
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 17 July 2023
Sec. Functional and Applied Plant Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1229870
 Nabieu Kamara1,2,3,4,5†
Nabieu Kamara1,2,3,4,5† Yamin Jiao1,2,3,4†
Yamin Jiao1,2,3,4† Weicong Huang1,2,3,4
Weicong Huang1,2,3,4 Lichong Cao1,2,3,4
Lichong Cao1,2,3,4 Lianjun Zhu1,2,3,4
Lianjun Zhu1,2,3,4 Chongchong Zhao1,2,3,4
Chongchong Zhao1,2,3,4 Xu Huang1,2,3,4
Xu Huang1,2,3,4 Fimanekeni Ndaitavela Shivute1,2,3,4,6
Fimanekeni Ndaitavela Shivute1,2,3,4,6 Xiangdong Liu1,2,3,4
Xiangdong Liu1,2,3,4 Jinwen Wu1,2,3,4*
Jinwen Wu1,2,3,4* Muhammad Qasim Shahid1,2,3,4*
Muhammad Qasim Shahid1,2,3,4*We aimed to investigate the genetic defects related to pollen development and infertility in NY2, a novel tetraploid rice germplasm known as Neo-tetraploid rice. This rice variety was created through the crossbreeding and selective breeding of various autotetraploid rice lines and has previously shown high fertility. Our previous research has revealed that the NY2 gene, encoding a eukaryotic translation initiation factor 3 subunit E, regulates pollen fertility. However, the underlying mechanism behind this fertility is yet to be understood. To shed light on this matter, we performed a combined cytological and transcriptome analysis of the NY2 gene. Cytological analysis indicated that ny2 underwent abnormal tapetal cells, microspore, and middle layer development, which led to pollen abortion and ultimately to male sterility. Genetic analysis revealed that the F1 plants showed normal fertility and an obvious advantage for seed setting compared to ny2. Global gene expression analysis in ny2 revealed a total of 7545 genes were detected at the meiosis stage, and 3925 and 3620 displayed upregulation and downregulation, respectively. The genes were significantly enriched for the gene ontology (GO) term “carbohydrate metabolic process. Moreover, 9 genes related to tapetum or pollen fertility showed down-regulation, such as OsABCG26 (ATP Binding Cassette G26), TMS9-1 (Thermosensitive Male Sterility), EAT1 (Programmed cell death regulatory), KIN14M (Kinesin Motor), OsMT1a (Metallothionein), and OsSTRL2 (Atypical strictosidine synthase), which were validated by qRT-PCR. Further analyses of DEGs identified nine down-regulated transcription factor genes related to pollen development. NY2 is an important regulator of the development of tapetum and microspore. The regulatory gene network described in this study may offer important understandings into the molecular processes that underlie fertility control in tetraploid rice.
Autotetraploid, as a new rice resource, has selective advantages over diploid rice, including high stable bulk density, high yield, resistance, nutrition, and larger and heavier grain size (Wang et al., 2009; Huang et al., 2009; Shahid et al., 2011; Koide et al., 2020; Chen et al., 2021; Gan et al., 2021; Ghouri et al., 2023). The relevance of autotetraploid rice for breeding is suggested by these studies. However, low fertility and low seed set have been frequently noted in autotetraploid rice throughout its known history compared to diploid rice. This problem has been a barrier preventing its utilization, leading to the slow progress of autotetraploid rice breeding research (Cai et al., 2007; Shahid et al., 2010; Chen et al., 2018; Li et al., 2018; Ghouri et al., 2019; Yang et al., 2019; Wu et al., 2019; Koide et al., 2020; Chen et al., 2021). After years of work, neo-tetraploid rice and tetraploid PMeS lines have recently been generated with high and stable seed setting characteristics (Chen et al., 2022). These rice materials are useful resources for the study of fertility-related molecular mechanisms and also the breeding application of tetraploid rice (Cai et al., 2007; He et al., 2011; Guo et al., 2017; Bei et al., 2019; Chen et al., 2019; Xiong et al., 2019; Ghaleb et al., 2020; Kamara et al., 2021; Liu et al., 2022).
The innermost anther cell layer, or tapetum, offers a secure environment, necessary nutrients, and components for developing microspores (Ariizumi and Toriyama, 2011). From male-sterile rice lines, several genes crucial for the growth of tapetum cells have been cloned. EAT1/DTD (Ji et al., 2013), TIP2/bHLH142 (Fu et al., 2014; Ko et al., 2014), UDT1 (Jung et al., 2005), TDR (Li et al., 2006), which encode bHLH transcription factor. R2R3-MYB family protein is encoded by OsTDF1 (Cai et al., 2015). A PHD-finger protein that plays a role in the late tapetum development stage is encoded by the gene PTC1 (Li et al., 2011), NY1, SUBSrP1 and DPS1, were needed for panicle and anther development of rice (Zafar et al., 2020; Ali et al., 2022; Kamara et al., 2022);. Early rice tapetum development is significantly influenced by these genes, and mutations in these genes showed abnormal tapetal cells and male sterility. The development of pollen in autotetraploid rice is thought to be significantly influenced by the transcription factor LOC_Os04g47890, a member of the myeloblastosis (MYB) family (Li et al., 2018). Recently, LOC_Os04g47890, a MORE FLORET 1 MYB transcription factor, was reported to control spikelet development and was named MOF1 (Ren et al., 2020). Understanding the regulatory mechanisms involving the tapetum and the expression levels of genes associated with the pollen wall is crucial for understanding pollen formation because the aforementioned genes are expressed there. All these genes were edited and investigated in diploid rice, but little is known in tetraploid rice.
RNA-Seq has the potential to identify differentially expressed genes with a greater expression level dynamic range (Wang et al., 2009). Many transcriptome investigations of plants, e.g., rice, maize, wheat, and Arabidopsis, etc., under various biotic and abiotic stresses, have been studied with efficacy (Ma et al., 2016; Vogel et al., 2016; Wang et al., 2016; Yadav et al., 2016). A reliable network of genes associated with pollen development has been utilized to learn more about how genes are regulated during the development of pollen activities of Arabidopsis, diploid, and polyploid rice, and insights from transcriptome analysis can be useful for identifying gene regulation during the development of rice pollen. Numerous studies have examined how autotetraploid rice develops its pollen (Wu et al., 2014; Wu et al., 2015; Li et al., 2016; Li et al., 2017). These RNA sequence data show the possibility of the genes controlling pollen development in tetraploid rice. However, the pollen fertility gene regulation network in tetraploid rice mutants remained largely unknown.
Our past investiagtion demostrated that the knock-out of NY2 (LOC_Os07g32040), which encodes a eukaryotic translation initiation factor 3 subunit E, resulted in defective pollen, increased sterility, and straggled chromosomes in meiosis (Kamara et al., 2021). Here, we used anther transverse section analysis to observe cytological defects in anther pollen development between ny2 and H1(wild type). The mutant ny2 underwent abnormal tapetal cells, abnormal microspores, and a defective pollen wall formation. Moreover, a global gene expression study revealed that ny2 alters the expression levels of pollen fertility-related genes, and qRT-PCR was employed to validate the expression patterns of these genes. Furthermore, between ny2 and H1, reciprocal crosses were created, and normal fertility and an obvious advantage for seed setting were detected in F1. This study provided a new understanding of the genetic basis for pollen fertility and development in neo-tetraploid rice.
Huaduo1 (H1), a neo-tetraploid line having high fertility that our lab developed, was used as a wild type. The ny2 mutant was developed by CRISPR/Cas9 knock-out technology. F1 hybrids were created by reciprocal crosses between H1 and ny2. Rice plants were grown at the farm of South China Agricultural University, Guangdong.
The pollen fertility was determined based on our earlier research (Kamara et al., 2021). Under a microscope, the mature pollens were stained with 1% I2-KI (Motic BA 200, Motic, Xiamen, China).
The anther transverse section analysis was carried out according to (Kamara et al., 2022). The anthers of ny2 mutants and wild-type (H1) plants were analyzed at various stages of anther development. First, the anthers were fixed in a solution of FAA (89% ethanol, 6% acetic acid, and 5% methyl aldehyde) for 48 hours at room temperature. After that, they were rinsed with 70% ethanol three times and dissected for semi-thin sectioning. A series of ethanol solutions were then used to dehydrate the anthers (80%, 90%, and 95%) for 30 minutes each. Finally, the tissues were processed according to the manufacturer’s protocol (Heraeus Kulzer) for Technovit 7100 Histological Analysis (Microtomy for Soft Tissues) at room temperature. Cross-sections of the anthers with a thickness of 3 μm were cut using a rotary microtome (Leica RM2235), stained with 1% toluidine blue, and sealed with neutral balsam. After that, the samples were examined and captured on camera with a microscope (Motic BA 200, China). The meiosis stage plays an important role in deciding the male fertility of rice. In this study, the frequency of tapetum abnormalities was observed during meiosis in ny2 mutant with an anther floret length range from 4.9 - 5.2 mm (Supplementary Table 1).
In order to analyze the seed set of F1 hybrids generated by crossing H1 with ny2, 10 plants of each population were investigated at maturity. The selfing of F1 populations of all hybrids developed the F2 generations. The criteria for evaluating these characteristics were based on Guo et al. (2017). Data analysis was performed according to Kamara et al. (2022).
The CRISPR/Cas9 editing system created a knock-out mutant of ny2 in the neo-tetraploid line Huaduo1 (H1). The mutant lines were grown in the field, and the T1 and T2 mutant plants were sequenced. We further selected the homozygous mutant of ny2 to observe the pollen development process. The anthers of H1 and T3 transgenic ny2 lines (homozygous mutant) were gathered at the meiotic stage and kept at -80°C in three biological replicates for RNA isolation. Total RNA of each sample was extracted according to Chen et al. (2018). The variations in gene expression between the samples were recorded according to Kamara et al. (2022).
A group of 13 DEGs (Differentially Expressed Genes) was chosen randomly for the validation of RNA-Seq data through qRT-PCR. Primer Premier 5.0 software was utilized to design gene-specific primers (Supplementary Table 2). The Roche Transcriptor First Strand cDNA Synthesis Kit was used to perform the reverse transcription. The qRT-PCR experiment was executed according to the procedure defined by Kamara et al. (2022). The gene expression patterns were estimated by 2-ΔΔCt method (Livak and Schmittgen, 2001). Every qRT-PCR test was run in triplicate.
The genomic sequence length of NY2 is 2327 base pairs (bp), and its CDS sequence is 1311 bp, and it encodes a eukaryotic translation initiation factor 3 subunit E, with 426 amino acids (aa) (Figure 1). A resequencing data set of 121 rice accession was used to analyze the diploid, autotetraploid, and neo-tetraploid rice’s DNA variants in the NY2 sequence. Seventeen mutations were identified in the aforementioned materials, and 13 mutant sites in CDS (Supplementary Table 3; Supplementary Figure 1). Numerous frameshift mutations were found in NY2, including mutants 5 and 6 (Supplementary Figure 2). The majority of neo-tetraploid lines clustered with their parent, T45, according to cluster analysis, suggesting that T45 might be associated with the high fertility rice gene (Supplementary Figure 3).
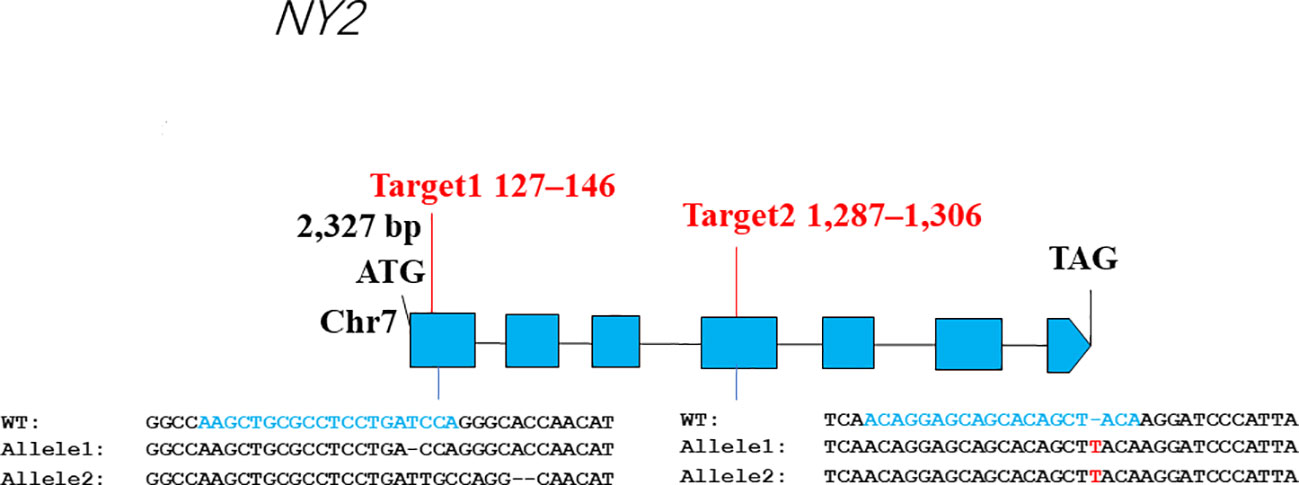
Figure 1 The gene structure and the CRISPR/Cas9 target sites of LOC_Os07g32040 (NY2) gene. The other side of the figure shows the alignment of ny2 sequences and wild-type (WT) having the CRISPR/Cas9 target sites.
To determine whether the ny2 mutation caused the transgenic lines’ male-sterile phenotype, reciprocal crosses were made between ny2 and H1. The F1 generation displayed normal plant types, pollen fertility, and obvious advantage for seed setting rate, just as the H1 (Figures 2A–H; Table 1), which is in contrast with the ny2 mutant (Figure 2D; Table 1). In F2 plants, the phenotypes were studied, and the ny2 mutant showed low seed set and pollen fertility (results not shown).
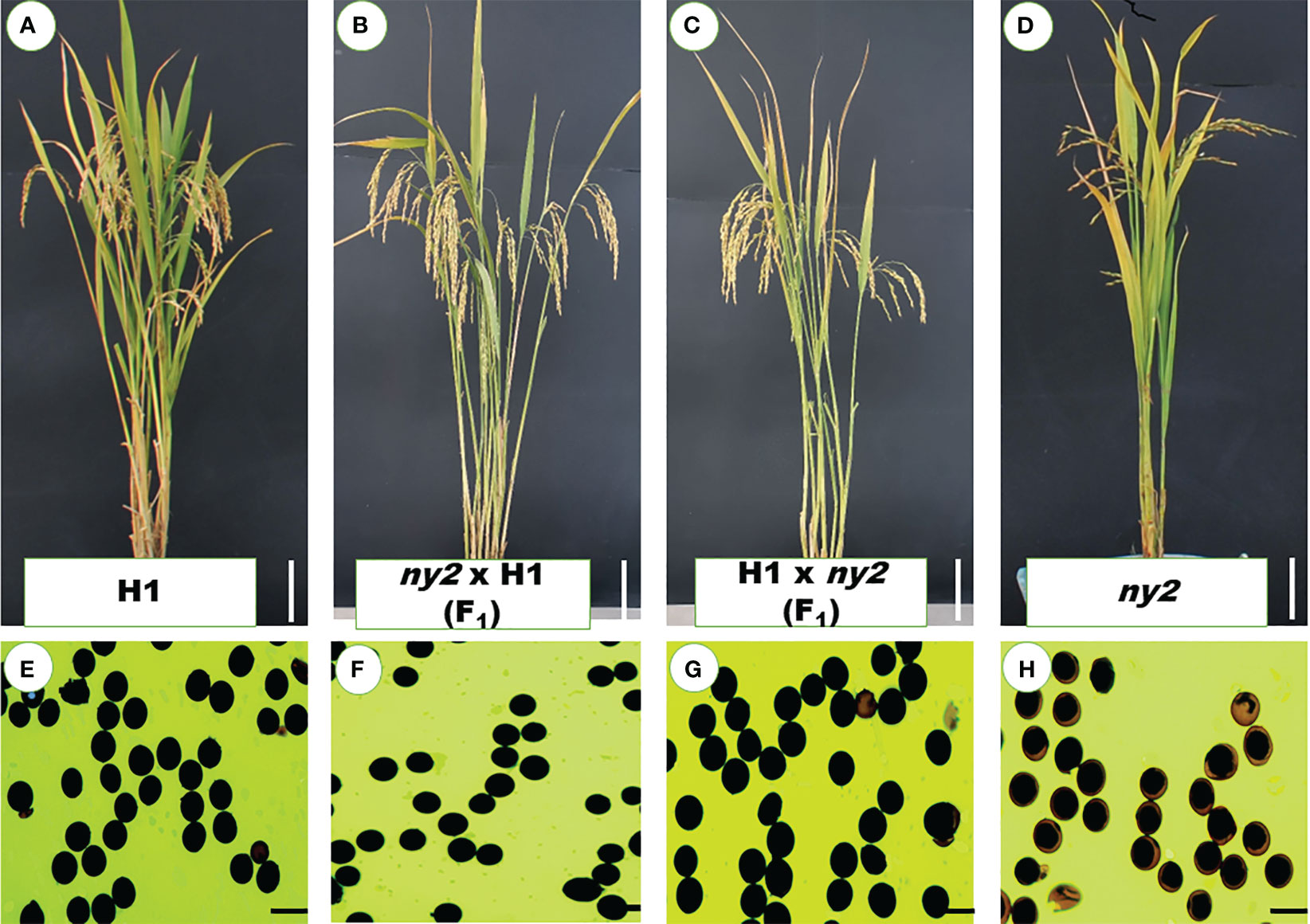
Figure 2 Hybrid lines’ and their parents’ phenotypic characteristics. (A–D) Plant morphology of H1, ny2 x H1, H1 x ny2 and ny2 at the mature stage. Bars = 10 cm. (E–H) Pollen fertility of H1, ny2 x H1, H1 x ny2 and ny2. Bars = 50 μm.
A cross-section of the ny2 and WT anthers at diverse pollen development phases was analyzed according to Kamara et al. (2022) to unveil the cytological abnormalities in anther pollen development. Stage 7 transverse sections of ny2 and WT anthers showed no discernible differences, and both showed a typical structure of all layers (Figures 3A, E). At stage S8a, the pollen mother cells were abnormal in the ny2 mutant compared to WT. The middle layer cells became narrower and more degenerated, and the tapetum cells of the WT exhibited high degeneration and became less vacuolated when compared with ny2 mutant anthers at stage S8a (Figures 3B, C, F, G). At stage S8b, middle layer cells and tapetum degenerated, and the pollen mother cells form tetrads. In contrast, the ny2 mutant anthers formed abnormal tetrads, the middle layer cells became more visible, and the tapetum became highly vacuolated (Figures 3D, H). During stage 9, WT tapetum cells turned thin, and the microspores were normal, but the tapetum of ny2 remained vacuolated, and the microspore cells were abnormal and significantly degenerated (Figures 3I, M). At stage 10, the WT tapetum cells shrunken further, the middle layer disappeared, and the microspores were fully developed in the WT circular anther locules. Still, the tapetum cell of ny2 was vacuolated, the middle layer remained obvious, and microspore cells were abnormal and degenerated (Figures 3J, N). During stage 11, the WT tapetum cell became less vacuolated, and the middle layer disappeared. In contrast, the microspore cells of ny2 were seriously affected and showed severe degeneration, the tapetum cell was normal, and the middle layer stayed visible at stage 11 (Figures 3K, O). At stage 12, the WT anther produced round, starchy pollen grains that had been fully stained. The pollen grains in ny2 were distorted and encircled by leftover tapetum fragments, which eventually caused male sterility and pollen abortion (Figures 3L, P).
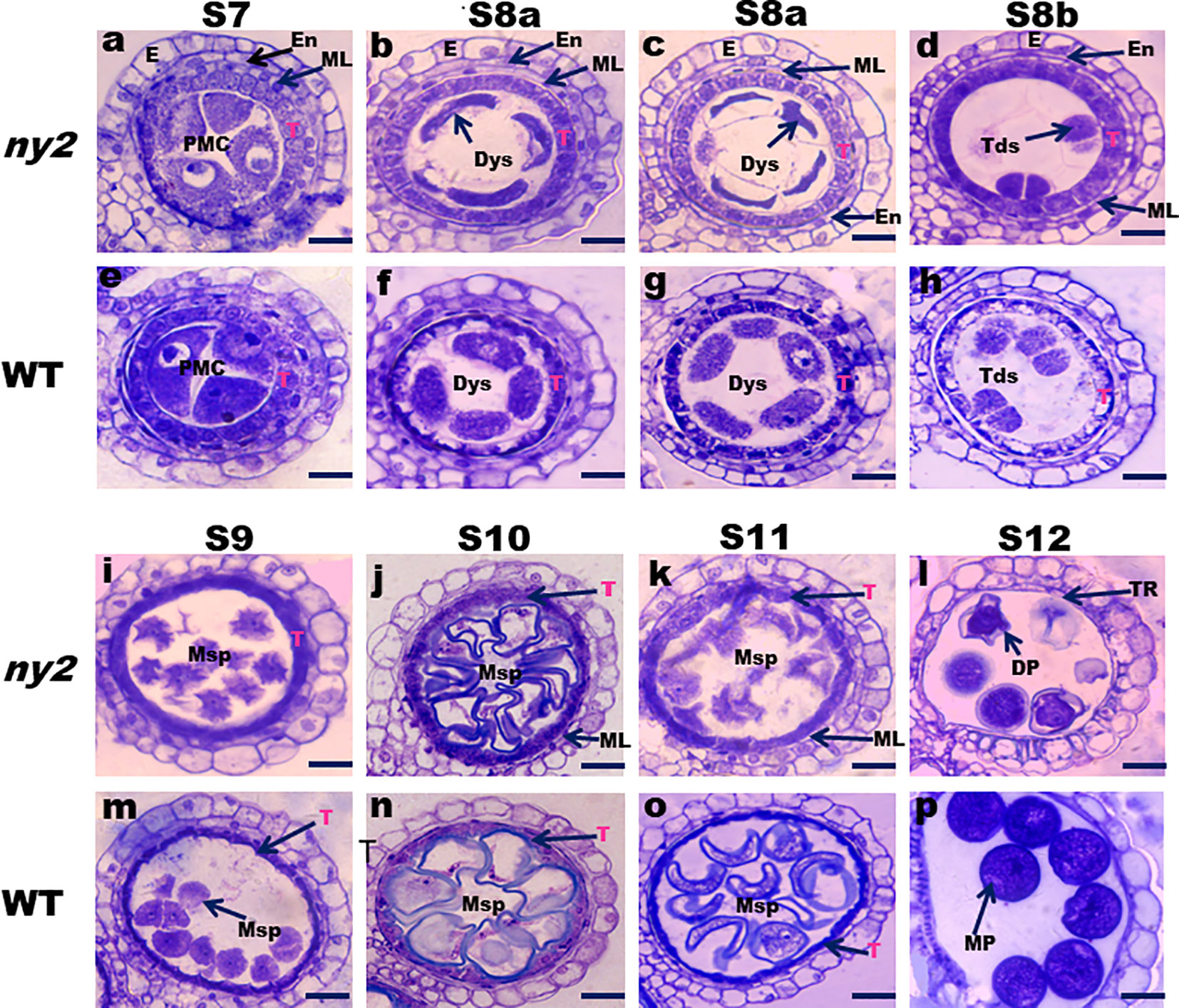
Figure 3 Transverse sections of anthers showing defective tapetal cell and microspore development in ny2 mutant. Stage 7 (A, E), Stage 8a (B, C, F, G), Stage 8b (D, H), Stage 9 (I, M), Stage 10 (J, N), Stage 11 (K, O), and Stage 12 (L, P). En, endothecium; E, epidermis; T, tapetum; PMC, pollen mother cells; ML, middle layer; Dys, dyad cells; DP, deformed pollen; Tds, tetrads; MP, mature pollen; TR, tapetal remnants Msp, microspore; Bars=20 μm (A–P).
The meiosis stage plays vital role in deciding the male fertility of rice. Hence, transcriptome analysis was used to investigate the patterns of gene expression during the meiosis stage in WT compared with ny2. Approximately 49,077,837 clean reads of high quality were found in each sample, which is 92.09% of the total 53,266,124 reads. When the clean reads were mapped onto the Nipponbare reference genome, 91.49 to 92.69% of the reference genome’s annotated transcripts were obtained in WT instead of ny2 (Table 2). The percentage of Q30 base was 93.47% or more, and the GC content ranged from 48.44% to 50.49% in all samples (Table 2). The correlation among the three replications of each sample varied from 0.77 to 0.98, suggesting the consistency of the three replicates (Supplementary Table 4).
A total of 7545 genes exhibited differential expression levels during meiosis in WT compared with ny2 (Figure 4B; Supplementary Table 5). Among these DEGs, 3925 and 3620 genes exhibited up- and down-regulation (Figure 4B; Supplementary Table 5). These results revealed that numerous genes were expressed during PMC meiosis in ny2. However, an abundance of key DEGs were highly expressed in H1 than ny2 mutant (Figure 4A). We also subjected the significant genes to the Kyoto encyclopedia of genes and genomes (KEGG) pathway and gene ontology (GO) enrichment analysis.
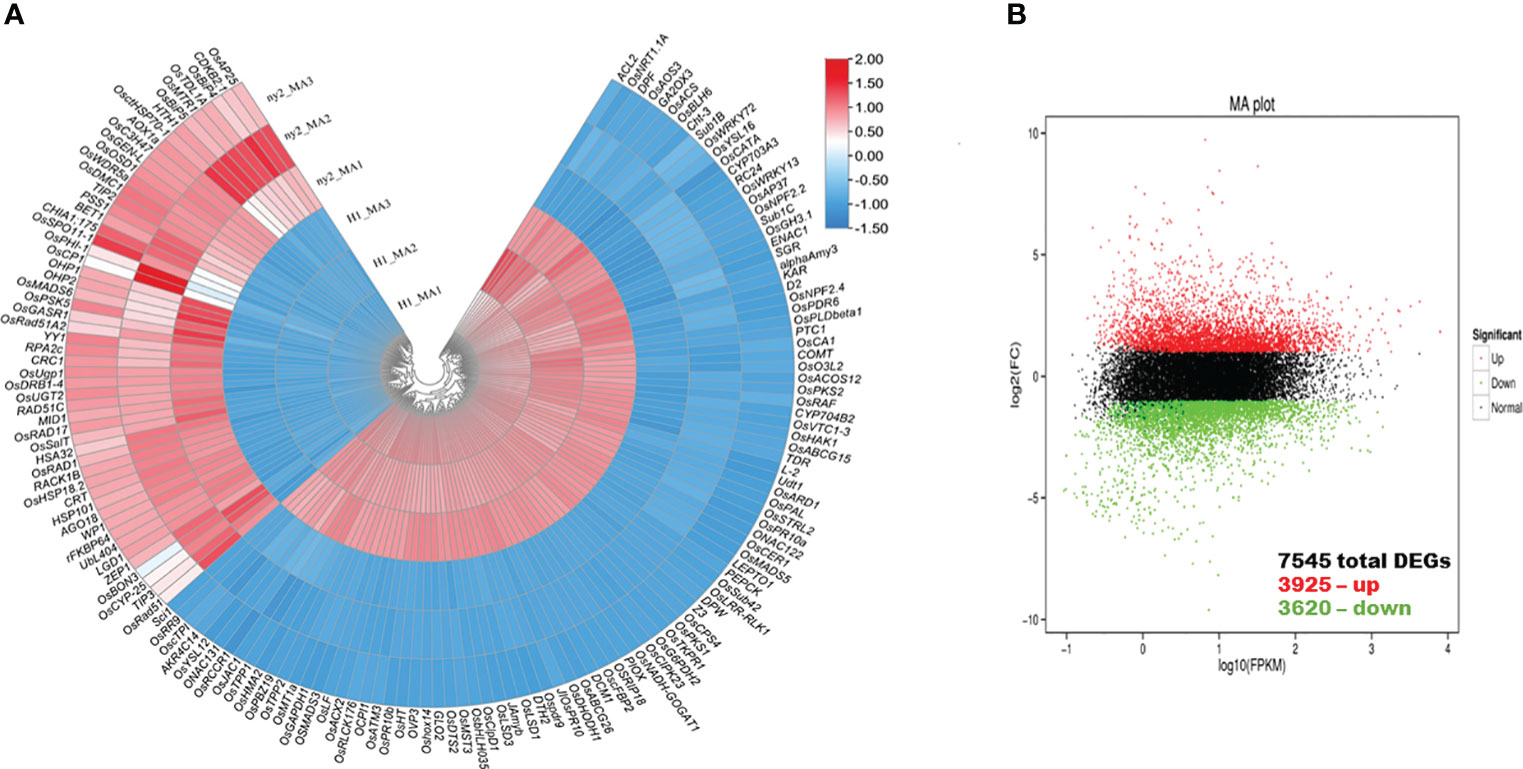
Figure 4 Sliding window and MA plot of the differential gene expression patterns in wild type (H1) compared with ny2. (A) The sliding window plot presenting the differentially expressed genes (DEGs) in wild type (H1) compared with ny2. (B) DEGs in wild type (H1) compared with ny2. The threshold used to assess the significance of gene expression differences was P<0.05.
Gene ontology (GO) enrichment analysis of the 7545 DEGs depicted 210 and 120 GO terms that were significantly down- and up-regulated in wild type (H1) compared with ny2, respectively. Cellular and metabolic processes had the highest percentage of representation in the biological process category. Binding and catalytic processes dominated the category of molecular function, while cells and cell parts dominated the category of cellular component (Supplementary Figure 4). We further characterized the GO terms related to down-regulated DEGs in ny2 in the biological process (Supplementary Table 6A). Of all these enrichment terms, some associated with anther development were known as significant ones. Of the down-regulated DEGs, disaccharide metabolic process, cellular carbohydrate metabolic process, and glycoside metabolic process were detected among the important enrichment terms, and the gene OsSSIIIa (LOC_Os08g09230) belonging to cellular carbohydrate metabolic process term participates in the glucan biosynthetic process and starch synthase activity. The gene RSUS2 (LOC_Os06g09450), belonging to the glycoside metabolic process term participated in the biosynthetic process, sucrose synthase and sucrose metabolic process, and the gene RSUS3 (LOC_Os07g42490), belonging to the disaccharide metabolic process term participated in the biosynthetic process, sucrose synthase and sucrose metabolic process (Supplementary Table 6A). It’s interesting to note that the GO term for the metabolism of carbohydrates was significantly enriched in both down- and up-regulated genes, and four DEGs Osg1 (LOC_Os01g71930), OsHXK7 (LOC_Os05g09500), OsHXK9 (LOC_Os01g52450) were down-regulated, and OsSUS7 (LOC_Os04g17650) was up regulated were identified to play a significant role in anther pollen development (Table 3; Supplementary Tables 6A, B). Except for the four important terms above, some other terms, including pollination, recognition of pollen, pollen-pistil interaction, reproduction, reproductive process, oligosaccharide metabolic process, carbohydrate biosynthetic process, oligosaccharide biosynthetic process, cellular lipid catabolic process, were represented in the down-regulated gene class transcripts (Supplementary Table 6A).
We mapped all DEGs to their various KEGG enrichment analyses to get a deeper perception of the specific pathways that control the biological processes mentioned above. The most significant biological pathways include protein processing in the endoplasmic reticulum, ribosome, and oxidative phosphorylation (Supplementary Figure 5). We further analyzed the down-regulated DEGs in WT compared with ny2. Among the top 20 most enriched down-regulated pathways, starch and sucrose metabolism (ko00500) and peroxisome (ko04146) were the most significantly down-regulated pathways in ny2 (Figure 5). Except for the two important pathways above, some other relevant biological pathways related to anther development, such as pyruvate metabolism (ko00620), fatty acid metabolism (ko01212), sphingolipid metabolism (ko00600) and fatty acid degradation (ko00071), were also recognized in the down-regulated DEGs. For example, starch and sucrose metabolism pathway DEGs were related to the synthase of sucrose, sucrose transporter, sucrose invertase, hexokinase, and starch. Sucrose and invertase are broken down into monosaccharides that are conveyed to the cells and utilized for starch biosynthesis (Ruan et al., 2010; Ruan, 2012). Two hexokinase genes (OsHXK7 and OsHXK9) and two sucrose synthase genes (RSUS2 and RSUS3) showed down-regulation in ny2. Saccharides are transported from source to sink by sucrose and monosaccharide transporters (Smeekens, 2000; Wang et al., 2008). OsSWEET4, a sucrose transporter gene and OsMST3, a monosaccharide transporter gene, exhibited down-regulation in ny2. In addition, a sugar degradation-related gene (Osg1) and a starch synthase gene (OsSSIIIa) were found to be down-regulated in ny2 (Table 3). LOC_Os03g07140, a DPW gene that belongs to fatty alcohol synthesis and peroxisome pathway, plays a vital role in pollen sporopollenin biosynthesis and anther cuticle in rice also showed down-regulation in ny2.

Figure 5 KEGG pathway analysis of the down-regulated DEGs in wild type (H1) compared with ny2. The abscissa represents the number of genes, the ordinate represents the enrichment pathways, the column length represents the number of genes, and color represents the p-value.
In previous studies, reduction of pollen fertility gene expression was frequently observed, which was thought to be the main cause of autotetraploid rice’s low pollen fertility (Wu et al., 2014; Li et al., 2018; Kamara et al., 2022). This study examined the genes at pollen mother cell (PMC) meiosis stage using comparative transcriptome profiling. We compared the pollen fertility-related genes with the DEGs detected in the present study (Fujita et al., 2010; Deveshwar et al., 2011; Yant et al., 2013; Wright et al., 2014; Wu et al., 2014) and found nine important down-regulated genes associated with tapetum or pollen fertility during meiosis in wild type (H1) compared with ny2, including LOC_Os10g35180, LOC_Os04g51070, LOC_Os09g27620, LOC_Os06g36080, LOC_Os11g47809, LOC_Os08g03682, LOC_Os02g02820, LOC_Os03g15710, and LOC_Os07g36460. Of these genes, OsABCG26 (LOC_Os10g35180) is an ABC transporter; its mutant displayed defects in tapetal cells and caused pollen sterility in rice (Chang et al., 2016). EAT1 (LOC_Os04g51070) is a programmed cell death regulatory gene, bHLH transcription factor, and delayed tapetum degradation during rice pollen development (Niu et al., 2013). A PHD-finger protein called TMS9-1 (LOC_Os09g27620), also referred to as PTC1 and OsMS1, is a thermosensitive male sterility gene that regulates pollen and tapetal cell development in rice (Qi et al., 2014; Yang et al., 2019; Wu et al., 2020). KIN14M (LOC_Os06g36080) is a kinesin motor domain-containing gene, and reduced pollen fertility and decreased seed setting in neo-tetraploid rice are the effects of this gene’s loss of function (Yu et al., 2020). OsMT1a (LOC_Os11g47809) is a metallothionein gene, and the knock-out of this gene causes pollen sterility (Lu et al., 2020). CYP703A3 (LOC_Os08g03682) cytochrome P450 hydroxylase gene that regulates tapetum and male fertility in rice (Yang et al., 2014). A tapetum development regulatory gene known as TDR (LOC_Os02g02820), its mutant plants showed tapetum and middle layer degeneration retardation and microspore collapse (Li et al., 2006). OsSTRL2 (LOC_Os03g15710), an atypical strictosidine synthase, which is highly expressed in microspores and tapetal cells, and its mutants cause male abortion (Zou et al., 2017) and UDT1 (LOC_Os07g36460) is a bHLH transcription factor, and a tapetum development regulatory, knock-out mutant showed defects in tapetal cells and caused male sterility in rice (Jung et al., 2005) (Table 4).
To confirm the gene expression levels of important genes in wild type (H1) and ny2, a subset of 13 tapetum or pollen fertility and transcription factor-associated genes were selected for qRT-PCR analysis. We contrasted the qRT-PCR results with the ones from the RNA-Seq analysis of these transcripts. The RNA-Seq analysis and expression trends were in agreement, with a correlation coefficient of R2 = 0.7949 (Figure 6; Supplementary Figure 6).
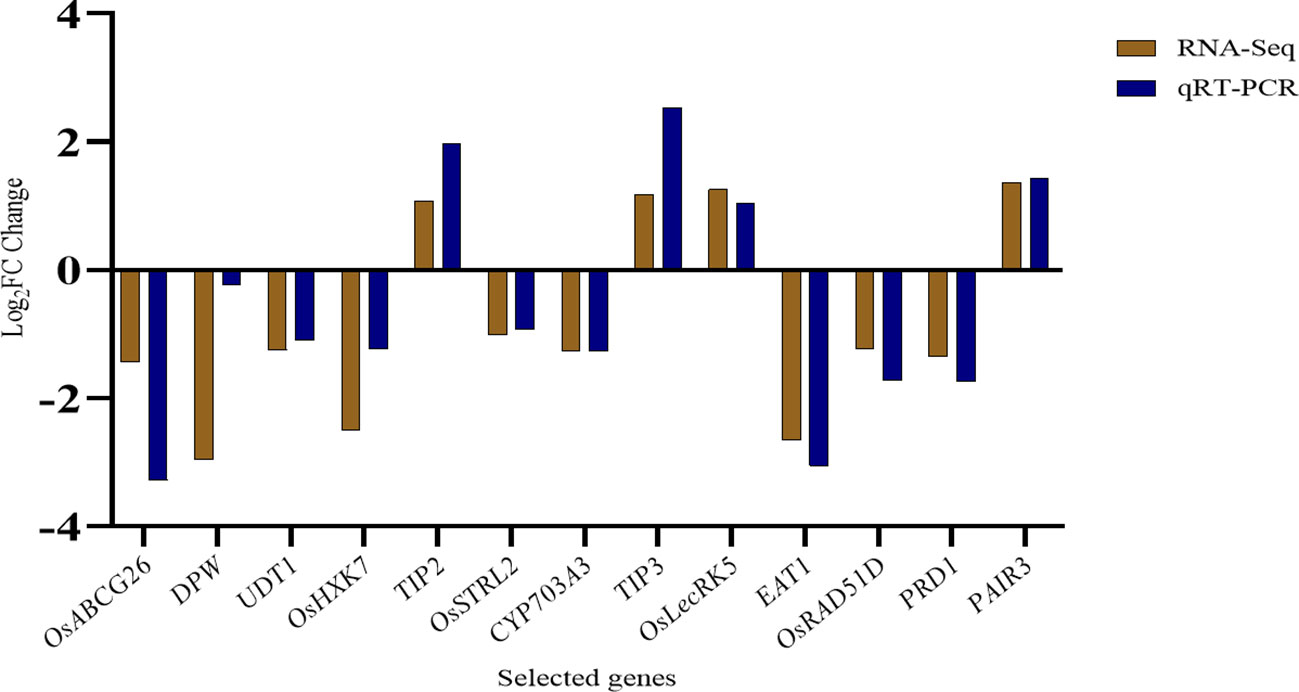
Figure 6 Comparison of the log2 (FC) of thirteen selected genes using qRT-PCR analysis in wild type (H1) compared with ny2.
Transcription factors are evident in controlling varied physiological activities in response to fertility. Transcription (GO: 0006350), regulation of transcription (GO: 0045449) and transcription regulator activity (GO: 0030528) were the significant GO terms enriched only in WT compared with ny2. In all, 212 DEGs were identified as transcription factors belonging to 34 (TFs) groups in wild type (H1) compared with ny2. Of the transcription factor related genes, 189 displayed down-regulation (Figure 7; Supplementary Table 7). The largest groups reported were transcription factor DEGs of NAC, bHLH, bZIP, MYB and ERF. Of these groups, bHLH, MYB family, MIKC_MADS and PHD-finger transcription factor were shown to participate in the pollen development process in previous studies (Zhang et al., 2010; Li et al., 2011; Sui et al., 2015; Zhang et al., 2015; Zhu et al., 2015; Ranjan et al., 2017; Xiang et al., 2020). Here, we identified 13 transcription factors related to genes that regulate pollen fertility in rice. Among these DEGs, EAT1, UDT1, TDR, TP2, OsbHLH138, and OsPIL16 encode the bHLH family transcription factor, PTC1 and TIP3 encode a PHD-finger transcription factor OSMADS3, OsMADS5 and OsMADS13 encode MIKC_MADS family protein, OsRR24 is an ARR-B transcription factor, and LOC_Os08g06370 is a MYB family transcription factor. Among these genes, EAT1, UDT1, TDR, OsbHLH138, OsPIL16, OsMADS3, OsMADS5, OsRR24 and PTC1 were down-regulated and LOC_Os08g06370, TIP, OsMADS13, and TIP3 were found to be up-regulated (Figure 7).
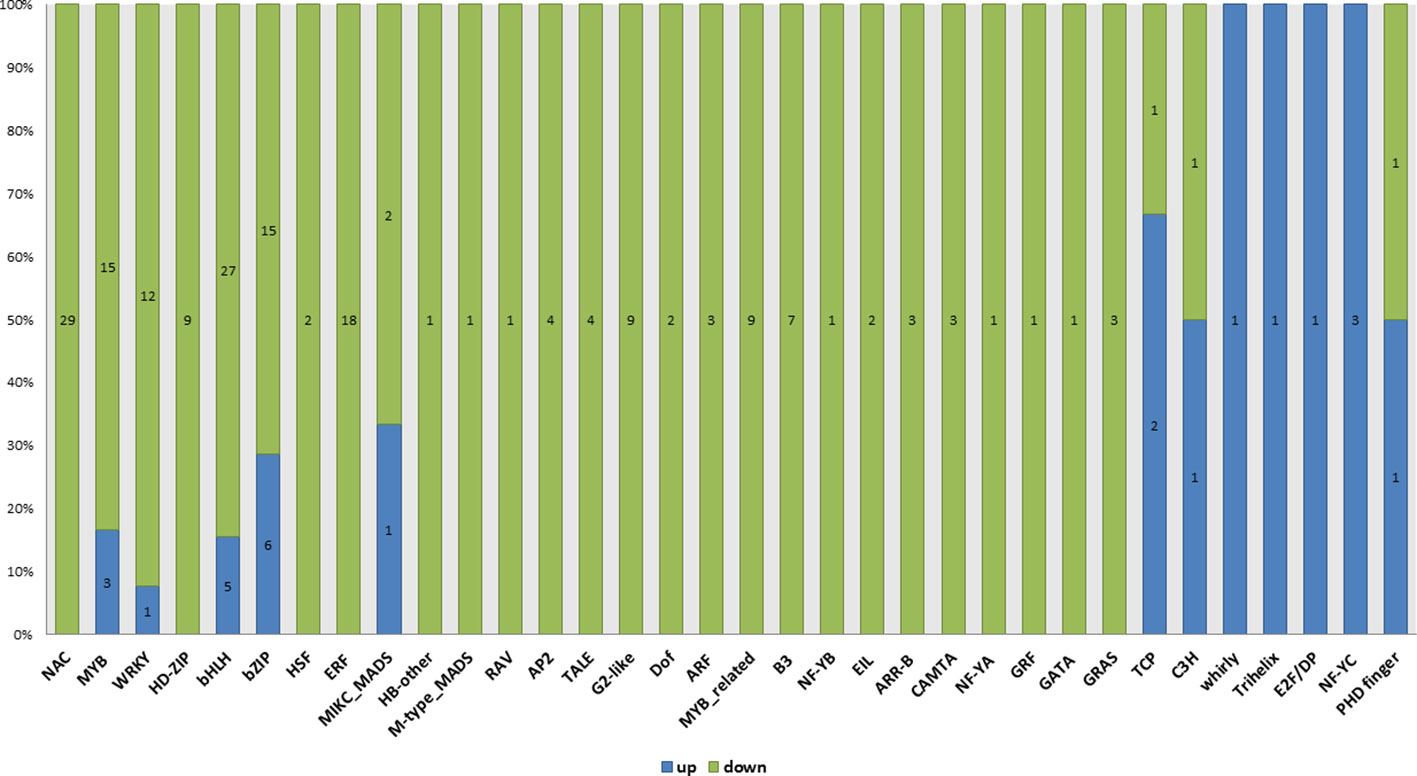
Figure 7 Differentially expressed genes associated with different transcription factor families discovered in WT compared with ny2. We discovered 212 putative TF DEGs, which were categorized into 34 groups.
The precise control of tapetal cell death is vital for anthers and pollen development in rice. Timely initiation of tapetal programmed cell death (PCD) is crucial for releasing pollen wall materials, such as carbohydrates, lipid molecules, and other related nutrients, from the tapetum cells and transporting them to the anther and microspore surface for deposition (Li et al., 2011). Many DEGs, including EAT1, UDT1, TDR, OsABCG26, PTC1, OsSTRL2, SUBSrP1, DPS1, and NY1 (Jung et al., 2005; Li et al., 2011; Niu et al., 2013; Chang et al., 2016; Zou et al., 2017; Zafar et al., 2020; Ali et al., 2022; Kamara et al., 2022), play critical controls over the development of the tapetum PCD and microspores. Silenced or loss of function of these genes delays tapetal cell degradation and promotes abnormal microspore development and pollen abortion. In autotetraploid rice, tapetal cells and microspore abnormalities were observed to be the principal causes of pollen abnormality (Li et al., 2018; Lu et al., 2020; Wu et al., 2020; Chen et al., 2022; Kamara et al., 2022). Similarly, in this study, the cytological analysis showed that the ny2 mutant displayed defects in tapetal cells, microspores and pollen wall development, which caused pollen sterility and male abortion.
The RNA-seq analysis revealed that 7545 genes displayed differential expression patterns during meiosis. Of these DEGs, 3925 and 3620 were up- and down-DEGs. Among the down-regulated genes, the expression levels of several key DEGs involved in tapetum or pollen fertility were significantly altered in the ny2 anthers. For example, OsABCG26 (LOC_Os10g35180) is an ABC transporter; its mutant plant displayed defects in tapetal cells and caused male sterility in rice (Chang et al., 2016). EAT1 (LOC_Os04g51070) is a programmed cell death regulatory gene, bHLH transcription factor; its mutant delayed tapetum degradation in rice (Niu et al., 2013). The rice gene TMS9-1 (LOC_Os09g27620), also referred to as PTC1 and OsMS1, is a PHD-finger protein that regulates tapetal cell and pollen fertility (Li et al., 2011; Qi et al., 2014; Yang et al., 2019; Wu et al., 2020). KIN14M (LOC_Os06g36080) is a kinesin motor gene, and the knock-out of KIN14M showed a decrease in pollen fertility and seed setting in neo-tetraploid rice (Yu et al., 2020). TDR (LOC_Os02g02820) is a tapetum development regulatory gene; its mutant plants exhibited abnormal tapetum, middle layer and collapse of microspores (Li et al., 2006). OsSTRL2 (LOC_Os03g15710) is an atypical strictosidine synthase highly expressed in microspores and tapetal cells. Its mutant showed abnormal fertility (Zou et al., 2017), and UDT1 (LOC_Os07g36460) is a tapetum development regulatory gene; its mutant showed defects in tapetal cells and ultimately resulted in pollen sterility in rice (Jung et al., 2005). Similar to these findings, a number of past studies showed that the down-regulated genes are the main cause of pollen abortion in tetraploid rice (Wu et al., 2014; Wu et al., 2017; Chen et al., 2019; Kamara et al., 2022). Our findings suggested that like the nine published tapetum or pollen fertility genes mention-above, NY2 is required for normal Pollen formation by regulating tapetal cells, microspore and pollen wall development in neo-tetraploid rice.
Energy from carbohydrates is needed to maintain the growth of the anthers and pollen, but they also serve as a growth indicator (Clement et al., 1996). The regulation of carbohydrates is actively regulated by many related proteins or genes. Numerous studies have shown that altered gene or protein expression in anthers impairs pollen development and adversely affects pollen fertility. Plant anthers possess the greatest sink capacity in flower, to support early development, enormous amounts of sugars are arranged on the anthers (Clement et al., 1996; Castro and Clément, 2007). Sucrose transporters (SUTs) are vital for sucrose uptake in diverse stages of sugar translocation and uphold a link-up relationship between pollen development and sucrose transport (Srivastava et al., 2008; Hirose et al., 2010). In plants, sugars are manufactured in the mesophyll cells of developed leaves that are best known as source organs. Hence, precise sugar production, storage, and transportation are essential for sustaining growth and development in plants. Due to decreased sucrose transport in the phloem, mutations in the AtSUC2 gene in the plant Arabidopsis thaliana cause growth and fertility reductions (Gottwald et al., 2000; Gould et al., 2012). OsSWEET4, OsSWEET11, or OsSWEET14 mutants that were silenced or lost their ability to function exhibited low fertility, delayed reproductive traits, smaller seeds, and shriveled caryopses (Chu et al., 2006; Antony et al., 2010; Li et al., 2012; Ma et al., 2017). The suppression of Osg1 causes male sterility (Wan et al., 2010). Therefore, all these results showed that sugar transporter or degradation and sucrose play an essential role in pollen formation. Here, the same sugar transporter gene (OsSWEET4) and (Osg1) were identified to be down-regulated in ny2. Sucrose synthase and sucrose invertase are important enzymes in sugar metabolism in plants (Ruan, 2012). Sucrose synthase genes (RSUS3) and (RSUS2) were significantly down-regulated, while the sucrose invertase gene (OSINV4) showed upregulation in ny2. Hexose, an essential energy source, plays an essential role in pollen development and forms an important component for cell wall synthesis. A deficiency in hexokinase HXK5 disturbed the usage of starch in pollen grains, which resulted in pollen sterility (Lee et al., 2020). Here, hexokinase genes (OsHXK7 and OsHXK9) showed down-regulation in ny2. In addition, Starch, a polymer of glucose residues, serves as the main energy source and carbon skeleton in developed pollen grains, enabling pollen germination and the development of pollen tubes for proper fertilization. Consequently, it is believed that deficiency of starch synthesis in pollen grains is the root of male sterility (Datta et al., 2002; Wu et al., 2016). As a result, modifying the starch contents of pollen grains has proven to be a new method of causing male abortion (Wu et al., 2016). In our study, a starch synthase gene (OsSSIIIa) was found to be down-regulated in ny2.
In pollen development, monosaccharide transporters play a crucial role (Wang et al., 2008). The development of rice pollen is evidently affected by the expressions of OsMST5 and OsMST8 (Ngampanya et al., 2003; Mamun et al., 2006). For example, cas mutant displayed remarkably lower expression levels of OsMST8, which reduced levels of carbohydrates and sugar in flower organs and led to pollen abortion (Zhang et al., 2010). Here, OsMST3, a monosaccharide transporter, was discovered to have decreased expression in ny2. Therefore, we speculated that the down-regulation of sucrose synthase, hexose, starch, and monosaccharide genes might be accountable for the defective pollen development in the ny2 mutant.
In higher plants, male gametophytes play an important role in pollen fertility. The complex and precise control of a large number of transcription factors is required for the tight regulation of pollen grain development (Guan et al., 2014; Lei et al., 2017).
A complex regulatory network of various transcription factors (TFs) working in a tissue-specific manner is essential to the development of anthers. Given that one of the key applications of plant biotechnology is the genetic changes in the male reproductive system, understanding the TFs related to the regulatory network of anther development is crucial. During the growth and development of plants, changes in the expression levels of some transcription factors may result in pollen defects and male sterility. For instance, overexpressing WRKY27 in Arabidopsis plants resulted in defects with pollen cracking, growth issues, and male sterility (Mukhtar et al., 2018). According to reports, OsTDF1 and OsMYB103 restored fertility in the tdf1 and ms188 mutants of Arabidopsis, respectively, and their gain- or loss-of-function induced male abortion in rice (Phan et al., 2012; Cai et al., 2015). OsMYB103 (OsMYB80) controls the development of anther by regulating the middle layer development and tapetum in rice (Zhang et al., 2010). The down-regulation of OsMYB106 causes pollen sterility in rice (Chen et al., 2014). Past studies also reported that knock-down and knock-out mutants of bHLH142 resulted in male sterile plants in rice (Fu et al., 2014; Ko et al., 2014). Additionally, the unique rice BHLH transcription factor DTD works in tandem with TDR to control the development of the tapetum and pollen in rice (Ji et al., 2013). Here, we identified 9 down-regulated transcription factor DEGs associated with rice pollen fertility. Of these genes, OsbHLH138 (LOC_Os03g27390) is a basic helix-loop-helix transcription factor, which controls thermos-sensitive male infertility in rice via activation of TMS5 (Wen et al., 2019; Chen et al., 2022). OsMADS3 (LOC_Os01g10504) is a MADS box gene, and the loss of function of its mutant causes male abortion in rice (Zhang et al., 2015). OsRR24 (LOC_Os02g08500) is a Type B response regulator that encodes an ARR-B transcription factor; its mutant showed abnormal meiosis and tapetal cells in rice (Zhao et al., 2018). Mutant plants of these three genes showed abnormal tapetum development and pollen defects. In the ny2 mutant, male sterility and a similar defect in the tapetum were also seen.
In this study, the cytological analysis indicated that ny2 underwent abnormal tapetal cells, microspores and middle layer development. Genetic analysis revealed that the F1 plants showed normal fertility and an obvious advantage for the seed set. Also, a large number of tapetum or pollen fertility, sucrose and starch metabolism and transcription factors related genes showed down-regulation. The regulatory gene network reported in this study may provide valuable information about the molecular mechanism related to fertility regulation in tetraploid rice
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
MQS and JW. designed and conceived the experiments. NK, YJ, WH, L C, LZ, CZ, XH and FNS. performed the experiment and analyzed the data. NK, YJ, MQS and XL wrote the paper. XL. generated the neo-tetraploid and autotetraploid rice lines. All authors contributed to the article and approved the submitted version.
This work was supported by the Base Bank of Lingnan Rice Germplasm Resources Project (2023), the National Natural Science Foundation of China (NSFC) (32050410294) and the Guangdong Basic and Applied Basic Research Foundation (2021A1515010748).
The authors thank Dr. Hang Yu, Lan Wang, Zhixiong Chen, and Ms. Shuhong Yu for assistance in experiment.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1229870/full#supplementary-material
Ali, A., Wu, T., Zhang, H., Xu, P., Zafar, S. A., Liao, Y., et al. (2022). A putative SUBTILISIN-LIKE SERINE PROTEASE 1 (SUBSrP1) regulates anther cuticle biosynthesis and panicle development in rice. J. Advanced Res. 42, 273–287. doi: 10.1016/j.jare.2022.01.003
Antony, G., Zhou, J., Huang, S., Li, T., Liu, B., White, B., et al. (2010). Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell. 22, 3864–3876. doi: 10.1105/ptc.110.078964
Ariizumi, T., Toriyama, K. (2011). Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 62, 437–460. doi: 10.1146/annurev-arplant-042809-112312
Bei, X. J., Shahid, M. Q., Wu, J. W., Chen, Z. X., Wang, L., Liu, X. D. (2019). Re-sequencing and transcriptome analysis reveal rich DNA variations and differential expressions of fertility-related genes in neo-tetraploid rice. PloS One 14, e0214953. doi: 10.1371/journal.pone.0214953
Cai, D. T., Chen, J. G., Chen, D. L., Dai, B. C., Zhang, W., Song, Z. J., et al. (2007). The breeding of two polyploid rice lines with the characteristic of polyploid meiosis stability. Sci. China C Life Sci. 50, 356–366. doi: 10.1007/s11427-007-0049-6
Cai, C., Zhu, J., Lou, Y., Guo, Z., Xiong, S., Wang, K., et al. (2015). The functional analysis of OsTDF1 reveals a conserved genetic pathway for tapetal development between rice and Arabidopsis. Sci. Bull. 60 (12), 1073–1082. doi: 10.1007/s11434-015-0810-3
Castro, A. J., Clément, C. (2007). Sucrose and starch catabolism in the anther of lilium during its development: a comparative study among the anther wall, locular fluid and microspore/pollen fractions. Planta. 225, 1573–1582. doi: 10.1007/s00425-008-0443-5
Chang, Z., Chen, Z., Yan, W., Xie, G., Lu, J., Lu, N. W. Q., et al. (2016). An ABC transporter, OsABCG26, is required for anther cuticle and pollen exine formation and pollen-pistil interactions in rice. Plant Science. 253, 21–30. doi: 10.1016/j.plantsci.2016.09.006
Chen, R., Feng, Z., Zhang, X., Song, Z., Cai, D. (2021). A new way of rice breeding: polyploid rice breeding. Plants. 10, 422. doi: 10.3390/plants10030422
Chen, L., Shahid, M. Q., Wu, J. W., Chen, Z. X., Wang, L., Liu, X. D. (2018). Cytological and transcriptome analyses reveal abrupt gene expression for meiosis and saccharide metabolisms that associated with pollen abortion in autotetraploid rice. Mol. Genet. Genomics 293 (6), 1407–1142. doi: 10.1007/s00438-018-1471-0
Chen, Y., Shahid, M. Q., Wu, J., Deng, R., Chen, Z., Wang, L., et al. (2022). Thermo-sensitive genic Male sterile lines of neo-tetraploid rice developed through gene editing technology revealed high levels of hybrid vigor. Plants. 11, 1390. doi: 10.3390/plants11111390
Chen, L., Wang, C., Yu, G., Xu, X., Qin, Y. (2014). Preliminary study of function of a MYB gene in rice. J. Huazhong Agric. Univ. 33, 7–12.
Chen, L., Yuan, Y., Wu, J. W., Chen, Z. X., Wang, L., Shahid, M. Q., et al. (2019). Carbohydrate metabolism and fertility related genes high expression levels promote heterosis in autotetraploid rice harboring double neutral genes. Rice. 12, 34. doi: 10.1186/s12284-019-0294-x
Chu, Z., Yuan, M., Yao, J., Ge, X., Yuan, B., Xu, C., et al. (2006). Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 20, 1250–1255. doi: 10.1101/gad.1416306
Clement, C., Burrus, M., Audran, J. C. (1996). Floral organ growth and carbohydrate content during pollen development in lilium. Am. J. Bot. 83, 459–469. doi: 10.2307/2446215
Datta, R., Chamusco, K. C., Chourey, P. S. (2002). Starch biosynthesis during pollen maturation is associated with altered patterns of gene expression in maize. Plant Physiol. 130, 1645–1656. doi: 10.1104/pp.006908
Deveshwar, P., Bovill, W. D., Sharma, R., Able, J. A., Kapoor, S. (2011). Analysis of anther transcriptomes to identify genes contributing to meiosis and male gametophyte development in rice. BMC Plant Biol. 11, 78. doi: 10.1186/1471-2229-11-78
Fu, Z. Z., Yu, J., Cheng, X. W., Zong, X., Xu, J., Chen, M. J., et al. (2014). The rice basic helix-loop-helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell. 26 (4), 1512–1524. doi: 10.1105/tpc.114.123745
Fujita, M., Horiuchi, Y., Ueda, Y., Mizuta, Y., Kubo, T., Yano, K., et al. (2010). Rice expression atlas in reproductive development. Plant Cell Physiol. 51, 2060–2081. doi: 10.1093/pcp/pcq165
Gan, L., Huang, B. S., Song, Z. J., Zhang, Y. C., Zhang, Y. J., Chen, S., et al. (2021). Unique glutelin expression patterns and seed endosperm structure facilitate glutelin accumulation in polyploid rice seed. Rice. 14, 61. doi: 10.1186/s.12284-021-00500-0
Ghaleb, M. A. A., Li, C., Shahid, M. Q., Yu, H., Liang, J. H., Chen, R. X., et al. (2020). Heterosis analysis and underlying molecular regulatory mechanism in a wide-compatible neo-tetraploid rice line with long panicles. BMC Plant Biol. 20, 83. doi: 10.1186/s12870-020-2291-z
Ghouri, F., Shahid, M. J., Liu, J., Lai, M., Sun, L., Wu, J., et al. (2023). Polyploidy and zinc oxide nanoparticles alleviated cd toxicity in rice by modulating oxidative stress and expression levels of sucrose and metal-transporter genes. J. Hazard. Mat., 130991. doi: 10.1016/j.jhazmat.2023.130991
Ghouri, F., Zhu, J., Yu, H., Wu, J., Baloch, F. S., Liu, X., et al. (2019). Deciphering global DNA variations and embryo sac fertility in autotetraploid rice line. Turk J. Agric. For. 43, 554–568. doi: 10.3906/tar-1901-13
Gottwald, J. R., Krysan, P. J., Young, J. C., Evert, R. F., Sussman, M. R. (2000). Genetic evidence for the in-planta role of phloem-specific plasma membrane sucrose transporters. Proc. Natl. Acad. Sci. U.S.A. 97, 1397. doi: 10.1073/pnas.250473797
Gould, N., Thorpe, M. R., Pritchardc, J., Christeller, J. T., Williams, L. E., Roebb, G., et al. (2012). AtSUC2 has a role for sucrose retrieval along the phloem pathway: evidence from carbon-11 tracer studies. Plant Sci. 188–189, 97–101. doi: 10.1016/j.plantsci.2011.12.018
Guan, Y., Meng, X., Khanna, R., LaMontagne, E., Liu, Y., Zhang, S. (2014). Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in arabidopsis. PloS Genet. 10, e1004384. doi: 10.1371/journal.pgen.1004384
Guo, H. B., Mendrikahy, J. N., Xie, L., Deng, J. F., Lu, Z. J., Wu, J. W., et al. (2017). Transcriptome analysis of neo-tetraploid rice reveals specific differential gene expressions associated with fertility and heterosis. Sci. Rep. 7, 40139. doi: 10.1038/srep40139
He, Y., Ge, J., Wei, Q., Jiang, A., Cai, D. (2011). Using a polyploid meiosis stability (PMeS) line as a parent improves embryo development and the seed set rate of a tetraploid rice hybrid. Can. J. Plant Sci. 91, 325–335. doi: 10.4141/CJPS09190
Hirose, T., Zhang, Z., Miyao, A., Hirochika, H., Ohsugi, R., Terao, T. (2010). Disruption of a gene for rice sucrose transporter, OsSUT1, impairs pollen function but pollen maturation is unaffected. J. Exp. Bot. 61 (13), 3639–3646. doi: 10.1093/jxb/erq175
Huang, Y., Huang, Q., Zhao, S. (2009). Comparative study on difference of grain characters of rice with different chromosome sets. J. Henan Agric. Sci. 10, 57–61.
Ji, C. H., Li, H. Y., Chen, L. B., Xie, M., Wang, F. P., Chen, Y. L., et al. (2013). A novel rice bHLH transcription factor, DTD, acts coordinately with TDR in controlling tapetum function and pollen development. Mol. Plant 6 (5), 1715–1718. doi: 10.1093/mp/sst046
Jung, K. H., Han, M. J., Lee, Y. S., Kim, Y. W., Hwang, I., Kim, M. J., et al. (2005). Rice undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell. 17 (10), 2705–2722. doi: 10.1105/tpc.105.034090
Kamara, N., Jiao, Y., Lu, Z., Aloryi, K. D., Wu, J., Liu, X., et al. (2021). Cytological observations and bulked-segregant analysis coupled global genome sequencing reveal two genes associated with pollen fertility in tetraploid rice. Int. J. Mol. Sci. 22, 841. doi: 10.3390/ijms22020841
Kamara, N., Lu, Z., Jiao, Y., Zhu, L., Wu, J., Chen, Z., et al. (2022). An uncharacterized protein NY1 targets EAT1 to regulate anther tapetum development in polyploid rice. BMC Plant Biol. 22, 582. doi: 10.1186/s12870-022-03976-0
Ko, S. S., Li, M. J., Sun-Ben Ku, M., Ho, Y. C., Lin, Y. J., Chuang, M. H., et al. (2014). The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice. Plant Cell. 26 (6), 2486–2504. doi: 10.1105/tpc.114.126292
Koide, Y., Kuniyoshi, D., Kishima, Y. (2020). Fertile tetraploids: new resources for future rice breeding. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01231
Lee, S. K. H., Kim, J. I., Cho, C. D., Nguyen, S., Moon, J. E., Park, H. R., et al. (2020). Deficiency of rice hexokinase HXK5 impairs synthesis and utilization of starch in pollen grains and causes male sterility. J. Exp. Bot. 71 (1), 116–125. doi: 10.1093/jxb/erz436
Lei, R., Li, X., Ma, Z., Lv, Y., Hu, Y., Yu, D. (2017). Arabidopsis WRKY2 and WRKY34 transcription factors interact with VQ20 protein to modulate pollen development and function. Plant J. 91 (6), 962–976. doi: 10.1111/ptj.13619
Li, X., Shahid, M. Q., Wu, J., Wang, L., Liu, X., Lu, Y. G. (2016). Comparative small RNA analysis of pollen development in autotetraploid and diploid rice. Int. J. Mol. Sci. 17, 499. doi: 10.3390/ijms17040499
Li, X., Shahid, M. Q., Xia, J., Lu, Z. J., Fang, N., Wang, L., et al. (2017). Analysis of small RNAs revealed differential expressions during pollen and embryo sac development in autotetraploid rice. BMC Genomics 18, 1–18. doi: 10.1186/s12864-017-3526-8
Li, C., Wei, J., Lin, Y., Chen, H. (2012). Gene silencing using the recessive rice bacterial blight resistance gene xa13 as a new paradigm in plant breeding. Plant Cell Rep. 31, 851–862. doi: 10.1007/s00299-011-1206-8
Li, X., Yu, H., Jiao, Y., Shahid, M. Q., Wu, J., Liu, X. D. (2018). Genome-wide analysis of DNA polymorphisms, the methylome and transcriptome revealed that multiple factors are associated with low pollen fertility in autotetraploid rice. PloS One 13, e0201854. doi: 10.1371/journal.pone.0201854
Li, H., Yuan, Z., Vizcay-Barrena, G., Yang, C., Liang, W., Zong, J., et al. (2011). PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol. 156, 615–630. doi: 10.1104/pp.111.175760
Li, N., Zhang, D. S., Liu, H. S., Yin, C. S., Li, X., Liang, W., et al. (2006). The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell. 18, 2999–3014. doi: 10.1105/tpc.106.044107
Liu, X., Wu, J., Shahid, M. Q. (2022). Development of neo-tetraploid rice and research progress on its heterosis mechanism. Biotechnol. Bulletin. 38 (1), 44–50. doi: 10.13560/j.cnki
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-DELTADELTACT method. Methods. 25, 402–408.
Lu, Z. J., Guo, X. T., Huang, Z. Y., Xia, J., Li, X., Wu, J. W., et al. (2020). Transcriptome and gene editing analyses reveal MOF1a defect alters the expression of genes associated with tapetum development and chromosome behavior at meiosis stage resulting in low pollen fertility of tetraploid rice. Int. J. Mol. Sci. 21, 7489. doi: 10.3390/ijms21207489
Ma, X., Xia, H., Liu, Y., Wei, H., Zheng, X., Song, C., et al. (2016). Transcriptomic and metabolomics studies disclose key metabolism pathways contributing to well-maintained photosynthesis under the drought and the consequent drought-tolerance in rice. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01886
Ma, L., Zhang, D., Miao, Q., Yang, J., Xuan, Y., Hu, Y. (2017). Essential role of sugar transporter OsSWEET11 during the early stage of rice grain filling. Plant Cell Physiol. 58, 863–873. doi: 10.1093/pcp/pcx040
Mamun, E. A., Alfred, S., Cantrill, L. C., Overall, R. L., Sutton, B. G. (2006). Effects of chilling on male gametophyte development in rice. Cell Biol. Int. 30, 583–591. doi: 10.1016/j.cellbi.2006.03.004
Mukhtar, M. S., Liu, X., Somssich, I. E. (2018). Elucidating the role of WRKY27 in male sterility in arabidopsis. Plant Signal Behav. 12 (9), e1363945. doi: 10.1080/15592324.2017.1363945
Ngampanya, B., Sobolewska, A., Takeda, T., Toyofuku, K., Narangajavana, J., Ikeda, A., et al. (2003). Characterization of rice functional monosaccharide transporter, OsMST5. Biosci. Biotechnol. Biochem. 67 (3), 556–562. doi: 10.1271/bbb.67.556
Niu, N., Liang, W., Yang, X., Jin, W., Wilson, Z. A., Hu, J., et al. (2013). EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. Commun. 4, 1445. doi: 10.1038/ncomms2396
Phan, H. A., Li, S., Parish, R. W. (2012). MYB80, a regulator of tapetal and pollen development, is functionally conserved in crops. Plant Mol. Biol. 78, 171–183. doi: 10.1007/s11103-011-9855-0
Qi, Y., Liu, Q., Zhang, L., Mao, B., Yan, D., Jin, Q., et al. (2014). Fine mapping and candidate gene analysis of the novel thermo-sensitive genic male sterility tms9-1 gene in rice. Theor. Appl. Genet. 127, 1173. doi: 10.1007/s00122-014-22890-8
Ranjan, R., Khurana, R., Malik, N., Badoni, S., Parida, S. K., Kapoor, S., et al. (2017). bHLH142 regulates various metabolic pathway-related genes to affect pollen development and anther dehiscence in rice. Sci. Rep. 7, 43397. doi: 10.1038/srep43397
Ren, D., Rao, Y., Yu, H., Xu, Q., Cui, Y., Xia, S., et al. (2020). MORE FLORET 1 encodes a MYB transcription factor that regulates spikelet development in rice. Plant Physiol. 184 (1), 251–265. doi: 10.1104/pp.20.00658
Ruan, Y. (2012). Signaling role of sucrose metabolism in development. Mol. Plant 5, 763–765. doi: 10.1093/mp/ssq046
Ruan, Y. L., Jin, Y., Yang, Y., Li, G., Boyer, J. S. (2010). Sugar input, metabolism, and signaling mediated by invertase: roles in development, yield potential, and response to drought and heat. Mol. Plant 3, 942–955. doi: 10.1093/mp/ssq044
Shahid, M. Q., Liu, G., Li, J., Naeem, M., Liu, X. D. (2011). Heterosis and gene action study of agronomic traits in diploid and autotetraploid rice. Acta Agric. Scand. Sect. B Soil Plant Sci. 61, 23–32.
Shahid, M. Q., Sun, J. F., Wei, C. M., Zhang, P., Liu, X. D. (2010). Studies on the abnormality of embryo sac and pollen fertility in autotetraploid rice during different growing seasons. Pak. J. Bot. 42, 7–19.
Smeekens, S. (2000). Sugar-induced signal transduction in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 49–81. doi: 10.1146/annurev.arplant.51.1.49
Srivastava, A. C., Ganesan, S., Ismail, I. O., Ayre, B. G. (2008). Functional characterization of the arabidopsis AtSUC2 sucrose/H+ symporter by tissue-specific complementation reveals an essential role in phloem loading but not in long-distance transport. Plant Physiol. 148, 200–211. doi: 10.1104/pp.108.124776
Sui, J., He, J., Wu, J., Gong, B., Cao, X., Seng, S., et al. (2015). Characterization and functional analysis of transcription factor LoMYB80 related to anther development in lily (Lilium oriental hybrids). J. Plant Growth Regulation. 34, 545–557. doi: 10.1007/s00344-015-9489-6
Vogel, C., Bodenhausen, N., Gruissem, W., Vorholt, J. A. (2016). The arabidopsis leaf transcriptome reveals distinct but also overlapping responses to colonization by phyllosphere commensals and pathogen infection with impact on plant health. New Phytol. 212, 192–207. doi: 10.1111/nph.14036
Wan, L., Zha, W., Chen, X., Liu, C., Lv, L., Liu, C., et al. (2010). A rice -1,3-glucanase gene Osg1 is required for callose degradation in pollen development. Planta. 233, 309–323. doi: 10.1007/s00425-010-1301-z
Wang, Z., Gerstein, M., Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Wang, X., Liu, H. N., Chen, Y., Qiu, L., Luan, L., Long, W. B., et al. (2009). Genetic improvement of the major agronomic traits in the parent materials of autotetraploid three-line rice. J. Southwest Univ. 31, 137–142.
Wang, Y., Xiao, Y., Zhang, Y., Chai, C., Wei, G., Wei, X., et al. (2008). Molecular cloning, functional characterization and expression analysis of a novel monosaccharide transporter gene OsMST6 from rice (Oryza sativa l.). Planta. 228, 525–535. doi: 10.1007/s00425-008-0755-8
Wang, Y., Zhou, Z., Gao, J., Wu, Y., Xia, Z., Zhang, H., et al. (2016). The mechanisms of maize resistance to fusarium verticillioides by comprehensive analysis of RNA-seq data. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01654
Wen, J., Wang, L., Wang, J., Zeng, Y., Xu, Y., Li, S. (2019). The transcription factor OsbHLH138 regulates thermosensitive genic male sterility in rice via activation of TMS5. Theor. Appl. Genet. 132, 1721–1732. doi: 10.1007/s00122-019-03310-7
Wright, K. M., Arnold, B., Xue, K. S., Šurinová, M., O’Connell, J., Bomblies, K. (2014). Selection on meiosis genes in diploid and tetraploid arabidopsis arenosa. Mol. Boil. Evol. 32, 944–955. doi: 10.1093/molbev/msu398
Wu, J., Chen, Y., Lin, H., Chen, Y., Yu, H., Lu, Z., et al. (2020). Comparative cytological and transcriptome analysis revealed the normal pollen development process and up-regulation of fertility-related genes in newly developed tetraploid rice. Int. J. Mol. Sci. 21, 7046. doi: 10.3390/ijms21197046
Wu, J., Chen, L., Shahid, M. Q., Chen, M., Dong, Q., Li, J., et al. (2017). Pervasive interactions of sa and Sb loci cause high pollen sterility and abrupt changes in gene expression during meiosis that could be overcome by double neutral genes in autotetraploid rice. Rice. 10, 49. doi: 10.1186/s12284-017-0188-8
Wu, Y., Fox, T. W., Trimnell, M. R., Wang, L., Xu, R., Cigan, A. M., et al. (2016). Development of a novel recessive genetic male sterility system for hybrid seed production in maize and other cross-pollinating crops. Plant Biotechnol. J. 14, 1046–1054. doi: 10.1111/pbi.12477
Wu, J. W., Shahid, M. Q., Chen, L., Chen, Z. X., Wang, L., Liu, X. D., et al. (2015). Polyploidy enhances F1 pollen sterility loci interactions that increase meiosis abnormalities and pollen sterility in autotetraploid rice. Plant Physiol. 169 (4), 2700–2717. doi: 10.1104/pp.15.00791
Wu, J., Shahid, M. Q., Chen, M., Li, X., Li, J., Xu, X., et al. (2019). Cytological and transcriptome analysis reveal that interaction at Sb pollen sterility locus cause down-regulation of important meiosis-related genes associated with high pollen sterility in autotetraploid rice hybrids. Plant Physiol. Biochem. 141, 73–82. doi: 10.1016/j.plaphy.2019.05.019
Wu, J., Shahid, M. Q., Guo, H., Yin, W., Chen, Z., Wang, L., et al. (2014). Comparative cytological and transcriptomic analysis of pollen development in autotetraploid and diploid rice. Plant Reprod. 27, 181–196. doi: 10.1007/s00497-014-0250-2
Xiang, X. J., Sun, L. P., Yu, P., Yang, Z. F., Zhang, P. P., Zhang, Y. X., et al. (2020). The MYB transcription factor Baymax1plays a critical role in rice male fertility. Theor. Appl. Genet. 134, 453–471. doi: 10.1007/s00122-020-03706-w
Xiong, Y., Gan, L., Hu, Y., Sun, W., Zhou, X., Song, Z., et al. (2019). OsMND1 regulates early meiosis and improves the seed set rate in polyploid rice. Plant Growth Reg. 87, 341–356. doi: 10.1007/s10725-019-00476-4
Yadav, I. S., Sharma, A., Kaur, S., Nahar, N., Bhardwaj, S. C., Sharma, T. R., et al. (2016). Comparative temporal transcriptome profiling of wheat near isogenic line carrying Lr57 under compatible and incompatible interactions. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01943
Yang, Z. F., Liu, L., Sun, L. P., Yu, P., Zhang, P. P., Abbas, A., et al. (2019). OsMS1 functions as a transcriptional activator to regulate programmed tapetum development and pollen exine formation in rice. Plant Mol. Biol. 99, 175–191. doi: 10.1007/s11103-018-0811-0
Yang, X., Liu, X., Zeng, X., Fang, Y., Sun, Y. (2019). Studies on the induction of autotetraploid rice. Nroth Rice. 6, 27–29.
Yang, X., Wu, D., Shi, J., He, Y., Pinot, F., Grausem, B., et al. (2014). Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J. Integr. Plant Biol. 56, 979–994. doi: 10.1111/jipb.12212
Yant, L., Hollister, J. D., Wright, K. M., Arnold, B. J., Higgins, J. D., Franklin, F. C. H., et al. (2013). Meiotic adaptation to genome duplication in arabidopsis arenosa. Curr. Biol. 23, 2151–2156. doi: 10.1016/j.cub.2013.08.059
Yu, H., Shahid, M. Q., Li, Q. H., Li, Y. D., Li, C., Lu, Z. J., et al. (2020). Production assessment and genome comparison revealed high yield potential and novel specific alleles associated with fertility and yield in neo-tetraploid rice. Rice. 13, 32. doi: 10.1186/s12284-020-00387-3
Zafar, S. A., Patil, S. B., Uzair, M., Fang, J., Zhao, J., Guo, T., et al. (2020). DEGENERATED PANICLE AND PARTIAL STERILITY 1 (DPS1) encodes a cystathionine β-synthase domain containing protein required for anther cuticle and panicle development in rice. New Phytologist. 225, 356–375.
Zhang, S., Fang, Z. J., Zhu, J., Gao, J. F., Yang, Z. N. (2010). OsMYB103 is required for rice anther development by regulating tapetum development and exine formation. Chin. Sci. Bulletin. 55 (29), 3288–3297. doi: 10.1007/s11434-010-4087-2
Zhang, H., Liang, W., Yang, X., Luo, X., Jiang, N., Ma, H., et al. (2010). Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. Plant Cell. 22, 672–689. doi: 10.1105/tpc.109.073668
Zhang, L., Mao, D., Xing, F., Bai, X., Zhao, H., Yao, W., et al. (2015). Loss of function of OsMADS3 via the insertion of a novel retrotransposon leads to recessive male sterility in rice (Oryza sativa l). Plant Science. 238, 188–197. doi: 10.1016/plantsci.2015.06.007
Zhao, T., Ren, L., Chen, X., Yu, H., Liu, C., Shen, Y., et al. (2018). The OsRR24/LEPTO1 type-b response regulator is essential for the organization of leptotene chromosomes in rice meiosis. Plant Cell. 30, 3024–3037. doi: 10.1105/tpc.18.00479
Zhu, E., You, C., Wang, S., Cui, J., Niu, B., Wang, Y., et al. (2015). The DYT1-interacting proteins bHLH010, bHLH089, and bHLH091 are redundantly required for arabidopsis anther development and transcriptome. Plant J. 83 (6), 976–990. doi: 10.1111/tpj.12942
Keywords: rice, RNA-Seq, pollen sterility, carbohydrate metabolism, tapetum
Citation: Kamara N, Jiao Y, Huang W, Cao L, Zhu L, Zhao C, Huang X, Shivute FN, Liu X, Wu J and Shahid MQ (2023) Comparative cytological and transcriptome analyses of ny2 mutant delayed degeneration of tapetal cells and promotes abnormal microspore development in neo-tetraploid rice. Front. Plant Sci. 14:1229870. doi: 10.3389/fpls.2023.1229870
Received: 27 May 2023; Accepted: 26 June 2023;
Published: 17 July 2023.
Edited by:
Gregory Thyssen, Agricultural Research Service (USDA), United StatesReviewed by:
Xiangdong Luo, Jiangxi Normal University, ChinaCopyright © 2023 Kamara, Jiao, Huang, Cao, Zhu, Zhao, Huang, Shivute, Liu, Wu and Shahid. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinwen Wu, and3dUBzY2F1LmVkdS5jbg==; Muhammad Qasim Shahid, cWFzaW1Ac2NhdS5lZHUuY24=
†These authors contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.