- Ministry of Education Key Laboratory of Cell Activities and Stress Adaptations, School of Life Sciences, Lanzhou University, Lanzhou, Gansu, China
Medicago truncatula has been selected as one of the model legume species for gene functional studies. To elucidate the functions of the very large number of genes present in plant genomes, genetic mutant resources are very useful and necessary tools. Fast Neutron (FN) mutagenesis is effective in inducing deletion mutations in genomes of diverse species. Through this method, we have generated a large mutant resource in M. truncatula. This mutant resources have been used to screen for different mutant using a forward genetics methods. We have isolated and identified a large amount of symbiotic nitrogen fixation (SNF) deficiency mutants. Here, we describe the detail procedures that are being used to characterize symbiotic mutants in M. truncatula. In recent years, whole genome sequencing has been used to speed up and scale up the deletion identification in the mutant. Using this method, we have successfully isolated a SNF defective mutant FN007 and identified that it has a large segment deletion on chromosome 3. The causal deletion in the mutant was confirmed by tail PCR amplication and sequencing. Our results illustrate the utility of whole genome sequencing analysis in the characterization of FN induced deletion mutants for gene discovery and functional studies in the M. truncatula. It is expected to improve our understanding of molecular mechanisms underlying symbiotic nitrogen fixation in legume plants to a great extent.
Introduction
Fast neutron produced from nuclear fission reactors or particle accelerator spallation sources are a type of high-energy ionizing irradiation effective in generating mutations in genomes of diverse organisms (Men et al., 2002). The nature of mutations induced by FN mutagenesis includes deletions of various sizes and other mutation types such as inversion, translocation and duplication (Bruggemann et al., 1998). Deletions cause gene loss or disruption and are especially valuable in functional studies of individual and tandemly-duplicated genes. Several large FN mutant populations have been developed in different plant species including Arabidopsis thaliana, Oryza sativa L., Glycine max, Lotus japonicus, barley and M. truncatula (Li et al., 2002; Alonso et al., 2003; Mitra et al., 2004; Rostoks et al., 2006; Wang et al., 2006; Hoffmann et al., 2007; Bolon et al., 2011).
M. truncatula is also an ideal model species for studying legume nodule development and symbiotic nitrogen fixation (SNF) (Cook, 1999). The symbiotic interaction between M. truncatula and rhizobia forms indeterminate nodules, which can be divided into five main zones: meristem, infection, intermediate transition, nitrogen fixation, and senescence. The apical meristem zone is mainly responsible for cell division and new cell supply for nodule development. The infection zone is the entry point for rhizobia into nodule cells through infection threads, and the transition zone is where rhizobia enter nodule cells and form symbiont. After entering the nitrogen fixation zone, bacteroids play a role in converting atmospheric nitrogen into ammonia, which can be used directly by plants. Finally, the senescence zone is where symbiont and bacteroids degrade and lose their nitrogen-fixing activity. Clearly, the symbiotic nitrogen fixation process is very delicate and complex, and depends on the mutual recognition between plant and rhizobia at the molecular level for signaling and metabolite exchange. The symbiotic relationship will cease if there are problems in any of the above areas. Loss of symbiotic nitrogen fixation in legumes can lead to the formation of different types of mutants, which are characterized as the three main categories: (1) non-nodulating mutants (Nod-), (2) nodulating but not nitrogen-fixing mutants (Nod+/Fix-), and (3) mutants with excessive nodulation (Nod+++) (Oldroyd, 2013).
Years of mutant analyses have led to identification of genes and molecular mechanisms involved in nodule initiation and development, including mutual recognition of rhizobia and root hairs. Mutants from the early recognition process mainly affect nodule formation, but not the nitrogen fixation (Mergaert et al., 2020). However, there are relatively less studies on genes and their mechanisms involved in rhizobia release to infected nodule cells and nodule senescence. Therefore, studies on the late development of legume-rhizobia symbiotic nitrogen fixation have become a major direction in legume research (Roy et al., 2020).
The establishment of an FN mutant library in M. truncatula provides opportunities for studying gene functions, and a large number of mutants with symbiotic nitrogen fixation deficiency phenotypes provide a reliable guarantee for identifying gene functions and studying molecular regulatory mechanisms (Chen and Chen, 2018). However, molecular cloning of genes from FN mutants traditionally relies on the map-based cloning approach, which is a time-consuming technique that limits the amount of mutants to be analyzed at the molecular level. Several complementary approaches, such as transcript-based deletion detection and DNA copy number variation detection using a whole genome tiling array comparative genomic hybridization (CGH) platform, have been developed to facilitate the characterization of deletion mutants in diverse organisms including animals and plants (Geschwind et al., 1998; Infante et al., 2003; Gong et al., 2004; Werner et al., 2005; Bejjani and Shaffer, 2006; Lakshmi et al., 2006; Emerson et al., 2008; Perry et al., 2008).
The array-based CGH (aCGH) method is a powerful tool for the analysis of copy number variation, polymorphisms, and structural variations in the genome (Bolon et al., 2011; Haun et al., 2011; Horvath et al., 2015; Kim et al., 2015). However, low accuracy and low sensitivity for small deletions by the CGH approach are apparent. It has a higher probe density in exons than in introns and 5’ and 3’ UTRs, which limits the detection of sequence variations to only covered regions (Chen et al., 2017).
Recently, the whole genome sequencing (WGS) platform has delivered comprehensive genetic analyses in humans, plants and animals (Sun et al., 2018). Paired-end whole genome sequencing involves sequencing both ends of a DNA fragment, which increases the likelihood of alignment to the reference genome and facilitates detection of genomic rearrangements, repetitive sequences, and gene fusions. Unlike focused approaches such as exome or targeted resequencing, which analyze a limited portion of the genome, whole genome sequencing conveys a comprehensive view of the entire genome. It is ideal for discovery applications, such as identifying genome deletions and duplications. Whole-genome sequencing can detect single nucleotide variants, insertions, deletions, copy number changes, and large structural variants. The application of WGS platforms to FN mutant populations can identify and locate genomic changes at the whole genome level (Du et al., 2021).
Here, we isolated and characterized of the M. truncatula FN007 mutant from fast neutron deletion mutant library. The FN007 mutant showed defective in biological nitrogen fixation and nodule development associated with early senescence phenotype. Detailed phenotypic analysis was carried out in the FN007 mutant and would provide new insight of symbiotic nitrogen fixation processes in leguminous plants. Furthermore, the FN007 mutant was analyzed by WGS platform to detect genomic alterations. Our results confirmed that the WGS platform was efficient in mapping genome deletions and would greatly facilitate characterization of a large number of FN mutants for gene functional studies in M. truncatula. Using this mutant screening and identification method should improve our understanding of molecular mechanisms underlying symbiotic nitrogen fixation.
Materials and methods
Plant materials and growth condition
M. truncatula seeds were scarified with concentrated sulfuric acid for 5 to 8 min, rinsed with deionized water for five times, surface sterilized with 5% sodium hypochlorite solution and rinsed with autoclaved deionized water for several times. After vernalization at 4°C for 3 days, seedlings were transplanted to pots and grown in greenhouse with 16 hr/8 hr light/dark cycle and 150 E/m2/sec light intensity. Perlite and sand (volume ratio of 3:1) were used as the planting medium. The perlite and sand were separately rinsed with deionized water, and then mixed in proportion for use. The germinated seeds were transferred to a 72-cell tray (530 mm × 270 mm, with a single well volume of 40 mL), watered with 1 L of 0.125 mM KNO3 (low-nitrogen nutrient) solution. Rhizobia were inoculated after one week of growth in the greenhouse.
Inoculation with the rhizobial: Sinorhizobium medicae ABS7 strain and Sinorhizobium meliloti Sm2011-GFP strain
The rhizobia strains were removed from a -80°C refrigerator. Two mL of the rhizobia were inoculated into 50 mL of TY liquid medium containing tetracycline and cultured at 28°C for 2 days. Then, the bacterial cultures were centrifuged at 4,000 rpm for fifteen min in a 50 mL centrifuge tube, re-suspended and diluted to OD600≈0.05 with the low nitrogen nutrient solution. Five mL of the above bacterial suspension were inoculated to each plant.
Microscopic observations
Three to five plants inoculated with S. medicae ABS7 strain carrying the hemA::LacZ reporter were harvested at 6 dpi, 8 dpi, 10 dpi, 13 dpi and 16 dpi (days post inoculation). Roots were washed with deionized water. The root nodules were fixed as previously described (Xi et al., 2013). For GFP studies, nodules from plants inoculated with the S. meliloti Sm2011 strain harboring hemA::mGFP reporter gene were washed with deionized water, embedded in 6% agarose, and sectioned with Vibratome™ 1000 Plus (Leica Biosystems, Germany). Nodule sections (50-70 μm) were mounted on glass slides and imaged using MVX10+DP74 microscope (Olympus).
Live/dead staining of nodule cells and potassium iodide staining
Plants inoculated with S. medicae ABS7 strain were used for 50-70 μm nodule section preparation. 5 μM SYTO9 and 30 μM PI were added to 50 mM Tris (pH7.0) buffer to make a staining solution. Nodule sections were stained for 20 min at room temperature. Sections were placed on slides covered with coverslips and observed with a confocal microscope [Nikon Confocal laser scanning microscope (A1R+ Ti2-E)].
Five-micrometer semi-thin sections were placed on glass slides and stained with 1% potassium iodide phosphate buffer in dark condition for 10 min. Then, the image was observed with microscope [Axio Imager.Z2 (ZEISS)].
Semithin section of root nodules and toluidine blue staining
The nodules were placed in the fixing solution, vacuumed for 10 min, and fixed overnight at 4°C. The fixed nodules were dehydrated in 70%, 85% and 95% ethanol series for 30 min each, and in 100% ethanol for 2 h. Finally, 0.1% eosin Y3 was added to 100% ethanol to dye for half an hour. After the nodules turned red, samples were replaced in a mixture of 100% ethanol and base liquid Technovit 7100 resin (v/v=1:1) for 2 h. The nodule samples were transferred to the infiltration solution overnight (infiltration solution: 1 g hardener I to 100 mL base liquid Technovit 7100 resin. Allow approximately 5 min for complete dissolution). The nodule samples were embedded after the permeation treatment. The temperature was set to 40°C and the embedding mould was dried beforehand. The infiltrate and hardener II (v/v=15:1) were mixed gently, and 200 μL of the mixture was added to each well of the embedding mould. The nodule materials were carefully picked up with forceps and placed on the tip of the embedding mould, with materials placed vertically to facilitate sectioning. The samples were arranged neatly and then polymerized overnight in a 40°C incubator. The Leica RM2265 rotary microtome was used to cut sections with a thickness of 3-5 μm. The semithin sections were placed on glass slides and stained with 1% toluidine blue for 5 min. Finally, the stained sections were observed and imaged with microscope [Axio Imager.Z2 (ZEISS)].
Whole genome sequencing analysis
The whole genome sequencing (WGS) analysis was performed by Shanghai Oebiotech Co. The compound leaf samples were collected from wild type (WT) A17 and FN007 mutant, respectively. The genomic DNA was extracted using the DNeasy plant mini kit (Qiagen Shanghai, China), following the supplier’s instructions. The library was constructed after the DNA quality was passing the bioanalyzer test. The DNA samples were randomly sheared into 350 bp-500 bp fragments, and the TruSeq DNA LT Sample Prep Kit was used for library construction. The DNA fragments were treated with the following steps to complete library construction: end repair, addition of ployA tails, addition of sequencing connectors, purification, and PCR amplification. After library construction and qualification, pair-end sequencing was performed on the sequencer (HiSeq X Ten sequencer).
The raw sequencing data were processed in two stages. Sequencing data quality was mainly examined through the sequencing error rate, data volume, comparison rate, coverage and other statistics to assess whether the sequenced library meet the standard for subsequent analysis. Then, the high-quality sequences were compared to the reference genome to detect the variation information in the samples. SNP (single nucleotide polymorphisms) and InDel (insertion or deletion) detection was performed using the GVCF mode of the Haplotypecaller module of the GATK (Version: 4.1.0.0) software, based on the results of the sample comparison with the reference genome. To reduce the error rate of SNP and InDel assays, a criterion of QD more than 2.0 was chosen for filtering, and only mutant loci that met this criterion were retained. QD is the ratio of the quality of variation divided by the depth of coverage, which is actually the quality of variation per unit depth. The SNP and InDel results were annotated using SnpEff (Version: 4.4) software. Finally, the genomic variation information detected was visualized using Circos plots.
To eliminate the effect of sequencing errors on the results, the original sequencing data were cleaned using the pre-processing software Fastp. Clean reads were compared to reference genome using BWA, and the comparison results were converted to SAMtools, and de-redundant using Picard (http://broadinstitute.github.io/picard/), and the results were analyzed using the Qualimap software (Li and Durbin, 2009; Li et al., 2009; Chen et al., 2018). M. truncatula reference genome can download at the website: https://medicago.toulouse.inra.fr/MtrunA17r5.0-ANR/(MtrunA17r5.0).
TAIL PCR
Thermal asymmetric interlaced PCR (TAIL PCR) assay was carried out essentially as previously described (Liu and Chen, 2007). The specific primer P1 adjacent to the proposed deletion border was combined with five short randomly primers (AD1, AD2, AD3, AD5, AD6) with low Tm values and amplified by PCR with thermally asymmetric temperature cycling, using the FN007 mutant genomic DNA as template.
The resulting PCR product was amplified again to obtain a flanking sequence using the nested primer P2 of P1 (Table S1) and the five random primers mentioned above. The PCR product was sequenced and compared with the genome sequence of M. truncatula A17 to confirm the deletion junctions in FN007 mutant.
Results
Phenotypes of FN007 mutant under low nitrogen condition
M. truncatula FN007 mutant was isolated from a Fast Neutron (FN) induced deletion mutant collection in the Jemalong A17 background. Under Nitrogen replete condition, the FN007 mutant is comparable with Wild Type (WT) plants in growth and development processes, including leaf, seed pod, flower and root development (data not shown). But under Nitrogen-limited and symbiotic condition, the FN007 mutant plant develop nitrogen starvation symptoms including stunted growth and yellowish leaves at 16 days post inoculation (dpi) with S. medicae ABS7 strain, indicating that these phenotypes were caused by defective symbiotic nitrogen fixation (Figures 1A, B). Nodules developed on the FN007 mutant roots were small and spherical in shape, most with white colors (due to degradation of leghemoglobin), whereas in the WT plant, the pink cylindrical nodules were developed (Figures 1C–F). Moreover, nodule distribution was concentrated in WT plants but scattered in FN007 mutant and the WT plant had fewer nodule numbers than the FN007 mutant.
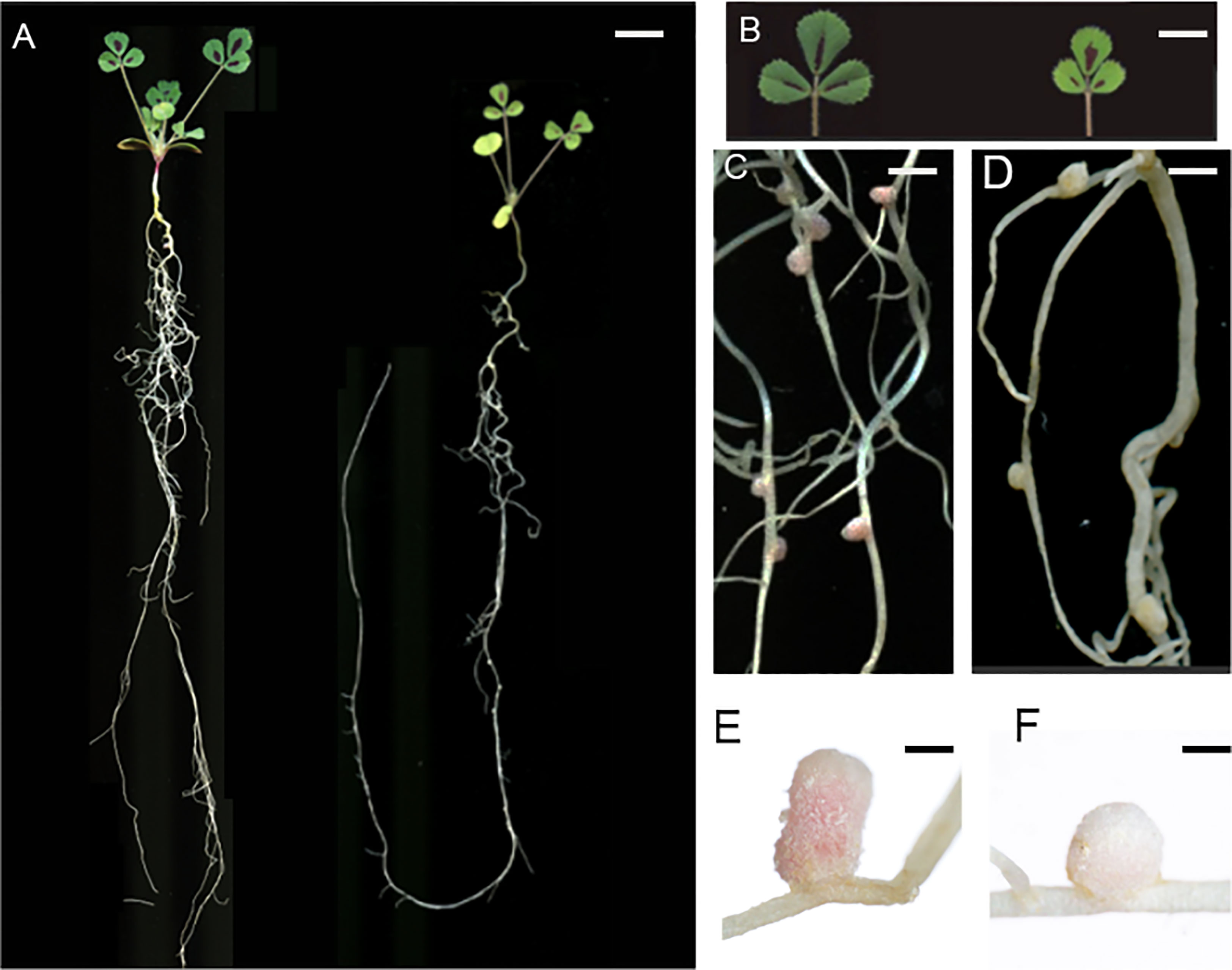
Figure 1 M. truncatula FN007 mutant phenotype. (A) Growth of FN007 (right) and wild type (WT, left) seedlings 16 days post inoculation (dpi) with S. medicae ABS7 strain, showing dwarf phenotype of FN007 mutant. (B) leaf phenotype of FN007 mutant (right) and WT (left), yellowish leaves are the sign of nitrogen deficiency phenotype. Nodules developed on (C) (WT) and (D) (FN007 mutant) were pink and white color, respectively. Close-up view of (E), WT and (F), FN007 mutant nodules. Scale bar: (A), 1 cm; (B), 50 mm; (C, D), 10 mm; (E, F), 1 mm.
Time-course observation of nodule phenotype
To further characterize the nodule development processes of the FN007 mutant, time-course experiments were carried out. Under low nitrogen condition (0.125 mM KNO3), the growth and development of root nodules in the FN007 mutant and WT plant inoculated with S. medicae ABS7 strain was continuously monitored and recorded. Morphological and color differences between FN007 mutant and WT nodules became apparent over time (Figure 2). At 6 dpi, WT (Figure 2A) and FN007 mutant (Figure 2F) nodules appeared to be similar, both white and spherical shape. At 9 dpi, WT nodules (Figure 2B) became pale pink color and conical in shape, while FN007 mutant nodules (Figure 2G) were still white color and cone-shaped. At 12dpi, WT nodules (Figure 2C) were pink and cylindrical shape, but FN007 mutant nodules (Figure 2H) gradually increased in size with increasing growth time, and remained white color. At 15 dpi, WT nodules (Figure 2D) continued to grow, almost entirely pink, and were already mature and capable of efficient nitrogen fixation. FN007 mutant nodules (Figure 2I) grew slowly, with one or two white and rod-like, and the remainder were basically white spheres or cones shape. At 19 dpi, WT nodules (Figure 2E) were generally the same as those at 15 dpi, but the number of mature nodules increased, while FN007 mutant nodules (Figure 2J) still appeared as white rod shape. Time-course experiments showed that early nodule development (6 dpi) appears to be normal in the FN007 mutant. However, differences began apparent at 6-9 dpi with leghemoglobin accumulation in WT nodules. Even though the FN007 nodules did not show the pink functional leghemoglobin, some nodules were able to elongate and grow into rod-like shape during later development stage, indicating that the nodule meristem still had normal developmental functions.
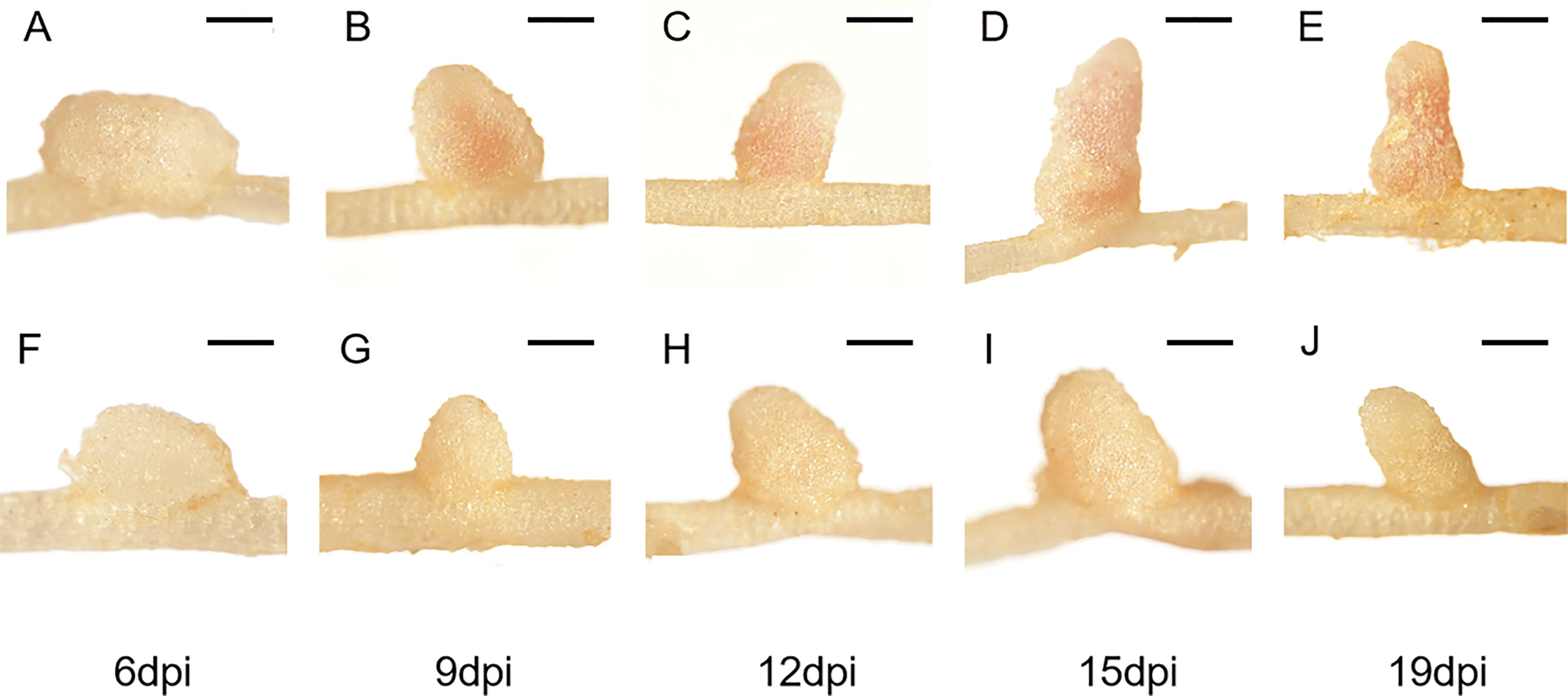
Figure 2 Time-course observations of nodule phenotypes. M.truncatula WT and FN007 mutant seedlings were inoculated with S. medicae ABS7 strain and inoculated root nodules were observed in a time-course assay. Representative images of (A), WT and (F), FN007 mutant roots 6 dpi; (B), WT and (G), FN007 mutant 9 dpi; (C), WT and (H), FN007 mutant 12 dpi; (D), WT and (I), FN007 mutant 15 dpi; (E), WT and (J), FN007 mutant 19 dpi. These observations suggest that FN007 mutant has an abnormal nodule development. Scale bar: 500 μm.
Under low nitrogen condition (0.125 mM KNO3), the FN007 mutant and WT seedlings grown for one week in greenhouse were inoculated with S. medicae ABS7 strain containing the hemA::LacZ reporter gene. Nodules were harvested at different development stages after inoculation and stained with LacZ staining buffer. At 12 dpi, in FN007 mutant nodules (Figure 3B), there were none or few rhizobia in nodule cells. For WT plant at the same time (Figure 3A), rhizobia filled the entire nodule cells. At 15 dpi, the number of rhizobia-containing nodule cells in FN007 mutant (Figure 3D) was decreased, and nodule cell debris was observed, at which point a distinct senescence zone (rather than a nitrogen-fixing zone) had formed, which was significantly different from the WT (Figure 3C). We speculated that between 6-7 dpi, the FN007 mutant failed to form a nitrogen-fixing zone and went directly into senescence, forming a region that accounted for an increasing proportion of nodules over time. To confirm this result, another rhizobia strain with GFP reporter gene, S. meliloti Sm2011-GFP, was inoculated. At 11 dpi, the fluorescence signal in wild type nodules was very strong, indicating that rhizobia released and filled with nodule cells (Figure 4A). However, in FN007 mutant nodules, though the fluorescence intensity of zones I and II was no different from the WT, zone III fluorescence was distributed in a punctiform pattern and no mature symbiont was formed (Figure 4B), indicating that rhizobia release into nodule cells during infection was normal, which was consistent with the results of nodule sections inoculated with S. medicae ABS7 strain. At 14 dpi, the WT nodules showed characteristic partitioning and a dominant green fluorescent signal in mature symbiotic cells completely filled with bacteroids (Figure 4C), whereas in the FN007 mutant nodules an increasingly severe degradation of the bacterial cycle in the nitrogen-fixing zone was observed (Figure 4D). This observation suggests the FN007 mutant nodules may undergo early senescence.
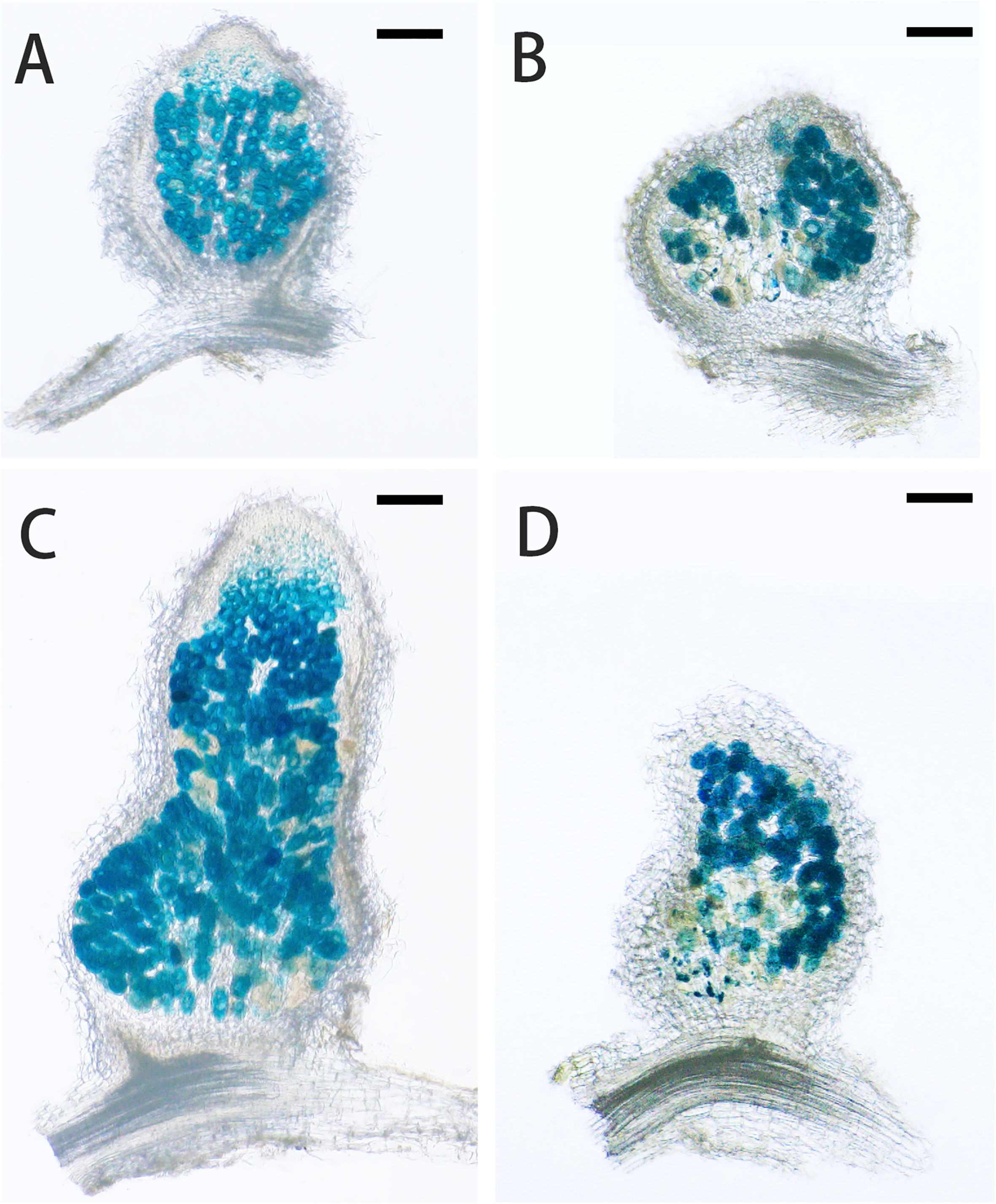
Figure 3 LacZ stained sections observation of nodule development. M.truncatula WT and FN007 mutant seedlings were inoculated with S. medicae ABS7 strain harboring hemA::LacZ reporter gene. Longitudinal sections of nodules were observed under a MVX10+DP74 microscope and representative images were shown. (A), WT and (B), FN007 mutant nodules 12 dpi; (C), WT and (D), FN007 mutant nodules 15 dpi. Scale bar: 200 μm.
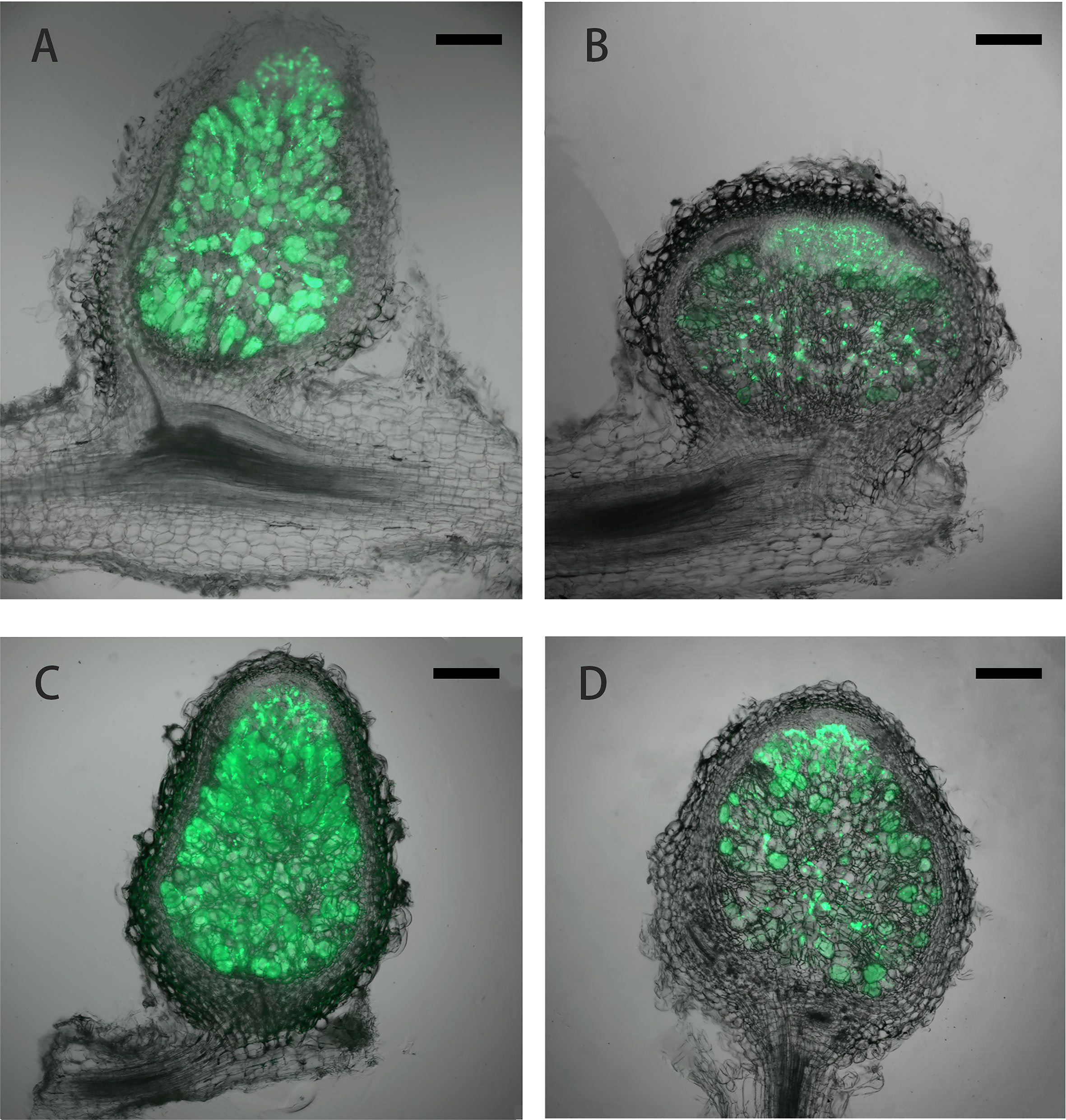
Figure 4 Laser confocal microscopic analysis of nodule development. M.truncatula WT and FN007 mutant seedlings were inoculated with S. meliloti Sm2011 strain harboring hemA::mGFP reporter gene. Longitudinal sections of nodules were observed under a MVX10+DP74 microscope and representative images were shown. (A), WT and (B), FN007 mutant nodules 11 dpi; (C), WT and (D), FN007 mutant nodules 14 dpi. Scale bar: 200 μm.
M. truncatula FN007 mutant is defective in symbiotic nitrogen fixation
In indeterminate nodules, rhizobia released into nodule cells differentiate before nitrogen fixation begins, and the differentiated rhizobia are morphologically longer than free-state rhizobia and have endoreduplication of their chromosomes (Mergaert et al., 2006). From GFP and LacZ stained sections experiments, we found that the FN007 mutant was normal in early nodule development stage including infection thread development and rhizobia release. To further determine the development and differentiation of rhizobia released into nodule cells, 5 μm semithin longitudinal nodule sections were stained with Toluidine blue for microscopic observation. In healthy symbionts, rhizobia undergo intracellular replication and differentiation into rod-like structures evenly distributed around a central vacuole, but rhizobia in infected cells need to differentiate to exert nitrogen-fixing function properly. At 13 dpi, WT nodules (Figure 5A) had formed four distinct zones: zone I (zones of meristem, ZI), zone II (infection, ZII), zone II-III (a transition zone between ZII and ZIII, IZ), and zone III (nitrogen fixation, ZIII). Zone II-III was a transition zone from zone II to III, when rhizobia had already entered into infected cells, and with a compartment area of about one-fifth of the total rhizobia area. Infected cells were filled with intact symbiosomes in WT. In this period, the FN007 mutant nodules appeared to have four zones in terms of cell morphology: I, II, IZ, and IV (senescence zone), but did not appear to have the nitrogen-fixing zone (ZIII) as observed in WT nodules, entering directly into the senescence zone. Symbiosomes in most infected cells were disintegrated at the proximal end of the root in the FN007 mutant (Figure 5D). In the WT, the senescence zone was formed at 35 dpi in the absence of environmental stress, but in the FN007 mutant, the senescence zone was formed at 13 dpi, which clearly showed an early senescence phenotype. Therefore, it was assumed that symbiotic nitrogen fixation defect occurs in the II-III transition zone, and rhizobia cannot complete the transition after its release into the infected cells in the FN007 nutant. In WT nodules (Figures 5B, C), infected cells were morphologically normal, mostly subrounded, with a large number of symbiosomes containing rod-shaped bodies regularly arranged around a large central vacuole. In the FN007 mutant, infected cells (Figures 5E, F) were smaller and irregularly shaped compared to that in WT nodules, with less symbiosomes and most were not rod-shaped, resembling the free-living cells.
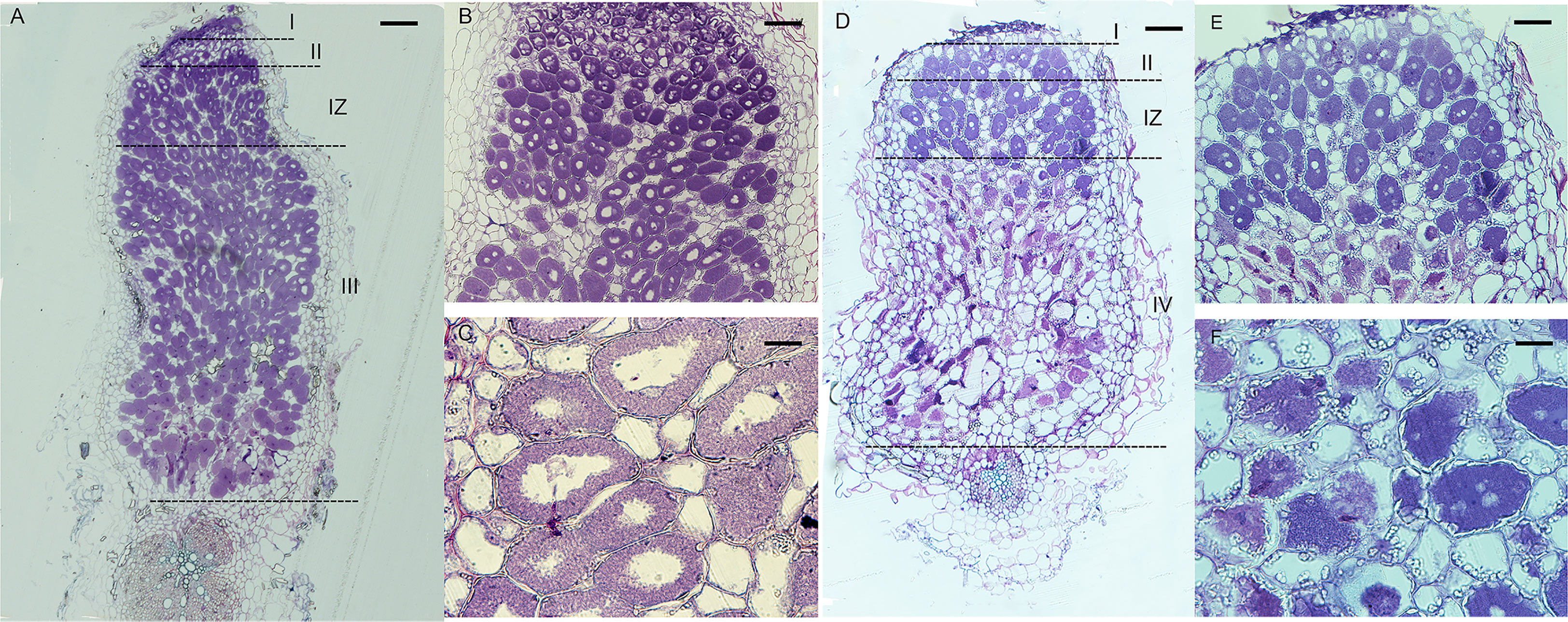
Figure 5 Semithin sections of WT and FN007 mutant nodules. M. truncatula, (A), WT and D, FN007 mutant nodules 13 days post inoculation (dpi) were sectioned longitudinally and stained with toluidine blue, showing development of meristem (I), infection (II) and transition zones (IZ), and infected cells in both WT and FN007 mutant. Compared with WT nodules, there was no fixation zone (III) in FN007 mutant, but a clearly senescence zone (IV) was emerged. Close-up view of (B), WT and (E), FN007 infected cells. Magnification view of (C), WT and (F), FN007 infected cells, showing degradation of symbiosomes in FN007 infected cells. Scale Bar: (A, D): 100 μm; (B, E): 20 μm; (C, F): 10 μm.
To further characterize the symbiotic phenotype of the FN007 mutant, semithin sections of nodules at 13 dpi were stained for starch granules with potassium iodide (KI). The WT nodules had almost no accumulation of starch granules at the distal root end (Figures 6A, B), but in the nitrogen fixation zone (Figure 6C), there was a small number of starch granules deposition around the cell membrane. Conversely, starch granules in the distal end of the FN007 nodules were evenly distributed and surrounded the cell membrane (Figure 6D), where clearly cell contours were observed and the starch granules were similar in size. At the proximal root end of the FN007 mutant nodule (Figures 6E, F), the number of deposited amyloplasts was increased and distributed in a disorganized manner. Overall, the number of starch granules in FN007 mutant nodules were greatly exceeded that of the wild type. These results indicated that the FN007 mutant cannot form a functional fixation zone, and thus many metabolic activities can not be carried out in infected cells, which accumulating more energy material (starch grains).
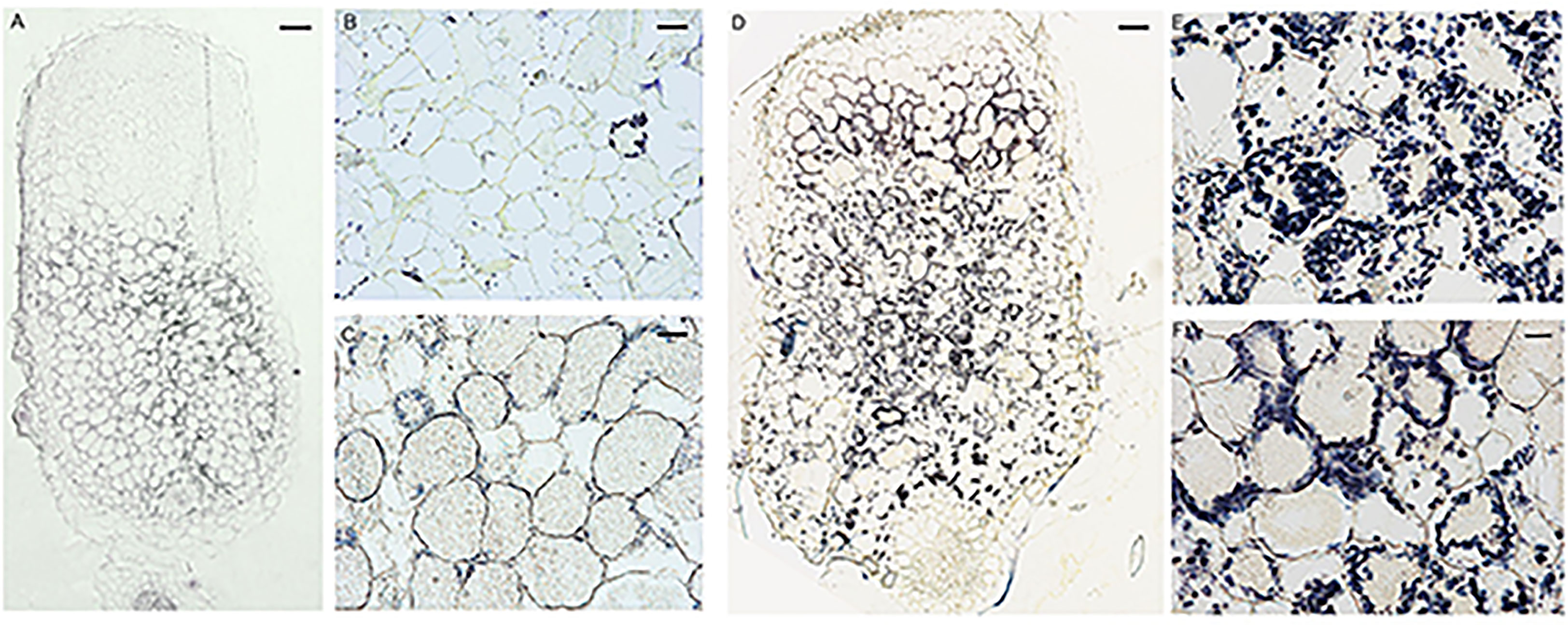
Figure 6 Accumulation of starch granules in FN007 mutant nodules. Potassium iodide staining of starch granules in infected cells in WT nodules (A–C) and FN007 mutant nodules (D–F). In WT nodules, starch granules was deposited in large amounts only in the II-III interval zone and in small amounts in the IV zone, while in FN007 mutant nodules, the starch granules deposition was greatly expanded. Scale bar: (A): 100 μm; (D):200 μm; (B, E): 20 μm; (C, F): 10 μm.
To confirm whether the FN007 mutant nodules undergo early senescence, we performed live/dead staining experiment, using fluorescence dyes SYTO9, which stains live bacteroids and nucleic acids of plant cells and propidium iodide (PI), which stains dead bacteroids. The FN007 mutant and WT plants were inoculated with S. medicae ABS7 strain. A large green fluorescence was observed within the WT nodule (Figures 7A–C), and clearly visible symbiosomes were formed within infected cells and regularly arranged around the central vacuole. At 13 dpi, the FN007 mutant nodules were mostly stained red fluorescence compared with the WT (Figures 7D–F), and high magnification images showed that infected cells contained both red and green scattered bacteroids, indicating that some bacteroids in the FN007 mutant were dead at this time. These observations suggest that nodule cells of the FN007 mutant exhibited signs of an early senescence phenotype (12 dpi).
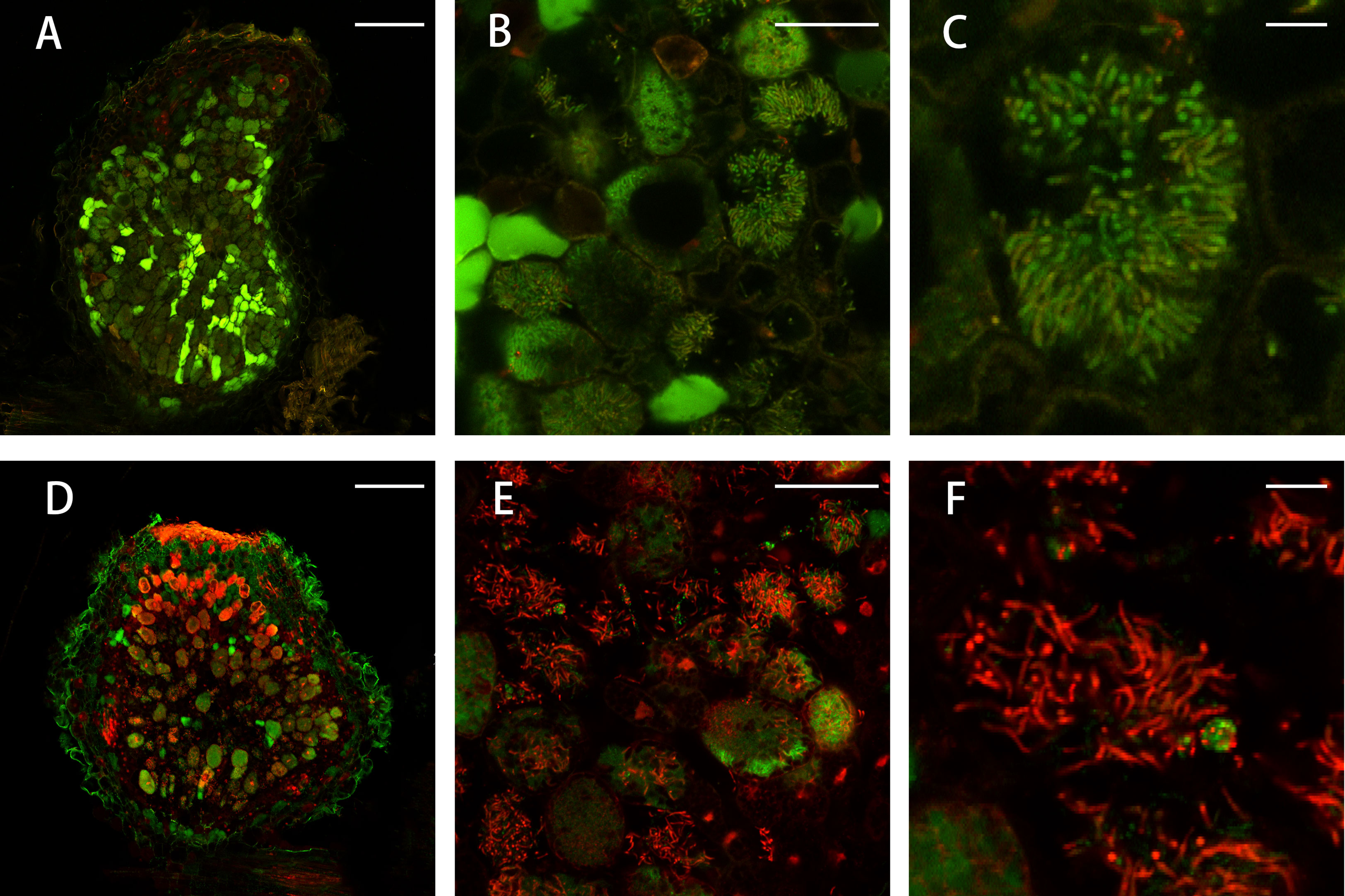
Figure 7 Live/dead staining of nodule cells. Live/dead staining of bacteroids in FN007 mutant compared with WT nodules (13 dpi). (A–C), wild type (WT) and (D–F), FN007 mutant. Live bacteroids have a green signal (GFP) and dying bacteroids have a red signal from propidium iodide (PI). PI can only enter bacteroids with compromised membranes. FN007 mutant nodules fail to sustain differentiated bacteroids. Scale bar: (A, D): 200 μm; (B, E): 50 μm; (C, F): 10 μm.
Whole genome sequencing analysis
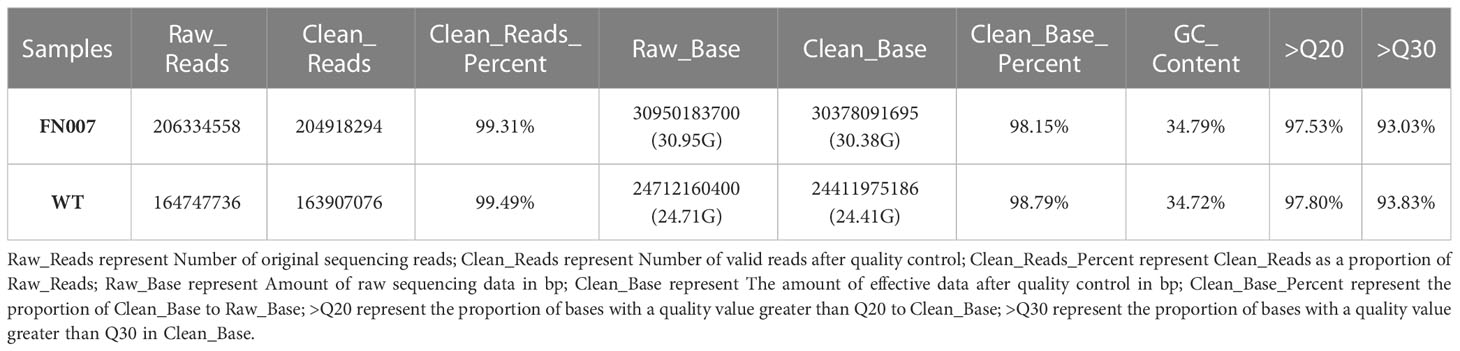
To identify mutations in the FN007 mutant, whole genome resequencing was carried out. Raw reads were cleaned after quality control and QC results were obtained (Table 1). Normally, for next generation sequencing values, Q20 should be over 95% (at worst, not below 90%), and Q30 is required to be more than 85% (at worst, not below 80%). The sequencing quality of the FN007 mutant and WT was high, with both Q20 values over 97% and both Q30 values over 93% (Table 1). The distribution of base error rates for pair-end reads in length range of 1 to 150 bp is shown in Figure 8. Regardless of read length, the average base error rate was less than 0.05%, indicating high sequencing quality.
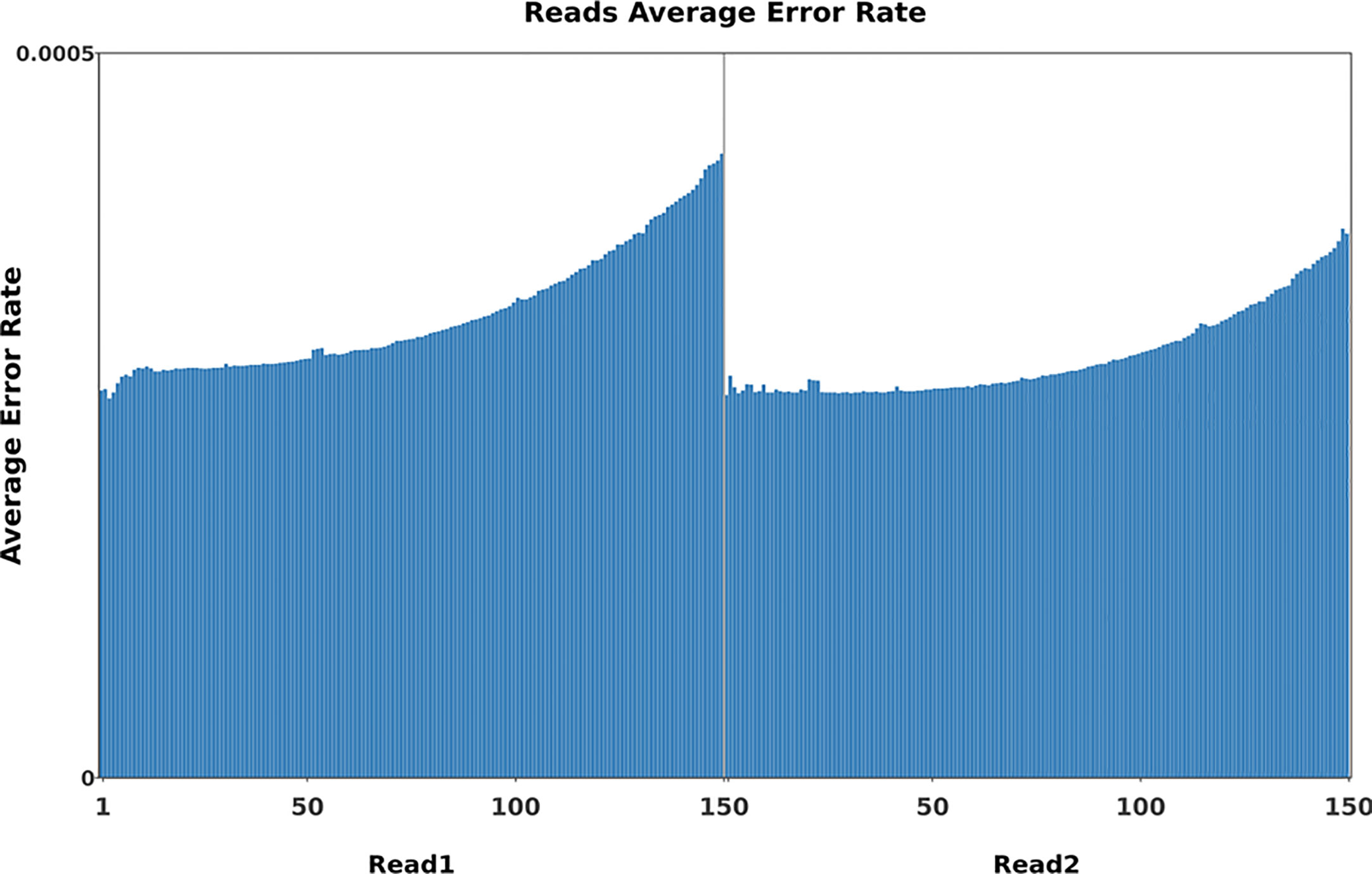
Figure 8 Base error rate distribution graph. Horizontal coordinates represent the base positions on reads, and vertical coordinates represent the average error rate of single bases; the left side shows the base quality distribution of R1 Reads in pair-end sequencing, and the right side shows the base quality distribution of R2 Reads.
Whole-genome sequencing uses non-strand-specific library building. Theoretically, G and C, and A and T bases should be equal in each sequencing cycle, and the whole sequencing process should be stable. The FN007 mutant base distribution sequencing data is shown in Figure 9. The start position of the sequence is linked to the primer junction during sequencing, so A, C, G and T will fluctuate at the beginning but stabilize later.
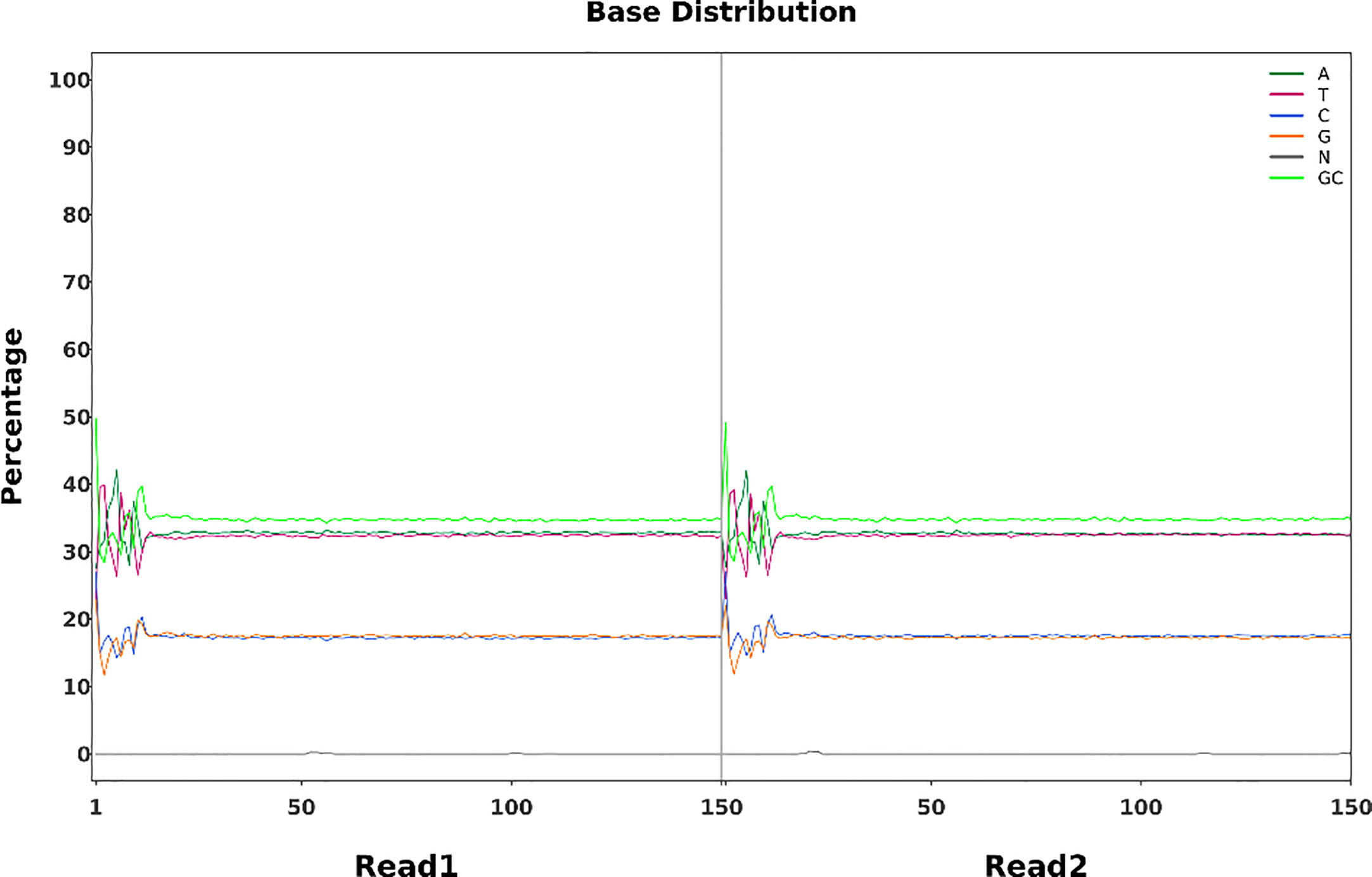
Figure 9 Base percentage distribution. Horizontal coordinates represent base position on the reads, and vertical coordinates represent the proportion of single bases, A,T,C,G,N, and GC, respectively.
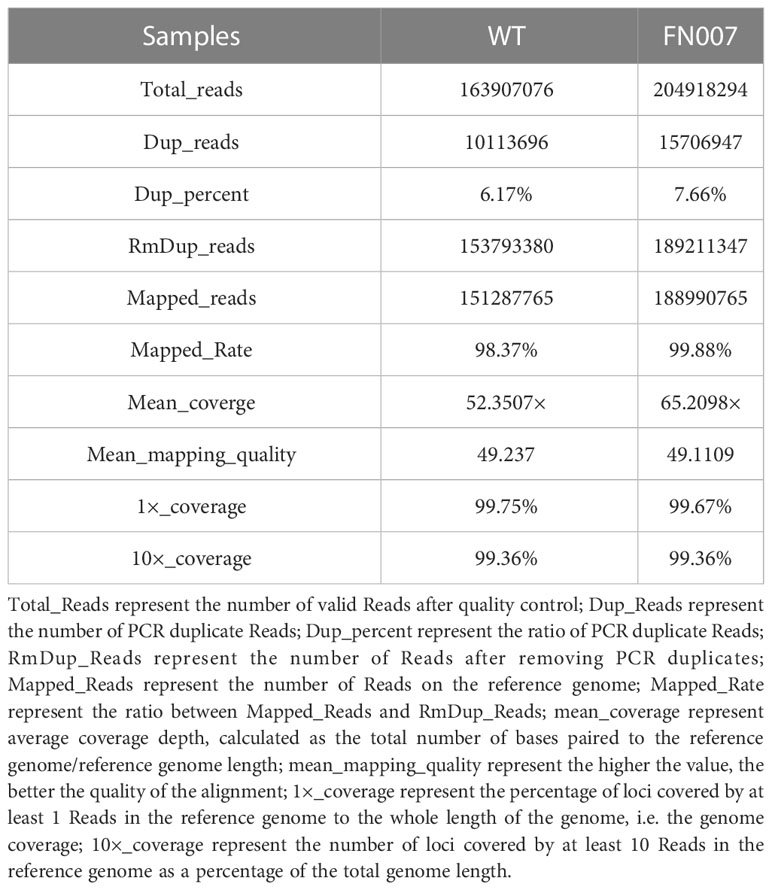
For species with a reference genome, their genomic information or complete transcript information can be downloaded from public databases, and then the sequenced reads can be compared to the reference genome. By quickly comparing each read to the reference genome, information about read position and match quality on the compared genome or other reference sequences can be obtained, and then the genes or transcripts can be annotated and quantified. After comparing the clean reads of the FN007 mutant and WT with the reference genome, the statistical results are shown in Table 2. It was suggested that reads from the FN007 mutant is comparable with WT. For the FN007 mutant, more than 189 million pair-end reads (2×150 bp) were produced, which represented a 65.2× sequencing depth. For the WT plant, 151 million reads were produced, representing 52.3× sequencing depth (Table 2). These reads were mapped to the latest reference genome of M. truncatula (MtrunA17r5.0). The total mean Mapping quality is nearly 50% in both mutant and WT. These result indicated that the WGS data has higher quality of the alignment.
SNP and InDel detection were performed using the GATK4 software Haplotypecaller module, based on the comparison of samples with the reference genome. QD is the ratio of variant quality divided by the depth of coverage, which is actually variant quality per unit depth, and most false positive variants have a QD of less than 2. Compared to transversion, transition is the most common mutation type, with T to C and A to G being the most common substitution types (Figure 10A). Of all the InDels obtained, deletions less than 4 bp accounted for 85.28% and insertions less than 4 bp accounted for 94.53% (Figure 10B). All the mutations detected were integrated into a Circos plot to visualize the genomic data (Figure 10C).
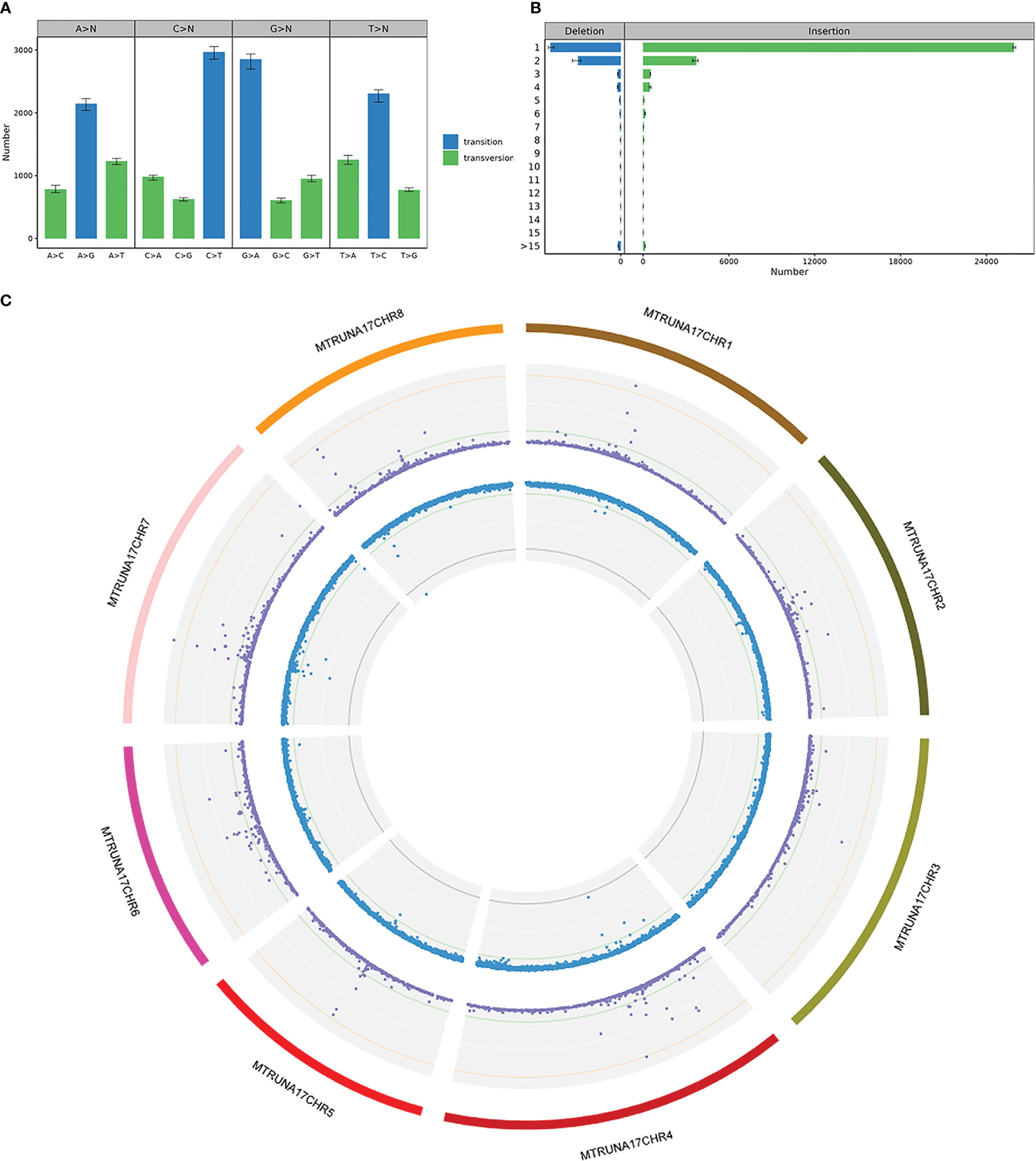
Figure 10 SNP & InDel Variant Detection in FN007 mutant (A) SNP substitution type statistics in FN007 mutant. Horizontal coordinates represent SNP substitution type, vertical coordinates are the mean, maximum and minimum values for each sample, and colors indicate whether SNP substitution type is a transition or a transversion. (B) InDel substitution type length statistics in FN007 mutant. Horizontal coordinates show the mean, maximum and minimum values of the InDel numbers, vertical coordinates show substitution base lengths, and colors indicate whether the substitution type is insertion or deletion. (C) Circos visualization of identified deletions in FN007 mutant. SNP and InDel distributions on individual chromosomes, where the first circle represents the FN007 chromosome, the purple dots in the second circle represent SNP density, and blue dots in the third circle represent InDel density.
The FN007 mutant resequencing data revealed that there was a copy number variation (CNV) in large segment of chromosome 3. CNVs may be biologically important for species-specific genome composition, species evolution and phylogeny, as well as for the expression and regulation of genes in specific genome regions. We used IGV software to visualize detection CNVs in the FN007 mutant, and found a possible large deletion on chromosome 3 from location 4,518,720 to 4,871,880 (Figure 11A). To determine the specific interval of the mutant deletion, four pairs of primer (Table S1) were first designed on both sides of each proposed boundary provided by the IGV data, and these primers were amplified by PCR in the FN007 mutant and WT (Figure 11B). The results showed that primers 3 and 4 could amplify the target bands in the FN007 mutant, but in contrast, primers 1 and 2 could not (Figure 11C). This suggests that the IGV data matches the actual deletion junction in the FN007 mutant. To identify the exactly deletion site, TAIL PCR was performed to identify the deletion regions in the FN007 mutant (Figure 11D). Results showed that the deletion border in the FN007 mutant was located at position 4,519,828-4,872,290 on chromosome 3. Based on the TAIL PCR results, we designed left and right border primers and verified the results, conformed that there was a deletion region in the FN007 mutant compared with WT (Figure 11E).
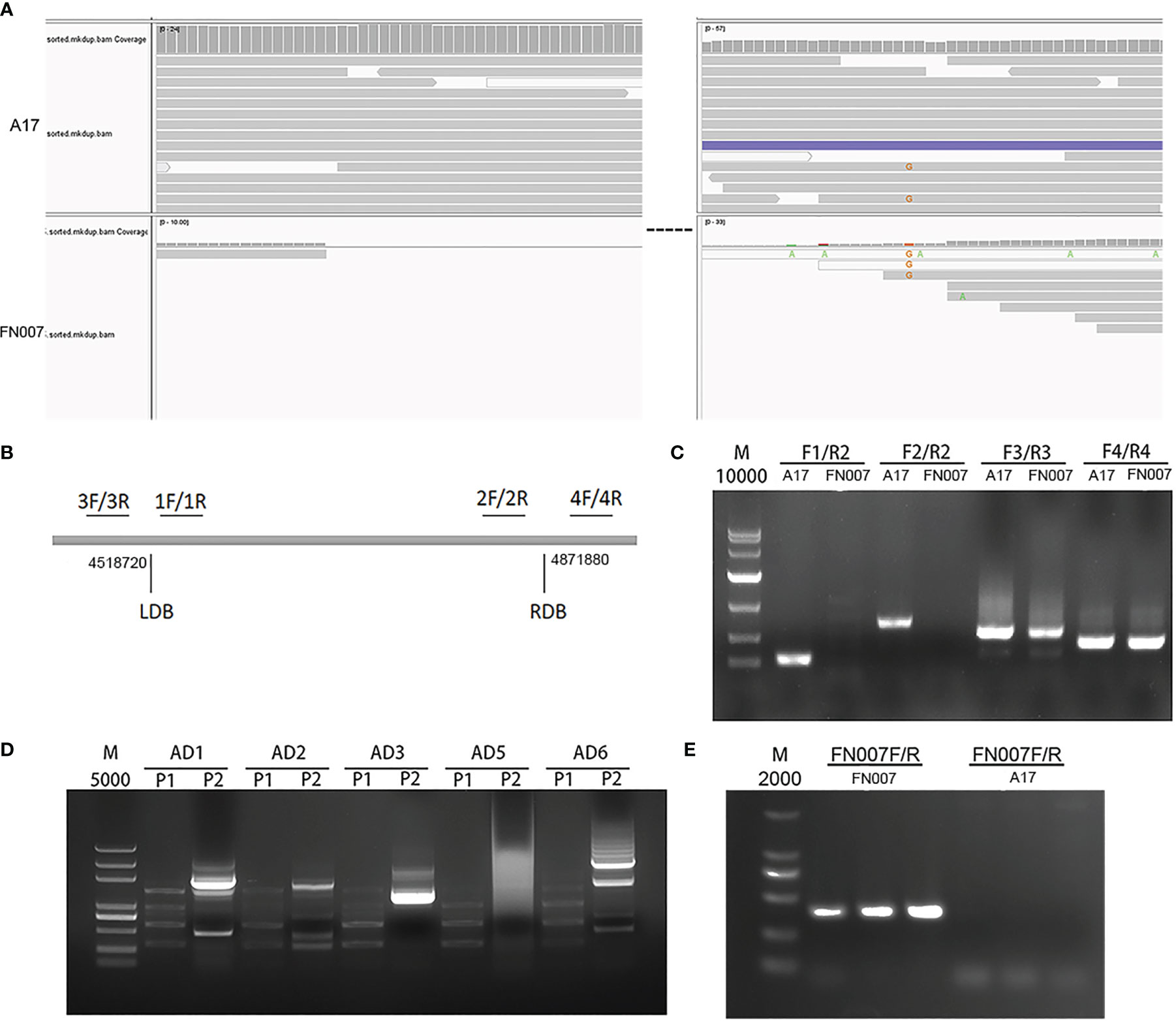
Figure 11 Identification of copy number variation (CNV) in FN007 mutant. (A) Visualization of the causal deletion using IGV. The reads alignment for WT and FN007 mutant. The sequencing showed a large deletion at the corresponding position in FN007 mutant; (B) Coordinate positions between four primers are designed for the identification of deletion border in FN007 mutant; (C) PCR detection for deletion junction through the flanking sequencing primers in FN007 mutant; (D) Tail-PCR was carried out, and the corresponding band were extracted for sequencing; (E) Deletion border confirmed by PCR in FN007 mutant. A pair of primer (FN007F/FN007R) was designed at deletion junction and amplified in FN007 mutant and WT. The results showed that FN007F/R could amplify the target band in FN007 mutant but not in WT.
Genetic linkage analysis
Whole genome sequencing analysis revealed that the FN007 mutant has a large deletion of approximately 348 kb on chromosome 3. To characterize the nature of the FN007 mutation, we generated F2 populations from cross-pollination between the FN007 mutant and its WT parent (Jemalong A17).
We analyzed 163 individual F2 plants and observed a ratio of 3.075:1 segregation of WT and mutant nodulation phenotypes in F2 populations, indicating a single recessive locus for the FN007 mutant phenotype. Next, we performed a linkage analysis, using pools of DNA samples isolated from individual F2 plants. The 123 plants with the WT phenotype were divided into 12 groups and 40 plants with the mutant phenotype were divided into 4 groups (the DNA from each group was pooled together). The deletion regions were identified by PCR using the corresponding primers (Figure 12A). The results showed that all the mutants plant could amplify the deletion border by MDB-F and MDB-R primer (data not shown), but could not get the corresponding band by the MDB-F and DB-R primer, while the WT plants can amplify the correct bands in the deletion region (Figure 12B). These results indicated that the fix- phenotype of the FN007 mutant was tightly linked to the deleted region on chromosome 3.
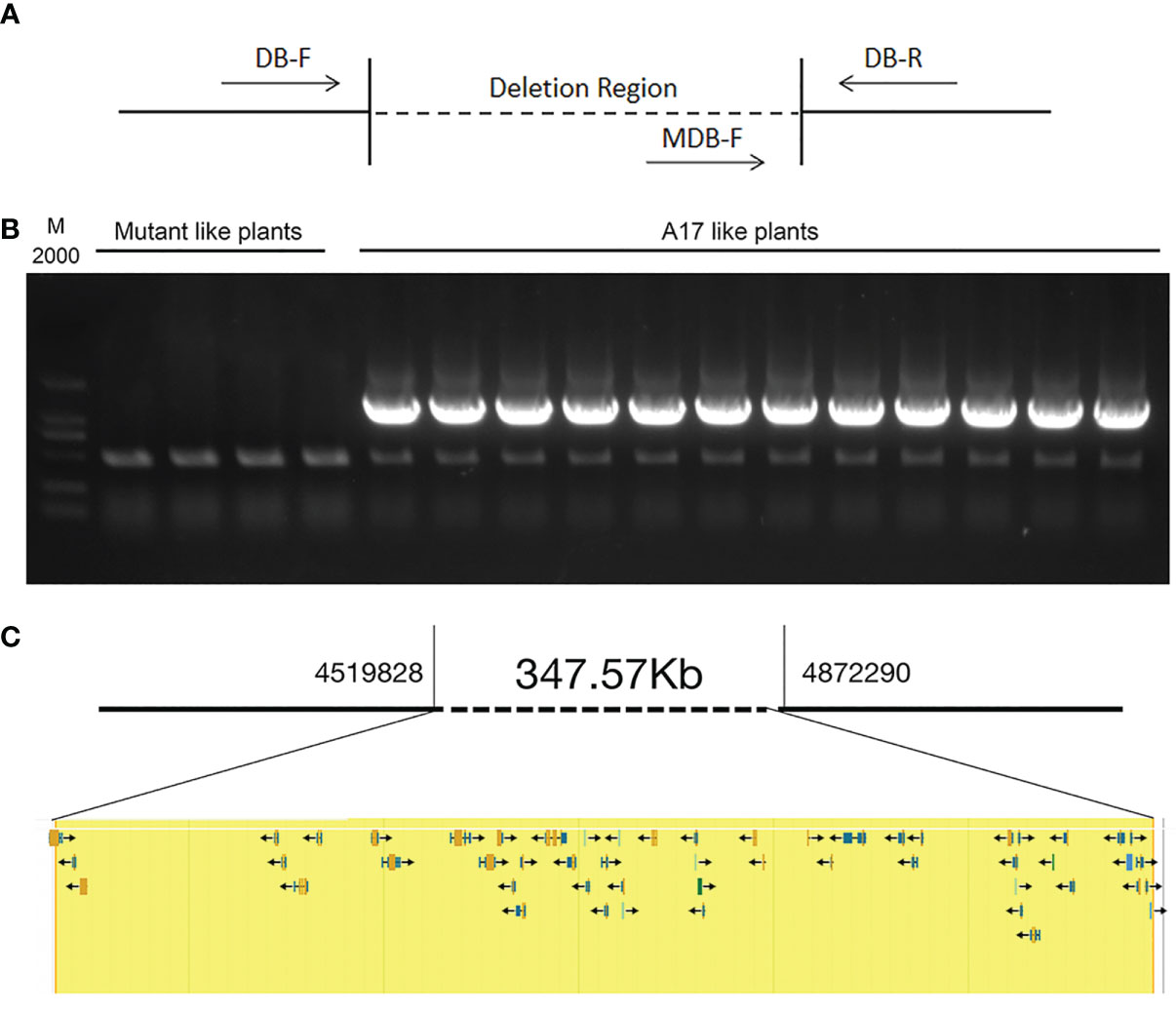
Figure 12 Genetic linkage analysis of FN007 mutant A genetic linkage analysis of FN007 F2 segregating population. (A) PCR analysis was carried out using primers spanning the deletion border in FN007 F2 population; (B) Shown are PCR amplification of deletion border in F2 mutant pools (mutant like plants, 10 plants per pool) and in wild type-like pools (WT like plants, 10 plants per pool). M, DL2000, molecular weight markers. There were no specific bands in F2 mutant pool by MDB-F and DB-R primer sets, while in WT like pools, all of pools could amplify the correct band. These results indicate that the deletion region has linkage with FN007 mutant phenotype; (C) A putative 348 kb deletion segment in FN007 mutant.
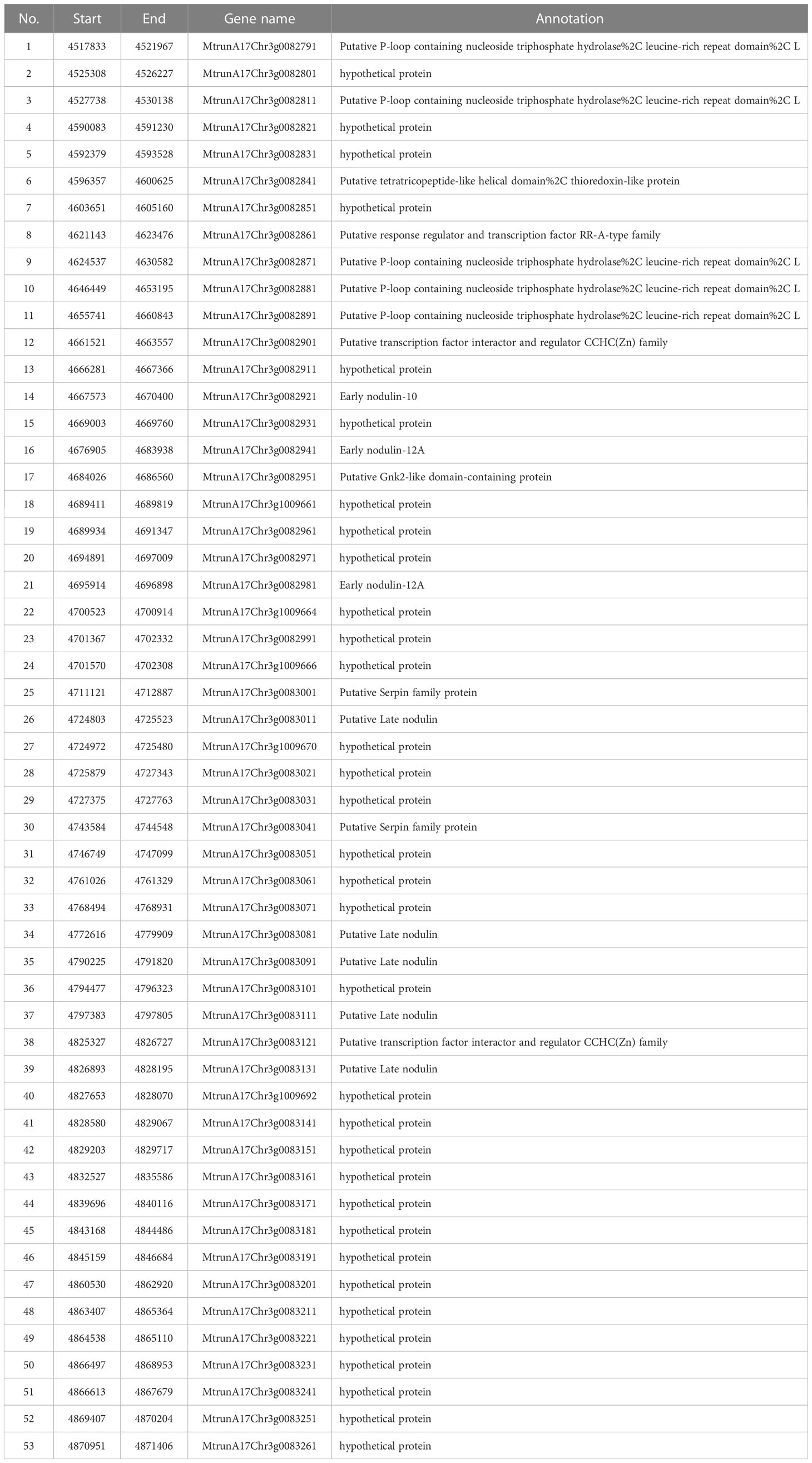
Within the deletion region on chromosome 3, there are 53 annotated genes base on M. truncatula A17 r5.0 genome (Figure 12C; Table 3). This, together with the results of the genetic linkage analysis, suggests that these genes are candidates for the cause of the phenotype of the FN007 mutant. Though we have successfully identified the deletion region in the FN007 mutant, it will take more effort to pinpoint the causal gene. To identify which is the target gene, RNA-seq technology would be used to narrow down the gene list in the deletion region. Finally, genetic complementation assay should be carried out, using Agrobacterium rhizogenes-mediated root transformation.
Discussion
Leguminous plants are able to establish symbiotic relationships with the soil bacteria rhizobia and as a result, develop nodules on their roots, in which biological nitrogen fixation take place. The development of nodules on roots of host plants is triggered by the perception of Nod factors by receptor-like kinase complexes of compatible host plants (Oldroyd et al., 2011; Zipfel and Oldroyd, 2017). Within cytoplasm of plant cells, rhizobia are enclosed by the plant-derived membrane, symbiosome membrane and differentiate into nitrogen fixing bacteroids, and symbiotic nitrogen fixation occurs. Later on, nodule cells senesce and nitrogen fixation stop (Puppo et al., 2005). In M. truncatula, nodules formed on roots are indeterminate. They are cylindrical and contain a series of development zones of different cell types. In mature nodules, there is a senescence zone, where the host cells and bacteroids degenerate. For most leguminous plants, significant nodule senescence is observed during pod filling, when nutrient reallocation and remobilization occur to ensure yield and seed quality. Therefore, increasing active nitrogen fixation in nodules by delaying their senescence might be a powerful strategy to improve nitrogen-use efficiency during seed development (Yu et al., 2023). However, the key factors that orchestrate nodule senescence have not been identified (Wang et al., 2022).
In this study, we isolated and characterized a new nodulation mutant from M. truncatula, FN007. The FN007 mutant was isolated as being defective in symbiotic nitrogen fixation from a M. truncatula deletion mutant collection generated by fast neutron bombardment. Phenotypic characterization showed that the FN007 mutant was defective in later stage of nodule development. In contrast to WT plants, which develop rod shape nodules, the FN007 mutant plants develop round, spherical nodules. A possible explanation for this could be that the deletion of the target gene affects the expression of the leghemoglobin gene in FN007 mutant (Jiang et al., 2021), resulting in white root nodules at all stages of development, and that the expression of genes involved in root nodule development is restricted (Li et al., 2022).
Microscopic observations of the FN007 mutant nodules showed the presence of the meristem zone, infection zone, transition zone, and senescence zone without a functional fixation zone. The symbiosomes and infected cells were degraded starting about 12 dpi. The number of nodules was much higher and the distribution of nodules was scattered in the FN007 mutant than WT plants after inoculation with rhizobia. These observations suggest that nodules developed on the FN007 mutant roots undergo early senescence.
Mutagenesis is a powerful tool for gene functional studies. Fast neutron, a high energy ionization mutagen, has been used in generating mutants in many plant species. A large portion of mutations derived from FN mutagenesis are due to DNA deletions that range in size from a few base pairs to mega base pairs. Many phenotype-associated genes have been successfully identified and characterized. Previously, molecular cloning of the underlying genes from FN mutants relied on a map-based approach, which is time -consuming and limits the number of mutants to be characterized at the molecular level. Recently, several complimentary approaches including genome tiling array-based comparative genomic hybridization (CGH) for DNA copy number variation detection, has been employed to facilitate the characterization of deletion mutants in diverse organisms including animals and plants. These approaches provided new solutions in characterizing with FN mutants. However, low resolution and low accuracy, especially for small deletions limited these methods to apply in identifying mutations for FN mutants.
With the rapid development of next generation sequencing (NGS) technologies, whole genome sequencing (WGS) has become more affordable, providing a new means of identifying causal deletions in FN mutants (Sun et al., 2018). WGS for FN mutants not only increases the sensitivity in detecting mutations on chromosomes, but also improves the reliability and resolution in identifying deletions (Du et al., 2021). In this study, we used it to analyze symbiotic nitrogen fixation deficiency mutant FN007 in M. truncatula and identified the deletion locus that was tightly linked with this phenotype. The causal deletion in the FN007 mutant was confirmed by Tail-PCR amplification, sequencing and genetic linkage analyses.
WGS can be used for large and small deletion identification in many species with reference genome sequences. WGS uses high coverage short reads to examine the large deletions or indel between the reference genome and short reads. In this study, we performed a 30×sequencing depth for mutation detection. Given sufficient sequencing depth, WGS could theoretically detect all potential deletions or indel caused by different mutagenesis. WGS provided a powerful platform to rapidly identify the candidate deletions for FN mutants.
In this study, the FN007 mutant showed a clearly different phenotype from WT under nitrogen-limited and symbiotic condition, so further use of the hairy root transformation technique to rescue the phenotype of the mutant and find the target gene is very important to elucidate the mechanism of symbiotic nitrogen fixation in leguminous plants. This work will provide a reference for most researchers to investigate similar mutant and explore the molecular mechanism of symbiotic nitrogen fixation in leguminous plants.
Conclusions
M. truncatula has been selected as model species for legume-rhizobial symbiosis and nodule development studies. Here, we describe the detail procedures that are being used to characterize SNF mutants in M. truncatula, which provide a reliable solutions to utilize FN mutants for SNF research. Our results demonstrate that the methods used here is feasible in screening and identifying the mutation sites for FN mutants in M. truncatula.
Data availability statement
The data presented in the study are deposited in the NCBI Sequence Read Archive (SRA) repository, accession number is PRJNA983295. The release date is: 2023-06-14. The SRA records will be accessible with the following link: https://www.ncbi.nlm.nih.gov/sra/PRJNA983295.
Author contributions
YC, YS and YM conceived the original research plans. YM, YS, DL, YP and MK performed all the phenotype analysis. YM, YS, DL, WT, RZ, SH, WS and YL performed all bioinformatics analysis. WH and DW performed most of the experiments. YC, YS and YM drafted, wrote, and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the foundation of the Ministry of Education Key Laboratory of Cell Activities and Stress Adaptations (lzujbky-2019-kb05; lzujbky-2021-kb05).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1209664/full#supplementary-material
References
Alonso, J. M., Stepanova, A. N., Solano, R., Wisman, E., Ferrari, S., Ausubel, F. M., et al. (2003). Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in arabidopsis. Proc. Natl. Acad. Sci. U S A. 100 (5), 2992–2997. doi: 10.1073/pnas.0438070100
Barker, D., Bianchi, S., Blondon, F., Dattee, Y., Duc, G., Essad, S., et al. (1990). Medicago truncatula, a model plant for studying the molecular genetics of the rhizobium-legume symbiosis. Plant Mol. Biol. Rep. 8, 40–49. doi: 10.1007/BF02668879
Bejjani, B. A., Shaffer, L. G. (2006). Application of array-based comparative genomic hybridization to clinical diagnostics. J. Mol. Diagn. 8 (5), 528–533. doi: 10.2353/jmoldx.2006.060029
Bolon, Y. T., Haun, W. J., Xu, W. W., Grant, D., Stacey, M. G., Nelson, R. T., et al. (2011). Phenotypic and genomic analyses of a fast neutron mutant population resource in soybean. Plant Physiol. 156 (1), 240–253. doi: 10.1104/pp.110.170811
Bruggemann, E. P., Doan, B., Handwerger, K., Storz, G. (1998). Characterization of an unstable allele of the arabidopsis HY4 locus. Genetics. 149 (3), 1575–1585. doi: 10.1093/genetics/149.3.1575
Chen, Y., Chen, R. (2018). Physical mutagenesis in medicago truncatula using fast neutron bombardment (FNB) for symbiosis and developmental biology studies. Methods Mol. Biol. 1822, 61–69. doi: 10.1007/978-1-4939-8633-0_4
Chen, Y., Wang, X., Lu, S., Wang, H., Li, S., Chen, R. (2017). An array-based comparative genomic hybridization platform for efficient detection of copy number variations in fast neutron-induced medicago truncatula mutants. J. Vis. Exp. 129), 56470. doi: 10.3791/56470
Chen, J., Yu, J., Ge, L., Wang, H., Berbel, A., Liu, Y., et al. (2010). Control of dissected leaf morphology by a Cys(2)His(2) zinc finger transcription factor in the model legume medicago truncatula. Proc. Natl. Acad. Sci. U S A. 107 (23), 10754–10759. doi: 10.1073/pnas.1003954107
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34 (17), i884–ii90. doi: 10.1093/bioinformatics/bty560
Cook, D. R. (1999). Medicago truncatula–a model in the making! Curr. Opin. Plant Biol. 2 (4), 301–304. doi: 10.1016/s1369-5266(99)80053-3
Du, H., Jiao, Z., Liu, J., Huang, W., Ge, L. (2021). Rapid identification of mutations caused by fast neutron bombardment in medicago truncatula. Plant Methods 17 (1), 62. doi: 10.1186/s13007-021-00765-y
Emerson, J. J., Cardoso-Moreira, M., Borevitz, J. O., Long, M. (2008). Natural selection shapes genome-wide patterns of copy-number polymorphism in drosophila melanogaster. Science. 320 (5883), 1629–1631. doi: 10.1126/science.1158078
Geschwind, D. H., Gregg, J., Boone, K., Karrim, J., Pawlikowska-Haddal, A., Rao, E., et al. (1998). Klinefelter's syndrome as a model of anomalous cerebral laterality: testing gene dosage in the X chromosome pseudoautosomal region using a DNA microarray. Dev. Genet. 23 (3), 215–229. doi: 10.1002/(sici)1520-6408(1998)23:3%3C215::aid-dvg7%3E3.0.co;2-x
Gong, J. M., Waner, D. A., Horie, T., Li, S. L., Horie, R., Abid, K. B., et al. (2004). Microarray-based rapid cloning of an ion accumulation deletion mutant in arabidopsis thaliana. Proc. Natl. Acad. Sci. U S A. 101 (43), 15404–15409. doi: 10.1073/pnas.0404780101
Haun, W. J., Hyten, D. L., Xu, W. W., Gerhardt, D. J., Albert, T. J., Richmond, T., et al. (2011). The composition and origins of genomic variation among individuals of the soybean reference cultivar williams 82. Plant Physiol. 155 (2), 645–655. doi: 10.1104/pp.110.166736
Hoffmann, D., Jiang, Q., Men, A., Kinkema, M., Gresshoff, P. M. (2007). Nodulation deficiency caused by fast neutron mutagenesis of the model legume lotus japonicus. J. Plant Physiol. 164 (4), 460–469. doi: 10.1016/j.jplph.2006.12.005
Horvath, B., Domonkos, A., Kereszt, A., Szucs, A., Abraham, E., Ayaydin, F., et al. (2015). Loss of the nodule-specific cysteine rich peptide, NCR169, abolishes symbiotic nitrogen fixation in the medicago truncatula dnf7 mutant. Proc. Natl. Acad. Sci. U S A. 112 (49), 15232–15237. doi: 10.1073/pnas.1500777112
Infante, J. J., Dombek, K. M., Rebordinos, L., Cantoral, J. M., Young, E. T. (2003). Genome-wide amplifications caused by chromosomal rearrangements play a major role in the adaptive evolution of natural yeast. Genetics. 165 (4), 1745–1759. doi: 10.1093/genetics/165.4.1745
Jiang, S., Jardinaud, M.-F., Gao, J., Pecrix, Y., Wen, J., Mysore, K., et al. (2021). NIN-like protein transcription factors regulate leghemoglobin genes in legume nodules. Science. 374 (6567), 625–628. doi: 10.1126/science.abg5945
Kim, M., Chen, Y., Xi, J., Waters, C., Chen, R., Wang, D. (2015). An antimicrobial peptide essential for bacterial survival in the nitrogen-fixing symbiosis. Proc. Natl. Acad. Sci. U S A. 112 (49), 15238–15243. doi: 10.1073/pnas.1500123112
Lakshmi, B., Hall, I. M., Egan, C., Alexander, J., Leotta, A., Healy, J., et al. (2006). Mouse genomic representational oligonucleotide microarray analysis: detection of copy number variations in normal and tumor specimens. Proc. Natl. Acad. Sci. U S A. 103 (30), 11234–11239. doi: 10.1073/pnas.0602984103
Li, H., Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics. 25 (14), 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence Alignment/Map format and SAMtools. Bioinformatics. 25 (16), 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, X., Lassner, M., Zhang, Y. (2002). Deleteagene: a fast neutron deletion mutagenesis-based gene knockout system for plants. Comp. Funct. Genomics 3 (2), 158–160. doi: 10.1002/cfg.148
Li, Y., Pei, Y., Shen, Y., Zhang, R., Kang, M., Ma, Y., et al. (2022). Progress in the self-regulation system in legume nodule development-AON (Autoregulation of nodulation). Int. J. Mol. Sci. 23 (12). doi: 10.3390/ijms23126676
Liu, Y. G., Chen, Y. L. (2007). High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. BioTechniques. 43 (5), 649–650. doi: 10.2144/000112601
Men, A., Laniya, T., Searle, I., Iturbe-Ormaetxe, I., Gresshoff, I., Jiang, Q., et al. (2002). Fast neutron mutagenesis of soybean ( glycine soja l.) produces a supernodulating mutant containing a Large deletion in linkage group h. Gen. Lett. 3, (3). doi: 10.1166/gl.2002.017
Mergaert, P., Kereszt, A., Kondorosi, E. (2020). Gene expression in nitrogen-fixing symbiotic nodule cells in medicago truncatula and other nodulating plants. Plant Cell. 32 (1), 42–68. doi: 10.1105/tpc.19.00494
Mergaert, P., Uchiumi, T., Alunni, B., Evanno, G., Cheron, A., Catrice, O., et al. (2006). Eukaryotic control on bacterial cell cycle and differentiation in the rhizobium-legume symbiosis. Proc. Natl. Acad. Sci. U S A. 103 (13), 5230–5235. doi: 10.1073/pnas.0600912103
Mitra, R. M., Gleason, C. A., Edwards, A., Hadfield, J., Downie, J. A., Oldroyd, G. E., et al. (2004). A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. U S A. 101 (13), 4701–4705. doi: 10.1073/pnas.0400595101
Oldroyd, G. E. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11 (4), 252–263. doi: 10.1038/nrmicro2990
Oldroyd, G. E., Murray, J. D., Poole, P. S., Downie, J. A. (2011). The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 45, 119–144. doi: 10.1146/annurev-genet-110410-132549
Pecrix, Y., Staton, S. E., Sallet, E., Lelandais-Briere, C., Moreau, S., Carrere, S., et al. (2018). Whole-genome landscape of medicago truncatula symbiotic genes. Nat. Plants. 4 (12), 1017–1025. doi: 10.1038/s41477-018-0286-7
Penmetsa, R. V., Cook, D. R. (2000). Production and characterization of diverse developmental mutants of medicago truncatula. Plant Physiol. 123 (4), 1387–1398. doi: 10.1104/pp.123.4.1387
Perry, G. H., Yang, F., Marques-Bonet, T., Murphy, C., Fitzgerald, T., Lee, A. S., et al. (2008). Copy number variation and evolution in humans and chimpanzees. Genome Res. 18 (11), 1698–1710. doi: 10.1101/gr.082016.108
Puppo, A., Groten, K., Bastian, F., Carzaniga, R., Soussi, M., Lucas, M. M., et al. (2005). Legume nodule senescence: roles for redox and hormone signalling in the orchestration of the natural aging process. New Phytol. 165, 683–701. doi: 10.1111/j.1469-8137.2004.01285.x
Rogers, C., Wen, J., Chen, R., Oldroyd, G. (2009). Deletion-based reverse genetics in medicago truncatula. Plant Physiol. 151 (3), 1077–1086. doi: 10.1104/pp.109.142919
Rostoks, N., Schmierer, D., Mudie, S., Drader, T., Brueggeman, R., Caldwell, D. G., et al. (2006). Barley necrotic locus nec1 encodes the cyclic nucleotide-gated ion channel 4 homologous to the arabidopsis HLM1. Mol. Genet. Genomics 275 (2), 159–168. doi: 10.1007/s00438-005-0073-9
Roy, S., Liu, W., Nandety, R. S., Crook, A., Mysore, K. S., Pislariu, C. I., et al. (2020). Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell. 32 (1), 15–41. doi: 10.1105/tpc.19.00279
Stacey, G., Libault, M., Brechenmacher, L., Wan, J., May, G. D. (2006). Genetics and functional genomics of legume nodulation. Curr. Opin. Plant Biol. 9 (2), 110–121. doi: 10.1016/j.pbi.2006.01.005
Sun, L., Ge, Y., Bancroft, A. C., Cheng, X., Wen, J. (2018). FNBtools: a software to identify homozygous lesions in deletion mutant populations. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00976
Tadege, M., Wen, J., He, J., Tu, H., Kwak, Y., Eschstruth, A., et al. (2008). Large-Scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume medicago truncatula. Plant J. 54 (2), 335–347. doi: 10.1111/j.1365-313X.2008.03418.x
Tang, H., Krishnakumar, V., Bidwell, S., Rosen, B., Chan, A., Zhou, S., et al. (2014). An improved genome release (version Mt4.0) for the model legume medicago truncatula. BMC Genomics 15, 312. doi: 10.1186/1471-2164-15-312
Wang, H., Chen, J., Wen, J., Tadege, M., Li, G., Liu, Y., et al. (2008). Control of compound leaf development by FLORICAULA/LEAFY ortholog SINGLE LEAFLET1 in medicago truncatula. Plant Physiol. 146 (4), 1759–1772. doi: 10.1104/pp.108.117044
Wang, D., Dong, W., Murray, J., Wang, E. (2022). Innovation and appropriation in mycorrhizal and rhizobial symbioses. Plant Cell. 34 (5), 1573–1599. doi: 10.1093/plcell/koac039
Wang, H., Li, G., Chen, R. (2006). Fast neutron bombardment (FNB) mutagenesis for forward and reverse genetic studies in plants. In Floriculture, ornamental and plant biotechnology: advances and topical issues, ed. Teixeira da Silva, J. A. (Isleworth: Global Science Books), 629–639.
Werner, J. D., Borevitz, J. O., Warthmann, N., Trainer, G. T., Ecker, J. R., Chory, J., et al. (2005). Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc. Natl. Acad. Sci. U S A. 102 (7), 2460–2465. doi: 10.1073/pnas.0409474102
Xi, J., Chen, Y., Nakashima, J., S-m, W., Chen, R. (2013). Medicago truncatula esn1 defines a genetic locus involved in nodule senescence and symbiotic nitrogen fixation. Mol. Plant Microbe Interact. 26 (8), 893–902. doi: 10.1094/MPMI-02-13-0043-R
Young, N. D., Debelle, F., Oldroyd, G. E., Geurts, R., Cannon, S. B., Udvardi, M. K., et al. (2011). The medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 480 (7378), 520–524. doi: 10.1038/nature10625
Yu, H. X., Xiao, A. F., Wu, J. S., Li, H. X., Duan, Y., Chen, Q. S., et al. (2023). GmNAC039 and GmNAC018 activate the expression of cysteine protease genes to promote soybean nodule senescence. Plant Cell, koad129. doi: 10.1093/plcell/koad129
Keywords: Medicago truncatula (barrel medic), symbiotic nitrogen fixation (SNF), nodule development, legume, whole genome sequencing (WGS)
Citation: Shen Y, Ma Y, Li D, Kang M, Pei Y, Zhang R, Tao W, Huang S, Song W, Li Y, Huang W, Wang D and Chen Y (2023) Biological and genomic analysis of a symbiotic nitrogen fixation defective mutant in Medicago truncatula. Front. Plant Sci. 14:1209664. doi: 10.3389/fpls.2023.1209664
Received: 21 April 2023; Accepted: 13 June 2023;
Published: 28 June 2023.
Edited by:
Jiejun Xi, Northwest A&F University, ChinaReviewed by:
Jiangqi Wen, Oklahoma State University, United StatesTianzuo Wang, Chinese Academy of Sciences (CAS), China
Huairong Pan, Hunan University, China
Copyright © 2023 Shen, Ma, Li, Kang, Pei, Zhang, Tao, Huang, Song, Li, Huang, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuhui Chen, cyh@lzu.edu.cn
†These authors have contributed equally to this work and share first authorship
 Yitong Shen
Yitong Shen