
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 25 September 2023
Sec. Plant Breeding
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1221537
This article is part of the Research Topic Enhancing Salinity Tolerance in Crops - Molecular and Practical Perspectives View all 6 articles
 Raj Kumar Gautam1,2*
Raj Kumar Gautam1,2* Pankaj Kumar Singh1
Pankaj Kumar Singh1 Krishnan Sakthivel1,3
Krishnan Sakthivel1,3 K. Venkatesan1,4
K. Venkatesan1,4 Shyam S. Rao1
Shyam S. Rao1 M. Srikumar1
M. Srikumar1 Joshitha Vijayan1,5
Joshitha Vijayan1,5 B. Rakesh1
B. Rakesh1 Soham Ray6
Soham Ray6 Jameel Akhtar2
Jameel Akhtar2 Bharat Raj Meena2
Bharat Raj Meena2 Sapna Langyan2*
Sapna Langyan2* Sharik Ali2
Sharik Ali2 S. L. Krishnamurthy7
S. L. Krishnamurthy7Introduction: Bacterial blight (BB) caused by Xanthomonas oryzae pv. oryzae is a major disease of rice, specially in the tropical regions of the world. Developing rice varieties with host resistance against the disease is the most effective and economical solution for managing the disease.
Methods: Pyramiding resistance genes (Xa4, xa5, xa13,and Xa21) in popular rice varieties using marker-assisted backcross breeding (MABB) has been demonstrated as a cost-effective and sustainable approach for establishing durable BB resistance. Here, we report our successful efforts in introgressing four resistance genes (Xa4, xa5, xa13, and Xa21) from IRBB60 to CARI Dhan 5, a popular salt-tolerant variety developed from a somaclonal variant of Pokkali rice, through functional MABB.
Results and discussion: Both BB and coastal salinity are among the major challenges for rice production in tropical island and coastal ecosystems. Plants with four, three, and two gene pyramids were generated, which displayed high levels of resistance to the BB pathogen at the BC3F2 stage. Under controlled salinity microplot environments, the line 131-2-175-1223 identified with the presence of three gene pyramid (Xa21+xa13+xa5) displayed notable resistance across locations and years as well as exhibited a salinity tolerance comparable to the recurrent parent, CARI Dhan 5. Among two BB gene combinations (Xa21+xa13), two lines, 17-1-69-334 and 46-3-95-659, demonstrated resistance across locations and years, as well as salt tolerance and grain production comparable to CARI Dhan 5. Besides salinity tolerance, five lines, 17-1-69-179, 46-3-95-655, 131-2-190-1197, 131-2-175-1209, and 131-2-175-1239, exhibited complete resistance to BB disease. Following multilocation testing, potential lines have been identified that can serve as a prospective candidate for producing varieties for the tropical Andaman and Nicobar Islands and other coastal locations, which are prone to BB and coastal salinity stresses.
Rice (Oryza sativa L.) is the world’s primary carbohydrate source, feeding nearly half of the global population. India must produce 135–140 million tons of rice by 2030 to maintain self-sufficiency and meet future food grain demands (Gujja and Thiyagarajan, 2009). This goal must be achieved while considering the challenges posed by limited agricultural lands, depleting water supplies, declining soil productivity, emerging pests and pathogens, and the potential adverse effects of climate change. Historically, rice has also been the principal grain crop of the Andaman and Nicobar Islands, situated in the Bay of Bengal, despite its poor average yield. The farmers of the Andaman and Nicobar Islands belonging to diverse socio-ethnic groups and tribes mostly cultivate traditional rice varieties. The warm and humid conditions prevalent almost throughout the year in tropical islands facilitate the growth and multiplication of biotic stress factors, including diseases and insect pests. Among the biotic stresses, bacterial blight (BB) caused by Xanthomonas oryzae pv. oryzae is one of the most important diseases of rice, which spreads epidemically in susceptible cultivars and causes yield losses ranging from 20% to 50%, depending on weather conditions (Singh et al., 2011). The favorable temperature for the development of disease is between 25 and 34°C, with a relative humidity above 70%, which commonly coincides with the weather conditions of the islands. The problem is further exacerbated by susceptible varieties, poor management practices, and closed farming system scattered across different islands. Chemical disease management is not a recommended practice in these islands due to its high cost and negative environmental impact on terrestrial and marine ecosystems. The prevalence of BB has a visible and direct detrimental effect on the Paddy-cum-fish and duck-type farming systems, as well as an indirect impact on the coastal mangrove ecology (Chukwu et al., 2019). The most effective approach to minimizing losses is to develop resistant/tolerant rice varieties. Additionally, the Andaman and Nicobar Islands face challenges from coastal salinity and tsunami effects, which affect about one-third of the total area and are compounded by the region’s vulnerability to climate change. While salt stress poses a worldwide threat to agricultural production (Hopmans et al., 2021), the effects of salinity are exacerbated in high rainfall coastal and islands’ ecosystems due to the presence of acid sulfate soils (Sarangi et al., 2022).
The development of resistant cultivars carrying resistant genes has been the most effective and economical strategy for controlling BB disease without causing environmental pollution (Huang et al., 1997; Singh et al., 2001; Jena and Mackill, 2008; Sundaram et al., 2008; Rajpurohit et al., 2011; Dokku et al., 2013; Suh et al., 2013). To confer resistance against BB, more than 46 disease-resistance (R) genes have been identified and designated with a series from Xa1 to Xa46 (Fiyaz et al., 2022). Many of these genes are tagged with DNA-based markers, and some have been cloned and characterized (Kumar et al., 2020). The marker-assisted introgression of multiple resistance genes into a single genetic background is invariably preferred because the probability of simultaneous mutation at multiple loci to overcome the resistance is low (Koide et al., 2010).
Pokkali is a traditionally popular rice variety cultivated in the coastal saline areas of Kerala, India (Gregorio et al., 2002). It has been identified as a universal donor of salt tolerance genes, of which Saltol is the most prominent (Thomson et al., 2010). The analysis of phenotypic responses, genomic composition, and QTLs present in the salt-tolerant introgressed lines (ILs) developed from a salt-tolerant rice landrace “Pokkali” using SSR and SNP markers has revealed potential salt tolerance mechanisms in these ILs, including Na+ dilution in leaves, vacuolar Na+ compartmentalization, and possibly the synthesis of compatible solutes (De Leon et al., 2017). Besides salt tolerance, Pokkali has also been found to be nutritionally rich in various micronutrients, bran oil, and antioxidants such as oryzanols, tocopherols, and tocotrienols (Shylaraj et al., 2018). In a quest to develop an agronomically desirable plant type, induction of somaclonal variation in Pokkali led to the development of an elite variety called CARI Dhan 5 (Mandal et al., 1999), which is now an increasingly popular and successful salt-tolerant variety in Andaman and Nicobar Islands due to its wide seed dissemination and larger-scale adoption (Singh et al., 2020). However, it is important to note that CARI Dhan 5 is susceptible to BB, which remains a major disease of rice in the tropical Andaman Islands.
Through our systematic research, we have evaluated the relative efficacy of various resistance genes for managing BB disease, further complemented by analyzing the diversity, distribution, and virulence of the pathogen. This has provided a deeper understanding of the disease dynamics and has opened up new opportunities through the host pathogen approach under Bay Islands (Gautam et al., 2015; Gautam et al., 2016a; Gautam et al., 2016b; Sakthivel et al., 2016; Sakthivel et al., 2017; Sakthivel et al., 2021). Our previous study proved the effectiveness of four gene combinations (Xa4, xa5, xa13, and Xa21) carried by IRBB60 under Andaman Islands conditions (Gautam et al., 2013). This led us to pyramid these resistance genes into the popular salt-tolerant variety CARI Dhan 5 using marker-assisted backcross breeding, considering the effectiveness of these genes and the susceptibility of CARI Dhan 5 to BB.
The donor parent, IRBB 60, possessed an impressive lineage of four BB resistance genes, namely, Xa4, xa5, xa13, and Xa21, which were incorporated into the background of IR24 through development at the International Rice Research Institute (IRRI), Philippines. On the other hand, the recurrent parent, salt-tolerant CARI Dhan5, was developed from the somaclonal variant selection from Pokkali (Mandal et al., 1999) and successfully notified for commercial cultivation in coastal saline and acid sulfate soils of the Andaman and Nicobar Islands. The crossing scheme for this hybridization was executed at Bloomsdale Research Farm, ICAR-CIARI, Port Blair, which is illustrated in Figure 1.
Following 40 days of planting, both the parents and hybridization-derived plants were inoculated with a virulent isolate from the Andaman Islands using the clipping procedure as outlined by Kauffman (1973). After a period of 15 days post-inoculation, the lesion length of the inoculated leaves was meticulously measured. Disease scoring was carried out as per the standard evaluation system (SES, IRRI, 1996; Sakthivel et al., 2017). Based on the mean lesion length, the lines were categorized into four distinct groups, namely, resistant (<5 cm), moderately resistant (5–10 cm), moderately susceptible (10–15 cm), and susceptible (>15 cm). A BB-resistant and -susceptible reaction after inoculation is presented in Figure 2.

Figure 2 BB resistance testing in resistant introgression lines (A) and in the recurrent parent (B).
In the study, a total of 35 pyramided lines (BC3F3/4) and a single check (CARI Dhan5) were transplanted in both normal and saline environments in microplots and experimental fields located at ICAR-CIARI, Port Blair, in the Andaman and Nicobar Islands. Various agronomic traits were recorded, including plant height, days to 50% flowering, number of tillers per plant, panicle length, leaf length, leaf width, grain length, grain width, 100-seed weight, and phenotypically acceptability score (Table S4). To evaluate the distinctness, uniformity, and stability (DUS) of the 15 pyramided lines in normal soil, 41 agro-morphological traits were recorded at Bloomdale Farm of ICAR-CIARI, Port Blair, in the Andaman and Nicobar Islands, during the kharif season of 2020. The chosen advanced generation pyramided lines were phenotyped for their grain yield and related traits performance in both salinity stress (EC 5.0 dSm−1) and normal soil conditions in a controlled micro-plot facility at ICAR-CIARI, Port Blair, Andaman and Nicobar Islands. Here, it is pertinent to mention that the 30-day-old nursery seedlings were transplanted in microplots. After 7 days of seedling establishment, artificially prepared saline water (EC ~ 5.00 dS/m) was applied throughout the crop season to maintain this level of salt stress until maturity. On the other hand, only normal or fresh water (EC ~ 0.00 dS/m) was applied to the normal microplots. The pyramided lines were also screened for BB resistance along with their evaluation for grain yield and related traits.
The CTAB method described by Doyle and Doyle (1990) was used to isolate high-quality genomic DNA from rice. In brief, this method involves the use of CTAB, a cationic detergent, to extract DNA from the plant tissues, followed by precipitation and washing steps to purify the DNA. The resulting DNA was quantified and used as a template for PCR amplification. The PCR reaction mixture consisted of 50 ng of template DNA, 10 pmol of each of the primers, 200 μM of each dNTP, 1 × PCR buffer (10 mM Tris–HCl, pH 8.3, 50 mM KCl), 1.5 mM MgCl2, and 0.5 units of Taq DNA polymerase in a volume of 20 μl. The cycling conditions for the PCR reaction involved an initial denaturation step at 94°C for 5 min, followed by 35 cycles of amplification at 95°C for 30 s, 55°C for 30 s for primer annealing (Chen et al., 1997; Panaud et al., 1996), and 72°C for 1 min for extension. The final extension was carried out at 72°C for 5 min. Template DNA was followed with the following cycling conditions: 30 s primers annealing at 55°C. The PCR products and the DNA fragments produced were separated by gel electrophoresis using an agarose gel, and gel images were analyzed on an Alpha Imager (Alpha Innotech, USA).
Sequence tagged site (STS) markers as well as functional PCR-based markers, Xa4F/Xa4R, for the Xa4 gene (Ma et al., 1999; Sundaram et al., 2011); RG556 and xa5SF/xa5SR/xa5FM-RF/xa5FM-RR for the xa5 gene (Hajira et al., 2016); RG136 and xa13-prom for xa13 gene (Chu et al., 2006); and pTA248 for the Xa21 gene (Song et al., 1995) were used for the foreground selection of resistance allele of each gene during each backcross generation (Table 1). In case of Xa4, the resistant allele is of size 150 bp while the susceptible allele is of size 120 bp. We have marked the identity of these bands in both the susceptible parent (SP) and the resistant parent (RP) to prevent any ambiguity. In case of the xa5 gene, the 424-bp band is common to all while a 313-bp band marks susceptible allele and a 134-bp band marks the resistant allele. We have marked the identity of these bands in both the susceptible parent (SP) and the resistant parent (RP) to prevent any ambiguity. The detection of Xa13 and Xa21 was performed as per Chu et al. (2006) and Song et al. (1995), respectively. Parental polymorphism survey was done between donor (IRBB60) and recipient (CARI Dhan 5) varieties using 200 HvSSR markers to select polymorphic markers associated with the recipient parents for background selection in later generations. The genomic contribution of the parent was determined by the assessment of SSR marker data in the selected recombinants. The marker-assisted backcross breeding approach was implemented from the F1 stage up to the BC3F1 generation. At each stage, foreground selection was carried out to identify plants carrying the BB resistance genes. Only progenies with the resistance alleles were advanced to the next generation (Figure 1).
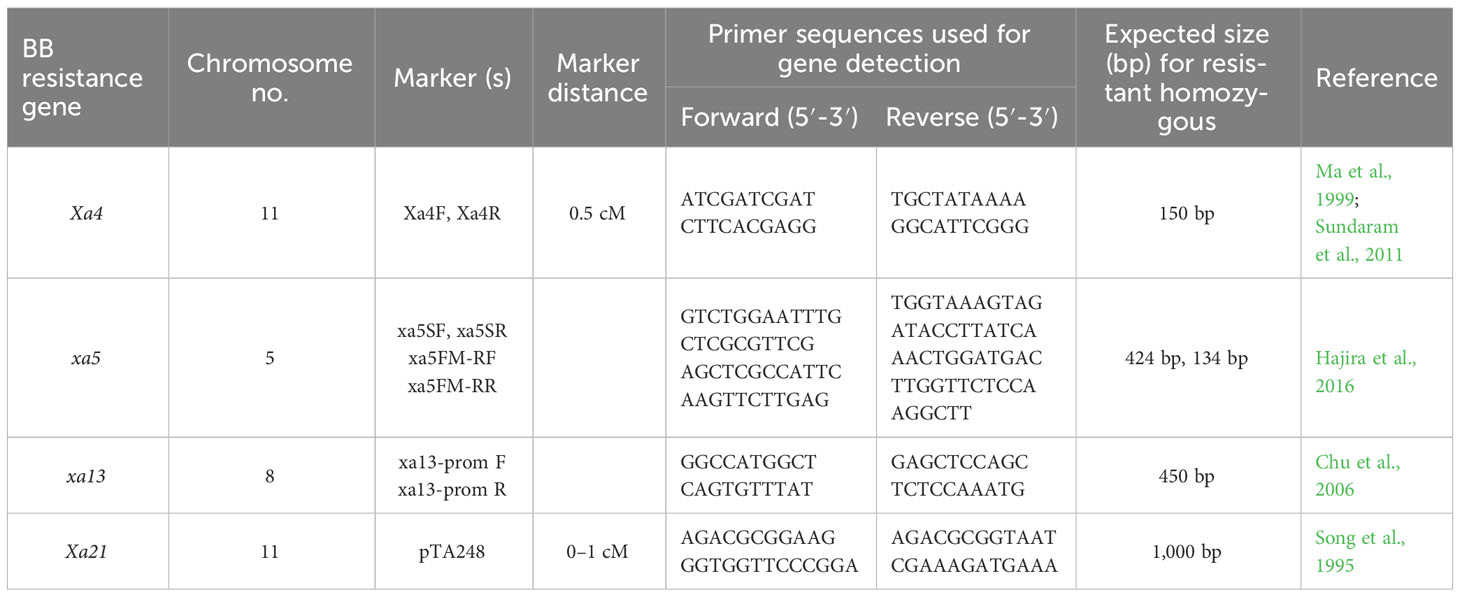
Table 1 Microsatellite markers used between CARI Dhan 5 and IRBB60 for BB resistance genes in foreground selection under study.
The experiments were performed in two to three replications and repeated twice for confirmation. The results were expressed as the mean ± SE of different independent replicates. An analysis of variance was performed in WASP software (version 2.0). P values of ≤0.05 were considered as statistically significant. The PCR amplification was performed in a Bio-Rad thermocycler (C1000 Thermal Cycler, Hong Kong, China). The images of the gel were analyzed on a Bio-Rad Gel Doc XR+System (California, USA).
The introgression of four BB resistance genes into CARI Dhan 5 was achieved via a crossing scheme (Figure 1) wherein IRBB60 (Xa4, xa5, xa13, and Xa21) was used as the donor parent. The results of the parental polymorphism survey revealed a significant level of polymorphism (23.5%) between CARI Dhan 5 (recurrent) and IRBB60 (non-recurrent) based on the analysis of 200 microsatellite markers. Among the markers surveyed, 47 were found to exhibit parental polymorphism. In addition, parental polymorphism was observed for all four target resistance genes, which was confirmed through the use of linked markers pTA248 (Xa21); RG136 (xa13); RG556, xa5SF/xa5SR (xa5); and Xa4F/Xa4R (Xa4).
In 2013–2014, F1 seeds were generated by crossing CARI Dhan 5 with IRBB 60. The BB-resistant F1 plants were confirmed based on hybridity for morphological and molecular markers of four resistance genes and were then pollinated by CARI Dhan 5 during the period 2014–2015 and 356 seeds of BC1F1 were obtained (Table 2). The progenies were thoroughly screened for BB resistance and the presence of linked genes through foreground selection, and BC2F1 crosses were attempted accordingly. In the Kharif season of 2015–2016, a total of 79 BC2F1 plants were screened using foreground selection, and only the plants exhibiting the presence of BB resistance genes were again pollinated by CARI Dhan 5 to obtain BC3 F1 seeds. In the year 2016–2017, a total of 3,505 BC3F1 plants were transplanted in the field, where foreground selection was carried out in conjunction with selection for CARI Dhan 5 phenotypes. Subsequently, the selected plants were self-pollinated to develop BC3F2 seeds.
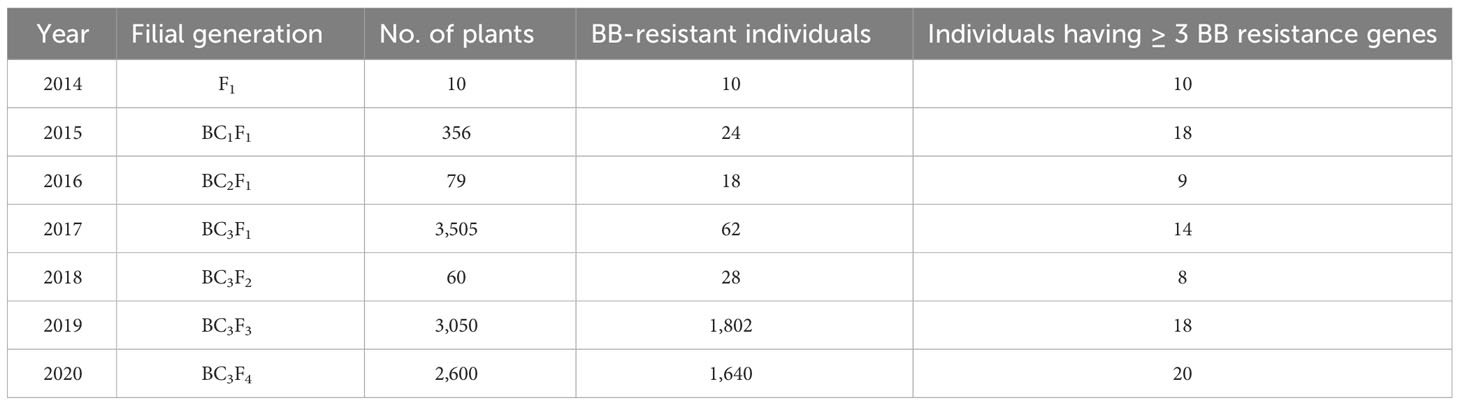
Table 2 Number and nature of breeding lines developed in a cross CARI Dhan 5 x IRBB60 over different filial generations.
To introgress BB resistance genes into CARI Dhan 5, single F1 plants were selected and further backcrossed with the recurrent parent. A total of 356 BC1F1 plants were produced and artificially inoculated with virulent Xoo isolates. Out of these, 24 plants scored highly for resistance to BB disease (Table 2). These twenty-four plants were subjected to foreground selection for the presence of BB resistance genes, namely, Xa4, xa5, xa13, and Xa21, by using corresponding linked markers. The molecular detection of the respective BB resistance genes, Xa4, xa5, xa13, and Xa21, in the BC1F1 plants was carried out, and specific bands were obtained as 19 (150 bp), 24 (300 bp), 16 (490 and 530 bp), and 18 (1,000 bp), respectively. Furthermore, the number of BC1F1 plants with different target resistance gene combinations obtained were as follows: (1) xa5, xa13, and Xa21 (4 lines viz., 21, 54, 80, and 93); (2) Xa4, xa5, and xa13 (1 line viz., 19); (3) Xa4, xa5, and Xa21 (4 lines viz., 43, 53, 57, and 79); and (4) Xa4, xa5, xa13, and Xa21(9 lines viz., 15, 17, 46, 86, 88, 113, 118, 131, and 134). Out of the nine BC1F1 plants carrying all four target resistance genes, three plants (17, 46, and 131) exhibited the highest similarity with CARI Dhan 5 and were selected for further back-crossing with the recurrent parent to obtain BC2F1 seeds.
Out of 79 BC2F1 progenies produced, 18 plants showed a highly resistant reaction to bacterial leaf blight with a score of 1, while 14 plants showed resistant to moderate resistant reaction with a score of 3, as shown in Table 2. The 18 highly resistant plants were subjected to foreground selection for the presence of Xa4, xa5, xa13, and Xa21 genes by using corresponding linked markers. In BC2F1 generation, the number of plants obtained with individual and different combinations of BB resistance genes were as [15 (Xa4), 17 (xa5), 12 (xa13), 4 (Xa21)] and [5 (Xa4, xa5, and xa13), 4 (Xa21, xa13, xa5 and Xa4)] (Figure S1). Out of four BC2F1 plant progenies, three progenies exhibited the amplification of all four resistance genes, and therefore, BC2F1 plants were pollinated by CARI Dhan 5 to generate BC3F1 seeds.
A total of 3,505 BC3F1 backcross derivative progenies were produced by backcrossing the plants showing maximum recurrent genomes with the recipient parent, CARI Dhan5. Out of these, 62 plants showed high resistance to bacterial leaf blight with a score of 1 and were subjected to foreground selection for the presence of Xa4, xa5, xa13, and Xa21 genes to identify plants that were homozygous for different resistant genes or their combinations using resistant gene linked markers (Table 2). In the case of individual R genes, 21 BC3F1 plants were positive for Xa21, 28 for xa5, 18 for xa13, and 21 for Xa4, whereas for different R gene combinations, the number of BC3F1 plants obtained for three and four gene combinations were as follows: three (lines 17-1-51, 17-1-55, and 46-5-143 with Xa4+xa5+xa13 genes), two (lines 131-2-189 and 46-5-94 with Xa4+xa13+Xa21 genes), two (lines 46-5-128 and 46-5-149 with Xa4+xa5+Xa21 genes), one (line 131-2-182 with xa5+xa13+Xa21 genes), and six (lines as 17-1-69, 131-2-175, 131-2-190, 46-3-95, 46-5-139, and 46-5-148 with Xa21+xa13+xa5+Xa4 genes), respectively (Supplementary Table 1). The BC3F1 derivative progenies carrying all four resistance genes, i.e., 17-1-69, 131-2-175, 131-2-190, 46-3-95, 46-5-139, and 46-5-148, were selfed to obtain BC3F2 lines. Then, the line 46-5-148 was backcrossed to obtain 75 BC4F1 seeds with four resistance genes (Xa4, xa5, xa13, and xa21).
Foreground selection was performed on 41 pyramided lines (BC3F4), as well as the parents (CARI Dhan 5 and IRBB 60) and a check (CSR 36), which were obtained by selfing lines 17-1-69, 131-2-175, 131-2-190, and 46-3-95. The selection was carried out using resistant gene linked markers to identify plants that were homozygous for different resistance genes or their combinations (Figures 3, 4).
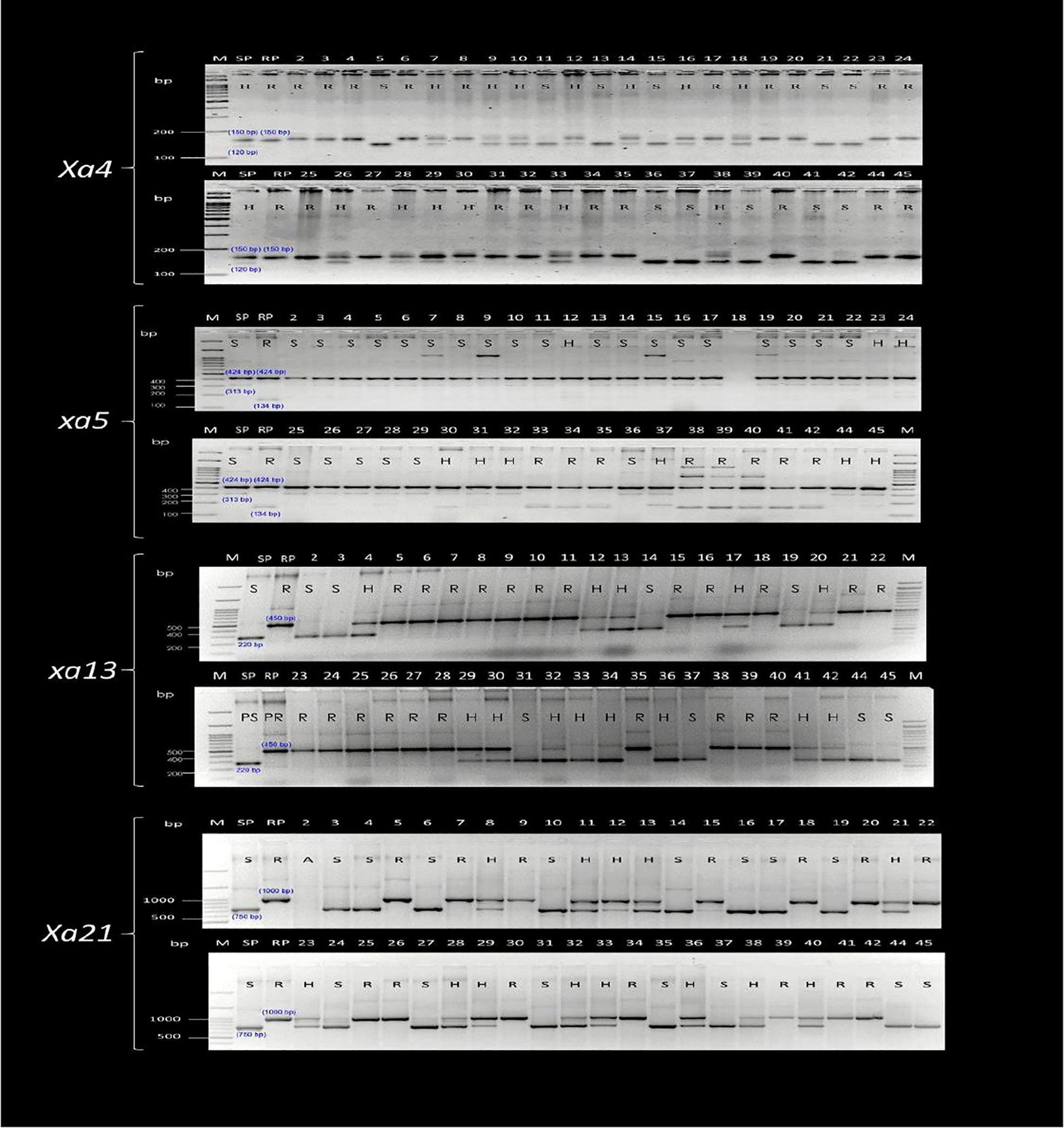
Figure 3 Marker profiling of the selected advanced breeding lines (ABLs) for foreground selection for Xa21, xa13, xa5, and Xa4 genes. M, Marker lane; SP/1, CARI Dhan-5; RP/43, IRBB 60; 2, 17-1-69-9; 3, 17-1-69-34: 4, 17-1-69-43; 5, 17-1-69-55; 6, 17-1-69-60; 7, 17-1-69-72; 8, 17-1-69-159; 9, 17-1-69-179; 10, 17-1-69-181; 11, 17-1-69-204; 12, 17-1-69-215; 13, 17-1-69-316; 14, 17-1-69-324; 15, 17-1-69-334; 16, 17-1-69-337; 17, 17-1-69-346; 18, 17-1-69-375; 19, 17-1-69-384; 20, 17-1-69-392; 21, 46-3-95-640; 22, 46-3-95-647; 23, 46-3-95-648; 24, 46-3-93-652; 25, 46-3-95-655; 26, 46-3-95-659; 27, 46-3-95-683; 28, 46-3-95-694; 29, 46-3-95-917; 30, 46-5-149-1185; 31, 131-2-190-785; 32, 131-2-190-795; 33, 131-2-190-1190; 34, 131-2-190-1196; 35, 131-2-190-1197; 36, 131-2-175-1207; 37, 131-2-175-1205; 38, 131-2-175-1209; 39, 131-2-175-1223; 40, 131-2-175-1224; 41, 131-2-175-1208, 42, 131-2-175-1239, 44, AVT-CSTVT-2020-1; 45, CSR 36.
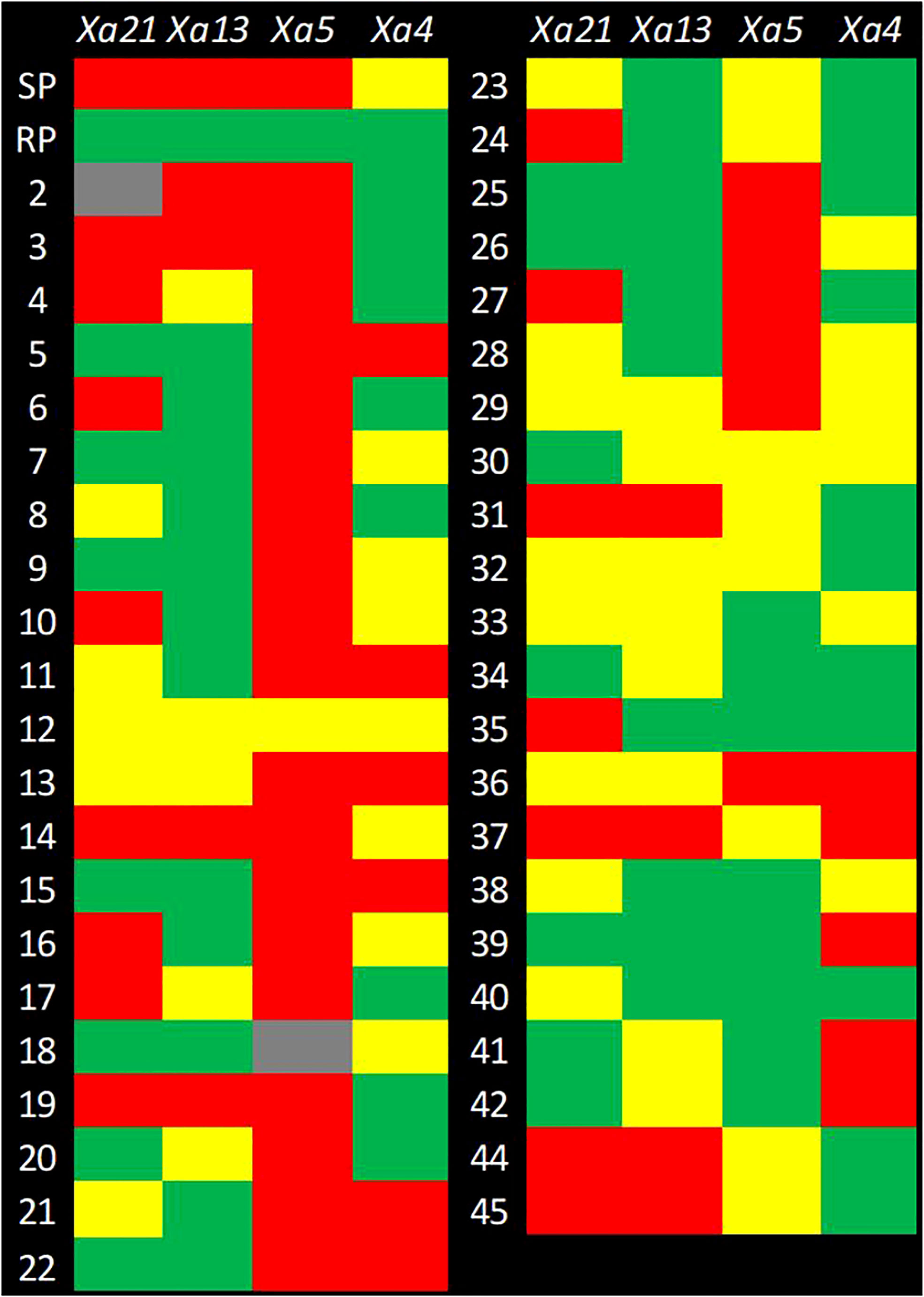
Figure 4 Genotyping of the selected advanced breeding lines (ABLs) for foreground selection. The checkerboard depicts the genotyping result for foreground selection. The rows represent genotypes and columns represent the genes for which genotyping has been performed. SP, Susceptible Parent; RP, Resistant Parent, 1–45*: Code number of SP/RP/selected ABLs for foreground selection. The Color Code: Green = Homozygous Resistant; Red = Homozygous Susceptible, Yellow = Heterozygous; and Gray = Missing Data. *SP/1, CARI Dhan-5; RP/43, IRBB 60; 2, 17-1-69-9; 3, 17-1-69-34: 4, 17-1-69-43; 5, 17-1-69-55; 6, 17-1-69-60; 7, 17-1-69-72; 8, 17-1-69-159; 9, 17-1-69-179; 10, 17-1-69-181; 11, 17-1-69-204; 12, 17-1-69-215; 13, 17-1-69-316; 14, 17-1-69-324; 15, 17-1-69-334; 16, 17-1-69-337; 17, 17-1-69-346; 18, 17-1-69-375; 19, 17-1-69-384; 20, 17-1-69-392; 21, 46-3-95-640; 22, 46-3-95-647; 23, 46-3-95-648; 24, 46-3-93-652; 25, 46-3-95-655; 26, 46-3-95-659; 27, 46-3-95-683; 28, 46-3-95-694; 29, 46-3-95-917; 30, 46-5-149-1185; 31, 131-2-190-785; 32, 131-2-190-795; 33, 131-2-190-1190; 34, 131-2-190-1196; 35, 131-2-190-1197; 36, 131-2-175-1207; 37, 131-2-175-1205; 38, 131-2-175-1209; 39, 131-2-175-1223; 40, 131-2-175-1224; 41, 131-2-175-1208, 42, 131-2-175-1239, 44, AVT-CSTVT-2020-1; 45, CSR 36.
Thirty-five promising pyramided (BC3F4) lines were evaluated for yield-related parameters under two different conditions, viz., salinity and normal, under controlled micro-plot facility at ICAR-CIARI, Port Blair. Based on disease score and genotyping results, the lines containing bacterial leaf blight (BB) resistance genes along with other desirable characteristics of CARI Dhan 5 were identified (Table S1). Under saline conditions, line 17-1-69-334 showed a score of 1 (resistant) for BB disease, while lines 17-1-69-316, 17-1-69-324, 17-1-69-334, and 17-1-69-337 exhibited a score of 3 (moderately resistant). On the other hand, under normal conditions, the pyramided lines 17-1-69-375, 17-1-69-334, 17-1-69-215, 131-2-190-1196, and 131-2-175-1223 all scored 1, indicating resistance against BB disease (Table S2). The elite 17 pyramided lines selected based on genotyping and phenotyping of advanced breeding lines (ABLs) were further subjected to confirmation for BB resistance and salinity tolerance (Ece ~ 5.0 dSm−1) in micro-plots over the years (Table 3). Consequently, five lines, viz., 17-1-69-55, 17-1-69-72, 17-1-69-179, 17-1-69-334, and 17-1-69-375 [all carrying two BB resistance genes (Xa21 and xa13)], were found to retain the original CARI Dhan 5 salinity tolerance with a salinity score of 1 (Table S2). A total of seven derivatives, viz., 17-1-69-55, 17-1-69-179, 17-1-69-334, 46-3-95-659 (Xa21 and xa13), 131-2-190-1196 (Xa21, xa5, and Xa4), 131-2-190-1197 (xa13, xa5, and Xa4), and 131-2-175-1223 (Xa21, xa13, and xa5), were found to have complete resistance as shown in Table S3. Interestingly, five lines, viz., 17-1-69-179 (Xa21 and xa13), 46-3-95-655 (Xa21, xa13 and Xa4), 131-2-190-1197 (xa13, xa5, and Xa4), 131-2-175-1209 (xa13 and xa5), and 131-2-175-1239 (Xa21 and xa5), were identified to possess desirable dual attributes of salinity tolerance and BB resistance, as shown in Table S2. Yield and yield-related traits of the above-mentioned pyramided lines at the micro-plot as well as field levels are presented in Table S2. In the saline micro-plots (EC 5.0 dSm−1), five lines viz., 131-2-175-1239 (146.9), 17-1-69-72 (145.5), 17-1-69-375 (131.8), 46-3-95-647 (130.6), and 46-3-95-659 (124.6) demonstrated grain yield (expressed in g per 20 plants) greater than or equal to that of CARI Dhan 5 (123.4). Meanwhile, under normal soil conditions, lines, viz., 17-1-69-334 (300.2), 17-1-69-179 (272.9), 46-3-95-655 (265.1), 17-1-69-375 (232.8), and 46-3-95-647 (232.8), exhibited higher grain yield than CARI Dhan 5 (187.6). The lines 46-3-95-659 (6,183.1), 46-3-95-647 (5,997.9), 17-1-69-375 (5,838.5), 131-2-175-1209 (4,629.6), and 131-2-175-1223 (4,526.8) demonstrated grain yield (kg ha−1) above or comparable to that of CARI Dhan 5 (4434.2) under natural field-level conditions (Table S3). It is also noteworthy to mention that the 17 pyramided BC3F3 lines were also subjected to DUS characterization using 41 rice-descriptor traits. The results showed that the BB-resistant and salinity-tolerant derivatives exhibited agro-morphological similarity with the recurrent parent, CARI Dhan 5, indicating the retention of the host background in these agronomically targeted lines. The details of this analysis are presented in Table S4. Here, lines 17-1-69-375 and 46-3-95-647 performed well for the grain quality and yield-traits.
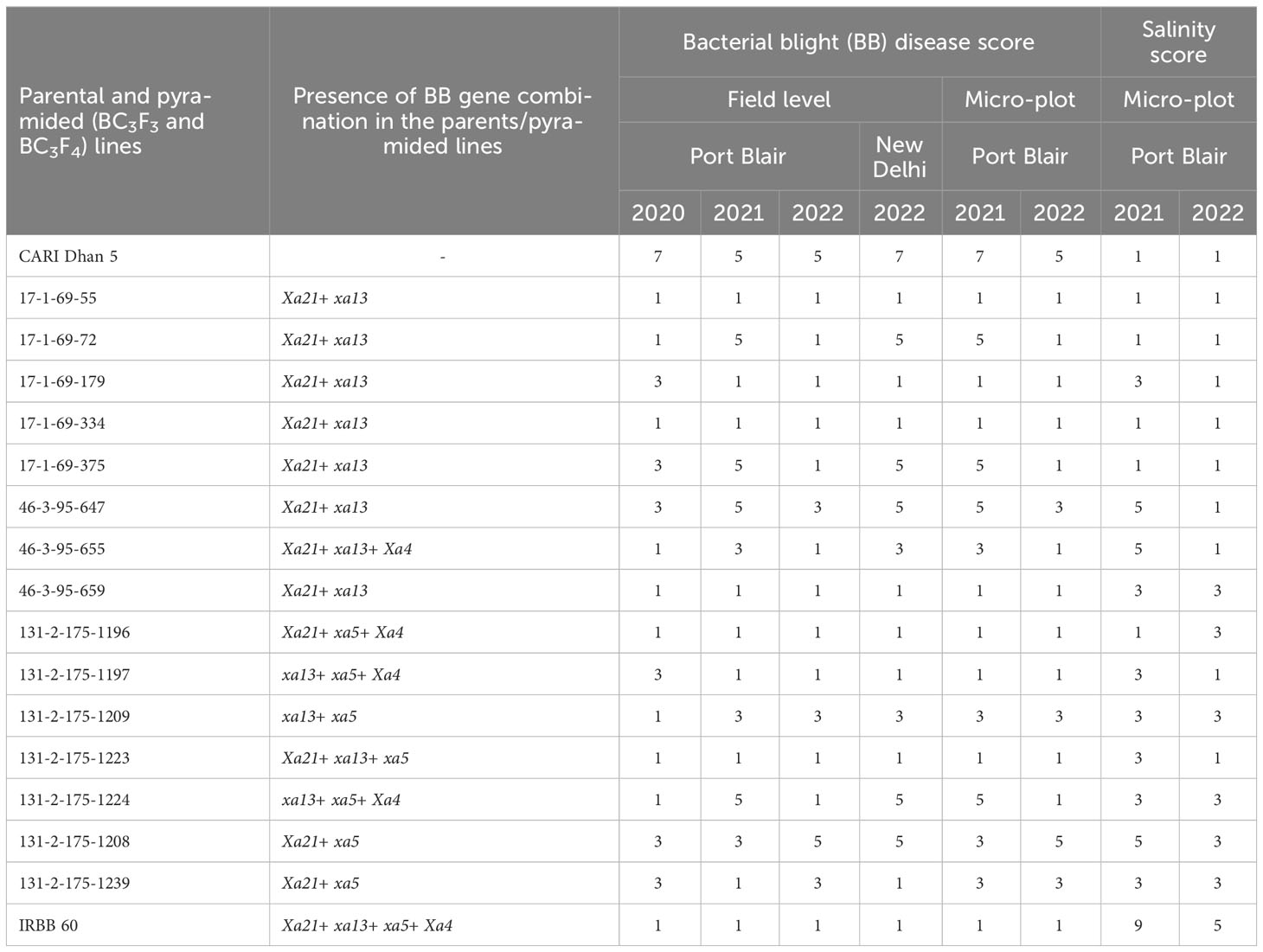
Table 3 Bacterial blight disease reaction of parental and pyramided lines against Xoo strains over the years and locations along with salinity score.
Salinity is a major abiotic stress for rice crops in coastal regions and the situation is expected to worsen due to climate change and the increasing number of irrigation networks. CARI Dhan 5, a somaclonal variant of the legendary rice landrace Pokkali, is a widely cultivated rice variety in the Andaman and Nicobar Islands due to its high yield and remarkable tolerance to coastal saline and acid sulfate soils (Mandal et al., 1999; Singh et al., 2020). However, CARI Dhan 5 is susceptible to BB disease caused by X. oryzae pv. oryzae, which is one of the most devastating diseases of rice. Being a tropical island, the hot and humid climate of the Andaman and Nicobar Islands provides congenial conditions for the completion of the disease triangle. In our previous studies, we conducted virulence profiling of Xoo isolates from the Andaman Islands, which revealed the prevalence of seven pathotypes of X. oryzae pv. oryzae belonging to two different clonal complexes (Sakthivel et al., 2021). Among these, pathotypes VI and VII were found to be highly virulent. Our analysis using multilocus sequence typing based on nucleotide sequence polymorphism in nine housekeeping genes, dnaK, fyuA, gyrB (two loci), rpoD, fusA, gapA, gltA, and lepA, further revealed that Xoo strains infecting rice in the Andaman and Nicobar Islands comprise a mixture of isolates representing mainland India and the islands, which might be due to trans-boundary movement of the BB pathogen through rice seeds (Sakthivel et al., 2021).
A combination of Xa4+xa5+xa13+Xa21 has been found to confer broad-spectrum resistance to BB disease caused by different strains of Xoo reported from different parts of the world. Moreover, studies conducted over the years and in multiple locations have demonstrated that the resistance imparted by this gene combination does not depict any sign of breakdown, making it a promising approach to combating the disease (Shanti and Shenoy, 2005; Pradhan et al., 2015). Therefore, in this study, we aimed to introgress these four BB resistance genes (Xa21, xa13, xa5, and Xa4) into the genetic background of CARI Dhan 5, a coastal salinity-tolerant rice cultivar, using marker-assisted breeding.
The genetic background of the recipient parent is known to play a significant role in the successful integration and expression of gene pyramids. In this context, our previous research on five distinct genetic backgrounds carrying xa5+xa13+Xa21 combinations revealed that the rice lines with IMP ASD16/60 and Improved Samba Mahsuri imparted complete resistance against all pathotypes of Andaman (Sakthivel et al., 2017). Fortunately, there are several resistance genes that confer resistance against Xoo. Notably, Xa21, xa13, and xa5 have been reported to impart strong levels of resistance against Xoo strains prevalent in India (Goel et al., 1998; Shanti et al., 2001; Joseph et al., 2004). Xa21 and Xa4 are well-studied dominant resistance genes encoding receptor-like kinases (RLKs) (Song et al., 1995; Leach et al., 2001), while the Xa21 protein contains a nucleotide-binding site and leucine-rich repeat (NBS-LRR) domain (Song et al., 1995) and Xa4 protein belongs to the category of wall-associated kinases (WAK) family (Leach et al., 2001). These two genes exhibit complete dominance, function independently, and are cumulative in nature, indicating their crucial role in two distinct pathways of resistance (Li et al., 2001). On the other hand, the xa13 gene is a recessive resistance allele of the susceptibility gene Os-8N3 (the rice homolog of the nodulin MtN3) (Chu et al., 2006; Antony et al., 2010). The allele xa13 contains a mutation in the promoter region that inhibits binding of the transcription factor and subsequent expression of the susceptibility gene (Antony et al., 2010). Similar to xa13, xa5 is also a recessive resistance gene that encodes a variant form of the basal transcription factor TFIIAγ. This variant form, resulting from a missense mutation, cannot bind to the promoter region of the susceptibility locus, providing resistance to Xoo (Han et al., 2020). Together, the two dominant resistance genes Xa4 and Xa21 confer vertical resistance, making the plant nearly immune, while the two recessive genes xa13 and xa5 offer strong horizontal resistance, which is durable in nature.
Previous studies have successfully generated four- or three-gene pyramids in popular rice cultivars including Mahsuri (Xa4+xa5+xa13+Xa21) (Shanti et al., 2010); Tapaswini (Xa4+xa5+xa13+Xa21) (Dokku et al., 2013); Swarna (Xa4+xa5+xa13+Xa21) (Pradhan et al., 2019); Putra-1 (Xa4+xa5+xa13+Xa21) (Chukwu et al., 2020); Ranidhan (Xa4+xa5+xa13+Xa21) (Pradhan et al., 2022); Basmati-385 (Xa4+xa5+xa13+Xa21) (Ullah et al., 2023); PR106 (xa5+xa13+Xa21) (Singh et al., 2001); Jalmagna (xa5+xa13+Xa21) (Pradhan et al., 2015); Samba Mahsuri (xa5+xa13+Xa21) (Sundaram et al., 2008); ASD 16 (xa5+xa13+Xa21) (Ramalingam et al., 2020); and ADT 43 (xa5+xa13+Xa21) (Ramalingam et al., 2020). Recent reports and reviews further suggest that the use of Xa4 might be redundant and that the gene combination of xa5+xa13+Xa21 is sufficient for providing resistance, which has been extensively used in marker-assisted backcross breeding in rice (Gautam et al., 2015; Fiyaz et al, 2022). It is worth noting that more than 70 rice varieties or hybrid parental lines have been improved for their BB resistance alone or in combination with genes/QTLs conferring tolerance to other biotic and abiotic stresses by the team.
In our previous study, we conducted a screening of 21 IRBB differentials that possessed individual Xa1 to Xa21 gene(s) as well as different gene combinations (Gautam et al., 2015). Based on these findings, it is concluded that three individual genes, namely, Xa4, Xa7, and Xa21, along with four combinations, viz. Xa4+xa5 (IRBB50), Xa4+Xa21 (IRBB52), xa5+xa13+Xa21 (IRBB59), and Xa4+xa5+xa13+Xa21 (IRBB60), conferred resistance against all the tested isolates from the Andaman Islands. IRBB60, in particular, has been a successful donor in rice breeding programs for conditioning BB resistance, as reported in several studies (Shanti et al., 2010; Dokku et al., 2013; Chukwu et al., 2019; Pradhan et al., 2019). Therefore, we employed a breeding strategy using IRBB60 as the donor parent and CARI Dhan 5 as the salt-tolerant recurrent parent to introgress at least three target Xa/xa genes into CARI Dhan 5. As a result, we successfully obtained a total of 14 BC3F1 plants with the desired gene combination. According to our findings, six BC3F1 progenies (17-1-69, 131-2-175, 131-2-190, 46-3-95, 46-5-139, and 46-5-148) were identified to harbor four gene pyramids (Xa21+xa13+xa5+Xa4). Additionally, three lines (17-1-51, 17-1-55, and 46-5-143) possessed Xa4+xa5+xa13 genes, while two lines (131-2-189 and 46-5-94) contained Xa4+xa13+Xa21 genes. The gene combination Xa4+xa5+Xa21 was identified in two lines (46-5-128 and 46-5-149), while the derivative 131-2-182 possessed the xa5+xa13+Xa21 combination. Therefore, these pyramided lines hold great promise as potential candidates for developing BB-resistant versions of CARI Dhan 5 for commercial release and cultivation in the coastal saline and acid sulfate soils of the Andaman and Nicobar Islands. In this study, we identified five resistant derivatives having three gene combinations, viz, 131-2-175-1223 (xa5+xa13+Xa21), 46-3-95-655 (Xa21+xa13+Xa4), 131-2-190-1196 (Xa21+xa5+Xa4), 131-2-190-1197 (xa13+xa5+Xa4), and 131-2-175-1224 (xa13+xa5+Xa4). Similar to 131-2-175-1223, the xa5+xa13+Xa21 combination has been proven to confer durable and broad-spectrum resistance in many popular rice varieties in India (Singh et al., 2001; Sundaram et al., 2008; Pradhan et al., 2015; Ramalingam et al., 2020). The disease reaction scoring over the years and locations established the resistance of the progenies in the following order: 131-2-175-1223 (xa5+xa13+Xa21) > 17-1-69-55, 17-1-69-72, 17-1-69-179, 17-1-69-334, 17-1-69-375, 46-3-95-647, 46-3-95-659 (xa13+Xa21) > 131-2-175-1208, 131-2-175-1239 (xa5+Xa21) > 131-2-175-1209 (xa5+xa13). It is evident from our gene-pyramided lines that an enhanced level of BB resistance is conditioned when the Xa21 gene (dominant allele) complements an allele of the xa5 or xa13 or both (recessive alleles). This finding is in line with several other earlier observations (Sanchez et al., 2000; Singh et al., 2001; Ramalingam et al., 2017) and is probably achieved due to the sword-shield effect mediated by resistance gene expression (the sword) coupled with non-expression of susceptibility gene (the shield). Four pyramided lines showed the presence of the Xa4 gene with other genes (Xa21/xa13/xa5) in combination as 46-3-95-655 (Xa21+xa13+Xa4), 131-2-190-1196 (Xa21+xa5+Xa4), 131-2-190-1197 (xa13+xa5+Xa4), and 131-2-175-1224 (xa13+xa5+Xa4) with a BB disease reaction score of 1 or 3 over the years and locations. We also observed that though the recurrent susceptible parent CARI Dhan 5 is heterozygous for the Xa4 gene, Xa4 gene is reported as a weak gene imparting incomplete resistance against very few isolates in India (Mishra et al., 2013). Incomplete resistance of Xa4 was also reported by our group in an earlier study under Andaman and Nicobar Island conditions (Gautam et al., 2015). Therefore, although CARI Dhan 5 harbors the Xa4 allele, it behaves as a susceptible genotype to the Xoo races prevalent in these islands. Hence, it is not unusual for the susceptible parent CARI Dhan 5 to contain both resistant alleles (150 bp) and susceptible alleles (120 bp) and possess heterozygous genetic constitution for the said locus.
The DUS characterization of pyramided lines for 41 agro-morphological and descriptor traits confirmed that the advanced elite lines recovered the genetic background of CARI Dhan 5 in addition to BB resistance (refer to Table S6 for details). It was imperative to confirm the retention of salinity tolerance in the ILs as compared to CARI Dhan 5. Based on the phenotypic scoring method, we found the ILs, viz., 17-1-69-55, 17-1-69-179, 17-1-69-334,131-2-175-1196, and 131-2-175-1197, as BB resistant (disease score 1) as well as salt tolerant (salinity score 1). Further evaluation under the saline micro-plot facility (EC 5.0 dSm−1) revealed that the pyramided line, viz., 17-1-69-334, exhibited a consistent score of 1 (resistant/tolerant) for both BB and salinity stresses across years and locations, with comparable grain yield performance of CARI Dhan 5. Overall, the results indicate that the salt-tolerant lines, viz., 17-1-69-179, 46-3-95-655, 131-2-190-1197, 131-2-175-1209, and 131-2-175-1239, also demonstrated complete resistance to BB disease. In view of their potential for dual attributes, these promising derivatives are being nominated for multi-location yield trials, under the All-India Co-ordinated Rice Improvement Program with their potential for release as BB-resistant and salt-tolerant varieties for cultivation in BB-prone and coastal saline conditions.
High salt concentrations in soils can cause a significant decline in rice yield and quality, and in some cases, it may even lead to complete crop failure (Xiao et al., 2020). CARI Dhan 5, a somaclonal variation-derived semi-dwarf variety from salt-tolerant rice landrace Pokkali in ICAR-CIARI, Port Blair, is a preferred rice variety in Andaman and Nicobar Islands due to its high yield, favorable grain quality, and high salinity tolerance. However, it is susceptible to BB disease, which is prevalent in the island’s environmental conditions. The present study endeavored to use marker-assisted backcross breeding to pyramid four BB resistance genes (Xa4, xa5, xa13, and Xa21) in the background of CARI Dhan 5. Consequently, five stabilized rice lines combining salt tolerance with BB resistance have been developed in the genetic background of CARI Dhan 5. The development of these improved CARI Dhan 5 lines is expected to have a strong and favorable impact on eco-friendly rice cultivation in the geographically remote Andaman and Nicobar Islands, which are affected by coastal salinity and BB disease.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
RG: Conceptualization, methodology, project execution, administration, primary draft, and editing. PS: Methodology (designed and conducted field experiment) and data compilation. KS: Bacterial inoculation, disease scoring, and data analysis using spatial software. KV: Methodology (designed and conducted field experiment) and data compilation. SSR: Genotyping and conducted field experiment. MS: Genotyping (molecular analysis using SSR markers). JV: Field experiments, data tabulation, and curation. BR: Field experiments, data tabulation, and genotyping. SR: Genotyping and draft preparation. JA: Disease scoring. BM: Disease scoring. SL: Writing the original draft, and review and editing. SA: Preparing tables and figures. SK: Phenotyping for salt tolerance. All authors contributed to the article and approved the submitted version.
The authors acknowledge the funding support from the Department of Biotechnology (DBT), Government of India, for the financial grant (BT/PR6575/AG11/106/892/2012) to the project “Marker assisted introgression of BB resistance in popular rice cultivars of Andaman and Nicobar Islands” under “Application of molecular markers under accelerated crop improvement programme”. We also acknowledge the funding support from the Bill and Melinda Gates Foundation and IRRI, Philippines, for the “Stress tolerant rice for poor farmers in Africa and South Asia” project.
The authors acknowledge the Indian Council of Agricultural Research for all-time support. The authors also thank the Director, ICAR-Central Island Agricultural Research Institute, Port Blair, for providing research facilities.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1221537/full#supplementary-material
Antony, G., Zhou, J., Huang, S., Li, T., Liu, B, White, F., et al. (2010). Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene os-11N3. Plant Cell 22 (11), 3864–3876. doi: 10.1105/tpc.110.078964
Chen, X., Temnykh, S., Xu, Y., Cho, Y. G., McCouch, S. R. (1997). Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Theor. Appl. Genet. 95, 553–567. doi: 10.1007/s001220050596
Chu, Z., Yuan, M., Yao, J., Ge, X., Yuan, B., Xu, C., et al. (2006). Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 20, 1250–1255. doi: 10.1101/gad.1416306
Chukwu, S. C., Rafii, M. Y., Ramlee, S. I., Ismail, S. I., Oladosu, Y., Kolapo, K., et al. (2020). Marker-assisted introgression of multiple resistance genes confers broad spectrum resistance against bacterial leaf blight and blast diseases in PUTRA-1 rice variety. Agronomy 10 (1), 42. doi: 10.3390/agronomy10010042
Chukwu, S. C., Rafii, M. Y., Ramlee, S. I., Ismail, S. I., Oladosu, Y., Okporie, E., et al. (2019). Marker-assisted selection and gene pyramiding for resistance to bacterial leaf blight disease of rice (Oryza sativa L.). Biotechnol. Biotechnol. Equip. 33, 440–455. doi: 10.1080/13102818.2019.1584054
Chukwu, S. C., Rafii, M. Y., Ramlee, S. I., Ismail, S. I., Hasan, M. M., Oladosu, Y. A., et al. (2019). Bacterial leaf blight resistance in rice: a review of conventional breeding to molecular approach. Mol. Biol. Rep. 46 (1), 1519–1532. doi: 10.1007/s11033-019-04584-2
De Leon, T. B., Linscombe, S., Subudhi, P. K. (2017). Identification and validation of QTLs for seedling salinity tolerance in introgression lines of a salt tolerant rice landrace `Pokkali'. PloS One 12 (4), e0175361. doi: 10.1371/journal.pone.0175361
Dokku, P., Das, K. M., Rao, G. J. N. (2013). Pyramiding of four resistance genes of bacterial blight in Tapaswini, an elite rice cultivar, through marker-assisted selection. Euphytica 192, 87–96. doi: 10.1007/s10681-013-0878-2
Fiyaz, R. A., Shivani, D., Chaithanya, K., Mounika, K., ChIranjeevi, M., Laha, G. S., et al. (2022). Genetic improvement of rice for bacterial blight resistance: present status and future prospects. Rice Sci. 29 (2), 118–132. doi: 10.1016/j.rsci.2021.08.002
Gautam, R. K., Dam Roy, S., Singh, P. K., Singh, S., Singh, A. K., Ahmed, S. K. Z., et al. (2016a). Improved varieties of field and horticultural crops for Andaman & Nicobar Islands (Port Blair, Andaman and Nicobar Islands: Technical Bulletin, ICAR-Central Island Agricultural Research Institute), 36.
Gautam, R. K., Singh, P. K., Birah, A., Kumar, K., Singh, A. K., Kumar, N., et al. (2013). Agro-technology for higher rice productivity in Andaman & Nicobar Islands (Port Blair, Andaman and Nicobar Islands: Technical Bulletin, ICAR-Central Island Agricultural Research Institute), 42.
Gautam, R. K., Singh, P. K., Sakthivel, K., Singh, A. K., Rajeni, E., Rao, S. S., et al. (2016b). “Development of bacterial blight (Xanthomonas oryzaepv.oryzae) resistant varieties of rice for Andaman & Nicobar Islands thought molecular markers technology: progress and prospects,” in Souvenir. Ed. Dam Roy, S., et al, 47–48. International Conference on Climate Change Adaptation and Biodiversity: Ecological sustainability and resource management for livelihood security. (ICAR-CIARI: A&N Islands, India).
Gautam, R. K., Singh, P. K., Sakthivel, K., Srikumar, M., Kumar, N., Kumar, K., et al. (2015). Analysis of pathogenic diversity of the rice bacterial blight pathogen (Xanthomonas oryzaepv. oryzae) in the Andaman Islands and identification of effective resistance genes. J.Phytopath. 16, 423–432. doi: 10.1111/jph.12338
Goel, R. K., Kaur, L., Saini, R. G. (1998). Effectiveness of different Xa genes against Xanthomonas oryzaepv. oryzae population causing bacterial blight of rice in Punjab (India). Rice Genet. Newsl. 15, 131–133.
Gregorio, G. B., Senadhira, D., Mendoza, R. D., Manigbas, N. L., Roxas, J. P., Cuerta, C. Q. (2002). Progress in breeding for salinity tolerance and associated abiotic stresses in rice. Field Crops Res. 76, 91–101. doi: 10.1016/S0378-4290(02)00031-X
Gujja, B., Thiyagarajan, T. M. (2009). New hope for Indian food security? the system of rice intensification. Gatekeeper 143, 3–18. Available at: https://hdl.handle.net/10535/5852.
Gupta, V. S., Raje Bhosale, M. D., Sodhi, M. S., Singh, S., Gnanamanickam, S. S., Dhaliwal, H. S., et al. (2001). Assessment of genetic variability and strain identification of Xanthomonas oryzaepv. oryzae using RAPD-PCR and IS1112-based PCR. Curr. Sci. 80, 1043–1049. Available at: https://www.jstor.org/stable/24105815.
Hajira, S. K., Sundaram, R. M., Laha, G. S., Yugander, A., Balachandran, S. M., Viraktamath, B. C., et al. (2016). A Single-Tube, Functional Marker-Based Multiplex PCR Assay for Simultaneous Detection of Major Bacterial Blight Resistance Genes Xa21, xa13 and xa5 in Rice. Rice Sci. 23 (3), 144–151. doi: 10.1016/j.rsci.2015.11.004
Han, J., Xia, Z., Liu, P., Li, C., Wang, Y., Guo, L., et al. (2020). TALEN-based editing of TFIIAy5 changes rice response to Xanthomonas oryzae pv. Oryzae. Sci. Rep. 10 (1), 2036. doi: 10.1038/s41598-020-59052-w
Hopmans, J. W., Qureshi, A. S., Kisekka, I., Munns, R., Grattan, S. R., Rengasamy, P., et al. (2021). Critical knowledge gaps and research priorities in global soil salinity. Adv. Agron. 169, 1–191. doi: 10.1016/bs.agron.2021.03.001
Huang, N., Angeles, E. R., Domingo, J., Magpantay, G., Singh, S., Zhang, G., et al. (1997). Pyramiding of bacterial blight resistance genes in rice: marker assisted selection using RFLP and PCR. Theor. Appl. Genet. 95, 313–320. doi: 10.1007/s001220050565
Jena, K. K., Mackill, D. J. (2008). Molecular markers and their use in marker-assisted selection in rice. Crop Sci. 48, 1266–1276. doi: 10.2135/cropsci2008.02.0082
Joseph, M., Gopalakrishnan, S., Sharma, R. K., Singh, V. P., Singh, A. K., Singh, N. K., et al. (2004). Combining bacterial blight resistance and basmati quality characteristics by phenotypic and molecular marker-assisted selection in rice. Mol. Breed. 13, 377–387. doi: 10.1023/B:MOLB.0000034093.63593.4c
Kauffman, H. E. (1973). An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep. 57, 537–541.
Koide, Y., Kawasaki, A., Telebanco-Yanoria, M. J., Hairmansis, A., Nguyet, N. T. M., Bigirimana, J., et al. (2010). Development of pyramided lines with two resistance genes, Pish and Pib, for blast disease (Magnaportheoryzae B. Couch) in rice (Oryza sativa L.). Plant Breed. 129 (6), 670–675. doi: 10.1111/j.1439-0523.2010.01781.x
Kumar, A., Kumar, R., Sengupta, D., Das, S. N., Pandey, M. K., Bohra, A., et al. (2020). Deployment of genetic and genomic tools toward gaining a better understanding of rice-Xanthomonas oryzaepv. oryzae interactions for development of durable bacterial blight resistant rice. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01152
Leach, J. E., Vera Cruz, C. M., Bai, J., Leung, H. (2001). Pathogen fitness penalty as a predictor of durability of disease resistance genes. Annu. Rev. Phytopathol. 39, 187–224. doi: 10.1146/annurev.phyto.39.1.187
Li, Z.-K., Sanchez, A., Angeles, E., Singh, S., Domingo, J., Huang, N., et al. (2001). Are the Dominant and Recessive Plant Disease Resistance Genes Similar? A Case Study of Rice R Genes and Xanthomonas oryzae pv. oryzae Races. Genetics 159 (2), 757–765. doi: 10.1093/genetics/159.2.757
Ma, B. J., Wenming, W., Bin, Z., Yongli, Z., Lihuang, Z., Wenxue, Z. (1999). Studies of PCR marker for the rice bacterial blight resistance gene Xa-4. Hereditas 21, 9–12.
Mandal, A. B., Pramanik, S. C., Chowdhury, B., Bandyopadhyay, A. K. (1999). Salt-tolerant Pokkalisomaclones: performance under normal and saline soils in Bay Islands. Field Crops Res. 61 (1), 13–21. doi: 10.1016/S0378-4290(98)00145-2
Mishra, D., Vishnupriya, M. R., Anil, M. G., Konda, K., Raj, Y., Sonti, R. V. (2013). Pathotype and genetic diversity amongst Indian isolates of Xanthomonas oryzae pv. oryzae. PloS One 8 (11), e81996. doi: 10.1371/journal.pone.0081996
Panaud, O., Chen, X., McCouch, S. R. (1996). Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol. Gen. Genet. 252, 597–607. doi: 10.1007/BF02172406
Pradhan, S. K., Nayak, D. K., Mohanty, S., Behera, L., Barik, S. R., Pandit, E., et al. (2015). Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety, Jalmagna. Rice 8, 19. doi: 10.1186/s12284-015-0051-8
Pradhan, K. C., Pandit, E., Mohanty, S. P., Moharana, A., Sanghamitra, P., Meher, J., et al. (2022). Development of broad spectrum and durable bacterial blight resistant variety through pyramiding of four resistance genes in rice. Agronomy 12, 1903. doi: 10.3390/agronomy12081903
Pradhan, S. K., Pandit, E., Pawar, S., Baksh, S. Y., Mukherjee, A. K., Mohanty, S. P. (2019). Development of flash-flood tolerant and durable bacterial blight resistant versions of mega rice variety ‘Swarna’ through marker-assisted backcross breeding. Sci. Rep. 9 (1), 12810. doi: 10.1038/s41598-019-49176-z
Rajpurohit, D., Kumar, R., Kumar, M., Paul, P., Awasthi, A. A., Basha, P. O., et al. (2011). Pyramiding of two bacterial blight resistance and a semi dwarfing gene in Type 3 Basmati using marker-assisted selection. Euphytica 178, 111–126. doi: 10.1007/s10681-010-0279-8
Ramalingam, J., Raveendra, C., Savitha, P., Vidya, V., Chaithra, T. L., Velprabakaran, S., et al. (2020). Gene pyramiding for achieving enhanced resistance to bacterial blight, blast, and sheath blight diseases in rice. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.591457
Ramalingam, J., Savitha, P., Alagarasan, G., Saraswathi, R., Chandrababu, R. (2017). Functional marker assisted improvement of stable cytoplasmic male sterile lines of rice for bacterial blight resistance. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01131
Sakthivel, K., Gautam, R. K., Manigundan, K., Singh, P. K., La Ha, G. S., Velazhahan, R. (2016). Isolation and molecular characterization of rice bacterial blight pathogen Xanthomonas oryzaepv. Oryzaefrom Andaman Islands, India. Plant Dis. Res. 30 (2), 131–146. Available at: https://api.semanticscholar.org/CorpusID:89000067.
Sakthivel, K., Gautam, R. K., Manigundan, K., Singh, R., RaMalingam, J., Laha, G. S., et al. (2017). The host background of rice influences the resistance expression of a three gene pyramid (xa5+xa13+Xa21) to bacterial blight (Xanthomonas oryzaepv. oryzae) pathotypes of Indian mainland and Bay islands. Plant Breed 136 (3), 357–364. doi: 10.1111/pbr.12472
Sakthivel, K., Kumar, A., Gautam, R. K., Manigundan, K., Laha, G. S., Velazhahan, R., et al. (2021). Intra-regional diversity of rice bacterial blight pathogen, Xanthomonas oryzae pv. oryzae, in the Andaman Islands, India: revelation by pathotyping and multilocus sequence typing. J. Appl. Microbiol. 130 (4), 1259–1272. doi: 10.1111/jam.14813
Sanchez, A. C., Brar, D. S., Huang, N., Li, Z., Khush, G. S. (2000). Sequence tagged site marker-assisted selection for three bacterial blight resistance genes in rice. Crop Sci. 40, 792–797. doi: 10.2135/cropsci2000.403792x
Sarangi, S. K., Mainuddin, M., Maji, B. (2022). Problems, management, and prospects of acidSulphate soils in the ganges delta. Soil Syst. 6, 95. doi: 10.3390/soilsystems6040095
SES, IRRI (1996). Standard evaluation system of rice (Philippines: International Rice Research Institute).
Shanti, M. L., George, M. L. C., Vera Cruz, C. M., Bernardo, M. A., Nelson, R. J., Leung, H., et al. (2001). Identification of resistance genes effective against bacterial leaf blight pathogen in eastern India. Plant Dis. 85, 506–512. doi: 10.1094/PDIS.2001.85.5.506
Shanti, M. L., Shenoy, V. V. (2005). Evaluation of resistance genes and their pyramids against rice bacterial leaf blight pathogen Xanthomonas oryzaepv. oryzae. Oryza 42 (3), 169–173.
Shanti, M., Shenoy, V., Devi, G. L., Kumar, V. M., Premalatha, P., Kumar, G. N., et al. (2010). Marker assisted breeding for resistance to bacterial leaf blight in popular cultivar and parental lines of hybrid rice. J. Plant Pathol. 92, 495–501.
Shylaraj, K. S., Nadh, S. G., Chandran, S. K. (2018). Comparative analysis of grain quality and nutraceutical properties of selected rice varieties from Kerala. Curr. Sci., 1088–1094. doi: 10.18520/cs/v114/i05/1088-1094
Singh, P. K., Gautam, R. K., Sakthivel, K., Ahmed, S. Z., Venkatesan, K., Varadan, R. J. (2020). Augmenting rice productivity in Andaman and Nicobar Islands through improved varieties and seed replacement rate. J. Indian Soc. Coast. Agric. Res. 38 (1), 1–10.
Singh, U. D., Gogoi, R., Mondal, K. K. (2011). Molecular approach for augmenting disease resistance in cereals: Rice and maize. Plant Pathol. Ind. 20, 21–30.
Singh, S., Sidhu, J. S., Huang, N., Vikal, Y., Li, Z., Brar, D. S., et al. (2001). Pyramiding three bacterial blight resistance genes (xa-5, xa-13 and Xa-21) using marker-assisted selection into indica rice cultivar PR-106. Theor. Appl. Genet. 102, 1011–1015. doi: 10.1007/s001220000495
Singh, S., Sodhi, M., Vikal, Y., George, M. L. C., Bala, G. S., Mangat, G. S., et al. (2003). DNA fingerprinting and virulence analysis of Xanthomonas oryzapv. oryzae isolates from Punjab, northern India. Euphytica 130, 107–115. doi: 10.1023/A:1022329024651
Song, W. Y., Wang, G. L., Chen, L. L., Kim, H. S., Pi, L. Y., Holsten, T., et al. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806. doi: 10.1126/science.270.5243.1804
Suh, J. P., Jeung, J. U., Noh, T. H., Cho, Y. C., Park, S. H., Park, H. S., et al. (2013). Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice 6, 5. doi: 10.1186/1939-8433-6-5
Sundaram, R., Laha, G., Viraktamath, B., Sujatha, K., Natarajkumar, P., Hari, Y., et al. (2011). Marker assisted breeding for development of bacterial blight resistant rice. J. Plant Pathol. 500, 154–182.
Sundaram, R. M., Vishnupriya, M. R., Biradar, S. K., Laha, G. S., Reddy, G. A., Rani, N. S., et al. (2008). Marker assisted introgression of bacterial blight resistance in Samba Mahsuri, an elite indica rice variety. Euphytica 160, 411–422. doi: 10.1007/s10681-007-9564-6
Thomson, M. J., de Ocampo, M., Egdane, J., Rahman, M. A., Sajise, A. G., Adorada, D. L., et al. (2010). Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice 3, 148–160. doi: 10.1007/s12284-010-9053-8
Ullah, I., Ali, H., Mahmood, T., Khan, M. N., Haris, M., Shah, H., et al. (2023). Pyramiding of four broad spectrum bacterial blight resistance genes in cross breeds of basmati rice. Plants 12, 46. doi: 10.3390/plants12010046
Keywords: bacterial blight, marker-assisted backcross breeding, pyramiding, resistance, rice, salinity tolerance, tropical islands
Citation: Gautam RK, Singh PK, Sakthivel K, Venkatesan K, Rao SS, Srikumar M, Vijayan J, Rakesh B, Ray S, Akhtar J, Meena BR, Langyan S, Ali S and Krishnamurthy SL (2023) Marker-assisted enhancement of bacterial blight (Xanthomonas oryzae pv. oryzae) resistance in a salt-tolerant rice variety for sustaining rice production of tropical islands. Front. Plant Sci. 14:1221537. doi: 10.3389/fpls.2023.1221537
Received: 12 May 2023; Accepted: 21 August 2023;
Published: 25 September 2023.
Edited by:
Hanwei Mei, Shanghai Agrobiological Gene Center, ChinaReviewed by:
Ramalingam Jegadeesan, Tamil Nadu Agricultural University, IndiaCopyright © 2023 Gautam, Singh, Sakthivel, Venkatesan, Rao, Srikumar, Vijayan, Rakesh, Ray, Akhtar, Meena, Langyan, Ali and Krishnamurthy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raj Kumar Gautam, UmFqLmdhdXRhbUBpY2FyLmdvdi5pbg==; Sapna Langyan, c2luZ2guc2FwbmEwNkBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.