- Department of Biology, Carleton University, Ottawa, ON, Canada
BLADE-ON-PETIOLE (BOP) genes are essential regulators of vegetative and reproductive development in land plants. First characterized in Arabidopsis thaliana (Arabidopsis), members of this clade function as transcriptional co-activators by recruiting TGACG-motif binding (TGA) basic leucine zipper (bZIP) transcription factors. Highly expressed at organ boundaries, these genes are also expressed in vascular tissue and contribute to lignin biosynthesis during secondary growth. How these genes function in trees, which undergo extensive secondary growth to produce wood, remains unclear. Here, we investigate the functional conservation of BOP orthologs in Populus trichocarpa (poplar), a widely-used model for tree development. Within the poplar genome, we identified two BOP-like genes, PtrBPL1 and PtrBPL2, with abundant transcripts in stems. To assess their functions, we used heterologous assays in Arabidopsis plants. The promoters of PtrBPL1 and PtrBPL2, fused with a β-glucuronidase (GUS) reporter gene showed activity at organ boundaries and in secondary xylem and phloem. When introduced into Arabidopsis plants, PtrBPL1 and PtrBPL2 complemented leaf and flower patterning defects in bop1 bop2 mutants. Notably, Arabidopsis plants overexpressing PtrBPL1 and PtrBPL2 showed defects in stem elongation and the lignification of secondary tissues in the hypocotyl and stem. Finally, PtrBPL1 and PtrBPL2 formed complexes with TGA bZIP proteins in yeast. Collectively, our findings suggest that PtrBPL1 and PtrBPL2 are orthologs of Arabidopsis BOP1 and BOP2, potentially contributing to secondary growth regulation in poplar trees. This work provides a foundation for functional studies in trees.
Introduction
Populus trichocarpa is a deciduous tree species that is commonly known as black cottonwood or western balsam poplar. A long-lived, mostly diploid species that is native to western North America, it is widely distributed in temperate and cold temperate regions (Cooke and Rood, 2007). P. trichocarpa and companion species in the genus Populus (poplar) are valuable model organisms for tree research (Taylor, 2002; Jansson and Douglas, 2007). The genus is closely related to the model plant Arabidopsis thaliana (Arabidopsis) and many poplar gene functions are conserved. An available variety of molecular tools, such as genetic transformation methods, CRISPR-based gene editing systems, and a growing number of sequenced genomes, make poplar species ideal for molecular studies (Jansson and Douglas, 2007; Bryant et al., 2020).
BLADE-ON-PETIOLE (BOP) co-transcriptional regulators, first described in Arabidopsis, belong to a family in land plants that have a BTB/POZ (Broad-complex, Tramtrack, and Bric-a-brac/POX virus and zinc finger) domain and ankyrin repeats for interaction with other proteins (Khan et al., 2014; Backer et al., 2019). Family members are classified into two phylogenetic subclades (Khan et al., 2014; Backer et al., 2019). The first subclade contains NPR1-type proteins involved in plant defense whereas BOP-type proteins primarily regulate plant development (Khan et al., 2014; Backer et al., 2019). Proteins from both subclades lack a DNA binding domain and interact TGACG-motif binding (TGA) basic leucine zipper (bZIP) transcription factors for the co-activation or co-repression of target genes. BTB-ankyrin proteins from both subclades can also function as E3 ubiquitin ligase adaptors involved in regulating protein abundance (Fu et al., 2012; Zhang et al., 2017; Chahtane et al., 2018; He et al., 2020). In monocots and dicots, BOPs contribute to a surprisingly large number of developmental processes, including lignin deposition as a defense (Zhang et al., 2019) and during secondary growth (Khan et al., 2012b; Woerlen et al., 2017; Shen et al., 2021; Liu et al., 2022).
In Arabidopsis, BOP1 and BOP2 genes are strongly expressed at organ boundaries, zones that connect organs to the plant body (Žádníková and Simon, 2014; Hepworth and Pautot, 2015). These areas regulate growth at the base of organs and produce axillary meristems for the development of lateral branches, flowers, and appendages such as stipules or nectaries. Boundaries are also sites where organs separate by abscission or dehiscence to release foliage, fruits, or seeds (Hepworth and Pautot, 2015). Phenotypic defects in bop1 bop2 double mutants are concentrated at lateral organ boundaries resulting in elongated leafy petioles, five-petalled flowers with a subtending bract, and defects in abscission (Hepworth et al., 2005; Norberg et al., 2005; McKim et al., 2008). In monocots and dicots, BOP1 and BOP2 paralogs participate in many of the same developmental processes (Wu et al., 2012; Tavakol et al., 2015; Couzigou et al., 2016; Jost et al., 2016; Xu et al., 2016; Toriba et al., 2019; Magne et al., 2020; Liu et al., 2022).
In Arabidopsis, BOP1 and BOP2 genes are also expressed in vascular tissues (Hepworth et al., 2005; Khan et al., 2012a). The overexpression of either gene inhibits stem elongation and disrupts secondary growth in stems and the root-hypocotyl (Khan et al., 2012a; Woerlen et al., 2017). In the inflorescence stem, lignified phloem and interfascicular fibers are completed earlier and the central pith becomes lignified in severe lines (Khan et al., 2012a). Secondary growth in the root-hypocotyl is also disrupted, with loss of xylem fiber differentiation (Liebsch et al., 2014; Woerlen et al., 2017). Unlike trees, secondary growth in the majority of the Arabidopsis stem is limited to the secondary thickening of xylem and phloem cell walls and the lignification of interfascicular fibers spaced between the primary vascular bundles. These fibers complete the vascular ring and provide mechanical support (Rogers and Campbell, 2004; Ehlting et al., 2005).
In woody dicot plants, cambium activity initiates in the primary vascular bundles (fascicular cambium) and spreads out to the interfascicular regions where differentiated cells regain the ability to divide (interfascicular cambium). In Arabidopsis, continuous vascular cambium only forms in the root-hypocotyl and at the base of the primary inflorescence 1-2 mm above the rosette (Sehr et al., 2010; Sanchez et al., 2012). The end result is a tube-like sheath of meristematic activity. Daughter cells on the outer rim of the cambium produce secondary phloem (inner bark) whereas daughter cells on the inside make secondary xylem (wood). In forest trees, this activity results in the radial thickening of stems and supports the development of large body architectures (Groover et al., 2006; Sehr et al., 2010; Sanchez et al., 2012).
P. trichocarpa and its relatives have emerged as an excellent platform for the study of trees (Jansson and Douglas, 2007; Bryant et al., 2020). The industrial impact of poplar is significant as hybrid poplars are among the fastest growing temperate forest trees in the world, ready for harvesting at 10 to 20 years. Trees can be grown on forest lands or marginal crop lands. Plantations, totalling some 9.6 million hectares worldwide, contribute to the storage of atmospheric CO2 as biomass, which is harvested for the production of wood chips, plywood, biofuels, and other materials. Lignin and wood engineering in forest trees can benefit from a detailed understanding of the molecular mechanisms controlling tree architecture and wood development (Sannigrahi et al., 2010; Chanoca et al., 2019).
Here, we investigate the function of two BOP-like genes in P. trichocarpa. Highly expressed in poplar xylem and phloem, we provide evidence that these genes when heterologously expressed in Arabidopsis can function at organ boundaries and regulate lignin deposition in tissues undergoing secondary growth. We also provide evidence of complex formation with TGA bZIP proteins indicating that gene functional networks might be conserved in poplar trees. The potential evolutionary implications of these findings are discussed.
Materials and methods
Plant material and growth conditions
The P. trichocarpa female clone “Nisqually-1” was used (Song et al., 2006). Cuttings from a tree grown on the University of British Columbia campus (a gift of Sean Mansfield) were rooted in soil and grown under natural lighting in a greenhouse. The Arabidopsis thaliana Columbia (Col-0) accession was used as the wild type. The double mutant bop1 bop2 (Hepworth et al., 2005), activation-tagged overexpression line bop1-6D and transgenic line 35S:BOP2 (Norberg et al., 2005) were previously described. Surface-sterilized seeds were sown on minimal media agar plates (Haughn and Somerville, 1986). Ten-day-old seedlings were planted in sterilized soil (Promix BX, Premier Tech, Quebec) supplemented with 20-20-20 fertilizer (Plant Product Co. Ltd, Brampton, Ontario). The plants were grown to maturity in chambers at 21°C under long days (8 h dark/16 h light, intensity 100 µmol m-2 s-1).
PtrBPL1 and PtrBPL2 complementation of Arabidopsis bop1 bop2 mutant
PtrBPL1 and PtrBPL2 genes were expressed under the control of a BOP1 promoter in Arabidopsis bop1 bop2 mutant plants. This promoter has been previously used for complementation studies (Khan et al., 2015). The cDNA sequences of PtrBPL1 (Potri.016G040500) and PtrBPL2 (Potri.006G043400) were amplified by polymerase chain reaction (PCR) using cDNA from mixed poplar tissue as the template and high-fidelity iProof polymerase (Biorad, Hercules, CA). The primers annealed to the 5’ and 3’ untranslated regions of each gene to ensure that amplification was specific. The resulting products were cloned into pCR™-BluntII-TOPO™ (Invitrogen, ThermoFisher Scientific, Waltham, MA) and verified by DNA sequencing (Eurofins Genomics, Louisville, KY). Verified clones were used as template to amplify the coding regions of PtrBPL1 and PtrBPL2 using PtrBPL-XbaI-F and PtrBPL-Sac1-R as the primers. The resulting products were cloned into the corresponding sites of binary vector pBAR (a gift from the Dangl lab, University of North Carolina) downstream of the AtBOP1 promoter (McKim et al., 2008). The resulting constructs named BOP1p:PtrBPL1 and BOP1p:PtrBPL2 were introduced into Agrobacterium tumefaciens strain C58C1 pGV101 pMP90 (Koncz and Schell, 1986). Arabidopsis bop1 bop2 plants were transformed by floral dipping (Clough and Bent, 1998). Glufosinate-resistant primary transformant (T1) plants were selected on soil using the herbicide Finale (Bayer Environmental Sciences, Sacramento, CA). Complementation of bop1 bop2 leaf, flower, and abscission defects was scored in the T1 generation. The progeny of ten independent T1 lines showing strong complementation were genotyped to confirm that they were bop1 bop2 double mutants. Three to five independent transgenic lines were selected for further analysis. Primers used for cloning are listed in Supplementary Table S1.
PtrBPL1 and PtrBPL2 promoter GUS reporter constructs
To determine the expression pattern of PtrBPL1 and PtrBPL2 genes, the promoters were PCR-amplified from genomic DNA template extracted from young poplar leaves using a Genomic DNA Mini Kit (Plant) (FroggaBio Inc., Concord, Ontario). A 3938-bp PtrBPL1 promoter (nucleotides -3867 to +112) was assembled using an overlap PCR approach because the full-length sequence was difficult to amplify. A 2.9-kb PtrBPL1 promoter fragment (nucleotides -3867 to -2876) was PCR-amplified using ptBPL1-pro-F10 and ptBPL1-pro-R1 as the primers. A 1.1-kb PtrBPL1 promoter fragment (-2807 to +112) was PCR amplified using BPL1-pro-F8 and BPL1-May14-R as the primers. These two PtrBPL1 overlapping promoter fragments were gel purified, mixed, and used as template to amplify a full-length product using PtBPL1-pro-F10 and BPL1-May14-R as the primer pair. The resulting PtrBPL1 promoter (including 112-bp of PtrBPL1 coding sequence) was column-purified and cloned into a Zero Blunt™ TOPO™ vector (Invitrogen, ThermoFisher Scientific). DNA sequencing identified a 42-bp sequence deletion (nucleotides -3757 to -3715) compared to publicly available database sequence. To create the reporter gene, the PtrBPL1 promoter was amplified from the above plasmid using BPL1-pro-BamH1-F and BPL1-pro-Nco1-R as the primer pair. The resulting product was column-purified, digested with BamH1 and Nco1 restriction enzymes and ligated into the corresponding sites of the pGreen-based pTGA9pro:GUS plasmid (Hellens et al., 2000; Murmu et al., 2010). This ligation placed the PtrBPL1 promoter upstream of a GUS reporter gene as a translational fusion. Similarly, a 4092-bp PtrBPL2 promoter sequence (nucleotide -4059 to +33) was PCR-amplified using BPL2-4kb-F and BPL2-Promo-R as the primers and cloned into the Zero Blunt™ TOPO™ vector (Invitrogen, ThermoFisher Scientific). The PtrBPL2 promoter insert was then amplified using BPL2-pro-BamH1-F and BPL2-pro-Nco1-R as the primers, digested with BamHI and NcoI restriction enzymes, and cloned into the corresponding sites of pTGA9pro:GUS (Murmu et al., 2010). DNA fragments destined for cloning were amplified using iProof high-fidelity polymerase (Biorad, Hercules, CA). All clones were verified by DNA sequencing (Eurofins Genomics). The resulting GUS reporter constructs were co-transformed with pSOUP into Agrobacterium tumefaciens strain C58C1 pGV101 pMP90 (Koncz and Schell, 1986; Hellens et al., 2000). Wild-type plants were transformed by floral dipping (Clough and Bent, 1998). Glufosinate-resistant primary transformant (T1) plants were selected on soil using the herbicide Finale (Bayer Environmental Sciences). Multiple independent transgenic lines per construct were evaluated for reporter GUS activity. Three independent transgenic lines were selected for further analysis. Primers used for cloning are listed in Supplementary Table S1.
Constructs for overexpression of PtrBPL1 and PtrBPL2
The coding sequences of PtrBPL1 and PtrBPL2 were PCR-amplified from cloned cDNA template using iProof as the polymerase (Biorad) and Pt6s04010CDS-F and Pt6s04010CDS-R primer set, and Pt6s04190CDS-F and Pt6s04190CDS-R primer set, respectively. The resulting products were placed into the Gateway entry vector pCR™8/GW/TOPO™ (Invitrogen, ThermoFisher Scientific) and moved into the pSM3 binary vector (Unda et al., 2017) downstream of a double 35S CaMV promoter (D35S) using Gateway™ LR Clonase™ (Invitrogen, ThermoFisher Scientific). Wild-type plants were transformed as described above. Hygromycin-resistant primary transformants were selected on agar plates. Phenotypes were scored in the T1 generation. Three to five independent transformants were selected for further analysis. Primers used for cloning are listed in Supplementary Table S1.
Reverse transcription-quantitative PCR
Total RNA was isolated from dissected poplar tissues (a gift of the Carl Douglas lab). 2 µg of RNA was used for cDNA synthesis using Superscript III reverse transcriptase (Invitrogen, ThermoFisher Scientific). PCR reactions in triplicate containing 2 µl of 10-fold diluted cDNA, gene-specific primers (Supplementary Table S1) and Power SYBR™ Green Master mix (Invitrogen, ThermoFisher Scientific) were carried out using a StepOnePlus thermocycler (Applied Biosystems, ThermoFisher Scientific). Relative transcript levels were calculated according to Pfaffl (2001). Values were normalized to the poplar elongation factor reference gene C672 (Wang et al., 2014) and then to young leaf. Data are the average of four measurements for each of two biological replicates. Error bars show standard deviation.
Localization of lignin deposition
Tissues were analyzed for lignin deposition as previously described (Khan et al., 2012b; Woerlen et al., 2017). Arabidopsis stems were harvested from five-week-old plants. A razor blade was used to hand-cut sections from the base of the primary inflorescence about 1 cm above the rosette leaves (Khan et al., 2012b). Hypocotyls were harvested from seven-week-old plants. A razor blade was used to hand-cut sections about 1.5 mm below the rosette leaves. Samples were placed in 2% phloroglucinol dissolved in 95% ethanol for five minutes. Then, five drops of concentrated hydrochloric acid were added. Two minutes were allowed for color development. Immediately, samples were transferred onto a glass slide and a cover slip was added. Images were collected using a Discovery V20 stereomicroscope (Carl Zeiss Canada, North York, Ontario).
Localization of GUS activity
Tissues were analyzed for β-glucuronidase (GUS) activity as previously described (Woerlen et al., 2017) with minor changes. The staining solution contained 4 mM KFe(CN) and 2 mM of 5-bromo-4-chloro-3-indoxyl-β-D-glucuronide (X-Gluc). The samples were incubated at 37°C for 3 to 24 hours until a localized blue precipitate was visible. After clearing in 70% ethanol, the samples were imaged using a Discovery V20 stereomicroscope (Carl Zeiss). For sections, the stained tissue was embedded in Paraplast Plus® (Sigma-Aldrich, St. Louis, MO) and processed using tert-butanol instead of xylenes (Woerlen et al., 2017). Tissue sections (20 µm) were fixed onto glass slides and dewaxed with tert-butanol. The samples were imaged using an Axio Imager M2 compound microscope (Carl Zeiss).
Yeast two-hybrid assay
Protein-protein interactions were assayed using a Matchmaker™ GAL4-based yeast two-hybrid system (Clontech, Takara Bio USA Inc, San Jose, CA) and Gateway-compatible pGBKT7-DEST (bait) and pGADT7-DEST (prey) plasmids modified from pGBKT7 and pGADT7-Res vectors, respectively (Lu et al., 2009). PtrBPL1 and PtrBPL2 proteins fused to the DNA-binding domain of yeast GAL4 were used as bait. AtTGA1, AtTGA4, AtTGA3, AtTGA7 and AtTGA8/PAN proteins fused to the transcriptional activation domain of yeast GAL4 were used as prey. To prepare Gateway entry vectors, the full-length coding sequences of bait and prey genes were cloned into pCR™8/GW/TOPO™TA (Invitrogen, ThermoFisher Scientific). All entry vector inserts were sequenced to confirm authenticity. To make the final plasmids, recombination reactions were performed using Gateway™ LR Clonase™ II enzyme mix according to the manufacturer’s instructions (Invitrogen, ThermoFisher Scientific). Bait and prey plasmid were co-transformed into yeast AH109 strain (Gietz and Schiestl, 2007). Transformed yeast colonies were identified by selection on synthetic complete (SC) media plates lacking Leu and Trp (SC/-Leu/-Trp). Dilution series were spotted onto SC/-Leu/-Trp medium or SC/-Leu-Trp-His medium plus 10 mM 3-amino-1,2,4-triazole (3-AT; Sigma-Aldrich) for assessment of His reporter gene activity. A 3-AT concentration (10 mM) sufficient to distinguish positive growth from background was determined empirically.
Bioinformatics
P. trichocarpa homologs of Arabidopsis BTB-ankyrin and TGA bZIP proteins were identified using the plant homologs tool at The Arabidopsis Information Resource (TAIR) (www.arabidopsis.org). The corresponding P. trichocarpa protein sequences were retrieved from Phytozome (Goodstein et al., 2012; phytozome-next.jgi.doe.gov). MEGA version 11 was used for the alignment of protein sequences by MUSCLE (Edgar, 2004) using the default parameters (www.megasoftware.net). Maximum Likelihood trees were constructed based on 100 bootstrap replicates using the Jones-Taylor-Thornton Model. The BOXSHADE alignment and sequence logos were prepared using Geneious Prime 2022.1 (www.geneioius.com). The percent similarity of protein pairs was calculated using the Expasy SIM tool (www.expasy.org/sim).
Promoter analysis
The 500-bp promoter sequences upstream of the translational start sites of AtBOP1, AtBOP2, PtrBPL1, and PtrBPL2 genes were retrieved from their genome assemblies found at TAIR (https://www.arabidopsis.org/) or Phytozome (https://phytozome-next.jgi.doe.gov/). Scanning for Transcription Factor Binding Sites (TFBS) was carried out using PlantPAN 3.0 (http://plantpan.itps.ncku.edu.tw/index.html). Selected TFBS were aligned to statistically enriched 6-mers identified by the motif finder tool at TAIR. Binding site locations were visualized by using TBtools v1.123 (Chen et al., 2020).
Results
Identification of BTB-ankyrin and TGA bZIP gene families in poplar
In the P. trichocarpa (poplar) genome, six BTB-ankyrin proteins were identified. The phylogenetic relationship Arabidopsis and poplar BTB-ankyrin proteins was investigated (Figure 1A). This analysis revealed one NPR1 protein (Potri.006G148100), three NPR3/4 proteins (Potri.012G118300; Potri.012G118500; Potri.015G117200) and two poplar BOP-like proteins designated as PtrBPL1 (Potri.016G040500) and PtrBPL2 (Potri.006G043400), respectively. Multiple sequence alignment showed that PtrBPL1 and PtrBPL2 are 94.3% similar to each other at the amino acid level and 80.2% versus 75.6% similar to AtBOP1, respectively. The similarity is broadly distributed across the length of the proteins, not just within the BTB/POZ and ankyrin repeat domain (Figures 1B–D; Supplementary Figure S1). Less conserved regions are situated within the BTB/POZ domain and near the C-terminus. For example, PtrBPL1 contains a deletion in the C-terminus compared to AtBOP1/2 and PtrBPL2 proteins (Figure 1B; Supplementary Figure S1). In parallel, we investigated the phylogenetic relationship of poplar TGA bZIP proteins as potential functional partners. Representatives from all five TGA clades found in Arabidopsis are present (Figure 1E; Supplementary Figure S2). Given these overall similarities, we anticipated that PtrBPL1 and PtrBPL2 might have conserved activities similar to AtBOP1 and AtBOP2.
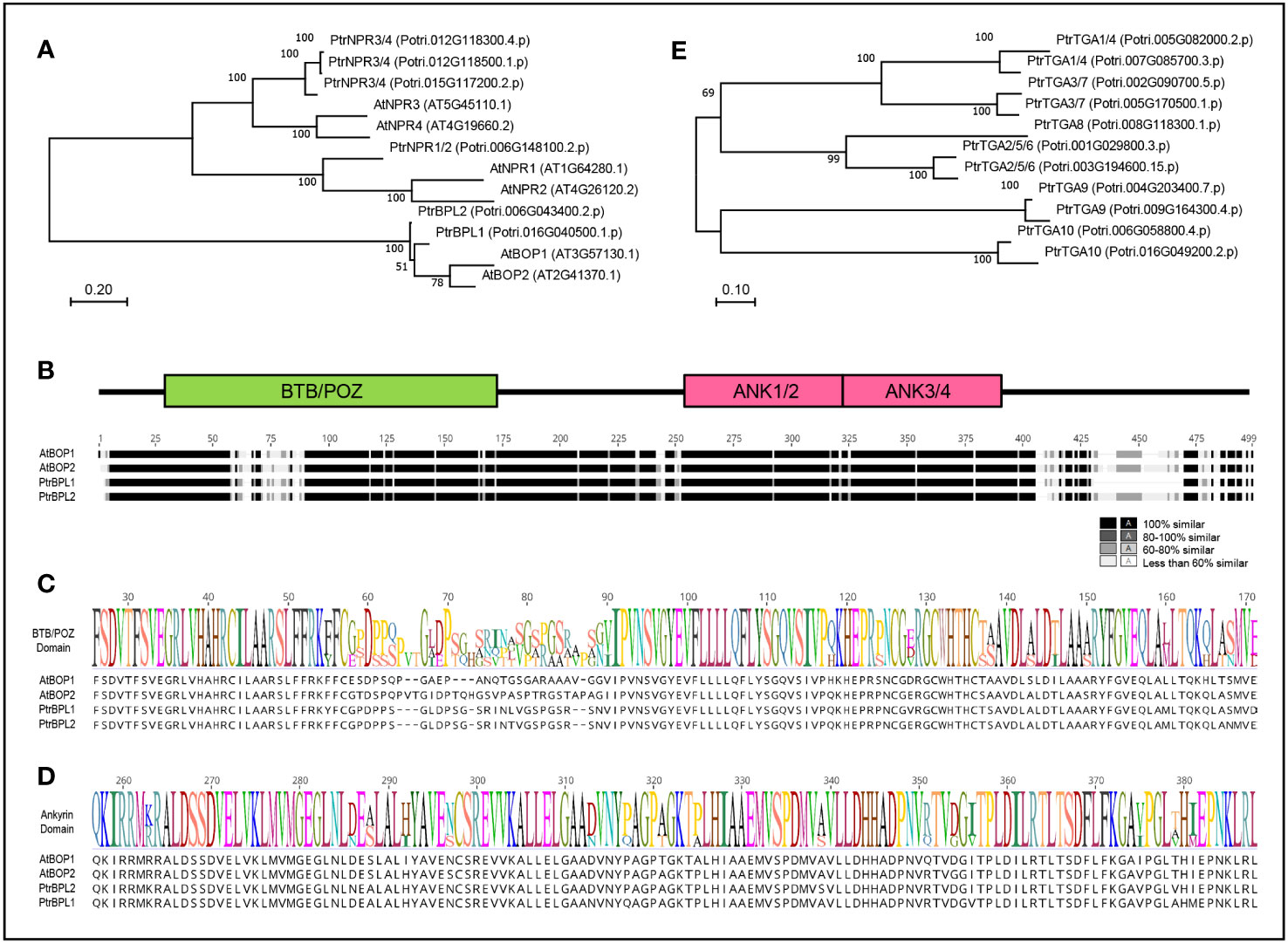
Figure 1 Proteins encoded by BTB-ankyrin and TGA bZIP genes from Arabidopsis and poplar. (A) Maximum Likelihood tree showing phylogenetic relationships between Arabidopsis (At) and poplar (Ptr) BTB-ankyrin proteins. (B) Multiple sequence alignment of AtBOP and PtrBPL proteins with corresponding domain map showing the relative position of conserved BTB/POZ and ankyrin (ANK) domains. Segments of highest similarity or identity are colored darkest and segments of lowest identity or similarity are colored lightest. (C) BTB/POZ domain sequence logo. (D) Ankyrin domain sequence logo. Protein sequences with listed identification numbers were retrieved from TAIR (www.arabidopsis.org) or Phytozome (phytozome-next.jgi.doe.gov). (E) Maximum Likelihood tree showing phylogenetic relationships between PtrTGA proteins.
PtrBPL1 and PtrBPL2 expression pattern
We next obtained expression data for PtrPBL1 and PtrBPL2 from the Bio-Analytic Resource for Plant Biology (BAR, www.utoronto.ca/bar). These data showed the wide expression of both genes in poplar tissues including seedlings, young leaves, catkins, roots, and xylem (Figures 2A, B). We then used RT-qPCR to monitor transcript abundance in selected tissues. These data showed that PtrBPL1 and PtrBPL2 transcripts are enriched in the petiole region of leaves compared to the blade. Significant expression was also observed in xylem and phloem tissues of young poplar stems (Figure 2C).
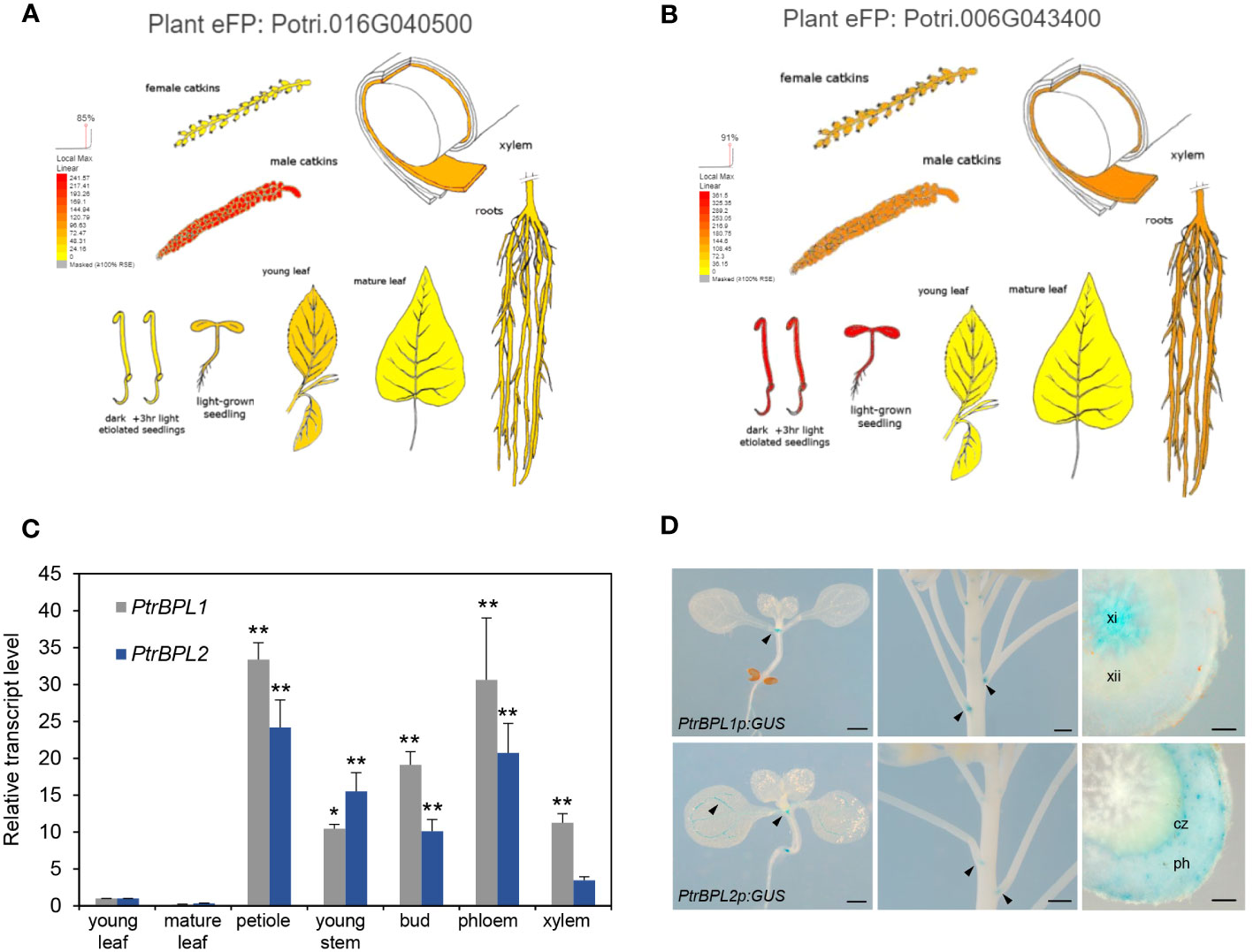
Figure 2 Expression of PtrBPL1 and PtrBPL2 in poplar and Arabidopsis. (A, B) Transcriptome data for P. trichocarpa compiled by the Bio-Analytic Resource for Plant Biology (www.bar.utoronto.ca). Red, high expression; orange, medium expression; yellow, lower expression. Plant eFP viewer output for (A) PtrBPL1 and (B) PtrBPL2. (C) The relative abundance of PtrBPL1 and PtrBPL2 transcript was independently measured using RT-qPCR in dissected poplar tissues. Data are the average ± SD of four measurements performed on each of two biological replicates. Values were normalized to young leaf. Asterisks indicate significant differences between tissues compared with young leaf (ANOVA with Tukey’s test, **P<0.01, *P<0.05). (D) Expression patterns of PtrBPL1p:GUS and PtrBPL2p:GUS in Arabidopsis seedlings, inflorescences, and sectioned hypocotyls. Arrows denote expression at organ boundaries. xi, secondary xylem I; xii, secondary xylem II; ph, secondary phloem; cz, cambium zone. Scale bars, 500 µm except 100 µm for hypocotyl sections.
We next investigated the spatial and temporal expression of PtrBPL1 and PtrBPL2 genes, by monitoring the expression of PtrBPL1p:GUS and PtrBPL2p:GUS reporter genes in Arabidopsis plants (Figure 2D). In seedlings, both genes were expressed in boundaries at the base of the cotyledons. During flowering, both genes were expressed at nodes in the stem where a boundary forms at the base of the flower pedicel. GUS expression was evident in floral organ boundaries and abscission zones at the base of young siliques for PtrBPL1 but not PtrBPL2 (Supplementary Figure S3). The vasculature of cotyledons, leaves, and the hypocotyl also showed differential expression. PtrBPL2 but not PtrBPL1 was expressed in veins of leaves (Figure 2D; Supplementary Figure S3). At the seedling stage, both genes were expressed at the root tip and at the base of lateral roots (Supplementary Figure S3). In the hypocotyl, PtrBPL1 was strongly expressed in early secondary xylem (xylem I) whereas PtrBPL2 was expressed at the outer edge of the cambial zone and in secondary phloem (Figure 2D; Supplementary Figure S3). Neither AtBOP1 nor AtBOP2 are expressed in secondary xylem (Liebsch et al., 2014; Woerlen et al., 2017) indicating a possible difference in gene regulation between the two species. To investigate this difference, a statistical motif analysis was carried out using the promoter regions (500 base pairs upstream of the start codon) of all four genes. This analysis showed an enrichment of MYB-related and NAC transcription factor binding sites in the promoters of PtrBPL1 and PtrBPL2 compared to AtBOP1 and AtBOP2 (Supplementary Figure S4) implicating these factors in the differential regulation of poplar orthologs.
PtrPBL1 and PtrBPL2 can complement bop1 bop2 leaf and flower patterning defects
To investigate to what extent PtrBPL1 and PtrBPL2 can substitute for AtBOP1 and AtBOP2, we expressed PtrBPL1 and PtrBPL2 under the control of an AtBOP1 promoter in bop1 bop2 mutants and tested for complementation. In total, we obtained 203 primary (T1) transformants for BOP1p:PtrBPL1 and 212 primary transformants for BOP1p:PtrBPL2 (Supplementary Table S2). Characteristic defects in bop1 bop2 mutants include leafy petioles, loss of floral organ abscission, and flowers with a bract and extra petals on the abaxial side (Hepworth et al., 2005). Strong or medium complementation of the leaf phenotype was observed in 54% and 52% of PtrBPL1 and PtrBPL2 transformants, respectively (Figure 3; Supplementary Table S2). Floral patterning defects were significantly restored in 65% and 76% of PtrBPL1 and PtrBPL2 transformants scored, respectively (Supplementary Table S2). Floral organ abscission was significantly restored in 76% and 62% of PtrBPL1 and PtrBPL2 transformants scored, respectively (Figure 3; Supplementary Table S2). These data confirm that heterologous expression of PtrBPL1 and PtrBPL2 can complement bop1 bop2 mutant phenotypes.
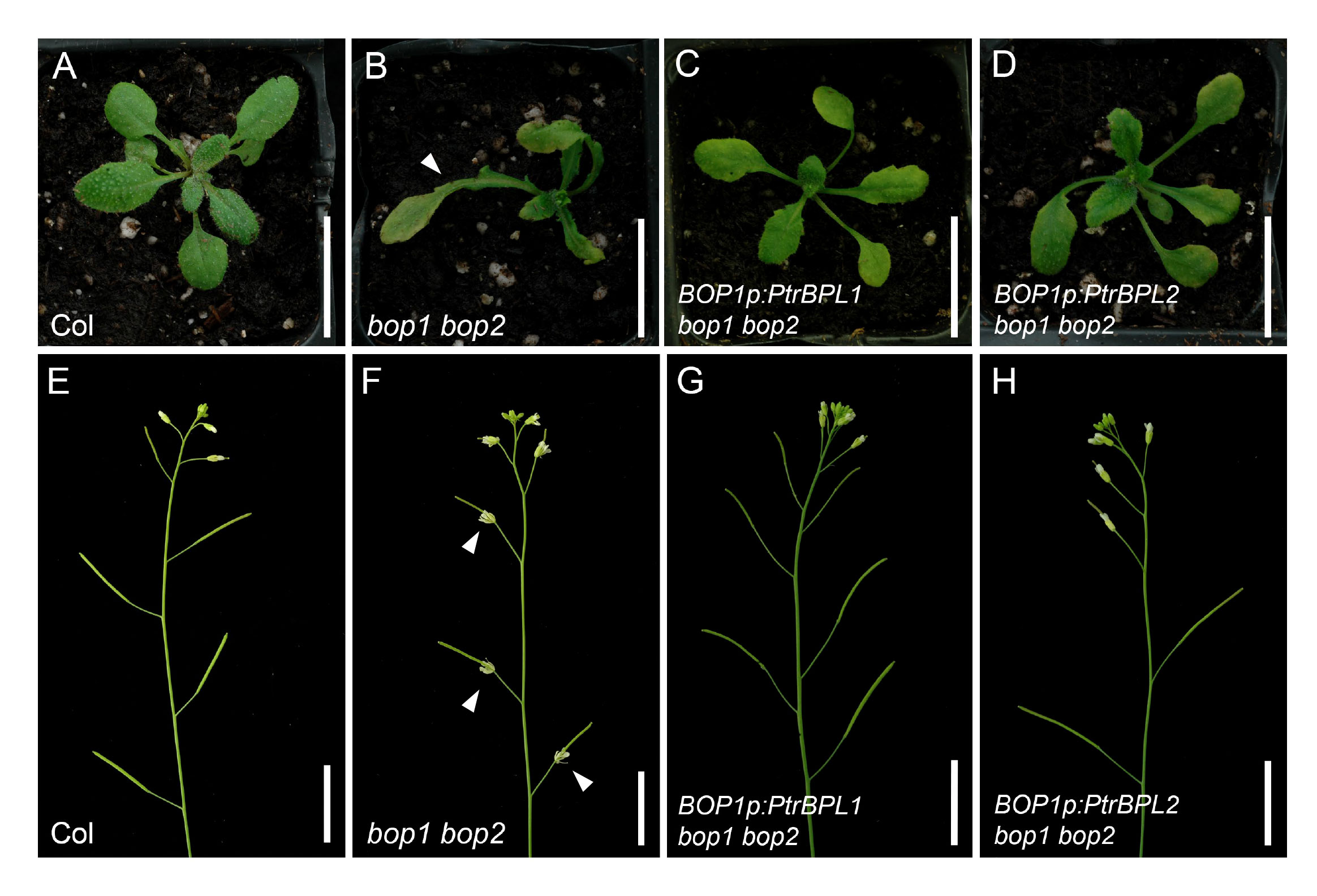
Figure 3 Complementation of Arabidopsis bop1 bop2 leaf and flower defects with PtrBPL1 and PtrBPL2. PtrBPL1 and PtrBPL2 coding regions were expressed under the control of the AtBOP1 promoter in bop1 bop2 plants. Representative plants are shown in the T2 generation. See Supplementary Table S2 for quantitative analysis of complementation for bop1 bop2 defects in leaf morphology, floral patterning, and abscission. (A, B) Wild-type plant showing (A) smooth leaf petioles and (B) inflorescence with floral organ abscission. (C, D) A bop1 bop2 mutant showing (C) leaves with a blade-on-petiole phenotype (arrow) and (D) inflorescence with asymmetric flowers and loss of floral organ abscission (arrows). (E, F) A BOP1p:PtrBPL1 bop1 bop2 plant showing (E) smooth leaf petioles and (F) inflorescence with floral organ abscission restored. (G, H) A BOP1p:PtrBPL2 bop1 bop2 plant showing (G) smooth leaf petioles and (H) inflorescence with floral organ abscission restored. Scale bars, 1.5 cm.
Overexpression of PtrBPL1 and PtrBPL2 alters secondary growth
Plants that overexpress AtBOP1 or AtBOP2 are short and bushy, with a wider pattern of secondary lignin deposition in stems (Khan et al., 2012b). Unlike trees, a continuous vascular cambium does not form except at the base of the stem. Instead, secondary growth is evident as the differentiation of abundant lignified interfascicular fibers (Ehlting et al., 2005).
To test the effect of PtrBPL1 and PtrBPL2 on stem development, Arabidopsis plants were transformed with a construct driving PtrBPL1 and PtrBPL2 expression from a strong, constitutive double cauliflower mosaic virus 35S promoter (D35S). Flowering D35S:PtrBPL1 and D35S:PtrBPL2 plants showed a loss of apical dominance and reduced stature in comparison to wild-type plants (Figure 4; Supplementary Table S3). The stems of 5-6 representative D3SS : PtrBPL1 and D35S:PtrBPL2 plants were hand-sectioned and stained with phloroglucinol to test for changes in lignin deposition. Several plants in this population showed an expanded pattern of lignin deposition in stems.
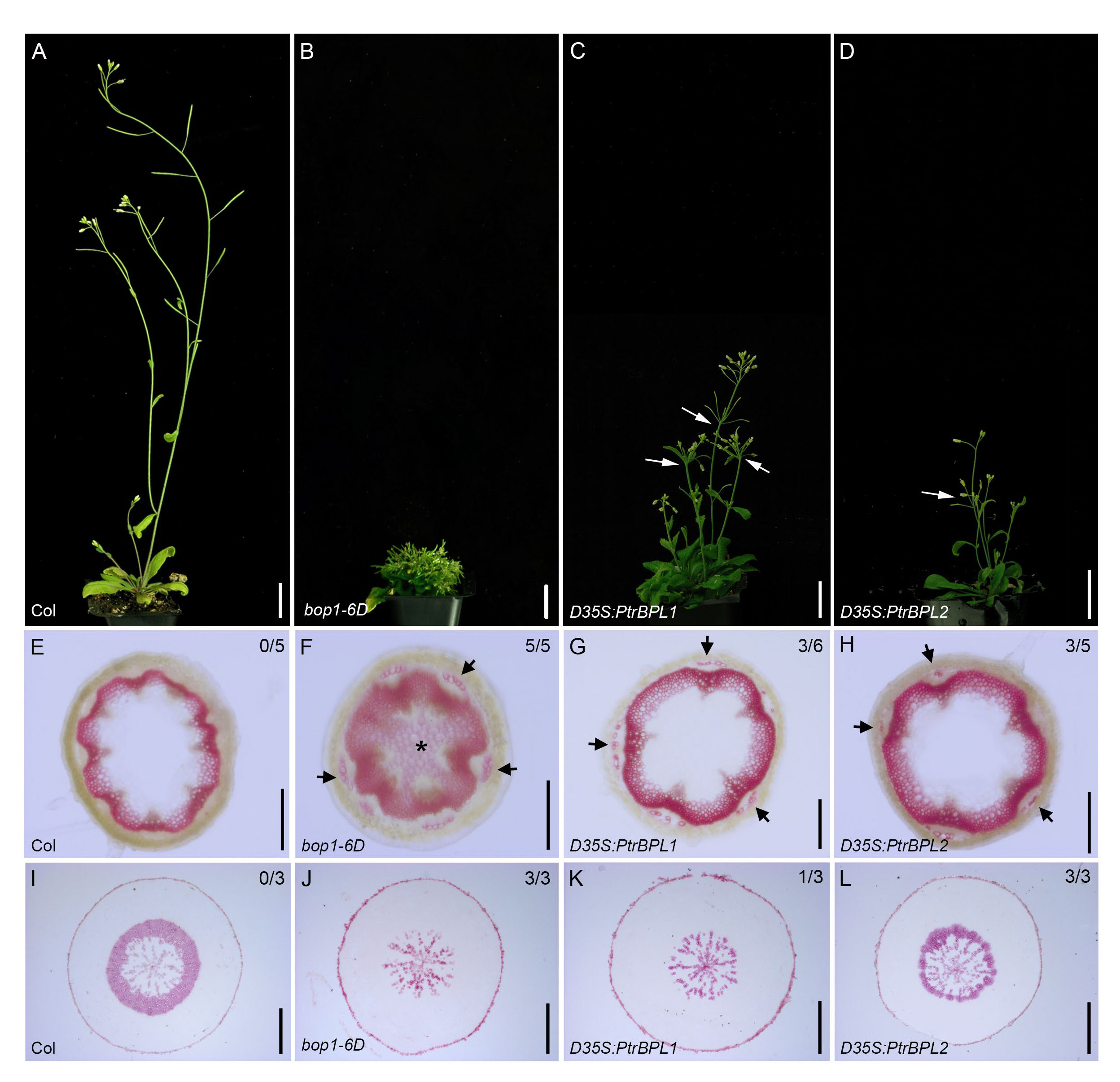
Figure 4 Overexpression of PtrBPL1 and PtrBPL2 in Arabidopsis plants. PtrBPL1 and PtrBPL2 coding regions were expressed under the control of a double 35S (D35S) cauliflower mosaic virus promoter in wild-type Arabidopsis plants. (A-D) Representative flowering plants are shown in the T1 generation. See Supplementary Table S3 for quantitative analysis. (A) Wild type plant, showing strong apical dominance and elongated internodes. (B) bop1-6D plant, showing a bushy, dwarf stature. (C) D35S:PtrBPL1 plant, showing a bushy, semi-dwarf stature. Arrows, clustered flowers and siliques. (D) D35S:PtrBPL2 plant, showing a bushy, semi-dwarf stature. Arrows, clustered flowers and siliques. (E-H) Transverse sections from the base of fully elongated stems were stained phloroglucinol-HCl to reveal lignin (pink). Top left, number of independent transgenic lines showing abnormal lignin deposition. Representative sections are shown for: (E) wild-type stem, showing a continuous vascular ring. (F) bop1-6D stem, showing a thicker vascular ring, lignified pith (asterisk) and lignified phloem fibers (arrows) (G) D35S:PtrBPL1 stem, showing a thick vascular ring and lignified phloem fibers (arrows). (H) D35S:PtrBPL2 stem, showing a thick vascular ring and lignified phloem fibers (arrows). (I-L) Transverse sections from the middle of the hypocotyl (1.5 mm below the rosette leaves) were stained phloroglucinol-HCl to reveal lignin (pink). Top left, number of independent transgenic lines showing abnormal lignin deposition. Representative sections are shown for: (I) Wild-type hypocotyl, showing a thick ring of xylem II. (J) bop1-6D hypocotyl, showing a lack of xylem II fibers and vessels. (K) D35S:PtrBPL1 hypocotyl, showing a lack of xylem II fibers and vessels. (L) D35S:PtrBPL2 hypocotyl, showing a xylem II ring of reduced thickness, compared to the wild type. Scale bars: (A-D) 1.5 cm; (E-H) 0.25 mm; (I-L) 0.5 mm.
In the root-hypocotyl where a continuous vascular cambium forms, two phases of secondary growth take place (Chaffey et al., 2002; Nieminen et al., 2015). During the first phase, the xylem I is composed of lignified vessel cells in a matrix of non-lignified parenchyma. Flowering triggers the second phase, in which xylem II forms as a thick ring composed of lignified fibers interspersed with lignified vessels (Chaffey et al., 2002; Nieminen et al., 2015). Arabidopsis plants that overexpress AtBOP1 and AtBOP2 lack xylem II features in the upper part of the root (Woerlen et al., 2017) and hypocotyl (Liebsch et al., 2014).
Analysis of secondary growth in the hypocotyl of D35S:PtrBPL1 and D35S:PtrBPL2 lines showed at least one D35S:PtrBPL1 line (n=3) missing xylem II fibers and vessels similar to bop1-6D and 35S:BOP2 control plants (Figure 4; Supplementary Figure S5). The xylem II ring in all D35S:PtrBPL2 lines (n=3) was reduced in thickness compared to wild-type control plants. These data provide further evidence that PtrBPL1 and PtrBPL2 can functionally substitute in Arabidopsis plants.
PtrBPL1 and PtrBPL2 interact with TGA bZIP factors
BTB-ankyrin proteins perform a variety of function by utilizing TGA bZIP factors as interaction partners (Backer et al., 2019). For example, Arabidopsis BOP1 and BOP2 interact with clade I and clade III TGAs in yeast (Hepworth et al., 2005). Such complexes are implicated in the regulation of stem development and lignin deposition in plants (Wang et al., 2019; Zhang et al., 2019). Arabidopsis BOP1 and BOP2 also interact with PAN/TGA8 for patterning functions in the flower (Hepworth et al., 2005). A highly sensitive method for detecting protein-protein interactions, we used the yeast two-hybrid assay to assess PtrBPL interactions with AtTGA factors. PtrBPL1 and PtrBPL2 fused to the DNA-binding domain of the yeast transcription activator protein GAL4 were used as bait. AtTGA1, AtTGA4, AtTGA3, and AtTGA7 fused to the activation domain of GAL4 were used as prey. Figure 5 shows that PtrBPL1 and PtrBPL2 interact with AtTGA1, AtTGA4, AtTGA3, and AtTGA7 providing evidence of conserved functional partners in Arabidopsis and poplar.
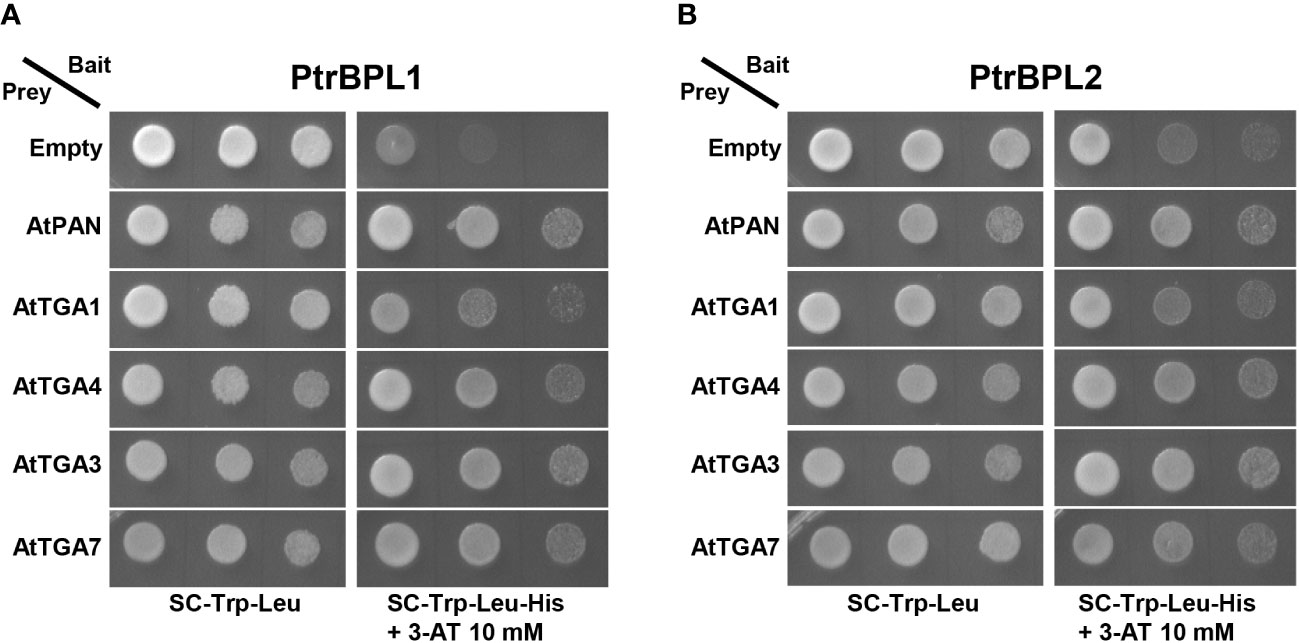
Figure 5 Pair-wise yeast two-hybrid assays showing (A) PtrBPL1 and (B) PtrBPL2 interaction with AtTGAs. PtrBPL1 and PtrBPL2 baits were fused to the DNA-binding domain of yeast GAL4. AtTGA1, AtTGA4, AtTGA3, and AtTGA7 preys were fused to the transcriptional activation domain of yeast GAL4. Plasmid constructs were co-transformed into yeast AH109 strain before serial dilutions (10-1, 10-2, 10-3) were plated onto SC/-Trp/-Leu medium with or without histidine + 10 mM 3-AT. Growth on SC/-Trp/-Leu/-His/+3-AT above background confirms a protein-protein interaction. Dilutions were spotted in replicate onto SC/-Trp/-Leu/-His/+3-AT medium. Photos on SC/-Trp/-Leu were taken three days after plating. For all assays, interaction with AtPAN/TGA8 was used as a positive control and an empty prey vector was used as a negative control.
Discussion
Trees display prominent radial growth in the stem in which secondary xylem and phloem tissues are produced by the vascular cambium. Current knowledge regarding the genetic regulation of cambium activity and secondary growth is still incomplete (Wang et al., 2021). Therefore, understanding gene families potentially involved in this process is desirable.
The BOP family of co-transcriptional regulators are conserved in dicots and monocots and contribute to numerous developmental processes. Studies in dicots such as tobacco (Wu et al., 2012), legumes (Frankowski et al., 2015; Couzigou et al., 2016; Magne et al., 2018), tomato (Xu et al., 2016; Izhaki et al., 2018) and monocots such as rice (Toriba et al., 2020), barley (Tavakol et al., 2015; Jost et al., 2016) and Brachypodium (Magne et al., 2020; Liu et al., 2022) reveal highly conserved functions, comparable to Arabidopsis, that influence organ patterning, inflorescence architecture, and abscission (Khan et al., 2014; Hepworth and Pautot, 2015). Taxon-specific roles in the maintenance of symbiotic nodule identity (Couzigou et al., 2012; Magne et al., 2018) and rhizome development (Toriba et al., 2020) have also been identified.
In flowering plants, BOPs are involved in boundary formation. These specific domains are important for the separation of meristematic regions from lateral organs (Aida and Tasaka, 2006; Žádníková and Simon, 2014). Compared to the shoot apical meristem, our knowledge of boundaries and their involvement in cambium meristems is limited. The conservation of BOPs across the plant kingdom (Khan et al., 2014) makes it plausible that orthologs in trees have roles in the vascular cambium and secondary growth.
The results shown here indicate that PtrBPL1 and PtrBPL2 play a role in secondary growth when overexpressed in Arabidopsis. D35S:PtrBPL1 and D35S:PtrBPL2 transgenic plants were characterized by dwarf stems with a thicker vascular ring and more developed phloem fibers whereas the increased activity of these genes in the hypocotyl interfered with xylem II production. Overexpression of either PtrBPL1 or PtrBPL2 resulted in phenotypes that were highly similar to those observed in Arabidopsis plants with overexpression of AtBOP1 or AtBOP2 (Khan et al., 2012b; Woerlen et al., 2017). Furthermore, the expression of PtrBPL1 or PtrBPL2 transgenes in a bop1 bop2 mutant background was sufficient to rescue leaf and floral patterning defects. Abscission defects in the bop1 bop2 mutant were also corrected. The PtrBPL1p:GUS and PtrBPL2p:GUS expression patterns in Arabidopsis plants were consistent with a role in boundary patterning and secondary growth. Finally, we identified a family of TGA bZIP factors in poplar with a clade structure similar that in Arabidopsis plants. Yeast two hybrid assays indicated that PtrBPL1 and PtrBPL2 can interact with AtTGAs known to mediate roles in plant development and lignin deposition. All of these results strongly suggest that BOP1/2-TGA modules characterized in Arabidopsis are conserved in P. trichocarpa. Therefore, it is feasible they contribute to stem development in trees.
In Arabidopsis plants, BOP1 and BOP2 genes are repressed by the class I KNOX homeodomain transcription factors SHOOT MERISTEMLESS (STM) and BREVIPEDICELLUS (BP) during meristem maintenance, stem development, and secondary xylem formation (Jun et al., 2010; Khan et al., 2012b; Liebsch et al., 2014; Khan et al., 2015; Woerlen et al., 2017). Consistent with these findings, PtrBPL1 is significantly downregulated in the stem of hybrid poplar overexpressing the STM ortholog ARBORKNOX1 (ARK1) (Liu et al., 2015). Both ARK1 and ARK2 (ortholog of BP) are strongly expressed in the cambial zone (Groover et al., 2006; Du et al., 2009). In the ARK1 overexpressor, the boundary between the cambium and the secondary xylem was uneven and phloem fibers were reduced in number (Groover et al., 2006). In the ARK2 overexpressor, extra secondary phloem was produced at the expense of phloem fibers and secondary xylem leading to an overall decrease in the differentiation of lignified cell-types. Conversely, an ark2 mutant generated by artificial miRNA silencing showed a premature differentiation of secondary xylem and phloem fibers, similar to the bp mutant in Arabidopsis (Du et al., 2009).
Studies in the tropical Cannabaceae tree Parasponia andersonii species complement these findings (Shen et al., 2021). CRISPR-Cas9 loss-of-function mutants in the AtBOP1 ortholog PanNODULE ROOT1 (PanNOOT1) were altered in the development of xylem and phloem tissues without any obvious difference in cambium organization and size. Compared to the wild-type, the differentiation of secondary xylem and phloem was inhibited resulting in a reduced stem diameter. Transcriptomic data showed a reduction in the expression of lignin metabolism-related genes (Shen et al., 2021). In both Arabidopsis and cotton (Gossypium hirsutum) and the grass Brachypodium distachyon model, BOPs positively regulate the expression of lignin biosynthesis genes and lignin deposition in stems (Khan et al., 2012b; Zhang et al., 2019; Liu et al., 2022).
In Arabidopsis plants, BOP1 and BOP2 promote the expression of two homeobox genes to influence lignin deposition (Khan et al., 2012a; Khan et al., 2012b; Woerlen et al., 2017). Poplar plants contain homeobox genes that are highly similar to ARABIDOPSIS HOMEOBOX GENE 1 (ATH1) (Potri.018G054700/Potri.006G230700) and KNOTTED-LIKE FROM ARABIDOPSIS THALIANA 6 (KNAT6) (Potri.010G043500/Potri.008G188700) suggesting that downstream pathway components are conserved. The homologous genes in P. andersonii were significantly downregulated in Pannoot1 stems showing a similar regulation as in Arabidopsis plants (Shen et al., 2021). The PagKNAT2/6b gene from a hybrid poplar clone (P. alba X P. glandulosa) is highly expressed in xylem and phloem. Compared to controls plants, transgenic poplar clones overexpressing PagKNAT2/6b showed a reduction in xylem formation by negatively regulating the expression of NAC domain transcription factor genes including PagXYLEM NAC DOMAIN 1 as a direct target (Zhao et al., 2020).
Spatial and temporal differences between PtrBPL1p:GUS and PtrBPL2p:GUS expression were observed. In Arabidopsis plants, the PtrBPL1 promoter was active in xylem I whereas PtrBPL2 the promoter was active at the boundary of the cambial zone and secondary phloem (Figures 2A, B, D). By contrast, a PanNOOT1p:GUS reporter was expressed in all three tissues during secondary growth (Shen et al., 2021). A similar pattern to PanNOOT1 was detected in aspen (P. tremula) and birch (Betula pendula) using high-spatial-resolution gene expression mapping (Sundell et al., 2017; Alonso-Serra et al., 2019; Shen et al., 2021). By contrast, AtBOP1p:GUS and AtBOP2p:GUS expression is normally repressed by BP in the cambial zone and secondary xylem of the root and hypocotyl (Liebsch et al., 2014; Khan et al., 2015; Woerlen et al., 2017). Thus, divergent transcriptional regulation may account for phenotypic differences in secondary growth between species. Compared to AtBOP1 and AtBOP2, the PtrBPL1 and PtrBPL2 promoters had a higher number of predicted binding sites for MYB-related and NAC transcription factors, which as a group contribute strongly to secondary growth in plants (Nakano et al., 2015).
Considering all of the above, BOP-TGA modules characterized in Arabidopsis are likely conserved in poplar. Information gained from this study forms a basis for further exploration of networks that regulate cambium development and secondary growth in trees.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
SH and EL designed the research. SL, BD, GA, JM, and EL performed the research. AB carried out bioinformatics analysis. SL, JM, and SH wrote the article. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada (RGPIN-2016-06193).
Acknowledgments
We thank Carl Douglas and Shawn Mansfield at the University of British Columbia for the gift of poplar cuttings, vectors, and poplar tissue cDNA. We thank Ying Wang and Kevin Xiong for help with cloning and Jenna O’Neill for images of GUS-stained seedlings.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1244583/full#supplementary-material
References
Aida, M., Tasaka, M. (2006). Genetic control of shoot organ boundaries. Curr. Opin. Plant Biol. 9, 72–77. doi: 10.1016/j.pbi.2005.11.011
Alonso-Serra, J., Safronov, O., Lim, K. J., Fraser-Miller, S. J., Blokhina, O. B., Campilho, A., et al. (2019). Tissue-specific study across the stem reveals the chemistry and transcriptome dynamics of birch bark. New Phytol. 222, 1816–1831. doi: 10.1111/nph.15725
Backer, R., Naidoo, S., van den Berg, N. (2019). The NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1) and related family: mechanistic insights in plant disease resistance. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00102
Bryant, N. D., Pu, Y., Tschaplinski, T. J., Tuskan, G. A., Muchero, W., Kalluri, U. C., et al. (2020). Transgenic poplar designed for biofuels. Trends Plant Sci. 25, 881–896. doi: 10.1016/j.tplants.2020.03.008
Chaffey, N., Cholewa, E., Regan, S., Sundberg, B. (2002). Secondary xylem development in Arabidopsis: a model for wood formation. Plant Physiol. 114, 594–600. doi: 10.1034/j.1399-3054.2002.1140413.x
Chahtane, H., Zhang, B., Norberg, M., Lemasson, M., Thévenon, E., Bakó, L., et al. (2018). LEAFY activity is post-transcriptionally regulated by BLADE ON PETIOLE2 and CULLIN3 in Arabidopsis. New Phytol. 220, 579–592. doi: 10.1111/nph.15329
Chanoca, A., de Vries, L., Boerjan, W. (2019). Lignin engineering in forest trees. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00912
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cooke, J. E. K., Rood, S. B. (2007). Trees of the people: the growing science of poplars in Canada and worldwide. Can. J. Bot. 85, 1103–1110. doi: 10.1139/b07-125
Couzigou, J. M., Magne, K., Mondy, S., Cosson, V., Clements, J., Ratet, P. (2016). The legume NOOT-BOP-COCH-LIKE genes are conserved regulators of abscission, a major agronomical trait in cultivated crops. New Phytol. 209, 228–240. doi: 10.1111/nph.13634
Couzigou, J. M., Zhukov, V., Mondy, S., Heba, G. A., Cosson, V., Noel Ellis, T. N., et al. (2012). NODULE ROOT and COCHLEATA maintain nodule development and are legume orthologs of Arabidopsis BLADE-ON-PETIOLE genes. Plant Cell 24, 4498–4510. doi: 10.1105/ypc.112.103747
Du, J., Mansfield, S. D., Groover, A. T. (2009). The Populus homeobox gene ARBORKNOX2 regulates cell differentiation during secondary growth. Plant J. 60, 1000–1014. doi: 10.1111/j.1365-313x.2009.04017.x
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Ehlting, J., Mattheus, N., Aeschliman, D. S., Li, E., Hamberger, B., Cullis, I. F., et al. (2005). Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J. 42, 618–640. doi: 10.1111/j.1365-313x.2005.02403.x
Frankowski, K., Wilmowicz, E., Kućko, A., Zienkiewicz, A., Zienkiewicz, K., Kopcewicz, J. (2015). Molecular cloning of the BLADE-ON-PETIOLE gene and expression analyses during nodule development in Lupinus luteus. Plant Physiol. 179, 35–39. doi: 10.1016/j.jplph.2015.01.019
Fu, Z. Q., Yan, S., Saleh, A., Wang, W., Ruble, J., Oka, N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232. doi: 10.1038/nature11162
Gietz, R. D., Schiestl, R. H. (2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34. doi: 10.1038/nprot.2007.13
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Groover, A. T., Mansfield, S. D., DiFazio, S. P., Dupper, G., Fontana, J. R., Millar, R., et al. (2006). The Populus homeobox gene ARBORKNOX1 reveals overlapping mechanisms regulating the shoot apical meristem and the vascular cambium. Plant Mol. Biol. 61, 917–932. doi: 10.1007/s11103-006-0059-y
Haughn, G. W., Somerville, C. (1986). Sulfonylurea-resistant mutants of Arabidopsis thaliana. Molec. Gen. Genet. 204, 430–434. doi: 10.1007/bf00331020
He, L., Lei, Y., Li, X., Peng, Q., Liu, W., Jiao, K., et al. (2020). Symmetric petals 1 encodes an ALOG domain protein that controls floral organ internal asymmetry in pea (Pisum sativum L.). Int. J. Mol. Sci. 21, 1–15. doi: 10.3390/ijms21114060
Hellens, R. P., Anne Edwards, E., Leyland, N. R., Bean, S., Mullineaux, P. M. (2000). pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. doi: 10.1023/a:1006496308160
Hepworth, S. R., Pautot, V. A. (2015). Beyond the divide: boundaries for patterning and stem cell regulation in plants. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01052
Hepworth, S. R., Zhang, Y., McKim, S., Li, X., Haughn, G. W. (2005). BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17, 1434–1448. doi: 10.1105/tpc.104.030536
Izhaki, A., Alvarez, J. P., Cinnamon, Y., Genin, O., Liberman-Aloni, R., Eyal, Y. (2018). The tomato BLADE ON PETIOLE and TERMINATING FLOWER regulate leaf axil patterning along the proximal-distal axes. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01126
Jansson, S., Douglas, C. J. (2007). Populus: a model system for plant biology. Annu. Rev. Plant Biol. 58, 435–458. doi: 10.1146/annrrev.arplant58.032806.103956
Jost, M., Taketa, S., Mascher, M., Himmelbach, A., Yuo, T., Shahinnia, F., et al. (2016). A homolog of BLADE-ON-PETIOLE 1 and 2 (BOP1/2) controls internode length and homeotic changes of the barley inflorescence. Plant Physiol. 171, 1113–1127. doi: 10.1104/pp.16.00124
Jun, J. H., Ha, C. M., Fletcher, J. C. (2010). BLADE-ON-PETIOLE1 coordinates organ determinacy and axial polarity in Arabidopsis by directly activating ASYMMETRIC LEAVES2. Plant Cell 22, 62–76. doi: 10.1105/tpc.109.070763
Khan, M., Ragni, L., Tabb, P., Salasini, B. C., Chatfield, S., Datla, R., et al. (2015). Repression of lateral organ boundary genes by PENNYWISE and POUND-FOOLISH is essential for meristem maintenance and flowering in Arabidopsis. Plant Physiol. 169, 2166–2186. doi: 10.1104/pp.15.00915
Khan, M., Tabb, P., Hepworth, S. R. (2012a). BLADE-ON-PETIOLE1 and 2 regulate Arabidopsis inflorescence architecture in conjunction with homeobox genes KNAT6 and ATH1. Plant Signal. Behav. 7, 788–792. doi: 10.4161/psb.20599
Khan, M., Xu, H., Hepworth, S. R. (2014). BLADE-ON-PETIOLE genes: setting boundaries in development and defense. Plant Sci. 215–216, 157–171. doi: 10.1016/j.plantsci.2013.10.019
Khan, M., Xu, M., Murmu, J., Tabb, P., Liu, Y., Storey, K., et al. (2012b). Antagonistic interaction of BLADE-ON-PETIOLE1 and 2 with BREVIPEDICELLUS and PENNYWISE regulates Arabidopsis inflorescence architecture. Plant Physiol. 158, 946–960. doi: 10.1104/pp.111.188573
Koncz, C., Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Molec. Gen. Genet. 204, 383–396. doi: 10.1007/bf00331014
Liebsch, D., Sunaryo, W., Holmlund, M., Norberg, M., Zhang, J., Hall, H. C., et al. (2014). Class I KNOX transcription factors promote differentiation of cambial derivatives into xylem fibers in the Arabidopsis hypocotyls. Development 141, 4311–4319. doi: 10.1242/dev.111369
Liu, L., Ramsay, T., Zinkgraf, M., Sundell, D., Street, N. R., Filkov, V., et al. (2015). A resource for characterizing genome-wide binding and putative target genes of transcription factors expressed during secondary growth and wood formation in Populus. Plant J. 82, 887–898. doi: 10.1111/tpj.12850
Liu, S., Magne, K., Daniel, S., Sibout, R., Ratet, P. (2022). Brachypodium distachyon UNICULME4 and LAXATUM-A are redundantly required for development. Plant Physiol. 188, 363–381. doi: 10.1093/plphys/kiab456
Lu, Q., Tang, X., Tian, G., Wang, F., Liu, K., Nguyen, V., et al. (2009). Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 61, 259–270. doi: 10.1111/j.1365-313X.2009.04048.x
Magne, K., Couzigou, J. M., Schiessl, K., Liu, S., George, J., Zhukov, V., et al. (2018). MtNODULE ROOT1 and MtNODULE ROOT2 are essential for indeterminate nodule identity. Plant Physiol. 178, 295–316. doi: 10.1104/pp.18.00610
Magne, K., Liu, S., Massot, S., Dalmais, M., Morin, H., Sibout, R., et al. (2020). Roles of BdUNICULME4 and BdLAXATUM-A in the non-domesticated grass Brachypodium distachyon. Plant J. 103, 645–659. doi: 10.1111/tpj.14758
McKim, S. M., Stenvik, G. E., Butenko, M. A., Kristiansen, W., Cho, S. K., Hepworth, S. R., et al. (2008). The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 135, 1537–1546. doi: 10.1242/dev.012807
Murmu, J., Bush, M. J., DeLong, C., Li, S., Xu, M., Khan, M., et al. (2010). Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol. 154, 1492–1504. doi: 10.1104/pp.110.159111
Nakano, Y., Yamaguchi, M., Endo, H., Rejab, N. A., Ohtani, M. (2015). NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 6, 288. doi: 10.3389/fpls.2015.00288
Nieminen, K., Blomster, T., Helariutta, Y., Mähönen, A. P. (2015). Vascular cambium development. Arabidopsis Book 13, e0177. doi: 10.1199/tab.0177
Norberg, M., Holmlund, M., Nilsson, O. (2005). The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development 132, 2203–2213. doi: 10.1242/dev.01815
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29, e45–e45. doi: 10.1093/nar/29.9.e45
Rogers, L. A., Campbell, M. M. (2004). The genetic control of lignin deposition during plant growth and development. New Phytol. 164, 17–30. doi: 10.1111/j.1469-8137.2004.01143.x
Sanchez, P., Nehlin, L., Greb, T. (2012). From thin to thick: major transitions during stem development. Trends Plant Sci. 17, 113–121. doi: 10.1016/j.tplants.2011.11.004
Sannigrahi, P., Ragauskas, A. J., Tuskan, G. A. (2010). Poplar as a feedstock for biofuels: A review of compositional characteristics. Biofuel. Bioprod. Biorefin. 4, 209–226. doi: 10.1002/bbb.206
Sehr, E. M., Agusti, J., Lehner, R., Farmer, E. E., Schwarz, M., Greb, T. (2010). Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation. Plant J. 63, 811–822. doi: 10.1111/j.1365-313x.2010.04283.x
Shen, D., Holmer, R., Kulikova, O., Mannapperuma, C., Street, N. R., Yan, Z., et al. (2021). The BOP-type co-transcriptional regulator NODULE ROOT1 promotes stem secondary growth of the tropical Cannabaceae tree Parasponia andersonii. Plant J. 106, 1366–1386. doi: 10.1111/tpj.15242
Song, J., Lu, S., Chen, Z. Z., Lourenco, R., Chiang, V. L. (2006). Genetic transformation of Populus trichocarpa genotype Nisqually-1: a functional genomic tool for woody plants. Plant Cell Physiol. 47, 1582–1589. doi: 10.1093/pcp/pcl018
Sundell, D., Street, N. R., Kumar, M., Mellerowicz, E. J., Kucukoglu, M., Johnsson, C., et al. (2017). AspWood: High-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell 29, 1585–1604. doi: 10.1105/tpc.17.00153
Tavakol, E., Okagaki, R., Verderio, G., Vahid, S. J., Hussien, A., Bilgic, H., et al. (2015). The barley Uniculme 4 gene encodes a BLADE-ON-PETIOLE-like protein that controls tillering and leaf patterning. Plant Physiol. 168, 164–174. doi: 10.1104/pp.114.252882
Taylor, G. (2002). Populus: Arabidopsis for forestry. Do we need a model tree? Ann. Bot. 90, 681–689. doi: 10.1093/aob/mcf255
Toriba, T., Tokunaga, H., Nagasawa, K., Nie, F., Yoshida, A., Kyozuka, J. (2020). Suppression of leaf blade development by BLADE-ON-PETIOLE orthologs is a common strategy for underground rhizome growth. Curr. Biol. 30, 509–516. doi: 10.1016/j.cub.2019.11.055
Toriba, T., Tokunaga, H., Shiga, T., Nie, F., Naramoto, S., Honda, E., et al. (2019). BLADE-ON-PETIOLE genes temporally and developmentally regulate the sheath to blade ratio of rice leaves. Nat. Commun. 10, 619. doi: 10.1038/s41467-019-08479-5
Unda, F., Kim, H., Hefer, C., Ralph, J., Mansfield, S. D. (2017). Altering carbon allocation in hybrid poplar (Populus alba × grandidentata) impacts cell wall growth and development. Plant Biotechnol. J. 15, 865–878. doi: 10.1111/pbi.12682
Wang, D., Chen, Y., Li, W., Li, Q., Lu, M., Zhou, G., et al. (2021). Vascular cambium: the source of wood formation. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.700928
Wang, S., Li, E., Porth, I., Chen, J. G., Mansfield, S. D., Douglas, C. J. (2014). Regulation of secondary cell wall biosynthesis by poplar R2R3 MYB transcription factor PtrMYB152 in Arabidopsis. Sci. Rep. 4, 5054. doi: 10.1038/srep05054
Wang, Y., Salasini, B. C., Khan, M., Devi, B., Bush, M., Subramaniam, R., et al. (2019). Clade I TGACG-motif binding basic leucine zipper transcription factors mediate BLADE-ON-PETIOLE-dependent regulation of development. Plant Physiol. 180, 937–951. doi: 10.1104/pp.18.00805
Woerlen, N., Allam, G., Popescu, A., Corrigan, L., Pautot, V., Hepworth, S. R. (2017). Repression of BLADE-ON-PETIOLE genes by KNOX homeodomain protein BREVIPEDICELLUS is essential for differentiation of secondary xylem in Arabidopsis root. Planta 245, 1079–1090. doi: 10.1007/s00425-017-2663-2
Wu, X. M., Yu, Y., Han, L. B., Li, C. L., Wang, H. Y., Zhong, N. Q., et al. (2012). The tobacco BLADE-ON-PETIOLE2 gene mediates differentiation of the corolla abscission zone by controlling longitudinal cell expansion. Plant Physiol. 159, 835–850. doi: 10.1104/pp.112.193482
Xu, C., Park, S. J., Van Eck, J., Lippman, Z. B. (2016). Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genes Dev. 30, 2048–2061. doi: 10.1101/gad.288415.116
Žádníková, P., Simon, R. (2014). How boundaries control plant development. Curr. Opin. Plant Biol. 17, 116–125. doi: 10.1016/j.pbi.2013.11.013
Zhang, B., Holmlund, M., Lorrain, S., Norberg, M., Bakó, L. S., Fankhauser, C., et al. (2017). BLADE-ON-PETIOLE proteins act in an E3 ubiquitin ligase complex to regulate PHYTOCHROME INTERACTING FACTOR 4 abundance. elife 6, e26759. doi: 10.7554/elife.26759
Zhang, Z., Wang, P., Luo, X., Yang, C., Tang, Y., Wang, Z., et al. (2019). Cotton plant defence against a fungal pathogen is enhanced by expanding BLADE-ON-PETIOLE1 expression beyond lateral-organ boundaries. Commun. Biol. 2, 238. doi: 10.1038/s42003-019-0468-5
Keywords: BLADE-ON-PETIOLE, TGA bZIP, Populus trichocarpa, secondary growth, lignin
Citation: Li S, Devi B, Allam G, Bhullar A, Murmu J, Li E and Hepworth SR (2023) Regulation of secondary growth by poplar BLADE-ON-PETIOLE genes in Arabidopsis. Front. Plant Sci. 14:1244583. doi: 10.3389/fpls.2023.1244583
Received: 22 June 2023; Accepted: 17 October 2023;
Published: 14 November 2023.
Edited by:
Verónica S. Di Stilio, University of Washington, United StatesReviewed by:
Ana Campilho, University of Porto, PortugalJesse L. Labbé, Technology Holding, United States
Copyright © 2023 Li, Devi, Allam, Bhullar, Murmu, Li and Hepworth. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shelley R. Hepworth, c2hlbGxleS5oZXB3b3J0aEBjYXJsZXRvbi5jYQ==
†Present address: Gamalat Allam, Department of Biology, Western University, London, ON, Canada
‡These authors have contributed equally to this work
 Sibei Li
Sibei Li Bhaswati Devi‡
Bhaswati Devi‡ Gamalat Allam
Gamalat Allam Armaan Bhullar
Armaan Bhullar Jhadeswar Murmu
Jhadeswar Murmu Shelley R. Hepworth
Shelley R. Hepworth