- 1State Key Laboratory of Wheat Breeding, Group of Wheat Quality and Molecular Breeding, College of Agronomy, Shandong Agricultural University, Tai’an, Shandong, China
- 2College of Biological and Chemical Engineering, Qilu Institute of Technology, Jinan, Shandong, China
- 3Taiyuan Agro-Tech Extension and Service Center, Taiyuan, Shanxi, China
- 4R&D Department, Shandong Huatian Agricultural Technology Co., Ltd, Feicheng, Shandong, China
Protein, starch, and their components are important for wheat grain yield and end-products, which are affected by wheat grain development. Therefore, QTL mapping and a genome-wide association study (GWAS) of grain protein content (GPC), glutenin macropolymer content (GMP), amylopectin content (GApC), and amylose content (GAsC) were performed on wheat grain development at 7, 14, 21, and 28 days after anthesis (DAA) in two environments using a recombinant inbred line (RIL) population of 256 stable lines and a panel of 205 wheat accessions. A total of 29 unconditional QTLs, 13 conditional QTLs, 99 unconditional marker−trait associations (MTAs), and 14 conditional MTAs significantly associated (p < 10−4) with four quality traits were found to be distributed on 15 chromosomes, with the phenotypic variation explained (PVE) ranging from 5.35% to 39.86%. Among these genomic variations, three major QTLs [QGPC3B, QGPC2A, and QGPC(S3|S2)3B] and SNP clusters on the 3A and 6B chromosomes were detected for GPC, and the SNP TA005876-0602 was stably expressed during the three periods in the natural population. The QGMP3B locus was detected five times in three developmental stages in two environments with 5.89%–33.62% PVE, and SNP clusters for GMP content were found on the 3A and 3B chromosomes. For GApC, the QGApC3B.1 locus had the highest PVE of 25.69%, and SNP clusters were found on chromosomes 4A, 4B, 5B, 6B, and 7B. Four major QTLs of GAsC were detected at 21 and 28 DAA. Most interestingly, both QTL mapping and GWAS analysis indicated that four chromosomes (3B, 4A, 6B, and 7A) were mainly involved in the development of protein, GMP, amylopectin, and amylose synthesis. Of these, the wPt-5870–wPt-3620 marker interval on chromosome 3B seemed to be most important because it played an important role in the synthesis of GMP and amylopectin before 7 DAA, in the synthesis of protein and GMP from 14 to 21 DAA, and in the development of GApC and GAsC from 21 to 28 DAA. Using the annotation information of IWGSC Chinese Spring RefSeq v1.1 genome assembly, we predicted 28 and 69 candidate genes for major loci from QTL mapping and GWAS, respectively. Most of them have multiple effects on protein and starch synthesis during grain development. These results provide new insights and information for the potential regulatory network between grain protein and starch synthesis.
1 Introduction
Wheat (Triticum aestivum L.) is one of the most important cereal crops in the world. It is the main source of carbohydrates, protein, and dietary fiber for humans (Ma et al., 2013). Protein and starch are important components of wheat grain and affect both the yield and processing quality of wheat. Wheat grains contain 8%–20% protein and 85% carbohydrates, most of which are starch (Anjum et al., 2007). In recent years, with the continuous improvement of human dietary habits and living standards, more attention has been given to the nutritional composition and flavor quality of wheat products. In addition, increasingly extreme weather conditions and an increasing world population impose a severe threat to wheat production (Zhang et al., 2021). Therefore, on the basis of ensuring high yield, breeding new wheat varieties with high quality and wide adaptability is the main goal of wheat quality breeding worldwide (Liu et al., 2018). It is of great significance to study and analyze the basic mechanism or important genes/loci affecting protein and starch content in wheat grain development.
The grain protein content (GPC) is important for dough and end-use quality because it influences the ratio of gliadin to glutenin. Therefore, improving GPC has become an important goal of wheat quality breeding, but previous studies showed that it was affected not only by genetics but also by the environment (Paull and Anderson, 1942). GPC is regarded as a quantitative trait. Several studies have reported quantitative trait loci (QTL) for GPC and flour protein content using different populations, which involved approximately 19 chromosomes. The majority of these QTLs were reported on chromosomes 6B and 7B. Of these, the marker Gpc-B1 on chromosome 6B has been used in wheat quality breeding. Two important QTLs, QGpc.ink-2B and QGpc.ink-6A, were detected for GPC by GWAS. The QGpc.ink-6A locus controlled both GPC and GSC (grain starch content) with opposite allelic effects (Muqaddasi et al., 2020). However, little is known about the genes determining the inheritance of GPC in wheat; the only identified causative gene in a GPC locus associated with high GPC yield thus far is an NAC transcription factor that originated from wild emmer wheat (Souer et al., 1996; Aida et al., 1997). In wheat, the endosperm-specific transcription factor TaNAC019 was found to affect grain quality by regulating glutenin and starch accumulation (Gao et al., 2021). Therefore, the identification of genetic loci or genes controlling GPC is helpful for the effective breeding of wheat varieties with high protein synthesis and lays a foundation for an in-depth understanding of the regulation between wheat protein and starch synthesis.
Polymeric glutenins are an important component of gluten proteins in wheat synthesized in starchy endosperm cells during grain filling. They contribute to the elasticity of gluten. The disulfide bonds of glutenin subunits are present in the glutenin macropolymer (GMP), allowing it to play an important role in the establishment of the gluten network structure when it achieves a certain size (Tao et al., 2022). It mainly comprises high- molecular-weight glutenin subunits (HMW-GS) and low- molecular-weight glutenin subunits (LMW-GS). The effects of the GMP, HMW-GS, and LMW-GS on dough and processing quality have been extensively studied, and most of them greatly contribute to wheat quality (Ma et al., 2005). Although genetic factors, such as the ratio of HMW-GS to LMW-GS and their allelic variations, affect the GMP and thus the viscoelastic properties of gluten, the GMP is also affected by the interaction between genotype and environment. The GMP is a quantitative trait. Few studies have been reported on the genes/loci for the GMP by QTL mapping or GWAS. Two major QTL clusters for glutenin have been reported, 1DL-2 and 6AS-3, at the chromosome regions 409.26–416.45 Mb and 44.02–50.35 Mb, respectively. HMW-GS and LMW-GS were detected, and Kompetitive Allele-Specific PCR (KASP) markers were developed using a recombinant inbred line population (Zhou et al., 2021). However, little is known about how many genes are involved in wheat grain development and how they work during that process.
Starch is a major storage component of wheat grain endosperm. Common wheat has an amylose content of 25%–30% and an amylopectin content of 70%–75%, both of which are very important for processing and end-use quality. Some studies showed that the GSC was positively correlated with the increase in grain size and eventually grain yield but was negatively correlated with the GPC, which indicated that a balance must be achieved to obtain the desired protein level and high yield (Muqaddasi et al., 2020). In addition, many key enzymes of starch synthesis, such as granule-bound starch synthase I (GBSS I), soluble starch synthase (SSS), branching enzyme (BE), and debranching enzyme (DBE), were found in wheat. Although some key enzymes were found, the genes determining the inheritance of GSC in bread wheat are still not clear. In addition to the known waxy genes, many other genes/QTLs should be further investigated. A few studies on QTL mapping of GSC have been reported, and the QTLs were mainly distributed on chromosomes 1A, 1D, 2A, 2D, 4A, 7A, 7B, and 7D (Tian et al., 2015). A major QTL for the B-type granule content was detected in wild Aegilops species (Muqaddasi et al., 2020). In addition, a genome-wide association study (GWAS) using the 90 K SNP assay identified 23 loci for percentage volumes of A- and B-granules and 25 loci for the ratio of A-/B-granule volumes, distributing on 15 chromosomes (Li et al., 2017). These findings support the hypothesis that wheat may alter grain starch content by inducing or inhibiting different genes.
The development of wheat grain is a complex process involving cellularization, grain filling, and maturation. Grain filling is a major stage for the synthesis and accumulation of storage molecules, such as starch and gluten proteins (Nadaud et al., 2010). Different gene expression patterns exist for the seed coat, endosperm, and embryo during the grain filling process (Brandizzi, 2021). The discovery of the conditional QTL mapping method laid a foundation for analyzing the dynamics of QTL/gene expression (Zhu, 1995; Bethke et al., 1996). The accumulations of protein and starch showed dynamic and orderly changes over a considerable time during the development of wheat grain. Therefore, previous researchers used this method to study the genetic basis of crops, such as cotton, rice, maize, soybeans, and wheat (Jiang et al., 2008; Wang et al., 2010; Cui et al., 2011; Wang et al., 2012; Zhang et al., 2017). In a few studies, conditional QTLs of GSC and GPC during grain development in DH populations were mapped. QGpc3A, QGpc4A-1, QGpc1D, and QGpc4A-2 were found to play the most important role in the accumulation of GPC, and QGsc4A was found to play an important role in the accumulation of starch (Tian et al., 2015). Two conditional QTLs related to protein content were detected at the grain filling stage. This is a good start for the study of the regulation of protein and starch content during wheat grain development by conditional QTLs (Li et al., 2015).
In addition to QTL mapping, genome-wide association studies (GWAS) based on high-density molecular markers are a powerful tool for determining the genetic anatomy of complex traits. They are an effective method to study the economic and biological values of germplasm resources (Alqudah et al., 2020). GWAS depend on the association between molecular markers and the phenotype of the target trait. The combined analysis of QTL mapping and GWAS can improve the accuracy of QTL/gene detection and has been used in rice (Famoso et al., 2011; Zhai et al., 2018; Pan et al., 2020; Chen et al., 2022) and maize (Wang et al., 2019; Li Z. et al., 2020). However, in wheat, few studies using this combination analysis have been reported (Chen et al., 2016), and the following genes were detected: TaFLL-5B1 for flag leaf length (Yan et al., 2020), qHR-1B for herbicide resistance (Shi et al., 2020), and S2B_26494801 for drought tolerance of winter wheat (Sallam et al., 2022). These results indicate that this combination analysis could be feasible for identifying QTLs/genes for protein, starch, and their components during wheat grain development.
Although genetic mapping of protein content, starch content, and their components in wheat grain has been performed, most studies used mature grain as the study material, not developing grains. Moreover, there are almost no reports on determining the development of grain protein, GMP, GAPC, and GASC using conditional and unconditional mapping by the combined analysis of QTL mapping and GWAS. Therefore, the objective of this study was to identify the important dynamic QTL/gene changes of wheat grain traits at the molecular level during continuous development under two environments at 7, 14, 21, and 28 DAA (days after anthesis) by combining conditional and unconditional QTL mapping and GWAS analysis. The results help explain the developmental genetic mechanism for regulating the contents of protein, starch, and their components and the GMP, which provides new insights into the potential regulatory network between grain quality and yield.
2 Materials and methods
2.1 Plant materials
The data presented in this study were derived from one recombinant inbred line (RIL) population and an association mapping panel comprising 205 wheat genotypes.
The RIL population of 256 lines was developed from a cross between the two winter wheat cultivars Nuomai 1 (female) (NM1) and Gaocheng 8901 (male) (GC8901) by the single seed descent method (Deng et al., 2015). NM1 (Jiangsu Baihuomai/Guandong107) has unique starch properties that are related to high-quality white salt noodles. GC8901 (77546-2/Lin zhang) exhibits high gluten strength and good bread-making qualities. The association mapping panel comprised 77 released cultivars, 55 landraces, and 73 breeding lines. Most of them are from 10 provinces that represent the major winter wheat production regions in China (Chen et al., 2016).
2.2 Growth conditions
The seeds of the RIL population and association mapping panel were planted in the experimental fields at Shandong Agricultural University, Tai’an location (36°57′N, 116°36′E) for two growing seasons, 2016–2017 and 2017–2018. The experiments were laid in a completely randomized block design with two replicates. Under each environment, the RILs, parents, and natural wheat lines were grown in 2-m-long four-row plots spaced 26 cm apart. During the growing season, field management was in accordance with local practices, and the plants were not damaged by disease or insects.
The four sampling stages are indicated by S1, S2, S3, and S4. S1 indicates 7 DAA, S2 indicates 14 DAA, S3 indicates 21 DAA, and S4 indicates 28 DAA.
2.3 Mill flour
The wheat spikes were tagged at the flowering stage and harvested at 7, 14, 21, and 28 DAA. Among them, 32 spikes were harvested at 7 DAA, 24 spikes at 14 DAA, 18 spikes at 21 DAA, and 28 spikes at 28 DAA. The harvested wheat spikes were faded at 105°C for 10 min and then dried at 80°C before threshing. The wheat grains were ground into whole flour by an LM3100 experimental mill and then stored at 4°C for use.
2.4 Quality trait measurements
Whole flour protein content (FPR) was measured by the Kjeldahl method according to GB/T 33862-2017.
Flour starch content of wheat samples was measured by the double-wave method (Chen et al., 2019).
The method from Sun’s study was used to determine the GMP content in grains with minor modifications (Sun et al., 1998). Whole wheat flour (0.1 g) was placed into a 10-ml centrifuge tube, and 5 ml of 1.5% SDS extract was added. The mixture was shaken for 2 h and centrifuged at 15,500 r/min at room temperature for 15 min. The supernatant was discarded, and the residual nitrogen content was determined by the biuret method. Four milliliters of biuret reagent diluted once and 1 ml of distilled water were added to the mixture, and it was shaken for 2 h and then centrifuged. The supernatant was used to measure the light absorption value at 540 nm, and the nitrogen content in the residue was used as the approximation of GMP.
2.5 The genetic linkage map
The linkage map of the RIL population was constructed in a previous study (Deng et al., 2016). A total of 501 markers were mapped, including 479 DArT markers, 17 SSR markers, 2 HMW-GS markers, and 3 Wx protein markers. It included 21 chromosomes comprising 25 linkage maps and covered 4,213.2 cM with an average marker distance of 8.4 cM.
SNP markers, genotyping, and the population structure of the association mapping panel have been previously reported with the high-density illumina wheat 90K SNP array (Chen et al., 2016). In total, 24,355 mapped SNP markers were used for the MTA analysis.
2.6 Statistical analyses
PASW Statistics 18 software was used for statistical analysis of population traits, QTL IciMapping 3.2 software (https://isbreeding.caas.cn/rj/qtllcmapping/294441.htm, released April 2012) was used for linkage analysis of RIL wheat lines, and a mixed linear model (MLM) in TASSEL3.0 software (Zhang et al., 2010) was used for identifying significant marker−trait associations (MTAs). The p-value was used to determine whether an MTA locus was associated with a marker. The R2 value was used to evaluate the magnitude of the MTA effects. The genome-wide significance threshold (p < 0.0001) was determined. When the MTA locus was detected in two or more environments, it was considered a stable association site. QGAStation 1.0 (http://ibi.zju.edu.cn/software/qga/) was used to calculate the conditional phenotypic values, which were subsequently used as input data for conditional QTL mapping.
To clarify the designations of the examined QTLs, the following rules were adopted: “Q” denotes “QTL”; the letter following “Q” is an abbreviation of its corresponding trait; a number followed by an upper case letter, “A,” “B,” or “D,” represents the chromosome in a given wheat genome where the corresponding QTL was detected; if there is more than one QTL on one chromosome, a serial number behind a hyphen is added (e.g., QGApC3B-1 stands for the second QTL for grain amylopectin content detected on chromosome 3B); and the growth stages are added before the chromosome number for conditional QTLs [e.g., QGApC (S4|S3)3B].
2.7 Forecasting candidate genes for major QTL/MTA loci during grain development
To identify the position of important QTLs on a physical map and possible candidate genes, significant markers detected in this study were used to identify putative candidate genes according to the method of Ji et al. (2021). A BLAST (Basic Local Alignment Search Tool) search was performed on the WheatOmics1.0 database (Ma et al., 2021) by Chinese Spring IWGSC RefSeq v1.1 genome assembly (http://wheatomics.sdau.edu.cn/blast/blast.html) using the sequence of the significant DArT markers and SNP markers identified by QTL mapping and GWAS. When a DArT marker or SNP marker sequence from the IWGSC was 100% identical to any wheat contig, the sequence was extended 2 Mb using the IWGSC BLAST results (Ji et al., 2021). Then, the extended sequence was used to run a BLAST search of the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov) and Ensembl Plants (http://plants.ensembl.org/Triticum_aestivum/Tools/Blast) to confirm possible candidate genes and functions.
3 Results
3.1 Phenotypic data and correlations
During grain development, the dynamic changes of GPC and GMP content in both the RIL population and the diversity panel showed basically consistent trends of “high–low–high” in the two environments (Tables S1, S2). However, the dynamic changes in amylopectin content (GApC) and amylose content (GAsC) in the RIL population and natural population were greatly affected by the environment and showed a gradual upwards trend (Tables S1, S2). All of the evaluated traits in the RIL population exhibited approximately continuous variation in different developmental stages in the two environments (Figures S1–S4), and transgressive segregation was observed for all the evaluated traits in these populations, indicating that alleles with positive effects were contributed by both parents.
By correlation analysis, a significantly positive correlation was observed between GPC and GMP content, but GPC was significantly negatively correlated with GApC and GAsC (Table S3). There was a significantly negative correlation between GMP content and GApC, but a significant positive correlation was found between GMP content and GAsC.
3.2 QTL mapping of four evaluated traits during grain development
A total of five unconditional QTLs for GPC were identified on chromosomes 1B, 2A, 3B, 3D, and 7A (Table 1). There were two major QTLs, QGPC3B and QGPC2A, with flanking markers of wPt-5870–wPt-3620 and wPt-2994–wPt-0071, accounting for 26.97% and 12.76% of the phenotypic variation explained (PVE), respectively. They were detected at 21 DAA and 28 DAA, respectively. The additive effects of the two loci were from the male parent Gaocheng 8901.
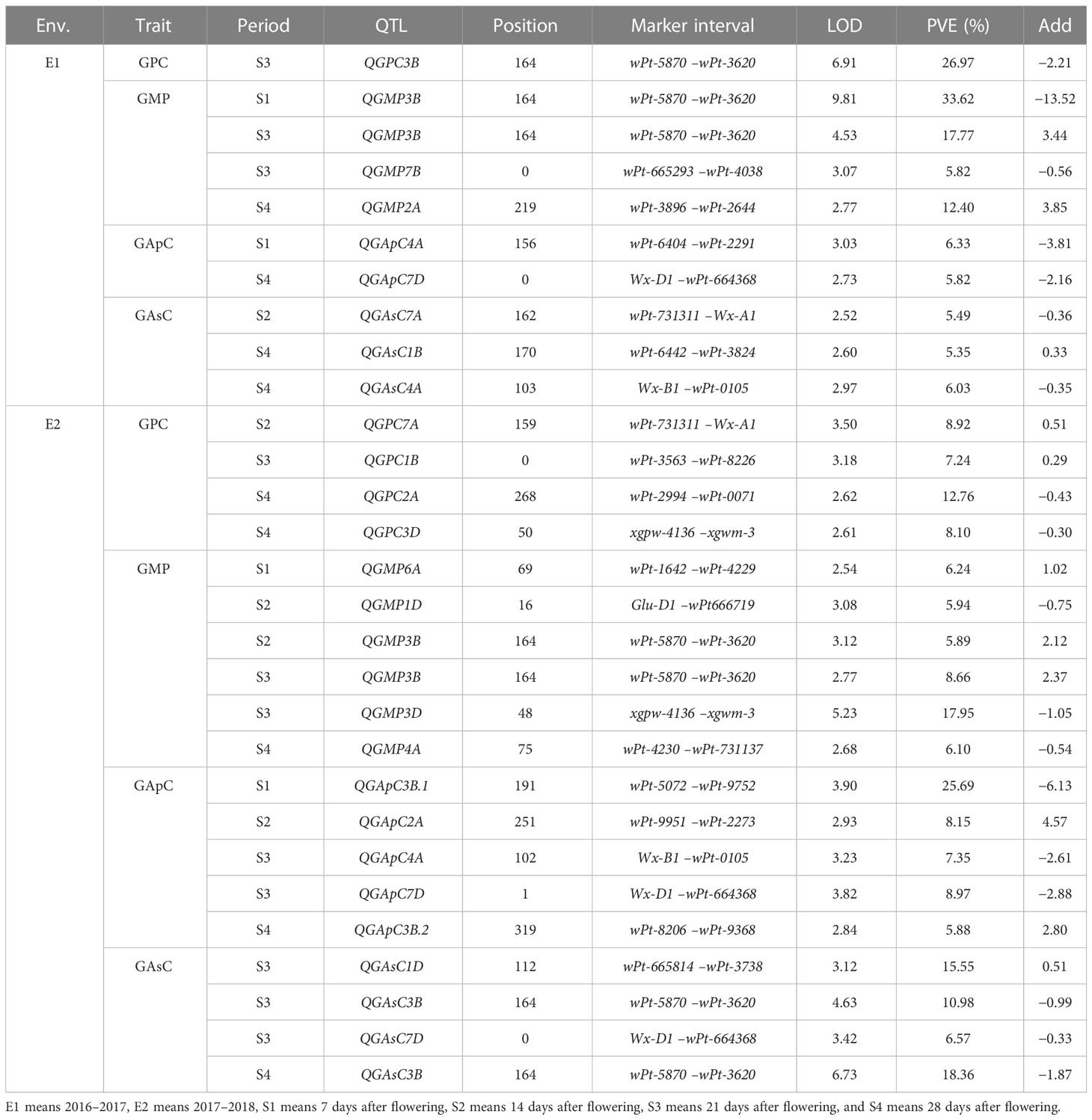
Table 1 Unconditional QTL analysis of the content of protein, GMP, amylopectin, and amylose during grain development in the RIL population.
There were 12 QTLs found for GMP content on chromosomes 1D, 2A, 3B, 4A, 6A, and 7B (Table 1). The major QTL, QGMP3B, was stably detected at three stages over 2 years, which indicated that this QTL was important for the development of GMP. The position of this locus was the same as that of QGPC3B, which indicated that this locus is important for protein synthesis.
Seven QTLs were identified for GApC on chromosomes 2A, 3B, 4A, and 7D (Table 1). Of these, QGApC7D was found with flanking markers between Wx-D1 and wPt-664368 at 28 DAA and 21 DAA in E1 and E2, respectively. Although there were two QTLs identified on chromosome 4A, they were not mapped to the same marker intervals. QGApC4A.2 was detected between markers Wx-B1 and wPt-0105 at 21 DAA in E2. On chromosome 3B, one major QTL, QGApC3B.1, was identified with 25.69% PVE.
A total of eight unconditional additive QTLs for GAsC were detected on chromosomes 1B, 1D, 3B, 4A, 7A, and 7D (Table 1; Figure 1). A major QTL, QGAsC3B, was found at 21 DAA and 28 DAA in the E2 environment, with flanking markers between wPt-5870 and wPt-3620, which indicated that this QTL was important for GAsC.
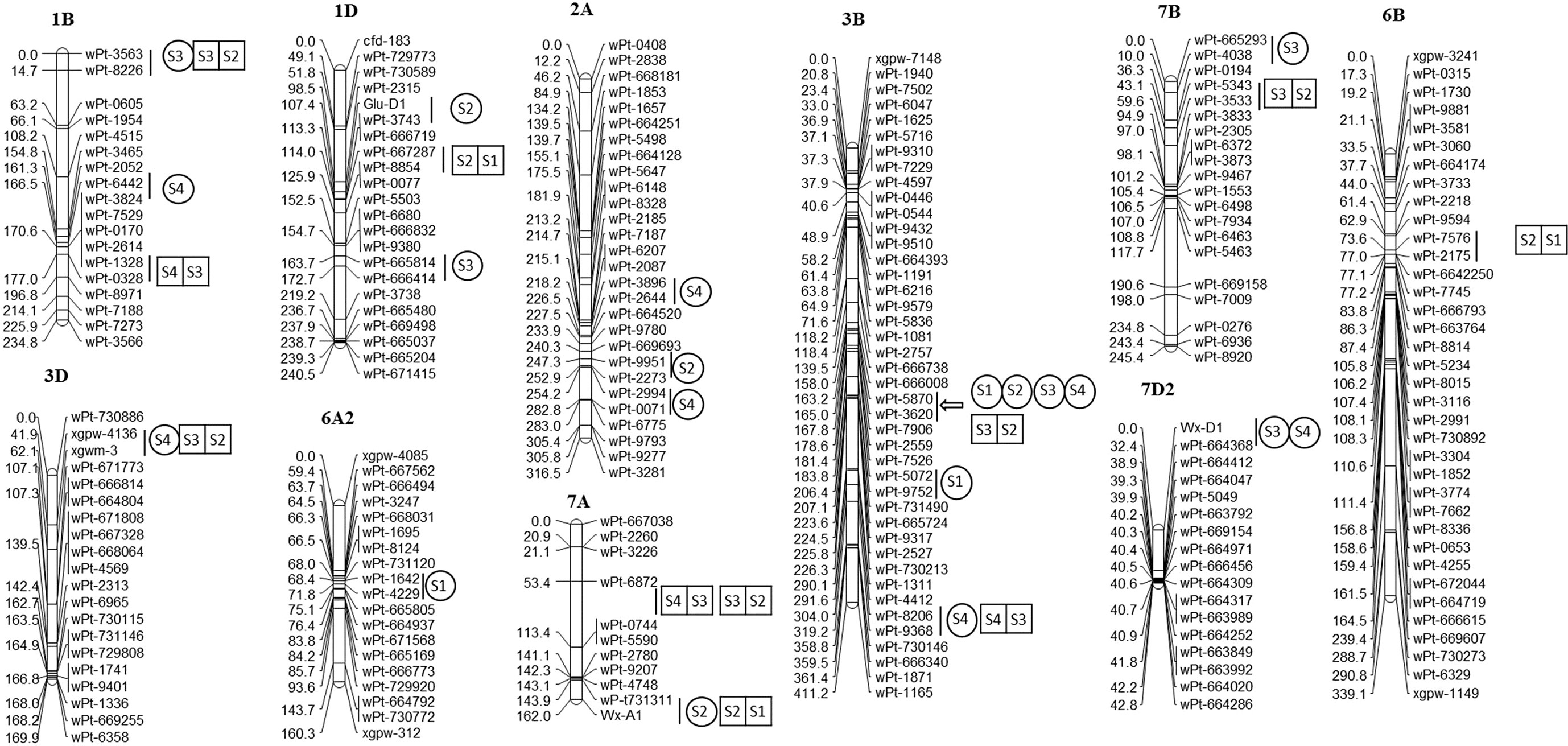
Figure 1 QTLs for four quality traits in the RIL population at four stages in 2 years.  represent unconditional QTLs at four periods, respectively;
represent unconditional QTLs at four periods, respectively;  represent conditional QTLs at four periods, respectively.
represent conditional QTLs at four periods, respectively.
A total of 13 conditional QTLs were identified in RIL wheat lines at four periods in 2 years (Table 2). Four QTLs for GPC were detected on chromosomes 1B, 3B, 7A, and 7B in 2 years. There were two major QTLs, QGPC(S3|S2)3B and QGPC(S3|S2)7A, with 29.17% and 14.42% PVE, respectively. Four conditional additive QTLs for GMP content were found on chromosomes 3B, 3D, and 7A, including three major QTLs, QGMP(S3|S2)3B, QGMP(S3|S2)3D, and QGMP(S4|S3)7A. For GApC, only one major QTL, QGApC (S4|S3)3B, was identified in E2, with 14.30% PVE. Four conditional additive QTLs for GAsC were found on chromosomes 1B, 1D, 6B, and 7A. For GAsC, there was one major QTL, QGAsC(S4|S3)7A, with a PVE of 21.86%.
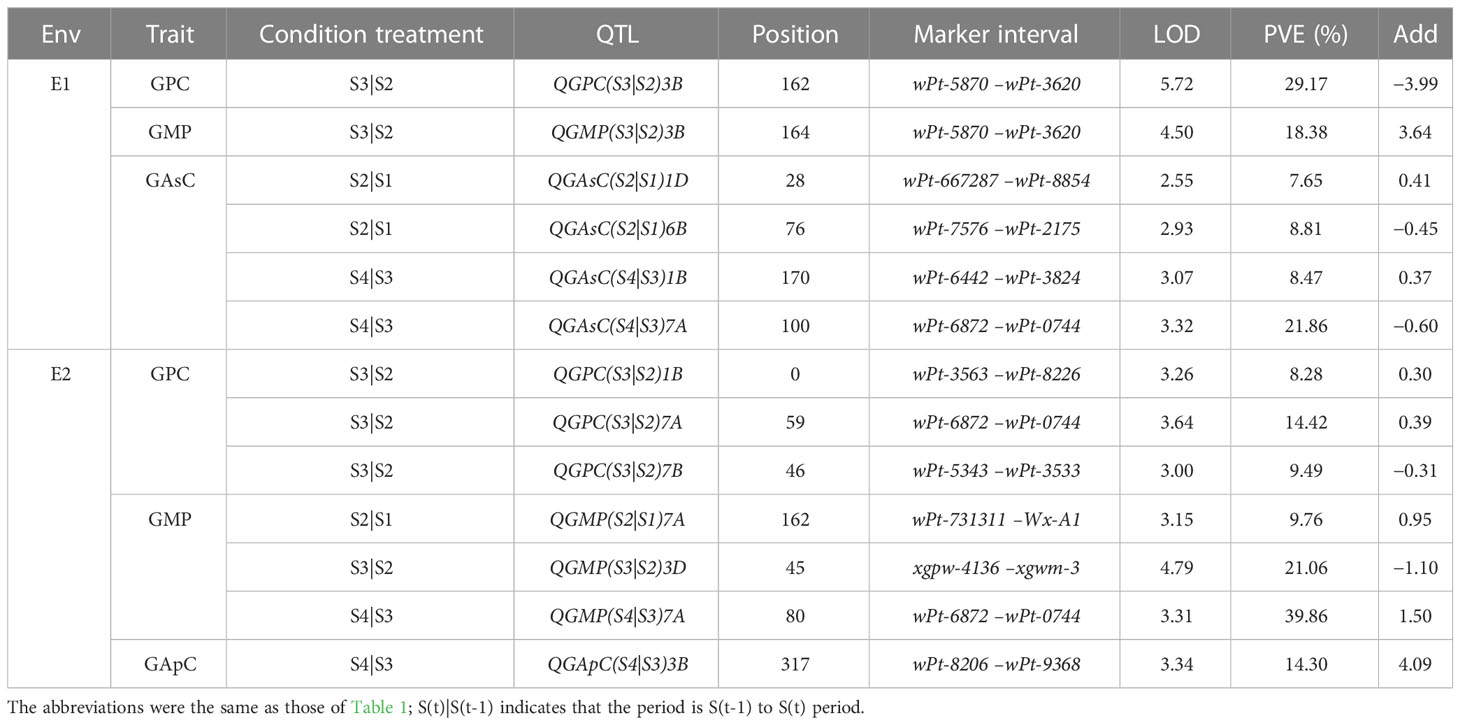
Table 2 Conditional QTL analysis of the content of protein, GMP, amylopectin, and amylose during grain development in the RIL population.
3.3 Comparative analysis of conditional and unconditional QTLs
During grain development, protein synthesis is affected by the cumulative effects of different QTLs/genes. No QTL was detected at all four stages (Table S5). From 14 DAA to 21 DAA, 10 QTLs explaining 7.24% to 29.71% of the phenotypic variation had high expression effects, which indicated that genes related to grain protein synthesis are actively expressed. At 21 DAA, two unconditional and conditional QTLs [QGPC3B, QGPC1B, QGPC(S3|S2)7A, and QGPC(S3|S2)7B] were significant for protein synthesis. This indicated that the two unconditional QTLs had cumulative genetic effects, while the two conditional QTLs had net genetic effects from 14 DAA to 21 DAA. Two unconditional QTLs, QGPC2A and QGPC3D, found only at 28 DAA, were not detected in the conditional QTL analysis, which indicated that their cumulative effects were important for protein synthesis during grain development. For GMP synthesis, there were 14 QTLs with high expression effects from 14 DAA to 21 DAA, explaining 5.82% to 39.86% of the variation. Of these, the loci with flanking markers between wPt-5870 and wPt-3620 on chromosome 3B included unconditional (QGMP3B) and conditional QTLs (QGMP(S3|S2)3B), which indicated that this region is important for GMP synthesis with a cumulative genetic effect, while QGMP(S3|S2)3D showed a net genetic effect. QGMP(S2|S1)7A and QGMP(S4|S3)7A also had net genetic effects from 7 DAA to 14 DAA and from 21 DAA to 28 DAA, respectively.
There were two stages with active expression of genes related to the synthesis of amylopectin from 7 DAA to 14 DAA and from 21 DAA to 28 DAA (Table S5). Seven unconditional QTLs had cumulative genetic effects, but one conditional QTL, QGApC(S4|S3)3B, showed a net genetic effect during GApC synthesis. The two active expression periods for GAsC are the same as those of amylopectin, but 11 QTLs were involved. From 7 DAA to 14 DAA, the unconditional QTL QGAsC7A showed a cumulative genetic effect, while QGAsC(S2|S1)1D and QGAsC(S2|S1)6B had a net genetic effect on GAsC synthesis. From 14 DAA to 21 DAA, there were three unconditional QTLs, QGAsC1D, QGAsC3B, and QGAsC7D, with cumulative genetic effects on GAsC development. However, from 21 DAA to 28 DAA, in addition to the cumulative genetic effects of QGAsC1B, QGAsC3B and QGAsC4A, net genetic effects existed for QGAsC(S4|S3)1B and QGAsC(S4|S3)7A. Therefore, we know that the expression of genes controlling grain protein and starch contents has obvious temporal and spatial characteristics.
3.4 Marker−trait associations of four evaluated traits during grain development
The MTAs at p < 10−4 were the focus of this study, so these loci are highlighted below. Thirty-one MTAs significantly associated with GPC (p < 10−4) (Table 3; Figures S5, S9) were identified; they explained 7.62% to 12.97% of the variation and were distributed on chromosomes 3A, 3B, 4A, 4B, 6A, 6B, 7A, and 7B. The SNP locus Excalibur_c46399_307 on chromosome 6B was detected in two or more environments. Meanwhile, the MTA locus RAC875_c7060_67 on chromosome 3A had the maximum PVE at 7 DAA in E2 (Figure S9).
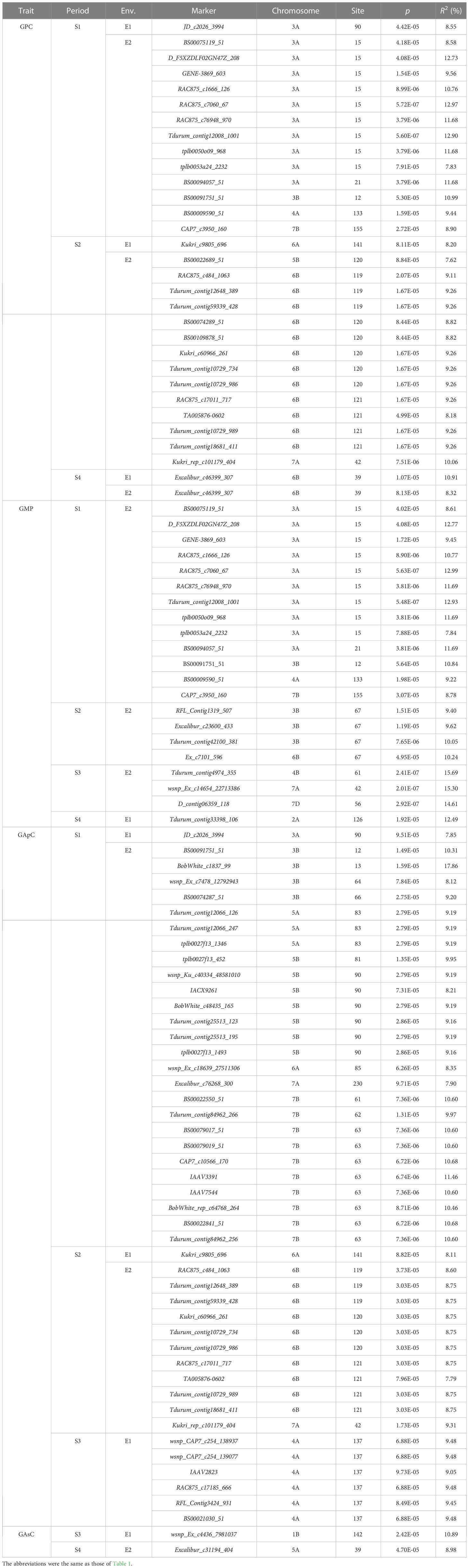
Table 3 Unconditional GWAS of GPC, GMP, GApC, and GAsC during grain development in the nature population (p < 10-4).
Twenty-one MTAs were significantly associated with GMP content (p < 10−4) (Figures S6, S10) on chromosomes 1A, 3A, 3B, 4A, 4B, 6B, 7A, 7B, and 7D, with 7.84% to 15.69% PVE. Of these, the MTA locus Tdurum_contig4974_355 on chromosome 4B had the highest PVE (15.69%) at 21 DAA in E2 (Figure S10).
Forty-five MTAs for GApC (p < 10−4) (Figures S7, S11) were found on chromosomes 3A, 3B, 4A, 5A, 5B, 6A, 6B, 7A, and 7B with 7.35% to 17.86% PVE. Of these, the MTA locus BobWhite_c1837_99 on chromosome 3B had the highest PVE at 7 DAA in E2 (Figure S11).
For GAsC, there were two MTAs (p < 10−4) (Figures S8, S12) identified on chromosomes 1B and 5A. Of these, the MTA locus wsnp_Ex_c4436_7981037 on chromosome 1B had a highest PVE of 10.89% at 21 DAA in E1.
By conditional analysis, at p < 10−4, a total of 14 MTAs were detected (Table 4). Of these, the MTA locus TA005876-0602 for GPC on chromosome 6B was found at four stages in E2 (Figure S13). The MTA locus Tdurum_contig76213_1156 for GApC on chromosome 4B was identified at four stages in E1 (Figure S14), while the MTA locus wsnp_Ex_c18639_27511306 on chromosome 6A was found at 14 DAA to 28 DAA in E2 (Figure S15). However, no MTAs at p < 10−4 were found under conditional analysis for GMP content and GAsC.
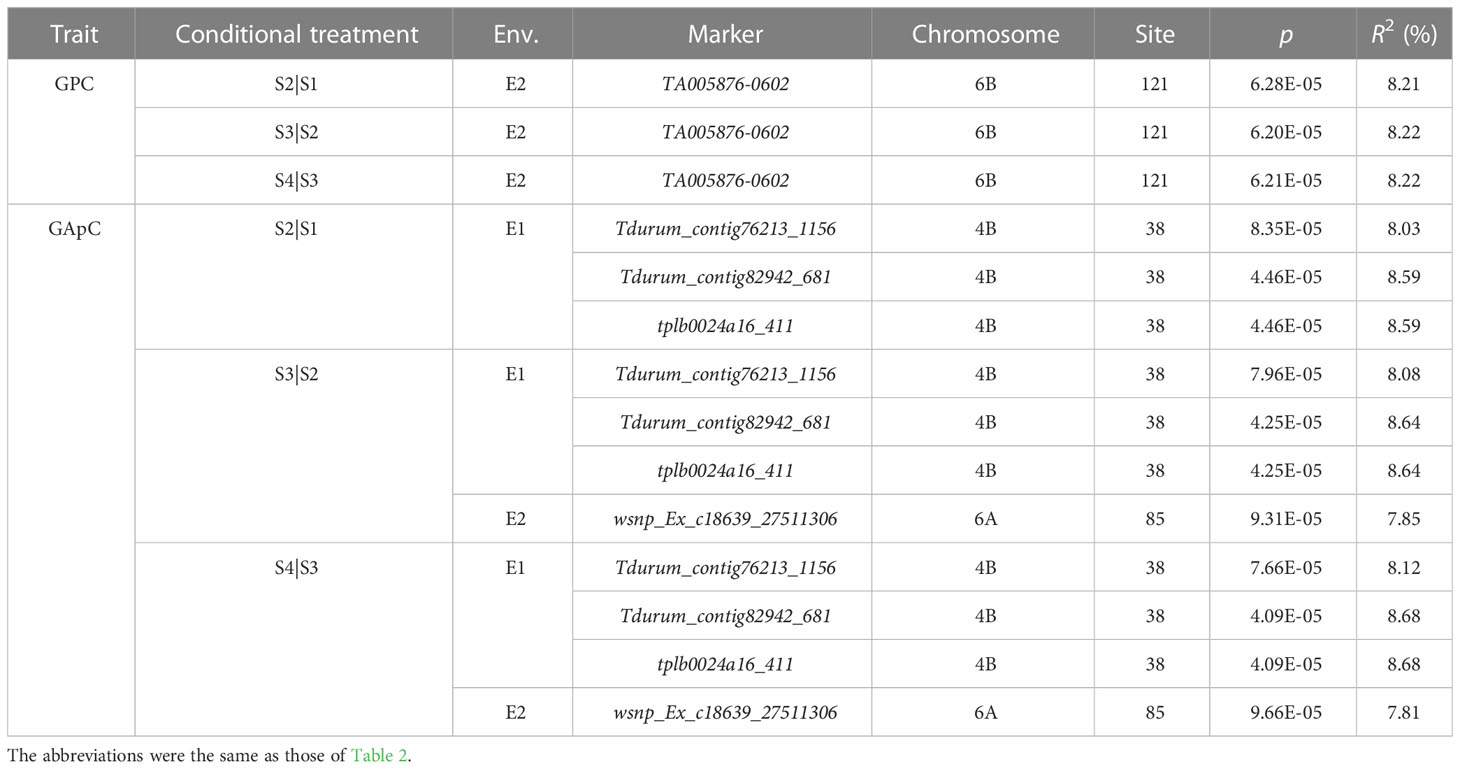
Table 4 Conditional GWAS of GPC, GMP, GApC, and GAsC during grain development in the nature population (p < 10-4).
3.5 Candidate genes predicted for four evaluated traits during grain development
Markers with high PVE values were selected from loci significantly associated with GMP content, GPC, amylose content, and amylopectin content. Twenty-eight and sixty-nine candidate genes were predicted in Triticum aestivum L. (Table S4) using QTL mapping and GWAS, respectively. Most of these genes have multiple effects during grain development. Two candidate genes (TraesCS3B02G159100 and TraesCS3B02G334400) (Li S. et al., 2020) for the loci linked with the marker wPt-5870 on chromosome 3B were found to be related to GMP content and GPC, and their functions are related to homeobox-leucine zipper protein 32 (HOX32) activity, DNA-binding transcription factor activity, ATP binding, and protein kinase activity, participating in the biological processes of protein phosphorylation and transcription regulation in T. aestivum L. Using the WheatOmics 1.0 (Ma et al., 2021) gene expression level database to screen these two genes (Table S7), we found that the expression levels of them were higher at 20 days after flowering, which is the same in our results (S3 development stage). For the conditional loci linked with the marker wPt-0744 related to GMP content, GPC, and GAsC, 13 candidate genes were predicted, but no genes were found on chromosome 7A. Two predictive genes, TraesCS6A02G298100 (Andleeb et al., 2023; Teng et al., 2022) and TraesCS5A02G326100, encode glutamine synthetase and cysteine protease, respectively, and influence protein synthesis during grain development. The functions of the residue candidate genes are mainly related to ATP binding, protein serine/threonine kinase activity, dipeptide or tripeptide transmembrane transporter activity, histone deacetylase activity, NAD+ binding, and transferase activity. These two genes have higher expression levels in S3 and S2 stages, respectively, by using the WheatOmics 1.0 gene expression level database analysis. The candidate gene TraesCS2A02G012600 is involved in carbohydrate metabolic processes. It had the highest expression level at 20 days after flowering (S3 development stage) using the WheatOmics 1.0 (Ma et al., 2021) gene expression level database. There are three candidate genes (TraesCS7A02G070100, TraesCS4A02G418200, and TraesCS7D02G064300) predicted for Wx-A1 on chromosomes 7A, 4A, and 7D (Liu et al., 2020; Tian et al., 2022; Kaushik et al., 2020; Guo et al., 2020) related to amylose and GPC, encoding starch synthase, chloroplastic/amyloplastic starch synthase, and granule-bound starch synthase. The expression levels of these three genes were higher at 10 days after flowering (S2 development stage) by using the WheatOmics 1.0 gene expression level database. Their functions are glycan biosynthesis, starch biosynthesis, and glycogen (starch) synthase activity, which confirms the accuracy of the results. In addition, one candidate gene was predicted for GMP content that is involved in cyclic pyranopterin monophosphate synthase activity. 999[JFATwo candidate genes (TraesCS2A02G238400 and TraesCS2A02G537100) (Grzesiak et al., 2019) for GPC are related to zeta-carotene desaturase and superoxide dismutase. The expression levels of these two genes were higher at 10 and 20 days after flowering (S2 development stage), respectively. Five candidate genes were predicted for GApC. Among the 69 candidate genes associated with important SNP sites, there were 39 candidate genes related to GMP content and GPC, mainly involving 11 chromosomes, especially 22 genes on 3A and 3B. Most of their functions are related to acid–amino acid ligase activity, myosin phosphatase activity, protein serine/threonine phosphatase activity, proton transmembrane transporter activity, GTPase activator activity, amino acid binding, aspartate carbamoyl transferase activity, phosphatidylinositol binding, and ubiquitin binding, participating in protein synthesis in wheat.
The SNP marker BS00091751_51 was associated with GApC, GMP content, and GPC during grain development. Eight candidate genes were predicted on chromosomes 3A, 3B, and 1B. The two genes TraesCS3A02G320800 (High expression level during S3 period) and TraesCS3A02G112600 (High expression level during S4 period), encode glucose-induced degradation protein 8 homologue and UBC core domain-containing protein, respectively. The candidate gene TraesCS1B02G368500 (High expression level during S4 period) has glycogen (starch) synthase activity and is involved in starch biosynthetic processes. Six candidate genes for SNP marker Kukri_rep_c101179_404 were also predicted to be associated with GApC, GMP content, and GPC. Of these, the candidate gene TraesCS3B02G345000 (High expression level around the S2 period) is related to BS00091751_51, not Kukri_rep_c101179_404. Although no genes were found on chromosome 7A, two genes (High expression level around the S3 period) with unknown functions (TraesCS7B02G305600 and TraesCS7B02G305500) were screened on chromosome 7B. The candidate gene TraesCS3B02G021900 is involved in glycosyltransferase activity and pentosyltransferase activity.
Five candidate genes for the Excalibur_c46399_307 marker related to GPC were found, and their functions were related to DNA binding, GTP binding, and GTPase activity; they encoded NAC transcription factors and ADP-ribosylation factors. There were 17 candidate genes predicted for GMP content mainly on chromosomes 3B, 2D, and 2A associated with SNP markers Tdurum_contig42100_381, Tdurum_contig4974_355, and wsnp_Ex_c14654_22713386. The gene TraesCS2D02G261300 (High expression level during the S1 or S2 period) has glutaminyl-tRNA synthase (glutamine-hydrolysing) activity, encoding glutamyl-tRNA (Gln) amidotransferase subunit B. The gene TraesCS3B02G254200 (High expression level during the S1 or S2 period) has glycosyltransferase activity, while the genes TraesCS2D02G308600 (Guo et al., 2020) (High expression level during the S1 or S2 period) and TraesCS1B02G368500 (High expression level during S4 period) are involved in starch biosynthesis. We found that some important genes on chromosome 3B were related to protein and starch synthesis during grain development using two mapping methods. These candidate genes may be important for GMP content, GPC, amylose content, and amylopectin content during grain development, and their functions will be further identified in future research.
4 Discussion
4.1 Accumulation changes of four quality characters during grain development
Previous studies showed that the accumulation changes in GPC presented a trend of “high–low–high” during grain filling (Zhang and Xu, 1995). In this study, the dynamic change in protein content also showed a “high–low–high” trend during the grain development of wheat, which was less affected by the environment and genetic background. The decrease in the total crude protein content observed during grain development might be due to extensive non- water-soluble carbohydrate accumulation, mainly starch. This decrease was followed by a vast accumulation of crude protein, which was mentioned before in other studies during the grain filling and maturation phases, which might result from the final desiccation prior to maturation (Panozzo et al., 2001; Verspreet et al., 2013). A study showed that GMP content changed from 12 DAA to the mature stage and that the accumulation amount remained relatively stable at a low level before 20 DAA, and then gradually increased from 20 DAA to the mature stage, during grain filling (Huang et al., 2007). In this study, the dynamic change in GMP content showed a trend of “high–low–high” from 7 DAA to the mature stage. This was similar to the accumulation trend of GPC. However, there have been few studies on the accumulation changes in GApC and GAsC during wheat grain development, and most of them focused on the key enzyme activity of starch synthesis in grain development. The activities of soluble starch synthase and starch granule-bound starch synthetase both first increased and then decreased during development (Wang et al., 2007), but they reached maximum activity at different times. This was similar to the accumulation changes in amylose content and amylopectin content. These results provide a reference for the study of protein content in wheat at certain developmental stages and is an important prerequisite for the study of how amylase affects starch content in wheat grains.
4.2 Major chromosomes for grain protein, GMP, amylose, and amylopectin synthesis during grain development
Previous studies have shown that most chromosomes are related to GPC. Using the RIL population, QTLs were identified on chromosomes 2A, 2B, 2D, 3D, 4A, 6B, 7A, and 7D (Prasad et al., 2003), and chromosomes 6B and 7A had major QTLs with flanking markers Xgwm133, Xgwm889, and Xgwm1171. Meanwhile, some important QTLs for GPC were detected on chromosomes 3B, 6B, and 7B (Chee et al., 2001; Sun et al., 2007; Bordes et al., 2011). In addition, previous researchers also found QTLs for GMP content on chromosomes 3B, 5B, 6B, and 7B (Li et al., 2007; Sun et al., 2007). In this study, both QTLs and MTAs for GPC were identified on chromosomes 3B, 7A, and 7B from 14 DAA to 21 DAA using QTL mapping and GWAS. The loci on chromosomes 3B, 7A, and 7B were closely linked to the markers wPt-5870–wPt-3620 and BS00091751_51, Wx-A1 and Kukri_rep_c101179_404, and wPt-5343 and CAP7_c3950_160, respectively. On chromosome 6B, there was an important SNP marker, TA005876-0602, linked to GPC at four stages. However, for GMP content during grain development, loci on chromosomes 2A, 3B, 4A, 7A, and 7B were found using both QTL mapping and GWAS. Some important linked markers were identified, such as wPt-3896 and Tdurum_contig33398_10 on chromosome 2A, wPt-3620 and BS00091751_51 on chromosome 3B, wPt-731137 and BS00009590_51 on chromosome 4A, Wx-A1 and wsnp_Ex_c14654_22713386 on chromosome 7A, and wPt-3533 and CAP7_c3950_160 on chromosome 7B. By comparison with previous studies, chromosomes 3B and 7B played important roles in protein and GMP synthesis during grain development. There was one important new locus on chromosome 3B involved in both protein and GMP synthesis.
Starch content affects not only wheat yield but also quality, which is influenced by amylose and amylopectin synthesis. Previous studies detected more than 20 QTLs for starch traits, most of which were found on chromosomes 3D, 6B, and 7B (Sun et al., 2007). The QTLs for GApC were identified on chromosomes 1B, 3A, 3B, and 5D (Tian et al., 2011). Although amylose synthesis is mainly affected by waxy genes, other chromosomes may also affect amylose content because it is a quantitative trait. There was one additive QTL found on chromosome 6B with flanking markers between Cau170 and xgwm234 (Wang et al., 2007). In the present study, there were 11 (2A, 3A, 3B, 4A, 5A, 5B, 6A, 6B, 7A, and 7B) and eight chromosomes (1B, 1D, 3B, 4A, 5A, 6B, 7A, and 7D) involved in amylopectin and amylose synthesis during grain development. For amylopectin content, the loci on chromosomes 3B and 4A were detected by both QTL mapping and GWAS, while for amylose, only the loci on chromosome 1B were detected by both methods. Most interestingly, the loci were identified on chromosomes 4A, 7A, and 7D close to three waxy genes. Of these, the loci on chromosome 4A were involved in not only amylose synthesis but also amylopectin synthesis during grain development. These results indicated that the results were accurate and reliable. However, by comparing the physical location of the important loci between QTL mapping GWAS, we unfortunately found that there was no loci on the same interval physical positions (Table S6).
4.3 The role of chromosome 3B in protein and starch synthesis during grain development
All four chromosomes (3B, 4A, 6B, and 7A) were simultaneously associated with protein, GMP, amylopectin, and amylose synthesis during grain development. In particular, the region of chromosome 3B with flanking markers between wPt-5870 and wPt-3620 was shown to be important for protein and starch synthesis. This chromosome region was involved in protein synthesis from 14 DAA to 21 DAA, GMP synthesis from 0 to 7 DAA, and amylose synthesis from 14 DAA to 21 DAA. In addition, from 0 to 7 DAA and from 21 DAA to 28 DAA, the other chromosome region of 3B was involved in amylopectin synthesis. Therefore, from 0 to 7 DAA, chromosome 3B was mainly involved in the synthesis of GMP and amylopectin; from 14 DAA to 21 DAA, it was mainly involved in protein and GMP synthesis; and from 21 DAA to 28 DAA, it was mainly involved in amylose and amylopectin synthesis.
4.4 Candidate genes during protein and starch accumulation in wheat grain filling
Previous studies have shown that protein accumulation during grain development is affected by regulation at the transcriptional level and post-transcriptional level, which involves some transcription factors (TFs) regulating prolamin genes, such as MYB TFs, AP2, and NAC TFs, and some important protein modifications, such as N-glycosylation, folding, and assembly of proteins, and protein bodies (Diaz et al., 2002; Rubio-Somoza et al., 2006; Zhang et al., 2019), while starch synthesis in wheat is affected by a series of enzymes, including ADP-glucose pyrophosphorylase, sucrose synthase, granule bound starch synthase, starch synthase, starch branching enzyme, and starch debranching enzyme (Kumar et al., 2018). Meanwhile, the interactions between opaque-2 and prolamin-box binding factor can regulate the gene networks for starch and protein biosynthesis in maize (Zhang et al., 2017). In rice, there is also a similar interaction between bZIP and DNA binding with one finger TFs (Kim et al., 2017). However, one storage protein activator, TaSPA-B, plays an important role in starch and protein biosynthesis by regulating a complex gene network in wheat (Guo et al., 2020). In our study, we also found some candidate genes related to not only protein synthesis but also starch synthesis during grain development. For example, TraesCS6A02G298100 (Andleeb et al., 2023) and TraesCS5A02G326100 function in ATP binding, glutamate-ammonia ligase activity, and cysteine-type peptidase activity, encoding glutamine synthetase and cysteine protease, respectively, which have been shown to play important roles in protein synthesis in previous studies (Gadaleta et al., 2011). Meanwhile, some identified genes have functions of glycosyltransferase activity, GTPase activator activity, 1,4-alpha-glucan branching enzyme activity, GTPase activity, etc., which are all involved in starch synthesis. Interestingly, we also identified genes that encode NAC transcription factors, TraesCS6B02G075200 (Saini et al., 2021), TraesCS5D02G059700 (Grzesiak et al., 2019), TraesCS5B02G054200 (Vranic et al., 2022), and TraesCS5A02G049100. The same two genes shown to be involved in starch synthesis in our study, TraesCS7D02G117800 (SSI) (Tian et al., 2022) and TraesCS2D02G308600 (GBE1, SBE), were also reported in Guo’s study (Guo et al., 2020). In addition, the gene network was constructed based on these predicted QTL/genes to verify the importance and connection among predicted genes (Table S7). We found that there were four clusters for predicted genes, which were related to grain hardness, grain length, grain size, grain weight, and starch grain. These indicated that they participated in the synthesis of protein and starch (Figure S16). However, a total of 67 candidate genes have been screened, and their multiple mechanisms of protein and starch synthesis during wheat grain development need to be studied further in the future.
5 Conclusion
We detected 10 unconditional major QTLs, 7 conditional major QTLs, and 36 unconditional major MTAs for four evaluated traits during grain development. Pleiotropic QTLs/MTAs have been identified. Four chromosomes were mainly involved in the development of protein, GMP, amylopectin, and amylose synthesis. Twenty-eight and 69 important genes for major loci were predicted, respectively, and most of them have multiple effects on protein and starch synthesis during grain development. Overall, our study provides novel insights that increase our understanding of the genetic information of protein and starch synthesis during grain development, and will help improve our understanding of the regulatory network between grain protein and starch in wheat breeding programs.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
ZD designed and revised this paper; JT constructed the populations; YG, GW, and DW analyzed the data and wrote the manuscript; XG, SC, and HY investigated and analyzed the phenotypic data; KJ, HH, and CW analyzed and screened the candidate genes; JC, YB, and WZ reviewed this paper; all authors have read and approved this manuscript.
Funding
This work was supported by the Key National Agricultural Science and Technology Project, the Agricultural Improved Variety Project of Shandong Province (No. 2022LZGCQY002), the Agricultural Improved Variety Project of Tai’an City (2022NYLZ06), the Natural Science Foundation of China (No. 31871613), Shandong Province Postgraduate Education Tutor Capacity Improvement Program (No. SDYY18114), and the project NK20220604.
Acknowledgments
We acknowledge Dr. Yongbin Zhuang for providing the gene network of predicted genes.
Conflict of interest
Author JT is employed by Shandong Huatian Agricultural Technology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1189887/full#supplementary-material
References
Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., Tasaka, M. (1997). Genes involved in organ separation in arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9 (6), 841–857. doi: 10.1105/tpc.9.6.841
Alqudah, A. M., Sallam, A., Baenziger, P. S., Börner, A. (2020). GWAS: fast-forwarding gene identification and characterization in temperate cereals: lessons from barley – a review. J. Advanced Res. 22 (C), 119–135. doi: 10.1016/j.jare.2019.10.013
Andleeb, T., Knight, E., Borell, P. (2023). Wheat NAM genes regulate the majority of early monocarpic senescence transcriptional changes including nitrogen remobilisation genes. G3 (Bethesda, Md.) 13 (2), jkac275. doi: 10.1093/G3JOURNAL/JKAC275
Anjum, F. M., Khan, M. R., Din, A., Saeed, M., Pasha, I., Arshad, M. U. (2007). Wheat gluten: high molecular weight glutenin subunits–structure, genetics, and relation to dough elasticity. J. Food Sci. 72 (3), R56–R63. doi: 10.1111/j.1750-3841.2007.00292.x
Bethke, P. C., Hillmer, S., Jones, R. L. (1996). Isolation of intact protein storage vacuoles from barley aleurone (Identification of aspartic and cysteine proteases). Plant Physiol. 110 (2), 521–529. doi: 10.1104/pp.110.2.521
Bordes, J., Ravel, C., Gouis, J. L., Lapierre, A., Charmet, G., Balfourier, F. (2011). Use of a global wheat core collection for association analysis of flour and dough quality traits. J. Cereal Sci. 54 (1), 137–147. doi: 10.1016/j.jcs.2011.03.004
Brandizzi, F. (2021). Adding to the understanding of grain filling processes through multiomics: high-throughput proteome and metabolome come into play. Plant J. 107 (3), 667–668. doi: 10.1111/tpj.15422
Chee, P. W., Elias, E. M., Anderson, J. A., Kianian, S. F. (2001). Evaluation of a high grain protein QTL from triticum turgidum l. var. dicoccoides in an adapted durum wheat background. Crop Sci. 41 (2), 295–301. doi: 10.2135/cropsci2001.412295x
Chen, X., Fang, W., Ji, M., Xu, S., Jiang, Y., Song, S., et al. (2019). Genome-wide association study of total starch and its components in common wheat. Euphytica 215 (12), 201. doi: 10.1007/s10681-019-2517-z
Chen, H., Zhai, L., Chen, K., Shen, C., Zhu, S., Qu, P., et al. (2022). Genetic background- and environment-independent QTL and candidate gene identification of appearance quality in three MAGIC populations of rice. Front. Plant Sci. 13. doi: 10.3389/FPLS.2022.1074106
Chen, G., Zhang, H., Deng, Z., Wu, R., Li, D., Wang, M., et al. (2016). Genome-wide association study for kernel weight-related traits using SNPs in a Chinese winter wheat population. Euphytica 212 (2), 173–185. doi: 10.1007/s10681-016-1750-y
Cui, F., Li, J., Ding, A., Zhao, C., Wang, L., Wang, X., et al. (2011). Conditional QTL mapping for plant height with respect to the length of the spike and internode in two mapping populations of wheat. Theor. Appl. Genet. 122 (8), 1517–1536. doi: 10.1007/s00122-011-1551-6
Deng, Z., Hu, S., Chen, F., Li, W., Chen, J., Sun, C., et al. (2015). Genetic dissection of interaction between wheat protein and starch using three mapping populations. Mol. Breed. 35 (1), 1–9. doi: 10.1007/s11032-015-0216-6
Deng, Z. Y., Li, W. J., Chen, F., Fang, W. Q., Chen, G. F., Sun, C. L., et al. (2016). Genetic dissection of flour whiteness by unconditional and conditional quantitative trait locus mapping in wheat. J. Agric. Sci. 155 (4), 544–555. doi: 10.1017/S0021859616000563
Diaz, I., Vicente-Carbajosa, J., Abraham, Z., Martínez, M., Isabel-La, M. I., Carbonero, P. (2002). The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J. 29 (4), 453–464. doi: 10.1007/0-306-46902-2_11
Famoso, A. N., Zhao, K., Clark, R. T., Tung, C.-W., Wright, M. H., Bustamante, C., et al. (2011). Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PloS Genet. 7 (8), 747–757. doi: 10.1371/journal.pgen.1002221
Gadaleta, A., Nigro, D., Giancaspro, A., Blanco, A. (2011). The glutamine synthetase (GS2) genes in relation to grain protein content of durum wheat. Funct. Integr. Genomics 11 (4), 665–670. doi: 10.1007/s10142-011-0235-2
Gao, Y., An, K., Guo, W., Chen, Y., Zhang, R., Zhang, X., et al. (2021). The endosperm-specific transcription factor TaNAC019 regulates glutenin and starch accumulation and its elite allele improves wheat grain quality. Plant Cell 33 (3), 603–622. doi: 10.1093/plcell/koaa040
Grzesiak, M. T., Hordyńska, N., Maksymowicz, A., Grzesiak, S., Szechyńska-Hebda, M. (2019). Variation among spring wheat (Triticum aestivum l.) genotypes in response to the drought stress. II-root system structure. Plants 8 (12), 584. doi: 10.3390/plants8120584
Guo, D. D., Hou, Q. L., Zhang, R. Q., Lou, H. Y., Li, Y. H., Zhang, Y. F., et al. (2020). Over-expressing TaSPA-b reduces prolamin and starch accumulation in wheat (Triticum aestivum l.) . grains. Int. J. Mol. Sci. 21 (9), 3257. doi: 10.3390/ijms21093257
Huang, Y., Yan, B. J., Ren, Z. L. (2007). Accumulation of grain protein components and glutenin macroaggregates in different wheat varieties. Southwest Agric. J. 04), 591–596. doi: 10.16213/j.cnki.scjas.2007.04.012
Ji, M., Fang, W., Li, W., Zhao, Y., Guo, Y., Wang, W., et al. (2021). Genome wide association study of the whiteness and colour related traits of flour and dough sheets in common wheat. Sci. Rep. 11 (1), 8790. doi: 10.1038/S41598-021-88241-4
Jiang, H., Jiang, L., Guo, L., Gao, Z., Zeng, D., Zhu, L., et al. (2008). Conditional and unconditional mapping of quantitative trait loci underlying plant height and tiller number in rice ( oryza sativa l.) grown at two nitrogen levels. Prog. Natural Sci. 18 (12), 1539–1547. doi: 10.1016/j.pnsc.2008.05.025
Kaushik, M., Rai, S., Venkadesan, S., Sinha, S. K., Mohan, S., Mandal, P. K. (2020). Transcriptome Analysis Reveals Important Candidate Genes Related to Nutrient Reservoir, Carbohydrate Metabolism, and Defence Proteins during Grain Development of Hexaploid Bread Wheat and Its Diploid Progenitors. Genes 11 (5), E509. doi: 10.3390/genes11050509
Kim, J. S., Chae, S., Jun, K. M., Pahk, Y.-M., Lee, T.-H., Chung, P. J., et al. (2017). Genome-wide identification of grain filling genes regulated by the OsSMF1 transcription factor in rice. Rice 10 (1), 16. doi: 10.1186/s12284-017-0155-4
Kumar, R., Mukherjee, S., Ayele, B. T. (2018). Molecular aspects of sucrose transport and its metabolism to starch during seed development in wheat: a comprehensive review. Biotechnol. Adv. 36 (4), 954–967. doi: 10.1016/j.biotechadv.2018.02.015
Li, S. M., Chen, N., Li, F. F., Mei, F. M., Wang, Z. X., Cheng, X. X., et al. (2020). Characterization of wheat homeodomain-leucine zipper family genes and functional analysis of TaHDZ5-6A in drought tolerance in transgenic arabidopsis. BMC Plant Biol. 20 (1), 50. doi: 10.1186/s12870-020-2252-6
Li, H. M., Liang, H., Li, Z., Tang, Z. X., Fu, S. L., Geng, Y. Y., et al. (2015). Dynamic QTL analysis of protein content and glutamine synthetase activity in recombinant inbred wheat lines. Genet. Mol. Res. 14 (3), 8706–8715. doi: 10.4238/2015.July.31.19
Li, Z., Liu, P., Zhang, X., Zhang, Y., Ma, L., Liu, M., et al. (2020). Genome-wide association studies and QTL mapping uncover the genetic architecture of ear tip-barrenness in maize. Physiologia plantarum 170 (1), 27–39. doi: 10.1111/ppl.13087
Li, Y., Tan, X., Wang, M., Li, B., Zhao, Y., Wu, C., et al. (2017). Exocyst subunit SEC3A marks the germination site and is essential for pollen germination in arabidopsis thaliana. Sci. Rep. 7 (1), 40279–40279. doi: 10.1038/srep40279
Li, H., Ye, G., Wang, J. (2007). A modified algorithm for the improvement of composite interval mapping. Genetics 175 (1), 361–374. doi: 10.1534/GENETICS.106.066811
Liu, Y., Tao, Y., Wang, Z., Guo, Q., Wu, F., Yang, X., et al. (2018). Identification of QTL for flag leaf length in common wheat and their pleiotropic effects. Mol. Breed. 38 (1), 1–11. doi: 10.1007/s11032-017-0766-x
Liu, H. Y., Wang, K., Jia, Z. M., Gong, Q., Lin, Z. S., Du, L. P., et al. (2020). Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized agrobacterium-mediated CRISPR system. J. Exp. Bot. 71 (4), 1337–1349. doi: 10.1093/jxb/erz529
Ma, W., Appels, R., Bekes, F., Larroque, O., Morell, M. K., Gale, K. R. (2005). Genetic characterisation of dough rheological properties in a wheat doubled haploid population: additive genetic effects and epistatic interactions. Theor. Appl. Genet. 111 (3), 410–422. doi: 10.1007/s00122-005-2001-0
Ma, S., Wang, M., Wu, J., Guo, W., Chen, Y., Li, G., et al. (2021). WheatOmics: a platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol. Plant 14, 1965–1968. doi: 10.1016/j.molp.2021.10.006
Ma, C., Yang, Y., Li, X., Pei, G., Yan, Y. (2013). Molecular cloning and characterization of six novel HMW-GS genes from aegilops speltoides and aegilops kotschyi. Plant Breed. 132 (3), 284–289. doi: 10.1111/pbr.12046
Muqaddasi, Q. H., Brassac, J., Ebmeyer, E., Kollers, S., Korzun, V., Argillier, O., et al. (2020). Prospects of GWAS and predictive breeding for European winter wheat's grain protein content, grain starch content, and grain hardness. Sci. Rep. 10 (1), 12541. doi: 10.1038/s41598-020-69381-5
Nadaud, I., Girousse, C., Debiton, C., Chambon, C., Bouzidi, M. F., Martre, P., et al. (2010). Proteomic and morphological analysis of early stages of wheat grain development. Proteomics 10 (16), 2901–2910. doi: 10.1002/pmic.200900792
Pan, X., Li, Y., Liu, W., Liu, S., Min, J., Xiong, H., et al. (2020). QTL mapping and candidate gene analysis of cadmium accumulation in polished rice by genome-wide association study. Sci. Rep. 10 (1), 11791. doi: 10.1038/s41598-020-68742-4
Panozzo, J. F., Eagles, H. A., Wootton, M. (2001). Changes in protein composition during grain development in wheat. Aust. J. Agric. Res. 52 (4), 485–493. doi: 10.1071/ar00101
Paull, A. E., Anderson, J. A. (1942). The effects of amount and distribution of rainfall on the protein content of western canadian wheat. Can. J. Biochem. Physiol. 20c 4), 212–227. doi: 10.1139/cjr42c-021
Prasad, M., Kumar, N., Kulwal, P. L., Röder, M. S., Balyan, H. S., Dhaliwal, H. S., et al. (2003). QTL analysis for grain protein content using SSR markers and validation studies using NILs in bread wheat. Theor. Appl. Genet. 106 (4), 659–667. doi: 10.1007/s00122-002-1114-y
Rubio-Somoza, I., Martinez, M., Abraham, Z., Diaz, I., Carbonero, P. (2006). Ternary complex formation between HvMYBS3 and other factors involved in transcriptional control in barley seeds. Plant J. 47 (2), 269–281. doi: 10.1111/j.1365-313X.2006.02777.x
Saini, D. K., Chopra, Y., Pal, N., Chahal, A., Srivastava, P., Gupta, P. K. (2021). Meta-QTLs, ortho-MQTLs and candidate genes for nitrogen use efficiency and root system architecture in bread wheat (Triticum aestivum l.). Physiol. Mol. Biol. Plants 27 (10), 2245–2267. doi: 10.1007/S12298-021-01085-0
Sallam, A., Eltaher, S., Alqudah, A. M., Belamkar, V., Baenziger, P. S. (2022). Combined GWAS and QTL mapping revealed candidate genes and SNP network controlling recovery and tolerance traits associated with drought tolerance in seedling winter wheat. Genomics 114 (3), 110358. doi: 10.1016/j.ygeno.2022.110358
Shi, C., Zheng, Y., Geng, J., Liu, C., Pei, H., Ren, Y., et al. (2020). Identification of herbicide resistance loci using a genome-wide association study and linkage mapping in Chinese common wheat. Crop J. 8 (04), 666–675. doi: 10.1016/j.cj.2020.02.004
Souer, E., Houwelingen, A., Kloos, D., Mol, J., Koes, R. (1996). The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85 (2), 159–170. doi: 10.1016/s0092-8674(00)81093-4
Sun, H., Lü, J., Fan, Y., Zhao, Y., Kong, F., Li, R., et al. (2007). Quantitative trait loci (QTLs) for quality traits related to protein and starch in wheat. Prog. Natural Sci. 18 (7), 825–831. doi: 10.1016/j.pnsc.2007.12.013
Sun, H., Yao, D. N., Li, B. Y., Liu, G. T., Zhang, S. Z. (1998). Correlation betwwen content of glutenin macropolymer (GMP) in wheat and baking quality. J. Chin. Cereals Oils Assoc. 06), 15–18.
Tao, Y., Qin, Z., Qi, W., Xiao, W., Jian, C., Mei, H., et al. (2022). Effects of nitrogen fertilizer on quality characteristics of wheat with the absence of different individual high-Molecular-Weight glutenin subunits (HMW-GSs). Int. J. Mol. Sci. 23 (4), 2178. doi: 10.3390/ijms23042178
Teng, W., He, X., Tong, Y. P. (2022). Genetic control of efficient nitrogen use for high yield and grain protein concentration in wheat: a review. Plants 11 (4), 492. doi: 10.3390/PLANTS11040492
Tian, B., Deng, Z., Xie, Q., Tian, J. (2015). Genetic dissection of the developmental behaviour of total starch content and its components in wheat grain. Crop Pasture Sci. 66 (5), 445–455. doi: 10.1071/CP14059
Tian, B., Liu, B., Zhu, Z. L., Xie, Q. G., Tian, J. C. (2011). Conditional and unconditioned QTL mapping of starch accumulation dynamics in wheat grain. Scientia Agricultura Sin. 44 (22), 4551–4559.
Tian, Y., Peng, K., Lou, G., Ren, Z., Sun, X., Wang, Z., et al. (2022). Transcriptome analysis of the winter wheat Dn1 in response to cold stress. BMC Plant Biol. 22 (1), 277. doi: 10.1186/s12870-022-03654-1
Tian, Y. S., Sang, W., Liu, P. P., Liu, J. D., Xiang, J. S., Cui, F. J., et al. (2022). Genome-wide association study for starch pasting properties in Chinese spring wheat. Front. Genet. 13. doi: 10.3389/fgene.2022.830644
Verspreet, J., Hemdane, S., Dornez, E., Cuyvers, S., Pollet, A., Delcour, J. A., et al. (2013). Analysis of storage and structural carbohydrates in developing wheat (Triticum aestivum l.) grains using quantitative analysis and microscopy. J. Agric. Food Chem. 61 (38), 9251–9259. doi: 10.1021/jf402796u
Vranic, M., Perochon, A., Benbow, H., Doohan, F. M. (2022). Comprehensive analysis of pathogen-responsive wheat NAC transcription factors: new candidates for crop improvement. G3 12 (11), 1–18. doi: 10.1093/G3JOURNAL/JKAC247
Wang, Y., Chen, J., Guan, Z., Zhang, X., Zhang, Y., Ma, L., et al. (2019). Combination of multi-locus genome-wide association study and QTL mapping reveals genetic basis of tassel architecture in maize. Mol. Genet. Genomics 294 (6), 1421–1440. doi: 10.1007/s00438-019-01586-4
Wang, L., Cui, F., Wang, J., Jun, L., Ding, A., Zhao, C., et al. (2012). Conditional QTL mapping of protein content in wheat with respect to grain yield and its components. J. Genet. 91 (3), 303–312. doi: 10.1007/S12041-012-0190-2
Wang, W. J., Wang, P., Wang, G. J. (2007). Study on enzyme activities related to starch synthesis in different types of winter wheat at filling stage. J. North China Agron. 02), 75–78.
Wang, Z., Wu, X., Ren, Q., Chang, X., Li, R., Jing, R. (2010). QTL mapping for developmental behavior of plant height in wheat (Triticum aestivum l.). Euphytica 174 (3), 447–458. doi: 10.1007/s10681-010-0166-3
Yan, X., Zhao, L., Ren, Y., Zhang, N., Dong, Z., Chen, F. (2020). Identification of genetic loci and a candidate gene related to flag leaf traits in common wheat by genome-wide association study and linkage mapping. Mol. Breed. 40 (2), 58. doi: 10.1007/s11032-020-01135-7
Zhai, L., Zheng, T., Wang, X., Wang, Y., Chen, K., Wang, S., et al. (2018). QTL mapping and candidate gene analysis of peduncle vascular bundle related traits in rice by genome-wide association study. Rice 11 (1), 13. doi: 10.1186/s12284-018-0204-7
Zhang, Z., Dong, J., Ji, C., Wu, Y., Messing, J. (2019). NAC-type transcription factors regulate accumulation of starch and protein in maize seeds. Proc. Natl. Acad. Sci. United States America 116 (23), 11223–11228. doi: 10.1073/pnas.1904995116
Zhang, Z. W., Ersoz, E., Lai, C. Q., Fodhunter, R. J., Tiwari, H. K., Gore, M. A., et al. (2010). Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42, 355–360. doi: 10.1038/ng.546
Zhang, S., Ghatak, A., Bazargani, M. M., Bajaj, P., Varshney, R. K., Chaturvedi, P., et al. (2021). Spatial distribution of proteins and metabolites in developing wheat grain and their differential regulatory response during the grain filling process. Plant J. 107 (3), 669–687. doi: 10.1111/tpj.15410
Zhang, Z., Liu, Z., Cui, Z., Hu, Y., Wang, B., Tang, J. (2017). Genetic analysis of grain filling rate using conditional QTL mapping in maize. PloS One 8 (2), e56344. doi: 10.1371/JOURNAL.PONE.0056344
Zhang, H. Y., Xu, Z. F. (1995). Dynamic study on protein accumulation in winter wheat during filling period. J. Triticeae Crops 01), 47–49.
Zhou, Z., Zhang, Z., Mason, A. S., Chen, L., Liu, C., Qin, M., et al. (2021). Quantitative traits loci mapping and molecular marker development for total glutenin and glutenin fraction contents in wheat. BMC Plant Biol. 21 (1), 455. doi: 10.1186/s12870-021-03221-0
Keywords: wheat, protein, starch, GMP, QTL mapping, GWAS
Citation: Guo Y, Wang G, Guo X, Chi S, Yu H, Jin K, Huang H, Wang D, Wu C, Tian J, Chen J, Bao Y, Zhang W and Deng Z (2023) Genetic dissection of protein and starch during wheat grain development using QTL mapping and GWAS. Front. Plant Sci. 14:1189887. doi: 10.3389/fpls.2023.1189887
Received: 20 March 2023; Accepted: 18 May 2023;
Published: 12 June 2023.
Edited by:
Yunfeng Xu, Kansas State University, United StatesCopyright © 2023 Guo, Wang, Guo, Chi, Yu, Jin, Huang, Wang, Wu, Tian, Chen, Bao, Zhang and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiying Deng, ZGVuZzg2OEAxNjMuY29t
†These authors have contributed equally to this work
 Yingxin Guo1,2†
Yingxin Guo1,2† Jichun Tian
Jichun Tian Jiansheng Chen
Jiansheng Chen Yinguang Bao
Yinguang Bao Zhiying Deng
Zhiying Deng