- 1Institute of Crop and Nuclear Technology Utilization, Zhejiang Academy of Agricultural Science, Hangzhou, China
- 2Central Laboratory, State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Zhejiang Academy of Agricultural Science, Hangzhou, China
- 3Institute of Biotechnology, Zhejiang Academy of Agricultural Science, Hangzhou, China
Effective weed control in the field is essential for maintaining favorable growing conditions and rapeseed yields. Sulfonylurea herbicides are one kind of most widely used herbicides worldwide, which control weeds by inhibiting acetolactate synthase (ALS). Molecular markers have been designed from polymorphic sites within the sequences of ALS genes, aiding marker-assisted selection in breeding herbicide-resistant rapeseed cultivars. However, most of them are not breeder friendly and have relatively limited application due to higher costs and lower throughput in the breeding projects. The aims of this study were to develop high throughput kompetitive allele-specific PCR (KASP) assays for herbicide resistance. We first cloned and sequenced BnALS1 and BnALS3 genes from susceptible cultivars and resistant 5N (als1als1/als3als3 double mutant). Sequence alignments of BnALS1 and BnALS3 genes for cultivars and 5N showed single nucleotide polymorphisms (SNPs) at positions 1676 and 1667 respectively. These two SNPs for BnALS1 and BnALS3 resulted in amino acid substitutions and were used to develop a KASP assay. These functional markers were validated in three distinct BC1F2 populations. The KASP assay developed in this study will be valuable for the high-throughput selection of elite materials with high herbicide resistance in rapeseed breeding programs.
Highlights
● Developed KASP assays for BnALS genes are high throughput, low-cost, and capable of screening for herbicide-resistant alleles for marker-assisted selection.
Introduction
Rapeseed (Brassica napus L., AACC) is one of the most important oil-producing crops worldwide, with an annual production of more than 28 million tons of vegetable oil globally (USDA ERS, 2021) and also provides important raw material for biofuel and other industrial products (Ohlrogge, 1994; Thelen and Ohlrogge, 2002). Weeds, especially the broad leaf cruciferous species, are well adapted to compete with rapeseed for sunlight, water, soil nutrients and physical space in the fields (Miki et al., 1990; Larue et al., 2019). Hence, weeds are a significant problem and greatly limit rapeseed yield. The development of herbicide-tolerant varieties is a high priority for varietal development and the most cost-effective tool to manage weeds (Tan et al., 2005; Green, 2014).
Acetolactate synthase (ALS) is the key enzyme for the biosynthesis of the branched chain amino acids, including valine, leucine, and isoleucine (Duggleby et al., 2008; Garcia et al., 2017). ALS has been proved to be the target site of several important herbicides, such as sulfonylurea (SU), imidazolinone (IMI), triazolopyrimidine (TP), pyrimidinyl-thiobenzoates (PTB) and sulfonyl- aminocarbonyl-triazolinone (SCT) (Yu and Powles, 2014). ALS harboring amino acid substitutions caused by gene editing or ethyl methane sulfonate (EMS) mutagenesis has been found to confer high resistance to sulfonylurea herbicides in crops including wheat, rapeseed and watermelon (Tian et al., 2018; Zhang et al., 2019; Guo et al., 2020). The genome information derived from Brassica napus cultivars Darmor-bzh and ZS11 shows that there are five copies in the BnALS gene family (Chalhoub et al., 2014; Sun et al., 2017). Of these, BnALS1 and BnALS3 are highly conserved, and constitutively expressed in all tissues (Wu et al., 2020). Thus, BnALS1 and BnALS3 are regarded to be essential ALS housekeeping genes and the ideal herbicide-resistance targets for genetic manipulation (Rutledge et al., 1991; Wu et al., 2020).
Single nucleotide polymorphism (SNP) is a kind of DNA polymorphism in a genome which results from a single nucleotide change in a DNA sequence (Drenkard et al., 2000; Vignal et al., 2002). Because amino acid substitution caused by single nucleotide mutation may change protein function to some extent, single nucleotide changes provide new insights into protein function (Henikoff and Comai, 2003). Specific single nucleotide change can alter protein function, which is closely related with agronomic traits, then was used as an important tool for crops genetic improvement (You et al., 2018; Zhang et al., 2018). Functional markers derived from polymorphic sites within genes causally affect phenotypic variation. Functional markers are superior to random DNA markers such as RFLPs, SSRs and AFLPs owing to complete linkage with trait locus alleles, and are considered to be more accurate and efficient for gene identification and marker-aided selection (Andersen and Lubberstedt, 2003; Varshney et al., 2005; Zhou et al., 2013; Li et al., 2022). Over the past few decades, allele-specific PCR (AS PCR) markers, cleaved amplified polymorphic sequences (CAPS) markers, derived CAPS (dCAPS) markers and loop-mediated isothermal amplification (LAMP) markers were developed in plants based on single nucleotide polymorphisms (Michaels and Amasino, 1998; Drenkard et al., 2000; Zhou et al., 2013; Pan et al., 2014; Guo et al., 2020; Wu et al., 2020; Wang et al., 2022). All these markers are used to detect and select interesting traits by differentiating between homozygous and heterozygous states of plants. However, these markers require fragments separation by electrophoresis and/or digestion with restriction enzyme after PCR amplification, making their application relatively limited due to higher costs and lower throughput.
The development of user-friendly tools and platforms makes the wide-scale use and application of SNP markers possible in breeding programs. The KASP (kompetitive allele-specific PCR) genotyping assay utilizes a unique form of competitive allele-specific PCR combined with a novel, homogeneous, fluorescence-based reporting system for the identification and measurement of genetic variation occurring at the nucleotide level to detect SNPs (He et al., 2014). With the advantages of being low-cost and high throughput for genotyping SNPs, the KASP technology has been extensively used in the fields of human, animal and plant genetics (He et al., 2014; Semagn et al., 2013).
In this study, we aimed to develop the KASP assays for high-throughput genotyping for herbicide resistance. The SNPs were identified on the basis of Sanger sequencing of cloned BnALS1 and BnALS3 genes from both the resistant 5N and susceptible cultivars of B. napus. Allele-specific assays were developed on the basis of SNPs at positions 1676 and 1667 bp from BnALS1 and BnALS3, respectively. The practical utility of the developed KASP assays was established by validating these in three segregating backcross progeny populations varying for herbicide resistance.
Materials and methods
Plant material
Three elite semi-winter B. napus cultivars (namely ZY50, ZY51 and ZS72) in Zhejiang province of China, and a double mutant 5N (als1als1/als3als3) with herbicide resistance (Guo et al., 2020) were used in this study. The seeds were sown usually in late September or early October and harvested around late May in Yangdu, Haining, Zhejiang Province.
Development of segregation populations with herbicide-resistant 5N
To obtain herbicide-resistant rapeseed lines with good agronomic and quality traits, three backcross progenies (BC1s) were developed from crosses of ZY50/5N//ZY50, ZY51/5N//ZY51 and ZS72/5N//ZS72. The heterozygous lines (ALS1als1/ALS3als3) from these three BC1F1 populations were then screened using newly developed KASP markers (Table 1; Figures 1A, B) and self-pollinated to produce three distinct BC1F2 populations for further analysis. The plants were cultivated in the experimental fields located in Yangdu, Haining, Zhejiang province.
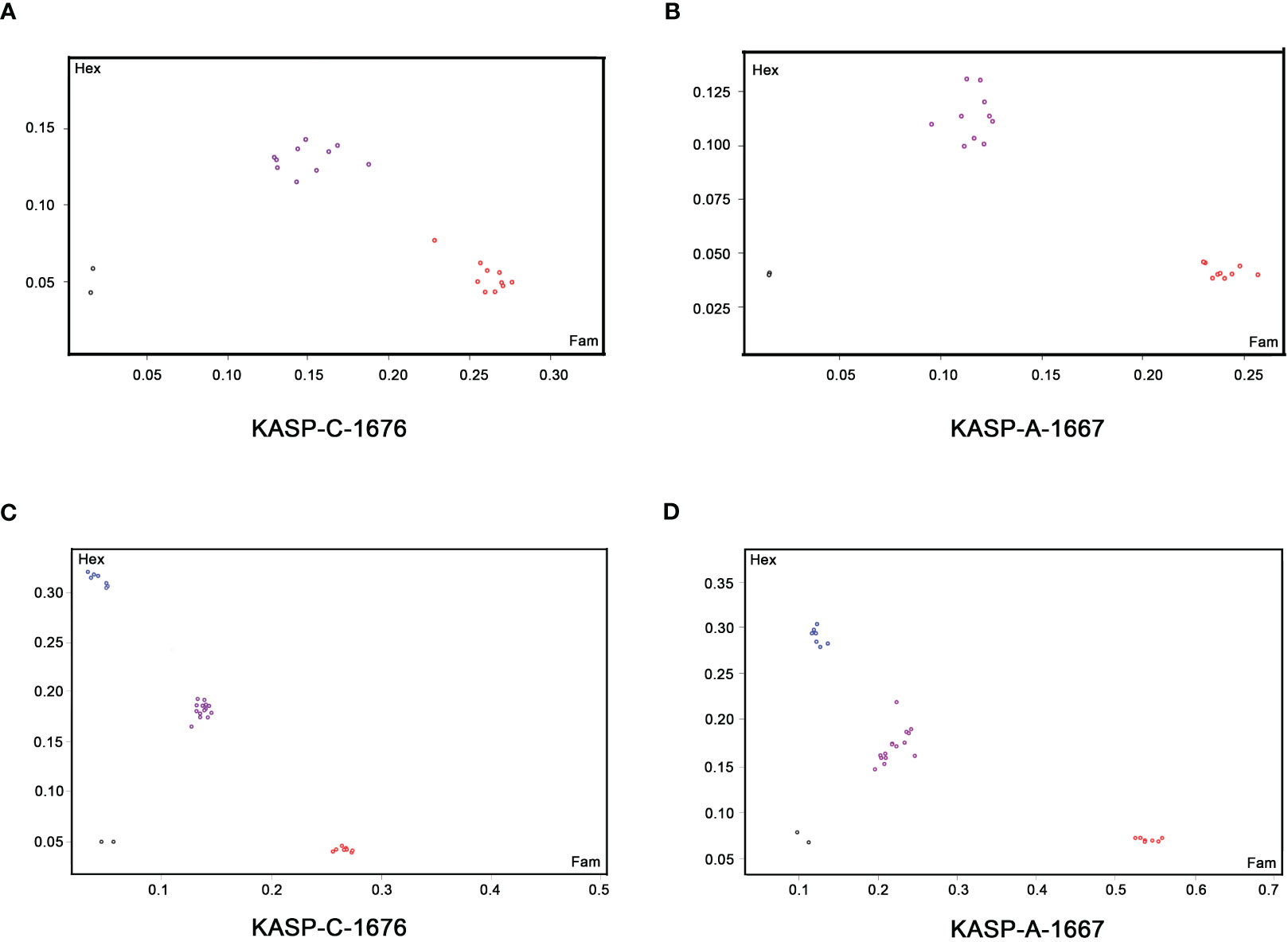
Figure 1 Kompetitive allele-specific PCR (KASP) genotyping using functional marker KASP-C-1676 and KASP-A-1667 in the segregating rapeseed populations for the selection of herbicide-resistant lines. (A, B) KASP genotyping of BnALS1 gene on BC1F1 lines (A) and BnALS3 gene on BC1F1 lines (B) respectively. The red, purple and black dots represent homozygous alleles (G/G), heterozygous alleles (G/T) and non-template control, respectively. (C, D) KASP genotyping of BnALS1 gene on BC1F2 lines (C) and BnALS3 gene on BC1F2 lines (D) respectively. The blue, red, purple and black dots represent homozygous alleles (T/T), homozygous alleles (G/G), heterozygous alleles (G/T) and non-template control, respectively.
Amplification and Sequence analysis of BnALS1 and BnALS3 Genes
Genomic DNA of young rapeseed leaves from each plant was extracted with a modified cetyltriethylammnonium bromide (CTAB) method (Shi et al., 2017). Full-length BnALS1 (2228 bp) and BnALS3 (2027 bp) genes were isolated and amplified separately from ZY50, ZY51, ZS72 and 5N using gene-specific primers as described (Guo et al., 2020). The resultant DNA fragments were sequenced by the Sanger dideoxy chain termination method on a capillary electrophoresis system (ABI 3730XL, Applied Biosystems, United States).
Nucleotide and amino acid multiple-sequence alignments were constructed using the CLUSTAL OMEGA program (Madeira et al., 2022) and colored by use of the GeneDoc 3.2 program with the default BLOSUM score. The sequence of the ALS gene from A. thaliana (GenBank accession no. NM_114714) was used as a reference. The nucleotide and amino acid sequences of BnALS1 and BnALS3 from three cultivars and 5N are listed in Supplementary data sheets 1, 2.
Primer design for BnALS1 and BnALS3 genes
BnALS1 and BnALS3 sequences from susceptible cultivars (ZY50, ZY51 and ZS72) and the double mutant 5N were amplified and analyzed as mentioned above. Two herbicide-resistant SNPs, G1676T for BnALS1 and G1667T for BnALS3, were used to develop functional markers. The KASP primers were designed according to the standard guidelines. Because BnALS1 and BnALS3 sequences are highly identical (97.6%), flanking sequence (including SNPs between BnALS1 and BnALS3) of different alleles at each locus were extracted and used for primer design (Supplementary Figure 1). For each gene, the KASP marker consisted of two SNP-specific primers and one common primer. Of these three primers, two G/T alleles were linked to the FAM and HEX fluorescent linker-specific sequence of the LGC KASP reagents at the 5’ end. The primer sequences are shown in Table 1.
Kompetitive allele-specific PCR genotyping
The genotyping assays of the developed KASP markers were performed on a 96-well plate. The KASP assay was performed in a 1.6 μL PCR reaction mix that consisted of 0.8 μL of KASP Master mix (LGC, Biosearch Technologies), approximately equal to 0.05 μL of primer, and 0.8 μL of DNA at a concentration of 10-20 ng/μL. The amplifications were performed using an IntelliQube (LGC, Biosearch Technologies) with the following cycling conditions: 94°C for 15min, 10 touchdown cycles (94°C for 20 s; touchdown at 61°C, dropping to -0.6°C per cycle 60 s) and followed by 26 cycles of amplification (94°C for 20 s, 55°C for 60 s).
Inheritance analysis
The susceptible rapeseed cultivars (ZY50, ZY51 and ZS72), 5N and the developed distinct BC1F2 populations were grown in the field, and seedlings at the 4-6 leaf stage were sprayed with tribenuron-methyl (TBM) at 20.25 g.a.i.ha-1. Resistance of the parents and their derived BC1F2 populations was evaluated 20 days after treatment. The response phenotypes were scored as resistant (R) if they showed no herbicide damage or only slight injury, or susceptible (S) if they died. The segregation of each population was assessed using a Chi square test.
Further herbicide resistance analysis of the homozygous genotypes
Three distinct BC1F2 populations were derived from the crosses ZY50/5N//ZY50, ZY51/5N//ZY51 and ZS72/5N//ZS72. For each population, the seedlings of BC1F2 populations was analyzed for the four homozygous genotypes (AABB, AAbb, aaBB and aabb) using the composite KASP markers. These homozygous lines were then self-pollinated to generate BC1F3 seeds.
These BC1F3 homozygous lines from the three distinct BC1F2 populations were sown and grown in plastic pots (diameter, 10cm) containing a 1:1:1 mixture of peat moss, perlite and vermiculite under natural light conditions.
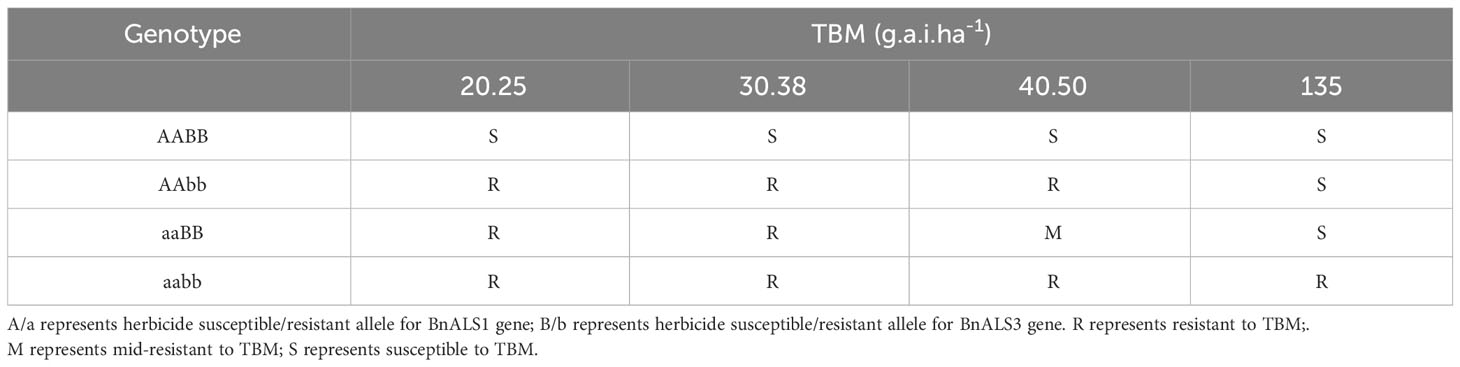
At least twenty BC1F3 seedlings from each of the four homozygous lines from the three distinct BC1F2 populations were sprayed with serial concentrations of 20.25, 30.38, 40.50 and 135 g.a.i.ha-1TBM at the 4-6 leaf stage. Symptoms were recorded as resistant (R - no herbicide damage or only slight injury), mid-resistant (M - chlorosis or necrosis on some leaves, but no death) or susceptible (S - dead plants) at 20 days after the treatment.
Results
Phenotypic symptom of herbicide resistance
To observe the resistance to herbicide, the seedlings of ZY50, ZY51, ZS72 and 5N were sprayed with TBM at a concentration of 20.25 g.a.i.ha-1. After exposure to TBM for 14 days, ZY50, ZY51 and ZS72 were growth injured with yellow or chlorotic leaves (Figures 2A, B). However, 5N, which harbored two resistant alleles, exhibited complete resistance, having no symptoms of chlorosis or necrosis (Figures 2A, B). Our results suggested that novel herbicide-resistant materials with good agronomic and quality traits could be developed through the crosses between the elite rapeseed cultivars and 5N.

Figure 2 Herbicide resistance phenotypes of three cultivars ZY50, ZY51 and ZS72 and 5N mutant. (A) Phenotypes of cultivars ZY50, ZY51 and ZS72 and 5N mutant grown under normal condition. (B) Symptoms of cultivars ZY50, ZY51 and ZS72 and 5N mutant 14 days after treatment with 20.25 g.a.i.ha-1 TBM.
Development of kompetitive allele-specific PCR marker for BnALS1 and BnALS3 genes
To detect single nucleotide polymorphisms (SNPs), BnALS1 and BnALS3 genes were separately cloned by PCR amplification from 5N and three cultivars, ZY50, ZY51 and ZS72 (Supplementary data sheet 1). Compared with 5N, these three cultivars have a common SNP at position G/T (1676) in BnALS1 and a common SNP at position G/T (1667) in BnALS3 (Supplementary Figure 1). A comparison of the amino acid sequences of susceptible cultivars/resistant 5N showed changes at W/L (474 in BnALS1; 471 in BnALS3) as compared to the changes at the two positions for the nucleotide sequence of BnALS1 and BnALS3 (Figure 3). A previous study proved that the substitutions of W/L in BnALS1 and BnALS3 could endow high herbicide resistance (Guo et al., 2020). Therefore, G1676T and G1667T were selected as the genotyping targets for BnALS1 and BnALS3 respectively. The KASP markers were designed for a SNP at position 1676 in BnALS1 and for a SNP at position 1667 in BnALS3 (Supplementary Figure 1). Both the marker KASP-C-1676 (specific to G1676T in BnALS1) and the marker KASP-A-1667 (specific to G1667 in BnALS3) could clearly distinguish type alleles GG, GT and TT among cultivars, cultivars/5N and 5N (Supplementary Figure 2). These two markers were also validated on BC1F1 populations, and formed separate clusters for heterozygous (GT) and homozygous (GG) alleles (Figures 1A, B).

Figure 3 Alignment of partial amino acid sequence of ALS proteins from Arabidopsis, ZY50, ZY51, ZS72 and herbicide-resistant mutant 5N (als1als1/als3als3). AtALS (GenBank accession no. NM_114714), BnALS1-ZY50, BnALS1-ZY51, BnALS1-ZS72, BnALS1-5N, BnALS3-ZY50, BnALS3-ZY51, BnALS3-ZS72 and BnALS3-5N (B. napus L. cv. ZY50, ZY51, ZS72 and 5N). The red arrow represents point mutations occurred in ALS1 and ALS3 of 5N. The shading of the alignment presents as follows: identical residues in black and different residues in dark gray. “*”indicates positions which have a single, fully conserved residue.
Validation of kompetitive allele-specific PCR assays on BC1F2 populations
To confirm the KASP assay on herbicide resistance, three distinct BC1F2 populations were genotyped using KASP-C-1676 and KASP-A-1667. DNA was extracted from the first true leaves of seedlings before the herbicide treatment. Seedlings at the 4-6 leaf stage were sprayed with TBM at a concentration of 20.25 g.a.i.ha-1. Phenotypic symptoms were observed at 20 days after the treatment. The KASP assays were used for specific amplification of BnALS1 and BnALS3. The frequency of the KASP alleles showed equivalence with the segregation expected for the BC1F2 populations (Figures 1C, D).
The combination of the KASP markers resulted in nine genotypes as shown by analysis of the seedlings in the BC1F2 populations (Table 2). These were AABB, AABb, AAbb, AaBB, AaBb, Aabb, aaBB, aaBb and aabb, and the ratio of isolation of these genotypes is 1:2:1:2:4:2:1:2:1 in the three distinct BC1F2 populations. In the three BC1F2 populations developed from crosses ZY50/5N//ZY50, ZY51/5N//ZY51 and ZS72/5N//ZS72, five, five and six homozygous plants with genotype AABB exhibited sensitivity to TBM treatment at 20 days after the treatment (Table 2). However, plants with other genotypes showed resistance to 20.25 g.a.i.ha-1 TBM treatment (Table 2). The ratio of susceptible lines to resistant lines is 1:15 in the three distinct BC1F2 populations (Table 2).
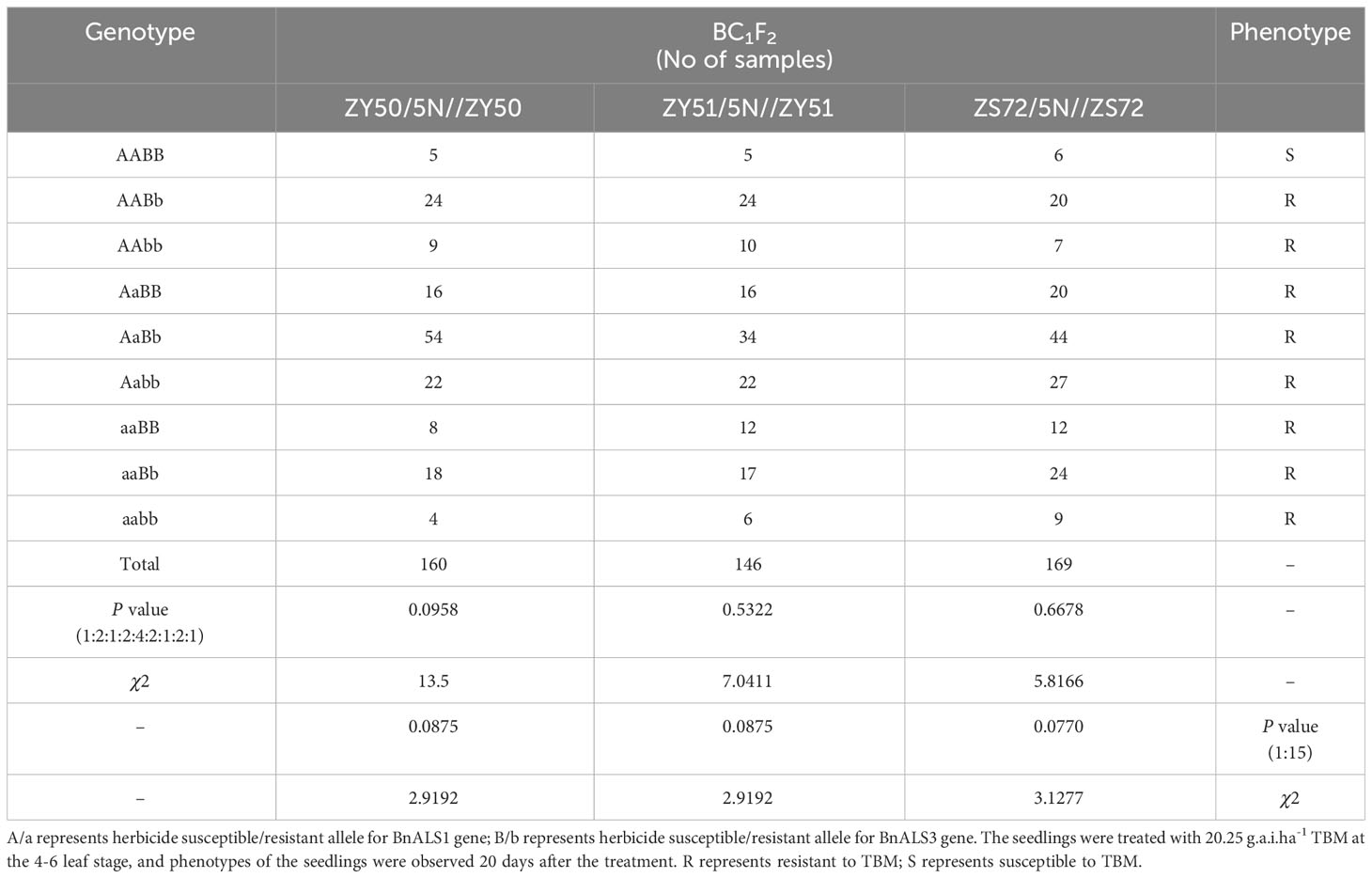
Table 2 Validation of the KASP assays for herbicide resistance in distinct BC1F2 populations of B. napus.
Further validation of genotype effect on herbicide resistance
To further validate the effect of genotype on herbicide resistance, we chose the BC1F3 plants with genotypes AABB, AAbb, aaBB and aabb developed from three distinct BC1F2 populations for resistance analysis. Plants with genotype AABB displayed serious damage with yellow leaves and eventual death within 20 days after treatment at all concentrations of TBM (Table 3). Plants with genotype AAbb exhibited resistance to 20.25-40.50 g.a.i.ha-1 TBM (Table 3). Plants with genotype aaBB showed resistance to 20.25-30.38 g.a.i.ha-1 TBM and mid-resistance to 40.50 g.a.i.ha-1 TBM (Table 3). However, at higher concentration of 135 g.a.i.ha-1 TBM, Plants with genotype AAbb and aaBB showed chlorotic stunting, destroyed apex and eventual death (Table 3). By contrast to plants with genotype AAbb and aaBB, plants with genotype aabb exhibited complete resistance, having no chlorosis or necrosis, even to the higher concentration of 135 g.a.i.ha-1 TBM (Table 3).
Discussion
Successful weed management helps to improve crop yield in modern agricultural production systems. Resistant cultivars are the most effective and environmentally responsible strategy for protecting crops from weeds. Thus, developing new cultivars with high resistance to herbicides is now a major breeding objective in rapeseed. Acetolactate synthase encoded by ALS gene is responsible for biosynthesis of the branched chain amino acids, including valine, leucine, and isoleucine (Haughn and Somerville, 1990). The mutation of ALS gene may result in amino acid substitutions of ALS and inhibit the binding of the ALS enzyme with herbicides, which endows the plants with resistance to herbicide (Duggleby et al., 2008; Murphy and Tranel, 2019; Guo et al., 2020; Wu et al., 2020). Functional markers, such as AS-PCR (Hu et al., 2015) and CAPS (Li et al., 2015; Hu et al., 2017; Guo et al., 2020; Huang et al., 2020), have been developed to discriminate the allelic variation for the ALS genes. However, all these are gel based markers, and have relatively limited potential for high-throughput application. Thus, the development of a high-throughput and relatively cost-efficient marker system is important and necessary for improving breeding strategies.
As a key enzyme for the biosynthesis of branched chain amino acids, improper mutation of ALS can destroy its function and result in plant death. However, ALS harboring point mutations could confer sufficient tolerance to some kinds of herbicides with little damage to plant growth (Yu et al., 2010; Zhao et al., 2020; Cheng et al., 2021). We independently cloned and sequenced BnALS1 and BnALS3 from three cultivars ZY50, ZY51 and ZS72, and from the 5N mutant. DNA sequence alignment showed that 5N contains a single-nucleotide mutation (G1676T) in BnALS1 and a single- nucleotide mutation (G1667T) in BnALS3 based on sequence comparison with the three herbicide-susceptible cultivars; ZY50, ZY51 and ZS72 (Supplementary Figure 1), resulting in amino acid alterations, W474L (W574L, numbered according to ALS sequence in Arabidopsis) in BnALS1 and W471L (W574L) in BnALS3 (Figure 2). The W574L substitution has been reported to confer resistance to ALS inhibitors in rapeseed, sunflower and cocklebur (Bernasconi et al., 1995; Hattori et al., 1995; Sala et al., 2012; Hu et al., 2017; Guo et al., 2020). Mutation at P197 also conferred good tolerance to sulfonylureas in Arabidopsis, rapeseed and wheat (Li et al., 2015; Chen et al., 2017; Zhang et al., 2019; Huang et al., 2020; Wu et al., 2020; Cheng et al., 2021; Guo et al., 2022). In addition, mutations at the sites of Ala122, Ala205 and Ser653 of ALS have been reported to confer resistance to ALS inhibitors (Tan et al., 2005; Murphy and Tranel, 2019). These SNPs in ALS genes can be used for marker-assisted breeding.
5N is an important herbicide-resistant material with simultaneous mutations in BnALS1 and BnALS3 genes (Guo et al., 2020). We planned to design KASP markers for BnALS1 and BnALS3 genes in the 5N double mutant. Considering the highly similar (97.6%) sequence of BnALS1 and BnALS3, it is difficult to develop high throughput markers capable of discriminating homozyous and heterozygous lines in these segregating populations. In this study, two KASP functional markers, KASP-C-1676 and KASP-A-1667, were successfully developed based on the specific characteristics of BnALS1 and BnALS3 genes from the cultivars and 5N (Table 1). These KASP markers will faciliate the use of 5N mutant for herbicide resistant rapeseed breeding.
The two KASP markers can clearly distinguish the genotypes of parents and hybrids (Supplementary Figure 2). Genotyping results performed by the two markers are highly consistent with the results of phenotypic evaluation (Figure 1; Table 2). Furthermore, these two KASP markers can distinguish the homozygous/heterozygous lines in three distinct segregated populations (BC1F2) developed from ZY50, ZY51, ZS72 and 5N (Table 2), which proved the high effectiveness of the KASP markers for genotyping under different genetic backgrounds. All these results suggested that the developed KASP markers are stable and effective to differentiate homozygous/heterozygous state of alleles in distinct populations and can be used for marker-assisted selection in rapeseed breeding projects.
In plants, synergistic effect is an important genetic phenomenon exhibited in the processes of hormone interaction, flower development and signal transduction (Poduska et al., 2003; Replogle et al., 2013; Yang et al., 2017). Synergistic effects have also been shown for herbicide resistance in crops. In B. napus, 5N (BnALS1-2R, W574L; BnALS3R, W574L) and DS3 (BnALS1-3R, P197L; BnALS3R, W574L) showed stronger herbicide resistance than mutants with single-point mutations (Guo et al., 2020 and Guo et al., 2022). In soybean, Als1 (P197S) and Als2 (W574L) exhibited synergistic resistant effects to ALS herbicides, and the combination of Als1 and Als2 conferred stronger tolerance to SU (Walter et al., 2014). In this study, four homozygous genotypes were characterized and selected using the developed KASP markers. We analyze the effects of four genotypes on herbicide resistance. Our results showed that the lines containing two mutated alleles exhibited relatively stronger TBM resistance compared with those lines with a single mutated allele (Table 3), which is consistent with the findings reported previously (Guo et al., 2020). Altogether, these results suggested that the developed KASP markers are valuable functional markers and could be used for the high throughput selection of superior herbicide resistant materials by providing precise genotypic information, which will expedite the process of breeding herbicide-resistant rapeseed.
Conclusion
In this study, two KASP markers for BnALS1 and BnALS3, KASP-C-1676 and KASP-A-1667, were successfully developed on the basis of SNPs in the ALS genes. These assays are highly gene specific and can effectively distinguish target genotype states. The developed KASP assays are high throughput and cost effective as compared to gel-based markers and can be used for marker-assisted selection of herbicide resistance.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
JS conceived and coordinated the study. TW cloned and aligned the genes. TZ, YF, and SL conducted the KASP assays. XN, HY, YZ, and JH performed the field experiments and phenotypic data collection. JS wrote the manuscript and JZ revised it. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (grant no. 31972875), the Project of Agriculture, Rural areas, Farmers and Nine Parties of Zhejiang Province (grant no. 2022SNJF010) and Key Laboratory of Digital Upland Crops of Zhejiang Province (grant no. 2022E10012).
Acknowledgments
We are grateful to Profs. Huiming Pu and Maolong Hu (Jiangsu Academy of Agricultural Science) for providing 5N material and to Dr. Yi Zhang (Molecular Marker (Wuhan) Biobreeding Co., LTD) for his assistance in marker designing. We are also grateful to Dr. Rebecca Horn at scientificproofreading.co.uk for her assistance in editing this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1213476/full#supplementary-material
References
Andersen, J. R., Lubberstedt, T. (2003). Functional markers in plants. Trends Plant Sci. 8 (11), 554–560. doi: 10.1016/j.tplants.2003.09.010
Bernasconi, P., Woodworth, A. R., Rosen, B. A., Subramanian, M. V., Siehl, D. L. (1995). A naturally occurring point mutation confers broad range tolerance to herbicides that target acetolactate synthase. J. Biol. Chem. 270 (29), 17381–17385. doi: 10.1074/jbc.270.29.17381
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345 (6199), 950–953. doi: 10.1126/science.1253435
Chen, Y., Wang, Z., Ni, H., Xu, Y., Chen, Q., Jiang, L. (2017). CRISPR/Cas9-mediated base-editing system efficiently generates gain-of-function mutations in Arabidopsis. Sci. China Life Sci. 60 (5), 520–523. doi: 10.1007/s11427-017-9021-5
Cheng, H., Hao, M., Ding, B., Mei, D., Wang, W., Wang, H., et al. (2021). Base editing with high efficiency in allotetraploid oilseed rape by A3A-PBE system. Plant Biotechnol. J. 19 (1), 87–97. doi: 10.1111/pbi.13444
Drenkard, E., Richter, B. G., Rozen, S., Stutius, L. M., Angell, N. A., Mindrinos, M., et al. (2000). A simple procedure for the analysis of single nucleotide polymorphisms facilitates map-based cloning in Arabidopsis. Plant Physiol. 124 (4), 1483–1492. doi: 10.1104/pp.124.4.1483
Duggleby, R. G., McCourt, J. A., Guddat, L. W. (2008). Structure and mechanism of inhibition of plant acetohydroxyacid synthase. Plant Physiol. Biochem. 46 (3), 309–324. doi: 10.1016/j.plaphy.2007.12.004
Garcia, M. D., Wang, J. G., Lonhienne, T., Guddat, L. W. (2017). Crystal structure of plant acetohydroxyacid synthase, the target for several commercial herbicides. FEBS J. 284 (13), 2037–2051. doi: 10.1111/febs.14102
Green, J. M. (2014). Current state of herbicides in herbicide-resistant crops. Pest Manag Sci. 70 (9), 1351–1357. doi: 10.1002/ps.3727
Guo, Y., Cheng, L., Long, W., Gao, J., Zhang, J., Chen, S., et al. (2020). Synergistic mutations of two rapeseed AHAS genes confer high resistance to sulfonylurea herbicides for weed control. Theor. Appl. Genet. 133 (10), 2811–2824. doi: 10.1007/s00122-020-03633-w
Guo, Y., Liu, C., Long, W., Gao, J., Zhang, J., Chen, S., et al. (2022). Development and molecular analysis of a novel acetohydroxyacid synthase rapeseed mutant with high resistance to sulfonylurea herbicides. Crop J. 10, 56–66. doi: 10.1016/j.cj.2021.05.006
Hattori, J., Brown, D., Mourad, G., Labbe, H., Ouellet, T., Sunohara, G., et al. (1995). An acetohydroxy acid synthase mutant reveals a single site involved in multiple herbicide resistance. Mol. Gen. Genet. 246 (4), 419–425. doi: 10.1007/BF00290445
Haughn, G. W., Somerville, C. R. (1990). A mutation causing imidazolinone resistance maps to the csr1 locus of arabidopsis thaliana. Plant Physiol. 92 (4), 1081–1085. doi: 10.1104/pp.92.4.1081
He, C., Holme, J., Anthony, J. (2014). SNP genotyping: the KASP assay. Methods Mol. Biol. 1145, 75–86. doi: 10.1007/978-1-4939-0446-4_7
Henikoff, S., Comai, L. (2003). Single-nucleotide mutations for plant functional genomics. Annu. Rev. Plant Biol. 54, 375–401. doi: 10.1146/annurev.arplant.54.031902.135009
Hu, M., Pu, H., Gao, J., Long, W., Chen, F., Zhou, X., et al. (2017). Inheritance and molecular characterization of resistance to AHAS-inhibiting herbicides in rapeseed. J. Integr. Agric. 16, 2421–2433. doi: 10.1016/S2095-3119(17)61659-9
Hu, M., Pu, H., Kong, L., Gao, J., Long, W., Chen, S., et al. (2015). Molecular characterization and detection of a spontaneous mutation conferring imidazolinone resistance in rapeseed and its application in hybrid rapeseed production. Mol. Breed 35, 46. doi: 10.1007/s11032-015-0227-3
Huang, Q., Lv, J., Sun, Y., Wang, H., Guo, Y., Qu, G., et al. (2020). Inheritance and molecular characterization of a novel mutated AHAS gene responsible for the resistance of AHAS-inhibiting herbicides in rapeseed (Brassica napus L.). Int. J. Mol. Sci. 21 (4), 1345–1362. doi: 10.3390/ijms21041345
Larue, C. T., Goley, M., Shi, L., Evdokimov, A. G., Sparks, O. C., Ellis, C., et al. (2019). Development of enzymes for robust aryloxyphenoxypropionate and synthetic auxin herbicide tolerance traits in maize and soybean crops. Pest Manag Sci. 75 (8), 2086–2094. doi: 10.1002/ps.5393
Li, H., Li, J., Zhao, B., Wang, J., Yi, L., Liu, C., et al. (2015). Generation and characterization of TBM herbicide-resistant rapeseed (Brasscia napus) for hybrid seed production using chemically induced male sterility. Theor. Appl. Genet. 128 (1), 107–118. doi: 10.1007/s00122-014-2415-7
Li, L., Sun, Z., Zhang, Y., Ke, H., Yang, J., Li, Z., et al. (2022). Development and utilization of functional kompetitive allele-specific PCR markers for key genes underpinning fiber length and strength in gossypium hirsutum L. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.853827
Madeira, F., Pearce, M., Tivey, A. R. N., Basutkar, P., Lee, J., Edbali, O., et al. (2022). Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 50 (W1), W276–W279. doi: 10.1093/nar/gkac240
Michaels, S. D., Amasino, R. M. (1998). A robust method for detecting single-nucleotide changes as polymorphic markers by PCR. Plant J. 14 (3), 381–385. doi: 10.1046/j.1365-313x.1998.00123.x
Miki, B. L., Labbe, H., Hattori, J., Ouellet, T., Gabard, J., Sunohara, G., et al. (1990). Transformation of Brassica napus canola cultivars with Arabidopsis thaliana acetohydroxyacid synthase genes and analysis of herbicide resistance. Theor. Appl. Genet. 80 (4), 449–458. doi: 10.1007/BF00226744
Murphy, B. P., Tranel, P. J. (2019). Target-site mutations conferring herbicide resistance. Plants (Basel) 8 (10). doi: 10.3390/plants8100382
Ohlrogge, J. B. (1994). Design of new plant products: engineering of fatty acid metabolism. Plant Physiol. 104 (3), 821–826. doi: 10.1104/pp.104.3.821
Pan, L., Li, J., Zhang, W. N., Dong, L. Y. (2014). Detection of the I1781L mutation in fenoxaprop-p-ethyl-resistant American sloughgrass (Beckmannia syzigachne Steud.), based on the loop-mediated isothermal amplification method. Pest Manag Sci. 71, 123–130. doi: 10.1002/ps.3777
Poduska, B., Humphrey, T., Redweik, A., Grbic, V. (2003). The synergistic activation of FLOWERING LOCUS C by FRIGIDA and a new flowering gene AERIAL ROSETTE 1 underlies a novel morphology in Arabidopsis. Genetics 163 (4), 1457–1465. doi: 10.1093/genetics/163.4.1457
Replogle, A., Wang, J., Paolillo, V., Smeda, J., Kinoshita, A., Durbak, A., et al. (2013). Synergistic interaction of CLAVATA1, CLAVATA2, and RECEPTOR-LIKE PROTEIN KINASE 2 in cyst nematode parasitism of Arabidopsis. Mol. Plant Microbe Interact. 26 (1), 87–96. doi: 10.1094/MPMI-05-12-0118-FI
Rutledge, R. G., Quellet, T., Hattori, J., Miki, B. L. (1991). Molecular characterization and genetic origin of the Brassica napus acetohydroxyacid synthase multigene family. Mol. Gen. Genet. 229 (1), 31–40. doi: 10.1007/BF00264210
Sala, C. A., Bulos, M., Altieri, E., Weston, B. (2012). Response to imazapyr and dominance relationships of two imidazolinone-tolerant alleles at the Ahasl1 locus of sunflower. Theor. Appl. Genet. 124 (2), 385–396. doi: 10.1007/s00122-011-1713-6
Semagn, K., Babu, R., Hearne, S., Olsen, M. (2013). Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement. Mol. Breed 33, 1–14. doi: 10.1007/s11032-013-9917-x
Shi, J., Lang, C., Wang, F., Wu, X., Liu, R., Zheng, T., et al. (2017). Depressed expression of FAE1 and FAD2 genes modifies fatty acid profiles and storage compounds accumulation in Brassica napus seeds. Plant Sci. 263, 177–182. doi: 10.1016/j.plantsci.2017.07.014
Sun, F., Fan, G., Hu, Q., Zhou, Y., Guan, M., Tong, C., et al. (2017). The high-quality genome of Brassica napus cultivar 'ZS11' reveals the introgression history in semi-winter morphotype. Plant J. 92 (3), 452–468. doi: 10.1111/tpj.13669
Tan, S., Evans, R. R., Dahmer, M. L., Singh, B. K., Shaner, D. L. (2005). Imidazolinone-tolerant crops: history, current status and future. Pest Manag Sci. 61 (3), 246–257. doi: 10.1002/ps.993
Thelen, J. J., Ohlrogge, J. B. (2002). Metabolic engineering of fatty acid biosynthesis in plants. Metab. Eng. 4 (1), 12–21. doi: 10.1006/mben.2001.0204
Tian, S., Jiang, L., Cui, X., Zhang, J., Guo, S., Li, M., et al. (2018). Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 37 (9), 1353–1356. doi: 10.1007/s00299-018-2299-0
USDA ERS. (2021). Available at: https://www.ers.usda.gov/data-products/oil-crops-yearbook/oil-crops-yearbook/.
Varshney, R. K., Graner, A., Sorrells, M. E. (2005). Genomics-assisted breeding for crop improvement. Trends Plant Sci. 10 (12), 621–630. doi: 10.1016/j.tplants.2005.10.004
Vignal, A., Milan, D., SanCristobal, M., Eggen, A. (2002). A review on SNP and other types of molecular markers and their use in animal genetics. Genet. Sel Evol. 34 (3), 275–305. doi: 10.1186/1297-9686-34-3-275
Walter, K. L., Strachan, S. D., Ferry, N. M., Albert, H. H., Castle, L. A., Sebastian, S. A. (2014). Molecular and phenotypic characterization of Als1 and Als2 mutations conferring tolerance to acetolactate synthase herbicides in soybean. Pest Manag Sci. 70 (12), 1831–1839. doi: 10.1002/ps.3725
Wang, M. L., Zhi Tang, Z., Liao, M., Cao, H. Q., Zhao, N. (2022). Loop-mediated isothermal amplification for detecting the Ile-2041-Asn mutation in fenoxaprop-P-ethyl-resistant Alopecurus aequalis. Pest Manag Sci. 79, 711–718. doi: 10.1002/ps.7239
Wu, J., Chen, C., Xian, G., Liu, D., Lin, L., Yin, S., et al. (2020). Engineering herbicide-resistant oilseed rape by CRISPR/Cas9-mediated cytosine base-editing. Plant Biotechnol. J. 18 (9), 1857–1859. doi: 10.1111/pbi.13368
Yang, Z. B., Liu, G., Liu, J., Zhang, B., Meng, W., Muller, B., et al. (2017). Synergistic action of auxin and cytokinin mediates aluminum-induced root growth inhibition in Arabidopsis. EMBO Rep. 18 (7), 1213–1230. doi: 10.15252/embr.201643806
You, Q., Yang, X., Peng, Z., Xu, L., Wang, J. (2018). Development and applications of a high throughput genotyping tool for polyploid crops: single nucleotide polymorphism (SNP) array. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00104
Yu, Q., Han, H. P., Vila-Aiub, M. M., Powles, S. B. (2010). AHAS herbicide resistance endowing mutations: effect on AHAS functionality and plant growth. J. Exp. Bot. 61 (14), 3925–3934. doi: 10.1093/jxb/erq205
Yu, Q., Powles, S. B. (2014). Resistance to AHAS inhibitor herbicides: current understanding. Pest Manag Sci. 70 (9), 1340–1350. doi: 10.1002/ps.3710
Zhang, R., Liu, J., Chai, Z., Chen, S., Bai, Y., Zong, Y., et al. (2019). Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat. Plants 5 (5), 480–485. doi: 10.1038/s41477-019-0405-0
Zhang, Y., Massel, K., Godwin, I. D., Gao, C. (2018). Applications and potential of genome editing in crop improvement. Genome Biol. 19 (1), 210. doi: 10.1186/s13059-018-1586-y
Zhao, N., Yan, Y. Y., Du, L., Zhang, X. L., Liu, W. T., Wang, J. J. (2020). Unravelling the effect of two herbicide resistance mutations on acetolactate synthase kinetics and growth traits. J. Exp. Bot. 71 (12), 3535–3542. doi: 10.1093/jxb/eraa120
Keywords: KASP assay, herbicide resistance, SNPs, ALS genes, marker-assisted selection
Citation: Shi J, Yu H, Fu Y, Wang T, Zhang Y, Huang J, Li S, Zheng T, Ni X and Zhao J (2023) Development and validation of functional kompetitive allele-specific PCR markers for herbicide resistance in Brassica napus. Front. Plant Sci. 14:1213476. doi: 10.3389/fpls.2023.1213476
Received: 28 April 2023; Accepted: 07 November 2023;
Published: 23 November 2023.
Edited by:
Baohua Wang, Nantong University, ChinaReviewed by:
Dengfeng Hong, Huazhong Agricultural University, ChinaNing Zhao, Anhui Agricultural University, China
Maolong Hu, Jiangsu Academy of Agricultural Sciences (JAAS), China
Copyright © 2023 Shi, Yu, Fu, Wang, Zhang, Huang, Li, Zheng, Ni and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiyuan Ni, bml4aXl1YW5AeWVhaC5uZXQ=; Tao Zheng, emhlbmd0YW9AemFhcy5hYy5jbg==
 Jianghua Shi
Jianghua Shi Huasheng Yu
Huasheng Yu Ying Fu
Ying Fu Tanliu Wang1
Tanliu Wang1 Yaofeng Zhang
Yaofeng Zhang Sujuan Li
Sujuan Li Tao Zheng
Tao Zheng Xiyuan Ni
Xiyuan Ni