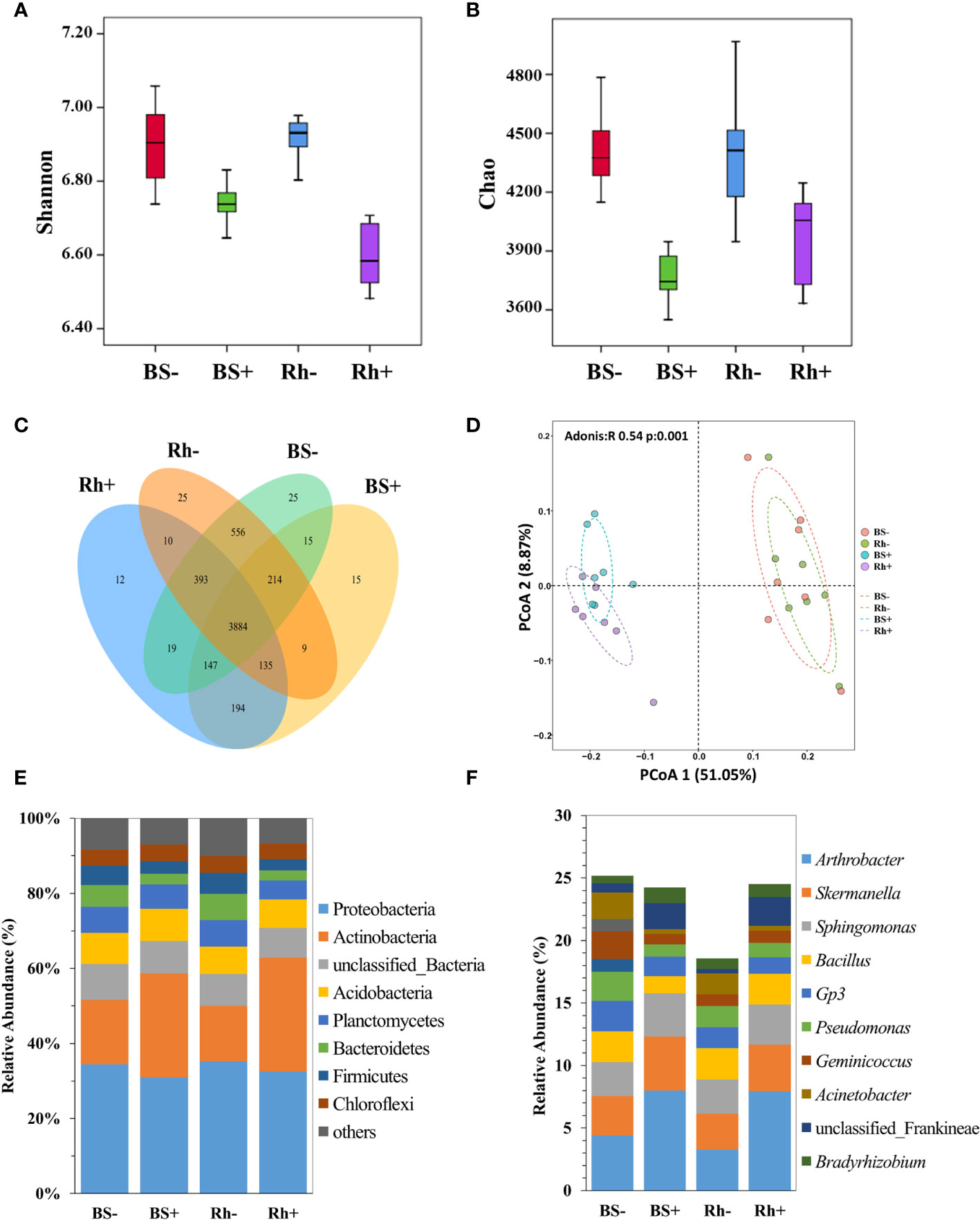
Figure 2 The effect of T. harzianum ST02 inoculation on the bacterial community in bulk soil and rhizosphere soil. (A) Comparison of Shannon index between groups. (B) Comparison of Chao index between groups. (C) Venn diagram showing the shared bacterial OTUs. (D) PCoA results based on Bray–Curtis distance metric; ellipses are 95% confidence. The relative abundance of bacteria at the (E) phylum level and (F) genus level. BS−, bulk soil without ST02 inoculation. BS+, bulk soil with ST02 inoculation. Rh−, rhizosphere soil without ST02 inoculation. Rh+, rhizosphere soil with ST02 inoculation. Different letters in (A, B) indicate significant differences among the treatments at the p < 0.05 level based on one-way ANOVA with Duncan’s multiple range test.
There were 25, 15, 25, and 12 unique OTUs observed in the bulk soil without Trichoderma inoculation (BS−), bulk soil with inoculation (BS+), rhizosphere soil without inoculation (Rh−), and rhizosphere soil with inoculation (Rh+) groups (Figure 2C), respectively. The PCoA based on the Bray–Curtis distance matrix showed a clear separation of treatments from inoculated with and without Trichoderma (Figure 2D). The first and second principal coordinates explained the variation of soil bacterial communities to 51.05% and 8.87%, respectively. The bacterial communities of bulk and rhizosphere soils were highly similar and clustered within the inoculation treatment.
According to the taxonomic annotation of OTUs, 12 dominant bacterial phyla (relative abundance >1.0%) accounted for 98.28%–99.12% of the total sequences in the four niches (Figure 2E). Proteobacteria, Actinobacteria, Bacteria, Acidobacteria, Planctomycetes, Bacteroidetes, and Firmicutes were the most predominant bacterial phyla with a mean relative abundance >5.0%. Trichoderma treatment significantly increased the relative abundances of Actinobacteria, Gemmatimonadota, Nitrospirae, and Acidobacteriota but decreased the relative abundances of Bacteroidetes, Proteobacteria, and Firmicutes in the rhizosphere and bulk soil (Figure 2E).
At the genus level, Arthrobacter had the highest relative abundance in Trichoderma-treated rhizosphere and bulk soil at 7.94% and 7.98%, respectively (Figure 2F). In rhizosphere soil, Trichoderma inoculation significantly increased the relative abundance of Arthrobacter, Nocardioides, unclassified Frankineae, and Bradyrhizobium by 142.07%, 152.61%, 550.26%, and 23.55%, respectively, but decreased the relative abundance of Planctomyces, Bacillus, and Gp3. In bulk soil, Trichoderma inoculation significantly increased the relative abundance of Arthrobacter, Nocardioides, unclassified Frankineae, and Bradyrhizobium by 81.36%, 126.83%, 184.52%, and 107.39%, respectively, but decreased the relative abundance of Geminicoccus and Bacillus.
3.4 Effects of Trichoderma inoculation on the abundance of N-cycling genes
The copy numbers of nifH, amoA, nirS, and nosZ genes were quantified by qPCR, which related to nitrogen fixation, nitrification, and denitrification, respectively. The results are shown in Figure 3. The abundance of nifH gene was significantly increased (Figure 3A, p < 0.05) in rhizosphere (from 8.88 × 105 to 2.80 × 106 copies) and bulk soil (from 1.35 × 106 to 3.76 × 106 copies) after inoculation with Trichoderma, which was 2.15 and 1.78 times greater than control treatments, respectively. The effects of Trichoderma inoculation on bacterial amoA gene abundance were similar to those of nifH, and the copy number of bacterial amoA increased significantly in both rhizosphere soil and bulk soil (Figure 3B), whereas the archaeal amoA was higher in inoculation treatment than the control only in the rhizosphere soil but not the bulk soil (Figure 3C). There was no rhizosphere effect or inoculation effect on the abundance of nirS (Figure 3D). The abundance of nosZ gene was greater in the rhizosphere soil than in bulk soil but not influenced by the inoculation effect (Figure 3E).
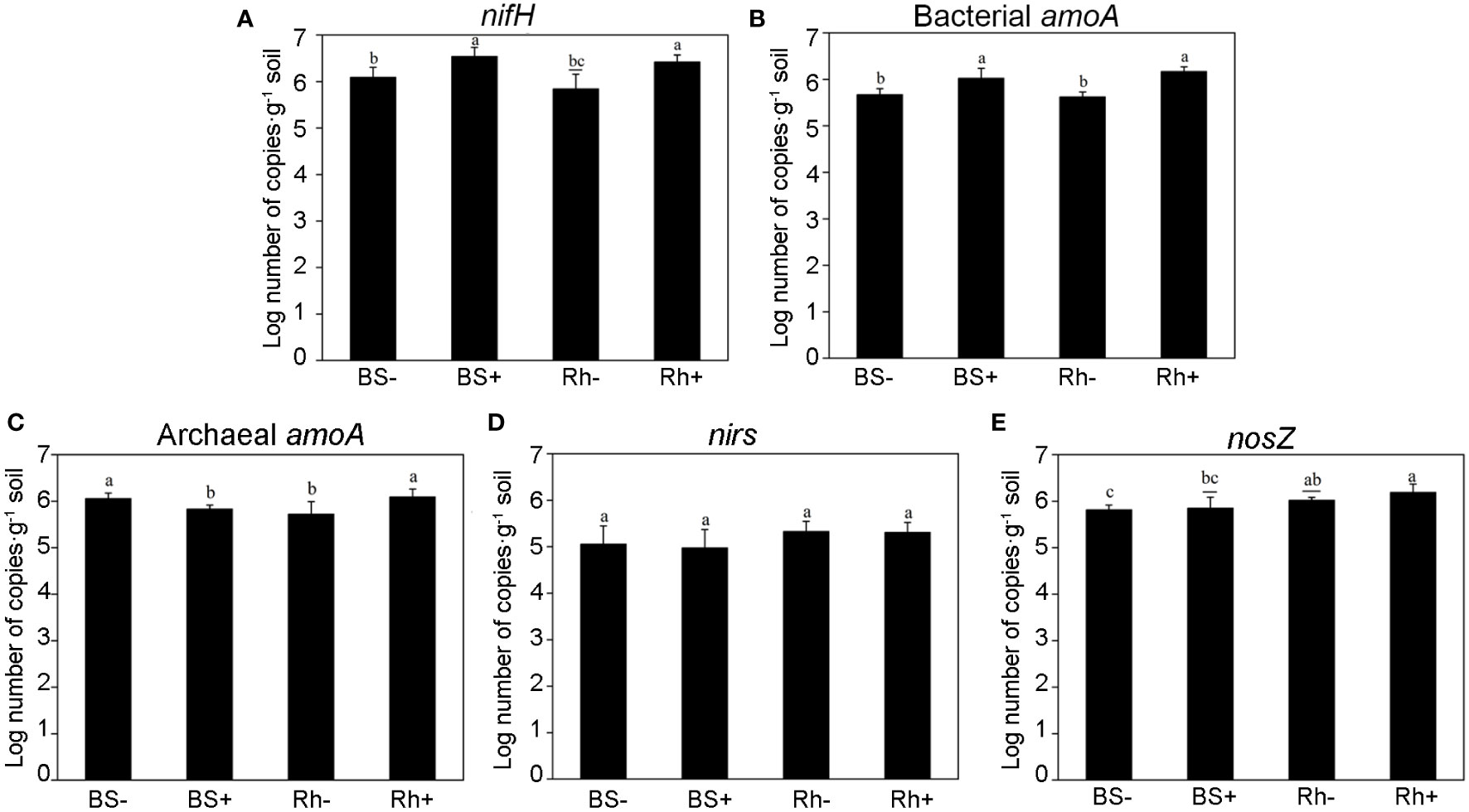
Figure 3 The abundances of nitrogen-cycling genes measured by quantitative PCR. The abundance of five N-cycling genes involved in nitrogen fixation (A, nifH), ammonia oxidation by bacteria (B, bacterial amoA) and archaea (C, archaeal amoA), and denitrification (D, E, nirS, nosZ), in different treatment groups. BS−, bulk soil without inoculation. BS+, bulk soil with inoculation. Rh−, rhizosphere soil without inoculation. Rh+, rhizosphere soil with inoculation. Different letters indicate significant differences among the treatments at the p < 0.05 level based on one-way ANOVA with Duncan’s multiple range test.
3.5 The relationship between rhizosphere soil properties and the structure of rhizobacterial communities
Spearman’s correlation analysis was used to test the rhizosphere soil factors influencing the microbial structure and functional pathway. The top 10 dominant bacterial genera were significantly correlated with at least one rhizosphere soil properties (Figure 4). The abundances of Arthrobacter, Streptomyces, and Nocardioides were positively correlated with content of SOM, AN, AP, and AK and negatively correlated with EC (p < 0.01), whereas those of Euzebya and Ramlibacter were negatively correlated with contents of SOM, AN, AP, and AK (p < 0.01) and positively correlated with EC (p < 0.01).
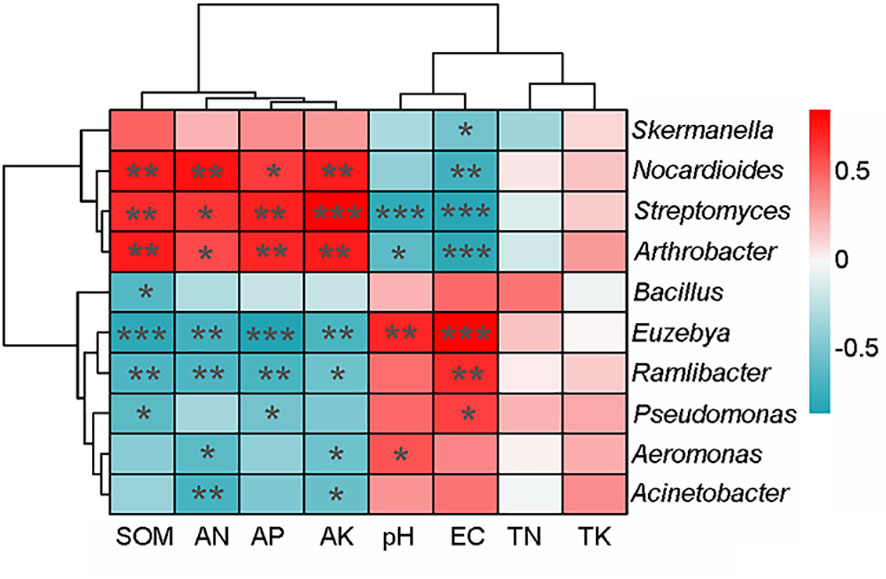
Figure 4 Correlation heatmap between the top 10 bacterial genera and soil properties. The result was based on Spearman’s correlation analysis. Positive relationships are represented in blue, whereas negative relationships are represented in red. The significant correlations are presented as asterisks (*p < 0.05; ** p < 0.01; *** p < 0.001).
The relationships between bacterial OTU composition and the soil properties were tested by redundancy analysis (Figure 5A). The first two axes explained 58.94% of the total variance. The soil SOM, AN, AK, and AP were positively corrected with the rhizosphere inoculated with Trichoderma, whereas pH and EC were correlated with non-inoculated control. For the relationships between the abundance of N-cycling genes and bacterial OTU composition (Figure 5B), the first two components accounted for 44.31% of the total variance and the inoculation of Trichoderma was positively corrected with the abundance of nifH and amoA in the rhizosphere soil.
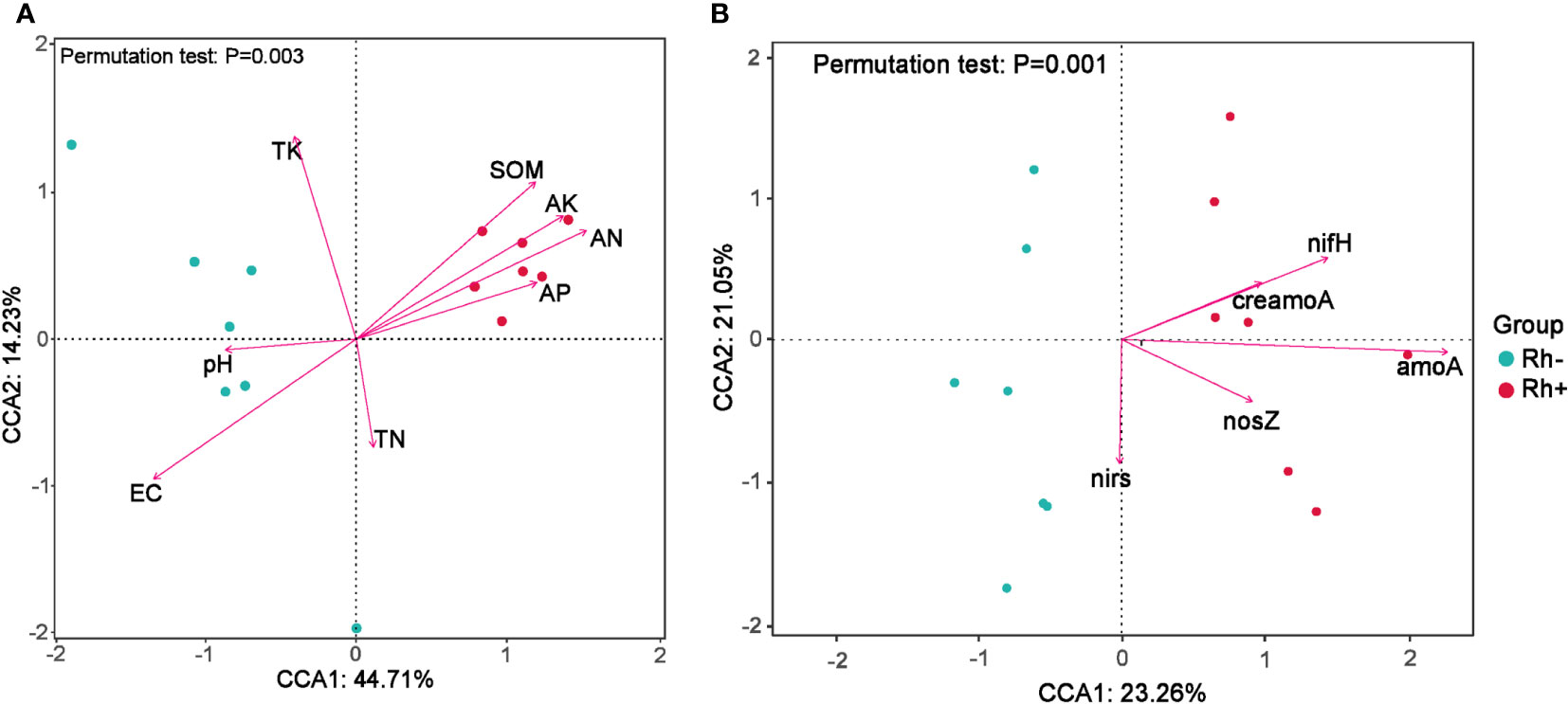
Figure 5 Redundancy analysis (RDA) to test the soil properties (A) and the abundance of N-cycling genes (B) influencing the bacterial OTU composition in the rhizosphere of sweet sorghum. Rh−, rhizosphere soil without ST02 inoculation. Rh+, rhizosphere soil with ST02 inoculation.
4 Discussion
Inoculation of beneficial microbes on agricultural crops is a cost-effective way to improve productivity. In this study, we revealed the beneficial effects and possible mechanism of T. harzianum ST02 on the growth of sweet sorghum under saline soil. We confirmed that inoculation with T. harzianum ST02 improved the shoot length, shoot dry weight, biomass, and total sugar content of sweet sorghum by modifying the rhizosphere soil, including increasing the available nutrients and changing the microbiome structure and function.
4.1 Trichoderma inoculation increased the growth and yield of sweet sorghum under saline conditions
Promotion of plant growth by Trichoderma under salt stress in wheat (Fu et al., 2019), cucumber (Zhao and Zhang, 2015), and barley (Gupta et al., 2021) has been reported. In our study, T. harzianum ST02 significantly increased sweet sorghum growth and grain yield in saline soil. These results were in agreement with our previous finding that T. harzianum ST02 increases the survival rate, plant height, and fresh weight the chlorophyll content and net photosynthetic rate of tomato seedlings under 200 mM NaCl stress (Zhao et al., 2021). Previous studies had found that T. longibrachiatum T6 increased IAA production and ACC-deaminase activity to enhance tolerance to NaCl stress and promote wheat growth (Zhang et al., 2019).
The main fermentable sugars in sweet sorghum stems are sucrose, fructose, and glucose (Almodares and Hadi, 2009); sucrose is the main sugar, accounting for 85% of the total content, and it is mainly accumulated through photosynthesis (Gowik et al., 2011; Sui et al., 2015). Under saline conditions, plants accumulate soluble sugars to promote osmotic regulation and thus improve dehydration tolerance (Farhangi-Abriz and Torabian, 2017). In our field trial, there was a significant increase in sugar content by inoculating ST02 than control (Figure 1C).
4.2 Effects of Trichoderma on soil properties and nutrient availability in the saline soil
Salinity stress affects the development of plant roots, thus affecting the utilization of soil nutrients and inhibiting plant growth (Zhao et al., 2020). Trichoderma can improve plant growth, especially when the conditions were unfavorable (Khomari and Davari, 2017), possibly by improving nutrient availability and photosynthesis efficiency (Asad, 2022). Previous studies had found that inoculated chickpea with Trichoderma increased nutrient availability and release of growth-promoting substances to enhance chickpea growth (Rudresh et al., 2004). In this study, we found that Trichoderma ST02 significantly decreased soil EC by 60.63% (Figure 1L, p < 0.01) to enhance soil nutrient availability and promote the growth of sweet sorghum. Additionally, compared with the control, Trichoderma enhanced the contents of SOM and AP by 30.6% and 44.77%, respectively, and the content of AN doubled in the rhizosphere soil, indicating that nutritional conditions of the sweet sorghum rhizosphere soil were improved. This might be attributed to that Trichoderma could release organic acid to solubilize nitrogen and phosphorus in soil and increase soil organic matter and soil fertility (Pang et al., 2017; Velmourougane et al., 2017; Ye et al., 2020). Similar results were also found in the AK; inoculation with Trichoderma could increase the AK content and restrain the Na+ uptake, which are beneficial for alleviating salt stress (Yu et al., 2019).
4.3 Trichoderma affected the composition of soil bacterial community
The composition and diversity of the soil microbial community in the rhizosphere are crucial for soil quality and plant health (Imane et al., 2021). In this study, compared with the control, Trichoderma inoculation significantly changed the structure and diversity of soil bacterial communities. It significantly reduced the bacterial diversity and richness and changed the bacterial taxonomic composition (Figure 2).
Trichoderma increased the relative abundance of Actinobacteria but decreased that of Bacteroidetes in both rhizosphere soil and bulk soil. Actinobacteria plays a vital role in the decomposition of organic matter, and they could also produce antibiotics to inhibit plant pathogens in soil (Zhang et al., 2017b), which might contribute to the growth and yield improvement of sweet sorghum. At the genus level, we found that Trichoderma treatment significantly increased the abundance of Arthrobacter, Streptomyces, Nocardioides, unclassified Frankineae, and Bradyrhizobium in both rhizosphere and bulk soil. Among them, Arthrobacter is the dominant bacteria in soil as the plant growth-promoting bacteria with the ability to degrade atrazine (Bazhanov et al., 2017). In addition, Jiang et al. (2004) had isolated Arthrobacter strain HS-G8 with nitrogen fixation activity from soil, which is in line with our results that nifH gene was higher in the soil with Trichoderma inoculation. Streptomyces, Nocardioides, and Frankineae were the dominant bacterial groups of Actinobacteria. Streptomyces can produce antibiotics and promote the decomposition of organic matter (Qin et al., 2011). Previous studies reported that Frankineae and Bradyrhizobium are linked to nitrogen fixation in plants (Sellstedt and Richau, 2013; Matos et al., 2021), which are related to the increase of nifH gene and available N in the soils with Trichoderma application in the present study. In addition, ST02 treatment decreased the abundance of Geminicoccus in both rhizosphere and bulk soil. Previous studies found that Gemmatimonadaceae (genus Gemmatimonas) was negatively correlated with the contents of AN and AP (Li et al., 2021). This was consistent with the result that the relative abundance of Gemmatimonadaceae decreased with higher AN and AP contents under Trichoderma ST02 treatment compared with control. These results suggested that the nitrogen cycle was enhanced in the Trichoderma treatment, thus promoting the uptake of nitrogen nutrients in sweet sorghum.
4.4 Trichoderma shift the abundance of N-cycling gene
Trichoderma can promote the nitrogen uptake and accumulation in tobacco and leafy vegetables (Singh et al., 2018; Visconti et al., 2020) and increased nitrate nitrogen in the soil (Cheng et al., 2020; Wu et al., 2022). In this study, we found that the mean abundance of nifH gene was greater than any other tested N cycling genes. Trichoderma ST02 increased the relative abundance of unclassified Frankineae and Bradyrhizobium, which were closely linked to nitrogen fixation and might explain the higher nifH gene abundance.
In addition, we also studied the effects of Trichoderma inoculation on the abundance of nitrification genes (bacterial amoA and archaea amoA) and denitrification genes (nirS and nosZ), which can change the contents of NH4+-N and NO3−-N in the soil (Wu et al., 2022). The first step of nitrification is ammonia oxidation, which is driven by ammonia-oxidizing archaea (AOA) and ammonia-oxidizing bacteria (AOB) (Shafiee et al., 2021). In this study, we found that the abundance of bacterial amoA and archaeal amoA genes increased significantly in the rhizosphere soil compared with control. However, in bulk soil, only bacterial amoA increased and the abundance of archaea amoA decreased significantly compared with control (Figures 3B, C). Previous studies have shown that the content of organic matter was the main factor affecting the growth of ammonia-oxidizing microorganisms and their ammonia-oxidizing function (Kirchman, 2012; Ouyang et al., 2018), and the lower AOA : AOB ratio in the fertilized treatments was associated with higher soil nitrification potential (Sun et al., 2015). Our results indicated that Trichoderma changed the abundance of amoA and promoted the conversion of ammonia nitrogen to nitrate nitrogen. For denitrification genes, ST02 had no significant effect on nirS and only increased the abundance of nosZ in rhizosphere soil.
5 Conclusion
This study demonstrated that T. harzianum ST02 significantly improved the growth and productivity of sweet sorghum. The promoting effect of Trichoderma was mainly attributed to the increase of the available nutrients and the reduction of EC in the rhizosphere soil, and the modified soil microbial community. Trichoderma inoculation of sweet sorghum remarkably increased the relative abundance of bacterial genera such as Arthrobacter, Nocardioides, unclassified Frankineae, and Bradyrhizobium, which were linked to nitrogen fixation. Therefore, the results of our study showed that T. harzianum ST02 had the potential to improve soil properties and enhance plant productivity in the saline soil.
Data availability statement
The data presented in the study are deposited in the CNGB Sequence Archive, accession number CNP0004730.
Author contributions
YLW: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. HY: Conceptualization, Data curation, Methodology, Writing – review & editing. JH: Data curation, Formal Analysis, Investigation, Methodology, Project administration, Software, Validation, Writing – review & editing. HL: Data curation, Investigation, Methodology, Resources, Writing – review & editing. ZZ: Conceptualization, Data curation, Formal Analysis, Methodology, Resources, Writing – review & editing. YZW: Conceptualization, Data curation, Investigation, Project administration, Software, Writing – review & editing. JL: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – review & editing. YZ: Conceptualization, Investigation, Methodology, Resources, Software, Supervision, Visualization, Writing – review & editing. KY: Data curation, Formal Analysis, Methodology, Writing – review & editing. HTY: Conceptualization, Formal Analysis, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – review & editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Provincial Key Research and Development Project (major scientific and technological innovation) from Shandong Province (Project ID: 2020CXGC010803), the Natural Science Foundation of Shandong Province (Project ID: ZR2023MC045), the International Cooperation Project of Science, Education and Industry Integration Innovation Pilot Project from Shandong Academy of Sciences (project ID: 2022GH011), the Independent training innovation team project from Jinan city (project ID: 2020GXRC003), and the major innovation projects of Qilu University of Technology (project ID: 2022JBZ02-05).
Acknowledgments
We gratefully acknowledge all the colleagues who have worked on this article and governments for their financial support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Almodares, A., Hadi, M. R. (2009). Production of bioethanol from sweet sorghum: a review. Afr. J. Agric. Res. 4, 772–780. doi: 10.5897/AJAR.9000567
Asad, S. A. (2022). Mechanisms of action and biocontrol potential of Trichoderma against fungal plant diseases -a review. Ecol. Complexity. 49, 100978. doi: 10.1016/j.ecocom.2021.100978
Bazhanov, D. P., Yang, K., Li, H., Li, C., Li, J., Chen, X., et al. (2017). Colonization of plant roots and enhanced atrazine degradation by a strain of Arthrobacter ureafaciens. Appl. Microbiol. Biotechnol. 101, 6809–6820. doi: 10.1007/s00253-017-8405-3
Bhattacharyya, D., Yu, S. M., Lee, Y. H. (2015). Volatile compounds from Alcaligenes faecalis JBCS1294 confer salt tolerance in Arabidopsis thaliana through the auxin and gibberellin pathways and differential modulation of gene expression in root and shoot tissues. Plant Growth Regul. 75 (1), 297–306. doi: 10.1007/s10725-014-9953-5
Calviño, M., Messing, J. (2012). Sweet sorghum as a model system for bioenergy crops. Curr. Opin. Biotechnol. 23 (3), 323–329. doi: 10.1016/j.copbio.2011.12.002
Cheng, H., Zhang, D., Huang, B., Song, Z., Ren, L., Hao, B., et al. (2020). Organic fertilizer improves soil fertility and restores the bacterial community after 1,3-dichloropropene fumigation. Sci. Total Environment. 738, 140345. doi: 10.1016/j.scitotenv.2020.140345
Contreras-Cornejo, H., Macias-Rodríguez, L., Alfaro-Cuevas, R., Lopez-Bucio, J. (2014). Trichoderma spp. improve growth of Arabidopsis seedlings under salt stress through enhanced root development, osmolite production, and Na+ elimination through root exudates. Mol. Plant-Microbe Interactions. 27 (6), 503–514. doi: 10.1094/MPMI-09-13-0265-R
Contreras-Cornejo, H., Macías-Rodríguez, L., Del-Val, E. K., Larsen, J. (2016). Ecological functions of Trichoderma spp. and their secondary metabolites in the rhizosphere: interactions with plants. FEMS Microbiol. Ecology. 92 (4), 36. doi: 10.1093/femsec/fiw036
Etesami, H., Maheshwari, D. K. (2018). Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: Action mechanisms and future prospects. Ecotoxicology Environ. Safety. 156, 225–246. doi: 10.1016/j.ecoenv.2018.03.013
Farhangi-Abriz, S., Torabian, S. (2017). Antioxidant enzyme and osmotic adjustment changes in been seedlings as affected by biochar under salt stress. Ecotoxicology Environ. Safety. 137, 64–70. doi: 10.1016/j.ecoenv.2016.11.029
Fu, J., Xiao, Y., Wang, Y., Liu, Z., Yang, K. (2019). Trichoderma affects the physiochemical characteristics and bacterial community composition of saline-alkaline maize rhizosphere soils in the cold region of Heilongjiang Province. Plant Soil. 436, 211–227. doi: 10.1007/s11104-018-03916-8
Gowik, U., Braeutigam, A., Weber, K. L., Weber, A. P., Westhoff, P. (2011). Evolution of C4 photosynthesis in the genus Flaveria: how many and which genes does it take to make C4? Plant Cell. 23 (6), 2087–2105. doi: 10.1105/tpc.111.086264
Guo, Y., Song, Y., Zheng, H., Zhang, Y., Sui, N. (2018). NADP-malate dehydrogenase of sweet sorghum improves salt tolerance of Arabidopsis thaliana. J. Agric. Food Chem. 66, 5992–6002. doi: 10.1021/acs.jafc
Gupta, S., Smith, P. M. C., Boughton, B. A., Rupasinghe, T. W. T., Natera, S. H. A., Roessner, U. (2021). Inoculation of barley with Trichoderma harzianum T-22 modifies lipids and metabolites to improve salt tolerance. J. Exp. Botany. 72 (20), 7229–7246. doi: 10.1093/jxb/erab335
Hassani, M. A., Durán, P., Hacquard, S. (2018). Microbial interactions within the plant holobiont. Microbiome. 6, 58. doi: 10.1186/s40168-018-0445-0
Imane, C., Nasreddine, E. O., Abdelaali, B., Naoual, E. M., Taoufiq, B., Cherki, G. (2021). Is the rhizosphere a source of applicable multi-beneficial microorganisms for plant enhancement? Saudi J. Biol. Sci. 29, 1246–1259. doi: 10.1016/j.sjbs.2021.09.032
Jiang, Y., Zhou, J., Zou, Y., Liu, D. (2004). Isolation and primary identification of a new nitrogen-fixation Arthrobacter strain. J. Cent. China Normal University. 38, 210–214. doi: 10.19603/j.cnki.1000-1190.2004.02.022
Kandeler, E., Deiglmayr, K., Tscherko, D., Bru, D., Philippot, L. (2006). Abundance of narG, nirS, nirK and nosZ genes of denitrifying bacteria during primary successions of a glacier foreland. Appl. Environ. Microbiol. 72, 5957–5962. doi: 10.1128/AEM.00439-06
Khomari, S., Davari, M. (2017). Trichoderma-Induced enhancement of soybean seedling performance in response to salt stress. J. Plant Physiol. Breeding. 7 (1), 27–39.
Kirchman, D. L. (2012). Marine archaea take a short cut in the nitrogen cycle. PNAS. 109 (44), 17732–17733. doi: 10.1073/pnas.1215654109
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2012). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1. doi: 10.1093/nar/gks808
Li, J., Cheng, B., Zhang, R., Li, W., Shi, X., Han, Y., et al. (2021). Nitrogen and phosphorus additions accelerate decomposition of slow carbon pool and lower total soil organic carbon pool in alpine meadows. Land Degradation Dev. 32 (4), 1761–1772. doi: 10.1002/ldr.3824
Li, H., Ruey, T., Wei, Y., Wang, Y., Hu, J., An, S., et al. (2022). Microbiomes across root compartments are shaped by inoculation with a fungal biological control agent. Appl. Soil Ecology. 170, 104230. doi: 10.1016/j.apsoil.2021.104230
Li, J., Wu, Y., Chen, K., Wang, Y., Hu, J., Wei, Y., et al. (2018). Trichoderma cyanodichotomus sp. nov., a new soil-inhabiting species with a potential for biological control. Can. J. Microbiol. 64 (12), 1020–1029. doi: 10.1139/cjm-2018-0224
Lozupone, C., Lladser, M. E., Knights, D., Stombaugh, J., Knight, R. (2010). UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172. doi: 10.1038/ismej.2010.133
Mastouri, F., Björkman, T., Harman, G. E. (2010). Seed treatment with Trichoderma harzianum alleviates biotic, abiotic, and physiological stresses in germinating seeds and seedlings. Phytopathology. 100 (11), 1213–1221. doi: 10.1094/PHYTO-03-10-0091
Matos, G., Rouws, L., Simes-Araújo, J. L., Baldani, J. I. (2021). Evolution and function of nitrogen fixation gene clusters in sugarcane associated Bradyrhizobium strains. Environ. Microbiol. 23 (10), 6148–6162. doi: 10.1111/1462-2920.15533
Olsen, S. R., Sommers, L. E. (1982). “Phosphorus,” in Methods of soil analysis part 2. Eds. Page, A. L., Miller, R. H., Keeney, D. R. (Madison, WI: ASASSSA), 403–427.
Ouyang, Y., Evans, S. E., Friesen, M. L., Tiemann, L. K. (2018). Effect of nitrogen fertilization on the abundance of nitrogen cycling genes in agricultural soils: a meta-analysis of field studies. Soil Biol. Biochem. 127, 71–78. doi: 10.1016/j.soilbio.2018.08.024
Pang, G., Cai, F., Li, R., Zhao, Z., Li, R., Gu, X., et al. (2017). Trichoderma-enriched organic fertilizer can mitigate microbiome degeneration of monocropped soil to maintain better plant growth. Plant Soil. 416, 181–192. doi: 10.1007/s11104-017-3178-0
Poly, F., Ranjard, L., Nazaret, S., Gourbiere, F., Monrozier, L. J. (2001). Comparison of nifH gene pools in soils and soil microenvironments with contrasting properties. Appl. Environ. Microbiol. 67, 2255–2262. doi: 10.1128/AEM.67.5.2255-2262.2001
Qin, Y., Druzhinina, I. S., Pan, X., Yuan, Zh. (2016). Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnol. Advances. 34 (7), 1245–1259. doi: 10.1016/j.bioteChadv.2016.08.005
Qin, S., Xing, K., Jiang, J., Xu, L., Li, W. (2011). Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic Actinobacteria. Appl. Microbiol. Biotechnol. 89, 457–473. doi: 10.1007/s00253-010-2923-6
Qiu, L., Kong, W., Zhu, H., Zhang, Q., Banerjee, S., Ishii, S., et al. (2022). Halophytes increase rhizosphere microbial diversity, network complexity and function in inland saline ecosystem. Sci. Total Environment. 831, 154944. doi: 10.1016/j.scitotenv.2022.154944
Rojas-Tapias, D., Moreno-Galván, A., Pardo-Díaz, S., Obando, M., Rivera, D., Bonilla, R. (2012). Effect of inoculation with plant growth-promoting bacteria (PGPB) on amelioration of saline stress in maize (Zea mays). Appl. Soil Ecology. 61, 264–272. doi: 10.1016/j.apsoil.2012.01.006
Rosier, A., Bishnoi, U., Lakshmanan, V., Sherrier, D. J., Bais, H. P. (2016). A perspective on inter-kingdom signaling in plant–beneficial microbe interactions. Plant Mol. Biol. 90, 537–548. doi: 10.1007/s11103-016-0433-3
Rudresh, D. L., Shivaprakash, M. K., Prasad, R. D. (2004). Effect of combined application of Rhizobium, phosphate solubilizing bacterium and Trichoderma spp. on growth, nutrient uptake and yield of chickpea (Cicer aritenium L.). Appl. Soil Ecol. 28 (2), 139–146. doi: 10.1016/j.apsoil.2004.07.005
Sellstedt, A., Richau, K. H. (2013). Aspects of nitrogen-fixing Actinobacteria, in particular free-living and symbiotic Frankia. FEMS Microbiol. Letters. 342, 179–186. doi: 10.1111/1574-6968.12116
Shafiee, R., Diver, P., Snow, J., Zhang, Q., Rickaby, R. (2021). Marine ammonia-oxidising archaea and bacteria occupy distinct iron and copper niches. ISME Commun. 1 (1). doi: 10.1038/s.43705-021-0001-7
Singh, B. N., Padmanabh, D., Sarma, B. K., Singh, G. S. (2018). Trichoderma asperellum T42 reprograms tobacco for enhanced nitrogen utilization efficiency and plant growth when fed with N nutrients. Front. Plant Science. 9. doi: 10.3389/fpls.2018.00163
Sui, N., Yang, Z., Liu, M., Wang, B. (2015). Identification and transcriptomic profiling of genes involved in increasing sugar content during salt stress in sweet sorghum leaves. BMC Genomics 16 (1). doi: 10.3389/fpls.2018.00007
Sun, R., Guo, X., Wang, D., Chu, H. (2015). Effects of long-term application of chemical and organic fertilizers on the abundance of microbial communities involved in the nitrogen cycle. Appl. Soil Ecology. 95, 171–178. doi: 10.1016/j.apsoil.2015.06.010
Tourna, M., Freitag, T. E., Nicol, G. W., Prosser, J. I. (2008). Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ. Microbiol. 10, 1357–1364. doi: 10.1111/j.1462-2920.2007.01563.x
Velmourougane, K., Prasanna, R., Singh, S., Chawla, G., Kumar, A., Saxena, A. K. (2017). Modulating rhizosphere colonisation, plant growth, soil nutrient availability and plant defense enzyme activity through Tichoderma viride-Azotobacter chroococcum, biofilm inoculation in chickpea. Plant Soil 421, 157–174. doi: 10.1007/s11104-017-3445-0
Visconti, D., Fiorentino, F., Cozzolino, E., Woo, S. L., Fagnano M. and Rouphael, Y. (2020). Can Trichoderma-based biostimulants optimize N use efficiency and stimulate growth of leafy vegetables in greenhouse intensive cropping systems? Agronomy. 10, 121–121. doi: 10.3390/agronomy10010121
Woo, S. L., Ruocco, M., Vinale, F., Nigro, M., Marra, R., Lombardi, N., et al. (2014). Trichoderma-based products and their widespread use in agriculture. Open Mycology J. 8 (1), 71–126. doi: 10.2174/1874437001408010071
Wu, J., Zhu, J., Zhang, D., Cheng, H., Hao, B., Cao, A., et al. (2022). Beneficial effect on the soil microenvironment of Trichoderma applied after fumigation for cucumber production. PloS One 17 (8), e0266347. doi: 10.1371/journal.pone.0266347
Xiong, Y., Gong, Y., Li, X., Chen, P., Ju, X., Zhang, C., et al. (2019). Enhancement of growth and salt tolerance of tomato seedlings by a natural halotolerant actinobacterium Glutamicibacter halophytocola KLBMP5180 isolated from a coastal halophyte. Plant Soil. 445, 307–322. doi: 10.1007/s11104-019-04310-8
Ye, L., Zhao, X., Bao, E., Li, J., Zhou, Z., Cao, K. (2020). Bio-organic fertilizer with reduced rates of chemical fertilization improves soil fertility and enhances tomato yield and quality. Sci. Rep. 10, 177. doi: 10.1038/s41598-019-56954-2
Yu, H., Zou, W., Chen, J., Chen, H., Yu, Z., Huang, J., et al. (2019). Biochar amendment improves crop production in problem soils: a review. J. Environ. Management. 232, 8–21. doi: 10.1016/j.jenvman.2018.10.117
Yuan, F., Leng, B., Wang, B. (2016). Progress in studying salt secretion from the salt glands in recretohalophytes: How do plants secrete salt? Front. Plant Science. 7. doi: 10.3389/fpls.2016.00977
Zhang, S., Gan, Y., Xu, B. (2019). Mechanisms of the IAA and ACC-deaminase producing strain of Trichoderma Longibrachiatum T6 in enhancing wheat seedling tolerance to NaCl stress. BMC Plant Biol. 19, 22. doi: 10.1186/s12870-018-1618-5
Zhang, R., Vivanco, J. M., Shen, Q. (2017b). The unseen rhizosphere root–soil–microbe interactions for crop production. Curr. Opin. Microbiol. 37, 8–14. doi: 10.1016/j.mib.2017.03.008
Zhang, H., Wang, R., Chen, S., Qi, G., He, Z., Zhao, X. (2017a). Microbial taxa and functional genes shift in degraded soil with bacterial wilt. Sci. Rep. 7, 39911. doi: 10.1038/srep39911
Zhao, Z., Hu, J., Chen, K., Wei, Y., Li, J. (2021). Effect of salt-tolerant Trichoderma ST02 on salt tolerance of tomato seed and seedling. Sci. Technol. Eng. 21 (7), 2632–2639. doi: 10.3969/j.issn.1671-1815.2021.07.010
Zhao, L., Zhang, Y. (2015). Effects of phosphate solubilization and phytohormone production of Trichoderma asperellum Q1 on promoting cucumber growth under salt stress. J. Integr. Agriculture. 14 (8), 1588–1597. doi: 10.1016/S2095-3119(14)60966-7
Zhao, W., Zhou, Q., Tian, Z., Cui, Y., Liang, Y., Wang, H. (2020). Apply biochar to ameliorate soda saline-alkali land, improve soil function and increase corn nutrient availability in the Songnen Plain. Sci. Total Environment. 722, 137428. doi: 10.1016/j.scitotenv.2020.137428
Keywords: Trichoderma harzianum, sweet sorghum, available nutrients, bacterial community, N-cycling genes
Citation: Wei Y, Yang H, Hu J, Li H, Zhao Z, Wu Y, Li J, Zhou Y, Yang K and Yang H (2023) Trichoderma harzianum inoculation promotes sweet sorghum growth in the saline soil by modulating rhizosphere available nutrients and bacterial community. Front. Plant Sci. 14:1258131. doi: 10.3389/fpls.2023.1258131
Received: 13 July 2023; Accepted: 21 August 2023;
Published: 12 September 2023.
Edited by:
Petronia Carillo, University of Campania Luigi Vanvitelli, ItalyReviewed by:
Giovanna Marta Fusco, University of Campania Luigi Vanvitelli, ItalyChiara Cirillo, University of Naples Federico II, Italy
Copyright © 2023 Wei, Yang, Hu, Li, Zhao, Wu, Li, Zhou, Yang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jishun Li, eWV3dTJAc2Rhcy5vcmc=; Yi Zhou, eWkuemhvdUBhZGVsYWlkZS5lZHUuYXU=
 Yanli Wei
Yanli Wei Han Yang
Han Yang Jindong Hu1
Jindong Hu1 Yuanzheng Wu
Yuanzheng Wu Yi Zhou
Yi Zhou