- 1Institute of Crop Sciences, National Wheat Improvement Center, Chinese Academy of Agricultural Sciences (CAAS), Beijing, China
- 2CAAS-IRRI Joint Laboratory for Genomics-Assisted Germplasm Enhancement, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, China
- 3School of Agriculture, Sun Yat-sen University, Shenzhen, China
- 4Sanya Nanfan Research Institute of Hainan University, Hainan University, Sanya, China
- 5Strategic Innovation Platform, International Rice Research Institute, Manila, Philippines
Mesocotyl is an essential organ of rice for pushing buds out of soil and plays a crucial role in seeding emergence and development in direct-seeding. Thus, identify the loci associated with mesocotyl length (ML) could accelerate breeding progresses for direct-seeding cultivation. Mesocotyl elongation was mainly regulated by plant hormones. Although several regions and candidate genes governing ML have been reported, the effects of them in diverse breeding populations were still indistinct. In this study, 281 genes related to plant hormones at the genomic regions associated with ML were selected and evaluated by single-locus mixed linear model (SL-MLM) and multi-locus random-SNP-effect mixed linear model (mr-MLM) in two breeding panels (Trop and Indx) originated from the 3K re-sequence project. Furthermore, superior haplotypes with longer mesocotyl were also identified for marker assisted selection (MAS) breeding. Totally, LOC_Os02g17680 (explained 7.1-8.9% phenotypic variations), LOC_Os04g56950 (8.0%), LOC_Os07g24190 (9.3%) and LOC_Os12g12720 (5.6-8.0%) were identified significantly associated with ML in Trop panel, whereas LOC_Os02g17680 (6.5-7.4%), LOC_Os04g56950 (5.5%), LOC_Os06g24850 (4.8%) and LOC_Os07g40240 (4.8-7.1%) were detected in Indx panel. Among these, LOC_Os02g17680 and LOC_Os04g56950 were identified in both panels. Haplotype analysis for the six significant genes indicated that haplotype distribution of the same gene varies at Trop and Indx panels. Totally, 8 (LOC_Os02g17680-Hap1 and Hap2, LOC_Os04g56950-Hap1, Hap2 and Hap8, LOC_Os07g24190-Hap3, LOC_Os12g12720-Hap3 and Hap6) and six superior haplotypes (LOC_Os02g17680-Hap2, Hap5 and Hap7, LOC_Os04g56950-Hap4, LOC_Os06g24850-Hap2 and LOC_Os07g40240-Hap3) with higher ML were identified in Trop and Indx panels, respectively. In addition, significant additive effects for ML with more superior haplotypes were identified in both panels. Overall, the 6 significantly associated genes and their superior haplotypes could be used to enhancing ML through MAS breeding and further promote direct-seedling cultivation.
Introduction
Rice (Oryza sativa) is one of the most important food crops in the world. Maintaining a higher and stable grain yield is crucial for food security especially in developing countries of Asia, such as China, Philippines, Vietnam and Malaysia. Traditional transplanting and direct-seeding are two major patterns for rice. Direct-seeding without transplanting process is labor-saving and water-efficient (Kumar and Ladha, 2011; Kato and Katsura, 2014; Liu et al., 2015; Ohno et al., 2018; Zhan et al., 2020). However, there are many disadvantages for direct-seeding, such as low seeding emergence rate, poor seeding establishment, weed infestation and high crop lodging rate (Mahender et al., 2015; Lee et al., 2017). Mesocotyl, an organ developed during rice seed germination in the dark and connects the coleoptile node and the basal part of seminal root, plays a key role in pushing buds out of deep water for successful seeding establishment (Zhan et al., 2020). Therefore, varieties with longer mesocotyl could be used to solve the problems induced by direct seeding cultivation (Lee et al., 2017; Zhan et al., 2020).
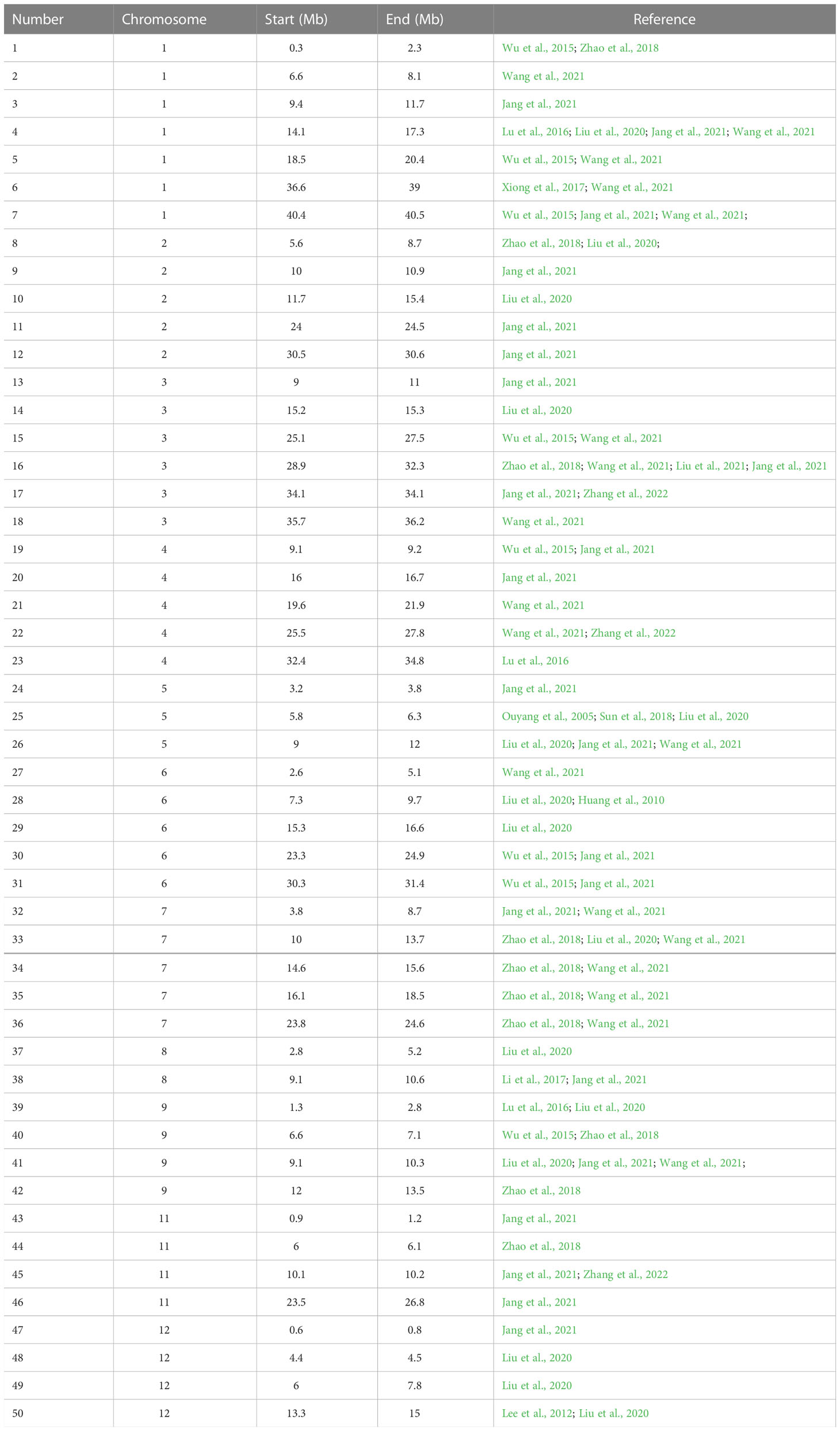
ML is a typical quantitative trait controlled by minor genes (Wu et al., 2015; Sun et al., 2018; Liu et al., 2020; Zhan et al., 2020; Jang et al., 2021; Zhang et al., 2022). Up to now, over 40 ML related Quantitative trait loci (QTLs) have been identified on 12 chromosomes and explain 5.7-27.8% of the phenotypic variations (Liu et al., 2020; Rohilla et al., 2020; Zhan et al., 2020). Recent advances in rice functional genomics facilitated the cloning and functional characterization of ML related genes, including GY1 (Xiong et al., 2017), OsGSK2 (Sun et al., 2018), OsSMAX1 (Zheng et al., 2020) and OsPAO5 (Lv et al., 2021). All the above cloned genes are involved in the plant hormone regulation. Also, previously reports showed that the mesocotyl elongation is regulated by various plant hormones, including Auxin (IAA), gibberellins (GA), ethylene (ETH), cytokinin (CTK) (Yuldashev et al., 2012), abscisic acid (ABA) (Watanabe and Takahashi, 1999; Watanabe et al., 2001; Wu et al., 2002), Jasmonic acid (JA), Strigolactones (SL) (Hu et al., 2014) and Brassinolide (BR). The mutual regulation of various plant hormones jointly regulates the elongation of rice mesocotyl (Xiong et al., 2017). Of these, IAA, GA, ETH, CTK and ABA can promote mesocotyl elongation; whereas JA and SL plays an inhibitory role. Lower concentration BR promotes mesocotyl elongation, whereas higher concentration inhibits. GA promotes cell elongation by changing the arrangement direction of cell microtubules and enhancing pectin methylation (Watanabe et al., 2001), whereas IAA mainly upregulates the activity of cell wall relaxant enzyme and promote cell growth (Zhan et al., 2020). ABA promotes the elongation of mesocotyl by inhibiting BR signaling pathway and then enhancing cell division near coleoptile node (Wu et al., 2002); whereas ETH promotes mesocotyl elongation by inhibiting JA synthesis (Xiong et al., 2017).
Association analysis is a powerful approach to understand clearly the genetic mechanism for complex traits (Flint-Garcia et al., 2003; Zhu et al., 2008; Wen et al., 2018). Single-locus mixed linear model (SL-MLM) is the most commonly used association analysis method, which was influenced seriously by polygenic background, including population structure and kinship (Zhu et al., 2008; Cui et al., 2018; Zhang et al., 2020a). Multi-locus association analysis (ML-AA), a method solves the SL-MLM induced shortcomings by estimating all the genetic effects across all the whole genome (Wen et al., 2018; Zhang et al., 2020b). ML-AA outperformed single locus-based methods in identify the minor effects loci of quantitative inheritance crop complex traits (Tamba and Zhang, 2018; Wen et al., 2018; Yang et al., 2020). Candidate gene association study (CAS) based on the target genes at the functional regions further increased the mapping resolution (Flint-Garcia et al., 2003; Zhu et al., 2008). Identifying minor genes of complex traits by CAS were conducted in Arabidopsis, rice, maize and common wheat (Zhao et al., 2015). Haplotype is the combination of alleles at different position on the same genomic regions for common inheritance (Liu et al., 2021), effective than SNP (Single nucleotide polymorphism) and InDel (Insertion-deletion) in crop marker-assisted selection (MAS) breeding (Li et al., 2017; Resende et al., 2017; Prodhomme et al., 2020). Superior haplotype identification has been proven to be an effective way to identify genes associated with complex traits and availability for crop breeding (Bevan et al., 2017; Abbai et al., 2019; Sinha et al., 2020). Previous approaches for genetic studies of ML were mainly focused on traditional linkage or association mapping, which hardly evaluate the existence and effects of natural variants and haplotypes. Thus, evaluating the genetic effects of the candidate gene for ML by CAS and identifying its correspondence superior haplotype in natural populations will accelerate the genetic improvement of ML.
Until now, substantial MAS breeding practices have been conducted to disease resistance, abiotic stress tolerance and yield related traits (Wang et al., 2020). However, MAS for ML is hindered due to the rare details of ML related genes and their haplotypes. To promote the progress of rice higher ML breeding, the effects of 281 selected genes related to plant hormones at reported ML genomic regions were evaluated in two breeding panels and the corresponding superior haplotypes were identified (Tables 1, S1).
Materials and methods
Plant materials
Two breeding populations (Trop and Indx) originated from the 3K re-sequence projects were employed in this study (Li et al., 2014; Alexandrov et al., 2015; Wang et al., 2018). The Trop panel including 331 Japonica (Geng) accessions mainly from Malaysia, United States, Philippines and Indonesia; whereas Indx panel including 470 Indica (Xian) accessions mainly from India, Philippines, China, Myanmar and Indonesia. The selected accessions have higher genetic polymorphism with various background.
Genotyping, population structure and haplotype analysis
All the genotypes of Trop and Indx panels were obtained from the 3K-resequence projects (https://snp-seek.irri.org/_snp.zul) (https://www.rmbreeding.cn/) (Alexandrov et al., 2015; Wang et al., 2018). Sequence reads (nearly 12×) were aligned to the Nipponbare RefSeq (IRGSP-1.0) (http://rice.plantbiology.msu.edu/index.shtml). The variants for each accession were called by the GATK V3.2.2. Stringent filtering strategy was conducted (QUAL < 30.0, QD < 10.0, FS > 200.0, MQRankSum < -12.5 and ReadPosRankSum < -8.0). In the present study, markers with minor allele frequency (MAF) < 0.05 and missing rate > 0.05 were removed. SNPs and InDels were annotated by ANNOVAR (Wang et al., 2010). The SNPs and Indels located in the CDS region and the promoter (-1500 bp) of the 281 selected genes were extracted and used for further CAS and haplotype analysis. Haplotype analysis was conducted by considering the nonsynonymous variations at RFGB database (https://www.rmbreeding.cn/). The SNPs for haplotype analysis were filtered according to the following requirements: (1) only two alleles; (2) missing data < 0.1; (3) MAF ≥ 0.05; (4) exclude the correlated markers (r2 = 1.0).
Phenotyping of ML and seedling height
The ML were measured according to Wang et al. (2021). In short, 15 plump seeds for each accession were sown in a plastic tray with nutrient soil at 6 cm), then the plastic tray was placed in a pallet with nutrient soil at 3 cm. The whole devices were then kept in a dark incubator (30°C/65% RH) for about 10 days after all seeds germinated. Seedings were carefully excavated and washed with ddH2O for ML measurement by Image J (https://imagej.en.softonic.com/). The mean of two replications was calculated as the phenotype data for further CAS analysis. The seedling height for the accessions from Trop and Indx were originated from the RFGB database (Wang et al., 2018) (https://www.rmbreeding.cn/).
Candidate gene study and superior haplotype identification
CAS was carried out using the Tassel V5.1 with a mixed linear model accounting for both PCA and kinship (Bradbury et al., 2007; Lipka et al., 2012). The Manhattan and QQ plots were drawn by CMplot (https://github.com/YinLiLin/CMplot) based on R v3.6.4. Mr-MLM V2.1, was used to perform the mr-MLM algorithm (Wang et al., 2020). The threshold for marker-trait association (MTA) was set as P>10-4 in SL-MLM and at a LOD value of 3.0 in mr-MLM. The significant genes were further used to identify superior haplotypes by Duncan analysis of ML means at Trop and Indx. Furthermore, only haplotypes existed at least five accessions in each panel were included for statistical analysis to ensure the accuracy of the results.
Results
Phenotype and genotype analysis
Continuous variation with transgressive segregation on both sides for ML and seedling height were observed across both Trop and Indx panels with approximately normal distributions (Figure S1). The ML for the Trop panel ranged from 0.20 to 4.40 cm with an average of 1.81 cm (Table S2), whereas the data ranged from 0 to 4.60 cm with an average of 1.40 cm for Indx panel (Table S3). The standard deviation and coefficient of variation of ML were 0.899 cm (coefficient of variation 0.50) and 0.893 cm (coefficient of variation 0.659) of Trop and Indx panel, respectively. The seedling height for the Trop panel ranged from 12.0 to 61.0 cm with an average of 34.7 cm (Table S2), whereas the data ranged from 16.0 to 74.0 cm with an average of 40.9 cm for Indx panel (Table S3). The standard deviation and coefficient of variation of seedling height were 10.4 cm (coefficient of variation 0.30) and 11.8 cm (coefficient of variation 0.29) of Trop and Indx panel, respectively.
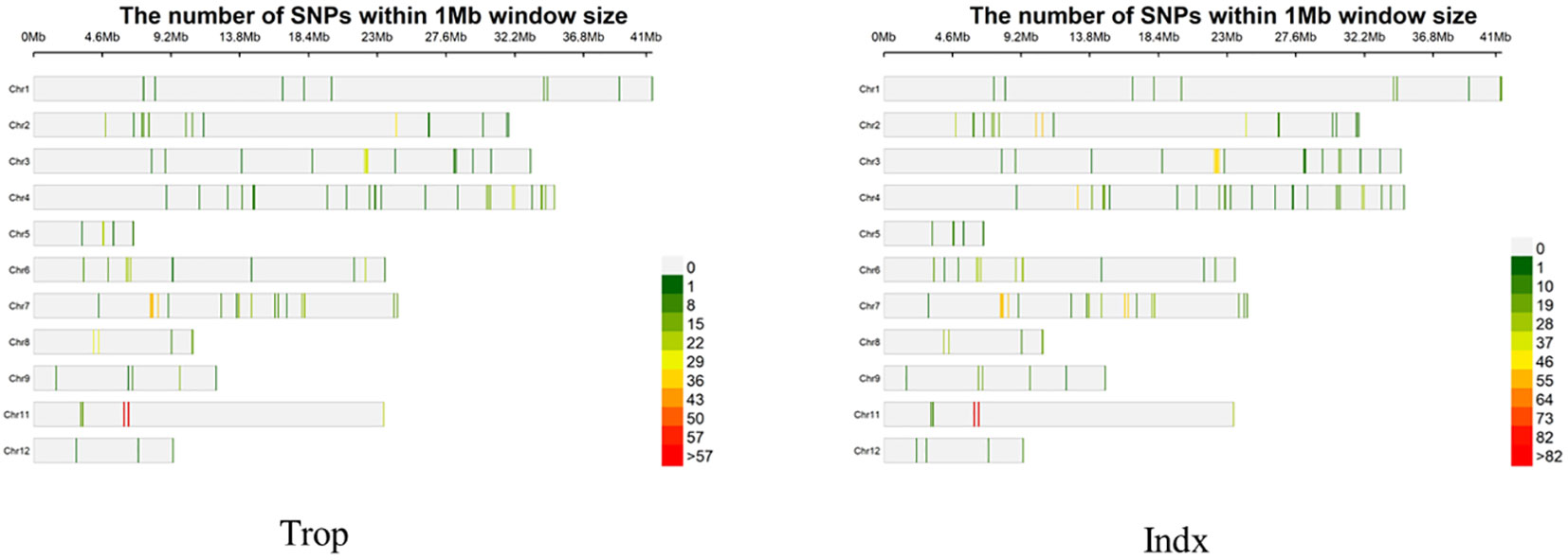
A total of 2277 SNPs and 1414 Indels were identified in the CDS and promoter regions of 281 selected genes in both panels. The SNPs and Indels for each gene ranged from 5 to 25 with the mean at 8.10 and 0 to 14 with the mean at 5.03 (Tables S4, S5; Figure 1). Trop and Indx subpopulation were classified in the 3K Rice Genomes Project and could be related to the geographic origins (Wang et al., 2018). Thus, population structure analysis was not conducted in this study and the MLM model were used for further CAS analysis. Principal component analysis indicated that the total variation explained by the top three PCs were 28.5%, 8.2% and 3.2% in Trop panel, whereas 24.5%, 9.2% and 7.3% in Indx panel.
SL-AA and ML-AA analysis
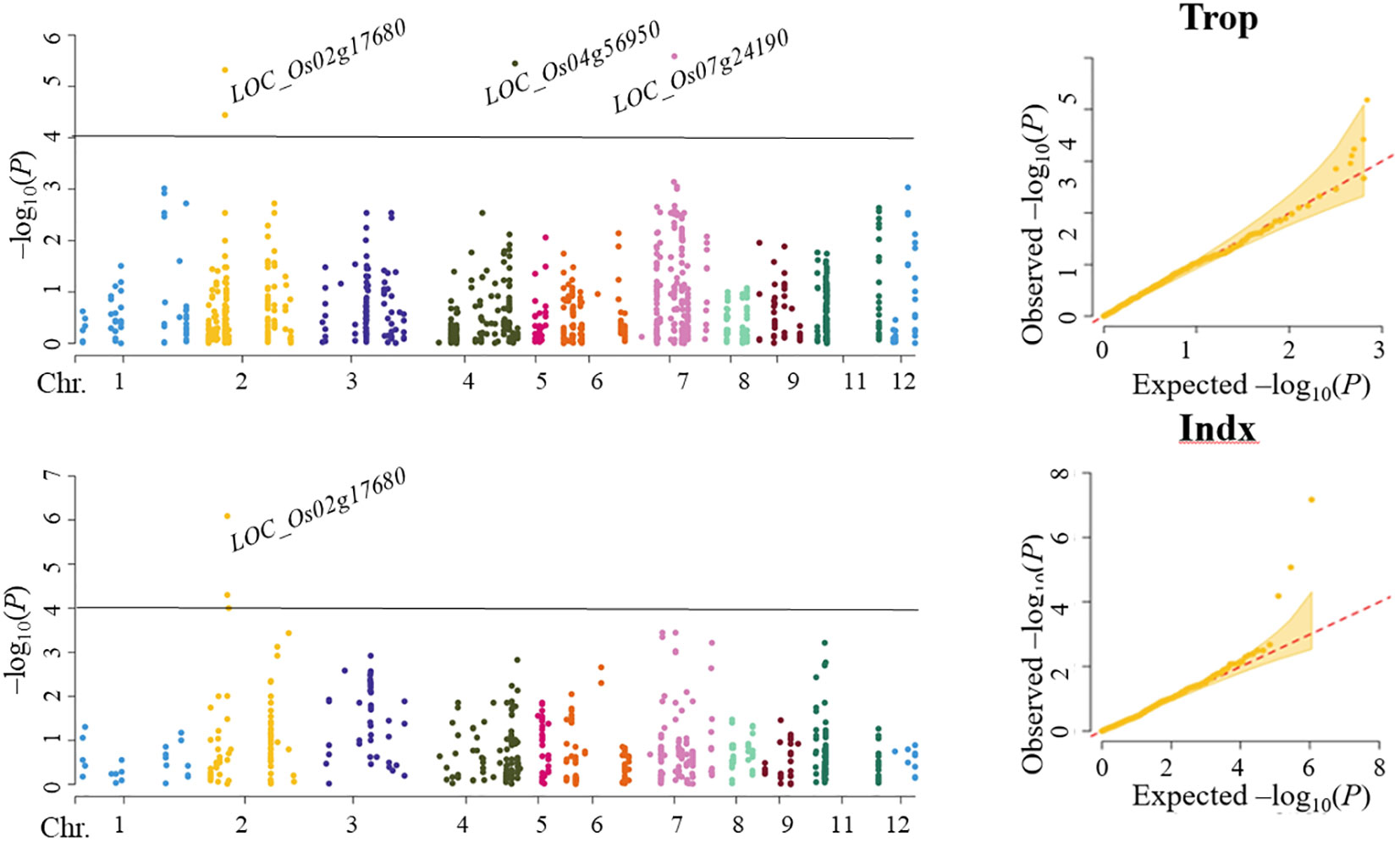
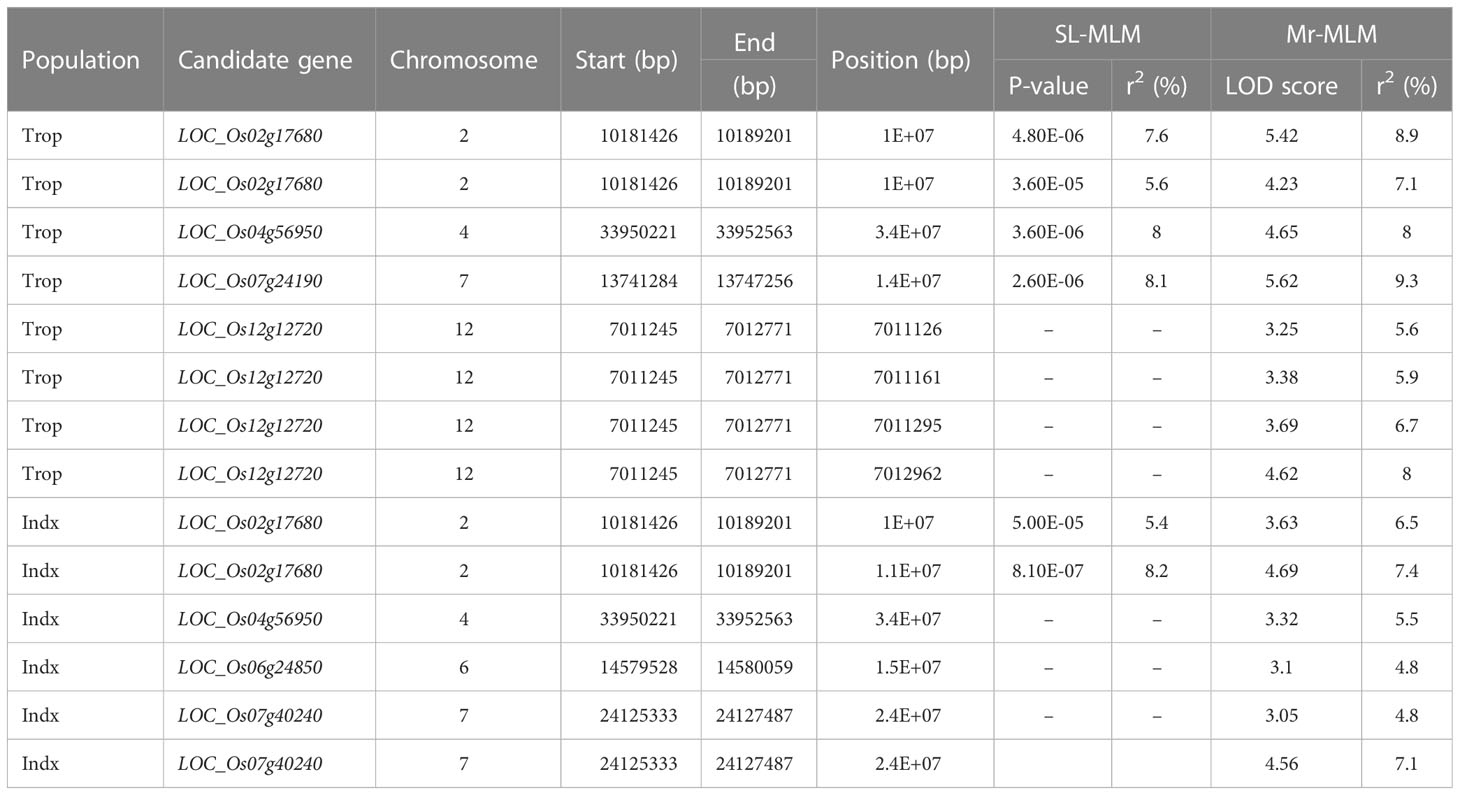
In Trop, four SNPs corresponding to LOC_Os02g17680 (including 2 SNPs), LOC_Os04g56950 and LOC_Os07g24190 were found to be significantly associated with ML by SL-MLM, and each explained the phenotypic variation of 5.6-7.6%, 8.0% and 8.1%, respectively (Figure 2; Table 2). As shown by SL-MLM, only LOC_Os02g17680 was significantly associated with ML in the Indx and explained phenotypic variations of 5.4-8.2%, respectively (Figure 2; Table 2). For mr-MLM, eight significant SNPs (LOD ≥ 3.0) corresponding to four candidate genes (LOC_Os02g17680, LOC_Os04g56950, LOC_Os07g24190 and LOC_Os12g12720) were simultaneously found to be significantly associated with the ML in Trop and explained phenotypic variation ranging from 5.6-9.3% (Table 2). Mr-MLM showed that six significant SNPs corresponding to four genes (LOC_Os02g17680, LOC_Os04g56950, LOC_Os06g24850 and LOC_Os07g40240) were significantly associated with ML in the Indx panel and explained phenotypic variations of 6.5-7.4% (2 SNPs), 5.5%, 4.8% and 4.8-7.1% (2 SNPs), respectively (Table 2).
Haplotype analysis for the significant genes
Haplotype analysis was performed for the six genes significantly associated with ML in Trop and Indx panel (Table 3). A total of 9 haplotypes of LOC_Os02g17680 were identifiedin Trop and Indx panel, and named as LOC_Os02g17680-Hap1-Hap9. Of these, only LOC_Os02g17680-Hap1, Hap2 and Hap4 existed in Trop, whereas LOC_Os02g17680-Hap1, Hap2, Hap3, Hap4, Hap5, Hap6, Hap7, Hap8 and Hap9 l existed in Indx. A total of 9 haplotypes of LOC_Os04g56950 were identified and named as LOC_Os04g56950-Hap1-Hap9. Of these, LOC_Os04g56950-Hap1, Hap2, Hap3, Hap6 and Hap8 distributed in Trop panel, whereas LOC_Os04g56950-Hap1, Hap3, Hap4, Hap5, Hap7 and Hap9 were existed in Indx panel. Totally, 3 haplotypes of LOC_Os06g24850 were identified and named as LOC_Os06g24850-Hap1-Hap3. LOC_Os06g24850-Hap1 and Hap2 distributed in Trop panel, whereas LOC_Os Os06g24850-Hap1, Hap2 and Hap3 existed in Indx panel. LOC_Os07g40240 including 3 haplotypes, e.g., LOC_Os07g40240-Hap1~Hap3. Of these, LOC_Os07g40240-Hap1, Hap2 and Hap3 distributed in Trop panel, whereas only LOC_Os07g40240-Hap1 and Hap3 were detected in Indx. Totally, LOC_Os07g24190 including six haplotypes and named as LOC_Os07g24190-Hap1-6. Among these, only Hap3 and Hap6 distributed in Trop panel, whereas Hap1-5 were identified in Indx panel. A total of 8 haplotypes of LOC_Os12g12720 were identified in all accessions and named LOC_Os12g12720-Hap1-Hap8. Of these, Hap1, Hap2, Hap3, Hap6 and Hap7 of LOC_ Os12g12720 distributed in Trop, whereas LOC_Os12g12720-Hap1, Hap3, Hap4, Hap5 and Hap8 were existed in Indx panel.
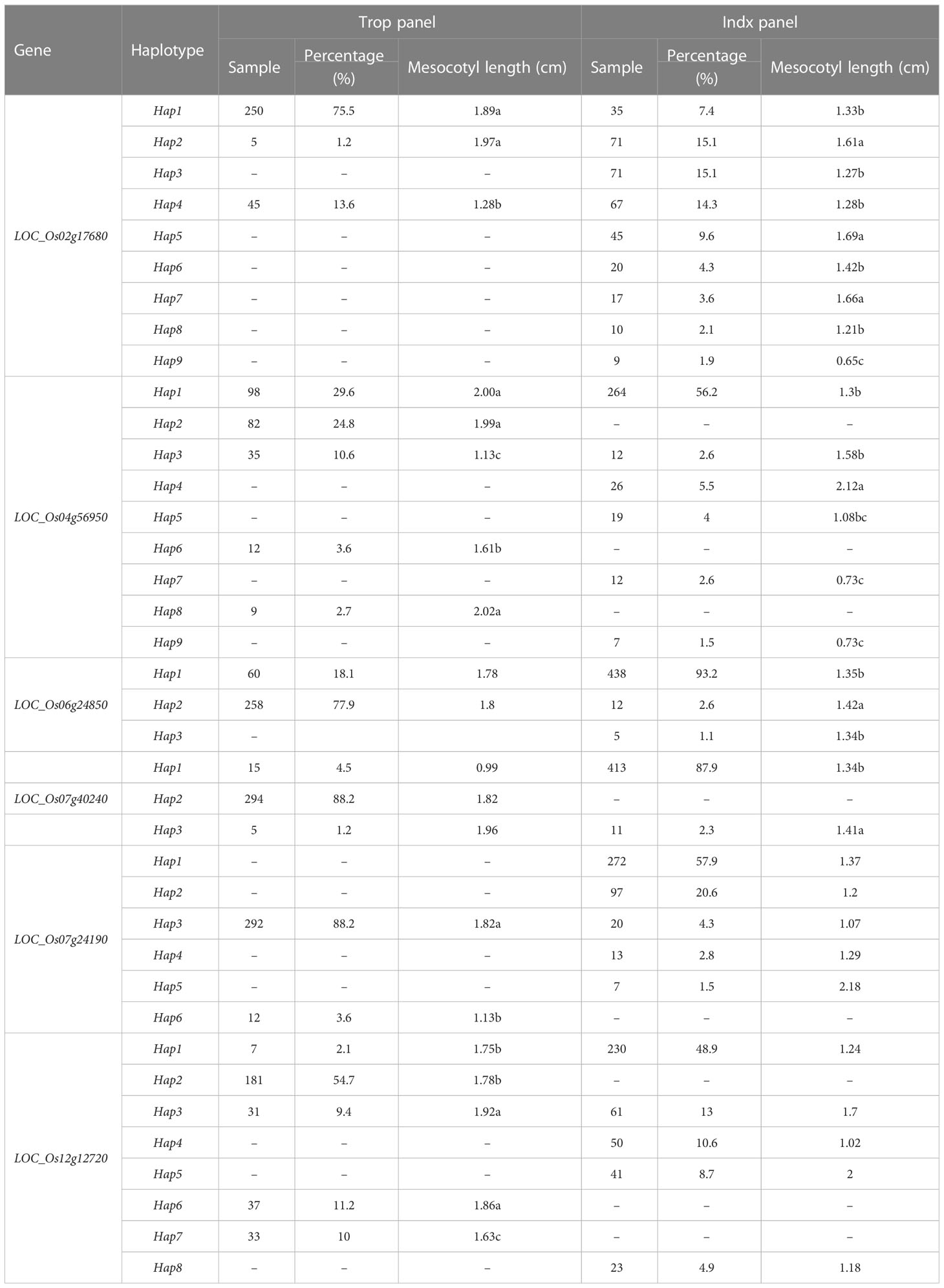
Table 3 The haplotype analysis and the superior haplotype for mesocotyl length in Trop and Indx panel.
The highest haplotype frequency in Trop were recorded in LOC_Os02g17680-Hap1 (75.5%), LOC_Os04g56950-Hap2 (29.6%), LOC_Os07g24190-Hap3 (88.2%) and LOC_Os12g12720-Hap2 (54.7%); whereas the lowest in Trop were recorded in LOC_Os02g17680-Hap2 (1.2%), LOC_Os04g56950-Hap8 (2.7%), LOC_Os07g24190-Hap6 (3.6%) and LOC_Os12g12720-Hap1 (2.1%). The highest haplotype frequency in Indx were recorded in LOC_Os02g17680-Hap2 (15.1%) and Hap3 (15.1%), LOC_Os04g56950-Hap1 (56.2%), LOC_Os06g24850-Hap1 (93.2%), LOC_Os07g40240-Hap1 (87.9%); whereas the lowest in Indx were recorded in LOC_Os02g17680-Hap9 (1.9%), LOC_Os04g56950-Hap9 (1.5%), LOC_Os06g24850-Hap3 (1.1%) and LOC_Os07g40240-Hap3 (2.3%).
The identification of superior haplotypes
To reduce the noise originated from population structure, the Duncan’s-test was established to identify the superior haplotypes of Trop and Indx separately (Table 3). According to the Table 3 and Figure S2, 8 superior haplotypes were identified in Trop panel (Figure S2), including LOC_Os02g17680-Hap1 (1.89 cm) and Hap2 (1.97 cm), which significantly long than Hap4 (1.28 cm); LOC_Os04g56950-Hap1 (2.00 cm), Hap2 (1.99 cm) and Hap8 (2.02 cm) were significantly longer than Hap3 (1.13 cm) and Hap6 (1.61 cm); LOC_Os07g24190-Hap3 (1.82 cm) with longest mesocotyl relative to the Hap6 (1.13 cm), Hap1 (1.75 cm), Hap2 (1.78 cm) and Hap7 (1.63 cm) of LOC_Os12g12720 were significantly shorter than Hap3 (1.92 cm) and Hap6 (1.86 cm) (P<0.05). Six superior haplotypes were identified in Indx panel, including the LOC_Os02g17680-Hap2 (1.61 cm), Hap5 (1.69 cm) and Hap7 (1.66 cm) were significantly longer than Hap1 (1.33 cm), Hap3 (1.27 cm), Hap4 (1.28 cm), Hap6 (1.42 cm), Hap8 (1.21 cm) and Hap9 (0.65 cm). LOC_Os04g56950-Hap4 (2.12 cm), which were significantly longer than Hpa1 (1.30 cm), Hap3 (1.58 cm), Hap5 (1.08 cm), Hap7 (0.73 cm) and Hap9 (0.73 cm). For LOC_Os06g24850-Hap2 (1.42 cm), Hap2 (1.42 cm) showed longer mesocotyl than Hap1 (1.35 cm) and Hap3 (1.34 cm). Furthermore, LOC_Os07g40240-Hap3 (1.45 cm) is higher than Hap1 (1.34 cm) (Figure S2) (P<0.05).
According to the data from RFGB database, the seedling height of superior haplotypes LOC_Os02g17680-Hap2 (36.4 cm) is higher than that of LOC_Os02g17680-Hap1 (35.6 cm) and LOC_Os02g17680-Hap4 (35.3 cm); LOC_Os04g56950-Hap1 (36.4 cm), Hap2 (35.2 cm) and Hap8 (35.6 cm) is higher than other haplotypes (32.0-35.5 cm) in Trop. However, the seedling height of superior haplotypes LOC_Os07g24190-Hap3 (34.5 cm), LOC_Os12g12720-Hap3 (34.1 cm) and Hap6 (33.9 cm) is lower than other haplotypes. In Trop, the seedling height of superior haplotypes LOC_Os02g17680-Hap2 (43.2 cm), Hap5 (41.9 cm) and Hap7 (39.3 cm), LOC_Os04g56950-Hap4 (37.69 cm) and LOC_Os07g40240-Hap3 (2.30%, 1.45 cm) is higher than other correspondence un-superior haplotypes; whereas LOC_Os06g24850-Hap2 (37.3 cm) is lower than correspondence un-superior haplotypes.
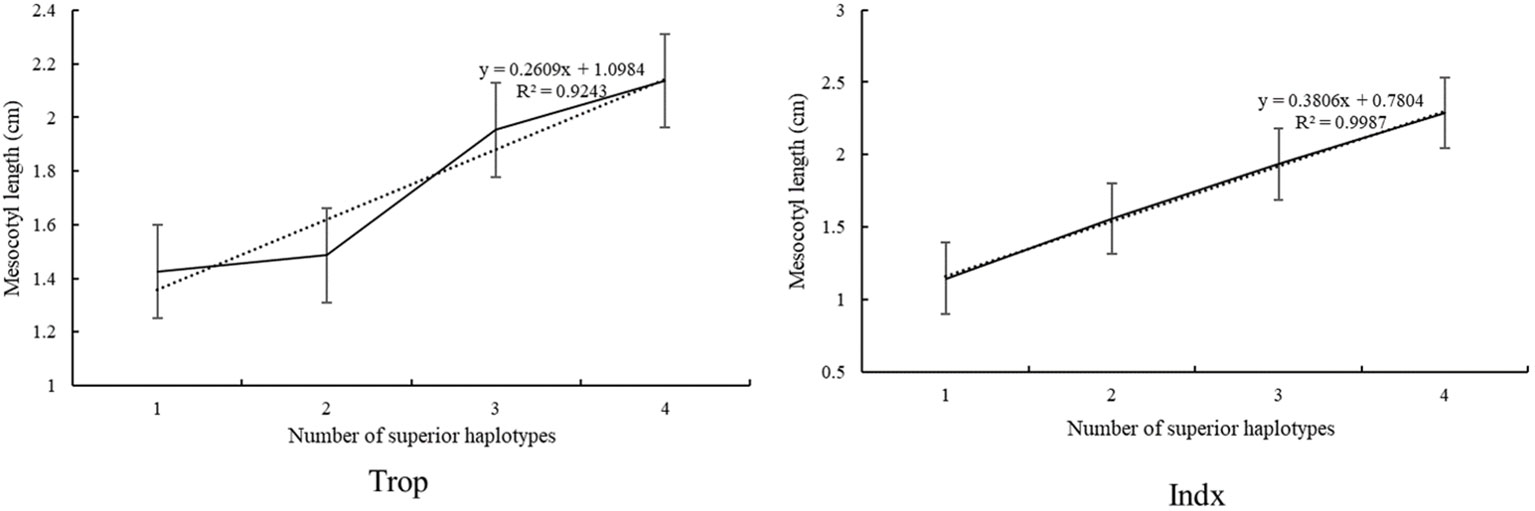
To further understand the additive effects of haplotypes on ML, we examined the number of superior haplotypes in each accession of Trop and Indx panel. The ML ranged from 1.41 cm to 2.15 cm with the superior haplotypes ranged from 0 to 3 in the Trop, whereas the ML ranged from 1.15 cm to 2.34 cm with the number of superior haplotypes ranged from 0 to 3 in the Indx. The relationships between ML and the numbers of superior haplotypes estimated by linear regression showed a dependence of ML on the number of superior haplotypes in both panels (Figure 3).
Discussion
The existence of considerable genetic and phenotype variations for ML has been observed in this study (Wu et al., 2005; Wu et al., 2015; Liu et al., 2020). Thus, evaluate the genetic effects and identify superior haplotypes is urgent and important for ML improvement. Long mesocotyl breeding is feasible and has great potential. Although a series of genomic regions and candidate genes for ML have been reported, their availability in rice breeding remains unclear. Deeper insights into the complex relationship among ML and identify corresponding candidate genes would greatly aid in the selection of appropriate genes and superior haplotypes. In this study, CAS based on 281 selected genes were separately conducted to identify the genes and corresponding superior haplotypes for ML.
Conventional SL-MLM have been widely applied to identify genetic variants in crops (Liu et al., 2017; Liu et al., 2020). However, SL-MLM have disadvantages as they ignore the overall effects of multiple minor loci, and suffer from multiple test corrections for critical values (Wen et al., 2018; Wang et al., 2020; Zhang et al., 2020). Differing from SL-MLM, all the potentially associated markers are selected by a random-SNP-effect MLM with a modified Bonferroni correction for significance test by mr-MLM. In this study, more loci for ML were identified by mr-MLM in both Trop and Indx panels. For example, LOC_Os12g12720 was only detected by the mr-MLM in Trop; LOC_Os04g56950, LOC_Os06g24850 and LOC_Os07g40240 were only detected by mr-MLM in Indx. These data illustrate that mr-MLM is more effective and powerful to detect minor gene/loci for quantitative inheritance complex traits (Segura et al., 2012; Cui et al., 2018; Wen et al., 2018; Zhang et al., 2020b). The reason for the higher effective of mr-MLM maybe the two-step association analysis of statistical model and the relatively loose threshold (Zhang et al., 2020b).
The distributions of haplotypes for the same gene were different in the Trop and Indx. Previous studies have reported that haplotype distributions differ across various populations (Qian et al., 2017; Wang et al., 2019; Liu et al., 2021). For example, in the present study, all the 9 haplotypes of LOC_Os02g17680 were existed in the Indx, whereas Hap1, Hap2 and Hap4 were only identified in the Trop; for LOC_Os04g56950, Hap1 and Hap3 distributed in both two panels, Hap2, Hap6 and Hap8 were only detected in Trop, whereas Hap4, Hap5, Hap7 and Hap9 detected in Indx panel. The frequencies of haplotype distribution in Trop and Indx were different. The frequency of LOC_Os02g17680-Hap1 accounted for 75.5% in the Trop, whereas 7.4% in Indx; the frequency of LOC_Os04g56950-Hap1 was about 29.6% in the Trop panel, whereas about 56.2% in Indx.
Plant hormones, such as SLs, CTK, ABA, BR, IAA and JAS have direct influence on mesocotyl elongation by affecting cell division or elongation. LOC_Os06g24850 on chromosome 6 belonged to OsIAA22-Auxin-responsive gene family. Auxin based on the indole ring plays crucial roles in plant growth and development, such as the cell differentiation, division and elongation (Xu and Xue, 2012). Feng et al. (2017) have reported that the exogenous IAA could promote mesocotyl elongation of rice seedings after germination under darkness. LOC_Os07g40240 on chromosomes 7 encodes the GASR9-Gibberellin-regulated GASA/GAST/Snakin family protein precursor. Liang et al. (2016) reported that the destabilization of cortical microtubules (CMTs) increased the GA level and further promote the mesocotyl elongation, while polymerization of CMT showed opposite effect by influencing the expression of GA20ox2, GA3ox2 and GID1 in GA biosynthesis. LOC_Os02g17680 on chromosomes 2 is an ethylene-responsive related protein. LOC_Os04g56950 and LOC_Os12g12720 on chromosome 4 and 12 encoding jasmonate O-methyltransferase and jasmonate-induced protein, respectively. ETH works as a signal to regulate cell elongation through JA biosynthesis pathway. Xiong et al. (2017) reported that the GY1 functions at the initial step of JA biosynthesis to repress mesocotyl and coleoptile elongation in etiolated rice seedings. ETH inhibits the expression of GY1 in the JA biosynthesis pathway and enhance mesocotyl and coleoptile growth by promoting cell elongation (Xiong et al., 2017). LOC_Os07g24190 on chromosome 7 encoding the CESA3-cellulose synthase, plays crucial roles in the roots, stems, and the elongation of root hair (Li et al., 2019; Moon et al., 2019).
Several studies have shown that mesocotyl has a significant impact on seedling height, and long mesocotyl accessions tend to with higher seedling height (Kumar and Ladha, 2011; Lee et al., 2017). This study verified the above results. Most of the superior haplotypes with higher seedling height, such as LOC_Os02g17680-Hap2, LOC_Os04g56950-Hap1, Hap2 and Hap8 in Trop panel, LOC_Os02g17680-Hap2, Hap5 and Hap7, LOC_Os04g56950-Hap4 and LOC_Os07g40240-Hap3 in Indx panel. However, we also identified few ML superior haplotypes with shorter seedling height, such as the LOC_Os07g24190-Hap3, LOC_Os12g12720-Hap3 and Hap6 in Trop panel and LOC_Os06g24850-Hap2 in the Indx. In rice breeding, seedling height selection is time-consuming and laborious, while mesocotyl phenotype evaluation can be carried out rapidly with high throughput. From the above results, in future rice breeding, superior haplotype accessions can be selected based on mesocotyl length, and then accessions with higher seedling height can be selected indirectly although seedling height is influenced by various factors besides mesocotyl length. However, these ML superior haplotypes with lower seedling height need to be specifically selected according to the breeding goal.
We examined the number of superior haplotypes in each accession to further understand the combined effects of alleles on ML. The ML ranged from 1.41 to 2.15 cm with the superior haplotypes ranged from 0 to 3 in the Trop panel, whereas the ML ranged from 1.15 to 2.34 cm with the superior haplotypes ranged from 0 to 3 in the Indx. A significant additive effect was identified from the linear regression between ML and the number of superior haplotypes, indicating that pyramiding of superior haplotypes will accelerate the genetic improvement of ML. As the distribution of superior haplotypes are different, genes and corresponding haplotypes should be selected specific for Trop and Indx. LOC_Os02g17680 (Hap1 (1.89 cm) and Hap2 (1.97 cm) of Trop; Hap2 (1.61 cm), Hap5 (1.69 cm), and Hap7 (1.66 cm) of Indx) and LOC_Os04g56950 (Hap1 (2.00 cm), Hap2 (1.99 cm) and Hap8 (2.02) cm of Trop; Hap4 (2.12 cm) of Indx) were detected in both Trop and Indx, implying that these genes play a stabilizing role in diverse accessions and could be widely used in rice breeding. LOC_Os07g24190 (Hap3 1.82 cm) and LOC_Os02g17680 (Hap2 (1.61 cm), Hap5 (1.69 cm), and Hap7 (1.66 cm)) explained the highest phenotypic variations and is the best choice for higher ML breeding in Trop and Indx panels, respectively. Furthermore, LOC_Os07g24190 (Hap3 1.82 cm) and LOC_Os12g12720 (Hap3 1.70 cm) could be applied in the Trop panel specifically, whereas the LOC_Os06g24850 (1.42 cm) and LOC_Os07g40240 (1.41 cm) could be used in Indx panel specifically. LOC_Os02g17680-Hap1 (1.89 cm) and Hap2 (1.97 cm), LOC_Os04g56950-Hap1 (2.00 cm), Hap2 (1.99 cm) and Hap8 (2.02 cm), LOC_Os07g24190-Hap3 (1.82 cm), LOC_Os12g12720-Hap3 (1.92 cm) and Hap6 (1.86 cm) are recommended for ML improvement in Trop, whereas the LOC_Os02g17680-Hap2 (1.61 cm), Hap5 (1.69 cm) and Hap7 (1.66 cm), LOC_Os04g56950-Hap4 (2.12 cm), LOC_Os06g24850-Hap2 (1.42 cm) and LOC_Os07g40240-Hap3 (1.41 cm) are suitable in Indx. Lines with higher ML and carrying multiple superior haplotypes, such as SUNGKAI, RIMBUN, SINAPLED, YAH YAW, IRAT 104/PALAWAN, BIKYAT and BUNTU DOMBA 1 in Trop, ARC 14064, CHNNOR, ARC 11857, BAIANG 6, LANJALI and JIA GEN in Indx could be used to rapidly combine several superior target haplotypes into one background.
Conclusion
In the present study, 281 ML related genes were selected to evaluate their effects for ML and identify superior haplotypes in two different populations. Totally, six unique genes were identified for ML. Of these, LOC_Os02g17680 and LOC_Os04g56950 were identified in both two panels. Totally, 8 and 6 superior haplotypes for ML were identified in Trop and Indx panel, respectively. A significant additive effect was identified from the linear regression between ML and the number of superior haplotypes. Introgression of these superior haplotypes by the haplotype‐based breeding is a promising strategy. The associated genes and superior haplotypes may pave the way for future rice ML breeding.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
JL and GY designed the research, analyzed the physiology data, YW and JL drafted the manuscript. YM and HL performed the experiment. JL revised the manuscript. All authors have read, edited and approved the current version of the manuscript.
Funding
This study was financially supported by the Young Elite Scientists Sponsorship Program by CAST (YESS) (2020QNRC001), General Project of Natural Science Foundation of Guangdong Province (2023A1515012040), China Postdoctoral Science Foundation (2022M723659), Guangdong Basic and Applied Basic Research Foundation (2022A1515111057).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1194119/full#supplementary-material
Supplementary Figure 1 | The distribution of mesocotyl length and seedling height in Trop and Indx panels.
Supplementary Figure 2 | Comparison of mesocotyl length between superior haplotypes and other haplotypes for the significant genes in the Trop and Indx panel. Different letters represent significant differences at the P=0.05 level.
References
Abbai, R., Singh, V. K., Nachimuthu, V. V., Sinha, P., Selvaraj, R., Vipparla, A. K., et al. (2019). Haplotype analysis of key genes governing grain yield and quality traits across 3K RG panel reveals scope for the development of tailor-made rice with enhanced genetic gains. Plant Biotechnol. J. 17, 1612–1622. doi: 10.1111/pbi.13087
Alexandrov, N., Tai, S., Wang, W., Mansueto, L., Palis, K., Fuentes, R. R., et al. (2015). SNP-seek database of SNPs derived from 3000 rice genomes. Nucleic. Acid. Res. 43, 1023–1027. doi: 10.1093/nar/gku1039
Bevan, M. W., Uauy, C., Wulff, B. B., Zhou, J., Krasileva, K., Clark, M. D. (2017). Genomic innovation for crop improvement. Nature 543, 346–354. doi: 10.1038/nature22011
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Cui, Y., Zhang, F., Zhou, Y. (2018). The application of multi-locus GWAS for the detection of salt-tolerance loci in rice. Front. Plant Sci. 9, 1464. doi: 10.3389/fpls.2018.01464
Feng, F. J., Mei, H. W., Fan, P. Q., Li, Y. N., Xu, X. Y., Wei, H. B., et al. (2017). Dynamic transcriptome and phytohormone profiling along the time of light exposure in the mesocotyl of rice seedling. Sci-Rep 7, 11961. doi: 10.1038/s41598-017-12326-2
Flint-Garcia, S. A., Thornsberry, J. M., Buckler, E. S. (2003). Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 54, 357–374. doi: 10.1146/annurev.arplant.54.031902.134907
Hu, Z. Y., Yamauchi, T., Yang, J. H., Jikumaru, Y., Tsuchida-Mayama, T., Ichi-kawa, H., et al. (2014). Strigolactone and cytokinin act antagonistically in regulating rice mesocotyl elongation in darkness. Plant Cell Physiol. 55, 30–41. doi: 10.1093/pcp/pct150
Huang , C., Jiang, S. K., Feng, L. L., Xu, J. Z., Chen, W. F. (2010). QTL Analysis for mesocotyl length in rice (Oryza sativa L.). Acta Agronomica Sinica 36, 1108–1113.
Jang, S. G., Park, S. Y., Lar, S. M., Zhang, H., Lee, A. R., Cao, F. Y., et al. (2021). Genome-wide association study (GWAS) of mesocotyl length for direct seeding in rice. Agronomy 11, 2527. doi: 10.3390/agronomy11122527
Kato, Y., Katsura, K. (2014). Rice adaptation to aerobic soils: physiological considerations and implications for agronomy. Plant Prod. Sci. 17, 1–12. doi: 10.1626/pps.17.1
Kumar, V., Ladha, J. K. (2011). Direct seeding of rice: recent developments and future research needs. Adv. Agron. 111, 297–413. doi: 10.1016/B978-0-12-387689-8.00001-1
Lee, H. S., Kang, J. W., Chung, N. J., Choi, K. S., Ahn, S. N. (2012). Identification of molecular markers for mesocotyl elongation in weedy rice. Kor. J. Breed. Sci. 44, 238–244.
Lee, H. S., Sasaki, K., Kang, J. W., Sato, T., Song, W. Y., Ahn, S. N. (2017). Mesocotyl elongation is essential for seedling emergence under deep-seeding condition in rice. Rice 10, 32. doi: 10.1186/s12284-017-0173-2
Li, Y., Cheng, X., Fu, Y., Wu, Q., Guo, Y., Peng, J., et al. (2019). A genome-wide analysis of the cellulose synthase-like (Csl) gene family in maize. Biologia. Plantarum 63, 721–732. doi: 10.32615/bp.2019.081
Li, J., Wang, J., Zeigler, R. S. (2014). The 3,000 rice genomes project: new opportunities and challenges for future rice research. Giga Sci. 3, 8. doi: 10.1186/2047-217X-3-8
Liang, Q., Wang, C., Ma, D. R., Li, L., Cui, Z. B., Wang, X. X., et al. (2016). Cortical microtubule disorganized related to an endogenous gibberellin increase plays an important role in rice mesocotyl elongation. Plant Biotechnol. J. 33, 59–69. doi: 10.5511/plantbiotechnology.16.0205a
Lipka, A. E., Tian, F., Wang, Q., Peiffer, J., Li, M., Bradbury, P. J., et al. (2012). GAPIT: genome association and prediction integrated tool. Bioinformatics 28, 2397–2399. doi: 10.1093/bioinformatics/bts444
Liu, J. D., He, Z. H., Rasheed, A., Wen, W. E., Yan, J., Zhang, P. Z., et al. (2017). Genome-wide association mapping of black point reaction in common wheat (Triticum aestivum l.). BMC Plant Biol. 17, 220. doi: 10.1186/s12870-017-1167-3
Li, Y. N., Yan, M., Feng, F. J., Wu, J. H., Xu, X. Y., Fan, P. Q., et al. (2017). Identification of chromosomal regions influencing mesocotyl elongation by bulked segregation analysis based on genome re-sequencing in rice. Acta Agric. Shanghai. 33, 10–15. (in Chinese with English abstract).
Liu, H., Hussain, S., Zheng, M., Peng, S., Huang, J., Cui, K., et al. (2015). Dry direct-seeded rice as an alternative to transplanted-flooded rice in central China. Agron. Sustain. Dev. 35, 285–294. doi: 10.1007/s13593-014-0239-0
Liu, H., Zhan, J., Li, J., Lu, X., Liu, J., Wang, Y., et al. (2020). Genome-wide association study (GWAS) for mesocotyl elongation in rice (Oryza sativa l.) under multiple culture conditions. Genes 11, 49.
Liu, J., Zhan, J., Chen, J., Lu, X., Zhi, S., Ye, G. (2021). Validation of genes affecting rice grain zinc content through candidate gene-based association analysis. Front. Genet. 12, 701658.
Lu, Q., Zhang, M. C., Niu, X. J., Wang, C. H., Xu, Q., Feng, Y. (2016). Uncovering novel loci for mesocotyl elongation and shoot length in indica rice through genome-wide association mapping. Planta 243, 645–657. doi: 10.1007/s00425-015-2434-x
Lv, Y., Shao, G., Jiao, G., Sheng, Z., Xie, L., Hu, S., et al. (2021). Targeted mutagenesis of POLYAMINE OXIDASE 5 that negatively regulates mesocotyl elongation enables the generation of direct-seeding rice with improved grain yield. Mol. Plant 14, 344–351. doi: 10.1016/j.molp.2020.11.007
Mahender, A., Anandan, A., Pradhan, S. K. (2015). Early seedling vigour, an imperative trait for direct-seeded rice: an overview on physio-morphological parameters and molecular markers. Planta 241, 1027–1050. doi: 10.1007/s00425-015-2273-9
Moon, S., Chandran, A. K. N., Kim, Y. J., Gho, Y., Hong, W. J., An, G., et al. (2019). Rice RHC encoding a putative cellulase is essential for normal root hair elongation. J. Plant Biol. 62, 82–91. doi: 10.1007/s12374-018-0393-z
Ohno, H., Banayo, N. P., Bueno, C. S., Kashiwagi, J. I., Nakashima, T., Corales, A. M., et al. (2018). Longer mesocotyl contributes to quick seedling establishment, improved root anchorage, and early vigor of deep-sown rice. Field Crop Res. 228, 84–92. doi: 10.1016/j.fcr.2018.08.015
Ouyang, Y. N., Zhang, Q. Y., Zhang, K. Q., Yu, S. M., Zhuang, J. Y., Jin, Q. Y. (2005). QTL mapping and interaction analysis of genotype/environment (Fe2+-concentrations) for mesocotyl length in rice (Oryza sativa l.). Acta Genet. Sin. 32, 712–718. (In Chinese with English abstract)
Prodhomme, C., Vos, P. G., Paulo, M. J., Tammes, J. E., Visser, R. G., Vossen, J. H., et al. (2020). Distribution of P1 (D1) wart disease resistance in potato germplasm and GWAS identification of haplotype-specific SNP markers. Theor. Appl. Genet. 6, 133. doi: 10.1007/s00122-020-03559-3
Qian, L., Hickey, L. T., Stahl, A., Werner, C. R., Hayes, B., Snowdon, R. J., et al. (2017). Exploring and harnessing haplotype diversity to improve yield stability in crops. Front Plant Sci. 8, 1534. doi: 10.3389/fpls.2017.01534
Resende, R. T., Resende, M. D. V., Silva, F. F., Azevedo, C. F., Takahashi, E. K., Silva-Junior, O. B., et al. (2017). Regional heritability mapping and genome-wide association identify loci for complex growth, wood and disease resistance traits in eucalyptus. New Phytol. 213, 1287–1300. doi: 10.1111/nph.14266
Rohilla, M., Singh, N., Mazumder, A., Sen, P., Roy, P., Chowdhury, D., et al. (2020). Genome-wide association studies using 50 K rice genic SNP chip unveil genetic architecture for anaerobic germination of deep-water rice population of Assam, India. Mol. Genet. Genomics 295, 1211–1226. doi: 10.1007/s00438-020-01690-w
Segura, V., Vilhjálmsson, B. J., Platt, A., Korte, A., Seren, Ü., Long, Q., et al. (2012). An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat. Genet. 44, 825–830. doi: 10.1038/ng.2314
Sinha, P., Singh, V. K., Saxena, R. K., Khan, A. W., Abbai, R., Chitikineni, A., et al. (2020). Superior haplotypes for haplotype-based breeding for drought tolerance in pigeonpea (Cajanus cajan l.). Plant Biotechnol. J. 18, 2482–2490. doi: 10.1111/pbi.13422
Sun, S. Y., Wang, T., Wang, L. L., Li, X. M., Jia, Y. C., Liu, C., et al. (2018). Natural selection of a GSK3 determines rice mesocotyl domestication by coordinating strigolactone and brassinosteroid signaling. Nat. Commun. 9, 2523. doi: 10.1038/s41467-018-04952-9
Tamba, C. L., Zhang, Y. M. (2018). A fast mr-MLM algorithm for multi-locus genome-wide association studies. Biorxiv 341784. doi: 10.1101/341784
Wang, S. B., Feng, J. Y., Ren, W. L., Huang, B., Zhou, L., Wen, Y. J., et al. (2019). Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci. Rep-UK. 6, 19444. doi: 10.1038/srep19444
Wang, K., Li, M., Hakonarson, H. (2010). ANNOVAR: functional annotation of genetic variants from next-generation sequencing data. Nucleic. Acids Res. 38, e164. doi: 10.1093/nar/gkq603
Wang, Y. M., Liu, J. D., Meng, Y., Liu, H. Y., Liu, C., Ye, G. Y. (2021). Rapid identification of QTL for mesocotyl length in rice through combining QTL-seq and genome-wide association analysis. Front. Genet. 12. doi: 10.3389/fgene.2021.713446
Wang, W., Mauleon, R., Hu, Z., Chebotarov, D., Tai, S., Wu, Z., et al. (2018). Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557, 43–49. doi: 10.1038/s41586-018-0063-9
Wang, Q., Tang, J., Han, B., Huang, X. (2020). Advances in genome-wide association studies of complex traits in rice. Theor. Appl. Genet. 133, 1415–1425. doi: 10.1007/s00122-019-03473-3
Watanabe, H., Takahashi, K. (1999). Effects of abscisic acid and its related compounds on rice seedling growth. Plant Growth Regul. 25, 5–8. doi: 10.1023/A:1006270514827
Watanabe, H., Takahashi, K., Saigusa, M. (2001). Morphological and anatomical effects of abscisic acid (ABA) and fluridone (FLU) on the growth of rice mesocotyls. Plant Growth Regul. 34, 273–275. doi: 10.1023/A:1013333718573
Wen, Y. J., Zhang, H., Ni, Y. L., Huang, B., Zhang, J., Feng, J. Y., et al. (2018). Methodological implementation of mixed linear models in multi-locus genome-wide association studies. Brief. Bioinform. 19, 700–712. doi: 10.1093/bib/bbw145
Wu, S. Q., Ding, R., Li, X. S. (2002). Regulation of mesocotyl growth by gibberellic acid and abscisic acid in etiolated seedlings of black rice. Amino. Acids Biotic. Res. 24, 44–45. (In Chinese with English abstract)
Wu, J. H., Feng, F. J., Lian, X. M., Teng, X. Y., Wei, H. B., Yu, H., et al. (2015). Genome-wide association study (GWAS) of mesocotyl elongation based on re-sequencing approach in rice. BMC Plant Biol. 15, 218. doi: 10.1186/s12870-015-0608-0
Wu, M., Zhang, G., Lin, J., Cheng, S. (2005). Screening for rice germplasms with specially elongated mesocotyl. Rice Sci. 12, 226–228.
Xiong, Q., Ma, B., Lu, X., Huang, Y. H., He, S. J., Yang, C., et al. (2017). Ethylene inhibited jasmonic acid biosynthesis promotes mesocotyl/coleoptile elongation of etiolated rice seedlings. Plant Cell 29, 1053–1072. doi: 10.1105/tpc.16.00981
Xu, Z. H., Xue, H. W. (2012). Plant hormones: function and molecular mechanism (Shanghai: Shanghai Scientific and Technical Publishers).
Yang, Y., Chai, Y., Zhang, X., Lu, S., Zhao, Z., Wei, D., et al. (2020). Multi-locus GWAS of quality traits in bread wheat: mining more candidate genes and possible regulatory network. Front. Plant Sci. 11, 1091. doi: 10.3389/fpls.2020.01091
Yuldashev, R., Avalbaev, A., Bezrukova, M., Vysotskaya, L., Khripach, V., Shakirova, F. (2012). Cytokinin oxidase is involved in the regulation of cytokinin content by 24-epibrassinolide in wheat seedlings. Plant Physiol. Bioch. 55, 1–6. doi: 10.1016/j.plaphy.2012.03.004
Zhan, J., Lu, X., Liu, H., Zhao, Q., Ye, G. (2020). Mesocotyl elongation, an essential trait for dry-seeded rice (Oryza sativa l.): a review of physiological and genetic basis. Planta 251, 1–14. doi: 10.1007/s00425-019-03322-z
Zhang, X. J., Lai, Y. C., Meng, Y., Tang, A., Dong, W. J., Liu, Y. H., et al. (2022). Analyses and identifications of quantitative trait loci and candidate genes controlling mesocotyl elongation in rice. J. Integr. Agr 22, 325–340. doi: 10.1016/j.jia.2022.08.080
Zhang, Y., Ponce, K. S., Meng, L., Chakraborty, P., Zhao, Q., Guo, L., et al. (2020). QTL identification for salt tolerance related traits at the seedling stage in indica rice using a multi-parent advanced generation intercross (MAGIC) population. Plant Growth Regul. 92, 365–373. doi: 10.1007/s10725-020-00644-x
Zhang, Y. W., Tamba, C. L., Wen, Y. J., Li, P., Ren, W. L., Ni, Y. L., et al. (2020). Mr-MLM v4. 0: an r platform for multi-locus genome-wide association studies. Genom. Proteom. Bioinf. 18, 481–487. doi: 10.1016/j.gpb.2020.06.006
Zhao, Y., Zhao, W. P., Jiang, C. H., Wang, X. N., Xiong, H. Y., Todorovska, E. G., et al. (2018). Genetic architecture and candidate genes for deep-sowing tolerance in rice revealed by non-syn GWAS. Front. Plant Sci. 9, 332. doi: 10.3389/fpls.2018.00332
Zhao, X. Q., Zhou, L. J., Ponce, K., Ye, G. Y. (2015). The usefulness of known genes/QTLs for grain quality traits in an indica population of diverse breeding lines tested using association analysis. Rice 8, 1–13. doi: 10.1186/s12284-015-0064-3
Zheng, J. S., Hong, K., Zeng, L. J., Wang, L., Kang, S. J., Qu, M. H., et al. (2020). Karrikin signaling acts parallel to and additively with strigolactone signaling to regulate rice mesocotyl elongation in darkness. Plant Cell 32, 2780–2805. doi: 10.1105/tpc.20.00123
Keywords: candidate gene association analysis, mesocotyl, haplotype, mr-MLM, Oryza sativa L
Citation: Wang Y, Liu H, Meng Y, Liu J and Ye G (2023) Validation of genes affecting rice mesocotyl length through candidate association analysis and identification of the superior haplotypes. Front. Plant Sci. 14:1194119. doi: 10.3389/fpls.2023.1194119
Received: 26 March 2023; Accepted: 02 May 2023;
Published: 30 May 2023.
Edited by:
Zhenyu Gao, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Joong Hyoun Chin, Sejong University, Republic of KoreaXiangjin Wei, China National Rice Research Institute (CAAS), China
Xiaoming Zheng, Chinese Academy of Agricultural Sciences, China
Copyright © 2023 Wang, Liu, Meng, Liu and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jindong Liu, bGl1amluZG9uZ0BjYWFzLmNu; Guoyou Ye, Zy55ZUBpcnJpLm9yZw==
 Yamei Wang1,2,3
Yamei Wang1,2,3 Hongyan Liu
Hongyan Liu Jindong Liu
Jindong Liu Guoyou Ye
Guoyou Ye