
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 13 July 2023
Sec. Plant Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1212979
The lignification of plant secondary walls is an important process that provides plants with mechanical support. However, the presence of lignin in the secondary walls affects the readily availability of cellulose required in various industries, including the biofuel, paper, and textile industries. Thus, plants with less lignin are ideal for usage in such industries. Molecular studies have identified genes that regulate plant lignification, including group III plant-specific patatin-related phospholipase genes. Recent studies have reported decreased lignin content when pPLAIIIα, pPLAIIIγ (from Arabidopsis thaliana), and pPLAIIIβ (from Panax ginseng) were overexpressed in Arabidopsis. However, the role played by a closely related gene pPLAIIIδ in lignin biosynthesis has not yet been reported. In this study, we found that overexpression of the pPLAIIIδ significantly reduced the lignin content in secondary cell walls, whereas the silencing of the gene increased secondary walls lignification. Transcript level analysis showed that the key structural and regulatory genes involved in the lignin biosynthesis pathway decreased in overexpression, and increased in plants with silenced pPLAIIIδ. Further analysis revealed that pPLAIIIδ played an influential role in several physiological processes including seed germination, and chlorophyll accumulation. Moreover, the gene also influenced the size of plants and plant organs, including leaves, seeds, and root hairs. Generally, our study provides important insights toward the use of genetic engineering for lignin reduction in plants and provides information about the agronomical and physiological suitability of pPLAIIIδ transgenic plants for utilization in biomass processing industries.
Secondary wall growth is an important developmental feature in plants. It provides the plants with the needed mechanical strength for vertical growth extension. Secondary walls are characterized by the lignification of vascular tissues, which help plants to resist the negative pressure resulting from transpiration and endow the plants with rigidity and hydrophobicity (Zhong and Ye, 2015). However, the presence of lignin affects the utilization of plant biomass in various industries. The separation of cellulose from lignin during the production of bio-products such as bioethanol, textiles, and paper requires mechanical or chemical pretreatment processes, which are costly. Moreover, the methods used to remove the lignin can affect the quality and commercial value of the final bio-product (Liu et al., 2017). Plants that accumulate less lignin would therefore be ideal in bioenergy industries, because using such plants would reduce production costs and increase the quality of the manufactured bio-products. Several research groups have explored the use of molecular techniques as an alternative approach for reducing the lignin content of plants. For example, the mutation of two genes in the lignin biosynthesis pathway, caffeic acid 3-O-methyltransferase (COMT) and caffeoyl-CoA 3-O-methyltransferase (CCoAOMT), reduced the lignin biosynthesis yield in Arabidopsis (Xie et al., 2019). In another study, the downregulation of genes coding for cinnamyl alcohol dehydrogenase in Morus alba (MaCADs) reduced lignification in stems and leaves (Chao et al., 2022). Similarly, our group has reported the involvement of several members of group III patatin-related phospholipase genes (pPLAIIIs) during lignin biosynthesis processes (Jang et al., 2019; Jang and Lee, 2020a; Jang et al., 2022). We have reported that the independent overexpression of pPLAIIIα and pPLAIIIγ in Arabidopsis caused a significant decrease in lignin content (Jang and Lee, 2020a; Jang et al., 2022). Moreover, a decrease in lignin accumulation was observed when a homolog from Panax ginseng, PgpPLAIIIβ, was overexpressed in poplar and Arabidopsis (Jang et al., 2019; Jang and Lee, 2020b). The discovery of the role played by other members of the pPLAIII group in plant lignin biosynthesis prompted a study to determine if the remaining member of the group, pPLAIIIδ, also influences lignification. The Arabidopsis pPLAIIIδ is closely related to the rest members of pPLAIII group. Apart from hydrolyzing phospho/galactolipids, pPLAIII enzymes (pPLAIIIα, β, γ, and δ) are also involved in plant responses to abiotic stress, auxin signaling (Scherer et al., 2010), and in resistance to viral diseases (Jang et al., 2020). Also, pPLAIIIs influence the development of vegetative tissues and seeds (Li et al., 2011; Liu et al., 2015; Jang et al., 2021). Previous studies have indicated that pPLAIIIδ is specifically involved in several cellular functions, including growth, organ development, and signal transduction (Li et al., 2013; Dong et al., 2014; Li et al., 2015). For example, the overexpression of pPLAIIIδ was reported to enhance oil content and cause a dwarf phenotype in Arabidopsis, Camelina sativa, and Brassica napus (Li et al., 2013; Dong et al., 2014; Li et al., 2015). Additionally, pPLAIIIδ was reported to cause a decrease in cellulose content when overexpressed in C. sativa (Li et al., 2015). However, no study has reported the role played by pPLAIIIδ in the lignification of plant secondary walls. In this study, we found that the overexpression of pPLAIIIδ in Arabidopsis resulted in a significant decrease in lignification, while the silencing of pPLAIIIδ caused an increase in the lignin content. We also found that pPLAIIIδ played a significant role in plant growth, chlorophyll production, and its overexpression decreased the initial seed germination rate.
The Arabidopsis thaliana Colombia ecotype (Col-0) was used as a wild type (WT) and background for the pPLAIIIδ gene overexpression and silenced lines. The pplalllδ (SALK_029470 and SALK_105929) SALK T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/). Arabidopsis seeds were sown in half-strength Murashige and Skoog (MS) medium (Duchefa Biochemie, Haarlem, The Netherlands) containing 1% sucrose, 0.5 g/L 2-(N-morpholino) ethanesulfonic acid, 0.8% phytoagar, and the pH was adjusted to pH 5.7. Vernalization treatment was conducted for 2 days at 4°C under dark conditions, after which seeds were grown under long-day light (16 h light/8 h dark) at 23°C. Arabidopsis seedlings grown in vitro for 8 days were transferred to sterilized soil mixed with vermiculite and perlite at a 3:2:1 ratio.
For the overexpression of pPLAIIIδ (pPLAIIIδ-OE), the full-length genomic sequence of pPLAIIIδ was obtained from wild-type Arabidopsis (Col-0) by polymerase chain reaction (PCR) using primers listed in Supplementary Table 1. Enzyme-digested PCR products were then cloned into the pCAMBIA 1390 vector driven by a native PLAIIIδ promoter and tagged with an enhanced cyan fluorescence protein at the C-terminus. To generate the promoterPLAIIIδ::GUS fusion construct (PrompPLAIIIδ::GUS), a pPLAIIIδ full promoter region was amplified from the upstream intergenic region using primers listed in Supplementary Table 1. The PCR-amplified product was cloned into a vector pCAMBIA1390 which contained a gusA reporter gene. RNA interference (RNAi) was used to silence the pPLAIIIδ gene. A pPLAIIIδ:RNAi construct was generated through the amplification of a 164 bp of pPLAIIIδ gene coding sequence (sense) and its complementary sequence (antisense) using primers listed in Supplementary Table 1. The amplicons were cloned into the pHANNIBAL vector at XhoI/KpnI restriction enzyme sites (sense orientation) and HindIII/XbaI restriction enzyme sites (antisense orientation). The dsRNA cassette under the 35S cauliflower mosaic virus promoter was then recloned into the pART27 vector. The generated constructs were introduced into Agrobacterium tumefaciens C58C1 (pMP90), which were later transformed into Arabidopsis by the floral dipping method (Clough and Bent, 1998).
The β-glucuronidase (GUS) activity was visualized in PrompPLAIIIδ::GUS expressing transgenic plants by incubating them in the staining buffer containing 1 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid cyclohexylammonium salt (X-Gluc, Duchefa Biochemie, Haarlem, The Netherlands), 0.1 M NaH2PO4, 0.01 M EDTA, 0.1% Triton-X, and 0.5 mM potassium ferri- and ferrocyanide in the dark at 37°C until blue color appeared. The stained plants were then washed in 70% (v/v) and 100% (v/v) ethanol sequentially for 2 hours each. Then, the plants were treated with a mixture of 10% (v/v) glycerol/50% (v/v) ethanol and 30% (v/v) glycerol/30% (v/v) ethanol to completely decolorize all non-stained tissues. A digital single-lens reflex (DSLR) camera (D8, Nikon, Tokyo, Japan) and a microscope (M165FC and DM3000 LED, Leica, Wetzlar, Germany) were used for imaging the GUS-stained plants.
Arabidopsis tissues from Col-0, vector control, pPLAIIIδOE, and pPLAIIIδ:RNAi were used for total RNA extraction following the TaKaRa MiniBEST plant RNA extraction kit (Takara, Shiga, Japan) user manual. The extracted total RNA was quantified using an ultraviolet (UV) spectrophotometer (Nano-MD, Scinco, Seoul, Korea). A 20 μL reaction volume was used to synthesize the complementary DNA (cDNA) using RevertAid reverse transcriptase (Thermo, Waltham, MA, USA). Quantitative PCR (qPCR) was conducted using TB Green™ Premix Ex Taq™ (Takara, Shiga, Japan) and a Thermal Cycler Dice real-time PCR system (Takara, Shiga, Japan). This was followed by standardization of the derived Ct value using the Ct value of a house-keeping gene β-actin. The expression level of the target gene was calculated through comparison with the control value using the formula 2-ΔΔCt. The specific primer sequences of the target genes analyzed by qPCR are listed in Supplementary Table 1.
Stem cross sections of 6-week-old Col-0, vector control, pPLAIIIδ-OE, and pPLAIIIδ:RNAi plants were used for lignin visualization. A sharp razor blade was used to cut 5 mm-thick sections from the stem taken at distance between 5 cm (basal sections) and 10 cm (apical sections) from the ground. The sections were treated with saturated phloroglucinol in HCl and immediately visualized and imaged under a microscope (M165FC and DM3000 LED, Leica, Wetzlar, Germany).
The lignin content was determined using an acetyl bromide assay (Yan et al., 2012). Stem segments from 6-week-old plants were ground into fine powder in liquid nitrogen and then freeze-dried for 48 hours. Approximately 10 mg of powder was rinsed with 95% ethanol four times and twice with distilled water. The samples were then dried at 60°C after which they were suspended in 2 mL of 25% acetylbromide (v/v in glacial acetic acid). The samples were then incubated for 30 minutes at 70°C, followed by the addition of 0.9 mL of 2 M NaOH, 3 mL glacial acetic, and 0.1 mL 7.5 M hydroxylamine hydrochloride. The samples were centrifuged at 4000 g for 10 minutes, and the supernatant was diluted 20-fold with glacial acetic acid. Finally, the absorbance was determined at 280 nm using a UV spectrophotometer (Scinco, Seoul, Korea).
For chlorophylls extraction, rosette leaf samples taken from 4-week-old plants were ground in liquid nitrogen, and 50 mg of each sample was then mixed with 5 mL of 80% acetone. The mixture was well mixed and centrifuged at 3000 g for 10 minutes. The supernatant was pooled and measured for absorbance at 480, 663, and 650 nm using a Nano-MD UV–vis spectrophotometer (Scinco, Seoul, Korea). The chlorophyll and carotenoid contents were calculated using the standard equations (Wintermans and De Mots, 1965). The experiment was repeated at least three times.
Arabidopsis seeds were sterilized using 70% ethanol and washed three times with autoclaved distilled water. The seeds were then sown and considered germinated when a radicle completely sprouted from a seed. Germinated seeds were counted at 20, 24, and 30 hours after exposure to light in the growth chamber. To analyze the effect of exogenous abscisic acid (ABA) on seed germination, 100 seeds (in triplicate) were sown in 1/2 MS media containing 0.1 and 0.5 μM ABA (Sigma-Aldrich, USA). The percent of germinated seeds was calculated after 24, 36, and 48 hours.
The lipid content of rosette leaves of 3-week-old plants was extracted. Lipidomic analysis was conducted via ultra-performance liquid chromatography (UPLC)-tandem mass spectrometry using an Ultimate RS 3000 UPLC system (Dionex) connected to a quadrupole-time-of-flight 5600 mass spectrometer (AB Sciex). Lipid extraction and lipidomic analysis procedures were conducted following the methods described by Jang et al. (2020).
The Arabidopsis pPLAIIIδ is closely related to the rest members of group III pPLAs (Li et al., 2011; Jang et al., 2019; Jang and Lee, 2020a; Jang and Lee, 2020b; Jang et al., 2020). Its amino acid sequence shares the key conserved domains found in other pPLAIIIs. Among four isoforms of pPLAIIIs, promoter::GUS constructs of pPLAIIIα and pPLAIIIγ were expressed highly in xylem and phloem cells (Jang et al., 2020; Jang et al., 2022), and their overexpression under the 35S promoter reduced cell lignification (Jang and Lee, 2020a; Jang et al., 2022). To determine the functional redundancy of pPLAIIIδ with other homologs, PrompPLAIIIδ::GUS expression was studied. Our study aimed to elucidate where pPLAIIIδ is expressed, especially within tissues such as the stem vascular bundles, for which pPLAIIIδ expression has not yet been studied. Our results showed pPLAIIIδ expressed throughout a young sprouting seedling with a stronger GUS signal in the cotyledons (Figures 1A, B). The seedlings grown under long-day light conditions (Figure 1C) exhibited a different pPLAIIIδ expression distribution from those grown under dark conditions; the distribution became stronger in the hypocotyls of etiolated seedlings grown under dark conditions (Figure 1D). In a mature plant, pPLAIIIδ was highly expressed in roots and rosette leaves, while cauline leaves, siliques, and inner floral parts exhibited weaker GUS signals (Figures 1E–G). Furthermore, the cross sections of the apical and basal parts of the stem showed high expression in their vascular bundles (Figure 1H).
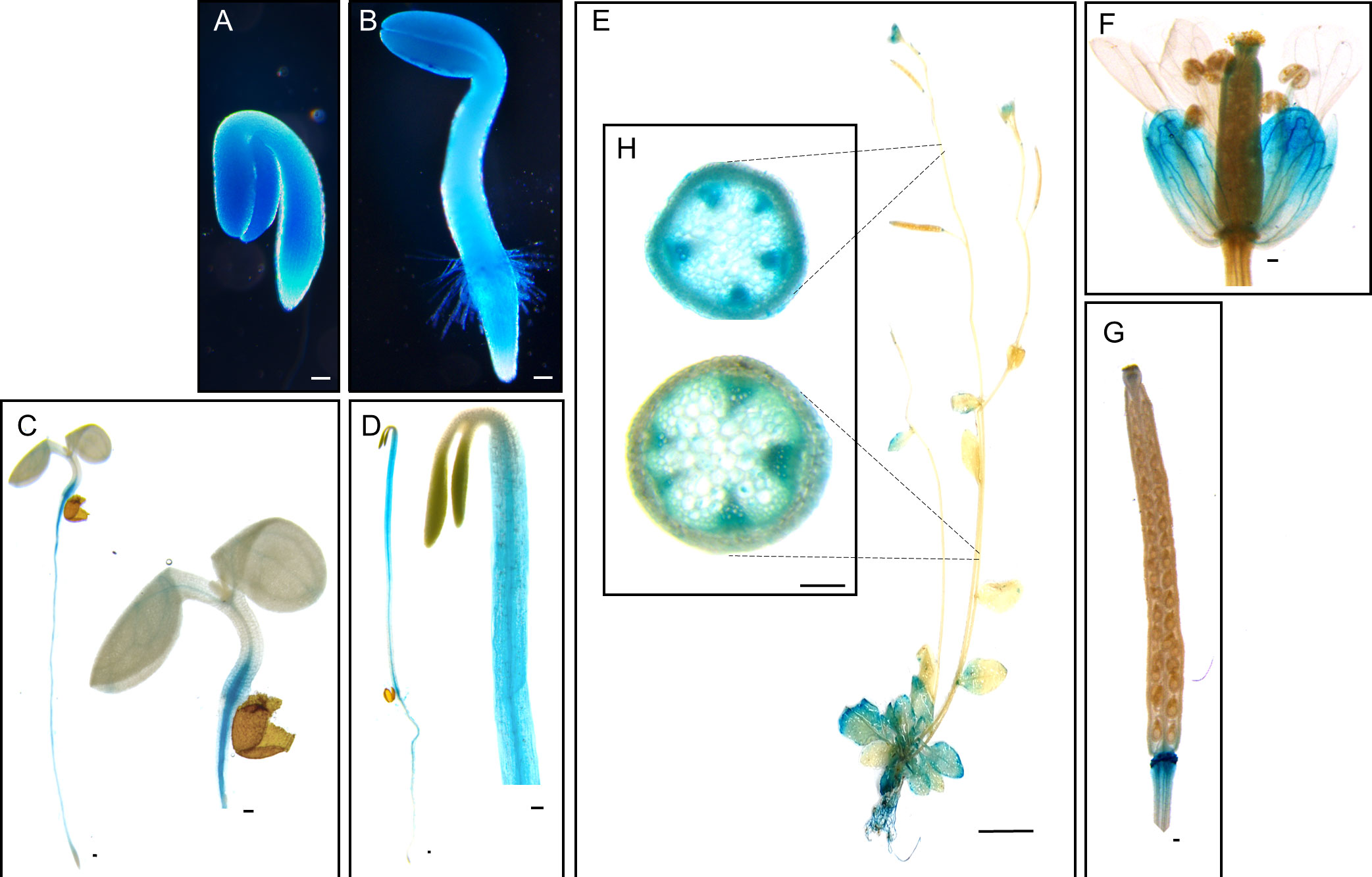
Figure 1 Histochemical analysis of GUS expression harboring PrompPLAIIIδ::GUS in different tissues at different stages of development. (A) 1-day-old seedling, (B) 2-day-old seedling, (C) 4-day-old seedling grown under long-day (16 h light/8 h dark) conditions, and (D) 4-day-old seedling grown under dark conditions. (E) 5-week-old plant. (F) Floral parts, and (G) silique. (H) Cross-sectional image of the apical and basal parts of the stem showing GUS expression in vascular bundles. Scale bars = 100 μm (A–D, F–H) and 1 cm (E).
Studies have reported that the overexpression (OE) of pPLAIII members (α, β, γ, δ) caused a dwarf phenotype under the 35S promoter (Li et al., 2011; Li et al., 2013; Dong et al., 2014; Liu et al., 2015; Jang and Lee, 2020a; Jang and Lee, 2020b; Jang et al., 2020; Jang et al., 2022). A study using a T-DNA insertional pplalllδ mutant (SALK_029470) revealed few phenotypic differences (Li et al., 2013; Dong et al., 2014), because T-DNA insertions were not within exon, and the mutant was not a knockout (Li et al., 2013, Supplementary Figure 1). Another available T-DNA insertional pplalllδ mutant (SALK_105929) was also found to be an incomplete knockout (Supplementary Figure 1). For this reason, RNA interference (pPLAIIIδ:RNAi) was constructed to silence the pPLAIIIδ gene, which resulted in an average of 0.13× downregulation (Figure 2A; Supplementary Figure 1). The pPLAIIIδ:RNAi lines showed a significant increase in plant height, leaf surface area, and seed length (Figures 2B–F; Supplementary Figure 2A). However, the growth of the overexpression plants of pPLAIIIδ under native promoter (pPLAIIIδ-OE) was stunted, with reduced leaf surface area, reduced seed length, and increased seed width (Figures 2B–G). Moreover, pPLAIIIδ-OE lines showed shorter root hairs and reduced root hair density (Figures 2H–J). The root hair length was not significantly changed in pPLAIIIδ:RNAi lines, but root hair density increased in pPLAIIIδ:RNAi#11 (Figures 2H–J). The observed phenotypic results support previously reported effect of pPLAIII genes OE in impeding anisotropic cell expansion (Li et al., 2011; Jang and Lee, 2020a; Jang and Lee, 2020b; Jang et al., 2020; Jang et al., 2021; Jang et al., 2022). The decreased size of pPLAIIIδ-OE plants coincided with the downregulation of genes regulating active gibberellic acid (GA) biosynthesis, GA20ox1 and GA3ox1 (Supplementary Figure 2B). Expression of a GA catabolism gene GA2ox1 was increased in OE line #10. In contrast, GA2ox1 was downregulated in pPLAIIIδ:RNAi #11 (Supplementary Figure 2C). Another GA catabolism gene GA2ox2 was unchanged in all transgenic lines. Surprisingly, the GA3ox1 gene was downregulated in pPLAIIIδ:RNAi lines. Similar observations were reported in OspPLAIIIα-OE which caused dwarf rice plants by decreasing gibberellic acid sensitivity (Liu et al., 2015).
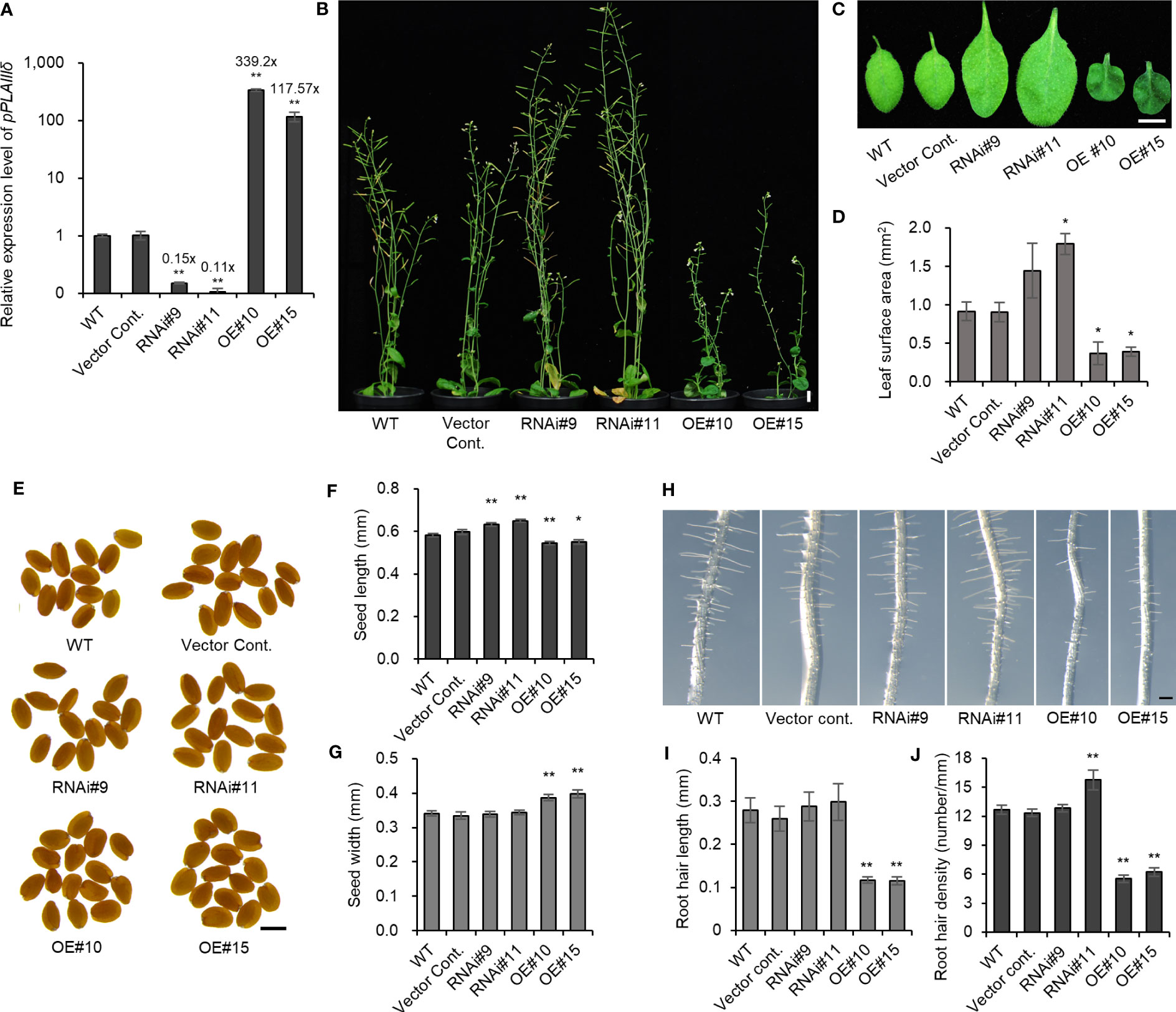
Figure 2 Overexpression of pPLAIIIδ alters the growth of plants. (A) pPLAIIIδ expression level in transgenic lines and in the controls. (B) 8-week-old plants. (C) Leaf phenotype and (D) leaf surface area. (E) Seed phenotype and statistical analysis of (F) seed length and (G) seed width. (H) 4-day-old root hair phenotype and quantification of (I) root hair length and (J) root hair density. Mean ± SE of three independent replicates. Asterisks indicate significant differences, obtained using the Student’s t-test (*P < 0.05 and **P < 0.01) compared with the wild type. Scale bar = 1 cm (B, C), 500 μm (E), and 0.2 mm (H). WT, wild type; Vector Cont., vector control; RNAi, pPLAIIIδ:RNAi; OE, pPLAIIIδ-OE.
The discovery of the role played by other members of the pPLAIII family in plant lignin biosynthesis prompted a study to determine if pPLAIIIδ also influences lignification. The expression of PrompPLAIIIδ::GUS in stem vascular cells (Figure 1) also indicates the possible involvement of pPLAIIIδ in lignin biosynthesis. Phloroglucinol-HCl staining is often used to visualize cell wall lignification as it can react with cinnamyl aldehydes to yield a pink or red-brown coloration (Pomer et al., 2002). Phloroglucinol-HCl staining was stronger in pPLAIIIδ:RNAi lines and weaker in pPLAIIIδ-OE lines compared with the controls (Figure 3A). The phloroglucinol-HCl staining results were consistent with results of the direct quantification of soluble lignin, which showed that the lignin content was significantly increased in pPLAIIIδ:RNAi lines but decreased in pPLAIIIδ-OE lines (Figure 3B). To identify the underlining mechanism through which pPLAIIIδ influences lignification, we checked the expression level of key transcription factors involved in the regulation of lignin biosynthesis, namely MYB58 and MYB63 (Zhou et al., 2009). Both transcription factors were upregulated in pPLAIIIδ:RNAi lines and downregulated in pPLAIIIδ-OE lines (Figure 3C). These results suggest that pPLAIIIδ functions upstream of the two lignin transcription factors. The structural genes in lignin biosynthesis are directly regulated by MYB58 and MYB68, mostly through interactions at the AC-rich elements (Zhou et al., 2009; Zhao and Dixon, 2011). Thus, the expression levels of phenylalanine ammonia-lyase (PAL), cinnamoyl CoA reductase (CCR), hydroxylcinnamoyl transferase (HCT), and caffeic acid 3-O-methyltransferase (COMT) structural genes involved in lignin biosynthesis (Zhao and Dixon, 2011) were analyzed (Figure 3D). The qPCR analysis revealed an increased expression of CCR1 (2.4-fold), PAL (1.9-fold), and HCT (1.9-fold) in pPLAIIIδ:RNAi lines, whereas the expressions of the genes decreased significantly in OE lines, except for PAL. The expression of the CCR1 gene in OE lines decreased by an average of 1.25-folds, whereas that of COMT decreased by 2.5-folds, and that of HCT1 decreased by 1.5-folds. Taken together, these results reveal that pPLAIIIδ negatively regulates lignin biosynthesis by influencing the expressions of MYB58 and MYB68, which consequently affects the structural gene expression in the lignin biosynthesis pathway.
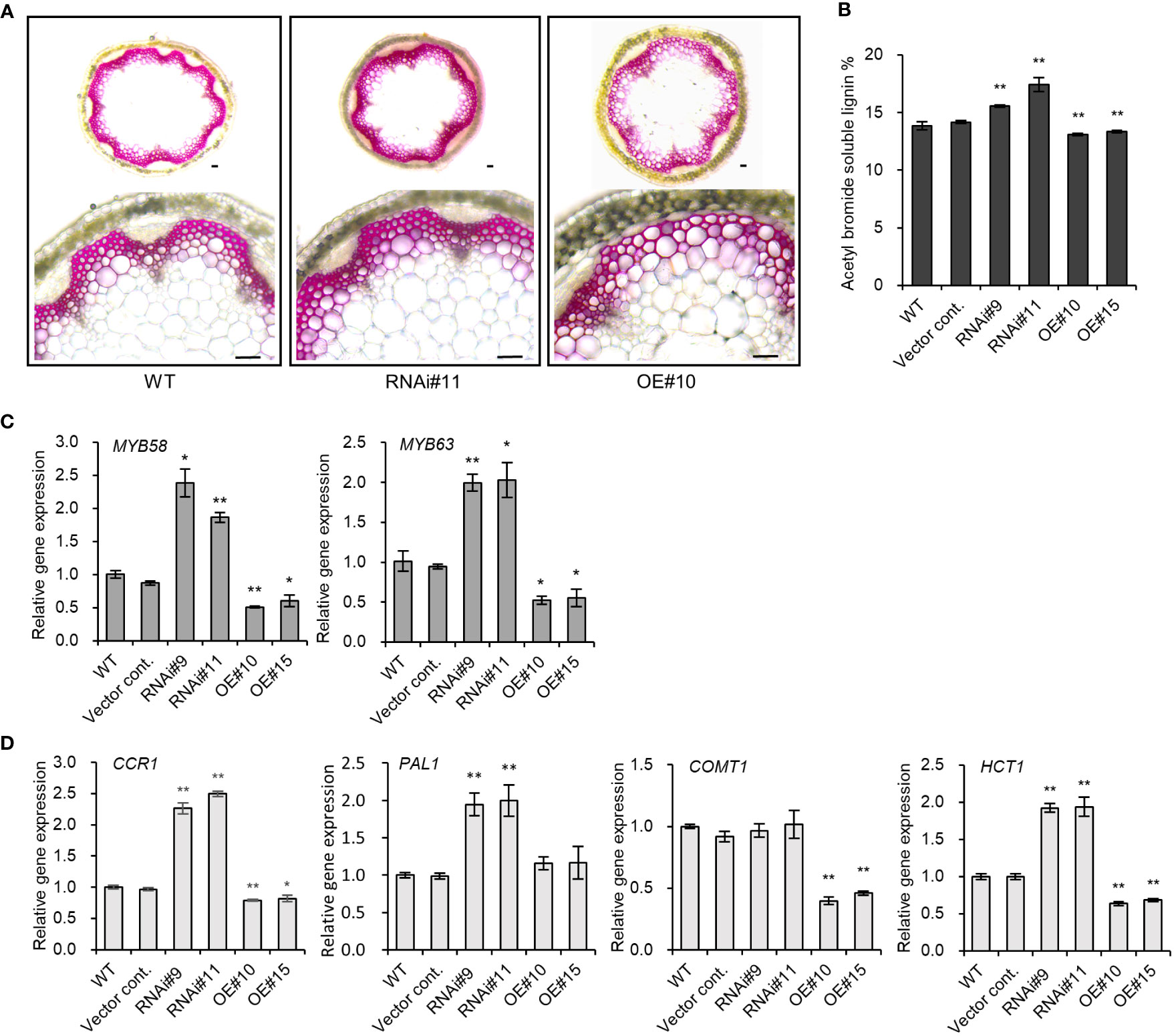
Figure 3 pPLAIIIδ alters the lignification of secondary walls. (A) Cross section of phloroglucinol-HCl stained Arabidopsis stem. Scale bar = 50 μm. (B) Quantification of the acetyl bromide soluble lignin content in RNAi, OE, and controls. (C) Transcription factors involved in the lignin biosynthesis pathway, and (D) the expression level of structural genes quantified by qPCR. Mean ± SE of the three independent replicates. Asterisks indicate significant differences, obtained using the Student’s t-test (*P < 0.05 and **P < 0.01) compared with the wild type.
The altered seed dimensions in pPLAIIIδ transgenic lines indicated a possible role of the gene in regulating seed germination. After 20 hours under light, the germination rate decreased by 24% in pPLAIIIδ-OE (Figure 4A). However, this initial delay in germination was not observed 30 hours after exposure to light because all seeds germinated (Figure 4A). pPLAIIIδ:RNAi showed no significant effect on the rate of seed germination. To confirm the delay of germination caused by pPLAIIIδOE, we used 24-hour imbibed seeds to check the expression level of genes involved in ethylene or gibberellin production. Studies have observed that seed germination depends on the accumulation of ethylene, which is produced by 1-aminocyclopropane-1-carboxylic acid (ACC) synthases (ACS4, ACS5, and ACS11) and oxidases (ACO1 and ACO2) (Corbineau et al., 2014; Dong et al., 2014). The activities of ACO1 and ACO2 play a more significant role than the ACS activity during seed germination (Corbineau et al., 2014). No significant change in both the ACO1 and ACO2 expression levels occurred (Figure 4B). Three ACS genes were also not significantly changed compared with controls (Supplementary Figure 3). Thus, we checked if the expression level of the genes involved in gibberellic acid (GA) production changed in pPLAIIIδ-OE seeds (Figure 4C). GA plays an important role in improving germination by working antagonistically with abscisic acid (ABA), which is known as a germination inhibitor (Koornneef et al., 2002). The expression levels of GA20ox1 and GA3ox1, which are the GA oxidases responsible for synthesizing bioactive gibberellins were downregulated 0.7-fold and 0.4-fold, respectively, in pPLAIIIδ-OE. Moreover, the expression levels of GA oxidases GA2ox1 and GA2ox2 in pPLAIIIδ-OE lines were increased, by 1.7- and 1.4-folds, respectively. GA2ox1 and GA2ox2 are known to inhibit active GA biosynthesis in plants (Li et al., 2021). The decrease of GA biosynthesis genes and increase of GA catabolism genes in pPLAIIIδ-OE germinating seeds, indicated an enhanced ABA sensitivity during seed germination. Germination test in media containing ABA showed pPLAIIIδ-OE seeds were more sensitive to ABA compared to the control seeds (Figures 4D–F). For example, the average germination of pPLAIIIδ-OE seeds in media containing 0.1 μM, and 0.5 μM ABA was reduced by 52.1%, and 38.2% respectively, 48 hours after exposure to light (Figures 4E, F). The germination rate of pPLAIIIδ:RNAi lines was not significantly changed. These results indicate that pPLAIIIδ-OE delays seed germination by affecting the ratio of GA to ABA.
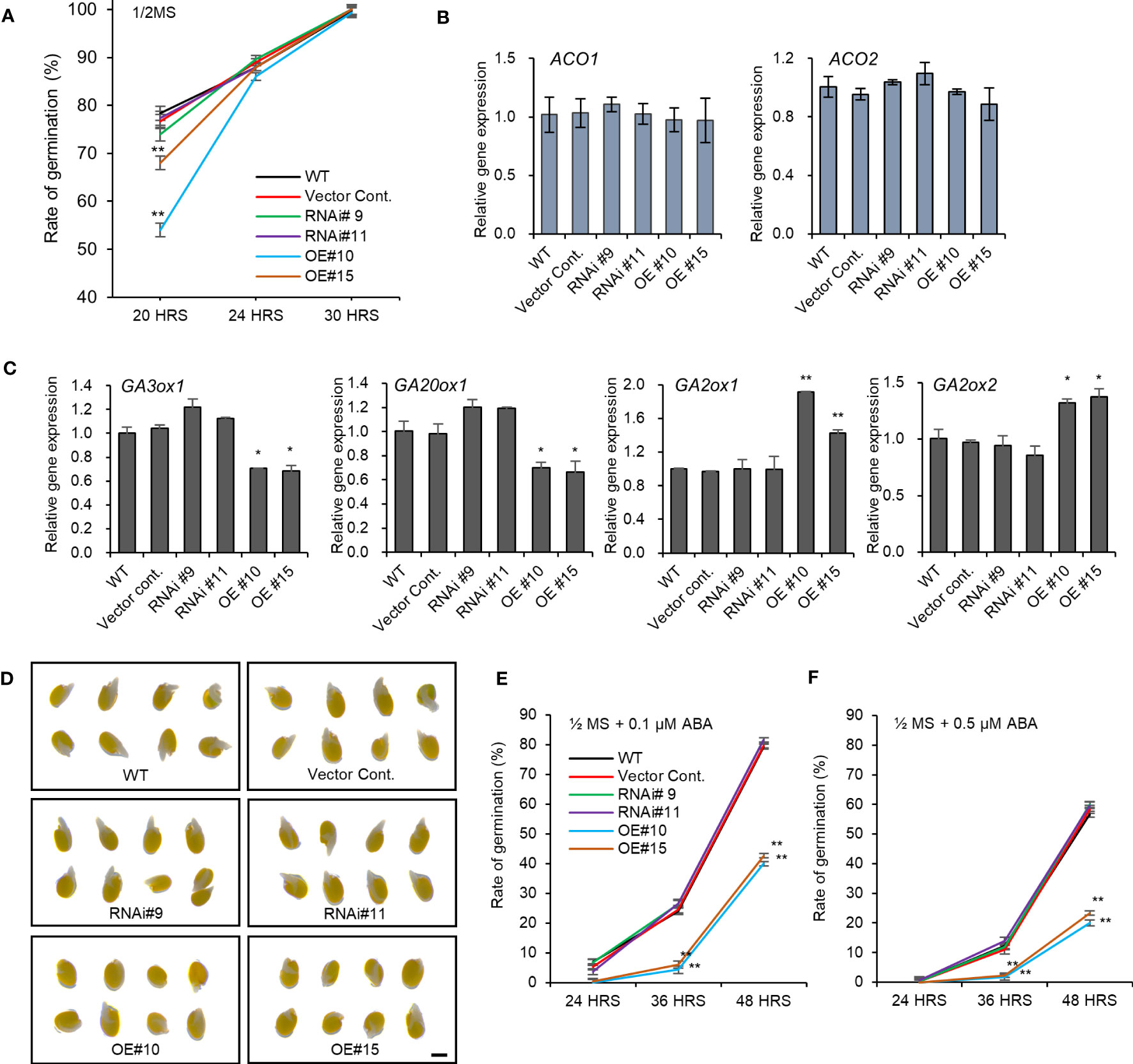
Figure 4 Seed germination rate altered by pPLAIIIδ-OE. (A) Seed germination rate. Expression levels of (B) ACC oxidases genes involved in ethylene biosynthesis. (C) Expression level of genes involved in active gibberellin biosynthesis (GA3ox1, GA20ox1), and genes involved in inactive gibberellin synthesis (GA2ox1, GA2ox2). (D) Seed germination with 0.1 μM ABA after 48 hours. (E) Seed germination rate with 0.1 μM ABA, and (F) 0.5 μM ABA. Mean ± SE of three independent replicates. Asterisks indicate significant differences, obtained using the Student’s t-test (*P < 0.05 and **P < 0.01). Scale bar= 0.5 mm.
The two galactolipids, monogalactosyldiacylglycerol (MGDG) and digalactosyldiacylglycerol (DGDG), constitute 75% of chloroplast lipidsthat play a key role in the structure and function of thylakoid membranes (Kobayashi et al., 2013). pplaIIIδ knockout mutant lines were reported to exhibit reduced total DGDG and MGDG levels compared to WT (Dong et al., 2014). Our study further identifies the individual molecular species of galactolipids that are affected by the silencing pPLAIIIδ, particularly because different molecular species may play different physiological roles (Wang et al., 2020). Using the pPLAIIIδ:RNAi line#11, we found that the significantly reduced MGDG molecular species were 34:1, 34:2-1, 34:2-2, 34:3, 34:4, 36:3, and 38:5 (Figure 5A). We did not find any significant difference in the amount of individual molecular species of DGDG (Figure 5A). To further elucidate the mechanism underlying the influence of pPLAIIIδ in MGDG production, we analyzed the expression levels of three MGDG synthase paralogs: MGD1, MGD2, and MGD3. MGD1 is the major isoform involved in the synthesis of MGDG in chloroplasts, whereas MGD2 and MGD3 function to supply MGDG as a substrate for DGDG biosynthesis, and are activated during phosphate deficiency (Kobayashi et al., 2009). The expression level of MGD1 was increased by 2.5-fold in pPLAIIIδ-OE lines, while the expression level of MGD2 did not significantly change in all lines, and MGD3 expression decreased by 0.3-fold in pPLAIIIδ-OE #10 (Figure 5B). The significant decrease in several molecular species of the chlorophyll-based lipid (MGDG) in pPLAIIIδ:RNAi and the increased expression of MGD1 in pPLAIIIδ-OE lines indicated a functional role of pPLAIIIδ in chlorophyll synthesis and function. To prove this hypothesis, we analyzed the chlorophyll content in the transgenic lines. Quantification analysis revealed that pPLAIIIδ-OE lines had higher total chlorophyll and carotenoid contents than the controls (Figures 5C, D). Moreover, the total chlorophyll content decreased in pPLAIIIδ:RNAi #11, while the carotenoid level increased slightly in pPLAIIIδ:RNAi lines compared to the controls. The leaf phenotype of pPLAIIIδ-OE lines also showed a relatively darker greenish color compared with the color of the pPLAIIIδ:RNAi and control lines (Figure 2C). These results are similar to those found when a PgpPLAIIIβ was overexpressed in hybrid poplar plants (Jang et al., 2019), which signifies that pPLAIIIs play a key role in chlorophyll accumulation.
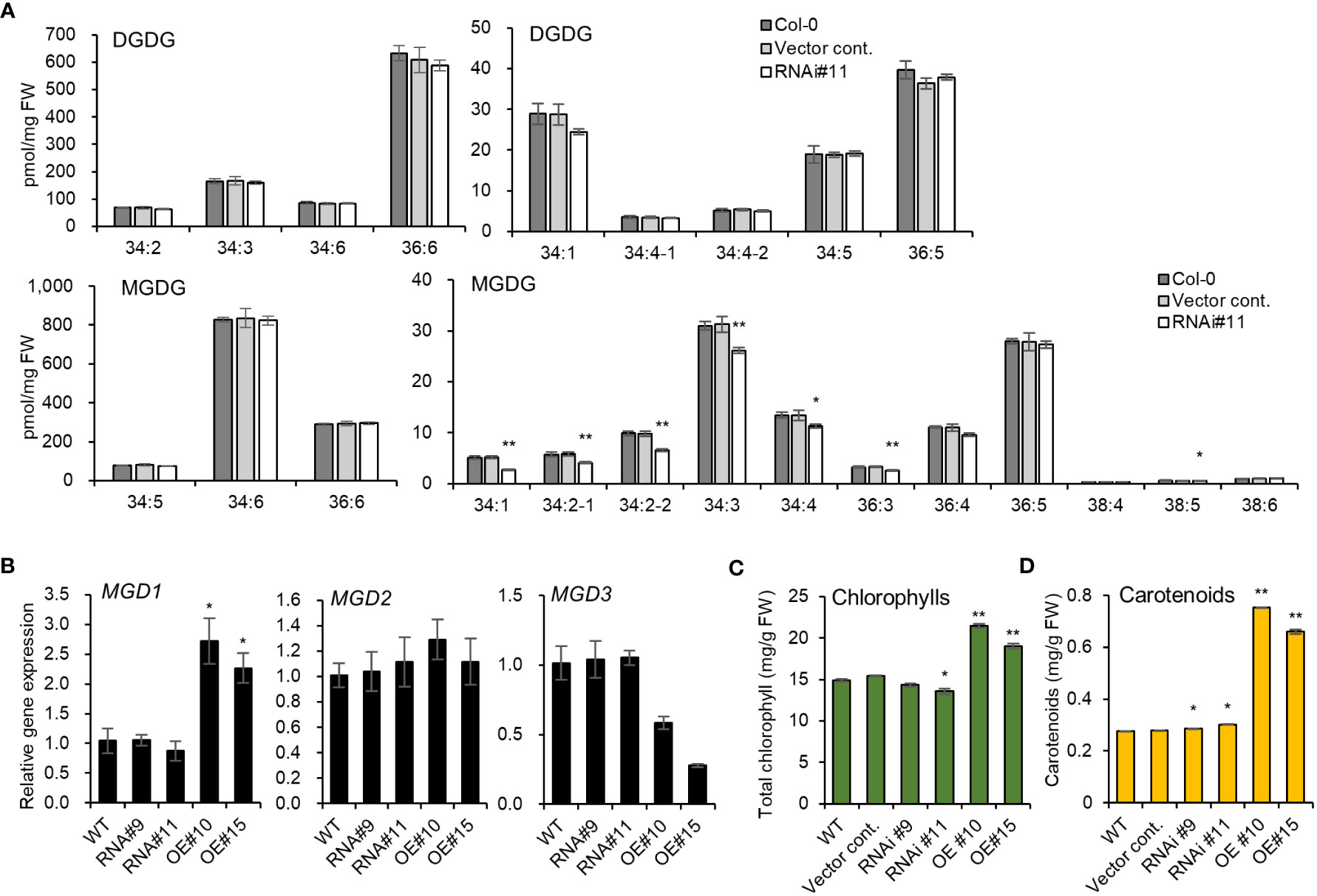
Figure 5 pPLAIIIδ plays a role in chlorophyll production. (A) The abundance of individual molecular species of galactolipids in pPLAIIIδ:RNAi and the controls. (B) The expression level of MGDG synthase genes, which codes for enzymes catalyzing MGDG synthesis. (C) Total chlorophylls, and (D) carotenoid contents. Mean ± SE of three independent replicates. Asterisks indicate significant differences, obtained using the Student’s t-test (*P < 0.05 and **P < 0.01) compared with the wild type.
Studying the overexpression and downregulation of pPLAIIIδ confirmed and elucidated the functional characteristics of the pPLAIII family genes. Specifically, the temporal and spatial expression patterns of pPLAIIIδ in roots and leaves were confirmed. In addition, the gene was well expressed in stem vascular bundles, where it played a significant role in secondary wall lignification. Our results also showed that the overall size of plants was reduced when pPLAIIIδ was overexpressed but enlarged when it was silenced. The altered seed size in pPLAIIIδ-OE led to delays in germination through the modulation of gibberellin-biosynthetic genes. This study also provides a link between pPLAIIIδ, MGDG levels, and chlorophyll contents.
The pPLAIIIδ expression was previously observed in roots, hypocotyl, the vascular bundles of leaves, and the meristem of the stem (Dong et al., 2014). Our study confirmed this expression pattern and also elucidated pPLAIIIδ expression in the stem xylem and phloem tissues (Figure 1). Expression in the vascular tissues of stems has also been observed in pPLAIIIα (Jang et al., 2020), and pPLAIIIγ (Jang et al., 2022). The two genes also played a role in the lignification of stem vascular tissues when overexpressed. Similarly, the phloroglucinol staining and the direct lignin quantification of stem tissues confirmed the role of pPLAIIIδ in stem lignification (Figure 3). Our results confirm the functional role of pPLAIIIs in plant lignification. Moreover, the increased lignin content in pPLAIIIδ:RNAi plants indicates that the gene played a more influential role in lignin formation than other members of the pPLAIIIs. In the case of pplaIIIα knockout mutants, the lignin level was not altered (Jang and Lee, 2020a). However, the downregulation or knockout effects of pPLAIIIβ and pPLAIIIγ genes on lignification needs to be studied further to confirm this notion.
The overexpression of the pPLAIII members has been reported to cause stunted growth (Li et al., 2011; Dong et al., 2014; Jang and Lee, 2020a; Jang and Lee, 2020b; Jang et al., 2022), limiting their potential use in bio-products industries. However, the effects of pPLAIIIs downregulation on the size of plants have not been fully revealed. Thus far, only the pplalllβ mutant has been reported to influence plant growth by increasing leaf size and root length (Li et al., 2011). The knockout mutant of pplalllα did not show any phenotypic difference (Jang et al., 2020), while that of pplalllγ is yet to be identified (Jang et al., 2021). An incomplete knockout mutant of pplalllδ (SALK_029470) revealed few phenotypic differences such as longer hypocotyls and trichomes (Li et al., 2013; Dong et al., 2014). Owing to the lack of knockout mutants of the pplalllδ gene, this study generated the first RNAi silenced lines, which displayed larger leaves, taller plants, and longer seeds, with opposite results in pPLAIIIδ-OE (Figure 2). Overall, depending on the threshold level of the mRNA expression of pPLAIIIs, overexpression drives reduced longitudinal growth, while silencing promotes overall growth. The pPLAIIIs-influcenced growth alterations have been linked to several internal processes such as changes in macrotubule-associated proteins (Jang et al., 2021), and altered GA metabolism (Liu et al., 2015). This study has shown that decreased transcript level of GA biosynthesis genes could be another underlying mechanism for dwarf phenotype seen in OE lines. Generally, the increased size observed in pPLAIIIδ:RNAi lines, and reduced lignification reported in pPLAIIIα and pPLAIIIγ-OE indicate a possibility of co-regulation of pPLAIII genes to obtain a phenotypically suitable transgenic plant for bio-products industries.
Although plant size and lignin amount are important traits in biomass processing industries, seed vigor is equally necessary for selecting suitable lines to be propagated for bioproducts manufacturing. Plants with rapid germination rate are considered more suitable, especially in the era of increased global temperatures (Reed et al., 2022). While the pPLAIIIδ-OE showed delayed seed germination, the pPLAIIIα-OE and the PgpPLAIIIβ-OE plants were reported to have increased the initial seed germination rate (Jang and Lee, 2020b; Jang et al., 2020). We further found that pPLAIIIδ-OE seeds were more sensitive to ABA (Figures 4E, F). The delayed germination in pPLAIIIδ-OE indicates that the gene behaves differently compared with pPLAIIIα-OE which increased seed germination by altering the biosynthesis of both ethylene and GA (Jang et al., 2020). These results suggest the existence of specific differential regulatory mechanisms in pPLAIIIs, underlined by changes in seed phenotype and initial germination rate. The antagonistic effects on the seed germination rate between pPLAIIIδ-OE and pPLAIIIα-OE suggest a possible gene suppression by ectopic gene expression. When we analyzed the transcript levels of the other pPLAIII genes in the pPLAIIIδ mutant lines (Supplementary Figure 4), the expression of pPLAIIIα was decreased in the pPLAIIIδ-OE lines, while the expression of pPLAIIIγ was increased in the pPLAIIIδ:RNAi lines. In the case of pPLAIIIγOE, the expression level of pPLAIIIδ was decreased (Jang et al., 2022), which also suggests the gene redundancy between pPLAIIIγ and pPLAIIIδ. Therefore, we can speculate that transgenic plants with silenced pPLAIIIδ and overexpressed pPLAIIIα could potentially have a good seed vigor as well as reduced lignin content making them suitable in biomass industries.
Silencing the pPLAIIIδ gene decreased key MGDG molecular species, including 34:3 (Figure 5A) which is required for photosynthesis (Wang et al., 2020), reducing the chlorophyll content (Figure 5C). Furthermore, the high level of photosynthetic pigments in the pPLAIIIδ-OE lines was a result of the increased expression of MDG1 (Figure 5B). Similar to our results, overexpression ofpPLAIIIβ increased chlorophylls content (Jang et al., 2019), and also increased the levels of MGDG and DGDG (Li et al., 2011). Moreover, similar to the effects of pPLAIIIδ:RNAi (Figure 5A), the pplaIIIβ and pplalllδ mutants also showed reduced galactolipids molecular species (Li et al., 2011; Dong et al., 2014). This suggests a functional redundancy between pPLAIIIβ and pPLAIIIδ during lipid biosynthesis. However, while silencing pPLAIIIδ resulted in reduced MGDG molecules (Figure 5A) and DGDGs (Dong et al., 2014), pplaIIIα knockout mutants did not change the levels of the two galactolipids (Jang et al., 2020). Nevertheless, the levels of MGDG and DGDG in pPLAIIIα-OE lines were decreased (Jang et al., 2020), indicating that another antagonistic function might exist between pPLAIIIδ and pPLAIIIα.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
OL conceived the project and designed the experiments. DS and JJ performed the experiments. OL, DS, and JJ analyzed the data and wrote the paper. All authors contributed to the article and approved the submitted version.
This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korea government (MSIT) (2019R1A2C1004140 and RS-2023-00217064); a grant from the New Breeding Technologies Development program (No. PJ01652301), Rural Development Administration, Republic of Korea.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1212979/full#supplementary-material
Chao, N., Huang, S., Kang, X., Yidilisi, K., Dai, M., Liu, L. (2022). Systematic functional characterization of cinnamyl alcohol dehydrogenase family members revealed their functional divergence in lignin biosynthesis and stress responses in mulberry. Plant Physiol. Biochem. 186, 145–156. doi: 10.1016/j.plaphy.2022.07.008
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 (6), 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Corbineau, F., Xia, Q., Bailly, C., El-Maarouf-Bouteau, H. (2014). Ethylene, a key factor in the regulation of seed dormancy. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00539
Dong, Y., Li, M., Zhang, P., Wang, X., Fan, C., Zhou, Y. (2014). Patatin-related phospholipase pPLAIIIδ influences auxin-responsive cell morphology and organ size in Arabidopsis and Brassica napus. BMC Plant Biol. 14, 332. doi: 10.1186/s12870-014-0332-1
Jang, J. H., Bae, E. K., Choi, Y. I., Lee, O. R. (2019). Ginseng-derived patatin-related phospholipase PgpPLAIIIβ alters plant growth and lignification of xylem in hybrid poplars. Plant Sci. 288, 110224. doi: 10.1016/j.plantsci.2019.110224
Jang, J. H., Lee, O. R. (2020a). Patatin-related phospholipase AtpPLAIIIα affects lignification of xylem in Arabidopsis and hybrid poplars. Plants 9 (4), 451. doi: 10.3390/plants9040451
Jang, J. H., Lee, O. R. (2020b). Overexpression of ginseng patatin-related phospholipase pPLAIIIβ alters the polarity of cell growth and decreases lignin content in Arabidopsis. J. Ginseng Res. 44 (2), 321–331. doi: 10.1016/j.jgr.2019.01.004
Jang, J. H., Nguyen, N. Q., Légeret, B., Beisson, F., Kim, Y. J., Sim, H. J., et al. (2020). Phospholipase pPLAIIIα increases germination rate and resistance to turnip crinkle virus when overexpressed. Plant Physiol. 184 (3), 1482–1498. doi: 10.1104/pp.20.00630
Jang, J. H., Seo, H. S., Lee, O. R. (2021). The reduced longitudinal growth induced by overexpression of pPLAIIIγ is regulated by genes encoding microtubule-associated proteins. Plants 10 (12), 2615. doi: 10.3390/plants10122615
Jang, J. H., Seo, H. S., Lee, O. R. (2022). Overexpression of pPLAIIIγ in Arabidopsis reduced xylem lignification of stem by regulating peroxidases. Plants 11 (2), 200. doi: 10.3390/plants11020200
Kobayashi, K., Nakamura, Y., Ohta, H. (2009). Type A and type B monogalacto syldiacylglycerol synthases are spatially and functionally separated in the plastids of higher plants. Plant Physiol. Biochem. 47 (6), 518–525. doi: 10.1016/j.plaphy.2008.12.012
Kobayashi, K., Narise, T., Sonoike, K., Hashimoto, H., Sato, N., Kondo, M., et al. (2013). Role of galactolipid biosynthesis in coordinated development of photosynthetic complexes and thylakoid membranes during chloroplast biogenesis in Arabidopsis. Plant J. 73 (2), 250–261. doi: 10.1111/tpj.12028
Koornneef, M., Bentsink, L., Hilhorst, H. (2002). Seed dormancy and germination. Curr. Opin. Plant Biol. 5 (1), 33–36. doi: 10.1016/s1369-5266(01)00219-9
Li, M., Bahn, S. C., Fan, C., Li, J., Phan, T., Ortiz, M., et al. (2013). Patatin-related phospholipase pPLAIIIδ increases seed oil content with long-chain fatty acids in Arabidopsis. Plant Physiol. 162 (1), 39–51. doi: 10.1104/pp.113.216994
Li, M., Bahn, S. C., Guo, L., Musgrave, W., Berg, H., Welti, R., et al. (2011). Patatin-related phospholipase pPLAIIIβ-induced changes in lipid metabolism alter cellulose content and cell elongation in Arabidopsis. Plant Cell 23 (3), 1107–1123. doi: 10.1105/tpc.110.081240
Li, Y., Shan, X., Jiang, Z., Zhao, L., Jin, F. (2021). Genome-wide identification and expression analysis of the GA2ox gene family in maize (Zea mays l.) under various abiotic stress conditions. Plant Physiol. Biochem. 166, 621–633. doi: 10.1016/j.plaphy.2021.06.043
Li, M., Wei, F., Tawfall, A., Tang, M., Saettele, A., Wang, X. (2015). Overexpression of patatin-related phospholipase AIIIδ altered plant growth and increased seed oil content in camelina. Plant Biotechnol. J. 13 (6), 766–778. doi: 10.1111/pbi.12304
Liu, M., Thygesen, A., Summerscales, J., Meyer, A. S. (2017). Targeted pre-treatment of hemp bast fibres for optimal performance in biocomposite materials: a review. Ind. Crops Prod. 108, 660–683. doi: 10.1016/j.indcrop.2017.07.027
Liu, G., Zhang, K., Ai, J., Deng, X., Hong, Y., Wang, X. (2015). Patatin-related phospholipase A, pPLAIIIα, modulates the longitudinal growth of vegetative tissues and seeds in rice. J. Exp. Bot. 66 (21), 6945–6955. doi: 10.1093/jxb/erv402
Pomer, E., Merino, F., Ros Barcelo, A. (2002). O-4-Linked conifeyl and sinapyl aldehydes in lignifying cell walls are the main targets of the wiesner (phloroglucinol-HCl) reaction. Protoplasma 220, 17–28. doi: 10.1007/s00709-002-0030-y
Reed, C. R., Bradford, K. J., Khanday, I. (2022). Seed germination and vigor: ensuring crop sustainability in a changing climate. Heredity 128, 450–459. doi: 10.1038/s41437-022-00497-2
Scherer, G. F. E., Ryu, S. B., Wang, X., Matos, A. R., Heitz, T. (2010). Patatin-related phospholipase A: nomenclature, subfamilies and functions in plants. Trends Plant Sci. 15 (12), 693–700. doi: 10.1016/j.tplants.2010.09.005
Wang, Y., Zhang, X., Huang, G., Feng, F., Liu, X., Guo, R., et al. (2020). Dynamic changes in membrane lipid composition of leaves of winter wheat seedlings in response to PEG-induced water stress. BMC Plant Biol. 20 (1), 84. doi: 10.1186/s12870-020-2257-1
Wintermans, J. F. G. M., De Mots, A. S. (1965). Spectrophotometric characteristics of chlorophylls a and b and their phenophytins in ethanol. Biochim. Biophys. Acta 109 (2), 448–453. doi: 10.1016/0926-6585(65)90170-6
Xie, H., Engle, N. L., Venketachalam, S., Yoo, C. G., Barros, J., Lecoultre, M., et al. (2019). Combining loss of function of FOLYLPOLYGLUTAMATE SYNTHETASE1 and CAFFEOYL-COA 3-O-METHYLTRANSFERASE1 for lignin reduction and improved saccharification efficiency in Arabidopsis thaliana. Biotechnol. Biofuels 12 (1), 108. doi: 10.1186/s13068-019-1446-3
Yan, L., Liu, S., Zhao, S., Kang, Y., Wang, D., Gu, T., et al. (2012). Identification of differentially expressed genes in sorghum (Sorghum bicolor) brown midrib mutants. Physiol. Plant 146 (4), 375–387. doi: 10.1111/j.1399-3054.2012.01646.x
Zhao, Q., Dixon, R. A. (2011). Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci. 16 (4), 227–233. doi: 10.1016/j.tplants.2010.12.005
Zhong, R., Ye, Z. (2015). Secondary cell walls: biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol. 56 (2), 195–214. doi: 10.1093/pcp/pcu140
Keywords: phospholipases, lignin, chlorophylls, pPLAIIIδ, MGDG
Citation: Simiyu DC, Jang JH and Lee OR (2023) A group III patatin-like phospholipase gene pPLAIIIδ regulates lignin biosynthesis and influences the rate of seed germination in Arabidopsis thaliana. Front. Plant Sci. 14:1212979. doi: 10.3389/fpls.2023.1212979
Received: 27 April 2023; Accepted: 19 June 2023;
Published: 13 July 2023.
Edited by:
Serena Varotto, University of Padua, ItalyCopyright © 2023 Simiyu, Jang and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ok Ran Lee, bXBpemxlZUBqbnUuYWMua3I=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.