- Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) OT Gatersleben, Seeland, Germany
Genome editing strategies in barley (Hordeum vulgare L.) typically rely on Agrobacterium-mediated genetic transformation for the delivery of required genetic reagents involving tissue culture techniques. These approaches are genotype-dependent, time-consuming, and labor-intensive, which hampers rapid genome editing in barley. More recently, plant RNA viruses have been engineered to transiently express short guide RNAs facilitating CRISPR/Cas9-based targeted genome editing in plants that constitutively express Cas9. Here, we explored virus-induced genome editing (VIGE) based on barley stripe mosaic virus (BSMV) in Cas9-transgenic barley. Somatic and heritable editing in the ALBOSTRIANS gene (CMF7) resulting in albino/variegated chloroplast-defective barley mutants is shown. In addition, somatic editing in meiosis-related candidate genes in barley encoding ASY1 (an axis-localized HORMA domain protein), MUS81 (a DNA structure-selective endonuclease), and ZYP1 (a transverse filament protein of the synaptonemal complex) was achieved. Hence, the presented VIGE approach using BSMV enables rapid somatic and also heritable targeted gene editing in barley.
1 Introduction
RNA-guided clustered regularly interspaced short palindromic repeats (CRISPR)-associated (Cas) endonucleases emerged as a versatile tool for targeted genetic engineering in plants (Bao et al., 2019; Chen et al., 2019; Koeppel et al., 2019; Hinge et al., 2021; Hisano et al., 2021). In barley, the transfer of genetic reagents required to elicit Cas9-mediated targeted genetic engineering relies on stable genetic transformation (Lawrenson et al., 2015; Gerasimova et al., 2020). However, only a limited number of genotypes is amenable to efficient genetic transformation (Hensel et al., 2008; Yeo et al., 2014; Hoffie et al., 2021) and the isolation of stable genetic transformants is time- and labor-consuming. Hence, the rapid application of targeted genetic engineering in barley is hampered.
More recently, virus-induced genome editing (VIGE) emerged as a targeted genome editing tool for plants (Cody and Scholthof, 2019; Oh et al., 2021; Varanda et al., 2021; Gentzel et al., 2022; Uranga and Daròs, 2022). Plant viruses are engineered to deliver either guide RNAs into plants stably expressing endonucleases or even the complete genome engineering components (Cody and Scholthof, 2019; Metje-Sprink et al., 2019; Tsanova et al., 2021; Varanda et al., 2021; Gentzel et al., 2022). VIGE has been applied in dicot plants including Nicotiana benthamiana, Arabidopsis thaliana, and Glycine max using plant RNA viruses such as Tobacco rattle virus (TRV) (Ellison et al., 2020; Nagalakshmi et al., 2022), Pea early browning virus (PEBV) (Ali et al., 2018), Beet necrotic yellow vein virus (BNYVV) (Jiang et al., 2019), Potato virus X (PVX) (Ariga et al., 2020), Barley yellow striate mosaic virus (BYSMV) (Gao et al., 2019) or Sonchus yellow net rhabdovirus (SYNV) (Ali et al., 2015; Zaidi and Mansoor, 2017; Cody and Scholthof, 2019; Ellison et al., 2020; Ma et al., 2020; Nagalakshmi et al., 2022). In monocots, Foxtail mosaic virus in maize (FoMV) (Mei et al., 2019) or BSMV in wheat, cotton, and maize (Hu et al., 2019; Li et al., 2021; Chen et al., 2022a; Chen et al., 2022b; Wang et al., 2022) were employed for VIGE.
BSMV is a positive-sense RNA hordeivirus with a tripartite genome consisting of RNA α, β, and γ (Petty et al., 1990; Jackson et al., 2009). BSMV was harnessed to deliver sgRNAs into plants that ectopically express Cas9, such as monocots (wheat and maize) and the dicot N. benthamiana, for eliciting Cas9-mediated targeted genome editing in somatic tissues (Hu et al., 2019; Li et al., 2021; Chen et al., 2022a; Wang et al., 2022). In wheat, different frequencies of BSMV-mediated heritable gene editing were observed, ranging from 0.8 to 3.0% (Chen et al., 2022a; Chen et al., 2022b), 12.9 to 100% (Li et al., 2021), and 0 to 19% (Wang et al., 2022), depending on the genotype, type of sgRNA (with/without mobile RNA elements), and the target site (Notaguchi et al., 2014; Zhang et al., 2016; Ellison et al., 2020; Nagalakshmi et al., 2022).
BSMV also infects several other agronomically important cereal crops such as oats (Avena sativa) (Pacak et al., 2010), culinary ginger (Zingiber officinale) (Renner et al., 2009), rye (Secale cereale) (Groszyk et al., 2017), millet (Setaria italica), and barley (Jackson et al., 2009). In barley, BSMV infects several genotypes/cultivars (Edwards and Steffenson, 1996; Holzberg et al., 2002; Bruun-Rasmussen et al., 2007), has been harnessed for virus-induced gene silencing (VIGS) (Holzberg et al., 2002; Yuan et al., 2011; Lee et al., 2012; Dommes et al., 2019), enters the germline (Carroll, 1972; Carroll and Mayhew, 1976; Brlansky et al., 1986), and can be transmitted via grains (Carroll, 1972). Hence, these features offer an opportunity for efficient heritable editing in barley based on BSMV-mediated VIGE (BSMVIGE).
We explored BSMV as a VIGE tool in barley cv. Golden Promise plants expressing Cas9 by targeting, as proof of principle, the ALBOSTRIANS gene, CMF7, involved in chloroplast development (Li et al., 2019). Somatic cmf7 mutations, induced in virus-infected plants, can be transmitted to subsequent generations resulting in albino or variegated plants defective for CMF7. In addition, somatic mutations were induced in three meiosis-related genes in barley: ASY1, encoding for a meiotic chromosome axis-localized HORMA domain protein (Armstrong et al., 2002; Steckenborn et al., 2023), MUS81, encoding for a DNA structure-selective endonuclease involved in the formation of meiotic class II crossover (CO) (Hartung et al., 2006; Berchowitz et al., 2007; Higgins et al., 2008) as well as ZYP1, encoding for a transverse filament protein of the synaptonemal complex (SC) (Barakate et al., 2014; Steckenborn et al., 2023). Hence, BSMVIGE enables rapid somatic and heritable targeted gene editing in barley.
2 Materials and methods
2.1 Plant material and growing conditions
N. benthamiana plants were grown in a greenhouse at 22°C, relative humidity of 65%, and a photoperiod of 16/8h light/dark at 80-100 µmol m-2 s-1. Within a containment facility of biosafety level S2, after BSMV infection, plants were incubated at constant 24.5°C, relative humidity of 65%, and a photoperiod of 16/8h light/dark at 80-100 µmol m-2 s-1 in a growth cabinet (Polyklima). Barley plants expressing Cas9 of Streptococcus pyogenes were grown in a greenhouse at constant 19°C, 65% relative humidity, and a photoperiod of 16/8h light/dark at 160-250 µmol m-2 s-1. Five days before and after the BSMV infection, plants were incubated in a controlled growth chamber (at constant 24.5°C, 65% relative humidity, and a photoperiod of 16/8h light/dark at 160-250 µmol m-2 s-1) within a containment facility of biosafety level S2. Offspring from virus-infected plants were grown under the same controlled conditions except at constant 21°C. Barley plants were supplemented with 0.2% Wuxal fertilizer (MANNA GmbH, Germany) once per week.
2.2 Construction of transformation vectors
An expression unit consisting of the maize Ubi1 promoter and 5’-UTR including its intron 1, Streptomyces pyogenes Cas9 with maize codon usage preceded by a 4x FLAG tag and bordered by two copies of the Simian virus 40 nuclear localization signal (NLS) was assembled in front of the nopalin synthase polyadenylation signal of Agrobacterium tumefaciens in plasmid pUbi-ABM (DNA Cloning Service, Germany). This unit was then transferred as a compatible SfiI fragment to the generic binary vector p7intUbi (DNA Cloning Service, Germany) to generate pSH151.
2.3 Generation of transgenic plants ubiquitously expressing Cas9
Transgenic barley plants were generated as described (Hensel et al., 2009; Marthe et al., 2015). Briefly, dissected immature embryos were inoculated and co-cultivated with the A. tumefaciens strain AGL1 (Lazo et al., 1991) carrying binary vector pSH151. After callus induction and plant regeneration under selective conditions using timentin to remove residual agrobacteria and bialaphos for transgenicity, plantlets with developed roots were transferred to the soil substrate. Transgenicity of regenerants (presence of Cas9) and Cas9 expression were confirmed (for primer sequences see Supplementary Table 1). From two selected T0 plants based on Cas9 expression and being phenotypically indistinguishable from the WT, homozygous progeny were produced by microspore-derived plant regeneration (Kapusi et al., 2013; Lippmann et al., 2015). From the doubled haploids obtained, progeny of line BG710-DH62 referred to as ZmUBI::Cas9 plants (positive for Cas9 expression) were used for BSMV infections.
2.4 Design of SpCas9-compatible guide RNAs
According to criteria defined by (Schindele et al., 2020), using CRISPOR (Haeussler et al., 2016) and RNAfold (Gruber et al., 2008), single guide (sg)RNAs with high target specificity, low off-target scores, and appropriate secondary structure were selected: CTCCTGGATTCAGGATCCAT(GGG), GCAGACGTTGCGGTAGGCGT(TGG), and TTCTAGATCAGACTTCACCG(AGG) complementary to a target region within exon 4 of HvASY1 (HORVU.MOREX.r3.5HG0494140), exon 1 of HvMUS81 (HORVU.MOREX.r3.3HG0257160) and exon 2 of HvZYP1 (HORVU.MOREX.r3.2HG0172550), respectively. As controls, sgRNAs CTCAAGGCGTGGTATGACAG(AGG) and GGCGAGGGCGATGCCACCTA(CGG) specific to HvCMF7 (HORVU.MOREX.r3.7HG0728080) (Li et al., 2019) and AvGFP (Wang et al., 2017) were used, respectively.
2.5 Generation of the generic guide RNA expression vector BSMV-γ-sg and its specification for Cas9 target motifs
To generate BSMV-γ-sg enabling the expression of sgRNAs downstream of the γb ORF in RNAγ, a 359 bp sequence flanking the ligation-independent cloning (LIC) site in the pCa-LICγb plasmid (Yuan et al., 2011) was replaced with a 1065 bp custom synthesized sequence via HpaI/BamHI. This sequence consists of the BSMV-γb cDNA sequence, the ccdB gene flanked on both sites by AarI for insertion of target sequences, and the sgRNA scaffold sequence compatible with SpCas9 (Figure 1A, Supplementary Table 2). Annealed complementary sgRNA oligonucleotides were cloned into BSMV-γ-sg via Golden Gate cloning (Engler et al., 2008) using AarI and confirmed by Sanger-Sequencing employing BSMV-γ-sg insert-specific primers (Supplementary Table 1).
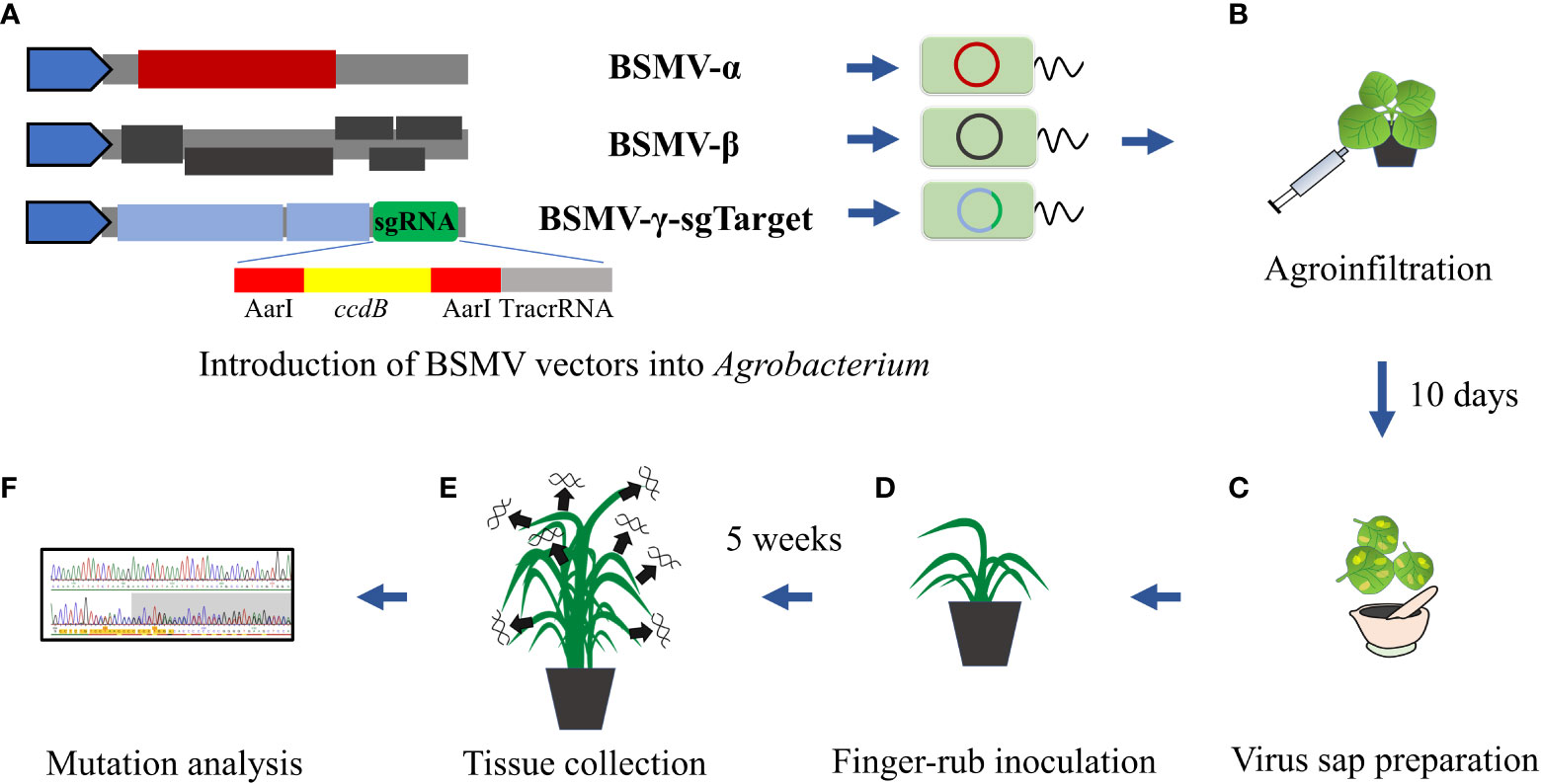
Figure 1 Schematic BSMVIGE workflow in barley. (A) Introduction of BSMV-α, BSMV-β, and BSMV-γ-sg vectors into Agrobacterium; (B) agroinfiltration using resuspended Agrobacterium mixtures harboring BSMVIGE vectors into N. benthamiana leaves; (C) virus leaf sap preparation 10 dpi; (D) finger-rub inoculation of virus sap into 3-4 week old ZmUBI::Cas9 plants; (E) tissue collection from multiple somatic structures 5 wpi; (F) mutation analysis of Sanger reads using ICE synthego tool (Conant et al., 2022).
2.6 Plant infection
BSMV-α, BSMV-β, and BSMV-γ-sg vectors (Yuan et al., 2011) were introduced individually into A. tumefaciens strain AGL1 (Figure 1A). An equal mixture (optical density at 600 nm of 1.2 of each culture) of AGL1 carrying the three BSMV components was infiltrated into leaves of 21 days old N. benthamiana plants to generate a high titer of functional BSMV (Figure 1B). Virus-symptom spotted leaves were harvested after 10-12 days post-infection (dpi) and ground in 10 mM phosphate buffer (pH 7) containing 0.5% of each celite 545 (Roth) and silicon carbide (400 mesh particle size, Sigma-Aldrich) (Figure 1C) on ice. The virus sap was used to inoculate 3 to 4 weeks-old barley plants by finger-rub inoculation of fully-emerged second and third leaves (Figure 1D).
2.7 Confirmation of BSMV infection
OneTaq one-step RT-PCR kit (NEB) and target-specific primers (Supplementary Table 1) were employed to amplify a region spanning the sgRNA in BSMV-γ-sg using 10 ng of extracted total RNA from leaf tissue.
2.8 Detection of targeted genome editing
Target regions were amplified using target-specific primers (Supplementary Table 1) and purified amplicons were Sanger-sequenced (Eurofins Genomics) (Figure 1E). Mutation efficiency (InDel percentage at target sites) and mutation frequency (frequency of plants showing mutation efficiency of at least 5% for each target) were estimated from Sanger reads using the ICE Synthego web tool (Conant et al., 2022) (Figure 1F). Sanger chromatograms of selected M1 plants (examples for high, medium or no editing at corresponding target site) are shown in Supplementary Figure 1.
3 Results
3.1 BSMVIGE mediates somatic editing at the ALBOSTRIANS locus in barley
To investigate the feasibility of BSMV-mediated genome editing in barley, initially, barley cv. Golden Promise plants that constitutively express Cas9 driven by the ZmUBI1 (maize ubiquitin 1) promotor were isolated. For a proof-of-principle of BSMVIGE in barley, a Cas9 target motif within the ALBOSTRIANS gene, CMF7, that had been successfully used for targeted gene editing, was selected (Li et al., 2019). As a negative control, a target motif specific to the AvGFP gene (Wang et al., 2017) that is absent in WT barley was chosen. For both target sequences, cognate gRNA 5’-ends were inserted into the BSMV-γ-sg vector resulting in BSMV-γ-sgCMF7 or BSMV-γ-sgGFP. The BSMV infection was performed according to (Yuan et al., 2011). For virus sap production, i.e. BSMVIGE-CMF7 or -GFP sap, N. benthamiana was infected with Agrobacterium harboring BSMV-α, BSMV-β, and BSMV-γ-sgCMF7 or BSMV-γ-sgGFP, respectively (Figure 1B). Then, virus sap was used for rub-infection of fully-emerged second and third leaves of 3-4 weeks old ZmUBI::Cas9 barley plants (Figures 1C, D). Assuming that systemic movement of the virus may lead to diverse Cas9-induced mutations in different plant tissues, potential presence of BSMV-mediated editing at the CMF7 target site was checked across multiple leaf tissues and emerging awns of different tillers in BSMVIGE-CMF7 or -GFP inoculated ZmUBI::Cas9 transgenic barley plants. To do so, pooled tissue samples, collected at 5 weeks post-infection (wpi) from individual plants, were used for Sanger read analysis using the ICE synthego tool to estimate the editing efficiency/types (Figures 1E, F) (Conant et al., 2022). Somatic editing was detected at the CMF7 target site in 8 out of 14 ZmUBI::Cas9 plants inoculated with BSMVIGE-CMF7. Mutation efficiencies of up to 94% in an individual plant and a mean mutation efficiency of 35% across all infected plants in two independent experiments were found (Figure 2, Supplementary Table 3). No editing in the target region was found in any ZmUBI::Cas9 barley plant infected with BSMVIGE-GFP, confirming the specificity of BSMVIGE-induced mutations for the CMF7 target motif (Supplementary Table 3). Given successful somatic editing in 8 out of 14 plants, we asked whether the presence of viral RNA in a given plant positively correlates with somatic editing or whether in turn absence of somatic editing in some plants was due to the absence of the virus. Note, while in wheat or other barley cultivars (Holzberg et al., 2002; Hu et al., 2019) infected with BSMV typical BSMV symptoms including yellow stripes/mosaic leaves are found, neither in BSMV- nor in rub-inoculated (no virus) ZmUBI::Cas9 plants any obvious phenotypic differences compared to uninoculated ZmUBI::Cas9 plants were found. Hence, absence of detectable BSMV symptoms in our materials inhibits visual identification of infected versus non-infected plants. Accordingly, seven randomly ZmUBI::Cas9 plants infected with BSMVIGE-CMF7 with mutation efficiencies ranging from zero to 94% were selected and analyzed for the presence of BSMV RNA 5 wpi. Except for a single plant that showed no signs of editing, BSMV was found in all further analyzed plants which include five plants with editing and one plant without editing. Hence, viral presence is required for editing at the target site but it does not assure in planta editing at detectable levels across various parts of a plant 5 wpi.
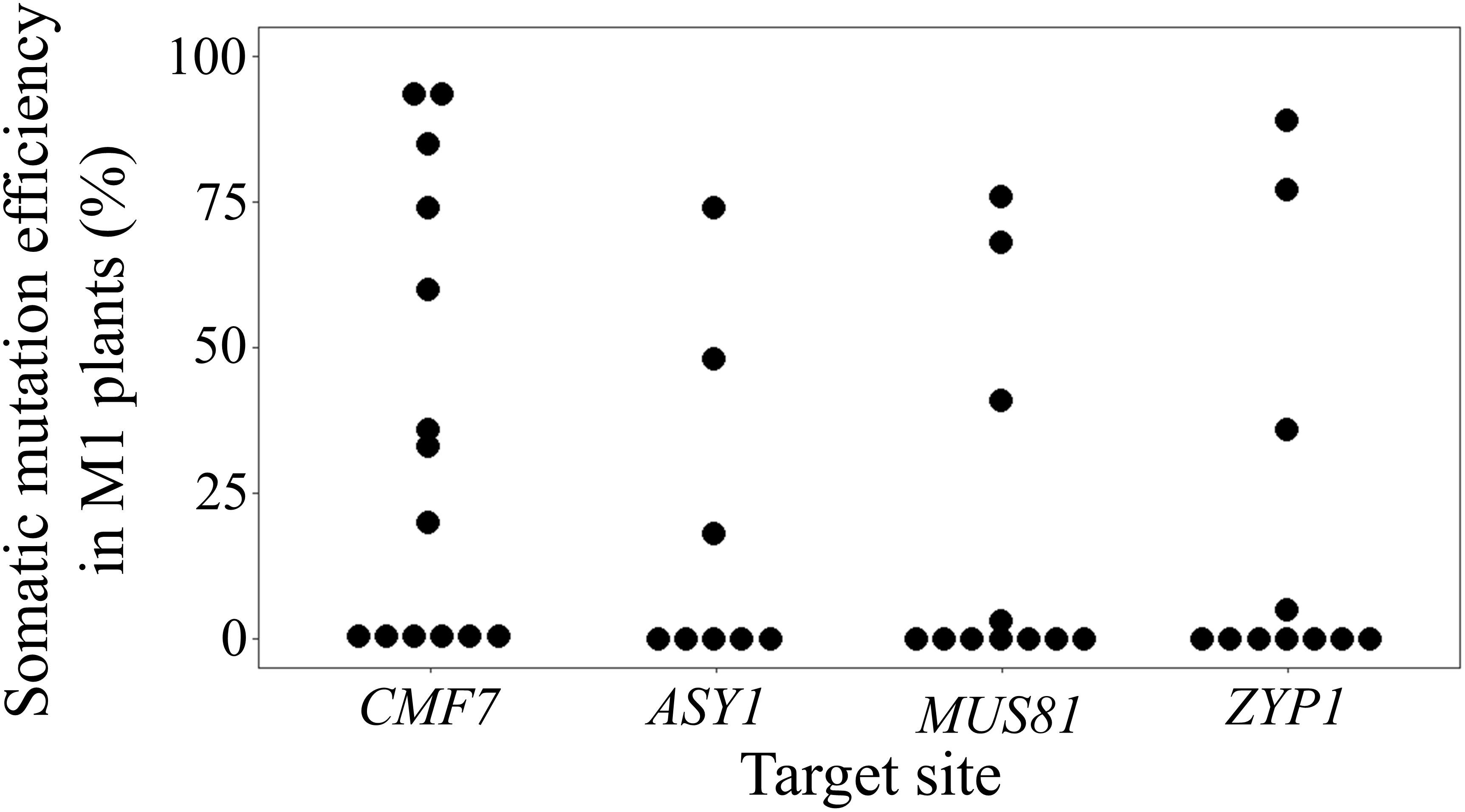
Figure 2 BSMVIGE mediated somatic gene editing in barley. Somatic mutation efficiency at CMF7, ASY1, MUS81, and ZYP1 target sites 5 wpi in ZmUBI::Cas9 plants inoculated with BSMVIGE-CMF7, -ASY1, -MUS81, and -ZYP1 virus saps, respectively.
3.2 Heritable editing at the CMF7 target site
Next, we asked whether induced somatic edits were transmitted into offspring plants. Three BSMV-positive primary mutant plants (M1) with variable editing efficiencies of 0, 33, and 94% termed M1-Null, M1-Medium, and M1-Highest, respectively, were selected. The CMF7 target region was sequenced in at least 20 offspring plants from each of the three selected M1 plants. We speculated that somatic mutation efficiency might aid in setting a selection criterion for choosing the appropriate M1 plant to be mined for heritable editing in offspring. Surprisingly, 33 of 62 analyzed M2 plants (~55%), comprising 8 (n=20), 9 (n=21), and 16 (n=21) M2 offspring from M1-Null, M1-Medium, and M1-Highest, had mutations at the target site (Figure 3A). Editing in offspring from M1-Null, being virus-positive five wpi without detectable somatic editing, suggested that BSMV-mediated editing might have occurred after tissue sampling e.g. in reproductive tissues such as the germline or the developing embryos. Hence, our Sanger-based analysis restricted to pooled somatic tissues hampered the identification of BSMV-dependent edits in M1-Null that likely occurred later than five wpi. Among 33 M2 cmf7 mutants, 17, 8, and 8 offspring plants had complex (bi-allelic or chimeric), heterozygous, and homozygous mutations at the target site, respectively (Figure 3A, Supplementary Table 4). Three M2 homozygous mutants showed a complete albino phenotype, reflecting a complete CMF7 loss-of-function phenotype (defective chloroplast development), while 5 M2 homozygous or heterozygous mutants showed a variegated phenotype likely based on residual/hypomorphic CMF7 activity (Figure 3B) (Li et al., 2019). Together, BSMVIGE induces somatic mutations at the CMF7 locus in BSMV-infected ZmUBI::Cas9 barley plants, which can be transmitted to offspring plants.
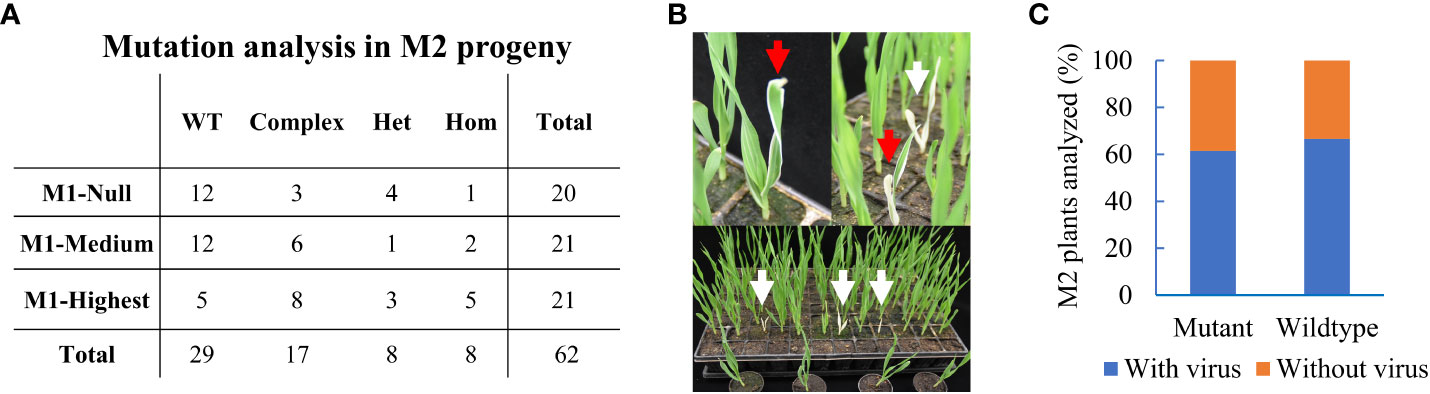
Figure 3 Heritable editing of the ALBOSTRIANS locus in barley. (A) M2 offspring plants from three selected BSMVIGE-CMF7 M1 plants (M1-Null, M1-Medium, and M1-Highest with somatic mutation efficiencies of 0%, 33%, and 94%, respectively): no mutation at the target locus (wild-type, WT), two or more mutations (complex), one mutant allele (heterozygous, Het) or similar mutation at both alleles (homozygous, Hom); (B) M2 progeny showing albino (white arrows) and variegated (red arrows) phenotypes; (C) M2 progeny analyzed for mutation at the CMF7 target site and presence of BSMV. Frequency of mutant or wild-type with presence/absence of BSMV RNA.
3.3 Virus-free offspring with heritable edits in M2 and M3 generations
BSMV transmission via grains depends on the BSMV strain and/or on the host genotype (Carroll, 1972). The generative transmission rate of the BSMVIGE-CMF7 virus (viral RNA presence/absence) was examined in randomly selected M2 offspring plants screened for heritable editing at the CMF7 target site. Among 38 analyzed M2 plants, 14 were virus-free, comprising 10 mutant and 4 WT plants for the CMF7 target site, and 24 plants were virus-positive, comprising 16 mutant and 8 WT plants for the CMF7 target site (Figure 3C, Supplementary Table 5). Hence, BSMVIGE-CMF7 can be generatively transmitted at a high rate in cv. Golden Promise, while virus-free mutant offspring plants can be recovered.
To determine whether the mutations found in cmf7 M2 plants will segregate in the M3 generation, a mutation analysis at the target region in M3 progeny from a virus-free M2 mutant plant being heterozygous for an ‘A’ insertion (cmf7) was performed. Among nine analyzed M3 offspring, three WT, one homozygous, and five heterozygous plants for cmf7 were found, suggesting the segregation of the cmf7 allele in the M3 generation (Supplementary Table 6). In addition, whether virus-free M3 cmf7 mutant progenies can be recovered, for both editing at the CMF7 target site and BSMV presence/absence in M3 progenies from a virus-positive M2 cmf7 mutant with multiple editing events (complex) was screened. Four virus-free and four virus-positive M3 offspring plants (n=8) were recovered. Among all these progeny plants, diverse editing events including complex and homozygous mutations at the CMF7 target site, which were transmitted from the M2 parent, were detected (Supplementary Table 6). Thus, regardless of virus transmission, BSMVIGE-induced cmf7 mutations are inherited into M3 progenies from M2 plants.
3.4 BSMVIGE induces somatic editing in putative meiotic genes in barley
To explore the reliability of BSMVIGE in barley, three putative meiosis-related genes that encode for the meiotic chromosome axis-associated protein ASY1 (Armstrong et al., 2002), a dHJ resolvase involved in the class II CO pathway, MUS81 (Hartung et al., 2006), and the transverse filament protein of the SC, ZYP1 (Barakate et al., 2014), were additionally addressed. Cas9 target motifs residing within exon 4 of ASY1, exon 1 of MUS81, or exon 2 of ZYP1 were selected and cognate gRNA 5’-ends cloned into BSMV-γ-sg to generate BSMV-γ-sgASY1, BSMV-γ-sgMUS81, and BSMV-γ-sgZYP1 (Figure 2, Supplementary Table 1). ZmUBI::Cas9 barley plants were infected using BSMVIGE-ASY1, BSMVIGE-MUS81, and BSMVIGE-ZYP1 virus saps. At 5 wpi, 3 out of 8 plants showed mutations at the ASY1 target site with a single plant showing a maximum mutation efficiency of 74%. In the case of BSMVIGE-MUS81, 3 out of 11 showed editing at the MUS81 target site while in the case of BSMVIGE-ZYP1, 4 out of 11 plants showed editing at the ZYP1 target site. The highest mutation efficiency observed in an individual plant at the target site was 76% and 89% for MUS81 and ZYP1 target sites, respectively. The mean mutation efficiencies at ASY1, MUS81, and ZYP1 target sites were 18%, 17%, and 19%, respectively. In summary, successful somatic editing at three additional target loci in barley suggests BSMV-VIGE as a rapid and reliable genome editing tool in barley.
4 Discussion
The use of RNA viruses to deliver gRNAs into plants expressing Cas9 for genome editing eliminates the need for further stable genetic transformation. Hence, it represents a rather rapid and easy-to-adopt plant genome editing approach, particularly in cereal crops. Therefore, we adapted a robust BSMVIGE workflow in barley that includes Golden Gate-based cloning of a single gRNA into a generic BSMVIGE vector followed by conventional tobacco leaf sap rub-inoculation of ZmUBI::Cas9 barley plants. To estimate somatic editing frequencies throughout infected plants, a rapid Sanger sequencing-based method is used. Accordingly, among a limited number of primary mutant plants, individuals are chosen for heritable editing screening in their offspring. By recreating the ALBOSTRIANS barley mutant, initially isolated through X-ray mutagenesis (Hagemann and Scholz, 1962) and CMF7 being recently identified as the causative gene in barley (Li et al., 2019), we show that virus-mediated heritable targeted gene editing based on BSMVIGE is possible in barley.
Somatic editing at the CMF7 target site was found 5 wpi in first-generation BSMVIGE-CMF7 virus-infected plants with an average frequency of 57% (n=14; mean mutation efficiency of 35%, ranging from zero to 94%). To possibly define somatic editing frequency as a criterion to choose plants for heritable editing screening in their offspring, M1 plants being positive for BSMV infection with no, intermediate (33%), and highest (94%) mutation efficiencies were selected to screen for heritable editing at the target site in their M2 progeny. Unexpectedly, not only heritable editing in the case of M1-Medium (43% frequency) and M1-Highest (76% frequency) offspring but also in the case of M1-Null (40% frequency) offspring was found. This suggests two non-exclusive possibilities: editing found in M1-Null offspring occurred only after 5 wpi analysis, e.g. in reproductive tissues, embryos or offspring plants, and/or the Sanger-based method used was not sensitive enough to detect the presence of low-frequency mutations. In any case, the highest heritable editing frequency with diverse editing events was found in the M1-Highest offspring. Whether a similar situation is found at other target sites, is unclear. However, M1 plants with rather high mutation efficiencies, represent obvious choices to screen for desired mutants in their offspring.
Notably, BSMV transmission into the next generation was found in barley, similar to wheat (Li et al., 2021), but at a higher frequency. However, virus-free plants with inherited mutations were obtained in both M2 and M3 generations and thus desired materials can be rapidly isolated by screening both for mutations at the target site(s) and viral presence/absence. In addition to our initial target site at the CMF7 gene, induction of somatic editing was also achieved at three putative meiosis-related genes, ASY1, MUS81, and ZYP1, with frequencies of 38%, 27%, and 36% (n=11), respectively, suggesting broad applicability of BSMVIGE for targeted genome engineering in barley.
The fusion of tRNAs or mobile RNA elements such as flowering locus T (FT) to gRNAs delivered by TRV can increase the heritable editing frequency in A. thaliana (Nagalakshmi et al., 2022) and N. benthamiana (Ellison et al., 2020). In wheat, both increased and decreased heritability of induced mutations upon fusion of RNA elements to gRNAs in BSMV were reported (Li et al., 2021; Chen et al., 2022a; Wang et al., 2022). These contradictory findings might be attributed to the different lengths and types of fusion RNAs as well as the different wheat genotypes employed. Whether in barley cv. Golden Promise the addition of tRNA or mobile RNA sequences to gRNAs improves BSMV-mediated (heritable) editing at target sites, should be addressed in future studies.
Multiple gRNAs addressing different targets can be expressed in a single virus such as TRV (Ellison et al., 2020) or Potato virus X (Uranga et al., 2021). Considering the limited cargo capacity of BSMV (Bruun-Rasmussen et al., 2007), stacking up gRNA arrays seems unfeasible. However, successful multiplexed gene editing in wheat using a mixed Agrobacterium pool strategy based on BSMVIGE (Li et al., 2021; Chen et al., 2022a; Wang et al., 2022), paves the way to test in the future, whether a similar strategy could also be adopted for barley. Furthermore, in existing mutant plants isolated either by conventional transformation strategies or by BSMVIGE being virus-free, likely BSMVIGE could be harnessed to edit independent genes of interest.
The current BSMVIGE approach is restricted to available barley genotypes that stably express Cas9, which is cv. Golden Promise in the present investigation. However, in addition to the possibility of generating further barley cultivars stably expressing Cas9, the current Cas9 transgene may be introgressed into other cultivars of interest (Chen et al., 2022a; Wang et al., 2022). Further, as demonstrated for wheat (Budhagatapalli et al., 2020), the development of Cas9 expressing haploid inducer barley lines infected with BSMV carrying gRNA(s) of interest could be utilized to induce mutations in the genome of the desired genotypes. Recently, a modified four-component BSMV system with increased cargo capacity (Cheuk and Houde, 2018) enabled transient somatic editing in cotton using split SpCas9 (Chen et al., 2022b). Whether reliable and frequent heritable editing based on a similar strategy can be achieved, needs to be addressed. Given the broad host range of BSMV and the application of BSMVIGE in barley (this study) or wheat (Li et al., 2021; Chen et al., 2022a; Wang et al., 2022), likely BSMVIGE can be adopted for other crops such as oat (Cho et al., 1999; Zhang et al., 1999) or rye (Popelka and Altpeter, 2003), where genetic transformation procedures are available.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
SHe acquired funding and designed this study. ST-N-A supported by FH, JL, RD and SD conducted most of the experiments and analyzed the data. SHi, GH, and JK generated barley Cas9-expressing lines. ST-N-A and SHe drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was supported by funding from the IPK (flagship project) and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No 949618). Costs for open-access publishing were partially funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation, grant 491250510).
Acknowledgments
We are grateful to Sabine Sommerfeld, Ingrid Otto, Carola Bollmann, Marius Dölling and Sylvia Swetik for excellent technical assistance, to the IPK gardeners for horticultural help, to Kostya Kanyuka for sharing materials and advice on BSMV work, to Manikandan Kalidass for critical reading of the manuscript, to all our lab members, IPK colleagues and Andreas Houben for fruitful discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1201446/full#supplementary-material
Supplementary Figure 1 | Sanger chromatogram analysis using ICE synthego tool (Conant et al., 2022) reveals somatic editing at target sites. Sanger traces at target sites within CMF7 (A), ASY1 (B), MUS81 (C) and ZYP1 (D) of selected BSMV-inoculated (top panel) compared with BSMV-uninoculated (bottom panel) M1 plants showing high, intermediate or no somatic editing 5 wpi analysed by ICE synthego tool. Details on plant materials indicated are given in Supplementary Table 3. In each case, the 20 bp Cas9 target motif is underlined in bold lines, while PAM is highlighted using dotted lines.
References
Ali, Z., Abulfaraj, A., Idris, A., Ali, S., Tashkandi, M., Mahfouz, M. M. (2015). CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 16. doi: 10.1186/s13059-015-0799-6
Ali, Z., Eid, A., Ali, S., Mahfouz, M. M. (2018). Pea early-browning virus-mediated genome editing via the CRISPR/Cas9 system in nicotiana benthamiana and arabidopsis. Virus Res. 244, 333–337. doi: 10.1016/j.virusres.2017.10.009
Ariga, H., Toki, S., Ishibashi, K. (2020). Potato virus X vector-mediated DNA-free genome editing in plants. Plant Cell Physiol. 61, 1946–1953. doi: 10.1093/pcp/pcaa123
Armstrong, S. J., Caryl, A. P., Jones, G. H., Franklin, F. C. H. (2002). Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in arabidopsis and brassica. J. Cell Sci. 115, 3645–3655. doi: 10.1242/jcs.00048
Bao, A., Burritt, D. J., Chen, H., Zhou, X., Cao, D., Tran, L.-S. P. (2019). The CRISPR/Cas9 system and its applications in crop genome editing. Crit. Rev. Biotechnol. 39, 321–336. doi: 10.1080/07388551.2018.1554621
Barakate, A., Higgins, J. D., Vivera, S., Stephens, J., Perry, R. M., Ramsay, L., et al. (2014). The synaptonemal complex protein ZYP1 is required for imposition of meiotic crossovers in barley. Plant Cell 26, 729–740. doi: 10.1105/tpc.113.121269
Berchowitz, L. E., Francis, K. E., Bey, A. L., Copenhaver, G. P. (2007). The role of AtMUS81 in interference-insensitive crossovers in a. thaliana. PloS Genet. 3, e132. doi: 10.1371/journal.pgen.0030132
Brlansky, R. H., Carroll, T. W., Zaske, S. K. (1986). Some ultrastructural aspects of the pollen transmission of barley stripe mosaic virus in barley. Can. J. Bot. 64, 853–858. doi: 10.1139/b86-111
Bruun-Rasmussen, M., Madsen, C. T., Jessing, S., Albrechtsen, M. (2007). Stability of barley stripe mosaic virus–induced gene silencing in barley. Mol. Plant-Microbe Interactions® 20, 1323–1331. doi: 10.1094/MPMI-20-11-1323
Budhagatapalli, N., Halbach, T., Hiekel, S., Büchner, H., Müller, A. E., Kumlehn, J. (2020). Site-directed mutagenesis in bread and durum wheat via pollination by cas9 /guide RNA-transgenic maize used as haploidy inducer. Plant Biotechnol. J. 18, 2376–2378. doi: 10.1111/pbi.13415
Carroll, T. W. (1972). Seed transmissibility of two strains of barley stripe mosaic virus. Virology 48, 323–336. doi: 10.1016/0042-6822(72)90043-8
Carroll, T. W., Mayhew, D. E. (1976). Occurrence of virions in developing ovules and embryo sacs of barley in relation to the seed transmissibility of barley stripe mosaic virus. Can. J. Bot. 54, 2497–2512. doi: 10.1139/b76-268
Chen, W., Huang, C., Luo, C., Zhang, Y., Zhang, B., Xie, Z., et al. (2022b). A new method for rapid subcellular localization and gene function analysis in cotton based on barley stripe mosaic virus. Plants 11, 1765. doi: 10.3390/plants11131765
Chen, H., Su, Z., Tian, B., Liu, Y., Pang, Y., Kavetskyi, V., et al. (2022a). Development and optimization of a Barley stripe mosaic virus -mediated gene editing system to improve fusarium head blight resistance in wheat. Plant Biotechnol. J. 20, 1018–1020. doi: 10.1111/pbi.13819
Chen, K., Wang, Y., Zhang, R., Zhang, H., Gao, C. (2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. doi: 10.1146/annurev-arplant-050718-100049
Cheuk, A., Houde, M. (2018). A new barley stripe mosaic virus allows Large protein overexpression for rapid function analysis. Plant Physiol. 176, 1919–1931. doi: 10.1104/pp.17.01412
Cho, M.-J., Jiang, W., Lemaux, P. G. (1999). High-frequency transformation of oat via microprojectile bombardment of seed-derived highly regenerative cultures. Plant Sci. 148, 9–17. doi: 10.1016/S0168-9452(99)00082-5
Cody, W. B., Scholthof, H. B. (2019). Plant virus vectors 3.0: transitioning into synthetic genomics. Annu. Rev. Phytopathol. 57, 211–230. doi: 10.1146/annurev-phyto-082718-100301
Conant, D., Hsiau, T., Rossi, N., Oki, J., Maures, T., Waite, K., et al. (2022). Inference of CRISPR edits from Sanger trace data. CRISPR J. 5, 123–130. doi: 10.1089/crispr.2021.0113
Dommes, A. B., Gross, T., Herbert, D. B., Kivivirta, K. I., Becker, A. (2019). Virus-induced gene silencing: empowering genetics in non-model organisms. J. Exp. Bot. 70, 757–770. doi: 10.1093/jxb/ery411
Edwards, M. C., Steffenson, B. J. (1996). Genetics and mapping of barley stripe mosaic virus resistance in barley. Phytopathology 86, 184–187. doi: 10.1094/Phyto-86-184
Ellison, E. E., Nagalakshmi, U., Gamo, M. E., Huang, P.-J., Dinesh-Kumar, S., Voytas, D. F. (2020). Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat. Plants 6, 620–624. doi: 10.1038/s41477-020-0670-y
Engler, C., Kandzia, R., Marillonnet, S. (2008). A one pot, one step, precision cloning method with high throughput capability. PLoS One 3, e3647. doi: 10.1371/journal.pone.0003647
Gao, Q., Xu, W.-Y., Yan, T., Fang, X.-D., Cao, Q., Zhang, Z.-J., et al. (2019). Rescue of a plant cytorhabdovirus as versatile expression platforms for planthopper and cereal genomic studies. New Phytol. 223, 2120–2133. doi: 10.1111/nph.15889
Gentzel, I. N., Ohlson, E. W., Redinbaugh, M. G., Wang, G.-L. (2022). VIGE: virus-induced genome editing for improving abiotic and biotic stress traits in plants. Stress Biol. 2. doi: 10.1007/s44154-021-00026-x
Gerasimova, S. V., Hertig, C., Korotkova, A. M., Kolosovskaya, E. V., Otto, I., Hiekel, S., et al. (2020). Conversion of hulled into naked barley by cas endonuclease-mediated knockout of the NUD gene. BMC Plant Biol. 20, 255. doi: 10.1186/s12870-020-02454-9
Groszyk, J., Kowalczyk, M., Yanushevska, Y., Stochmal, A., Rakoczy-Trojanowska, M., Orczyk, W. (2017). Identification and VIGS-based characterization of Bx1 ortholog in rye (Secale cereale l.). PLoS One 12, e0171506. doi: 10.1371/journal.pone.0171506
Gruber, A. R., Lorenz, R., Bernhart, S. H., Neubock, R., Hofacker, I. L. (2008). The Vienna RNA websuite. Nucleic Acids Res. 36, W70–W74. doi: 10.1093/nar/gkn188
Haeussler, M., Schönig, K., Eckert, H., Eschstruth, A., Mianné, J., Renaud, J.-B., et al. (2016). Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148. doi: 10.1186/s13059-016-1012-2
Hagemann, R., Scholz, F. (1962). A case of gene induced mutations of the plasmotype in barley. Theor. Appl. Genet. 32, 50–59.
Hartung, F., Suer, S., Bergmann, T., Puchta, H. (2006). The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRecQ4A. Nucleic Acids Res. 34, 4438–4448. doi: 10.1093/nar/gkl576
Hensel, G., Kastner, C., Oleszczuk, S., Riechen, J., Kumlehn, J. (2009). Agrobacterium-mediated gene transfer to cereal crop plants: current protocols for barley, wheat, triticale, and maize. Int. J. Plant Genomics 2009, 71–82. doi: 10.1155/2009/835608
Hensel, G., Valkov, V., Middlefell-Williams, J., Kumlehn, J. (2008). Efficient generation of transgenic barley: the way forward to modulate plant–microbe interactions. J. Plant Physiol. 165, 71–82. doi: 10.1016/j.jplph.2007.06.015
Higgins, J. D., Buckling, E. F., Franklin, F. C. H., Jones, G. H. (2008). Expression and functional analysis of AtMUS81 in arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 54, 152–162. doi: 10.1111/j.1365-313X.2008.03403.x
Hinge, V. R., Chavhan, R. L., Kale, S. P., Suprasanna, P., Kadam, U. S. (2021). Engineering resistance against viruses in field crops using CRISPR- Cas9. Curr. Genomics 22, 214–231. doi: 10.2174/1389202922666210412102214
Hisano, H., Abe, F., Hoffie, R. E., Kumlehn, J. (2021). Targeted genome modifications in cereal crops. Breed. Sci. 71, 405. doi: 10.1270/jsbbs.21019
Hoffie, R. E., Otto, I., Perovic, D., Budhagatapalli, N., Habekuß, A., Ordon, F., et al. (2021). Targeted knockout of eukaryotic translation initiation factor 4E confers bymovirus resistance in winter barley. Front. Genome Editing 34. doi: 10.3389/fgeed.2021.784233
Holzberg, S., Brosio, P., Gross, C., Pogue, G. P. (2002). Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 30, 315–327. doi: 10.1046/j.1365-313X.2002.01291.x
Hu, J., Li, S., Li, Z., Li, H., Song, W., Zhao, H., et al. (2019). A barley stripe mosaic virus-based guide RNA delivery system for targeted mutagenesis in wheat and maize. Mol. Plant Pathol. 20, 1463–1474. doi: 10.1111/mpp.12849
Jackson, A. O., Lim, H.-S., Bragg, J., Ganesan, U., Lee, M. Y. (2009). Hordeivirus replication, movement, and pathogenesis. Annu. Rev. Phytopathol. 47, 385–422. doi: 10.1146/annurev-phyto-080508-081733
Jiang, N., Zhang, C., Liu, J. Y., Guo, Z. H., Zhang, Z. Y., Han, C. G., et al. (2019). Development of beet necrotic yellow vein virus-based vectors for multiple-gene expression and guide RNA delivery in plant genome editing. Plant Biotechnol. J. 17, 1302–1315. doi: 10.1111/pbi.13055
Kapusi, E., Hensel, G., Coronado, M.-J., Broeders, S., Marthe, C., Otto, I., et al. (2013). The elimination of a selectable marker gene in the doubled haploid progeny of co-transformed barley plants. Plant Mol. Biol. 81, 149–160. doi: 10.1007/s11103-012-9988-9
Koeppel, I., Hertig, C., Hoffie, R., Kumlehn, J. (2019). Cas endonuclease technology–a quantum leap in the advancement of barley and wheat genetic engineering. Int. J. Mol. Sci. 20, 2647. doi: 10.3390/ijms20112647
Lawrenson, T., Shorinola, O., Stacey, N., Li, C., Østergaard, L., Patron, N., et al. (2015). Induction of targeted, heritable mutations in barley and brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 16. doi: 10.1186/s13059-015-0826-7
Lazo, G. R., Stein, P. A., Ludwig, R. A. (1991). A DNA transformation–competent arabidopsis genomic library in agrobacterium. Bio/technology 9, 963–967. doi: 10.1038/nbt1091-963
Lee, W. S., Hammond-Kosack, K. E., Kanyuka, K. (2012). Barley stripe mosaic virus-mediated tools for investigating gene function in cereal plants and their pathogens: virus-induced gene silencing, host-mediated gene silencing, and virus-mediated overexpression of heterologous protein. Plant Physiol. 160, 582–590. doi: 10.1104/pp.112.203489
Li, M., Hensel, G., Mascher, M., Melzer, M., Budhagatapalli, N., Rutten, T., et al. (2019). Leaf variegation and impaired chloroplast development caused by a truncated CCT domain gene in albostrians barley. Plant Cell. 31, 1430–1445. doi: 10.1105/tpc.19.00132
Li, T., Hu, J., Sun, Y., Li, B., Zhang, D., Li, W., et al. (2021). Highly efficient heritable genome editing in wheat using an RNA virus and bypassing tissue culture. Mol. Plant 14, 1787–1798. doi: 10.1016/j.molp.2021.07.010
Lippmann, R., Friedel, S., Mock, H.-P., Kumlehn, J. (2015). The low molecular weight fraction of compounds released from immature wheat pistils supports barley pollen embryogenesis. Front. Plant Sci. 6, 498. doi: 10.3389/fpls.2015.00498
Ma, X., Zhang, X., Liu, H., Li, Z. (2020). Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nat. Plants 6, 773–779. doi: 10.1038/s41477-020-0704-5
Marthe, C., Kumlehn, J., Hensel, G. (2015). “Barley (Hordeum vulgare l.) transformation using immature embryos,” in Agrobacterium protocols, vol. 1 . Ed. WANG, K. (New York, NY: Springer New York).
Mei, Y., Beernink, B. M., Ellison, E. E., Konečná, E., Neelakandan, A. K., Voytas, D. F., et al. (2019). Protein expression and gene editing in monocots using foxtail mosaic virus vectors. Plant Direct 3, e00181. doi: 10.1002/pld3.181
Metje-Sprink, J., Menz, J., Modrzejewski, D., Sprink, T. (2019). DNA-Free genome editing: past, present and future. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01957
Nagalakshmi, U., Meier, N., Liu, J.-Y., Voytas, D. F., Dinesh-Kumar, S. P. (2022). High-efficiency multiplex biallelic heritable editing in arabidopsis using an RNA virus. Plant Physiol. 189, 1241–1245. doi: 10.1093/plphys/kiac159
Notaguchi, M., Higashiyama, T., Suzuki, T. (2014). Identification of mRNAs that move over long distances using an RNA-seq analysis of Arabidopsis/Nicotiana benthamiana heterografts. Plant Cell Physiol. 56, 311–321. doi: 10.1093/pcp/pcu210
Oh, Y., Kim, H., Kim, S.-G. (2021). Virus-induced plant genome editing. Curr. Opin. Plant Biol. 60, 101992. doi: 10.1016/j.pbi.2020.101992
Pacak, A., Geisler, K., Jørgensen, B., Barciszewska-Pacak, M., Nilsson, L., Nielsen, T. H., et al. (2010). Investigations of barley stripe mosaic virus as a gene silencing vector in barley roots and in brachypodium distachyon and oat. Plant Methods 6, 26. doi: 10.1186/1746-4811-6-26
Petty, I. T., French, R., Jones, R. W., Jackson, A. O. (1990). Identification of barley stripe mosaic virus genes involved in viral RNA replication and systemic movement. EMBO J. 9, 3453–3457. doi: 10.1002/j.1460-2075.1990.tb07553.x
Popelka, J. C., Altpeter, F. (2003). Agrobacterium tumefaciens-mediated genetic transformation of rye (Secale cereale L.). Molecular Breeding 11, 203–211. doi: 10.1023/A:1022876318276
Renner, T., Bragg, J., Driscoll, H. E., Cho, J., Jackson, A. O., Specht, C. D. (2009). Virus-induced gene silencing in the culinary ginger (Zingiber officinale): an effective mechanism for down-regulating gene expression in tropical monocots. Mol. Plant 2, 1084–1094. doi: 10.1093/mp/ssp033
Schindele, P., Wolter, F., Puchta, H. (2020). “CRISPR guide RNA design guidelines for efficient genome editing,” in RNA Tagging: methods and protocols. Ed. HEINLEIN, M. (New York, NY: Springer US).
Steckenborn, S., Cuacos, M., Ayoub, M. A., Feng, C., Schubert, V., Hoffie, I., et al. (2023). The meiotic topoisomerase VI b subunit (MTOPVIB) is essential for meiotic DNA double-strand break formation in barley (Hordeum vulgare l.). Plant Reprod. 36, 1–15. doi: 10.1007/s00497-022-00444-5
Tsanova, T., Stefanova, L., Topalova, L., Atanasov, A., Pantchev, I. (2021). DNA-Free gene editing in plants: a brief overview. Biotechnol. Biotechnol. Equip. 35, 131–138. doi: 10.1080/13102818.2020.1858159
Uranga, M., Aragonés, V., Selma, S., Vázquez-Vilar, M., Orzáez, D., Daròs, J. A. (2021). Efficient Cas9 multiplex editing using unspaced sgRNA arrays engineering in a potato virus X vector. Plant J. 106, 555–565. doi: 10.1111/tpj.15164
Uranga, M., Daròs, J. A. (2022). Tools and targets: the dual role of plant viruses in CRISPR–cas genome editing. Plant Genome, e20220. doi: 10.1002/tpg2.20220
Varanda, C. M. R., Félix, M. D. R., Campos, M. D., Patanita, M., Materatski, P. (2021). Plant viruses: from targets to tools for CRISPR. Viruses 13, 141. doi: 10.3390/v13010141
Wang, B., Krall, E. B., Aguirre, A. J., Kim, M., Widlund, H. R., Doshi, M. B., et al. (2017). ATXN1L, CIC, and ETS transcription factors modulate sensitivity to MAPK pathway inhibition. Cell Rep. 18, 1543–1557. doi: 10.1016/j.celrep.2017.01.031
Wang, W., Yu, Z., He, F., Bai, G., Trick, H. N., Akhunova, A., et al. (2022). Multiplexed promoter and gene editing in wheat using a virus-based guide RNA delivery system. Plant Biotechnol. J. 20, 2332–2341. doi: 10.1111/pbi.13910
Yeo, F., Hensel, G., Vozábová, T., Martin-Sanz, A., Marcel, T., Kumlehn, J., et al. (2014). Golden SusPtrit: a genetically well transformable barley line for studies on the resistance to rust fungi. Theor. Appl. Genet. 127, 325–337. doi: 10.1007/s00122-013-2221-7
Yuan, C., Li, C., Yan, L., Jackson, A. O., Liu, Z., Han, C., et al. (2011). A high throughput barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS One 6, e26468. doi: 10.1371/journal.pone.0026468
Zaidi, S. S.-E.-A., Mansoor, S. (2017). Viral vectors for plant genome engineering. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00539
Zhang, S., Cho, M. J., Koprek, T., Yun, R., Bregitzer, P., Lemaux, P. G. (1999). Genetic transformation of commercial cultivars of oat (Avena sativa l.) and barley (Hordeum vulgare l.) using in vitro shoot meristematic cultures derived from germinated seedlings. Plant Cell Rep. 18, 959–966. doi: 10.1007/s002990050691
Keywords: CRISPR/Cas9, BSMV, VIGE, albostrians, barley, genome editing
Citation: Tamilselvan-Nattar-Amutha S, Hiekel S, Hartmann F, Lorenz J, Dabhi RV, Dreissig S, Hensel G, Kumlehn J and Heckmann S (2023) Barley stripe mosaic virus-mediated somatic and heritable gene editing in barley (Hordeum vulgare L.). Front. Plant Sci. 14:1201446. doi: 10.3389/fpls.2023.1201446
Received: 06 April 2023; Accepted: 02 June 2023;
Published: 19 June 2023.
Edited by:
Magdalena Klimek-Chodacka, University of Agriculture in Krakow, PolandReviewed by:
Xingliang Ma, University of Saskatchewan, CanadaUlhas S. Kadam, Gyeongsang National University, Republic of Korea
Copyright © 2023 Tamilselvan-Nattar-Amutha, Hiekel, Hartmann, Lorenz, Dabhi, Dreissig, Hensel, Kumlehn and Heckmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefan Heckmann, aGVja21hbm5AaXBrLWdhdGVyc2xlYmVuLmRl
†Present address: Goetz Hensel, Centre for Plant Genome Engineering, Institute of Plant Biochemistry, Heinrich Heine University, Düsseldorf, Germany
‡ORCID: Suriya Tamilselvan-Nattar-Amutha, orcid.org/0000-0002-2940-4354
Steven Dreissig, orcid.org/0000-0002-4766-9698
Goetz Hensel, orcid.org/0000-0002-5539-3097
Jochen Kumlehn, orcid.org/0000-0001-7080-7983
Stefan Heckmann, orcid.org/0000-0002-0189-8428
 Suriya Tamilselvan-Nattar-Amutha‡
Suriya Tamilselvan-Nattar-Amutha‡ Steven Dreissig
Steven Dreissig Jochen Kumlehn
Jochen Kumlehn Stefan Heckmann
Stefan Heckmann