- 1Department of Breeding and Genomics, The New Zealand Institute for Plant and Food Research Limited, Christchurch, New Zealand
- 2Department of Plant and Microbial Biology, University of Zürich, Zurich, Switzerland
- 3Department of Plant Science, McGill University, Lincoln, QC, Canada
- 4Department of Wine, Food and Molecular Biosciences, Lincoln University, Canterbury, New Zealand
Pilosella piloselloides var. praealta (syn. P. praealta; Hieracium praealtum) is a versatile model used to study gametophytic apomixis. In this system apomixis is controlled by three loci: one that controls the avoidance of meiosis (LOA), one that controls the avoidance of fertilization (LOP) and a third that controls autonomous endosperm formation (AutE). Using a unique polyhaploid mapping approach the LOP locus was mapped to a 654 kb genomic interval syntenic to linkage group 8 of Lactuca sativa. Polyhaploids form through the gametophytic action of a dominant determinant at LOP, so the mapped region represents both a functional and a physical domain for LOP in P. piloselloides. Allele sequence divergence (ASD) analysis of the PARTHENOGENESIS (PAR) gene within the LOP locus revealed that dominant PAR alleles in Pilosella remain highly similar across the genus, whilst the recessive alleles are more divergent. A previous report noted that dominant PAR alleles in both Pilosella and Taraxacum are modified by the presence of a class II transposable element (TE) in the promoter of the gene. This observation was confirmed and further extended to the related genus Hieracium. Sufficient differences were noted in the structure and location of the TE elements to conclude that TE insertional events had occurred independently in the three genera. Measures of allele crossover amongst the polyhaploids revealed that P. piloselloides is an autopolyploid species with tetrasomic inheritance. It was also noted that the dominant determinant of LOP in P. piloselloides could transmit via a diploid gamete (pollen or egg) but not via a haploid gamete. Using this information, a model is presented of how gametophytic apomixis may have evolved in several members of the Lactuceae, a tribe of the Asteraceae.
Introduction
Pilosella piloselloides var. praealta (syn. P. praealta; Hieracium praealtum) (Asteraceae, Cichorieae) is a model plant used to study gametophytic apomixis, the asexual formation of seeds (Catanach et al., 2006; Rodrigues et al., 2008; Koltunow et al., 2011; Bicknell et al., 2016; Bräuning et al., 2018; Underwood et al., 2022). In this system, apomixis is conferred by the inheritance of dominant determinants at two, unlinked loci: one associated with the avoidance of meiosis (the LOSS OF APOMEIOSIS locus; LOA) and another with spontaneous embryo formation (the LOSS OF PARTHENOGENESIS locus; LOP) (Catanach et al., 2006; Koltunow et al., 2011; Shirasawa et al., 2014). In apomictic species, as in almost all angiosperms, embryo maturation requires the support of nutritive endosperm tissue. Apomictic species differ in the origin of this tissue. In some apomicts the endosperm is derived sexually (‘pseudogamous apomixis’) while in others it arises asexually (‘autonomous apomixis’). Apomictic species in the genus Pilosella feature autonomous apomixis, the consequence of which is that fertilization is neither required for initiation of the embryo nor for the formation of the endosperm. As a result, seed formation in apomictic Pilosella proceeds without pollination and often does so precociously, being well advanced before the flower opens (Koltunow et al., 1998). Autonomous endosperm in Pilosella piloselloides var. praealta (syn. H. praealtum; syn. Hieracium caespitosum) was recorded by Catanach et al. (2006) to be associated with the inheritance of a dominant allele at the LOP locus. Ogawa et al. (2013), however, described two segregants from amongst the progeny of related Pilosella species in which autonomous endosperm was expressed without parthenogenesis, suggesting the presence of an ‘AUTONOMOUS ENDOSPERM’ locus (AutE) in this genus. The structure, function and location of the AutE locus, however, remains unclear.
The dominant actions of the LOA and LOP loci are inferred by their influence when present as simplex copies. In Pilosella piloselloides var. piloselloides (syn. P. piloselloides; Hieracium piloselloides) and Pilosella piloselloides var. praealta (syn. Hieracium praealtum) the loss of LOA by deletion resulted in plants exhibiting a meiotic phenotype, while the loss of LOP resulted in plants requiring fertilization to initiate both embryogenesis and endosperm formation (Catanach et al., 2006; Koltunow et al., 2011) The loss of both loci resulted a ‘mutant’ phenotype of sexual reproduction, in this otherwise asexually reproducing plant. The uncovering of sexual reproduction following the removal of these loci supports the proposal that sexual reproduction is the base state with apomixis acting as an overlay process in this species (Koltunow et al., 2011). Further support for this hypothesis comes from studies of gene expression in apomictic and sexual biotypes of Pilosella. Tucker et al. (2003) reported that the same genes appear to act in both biotypes throughout most of gametogenesis, indicating that the sexual and apomictic pathways of reproduction in Pilosella (syn. Hieracium) differ in the expression of only a small percentage of the genes and/or alleles necessary to achieve the formation of a new sporophytic generation. Bräuning et al. (2018) further noted that the transcriptome of an LOP mutant of Pilosella piloselloides var. praealta surveyed over the period of parthenogenesis was substantially similar to that of the corresponding wild-type.
Shirasawa et al. (2014) constructed genetic linkage maps for two apomictic species, Pilosella piloselloides var. piloselloides (syn. Hieracium piloselloides) and Pilosella piloselloides var. praealta (syn. Hieracium praealtum) using SSR markers associated with expressed transcripts. In Pilosella piloselloides var. praealta the LOA and LOP loci were mapped to linkage groups LGR15 and LGR08 respectively, supporting previous findings that the two loci are unlinked in this plant. The partial structure of the LOA locus was described by Kotani et al. (2014). It was found to be a hemizygous region of low recombination, containing extensive, often complex repeat sequences. This is similar to the reported structure of the apospory-specific genomic region (ASGR) in the apomictic grass species Pennisetum squamulatum and Cenchrus ciliaris, which has an analogous function to LOA and LOP, combined (Roche et al., 1999; Akiyama et al., 2004; Zeng et al., 2011). Kotani et al. (2014) addressed the potential role of repeat sequence at the LOA locus, noting that LOA remained functional when it was separated from the majority of the surrounding repeat sequence through recombination. They concluded that the association between hemizygosity, repeat sequence and apomixis loci is more likely to be a consequence of the action of these loci in suppressing recombination, rather than a requirement for their function.
The complete structure of the LOP locus of Pilosella piloselloides var. praealta is currently unreported, although one gene within the locus, PAR, is now known. Underwood et al. (2022) reported the structure and function of the PAR gene of the apomict Taraxacum officinale (ToPAR), and also the orthologous sequence (PpPAR) from Pilosella piloselloides var. praealta. In both systems PAR acts as an initiator of parthenogenesis, activating the egg cell to enter into the first mitotic division of embryogenesis. The presence of a transposable element was noted in the promoters of the dominant alleles of PAR in both species and it was postulated that this element is responsible for an alteration in the expression of these alleles in apomictic plants.
Apomixis is a form of asexual reproduction. Consequently, it promotes the establishment of uniform populations, and enables the perpetuation of clonal lineages over multiple seedling generations. As with all forms of clonal reproduction, apomixis restricts potential genetic advance through recombination (Van Dijk et al., 2009; Hörandl, 2011; Mau et al., 2015). It has, therefore, been described as an ‘evolutionary dead-end’ (Darlington, 1939; Stebbins, 1950) but this viewpoint is now generally regarded as too simplistic (Van Dijk and Vijverberg, 2005). Apomixis is very seldom expressed as an obligate, or exclusive, trait. Typically, it co-exists with sexual reproduction, both through the perpetuation of populations of inter-fertile, mixed biotypes and/or through the expression of sexual and asexual developmental pathways on the same plant (Matzk et al., 2000; Bicknell et al., 2003; Hörandl and Hojsgaard, 2012; Hand and Koltunow, 2014; Hojsgaard and Hörandl, 2019). In this way ‘facultatively apomictic’ species retain the ability to exploit allelic crossover and recombination, facilitating their advance under selection. The genus Pilosella provides a good example. Both sexual and apomictic biotypes are commonly found in close proximity, and they are known to readily hybridize in natural habitats (Krahulcová and Krahulec, 1999; Houliston and Chapman, 2001; Krahulcová et al., 2001; Fehrer et al., 2007; Mráz et al., 2009). Also, apomixis in this genus is facultative, with progeny forming from both sexual and asexual developmental pathways on the same plant, and occasionally from a combination of both Bicknell et al. (2003). One expected consequence of facultative apomixis is that, while allele diversity is expected to occur in these plants, patterns of ‘Allele Sequence Divergence’ (ASD) will differ between genomic intervals associated with the expression of apomixis and regions that are not critical to the inheritance of the trait. This effect arises because alleles which confer elements of apomixis are more likely to reside within clonal lineages. Conversely, alleles that are not involved in the inheritance of apomixis will more frequently reside in sexual biotypes and be subject to greater rates of recombination. Studies of ASD in apomicts, however, are uncommon, principally because so little is known of the loci that control apomixis.
The aims of this study were to; define the limits of the functional domain of the LOP locus in Pilosella piloselloides var. praealta; compare rates of recombination around LOP against the physical map of the locus; measure allelic exchange near LOP within this autopolyploid species; characterize ASD amongst a group of related species at LOP, and finally to measure LOP allele exchange through gametes of different ploidy. We propose a model of how gametophytic apomixis may have arisen in the genus Pilosella and other members of the Lactuceae.
Materials and methods
Plant material
Throughout the study, the accession Pilosella piloselloides var. praealta R35 was used as the apomictic, wild-type, control. A taxonomic re-assessment of the genera Hieracium and Pilosella (Bräutigam and Greuter, 2007) resulted in the re-organization of these taxa, and also in name changes for several of the species. In a previous deletion mapping study (Catanach et al., 2006), R35 was given the name H. caespitosum C4D. This plant was later reassigned as Hieracium praealtum R35 and is now designated as Pilosella piloselloides var. praealta R35. This is the name used throughout this publication; however, as earlier references use alternative names, synonyms are provided in the introduction and discussion sections to aid understanding. Eleven of the mutants described by Catanach et al. (LOP γ115, LOP γ116, LOP γ136, LOP γ138, LOP γ143, LOP γ144, LOP γ161 and LOP γ179; LOA γ165, LOA γ134 and LOA γ186) were used in the current study to construct tiles of Bacterial Artificial Chromosome (BAC) clones and to facilitate construction of the physical map of LOP. Other wild-type plants (as listed in Table 1) were obtained from European botanical gardens or were collected from natural habitats.
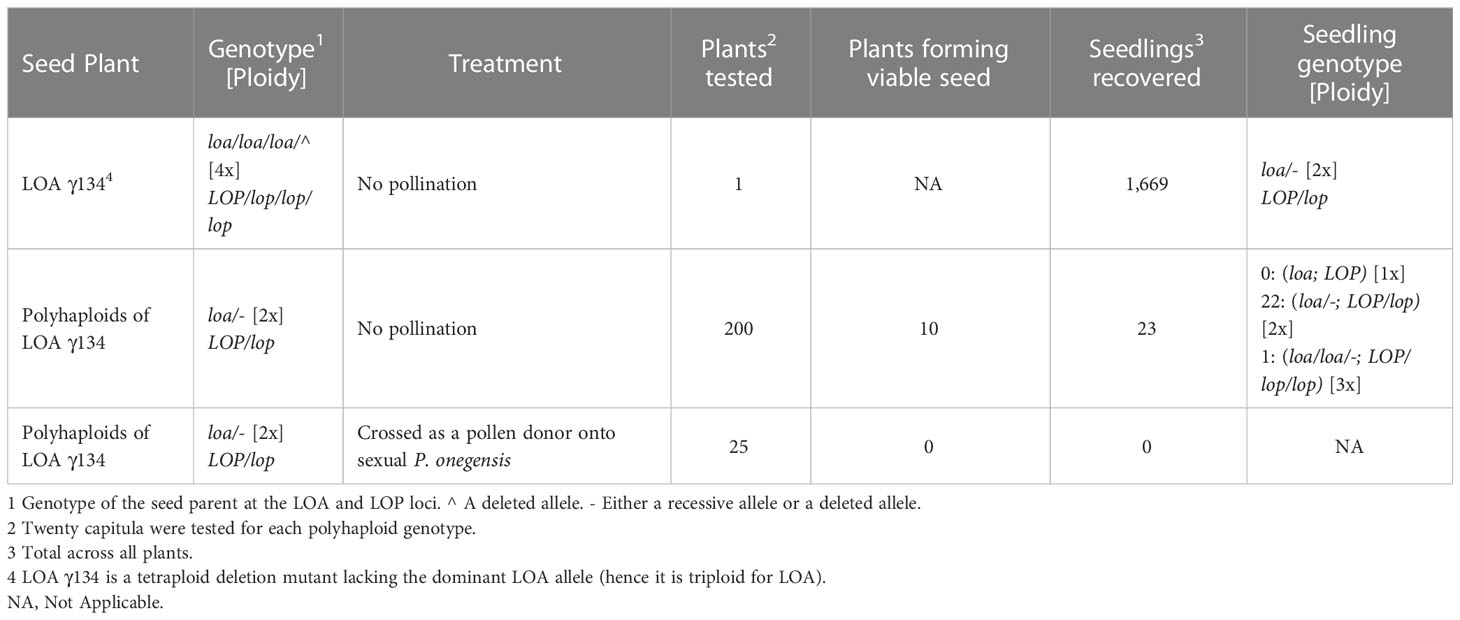
Two polyhaploid populations were produced from R35. Polyhaploids are the product of the parthenogenic development of a reduced egg cell and therefore contain half the genetic material of the maternal plant (Asker and Jerling, 1992). In this case, as the maternal plant was tetraploid at LOP, the recovered polyhaploids were diploids. The first population of 500 individuals was produced using the method of Bicknell et al. (2003). Seed from a plant of R35, transgenic for a simplex copy the codA negative selectable marker, was sown onto hormone-free tissue culture medium containing 200 mg/L 5-fluorocytosine. Any polyhaploid, lacking the marker gene following meiotic reduction, grew on the selective medium and was recovered. This method provides a simple way of producing polyhaploid populations for mapping studies without significant genome modification, providing the wild-type produces polyhaploids at a favorable rate and the transgene is not in linkage with a genetic region of interest. Approximately 1% of the progeny of R35 are polyhaploids (Bicknell et al., 2003). As 50% of segregants inherit the codA marker and do not grow, this method produced polyhaploids at a rate of approximately 0.5% of the seed sown. To provide more polyhaploids, a second method was also used. LOA γ134 is a mutant of R35 with a deletion across the LOA locus (Catanach et al., 2006). Consequently, LOA γ134 lacks LOA function and produces reduced egg cells following meiosis. Mutant LOA γ134, however, retains the LOP locus so egg cells carrying the dominant LOP determinant are capable of parthenogenesis, the product of which is a polyhaploid. The mutant LOA γ134 therefore generates polyhaploids at a rate of approximately 50% of the seed sown. A second population of 1,669 polyhaploids was produced by this method, bringing the total number of polyhaploids used in the study to 2,169. All the polyhaploids were tested by flow cytometry to confirm that they were diploids. Data from the two populations were initially analysed separately to be sure that the method of polyhaploid formation did not influence the result, particularly since mutant LOA γ134 was generated via gamma-induced mutagenesis (Catanach et al., 2006). No population effect was seen, so the data are presented collectively. PH70, the polyhaploid plant used for genome sequencing, was derived by the negative selectable marker technique to avoid potential difficulties with mutation. PH70 was chosen for sequencing as: it had dominant determinants at both the LOA and LOP loci; it formed apomictic seed at a high rate; it was diploid and it was also comparatively vigorous.
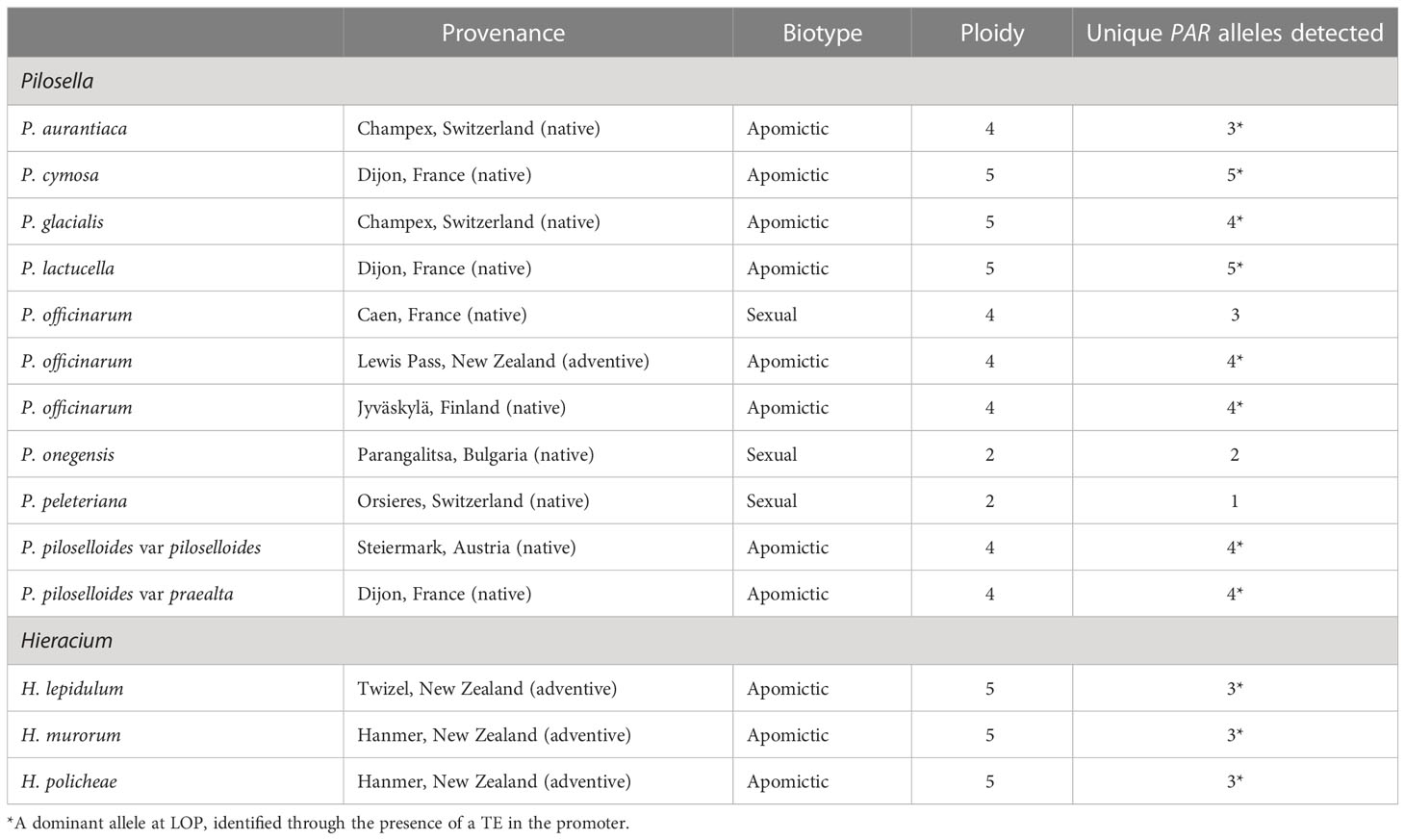
For the study of allele sequence divergence genomic DNA was isolated from the following species: P. aurantiaca, P. lactucella, P. cymosa, P. glacialis, P. officinarum, P. onegensis, P. peleteriana, P. piloselloides var. piloselloides, P. piloselloides var. praealta, Hieracium lepidulum, Hieracium murorum, and Taraxacum officinarum. The DNA was extracted using a Qiagen DNeasy Plant Mini Kit® in accordance with the supplier’s instructions and normalised to a concentration of 2ng/µL prior to use. For the species P. officinarum, four accessions were used, varying in geographic origin and biotype. The ploidy of each plant was determined by flow cytometry, using the staining method of Otto (1990) and measured on a Partec Space flow cytometer, operating Flow Max software (v2.9). Plants of known ploidy were available for the species P. aurantiaca, P. piloselloides var. piloselloides, P. piloselloides var. praealta and P. onegensa. These were used as ploidy references to estimate the ploidy of the remaining samples (Table 1).
BAC library construction and the identification of seed BACs
A Bacterial Artificial Chromosome (BAC) library of the R35 genome was constructed according to the method of Luo and Wing (2003), utilizing the vector pIndigoBAC536, a derivative of pBeloBAC. The final picked library included 258,048 clones. The average insert size was estimated to be 141kb using 117 randomly selected clones. End sequencing of a random selection of 1,536 clones indicated that the representations of mitochondrial and chloroplastic sequences were 0.13% and 0.33% respectively. The Pilosella haploid genome is estimated at 1.9 x 109 bp (http://data.kew.org/cvalues), so the library represents an estimated 21-fold coverage of the haploid genome and a 5.4-fold coverage of the complete genome of the plant R35. The library and printed filters are available from the resource center of the Arizona Genomics Institute under the species name Hieracium caespitosum (http://www.genome.arizona.edu).
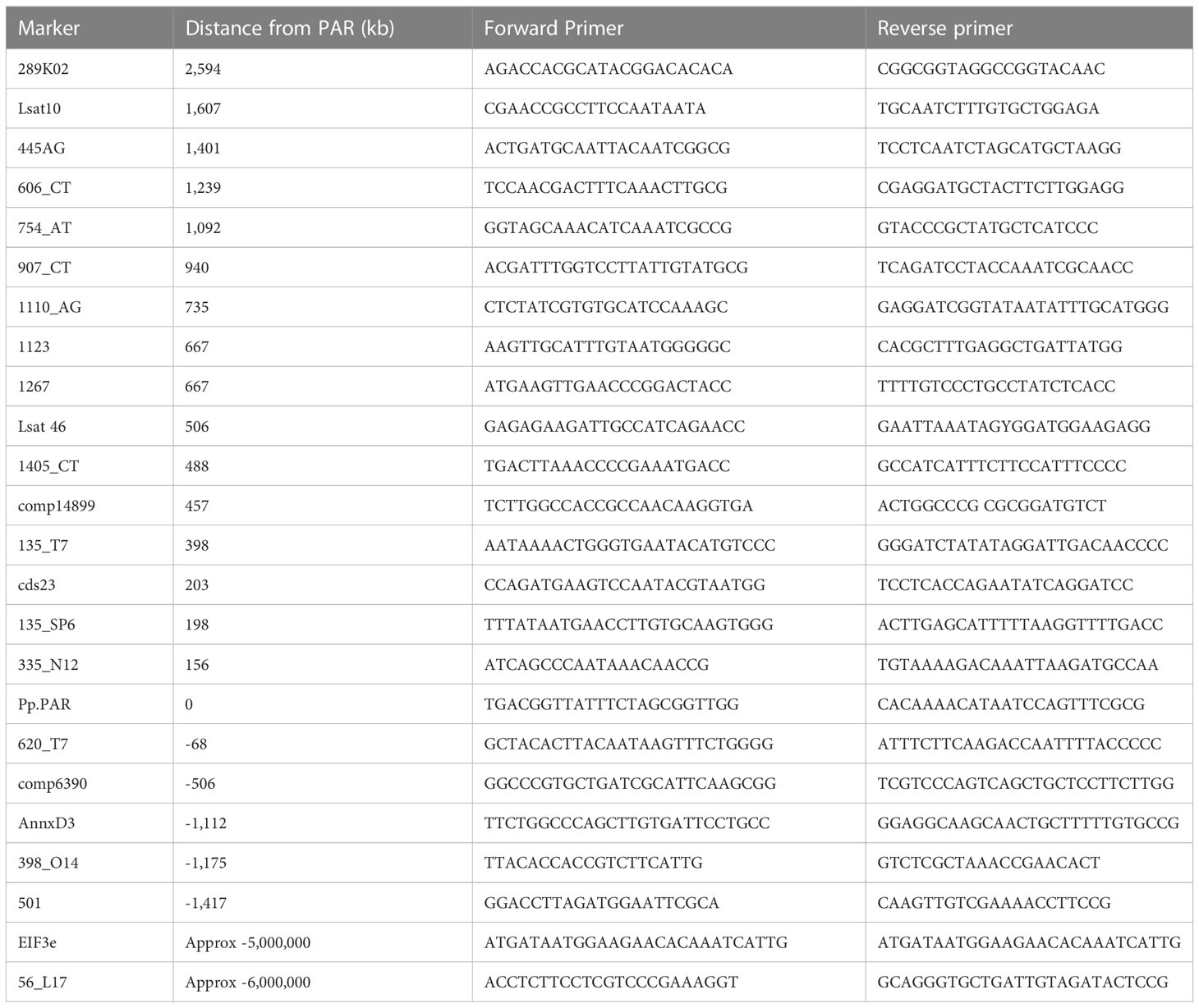
Secondary digest AFLP (sdAFLP) was conducted using markers described by Catanach et al. (2006) according to the method of Knox and Ellis (2001). The identified bands were used to design primers for seed-BAC identification. BAC clones associated with four LOP-linked makers (LOP-516, LOP-93, LOP-110 and LOP-380) were identified and sequenced by Sanger sequencing. Within the BAC library the corresponding BAC addresses are 56L17, 192I22, 501A19 and 289K02, respectively. Linkage to the dominant LOP allele was confirmed using a panel of plants (the wild-type R35 and the LOP deletion mutants: γ115, γ116, γ136, γ138, γ143, γ144, γ161 and γ179). The BAC sequences were later used for proofing the genomic assembly and for haplotyping genomic scaffolds.
Genomic sequencing
DNA was isolated from polyhaploid PH70 using a NucleoMag Plant Kit from Machery-Nagel according to the manufacturer’s instructions. DNA samples were then purified by chloroform (1:1 v/v) extraction using Qiagen MaXtract High Density tubes and concentrated using magnetic bead separation (Kappa HyperPure beads). Twenty micrograms of DNA were size-selected to a lower cutoff of 17kb using a Sage Science Blue Pippin as per the manufacturer’s instructions. Size-separated DNA was concentrated using Kappa Hyper beads and DNA purity, concentration and integrity determined using a Qubit Fluorometer and an Agilent S200 series fragment analyzer.
Genome sequencing was carried out by using the GENEWIZ commercial Next Generation Sequencing service to generate short-read (Illumina) sequencing data, and by Oxford Nanopore MinION sequencing to generate long-read sequence data. Approximately 450GB of raw short-read sequence data (150bp paired-end reads, providing approximately 140x coverage of the diploid genome), were generated. In addition, a further 42x of the diploid genome was generated using ONT Minion sequencing. Sequencing libraries were generated using a LSK-SQK109 sequencing Kit (ONT, UK) and the prepared libraries were sequenced on MIN FLO109 flow cells. Data collection was conducted with ONT’s MinKnow (Oxford Nanopore, Windows version 20.06.5) and base calling of the raw data performed off-line using the NeSI compute cluster and the GUPPY base caller (Oxford Nanopore, LINUX version 4.2.2) using default settings recommended by ONT.
Hybrid assembly of the Hieracium genome was carried using the MaSuRCA (Zimin et al., 2013; Luo et al., 2017) and FLYE (Kolmogorov et al., 2019) assembly tools and default conditions. The resulting output files were then used to generate a local BLAST database and screened for homology with Pilosella BAC sequences using BLAST (Altschul et al., 1990; Camacho et al., 2009) to identify contigs and scaffolds corresponding to the LOP locus.
Polyhaploid mapping of the LOP locus
Polyhaploids arise from the parthenogenic development of meiotically-reduced egg cells and can, therefore, be used to generate uni-parental mapping populations in facultatively apomictic Pilosella species (Bicknell et al., 2003; McGee, 2013; Shirasawa et al., 2014). In Pilosella piloselloides var. praealta, LOP acts as a gametophytic locus (Catanach et al., 2006; Koltunow et al., 2011) with the dominant determinant present in every polyhaploid (Koltunow et al., 2011). As a functional copy of LOP is required, the functional/structural bounds of the locus can be defined by the limits of recombination within a polyhaploid population. In keeping with other known apomixis loci, the frequency of recombination about LOP is low (Shirasawa et al., 2014), so the mapping population needed to be correspondingly large. As mentioned above, 2,169 polyhaploids were used in this study. PCR-based markers were developed, using sequence from the seed-BAC clones, from a syntenic region on LG8 of the lettuce genome (http://lgr.genomecenter.ucdavis.edu) and from the de novo sequencing of the PH70 genome. Earlier deletion mapping (Catanach et al., 2006) determined that the LOP locus lay between the flanking markers LOP-516 and LOP-380. Allele-specific markers were developed for LOP-516 and LOP-380 and used to identify a group of 52 polyhaploids recombinant between the flanking markers. Based on synteny to lettuce the genomic interval between these markers, which includes the entire LOP locus, is estimated at 8 Mb. The 52 recombinants were then used to map the limits of LOP to an approximate resolution of 0.15 cM. The primers used in this study are listed in Table 2.
Patterns of allele inheritance
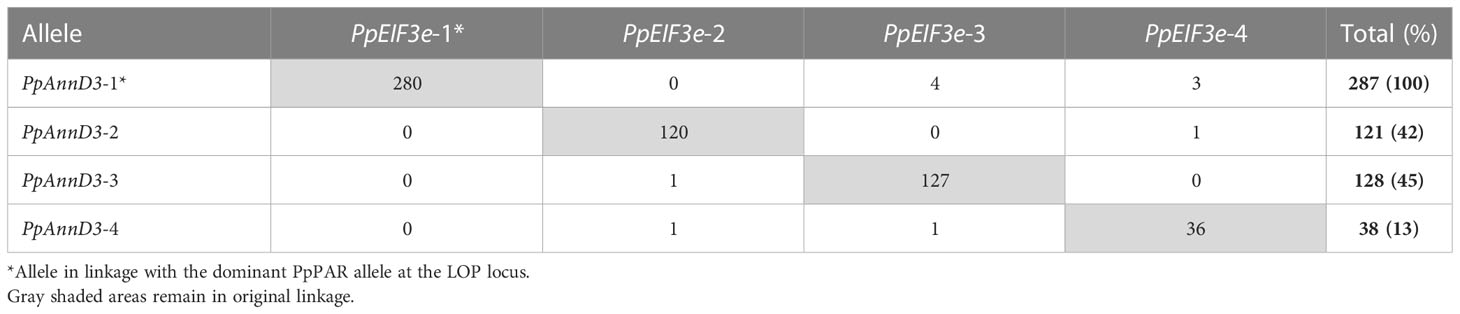
Two linked genes were selected to determine patterns of homologue exchange close to the LOP locus of P. piloselloides var. praealta: Annexin D3 (PpAnnD3) which is located 1.1Mb from the PAR gene and Eukaryotic translation Initiation Factor 3e (PpeIF3e) which is located approximately a further 4Mb from PAR (estimate based on synteny with lettuce) (Reyes-Chin-Wo et al., 2017). Using sequence from cloned AFLP markers 110 and 93 (Catanach et al., 2006), BACs were identified that included alleles of these genes linked to the dominant allele of the PAR gene. Redundant primers and low-stringency hybridization were then used to develop intron-spanning allele-specific markers, for all four alleles of PpAnnD3 and PpeIF3e.
Allele segregation was then assessed on a population of 287 polyhaploids, generated by the marker segregation approach and 109 from the deletion mutant approach. For each diploid plant the two alleles at PpAnnD3 and PpeIF3e were scored and the rates of recombination calculated. Alleles in linkage with the dominant determinant at PpPAR were designated as ‘Allele 1’. As the PpPAR allele is gametophytic in action and necessary for polyhaploid formation, it was present in all polyhaploids. Alleles in linkage with LOP-PAR, therefore, were also commonly present and easily identified. The remaining alleles for each gene were arbitrarily designated as ‘Allele 2, 3 and 4’.
Allele sequence divergence at PAR
The PARTHENOGENESIS (PpPAR) gene is located within the LOP locus of P. piloselloides var. praealta (Underwood et al., 2022). PAR encodes a K2-2 zinc finger protein, known to be necessary for the expression of parthenogenesis in the related genus Taraxacum (Underwood et al., 2022). Using alignments with published genomes, the genomic sequence of PH70 and data from the MinIon sequencing of Hieracium policheae genomic DNA, primers were designed that amplified exonic sequence for this gene in a range of plant species. The sequences of the primers used are listed in Table 2. Established ribosomal ITS primers (Gardes and Bruns, 1993) were also used to phylogenetically order the species under study.
For each species/accession, amplicons for PAR and rRNA ITS region were separately generated, barcoded, normalized for concentration, combined and sequenced using the Oxford nanopore MinION sequencing platform. The sequenced amplicons were then processed using the Geneious Prime software suite (Version 2022.0.1) For each gene/species combination at least 2,000 reads were used. Consensus allele sequences were derived using the ‘De Novo Assembly’ function and proof-checked by eye using the read alignment tool. Tree assemblies were conducted with the ‘Geneious Tree Builder’ tool using a HKY genetic distance model, a cost matrix of 65% similarity and bootstrap resampling with 5,000 replicates. Published sequence for Lactuca sativa was used to provide an outgroup. For each of the apomictic species studied, the dominant allele at PpPAR was readily identifiable, due to the presence of transposon-derived sequence in the promoter of the gene, as described by Underwood et al. (2022). Only PAR coding sequence was considered in the analysis.
Allele inheritance in haploid and diploid gametes
As described above, the deletion mutant LOA γ134 has a deletion across the dominant LOA allele associated with apomeiosis (genotype */loa/loa/loa). Therefore, LOA γ134 only produces reduced egg cells following the action of meiosis (Catanach et al., 2006). Egg cells carrying the dominant LOP allele can then give rise to a diploid seedling through the action of parthenogenesis (a polyhaploid). As LOA γ134 is simplex for the dominant LOP allele, all its polyhaploids are heterozygous for LOP (LOP/lop) and they are also meiotic (loa/loa or loa/-), (see Table 3). We therefore reasoned that the polyhaploids of LOA γ134 should, in turn, predominantly form haploid egg cells through the action of meiosis. Any egg cell inheriting the dominant LOP allele might then be capable of development into a haploid seedling, provided the dominant LOP allele was capable of transmission through a haploid female gamete and/or that haploid plant formation was possible with only the dominant LOP allele present. A total of 200 polyhaploids were tested. For each polyhaploid, seed was collected from at least 20 capitula, sown onto hormone-free tissue culture medium and maintained under sterile conditions to ensure the survival of weak plants. Germinated seedlings were ploidy tested using a Partec Space flow cytometer and the PpPAR alleles were determined using primers listed in Table 2. Similarly, to test allele transfer through haploid male gametes, 25 polyhaploids were crossed, as the pollen parent, with a sexual, diploid accession of Pilosella onegensis. For each cross-combination, 15 capitula were tested and the seed sown in tissue culture.
Results
The Pilosella genome
Genomic sequence of the diploid apomict Pilosella piloselloides var. praealta PH70, assembled using the hybrid assembler MaSuRCA (Zimin et al., 2013; Luo et al., 2017) resolved the diploid genome into 21,000 contigs, with a predicted haploid genome size of 1.6 Gbp. Analysis of the completeness of the genome gave a BUSCO score of 94%. To identify and interrogate the LOP locus, a BLAST library was prepared from these contigs in GeneiousPrime Version 2023.1.1 (www.geneious.com). Sequences of predicted genes, marker sequences and BAC sequences were used to identify contigs with suitable sequence identity to sequences previously associated with the LOP locus. To resolve haplotigs associated with the dominant and recessive alleles of LOP, BAC sequences and marker sequences were used to manually assemble and resolve individual contigs across 1.6 Mbp. All genome and amplicon sequences are available under the BioProject accession code: PRJNA982544: Pilosella piloselloides Genome sequencing and assembly (TaxID: 1628030).
Size of the LOP locus in Pilosella piloselloides var. praealta
Figure 1 illustrates the relationship between the physical and recombination-based maps of the LOP locus of P. piloselloides var. praealta PH70, approximately centered on the PpPAR gene and extending over a total genomic interval of approximately 3 Mb. The illustrated physical distances are based primarily on dominant LOP allele sequence, resolved by combining the outputs of the MaSuRCA (Zimin et al., 2013) and FLYE (Kolmogorov et al., 2019) assembly tools. Estimations of genetic distance are also based on a single recombinant class (those inheriting the dominant LOP allele) and the total population size (2,169 polyhaploids) is based only on plants which inherited that allele, as polyhaploids do not form without it. Consequently, both the physical and genetic maps presented in Figure 1 represent the dominant allele of the LOP locus in P. piloselloides var. praealta.
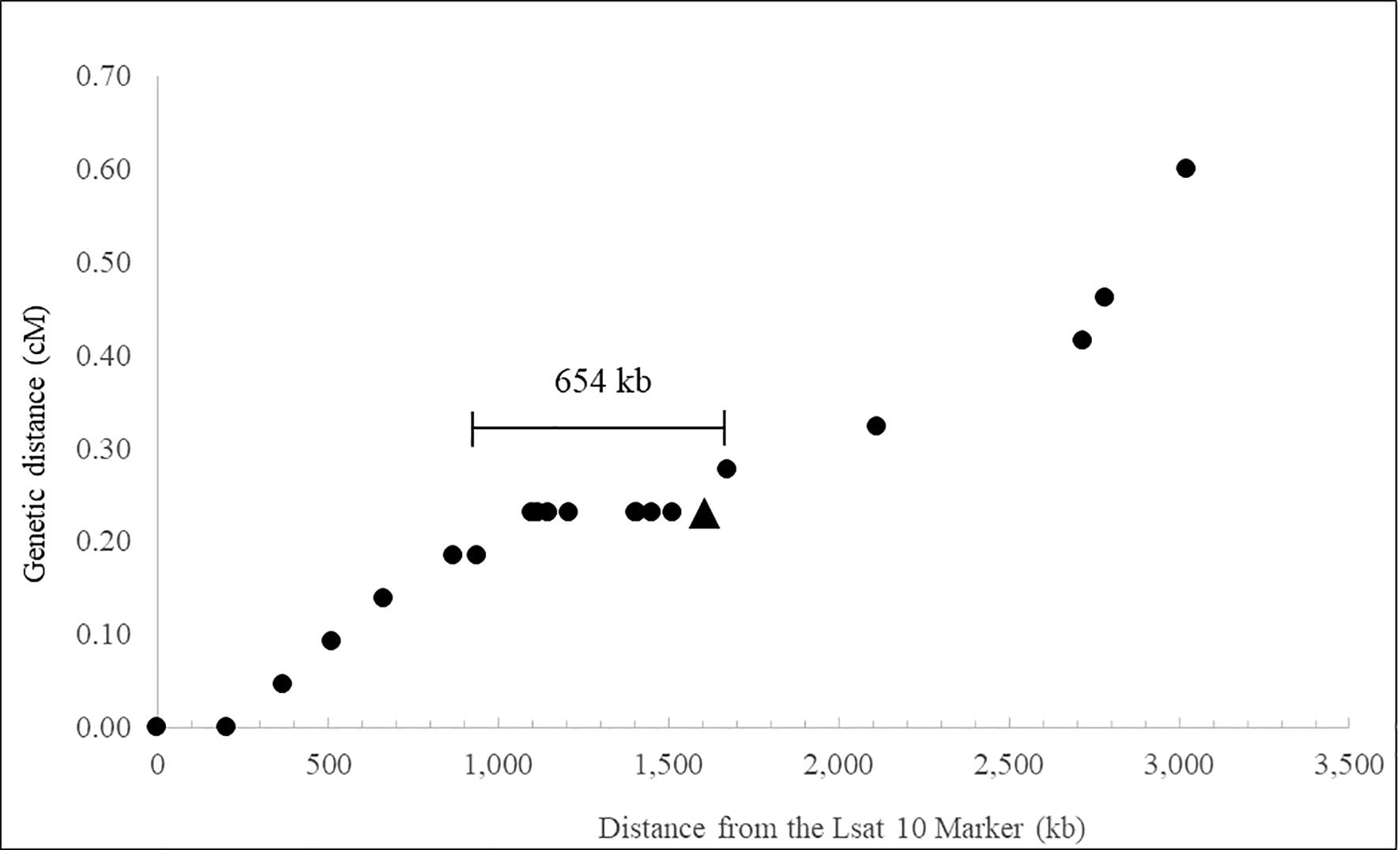
Figure 1 Recombination at the LOP locus of Pilosella piloselloides. ● A SCAR or SSR marker. ▲ The PpPAR gene.
Polyhaploid mapping defined the functional limits of the LOP locus to a region between markers LOP_1267 and 620_T7, a genomic interval of 654 kb (Figure 1). Deletion mutant LOP γ138, which lacks LOP function, has a break point within this interval, suggesting that the LOP locus may be as small as 527 kb. Alternatively, this result may indicate that LOP includes more than one gene critical to its function. As previously noted (Underwood et al., 2022) the PAR gene lies within the mapped interval and mutant LOP γ138 lacks LOP function (Catanach et al., 2006). The total genetic distance of the 3 Mbp genomic region studied was estimated as 0.6 cM.
Patterns of allele inheritance
Table 4 records the observed distribution of allelic inheritance, for two loci: Annexin D3 (PpAnnD3), which is located 1.1 Mb from the PAR gene, and Eukaryotic translation Initiation Factor 3e (PpeIF3e), which is located a further 4 Mb from PAR, in a population of 287 polyhaploids. All 287 polyhaploids carried the PpAnnD3-1 allele, which is tightly linked to the dominant LOP locus, but they varied with respect to the other allele present. No significant difference was observed between the frequency of inheritance of alleles PpAnnD3-2 and PpAnnD3-3 (χ2 p > 0.1); however, a highly significant bias against the inheritance of allele PpAnnD3-4 was apparent (χ2 p < 0.001). Crossover events between the loci detected all possible allele combinations, except between alleles 1 and 2.
Phylogenetic relationships
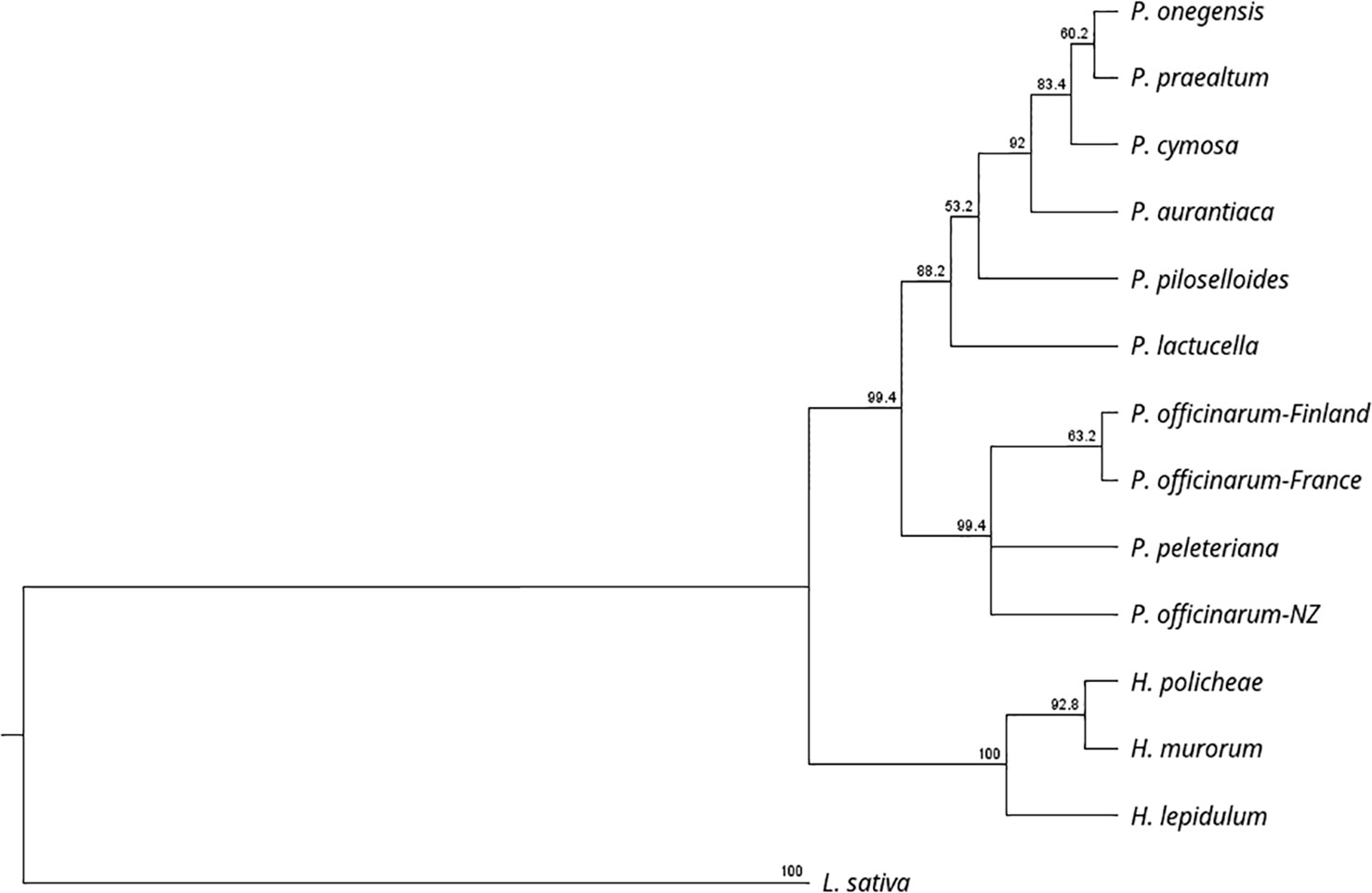
Table 1 lists the plants used in this study, their biotype, ploidy, and the numbers of unique allelic sequences at PAR determined by our analysis. For some of the accessions the number of unique allelic sequences equaled that predicted from the ploidy analysis; however, in many cases a lesser number was determined. It is unclear whether this was due to similarities between alleles and/or whether it resulted from differential-PCR amplification because of differences in primer specificity. Figure 2 illustrates the taxonomic relationship of the plants studied, as defined by rRNA-ITS sequence, with the inclusion of Lactuca sativa as a related outgroup. Species within the two, closely related genera, Pilosella and Hieracium, grouped into distinct clades, in accordance with their reported taxonomic relationship (Bräutigam and Greuter, 2007).
Allele sequence divergence at PAR
The cladogram of allele sequence relationships at the PAR gene is illustrated in Figure 3. As observed with rRNA, sequences from the two genera, Pilosella and Hieracium, clearly separated. Dominant alleles at PAR, which associate with the expression of parthenogenesis (labeled PAR-1*) were identified in the three apomictic accessions of Hieracium and the nine apomictic accessions of Pilosella studied. For Pilosella, dominant PAR alleles clustered into a single, invariate clade. Dominant alleles in the Hieracium species also clustered but were more variable than those of Pilosella.
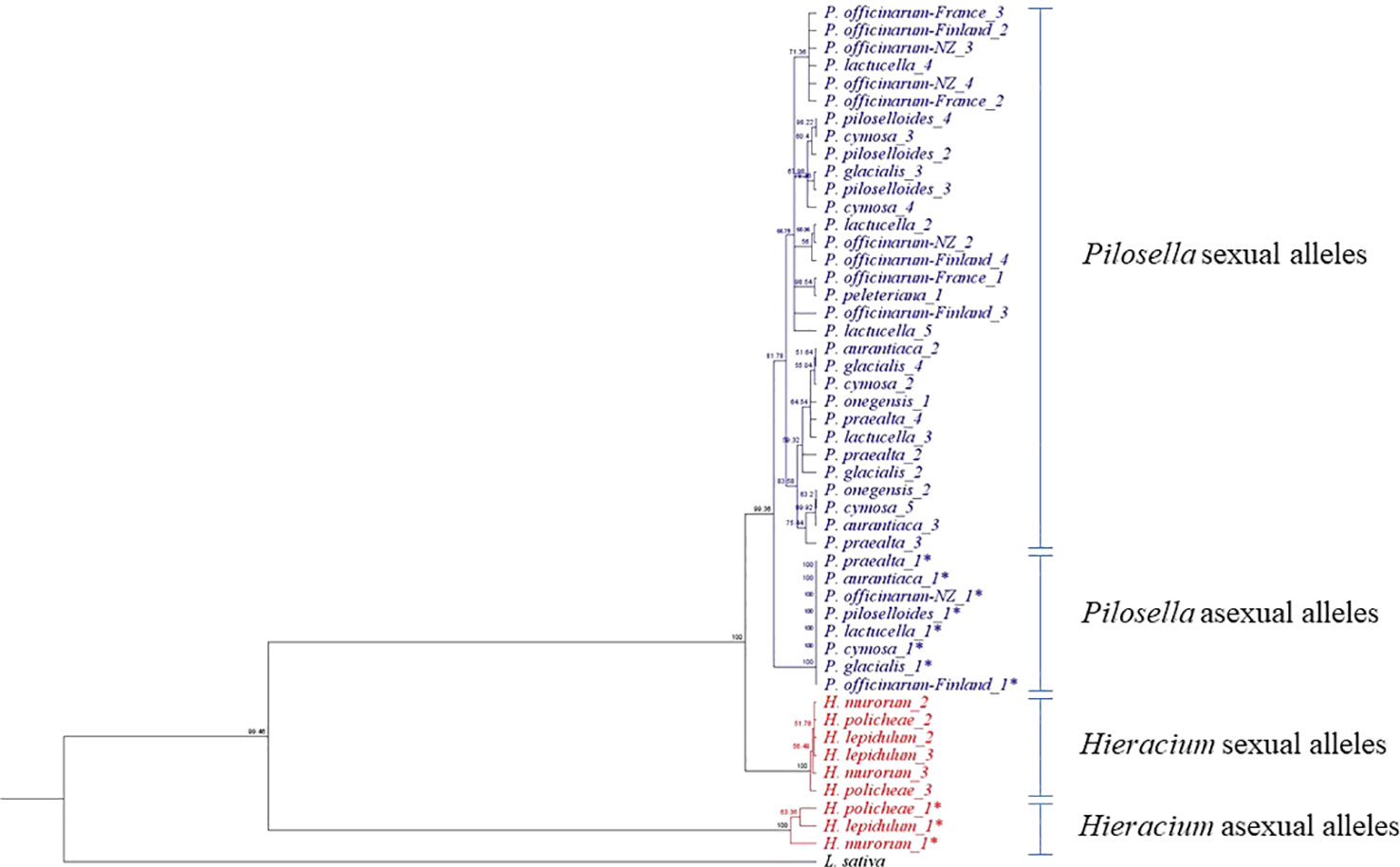
Figure 3 Allele Diversity at PAR. * A dominant allele, identified by the presence of a TE in the promoter.
Structure of dominant PAR alleles
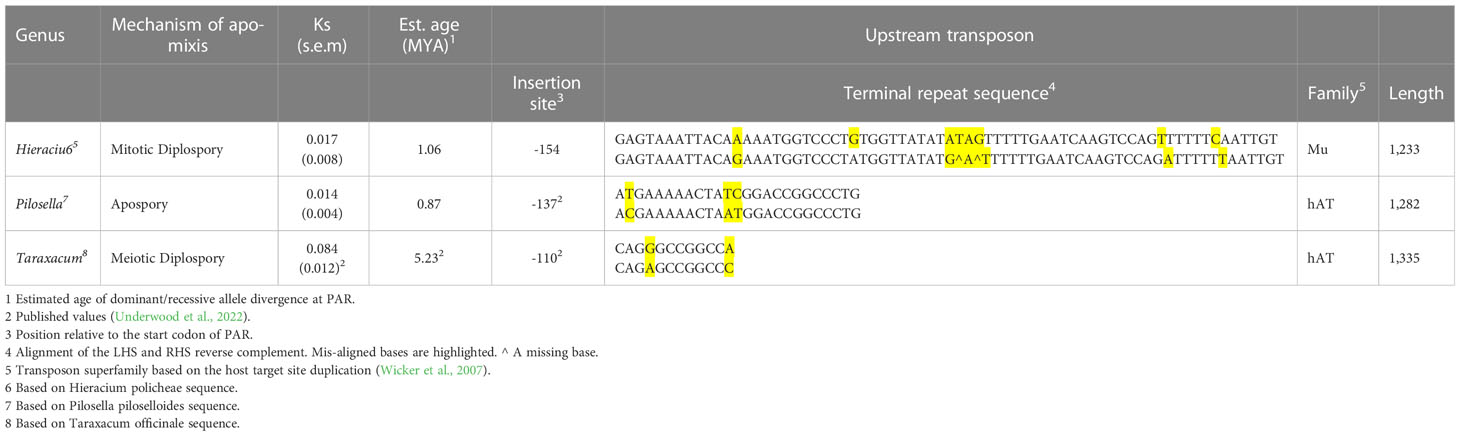
Table 5 lists the observed characteristics of dominant PAR alleles in Hieracium policheae, Pilosella piloselloides and Taraxacum officinale. In all cases a class II transposable element was present, positioned in the promoter of PAR within a conserved 200 bp interval, previously described by Underwood et al. (2022). The location, length, terminal repeat sequences, TE superfamily and estimated age of the insertion events, however, differed significantly between the genera studied. The presented Ks values and predicted ages of allele separation are based solely on PAR exonic sequence and therefore need to be considered with caution. PAR is a gene that resides in a region of suppressed recombination and appears to be under strong stabilizing selection. The presented values, therefore, are best viewed in relation to each other rather than as absolute estimates of the age of genomic divergence.
Allele inheritance in haploid and diploid gametes
Table 3 records the frequency, ploidy and genotype of progeny derived from the use of LOA γ134 and its polyhaploid progeny. The genotype of LOA γ134, with respect to the LOP locus, is LOP/lop/lop/lop. The polyhaploid progeny of that plant, therefore, were expected to have the genotype LOP/lop, as was observed (Table 3). Using similar reasoning, the diploid polyhaploids were expected to produce haploid progeny by parthenogenesis with the genotype LOP; however, none was recovered, despite the large number of plants tested. Similarly, no evidence was seen of any haploid pollen function in the diploid polyhaploids plants. Interestingly, viable seedlings were recovered from ten of the polyhaploids. Of the 23 seedlings recovered, 22 were diploid and heterozygous for LOP and one was triploid and heterozygous for LOP.
Discussion
LOP maps to a 654 kb interval
Shirasawa et al. (2014) constructed a genetic map for the accession P. piloselloides var. praealta R35, positioning 418 markers into 18 linkage groups. The location of the LOP locus, however, could not be defined for this accession but was defined in a study using a smaller population segregating for LOP transferred from the closely related parental line D36 (Pilosella piloselloides var. piloselloides). In that population LOP mapped to a single linkage group, but only to within a region with low marker density. In the current study, using polyhaploid mapping, the LOP locus of P. piloselloides var. praealta was mapped to a 654 kb genomic interval syntenic to linkage group 8 of Lactuca sativa (Reyes-Chin-Wo et al., 2017) with a mapping resolution of approximately 0.15 cM (Figure 1). The LOP locus is, therefore, significantly smaller than the analogous ‘Apospory Specific Genomic Region’ (ASGR) of the apomictic grass Pennisetum squamulatum and closely related Cenchrus ciliaris (Sherwood et al., 1994; Ozias-Akins et al., 1998). The ASGR is estimated to be approximately 50 Mbp in size and is believed to be composed largely of repeat sequence (Ozias-Akins et al., 1993; Ozias-Akins et al., 1998; Roche et al., 2001; Roche et al., 2002).
Polyhaploids are known to form in P. piloselloides var. praealta through the gametophytic action of LOP (Bicknell et al., 2003; Koltunow et al., 2011). The mapped region, therefore, represents both a functional and a physical domain for LOP. Whether the bounds of the mapped region are defined by suppressed recombination and/or due to the presence of more than one critical element within the locus remains unclear. The deletion mutant LOP γ138, which lacks LOP function (Catanach et al., 2006), has a break point within the 654 kb interval. If the bounds of the locus are defined by suppressed recombination alone, then this suggests that the LOP locus may be as small as 527 kb. However, as described above the phenotype of LOP γ138 could also be the result of a partial loss of LOP functionality. The PpPAR gene, which has been identified as a critical determinant in the expression of parthenogenesis in P. piloselloides var. praealta (Underwood et al., 2022) maps within the 654kb interval (Figure 1) but is positioned 68kb from a recombinant flanking marker, placing it very near the edge of the mapped interval. This suggests that either a mechanism is acting to suppress recombination on only one side of PAR, or that other genes or regulatory elements are present in the remaining non-recombinant region of the LOP locus which are necessary for the formation of polyhaploids through parthenogenesis.
Tetrasomic allelic exchange indicates an autoploid origin for P. piloselloides var. praealta R35
The accession P. piloselloides var. praealta R35 is tetraploid at the LOP locus, as evidenced by flow cytometric analysis (Table 4) and the presence of four distinct allelic sequences amongst the genes sequenced. Tetraploid plants can arise either through the duplication of a single genome (auto-polyploidization) or by the fusion of two distinct species, followed by genome duplication (allo-polyploidization) (Harlan and De Wet, 1975; Ramsey and Schemske, 2002; Ranney, 2006). Tetrasomic patterns of crossover, where allelic exchange can occur between any of the four alleles present, is characteristic of autopolyploidy, whilst disomic inheritance, where an allele will exchange with only one other, is characteristic of allopolyploidy. Table 4 clearly illustrates the action of tetrasomic exchange in the formation of polyhaploids amongst the progeny of R35, indicating that this an autopolyploid species.
Because of the gametophytic action of LOP, every polyhaploid inherited allele 1 (linked to the dominant allele at PAR) but polyhaploids differed in the second allele present. Although alleles 2 and 3 were similarly represented in the polyhaploid population, there was a significant bias against the inheritance of allele 4. Evidence of crossover between allele 4 and the remaining alleles (Table 4) indicates similarity in sequence, yet allele 4 was clearly either less frequently transmitted through the egg cell following meiosis and/or the subsequent survival of the allele 1: allele 4 genotype was disadvantaged. Either possibility indicates that recessive (sexual) allele inheritance at LOP is influenced by selection and, therefore, that recessive alleles play a functional role in this system. Underwood et al. (2022) proposed such a mechanism for alleles at the PAR locus of Taraxacum and Pilosella, hypothesizing that recessive alleles may direct expression of the gene in the pollen grain, while the dominant allele directs expression in the egg cell, placing both under natural selection. Our data suggest that recessive par alleles are also critical to the development and/or function of the egg cell as well.
Allele inheritance in haploid and diploid gametes
It is reported that dominant factors governing gametophytic apomixis seldom, if ever, transmit through haploid gametes, yet they will transmit through diploid or polyploid gametes (Asker and Jerling, 1992; Van Dijk et al., 2009). Nogler (1984) appears to have been the first to have noted this phenomenon, following a study of the pollen-mediated transmission of apomixis in Ranunculus auricomus. He proposed that an ‘apomixis’ allele may be haploid-lethal, requiring the co-inheritance of another, complementing ‘sexual’ allele to ensure transfer. An expected consequence of this mechanism would be to establish apomixis in polyploid lineages, as is commonly observed (Nogler, 1986). Underwood et al. (2022) proposed that dominant and recessive alleles of the PAR gene, the dominant allele of which confers parthenogenesis in Taraxacum and Pilosella, may be expressed in different tissues of the flower. If both functions are essential for female gametophyte development and/or function, then this would meet the conditions of Nogler’s hypothesis, limiting transmission of the dominant allele to heterozygotic gametes. Similarly, Van Dijk et al. (2009) demonstrated haploid pollen lethality was linked to the DIP locus of the apomict Taraxacum officinale. The dominant DIP allele confers apomeiosis in this system, which is a separate component of apomixis from parthenogenesis. It is intriguing, therefore, to note that both apomeiosis and parthenogenesis-conferring alleles appear to confer haploid lethality and can, therefore, only transmit in gametes also carrying at least one recessive allele.
In the current study the LOP locus of Pilosella piloselloides var. praealta transmitted readily through the diploid egg cells of the plant LOA γ134 but not through the haploid egg cells of the diploid derivatives of LOA γ134. Furthermore, the pollen of the polyhaploids also appeared to be non-functional. We conclude, therefore, that the dominant allele of LOP cannot transmit through either female or male haploid gametes in this system, in agreement with Nogler’s hypothesis. The failure of all pollen function, however, suggests that pollen malfunction may be a general problem with these diploid plants, probably associated with the greater extent of exposed genetic load in polyhaploids. Interestingly, 22 diploid seedlings and one triploid seedling were recovered from the diploid polyhaploid plants tested (Table 3). The triploid plant was probably the product of self-fertilization involving the union of a reduced and an unreduced gamete. All the diploid seedlings were heterozygous for LOP, indicating that they were either derived following self-fertilization and or through the action of apomixis. The expression of apomixis in these plants, however, would require a mechanism for apomeiosis, yet the LOA locus is known to be absent in this plant material. This finding will be further discussed below, with respect to its implications for the evolution of apomixis in this system.
The structure of PAR and allelic sequence divergence at this locus
Genomic regions subject to reduced amounts of homologous recombination are known to accumulate mutational differences, including the accumulation of sequence derived from transposable elements (Kimura et al., 1963; Muller, 1964; Corral et al., 2009). As a consequence, these loci tend to increase in size over time and the allelic sequences of genes within them become increasingly divergent. This effect has been recorded for specific genomic regions within individuals (such as the sex chromosomes of mammals) and at the population level between related sexual and clonal lineages (Mark Welch and Meselson, 2000). The presence of ASD has, therefore, been proposed as an indicator of ancestral asexuality (Corral et al., 2009). As described above, the LOP locus of Pilosella is a genomic region of suppressed recombination (Figure 1) and the perpetuation of apomictic lineages would have further isolated genomes from the influences of meiotic recombination. We, therefore, reasoned that ASD was likely to be measurable in this system, and that the pattern of ASD might inform our understanding of the evolution of apomixis in this plant group. The genus Pilosella is a member of the Lactuceae, a tribe of the Asteraceae. Pilosella is closely related to the genus Hieracium (Bräutigam and Greuter, 2007) and more distantly related to the comprehensively characterized genera Lactuca (lettuce) and Cichorium (chicory) (Tutin et al., 1976). Most species and clones of Pilosella are polyploid, facultative, gametophytic apomicts, using the mechanism of apospory to generate clonal seeds. During apospory, an unreduced embryo sac arises from a nucellar cell, alongside similar meiotically-derived structures within the ovule. Competition between the meiotic and apo-meiotic structures leads to the formation of a mixture of sexually and asexually derived seed. Typically, asexually derived seed predominates. In P. aurantiaca (syn. H. aurantiacum) and P. piloselloides var. piloselloides (syn. H. piloselloides) meiotically derived seed was recorded at rates of 2.6% and 2.8% respectively (Bicknell et al., 2003). Furthermore, obligate sexual accessions are known to occur in some Pilosella species (Gadella, 1972; Gadella, 1984; Gadella, 1991) including P. officinarum (as used in this study), and entirely obligate sexual species are also known, such as the diploid P. onegensis. For Pilosella, therefore, sexual reproduction continues to operate and to contribute to the formation of seed in most plants and in most populations, albeit often at a low rate.
In contrast, plants in the genus Hieracium are almost entirely polyploid, obligate gametophytic apomicts, generating seeds by mitotic diplospory of the Antennaria type (Crane, 2001; Mráz et al., 2009; Mráz and Zdvořák, 2018). In this form of gametophytic apomixis, an unreduced embryo sac forms directly from a nucellus cell without the mediation of any meiotic event. Meiotic products, therefore, do not form in the ovule, as seen during apospory, so the resulting seed is solely of clonal origin (Asker and Jerling, 1992). Sexual biotypes of Hieracium are known, but amongst the hundreds of described species they appear to be restricted to a small number of diploids of restricted distribution (Mráz and Paule, 2006; Mráz et al., 2011; Mráz and Zdvořák, 2018).
Figure 2 illustrates the taxonomic relationship between Pilosella and Hieracium, as defined by rRNA ITS sequence diversity. It supports a recent separation of Pilosella and Hieracium into two distinct clades (Bräutigam and Greuter, 2007) and illustrates their association with the closely related model species Lactuca sativa. Figure 3 illustrates allelic diversity for the PAR coding sequence in the accessions studied. As detailed above, PAR lies within the region of suppressed recombination that defines the LOP locus. The separation of Pilosella and Hieracium is again clearly evident in Figure 3, which includes the dominant PAR alleles, suggesting they have different origins, as confirmed by sequence analysis of the transposon insertion sites and internal transposon sequence (Table 5). Notably, the dominant PAR alleles of Pilosella cluster into a distinct clade, separately from the recessive alleles of the individual accessions tested. This indicates that the dominant alleles share a common and recent origin, supporting the theory that apomixis can radiate rapidly through pollen transfer of the enabling alleles (Van Dijk et al., 2009; Hojsgaard and Hörandl, 2019).
The expression of apomixis in the Lactuceae
Gametophytic apomixis is a comparatively common trait in the Lactuceae. Of the 70 genera assigned to this tribe, seven (Taraxacum, Hieracium, Pilosella, Ixeris, Chrondrilla, Cichorium and Crepis) are known to include apomictic species (Noyes, 2007). An incidence rate of 10% of genera is well above the estimated rate amongst all flowering plant genera of 0.8% (Carman, 1997). The factor(s) that predispose members of this taxon to the expression of gametophytic apomixis are incompletely understood, but available data indicate that apomixis evolved from sexual reproduction many times in this group, rather than being co-inherited from a common apomictic ancestor. One line of evidence is the diversity of apomictic mechanisms found in the Lactuceae. Of the genera listed above, two (Crepis and Pilosella) form asexual seeds by apospory, three (Chondrilla, Ixeris and Taraxacum) by meiotic diplospory, and one (Hieracium) by mitotic diplospory (Carman, 1997; Crane, 2001; Noyes, 2007). The defining differences between these mechanisms relate to events within the ovule at the time of meiosis. Unlike sexual plants, apomictic species either avoid, or utilize a truncated form of meiosis to generate a cell competent to enter gametogenesis, the subsequent product of which is an unreduced embryo sac (Asker and Jerling, 1992). The collective term for the avoidance of the reduction/division step of meiosis in apomicts is ‘apomeiosis’. As apospory, meiotic diplospory and mitotic diplospory differ significantly in the developmental processes involved in apomeiosis, it appears likely that they are controlled by different genetic elements.
Conversely, all the apomictic members of the Lactuceae spontaneously initiate both embryogenesis (parthenogenesis) and endosperm formation (autonomous endospermy) without pollination, indicating a possible commonality in these component processes of apomixis across the Lactuceae. This conclusion was recently proposed by Underwood et al. (2022), who reported that the gene PAR was a key determinant in the expression of parthenogenesis in both T. officinale and P. piloselloides and that the dominant (asexual) alleles conferring parthenogenesis differed from the corresponding recessive (sexual) alleles by the presence of a TE in a highly conserved 200 bp region of the PAR promoter. Critically, the insertion site, length and internal sequence of the TE’s were different, implying that they arose from independent insertion events. Taraxacum officinale uses meiotic diplospory to form asexual seeds, while P. piloselloides uses apospory. In the current study, this observation was further extended to include Hieracium policheae, a species using mitotic diplospory, the third mechanism of gametophytic apomixis found in the Lactuceae. Again, the PAR gene was found to be modified by a non-autonomous type II transposon, the insertion site of which also located to the conserved promoter region of PAR but differed from those of T. officinale and P. piloselloides (Table 5). Furthermore, analysis of synonymous SNP variation also indicates that the Taraxacum insertion event occurred earlier than the Hieracium and Pilosella events (Table 5).
Model for the evolution of apomixis in the Lactuceae
Gametophytic apomixis in flowering plants requires three key elements: apomeiosis, parthenogenesis, and the development of an endosperm to ensure the nourishment of the developing embryo. Endosperm formation in apomicts often occurs by a sexual process, but apomeiosis and parthenogenesis represent significant departures from sexuality. Notably, both apomeiosis and parthenogenesis act as deleterious traits when operating alone. Apomeiosis, when expressed without parthenogenesis, leads to the formation of unreduced eggs that require fertilization to initiate embryogenesis. Fertilization then leads to an increase in ploidy with each generation and ultimately to the isolation of the trait in genotypes with reduced vegetative fitness and reduced fertility (Bicknell and Koltunow, 2004). Conversely, parthenogenesis, when expressed without apomeiosis, leads to the production of progeny with successively decreasing ploidy, also resulting in reduced fitness and lost fertility. There is, therefore, a question as to how gametophytic apomixis evolved in seven genera of the Lactuceae, when it requires the coordinated establishment of two independently deleterious traits.
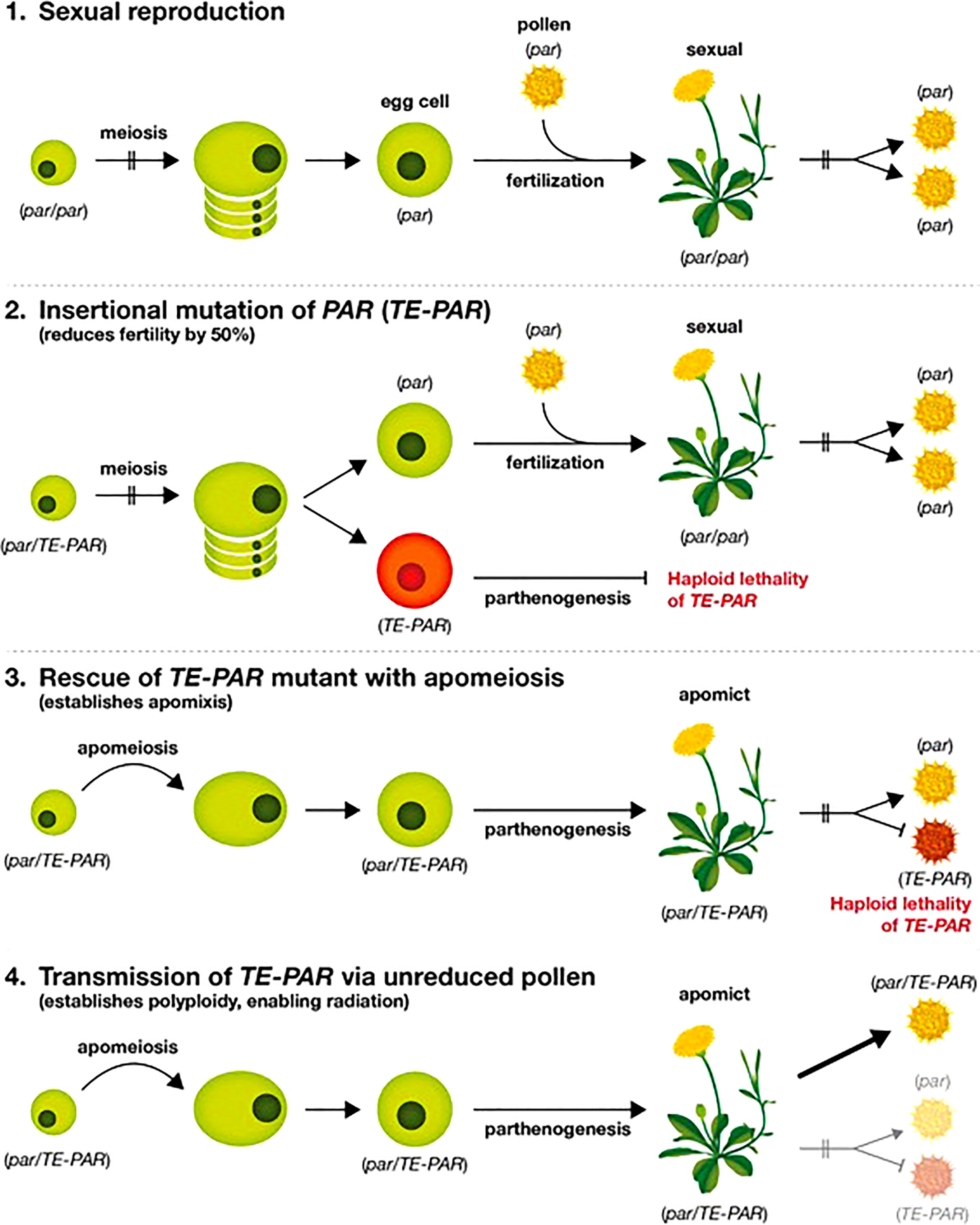
Based on the data presented above and on observations within the literature we present a model for the evolution of gametophytic apomixis in the Lactuceae (Figure 4). We propose that the primary event in the evolution of apomixis is the insertion of a transposable element within a 200 bp conserved region of the PAR gene promoter, immediately upstream of the start codon (Underwood et al., 2022) as illustrated in step 2 of Figure 4. This establishes a dominant allele for PAR, the expression of which promotes spontaneous egg cell activation without fertilization (parthenogenesis) as described by Underwood et al. (2022). We have evidence that this event occurred independently in at least three separate apomictic genera within the Lactuceae: Taraxacum, Pilosella and Hieracium (Underwood et al., 2022) (see also Table 5, Figure 3). Given the differences noted between these elements (Table 5), it appears that the precise identity of the transposon is not critical, however the three elements do share a number of common characteristics that may be of importance. All are class II, DNA-mediated elements, although they differ in their internal sequence, terminal repeat sequence and TE superfamily. All are non-autonomous elements, and they all have imperfectly matched terminal repeat sequences that are likely to prevent their excision. Also, although they all resemble MITE elements in their structure (Wicker et al., 2007), they are all longer than a typical MITE element. Notably, they are all of a similar length (1,233 – 1,335 bp) indicating either the possible presence of regulatory sequences within them or the critical separation of regulatory elements within the native promoter of PAR, either of which may direct the ectopic expression of PAR.
Meiosis will remain functional following such a TE insertion event, therefore 50% of gametes will carry this modified allele and 50% will not. Our data suggest that PAR alleles modified by a TE insertion cannot transmit through haploid gametes, either via eggs or pollen (Table 3) so an immediate outcome of the TE-PAR mutation would be to reduce both seed and pollen fertility by 50%. While this would restrain the spread of the TE-PAR mutation, it is unlikely to prevent it altogether, as unreduced pollen grains are known to form in a wide variety of flowering plant species (Brownfield and Köhler, 2011). The mediation of unreduced pollen promotes the formation of polyploid progeny, so it is interesting to note that gametophytic apomixis is almost exclusively found in polyploid plants, often existing alongside diploid, sexual relatives (Asker and Jerling, 1992). At this step in the evolution of apomixis, therefore, modified PAR alleles would be restricted either to single individuals or to a limited number of polyploid (probably triploid) offspring derived from outcrossing. We predict, therefore, that most TE-PAR mutations are lost before a more durable form of apomixis is achieved.
If, however, a mechanism of apomeiosis subsequently evolves, it would act to ‘rescue’ a TE-PAR mutation by enabling reproduction through apomixis rather than sexual reproduction (step 3 in Figure 4). Such mutations are likely to occur rarely, so it is notable that gametophytic apomixis is almost exclusively associated with a perennial habit; it is often seen in plants with other forms of vegetative increase; and it typically occurs in polycarpic species that produce abundant seed (Asker and Jerling, 1992). These are all traits that increase the numbers of gametes produced and/or extend the life of a clone, increasing the likelihood that a form of apomeiosis will establish before a modified TE-PAR allele is lost. In the Lactuceae, for example, apomicts are restricted to perennial species, whilst annuals such as lettuce (Lactuca sativa) are well represented amongst the sexual members of the group. The form of newly installed apomeiosis does not appear to be critical for the rescue of dominant PAR alleles, as apospory, meiotic diplospory and mitotic diplospory are all found in closely related genera within the Lactuceae. It is also notable that, in the two cases studied: diplospory in Taraxacum (Vijverberg et al., 2004) and apospory in Pilosella (Koltunow et al., 2011), apomeiosis is conferred by dominant alleles, which are the most likely to establish apomixis in a mutant background where TE-PAR alleles are not participating in fertilization.
Finally, the collective instalment of apomeiosis and parthenogenesis would only lead to the perpetuation of a single clone, unless there was a mechanism enabling the dispersal of the critical alleles into a wider germplasm pool. Pollen transfer is the most likely mechanism to mediate this as apomixis restricts access to the female gamete. As described above, dominant alleles conferring both apomeiosis and parthenogenesis appear to transfer only in diploid or polyploid gametes, so the mediation of unreduced pollen grains is the most likely mechanism for their transfer. Unreduced pollen promotes the production of polyploid progeny following the fertilization of a sexual recipient (step 4 in Figure 4). In this way polyploid apomictic clones would rapidly establish and hybridize, facilitating their diversification and adaptation over time as previously proposed (Van Dijk et al., 2009; Hojsgaard and Hörandl, 2019). This process, presumably, underlies the observation that the apomicts of Pilosella are autopolyploid species (Table 4) and that the dominant alleles of Pilosella share a very high degree of similarity (Figure 3), indicating a common origin and recent exchange between interfertile plants within this genus.
The evolution of the remaining component of apomixis, endosperm formation, remains unclear. Apomicts in the Asteraceae typically develop an endosperm through the spontaneous fusion and subsequent division of the polar nuclei (‘autonomous endosperm’). Conversely, most apomicts beyond the Asteraceae develop an endosperm following the fertilization of polar nuclei, in a manner similar to sexual seed formation (‘pseudogamous apomixis’) (Asker and Jerling, 1992; Crane, 2001). This suggests a predisposition in the Asteraceae towards autonomous endosperm formation, but the nature of that predisposition remains speculative. In Pilosella piloselloides var. praealta R35, autonomous endosperm formation was lost when the LOP locus was deleted (Catanach et al., 2006), indicating that it is either encoded by genetic elements within the LOP locus and/or that it requires other genes but is dependent on the action of LOP to be expressed. Ogawa et al. (2013) however, reported the genetic uncoupling of autonomous endosperm formation from both apospory and parthenogenesis in Pilosella piloselloides var. piloselloides (syn. Hieracium piloselloides D3), a relative of Pilosella piloselloides var. praealta R35. This indicates the action of another locus (AutE) controlling autonomous endosperm and therefore a further evolutionary step in the development of apomixis in this system, but the location and structure of the AutE locus are unknown.
Conclusions
The LOSS OF PARTHENOGENESIS (LOP) locus, which mediates spontaneous embryo formation in P. piloselloides, acts as a non-recombinant genomic interval with an estimated size of 654 kb. The PAR gene lies near a limit of that interval, within 68 kb of the nearest recombinant marker. Beyond the LOP locus, allelic exchange occurs between all four alleles in this system, indicating tetrasomic inheritance in an autopolyploid system. Allele sequence divergence was assessed for the PAR genes of Pilosella and Hieracium. In Pilosella, dominant PAR alleles appear to share a recent co-ancestry, whilst recessive alleles clustered in a manner suggestive of reticulate exchange between species in the genus. In Hieracium, dominant alleles also clustered separately to the recessive alleles, but both were well separated from the Pilosella clades, indicating a separate origin. Further evidence of this separation came from sequence analysis of the Hieracium dominant alleles. Whilst both Hieracium and Pilosella dominant PAR alleles have a transposon insertion immediately upstream of the PAR start codon, the length, location, terminal repeat and internal sequences of the different transposons differed significantly, indicating they represent separate ancestral mutations of PAR. The importance of ploidy on LOP allele transfer was then tested. Dominant ‘parthenogenesis’ alleles of LOP could only transfer through egg cells, when they were accompanied by a recessive lop allele. Haploid transfer of a dominant LOP allele was never observed, neither through the egg cell nor pollen. Interestingly, biased inheritance was observed between the recessive alleles present, indicating a functional role for both dominant and recessive alleles in reproduction in Pilosella. Using the information outlined above, a model for the establishment of apomixis in the Lactuceae is presented.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, BioProject PRJNA982544.
Author contributions
RB, MG, AC, CW: Contributed to the research plan. RB, MG, AC, RM, SE, BF, CW: Contributed to the data collection. RB, MG, AC, RM, SE, BF, CW: Contributed to writing the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The authors would like to thank the former New Zealand Foundation for Science and Technology (now the New Zealand Ministry of Business Innovation and Employment), and the New Zealand Ministry for Primary Industries for their financial support of this study.
Acknowledgments
The authors thank Dr R. Michelmore for providing access to the draft lettuce genomic sequence and Dr R. Wing and members of his lab for their help with the construction of the BAC library. Our thanks also to Donna Gibson for her help with the figures.
Conflict of interest
Authors RB, AC, SE, and BF were employed by the company The New Zealand Institute for Plant and Food Research Limited.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Akiyama, Y., Conner, J. A., Goel, S., Morishige, D. T., Mullet, J. E., Hanna, W. W., et al. (2004). High-resolution physical mapping in Pennisetum squamulatum reveals extensive chromosomal heteromorphism of the genomic region associated with apomixis. Plant Physiol. 134, 1733–1741. doi: 10.1104/pp.103.033969
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Bicknell, R., Catanach, A., Hand, M., Koltunow, A. (2016). Seeds of doubt: Mendel’s choice of Hieracium to study inheritance, a case of right plant, wrong trait. Theor. Appl. Genet. 129, 2253–2266. doi: 10.1007/s00122-016-2788-x
Bicknell, R. A., Koltunow, A. M. (2004). Understanding apomixis: Recent advances and remaining conundrums. Plant Cell 16, S228–S245. doi: 10.1105/tpc.017921
Bicknell, R. A., Lambie, S. C., Butler, R. C. (2003). Quantification of progeny classes in two facultatively apomictic accessions of Hieracium. Hereditas 138, 11–20. doi: 10.1034/j.1601-5223.2003.01624.x
Bräuning, S., Catanach, A., Lord, J. M., Bicknell, R., Macknight, R. C. (2018). Comparative transcriptome analysis of the wild-type model apomict Hieracium praealtum and its loss of parthenogenesis (lop) mutant. BMC Plant Biol. 18, 206. doi: 10.1186/s12870-018-1423-1
Bräutigam, S., Greuter, W. (2007). A new treatment of Pilosella for the Euro-Mediterranean flora. Willdenowia, 123–137. doi: 10.3372/wi.37.37106
Brownfield, L., Köhler, C. (2011). Unreduced gamete formation in plants: mechanisms and prospects. J. Exp. Bot. 62, 1659–1668. doi: 10.1093/jxb/erq371
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinf. 10, 1–9. doi: 10.1186/1471-2105-10-421
Carman, J. G. (1997). Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol. J. Linn Soc. 61, 51–94. doi: 10.1111/j.1095-8312.1997.tb01778.x
Catanach, A. S., Erasmuson, S. K., Podivinski, E., Jordan, B. R., Bicknell, R. A. (2006). Deletion mapping of genetic elements associated with apomixis in Hieracium. Proc. Natl. Acad. Sci. U.S.A. 103, 18650–18655. doi: 10.1073/pnas.0605588103
Corral, J. M., Piwczynski, M., Sharbel, T. F. (2009). Allelic sequence divergence in the apomictic Boechera holboellii complex. Lost sex: evolution. Biol. parthenogen., 495–516. doi: 10.1007/978-90-481-2770-2_23
Crane, C. F. (2001). “Classification of apomictic mechanisms,” in Flowering of apomixis: From mechanisms to genetic engineering. Eds. Savidan, Y., Carman, J. G., Dresselhaus, T. (Mexico: CIMMYT, IRD, European Commission DG VI), 24–34.
Fehrer, J., Gemeinholzer, B., Chrtek, J. J., Bräutigam, S. (2007). Incongruent plastid and nuclear DNA phylogenies reveal ancient intergeneric hybridization in Pilosella hawkweeds (Hieracium, Cichorieae, Asteraceae). Mol. Phylogenet. Evol. 42, 347–361. doi: 10.1016/j.ympev.2006.07.004
Gadella, T. W. (1972). Biosystematic studies in Hieracium pilosella L. and some related species of the subgenus Pilosella. Botaniska notiser 125, 361–369.
Gadella, T. W. J. (1984). Cytology and the mode of reproduction of some taxa of Hieracium subgnenus Pilosella. Proc. Kon Ned Acad. Wetensch C 87, 387–399.
Gadella, T. W. J. (1991). Variation, hybridization and reproductive biology of Hieracium pilosella L. Proc. Kon. Ned. Akad. V. Wetensch. 94, 455–488.
Gardes, M., Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Hand, M. L., Koltunow, A. M. G. (2014). The genetic control of apomixis: Asexual seed formation. Genetics 197, 441–450. doi: 10.1534/genetics.114.163105
Harlan, J. R., De Wet, J. M. J. (1975). On Ö. Winge and a prayer: The origins of polyploidy. Bot. Review. 41, 361–390.
Hojsgaard, D., Hörandl, E. (2019). The rise of apomixis in natural plant populations. Front. Plant Sci. 10, 358. doi: 10.3389/fpls.2019.00358
Hörandl, E. (2011). Evolution and biogeography of alpine apomictic plants. Taxon 60, 390–402. doi: 10.1002/tax.602009
Hörandl, E., Hojsgaard, D. (2012). The evolution of apomixis in angiosperms: a reappraisal. Plant Biosystems-An Int. J. Dealing all Aspects Plant Biol. 146, 681–693. doi: 10.1080/11263504.2012.716795
Houliston, G. J., Chapman, H. M. (2001). Sexual reproduction in field populations of the facultative apomict, Hieracium pilosella. New Zeal J. Bot. 39, 141–146. doi: 10.1080/0028825X.2001.9512722
Kimura, M., Maruyama, T., Crow, J. F. (1963). The mutation load in small populations. Genetics 48, 1303–1312. doi: 10.1093/genetics/48.10.1303
Knox, M. R., Ellis, T. H. N. (2001). Stability and inheritance of methylation states at PstI sites in Pisum. Mol. Genet. Genomics 265, 497–507. doi: 10.1007/s004380000438
Kolmogorov, M., Yuan, J., Lin, Y., Pevzner, P. A. (2019). Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546. doi: 10.1038/s41587-019-0072-8
Koltunow, A., Johnson, S. D., Bicknell, R. A. (1998). Sexual and apomictic development in Hieracium. Sex Plant Reprod. 11, 213–230. doi: 10.1007/s004970050144
Koltunow, A. M. G., Johnson, S. D., Rodrigues, J. C. M., Okada, T., Hu, Y., Tsuchiya, T., et al. (2011). Sexual reproduction is the default mode in apomictic Hieracium subgenus Pilosella where two dominant loci function to enable apomixis. Plant J. 66, 890–902. doi: 10.1111/j.1365-313X.2011.04556.x
Kotani, Y., Henderson, S. T., Suzuki, G., Johnson, S. D., Okada, T., Siddons, H., et al. (2014). The LOSS OF APOMEIOSIS (LOA) locus in Hieracium praealtum can function independently of the associated large-scale repetitive chromosomal structure. New Phytol. 201, 973–981. doi: 10.1111/nph.12574
Krahulcová, A., Krahulec, F. (1999). Chromosome numbers and reproductive systems in selected representatives of Hieracium subgen. Pilosella in the Krkonose Mts (the Sudeten Mts). Preslia 71, 217–234.
Krahulcová, A., Krahulec, F., Chrtek, J. (2001). Chromosome numbers and reproductive systems in selected representatives of Hieracium subgen. Pilosella in the Krkonose Mts (the Sudeten Mts): 2. Preslia 73, 193–211.
Luo, M. C., Gu, Y. Q., Puiu, D., Wang, H., Twardziok, S. O., Deal, K. R., et al. (2017). Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nat. Biotechnol. 551, 498–502. doi: 10.1038/nature24486
Luo, M., Wing, R. A. (2003). “An improved method for plant BAC library construction,” in Plant functional genomics (New York, NY: Springer New York), 3–19. doi: 10.1385/1-59259-413-1:3
Mark Welch, D. B., Meselson, M. (2000). Evidence for the evolution of bdelloid rotifers without sexual reproduction or genetic exchange. Science 288, 1211–1215. doi: 10.1126/science.288.5469.1211
Matzk, F., Meister, A., Schubert, I. (2000). An efficient screen for reproductive pathways using mature seeds of monocots and dicots. Plant J. 21, 97–108. doi: 10.1046/j.1365-313x.2000.00647.x
Mau, M., Lovell, J. T., Corral, J. M., Kiefer, C., Koch, M. A., Aliyu, O. M., et al. (2015). Hybrid apomicts trapped in the ecological niches of their sexual ancestors. Proc. Natl. Acad. Sci. 112, E2357–E2365. doi: 10.1073/pnas.1423447112
McGee, R. (2013). Allelism and allele sequence divergence of LOP, the locus of parthenogenesis in the model apomict Hieracium praealtum (Asteraceae) (New Zealand: University of Canterbury). Thesis.
Mráz, P., Chrtek, J., Fehrer, J. (2011). Interspecific hybridization in the genus Hieracium s. str.: evidence for bidirectional gene flow and spontaneous allopolyploidization. Plant Sys. Evol. 293, 237–245. doi: 10.1007/s00606-011-0441-3
Mráz, P., Chrtek, J., Šingliarová, B. (2009). Geographical parthenogenesis, genome size variation and pollen production in the arctic-alpine species. Hieracium alpinum. Botanica Helv. 119, 41–51. doi: 10.1007/s00035-009-0055-3
Mráz, P., Paule, J. (2006). Experimental hybridization in the genus Hieracium s. str.: crosses between diploid taxa. Preslia 78, 1–26.
Mráz, P., Zdvořák, P. (2018). Reproductive pathways in Hieracium s.s. (Asteraceae): strict sexuality in diploids and apomixis in polyploids. Ann. Bot. 123, 391–403. doi: 10.1093/aob/mcy137
Muller, H. J. (1964). The relation of recombination to mutational advance. Mutat. Res. 1, 2–9. doi: 10.1016/0027-5107(64)90047-8
Nogler, G. A. (1984). Genetics of apospory in apomictic Ranunculus auricomus. V. Conclusion. Botanica Helv. 94, 411–422.
Noyes, R. D. (2007). Apomixis in the Asteraceae: diamonds in the rough. Funct. Plant Sci. Biotechnol. 1, 207–222.
Ogawa, D., Johnson, S. D., Henderson, S. T., Koltunow, A. M. G. (2013). Genetic separation of autonomous endosperm formation (AutE) from the two other components of apomixis in Hieracium. Plant Reprod. 26, 113–123. doi: 10.1007/s00497-013-0214-y
Otto, F. (1990). DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. Methods Cell Biol. 33, 105–110. doi: 10.1016/S0091-679X(08)60516-6
Ozias-Akins, P., Lubbers, E. L., Hanna, W. W., Mcnay, J. W. (1993). Transmission of the apomictic mode of reproduction in pennisetum - co-inheritance of the trait and molecular markers. Theor. Appl. Genet. 85, 632–638. doi: 10.1007/BF00220923
Ozias-Akins, P., Roche, D., Hanna, W. W. (1998). Tight clustering and hemizygosity of apomixis-linked molecular markers in Pennisetum squamulatum genetic control of apospory by a divergent locus that may have no allelic form in sexual genotypes. Proc. Natl. Acad. Sci. U.S.A 95, 5127–5132. doi: 10.1073/pnas.95.9.5127
Ramsey, J., Schemske, D. W. (2002). Neopolyploidy in flowering plants. Annu. Rev. Ecol. sys. 33, 589–639. doi: 10.1146/annurev.ecolsys.33.010802.150437
Ranney, T. G. (2006). “Polyploidy: From evolution to new plant development,” in Combined Proceedings International Plant Propagators’ Society. 137–142.
Reyes-Chin-Wo, S., Wang, Z., Yang, X., Kozik, A., Arikit, S., Song, C., et al. (2017). Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 8, 14953. doi: 10.1038/ncomms14953
Roche, D., Chen, Z. B., Hanna, W. W., Ozias-Akins, P. (2001). Non-Mendelian transmission of an apospory-specific genomic region in a reciprocal cross between sexual pearl millet (Pennisetum glaucum) and an apomictic F1 (P. glaucum x P. squamulatum). Sex Plant Reprod. 13, 217–223. doi: 10.1007/s004970000060
Roche, D., Cong, P. S., Chen, Z. B., Hanna, W. W., Gustine, D. L., Sherwood, R. T., et al. (1999). An apospory-specific genomic region is conserved between Buffelgrass (Cenchrus ciliaris L.) and Pennisetum squamulatum Fresen. Plant J. 19, 203–208. doi: 10.1046/j.1365-313X.1999.00514.x
Roche, D., Conner, J. A., Budiman, M. A., Frisch, D., Wing, R., Hanna, W. W., et al. (2002). Construction of BAC libraries from two apomictic grasses to study the microcolinearity of their apospory-specific genomic regions. Theor. Appl. Genet. 104, 804–812. doi: 10.1007/s00122-001-0795-y
Rodrigues, J. C. M., Tucker, M. R., Johnson, S. D., Hrmova, M., Koltunow, A. M. G. (2008). Sexual and apomictic seed formation in hieracium requires the plant polycomb-group gene FERTILIZATION INDEPENDENT ENDOSPERM. Plant Cell Online 20, 2372–2386. doi: 10.1105/tpc.108.059287
Sherwood, R. T., Berg, C. C., Young, B. A. (1994). Inheritance of apospory in buffelgrass. Crop Sci. 34, 1490–1494. doi: 10.2135/cropsci1994.0011183X003400060014x
Shirasawa, K., Hand, M. L., Henderson, S. T., Okada, T., Johnson, S. D., Taylor, J. M., et al. (2014). A reference genetic linkage map of apomictic Hieracium species based on expressed markers derived from developing ovule transcripts. Ann. Bot. 115, 567–580. doi: 10.1093/aob/mcu249
Tucker, M. R., Araujo, A. C. G., Paech, N. A., Hecht, V., Schmidt, E. D. L., Rossell, J. B., et al. (2003). Sexual and apomictic reproduction in Hieracium subgenus Pilosella are closely interrelated developmental pathways. Plant Cell 15, 1524–1537. doi: 10.1105/tpc.011742
Tutin, T. G., Heywood, V. H., Burges, N. A., Moore, D. M., Valentine, D. H., Walters, S. M., et al. (1976). Flora Europaea 4, 505 p. Plantaginaceae to Compositae (and Rubiaceae) Cambridge: Cambridge University Press.
Underwood, C., Vijverberg, K., Rigola, D., Okamoto, S., Oplaat, C., Camp, R., et al. (2022). A PARTHENOGENESIS allele from apomictic dandelion can induce egg cell division without fertilization in lettuce. Nat. Genet. 54, 84–93. doi: 10.1038/s41588-021-00984-y
Van Dijk, P., De Jong, H., K., V., Biere, A. (2009). “An apomixis-gene’s view on dandelions,” in Lost Sex. Eds. Schön, I., Martens, K., Dijk, P.V. (Dordrecht: Springer), 475–494.
Van Dijk, P. J., Vijverberg, K. (2005). “The significance of apomixis in the evolution of the angiosperms: a reappraisal,” in Plant Species-Level Systematics: New Perspectives on Pattern and Process. Eds. Bakker, F., Chatrou, L., Gravendeel, B., Pelser, P. (Lichtenstein: Gantner Verlag, Ruggell), 101–116.
Vijverberg, K., van der Hulst, R. G. M., Lindhout, P., Van Dijk, P. J. (2004). A genetic linkage map of the diplosporous chromosomal region in Taraxacum officinale (common dandelion; Asteraceae). Theor. Appl. Genet. 108, 725–732. doi: 10.1007/s00122-003-1474-y
Wicker, T., Sabot, F., Hua-Van, A., Bennetzen, J. L., Capy, P., Chalhoub, B., et al. (2007). A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8, 973–982. doi: 10.1038/nrg2165
Zeng, Y., Conner, J., Ozias-Akins, P. (2011). Identification of ovule transcripts from the Apospory-Specific Genomic Region (ASGR)-carrier chromosome. BMC Genomics 12, 206. doi: 10.1186/1471-2164-12-206
Keywords: parthenogenesis (asexual reproduction), apomixis, pilosella, allele sequence divergence, egg cell activation
Citation: Bicknell R, Gaillard M, Catanach A, McGee R, Erasmuson S, Fulton B and Winefield C (2023) Genetic mapping of the LOSS OF PARTHENOGENESIS locus in Pilosella piloselloides and the evolution of apomixis in the Lactuceae. Front. Plant Sci. 14:1239191. doi: 10.3389/fpls.2023.1239191
Received: 13 June 2023; Accepted: 01 August 2023;
Published: 25 August 2023.
Edited by:
Shuqing Xu, Johannes Gutenberg University Mainz, GermanyReviewed by:
Diego Hojsgaard, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), GermanyFulvio Pupilli, National Research Council (CNR), Italy
Copyright © 2023 Bicknell, Gaillard, Catanach, McGee, Erasmuson, Fulton and Winefield. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ross Bicknell, cm9zcy5iaWNrbmVsbEBwbGFudGFuZGZvb2QuY28ubno=
 Ross Bicknell
Ross Bicknell Marion Gaillard
Marion Gaillard Andrew Catanach
Andrew Catanach Robert McGee3
Robert McGee3 Beatrice Fulton
Beatrice Fulton