
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 29 September 2023
Sec. Plant Membrane Traffic and Transport
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1250588
This article is part of the Research Topic Intracellular Transition Metal Homeostasis in Plants and Algae View all 6 articles
Iron (Fe) is an essential micronutrient for plant growth and development due to its role in crucial processes such as photosynthesis and modulation of the redox state as an electron donor. While Fe is one of the five most abundant metals in the Earth’s crust, it is poorly accessible to plants in alkaline soils due to the formation of insoluble complexes. To limit Fe deficiency symptoms, plant have developed a highly sophisticated regulation network including Fe sensing, transcriptional regulation of Fe-deficiency responsive genes, and post-translational modifications of Fe transporters. In this mini-review, we detail how plants perceive intracellular Fe status and how they regulate transporters involved in Fe uptake through a complex cascade of transcription factors. We also describe the current knowledge about intracellular trafficking, including secretion to the plasma membrane, endocytosis, recycling, and degradation of the two main Fe transporters, IRON-REGULATED TRANSPORTER 1 (IRT1) and NATURAL RESISTANCE ASSOCIATED MACROPHAGE PROTEIN 1 (NRAMP1). Regulation of these transporters by their non-Fe substrates is discussed in relation to their functional role to avoid accumulation of these toxic metals during Fe limitation.
Iron (Fe) is a crucial micronutrient for most living organism due to its redox properties. Thanks to its ability to easily transfer electrons (gain or lose), Fe serves as co-factor in many vital chemical reactions such as cellular respiration, DNA synthesis and photosynthesis (Hänsch and Mendel, 2009; Palmer and Guerinot, 2009; Rengel et al., 2023). Consequently, perturbation of Fe homeostasis leads to several growth, developmental or health symptoms (Briat et al., 1995). According to the World Health Organization, Fe deficiency in humans is the most severe nutritional issue in the world, with anemia caused by dietary Fe deficiency affecting around 15% of the world’s population (World Health Organization, 2016; Safiri et al., 2021). In plants, Fe starvation is associated with both reduced growth and chlorosis (Briat et al., 1995). Although Fe is the second most abundant metal in the Earth’s crust, its poor solubility renders Fe limiting for plant growth (Rengel et al., 2023). In particular, alkaline and aerobic soil conditions favor Fe oxidation and precipitation of insoluble ferric (Fe3+) complexes. To counteract the Fe unavailability, plants have developed two different Fe acquisition strategies.
The first one, referred to as strategy I, is a reduction-based strategy used by all plant species except grasses to absorb Fe. It consists in a multistep process depending on a local rhizosphere acidification by root-released protons, a subsequent Fe reduction (Fe3+ to Fe2+) and the ferrous iron (Fe2+) uptake into root cells (Briat et al., 1995; Schmidt, 2003; Briat et al., 2015). In the model plant Arabidopsis thaliana, strategy I is driven by a complex of three proteins localized to the plasma membrane of root epidermal cells. First, the ARABIDOPSIS H+-ATPase 2 (AHA2) proton pump actively exports protons into the rhizosphere to reduce the pH, hence increasing Fe3+ solubility. Then, the FERRIC REDUCTION OXIDASE 2 (FRO2) reduces ferric iron into Fe2+ at the root surface. Finally, the high-affinity Fe transporter IRON-REGULATED TRANSPORTER 1 (IRT1) transports Fe2+ into root epidermal cells (Robinson et al., 1999; Vert et al., 2002; Santi and Schmidt, 2009). Upon Fe starvation, IRT1 is the major Fe transporter. In Fe-replete conditions however, the metal transporter NATURAL RESISTANCE-ASSOCIATED MACROPHAGE PROTEIN 1 (NRAMP1) cooperates with IRT1 and takes part in the Fe2+ uptake process as a low-affinity transporter (Castaings et al., 2016). Plants using strategy I also secrete secondary metabolites such as coumarins through the ATP-binding cassette (ABC) transporter ABCG37 (also named PLEIOTROPIC DRUG RESISTANCE 9: PDR9) to increase Fe mobility in the rhizosphere (Fourcroy et al., 2014). As coumarins play an important role in alkaline soils, it has been hypothesized that they might also form complexes with Fe3+ that could be directly taken up into the root to optimize Fe uptake (Robe et al., 2021). Graminaceous plants use a chelation-based strategy for Fe acquisition, named strategy II. They massively excrete phytosiderophores such as mugineic acids (MAs) (Takagi et al., 1984; Schmidt, 2003). MAs are secreted in the root vicinity through the TRANSPORTER OF MUGINEIC ACID 1 (TOM1) efflux transporter, to chelate and solubilize Fe3+ (Takagi et al., 1984; Nozoye et al., 2011; Nozoye et al., 2015). Fe-MA complexes are then taken up into root cells by the YELLOW STRIPE 1 (YS1)/YS1-LIKE (YSL) transporter (Inoue et al., 2009; Lee et al., 2009).
Mechanisms involved in strategy II will not be further discussed in this mini-review which focusses on the regulation of Fe uptake processes dedicated to strategy I. First, we will give a brief overview of Fe status sensing and the following steps regulating Fe homeostasis in plants, including the rapid transcriptional and post-transcriptional events occurring upon Fe deficiency. In a second part, we will summarize the current knowledge on the intracellular trafficking and post-translational regulation of the two main Arabidopsis Fe2+ transporters.
Fe homeostasis is controlled by a transcriptional cascade involving an intricate network of transcription factors (TFs) regulating the expression of genes encoding proteins involved in Fe uptake, distribution and storage (Figure 1). Fe depletion at the vicinity of plant roots induces the phosphorylation of the basic helix-loop-helix (bHLH) transcription factor bHLH121/UPSTREAM REGULATOR OF IRT1 (URI) and its root cellular relocalization from the central cylinder and the endodermis to the cortex and the epidermis (Gao et al., 2020a). Phosphorylated URI then forms heterodimers with members of the bHLH subgroup IVc (bHLH34, bHLH104, bHLH105/IAA-LEUCINE RESISTANT 3 (ILR3) and bHLH115) (Zhang et al., 2015; Li et al., 2016b; Liang et al., 2017; Kim et al., 2019; Tissot et al., 2019; Gao et al., 2020a; Lei et al., 2020). Cooperatively, these TFs acts as direct transcriptional activators of key genes involved in the Fe regulatory network, including four bHLH Ib genes (bHLH38, bHLH39, bHLH100, bHLH101), POPEYE (PYE/bHLH47), as well as BRUTUS (BTS) and BRUTUS LIKE 1 (BTSL1), which both encode E3 ubiquitin ligases (Kim et al., 2019; Gao et al., 2020a; Lei et al., 2020). In turn, members of the bHLH Ib subgroup associate with bHLH29/FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR (FIT) to form heterodimers that promote the expression of the Fe uptake machinery (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2008). PYE, a bHLH of the subgroup IVb, interacts with bHLH105/ILR3, to act as a transcriptional repressor of its own expression and of the expression of FERRITIN (FER) genes involved in Fe storage (Long et al., 2010; Samira et al., 2018; Tissot et al., 2019; Akmakjian et al., 2021), which are otherwise induced by bHLH121/URI (Gao et al., 2020b). PYE, likely in interaction with ILR3, also controls Fe redistribution, especially regarding its mobility, through the negative control imposed on the expression of NAS4 (NICOTIANAMINE SYNTHASE 4) involved in the synthesis of nicotianamine (NA), a Fe chelator important for intercellular Fe transport, and on the expression of ZINC-INDUCED FACILITATOR 1 (ZIF1) involved in the vacuolar sequestration of NA in roots (Long et al., 2010; Haydon et al., 2012; Schuler et al., 2012; Tissot et al., 2019). PYE was also found to repress the transcription of bHLH Ib genes (bHLH38, bHLH39, bHLH100, and bHLH101) by directly binding to their promoters, hence triggering the subsequent downregulation of IRT1 and FRO2 (Pu and Liang, 2023). Moreover, two transcription factors of the myeloblastosis (MYB) family, MYB10 and MYB72, cooperate to positively regulate NAS2 and NAS4 expression (Palmer et al., 2013), and the expression of both MYB is induced upon Fe deficiency under the control of heterodimeric complexes FIT/bHLH Ib and might also be dependent on bHLH121/URI (Palmer et al., 2013; Gao et al., 2020a; Lei et al., 2020).
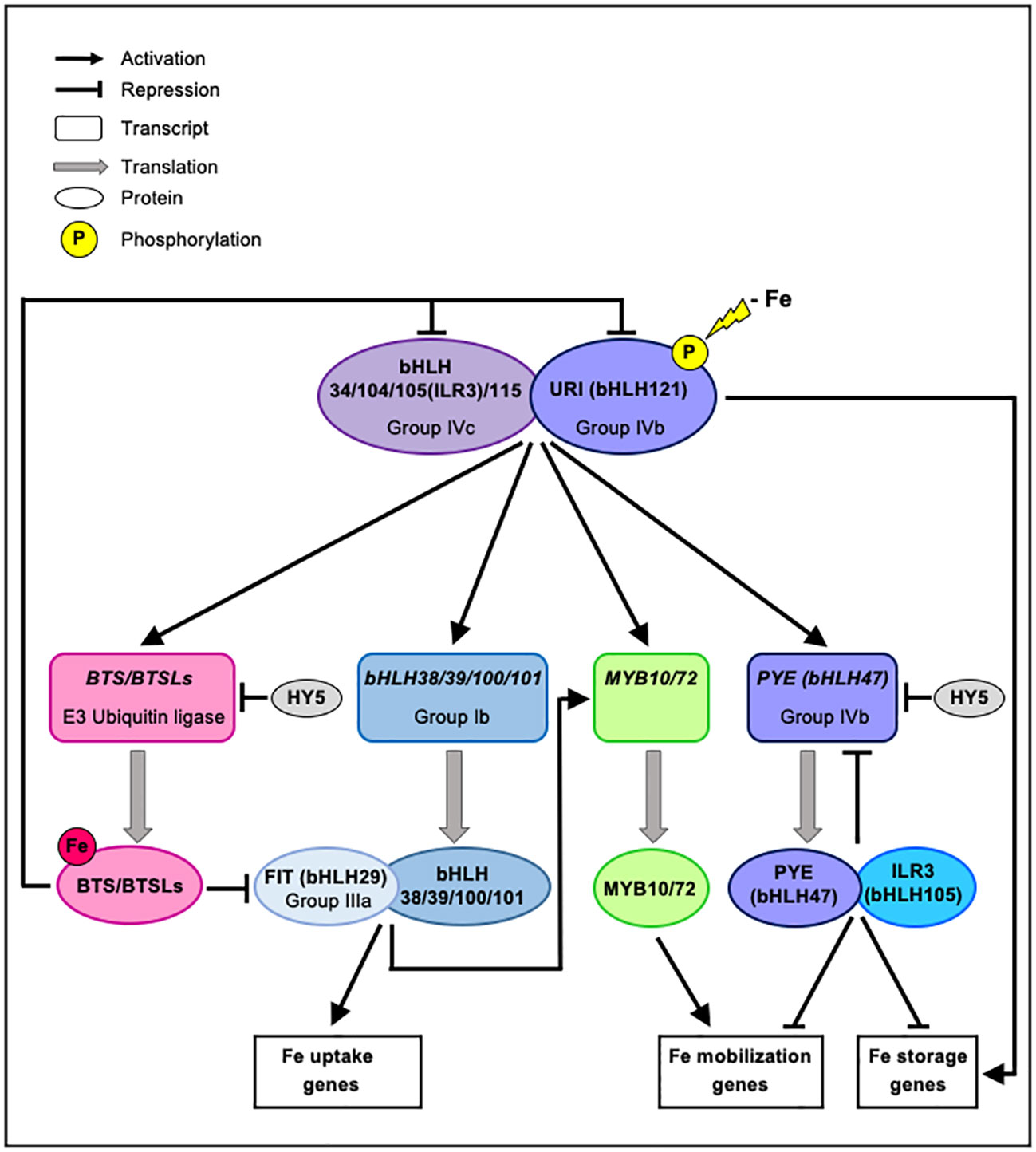
Figure 1 Model for transcriptional regulation of Fe homeostasis. Upon low-Fe conditions, URI (bHLH121) is phosphorylated and interacts with subgroup IVc bHLH transcription factors (bHLH34/104/105(ILR3)/115). These heterodimers activate the expression of several genes involved in the regulation of Fe homeostasis, including genes encoding the E3 ubiquitin ligases BTS/BTSLs, and the MYB10, MYB72, PYE (bHLH47) and subgroup Ib bHLH (bHLH38/39/100/101) transcription factors. In contrast, the bZIP transcription factor HY5 negatively regulate the expression of BTS and PYE. Members of the bHLH Ib subgroup form heterodimers with FIT (bHLH29) to promote the expression of MYB10/72 and of genes involved in Fe uptake. PYE (bHLH47), a subgroup IVb bHLH, interacts with ILR3 (bHLH105) to negatively regulates its own expression and also to act as a transcriptional repressor of genes involved in Fe storage which are otherwise induced by URI (bHLH121). PYE (bHLH47) with ILR3 (bHLH105) likely represses the expression of genes involved in Fe mobilization that are induced under the control of MYB10/72. PYE also negatively regulates the expression of bHLH Ib genes (not shown). When Fe becomes available, E3 ubiquitin ligases of the BTS/BTSLs family target bHLH transcription factors (bHLH29(FIT)/104/105(ILR3)/115/121(URI)) leading to their degradation via the 26S proteasome to turn off Fe-deficiency signaling and prevent Fe overload. For clarity, the involvement of IMA peptides in the transcriptional cascade controlling Fe homeostasis is not shown, but is discussed in the text.
Fine tuning of this complex transcriptional regulatory cascade may be achieved by the Fe-deficiency-dependent phosphorylation of bHLH121/URI and its subsequent phospho-dependent degradation by BTS (Kim et al., 2019). It is noteworthy, however, that BTS expression pattern is restrained to the stele (Selote et al., 2015; Rodríguez-Celma et al., 2019), where BTS would coexist with an unphosphorylated, hence undegradable bHLH121/URI protein (Kim et al., 2019). Furthermore, no interaction was uncovered between bHLH121/URI and BTS or BTSL proteins in yeast two-hybrid assays (Gao et al., 2020a). BTS and its homologs BTSL1 and BTSL2 are all E3 ubiquitin ligases that are able to bind Fe and to adjust their biological function accordingly to negatively regulate Fe homeostasis (Selote et al., 2015; Hindt et al., 2017; Kim et al., 2019). The negative control imposed by BTS and BTSLs results from the proteasomal degradation of their targets, among which are found several bHLHs especially important for Fe homeostasis (e.g. bHLH29/FIT, bHLH104, bHLH105/ILR3 and bHLH115) (Selote et al., 2015; Rodríguez-Celma et al., 2019). BTS and BTSL proteins all possess hemerythrin/HHE Fe-binding domains at their N-terminus (Hindt et al., 2017), and the BTS protein is stabilized in the absence of Fe bound to glutamic acid residues located within HHE domains (Selote et al., 2015). At the transcriptional level, BTS and BTSL genes are upregulated by Fe deficiency at least through the control of three bHLH transcription factors (bHLH105/ILR3, bHLH115 and bHLH121/URI) (Long et al., 2010; Liang et al., 2017; Rodríguez-Celma et al., 2019; Gao et al., 2020a). In contrast, ELONGATED HYPOCOTYL 5 (HY5), a light signaling basic leucine zipper (bZIP) transcription factor, has been identified as a negative regulator of BTS and PYE expression to modulate Fe-deficiency responses (Mankotia et al., 2023). It was recently shown that IRON MAN (IMA) peptides interact with and are ubiquitinated by BTS, which inhibits the degradation of bHLH105 and bHLH115 through binding competition (Li et al., 2021). In the same line of evidence, the interaction of BTSL1 with PYE and bHLHs of the subgroup IVc is attenuated by the peptide IMA1 (Lichtblau et al., 2022). Interestingly, mutation applied to the bHLH121/URI gene leads to an up-regulation of IMA1/2/3 and BTS genes under Fe deficiency (Lei et al., 2020).
Interestingly, crosstalks exist between Fe homeostasis and phytohormone signaling pathways. Indeed, FIT interacts with ETHYLENE INSENSITIVE 3 (EIN3) and ETHYLENE INSENSITIVE 3-LIKE 1 (EIL1) (Lingam et al., 2011). Two components of the jasmonic acid (JA) pathway, MYC2 and JAR1, negatively regulate the expression of FIT and bHLH Ib genes in response to JA (Cui et al., 2018). Moreover, FIT interactions with four IVa bHLHs (bHLH18, bHLH19, bHLH20 and bHLH25) trigger JA-mediated FIT degradation by the 26S proteasome (Cui et al., 2018). FIT, bHLH38 and bHLH39, as well as FRO2, IRT1, expression is also positively regulated by two key enzymes of the gibberellin (GA) biosynthetic pathway (gibberellin 3-oxidase 1 and 2, GA3ox1 and GA3ox2) (Matsuoka et al., 2014). DELLA proteins, which are negatively regulated by bioactive GA, associate with FIT, bHLH38 and bHLH39 to inhibit their activity (Wild et al., 2016).
FIT and Ib bHLH genes, hence their direct targets (at least IRT1 (Schwarz and Bauer, 2020) and putatively NRAMP1, as they are known to be co-regulated (Colangelo and Guerinot, 2004)) are positively regulated by a wide range of metal excess (zinc (Zn2+), cobalt (Co2+), nickel (Ni2+), cadmium (Cd2+) and manganese (Mn2+)) (Wu et al., 2012; Lešková et al., 2017). The effect of metal excess on the expression of aforementioned genes likely results from competition with Fe uptake as it was demonstrated for cadmium (Wu et al., 2012).
Upon Fe deficiency, many genes mentioned above undergo changes not only at the transcriptional level but also at the post-transcriptional level through modification of their splicing patterns (Lan et al., 2013; Li et al., 2013; Li et al., 2016a; Hsieh et al., 2022). Alternative splicing events occurring upon Fe deficiency might be regulated by many splicing factors, including members of the serine/arginine-rich (SR) family (Zhang et al., 2014; Dong et al., 2018; Fanara et al., 2022). Although an alternatively spliced isoform of IRT1 mRNA retaining an intron has been previously described (Li et al., 2013), no significant changes appear in the IRT1 splicing profile in a wild-type plant upon long- or short-term Fe deficiency (Fanara et al., 2022; Hsieh et al., 2022). Among the genes of the bHLH Ib subgroup, only bHLH100 and bHLH101 undergo differential intron retention (DIR) upon short-term Fe deficiency (Hsieh et al., 2022). While bHLH29/FIT intensively undergoes differential donor or acceptor (DDA) splice sites events (Hsieh et al., 2022), changes in the NRAMP1 splicing profile occur through both DIR (Lan et al., 2013; Li et al., 2013) and DDA (Lan et al., 2013; Li et al., 2016a). DDA events define new borders between exon and intron at the 5’ and 3’ ends of the intron, respectively, and might result in frameshifts and subsequent premature termination codons. Alternatively, if in-frame, DDA could modify the length of exonic regions (English et al., 2010). Alternative splicing therefore has the potential to partially modify important domain of a protein, hence modifying its interaction network, function, stability, or even its intracellular localization (English et al., 2010; Kalyna et al., 2012; Drechsel et al., 2013; Remy et al., 2013).
As previously mentioned, Fe2+ is taken up by the cooperation of two transporters, NRAMP1 and IRT1 in plants (Curie et al., 2000; Vert et al., 2002; Castaings et al., 2016). The low affinity Fe transporter NRAMP1 plays an important role in Fe-replete conditions to support Fe homeostasis. However, upon Fe deficiency, the high affinity Fe transporter IRT1 is rapidly and strongly induced to ensure efficient Fe uptake.
Membrane proteins are synthesized in the endoplasmic reticulum (ER) (High and Laird, 1997), and use well-characterized secretion signals and pathways to traffic in the cell to their destination (Peer, 2011; Chung et al., 2016). Exit from the ER requires the direct interaction with the coat protein complex II (COPII) using specific motifs, which allows their transport to the Golgi apparatus (Barlowe et al., 1994; Matheson et al., 2006; Lord et al., 2013). Then, membrane proteins follow the secretory pathway through the trans-Golgi network (TGN) and reach their final destination using various vesicles such as clathrin-coated vesicles or exocysts (Matheson et al., 2006; Peer, 2011; Chung et al., 2016). The mechanisms by which IRT1 and NRAMP1 reach the plasma membrane is still elusive and lacks experimental evidence. However, one of the first characterized ER export motifs, the diacidic motif (D/E)X(D/E), is present in both transporters (Hanton et al., 2005). NRAMP1 harbors a diacidic motif (DVD) in its cytoplasmic C-terminal part, as many others membrane proteins, and IRT1 possesses the diacidic motif (EDD) in its largest cytoplasmic loop. Besides, mutation of this motif in IRT1 from Malus xiaojinesis leads to its retention into the ER (Tan et al., 2018). In addition, two of the five COPII core components, SEC13a and SEC31b, were recently identified in the Arabidopsis IRT1 interactome (Matheson et al., 2006; Lord et al., 2013; Martìn-Barranco et al., 2020). Altogether, these findings suggest that IRT1 is exported from the ER to the Golgi apparatus using the COPII machinery. Then, during the post-Golgi trafficking, the CHOLINE TRANSPORTER-LIKE 1 (CTL1) plays an essential role to allow the delivery of NRAMP1 and probably IRT1 at the plasma membrane (Gao et al., 2017).
Interestingly, both transporters have low Fe specificity allowing the entry, in plant cells, of others divalent metals (hereafter named non-Fe metals), such as Zn2+, Mn2+, Co2+, and Cd2+ (Vert et al., 2002; Cailliatte et al., 2010; Barberon et al., 2011). In order to fine tune Fe acquisition and hence limit toxicity of their non-Fe metal substrates, both transporters are regulated at the post-translational level leading to a complex intracellular trafficking (Figure 2) (Barberon et al., 2011; Barberon et al., 2014; Agorio et al., 2017; Dubeaux et al., 2018; Castaings et al., 2021; Spielmann et al., 2022). Besides Fe transport, NRAMP1 is also essential for manganese uptake (Curie et al., 2000; Cailliatte et al., 2010; Castaings et al., 2016). Indeed, manganese availability impacts NRAMP1 subcellular localization. In control condition, NRAMP1 is shared between plasma membrane and endosomal vesicles of the trans-Golgi network/early endosome (TGN/EE) and the late endosome/multivesicular body (LE/MVB) (Agorio et al., 2017; Castaings et al., 2021). Increasing manganese availability gradually reduces the amount of NRAMP1 at the plasma membrane while enhancing its presence in the endosomal fraction (Castaings et al., 2021). This manganese-induced NRAMP1 endocytosis involves the clathrin-mediated endocytic pathway and occurs after phosphorylation of NRAMP1 (Castaings et al., 2021). The increase of intracellular manganese concentration seems to induce an intracellular transient calcium signal which is perceived by two calcium sensors, the CALCINEURIN-B-LIKE 1 (CBL1) and CALCINEURIN-B-LIKE 9 (CBL9). CBL1/9 interact with and activate the CBL-INTERACTING PROTEIN KINASE 23 (CIPK23) which phosphorylates NRAMP1 at serine 20 (S20), thereby inducing endocytosis of NRAMP1 to reduce its distribution on the plasma membrane and enhance plant tolerance to manganese toxicity. Indeed, in cipk23 and cbl1/cbl9 mutant backgrounds, NRAMP1 manganese-mediated endocytosis is abolished (Zhang et al., 2023b). Additionally, a nonphosphorylatable NRAMP1 mutant on S20 is blocked at the plasma membrane unlike the corresponding phosphomimic variant which is constitutively endocytosed, thus suggesting that phosphorylation is a pivotal step for NRAMP1 trafficking (Castaings et al., 2021). Finally, NRAMP1 undergoes vacuolar degradation through an unclear process depending on the PLECKSTRIN HOMOLOGY (PH) DOMAIN-CONTAINING PROTEIN 1 (PH1), but independent of manganese availability (Agorio et al., 2017; Castaings et al., 2021). Even if perturbation of NRAMP1 endocytosis was found to cause manganese toxicity (Castaings et al., 2021), most factors and mechanisms driving NRAMP1 trafficking and degradation remain to be identified and characterized.
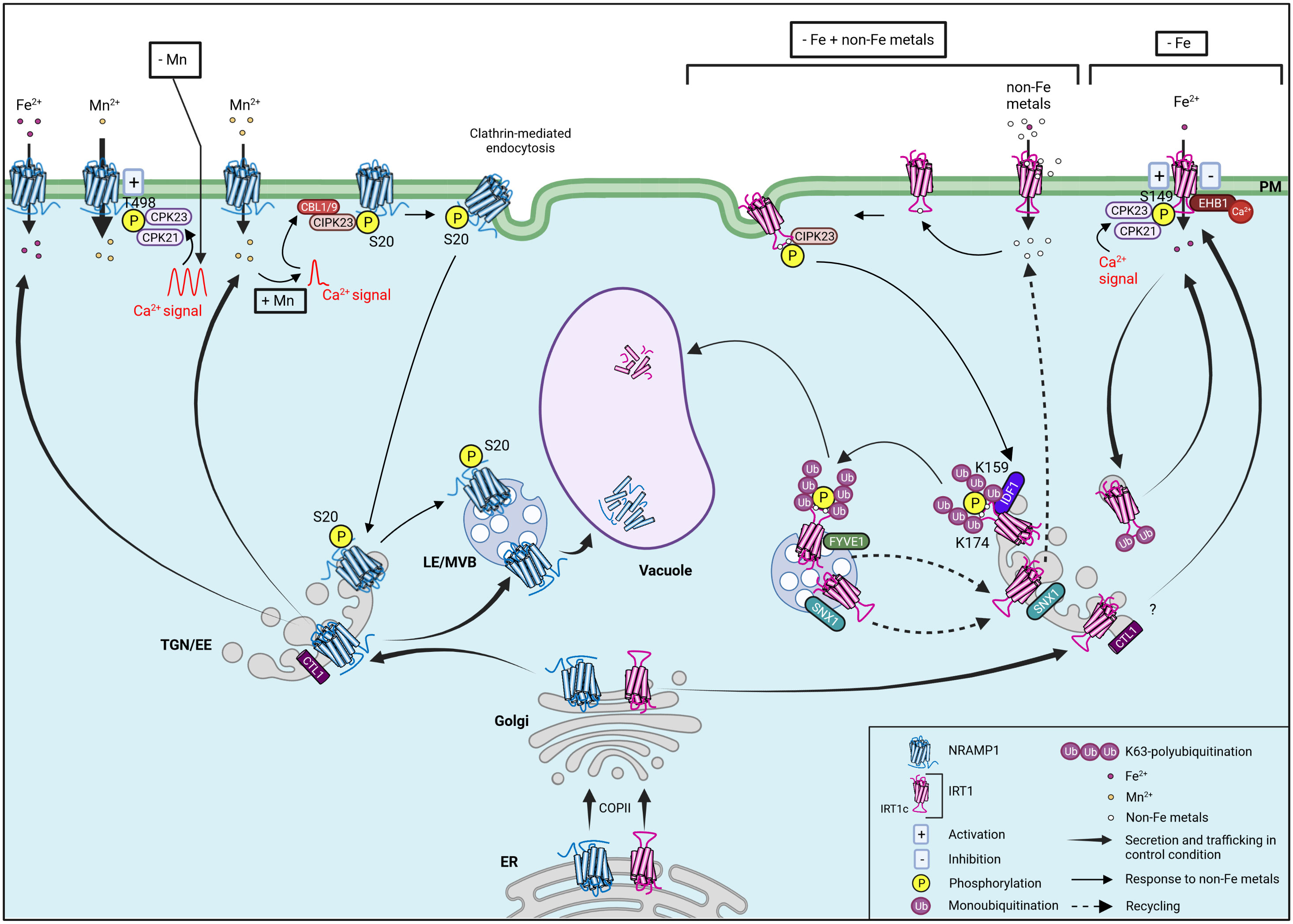
Figure 2 Model summarizing intracellular trafficking and post-translational regulation of NRAMP1 and IRT1. The two Fe transporters NRAMP1 and IRT1 are synthesized in the ER and exit this compartment by interacting with COPII. Then, the proteins follow the secretory pathway through the EE/TGN and reach the PM by a process involving CTL1 for NRAMP1 and probably also for IRT1. At the PM, the low affinity Fe2+ transporter NRAMP1 plays an important role in Fe-replete conditions, whereas the high affinity Fe2+ transporter IRT1 is induced upon Fe deficiency to ensure efficient Fe uptake. In control condition, NRAMP1 is found at the PM, the TGN/EE, the LE/MVB and also undergoes vacuolar degradation. NRAMP1 steady-state localization pattern is dependent on endocytosis (not shown). Besides Fe2+ transport, NRAMP1 is also essential for Mn2+ uptake. Mn2+ depletion (- Mn) induces intracellular Ca2+ oscillations decoded by CPK21 and CPK23 which phosphorylate NRAMP1 at T498 leading to NRAMP1 activation and increased Mn2+ uptake. The increase in intracellular Mn2+ concentration (+ Mn) induces a transient Ca2+ signal received by CBL1 and CBL9 located on the PM which activate CIPK23 to interact with and phosphorylate NRAMP1 at S20. This phosphorylation induces clathrin-mediated endocytosis of NRAMP1 reducing NRAMP1 distribution on the PM while enhancing its presence in the endosomal fraction to prevent Mn2+ toxicity. In addition to Fe, IRT1 also transports non-Fe metals (Zn2+, Mn2+, Co2+, Cd2+). In the combined absence of Fe and non-Fe metals, IRT1 is localized in the outer PM domain facing the rhizosphere in root epidermal cells. When Fe is limited and non Fe-metals are present at physiological concentration (- Fe), IRT1 is multimonoubiquinated and cycles between the PM and TGN/EE. In these conditions, the Fe transport activity of IRT1 is negatively regulated by Ca2+-promoted interaction of EHB1 with IRT1. In contrast, the phosphorylation of S149 in IRT1c by Ca2+-activated CPK21 and CPK23 stimulates IRT1 transport activity to promote Fe uptake. In Fe-depleted and non-Fe metal excess conditions (- Fe + non-Fe metals), IRT1 is removed from the PM. First, the high influx rate of non-Fe metals is directly sensed by IRT1 through non-Fe metal binding to histidine residues within IRT1c. This interaction triggers the recruitment of CIPK23 which phosphorylates S/T residues within IRT1c, creating a docking site for the E3 ubiquitin ligase IDF1, which decorates K159 and K174 in IRT1c with K63-linked polyubiquitin chains. IRT1 is then sorted toward LE/MVB for subsequent degradation in the vacuole. IRT1 may be recycled to earlier endocytic compartments or the PM through pathways involving SNX1 and FYVE1. IRT1 is also thought to be degraded via the 26S proteasome and autophagy pathways in response to Cd2+ stress (not shown). To simplify the scheme, only key proteins are shown (SEC13a, SEC31b, PH1, UBC35, UBC36, ESCRT complex, ATL31 and PATL2 are not represented). ER endoplasmic reticulum, TGN/EE trans-Golgi network/early endosome, LE/MVB late endosome/multivesicular body.
In the absence of Fe and non-Fe metals, IRT1 is mainly localized at the plasma membrane of root epidermal cells in a soil-facing polar fashion to optimize Fe uptake (Barberon et al., 2014; Dubeaux et al., 2018). When plants are exposed to non-Fe metals, IRT1 is subjected to a rapid and complex post-translational regulation to limit accumulation and toxicity of non-Fe metals. IRT1 acts as a transceptor, combining transporter and receptor activities, enabling sensing of intracellular metal status and regulation of its cell surface levels (Cointry and Vert, 2019). Increasing the concentration of non-Fe metals gradually removes IRT1 from the cell surface, allowing IRT1 to traffic through endosomal compartments on its way to vacuolar degradation (Dubeaux et al., 2018). Thanks to a histidine-rich motif located in the largest cytosolic loop (thereafter called IRT1c), IRT1 directly binds non-Fe metals (Dubeaux et al., 2018; Spielmann et al., 2022). This interaction triggers the recruitment of the CIPK23 kinase that phosphorylates IRT1c and thus creates a docking site for the E3 ubiquitin ligase IRON DEGRADATION FACTOR 1 (IDF1) (Shin et al., 2013; Dubeaux et al., 2018). Through the cooperation with two E2 UBIQUITIN CONJUGATING ENZYMES 35 and 36 (UBC35 and UBC36), IDF1 decorates two IRT1c lysine residues (K159 and K174) with K63-linked polyubiquitin chains (Shin et al., 2013; Dubeaux et al., 2018; Spielmann et al., 2022). IRT1 is then sorted toward late endosomes (LEs) and the vacuole for degradation, but the exact underlying mechanisms remain unclear (Barberon et al., 2011; Dubeaux et al., 2018; Spielmann et al., 2022). Polyubiquitinated IRT1 is probably recognized by the ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT (ESCRT) complex to be sent to the vacuole for degradation unless recycled (Dubeaux and Vert, 2017). In fact, two recycling mechanisms have been identified to exit the route to the vacuole (Barberon et al., 2014; Ivanov et al., 2014). One of these pathway preventing IRT1 degradation involves the SORTING NEXIN 1 (SNX1) protein which partly co-localise with IRT1 in TGN/EE and to a lesser extent in LE. A snx1 loss-of-function plant displays lower IRT1 levels than wild-type plants due to enhanced degradation rate. This suggests a role for SNX1 in the recycling of IRT1 (Ivanov et al., 2014). Secondly, IRT1 interacts with the phosphatidylinositol-3-phosphate-binding protein FYVE1 in LE, which alters its fate. Indeed, plants with enhanced expression of FYVE1 not only show an accumulation of IRT1 at the plasma membrane, but also display a loss of IRT1 lateral polarity (Barberon et al., 2014). Considering that FYVE1 is part of the ESCRT complex and mediates MVB sorting (Gao et al., 2014), the mechanisms by which FYVE1 overexpression reroutes IRT1 to the plasma membrane are still unclear.
IRT1 was also recently demonstrated to be degraded via the 26S proteasome and autophagy pathways. Indeed, IRT1 directly interacts with the plasma membrane-localized RING-type E3 ubiquitin ligase ARABIDOPSIS TOXICOS EN LEVADURA 31 (ATL31), which enhances IRT1 ubiquitination and leads to both proteasome- and autophagy-dependent degradation in response to cadmium stress. In addition, the WRKY33 transcription factor directly activates ATL31 expression in response to cadmium stress, suggesting a WRKY33-ATL31-IRT1 regulatory module involved in cadmium-specific plant tolerance (Zhang et al., 2023a). It remains to be determined how the proteasome may convey the degradation of a highly hydrophobic membrane protein like IRT1.
Finally, IRT1 post-translational regulatory mechanisms have been proposed to modulate Fe uptake. The peripheral membrane protein ENHANCED BENDING 1 (EHB1), a calcium-binding protein, acts as an Fe uptake inhibitor even during Fe starvation. EHB1 binds to IRT1c and thus interacts with IRT1 in a calcium-dependent manner, reducing Fe acquisition by a still unknown process. The IRT1-EHB1 interaction might represent a rapid mechanism to switch off Fe import once its optimal intracellular concentration has been reached, avoiding Fe overaccumulation and toxicity (Khan et al., 2019). The peripheral plasma membrane SEC14-Golgi dynamics (SEC14-GOLD) protein PATELLIN 2 (PATL2) also binds IRT1c to reduce membrane oxidative damage during Fe import via IRT1 (Hornbergs et al., 2023). In contrast, Fe deficiency leads to phosphorylation of serine 149 (S149) in IRT1c by the CALCIUM-DEPENDENT PROTEIN KINASE 21 and 23 (CPK21 and CPK23) calcium-regulated kinases to increase IRT1 transport activity to optimize Fe uptake (Wang et al., 2023). In response to manganese depletion, these two kinases also catalyze the phosphorylation of NRAMP1 at threonine 498 (T498), leading to NRAMP1 activation and optimal manganese uptake (Fu et al., 2022).
The control of cellular Fe relies on a complex pathway regulated at several levels and highly integrated. Fe deficiency quickly leads to an extensive cascade of transcriptional regulation mainly controlled by key members of the bHLH family and Fe-dependent protein degradation governed by BTS/BTSLs. Post-transcriptional events, including splicing, add a new level of complexity leading to a plethora of mRNAs rapidly responding to Fe deficiency and creating a diverse proteome controlling Fe uptake, as well as its intra- and intercellular Fe distribution. The trafficking and post-translational regulation of both NRAMP1 and IRT1 emerge as crucial for proper metal uptake and reveal some interesting parallels between the two transporters. Indeed, many post-translational modifications may affect the activity of these transporters, such as the phosphorylation of critical residues leading to their activation in limiting conditions or to their removal from the plasma membrane to avoid toxicity under metal stress. Finally, the polarity of membrane transporters is also inherently connected to their ability to drive directional Fe and metal uptake. Mechanisms controlling polar localization and maintenance of transporters therefore deserve more attention.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
This work was supported by research grants from the French National Research Agency (ANR-21-CE20-0046) and the French Laboratory of Excellence (project “TULIP” grant nos. ANR-10-LABX-41 and ANR-11-IDEX-0002-02) to GV.
We apologize to colleagues whose work could not be cited due to space limitation. We acknowledge the use of the BioRender software (https://biorender.com) for creating Figure 2.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Agorio, A., Giraudat, J., Bianchi, M. W., Marion, J., Espagne, C., Castaings, L., et al. (2017). Phosphatidylinositol 3-phosphate-binding protein AtPH1 controls the localization of the metal transporter NRAMP1 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114, E3354–E3363. doi: 10.1073/pnas.1702975114
Akmakjian, G. Z., Riaz, N., Guerinot, M. L. (2021). Photoprotection during iron deficiency is mediated by the bHLH transcription factors PYE and ILR3. Proc. Natl. Acad. Sci. U.S.A. 118, e2024918118. doi: 10.1073/pnas.2024918118
Barberon, M., Dubeaux, G., Kolb, C., Isono, E., Zelazny, E., Vert, G. (2014). Polarization of IRON-REGULATED TRANSPORTER 1 (IRT1) to the plant-soil interface plays crucial role in metal homeostasis. Proc. Natl. Acad. Sci. U.S.A. 111, 8293–8298. doi: 10.1073/pnas.1402262111
Barberon, M., Zelazny, E., Robert, S., Conéjéro, G., Curie, C., Friml, J., et al. (2011). Monoubiquitin-dependent endocytosis of the IRON-REGULATED TRANSPORTER 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. U.S.A. 108, E450–E458. doi: 10.1073/pnas.1100659108
Barlowe, C., Orci, L., Yeung, T., Hosobuchi, M., Hamamoto, S., Salama, N., et al. (1994). COPII: A membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77, 895–907. doi: 10.1016/0092-8674(94)90138-4
Briat, J.-F., Dubos, C., Gaymard, F. (2015). Iron nutrition, biomass production, and plant product quality. Trends Plant Sci. 20, 33–40. doi: 10.1016/j.tplants.2014.07.005
Briat, J.-F., Fobis-Loisy, I., Grignon, N., Lobréaux, S., Pascal, N., Savino, G., et al. (1995). Cellular and molecular aspects of iron metabolism in plants. Biol. Cell 84, 69–81. doi: 10.1016/0248-4900(96)81320-7
Cailliatte, R., Schikora, A., Briat, J.-F., Mari, S., Curie, C. (2010). High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell 22, 904–917. doi: 10.1105/tpc.109.073023
Castaings, L., Alcon, C., Kosuth, T., Correia, D., Curie, C. (2021). Manganese triggers phosphorylation-mediated endocytosis of the Arabidopsis metal transporter NRAMP1. Plant J. 106, 1328–1337. doi: 10.1111/tpj.15239
Castaings, L., Caquot, A., Loubet, S., Curie, C. (2016). The high-affinity metal transporters NRAMP1 and IRT1 team up to take up iron under sufficient metal provision. Sci. Rep. 6, 37222. doi: 10.1038/srep37222
Chung, K. P., Zeng, Y., Jiang, L. (2016). COPII paralogs in plants: functional redundancy or diversity? Trends Plant Sci. 21, 758–769. doi: 10.1016/j.tplants.2016.05.010
Cointry, V., Vert, G. (2019). The bifunctional transporter-receptor IRT1 at the heart of metal sensing and signalling. New Phytol. 223, 1173–1178. doi: 10.1111/nph.15826
Colangelo, E. P., Guerinot, M. L. (2004). The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16, 3400–3412. doi: 10.1105/tpc.104.024315
Cui, Y., Chen, C.-L., Cui, M., Zhou, W.-J., Wu, H.-L., Ling, H.-Q. (2018). Four IVa bHLH transcription factors are novel interactors of FIT and mediate JA inhibition of iron uptake in Arabidopsis. Mol. Plant 11, 1166–1183. doi: 10.1016/j.molp.2018.06.005
Curie, C., Alonso, J. M., Le Jean, M., Ecker, J. R., Briat, J.-F. (2000). Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem. J. 347, 749–755. doi: 10.1042/0264-6021:3470749
Dong, C., He, F., Berkowitz, O., Liu, J., Cao, P., Tang, M., et al. (2018). Alternative splicing plays a critical role in maintaining mineral nutrient homeostasis in rice (Oryza sativa). Plant Cell 30, 2267–2285. doi: 10.1105/tpc.18.00051
Drechsel, G., Kahles, A., Kesarwani, A. K., Stauffer, E., Behr, J., Drewe, P., et al. (2013). Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 25, 3726–3742. doi: 10.1105/tpc.113.115485
Dubeaux, G., Neveu, J., Zelazny, E., Vert, G. (2018). Metal sensing by the IRT1 transporter-receptor orchestrates its own degradation and plant metal nutrition. Mol. Cell 69, 953–964. doi: 10.1016/j.molcel.2018.02.009
Dubeaux, G., Vert, G. (2017). Zooming into plant ubiquitin-mediated endocytosis. Curr. Opin. Plant Biol. 40, 56–62. doi: 10.1016/j.pbi.2017.07.005
English, A. C., Patel, K. S., Loraine, A. E. (2010). Prevalence of alternative splicing choices in Arabidopsis thaliana. BMC Plant Biol. 10, 102. doi: 10.1186/1471-2229-10-102
Fanara, S., Schloesser, M., Hanikenne, M., Motte, P. (2022). Altered metal distribution in the sr45-1 Arabidopsis mutant causes developmental defects. Plant J. 110, 1332–1352. doi: 10.1111/tpj.15740
Fourcroy, P., Sisó-Terraza, P., Sudre, D., Savirón, M., Reyt, G., Gaymard, F., et al. (2014). Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol. 201, 155–167. doi: 10.1111/nph.12471
Fu, D., Zhang, Z., Wallrad, L., Wang, Z., Höller, S., Ju, C., et al. (2022). Ca2+-dependent phosphorylation of NRAMP1 by CPK21 and CPK23 facilitates manganese uptake and homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 119, e2204574119. doi: 10.1073/pnas.2204574119
Gao, Y.-Q., Chen, J.-G., Chen, Z.-R., An, D., Lv, Q.-Y., Han, M.-L., et al. (2017). A new vesicle trafficking regulator CTL1 plays a crucial role in ion homeostasis. PloS Biol. 15, e2002978. doi: 10.1371/journal.pbio.2002978
Gao, C., Luo, M., Zhao, Q., Yang, R., Cui, Y., Zeng, Y., et al. (2014). A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr. Biol. 24, 25562563. doi: 10.1016/j.cub.2014.09.014
Gao, F., Robe, K., Bettembourg, M., Navarro, N., Rofidal, V., Santoni, V., et al. (2020a). The transcription factor bHLH121 interacts with bHLH105 (ILR3) and its closest homologs to regulate iron homeostasis in Arabidopsis. Plant Cell 32, 508–524. doi: 10.1105/tpc.19.00541
Gao, F., Robe, K., Dubos, C. (2020b). Further insights into the role of bHLH121 in the regulation of iron homeostasis in Arabidopsis thaliana. Plant Signal. Behav. 15, 1795582. doi: 10.1080/15592324.2020.1795582
Hänsch, R., Mendel, R. R. (2009). Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr. Opin. Plant Biol. 12, 259–266. doi: 10.1016/j.pbi.2009.05.006
Hanton, S. L., Renna, L., Bortolotti, L. E., Chatre, L., Stefano, G., Brandizzi, F. (2005). Diacidic motifs influence the export of transmembrane proteins from the endoplasmic reticulum in plant cells. Plant Cell 17, 3081–3093. doi: 10.1105/tpc.105.034900
Haydon, M. J., Kawachi, M., Wirtz, M., Hillmer, S., Hell, R., Krämer, U. (2012). Vacuolar nicotianamine has critical and distinct roles under iron deficiency and for zinc sequestration in Arabidopsis. Plant Cell 24, 724–737. doi: 10.1105/tpc.111.095042
High, S., Laird, V. (1997). Membrane protein biosynthesis - all sewn up? Trends Cell Biol. 7206–210. doi: 10.1016/S0962-8924(97)01035-0
Hindt, M. N., Akmakjian, G. Z., Pivarski, K. L., Punshon, T., Baxter, I., Salt, D. E., et al. (2017). BRUTUS and its paralogs, BTS LIKE1 and BTS LIKE2, encode important negative regulators of the iron deficiency response in Arabidopsis thaliana. Metallomics 9, 876–890. doi: 10.1039/c7mt00152e
Hornbergs, J., Montag, K., Loschwitz, J., Mohr, I., Poschmann, G., Schnake, A., et al. (2023). SEC14-GOLD protein PATELLIN2 binds IRON-REGULATED TRANSPORTER1 linking root iron uptake to vitamin E. Plant Physiol. 192, 504–526. doi: 10.1093/plphys/kiac563
Hsieh, E.-J., Lin, W.-D., Schmidt, W. (2022). Genomically hardwired regulation of gene activity orchestrates cellular iron homeostasis in Arabidopsis. RNA Biol. 19, 143–161. doi: 10.1080/15476286.2021.2024024
Inoue, H., Kobayashi, T., Nozoye, T., Takahashi, M., Kakei, Y., Suzuki, K., et al. (2009). Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J. Biol. Chem. 284, 3470–3479. doi: 10.1074/jbc.M806042200
Ivanov, R., Brumbarova, T., Blum, A., Jantke, A.-M., Fink-Straube, C., Bauer, P. (2014). SORTING NEXIN1 is required for modulating the trafficking and stability of the Arabidopsis IRON-REGULATED TRANSPORTER1. Plant Cell 26, 1294–1307. doi: 10.1105/tpc.113.116244
Jakoby, M., Wang, H.-Y., Reidt, W., Weisshaar, B., Bauer, P. (2004). FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett. 577, 528–534. doi: 10.1016/j.febslet.2004.10.062
Kalyna, M., Simpson, C. G., Syed, N. H., Lewandowska, D., Marquez, Y., Kusenda, B., et al. (2012). Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 40, 2454–2469. doi: 10.1093/nar/gkr932
Khan, I., Gratz, R., Denezhkin, P., Schott-Verdugo, S. N., Angrand, K., Genders, L., et al. (2019). Calcium-promoted interaction between the C2-domain protein EHB1 and metal transporter IRT1 inhibits Arabidopsis iron acquisition. Plant Physiol. 180, 1564–1581. doi: 10.1104/pp.19.00163
Kim, S. A., LaCroix, I. S., Gerber, S. A., Guerinot, M. L. (2019). The iron deficiency response in Arabidopsis thaliana requires the phosphorylated transcription factor URI. Proc. Natl. Acad. Sci. U.S.A. 116, 24933–24942. doi: 10.1073/pnas.1916892116
Lan, P., Li, W., Lin, W.-D., Santi, S., Schmidt, W. (2013). Mapping gene activity of Arabidopsis root hairs. Genome Biol. 14, R67. doi: 10.1186/gb-2013-14-6-r67
Lee, S., Chiecko, J. C., Kim, S. A., Walker, E. L., Lee, Y., Guerinot, M. L., et al. (2009). Disruption of OsYSL15 leads to iron inefficiency in rice plants. Plant Physiol. 150, 786–800. doi: 10.1104/pp.109.135418
Lei, R., Li, Y., Cai, Y., Li, C., Pu, M., Lu, C., et al. (2020). bHLH121 functions as a direct link that facilitates the activation of FIT by bHLH IVc transcription factors for maintaining Fe homeostasis in Arabidopsis. Mol. Plant 13, 634–649. doi: 10.1016/j.molp.2020.01.006
Lešková, A., Giehl, R. F. H., Hartmann, A., Fargašová, A., von Wirén, N. (2017). Heavy metals induce iron deficiency responses at different hierarchic and regulatory levels. Plant Physiol. 174, 1648–1668. doi: 10.1104/pp.16.01916
Li, W., Lin, W.-D., Ray, P., Lan, P., Schmidt, W. (2013). Genome-wide detection of condition-sensitive alternative splicing in Arabidopsis roots. Plant Physiol. 162, 1750–1763. doi: 10.1104/pp.113.217778
Li, Y., Lu, C. K., Li, C. Y., Lei, R. H., Pu, M. N., Zhao, J. H., et al. (2021). IRON MAN interacts with BRUTUS to maintain iron homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 118, e2109063118. doi: 10.1073/pnas.2109063118
Li, S., Yamada, M., Han, X., Ohler, U., Benfey, P. N. (2016a). High-resolution expression map of the Arabidopsis root reveals alternative splicing and lincRNA regulation. Dev. Cell 39, 508–522. doi: 10.1016/j.devcel.2016.10.012
Li, X., Zhang, H., Ai, Q., Liang, G., Yu, D. (2016b). Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana. Plant Physiol. 170, 2478–2493. doi: 10.1104/pp.15.01827
Liang, G., Zhang, H., Li, X., Ai, Q., Yu, D. (2017). bHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis thaliana. J. Exp. Bot. 68, 1743–1755. doi: 10.1093/jxb/erx043
Lichtblau, D. M., Schwarz, B., Baby, D., Endres, C., Sieberg, C., Bauer, P. (2022). The iron deficiency-regulated small protein effector FEP3/IRON MAN1 modulates interaction of BRUTUS-LIKE1 with bHLH Subgroup IVc and POPEYE transcription factors. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.930049
Lingam, S., Mohrbacher, J., Brumbarova, T., Potuschak, T., Fink-Straube, C., Blondet, E., et al. (2011). Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 23, 1815–1829. doi: 10.1105/tpc.111.084715
Long, T. A., Tsukagoshi, H., Busch, W., Lahner, B., Salt, D. E., Benfey, P. N. (2010). The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22, 2219–2236. doi: 10.1105/tpc.110.074096
Lord, C., Ferro-Novick, S., Miller, E. A. (2013). The highly conserved COPII coat complex sorts cargo from the endoplasmic reticulum and targets it to the Golgi. Cold Spring Harb. Perspect. Biol. 5, a013367. doi: 10.1101/cshperspect.a013367
Mankotia, S., Singh, D., Monika, K., Kalra, M., Meena, H., Meena, V., et al. (2023). ELONGATED HYPOCOTYL 5 regulates BRUTUS and affects iron acquisition and homeostasis in Arabidopsis thaliana. Plant J. 114, 1267–1284. doi: 10.1111/tpj.16191
Martìn-Barranco, A., Spielmann, J., Dubeaux, G., Vert, G., Zelazny, E. (2020). Dynamic control of the high-affinity iron uptake complex in root epidermal cells. Plant Physiol. 184, 1236–1250. doi: 10.1104/pp.20.00234
Matheson, L. A., Hanton, S. L., Brandizzi, F. (2006). Traffic between the plant endoplasmic reticulum and Golgi apparatus: to the Golgi and beyond. Curr. Opin. Plant Biol. 9, 601–609. doi: 10.1016/j.pbi.2006.09.016
Matsuoka, K., Furukawa, J., Bidadi, H., Asahina, M., Yamaguchi, S., Satoh, S. (2014). Gibberellin-induced expression of Fe uptake-related genes in Arabidopsis. Plant Cell Physiol. 55, 87–98. doi: 10.1093/pcp/pct160
Nozoye, T., Nagasaka, S., Kobayashi, T., Sato, Y., Uozumi, N., Nakanishi, H., et al. (2015). The phytosiderophore efflux transporter TOM2 is involved in metal transport in rice. J. Biol. Chem. 290, 27688–27699. doi: 10.1074/jbc.M114.635193
Nozoye, T., Nagasaka, S., Kobayashi, T., Takahashi, M., Sato, Y., Sato, Y., et al. (2011). Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J. Biol. Chem. 286, 5446–5454. doi: 10.1074/jbc.M110.180026
Palmer, C. M., Guerinot, M. L. (2009). Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nat. Chem. Biol. 5, 333–340. doi: 10.1038/nchembio.166
Palmer, C. M., Hindt, M. N., Schmidt, H., Clemens, S., Guerinot, M. L. (2013). MYB10 and MYB72 are required for growth under iron-limiting conditions. PloS Genet. 9, e1003953. doi: 10.1371/journal.pgen.1003953
Peer, W. A. (2011). “Plasma Membrane Protein Trafficking,” in The Plant Plasma Membrane. Plant Cell Monographs, vol. 19 . Eds. Murphy, A., Schulz, B., Peer, W. (Berlin, Heidelberg: Springer), 31–56. doi: 10.1007/978-3-642-13431-9_2
Pu, M. N., Liang, G. (2023). The transcription factor POPEYE negatively regulates the expression of bHLH Ib genes to maintain iron homeostasis. J. Exp. Bot. 74, 2754–2767. doi: 10.1093/jxb/erad057
Remy, E., Cabrito, T. R., Baster, P., Batista, R. A., Teixeira, M. C., Friml, J., et al. (2013). A major facilitator superfamily transporter plays a dual role in polar auxin transport and drought stress tolerance in Arabidopsis. Plant Cell 25, 901–926. doi: 10.1105/tpc.113.110353
Rengel, Z., Cakmak, I., White, P. J. (2023). Marschner’s mineral nutrition of higher plants. Elsevier/Academic Press. doi: 10.1016/C2019-0-00491-8
Robe, K., Izquierdo, E., Vignols, F., Rouached, H., Dubos, C. (2021). The coumarins: secondary metabolites playing a primary role in plant nutrition and health. Trends Plant Sci. 26, 248–259. doi: 10.1016/j.tplants.2020.10.008
Robinson, N. J., Procter, C. M., Connolly, E. L., Guerinot, M. L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397, 694–697. doi: 10.1038/17800
Rodríguez-Celma, J., Connorton, J. M., Kruse, I., Green, R. T., Franceschetti, M., Chen, Y.-T., et al. (2019). Arabidopsis BRUTUS-LIKE E3 ligases negatively regulate iron uptake by targeting transcription factor FIT for recycling. Proc. Natl. Acad. Sci. U.S.A. 116, 17584–17591. doi: 10.1073/pnas.1907971116
Safiri, S., Kolahi, A.-A., Noori, M., Nejadghaderi, S. A., Karamzad, N., Bragazzi, N. L., et al. (2021). Burden of anemia and its underlying causes in 204 countries and territories 1990-2019: results from the global burden of disease study 2019. J. Hematol. Oncol. 14, 185. doi: 10.1186/s13045-021-01202-2
Samira, R., Li, B., Kliebenstein, D., Li, C., Davis, E., Gillikin, J. W., et al. (2018). The bHLH transcription factor ILR3 modulates multiple stress responses in Arabidopsis. Plant Mol. Biol. 97, 297–309. doi: 10.1007/s11103-018-0735-8
Santi, S., Schmidt, W. (2009). Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 183, 1072–1084. doi: 10.1111/j.1469-8137.2009.02908.x
Schmidt, W. (2003). Iron solutions: acquisition strategies and signaling pathways in plants. Trends Plant Sci. 8, 188–193. doi: 10.1016/S1360-1385(03)00048-7
Schuler, M., Rellán-Álvarez, R., Fink-Straube, C., Abadía, J., Bauer, P. (2012). Nicotianamine functions in the phloem-based transport of iron to sink organs, in pollen development and pollen tube growth in Arabidopsis. Plant Cell 24, 2380–2400. doi: 10.1105/tpc.112.099077
Schwarz, B., Bauer, P. (2020). FIT, a regulatory hub for iron deficiency and stress signaling in roots, and FIT-dependent and -independent gene signatures. J. Exp. Bot. 71, 1694–1705. doi: 10.1093/jxb/eraa012
Selote, D., Samira, R., Matthiadis, A., Gillikin, J. W., Long, T. A. (2015). Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiol. 167, 273–286. doi: 10.1104/pp.114.250837
Shin, L.-J., Lo, J.-C., Chen, G.-H., Callis, J., Fu, H., Yeh, K.-C. (2013). IRT1 DEGRADATION FACTOR1, a RING E3 ubiquitin ligase, regulates the degradation of IRON-REGULATED TRANSPORTER1 in Arabidopsis. Plant Cell 25, 3039–3051. doi: 10.1105/tpc.113.115212
Spielmann, J., Cointry, V., Devime, F., Ravanel, S., Neveu, J., Vert, G. (2022). Differential metal sensing and metal-dependent degradation of the broad spectrum root metal transporter IRT1. Plant J. 112, 1252–1265. doi: 10.1111/tpj.16010
Takagi, S.-I., Nomoto, K., Takemoto, T. (1984). Physiological aspect of mugineic acid, a possible phytosiderophore of graminaceous plants. J. Plant Nutr. 7, 469–477. doi: 10.1080/01904168409363213
Tan, S., Zhang, P., Xiao, W., Feng, B., Chen, L.-Y., Li, S., et al. (2018). TMD1 domain and CRAC motif determine the association and disassociation of MxIRT1 with detergent-resistant membranes. Traffic 19, 122–137. doi: 10.1111/tra.12540
Tissot, N., Robe, K., Gao, F., Grant-Grant, S., Boucherez, J., Bellegarde, F., et al. (2019). Transcriptional integration of the responses to iron availability in Arabidopsis by the bHLH factor ILR3. New Phytol. 223, 1433–1446. doi: 10.1111/nph.15753
Vert, G., Grotz, N., Dédaldéchamp, F., Gaymard, F., Guerinot, M. L., Briat, J.-F., et al. (2002). IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14, 1223–1233. doi: 10.1105/tpc.001388
Wang, Z., Zhang, Y., Liu, Y., Fu, D., You, Z., Huang, P., et al. (2023). Calcium-dependent protein kinases CPK21 and CPK23 phosphorylate and activate the iron-regulated transporter IRT1 to regulate iron deficiency in Arabidopsis. Sci. China Life Sci. doi: 10.1007/s11427-022-2330-4
Wild, M., Davière, J. M., Regnault, T., Sakvarelidze-Achard, L., Carrera, E., Lopez Diaz, I., et al. (2016). Tissue-specific regulation of gibberellin signaling fine-tunes Arabidopsis iron-deficiency responses. Dev. Cell 37, 190–200. doi: 10.1016/j.devcel.2016.03.022
World Health Organization (2016) Guideline: daily iron supplementation in infants and children. Available at: https://www.who.int/publications/i/item/9789241549523.
Wu, H., Chen, C., Du, J., Liu, H., Cui, Y., Zhang, Y., et al. (2012). Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol. 158, 790–800. doi: 10.1104/pp.111.190983
Yuan, Y., Wu, H., Wang, N., Li, J., Zhao, W., Du, J., et al. (2008). FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 18, 385–397. doi: 10.1038/cr.2008.26
Zhang, W., Du, B., Liu, D., Qi, X. (2014). Splicing factor SR34b mutation reduces cadmium tolerance in Arabidopsis by regulating iron-regulated transporter 1 gene. Biochem. Biophys. Res. Commun. 455, 312–317. doi: 10.1016/j.bbrc.2014.11.017
Zhang, Z., Fu, D., Xie, D., Wang, Z., Zhao, Y., Ma, X., et al. (2023b). CBL1/9-CIPK23-NRAMP1 axis regulates manganese toxicity. New Phytol. 239, 660–672. doi: 10.1111/nph.18968
Zhang, J., Liu, B., Li, M., Feng, D., Jin, H., Wang, P., et al. (2015). The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell 27, 787–805. doi: 10.1105/tpc.114.132704
Keywords: iron, Fe, iron homeostasis, iron uptake, transcriptional and post-translational regulations, transcription factors, iron transporters, intracellular trafficking
Citation: Spielmann J, Fanara S, Cotelle V and Vert G (2023) Multilayered regulation of iron homeostasis in Arabidopsis. Front. Plant Sci. 14:1250588. doi: 10.3389/fpls.2023.1250588
Received: 30 June 2023; Accepted: 07 September 2023;
Published: 29 September 2023.
Edited by:
Marie-Theres Hauser, University of Natural Resources and Life Sciences Vienna, AustriaReviewed by:
Jeeyon Jeong, Amherst College, United StatesCopyright © 2023 Spielmann, Fanara, Cotelle and Vert. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Valérie Cotelle, dmFsZXJpZS5jb3RlbGxlQHVuaXYtdGxzZTMuZnI=; Grégory Vert, Z3JlZ29yeS52ZXJ0QHVuaXYtdGxzZTMuZnI=
†These authors have contributed equally to this work and share first authorship
‡These authors have contributed equally to this work and share last authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.