
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 28 August 2023
Sec. Plant Biotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1226911
This article is part of the Research Topic СRISPR tools, technology development, and application View all 9 articles
Introduction: Clustered regularly interspaced short palindromic repeats (CRISPR)/ CRISPR-associated protein 9 (Cas9) is a precise genome editing tool used to introduce genetic modifications in a wide range of crop species. Thus far, there is no report of CRISPR/Cas9-mediated genome editing in onions (Allium cepa L.).
Methods: In the present study, we targeted two exons of the gene coding for Phytoene desaturase (AcPDS) in onion cv. Bhima Super. The sgRNA-carrying constructs were co-cultivated with 8-week-old embryogenic calli using an Agrobacterium-mediated transformation protocol and incubated on the media without hygromycin B selection.
Results and discussion: Out of the total 617 co-cultivated calli, 21 (3.4%) regenerated shoots exhibited three distinct phenotypes: albino, chimeric, and pale green; in comparison to the wild-type non-transformed regenerated shoots. Total chlorophyll content was drastically reduced in albino shoots and significantly decreased in chimeric shoots. Out of the six Cas9 gene PCR-confirmed regenerated shoots, two exhibited the albino phenotype due to insertions/deletions (InDels) and substitution-based mutations in and around the AcPDS target sites. Deep amplicon sequencing revealed a significantly variable InDel frequency between two sgRNAs, ranging from 1.2% to 63.4%, along with a 53.4% substitution frequency. The mutation of the AcPDS gene generated a visually detectable albino phenotype, thus confirming the successful editing of the AcPDS gene. This is the first time a CRISPR/Cas9-mediated genome editing protocol has been successfully established in onion, with the AcPDS gene serving as an example. This study will provide the necessary momentum for researchers to further basic and applied research on onions.
Onion (Allium cepa L.) is one of the important horticultural crops cultivated throughout the world. It is known for its distinct flavor and pungency, which are attributed to a variety of sulfur compounds. Onions are beneficial in terms of offering health benefits due to the presence of flavonoids, anthocyanins, fructooligosaccharides, and organosulfur compounds (Goldman, 2011; Bisen and Emerald, 2016; Putnik et al., 2019). Since domestication, productivity, quality, and resilience to biotic and abiotic stresses have been improved in onions using several conventional breeding practices. However, conventional approaches have not produced substantial progress in the genetic improvement of onions and have been hampered by the biennial life cycle, cross-pollinated nature, and severe inbreeding depression (McCallum, 2007; Sudha et al., 2019; Lee et al., 2021). Despite the fact that onions have a large genetic variability, crop improvement has lagged behind other monocot crops (McCallum, 2007; Varshney et al., 2012). The burden of the current global challenges involving food security, increasing human population and changes in climate experienced worldwide lies in the need to improve horticultural crops with high yield and enhanced adaptation to the environment, for which conventional breeding is unlikely to meet the demand (Brewster, 2008; Van den Broeck and Maertens, 2016). To overcome these obstacles, advanced and innovative molecular approaches should be strategically employed to strengthen onion genetic improvement programs.
Several genome editing tools, such as transcription activator-like effector nucleases (TALENs), zinc-finger nucleases (ZFNs), and meganucleases (MNs), have facilitated targeted manipulation of a desired gene in crop plants. However, these methods are expensive, labor-intensive, and require intricate procedures to achieve successful editing (Razzaq et al., 2019). In contrast, CRISPR/Cas9 has emerged as a promising and versatile approach for precise and efficient plant genome editing. It offers an appealing advantage due to its user-friendly design, cost-effectiveness, and adaptability, making it an attractive tool for researchers in the field (Razzaq et al., 2019). The advent of CRISPR/Cas9-targeted genome editing has paved the way toward incorporating beneficial genetic changes for addressing limitations affecting production and utilization. CRISPR/Cas9 genome editing, as a popular genome engineering tool useful for crop improvement, has drawn the attention of breeders and stands out as an essential tool to generate plants with the ability to cope with challenges. The CRISPR/Cas9 system has been applied to a large number of plant species. Still, there are several horticultural crops where the applicability of this technology has not been demonstrated (Koltun et al., 2018), and onion is one among them. A reproducible system for the design, construction, and application of Cas9/guide RNA (gRNA) must be established and evaluated in onions in order to assess the potential of the CRISPR/Cas9 system. The strategy to knock out the Phytoene desaturase (PDS) gene has been widely used to validate the practicability of the CRISPR/Cas9 system in several plants, including cassava (Manihot esculenta) (Odipio et al., 2017), banana cv. Rasthali (Musa acuminata cv. Rasthali) (Kaur et al., 2018), and Cavendish banana (Musa acuminata subgroup Cavendish) (Naim et al., 2018). Only recently has the onion genome sequence become available (Finkers et al., 2021), thereby enabling the exploration of the potential for CRISPR/Cas9-based genome editing in applied research for this crop.
In the present study, for the first time, we report the successful establishment of a CRISPR/Cas9-based genome editing system by targeting the gene coding for PDS of the Indian short-day onion. The insights presented here lay the groundwork for future investigations into gene function and trait development in this important vegetable crop, leveraging the capabilities of the CRISPR/Cas9-based gene editing system.
The annotated PDS sequence of Asparagus officinalis (Accn. No. XP_020273319.1) was retrieved from the National Centre for Biotechnology Information (NCBI) database and used as a query for the Basic Local Alignment Search Tool-Nucleotide (BLASTn) search against the draft genome sequence of onion (https://www.oniongenome.wur.nl/; Finkers et al., 2021) to retrieve PDS gene sequence of onion. Primers were designed in the coding region (Supplementary Table S1) of retrieved PDS sequences of double haploid onion line DHCU066619 PDS (accession: GBRN01009640.1) for the isolation of the full-length PDS gene from the Indian short-day onion cv. Bhima Super (B. Super). Genomic DNA was isolated from onion cv. B. Super leaf tissue following GenElute Plant Genomic DNA Miniprep Kit protocol (Sigma, USA). PCR amplification was performed using 100 ng of genomic DNA as a template and a gene-specific primer set (AcPDS F/AcPDS R) with the help of the PrimeSTAR® GXL DNA Polymerase (Takara, Japan), as per the PCR condition described in Supplementary Table S1. The amplified PCR product was cloned into a pJET1.2 vector (ThermoScientific, USA) as per the manufacturer’s instructions. Clones were sequenced by Sanger’s platform at Eurofins Genomics India Pvt. Ltd., India and the sequence was used for BLAST search against the non-redundant database.
The exon–intron pattern of the AcPDS gene of cv. B. Super was analyzed using Gene Structure Display Server 2.0 (http://gsds.gao-lab.org/). The physicochemical properties, such as molecular weight and isoelectric point (pI), were calculated using the protparam tool (http://web.expasy.org/protparam/). The online Pfam tool (https://pfam.xfam.org/) was used to detect the presence of a conserved domain in the AcPDS.
The protein sequences of AcPDS of onion cv. B. Super and PDS from 18 different plant species from monocots and dicots were aligned using the MUSCLE program, and the aligned file was subjected to phylogenetic tree analysis. A phylogenetic tree was reconstructed in MEGA 11 (Tamura et al., 2021) using the maximum likelihood method, the Jones–Taylor–Thornton (JTT) model with a 1,000 bootstrap value.
Based on onion cv. B. Super AcPDS gene sequences, we targeted exon 3 and exon 4 regions of the AcPDS gene for CRISPR/Cas9-based genome editing, as they are located in the N-terminal region of the conserved amino-oxidase domain. Two different guide RNAs targeting exon 3 (AcPDS exon 3 gRNA-GAAGCTAGAGATGTTCTGGGAGG) and exon 4 (AcPDS exon 4 gRNA-GGTTGCTGCTTGGAAAGACAAGG) were predicted on the positive strand by the CRISPR-P2.0 (http://crispr.hzau.edu.cn/) web-based tool with highest efficacy. To elucidate the conserved sequence of target regions, a primer set was designed in the flanking regions spanning both target sites and used for PCR amplification of the AcPDS fragment using genomic DNA from 20 individual plants of onion cv. B. Super as a template and as per the PCR conditions described in Supplementary Table S1. After confirming the conserved target sequences, BsaI adaptors (5′GGCA in the top strand and 5′AAAC in the bottom strand) were added to the predicted gRNAs, and oligomers of both gRNAs were synthesized. These oligomers with BsaI overhangs were duplexed and cloned independently at the BsaI site of the CRISPR/Cas9-based binary vector pRGEB31, under the control of the OsU3 promoter and PolIII terminator. The insertion and stability of the final binary vector constructs were confirmed by Sanger sequencing. The resulting constructs were named as pRGEB31-AcPDS E3 target and pRGEB31-AcPDS E4 target. Both binary vectors were independently introduced into the Agrobacterium tumefaciens strain LBA4404 by employing the tri-parental mating method.
Agrobacterium LBA4404 harboring AcPDS gene editing constructs, viz. pRGEB31-AcPDS E3 target and the pRGEB31-AcPDS E4 target separately were used for co-transformation. A total of 617 eight-week-old embryogenic calli of onion were co-transformed using Agrobacterium-mediated transformation as described by Manape et al. (2022) in seven independent batches. An Agrobacterium-infected calli was blot-dried on sterile filter paper, transferred to the co-cultivation medium for 3 days, and then transferred to the resting medium for 15 days. Later, these calli were directly transferred to a shooting medium (devoid of hygromycin B) and subcultured regularly on the fresh shooting medium at intervals of 15 days. After shooting induction, the regenerated shoots were transferred to the rooting medium (½ strength Murashige and Skoog medium) to develop into complete plantlets. In each batch, 30–34 calli were also maintained without co-cultivation as an experimental control and subcultured routinely as per co-cultivated calli.
The morphology of the regenerated shoots from co-cultivated calli on the shooting medium was visually observed 8-weeks after co-cultivation. The genomic DNA was extracted from phenotypically distinct putatively transformed shoots and non-transformed shoots using the GenElute Plant Genomic DNA Miniprep Kit (Sigma) as per the manufacturer’s instructions. To investigate the presence of the Cas9 gene in phenotypically distinct shoots, the Cas9 F/Cas9 R primer set was used for amplification of the internal sequence of the Cas9 gene using 100 ng of DNA as a template as per the PCR conditions described in Supplementary Table S1. Whereas, to investigate the mutation induced in the exon 3 or exon 4 regions of AcPDS in the phenotypically distinct calli, PCR amplification of the AcPDS fragment harboring both target sites was carried out by using the DNA of these calli as a template and PrimeSTAR® GXL DNA Polymerase (DSS Takara) with the help of another primer set (AcPDSg F/AcPDSg R). PCR-amplified products from Cas9-positive shoots were cloned into the pJET1.2 vector using a CloneJET PCR Cloning Kit (ThermoFisher Scientific), and 10 clones of each were sequenced using Sanger’s platform at Eurofins Genomics India Pvt. Ltd. The sequences of amplified fragments were aligned using the Clustal Omega online tool (https://www.ebi.ac.uk/Tools/msa/clustalo/). The identified InDels and substitutions were further validated by analyzing chromatograms using the Synthego ICE analysis online tool (https://ice.synthego.com/). In addition, we used purified PCR products, each with an equal concentration of 100 ng, from Cas9-positive plants. These products were amplified using flanking primers and then pooled together for sequencing using the Illumina Novaseq6000 platform to validate the results from Sanger sequencing. The raw reads were analyzed using the Cas-analyzer tool (Park et al., 2017). To estimate the frequency and type of mutation induced by each target, we used a 65-bp comparison range around the target CRISPR RNA (crRNA) sequences.
Total chlorophyll was extracted from 30 mg of shoot tissues from phenotypically distinct regenerated shoots and nontransformed green shoots as controls by following the protocol described by Arnon (1949). The chlorophyll content was estimated by the SPECTROstar Nano microplate reader (BMG Labtech, Germany) by recording the absorbance at 645 nm and 663 nm. Total chlorophyll content was estimated by using Arnon’s equation.
The chlorophyll estimation experiment was carried out in triplicates, and the results of phenotypically distinct regenerated shoots were compared with control non-transformed green shoots. The average reduction in the chlorophyll content was calculated in percentage based on a mean value of distinct phenotype compared with the control regenerated shoots. The data were presented in the mean ± SD format and analyzed by one-way ANOVA. The statistical significance was checked at p ≤ 0.05–0.001 (*p ≤ 0.001) with respect to control. The statistical analysis was performed using Analysis ToolPak in Excel (2021).
The nucleotide sequence of Asparagus officinalis PDS (XP_020273319.1) was used as a query to search for similar sequences in the draft genome sequence of onion (Allium cepa L.), and a single copy of AcPDS on chromosome 4 was identified. The coding sequence (CDS) of size 4,278 bp was amplified by PCR from the genomic DNA of onion cv. B. Super and submitted to the NCBI database (Accn. No. OP004915). The BLASTn analysis of the AcPDS of onion cv. B. Super revealed 23 mismatches and 4 gaps when compared to the draft genome sequence of the double haploid onion line DHCU066619 PDS (GBRN01009640.1) (https://www.oniongenome.wur.nl/). The sequence was confirmed by independent sequencing three times and was later predicted to be translated into a functional protein. The genomic AcPDS gene sequence of 4,278 bp size comprises 14 exons and 13 introns and has an open reading frame of 1,719 bp that encodes a protein of 572 amino acids with a predicted molecular weight of 63.9 kDa. Pfam database analysis of the AcPDS protein has shown the presence of a conserved amino-oxidase domain of the PDS family (Figure 1A). The AcPDS sequence showed 85.17% and 87.32% identity with the annotated PDS of Narcissus (AFH53812.1) and Asparagus (XP_020273319.1), respectively, suggesting a conserved function in the onion. The multiple sequence alignment analysis of the AcPDS protein had shown three characteristic conserved motifs of the PDS family, viz., the dinucleotide binding motif, the putative carrier motif, and the carotenoid-binding motif responsible for the catalytic activity of AcPDS (Supplementary Figure S1). The phylogenetic analysis of the AcPDS protein along with the PDS proteins from 18 different monocot and eudicot species showed that AcPDS clustered with the PDS proteins from monocot species and formed a separate clade, whereas PDS from dicot species formed a clade distinct from monocot plants (Figure 1B).
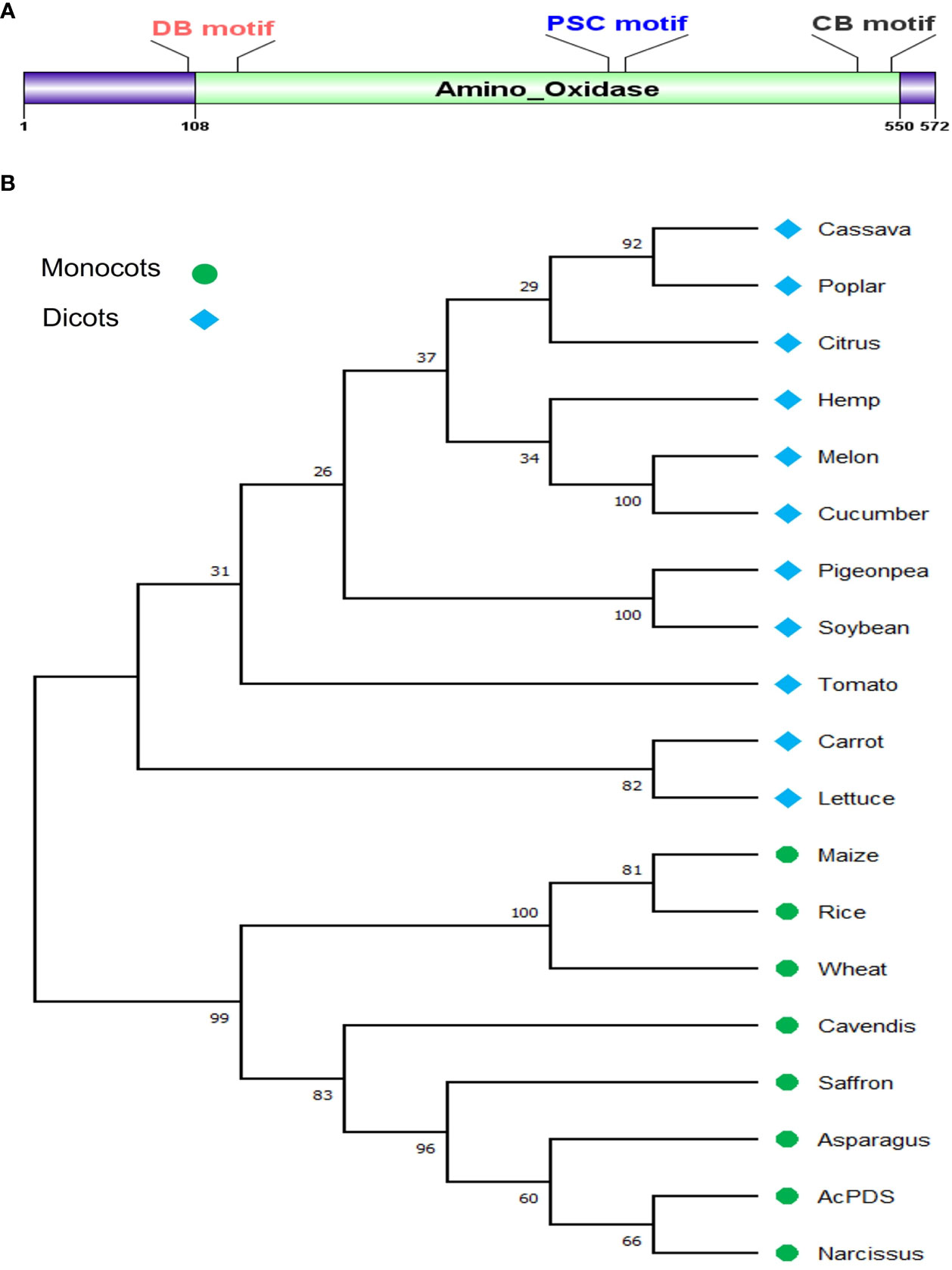
Figure 1 In silico analysis of AcPDS gene, (A) Schematic representation of conserved amino oxidase domain in AcPDS protein. (B) Phylogenetic relationship of AcPDS protein of Allium cepa L. cv. B. Super with PDS protein sequences from 11 dicot and seven monocot species was constructed using the MEGA 11 tool.
A total of 617 embryogenic calli were co-transformed with the pRGEB31-AcPDS E3 target (Figure 2A), harboring exon 3 sgRNA (Figure 2C), and the pRGEB31-AcPDS E4 target (Figure 2B), harboring exon 4 sgRNA (Figure 2D), using Agrobacterium-mediated transformation in seven independent batches. As mutations in the PDS gene resulted in visible albino or chimeric phenotypes, the calli were not screened on selection steps (MS medium supplemented with 50 mg/L of hygromycin B) of Manape et al. (2022) to reduce the time of regeneration protocol and were directly regenerated on the shooting medium devoid of hygromycin B. After 8-weeks of co-cultivation, the co-cultivated and non-transformed calli had regenerated into shoots. Shoots regenerated from non-transformed calli with shooting efficiency of 76%, and all were normal green (Figure 2F). On the contrary, out of 617 co-transformed calli, 456 shoots were regenerated, of which 21 shoots had three additional different phenotypes along with the normal green phenotype, viz., 9.52% complete albino (two of 21; callus #3 and #41), 19.04% chimeric (four of 21; callus #1, #2, #8, and #48), and 71.42% pale green (15/21) (Figure 2E). The regenerated, phenotypically distinct shoots were rooted on rooting media. On rooting media, the pale green shoots were turned into normal green plantlets, whereas chimeric shoots were dried off quickly, and newly emerged leaves were not chimeric and looked more like leaves of the wild-type. The albino plants retained the albino phenotype throughout the life cycle but showed a slower growth rate than non-transformed control plants (Figure 2G).
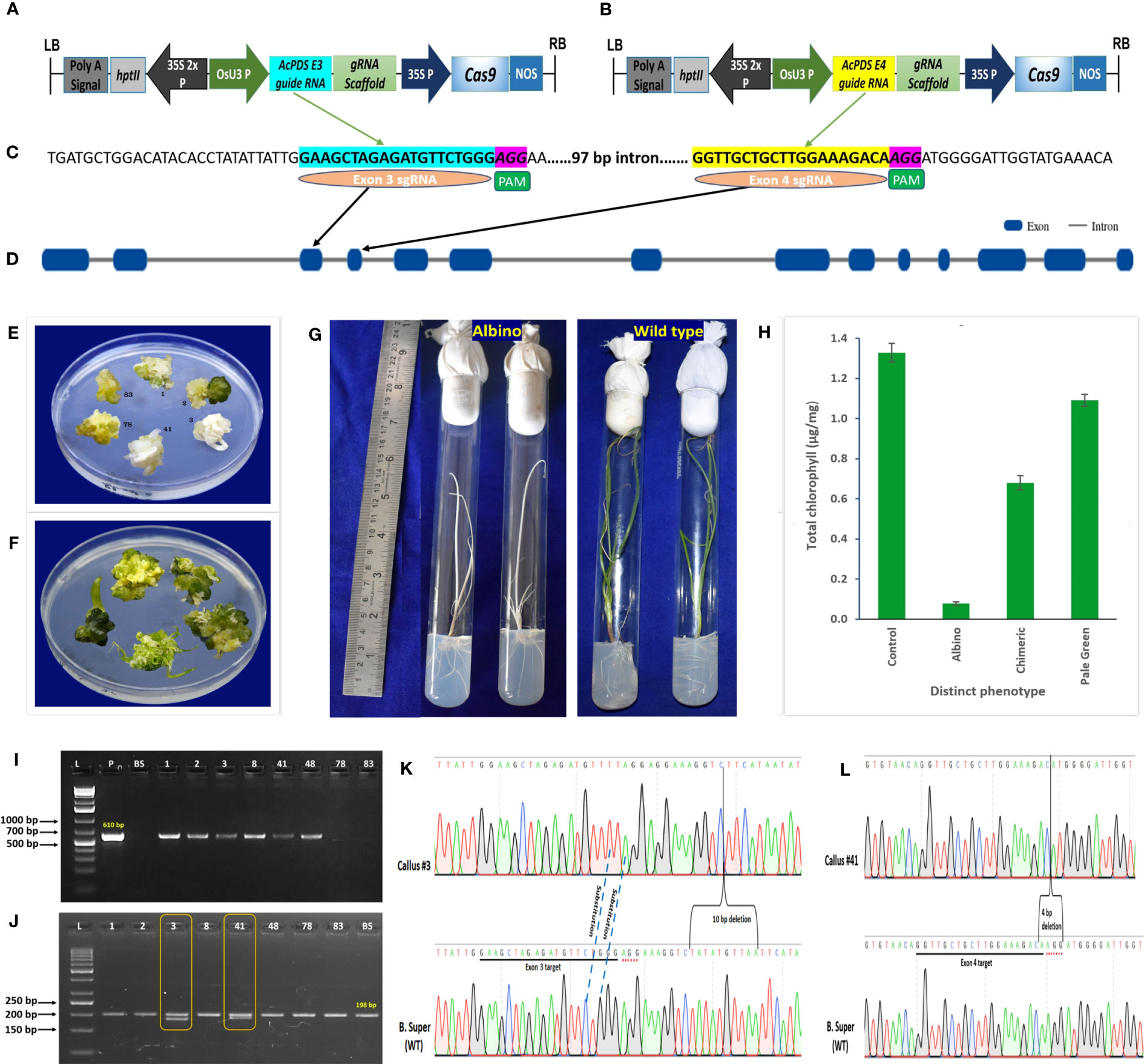
Figure 2 CRISPR/Cas9-mediated genome-editing of onion by targeting phytoene desaturase (AcPDS) gene. (A) Schematic presentation of binary vector pRGEB31-AcPDS E3 target. (B) Schematic presentation of binary vector pRGEB31-AcPDS E4 target. (C) Protospacer sequences for exon 3 target (cyan highlighted) and for exon 4 target (yellow highlighted); PAM sequences in pink highlighted. (D) Schematic representation of gene structure of AcPDS isolated from onion cv. B. Super, showing the gRNA targets. Blue blocks indicate exons of the gene. (E) Co-cultivated regenerated shoots showing three distinct phenotypes, viz. albino (regenerated shoots #3 and #41), chimeric (regenerated shoots #1 and #2), and pale green (regenerated shoots #78 and #83). (F) Nontransformed control regenerated shoots showing dark green shoots. (G) Regenerated plantlets showing albino and normal green wild-type phenotypes. (H) Chlorophyll content estimation in the three phenotypically distinct regenerated shoots. Bars represent the mean ± SD of the three independent replicates (n = 3). (I) Representative PCR analysis of phenotypically distinct shoots to confirm the presence of Cas9 gene. (J) PCR analysis of albino and chimeric shoots showing varying sizes of amplicons in the target regions. (L, 1 kb plus ladder in (H) and 50 bp ladder in (I); P, pRGEB31-plasmid (positive control); BS: onion cv. B. Super (negative control); 1, 2, 3, 8, 41, 48, 78, and 83: phenotypically distinct co-cultivated regenerated shoots). (K, L) Schematic chromatogram presentation of site-specific mutations of AcPDS induced by exon 3 and exon 4 sgRNAs in albino shoots regenerated from callus #3 and #41, respectively. Dot lines specify the type of mutation, and parentheses indicates large (4 nt and 10 nt) deletions. PAM is in red dots, and protospacers are underlined.
The chlorophyll content also varied in these different phenotypes, viz. 0.073 ± 0.008 μg/mg to 0.085 ± 0.012 μg/mg in albino, 0.644 ± 0.071 μg/mg to 0.724 ± 0.033 μg/mg in chimeric, and 1.027 ± 0.026 μg/mg to 1.187 ± 0.019 μg/mg in the pale green phenotypes, whereas the chlorophyll content in the non-transformed control regenerated shoots was 1.328 ± 0.046 μg/mg (Figure 2H). There was significant reduction of the chlorophyll content in the albino (94.07%) and chimeric (48.79%) and a nonsignificant reduction in the pale green (17.80%) shoots as compared to the control non-transformed green shoots.
The PCR analysis of these 21 phenotypically different shoots showed the presence of a 610-bp-sized Cas9 gene fragment in six regenerated shoots (Figure 2I). Out of six, two had complete albino and four had chimeric (mixture of albino and green) shoot morphology, whereas the Cas9 gene was absent in the pale green transformed and non-transformed control regenerated shoots. With the help of the AcPDSg F/AcPDSg R primer set, DNA fragments of size 198 bp spanning both the target sites were PCR amplified from these six transgenic shoots and from non-transgenic control shoots. Moreover, an additional 188 bp and 194 bp fragments were amplified in albino shoots #3 and #41, respectively (Figure 2J). The sequence alignment of 188 bp and 194 bp fragments with the reference sequence of onion cv. B. Super revealed that shoot #3 had 2 nucleotide substitutions in the exon 3 target site along with 10 nucleotide deletions (+8 to +17 bases from exon 3 PAM site) in the downstream region of the exon 3 target site (Figure 2K). Shoot #41 showed a 4-bp deletion within the exon 4 target site, including the PAM sequence (Figure 2L).
A comparative analysis was conducted using the Synthego ICE analysis tool on the Sanger view plot of the wild-type B. Super chromatogram, alongside the chromatograms of shoots #3 and #41, to validate multiple sequence alignment results. Synthego ICE tool analysis of shoot #3 revealed the cut site in the exon 3 gRNA target sequence along with 2 bp substitutions, and no cut site was predicted in the exon 4 gRNA sequence. Additionally, a 10-bp deletion was observed in the downstream region of the exon 3 PAM site of shoot #3, accounting for 30% of the total InDel mixture (Supplementary Figure S2B). On the other hand, the analysis of shoot #41 using the Synthego ICE tool revealed the cut site in the exon 4 gRNA target sequence, while no cut site was predicted in the exon 3 gRNA target site. An InDel plot of shoot #41 demonstrated a 4 bp deletion within the exon 4 PAM site, constituting 58% of the total InDel mixture (Supplementary Figure S3B). These findings clearly indicated that the observed InDels in shoots #3 and #41 resulted from editing due to the action of exon 3 and exon 4 sgRNAs, respectively.
The chimeric transgenic onion shoots did not show any editing in and around both target sites and had a similar sequence as that of the wild-type. This could be due to the limited number of clones (i.e., 10 clones per sample) were sequenced by Sanger sequencing. Thus, the genome editing efficiency of 33.33% (two of six) was observed among the PCR-positive regenerated onion shoots.
We performed deep amplicon sequencing of the AcPDS target region on six Cas9-positive shoots using the Illumina NovaSeq6000 platform with 2 × 150 bp chemistry. This yielded 5.96 million paired-end reads, corresponding to 2.54 Gb of sequence data. Using the Cas-analyzer tool, we analyzed the AcPDS Illumina amplicons and identified insertions, deletions, and substitution-type mutations. Our next-generation sequencing (NGS) data indicated InDel frequencies of 63.4% and 1.237% in and around exon 3 and exon 4 sgRNAs, respectively (Table 1). Among the 63.4% InDel frequency of exon 3 sgRNA, 60.21% were deletions, primarily of size 10 bp downstream to the target site of exon 3 sgRNA (58.75%), while 3.19% were insertions, mostly of size 3 bp and 8 bp downstream to the target site of exon 3 sgRNA (2.86%) (Supplement S2; Supplementary Figure S4). We also observed 3.04% substitutions, of which 0.22% were at the exon 3 sgRNA target, featuring specific base changes (i.e., C is replaced by T at the 16th position and G is replaced by A at the 18th position of exon 3 gRNA). These same substitutions also appeared in deletion counts (Supplementary Figure S6). After combining these counts manually, the substitution frequency for these specific bases in exon 3 sgRNA increased from 0.22% to 54.84%. The remaining fraction of InDels (1.46% deletions and 0.33% insertions of 1 to 2 bp) and substitutions at various positions (2.8%) were around the exon 3 sgRNA, possibly from chimeric shoots. As deep sequencing could be able to detect the mutants other than editing identified by the Sanger platform, therefore, these edits could be possibly derived from chimeric shoots. Regarding the exon 4 target, the overall InDel frequency was 1.237%, primarily deletions of size 4 bp at the PAM site (1.1%), including the last nucleotide of exon 4 sgRNA, with negligible insertion frequency (Supplement S3; Supplementary Figure S5). We also observed negligible (0.54%) substitutions in exon 4 sgRNA. Overall, our NGS data were consistent with Sanger sequencing data. Additionally, we detected low-frequency InDels and substitutions in the NGS data, potentially from chimeric shoots (Supplement S2, S3).
The CRISPR/Cas9 technology has so far been used in numerous plant species for basic research and trait development. Apart from the evaluation of gene functions, the application of CRISPR/Cas9 technology has great potential in onion for manipulation of a metabolic pathway to alter flavor profile, flowering behavior, quality, and traits of agronomic importance for increasing yield potential. Though onion is one of the most important vegetable crops throughout the world, the study for CRISPR/Cas9-mediated gene editing in onion has not been conducted yet. To the best of our knowledge, the present study is the first report on CRISPR/Cas9-mediated genome editing in onion.
The PDS gene is involved in the carotenoid biosynthesis pathway, and its loss-of-function mutation leads to the development of the photobleached or albino phenotype, which can be easily and quickly detected by naked eye and does not require any biochemical, physiological, or microscopic assays. Therefore, PDS has been widely used as a common visual marker to validate the practicability of genome editing by CRISPR/Cas9 in several plant species, viz. populus (Populus angustifolia) (Fan et al., 2015), apple (Malus prunifoli) (Nishitani et al., 2016), grape (Vitis vinifera) (Nakajima et al., 2017), cassava (Manihot esculenta) (Odipio et al., 2017), banana cv. Rasthali (Musa acuminata cv. Rasthali) (Kaur et al., 2018), cavendish banana (Musa acuminata subgroup Cavendish) (Naim et al., 2018), melon (Cucumis melo) (Hooghvorst et al., 2019), strawberry (Fragaria ananassa) (Wilson et al., 2019), lettuce (Lactuca sativa) (Henderson et al., 2020), poplar (Populus tomentosa Carr) (Fan et al., 2015; Wang et al., 2020), and yam (Dioscorea rotunda) (Syombua et al., 2021). Thus, we targeted the AcPDS gene to evaluate the editing efficiency induced by the CRISPR/Cas9 system.
A detailed analysis of the targeted gene and its putative functions is a prerequisite for any genome editing studies. We retrieved the AcPDS sequence from the draft onion genome based on a homology search. As the B. Super cultivar was used for the genome editing study, the gene was additionally isolated from the Indian onion cv. B. Super and afterward employed for designing and constructing targets for genome editing studies. PDS has been reported as a single copy gene in many crop species like melon (Hooghvorst et al., 2019), cassava (Odipio et al., 2017), and cavendish banana (Naim et al., 2018). Likewise, BLASTn analysis also showed a single copy of the PDS gene in the draft genome sequence of onion. Furthermore, in-silico analysis of the AcPDS protein sequence from B. Super cultivar showed that it contained the characteristic signatures of the conserved amino-oxidase domain known from other crops (Li et al., 2013b; Kaur et al., 2017). Therefore, we targeted the 5′ region of a conserved amino-oxidase domain of AcPDS to ensure functional knockdown upon mutation. Moreover, we selected two gRNA targets in the amino-oxidase domain, i.e., in exon 3 and exon 4, to improve our chances of success and to evaluate any large deletions in addition to InDels, as they were used in other crops (Odipio et al., 2017; Ntui et al., 2020; Lu and Tian, 2022).
Onion is recalcitrant to transformation, has very low transformation efficiency (i.e., 0.2% to 2.5%), and transformation is highly genotype-dependent (Eady et al., 2000; Zheng et al., 2001; Eady et al., 2003; Zheng et al., 2005; Kamata et al., 2011; Naini et al., 2019; Manape et al., 2022). There are two reported techniques for the genetic transformation of onions: i.e., biolistic transformation and Agrobacterium-mediated transformation. A biolistic method is challenging to carry out within crop plants and requires expensive consumables such as gold particles or specialized machinery (e.g., gene gun). On the other hand, Agrobacterium-mediated is inexpensive, does not require any specialized equipment, and has been standardized in our laboratory for the onion cv. B. Super with 1% transformation efficiency (Manape et al., 2022). Therefore, we employed the same protocol to study the CRISPR/Cas9-based genome editing in the same onion cultivar and regenerated six Cas9 PCR-positive shoots out of 617 co-cultivated calli with a comparable transformation efficiency of 0.97%.
In general, putatively transformed calli are selected on growth medium by using either negative agents, namely, kanamycin, hygromycin B, and spectinomycin, or positive ones like mannose (Bibi et al., 2013; Rastogi et al., 2018). Negative selection can inhibit and delay shoot growth, cause chlorosis in shoots, and produce pale green phenotypes, as observed in crops like sugarcane (Rastogi et al., 2018) and cotton (Bibi et al., 2013). The pale green phenotypes were also observed due to Cas9-induced InDels in the PDS gene of the Cavendish banana (Naim et al., 2018) and hybrid poplar (Wang et al., 2020). This necessitates the screening of pale green shoots to differentiate between non-transformed and transformed shoots, which is a laborious and time-consuming process. In the onion variety B. Super, embryogenic calli induce shoots within 3 weeks on a shooting medium devoid of hygromycin B, whereas transformed calli take 6–8 weeks in the presence of hygromycin B, occasionally producing shoots with chlorotic effects (unpublished data). However, using a positive selection method requires optimization of the selection agent concentration, which is also labor- and time-intensive. Given that the regeneration protocol was already standardized for onion cv. B. Super, we transferred co-cultivated calli directly to a shooting medium without hygromycin B. This approach eliminated confusion over pale green shoots and expedited the regeneration protocol for quick validation of CRISPR/Cas9-mediated genome editing. Our study successfully regenerated 21 shoots with varying degrees of albino or chlorophyll loss within 7–8 weeks using Agrobacterium-mediated co-transformation of embryogenic calli of onion with two independent AcPDS targeting CRISPR/Cas9 constructs. This is significantly faster than the 22–24 weeks required by the transformation protocol that we had developed earlier (Manape et al., 2022). Of these 21 shoots, two were complete albinos, four were chimeric, and 15 were pale green. Similar phenotypes have been reported in PDS knockout experiments in various plant species (Pan et al., 2016; Naim et al., 2018; Hooghvorst et al., 2019; Henderson et al., 2020; Wang et al., 2020; Lu and Tian, 2022).
To ascertain genome editing in onion, PCR analysis was performed with a flanking primer set in six Cas9-positive regenerated shoots, and multiple clones of each were sequenced on Sanger’s platform to investigate mutations within and around one or both target sites. Sanger sequencing results revealed InDels and substitutions in and around the target sites of both sgRNAs in the albino phenotypes. A similar type of mutation has also been reported in other plants, including Arabidopsis (Arabidopsis thaliana) (Feng et al., 2014), apple (Malus prunifoli) (Nishitani et al., 2016), and Rasthali banana (Musa acuminata cv. Rasthali) (Kaur et al., 2018). The InDels recorded here could have resulted from a nonhomologous end-joining repair (NHEJ) pathway following the integration of T-DNA (Fan et al., 2015; Nishitani et al., 2016).
InDels are the most common mutations in CRISPR/Cas9-mediated gene editing, induced by NHEJ. The evaluation of the Sanger sequencing data, using multiple sequence alignment and the Synthego ICE Analysis tool, was found to be consistent with the results from NGS data. Both sets of data indicated similar InDels in the vicinity of the target sgRNAs. NGS data showed a high level (63.4% in exon 3 sgRNA) to a low level (1.1% in exon 4 sgRNA) of InDel frequencies. This clearly demonstrated a significant difference in the editing efficiency of exon 3 and exon 4 sgRNAs. The GC content of the target sgRNA sequence and secondary structure of targeted sgRNAs affect the editing efficiency of Cas9 (Wang et al., 2014; Zhang et al., 2014; Ma et al., 2015). Both the sgRNAs had 50% GC content. Therefore, we analyzed the secondary structures of both target sgRNAs. Analyzing the secondary structure revealed that exon 3 target sgRNA had pairings of only 2 to 4 bp (i.e., continuous pairing less than 5 bp) in the target sgRNA sequence. However, exon 4 target sgRNA had a typical stem-loop structure with a pairing of continuous 7 and 4 bp of the target sgRNA sequence (Supplementary Figure S7). Secondary structures of target-sgRNAs with more than continuous 6 bp pairing of the target sgRNA sequence with stem-loop structures had significantly low editing efficiency, possibly due to inhibition of binding of the sgRNA to the target strand attributed to the formation of stem-loop structures (Ma et al., 2015).
Along with InDels, we also found a high frequency (54.84%) of substitutions in the exon 3 sgRNA sequence. Similar findings on the high level of substitutions mediated by CRISPR/Cas9 have been made by other studies. For example, in cotton (Gossypium hirsutum L.) protoplasts, only substitutions were found (Chen et al., 2017); in melon protoplasts and plants, 91% of mutations were substitutions (Hooghvorst et al., 2019); in cassava plants, substitutions occurred more frequently than InDel mutations (Odipio et al., 2017); in soybean (Glycine max L.) protoplasts and plants, a high level of substitutions were reported in the Cas9-edited events (Sun et al., 2015); and in rice (Oryza sativa L.), 25%–45% of the examined mutations were substitutions (Macovei et al., 2018). Observation of nucleotide substitution downstream of the target region implies activation of the homologous recombination (HR) pathway to repair double-strand breaks created in AcPDS. Simultaneous triggering of HR and NHEJ repair pathways proceeding integration of CRISPR/Cas9 gene T-DNA was documented earlier in apple (Malus prunifoli), Arabidopsis (Arabidopsis thaliana), tobacco (N. benthamiana), cassava (Manihot esculenta), and cotton (Gossypium hirsutum) (Li et al., 2013a; Nishitani et al., 2016; Gao et al., 2017; Odipio et al., 2017).
Four regenerated chimeric shoots showed the presence of the Cas9 gene, but no editing was detected in and around the target sites when Sanger sequencing results were analyzed. However, NGS data revealed additional InDels and substitutions, albeit at very low frequencies. As we sequenced only a few clones from each chimeric shoot using the Sanger platform, it is likely that we were not able to detect these very low frequencies of InDels and substitutions, which potentially could have been induced by transient expression of the Cas9 gene and sgRNA from the transformed construct. Bleached patches of tissue, similar to our observations, were reported in a study on yam, and they were attributed to the transient expression of Cas9 and PDS-targeted sgRNA (Syombua et al., 2021). While this is a possibility in our study, another plausible explanation could stem from the use of a non-selective medium leading to the regeneration of chimeric transformants.
In our study, out of the six Cas9-positive shoots identified, two demonstrated CRISPR/Cas9-mediated genome editing in the AcPDS gene, yielding an editing efficiency of 33.33%. Mutation efficiency in monocotyledons using 35S promoter-driven Cas9 constructs is 1.8%–81% (Basu et al., 2023). Therefore, mutation efficiency is moderate and can be improved in future studies. We used the pRGEB31 vector, which contains the cauliflower mosaic virus (CaMV35S) and Oryza sativa U3 promoter for driving the expression of Cas9 and sgRNAs, respectively. Optimal CRISPR/Cas9 function requires high transcription levels of both the sgRNA and Cas9 proteins (Mao et al., 2016). Editing efficiency is significantly influenced by the choice of promoter driving Cas9, with endogenous constitutive promoters yielding high transgene expression and, consequently, higher mutation efficiencies (Hassan et al., 2021). Similarly, the endogenous U6 promoter has been shown to enhance sgRNA expression and thus the overall CRISPR/Cas9-mediated editing efficiency by up to 87% (Yan et al., 2015; Zhang et al., 2018). As the promoters used in our study were not endogenous, it may have contributed to the less effective performance of the CRISPR/Cas9 system. Therefore, the utilization of endogenous promoters for Cas9 and sgRNA expression could be a more effective strategy for boosting CRISPR/Cas9 efficiency in onions.
We estimated the chlorophyll content in the regenerated albino, chimeric, pale green, and non-transformed control shoots to investigate the impact of AcPDS mutations. We observed a significant reduction in chlorophyll content in albino and chimeric shoots. InDels produced frameshift mutations and ectopic stop codons in the AcPDS gene of albino shoots. These alterations led to the early termination of AcPDS protein, leading to albinism; consistent with findings in Rasthali banana (Kaur et al., 2018). The variegation and reduced chlorophyll content in chimeric phenotypes could be the result of a transient expression of Cas9-mediated alterations in AcPDS. Similar observations were reported in yam, where bleached patches on shoots were linked to Agrobacterium-mediated transient expression of Cas9 and PDS sgRNA (Syombua et al., 2021). The nonsignificant reduction in chlorophyll content in the 15 pale green calli may be attributed to somaclonal variation induced by frequent culturing on the shooting medium. Further detailed analysis is required to confirm this hypothesis. In summary, the substantial decrease in chlorophyll content in albino shoots indicates a functional knockout of the AcPDS gene. This disruption likely affects both the chlorophyll and gibberellic acid biosynthesis pathways. The slow growth rate observed in albino plants may be due to the disruption of gibberellic acid biosynthesis, which hinders their growth and development. This is supported by a study in Arabidopsis, where the application of exogenous GA3 partially reversed the dwarf phenotypes of AcPDS mutants (Qin et al., 2007).
In conclusion, the CRISPR/Cas9-mediated AcPDS knockout showcased in this study produced a readily observable mutant phenotype within a rapid timeframe of 8-weeks. This strategy provides a valuable tool for accelerating the evaluation and optimization of CRISPR/Cas9-mediated genome editing in onions, ultimately contributing to the improvement of their traits. While the observed editing efficiency in our study was moderate, the use of an endogenous promoter-driven Cas9 and sgRNA could potentially enhance it in future applications.
The data presented in the study are deposited in the NCBI database repository, accession number PRJNA976681 (http://www.ncbi.nlm.nih.gov/bioproject/976681). The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
PM, TKM, and SA conceived and designed the experiments. PM and TKM conducted all the experiments. PM, TKM, and SA drafted the manuscript. SA and VS carried out the data analysis and manuscript editing. SA supervised the entire work. All authors contributed to the article and approved the submitted version.
The research work was supported by a grant from the Indian Council of Agricultural Research (ICAR)-National Fellow Project (Project Code: HORTDOGRSOL201700800088).
Financial support and fellowship to PM and TKM provided by the Indian Council of Agricultural Research (ICAR)-National Fellow Project, Project Code: HORTDOGRSOL201700800088) is gratefully acknowledged. The authors thank Dr. Major Singh, Former Director, ICAR-DOGR, for his support during the research work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1226911/full#supplementary-material
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts. polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–15. doi: 10.1104/pp.24.1.1
Basu, U., Riaz Ahmed, S., Bhat, B. A., Anwar, Z., Ali, A., Ijaz, A., et al. (2023). A CRISPR way for accelerating cereal crop improvement: Progress and challenges. Front. Genet. 13. doi: 10.3389/FGENE.2022.866976
Bibi, N., Fan, K., Yuan, S., Ni, M., Ahmed, I. M., Malik, W., et al. (2013). An efficient and highly reproducible approach for the selection of upland transgenic cotton produced by pollen tube pathway method. Aust. J. Crop Sci. 7, 1714–1722.
Bisen, S. P., Emerald, M. (2016). Nutritional and therapeutic potential of garlic and onion (Allium sp.). Current Nutrition & Food Science 12 (3), 190–199. doi: 10.2174/1573401312666160608121954
Brewster, J. L. (2008). Onions and other vegetable alliums. Onions other Veg. alliums. 15. doi: 10.1079/9781845933999.0000
Chen, X., Lu, X., Shu, N., Wang, S., Wang, J., Wang, D., et al. (2017). Targeted mutagenesis in cotton (Gossypium hirsutum L.) using the CRISPR/Cas9 system. Sci. Rep. 7, 1–7. doi: 10.1038/srep44304
Eady, C. C., Davis, S., Farrant, J., Reader, J., Kenel, F. (2003). Agrobacterium tumefaciens-mediated transformation and regeneration of herbicide resistant onion (Allium cepa) plants. Ann. Appl. Biol. 142, 213–217. doi: 10.1111/j.1744-7348.2003.tb00243.x
Eady, C. C., Weld, R. J., Lister, C. E. (2000). Agrobacterium tumefaciens-mediated transformation and transgenic-plant regeneration of onion (Allium cepa L.). Plant Cell Rep. 19, 376–381. doi: 10.1007/s002990050743
Fan, D., Liu, T., Li, C., Jiao, B., Li, S., Hou, Y., et al. (2015). Efficient CRISPR/cas9-mediated targeted mutagenesis in populus in the first generation. Sci. Rep. 5, 1–7. doi: 10.1038/srep12217
Feng, Z., Mao, Y., Xu, N., Zhang, B., Wei, P., Yang, D. L., et al. (2014). Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 111, 4632–4637. doi: 10.1073/PNAS.1400822111
Finkers, R., van Kaauwen, M., Ament, K., Burger-Meijer, K., Egging, R., Huits, H., et al. (2021). Insights from the first genome assembly of Onion (Allium cepa). G3, 11 (9) 1–8. Genes|Genomes|Genetics 11. doi: 10.1093/G3JOURNAL/JKAB243
Gao, W., Long, L., Tian, X., Xu, F., Liu, J., Singh, P. K., et al. (2017). Genome editing in cotton with the CRISPR/Cas9 system. Front. Plant Sci. 8. doi: 10.3389/FPLS.2017.01364
Goldman, I. L. (2011). Molecular breeding of healthy vegetables. EMBO Rep. 12, 96–102. doi: 10.1038/EMBOR.2010.215
Hassan, M. M., Zhang, Y., Yuan, G., De, K., Chen, J. G., Muchero, W., et al. (2021). Construct design for CRISPR/Cas-based genome editing in plants. Trends Plant Sci. 26, 1133–1152. doi: 10.1016/J.TPLANTS.2021.06.015
Henderson, S. W., Henderson, S. T., Goetz, M., Koltunow, A. M. G. (2020). Efficient CRISPR/cas9-mediated knockout of an endogenous PHYTOENE DESATURASE gene in T1 progeny of apomictic hieracium enables new strategies for apomixis gene identification. Genes 11, 1064. doi: 10.3390/GENES11091064
Hooghvorst, I., López-Cristoffanini, C., Nogués, S. (2019). Efficient knockout of phytoene desaturase gene using CRISPR/Cas9 in melon. Sci. Rep. 9, 1–7. doi: 10.1038/s41598-019-53710-4
Kamata, Y., Masamura, N., Miyazaki, A., Nagata, T. (2011). A novel autofluorescence-based selection of calli amenable to Agrobacterium-mediated transformation in onion (Allium cepa L.). Plant Biotechnol. 28, 361–371. doi: 10.5511/plantbiotechnology.11.0513a
Kaur, N., Alok, A., Shivani, Kaur, N., Pandey, P., Awasthi, P., et al. (2018). CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Funct. Integr. Genomics 18, 89–99. doi: 10.1007/S10142-017-0577-5
Kaur, N., Pandey, A., Shivani, Kumar, P., Pandey, P., Kesarwani, A. K., et al. (2017). Regulation of banana phytoene synthase (MaPSY) expression, characterization and their modulation under various abiotic stress conditions. Front. Plant Sci. 8. doi: 10.3389/FPLS.2017.00462
Koltun, A., Corte, L. E. D., Mertz-Henning, L. M., Gonçalves, L. S. A. (2018). Genetic improvement of horticultural crops mediated by CRISPR/Cas: a new horizon of possibilities. Hortic. Bras. 36, 290–298. doi: 10.1590/S0102-053620180302
Lee, Y. R., Kim, C. W., Han, J., Choi, H. J., Han, K., Lee, E. S., et al. (2021). Genotyping-by-sequencing derived genetic linkage map and quantitative trait loci for sugar content in onion (Allium cepa L.). Plants 10, 2267. doi: 10.3390/PLANTS10112267
Li, M., Gan, Z., Cui, Y., Shi, C., Shi, X. (2013b). Structure and function characterization of the phytoene desaturase related to the lutein biosynthesis in Chlorella protothecoides CS-41. Mol. Biol. Rep. 40, 3351–3361. doi: 10.1007/S11033-012-2410-5
Li, J. F., Norville, J. E., Aach, J., McCormack, M., Zhang, D., Bush, J., et al. (2013a). Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691. doi: 10.1038/nbt.2654
Lu, Q. S. M., Tian, L. (2022). An efficient and specific CRISPR-Cas9 genome editing system targeting soybean phytoene desaturase genes. BMC Biotechnol. 22, 1–16. doi: 10.1186/S12896-022-00737-7
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/J.MOLP.2015.04.007
Macovei, A., Sevilla, N. R., Cantos, C., Jonson, G. B., Slamet-Loedin, I., Čermák, T., et al. (2018). Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol. J. 16, 1918–1927. doi: 10.1111/PBI.12927
Manape, T. K., Satheesh, V., Singh, S., Singh, M., Anandhan, S. (2022). Improved method for regeneration and Agrobacterium-mediated transformation of Indian short-day onion (Allium cepa L.). Plant Cell. Tissue Organ Cult. 148, 61–72. doi: 10.1007/S11240-021-02161-9
Mao, Y., Botella, J. R., Zhu, J. K. (2016). Heritability of targeted gene modifications induced by plant-optimized CRISPR systems. Cell. Mol. Life Sci. 74, 1075–1093. doi: 10.1007/S00018-016-2380-1
Naim, F., Dugdale, B., Kleidon, J., Brinin, A., Shand, K., Waterhouse, P., et al. (2018). Gene editing the phytoene desaturase alleles of Cavendish banana using CRISPR/Cas9. Transgenic Res. 27, 451–460. doi: 10.1007/S11248-018-0083-0
Naini, R., Pavankumar, P., Prabhakar, S., Kancha, R. K., Rao, K. V., Reddy, V. D. (2019). Evolvement of nutraceutical onion plants engineered for resveratrol biosynthetic pathway. Plant Cell Rep. 38, 1127–1137. doi: 10.1007/s00299-019-02432-y
Nakajima, I., Ban, Y., Azuma, A., Onoue, N., Moriguchi, T., Yamamoto, T., et al. (2017). CRISPR/Cas9-mediated targeted mutagenesis in grape. PloS One 12, e0177966. doi: 10.1371/JOURNAL.PONE.0177966
Nishitani, C., Hirai, N., Komori, S., Wada, M., Okada, K., Osakabe, K., et al. (2016). Efficient genome editing in apple using a CRISPR/cas9 system. Sci. Rep. 6, 1–8. doi: 10.1038/srep31481
Ntui, V. O., Tripathi, J. N., Tripathi, L. (2020). Robust CRISPR/Cas9 mediated genome editing tool for banana and plantain (Musa spp.). Curr. Plant Biol. 21, 100128. doi: 10.1016/J.CPB.2019.100128
Odipio, J., Alicai, T., Ingelbrecht, I., Nusinow, D. A., Bart, R., Taylor, N. J. (2017). Efficient CRISPR/cas9 genome editing of phytoene desaturase in cassava. Front. Plant Sci. 8. doi: 10.3389/FPLS.2017.01780
Pan, C., Ye, L., Qin, L., Liu, X., He, Y., Wang, J., et al. (2016). CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 6, 1–9. doi: 10.1038/srep24765
Park, J., Lim, K., Kim, J. S., Bae, S. (2017). Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics 33, 286–288. doi: 10.1093/BIOINFORMATICS/BTW561
Putnik, P., Gabrić, D., Roohinejad, S., Barba, F. J., Granato, D., Lorenzo, J. M., et al. (2019). Bioavailability and food production of organosulfur compounds from edible Allium species. Innov. Therm. Non-Thermal Process. Bioaccessibility Bioavailab. Nutr. Bioact. Compd., 293–308. doi: 10.1016/B978-0-12-814174-8.00010-X
Qin, G., Gu, H., Ma, L., Peng, Y., Deng, X. W., Chen, Z., et al. (2007). Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 17, 471–482. doi: 10.1038/cr.2007.40
Rastogi, J., Jyoti Rastogi, C., Bubber, P., Singh, R., Singh, R. (2018). Determination of minimal inhibitory concentration of kanamycin as selective agents and marker genes for use in Agrobacterium mediated transformation in sugarcane. J. Pharmacogn. Phytochem. 7, 1861–1864.
Razzaq, A., Saleem, F., Kanwal, M., Mustafa, G., Yousaf, S., Arshad, H. M. I., et al. (2019). Modern trends in plant genome editing: an inclusive review of the CRISPR/cas9 toolbox. Int. J. Mol. Sci. 20, 4045. doi: 10.3390/IJMS20164045
Sudha, G. S., Ramesh, P., Sekhar, A. C., Krishna, T. S., Bramhachari, P. V., Riazunnisa, K. (2019). Genetic diversity analysis of selected Onion (Allium cepa L.) germplasm using specific RAPD and ISSR polymorphism markers. Biocatal. Agric. Biotechnol. 17, 110–118. doi: 10.1016/J.BCAB.2018.11.007
Sun, X., Hu, Z., Chen, R., Jiang, Q., Song, G., Zhang, H., et al. (2015). Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci. Rep. 5 (1), 10342. doi: 10.1038/SREP10342
Syombua, E. D., Zhang, Z., Tripathi, J. N., Ntui, V. O., Kang, M., George, O. O., et al. (2021). A CRISPR/Cas9-based genome-editing system for yam (Dioscorea spp.). Plant Biotechnol. J. 19, 645–647. doi: 10.1111/PBI.13515
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/MOLBEV/MSAB120
Van den Broeck, G., Maertens, M. (2016). Horticultural exports and food security in developing countries. Glob. Food Sec. 10, 11–20. doi: 10.1016/J.GFS.2016.07.007
Varshney, R. K., Ribaut, J. M., Buckler, E. S., Tuberosa, R., Rafalski, J. A., Langridge, P. (2012). Can genomics boost productivity of orphan crops? Nat. Biotechnol. 30, 1172–1176. doi: 10.1038/nbt.2440
Wang, T., Wei, J. J., Sabatini, D. M., Lander, E. S. (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84. doi: 10.1126/SCIENCE.1246981
Wang, J., Wu, H., Chen, Y., Yin, T. (2020). Efficient CRISPR/cas9-mediated gene editing in an interspecific hybrid poplar with a highly heterozygous genome. Front. Plant Sci. 11. doi: 10.3389/FPLS.2020.00996
Wilson, F. M., Harrison, K., Armitage, A. D., Simkin, A. J., Harrison, R. J. (2019). CRISPR/Cas9-mediated mutagenesis of phytoene desaturase in diploid and octoploid strawberry. Plant Methods 15, 1–13. doi: 10.1186/S13007-019-0428-6
Yan, L., Wei, S., Wu, Y., Hu, R., Li, H., Yang, W., et al. (2015). High-efficiency genome editing in arabidopsis using YAO promoter-driven CRISPR/cas9 system. Mol. Plant 8, 1820–1823. doi: 10.1016/j.molp.2015.10.004
Zhang, J., Zhang, H., Botella, J. R., Zhu, J. K. (2018). Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J. Integr. Plant Biol. 60, 369–375. doi: 10.1111/JIPB.12620
Zhang, H., Zhang, J., Wei, P., Zhang, B., Gou, F., Feng, Z., et al. (2014). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12, 797–807. doi: 10.1111/PBI.12200
Zheng, S. J., Henken, B., De Maagd, R. A., Purwito, A., Krens, F. A., Kik, C. (2005). Two different Bacillus thuringiensis toxin genes confer resistance to beet armyworm (Spodoptera exigua Hübner) in transgenic Bt-shallots (Allium cepa L.). Transgenic Res. 14, 261–272. doi: 10.1007/s11248-005-0109-2
Keywords: PDS, onion transformation, CRISPR/Cas9, genome editing, chlorophyll
Citation: Mainkar P, Manape TK, Satheesh V and Anandhan S (2023) CRISPR/Cas9-mediated editing of PHYTOENE DESATURASE gene in onion (Allium cepa L.). Front. Plant Sci. 14:1226911. doi: 10.3389/fpls.2023.1226911
Received: 22 May 2023; Accepted: 28 July 2023;
Published: 28 August 2023.
Edited by:
Suprasanna Penna, Amity University, Mumbai, IndiaReviewed by:
Jitesh Kumar, University of Minnesota Twin Cities, United StatesCopyright © 2023 Mainkar, Manape, Satheesh and Anandhan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sivalingam Anandhan, YW5hbmRoYW5zQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work and share first authorship
‡ORCID: Sivalingam Anandhan, orcid.org/0000-0002-1310-0807
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.