
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 18 July 2023
Sec. Plant Symbiotic Interactions
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1205894
Bacillus species and their metabolites have potential alternative uses as chemical pesticides that can limit the growth of potential plant pathogens and enhance crop productivity. The aim of this study was to investigate the potential of Bacillus subtilis FJ3 for promoting plant growth and controlling fungal plant pathogens. The study evaluated the ability of the strain to promote plant growth in vitro by characterizing its growth-promoting traits, which included the production of hydrolytic enzymes, indole acetic acid, siderophores, biofilm formation, and phosphate solubilization. Polymerase Chain Reaction (PCR) testing revealed that strain FJ3 has the potential to produce lipopeptides such as fengycin, surfactin, mycosubtilin, and pilpastatin. Through in vitro antagonism testing it was demonstrated that strain FJ3 is able to inhibit Fusarium oxysporum by 52% compared to the untreated control and was antagonistic against Aspergillus flavus, Aspergillus niger, and Rhizopus oryzae using a dual method. The minimum inhibitory concentration of Bacillus crude extract resulted in a 92%, 90%, 81.5%, and 56% growth inhibition of Fusarium oxysporum, A. niger, A. flavus, and Rhizopus oryzae, respectively. In FT-IR and GC-MS analysis of crude LPs extract, the transmission and mass spectrum confirmed the existence of aforesaid lipopeptides containing β-fatty acids with chain lengths ranging from C14 to C21 in which the majority were saturated fatty acids. Greenhouse experimentation revealed that Bacillus strain FJ3 and its metabolites significantly diminished the disease incidence with an average reduction of 31.56%. In sterilized soil, FJ3 and its metabolites caused 24.01% and 10.46% growth promotion, respectively, in chickpea. The results demonstrated that Bacillus strain FJ3 has broad-spectrum antifungal and plant growth-promoting applications and could be a promising candidate for development into a commercialized biobased product for use in sustainable agriculture practice.
With the increasing human population the need for increased food production and to ensure global food security will continue to mount. Discrepancies between food supply and demand create an imbalance that has dramatically worsened during the past few decades (Pinstrup-Andersen and Pandya-Lorch, 1998). During the next 35 years, it seems likely that food security, water scarcity, and environmental pollution will become increasingly important issues with regard to crop agriculture (Shiferaw et al., 2013). Numerous biotic and abiotic factors are responsible for productivity losses each year. Reductions in crop yield occur both qualitatively and quantitatively either in the field (pre-harvest) or during storage (post-harvest) due to pests and pathogens. Plant pathogens are major threats to agricultural outputs that range in complexity from the simplest viroid to more sophisticated pathogens such as bacteria, viruses, fungi, nematodes, and oomycetes that cause a wide range of plant infections (Seyi-Amole and Onilude, 2021). Recent research indicated that phytopathogenic fungi are responsible for 40% of crop loss worldwide at pre- and post-harvesting stages (De Angelis et al., 2022). Fungal pathogens are very problematic because of their adaptability patterns and capacity to persist in low-nutrient environments (Paulussen et al., 2017). Being an agricultural economy, Pakistan faces huge losses due to plant infections, particularly by Fusarium and Aspergillus species. Fusarium spp. causes infections such as root rot, crown rot, and vascular wilts in more than 2000 plant species (Zehra et al., 2022). It causes postharvest loss of grains, pulses, dry fruits, and spices by secreting several types of mycotoxins (patulin, aflatoxins, ochratoxins, zearalenone, and fumonisin deoxynivalenol), that are not only food poisoning agents but can also cause cancer, kidney failure, liver damage, and paralysis in humans (Awuchi et al., 2021).
Existing agriculture management strategies chiefly rely on synthetic chemicals that are either recalcitrant or may create a significant ecological disturbance. Under these circumstances, promotion of green technologies and bio-based products need to be promoted (Gontard et al., 2018). Furthermore, excessive input of pesticides and antimicrobials affects the density and diversity of beneficial microbial flora. Therefore, there is an urgent need to shift from conventional agricultural methods to “Green” and sustainable processes/products (Singh et al., 2019).
Bacillus-based biocontrol products are considered essential as biopesticides against a wide variety of plant pathogens. Typically, Bacillus is given priority over other biocontrol agents due to some of its basic properties such as rapid replication, resistance to adverse environmental conditions, and development and longevity of spores (Albayrak, 2019). Bacillus species isolated from rhizospheric soil can act as an effective plant growth promoter, induce systemic resistance in the host plant, and produce a broad range of antimicrobial compounds such as antibiotics, lipopeptides, and enzymes (Miljaković et al., 2020). Moreover, they act as major competitors for space and nutrients with pathogens in the soil (Shafi et al., 2017). Currently, a number of registered products and those in the development stage are derived from Bacillus species. Reported biocontrol agents (BCAs) from Bacillus species such as Bacillus subtilis, Bacillus pumilis, and Bacillus thuriengiensis are characterized by their antagonist activity and manifest fungistatic and fungicidal effects (Rajkumari and Pandey, 2019). Bacillus spp. support plant growth through phosphate solubilization, nitrogen fixation, and phytohormone synthesis, in addition to being the most promising biocontrol agents (Khan et al., 2022).
Bacillus spp. antagonist mechanism is predicted to be either by the production of cyclic lipopeptides (LPs) such as fengycin or plipastatin, surfactin, iturin, bacillomycin, mycosubtilin or through production of lytic enzymes such as chitinases, cellulase, endoglucanase, or hemicellulase which suppresses the growth of pathogens (Dimkić et al., 2022). LPs in Bacillus are synthesized by multi-modular enzyme systems known as non-ribosomal peptide synthetases (NRPS) by the thio-template mechanism (Caulier et al., 2019). Rearrangement of the modular organization of these enzymes within the NRPS genetic clusters is responsible for the production of a wide variety of lipopeptides. Among all the diversity, LPs of the families surfactin, iturin, and fengycin or plipastatin are regarded as the most active. These lipopeptides not only manifest antifungal activities but also play crucial roles in root colonization and plant growth-promoting activities (Nahas et al., 2021).
Bacillus subtilis strains are potentially effective and environmentally friendly agents for disease control. Recent studies have shown that treating these strains with nanoparticles coated with gold, aluminum, and silver can promote plant growth and prevent the growth of harmful fungi in the rhizosphere, indicating their potential as nano-biofertilizers (do Espirito Santo Pereira et al., 2021). Bacillus subtilis strains can activate natural defense mechanisms in plants against a wide range of pathogens, making them useful biocontrol agents (Hashem et al., 2019). They are considered safe for use in the food industry and are easy to formulate and store because of their endospore formation ability as their endospores are resistant to heat, UV radiation, organic solvents, and desiccation. Furthermore, B. subtilis strains can maintain their ability to activate plant defense responses under adverse conditions (Zhang et al., 2020). Considering these issues, the current research was focused on the isolation and characterization of lipopeptide-producing Bacillus strains as biocontrol agents against some of the most devastating plant pathogens. The ALPs were characterized and tested in-vitro against selected fungal pathogens and their plant growth stimulatory role was estimated.
Fusarium oxysporum was isolated from chickpea plants, from Bakkar (Pakistan) and was used in the present study. The isolated fungus was stored on sabouraud dextrose agar (SDA) at 4°C. For further experimentation, the isolate was subcultured on an SDA medium at 28 ± 1°C for 7 days.
The 13 Bacillus strains scrutinized in this study were isolated from the soil samples of various agricultural fields of the Swat district. Preliminary screening was performed on a minimal salt medium, which was used for the enrichment of samples and 1 gram of soil was suspended in 10 mL of sterile distilled water and serially diluted. In total, 9 dilutions of each sample were made. An aliquot of 100 µL from 10−3 and 10−9 dilutions was distributed on nutrient agar and Hi-chrome Bacillus agar plates for culturing at 37°C for 24 h. Then, morphological and biochemical identification was carried out via microscopy and biochemical tests.
The bacterial strains were selected based on preliminary screening. The genomic DNA of selected bacterial strains and isolated fungal strains were extracted through the PCI (phenol, chloroform, isoamyl) method (Munir et al., 2018). Identification of fungal strains was validated by 18S rDNA and 16S rDNA for bacteria using universal primer (Chen et al., 2014) sequences. Sequencing was done at the Alpha genomics. The sequences were edited using the MEGA software version X. The nucleotide was compared to the NCBI database through the Basic Local Alignment Search Tool (NCBI) algorithm. The sequences were further submitted to GeneBank for accession number. In order to check the taxonomic placement of test organisms a phylogenetic tree was constructed on MEGA software version X (Dissanayake et al., 2020).
NBRIP agar medium was utilized to evaluate the potential of a selected bacterial strain to solubilize phosphate. The medium comprised the following: 10 g L−1 glucose, 5 g L−1 MgCl2.6H2O, 0.25 g L−1 MgSO4.7H2O, 5 g L−1 Ca3(PO4)2, 0.2 g L−1 KCL, 0.1 g L−1 (NH4)2SO4, 15% agar, and pH 7. Then, 5% tri-calcium phosphate was added to the medium, and bacterial strains were spot-inoculated on the NBRIP agar medium, followed by incubation at 30°C for 7 days (Mohamed et al., 2019).
Chrome azurol sulphonate agar medium was used to identify siderophore production by selected bacterial strains. After 5 days of incubation at 28°C, the change in medium color from green to yellow indicates positive results (Grobelak and Hiller, 2017).
Indole acetic acid production was determined by inoculating selected bacterial strains in 30 mL of LB broth supplemented with tryptophan and slowly shaken at 28°C for three to five days. After incubation, the broth was subjected to centrifugation at 10,000 rpm and 4°C for 10 minutes. A two mL portion of the cell-free supernatant was mixed with 2 mL of Salkowski reagent, which is composed of 35% HClO4 and 0.5 M FeCl3 (Ullah and Bano, 2019). The mixture was then kept in the dark at room temperature for 20 to 30 min; the appearance of a pink color indicated the production of IAA.
Isolated Bacillus strains were assayed for the production of protease, cellulase (endoglucanase), and amylase enzymes following the method of Durairaj et al. (2017). Qualitative protease activity was performed on Modified Basal Media (MM) containing 5g L−1 casein. The ability of the bacterial strains to produce cellulase was evaluated on plates containing 1% carboxymethylcellulose and 1M NaCl while the starch iodine method was employed for amylase assay.
The Congo Red assay, described by Freeman et al. (1989), was used for confirmation of biofilm formation. The selected bacterial strain was streaked onto Congo Red agar, which comprised brain heart infusion (BHI) agar, sucrose, and Congo Red dye. The plate was then incubated at 37°C for 48 h.
Polymerase Chain Reaction (PCR) testing was performed to identify the lipopeptide biosynthesis genes for fengycin, plipastatin, surfactin, and mycosubtilin. Primers were checked on the NRPS-producing Bacillus strain FJ3. Primers were ordered according to Tapi et al. (2010) from Macrogen. A list of the primers is given in Table 1. Genomic DNA concentration from each extract was evaluated from Nanodrop. The final DNA dilution used for PCR was between 10 and 30 ng µL−1.
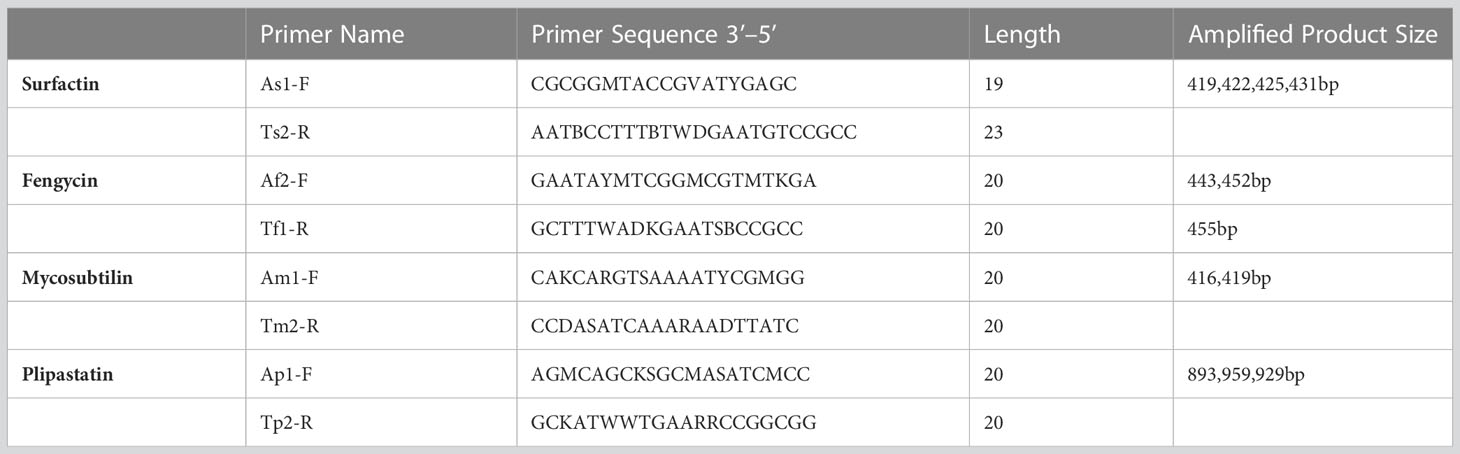
Table 1 List of primers and their characteristics ordered for detection of lipopeptide biosynthetic genes (Tapi et al., 2010).
PCR conditions for each set of primers were experimentally determined by running an annealing temperature gradient in a gradient thermocycler PCR (Bio-Rad). The thermal cycler was programmed for 30 cycles of PCR, including initial denaturation at 94°C for 3 min, followed by a denaturation step at 94°C for 1 min. The annealing temperature for the fengycin gene was set at 45°C, for plipastatin at 58°C, and surfactin at 43°C while for mycosubtilin it was set at 45°C. Extension for each set of primers was done at 72°C for 45 sec, (except for plipastatin for 75 sec at 72°C), followed by a final extension step of 72°C for 1 min (Caracciolo, 2018).
Three μL of amplified PCR product mixed with loading dye was loaded into the Agarose gel 2X stained with Et.Br. For band size estimation, a gene ladder of 1 kb was used. A DNA ladder of 1 kb was ordered from Solis Bio Dyne. The gels were observed under a UV illuminator at low and high resolutions to detect DNA bands, and photographs were captured using a Kodak DC 290 digital camera from New York, USA. Positive PCR results were indicated when bands of appropriate size were visualized under a UV illuminator (Weal Tax).
The antagonistic effect of a bacterial strain against isolated fungal strains of Fusarium oxysporum was assessed. The spore suspension method was employed for fungal culturing (Jangir et al., 2018). The pathogenic fungal strains were transferred to sabouraud dextrose agar (SDA) plates prior to use. The cell-free supernatants of bacterial strains were taken after centrifugation. The Bacillus strain (72 h growth) was inoculated at an equidistant central point via the agar well diffusion method. The plates were sealed with parafilm and incubated at 28 ± 1°C. The inhibition zones were recorded using a scale (Chopra et al., 2020). Moreover, growth inhibition of mycelium by the potentially best Bacillus sp. was documented from the 5th to the 10th day after confrontation.
Using a dual culture assay reported by Limtong et al. (2020) with slight alterations, 5 mm plugs of fungi from SDA plates were inoculated at 3 cm apart from the dish edge. Antagonistic Bacillus strains cultured in nutrient broth were streaked at 3 cm from the inoculated fungal plug. Control plates contained only fungal inoculum. After incubating the plates at 28 ± 2°C for 7 days, the percentage of inhibition of fungal growth by Bacillus strain was calculated using the following method of Limtong et al. (2020):
A solvent extraction technique was used for the initial isolation of lipopeptides from selected bacterial strains using ethyl acetate (Alajlani et al., 2016). Antimicrobial activity was checked by scratching and then dissolving the crude extract in methanol. Then, the antifungal activity was analyzed against selected phytopathogens by the cell diffusion method (Kaur et al., 2017).
The micro-dilution method was used for the determination of the antimicrobial susceptibility of the bacterial strains. The 96-well microtiter plates were inoculated with SDA broth (70 µL) crude extract (20 µL) and fungal spore suspension (10 µL) making the total mixture 100 µL. After inoculation, microtiter plates were incubated for 30 minutes at 27°C, and absorbance was noted using an ELISA plate reader at 630 nm. Then the development was computed after 48 h at 27°C, 630 nm using an ELISA plate reader. The experiment was run in triplicate. The formula for computing the susceptibility is (Juboi, 2017).
Where ΔC = Corrected absorbance after 48 h.
ΔT = Absorbance calculated after 30 min.
FT-IR is a fast and inexpensive technique to characterize and identify functional groups of lipopeptides produced by Bacillus species. The crude metabolite was extracted using solvent extraction and measurements were carried out in transmittance mode with a wavelength in the range of 400–4,000 cm (Biniarz et al., 2017).
GC-MS analysis was performed on the extracted metabolite to confirm the presence of both free and esterified fatty acids. The process involved hydrolytic cleavage of the links between the carbohydrate and peptide portion of the lipopeptide and the lipid portion, followed by derivatization of the resulting fatty acid chains into fatty acid methyl esters (FAME) to aid in GC-MS analysis. The samples were dissolved in methanol and filtered through a 0.22 µm pore size syringe filter (Yakimov et al., 1995).
Chickpea seeds of the “Bittal 2016” variety were acquired from the National Agriculture Research Center (NARC) in Islamabad, Pakistan. The seeds were treated by immersing them in a solution of 5% NaOCl for 4 min, followed by multiple rinses with autoclaved water to eliminate any remaining NaOCl residues. Seeds were further disinfected with 75% alcohol and dried in a biosafety cabinet on filter paper. The soil was mixed with sand at a 2:1 proportion and autoclaved at 121°C for 21 min. Respective sterilized and un-sterilized soil was inoculated with the pathogenic fungus according to the protocol of Manghwar et al. (2021) with a few minor changes. Two seeds were sown in each pot. Five mL each of selected antagonistic Bacillus species was cultured in nutrient broth at 37°C and the cell-free supernatant broth was used as a bioprimer at the time of sowing. The seeds without fungal and bioprimers were regarded as negative controls and with fungus only as positive controls. Each treatment consisted of three replications including controls. The pots were placed in a greenhouse and irrigated regularly to maintain soil humidity. The chlorophyll content was measured using a SPAD-502 meter, which gives readings without damaging the leaves and, during measurement, the leaf remains attached to the plant. Harvesting was done 6 weeks after sowing. The percentage of seed germination, seedling vigor index (SVI), disease incidence, and disease control were calculated as described by Limtong et al. (2020) with an additional variable, the percentage of growth increase.
XLSTAT® software was utilized for data analysis and ANOVA was performed. To compare means, Duncan’s multiple range test was applied, and statistical significance was determined at a significance level of p ≤ 0.05.
A total of 13 purified bacterial isolates were obtained after the enrichment of the soil. These isolates were subjected to initial screening after which only one strain FJ3 was selected, as it had the greatest ability to inhibit the growth of plant pathogens. The isolated bacterial strain FJ3 and fungal strain FJ81 were further identified as Bacillus subtilis and Fusarium oxysporum via 16s rRNA and 18s rRNA gene sequencing. The taxonomic tree indicated a relationship of both Bacillus and FOC with later formerly identified Bacillus strains and Fusarium oxysporum; these are highlighted in colored text. The gene sequence of both FJ3 and FJ81 was submitted to the GeneBank database and accession numbers were assigned (FJ3 OQ073531 and FJ81 OQ073532). Light microscopy results of FJ3 revealed Gram-positive rods and motile and single cells. Identification characteristics are presented in Figures 1A, B, 2A, B.

Figure 2 Neighborhood joining method showing the phylogenetic position of (A) FJ3 and (B) FJ81 other related taxa of the genera Bacillus and Fusarium based on 16s rRNA and 18sRNA gene sequences. Bootstrap value is > 50%.
To determine the phosphate solubilizing activity of selected bacterial strain FJ3, the strain was point inoculated at the center on NBRIP media. The results showed that strain FJ3 could solubilize tricalcium phosphate in large quantities. The halo zone around the colony indicated phosphate solubilization (Figure 3A).
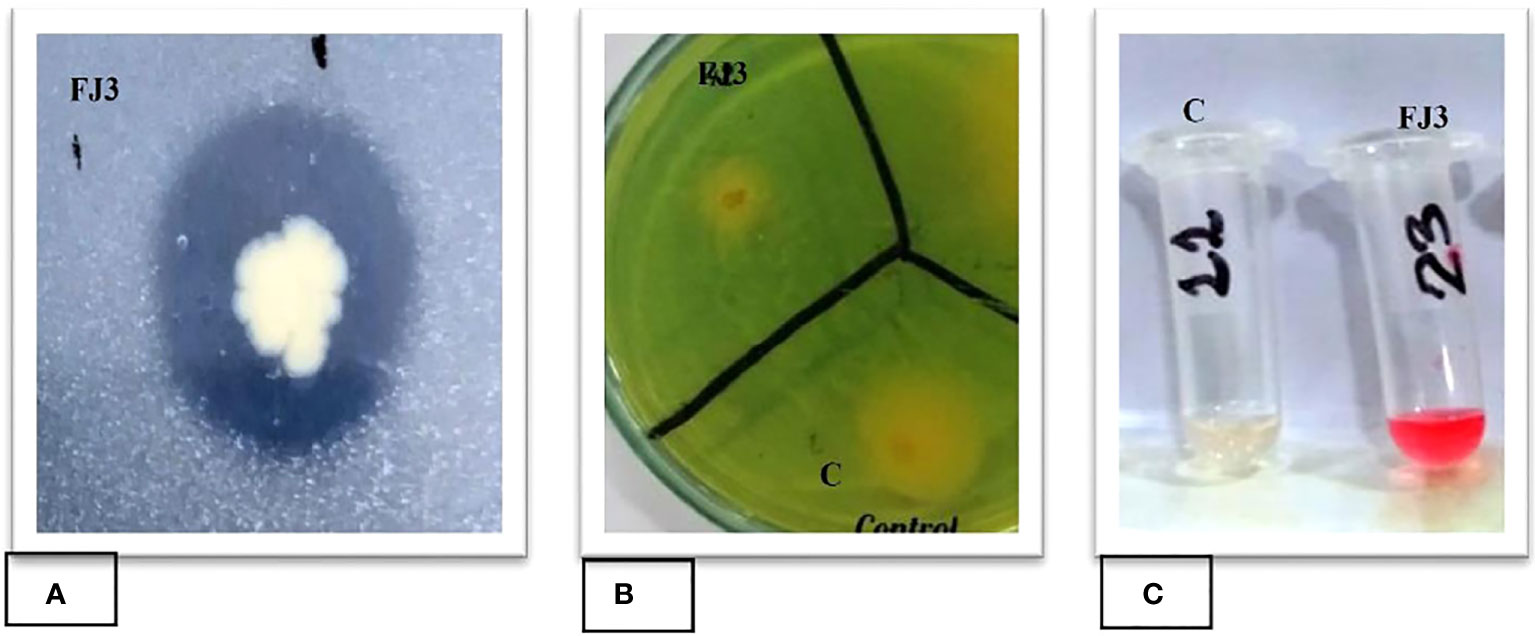
Figure 3 Plant growth promoting traits of strain FJ3: (A) Phosphate solubilization, (B) Siderophore production, and (C) IAA production.
Chrome Azurol Sulphonate (CAS) agar media was used to determine the siderophore production activity of both selected strains. The change in medium color from green to yellow indicates the siderophore ability of both strains (Figure 3B).
Bacillus subtilis strain FJ3 was assessed for its indole acetic acid production ability. The FJ3 strain was inoculated in Luria Bertani broth with tryptophan. The appearance of pink color after adding Salkowski reagent in cell-free supernatant indicates a positive result. Figure 3C showed that strain FJ3 secreted indole acetic acid.
The selected Bacillus subtilis strain FJ3 manifested positive results for protease, amylase, and endoglucanase. Results are depicted in Figure 4.
To determine the ability of Bacillus strains to form a biofilm, Congo Red agar was utilized. Wild-type Bacillus, having the gene for TasA amyloid protein, will appear red while mutated strains will not bind to the dye. The Bacillus subtilis strain FJ3 appeared red, indicating its potential to produce biofilm (Figure 5).
Primer pairs of As1-F/Ts2-R, Af2-F/Tf1-R, Ap1-F/Tp12-R, and Am1-F/Tm1-R were used to amplify surfactin, fengycin, plipastatin, and mycosubtilin genes, respectively. The NRPS gene clusters for mycosubtilin, surfactin, and fengycin in FJ3 were detected using multiplex PCR. In Bacillus strain FJ3 surfactin, mycosubtilin, and fengycin were detected in the range of 420–450 bp and for plipastatin in the range of 890–950 bp, suggesting a positive result for the presence aforesaid lipopeptides. The results of PCR testing are illustrated in Figure 6.
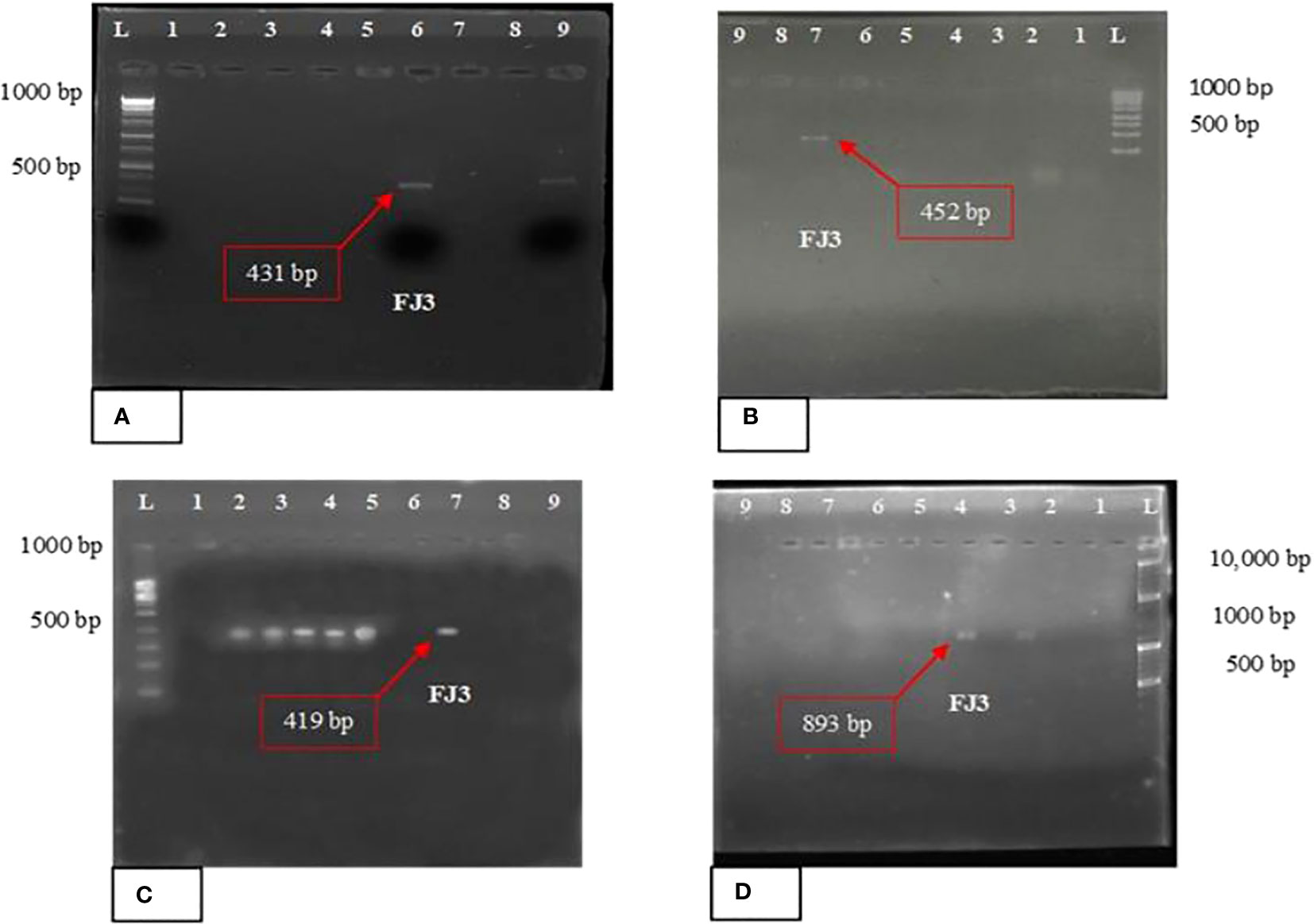
Figure 6 PCR amplification with different primer pairs of (A) surfactin, (B) fengycin, (C) mycosubtilin, and (D) plipastatin. Bacillus strain FJ3 L = Ladder of 1kb and 10kb.
Bacillus subtilis FJ3 showed broad-spectrum antifungal activity against F. oxysporum and other tested fungal phytopathogens. In a dual culture assay, FJ3 inhibited 52.92± 0.72% mycelial growth of F. oxysporum when incubated at 28 ± 1°C (Figure 7).
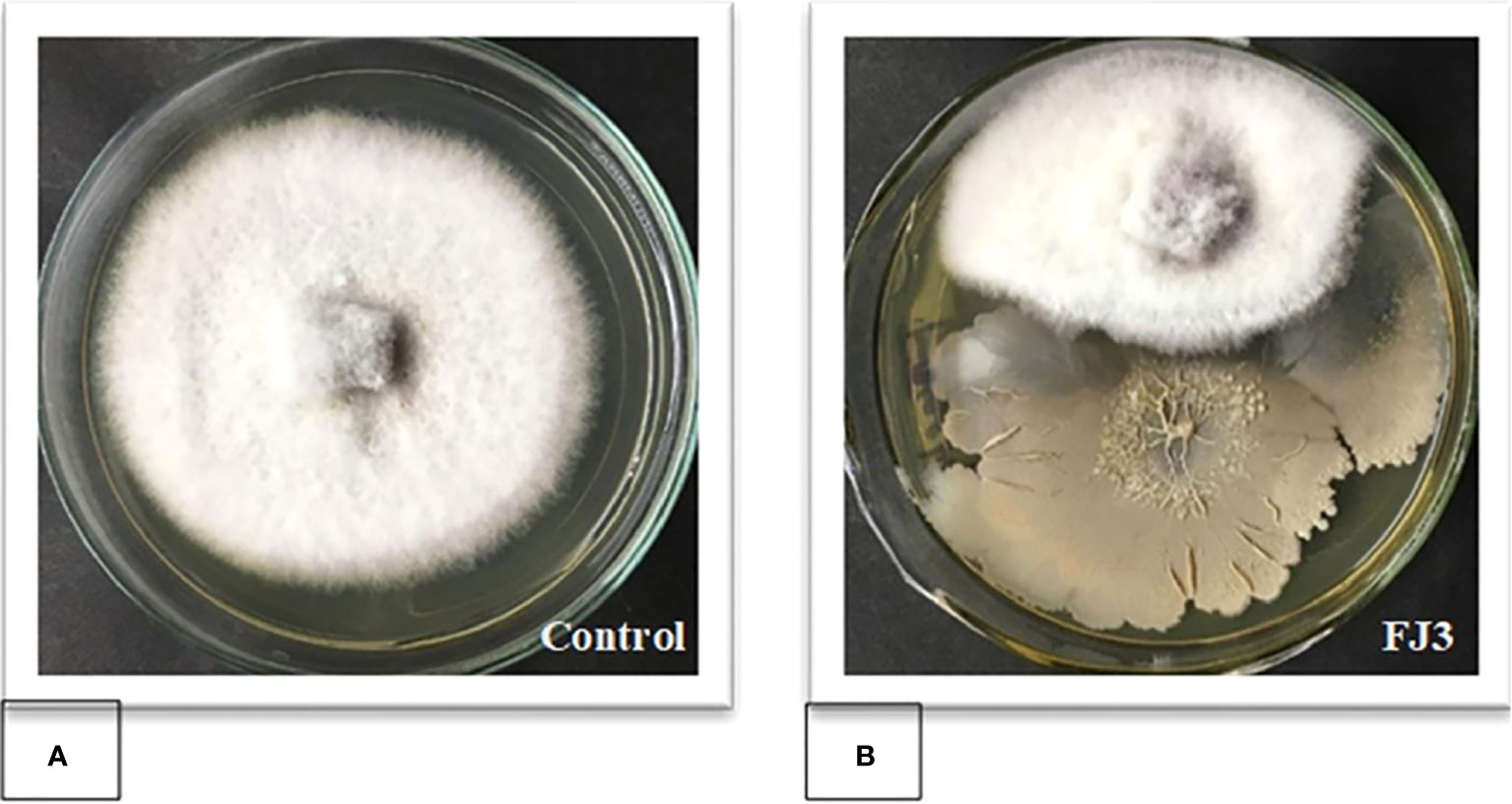
Figure 7 Dual culture assay of Bacillus strain FJ3 against F. oxysporum to measure the percentage of growth inhibition. (A) Control (F. oxysporum). (B) Bacillus subtilis FJ3 with F. oxysporum.
In an agar well diffusion assay B. subtilis FJ3 showed antifungal activity in an SDA medium against tested fungal pathogens A. niger, A. flavus, R. oryzae, and F. oxysporum. The results of in vitro antifungal activity indicated that the cell-free supernatant obtained from B. subtilis FJ3 had significantly higher inhibitory activity, 53 mm, against F. oxysporum, 40 mm for A. niger, 42 mm against A. flavus, and 38 mm against R. oryzae (Figure 8).
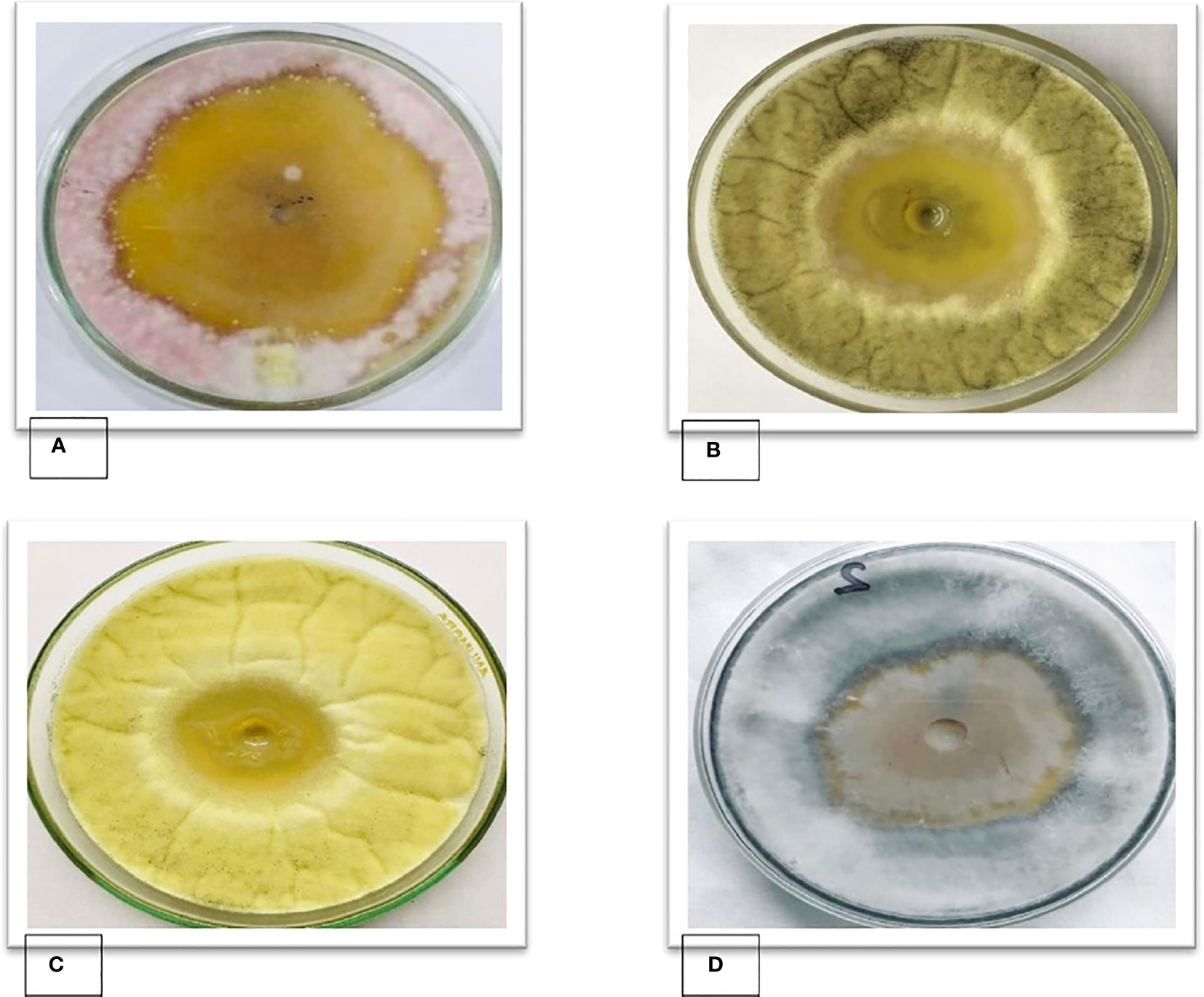
Figure 8 Zones of inhibition of cell-free supernatants of Bacillus strain FJ3 against (A) F. oxysporum, (B) A. niger, (C) A. flavus and (D) R. oryzae.
The minimum inhibitory concentration (MIC) of the crude lipopeptide extracts from strains Bacillus subtilis FJ3 illustrated broad anti-fungal activity with >40% inhibition against almost all tested pathogens. Growth inhibition was against Fusarium oxysporum (92%), Aspergillus flavus (81.5%), A. niger (90%), and Rhizopus oryzae (56%). The commercialized anti-fungal drug, Nystatin, showed maximum growth inhibition while no growth inhibition was manifested by methanol.
Crude metabolite FJ3 against which fungal pathogens were found susceptible (must have inhibition activity greater than 20%) was subjected to an MIC test. MIC was evaluated at a range of dilutions. The results of the calculated MIC are shown in Table 2. The control (nystatin) had the highest MIC value of 0.781 mg mL−1 against A. flavus, and 1.56 mg mL−1 against F. oxysporum, and P. oryzae and FJ3 exhibited excellent MIC against pathogenic fungal strains of F. oxysporum and A. flavus (Table 2).
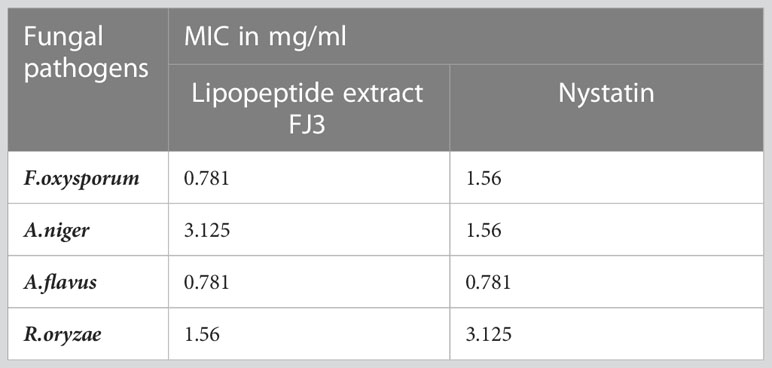
Table 2 Minimum inhibitory antifungal activity of lipopeptide extract from FJ3 at various dilutions.
The infra-red spectrum of crude lipopeptide extracts from FJ3 showed intense bands at 3,287.67 cm−1 and 3,287.94 cm−1, indicating the presence of an amino stretch in the metabolite architecture (Figure 9). Another band at 2,932.51 cm−1 and 2,931.49 cm−1 confirmed the presence of an aliphatic side chain stretch in the extract. Peaks at 1,640.26 cm−1 and 1,641.80 cm−1 corresponded to the peptide moiety while intense bands at 1,454.56 cm−1 and 1,455.01 cm−1 underlined the presence of methyl bonds and aliphatic (C-H) bonds respectively.
Mass spectrometry analysis showed the presence of saturated β-fatty acids with chain lengths ranging from C14 to C21, and most of the fatty acids were saturated. (Table 3). Among the detected fatty acids, hexadecanoic acid is found to be in the majority proportion. Other detected fatty acids include oleic acid, ascorbic acids, pentadecanoic acid, 6-octadecenoic acid, and 10-heptadecenoic acid (Figures 10, 11). The result is consistent with the reports of Peng et al. (2008) and Ibrahim et al. (2013).
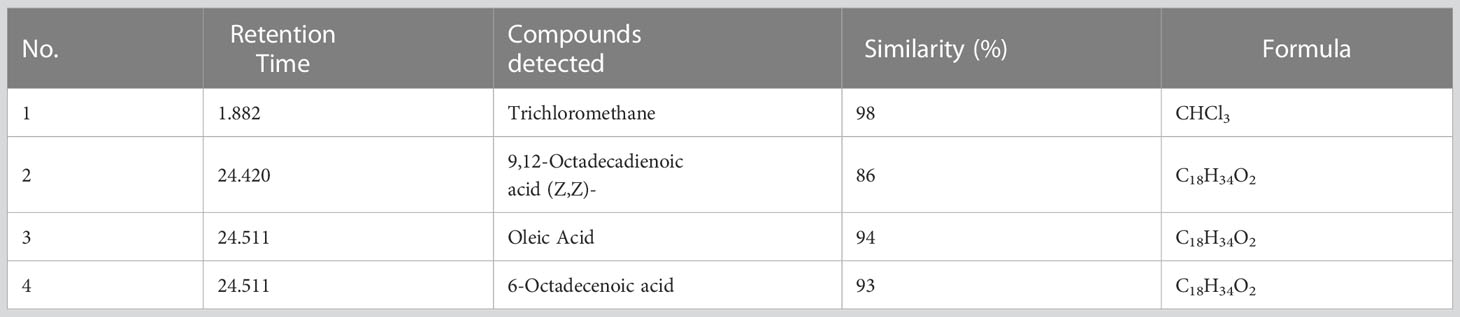
Table 3 GC-MS analysis of the fatty acid composition of lipopeptide produced by Bacillus subtilis FJ3.
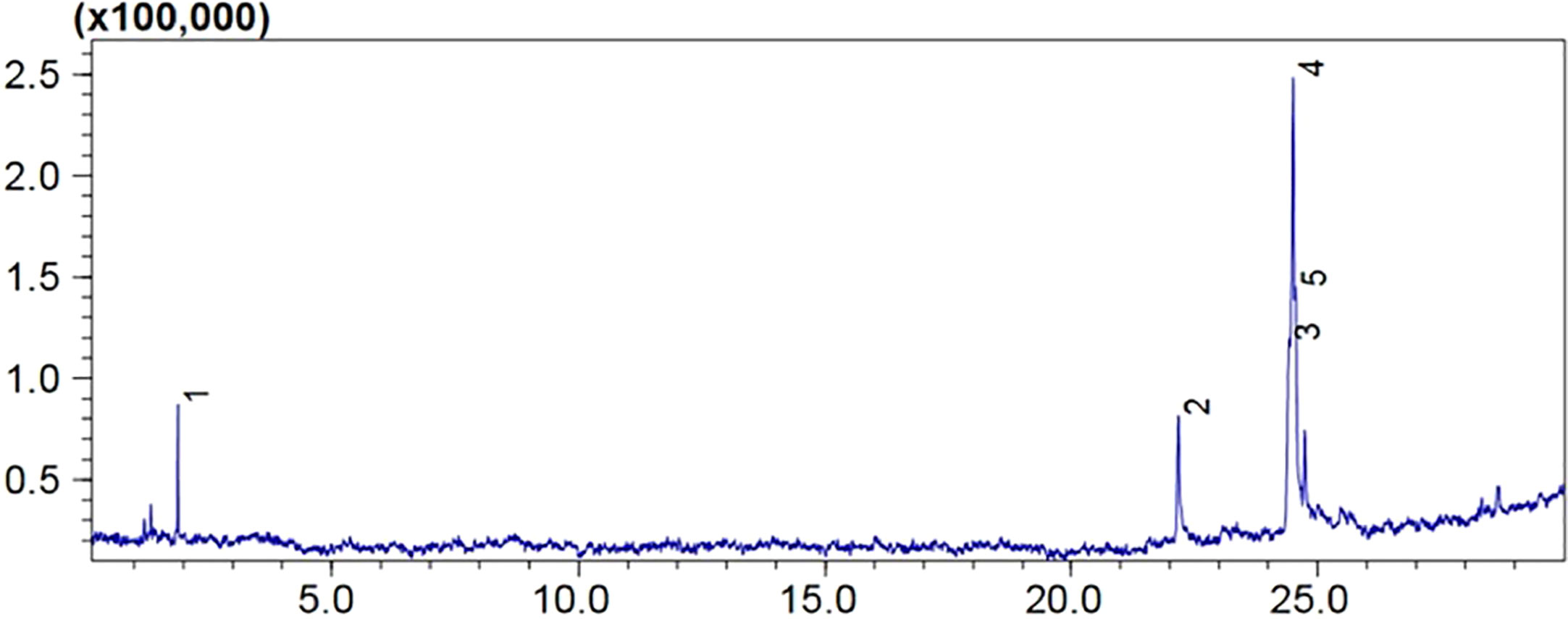
Figure 10 The GC-MS total ion chromatogram of fatty acids in lipopeptide isolate of FJ3. Peaks are numbered 1 to 5 as shown in the table.
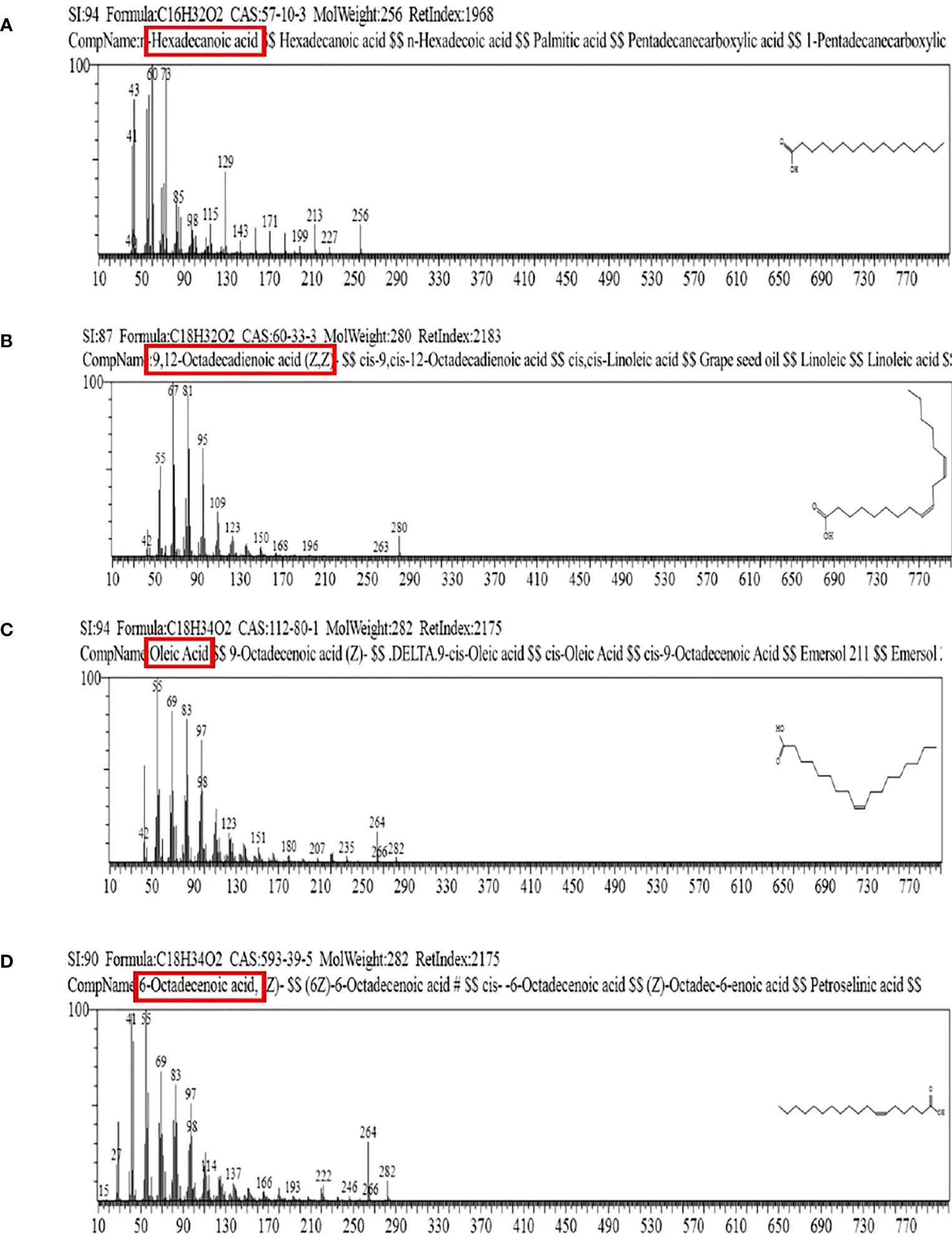
Figure 11 Mass spectrogram of methyl esterified fatty acid chain in lipopeptide extract of FJ3. (A–D) Correspond to peaks 1, 2, 3, and 4 as depicted in the figure.
In sterilized soil, Bacillus subtilis FJ3 and its cell-free supernatant significantly controlled disease and resulted in enhanced growth as compared to the negative control. It promoted root and shoot growth appreciably with increased chlorophyll content. Although Bacillus subtilis and its cell-free supernatant considerably lowered the pathogen incidence in un-sterilized soil, it did not result in the promotion of growth as did the SVI for all treatments in un-sterilized soil, including controls, that were not significantly different according to Duncan multiple range test. The results are presented in Figure 12 and Table 4 and Figure 13 and Table 5, respectively.

Figure 12 Effect of Bacillus subtilis FJ3 and its cell-free supernatant (CFS) treatments on Fusarium wilt of chickpea.
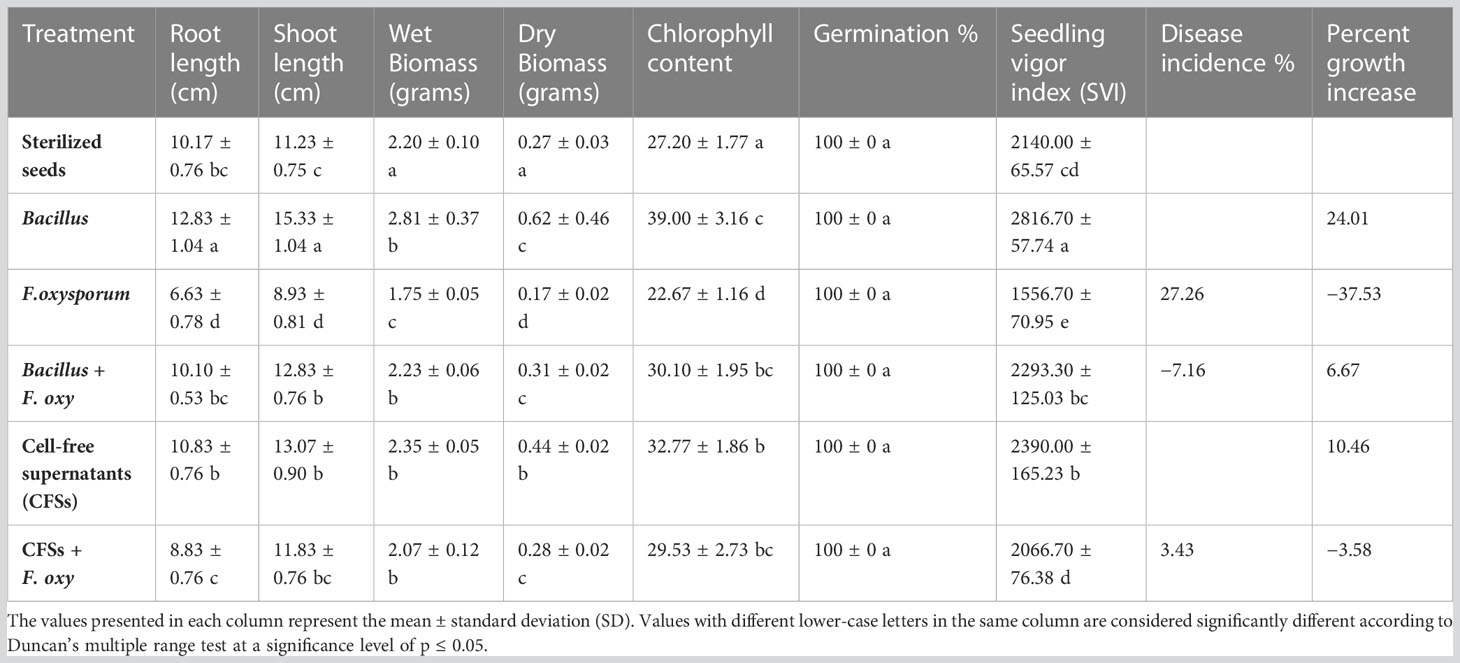
Table 4 Control of Fusarium wilt in chickpea caused by Fusarium oxysporum by Bacillus subtilis FJ3 and its cell-free supernatant in sterilized soil.
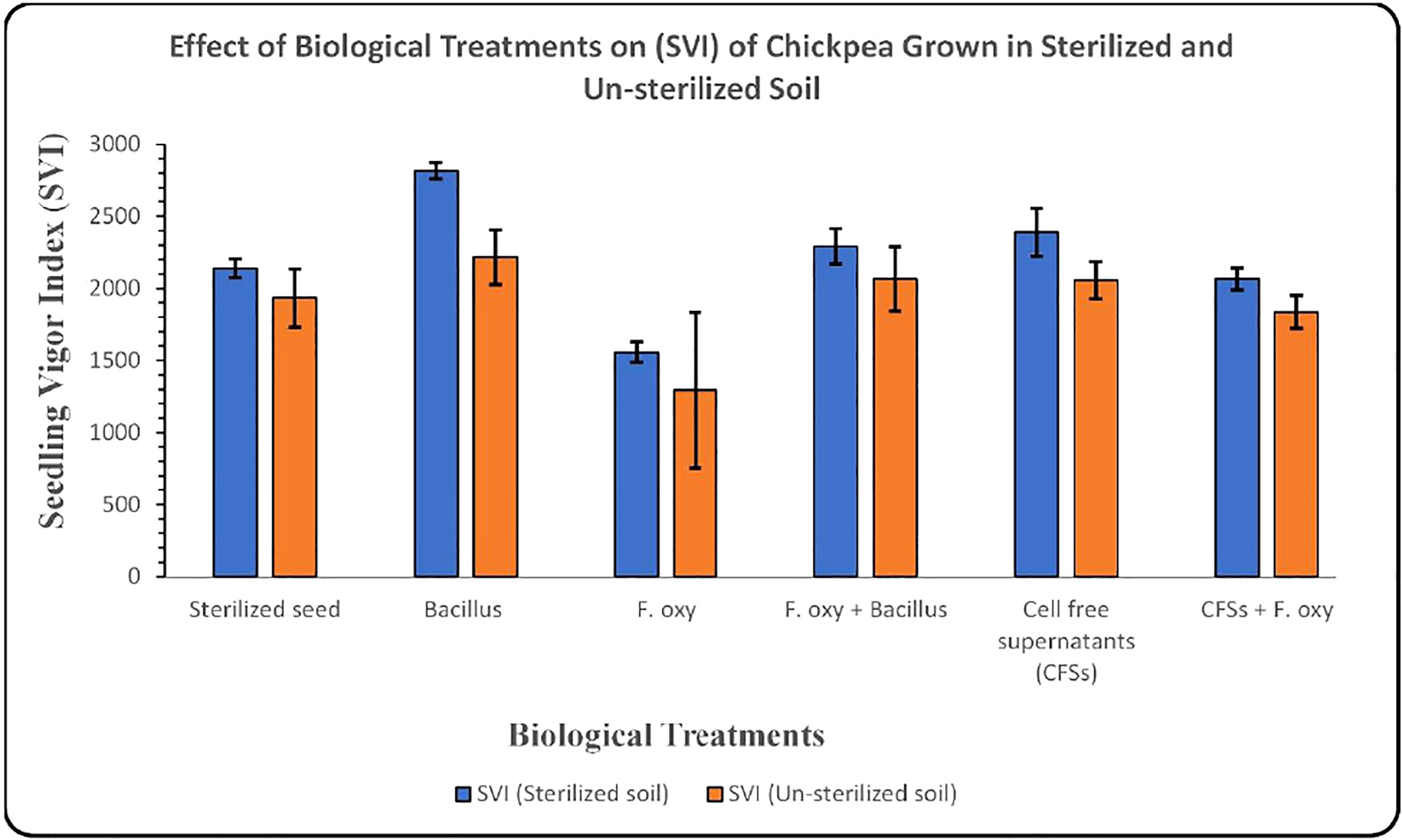
Figure 13 Graph showing the effect of biological treatments on the SVI of chickpea grown in sterilized and un-sterilized soil (Error bars represent standard deviations).
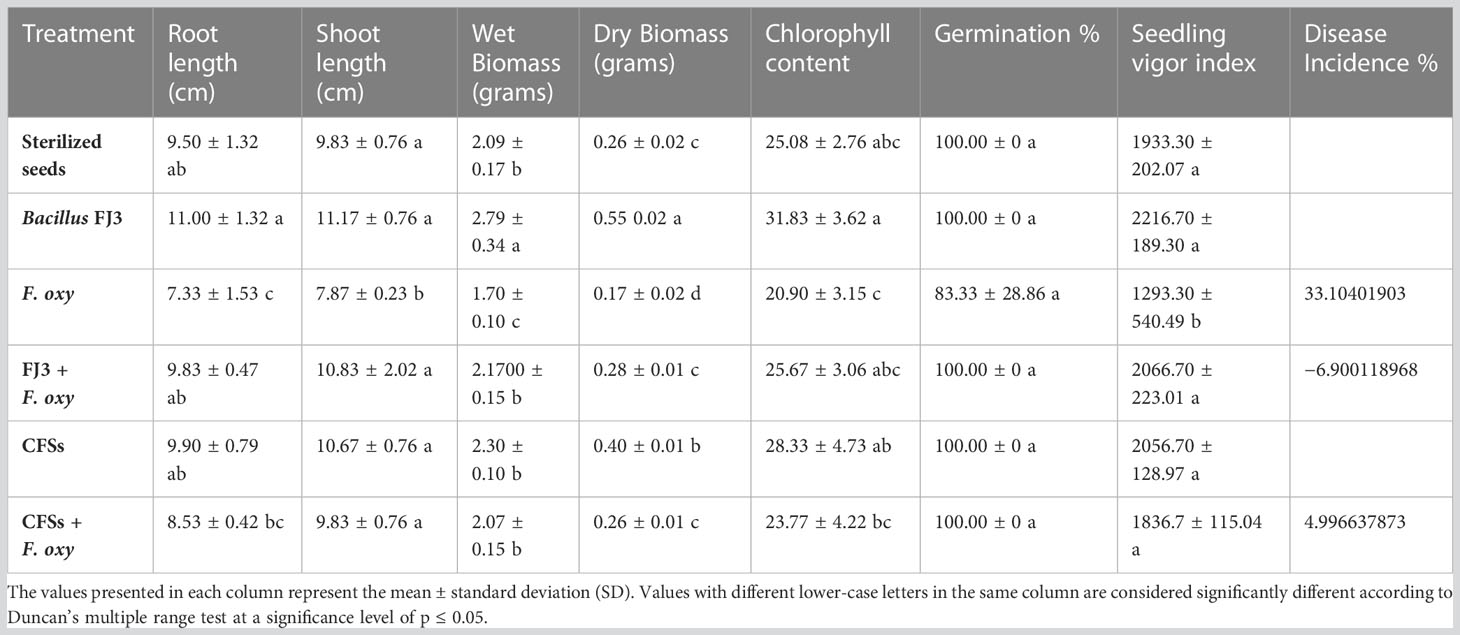
Table 5 Control of Fusarium wilt in chickpea caused by Fusarium oxysporum by Bacillus subtilis FJ3 and its cell-free supernatant in un-sterilized soil.
The utilization of microorganisms and their metabolic products to enhance plant productivity by mitigating the effects of phytopathogens has been increasing since the beginning of this millennium. The innate ability of microbial agents to control plant pathogens, with the added advantage of enhancing agricultural productivity, has been gaining appreciation among those working in this area (He et al., 2021). The genus Bacillus is often more effective than other potential biocontrol microorganisms owing to its unique metabolic attributes such as the fabrication of a wide range of antimicrobial metabolites and its endospore-forming capability. Bacillus can accomplish the work of disease suppression through a variety of mechanisms, such as antibiosis, parasitism, competition for space and nutrients with the pathogen, or by directly inducing systemic resistance in host plants. Thus, there is a compelling demand for seeking an active bioagent that not only aids in combating disease-causing pathogens but also causes no environmental hazard (Fira et al., 2018).
The intent of the current study was to find microorganisms with highly effective anti-fungal potential and plant growth-promoting activities. This was accomplished by the isolation of soil-borne Bacillus strains from agricultural soils of Swat, KPK. In total, 4 out of the 13 isolates manifested good levels of anti-fungal potential against three selected test pathogens and Fusarium oxysporum FJ81 isolated from chickpea. Based on the zone of inhibition obtained in the preliminary spot inoculation tests, four strains FJ3, FJ9, FJ26, and FJ55 were selected as efficient strains of Bacillus with clear antagonism against the tested pathogens. Isolate FJ3 manifested the greatest antagonistic activity and was subjected to detailed characterization. The strain was identified as Bacillus subtilis by the 16S rRNA technique.
In preliminary screening, the strain Bacillus subtilis FJ3 displayed numerous plant growth-promoting traits such as phosphate solubilization, IAA production, siderophore production, and biofilm formation. The Bacillus subtilis strain FJ3 was able to solubilize phosphorus, an important feature for increasing phosphorus availability in soil for plants, to enhance crop yield. On NBRIP agar plates, the selected strain FJ3 converted insoluble tricalcium phosphate into the soluble form with a solubility index greater than 3 mm comparable with a recent study in which Javoreková et al. (2021) reported a medium solubility index value of approximately 2.96 mm for phosphate solubilization. PSBs increase phosphorus availability by secreting phosphatases and organic acids which convert phosphate to plant-available forms (Breedt et al., 2017).
The strain FJ3 was shown to produce the phytohormone IAA during an in-vitro assay which is involved in cell enlargement, cell division, and root growth and development, resulting in a larger root surface area allowing the plant to acquire more nutrients from the soil.
Strain FJ3 tested positive for siderophore production, indicated by the appearance of a yellowish-orange halo zone on CAS agar media (Figure 3B). Javoreková et al. (2021) reported that Gram-negative bacteria are primarily responsible for the formation of siderophores. Siderophores excreted by rhizosphere bacteria may promote plant growth by enhancing Fe nutrition and protecting plants against a variety of fungal and bacterial infections. Bacteria that produce siderophores can play a significant role in the biocontrol of several phytopathogens (Javorekova et al., 2020).
Bacillus subtilis strain FJ3 formed dry reddish colonies on Congo Red agar. Romero et al. (2010) indicated that the integrity of the extracellular matrix is strengthened by amyloid fibers formed of an amyloid protein TasA. TasA also helps in biofilm formation. TasA gives the colony a red appearance on binding with Congo Red dye. Bacillus species having a mutated form of the gene for TasA cannot form biofilm and colonies do not appear red. Anckaert et al. (2021) reported that biofilm formation helps Bacillus strains to occupy space and acquire nutrients as the mobility of nutrients is enhanced within the rhizosphere by the extracellular matrix, resulting in a reduction in the colonization of plant pathogens. According to Chen et al. (2013), restricted production of the matrix by Bacillus subtilis (3,610) resulted in decreased biocontrol efficacy against the Rhizoctonia solanacearum pathogen of tomato. Our Bacillus species was a biofilm producer with capacity to occupy space by inhibiting the mycelial growth of isolated fungi in vitro and to inhabit plant surfaces for disease prevention.
Plant growth-promoting rhizobacteria (PGPR) have been recognized for their potential to control soil-borne plant pathogens through the production of antibiotics and hydrolytic enzymes, which are recognized as key traits for biocontrol. In particular, PGPR can prevent the growth of phytopathogens by producing enzymes that can degrade fungal cell walls. A recent study has shown that strain FJ3, like other biocontrol bacteria, can synthesize enzymes such as protease, glucanase, cellulase, and pectinase. These enzymes break down specific components of the cell wall of several pathogenic fungi, such as Rhizoctonia solani, Sclerotium rolfsii, Fusarium oxysporum, Phytophthora spp., Pythium ultimum, and Botrytis cinerea. This enzymatic activity can efficiently control the spread of diseases by inhibiting the growth of these fungi (Beneduzi et al., 2012).
Bacillus subtilis FJ3 was also screened in specific media to produce various hydrolytic enzymes (protease, endoglucanases, and amylase). Production of these enzymes was obvious from the zone of hydrolysis during enzyme assays. Chitinases and endoglucanases destroy the basic building block of fungal cell walls, i.e., chitin and β 1,3 glucanase and β 1,4 glucanase, resulting in the loss of cell wall integrity by fungal pathogens. Production of these enzymes in Bacillus species was also confirmed in several recent papers (Thakaew and Niamsup, 2013; Anabella et al., 2013; Mnif and Ghribi, 2015).
PGPR primarily employ the production of antifungal metabolites and antibiotics as their key mechanisms to restrict the invasion of pathogens in host plant tissues. To investigate the antibiosis mechanisms of the FJ3 strain, PCR analysis was performed to screen four non-ribosomal peptide (NRP) genes that are responsible for synthesizing lipopeptides. Primer pairs of As1-F/Ts2-R, Af2-F/Tf1-R, Ap1-F/Tp12-R, and Am1-F/Tm1-R were able to amplify surfactin, fengycin, mycosubtilin, and plipastatin genes respectively. Fengycins and iturins exhibit potent antifungal properties and effectively inhibit the growth of various plant pathogens. Although surfactins alone do not possess antifungal properties, they have a synergistic effect with the antifungal activity of iturins (Ongena and Jacques, 2008). Results were considered positive when bands of appropriate size were obtained. The presence of surfactin genes in Bacillus subtilis and Bacillus cereus has been described by many authors; it was surprising to detect surfactin biosynthetic genes in other Bacillus species, i.e., Bacillus tropicus, Bacillus velenzensis, Bacillus proteolyticus, and Bacillus paramycoides. Similarly, the presence of plipastatin biosynthetic genes was observed in Bacillus subtilis, Bacillus tropicus, Bacillus velenzensis, and Bacillus proteolyticus and the fengycin biosynthetic gene were observed in Bacillus subtilis, Bacillus tropicus, Bacillus velenzensis, Bacillus proteolyticus, and Bacillus paramycoides. Bands of various intensities were discerned with Bacillus strains for each pair of primers. A band of high intensity corresponds to the amplification of many fragments of similar size. The selected Bacillus strain FJ3 showed positive results for surfactin, fengycin, mycosubtilin, and plipastatin genes. The amplification of expected gene size is in accordance with the work of Tapi et al. (2010). This method is highly effectual in multiplex PCR and in metagenomics.
To date, to our knowledge, this is the first report that confirms the presence of NRPS genes in strains of Bacillus tropicus, Bacillus velenzensis, Bacillus proteolyticus, and Bacillus paramycoides. B. velezensis is known to be a lipopeptide producer, i.e., B. velezensis FZB2 is known to be a PGPR (Antil et al., 2023).
The present study conducted in vitro inhibition assays against four fungal phytopathogens to determine the broad antagonistic ability of B. subtilis FJ3. The use of the in vitro dual culture method to evaluate potential biocontrol activities has been widely employed in previous studies. Bacillus strain FJ3 inhibited the mycelial growth of Fusarium by up to 52.92%. Mejía-Bautista et al. (2016) reported that Bacillus strains isolated from soil showed growth inhibition of Fusarium solani (3.76% to 69.16%) and Fusarium equiseti (2.15% to 71.55%). Using the agar well diffusion method, FJ3 showed broad-spectrum antifungal activities with maximum inhibition against Aspergillus flavus, Aspergillus niger, Fusarium oxysporum, and Rhizopus oryzae. Antifungal lipopeptides, extracted with solvents, illustrated broad anti-fungal activity with >40% inhibition against almost all tested pathogens.
Growth inhibition against Aspergillus flavus was 81.5%, Fusarium oxysporum was 92%, Aspergillus niger was 90%, and Rhizopus oryzae was 56%. A distinctive novel compound manifested antifungal activity. Previous in vitro studies have shown that Bacillus subtilis (strain RMB7) can inhibit the mycelial growth of Fusarium oxysporum (71%), Rhizoctonia solani (70%), Pythium ultimum (83%), and Aspergillus flavus (75%) (Ali et al., 2014). Similarly, another B. subtilis (strain BP9) was evaluated for potential biocontrol against F. oxysporum and was shown to inhibit mycelial growth by 62.5% (Gajbhiye et al., 2010). The high anti-fungal activity shown by FJ3 in this study was subjected to the collective effect of anti-fungal metabolites including lytic enzymes, diffusible antibiotics, and siderophores, or by high production of cyclic lipopeptides. The lipopeptides extracted showed good results with MIC values ranging from 0.78 to 3.125 mg mL−1 and exerted a fungicidal effect on all tested fungal pathogens. The best results were obtained against A. flavus and Fusarium oxysporum species (MIC = 0.781).
The FTIR results of crude extract evinced peaks with characteristics of cyclic lipopeptides encompassing surfactin, fengycin, and iturin. The infra-red spectrum of FJ3 crude lipopeptide content indicated intense bands at 2,853.51 cm−1 and 2,923.01 cm−1, confirming the presence of aliphatic side chain stretch in the metabolite architecture. Peaks at 1,710.18 cm−1 and 1,739.91 cm−1 indicate lactone – carbonyl absorption, while intense bands at 1,457.48 cm-1and 1,378.14 cm−1 confirmed the presence of methyl bonds and aliphatic (C-H) bonds, respectively. These bands show a chemical structure resembling lipopeptides with aliphatic chains (hydrophobic domain) and peptide moieties (hydrophilic domain). Comparison with published literature indicates the presence of surfactin (Sivapathasekaran et al., 2010; Veshareh and Nick, 2018), fengycin (Wei and Yang, 2010), and iturin (Narendrakumar et al., 2016). Based on the results of this study and the comparison of these results with the lipopeptide spectra published in the published literature, it is speculated that the compound could be lipopeptide in nature (Wu et al., 2019).
GC-MS results of isolated lipopeptides evinced various types of free and bound fatty acids. The fatty acids detected are similar to those described by Peng et al. (2008) and Ibrahim et al. (2013). Representative lipopeptides, i.e., surfactin, fengycin, and iturin, are characterized by the presence of β-hydroxy fatty acids which are crucial for the surface activity of biosurfactants. The composition of fatty acids integrated into lipopeptides relies on medium composition. Thus, the presence of the predominant hexadecanoic acid C16H32O2 at maximum concentration indicates the importance of this substrate for bacterial growth and proliferation (Guo et al., 2012). Nonetheless, owing to the vast structural diversity of the lipopeptides produced by Bacillus species, the determination of the exact fatty acid structure in cyclic lipopeptides is not well understood yet.
Although in vitro assays are significant in determining the antifungal properties of B. subtilis, it is equally vital to assess its biocontrol efficacy in vivo. In vivo experiments carried out on chickpea crops revealed that the FJ3 strain is highly effective in suppressing root rot caused by Fusarium spp. In autoclaved soil, our isolated Bacillus strain FJ3 and its metabolites (cell-free supernatant) significantly lowered the disease incidence and resulted in 24% and 10.46% increases in plant growth compared with controls, respectively. However, seed germination did not vary meaningfully. Souad et al. (2013) also demonstrated that Bacillus species significantly control Fusarium wilt in chickpea and enhanced plant growth in sterilized soil. They further noticed that seed germination was better when inoculated with only Fusarium oxysporum f. sp. ciceris. Posada et al. (2016) stated that cell-free supernatants either derived from endospore or vegetative cells substantially enhanced dry weight and increased shoot length of Musa plants as compared to the control. The higher plant growth promotion in the case of Bacillus treated seeds, as compared to metabolites, indicates possible production of plant growth-promoting substances or helping the plants in nutrient acquisition by establishing an association via biofilm production during seedling establishment. Moreover, metabolite lability (sensitive to temperature, strong acids, and bases) limits CFSs as biofertilizers used in soil (Pellegrini et al., 2020). The reduced plant growth in Fusarium-infested soil might be due to stress induction in the seedlings as indicated by the significantly reduced biomass and chlorophyll contents. Shoaib et al. (2018) also reported that onion infected with the soil-borne fungus Fusarium oxysporum f. sp. cepa showed significantly decreased (30–70%) plant growth, biomass, and chlorophyll content. Das and Roychoudhury (2014) stated that developmental and physiological variables are affected by the overproduction of ROS, leading to reduced pigments and root growth. Our study is in agreement with Baghbani et al. (2019), who noted reduced chlorophyll concentrations in maize infected with Fusarium verticilloides. In non-autoclaved soil, our Bacillus species and its CFSs significantly lowered the disease incidence by up to 39% and 29%, respectively, however, SVI, a parameter of plant growth, was not enhanced when compared with the negative control. Martínez-Viveros et al. (2010) stated that to produce a significant change in plant growth, a critical population density of beneficial microbes is needed, however, in un-sterilized soil, there is a rapid decline in bacterial population compared with sterilized soil due to competition with indigenous microbiota and predation by nematodes and protozoa. Further rapid re-colonization of bacterial communities was also observed by Marschner and Rumberger (2004) in sterilized soil. These studies justify the observed differences in plant growth in sterilized and non-sterilized soils. To concisely summarize the results of this research project, Bacillus species are one of the most interesting and promising options for use as biocontrol agents, demonstrating wide inhibitions against plant pathogens. Moreover, it is desirable to consider bio-controls as a green alternative to chemical pesticides to be used in formulations of commercial biocontrol products. In this way, this novel concept of bio-controls will eventually have a space outside the laboratory, and we will be able to see its fruits in present and future crop production systems.
The presented research has revealed that B. subtilis FJ3 exhibits multiple plant growth-promoting traits, including phosphate solubilization, IAA production, siderophore synthesis, biofilm formation, hydrolytic enzyme production, and lipopeptide synthesis, indicating its potential as a microbial inoculant. In vitro and in vivo investigations have also indicated that B. subtilis FJ3 could be a promising biocontrol agent for managing Fusarium wilt caused by Fusarium species in chickpea plants. Therefore, future studies should focus on evaluating the application of FJ3 in field trials for chickpea and other crops to facilitate the development of a commercialized product.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
FJ established the experiments, collected data, and wrote the manuscript. HA and MA were involved in the experimental setup and data collection. AJ provided intellectual guidance and reviewed the manuscript. DS provided funding, intellectual guidance, editorial input, and review of the manuscript. All authors contributed to the article and approved the submitted version.
We are grateful to Department of Microbiology, Quaid-i-Azam University Islamabad, Pakistan and DS for financial support to this research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alajlani, M., Shiekh, A., Hasnain, S., Brantner, A. (2016). Purification of bioactive lipopeptides produced by Bacillus subtilis strain BIA. Chromatographia 79, 1527–1532. doi: 10.1007/s10337-016-3164-3
Albayrak, Ç. B. (2019). Bacillus species as biocontrol agents for fungal plant pathogens. Bacilli Agrobiotechnol.: Phytostimulation Biocontrol 2, 239–265. doi: 10.1007/978-3-030-15175-1_13
Ali, S., Hameed, S., Imran, A., Iqbal, M., Lazarovits, G. (2014). Genetic, physiological and biochemical characterization of bacillus sp. strain RMB7 exhibiting plant growth promoting and broad spectrum antifungal activities. Microb. Cell Factories 13 (1), 144.
Anabella, I., Jacylyn, V., Ben, W., Ignacio, F. (2013). Spectroscopic and kinetic investigation of the reactions of peroxyacetic acid with burkholderia pseudomallei catalase-peroxidase. KatG.
Anckaert, A., Arias, A. A., Hoff, G., Calonne-Salmon, M., Declerck, S., Ongena, M. (2021). The use of Bacillus spp. as bacterial biocontrol agents to control plant diseases. Burleigh Dodds Series in Agricultural Science 247–300. doi: 10.19103/AS.2021.0093.10
Antil, S., Kumar, R., Pathak, D. V., Kumari, A. (2023). Recent advances in utilizing bacteria as biocontrol agents against plant parasitic nematodes emphasizing Meloidogyne spp. Biol. Control, 105244. doi: 10.1016/j.biocontrol.2023.105244
Awuchi, C. G., Ondari, E. N., Ogbonna, C. U., Upadhyay, A. K., Baran, K., Okpala, C. O. R., et al. (2021). Mycotoxins affecting animals, foods, humans, and plants: types, occurrence, toxicities, action mechanisms, prevention, and detoxification strategies–a revisit. Foods 10 (6), 1279. doi: 10.3390/foods10061279
Baghbani, F., Lotfi, R., Moharramnejad, S., Bandehagh, A., Roostaei, M., Rastogi, A., et al. (2019). Impact of fusarium verticillioides on chlorophyll fluorescence parameters of two maize lines. Eur. J. Plant Pathol. 154, 337–346. doi: 10.1007/s10658-018-01659-x
Beneduzi, A., Ambrosini, A., Passaglia, L. M. (2012). Plant growth-promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35, 1044–1051. doi: 10.1590/S1415-47572012000600020
Biniarz, P., Łukaszewicz, M., Janek, T. (2017). Screening concepts, characterization and structural analysis of microbial-derived bioactive lipopeptides: a review. Crit. Rev. Biotechnol. 37 (3), 393–410. doi: 10.3109/07388551.2016.1163324
Breedt, G., Labuschagne, N., Coutinho, T. A. (2017). Seed treatment with selected plant growth-promoting rhizobacteria increases maize yield in the field. Ann. Appl. Biol. 171 (2), 229–236. doi: 10.1111/aab.12366
Caracciolo, R. (2018). Evaluation of the antagonistic activity of Bacillus amyloliquefaciens against fungal pathogens of wheat.
Caulier, S., Nannan, C., Gillis, A., Licciardi, F., Bragard, C., Mahillon, J. (2019). Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front. Microbiol. 10, 302. doi: 10.3389/fmicb.2019.00302
Chen, L., Cai, Y., Zhou, G., Shi, X., Su, J., Chen, G., et al. (2014). Rapid Sanger sequencing of the 16S rRNA gene for identification of some common pathogens. PloS One 9 (2), e88886. doi: 10.1371/journal.pone.0088886
Chen, Y., Yan, F., Chai, Y., Liu, H., Kolter, R., Losick, R., et al. (2013). Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ. Microbiol. 15 (3), 848–864. doi: 10.1111/j.1462-2920.2012.02860.x
Chopra, A., Bobate, S., Rahi, P., Banpurkar, A., Mazumder, P. B., Satpute, S. (2020). Pseudomonas aeruginosa RTE4: a tea rhizobacterium with potential for plant growth promotion and biosurfactant production. Front. Bioeng. Biotechnol. 8, 861. doi: 10.3389/fbioe.2020.00861
Das, K., Roychoudhury, A. (2014). Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2, 53. doi: 10.3389/fenvs.2014.00053
De Angelis, G., Simonetti, G., Chronopoulou, L., Orekhova, A., Badiali, C., Petruccelli, V., et al. (2022). A novel approach to control Botrytis cinerea fungal infections: uptake and biological activity of antifungals encapsulated in nanoparticle based vectors. Sci. Rep. 12 (1), 7989.
Dimkić, I., Janakiev, T., Petrović, M., Degrassi, G., Fira, D. (2022). Plant-associated Bacillus and Pseudomonas antimicrobial activities in plant disease suppression via biological control mechanisms-a review. Physiol. Mol. Plant Pathol. 117, 101754. doi: 10.1016/j.pmpp.2021.101754
Dissanayake, A. J., Bhunjun, C. S., Maharachchikumbura, S. S. N., Liu, J. K. (2020). Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere 11 (1), 2652–2676. doi: 10.5943/mycosphere/11/1/18
do Espirito Santo Pereira, A., Caixeta Oliveira, H., Fernandes Fraceto, L., Santaella, C. (2021). Nanotechnology potential in seed priming for sustainable agriculture. Nanomaterials 11 (2), 267. doi: 10.3390/nano11020267
Durairaj, K., Velmurugan, P., Park, J. H., Chang, W. S., Park, Y. J., Senthilkumar, P., et al. (2017). Potential for plant biocontrol activity of isolated Pseudomonas aeruginosa and Bacillus stratosphericus strains against bacterial pathogens acting through both induced plant resistance and direct antagonism. FEMS Microbiol. Lett. 364 (23), fnx225. doi: 10.1093/femsle/fnx225
Fira, D., Dimkić, I., Berić, T., Lozo, J., Stanković, S. (2018). Biological control of plant pathogens by Bacillus species. J. Biotechnol. 285, 44–55. doi: 10.1016/j.jbiotec.2018.07.044
Freeman, D., Falkiner, F., Keane, C. T. (1989). New method for detecting slime production by coagulase negative staphylococci. JCP BMJ Com. 42, 872–874. doi: 10.1136/jcp.42.8.872
Gajbhiye, A., Rai, A. R., Meshram, S. U., Dongre, A. B. (2010). Isolation, evaluation and characterization of Bacillus subtilis from cotton rhizospheric soil with biocontrol activity against Fusarium oxysporum. World J. Microbiol. Biotechnol. 26, 1187–1194. doi: 10.1007/s11274-009-0287-9
Gontard, N., Sonesson, U., Birkved, M., Majone, M., Bolzonella, D., Celli, A., et al. (2018). A research challenge vision regarding management of agricultural waste in a circular bio-based economy. Crit. Rev. Environ. Sci. Technol. 48 (6), 614–654. doi: 10.1080/10643389.2018.1471957
Grobelak, A., Hiller, J. (2017). Bacterial siderophores promote plant growth: screening of catechol and hydroxamate siderophores. Int. J. Phytoremediation 19 (9), 825–833. doi: 10.1080/15226514.2017.1290581
Guo, Y., Huang, E., Yuan, C., Zhang, L., Yousef, A. E. (2012). Isolation of a Paenibacillus sp. strain and structural elucidation of its broad-spectrum lipopeptide antibiotic. Appl. Environ. Microbiol. 78 (9), 3156–3165.
Hashem, A., Tabassum, B., Abd_Allah, E. F. (2019). Bacillus subtilis: a plant-growth promoting rhizobacterium that also impacts biotic stress. Saudi J. Biol. Sci. 26 (6), 1291–1297. doi: 10.1016/j.sjbs.2019.05.004
He, D. C., He, M. H., Amalin, D. M., Liu, W., Alvindia, D. G., Zhan, J. (2021). Biological control of plant diseases: an evolutionary and eco-economic consideration. Pathogens 10 (10), 1311. doi: 10.3390/pathogens10101311
Ibrahim, M. L., Ijah, U. J. J., Manga, S. B., Bilbis, L. S., Umar, S. (2013). Production and partial characterization of biosurfactant produced by crude oil degrading bacteria. Int. Biodeterior. Biodegrad. 81, 28–34. doi: 10.1016/j.ibiod.2012.11.012
Ivancich, A., Donald, L. J., Villanueva, J., Wiseman, B., Fita, I., Loewen, P. C (2013). Spectroscopic and kinetic investigation of the reactions of peroxyacetic acid with burkholderia pseudomallei catalase-peroxidase, KatG. Biochemistry 52 (41), 7271–7282. doi: 10.1021/bi400963j
Jangir, M., Pathak, R., Sharma, S., Sharma, S. (2018). Biocontrol mechanisms of Bacillus sp., isolated from tomato rhizosphere, against fusarium oxysporum f. sp. lycopersici. Biol. Control 123, 60–70. doi: 10.1016/j.biocontrol.2018.04.018
Javorekova, S., Cinkocki, R., Maková, J., Hricáková, N. (2020). Isolation and identification of rhizobacteria from maize (Zea mays L.) in luvisols and documentation their plant growth promoting traits. J. Microbiol. Biotechnol. Food Sci. 10 (3), 505–510.
Javoreková, S., Kovácsová, S., Medo, J., Maková, J., Petrová, J., Hleba, L., et al. (2021). Soil amended with organic fertilizers as a source of actinomycetes with high potential as biocontrol agents. J. Microbiol. Biotechnol. Food Sci. 2021, 1352–1359.
Juboi, H. A. (2017). Evaluation of the antibiotics and secondary metabolites biosynthetic potential of Plantactinospora sp. kbS50, a rare marine-derived actinomycete (Doctoral dissertation, Thesis (biology) published. Corpus ID: 132381879).
Kaur, P. K., Joshi, N., Singh, I. P., Saini, H. S. (2017). Identification of cyclic lipopeptides produced by Bacillus vallismortis R2 and their antifungal activity against Alternaria alternata. J. Appl. Microbiol. 122 (1), 139–152. doi: 10.1111/jam.13303
Khan, A. R., Mustafa, A., Hyder, S., Valipour, M., Rizvi, Z. F., Gondal, A. S., et al. (2022). Bacillus spp. as bioagents: uses and application for sustainable agriculture. Biology 11 (12), 1763.
Limtong, S., Into, P., Attarat, P. (2020). Biocontrol of rice seedling rot disease caused by Curvularialunata and Helminthosporiumoryzae by epiphytic yeasts from plant leaves. Microorganisms 8 (5), 647. doi: 10.3390/microorganisms8050647
Manghwar, H., Hussain, A., Ali, Q., Saleem, M. H., Abualreesh, M. H., Alatawi, A., et al. (2021). Disease severity, resistance analysis, and expression profiling of pathogenesis-related protein genes after the inoculation of Fusarium equiseti in wheat. Agronomy 11 (11), 2124. doi: 10.3390/agronomy11112124
Marschner, P., Rumberger, A. (2004). Rapid changes in the rhizosphere bacterial community structure during re-colonization of sterilized soil. Biol. Fertil. Soils 40, 1–6. doi: 10.1007/s00374-004-0736-4
Martínez-Viveros, O., Jorquera, M. A., Crowley, D. E., Gajardo, G. M. L. M., Mora, M. L. (2010). Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. J. Soil Sci. Plant Nutr. 10 (3), 293–319. doi: 10.4067/S0718-95162010000100006
Mejía-Bautista, M. Á., Reyes-Ramírez, A., Cristóbal-Alejo, J., Tun-Suárez, J. M., Borges-Gómez, L. D. C., Pacheco-Aguilar, J. R. (2016). Bacillus spp. en el control de la marchitez causada por Fusarium spp. en Capsicum chinense. Rev. Mexicana Fitopatología 34 (3), 208–222.
Miljaković, D., Marinković, J., Balešević-Tubić, S. (2020). The significance of Bacillus spp. in disease suppression and growth promotion of field and vegetable crops. Microorganisms 8 (7), 1037.
Mnif, I., Ghribi, D. (2015). Review lipopeptides biosurfactants: mean classes and new insights for industrial, biomedical, and environmental applications. Pept. Sci. 104 (3), 129–147. doi: 10.1002/bip.22630
Mohamed, A. E., Nessim, M. G., Abou-el-seoud, I. I., Darwish, K. M., Shamseldin, A. (2019). Isolation and selection of highly effective phosphate solubilizing bacterial strains to promote wheat growth in Egyptian calcareous soils. Bull. Natl. Res. Centre 43 (1), 1–13. doi: 10.1186/s42269-019-0212-9
Munir, S., Li, Y., He, P., He, P., He, P., Cui, W., et al. (2018). Bacillus subtilis L1-21 possible assessment of inhibitory mechanism against phytopathogens and colonization in different plant hosts. Pakistan J. Agric. Sci. 55 (4), 996–1002.
Nahas, H. H. A., Nouh, F. A. A., Husien, S., Gezaf, S. A., Mansour, S. A., Abdel-Azeem, A. M., et al. (2021). “Lipopeptides biosurfactants production, and applications in bioremediation and health,” in Microbial surfactants (CRC Press), 63–84.
Narendrakumar, P., Nageshwar, L., Parameshwar, J., Khan, M. Y., Rayulu, J., Hameeda, B. (2016). In silico and in vitro studies of fungicidal nature of lipopeptide (Iturin a) from bacillus amyloliquefaciens RHNK 22 and its plant growth-promoting traits. Indian Phytopathol. 69 (4s), 569–574.
Ongena, M., Jacques, P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16 (3), 115–125. doi: 10.1016/j.tim.2007.12.009
Paulussen, C., Hallsworth, J. E., Álvarez-Pérez, S., Nierman, W. C., Hamill, P. G., Blain, D., et al. (2017). Ecology of aspergillosis: insights into the pathogenic potency of Aspergillus fumigatus and some other Aspergillus species. Microb. Biotechnol. 10 (2), 296–322. doi: 10.1111/1751-7915.12367
Pellegrini, M., Pagnani, G., Bernardi, M., Mattedi, A., Spera, D. M., Gallo, M. D. (2020). Cell-free supernatants of plant growth-promoting bacteria: a review of their use as biostimulant and microbial biocontrol agents in sustainable agriculture. Sustainability 12 (23), 9917. doi: 10.3390/su12239917
Peng, F., Wang, Y., Sun, F., Liu, Z., Lai, Q., Shao, Z. (2008). A novel lipopeptide produced by a pacific ocean deep-sea bacterium, rhodococcus sp. TW53. J. Appl. Microbiol. 105 (3), 698–705.
Pinstrup-Andersen, P., Pandya-Lorch, R. (1998). Food security and sustainable use of natural resources: a 2020 vision. Ecol. Econ. 26 (1), 1–10. doi: 10.1016/S0921-8009(97)00067-0
Posada, L. F., Ramírez, M., Ochoa-Gómez, N., Cuellar-Gaviria, T. Z., Argel-Roldan, L. E., Ramírez, C. A., et al. (2016). Bioprospecting of aerobic endospore-forming bacteria with biotechnological potential for growth promotion of banana plants. Scientia Hortic. 212, 81–90. doi: 10.1016/j.scienta.2016.09.040
Rajkumari, J., Pandey, P. (2019). Genomic insights and comparative genomics of Bacillus species having diverse mechanisms of biocontrol against fungal phytopathogens. Bacilli Agrobiotechnol.: Phytostimulation Biocontrol 2, 217–237. doi: 10.1007/978-3-030-15175-1_12
Romero, D., Aguilar, C., Losick, R., Kolter, R. (2010). Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U. States America 107 (5), 2230–2234. doi: 10.1073/pnas.0910560107
Seyi-Amole, D. O., Onilude, A. A. (2021). “Microbiological control: a new age of maize production,” in Cereal grains, vol. 2. (IntechOpen).
Shafi, J., Tian, H., Ji, M. (2017). Bacillus species as versatile weapons for plant pathogens: a review. Biotechnol. Biotechnol. Equip. 31 (3), 446–459.
Shiferaw, B., Smale, M., Braun, H. J., Duveiller, E., Reynolds, M., Muricho, G. (2013). Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Secur. 5, 291–317. doi: 10.1007/s12571-013-0263-y
Shoaib, A., Meraj, S., Khan, K. A., Javaid, M. A. (2018). Influence of salinity and Fusarium oxysporum as the stress factors on morpho-physiological and yield attributes in onion. Physiol. Mol. Biol. Plants 24, 1093–1101. doi: 10.1007/s12298-018-0570-z
Singh, R., Singh, H., Raghubanshi, A. S. (2019). Challenges and opportunities for agricultural sustainability in changing climate scenarios: a perspective on Indian agriculture. Trop. Ecol. 60, 167–185. doi: 10.1007/s42965-019-00029-w
Sivapathasekaran, C., Das, P., Mukherjee, S., Saravanakumar, J., Mandal, M., Sen, R. (2010). Marine bacterium derived lipopeptides: characterization and cytotoxic activity against cancer cell lines. Int. J. Pept. Res. Ther. 16 (4), 215–222.
Souad, Z., Belabid, L., Bellahcene, M. (2013). Biocontrol of chickpea fusarium wilt by bacillus spp. rhizobacteria. J. Plant Prot. Res. 53, 177 –1183. doi: 10.2478/jppr-2013-0027
Tapi, A., Chollet-Imbert, M., Scherens, B., Jacques, P. (2010). New approach for the detection of non-ribosomal peptide synthetase genes in Bacillus strains by polymerase chain reaction. Appl. Microbiol. Biotechnol. 85 (5), 1521–1531. doi: 10.1007/s00253-009-2176-4
Thakaew, R., Niamsup, H. (2013). Inhibitory activity of bacillus subtilis BCC 6327 metabolites against growth of aflatoxigenic fungi isolated from bird chili powder. Int. J. Biosci. Biochem. Bioinforma. 3 (1), 27.
Ullah, A., Bano, A. (2019). Role of PGPR in the reclamation and revegetation of saline land. Pak. J. Bot. 51 (1), 27–35. doi: 10.30848/PJB2019-1(43)
Veshareh, M. J., Nick, H. M. (2018). “Learnings from reservoir souring treatment by nitrate injection in the Halfdan oil field,” in 80th EAGE Conference and Exhibition 2018, Vol. 2018. 1–5 (European Association of Geoscientists & Engineers).
Wei, B., Yang, L. (2010). A review of heavy metal contaminations in urban soils, urban road dusts and agricultural soils from China. Microchem. J. 94 (2), 99–107. doi: 10.1016/j.microc.2009.09.014
Wu, Y., Zhou, L., Lu, F., Bie, X., Zhao, H., Zhang, C., et al. (2019). Discovery of a novel antimicrobial lipopeptide, brevibacillin V, from Brevibacillus laterosporus fmb70 and its application on the preservation of skim milk. J. Agric. Food Chem. 67 (45), 12452–12460. doi: 10.1021/acs.jafc.9b04113
Yakimov, M. M., Timmis, K. N., Wray, V., Fredrickson, H. L. (1995). Characterization of a new lipopeptide surfactant produced by thermotolerant and halotolerant subsurface Bacillus licheniformis BAS50. Appl. Environ. Microbiol. 61 (5), 1706–1713. doi: 10.1128/aem.61.5.1706-1713.1995
Zehra, R., Moin, S., Enamullah, S. M., Ehteshamul-Haque, S. (2022). Detection and identification of quarantine bacteria and fungi associated with imported and local potato seed tubers. Pak. J. Bot. 54 (3), 1157–1161. doi: 10.30848/PJB2022-3(25)
Keywords: Bacillus subtilis FJ3, lipopeptides, antifungal, plant growth promoting traits and agriculture, biocontrol
Citation: Jan F, Arshad H, Ahad M, Jamal A and Smith DL (2023) In vitro assessment of Bacillus subtilis FJ3 affirms its biocontrol and plant growth promoting potential. Front. Plant Sci. 14:1205894. doi: 10.3389/fpls.2023.1205894
Received: 14 April 2023; Accepted: 23 June 2023;
Published: 18 July 2023.
Edited by:
Estibaliz Sansinenea, Meritorious Autonomous University of Puebla, MexicoReviewed by:
Parul Chaudhary, Graphic Era Hill University, IndiaCopyright © 2023 Jan, Arshad, Ahad, Jamal and Smith. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Asif Jamal, YXNpZmphbWFsQHFhdS5lZHUucGs=; Donald L. Smith, ZG9uYWxkLnNtaXRoQG1jZ2lsbC5jYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.