
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 23 October 2023
Sec. Plant Development and EvoDevo
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1227859
This article is part of the Research TopicAdvanced Imaging in Plants: Exploring Development and FunctionView all 4 articles
Bioenergy sorghum is a drought-tolerant high-biomass C4 grass targeted for production on annual cropland marginal for food crops due primarily to abiotic constraints. To better understand the overall contribution of stem wax to bioenergy sorghum’s resilience, the current study characterized sorghum stem cuticular wax loads, composition, morphometrics, wax pathway gene expression and regulation using vegetative phase Wray, R07020, and TX08001 genotypes. Wax loads on sorghum stems (~103-215 µg/cm2) were much higher than Arabidopsis stem and leaf wax loads. Wax on developing sorghum stem internodes was enriched in C28/30 primary alcohols (~65%) while stem wax on fully developed stems was enriched in C28/30 aldehydes (~80%). Scanning Electron Microscopy showed minimal wax on internodes prior to the onset of elongation and that wax tubules first appear associated with cork-silica cell complexes when internode cell elongation is complete. Sorghum homologs of genes involved in wax biosynthesis/transport were differentially expressed in the stem epidermis. Expression of many wax pathway genes (i.e., SbKCS6, SbCER3-1, SbWSD1, SbABCG12, SbABCG11) is low in immature apical internodes then increases at the onset of stem wax accumulation. SbCER4 is expressed relatively early in stem development consistent with accumulation of C28/30 primary alcohols on developing apical internodes. High expression of two SbCER3 homologs in fully elongated internodes is consistent with a role in production of C28/30 aldehydes. Gene regulatory network analysis aided the identification of sorghum homologs of transcription factors that regulate wax biosynthesis (i.e., SbSHN1, SbWRI1/3, SbMYB94/96/30/60, MYS1) and other transcription factors that could regulate and specify expression of the wax pathway in epidermal cells during cuticle development.
The cuticle covers the aerial surfaces of most terrestrial plants serving an essential role in the adaptation of plants to the environment. The biogenesis, chemistry, morphology, and function of the plant cuticle has been the focus of extensive research (Jenks et al., 1992; Koch and Ensikat, 2008; Samuels et al., 2008; Bernard and Joubes, 2013; Yeats and Rose, 2013; Lee and Suh, 2015; Busta et al., 2017; Lewandowska et al., 2020; Batsale et al., 2021). The cuticle aids in reducing water loss from plant surfaces, dissipates harmful solar radiation, and helps protect plants from pathogens and insects in part by mediating signaling between plants and beneficial microbes/pathogens (Nwanze et al., 1992; Lewandowska et al., 2020; Cardona et al., 2022). The cuticle is composed of cutin, a polyester comprised primarily of cross-linked C16-C18 fatty acids, intracuticular wax and oils embedded in the cutin layer (Buschhaus and Jetter, 2011), and epicuticular wax located on the external surface of the cutin matrix. Intracuticular and epicuticular waxes are composed of mixtures of hydrophobic aliphatic compounds that include long chain fatty acids, primary and secondary alcohols, esters, aldehydes, ketones and lower amounts of triterpenoids, sterols, and polyketides. Epicuticular wax can form plates, crystals, rods, needles, and tubules on surfaces depending in part on the chemistry of secreted wax (Wettstein-Knowles, 1972; Chambers et al., 1975; Koch and Ensikat, 2008). The composition and thickness of epicuticular wax varies depending on species, organ, surface (adaxial, abaxial), genotype and environmental conditions. The molecular basis of extensive variation in wax composition and load and the functional impact of variation has been the subject of numerous studies.
The genetic and biochemical pathways involved in epicuticular wax formation have been characterized in Arabidopsis thaliana aided by an extensive collection of eceriferum (cer) mutants that have altered cuticle appearance and chemical composition (Bernard and Joubes, 2013; Busta et al., 2017). These studies showed that the main components of wax are derived from C16 and C18 fatty acid thioesters that are synthesized in plastids then hydrolyzed and transported to the endoplasmic reticulum (ER) for conversion to long chain acyl-CoAs by long chain acyl-CoA synthases (LACSs) (Busta et al., 2017). The resulting acyl-CoAs are converted to very-long-chain fatty acids (VLCFAs) with chain lengths of 24-34 carbons by a fatty acid elongase complex consisting of KCS/CER6, KCR, HDC/PAS2, and ECR. Proteins encoded by CER2, CER26, and CER2-like are involved in elongation that produces C30-C34 acyl-CoAs (Negruk et al., 1996; Pascal et al., 2013; Haslam et al., 2015; Haslam et al., 2017; Xue et al., 2017). The long chain acyl-CoAs (Yeats and Rose, 2013) are directed into alcohol-forming or alkane-forming wax pathways prior to export or are oxidized into a VLCFA. The alcohol-forming pathway, mediated by proteins encoded by CER4 and WSD1, produces primary alcohols and esters, whereas the alkane-forming pathway, involving the gene products of CER3, CER1, and MAH1, produces aldehydes, alkanes, secondary alcohols, and ketones. The wax components are secreted through the plasmalemma via ABCG-transporters (ABCG12, ABCG11) and carried across the wall space in association with lipid transfer proteins (LTPs) (Bernard and Joubes, 2013; Lewandowska et al., 2020).
The regulation of wax biosynthesis is complex in part because cuticle synthesis and organ growth are coordinated and wax load and composition varies depending on stage of development, organ type, and abiotic and biotic factors (Avato et al., 1984; Medeiros et al., 2017; Bourgault et al., 2020; Lykholat et al., 2020). Transcription factors that regulate wax biosynthesis have been identified through analysis of mutants, association studies, and co-expression analysis. For example, analysis of Arabidopsis wax mutants and biochemical studies led to the identification of MYB94 and MYB96 as direct regulators of KCS1/2/6, KCR1, CER1/2/3 and WSD1 expression (Seo et al., 2011; Lee et al., 2016). Additional R2R3-MYB factors including MYB30 and MYB41 also modulate cuticle and wax formation (Cominelli et al., 2008; Raffaele et al., 2008). Diel regulation of wax biosynthesis in leaves is regulated by MYS1/2, DEWAX, SPL9, CER7 and miR156 (Klein et al., 1996; Hooker et al., 2007; Li et al., 2019; Liu et al., 2022). In addition, the AP2/ERF factors WIN1/SHN1 and WRI1 regulate cutin and wax biosynthesis and in Arabidopsis, WRI1 directly modulates expression of LACS1, KCR1, PAS2, ECR, and WSD1 (Park et al., 2016). Increased wax accumulation in response to water deficit is regulated by induction of RAP2.4 which encodes an AP2/DREB transcription factor (Yang et al., 2020), and increased expression of MYB94/96 (Seo et al., 2011).
Sorghum is a genetically diverse drought tolerant C4 grass used for production of grain, forage, and biomass for bioenergy (Rooney et al., 2007; Mullet et al., 2014; Boyles et al., 2019; Tao et al., 2021).The heavy wax loads that accumulate on aerial surfaces of Sorghum bicolor have been analyzed to better understand the contribution of wax load/chemistry to sorghum’s exceptional tolerance to drought and heat. Early studies found that higher leaf wax loads on sorghum leaves were correlated with reduced cuticular transpiration (Nobel and Jordan, 1983; Jordan et al., 1984) and that leaf wax load and water use efficiency increased in response to water deficit (Saneoka and Ogata, 1987). Variation in wax loads were also correlated with differences in susceptibility to fungal pathogens (Jenks et al., 1994) and insect-plant interactions (Bernays, 1972; Nwanze et al., 1992). A recent study of the sorghum wax phyllosphere found that Sphingomonadaceae and Rhizobiaceae families were the major taxa associated with stem wax (Mechan-Llontop et al., 2023). SEM analysis revealed that sorghum leaf sheath wax exhibits a wide range of morphologies including plates, crystals, and tubules (McWhorter and Rex, 1989; McWhorter et al., 1990; Jenks et al., 1992). Analysis of sorghum wax chemistry showed that triterpenoids could have an important impact on cuticular transpiration at high temperatures (Busta et al., 2021). Differences in the wax composition of sorghum leaf blades, leaf sheaths, stems and grain has been documented although the functional significance of these differences requires further investigation (Hwang et al., 2002; Harron et al., 2017; Xiao et al., 2020; Busta et al., 2021; Tang et al., 2022). The availability of variation in wax load among sorghum accessions and the generation and characterization of sorghum wax mutants (Peters et al., 2009; Xin et al., 2009; Xin et al., 2021) has enabled QTL, association, and map-based identification of several sorghum genes involved in cuticle and wax biosynthesis (Burow et al., 2008; Burow et al., 2009; Awika et al., 2017; Punnuri et al., 2017; Uttam et al., 2017; Elango et al., 2020). These studies characterized WBC11, an ABC transporter that is involved in wax transport (Mizuno et al., 2013) and a GDSL-lipase homolog of cutin synthase that reduces wax accumulation when mutated (Jiao et al., 2017).
Drought and heat tolerance are especially important traits in bioenergy sorghum because this crop is being developed for biomass production on marginal annual cropland suitable for bioenergy crops. Moreover, bioenergy sorghum’s long vegetative growth duration (>150 days) typically results in exposure to abiotic stress during a significant portion of the growing season. Most of the previous studies of sorghum wax have utilized dwarfed grain sorghum genotypes and focused on wax accumulation on leaf blades and leaf sheaths. In contrast, bioenergy sorghum stems are 4-5 m in length by the end of the growing season and stems, like leaf sheaths, are covered with a heavy wax load. Therefore, the current study focused on characterizing the developmental timing and extent of stem wax accumulation, morphology and composition during plant development and the identification of genes involved in stem wax biosynthesis. This analysis revealed that stem wax loads are very high and that wax composition changes during stem development resulting in the accumulation of high levels of long chain waxy aldehydes on the surface of older stem internodes. Transcriptome and gene regulatory network analysis enabled the identification of transcription factors that are predicted to regulate stem wax biosynthesis.
Stem tissues for wax analysis were obtained from TX08001, a photoperiod sensitive late flowering bioenergy sorghum hybrid developed at Texas A&M University, R07020, a photoperiod sensitive bioenergy sorghum inbred, and Wray, a photoperiod sensitive sweet sorghum inbred. Seeds were obtained from the Sorghum Breeding Lab at Texas A&M University in College Station, Texas and planted in the field or in a greenhouse in 6 L pots filled with Jolly Gardner Pro-Line C25 soil or in rhizotrons (20 cm diameter x 75cm long) filled with field soil. TX08001, R07020, and Wray were grown in a greenhouse for 90 days under long day conditions to maintain plants in the vegetative phase. Stem internode tissue was harvested from Wray (74 DAE) and R07020(60 DAE) plants (DAE -Days after Emergence) (Casto et al., 2018; Fu et al., 2023). Stem tissues were harvested from TX08001 at 90 DAE for SEM analysis. TX08001 plants were also grown in rhizotrons for 120 days in an automated phenotyping greenhouse with tall side walls in 2022 and harvested for wax composition analysis. Stem samples were also obtained from field grown TX08001 in 2021 harvested at 125 DAE. Wax was extracted for wax load and composition analysis from TX08001 grown at the Texas A&M University Farm in Burleson County, TX in 2019 and 2020. Field grown plants were fertilized at planting with a solution of liquid ammonium polyphosphate (11-37-0), UAN 32% and zinc sulfate. Field soil is designated Roetex Clay. TX08001 plants were thinned at 21 DAE to 15 cm spacing between plants in rows, with 76 cm spacing between rows. Seeds were treated with Concep III, Nugro, and Apron X. Field plots were not irrigated after planting.
Internode epidermal tissue was excised and placed onto Kimwipes and then put under a low-pressure nitrogen stream for two hours to pre-dry the tissue. The samples were then transferred to a desiccation globe and dried by exposure to low pressure for 48 hours. Samples were attached to 1 cm pin mount stands with Electron Microscopy Sciences conductive double sided carbon tape (8mm). The samples were sputter coated with 7nm of palladium, then analyzed with a FEI Quanta 600 FE-SEM at 10kV under low pressure.
Plant material (TX08001) was harvested by cutting plants at the base of the stem for deconstruction and wax extraction. Stems were divided into node-internode sections and labeled based on phytomer number (P1 corresponds to the youngest phytomer located at the top of the plant). The surface area of each tissue type targeted for wax extraction was measured while samples were still fresh. Wax extraction was carried out by immersing samples in hexane for 40 seconds followed by the transfer of hexane extracts to preweighed glass autosampler tubes. The sample was then subjected to a second and third hexane extraction of 40 sec each. The three hexane washes were obtained, each was dried to remove hexane, and the weight of extracted wax was summed to calculate wax load. This protocol was developed to optimize the extraction of epicuticular and intracuticular wax while minimizing the extraction of C16 and C18 fatty acids and chlorophyll. The yield of wax decreased and C16/C18 fatty acids increased when more than three 40 sec washes were utilized for extraction. Following three hexane extractions of older stem internodes (>P9) residual wax was still visible on stem surfaces and could be removed with a hexane-soaked cloth. This additional wax fraction was not included in the wax yields presented in this paper.
TX08001 internode diameters were measured with calipers to a tenth of a millimeter above the pulvinus (Base of the Internode), below the nodal plexus (Top of Internode) and at the mid-point of the internode (Middle of the Internode) which typically has the smallest diameter. The length (h) of the internode was also measured with calipers to a tenth of a millimeter. Surface area was then estimated based on a model of stem internodes as the walls of two intersecting truncated cones where the largest diameter of each cone corresponded to the ends of the internode (resulting in two trapezoidal calculations). + (h/2)(Middle of internode+Base of Internode)
TX08001 waxes were transferred in hexane to autosampler vials for analysis in 500 µg aliquots, then dried under a nitrogen stream before adding 100 uL of BSTFA (with 1% TCMS) and incubating at 85°C for 12 hours. Excess BSTFA was evaporated under nitrogen and chloroform was used to suspend the sample at 500 µg/mL. Samples were then analyzed on an Agilent 7890A/7693A/5975C XL GC-MS equipped with a 30-m DB-5MS column (0.25-mm film) using pulsed splitless injection (290°C inlet). Helium was used as the carrier gas at 105.75 mL/min. The oven was held at 70°C for 1 min, then ramped at 10°C/min to 200°C, then ramped at 4°C/min to 295°C and maintained at 295°C for 20 min (total run time 57.75 minutes). The ion source was maintained at 230°C and the quadrupole at 150°C. Mass charge selection for the quadrupole was set to 198-552 m/z (Yang et al., 2017).
High temperature GC-MS was performed by the Thermofisher Demo Lab on a Thermo Scientific ISQ 7000. Bulk wax was resuspended in hexane and transferred to autosampler vials in 1 mg aliquots, then dried under a nitrogen stream. Samples were resuspended in chloroform at a concentration of 1mg/mL and separated on a RESTEK Rxi-5HG column (15m x.25mm x.1µm). The inlet and detector were set to 390°C, and the oven was set to ramp from 120°C to 240°C at a rate of a of 15°C/min and then from 240°C-390°C at 8°C/min and maintained at 390°C for 6 minutes. Samples were injected at a volume of 1µL at concentrations ranging from 400-2000µg/mL (in chloroform). Mass charge selection for the quadrupole was set to 48-952 m/z (Tada et al., 2014).
Three replicates of stem plant tissues were harvested for RNAseq analysis. Some of the R07020 stem samples were cored to separate rind and stem core tissues prior to RNA extraction. The stem tissue samples were placed in 50 mL conical tubes, frozen in liquid nitrogen and stored at -80 C. The tissue was then ground into a fine powder using a mortar and pestle cooled with liquid nitrogen before RNA was extracted using the Zymo Research Direct-zol RNA MiniPrep Plus kit. RNA quality was checked using an Aglient Bioanalyzer then sent to the Joint Genome Institute for library construction and sequencing at a read depth of 30-50 M reads/replicate. Sequenced reads were aligned to the Sorghum bicolor V3.1 genome using HISAT2 aligner. The transcriptome assembly and TPM normalization were conducted using String Tie version 1.3 and relative expression used TPM normalized data. RNAseq data from two prior studies was used in the current study (Casto et al., 2018; Fu et al., 2023). Four replicates of Wray stem internode tissue were harvested from vegetative stage plants at 74 DAE, frozen, sent to the Pacific Northwest National Laboratory where stem cell types were isolated using laser capture microdissection (LCM) and RNAseq analysis was done to identify cell-type transcriptomes. The details of LCM methodology, RNA extraction and library construction are described in (Fu et al., 2023). RNAseq data derived from LCM analysis was processed as described above and used to examine the expression of wax pathway genes in stem cell types and for gene regulatory network analysis.
Sorghum genes involved in stem wax biosynthesis were identified in a multi-step process. Sorghum homologs of validated wax pathway genes characterized in Arabidopsis thaliana, Oyrza sativa, and Zea mays were identified initially through Blast X analysis. Sorghum homologs with an E-value of 1e-20 or lower that encoded proteins with the same PFAM domains as the protein encoded by a validated target wax pathway gene were used for phylogenetic analysis using MEGA X (Tamura et al., 2011; Kumar et al., 2018; Tamura et al., 2021). Alignments were performed with Clustal-W and were co-validated with MUSCLE. The phylogenetic tree was constructed using maximum likelihood with the Jones-Taylor-Thornton amino acid substitution model and all families were bootstrapped with at least 500 replicates. The MYB phylogenetic tree was produced as described in (Singh et al., 2020). The phylogenetic analysis conducted in the current study extended a prior analysis of sorghum MYB factors carried out by (Singh et al., 2020) by including SbMYB86, a gene is relevant to this study. During review, it was noted that the results of our sorghum MYB phylogenetic analysis were consistent with a prior multi-species phylogenetic analysis of MYB factors (Hennet et al., 2020). Sorghum genes that clustered with the validated wax pathway gene at nodes with >80% bootstrap support (SbLACS9 and SbWSD1 failed this threshold Supplemental Figure 3A) were analyzed further to identify genes that were differentially expressed in the stem epidermis of internode 4 (Wray) compared to other stem cell types (pith parenchyma, xylem, phloem, vascular parenchyma) (Fu et al., 2023). The subset of sorghum wax pathway genes that were differentially expressed in the epidermis were used for wax pathway gene regulatory network (GRN) analysis and to investigate the expression of these genes during bioenergy sorghum internode development.
Stem cell type transcriptome data obtained by laser capture microdissection (LCM) (Fu et al., 2023) was used to construct a wax pathway gene regulatory network (GRN). The general approach and methods used for gene regulatory analysis have been previously described (Varala et al., 2018; Yu et al., 2022; Fu et al., 2023). Genes involved in wax biosynthesis are differentially expressed at higher levels in the stem epidermis compared to other stem cell types. Transcriptome profiles of stem epidermal cells, pith cells, vascular bundle sclerenchyma, phloem, and vascular bundle parenchyma were obtained in a separate study using laser-capture microdissection and RNAseq analysis (Fu et al., 2023). The stem cell type transcriptome profiles were used to identify genes encoding transcription factors that were differentially expressed in epidermal cells (vs. the other stem cell types), that are co-expressed with wax pathway genes, and that have predicted binding sites in the promoters of the wax pathway genes and TFs that are part of the wax pathway gene regulatory network. To do this, data from each cell type replicate was summed across transcript-level TPMs and counts to obtain gene-level TPMs and counts. Next, an expression threshold was applied to retain genes that exhibited expression of TPM ≥ 5 in at least one sample (one sample being the mean of three biological replicates). TPM normalized expression was used instead of count data for expression thresholding because TPM values allow for comparison across samples and genes. Next, read counts for the same gene set were used to calculate differential expression (DE) with the edgeR package. To calculate DE, gene-level read counts were normalized using Trimmed Mean of M-values normalization. DE is calculated using two groups. The first group was comprised of the epidermis of the LCM dataset. The second group was comprised of bundle sheath, phloem, pith parenchyma, xylem/vascular parenchyma, and xylem cell types of the LCM dataset. We contrasted the epidermal cells relative to the other cell types and defined the genes that were positively or negatively differentially expressed in the epidermis as upregulated or down-regulated, respectively. Following DE analysis, the dataset was further constrained to genes that exhibited DE ≥ 5, FDR < 0.05 (in addition to being expressed > 5 TPM in one triplicate sample). The TPM data of the genes that were DE were used to construct the GRN by integrating three different metrics: (i) Pearson’s correlation coefficient (PCC); (ii) Mutual Rank (calculated as the geometric mean of the ranking of each gene in the PCC rank of the other gene of the pair); and (iii) the capacity of one of the genes of the pair to bind to the promoter of the other gene (i.e., one of the genes being a TF and the promoter of the other gene possessing an enriched conserved regulatory element (CRE) that could be bound by the TF). To facilitate the third criterion, the putative promoter sequences of all genes in the sorghum genome were subjected to DNA pattern analysis using the position weight matrix available in Plant Promoter Analysis Navigator–PLANTPAN3.0; (http://plantpan.itps.ncku.edu.tw/index.html) (Chow et al., 2019) to match known CREs with sequences in sorghum promoters. Promoter sequences annotated spanned 1 kb upstream of the transcription start site. Transcription start site locations were obtained from the Morokoshi sorghum transcriptome database (Makita et al., 2015). As construction of the network considers the potential for promoter binding as described above, every edge in the network consists of at least one TF. The GRN was constructed by selecting all the edges that had PCC ≥ 0.9, FDR ≤ 0.05 and where at least one of the two genes was a TF capable of binding to an enriched CRE in the promoter of the other gene in the pair. When a PCC of -0.9 was used to analyze down-regulated genes, no down-regulated TFs were part of the GRN. If a PCC of -0.7 was used, the down-regulated TFs incorporated into the GRN did not have direct connections to wax pathway genes. Therefore, GRN analysis focused only on genes that were upregulated in stem epidermal cells. The GRN that included genes >5-fold upregulated in stem epidermal cells consisted of 626 genes, and 1416 edges. These statistical thresholds were selected because they resulted in a GRN that included genes that, based on previous knowledge, are likely involved in wax biosynthesis. Further analysis and rendering of the network were conducted in Cytoscape. The GRN network file, the script used to calculate the GRN, and the Cytoscape session file are included in the Supplemental Materials and are hosted at https://github.com/brianamckinley/Chemelewski_Wax_2023. In depth analysis of sorghum stem cell type specific gene regulatory networks is described in (Fu et al., 2023).
During sorghum’s adult vegetative phase, a new phytomer (P) is formed immediately below the shoot apical meristem approximately every 3-4 days in good growing conditions. During phytomer development, a leaf blade-sheath grows out from the stem nodal plexus, followed by elongation of the stem internode below the nodal plexus. Stem tissue associated with the youngest 3-4 apical phytomers (all following phytomer (P) numbers are assuming 3 apical phytomers) are short, contain minimal internode tissue, and increase in size primarily through cell division (Kebrom et al., 2017; Yu et al., 2022) (Supplementary Figure 1). The onset of stem internode growth is observed in P4 and continues for 9-12 days through the generation of additional cells by the intercalary meristem located at the base of the internode and the elongation of cells in a region immediately above the intercalary meristem (Yu et al., 2022) (Supplementary Figure 1). Cessation of internode cell elongation is followed by secondary cell wall formation in a zone of maturation at the upper end of a developing internode. The zone of maturation increases in length during internode growth, eventually spanning the entire internode once cell division ceases (Supplementary Figure 1). In a prior study (Kebrom et al., 2017; Yu et al., 2022), the first short internode that became visible below the shoot apex was derived from P4 and was designated Internode #1 (Int#1); internodes at later stages of development were numbered sequentially (Int#2, P5; Int#3, P6, etc.). In the current study, visual inspection of the external surface of stem tissue associated with P1-P4 showed minimal wax accumulation, however, by P7 (Int#4), a white wax layer was visible covering the majority of the internode.
SEM has been used to visualize wax accumulation and morphology on sorghum surfaces (McWhorter and Rex, 1989; Jenks et al., 1992). In the current study, SEM was used to examine the deposition of wax on the external surface of stem internodes during internode development (Figures 1–3). Consistent with visual inspection, SEM analysis showed minimal wax on Int#1 (P4) and Int#2 (P5) (Figure 1, Int#1 & 2, A-C) (Supplementary Figure 1, locations of SEM sampling). However, stomata (St), putative silica cells (Sc) and nascent papillae (P) that later are associated with wax extrusion were observed on the epidermis of Int#2 (Figure 1C). The apical stem tissues are soft and following SEM sample preparation the external cell layer of stem Int#1 (P4) and Int#2 (P5) showed a range of contours and folds. Epicuticular wax was visible at the apical end of Int#3 (P6) distal to the zone of cell elongation (Figures 1D–F). The short wax tubules observed on the upper end of Int#3 were associated with the apical end of cells shaped like a clover leaf that have been previously identified as silica cells (Sc) (Figures 1D–F) (Jenks et al., 1992). The wax forming on these cells had a tube-like structure (wax tubules) and was associated with papillae (P) (Figures 1E, F). Multiple papillae were almost always located at one end of the silica cells (Jenks et al., 1992). SEM analysis of wax accumulation at the middle and upper end of Int#4 is shown in Figure 2. Minimal wax was observed on epidermal tissue from the middle of Int#4 (Figure 2C), a region of the internode just above the growing zone. This is contrasted by the apical end of the internode spanning fully elongated cells that were covered with epicuticular wax (Figure 2A). The morphology of wax that accumulates on fully expanded internodes of older phytomers was also analyzed (Figure 3, Int#6, Int#25). Portions of Int#6 (P9) were covered with long wax tubes (Figure 3A) which appeared to be extruded through the cutin layer possibly from rows of papillae (Figure 3C). Wax crystals and plate-like wax was also observed on the surface (Figure 3B). Hexane removal of wax from the surface of Int#6 revealed cloverleaf shaped silica cells (Sc) and associated papillae (P) (Figure 3C). SEM analysis of Int#25 from an older plant showed the presence of a dense mat of wax tubes of different diameters (Figure 3D). Removal of the wax plate through mechanical lifting revealed a near perfect cast of the underlying epidermal cells (Figure 3E) and clover leaf shaped silica cells and associated papillae (Figures 3E, F). On occasion, papillae unassociated with a silica cell were observed on the external surface (Figure 3F).
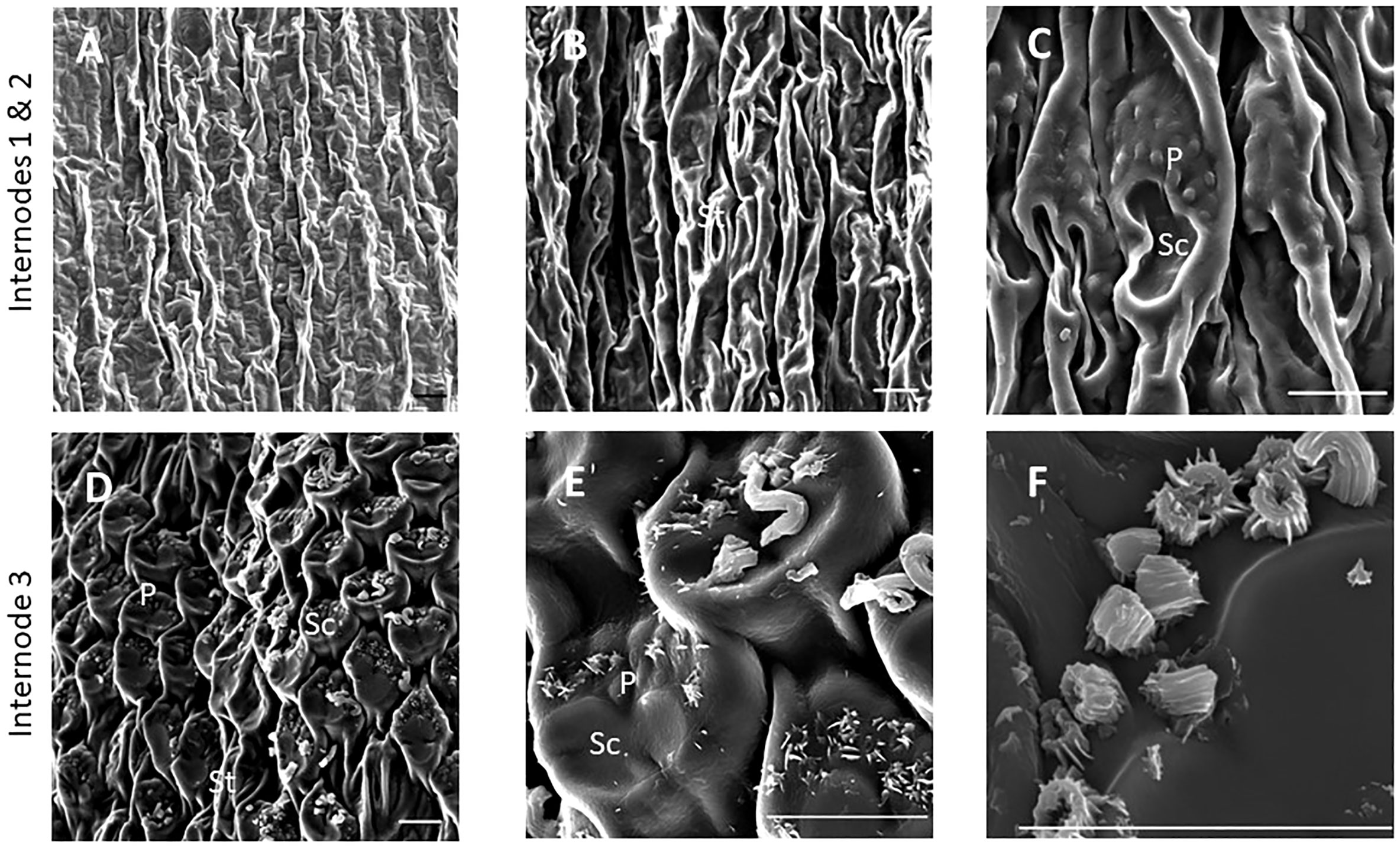
Figure 1 SEM micrographs of the epidermis of TX08001 stem internodes at various stages of development (scale bar = 10 μm). (A) Internode 1, the youngest internode analyzed shows minimal wax accumulation. (B, C) Stem epidermal tissue taken from the upper end of internode 2 showing stomata [St], silica cells [Sc], and papillae [P]. (D–F) Stem epidermal tissue taken from the upper end of the internode 3 showing the onset of wax tubule formation associated with silica cells and papillae. A diagram showing the location of SEM stem sample collection is provided in Supplementary Figure 1.
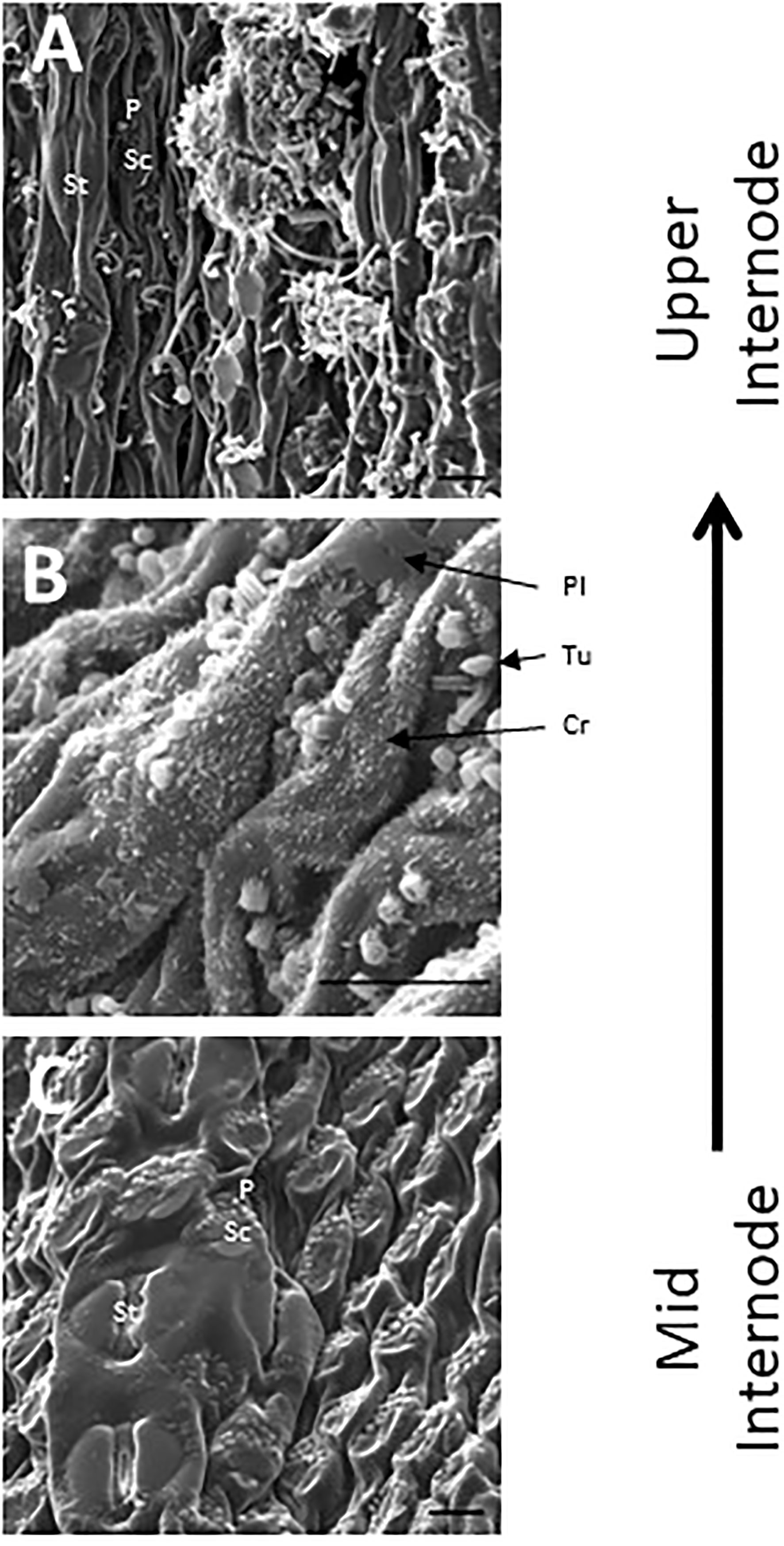
Figure 2 SEM micrographs of the stem epidermis of TX08001 across the upper portion of internode 4 (scale bar = 10 μm). (A) SEM of the upper end of internode 4 showing long wax tubules, globular and wax crystals accumulating on epidermal tissue. (B) Accumulation of short wax tubules and wax crystals on the surface of less developed internode tissue. Plate wax (Pl), Wax Crystals (Cr), Tubular Wax (Tu). (C) SEM aided visualization of silica cells and papillae and minimal wax accumulation on stem tissue taken from the middle of internode 4. A diagram showing the location of SEM stem sample collection across internode 4 is provided in Supplementary Figure 1.
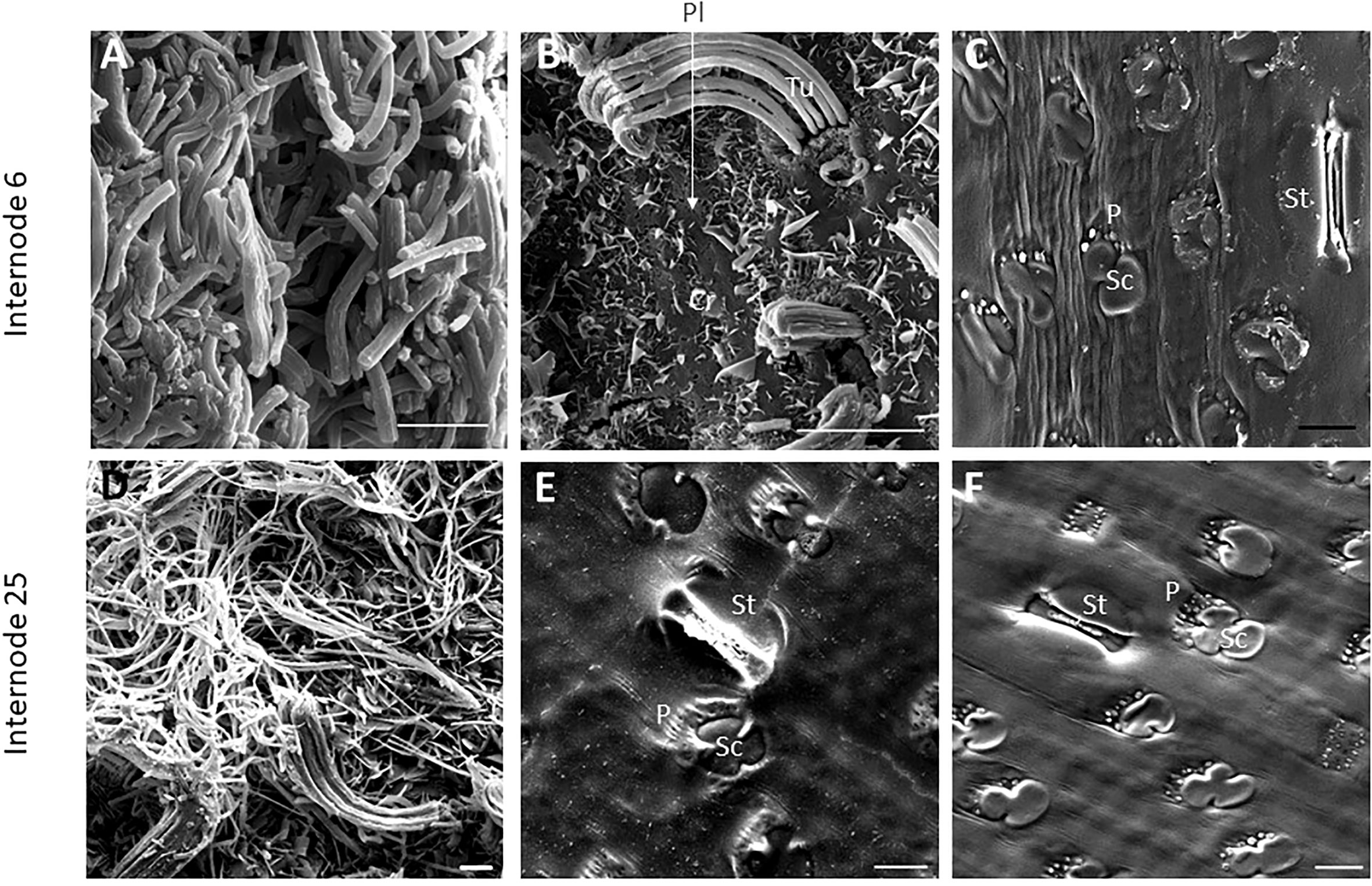
Figure 3 SEM micrographs of fully elongated TX08001 internodes (scale bar= 10μm). (A, B) SEM images of Internode 6 showing dense mats of wax tubules (A) and regions with fewer wax tubules that appear to be secreted in rows through the cutin layer (B). (B) Plate wax (Pl), Wax Crystals (Cr), Tubular Wax (Tu). (C) SEM images of a hexane washed section of internode 6 showing epidermal cells including stomata (St), silica cells (Sc) and papillae (P) (image Supplementary Figure 7 is the non-contrast adjusted image). SEM images of the epidermis of internode 25 (D–F) showing wax tubules, filaments, and scales. (E) SEM image of the underside of the wax plate on internode 25 showing a ‘negative’ impression of the surface of the differentiated structures. (F) The cuticular and epidermal surface of internode 25. To evaluate if the same patterns of secretion were occurring, the wax was peeled to view the back side of the wax mat showing that it coats the surface of the cutin.
Wax extraction from bioenergy sorghum stems was optimized to maximize recovery of wax components while minimizing extraction of C16 and C18 fatty acids and chlorophyll (see methods). Previous estimates of wax loads on grain sorghum leaf sheaths ranged from ~38-206 ug/cm2 (McWhorter et al., 1990; Burow et al., 2008; Xiao et al., 2020). In the current study, the wax load from a pioneering survey in 2019 on internode #8 of field grown TX08001 at 90 DAE was ~200 ug/cm2 and the wax load on leaf sheath #8 was ~42 ug/cm2. In 2020, the wax load on internode #8 of field grown TX08001 plants at 60 DAE was ~103 ug/cm2 and the leaf sheath wax load was ~51 ug/cm2. Internode #8 (60 DAE) became internode #23 at 120 DAE due to the production of additional phytomers by the shoot apical meristem. By 120 DAE, the wax load on internode #23 was ~215 ug/cm2 (Table 1). High variance in wax load estimates from internode #23 could be due to loss of wax flakes during plant collection/deconstruction prior to wax extraction and the wax flaking into solution during extraction. However, when the removal of residual wax with a cloth was factored in (see Stem Wax Extraction in Methods), the variance was greatly reduced suggesting this is likely variance associated with the hexane-based wax extraction method.
Preliminary analyses of wax composition on internodes at various stages of development were conducted in 2019/2020. In 2022, TX08001 plants were grown in rhizotrons in a new automated phenotyping greenhouse with high side walls that enabled vegetative growth of tall bioenergy sorghum for 120 days prior to wax composition analysis. Plant shoots were harvested, leaves and leaf sheaths removed from stems and wax was extracted from stem segments associated with P1-7, P8-15, P16-P23 and P24+. GC-MS analysis of wax extracts collected from P1-7 revealed that C28/30 primary alcohols were the most abundant wax component during early stem development (Figure 4; Supplementary Figure 2). The wax alcohols consisted primarily of octacosanol and melyssil alcohol (C28 and C30 primary alcohols, respectively), with small amounts of octacosanal and triacotonal (Figure 4). In contrast, wax from older more developed internodes was enriched in long chain fatty aldehydes (C28, C30) (Figure 4). Wax esters and alkanes were relatively minor wax components that did not change significantly in relative abundance during internode development.
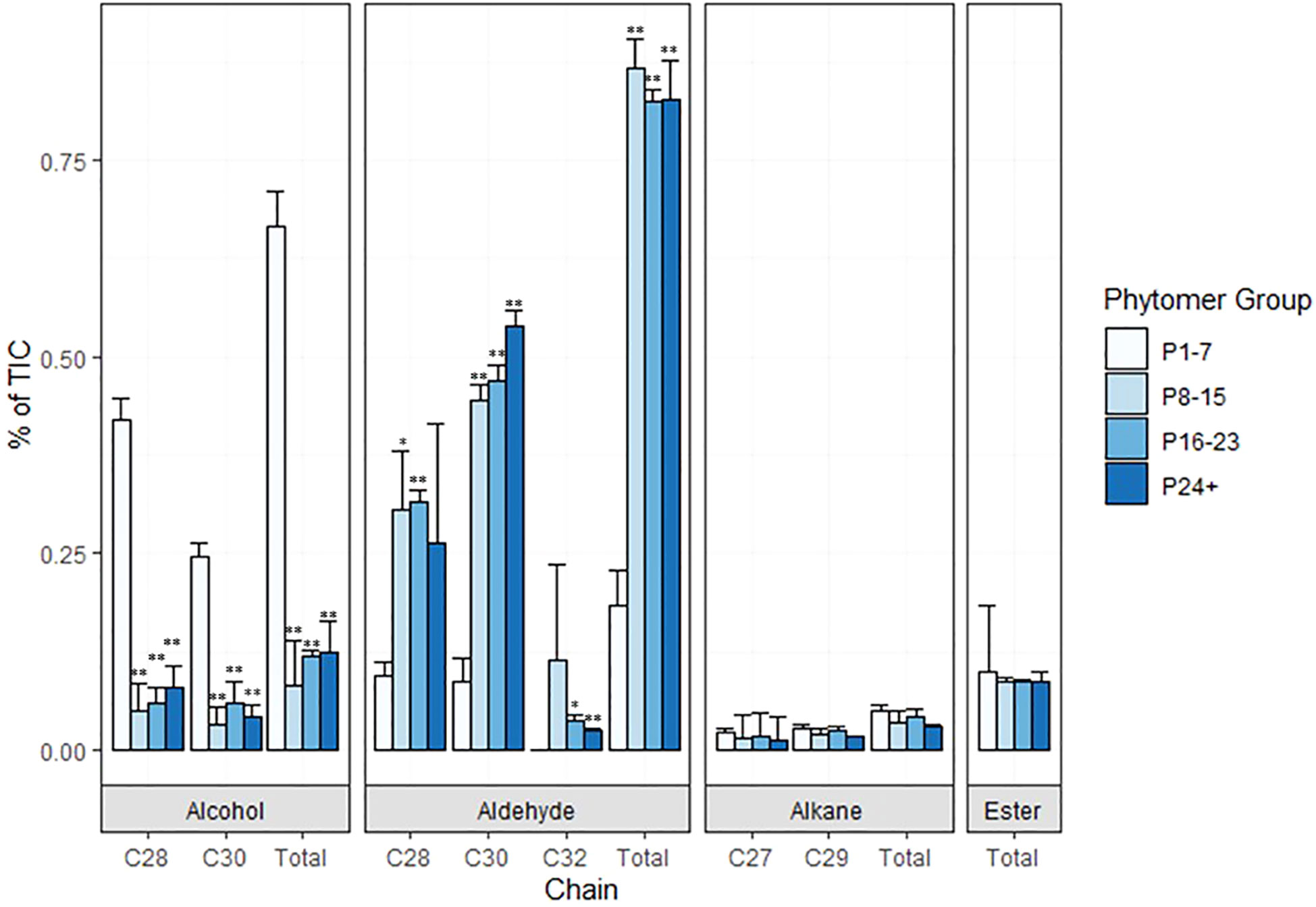
Figure 4 Composition of sorghum stem wax on phytomers of TX08001 120 DAE plants. Wax was extracted from groups of phytomers of increasing age and development (1-7, 8-15, 16-23, and 24+) for composition analysis. The relative abundance of wax components (C28/C30 alcohols, aldehydes, alkanes, esters) extracted from stems of each set of phytomers is represented by bar graphs (P-values designated by asterisks as follows: *<.01, **<.001, P1-7). % of TIC (Total Ion Current) is calculated from area under the peak with local background subtraction.
Sorghum homologs of genes involved in stem wax biosynthesis are annotated in the BTx623 reference genome (Phytozome v3.1) (McCormick et al., 2018). To confirm and refine the annotations, sorghum homologs of validated Arabidopsis and maize wax pathway genes were identified and used as the starting point for phylogenetic analysis. Sorghum homologs that contained the same PFAM domains as the validated wax pathway genes and that had BLAST X score e-values <1e-20 were included in the analysis. The phylogenetic relationships among the sorghum, Arabidopsis, rice, and maize wax pathway homologs were analyzed using Mega-X (Tamura et al., 2011; Kumar et al., 2018; Tamura et al., 2021) (Supplementary Figures 3A–Z). This approach identified sorghum gene homologs encoding long chain acyl-CoA synthetases (LACS1/2/4/9), subunits of the fatty acid elongase (CER6, KCR1, PAS2, ECR/CER10, CER2), the wax alcohol forming pathway (CER4, WSD1), the wax alkane pathway (CER3, CER1, MAH1, WSD1) and wax transporters (CER5/ABCG12, ABCG11, LPTG) (Supplementary Table 1). This method identified Sobic.006G155700 as a wax transporter (WBC11/ABCG11) consistent with sorghum mutant analysis (Mizuno et al., 2013).
One goal of this study was to identify wax pathway genes involved in stem epicuticular wax formation. Therefore, the sorghum homologs of genes involved in wax biosynthesis and transport were analyzed to identify gene family members that were differentially expressed in stem epidermal cells. This was accomplished by comparing the expression of each wax pathway gene family member in sorghum stem internode epidermal cells (Ep) to non-epidermal cell types [i.e., pith parenchyma (PP), xylem fibers (XF), xylem/vascular parenchyma (XP) and the phloem (Ph)] using data derived from LCM-based RNAseq analysis (Fu et al., 2023). The subset of the sorghum wax pathway gene homologs in each gene family that were differentially expressed in stem epidermal cells was identified and targeted for further analysis (Supplementary Table 2). The sorghum triterpene synthase gene (Sobic.008G142400) that produces cuticular triterpenoids (Busta et al., 2021) and the sorghum gene (Sobic.001G228100) encoding a GDSL-lipase required for wax accumulation on the leaf sheath (Jiao et al., 2017) were also differentially expressed in epidermal cells (Supplementary Table 2).
Expression of the sorghum wax pathway genes during stem development was characterized using RNAseq data derived from developing vegetative phase stems of R07020 (Casto et al., 2018). R07020 stem tissue was collected from two apical internode sections (Int#1, Int#2) that had not yet initiated rapid internode elongation, apical to basal sections from Int#3 (i.e., 3-1, 3-2, 3-3, 3-4, 3-5), an internode that was undergoing rapid elongation, and apical to basal sections from Int#4, an internode that was fully elongated (Casto et al., 2018). SbLACS1 was expressed at ~16 TPM in Int#1/2 and at somewhat higher levels in most sections of Int#3 and Int#4 (~29-53 TPM) (Supplementary Table 3). SbLACS4 expression was also relatively high in Int#1-4 (19-87 TPM). In contrast, SbLACS2 and SbLACS9 were expressed at 5-21 TPM in Int#1-3, then at lower levels in the upper older sections of Int#4 (1-5 TPM). SbPAS2 and SbECR were expressed at relatively high levels in Int#1-4 (53-272 TPM, 134-273 TPM respectively) (Supplementary Table 3).
Several genes in the wax pathway showed more extensive variation in expression during stem internode development (Figure 5). For example, expression of SbKCS1, SbKCS6 and SbCER2 was <1 TPM in Int#1/2, then expression increased in the upper more fully developed sections of Int#3 and Int#4. SbKCR1 was expressed at ~2 TPM in Int#1/2 and at 4-fold higher levels in the upper more fully developed portion of Int#4. SbCER3-1, SbWSD1, SbABCG12, SbABC11 had a similar developmental pattern of expression with low expression in Int#1/2 and 5-20-fold higher expression in the fully elongated, older, more developed portions of Int#3/4. Other genes in the alkane and alcohol wax pathways had more complex patterns of expression. For example, SbCER3-2 was highly expressed in Int#1/2 and the basal section of Int#3 that spans the intercalary meristem, but also in the upper more fully developed portion of Int#4 where SbCER3-1 is also highly expressed in conjunction with SbCER1-2. In contrast, SbCER1-1 and SbCER1-2 were expressed at high levels in Int#1/2 and the basal growing zone sections of Int#3 and Int#4 but at lower levels in the upper more developed portions of Int#3/4. SbCER4 was expressed in Int#1/2, throughout most of Int#3, then at the base and top of Int#4 (stem node tissue).
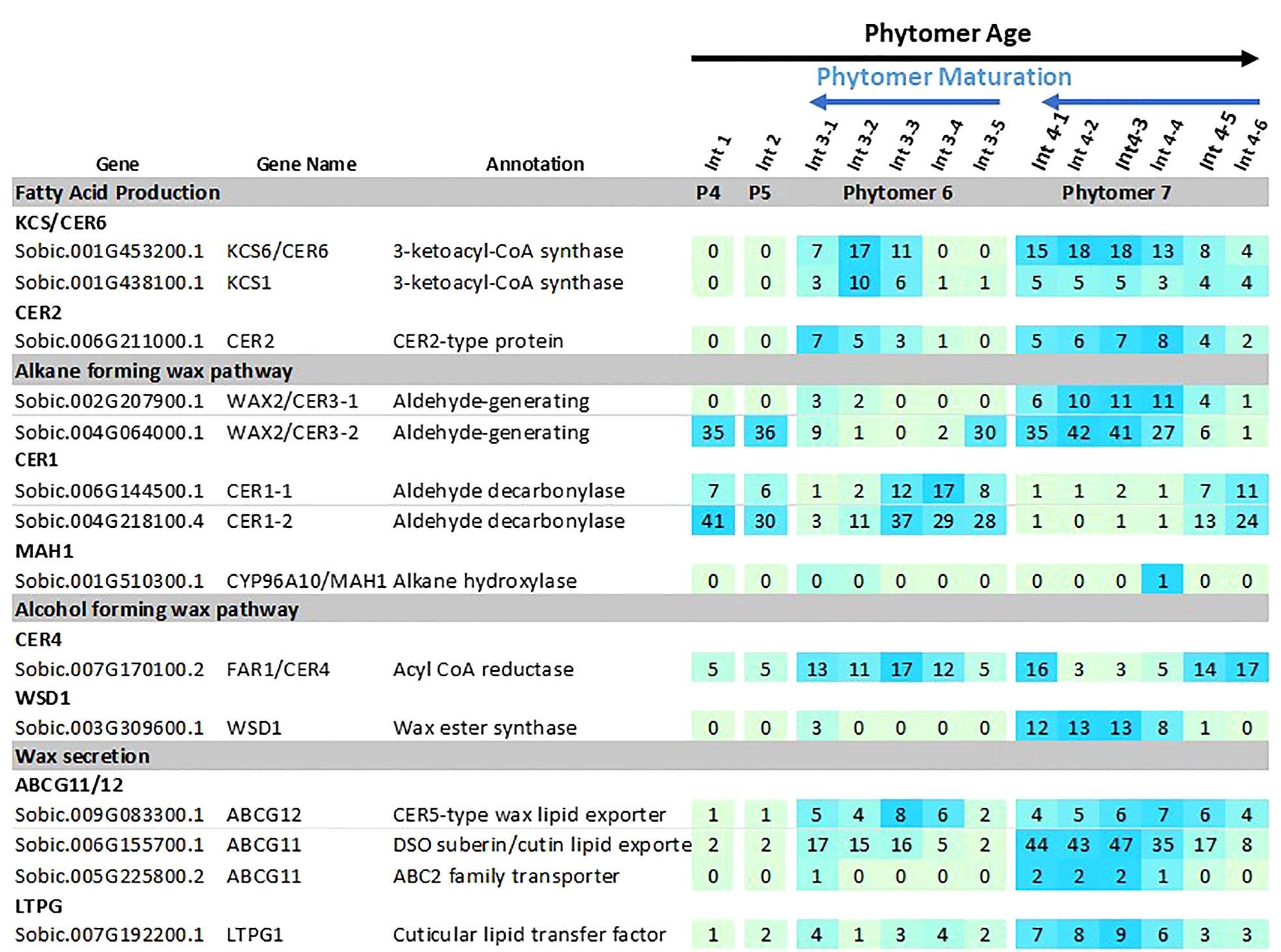
Figure 5 Expression of sorghum wax pathway genes during stem internode development of R07020. Expression (TPM) of wax pathway genes in R07020 stem internodes 1-4 (Casto et al., 2018). Stem internodes 3 and 4 were divided into sections where Int#3-1 and Int#4-1 are from the upper end of the internode and Int3-5 and Int4-6 were from the lower end of the internode, a region spanning the growing zone. Cells are shaded in light green>blue (low to high) scale as relative expression within the data set. Additional information on wax pathway gene expression is found in Supplementary Tables 1–3).
Transcription factors expressed in the stem epidermis that potentially regulate wax pathway gene expression were identified by gene regulatory network analysis (Varala et al., 2018; Arachchilage et al., 2020; Yu et al., 2022). Sorghum stem cell type transcriptome data (Casto et al., 2018) was used to determine that approximately 1,669 genes were differentially expressed at >5-fold higher levels in the stem epidermis compared to other stem cell types (cutoffs = 5 transcripts per million [TPM], 0.05 false discovery rate [FDR]). After applying a Pearson Correlation Coefficient (PCC) threshold of > 0.9, 626 genes with 1416 edges were retained in the network. Sorghum wax pathway genes and genes encoding many transcription factors (TFs) were differentially expressed in sorghum stem epidermal cells. Potential connections between the TFs and wax pathway gene promoters were identified through gene regulatory network (GRN) analysis (Supplementary Figure 4). The sorghum stem epidermis wax pathway GRN included homologs of TFs that have previously been identified as regulators of wax biosynthesis in Arabidopsis (i.e., MYB96/94, WRI1, SHN1, WIN1) (Broun et al., 2004; Kannangara et al., 2007; Seo et al., 2011; Lee and Suh, 2015; Lee et al., 2016; Park et al., 2016). Connections between these TFs and their target genes in the stem wax pathway are shown in Figure 6 (left side). Other TFs (i.e., WRI3, MYS1, MYB30) that are involved directly or indirectly in wax biosynthesis (Raffaele et al., 2008; Park et al., 2016; Liu et al., 2022) were part of more complex sub-networks that had 47 predicted connections to genes in the wax pathway (Figure 6, right). Four sub-networks were tentatively identified that included specific subsets of TFs; (A) WRI3/MYB86/HB16, (B) MYS1/G2-GARP/FMA/VRN1, (C) NAC034, which is potentially regulated by HB16/MYS1/MYB36 and connected to genes encoding MYB30/MYB60/WRKY42, and (D) MYB30/MYB60/EGL3, with predicted regulation by PDF2, NAC034, and FMA. The network of TFs was connected to every gene in the wax pathway except CER1 and LACS9. Most wax pathway genes were regulated by several TFs and CER3-2 (8 connections) and ABCB11-1 (10 connections) were most highly connected to the TF regulatory network. The specific wax pathway genes that could potentially be regulated through each of the four subnetworks were identified (Supplementary Figures 5A–4D). Since MYB-factors play a central role in the sorghum stem wax GRN (MYB30/32/36/60/86/94/96/MYS1) the phylogenetic relationships among the sorghum proteins and Arabidopsis MYB proteins were analyzed using MEGA-X (Supplementary Figure 6) (Singh et al., 2020). This analysis showed that sorghum and Arabidopsis MYB30/60/94/96 clustered in the same clade whereas SbMYB32, SbMYB36, and SbMYB86 clustered with their Arabidopsis homologs in other clades.
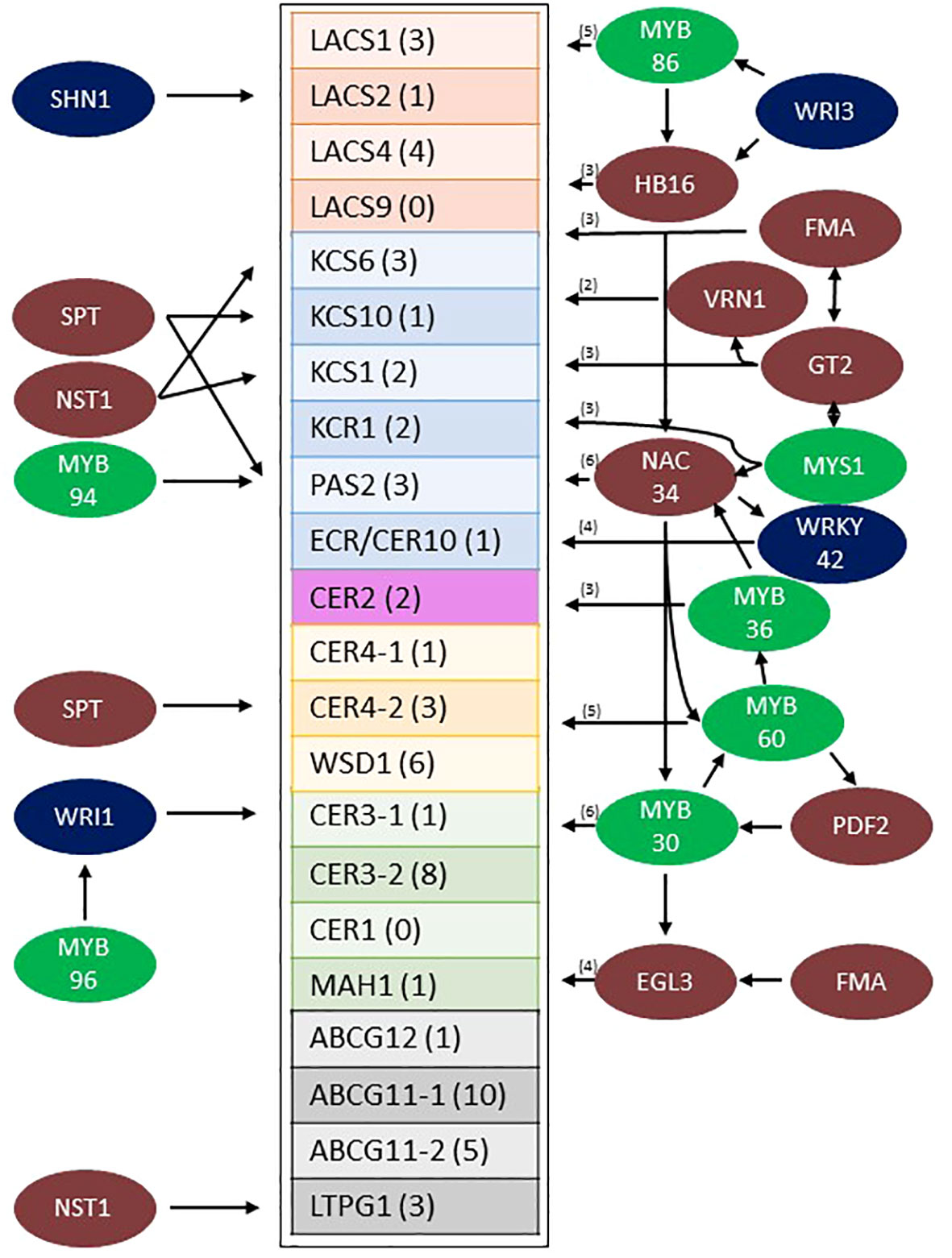
Figure 6 Model of the predicted sorghum stem wax pathway gene regulatory network from Wray LCM data (Fu et al., 2023). Sorghum wax pathway genes that are differentially expressed at higher levels in the stem epidermis are boxed (the number of TF connections to each gene is shown in parentheses). TFs (colored circles, left side) are shown with arrows indicating connections to specific wax pathway genes. TFs with 2-6 predicted connections to wax pathway genes are shown on the right (the number of connections in parentheses). Specific connections between TFs shown to the right and their target wax pathway genes are shown in Supplementary Figures 5A–D. MYB factors are color coded green, previously identified EW associated TFs are colored blue and other TFs are colored brown. EW, Epicuticular Wax; TF, Transcription Factor.
Most prior studies of sorghum wax focused on grain sorghum to better understand the contribution of leaf wax to sorghum’s drought tolerance. Early studies correlated higher leaf blade wax load to reduced non-stomatal transpiration from leaf surfaces (Nobel and Jordan, 1983; Jordan et al., 1984). Sorghum leaf sheath wax has also been characterized in part because the external surface of the sheath has a heavy wax load and diverse wax morphologies (Jenks et al., 1994; Jenks et al., 2000). Grain wax has also been analyzed to determine its potential as a co-product of grain production (Hwang et al., 2002; Xiao et al., 2020). However, sorghum stem wax has been studied to a limited extent most likely because dwarf grain sorghum genotypes have short stems that are covered by leaf sheaths. In contrast, bioenergy sorghum hybrids developed for biomass/bioenergy production have 4-5 m long stems (x 20-30 mm diameter) that account for ~25% of the above ground plant surface area. Stems of bioenergy sorghum plants grow continuously by the sequential addition of >40 apical phytomers and internodes during a long growing season (150-200 days). Following canopy closure, leaf blades and leaf sheaths lower in the canopy senesce exposing wax covered stems to the environment. Exposed stem surfaces accumulate a thick wax plate that often covers internode stomata potentially reducing water loss from internode surfaces and contributing to biotic resistance by blocking stomatal pores. Given the potential importance of stem wax to biotic/abiotic resilience and its utility as a valuable co-product, the current study was undertaken to characterize sorghum stem wax and the regulation of stem wax pathway gene expression.
In Arabidopsis, wax loads on stems (13-32 ug/cm2) are >10-fold higher than on leaf blades (0.7-1.5 ug/cm2) (Suh et al., 2005; Lee and Suh, 2015). In sorghum, leaf blade wax loads have been reported to vary from 2-25 ug/cm2 depending on genotype and stage of development (Nobel and Jordan, 1983; McWhorter et al., 1990; Burow et al., 2008; Burow et al., 2009; Xiao et al., 2020; Busta et al., 2021; Tang et al., 2022). Higher wax loads are reported for sorghum leaf sheaths (~36-206 ug/cm2) (McWhorter et al., 1990; Burow et al., 2008; Burow et al., 2009; Xiao et al., 2020). In the current study, wax loads on the leaf sheath associated with internode #8 of the bioenergy sorghum hybrid TX08001 ranged from 41-51 ug/cm2 between 60-90 DAE. Stems of TX08001 had a wax load of ~100 ug/cm2 at 60 DAE (internode #8), ~197 ug/cm2 at 90 DAE (internode #8) and ~215 ug/cm2 at 120 DAE (internode #23). Therefore, sorghum wax loads are much higher than Arabidopsis, but like Arabidopsis, sorghum stem wax loads are higher than leaf wax loads. Stems are not thought to be large contributors to non-stomatal transpiration because they are typically shaded by leaf blades and often covered by leaf sheaths that have heavy wax loads. This suggests that stem wax may play an important role in biotic stress resistance in addition to reducing water loss from cuticular surfaces. Of the total shoot surface area at bioenergy sorghum harvest, stem surface area accounts for ~24%, leaf sheaths ~30%, and leaf blades ~46%. High wax loads on bioenergy sorghum stem and leaf sheaths could potentially be extracted at biorefineries to provide high value wax co-products (Cominelli et al., 2008). Recovery of wax from harvested bioenergy sorghum stems, leaf sheaths and leaf blades could allow recovery of 100-200 kg of wax per hectare when the crop is planted at a standard planting density of 132,000 plants/ha (Olson et al., 2012). Sugarcane, a related C4 grass, was estimated to accumulate 40-240 kg of wax per hectare assuming average crop yields (Jetter and Kunst, 2008). Sugarcane stem wax contains higher levels of short chain wax components relative to bioenergy sorghum (Inarkar and Lele, 2012).
The current study found that sorghum stem wax on younger more apical internodes (P1-7, Int#1-4) contained high levels of C28/30 primary alcohols (~65%) and lower levels of C28/30 aldehydes (~18%) and esters (~7%). In contrast, stem wax on older sorghum internodes (P8+) was highly enriched in C28/30 aldehydes (~80%) and lower amounts of primary alcohols (6-12%) and esters (~8%). Stem wax on sorghum grain genotypes was also reported to be enriched in C28/30 aldehydes (Xiao et al., 2020). The functional significance of accumulation of C28/30 wax aldehydes during stem development is unknown, however, variation in wax composition during organ development is common. For example, the composition of sorghum leaf wax changes during the juvenile/adult phase transition (Avato et al., 1984; Busta et al., 2021). Sorghum juvenile leaf wax is enriched in C28/C30 primary alcohols (80%) whereas wax on adult phase leaves is a mixture of fatty acids, primary alcohols, aldehydes, alkanes, and triterpenoids (Busta et al., 2021). In maize, an increase in the ratio of wax esters/alkanes during leaf development coincided with changes in the water barrier properties of the cuticle (Bourgault et al., 2020). Variation in wax composition during development, on different organ surfaces and species is extensive (Lee and Suh, 2015). Changes in wax chemistry during organ development and variation among organs and plant species could result in differences in wax morphology (Chambers et al., 1975) and the functional properties of wax. As in Zea mays (Bourgault et al., 2020), we observed a shift from alkanes to primary alcohols and then aldehydes during stem internode epidermal cell development. The variation in wax composition during development were correlated with changes in the morphology of the SEM visualized epicuticular surface from a smooth wax surface, followed by the appearance of wax tubules on the surface, and finally to a surface with tubules and wax plates.
A developmental study of wax accumulation and gene expression in the Arabidopsis stem epidermis was highly successful in identifying genes involved in epicuticular wax biosynthesis (Suh et al., 2005). Therefore, in the current study a similar approach was used to identify sorghum wax pathway genes that contribute to stem wax accumulation. Initially, sorghum homologs of known wax pathway genes were identified using BLAST X, PFAM domain information, and phylogenetic analysis. This approach identified small sorghum gene families for nearly every step in the wax pathway. Wax pathway gene family members that were differentially expressed in stem internode epidermal cells were identified using transcriptome profiles of sorghum stem internode cell types (Fu et al., 2023). Many of the sorghum wax pathway genes identified using this method were homologs of Arabidopsis genes expressed in the stem epidermis (Suh et al., 2005). For example, in Arabidopsis, AtLACS1, AtLACS2 and AtLACS3 were expressed in the stem epidermis. In sorghum, homologs of AtLACS1, AtLACS2, AtLACS4 and AtLACS9 were expressed in the stem epidermis where they could contribute to cutin and wax formation. AtLACS1 and AtLACS2 synthesize long-chain acyl-CoAs for wax and cutin biosynthesis (Lu et al., 2009; Weng et al., 2010) and AtLACS1 and AtLACS9 contribute to the synthesis of tryphine lipids in the pollen coat (Jessen et al., 2011). SbKCS1, SbKCS6 and SbKCS10 are also differentially expressed in the stem epidermis as are the corresponding Arabidopsis homologs (Suh et al., 2005). KCS is considered the rate limiting enzyme in the FAE complex and KCS gene family members influence the type of VLCFAs that are produced (Guo et al., 2016). KCS1, for example, primarily catalyzes the elongation of C20-C26 VLCFAs, whereas KCS6, encoded by the KCS-gene with the highest expression in the sorghum stem epidermis, generates VLCFAs longer than C24, consistent with its role in wax biosynthesis (Trenkamp et al., 2004; Huang et al., 2022). SbKCS10 is a homolog of FIDDLEHEAD, a gene that impacts the development of the adaxial epidermis of rosette leaves and trichomes (Yephremov et al., 1999). Sorghum genes encoding the other subunits of the FAE (SbKCR1, SbPAS2, SbECR) and SbCER2, a gene that increases the production of VLCFA with chain lengths greater than C28 (Tacke et al., 1995; Pascal et al., 2013; Haslam et al., 2015; Xue et al., 2017; Alexander et al., 2020), were also differentially expressed at higher levels in the stem epidermis compared to non-epidermal cells. In addition, another SbKCR1 homolog (Sobic.004G203900) was highly expressed throughout the stem in Int#1-Int#4.
In the wax alkane pathway, CER3 generates aldehydes, CER3:CER1 complexes convert aldehydes to alkanes, and MAH1 converts wax alkanes to secondary alcohols and ketones (Bernard and Joubes, 2013). In Arabidopsis, AtCER3 and AtCER1 were expressed at similar levels in the stem epidermis consistent with accumulation of high levels of C29 wax alkanes (32%) relative to C28/30 aldehydes (5%) (Lee and Suh, 2015). In sorghum two SbCER3 and two SbCER1 homologs were differentially expressed in the sorghum stem epidermis. However, unlike Arabidopsis, SbCER3 expression was 58-fold higher than SbCER1 consistent with elevated accumulation of C28/30 aldehydes in the sorghum stem epidermis relative to wax alkanes. Sorghum stem wax also accumulated only low levels of wax secondary alcohols and ketones consistent with very low expression of the sorghum homolog of AtMAH1, a gene involved in generating secondary wax alcohols and ketones (Greer et al., 2007; Wen and Jetter, 2009). The formation of wax primary alcohols is mediated by an NADH-dependent fatty acyl-coA reductase (FAR) (Kolattukudy, 1971). In Arabidopsis AtCER4/FAR3 mediates wax primary alcohol formation whereas in Triticeae, TaFAR1 encodes the enzyme that mediates production of primary alcohols in the cuticle (Wang et al., 2015). The sorghum homolog (Sobic.005G063300) of AtCER4/FAR3 was expressed at very low levels in the sorghum stem epidermis. In contrast, the sorghum homolog (Sobic.007G170100) of TaFAR1 (Wang et al., 2015) was differentially expressed in the epidermis of sorghum stems consistent with a role in stem wax primary alcohol formation. A sorghum homolog (Sobic.003G309600) of AtWSD1 (Li et al., 2008), ZmWSD1, OsWSD1 and ShWSD1 (Kawahara et al., 2013; Garsmeur et al., 2018; Liu et al., 2021; Li et al., 2022) was differentially expressed in the stem epidermis. Another SbWSD1 homolog (Sobic.009G226600) that was most closely related to the grass WSD1 genes was not differentially expressed in stem epidermal cells. The homolog discovery approach employed also led to the identification of sorghum homologs of ABCG12/11 cutin/wax transporters and LTPG that are differentially expressed at higher levels in the stem epidermis.
SEM analysis of stem surfaces showed minimal wax accumulation on Int#1/2, young non-elongated stem segments located just below the stem apex. SEM analysis first detected wax accumulation on the upper portion of Int#3, a region of the internode that contains fully elongated cells, but not on the lower portion of internode 3 that spans the intercalary meristem and zone of elongation (Kebrom et al., 2017; Yu et al., 2022). A similar distribution of wax accumulation was observed on internode #4, although larger wax deposits were present on the upper portion of this internode, and wax was visible on a large portion of the internode. Wax on Int#6 and Int#25 was more profuse and wax tubules completely covered the surface of these internodes. Expression of SbKCS6, SbKSC1, SbCER3-1, SbWSD1 and SbABCG12/11-1 was low in Int#1/2 followed by increased expression in the upper portion of Int#3 and Int#4, a pattern of wax pathway gene expression correlated with the appearance of wax on the surface of internodes. In contrast, SbCER1-1 and SbCER1-2 were expressed at higher levels in Int#1 and Int#2 and the lower portion (growing zone) of Int#3 and Int#4 during development. SbCER3-2 also showed relatively high expression in Int#1, Int#2, and the lower portion of Int#3. This suggests that SbCER3-2 and SbCER1-1/1-2 could generate alkanes that accumulate on Int#1/2 prior to the onset of internode elongation and in the growing zone of Int#3/4. Preliminary MS data showing the presence of alkanes on Int#1 and Int#2 supports this hypothesis and Int#4 during development (Supplementary Figure 8). SbCER3-2 also showed relatively high expression in Int#1, Int#2, and the lower portion of Int#3. There was no visible epicuticular wax accumulation on these nascent internodes suggesting alkane may be a component of intracuticular wax. SbCER4 was expressed in Int#2/3, then increased in Int#3 followed by decreased expression in Int#4 but with retention of expression in nodal tissues (pulvinus, nodal plexus). SbCER4 expression in Int#1-3 is consistent with accumulation of primary alcohols in wax early in internode development. The induction of SbCER3-2 expression (but not SbCER1) in the upper portion of Int#4 could contribute to the generation of C28/30 wax aldehydes that become the predominant component of sorghum stem wax on fully developed stem internodes. Taken together, the timing of elevated expression of SbCER4 before high levels of SbCER3-2 during internode development is consistent with early accumulation of wax alcohols followed by wax aldehydes (Figure 4; Supplementary Figure #). Assessment of the impact of continued expression of SbCER1 and SbCER4 expression in stem nodes will require targeted analysis of wax composition on this stem tissue to determine if wax alkanes and primary alcohols accumulate preferentially on stem nodal tissues.
Several TFs that regulate wax biosynthesis have been identified in Arabidopsis (i.e., SHN1/WIN1, WRI4, MYB30/94/96, MYS1) (Aharoni et al., 2004; Kannangara et al., 2007; Bernard and Joubes, 2013; Liu et al., 2022). Sorghum homologs of known wax pathway TFs were identified through phylogenetic analysis and then screened to identify gene family members with high expression in the stem epidermis. This approach identified sorghum homologs of several TFs that are known to regulate cuticle/epidermal cell development (i.e., SbPDF2, SbHDG1, SbEGL3) (Bernhardt et al., 2003; Nakamura et al., 2006; Singh et al., 2020; Nagata and Abe, 2023) and/or biosynthesis of epicuticular wax (i.e., SbSHN1.WIN1, SbMYB30/94/96, SbWRI1, SbWRI3, SbMYS1). Gene regulatory network analysis of genes co-expressed in wax producing stem epidermal cells identified predicted TF binding sites in the promoters of genes in the sorghum stem wax pathway as well as among TFs in the network. For example, SbSHN1/WIN1 was differentially expressed in stem epidermal cells and GRN analysis predicted SbSHN1.WIN1 could regulate the expression of SbLACS2, consistent with studies showing AtWIN1 regulates the expression of AtLACS2 (Kannangara et al., 2007). A different member of the sorghum SHN/WIN gene family, SbWINL is highly expressed in leaves, and when over-expressed in Arabidopsis wax crystals accumulated (Bao et al., 2016). Arabidopsis WRINKLED (WRI) TFs regulate tissue specific fatty acid biosynthesis (To et al., 2012) and AtWRI4 was found to activate wax biosynthesis in stems (Park et al., 2016). In the current study, SbWRI1 and SbWRI3 were differentially expressed in the stem epidermis and predicted to activate CER3 (Sobic.002G207900) and SbMYB86/SbHB16, TFs that have predicted connections to eight genes in the stem wax pathway. In Arabidopsis, AtMYB86 is involved in secondary cell wall formation (Taylor-Teeples et al., 2015); in tea plants, light regulated flavonoid synthesis (Ye et al., 2021); and in black cottonwood with xylem formation (Wessels et al., 2019). SbMYB86 was not identified in the sorghum stem secondary cell wall gene regulatory network (Fu et al., 2023), however, a role in coordinating wax and flavonoid synthesis in sorghum stem epidermal cells is possible. SbMYB86 was also connected to SbGlossy6 (Sobic.008G131299), a gene implicated in wax transport (Li L. et al., 2019). SbGlossy6 is a homolog of ZmGlossy6, a gene that encodes a DUF538 containing protein that influences wax secretion (Li L. et al., 2019). The Arabidopsis Glossy6 homologs AtSVB and AtSVB2 (ZmGlossy6 homologs) are modulated by ABA and involved in trichome formation (Hussain et al., 2021).
AtMYB94/96 induce the expression of genes involved in cuticular wax biosynthesis and activate wax accumulation under conditions of water deficit (Koch and Ensikat, 2008; Lee and Suh, 2015; Lee et al., 2016). In the current study, SbMYB94 and SbMYB96 were differentially expressed in the sorghum stem epidermis and network analysis indicated these TFs could regulate PAS2 and WRI1 expression. Further studies will be needed to determine if SbMYB94/96 play a role in increasing stem wax accumulation in response to drought stress. Phylogenetic analysis showed that SbMYB30 is closely related to SbMYB94/96. SbMYB30 was differentially expressed in the sorghum stem epidermis and GRN analysis predicted interactions with the promoters of six genes in the wax pathway and SbGlossy6. In Arabidopsis, AtMYB30 regulates VLCFA and wax biosynthesis (Raffaele et al., 2008) and also modulates ROS-signaling, PIF-signaling, and hypersensitive responses (Raffaele et al., 2008; Fichman and Mittler, 2020; Yan et al., 2020). GRN analysis predicted SbMYB30 expression could be regulated by SbPDF2, which could contribute to differential expression in the sorghum stem epidermis. The corresponding AtPDF2 homolog (AT4G04890) is also differentially expressed in the Arabidopsis stem epidermis (Suh et al., 2005) and is known to help specify shoot epidermal cell differentiation (Ogawa et al., 2015). GRN analysis predicted that SbMYB30 could regulate SbMYB60 and SbEGL3 expression and that SbMYB60 could potentially regulate the expression of 5 genes and SbELG3 could potentially regulate the expression of four genes in the wax pathway. In Arabidopsis, AtMYB60 is expressed in stomatal guard cells (Rusconi et al., 2013) and represses anthocyanin biosynthesis (Park et al., 2008). In sorghum SbMYB60 regulates secondary cell wall formation (Scully ED et al., 2016; Hennet et al., 2020). If SbMYB60 expression occurs in sorghum epidermal guard cells, the results suggest that SbMYB60 could modulate wax biosynthesis in that cell type in addition to its other functions. SbEGL3 is a homolog of AtELG3, a gene involved in trichome development (Hung et al., 2020), so it is possible that SbEGL3 modulates wax pathway expression in this specialized epidermal cell type. In Arabidopsis leaves, AtMYS1/2 regulates wax production by repressing DEWAX (Liu et al., 2021), a diel regulated gene that represses wax synthesis in conjunction with SPL9 (Li RJ. et al., 2019). SbMYS1 was expressed in sorghum stem epidermal cells, but SbDEWAX expression was not detected. GRN analysis indicated that SbMYS1 could interact with the promoters of three genes in the epicuticular wax biosynthesis pathway (SbCER3-2, SbABCG11-1/WBC11, SbABCG11-2). SbMYS1 also was predicted to interact with two TFs, SbNAC034, a gene with predicted connections to 6 genes in the wax pathway, and SbG2-GARP which encodes a TF with three connections to wax pathway genes. In addition, sorghum homologs of AtNST1, a gene involved in secondary cell wall formation/lignin biosynthesis (Scully ED et al., 2016; Hennet et al., 2020) and AtMYB36, a gene involved in suberin biosynthesis in root casparian strips in Arabidopsis, were differentially expressed in the sorghum stem epidermis and connected to the wax GRN. Sorghum epidermal cells accumulate secondary cell walls (Fu et al., 2023) and suberin requires synthesis of VLCFA, so there may be regulatory connections between these co-expressed pathways in stem epidermal cells.
The gene regulatory network analysis predictions reported here merit extensive testing using DAPseq, CHIPseq, and other methods to validate the predictions. The analysis could also be refined by collecting transcriptome profiles from specific cell types that comprise the sorghum stem epidermis (i.e., guard cells, silica cells, cork cells, long cells). The current study provides a starting point for an analysis of the regulation of wax pathway expression during sorghum stem development, the basis of sorghum’s heavy wax loads on stems relative to other plant species and among sorghum genotypes, and on different sorghum organ surfaces (i.e., stem wax, adaxial/abaxial leaf and sheath surfaces).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Table 6.
RC conducted all experiments and analysis. RC and BM contributed to the bioinformatics experiments. SF contributed machine time for initial MS experiments and training. All authors (RC, BM, SF, and JM) contributed to writing the manuscript.
This work was funded by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Science Grant/Award: DE-SC0018409).
The authors thank Kerrie Barry and the Joint Genome Institute for contributing to RNA-seq analysis. The work (proposal: 10.46936/10.25585/60001026 and 10.46936/10.25585/60000987) conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, supported by the Office of Science of the US Department of Energy operated under contract no. DE-AC02-05CH11231. The authors acknowledge the Texas A&M Microscopy and Imaging Center’s help with microscopic analyses. High temperature mass spectrometry was performed in collaboration with Amit Gujar and Kenneth Free at Thermo Fisher Scientific in Austin, Texas. Additional mass spectrometry validation was done with machine time donated by Tim Devarenne. Supplemental Figure 1 is an artist rendering of the developing phytomers (as 3 precursors to the first internode) by Veronica Chemelewski.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1227859/full#supplementary-material
Supplementary Figure 1 | Sorghum stem development diagram. The youngest phytomers are located immediately below the shoot apex (i.e., Phytomers 1-3, Nascent Leaf) with older and more developed phytomers are located further from the stem apex (i.e., Phytomer 7) (right, phytomer developmental arrow). Internodes associated with each phytomer are comprised of three stem tissues, the nodal plexus, internode and pulvinus. Internodes (Int), the last tissue produced during phytomer development, is formed by the action of an intercalary meristem (IM) (cell division), followed by cell elongation above the IM, and then cell maturation (accumulation of secondary cell walls). The locations where stem samples for SEM analysis (, , ) were taken is shown to the right of the stem diagram. SEM samples shown in were taken from older internodes (Int8, Int25) not shown in Supplementary Figure 1.
Supplementary Figure 2 | Representative GC-MS spectra used to generate wax composition data that are shown in. (A) GC-MS profiles of wax extracted from internodes associated with phytomers 1-7 (P1-7), P8-15, P16-23, and P24+. (B) Portion of the GC-MS spectra showing wax esters including Carnauba wax as a reference.
Supplementary Figure 3 | Phylogenetic analysis of sorghum wax pathway gene homologs (e-values <-20). (A) Bootstrap (BS) Arabidopsis E-Score and BS values for Arabidopsis and Zea mays. Values contained in [] is the BS value without added Zea mays genes, † delineates predictive Zea mays gene in literature by expression, ‡ delineates predictive gene from Saccharum officinarum, () delineates which supplemental figure panel specifies the tree for the gene family. Green highlighted genes designate where single candidates were selected and are green highlighted in the associated tree. (B) Phylogenetic tree of CER6 gene family. (C) Phylogenetic tree of KCR1 gene family. (D) Phylogenetic tree of PAS2 gene family. (E) Phylogenetic tree of CER10 gene family. (F) Phylogenetic tree of CER2 gene family. (G) Phylogenetic tree of LACS1 gene family. (H) Phylogenetic tree of CER1&3 gene family. (I) Phylogenetic tree of MAH1 gene family. (J) Phylogenetic tree of CER4 gene family. (K) Phylogenetic tree of WSD1 gene family. (L) Phylogenetic tree of WSD1 gene family that includes Saccharum officinarum. (M) Phylogenetic tree of ABCG11&12 gene family. (N) Phylogenetic tree of LTPG1 gene family.
Supplementary Figure 4 | Cytoscape figure of the wax pathway gene regulatory network (GRN). Transcription factor (blue nodes) connections to target gene promoters are denoted by lines. Selected genes in the wax pathway are noted.
Supplementary Figure 5 | (A-D): Sub-networks of the wax pathway GRN showing connections between TFs and wax pathway genes. The four networks contain the following TFs: (A) WRI3/MYB86/HB16/NAC34, (B) GT2/VRN1/EGL3/FMA/MYS1, (C) NAC34/WRKY42/MYB30/HB16/MYS1/MYB36/MYB60, (D) MYB30/MYB60/EGL3/NAC34/PDF2. All depicted connections meet the threshold of PCC>.9 and FDR<.05.
Supplementary Figure 6 | Phylogenetic analysis of sorghum MYB factors. Same color arrows denotes homologous Arabidopsis and Sorghum MYB factors. Arabidopsis MYB factors MYB30, MYB32, MYB36, MYB60, MYB60-2, MYB86, MYB94 and MYB96 were used in the analysis.
Supplementary Figure 7 | Unedited SEM (no contrast adjustment).
Supplementary Figure 8 | GC-MS of BSTFA derivatized wax extracted from sorghum stems on DB-5MS. (A) Internode 2, (B) Internode 3, (C) Internode 4, (D) Internode 25. Red arrows indicate C28/30 wax alcohols and blue arrows point to the derivatized C28/30 aldehydes. Purple arrow indicates Nonacosane.
Supplementary Figure 9 | Bargraph of selected genes from Supplementary Table 3. (A) CER6 (Sobic.001G453200.1) & (B) WSD1 (Sobic.003G309600.1) the bargraphs on the left show the TPM as a function of internode number and position. The bargraphs on the right show core vs rind expression of the gene within the Internode #4.
Supplementary Table 1 | List of sorghum gene family homologs of wax biosynthetic genes identified in other species. Genes that are differentially expressed in the stem epidermis are highlighted in green. The sorghum to Arabidopsis gene Blast X E-values are listed. The fold change of RNA expression in rind/core of internode 4 of R07020 (red means additive expression, due to no core expression). Gene expression (TPM) of wax pathway genes in stem cell types (epidermis, pith, xylem, vascular bundle parenchyma, phloem, fibers) is shown to the right (based on LCM analysis, see methods) (Casto et al., 2018; Fu et al., 2023).
Supplementary Table 2 | List of sorghum gene homologs of wax biosynthetic genes that are differentially expressed in the stem epidermis (Fu et al., 2023).
Supplementary Table 3 | Expression of sorghum homologs of wax pathway genes during stem development. Relative gene expression (TPM) in internode 1 (Int1), internode 2 (Int2), sections of internode 3 (3-1 (top) to 3-5 (lower portion of Int3) and internode 4 (4-1 to 4-6). Intensity of blue highlighting indicates relative expression. The transcriptomic data for this analysis was generated in a previous study (Casto et al., 2018).
Supplementary Table 4 | GRN analysis identifying first tier connections between transcription factors and the wax biosynthetic genes that are predicted to interact with. Grey boxes delineate wax biosynthetic gene families, a blue box designates a predicted wax biosynthetic gene, with the white boxes are TFs with a PCC of.95 to the biosynthetic gene.
Supplementary Table 5 | Table showing predicted interactions between TFs that are part of the wax pathway GRN (listed at the top of the Table) and wax pathway genes (on the left) that the TFs are connected to (blue highlighted boxes indicate a predicted interaction).
Supplementary Table 6 | SRA information used in this study. (A) R07020 SRA information (Casto et al., 2018). (B) LCM SRA information (Fu et al., 2023).
Aharoni, A., Dixit, S., Jetter, R., Thoenes, E., Arkel van, G., Pereira, A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16 (9), 2463–2480. doi: 10.1105/tpc.104.022897
Alexander, L. E., Okazaki, Y., Schelling, M. A., Davis, A., Zheng, X., Rizhsky, L., et al, et al. (2020). Maize glossy2 and glossy2-like genes have overlapping and distinct functions in cuticular lipid deposition. Plant Physiol. 183 (3), 840–853. doi: 10.1104/pp.20.00241
Arachchilage, M. H., Mullet, J. E., Marshall-Colon, A. (2020). Sorghum bicolor cultivars have divergent and dynamic gene regulatory networks that control the temporal expression of genes in stem tissue. bioRxiv. doi: 10.1101/2020.06.17.158048
Avato, P., Giorgio, B., Mariani, G. (1984). Epicuticular waxes of Sorghum and some compositional changes with plant age. Phytochemistry 23 (12), 2843–2846. doi: 10.1016/0031-9422(84)83026-5
Awika, H. O., Hays, D. B., Mullet, J. E., Rooney, W. L., Weer, B. D. (2017). QTL mapping and loci dissection for leaf epicuticular wax load and canopy temperature depression and their association with QTL for staygreen in Sorghum bicolor under stress. Euphytica 213 (9). doi: 10.1007/s10681-017-1990-5
Bao, S.-G., Shi, J.-X., Luo, F., Ding, B., Hao, J.-Y., Xie, X.-D., et al. (2016). Overexpression of Sorghum WINL1 gene confers drought tolerance in Arabidopsis thaliana through the regulation of cuticular biosynthesis. Plant Cell Tissue Organ Culture (PCTOC) 128 (2), 347–356. doi: 10.1007/s11240-016-1114-2
Batsale, M., Bahammou, D., Fouillen, L., Mongrand, S., Joubes, J., Domergue, F. (2021). Biosynthesis and functions of very-long-chain fatty acids in the responses of plants to abiotic and biotic stresses. Cells 10 (6), 1284. doi: 10.3390/cells10061284
Bernard, A., Joubes, J. (2013). Arabidopsis cuticular waxes: advances in synthesis, export and regulation. Prog. Lipid Res. 52 (1), 110–129. doi: 10.1016/j.plipres.2012.10.002
Bernays, E. (1972). Changes in the first instar cuticle of Schistocerca gregaria before and associated with hatchin. J. Insect Physiol. 18 (5), 897–912. doi: 10.1016/0022-1910(72)90028-5
Bernhardt, C., Lee, M.M., Gonzalez, A., Zhang, F., Lloyd, A., Schiefelbein, J. (2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130 (26), 6431–6439. doi: 10.1242/dev.00880
Bourgault, R., Vasquez, M., Qiao, P., Sonntag, A., Charlebois, C., Mohammadi, M., et al. (2020). Constructing functional cuticles: analysis of relationships between cuticle lipid composition, ultrastructure and water barrier function in developing adult maize leaves. Ann. Bot. 125 (1), 79–91. doi: 10.1093/aob/mcz143
Boyles, R. E., Brenton, Z. W., Kresovich, S. (2019). Genetic and genomic resources of sorghum to connect genotype with phenotype in contrasting environments. Plant J. 97 (1), 19–39. doi: 10.1111/tpj.14113
Broun, P., Poindexter, P., Osborne, E., Jiang, C. Z., Riechmann, J. L. (2004). WIN1, a transcriptional activator of epidermal waxaccumulation inArabidopsis. PNAS 101 (13), 4706–4711. doi: 10.1073/pnas.0305574101
Burow, G. B., Franks, C.D., Acosta-Martinez, V., Xin, Z. (2009). Molecular mapping and characterization of BLMC, a locus for profuse wax (bloom) and enhanced cuticular features of Sorghum (Sorghum bicolor (L.) Moench.). Theor. Appl. Genet. 118 (3), 423–431. doi: 10.1007/s00122-008-0908-y
Burow, G. B., Franks, C. D., Xin, Z. (2008). Genetic and physiological analysis of an irradiated bloomless mutant (Epicuticular wax mutant) of sorghum. Crop Sci. 48 (1), 41–48. doi: 10.2135/cropsci2007.02.0119
Buschhaus, C., Jetter, R. (2011). Composition differences between epicuticular and intracuticular wax substructures: how do plants seal their epidermal surfaces? J. Exp. Bot. 62 (3), 841–853. doi: 10.1093/jxb/erq366
Busta, L., Hegebarth, D., Kroc, E., Jetter, R. (2017). Changes in cuticular wax coverage and composition on developing Arabidopsis leaves are influenced by wax biosynthesis gene expression levels and trichome density. Planta 245 (2), 297–311. doi: 10.1007/s00425-016-2603-6
Busta, L., Schmitz, E., Kosma, D. K., Schnable, J. C., Cahoon, E. B. (2021). A co-opted steroid synthesis gene, maintained in sorghum but not maize, is associated with a divergence in leaf wax chemistry. Proc. Natl. Acad. Sci. U. S. A. 118 (12), e2022982118. doi: 10.1073/pnas.2022982118
Cardona, J. B., Grover, S., Busta, L., Sattler, S. E., Louis, J. (2022). Sorghum cuticular waxes influence host plant selection by aphids. Planta 257 (1), 22. doi: 10.1007/s00425-022-04046-3
Casto, A. L., McKinley, B. A., Yu, K. M. J., Rooney, W. L., J., Mullet, E. (2018). Sorghum stem aerenchyma formation is regulated by SbNAC_D during internode development. Plant Direct 2 (11), e00085. doi: 10.1002/pld3.85
Chambers, T. C., Ritchie, I. M., Booth, M. A. (1975). Chemical models for plant wax morphogenesis. New Phytol. 77 (1), 43–49. doi: 10.1111/j.1469-8137.1976.tb01499.x
Chow, C. N., Lee, T. Y., Hung, Y. C., Li, G. Z., Tseng, K. C., Liu, Y. H., et al. (2019). PlantPAN3.0: a new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 47 (D1), D1155–D1163. doi: 10.1093/nar/gky1081
Cominelli, E., Sala, T., Calvi, D., Gusmaroli, G., Tonelli, C. (2008). Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J. 53 (1), 53–64. doi: 10.1111/j.1365-313X.2007.03310.x
Elango, D., Xue, W., Chopra, S., et al. (2020). Genome wide association mapping of epi-cuticular wax genes in Sorghum bicolor. Physiol. Mol. Biol. Plants 26 (8), 1727–1737. doi: 10.1007/s12298-020-00848-5
Fichman, Y., Mittler, R. (2020). Rapid systemic signaling during abiotic and biotic stresses: is the ROS wave master of all trades? Plant J. 102 (5), 887–896. doi: 10.1111/tpj.14685
Fu, J., McKinley, B., James, B., Chrisler, W., Markillie, L. M., Gaffrey, M. J., et al. (2023). The stem cell-type transcriptome of bioenergy sorghum reveals the spatial regulation of secondary cell wall networks. bioRxiv. doi: 10.1101/2023.04.22.537921
Garsmeur, O., Droc, G., Antonise, R., Grimwood, J., Potier, B., Aitken, K., et al. (2018). A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat. Commun. 9 (1), 2638. doi: 10.1038/s41467-018-05051-5
Greer, S., Wen, M., Bird, D., Wu, X., Samuels, L., Kunst, L., et al. (2007). The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol. 145 (3), 653–667. doi: 10.1104/pp.107.107300
Guo, H. S., Zhang, Y. M., Sun, X. Q., Li, M. M., Hang, Y. Y., Xue, J. Y., et al. (2016). Evolution of the KCS gene family in plants: the history of gene duplication, sub/neofunctionalization and redundancy. Mol. Genet. Genomics 291 (2), 739–752. doi: 10.1007/s00438-015-1142-3
Harron, A. F., Powell, M. J., Nunez, A., Moreau, R. A. (2017). Analysis of sorghum wax and carnauba wax by reversed phase liquid chromatography mass spectrometry. Ind. Crops Prod. 98, 116–129. doi: 10.1016/j.indcrop.2016.09.015
Haslam, T. M., Gerelle, W. K., Graham, S. W., Kunst, L. (2017). The unique role of the ECERIFERUM2-LIKE clade of the BAHD acyltransferase superfamily in cuticular wax metabolism. Plants (Basel) 6 (2), 23. doi: 10.3390/plants6020023
Haslam, T. M., Haslam, R., Thoraval, D., Pascal, S., Delude, C., Domergue, F., et al. (2015). ECERIFERUM2-LIKE proteins have unique biochemical and physiological functions in very-long-chain fatty acid elongation. Plant Physiol. 167 (3), 682–692. doi: 10.1104/pp.114.253195
Hennet, L., Berger, A., Trabanco, N., Ricciuti, E., Dufayard, J.-F., Bocs, S., et al. (2020). Transcriptional regulation of sorghum stem composition: key players identified through co-expression gene network and comparative genomics analyses. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00224
Hooker, T. S., Lam, P., Zheng, H., Kunst, L. (2007). A core subunit of the RNA-processing/degrading exosome specifically influences cuticular wax biosynthesis in Arabidopsis. Plant Cell 19 (3), 904–913. doi: 10.1105/tpc.106.049304
Huang, H., Ayaz, A., Zheng, M., Yang, X., Zaman, W., Zhao, H., et al. (2022). ArabidopsisKCS5 and KCS6 play redundant roles in wax synthesis. Int. J. Mol. Sci. 23 (8), 4450. doi: 10.3390/ijms23084450
Hung, F. Y., Chen, J. H., Feng, Y. R., Lai, Y. C., Yang, S., Wu, K. (2020). Arabidopsis JMJ29 is involved in trichome development by regulating the core trichome initiation gene GLABRA3. Plant J. 103 (5), 1735–1743. doi: 10.1111/tpj.14858
Hussain, S., Zhang, N., Wang, W., Ahmed, S., Cheng, Y., Chen, S., et al. (2021). Involvement of ABA responsive SVB genes in the regulation of trichome formation in arabidopsis. Int. J. Mol. Sci. 22 (13), 6790. doi: 10.3390/ijms22136790
Hwang, K., Weller, C. L., Hanna, M. A. (2002). HPLC of grain sorghum wax classes highlighting seperation of aldehydes from wax esters and steryl esters. J. Separation Sci. 25 (9), 619–623. doi: 10.1002/1615-9314(20020601)25:9<619::AID-JSSC619>3.0.CO;2-J
Inarkar, M. B., Lele, S. S. (2012). Extraction and characterization of sugarcane peel wax. ISRN Agron. 2012, 1–6. doi: 10.5402/2012/340158
Jenks, M. A., Rich, P. J., Rhodes, D., Ashworth, E. N., Axtell, J. D., Ding, C. K. (2000). Leaf sheath cuticular waxes on bloomless and sparse-bloom mutants of Sorghum bicolor. Phytochemistry 54 (6), 577–584. doi: 10.1016/S0031-9422(00)00153-9
Jenks, M. A., Joly, R. J., Peters, P. J., Rich, P. J., Axtell, J. D., Ashworth, E. N. (1994). Chemically induced cuticle mutation affecting epidermal conductance to water vapor and disease susceptibility in sorghum bicolor (L.) moench. Plant Physiol. 105 (4), 1239–1245. doi: 10.1104/pp.105.4.1239
Jenks, M. A. R., Peters, P. J., Axtell, J. D., Ashworth, E. N. (1992). Epicuticular wax morphology of bloomless (bm) mutants in sorghum bicolor. Int. J. Plant Sci. 153 (3), 311–319. doi: 10.1086/297034
Jessen, D., Olbrich, A., Knufer, J., Kruger, A., Hoppert, M., Polle, A., et al. (2011). Combined activity of LACS1 and LACS4 is required for proper pollen coat formation in Arabidopsis. Plant J. 68 (4), 715–726. doi: 10.1111/j.1365-313X.2011.04722.x
Jetter, R., Kunst, L. (2008). Plant surface lipid biosynthetic pathways and their utility for metabolic engineering of waxes and hydrocarbon biofuels. Plant J. 54 (4), 670–683. doi: 10.1111/j.1365-313X.2008.03467.x
Jiao, Y., Burow, G., Gladman, N., Acosta-Martinez, V., Chen, J., Burke, J., et al. (2017). Efficient Identification of Causal Mutations through Sequencing of Bulked F (2) from Two Allelic Bloomless Mutants of Sorghum bicolor. Front. Plant Sci. 8, 2267. doi: 10.3389/fpls.2017.02267
Jordan, W. R., Blum, A., Miller, F. R., Monk, R. L. (1984). Environmental physiology of sorghum. II. Epicuticular wax load and cuticular transpiration. Crop Sci. 24 (6), 1168–1173. doi: 10.2135/cropsci1984.0011183X002400060038x
Kannangara, R., Burow, G., Gladman, N., Acosta-Martinez, V., Chen, J., Burke, J., et al. (2007). The transcription factor WIN1/SHN1 regulates Cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19 (4), 1278–1294. doi: 10.1105/tpc.106.047076
Kawahara, Y., de la Bastide, M., Hamilton, J. P., Kanamori, H., McCombie, W. R., Ouyang, S., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (NY) 6, 4. doi: 10.1186/1939-8433-6-4
Kebrom, T. H., McKinley, B., Mullet, J. E. (2017). Dynamics of gene expression during development and expansion of vegetative stem internodes of bioenergy sorghum. Biotechnol. Biofuels 10, 159. doi: 10.1186/s13068-017-0848-3
Klein, J., Saedler, H., Huijser, P. (1996). A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 250, 7–16. doi: 10.1007/BF02191820
Koch, K., Ensikat, H. J. (2008). The hydrophobic coatings of plant surfaces: epicuticular wax crystals and their morphologies, crystallinity and molecular self-assembly. Micron 39 (7), 759–772. doi: 10.1016/j.micron.2007.11.010
Kolattukudy, P. (1971). Enzymatic Synthesis of fatty alcohols in Brassica oleracea. Arch. Biochem. Biophys. 142 (2), 701–709. doi: 10.1016/0003-9861(71)90536-4
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi: 10.1093/molbev/msy096
Lee, S. B., Kim, H. U., Suh, M. C. (2016). MYB94 and MYB96 additively activate cuticular wax biosynthesis in arabidopsis. Plant Cell Physiol. 57 (11), 2300–2311. doi: 10.1093/pcp/pcw147
Lee, S. B., Suh, M. C. (2015). Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species. Plant Cell Rep. 34 (4), 557–572. doi: 10.1007/s00299-015-1772-2
Lewandowska, M., Keyl, A., Feussner, I. (2020). Wax biosynthesis in response to danger: its regulation upon abiotic and biotic stress. New Phytol. 227 (3), 698–713. doi: 10.1111/nph.16571
Li, F., Wu, X., Lam, P., Bird, D., Zheng, H., Samuels, L., et al. (2008). Identification of the wax ester synthase/acyl-coenzyme A: diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol. 148 (1), 97–107. doi: 10.1104/pp.108.123471
Li, R. J., Li, L. M., Liu, X.L., Kim, J.C., Jenks, M. A., Lu, S., et al. (2019). Diurnal regulation of plant epidermal wax synthesis through antagonistic roles of the transcription factors SPL9 and DEWAX. Plant Cell 31 (11), 2711–2733. doi: 10.1105/tpc.19.00233
Li, L., Du, Y., He, C. C., Dietrich, R., Li, J., Ma, X., et al. (2019). Maize glossy6 is involved in cuticular wax deposition and drought tolerance. J. Exp. Bot. 70 (12), 3089–3099. doi: 10.1093/jxb/erz131
Li, P., Lin, P., Zhao, Z., Li, Z., Liu, Y., Huang, C., et al. (2022). Gene co-expression analysis reveals transcriptome divergence between wild and cultivated sugarcane under drought stress. Int. J. Mol. Sci. 23 (1), 569. doi: 10.3390/ijms23010569
Liu, X., Bourgault, R., Galli, M., Strable, J., Chen, Z., Feng, F., et al. (2021). The FUSED LEAVES1-ADHERENT1 regulatory module is required for maize cuticle development and organ separation. New Phytol. 229 (1), 388–402. doi: 10.1111/nph.16837
Liu, Q., Huang, H., Chen, Y., Yue, Z., Wang, Z., Qu, T., et al. (2022). Two Arabidopsis MYB-SHAQKYF transcription repressors regulate leaf wax biosynthesis via transcriptional suppression on DEWAX. New Phytol. 236 (6), 2115–2130. doi: 10.1111/nph.18498
Lu, S., Song, T., Kosma, D. K., Parsons, E. P., Rowland, O., Jenks, M. A. (2009). Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 59 (4), 553–564. doi: 10.1111/j.1365-313X.2009.03892.x
Lykholat, Y. V., Didur, O. O., Alexeyeva, A. A., Lykholat, T. Y. (2020). Modification of the epicuticular waxes of plant leaves due to increased sunlight intensity. Biosys. Diversity 28 (1), 29–33. doi: 10.15421/012005
Makita, Y., Shimada, S., Kawashima, M., Kondou-Kuriyama, T., Toyoda, T., Matsui, M. (2015). MOROKOSHI: transcriptome database in Sorghum bicolor. Plant Cell Physiol. 56 (1), e6. doi: 10.1093/pcp/pcu187
McCormick, R. F., Truong, S. K., Sreedasyam, A., Jenkins, J., Shu, S., Sims, D., et al. (2018). The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 93 (2), 338–354. doi: 10.1111/tpj.13781
McWhorter, C., Paul, R., Barrentine, W. (1990). Morphology, development, and recrystallization of epicuticular waxes of johnsongrass (Sorghum halepense). Weed Sci. 38 (1), 22–33. doi: 10.1017/S004317450005606X
McWhorter, C. G. P., Rex, N. (1989). The involvement of cork-silica cell pairs in the production of wax filaments in johnsongrass (Sorghum halepense) leaves. Weed Sci. 37 (3), 458–470. doi: 10.1017/S0043174500072222
Mechan-Llontop, M. E., Mullet, J., Shade, A. (2023). Phyllosphere exudates select for distinct microbiome members in sorghum epicuticular wax and aerial root mucilage. Phytobiomes J. 1–14. doi: 10.1094/PBIOMES-08-22-0046-FI
Medeiros, C. D., Almeida-Cortez, J., Santos, D. Y. A. C., Oliveira, A. F. M., Santos, M. G. (2017). Leaf epicuticular wax content changes under different rainfall regimes, and its removal affects the leaf chlorophyll content and gas exchanges of Aspidosperma pyrifolium in a seasonally dry tropical forest. South Afr. J. Bot. 111, 267–274. doi: 10.1016/j.sajb.2017.03.033
Mizuno, H., Kawahigashi, H., Ogata, J., Minami, H., Kanamori, H., Nakagawa, H., et al. (2013). Genomic inversion caused by gamma irradiation contributes to downregulation of a WBC11 homolog in bloomless sorghum. Theor. Appl. Genet. 126 (6), 1513–1520. doi: 10.1007/s00122-013-2069-x
Mullet, J., Morishige, D., McCormick, R., Truong, S., Hilley, J., McKinley, B., et al. (2014). Energy sorghum–a genetic model for the design of C4 grass bioenergy crops. J. Exp. Bot. 65 (13), 3479–3489. doi: 10.1093/jxb/eru229
Nagata, K., Abe, M. (2023). A conserved mechanism determines the activity of two pivotal transcription factors that control epidermal cell differentiation in Arabidopsis thaliana. J. Plant Res. 136 (3), 349–358. doi: 10.1007/s10265-023-01439-7
Nakamura, M., Katsumata, H., Abe, M., Yabe, N., Komeda, Y., Yamamoto, K. T., et al. (2006). Characterization of the class IV homeodomain-Leucine Zipper gene family in Arabidopsis. Plant Physiol. 141 (4), 1363–1375. doi: 10.1104/pp.106.077388
Negruk, V., Yang, P., Subramanian, M., McNevin, J. P., Lemieux, B. (1996). Molecular cloning and characterization of the CER2 gene of Arabidopsis thaliana. Plant J. 9 (2), 137–145. doi: 10.1046/j.1365-313X.1996.09020137.x
Nobel, P. S., Jordan, P. W. (1983). Transpiration stream of desert species: resistances and capacitances for a C3, a C4, and a CAM plant. J. Exp. Bot. 34 (147), 1379–1391. doi: 10.1093/jxb/34.10.1379
Nwanze, K. F. S., Butler, D. R., Reddy, D. D. R., Reddy, Y. V. R., Soman, P. (1992). The dynamics of leaf surface wetness of sorghum seedlings in relation to resistance to the shoot fly, Atherigona soccata. Entomol. Experimentalis Appl. 64 (2), 151–160. doi: 10.1111/j.1570-7458.1992.tb01604.x
Ogawa, E., Yamada, Y., Sezaki, N., Kosaka, S., Kondo, H., Kamata, N., et al. (2015). ATML1 and PDF2 play a redundant and essential role in arabidopsis embryo development. Plant Cell Physiol. 56 (6), 1183–1192. doi: 10.1093/pcp/pcv045
Olson, S. N., Ritter, K., Rooney, W., Kemanian, A., McCarl, B. A., Zhang, Y., et al. (2012). High biomass yield energy sorghum: developing a genetic model for C4 grass bioenergy crops. Biofuels Bioprod. Bioref. 6 (6), 640–655. doi: 10.1002/bbb.1357
Park, J. S., Kim, J. B., Cho, K. J., Cheon, C. I., Sung, M. K., Choung, M. G., et al. (2008). Arabidopsis R2R3-MYB transcription factor AtMYB60 functions as a transcriptional repressor of anthocyanin biosynthesis in lettuce (Lactuca sativa). Plant Cell Rep. 27 (6), 985–994. doi: 10.1007/s00299-008-0521-1
Park, C. S., Go, Y. S., Suh, M. C. (2016). Cuticular wax biosynthesis is positively regulated by WRINKLED4, an AP2/ERF-type transcription factor, in Arabidopsis stems. Plant J. 88 (2), 257–270. doi: 10.1111/tpj.13248
Pascal, S., Sorel, M., Pervent, M., Vile, D., Haslam, R. P., Napier, J. A., et al. (2013). The Arabidopsis cer26 mutant, like the cer2 mutant, is specifically affected in the very long chain fatty acid elongation process. Plant J. 73 (5), 733–746. doi: 10.1111/tpj.12060
Peters, P. J., Rich, P. J., Axtell, J. D., Ejeta, G. (2009). Mutagenesis, selection, and allelic analysis of epicuticular wax mutants in sorghum. Crop Sci. 49 (4), 1250–1258. doi: 10.2135/cropsci2008.08.0461
Punnuri, S., Harris-Shultz, K., Knoll, J., Ni, X., Wang, H. (2017). The genes bm2 and blmc that affect epicuticular wax deposition in sorghum are allelic. Crop Sci. 57 (3), 1552–1556. doi: 10.2135/cropsci2016.11.0937
Raffaele, S., Vailleau, F., Leger, A., Joubes, J., Miersch, O., Huard, C., et al. (2008). A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20 (3), 752–767. doi: 10.1105/tpc.107.054858
Rooney, W. L., Blumenthal, J., Bean, B., Mullet, J. E. (2007). Designing sorghum as a dedicated bioenergy feedstock. Biofuels Bioprod. Bioref. 1 (2), 147–157. doi: 10.1002/bbb.15
Rusconi, F., Simeoni, F., Francia, P., Cominelli, E., Conti, L., Riboni, M., et al. (2013). The Arabidopsis thaliana MYB60 promoter provides a tool for the spatio-temporal control of gene expression in stomatal guard cells. J. Exp. Bot. 64 (11), 3361–3371. doi: 10.1093/jxb/ert180
Samuels, L., Kunst, L., Jetter, R. (2008). Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 59, 683–707. doi: 10.1146/annurev.arplant.59.103006.093219
Saneoka, H., Ogata, S. (1987). Relationship between Water use Efficiency and Cuticular Wax Deposition in Warm Season Forage Crops Grown under Water Deficit Conditions. Soil Sci. Plant Nutr. 33 (3), 439–448. doi: 10.1080/00380768.1987.10557590
Scully ED, G. T., Sarath, G., Palmer, N. A., Baird, L., Serapiglia, M. J., Dien, B. S., et al. (2016). Overexpression of SbMyb60 impacts phenylpropanoid biosynthesis and alters secondary cell wall composition in Sorghum bicolor. Plant J. 85 (3), 378–395. doi: 10.1111/tpj.13112
Seo, P. J., Lee, S. B., Suh, M. C., Park, M. J., Go, Y. S., Park, C. M. (2011). The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23 (3), 1138–1152. doi: 10.1105/tpc.111.083485
Singh, V., Kumar, N., Dwivedi, A.K., Sharma, R., Sharma, M.K. (2020). Phylogenomic analysis of R2R3 MYB transcription factors in sorghum and their role in conditioning biofuel syndrome. Curr. Genomics 21 (2), 138–154. doi: 10.2174/1389202921666200326152119
Suh, M. C., Samuels, A. L., Jetter, R., Kunst, L., Pollard, M., Ohlrogge, J., et al. (2005). Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 139 (4), 1649–1665. doi: 10.1104/pp.105.070805
Tacke, E., Korfhage, C., Michel, D., Maddaloni, M., Motto, M., Lanzini, S., et al. (1995). Transposon tagging of the maize Glossy2 locus with the transposable element En/Spm. Plant J. 8 (6), 907–917. doi: 10.1046/j.1365-313X.1995.8060907.x
Tada, A., Ishizuki, K., Yamazaki, T., Sugimoto, N., Akiyama, H. (2014). Method for the determination of natural ester-type gum bases used as food additives via direct analysis of their constituent wax esters using high-temperature GC/MS. Food Sci. Nutr. 2 (4), 417–425. doi: 10.1002/fsn3.117
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 (10), 2731–2739. doi: 10.1093/molbev/msr121
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi: 10.1093/molbev/msab120
Tang, W., Liao, L., Xiao, Y., Zhai, J., Su, H., Chen, Y., et al. (2022). Epicuticular wax of sweet sorghum influenced the microbial community and fermentation quality of silage. Front. Microbiol. 13, 960857. doi: 10.3389/fmicb.2022.960857
Tao, Y., Luo, H., Xu, J., Cruickshank, A., Zhao, X., Teng, F., et al. (2021). Extensive variation within the pan-genome of cultivated and wild sorghum. Nat. Plants 7 (6), 766–773. doi: 10.1038/s41477-021-00925-x
Taylor-Teeples, M., Lin, L., de Lucas, M., Turco, G., Toal, T. W., Gaudinier, A., et al (2015). An Arabidopsis gene regulatory networkfor secondary cell wall synthesis. Nature 2015 (517), 571–575. doi: 10.1038/nature14099
To, A., Joubes, J., Barthole, G., Lecureuil, A., Scagnelli, A., Jasinski, S., et al. (2012). WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell 24 (12), 5007–5023. doi: 10.1105/tpc.112.10612010
Trenkamp, S., Martin, W., Tietjen, K. (2004). Specific and differential inhibition of very-long-chainfatty acid elongases fromArabidopsis thalianabydifferent herbicides. PNAS 101 (32), 11903–11908. doi: 10.1073/pnas.0404600101
Uttam, G. A., Praveen, M., Rao, Y. V., Tonapi, V. A., Madhusudhana, R. (2017). Molecular mapping and candidate gene analysis of a new epicuticular wax locus in sorghum (Sorghum bicolor L. Moench). Theor. Appl. Genet. 130 (10), 2109–2125. doi: 10.1007/s00122-017-2945-x
Varala, K., Marshall-Colon, A., Cirrone, J., Brooks, M.D., Pasquino, A.V., Leran, S., et al. (2018). Temporal transcriptional logic of dynamic regulatory networks underlying nitrogen signaling and use in plants. Proc. Natl. Acad. Sci. U.S.A. 115 (25), 6494–6499. doi: 10.1073/pnas.1721487115
Wang, Y., Wang, J., Chai, G., Li, C., Hu, Y., Chen, X., et al. (2015). Developmental changes in composition and morphology of cuticular waxes on leaves and spikes of glossy and glaucous wheat (Triticum aestivum L.). PloS One 10 (10), e0141239. doi: 10.1371/journal.pone.0141239
Wen, M., Jetter, R. (2009). Composition of secondary alcohols, ketones, alkanediols, and ketols in Arabidopsis thaliana cuticular waxes. J. Exp. Bot. 60 (6), 1811–1821. doi: 10.1093/jxb/erp061
Weng, H., Molina, I., Shockey, J., Browse, J. (2010). Organ fusion and defective cuticle function in a lacs1 lacs2 double mutant of Arabidopsis. Planta 231 (5), 1089–1100. doi: 10.1007/s00425-010-1110-4
Wessels, B., Seyfferth, C., Escamez, S., Vain, T., Antos, K., Vahala, J., et al. (2019). An AP2/ERF transcription factor ERF139 coordinates xylem cell expansion and secondary cell wall deposition. New Phytol. 224 (4), 1585–1599. doi: 10.1111/nph.15960
Wettstein-Knowles, P. (1972). Genetic control of β-diketone and hydroxy-β-diketone synthesis in epicuticular waxes of barley. Planta 106, 113–130. doi: 10.1007/BF00383991
Xiao, Y., Li, X., Yao, L., Xu, D., Li, Y., Zhang, X., et al. (2020). Chemical profiles of cuticular waxes on various organs of Sorghum bicolor and their antifungal activities. Plant Physiol. Biochem. 155, 596–604. doi: 10.1016/j.plaphy.2020.08.026
Xin, Z., Wang, M. L., Burow, G., Burke, J. (2009). An induced sorghum mutant population suitable for bioenergy research. Bioenergy Res. 2 (1-2), 10–16. doi: 10.1007/s12155-008-9029-3
Xin, Z., Wang, M., Cuevas, H. E., Chen, J., Harrison, M., Pugh, N. A., et al. (2021). Sorghum genetic, genomic, and breeding resources. Planta 254 (6), 114. doi: 10.1007/s00425-021-03742-w
Xue, D., Zhang, X., Lu, X., Chen, G., Chen, Z.H. (2017). Molecular and evolutionary mechanisms of cuticular wax for plant drought tolerance. Front. Plant Sci. 8, 621. doi: 10.3389/fpls.2017.00621
Yan, Y., Li, C., Dong, X., Li, H., Zhang, D., Zhou, Y., et al. (2020). MYB30 is a key negative regulator of arabidopsis photomorphogenic development that promotes PIF4 and PIF5 protein accumulation in the light. Plant Cell 32 (7), 2196–2215. doi: 10.1105/tpc.19.00645
Yang, Y., Zhou, B., Zhang, J., Wang, C., Liu, C., Liu, Y., et al. (2017). Relationships between cuticular waxes and skin greasiness of apples during storage. Postharvest Biol. Technol. 131, 55–67. doi: 10.1016/j.postharvbio.2017.05.006
Yang, X., Feng, T., Li, S., Zhao, H., Zhao, S., Ma, C., et al. (2020). CER16 inhibits post-transcriptional gene silencing of CER3 to regulate alkane biosynthesis. Plant Physiol. 182 (3), 1211–1221. doi: 10.1104/pp.19.01002
Ye, J. H., Lv, Y. Q., Liu, S. R., Jin, J., Wang, Y. F., Wei, C. L., et al. (2021). Effects of light intensity and spectral composition on the transcriptome profiles of leaves in shade grown tea plants (Camellia sinensis L.) and regulatory network of flavonoid biosynthesis. Molecules 26 (19), 5836. doi: 10.3390/molecules26195836
Yeats, T. H., Rose, J. K. (2013). The formation and function of plant cuticles. Plant Physiol. 163 (1), 5–20. doi: 10.1104/pp.113.222737
Yephremov, A., Wisman, E., Huijser, P., Huijser, C., Wellesen, K., Saedler, H. (1999). Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11 (11), 2187–2201. doi: 10.1105/tpc.11.11.2187
Keywords: Sorghum, bioenergy, stem cuticular wax, scanning electron microscopy, wax load, gene regulatory network analysis, policosanol
Citation: Chemelewski R, McKinley BA, Finlayson S and Mullet JE (2023) Epicuticular wax accumulation and regulation of wax pathway gene expression during bioenergy Sorghum stem development. Front. Plant Sci. 14:1227859. doi: 10.3389/fpls.2023.1227859
Received: 26 May 2023; Accepted: 11 September 2023;
Published: 23 October 2023.
Edited by:
Laura Rossini, University of Milan, ItalyReviewed by:
David Pot, Institut National de la Recherche Agronomique (INRA), FranceCopyright © 2023 Chemelewski, McKinley, Finlayson and Mullet. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John E. Mullet, am9obi5tdWxsZXRAYWcudGFtdS5lZHU=
†ORCID: Robert Chemelewski, orcid.org/0000-0001-7739-2741
John E. Mullet, orcid.org/0000-0003-2502-2671
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.