
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 30 August 2023
Sec. Plant Development and EvoDevo
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1209834
 Zeng-Yu Lin1,2
Zeng-Yu Lin1,2 Gen-Fa Zhu1
Gen-Fa Zhu1 Chu-Qiao Lu1
Chu-Qiao Lu1 Jie Gao1
Jie Gao1 Jie Li1
Jie Li1 Qi Xie1
Qi Xie1 Yong-Lu Wei1
Yong-Lu Wei1 Jian-Peng Jin1
Jian-Peng Jin1 Feng-Lan Wang2*
Feng-Lan Wang2* Feng-Xi Yang1*
Feng-Xi Yang1*Cymbidium sinense is one of the most important traditional Chinese Orchids due to its unique and highly ornamental floral organs. Although the ABCDE model for flower development is well-established in model plant species, the precise roles of these genes in C. sinense are not yet fully understood. In this study, four SEPALLATA-like genes were isolated and identified from C. sinense. CsSEP1 and CsSEP3 were grouped into the AGL9 clade, while CsSEP2 and CsSEP4 were included in the AGL2/3/4 clade. The expression pattern of CsSEP genes showed that they were significantly accumulated in reproductive tissues and expressed during flower bud development but only mildly detected or even undetected in vegetative organs. Subcellular localization revealed that CsSEP1 and CsSEP4 were localized to the nucleus, while CsSEP2 and CsSEP3 were located at the nuclear membrane. Promoter sequence analysis predicted that CsSEP genes contained a number of hormone response elements (HREs) and MADS-box binding sites. The early flowering phenotype observed in transgenic Arabidopsis plants expressing four CsSEP genes, along with the expression profiles of endogenous genes, such as SOC1, LFY, AG, FT, SEP3 and TCPs, in both transgenic Arabidopsis and C. sinense protoplasts, suggested that the CsSEP genes played a regulatory role in the flowering transition by influencing downstream genes related to flowering. However, only transgenic plants overexpressing CsSEP3 and CsSEP4 caused abnormal phenotypes of floral organs, while CsSEP1 and CsSEP2 had no effect on floral organs. Protein-protein interaction assays indicated that CsSEPs formed a protein complex with B-class CsAP3-2 and CsSOC1 proteins, affecting downstream genes to regulate floral organs and flowering time. Our findings highlighted both the functional conservation and divergence of SEPALLATA-like genes in C. sinense floral development. These results provided a valuable foundation for future studies of the molecular network underlying floral development in C. sinense.
Orchidaceae is one of the largest families of flowering plants, comprising over 27,000 species grouped into approximately 900 genera. Renowned for their distinct floral patterns, fragrances, colors, and leaf art, Cymbidium species are highly sought after across the globe, especially in Asia (Roberts and Dixon, 2008; Sun et al., 2021; Yang et al., 2021; Li et al., 2022). Cymbidium sinense, one of the eight traditional Chinese Orchids, is a significant ornamental plant and cash crop (Motomura et al., 2010; Zhu et al., 2015; Yang et al., 2021). As an epiphytic plant, it thrives in well-ventilated and cool, moist environments, typically found at low latitudes. The flowering season of C. sinense occurs from January to March, coinciding with the Chinese Spring Festival. However, C. sinense has a lengthy juvenile phase and requires over three years of cultivation before blooming. The conditions required for flowering include a prolonged photoperiod that triggers spike formation, a period of approximately 6 months of semi-dormancy, and finally a phase of low temperature that promotes inflorescence elongation and flowering (Huang et al., 2012; Yang et al., 2019; Ahmad et al., 2022).
A typical angiosperm flower consists of four whorls of floral organs, namely sepals, petals, stamens, and carpels (Robles and Pelaz, 2005). The ABCDE model (Fan et al., 1997) explains that the combination of A- and E-class genes controls sepal identity, while petal identity is regulated by the protein complexes of A, B, and E-class genes. Similarly, male stamen identity is specified by the expression of B, C, and E-class genes, and female carpel identity by C and E-class genes. Ovule development, on the other hand, is regulated by C, D, and E-class genes (Theissen, 2001; Theissen and Saedler, 2001; Ditta et al., 2004). Given their role in the MADS protein-protein interaction network, SEP-like proteins are essential “hub” proteins (Ruokolainen et al., 2010; Hugouvieux and Zubieta, 2018).
The E-class MADS-box genes, which include SEP1, SEP2, SEP3, and SEP4, are essential for determining floral organ identity and are involved in floral meristem identity (Pelaz et al., 2000; Pelaz et al., 2001). The SEP genes have undergone multiple duplications, leading to two clades known as SEP3 (AGL9) and LOFSEP (AGL2/3/4 or SEP1/2/4). Subsequent duplications have occurred independently within these clades after the divergence of eudicots and monocots. The LOFSEP clades are further divided into Petunia PhFBP9/23, Arabidopsis AtSEP1/2/4 subclades, and rice SEP homologous genes LHS1 and OsMADS5/34 (Malcomber and Kellogg, 2005; Zahn et al., 2005; Shan et al., 2009). SEP-like genes are widely reported in both monocots and eudicots due to their crucial role in regulating floral meristem, floral organs, and flowering period. In eudicots, the sep1 sep2 sep3 triple mutants and sep1 sep2 sep3 sep4 quadruple mutants result in sepal-like organs and leaf-like structures in Arabidopsis, respectively (Pelaz et al., 2000; Ditta et al., 2004; Castillejo et al., 2005). A mutation in the fbp2 gene and a double mutation in the fbp2 and fbp5 genes in petunia cause the transformation of floral organs (Vandenbussche et al., 2003; Matsubara et al., 2008). In monocots, overexpression of the OsMADS5 gene in rice only promotes early flowering (Jeon et al., 2000), and knockdown of OsMADS1, OsMADS5, OsMADS7, and OsMADS8 genes simultaneously results in the formation of leaf-like organs (Cui et al., 2010). Additionally, the SEP homologous genes have been shown to be crucial for floral organ and floral meristem development in Gerbera (Kotilainen et al., 2000), Lilium (Tzeng et al., 2003), maize (Lid et al., 2004), Malus (Ireland et al., 2013), cucumber (Wang et al., 2016), Prunus (Li et al., 2017), and tomato (Zhang et al., 2018), making it an excellent example of duplication and divergence in the evolution of the function of the SEP genes in angiosperms (Uimari et al., 2004).
Orchidaceae is the largest family of flowering plants and relies on SEPALLATA-like genes to maintain floral organ identity (Tsai and Chen, 2006; Salemme et al., 2013). The first E-class gene to be identified in orchids was OM1, which was isolated from the hybrid Aranda Deborah (Lu et al., 1993). Later, three additional E-class genes (DOMADS1, DOMADS3, and DcOSEP1) were identified in Dendrobium, and these genes are sequentially activated during floral transition and persist in mature flowers (Yu and Goh, 2000; Yu et al., 2002). Higher-order complexes resulting from the formation of DcOAP3-DcOPI-DcOSEP1 have been detected via yeast three-hybrid experiments (Xu et al., 2006). Recently, diverse functions of E-class genes have been reported in various orchid species. For example, in P. equestris, the PeSEP3 gene can cause tepals to become leaf-like organs (Pan et al., 2014). Additionally, the expression of SEP-like genes has been detected in all floral organs and plays a vital role in the development of the column, lip, and petals in Habenaria radiata and P. henryanum (Mitoma and Kanno, 2018; Valoroso et al., 2019; Cheng et al., 2022). Defects in CeSEP1/3-clade genes also contribute to the leaf-like flower phenotype in Cymbidium ensifolium mutants, consistent with research from Phalaenopsis (Wei et al., 2020). In contrast, CeSEP-2 is crucial for the specialized lip and causes the formation of a peloric flower shape in C. ensifolium (Ai et al., 2021), unlike in P. equestris. Furthermore, the E-class MADS-box PeMADS8 gene may regulate ovule development and form higher-order complexes with other MADS-boxes (Shen et al., 2021). In E. pusilla, SEP-like genes EpMADS8 and EpMADS9 are involved in orchid fruit development (Lin et al., 2016; Dirks-Mulder et al., 2019).
C. sinense is one of the eight traditional Chinese orchid types, valued for its unique floral organ development and flowering time. In this study, we cloned and characterized four SEPALLATA-like genes from C. sinense and found that these genes exhibited divergent expression patterns. Transgenic Arabidopsis plants expressing all four CsSEP genes showed a common phenotype of early flowering, suggesting functional conservation. However, only transgenic plants overexpressing CsSEP3 and CsSEP4 displayed abnormal floral organ phenotypes, indicating divergence in SEPALLATA-like genes in C. sinense. The expression profile of endogenous genes in transgenic Arabidopsis, transient expression in C. sinense protoplasts, and protein interaction assays suggested that CsSEP genes could affect downstream-related genes to regulate floral organs and flowering time in C. sinense. Our study suggested that CsSEP genes played a critical role in regulating floral development in C. sinense.
The wild-type plants of C. sinense ‘Baimo’ utilized in this study were artificially cultivated and collected from the cultivation base of the Environmental Horticulture Research Institute at Guangdong Academy of Agricultural Sciences, China. The plants were grown and maintained in pots inside a greenhouse, with day/night temperatures of 26/23 °C and a 16-h light/8-h dark photoperiod.
Based on the annotated coding sequence (CDS) in the genome database (Yang et al., 2021), we cloned the CDS of CsSEP1, CsSEP2, CsSEP3, and CsSEP4 using specific primers designed by PrimerPremier 5.0. Total RNA was extracted from natural flowers of C. sinense ‘Baimo’ using the RNAprep Pure Plant Kit (TIANGEN, China) following the manufacturer’s instructions. RNA content was measured with a Nano-Drop 2000 Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE), and 1 μg of RNA was used to synthesize first-strand complementary DNA (cDNA) with the HiScript III 1st Strand cDNA Synthesis Kit (Vazyme, China). The cDNA was then used as a template to clone the CDS of CsSEP genes with high-fidelity Taq DNA polymerase (Vazyme, China), and the gene-specific primers used are listed in Supplementary Table S3. The PCR products were purified using the FastPure® Gel DNA Extraction Mini Kit (Vazyme, China) and cloned into the pCE2 TA/Blunt-Zero Vector (Vazyme, China) for transformation into DH5α (Tiangen, China). Positive clones (8-10) were selected for identification and sequenced by the Sangon Company in Shanghai. The plasmids were extracted with the FastPure® Plasmid Mini Kit (Vazyme, China).
We retrieved partial SEP-like genes from previously published studies (Yang et al., 2021; Cheng et al., 2022) and obtained others from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) for phylogenetic analysis. The registration numbers for these genes are listed in Supplementary Table S2. We aligned full-length amino acid sequences using the default settings in ClustalW implemented in MEGA v5.2 (Kumar et al., 2004) and manually adjusted the alignment with the reference alignment provided by Zahn et al. (2005). We used IQ-TREE v1.6.12 (Nguyen et al., 2015) to reconstruct the phylogenetic tree from 1,000 ultrafast bootstrap maximum likelihood (ML) tree replicates. The resulting tree was visualized using Evolview v3 (Subramanian et al., 2019). We performed multiple sequence alignments of CsSEP genes against SEP-like genes of other known orchids and selected angiosperm species using DNAMAN (v.6.0) software.
To verify the spatiotemporal expression patterns of the four CsSEP genes, we analyzed different stages of flower buds (S1, S2, S3, S4, S5), root, stem, leaf, flower, pod, sepals, petals, lips, and column (Yang et al., 2021) from C. sinense by qRT-PCR. Gene-specific primers were designed for each gene within the non-conservative C-terminal region (Supplementary Table S3). qRT-PCR was conducted on a qTOWER 2.0 Real-Time PCR System (Jena, Germany) using ChamQ™ Universal SYBR® qPCR Master Mix (Vazyme) with three biological replications. The qRT-PCR products were amplified in a 20-μL reaction mixture containing 2 μL cDNA, 2 μM of each primer, 10 μL 2×ChamQ Universal SYBR qPCR Master Mix* (Vazyme), and double-distilled water to 20 μL. Briefly, after an initial denaturation step at 95°C for 5 min, the amplifications were carried out with 40 cycles at a melting temperature of 95°C for 15 s, an annealing temperature of 60°C for 30 s, and an extension temperature of 72°C for 30 s, followed by an extra extension step at 72°C for 5 min. In addition, β-actin (Mol013347) was selected as an internal reference gene in C. sinense. The relative expressions of CsSEP genes at the mRNA level were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
The full-length CDS of CsSEPs were cloned into the control vector PAN580-GFP using the ClonExpress® II One Step Cloning Kit (Vazyme, China) in accordance with the manufacturer’s instructions. Specific primers with overlapping homologous ends were designed and are listed in Supplementary Table S3. The plasmids were extracted using the GoldHi EndoFree Plasmid Maxi Kit (CWBIO, China). The protoplast-based transient expression system (PTES) was used to transform the plasmid DNA into C. sinense protoplasts, following the method described by Ren et al. (2020). After incubation for 12-16 h under conditions of 23 °C and darkness, the LSM710 confocal laser scanning microscope was used to visualize the fluorescence of GFP-proteins. To detect cell nuclei, the transfected protoplasts were stained with 50 µg/mL 4’-6’-diamidino-2-phenylindole (DAPI) (Sigma, Germany) at 37 °C for 10 min. DAPI was illuminated by UV light and detected by blue or cyan filters in fluorescence microscopy, and the red self-luminescence of chloroplasts was visualized through the green light of a fluorescence microscope. In addition, three to five images were randomly taken under an LSM710 confocal laser scanning microscope.
Using the genome sequence of C. sinense as a reference, we selected 2,000 bp of the promoter sequence upstream of the ATG start codon for CsSEP1, CsSEP2, CsSEP3, and CsSEP4. We predicted the cis-acting elements and binding sites using the online tools PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and PlantPan3.0 (http://plantpan.itps.ncku.edu.tw/promoter.php) and then generated a visual representation using Tbtools.
Arabidopsis thaliana ecotype Columbia (Col-0) plants were grown under standard glasshouse conditions for use in the functional analysis. We cloned the full-length CDS of CsSEPs into the vector pOCA30 using homologous recombination, resulting in four recombinant plasmids: 35S: CsSEP1, 35S: CsSEP2, 35S: CsSEP3, and 35S: CsSEP4. These plasmids were introduced into A. tumefaciens GV3101 Chemically Competent Cells (WEIDI, China) by the freeze-thaw method and then transformed into wild-type Arabidopsis using the floral dip method (Clough and Bent, 1998). The seeds collected from the infiltrated plants were sterilized and germinated on 1/2 Murashige & Skoog select medium containing 50 μg/mL kanamycin. We analyzed the expressions of the four CsSEP exogenous genes in T2 transgenic plants and wild-type Arabidopsis plants by RT-PCR and normalized the template amounts using the Arabidopsis Actin2 gene (AT3G18780.2). We evaluated the flowering time in T3 transgenic plants using two reference points. We counted the total number of days required to bolt and the number of rosette leaves at bolting, with no fewer than 10 Arabidopsis plants counted each time.
Leaves were harvested from 2-week-old seedlings of 35S: CsSEPs transgenic Arabidopsis plants (T2) for total RNA extraction. The expressions of endogenous flowering-related and leaf-related genes, including AtFT, AtSOC1, AtSEP3, AtTCP20, AtGRF2, AtGRF1, AtTCP3, AtARF2, AtAG, and AtLFY (Kaufmann et al., 2009; Pajoro et al., 2014), were analyzed by qRT-PCR with three biological replications. The regulations to the downstream genes were verified by overexpressing CsSEP genes in C. sinense protoplasts. Fluorescein diacetate (FDA) was used to detect the activity of isolated protoplasts under a fluorescent microscope, and those with transfection efficiency exceeding 50% were chosen for subsequent qRT-PCR expression analysis. The regulations to the downstream flowering-related genes CsSOC1, CsFT, CsLFY, CsAP3-2 and CsAG1were analyzed by qRT-PCR with three biological replications.
The full-length CDSs of CsSEP1, CsSEP2, CsSEP3, CsSEP4, CsAP3-2, and CsSOC1 were cloned and recombined into the pGBKT7 and pGADT7 vectors, resulting in the creation of pGBK-CsSEP1, pGBK-CsSEP2, pGBK-CsSEP3, pGBK-CsSEP4, pGAD-CsSAP3-2, and pGAD-CsSOC1, respectively. Each pair of the eight constructs was co-transformed into Y187 and Y2HGold yeast strains, respectively. Diploids containing pGAD-CsAP3-2, pGAD-CsSOC1, and pGBK-CsSEP1/2/3/4 were obtained using a small-scale protocol (Clontech, Palo Alto, USA). After selecting co-transformants in DDO (SD/-Trp/-Leu) medium, they were transferred to QDO/X/A (SD/-Trp/-His/-Trp/-Ade/X-α-gal/AbA) medium. Yeast cells carrying the pGBKT7-53 and pGADT7-T plasmids were used as positive controls, while pGBKT7-Lam and pGADT7-T were co-transformed and served as negative controls. Blue colonies were observed on QDO/X/A plates within 3-5 days, indicating protein interaction between the two proteins.
Four SEP-like genes are isolated from C. sinense ‘Baimo’ and named CsSEP1, CsSEP2, CsSEP3, and CsSEP4. Sequence analysis showed that the CDS of these genes contained open read frames (ORFs) of 732, 504, 753, and 744 base pairs, encoding putative proteins of 266, 168, 251, and 248 amino acids, respectively (Supplementary Figure S1). The predicted amino acid sequences of these genes shared 34.07% to 60.44% identity (Supplementary Table S1). CsSEP1 and CsSEP3 showed high homology to CgSEP1 and CgSEP3 of C. goeringii, with 75.46% and 90.48% identity, respectively. CsSEP2 shared 52.01% identity with its homolog in C. ensifolium, CeSEP2. The amino acid sequence of CsSEP4 was found to be identical to those of C. ensifolium and C. goeringii, while shared 86.23% identity with PeSEP4 of P. equestris. Based on the amino acid sequences, we constructed a phylogenetic tree to determine the evolutionary relationships of these SEP-like genes, which included a number of available orchids SEP homologs and other selected angiosperm species (Supplementary Table S2). Phylogenetic analysis showed that all the SEP genes fell within the SEP clade and formed two separate monophyletic clades, with AGL6 and SQUA family members used as outgroups (Figure 1). The angiosperm SEP-like genes formed two major subclades, the LOFSEP clade (also known as the SEP1/2/4 or AGL2/3/4 clade) and the SEP3 (or AGL9) clade (Zahn et al., 2005). Based on this analysis, the phylogenetic tree was divided into two major clades (SEP1/2/4 and SEP3), with CsSEP1 and CsSEP3 grouped into the orchid SEP3 (in the SEP3 clade), and CsSEP2 and CsSEP4 included in the orchid SEP1/2 (in the SEP1/2/4 clade) (Figure 1). Like other monocot lineages, the multiple sequence alignment of the SEP genes from orchids showed that CsSEP proteins possessed the conserved MIK domain. The C-terminal domain of CsSEP proteins had a divergent C-terminal domain with the conserved SEP I motif and SEP II motif (Figure 2). Interestingly, both the SEP I motif and SEP II motif of CsSEP2 were lost together, which is similar to the SEPALLATA-like gene DcMADS26 identified by Zhang et al. (2016) in D. catenatum (Supplementary Figure S2).
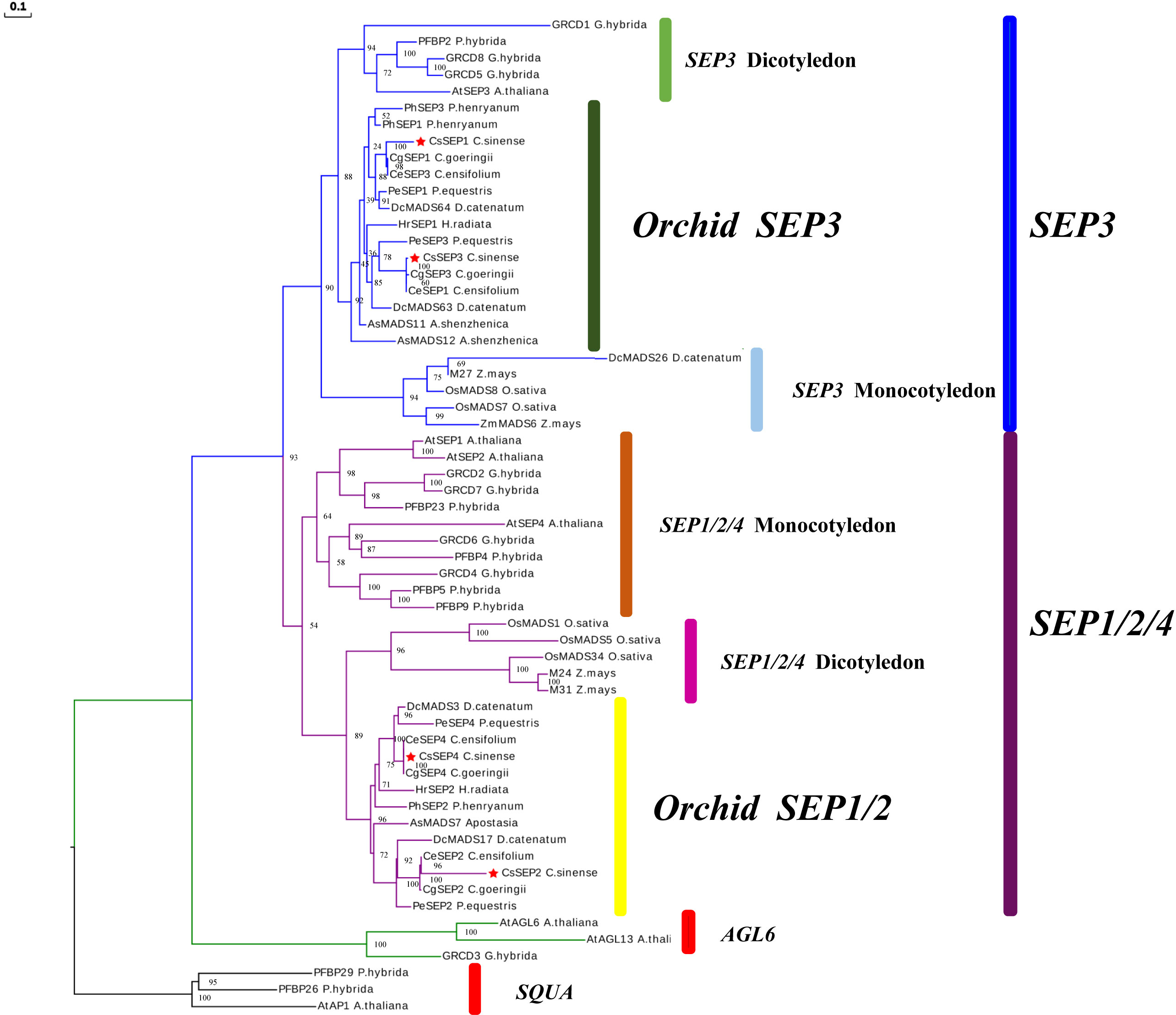
Figure 1 ML-based phylogenetic tree of C. sinense, Phalaenopsis equestris, Cymbidium ensifolium, Paphiopedilum henryanum, Cymbidium goeringii, Apostasia shenzhenica, Dendrobium catenatum, and other selected angiosperm species. The CsSEP1, CsSEP2, CsSEP3, and CsSEP4 genes were marked with red stars.
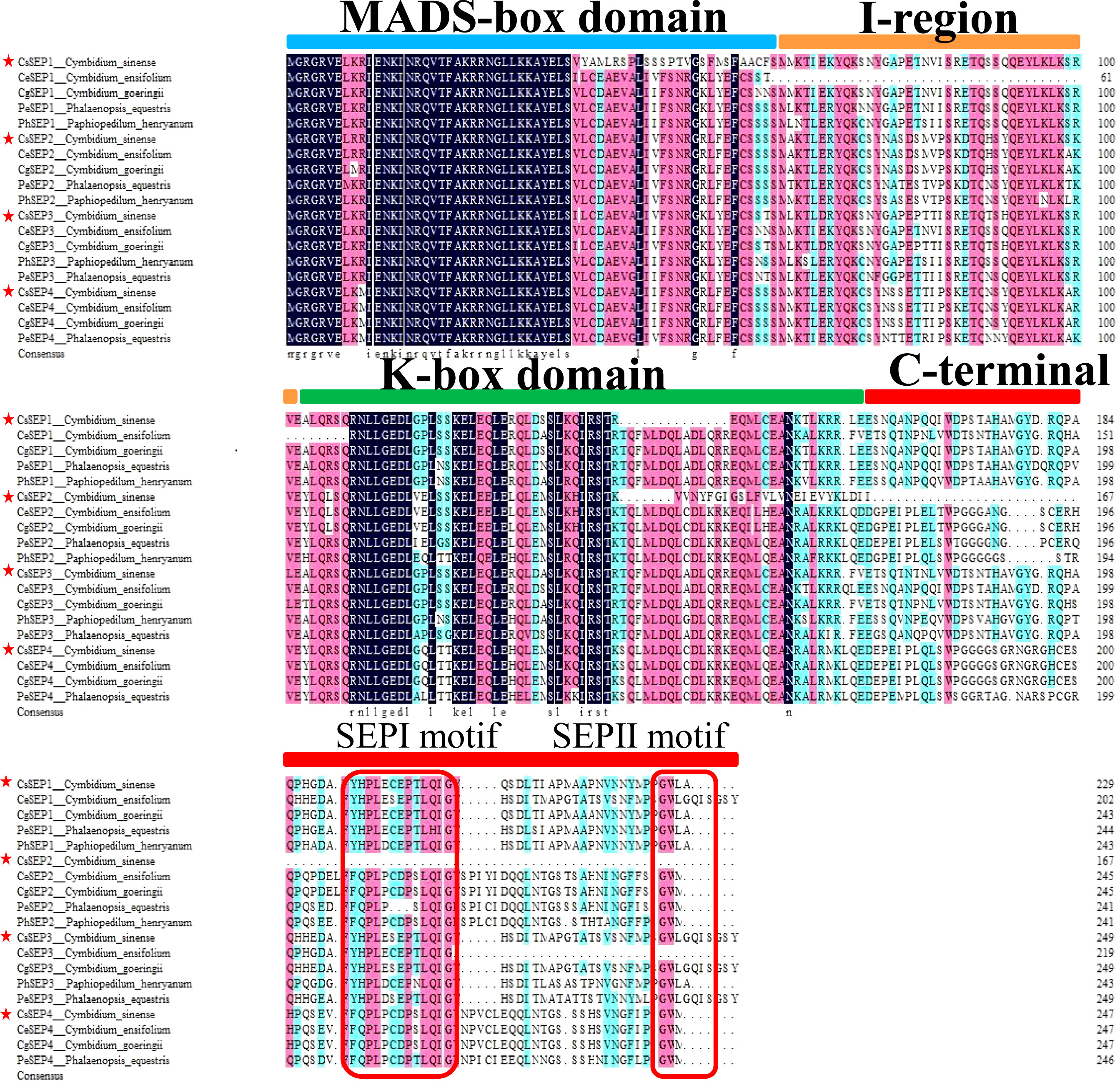
Figure 2 Multiple alignments of the amino acid sequence of CsSEP1, CsSEP2, CsSEP3, and CsSEP4. CsSEP proteins of C. sinense were marked by red stars, which possess the MADS-box domain (light blue line), I-region (orange line), K-box domain (green line), and C-terminal domains (red line). The SEP I and SEP II motifs are indicated with red boxes.
To investigate the role of CsSEP genes in floral development, we examined the expression patterns of four CsSEPs in various tissues and organs (root, stem, leaf, flower, and pod), as well as floral organs and different stages of developing flower buds (Figure 3A). Our findings, as shown in Figures 3B-E, indicated that CsSEP mRNA were significantly accumulated in reproductive tissues, such as flowers and pods. By contrast, the mRNA of all CsSEP genes was only mildly detected or undetected in vegetative organs, such as the root, stem, and leaf. In particular, transcripts of all CsSEP genes were highest in flowers, suggesting an essential role in flower development. In terms of the expression pattern of the five flower bud development stages, CsSEP1, CsSEP2, CsSEP3, and CsSEP4 were expressed throughout all stages of flower bud development (Figures 3F–I). Notably, the expression of CsSEP1 was significantly upregulated in S5 (Figure 3F), while transcripts of CsSEP2 and CsSEP4 were highest in S1 and gradually downregulated (Figures 3G, I). On the other hand, no significant changes were observed in the expression of CsSEP3 (Figure 3H). In the expression profile of floral organs, the expression levels of CsSEP1 and CsSEP2 genes in sepals and petals were higher than those in lips and columns (Figures 3J, K), while CsSEP3 mRNA in sepals, petals, and lips was nearly the same, but higher than those in the column (Figure 3L). Interestingly, the transcripts of the CsSEP4 gene were specifically highly expressed only in the column (Figure 3M). Based on these results, we concluded that CsSEP genes played an indispensable role in floral development, consistent with previous studies, although there were some differences in our findings.
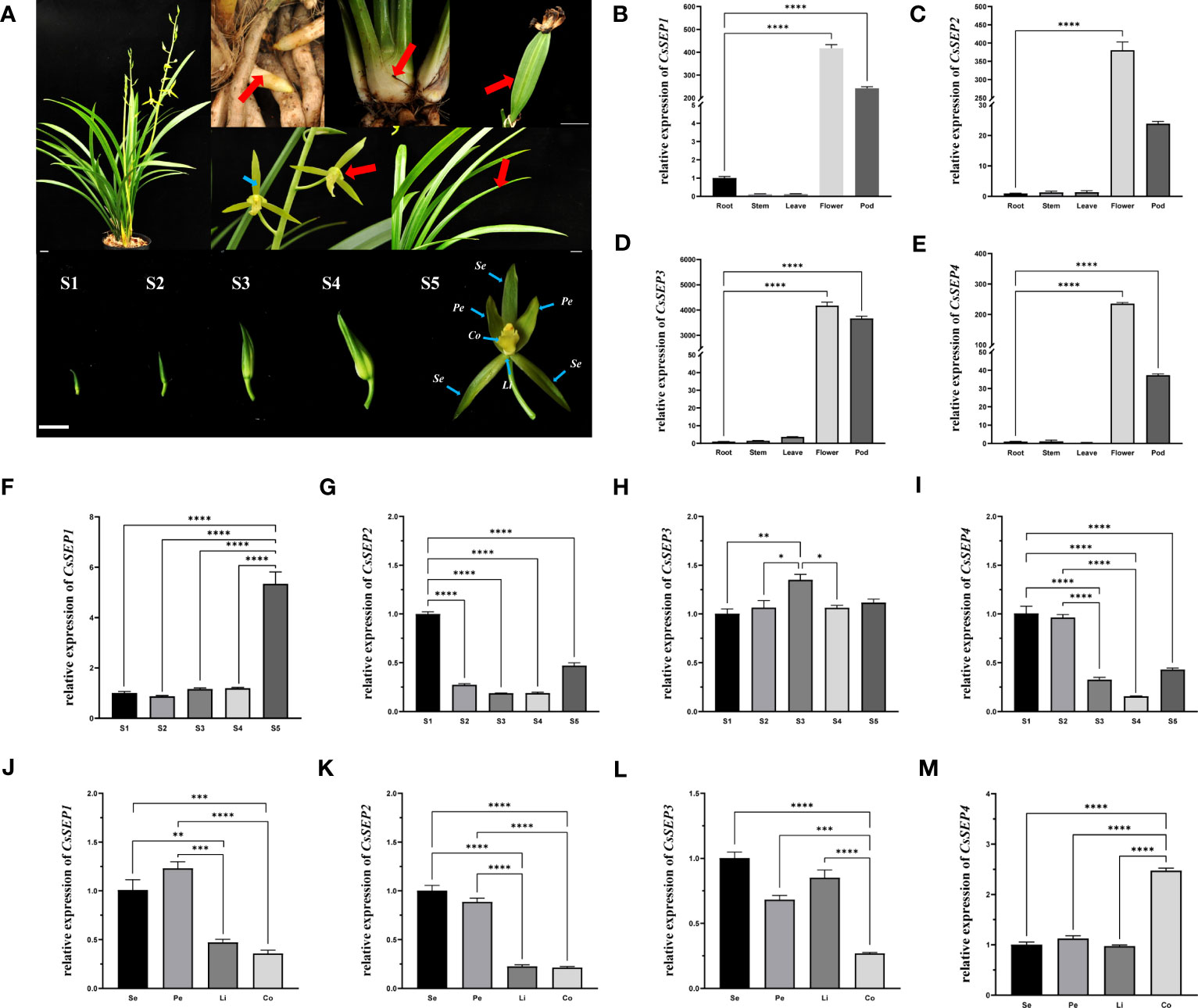
Figure 3 Expression patterns of CsSEP genes in C. sinense. (A) root, stem, leaf, flower, and pod, different stages of flower buds (S1-5) and floral organs in nature C. sinense plants. Se: Sepals, Pe: Petals, Li: Lip, Co: Column. (B–E) Relative expression patterns of CsSEP1/2/3/4 in different tissues and organs were examined by qRT-PCR. (F–I) Relative expression patterns of CsSEP1/2/3/4 at five stages of flower buds. (J–M) Relative expression patterns of CsSEP1/2/3/4 in the floral organ. Scale bars: (A) 1 cm; Error bars (B–M): ± SD. Significant difference was assessed by One-Way ANOVA and indicated by asterisks; (*)p ≤ 0.05, (**) p ≤ 0.01, (***)p ≤ 0.001, (****) p ≤ 0.0001. Data are expressed as the mean of three biological replicates, with error bars indicating the SD values.
To investigate the function of CsSEP proteins, we transformed recombinant vectors expressing fusion proteins PAN580-CsSEP1:GFP, PAN580-CsSEP2:GFP, PAN580-CsSEP3:GFP, PAN580-CsSEP4:GFP, and an empty control vector PAN580-GFP into C. sinense protoplasts, using a highly efficient PTES (Ren et al., 2020) (Figure 4A). Subcellular localization analysis showed that strong green fluorescence from PAN580-CsSEP1:GFP and PAN580-CsSEP4:GFP was observed in the nucleus, external to the chloroplast signal, and colocalized with the DAPI signal (Figure 4B). This finding indicated that CsSEP1 and CsSEP2 were nuclear proteins and consistent with their role as transcription factors (TFs). In contrast, green fluorescence from the fusion proteins PAN580-CsSEP2:GFP and PAN580-CsSEP3:GFP was distributed in both the nucleus and membrane, with the nucleus and chloroplasts being highlighted by blue and spontaneous red light, respectively (Figure 4B). Finally, the green fluorescence from the empty control vector PAN580-GFP was distributed throughout the entire cell.
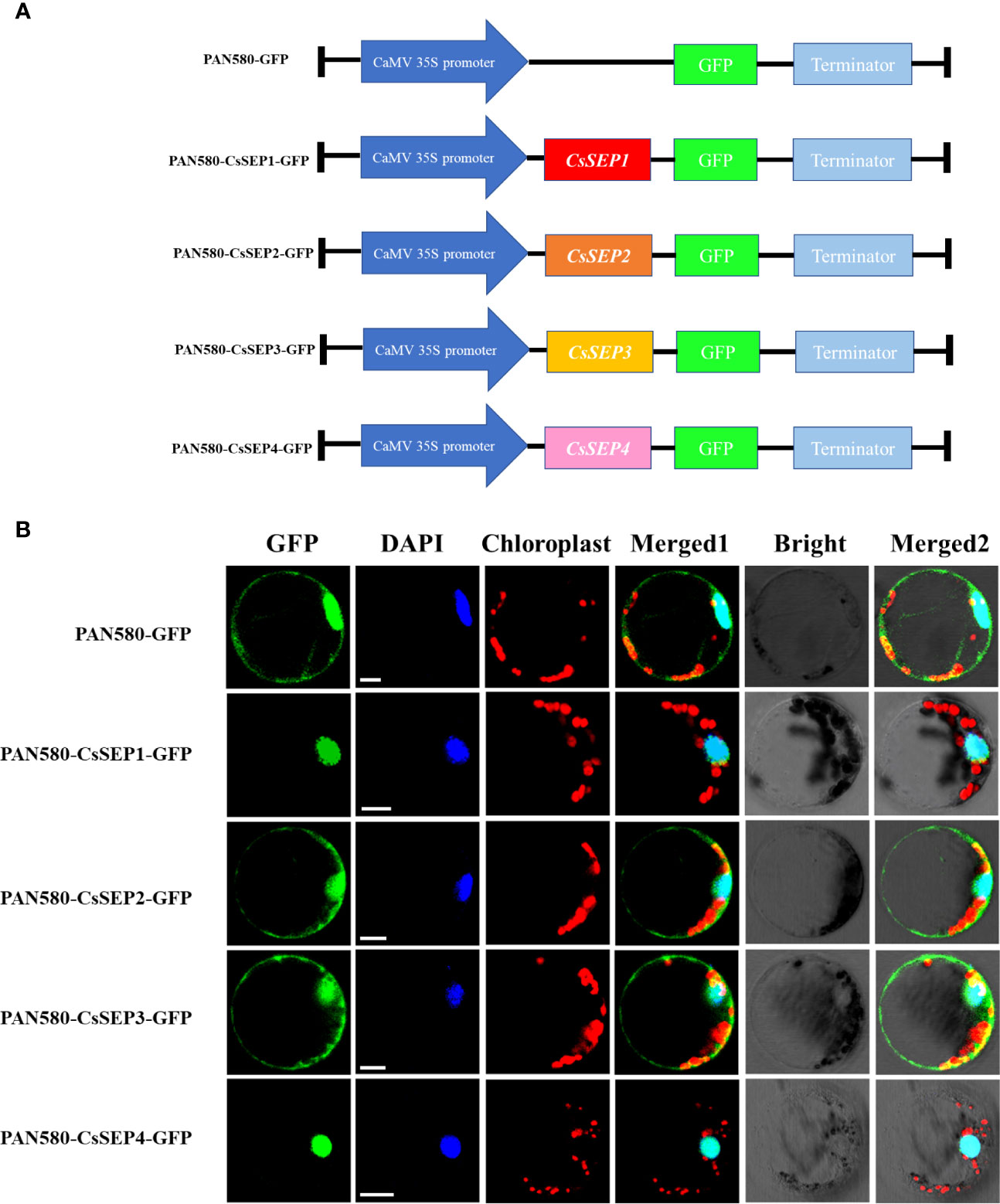
Figure 4 Subcellular localization of CsSEPs in C. sinense protoplasts. (A) Vectors for transient expression were obtained by cloning the full-length CDSs into the vector PAN580-GFP. (B) Transient expression of CsSEP fusion protein and the PAN580-GFP control in protoplasts of C. sinense, and the fluorescences were visualized through LSM 710 confocal laser microscope. GFP: represents gene localization; DAPI: represents nuclear staining; Chloroplast: represents chloroplast self-luminescence; Scale bars: 10 μm.
In order to gain a better understanding of the upstream regulation of CsSEP genes, we obtained the 2,000 bp promoter region upstream of the coding region based on the genome reference sequence. Using the PlantCARE online website, we predicted the cis-elements in the CsSEP promoter region and found that the promoter region contained a large number of core promoter elements. Different types of light response elements (G-box, Box4, GT1-motif, chs-CMA1a, TCT-motif, GA-motif, and TCCC-motif) and HREs (GARE-motif, P-box, TGACG-motif, ABRE, TGA-element, and TCA-element) were widely distributed in the CsSEP promoter (Figure 5). Notably, the promoter region of CsSEP4 contained circadian (circadian control), HD-Zip 1 (differentiation of the palisade mesophyll cells), and CCAAT-box (MYBHv1 binding site) motifs. The defense and stress responsiveness element (TC-rich repeats) and auxin-responsive element (TGA-element) were only found in the promoter region of CsSEP1, while the drought-inducibility element (MBS) was only present in the promoter region of CsSEP2. The meristem expression element (CAT-box) was predicted in the promoter regions of CsSEP1, CsSEP2, and CsSEP4 but not in CsSEP3. Furthermore, numerous MADS-box transcription factor binding sites were uniformly distributed in the CsSEP promoter region predicted from the PlantPan3.0 online website. Specifically, we calculated 16, 22, 12, and 13 MADS-box binding sites in the promoter regions of CsSEP1, CsSEP2, CsSEP3, and CsSEP4, respectively (Supplementary Figure S3).
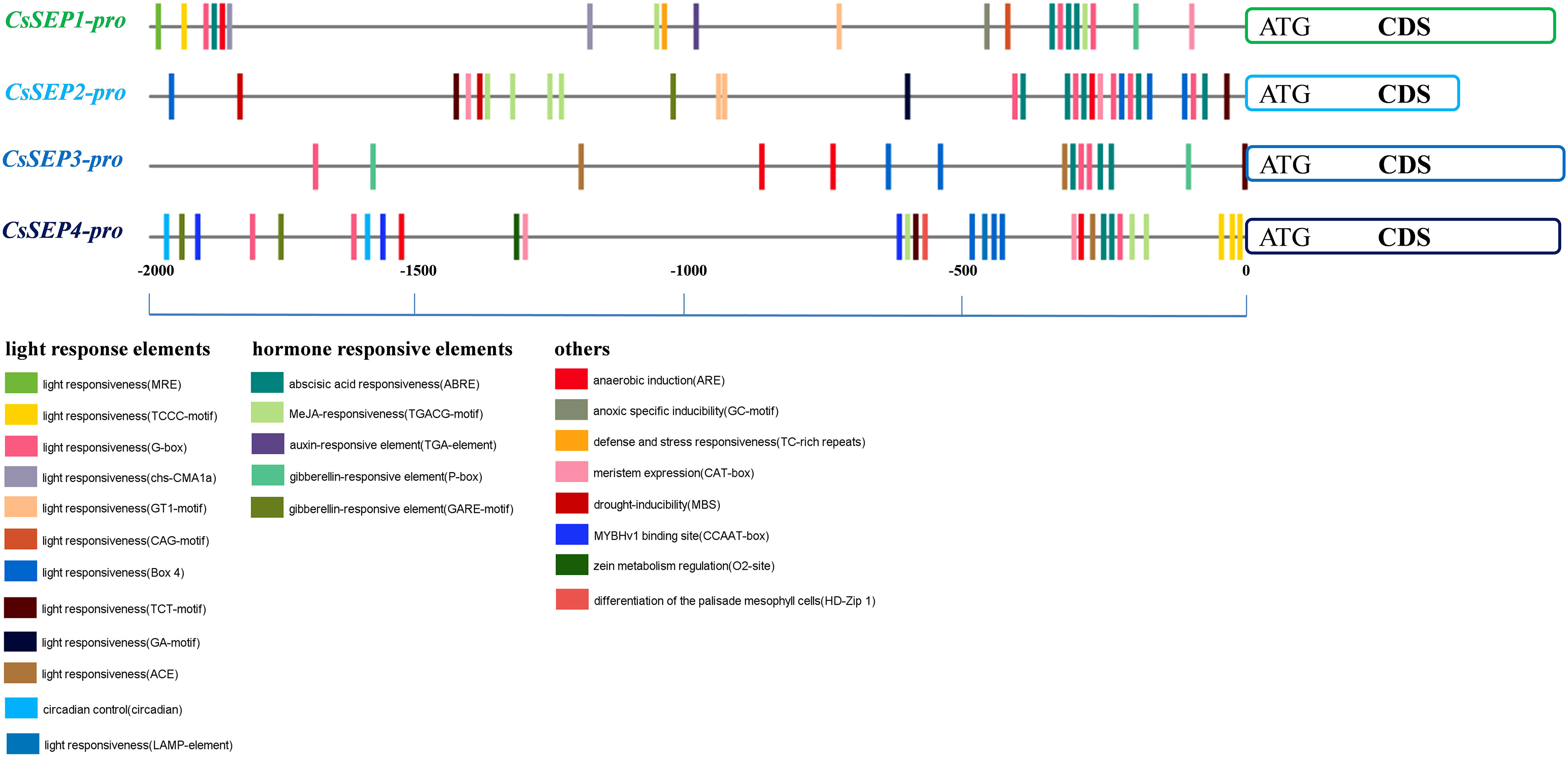
Figure 5 Horizontal lines and short vertical bars of different colors represent promoter regions of CsSEP1, CsSEP2, CsSEP3, and CsSEP4 genes and the position of putative cis-elements, respectively.
To further investigate the function of C. sinense CsSEP genes in flower development, we ectopically expressed CsSEP1, CsSEP2, CsSEP3, and CsSEP4 genes in Arabidopsis using the cauliflower mosaic virus 35S promoter. After screening transgenic plants, we used three independent, stably overexpressing transgenic lines for further analysis. We found that the four CsSEP genes of C. sinense produced early flowering in transgenic A. thaliana, which could be divided into strong phenotype transgenic lines and weak phenotype transgenic lines based on the curling degree of rosette leaves, flowering time, and plant type (Figures 6A–H). The expression levels of the heterologous gene CsSEP1/2/4 in the strong phenotype transgenic lines (35S:CsSEP-S) were higher than those in the weak phenotype transgenic lines (35S:CsSEP-W), as detected by RT-PCR (Figure 6K).

Figure 6 Phenotypic analysis of transgenic Arabidopsis ectopically expressing CsSEP1, CsSEP2, CsSEP3, and CsSEP4 genes. (A) Transgenic lines of 35S:CsSEPs strongly affected the flowering time. (B) Wild-type Col-0. (C-H) Weak and strong phenotypes of 35S:CsSEP2, 35S:CsSEP3, and 35S:CsSEP4 transgenic Arabidopsis thaliana, W: weak phenotype transgenic lines; S: strong phenotype transgenic lines. (I) Distribution of trichomes in wild-type Col-0 and transgenic Arabidopsis thaliana adaxial leaves. (J) Abnormal phenotypes of floral organs in 35S:CsSEPs transgenic Arabidopsis thaliana. (K) RT-PCR analysis of wild-type Col-0 and 35S:CsSEPs transgenic Arabidopsis lines. An ACTIN (ACT) gene fragment was amplified as an internal control. (L) The number of rosette leaves and bolting time were compared between wild-type Col-0 and 35S:CsSEPs transgenic Arabidopsis plants. Scale bars: 1 cm (A); 0.5 mm (I).
All four CsSEPs transgenic A. thaliana showed early flowering under long and short days, the bolting time of Wild-type Col-0 was about 46 days, while the bolting time of 35S:CsSEPs was about 20 to 35 days. Among them, the earliest bolting time were 35S:CsSEP3-W and 35S:CsSEP4-W about 20 days, which were 26 days earlier than wild type Col-0; the next was 35S:CsSEP3-S about 22 days; and the bolting time of 35S:SEP1-S was about 35 days, which only 11 days earlier than Wild type Col-0 (Figure 6L).
Ectopic expression of CsSEPs gene also effected the development of leaves in A. thaliana. The number of rosette leaves of wild type Col-0 at bolting were about 17, while the 35S:SEPs transgenic lines only had 5 to7 rosette leaves (Figure 6L). And the different degrees of curled leaves were found in 35S: CsSEP2, 35S: CsSEP3, and 35S: CsSEP4, the degrees of curled leaves in the strong phenotype transgenic lines were more severe than that of weak phenotype transgenic lines, but curled leaves were undetected in the 35S:CsSEP1 transgenic line (Figures 6A–H). Additionally, curly leaves on strong phenotypic lines of 35S: CsSEP2, 35S: CsSEP3, and 35S: CsSEP4 were exhibited smooth compared with the normal leaves of wild-type Col-0 (Figures 6B–I). Microscopic examination revealed that the distribution density of trichomes on the adaxial epidermis of these leaves was much lower than that of wild-type plants (Figure 6I). Interestingly, the phenotypes of cauline leaves, terminal flowers and smaller plant size were also found in 35S: SEPs transgenic lines compared to the wild type Col-0 (Figures 6A–H).
CsSEP genes not only affected flowering but also played a crucial role in regulating the development of floral organs (Figure 6J). In 35S: CsSEP3 had an abnormal stamen and ovule, and the carpel was dehiscent, rendering it unable to develop normal seeds is similar to Phalaenopsis (Pan et al., 2014), Particularly, 35S: CsSEP4 transgenic Arabidopsis plants was divergent, the petals transitioned into carpel-like structures, and the floral organs failed to develop in the first and second whorls. However, the phenotypes of 35S: CsSEP1 and 35S: CsSEP2 were consistent with those of wild-type plants, showing no phenotypic changes in floral organ development.
We examined the expression profile of endogenous genes involved in floral and leaf development to elucidate the underlying mechanisms of the observed phenotypes resulting from the ectopic expression of CsSEPs in Arabidopsis. Marker genes including AtFT, AtLFY, AtSEP3, AtSOC1, AtAG, AtARF2, AtGRF1, AtGRF2, AtTCP3, and AtTCP20 were chosen for further analysis. Our results (Figure 7) from qRT-PCR assays showed that the expression levels of AtFT, AtLFY, AtSEP3, AtSOC1, and AtAG, which are related to flowering, were significantly upregulated in 35S:CsSEPs transgenic plants. The highest increase in mRNA transcription level was observed for AtSEP3 among these genes, increased by hundreds of times (Figure 7E); the second was AtLFY, AtFT and AtAG, increased dozens of times (Figures 7A, C, D); the expression of AtSOC1 also exhibited significant up-regulation, but not as high as AtFT, AtLFY, AtSEP3 and AtAG (Figure 7B). We also examined leaf development-related genes in 35S:CsSEPs transgenic plants. The transcriptional activation levels of AtTCP3 and AtTCP20 in 35S:CsSEP2/34 transgenic lines with curled leaves phenotype were found an upregulated expression, which were not upregulated in 35S:CsSEP1 transgenic lines without curled leaves phenotype (Supplementary Figures S4A, B). However, for the transcript levels of AtARF2, AtGRF1 and AtGRF2, there were not apparent changed in 35S:CsSEPs transgenic lines (Supplementary Figures S4C-E).
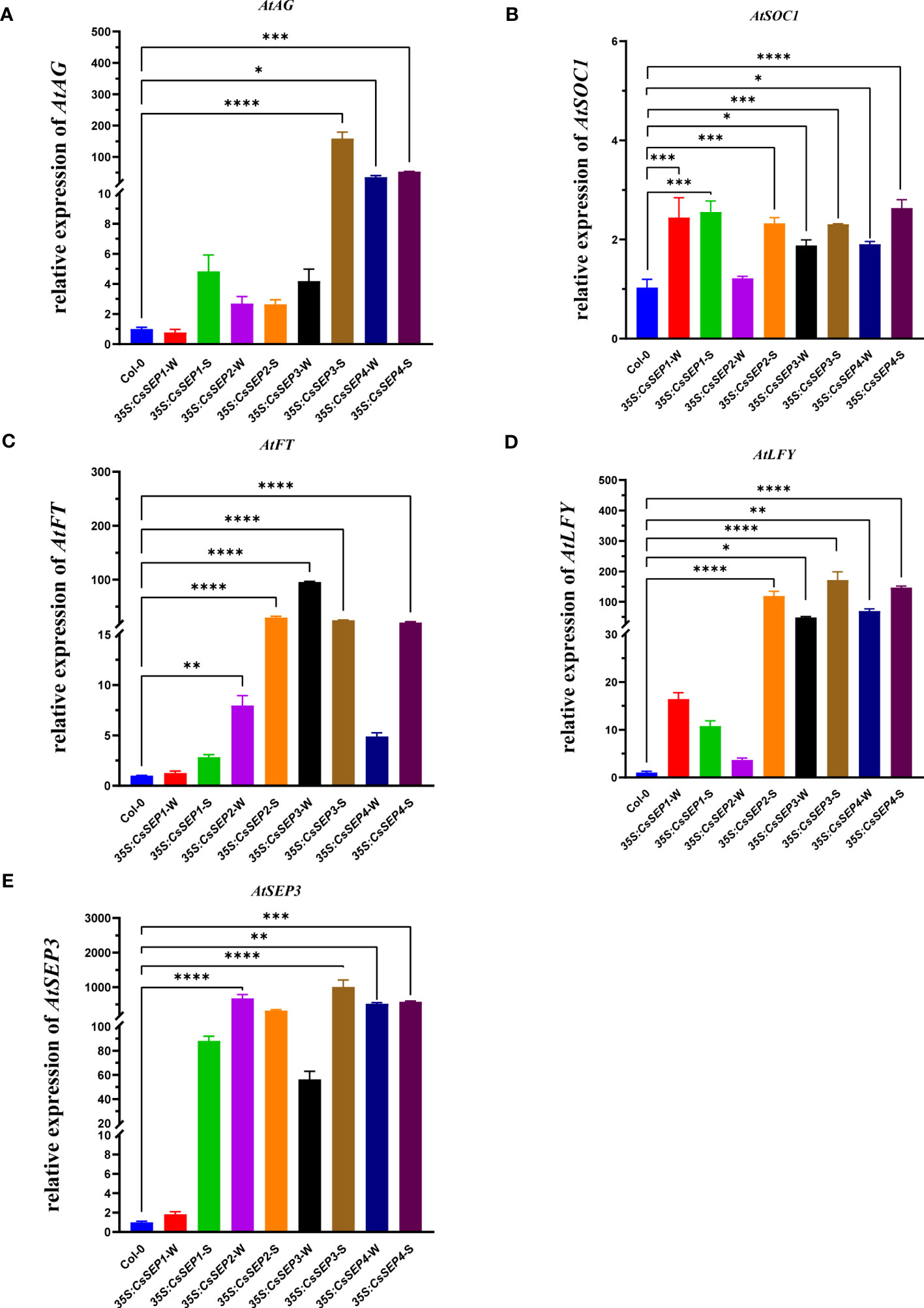
Figure 7 The expressions of (A) AtAG, (B) AtSOC1, (C) AtFT, (D) AtLFY and (E) AtSEP3 endogenous genes related to floral development were examined with qRT-PCR in wild-type Col-0, 35S:CsSEP1, 35S:CsSEP2, 35S:CsSEP3, and 35S:CsSEP4 transgenic Arabidopsis. Significant difference was assessed by One-Way ANOVA and indicated by asterisks; (*)p ≤ 0.05, (**) p ≤ 0.01, (***)p ≤ 0.001, (****) p ≤ 0.0001. Data are expressed as the mean of three biological replicates, with error bars indicating the SD values.
To gain a deeper understanding of the regulatory mechanism of CsSEPs in vivo, we transiently overexpressed CsSEPs genes in C. sinense protoplasts using a highly efficient PTES. We were able to obtain intact and clean protoplasts with an activity of over 95%, which could be used for further transfection (Figure 8A). We successfully transfected the recombinant plasmids into C. sinense protoplasts, as detected by fluorescence microscope (Figure 8B), and examined the expression of flowering-related genes using qRT-PCR. As expected, the expression levels of CsSEP1, CsSEP2, CsSEP3, and CsSEP4 were significantly elevated after transfection (Figure 8C). We also detected the transcriptional levels of endogenous floral development related genes CsSOC1, CsFT, CsLFY, CsAP3-2 and CsAG1 in C. sinense protoplasts and found that the expression of CsSOC1, CsAP3-2 and CsAG1 were significantly upregulated after transfection (Figures 8F–H). Moreover, the other two flowering-related genes also showed a trend of up-regulation (Figures 8D, E). These results suggested that CsSEP genes might regulate floral development by affecting the expression of downstream related genes in C. sinense.
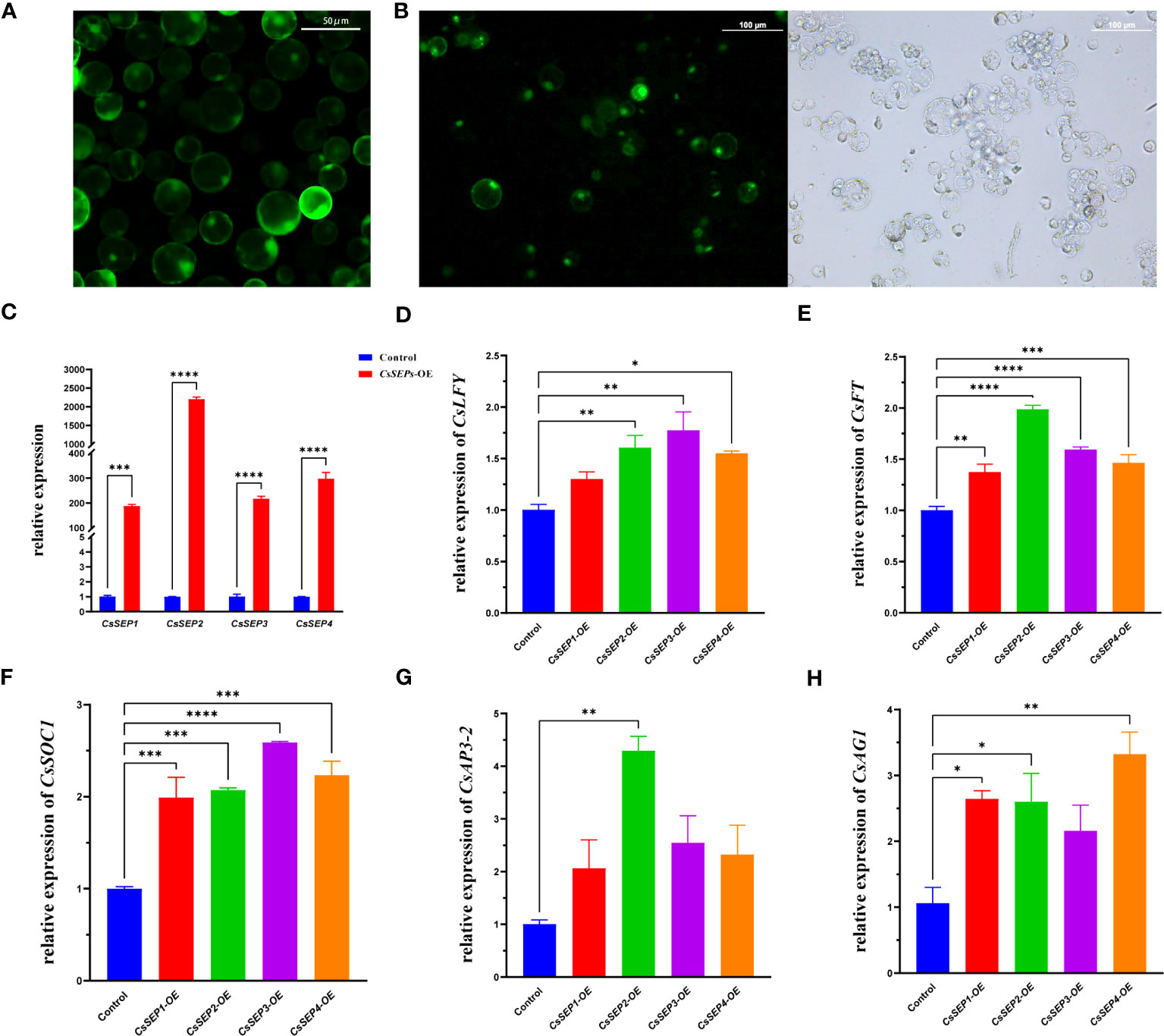
Figure 8 Regulation analysis of CsSEP1, CsSEP2, CsSEP3, and CsSEP4 proteins to express floral development related genes with qRT-PCR. (A) Isolated highly active protoplasts; (B) The transfection efficiency of protoplasts; (C) The expression level of CsSEP genes in C. sinense protoplasts, (D-H) The expressions of CsAP3-2, CsAG1, CsFT, CsSOC1, and CsLFY endogenous flower development genes were examined with qRT-PCR in C. sinense. Significant difference was assessed by One-Way ANOVA and indicated by asterisks; (*)p ≤ 0.05, (**) p ≤ 0.01, (***)p ≤ 0.001, (****) p ≤ 0.0001. Data are expressed as the mean of three biological replicates, with error bars indicating the SD values.
In our study, we observed that overexpression of the CsSEP genes in Arabidopsis resulted in widespread early flowering and floral organ transformation. To analyze the relationship between CsSEP proteins and other MADS-box proteins, we conducted yeast two-hybrid assays (Figure 9). Specifically, we investigated the protein-protein interactions within E-class SEPALLATA-like proteins, B-class protein AP3-2, and MADS-box flowering-related protein SOC1 in C. sinense. Our results indicated that yeast cells cotransformed with recombinant vectors pGBK-CsSEP1/2/3/4+pGAD-CsAP3-2 and pGBK-CsSEP1/2/3/4+pGAD-CsSOC1 yielded blue colonies on QDO/X/A medium, consistent with the positive control. This finding suggested that CsSEP1, CsSEP2, CsSEP3, and CsSEP4 showed interactions with CsAP3-2 and CsSOC1 at the protein level. Moreover, the intensity of interaction among these MADS-box proteins varied. Specifically, CsSEP4 showed the strongest interaction with CsAP3-2 and CsSOC1, while the interaction between CsAP3-2, CsSOC1, and CsSEP2 was also very strong. The intensity of interaction between CsSEP1 and CsAP3-2 and between CsSEP2 and CsSOC1 was strong. However, the interaction ability of the CsSEP3 protein showed only moderate intensity to CsAP3-2 and CsSOC1 proteins.
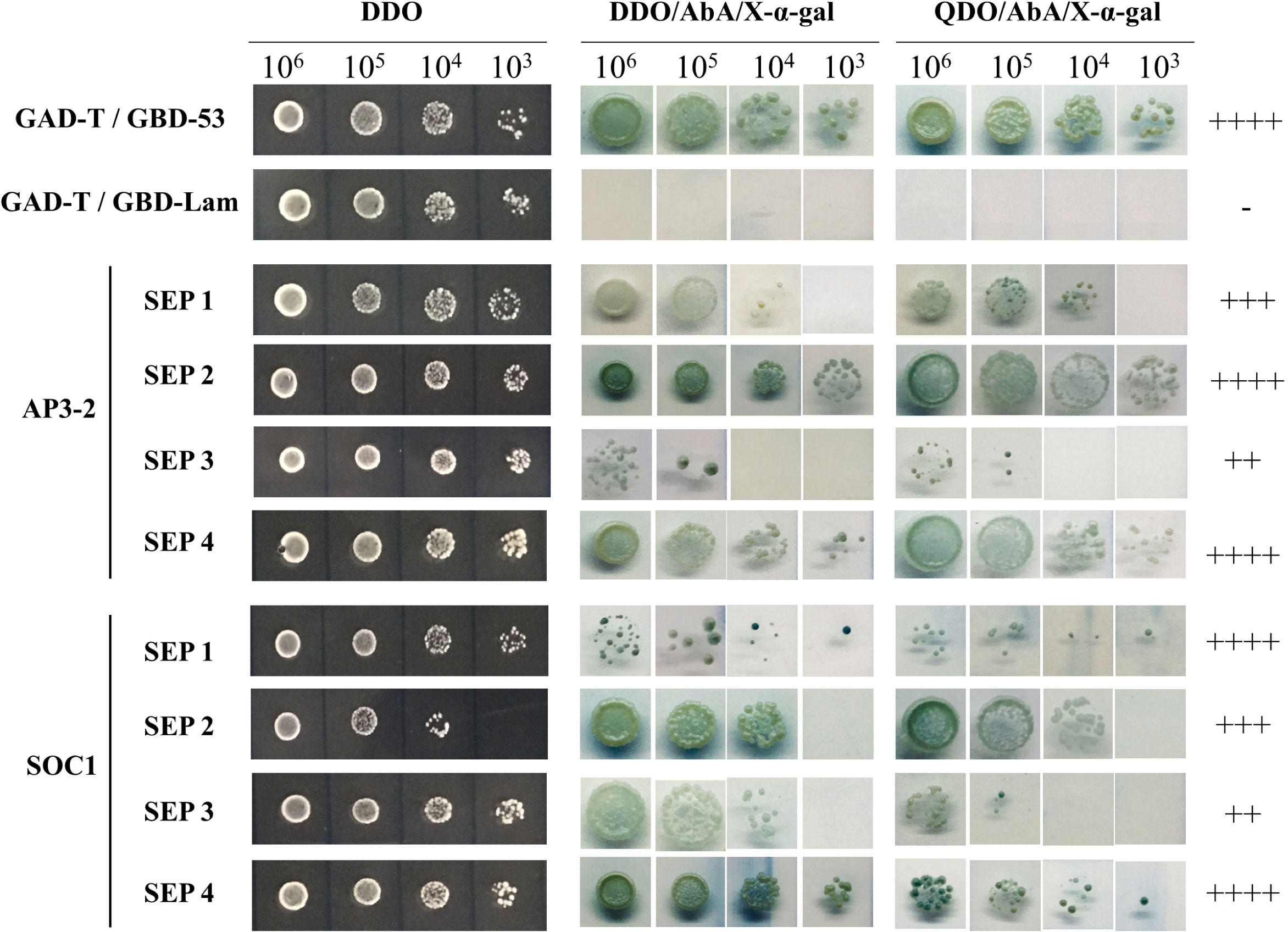
Figure 9 Analysis of protein–protein interactions between CsAP3-2, CsSOC1, and CsSEP1/2/3/4 of C. sinense by the yeast two-hybrid system. ‘++++’, ‘+++’, ‘++’ and ‘+’ are very strong, strong, moderate and weak interactions, respectively, on selective plates. ‘-’, no interaction.
In previous studies of model plants including Arabidopsis (Coen and Meyerowitz, 1991) and Oryza sativa (Cui et al., 2010), four SEP genes exhibit redundant functions in determining floral development and meristem. In this study, we cloned and identified four SEP genes in C. sinense. Phylogenetic analysis revealed that CsSEP2 and CsSEP4 belonged to the SEP1/2/4 clade, forming an Orchid SEP1/2 clade with C. ensifolium, C. goeringii, and other Orchids, while CsSEP1 and CsSEP3 belonged to the SEP3 clade (Figure 1). This finding was consistent with previous reports on the phylogeny of SEP1/3-like genes in P. henryanum and P. equestris. In Orchids, four clades of SEP genes have been generated from orchid-specific duplications within monocot SEP clades (Pan et al., 2014; Ai et al., 2021; Sun et al., 2021; Yang et al., 2021; Cheng et al., 2022). Therefore, we speculated that the endemic Orchid SEP1/2 and Orchid SEP3 branches were formed during plant evolution. The predicted amino acid sequences of CsSEP proteins exhibited a high degree of identity with homologous proteins from P. equestris, C. goeringii, and C. ensifolium. As expected, CsSEP proteins possessed the conserved MIK domain, and the C-terminal domain of CsSEP proteins had a divergent C-terminal domain with the conserved SEP I motif and SEP II motif, supporting their characterization as E-class floral meristem identity genes and suggesting a similar functionality (Figure 2). However, both the SEP I motif and SEP II motif of CsSEP2 were lost together, similar to the SEPALLATA-like gene DcMADS26 identified by Zhang et al. (2016) in D. catenatum (Supplementary Figure S2). This may be due to the loss of SEP genes caused by frequent replication during plant evolution and leading to functional divergence. Hence, the extensive duplication of MADS-box genes and the resulting subfunctional and expressional differentiation were associated with the regulation of species-specific flower traits (Elomaa et al., 2018).
In eudicots, SEPALLATA homologous genes are found to accumulate to high levels in flower buds while being low in shoot tips and undetectable in leaves and roots (Ampomah-Dwamena et al., 2002; Ma et al., 2022; Zhang et al., 2022). In this study, the expression pattern of CsSEP genes in reproductive organs was also found to be conserved. Specifically, the mRNA of CsSEPs showed significant accumulation in flowers and pods, but was only mildly detected or even undetected in roots, stems, and leaves (Figures 3B–E). These results could be verified in other orchids, such as C. goeringii, E. pusilla, and P. henryanum, where the same expression pattern of SEP homologous genes is found to be expressed in floral organs, with low or no expression in vegetative organs (Lin et al., 2016; Dirks-Mulder et al., 2019; Sun et al., 2021; Cheng et al., 2022). However, in the flower bud development process (Figures 3F–I), the expression of CsSEP1 was significantly upregulated in S5. In contrast, the transcripts of CsSEP2 and CsSEP4 were highest in S1. No significant change was observed in the expression of CsSEP3 during the flower bud development stages. These results showed non-overlapping expression profiles temporally. Similarly, the expression levels of SEP homologous genes are found to be very different at different floral bud developmental stages (Xiang et al., 2020). Regarding the expression profile of floral organs (Figures 3J–M), CsSEP1 and CsSEP2 genes in sepals and petals were expressed more highly than those in lips and columns. The CsSEP4 gene was specifically highly expressed in the column, implying that it played a role in determining the column. In contrast, in Phalaenopsis, PeSEP2 is highly expressed in the column, PeSEP3 expression is dominant in the petal, and PeSEP4 expression is extremely low in floral organs (Pan et al., 2014). SEP3 homologous and SEP4 homologous genes are mainly highly expressed in the outer two whorls of marigold and gerbera (Zhang T. et al., 2017; Zhang et al., 2021). The non-ubiquitous expression patterns of SEPALLATA-like genes offered a rare opportunity to investigate their functional conservation and divergence in C. sinense.
In both model and non-model plants, including Arabidopsis, rice, tomato, Gerbera and Prunus, SEP-like genes are well-conserved floral organ identity genes and key regulators of flower development (Ferrario et al., 2003; Teo et al., 2014; Zhang T. et al., 2017; Zhou et al., 2017). Overexpression of SEPALLATA-like genes in Arabidopsis, which is commonly observed, leads to early flowering (Ditta et al., 2004), and this phenomenon is well-conserved in distant angiosperm species. In this study, the four CsSEP genes of C. sinense produced early flowering in transgenic A. thaliana (Figure 6A). However, additional phenotypes including smooth curled leaves, reduced plant size and terminal flowers (Figures 6B–H) were also observed. However, only one or more SEP homologous in other species can cause early flowering phenotype, such as in Phalaenopsis, only PeSEP3 gene can lead to early flowering phenotype in Arabidopsis (Pan et al., 2014), differently, four CsSEP genes all showed early flowering in transgenic Arabidopsis, indicating that the functions of SEPs genes are not only conserved but also divergent in C. sinense.
Considering that the SEP homologous genes in different angiosperms are diverse, leading to the homeotic conversion of floral organs and even affecting fruit development (Zhang S. et al., 2017; Qi et al., 2020; Zhang et al., 2021; Zhang et al., 2022). For example, SEP homologous genes in Isatis indigotica, Marigold, and Gerbera can affect the structure of sepals, petals, stamens and carpels (Uimari et al., 2004; Ma et al., 2019; Zhang et al., 2021). Additionally, SEP homologous genes can effect fruit and seed also have reported in grapevine (Zhang et al., 2022). It is reasonable that these homologous genes showed separation of phenotypic traits. In orchids, C. ensifolium CeSEP2 gene can affect floral organ development (Ai et al., 2021), and the function of Phalaenopsis PeSEP3 gene affecting the floral organ structure is consistent with the CsSEP3 gene in this study (Pan et al., 2014), which exhibited an abnormal stamen and ovule, with the carpel being dehiscent and unable to develop normal seeds (Figure 6J). These conclusions further reveal that the effection of SEP gene on floral organs was conserved. Conversely, the effection of four CsSEPs genes on floral organs were divergent from each other in C. sinense. In this study, the overexpression of CsSEP3 and CsSEP4 caused morphological changes of floral organs, however, the 35S:CsSEP1 and 35S:CsSEP2 showed no phenotypic changes in floral organ development (Figure 6J). In particular, 35S:CsSEP4 transgenic plants resulted in the transformation of petals into carpel-like structures, with floral organs failing to develop in the first and second whorls, which has not been reported before. In species that have undergone recent evolution, the impact of SEP homologs on flower organs appears to be divergent in function. While the relationship between the four SEP genes remains unclear, several studies have suggested that the functions of SEP1 and SEP2 are more similar to those of SEP4. On the other hand, other reports have supported that the function of SEP3 is more similar to that of SEP1 and SEP2 (Zhang et al., 2021).
The phenotypic changes in transgenic plants were verified to be related to the expression levels of their endogenous genes regulated by SEP genes. For instance, in marigold and Platanus acerifolia, ectopic expression of SEP homologous genes regulates the expression of endogenous genes in A. thaliana (Zhang S. et al., 2017; Zhang et al., 2021). Consistent with those conclusions, the expression levels of endogenous floral development genes, such as AtFT, AtLFY, AtSEP3, AtSOC1 and AtAG, were significantly upregulated in 35S: CsSEP transgenic plants (Figure 7), and the phenotypic differences might be due to the different capabilities of activating downstream target genes in Arabidopsis. In Arabidopsis, FT and SOC1 were the main flowering signal pathway integrators. SOC1 can, together with AGL24, directly activate LFY (Parcy, 2005; Lee and Lee, 2010). These homologous genes in orchids also play a conserved role in influencing flowering. For instance, overexpression of DOFT in Dendrobium results in increased transcript levels of DOSOC1 (Wang et al., 2017). Moreover, when the CsSEP genes were transiently overexpressed in C. sinense protoplasts, the expression levels of CsSOC1, CsFT, CsLFY, CsAP3-2 and CsAG1 were shown to have an upregulated trend (Figures 8D–H), respectively, which was consistent with the promotive effect of CsSEPs on endogenous floral development genes in Arabidopsis. These results indicated that activation of floral development genes by SEPALLATA-like genes was well-conserved in C. sinense.
SEP proteins act as “glue,” facilitating interactions within MADS proteins and driving the formation of distinct tetrameric complexes (Honma and Goto, 2001; Ditta et al., 2004; Zahn et al., 2005; Immink et al., 2009). In Arabidopsis, B-class genes are direct targets of SEP proteins, and SEP3 displays extensive interactions with other MADS-box proteins (de Folter et al., 2005). Similarly, in orchids, SEP-like proteins bind to CArG-box sequences and interact with AP1/FUL, B, C, D, and other MADS-box proteins during flower development (Tilly et al., 1998; Pan et al., 2014; Mitoma and Kanno, 2018; Cheng et al., 2022). In the evolution of the orchid family, B-class MADS-box genes is diverged into different subclades, and involved with E-class SEP genes in the development of a highly modified perianth (Pan et al., 2011; Lucibelli et al., 2021). For example, the loss of class B and E genes leads to the adaxial petal not differentiating into a specialized lip in Apostasia shenzhenica (Zhang G. Q. et al., 2017); in Phalaenopsis, PeSEP1 gene plays a role in the control of floral morphogenesis directly or indirectly via interact with two AP3 homologous genes PeMADS2 and PeMADS4 (Pan et al., 2014). Similarly in C. sinense, E-class genes were still conserved interact with B-class gene, that was consistent with Phalaenopsis. However, different from Phalaenopsis, the CsSEP1, CsSEP2, CsSEP3 and CsSEP4 proteins all had different degrees of interaction with only one B-class CsAP3-2 protein (Figure 9), this finding further supported the conservative and divergent role of class B and E proteins in regulating floral development in C. sinense. Furthermore, protein interaction analysis also detected that CsSEP1, CsSEP2, CsSEP3, and CsSEP4 interacted with CsSOC1. Previous studies only confirmed that SEP genes can interact with ABCDE-class genes and have not reported interactions with SOC1. This new discovery shows that the CsSEPs genes had produce functional divergence and may recruit CsSOC1 protein to regulate the flowering transition in C. sinense.
In this study, we isolated and identified four SEPALLATA-like genes from C. sinense and conducted a systematic analysis of their phylogenetic relationships, protein sequences, and expression patterns. Our findings revealed that CsSEP genes were both conserved and divergent in floral initiation and development. Based on our analysis of transgenic phenotypes in Arabidopsis, expression profiles of endogenous genes, and protein interaction patterns, we suggested that CsSEPs genes played an important role in regulating flowering time and floral organ development in C. sinense. The results of our study on the early flowering phenotype of transgenic plants overexpressing CsSEP genes demonstrated their conservative functions in regulating flowering transition. However, the regulation of floral organ development by CsSEPs was divergent, suggesting that CsSEP3 and CsSEP4 could be potential candidate genes for further research. Furthermore, our study on the different interaction patterns of CsSEP proteins with MADS-box genes shed new light on the conservative and divergent functions of these proteins. We predicted that CsSEPs might form a protein complex with B-class and CsSOC1 proteins to affect downstream genes and regulate floral development. Although the study on the mechanism of orchid flower development is still in its preliminary stage, our findings on the role of SEPALLATA genes in regulating floral development in C. sinense will lay a foundation for further study of the molecular network.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
F-XY, G-FZ, and F-LW designed the experiments and edited the manuscript, Z-YL, C-QL, and JG executed the experiments and assembled the figures, Y-LW, J-PJ, JL and QX conducted the qRT-PCR, F-XY, F-LW, and Z-YL wrote the paper with inputs from other authors. All authors read and approved the final manuscript.
This research was funded by grants from the National Natural Science Foundation of China (31872151, 31672184), the Natural Science Foundation of Guangdong province (2017A030312004), Guangzhou Science and Technology Project (2022B03J703), Innovation Team of Modern Agricultural Industry Technology System in Guangdong Province (2023KJ121), the Guangdong Academy of Agricultural Sciences Discipline Team Construction Project (202127TD, R2020PY-JX018), Yunfu scientific research project (2021020606), Guangzhou Science and Technology Project (2023B03J1322) and Guangzhou scientific research Project (202102020809).
We are thankful to funding agencies for funding and Plant Editors for providing professional services for language editing and the final polishing of our manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1209834/full#supplementary-material
Ahmad, S., Chen, J., Chen, G., Huang, J., Hao, Y., Shi, X., et al. (2022). Transcriptional proposition for uniquely developed protocorm flowering in three orchid species: resources for innovative breeding. Front. Plant Sci. 13, 942591. doi: 10.3389/fpls.2022.942591
Ai, Y., Li, Z., Sun, W.-H., Chen, J., Zhang, D., Ma, L., et al. (2021). The Cymbidium genome reveals the evolution of unique morphological traits. Horticulture Res. 8 (1), 255. doi: 10.1038/s41438-021-00683-z
Ampomah-Dwamena, C., Morris, B. A., Sutherland, P., Veit, B., Yao, J. L. (2002). Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol. 130 (2), 605–617. doi: 10.1104/pp.005223
Castillejo, C., Romera-Branchat, M., Pelaz, S. (2005). A new role of the Arabidopsis SEPALLATA3 gene revealed by its constitutive expression. Plant J. 43 (4), 586–596. doi: 10.1111/j.1365-313X.2005.02476.x
Cheng, H., Xie, X., Ren, M., Yang, S., Zhao, X., Mahna, N., et al. (2022). Characterization of three SEPALLATA-like MADS-box genes associated with floral development in paphiopedilum henryanum (Orchidaceae). Front. Plant Sci. 13, 916081. doi: 10.3389/fpls.2022.916081
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 (6), 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Coen, E. S., Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353 (6339), 31–37. doi: 10.1038/353031a0
Cui, R., Han, J., Zhao, S., Su, K., Wu, F., Du, X., et al. (2010). Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J. 61 (5), 767–781. doi: 10.1111/j.1365-313X.2009.04101.x
de Folter, S., Immink, R. G., Kieffer, M., Parenicová, L., Henz, S. R., Weigel, D., et al. (2005). Comprehensive interaction map of the Arabidopsis MADS Box transcription factors. Plant Cell 17 (5), 1424–1433. doi: 10.1105/tpc.105.031831
Dirks-Mulder, A., Ahmed, I., Uit Het Broek, M., Krol, L., Menger, N., Snier, J., et al. (2019). Morphological and molecular characterization of orchid fruit development. Front. Plant Sci. 10, 137. doi: 10.3389/fpls.2019.00137
Ditta, G., Pinyopich, A., Robles, P., Pelaz, S., Yanofsky, M. F. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14 (21), 1935–1940. doi: 10.1016/j.cub.2004.10.028
Elomaa, P., Zhao, Y., Zhang, T. (2018). Flower heads in Asteraceae-recruitment of conserved developmental regulators to control the flower-like inflorescence architecture. Hortic. Res. 5, 36. doi: 10.1038/s41438-018-0056-8
Fan, H. Y., Hu, Y., Tudor, M., Ma, H. (1997). Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant J. 12 (5), 999–1010. doi: 10.1046/j.1365-313x.1997.12050999.x
Ferrario, S., Immink, R. G., Shchennikova, A., Busscher-Lange, J., Angenent, G. C. (2003). The MADS box gene FBP2 is required for SEPALLATA function in petunia. Plant Cell 15 (4), 914–925. doi: 10.1105/tpc.010280
Honma, T., Goto, K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409 (6819), 525–529. doi: 10.1038/35054083
Huang, W., Fang, Z., Zeng, S., Zhang, J., Wu, K., Chen, Z., et al. (2012). Molecular cloning and functional analysis of Three FLOWERING LOCUS T (FT) homologous genes from Chinese Cymbidium. Int. J. Mol. Sci. 13 (9), 11385–11398. doi: 10.3390/ijms130911385
Hugouvieux, V., Zubieta, C. (2018). MADS transcription factors cooperate: complexities of complex formation. J. Exp. Bot. 69 (8), 1821–1823. doi: 10.1093/jxb/ery099
Immink, R. G., Tonaco, I. A., de Folter, S., Shchennikova, A., van Dijk, A. D., Busscher-Lange, J., et al. (2009). SEPALLATA3: the 'glue' for MADS box transcription factor complex formation. Genome Biol. 10 (2), R24. doi: 10.1186/gb-2009-10-2-r24
Ireland, H. S., Yao, J. L., Tomes, S., Sutherland, P. W., Nieuwenhuizen, N., Gunaseelan, K., et al. (2013). Apple SEPALLATA1/2-like genes control fruit flesh development and ripening. Plant J. 73 (6), 1044–1056. doi: 10.1111/tpj.12094
Jeon, J.-S., Lee, S., Jung, K.-H., Yang, W.-S., Yi, G.-H., Oh, B.-G., et al. (2000). Production of transgenic rice plants showing reduced heading date and plant height by ectopic expression of rice MADS-box genes. Mol. Breed. 6 (6), 581–592. doi: 10.1023/A:1011388620872
Kaufmann, K., Muino, J. M., Jauregui, R., Airoldi, C. A., Smaczniak, C., Krajewski, P., et al. (2009). Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PloS Biol. 7 (4), e1000090. doi: 10.1371/journal.pbio.1000090
Kotilainen, M., Elomaa, P., Uimari, A., Albert, V. A., Yu, D., Teeri, T. H. (2000). GRCD1, an AGL2-like MADS box gene, participates in the C function during stamen development in Gerbera hybrida. Plant Cell 12 (10), 1893–1902. doi: 10.1105/tpc.12.10.1893
Kumar, S., Tamura, K., Nei, M. (2004). MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 5 (2), 150–163. doi: 10.1093/bib/5.2.150
Lee, J., Lee, I. (2010). Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61 (9), 2247–2254. doi: 10.1093/jxb/erq098
Li, J., Li, F., Qian, M., Han, M., Liu, H., Zhang, D., et al. (2017). Characteristics and regulatory pathway of the PrupeSEP1 SEPALLATA gene during ripening and softening in peach fruits. Plant Sci. 257, 63–73. doi: 10.1016/j.plantsci.2017.01.004
Li, Y., Zhang, B., Yu, H. (2022). Molecular genetic insights into orchid reproductive development. J. Exp. Bot. 73 (7), 1841–1852. doi: 10.1093/jxb/erac016
Lid, S. E., Meeley, R. B., Min, Z., Nichols, S., Olsen, O.-A. (2004). Knock-out mutants of two members of the AGL2 subfamily of MADS-box genes expressed during maize kernel development. Plant Sci. 167 (3), 575–582. doi: 10.1016/j.plantsci.2004.04.031
Lin, C. S., Hsu, C. T., Liao, D. C., Chang, W. J., Chou, M. L., Huang, Y. T., et al. (2016). Transcriptome-wide analysis of the MADS-box gene family in the orchid Erycina pusilla. Plant Biotechnol. J. 14 (1), 284–298. doi: 10.1111/pbi.12383
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Lu, Z. X., Wu, M., Loh, C. S., Yeong, C. Y., Goh, C. J. (1993). Nucleotide sequence of a flower-specific MADS box cDNA clone from orchid. Plant Mol. Biol. 23 (4), 901–904. doi: 10.1007/bf00021545
Lucibelli, F., Valoroso, M. C., Theissen, G., Nolden, S., Mondragon-Palomino, M., Aceto, S. (2021). Extending the toolkit for beauty: differential co-expression of DROOPING LEAF-like and class B MADS-box genes during Phalaenopsis flower development. Int. J. Mol. Sci. 22 (13):7025. doi: 10.3390/ijms22137025
Ma, Y. Q., Pu, Z. Q., Tan, X. M., Meng, Q., Zhang, K. L., Yang, L., et al. (2022). SEPALLATA–like genes of Isatis indigotica can affect the architecture of the inflorescences and the development of the floral organs. PeerJ 10, e13034. doi: 10.7717/peerj.13034
Ma, Y. Q., Pu, Z. Q., Zhang, L., Lu, M. X., Zhu, Y., Hao, C. Y., et al. (2019). A SEPALLATA1-like gene of Isatis indigotica Fort. regulates flowering time and specifies floral organs. Gene 713, 143974. doi: 10.1016/j.gene.2019.143974
Malcomber, S. T., Kellogg, E. A. (2005). SEPALLATA gene diversification: brave new whorls. Trends Plant Sci. 10 (9), 427–435. doi: 10.1016/j.tplants.2005.07.008
Matsubara, K., Shimamura, K., Kodama, H., Kokubun, H., Watanabe, H., Basualdo, I. L., et al. (2008). Green corolla segments in a wild Petunia species caused by a mutation in FBP2, a SEPALLATA-like MADS box gene. Planta 228 (3), 401–409. doi: 10.1007/s00425-008-0744-y
Mitoma, M., Kanno, A. (2018). The greenish flower phenotype of Habenaria radiata (Orchidaceae) is caused by a mutation in the SEPALLATA-like MADS-box gene HrSEP-1. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00831
Motomura, H., Selosse, M. A., Martos, F., Kagawa, A., Yukawa, T. (2010). Mycoheterotrophy evolved from mixotrophic ancestors: evidence in Cymbidium (Orchidaceae). Ann. Bot. 106 (4), 573–581. doi: 10.1093/aob/mcq156
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32 (1), 268–274. doi: 10.1093/molbev/msu300
Pajoro, A., Madrigal, P., Muiño, J. M., Matus, J. T., Jin, J., Mecchia, M. A., et al. (2014). Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 15 (3), R41. doi: 10.1186/gb-2014-15-3-r41
Pan, Z. J., Chen, Y. Y., Du, J. S., Chen, Y. Y., Chung, M. C., Tsai, W. C., et al. (2014). Flower development of Phalaenopsis orchid involves functionally divergent SEPALLATA-like genes. New Phytol. 202 (3), 1024–1042. doi: 10.1111/nph.12723
Pan, Z. J., Cheng, C. C., Tsai, W. C., Chung, M. C., Chen, W. H., Hu, J. M., et al. (2011). The duplicated B-class MADS-box genes display dualistic characters in orchid floral organ identity and growth. Plant Cell Physiol. 52 (9), 1515–1531. doi: 10.1093/pcp/pcr092
Parcy, F. (2005). Flowering: a time for integration. Int. J. Dev. Biol. 49 (5-6), 585–593. doi: 10.1387/ijdb.041930fp
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405 (6783), 200–203. doi: 10.1038/35012103
Pelaz, S., Gustafson-Brown, C., Kohalmi, S. E., Crosby, W. L., Yanofsky, M. F. (2001). APETALA1 and SEPALLATA3 interact to promote flower development. Plant J. 26 (4), 385–394. doi: 10.1046/j.1365-313x.2001.2641042.x
Qi, X., Liu, C., Song, L., Li, M. (2020). PaMADS7, a MADS-box transcription factor, regulates sweet cherry fruit ripening and softening. Plant Sci. 301, 110634. doi: 10.1016/j.plantsci.2020.110634
Ren, R., Gao, J., Lu, C., Wei, Y., Jin, J., Wong, S. M., et al. (2020). Highly efficient protoplast isolation and transient expression system for functional characterization of flowering related genes in Cymbidium orchids. Int. J. Mol. Sci. 21 (7). doi: 10.3390/ijms21072264
Roberts, D. L., Dixon, K. W. (2008). Orchids. Curr. Biol. 18 (8), R325–R329. doi: 10.1016/j.cub.2008.02.026
Robles, P., Pelaz, S. (2005). Flower and fruit development in Arabidopsis thaliana. Int. J. Dev. Biol. 49 (5-6), 633–643. doi: 10.1387/ijdb.052020pr
Ruokolainen, S., Ng, Y. P., Albert, V. A., Elomaa, P., Teeri, T. H. (2010). Large scale interaction analysis predicts that the Gerbera hybrida floral E function is provided both by general and specialized proteins. BMC Plant Biol. 10, 129. doi: 10.1186/1471-2229-10-129
Salemme, M., Sica, M., Gaudio, L., Aceto, S. (2013). The OitaAG and OitaSTK genes of the orchid Orchis italica: a comparative analysis with other C- and D-class MADS-box genes. Mol. Biol. Rep. 40 (5), 3523–3535. doi: 10.1007/s11033-012-2426-x
Shan, H., Zahn, L., Guindon, S., Wall, P. K., Kong, H., Ma, H., et al. (2009). Evolution of plant MADS box transcription factors: evidence for shifts in selection associated with early angiosperm diversification and concerted gene duplications. Mol. Biol. Evol. 26 (10), 2229–2244. doi: 10.1093/molbev/msp129
Shen, C.-Y., Chen, Y.-Y., Liu, K.-W., Lu, H.-C., Chang, S.-B., Hsiao, Y.-Y., et al. (2021). Orchid Bsister gene PeMADS28 displays conserved function in ovule integument development. Sci. Rep. 11 (1). doi: 10.1038/s41598-020-79877-9
Subramanian, B., Gao, S., Lercher, M. J., Hu, S., Chen, W. H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47 (W1), W270–w275. doi: 10.1093/nar/gkz357
Sun, Y., Chen, G.-Z., Huang, J., Liu, D.-K., Xue, F., Chen, X.-L., et al. (2021). The Cymbidium goeringii genome provides insight into organ development and adaptive evolution in orchids. Ornamental Plant Res. 1 (1), 1–13. doi: 10.48130/opr-2021-0010
Teo, Z. W., Song, S., Wang, Y. Q., Liu, J., Yu, H. (2014). New insights into the regulation of inflorescence architecture. Trends Plant Sci. 19 (3), 158–165. doi: 10.1016/j.tplants.2013.11.001
Theissen, G. (2001). Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 4 (1), 75–85. doi: 10.1016/s1369-5266(00)00139-4
Theissen, G., Saedler, H. (2001). Plant biology. Floral quartets. Nature 409 (6819), 469–471. doi: 10.1038/35054172
Tilly, J. J., Allen, D. W., Jack, T. (1998). The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development 125 (9), 1647–1657. doi: 10.1242/dev.125.9.1647
Tsai, W. C., Chen, H. H. (2006). The orchid MADS-box genes controlling floral morphogenesis. Sci. World J. 6, 1933–1944. doi: 10.1100/tsw.2006.321
Tzeng, T. Y., Hsiao, C. C., Chi, P. J., Yang, C. H. (2003). Two lily SEPALLATA-like genes cause different effects on floral formation and floral transition in Arabidopsis. Plant Physiol. 133 (3), 1091–1101. doi: 10.1104/pp.103.026997
Uimari, A., Kotilainen, M., Elomaa, P., Yu, D., Albert, V. A., Teeri, T. H. (2004). Integration of reproductive meristem fates by a SEPALLATA-like MADS-box gene. Proc. Natl. Acad. Sci. U.S.A. 101 (44), 15817–15822. doi: 10.1073/pnas.0406844101
Valoroso, M. C., Censullo, M. C., Aceto, S. (2019). The MADS-box genes expressed in the inflorescence of Orchis italica (Orchidaceae). PloS One 14 (3), e0213185. doi: 10.1371/journal.pone.0213185
Vandenbussche, M., Zethof, J., Souer, E., Koes, R., Tornielli, G. B., Pezzotti, M., et al. (2003). Toward the analysis of the petunia MADS box gene family by reverse and forward transposon insertion mutagenesis approaches: B, C, and D floral organ identity functions require SEPALLATA-like MADS box genes in petunia. Plant Cell 15 (11), 2680–2693. doi: 10.1105/tpc.017376
Wang, X., Gao, D., Sun, J., Liu, M., Lun, Y., Zheng, J., et al. (2016). An exon skipping in a SEPALLATA-Like gene is associated with perturbed floral and fruits development in cucumber. J. Integr. Plant Biol. 58 (9), 766–771. doi: 10.1111/jipb.12472
Wang, Y., Liu, L., Song, S., Li, Y., Shen, L., Yu, H. (2017). DOFT and DOFTIP1 affect reproductive development in the orchid Dendrobium Chao Praya Smile. J. Exp. Bot. 68 (21-22), 5759–5772. doi: 10.1093/jxb/erx400
Wei, Y., Jin, J., Yao, X., Lu, C., Zhu, G., Yang, F. (2020). Transcriptome Analysis Reveals Clues into leaf-like flower mutant in Chinese orchid Cymbidium ensifolium. Plant Divers. 42 (2), 92–101. doi: 10.1016/j.pld.2019.12.001
Xiang, Y., Huang, Y., He, H., Xu, Q. (2020). Phylogenetic and expression analysis of SEPALLATA-like gene in Brassica oleracea L. var. acephala. Sheng Wu Gong Cheng Xue Bao 36 (11), 2398–2412. doi: 10.13345/j.cjb.200422
Xu, Y., Teo, L. L., Zhou, J., Kumar, P. P., Yu, H. (2006). Floral organ identity genes in the orchid Dendrobium crumenatum. Plant J. 46 (1), 54–68. doi: 10.1111/j.1365-313X.2006.02669.x
Yang, F. X., Gao, J., Wei, Y. L., Ren, R., Zhang, G. Q., Lu, C. Q., et al. (2021). The genome of Cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol. J. 19 (12), 2501–2516. doi: 10.1111/pbi.13676
Yang, F., Zhu, G., Wei, Y., Gao, J., Liang, G., Peng, L., et al. (2019). Low-temperature-induced changes in the transcriptome reveal a major role of CgSVP genes in regulating flowering of Cymbidium goeringii. BMC Genomics 20 (1), 53. doi: 10.1186/s12864-019-5425-7
Yu, H., Goh, C. J. (2000). Identification and characterization of three orchid MADS-box genes of the AP1/AGL9 subfamily during floral transition. Plant Physiol. 123 (4), 1325–1336. doi: 10.1104/pp.123.4.1325
Yu, H., Yang, S. H., Goh, C. J. (2002). Spatial and temporal expression of the orchid floral homeotic gene DOMADS1 is mediated by its upstream regulatory regions. Plant Mol. Biol. 49 (2), 225–237. doi: 10.1023/a:1014958118852
Zahn, L. M., Kong, H., Leebens-Mack, J. H., Kim, S., Soltis, P. S., Landherr, L. L., et al. (2005). The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169 (4), 2209–2223. doi: 10.1534/genetics.104.037770
Zhang, J., Hu, Z., Yao, Q., Guo, X., Nguyen, V., Li, F., et al. (2018). A tomato MADS-box protein, SlCMB1, regulates ethylene biosynthesis and carotenoid accumulation during fruit ripening. Sci. Rep. 8 (1), 3413. doi: 10.1038/s41598-018-21672-8
Zhang, G. Q., Liu, K. W., Li, Z., Lohaus, R., Hsiao, Y. Y., Niu, S. C., et al. (2017). The Apostasia genome and the evolution of orchids. Nature 549 (7672), 379–383. doi: 10.1038/nature23897
Zhang, S., Lu, S., Yi, S., Han, H., Liu, L., Zhang, J., et al. (2017). Functional conservation and divergence of five SEPALLATA-like genes from a basal eudicot tree, Platanus acerifolia. Planta 245 (2), 439–457. doi: 10.1007/s00425-016-2617-0
Zhang, C., Wei, L., Yu, X., Li, H., Wang, W., Wu, S., et al. (2021). Functional conservation and divergence of SEPALLATA-like genes in the development of two-type florets in marigold. Plant Sci. 309, 110938. doi: 10.1016/j.plantsci.2021.110938
Zhang, G. Q., Xu, Q., Bian, C., Tsai, W. C., Yeh, C. M., Liu, K. W., et al. (2016). The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 6, 19029. doi: 10.1038/srep19029
Zhang, S., Yao, J., Wang, L., Wu, N., van Nocker, S., Li, Z., et al. (2022). Role of grapevine SEPALLATA-related MADS-box gene VvMADS39 in flower and ovule development. Plant J. 111 (6), 1565–1579. doi: 10.1111/tpj.15907
Zhang, T., Zhao, Y., Juntheikki, I., Mouhu, K., Broholm, S. K., Rijpkema, A. S., et al. (2017). Dissecting functions of SEPALLATA-like MADS box genes in patterning of the pseudanthial inflorescence of Gerbera hybrida. New Phytol. 216 (3), 939–954. doi: 10.1111/nph.14707
Zhou, Y., Xu, Z., Yong, X., Ahmad, S., Yang, W., Cheng, T., et al. (2017). SEP-class genes in Prunus mume and their likely role in floral organ development. BMC Plant Biol. 17 (1), 10. doi: 10.1186/s12870-016-0954-6
Zhu, G., Yang, F., Shi, S., Li, D., Wang, Z., Liu, H., et al. (2015). Transcriptome characterization of cymbidium sinense 'Dharma' Using 454 pyrosequencing and its application in the identification of genes associated with leaf color variation. PloS One 10 (6), e0128592. doi: 10.1371/journal.pone.0128592
Keywords: C. sinense, SEPALLATA-like genes, expression pattern, ectopic overexpression, transient expression, protein interaction, floral development
Citation: Lin Z-Y, Zhu G-F, Lu C-Q, Gao J, Li J, Xie Q, Wei Y-L, Jin J-P, Wang F-L and Yang F-X (2023) Functional conservation and divergence of SEPALLATA-like genes in floral development in Cymbidium sinense. Front. Plant Sci. 14:1209834. doi: 10.3389/fpls.2023.1209834
Received: 21 April 2023; Accepted: 08 August 2023;
Published: 30 August 2023.
Edited by:
Xiaozeng Yang, Beijing Academy of Agricultural and Forestry Sciences, ChinaReviewed by:
Xu Xiaozhao, China Agricultural University, ChinaCopyright © 2023 Lin, Zhu, Lu, Gao, Li, Xie, Wei, Jin, Wang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feng-Lan Wang, d2FuZ2ZsMjAwMkAxNjMuY29t; Feng-Xi Yang, ZmVuZ3hpX3dlaUBzaW5hLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.