
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 21 July 2023
Sec. Plant Metabolism and Chemodiversity
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1232880
This article is part of the Research Topic Contribution of Phenylpropanoid Metabolism to Plant Development and Stress Responses View all 8 articles
Jasmonic acid (JA) is a phytohormone involved in plant defense, growth, and development, etc. However, the regulatory mechanisms underlying JA-mediated lignin deposition and secondary cell wall (SCW) formation remain poorly understood. In this study, we found that JA can inhibit lignin deposition and SCW thickening in poplar trees through exogenous MeJA treatment and observation of the phenotypes of a JA synthesis mutant, opdat1. Hence, we identified a JA signal inhibitor PtoJAZ5, belonging to the TIFY gene family, which is involved in the regulation of secondary vascular development of Populus tomentosa. RT-qPCR and GUS staining revealed that PtoJAZ5 was highly expressed in poplar stems, particularly in developing xylem. Overexpression of PtoJAZ5 inhibited SCW thickening and down-regulated the expression of SCW biosynthesis-related genes. Further biochemical analysis showed that PtoJAZ5 interacted with multiple SCW switches NAC/MYB transcription factors, including MYB3 and WND6A, through yeast two-hybrid and bimolecular fluorescent complementation experiments. Transcriptional activation assays demonstrated that MYB3-PtoJAZ5 and WND6A-PtoJAZ5 complexes regulated the expression of lignin synthetic genes. Our results suggest that PtoJAZ5 plays a negative role in JA-induced lignin deposition and SCW thickening in poplar and provide new insights into the molecular mechanisms underlying JA-mediated regulation of SCW formation.
The secondary cell wall (SCW) is a crucial component of wood, composed of lignin, cellulose, and hemicelluloses. It provides mechanical support for water transport and physical defense against pathogenic bacteria in woody plants. Lignin, a phenolic polymer, exists in a highly crosslinked state in the SCW of plants and is one of the most important substances for plants to resist pathogenic microorganisms and abiotic stresses (Vanholme et al., 2010).
Lignin is composed of three main types of monolignols: p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units. The biosynthesis of monolignols occurs through the phenylpropanoid pathway and involves an 11-step enzymatic reaction in the cytoplasm, requiring the participation of various enzymes, including phenylalanine ammonia lyase (PAL), 4-coumarate-CoA ligase (4CL), cinnamate 4-hydroxylase (C4H), cinnamoyl-CoA reductase (CCR), p-hydroxycinnamoyl-CoA: quinate/shikimate p-hydroxycinnamoyltransferase (HCT), cinnamyl alcohol dehydrogenase (CAD), p-coumaroyl shikimate 3-hydroxylase (C3’H), caffeoyl-CoA O-methyltransferase (CCoAOMT), caffeic acid O-methyltransferase (COMT), and ferulate-5-hydroxylase (F5H) (Vanholme et al., 2010). Finally, monolignols are transported across the plasma membrane to the apoplast, where they undergo oxidative polymerization and crosslinking by peroxidases and/or laccases to form the lignin polymer, which stabilizes the cell wall structure (Freudenberg, 1959; Vanholme et al., 2019). Cellulose is synthesized by a cellulose synthase complex (CSC) located in the plasma membrane, first forming β-1,4-D-glucan chains, and then 24 or 36 β-1,4-D-glucan chains are polymerized to form microfibrils, cellulose microfibres provide essential load-bearing properties to cell walls (Mutwil et al., 2008). Xylan is the main component of hemicellulose, it is mainly composed of linear chains of xylose linked by β-1,4 bonds, synthesized in the Golgi apparatus, and then transported to the cell membrane to participate in cell wall synthesis (Rennie and Scheller, 2014).
SCW biosynthesis is a complex and highly regulated process that involves multiple transcription factors. In model plants, such as Arabidopsis (Arabidopsis thaliana) and poplar, a multi-stage feedback regulatory network of SCW synthesis has been established, mainly composed of NAC/MYB transcription factors (Mccahill and Hazen, 2019). NAC transcription factors, including VND1-7, NST1, NST2, and NST3/SND1 in Arabidopsis (Kubo et al., 2005; Zhong et al., 2006; Mitsuda et al., 2007), and PtrWNDs in poplar (Zhong et al., 2010), act as primary switches to initiate the transcriptional regulation of SCWs. Additionally, MYB transcription factors, such as PtrMYB02/03/20/21 in poplar (Wilkins et al., 2009; Mccarthy et al., 2010; Chai et al., 2014), act as secondary switches to activate the transcription of SCW synthase genes. The activation of these switch factors is closely related to plant growth and development, and is precisely regulated by plant hormones.
Jasmonic acid (JA) and its derivatives are lipid-derived phytohormones that are widely involved in the regulation of plant growth, development, and defense response (Staswick and Tiryaki, 2004). Recent studies have revealed that JA is also involved in the lignification process of plant cells. For example, exogenous methyl jasmonate (MeJA) treatment enhances lignin deposition and SCW thickening in Arabidopsis (Sehr et al., 2010), and mechanical injury to plants causes a rapid increase in JA content and lignin synthesis (Denness et al., 2011). Additionally, researchers cultured pine seedlings at an angle and found that JA was consistently distributed in the upper part of the angled stems, where G-type lignin was predominantly present. (Salazar et al., 2019). These findings imply that there is a close relationship between JA and lignin synthesis and secondary cell wall thickening, although the molecular mechanisms involved in this relationship remain unclear.
The main signal transduction pathway of JA is mediated by JAZ (JASMONATE-ZIM-DOMAIN) proteins (Chini et al., 2007). In the absence of JA, JAZ proteins bind to transcription factors, such as MYC2 (Chen et al., 2011), inhibiting their transcriptional activity (Thines et al., 2007). When JA level is elevated, the bioactive derivative JA-Ile binds to the receptor COI1 protein, which then interacts with JAZ proteins, promoting their ubiquitination and degradation by the 26S proteasome, and ultimately leading to the activation of downstream gene expression (Katsir et al., 2008; Kramell et al., 2009). Previous research has demonstrated that MeJA treatment and injury can both increase the expression of the genes involved in lignin production (Tianpei et al., 2015; Hyde et al., 2018; Valenzuela-Riffo et al., 2020), but the regulatory network underlying this process remains to be fully elucidated.
In this study, we investigated the role of JA signaling in regulating lignin deposition and SCW thickening in poplar. We employed the opdat1 mutant, a poplar JA synthesis mutant obtained from our previous study (Zhao et al., 2021), and confirmed that JA deficiency leads to a reduction in secondary cell wall thickness. Treatment of wild-type poplar with exogenous methyl jasmonate (MeJA) induced the expression of lignin synthesis genes. Furthermore, PtoJAZ5-overexpression poplar inhibited JA signal transduction, resulting in a block in the biosynthesis of the SCW. We used yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) experiments to confirm that PtoJAZ5 interacts with WND and MYB transcription factors involved in secondary wall synthesis. In addition, our transient transactivation assays of GUS activity showed that PtoJAZ5 suppresses transcriptional activation of downstream SCW synthesis genes by WND6A and MYB3. These findings suggest that JA signaling can regulate the transcript levels of lignin and SCW synthesis genes through PtoJAZ proteins, thereby affecting lignin deposition and SCW thickening in poplar.
Poplar plants (Populus tomentosa Carr. clone 741) were grown in a greenhouse at 23-25°C, 16 h light/8 h dark, 5000 lux supplementary light, and humidity 60%. The Col-0 seedlings of Arabidopsis thaliana were grown in a incubator at 22°C, 16 h light/8h dark, 10,000 lux supplementary light, and humidity 80%. Arabidopsis seeds were germinated on MS medium (8.5 g/L Agar and 30 g/L sucrose) and placed in a lighted incubator. Arabidopsis seedlings were transferred to nutrient soil and continued to be incubated and growth in a light incubator. The culture conditions: 80% humidity, 20–23°CC, 16/8 h light/dark cycle and 555 mmol m−2 s−1 supplemental light (Murashige and Skoog, 1962). Arabidopsis transformation using 30-day-old plants by floral dip method (Zhang et al., 2006). For poplar transformation, the leaf disk method was used to the pCXSN-PtoJAZ5 vector according to the methods previously (Jia et al., 2010a).
Exogenous jasmonic acid (JA) treatment of wild-type (WT) poplars was performed by continuous watering with a 1 mM solution of methyl Jasmonate (MeJA) for one month, and the plants were cultured in a greenhouse and observed for phenotype.
Total RNA was extracted from poplar stems and reverse transcribed into cDNA using a kit (TAKARA), specific primers were designed and the full-length coding sequence of PtoJAZ5 (831 bp) was amplified by Polymerase Chain Reaction (PCR). The amplified fragments were then cloned into pCXSN vector (Chen et al., 2009). For construction of the PtoJAZ5pro::GUS reporter vector, the 1857-bp promoter fragment was amplified and assembled into the pCAMBIA1305 vector. All plant expression vectors were used to transform plant material using the Agrobacterium-mediated leaf disc method according to previous study (Jia et al., 2010b). The software DNAMAN9 (Lynnon Biosoft, San Ramon, CA, USA) was used for multiple sequence alignments, and the software MEGA7.0 was used for phylogenic analysis using the neighbor-joining (NJ) method.
Plant materials such as roots, stems, xylem, bast, old and young leaves in good growth condition were selected and ground thoroughly using liquid nitrogen, and then total RNA was extracted using the Trizol Reagent (Tiangen, China), followed by reverse transcription of cDNA using the PrimeScript™ RT reagent Kit (Takara, Shiga, Japan). Specific quantitative primers were designed as needed, and the quantitative RT-qPCR was performed using SYBR Premix ExTaq™ (Takara) in a TP800 Real-Time PCR machine (Takara). The UBQ11 gene was used as an internal control in poplar and AtUBC in Arabidopsis. The RT-qPCR reaction conditions were 95°C, 30 s; 40 cycles: 95°C, 5 s; 60°C, 1 min; 95°C, 15 s; 60°C, 30 s; 95°C, 15 s. cycles: 95°C, 5 s; 60°C, 1 min; 95°C, 15 s; 60°C, 30 s; 95°C, 15 s. Three biological replicates were required for each gene of the RT-qPCR, and the primers involved were listed in Table S1. To analyze their expression levels, the 2-ΔΔCt method (Livak and Schmittgen, 2001) was used in Figure 1F and the 2-ΔCt method (Chiba-Falek et al., 2006) was used in other figures.
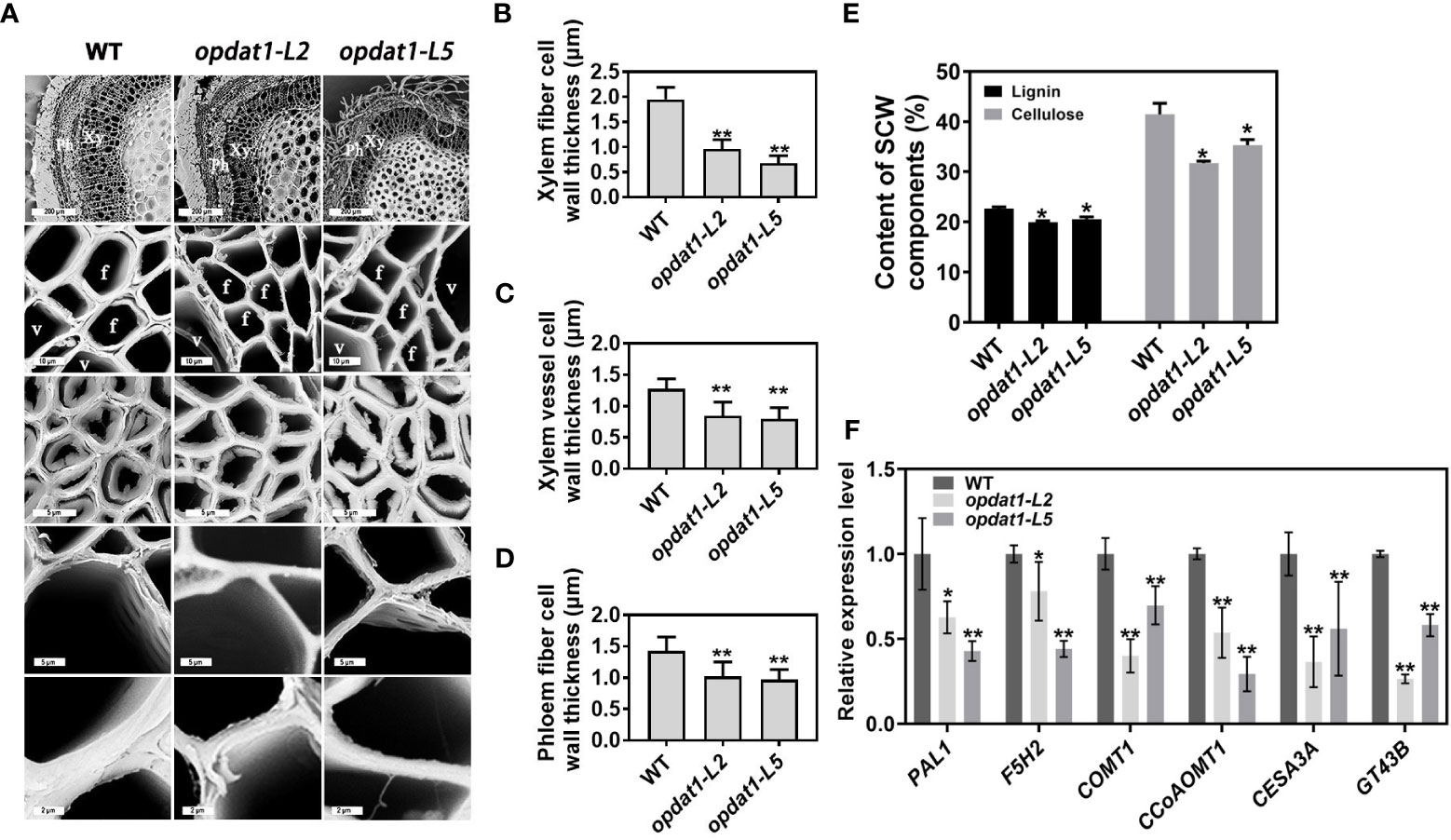
Figure 1 Secondary cell wall thinning in the opdat11 poplar mutant. (A) Scanning electron microscope observation of 5th internode stem section of transgenic poplar, from bottom to top, magnified xylem fiber cells, magnified xylem vessel cells, phloem fiber cells, xylem cells and stem cross-sectional anatomy. The scale bars from bottom to top = 5 μm, 2 μm, 5 μm, 10 μm, 200 μm. Xy, xylem; Ph, phloem; V, vessel cells; F, fiber cells. (B) Xylem fiber cell wall thickness statistics. (C) Xylem vessel cell wall thickness statistics. (D) Phloem fiber cell wall thickness statistics. (E) Determination of lignin and cellulose content of three-month-old poplar stems. (F) RT-qPCR was performed to detect the expression levels of SCW synthesis genes of three-month-old poplar stems. The error bars indicate the standard deviation of the data from the three biological replicates in the experiment. Asterisks indicate that the data from the experimental replicates show significant differences based on Student’s t-test (*P < 0.05; **P < 0.01).
The full-length coding sequence of PtoJAZ5 was amplified via the specific primer and assembled into the pCAMBIA1300 vector. Then the tobacco leaf flesh epidermal cells were transformed using A. tumefaciens mediated transformation method. Empty vector 35S-GFP was used as positive control. 4’,6-diamidino-2-phenylindole (DAPI) was used to stain the nucleus of cells. A confocal laser microscope (Olympus FV1200, Tokyo, Japan) was used to detected the DAPI and GFP fluorescent signals.
Full-length and mutant DNA sequences of PtoJAZ5 were amplified and constructed into the PGBKT7 (BD) vector, while full-length DNA sequences of PtoMYC2, PtoMYB3/2/20/21/74, and PtoWND1A/1B/2A/2B/3A/3B/4A/4B/5A/5B/6A/6B were amplified and constructed into the PGADT7 (AD) vector. Positive controls included AD-T and BD-53 vectors, while negative controls consisted of AD-T and BD-lam, BD and AD empty vectors. Y2H gold strain was used in yeast experiments, yeast transformation was performed using the PEG/LiAC method according to the Yeast Protocol Handbook (Clontech), and transformed yeast colonies were selected on nutrient-deficient medium. Transcriptional self-activation verification was performed using media lacking histidine, adenine, and tryptophan (SD-Ade-His-Trp) or only lacking tryptophan (SD-Trp), while protein interaction verification was performed using media lacking leucine and tryptophan (SD-Leu-Trp) or lacking tryptophan, leucine, histidine, and adenine (SD-Leu-Trp-Ade-His). Transcription activation activity was identified using X-α-gal, a chromogenic substrate for yeast galactosidase (MEL1). The transformed yeast cells were grown in a 30°C constant temperature incubator for 3-5 days.
The ORF of PtoJAZ5 was cloned into the PXY106 vector containing the N-terminal of YFP and formed into a complete fusion expression vector. The ORF of PtoMYB3 and PtoWND6A were cloned into the PXY104 vector containing the C-terminal of YFP and formed into a complete fusion expression vector. The resulting constructs were transfered into tobacco leaf flesh epidermal cells using A. tumefaciens mediated transformation method. A confocal laser microscope (Olympus FV1200, Tokyo, Japan) was used to detected the DAPI and YFP fluorescent signals after 2-3 days of incubation.
Poplar and Arabidopsis stems were cross-sectioned by hand or semi-automatic slicers (FINESSE 325, Thermo, Runcorn, UK). Histochemical staining including phloroglucinol-HCl, toluidine blue and GUS staining of stem cross sections was performed according to previously described (Li et al., 2015). Scanning electron microscope (Phenomtm Pure FEI, Eindhoven, Netherland) was used to observe the stem cross-sections according to the operation manual.
For immunocytochemical analysis of JAs, stem samples were fixed with 4% (w/v) 1-ethyl-3-(3-dimethyl aminopropyl)-carbodiimide hydrochloride (Merck KgaA) in PBS, embedded in polyethylene glycol (PEG) 1500, and immunostained with an anti-JA antibody as reported for immunocytochemical examination of JAs (Mielke et al., 2011). Refer to the previous description for the detailed operation steps (Fu et al., 2021).
Klason and acid-soluble lignin methods as previously described (Li et al., 2015) were used to measure the total lignin content of poplar stems. Cellulose, pectin and hemicellulose were determined using Peng method (Peng et al., 2000; Fan et al., 2020).
The ImageJ (https://imagej.nih.gov/ij/) (Collins, 2007) software was used for image data measurement, the GraphPad Prism 7 software (Mitteer et al., 2018) was used for data analysis and statistics and One-way ANOVA followed by Dunnett’s test analysis was performed to evaluate statistical significance. For pairwise comparisons, Dunnett’s test was utilized in the statistical analysis.
The promoter sequences of the lignin synthesis genes PtoCCR2, PtoCOMT2, PtoCCoAOMT1 and PtoMYB3 were amplified and fused to the GUS reporter gene into the PCXGUS-P vector. Agrobacterium cells carrying the 35S-PtoJAZ5, 35S-PtoMYB3 and 35S-PtoWND6A constructs were used as effectors. Empty vector as negative control. Infiltrating into 2-week-old tobacco leaf flesh epidermal cells with different mixes contain the effectors and reporters of Agrobacterium spp. After three days of cultivation, transgenic tobacco leaves were then collected and GUS activity was quantitatively measured by a spectrophotometry according to previously method (Jefferson et al., 1987).
To determine the distribution of JA in poplar stem, we conducted immunocytochemical detection experiments and observed that JA is predominantly located in the phloem and developing xylem regions (Figure S1), suggesting its potential role in the regulation of xylem development. To further explore the essential role of JA in SCW development, we observed sections of the JA synthesis mutant opdat1 (Zhao et al., 2021) under scanning electron microscope, which revealed significant reduction in SCW thickness of xylem fiber cells, xylem vessel cells and phloem fibers (Figures 1A-D). Furthermore, the lignin and cellulose content of SCW also significantly decreased in opdat1 mutants (Figure 1E), while the xylan content did not differ significantly. RT-qPCR analysis further demonstrated a significant down-regulation in the expression level of key SCW synthesis-related enzyme genes, including PAL1, F5H2, COMT1, CCoAOMT1, CESA3A, and GT43B in the opdat1 mutant poplar (Figure 1F). These findings suggest that the decreased JA content leads to delayed SCW development in poplar.
To investigate the effects of JA on lignin deposition and SCW thickening in poplar, we treated wild-type (WT) poplar with MeJA solution for one month. Methyl jasmonate (MeJA) treatment significantly inhibited the growth of poplar stems (Figure S2). Toluidine blue and phloroglucinol-HCl staining of sections from the sixth internode stem revealed that MeJA treatment significantly increased the depth of xylem staining and SCW thickness (Figure S3A). Scanning electron microscopy also showed that MeJA treatment promoted the thickening of SCW in vessel and fiber cells (Figures S3B-D), which was consistent with the increased lignin content in treated poplar (Figure S3E). Furthermore, RT-qPCR analysis revealed that the expression levels of lignin synthase genes (COMT2, CCoAOMT1, CCR2, CAld5H2, PAL1, CSE1, C4H2, and 4CL5), cellulose synthase gene (CESA3A), xylan synthase gene (GT43B), and some laccase genes (LAC29, LAC30, and LAC41) were significantly induced by MeJA treatment (Figure S3F). The application of MeJA to the opdat1 mutant also significantly restored the phenotype of SCW thinning (Figure 2), suggesting that MeJA treatment promotes SCW thickening by inducing the expression of genes related to secondary wall biosynthesis.
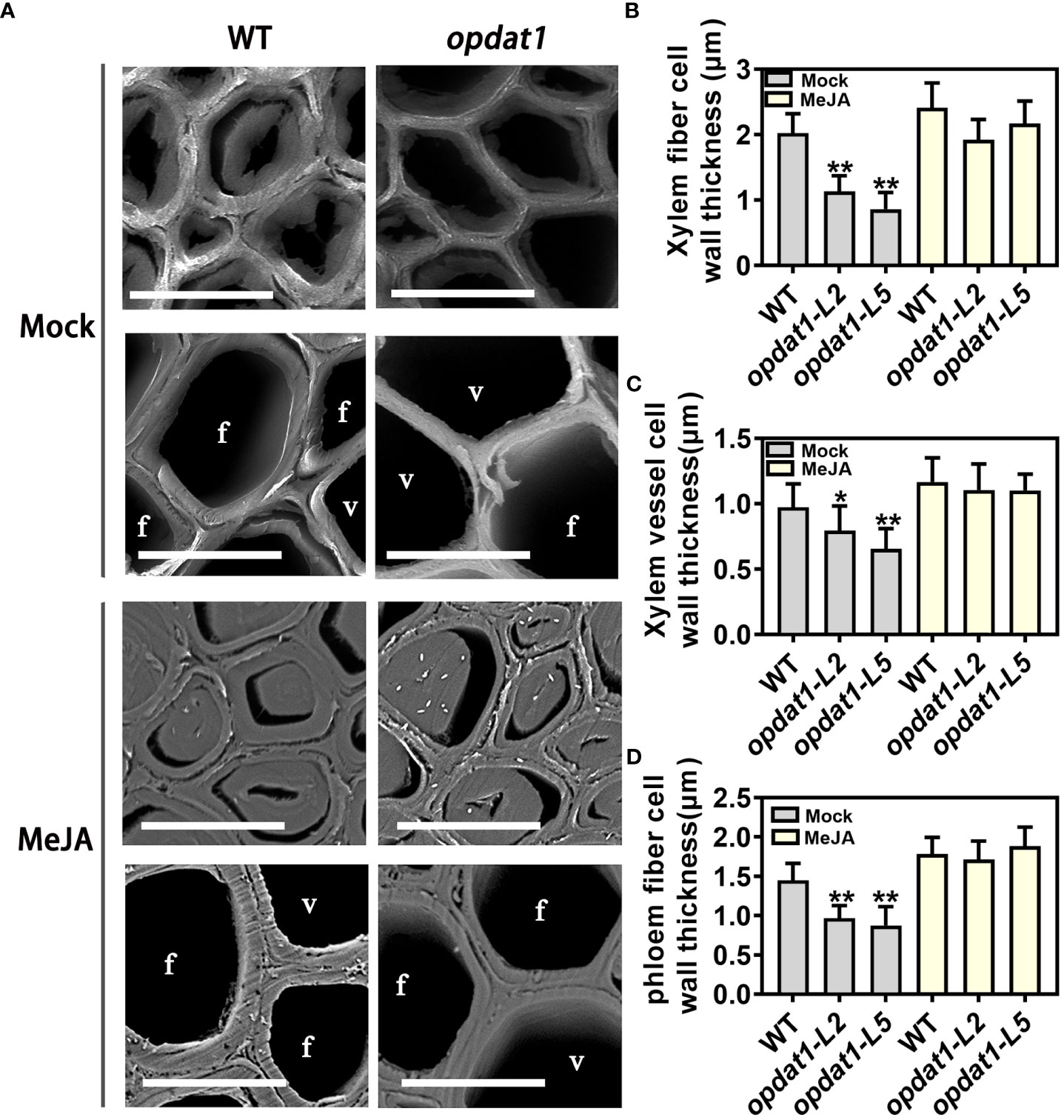
Figure 2 Section observation after MeJA treatment of opdat1 mutant poplars. (A) Observation of poplar 5th internode stem sections by scanning electron microscope. Phloem cells above and xylem cells below in the treatment and control groups. (B) Xylem fiber cell secondary wall measurement. fiber cells. (C) Xylem vessel cell secondary wall measurement. v, vessel cell. (D) Phloem fiber cell secondary wall measurement. The scale bars = 10 μm. The error bars indicate the standard deviation of the data from the three biological replicates in the experiment. Asterisks indicate that the data from the experimental replicates show significant differences based on Student’s t-test (*P < 0.05; **P < 0.01).
It is well known that the degradation of JAZ proteins is the main mechanism of JA signaling in plants (Thines et al., 2007). To investigate the role of JA signaling in lignin deposition and SCW synthesis, we aimed to reduce JA signaling by overexpressing JAZ genes. We identified 12 JAZ proteins in poplar and constructed a phylogenetic tree of Arabidopsis and poplar JAZ protein sequences (Figure S4A). Poplar JAZ5 has a 58.48% amino acid sequence similarity to Arabidopsis AtJAZ1/2, and contains the conserved Tify and Jas domains with a conserved LPIARRA amino acid sequence in the Jas domain (Figure S4B). Previous studies have demonstrated that AtJAZ1 is involved in plant growth, development, and response to freezing and senescence (Robson et al., 2010; Zhou et al., 2017; Valenzuela-Riffo et al., 2020). Gene expression profile data of poplar JAZ proteins showed that PtoJAZ5 is specifically expressed in the xylem (Figures S4C, D), indicating that PtoJAZ5 may play a role in the transcriptional regulation of JA signaling in the xylem region. Therefore, we cloned the poplar PtoJAZ5 gene for further investigation.
To investigate the subcellular localization of PtoJAZ5, we generated a PtoJAZ5-GFP fusion protein driven by the CaMV 35S promoter and transiently expressed it in tobacco epidermal cells. The localization of the fusion protein was visualized by observing the GFP fluorescence signal. The results showed that PtoJAZ5 is primarily localized in the nucleus and co-localized with the DAPI signal, confirming that PtoJAZ5 is a nuclear-localized protein (Figure 3A). To further explore the transcriptional activity of PtoJAZ5, we fused PtoJAZ5 to the GAL4 DNA binding domain (BD) and transformed the recombinant plasmids (BD-PtoJAZ5) into yeast cells. The transformed yeast cells were able to grow on selective medium lacking tryptophan (SD/-T), but could not grow on medium lacking tryptophan, histidine and adenine (SD/-ATH), indicating that PtoJAZ5 does not have transcriptional activation activity (Figure 3B).
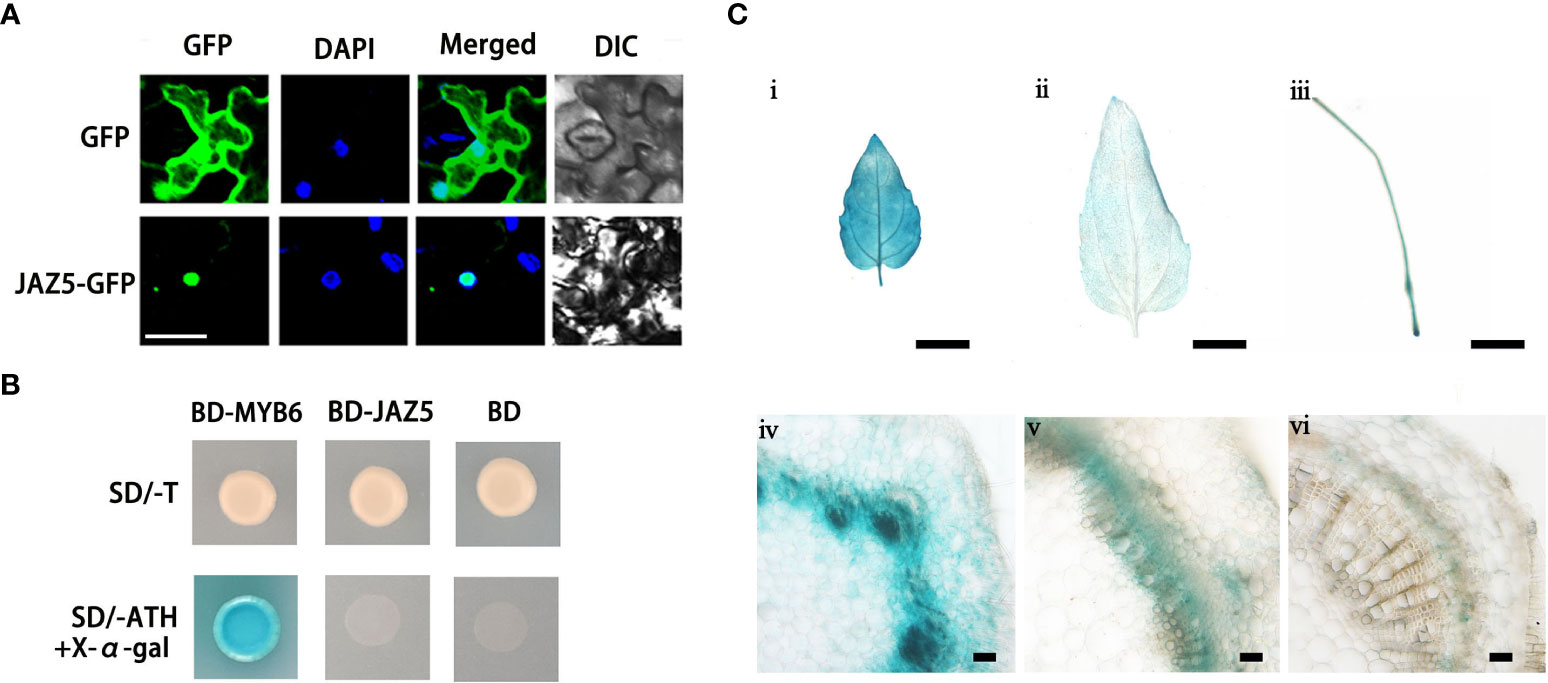
Figure 3 Analysis of subcellular localization, tissue expression and transcriptional activity of poplar JAZ5 protein. (A) Subcellular localization observation of JAZ5. DAPI is a nuclear dye; DIC indicates observation under white light. Scale bar: 50 μm. (B) Analysis of transcriptional activity of JAZ5. MYB6 is a transcriptional activator and was used as a positive control in this experiment (Wang et al., 2019) and BD is the empty vector for negative control. SD/-T represents one deficient yeast medium lacking tryptophan. SD/-ATH represents three deficient yeast medium lacking tryptophan, histidine and adenine. X-α-gal is a chromogenic substrate for yeast galactosidase (MEL1). (C) GUS staining for young leaves (і), old leaves (і), roots (ii), stem second internode (iii), stem fourth internode (iv), and stem base (v). scale bars: 0.5 cm (і-iii); 100 μm (iii-v).
To investigate the spatiotemporal expression pattern of PtoJAZ5, we constructed a PtoJAZ5 promoter-GUS fusion vector. GUS staining revealed that PtoJAZ5 is mainly expressed in the cambium layer and developing xylem region, young leaves, roots, and stems of poplar (Figure 3C). These results suggest that PtoJAZ5 may play a role in the growth and development of xylem.
To determine the role of PtoJAZ5 in SCW formation, we overexpressed PtoJAZ5 in poplar under the control of the CaMV 35S promoter. RT-qPCR assay was performed to select transgenic lines (Line 8 and Line 9) with high expression of PtoJAZ5 for further experiments (Figure S5D). No significant difference in the appearance phenotype was observed between transgenic and WT poplars, except for a slight increase in height of some transgenic plants (Figures S5A-C). RT-qPCR results showed that the expression levels of JA signaling marker genes (PR4, VSP1, VSP2) were significantly down-regulated in PtoJAZ5 overexpression plants (Figure S5E), indicating the suppression of JA signaling.
To further investigate the effect of PtoJAZ5 on SCW formation, we performed section staining and scanning electron microscopy on the sixth internode stem of transgenic poplar. In comparison to WT, a lighter staining color of the xylem region and a significant reduction in the thickness of vessel cells and fiber cells were observed in the PtoJAZ5 overexpression plants compared with WT. However, the area of vessel cells and fiber cells was significantly increased (Figures 4A-E). RT-qPCR results showed that the expression levels of SCW biosynthase genes including lignin synthase genes (C4H2, CCR2, CAD1, CCoAOMT1, and CAld5H2), cellulose synthase genes (CESA3A, CESA2B), and xylan synthase genes (GT8D and GT43B) were significantly down-regulated in the transgenic plants (Figures 4F, G). Additionally, exogenous MeJA treatment significantly restored the phenotype of SCW thinning in transgenic poplars (Figure S8). These results indicate that the SCW thinning in PtoJAZ5 overexpression poplar is mediated by a decrease in the expression level of SCW synthesis-related enzyme genes.
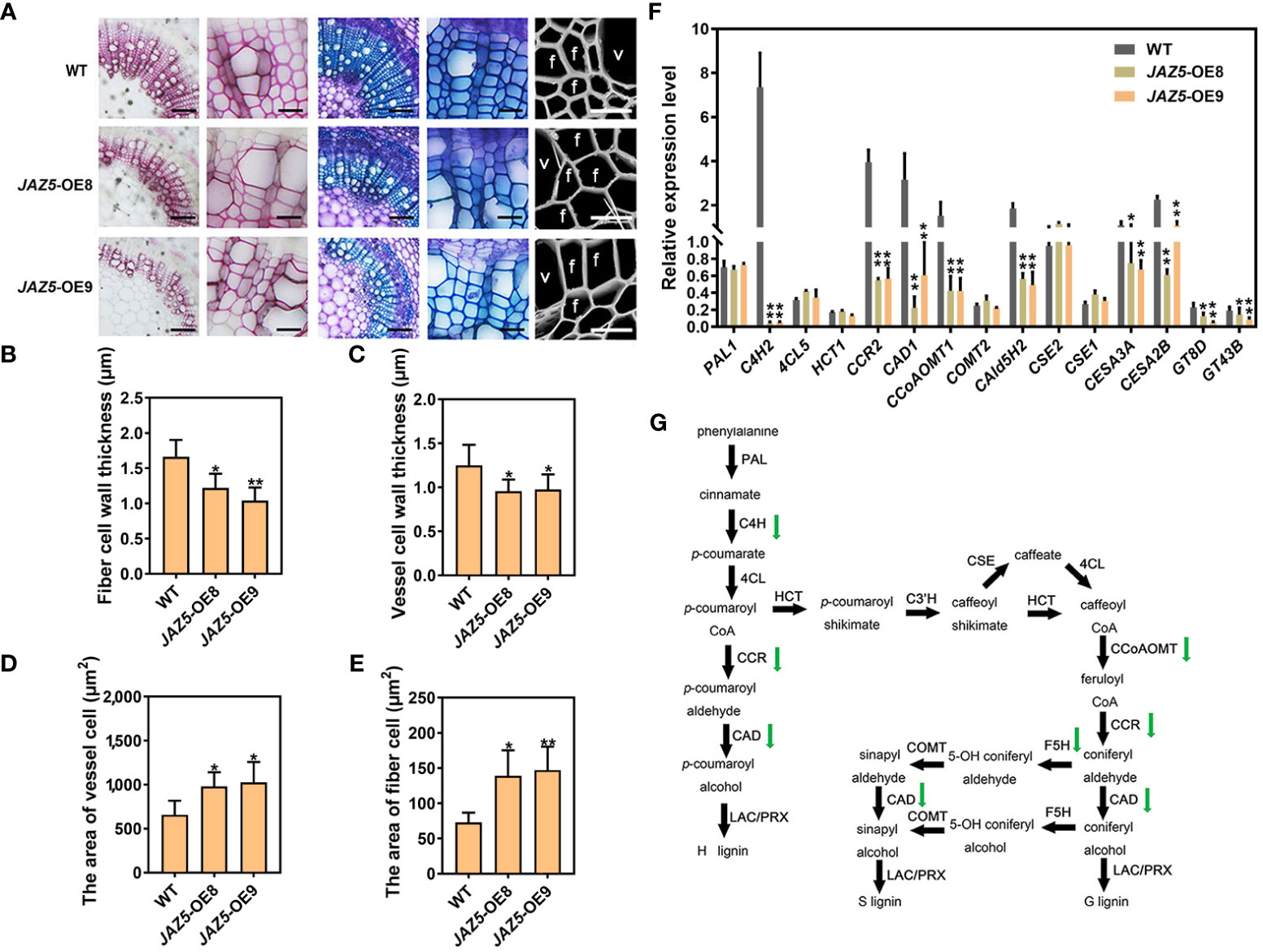
Figure 4 Reduced lignin deposition and secondary cell wall thinning during wood formation in JAZ5-OE transgenic poplar plants. (A) Section observation of wild-type and transgenic plants, from left to right, with Ph-cl, toluidine blue staining and scanning electron microscopy. (B-E) Statistical analysis of the stem region for fiber secondary wall, vessel cell secondary wall, vessel cell size and fiber cell size. Scale bars (Figure A) = 100 μm (columns 1, 3), 50 μm (columns 2, 4), 20 μm (columns 5). In the picture, f represents fiber cells and v represents vessel cells. OE8 and OE9 represent two different JAZ5 overexpression lines. (F) RT-qPCR of secondary cell wall synthesis-related enzyme genes of stem in JAZ5-OE poplar, including lignin synthase genes (PAL1, C4H2, 4CL5, HCT1, CCR2, CAD1, CCoAOMT1, COMT2, CAld5H2, CSE2 and CSE1), cellulose synthase genes (CESA3A and CESA2B) and xylan synthase genes (GT8D and GT43B). (G) Schematic diagram of the lignin synthesis pathway. The error bars indicate the standard deviation of the data from the three biological replicates in the experiment. Asterisks indicate that the data from the experimental replicates show significant differences based on Student’s t-test (*P < 0.05; **P < 0.01).
To examine the conservation of JAZ5 function, we also heterologously overexpressed PtoJAZ5 in Arabidopsis. Analysis of stem sections of transgenic Arabidopsis showed that the autofluorescence intensity of lignin was significantly reduced, and the color of toluidine blue staining and phloroglucinol-HCl staining in stem of Arabidopsis overexpressing PtoJAZ5 was lighter compared to WT. Scanning electron microscopy revealed significant thinning of SCW in transgenic Arabidopsis (Figure S6). Consistent with the phenotypic observation, RT-qPCR results revealed that transcript levels of SCW synthase genes in transgenic Arabidopsis were significantly reduced (Figure S7). These results suggest that PtoJAZ5 is able to inhibit lignin deposition and SCW thickening in Arabidopsis, indicating its conserved function.
Intensive studies have shown that JA signaling is mediated by the interaction of JAZ proteins with transcription factors, which results in the suppression or activation of downstream genes depending on JA levels (Chini et al., 2007; Thines et al., 2007; Katsir et al., 2008; Pauwels et al., 2010). Using yeast two-hybrid (Y2H) experiments, we searched for SCW synthesis-related transcription factors that interact with JAZ5. The Y2H results revealed that PtoJAZ5 interacted with several SCW synthesis switch masters, such as MYB2, MYB3, MYB74, WND3A, WND4A, WND6A, and WND6B (Figure 5A). Previous studies have shown that poplar MYB3/2/20/21, which are mainly expressed in the developing xylem region, act as secondary switches in the regulatory network of SCW synthesis (Wilkins et al., 2009; Mccarthy et al., 2010). The WNDs, belonging to the NAC gene family, are first-order switches in the regulatory network of SCW synthesis in poplar (Zhong et al., 2010; Zhong et al., 2011). Based on the gene expression analysis and Y2H experiment, we selected MYB3 and WND6A as representative transcription factors to further investigate the function of PtoJAZ5 (Figure 5A and Figure S9). We constructed PtoJAZ5-PXY104, a C-terminal fusion expression vector of PtoJAZ5 with yellow fluorescent protein (PtoJAZ5-cYFP), and PXY106-MYB3 and PXY106-WND6A, fusion expression vectors of the N-terminal of YFP with MYB3 and WND6A (nYFP-MYB3 and nYFP-WND6A), for fluorescent bimolecular complementation experiments (BiFC). The combination of PtoJAZ5/MYB3 and PtoJAZ5/WND6A showed a yellow fluorescent signal in tobacco leaves, while the other two negative controls showed no signal when observed under laser confocal electron microscopy (Figures 5B, C). These results suggest that PtoJAZ5 interacts with MYB3 and WND6A in plant cells.
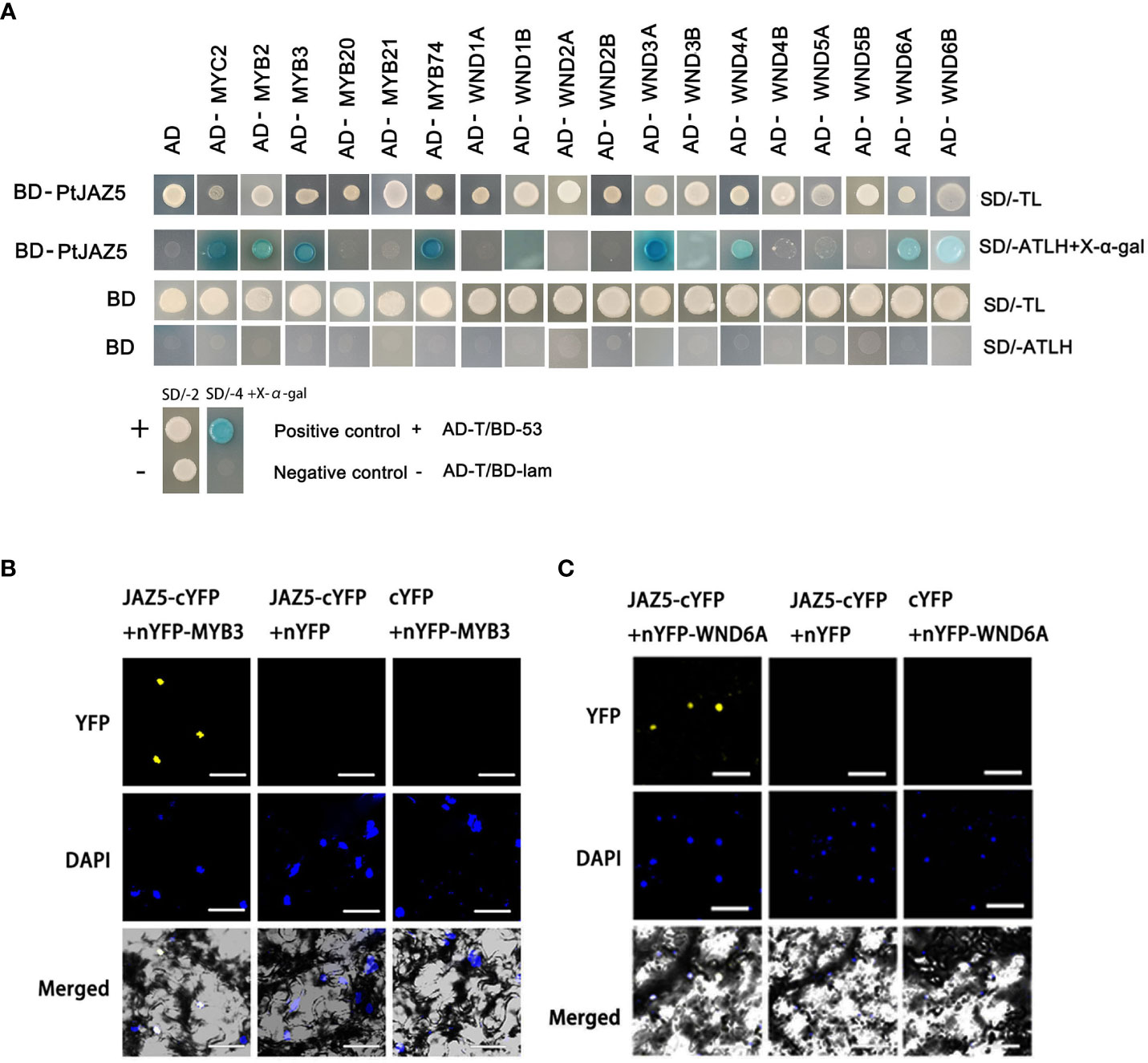
Figure 5 Interaction analysis of JAZ5 with NAC and MYB transcription factors from poplar via Y2H assay. (A) AD and BD denote PGADT7 and PGBKT7 vectors, respectively. The BD vector fusion protein JAZ5 was constructed, and the AD vector fusion protein included JA signaling transcription factor MYC2 and key regulators of secondary wall synthesis (MYB transcription factor MYB2/3/20/21/74 and NAC transcription factor WND1A/1B/2A/2B/3A/3B/4A/4B/5A/5B/6A/6B). AD-T and BD-53 vectors are positive controls; AD-T and BD-lam, BD and AD empty vectors are negative controls. SD/-TL represents a two-deficient yeast medium lacking tryptophan and leucine. SD/-ATLH represents a four-deficient yeast medium lacking tryptophan, leucine, histidine and adenine. X-α-gal is a chromogenic substrate for yeast galactosidase (MEL1). (B, C) Bimolecular Fluorescent Complimentary (BiFC), the C-terminal fusion protein JAZ5-cYFP expression vector of JAZ5-linked yellow fluorescent protein (YFP) and the N-terminal fusion proteins MYB3-nYFP and WND6A-nYFP expression vectors of MYB3- and WND6A-linked YFP were constructed, and the C-terminal and N-terminal empty expression vectors of YFP were used as negative controls to transform tobacco leaves for transient expression. The scale bars = 10 μm.
To identify the core amino acid sequence fragments of PtoJAZ5 that interact with MYB3 or WND6A, we constructed a yeast fusion expression vector of PGBKT7 by truncating PtoJAZ5 into different structural domains, including JAS domain, Tify domain, and N-terminal domain, based on the amino acid sequence specificity of PtoJAZ5. The Y2H assays showed that proteins containing the JAS domain could interact with MYB3 or WND6A, and yeast transformants grew normally on SD/-ATLH-deficient medium. Conversely, yeast transformants failed to grow when the JAS domain was absent (Figures S10A, B). Therefore, we conclude that the JAS domain is the necessary core sequence for PtoJAZ5 to interact with MYB3 or WND6A.
Previous studies have revealed that the JAS structural domain mediates the degradation of JAZ protein by JA receptor COI1 (Sheard et al., 2010). Based on amino acid sequence analysis of PtoJAZ5, we mutated the core sequence of the JAZ degradation domain, LPIAR, to LPIGA, to construct a PtoJAZ5 mutant protein (PtoJAZ5m). Using a Y2H assay to verify whether PtoJAZ5m protein could still interact with MYB3/WND6A, and found that this sequence mutation did not affect the interaction of PtoJAZ5 with MYB3 or WND6A (Figure S10C). Next, we investigated whether the LPIAR sequence mutation affected the degradation of PtoJAZ5. We constructed PtoJAZ5-GFP and PtoJAZ5m-GFP fusion protein expression vectors and transformed them into Arabidopsis using floral dip method. Transgenic Arabidopsis seedlings were treated with MeJA solution and observed under a fluorescence microscope. The results showed that the green fluorescence signal was diminished or even disappeared in transgenic Arabidopsis roots expressing PtoJAZ5 but not in those expressing PtoJAZ5m (Figures S11A, B). These findings indicate that LPIAR is the core sequence of the degradation domain of PtoJAZ5, which responds to JA signals.
To investigate whether the interaction between PtoJAZ5 and MYB3/WND6A affects downstream SCW synthase gene expression levels, we used the promoters of CCoAOMT1, CCR2, COMT2, and C4H2 to control the expression of GUS reporter genes. The generated reporter vectors were co-transformed into tobacco leaf epidermal cells with overexpression vectors, enabling transactivation analysis. Our findings demonstrate that MYB3 or WND6A alone significantly activated the expression of GUS genes under the control of the aforementioned promoters. However, this activation was inhibited by PtoJAZ5. Upon MeJA treatment, the repressive effect of PtoJAZ5 on MYB3 and WND6A was alleviated (Figure 6). Thus, our results suggest that PtoJAZ5 regulates SCW biosynthesis via interacting with MYB3 or WND6A, and that this regulatory mechanism is modulated by JA signaling.
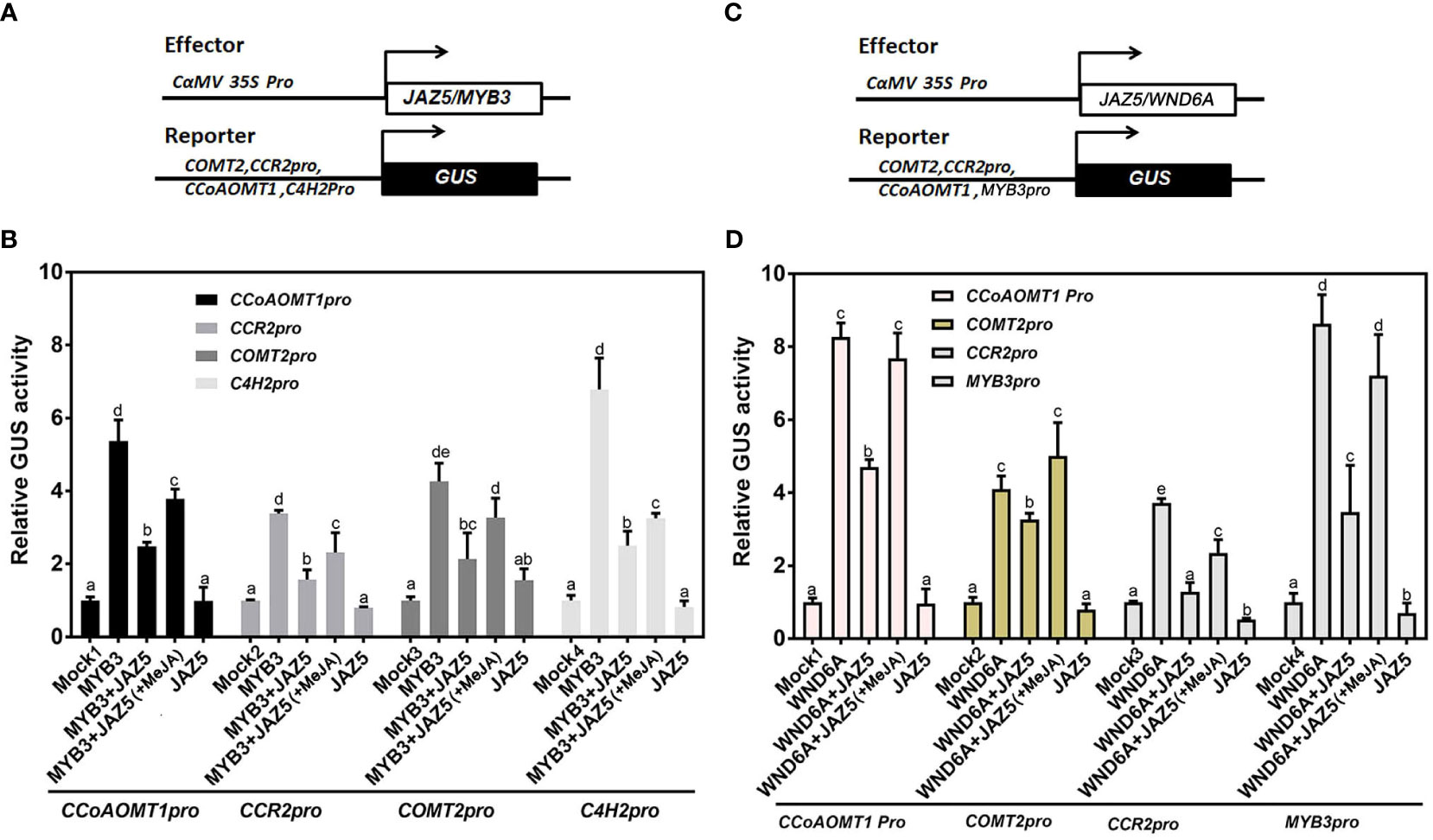
Figure 6 JAZ5 represses transcriptional activation of downstream genes by MYB3 and WND6A. (A, B) JAZ5 represses the transcriptional activation of lignin synthesis-related enzyme genes by MYB3. (C, D) JAZ5 represses the transcriptional activation of lignin synthesis-related enzyme genes and MYB3 via WND6A. Experiments were performed using tobacco leaf transient expression technology, in which the promoter-linked GUS reporter gene expression vectors of CCoAOMT1, CCR2, COMT2, C4H2, and MYB3 were co-transformed with the expression vectors of effector proteins JAZ5 and MYB3/WND6A to detect GUS protein activity. MeJA in parentheses indicates exogenous MeJA treatment. The error bars indicate the standard deviation of the data from the three biological replicates. Different lowercase letters represent significant differences, and one-way ANOVA was used for statistical analysis of differences.
In woody plants, lignin deposition and SCW thickening are crucial for providing mechanical support and physical defensive structures. Various plant hormones, such as auxin, gibberellin, ethylene, abscisic acid, and JA, play important roles in this process (Burger and Chory, 2019; Buttò et al., 2019; Jewaria et al., 2021). Despite their importance, the mechanism by which JA signaling regulates plant SCW development remains unclear.
Previous studies have shown that exogenous application of MeJA treatment promotes thickening of xylem SCWs and ectopic deposition of lignin in Arabidopsis stems (Sehr et al., 2010). However, its molecular mechanism remains unknown and has not been studied in woody plants. In this study, we provide evidence that MeJA treatment induces elevated expression levels of genes related to SCW synthesis, thereby promoting lignin deposition and SCW thickening in poplar stems (Figure S3). Additionally, we confirmed that lower JA levels in poplar led to thinning of the SCW using section observation of the opdat1 mutant and exogenous JA reversion experiments (Figures 1, 2). These results indicate that proper JA content is necessary for SCW development in poplar.
Numerous studies have confirmed that JA mainly functions through the regulation of the repressor JAZ protein and the core transcription factor MYC2 (Chini et al., 2007; Pauwels and Goossens, 2011; Breeze, 2019). For example, MYC2, MYC3, and MYC4 can bind to JAZs to regulate root growth, insect defense, leaf senescence and wounding response in plants (Sasaki-Sekimoto et al., 2013; Zhuo et al., 2020). JAZ1/8/11 regulates anthocyanin synthesis by inhibiting the action of MBW complex (Qi et al., 2011). Therefore, in order to further investigate the mechanism of JA in secondary cell wall development, we overexpressed PtoJAZs to inhibit the JA signaling pathway. We identified 12 poplar JAZ proteins by sequence alignment with Arabidopsis JAZ proteins, and selected PtoJAZ5, which was specifically and highly expressed in stems, as a representative for further study. A plant expression vector carrying PtoJAZ5-OE cassettes was constructed and transgenic poplar plants were generated (Figures S5, S6). Histochemical staining of toluidine blue and phloroglucinol-HCl, and scanning electron microscope observation showed that constitutive expression of PtoJAZ5 inhibited SCW thickening and lignin deposition in poplar (Figure 4) and Arabidopsis (Figures S6, S7). RT-qPCR analysis revealed that overexpression of PtoJAZ5 resulted in decreased expression levels of SCW biosynthetic genes for lignin, cellulose and xylan (Figures 4F, G). Similar results were obtained from PtoJAZ5-OE Arabidopsis (Figure S7). These results are consistent with the findings in Arabidopsis (Sehr et al., 2010) and demonstrate the regulatory role of JA in secondary cell wall development in poplar.
In conclusion, our study provides evidence that JA promotes lignin deposition and SCW thickening in poplar trees. Specifically, we found that JAZ proteins, such as PtoJAZ5, act as inhibitors of JA signal transduction and can suppress the expression and activation of SCW synthase genes by interacting with the SCW synthesis switch factor WND/MYB, such as WND6A/MYB3 (Figures 5, 6). This leads to a reduction in SCW synthesis and lignin deposition in poplar trees. JAZ proteins may not only interact with MYC2 to regulate growth and development and secondary wall synthesis (Zhang et al., 2018; Luo et al., 2022), but they also directly interact with secondary wall synthesis switch factors to influence secondary wall deposition. These findings extent our understanding of the regulatory network of JA in coordinating plant growth and development, as well as defense responses.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
XZ, CX, and KL designed the work. XZ, XJ, ZL and QS performed experiments and analyzed data. XZ, and KL drafted the manuscript. CX and KL revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from National Science Foundation of China (31870657, 31870175 and 32271835), and Chongqing Youth Top Talent Program (CQYC201905028).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1232880/full#supplementary-material
Breeze, E. (2019). Master MYCs: MYC2, the jasmonate signaling "Master switch. Plant Cell 31, 9–10. doi: 10.1105/tpc.19.00004
Burger, M., Chory, J. (2019). Stressed out about hormones: how plants orchestrate immunity. Cell Host Microbe 26, 163–172. doi: 10.1016/j.chom.2019.07.006
Buttò, V., Deslauriers, A., Rossi, S., Rozenberg, P., Shishov, V., Morin, H. (2019). The role of plant hormones in tree-ring formation. Trees 34, 315–335. doi: 10.1007/s00468-019-01940-4
Chai, G., Wang, Z., Tang, X., Yu, L., Qi, G., Wang, D., et al. (2014). R2R3-MYB gene pairs in Populus: evolution and contribution to secondary wall formation and flowering time. J. Exp. Bot. 65, 4255–4269. doi: 10.1093/jxb/eru196
Chen, S., Songkumarn, P., Liu, J., Wang, G. L. (2009). A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 150, 1111–1121. doi: 10.1104/pp.109.137125
Chen, Q., Sun, J., Zhai, Q., Zhou, W., Qi, L., Xu, L., et al. (2011). The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 23, 3335–3352. doi: 10.1105/tpc.111.089870
Chiba-Falek, O., Lopez, G. J., Nussbaum, R. L. (2006). Levels of alpha-synuclein mRNA in sporadic Parkinson disease patients. Mov Disord. 21, 1703–1708. doi: 10.1002/mds.21007
Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Denness, L., Mckenna, J. F., Segonzac, C., Wormit, A., Madhou, P., Bennett, M., et al. (2011). Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in Arabidopsis. Plant Physiol. 156, 1364–1374. doi: 10.1104/pp.111.175737
Fan, C., Yu, H., Qin, S., Li, Y., Alam, A., Xu, C., et al. (2020). Brassinosteroid overproduction improves lignocellulose quantity and quality to maximize bioethanol yield under green-like biomass process in transgenic poplar. Biotechnol. Biofuels 13, 9. doi: 10.1186/s13068-020-1652-z
Freudenberg, K. (1959). Biosynthesis and constitution of lignin. Nature 183, 1152–1155. doi: 10.1038/1831152a0
Fu, X., Su, H., Liu, S., Du, X., Xu, C., Luo, K. (2021). Cytokinin signaling localized in phloem noncell-autonomously regulates cambial activity during secondary growth of Populus stems. New Phytol. 230, 1476–1488. doi: 10.1111/nph.17255
Hyde, L. S., Pellny, T. K., Freeman, J., Michaelson, L. V., Simister, R., Mcqueen-Mason, S. J., et al. (2018). Response of cell-wall composition and RNA-seq transcriptome to methyl-jasmonate in Brachypodium distachyon callus. Planta 248, 1213–1229. doi: 10.1007/s00425-018-2968-9
Jefferson, R. A., Kavanagh, T. A., Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x
Jewaria, P. K., Yu, M., Li, X. (2021). Cell wall and hormone interplay controls growth asymmetry. Trends Plant Sci. 26, 665–667. doi: 10.1016/j.tplants.2021.04.003
Jia, Z., Gou, J., Sun, Y., Yuan, L., Tang, Q., Yang, X., et al. (2010a). Enhanced resistance to fungal pathogens in transgenic Populus tomentosa Carr. by overexpression of an nsLTP-like antimicrobial protein gene from motherwort (Leonurus japonicus). Tree Physiol. 30, 1599–1605. doi: 10.1093/treephys/tpq093
Jia, Z., Sun, Y., Yuan, L., Tian, Q., Luo, K. (2010b). The chitinase gene (Bbchit1) from Beauveria bassiana enhances resistance to Cytospora chrysosperma in Populus tomentosa Carr. Biotechnol. Lett. 32, 1325–1332. doi: 10.1007/s10529-010-0297-6
Katsir, L., Chung, H. S., Koo, A. J. K., Howe, G. A. (2008). Jasmonate signaling: a conserved mechanism of hormone sensing. Curr. Opin. Plant Biol. 11, 428–435. doi: 10.1016/j.pbi.2008.05.004
Kramell, R., Miersch, O., Wasternack, C., Solano, R., Fonseca, S., Chini, A., et al. (2009). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350. doi: 10.1038/nchembio.161
Kubo, M., Udagawa, M., Nishikubo, N., Horiguchi, G., Yamaguchi, M., Ito, J., et al. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860. doi: 10.1101/gad.1331305
Li, C., Wang, X., Ran, L., Tian, Q., Fan, D., Luo, K. (2015). PtoMYB92 is a transcriptional activator of the lignin biosynthetic pathway during secondary cell wall formation in populus tomentosa. Plant Cell Physiol. 56, 2436–2446. doi: 10.1093/pcp/pcv157
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, F., Zhang, Q., Xin, H., Liu, H., Yang, H., Doblin, M. S., et al. (2022). A Phytochrome B-PIF4-MYC2/MYC4 module inhibits secondary cell wall thickening in response to shaded light. Plant Commun. 3 (6), 100416. doi: 10.1016/j.xplc.2022.100416
Mccahill, I. W., Hazen, S. P. (2019). Regulation of cell wall thickening by a medley of mechanisms. Trends Plant Sci. 24, 853–866. doi: 10.1016/j.tplants.2019.05.012
Mccarthy, R. L., Zhong, R., Fowler, S., Lyskowski, D., Piyasena, H., Carleton, K., et al. (2010). The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol. 51, 1084–1090. doi: 10.1093/pcp/pcq064
Mielke, K., Forner, S., Kramell, R., Conrad, U., Hause, B. (2011). Cell-specific visualization of jasmonates in wounded tomato and Arabidopsis leaves using jasmonate-specific antibodies. New Phytol. 190, 1069–1080. doi: 10.1111/j.1469-8137.2010.03638.x
Mitsuda, N., Iwase, A., Yamamoto, H., Yoshida, M., Seki, M., Shinozaki, K., et al. (2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19, 270–280. doi: 10.1105/tpc.106.047043
Mitteer, D. R., Greer, B. D., Fisher, W. W., Cohrs, V. L. (2018). Teaching behavior technicians to create publication-quality, single-case design graphs in graphpad prism 7. J. Appl. Behav. Anal. 51, 998–1010. doi: 10.1002/jaba.483
Murashige, T., Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia plantarum 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Mutwil, M., Debolt, S., Persson, S. (2008). Cellulose synthesis: a complex complex. Curr. Opin. Plant Biol. 11, 252–257. doi: 10.1016/j.pbi.2008.03.007
Pauwels, L., Barbero, G. F., Geerinck, J., Tilleman, S., Grunewald, W., Pérez, A. C., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791. doi: 10.1038/nature08854
Pauwels, L., Goossens, A. (2011). The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23, 3089–3100. doi: 10.1105/tpc.111.089300
Peng, L., Hocart, C. H., Redmond, J. W., Williamson, R. E. (2000). Fractionation of carbohydrates in Arabidopsis root cell walls shows that three radial swelling loci are specifically involved in cellulose production. Planta 211, 406–414. doi: 10.1007/s004250000301
Qi, T., Song, S., Ren, Q., Wu, D., Huang, H., Chen, Y., et al. (2011). The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23, 1795–1814. doi: 10.1105/tpc.111.083261
Rennie, E. A., Scheller, H. V. (2014). Xylan biosynthesis. Curr. Opin. Biotechnol. 26, 100–107. doi: 10.1016/j.copbio.2013.11.013
Robson, F., Okamoto, H., Patrick, E., Harris, S. R., Wasternack, C., Brearley, C., et al. (2010). Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22, 1143–1160. doi: 10.1105/tpc.109.067728
Salazar, R., Pollmann, S., Morales-Quintana, L., Herrera, R., Caparros-Ruiz, D., Ramos, P. (2019). In seedlings of Pinus radiata, jasmonic acid and auxin are differentially distributed on opposite sides of tilted stems affecting lignin monomer biosynthesis and composition. Plant Physiol. Biochem. 135, 215–223. doi: 10.1016/j.plaphy.2018.12.008
Sasaki-Sekimoto, Y., Jikumaru, Y., Obayashi, T., Saito, H., Masuda, S., Kamiya, Y., et al. (2013). Basic helix-loop-helix transcription factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate responses in Arabidopsis. Plant Physiol. 163, 291–304. doi: 10.1104/pp.113.220129
Sehr, E. M., Agusti, J., Lehner, R., Farmer, E. E., Schwarz, M., Greb, T. (2010). Analysis of secondary growth in the Arabidopsis shoot reveals a positive role of jasmonate signalling in cambium formation. Plant J. 63, 811–822. doi: 10.1111/j.1365-313X.2010.04283.x
Sheard, L. B., Tan, X., Mao, H., Withers, J., Ben-Nissan, G., Hinds, T. R., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405. doi: 10.1038/nature09430
Staswick, P. E., Tiryaki, I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16, 2117–2127. doi: 10.1105/tpc.104.023549
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Tianpei, X., Li, D., Qiu, P., Luo, J., Zhu, Y., Li, S. (2015). Scorpion peptide LqhIT2 activates phenylpropanoid pathways via jasmonate to increase rice resistance to rice leafrollers. Plant Sci. 230, 1–11. doi: 10.1016/j.plantsci.2014.10.005
Valenzuela-Riffo, F., Zuniga, P. E., Morales-Quintana, L., Lolas, M., Caceres, M., Figueroa, C. R. (2020). Priming of Defense Systems and Upregulation of MYC2 and JAZ1 Genes after Botrytis cinerea Inoculation in Methyl Jasmonate-Treated Strawberry Fruits. Plants (Basel) 9 (4), 447. doi: 10.3390/plants9040447
Vanholme, R., Demedts, B., Morreel, K., Ralph, J., Boerjan, W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153, 895–905. doi: 10.1104/pp.110.155119
Vanholme, R., De Meester, B., Ralph, J., Boerjan, W. (2019). Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 56, 230–239. doi: 10.1016/j.copbio.2019.02.018
Wang, L., Lu, W., Ran, L., Dou, L., Yao, S., Hu, J., et al. (2019). R2R3-MYB transcription factor MYB6 promotes anthocyanin and proanthocyanidin biosynthesis but inhibits secondary cell wall formation in Populus tomentosa. Plant J. 99, 733–751. doi: 10.1111/tpj.14364
Wilkins, O., Nahal, H., Foong, J., Provart, N. J., Campbell, M. M. (2009). Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 149, 981–993. doi: 10.1104/pp.108.132795
Zhang, X., Henriques, R., Lin, S.-S., Niu, Q.-W., Chua, N.-H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Zhang, Q., Xie, Z., Zhang, R., Xu, P., Liu, H., Yang, H., et al. (2018). Blue light regulates secondary cell wall thickening via MYC2/MYC4 activation of the NST1-directed transcriptional network in arabidopsis. Plant Cell 30, 2512–2528. doi: 10.1105/tpc.18.00315
Zhao, X., Li, N., Song, Q., Li, X., Meng, H., Luo, K. (2021). OPDAT1, a plastid envelope protein involved in 12-oxo-phytodienoic acid export for jasmonic acid biosynthesis in Populus. Tree Physiol. 41, 1714–1728. doi: 10.1093/treephys/tpab037
Zhong, R., Demura, T., Ye, Z. H. (2006). SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18, 3158–3170. doi: 10.1105/tpc.106.047399
Zhong, R., Lee, C., Ye, Z. H. (2010). Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol. 152, 1044–1055. doi: 10.1104/pp.109.148270
Zhong, R., Mccarthy, R. L., Lee, C., Ye, Z. H. (2011). Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol. 157, 1452–1468. doi: 10.1104/pp.111.181354
Zhou, Y., Zhou, X., Li, Q., Chen, J., Xiao, Y., Zhang, L., et al. (2017). Molecular cloning, bioinformatics analysis, and transcriptional profiling of JAZ1 and JAZ2 from Salvia miltiorrhiza. Biotechnol. Appl. Biochem. 64, 27–34. doi: 10.1002/bab.1454
Keywords: JAZ protein, secondary cell wall, lignin deposition, poplar, jasmonic acid
Citation: Zhao X, Jiang X, Li Z, Song Q, Xu C and Luo K (2023) Jasmonic acid regulates lignin deposition in poplar through JAZ5-MYB/NAC interaction. Front. Plant Sci. 14:1232880. doi: 10.3389/fpls.2023.1232880
Received: 01 June 2023; Accepted: 10 July 2023;
Published: 21 July 2023.
Edited by:
Yingzhen Kong, Qingdao Agricultural University, ChinaReviewed by:
Liang Zhang, University of Georgia, United StatesCopyright © 2023 Zhao, Jiang, Li, Song, Xu and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keming Luo, bHVva2VtaW5nQGhvdG1haWwuY29t; Changzhen Xu, eHVjekBzd3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.