
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 05 July 2023
Sec. Functional and Applied Plant Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1214732
 Qi Wang1†
Qi Wang1† Zhenbiao Zhang2†
Zhenbiao Zhang2† Cun Guo3†
Cun Guo3† Xiaobo Zhao1
Xiaobo Zhao1 Zhiyuan Li2
Zhiyuan Li2 Yifei Mou1
Yifei Mou1 Quanxi Sun1
Quanxi Sun1 Juan Wang1
Juan Wang1 Cuiling Yuan1
Cuiling Yuan1 Chunjuan Li1
Chunjuan Li1 Ping Cong2*
Ping Cong2* Shihua Shan1*
Shihua Shan1*Heat shock transcription factors (Hsfs) play important roles in plant developmental regulations and various stress responses. In present study, 46 Hsf genes in peanut (AhHsf) were identified and analyzed. The 46 AhHsf genes were classed into three groups (A, B, and C) and 14 subgroups (A1-A9, B1-B4, and C1) together with their Arabidopsis homologs according to phylogenetic analyses, and 46 AhHsf genes unequally located on 17 chromosomes. Gene structure and protein motif analysis revealed that members from the same subgroup possessed similar exon/intron and motif organization, further supporting the results of phylogenetic analyses. Gene duplication events were found in peanut Hsf gene family via syntenic analysis, which were important in Hsf gene family expansion in peanut. The expression of AhHsf genes were detected in different tissues using published data, implying that AhHsf genes may differ in function. In addition, several AhHsf genes (AhHsf5, AhHsf11, AhHsf20, AhHsf24, AhHsf30, AhHsf35) were induced by drought and salt stresses. Furthermore, the stress-induced member AhHsf20 was found to be located in nucleus. Notably, overexpression of AhHsf20 was able to enhance salt tolerance. These results from this study may provide valuable information for further functional analysis of peanut Hsf genes.
The growth and development of plants is significantly constrained by abiotic stresses, such as salt, drought, cold and heat. These abiotic stresses have a negative effect on the quality and yield of crops (Umezawa et al., 2006). In order to cope with environmental stresses, plants form various regulatory mechanisms in the course of long-term evolution (Shinozaki et al., 2003). With the global warming, the effect of heat stress on plants has been paid more and more attention. The heat shock response (HSR), as a universal protection mechanism, was activated when faced with heat stress and other stimulating factors (Schoffl et al., 1998). Meanwhile, the heat shock proteins (HSPs), as molecular chaperone, accumulated rapidly to protect the plant from heat stress by maintaining protein homeostasis and repairing damaged proteins under HSR (Wang et al., 2004; Zhang et al., 2013). Furthermore, expression of HSP genes is mainly regulated by the Hsfs on a transcriptional level, which can combine with heat shock elements (HSEs:5’-nGAAnnTTCn-3’) in HSP gene promoters (Bienz and Pelham, 1987; Lin et al., 2011). The heat shock transcription factors (Hsfs) are the terminal component of the signal transduction chain, which can mediate the activation of the response to heat stress and/or other stress-stimulated genes (Nover et al., 2001).
Previous studies have reported that the Hsfs have several conserved domains, including DNA binding domain (DBD), oligomerization domain (OD), nuclear localization signal (NLS), nuclear export signal (NES), C-terminal activation domain (CTAD), and repressor domain (RD) (Nover et al., 2001; Kotak et al., 2004; Guo et al., 2008). Among them, DBD is the most conserved domain in Hsf proteins, which is characterized by its hydrophobic core containing a typical helix-turn-helix conserved motif. It is essential for recognizing and binding to the conserved motif of the heat shock element (HSE) in the promoter of the target genes (von Koskull-Doring et al., 2007; Huang et al., 2015). OD (HR-A/B) is composed of two hydrophobic seven peptide repeat regions, and the domain can promote Hsf effectively bind to the HSE in HSP promoter by forming a specific structure (Scharf et al., 2012). CTAD is the least conserved region in Hsf protein sequence, which contains a short peptide AHA motif and is a necessary condition for Hsfs transcriptional activity (Doring et al., 2000). According to the number of amino acid residues connected between HR-A/B, plant Hsf protein families can be divided into three groups: A, B, and C. The number of amino acid residues insertion between HR-A/B is 21 and 7 in group A and group C Hsfs, respectively, whereas group B Hsfs have no amino acid insertion between HR-A/B (Scharf et al., 2012). It is worth noting that only group A Hsfs contain the C-terminal activation motifs (AHA motifs).
To date, a large number of heat shock transcription factors have been identified in different plants, including 21 genes in Arabidopsis, 25 in rice (Guo et al., 2008), 27 in potato (Tang et al., 2016), 24 in tomato (Yang et al., 2016), 82 in wheat (Duan et al., 2019), 25 in maize (Lin et al., 2011), and 38 in soybean (Li et al., 2014). In plants, the class A Hsfs consist of majority of the Hsf proteins. Members of class A play important roles in regulating plant response to abiotic stresses (Scharf et al., 2012). In Arabidopsis, the expression levels of most heat-stress response genes are regulated by AtHsfA1 (AtHsfA1a, AtHsfA1b, AtHsfA1d and AtHsfA1e). The expression of HS-responsive genes, including transcription factors and molecular chaperones, was significantly decreased in hsfa1a/b/d triple mutants. Moreover, the hsfa1a/b/d/e quadruple mutant exhibited growth retardation and more sensitive to temperature compared with wide type (Lohmann et al., 2004; Nishizawa-Yokoi et al., 2011; Yoshida et al., 2011). In subgroup A1, AtHsfA1b and AtHsfA1d were reported to enhance drought and heat tolerance, respectively (Bechtold et al., 2013; Higashi et al., 2013). AtHsfA2 in subgroup A2 and AtHsfA3 in subgroup A3 have been reported to function in heat tolerance and AtHsfA2 also enhances anoxia tolerance (Schramm et al., 2006; Charng et al., 2007; Banti et al., 2010; Chen et al., 2010; Friedrich et al., 2021). Meanwhile, overexpression of AtHsfA3 in Arabidopsis increased galactinol level and oxidative stress tolerance (Song et al., 2016). Besides, AtHsfA6b in subgroup A6 was highly induced by salt and drought conditions, and positively regulated Arabidopsis tolerance to ABA-mediated salt, drought, and heat stresses (Huang et al., 2016).
In addition, Hsf members have been reported to be involved in plant growth and development. For example, in subgroup A9, AtHsfA9 was proved to sever as a seed-specific transcription factor showing to function in regulating expression of heat stress protein-encoding genes during seed development of Arabidopsis (Kotak et al., 2007). AtHsfB2a from subgroup B2, was proven to be involved in regulating gametophyte development of Arabidopsis (Wunderlich et al., 2014). Besides, AtHsfB4-overexpressing transgenic lines displayed short root length phenotype compared with wild type, indicating AtHsfB4 in subgroup B4 was involved in the negative regulation of root development (Begum et al., 2013).
Peanut (Arachis hypogaea L.) is an important economic and oil crop in the world, suppling oils and proteins for the human nutrition. The production of peanut was affected by several abiotic stresses, including extreme temperatures, high salinity, and drought, during the growing stage. Utilization of resistant varieties is the most cost-efficient measure to control these stresses. With the availability of the peanut genome database, studies of a number of peanut gene families have been reported, but our understanding of the peanut Hsf gene family is very limited. Here, a total of 46 peanut Hsf genes were identified from the peanut. Comprehensive analyses on the gene structures, cis-acting element composition, chromosome distribution, phylogenetic relationships, and expression patterns were performed. The results of which suggested that the peanut Hsf members might play important roles in peanut development and in response to stresses.
The peanut genome data and the Arabidopsis genome data were downloaded from the Peanut Base (https://www.peanutbase.org/) and TAIR (https://www.arabidopsis.org/), respectively. Arabidopsis Hsf full-length protein sequences were used as queries to perform BLASTP program against the peanut genome database with an E-value of 0.0001. The resulting sequences were then subjected to Pfam (El-Gebali et al., 2019) and SMART (Letunic and Bork, 2018) analyses to detect the presence of the Hsf-type DBD and OD domain. The ProtParam (Wilkins et al., 1999) was used to calculate Mw (molecular weight) and theoretical pI (isoelectric point) of the putative Hsf proteins from peanut.
MAFFT (Katoh et al., 2019) under the default parameters was applied in protein sequences alignment analyses of newly identified peanut Hsf members and previously reported Arabidopsis Hsf members. A neighbor-joining (NJ) tree was constructed by MEGA 11 with the following parameters: Poisson correction, 1000 bootstrap values, and pairwise deletion (Tamura et al., 2021).
The structure of peanut Hsf genes was analyzed using the gene structure display server (Hu et al., 2015) by comparing the coding sequence (CDS) and genomic sequence obtained from the peanut genome database. The 2000-bp sequences upstream of peanut Hsf genes were obtained from peanut genome database as the promoter (Wang et al., 2021). And then the cis-regulatory elements of these promoters were identified by PlantCARE (Lescot et al., 2002).
The typical functional structure domains of peanut Hsf protein were analyzed by Pfam and SMART. The nuclear localization signal (NLS) prediction was done by using the online tool (Nguyen Ba et al., 2009). MEME tools (Bailey et al., 2009) was applied to identify conserved motifs of the AhHsfs full-length proteins.
The chromosomal location image of peanut Hsf genes was generated by TBtools (Chen et al., 2020), according to the data obtained from the peanut genome database. The tandem and segmental duplications were identified by using MCScanX program (Wang et al., 2012) and the results were visualized by Circos (Krzywinski et al., 2009). The syntenic analysis of the orthologous genes obtained from peanut and other three plant species were investigated by TBtools (Chen et al., 2020). Subsequently, the synonymous substitution (Ks) and non-synonymous substitution (Ka) rates were calculated using DnaSP 5.0 software (Librado and Rozas, 2009).
The peanut cultivar HY9312 plants were used to analyze the expression of AhHsf genes in this study. The peanut seeds were germinated on MS medium in a light incubator at 25 °C for two weeks. The seedlings were then transferred to 20% PEG 6000 and 200 mM NaCl for drought and salt stresses, respectively. After treatments of 0, 6, 24, and 48 h, the leaves were harvested for RNA extraction and qRT-PCR analysis. The samples were frozen in liquid nitrogen immediately and then transferred to -80 °C for RNA extraction. Three biological replicates were used for each sample.
Total RNA was extracted by Trizol Reagent according to the manufacturer’s instructions. RNA quality and concentration were tested by using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, United States). The synthesis of fist-strand cDNA by using the PrimeScript ™ RT reagent Kit (TaKaRa, Shiga, Japan) with Oligo (dT) primers. The qRT-PCR was performed on an ABI7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, United States) with 2 µL template cDNA. The peanut Actin11 gene was adopted as the internal control (Li et al., 2021). The relative primer sequences were listed in Supplementary Table 1. All reactions were performed with three biological replications and the resulting data were analyzed by using the 2 −ΔΔCT method (Livak and Schmittgen, 2001).
The coding regions (without stop codon) of AhHsf20 were amplified from the cDNA of peanut leaves and inserted into the pCHF3-cGFP vector by Infusion (Invitrogen), generating the AhHsf20-GFP fusion fragment under control of the CaMV-35S promoter. The 35S::AhHsf20-GFP plasmid was introduced into Agrobacterium competent cell GV3101 for transient expression in the leaf of Nicotiana benthamiana. After growth of three days in light chamber, these leaves were subjected to 4,6-diamidino-2-phenylindole (DAPI) staining to confirm the position of the nucleus. The fluorescence signals were monitored by using the confocal microscope (TCS-SP8 Leica, Wetzlar, Germany), as previously reported (Li et al., 2019).
The coding sequence of AhHsf20 was inserted into the SacI-digested pCHF3 vector by Infusion (Clontech) to produce 35S::AhHsf20, which was then transformed into Agrobacterium tumefaciens GV3101 and used to transform Arabidopsis Col-0 plants by the floral dip method (Zhang et al., 2006). T3 homozygous seedlings were used for further analyses. The sterilized wild-type and transgenic Arabidopsis seeds were evenly sown on 1/2 MS media. The seeds were stratified at 4 °C in darkness for two days, and then transferred to culture room (23 °C, continuous light). Wild-type and transgenic Arabidopsis seedlings growing normally for seven days were transferred to 1/2 MS media containing 100 and 150 mM NaCl. The primary root lengths were measured after seven days of upright growth in 1/2 MS media. Three biological replicates.
The GraphPad Prism 8 (t test) was used to analyze significant differences (P < 0.05, P < 0.01, P < 0.001). All data were obtained from three replicates.
To identify Hsf genes in peanut, the BLASTP search was performed using the Pfam Hsf-type DBD domain (PF00447) as query. After deleting redundant genes, we identified a total of 46 Hsf family proteins in peanut, designating the newly identified peanut Hsf genes AhHsf1 to AhHsf46 according to their position on chromosome. The Chr5, Chr13, and Chr15 contained the largest number of AhHsf genes, with six genes. There was followed by Chr3, which had five AhHsf genes. The Chr6 and Chr16 contained four AhHsf genes, respectively. The Chr8, Chr9, Chr17, and Chr19 contained two AhHsf genes, respectively. Additionally, Chr1, Chr2, Chr7, Chr10, Chr11, Chr12, and Chr20 contained only one AhHsf gene (Supplementary Table 2). The lengths of the predicted AhHsf proteins ranged from 207 amino acids (AhHsf29) to 599 amino acids (AhHsf44). The molecular weights (Mw) of Hsf proteins ranged from 23.6 (AhHsf29) to 66.7 (AhHsf44) kDa, and the theoretical isoelectric points (pI) ranged from 4.82 (AhHsf8 and AhHsf32) to 8.81 (AhHsf4) (Supplementary Table 2).
In order to further study the evolutionary relationship among the peanut Hsf proteins, we constructed a neighbor-joining phylogenetic tree from alignments of 46 peanut Hsf proteins and 21 Arabidopsis Hsf proteins (Figure 1). The results showed that all the Hsf proteins from peanut and Arabidopsis were divided into three groups: A, B, and C. Furthermore, 28 AhHsfs in group A were divided into nine subgroups comprising A1-A9; 16 AhHsfs in group B were divided into four subgroups comprising B1, B2, B3, and B4; and 2 AhHsfs in group C were divided into C1 subgroup. The A1 and B4 subgroups contained the largest number of AhHsf members with six members. There were followed by subgroup A2, A4, A5, B2, and B3 subgroups, all of which had four members of the AhHsf family. Then subgroup A3, A6, A7, A8, A9, B1, and C1 subgroups all contained two members of the AhHsf family (Figure 1).
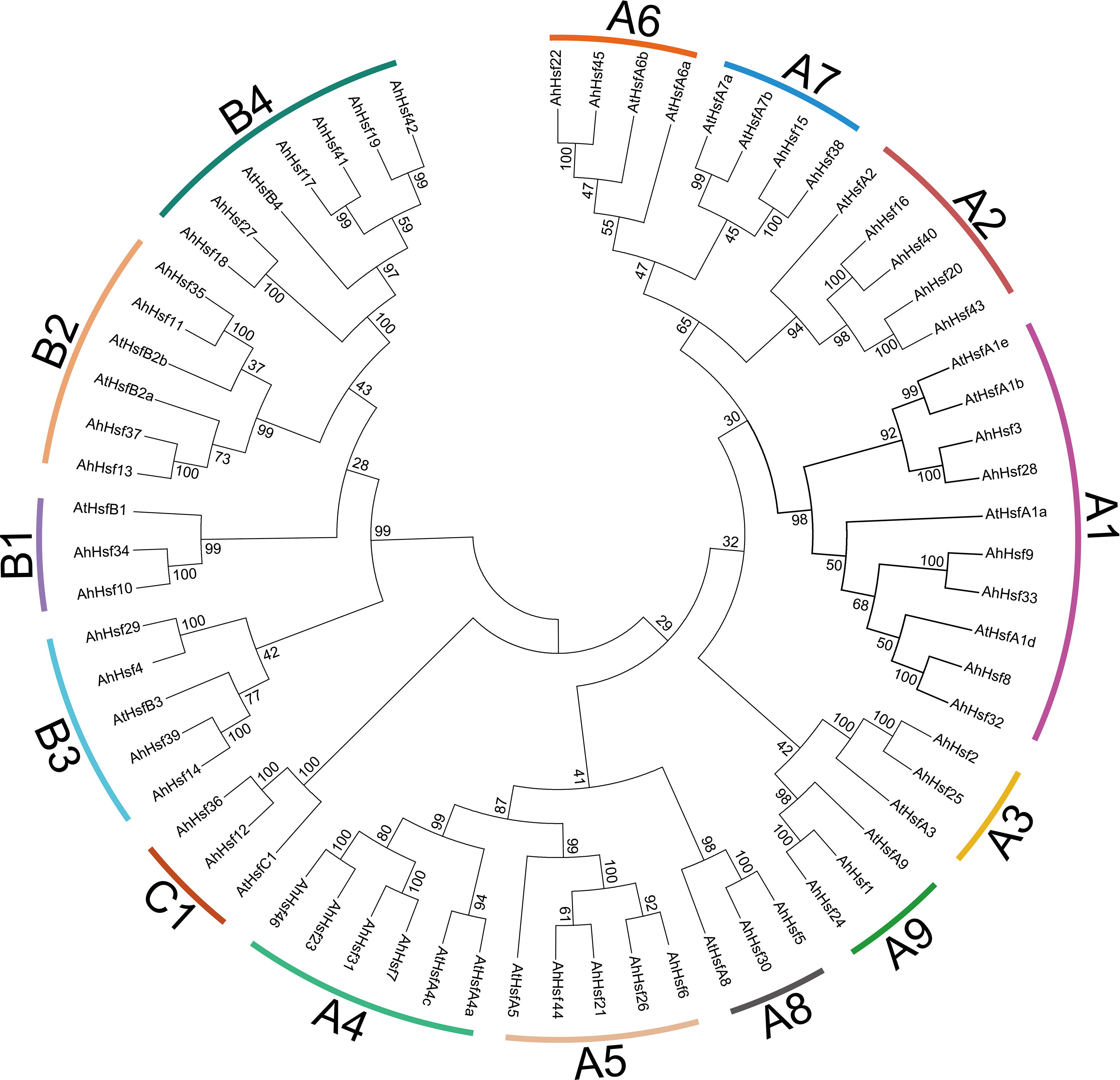
Figure 1 Phylogenetic analysis of peanut Hsf family members. The phylogenetic tree was generated from the alignment of peanut and Arabidopsis Hsf proteins with 1000 bootstrap replicates using the neighbor-joining (NJ) method. The peanut Hsf members together with their Arabidopsis homologs were classified into 14 subgroups.
We identified five typical conserved domains in peanut Hsf proteins, including DBD, OD, NLS, AHA, and NES domains (Supplementary Table 3). All of the AhHsf proteins possessed a highly conserved section, DBD domain, which contained three α-helices and four β-sheets (α1-β1-β2-α2-α3-β3-β4) in N-terminal region (Supplementary Figure 1A). Adjacent to the DBD domain is the HR-A/B domain, which is characterized by a coiled-coil structure in C-terminal region of the AhHsf proteins. We found that group A and group C Hsfs have different amounts of amino acid residues are respectively inserted between the HR-A and HR-B, whereas it is not found in the group B Hsfs (Supplementary Figure 1B). We also found that most of AhHsf proteins have NLS and NES domain, which are essential for maintaining the changes dynamically of Hsf proteins between nucleus and cytoplasm (Scharf et al., 1998; Heerklotz et al., 2001). 32 AhHsf proteins have NLS domain and NES was detected in 16 AhHsf members. Notably, the AHA motifs exist only in class A Hsfs. Among of them, seven AhHsf members (AhHsf8, AhHsf9, AhHsf23, AhHsf25, AhHsf32, AhHsf33, and AhHsf46) contained two AHA motif (Supplementary Table 3).
We analyzed the gene structure of all the AhHsf genes by GSDS, including introns and exons. The results showed that all AhHsf genes contained introns, but the number and length of introns were different (Figure 2B). In group A, there were four introns in AhHsf3 gene, two introns in four AhHsf genes (AhHsf6, AhHsf16, AhHsf40, and AhHsf44), only one intron in others. In group B and group C, all AhHsf genes contained one intron. AhHsf genes had similar gene structures in the same subgroup.
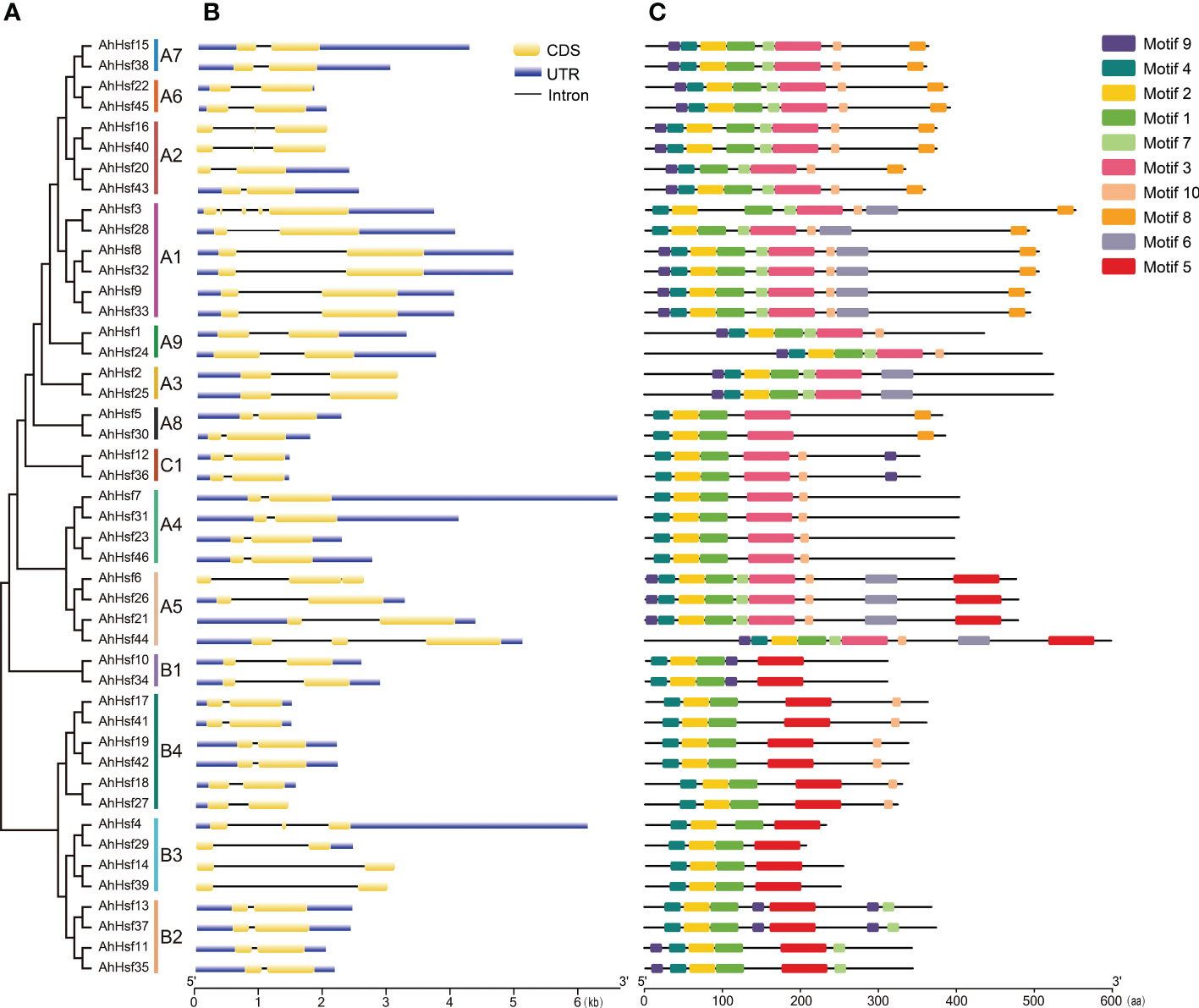
Figure 2 The motif and gene structure organizations of AhHsf members. (A) The evolutionary tree peanut Hsf family members. (B) Gene structure of AhHsf genes. (C) Protein motif of peanut Hsf members.
We used MEME to analyze the motif composition of AhHsf proteins (Figure 2C). A total of 10 motifs (named motif 1-10), ranging from 6 to 50 amino acids, were predicted based on the feature of AhHsf protein sequences, and all sequences were listed in Supplementary Figure 2. Motif 1, motif 2, and motif 4 correspond to the DBD. Motif 3 and motif 5 correspond to the OD. Motif 10 correspond to the NLS. Motif 8 correspond to the NES.
To further understand about the phylogeny of the peanut Hsf family members, syntenic analysis was performed between the Hsf genes of peanut and the Hsf genes of the other four plant species, including Arabidopsis, soybean (Glycine max), tomato (Solanum lycopersicum), and rice (Oryza sativa) (Figure 3). The results revealed that 39, 38, 27, and eight AhHsf genes were synchronized with Hsf genes in soybean, tomato, Arabidopsis, and rice, respectively. The number of collinear pairs between peanut and other four plant species (soybean, tomato, Arabidopsis, and rice), were 121, 52, 36, and 14, respectively, suggesting that the genetic relationship between peanut Hsf genes and soybean Hsf genes was close. Meanwhile, we found that the AhHsf45 of peanut was associated with six, three, two, and two Hsf genes in soybean, Arabidopsis, tomato, and rice, respectively, suggesting that AhHsf45 may play an important role during the evolution in peanut. Notably, a total of six Hsf genes formed collinear pairs with Hsf genes from all of other four plants, suggesting that these Hsf genes may have existed before the divergence of these plant species. The detailed information of syntenic gene pairs is provided in Supplementary Table 4.
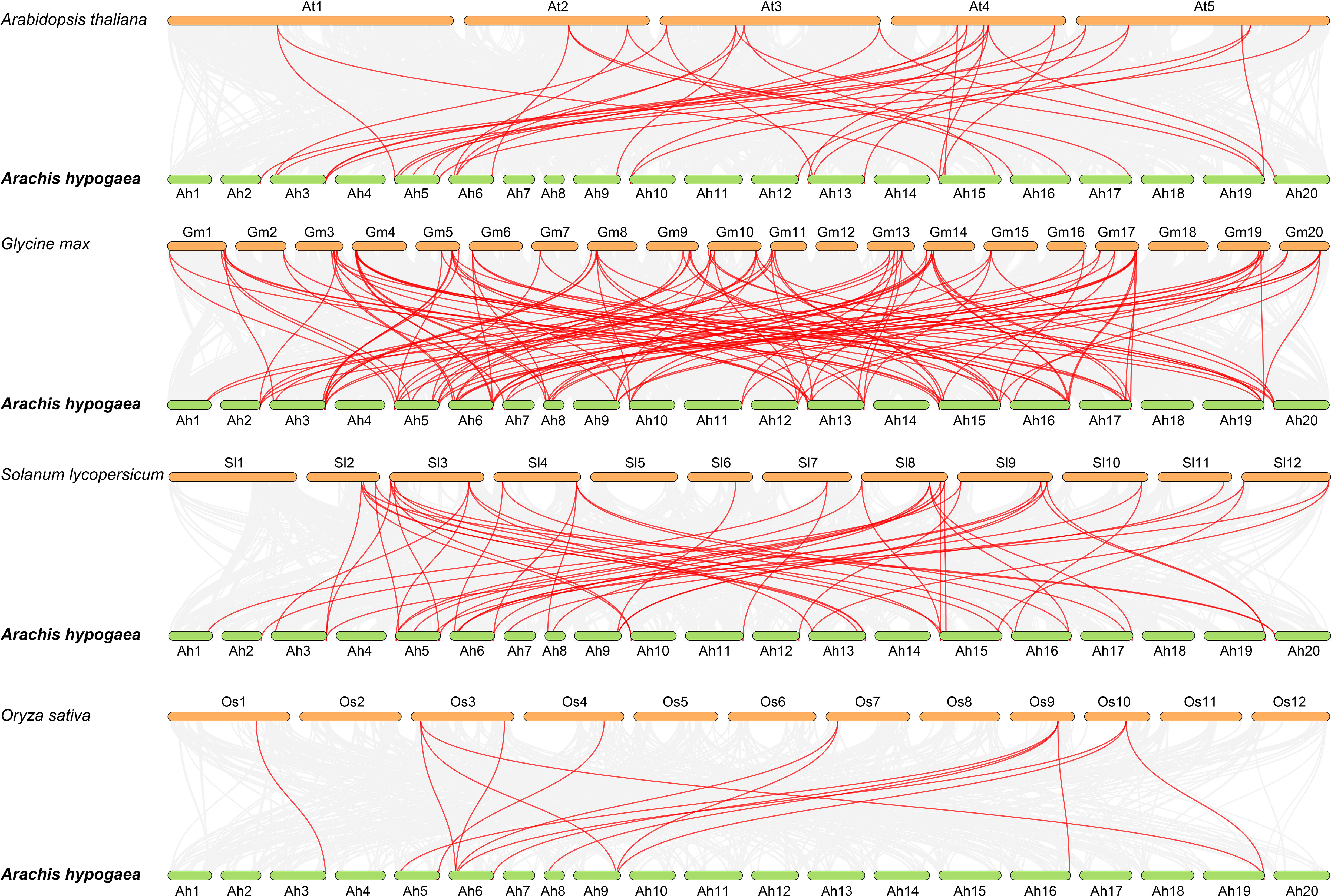
Figure 3 Synteny analysis of Hsf genes between peanut and four other plant species. The gray line in the background represented the collinear blocks between peanut and four other plant species, while the red line exhibited the syntenic Hsf gene pairs.
In this study, 46 AhHsf genes were mapped on 17 peanut chromosomes, and each of which contains different number of the AhHsf genes (Figure 4A). The most peanut Hsf genes (six) were found on Ah5, Ah13, and Ah15, while Ah1, Ah2, Ah7, Ah10, Ah11, Ah12, and Ah20 only have one peanut Hsf gene. It has been reported that when a chromosome region within 200 kb possesses two or more genes of the same family, these genes are defined to be a gene cluster and genes sharing an identity of more than 70% in a cluster are considered to be tandem duplication genes (Holub, 2001). Homology analysis showed that there were two tandem duplication pairs (AhHsf8/AhHsf9 and AhHsf32/AhHsf33) in peanut Hsf gene family (Figure 4A).
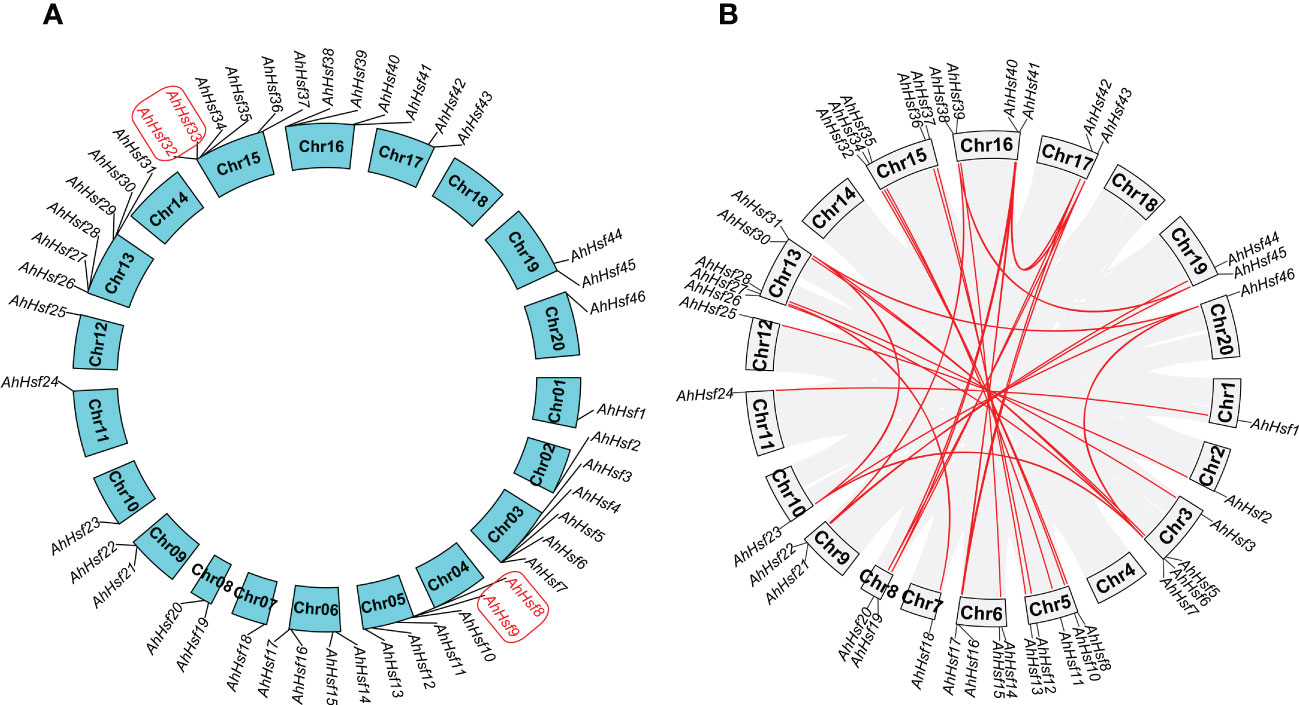
Figure 4 Chromosomal distribution and duplication events. (A) In peanut, 46 AhHsf genes were mapped on 17 peanut chromosomes. The tandem duplication pairs were featured by the red color. (B) The 35 putative segmental duplication pairs of AhHsf genes were investigated using MCScanX and linked by the colored lines, respectively. The gray lines indicate all putative segmental duplication pairs in the peanut genome, while the AhHsf segmental duplication pairs were linked by the red line.
We used the MCScanX to analyze segmental duplication or whole genome duplication of the AhHsf genes. In a total, 35 segmental duplication pairs were identified in 42 AhHsf genes (Figure 4B). 23 pairs of segmental duplication occurred in group A Hsf genes. 11 pairs of segmental duplication occurred in group B Hsf genes. However, only one pair of segmental duplication (AhHsf12/AhHsf36) occurred in group C Hsf genes. These results suggested that the generation of some Hsf genes may be due to gene duplication events. All of the tandem and segmental duplication genes were listed in Supplementary Table 5.
Ka/Ks refers to the ratio between non-synonymous and synonymous substitutions, which can estimate whether the selective pressure acts on the protein-coding gene. There are three selection roles in evolutionary analysis, including the positive selection, neutral selection, and purifying selection. All Ka/Ks ratios of the 35 segmental and two tandem duplication pairs were less than one, suggesting that these Hsf genes may have undergone purifying selective pressure in process of evolution (Supplementary Table 5).
In order to study the potential function of AhHsf genes in stress responses, plantCARE was used to predict the cis-elements in promoter regions (Figure 5). Four hormone-responsive elements were identified in most AhHsf gene promoters, including ABRE, GARE, TCA-element, and CGTCA-motif, which regulate the plant responses to ABA, gibberellin, salicylic acid (SA), and methyl jasmonate (MeJA), respectively. The result suggested that most AhHsf genes may be involved in ABA-mediated and MeJA-mediated stress responses. Notably, the AuxRR-core element of regulating the plant responses to auxin was only detected in promoter regions of certain AhHsf genes, including AhHsf2, AhHsf3, AhHsf5, AhHsf12, and AhHsf25. Six stress-responsive elements including anaerobic induction element (ARE), wound-responsive element (WUN-motif), stress-responsive element (TC-rich repeats), WRKY binding site (W-box), MYB binding site (MBS), and low-temperature-responsive element (LTR) were also detected in the promoter regions. These AhHsf genes may be involved in responses to different stress conditions. Furthermore, a cis-element related to flavonoid biosynthetic (MBSI) was found in promoter regions of five AhHsf genes.
To determine the expression patterns of the AhHsf genes in different tissues, the corresponding transcriptome data with 22 tissues were accessed in the Peanut Base. It was found that the expressions of the AhHsf genes were significant differences in different tissues. As shown in Figure 6, AhHsf5 from group A was highly expressed in the all-tested tissues, whereas the expression of its homologue AhHsf30 was barely detectable. AhHsf1 and AhHsf24 were highly expressed in seed Pat. The expression levels of AhHsf23 and AhHsf46 in leaves were higher than other tissues. In group B, AhHsf14 and AhHsf39 were highly expressed in root and nodule. However, the expression of these two genes were lower in other tissues. Additionally, in group C, the expression levels of AhHsf12 and AhHsf36 were similar in the all-tested tissues. The various expression patterns suggested that AhHsf genes have different functions in peanut growth and development.
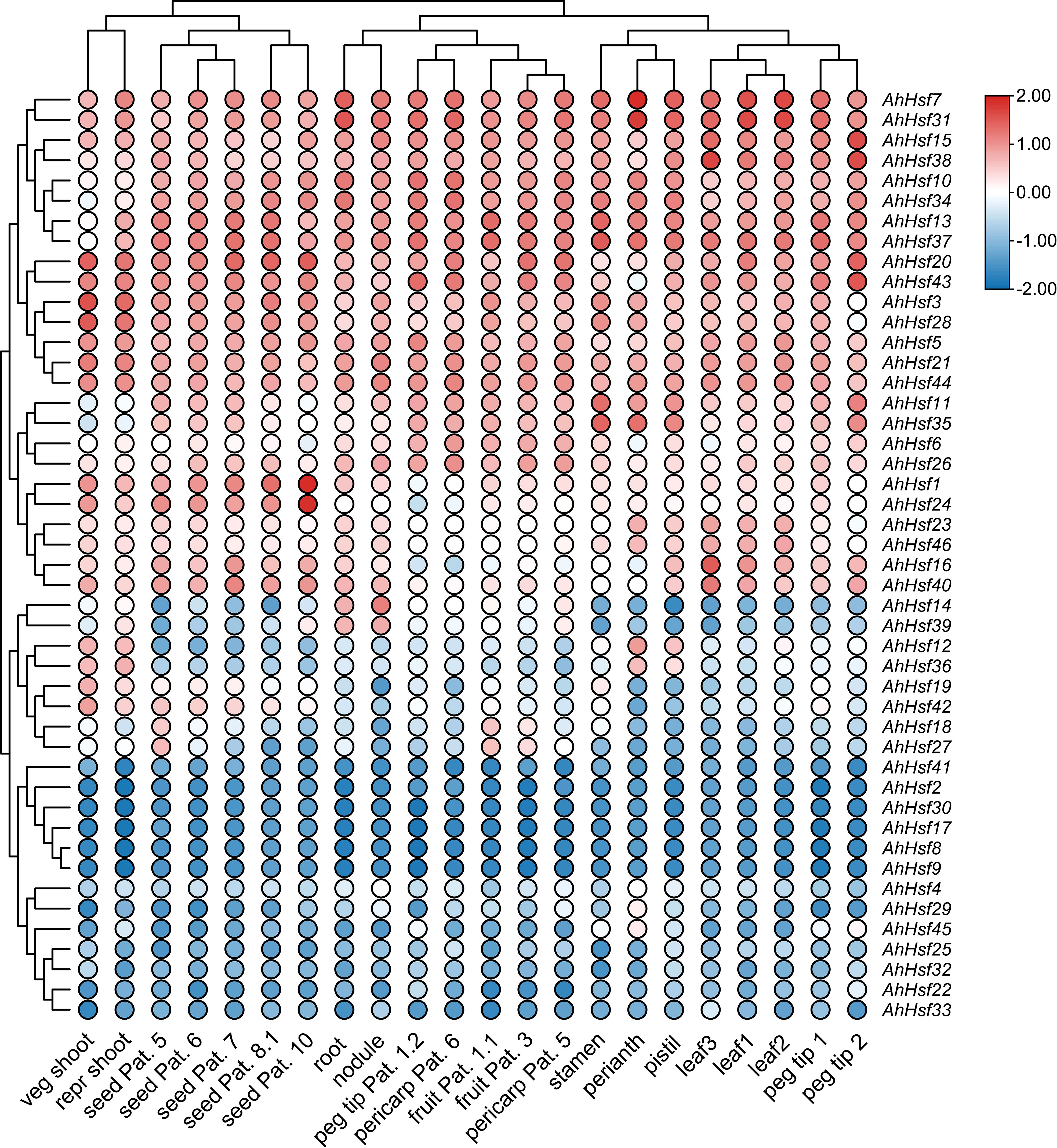
Figure 6 The expression patterns of AhHsf genes in 22 tissues. The FPKM values of each AhHsf gene were normalized and clustered using TBtools.
To reveal peanut Hsf genes expression patterns under abiotic stresses, we utilized transcriptome data after drought and salt stresses from NCBI (Zhao et al., 2018; Zhang et al., 2020). As shown in Figure 7, most peanut Hsf genes exhibited several different expression patterns. Some genes, such as AhHsf11, AhHsf24, and AhHsf35, were upregulated under drought and salt stresses. Several peanut Hsf genes showed conflicting expression patterns under the two stress conditions. For instance, AhHsf1 was insensitive to salt stress, whereas it was significantly induced by drought stress. Additionally, some genes did not show significant expression changes after drought and salt stress treatments, such as AhHsf46. The various expression patterns reflected the different roles of peanut Hsf genes in abiotic stress-response pathways.
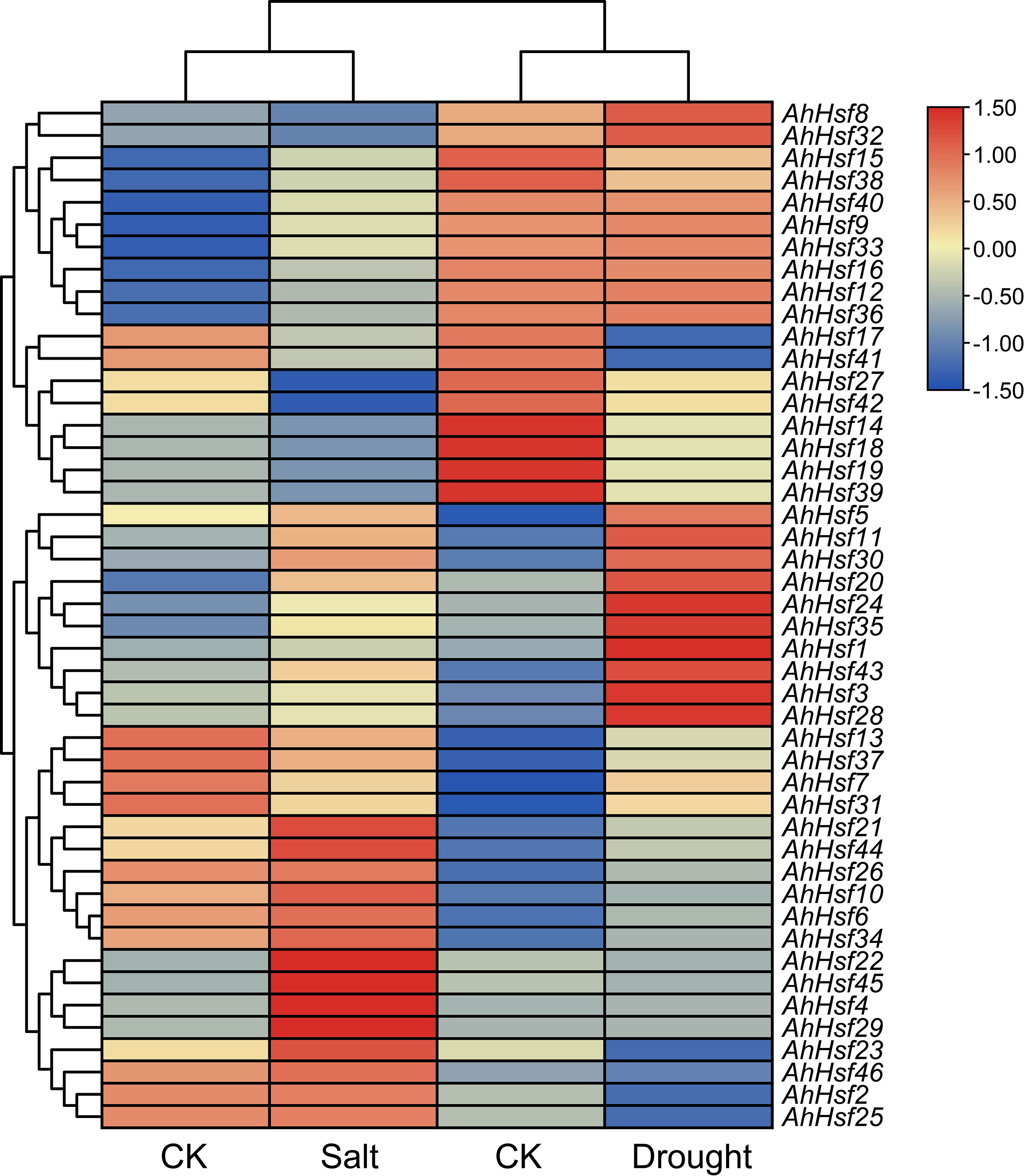
Figure 7 Heatmap of AhHsf genes under drought and salt stresses. The FPKM values of each AhHsf gene were normalized and clustered using TBtools.
qRT-PCR was used to verify the expression pattern of peanut Hsf genes in response to abiotic stresses, including drought and salt. According to the results, we found that several AhHsf genes were induced by drought and salt stresses (Figure 8). For example, the expression levels of AhHsf1, AhHsf20, and AhHsf24 were continually up-regulated under drought stress, AhHsf4 and AhHsf24 were continually up-regulated under salt stress. Moreover, expression levels of AhHsf1/4/20/24/29/35/43 were significantly increased (>10-fold) in drought stress condition. AhHsf4, AhHsf20, and AhHsf29 were significantly increased (>7-fold) in salt stress condition. Notably, seven AhHsf genes, including AhHsf1, AhHsf4, AhHsf20, AhHsf24, AhHsf29, AhHsf35, and AhHsf43, were induced under both drought and salt stresses. These results suggested that peanut Hsf genes might be involved in response to drought and salt stresses.
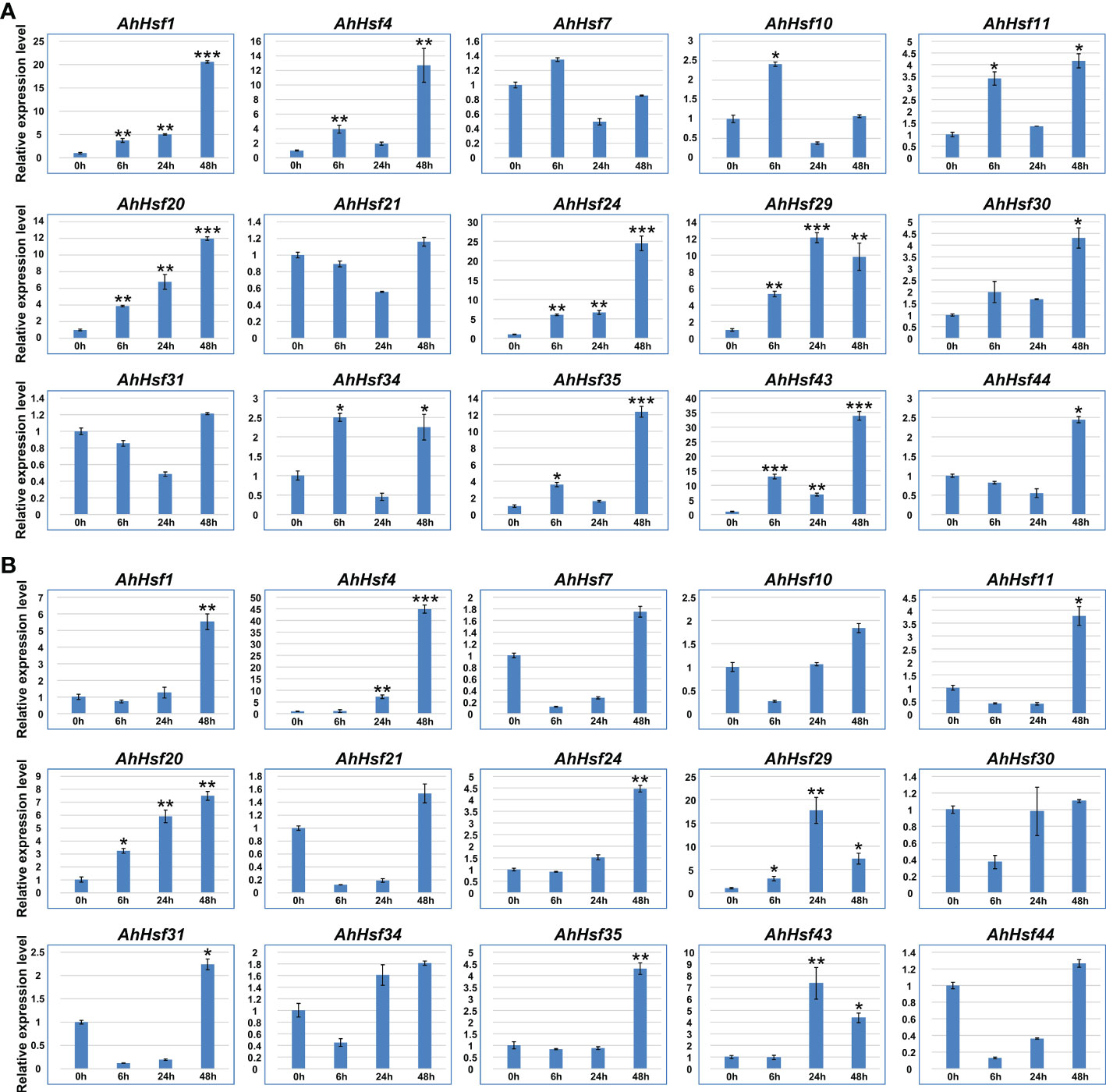
Figure 8 The expression patterns of AhHsf genes under drought and salt conditions. (A) The expression pattern of selected AhHsf genes in response to drought stress treatments, which was calculated as folds relative to the untreated control. (B) The expression pattern of selected AhHsf genes in response to salt stress treatments, which was calculated as folds relative to the control. The data were means ± SD of three biological repeats. *p < 0.05, **p < 0.01, ***p < 0.001 (t-tests).
In order to further explore the potential function of the AhHsf genes. The stress-induced AhHsf20 gene was selected for subcellular localization analysis (Figure 9). The coding region of AhHsf20 without the stop codon was fused to the GFP reporter gene under control of the CaMV35S promoter. The Agrobacterium cultures with the recombinant construct were used to inject the tobacco leaf epidermal cells, using 35S::GFP vector as the control. Subsequently, the signal of GFP protein was observed by confocal microscopy. It can be seen from Figure 9 that AhHsf20 fusion protein was specifically located in the nucleus, whereas the control group was uniformly distributed throughout the whole cell. These results showed that AhHsf20 protein was located in nucleus.
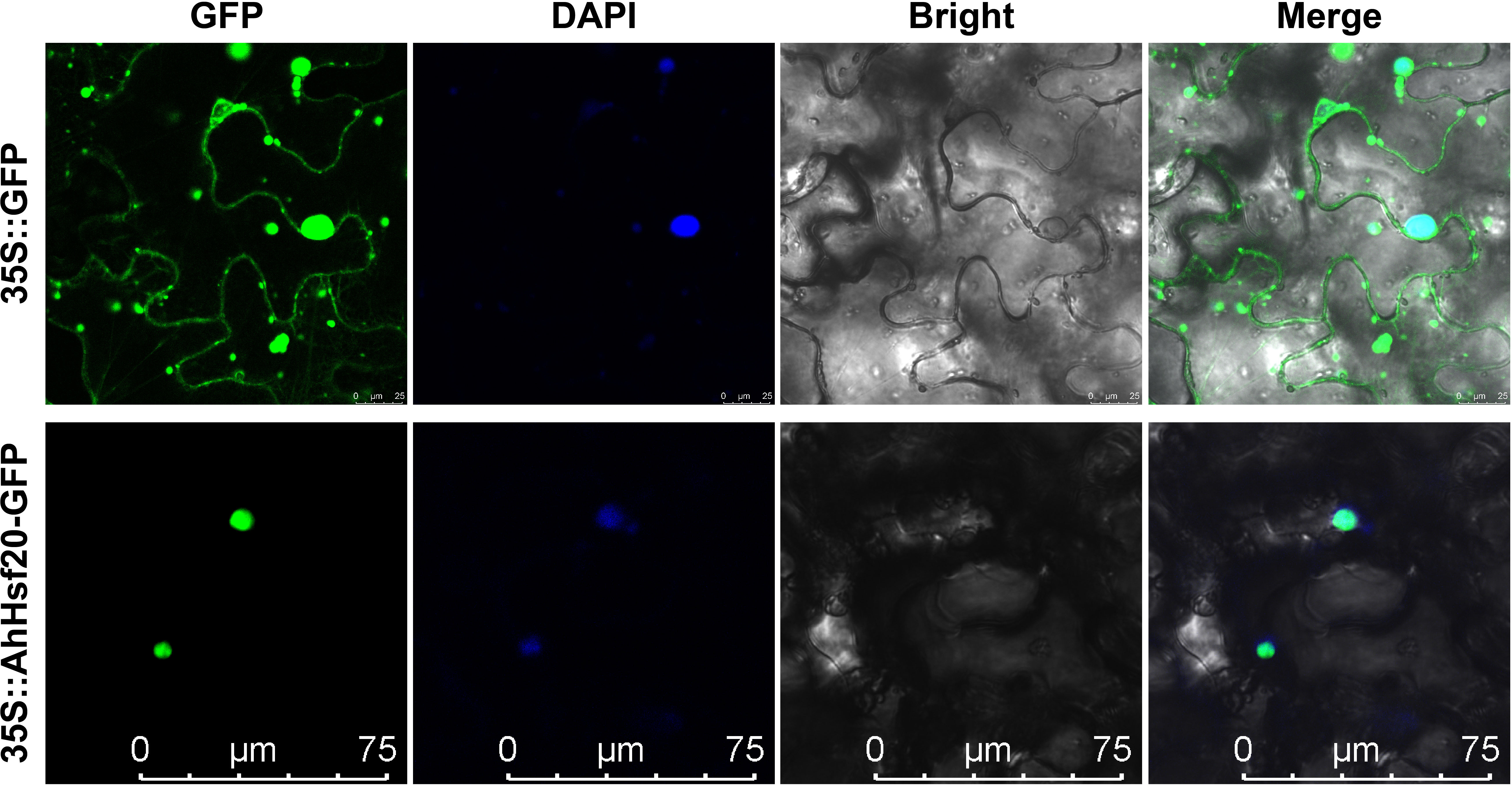
Figure 9 The subcellular localization of AhHsf20 in tobacco epidermal cells. The AhHsf20-GFP fusion construct and GFP gene driven by the CaMV-35S promoter were transiently expressed in tobacco, respectively. DAPI (dye 4,6-diamidino-2-phenylindole) staining indicted the nucleus.
To explore the underlying mechanisms of action of AhHsf members, interaction networks of AhHsfs in peanut were predict using STRING online tool (https://cn.string-db.org/). We found that protein-protein interaction occurred between 18 peanut Hsf members (Supplementary Figure 3). There were four hub nodes in the AhHsf protein interaction network, including AhHsf13, AhHsf10, AhHsf2, and AhHsf18. Among them, AhHsf13 was found to interact with ten other Hsf members, followed by AhHsf2 (4), AhHsf10 (3), AhHsf18 (3). The results showed that these interacting proteins might co-exert specific biological functions in peanut.
To further investigate the function of AhHsf20 in response to abiotic stresses, we generated transgenic lines overexpressing AhHsf20 under the control of the 35S promoter in Arabidopsis Col-0 plants (AhHsf20-OE). Two overexpression lines AhHsf20-OE1 and AhHsf20-OE3 were selected for further studies (Figure 10B). Compared with wild-type, AhHsf20-OE1 and AhHsf20-OE3 plants displayed a longer root phenotype under 100 and 150 mM NaCl (Figures 10A, C). The results revealed that the overexpression of AhHsf20 enhanced the salt tolerance in transgenic Arabidopsis.
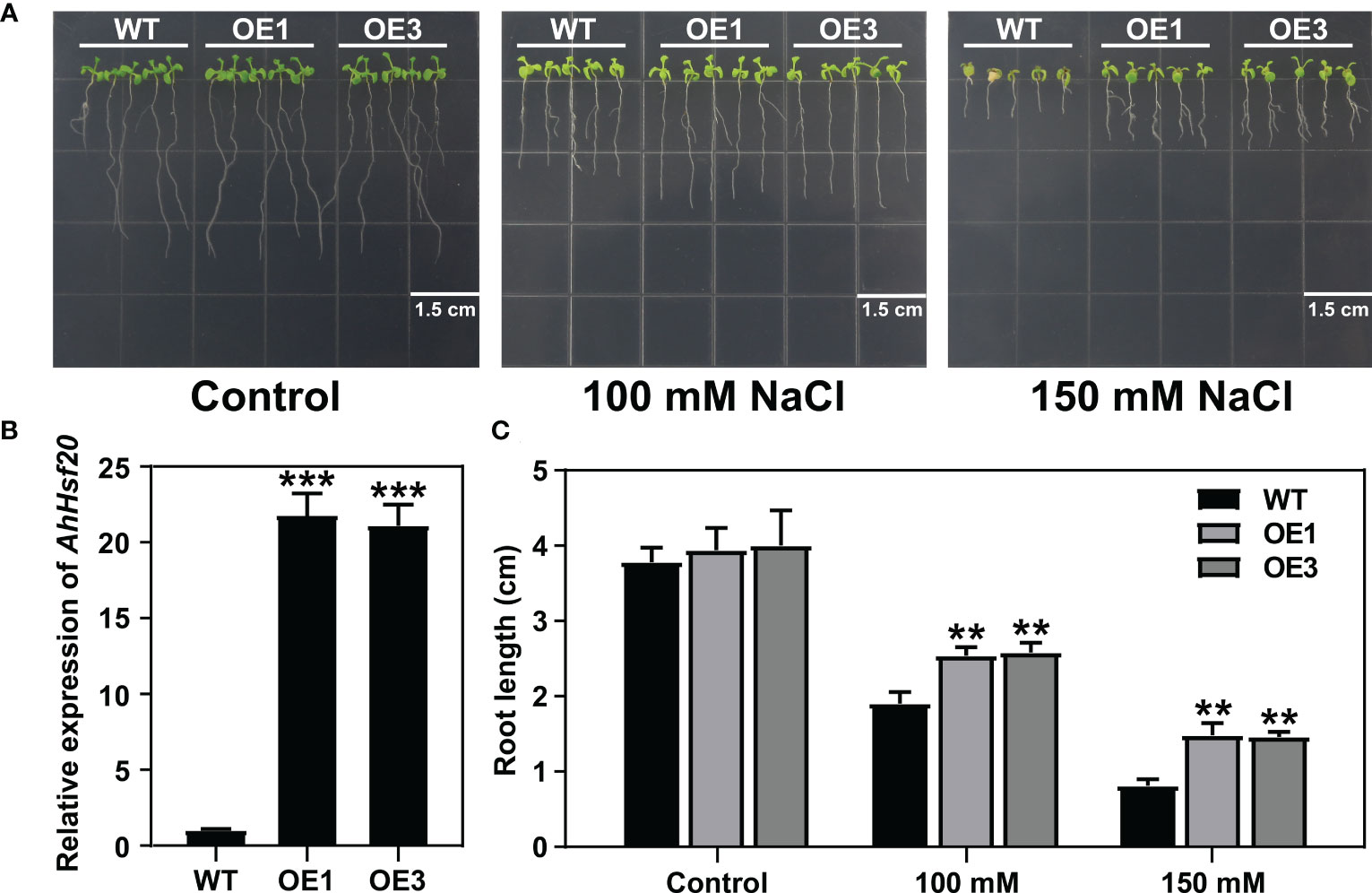
Figure 10 Effects of salt stress treatments on the root growth of the AhHsf20 gene overexpressed in Arabidopsis. (A) Primary root lengths of the WT and AhHsf20 overexpression lines under salt treatments in transgenic Arabidopsis. (B) The expression levels of AhHsf20 gene in WT and two overexpression lines. The ratios of gene expression were calculated relative to the WT. (C) Quantification of the primary root lengths under normal condition and 100 and 150 mM NaCl treatments. The data were retrieved from three biological replicates. WT, wild type. Data are the mean ± SD of three biological repeats. **p < 0.01, ***p < 0.001 (t tests).
It has been reported that Hsf genes are heat shock transcription factors and play important roles in response to abiotic stresses and plant development processes (von Koskull-Doring et al., 2007). Cultivated peanut (Arachis hypogaea L.) is not only an important economic crop but also an oil crop. Thus, identification and analysis Hsf genes in peanut will be great significance on various stress responses. Previous study found that 16 and 17 Hsf genes were identified from Arachis duranensis and A. ipaensis, respectively (Wang et al., 2017). However, the Hsf gene family in cultivated peanut has not been comprehensive analyses so far. There are only 21 Hsf genes in Arabidopsis (Guo et al., 2008). In the present study, a total of 46 Hsf genes were identified from peanut, probably due to peanut is an allopolytraploid. Consistent with the classification of Hsf genes in other plant species including Arabidopsis, rice (Guo et al., 2008), potato (Tang et al., 2016), and tomato (Yang et al., 2016), peanut Hsf genes were also divided into three groups: A, B, and C (Figure 1). There are 28 Hsf genes belong to group A (A1-A9), 16 in group B (B1-B4), and two in group C (C1).
Conserved motif analysis suggested that all of 46 AhHsf proteins contain two necessary domains (DBD and OD) and/or three specific domains (NLS, AHA, and NES). The transcriptional activation activities of Hsf from group A were worked by the AHA motif in the C-terminal region (Doring et al., 2000). In the peanut Hsf family, 27 of 28 AhHsfA members with AHA motif (Supplementary Table 3), suggesting that these AhHsf members may have self-transcriptional activation activity, whereas the AhHsf16 from subgroup A2 without AHA motif. It has been reported that HsfA members with no AHA motif might be activated by forming hetero-polymers with other group A Hsf (Scharf et al., 2012). Furthermore, the gene structures of peanut Hsf genes are highly conserved within the same subgroup (Figure 2), further supporting the result of evolutionary analysis.
Segmental duplication and tandem duplication events contribute to the expansion of the gene families in plant genomes (Vision et al., 2000; Cannon et al., 2004). In this study, we found that the expansion of the AhHsf genes in peanut is mainly caused by segmental duplication, not tandem duplication. A total of 35 segmental and two tandem duplication pairs were identified (Figure 4), with all the Ka/Ks ratios of these duplication pairs were less than one (Supplementary Table 5), suggesting that these duplicated peanut Hsf genes might have undergone purifying selective pressure in process of peanut evolution. In addition, duplicated peanut Hsf genes belonged to the same subgroup, such as AhHsf8/AhHsf9 in subgroup A1 and AhHsf10/AhHsf34 in subgroup B1. Notably, AhHsf10 and AhHsf34, as a segmental duplication pair, exhibited consistent organ/tissue expression patterns (Figure 6).
Most of the Hsf transcription factors have been reported to be involved in the regulation of Arabidopsis developmental processes. For example, AtHsfA9 from subgroup A9 was reported to be involved in regulation of seed development in Arabidopsis (Kotak et al., 2007). A segmental duplication pair AhHsf1 and AhHsf24 was clustered together with AtHsfA9 in subgroup A9, and these two genes both highly expressed in seeds (Figure 6), suggesting that AhHsf1 and AhHsf24 might also be involved in regulating seed development. In addition, AtHsfB2a was shown to play important role in regulating gametophyte development of Arabidopsis (Wunderlich et al., 2014). Its homolog AhHsf13 showed high expression level in stamen (Figures 3, 6), which suggested that AtHsfB2a and AhHsf13 might have similar biological functions.
Drought and salt stresses are the two major abiotic stresses adversely affecting the growth development and yields of crops (Suzuki et al., 2014). The plant cells will exhibit consistent changes in water loss under drought or salt stress. Osmotic adjusting can enhance water absorption to cope with salt and drought stresses, simultaneously (Munns et al., 2020; Ozturk et al., 2021). For example, under salt and drought stresses, a Chloride (Cl-)-tolerant species Pugionium cornutum that can accumulate Cl- and enhance osmotic adjustment capacity to improve growth (Cui et al., 2020). Notably, the accumulation of osmotic substances can be realized by altering relative gene expression levels. In tomato, transgenic SlWRKY8 plants exhibit enhanced salt and drought tolerance, with higher osmotic substances like proline (Gao et al., 2020). It is worth noting that a number of Hsf members have been reported to be involved in response to plant environment stresses. In subgroup A1, AtHsfA1b has been reported to enhance drought resistance (Bechtold et al., 2013). Its peanut homologs AhHsf3 and AhHsf28 were found to be induced by drought stress (Figures 3, 7), implying that these AhHsf genes might be involved in regulating drought stress response. Furthermore, AhHsf3 and AhHsf28 were predicted to have arisen from segmental duplication events (Figure 4B). In subgroup A6, AtHsfA6b was demonstrated to induce stress-responsible genes under salt or drought stresses via the ABA dependent signaling pathway (Huang et al., 2016). AhHsf22 and AhHsf45 were clustered together with AtHsfA6b in subgroup A6, and both were induced by salt stress (Figure 7). Furthermore, the promoter regions of AhHsf22 and AhHsf45 have one ABA-responsive element (ABRE) respectively (Figure 5), suggesting that these peanut Hsf genes might be involved in response to salt stress through the ABA signaling pathway. Notably, salt and drought stress treatments can significantly induce the expression of AhHsf20 (Figures 7, 8), implying that AhHsf20 may be involved in salt- and drought-stress responses. Further experiments indicated that the overexpression of AhHsf20 can enhance salt tolerance in transgenic Arabidopsis (Figure 10), supporting the potential roles of more AhHsf proteins in stress responses.
In this study, a systematic analysis of the peanut Hsf gene family was performed, and a total of 46 AhHsf members were identified. They were divided into three groups and 14 subgroups together with their Arabidopsis homologs. Our analyses of Hsf genes of peanut and Arabidopsis revealed their diversity in the number of members, evolutionary relationship, gene structure, chromosome locations, gene duplication, collinearity, tissue expression patterns and expression patterns in response to salt and drought stresses. The results indicate that the AhHsf genes may be important for regulating peanut responses to abiotic stresses and development. Notably, AhHsf20 was induced by salt and drought stresses and was able to enhance salt tolerance in transgenic Arabidopsis. Overall, these results from this study can help us to further examine the specific functions of the AhHsf genes and provide some new genes resource for peanut stress resistance breeding.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
PC and SS conceived this research, designed the experiments. QW, Z, and CG conducted the research and drafted the manuscript. XZ, ZL, YM, QS, JW, CY, and CL assisted in data collection and analysis. All authors have read and approved the final version of the manuscript.
This work was supported by the National Key Research and Development Program of China (2022YFD1200400), the Innovation Project of SAAS (CXGC2022E20, CXGC2023F20), the Agro-industry Technology Research System of Shandong Province (SDAIT-04-02), and the Shandong Elite Variety Project (2020LZGC001).
CG was employed by Kunming Branch of Yunnan Provincial Tobacco Company.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1214732/full#supplementary-material
Supplementary Figure 1 | Multiple sequence alignment of the DBD domains (A) and HR-A/B regions (B) of the Hsf protein family in peanut.
Supplementary Figure 2 | The putative conserved motifs in AhHsf proteins.
Supplementary Figure 3 | Interaction networks of AhHsf in peanut.
Supplementary Table 1 | The qRT-PCR primers used in this study.
Supplementary Table 2 | The detail information of identified peanut Hsf family members.
Supplementary Table 3 | The functional domains and motifs of peanut Hsf members.
Supplementary Table 4 | The syntenic pairs between peanut and other four plant species.
Supplementary Table 5 | The detail information of segmental and tandem duplication gene pairs.
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Banti, V., Mafessoni, F., Loreti, E., Alpi, A., Perata, P. (2010). The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol. 152, 1471–1483. doi: 10.1104/pp.109.149815
Bechtold, U., Albihlal, W. S., Lawson, T., Fryer, M. J., Sparrow, P. A., Richard, F., et al. (2013). Arabidopsis HEAT SHOCK TRANSCRIPTION FACTORA1b overexpression enhances water productivity, resistance to drought, and infection. J. Exp. Bot. 64, 3467–3481. doi: 10.1093/jxb/ert185
Begum, T., Reuter, R., Schoffl, F. (2013). Overexpression of AtHsfB4 induces specific effects on root development of Arabidopsis. Mech. Dev. 130, 54–60. doi: 10.1016/j.mod.2012.05.008
Bienz, M., Pelham, H. R. (1987). Mechanisms of heat-shock gene activation in higher eukaryotes. Adv. Genet. 24, 31–72. doi: 10.1016/s0065-2660(08)60006-1
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10
Charng, Y. Y., Liu, H. C., Liu, N. Y., Chi, W. T., Wang, C. N., Chang, S. H., et al. (2007). A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 143, 251–262. doi: 10.1104/pp.106.091322
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, H., Hwang, J. E., Lim, C. J., Kim, D. Y., Lee, S. Y., Lim, C. O. (2010). Arabidopsis DREB2C functions as a transcriptional activator of HsfA3 during the heat stress response. Biochem. Biophys. Res. Commun. 401, 238–244. doi: 10.1016/j.bbrc.2010.09.038
Cui, Y. N., Li, X. T., Yuan, J. Z., Wang, F. Z., Guo, H., Xia, Z. R., et al. (2020). Chloride is beneficial for growth of the xerophyte Pugionium cornutum by enhancing osmotic adjustment capacity under salt and drought stresses. J. Exp. Bot. 71, 4215–4231. doi: 10.1093/jxb/eraa158
Doring, P., Treuter, E., Kistner, C., Lyck, R., Chen, A., Nover, L. (2000). The role of AHA motifs in the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell 12, 265–278. doi: 10.2307/3870927
Duan, S., Liu, B., Zhang, Y., Li, G., Guo, X. (2019). Genome-wide identification and abiotic stress-responsive pattern of heat shock transcription factor family in Triticum aestivum l. BMC Genomics 20, 257. doi: 10.1186/s12864-019-5617-1
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Friedrich, T., Oberkofler, V., Trindade, I., Altmann, S., Brzezinka, K., Lamke, J., et al. (2021). Heteromeric HSFA2/HSFA3 complexes drive transcriptional memory after heat stress in Arabidopsis. Nat. Commun. 12, 3426. doi: 10.1038/s41467-021-23786-6
Gao, Y. F., Liu, J. K., Yang, F. M., Zhang, G. Y., Wang, D., Zhang, L., et al. (2020). The WRKY transcription factor WRKY8 promotes resistance to pathogen infection and mediates drought and salt stress tolerance in Solanum lycopersicum. Physiol. Plant 168, 98–117. doi: 10.1111/ppl.12978
Guo, J., Wu, J., Ji, Q., Wang, C., Luo, L., Yuan, Y., et al. (2008). Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genomics 35, 105–118. doi: 10.1016/S1673-8527(08)60016-8
Heerklotz, D., Doring, P., Bonzelius, F., Winkelhaus, S., Nover, L. (2001). The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Mol. Cell Biol. 21, 1759–1768. doi: 10.1128/MCB.21.5.1759-1768.2001
Higashi, Y., Ohama, N., Ishikawa, T., Katori, T., Shimura, A., Kusakabe, K., et al. (2013). HsfA1d, a protein identified via FOX hunting using Thellungiella salsuginea cDNAs improves heat tolerance by regulating heat-stress-responsive gene expression. Mol. Plant 6, 411–422. doi: 10.1093/mp/sst024
Holub, E. B. (2001). The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2, 516–527. doi: 10.1038/35080508
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Huang, Y., Li, M. Y., Wang, F., Xu, Z. S., Huang, W., Wang, G. L., et al. (2015). Heat shock factors in carrot: genome-wide identification, classification, and expression profiles response to abiotic stress. Mol. Biol. Rep. 42, 893–905. doi: 10.1007/s11033-014-3826-x
Huang, Y. C., Niu, C. Y., Yang, C. R., Jinn, T. L. (2016). The heat stress factor HSFA6b connects ABA signaling and ABA-mediated heat responses. Plant Physiol. 172, 1182–1199. doi: 10.1104/pp.16.00860
Katoh, K., Rozewicki, J., Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kotak, S., Port, M., Ganguli, A., Bicker, F., von Koskull-Doring, P. (2004). Characterization of c-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class a hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. 39, 98–112. doi: 10.1111/j.1365-313X.2004.02111.x
Kotak, S., Vierling, E., Baumlein, H., von Koskull-Doring, P. (2007). A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell 19, 182–195. doi: 10.1105/tpc.106.048165
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi: 10.1093/nar/gkx922
Li, X., Ahmad, S., Ali, A., Guo, C., Li, H., Yu, J., et al. (2019). Characterization of somatic embryogenesis receptor-like kinase 4 as a negative regulator of leaf senescence in Arabidopsis. Cells 8, 50. doi: 10.3390/cells8010050
Li, C., Yan, C., Sun, Q., Wang, J., Yuan, C., Mou, Y., et al. (2021). The bHLH transcription factor AhbHLH112 improves the drought tolerance of peanut. BMC Plant Biol. 21, 540. doi: 10.1186/s12870-021-03318-6
Li, P. S., Yu, T. F., He, G. H., Chen, M., Zhou, Y. B., Chai, S. C., et al. (2014). Genome-wide analysis of the hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genomics 15, 1009. doi: 10.1186/1471-2164-15-1009
Librado, P., Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lin, Y. X., Jiang, H. Y., Chu, Z. X., Tang, X. L., Zhu, S. W., Cheng, B. J. (2011). Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genomics 12, 76. doi: 10.1186/1471-2164-12-76
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lohmann, C., Eggers-Schumacher, G., Wunderlich, M., Schoffl, F. (2004). Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol. Genet. Genomics 271, 11–21. doi: 10.1007/s00438-003-0954-8
Munns, R., Passioura, J. B., Colmer, T. D., Byrt, C. S. (2020). Osmotic adjustment and energy limitations to plant growth in saline soil. New Phytol. 225, 1091–1096. doi: 10.1111/nph.15862
Nguyen Ba, A. N., Pogoutse, A., Provart, N., Moses, A. M. (2009). NLStradamus: a simple hidden Markov model for nuclear localization signal prediction. BMC Bioinf. 10, 202. doi: 10.1186/1471-2105-10-202
Nishizawa-Yokoi, A., Nosaka, R., Hayashi, H., Tainaka, H., Maruta, T., Tamoi, M., et al. (2011). HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the hsf signaling network in response to environmental stress. Plant Cell Physiol. 52, 933–945. doi: 10.1093/pcp/pcr045
Nover, L., Bharti, K., Doring, P., Mishra, S. K., Ganguli, A., Scharf, K. D. (2001). Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6, 177–189. doi: 10.1379/1466-1268(2001)006<0177:aathst>2.0.co;2
Ozturk, M., Turkyilmaz Unal, B., Garcia-Caparros, P., Khursheed, A., Gul, A., Hasanuzzaman, M. (2021). Osmoregulation and its actions during the drought stress in plants. Physiol. Plant 172, 1321–1335. doi: 10.1111/ppl.13297
Scharf, K. D., Berberich, T., Ebersberger, I., Nover, L. (2012). The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim. Biophys. Acta 1819, 104–119. doi: 10.1016/j.bbagrm.2011.10.002
Scharf, K. D., Heider, H., Hohfeld, I., Lyck, R., Schmidt, E., Nover, L. (1998). The tomato hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol. Cell Biol. 18, 2240–2251. doi: 10.1128/MCB.18.4.2240
Schoffl, F., Prandl, R., Reindl, A. (1998). Regulation of the heat-shock response. Plant Physiol. 117, 1135–1141. doi: 10.1104/pp.117.4.1135
Schramm, F., Ganguli, A., Kiehlmann, E., Englich, G., Walch, D., von Koskull-Doring, P. (2006). The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol. Biol. 60, 759–772. doi: 10.1007/s11103-005-5750-x
Shinozaki, K., Yamaguchi-Shinozaki, K., Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417. doi: 10.1016/s1369-5266(03)00092-x
Song, C., Chung, W. S., Lim, C. O. (2016). Overexpression of heat shock factor gene HsfA3 increases galactinol levels and oxidative stress tolerance in Arabidopsis. Mol. Cells 39, 477–483. doi: 10.14348/molcells.2016.0027
Suzuki, N., Rivero, R. M., Shulaev, V., Blumwald, E., Mittler, R. (2014). Abiotic and biotic stress combinations. New Phytol. 203, 32–43. doi: 10.1111/nph.12797
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tang, R., Zhu, W., Song, X., Lin, X., Cai, J., Wang, M., et al. (2016). Genome-wide identification and function analyses of heat shock transcription factors in potato. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00490
Umezawa, T., Fujita, M., Fujita, Y., Yamaguchi-Shinozaki, K., Shinozaki, K. (2006). Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 17, 113–22. doi: 10.1016/j.copbio.2006.02.002
Vision, T. J., Brown, D. G., Tanksley, S. D. (2000). The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117. doi: 10.1126/science.290.5499.2114
von Koskull-Doring, P., Scharf, K. D., Nover, L. (2007). The diversity of plant heat stress transcription factors. Trends Plant Sci. 12, 452–457. doi: 10.1016/j.tplants.2007.08.014
Wang, Q., Guo, C., Li, Z., Sun, J., Wang, D., Xu, L., et al. (2021). Identification and analysis of bZIP family genes in potato and their potential roles in stress responses. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.637343
Wang, P., Song, H., Li, C., Li, P., Li, A., Guan, H., et al. (2017). Genome-wide dissection of the heat shock transcription factor family genes in Arachis. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00106
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, W., Vinocur, B., Shoseyov, O., Altman, A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. doi: 10.1016/j.tplants.2004.03.006
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552. doi: 10.1385/1-59259-584-7:531
Wunderlich, M., Gross-Hardt, R., Schoffl, F. (2014). Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol. Biol. 85, 541–550. doi: 10.1007/s11103-014-0202-0
Yang, X., Zhu, W., Zhang, H., Liu, N., Tian, S. (2016). Heat shock factors in tomatoes: genome-wide identification, phylogenetic analysis and expression profiling under development and heat stress. Peer J 4, e1961. doi: 10.7717/peerj.1961
Yoshida, T., Ohama, N., Nakajima, J., Kidokoro, S., Mizoi, J., Nakashima, K., et al. (2011). Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Genet. Genomics 286, 321–332. doi: 10.1007/s00438-011-0647-7
Zhang, J., Li, J., Liu, B., Zhang, L., Chen, J., Lu, M. (2013). Genome-wide analysis of the populus Hsp90 gene family reveals differential expression patterns, localization, and heat stress responses. BMC Genomics 14, 532. doi: 10.1186/1471-2164-14-532
Zhang, X., Rossana, H., Lin, S. S., Niu, Q. W., Chua, N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Zhang, H., Zhao, X., Sun, Q., Yan, C., Wang, J., Yuan, C., et al. (2020). Comparative transcriptome analysis reveals molecular defensive mechanism of Arachis hypogaea in response to salt stress. Int. J. Genomics 2020, 6524093. doi: 10.1155/2020/6524093
Keywords: peanut, Hsf, abiotic stress, transcription factor, gene family
Citation: Wang Q, Zhang Z, Guo C, Zhao X, Li Z, Mou Y, Sun Q, Wang J, Yuan C, Li C, Cong P and Shan S (2023) Hsf transcription factor gene family in peanut (Arachis hypogaea L.): genome-wide characterization and expression analysis under drought and salt stresses. Front. Plant Sci. 14:1214732. doi: 10.3389/fpls.2023.1214732
Received: 30 April 2023; Accepted: 07 June 2023;
Published: 05 July 2023.
Edited by:
Yunpeng Cao, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Muhammad Anwar, Hainan University, ChinaCopyright © 2023 Wang, Zhang, Guo, Zhao, Li, Mou, Sun, Wang, Yuan, Li, Cong and Shan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ping Cong, Y29uZ3BpbmdAY2Fhcy5jbg==; Shihua Shan, c2hhbnNoX3NwcmlAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.