
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 17 October 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1217193
Salt and alkaline stresses often occur together, severely threatening plant growth and crop yields. Salt stress induces osmotic stress, ionic stress, and secondary stresses, such as oxidative stress. Plants under saline-alkali stress must develop suitable mechanisms for adapting to the combined stress. Sustained plant growth requires maintenance of ion and pH homeostasis. In this review, we focus on the mechanisms of ion and pH homeostasis in plant cells under saline-alkali stress, including regulation of ion sensing, ion uptake, ion exclusion, ion sequestration, and ion redistribution among organs by long-distance transport. We also discuss outstanding questions in this field.
Plants face many biotic and abiotic stresses from their environment. Soil salinization and alkalization cause widespread abiotic stress in plants, adversely affecting global crop productivity. Saline-alkali stress includes the combined effects of salt and alkali stresses.
Salt stress is a major threat limiting plant growth and crop yield (Yang and Guo, 2018a; Kotula et al., 2020). More than 800 million hectares of the world’s land are affected by salinity (Munns and Tester, 2008). One-fifth of irrigated lands are already affected (Morton et al., 2019), and this area is growing larger as a result of overirrigation and climate changes. Soils are classified as saline when the electrical conductivity of a saturated soil extract is above 4 dS·m–1. Halophytic plants tolerate high salt concentrations, while glycophytes are sensitive to saline soils (Munns and Tester, 2008; Flowers et al., 2015). Most crop species are glycophytes, and their growth and reproduction are severely affected by salt stress, which is typically caused by sodium chloride, sodium sulfate, and other neutral salts in soil. Sodium ions (Na+) are generally considered to be the major toxic ions at high concentrations. Three stress signaling pathways are induced under salt stress: osmotic stress, ionic stress, and secondary stress (Sewelam et al., 2014; Yang and Guo, 2018a). High salt concentrations increase the osmotic pressure and lower the water potential of soil solutions, leading to water deficiency in plants (Hasegawa et al., 2000). Na+ in plant cells affect the absorption of other ions necessary for normal physiological processes (Yang and Guo, 2018b). Osmotic stress and ionic stress cause long-term secondary stresses, including oxidative stress. Salt-induced oxidative stress leads to accumulation of reactive oxygen species (ROS) in plant cells, including singlet oxygen (1O2), superoxide anion (O2–), hydrogen peroxide (H2O2), and hydroxyl radicals (HO·). Excess ROS cause oxidative damage to DNA, proteins, and lipids, affecting the integrity of plant cell organelles.
Alkali stress is often concomitant with salt stress owing to the abundance of sodium carbonate and sodium bicarbonate in soil. High pH in soil causes deposition of metal ions, such as iron (Fe2+), magnesium (Mg2+), and calcium (Ca2+). The uptake of mineral elements is blocked in plants, leading to nutrient deficiency. Meanwhile, many plant metabolic activities are severally interrupted, such as root vitality, cell pH stability, cell membrane integrity, and photosynthesis.
The combination of saline and alkali stresses affects plant growth and development more severely than salt stress or alkali stress alone. Most studies have focused mainly on either salt stress or alkali stress, with only a few paying attention to mixed saline-alkali stress. With an increasing world population, a 50% increase in food production by 2050 will be required (FAO, 2017). Understanding the mechanisms of plant responses to saline-alkali stress will provide information for genetically engineering saline-alkali-tolerant crops to meet this demand. Here, we discuss recent studies on the mechanisms underlying plant responses to saline-alkali stress.
High concentrations of salt and high soil pH induce multiple stresses in plant cells. Plants must sense saline-alkali stress rapidly and trigger precise responses. Sensing Na+, osmotic stress, and high pH is the first step in initiating such responses. Salt-induced Na+ accumulation occurs within 2 minutes after exposure to salt stress, while Na+ begins to efflux from roots within 10 minutes (Essah et al., 2003; Bose et al., 2015). This indicates that high Na+ concentrations are rapidly perceived, triggering specific downstream responses.
The mechanism underlying salt stress sensing has been an open question for a long time. High salt induces both ionic and osmotic stresses in plants. It was thought that plants perceive osmotic stress rather than Na+ (Munns and Tester, 2008). However, rapid sodium-specific calcium waves have since been identified (Dodd et al., 2010; Hashimoto and Kudla, 2011; Martí et al., 2013). Salt (NaCl) induces an increase in the cytosolic free calcium concentration ([Ca2+]cyt) in cortical and endodermal cells of roots, while osmotic stress (mannitol) induces an increase in [Ca2+]cyt in epidermal cells (Kiegle et al., 2000; Choi et al., 2014). Increased [Ca2+]cyt activates Ca2+-binding proteins and upregulates the Na+/H+ antiporter to efflux Na+. Accordingly, plants may perceive salt stress through Na+ sensors and osmotic sensors in different cell types.
Osmotic sensor contributes to the perception of salt stress. The Arabidopsis (Arabidopsis thaliana) reduced hyperosmolality-induced [Ca2+] increase 1 (osca1) mutant was isolated through Ca2+-imaging-based forward genetic screening (Yuan et al., 2014). Under osmotic stress, OSCA1 induces a rapid increase in [Ca2+]cyt. OSCA1 is a Ca2+-permeable channel on the plasma membrane identified as a putative sensor of hyperosmotic stress and required for osmotic-stress-induced Ca2+ signaling (Yuan et al., 2014). The osca1 mutant shows impaired Ca2+ signaling in response to osmotic stress but not salt stress (Yuan et al., 2014). Recent mathematical fitting/modeling analysis in wild-type Arabidopsis and the osca1 mutant revealed that OSCA1 is responsible for the increase of [Ca2+]cyt induced by osmotic stress rather than ionic stress (Pei et al., 2022).
The plant cell wall is involved in perceiving and responding to salt stress (Endler et al., 2015; Van der Does et al., 2017; Parrotta et al., 2015). It is important for cells to sense salt-induced changes in the cell wall and maintain cell wall integrity. Plasma membrane-located receptor-like kinases (RLKs), MALE DISCOVERER 1-INTERACTING RECEPTOR LIKE KINASE 2 (MIK2), FEI1 and FEI2 (named after the Chinese word for fat), and FERONIA (FER) are involved in cell wall sensing and the response to salt stress (Xu et al., 2008; Van der Does et al., 2017; Feng et al., 2018). Salt-specific ionic stress causes cell wall softening, which is sensed by FER (Feng et al., 2018). FER interacts with pectin in the cell wall and elicits salt-induced Ca2+ transients to maintain cell wall integrity under high salinity. Thus, FER senses cell wall integrity under salt stress. Salt stress also induces the secretion of mature RAPID ALKALINIZATION FACTORs (RALFs), which interact with FER to trigger a series of physiological process such as deactivation of plasma membrane H+-ATPases (PM H+-ATPases) and apoplastic alkalinization (Haruta et al., 2014).
The Arabidopsis monocation-induced Ca2+ increases 1 (moca1) mutant was identified through a Ca2+-imaging-based forward genetic screen for mutants with disturbed [Ca2+]cyt induced by salt. Increases in [Ca2+]cyt in response to Na+, potassium ions (K+), or lithium ions (Li+) are reduced in the moca1 mutant than in wild-type plants (Jiang et al., 2019). Phenotype analysis revealed that the moca1 mutant is hypersensitive to salt stress. MOCA1 is a glucuronosyltransferase for glycosylinositol phosphorylceramide (GIPC) sphingolipids in the plasma membrane. Na+ binds to GIPC to potentially gate Ca2+ channels mediating Ca2+ influx and induce downstream responses. Therefore, MOCA1-dependent GIPC sphingolipids are likely involved in sensing salt stress through unidentified Ca2+ transporters in the plasma membrane (Jiang et al., 2019). Arabidopsis ANNEXIN1 (ANN1) and AtANN4 are Ca2+-permeable transporters (Laohavisit et al., 2013; Ma et al., 2019) required for salt-induced increases in [Ca2+]cyt. AtANN4 interacts with SALT OVERLY SENSITIVE 2 (SOS2) and SOS3-LIKE CALCIUM-BINDING PROTEIN 8 (SCaBP8) to generate Ca2+ signals that activate the salt overly sensitive (SOS) pathway under salt stress. Meanwhile, AtANN4 is repressed by the SOS pathway, forming a negative feedback loop to fine-tune Ca2+ signals for optimizing the plant salt response (Ma et al., 2019). It remains to be confirmed whether the MOCA1-dependent GIPC sphingolipids gate operates through AtANN1 and AtANN4 to trigger downstream responses to salt -specific ionic stress.
Different from animals, plants might sense salt through a multicomponent ion channel complex. In addition, this ion channel complex might not be controlled directly by salt (Jiang et al., 2019; van Zelm et al., 2020; Pei et al., 2022). Identifying the salt sensor remains a problem that scientists are eager to solve.
Alkali stress is often associated with salt stress. However, high pH may be sensed through a different mechanism in plants. The cell-surface peptide-receptor complexes RGF1-RGFR (involving ROOT MERISTEM GROWTH FACTOR 1 [RGF1] and its receptors [RGFRs]) and Pep1-PEPR (involving plant elicitor peptides [Peps] and their receptors [PEPRs]) function as pH sensors to regulate extracellular pH-mediated growth and immunity in root apical meristem regions (Liu et al., 2022). Extracellular alkalization inhibits the interaction between RGF1 and RGFRs through sulfotyrosine, thereby inhibiting root meristem growth. Extracellular alkalization also facilitates alkaline-dependent binding of Peps to PEPRs through the pH sensor Glu/Asp, thereby promoting immunity (Liu et al., 2022). Further investigation is needed to reveal whether high pH during alkali stress is also sensed by plant membrane RLKs.
In addition to high pH, excess bicarbonate or carbonate caused by saline-alkali stress may injury plants in unique way. SLOWLY ACTIVATING ANION CONDUCTANCE HOMOLOGUE 3 in Glycine soja (GsSLAH3) was found to be induced be NaHCO3 treatment. Overexpression of GsSLAH3 in Arabidopsis improves alkaline tolerance under treatments of NaHCO3 or KHCO3, rather than high pH treatment, which indicates that GsSLAH3 is a positive alkali responsive gene that increases bicarbonate resistance specifically (Duan et al., 2018). Similarly, GsERF6 and GsBOR2 also exhibit specific response to bicarbonate stress (Yu et al., 2016; Duan et al., 2018). These results suggest that plants response to bicarbonate stress in a unique way. It is still indistinct whether plants also possess unique sensors to bicarbonate stress.
It is important to maintain ion homeostasis in plant cells under saline-alkali stress. Under normal conditions, plants maintain relatively high K+ concentrations and low Na+ concentrations in cells (Binzel et al., 1988). A high ratio of cytosolic K+ to Na+ is necessary for plant growth and development (Hussain et al., 2021). However, under saline-alkali stress, excess Na+ is transported into the cytoplasm, which disturbs ion homeostasis.
Plants maintain Na+ homeostasis under saline-alkali stress by restricting Na+ import, compartmentalizing Na+, and increasing Na+ efflux (Yamaguchi et al., 2013; Yang and Guo, 2018a). The specific import system for Na+ has not been identified, but Na+ is known to be transported into cells through K+ carriers. Electrophysiological evidence shows that Na+ is transported across the plasma membrane through nonselective cation channels (NSCCs), high-affinity K+ transporters (HKTs), and low-affinity K+ transporters (such as ARABIDOPSIS K+ TRANSPORTER 1 [AKT1]) (Blumwald et al., 2000; Tuteja, 2007).
HKTs are key determinants of plant responses to salt stress (Horie et al., 2006; Platten et al., 2006; Horie et al., 2007). HKT1 from wheat (Triticum aestivum) was first identified as an H+/K+ symporter and then confirmed as a Na+/K+ symporter (Schachtman and Schroeder, 1994; Rubio et al., 1995). High-affinity K+ uptake by HKT1 is activated by micromolar concentrations of Na+ and inhibited by high concentrations of Na+, which initiate low-affinity Na+ uptake (Rubio et al., 1995). AtHKT1 from Arabidopsis and OsHKT1 from rice (Oryza sativa) display selective Na+ transport activity rather than K+ transport activity in Xenopus (Xenopus laevis) oocytes (Uozumi et al., 2000; Horie et al., 2001; Garciadeblás et al., 2003). OsHKT1 plays an exclusive role in Na+ uptake by rice roots under K+ starvation. However, under salt stress, OsHKT1 activity is rapidly downregulated to restrict excess Na+ influx (Horie et al., 2007). Arabidopsis hkt1 mutants accumulate more Na+ than wild-type plants and are sensitive to salt stress (Mäser et al., 2002; Rus et al., 2004; Davenport et al., 2007). Excess Na+ influx into the cytoplasm can decrease membrane potential. Therefore, K+ outflow channels (NSCC, STELAR K+ OUTWARD RECTIFIER [SKOR], and GATED OUTWARDLY-RECTIFYING K+ CHANNEL [GORK]) are activated and the ratio of K+ to Na+ decreased, weakening plant adaption to salt stress (Falhof et al., 2016). Recently, AKT1 is found to mediates K+ uptake under salt stress (Li et al., 2023). SCaBP8 binds to AKT1 to reduce its activity (Ren et al., 2013); under salt stress, phosphorylation of SCaBP8 by SOS2, promoted by phosphatidic acid (PA), releases the inhibition of AKT1, enhancing K+ influx (Li et al., 2023). Meanwhile, PA also promotes Na+ efflux through the SOS2–SOS1 module to maintain Na+/K+ ratios under salt stress (Li et al., 2023).
In addition to salt-induced Na+ influx and changes in the Na+/K+ ratio under saline-alkali stress, high pH also causes nutrient ions, such as Ca2+, Mg2+, Fe2+, zinc (Zn2+), and copper (Cu2+), to precipitate in soil. Under saline-alkali stress, nutrient recycling and availability for root cells are decreased, further leading to nutrient deficiency in plants (Jia et al., 2020). Saline-alkali stress induces plants to generate and secrete organic acids such as oxalate, acetate, and malate, which function as buffering substances to maintain intracellular pH homeostasis and ion balance (Fang et al., 2021). The key enzyme in this biochemical process is phosphoenolpyruvate carboxylase, which is regulated by phosphoenolpyruvate carboxylase kinases. These kinases are dramatically upregulated by NaHCO3 stress (Xiang et al., 2019).
Excess Na+ accumulates in plant cytoplasm under saline-alkali stress. Plants transport Na+ from the cytoplasm to the apoplast or vacuoles mainly through Na+/H+ antiporters, which transport Na+ in exchange for H+ (Mansour and Hassan, 2022).
The signaling pathways for Na+ efflux have been well described, with the most important being the SOS regulatory pathway (Yang and Guo, 2018a; Yang and Guo, 2018b; Ismail et al., 2020; van Zelm et al., 2020). The SOS pathway has three major components: SOS1, SOS2, and SOS3 or SCaBP8. SOS1 is a Na+/H+ antiporter belonging to the Na+/H+ exchangers (NHX) family. SOS2 is a serine/threonine-protein kinase. SOS3 and SCaBP8 are EF-hand calcium-binding proteins. SOS3 mainly functions in roots while SCaBP8 mainly functions in shoots (Quan et al., 2007; Monihan et al., 2016; Yang and Guo, 2018a). SCaBP8 overexpression in sos3 mutants partially rescues the sos3 sensitive phenotype under salt stress. However, SOS3 overexpression fails to complement the scabp8 mutant. These results indicate that SCaBP8 and SOS3 are only partially redundant in their function, with each playing additional and unique roles in the plant response to salt stress (Quan et al., 2007). Under salt stress, SOS3 and SCaBP8 perceive salt-induced [Ca2+]cyt to release self-inhibition of SOS2, recruiting SOS2 to the plasma membrane and activating SOS2 (Halfter et al., 2000; Kim et al., 2007; Lin et al., 2009). SOS2 then phosphorylates SOS1 at serine 1044 in the C-terminal domain to relieve SOS1 autoinhibition. Activated SOS1 transports Na+ from the cytoplasm to the apoplast to relieve salt stress (Quintero et al., 2011).
The Na+/H+ antiporter SOS1 is a key component of Na+ efflux from the cytoplasm to the apoplast. The knockout mutant sos1-1 is highly sensitive to salt stress (Shi et al., 2000; Qiu et al., 2002; Quintero et al., 2002). In Arabidopsis, salt induces phospholipase D activity to hydrolyze phospholipids into PA. PA then binds to MITOGEN-ACTIVATED PROTEIN KINASE KINASE 7 (MKK7) and MKK9, increasing their kinase activity. Downstream MITOGEN-ACTIVATED PROTEIN KINASE 6 (MPK6) is then phosphorylated, and activated MPK6 phosphorylates and activates the SOS1 antiporter to export Na+ from cells (Shen et al., 2019). Under normal conditions, SOS1 activity is inhibited by the clade D protein phosphatase 2Cs (PP2C) PP2C.D6 and PP2C.D7. Under salt stress, however, SCaBP8 locates to the plasma membrane and inhibits PP2C.D6 and PP2C.D7 phosphatase activity. PP2C.D6 is released into the cytoplasm, and inhibition of SOS1 by PP2C.D6 and PP2C.D7 is abolished, allowing SOS1 to export Na+ from the cytoplasm (Fu et al., 2022). The Ca2+ sensor SOS3/CALCINEURIN B-LIKE PROTEIN 4 (CBL4) triggers basal salt tolerance through the SOS3/CBL4–SOS2/CBL-INTERACTING PROTEIN KINASE 24 (CIPK24)–SOS1 pathway, while another Ca2+ sensor, CBL8, triggers the CBL8–SOS2/CIPK24–SOS1 module in response to severe salt stress (Steinhorst et al., 2022). The mechanism for sensing mild salt stress may differ from that for sensing severe salt stress in plant cells.
The kinase activity of SOS2 is controlled by several key regulators, such as the PP2C ABA INSENSITIVE 2 (ABI2), GIGANTEA (GI), 14-3-3 proteins, SOS2-LIKE PROTEIN KINASE 5 (PKS5), and BRASSINOSTEROID-INSENSITIVE 2 (BIN2) (Kim et al., 2013; Zhou et al., 2014; Yang Z. et al., 2019; Duscha et al., 2020; Li et al., 2020). ABI2 interacts with SOS2 through the protein phosphatase interaction motif of SOS2. This interaction is disrupted in abi2-1 mutants, which are more tolerant to salt stress than wild-type plants (Ohta et al., 2003). Interaction of ABI2 with SOS2 therefore negatively regulates salt tolerance. The flowering-time protein GI also negatively regulates salt tolerance. In the absence of salt stress, GI confines SOS2 to the cytoplasm and nucleoplasm, inhibiting its activation of SOS1. Under salt stress, GI is degraded by the 26S proteasome, releasing SOS2 to activate SOS1 to export Na+. Meanwhile, flowering is also delayed. GI therefore balances floral transition and salt stress adaption in Arabidopsis (Kim et al., 2013). Similarly, 14-3-3 proteins interact with SOS2, inhibiting SOS2 activity. Salt stress decreases the interaction between 14-3-3 proteins and SOS2, releasing SOS2 and thus activating SOS1 activity (Zhou et al., 2014). Interaction between 14-3-3 proteins and SOS2 is promoted by phosphorylation of SOS2 at serine 294 by PKS5. Salt stress also induces an interaction between 14-3-3 proteins and PKS5, which inhibits the kinase activity of PKS5 and retards the inhibition of SOS2 (Yang Z. et al., 2019). These regulators are involved in the salt stress response phase. Li et al. (2020) identified a molecular switch in the recovery phase following salt stress. After salt stress, SOS3 and SCaBP8 perceive a Ca2+ signal and promote BIN2 relocation to the plasma membrane. BIN2 then phosphorylates and inhibits SOS2 to repress the salt stress response and enhances plant growth by releasing the transcriptional activity of BRASSINAZOLE RESISTANT 1 (BZR1) and BRI1-EMS-SUPPRESSOR 1 (BES1). BIN2 therefore functions as a molecular switch between the salt stress response and recovery of growth after salt stress (Li et al., 2020). Recently, evidences showed that light regulates plants’ response to salt stress. Arabidopsis are more tolerant to salt stress under light conditions than under dark conditions. In the light, phytochrome A and phytochrome B (phyA and phyB) translocate into nucleus, interacts with SOS2 and promote SOS2 kinase activity. In nucleus, highly activated SOS2 mediates phosphorylation and degradation of PIF1 and PIF3, leading to alleviate their inhibition of salt tolerance. Besides, light-activated phyA and phyB also enhance SOS2 activity in cytosol, thus enhancing SOS1 activity on the plasma membrane (Ma et al., 2023).
PM H+-ATPase is another key regulator of Na+ efflux. PM H+-ATPase belongs to the P-type subfamily of H+-ATPase and export cytoplasmic protons into the apoplast, generating proton gradients and electrical potential differences (Palmgren, 2001; Falhof et al., 2016). SOS1-mediated Na+ transport is driven by such proton gradients (Qiu et al., 2002; Yang Y. et al., 2019; Li et al., 2022). Na+ exclusion capability of plants generally attenuated under saline-alkali stress due to the influence of high pH on the proton motive force. Thus, H+-ATPase activity is a primary determinant of plant saline-alkali tolerance. The Arabidopsis PM H+-ATPase PLASMA MEMBRANE PROTON ATPASE2 (AHA2) interacts with the 14-3-3 class of chaperones, which alleviates AHA2 autoinhibition and activates AHA2 (Fuglsang et al., 1999; Fuglsang et al., 2007). Under normal conditions, the SCaBP1-PKS5, SCaBP1-PKS24, and SCaBP3-PKS5 modules phosphorylate Ser-931 of AHA2, which inhibits the interaction between AHA2 and 14-3-3 proteins, thereby repressing its H+-ATPase activity, and consequently inhibiting SOS1 activity (Fuglsang et al., 2007; Lin et al., 2014; Zhou et al., 2014; Yang Y. et al., 2019). Under salt stress, ([Ca2+]cyt) increases. This Ca2+ signal is decoded by 14-3-3 proteins, which interact with and repress PKS5 to activate PM H+-ATPases, improving SOS1 activity (Yang Z. et al., 2019). DNAJ HOMOLOG 3 (J3) also interacts with PKS5 under salt stress to inhibit its kinase activity and relieve repression of PM H+-ATPases. J3 loss-of-function mutants have lower PM H+-ATPase activity than wild-type plants and display hypersensitivity to saline and alkaline stresses (Yang et al., 2010). SCaBP3 separates from PM H+-ATPases when it senses Ca2+ signals triggered by saline-alkali stress. Activity of the SCaBP3-PKS5 complex and phosphorylation at Ser-931 of AHA2 decrease, allowing AHA2 to be activated by 14-3-3 proteins, subsequently enhancing PM H+-ATPase activity (Yang Y. et al., 2019). Acetylation of lysine 56 of GENERAL REGULATORY FACTOR 6 (GFR6)/14-3-3λ also inhibits AHA2 activity, making plants hypersensitive to alkali stress (Guo et al., 2022). Moreover, lipids dynamically change under saline-alkali stress (Tasseva et al., 2004). Phosphatidylinositol (PI) inhibits PM H+-ATPase activity under normal conditions. Under salt stress, PHOSPHOINOSITIDE 4-KINASE (PI4K) converts PI to phosphatidylinositol 4-phosphate (PI4P), which relieves the inhibition of PM H+-ATPase and activates the PM Na+/H+ antiporter SOS1 to transport Na+ out of cells and maintain ion homeostasis (Yang et al., 2021). Recently, a Ca2+-dependent signaling pathway to enhance H+-ATPase activity and saline-alkali stress tolerance is found in wheat. The researchers found that PM H+-ATPase activity is highly related to alkali tolerance in the wheat Shanrong 4 (SR4) variety. Alkali stress induces the expression of Ca2+-binding protein C-TERMINAL CENTRIN-LIKE DOMAIN CONTAINING PROTEIN 1 of Triticum aestivum (TaCCD1), TaCCD1 interacts with SMALL AUXIN UP RNA215 (TaSAUR215) and enhances the inhibitory effect of TaSAUR215 on two D-CLADE TYPE 2C PROTEIN PHOSPHATASES (TaPP2C.D1 and TaPP2C.D8), which can directly dephosphorylate Thr947 in H+-ATPase. Ultimately, the activity of PM H+-ATPase TaHA2 is enhanced and alkali stress tolerance is promoted (Cui et al., 2023). Besides, transcriptional regulation also plays important roles in stimulating PM H+-ATPase activity under salt stress (Wang et al., 2013; Yao et al., 2019). Populus euphratica transcription factor WRKY1 was found to bind the promotor of PM H+-ATPase gene PeHA1 to enhance the PeHA1 gene expression and salt tolerance (Yao et al., 2019). Phospholipase Dδ in Populus euphratica (PePLDδ) is a key enzyme in producing signaling molecules phosphatidic acid (PA). Overexpression of PePLDδ in Arabidopsis increases the transcription of AtSOS1 and PM H+-ATPase AtAHA2, leading to enhanced Na+ exclusion under salt stress (Zhang et al., 2022). Transcriptional regulation of PM H+-ATPases provides an important method for saline-alkaline tolerant crop breeding.
Compartmentalizing Na+ into vacuoles is an important mechanism for reducing the cytoplasmic Na+ concentration under saline-alkali stresses. Vacuolar Na+/H+ exchangers (NHXs), driven by the proton gradient generated by vacuolar H+-ATPase and H+-pyrophosphatase, are key components for Na+ compartmentation in vacuoles (Drozdowicz and Rea, 2001; Brini and Masmoudi, 2012). The NHX family is divided into two classes. Class I comprises NHX1–NHX4 in Arabidopsis, all localized in vacuoles (Yokoi et al., 2002; Pardo et al., 2006). Overexpressing tonoplast-localized NHX1 in Arabidopsis increases salt tolerance and Na+ accumulation under salt stress (Apse et al., 2003). Similar to the plasma membrane Na+/H+ exchanger SOS1, tonoplast NHXs in Arabidopsis are activated by SOS2 (Qiu et al., 2004). SCaBP8 interacts with SOS2 to activate Na+ sequestration (Kim et al., 2007). However, the Na+ content in leaf tissue of nhx1 nhx2 double mutants under salt stress is higher than that in wild-type plants, while the vacuolar K+ content is lower. These results indicate that NHX1 and NHX2 mainly function as K+/H+ exchangers in the Arabidopsis vacuole (Barragán et al., 2012; Bassil et al., 2012; Bassil and Blumwald, 2014). It seems that other Na+ transporters mediate Na+ influx into the vacuole from the cytoplasm in Arabidopsis, or that the NHX1 and NHX2 K+ transport activity might be mediated by Na+ concentration in the cytosol.
Vacuolar H+-ATPase is the major H+ pump that transports protons, generating a proton gradient and electrical potential to drive secondary active transport of various ions. Under salt stress, SOS2 directly interacts with vacuolar H+-ATPases and enhances their H+ transport activity to increase the driving force for Na+ compartmentation into the vacuole (Batelli et al., 2007).
In addition to Na+, Ca2+ is also transported into vacuoles under salt stress. CALCIUM EXCHANGER 1 (CAX1) and CAX3 are vacuolar H+/Ca2+ transporters localized on the tonoplast. CAX1 is mainly expressed in leaves, while CAX3 is mainly expressed in roots (Cheng et al., 2005). CAX1-like chimeric transporters are activated by SOS2. Biochemical analysis of Ca2+ transport in cax1 mutants reveals that CAX1 mediates approximately 50% of H+/Ca2+ transport between the cytoplasm and vacuole; cax1 and cax3 mutants display a 38% and 22% reduction in vacuolar H+-ATPase activity, respectively. Overexpressing a deregulated version of CAX1 led to salt sensitivity (Cheng et al., 2005). These results indicate a link between Na+ and Ca2+ homeostasis in Arabidopsis under salt stress.
At the whole-plant level, inorganic ions, including Na+, are absorbed through the root hairs, epidermis, and cortex, then loaded into xylem vessels for transport upward throughout the plant driven by the transpiration pull (Boer and Volkov, 2003; Raddatz et al., 2020). Thereafter, ions are unloaded by shoot or leaf xylem parenchyma cells and transported into phloem sieves (Sunarpi et al., 2005). SOS1 activity during xylem loading and HKT1 activity during xylem unloading might coordinately control the amount of Na+ that is eventually exported from roots to shoots.
In addition to Na+ exclusion, SOS1 also functions in controlling long-distance Na+ transport from roots to shoots to protect roots from salt-induced damage. In Arabidopsis, SOS1 is preferentially expressed in epidermal cells at the root tip and in parenchyma cells at the xylem/symplast boundary of roots, stems, and leaves (Shi et al., 2002). Na+ content analysis of shoots in the sos1 mutant under mild salt stress (25 mM NaCl) and under severe salt stress (100 mM NaCl) indicates a role for SOS1 in retrieving Na+ from xylem sap under severe salt stress, whereas it may function in loading Na+ into the xylem under mild salt stress (Shi et al., 2002). The rice sos1 loss-of-function mutant also shows sensitivity to salt, caused by excessive Na+ intake and disturbed Na+ loading into the xylem (El Mahi et al., 2019).
Arabidopsis HKT1 in xylem parenchyma cells has a key role in Na+ unloading from xylem vessels, thus protecting Arabidopsis leaves from Na+ stress. hkt1 mutants have increased Na+ concentration in the xylem sap and reduced Na+ concentration in phloem sap. However, the K+ content of the xylem is not dramatically influenced (Sunarpi et al., 2005). Under salt stress, hkt1 mutants show hypersensitive leaves and Na+ overaccumulation in xylem sap and shoots, with concomitant Na+ reduction in roots. These results indicate that HKT1 selectively transports Na+ from xylem vessels to xylem parenchyma cells. Overexpressing HKT1 in Arabidopsis root stele reduces Na+ accumulation in the shoot and enhances salt tolerance (Berthomieu et al., 2003; Møller et al., 2009). In line with this, salt-tolerant rice cultivars accumulate less Na+ in leaves and shoots compared with salt-sensitive rice cultivars (Ren et al., 2005), and Na+ accumulation in shoots is negatively correlated with salt tolerance in rice (Coskun et al., 2013).
In summary, to protect roots under salt stress, SOS1 exports Na+ out of roots and improves Na+ loading into the xylem. Meanwhile, HKT1 transports Na+ from xylem vessels into xylem parenchyma cells to protect leaves and shoots (Ali et al., 2021). Therefore, SOS1 and HKT1 antagonistically control the amount of Na+ in roots and shoots, and their activities are fine-tuned to avoid futile cycles of Na+ loading and unloading (Figure 1).
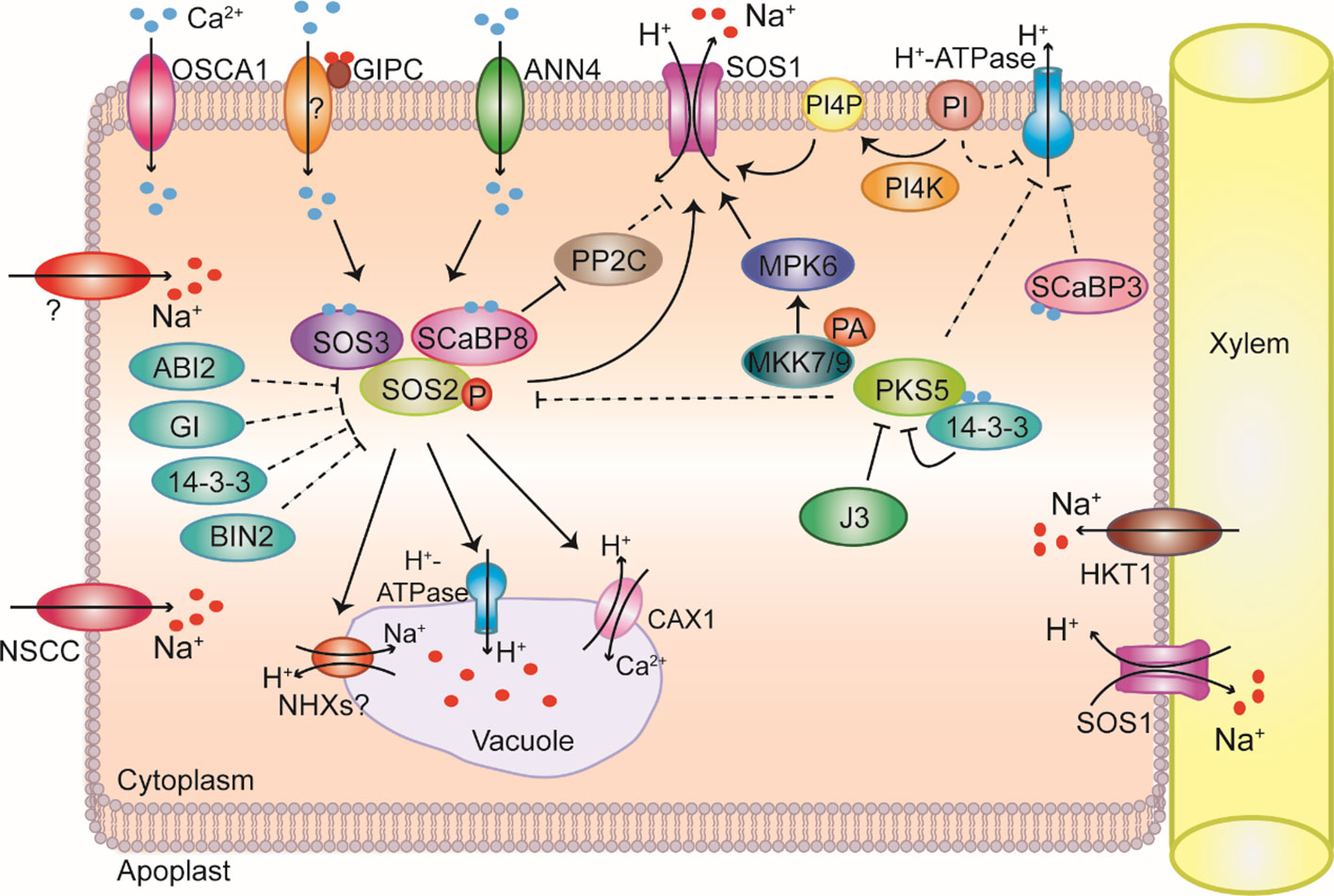
Figure 1 Overview of Na+ homeostasis in plant cells under salt stress. Na+ (red dots) enters plant cells through nonselective cation channels (NSCC) and unknown transporters. Na+ is sensed via an unknown sensing mechanism. The REDUCED HYPEROSMOLALITY-INDUCED [CA2+] INCREASE 1 (OSCA1) calcium-permeable channel on the plasma membrane is required for osmotic-stress-induced Ca2+ (blue dots) signaling. Na+ binds to glycosylinositol phosphorylceramide (GIPC) to gate Ca2+ influx channels and induce downstream responses to salt stress. The Ca2+-permeable transporter ANNEXIN4 (ANN4) generates Ca2+ signals that activate the salt overly sensitive pathway under salt stress. SALT OVERLY SENSITIVE 3 (SOS3) and SOS3-LIKE CALCIUM-BINDING PROTEIN 8 (SCaBP8) perceive salt-induced Ca2+ signals to release self-inhibition of SOS2 and recruit SOS2 to the plasma membrane where it becomes active. SOS2 then phosphorylates SOS1, relieving SOS1 autoinhibition. Activated SOS1 transports Na+ from the cytoplasm to the apoplast. SCaBP8 locates to the plasma membrane and inhibits the activities of the protein phosphatase 2Cs (PP2C) PP2C.D6 and PP2C.D7 under salt stress, thereby abolishing inhibition of SOS1 by PP2C.D6 and PP2C.D7. Phosphatidic acid (PA) binds to MITOGEN-ACTIVATED PROTEIN KINASE KINASE 7 (MKK7) and MKK9, increasing their kinase activity. MKK7 and MKK9 then activate MITOGEN-ACTIVATED PROTEIN KINASE 6 (MPK6), which phosphorylates and activates the SOS1 antiporter. SOS2 kinase activity is regulated by the PP2C ABA INSENSITIVE 2 (ABI2), GIGANTEA (GI), 14-3-3 proteins, BRASSINOSTEROID-INSENSITIVE 2 (BIN2), and SOS2-LIKE PROTEIN KINASE 5 (PKS5). Increased Ca2+ signals are decoded by 14-3-3 proteins, which interact with and repress PKS5 to activate plasma membrane H+-ATPases (PM H+-ATPases), improving SOS1 activity. DNAJ HOMOLOG 3 (J3) also interacts with PKS5 under salt stress to inhibit its kinase activity and relieve repression of PM H+-ATPases. SCaBP3 separates from PM H+-ATPases upon sensing Ca2+ signals triggered by saline-alkali stress, allowing PM H+-ATPases to be activated by 14-3-3 proteins. Na+ is compartmentalized in the vacuole through as-yet-unidentified transporters. Na+/H+ exchanger (NHX) proteins might function as Na+/H+ exchangers, but this has not been confirmed. In addition to Na+, Ca2+ ions are also transported into vacuoles through CALCIUM EXCHANGER 1 (CAX1) under salt stress. SOS2 interacts with vacuolar H+-ATPases to generate an increased driving force for compartmentalizing Na+ into the vacuole under salt stress through putative vacuolar H+/Na+ and H+/Ca2+ transporters. SOS1 also functions in controlling long-distance Na+ transport from roots to shoots to protect roots from salt-induced damage by loading Na+ into the xylem. HIGH-AFFINITY K+ TRANSPORTER 1 (HKT1) in xylem parenchyma cells mediates Na+ unloading from xylem vessels, thus protecting Arabidopsis leaves from Na+ stress.
In order to improve saline-alkali tolerance in crops, many saline-alkali-tolerant genes or locus were identified through Genome-wide association study (GWAS) analysis. Na+ Content under Saline-Alkali Condition (ZmNSA1) was identified in by GWAS analysis in natural maize inbred lines with variations of sensitivity to saline-alkali stress (Cao et al., 2020). Under saline-alkaline condition, Ca2+ binds to EF-hand domain of ZmNSA1 and triggers its degradation, then increases the transcription level of PM-H+-ATPases. Increased abundance of PM-H+-ATPases enhance H+ efflux and SOS1-mediated Na+ efflux from roots, which finally promoting saline-alkali tolerance (Cao et al., 2020). Recently, Alkaline tolerance 1 (AT1) in Sorghum bicolor, has been identified as a negative regulator of alkali tolerance by GWAS analysis (Zhang et al., 2023). AT1 encodes an atypical G protein γ subunit, which inhibits the phosphorylation of aquaporins PIP2s (Plasma membrane intrinsic protein 2s) to regulate the export of hydrogen peroxide, and then modulate ROS homeostasis under alkaline stress (Zhang et al., 2023). Knockout of AT1 increased alkali tolerance and production of sorghum, millet (Setaria italica), rice (Oryza sativa), and maize (Zea mays) (Zhang et al., 2023). These results indicate AT1 with a great applicational potential in genetic engineering of alkali-tolerant crops.
Saline-alkali stress severely affects plant growth and crop yield, posing a huge threat to agriculture and world food security. Understanding the mechanisms of plant responses to saline-alkali stress is critical for genetically engineering tolerance to saline-alkali stress in crops. Key components that involve in saline-alkali stress responses have been identified over the past few decades, with the mechanisms of ion and pH homeostasis studied in depth. However, some important questions remain. Here we discuss three aspects that need further research.
1) The sodium-sensing mechanism needs to be identified. The cell-surface peptide-receptor complexes RGF1-RGFR and Pep1-PEPR have been identified as pH sensors that function in extracellular pH-mediated growth and immunity in the root apical meristem (Liu et al., 2022). However, it is unknown whether high pH during alkali stress is also sensed by plant membrane RLKs. 2) Studying the specific influence of Na+ on plant growth requires disentangling of osmotic and Na+ stresses. 3) The mechanisms of plant responses to mild salt stress may be different from those to severe salt stress. 4) Most studies have focused on salt stress or alkali stress alone, with only a few assessing saline-alkali mixed stress. Whether the combination of salt and alkali stresses necessitate distinct response mechanisms in plants is an interesting question.
As research progressed, more key regulators in plant responses to saline-alkaline stress have been identified. Overexpressing positive regulators or knockout negative regulators to saline-alkaline stress has been a common method to genetically improve plant tolerance. However, the balance between plant growth/development and saline-alkaline tolerance should be considered. TOR kinase and ABA receptor balances plant growth and stress response (Wang et al., 2018). Receptor-like kinase FERONIA (FER) coordinates plant growth and salt tolerance via the phosphorylation of phytochrome B (phyB) (Liu et al., 2023). Genes not only enhancing stress tolerance but also improving plant growth, or at least not impacting to plant growth would be an important selection criterion for genetic engineering. The AT1 nonfunctional mutants, either natural varieties or gene editing mutants in sorghum, millet, rice, and maize, can improve the alkali tolerance and production when cultivated on sodic lands (Zhang et al., 2023). Therefore, genes like AT1 could be enormous potential for application in crop breeding for salt and alkali tolerance.
Artificial Intelligence (AI) has been a revolutionary technology in many fields and it is also hopeful for promoting crop breeding. The main possible applications of AI in crop breeding contains two major fields, phenomics and genomics. Smart breeding requires gene-phenotype-genomics big data which could be obtained by automatic equipment. Traditional phenotype identification requires a large amount of manpower, funding, time and plentiful breeding experience. AI may contribute in phenotype collecting, phenotype analyzing and phenotype assessment. Although there are many technical difficulties in image acquisition under complex environment and in AI algorithm optimization to analyze massive phenotypic elements, significant achievements have been achieved. Breeding robots and data analysis platforms have been established by several groups or companies, but the scope of application needs to be expanded to more plant species. Genotype-phenotype-envirotype big data is analyzed by AI algorithm to predict possible gene function and phenotype. Saline-alkali tolerant crops would be obtained by experimental methods, such as marker-assisted breeding, transgenic breeding, CRISPR-Cas9 based genome editing (Figure 2). As more and more high-throughput data has been obtained through plant multi-omics analysis, AI is more efficient to handle these massive data and screen functional genes to accelerate crop breeding for saline-alkali tolerance.
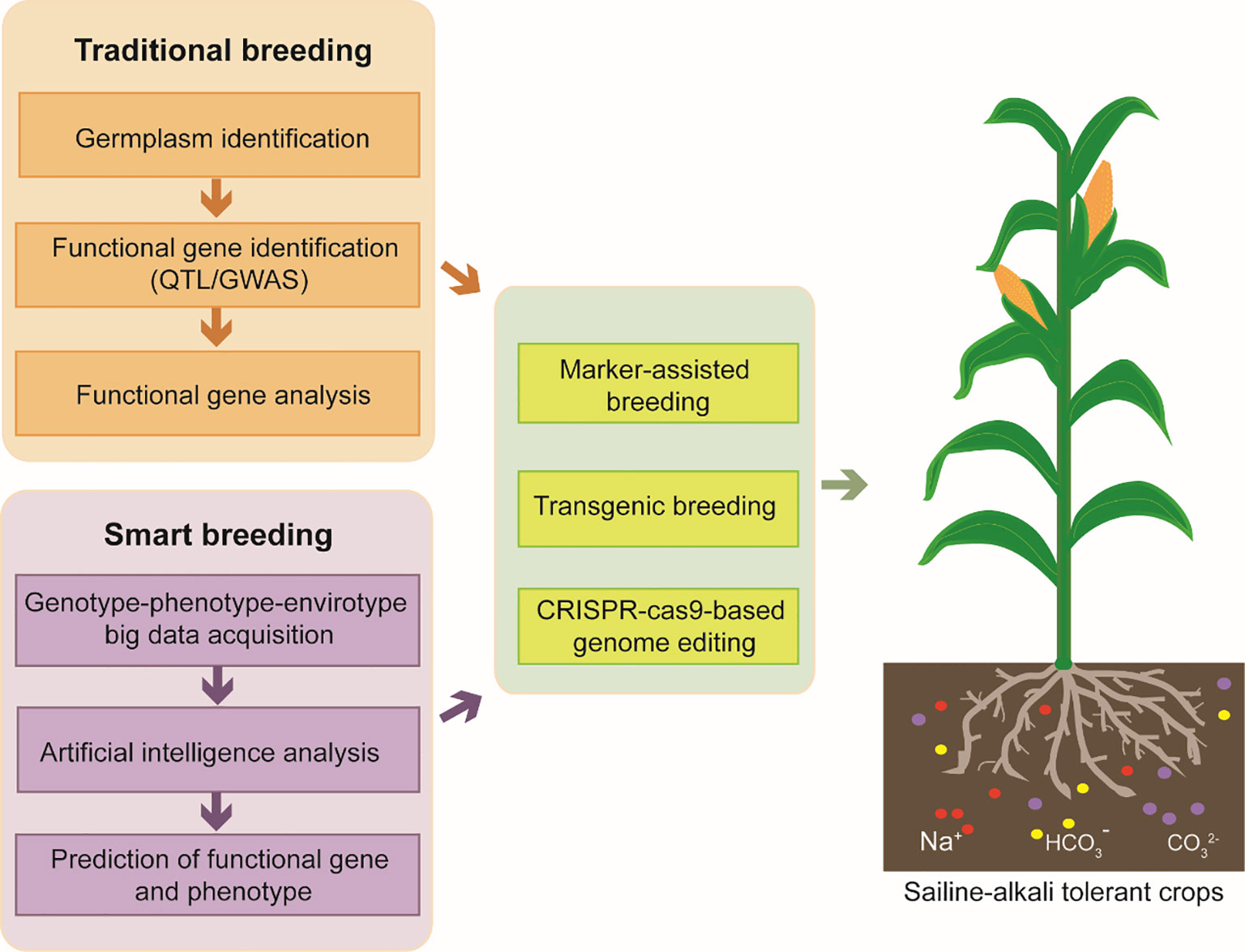
Figure 2 The strategies for improving plant resistance of saline-alkali stress. Traditional breeding requires tolerant/sensitive germplasm identification for saline-alkali stress, key gene identification and gene function analysis. In developing smart breeding, AI could be used in phenotype collecting, phenotype analyzing and phenotype assessment. Genotype-phenotype-envirotype big data could be analyzed by AI algorithm to predict possible gene function and phenotype. Finally, Saline-alkali tolerant crops would be obtained by modern experimental methods, such as marker-assisted breeding, transgenic breeding, CRISPR-Cas9 based genome editing.
YY and JL proposed the concept and content. JL drafted the manuscript. YY revised the manuscript. All authors contributed to the article and approved the submitted version.
This work is supported by the National Natural Science Foundation of China (NSFC) (Grant No.31800220 to JL; Grant No. 32070301 to YY) and Scientific Research Project of Beijing Municipal Commission of Education (Grant No. KM201910020014 to JL). Young Teacher Scientific Research Project of Beijing University of Agriculture (Grand No. 17ZK002 to JL).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ali, A., Raddatz, N., Pardo, J. M., Yun, D. J. (2021). HKT sodium and potassium transporters in Arabidopsis thaliana and related halophyte species. Physiol. Plant 171 (4), 546–558. doi: 10.1111/ppl.13166
Apse, M. P., Sottosanto, J. B., Blumwald, E. (2003). Vacuolar cation H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J. 36 (2), 229–239. doi: 10.1046/j.1365-313x.2003.01871.x
Barragán, V., Leidi, E. O., Andrés, Z., Rubio, L., De Luca, A., Fernández, J. A., et al. (2012). Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 24 (3), 1127–1142. doi: 10.1105/tpc.111.095273
Bassil, E., Blumwald, E. (2014). The ins and outs of intracellular ion homeostasis: NHX-type cation/H+ transporters. Curr. Opin. Plant Biol. 22, 1–6. doi: 10.1016/j.pbi.2014.08.002
Bassil, E., Coku, A., Blumwald, E. (2012). Cellular ion homeostasis: emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J. Exp. Bot. 63 (16), 5727–5740. doi: 10.1093/jxb/ers250
Batelli, G., Verslues, P. E., Agius, F., Qiu, Q., Fujii, H., Pan, S., et al. (2007). SOS2 promotes salt tolerance in part by interacting with the vacuolar H+-ATPase and upregulating its transport activity. Mol. Cell Biol. 27 (22), 7781–7790. doi: 10.1128/MCB.00430-07
Berthomieu, P., Conéjéro, G., Nublat, A., Brackenbury, W. J., Lambert, C., Savio, C., et al. (2003). Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J. 22 (9), 2004–2014. doi: 10.1093/emboj/cdg207
Binzel, M. L., Hess, F. D., Bressan, R. A., Hasegawa, P. M. (1988). Intracellular compartmentation of ions in salt adapted tobacco cells. Plant Physiol. 86 (2), 607–614. doi: 10.1104/pp.86.2.607
Blumwald, E., Aharon, G. S., Apse, M. P. (2000). Sodium transport in plant cells. Biochim. Biophys. Acta 1465 (1-2), 140–151. doi: 10.1016/S0005-2736(00)00135-8
Boer, A., Volkov, V. (2003). Logistics of water and salt transport through the plant: structure and functioning of the xylem. Plant Cell Environ. 26 (1), 87–101. doi: 10.1046/j.1365-3040.2003.00930.x
Bose, J., Rodrigo-Moreno, A., Lai, D., Xie, Y., Shen, W., Shabala, S. (2015). Rapid regulation of the plasma membrane H+-ATPase activity is essential to salinity tolerance in two halophyte species, Atriplex lentiformis and Chenopodium quinoa. Ann. Bot. 115 (3), 481–494. doi: 10.1093/aob/mcu219
Brini, F., Masmoudi, K. (2012). Ion transporters and abiotic stress tolerance in plants. ISRN Mol. Biol. 2012, 927436. doi: 10.5402/2012/927436
Cao, Y., Zhang, M., Liang, X., Li, F., Shi, Y., Yang, X., et al. (2020). Natural variation of an EF-hand Ca2+-binding-protein coding gene confers saline-alkaline tolerance in maize. Nat. Commun. 11 (1), 186. doi: 10.1038/s41467-019-14027-y
Cheng, N. H., Pittman, J. K., Shigaki, T., Lachmansingh, J., LeClere, S., Lahner, B., et al. (2005). Functional association of Arabidopsis CAX1 and CAX3 is required for normal growth and ion homeostasis. Plant Physiol. 138 (4), 2048–2060. doi: 10.1104/pp.105.061218
Choi, W. G., Toyota, M., Kim, S. H., Hilleary, R., Gilroy, S. (2014). Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. U.S.A. 111 (17), 6497–6502. doi: 10.1073/pnas.1319955111
Coskun, D., Britto, D. T., Jean, Y. K., Kabir, I., Tolay, I., Torun, A. A., et al. (2013). K+ efflux and retention in response to NaCl stress do not predict salt tolerance in contrasting genotypes of rice (Oryza sativa L.). PloS One 8 (2), e57767. doi: 10.1371/journal.pone.0057767
Cui, R. J., Li, A., Li, D., Yin, P. A., Chen, A., Qin, M., et al (2023). Ca2+-dependent TaCCD1 cooperates with TaSAUR215 to enhance plasma membrane H+-ATPase activity and alkali stress tolerance by inhibiting PP2C-mediated dephosphorylation of TaHA2 in wheat. Mol. Plant 16 (3), 571–587. doi: 10.1016/j.molp.2023.01.010
Davenport, R. J., Muñoz-Mayor, A., Jha, D., Essah, P. A., Rus, A., Tester, M. (2007). The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ. 30 (4), 497–507. doi: 10.1111/j.1365-3040.2007.01637.x
Dodd, A. N., Kudla, J., Sanders, D. (2010). The language of calcium signaling. Annu. Rev. Plant Biol. 61, 593–620. doi: 10.1146/annurev-arplant-070109-104628
Drozdowicz, Y. M., Rea, P. A. (2001). Vacuolar H+ pyrophosphatases: from the evolutionary backwaters into the mainstream. Trends Plant Sci. 6 (5), 206–211. doi: 10.1016/S1360-1385(01)01923-9
Duan, X., Yu, Y., Zhang, Y., Chen, C., Duanmu, H., Cao, L., et al. (2018). A potential efflux boron transporter gene GsBOR2, positively regulates Arabidopsis bicarbonate tolerance. Plant Sci. 274, 284–292. doi: 10.1016/j.plantsci.2018.05.032
Duscha, K., Martins Rodrigues, C., Müller, M., Wartenberg, R., Fliegel, L., Deitmer, J. W., et al. (2020). 14-3-3 Proteins and other candidates form protein-protein interactions with the cytosolic c-terminal end of SOS1 affecting its transport activity. Int. J. Mol. Sci. 21 (9), 3334. doi: 10.3390/ijms21093334
El Mahi, H., Perez-Hormaeche, J., De Luca, A., Villalta, I., Espartero, J., Gamez-Arjona, F., et al. (2019). A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice. Plant Physiol. 180 (2), 1046–1065. doi: 10.1104/pp.19.00324
Endler, A., Kesten, C., Schneider, R., Zhang, Y., Ivakov, A., Froehlich, A., et al. (2015). A mechanism for sustained cellulose synthesis during salt stress. Cell 162 (6), 1353–1364. doi: 10.1016/j.cell.2015.08.028
Essah, P. A., Davenport, R., Tester, M. (2003). Sodium influx and accumulation in Arabidopsis. Plant Physiol. 133 (1), 307–318. doi: 10.1104/pp.103.022178
Falhof, J., Pedersen, J. T., Fuglsang, A. T., Palmgren, M. (2016). Plasma membrane H+-ATPase regulation in the center of plant physiology. Mol. Plant 9 (3), 323–337. doi: 10.1016/j.molp.2015.11.002
Fang, S., Hou, X., Liang, X. (2021). Response mechanisms of plants under saline-alkali stress. Front. Plant Sci. 12, 667458. doi: 10.3389/fpls.2021.667458
Feng, W., Kita, D., Peaucelle, A., Cartwright, H. N., Doan, V., Duan, Q., et al. (2018). The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr. Biol. 28 (5), 666–675.e665. doi: 10.1016/j.cub.2018.01.023
Flowers, T. J., Munns, R., Colmer, T. D. (2015). Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Ann. Bot. 115 (3), 419–431. doi: 10.1093/aob/mcu217
Fu, H., Yu, X., Jiang, Y., Wang, Y., Yang, Y., Chen, S., et al. (2022). SALT OVERLY SENSITIVE 1 is inhibited by clade D Protein phosphatase 2C D6 and D7 in Arabidopsis thaliana. Plant Cell 35 (1), 279–297. doi: 10.1093/plcell/koac283
Fuglsang, A. T., Guo, Y., Cuin, T. A., Qiu, Q., Song, C., Kristiansen, K. A., et al. (2007). Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 19 (5), 1617–1634. doi: 10.1105/tpc.105.035626
Fuglsang, A. T., Visconti, S., Drumm, K., Jahn, T., Stensballe, A., Mattei, B., et al. (1999). Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr(946)-Thr-Val and requires phosphorylation of Thr(947). J. Biol. Chem. 274 (51), 36774–36780. doi: 10.1074/jbc.274.51.36774
Garciadeblás, B., Senn, M. E., Bañuelos, M. A., Rodríguez-Navarro, A. (2003). Sodium transport and HKT transporters: the rice model. Plant J. 34 (6), 788–801. doi: 10.1046/j.1365-313X.2003.01764.x
Guo, J., Chai, X., Mei, Y., Du, J., Du, H., Shi, H., et al. (2022). Acetylproteomics analyses reveal critical features of lysine-ϵ-acetylation in Arabidopsis and a role of 14-3-3 protein acetylation in alkaline response. Stress Biol. 2 (1), 19. doi: 10.1007/s44154-021-00024-z
Halfter, U., Ishitani, M., Zhu, J. K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. U.S.A. 97 (7), 3735–3740. doi: 10.1073/pnas.97.7.3735
Haruta, M., Sabat, G., Stecker, K., Minkoff, B. B., Sussman, M. R. (2014). A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343 (6169), 408–411. doi: 10.1126/science.1244454
Hasegawa, P. M., Bressan, R. A., Zhu, J. K., Bohnert, H. J. (2000). Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 463–499. doi: 10.1146/annurev.arplant.51.1.463
Hashimoto, K., Kudla, J. (2011). Calcium decoding mechanisms in plants. Biochimie 93 (12), 2054–2059. doi: 10.1016/j.biochi.2011.05.019
Horie, T., Costa, A., Kim, T. H., Han, M. J., Horie, R., Leung, H. Y., et al. (2007). Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J. 26 (12), 3003–3014. doi: 10.1038/sj.emboj.7601732
Horie, T., Horie, R., Chan, W. Y., Leung, H. Y., Schroeder, J. I. (2006). Calcium regulation of sodium hypersensitivities of sos3 and athkt1 mutants. Plant Cell Physiol. 47 (5), 622–633. doi: 10.1093/pcp/pcj029
Horie, T., Yoshida, K., Nakayama, H., Yamada, K., Oiki, S., Shinmyo, A. (2001). Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J. 27 (2), 129–138. doi: 10.1046/j.1365-313x.2001.01077.x
Hussain, S., Hussain, S., Ali, B., Ren, X., Chen, X., Li, Q., et al. (2021). Recent progress in understanding salinity tolerance in plants: Story of Na+/K+ balance and beyond. Plant Physiol. Biochem. 160, 239–256. doi: 10.1016/j.plaphy.2021.01.029
Ismail, A., El-Sharkawy, I., Sherif, S. (2020). Salt stress signals on demand: Cellular events in the right context. Int. J. Mol. Sci. 21 (11), 3918. doi: 10.3390/ijms21113918
Jia, X., Zhu, Y., Zhang, R., Zhu, Z., Zhao, T., Cheng, L., et al. (2020). Ionomic and metabolomic analyses reveal the resistance response mechanism to saline-alkali stress in Malus halliana seedlings. Plant Physiol. Biochem. 147, 77–90. doi: 10.1016/j.plaphy.2019.12.001
Jiang, Z., Zhou, X., Tao, M., Yuan, F., Liu, L., Wu, F., et al. (2019). Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 572 (7769), 341–346. doi: 10.1038/s41586-019-1449-z
Kiegle, E., Moore, C. A., Haseloff, J., Tester, M. A., Knight, M. R. (2000). Cell-type-specific calcium responses to drought, salt and cold in the Arabidopsis root. Plant J. 23 (2), 267–278. doi: 10.1046/j.1365-313x.2000.00786.x
Kim, W. Y., Ali, Z., Park, H. J., Park, S. J., Cha, J. Y., Perez-Hormaeche, J., et al. (2013). Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 4, 1352. doi: 10.1038/ncomms2357
Kim, B. G., Waadt, R., Cheong, Y. H., Pandey, G. K., Dominguez-Solis, J. R., Schültke, S., et al. (2007). The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J. 52 (3), 473–484. doi: 10.1111/j.1365-313X.2007.03249.x
Kotula, L., Garcia Caparros, P., Zorb, C., Colmer, T. D., Flowers, T. J. (2020). Improving crop salt tolerance using transgenic approaches: An update and physiological analysis. Plant Cell Environ. 43 (12), 2932–2956. doi: 10.1111/pce.13865
Laohavisit, A., Richards, S. L., Shabala, L., Chen, C., Colaço, R. D., Swarbreck, S. M., et al. (2013). Salinity-induced calcium signaling and root adaptation in Arabidopsis require the calcium regulatory protein annexin1. Plant Physiol. 163 (1), 253–262. doi: 10.1104/pp.113.217810
Li, J., Guo, Y., Yang, Y. (2022). The molecular mechanism of plasma membrane H+-ATPases in plant responses to abiotic stress. J. Genet. Genomics 49 (8), 715–725. doi: 10.1016/j.jgg.2022.05.007
Li, J., Shen, L., Han, X., He, G., Fan, W., Li, Y., et al. (2023). Phosphatidic acid–regulated SOS2 controls sodium and potassium homeostasis in Arabidopsis under salt stress. EMBO J. 42 (8), e112401. doi: 10.15252/embj.2022112401
Li, J., Zhou, H., Zhang, Y., Li, Z., Yang, Y., Guo, Y. (2020). The GSK3-like kinase BIN2 is a molecular switch between the salt stress response and growth recovery in Arabidopsis thaliana. Dev. Cell 55 (3), 367–380.e366. doi: 10.1016/j.devcel.2020.08.005
Lin, H., Du, W., Yang, Y., Schumaker, K. S., Guo, Y. (2014). A calcium-independent activation of the Arabidopsis SOS2-like protein kinase24 by its interacting SOS3-like calcium binding protein1. Plant Physiol. 164 (4), 2197–2206. doi: 10.1104/pp.113.232272
Lin, H., Yang, Y., Quan, R., Mendoza, I., Wu, Y., Du, W., et al. (2009). Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 21 (5), 1607–1619. doi: 10.1105/tpc.109.066217
Liu, X., Jiang, W., Li, Y., Nie, H., Cui, L., Li, R., et al. (2023). FERONIA coordinates plant growth and salt tolerance via the phosphorylation of phyB. Nat. Plants 9, 645–660. doi: 10.1038/s41477-023-01390-4
Liu, L., Song, W., Huang, S., Jiang, K., Moriwaki, Y., Wang, Y., et al. (2022). Extracellular pH sensing by plant cell-surface peptide-receptor complexes. Cell 185 (18), 3341–3355.e3313. doi: 10.1016/j.cell.2022.07.012
Ma, L., Ye, J., Yang, Y., Lin, H., Yue, L., Luo, J., et al. (2019). The SOS2-SCaBP8 complex generates and fine-tunes an AtANN4-dependent calcium signature under salt stress. Dev. Cell 48 (5), 697–709 e695. doi: 10.1016/j.devcel.2019.02.010
Ma, L., Han, R., Yang, Y., Liu, X., Li, H., Zhao, X., et al (2023). Phytochromes enhance SOS2-mediated PIF1 and PIF3 phosphorylation and degradation to promote Arabidopsis salt tolerance. Plant Cell 35 (8), 2997–3020. doi: 10.1093/plcell/koad117
Mansour, M. M. F., Hassan, F. A. S. (2022). How salt stress-responsive proteins regulate plant adaptation to saline conditions. Plant Mol. Biol. 108 (3), 175–224. doi: 10.1007/s11103-021-01232-x
Martí, M. C., Stancombe, M. A., Webb, A. A. (2013). Cell- and stimulus type-specific intracellular free Ca2+ signals in Arabidopsis. Plant Physiol. 163 (2), 625–634. doi: 10.1104/pp.113.222901
Mäser, P., Eckelman, B., Vaidyanathan, R., Horie, T., Fairbairn, D. J., Kubo, M., et al. (2002). Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 531 (2), 157–161. doi: 10.1016/S0014-5793(02)03488-9
Møller, I. S., Gilliham, M., Jha, D., Mayo, G. M., Roy, S. J., Coates, J. C., et al. (2009). Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21 (7), 2163–2178. doi: 10.1105/tpc.108.064568
Monihan, S. M., Magness, C. A., Yadegari, R., Smith, S. E., Schumaker, K. S. (2016). Arabidopsis CALCINEURIN B-LIKE10 functions independently of the SOS pathway during reproductive development in saline conditions. Plant Physiol. 171 (1), 369–379. doi: 10.1104/pp.16.00334
Morton, M. J. L., Awlia, M., Al-Tamimi, N., Saade, S., Pailles, Y., Negrão, S., et al. (2019). Salt stress under the scalpel - dissecting the genetics of salt tolerance. Plant J. 97 (1), 148–163. doi: 10.1111/tpj.14189
Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Ohta, M., Guo, Y., Halfter, U., Zhu, J. K. (2003). A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. U.S.A. 100 (20), 11771–11776. doi: 10.1073/pnas.2034853100
Palmgren, M. G. (2001). PLANT PLASMA MEMBRANE H+-ATPases: Powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 817–845. doi: 10.1146/annurev.arplant.52.1.817
Pardo, J. M., Cubero, B., Leidi, E. O., Quintero, F. J. (2006). Alkali cation exchangers: roles in cellular homeostasis and stress tolerance. J. Exp. Bot. 57 (5), 1181–1199. doi: 10.1093/jxb/erj114
Parrotta, L., Guerriero, G., Sergeant, K., Cai, G., Hausman, J. F. (2015). Target or barrier? The cell wall of early- and later-diverging plants vs cadmium toxicity: differences in the response mechanisms. Front. Plant Sci. 6, 133. doi: 10.3389/fpls.2015.00133
Pei, S., Liu, Y., Li, W., Krichilsky, B., Dai, S., Wang, Y., et al. (2022). OSCA1 is an osmotic specific sensor: a method to distinguish Ca2+-mediated osmotic and ionic perception. New Phytol. 235 (4), 1665–1678. doi: 10.1111/nph.18217
Platten, J. D., Cotsaftis, O., Berthomieu, P., Bohnert, H., Davenport, R. J., Fairbairn, D. J., et al. (2006). Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci. 11 (8), 372–374. doi: 10.1016/j.tplants.2006.06.001
Qiu, Q. S., Guo, Y., Dietrich, M. A., Schumaker, K. S., Zhu, J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U.S.A. 99 (12), 8436–8441. doi: 10.1073/pnas.122224699
Qiu, Q. S., Guo, Y., Quintero, F. J., Pardo, J. M., Schumaker, K. S., Zhu, J. K. (2004). Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J. Biol. Chem. 279 (1), 207–215. doi: 10.1074/jbc.M307982200
Quan, R., Lin, H., Mendoza, I., Zhang, Y., Cao, W., Yang, Y., et al. (2007). SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell 19 (4), 1415–1431. doi: 10.1105/tpc.106.042291
Quintero, F. J., Martinez-Atienza, J., Villalta, I., Jiang, X., Kim, W. Y., Ali, Z., et al. (2011). Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. U.S.A. 108 (6), 2611–2616. doi: 10.1073/pnas.1018921108
Quintero, F. J., Ohta, M., Shi, H., Zhu, J. K., Pardo, J. M. (2002). Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. U.S.A. 99 (13), 9061–9066. doi: 10.1073/pnas.132092099
Raddatz, N., Morales de Los Ríos, L., Lindahl, M., Quintero, F. J., Pardo, J. M. (2020). Coordinated transport of nitrate, potassium, and sodium. Front. Plant Sci. 11, 247. doi: 10.3389/fpls.2020.00247
Ren, Z. H., Gao, J. P., Li, L. G., Cai, X. L., Huang, W., Chao, D. Y., et al. (2005). A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 37 (10), 1141–1146. doi: 10.1038/ng1643
Ren, X. L., Qi, G. N., Feng, H. Q., Zhao, S., Zhao, S. S., Wang, Y., et al. (2013). Calcineurin B-like protein CBL10 directly interacts with AKT1 and modulates K+ homeostasis in Arabidopsis. Plant J. 74 (2), 258–266. doi: 10.1111/tpj.12123
Rubio, F., Gassmann, W., Schroeder, J. I. (1995). Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270 (5242), 1660–1663. doi: 10.1126/science.270.5242.1660
Rus, A., Lee, B. H., Muñoz-Mayor, A., Sharkhuu, A., Miura, K., Zhu, J. K., et al. (2004). AtHKT1 facilitates Na+ homeostasis and K+ nutrition in planta. Plant Physiol. 136 (1), 2500–2511. doi: 10.1104/pp.104.042234
Schachtman, D. P., Schroeder, J. I. (1994). Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 370 (6491), 655–658. doi: 10.1038/370655a0
Sewelam, N., Oshima, Y., Mitsuda, N., Ohme-Takagi, M. (2014). A step towards understanding plant responses to multiple environmental stresses: a genome-wide study. Plant Cell Environ. 37 (9), 2024–2035. doi: 10.1111/pce.12274
Shen, L., Zhuang, B., Wu, Q., Zhang, H., Nie, J., Jing, W., et al. (2019). Phosphatidic acid promotes the activation and plasma membrane localization of MKK7 and MKK9 in response to salt stress. Plant Sci. 287, 110190. doi: 10.1016/j.plantsci.2019.110190
Shi, H., Ishitani, M., Kim, C., Zhu, J. K. (2000). The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. U.S.A. 97 (12), 6896–6901. doi: 10.1073/pnas.120170197
Shi, H., Quintero, F. J., Pardo, J. M., Zhu, J. K. (2002). The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14 (2), 465–477. doi: 10.1105/tpc.010371
Steinhorst, L., He, G., Moore, L. K., Schültke, S., Schmitz-Thom, I., Cao, Y., et al. (2022). A Ca2+-sensor switch for tolerance to elevated salt stress in Arabidopsis. Dev. Cell 57 (17), 2081–2094.e2087. doi: 10.1016/j.devcel.2022.08.001
Sunarpi, Horie, T., Motoda, J., Kubo, M., Yang, H., Yoda, K., et al. (2005). Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J. 44 (6), 928–938. doi: 10.1111/j.1365-313X.2005.02595.x
Tasseva, G., Richard, L., Zachowski, A. (2004). Regulation of phosphatidylcholine biosynthesis under salt stress involves choline kinases in Arabidopsis thaliana. FEBS Lett. 566 (1-3), 115–120. doi: 10.1016/j.febslet.2004.04.015
Tuteja, N. (2007). Mechanisms of high salinity tolerance in plants. Methods Enzymol. 428, 419–438. doi: 10.1016/S0076-6879(07)28024-3
Uozumi, N., Kim, E. J., Rubio, F., Yamaguchi, T., Muto, S., Tsuboi, A., et al. (2000). The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 122 (4), 1249–1259. doi: 10.1104/pp.122.4.1249
Van der Does, D., Boutrot, F., Engelsdorf, T. (2017). The Arabidopsis leucine-rich repeat receptor kinase MIK2/LRR-KISS connects cell wall integrity sensing, root growth and response to abiotic and biotic stresses. PloS Genet. 13 (6), e1006832. doi: 10.1371/journal.pgen.1006832
van Zelm, E., Zhang, Y., Testerink, C. (2020). Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 71, 403–433. doi: 10.1146/annurev-arplant-050718-100005
Wang, M., Wang, Y., Sun, J., Ding, M., Deng, S., Hou, P., et al. (2013). Overexpression of PeHA1 enhances hydrogen peroxide signaling in salt-stressed Arabidopsis. Plant Physiol. Biochem. 71, 37–48. doi: 10.1016/j.plaphy.2013.06.020
Wang, P., Zhao, Y., Li, Z., Hsu, C.-C., Liu, X., Fu, L., et al. (2018). Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell 69 (1), 100–112.e106. doi: 10.1016/j.molcel.2017.12.002
Xiang, G., Ma, W., Gao, S., Jin, Z., Yue, Q., Yao, Y. (2019). Transcriptomic and phosphoproteomic profiling and metabolite analyses reveal the mechanism of NaHCO3-induced organic acid secretion in grapevine roots. BMC Plant Biol. 19 (1), 383. doi: 10.1186/s12870-019-1990-9
Xu, S. L., Rahman, A., Baskin, T. I., Kieber, J. J. (2008). Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 20 (11), 3065–3079. doi: 10.1105/tpc.108.063354
Yamaguchi, T., Hamamoto, S., Uozumi, N. (2013). Sodium transport system in plant cells. Front. Plant Sci. 4, 410. doi: 10.3389/fpls.2013.00410
Yang, Y., Guo, Y. (2018a). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217 (2), 523–539. doi: 10.1111/nph.14920
Yang, Y., Guo, Y. (2018b). Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 60 (9), 796–804. doi: 10.1111/jipb.12689
Yang, Y., Han, X., Ma, L., Wu, Y., Liu, X., Fu, H., et al. (2021). Dynamic changes of phosphatidylinositol and phosphatidylinositol 4-phosphate levels modulate H+-ATPase and Na+/H+ antiporter activities to maintain ion homeostasis in Arabidopsis under salt stress. Mol. Plant 14 (12), 2000–2014. doi: 10.1016/j.molp.2021.07.020
Yang, Y., Qin, Y., Xie, C., Zhao, F., Zhao, J., Liu, D., et al. (2010). The Arabidopsis chaperone J3 regulates the plasma membrane H+-ATPase through interaction with the PKS5 kinase. Plant Cell 22 (4), 1313–1332. doi: 10.1105/tpc.109.069609
Yang, Z., Wang, C., Xue, Y., Liu, X., Chen, S., Song, C., et al. (2019). Calcium-activated 14-3-3 proteins as a molecular switch in salt stress tolerance. Nat. Commun. 10 (1), 1199. doi: 10.1038/s41467-019-09181-2
Yang, Y., Wu, Y., Ma, L., Yang, Z., Dong, Q., Li, Q., et al. (2019). The Ca2+ sensor SCaBP3/CBL7 modulates plasma membrane H+-ATPase activity and promotes alkali tolerance in Arabidopsis. Plant Cell 31 (6), 1367–1384. doi: 10.1105/tpc.18.00568
Yao, J., Shen, Z., Zhang, Y., Wu, X., Wang, J., Sa, G., et al. (2019). Populus euphratica WRKY1 binds the promoter of H+-ATPase gene to enhance gene expression and salt tolerance. J. Exp. Bot. 71 (4), 1527–1539. doi: 10.1093/jxb/erz493
Yokoi, S., Quintero, F. J., Cubero, B., Ruiz, M. T., Bressan, R. A., Hasegawa, P. M., et al. (2002). Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J. 30 (5), 529–539. doi: 10.1046/j.1365-313X.2002.01309.x
Yu, Y., Liu, A., Duan, X., Wang, S., Sun, X., Duanmu, H., et al. (2016). GsERF6, an ethylene-responsive factor from Glycine soja, mediates the regulation of plant bicarbonate tolerance in Arabidopsis. Planta 244 (3), 681–698. doi: 10.1007/s00425-016-2532-4
Yuan, F., Yang, H., Xue, Y., Kong, D., Ye, R., Li, C., et al. (2014). OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 514 (7522), 367–371. doi: 10.1038/nature13593
Zhang, H., Yu, F., Xie, P., Sun, S., Qiao, X., Tang, S., et al. (2023). A Gγ protein regulates alkaline sensitivity in crops. Science 379 (6638), doi: eade8416. doi: 10.1126/science.ade8416
Zhang, Y., Yao, J., Yin, K., Liu, Z., Zhang, Y., Deng, C., et al. (2022). Populus euphratica Phospholipase Dδ increases salt tolerance by regulating K+/Na+ and ROS homeostasis in Arabidopsis. Int. J. Mol. Sci. 23 (9), 4911. doi: 10.3390/ijms24098208
Keywords: saline-alkali stress, ion homeostasis, pH homeostasis, plant cell, abiotic stress
Citation: Li J and Yang Y (2023) How do plants maintain pH and ion homeostasis under saline-alkali stress? Front. Plant Sci. 14:1217193. doi: 10.3389/fpls.2023.1217193
Received: 05 May 2023; Accepted: 25 September 2023;
Published: 17 October 2023.
Edited by:
Habib-ur-Rehman Athar, Bahauddin Zakariya University, PakistanReviewed by:
Jian Sun, Jiangsu Normal University, ChinaCopyright © 2023 Li and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongqing Yang, eWFuZ3lvbmdxaW5nQGNhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.