
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 23 August 2023
Sec. Plant Biotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1249760
 Hengzuo Xiang1,2,3,4,5†
Hengzuo Xiang1,2,3,4,5† Sida Meng1,2,3,4,5†
Sida Meng1,2,3,4,5† Yunzhu Ye1,2,3,4,5
Yunzhu Ye1,2,3,4,5 Leilei Han1,2,3,4,5
Leilei Han1,2,3,4,5 Yi He2,3
Yi He2,3 Yiqing Cui2,3
Yiqing Cui2,3 Changhua Tan1,2,3,4,5
Changhua Tan1,2,3,4,5 Jian Ma1,2,3,4,5
Jian Ma1,2,3,4,5 Mingfang Qi1,2,3,4,5*
Mingfang Qi1,2,3,4,5* Tianlai Li1,2,3,4,5*
Tianlai Li1,2,3,4,5*Malformed tomato fruit with multiple locules is a common physiological disorder that significantly affects the quality of tomatoes. Research has shown that the occurrence of malformed fruit in tomatoes is closely linked to the number of locules, and two key QTLs, lc and fas, are involved in controlling this trait. It has been observed that lc has a relatively weaker effect on increasing locule number, which is associated with two SNPs in the CArG repressor element downstream of the SlWUS. However, the precise molecular mechanism underlying lc is not yet fully understood. In this study, we investigated the role of lc in tomato locule development. We found that the number of floral organs and fruit locules significantly increased in tomato lc knockout mutants. Additionally, these mutants showed higher expression levels of the SlWUS during carpel formation. Through cDNA library construction and yeast one-hybrid screening, we identified the MADS-box transcription factor SlSEP3, which was found to bind to lc. Furthermore, we observed an increase in floral organs and fruit locules similar to the lcCR plant on SlSEP3 silencing plants. However, it should be noted that the lc site is located after the 3′ untranslated region (UTR) of SlWUS in the tomato genome. As a result, SlSEP3 may not be able to exert regulatory functions on the promoter of the gene like other transcription factors. In the yeast two-hybrid assay, we found that several histone deacetylases (SlHDA1, SlHDA3, SlHDA4, SlHDA5, SlHDA6, SlHDA7, and SlHDA8) can interact with SlSEP3. This indicated that SlSEP3 can recruit these proteins to repress nucleosome relaxation, thereby inhibiting SlWUS transcription and affecting the number of locules in tomato fruit. Therefore, our findings reveal a new mechanism for lc playing a significant role in the genetic pathway regulating tomato locule development.
Fruit size and shape are crucial agronomic traits that have greatly influenced the domestication and improvement of tomato varieties. Compared with wild tomatoes, such as Solanum pimpinellifolium, the number of floral organs and fruit locules in cultivated tomatoes (Solanum lycopersicum) increases by two to five times, or even more, which directly results in the incidence of malformed fruits with multiple locules increasing with fruit size (Lippman and Tanksley, 2001; Tanksley, 2004). In addition to environmental factors such as light, water, and nutrients, the occurrence of malformed fruit is also affected by genetic factors. Fasciated (fas) and locule number (lc) are two key QTLs that control tomato locule number and fruit size (Tanksley, 2004; van der Knaap et al., 2014). The development of tomato locules is closely associated with the differentiation of stem cells in the shoot apical meristem (SAM) and floral meristem (FM).
The mutation of CLAVATA3 (CLV3) and its ligands in the CLV-WUS pathway can lead to abnormal plant development in Arabidopsis and tomatoes. These mutations result in excessive proliferation of stem cells, expansion of the meristem, and the production of additional organs such as flowers and fruits (Xu et al., 2015; Somssich et al., 2016). SlCLV3, which is identified as a candidate gene for fas, regulates the differentiation of tomato stem cells through the CLV-WUS pathway (Somssich et al., 2016; Chu et al., 2019). The phenotype of the slclv3 mutant is similar to that of fas, but most slclv3pro lines after editing the cis-regulatory elements (CRE) of the SlCLV3 promoter show more locules than fas, with some lines reaching 15–16 locules (Wang et al., 2021). In the absence of SlCLV3, SlCLE9 initiates a weak and partial compensatory effect. The phenotype of slcle9 is similar to the wild type, but the slcle9-slclv3 double-mutant shows a more severe phenotype than the slclv3 mutant (Rodríguez-Leal et al., 2019).
The inversion at fas results in a partial loss of function of SlCLV3, while two SNP mutations at lc lead to a gain of function of SlWUS (Huang and van der Knaap, 2011; Xu et al., 2015). Compared with fas, lc has a weaker effect on increasing locule number, which is associated with two SNPs in the CArG repressor element (similar to CRE) downstream of SlWUS (Muños et al., 2011; Somssich et al., 2016). SlWUS plays a direct role in regulating locule numbers. Its expression is increased after mutation of the CArG repressor element, suggesting that lc affects tomato locule number through SlWUS (Li et al., 2017). SlWUS is highly expressed in shoot apical and young flower buds, and gene silencing results in significantly smaller flowers and fruits with fewer locules (van der Knaap et al., 2014; Li et al., 2017). Furthermore, the regulatory factors in the plant hormone pathway that interact with SlWUS can also have a significant impact on tomato locule number (Su et al., 2022). In homozygous slwus mutants, SAM activity is terminated, and the transition to reproductive growth is disrupted, resulting in the absence of locules. Interestingly, when the CREs of the SlWUS promoter are edited, the floral organs and fruit shape of slwuspro mutants develop normally. However, only the slwuspro-6 mutant, which carries an inserted 223-bp fragment and an inverted 554-bp fragment of CRE, shows a phenotype similar to lc (Wang et al., 2021). This suggests that the role of lc in regulating tomato locule number is primarily mediated through WUS in the CLV-WUS regulatory module. However, it remains unknown whether lc has other regulatory pathways besides SlWUS through CRE elements.
In summary, the occurrence of malformed tomato fruit has a significant impact on yield and quality, which are primarily closely related to locule number. The regulation of locule number in tomatoes is influenced by two key factors: lc and fas. While the role of fas in this process has been well-established through the CLV-WUS module, the mechanism of how lc participates in tomato locule numbers remains unclear. In this study, we utilized CRISPR/Cas9 technology to generate a tomato lc knockout mutant. The homozygous mutant, which had a normal fas and a mutated lc, exhibited an increase in floral organs and fruit locules. Additionally, SlWUS expression, which is known to be associated with locule development, was significantly increased in the CR-lc#11 mutant. It indicated that lc affected SlWUS by some unknown pathway. Therefore, exploring the regulatory network between lc and SlWUS will further improve the molecular mechanism of tomato locule development.
Tomato AC (S. lycopersicum cultivar “Ailsa Craig”) with two locules was used as material for constructing lcCR mutants, lcCR T1 generation was grown in a greenhouse (16 h of light at 25°C, 8 h of dark at 15°C), and conventional cultivation management was carried out.
Young leaves from the lcCR T1 generation were sampled for target detection. The shoot apicals from both lcCR T1 generation and AC were taken during the carpellary formation stage for qRT-PCR analysis; three biological replicates were set up for each sample. The shoot apical from AC were taken at different stages of flower bud differentiation, including early flower bud differentiation (S1), middle flower bud differentiation (sepal and petal formation stage (S2)), and late flower bud differentiation stage (carpel formation stage (S3)). Every 15–20 shoot apical samples were collected for library construction and qRT-PCR analysis. Both samples were stored in an ultra-low-temperature refrigerator to be determined.
One single guide RNA (sgRNA) was designed to target lc for obtaining genome-edited mutants. The sgRNA sequence is listed in the Supplementary Table. The U6-lc-sgRNA was cloned into CRISPR/Cas9 binary vector pCBC-DT1T2-tomatoU6 to generate pCBC-DT1T2-tomatoU6-lc. This construct was then transformed into competent cells of Agrobacterium (Agrobacterium tumefaciens) strain LBA4404 using electroporation. Positive colonies obtained were used to transform the wild-type tomato AC through leaf disc cocultivation. Each primary transformant (T0) was genotyped by sequencing PCR products amplified from the relevant target region.
The plant genomic DNA extraction kit (Tiangen, Beijing, China) was used to extract genomic DNA, following the manufacturer’s instructions (Code No. DP305, Tiangen). The extracted products were sequenced and compared with Cas9/gRNA using DNAMAN9.0 to determine whether the target was mutated and the specific mutation mode. Primers used are listed in the Supplementary Table.
lcCR T1 generation mutants and wild-type plants were selected for 10 strains, respectively, and the number of sepals, stamens, petals, and locules in the first inflorescence flowers and fruit were investigated. Additionally, the number of sepals, petals, and fruit locules of flowers in the first and second inflorescence for SlSEP3 silenced strains and wild-type plants were investigated. Data statistics and significant differences were calculated and analyzed by Excel software.
Total RNA from shoot apical of lcCR mutants and AC was extracted using TRIzol reagent (Invitrogen, Carlsbad, California, USA). The cDNA was synthesized using PrimeScript RT Master Mix (TaKaRa, Kusatsu, Shiga, Japan) and diluted fourfold in ddH2O. Gene-specific primers were designed using the National Center for Biotechnology Information (NCBI) website (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi). Real-time quantitative analysis was performed using TB Green® Premix Ex Taq II (TaKaRa, Japan). The 20-μL reaction system contained 2 μL of diluted cDNA, 10 µL of 2 × TB Green Premix Ex Taq, and 0.3 μM of each primer with a qTOWER3/G real-time system (Analytik Jena, Jena, Thuringia, Germany). The reaction program consisted of 95°C for 30 s, 95°C for 5 s (45 cycles), 60°C for 34 s (45 cycles), and Melt for 15 s. The tomato actin (NCBI: NM_001330119.1) was used as an internal control. Primers are listed in the Supplementary Table.
Total RNA was extracted from shoot apicals at different stages of flower bud differentiation using TRIzol reagent (Invitrogen, USA). The extracted RNA was then evaluated for integrity, purity, and concentration. An equal amount of RNA from each stage was mixed, and 1 μg was used as a template. The first strand of cDNA was synthesized using SMART cDNA Library Construction Kit (Taraka, Japan) according to the manufacturer’s instructions (Code No. 634901, Taraka). The second strand cDNA was amplified using Advantage 2 PCR Kit (Taraka, Japan) following the manufacturer’s instructions (Code No. 639206, Taraka). Double-stranded cDNA was purified using the DNA Fragment Kit (Taraka, Japan) according to the manufacturer’s instructions (Code No. 9761, Taraka), and SfiI was used for enzyme digestion. Short fragments were purified using CHROMA SPIN-1000-TE (Taraka, Japan) according to the manufacturer’s instructions (Code No. 636079, Taraka). The cDNA sample with the removed short fragment was attached to three pGADT7-SfiI containing different reading frames using a DNA ligation Kit (Taraka, Japan) according to the manufacturer’s instructions (Code No. 6022, Taraka). Three primary cDNA libraries were transformed into Escherichia coli HST08 premium electro-cells by electric shock according to the manufacturer’s instructions (Code No. 9028, Taraka). The capacity of the library was calculated by diluting the transformed products 103-fold and culturing them on an LB agar medium. The number of single colonies obtained was multiplied by dilution times and the total volume of solution (mL) and then divided by the volume of culture medium (mL) to calculate the capacity of the library. To use 2×Taq PCR Mastermix (Tiangen, Beijing) for PCR amplification, 48 single colonies were randomly selected. The primary cDNA library was transformed into HST08 again, and all single colonies were collected for germiculture. The Plasmid Purification Kit (Taraka, Japan) was used to extract the plasmid according to the manufacturer’s instructions (Code No. 9760, Taraka). Finally, the concentration of the final cDNA library was detected.
The bait fragment used for yeast one-hybrid library screening was designed based on the CArG element found in the wild-type lc. It consisted of three tandem copies of a specific nucleotide sequence, which is listed in the Supplementary Table. Two oligonucleotide chains of the bait fragment were synthesized using the gene-synthesis technique and mixed in a 1:1 ratio. The reaction program consisted of 95°C for 30 s, 72°C for 2 min, 37°C for 2 min, 25°C for 2 min, and storage at 4°C. To generate the bait vector, the bait sequence was cloned into the pAbAi vector, which was digested with XhoI and SmaI restriction enzymes. The resulting construct was named pAbAi-lc. Next, the bait vector pAbAi-lc was digested with BbsI and BstbI enzymes. The p53 vector was used as a positive control. The linearized pAbAi-lc and p53 vectors were then transformed into Y1HGold yeast-competent cells using Yeast Transformation System 2 (Taraka, Japan) according to the manufacturer’s instructions (Code No. 630439, Taraka). The transformed products were detected using Matchmaker Insert Check PCR Mix 1 (Taraka, Japan). The concentration of aureobasidin A (AbA) in the bait and positive control yeast strains was determined using Matchmaker Gold Yeast One-Hybrid Library Screening System Kit (Clontech, Japan, Mountain View, CA, USA) according to the manufacturer’s instructions (Code No. 630491, Clontech). For library screening, 15 μg of the cDNA library was transformed into Y1HGold[pAbAi-lc] yeast-competent cells. The resulting product was diluted 10-, 100-, and 103-fold, respectively, and then cultured on SD/-Leu and SD/-Leu/AbA medium (90 mm). The number of colonies was counted on the 100-fold dilution medium, and the rest of the reaction liquid was cultured on SD/-Leu/AbA medium (150 mm). The number of colonies obtained was calculated using the formula: number of single colonies × dilution times × suspension volume (mL) ÷ volume on medium (mL). Finally, the colonies on the 150-mm medium were detected using Matchmaker Insert Check PCR Mix 2 (Taraka, Japan).
The coding sequence of SlSEP3 was inserted into the pGADT7 vector (Clontech), while the CArG element in the wild-type lc was cloned into the pAbAi vector (Clontech) individually. The assay was performed with the Matchmaker Gold Yeast One-Hybrid Library Screening System Kit (Clontech, Japan) according to the manufacturer’s instructions (Code No.630491, Clontech). Primers used are listed in the Supplementary Table.
The coding sequence of SlSEP3 was cloned into the pGADT7 vector and transformed into Y1HGold yeast-competent cells. Positive colonies were cultured in a YPDA liquid medium containing 50 mg mL−1 ampicillin at 30°C until the OD600 value reached 1.0. Yeast protein was extracted using a yeast protein extraction kit (Solarbio, Beijing, China) following the manufacturer’s instructions (Code No. BC3780, Solarbio). The extracted protein was purified using an Amicon® Ultra-4 centrifugal filter (Merck, Burlington, MA, USA) to remove small molecule salts and heteroproteins according to the manufacturer’s instructions (Code No. UFC8010, Merck). The purified protein was dissolved in 1× PBS buffer and stored at −20°C in a refrigerator.
Electrophoretic mobility shift assay (EMSA) assay was performed using a Chemiluminescent EMSA Kit (Beyotime, Shanghai, China) according to the manufacturer’s instructions (Code No. GS009, Beyotime). The biotin-labeled probe was prepared by SBS Genetech Co. Ltd. (Beijing, China). All oligonucleotide probes used are listed in the Supplementary Table.
Protein sequences of E-class genes involved in flower development in Arabidopsis and tomatoes were retrieved from the NCBI website. Multiple sequence alignment was conducted using Clustalw, and the neighbor-joining model was employed for constructing the phylogenetic tree.
SlSEP3 silencing fragments were designed using the virus induced gene silencing (VIGS) tool on the Sol Genomics Network (https://solgenomics.net/) and amplified by PCR, then ligated into the pTRV2 vector to generate the pTRV2-SlSEP3 constructs. The pTRV2-SlSEP3, pTRV2 empty vector, and pTRV1 empty vectors were transformed into Agrobacterium (Agrobacterium tumefaciens) strain GV3101 individually. A mixture of cultures containing pTRV1 and pTRV2-SlSEP3 was used for infection as previously described (Liu et al., 2002). Two infections were performed, one on germinated seeds and the other on stems. Silencing efficiency was determined using qRT-PCR on both young buds and leaves below the infected site. The primers used are listed in the Supplementary Table.
The coding sequence of the histone deacetylase SlHDAs was cloned into the pGADT7 vector (Clontech, Japan), while the coding sequence of SlSEP3 was cloned into the pGBKT7 vector (Clontech, Japan). The assay was performed with the Matchmaker Gold Yeast Two-Hybrid System Kit (Clontech, Japan) according to the manufacturer’s instructions (Code No. 630489, Clontech, Japan). Primers used are listed in the Supplementary Table.
To investigate the molecular function of lc in affecting tomato locule number, four lc mutants (#11, #12, #9, and #22) were generated using the CRISPR/Cas9 system under AC background. The lc target region in T1 generation plants was amplified and sequenced, revealing a 5-bp deletion, 1-bp insertion, 1-bp deletion, and 2-bp deletion in the CArG motif of the target sequences of T1 generation lines (Figure 1A). Compared with WT, the homozygous CR-lc#11 mutant showed an increase in floral organ number (Figures 1B, C; Supplemental Dataset), with some partial flowers composed of five to seven sepals, five to six stamens, and five to seven petals (Figure 1C). Additionally, the locule number increased from two to three (Figure 1D), with 14.2% of the fruits from the CR-lc#11 mutant exhibiting three locules (Figure 1E; Supplemental Dataset). However, most flowers in the CR-lc#11 mutant remained unchanged from the WT, with flowers composed of four to five sepals, four to five stamens, and four to five petals. In addition, we also analyzed the phenotype of flower organ number and locule number in homozygous CR-lc#12 mutant (Supplementary Figure S1, Supplemental Dataset). Similar to the CR-lc#11 mutant, the number of petals, sepals, and stamens increased compared with WT in the CR-lc#12 mutant. Furthermore, 10.7% of the fruits exhibited three locules. These findings suggest that the lc mutation affects the differentiation of stem cells in partial flowers.
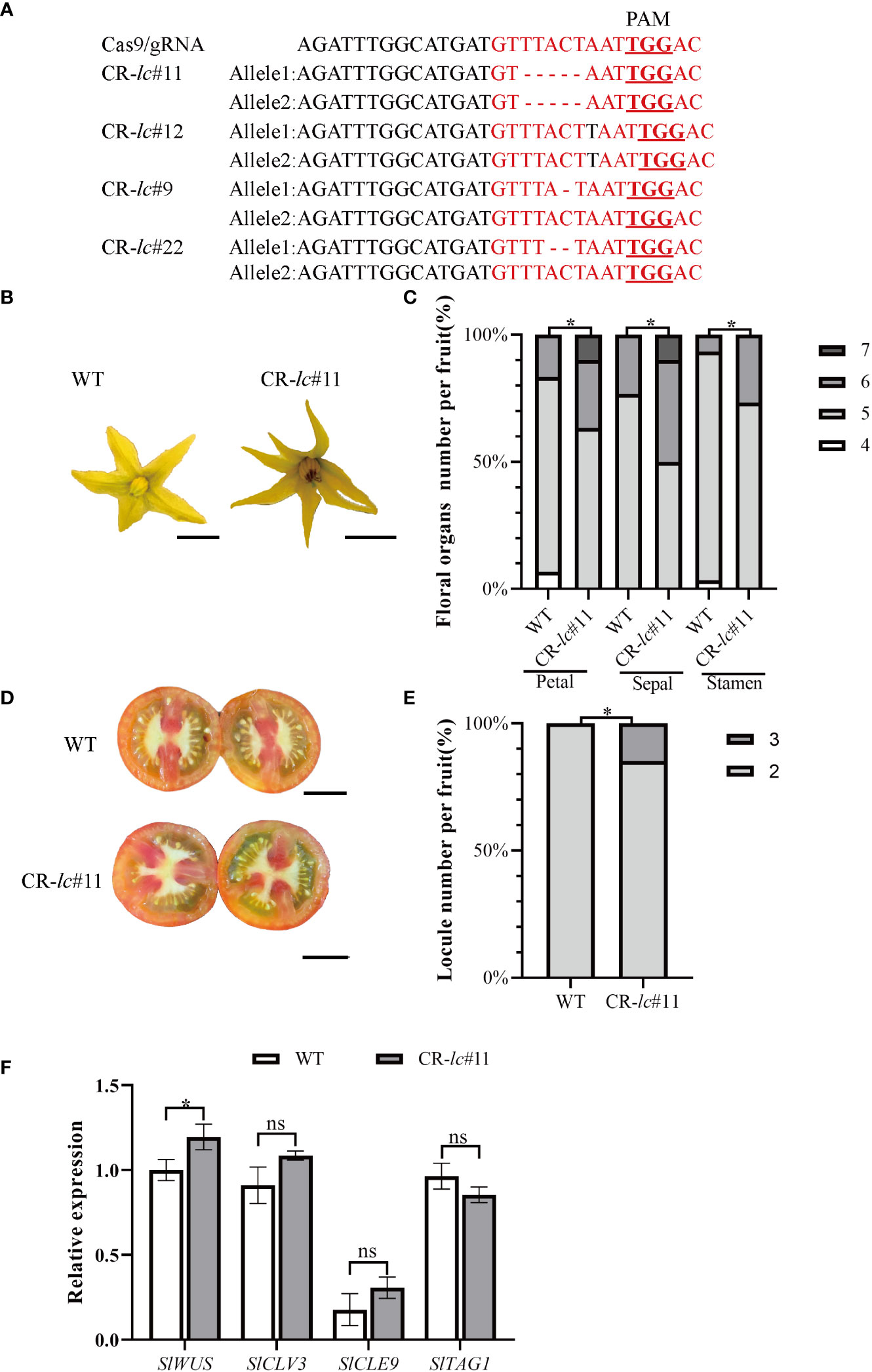
Figure 1 Tomato lc mutation increases the number of floral organs and fruit locules by altering the expression of the SlWUS. (A) Identification of CArG-motif (red font) editing in tomato lcCR mutants: the red font with underline indicates PAM sequences. (B) Comparison of flower phenotypes between the lcCR mutant and WT. Bars represent 0.5 cm. (C) Percentage of floral organ count statistics between the lcCR mutant and WT. (D) Comparison of lcCR and WT fruit cross-sectional phenotypes. Bars represent 1.0 cm. (E) The percentage of single fruit locule number count statistics between the lcCR mutant and WT. (F) Analysis of gene expression related to meristematic differentiation in tomato lcCR T1 generation. The data are presented as means ± SD of three independent tests. Significant differences were determined using an unpaired two-tailed Student’s t-test: *p-value < 0.05; ns, no significant difference.
In the CR-lc#11 mutant, we found that SlWUS expression was significantly increased during the differentiation of stem cells, while the expression of SlCLV3, SlTAG1, and SlCLE9 was not changed (Figure 1F; Supplemental Dataset). It suggested that lc might change locule number by affecting SlWUS transcription.
To investigate the upstream regulation of tomato lc, we first constructed a shoot apical cDNA library for screening proteins that bind to lc. We extracted shoot apical RNA from different stages of flower bud differentiation in tomato AC (Supplementary Figure S2A) and synthesized double-stranded cDNA from equal amounts of total RNA. The resulting electrophoretic bands of double-stranded cDNA fragments were dispersed, with a majority concentrated in the 500- to 3,000-bp range (Supplementary Figure S2B). We purified the cDNA by removing highly expressed components and retaining the effective fragments within the enriched interval (Supplementary Figure S2C). The normalized full-length cDNA was then digested with SfiI (Supplementary Figure S2D) and ligated to a three-frame pGADT7-SfiI vector to generate the primary cDNA library. We ultimately transformed the primary cDNA library into competent cells from Escherichia coli and measured its capacity, which was calculated based on the number of colonies. The primary cDNA library capacity of the three reading frame vectors was 2.6 × 106 cfu, 1.6 × 106 cfu, and 2.4 × 106 cfu, respectively. Finally, we detected the length of inserted fragments, which was mainly distributed in the 500- to 3,000-bp range (Supplementary Figure S2E). We mixed the colonies for germiculture in an LB liquid medium, the concentration of the final cDNA library plasmid was 1 mg mL−1.
Secondly, we performed a yeast one-hybrid screen. We selected an OD600 value of 0.002 to screen for the appropriate concentration of AbA (Supplementary Figure S3A). We found that 150 ng mL−1 of AbA completely inhibited yeast growth. To ensure the reliability of subsequent library screens, we selected a concentration of 200 ng mL−1 of AbA. We transformed the final cDNA library into a bait yeast strain and used the P53 positive yeast strain as a control. The number of positive control colonies on SD/-Leu and SD/-Leu/AbA200 agar mediums was roughly the same, but the number of bait yeast strain colonies on SD/-Leu/AbA200 agar medium was significantly lower than that on SD/-Leu agar medium. We counted 137 single colonies on SD/-Leu/AbA200 agar medium (Supplementary Figure S3B). The number of obtained clones was calculated to be 137 cfu/0.1 mL × 100 mL × 15 mL = 2.055 × 106, which was greater than 106, indicating that the library screen test was successful.
Finally, we selected every single colony on the 150-mm agar medium for PCR detection and sequencing. The obtained results were analyzed on the Tomato Genomics Network, and we obtained statistical information about the function of the screened factors (Table 1). Eventually, two candidate transcription factors were identified: Solyc05g015750, which encodes SEPALLATA3 (SEP3) of the tomato MADS-box transcription factor, and Solyc02g092370, which encodes GARS11 of the tomato GRAS family transcription factor. Based on the function of transcription factors, we considered SlSEP3 to be a potential candidate for binding to lc.
To ensure the reliability of yeast one-hybrid screening results, we implemented a yeast one-hybrid assay and an electrophoretic mobility shift assay. We recombined the coding region of SlSEP3 with the pGADT7 vector and performed a yeast one-hybrid assay with the CArG element in wild-type lc. We used an empty pGADT7 vector as the negative control and mutant lc as a control (Figure 2A). The results demonstrated that the SlSEP3 transcription factor interacted with the wild-type lc but not with the mutant lc. Furthermore, we transferred the recombinant expression vector pGADT7-SlSEP3 into yeast Y1HGold-competent cells for cultivation and obtained a purified recombinant HA-tagged SlSEP3 protein from yeast for the EMSA. To exclude any influence from HA-tagged yeast protein, it was set as the experimental control. We confirmed that HA-SlSEP3 yeast protein bound to the wild-type lc, and the binding ability decreased when the concentration of the cold competition probe was increased to 200 times (Figure 2B). Taken together, the results from the yeast one-hybrid assay and EMSA provided strong evidence supporting the reliability of the yeast one-hybrid screening.

Figure 2 In vivo yeast one-hybrid (A) and in vitro EMSA validation (B) of the SlSEP3 transcription factor bound to the wild-type lc.
In order to elucidate the role of SlSEP3 in regulating tomato locule number, we performed qRT-PCR analysis to examine the expression level of SlSEP3 and SlWUS in the shoot apical (Figure 3). Our findings revealed that SlWUS expression was downregulated during flower bud differentiation, particularly after the early stage, whereas the expression of SlSEP3 was upregulated, especially after the sepal and petal formation. Notably, the expression patterns of SlSEP3 and SlWUS were found to be opposite to each other. Given that SlSEP3 functions as a transcription factor that interacts with lc, it is plausible to speculate that SlSEP3 may serve as an inhibitor of SlWUS. In addition, we performed a phylogenetic analysis of the E-class genes that influence tomato flower development. There are five E-class genes in tomato, and upon comparing them with the AtSEP in Arabidopsis, it was observed that tomato SlSEP3 clustered preferentially with AtSEP3. This suggests that tomato SlSEP3 shares a similar function with AtSEP3 in Arabidopsis. By analyzing the expression patterns of other E-class genes at different stages of floral bud differentiation in wild-type tomatoes, we found that the SlMADS1 and SlSEP3 might have similar expression patterns. However, further research is needed to determine whether there is functional redundancy between these two genes in regulating fruit locule number.
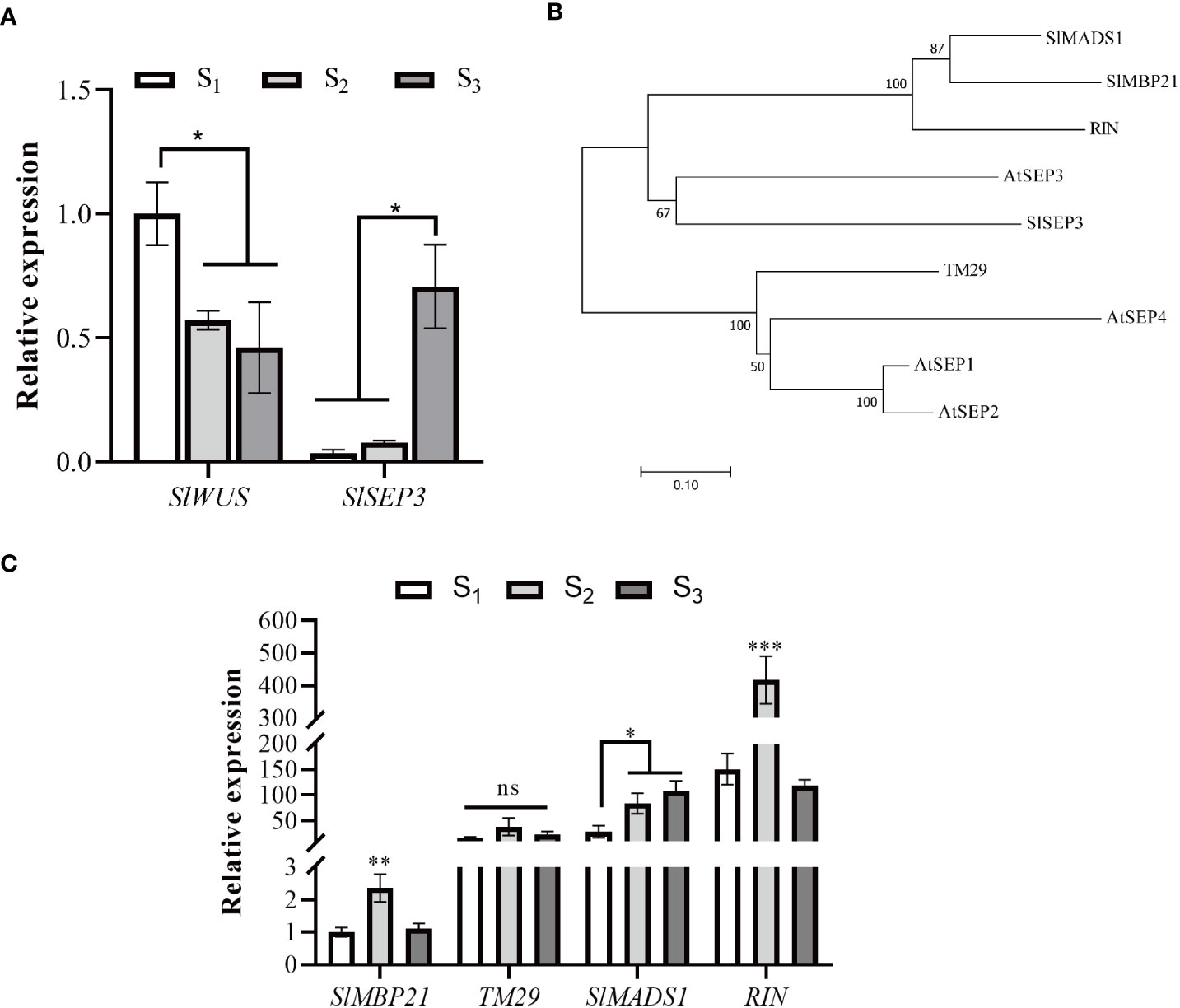
Figure 3 Tomato SlSEP3 and SlWUS have opposite effects on flower bud differentiation (A). Expression analysis of SlSEP3 and SlWUS in tomato flower bud differentiation stages. S1, S2, and S3 respectively represent early flower bud differentiation, middle flower bud differentiation (sepal and petal formation stage), and late flower bud differentiation stages (carpel formation stage). The data are presented as the means ± SD of three independent tests. Significant differences were determined using one-way ANOVA with Dunnett’s test: *p-value < 0.05. (B) Systematic phylogenetic analysis of E-class genes involved in tomato and Arabidopsis flower development. The evolutionary history was inferred using the neighbor-joining method. (C) Expression analysis of E-class genes involved in tomato flower bud differentiation stages. The data are presented as the means ± SD of three independent tests. Significant differences were determined using one-way ANOVA with Tukey’s multiple comparisons test: *p-value < 0.05; **p-value < 0.01; ***p-value < 0.0005.
To further investigate whether SlSEP3 is a negative regulator of fruit locule number, we generated a set of tomato lines with individually silenced SlSEP3 using virus-induced gene silencing and analyzed SlSEP3 expression in young buds and leaves by qRT-PCR (Figures 4A, B; Supplemental Dataset). Compared with wild-type plants, pTRV-SlSEP3 plants displayed an increase in the number of floral organs in both the first and second inflorescence (Figures 4C, 5A, B; Supplemental Dataset). In the first inflorescence, only 20% of the flowers in wild-type plants had six petals and six sepals, whereas 60% of flowers in pTRV-SlSEP3 plants had six petals and 30% of the flowers had six sepals (Figure 5A). Moreover, the number of petals in the second inflorescence of pTRV-SlSEP3 plants was significantly increased (Figure 5B), particularly in pTRV-SlSEP3-3 plants where seven and eight petals were observed (Figure 4C). Furthermore, there was an increase in the number of locules in both the first and second inflorescence of pTRV-SlSEP3 plants, with 20% of the fruits in the first inflorescence and 30% of the fruits in the second inflorescence of pTRV-SlSEP3 plants having three locules (Figures 4D, 5C, D). These results suggested that SlSEP3 acts as a negative regulator of tomato locule number.
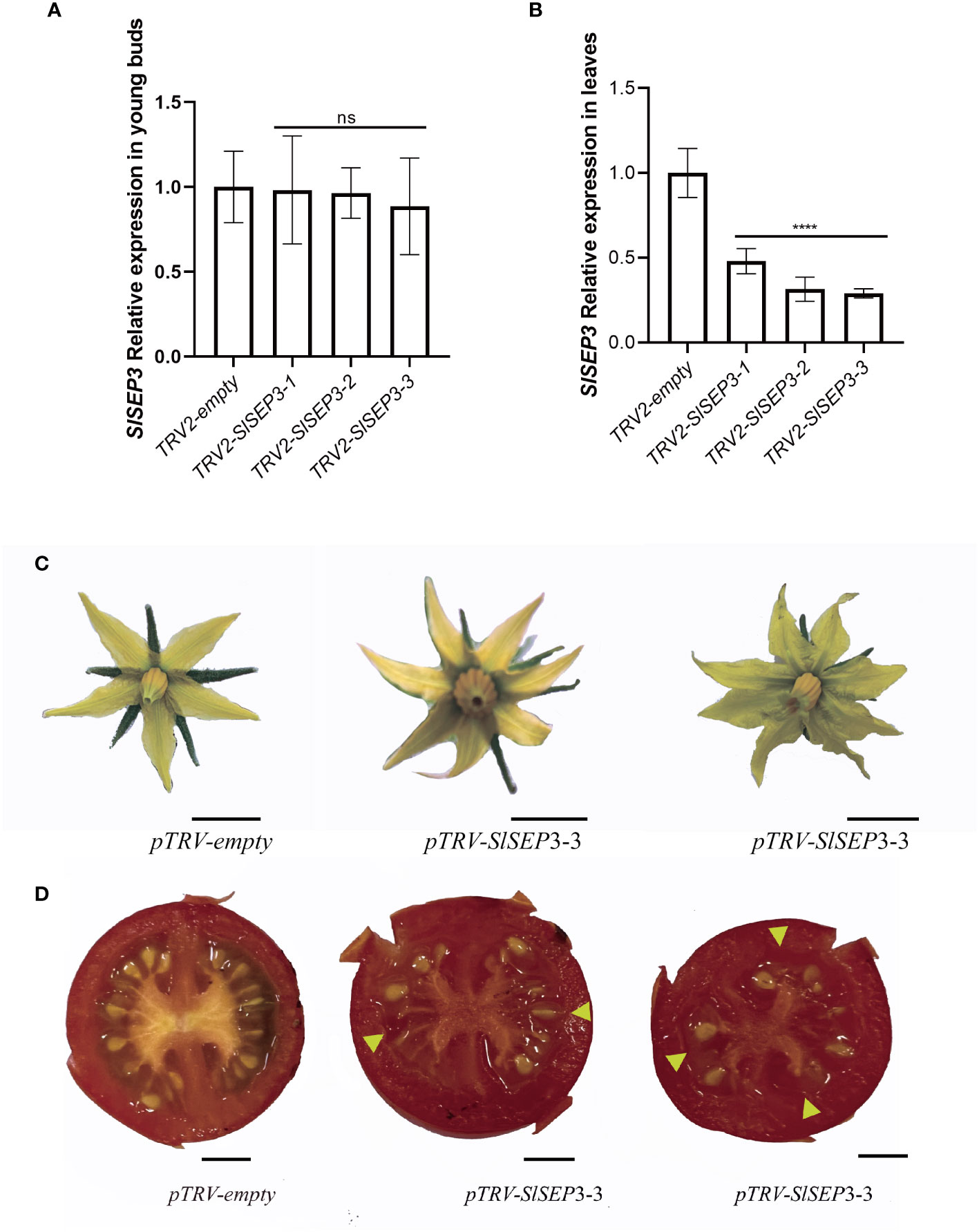
Figure 4 The number of petals and locules in partially silenced SlSEP3 plants increased. (A) Detection of the transient silencing efficiency of SlSEP3 in young buds. (B) Detection of the transient silencing efficiency of SlSEP3 in leaves. Data are the means ± SD of three independent tests. Significant differences were performed with a two-way ANOVA with Dunnett’s multiple comparisons test: ****p-value < 0.0001; ns, no significant difference. (C, D) Comparison of flower phenotypes (C) and fruit cross-sectional phenotypes (D) between the SlSEP3-3-silenced plant line and the control plant line. Bars represent 0.5 cm.
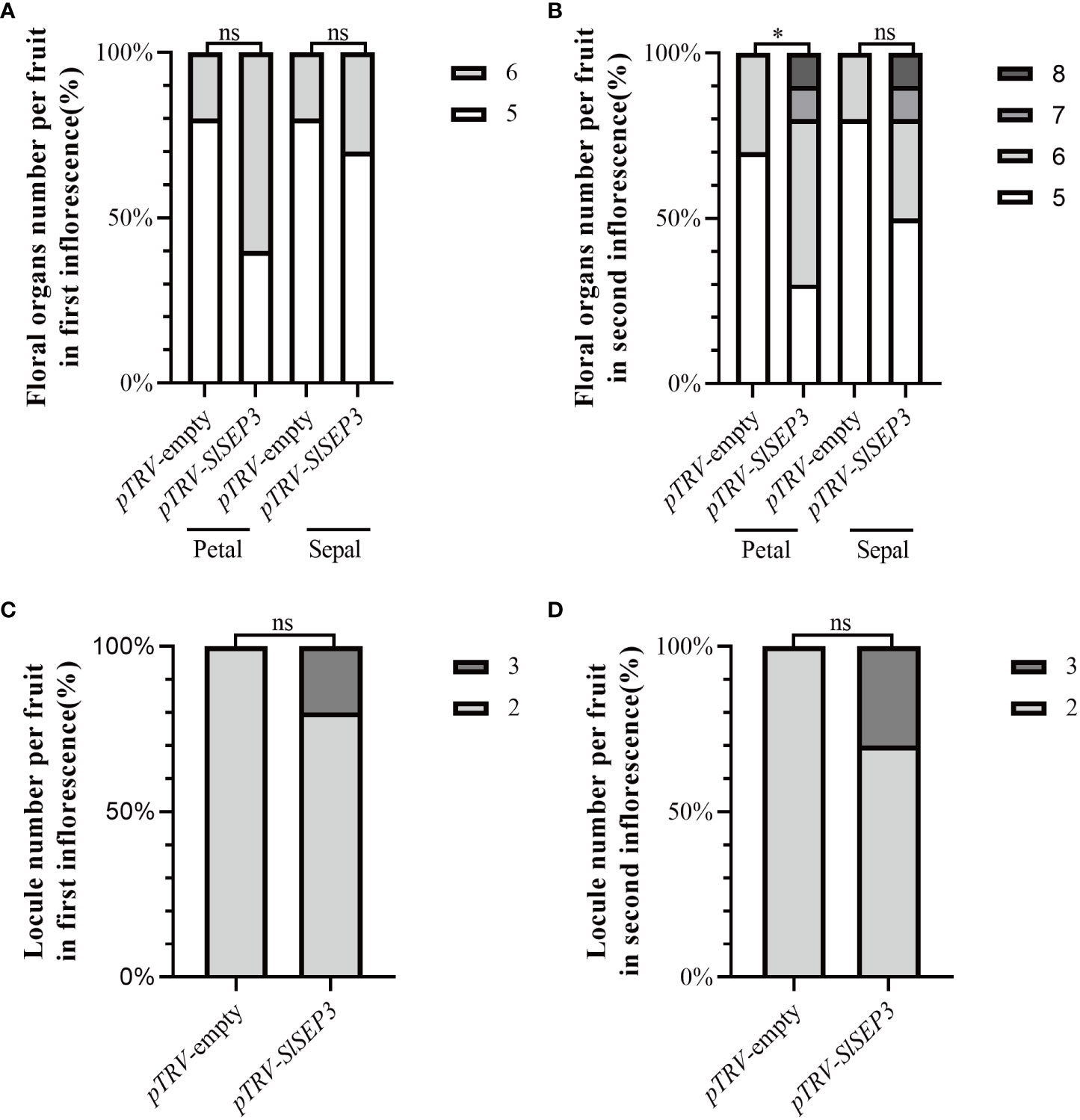
Figure 5 Silencing of SlSEP3 results in an increase in the number of floral organs and fruit locules. Percentage of the floral organ (A, B) count statistics and single fruit locule number (C, D) count statistics between the SlSEP3-silenced plants and the control plant line. Significant differences were determined using a two-way ANOVA with Sidak’s multiple comparisons test: *p-value < 0.05; ns, no significant difference.
To further investigate the regulation of SlWUS expression by the tomato SlSEP3 transcription factor after binding to lc, we conducted an analysis of the interaction between SlSEP3 and tomato histone deacetylase, drawing upon the functions of homologous genes in Arabidopsis (Figure 6). We cloned the coding sequences of SlHDA1–9 and SlHDT1–3 from the tomato histone deacetylase family and performed yeast two-hybrid assays. The results provide preliminary evidence that SlSEP3 could interact with several tomato histone deacetylases, including SlHDA1, SlHDA3, SlHDA4, SlHDA5, SlHDA6, SlHDA7, and SlHDA8 in yeast cells.
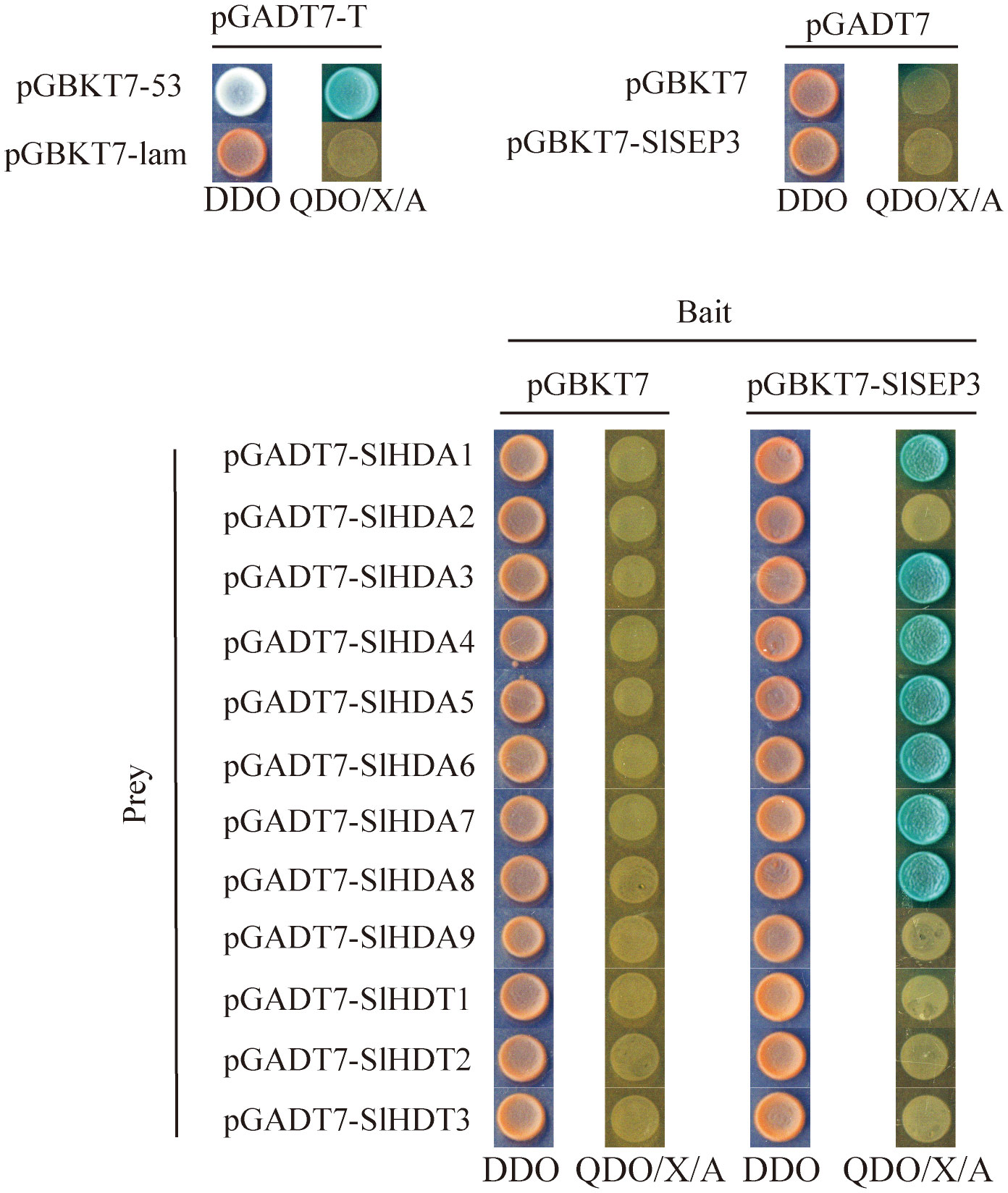
Figure 6 Yeast two-hybrid analysis of the interaction between tomato SlSEP3 transcription factor and the histone deacetylase family.
In wild-type tomatoes, the CArG-motif of lc is mainly composed of “GTTTACTAATTGGAC”. However, in the lc mutant, there is a mutation at the second and eighth nucleotides. This mutation has been shown to increase the fruit locule number to three to four (Barrero et al., 2006; Muños et al., 2011). In this study, we deleted a 5-bp fragment of the CArG motif in the lcCR under AC background and found that 14.2% of fruits had locule numbers up to 3 (Figures 1A–E; Supplemental Dataset). This is consistent with the phenotype of 11% fruits in S.pim-lcNIL with three to four locules (Rodríguez-Leal et al., 2017). Additionally, we also observed a slight increase in floral organ number and fruit locule number in the lcCR with a 1-bp insertion in the CArG-motif, with 10.7% of fruits displaying three locules (Supplementary Figure S1, Supplemental Dataset). Furthermore, since the lc does not encode any protein, we are particularly interested in obtaining strains with a complete deletion of the cArG motif. Considering that the CR-lc#12 strain only exhibited a 1-bp insertion at the target site, we believe that the CR-lc #11 strain is more suitable. These results suggest that the CArG motif in lc is involved in the differentiation of stem cells during floral development. Previous studies have shown that SlWUS is upregulated after natural variation in lc and may be the target of lc (Li et al., 2017). In the CR-lc#11 mutant, only SlWUS expression was significantly increased, while other genes related to stem cell differentiation, such as SlCLV3, SlTAG1, and SlCLE9, were unaffected (Figure 1F, Supplemental Dataset). This suggests that the lc site may directly affect SlWUS transcription to change locule number, although the specific mechanism remains unclear. It is worth noting that although the upregulation of SlWUS expression reaches a significant level, the magnitude of change is only around 20%. Therefore, in order to accurately clarify the tissue-specific SlWUS expression in lcCR mutants, we plan to utilize transgenic plants with a SlWUS promoter fused with GFP or GUS tags for further in-depth analysis in our future studies.
In Arabidopsis, the MADS-box transcription factor AtSEP is involved in floral organ formation and the establishment of whorled organ patterns (Theissen, 2001; Becker and Theissen, 2003). Loss-of-function mutations in the sep3 gene lead to a reduced number and size of floral organs, resulting in defective flower development with no petals, stamens, or carpels. These phenotypes are similar to those observed in pin1 and arf3 (Kaufmann et al., 2009). To identify potential regulators of the lc site, a Y1H library screening was performed, which identified 23 proteins that bind to the lc and activate GAL4 transcription (Table 1). Among these proteins, a MADS-box transcription factor known as SlSEP3 (Solyc05g015750) was found. The Y1H and EMSA results confirmed the binding of SlSEP3 to the lc site (Figures 2A, B). This suggests that SlSEP3 may be involved in regulating SlWUS expression. To further investigate the function of SlSEP3, VIGS was used to silence SlSEP3 expression. The floral organ and fruit phenotype in SlSEP3-silenced plants resembled those of the lc mutant (Figures 1B, D, 4C, D). Additionally, the number of sepals, stamens, and petals were similarly affected in lc mutant and SlSEP3-silenced plants (Figures 1C, E, 5A, B). Moreover, SlSEP3 expression was up-regulated during flower bud differentiation, particularly in the carpel formation stage (Figure 3A). These results suggest that SlSEP3 affects locule formation by binding to the lc site. However, due to the limitations of the VIGS method, it is important to obtain CRISPR-edited SlSEP3 mutants to better elucidate the role of SlSEP3 in tomato locules. Additionally, as the lc site is located after the 3′UTR of SlWUS in the genome, the mechanism of its function remains unclear.
Studies have indicated that AtSEP3 can activate the expression of class B and C genes by recruiting BRM and SYD to their promoter regions, thus overcoming PRC2-mediated inhibition (Wu et al., 2012). AtSEP3 can also form tetramers with AG to inhibit H3K27me3 and activate the expression of CRC and KNU (Hugouvieux et al., 2018). AG tetramers bind to Polycomb response elements in KNU promoter and compete for binding to histone H3, ultimately ensuring the normal differentiation of meristem by inhibiting WUS expression (Sun et al., 2014). In tomatoes, MADS-box transcription factors TAG1 and tomato MADS-box 29 (TM29) jointly regulate reproductive development by recruiting histone deacetylases PRD3/HDA1 subfamily genes SlHDA1 and SlHDA4. TM29 is highly expressed in the primordium of all four-wheel floral organs, and downregulated expression of TM29 leads to parthenogenesis and abnormalities in the second and third-round floral organs (Ampomah-Dwamena et al., 2002). In this study, it was found that SlSEP3 could recruit some PRD3/HDA1 subfamily members such as SlHDA1, SlHDA3, SlHDA4, SlHDA5, SlHDA6, SlHDA7, and SlHDA8 (Figure 6). However, a BIFC or pull-down assay is needed to further investigate the interaction between SlHDA and SlSEP3. Additionally, the increase in SlSEP3 expression also promoted the recruitment of SlHDA proteins near the lc site during flower bud differentiation (Figure 3). This suggests that the nucleosome near the lc site may not be easily opened, leading to the inhibition of transcription of nearby genes. Moreover, the lc site is located after the 3’UTR of SlWUS in the genome, so the inhibitory effect on SlWUS transcription is relatively weak. This also explains why only some fruits have an increase in locule number after lc mutation and SlSEP3 silencing.
In conclusion, the lc site has a significant impact on the development of tomato locules, and its mutation can increase the number of locules in the fruit. The SlSEP3 protein initially binds to the lc site and then recruits several histone deacetylases, including SlHDA, to inhibit nucleosome relaxation. This ultimately affects the transcription of SlWUS, which regulates the locule number in the fruit (Figure 7). Therefore, the SlSEP3-SlHDA-lc-SlWUS module is a crucial addition to enhancing the molecular network of tomato locule formation.
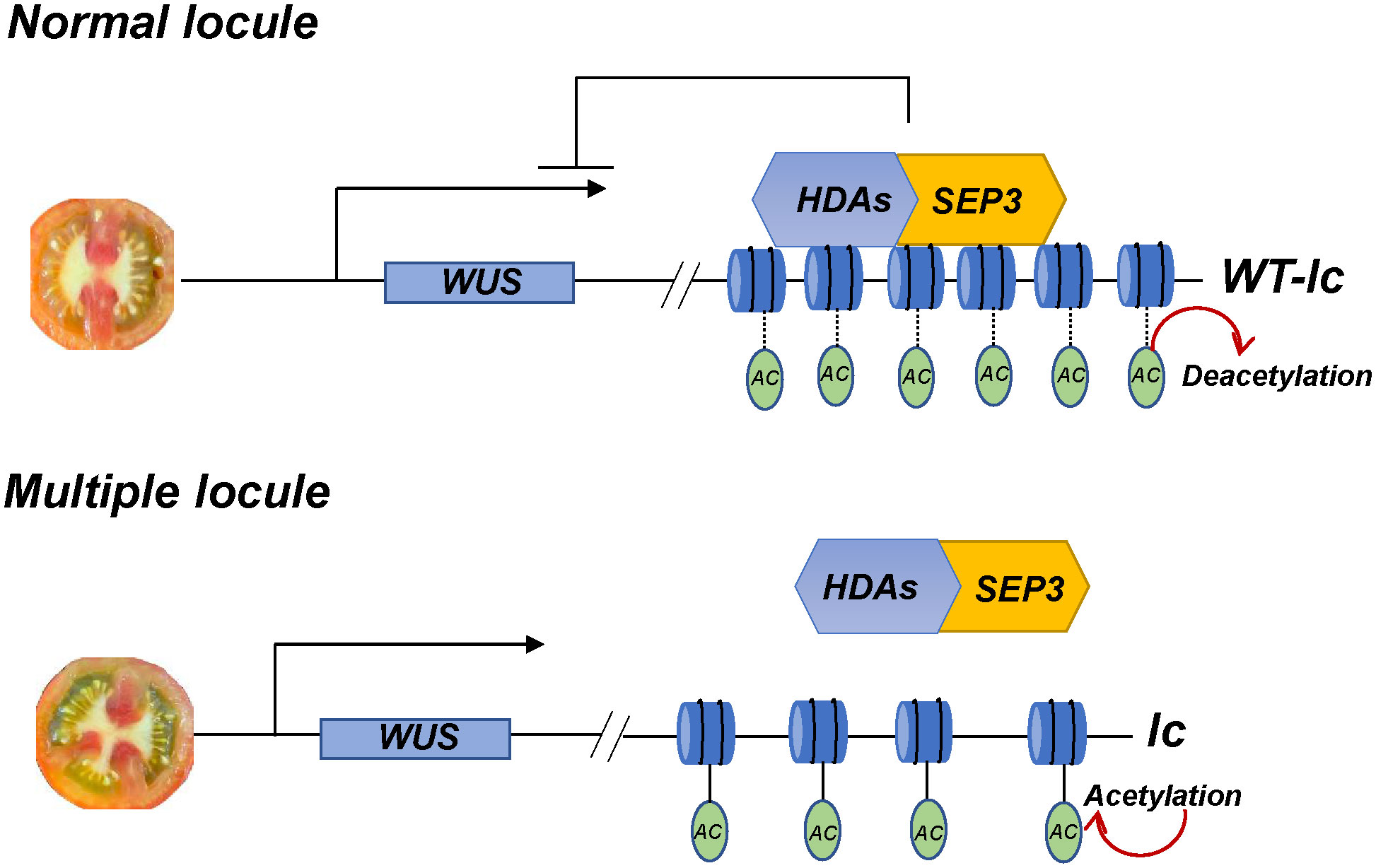
Figure 7 The module of lc QTL is responsible for regulating tomato locule formation. In normal locule tomato plants, the MADS-box transcription factor SlSEP3 binds to lc and recruits histone deacetylase SlHDAs. This process removes the charge and acetyl (Ac) on histone lysine residues, resulting in DNA tightly binding to histone nucleosomes, which limits the effects of transcription factors, polymerase, and DNA. Consequently, the transcription of SlWUS near lc is inhibited, and the locule number is maintained at a normal level. In multiple locule tomato plants, the relevant inhibitory effect is relieved, and the chromatin nucleosome structure will relax, maintaining normal transcription of SlWUS and resulting in an increase in the locule number.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
HX and SM designed and drafted the manuscript. HX and SM conducted experiments and data analysis. YY and LH participated in some data analysis. JM and CT conducted a field data survey. YH and YC help review and edit manuscripts. MQ and TL were involved in the critical revision of important intellectual content and project management. All authors contributed to the manuscript revision and read and approved the submitted version.
This research was funded by the National Natural Science Foundation of China (Grant Nos. 31972397, 31991184, and 31861143045), the Major Agricultural Project of Liaoning Province (2022020768-JH1/102-01), and the 2021 Scientific Research Funding Project of Liaoning Provincial Department of Education (Grant No. LJKZ0639).
The authors are grateful to the National and Local Joint Engineering Research Center of Northern Horticultural Facilities Design and Application Technology (Liaoning), the Key Laboratory of Protected Horticulture (Shenyang Agricultural University), the Ministry of Education, the Key Laboratory of Horticultural Equipment, the Ministry of Agriculture, and the Rural and Collaborative Innovation Center of Protected Vegetable Provincial Co-construction Surrounds Bohai Gulf Region for supporting the project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1249760/full#supplementary-material
Ampomah-Dwamena, C., Morris, B. A., Sutherland, P., Veit, B., Yao, J. L. (2002). Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol. 130, 605–617. doi: 10.1104/pp.005223
Barrero, L. S., Cong, B., Wu, F., Tanksley, S. D. (2006). Developmental characterization of the fasciated locus and mapping of Arabidopsis candidate genes involved in the control of floral meristem size and carpel number in tomato. Genome. 49, 991–1006. doi: 10.1139/g06-059
Becker, A., Theissen, G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet Evol. 29, 464–489. doi: 10.1016/s1055-7903(03)00207-0
Chu, Y. H., Jang, J. C., Huang, Z. J., van der Knaap, E. (2019). Tomato locule number and fruit size controlled by natural alleles of lc and fas. Plant Direct. 3, e00142. doi: 10.1002/pld3.142
Huang, Z. J., van der Knaap, E. (2011). Tomato fruit weight 11.3 maps close to fasciated on the bottom of chromosome 11. Theor. Appl. Genet. 123, 465–474. doi: 10.1007/s00122-011-1599-3
Hugouvieux, V., Silva, C. S., Jourdain, A., Stigliani, A., Charras, Q., Conn, V., et al. (2018). Tetramerization of MADS family transcription factors SEPALLATA3 and AGAMOUS is required for floral meristem determinacy in Arabidopsis. Nucleic. Acids Res. 46, 4966–4977. doi: 10.1093/nar/gky205
Kaufmann, K., Muino, J. M., Jauregui, R., Airoldi, C. A., Smaczniak, C., Krajewski, P., et al. (2009). Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PloS Biol. 7, e1000090. doi: 10.1371/journal.pbio.1000090
Li, H., Qi, M. F., Sun, M. H., Liu, Y., Liu, Y. D., Xu, T., et al. (2017). Tomato transcription factor slWUS plays an important role in tomato flower and locule development. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00457
Lippman, Z., Tanksley, S. D. (2001). Dissecting the genetic pathway to extreme fruit size in tomato using a cross between the small-fruited wild species Lycopersicon pimpinellifolium and L. esculentum var. Giant Heirloom. Genetics. 158, 413–422. doi: 10.1093/genetics/158.1.413
Liu, Y., Schiff, M., Dinesh-Kumar, S. P. (2002). Virus-induced gene silencing in tomato. Plant J. 31, 777–786. doi: 10.1046/j.1365-313X.2002.01394.x
Muños, S., Ranc, N., Botton, E., Bérard, A., Rolland, S., Duffé, P., et al. (2011). Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol. 156, 2244–2254. doi: 10.1104/pp.111.173997
Rodríguez-Leal, D., Lemmon, Z. H., Man, J., Bartlett, M. E., Lippman, Z. B. (2017). Engineering quantitative trait variation for crop improvement by genome editing. Cell. 171, 470–480. doi: 10.1016/j.cell.2017.08.030
Rodríguez-Leal, D., Xu, C., Kwon, C. T., Soyars, C., Demesa-Arevalo, E., Man, J., et al. (2019). Evolution of buffering in a genetic circuit controlling plant stem cell proliferation. Nat. Genet. 51, 786–796. doi: 10.1038/s41588-019-0389-8
Somssich, M., Je, B., Simon, R., Jackson, D. (2016). CLAVATA-WUSCHEL signaling in the shoot meristem. Development. 143, 3238–3248. doi: 10.1242/dev.133645
Su, D. D., Wen, L., Xiang, W., Shi, Y., Lu, W., Liu, Y. D., et al. (2022). Tomato transcriptional repressor SlBES1.8 influences shoot apical meristem development by inhibiting the DNA binding ability of SlWUS. Plant J. 110, 482–498. doi: 10.1111/tpj.15683
Sun, B., Looi, L. S., Guo, S. Y., He, Z. M., Gan, E. S., Huang, J. B., et al. (2014). Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science. 343, 1248559. doi: 10.1126/science.1248559
Tanksley, S. D. (2004). The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell. 16suppl, S181–S189. doi: 10.1105/tpc.018119
Theissen, G. (2001). Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 4, 75–85. doi: 10.1016/s1369-5266(00)00139-4
van der Knaap, E., Chakrabarti, M., Chu, Y. H., Clevenger, J. P., Illa-Berenguer, E., Huang, Z. J., et al. (2014). What lies beyond the eye: The molecular mechanisms regulating tomato fruit weight and shape. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00227
Wang, X. G., Aguirre, L., Rodríguez-Leal, D., Hendelman, A., Benoit, M., Lippman, Z. B. (2021). Dissecting cis-regulatory control of quantitative trait variation in a plant stem cell circuit. Nat. Plants. 7, 419–427. doi: 10.1038/s41477-021-00898-x
Wu, M. F., Sang, Y., Bezhani, S., Yamaguchi, N., Han, S. K., Li, Z. T., et al. (2012). SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc. Natl. Acad. Sci. 109, 3576–3581. doi: 10.1073/pnas.1113409109
Keywords: tomato, lc, locule number, SlWUS, SlSEP3 transcription factor
Citation: Xiang H, Meng S, Ye Y, Han L, He Y, Cui Y, Tan C, Ma J, Qi M and Li T (2023) A molecular framework for lc controlled locule development of the floral meristem in tomato. Front. Plant Sci. 14:1249760. doi: 10.3389/fpls.2023.1249760
Received: 29 June 2023; Accepted: 02 August 2023;
Published: 23 August 2023.
Edited by:
Jia-Long Yao, The New Zealand Institute for Plant and Food Research Ltd, New ZealandReviewed by:
Chunying Kang, Huazhong Agricultural University, ChinaCopyright © 2023 Xiang, Meng, Ye, Han, He, Cui, Tan, Ma, Qi and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingfang Qi, cWltaW5nZmFuZ0BzeWF1LmVkdS5jbg==; Tianlai Li, bHRsQHN5YXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.