- 1Department of Horticulture, Zijingang Campus, Zhejiang University, Hangzhou, China
- 2Key Laboratory of Horticultural Plants Growth and Development, Agricultural Ministry of China, Hangzhou, China
Plant synthetic biology has emerged as a powerful and promising approach to enhance the production of value-added metabolites in plants. Flavonoids, a class of plant secondary metabolites, offer numerous health benefits and have attracted attention for their potential use in plant-based products. However, achieving high yields of specific flavonoids remains challenging due to the complex and diverse metabolic pathways involved in their biosynthesis. In recent years, synthetic biology approaches leveraging transcription factors and enzyme diversity have demonstrated promise in enhancing flavonoid yields and expanding their production repertoire. This review delves into the latest research progress in flavonoid metabolic engineering, encompassing the identification and manipulation of transcription factors and enzymes involved in flavonoid biosynthesis, as well as the deployment of synthetic biology tools for designing metabolic pathways. This review underscores the importance of employing carefully-selected transcription factors to boost plant flavonoid production and harnessing enzyme promiscuity to broaden flavonoid diversity or streamline the biosynthetic steps required for effective metabolic engineering. By harnessing the power of synthetic biology and a deeper understanding of flavonoid biosynthesis, future researchers can potentially transform the landscape of plant-based product development across the food and beverage, pharmaceutical, and cosmetic industries, ultimately benefiting consumers worldwide.
1 Introduction
Flavonoids are natural products derived from plants, characterized by a basic C6-C3-C6 structure formed by linking two benzene rings (A ring and B ring in Figure 1) via a heterocyclic pyrane ring (C ring). These compounds are widely distributed in plant tissues such as leaves, flowers, and fruits, and play a crucial role in plant-environment interactions. They protect plants from adverse environmental factors, such as UV radiation, and attract animals for plant pollination and seed dispersal (Grotewold, 2006; Falcone Ferreyra et al., 2012). Additionally, flavonoids have significant beneficial effects on human health (Winkel-Shirley, 2002; Grotewold, 2006; Falcone Ferreyra et al., 2012), acting as major bioactive secondary metabolites in medicinal herbs. They possess excellent free radical scavenging and antioxidant activities, reducing inflammation and the risk of chronic diseases such as cancer, diabetes, and cardiovascular diseases (Ravishankar et al., 2013; Lin et al., 2018; Perez-Vizcaino and Fraga, 2018; López, 2019). Diverse classes of flavonoids are currently manufactured and consumed in the food, pharmaceutical, and nutraceutical industries (Tian et al., 2017). However, the primary source of industrialized flavonoids relies on phytoextraction, which is constrained by limited supplies of raw plant materials containing the targeted compounds. In some cases, the desired flavonoids are present at low abundance in plants, further decreasing the product yields (Pyne et al., 2019; Sheng et al., 2020).
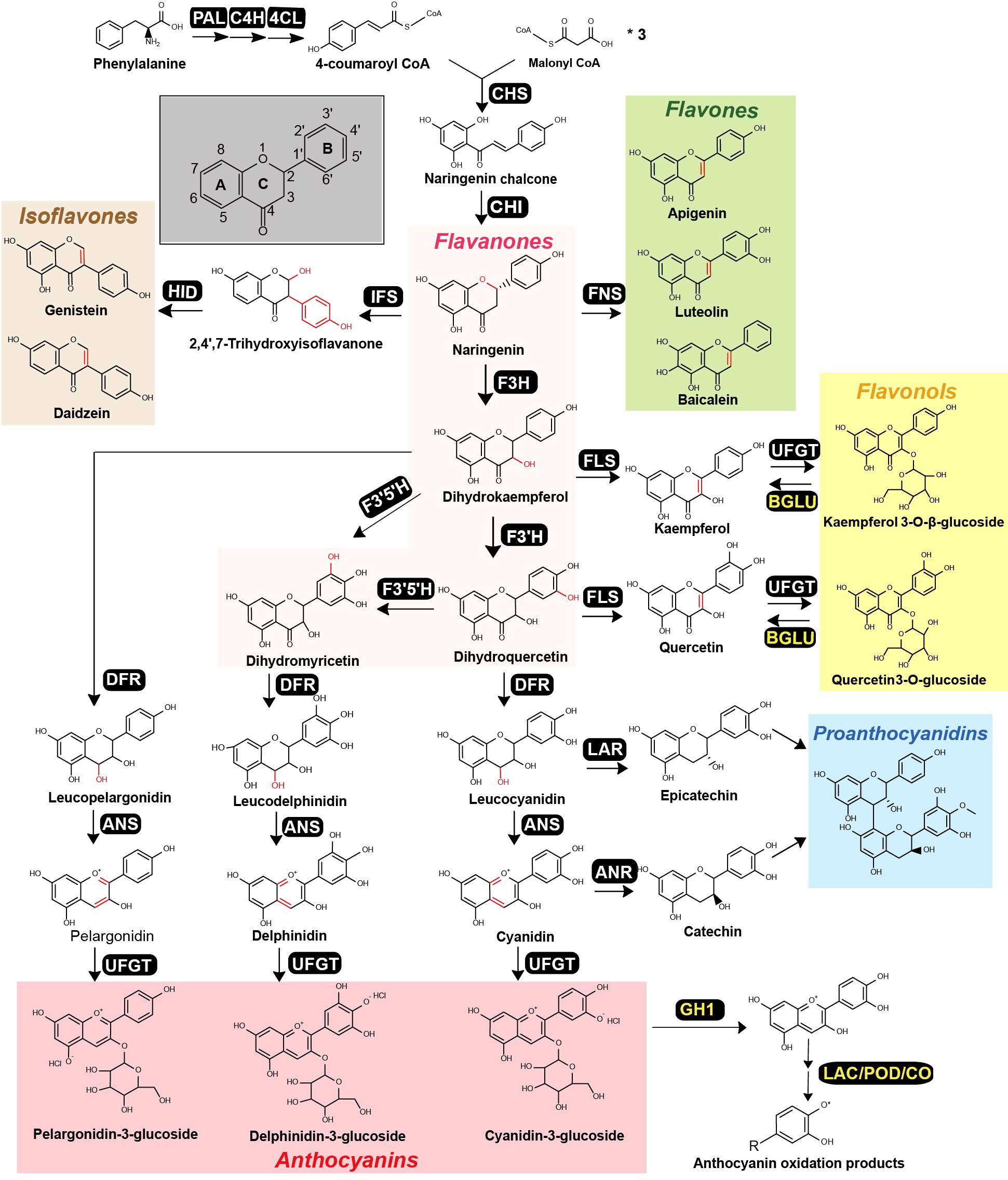
Figure 1 Canonical flavonoid metabolic pathway that produces a diverse range of flavonoids in plants. The pathway, beginning with phenylalanine, involves multiple enzymatic steps, yielding various flavonoid classes: flavanones (highlighted in pink), flavones (green), isoflavones (orange), flavonols (yellow), anthocyanins (red) and proanthocyanidins (blue). Each distinct enzymatic step is catalyzed by a specific enzyme, with abbreviation provided next to the corresponding product. These abbreviations include phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumaroyl-CoA ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavone synthase (FNS), isoflavone synthase (IFS), flavanone 3-hydroxylase (F3H), flavonoid 3’-hydroxylase (F3’H), flavonoid 3’, 5’-hydroxylase (F3’5’H), flavonol synthase (FLS), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS), leucoanthocyanin reductase (LAR), anthocyanidin reductase (ANR), UDP-flavonoid glycosyltransferases (UFGT), β- glucosidase (BGLU), GH (glycoside hydrolase), Laccase (LAC), Peroxidases (POD), Catechol oxidases (CO).The chemical modifications mediated by each enzyme are highlighted in red within the respective chemical structures. The enzymes colored in yellow indicate that they are involved in flavonoid degradation steps.
Modern biotechnology methods, featured by metabolic engineering and synthetic biology, are capable of introducing new biochemical pathways or rewiring existing metabolic networks in organisms to produce desired compounds. These techniques provide effective systems for developing biofortify plants with enhanced health-promoting chemicals (Martin, 2013; Lau et al., 2014; Fu et al., 2018). Historically, microorganisms such as Escherichia coli and yeast Saccharomyces cerevisiae have served as major hosts for metabolic engineering to produce the value-added natural products. For example, many flavonoids with nutraceutical and pharmaceutical interests have been produced by microorganisms, including flavanones (silybin and isosilybin, etc.) (Lv et al., 2019; Yang et al., 2020), flavones (luteolin and apigenin, etc.) (Pei et al., 2020; Zhu et al., 2020b), flavonols (quercetin and kaempferol, etc.) (Sultana and Anwar, 2008; Sheng et al., 2020). In recent years, plants have emerged as an alternative choice of synthetic biology chassis, particularly for producing plant-derived compounds. Although selecting plants as hosts poses challenges for metabolic engineering due to low transformation efficiency and long-life cycles, they offer several advantages, such as photosynthesis-driven growth using CO2 as the carbon source and the ability to expand production using existing agricultural or horticultural infrastructure (Zhu et al., 2017; Fu et al., 2018). Furthermore, plant chassis provides a promising platform to effectively generate plant-derive nutrients or bioactive chemicals that are difficult to produce in microbes, but can be more easily manipulated in plants by altering existing regulatory or biosynthetic enzymes (Zhu et al., 2017; Garg et al., 2018; Pyne et al., 2019).
Flavonoids are highly desirable compounds for plant metabolic engineering, and the flavonoid metabolic network has been successfully engineered in many plants to optimize the biosynthesis of health-promoting anthocyanins, resulting in anthocyanin-rich crops such as tomatoes, rice, and maize (Ogo et al., 2013; Zhang et al., 2015; Zhu et al., 2017; Liu et al., 2018a). Efforts to engineer desired flavonoids in plants have been pursued along two main directions: boosting flavonoid content by manipulating transcription regulatory components and introducing novel flavonoids through engineering enzyme diversity (Bovy et al., 2007; Nabavi et al., 2020). Simultaneously overexpressing multiple genes encoding biosynthetic enzymes efficiently promotes plant flavonoid contents, which can be achieved through manipulating master regulators, such as transcription factors (Hichri et al., 2011; Nabavi et al., 2020). Multiple transcription factors regulating flavonoids biosynthesis have been identified in various plant species, have been served as targets for plant flavonoid engineering. Concurrently, the natural variation of plant flavonoid biosynthetic enzymes, which exhibit promiscuous catalytic specificity, can be leveraged to expand chemical diversity and engineer flavonoids in selected plant hosts (Su et al., 2017; Jia et al., 2019; Shang and Huang, 2020; Jiang et al., 2021).
Overall, plant flavonoid metabolic engineering offers a valuable approach to modify the flavor, aroma, and color of plant-based products, ultimately enhancing their appeal to consumers (Zhu et al., 2020b). This can benefit the food and beverage industries, as well as for the pharmaceutical and cosmetic industries, where the demand for flavonoid-rich products is on the rise. In this review, we provide a comprehensive summary of recent research advancements in flavonoid metabolic engineering, encompassing the diverse benefits of flavonoids across various plant families, the transcription factors that govern flavonoid biosynthesis, and the enzyme promiscuity that can modulate flavonoid substrate specificity. Specifically, we emphasize the potential of utilizing transcription factors to increase plant flavonoid production and leveraging enzyme natural variation to expand flavonoid diversity as synthetic biology strategies.
2 Flavonoid biosynthesis and degradation
Flavonoids are a diverse group of secondary metabolites found in various plant families and can be categorized into several classes, including flavanones, flavones, isoflavones, flavonols, anthocyanins, and proanthocyanidins (Brodowska, 2017; Tohge et al., 2017; Dias et al., 2021). The canonical pathway for flavonoid biosynthesis begins with phenylalanine, which undergoes sequential catalysis by three enzymes, phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-coumaroyl-CoA ligase (4CL), to produce 4-coumaroyl-CoA (Winkel-shirley, 2001; Falcone Ferreyra et al., 2012; Dias et al., 2021) (Figure 1). Chalcone synthase (CHS) then converts one molecule of 4-coumaroyl-CoA and three molecules of malonyl-CoA to naringenin chalcone, the essential precursor of flavonoids. Naringenin chalcone is subsequently converted to naringenin, a ternary ring structure, by the enzyme chalcone isomerase (CHI). Naringenin serves as a central metabolic node for various flavonoid classes (Figure 1), and can be converted to flavones (with a double bond between C-2 and C-3) by flavone synthase (FNS), to Fabaceae-specific isoflavones (with the benzene ring on C-3) by isoflavone synthase (IFS), or to dihydroflavonols (with C-3 hydroxylation) by flavanone 3-hydroxylase (F3H). Dihydroflavonols are further classified into dihydrokaempferol, dihydroquercetin and dihydromyricetin, depending on the position of B-ring hydroxylation at B-3’ or B-5’, which is catalyzed by flavonoid 3’-hydroxylase (F3’H) or flavonoid 3’, 5’-hydroxylase (F3’5’H) (Figure 1). Dihydroflavonols can also be converted to flavonols by flavonol synthase (FLS) similarly to FNS, forming a double bond between C-2 and C-3 positions. Furthermore, dihydroflavonols can be converted to anthocyanidins, a major group of flower pigments, by the sequential actions of dihydroflavonol 4-reductase (DFR) and anthocyanidin synthase (ANS). In some plants, leucocyanidin and cyanidin can be transformed to epicatechin by leucoanthocyanins reductase (LAR) and catechin by anthocyanidin reductase (ANR), respectively, which are further condensed to the polyphenol proanthocyanidins (Figure 1). Finally, UDP-flavonoid glycosyltransferases (UFGTs) catalyze the final glycosylation steps and are the major contributor to flavonoid chemical diversity.
Flavonoid catabolism is influenced by multiple factors including developmental signals and environmental conditions, including light, temperature, pH, etc. Current research exploring the catabolic enzymes responsible for flavonoid degradation mostly focusing on anthocyanins. These enzymes originate from various protein families including glycoside hydrolase (GH1) and oxidation related enzymes (LAC, POD, CO) (Figure 1). Enzymes in the glycoside hydrolase1 (GH1) family are primarily associated with the processing of plant defense-related glycosylated specialized metabolites, such as saponins, coniferin, flavonoids, and inactive glycosylated forms of plant hormones (Minic, 2008; Cao et al., 2017; Dong et al., 2019). Recent studies have found that GH1 plays a role in flavonoid catabolism, specifically targeting the β-glycoside linkage between glucose and the aglycone component of the flavonol glucosides (Bozzo and Unterlander, 2021). For instance, AtBGLU15 catalyzes the hydrolysis quercetin 3-O-β-glucoside to quercetin and glucose (Roepke and Bozzo, 2015; Roepke et al., 2017), while CsBGLU12 specializes in the hydrolysis of flavonol 3-O-β-glucosides, such as quercetin 3-O-β-glucoside (Baba et al., 2017). Laccases (LACs), copper-containing polyphenol oxidases, utilize oxygen to oxidize a broad spectrum of aromatic and non-aromatic compounds. Though primarily associated with lignin accumulation, certain LACs also contribute to the color fading of plant fruits, a process involving the breakdown of anthocyanins (Claus, 2004; Pourcel et al., 2007). Notably, a PbLAC4-like gene, identified in pears and regulated by the transcription factor PbMYB26, participates in anthocyanin degradation, causing a fading in the red hue of various pear tissues (Zhao et al., 2021b). In Arabidopsis seeds, the TT10 laccase in uniquely involved in flavonoid degradation through oxidation reactions among the 17 identified LACs (Pourcel et al., 2005). Flavonoid-peroxidases (PODs), primarily function as H2O2-scavengers, play a crucial role in plant cell detoxification (Yamasaki et al., 1997; Pourcel et al., 2007). In the case of Brunfelsia calycina, BcPrx01, a vacuolar type Class III peroxidase, was integral in the degradation of anthocyanins, accelerating the color transition of calyx flowers from deep purple to white after blooming (Zipor et al., 2015). In a separate study, Liu (Liu et al., 2022) observed an increase in POD activity coinciding with color fading, which is due to the anthocyanin degradation catalyzed by POD.
In summary, the multifaceted enzymatic machinery participating in flavonoid metabolism forms a complex metabolic network (Figure 1). This network serves as a blueprint for metabolic engineering strategies aimed at enhancing the production and modification of flavonoids in plants. By manipulating the key enzymes and regulatory factors within this system, researchers can unlock the potential for tailored production of specific flavonoids with desired properties, paving the way for the development of novel therapeutics, nutraceuticals, and functional food ingredients.
3 Chemical diversity, and potential health benefits of the flavonoid found across plant families
Flavonoids are widely distributed among different plant families and species, offering a range of benefits to humans (Figure 2). These flavonoid-rich plant species can be grouped into three categories based on their perceived benefits: medicinal plants with antibacterial or anti-inflammatory flavonoids, edible plants with health-promoting flavonoids, and ornamental plants with flavonoid pigments used for their floral coloration (Figure 2). Flavonoids have been identified as the primary bioactive compounds in various medicinal plants. These compounds have been extensively studied for their potential health benefits, demonstrating effectiveness in preventing and treating a variety of diseases. Baicalin and wogonin, found in the Scutellaria genus, have been shown to be effective in the prevention and treatment of cancer (Pei et al., 2022). Numerous studies have revealed their anti-tumor properties, inhibiting the growth and proliferation of cancer cells (Singh et al., 2021; Banik et al., 2022). Isoliquiritigenin and liquiritigenin, found mainly in the Glycyrrhiza genus, possess potent antioxidant and anti-aging properties. These compounds have been shown to protect against oxidative stress and inflammation, which are key factors in the development of aging-related diseases such as Alzheimer’s disease (AD) and Parkinson’s disease (PD) (Ramalingam et al., 2018). Silybin and isosilybin, discovered primarily in Silybum marianum, have a long history of use in traditional medicine for the treatment of liver disease. Modern research has confirmed the efficacy of these compounds in protecting the liver and promoting liver health (Lv et al., 2019). In addition to these specific compounds, other flavonoids, such as xanthohumol, apigenin, taxifolin, luteolin, and acacetin, have been found in various medicinal plants and have demonstrated diverse medicinal properties, including anti-inflammatory, antioxidant, and anti-cancer properties, among other health benefits (Tungmunnithum et al., 2018; Wang et al., 2018c; Ullah et al., 2020).
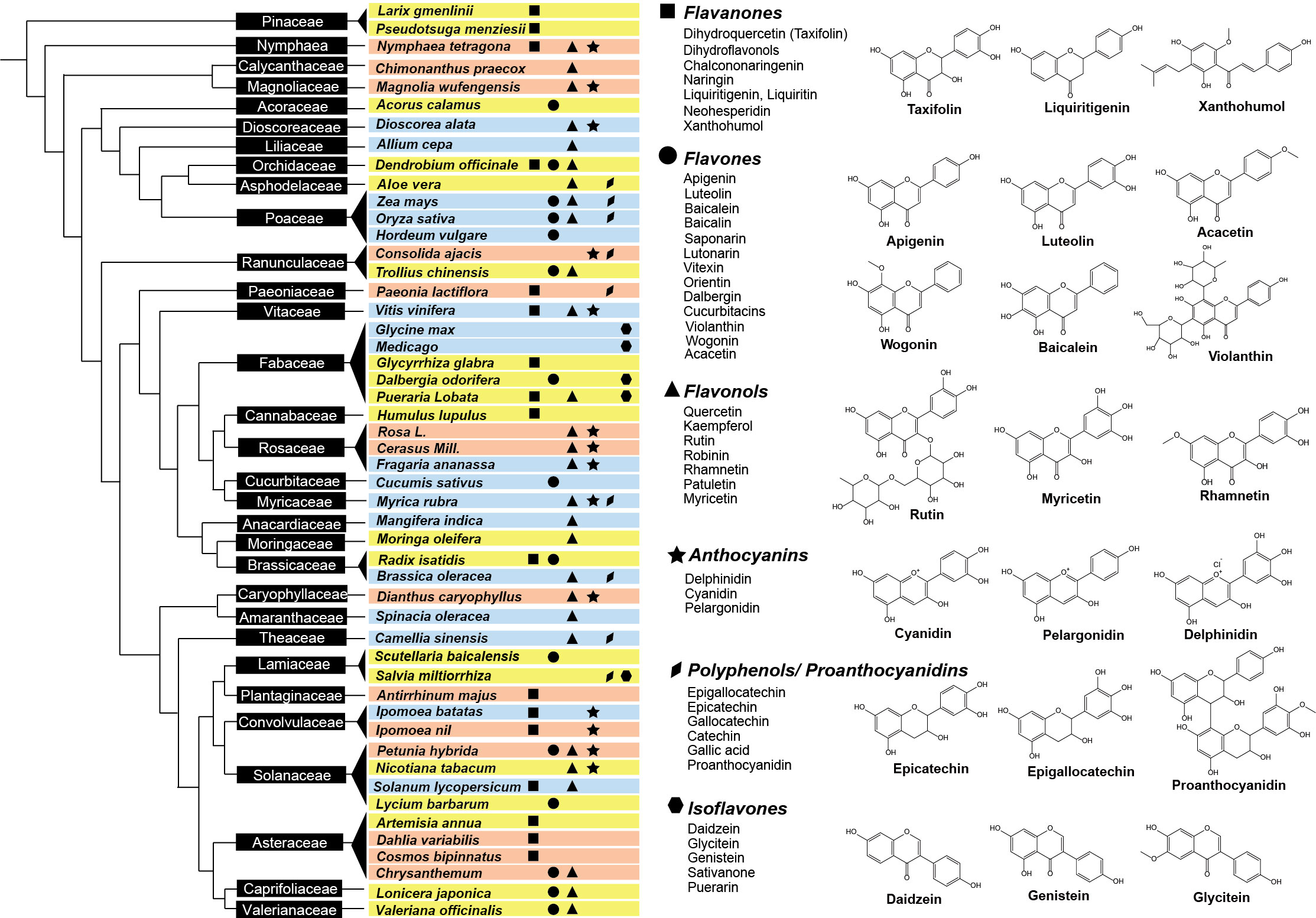
Figure 2 Distribution of flavonoid classes across plant phylogenetic tree. The left panel illustrates the phylogenetic distribution of representative plant species producing flavonoids with pharmaceutical (the corresponding plants are highlighted in yellow), nutritional (blue), or ornamental value (red). Plant families are displayed in black boxes. The right panel shows the major flavonoid classes produced by different plants, represented by unique shapes – squares for flavanones, circles for flavones, triangles for flavonols, stars for anthocyanins, diamonds for proanthocyanidins, and hexagons for isoflavones. Chemical structures of representative flavonoids from each class are also provided.
Plants have long been recognized as excellent sources of flavonoids, which not only aid in food preservation, but also promote a healthy diet (Sultana and Anwar, 2008). These compounds are commonly found in daily food and beverage consumption including fruits, vegetables, and tea, and have gained widespread attention due to their potent antioxidant properties – with some dietary flavonoids possessing stronger antioxidant effects even surpassing vitamin C and vitamin E (Sokół-Łętowska et al., 2007; Sultana and Anwar, 2008). Studies have shown that different plant species are abundant in diverse types of nutritional flavonoids. For example, soybeans are rich in isoflavones (Rochfort and Panozzo, 2007; Chen and Chen, 2021), yams are high in catechins (Dragovic-Uzelac et al., 2007), citrus fruits contain large amounts of flavones and flavanones (Dias et al., 2021), and red or purple fruits such as grapes, waxberries, and blueberries are abundant in anthocyanins (Sultana and Anwar, 2008; Terahara, 2015). Consuming these dietary flavonoids is not only beneficial for providing daily nutritional supplementation, but also helps regulate various body functions and maintain overall health (Yao et al., 2004; Sultana and Anwar, 2008; Terahara, 2015).
Flavonoids also play a role in adding color to various parts of plants, including flowers and leaves. Anthocyanin, one of the most well-known pigments, is estimated to contribute to the color of approximately 80% of flowers in angiosperms (Kong et al., 2003; Castañeda-Ovando et al., 2009). Chalcones and flavonols are primarily responsible for providing plants with a yellow color, while differences in the substituents present at various positions on the basic skeleton structure of flavonoids result in a diverse range of anthocyanins, generating colors such as red, purple, blue-purple, and blue (Castañeda-Ovando et al., 2009; Pasam et al., 2017). Flavonoids play a vital role in coloring various cut flower plants. For example, yellow color flower in common ornamental plants like rose, chrysanthemum, and hydrangeas are primarily due to the presence of chalcones, flavones, and flavonols, while orange, red, purple, and blue flowers, such as roses, tulips, commerinas, delphiniums, and violets are primarily due to the anthocyanin pigments (Nishihara and Nakatsuka, 2011; Iwashina, 2015; Iwashina and Mizuno, 2020; Stavenga et al., 2021). The so-called “black flowers”, such as Fritillaria camtschatcensis (Liliaceae) are also a result of the anthocyanins (Iwashina, 2003; Deguchi et al., 2016). It is worth noting that the role of flavonoids in plant coloration is not limited to visual appeal, but also contributes to attracting pollinators and providing protection against UV radiation.
The role of flavonoids in human daily life is multifaceted, and plants serve as a crucial and convenient source of these compounds. Although different plants contain varying amounts and compositions of flavonoid, with some plants generating limited types or producing low quantities overall, plant metabolic engineering has been used to increase the production of target components or broaden the chemical space of plant flavonoids (Pandey et al., 2016; Marsafari et al., 2020; Zhu et al., 2020b). By utilizing this approach, researchers have been able to enhance the content of existing flavonoids or create novel flavonoids not previously found in plants, providing a more efficient means to meet the nutritional and health needs of humans.
4 Transcriptional regulation of flavonoid biosynthesis pathway for metabolic engineering in plants
Plant flavonoid metabolism represents a complex and highly regulated process controlled by multiple transcription factors, which govern the gene expression of multiple steps within the flavonoid biosynthesis pathway. These transcriptional regulators can be harnessed for plant metabolic engineering, enabling the fine-tuning of gene expression to activate or repress metabolic pathway genes and enhance flavonoid content in plants (Broun, 2004; Grotewold, 2008; Hichri et al., 2011). In recent years, significant progress has been made in uncovering the biosynthesis-related transcription factors for flavonoids in numerous plant species as listed in Table 1. This offers promising opportunities for metabolic engineering to develop new and improved plant varieties with elevated flavonoid levels for human consumption.
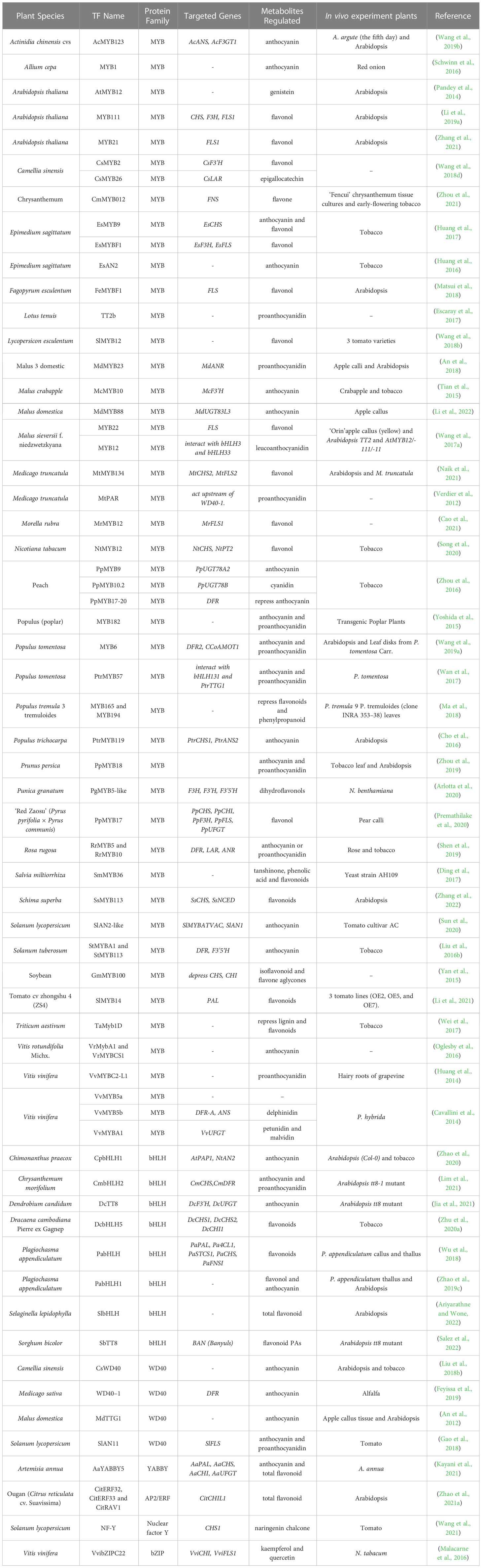
Table 1 Recently identified transcription factors that regulate the expression of genes involved in plant flavonoid biosynthesis.
4.1 R2R3-type MYB transcription factors and their roles in regulating flavonoid biosynthesis
The regulation of flavonoid biosynthesis-related genes involves several well-characterized transcription factors, including the R2R3 MYB family, basic helix-loop-helix (bHLH) family, and WD40 proteins. These transcription factors are responsible for regulating the expression of genes in two primary groups (Figure 3A): the early biosynthetic genes (EBGs) and the late biosynthetic genes (LBGs) (Petroni and Tonelli, 2011; Falcone Ferreyra et al., 2012; Xu et al., 2015). EBGs, such as CHS, FNS, and FLS, encode enzymes involved in the biosynthesis of compounds upstream of dihydrokaempferol, including flavones and flavonols. LBGs, including DFR, ANS, LAR, and ANR, encode enzymes involved in the downstream pathway leading to the production of anthocyanins and proanthocyanidins (Dixon et al., 2005; Dixon and Sarnala, 2020) (Figure 3A). MYB proteins primarily regulate the EBGs, while LBGs are regulated by the MBW complex, comprising MYB, bHLH, and WD40 proteins (Li, 2014).
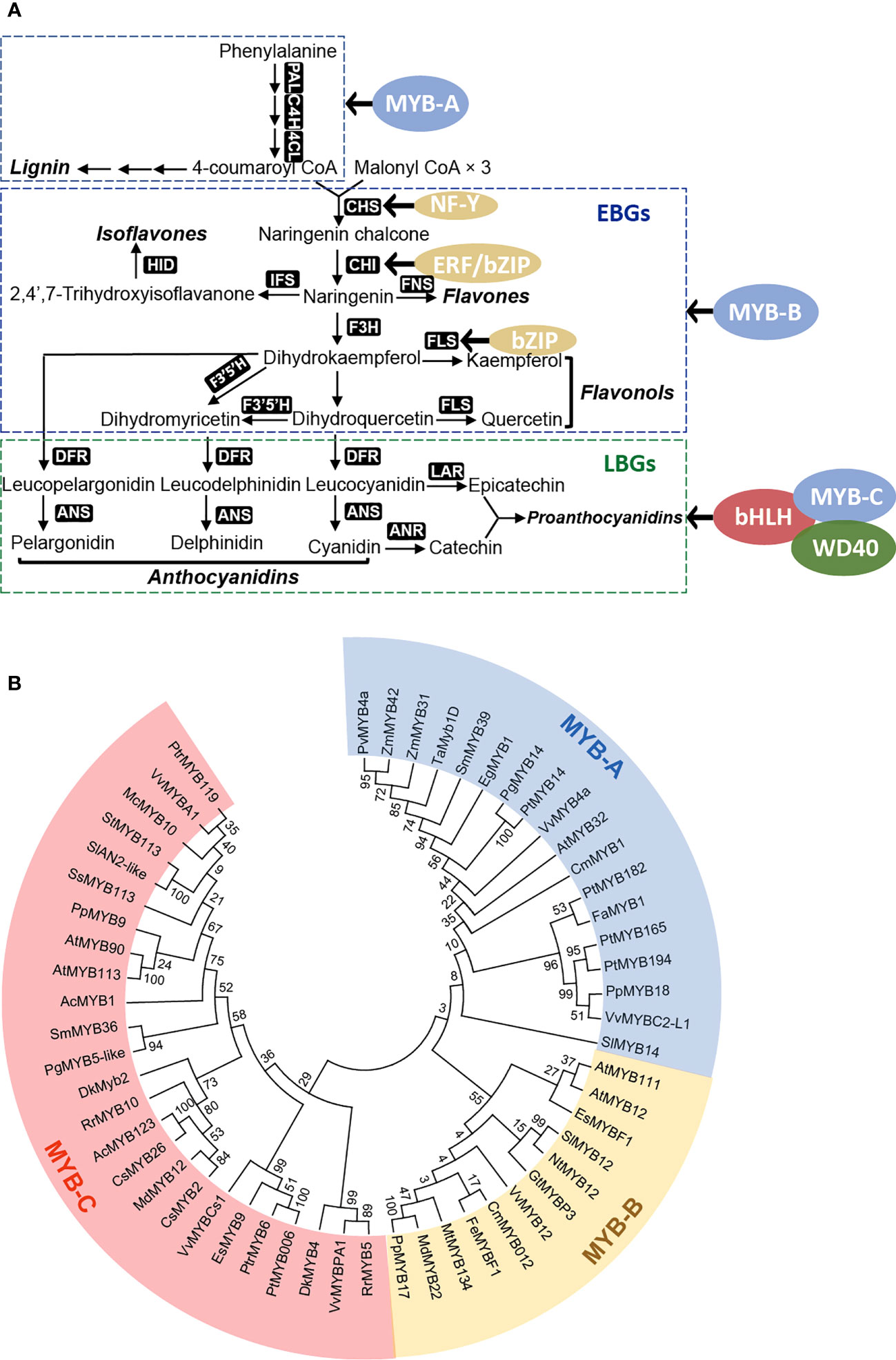
Figure 3 Transcriptional regulatory network in plant flavonoid biosynthesis. (A) Transcription factors critical for regulating genes in flavonoid biosynthesis, can be categorized into early biosynthetic genes (EBGs) and late biosynthetic genes (LBGs). Transcription factors belong to several protein families including MYB (blue), bHLH (red), WD40 (green), and other sporadically identified regulators (yellow); (B) Phylogenetic analysis reveals distinct groups of MYB proteins in regulating flavonoid biosynthesis. These recently identified R2R3-MYBs can been phylogenetically and functionally categorized into three groups: MYB-A (blue), MYB-B (yellow), and MYB-C (red), serving as the major regulators of flavonoid biosynthesis.
R2R3-type MYB transcription factors are the most frequently identified regulators of flavonoid biosynthesis. These factors bind to the cis-elements of promoters to regulate flavonoid metabolism (Dubos et al., 2010; Xu et al., 2015). Recently identified flavonoid biosynthesis-related R2R3-MYBs are phylogenetically divided into three groups that control the biosynthesis of specific flavonoid products (Figure 3B). The MYB-A group predominantly regulates genes involved in the upstream phenylpropanoid pathway and those associated with lignin biosynthesis. The MYB-B group mainly regulates the expression of EBGs, thus controlling the biosynthesis of flavonols or flavones, while the MYB-C group regulates the LBGs expression, determining the biosynthesis of anthocyanins or proanthocyanidins (Figure 3B). MYB-C group transcription factors often form a protein complex with bHLH and WD40 to execute their regulatory functions.
The MYB-B group, comprising MYB11, MYB12 and MYB111, is responsible for up-regulating the expression of essential EBGs, including CHS, F3H and FLS1. This leads to the accumulation of specific flavonols (Stracke et al., 2007; Stracke et al., 2010; Pandey et al., 2014; Pandey et al., 2015). AtMYB11, AtMYB12, AtMYB111 directly regulate the expression of AtF3H and AtFLS, impacting flavonol biosynthesis (Stracke et al., 2007; Li et al., 2019a). MYB12 has been extensively studied and primarily directs the flavonol biosynthesis. In tobacco, NtMYB12 regulates the expression of NtCHS and NtPT2, thereby increasing flavonol accumulation and enhancing tolerance to low phosphorus stress (Song et al., 2020). In the traditional medicinal plant Scutellaria baicalensis, the nuclear localized transcription factor SbMYB12 activates the expression of SbCCL7-4, SbCH-2, and SbF6H-1 by binding to their promoters, resulting in increased baicalin and wogonoside content in the SbMYB12-overexpressed hairy roots of S. baicalensis (Wang et al., 2022). Ectopic overexpression of MrMYB12 from Morella rubra in transgenic tobacco induced the expression of NtCHS, NtF3H and NtFLS, resulting in a significant increase in flavonol content and a pale pink or pure white flower color due to a decrease in anthocyanin content (Cao et al., 2021). Overexpression of SlMYB12 in tomato fruit, using the fruit-specific E8 promoter, significantly increased flavonol content (Wang et al., 2018b). In Asiatic hybrid lily flowers, an ornamental plant, MYB12 upregulation under high temperature conditions enhances anthocyanin content and improves flower coloring (Yamagishi and Sakai, 2020; Yamagishi, 2022).
Several MYB variants in the MYB-B group, besides MYB11, MYB12, and MYB111, have also been identified to regulate the expression of EBGs in various plants. For instance, MtMYB134 in Medicago truncatula enhances flavonol biosynthesis through the activation of CHS and FLS genes (Naik et al., 2021). The MYB17 transcription factor PpMYB17 from pear binds to the promoters of PpCHS, PpCHI, PpF3H, PpFLS and PpUFGT, increasing their expression and leading to higher levels of flavonols, flavanones, and flavones in PpMYB17-overexpressing pear calli (Premathilake et al., 2020). MYB22 in apple (Malus sieversii) stimulates the flavonol production by directly binding to the FLS promoter. In this case, MdMYB22 was able to alleviate the flavonol deficiency phenotype in the Arabidopsis AtMYB11/-12/-111 triple mutant, suggesting that MYB22 function is conserved with other MYB-B group members (Wang et al., 2017a).
The MYB-C group of transcription factors regulates flavonoid biosynthesis by forming a ternary protein complex with bHLH and WD40, referred to as the MBW complex. In A. thaliana, MYB-C proteins, such as MYB75, MYB90, and MYB113, along with bHLH family proteins TT8, GL3, and EGL3, have been identified as participants in leaf anthocyanin pigmentation (Borevitz et al., 2000; Gonzalez et al., 2008; Zuluaga et al., 2008). Only one WD40 protein, TTG1, has been identified in A. thaliana (Nesi et al., 2000; Zhang et al., 2003; Dubos et al., 2010), and it interacts with MYB and bHLH in various ways to jointly regulate anthocyanin biosynthesis (Xu et al., 2014; Xu et al., 2015). The same mechanism was also observed in the tea plant Camellia sinensis, where CsWD40 interacts with bHLH and MYB to form the MBW complex, regulating the biosynthesis of anthocyanin and proanthocyanidin (Liu et al., 2018b). In the edible plant mulberry, the transcriptional regulation of fruit flavonoids involves the cooperation of two MBW complexes, one regulating anthocyanin biosynthesis (MYBA-bHLH3-TTG1 complex) and another regulating proanthocyanidin biosynthesis (TT2L1/TT2L2-bHLH3/GL3-TTG1 complex). Both complexes activate the expression of MYB4, which fine-tunes the content of anthocyanins and proanthocyanidins in the plant (Li et al., 2020). In ornamental plants, the interaction between MYB-C group transcription factors and other components, such as bHLH and WD40, to form MBW complexes is frequently observed for the regulation of flavonoid biosynthesis. For instance, in coleus (Solenostemon scutellarioides), the MYB-C factor SsMYB3 forms an MBW complex with AtTT8 and AtTTG1 of Arabidopsis to regulate flavonoid biosynthesis (Zhu et al., 2015). Additionally, when SsMYB3 was expressed ectopically in tobacco, it interacted with two tobacco bHLH proteins (NtAn1a and NtJAF13-1) and a WD40 protein (NtAn11-1) to form an MBW complex that regulates the expression of anthocyanin-related genes NtDFR, NtLAR, and NtANS (Zhu et al., 2015). This suggests that MYB-C may be conserved across different plant lineages. Studies also showed that MYB-C group transcription factors may form complexes with only bHLH proteins, as not all plants contain WD40 proteins (Li et al., 2014; Xu et al., 2015). For example, in the medicinal plant Epimedium sagittatum, co-expression of EsMYB9 with EsTT8, a bHLH gene, strongly induced the expression of LBGs, such as DFR and ANS, leading to increased anthocyanin accumulation (Huang et al., 2017; Jiang et al., 2020). Similarly, in the ornamental plant Chrysanthemum x morifolium, the interaction between CmMYB6 and CmbHLH2 enhances the expression of CmDFR and promotes anthocyanin accumulation (Lim et al., 2021).
On the other hand, some MBW complexes may also function as negative regulators in plant flavonoid biosynthesis, offering opportunities for fine-tuning flavonoid production in plant metabolic engineering. For example, in poplar trees, PtrMYB57 interacts with bHLH131 and PtrTTG1 to form an MBW complex that binds to the promoters of flavonoid genes, inhibiting gene expression and negatively impacting anthocyanin and proanthocyanidin biosynthesis (Wan et al., 2017). Similarly, in peach (Prunus persica), PpMYB18 interacts with PpbHLH3 and PpbHLH33 to inhibit the production of anthocyanin glycosides (Zhou et al., 2019). In the ornamental plant Japanese gentian (Gentiana triflora), GtMYB1R1 interacts with GtbHLH1 protein to form a GtMYB3-GtbHLH1 complex that controls floral anthocyanin biosynthesis. However, expressing GtMYB1R heterologously in tobacco flowers decreases the expression of CHI, DFR and ANS and reduces anthocyanin pigmentation (Nakatsuka et al., 2013).
In some instances, MYB transcription factors in the MYB-C group can regulate anthocyanidin biosynthesis even without a well-defined partnership with bHLH or WD40 proteins. For example, in peach, PpMYB9 or PpMYB10 have been found to bind to the promoters of various UFGT genes in petal flowers, influencing anthocyanin accumulation and, ultimately, petal color (Zhou et al., 2016). Similarly, in crabapple, McMYB10 has been shown to bind to the promoter of McF3’H and increase its expression, leading to enhanced anthocyanin accumulation in stably transformed crabapple plants over-expressing McMYB10 (Tian et al., 2015). In grapes, the accumulation of anthocyanin pigments is regulated by a number of MYB-R2R3 transcription factors such as VvMYBC2-L1, VvMYB5a, VvMYB5b, and VvMYBA1. However, their relationships with bHLH and WD40 proteins have yet to be fully understood (Cavallini et al., 2014; Huang et al., 2014).
4.2 Regulation of flavonoid biosynthesis by bHLH and WD40 proteins of the MYB-bHLH-WD40 complex
The bHLH and WD40 proteins, integral components of the MYB-bHLH-WD40 (MBW) complex, play pivotal roles in regulating flavonoid biosynthesis in plants. The bHLH transcription factor family, encompassing various growth, developmental, and environmental adaptation processes in plants, has been found to be instrumental in the regulation of flavonoid biosynthesis (Samira et al., 2018; Zhu et al., 2019; Zhu et al., 2020a). Numerous studies have demonstrated the potential of leveraging bHLH proteins in modulating plant flavonoid production. For example, DcTT8, a bHLH protein in Dendrobium candidum, promotes the expression of F3’H and UFGT, leading to increased plant anthocyanin accumulation (Jia et al., 2021). Similarly, the bHLH protein DcbHLH5 from Dracaena cambodiana activates the expression of CHS1, CHS2, and CHI1, key genes involved in flavonoid biosynthesis, resulting in increased leaf flavonoid content and enhanced UV-B resistance (Zhu et al., 2020a). In the liverworts plant Plagiochasma appendiculatum, PabHLH1 positively regulates flavonol and anthocyanin biosynthesis by activating multiple genes involved in flavonoid biosynthesis, such as PAL, CHS, CHI, F3H, DFR, and FLS, leading to increased flavonoid and anthocyanin accumulation (Zhao et al., 2019c). PabHLH1’s functions appear evolutionarily conserved, as its heterologous expression in Arabidopsis also promotes the expression of EBGs and LBGs involved in flavonoid biosynthesis and results in increased flavonoid and anthocyanin accumulation (Wu et al., 2018; Zhao et al., 2019c). In another example, the sorghum bHLH transcription factor SbTT8 regulates the expression of genes related to proanthocyanidin biosynthesis. Heterologous overexpression of SbTT8 in the Arabidopsis tt8 mutant rescues the abnormal phenotype of seed dormancy and seed coat color by reestablishing proanthocyanidin biosynthesis (Salez et al., 2022).
WD40 proteins, composed of four or more highly conserved amino acid repeating units terminated by a tryptophan-aspartic acid (WD) dipeptide, are a widespread protein family in eukaryotes, participating in various biological processes such as signal transduction, transcriptional regulation, and cell cycle regulation (Neer et al., 1994; Stirnimann et al., 2010; Gao et al., 2018). In plants, WD40 proteins involved in anthocyanin biosynthesis typically form an MBW complex with bHLHs or MYBs to perform their functions (Xu et al., 2015; Lloyd et al., 2017). However, manipulation of the WD40 protein alone can also effectively control plant flavonoid biosynthesis. For instance, overexpression of the FtWD40 protein from Fagopyrum tataricum in tobacco enhances the expression of DFR and ANS, resulting in increased anthocyanin accumulation and pigmentation in petals (Yao et al., 2017). Similarly, overexpression of the tomato WD40 gene SlAN11 in tobacco results in higher leaf anthocyanin and seed proanthocyanidin contents, while reducing flavonol accumulation as dihydroflavonol precursors are redirected to anthocyanin biosynthesis (Gao et al., 2018). In the ornamental plant Freesia hybrida, FhTTG1 interacts with FhbHLH proteins (FhTT8L and FhGL3L) to form the MBW complex. When FhbHLH is co-expressed with MYB and bHLH partners, significant activation of anthocyanin and proanthocyanidin biosynthesis is observed (Shan et al., 2019). Furthermore, WD40 proteins can regulate flavonoid biosynthesis through interaction with bHLH proteins, without the involvement of MYB. For example, the apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin biosynthesis (An et al., 2012). Similarly, SlAN11, a tomato WD40 protein, interacts with bHLH proteins but not MYB to promote leaf anthocyanin and seed proanthocyanidin accumulation, while reducing flavonol content (Gao et al., 2018).
4.3 The regulatory network of flavonoid biosynthesis beyond MYB, bHLH, and WD40 proteins
In addition to MYB, bHLH, and WD40 proteins, other transcription factors also play a role in regulating flavonoid biosynthesis. For example, a bZIP transcription factor, VvibZIPC22, was identified in grape to activate the promoters of flavonoid pathway genes (VviCHI and VviFLS1), leading to increased flavonol production (Malacarne et al., 2016). Ectopic overexpression of VvibZIPC22 in tobacco promotes the expression of several floral genes involved in flavonoid biosynthesis, resulting in a significant increase in flavonoid content in the flowers (Malacarne et al., 2016). YABBY transcription factors, exclusive to seed plants, also play crucial roles in regulating primary and secondary metabolism, including flavonoid biosynthesis. In Artemisia annua, the YABBY5 transcription factor activates multiple genes of the flavonoid pathway, including AaPAL, AaCHS, AaCHI, and AaUFGT, which led to a significant increase in total flavonoid accumulation and an increase in anthocyanin production that causes the stem to turn deep purple (Kayani et al., 2021). Nuclear factor Y (NF-Y) proteins, evolutionarily conserved transcription factors that bind to the CAAT box of many genes, have also been found to play a role in flavonoid regulation. In tomato, an NF-Y factor binds to the CHS1 promoter and inhibits its expression, leading to a decrease in overall flavonoid content that typically imparts a yellowish color to the fruit during ripening, resulting in the production of pink-colored fruit with colorless peels (Wang et al., 2021). In citrus, the AP2/ERF transcription factors affect flavonoid biosynthesis. CitERF32 and CitERF33, two citrus AP2/ERF proteins, enhance the expression of CitCHIL1, enabling the substrate to flow more efficiently into CHS and thereby increasing overall flavonoid content (Zhao et al., 2021a).
In summary, transcription factors play a vital role in the regulation of flavonoid biosynthesis in plants, and their use in plant metabolic engineering holds great potential. By expressing selected transcription factors in specific organs, it is possible to manipulate the expression of key target genes involved in flavonoid production and thus control the levels of specific flavonoid components. This approach opens up a promising avenue to create plants with elevated flavonoid content, which are increasingly in demand for their health-promoting properties. However, it is important to note that while transcription factors are effective regulators of flavonoid biosynthesis, they mainly regulate certain classes of flavonoids. To specifically regulate the synthesis of a particular flavonoid, it is necessary to perform plant metabolic engineering experiments in combination with key structural genes.
5 Explore the natural diversity of flavonoid biosynthesis for synthetic biology
Plant-derived natural products have historically served as therapeutic agents, with their pharmacological properties largely determined by their core structures. However, to develop innovative drugs, it is often necessary to modify these structures (Guo, 2017). Such innovations can improve drug metabolic stability, increase activity or selectivity, adjust physicochemical properties, or simplify structures for easier production (Guo, 2017; Atanasov et al., 2021). The natural diversity of plant chemicals offers a reservoir of structural variations that can be exploited for drug innovation through metabolic engineering and synthetic biology (King et al., 2016; Lautié et al., 2020). Flavonoids are a class of plant natural products with potential for drug innovation, which are widely distributed throughout the plant kingdom and exhibit various structural modifications on their central structures, such as hydroxylation, methylation, and glycosylation. These modifications can significantly affect the bioactivity and bioavailability of flavonoid compounds used in drugs or functional foods. For example, methylation of the hydroxyl group in flavonoids can drastically improve their metabolic stability and membrane permeability, facilitating absorption and bioavailability (Wen and Walle, 2006; Koirala, 2016). Glycosylation of flavonoids can improve drug solubility and enhance uptake (Fernández et al., 2000). Metabolic engineering of flavonoid biosynthesis offers an opportunity to exploit the natural chemical diversity of flavonoids for drug innovation. By identifying and manipulating enzyme variants that can catalyze desired structural modifications, biosynthetic pathways can be simplified and enzyme performance improved. Such synthetic biology strategies hold great promise for developing new drug candidates and functional food ingredients, and for enabling more efficient and sustainable production processes.
5.1 Enzyme divergence at early flavonoid biosynthetic steps determines B-ring hydroxylation and methylation patterns
Naturally, the flavonoid core structure is hydroxylated and O-methylated at various positions. The site and number of these substitution groups vary across plant species and have diverse physiological relevance, particularly in terms of flavonoid B-ring modification patterns. For example, six anthocyanidins, including cyanidin, pelargonidin, delphinidin, peonidin, petunidin, and malvidin, are differentiated by B-ring modifications and exist widely in flowers with colors of pink, scarlet, red, or magenta. The colors of these anthocyanidins are determined by the number of hydroxylations - a single hydroxylation on the 4’ position results in scarlet, while multiple hydroxylations on the 3’, 4’, and 5’ position give blue (Iwashina, 2015). In an extreme case, the Scutellaria genus of the Lamiaceae family produces unique 4’-deoxyflavones that lack any decoration on the B-ring, such as wogonin, baicalein, and chrysin, in addition to the 4’-hydroxyflavones apigenin and scutellarein (Figure 4) (Zhao et al., 2019a; Pei et al., 2022). Medical research has revealed that the 4’-deoxyflavones extracted from the roots of the Chinese herb S. baicalensis exhibit higher pharmacological activities than typical 4’-hydroxyflavones (Himeji et al., 2007; Zhao et al., 2019b).
The functional divergence of multiple enzymes in the early steps of flavonoid biosynthesis influences flavonoid B-ring diversity. In the canonical flavonoid biosynthetic pathway, the B-ring 4’-hydroxyl group is derived from p-coumaroyl-CoA, which is condensed with three malonyl-CoAs to produce the backbone naringenin chalcone that is catalyzed by CHS (Figure 1). The 3’- and 5’-hydroxylation are then generated by F3’Hs and F3’5’Hs. However, the 4’-deoxyflavones are less likely to be synthesized by the canonical pathway, which requires an as-yet unidentified dehydroxylase to remove the B-ring 4’-hydroxyl. Recent studies on S. baicalensis flavonoid biosynthesis have shown that enzyme divergence of p-coumaroyl-CoA ligase (4CL), CHS, and flavone synthase II (FNS II) leads to the production of unique 4’-deoxyflavones in the roots (Zhao et al., 2016; Zhao et al., 2019a). The divergence is initiated by a cinnamate-CoA-like enzyme, which prefers to activate cinnamic acid to yield cinnamoyl-CoA, in contrast to 4CL that generates p-coumaroyl-CoA using p-coumaric acid (Zhao et al., 2016). Then, a root-specific CHS2, separated from CHS1 after the divergence of the Lamiaceae family, evolved the function to produce pinocembrin chalcone, a flavanone precursor without B ring hydroxylation (Zhao et al., 2016). A regular CHI catalyzes pinocembrin chalcone to form pinocembrin, which is further converted to chrysin – the precursor of all other 4’-deoxyflavones – by a recently evolved, root-specific FNS II-2 (Zhao et al., 2016). Chrysin is then decorated on the A-ring by F6H to yield baicalein or by F8H and O-methyltransferase (PFOMT) to yield wogonin (Zhao et al., 2018; Zhao et al., 2019a). In the above case, the expanded substrate specificity of 4CL to activate cinnamic acid and CHS2 to accept cinnamate-CoA leads to the unique non-decorated flavonoid B-ring. In angiosperms, CHSs often undergo gene duplication and are expressed in different tissues or cell types with conserved biochemical functions (Huang et al., 2004; Zavala and Opazo, 2015). A study on the flavonoid biosynthetic pathway of the fern plant Stenoloma chusanum identified promiscuous 4CL and CHS that incorporate p-coumaroyl, cinnamoyl, caffeoyl, and feruloyl groups into flavonoids (Ni et al., 2022). The enzymes Sc4CL1 and Sc4CL2 can activate cinnamic acid and its three derivatives, p-coumaric acid, caffeic acid, and ferulic acid, to form the CoA-thioesters (Ni et al., 2022). ScCHS1 showed extraordinary substrate promiscuity towards these acyl-CoAs, which are condensed with malonyl-CoA to produce pinocembrin chalcone, naringenin chalcone, eriodictyol chalcone, and homoeriodictyol chalcone (Ni et al., 2022) (Figure 4).
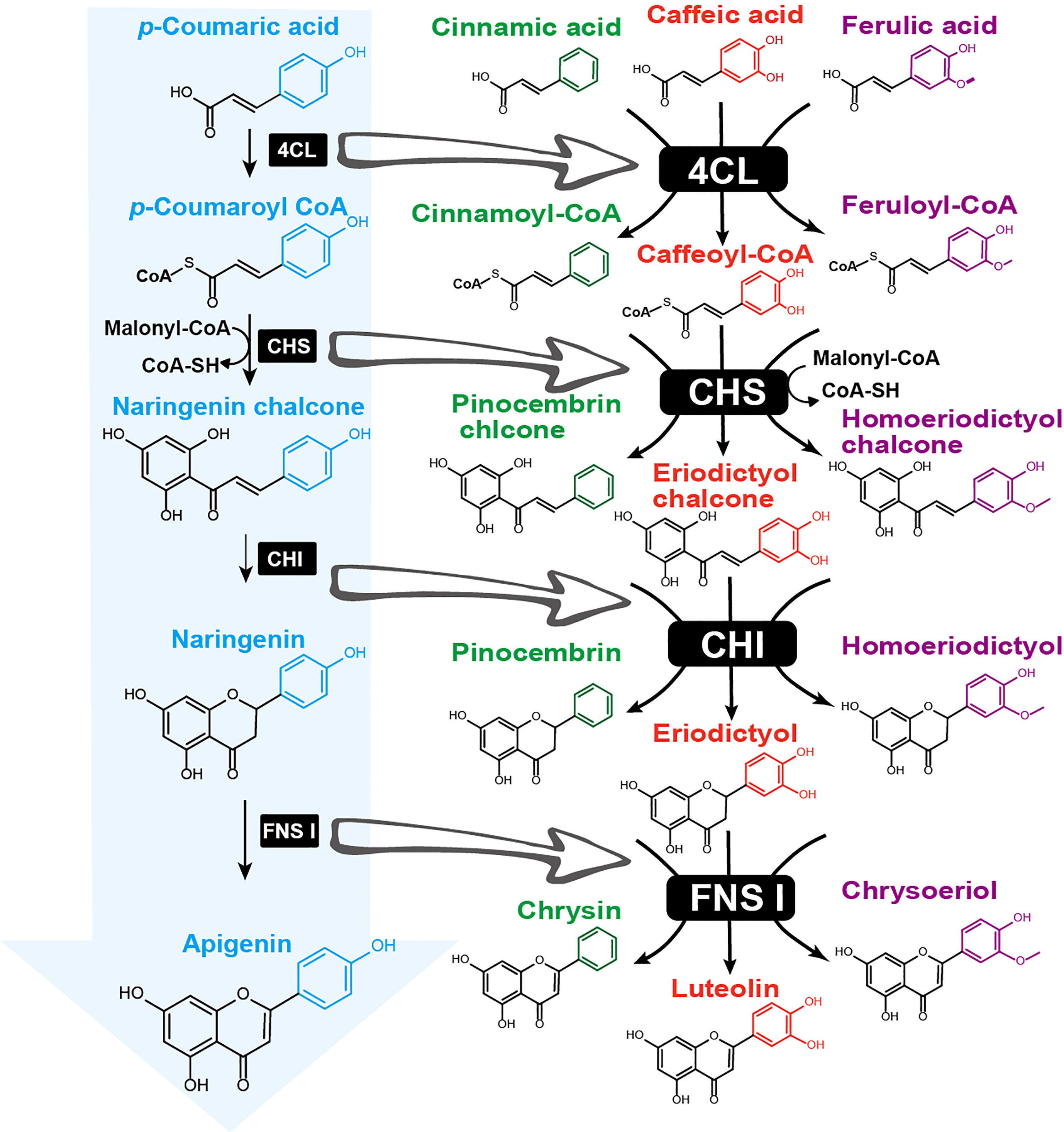
Figure 4 Enzyme promiscuity diversifies flavonoid B-ring hydroxylation and methylation patterns during early biosynthetic steps. The canonical flavonoid biosynthetic pathway (indicated by a blue arrow) begins with p-coumaric acid, which is sequentially catalyzed by 4-coumaroyl-CoA ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), and flavone synthase I (FNS I) to produce apigenin. Enzyme divergence led to promiscuous 4CL accepting multiple substrates with varying hydroxylation and methylation patterns on the aromatic ring (highlighted with different colors), including cinnamic acid (green), caffeic acid (red), and ferulic acid (purple). Acyl-CoA products are further converted by promiscuous CHS, CHI, and FNS to generate diverse flavones, such as chrysin, luteolin, and chrysoeriol.
In canonical flavonoid biosynthesis, hydroxylases and O-methyltransferases are required for decorating all positions on the B-ring except for the 4’ position (Figure 1). However, promiscuous 4CL and CHS enzymes can shorten the biosynthetic routes and generate flavonoid C6-C3-C6 core structures with predetermined B-ring decorations directly from phenylpropanoic acid. This is especially useful for metabolic engineering of flavonoids with desired B-ring modifications in microorganisms such as Escherichia coli and Saccharomyces cerevisiae. By feeding appropriate phenylpropanoic acids such as ferulic acid, engineered E. coli harboring 4CL1 and CHS1 can efficiently produce homoeriodictyol (Cui et al., 2019; Dunstan et al., 2020; Ni et al., 2022), circumventing the need for hydroxylases and O-methyltransferases. When two additional genes, CHI and FNS I, are introduced, a diverse group of flavanones and flavones can be produced, further demonstrating the power of leveraging enzyme diversity for flavonoid metabolic engineering with synthetic biology (Li et al., 2019b; Ni et al., 2022).
5.2 Bifunctional enzymes in flavonoid biosynthesis: potential applications in metabolic engineering
Bifunctional enzymes are rare yet intriguing multifunctional proteins that catalyze two distinct biochemical reactions, often part of consecutive steps in a metabolic pathway (Moore, 2004). These enzymes have evolved through gene fusion, neofunctionalization following gene duplication, or retention of ancestral enzymes’ promiscuous activity (Moore, 2004; Copley, 2015; Seelig, 2017; Siddiq et al., 2017). The development of bifunctional enzymes was likely driven by the need for enhanced metabolic catalytic efficiency and improved coordination of biosynthetic steps. Consequently, bifunctional enzymes hold considerable potential for synthetic biology applications, simplifying biosynthetic steps and generating desired compounds more efficiently. In this section, we present two examples of bifunctional enzymes involved in flavonol and proanthocyanidin biosynthesis and discuss their potential applications in metabolic engineering.
Flavonols, a major flavonoid subgroup, are characterized by a hydroxyl group at the C3 position and a double bond between C2 and C3 positions of the C-ring. Known for their anti-inflammatory and antioxidant properties, flavonols have become important sources of drugs to treat diseases like cancer and trypanosomosis (Borsari et al., 2016; Salehi et al., 2020; Holland et al., 2023; Oo et al., 2022). Flavonol biosynthesis involves two critical modifications of the C-ring, namely C3 hydroxylation by F3H and C2-C3 bond desaturation by flavone synthase I (FNS I) or flavonol synthase (FLS). All three enzymes, FNS I, F3H, and FLS, belong to the 2-oxoglutarate-dependent dioxygenase (2-ODD) family (Farrow and Facchini, 2014), with FNS I and F3H exhibiting a closer phylogenetic relationship than FLS (Li et al., 2020). Bifunctional FLS enzymes have been identified in various plant species, including Oryza sativa (Park et al., 2019), Brassica napus (Schilbert et al., 2021), Citrus unshiu (Lukačin et al., 2003), Ginkgo bilobaand (Xu et al., 2012), and Populus deltoides (Kim et al., 2010), which can catalyze consecutive reactions of C3 hydroxylation and C2-C3 desaturation, producing flavonols from flavanones (Figure 5A). Although the evolutionary mechanisms leading to bifunctional FLS remain unclear, it is hypothesized that the dual-functional enzyme evolved early after plant terrestrialization, as promiscuous FNS I/F3H were observed in basal land plants such as liverworts and mosses (Li et al., 2020). From synthetic biology perspective, bifunctional FLS enzymes are useful for efficiently generating desired flavonols. Indeed, a study aiming to produce the plant flavonol kaempferol using yeast as a cell factory found that the bifunctional PdFLS or CitFLS alone yielded more kaempferol than co-expressing single-function enzymes MdFLS and ZmFLS (Duan et al., 2017).
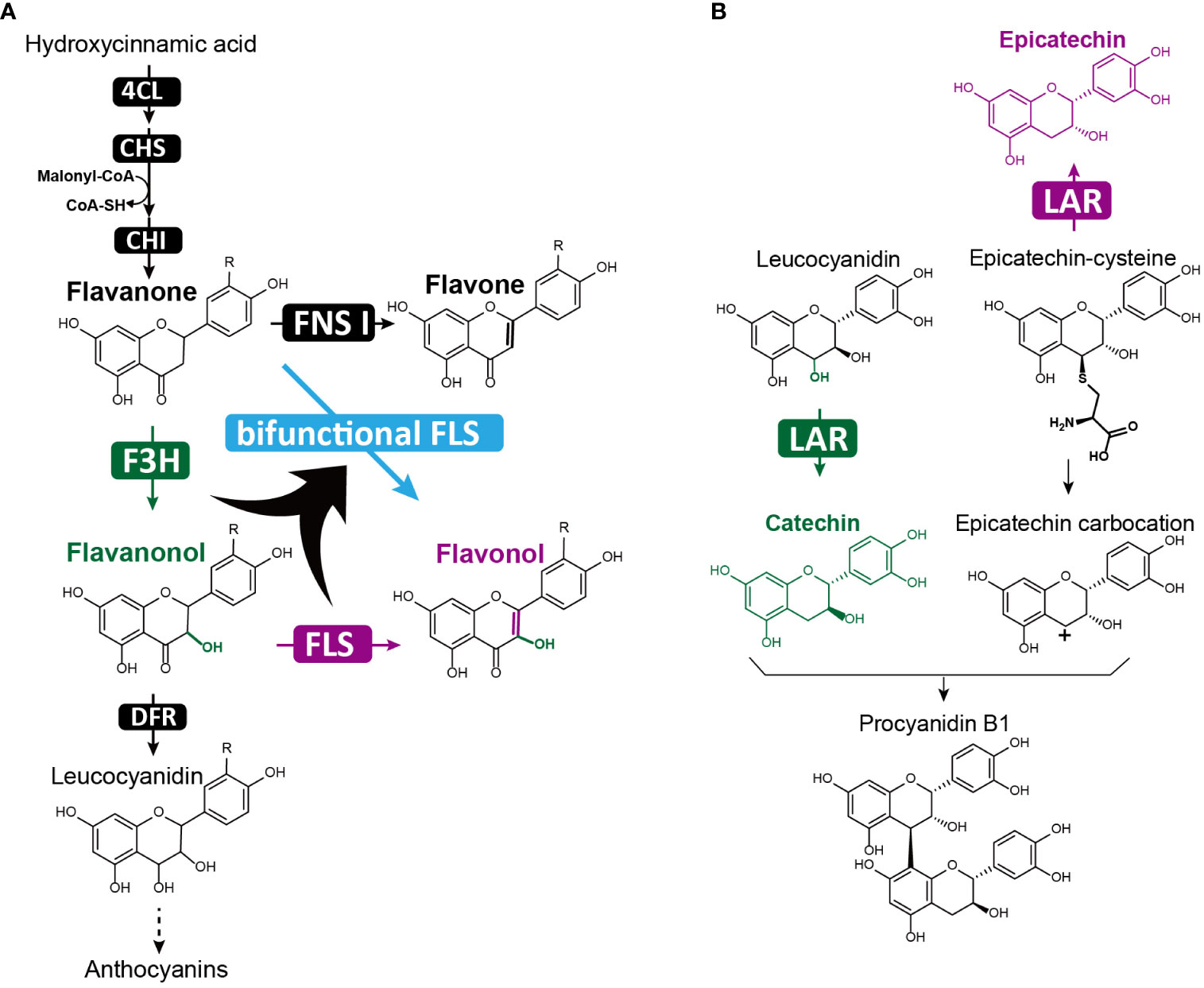
Figure 5 Bifunctional enzymes in flavonol and proanthocyanidin biosynthesis. (A) The bifunctional FLS enzyme (highlighted in blue) catalyzes two sequential reactions: adding a hydroxyl group at the C3 position and forming a double bond between the C2 and C3 positions of the flavonoid C-ring. In the canonical flavonoid biosynthetic pathway, these reactions are catalyzed separately by two single functional enzymes: F3H (highlighted in green) and FLS (highlighted in purple); (B) The bifunctional LAR performs dual functions in proanthocyanidin biosynthesis, converting leucoanthocyanidins to (+)-catechin (highlighted in green), and 4β-(S-cysteinyl)-epicatechin (epicatechin–cysteine) – a specialized form of the proanthocyanidin carbocation unit pool – into (-)-epicatechin (highlighted in purple).
Proanthocyanidins (PAs), or condensed tannins, are oligomeric flavonoids composed of flavan-3-ol units and can be found in numerous plants, particularly in berries such as cranberries, blueberries, and grape seeds. PAs and their oligomers serve as plant protectants against stress (Dixon et al., 2005), and are being investigated for their potential health benefits as therapeutics for cancer, cardiovascular diseases, and lipid metabolic disorders (Wang et al., 2020). The biosynthesis of PAs involves the polymerization of flavan-3-ol monomers, primarily (-)-epicatechin and (+)-catechin, with the flavan-3-ol carbocations attacking the C8 position of another monomer to extend the polymer (Dixon et al., 2005). Among the enzymes that generate flavan-3-ol monomers, leucoanthocyanidin reductase (LAR) play a crucial role in converting leucoanthocyanidins to (+)-catechin (Tanner et al., 2003) (Figure 5B). However, transgenic experiments have shown that LAR overexpression does not increase (+)-catechin production, but instead results in higher accumulation of (-)-epicatechin (Liu et al., 2013; Pang et al., 2013; Wang et al., 2018a). This led to the discovery of the second function of LAR, which converts 4β-(S-cysteinyl)-epicatechin(epicatechin–cysteine) – a special form of the PA carbocation unit pool – into (-)-epicatechin (Liu et al., 2016a; Yu et al., 2019) (Figure 5B). As a result, bifunctional LAR acts as a regulator of PA extension, fine-tuning the balance of insoluble long PA chains with soluble short PA chains and monomers (Liu et al., 2016a) (Figure 5B). Knocking out LAR in the legume Medicago truncatula disrupted this balance, cause an accumulation of insoluble PAs and a loss of soluble epicatechin-derived PAs (Liu et al., 2016a). In plant metabolic engineering, bifunctional LAR could be employed to manipulate the ratio of shorter and longer PA chains, as well as their biosynthetic monomeric precursors. This fine-tuning of PA composition by LAR provides an crucial approach for engineering health-promoting PAs in fruits, particularly optimizing PAs in grapevines, which directly affects red wine quality (Yu et al., 2019) (Figure 5).
5.3 Enzyme promiscuity and structure-guided glycosyltransferase engineering for flavonoid C-glycosylation
Flavonoids are commonly found as glycosides, including glucosides, glucuronsides, rhamnosides, and galactosides. The addition of sugar moieties to the flavonoid core significantly increases solubility and enhances bioavailability, influencing their pharmacological activity (Kren and Martinkova, 2001). Most glycosyl flavonoids are O-glycosylated with sugar moieties attached to hydroxyl groups. However, a survey of plant flavonoid chemical diversity revealed the presence of C-glycosides, especially in cereals (Brazier-Hicks et al., 2009). C-glycosylflavonoids are unique compounds with sugar groups directly conjugated to the aglycone via a C-C glycosidic bond. These flavonoids are highly stable against spontaneous and gastrointestinal hydrolysis due to the rigid C-C bond, resulting in a long in vivo half-life and remarkable druggability (Langenhan et al., 2005; Williams et al., 2007; Oualid and Silva, 2012).
Glycosyltransferases catalyze flavonoid glycosylation using activated sugar nucleotides as the glycosyl donor, such as UDP-glucose (UDP-Glc), UDP-galactose, and UDP-xylose (Slámová et al., 2018). However, compared to the relatively larger and well-studied O-glycosyltransferases (OGTs) family, only a few examples of C-glycosyltransferases (CGTs) have been reported in plants, such as Oryza sativa (Hao et al., 2016), Glycine max (Hirade et al., 2015), Gentiana triflora (Sasaki et al., 2015), Fagopyrum esculentum (Nagatomo et al., 2014), and Pueraria lobata (Wang et al., 2017b). The catalytic mechanism underlying C-glycosyl transfer remains unclear. However, recent comprehensive studies on promiscuous CGTs, capable of catalyzing O-, C-, and di-C-glycosylation, shed light on the mechanistic basis of CGT-mediated C-glycosylation (Gutmann and Nidetzky, 2012; Chen et al., 2018; He et al., 2019; Zhang et al., 2020; Dai et al., 2021). These findings have paved the way for developing efficient and versatile CGTs through protein engineering.
Recent studies have identified key amino acid residues in glycosyltransferases (GTs) that contribute to the switch between O- and C-glycosylation and have applied them for enzyme engineering (Gutmann and Nidetzky, 2012; He et al., 2019). For instance, a notably promiscuous TcCGT1 from Trollius chinensis was found to catalyze C-glycosylation or O-glycosylation using 41 out of 114 tested structurally diverse flavonoid substrates with UDP-Glc as the sugar donor(He et al., 2019). Using apigenin and wogonin as the respective C- and O-glycosylation acceptors (Figure 6), protein structural analysis coupled with molecular docking revealed that the binding pose of the C-glycosyl acceptor is almost vertical to the O-glycosyl acceptor (He et al., 2019), thus necessitating a more spacious acceptor binding pocket, partially determined by the key residue G284. Mutating G284 to bulkier residues such as Phe, Gln, Tyr, or Lys significantly reduced C-glycosylation activity of TcCGT1. Introducing a salt-bridge between G284 and I94 through site-directed mutagenesis of G284F, G284Q, G284Y, and I94E/G284K completely blocked C-glycosylation and turned TcCGT1 into a single functional OGT (He et al., 2019). In another study, Gutmann and Nidetzky identified the active motif in the single-functional enzymes OsCGT and PcOGT using structural-functional analysis (Gutmann and Nidetzky, 2012). They found that reciprocally switching the key residues in the active motif (I117 and D118) in PcOGT to (D120 and I121) in OsCGT created promiscuous enzymes capable of catalyzing both O- and C-glycosylation with the dihydrochalcone phloretin as the acceptor (Gutmann and Nidetzky, 2012).
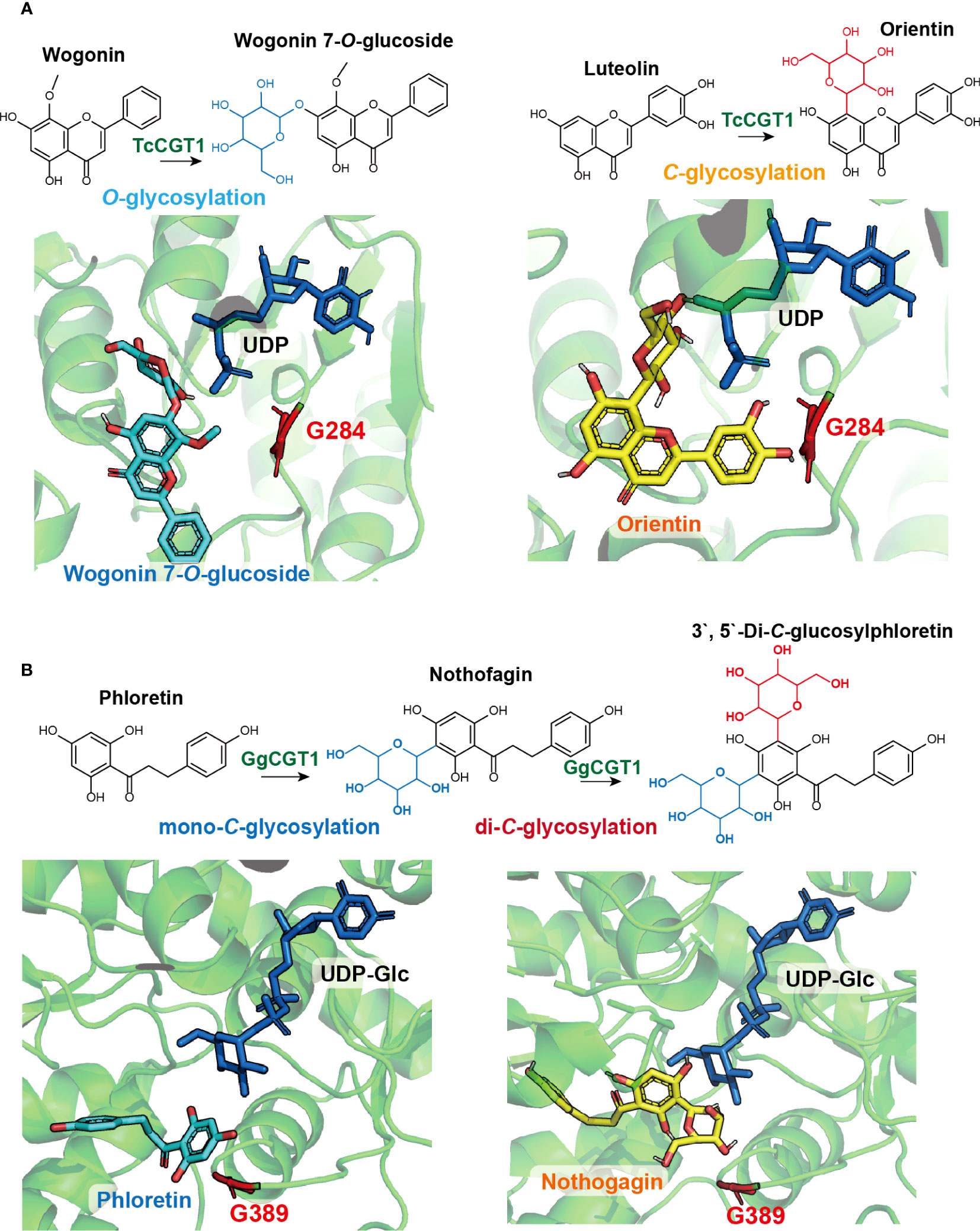
Figure 6 Natural variation and structure-function-guided exploration of key amino acid residues affecting flavonoid C-glycosylation. (A) TcCGT1 from Trollius chinensis is a promiscuous enzyme catalyzing both C- and O-glycosylation using apigenin and wogonin as acceptor substrates, respectively. Protein structural analysis and molecular docking reveal the binding pose of the C-glycosylation product, wogonin 7-O-glucoside (blue), is nearly perpendicular to the C-glycosylation product, orientin. The key residue G284 (red) allows for a more spacious acceptor binding pocket for the C-glycosyl acceptor; (B) GgCGT from Glycyrrhiza glabra can perform consecutive C-glycosylation to produce di-C-glycosylflavonoid, 3’,5’-Di-C-glucosylphloretin. Protein structural analysis and molecular docking suggested that after the first round of C-glycosylation, the acceptor substrate, phloretin (blue), may rotate in the binding pocket, exposing the second C-glycosylation site to UDP-Glc for the subsequent glycosyl transfer. The key residue G389 (red) provides a spacious pocket to accommodate nothofagin (yellow), the mono-C-glycosylation product from the first round of glycosylation.
Among the diverse plant CGTs, some are capable of performing consecutive C-glycosylation to produce di-C-glycosylflavonoids (Chen et al., 2018; Zhang et al., 2020; Dai et al., 2021). Zhang et al. identified a promiscuous GgCGT from Glycyrrhiza glabra, which could catalyze the C-glycosylation of 33 out of 51 tested phenolic compounds as the acceptor substrates, with six compounds converted to di-C-glucosides (Zhang et al., 2020). The crystal structure of GgCGT was solved using the proteins in complex with sugar donor UDP-Glc and acceptor phloretin. The binding poses of UDP-Glc, phloretin, and its mono-C-glycoside nothofagin in the enzyme reaction center were investigated (Zhang et al., 2020). This analysis revealed that the acceptor phloretin possibly rotates in the binding pocket after the first round of C-glycosylation, exposing the second site of C-glycosylation to UDP-Glc for the glycosyl transfer reaction. The binding pose of nothofagin requires a spacious pocket that is determined by the key residue G389. Replacing G389 with bulkier residues Arg or Trp hindered the enzymatic reaction of the second round of C-glycosylation, whereas mutating G389 into Lys almost completely turned the enzyme into a mono-C-glycosyltransferase (Zhang et al., 2020). In a similar study, enzyme promiscuity was leveraged to examine two recently diverged CGTs from Mangifera indica and to uncover key residues contributing to di-C-glycosylation (Chen et al., 2018). Although MiCGT and MiCGTb share 90% amino acid sequence similarity, MiCGTb could transfer two C-glycosyl groups to phloretin, while MiCGT only catalyzes mono-C-glycosylation. A single amino acid – either I152 or E152 – located in the enzyme binding pocket was pinpointed as influencing donor-acceptor substrate permissiveness and determining the transition between mono-C and di-C-glycosylation (Chen et al., 2018). These catalytic residues that switch flavonoid O-, mono-C-, or di-C-glycosylation further expand the toolbox for metabolic engineering of structurally diverse and pharmacologically active flavonoid C-glycosides. The identification of key amino acid residues of GTs contributing to the switch between O- and C-glycosylation, such as those identified in GgCGT and Mangifera indica CGTs, can be applied for enzyme engineering in the production of valuable C-glycosides.
Conclusion
Recent advancements in plant synthetic biology have demonstrated the significant potential of utilizing plants as alternative hosts for metabolic engineering. This approach offers a promising platform to produce plant-derived nutrients or bioactive chemicals that are difficult to produce in microbes but can be more easily manipulated in plants by modifying existing regulatory or biosynthetic enzymes. In particular, flavonoid metabolic engineering in plants has been successfully pursued in two main directions: boosting flavonoid content by manipulating transcription regulatory components and introducing novel flavonoids through engineering enzyme diversity discovered by exploring natural plant variation. By leveraging these two strategies, future plant flavonoid metabolic engineering will provide a variety of approaches to alter the flavor, aroma, and color of plant-based products, making them more appealing to consumers and benefiting the food, beverage, and pharmaceutical industries.
Author contributions
PF, LJ, YG, WZ, and LH contributed to the conceptualization of the review. LJ and PF wrote the first draft of the manuscript. YG, WZ, and LH reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study is supported by Natural Science Foundation of Zhejiang province, China (Grant No. LZ22C150005) and the Starry Night Science Fund of Zhejiang University Shanghai Institute for Advanced Study (SN-ZJU-SIAS-0011).
Acknowledgments
The authors acknowledge all the reviewers for their critical comments and valuable suggestions to improve the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
An, J. P., Li, R., Qu, F. J., You, C. X., Wang, X. F., Hao, Y. J. (2018). R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 96, 562–577. doi: 10.1111/tpj.14050
An, X. H., Tian, Y., Chen, K. Q., Wang, X. F., Hao, Y. J. (2012). The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J. Plant Physiol. 169, 710–717. doi: 10.1016/j.jplph.2012.01.015
Ariyarathne, M. A., Wone, B. W. M. (2022). Overexpression of the Selaginella lepidophylla bHLH transcription factor enhances water-use efficiency, growth, and development in arabidopsis. Plant Sci. 315, 111129. doi: 10.1016/j.plantsci.2021.111129
Arlotta, C., Puglia, G. D., Genovese, C., Toscano, V., Karlova, R., Beekwilder, J., et al. (2020). MYB5-like and bHLH influence flavonoid composition in pomegranate. Plant Sci. 298, 110563. doi: 10.1016/j.plantsci.2020.110563
Atanasov, A. G., Zotchev, S. B., Dirsch, V. M., Supuran, C. T. (2021). Natural products in drug discovery: advances and opportunities. Nat. Rev. Drug Discovery 20, 200–216. doi: 10.1038/s41573-020-00114-z
Baba, S. A., Vishwakarma, R. A., Ashraf, N. (2017). Functional characterization of CsBGlu12, a β-glucosidase from Crocus sativus, provides insights into its role in abiotic stress through accumulation of antioxidant flavonols. J. Biol. Chem. 292, 4700–4713. doi: 10.1074/jbc.M116.762161
Banik, K., Khatoon, E., Harsha, C., Rana, V., Parama, D., Thakur, K. K., et al. (2022). Wogonin and its analogs for the prevention and treatment of cancer: a systematic review. Phytother. Res. 36, 1854–1883. doi: 10.1002/ptr.7386
Borevitz, J. O., Xia, Y., Blount, J., Dixon, R. A., Lamb, C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2393. doi: 10.1105/tpc.12.12.2383
Borsari, C., Luciani, R., Pozzi, C., Poehner, I., Henrich, S., Trande, M., et al. (2016). Profiling of flavonol derivatives for the development of antitrypanosomatidic drugs. J. Med. Chem. 59, 7598–7616. doi: 10.1021/acs.jmedchem.6b00698
Bovy, A., Schijlen, E., Hall, R. D. (2007). Metabolic engineering of flavonoids in tomato (Solanum lycopersicum): the potential for metabolomics. Metabolomics 3, 399–412. doi: 10.1007/s11306-007-0074-2
Bozzo, G. G., Unterlander, N. (2021). In through the out door: biochemical mechanisms affecting flavonoid glycoside catabolism in plants. Plant Sci. 308, 110904. doi: 10.1016/j.plantsci.2021.110904
Brazier-Hicks, M., Evans, K. M., Gershater, M. C., Puschmann, H., Steel, P. G., Edwards, R. (2009). The C-glycosylation of flavonoids in cereals. J. Biol. Chem. 284, 17926–17934. doi: 10.1074/jbc.M109.009258
Brodowska, K. M. (2017). Natural flavonoids: classification, potential role, and application of flavonoid analogues. Eur. J. Biol. Res. 7, 108–123. doi: 10.5281/zenodo.545778
Broun, P. (2004). Transcription factors as tools for metabolic engineering in plants. Curr. Opin. Plant Biol. 7, 202–209. doi: 10.1016/j.pbi.2004.01.013
Cao, Y. Y., Yang, J. F., Liu, T. Y., Su, Z. F., Zhu, F. Y., Chen, M. X., et al. (2017). A phylogenetically informed comparison of GH1 hydrolases between arabidopsis and rice response to stressors. Front. Plant Sci. 8, 350. doi: 10.3389/fpls.2017.00350
Cao, Y., Jia, H., Xing, M., Jin, R., Grierson, D., Gao, Z., et al. (2021). Genome-wide analysis of MYB gene family in Chinese bayberry (Morella rubra) and identification of members regulating flavonoid biosynthesis. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.691384
Castañeda-Ovando, A., de Lourdes Pacheco-Hernández, M., Páez-Hernández, M. E., Rodríguez, J. A., Galán-Vidal, C. A. (2009). Chemical studies of anthocyanins: a review. Food Chem. 113, 859–871. doi: 10.1016/j.foodchem.2008.09.001
Cavallini, E., Zenoni, S., Finezzo, L., Guzzo, F., Zamboni, A., Avesani, L., et al. (2014). Functional diversification of grapevine MYB5a and MYB5b in the control of flavonoid biosynthesis in a petunia anthocyanin regulatory mutant. Plant Cell Physiol. 55, 517–534. doi: 10.1093/pcp/pct190
Chen, L. R., Chen, K. H. (2021). Utilization of isoflavones in soybeans for women with menopausal syndrome: an overview. Int. J. Mol. Sci. 22, 3212. doi: 10.3390/ijms22063212
Chen, D., Fan, S., Chen, R., Xie, K., Yin, S., Sun, L., et al. (2018). Probing and engineering key residues for bis- C -glycosylation and promiscuity of a C -glycosyltransferase. ACS Catal. 8, 4917–4927. doi: 10.1021/acscatal.8b00376
Cho, J. S., Nguyen, V. P., Jeon, H. W., Kim, M. H., Eom, S. H., Lim, Y. J., et al. (2016). Overexpression of PtrMYB119, a R2R3-MYB transcription factor from Populus trichocarpa, promotes anthocyanin production in hybrid poplar. Tree Physiol. 36, 1162–1176. doi: 10.1093/treephys/tpw046
Claus, H. (2004). Laccases: structure, reactions, distribution. Micron 35, 93–96. doi: 10.1016/j.micron.2003.10.029
Copley, S. D. (2015). An evolutionary biochemist’s perspective on promiscuity. Trends Biochem. Sci. 40, 72–78. doi: 10.1016/j.tibs.2014.12.004
Cui, H., Song, M. C., Lee, J. Y., Yoon, Y. J. (2019). Microbial production of O -methylated flavanones from methylated phenylpropanoic acids in engineered Escherichia coli. J. Ind. Microbiol. Biotechnol. 46, 1707–1713. doi: 10.1007/s10295-019-02239-6
Dai, L., Hu, Y., Chen, C., Ma, L., Guo, R. (2021). Flavonoid C -glycosyltransferases: function, evolutionary relationship, catalytic mechanism and protein engineering. ChemBioEng Rev. 8, 15–26. doi: 10.1002/cben.202000009
Deguchi, A., Tatsuzawa, F., Hosokawa, M., Doi, M., Ohno, S. (2016). Quantitative evaluation of the contribution of four major anthocyanins to black flower coloring of dahlia petals. Hortic. J. 85, 340–350. doi: 10.2503/hortj.MI-121
Dias, M. C., Pinto, D. C. G. A., Silva, A. M. S. (2021). Plant flavonoids: chemical characteristics and biological activity. Molecules 26, 5377. doi: 10.3390/molecules26175377
Ding, K., Pei, T., Bai, Z., Jia, Y., Ma, P., Liang, Z. (2017). SmMYB36, a novel R2R3-MYB transcription factor, enhances tanshinone accumulation and decreases phenolic acid content in Salvia miltiorrhiza hairy roots. Sci. Rep. 7, 1–15. doi: 10.1038/s41598-017-04909-w
Dixon, R. A., Sarnala, S. (2020). Proanthocyanidin biosynthesis–a matter of protection. Plant Physiol. 184, 579–591. doi: 10.1104/pp.20.00973
Dixon, R. A., Xie, D., Sharma, S. B. (2005). Proanthocyanidins – a final frontier in flavonoid research? New Phytol. 165, 9–28. doi: 10.1111/j.1469-8137.2004.01217.x
Dong, X., Jiang, Y., Hur, Y. (2019). Genome-wide analysis of glycoside hydrolase family 1 β-glucosidase genes in Brassica rapa and their potential role in pollen development. Int. J. Mol. Sci. 20, 1663. doi: 10.3390/ijms20071663
Dragovic-Uzelac, V., Levaj, B., Mrkic, V., Bursac, D., Boras, M. (2007). The content of polyphenols and carotenoids in three apricot cultivars depending on stage of maturity and geographical region. Food Chem. 102, 966–975. doi: 10.1016/j.foodchem.2006.04.001
Duan, L., Ding, W., Liu, X., Cheng, X., Cai, J., Hua, E., et al. (2017). Biosynthesis and engineering of kaempferol in Saccharomyces cerevisiae. Microb. Cell Factories 16, 165. doi: 10.1186/s12934-017-0774-x
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., Lepiniec, L. (2010). MYB transcription factors in arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Dunstan, M. S., Robinson, C. J., Jervis, A. J., Yan, C., Carbonell, P., Hollywood, K. A., et al. (2020). Engineering Escherichia coli towards de novo production of gatekeeper (2S)-flavanones: naringenin, pinocembrin, eriodictyol and homoeriodictyol. Synth. Biol. 5, ysaa012. doi: 10.1093/synbio/ysaa012
Escaray, F. J., Passeri, V., Perea-García, A., Antonelli, C. J., Damiani, F., Ruiz, O. A., et al. (2017). The R2R3-MYB TT2b and the bHLH TT8 genes are the major regulators of proanthocyanidin biosynthesis in the leaves of Lotus species. Planta 246, 243–261. doi: 10.1007/s00425-017-2696-6
Falcone Ferreyra, M. L., Rius, S. P., Casati, P. (2012). Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front. Plant Sci. 3, 222. doi: 10.3389/fpls.2012.00222
Farrow, S. C., Facchini, P. J. (2014). Functional diversity of 2-oxoglutarate/Fe(II)-dependent dioxygenases in plant metabolism. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00524
Fernández, C., Nieto, O., Rivas, E., Montenegro, G., Fontenla, J. A., Fernández-Mayoralas, A. (2000). Synthesis and biological studies of glycosyl dopamine derivatives as potential antiparkinsonian agents. Carbohydr. Res. 327, 353–365. doi: 10.1016/s0008-6215(00)00073-2
Feyissa, B. A., Arshad, M., Gruber, M. Y., Kohalmi, S. E., Hannoufa, A. (2019). The interplay between miR156/SPL13 and DFR/WD40-1 regulate drought tolerance in alfalfa. BMC Plant Biol. 19, 1–19. doi: 10.1186/s12870-019-2059-5
Fu, R., Martin, C., Zhang, Y. (2018). Next-generation plant metabolic engineering, inspired by an ancient Chinese irrigation systemu. Mol. Plant 11, 47–57. doi: 10.1016/j.molp.2017.09.002
Gao, Y., Liu, J., Chen, Y., Tang, H., Wang, Y., He, Y., et al. (2018). Tomato SlAN11 regulates flavonoid biosynthesis and seed dormancy by interaction with bHLH proteins but not with MYB proteins. Hortic. Res. 5, 27. doi: 10.1038/s41438-018-0032-3
Garg, M., Sharma, N., Sharma, S., Kapoor, P., Kumar, A., Chunduri, V., et al. (2018). Biofortified crops generated by breeding, agronomy, and transgenic approaches are improving lives of millions of people around the world. Front. Nutr. 12. doi: 10.3389/fnut.2018.00012
Gonzalez, A., Zhao, M., Leavitt, J. M., Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Grotewold, E. (2008). Transcription factors for predictive plant metabolic engineering: are we there yet? Curr. Opin. Biotechnol. 19, 138–144. doi: 10.1016/j.copbio.2008.02.002
Guo, Z. (2017). The modification of natural products for medical use. Acta Pharm. Sin. B 7, 119–136. doi: 10.1016/j.apsb.2016.06.003
Gutmann, A., Nidetzky, B. (2012). Switching between O - and C -glycosyltransferase through exchange of active-site motifs. Angew. Chem. Int. Ed. 51, 12879–12883. doi: 10.1002/anie.201206141
Hao, B., Caulfield, J. C., Hamilton, M. L., Pickett, J. A., Midega, C. A. O., Khan, Z. R., et al. (2016). Biosynthesis of natural and novel C-glycosylflavones utilising recombinant Oryza sativa c-glycosyltransferase (OsCGT) and Desmodium incanum root proteins. Phytochemistry 125, 73–87. doi: 10.1016/j.phytochem.2016.02.013
He, J., Zhao, P., Hu, Z., Liu, S., Kuang, Y., Zhang, M., et al. (2019). Molecular and structural characterization of a promiscuous C -glycosyltransferase from Trollius chinensis. Angew. Chem. Int. Ed. 58, 11513–11520. doi: 10.1002/anie.201905505
Hichri, I., Barrieu, F., Bogs, J., Kappel, C., Delrot, S., Lauvergeat, V. (2011). Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 62, 2465–2483. doi: 10.1093/jxb/erq442
Himeji, M., Ohtsuki, T., Fukazawa, H., Tanaka, M., Yazaki, S., Ui, S., et al. (2007). Difference of growth-inhibitory effect of Scutellaria baicalensis -producing flavonoid wogonin among human cancer cells and normal diploid cell. Cancer Lett. 245, 269–274. doi: 10.1016/j.canlet.2006.01.011
Hirade, Y., Kotoku, N., Terasaka, K., Saijo-Hamano, Y., Fukumoto, A., Mizukami, H. (2015). Identification and functional analysis of 2-hydroxyflavanone C -glucosyltransferase in soybean (Glycine max). FEBS Lett. 589, 1778–1786. doi: 10.1016/j.febslet.2015.05.010
Holland, T. M., Agarwal, P., Wang, Y., Dhana, K., Leurgans, S. E., Shea, K., et al. (2023). Association of dietary intake of flavonols with changes in global cognition and several cognitive abilities. Neurology 100, e694–e702. doi: 10.1212/WNL.0000000000201541
Huang, W., Khaldun, A. B. M., Lv, H., Du, L., Zhang, C., Wang, Y. (2016). Isolation and functional characterization of a R2R3-MYB regulator of the anthocyanin biosynthetic pathway from Epimedium sagittatum. Plant Cell Rep. 35, 883–894. doi: 10.1007/s00299-015-1929-z
Huang, W., Lv, H., Wang, Y. (2017). Functional characterization of a novel R2R3-MYB transcription factor modulating the flavonoid biosynthetic pathway from Epimedium sagittatum. Front. Plant Sci. 8, 1274. doi: 10.3389/fpls.2017.01274
Huang, J. X., Qu, L., Yang, J., Yin, H., Gu, H. (2004). A preliminary study on the origin and evolution of chalcone synthase (CHS) gene in angiosperms. Acta Bot. Sin. 46, 10–19. Available at: https://www.jipb.net/EN/Y2004/V46/I1/10.
Huang, Y. F., Vialet, S., Guiraud, J. L., Torregrosa, L., Bertrand, Y., Cheynier, V., et al. (2014). A negative MYB regulator of proanthocyanidin accumulation, identified through expression quantitative locus mapping in the grape berry. New Phytol. 201, 795–809. doi: 10.1111/nph.12557
Iwashina, T. (2003). Flavonoid function and activity to plants and other organisms. Biol. Sci. Space Uchū Seibutsu Kagaku 17, 24–44. doi: 10.2187/bss.17.24
Iwashina, T. (2015). Contribution to flower colors of flavonoids including anthocyanins: a review. Nat. Prod. Commun. 10, 529–544. doi: 10.1177/1934578X1501000335
Iwashina, T., Mizuno, T. (2020). Flavonoids and xanthones from the genus Iris: phytochemistry, relationships with flower colors and taxonomy, and activities and function. Nat. Prod. Commun. 15, 1–15. doi: 10.1177/1934578X20937151
Jia, Y., Li, B., Zhang, Y., Zhang, X., Xu, Y., Li, C. (2019). Evolutionary dynamic analyses on monocot flavonoid 3′-hydroxylase gene family reveal evidence of plant-environment interaction. BMC Plant Biol. 19, 1–16. doi: 10.1186/s12870-019-1947-z
Jia, N., Wang, J. J., Liu, J., Jiang, J., Sun, J., Yan, P., et al. (2021). DcTT8, a bHLH transcription factor, regulates anthocyanin biosynthesis in Dendrobium candidum. Plant Physiol. Biochem. 162, 603–612. doi: 10.1016/j.plaphy.2021.03.006
Jiang, L., Fan, Z., Tong, R., Yin, H., Li, J., Zhou, X. (2021). Flavonoid 3′-hydroxylase of Camellia nitidissima chi. promotes the synthesis of polyphenols better than flavonoids. Mol. Biol. Rep. 48, 3903–3912. doi: 10.1007/s11033-021-06345-6
Jiang, L., Fan, Z., Tong, R., Zhou, X., Li, J., Yin, H. (2020). Functional diversification of the dihydroflavonol 4-reductase from Camellia nitidissima chi. in the control of polyphenol biosynthesis. Genes 11, 1341. doi: 10.3390/genes11111341
Kayani, S. I., Shen, Q., ur Rahman, S., Fu, X., Li, Y., Wang, C., et al. (2021). Transcriptional regulation of flavonoid biosynthesis in Artemisia annua by AaYABBY5. Hortic. Res. 8, 257. doi: 10.1038/s41438-021-00693-x
Kim, B.-G., Joe, E. J., Ahn, J.-H. (2010). Molecular characterization of flavonol synthase from poplar and its application to the synthesis of 3-O-methylkaempferol. Biotechnol. Lett. 32, 579–584. doi: 10.1007/s10529-009-0188-x
King, J. R., Edgar, S., Qiao, K., Stephanopoulos, G. (2016). Accessing nature’s diversity through metabolic engineering and synthetic biology. F1000Research 5, 397. doi: 10.12688/f1000research.7311.1
Koirala, N. (2016). Methylation of flavonoids: chemical structures, bioactivities, progress and perspectives for biotechnological production. Enzyme Microb. Technol. 86, 103–116. doi: 10.1016/j.enzmictec.2016.02.003
Kong, J.-M., Chia, L.-S., Goh, N.-K., Chia, T.-F., Brouillard, R. (2003). Analysis and biological activities of anthocyanins. Phytochemistry 64, 923–933. doi: 10.1016/S0031-9422(03)00438-2
Kren, V., Martinkova, L. (2001). Glycosides in medicine: “The role of glycosidic residue in biological activity”. Curr. Med. Chem. 8, 1303–1328. doi: 10.2174/0929867013372193
Langenhan, J. M., Griffith, B. R., Thorson, J. S. (2005). Neoglycorandomization and chemoenzymatic glycorandomization: two complementary tools for natural product diversification. J. Nat. Prod. 68, 1696–1711. doi: 10.1021/np0502084
Lau, W., Fischbach, M. A., Osbourn, A., Sattely, E. S. (2014). Key applications of plant metabolic engineering. PloS Biol. 12, e1001879. doi: 10.1371/journal.pbio.1001879
Lautié, E., Russo, O., Ducrot, P., Boutin, J. A. (2020). Unraveling plant natural chemical diversity for drug discovery purposes. Front. Pharmacol. 11, 37. doi: 10.3389/fphar.2020.00397
Li, S. (2014). Transcriptional control of flavonoid biosynthesis: fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal. Behav. 9, e27522. doi: 10.4161/psb.27522
Li, B., Fan, R., Guo, S., Wang, P., Zhu, X., Fan, Y., et al. (2019a). The arabidopsis MYB transcription factor, MYB111 modulates salt responses by regulating flavonoid biosynthesis. Environ. Exp. Bot. 166, 103807. doi: 10.1016/j.envexpbot.2019.103807
Li, Y., Li, P., Zhang, L., Shu, J., Court, M. H., Sun, Z., et al. (2022). Genome-wide analysis of the apple family 1 glycosyltransferases identified a flavonoid-modifying UGT, MdUGT83L3, which is targeted by MdMYB88 and contributes to stress adaptation. Plant Sci. 321, 111314. doi: 10.1016/j.plantsci.2022.111314
Li, D. D., Ni, R., Wang, P. P., Zhang, X. S., Wang, P. Y., Zhu, T. T., et al. (2020). Molecular basis for chemical evolution of flavones to flavonols and anthocyanins in land plants. Plant Physiol. 184, 1731–1743. doi: 10.1104/pp.20.01185
Li, Z., Peng, R., Yao, Q. (2021). SlMYB14 promotes flavonoids accumulation and confers higher tolerance to 2,4,6-trichlorophenol in tomato. Plant Sci. 303, 110796. doi: 10.1016/j.plantsci.2020.110796
Li, J., Tian, C., Xia, Y., Mutanda, I., Wang, K., Wang, Y. (2019b). Production of plant-specific flavones baicalein and scutellarein in an engineered E. coli from available phenylalanine and tyrosine. Metab. Eng. 52, 124–133. doi: 10.1016/j.ymben.2018.11.008
Li, Q., Zhao, P., Li, J., Zhang, C., Wang, L., Ren, Z. (2014). Genome-wide analysis of the WD-repeat protein family in cucumber and arabidopsis. Mol. Genet. Genomics 289, 103–124. doi: 10.1007/s00438-013-0789-x
Lim, S.-H., Kim, D.-H., Jung, J.-A., Lee, J.-Y. (2021). Alternative splicing of the basic helix–loop–helix transcription factor gene CmbHLH2 affects anthocyanin biosynthesis in ray florets of Chrysanthemum (Chrysanthemum morifolium). Front. Plant Sci. 12, 669315. doi: 10.3389/fpls.2021.669315
Lin, M., Zhang, J., Chen, X. (2018). Bioactive flavonoids in Moringa oleifera and their health-promoting properties. J. Funct. Foods 47, 469–479. doi: 10.1016/j.jff.2018.06.011
Liu, Y., Hou, H., Jiang, X., Wang, P., Dai, X., Chen, W., et al. (2018b). A WD40 repeat protein from Camellia sinensis regulates anthocyanin and proanthocyanidin accumulation through the formation of MYB–bHLH–WD40 ternary complexes. Int. J. Mol. Sci. 19, 1686. doi: 10.3390/ijms19061686
Liu, Y., Lin-Wang, K., Espley, R. V., Wang, L., Yang, H., Yu, B., et al. (2016b). Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 67, 2159–2176. doi: 10.1093/jxb/erw014
Liu, Y., Shi, Z., Maximova, S., Payne, M. J., Guiltinan, M. J. (2013). Proanthocyanidin synthesis in Theobroma cacao: genes encoding anthocyanidin synthase, anthocyanidin reductase, and leucoanthocyanidin reductase. BMC Plant Biol. 13, 202. doi: 10.1186/1471-2229-13-202
Liu, C., Wang, X., Shulaev, V., Dixon, R. A. (2016a). A role for leucoanthocyanidin reductase in the extension of proanthocyanidins. Nat. Plants 2, 16182. doi: 10.1038/nplants.2016.182
Liu, J., Wang, Y., Zhang, M., Wang, Y., Deng, X., Sun, H., et al. (2022). Color fading in lotus (Nelumbo nucifera) petals is manipulated both by anthocyanin biosynthesis reduction and active degradation. Plant Physiol. Biochem. 179, 100–107. doi: 10.1016/j.plaphy.2022.03.021
Liu, X., Yang, W., Mu, B., Li, S., Li, Y., Zhou, X., et al. (2018a). Engineering of ‘Purple embryo maize’with a multigene expression system derived from a bidirectional promoter and self-cleaving 2A peptides. Plant Biotechnol. J. 16, 1107. doi: 10.1111/pbi.12883
Lloyd, A., Brockman, A., Aguirre, L., Campbell, A., Bean, A., Cantero, A., et al. (2017). Advances in the MYB-bHLH-WD repeat (MBW) pigment regulatory model: addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 58, 1431–1441. doi: 10.1093/pcp/pcx075
López, J. G.-E. (2019). Flavonoids in health and disease. Curr. Med. Chem. 26, 6972–6975. doi: 10.2174/092986732639191213095405
Lukačin, R., Wellmann, F., Britsch, L., Martens, S., Matern, U. (2003). Flavonol synthase from Citrus unshiu is a bifunctional dioxygenase. Phytochemistry 62, 287–292. doi: 10.1016/S0031-9422(02)00567-8
Lv, Y., Xu, S., Lyu, Y., Zhou, S., Du, G., Chen, J., et al. (2019). Engineering enzymatic cascades for the efficient biotransformation of eugenol and taxifolin to silybin and isosilybin. Green Chem. 21, 1660–1667. doi: 10.1039/C8GC03728K
Ma, D., Reichelt, M., Yoshida, K., Gershenzon, J., Constabel, C. P. (2018). Two R2R3-MYB proteins are broad repressors of flavonoid and phenylpropanoid metabolism in poplar. Plant J. 96, 949–965. doi: 10.1111/tpj.14081
Malacarne, G., Coller, E., Czemmel, S., Vrhovsek, U., Engelen, K., Goremykin, V., et al. (2016). The grapevine VvibZIPC22 transcription factor is involved in the regulation of flavonoid biosynthesis. J. Exp. Bot. 67, 3509–3522. doi: 10.1093/jxb/erw181
Marsafari, M., Samizadeh, H., Rabiei, B., Mehrabi, A. A., Koffas, M., Xu, P. (2020). Biotechnological production of flavonoids: an update on plant metabolic engineering, microbial host selection, and genetically encoded biosensors. Biotechnol. J. 15, 1–13. doi: 10.1002/biot.201900432
Martin, C. (2013). The interface between plant metabolic engineering and human health. Curr. Opin. Biotechnol. 24, 344–353. doi: 10.1016/j.copbio.2012.11.005
Matsui, K., Oshima, Y., Mitsuda, N., Sakamoto, S., Nishiba, Y., Walker, A. R., et al. (2018). Buckwheat R2R3 MYB transcription factor FeMYBF1 regulates flavonol biosynthesis. Plant Sci. 274, 466–475. doi: 10.1016/j.plantsci.2018.06.025
Minic, Z. (2008). Physiological roles of plant glycoside hydrolases. Planta. 227, 723–740. doi: 10.1007/s00425-007-0668-y
Moore, B. (2004). Bifunctional and moonlighting enzymes: lighting the way to regulatory control. Trends Plant Sci. 9, 221–228. doi: 10.1016/j.tplants.2004.03.005
Nabavi, S. M., Šamec, D., Tomczyk, M., Milella, L., Russo, D., Habtemariam, S., et al. (2020). Flavonoid biosynthetic pathways in plants: versatile targets for metabolic engineering. Biotechnol. Adv. 38, 107316. doi: 10.1016/j.biotechadv.2018.11.005
Nagatomo, Y., Usui, S., Ito, T., Kato, A., Shimosaka, M., Taguchi, G. (2014). Purification, molecular cloning and functional characterization of flavonoid C -glucosyltransferases from Fagopyrum esculentum m. (buckwheat) cotyledon. Plant J. 80, 437–448. doi: 10.1111/tpj.12645
Naik, J., Rajput, R., Pucker, B., Stracke, R., Pandey, A. (2021). The R2R3-MYB transcription factor MtMYB134 orchestrates flavonol biosynthesis in Medicago truncatula. Plant Mol. Biol. 106, 157–172. doi: 10.1007/s11103-021-01135-x
Nakatsuka, T., Yamada, E., Saito, M., Fujita, K., Nishihara, M. (2013). Heterologous expression of gentian MYB1R transcription factors suppresses anthocyanin pigmentation in tobacco flowers. Plant Cell Rep. 32, 1925–1937. doi: 10.1007/s00299-013-1504-4
Neer, E. J., Schmidt, C. J., Nambudripad, R., Smith, T. F. (1994). The ancient regulatory-protein family of WD-repeat proteins. Nature 371, 297–300. doi: 10.1038/371297a0
Nesi, N., Debeaujon, I., Jond, C., Pelletier, G., Caboche, M., Lepiniec, L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in arabidopsis siliques. Plant Cell 12, 1863–1878. doi: 10.1105/tpc.12.10.1863
Ni, R., Niu, M., Fu, J., Tan, H., Zhu, T., Zhang, J., et al. (2022). Molecular and structural characterization of a promiscuous chalcone synthase from the fern species Stenoloma chusanum. J. Integr. Plant Biol. 64, 1935–1951. doi: 10.1111/jipb.13335
Nishihara, M., Nakatsuka, T. (2011). Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotechnol. Lett. 33, 433–441. doi: 10.1007/s10529-010-0461-z
Oglesby, L., Ananga, A., Obuya, J., Ochieng, J., Cebert, E., Tsolova, V. (2016). Anthocyanin accumulation in muscadine berry skins is influenced by the expression of the MYB transcription factors, MybA1, and MYBCS1. Antioxidants 5, 1–14. doi: 10.3390/antiox5040035
Ogo, Y., Ozawa, K., Ishimaru, T., Murayama, T., Takaiwa, F. (2013). Transgenic rice seed synthesizing diverse flavonoids at high levels: a new platform for flavonoid production with associated health benefits. Plant Biotechnol. J. 11, 734–746. doi: 10.1111/pbi.12064
Oo, A. M., Mat Nor, M. N., Lwin, O. M., Simbak, N. (2022). Flavonol quercetin: immunomodulatory and anticancer properties. Asian J. Med. Biomed. 6, 17–31. doi: 10.37231/ajmb.2022.6.1.452
Oualid, O., Silva, M. S. (2012). Advances in C-glycosylflavonoid research. Curr. Org. Chem. 16, 859–896. doi: 10.2174/138527212800194791
Pandey, A., Misra, P., Khan, M. P., Swarnkar, G., Tewari, M. C., Bhambhani, S., et al. (2014). Co-Expression of arabidopsis transcription factor, AtMYB12, and soybean isoflavone synthase, GmIFS1, genes in tobacco leads to enhanced biosynthesis of isoflavones and flavonols resulting in osteoprotective activity. Plant Biotechnol. J. 12, 69–80. doi: 10.1111/pbi.12118
Pandey, A., Misra, P., Trivedi, P. K. (2015). Constitutive expression of arabidopsis MYB transcription factor, AtMYB11, in tobacco modulates flavonoid biosynthesis in favor of flavonol accumulation. Plant Cell Rep. 34, 1515–1528. doi: 10.1007/s00299-015-1803-z
Pandey, R. P., Parajuli, P., Koffas, M. A. G., Sohng, J. K. (2016). Microbial production of natural and non-natural flavonoids: pathway engineering, directed evolution and systems/synthetic biology. Biotechnol. Adv. 34, 634–662. doi: 10.1016/j.biotechadv.2016.02.012
Pang, Y., Abeysinghe, I. S. B., He, J., He, X., Huhman, D., Mewan, K. M., et al. (2013). Functional characterization of proanthocyanidin pathway enzymes from tea and their application for metabolic engineering. Plant Physiol. 161, 1103–1116. doi: 10.1104/pp.112.212050
Park, S., Kim, D.-H., Park, B.-R., Lee, J.-Y., Lim, S.-H. (2019). Molecular and functional characterization of Oryza sativa flavonol synthase (OsFLS), a bifunctional dioxygenase. J. Agric. Food Chem. 67, 7399–7409. doi: 10.1021/acs.jafc.9b02142
Pasam, V. R., Kiran, S., R., P., B., P. (2017). Flavonoid : a review on naringenin. J. Pharmacogn. Phytochem. 6, 2778–2783. Available at: https://www.phytojournal.com/archives/2017.v6.i5.2042/flavonoid-a-review-on-naringenin.
Pei, J., Sun, Q., Gu, N., Zhao, L., Fang, X., Tang, F., et al. (2020). Production of isoorientin and isovitexin from luteolin and apigenin using coupled catalysis of glycosyltransferase and sucrose synthase. Appl. Biochem. Biotechnol. 190, 601–615. doi: 10.1007/s12010-019-03112-z
Pei, T., Yan, M., Huang, Y., Wei, Y., Martin, C., Zhao, Q. (2022). Specific flavonoids and their biosynthetic pathway in Scutellaria baicalensis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.866282
Perez-Vizcaino, F., Fraga, C. G. (2018). Research trends in flavonoids and health. Arch. Biochem. Biophys. 646, 107–112. doi: 10.1016/j.abb.2018.03.022
Petroni, K., Tonelli, C. (2011). Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 181, 219–229. doi: 10.1016/j.plantsci.2011.05.009
Pourcel, L., Routaboul, J., Cheynier, V., Lepiniec, L., Debeaujon, I. (2007). Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends Plant Sci. 12, 29–36. doi: 10.1016/j.tplants.2006.11.006
Pourcel, L., Routaboul, J.-M., Kerhoas, L., Caboche, M., Lepiniec, L., Debeaujon, I. (2005). TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17, 2966–2980. doi: 10.1105/tpc.105.035154
Premathilake, A. T., Ni, J., Bai, S., Tao, R., Ahmad, M., Teng, Y. (2020). R2R3-MYB transcription factor PpMYB17 positively regulates flavonoid biosynthesis in pear fruit. Planta 252, 1–16. doi: 10.1007/s00425-020-03473-4
Pyne, M. E., Narcross, L., Martin, V. J. J. (2019). Engineering plant secondary metabolism in microbial systems. Plant Physiol. 179, 844–861. doi: 10.1104/pp.18.01291
Ramalingam, M., Kim, H., Lee, Y., Lee, Y.-I. (2018). Phytochemical and pharmacological role of liquiritigenin and isoliquiritigenin from radix glycyrrhizae in human health and disease models. Front. Aging Neurosci. 10, 348. doi: 10.3389/fnagi.2018.00348
Ravishankar, D., Rajora, A. K., Greco, F., Osborn, H. M. I. (2013). Flavonoids as prospective compounds for anti-cancer therapy. Int. J. Biochem. Cell Biol. 45, 2821–2831. doi: 10.1016/j.biocel.2013.10.004
Rochfort, S., Panozzo, J. (2007). Phytochemicals for health, the role of pulses. J. Agric. Food Chem. 55, 7981–7994. doi: 10.1021/jf071704w
Roepke, J., Bozzo, G. G. (2015). Arabidopsis thaliana β-glucosidase BGLU15 attacks flavonol 3-O-β-glucoside-7-O-α-rhamnosides. Phytochemistry 109, 14–24. doi: 10.1016/j.phytochem.2014.10.028
Roepke, J., Gordon, H. O. W., Neil, K. J. A., Gidda, S., Mullen, R. T., Freixas Coutin, J. A., et al. (2017). An apoplastic β-glucosidase is essential for the degradation of flavonol 3-O-β-glucoside-7-O-α-rhamnosides in arabidopsis. Plant Cell Physiol. 58, 1030–1047. doi: 10.1093/pcp/pcx050
Salehi, B., Machin, L., Monzote, L., Sharifi-Rad, J., Ezzat, S. M., Salem, M. A., et al. (2020). Therapeutic potential of quercetin: new insights and perspectives for human health. ACS Omega 5, 11849–11872. doi: 10.1021/acsomega.0c01818
Salez, M. A., González, J. M., Bernacchini, J. C., Rodriguez, M. V., Falcone, M. L., Paula, F., et al. (2022). Sb TT8, a new sorghum bHLH transcription factor that rescues brown seed coat phenotype in Arabidopsis tt8 mutant plants. J. Plant Biol. 65, 473–485. doi: 10.1007/s12374-022-09365-2
Samira, R., Li, B., Kliebenstein, D., Li, C., Davis, E., Gillikin, J. W., et al. (2018). The bHLH transcription factor ILR3 modulates multiple stress responses in arabidopsis. Plant Mol. Biol. 97, 297–309. doi: 10.1007/s11103-018-0735-8
Sasaki, N., Nishizaki, Y., Yamada, E., Tatsuzawa, F., Nakatsuka, T., Takahashi, H., et al. (2015). Identification of the glucosyltransferase that mediates direct flavone C -glucosylation in Gentiana triflora. FEBS Lett. 589, 182–187. doi: 10.1016/j.febslet.2014.11.045
Schilbert, H. M., Schöne, M., Baier, T., Busche, M., Viehöver, P., Weisshaar, B., et al. (2021). Characterization of the Brassica napus flavonol synthase gene family reveals bifunctional flavonol synthases. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.733762
Schwinn, K. E., Ngo, H., Kenel, F., Brummell, D. A., Albert, N. W., McCallum, J. A., et al. (2016). The onion (Allium cepa l.) R2R3-MYB gene MYB1 regulates anthocyanin biosynthesis. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01865
Seelig, B. (2017). Multifunctional enzymes from reduced genomes – model proteins for simple primordial metabolism? Mol. Microbiol. 105, 505–507. doi: 10.1111/mmi.13742
Shan, X., Li, Y., Yang, S., Gao, R., Zhou, L., Bao, T., et al. (2019). A functional homologue of Arabidopsis TTG1 from Freesia interacts with bHLH proteins to regulate anthocyanin and proanthocyanidin biosynthesis in both Freesia hybrida and Arabidopsis thaliana. Plant Physiol. Biochem. 141, 60–72. doi: 10.1016/j.plaphy.2019.05.015
Shang, Y., Huang, S. (2020). Engineering plant cytochrome P450s for enhanced synthesis of natural products: past achievements and future perspectives. Plant Commun. 1, 100012. doi: 10.1016/j.xplc.2019.100012
Shen, Y., Sun, T., Pan, Q., Anupol, N., Chen, H., Shi, J., et al. (2019). RrMYB5- and RrMYB10-regulated flavonoid biosynthesis plays a pivotal role in feedback loop responding to wounding and oxidation in Rosa rugosa. Plant Biotechnol. J. 17, 2078–2095. doi: 10.1111/pbi.13123
Sheng, H., Sun, X., Yan, Y., Yuan, Q., Wang, J., Shen, X. (2020). Metabolic engineering of microorganisms for the production of flavonoids. Front. Bioeng. Biotechnol. 8, 589069. doi: 10.3389/fbioe.2020.589069
Siddiq, M. A., Hochberg, G. K., Thornton, J. W. (2017). Evolution of protein specificity: insights from ancestral protein reconstruction. Curr. Opin. Struct. Biol. 47, 113–122. doi: 10.1016/j.sbi.2017.07.003
Singh, S., Meena, A., Luqman, S. (2021). Baicalin mediated regulation of key signaling pathways in cancer. Pharmacol. Res. 164, 105387. doi: 10.1016/j.phrs.2020.105387
Slámová, K., Kapešová, J., Valentová, K. (2018). “Sweet flavonoids”: glycosidase-catalyzed modifications. Int. J. Mol. Sci. 19, 2126. doi: 10.3390/ijms19072126
Sokół-Łętowska, A., Oszmiański, J., Wojdyło, A. (2007). Antioxidant activity of the phenolic compounds of hawthorn, pine and skullcap. Food Chem. 103, 853–859. doi: 10.1016/j.foodchem.2006.09.036
Song, Z., Luo, Y., Wang, W., Fan, N., Wang, D., Yang, C., et al. (2020). NtMYB12 positively regulates flavonol biosynthesis and enhances tolerance to low pi stress in Nicotiana tabacum. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01683
Stavenga, D. G., Leertouwer, H. L., Dudek, B., van der Kooi, C. J. (2021). Coloration of flowers by flavonoids and consequences of pH dependent absorption. Front. Plant Sci. 11, 600124. doi: 10.3389/fpls.2020.600124
Stirnimann, C. U., Petsalaki, E., Russell, R. B., Müller, C. W. (2010). WD40 proteins propel cellular networks. Trends Biochem. Sci. 35, 565–574. doi: 10.1016/j.tibs.2010.04.003
Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., et al. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Stracke, R., Jahns, O., Keck, M., Tohge, T., Niehaus, K., Fernie, A. R., et al. (2010). Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12-and MYB111-independent flavonol glycoside accumulation. New Phytol. 188, 985–1000. doi: 10.1111/j.1469-8137.2010.03421.x
Su, X., Shen, G., Di, S., Dixon, R. A., Pang, Y. (2017). Characterization of UGT716A1 as a multi-substrate UDP: flavonoid glucosyltransferase gene in Ginkgo biloba. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02085
Sultana, B., Anwar, F. (2008). Flavonols (kaempeferol, quercetin, myricetin) contents of selected fruits, vegetables and medicinal plants. Food Chem. 108, 879–884. doi: 10.1016/j.foodchem.2007.11.053
Sun, C., Deng, L., Du, M., Zhao, J., Chen, Q., Huang, T., et al. (2020). A transcriptional network promotes anthocyanin biosynthesis in tomato flesh. Mol. Plant 13, 42–58. doi: 10.1016/j.molp.2019.10.010
Tanner, G. J., Francki, K. T., Abrahams, S., Watson, J. M., Larkin, P. J., Ashton, A. R. (2003). Proanthocyanidin biosynthesis in plants. J. Biol. Chem. 278, 31647–31656. doi: 10.1074/jbc.M302783200
Terahara, N. (2015). Flavonoids in foods: a review. Nat. Prod. Commun. 10, 1934578X1501000334. doi: 10.1177/1934578X1501000334
Tian, L., Hu, Y., Chen, X.-Y. (2017). Advancing human health through exploration of plant metabolism and reaping the renefits of edible medicinal plants. Mol. Plant 10, 533–536. doi: 10.1016/j.molp.2017.01.009
Tian, J., Peng, Z., Zhang, J., Song, T., Wan, H., Zhang, M., et al. (2015). McMYB10 regulates coloration via activating McF3′H and later structural genes in ever-red leaf crabapple. Plant Biotechnol. J. 13, 948–961. doi: 10.1111/pbi.12331
Tohge, T., de Souza, L. P., Fernie, A. R. (2017). Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 68, 4013–4028. doi: 10.1093/jxb/erx177
Tungmunnithum, D., Thongboonyou, A., Pholboon, A., Yangsabai, A. (2018). Flavonoids and other phenolic compounds from medicinal plants for pharmaceutical and medical aspects: an overview. Medicines 5, 93. doi: 10.3390/medicines5030093
Ullah, A., Munir, S., Badshah, S. L., Khan, N., Ghani, L., Poulson, B. G., et al. (2020). Important flavonoids and their role as a therapeutic agent. Molecules 25, 5243. doi: 10.3390/molecules25225243
Verdier, J., Zhao, J., Torres-Jerez, I., Ge, S., Liu, C., He, X., et al. (2012). MtPAR MYB transcription factor acts as an on switch for proanthocyanidin biosynthesis in Medicago truncatula. Proc. Natl. Acad. Sci. U. S. A. 109, 1766–1771. doi: 10.1073/pnas.1120916109
Wan, S., Li, C., Ma, X., Luo, K. (2017). PtrMYB57 contributes to the negative regulation of anthocyanin and proanthocyanidin biosynthesis in poplar. Plant Cell Rep. 36, 1263–1276. doi: 10.1007/s00299-017-2151-y
Wang, S., Chu, Z., Jia, R., Dan, F., Shen, X., Li, Y., et al. (2018b). SlMYB12 regulates flavonol synthesis in three different cherry tomato varieties. Sci. Rep. 8, 1–12. doi: 10.1038/s41598-018-19214-3
Wang, W., Hu, S., Yang, J., Zhang, C., Zhang, T., Wang, D., et al. (2022). A novel R2R3-MYB transcription factor SbMYB12 positively regulates baicalin biosynthesis in Scutellaria baicalensis georgi. Int. J. Mol. Sci. 23, 10–23. doi: 10.3390/ijms232415452
Wang, T., Li, Q., Bi, K. (2018c). Bioactive flavonoids in medicinal plants: structure, activity and biological fate. Asian J. Pharm. Sci. 13, 12–23. doi: 10.1016/j.ajps.2017.08.004
Wang, J., Li, G., Li, C., Zhang, C., Cui, L., Ai, G., et al. (2021). NF-y plays essential roles in flavonoid biosynthesis by modulating histone modifications in tomato. New Phytol. 229, 3237–3252. doi: 10.1111/nph.17112
Wang, X., Li, C., Zhou, C., Li, J., Zhang, Y. (2017b). Molecular characterization of the C -glucosylation for puerarin biosynthesis in Pueraria lobata. Plant J. 90, 535–546. doi: 10.1111/tpj.13510
Wang, L., Lu, W., Ran, L., Dou, L., Yao, S., Hu, J., et al. (2019a). R2R3-MYB transcription factor MYB6 promotes anthocyanin and proanthocyanidin biosynthesis but inhibits secondary cell wall formation in Populus tomentosa. Plant J. 99, 733–751. doi: 10.1111/tpj.14364
Wang, L., Tang, W., Hu, Y., Zhang, Y., Sun, J., Guo, X., et al. (2019b). A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. hongyang. Plant J. 99, 359–378. doi: 10.1111/tpj.14330
Wang, W., Wang, Y., Li, H., Liu, Z., Cui, X., Zhuang, J. (2018d). Two MYB transcription factors (CsMYB2 and CsMYB26) are involved in flavonoid biosynthesis in tea plant [Camellia sinensis (L.) o. kuntze]. BMC Plant Biol. 18, 1–15. doi: 10.1186/s12870-018-1502-3
Wang, N., Xu, H., Jiang, S., Zhang, Z., Lu, N., Qiu, H., et al. (2017a). MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 90, 276–292. doi: 10.1111/tpj.13487
Wang, T. K., Xu, S., Li, S., Zhang, Y. (2020). Proanthocyanidins should be a candidate in the treatment of cancer, cardiovascular diseases and lipid metabolic disorder. Molecules 25, 5971. doi: 10.3390/molecules25245971
Wang, P., Zhang, L., Jiang, X., Dai, X., Xu, L., Li, T., et al. (2018a). Evolutionary and functional characterization of leucoanthocyanidin reductases from Camellia sinensis. Planta 247, 139–154. doi: 10.1007/s00425-017-2771-z
Wei, Q., Zhang, F., Sun, F., Luo, Q., Wang, R., Hu, R., et al. (2017). A wheat MYB transcriptional repressor TaMyb1D regulates phenylpropanoid metabolism and enhances tolerance to drought and oxidative stresses in transgenic tobacco plants. Plant Sci. 265, 112–123. doi: 10.1016/j.plantsci.2017.09.020
Wen, X., Walle, T. (2006). Methylated flavonoids have greatly improved intestinal absorption and metabolic stability. Drug Metab. Dispos. Biol. Fate Chem. 34, 1786–1792. doi: 10.1124/dmd.106.011122
Williams, G. J., Zhang, C., Thorson, J. S. (2007). Expanding the promiscuity of a natural-product glycosyltransferase by directed evolution. Nat. Chem. Biol. 3, 657–662. doi: 10.1038/nchembio.2007.28
Winkel-shirley, B. (2001). Flavonoid biosynthesis. a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. doi: 10.1104/pp.126.2.485
Winkel-Shirley, B. (2002). Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 5, 218–223. doi: 10.1016/S1369-5266(02)00256-X
Wu, Y. F., Zhao, Y., Liu, X. Y., Gao, S., Cheng, A. X., Lou, H. X. (2018). A bHLH transcription factor regulates bisbibenzyl biosynthesis in the liverwort Plagiochasma appendiculatum. Plant Cell Physiol. 59, 1187–1199. doi: 10.1093/pcp/pcy053
Xu, W., Dubos, C., Lepiniec, L. (2015). Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 20, 176–185. doi: 10.1016/j.tplants.2014.12.001
Xu, W., Grain, D., Bobet, S., Le Gourrierec, J., Thévenin, J., Kelemen, Z., et al. (2014). Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in arabidopsis seed. New Phytol. 202, 132–144. doi: 10.1111/nph.12620
Xu, F., Li, L., Zhang, W., Cheng, H., Sun, N., Cheng, S., et al. (2012). Isolation, characterization, and function analysis of a flavonol synthase gene from Ginkgo biloba. Mol. Biol. Rep. 39, 2285–2296. doi: 10.1007/s11033-011-0978-9
Yamagishi, M. (2022). High temperature enhances anthocyanin coloration in Asiatic hybrid lily flowers via upregulation of the MYB12 positive regulator. Hortic. Plant J. 8, 769–776. doi: 10.1016/j.hpj.2022.05.003
Yamagishi, M., Sakai, M. (2020). The microRNA828/MYB12 module mediates bicolor pattern development in Asiatic hybrid lily (Lilium spp.) flowers. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.590791
Yamasaki, H., Sakihama, Y., Ikehara, N. (1997). Flavonoid-peroxidase reaction as a detoxification mechanism of plant cells against H2O2. Plant Physiol. 115, 1405–1412. doi: 10.1104/pp.115.4.1405
Yan, J., Wang, B., Zhong, Y., Yao, L., Cheng, L., Wu, T. (2015). The soybean R2R3 MYB transcription factor GmMYB100 negatively regulates plant flavonoid biosynthesis. Plant Mol. Biol. 89, 35–48. doi: 10.1007/s11103-015-0349-3
Yang, J., Liang, J., Shao, L., Liu, L., Gao, K., Zhang, J.-L., et al. (2020). Green production of silybin and isosilybin by merging metabolic engineering approaches and enzymatic catalysis. Metab. Eng. 59, 44–52. doi: 10.1016/j.ymben.2020.01.007
Yao, L. H., Jiang, Y.-M., Shi, J., Tomas-Barberan, F. A., Datta, N., Singanusong, R., et al. (2004). Flavonoids in food and their health benefits. Plant Foods Hum. Nutr. 59, 113–122. doi: 10.1007/s11130-004-0049-7
Yao, H. P., Li, C. L., Zhao, H. X., Zhao, J. L., Chen, H., Bu, T. L., et al. (2017). Deep sequencing of the transcriptome reveals distinct flavonoid metabolism features of black tartary buckwheat (Fagopyrum tataricum garetn.). Prog. Biophys. Mol. Biol. 124, 49–60. doi: 10.1016/j.pbiomolbio.2016.11.003
Yoshida, K., Ma, D., Constabel, C. P. (2015). The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol. 167, 693–710. doi: 10.1104/pp.114.253674
Yu, K., Jun, J. H., Duan, C., Dixon, R. A. (2019). VvLAR1 and VvLAR2 are bifunctional enzymes for proanthocyanidin biosynthesis in grapevine. Plant Physiol. 180, 1362–1374. doi: 10.1104/pp.19.00447
Zavala, K., Opazo, J. C. (2015). Lineage-specific expansion of the chalcone synthase gene family in rosids. PLoS One 10, e0133400. doi: 10.1371/journal.pone.0133400
Zhang, Y., Butelli, E., Alseekh, S., Tohge, T., Rallapalli, G., Luo, J., et al. (2015). Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 6, 8635. doi: 10.1038/ncomms9635
Zhang, F., Gonzalez, A., Zhao, M., Payne, C. T., Lloyd, A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of arabidopsis. Development. 130, 4859–4869. doi: 10.1242/dev.00681
Zhang, X., He, Y., Li, L., Liu, H., Hong, G. (2021). Involvement of the R2R3-MYB transcription factor MYB21 and its homologs in regulating flavonol accumulation in arabidopsis stamen. J. Exp. Bot. 72, 4319–4332. doi: 10.1093/jxb/erab156
Zhang, M., Li, F.-D., Li, K., Wang, Z.-L., Wang, Y.-X., He, J.-B., et al. (2020). Functional characterization and structural basis of an efficient di- C -glycosyltransferase from Glycyrrhiza glabra. J. Am. Chem. Soc 142, 3506–3512. doi: 10.1021/jacs.9b12211
Zhang, G., Yu, Z., Yao, B., Teixeira da Silva, J. A., Wen, D. (2022). SsMYB113, a Schima superba MYB transcription factor, regulates the accumulation of flavonoids and functions in drought stress tolerance by modulating ROS generation. Plant Soil. 478, 427–444. doi: 10.1007/s11104-022-05466-6
Zhao, Q., Cui, M.-Y., Levsh, O., Yang, D., Liu, J., Li, J., et al. (2018). Two CYP82D enzymes function as flavone hydroxylases in the biosynthesis of root-specific 4′-deoxyflavones in Scutellaria baicalensis. Mol. Plant 11, 135–148. doi: 10.1016/j.molp.2017.08.009
Zhao, C., Liu, X., Gong, Q., Cao, J., Shen, W., Yin, X., et al. (2021a). Three AP2/ERF family members modulate flavonoid synthesis by regulating type IV chalcone isomerase in citrus. Plant Biotechnol. J. 19, 671–688. doi: 10.1111/pbi.13494
Zhao, R., Song, X., Yang, N., Chen, L., Xiang, L., Liu, X. Q., et al. (2020). Expression of the subgroup IIIf bHLH transcription factor CpbHLH1 from Chimonanthus praecox (L.) in transgenic model plants inhibits anthocyanin accumulation. Plant Cell Rep. 39, 891–907. doi: 10.1007/s00299-020-02537-9
Zhao, T., Tang, H., Xie, L., Zheng, Y., Ma, Z., Sun, Q., et al. (2019b). Scutellaria baicalensis georgi. (Lamiaceae): a review of its traditional uses, botany, phytochemistry, pharmacology and toxicology. J. Pharm. Pharmacol. 71, 1353–1369. doi: 10.1111/jphp.13129
Zhao, G., Xiang, F., Zhang, S., Song, J., Li, X., Song, L., et al. (2021b). PbLAC4-like, activated by PbMYB26, related to the degradation of anthocyanin during color fading in pear. BMC Plant Biol. 21, 469. doi: 10.1186/s12870-021-03220-1
Zhao, Q., Yang, J., Cui, M.-Y., Liu, J., Fang, Y., Yan, M., et al. (2019a). The reference genome sequence of Scutellaria baicalensis provides insights into the evolution of wogonin biosynthesis. Mol. Plant 12, 935–950. doi: 10.1016/j.molp.2019.04.002
Zhao, Y., Zhang, Y. Y., Liu, H., Zhang, X. S., Ni, R., Wang, P. Y., et al. (2019c). Functional characterization of a liverworts bHLH transcription factor involved in the regulation of bisbibenzyls and flavonoids biosynthesis. BMC Plant Biol. 19, 1–13. doi: 10.1186/s12870-019-2109-z
Zhao, Q., Zhang, Y., Wang, G., Hill, L., Weng, J.-K., Chen, X.-Y., et al. (2016). A specialized flavone biosynthetic pathway has evolved in the medicinal plant, Scutellaria baicalensis. Sci. Adv. 2, e1501780. doi: 10.1126/sciadv.1501780
Zhou, L. J., Geng, Z., Wang, Y., Wang, Y., Liu, S., Chen, C., et al. (2021). A novel transcription factor CmMYB012 inhibits flavone and anthocyanin biosynthesis in response to high temperatures in Chrysanthemum. Hortic. Res. 8, 248. doi: 10.1038/s41438-021-00675-z
Zhou, H., Lin-Wang, K., Wang, F., Espley, R. V., Ren, F., Zhao, J., et al. (2019). Activator-type R2R3-MYB genes induce a repressor-type R2R3-MYB gene to balance anthocyanin and proanthocyanidin accumulation. New Phytol. 221, 1919–1934. doi: 10.1111/nph.15486
Zhou, H., Peng, Q., Zhao, J., Owiti, A., Ren, F., Liao, L., et al. (2016). Multiple R2R3-MYB transcription factors involved in the regulation of anthocyanin accumulation in peach flower. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01557
Zhu, Z., Liang, H., Chen, G., Li, F., Wang, Y., Liao, C., et al. (2019). The bHLH transcription factor SlPRE2 regulates tomato fruit development and modulates plant response to gibberellin. Plant Cell Rep. 38, 1053–1064. doi: 10.1007/s00299-019-02425-x
Zhu, Q., Sui, S., Lei, X., Yang, Z., Lu, K., Liu, G., et al. (2015). Ectopic expression of the coleus R2R3 MYB-type proanthocyanidin regulator gene SsMYB3 alters the flower color in transgenic tobacco. PLoS One 10, 1–19. doi: 10.1371/journal.pone.0139392
Zhu, Q., Wang, B., Tan, J., Liu, T., Li, L., Liu, Y. G. (2020b). Plant synthetic metabolic engineering for enhancing crop nutritional quality. Plant Commun. 1, 1–18. doi: 10.1016/j.xplc.2019.100017
Zhu, J. H., Xia, D. N., Xu, J., Guo, D., Li, H. L., Wang, Y., et al. (2020a). Identification of the bHLH gene family in Dracaena cambodiana reveals candidate genes involved in flavonoid biosynthesis. Ind. Crops Prod. 150, 112407. doi: 10.1016/j.indcrop.2020.112407
Zhu, Q., Yu, S., Zeng, D., Liu, H., Wang, H., Yang, Z., et al. (2017). Development of “purple endosperm rice” by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system. Mol. Plant 10, 918–929. doi: 10.1016/j.molp.2017.05.008
Zipor, G., Duarte, P., Carqueijeiro, I., Shahar, L., Ovadia, R., Teper-Bamnolker, P., et al. (2015). In planta anthocyanin degradation by a vacuolar class III peroxidase in Brunfelsia calycina flowers. New Phytol. 205, 653–665. doi: 10.1111/nph.13038
Keywords: flavonoid, plant synthetic biology, transcription factor, enzyme diversity, plant chassis
Citation: Jiang L, Gao Y, Han L, Zhang W and Fan P (2023) Designing plant flavonoids: harnessing transcriptional regulation and enzyme variation to enhance yield and diversity. Front. Plant Sci. 14:1220062. doi: 10.3389/fpls.2023.1220062
Received: 11 May 2023; Accepted: 05 July 2023;
Published: 28 July 2023.
Edited by:
Claudia Renate Stange, University of Chile, ChileCopyright © 2023 Jiang, Gao, Han, Zhang and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pengxiang Fan, pxfan@zju.edu.cn
 Lina Jiang1
Lina Jiang1