- 1Key Laboratory of Tea Plant Biology and Resources Utilization, Ministry of Agriculture, Tea Research Institute, Chinese Academy of Agricultural Sciences, Hangzhou, China
- 2Graduate School of Chinese Academy of Agricultural Sciences, Beijing, China
- 3Xihu National Agricultural Experimental Station for Soil Quality, Hangzhou, China
Nitrogen is one of the most important nutrients for tea plants, as it contributes significantly to tea yield and serves as the component of amino acids, which in turn affects the quality of tea produced. To achieve higher yields, excessive amounts of N fertilizers mainly in the form of urea have been applied in tea plantations where N fertilizer is prone to convert to nitrate and be lost by leaching in the acid soils. This usually results in elevated costs and environmental pollution. A comprehensive understanding of N metabolism in tea plants and the underlying mechanisms is necessary to identify the key regulators, characterize the functional phenotypes, and finally improve nitrogen use efficiency (NUE). Tea plants absorb and utilize ammonium as the preferred N source, thus a large amount of nitrate remains activated in soils. The improvement of nitrate utilization by tea plants is going to be an alternative aspect for NUE with great potentiality. In the process of N assimilation, nitrate is reduced to ammonium and subsequently derived to the GS-GOGAT pathway, involving the participation of nitrate reductase (NR), nitrite reductase (NiR), glutamine synthetase (GS), glutamate synthase (GOGAT), and glutamate dehydrogenase (GDH). Additionally, theanine, a unique amino acid responsible for umami taste, is biosynthesized by the catalysis of theanine synthetase (TS). In this review, we summarize what is known about the regulation and functioning of the enzymes and transporters implicated in N acquisition and metabolism in tea plants and the current methods for assessing NUE in this species. The challenges and prospects to expand our knowledge on N metabolism and related molecular mechanisms in tea plants which could be a model for woody perennial plant used for vegetative harvest are also discussed to provide the theoretical basis for future research to assess NUE traits more precisely among the vast germplasm resources, thus achieving NUE improvement.
1 Introduction
Nitrogen is an essential mineral nutrient for plant growth and reproduction. Apart from being a fundamental building block of proteins and nucleic acids, N also participates in carbon fixation through photosynthesis as a component of chlorophyll (Bernard and Habash, 2009). In agricultural production, applying N fertilizers generally leads to significant yield increases (Suárez et al., 2002; Liu et al., 2021c), for which N fertilizers’ use is expected to increase up to 236 million metric tons to meet the global food demands by 2050 (Beatty and Good, 2018). However, less than 50% of the applied N as fertilizer is absorbed by plants and harvested in grains (Raun and Johnson, 1999; Camargo et al., 2005). Thus, a high amount of “unuse” N supplied as fertilizer is transferred to water and the atmosphere, resulting in energy waste, soil acidification, water eutrophication and greenhouse gas emissions (Godfray et al., 2010; Liu et al., 2010). This negative environmental consequence of nitrogen fertilization became a huge challenge for stable and sustainable agricultural production (Bodirsky et al., 2014). There is an urgent need for research advances on N metabolism in the ecosystem; in this context, we need to improve N use efficiency (NUE) by crops, for which the genetic potential for N uptake and assimilation must be further explored.
Tea is processed from the leaves of Camellia sinensis (L.) O. Kuntze and becoming one of the most widely non-alcoholic beverages consumed worldwide due to its unique taste and potential health benefits (Wei et al., 2018). Since 2011, the global planting area of tea have increased steadily and gradually, from 3.84 million hectares in 2011 to 5.09 million hectares in 2020 (Liu et al., 2023). This perennial evergreen woody plant is cultivated in over 30 countries, and China has the greatest cultivated area (Zhang et al., 2019b; Lei et al., 2022). In 2022, tea planting area of China reached 3.33 million hectares (Mei and Zhang, 2022). The geographic origin of the tea plant is assigned to Yunnan province and neighboring regions in southwestern China (Chen et al., 2005). China has traditionally been the largest tea producer worldwide with abundant germplasm resources, and China’s tea have been exported to more than 140 countries or regions (Wei et al., 2012). Currently, many cultivated tea varieties are extensively grown in tropical and subtropical regions across the world, and tea cultivation may increase the local smallholder income, especially in mountainous areas, contributing to local economic development (Yao et al., 2012). The N concentration in young buds and leaves is about 60-70 g·kg−1 (Ma et al., 2013). Tea plants form new shoots every season, and multiple picking and pruning have been done. In agricultural production, tea plants have a high demand for N, which is generally fulfilled through fertilization, active N uptake, assimilation and translocation, as well as remobilization processes. In China, the average annual N inputs reach 300-450 kg·hm−2 to cover tea N demand; an excessive N application rate has been reported in over 30% of the tea plantation area (Ma et al., 2013; Ni et al., 2019). These numerical data reinforce the crucial and urgent need for optimizing the NUE of tea plant. A series of interconnected processes, including N transport, assimilation and remobilization, are involved in NUE, thus the understanding on N metabolism at molecular level will provide the basis for a more rational application of N fertilizers during tea production.
Nitrogen is involved in many important metabolic pathways closely related to the synthesis of amino acids (AAs), caffeine, polyphenols, and other substances responsible for tea quality (Tang et al., 2020). Inorganic N sources, including ammonium (NH4+) and nitrate (NO3-), and small organic N-containing compounds can be uptaken from the soil by the tea plant roots and subsequently transported to the leaves by ammonium transporters (AMTs), nitrate transporters (NRTs), and amino acid transporters (AATs). The absorbed NO3− is first reduced into nitrite (NO2−) in the cytoplasm by nitrate reductase (NR) and further reduced to NH4+ in plastids by nitrite reductase (NiR). Ammonium assimilation involves the conversion of inorganic N to organic N, mainly through the glutamine-glutamate (GS-GOGAT) cycle, catalyzed by glutamine synthetase (GS) and glutamine-2-oxoglutarate aminotransferase/ glutamate synthase (GOGAT) (Bernard and Habash, 2009; Liu et al., 2022). It is noteworthy that glutamate and ethylamine are catalyzed by theanine synthetase (TS) to biosynthesize theanine (γ-glutamyl-L-ethylamide), a unique non-proteinogenic amino acid responsible for umami taste and healthy beneficial component in tea. Thus, the content of Thea is an important indicator for cultivar breeding and evaluating NUE. These processes are schematically illustrated in Figure 1. Further details on substrates, transporters, enzyme isoforms, and cell compartments relevant to the N cycle in tea plants are given in the following sections.
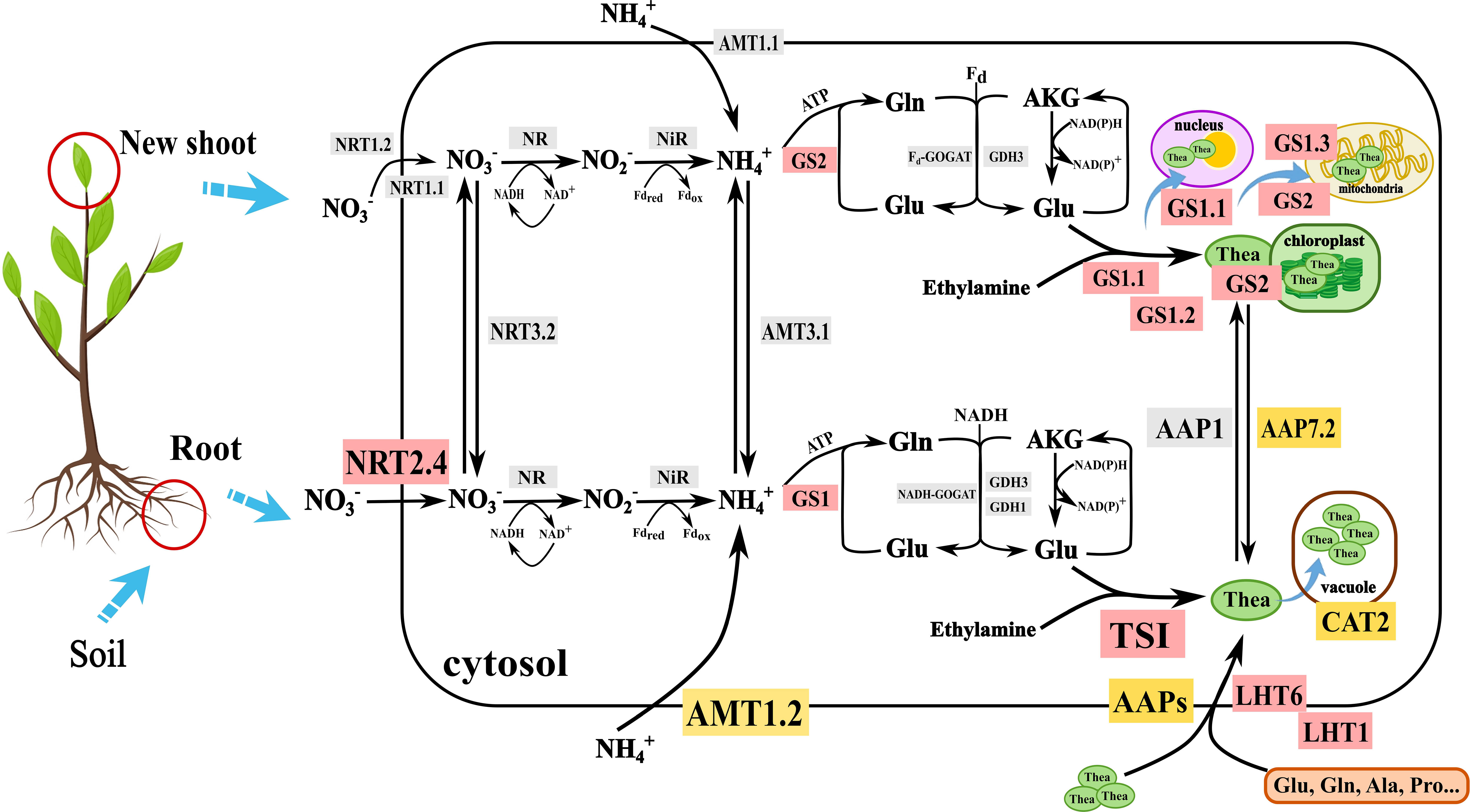
Figure 1 Molecular mechanism of nitrogen nutrient absorption and utilization in tea plant. NRT, nitrate transporter; NR, Nitrate reductase; NiR, Nitrite reductase; AMT, Ammonium transporter; GS, Glutamine synthetase; GOGAT, Glutamate synthase; GDH, Glutamate dehydrogenase; Gln, Glutamine; Glu, Glutamic acid; TS, Theanine synthetase; Thea, Theanine; LHT, Lysine and histidine transporter; CAT, Cationic amino acid transporter; Ala, Alanine; Pro: Proline; AKG, α-Ketoglutaric acid. Gray background represents the genes just were cloned in vitro; Yellow background represents functions of these proteins were validated in yeast; The red background represents functions of these proteins were validated in Arabidopsis, Nicotiana tabacum or Camellia sinensis.
Since the tea plant genome was sequenced (Xia et al., 2020), many enzymes involved in N metabolism and their encoding genes were identified. Nitrogen dynamic regulation and physiological function were widely investigated in tea plant, as these are all critical aspects to improve NUE. In this article, we outline the results of recent investigations about the mechanisms underlying: (1) N absorption and transport in the form of NH4+, NO3−, and AAs; (2) metabolic reduction of nitrate; (3) ammonia assimilation and theanine (γ-glutamyl-L-ethylamide) biosynthesis. We also discuss the use of genetic, genomic, and phenotyping technologies for improving NUE by tea plants and stress the relevance of understanding the genetic basis of tea plant adaptive responses to different N forms.
2 General nitrogen utilization traits by tea plants
2.1 Tea plants acquire N preferentially as NH4+
Tea plant shows a preferential uptake of N in the form of ammonium (NH4+-N). Using the scanning ion-selective electrode technique, Ruan et al. (2016) found that the NH4+ influx rate in the roots of tea plant was higher than that of NO3−, and the presence of NH4+ would promote NO3− influx rate. The yield of young shoots, total root length, N uptake rate, and the contents of caffeine, theanine (Thea), glutamine (Gln), glutamate (Glu) and aspartate (Asp) in tea leaves were significantly higher when NH4+ was the main N source, compared with NO3− (Ruan et al., 2007; Ruan et al., 2010; Ruan et al., 2019; Wang et al., 2022a). NH4+-N can also promote phosphorus (P) uptake (Chen et al., 2019) and increase iron (Fe) and chloride (Cl) contents in mature leaves and sulfur (S) content in the roots (Tang et al., 2019). As a signaling molecule, NH4+ could induce Thea and catechin biosynthesis in a short period (Liu et al., 2017b; Huang et al., 2018). By sensing NH4+ levels, lysine-acetylated and crotonylated proteins profoundly influenced some primary metabolic processes involved in amino acid metabolism, photosynthesis, glycolysis, and carbon fixation (Jiang et al., 2018; Sun et al., 2019).
2.2 Nitrogen concentration influences tea plant growth and biochemical profile
To obtain an appropriate amount of harvestable product, i.e., young buds and leaves, multiple tender shoots are picked from the plants every year. Adequate N nutrition is necessary to increase the formation of young shoots, enhance the growth vigor and maintain the C/N balance (Ruan et al., 2010). A balanced C/N ratio is also important to promote chlorophyll biosynthesis (Yang, 2011) and to ensure adequate availability of free AAs (Liu et al., 2020; Wang et al., 2022a), thus providing N reserve for reproductive growth (Fan et al., 2019).
N metabolism of tea plant is dynamically regulated by environmental factors. Likewise, the growth of lateral roots was regulated by N levels: their length and numbers decreased with increasing N concentrations (Chen et al., 2023; Hu et al., 2023). Under N deficiency, the content of N, L-Thea, and chlorophyll decreased significantly. The activity of many antioxidant enzymes and leaf CO2 assimilation capacity also diminished (Lin et al., 2016; Lin et al., 2019). However, low N levels positively regulated the expression of phosphate transporter genes and promoted flavonoids and polyphenols synthesis in tea leaves (Lin et al., 2023b). Appropriate N supply contributes to the aroma and flavor quality of tea infusion. The activity of the rate-limiting enzyme for N assimilation, GS, increased with N application level, and the content of total AAs, alcohols, and ketone compounds conferring aroma also increased, thus promoting tea products’ integrated quality (Ruan et al., 2010; Deng et al., 2012; Liu et al., 2021b).The accumulation of caffeine, a component of the bitter taste and a central nervous system stimulant in tea, can be increased with the increasing N supply (Ruan et al., 2010). Sufficient N also promotes flavonol glycoside biosynthesis through the expression of relevant genes and the accumulation of the corresponding substrate carbohydrates (Dong et al., 2019). Lipidomic studies revealed that the content of precursors for the formation of aroma-related substances such as monogalactosyl diaclyglycerol (36:6 MGDG) and digalactosyl diacylglycerol (36:6 DGDG) increased when the N fertilizer was applied at adequate amounts, while an excessive N application led to overaccumulation of hexenol and hexenal, compounds which cause an unpleasant grassy smell in tea (Liu et al., 2017a). With the increase in N supply, more C was allocated to N-containing compounds in mature tea leaves and roots, leading to a decrease in flavonoid concentration in the young shoots (Liu et al., 2021a). Long-term N overfertilization reduced significantly benzyl alcohol and 2-phenylethanol contents in tea leaves, as well as those of (E)-nerolidol and indoles in withering leaves, becoming not conducive to the generation of floral and fruity fragrances (Chen et al., 2021).
3 Nitrogen transport in plants
In a wide range of organisms, N transport as NH4+, NO3−, and soluble organic compounds across membranes is mediated by transporter proteins (Wirén et al., 1997). These transporters can be divided into high-affinity transporter systems (HATS) and low-affinity transporter systems (LATS), depending on the specific substrate affinity. The external N level also regulates the affinities of transporters. For instance, there are inducible high-affinity transporter systems (iHATS) and constitutive high-affinity transporter systems (cHATS) to accomplish NO3− transport (Crawford and Glass, 1998; Forde, 2000). These transport proteins play a vital role in both short- and long-distance translocation of N inorganic ions and N-organic compounds.
3.1 NH4+ transport
The membrane-localized ammonium transporter/methylammonium permease (AMT/MEP) facilitates the import and export of NH4+ (Howitt and Udvardi, 2000). In higher plants, AMT proteins can be divided into two types: AMT1 and AMT2. Most AMT1 proteins belong to the HATS group and are synergically involved in NH4+ transport through the apoplastic and the symplastic routes (Yuan et al., 2007). AMT2 plays a role in the translocation of NH4+ from roots to shoots (Giehl et al., 2017). The transcription of the gene encoding this protein is tightly controlled through multiple factors, including external N level, circadian rhythm, hormone contents, and mycorrhizal symbiosis (Couturier et al., 2007; Kobae et al., 2010; Li et al., 2012; Li et al., 2016).
To cope with elevated NH4+ concentrations, the AMT activity may be post-translationally modified via the reversible phosphorylation of the cytosolic C-terminal region, thus allowing rapid adaptation to variable environmental conditions (Yuan et al., 2013; Wu et al., 2019). In tea plants, CsAMTs expression seems to be tissue-specific: CsAMT1.2 reached the highest transcript abundance in roots, while CsAMT1.4 was mainly expressed in flower buds. However, CsAMT1.1 and CsAMT3.1 were highly expressed in all tissues, suggesting that these genes might have diverse functions in NH4+ transport (Zhang et al., 2018; Wang et al., 2022c; Zhang et al., 2022a). Likewise, AMTs expression levels are responsive to changes in NH4+ availability. In roots, CsAMT1.1 expression peaked at 12 h after the exogenous NH4+ resupply, while CsAMT3.1 showed an upward trend after 24 h and CsAMT1.2 expression level increased at 10 h, with a 2.5-fold change compared to 0 h, and then decreased again by 24 h. In NH4+-treated leaves, CsAMT1.1 expression was up-regulated only after 4 h, exhibiting a 4.75-fold increase, whereas CsAMT1.2 and CsAMT3.1 expression levels did not change until 24 h later. These data indicate that NH4+ transport in tea roots is mainly regulated by CsAMT1.2, while in leaves, the NH4+ induction is mainly controlled by CsAMT1.1 in the short term (Tang et al., 2020). Across different experimental NH4+ concentrations, most CsAMTs were expressed at higher levels in leaves than roots, except for CsAMT1.2, CsAMT1.4, and CsAMT2.1a. Remarkably, CsAMT1.2 expression was significantly higher in roots than leaves under NH4+ deficiency (0 mM NH4+) or at 4 mM NH4+, demonstrating the major role of this transporter in NH4+ uptake. Other genes involved in NH4+ transport, such as CsAMT2.1b, CsAMT3.3, CsAMT4.1a, CsAMT4.1b, CsAMT4.1c, and CsAMT4.1d, exhibited similar expression profiles, with a decreasing trend under low N supply and a notorious induction under high N supply (Wang et al., 2022c). Furthermore, this report indicates that CsAMTs expression in tea leaves is differentially regulated over time by abiotic stresses, including drought and salinity, as well as after methyl jasmonate treatments. Under these treatments, specific CsAMTs genes were up-regulated or down-regulated in different ways, suggesting different functions to cope with various stresses (Wang et al., 2022c).
Transcriptome data revealed that CsAMT1.2 expression could be highly induced by NH4+-resupply; weighted gene co-expression network analyses and the functional validation in an NH4+-uptake defective yeast line further corroborated that the high-affinity transporter CsAMT1.2 was a “hub gene” in the N metabolic network of tea plants, controlling NH4+ uptake from the soil to the roots (Zhang et al., 2020). Also, Wang et al. (Wang et al., 2022c) found that 11 yeast transformant lines grew well on 0.3 mM NH4+ as the sole N source, indicating their high affinity for NH4+ permeation. The transcriptional regulation of CsAMTs differed even at the cultivar level (Li et al., 2017). CsAMT1.1 and CsAMT1.5 expression levels were significantly higher in the roots of the FuDingDaBaiCha cultivar than Longjin43 cultivar (Zhang et al., 2022a). After NH4+ resupply, CsAMT1.2, CsAMT2.2, and CsAMT2.3 genes were differentially induced in tea cultivars with different NH4+-uptake efficiency, indicating the uneven NH4+ transport capacity among cultivars (Zhang et al., 2018; Zhang et al., 2022b).
3.2 NO3− transport
Membrane-bound nitrate transporters (NRTs) are required for NO3− uptake in plants. The members of the large NRT family can be divided into four subfamilies: nitrate transporter 1/peptide transporter (NRT1/PTR), collectively known as NPF, nitrate transporter 2 (NRT2), chloride channel (CLC), and slow anion channel associated/homologue (SLAC/ SLAH) (Krapp et al., 2014).
The NRT1 subfamily harbors many members, acting in NO3− transport from roots to shoots (Krapp et al., 2014). NRT transport activity is also regulated through phosphorylation. AtNRT1.1 is a dual-affinity protein: phosphorylation of the Thr101 residue by the CBL-interacting protein kinase 23 changes its substrate affinity (Sun et al., 2014). NRT2 are HATS proteins and belong to the nitrate/nitrite porter (NNP) family, mainly expressed in roots. These proteins have a role in both NO3− accumulation and NO3− transport (Chopin et al., 2007; Li et al., 2007; Kiba et al., 2012). To date, four CsNRT1 and four CsNRT2/3 genes have been isolated from tea plants. These genes show tissue-specific expression patterns and are differentially induced by exogenous NO3−. It was reported that CsNRT1.1 and CsNRT1.2 were mainly expressed in leaves. CsNRT1.7, CsNRT2.5, and CsNRT3.2 had higher expression levels in mature leaves than other tissues, while CsNRT1.5, CsNRT2.4, and CsNRT3.1 transcripts mainly accumulated in tea roots (Feng et al., 2014; Wang, 2014; Wang et al., 2014; Yang et al., 2016; Zhang et al., 2021). Further research showed that CsNRT2.4 expression was root-specific and strongly induced by N resupply. Arabidopsis seedlings overexpressing CsNRT2.4 had a significantly higher fresh weight and lateral roots length than wild-type seedlings, especially under low N availability (0.1 mM NO3−), pointing out CsNRT2.4 as a high-affinity nitrate transporter that might improve NO3− uptake rate (Zhang et al., 2021). Additionally, Wang et al. (2022b) identified a total of 109 CsNPF members by analyzing the tea genome; these proteins could be divided into 8 groups according to their phylogenetic relationships, and the transcription of most of these genes responded to NO3− supply. Similarly, CsNRTs expression profiles varied in tea cultivars with different NUE (Wang et al., 2014). The expression of CsNRT2.4 and CsNRT3.2 in the cultivar LongJin43 was higher than that observed in ZhongCha108, indicating higher responsiveness to external NO3− supply in the former (Su et al., 2020).
Initially, CLC proteins were thought to be specifically involved in chloride (Cl-) transport as channels or 2 Cl−/1 H+ antiporters (Jentsch, 2008). Further research showed that AtCLCa is a tonoplast-located 2 NO3−/1 H+ antiporter that drives NO3− accumulation in the vacuoles (Jentsch, 2008; Monachello et al., 2009). As anion channels, SLAC/SLAH proteins showed a strong preference for NO3− and have been associated with CO2 and abscisic acid-dependent stomatal closure (Negi et al., 2008; Vahisalu et al., 2008). In tea plants, Xing et al. (2020) identified eight CLC genes across the wide genome of this species and named them CsCLC1-8. Phylogenetic studies demonstrated that the proteins encoded by these genes belonged to two subclasses; further studies showed that CsCLC transporters might participate in the uptake and long-distance transport of Cl− and F−, as their expression levels varied in response to the addition of these two ions at different concentrations. However, the role of CsCLCs in NO3− transport has not been elucidated. Similarly, there are no published reports related to SLAC/SLAH proteins in tea plants.
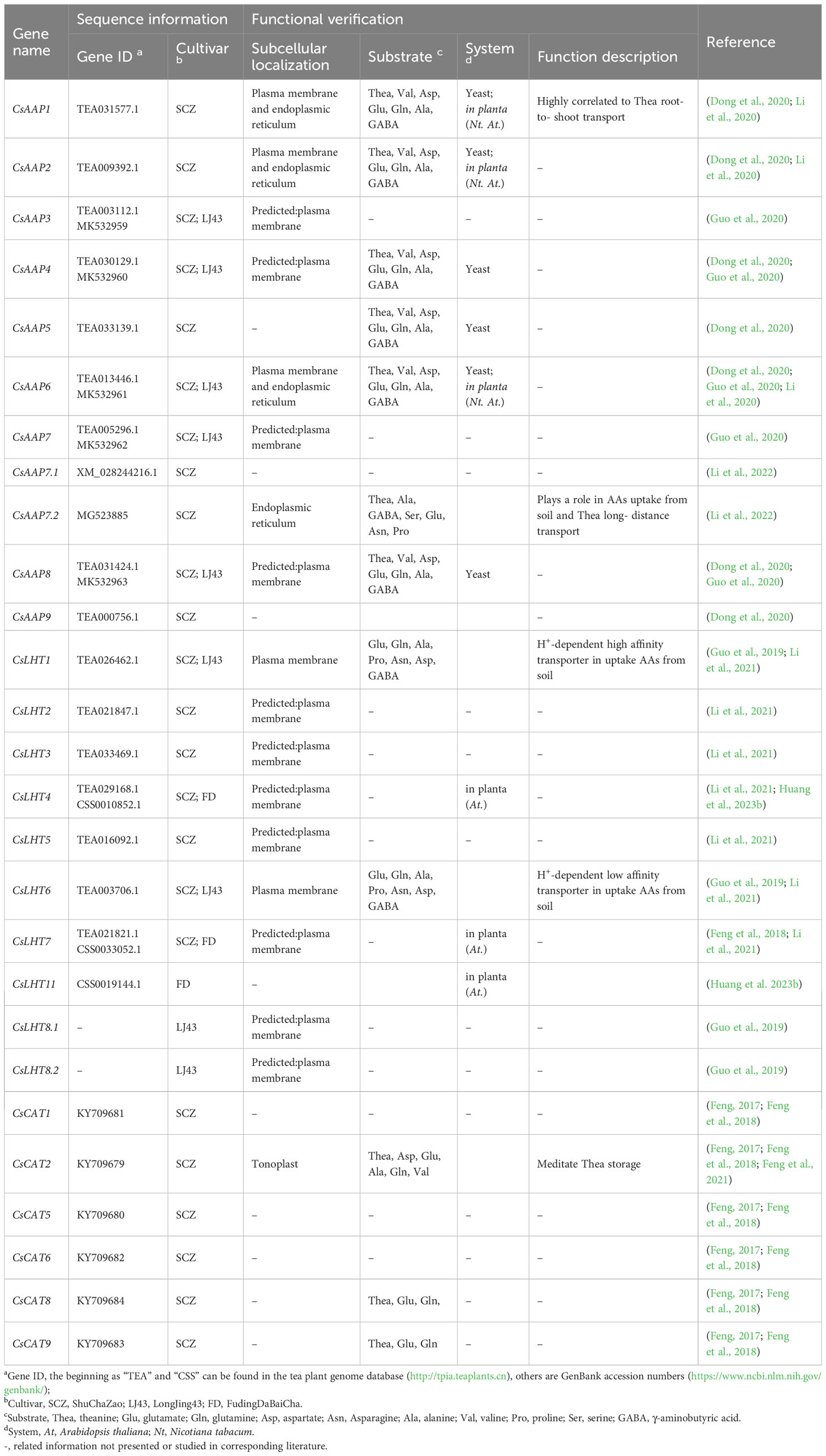
To summarize the precedent information, Table 1 lists genes involved in NH4+ and NO3− sensing, uptake, and transport in tea plants reported to date. Further information about the subcellular localization, sequence data, and functional corroboration experiments is also provided.
3.3 Amino acid- N transport
Tea plants can directly absorb organic N and transport it to actively growing parts. The amino acid theanine (Thea) is synthesized and stored in root cells and then transported from the root to the flush shoot in spring. These movements, which include xylem loading/unloading, xylem-to-phloem transfer, and post-vascular movements into the sink cells, are driven by plasmatic membrane-localized amino acid transporters (AATs) (Fischer et al., 1995; Dong et al., 2020; Lin et al., 2023a). Studies on tea plants AAT proteins have mostly focused on amino acid permeases (AAP), lysine and histidine transporters (LHT), and cationic amino acid transporters (CAT), which play important roles in AAs acquisition and long-distance transport from source to sink (Guo et al., 2019; Guo et al., 2020; Li et al., 2020; Liu, 2020).
Six CsAAPs members, CsAAP1, CsAAP2, CsAAP4, CsAAP5, CsAAP6, and CsAAP8, were identified in tea plants through the screening of a Saccharomyces cerevisiae mutant library. The expression of genes encoding these transporters was tissue-specific and regulated by the season and N levels. Thus, CsAAP1 expression in roots increased in March and decreased by mid-April and was highly correlated with root-to-bud Thea transport in seven tea cultivars (Dong et al., 2020; Li et al., 2020). Besides, shading promoted CsAAP2, CsAAP4, CsAAP5, and CsAAP8 expression in young stems and suppressed CsAAP1, CsAAP2, CsAAP4, CsAAP5, and CsAAP6 expression in the leaves, in accordance with Thea levels in these tissues. These findings indicate that CsAAP2, CsAAP4, CsAAP5, and CsAAP8 functions may be related to Thea movements in the xylem, leading to high Thea accumulation in the stem. The up-regulated genes might induce Thea transport into the companion cells in the sieve elements for phloem loading and Thea delivery to the terminal leaves (Yang et al., 2021).
LHT proteins were investigated more deeply. The CsLHTs family comprises multiple members, among which CsLHT1 and CsLHT6, highly expressed in roots, were identified as H+-dependent high- and low-affinity amino acid transporters in yeast heterologous systems. The overexpression of CsLHT1 and CsLHT6 in Arabidopsis lines significantly increased the root ability to uptake exogenous nitrogen supplied as 15N-Gln and 15N-Glu, suggesting that these transporters may contribute to the use of organic N from the soil (Guo et al., 2019; Li et al., 2021). Likewise, the heterologous expression of CsLHT4, CsLHT7, and CsLHT11 in Arabidopsis was associated with a decline in aerial parts biomass compared with WT plants, but CsLHT11 overexpressing plants had increased biomass in the rosette leaves, regardless the N levels. Therefore, this protein might have a regulatory function relevant to the development of harvestable, young shoots in tea plants (Huang et al., 2023b).
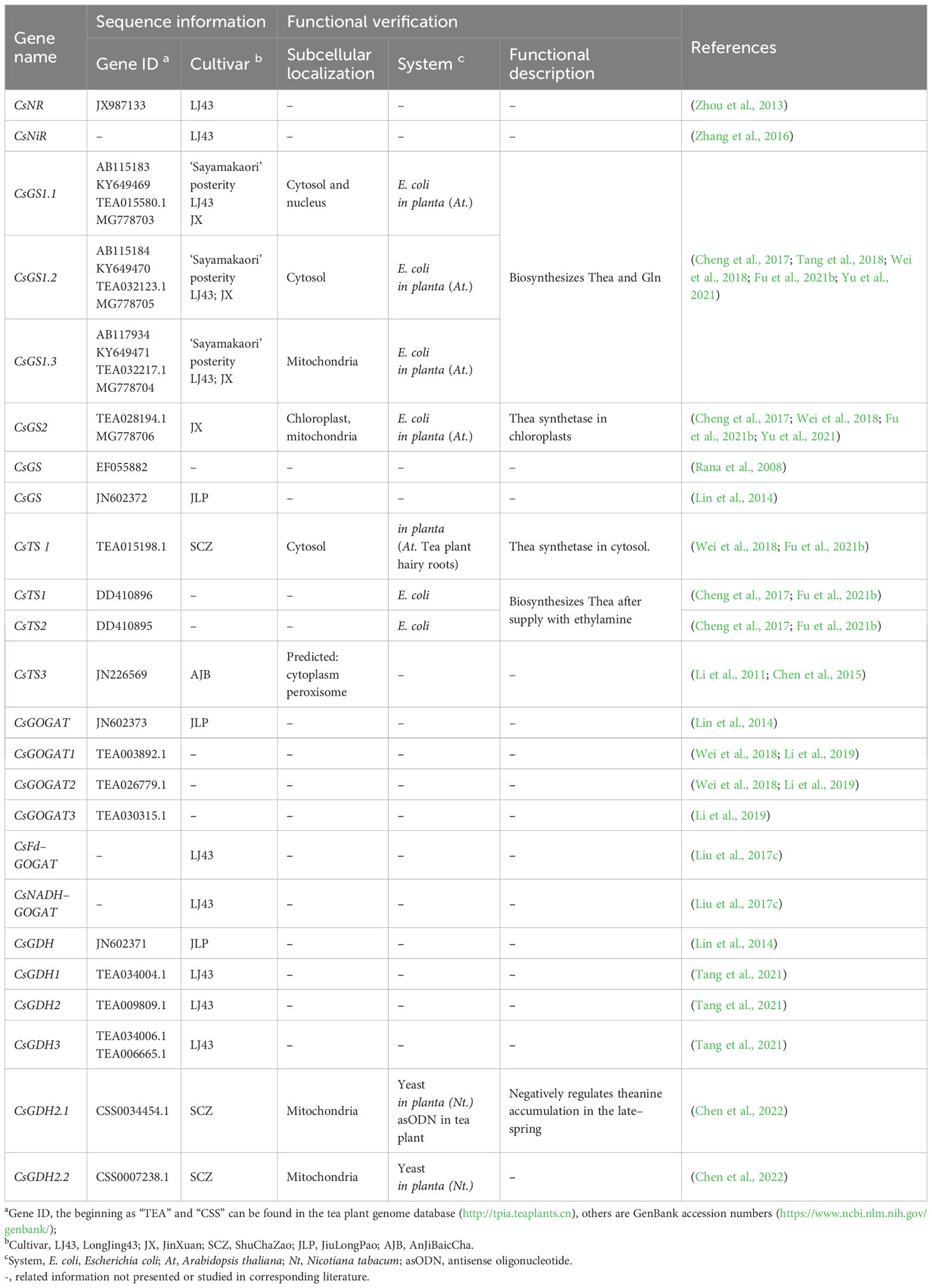
Regarding the cationic acid transporters, it was reported that the CsCAT gene family includes six members, mainly expressed in roots and stems. It was also found that some CsCATs modify their expression levels in response to abiotic stress and the exogenous application of Thea, Gln, and ethylamine hydrochloride, a precursor of Thea biosynthesis (Feng et al., 2018). CsCAT2 from tea plant was homologous to glutamine permease 1 (GNP1) from yeast, and it was found to be localized in the tonoplast as an H+-dependent amino acid transporter. CsCAT2 was highly expressed in the roots in winter, and this was negatively correlated with Thea root-to-shoot translocation, providing evidence that this transporter may meditate Thea storage in tea cell vacuoles (Feng et al., 2021). These findings enrich our understanding of N homeostasis in the form of AAs. Table 2 lists the genes involved in AAs transport in tea plants. When available, data on subcellular localization, sequencing, tea cultivars analyzed, specific substrates, and functional corroboration experiments are supplied.
4 N utilization
4.1 NO3− metabolic reduction
NO3− absorbed by plants is a nitrogen form in a highly oxidized state, which must be reduced to NH4+ through metabolic reduction to be further utilized. In this process, nitrate reductase (NR) is the rate-limiting enzyme (Jackson et al., 2008). Both NR and NiR are substrate-inducible enzymes; their function is to transfer electrons for NO3− reduction. NO3− taken up by roots was reduced into ammonium in mesophyll cells of shoots, and the metabolic reduction can also be catalyzed in roots (Miller and Cramer, 2005). In rice, the alleles of OsNR2 present differences between the two most common subspecies, indica and japonica. Thus, OsNR2 in indica rice promotes NO3− uptake through OsNRT1.1B, conferring to this subspecies increased yield and greater NUE compared with japonica rice (Gao et al., 2019).
In tea plants, studies have mainly focused on the activity and expression of CsNR and CsNiR. The activity of NR was related to NO3− content. Besides, this activity was lower in the less vigorously growing organs, such as the larger roots, older leaves and stems. In the new shoots, the in vitro NR activity decreased with the degree of leaf development, being highest in the first leaf and lowest in the fifth one (Wang and Su, 1990; Wu and Wu, 1993). NR activity was also responsive to trace elements including copper (Cu) and zinc (Zn). Foliar spraying of Cu and Zn increased the content of N-containing compounds and the activity of NR (Han and Wu, 1992). Under the same conditions, CsNR and CsNiR expression levels in tea roots were more strongly influenced by NH4+ than NO3− (Tang et al., 2020). CsNR expression in tea roots was higher than in other tissues and was up-regulated by environmental stresses (Zhou, 2014). However, the expression level of this gene significantly differed across various cultivars (Zhou et al., 2013). On the other hand, the expression of CsNiR was higher in mature leaves than in new shoots and roots, and in roots, this gene expression was up-regulated after a short period of N resupply. The change in gene expression was slower in leaves, and this responsiveness also varied in different cultivars representing diverse genotypes (Zhang et al., 2016). Most of these findings correspond to earlier experiments; the experimental evidence for CsNR and CsNiR functions in tea plants is still scarce.
4.2 Ammonia assimilation
Both NH4+ absorbed directly by plant roots and NH4+/NH3 formed through NO3- reduction can be derived to AAs synthesis using various keto acids generated through respiration; this process is known as ammonia assimilation. In higher plants, more than 95% of the NH4+/NH3 pool is assimilated via the GS-GOGAT cycle. Glutamine synthetase (GS) is the key enzyme in this pathway, playing a major role in fixing NH4+ to the δ-carboxyl group of Glu to form Gln (Thomsen et al., 2014). Tea plants have a particular ammonia-assimilation route; their roots can biosynthesize a unique amino acid, Theanine (Thea), a homolog of Gln (Lin et al., 2023a). Glutamate synthase (GOGAT) catalyzes the conversion of Gln and 2-oxoglutarate to Glu, thus providing Glu for ammonia assimilation (Bernard and Habash, 2009; Valderrama-Martín et al., 2022). When plants germinate, senesce, and begin to form seeds, glutamate dehydrogenase (GDH) can catalyze the reversible amination/deamination so that the GS-GOGAT cycle allows NH3 reuse, necessary for ammonia detoxification (Fontaine et al., 2012; Zhou et al., 2015). Through these pathways, N absorbed by roots is incorporated into proteins, nucleic acids, and other substances needed for plant growth.
4.2.1 Glutamine synthetase (GS) and theanine synthetase (TS)
Two isoforms of GS were first identified by ion exchange chromatography: cytosolic GS (GS1) and plastidic GS (GS2) (Bernard and Habash, 2009). GS1 is localized in the cytoplasm of non-photosynthetic tissues and is mainly involved in assimilating NH4+ absorbed from the soil and released from the plant N cycle. GS2 is localized in the chloroplast stroma and is the main isoform in chlorenchyma, having a major role in NH4+ assimilation within the photorespiratory pathway and NO3- reduction in plastids (Swarbreck et al., 2011; Thomsen et al., 2014).
GS has been studied in tea plants extensively. It may be noticed in the GenBank database that the Japanese researchers Tanaka and Taniguchi were the first to clone three CsGS1 genes from tea roots in 2011: CsGS1.1 (AB115183), CsGS1.2 (AB115184), and CsGS1.3 (AB117934), but the functions of these genes had not been reported (Lin et al., 2023a). Tang et al. (2018) cloned three CsGS1 genes from the leaf of the cultivar Longjing43; the information obtained from the sequence analysis showed that the 3’/5’-untranslated region differed from those obtained previously, and CsGS1.1 and CsGS1.3 were mainly expressed in roots, while CsGS1.2 was mainly expressed in mature leaves. NH4+ or NO3− supply also influences the expression levels of these genes. The expression of CsGS1.1 in leaves was up-regulated only by NO3− in a similar manner as AtGLN1.2, indicating that its role in ammonia assimilation originates from NO3− reduction (Lothier et al., 2011; Guan et al., 2014). However, under the NH4+ treatment, the expression of CsGS1.2 was induced in both leaves and roots, and CsGS1.3 expression was only significantly increased in leaves (Tang et al., 2018). Further research indicated that GS activity in tea plants was quickly inhibited upon methionine sulfoximine addition, leading to the reprogramming of AAs and nitrogenated lipids. This change involved a decrease in the biosynthesis of all other AAs and nitrogenated lipids, whereas the content of NH4+, Thea, and glycolysis and tricarboxylic acid cycle-related metabolites increased, indicating that the inhibition reduced N reutilization in the leaves (Liu et al., 2019).
L-theanine (γ-glutamyl-L-ethylamide), also known as L-Thea, is a distinctive non-proteinogenic amino acid that contributes an umami taste and exhibits anti-depression benefits (Liu et al., 2017d). Thea accumulation was dynamically regulated by developmental growth, and environmental factors, including N supply, temperature, light intensity, and salt stress (Ashihara, 2015). The synthesis of L-Thea is a unique and highly characteristic aspect of nitrogen assimilation in tea plants. Deciphering the underlying molecular mechanism of L-Thea synthesis will provide valuable guidance for fertilization and breeding strategies. Theanine synthetase (TS), an essential enzyme for Thea metabolism, catalyzes the biosynthesis of Thea from ethylamine and Glu, mainly in tea roots (Fu et al., 2021a). The structure and properties of L-Thea are similar to those of L-Gln, and some studies have confirmed that TS is highly homologous to GS (Cheng et al., 2017). As indicated before, CsTS1 (DD410895) and CsTS2 (DD410896) were firstly isolated through cDNA library screening. CsTS1 is mainly expressed in the new shoots, roots, and mature leaves, while CsTS2 reached higher expression levels in shoots (Deng et al., 2008). Both genes are involved in Thea biosynthesis; this was validated through a heterologous expression system (Lin et al., 2023a).
By performing genome studies, Wei et al. (Wei et al., 2018) found that the predicted CsGSⅠ sequence shared high homology with that of PtGS (Pseudomonas taetrolens), and PtGSI has been engineered for Thea production at high levels, for which CsGSⅠ was renamed as CsTSⅠ. The function of CsTS and CsGS was investigated in depth through the transient overexpression in Nicotiana benthamiana leaves or the stable expression in Arabidopsis and knockdown in tea plants. The expression pattern and distribution of CsTSⅠ correlated with Thea and Gln contents in different tissues. CsTS I mainly accumulated in root tip epidermal, pericycle, and procambium cells to form cytoplasmic proteins. When fed with 10 mM ethylamine, CsTSI-overexpressing Arabidopsis seedlings showed a significantly higher Thea content than wild-type seedlings. Further research allowed the construction of CsTSI RNAi and CsTSI overexpressing chimerical tea seedlings with transgenic hair roots; the results demonstrated that the content of Thea decreased and that of Gln increased, thus proving that CsTSI biosynthesized Gln and Thea used glutamate as an acceptor and ammonium or ethylamine as a donor, respectively (Wei et al., 2018; Fu et al., 2021b; She et al., 2022). Fu et al. (2021b) used a non-aqueous fractionation method and could determine that, in roots, L-Thea biosynthesis mainly occurred in the cytosol through the action of the key and cytosolic enzyme L-Thea synthetase CsTSI, whereas in shoots, both the cytosol and chloroplasts were the major sites for L-Thea biosynthesis, and CsGS1.1 and CsGS2 were, most likely, the fundamental L-theanine synthetase. CsGS2 was identified as a key enzyme regulating Thea biosynthesis in chloroplasts, L-Thea content and distribution in leaf tissues would be affected by light, as long-term shading treatment led to a decrease in the proportion of L-Thea in the plastids by reducing CsGS2 expression levels. Thus, new shoots could accumulate more L-Thea. In contrast, CsGS1.2 expression in albino new shoots was higher than that found in common cultivars as a way to compensate for the low CsGS2 expression in undeveloped chloroplasts. These findings indicate that the mechanism underlying Thea synthesis might differ across tea genotypes (Yu et al., 2021).
4.2.2 Glutamate synthase (GOGAT) and glutamate dehydrogenase (GDH)
There are two isoforms of GOGAT in plants, with different functions: ferredoxin-dependent GOGAT (Fd-GOGAT) and nicotinamide adenine dinucleotide-dependent GOGAT (NADH-GOGAT). Fd-GOGAT assimilates ammonia through photorespiration in leaves, while NADH-GOGAT accumulates in non-green tissues, playing a role in ammonia assimilation in root (Suzuki and Knaff, 2005; Konishi et al., 2014).
GDH is abundant in plant tissues; this enzyme catalyzes ammonia conversion to Glu and also deaminates Glu to α-ketoglutarate. GDH-mediated ammonia assimilation and as a stress-responsive enzyme, GDH detoxified the intracellular high ammonia and biosynthesize Glu (Lea and Miflin, 2003; Fontaine et al., 2012; Zhou et al., 2015). CsGOGAT was found to have significantly higher expression in the leaf than in the root (Chen et al., 2015). Under N starvation, CsGOGAT expression increased, and CsGDH expression decreased significantly; these changes were correlated with leaf N content (Lin et al., 2014). CsGOGAT also have a regulatory role in AAs changes in postharvest tea plant leaves. The Thea content changed in spreading tea leaves under different treatments, and CsGOGAT was involved in Thea metabolic pathway, regardless of external light and temperature. Also, CsGOGAT would interact with CsTS I and CsNiR during N metabolism (Liu et al., 2017c). In tea plant, all CsGDHs identified to date belong to the NADH-GDH group. Accumulation of CsGDH2 transcripts seemed to be flower-specific compared with the other five plant tissues analyzed; CsGDH1 was mainly expressed in mature leaves and roots, and CsGDH3 in new shoots and roots. Under high NH4+ supply, CsGS inhibition resulted in a significant up-regulation of CsGDH3 and CsGDH2 in roots and leaves, indicating the synergistic effect of CsGSs and CsGDHs in the process of ammonia assimilation (Tang et al., 2021). The expression of CsGDH2.1 in shoots increased greatly in the late spring; further investigation revealed that Glu was a signal for Thea hydrolysis, and CsGDH2.1-mediated Glu catabolism negatively regulated Thea accumulation in the new shoots in the late spring, improving green quality by targeting to reduce CsGDH2.1 expression (Chen et al., 2022).
Summing up, studies directed to analyze the genes related to N metabolism in tea plants mostly focused on their function in regulating AAs biosynthesis. Knowledge about the functioning and regulation of the enzymes involved in these processes is mostly based on transcript analyses. There are still many gaps in our understanding of their functions, especially for NR, NiR, and GOGAT, concerning NO3− reduction, N assimilation, remobilization, and reassimilation of photorespiratory NH3. It is noteworthy that, apart from the transcriptional regulation, post-translational modifications (PTMs) can also be critical for the regulation of many proteins relevant to N metabolism in plants (Liu et al., 2022). Therefore, more detailed studies will deepen our understanding of NUE determinants and allow further optimization of NUE under actual tea garden production scenarios.
Genes involved in ammonia assimilation by tea plants and their most relevant data are shown in Table 3.
5 An overview of nitrogen use efficiency assessment by tea plants
Nitrogen use efficiency (NUE) is a complex trait influenced by the interaction between environmental factors and intrinsic plant factors; this variable can be approached at different levels and calculated in different ways (Santa-María et al., 2015). Initially, NUE was defined as the crop yield per unit of applied N, a parameter also termed partial fertilizer productivity (PFP) (Moll et al., 1982). Under specific N supply conditions, NUE can be divided into two components: nitrogen uptake efficiency (NUpE) and nitrogen utilization efficiency (NUtE) or nitrogen physiological efficiency (NPE). NUpE may be defined as the total amount N absorbed and NUtE as the dry weight or grain yield per unit of absorbed N, accounting for the results at this growth stage (Williams et al., 2021). Tea germplasm resources are abundant in China; the genetic diversity of this plant, resulting from a long time of artificial domestication and cultivar-breeding improvement, has determined quite different N requirements (Zhang et al., 2018). Additionally, because tea production does not target grain yield, dissimilar NUE assessment criteria were adopted. Here, we integrate the results of several studies and present four approaches to assess NUE by tea plants.
5.1 Biomass accumulation
By the end of the 20th century, it was reported that the rate of increase in tea ground stem diameter and height and dry matter production in different cultivars varied under sufficient N supply compared to no N application (Ruan et al., 1993). Under low N supply, tea plants’ height, root and shoot dry weight, and leaf SPAD values were significantly decreased (Wang et al., 2015). Wang et al. (2004) measured the added-N content in the biomass and the growth of new shoots in six tea cultivars under four N levels (based on 15N isotope labeling techniques), and redefined five interdependent traits—nitrogen use efficiency (NE), nitrogen uptake efficiency (NUE), nitrogen physiological utilization efficiency (NPE), nitrogen economic efficiency (NEE) and N responsiveness—according to growth characters and harvesting organs. They found that the biomass increase was significantly correlated with NEE, the weight of the new shoots was significantly (positively) correlated with NE, NUE, and NEE, while NUE was the main determinant of NE. These authors indicated that by comparing the NE values of different cultivars, it is possible to detect which cultivar can achieve the highest NUE for a given level of N supply.
5.2 Root-related traits
The root is the main organ for nutrient uptake and plays a direct role in N acquisition (Lynch, 2007; Zhu et al., 2011). Root development and activity are responsive to soil N levels (Ju et al., 2015). Studies on plant response to N concentration gradients using different tea cultivars suggested that N concentration has a significant effect on root/shoot ratio, and this ratio could be used as a screening index to detect low-N-tolerant cultivars (Wang et al., 2015). On the other hand, the differences among cultivars in root-related parameters such as root dry weight, root volume, or root active uptake area were greater than those of root activity. Likewise, root volume and active uptake area varied significantly across N levels. Further correlation studies provided evidence that these parameters may be considered as promising indices for selecting and breeding tea cultivars with high NUE (Wang et al., 2005).
5.3 NH4+ influx kinetics
In the early 1950s, Epstein and Hagen (1952) applied the Michaelis-Menten equation for the first time to describe the absorption process of ionic nutrients by plants. In this equation, Vmax represents the maximum uptake rate; this value is directly proportional to the uptake rate for ions, and Km is inversely proportional to the affinity of the cell membrane for nutrient ions (Zhang et al., 2018). Because tea roots show a preference for NH4+ uptake as the nitrogen source, the kinetic parameters of this cation are usually used to define tea adaptability to N availability. According to current studies on NH4+ dynamics, tea cultivars may be classified into three categories: (1) cultivars with high Km and high Vmax can produce high yields in soils with elevated N contents; TeiGuanYin, HuangDan, and Yubukita cultivars belong to this type; (2) cultivars with low Km and low Vmax may display a good performance in soils with low N concentrations; YingShuang and MaoXie belong to this type; (3) cultivars with high Km and low Vmax are the most flexible concerning N levels, being appropriate for both high and low N conditions; ZhongCha#302 and FuDingDaBaiCha belong to this type (Wang et al., 2005; Liu, 2016; Zhang et al., 2018; Zhang et al., 2022b). Notably, N flux was calculated as the N content in the roots based on 15N labeling in most studies, and there are still many cultivars falling into different groups in different studies due to different number of tested cultivars and methodological approaches. Though NH4+ influx kinetics allowed a better understanding of N use by tea plants, more precise methods, such as non-invasive procedures based on micro-test technology, will be useful for future experiments (Ruan et al., 2016; Su et al., 2020).
5.4 Activity and gene expression of N-assimilation-related enzymes
The leaves are the main assimilation organs of inorganic N. The accumulation of N-assimilates and the enzymes and genes regulating these metabolic processes could indicate NUE-related sub-traits (Sun et al., 2019). Some studies showed that GS activity varied among cultivars and N levels and was positively correlated with N assimilation rate and NUpE (Wang et al., 2005; Du et al., 2015). Lin et al. (2017; 2018) examined the activity of some antioxidant enzymes and found increased activities in the low N-tolerant cultivar HuangDan in a nitrogen-deficient environment. This was linked to the maintenance of high photosynthetic rates and to the adequate output of N-assimilates in the leaves. Still, by combining genes, enzymes, and assimilates and exploring their affiliation links, it was possible to evaluate NUE traits comprehensively. Zhou (2012) measured soluble sugars, soluble proteins, total N content, N-related enzymatic activities, and the expression of AAs biosynthetic genes. Their results suggested that the differences in these indicators varied in the five cultivars tested as the N concentration increased and membership function could be used to evaluated the NUE of each cultivar synthetically. Also, CsAMTs expression profiles in response to NH4+ differed among cultivars (Zhang et al., 2018; Zhang et al., 2022b). Still, it was possible to detect that CsNRT2 participated in NO3− transport under low N conditions (Hu et al., 2023; Lin et al., 2023b). The AuTophaGy-related genes CsATG8e and CsATG3a were linked to an improved plant ability for N recycling and tolerance to low N levels (Huang et al., 2020; Huang et al., 2023a). These genes emerge as promising indicators and may contribute to identifying higher NUE among various germplasm resources.
Although multiple investigations have addressed NUE of tea plants, most NUE-related traits were identified based on individual morphology, physiological processes, relevant biochemical components, or gene expression patterns. Nevertheless, there are no universal standards for grading NUE in tea plants, and some cultivars have shown heterogeneous results. The measurement of biomass is time-consuming and susceptible to environmental changes. And only the processes of N uptake, transport or utilization not the comprehensive NUE have been measured in tea plants. The practicability of method also depends on the number of tested cultivars. Most importantly, NUE estimates are complicated and current evaluation methods are not comprehensive enough to cover and explain the meaning of NUE. The methodological limitations still resist our understanding of N metabolic mechanisms. Therefore, analyses combining omics data and molecular and genetic approaches will be useful to elucidate further heritability and inheritance in this species with a point of great value to improve NUE by tea plants.
6 Conclusions and perspectives
N is the driving factor for tea yield and quality. Facing the practical problem of the disproportionate amount of N fertilizers applied and the low N utilization rate by tea plantations, a comprehensive study on the process of N transport, absorption, and utilization is necessary to increase NUE, to improve quality features such as aroma and flavor, and, ultimately, to promote the sustainable development of the industry.
Currently, it is clear that tea plants show a preferential uptake and assimilation of NH4+ over NO3−, and more NH4+ availability allows tea plants to produce more AAs, which further act as signaling molecules involved in other metabolic pathways. In addition, great progress has been made in the elucidation of the N primary metabolism network. Genes contributing to N transport and assimilation have been cloned and sequenced, and the functions of many genes have been identified by transgenic experiments in yeast, Arabidopsis, and Nicotiana tabacum. However, the current methods to assess tea NUE under actual productive settings have limitations. For instance, some basic indices related to plant physiological performance and gene expression were proposed, but these approaches are time-consuming and inappropriate for large-scale field cultivar assessment. One drawback is that a stable transgenic system has not been established yet; hence, we cannot knock out or overexpress genes to provide functional evidence in homologous systems. Therefore, there is an urgent need to develop an efficient and stable gene transformation system for tea plants, even more considering that the N metabolism network is regulated by multiple genes. Future research should consider the following issues.
Firstly, most research on N uptake and utilization by tea plants has focused on ammonia assimilation and AAs biosynthesis. However, NH4+-based fertilizers and urea are widely applied in tea gardens, and these N forms are expected to be converted to NO3− by nitrification, entailing the risk of leaching. It has been reported that NO3- was the main chemical form of N loss by leaching: about 51%-63% of the added N is lost in this way (Zheng, 2022). Therefore, the biological significance of NRT, NR, and NiR in N utilization is not negligible. In rice, the nitrate sensor NRT1.1B could perceive NO3− signal at the plasma membrane and facilitated SPX4 degradation by recruiting NBIP1, resulting in the cytoplasm-to-nuclear shuttling of OsNLP3 to transduce NO3− response (Hu et al., 2019). Also, in Arabidopsis, the phosphorylation state of NRT1.1 regulates the nitrate signaling for lateral root growth, and the non-phosphorylable NRT1.1T101A would activate Ca2+-CPKs-NLPs signaling pathway by inducing its endocytosis under high NO3− concentration (Zhang et al., 2019a).
Secondly, although significant progress has been made in recent years regarding our understanding of the transcriptional regulation of the GS-GOGAT cycle, there are few reports on how transcription factors (TFs) regulate the expression of these downstream genes. The latest research revealed that the lateral organ boundaries domain gene CsLBD39 negatively regulated NO3− transduction (Teng et al., 2022). Functional studies on the regulation of N metabolism by TFs need to be further expanded in both scope and depth. Additionally, PTMs also influence NUE through their effects on relevant proteins in plants. Phosphorylation and dephosphorylation of NR are involved in regulating NR activity, and phosphorylation, oxidation, tyrosine nitration, and S-nitrosylation of GS protein are also key mechanisms for GS function in many crops, including wheat, rice, and maize (Liu et al., 2022). A recent study in tea plants found that CsALT, CsTSI, CsGS, and CsAlaDC, proteins involved in Thea synthesis, were modified through ubiquitination, implying that these enzymes’ stabilities were regulated by this modification (Wang et al., 2021b). Consequently, to establish a comprehensive N mechanism network for tea plant, N transport, reduction and assimilation requires precise regulation at both the transcriptional and post translational levels, many efforts need to be made to explore the PTMs, particularly to identify the modification sites that may be relevant for N use regulation by tea plants.
Furthermore, plants can respond to changes in N uptake by adjusting leaf expansion and photosynthetic rates, as well as chlorophyll content. In senescent leaves, N assimilation decreased; this was associated with the degradation of proteins and nucleic acids; the released N was remobilized to developing tissues. The expression of genes related to GS/GOGAT cycle during leaf senescence was widely investigated; most of these genes were expressed in phloem companion and parenchyma cells in cereals, suggesting that GS/GOGAT cycle plays a vital role in N remobilization from senescent organs to developing organs (Havé et al., 2017; Liu et al., 2022). In addition, NH4+, NO3−, AAs, and peptide transporters also can be up- or down-regulated during leaf senescence. Thus, many aspects of N metabolic pathways would be influenced by N recycling and remobilization (Breeze et al., 2011). It is reasonable to hypothesize that there are some other undiscovered factors and pathways, for example, components of C metabolism that regulate N remobilization. Tea production involves the pruning and picking of the tender leaves every season; this leads to a more active N turnover between the senescent leaves and the new shoots. Hence, for tea plants, an overview of the mechanisms involved in N recycling and remobilization is important to improve N resorption efficiency and also to reduce the use of N chemical fertilizers, which are responsible for a large part of greenhouse gas emissions.
Finally, along with the deciphering of the tea genome in multiple cultivars (Wang et al., 2021a), whole genome resequencing could provide more efficient single nucleotide polymorphisms (SNPs) markers to construct a high-density linkage map of tea populations. Such maps will lay a foundation for further investigations of quantitative trait loci (QTL) mapping and genome-wide association studies (GWAS) in order to reveal the molecular basis for important agronomic traits. In rice, forward genetics approaches revealed that allelic variation at OsNR2 and OsNRT1.1B resulted in the nitrate-use divergence between indica and japonica subspecies and were used to improve the NUE of rice (Hu et al., 2015; Gao et al., 2019). Multiple attempts have been made to detect relevant QTLs or variation sites and quality-related traits in tea plants, including biochemical components, leaf area (An et al., 2021), seed setting rate (Wei et al., 2021), bud flush timing (Tan et al., 2022), and AAs (Huang et al., 2022), caffeine (Ma et al., 2018), and flavonoid (Xu et al., 2018) contents. However, fewer attempts to unravel nutrient uptake and utilization traits in the context of genotype-to-phenotype mapping research have been reported. Nutrient-related traits are generally regulated by multiple genes and environmental factors, so it is difficult to quantify their phenotypes precisely. More attempts need to reveal the processes of N cycling, and to define the phenotypic indicators that reflect each step of N metabolism. For example, chlorate (ClO3−) is an analogic tracer for NO3− and the resistance ability to ClO3− is an efficient indicator for fast screening the process of NO3− transport and reduction divergency (Hu et al., 2015). How to apply this method in woody plants is a challenge that needs to be considered in future research. Population genetics can help us to explore better the gene regulatory loci affecting NUE-related traits and to identify the TFs or promoters which are able to regulate or activate the transcription of downstream structural genes. Exploiting interpopulation genetic variation in different germplasms will be instrumental for cultivar improvement. Thus, the use of precise phenotyping methods on population is challenging but necessary for future studies of discovering genetic variation associated with NUE-related traits.
Therefore, future studies should focus on the regulation mechanisms of NO3− uptake and reduction in tea plants to increase the utilization of NO3− from the soils and reduce leaching losses, a point of great significance for the genetic improvement directed to high NUE cultivars as well as for developing a sustainable tea plantations.
Author contributions
WZ and LL contributed to the conceptualization. WZ prepared the first draft and figures. LL and KN contributed with inputs and made revisions in the text and figures. LL and JR supervised the overall process. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-ASTIP-TRICAAS) and Zhejiang Provincial Natural Science Foundation of China under Grant No. LY23C150008.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
An, Y., Chen, L., Tao, L., Liu, S., Wei, C. (2021). QTL mapping for leaf area of tea plants (Camellia sinensis) based on a high-quality genetic map constructed by whole genome resequencing. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.705285
Ashihara, H. (2015). Occurrence, biosynthesis and metabolism of theanine (γ-glutamyl-L-ethylamide) in plants: a comprehensive review. Nat. Prod. Commun. 10, 803–810. doi: 10.1177/1934578X15010005
Beatty, P. H., Good, A. G. (2018). “Improving nitrogen use efficient in crop plants using biotechnology approaches,” in Engineering nitrogen utilization in crop plants. Eds. Shrawat, A., Zayed, A., Lightfoot, D. A. (Cham: Springer International Publishing), 15–35.
Bernard, S. M., Habash, D. Z. (2009). The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New. Phytol. 182, 608–620. doi: 10.1111/j.1469-8137.2009.02823.x
Bodirsky, B. L., Popp, A., Lotze-Campen, H., Dietrich, J. P., Rolinski, S., Weindl, I., et al. (2014). Reactive nitrogen requirements to feed the world in 2050 and potential to mitigate nitrogen pollution. Nat. Commun. 5, 3858. doi: 10.1038/ncomms4858
Breeze, E., Harrison, E., McHattie, S., Hughes, L., Hickman, R., Hill, C., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell. 23, 873–894. doi: 10.1105/tpc.111.083345
Camargo, J. A., Alonso, A., Salamanca, A. (2005). Nitrate toxicity to aquatic animals: a review with new data for freshwater invertebrates. Chemosphere 58, 1255–1267. doi: 10.1016/j.chemosphere.2004.10.044
Chen, L., Chen, J., Wang, N., Zhang, X. (2019). The role of plasma membrane H+-ATPase on nitrogen-regulated phosphorus uptake in tea plants. J. Tea Sci. 39, 723–730. doi: 10.13305/j.cnki.jts.2019.06.011
Chen, T., Lin, S., Chen, Z., Yang, T., Zhang, S., Zhang, J., et al. (2023). Theanine, a tea-plant-specific non-proteinogenic amino acid, is involved in the regulation of lateral root development in response to nitrogen status. Hortic. Res. 10, uhac267. doi: 10.1093/hr/uhac267
Chen, T., Ma, J., Li, H., Lin, S., Dong, C., Xie, Y., et al. (2022). CsGDH2.1 negatively regulates theanine accumulation in the late-spring tea plants (Camellia sinensis var. sinensis). Hortic. Res. 10, uhac245. doi: 10.1093/hr/uhac245
Chen, Q., Meng, X., Jiang, X., Yu, S., Wan, X. (2015). Tissue specificity expression analysis of theanine metabolism pathway related genes in Camellia Sinensis. J. Nucl. Agric. Sci. 29, 1285–1291.
Chen, Y., Wang, F., Wu, Z., Jiang, F., Yu, W., Yang, J., et al. (2021). Effects of long-term nitrogen fertilization on the formation of metabolites related to tea quality in subtropical China. Metabolites 11, 146. doi: 10.3390/metabo11030146
Chen, J., Wang, P., Xia, Y., Xu, M., Pei, S. (2005). Genetic diversity and differentiation of Camellia sinensis L. (cultivated tea) and its wild relatives in Yunnan province of China, revealed by morphology, biochemistry and allozyme studies. Genet. Resour. Crop Evol. 52, 41–52. doi: 10.1007/s10722-005-0285-1
Cheng, S., Fu, X., Wang, X., Liao, Y., Zeng, L., Dong, F., et al. (2017). Studies on the biochemical formation pathway of the amino acid L-Theanine in tea (Camellia sinensis) and other plants. J. Agric. Food Chem. 65, 7210–7216. doi: 10.1021/acs.jafc.7b02437
Chopin, F., Orsel, M., Dorbe, M. F., Chardon, F., Truong, H.-N., Miller, A. J., et al. (2007). The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell 19, 1590–1602. doi: 10.1105/tpc.107.050542
Couturier, J., Montanini, B., Martin, F., Brun, A., Blaudez, D., Chalot, M. (2007). The expanded family of ammonium transporters in the perennial poplar plant. New Phytol. 174, 137–150. doi: 10.1111/j.1469-8137.2007.01992.x
Crawford, N. M., Glass, A. D. M. (1998). Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 3, 389–395. doi: 10.1016/S1360-1385(98)01311-9
Deng, W.-W., Ogita, S., Ashihara, H. (2008). Biosynthesis of theanine (γ-ethylamino-l-glutamic acid) in seedlings of Camellia sinensis. Phytochem. Lett. 1, 115–119. doi: 10.1016/j.phytol.2008.06.002
Deng, M., Xu, Z., Hu, L., Yao, Y., Xu, M., Pi, L. (2012). Effect of different levels of nitrogen application on yield and quality of FudingDabaicha. Southwest Chin. J. Agric. Sci. 25, 1330–1333. doi: 10.16213/j.cnki.scjas.2012.04.051
Dong, F., Hu, J., Shi, Y., Liu, M., Zhang, Q., Ruan, J. (2019). Effects of nitrogen supply on flavonol glycoside biosynthesis and accumulation in tea leaves (Camellia sinensis). Plant Physiol. Biochem. 138, 48–57. doi: 10.1016/j.plaphy.2019.02.017
Dong, C., Li, F., Yang, T., Feng, L., Zhang, S., Li, F., et al. (2020). Theanine transporters identified in tea plants (Camellia sinensis L.). Plant J. 101, 57–70. doi: 10.1111/tpj.14517
Du, X., Peng, F., Jiang, J., Tan, P., Wu, Z. Z., Liang, Y. W., et al. (2015). Inorganic nitrogen fertilizers induce changes in ammonium assimilation and gas exchange in Camellia sinensis L. Turk. J. Agric. For. 39, 28–38. doi: 10.3906/TAR-1311-82
Epstein, E., Hagen, C. E. (1952). A kinetic study of the absorption of alkali cations by barley roots. Plant Physiol. 27, 457–474. doi: 10.1104/pp.27.3.457
Fan, K., Zhang, Q., Liu, M., Ma, L., Shi, Y., Ruan, J. (2019). Metabolomic and transcriptional analyses reveal the mechanism of C, N allocation from source leaf to flower in tea plant (Camellia sinensis. L). J. Plant Physiol. 232, 200–208. doi: 10.1016/j.jplph.2018.11.007
Feng, S. (2014). “Cloning an expressing of the nitrogen transporter Gene NRT1.2, NRT1.5 and NRT2.5 in tea plant,” in Dissertation (Beijing: Chinese Academy of Agricultural Sciences). Chinese dissertation
Feng, L. (2017). “Cloning and functional analysis of CAT amino acid transporter genes in tea plant (Camellia sinensis L.),” in Dissertation (Hefei, Anhui Province: Anhui Agricultural University).
Feng, S., Wang, L., Chen, C., Lin, Z., Cheng, H., Wei, K., et al. (2014). Cloning and expressing analysis of a nitrogen transporter 2.5 gene from tea plant [Camellia sinensis (L.)]. J. Tea Sci. 34, 364–370.
Feng, L., Yang, T., Zhang, Z., Li, F., Chen, Q., Sun, J., et al. (2018). Identification and characterization of cationic amino acid transporters (CATs) in tea plant (Camellia sinensis). Plant Growth Regul. 84, 57–69. doi: 10.1007/s10725-017-0321-0
Feng, L., Yu, Y., Lin, S., Yang, T., Chen, Q., Liu, L., et al. (2021). Tonoplast-localized theanine transporter CsCAT2 may mediate theanine storage in the root of tea plants (Camellia sinensis L.). Front. Plant Sci. 12. doi: 10.3389/fpls.2021.797854
Fischer, W.-N., Kwart, M., Hummel, S., Frommer, W. B. (1995). Substrate specificity and expression profile of amino acid transporters (AAPs) in Arabidopsis*. J. Biol. Chem. 270, 16315–16320. doi: 10.1074/jbc.270.27.16315
Fontaine, J.-X., Tercé-Laforgue, T., Armengaud, P., Clément, G., Renou, J. P., Pelletier, S., et al. (2012). Characterization of a NADH-dependent glutamate dehydrogenase mutant of Arabidopsis demonstrates the key role of this enzyme in root carbon and nitrogen metabolism. Plant Cell 24, 4044–4065. doi: 10.1105/tpc.112.103689
Forde, B. G. (2000). Nitrate transporters in plants: structure, function and regulation. BBA - Biomembranes 1465, 219–235. doi: 10.1016/S0005-2736(00)00140-1
Fu, X., Liao, Y., Cheng, S., Deng, R., Yang, Z. (2021a). Stable isotope-labeled precursor tracing reveals that L-alanine is converted to L-theanine via L-glutamate not ethylamine in tea plants In Vivo. J. Agric. Food Chem. 69, 15354–15361. doi: 10.1021/acs.jafc.1c06660
Fu, X., Liao, Y., Cheng, S., Xu, X., Grierson, D., Yang, Z. (2021b). Nonaqueous fractionation and overexpression of fluorescent-tagged enzymes reveals the subcellular sites of L-theanine biosynthesis in tea. Plant Biotechnol. J. 19, 98–108. doi: 10.1111/pbi.13445
Gao, Z., Wang, Y., Chen, G., Zhang, A., Yang, S., Shang, L., et al. (2019). The indica nitrate reductase gene OsNR2 allele enhances rice yield potential and nitrogen use efficiency. Nat. Commun. 10, 5207. doi: 10.1038/s41467-019-13110-8
Giehl, R. F. H., Laginha, A. M., Duan, F., Rentsch, D., Yuan, L., von Wirén, N. (2017). A critical role of AMT2;1 in root-to-shoot translocation of ammonium in Arabidopsis. Mol. Plant 10, 1449–1460. doi: 10.1016/j.molp.2017.10.001
Godfray, H. C. J., Beddington, J. R., Crute, I. R., Haddad, L., Lawrence, D., Muir, J. F., et al. (2010). Food security: the challenge of feeding 9 billion people. Science 327, 812–818. doi: 10.1126/science.1185383
Guan, P., Wang, R., Nacry, P., Breton, G., Kay, S. A., Pruneda-Paz, J. L., et al. (2014). Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 111, 15267–15272. doi: 10.1073/pnas.1411375111
Guo, L., Zhang, F., Cheng, H., Wei, K., Ruan, L., Wu, L., et al. (2020). Molecular cloning and expression analysis of CsAAPs gene subfamily in Camellia Sinensis. J. Tea Sci. 40, 454–464. doi: 10.13305/j.cnki.jts.20200612.002
Guo, L., Zhang, F., Zhang, Y., Cheng, H., Wei, K., Ruan, L., et al. (2019). Molecular cloning and expression analysis of CsLHTs gene subfamily in tea plants (Camellia sinensis). J. Tea Sci. 39, 280–288. doi: 10.13305/j.cnki.jts.2019.03.005
Han, W., Wu, X. (1992). Effect of copper and zinc on nitrate reductase activity in tea plant. J. Tea Sci. 12, 159–160. doi: 10.13305/j.cnki.jts.1992.02.010
Havé, M., Marmagne, A., Chardon, F., Masclaux-Daubresse, C. (2017). Nitrogen remobilization during leaf senescence: lessons from Arabidopsis to crops. J. Exp. Bot. 68, 2513–2529. doi: 10.1093/jxb/erw365
Howitt, S. M., Udvardi, M. K. (2000). Structure, function and regulation of ammonium transporters in plants. BBA - Biomembranes 1465, 152–170. doi: 10.1016/S0005-2736(00)00136-X
Hu, S., Hu, Y., Mei, H., Li, J., Xuan, W., Jeyaraj, A., et al. (2023). Genome-wide analysis of long non-coding RNAs (lncRNAs) in tea plants (Camellia sinensis) lateral roots in response to nitrogen application. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1080427
Hu, B., Jiang, Z., Wang, W., Qiu, Y., Zhang, Z., Liu, Y., et al. (2019). Nitrate–NRT1.1B–SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants 5, 401–413. doi: 10.1038/s41477-019-0384-1
Hu, B., Wang, W., Ou, S., Tang, J., Li, H., Che, R., et al. (2015). Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47, 834–838. doi: 10.1038/ng.3337
Huang, W., Ma, D. N., Liu, H. L., Luo, J., Wang, P., Wang, M. L., et al. (2020). Genome-wide identification of CsATGs in tea plant and the involvement of CsATG8e in nitrogen utilization. Int. J. Mol. Sci. 21, 7043. doi: 10.3390/ijms21197043
Huang, W., Ma, D., Xia, L., Zhang, E., Wang, P., Wang, M., et al. (2023a). Overexpression of CsATG3a improves tolerance to nitrogen deficiency and increases nitrogen use efficiency in arabidopsis. Plant Physiol. Biochem. 196, 328–338. doi: 10.1016/j.plaphy.2023.01.057
Huang, W., Ma, D., Zaman, F., Hao, X., Xia, L., Zhang, E., et al. (2023b). Identification of the lysine and histidine transporter family in Camellia sinensis and the characterizations in nitrogen utilization. Hortic. Plant J. doi: 10.1016/j.hpj.2023.01.009
Huang, R., Wang, J. Y., Yao, M. Z., Ma, C. L., Chen, L. (2022). Quantitative trait loci mapping for free amino acid content using an albino population and SNP markers provides insight into the genetic improvement of tea plants. Hortic. Res. 9, uhab029. doi: 10.1093/hr/uhab029
Huang, H., Yao, Q., Xia, E., Gao, L. (2018). Metabolomics and transcriptomics analyses reveal nitrogen influences on the accumulation of flavonoids and amino acids in young shoots of tea plant (Camellia sinensis L.) associated with tea flavor. J. Agric. Food Chem. 66, 9828–9838. doi: 10.1021/acs.jafc.8b01995
Jackson, L. E., Burger, M., Cavagnaro, T. R. (2008). Roots, nitrogen transformations, and ecosystem services. Annu. Rev. Plant Biol. 59, 341–363. doi: 10.1146/annurev.arplant.59.032607.092932
Jentsch, T. (2008). CLC chloride channels and transporters: from genes to protein structure, pathology and physiology. Crit. Rev. Biochem. Mol. 43, 3–36. doi: 10.1080/10409230701829110
Jiang, J., Gai, Z., Wang, Y., Fan, K., Sun, L., Wang, H., et al. (2018). Comprehensive proteome analyses of lysine acetylation in tea leaves by sensing nitrogen nutrition. BMC Genom. 19, 840. doi: 10.1186/s12864-018-5250-4
Ju, C., Buresh, R. J., Wang, Z., Zhang, H., Liu, L., Yang, J., et al. (2015). Root and shoot traits for rice varieties with higher grain yield and higher nitrogen use efficiency at lower nitrogen rates application. Field Crop Res. 175, 47–55. doi: 10.1016/j.fcr.2015.02.007
Kiba, T., Feria-Bourrellier, A.-B., Lafouge, F., Lezhneva, L., Boutet-Mercey, S., Orsel, M., et al. (2012). The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell 24, 245–258. doi: 10.1105/tpc.111.092221
Kobae, Y., Tamura, Y., Takai, S., Banba, M., Hata, S. (2010). Localized expression of arbuscular mycorrhiza-inducible ammonium transporters in soybean. Plant Cell Physiol. 51, 1411–1415. doi: 10.1093/pcp/pcq099
Konishi, N., Ishiyama, K., Matsuoka, K., Maru, I., Hayakawa, T., Yamaya, T., et al. (2014). NADH-dependent glutamate synthase plays a crucial role in assimilating ammonium in the Arabidopsis root. Physiol. Plant 152, 138–151. doi: 10.1111/ppl.12177
Krapp, A., David, L. C., Chardin, C., Girin, T., Marmagne, A., Leprince, A.-S., et al. (2014). Nitrate transport and signalling in Arabidopsis. J. Exp. Bot. 65, 789–798. doi: 10.1093/jxb/eru001
Lea, P. J., Miflin, B. J. (2003). Glutamate synthase and the synthesis of glutamate in plants. Plant Physiol. Biochem. 41, 555–564. doi: 10.1016/S0981-9428(03)00060-3
Lei, X., Wang, T., Yang, B., Duan, Y., Zhou, L., Zou, Z., et al. (2022). Progress and perspective on intercropping patterns in tea plantations. Bev. Plant Res. 2, 1–10. doi: 10.48130/BPR-2022-0018
Li, J., Deng, T., Wu, Y., Liu, S., Li, Q., Liu, Z., et al. (2011). Full-length cDNA cloning and sequence analysis of theanine synthease gene in Camellia Sinensis. J. Tea Sci. 31, 411–418. doi: 10.13305/j.cnki.jts.2011.05.012
Li, F., Dong, C., Yang, T., Bao, S., Fang, W., Lucas, W. J., et al. (2021). The tea plant CsLHT1 and CsLHT6 transporters take up amino acids, as a nitrogen source, from the soil of organic tea plantations. Hortic. Res. 8, 1–12. doi: 10.1038/s41438-021-00615-x
Li, F., Dong, C., Yang, T., Ma, J., Zhang, S., Wei, C., et al. (2019). Seasonal theanine accumulation and related gene expression in the roots and leaf buds of tea plants (Camellia Sinensis L.). Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01397
Li, H., Han, J.-L., Chang, Y. H., Lin, J., Yang, Q. S. (2016). Gene characterization and transcription analysis of two new ammonium transporters in pear rootstock (Pyrus betulaefolia). J. Plant Res. 129, 737–748. doi: 10.1007/s10265-016-0799-y
Li, F., Li, H., Dong, C., Yang, T., Zhang, S., Bao, S., et al. (2020). Theanine transporters are involved in nitrogen deficiency response in tea plant (Camellia sinensis L.). Plant Signal. Behav. 15, 1728109. doi: 10.1080/15592324.2020.1728109
Li, S. M., Li, B. Z., Shi, W. M. (2012). Expression patterns of nine ammonium transporters in rice in response to N status. Pedosphere 22, 860–869. doi: 10.1016/S1002-0160(12)60072-1
Li, F., Lv, C., Zou, Z., Duan, Y., Zhou, J., Zhu, X., et al. (2022). CsAAP7.2 is involved in the uptake of amino acids from soil and the long-distance transport of theanine in tea plants (Camellia sinensis L.). Tree Physiol. 42, 2369–2381. doi: 10.1093/treephys/tpac071
Li, W., Wang, Y., Okamoto, M., Crawford, N. M., Siddiqi, M. Y., Glass, A. D. M. (2007). Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol. 143, 425–433. doi: 10.1104/pp.106.091223
Li, W., Xiang, F., Zhong, M., Zhou, L., Liu, H., Li, S., et al. (2017). Transcriptome and metabolite analysis identifies nitrogen utilization genes in tea plant (Camellia sinensis). Sci. Rep. 7, 1693. doi: 10.1038/s41598-017-01949-0
Lin, S., Chen, Z., Chen, T., Deng, W., Wan, X., Zhang, Z. (2023a). Theanine metabolism and transport in tea plants (Camellia sinensis L.): advances and perspectives. Crit. Rev. Biotechnol. 43, 327–341. doi: 10.1080/07388551.2022.2036692
Lin, Z. H., Chen, C. S., Zhao, S. Q., Liu, Y., Zhong, Q. S., Ruan, Q. C., et al. (2023b). Molecular and physiological mechanisms of tea (Camellia sinensis (L.) O. Kuntze) leaf and root in response to nitrogen deficiency. BMC Genom. 24, 27. doi: 10.1186/s12864-023-09112-y
Lin, Z., Zhong, Q., Chen, C., Chen, Z., You, X. (2014). Expression analysis of key enzyme genes of nitrogen synthesis from tea tree leave under different nitrogen level. J. Nucl. Agric. Sci. 28, 985–989.
Lin, Z., Zhong, Q., Chen, C., Ruan, Q., Chen, Z., You, X. (2016). Carbon dioxide assimilation and photosynthetic electron transport of tea leaves under nitrogen deficiency. Bot. Stud. 57, 37. doi: 10.1186/s40529-016-0152-8
Lin, Z., Zhong, Q., Hao, Z., You, X., Chen, Z., Chen, C., et al. (2017). Effects of chlorophyll fluorescence parameters of different tea cultivars in response to low nitrogen. J. Tea Sci. 37, 363–372. doi: 10.13305/j.cnki.jts.2017.04.008
Lin, Z., Zhong, Q., You, X., Chen, Z., Chen, C., Shan, R., et al. (2018). Antioxidant enzyme activity of tea plant (Camellia sinensis) in response to low temperature stress. J. Tea Sci. 38, 363–371. doi: 10.13305/j.cnki.jts.2018.04.004
Lin, Z., Zhong, Q., You, X., Chen, Z., Chen, C., Shan, R., et al. (2019). Antioxidant enzyme activity and growth of tea plants (Camellia sinensis) as affected by low-nitrogen stress. Acta Tea Sin. 60, 57–63.
Liu, Y. (2016). “The altered expression of genes related to nitrogen absorption and utilization of tea cultivars with different nitrogen use efficiency,” in Dissertation (Beijing: Chinese Academy of Agricultural Sciences).
Liu, H. (2020). “Functional characterization of amino acid transporter genes CsVAT1.3, CsLHT8L, CsCAT9.1, CsAAP3.1 in Camellia sinensis,” in Dissertation (Wuhan, Hubei Province: HuangZhong Agricultural University).
Liu, M. Y., Burgos, A., Ma, L., Zhang, Q., Tang, D., Ruan, J. (2017a). Lipidomics analysis unravels the effect of nitrogen fertilization on lipid metabolism in tea plant (Camellia sinensis L.). BMC Plant Biol. 17, 165. doi: 10.1186/s12870-017-1111-6
Liu, M.-Y., Burgos, A., Zhang, Q., Tang, D., Shi, Y., Ma, L., et al. (2017b). Analyses of transcriptome profiles and selected metabolites unravel the metabolic response to NH4+ and NO3– as signaling molecules in tea plant (Camellia sinensis L.). Sci. Hortic. 218, 293–303. doi: 10.1016/j.scienta.2017.02.036
Liu, X., Hu, B., Chu, C. (2022). Nitrogen assimilation in plants: current status and future prospects. J. Genet. Genomics 49, 394–404. doi: 10.1016/j.jgg.2021.12.006
Liu, Z. W., Li, H., Liu, J. X., Wang, Y., Zhuang, J. (2020). Integrative transcriptome, proteome, and microRNA analysis reveals the effects of nitrogen sufficiency and deficiency conditions on theanine metabolism in the tea plant (Camellia sinensis). Hortic. Res. 7, 1–13. doi: 10.1038/s41438-020-0290-8
Liu, Z. W., Li, H., Wang, W. L., Wu, Z. J., Cui, X., Zhuang, J. (2017c). CsGOGAT is important in dynamic changes of theanine content in postharvest tea plant leaves under different temperature and shading spreadings. J. Agric. Food Chem. 65, 9693–9702. doi: 10.1021/acs.jafc.7b04552
Liu, J., Liu, M., Fang, H., Zhang, Q., Ruan, J. (2021a). Accumulation of amino acids and flavonoids in young tea shoots is highly correlated with carbon and nitrogen metabolism in roots and mature leaves. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.756433
Liu, M., Tang, D., Jiao, Z., Shi, Y., Ma, L., Zhang, Q., et al. (2021b). Suitable nitrogen fertilization rate effectively improve the quality of summer green tea. J. Plant Nutr. Fer. 27, 1407–1419. doi: 10.11674/zwyf.2021012
Liu, M. Y., Tang, D., Shi, Y., Ma, L., Li, Y., Zhang, Q., et al. (2019). Short-term inhibition of glutamine synthetase leads to reprogramming of amino acid and lipid metabolism in roots and leaves of tea plant (Camellia sinensis L.). BMC Plant Biol. 19, 425. doi: 10.1186/s12870-019-2027-0
Liu, Y., Wang, H., Jiang, Z., Wang, W., Xu, R., Wang, Q., et al. (2021c). Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 590, 600–605. doi: 10.1038/s41586-020-03091-w
Liu, Z. W., Wu, Z. J., Li, H., Wang, Y. X., Zhuang, J. (2017d). L-theanine content and related gene expression: novel insights into theanine biosynthesis and hydrolysis among different tea plant (Camellia sinensis L.) tissues and cultivars. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00498
Liu, J., You, L., Amini, M., Obersteiner, M., Herrero, M., Zehnder, A. J. B., et al. (2010). A high-resolution assessment on global nitrogen flows in cropland. Proc. Natl. Acad. Sci. U.S.A. 107, 8035–8040. doi: 10.1073/pnas.0913658107
Liu, S. X., Zhang, H. J., Liu, X. Q. (2023). Comparative analysis and improvement of international competitiveness of Chinese tea industry. Food Nutri China. doi: 10.19870/j.cnki.11-3716/ts.20230609.002
Lothier, J., Gaufichon, L., Sormani, R., Lemaître, T., Azzopardi, M., Morin, H., et al. (2011). The cytosolic glutamine synthetase GLN1;2 plays a role in the control of plant growth and ammonium homeostasis in Arabidopsis rosettes when nitrate supply is not limiting. J. Exp. Bot. 62, 1375–1390. doi: 10.1093/jxb/erq299
Lynch, J. (2007). Roots of the second green revolution. Aust. J. Bot. 55, 493–512. doi: 10.1071/BT06118
Ma, L., Chen, H., Shan, Y., Jiang, M., Zhang, G., Geng, M., et al. (2013). Status and suggestions of tea garden fertilization on main green tea-producing counties in Zhejiang province. J. Tea Sci. 33, 74–84. doi: 10.13305/j.cnki.jts.2013.01.010
Ma, J. Q., Jin, J. Q., Yao, M. Z., Ma, C. L., Xu, Y. X., Hao, W. J., et al. (2018). Quantitative trait loci mapping for theobromine and caffeine contents in tea plant (Camellia sinensis). J. Agric. Food Chem. 66, 13321–13327. doi: 10.1021/acs.jafc.8b05355
Mei, Y., Zhang, S. (2022). Analysis of China's tea production and domestic sales in 2022. China Tea. 45, 25–30.
Miller, A. J., Cramer, M. D. (2005). Root nitrogen acquisition and assimilation. Plant Soil 274, 1–36. doi: 10.1007/s11104-004-0965-1
Moll, R. H., Kamprath, E. J., Jackson, W. A. (1982). Analysis and interpretation of factors which contribute to efficiency of nitrogen utilization1. Agron. J. 74, 562–564. doi: 10.2134/agronj1982.00021962007400030037x
Monachello, D., Allot, M., Oliva, S., Krapp, A., Daniel-Vedele, F., Barbier-Brygoo, H., et al. (2009). Two anion transporters AtClCa and AtClCe fulfil interconnecting but not redundant roles in nitrate assimilation pathways. New Phytol. 183, 88–94. doi: 10.1111/j.1469-8137.2009.02837.x
Negi, J., Matsuda, O., Nagasawa, T., Oba, Y., Takahashi, H., Kawai-Yamada, M., et al. (2008). CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452, 483–486. doi: 10.1038/nature06720
Ni, K., Liao, W., Yi, X., Niu, S., Ma, L., Shi, Y., et al. (2019). Fertilization status and reduction potential in tea gardens of China. J. Plant Nutr. Fer. 25, 421–432. doi: 10.11674/zwyf.18078
Rana, N. K., Mohanpuria, P., Yadav, S. K. (2008). Cloning and characterization of a cytosolic glutamine synthetase from Camellia sinensis (L.) O. Kuntze that is upregulated by ABA, SA, and H2O2. Mol. Biotechnol. 39, 49–56. doi: 10.1007/s12033-007-9027-2
Raun, W. R., Johnson, G. V. (1999). Improving nitrogen use efficiency for cereal production. Agron. J. 91, 357–363. doi: 10.2134/agronj1999.00021962009100030001x
Ruan, J., Gerendás, J., Härdter, R., Sattelmacher, B. (2007). Effect of root zone pH and form and concentration of nitrogen on accumulation of quality-related components in green tea. J. Sci. Food Agr. 87, 1505–1516. doi: 10.1002/jsfa.2875
Ruan, J., Haerdter, R., Gerendás, J. (2010). Impact of nitrogen supply on carbon/nitrogen allocation: a case study on amino acids and catechins in green tea [Camellia sinensis (L.) O. Kuntze] plants*. Plant Biol. 12, 724–734. doi: 10.1111/j.1438-8677.2009.00288.x
Ruan, J., Wang, X., Cui, S., Yao, G. (1993). Study on the mechanism in the difference of nitrogen nutrition among different cultivars. China Tea, 35–37.
Ruan, L., Wei, K., Wang, L., Cheng, H., Wu, L., Li, H. (2019). Characteristics of free amino acids (the quality chemical components of tea) under spatial heterogeneity of different nitrogen forms in tea (Camellia sinensis) Plants. Molecules 24, 415. doi: 10.3390/molecules24030415
Ruan, L., Wei, K., Wang, L., Cheng, H., Zhang, F., Wu, L., et al. (2016). Characteristics of NH4+ and NO3– fluxes in tea (Camellia sinensis) roots measured by scanning ion-selective electrode technique. Sci. Rep. 6, 38370. doi: 10.1038/srep38370
Santa-María, G. E., Moriconi, J. I., Oliferuk, S. (2015). Internal efficiency of nutrient utilization: what is it and how to measure it during vegetative plant growth? J. Exp. Bot. 66, 3011–3018. doi: 10.1093/jxb/erv162
She, G., Yu, S., Li, Z., Peng, A., Li, P., Li, Y., et al. (2022). Characterization of CsTSI in the biosynthesis of theanine in tea plants (Camellia sinensis). J. Agric. Food Chem. 70, 826–836. doi: 10.1021/acs.jafc.1c04816
Song, B., Zhang, L., Chen, Z., Liu, M., Zhang, W., Zhao, J., et al. (2023). Effects of different fertilization treatments on the physiological activities of roots and the expression of CsAMT2s of potted tea cuttings of the different varieties. Chin. J. Appl. Environ. Biol., 1–11. doi: 10.19675/j.cnki.1006-687x.2022.09029
Su, J., Ruan, L., Wang, L., Wei, K., Wu, L., Bai, P., et al. (2020). Early identification of nitrogen absorption efficiency in tea plants. J. Tea Sci. 40, 576–587. doi: 10.13305/j.cnki.jts.2020.05.002
Suárez, M. F., Avila, C., Gallardo, F., Cantón, F. R., García-Gutiérrez, A., Claros, M. G., et al. (2002). Molecular and enzymatic analysis of ammonium assimilation in woody plants. J. Exp. Bot. 53, 891–904. doi: 10.1093/jexbot/53.370.891
Sun, J., Bankston, J. R., Payandeh, J., Hinds, T. R., Zagotta, W. N., Zheng, N. (2014). Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature 507, 73–77. doi: 10.1038/nature13074
Sun, J., Qiu, C., Qian, W., Wang, Y., Sun, L., Li, Y., et al. (2019). Ammonium triggered the response mechanism of lysine crotonylome in tea plants. BMC Genom. 20, 340. doi: 10.1186/s12864-019-5716-z
Suzuki, A., Knaff, D. B. (2005). Glutamate synthase: structural, mechanistic and regulatory properties, and role in the amino acid metabolism. Photosynth. Res. 83, 191–217. doi: 10.1007/s11120-004-3478-0
Swarbreck, S. M., Defoin-Platel, M., Hindle, M., Saqi, M., Habash, D. Z. (2011). New perspectives on glutamine synthetase in grasses. J. Exp. Bot. 62, 1511–1522. doi: 10.1093/jxb/erq356
Tan, L., Cui, D., Wang, L., Liu, Q., Zhang, D., Hu, X., et al. (2022). Genetic analysis of the early bud flush trait of tea plants (Camellia sinensis) in the cultivar ‘Emei Wenchun’ and its open-pollinated offspring. Hortic. Res. 9, uhac086. doi: 10.1093/hr/uhac086
Tang, D., Jiao, Z., Zhang, Q., Liu, M.-Y., Ruan, J. (2021). Glutamate dehydrogenase isogenes CsGDHs cooperate with glutamine synthetase isogenes CsGSs to assimilate ammonium in tea plant (Camellia sinensis L.). Plant Sci. 312, 111031. doi: 10.1016/j.plantsci.2021.111031
Tang, D., Liu, M., Zhang, Q., Fan, K., Shi, Y., Ma, L., et al. (2018). Isolation and expression profiles of cytosolic glutamine synthetase genes CsGS1s in tea plant (Camellia sinensis). Plant Physiol. J. 54, 71–80. doi: 10.13592/j.cnki.ppj.2017.0265
Tang, D., Liu, M.-Y., Zhang, Q., Ma, L., Shi, Y., Ruan, J. (2020). Preferential assimilation of NH4+ over NO3– in tea plant associated with genes involved in nitrogen transportation, utilization and catechins biosynthesis. Plant Sci. 291, 110369. doi: 10.1016/j.plantsci.2019.110369
Tang, D., Liu, M., Zhang, Q., Shi, Y., Ma, L., Ruan, J. (2019). Effects of nitrogen form and root-zone pH on nutrient uptake and concentrations of organic anions in tea plants (Camellia sinensis). J. Tea Sci. 39, 159–170. doi: 10.13305/j.cnki.jts.2019.02.005
Teng, R. M., Yang, N., Li, J. W., Liu, C. F., Chen, Y., Li, T., et al. (2022). Isolation and characterization of an LBD transcription factor CsLBD39 from tea plant (Camellia sinensis) and its roles in modulating nitrate content by regulating nitrate-metabolism-related genes. Int. J. Mol. Sci. 23, 9294. doi: 10.3390/ijms23169294
Thomsen, H. C., Eriksson, D., Møller, I. S., Schjoerring, J. K. (2014). Cytosolic glutamine synthetase: a target for improvement of crop nitrogen use efficiency? Trends Plant Sci. 19, 656–663. doi: 10.1016/j.tplants.2014.06.002
Vahisalu, T., Kollist, H., Wang, Y. F., Nishimura, N., Chan, W. Y., Valerio, G., et al. (2008). SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452, 487–491. doi: 10.1038/nature06608
Valderrama-Martín, J. M., Ortigosa, F., Ávila, C., Cánovas, F. M., Hirel, B., Cantón, F. R., et al. (2022). A revised view on the evolution of glutamine synthetase isoenzymes in plants. Plant J. 110, 946–960. doi: 10.1111/tpj.15712
Wang, J. (2014). “Cloning and expression analysis of genes related to nitrogen transporter in tea plants (Camellia sinesis),” in Dissertation (Hefei, Anhui Province: Anhui Agricultural University).
Wang, L., Chen, C., Lin, Z., Wei, K., Wu, L., Feng, S., et al. (2015). Growth characteristic of different cultivars of tea plant in response to nitrogen contents. J. Tea Sci. 35, 423–428. doi: 10.13305/j.cnki.jts.2015.05.003
Wang, Y., Cheng, X., Yang, T., Su, Y., Lin, S., Zhang, S., et al. (2021b). Nitrogen-regulated theanine and flavonoid biosynthesis in tea plant roots: protein-level regulation revealed by multiomics analyses. J. Agric. Food. Chem. 69, 10002–10016. doi: 10.1021/acs.jafc.1c02589
Wang, Y., Ouyang, J.-X., Fan, D.-M., Wang, S.-M., Xuan, Y.-M., Wang, X.-C., et al. (2022a). Transcriptome analysis of tea (Camellia sinensis) leaves in response to ammonium starvation and recovery. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.963269
Wang, X., Su, J. (1990). Relationship between nitrate reductase activity and yield traits and nitrogen nutrition in tea plants. J. Tea Sci. 64.
Wang, J., Tian, X., Jiang, C., Li, Y. (2014). Cloning and expression analysis of nitrate transporter gene in Camellia sinensis. Plant Physiol. J. 50, 983–988. doi: 10.13592/j.cnki.ppj.2014.0049
Wang, Y., Wei, K., Ruan, L., Bai, P., Wu, L., Wang, L., et al. (2022b). Systematic investigation and expression profiles of the nitrate transporter 1/peptide transporter family (NPF) in tea plant (Camellia sinensis). Int. J. Mol. Sci. 23, 6663. doi: 10.3390/ijms23126663
Wang, Y., Xuan, Y.-M., Wang, S. M., Fan, D. M., Wang, X. C., Zheng, X. Q. (2022c). Genome-wide identification, characterization, and expression analysis of the ammonium transporter gene family in tea plants (Camellia sinensis L.). Physiol. Plant 174, e13646. doi: 10.1111/ppl.13646
Wang, X., Yang, Y., Chen, L., Ruan, J. (2004). Genotypic difference of nitrogen efficiency in tea plant [Camellia sinensis (L.) O. Kuntze]. J. Tea Sci. 24, 93–98. doi: 10.13305/j.cnki.jts.2004.02.004
Wang, X., Yang, Y., Chen, L., Ruan, J. (2005). Preliminary study on physiological and biochemical indices related to nitrogen use efficiency in tea plant [Camelliasinensis(L.)O·Kuntze). Acta Agronomica Sin. 31, 926–931.
Wang, P., Yang, J., Zhang, X., Ye, N. (2021a). Research advance of tea plant genome and sequencing technologies. J. Tea Sci. 41, 743–752. doi: 10.13305/j.cnki.jts.2021.06.002
Wei, G., Huang, J., Yang, J. (2012). The impacts of food safety standards on China’s tea exports. China Econ. Rev. 23, 253–264. doi: 10.1016/j.chieco.2011.11.002
Wei, K., Wang, X., Hao, X., Qian, Y., Li, X., Xu, L., et al. (2021). Development of a genome-wide 200K SNP array and its application for high-density genetic mapping and origin analysis of Camellia sinensis. Plant Biotechnol. J. 20, pbi.13761. doi: 10.1111/pbi.13761
Wei, C., Yang, H., Wang, S., Zhao, J., Liu, C., Gao, L., et al. (2018). Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. U.S.A. 115, E4151–E4158. doi: 10.1073/pnas.1719622115
Williams, S., Arcand, M., Congreves, K. (2021). Nitrogen use efficiency definitions of today and tomorrow. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.637108
Wirén, N., Gazzarrini, S., Frommer, W. (1997). Regulation of mineral nitrogen uptake in plants. Plant Soil 196, 191–199. doi: 10.1023/A:1004241722172
Wu, X., Liu, T., Zhang, Y., Duan, F., Neuhäuser, B., Ludewig, U., et al. (2019). Ammonium and nitrate regulate NH4+ uptake activity of Arabidopsis ammonium transporter AtAMT1;3 via phosphorylation at multiple C-terminal sites. J. Exp. Bot. 70, 4919–4930. doi: 10.1093/jxb/erz230
Wu, B., Wu, X. (1993). Determination of nitrate reductase activity of tea pant leaves in vitro and in situ. China Tea 8–10.
Xia, E., Tong, W., Hou, Y., An, Y., Chen, L., Wu, Q., et al. (2020). The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation. Mol. Plant 13, 1013–1026. doi: 10.1016/j.molp.2020.04.010
Xing, A., Ma, Y., Wu, Z., Nong, S., Zhu, J., Sun, H., et al. (2020). Genome-wide identification and expression analysis of the CLC superfamily genes in tea plants (Camellia sinensis). Funct. Integr. Genomics 20, 497–508. doi: 10.1007/s10142-019-00725-9
Xu, L. Y., Wang, L. Y., Wei, K., Tan, L. Q., Su, J. J., Cheng, H. (2018). High-density SNP linkage map construction and QTL mapping for flavonoid-related traits in a tea plant (Camellia sinensis) using 2b-RAD sequencing. BMC Genom. 19, 955. doi: 10.1186/s12864-018-5291-8
Yang, Y. (2011). “Quality-related constituents in tea (Camellia sinensis (L.) O. Kuntze) leaves as affected by nitrogen,” in Dissertation (Nanjing, Jiangsu Province: Nanjing Agricultural University).
Yang, Y., Hu, Y., Wan, Q., Li, R., Wang, F., Ruan, J. (2016). Cloning and expression analysis of nitrate transporter NRT1.1 gene in tea plant (Camellia sinensis (L.)). J. Tea Sci. 36, 505–512. doi: 10.13305/j.cnki.jts.2016.05.009
Yang, T., Xie, Y., Lu, X., Yan, X., Wang, Y., Ma, J., et al. (2021). Shading promoted theanine biosynthesis in the roots and allocation in the shoots of the tea plant (Camellia sinensis L.) cultivar Shuchazao. J. Agric. Food Chem. 69, 4795–4803. doi: 10.1021/acs.jafc.1c00641
Yao, M. Z., Ma, C. L., Qiao, T. T., Jin, J. Q., Chen, L. (2012). Diversity distribution and population structure of tea germplasms in China revealed by EST-SSR markers. Tree. Genet. Genomes. 8, 205–220. doi: 10.1007/s11295-011-0433-z
Yu, Y., Kou, X., Gao, R., Chen, X., Zhao, Z., Mei, H., et al. (2021). Glutamine synthetases play a vital role in high accumulation of theanine in tender shoots of albino tea germplasm “Huabai 1.” J. Agric. Food Chem. 69, 13904–13915. doi: 10.1021/acs.jafc.1c04567
Yuan, L., Gu, R., Xuan, Y., Smith-Valle, E., Loqué, D., Frommer, W. B., et al. (2013). Allosteric regulation of transport activity by heterotrimerization of Arabidopsis ammonium transporter complexes in vivo. Plant Cell 25, 974–984. doi: 10.1105/tpc.112.108027
Yuan, L., Loqué, D., Kojima, S., Rauch, S., Ishiyama, K., Inoue, E., et al. (2007). The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-type transporters. Plant Cell 19, 2636–2652. doi: 10.1105/tpc.107.052134
Zhang, X., Cui, Y., Yu, M., Su, B., Gong, W., Baluška, F, et al. (2019a). Phosphorylation-mediated dynamics of nitrate transceptor NRT1.1 regulate auxin flux and nitrate signaling in lateral root growth. Plant Physiol. 181, 480–498. doi: 10.1104/pp.19.00346
Zhang, F., He, W., Yuan, Q., Wei, K., Ruan, L., Wang, L., et al. (2021). Transcriptome analysis identifies CsNRT genes involved in nitrogen uptake in tea plants, with a major role of CsNRT2.4. Plant Physiol. Biochem. 167, 970–979. doi: 10.1016/j.plaphy.2021.09.024
Zhang, X., Li, Y., Wang, Y., Cai, H., Zeng, H., Wang, Z. (2019b). Influence of future climate change in suitable habitats of tea in different countries. Biodivers. Sci. 27, 595–606. doi: 10.17520/biods.2019085
Zhang, W., Lin, L., Chen, M., Sun, W. (2022a). Molecular cloning and expression analysis of CsAMT1s gene subfamily in Camellia sinensis. Chin. J. Appl. Environ. Biol. 28, 57–66. doi: 10.19675/j.cnki.1006-687x.2020.10011
Zhang, W., Lin, L., Wang, T., Chen, M., Song, B., Sun, W. (2022b). Genome-wide identification of AMT2-type ammonium transporters reveal that CsAMT2.2 and CsAMT2.3 potentially regulate NH4+ absorption among three different cultivars of Camellia sinensis. Int. J. Mol. Sci. 23, 15661. doi: 10.3390/ijms232415661
Zhang, F., Liu, Y., Wang, L., Bai, P., Ruan, L., Zhang, C., et al. (2018). Molecular cloning and expression analysis of ammonium transporters in tea plants (Camellia sinensis (L.) O. Kuntze) under different nitrogen treatments. Gene 658, 136–145. doi: 10.1016/j.gene.2018.03.024
Zhang, F., Wang, L., Bai, P., Wei, K., Zhang, Y., Ruan, L., et al. (2020). Identification of regulatory networks and hub genes controlling nitrogen uptake in tea plants [Camellia sinensis (L.) O. Kuntze]. J. Agric. Food Chem. 68, 2445–2456. doi: 10.1021/acs.jafc.9b06427
Zhang, F., Wang, L., Cheng, H., Wei, K., Hu, J., Zhang, C., et al. (2016). Molecular cloning and expression analysis of nitrite reductase gene CsNiR in tea plant. Acta Hortica 43, 1348–1356. doi: 10.16420/j.issn.0513-353x.2016-0324
Zheng, S. (2022). “Study on the monitoring of nitrate leaching and nitrogen balance in tea plantation,” in Dissertation (Beijing: Chinese Academy of Agricultural Sciences).
Zhou, X. (2012). “Study on rapid diagnosis and physiology and biochemistry indexes of nitrogen nutrition in tea plants (Camellia sinensis),” in Dissertation (Hefei, Anhui Province: Anhui Agricultural University).
Zhou, Y. (2014). “Genotype difference study on nitrogen efficiency of tea plants (Camellia sinensis) and cloning, expression analysis of nitrate reductase,” in Dissertation (Hefei, Anhui Province: Anhui Agricultural University).
Zhou, Y., Pang, L., Li, Y., Jiang, C. (2013). Cloning and expression analysis nitrate reductase gene in Camellia sinensis. Acta Bot. Boreal-Occident Sin. 33, 1292–1297. Chinese dissertation
Zhou, Y., Zhang, C., Lin, J., Yang, Y., Peng, Y., Tang, D., et al. (2015). Over-expression of a glutamate dehydrogenase gene, MgGDH, from Magnaporthe grisea confers tolerance to dehydration stress in transgenic rice. Planta 241, 727–740. doi: 10.1007/s00425-014-2214-z
Keywords: nitrogen transport, nitrate reduction, ammonia assimilation, NUE, camellia sinensis, challenges and prospects
Citation: Zhang W, Ni K, Long L and Ruan J (2023) Nitrogen transport and assimilation in tea plant (Camellia sinensis): a review. Front. Plant Sci. 14:1249202. doi: 10.3389/fpls.2023.1249202
Received: 28 June 2023; Accepted: 04 September 2023;
Published: 22 September 2023.
Edited by:
Karl H. Mühling, University of Kiel, GermanyReviewed by:
Joska Gerendas, University of Kiel, GermanyWei-Wei Deng, Anhui Agricultural University, China
Copyright © 2023 Zhang, Ni, Long and Ruan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lizhi Long, bG9uZ2xpemhpQHRyaWNhYXMuY29t; Jianyun Ruan, anJ1YW5AbWFpbC50cmljYWFzLmNvbQ==
 Wenjing Zhang
Wenjing Zhang Kang Ni
Kang Ni Lizhi Long
Lizhi Long Jianyun Ruan
Jianyun Ruan