Abstract
Introduction:
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is an essential key enzyme in the glycolytic pathway and plays an important role in stress responses. Although GAPDH family genes have been found in different plant species, the determination of their gene family analysis and their functional roles in soybean are still unknown.
Methods:
In this study, gene sequence and expression data were obtained using online tools, and systematic evolution, expression profile analysis, and qRT-PCR analysis were conducted.
Results and Discussion:
Here a total of 16 GmGAPDH genes were identified on nine chromosomes, which were classified into three clusters. Additionally, all GmGAPDH genes harbor two highly conserved domains, including Gp_dh_N (PF00044) and Gp_dh_C (PF02800). The qRTPCR analysis also showed that most GmGAPDH genes significantly responded to multiple abiotic stresses, including NaHCO3, polyethylene glycol, cold, and salt. Among them, GmGAPDH14 was extraordinarily induced by salt stress. The GmGAPDH14 gene was cloned and overexpressed through soybean hair roots. The overexpressed transgenic soybean plants of the GmGAPDH14 gene have also shown better growth than that of control plants. Moreover, the overexpressed transgenic plants of GmGAPDH14 gene had higher activities of superoxide dismutase but lower malonaldehyde (MDA) content than those of control plants under salt stress. Meanwhile, a total of four haplotypes were found for the GmGAPDH14 gene, and haplotypes 2, 3, and 4 were beneficial for the tolerance of soybean to salt stress. These results suggest that the GmGAPDH14 gene might be involved in the process of soybean tolerance to salt stress. The results of this study will be valuable in understanding the role of GAPDH genes in the abiotic stress response of soybean.
Introduction
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a key enzyme in the glycolytic metabolic pathway, which widely exists in biological cells (Zhang et al., 2019). GAPDHs catalyzed glyceraldehyde-3-phosphate to form 1,3-biphosphoglycerate in the presence of NAD+ and inorganic phosphate (Sirover, 2011). The major functions of the GAPDH gene refer to immune response (Henry et al., 2015), expression regulation (Zhang et al., 2017), and autophagy (Colell et al., 2007).
In plants, GAPDH genes are involved in glycolytic or photosynthetic pathways (Plaxton, 1996). Meanwhile, GAPDH genes can be divided into three categories according to their functions in cells (Guo et al., 2012; Guo et al., 2014). In chloroplasts, NADP-specific GAPDHs (GAPA/B) were involved in photosynthetic CO2 fixation. In the cytoplasm, NAD-dependent GAPDH (GAPC) converted glyceraldehyde-3-P (Ga3P) to 1,3-bisphosphoglycerate. In plastids, GAPCp isoforms may be involved in glycolytic energy production. Moreover, all GAPDH proteins contained highly conserved domains, including the Gp_dh_N (PF00044) and Gp_dh_C (PF02800) domains (Jiao et al., 2011; Zeng et al., 2016; Miao et al., 2019). It was also found that some GAPDHs also contained CP12 (PF02672) domain.
To date, a series of GAPDH genes have been cloned and characterized, including Arabidopsis thaliana (Guo et al., 2014), Oryza sativa (Lim et al., 2021), Zea may (Bustos et al., 2007), and Cucumis sativus (Chaturvedi et al., 2016). Based on subcellular localization, it has been proven that GAPDH was divided into cytosolic (Cy) and plastic (P) isoforms (Miao et al., 2019; Wei et al., 2022). In A. thaliana, GAPDH genes distributed in different subcellular compartments: GAPC1 and GAPC2 were located in the cytosol, and the rest of the GAPDH genes were located in plastids (Rius et al., 2008; Anoman et al., 2015). Some researchers have revealed that GAPCs can regulate the accumulation of oil content in seeds (Guo et al., 2014). The seed oil content was reduced by 3% when GAPDH was knocked out of the cytoplasm in A. thaliana, suggesting that cytosolic GAPDH was vital for regulating the content of seed oil (Guo et al., 2014). Furthermore, the plastidic GAPCp has been shown to be involved in starch metabolism (Muñoz-Bertomeu et al., 2009). In soybean, the knockdown of GAPC1 decreased the nodule nitrogenase activity without affecting the nodule weight (Ke et al., 2022). Moreover, the key role of GAPDH genes in plant growth and development and responses to abiotic stresses has been extensively confirmed, including heat (Kim et al., 2020), cold (Liu et al., 2017), salinity (Cho et al., 2014), and drought (Li et al., 2019). Previous studies have shown that the overexpression of PsGAPDH can increased salt tolerance in potato (Jeong et al., 2001). In Arabidopsis, the overexpression of TaGApC gene from Chinese spring Triticum aestivum displayed improved drought tolerance by decreasing the reactive oxygen species (ROS) levels (Zhang et al., 2020). Furthermore, it was also found that salicylic acid restrains the GAPDH activity in vitro (Pokotylo et al., 2020).
Soybean was the main oil crop in the world (Holle and Damme, 2015). However, the yield and the quality of soybean were often affected by abiotic stresses such as low temperature, drought, and salinization (Feng et al., 2020). Therefore, it was significant to study the salt resistance mechanism of soybean and excavate the stress-resistant genes for improving the yield and quality of soybean. Although GAPDHs have been characterized and analyzed in many plant species, the characterization of the GAPDH gene family in soybean is still limited, and it is unknown how GAPDH regulates the molecular mechanism of salt stress in soybean.
Although most studies have described the biological and physiological functions of the GAPDH gene, few research were known in terms of the functional divergence of the GAPDH gene family in soybean. In this study, 16 of the GAPDH gene members in soybean were identified, and their phylogenetic relationships, gene structure, chromosomal localization, and stress responses were analyzed. Furthermore, the function of GmGAPDH14 gene in soybean tolerance to salt stress was tentatively verified, indicating the important role of GmGAPDH14 gene in salt stress.
Materials and methods
Identification of the GAPDH gene family in soybean
To identify the GAPDH gene sequence of soybean, systematic BLASTP was conducted against the soybean reference genome database (https://www.soybase.org/) and the Phytozome database (https://phytozome-next.jgi.doe.gov/) using the published Arabidopsis GAPDH as alignment sequence. The screening threshold was set to E-value (<10−10), and the protein length was greater than 200 aa. The candidate GAPDH genes were determined by SMART (http://smart.embl-heidelberg.de) and Pfam (http://pfam.xfam.org/) software with both Gp_dh_N and Gp_dh_C domains. The open reading frame length was obtained from the Phytozome database. The molecular weight and the isoelectric point values were downloaded from the ExPASy (Artimo et al., 2012) software (https://web.expasy.org/protparam/). CELLO 2.5 (Yu et al., 2004) was used to predict subcellular localization.
Phylogeny, gene structure, and conserved domain analysis
The protein sequences of GAPDHs from soybean (Glycine max), maize (Zea mays), rice (O. sativa), and Arabidopsis (A. thaliana) were used to construct a phylogenetic tree using the neighbor-joining method and bootstrap test set at 1,000 replications through the MEGA7.0 software (Kumar et al., 2016). The exon/intron structures of GmGAPDHs were demonstrated at the GSDS online server (Hu et al., 2015). The coding and genomic sequences of GmGAPDH were collected from the Phytozome database. The conserved domains of GAPDH were determined by SMART (http://smart.embl-heidelberg.de) (Letunic et al., 2015) and Pfam (http://pfam.xfam.org/) (Finn et al., 2016) software, including Gp_dh_N and Gp_dh_C domains, and the structure of GAPDH proteins was visualized using the IBS 6.0 software (Liu et al., 2015).
Promoter analysis of GmGAPDHs
To investigate the critical cis-acting elements in the promoter of GmGAPDH genes, the sequence at 2.0 kb upstream of the position of the ATG codon in these genes was obtained from the Phytozome database (https://phytozome-next.jgi.doe.gov/).The plant CARE database was used to predict the cis-acting regulatory elements, including motifs related to plant growth and development, plant hormone responses, and abiotic and biotic stress responses.
Expression analysis of GmGAPDHs during soybean development and response to abiotic stresses
The expression patterns of soybean GmGAPDHs at different tissues were obtained using the Phytozome database. The heat maps were generated by cluster analysis with the TBtools software (Chen et al., 2020), and the expression data were log2-transformed. To explore the expression patterns of GmGAPDHs in seeds at different developmental stages, soybean seeds (DN50) were collected at 10, 20, 30, and 40 days after flowering (DAF), and the total RNA extraction of each sample was performed to analyze the expression patterns of GmGAPDHs under abiotic stresses, including cold, salt, NaHCO3, and drought. Briefly, soybeans (DN50) were grown in a plant incubator. There were two different plant cultivation methods used: (a) for low temperature treatment, seeds of soybean were sown in soil and vermiculite (v:v/1:1) and (b) for the salinity, NaHCO3, and drought treatments, soybeans were grown in a hydroponic culture, and the growth condition was 24°C and a 16-h/8-h (day/night) daily photoperiod cycle. Second-trifoliolate-stage seedlings of uniform growth were subjected to cold treatment with a low temperature of 4°C, salt treatment with 150 mM salt, drought treatment with 20% polyethylene glycol (PEG, 6,000 g/M), and alkali treatment with 100 mM NaHCO3. The soybean leaves were sampled at 0, 6, 12, and 24 h after the treatments. The sample total RNA was extracted using Trizol reagent (Invitrogen). The expressions of GmGAPDH genes in soybean seed samples at 10 DAF were used as a calibrator. The expressions of GmGAPDH genes in soybean samples at 0 h were used as a calibrator. GmACTIN4 (GenBank accession no. AF049106) was used as an internal reference. Quantitative real-time RT-PCR (qRT-PCR) was conducted using the CFX Connect TMreal-time system (BIO-RAD) with SYBR Select Master Mix RT-PCR (SYBR Green, TOYOBO, Osaka, Japan). Three biological replicates with three technical replicates were applied to each sample. The expression levels of GmGAPDHs were calculated using the 2–ΔΔct method (Hong et al., 2010), and all primers used for the expression analysis were listed in Supplementary Table S1.
Agrobacterium-mediated transformation of GmGAPDH14 soybean hairy roots
Soybean cultivar DN50 was used for the Agrobacterium rhizogenes strain K599 transformation in soybean hypocotyls. The cDNA of GmGAPDH14 was directly ligated into the vector pCambia3300. The recombinant plasmid and empty pCambia3300 vector (EV) were transferred into Agrobacterium rhizogenes strain K599 and then injected into the hypocotyls following a previous report (Kereszt et al., 2007; Yu et al., 2021). The transgenic plants were identified by PCR amplification, and the non-transgenic hairy roots in the seedlings were removed.
Detection of physiological indicators after salt treatments
The hairy root soybean plants were grown in Hoagland nutrient solution, and the growth chamber was set at a 16/8-h light–dark daily photoperiodic cycle. The transgenic plants were treated with 0 and 150 mM salt for 3 days (d), respectively.
The leaves of overexpression GmGAPDH14 (OE-GmGAPDH14) and EV seedlings were analyzed to measure physiological indicators. The measurements of superoxide dismutase (SOD) and malonaldehyde (MDA) were conducted according to corresponding assay kit protocols (Cominbio, Suzhou, China). All measurements were obtained with three biological replicates.
Prediction and haplotype analysis of GmGAPDH14 gene for soybean salt resistance
A total of 131 soybean germplasms were collected and grown in Harbin for 2 consecutive years (2019–2020). All accessions were treated with 0 and 150 mM salt. The relative swelling rate was obtained according to Zhang’s method (Zhang et al., 2014). According to genomic re-sequencing data, the SNPs in genomic regions including the promoter, 5′UTRs, exon, intron, and 3′UTRs of GmGAPDH14 gene were analyzed in 131 soybean lines using the generalized linear model (GLM) method as conducted with Tassel version 5.0 software (Bradbury et al., 2017).
Statistical analysis
The statistical significance was evaluated using Student’s t-test as performed with the SPSS 22.0 software. The significance levels were *p < 0.05 and **p < 0.01. The standard deviation (mean ± SD) was calculated with at least three biological replicates.
Results
Characterization of the GAPDH gene families of soybean
To identify the GAPDH gene family of soybean, GAPDH genes were selected by BLASTp tool with Arabidopsis as the alignment sequence. A total of 16 GAPDH genes (GmGAPDH1–GmGAPDH16) were retrieved (Supplementary Table S2). The full-length CDS sequences of GmGAPDH1–GmGAPDH16 varied from 786 to 1,362 bp. The isoelectric points of GmGAPDHs ranged from 6.54 to 8.71, and the molecular weight of GmGAPDHs ranged from 28.2 to 48.4 kDa (Supplementary Table S2).
To understand the evolutionary relationship of GmGAPDHs in soybean, the amino acid sequences of GmGAPDHs from A. thaliana (seven), Z. mays (12), and O. sativa (seven) were obtained from the NCBI database (https://www.ncbi.nlm.nih.gov/), and a phylogenetic tree was built. The phylogenetic tree indicated that the GAPDH proteins of soybean were clearly divided into three clusters (I–III) (Figure 1A). Cluster I, consisting of GmGAPDH4–6, GmGAPDH8–10, and GmGAPDH14–15, corresponded with cytosol isoforms containing AtGAPC proteins. Cluster II, including GmGAPDH2 and GmGAPDH12, corresponded with plastid isoforms containing AtGAPCp1 and AtGAPCp2 proteins. Cluster III, covering GmGAPDH1/3/7/11/13/16, corresponded with AtGAPA1, AtGAPA2, and AtGAPB proteins (Figure 1A).
Figure 1
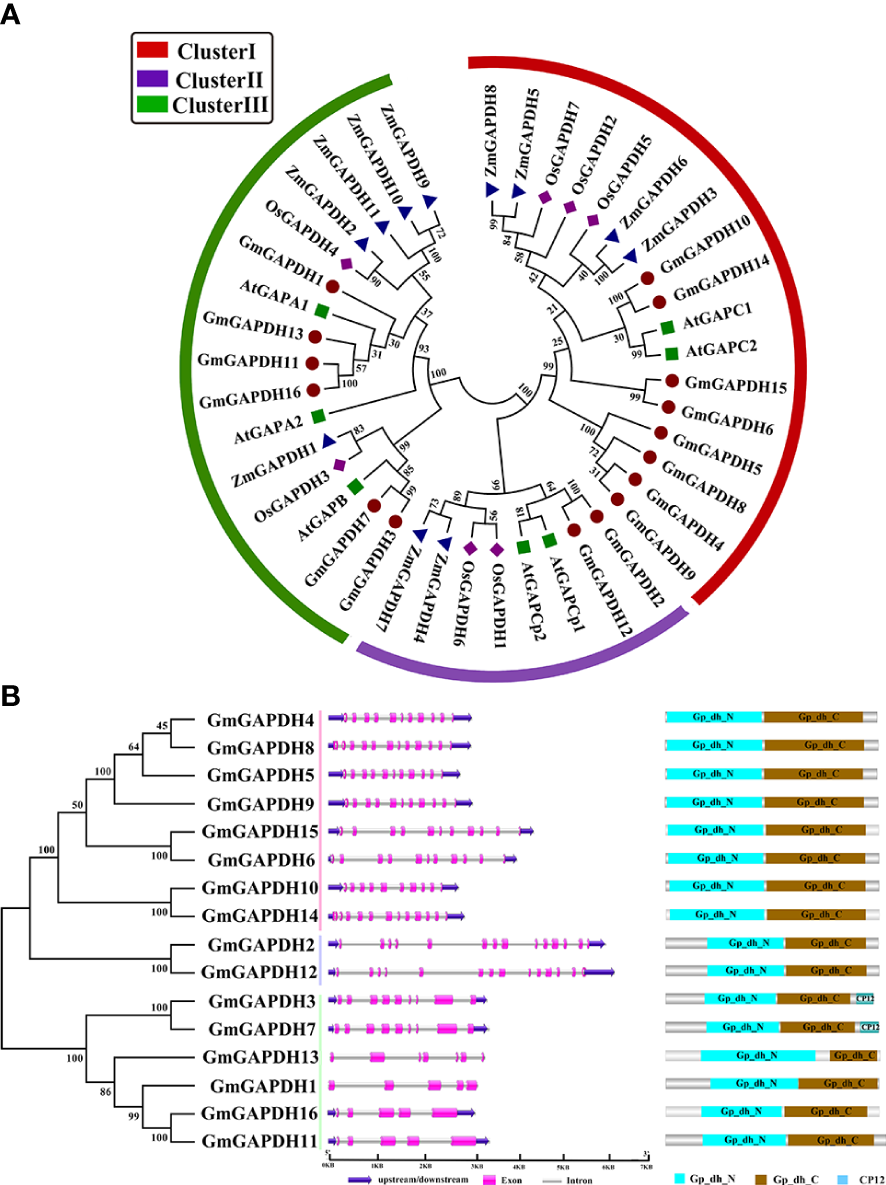
(A) Phylogenetic tree construction of GAPDH proteins from soybean (G. max), maize (Z. mays), Arabidopsis (A. thaliana), and rice (O. sativa). The phylogenetic tree was constructed through the neighbor-joining method based on MEGA7.0. The different colors of the rings represent different subfamilies: red, blue, and green represent clusters I, II, and III, respectively. (B) Exon–intron structure and domain analysis of GmGAPDHs of soybean. The untranslated region, exon, and intron are represented with blue, pink, and gray, respectively. Different-colored boxes were represented using different signal peptides.
Exon/intron structure and the conserved domain of soybean GAPDH genes
The structures of soybean GAPDH genes were characterized with the GSDS software. As shown in Figure 1, cluster II (GmGAPDH2 and GmGAPDH12) had the largest number of exons, including 12 exons. The exon number of cluster I ranged between 10 (GmGAPDH10) and 12 (GmGAPDH14); the remaining GmGAPDHs contained nine exons. The exon number of cluster III ranged from five to nine, and only GmGAPDH3/7 had nine exons; the remaining GmGAPDHs had five exons (Figure 1B).
The conserved domains analysis for 16 GmGAPDHs indicated that the GmGAPDHs revealed a multiple-domain protein, including Gp_dh_N (PF00044) and Gp_dh_C (PF02800) domains (Figure 1B). The Gp_dh_N domain (INGFGRIGR) and Gp_dh_C (GAAKAV) sequences were identified as highly conserved in the GmGAPDHs (Supplementary Figures S1, S2). A conserved active site (PS00071: ASCTTNCL) was found in most GmGAPDHs, except for GmGAPDH1 and GmGAPDH13 (Supplementary Figure S1). The similarity of the gene structure and conserved domains of soybean GAPDH genes implies that they have undergone gene duplication during evolution.
Analysis of regulatory elements in the promoter of GmGAPDHs
To obtain the cis-elements of GmGAPDHs, sequencing of 2,000 bp upstream of all GmGAPDHs gene was performed based on the PlantCARE software. As shown in Figure 2A, a total of 22 cis-elements were found with plant growth and development, phytohormone-responsive, and abiotic and biotic stresses in the upstreams of 16 GmGAPDH genes. The ERE and ARE cis-elements were found with almost all GmGAPDH genes. The GCN4_motif (endosperm expression) elements were discovered in GmGAPDH6-7 and GmGAPDH9-11. Meanwhile, GmGAPDH15 and GmGAPDH14 were analyzed only involving AuxRR-core (auxin-responsive) and GC-motif (anoxic-specific inducibility) elements, respectively. It was noteworthy that there were five GmGAPDHs that harbored low temperature responsiveness (LTR) elements while five GmGAPDHs contained MBS (drought-responsive) elements. In addition, O2 site (gliadin metabolic regulatory), CCGTCC box (specific activation), and CAT box (meristem expression) were found in the GmGAPDHs gene. TC-rich repeats (defense and stress responsiveness), GC motif (involved in anoxic-specific inducibility), and WUN motif (mechanical injury response) elements were observed in six, one, and 10 GmGAPDH genes, respectively (Figure 2B). These results showed that the GAPDH family may play an important role in growth and development and response to environmental stress in soybean.
Figure 2
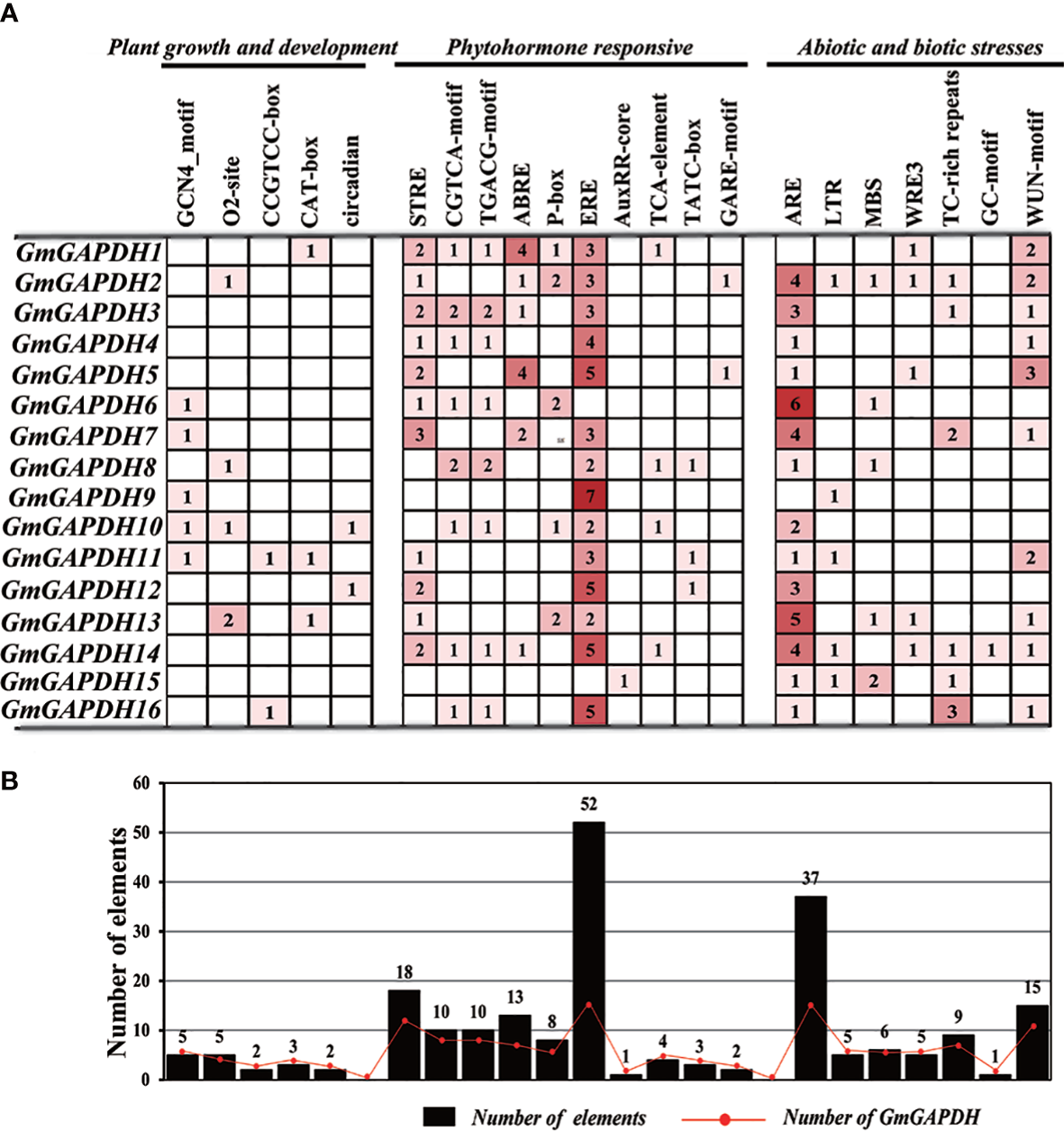
Analysis of cis-elements in the promoter of GAPDH genes. (A) Number of cis-elements in the 2.0-kb promoter region upstream of GmGAPDH genes. (B) Statistical analysis for the total number of GmGAPDH genes; the black box corresponds to the total number of cis-elements, and the red dot corresponds to the number of GmGAPDH genes.
Synteny analysis of GmGAPDHs
To further characterize duplicated events within the soybean genome, a synteny analysis of GmGAPDH genes was performed. As shown in Figure 3A, the GmGAPDH genes were scattered on nine of the 20 soybean chromosomes. The nine soybean chromosomes distributed one to three GmGAPDH genes (Figure 3A). The replication relationship with the soybean GAPDH genes was analyzed. A total of 10 duplicated gene pairs were identified within the soybean (Figure 3A). Meanwhile, a synteny analysis was conducted from G. max and A. thaliana. As shown in Figure 3B, GmGAPDHs had a replication relationship with AtGAPDHs, including nine replication relationship pairs between G. max and A. thaliana (GmGAPDH1/AtGAPC1, GmGAPDH10/AtGAPC2, GmGAPDH11/AtGAPA2, GmGAPDH11/AtGAPA1, GmGAPDH11/AtGAPA2, GAPDH13/AtGAPC1, GAPDH14/AtGAPC2, GAPDH16/AtGAPA1, and GAPDH16/AtGAPC1) (Figure 3B). The duplicated genes showed their common genomic origin and maybe functional similarity.
Figure 3
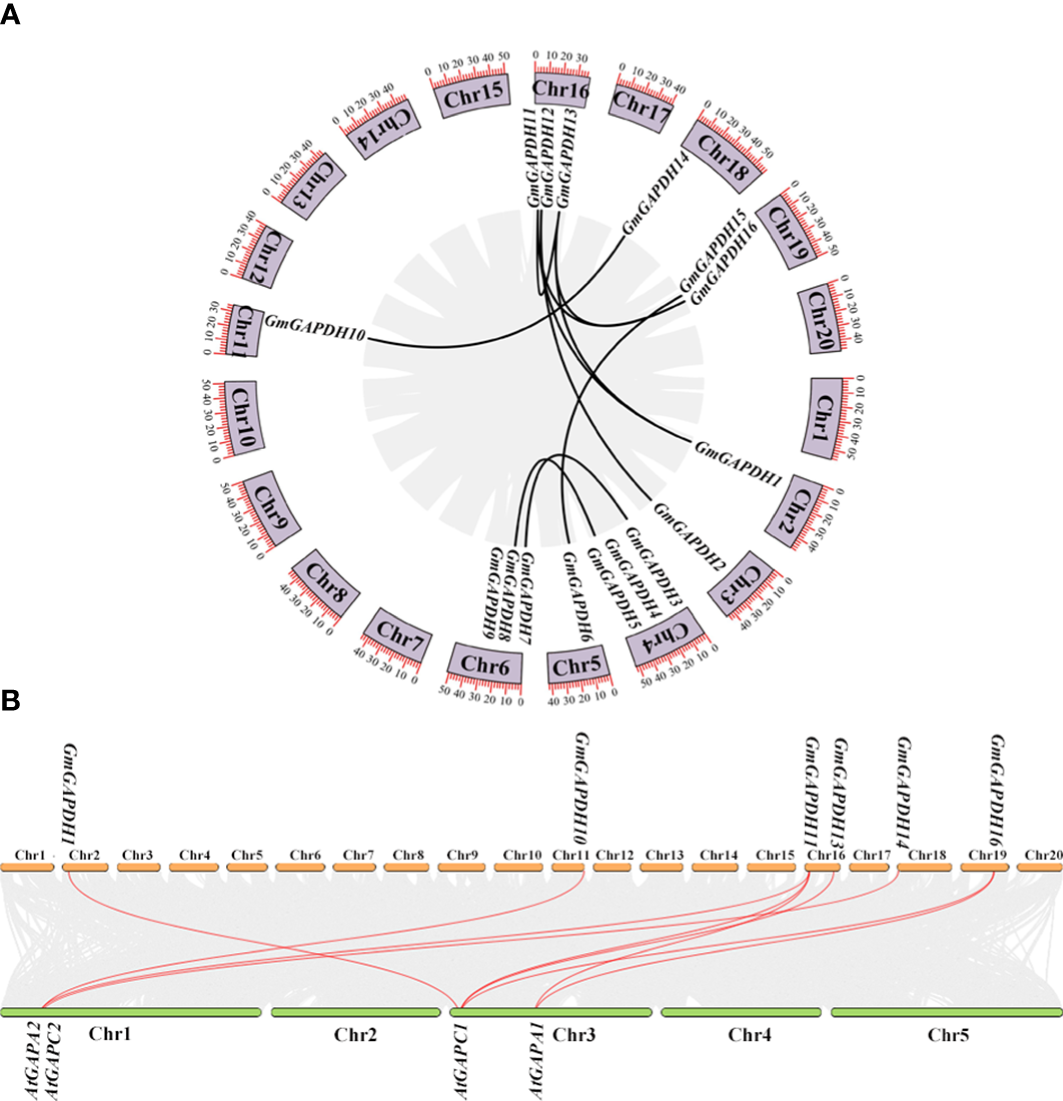
Syntenic analysis of GmGAPDH family genes. (A) Chromosome location and duplication of GmGAPDH genes on soybean genome. (B) Syntenic analysis of GAPDHs with the corresponding genes in G max and A thaliana.
Expression profiles of soybean GAPDHs in diverse tissues and developmental stages
To determine the expression pattern of soybean GAPDH genes in different development phases, we retrieved the high-throughput sequencing data of the Phytozome database and conducted an expression analysis. As demonstrated in Figure 4, the expression of GmGAPDHs was revealed in diverse tissues. GmGAPDH4, 5, 8, 9, and 14 were found to have a higher expression in different tissues. Meanwhile, the GmGAPDH1, GmGAPDH6, and GmGAPDH15 genes were feebly expressed in nine different tissues. Moreover, GmGAPDH3, 7, 11, and 16 were expressed only in the shoot apical meristem and leaf tissues. GmGAPDH2 and GmGAPDH12 were especially expressed only in nodules (Figure 4A). Moreover, the expression of GmGAPDHs during the development of soybean seed is shown in Figure 4B (10 to 40 DAF). The expression levels of GmGAPDH4, GmGAPDH5, GmGAPDH9, and GmGAPDH11 were found to be upregulated in seeds at 20 DAF. The expression of the GmGAPDH8 and GmGAPDH14 genes was significantly upregulated in seeds at 30 DAF.
Figure 4
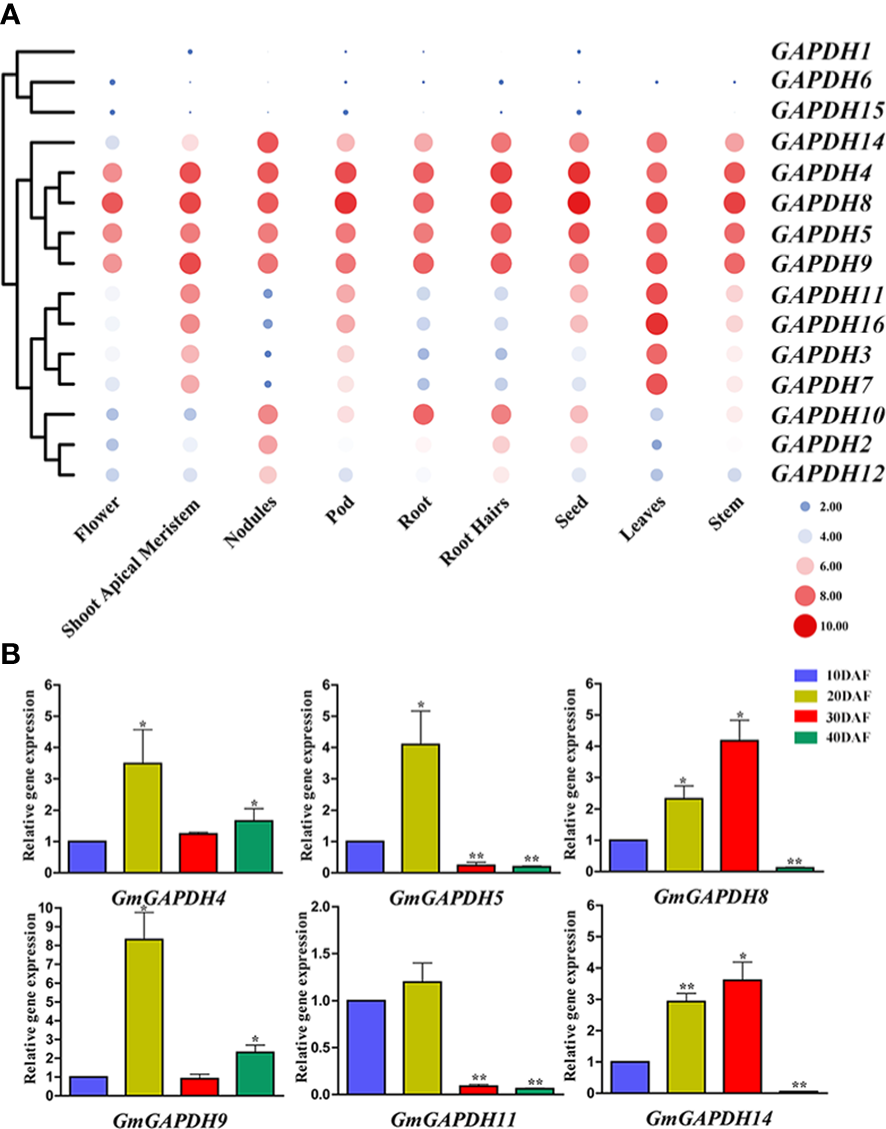
Expression levels of GmGAPDH genes during different tissue and developmental stages. (A) The expression profile analyses of soybean GAPDH genes were characterized in different tissues. The expressions of different tissues were displayed in heat maps. The color scale indicates the log2 expression level, the red circles indicate the high transcription levels, and the blue circles indicate the low transcription levels. (B) Relative expression level of GmGAPDHs in different developing seeds at 10, 20, 30, and 40 days after flowering in soybean (DN50). The expression of GmGAPDHs in developing seeds at 10 DAF was used as the internal reference. Student’s t-test was carried out to determine the significance levels (*P < 0.05, **P < 0.01).
Expression profiles of GmGAPDH genes under abiotic stresses
To confirm the role of GmGAPDH gene responses to various abiotic stresses, the transcription level of 16 GmGAPDH genes under NaHCO3 (100 mM), PEG (20%), Cold (4°C), and Slat (150 mM) stresses was determined by qRT-PCR. Under simulated alkali stress using NaHCO3, GmGAPDH 4, 5, 8, 10, 12, and 14 were extraordinarily upregulated (more than six folds) and peaked at 6 and 12 h, respectively (Figure 5A). After simulated drought stress using PEG, GmGAPDH14 was upregulated by more than 20 folds in 24 h after the treatment. In comparison, the expression of GmGAPDH16 significantly decreased in 6, 12, and 24 h, respectively (Figure 5B). In response to cold treatment, most of GmGAPDHs had upregulated expression, especially GmGAPDH4, which had a significantly higher level of expression at 24 h (Figure 5C). Furthermore, the GmGAPDH genes were shown to have a higher expression level at different timepoints of cold stress. The expression levels of GmGAPDHs were different under simulated salt stress with salt (Figure 5D). The GmGAPDH14 gene was shown to have the highest expression level at 6, 12, and 24, which was upregulated by more than 150 folds than that at 24 h. Meanwhile, the GmGAPDH4 gene exhibited a higher expression level at 6 and 12 h (20 and 40 folds). In contrast to other stresses, the whole expression of GmGAPDHs under NaHCO3 stress was found to be relatively low (Figure 5A). Remarkably, the expression of GmGAPDH14 was sharply induced under salt stress, indicating that this gene might play a role in salt stress resistance.
Figure 5
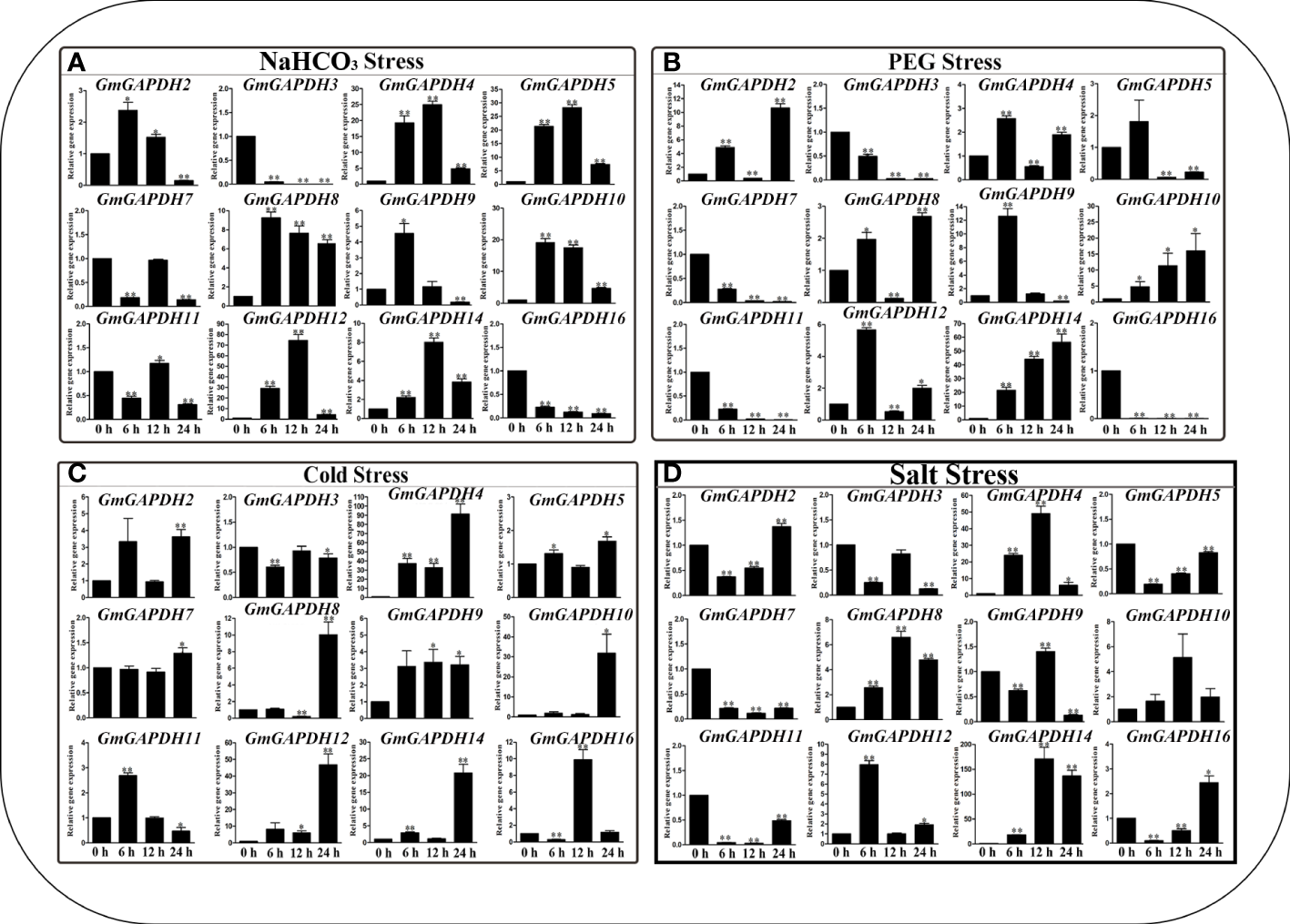
Expression level of GmGAPDHs in response to NaHCO3(A), polyethylene glycol (B), cold (C), and salt (D) stresses in the leaves of soybean. The expression of GmGAPDHs in a non-stress environment was used as a calibrator. Three technical replicates and three biological replicates were applied to each sample. The expression level of GmGAPDHs was calculated with the 2–ΔΔct method.
Overexpression of GmGAPDH14 improved the tolerance to salt stress
To deeply illustrate how GmGAPDH14 genes respond to salt stress, hairy roots with Agrobacterium rhizogenes K599 containing pCambia3300-GmGAPDH14 plasmid or the pCambia3300 empty vector were transformed. Eight soybeans were proven through the PCR method to be positive transgenic. Furthermore, it was found that the effect of overexpression of GmGAPDH14 is such that it can regulate soybean hair roots in response to salt tolerance. Furthermore, 2-week soybean positive lines, involving 0 or 150 mM salt for 3 days, were transferred.
Previous studies showed that plant GAPDHs are involved in functions such as response to oxidative stress (Guo et al., 2012). Hence, the overexpression of GmGAPDH14 may further increase the antioxidant level under salt stress in this study. Therefore, the SOD activity and the MDA content were tested in soybeans positive transgenic at 3 d after 150 mM salt treatment. After 3 d of 150 mM salt stress, the overexpression of GmGAPDH14 was exhibited to enhance the resistance to salt stress than those of the control (transformed by pCambia3300 EV) (Figures 6A, B). As shown in Figure 6, the OE-GmGAPDH14 lines had a higher activity of SOD than that of the EV lines (Figure 6C). The above-mentioned results showed that GmGAPDH14 could participate in regulating the ROS level. The MDA content showed that the contents of the OE-GmGAPDH14 lines were significantly lower than that of the EV lines after the salt treatment (Figure 6D).
Figure 6
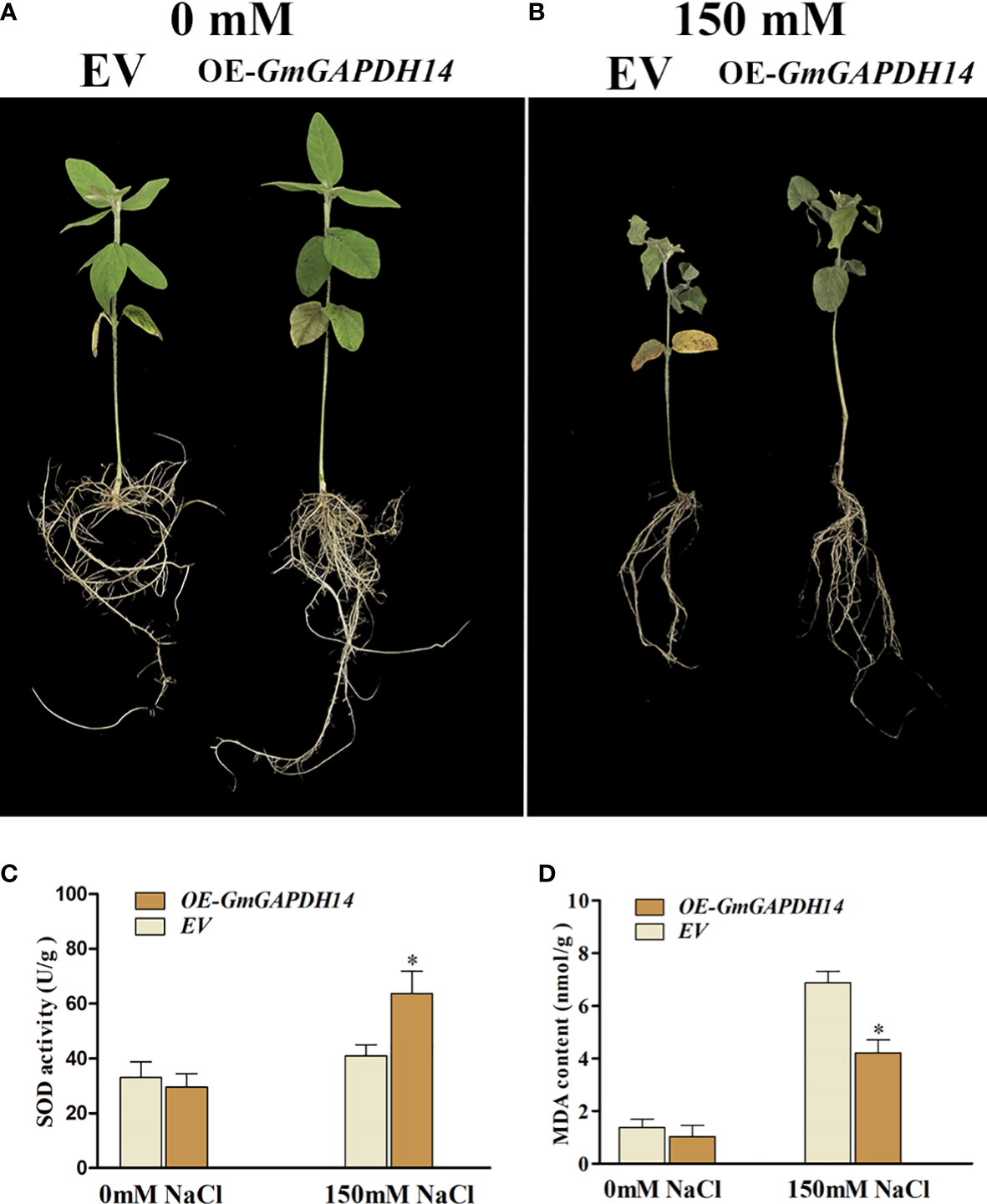
Effect of GmGAPDH14 overexpression: (A) 0 mM salt, (B) 150 mM salt, (C) superoxide dismutase (SOD) activity, and (D) malondialdehyde (MDA) content in transgenic lines under salt stress. EV represents empty vector pCambia3300 (transgenic soybean hairy roots, control), and OE-GmGAPDH14 represents recombinant vector pCambia3300–GmGAPDH14 (transgenic soybean hairy roots). The activity of SOD and the MDA content in soybean seedling at 3 days after 150 mM salt treatment. The asterisks represent significant differences between EV and OE-GmGAPDH14 by Student’s t-test (∗P < 0.05).
Haplotype analysis of GmGAPDH14 gene for salt stress
To further confirm the potential effects of GmGAPDH14 gene for salt stress, gene-based association analysis was applied through the GLM method. A total of five SNPs in the GmGAPDH14 gene were identified among 131 lines (Supplementary Table S3). All the five SNPs were significantly associated with salt stress, and they were located in the exon, intron, UTR region, and upstream regions of the GmGAPDH14 gene, respectively (Table 1). Four haplotypes of the GmGAPDH14 gene were defined by the five SNPs. Haplotypes 2, 3, and 4 were composed of the combination of TAG, TTT, and AAT alleles and were beneficial for the salt tolerance of soybean. The carriers of haplotype 1 were composed of a combination of AAG alleles which tended to be sensitive to salt stress (Figure 7, Supplementary Figure S3). The difference of salt tolerance between the carriers of two haplotypes reached a very significant level.
Table 1
| Chromosome | Position (bp) | Region | Alleles | Year | -log10(p) |
|---|---|---|---|---|---|
| 18 | 693402 | Exon | G/T | 2019 | 2.74 |
| 18 | 693891 | Intron | C/T | 2019 | 1.82 |
| 18 | 695147 | Intron | T/C | 2019 | 3.56 |
| 18 | 695420 | UTR | A/T | 2019 | 2.35 |
| 18 | 695922 | Promoter | A/T | 2019 | 2.74 |
| 18 | 693402 | Exon | G/T | 2020 | 1.08 |
| 18 | 693891 | Intron | C/T | 2020 | 2.6 |
| 18 | 695147 | Intron | T/C | 2020 | 2.14 |
| 18 | 695420 | UTR | A/T | 2020 | 1.53 |
| 18 | 695922 | Promoter | A/T | 2020 | 1.08 |
The association between SNP in GmGAPDH14 gene and soybean resistance to salt based on 131 soybean germplasms.
Figure 7
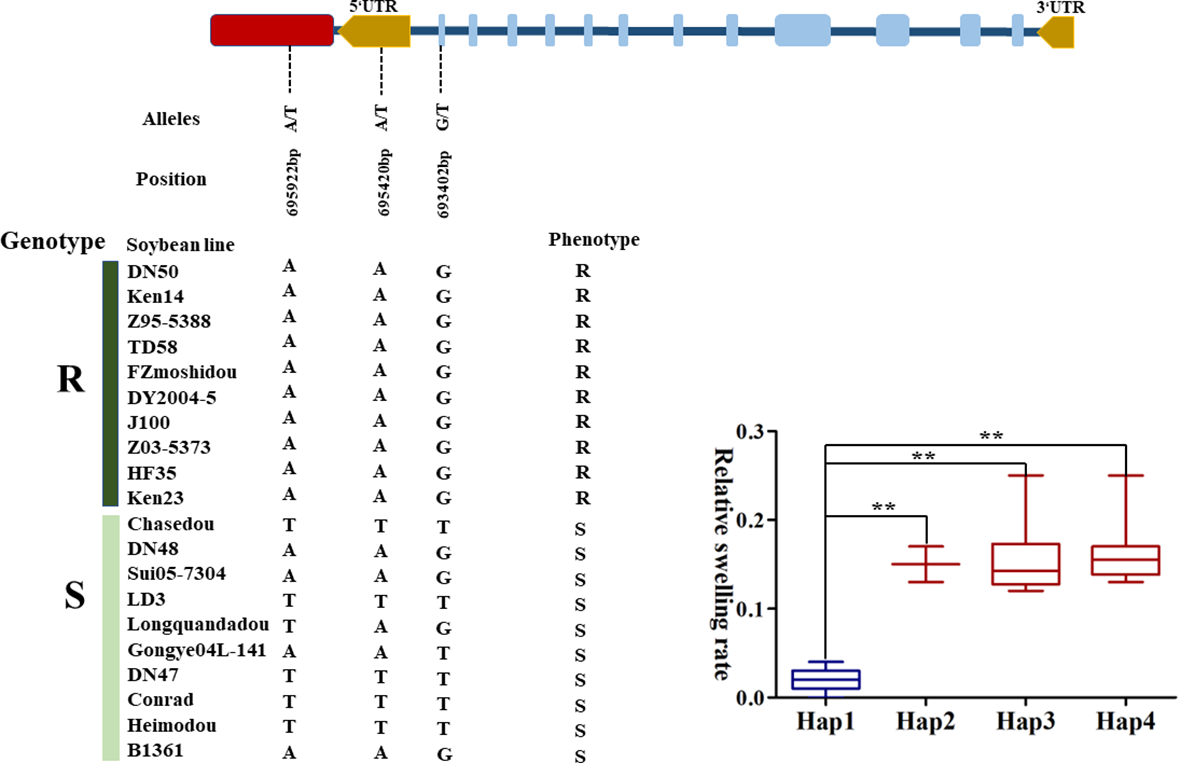
Haplotype analysis of GmGAPDH14 genes related to salt stress. The asterisks represent significant differences by Student’s t-test (∗∗P < 0.01).
Discussion
GAPDH is a pivotal enzyme in the glycolytic pathway. Previous studies have exhibited that GAPDHs play a crucial role in plant growth and response to various stresses (Guo et al., 2012; Liu et al., 2017; Kim et al., 2020). In this study, the candidate genes of soybean GAPDH family were identified, which were proven to regulate plant growth and stress. Although GmGAPDH genes have been analyzed in many plants, including Arabidopsis (Guo et al., 2014; Kim et al., 2020), potato (Liu et al., 2017), wheat (Li et al., 2019), and rice (Wang et al., 2021), nevertheless, finite information was found about the GAPDH molecular function of soybean.
In the present study, a total of 16 GmGAPDH genes were identified from the soybean genome. The phylogenetic analysis can clearly prove the evolutionary relationships between soybean GAPDH and those of other species. The result showed that 16 GAPDHs were segmented into three clusters based on their different subcellular locations. Previous studies showed that the different GAPDHs can target various regions (Rius et al., 2008; Muñoz-Bertomeu et al., 2009). For cluster I, eight GmGAPDHs isoforms were located on the cytosol of Arabidopsis GAPCs; for cluster II, two GmGAPDHs isoforms were located on the chloroplast; and for cluster III, six GmGAPDHs isoforms were located on the plastid (Figure 1) (Vescovi et al., 2013; Anoman et al., 2015; Marri et al., 2015). The result showed that the three distinct types of GmGAPDHs can implement corresponding functions in plants. Gene duplication was the key mechanism for the creation of unique evolutionary innovations, which mainly include segmental and tandem duplications (Jiao et al., 2011). The previous studies showed that a family gene will show a highly conserved duplication style in various species (Innan and Kondrashov, 2010). In this study, most of the GmGAPDH genes were found to carry out segmental duplication in the soybean genome (Figure 3). The result of this study indicated that soybean GAPDH did not maintain a conserved duplication. These duplicated genes proved a common genomic source and performed similar functions, and segmental duplication was advantageous to extend of GAPDH genes family in soybean.
In this study, to reveal the response of soybean GAPDHs to abiotic stresses, the expression pattern of GAPDH genes was tested via qRT-PCR analysis. The promoter sequences of the soybean GAPDHs involved a number of cis-elements—for example, ERE, ARE, and LTR (Figure 2). According to this result, NaHCO3, PEG, cold stress, and salt stress can significantly induce GmGAPDHs expression (Figure 5). AtGAPC was found to interact with phospholipase Dδ (PLDδ), transmit H2O2 signaling under drought stress, and increase the seed oil content (Guo et al., 2012; Kim et al., 2020). In wheat, GAPDH12 was found to be remarkably upregulated under salt, cold, high temperature, and drought stresses (Zeng et al., 2016). The overexpression of GAPC3 can improve salt tolerance in rice (Zhang et al., 2011). In Arabidopsis, the overexpression of TaGAPC1 enhanced the tolerance to drought stress (Zhang et al., 2019). In potato, StGAPC1, StGAPC2, and StGAPC3 were found to be cold-induced in the tubers (Liu et al., 2017). In Arabidopsis, the overexpression of GAPC improved its heat tolerance (Kim et al., 2020). In this study, GAPDH12 was found to be strongly under NaHCO3 stress at 12 h. In PEG stress, GAPDH14 was found to be strongly under stress at 24 h. In cold stress, GAPDH4 was found to be strongly under stress at 24 h. It is worth noting that GmGAPDH14 responded more strongly to salt stress than the other genes, and GmGAPDH14 reached a maximum expression level with 12 h to salt stress (about 150 folds). Therefore, GAPDH14 may play an important role in response to salt stress.
Previous studies showed that GAPDH proteins—PsGAPDH, NbGAPC, and AtGAPC—play key roles in growth and abiotic stress response in plants (Guo et al., 2014; Han et al., 2015; Lim et al., 2021). Next, to further understand the molecular function of GmGAPDH14 in response to salt stress, the overexpression of GmGAPDH14 significantly increased the salt tolerance of transgenic soybean lines (Figures 6A, B). In this study, the SOD activity of OE-GmGAPDH14 plants was significantly higher than that of EV plants. The above-mentioned data showed that GmGAPDH14 may play an important role in reducing ROS accumulation under salt stress, which was consistent with the results of previous studies (Zhang et al., 2011). The content of MDA was commonly considered as a marker of oxidative stress (Anjum et al., 2015). It was certified that the EV plants were found to have more serious damage than OE-GmGAPDH14 plants (Figure 6D). In addition, the five SNPs of the GmGAPDH14 gene were significantly associated with soybean tolerance to salt stress. Four haplotypes of the GmGAPDH14 gene were defined by the five SNPs, and the difference of salt tolerance between the carriers of the four haplotypes reached a very significant level. Meantime, the 693402 position SNP was found to be located in the Gp_dh_C domain. This result suggests that variations in the domain may enhance the salt tolerance of the GAPDH14 gene. In this study, the involvement of GmGAPDH14 gene in soybean salt tolerance was verified, and the salt tolerance effect of the gene related to the natural variation of the gene sequence was also proven. These results may provide ideas for exploring the beneficial salt-resistant SNPs.
In conclusion, a total of 16 soybean GAPDH genes were clearly divided into three clusters. These GmGAPDH genes had a different expression level under various abiotic stresses. The 16 GmGAPDH genes, including GmGAPDH14, had a markedly induced response to salinity.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
Conceptualization: YH, YL, and XueZ; methodology: XunZ, NX, and JW; software: YQ, YZ, and WT; formal analysis: XunZ and JW; investigation: XunZ, HL, and WL; resources: YH; data curation: YQ, JW, and YZ; writing—original: XunZ; writing—review and editing: YH and YL; supervision: YH and YL; funding acquisition: YH and XueZ. All authors All authors contributed to the article and approved the submitted version.
Funding
This study was conducted in the Key Laboratory of Soybean Biology of the Chinese Education Ministry, Soybean Research and Development Center, and the Key Laboratory of Northeastern Soybean Biology and Breeding/Genetics of the Chinese Agriculture Ministry and was financially supported by the Chinese National Natural Science Foundation (32001570), the Chinese National Natural Science Foundation (U22A20473, 31971967), the Key Fund of Heilongjiang Province (ZD2022C002), the National Project (2014BAD22B01 and 2016ZX08004001-007), the Youth Leading Talent Project of the Ministry of Science and Technology in China (2015RA228), the National Ten-Thousand Talents Program, Postdoctoral Fund in Heilongjiang Province (LBH-Q20004), and the National Project (CARS-04-PS04).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1193044/full#supplementary-material
Supplementary Figure 1Analysis of the conserved active site of GAPDH in soybean.
Supplementary Figure 2Analysis of the conserved motifs of GAPDH in soybean.
Supplementary Figure 3Haplotype analysis of GmGAPDH14 gene related to salt stress.
References
1
Anjum N. A. Sofo A. Scopa A. Roychoudhury A. Gill S. S. Iqbal M. et al . (2015). Lipids and proteins–major targets of oxidative modifications in abiotic stressed plants. Environ. Sci. pollut. Res. Int.22, 4099–4121. doi: 10.1007/s11356-014-3917-1
2
Anoman A. D. Muñoz-Bertomeu J. Rosa-Téllez S. Flores-Tornero M. Serrano R. Bueso E. et al . (2015). Plastidial glycolytic glyceraldehyde-3-phosphate dehydrogenase is an important determinant in the carbon and nitrogen metabolism of heterotrophic cells in Arabidopsis. Plant Physiol.169, 1619–1637. doi: 10.1104/pp.15.00696
3
Artimo P. Jonnalagedda M. Arnold K. Baratin D. Csardi G. de Castro E. et al . (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res.40, W597–W603. doi: 10.1093/nar/gks400
4
Bradbury P. J. Zhang Z. Kroon D. E. Casstevens T. M. Ramdoss Y. Buckler E. S. (2017). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics23, 2633–2635. doi: 10.1093/bioinformatics/btm308
5
Bustos D. M. Bustamante C. A. Iglesias A. A. (2007). Involvement of non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase in response to oxidative stress. J. Plant Physiol.165, 456–461. doi: 10.1016/j.jplph.2007.06.005
6
Chaturvedi S. Seo J. K. Rao A. L. (2016). Functionality of host proteins in cucumber mosaic virus replication: GAPDH is obligatory to promote interaction between replication-associated proteins. Virology494, 47–55. doi: 10.1016/j.virol.2016.04.001
7
Chen C. Chen H. Zhang Y. Thomas H. R. Frank M. H. He Y. et al . (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant3, 1194–1202. doi: 10.1016/j.molp.2020.06.009
8
Cho J. I. Lim H. M. Siddiqui Z. S. Park S. H. Kim A. R. Kwon T. R. et al . (2014). Over-expression of PsGPD, a mushroom glyceraldehyde-3-phosphate dehydrogenase gene, enhances salt tolerance in rice plants. Biotechnol. Lett.36, 1641–1648. doi: 10.1007/s10529-014-1522-1525
9
Colell A. Ricci J. E. Tait S. Milasta S. Maurer U. Bouchier-Hayes L. et al . (2007). GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell129, 983–997. doi: 10.1016/j.cell.2007.03.045
10
Feng Z. Ding C. Li W. Wang D. Cui D. (2020). Applications of metabolomics in the research of soybean plant under abiotic stress. Food Chem.25 (310), 125914. doi: 10.1016/j.foodchem.2019.125914
11
Finn R. D. Coggill P. Eberhardt R. Y. Eddy S. R. Mistry J. Mitchell A. L. et al . (2016). The pfam protein families database: towards a more sustainable future. Nucleic Acids Res.44, D279–D285. doi: 10.1093/nar/gkv1344
12
Guo L. Devaiah S. P. Narasimhan R. Pan X. Zhang Y. Zhang W. et al . (2012). Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase dδ to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell.24, 2200–2212. doi: 10.1105/tpc.111.094946
13
Guo L. Ma F. Wei F. Fanella B. Allen D. K. Wang X. (2014). Cytosolic phosphorylating glyceraldehyde-3-phosphate dehydrogenases affect Arabidopsis cellular metabolism and promote seed oil accumulation. Plant Cell.26, 3023–3035. doi: 10.1105/tpc.114.126946
14
Han S. Wang Y. Zheng X. Jia Q. Zhao J. Bai F. et al . (2015). Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in nicotiana benthamiana. Plant Cell.27, 1316–1331. doi: 10.1105/tpc.114.134692
15
Henry E. Fung N. Liu J. Drakakaki G. Coaker G. (2015). Beyond glycolysis: GAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLoS Genet.11, e1005199. doi: 10.1371/journal.pgen.1005199
16
Holle S. V. Damme E. V. (2015). Distribution and evolution of the lectin family in soybean (Glycine max). Molecules20, 2868–2891. doi: 10.3390/molecules20022868
17
Hong S. Bahn S. C. Lyu A. Jung H. S. Ahn J. H. (2010). Identification and testing of superior reference genes for a starting pool of transcript normalization in arabidopsis. Plant Cell Physiol.51, 1694–1706. doi: 10.1093/pcp/pcq128
18
Hu B. Jin J. Guo A. Y. Zhang H. Luo J. Gao G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics31, 1296–1297. doi: 10.1093/bioinformatics/btu817
19
Innan H. Kondrashov F. (2010). The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet.11, 97–108. doi: 10.1038/nrg2689
20
Jeong M. J. Park S. C. Byun M. O. (2001). Improvement of salt tolerance in transgenic potato plants by glyceraldehyde-3 phosphate dehydrogenase gene transfer. Mol. Cell.12, 185–189. doi: 10.14348/molcells.2001.0185
21
Jiao Y. Wickett N. J. Ayyampalayam S. Chanderbali A. S. Landherr L. Ralph P. E. et al . (2011). Ancestral polyploidy in seed plants and angiosperms. Nature473, 97–100. doi: 10.1038/nature09916
22
Ke X. Xiao H. Peng Y. Wang J. Lv Q. Wang X. (2022). Phosphoenolpyruvate reallocation links nitrogen fixation rates to root nodule energy state. Science378, 971–977. doi: 10.1126/science.abq8591
23
Kereszt A. Li D. Indrasumunar A. Nguyen C. D. Nontachaiyapoom S. Kinkema M. et al . (2007). Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc.2, 948–952. doi: 10.1038/nprot.2007.141
24
Kim S. C. Guo L. Wang X. (2020). Nuclear moonlighting of cytosolic glyceraldehyde-3-phosphate dehydrogenase regulates Arabidopsis response to heat stress. Nat. Commun.113439. doi: 10.1038/s41467-020-17311-4
25
Kumar S. Stecher G. Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol.33, 1870–1874.
26
Letunic I. Doerks T. Bork P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res.43, D257–D260. doi: 10.1093/nar/gku949
27
Li X. Wei W. Li F. Zhang L. Deng X. Liu Y. et al . (2019). The plastidial glyceraldehyde-3-phosphate dehydrogenase is critical for abiotic stress response in wheat. Int. J. Mol. Sci.20, 1104. doi: 10.3390/ijms20051104
28
Lim H. Hwang H. Kim T. Kim S. Chung H. Lee D. et al . (2021). Transcriptomic analysis of rice plants overexpressing PsGAPDH in response to salinity stress. Genes12, 641. doi: 10.3390/genes12050641
29
Liu T. Fang H. Liu J. Reid S. Hou J. Zhou T. et al . (2017). Cytosolic glyceraldehyde-3-phosphate dehydrogenases play crucial roles in controlling cold-induced sweetening and apical dominance of potato (Solanum tuberosum l.) tubers. Plant Cell Environ.40, 3043–3054. doi: 10.1111/pce.13073
30
Liu W. Xie Y. Ma J. Luo X. Nie P. Zuo Z. et al . (2015). IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics31, 3359–3361. doi: 10.1093/bioinformatics/btv362
31
Marri L. Sparla F. Pupillo P. Trost P. (2015). Co-Ordinated gene expression of photosynthetic glyceraldehyde-3-phosphate dehydrogenase, phosphoribulokinase, and CP12 in Arabidopsis thaliana. J. Exp. Bot.56, 73–80. doi: 10.1093/jxb/eri020
32
Miao L. Chen C. Yao L. Tran J. Zhang H. (2019). Genome-wide identification, characterization, interaction network and expression profile of GAPDH gene family in sweet orange (Citrus sinensis). Peer J.7, e7934. doi: 10.7717/peerj.7934
33
Muñoz-Bertomeu J. Cascales-Miñana B. Mulet J. M. Baroja-Fernández E. Pozueta-Romero J. Kuhn J. M. et al . (2009). Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiol.151, 541–558. doi: 10.1104/pp.109.143701
34
Plaxton W. C. (1996). The organization and regulation of plant glycolysis. Annu. Rev. Plant Biol.47, 185–214. doi: 10.1146/annurev.arplant.47.1.185
35
Pokotylo I. Hellal D. Bouceba T. Hernandez-Martinez M. Kravets V. Leitao L. et al . (2020). Deciphering the binding of salicylic acid to Arabidopsis thaliana chloroplastic GAPDH-A1. Int. J. Mol. Sci.21, 4678. doi: 10.3390/ijms21134678
36
Rius S. P. Casati P. Iglesias A. A. Gomez-Casati D. F. (2008). Characterization of Arabidopsis lines deficient in GAPC-1, a cytosolic NAD-dependent glyceraldehyde-3-phosphate dehydrogenase. Plant Physiol.148, 1655–1667. doi: 10.1104/pp.108.128769
37
Sirover M. A. (2011). On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim. Biophys. Acta1810, 741–751. doi: 10.1016/j.bbagen.2011.05.010
38
Vescovi M. Zaffagnini M. Festa M. Trost P. Lo Schiavo F. Costa A. (2013). Nuclear accumulation of cytosolic glyceraldehyde-3-phosphate dehydrogenase in cadmium-stressed arabidopsis roots. Plant Physiol.162, 333–346. doi: 10.1104/pp.113.215194
39
Wang Q. Lu L. Zeng M. Wang D. Zhang T. Xie Y. et al . (2021). Rice black-streaked dwarf virus P10 promotes phosphorylation of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) to induce autophagy in Laodelphax striatellus. Autophagy27, 1–20. doi: 10.1080/15548627.2021.1954773
40
Wei H. Movahedi A. Yang J. Zhang Y. Liu G. Zhu S. et al . (2022). Characteristics and molecular identification of glyceraldehyde-3-phosphate dehydrogenases in poplar. Int. J. Biol. Macromol.219, 185–198. doi: 10.1016/j.ijbiomac.2022.08.001
41
Yu C. Lin C. Hwang J. K. (2004). Predicting subcellular localization of proteins for gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci.13, 1402–1406. doi: 10.1110/ps.03479604
42
Yu Q. Liu Y. Sun G. Liu Y. Chen J. Zhou Y. et al . (2021). Genome-wide analysis of the soybean calmodulin-binding protein 60 family and identification of GmCBP60A-1 responses to drought and salt stresses. Int. J. Mol. Sci.22, 13501. doi: 10.3390/ijms222413501
43
Zeng L. Deng R. Guo Z. Yang S. Deng X. (2016). Genome-wide identification and characterization of glyceraldehyde-3-phosphate dehydrogenase genes family in wheat (Triticum aestivum). BMC Genomics17, 240. doi: 10.1186/s12864-016-2527-3
44
Zhang H. Zhao Y. Zhou D. (2017). Rice NAD+-dependent histone deacetylase OsSRT1 represses glycolysis and regulates the moonlighting function of GAPDH as a transcriptional activator of glycolytic genes. Nucleic Acids Res.45, 12241–12255. doi: 10.1093/nar/gkx825
45
Zhang J. Niu Y. Bu S. H. Li M. Feng Y. Zhang J. et al . (2014). Epistatic association mapping for alkaline and salinity tolerance traits in the soybean germination stage. PLoS One9, e84750. doi: 10.1371/journal.pone.0084750
46
Zhang L. Xu Z. Ji H. Zhou Y. Yang S. (2019). TaWRKY40 transcription factor positively regulate the expression of TaGAPC1 to enhance drought tolerance. BMC Genomics20, 795. doi: 10.1186/s12864-019-617
47
Zhang L. Zhang. H. Yang. S. (2020). Cytosolic TaGAPC2 enhances tolerance to drought stress in transgenic Arabidopsis plants. Int. J. Mol. Sci.21, 7499. doi: 10.3390/ijms21207499
48
Zhang X. Rao X. Shi H. Li R. Lu Y. (2011). Overexpression of a cytosolic glyceraldehyde-3-phosphate dehydrogenase gene OsGAPC3 confers salt tolerance in rice. Plant Cell Tissue Organ Cult.107, 1–11. doi: 10.1007/s11240-011-9950-6
Summary
Keywords
GAPDH protein, hairy roots, salt tolerance, soybean, gene family
Citation
Zhao X, Wang J, Xia N, Qu Y, Zhan Y, Teng W, Li H, Li W, Li Y, Zhao X and Han Y (2023) Genome-wide identification and analysis of glyceraldehyde-3-phosphate dehydrogenase family reveals the role of GmGAPDH14 to improve salt tolerance in soybean (Glycine max L.). Front. Plant Sci. 14:1193044. doi: 10.3389/fpls.2023.1193044
Received
24 March 2023
Accepted
26 April 2023
Published
06 June 2023
Volume
14 - 2023
Edited by
Baohua Wang, Nantong University, China
Reviewed by
Hengyou Zhang, Northeast Institute of Geography and Agroecology (CAS), China; Shiming Liu, Institute of Plant Protection (CAAS), China; Long Yan, Hebei Academy of Agriculture and Forestry Sciences (HAAFS), China; Xiangjin Wei, China National Rice Research Institute (CAAS), China
Updates
Copyright
© 2023 Zhao, Wang, Xia, Qu, Zhan, Teng, Li, Li, Li, Zhao and Han.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongguang Li, Yongguangli@neau.edu.cn; Xue Zhao, xuezhao@neau.edu.cn; Yingpeng Han, hyp234286@aliyun.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.