- 1Research and Development of Center, China Tobacco Yunnan Industrial Co., Ltd., Kunming, China
- 2School of Ethnic Medicine, Yunnan Minzu University, Kunming, China
- 3Technology Center, China Tobacco Yunnan Industrial Co., Ltd., Kunming, China
- 4Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, Beijing, China
- 5University of Chinese Academy of Sciences, Beijing, China
Background: Nicotiana tabacum is an important economic crop, which is widely planted in the world. Lignin is very important for maintaining the physiological and stress-resistant functions of tobacco. However, higher lignin content will produce lignin gas, which is not conducive to the formation of tobacco quality. To date, how to precisely fine-tune lignin content or composition remains unclear.
Results: Here, we annotated and screened 14 CCoAOMTs in Nicotiana tabacum and obtained homozygous double mutants of CCoAOMT6 and CCoAOMT6L through CRSIPR/Cas9 technology. The phenotype showed that the double mutants have better growth than the wild type whereas the S/G ratio increased and the total sugar decreased. Resistance against the pathogen test and the extract inhibition test showed that the transgenic tobacco has stronger resistance to tobacco bacterial wilt and brown spot disease, which are infected by Ralstonia solanacearum and Alternaria alternata, respectively. The combined analysis of metabolome and transcriptome in the leaves and roots suggested that the changes of phenylpropane and terpene metabolism are mainly responsible for these phenotypes. Furthermore, the molecular docking indicated that the upregulated metabolites, such as soyasaponin Bb, improve the disease resistance due to highly stable binding with tyrosyl-tRNA synthetase targets in Ralstonia solanacearum and Alternaria alternata.
Conclusions: CAFFEOYL-COA 3-O-METHYLTRANSFERASE 6/6L can regulate the S/G ratio of lignin monomers and may affect tobacco bacterial wilt and brown spot disease resistance by disturbing phenylpropane and terpene metabolism in leaves and roots of Nicotiana tabacum, such as soyasaponin Bb.
Introduction
Nicotiana tabacum is an important economic crop, which is planted worldwide. Tobacco production involves many processes such as planting and baking, among which resistance, growth, baking ability, quality, and other indicators are key traits for evaluating excellent varieties. In the field, the bacterial wilt and brown spot are bacterial and fungal diseases caused by the infection of roots and leaves by Ralstonia solanacearum and Alternaria alternata, respectively, which are the most serious threats to the quality of tobacco leaves (Tong et al., 2012; Lan et al., 2014). The application of resistant varieties is one of the effective approaches for disease control and key trait improvement, and these are also closely related to metabolism during tobacco cultivation (Lebeau et al., 2011).
Lignin is a kind of phenolic macromolecular polymer in plants, which is the main component of the secondary cell wall. It widely exists in tobacco and has important physiological functions. For example (1), it enhances the physical and mechanical properties of plants and improves the strength of stems (Jung and Ni, 1998); (2) it is conducive to the transportation of water and nutrients and improves the water retention capacity of plants (Bernard-Vailhé et al., 1996; Hopkins et al., 2001; Pedersen et al., 2005; Webster et al., 2005); and (3) it provides defense against biological invasion such as pathogens and nematodes, as well as abiotic stresses such as drought and low temperature (Pellegrini et al., 1993; Martz et al., 1998; Maury et al., 1999; Guillaumie et al., 2009). Generally, lowering the lignin content will cause abnormal growth or even death of plants, so it plays an important role in maintaining normal growth and development of plants. However, higher lignin content will produce lignin gas, which is not conducive to the formation of tobacco quality (Li et al., 2011; Zang et al., 2015; Jin et al., 2018). Therefore, the precise fine-tuning lignin content or composition will become a breakthrough to explore better-performance tobacco varieties.
Lignin is composed of three kinds of monomers, namely, guaiacyl lignin (G-monomer), syringyl lignin (S-monomer), and hydroxy-phenyl lignin (H-monomer). Its biosynthesis includes monomer synthesis and monomer polymerization (Rastogi and Dwivedi, 2008; Li et al., 2010; Vanholme et al., 2010). The biosynthesis of lignin monomer needs to go through the shikimic acid pathway, the phenylpropanoid pathway, and the specific lignin pathway in turn. Phenylalanine undergoes deamination reaction, hydroxylation, methylation reaction, redox reaction, and a series of reactions to finally produce three monomers (Li et al., 2010; Vanholme et al., 2010). Caffeoyl-CoA 3-O-methyltransferase (CCoAOMT) and caffeic acid 3-O-methyltransferase (COMT) are two key enzymes in the specific lignin pathway (Xie et al., 2019).
CCoAOMT can catalyze the phenol hydroxymethylation at the C3 position on the benzene ring of caffeoyl-CoA, that is, the formation of the G-monomer; COMT can catalyze C3 and C5 positions of the benzene ring at the same time, that is, the S-monomer (Li et al., 2010). The free C-5 position on the G-monomer can react with other monomers to form a stable C–C cross link, which is relatively difficult to separate and degrade, whereas the S-monomer is relatively easy to degrade, so the S/G ratio is often used as an important indicator to evaluate lignin (Jung and Ni, 1998; Christensen et al., 2000; Rastogi and Dwivedi, 2008; Verma and Dwivedi, 2014). In Arabidopsis, the mutation of AtCCoAOMT1 does not affect the growth and development of plants but can reduce the biosynthesis of G-monomers and thus increase the S/G ratio (Xie et al., 2019); however, the double mutant comtccomt will cause abnormal growth and development, leading to death (Do et al., 2007; Nelson et al., 2017).
In this study, we annotated and screened CCoAOMTs in Nicotiana tabacum and obtained homozygous double mutants of CCoAOMT6 and CCoAOMT6L through CRSIPR/Cas9 technology. It was found that the double mutants have better growth and disease resistance than the wild type whereas the S/G ratio increased and the total sugar decreased. Furthermore, through the multi-omics and molecular docking, we considered that S lignin and upregulated compounds, such as soyasaponin Bb, may enhance tobacco growth and resistance. CCoAOMT6/6L may be the potential genes for tobacco breeding.
Results
Identification of the CCoAOMT gene family in tobacco
There were 14 predicted NtCCoAOMT genes that were searched from the Nicotiana tabacum genome in the NCBI database, of which nine genes were explicitly annotated (Figure 1A). The NtCCoAOMT family generally contains around 240 amino acids, all of which contain the caffeoyl-CoA O-methyltransferase domain (PLN02589 or PLN02781). A phylogenetic tree was constructed using seven AtCCoAOMTs from Arabidopsis thaliana and the above 14 NtCCoAOMTs. The results showed that LOC107826706, LOC107792528, LOC107795104, LOC107799225, and LOC107832128 were clustered together with AtCCoAOMT7 (At4g26220), whereas NtCCoAOMT1-6, 3L, 5L, 6L clade with AtCCoAOMT1 (At4g34050), which was confirmed to be the key member involved in the monolignol pathway (Do et al., 2007) (Figure 1A). Furthermore, we analyze the tissue expression level of these nine NtCCoAOMTs by qRT-PCR and found that NtCCoAOMT6 was the most highly expressed in tobacco roots and leaves (Figures 1B, C), suggesting its critical position in the monolignol biosynthesis. NtCCoAOMT6L shares 95.7% sequence identity and similar expression patterns with NtCCoAOMT6. Because the common tobacco is an allotetraploid formed by combining different chromosomes of two wild species N. sylvestris and N. tomentosiformis (Sierro et al., 2014), we compared NtCCoAOMT6 and NtCCoAOMT6L in these two wild species, respectively. It shows that NtCCoAOMT6 originates from N. tomentosiformis (99% identities), whereas NtCCoAOMT6L originates from N. sylvestris (100% identities), which means they are orthologous genes from different subgenomes. Thus, we created loss-of-function mutants for these two genes in Nicotiana tabacum.
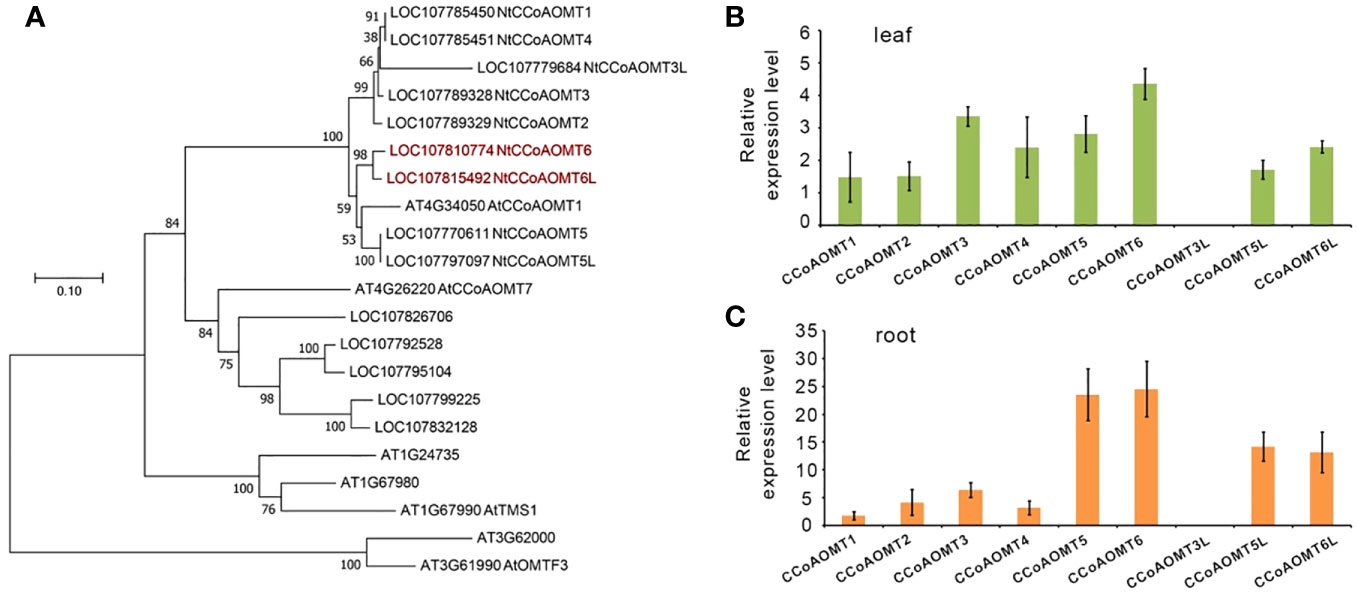
Figure 1 The CCoAOMT gene family in Nicotiana tabacum. (A) The phylogenetic tree of the CCoAOMT gene family in Nicotiana tabacum and Arabidopsis thaliana. A neighbor-joining (NJ) tree is reconstructed using the MEGA program version 7.0. Sequences are aligned by Muscle. Bootstrap values (expressed as percentages of 1,000 replications) are shown at branch points. (B) Relative expression analysis of the primary CCoAOMTs in the leaf. (C) Relative expression analysis of the primary CCoAOMTs in the root.
Generation and characterization of double mutant ccoaomt6 ccoaomt6l
We used CRISPR/Cas9 technology to generate a ccoaomt6 ccoaomt6l double mutant. A common sgRNA target (5′-TTGCCCGTAATCCAAAAGGCTGG-3′) in the two genes was selected (Figure 2A). The CRISPR construct was transformed into wild-type Nicotiana tabacum, and the homozygous double mutant was obtained. The double mutant contained a nucleotide insertion in the target sites of CCoAOMT6 and CCoAOMT6L, leading to frameshift mutation, separately (Figure 2B). Under natural conditions, compared with the wild type, the mutant has a longer squaring stage, darker green leaves, and vigorous growth and development (Figure 2C). Statistical analysis showed that the plant height, leaf length, and leaf width were significantly increased (p < 0.01) (Figure 2D).
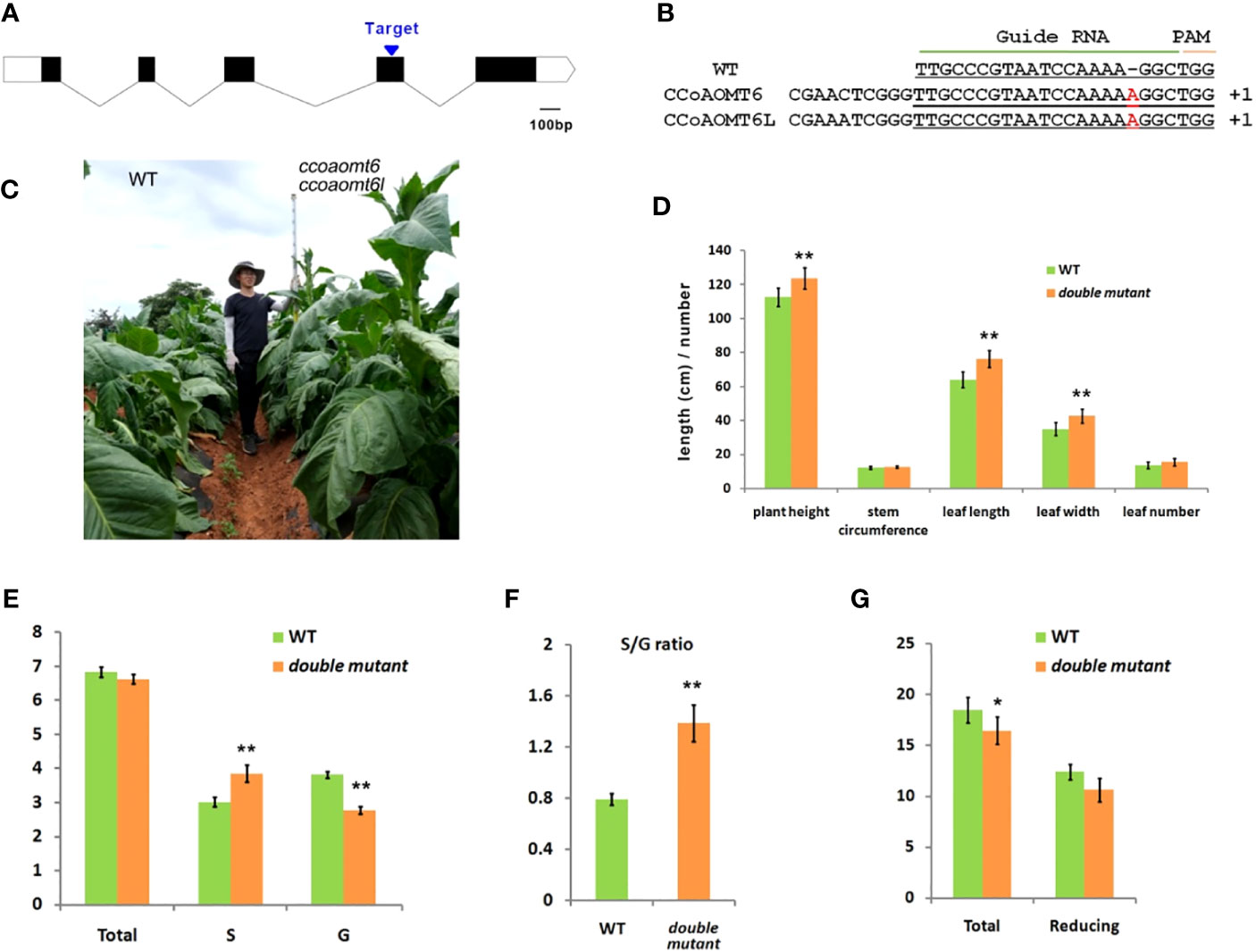
Figure 2 Sequence and phenotype of the CRISPR/Cas9 mutant. (A) Diagram of CCoAOMT6 and CCoAOMT6L showing the target site for CRISPR/Cas9 technology. The CDS and UTR regions are indicated by the black box and white box, separately. (B) The sequence in the ccoaomt6ccoaomt6l double mutant. (C) The appearance of the WT (left) and ccoaomt6ccoaomt6l double mutant (right). (D) The agronomic traits of WT and ccoaomt6ccoaomt6l double mutant. The data are presented as mean ± s.d., n = 10. (E) Determination of total lignin and lignin monomer content. (F) Determination of the S/G ratio between the wild-type and double mutant. (G) Determination of total sugar and reducing sugar content. The data are presented as mean ± s.d., n = 5. *P < 0.05, **P < 0.01, Student’s tests.
Knockout of CCoAOMT6 and CCoAOMT6L increases the S/G ratio of lignin monomers
CCoAOMT is the key methylation enzyme of caffeoyl-CoA, which produces feruloyl-CoA, and is the upstream component of G-lignin and S-lignin monomers. Therefore, we also detected the total lignin content and lignin monomer content in the wild type and the double mutant. The results showed that there was no significant difference in total lignin content between the mutant and the control; however, S-lignin content increased by around 27.5% and G-lignin content decreased by around 24.3% (Figure 2E), thus increasing the S/G ratio by nearly 68.4% (Figure 2F). The above results prove that tobacco CCoAOMT can control the type of lignin monomer by influencing the step of caffeoyl-CoA converting to feruloyl-CoA.
Furthermore, we detected the content of total sugar and reducing sugar that are important for plant growth and development. Compared with the wild type, the total sugar content in the double mutant leaves decreased by 11.0%, and the reducing sugar content decreased by 14.3% (Figure 2G), indicating that the decomposition of sugar may have promoted the synthesis of lignin monomers. In general, the mutation of CCoAOMT6 and CCoAOMT6L has improved the economic traits of tobacco.
Knockout of CCoAOMT6 and CCoAOMT6L improves tobacco disease resistance
Bacterial wilt and brown spot are the main diseases of tobacco plants, caused by Ralstonia solanacearum infection and Alternaria alternata infection, respectively. To further study the effect of the changes in lignin monomer composition caused by CCoAOMT6 and CCoAOMT6L knockout on tobacco disease resistance, Ralstonia solanacearum and Alternaria alternata strains were inoculated on the wild-type and double-mutant leaves, respectively, according to the methods of Guo et al. (2004)and Ma et al. (2017). The disease lesion diameter around the inoculation sites was measured after 72 h of inoculation. It was found that on the wild-type tobacco leaves, the bacterial wilt disease symptoms were obvious and the brown spot disease symptoms developed rapidly. For the mutant leaves, however, the bacterial wilt disease either did not appear or developed slowly, and the development of brown spot diseases disease symptoms is relatively slow (Figures 3A–C).
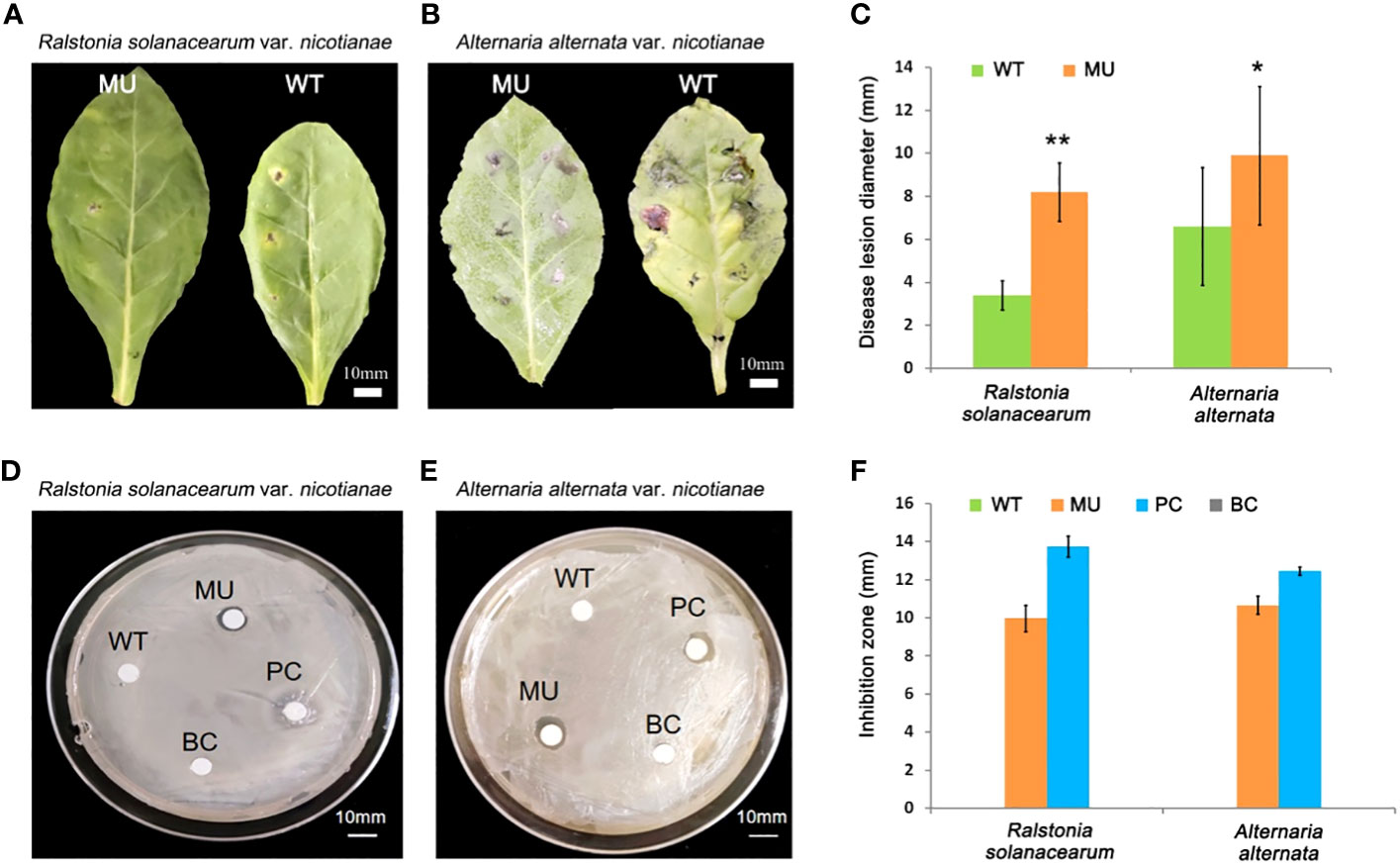
Figure 3 Disease resistance test of the CRISPR/Cas9 mutant. (A, B) Pathogenicity of tobacco bacterial wilt disease (A) and brown spot disease (B). Bars = 10 mm. (C) Statistics of the disease lesion diameter against tobacco bacterial wilt disease and brown spot disease. MU, the double mutant; WT, the wild type. (D, E) Inhibition test of Ralstonia solanacearum (D) and Alternaria alternata (E). MU, double-mutant tobacco leaf extract; WT, WT tobacco leaf extract; PC, 1 mg/mL cefotaxime sodium–95% ethanol solution (positive control); BC, 95% ethanol (blank control). (F) Statistics of the inhibition zone diameter against tobacco bacterial wilt disease and brown spot disease. Values show as mean ± s.d., n = 5. *P < 0.05, **P < 0.01, Student’s tests.
Similarly, we also investigated the inhibitory effect of transgenic tobacco leaf extract on the growth of Ralstonia solanacearum and Alternaria alternata, according to the protocol provided by Guo et al (Guo et al., 2004). The results showed that the ethanol extract of transgenic tobacco leaves indeed inhibited the growth of Ralstonia solanacearum and Alternaria alternata, whereas the extract of wild-type leaves did not (Figures 3D–F). This indicates that compounds in the mutant tobacco extract has a “chemical defense” response to Ralstonia solanacearum and Alternaria alternata pathogens. Taken together, it is proved that the knockout of CCoAOMT6 and CCoAOMT6L can enhance the disease resistance of tobacco.
Knockout of CCoAOMT6 and CCoAOMT6L altered the primary and secondary metabolites
To investigate the effects of S/G-lignin ratio changes on the whole plant metabolism, widely targeted metabolome sequencing was performed between the leaves and roots of the wild type and double mutants. A total of 266 metabolites were detected, including carbohydrates, amino acids and derivatives, organic acids and derivatives, lipids, terpenes and sterols, phenolic acids, nucleosides, flavonoid, and coumarin; different metabolites of roots and leaves were screened (Tables S1, S2).
Compared with the wild type, 56 metabolites were differently detected from leaves of the double mutant, i.e., 15 upregulated and 41 downregulated (Figure S2A). Among them, eight differential amino acid and derivatives (eight downregulated) were identified, namely, histidine, leucine, tyrosine, phenylalanine, targinine, methionine, arginine, and proline. Moreover, there were 17 differential metabolites in the fatty acid pathway, of which four were upregulated and 13 were downregulated. Also, six organic acids were identified and all of them downregulated in the mutant line. There were five differential terpenoids or steroids identified, including two downregulated (campesterol, scoparone) and upregulated (asiatic acid, epiandrosterone, soyasaponin Bb). It also included nine metabolites involved in the shikimate pathway (five downregulated and four upregulated), namely, cinnamic acid, 4-coumaric acid, coumaroylquinic acid, one coumarin, two lignin, and three flavonoids. Scopoletin belongs to coumarin, which was downregulated in the mutant. Lactose and alpha-D-glucose-1,6-diphosphate were involved in the carbohydrate metabolic pathway, both of which were upregulated in the mutant (Figure 4).
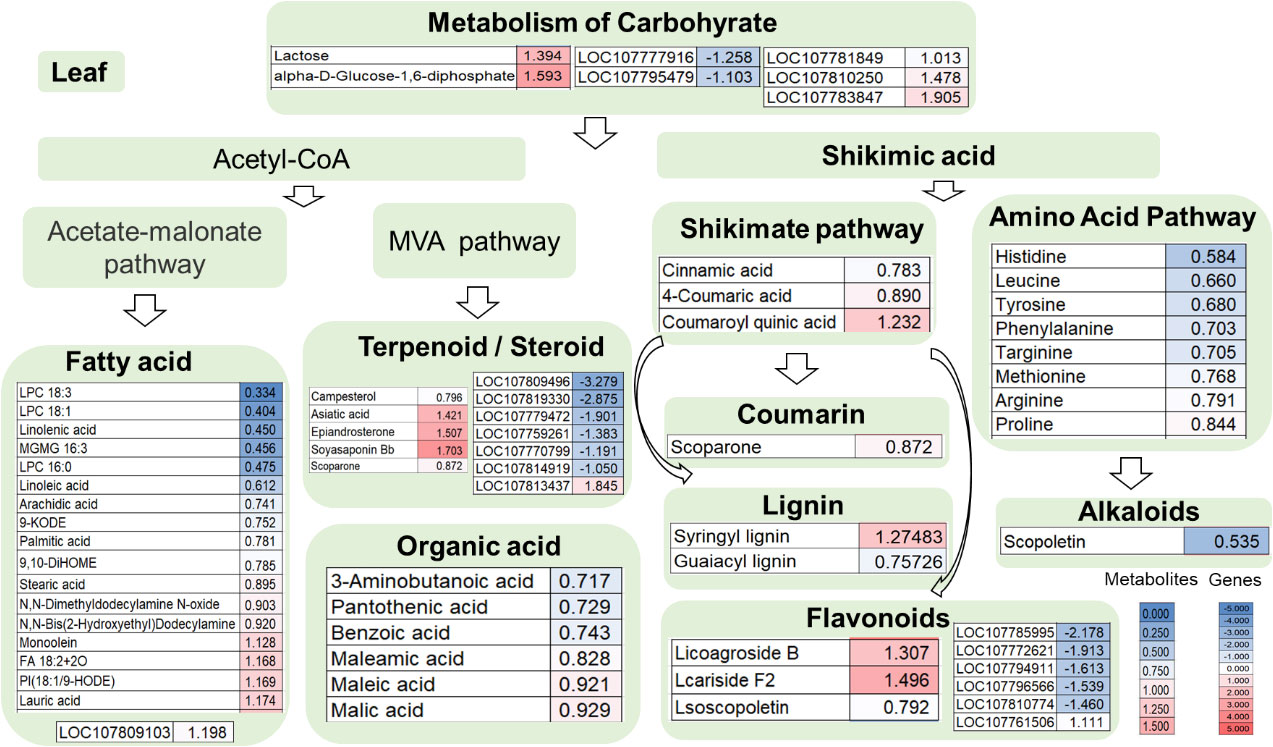
Figure 4 Leaf transcriptome combined with metabolome analysis in the double mutant. The heat map scale represents the relative level of metabolites and gene expression.
Meanwhile, 32 differential metabolites were identified from the roots between the double mutant and the wild type, of which 11 were upregulated and 21 were downregulated (Figure S2B). Among them, three differential flavonoid and derivatives (two down and one up) were identified, namely cyanidin-3-O-galactoside, hyperoside, and plantamajoside. Also, two metabolites were found in the fatty acid pathway, of which one was upregulated and one was down-regulated. Moreover, seven organic acids were identified of which three were upregulated and four were downregulated and 13 differential metabolites in the shikimate pathway (nine down and four up) were identified, one coumarin (caffeoylquinic acid up), two anthocyanin (two down), and five flavonoids (two down and three up). Nicotine was also identified, which was downregulated in the mutant line. N,N-Bis (2-hydroxyethyl)dodecylamine was involved in carbohydrate metabolism, which was downregulated in the mutant line (Figure 5).
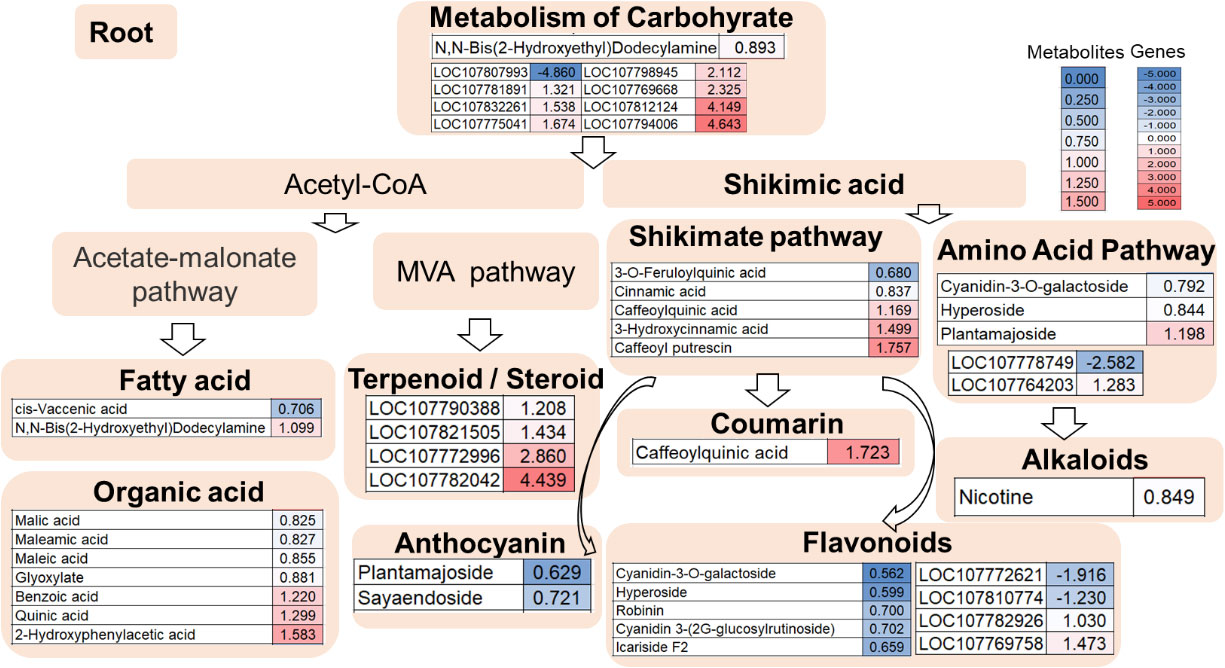
Figure 5 Root transcriptome combined with metabolome analysis in the double mutant. The heat map scale represents the relative level of metabolites and gene expression.
Knockout of CCoAOMT6 and CCoAOMT6L altered the gene expression in the primary and secondary metabolic pathways
To better analyze the potential roles of CCoAOMT6 and CCoAOMT6L in primary and secondary metabolic regulation, we conducted transcriptome profiling of the leaves and roots in the double mutant and the wild type. A total of 248 DEGs (differentially expressed genes) were detected in the leaf data, which were composed of 226 genes that were upregulated and 22 genes that were downregulated (Figures S3A, B). Moreover, 332 DEGs were detected in the root data, composed of 149 upregulated genes and 183 downregulated genes (Figures S3A, C). Only 38 DEGs were identified in both leaves and root, suggesting that the functions of CCoAOMT6 and CCoAOMT6L in different tissues were slightly different (Figure S3A).
All the DEGs were annotated according to GO, KEGG, TF, COG, uniProt, InterPro, and WoLF PSORT. The GO terms “protein folding,” “protein complex oligomerization,” “response to hydrogen peroxide,” and “response to reactive oxygen species” are the most highly enriched in both leave and root data (Figure S4). KEGG analysis of the leaf data showed that “protein processing in endoplasmic reticulum,” “vitamin B6 metabolism,” “stilbenoid, diarylheptanoid and gingerol biosynthesis,” “sesquiterpenoid and triterpenoid biosynthesis,” and “flavonoid biosynthesis” were significantly enriched (Figure S5A). Meanwhile, KEGG analysis of the root data showed that “protein processing in endoplasmic reticulum,” “galactose metabolism,” “diterpenoid biosynthesis,” “ascorbate and aldarate metabolism,” and “stilbenoid, diarylheptanoid and gingerol biosynthesis” were significantly enriched (Figure S5B).
In the leaf data shown in Figure 4, among primary metabolite pathway biosynthesis enzyme genes, the expression of transcripts coding for carbohydrate such as galactinol synthase 2-like (LOC107783847), galactinol-sucrose galactosyltransferase-like (LOC107810250), and 1,4-alpha-glucan-branching enzyme 3 (LOC107781849) were upregulated and mannan endo-1,4-beta-mannosidase 7-like (LOC107777916) and probable isoaspartyl peptidase/L-asparaginase 3 (LOC107795479) were reduced. Fatty acid elongation gene LOC107809103 in the fatty acid pathway was upregulated (Figure 4). Among flavonoid biosynthesis enzyme genes, the transcripts coding for feruloyl CoA ortho-hydroxylase 1-like (LOC107796566), caffeoyl-CoA O-methyltransferase 6-like (LOC107772621), agmatine coumaroyltransferase-2-like (LOC107794911), caffeoyl-CoA O-methyltransferase 6 (LOC107810774), and trans-resveratrol-di-O-methyltransferase-like (LOC107785995) were downregulated whereas the transcript coding for agmatine coumaroyltransferase-2-like (LOC107761506) was upregulated (Figure 4). In addition, seven genes involving the terpenoid/steroid biosynthesis enzyme gene were detected by transcriptome profiling. Dehydrodolichyl diphosphate synthase 6-like (LOC107814919), 3-hydroxy-3-methylglutaryl-coenzyme A reductase-like (LOC107770799), ent-copalyl diphosphate synthase, chloroplastic-like (LOC107809496), 5-epiaristolochene 1,3-dihydroxylase (LOC107759261), alpha-farnesene synthase-like (LOC107779472), and probable terpene synthase 3 (LOC107819330) were downregulated, while the transcript coding for gibberellin 2-beta-dioxygenase 1-like (LOC107813437) was upregulated (Figure 4).
In the root data shown in Figure 5, among primary metabolite pathway biosynthesis enzyme genes, the expressions of transcripts coding for carbohydrate such as galactinol synthase 1-like (LOC107812124), bifunctional UDP-glucose 4-epimerase and UDP-xylose 4-epimerase 1-like (LOC107775041), bifunctional UDP-glucose 4-epimerase and UDP-xylose 4-epimerase 1-like (LOC107781891), acid beta-fructofuranosidase-like (LOC107807993), beta-amylase 1, chloroplastic-like (LOC107832261), alpha-amylase-like (LOC107769668), and alpha-galactosidase 3-like (LOC107798945) were upregulated and acid beta-fructofuranosidase-like (LOC107807993) was reduced. 1-(5-Phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase (LOC107778749) and homogentisate 1,2-dioxygenase-like (LOC107764203) in the amino acid pathway were down- and upregulated, respectively (Figure 5). Among flavonoid biosynthesis enzyme genes, the transcripts coding for caffeoyl-CoA O-methyltransferase 6 (LOC107810774) and caffeoyl-CoA O-methyltransferase 6-like (LOC107772621) were downregulated whereas the transcript coding for a4-coumarate-CoA ligase 2-like (LOC107769758) and peroxidase P7-like (LOC107782926) were upregulated (Figure 5). In addition, four genes involving terpenoid/steroid biosynthesis enzyme genes were detected by transcriptome profiling. The expression of transcripts coding for 8-hydroxygeraniol dehydrogenase-like (LOC107772996), gibberellin 2-beta-dioxygenase 1-like (LOC107782042), gibberellin 2-beta-dioxygenase 1-like (LOC107821505), and gibberellin 2-beta-dioxygenase 1-like (LOC107790388) were upregulated (Figure 5).
Knockout of CCoAOMT6 and CCoAOMT6L upregulated metabolite docking study for possible chemical mechanism of tobacco disease resistance
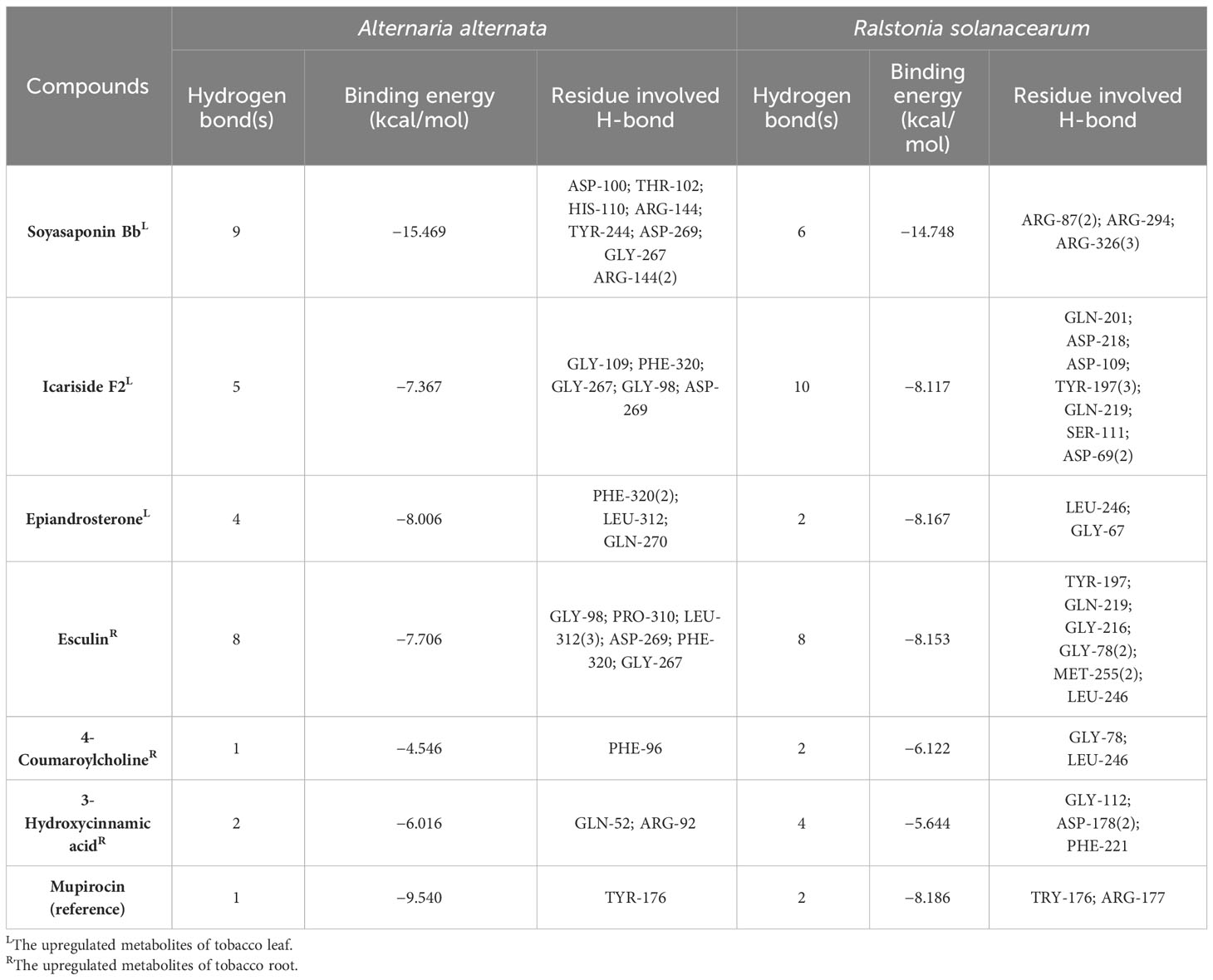
The above bacteriostatic experiments showed that there were some antimicrobial agents in transgenic tobacco leaves. Because phenolic acids, coumarins, terpenoids, and other compounds have good biological activities, we screened six compounds such as icariside F2, epiandrosterone, soyasaponin Bb, 3-hydroxycinnamic acid, esculin, and 4-coumaroylcholine from upregulated metabolites (the threshold is 1.5-fold). Aminoacyl-tRNA synthetases (aaRSs) play a key role in protein biosynthesis, and the inhibition of aaRSs affects cell growth (Pisano et al., 2019). The tyrosyl-tRNA synthetase 3D structures of Ralstonia solanacearum and Alternaria alternata were obtained from UniProt (https://www.uniprot.org/). Mupirocin, which target to tyrosyl-tRNA synthetase, is a good inhibitor of pathogens as a reference molecule (Francklyn and Mullen, 2019). Molecular docking results are shown in Table 1 and Figure 6. From Table 1, it can be found that the binding energies of A with tyrosyl-tRNA synthetase of Alternaria alternata and Ralstonia solanacearum are −15.469 and −14.748 kcal/mol, respectively, which are better than those of reference molecules mupirocin’s −9.540 and −8.186 kcal/mol, respectively. As shown in Figure 6, soyasaponin Bb can interact with tyrosyl-tRNA synthetase of Alternaria alternata residues ASP-100, THR-102, HIS-110, ARG-144, TYR-244, ASP-269, GLY-267, and ARG-144 form nine hydrogen bonds. It forms three hydrophobic interactions with ASP-100 and LYS-322. Moreover, soyasaponin Bb can form six hydrogen bonds with residues ARG-87, ARG-294, and ARG-326 and form four hydrophobic interactions with residues PHE-293, ARG-294, and ALA-325 in tyrosyl-tRNA synthetase of Ralstonia solanacearum (Table 1 and Figure 6). It indicates that soyasaponin Bb may have a good inhibitory effect on Ralstonia solanacearum and Alternaria alternata and upregulation of soyasaponin Bb may be a chemical reason for the improvement of disease resistance in the mutant tobacco. Of course, other upregulated metabolites have high binding energy with tyrosyl-tRNA synthetase (<−5 kcal/mol), which may also enhance the tobacco resistance (Table 1). It may also be that the synergism of upregulated metabolites improves the disease resistance of tobacco.
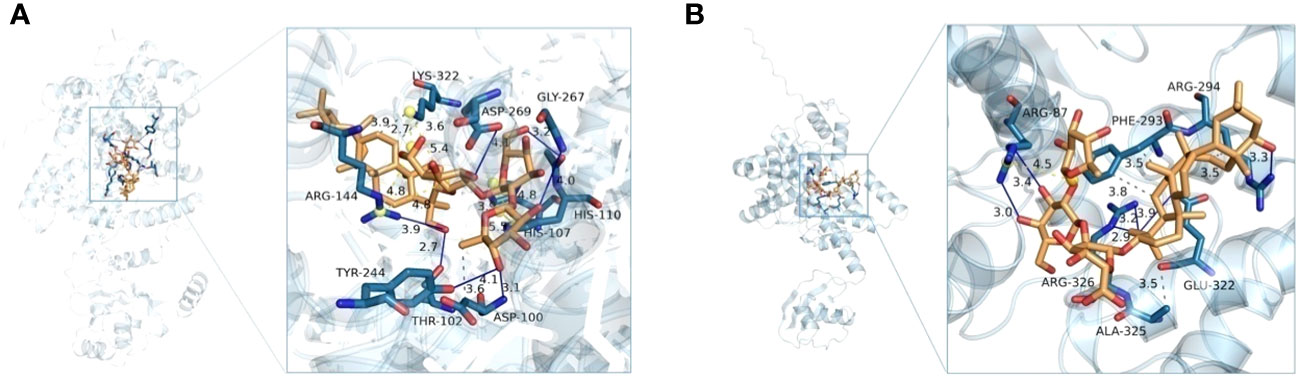
Figure 6 Soyasaponin Bb in the tyrosyl-tRNA synthetase active sites of Ralstonia solanacearum (A) and Alternaria alternata (B).
Discussion
In plants, COMT and CCoAOMT are methylation enzymes at two different substrate levels in the lignin biosynthetic pathway, giving rise to two parallel pathways. Compared with COMT, the role of CCoAOMT in the lignin biosynthesis pathway was studied relatively late. At present, CCoAOMT has been successfully cloned from pomelo, poplar, and other plants and the overexpression plants have been constructed (Liu et al., 2014; Xu et al., 2014). Some studies have found that COMT and CCoAOMT have relatively consistent high expression in cotton root, stem, and other vascular tissues (Lv et al., 2010). In this study, 14 CCoAOMT genes were identified in tobacco, of which CCoAOMT5/5L and CCoAOMT6/6L were highly expressed in leaves and roots, especially in roots, suggesting that they may play a key role in tobacco lignin biosynthesis (Figure 1).
In recent years, more and more studies have been conducted on the regulation of lignin content in plants by inhibition of CCoAOMT activity. Inhibiting the expression of CCoAOMT will inhibit the biosynthesis of S-monomer lignin or G-monomer lignin to a varying extent, especially G-monomer, so as to control the S/G ratio. It is found that the lignin content in transgenic poplars with inhibited CCoAOMT expression decreased by 12% and the S/G ratio increased by 11% (Meyermans et al., 2000). Zhong et al. transferred the antisense-CCoAOMT into poplar and found that the lignin content decreased significantly, and the loose structure was conducive to delignification (Zhong et al., 2000). The difference is that this study found that the knockout of CCoAOMT6/6L in tobacco does not affect the content of total lignin but increases and decreases the content of S-monomer and G-monomer to increase the S/G ratio by 68.4%, and the plant growth and development are better (Figures 2, 3). On the one hand, this may be due to its functional redundancy with CCoAOMT5/5L to ensure plant growth; on the other hand, it shows the importance of CCoAOMT6/6L in regulating the S/G ratio. However, the reduction of total sugar content may ensure the metabolic balance of this lignin fine-tuning (Figure 3).
The combined analysis of transcriptome and metabolism showed that the functions of CCoAOMT6/6L in roots and leaves were similar, both of which mainly affected the primary and secondary metabolism. However, the upregulation of primary metabolism and terpenoid metabolism genes in roots is more obvious whereas the phenylpropane metabolism pathway is mainly affected in leaves (Figures 4, 5, S5). This result is consistent with the phenotype. The increase of primary metabolism in roots may promote plant growth due to the sink–source relationship (Caretto et al., 2015). The phenylpropane metabolic pathway also plays an important role in plant–microorganism interaction (Naoumkina et al., 2010). Lignin biosynthesis of Verticillium dahliae-resistant tomato line LA3038 increased (Gayoso et al., 2010). After cotton hypocotyls were treated with V. dahliae, the biosynthesis of lignin and lignin phenolic polymers increased, and the expression of phenylpropanoid pathway genes, such as chalcone synthase (CHS) and phenylalanine ammonia-lyase (PAL), was upregulated (Smit and Dubery, 1997). In addition, terpenoid metabolic pathways are also closely related to plant growth, development, and resistance. The triterpene metabolism pathway in Arabidopsis thaliana can specifically regulate the composition of rhizosphere microorganisms to affect plant growth and development (Huang et al., 2019). Triterpenoids in rice tapetum can regulate the formation of pollen coat (Xue et al., 2018). The triterpene saponins in oat root have antibacterial function, which can prevent soil borne diseases, such as take-all disease (Li et al., 2021).
On the other hand, lignin plays an important role in the plant defense response to variant diseases. In previous reports, S lignin inhibits the infection and reproduction of nematodes in Arabidopsis and tobacco (Wuyts et al., 2006) and S lignin will accumulate in the process of wheat-hypersensitive resistance (Menden et al., 2007). In Ma’s work, total lignin and S lignin were considered to be the main factors contributing to the basic defense response of tobacco (Ma et al., 2017). Ma believes that S lignin is only developed in angiosperms and S lignin is evolved by plants to cope with more complex disease-resistant environments. Long et al. studied the defense effect of tobacco against Alternaria alternata and Phytophthora nicotianae pathogens and showed that the induced expression of phenylalanine biosynthesis and sesquiterpenoid biosynthesis pathway synthesis genes, resulting in phytoalexin scopoletin and capsidiol, played a chemical defense role (Long et al., 2021). A triterpenoid saponin, soyasaponin has anticancer, antioxidant, and antiviral activities and has a different influence on the oil bacterial genera (Sugiyama, 2019; Fujimatsu et al., 2020; Nakayasu et al., 2021). According to the results of molecular docking (Table 1 and Figure 6 and ), the upregulated compounds such as soybean soyasaponin Bb, as chemical defense, enhance the defense response to tobacco varieties. Considering the previous reports, in addition to S lignin, upregulated metabolisms like soyasaponin Bb are other important factors that contribute to the basic defense response in tobacco. Therefore, it is speculated that the growth phenotype of the double mutants may be caused by the perturbation of phenylpropanoid and terpenoid metabolisms by the fine-tuning of lignin monomers. In addition, S lignin features an extra methoxy functional group compared with G lignin, thereby leading to a heightened concentration of active moieties within the double mutant. This particular attribute holds the potential to play a role in enhancing the growth and developmental outcomes of the double mutant line (Parra et al., 2011).
Conclusions
In total, we have provided the physiological and molecular evidence to demonstrate that CCoAOMT6 and CCoAOMT6L are involved in growth development and disease resistance by adjusting the S/G ratio of lignin monomers in Nicotiana tabacum. Furthermore, the combined analysis of metabolome and transcriptome proved that these phenotypes may be caused by affecting phenylpropane and terpene metabolism in the leaves and roots. In addition to S lignin, upregulated compounds such as soyasaponin Bb may enhance the tobacco disease defense. Our research provides a theoretical basis for the improvement of tobacco varieties.
Methods
Identification and phylogenetic analysis
To identify potential members of the CCoAOMT gene family in Nicotiana tabacum, we performed NCBI database searches. Seven Arabidopsis CCoAOMT sequences were used as queries in BLAST searches against the Nicotiana tabacum genome sequence (https://www.ncbi.nlm.nih.gov/genome/?term=Nicotiana+tabacum). The exons and introns were analyzed and calculated by IMEter v2.0 (Wu et al., 2023). SMART toolkit (http://smart.embl-heidelberg.de/) was used for protein domain analysis.
Full-length protein sequences of 14 tobacco CCoAOMTs and 7 Arabidopsis CCoAOMTs were used for building the phylogenetic tree. Multiple-sequence alignments were performed using Muscle 3.52. A neighbor-joining (NJ) tree is reconstructed using the MEGA program version 7.0. Support for each node was tested with 1,000 bootstrap replicates.
RNA extraction and cDNA synthesis
The total RNA of the wild type and the double mutant was isolated respectively from finely ground tissue samples of leaves and roots of tobacco growing in the open field in Kunming (2021), China. We used the BGI-NGS-TQ-RNA-005 A0 Kit (BGI) for the extractions. The purity and integrity of the RNA were evaluated through gel electrophoresis and the ratios of A260/A230 and A260/A280. The qualified RNA is used for subsequent reverse transcription and transcriptome library construction. We used the TransScript II First-Strand cDNA Synthesis Kit (TransGen) for cDNA reverse transcription.
Real-time PCR
To analyze gene expression levels, a qRT-PCR assay was completed with SYBR Green (Takara) and a 7500 Real-Time PCR system (Life Technologies).The reaction mixture was 20 μL, including SYBR Premix Ex Taq 10 μL, 10 μM reverse-specific primer 0.4 μL, and 0.1 μM cDNA templates 2 μL. Actin was used as a reference gene. The 2−△△CT method was used to analyze the relative gene expression (Livak and Schmittgen, 2001). The experiment was repeated three times, and the primers are listed in Table S3.
Plasmid construction
The binary vector pORE-Cas9 was used as backbone like that previously described (Gao et al., 2015; Gao et al., 2018), which contains the CRISPR/Cas9 components. The 25-bp oligos (F: GATTGTTGCCCGTAATCCAAAAGGC; R: AAACGCCTTTTGGATTACGGGCAAC) with GAAT and AAAC overhands, respectively, were annealed and then inserted into the BsaI site of the pORE-Cas9 binary vector by T7 ligase.
Creation of the double mutant using CRISPR/Cas9 technology
The pORE-Cas9 binary vectors containing the specific guide RNA and Cas9 expression cassettes were transformed into Agrobacterium tumefaciens LBA4404 by the freeze–thaw method. The positive clones were then used to produce target gene mutant tobacco plants with the leaf discs method (Horsch et al., 1985). Kanamycin-resistant seedlings were obtained, and mutants were identified.
Lignin content analysis
The total lignin contents of dried tobacco leave and root tissues were quantitatively determined by using the Klason method (Eudes et al., 2006; Ma et al., 2017). Lignin monomer composition was measured using thioacidolysis (Zhao et al., 2010; Ma et al., 2017).
Total sugar and reducing sugar analysis
Determination methods of total sugar and reducing sugar followed protocols of Tobacco Trade Standards (China National Tobacco Quality Supervision&TestCenter, 2002).
Transcriptome sequencing
Extracted RNA was assessed for quality and quantity using an Agilent 2100 Bioanalyzer (Agilent Technologies), and mRNA library preparation refers to the construction process BGI-NGS-JK-RNA-001 (https://www.yuque.com/yangyulan-ayaeq/oupzan/gu8ls7). The amplified libraries were sequenced on a DNBSEQ sequencing machine at BGI company according to Qi et al (Qin et al., 2020). The sequencing data were filtered with SOAPnuke (Li et al., 2008). Bowtie2 (Langmead and Salzberg, 2012) was applied to align the clean reads to the gene set. The gene expression level was calculated by RSEM (v1.3.1) (Li and Dewey, 2011). Differential expression analysis was performed using the DESeq2 (v1.4.5) (Love et al., 2014) with log2FC ≥1 and Q value ≤0.05. All DEGs were annotated according to GO, KEGG, KOG, Swiss-Prot, and TrEMBL databases. There are six replicates in the leaf and root groups between WT and the mutant.
Tobacco disease resistance test
Resistance against pathogens test. The leaves were the fourth leaf from the top, taken from the 2-month planted WT tobacco and double-mutant tobacco. The wound sites were scratched by the toothpick back of the leaf, evenly distributed on both sides of the main leaf vein. The challenged leaves were put into a 150-mm culture dish with water-soaked filter paper at the bottom and incubated at 28°C for 72 h, then the photoperiod was set as 16-h light and 8-h darkness.
Tobacco leaf extract inhibition test. 100 mg of MU and WT tobacco leaf-lyophilized powder was ultrasonically extracted for 40 min by 10 mL of 95% ethanol and filtered, and MU and WT extracts were obtained after filtration. The 6-mm-diameter filter paper was put into the MU extract, WT extract, 95% ethanol, and 1 mg/mL cefotaxime sodium ethanol solutions and soaked for 10 min. 100 μL of Alternaria alternata and Ralstonia solanacearum bacterial fluids was taken and coated in PDA and LB plate culture medium, respectively, then kept in a 28°C incubator for 24 h.
Widely targeted metabolomic analysis
20.0 mg (accurate to 0.1 mg) sample powder was weighed, and 4 mL of methanol–water (v:v = 8:2) solution was added, shaken on a vortex mixer at 2,000 r/min for 30 s, sonicated in an ice-water bath for 30 min, centrifuged at 13,000 r/min for 15 min at 4°C, and passed through a 0.22-μm organic filter membrane, ready for analysis.
High-performance liquid chromatography (LC-20A, Shimadzu) tandem time-of-flight mass spectrometry (AB SCIEX 5600+, Shimadzu), and XSelect HSS T3 (4.6 × 150 mm, 3.5 µm) column chromatography were performed for the metabolomic analysis. The positive ionization mode mobile phase was 0.1% formic acid/water (A) and acetonitrile (B), and the negative ionization mode mobile phase was 5 mM ammonium formate (A) and acetonitrile (B). The mobile phase gradient elution program was: 0.00 min–3.00 min, 10%B; 3.00 min–21.00 min, 10%–95%B; 21.00 min–28.00 min, 95%B; 28.00 min–28.10 min, 10%B; 28.10 min–34.00 min, 10%B. The QTOF/MS scanning mode is an information-dependent (IDA) full-scan mode, the mass scanning range is m/z 50–1,000, the source voltages of the positive and negative ionization modes are 5,500 V and 4,500 V, respectively, and the curtain gas, atomizer (gas1), and heating gas (gas2) flow pressures are 25 psi, 50 psi, and 50 psi, respectively. The collision energies (CE) were 30 V and −30 V, respectively. Qualitative analysis of metabolites was based on the local database (Jiangsu Academy of Agricultural Sciences) and the public database of metabolite information, including NIST, METLIN (https://metlin.scripps.edu), and HMDB (https://hmdb.ca/). Metabolites with significant differences were set with thresholds of variable importance in projection (VIP) ≥1 and fold change ≥1 or ≤0.5. Eight biological replicates of each sample were performed in each group.
Molecular docking
In this study, AutoDock Vina (Vina, version 1.1.2) is running with a semi-flexible docking method, with docking accuracy up to 78%. PyMOL (version 4.3.0) software (https://pymol.org/) was used for separation of original ligand and protein structure, dehydration, and removal of organic matter. AutoDockTools (http://mgltools.scripps.edu/downloads) is used for hydrogenation, checking the charge, designating the atom type as AD4 type, calculating the Gasteiger, and constructing the docking grid box of the protein structure. In addition, the chemical composition (small-molecule ligand) should be determined as the root, and the reversible bond of the ligand should be selected in AutoDockTools.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
ML performed the experiments and contributed to the original manuscript. HL analyzed the data. JZ contributed to the plotting and data analysis. CL and YL contributed to the analysis and interpretation of the data and drafting and revision of the manuscript. GY, TX, and HH contributed to the analysis and interpretation of data. YX and WK contributed to the analysis and interpretation of data. BH, XQ, and JW contributed to the design and conduct of the study, and the drafting and revision of the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
We thank Prof. Qingyou Xia (Southwestern University, Chongqing, China) for providing the CRISPR/Cas9 vector.
Conflict of interest
Authors ML, HL, JZ, GY, HH, YX, JW, WK were employed by the company China Tobacco Yunnan Industrial Co., Ltd.
The authors declare that this study received funding from China Tobacco Yunnan Industrial (No. 2022JY02) and Yunnan Key Laboratory of Tobacco Chemistry (No. 2021539200340239). The funder had the following involvement in the study: They provided some of the experimental materials and instructions.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer JW declared a shared affiliation with the authors BH, XQ to the handling editor at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1216702/full#supplementary-material
References
Bernard-Vailhé, M. A., Cornu, A., Robert, D., Maillot, M. P., Besle, J. M. (1996). Cell wall degradability of transgenic tobacco stems in relation to their chemical extraction and lignin quality. J. Agric. Food Chem. 44 (4), 1164–1169. doi: 10.1021/jf950481j
Caretto, S., Linsalata, V., Colella, G., Mita, G., Lattanzio, V. (2015). Carbon fluxes between primary metabolism and phenolic pathway in plant tissues under stress. Int. J. Mol. Sci. 16, 26378–26394. doi: 10.3390/ijms161125967
China National Tobacco Quality Supervision&TestCenter (2002). Tobacco and tobacco products-Detelrmination of water soluble sugars-Continuous flow method:YC/T 159 —2002 (Beijing: State Tobacco Mono-poly Administration).
Christensen, J. H., Baucher, M., O’connell, A., Van Montagu, M., Boerjan, W. (2000). “Control of lignin biosynthesis.” Molecular biology of woody plants (Dordrecht: Springer) 1, 227–267.
Do, C.-T., Pollet, B., Thévenin, J., Sibout, R., Denoue, D., Barrière, Y., et al. (2007). Both cafeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, favonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta. 226 (5), 1117–1129.
Eudes, A., Pollet, B., Sibout, R., Do, C. T., Séguin, A., Lapierre, C., et al. (2006). Evidence for a role of AtCAD 1 in lignification of elongating stems of Arabidopsis thaliana. Planta 225 (1), 23–39. doi: 10.1007/s00425-006-0326-9
Francklyn, C. S., Mullen, P. (2019). Progress and challenges in aminoacyl-tRNAsynthetase-based therapeutics. J. Biol. Chem. 294 (14), 5365–5385. doi: 10.1074/jbc.REV118.002956
Fujimatsu, T., Endo, K., Yazaki, K., Sugiyama, A. (2020). Secretion dynamics of soyasaponins in soybean roots and effects to modify the bacterial composition. Plant Direct 4 (9), e00259. doi: 10.1002/pld3.259
Gao, J., Wang, G., Ma, S., Xie, X., Wu, X., Zhang, X., et al. (2015). CRISPR/Cas9-mediated targeted mutagenesis in Nicotianatabacum. Plant Mol. Biol. 87 (1-2), 99–110.
Gao, J., Zhang, T., Xu, B., Jia, L., Xiao, B., Liu, H., et al. (2018). CRISPR/cas9-mediated mutagenesis of carotenoid cleavage dioxygenase 8 (CCD8) in tobacco affects shoot and root architecture. Int. J. Mol. Sci. 19 (4), 1062. doi: 10.3390/ijms19041062
Gayoso, C., Pomar, F., Novo-Uzal, E., Merino, F., De Ilarduya, O. M. (2010). The Ve-mediated resistance response of the tomato to Verticillium dahlia involves H2O2, peroxidase and lignins and drives PAL gen expression. BMC Plant Biol. 10, 232. doi: 10.1186/1471-2229-10-232
Guillaumie, S., Mzid, R., Méchin, V., Léon, C., Hichri, I., Destrac-Irvine, A., et al. (2009). The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco. Plant Mol. Biol. 72 (1-2), 215–234.
Guo, Z. J., Chen, X. J., Wu, X. L., Ling, J. Q., Xu, P. (2004). Overexpression of the AP2/EREBP transcription factor OPBP1 enhances disease resistance and salt tolerance in tobacco. Plant Mol. Biol. 55, 607–618. doi: 10.1007/s11103-004-1521-3
Hopkins, D. W., Webster, E. A., Chudek, J. A., Halpin, C. (2001). Decomposition in soil of tobacco plants with genetic modifications to lignin biosynthesis. Soil Biol. Biochem. 33 (11), 1455–1462. doi: 10.1016/S0038-0717(01)00054-2
Horsch, R. B., Fry, J. E., Hoffmann, N. L., Wallroth, M., Eichholtz, D., Rogers, S. G., et al. (1985). A simple and general method for transferring genes into plants. Science 227 (4691), 1229–1231.
Huang, A. C., Jiang, T., Liu, Y. X., Bai, Y. C., Reed, J., Qu, B., et al. (2019). A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science, 364, (6440), eaau6389.
Jin, K. X., Wang, K., Cui, H. S., Yang, S. M., Tian, G. L., Liu, X. E., et al. (2018). Application of Raman spectroscopy in the study of lignin. Forestry Sci. 54 (03), 144–151.
Jung, H. J. G., Ni, W. (1998). Lignification of plant cell walls: impact of genetic manipulation. Proc. Natl. Acad. Sci. 95 (22), 12742–12743. doi: 10.1073/pnas.95.22.12742
Lan, T., Zheng, S., Yang, L., Wu, S., Wang, B., Zhang, S., et al. (2014). Mapping of quantitative trait loci conferring resistance to bacterial wilt in tobacco (Nicotiana tabacum L.). Plant Breed. 133, 672–677. doi: 10.1111/pbr.12202
Langmead, B., Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9 (4), 357–359. doi: 10.1038/nmeth.1923
Lebeau, A., Daunay, M. C., Frary, A., Palloix, A., Wang, J. F., Dintinger, J., et al. (2011). Bacterial wilt resistance in tomato, pepper, and eggplant: genetic resources respond to diverse strains in the Ralstonia solanacearum species complex. Phytopathology 101, 154—165. doi: 10.1094/PHYTO-02-10-0048
Li, X., Bonawitz, N. D., Weng, J. K., Chapple, C. (2010). The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22 (5), 1620–1632. doi: 10.1105/tpc.110.074161
Li, B., Dewey, C. N. (2011). RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 323. doi: 10.1186/1471-2105-12-323
Li, Y., Leveau, A., Zhao, Q., Feng, Q., Lu, H., Miao, J., et al. (2021). Subtelomeric assembly of a multi-gene pathway for antimicrobial defense compounds in cereals. Nat. Commun. 12 (1), 2563. doi: 10.1038/s41467-021-22920-8
Li, R., Li, Y., Kristiansen, K., Wang, J. (2008). SOAP: Short oligonucleotide alignment program. Bioinformatics. 24 (5), 713–714. doi: 10.1093/bioinformatics/btn025
Li, H. T., Yao, K., Jia, D. R., He, Q. (2011). Method for determination of lignin content. Leather Sci. Eng. 21 (02), 31–34.
Liu, H. M., Hu, S. L., Lu, X. Q., Cao, Y., Ren, P. (2014). Cloning and bioinformatics analysis of CCoAOMT gene in PopuluscathayanaRehd. Hubei Agric. Sci. 53 (11), 2670–2674.
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Long, J., Yang, M., Zuo, C., Song, N., He, J. M., Zeng, J., et al. (2021). Requirement of Jasmonate signaling for defense responses against Alternariaalternata and Phytophthoranicotiane in tobacco. Crop Sci. 61 (6), 4273–4283. doi: 10.1002/csc2.20625
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Lv, M., Ni, Z. Y., Wang, J., Li, B., Luo, S. P., Fan, L. (2010). Tissue specificity express analysis of methyltransferase gene COMT and CCoAOMT in cotton. J. Nucl. Agric. Sci. 24 (4), 713–719.
Ma, Q. H., Zhu, H. H., Qiao, M. Y. (2017). Contribution of both lignin content and sinapyl monomer to disease resistance in tobacco. Plant Pathol. 67 (3), 642–650.
Martz, F., Maury, S., Pinçon, G., Legrand, M. (1998). cDNA cloning, substrate specificity and expression study of tobacco caffeoyl-CoA 3-O-methyltransferase, a lignin biosynthetic enzyme. Plant Mol. Biol. 36 (3), 427–437. doi: 10.1023/A:1005969825070
Maury, S., Geoffroy, P., Legrand, M. (1999). Tobacco O-methyltransferases involved in phenylpropanoid metabolism. The different caffeoyl-coenzyme A/5-hydroxyferuloyl-coenzyme A 3/5-O-methyltransferase and caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase classes have distinct substrate specificities and expression patterns. Plant Physiol. 121 (1), 215–224.
Menden, B., Kohlhoff, M., Moerschbacher, B. M. (2007). Wheat cells accumulate a syringyl-rich lignin during the hypersensitive resistance response. Phytochemistry 68 (4), 513–520. doi: 10.1016/j.phytochem.2006.11.011
Meyermans, H., Morreel, K., Lapierre, C. (2000). Modification in lignin and accumulation of phenolic glucosides in poplar xylem upon down-regulation of caffeoyl-coenzyme A O-methyltransferase, an enzyme involved in lignin biosynthesis. J. Biol. Chem. 275, 36899–36909. doi: 10.1074/jbc.M006915200
Nakayasu, M., Yamazaki, S., Aoki, Y., Yazaki, K., Sugiyama, A. (2021). Triterpenoid and steroidal saponins differentially influence soil bacterial genera. Plants 10 (10), 2189. doi: 10.3390/plants10102189
Naoumkina, M. A., Zhao, Q., Gallego-Giraldo, L., Dai, X., Zhao, P. X., Dixon, R. A. (2010). Genome‐wide analysis of phenylpropanoid defence pathways. Mol. Plant Pathol. 11 (6), 829–846.
Nelson, R. S., Stewart, C. N., Gou, J., Holladay, S., Gallego-Giraldo, L., Flanagan, A., et al. (2017). Development and use of a switchgrass (Panicum virgatum L.) transformation pipeline by the BioEnergy Science Center to evaluate plants for reduced cell wall recalcitrance. Biotechnol. Biofuels 10 (1), 309.
Parra, G., Bradnam, K., Rose, A. B., Korf, I. (2011). Comparative and functionalanalysis of intron-mediated enhancement signals reveals conserved features among plants. Nucleic Acids Res. 39, 5328–5337. doi: 10.1093/nar/gkr043
Pedersen, J. F., Vogel, K. P., Funnell, D. L. (2005). Impact of reduced lignin on plant fitness. Crop Sci. 45 (3), 812–819. doi: 10.2135/cropsci2004.0155
Pellegrini, L., Geoffroy, P., Fritig, B., Legrand, M. (1993). Molecular cloning and expression of a new class of ortho-diphenol-O-methyltransferases induced in tobacco (Nicotianatabacum L) leaves by infection elicitor treatment. Plant Physiol. 103 (2), 509–517.
Pisano, M. B., Kumar, A., Medda, R., Gatto, G., Pal, R., Fais, A., et al. (2019). Antibacterial activity and molecular docking studies of a selected series of hydroxy-3-arylcoumarins. Molecules 24 (15), 2815. doi: 10.3390/molecules24152815
Qin, Y., Bai, S., Li, W., Sun, T., Galbraith, D. W., Yang, Z., et al. (2020). Transcriptome analysis reveals key genes involved in the regulation of nicotine biosynthesis at early time points after topping in tobacco (Nicotianatabacum L). BMC Plant Biol. 20, 1–15. doi: 10.1186/s12870-020-2241-9
Rastogi, S., Dwivedi, U. N. (2008). Manipulation of lignin in plants with special reference to O-methyltransferase. Plant Sci. 174 (3), 264–277. doi: 10.1016/j.plantsci.2007.11.014
Sierro, N., Battey, J. N., Ouadi, S., Bakaher, N., Bovet, L., Willig, A., et al. (2014). The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 5 (1), 3833. doi: 10.1038/ncomms4833
Smit, F., Dubery, I. A. (1997). Cell wall reinforcement in cotton hypocotyls in response to a Verticilliumdahliae elicitor. Phytochemistry 44 (5), 811–815. doi: 10.1016/S0031-9422(96)00595-X
Sugiyama, A. (2019). The soybean rhizosphere: Metabolites, microbes, and beyond—A review. J. Advanced Res. 19, 67–73. doi: 10.1016/j.jare.2019.03.005
Tong, Z., Jiao, T., Wang, F., Li, M., Leng, X., Gao, Y., et al. (2012). Mapping of quantitative trait loci conferring resistance to brown spot in flue-cured tobacco (Nicotiana tabacum L.). Plant Breed. 131, 335–339. doi: 10.1111/j.1439-0523.2011.01940.x
Vanholme, R., Demedts, B., Morreel, K., Ralph, J., Boerjan, W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153 (3), 895–905. doi: 10.1104/pp.110.155119
Verma, S. R., Dwivedi, U. N. (2014). Lignin genetic engineering for improvement of wood quality: Applications in paper and textile industries, fodder and bioenergy production. South Afr. J. Bot. 91, 107–125. doi: 10.1016/j.sajb.2014.01.002
Webster, E., Halpin, C., Chudek, J., Tilston, E. L., Hopkins, D. W. (2005). Decomposition in soil of soluble, insoluble and lignin-rich fractions of plant material from tobacco with genetic modifications to lignin biosynthesis. Soil Biol. Biochem. 37 (4), 751–760. doi: 10.1016/j.soilbio.2004.09.012
Wu, D., Wang, Y., Qi, S., Yuan, Y., Guo, J., Chen, G., et al. (2023). Effects of the structure and molecular weight of alkali-oxygen lignin isolated from rice straw on the growth of maize seedlings. Biomacromolecules 24 (3), 1377–1387. doi: 10.1021/acs.biomac.2c01392
Wuyts, N., Lognay, G., Swennen, R., De Waele, D. (2006). Nematode infection and reproduction in transgenic and mutant Arabidopsis and tobacco with an altered phenylpropanoid metabolism. J. Exp. Bot. 57 (11), 2825–2835. doi: 10.1093/jxb/erl044
Xie, H. L., Engle, N. L., Venketachalam, S., Yoo, C. G., Barros, J., Lecoultre, M., et al. (2019). Combining loss of function of FOLYLPOLYGLUTAMATE SYNTHETASE1 and CAFFEOTYL-COA 3-O-METHYLTRANSFERASE1 for lignin reduction and improved asccharification efficiency in Arabidopsis thaliana. BMC Plant Biol. 12, 108–128.
Xu, Y., Pan, T., Pang, D. (2014). Cloning of caffeoyl-coenzyme A O-methyltransferase gene from Citrus maxima (Burm.) Merr. And its plant expressing vector construction. Chin. Agric. Sci. Bull. 30 (25), 148–153.
Xue, Z., Xu, X, Zhou, Y., Wang, X., Zhang, Y., Liu, D., et al. (2018). Deficiency of a triterpene pathway results in humidity-sensitive genic male sterility in rice. Nat. Commun. 9, 604. doi: 10.1038/s41467-018-03048-8
Zang, Y. N., Kong, N. C., Li, D. Y., Dang, L. Z., Duan, Y. Q., Li, J., et al. (2015). Research overview of tobacco lignin. J. Yunnan Agric. Univ. (Natural Sciences) 30 (05), 822–828.
Zhao, Q., Gallego-Giraldo, L., Wang, H., Zeng, Y., Ding, S. Y., Chen, F., et al. (2010). An NAC transcription factor orchestrates multiple features of cell wall development in Medicago truncatula. Plant J. 63 (1), 100–114.
Keywords: CCoAOMT 6/6L, lignin, tobacco bacterial wilt, tobacco brown spot, transcriptome, metabolome, soyasaponin Bb, Nicotiana tabacum
Citation: Liu M, Liu H, Zhang J, Li C, Li Y, Yang G, Xia T, Huang H, Xu Y, Kong W, Hou B, Qi X and Wang J (2023) Knockout of CAFFEOYL-COA 3-O-METHYLTRANSFERASE 6/6L enhances the S/G ratio of lignin monomers and disease resistance in Nicotiana tabacum. Front. Plant Sci. 14:1216702. doi: 10.3389/fpls.2023.1216702
Received: 13 June 2023; Accepted: 13 September 2023;
Published: 05 October 2023.
Edited by:
Shaojun Dai, Shanghai Normal University, ChinaReviewed by:
Rui Shi, North Carolina State University, United StatesDaping Gong, Chinese Academy of Agricultural Sciences, China
Copyright © 2023 Liu, Liu, Zhang, Li, Li, Yang, Xia, Huang, Xu, Kong, Hou, Qi and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bingzhu Hou, aG91YnpAaWJjYXMuYWMuY24=; Xiaoquan Qi, eHFpQGliY2FzLmFjLmNu; Jin Wang, d2FuZ2ppbkBpY2Nhcy5hYy5jbg==
†These authors share first authorship
 Mingxin Liu1,2†
Mingxin Liu1,2† Bingzhu Hou
Bingzhu Hou Xiaoquan Qi
Xiaoquan Qi Jin Wang
Jin Wang