- State Key Laboratory of Southwestern Chinese Medicine Resources, Department of Pharmacy, Chengdu University of Traditional Chinese Medicine, Chengdu, China
Introduction: P. ternata is a perennial herb of the family Araceae that grows in China and has various medicinal properties and applications. At present, the artificial cultivation of P. ternata is constrained by seedling propagation. To address the problems of low seedling breeding propagation efficiency and high cost, our group has developed a highly efficient cultivation technology for "hydroponic cuttings of P. ternata "for the first time. P. ternata is used as the source material and is grown in a hydroponic system, increasing the seedling production rate 10-fold compared with the traditional cultivation mode. However, the callus formation mechanism in cuttings from hydroponic cultivation is still remains unclear.
Methods: In order to better understand the biological process of callus formation in cuttings from hydroponic P. ternata, anatomical characterization, endogenous hormone content determination and transcriptome sequencing were performed on five callus stages from early growth to early senescence.
Results: Regarding the four major hormones during the callus developmental stages of P. ternata hydroponic cuttings, cytokinins showed an increasing trend during callus formation. IAA(indole-3-acetic acid) and abscisic acid contents increased at 8d and then decreased, while jasmonic acid content gradually decreased. A total of 254137 unigenes were identified by transcriptome sequencing in five callus formation stages. Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of the differentially expressed genes (DEGs) that differentially expressed unigenes were involved in various plant hormone signaling and hormone synthesis-related pathways. The expression patterns of 7 genes were validated using quantitative real-time PCR.
Discussion: This study presented integrated transcriptomic and metabolic analysis approach to obtain insights into the underlying biosynthetic mechanisms and function of key hormones involved in the callus formation process from hydroponic P. ternata cuttings.
1 Introduction
The medicinal plant P. ternata is widely used in China, Korea, and Japan (Bai et al., 2022) and has been proven to be very effective in treating cough, vomiting, infections, and inflammatory diseases (Peng et al., 2019; Peng et al., 2022). Modern pharmacological studies have demonstrated that it possesses a variety of activities such as cough suppressant, expectorant, anti-vomiting, anti-tumor, anti-bacterial, hypnotic, and others (Kong et al., 2018). It is used in various aspects of Chinese medicine, including clinical formulations of Chinese medicine and raw materials of many types of patented Chinese medicines, among which more than 400 Chinese patented medicines such as Huoxiang Zhengqi Oral Liquid, Banxia Cough Tablets and Banxia Tianma Pills (Mao and He, 2020). The current market demand for P. ternata exceeds 8,000 tons annually, far exceeding the sum of the current cultivated and wild P. ternata production. The disparity between supply and demand is outstanding (Kumari et al., 2017). The shrinkage of wild resources and the low degree of mechanization seriously restrict the increase of production. The cultivation and management techniques are not in place, resulting in low yield and uneven quality of the herb, which seriously affects the quality and clinical efficacy of P. ternata.
Currently, the P. ternata market is in a state of oversupply. Appropriate propagation methods, including seeds, tubers, bulbil, and group culture, have been evaluated domestically and abroad. Seed propagation faces problems such as low reproduction coefficient, long reproduction cycle, and quality degradation. As the medicinal part of P. ternata is the tuber, there is a contradiction between seed retention and the commercial use of tuber propagation. Bulbil propagation generally takes 2 to 3 years before the bulbils are used for medicinal purposes, and there are issues with the long propagation cycle. Rapid propagation of P. ternata using seedlings in tissue culture has been reported under laboratory conditions. This method requires steps such as sowing, rooting, and transplanting, which are demanding and complicated. Therefore, there is an urgent need to explore more convenient breeding and propagation methods to expand the propagation resources of P. ternata.
To expand the P. ternata resources and explore a more convenient breeding method, our group has established a system of cuttings obtained from the hydroponic culture of P. ternata, in which we induced the formation of callus on the cut surface of leafy stems above the Bulbils of P. ternata. These were further differentiated to form multiple adventitious buds without using exogenous hormones and further developed and matured to form tubers. Plants exhibit multiple regeneration patterns after trauma to a body part (Birnbaum and Alvarado, 2008; Ikeuchi et al., 2016). Plant regeneration is characterized by the formation of new shoots and/or roots through organogenesis (de novo organogenesis) from plant tissues. Callus formation is the protective response after a plant is injured and regenerates new organs. The wound-induced callus is formed at the wound site, preventing water loss and serving as a source of cells for vascular differentiation and/or ab initio organogenesis (Ikeuchi et al., 2013; Ikeuchi et al., 2016). Cells near the wound proliferate and establish new shoots or root tip meristematic tissues, giving rise to new organs. These newly formed meristematic tissues can be derived directly from the parent plant or indirectly from the callus formed at the wound (Ikeuchi et al., 2013). Many plants can regenerate from shoots or root explants, and this property has contributed to the increased possibility of asexual reproduction of stem segments. Surprisingly, in some plant species, whole plants can develop through certain plant tissue regeneration (Nakayama et al., 2014). Currently, only a few species can regenerate whole plants when the cut damage occurs naturally. Still, in this study, we found that after cutting the stem, the stem segment of P. ternata can regenerate to form a complete plant in pure water cutting culture. Callus is a key link in the development and maturation of hydroponic cuttings of P. ternata to form tubers, so exploring the mechanism of callus formation is currently a key priority.
Callus proliferation and differentiation are regulated by the expression of multiple genes (Chu et al., 2017; Ferdausi et al., 2021). The growth of the model plant Arabidopsis thaliana is regulated by various genes in an auxin-rich media, such as the PLT family of lateral root regulators, the LBD family of root formation regulators (Fan et al., 2012), the WOX5 root meristem regulator genes (Sugimoto et al., 2010), and WOX11-LBD16, which promotes the callus totipotency acquisition. Moreover, several APETALA2/ethylene response factor (AP2/ERF) transcription factors termed trauma-induced dedifferentiation 1 (WIND1), WIND2, WIND3, and WIND4 play key roles in trauma-induced callus formation (Iwase et al., 2011). Recent studies have shown that WIND1 directly upregulates the expression of the Arabidopsis AP2/ERF transcription factor ENHANCER of SHOOT REGENERATION1 (ESR1) to promote callus formation and plant regeneration (Iwase et al., 2017). Auxin can stabilize the expression of AtbZIP59, and in AtbZIP59 mutants, auxin can maintain callus formation, while overexpression of AtbZIP59 can promote callus formation (Xu et al., 2018). In addition, MYB94 and MYB96 can directly control AtLBD29 expression, and plants overexpressing AtLBD29 showed significant phenotypic changes during growth and development, inhibiting callus inhibition (Dai et al., 2020). JA is a trauma-induced hormone (Creelman et al., 1992) and has a slight inhibitory effect on callus formation at wound sites (Ikeuchi et al., 2017). ABA transduces various stress responses, such as drought and heat stress, but only a few studies have reported its involvement in trauma-induced signaling (Pēna-Cortés et al., 1989; Pena-Cortes et al., 1991; Dammann et al., 1997). Cytokinins (CKs) are a large group of plant hormone promoting cell division and prevent senescence. Post-trauma–induced CKs responses have been found to regulate callus formation (Ikeuchi et al., 2017). IAA plays a key role in regulating root regeneration after injury (Chen et al., 2016), and it also promotes the formation of callus in tissue culture. Although the molecular mechanisms of callus differentiation have been reported in many plant species, the genes involved in the proliferation and differentiation of P. ternata callus have not yet been adequately studied, and the underlying molecular mechanisms are still unclear.
In order to elucidate the molecular mechanism underlying of the hydroponic cutting callus formation in P. ternata. Through analysis of the histological characteristics and hormonal metabolism of callus samples at different stages, we identified changes in four major classes of plant hormones during callus formation in P. ternata hydroponic cuttings. Our findings showed that the total CK and auxin contents are crucial for callus formation, highlighting their importance during callus formation. We also analyzed the transcriptome dynamics of five stages of P. ternata callus development and examined the expression of genes involved in phytohormone biosynthesis and signaling pathways. Our study revealed variations in gene expression at different callus stages, suggesting that callus formation is a complex process that involves multiple genes and interconnected mechanisms. This study investigated the molecular mechanism of callus formation in P. ternata hydroponic cuttings, laying the groundwork for further investigation of the molecular regulatory network underlying callus formation and providing a theoretical basis for expanding P. ternata resources.
2 Materials and methods
2.1 Plant material and experiment at conditions
P. ternata, originally harvested from Qingshan Village, Yingshan County, Nanchong City, Sichuan Province, was collected from the Chengdu University of Traditional Chinese Medicine. P. ternata were planted in small potting trials in river sand with humidity maintained at 85% and temperature at 28°C. When the bulbils were mature (brown), the three-leave stem above the bulbils were collected as cuttings. For disinfection, the cuttings were washed with sterile water. Afterward, the cuttings were inserted into a sterilized foam plate and immediately placed into a hydroponic tank. The basal cut of the stem cutting was immersed in 0.1 mg/liter of 6-BA (6-benzyladenine) solution for culture. The concentration of the 6-BA solution was determined according to agricultural practices. The culture was incubated in an air-conditioned room at 28°C, 14/10h light/dark and 75% relative humidity. The 6-BA culture solution was changed once a week.
At 0, 8, 15, 25, and 30 days, 120 stem cuttings from each period (40 samples per replicate) were collected from the base of stem segments (approximately 2.0 cm of callus growth). A part of the callus was preserved in 4% paraformaldehyde for histological morphological observation by staining with Saffron solid green (SSG) and Periodic acid-Schiff (PAS). The other sampled callus tissue was snap-frozen in liquid nitrogen and stored at −80°C in an ultra-low temperature refrigerator for determination of oxidase activity, transcriptome analysis, and targeted endogenous hormone content determination.
2.2 Histomorphological, histochemical, and biochemical characterization
Fresh callus was photographed under a camera for observation. Light microscopic observation was performed using cross-sectional dissection after fixation of the callus in 4% paraformaldehyde, and paraffin sections were embedded. Sections (5 μm in thickness) were cut using a Leica RM2235 (Leica, Shanghai, China) rotary sectioning machine. Cell proliferation was observed using staining with SSG. Insoluble polysaccharides and starch were detected by staining with PAS.
2.3 Analysis of peroxidase, phenylene oxide, and indoleacetic acid oxidase
The enzymatic activities of peroxidase (POD), polyphenol oxidase (PPO), and indoleacetic acid oxidase (IAAO) were assessed in the callus samples according to the method used by Zhang et al1. The sample (0.5 g) was ground into a homogenate in 1 ml of cold phosphate buffer (50 mM, pH 6.5) and a small amount of quartz sand in a mortar in an ice bath. After diluting the homogenate to 2 ml, the supernatant was collected by centrifugation (1790 × g, 20 min, 4°C) and readjusted to a constant volume. For analysis of IAAO, 1 ml of enzyme solution was added to the reaction solution (1 ml 1×10−3 MnCl2, 1 ml 1×10−3 M 2,4-dichlorophenol, 2 ml 1×10−3 M IAA (indole-3-acetic acid) and 5 ml of phosphate buffer and mixed in tube 1. The same volume of phosphate buffer was added to the same reaction system in tube 2. Distilled water was added to tube 3 as a control. Then, 1 ml of each mixed solution was added to the addition of 2 ml of FeCl3 perchlorate reagent, shaken, and incubated for 30 min at 30°C in the dark. Absorbance values were measured at 530 nm. For analysis of PPO, after a 1-min reaction in 2.5 ml of phosphate buffer (pH 6.0) 0.5 ml enzyme solution, 1 ml catechol (10 M) was added and shaken. The absorbance was measured at 420 nm. For POD analysis, 0.1 ml of enzyme solution was added to the reaction solution, 0.1 ml of enzyme solution was added to the reaction solution (2.9 ml of 0.05 M phosphate buffer, 1.0 ml of 2% H2O2, 1.0 ml of 0.05 M guaiacol) and incubated in a water bath at 30°C for 3 min. Then, the absorbance values were measured at 470 nm.
2.4 Plant hormone determination
When the cuttings were obtained, there was no callus formation, and the 2 cm of tissue at the base of the stem was used for the phytohormones determination. Afterward, when the callus was formed, the callus’s phytohormone content at the stem’s base was measured in five time periods. The harvested callus from the five-time periods was immediately frozen in liquid nitrogen, ground into powder (30 Hz, 1 min) using a ball mill (MM400, Retsch, Germany), and stored at −80°C until needed. 50 mg of plant sample was weighed into a 2 ml of plastic microtube and frozen in liquid nitrogen, dissolved in 1 ml of methanol/water/formic acid (15:4:1, V/V/V). Ten microliter internal standard (IS) mixed solution (100 ng/ml) was added into the extract as IS for the quantification. The mixture was vortexed for 10 min, then centrifugation for 5 min (12000 r/min and 4°C), the supernatant was transferred to clean plastic microtubes, followed by evaporation to dryness and dissolved in 100 μl of 80% methanol (V/V), and filtered through a 0.22-μm membrane filter for further LC-MS/MS analysis (Floková et al., 2014; Li et al., 2016). Phytohormones contents were determined by MetWare2based on the AB Sciex QTRP6500+LC-MS/MS platform. All assays were performed with three biological replicates.
2.5 RNA extraction, library preparation, and RNA-seq
Total RNA was extracted using the Plant RNA Extraction Kit (Tiangen, Beijing, China). RNA concentration and integrity were assessed using a Qubit 2.0 fluorometer (Life Technologies, California, USA) and an Agilent 2100 Bioanalyzer (Agilent Technologies, California, USA). Each sample (≥1 μg) was used for sequencing library preparation using the NEBNext® Ultra™ RNA Library Preparation Kit (NEB, USA) according to the manufacturer’s instructions. Library quality was subsequently assessed by an Agilent 2100 Bioanalyzer system. The Illumina NovaSeq 6000 platform was used for subsequent sequencing.
2.6 Data filtering and transcriptome analysis
After removing adapter sequences, ambiguous reads, and low-quality reads, the remaining high-quality clean reads were used for subsequent analysis. Transcript assembly was performed on clean reads using Trinity, and the transcripts obtained from the assembly were clustered and de-redundant using Corset3. ClusterProfile software was used for GO (Gene Ontology) functional analysis, and the KEGG (Kyoto Encyclopedia of Genes and Genomes)4 database was used for pathway enrichment analysis. OmicShare tool5 was used to graphically display the results of GO and KEGG functional enrichment analysis with adjusted p values.
2.7 Identification of differentially expressed genes
Fragment per kilobase fragment per million fragment transcripts (FPKM) values were used to normalize gene expression. The DESeq2 R software (1.16.1) was used to identify differentially expressed genes (DEGs, 0d vs. 8d, 15d, 25d, and 30d) according to the following threshold criteria:|log2 (fold change) |>1, p adjusted < 0.05. Pearson correlation coefficients were used to assess correlation of the biological replicates.
2.8 Quantitative real-time polymerase chain reaction assay
For expression validation of differentially expressed genes, special primers were designed for seven differentially expressed genes were selected by using Premier 5.0 software. The specificity of each primer set was assessed with polymerase chain reaction (PCR) amplicon sequencing, and the efficiencies of the different primer sets were similar. Details of all primers are described in Supplementary Table S9. Total RNA, initially treated with DNase I Amplification Grade enzyme (Tsingke, Beijing, China), was used as the template for the reverse transcriptase reactions. An aliquot of treated RNA was used in quantitative polymerase chain reaction (qPCR) to rule out DNA contamination. Complementary DNA (cDNA) synthesis was done using Super Script First-Strand Synthesis System for RT-PCR (Tsingke, Beijing, China) with random hexamers and oligo (dT) primers. qPCR reaction was performed in a volume of 20 μl containing 10 μl 2 × All-in- One TM (Gene Copoeia, Los Angeles, USA) qPCR Mix, 2 μl of cDNA, 1 μl of each 4 μM primer, and 6 μl RNase-free sterile water. To normalize the relative expression of selected genes, the GAPDH gene was used as a reference (Zhu et al., 2013). qPCR was performed using an iQ5 Real-Time PCR Detection System (Bio-Rad, Hercules, USA). PCR reactions were performed at 95°C for 2 min, followed by 40 cycles of 95°C for 15 s, 60°C for 20 s, and 72°C for 20 s. Melting curve analysis was conducted for each reaction to confirm the specificity of the reaction, and all the cDNA samples were analyzed in triplicate. The relative expression levels of candidate genes were calculated using the 2−ΔΔCt method.
2.9 Statistical analysis
Statistical processing of plant hormone content and qRT-PCR data was performed in Excel. The experiments were conducted in triplicate, and data were analyzed using standard analysis of variance (ANOVA) followed by Duncan’s multiple range test (DMRT) through Origin software.
3 Results
3.1 Morphological and histological analysis
The first morphological change in the process of callus formation occurred in the first eight days of culture(8d), when the tissue at the base of the cuttings expanded slightly and strong cell proliferation was observed on its surface. Callus cell were produced that were larger in size and grew more vigorously than those in the control group(0d) (Figure 1). The callus continued to grow, and after 15 days of culture (15d), instead of the initially loosely arranged cells, the callus cells at the outermost layer became relatively small and showed a high capacity to divide, thus enabling the outer layer expansion. The cells at the inner callus layers were uniformly colored and arranged without cell gaps. The callus continued to differentiate and formed shoot primordia (Figures 1A, C). After 25 days of culture (25d), the shoot primordium cells continued to divide, resulting in a significant increase in the number of cells and the formation of one or more shoots. After thirty days of culture (30d), multiple shoots gradually differentiated and formed stems and leaves. Histological callus observation was carried out at different time periods using a light microscope. Starch reserves were observed by staining with a periodic acid Schiff (PAS) reagent. As the time and callus growth progressed, starch accumulated in the callus, indicating that the cells were metabolically competent.
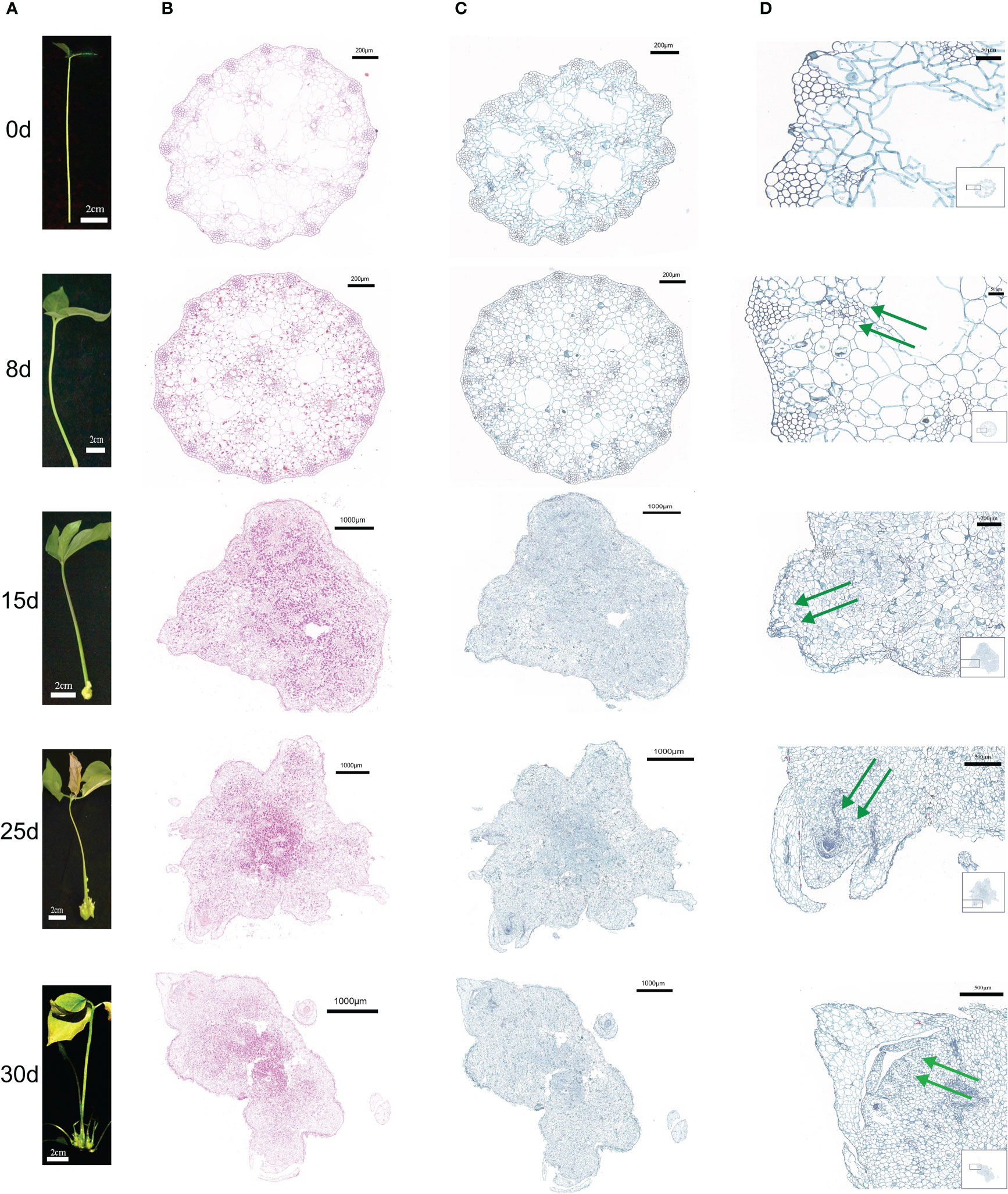
Figure 1 Morphological and histological characteristics of callus from P. ternata hydroponic cuttings cultured in water at different induction stages (0d, 8d, 15d, 25d, and 30d). (A) External morphology of different callus in order from top to bottom. (B) PAS staining of transverse callus sections at different points in order from top to bottom. (C) Staining of callus cross sections at different periods of time in order from top to bottom. (D) Local organization of saffron green in cross sections of callus at different induction stages periods of time in order from top to bottom, with green arrows indicating the process of bud formation at different induction stages. The scale bars are 2 cm, 200 μm, 1000 μm, and 500 μm, in that order.
3.2 Variations in oxidative enzyme activity during callus formation
The callus POD, polyphenol oxidase (PPO), and indoleacetic acid oxidase (IAAO) activities at five time points of callus growth are shown in Figure (Supplementary Figure S1). The POD and PPO activities consistently increased, reaching their highest values on the 25th day after explant cutting (Supplementary Figures S1A, B). The increase in POD activity may be a result of the damage to the cuttings during excision. POD activity remained high throughout the callus formation process, probably due to the effects of exogenous hormones, that promoted the growth and development of the callus. PPO affects plant growth mainly by catalyzing the formation of IAA-phenolic acid complex. It increased rapidly during callus formation and differentiation to form buds in the early stage, producing high concentrations of the IAA-phenolic acid compounds to promote callus growth, differentiation, and bud formation. The high IAA-phenolic acid compound concentration is produced to promote callus growth and buds’ differentiation. However, after the plant is damaged or during virus infestation, PPO catalyzes the oxidation of phenols and O2 to form quinone, resulting in tissue browning restoring the damage, and preventing infection. In the early stage of cuttings, IAAO activity is relatively low, facilitating callus formation. At the later stage of P. ternata callus development, IAAO activity increased and resulted in a reduction in endogenous IAA levels, which was favorable to the differentiation of callus and the formation of shoot primordia (Supplementary Figure S1B).
3.3 Targeted metabolomic analysis of callus formation
A total of 73 metabolites of indole-3-acetic acid (IAA), CK, jasmonic acid (JA), abscisic acid (ABA), and their intermediates were measured by liquid chromatography–mass spectrometry (LC-MS) used for targeted metabolome determination at 0d, 8d, 15d, 25d, and 30d of callus formation. The results revealed 55 hormone metabolites, including 20 auxin analogs and their derivative. PCA was performed, with the first principal component explaining 48.78% of the variation and the second principal component explaining 21.58% of the variation (Supplementary Figure S2A). There was a clear separation between the samples of different time points in these two dimensions. The clustering of QC samples also indicated that the experimental results were reliable. All samples were within the 95% confidence interval, indicating significant differences in metabolites in different periods of callus formation in P. ternata hydroponic cuttings.
3.3.1 Analysis of differential metabolites during the different callus formation stages
In order to evaluate metabolite changes between the different P. ternata callus stages, callus samples at 0d, 8d, 15d, 25d, and 30d were assessed. The OPLA-DA model was used to test for differences to eliminate the interference of irrelevant variables. Metabolites with fold change ≥ 2 and fold change ≤ 0.5 in the control (0d) and experiment groups (8d, 15d, 25d, and 30d) were selected were considered significantly different (Supplementary Figure S2). Compared with 0d (CK), 44, 44, 38, and 14 metabolites were differentially accumulated in 8d, 15d, 25d, and 30d callus (Supplementary Figures S2B, C), and a total of 29 metabolites were differed in the four comparison groups, mainly corresponding to auxin, CK, and JA metabolites (Supplementary Figure S2D; Supplementary Table S1).
3.3.2 Variations in phytohormone contents during the callus formation process
3.3.2.1 Cytokinins
CKs are a class of adenine derivatives. Natural CKs are divided into free-state CKs and bound-state CKs (Záveská Drábková et al., 2021). The natural free-state CKs in plants are zeatin (ZT), zeatin riboside (ZR), and dihydro zeatin (DZ), dihydrozeatin ribonucleoside (DHZR), and N6-isopentenyladenine (IP). The bound state CKs are methionyl zeatin, methionyl isopentenyl adenosine, and 3-Indolepropionic acid (iPA).
The content of various forms of CKs varied in the callus during the different time periods. The tZOG (trans-Zeatin-O-glucoside) was initially significantly increased and then significantly decreased (P < 0.05), with the highest level measured at 8d and 15d, indicating that tZOG plays a vital role in the early callus formation stages (Figure 2A). The concentration decreased in the later period of differentiation during shoot formation indicating that the level of cZROG (cis-Zeatin-O-glucoside riboside) can promote the formation of shoots in the late differentiation of callus (Figure 2A). Both IP7G (N6-Isopentenyl-adenine-7-glucoside) and IP9G (N6-Isopentenyl-adenine-7-glucoside) in IP showed an initial increasing trend, and then they both decreased. The level of IP7G increased sharply at 15d, indicating a role in callus formation. However, IP9G showed high levels at 8d but decreased after that time point (Supplementary Figure S3B). Moreover, 2MeScZR (2-Methylthio-cis-zeatin riboside) and 2MeSiP (2-Methylthio-N6-isopentenyladenine) were not detected at 8d (Figure 2D), suggesting they do not play a significant role in callus formation process. K9G content reached its highest levels at 15d (Figure 2E). BAP9G (N6-Benzyladenine -9-glucoside) levels also remained high after an initial increase at 8d (Figure 2F). mT (meta-Topolin) decreased gradually after a significant increase at 8d, while mT9G (meta-Topolin-9-glucoside) gradually increasing (Figure 2G). oT9G (ortho-Topolin-9-glucoside) also showed the same trend (Figure 2I).
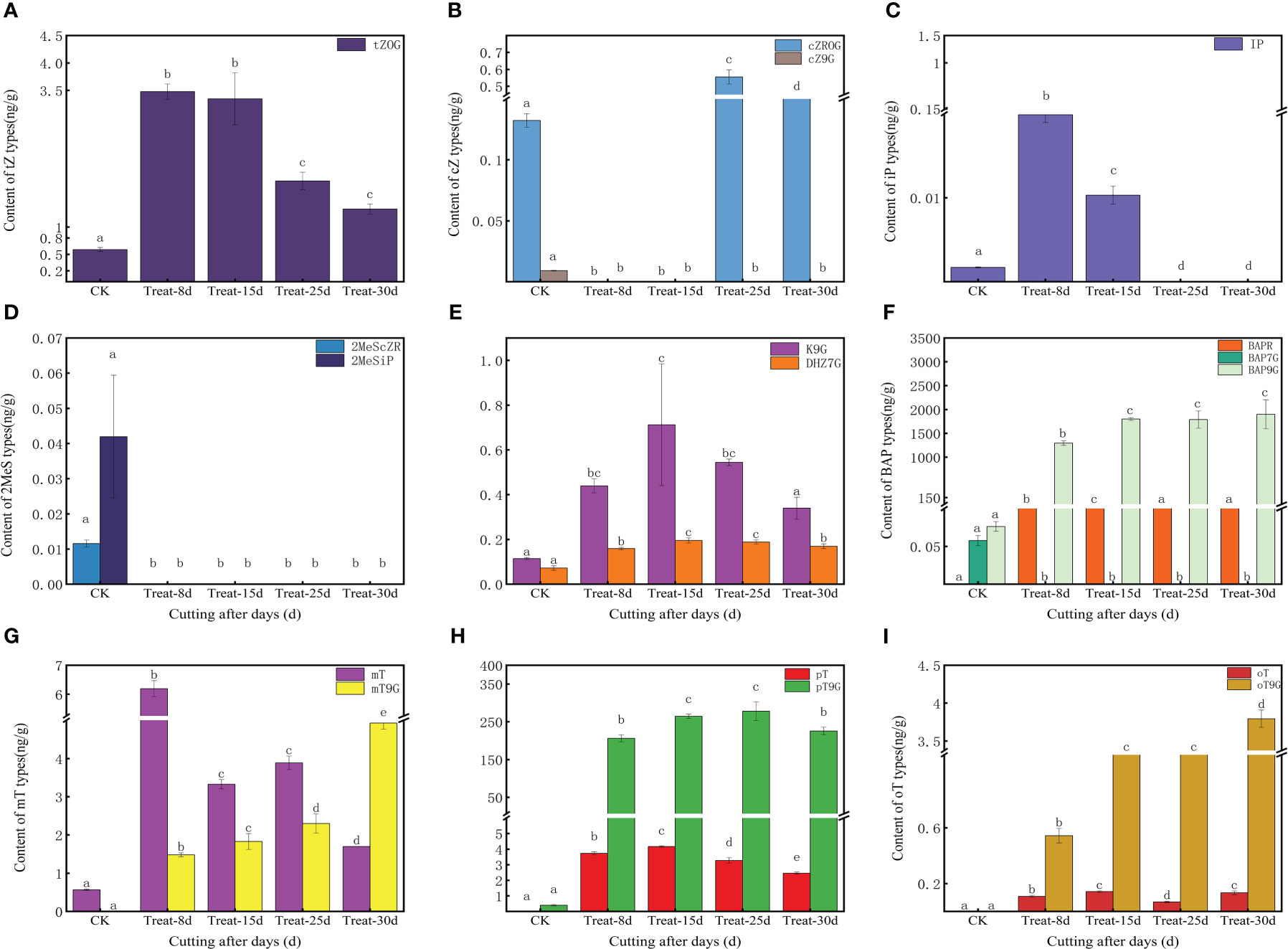
Figure 2 Changes in endogenous cytokinins (CKs) content at five stages of callus formation from hydroponic cuttings. (A) trans-zeatin (tZ) type; (B) cis-zeatin (cZ) type; (C) N6-(Δ2-prenyl) adenine (iP) type; (D) methyl mercaptan type (2MeScZR, 2MeSiP); (E) dihydrozeatin; (F) 6- benzylaminopurine (BAP) type; (G) 3-[(9H-purin-6-ylamino) methyl] phenol (mT) type; (H) 2-[(9H-purin-6-ylamino) methyl] phenol (pT); (I) 4-[(9H-purin-6-ylamino) methyl] phenol (oT) type. The bars with different letters are significantly different (p < 0.05). Values are means of three replicates ± SE.
While 17 CKs were identified in the shared differentially accumulated metabolite, the clustering heat map results indicated that cZ9G, BAP9G, 2MeScZR, and 2MesiP were present in high levels only in control at 0d (Supplementary Figure S2D). A significant elevation of IP and mT at 8d, suggesting a potentially important role in triggering callus induction. BAPR was significant expression elevated at 15d, suggesting a role in callus growth and expansion, while cZROG was elevated at 25d, and mT9G and oT9G were elevated at 30d (Supplementary Figure S3).
3.3.2.2 Auxin
The endogenous auxin content changed significantly during the formation of callus in P. ternata. The level of IAA (Indole-3-acetic acid) decreased significantly (P < 0.05) from the cuttings excision date and remained low during the formation of the callus (Figure 3A). The level of MEIAA (methyl indole-3-acetate) increased significantly (P < 0.05) at 8d and remained elevated, indicating that MEIAA was beneficial for callus induction (Supplementary Figure S4A). IBA (3-Indolebutyric acid) is another form of auxin in plants, which is more stable than IAA, but its levels were low in this experiment (Supplementary Figure S4A). IAN (indole-3-acetonitrile) and IAM (indole-3-acetamide) act as important precursors for IAA synthesis and determine the amount of IAA synthesized. The level of IAM increased sharply at 8d (P < 0.05) during callus. Still, its level was 0 at 15d, 25d, and 30d, indicating that IAM facilitates callus initiation (Supplementary Figure S4C). In contrast, the content of IAN decreased at 8d and was maintained at these levels at 15d but increased again at 25d and eventually decreased at 30d (P < 0.05) (Supplementary Figure S4D). This indicates that IAN carries an important role in the late differentiation callus and in shoots formation. IAA synthesis includes both tryptophan-dependent and tryptophan-independent pathways. TRP (tryptophan) is rate-limiting for auxin biosynthesis and its derivatives. TRP content increased significantly (P < 0.05) on the date of cutting excision and remained high (Figure 3B), indicating that TRP is beneficial for callus formation. TRA (Tryptamine) is a TRP catabolism product, a precursor of indole-3-glyoxime by the action of YUCC enzyme, and of IAN synthesis catalyzed by indole-3-glyoxime dehydratase. In this study, the TRA content exhibited significant differences at 8d and 25d (P < 0.05), which are critical periods for callus formation (Figure 3C), indicating that TRA could positively affect the process. During auxin degradation, intermediates are formed with amino acids, which are storage forms of IAA and can have direct signaling functions. Among them, the level of amino acid conjugate of IAA-Asp (Indole-3-acetyl-L-aspartic acid) was significantly higher at 8d (P < 0.05), suggesting that its accumulation facilitates the initial callus formation process (Figure 3D).
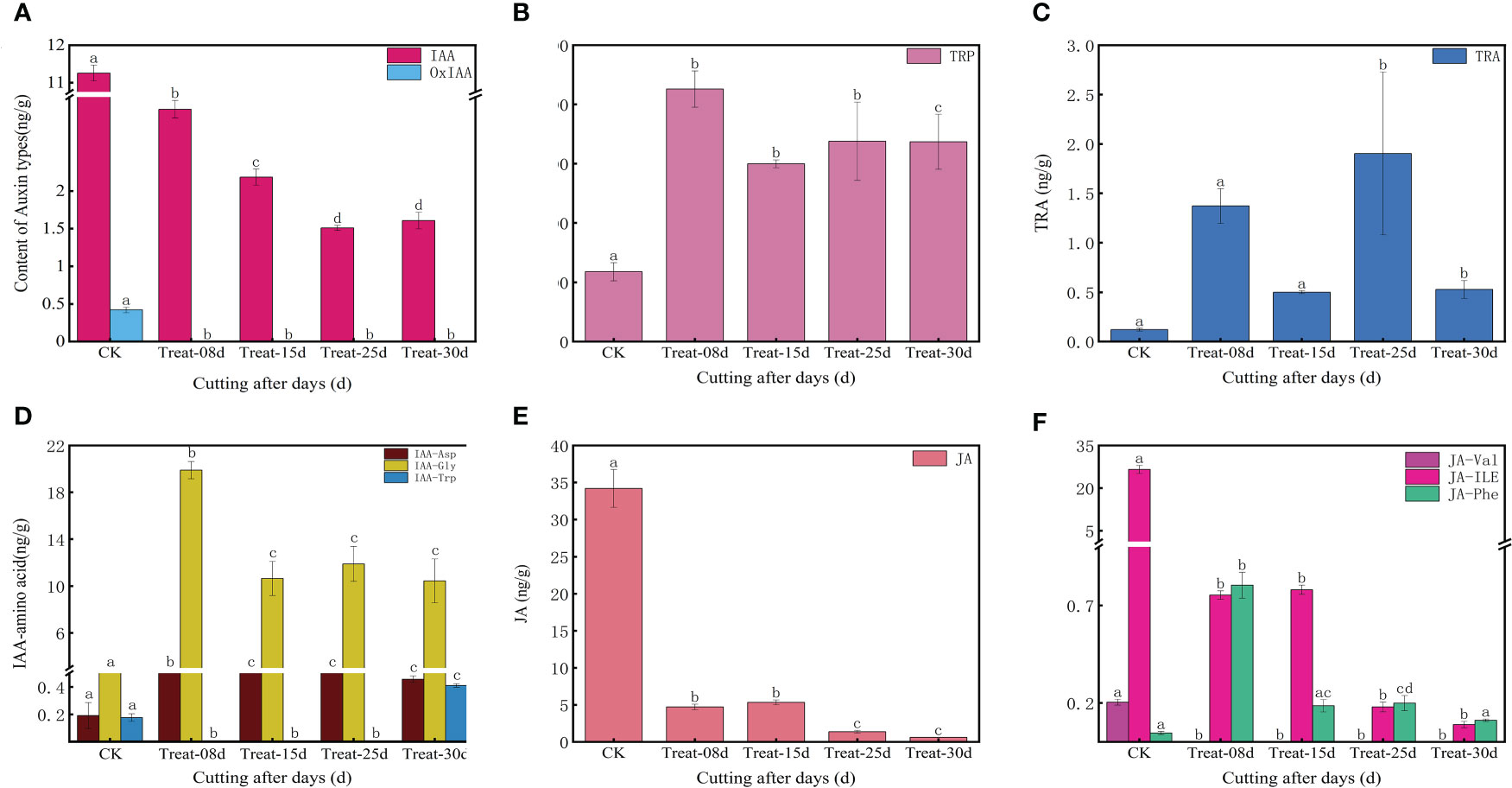
Figure 3 Changes in endogenous auxin, jasmonic acid, and abscisic acid contents in five stages of callus formation in P. ternata hydroponic cuttings. (A) IAA active compound; (B) tryptophan, a key compound for IAA synthesis; (C) tryptamine; (D) amino acid coupling substance of IAA; (E) JA; (F) amino acid conjugates of JA. The bars with different letters are significantly different from each treatment (p < 0.05). Values are means of three replicates ± SE.
Based on the cluster heat map of the shared differentially accumulated metabolites at different periods (Supplementary Figure S2D), the levels of IAA and OxIAA (2-oxindole-3-acetic acid) were highest at 0d. They decreased sharply during callus formation, while MEIAA, IAA-Gly (Indole-3-acetyl glycine), and IAA-Asp levels were highest at 8d and TRA at 25d, and IAA-Trp (Indole-3-acetyl-L-tryptophan) accumulated significantly at 30d, suggesting that the different auxin types may have different mechanisms of action at different stages of callus formation (Supplementary Figure S4).
3.3.2.3 Stress hormones (jasmonic acid and abscisic acid)
JA (Figure 3E) and jasmonic acid isoleucine content (Figure 3F) showed a gradual decline (P < 0.05) during the callus formation. This indicates that JA and jasmonic acid isoleucine may not be directly involved in the process. In contrast, OPC-4 (3-oxo-2-(2-(Z)-Pentenyl) cyclopentane-1-butyric acid) levels were elevated at 8d and maintained at high levels at 15d. However, its levels were not detected at 25d and 30d, suggesting that OPC-4 plays a role in the developmental process of callus expansion. ABA was elevated at 8d and later returned to its initial level, suggesting a role for ABA in the early stages of callus formation at 8d (Supplementary Figure S4D). Among the shared differentially accumulated metabolites, only JA-Phe (N-[(-)-Jasmonoyl]-(l)-phenalanine) showed show significant differences at 8d (Figure 3F).
3.4 Transcriptomic analysis of callus formation
3.4.1 Transcriptome sequencing quality testing, assembly, and differentially expressed gene analysis
Five callus formation stages at 0d, 8d, 15d, 25d, and 30d were selected for transcriptome sequencing. The reads were filtered and used for de novo transcriptome assembly in the absence of a reference genome. A total of 121.74 Gb Clean Data were obtained, with approximately 5 Gb of clean data per sample and a Q30 bases percentage greater than 91%. The 15 samples had GC content between 53.7% and 54.82%. The results, indicating that the nucleic acid sequence results had met the requirements for subsequent quality assessment of the analysis (Supplementary Table S2).
A total of 132,435 (52.11%) unigenes were matched to the Nr database by BLAST analysis, while 111,436 (43.85%), 98,381 (38.71%), 78766 (30.99%), and 92,351 (36.34%) (Supplementary Table S3). Also, a high percentage of annotated sequences in P. ternata callus were highly similar to Elaeis guineensis (16.03%) and Phoenix dactylifera (13.95%) (Supplementary Figure S5).
The raw data were analyzed using the negative binomial distribution using DESeq26 software. The screening criteria were P-value < 0.05, |log2 FC| ≥ 1, and genes differentially expressed between the two groups were screened. 12,743 DEGs were upregulated, and 12,589 DEGs were downregulated in CK at 0d compared with the callus at 8d. In CK compared with the callus at 15d, 25,150 genes were differentially expressed, with 12,862 upregulated and 12,288 downregulated transcripts were detected. When comparing the CK versus callus at 25d, 27,509 differentially expressed, 12,671 upregulated and 14,838 downregulated transcripts were identified. In the CK versus the callus at 30d comparison, 24,332 differentially expressed genes, 12,397 upregulated and 11,935 downregulated transcripts were identified (Supplementary Figure S6). Meanwhile, a total of 11,067 co-expressed genes were detected in the four comparison groups, of which 4,309 were co-upregulated and 6,702 co-downregulated (Supplementary Figures S6A, B).
To visualize the DEGs in metabolic pathways, we classified these DEGs according to KEGG pathway enrichment analysis. The DEGs in the CK versus 8d callus comparison were mainly associated with metabolic pathways (3,978), biosynthesis of secondary metabolites (2,181), plant-pathogen interactions (596), and phytohormone signaling (521) (Supplementary Table S5). DEGs in the CK versus 15d callus were mainly associated with metabolic pathways (3,861), biosynthesis of secondary metabolites (2,145), plant-pathogen interactions (595), and plant hormone signaling transduction (558) (Supplementary Table S6). DEGs in the CK versus 25d callus were mainly associated with metabolic pathways (4,103), biosynthesis of secondary metabolites (2,246), ribosomes (909), and plant hormone signaling transduction (557) (Supplementary Table S7). Finally, the DEGs in the CK versus 30d callus comparison were mainly enriched in metabolic pathways (3,558), secondary metabolite biosynthesis (2,013), plant-pathogen interactions (575), and plant hormone signaling transduction (551) (Supplementary Table S8). In this study, we focused on signaling pathways such as “plant hormone signaling transduction” and metabolic pathways related to hormone biosynthesis.
3.4.2 Analysis of key differentially expressed genes involved in the plant hormone signaling transduction pathways
A clustering heat map was used to illustrate changes in the expression of genes associated with plant hormone signaling transduction pathways and to classify genes with similar expression patterns (Figure 4). A total of 253 shared DEGSs were identified as hormone related during callus development in P. ternata, including DEGs of auxin, CK, JA, and ABA signaling pathways. Most of the DEGSs tended to be downregulated during callus formation from P. ternata hydroponic cuttings, while a few showed an upregulation trend.
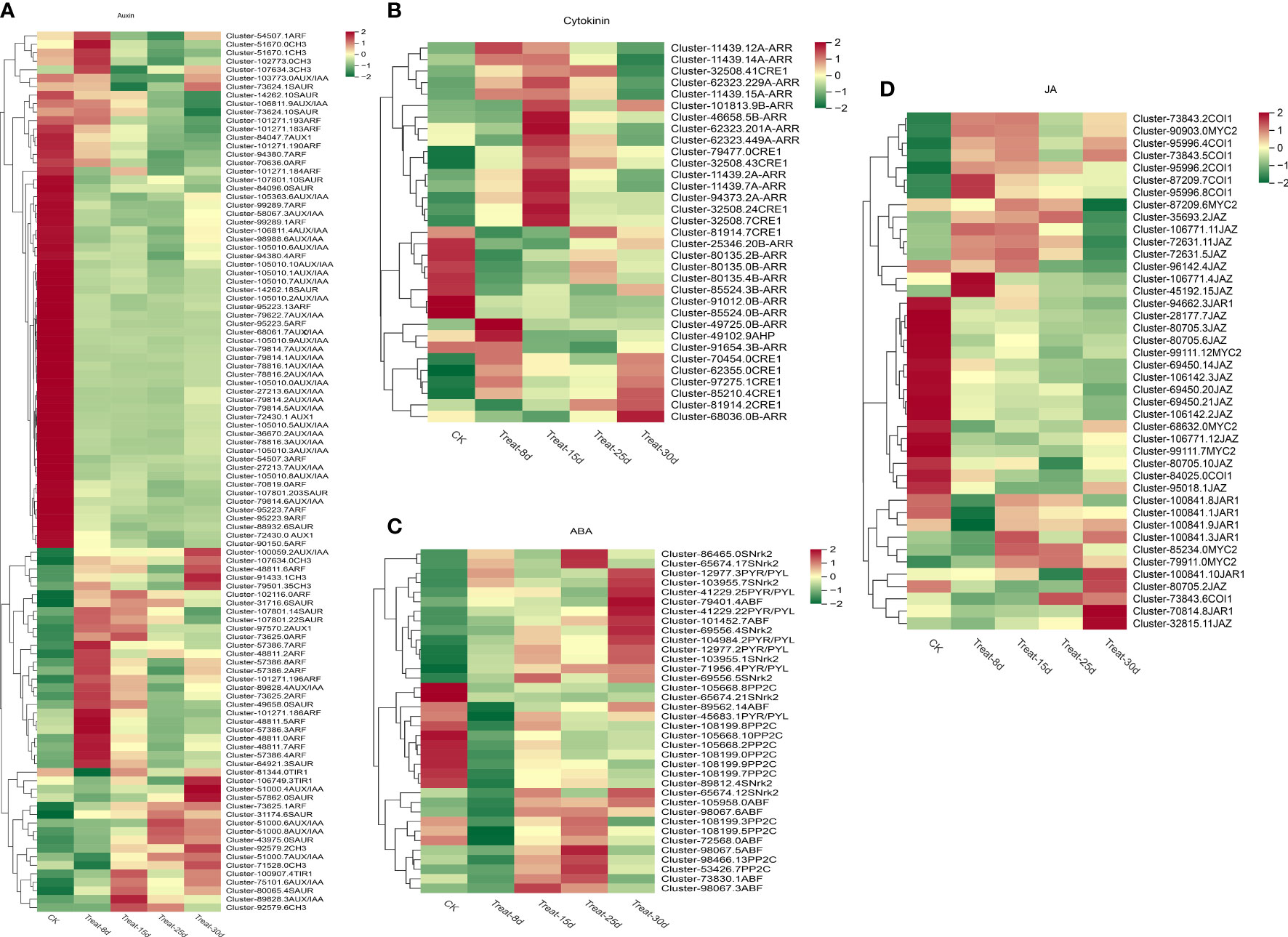
Figure 4 Changes in the expression levels of differentially expressed genes (DEGs) involved in auxin, cytokinin, jasmonic acid, and abscisic acid signaling pathways during the five stages of P. ternata callus formation. (A) Expression levels of DEGs in the auxin signaling pathway. (B) Expression levels of DEGs involved in cytokinin signaling. (C) Expression levels of DEGs of the abscisic acid signaling pathway. (D) Expression levels of DEGs of the jasmonic acid signaling pathway.
The auxin signaling network regulation during callus differentiation is very complex. One hundred four DEGs were involved in the auxin signal transduction pathway, accounting for the largest proportion of all hormone-related DEGs. Auxin-related DEGs involved four AUX1 [auxin influx vector (AUX1 LAX series)], three TIR1 (transport inhibitor response 1), 37 AUX/IAA (auxin-responsive protein IAA), 33 ARF (auxin-responsive factor), 10 CH3 (auxin-responsive CH3 gene family), and 17 SAUR (SAUR family protein), and so forth (Figure 4A). Among them, AUX1, AUX/IAA, and ARF DEGs mainly showed a decreasing expression trend, while an increasing expression trend was observed with TIR1, CH3, and SAUR DEGs. Notably, four CH3 genes and eight ARF genes were significantly expressed only at 8d, suggesting that these two genes may be key regulators for the initiation of the callus regeneration system. The AUX/IAA genes (Cluster-51000.4), TIR1 gene (Cluster106749.3), and SAUR gene (Cluster- 57862.0) were significantly upregulated only at 30d, which may be a key regulatory gene for shoot differentiation after callus formation. Further experimental studies are needed to confirm the function of these genes.
A total of 33 DEGs were involved in the CK signaling transduction pathway, and the related DEGs included 11 CRE1 [Arabidopsis histidine kinase 2/3/4 (cytokinin receptor)], one AHP (histidine-containing phosphotransfer protease), 12 B-ARR (two-component response regulator ARR-B series), eight A-ARR (two-component response regulator ARR-A series), and others (Figure 4B). Most of these genes were activated at 15d callus stage, including two CRE1 genes, one B-ARR gene, and six A-ARR genes. CK rapidly activates the expression of A-type ARRs, a family of genes considered to be negative feedback inhibitors of the CK response. It is worth emphasizing that the expression of AHP was significantly upregulated at 8d but was then downregulated. B-type ARR can promote the transduction of CK response. The gene Cluster-49725.0 of B-ARR was strongly activated only at 8d, while Cluster-68036.0 was significantly activated only at 30d.
A total of 36 DEGs were involved in the ABA signaling pathway. The related DEGs included seven PYR/PYL (abscisic acid receptor PYR/PYL family), 11 PP2C (protein phosphatase 2C), eight SNrk2 (serine/threonine protein kinase SRK2), and eight ABF (ABA response element binding factor) (Figure 4C). Among them, two SNrk2 genes and two PP2C genes were significantly upregulated at 25d. Most of the PYR/PYL family genes were significantly upregulated at 30d. Only one ABF family gene (Cluster-98067.3) showed q higher expression at 15d.
A total of 42 DEGs were involved in the JA signaling pathway, and the related DEGs involved seven JAR1 (jasmonic acid-amino synthetase), seven COI1 (jasmonic acid signaling receptor and coronin insensitive protein 1), 19 JAZ (jasmonate ZIM structural domain protein), seven MYC2 (transcription factor MYC2), and others (Figure 4D). Most genes exhibited their highest expression at 0d and showed a gradually decreasing trend. Among the JAR1 family genes, Cluster-70814.8 was significantly upregulated at 30d, the JAZ gene Cluster-45192.15 was most significantly expressed at 8d, and Cluster-32815.11 was most abundantly expressed at 30d.
3.4.3 Analysis of key differentially expressed genes involved in plant hormone biosynthesis pathways
Gene-level analysis of plant hormone metabolic pathways revealed 487 shared DEGs associated with hormone synthesis pathways during callus development of P. ternata in hydroponically grown cuttings of P. ternata. These included DEGs involved in the auxin, CK, JA, and ABA biosynthesis pathways.
A total of 126 DEGs were involved in the tryptophan metabolic pathway of auxin biosynthesis, accounting for the largest proportion of all hormone synthesis-related DEGs. The auxin-related DEGs included TDC (L-tryptophan decarboxylase), TAA1 (L-tryptophan–pyruvate aminotransferase), YUCCA (indole-3-pyruvate monooxygenase), ALDH [aldehyde dehydrogenase (NAD+)], AMIE (amidase), and others (Figure 5A). Among them, three TDC genes, four TAA1 genes, and one YUCCA gene were significantly upregulated at the 8d callus stage. One TDC gene, two TAA1 genes, and four AMIE genes were significantly expressed at 15d compared with 0d. Five AMIE genes, one YUCCA gene, and two ALDH genes were highly upregulated at the 25d callus stage.one TDC gene, one TAA1 gene, one YUCCA gene, and two ALDH genes were highly upregulated at 30d.
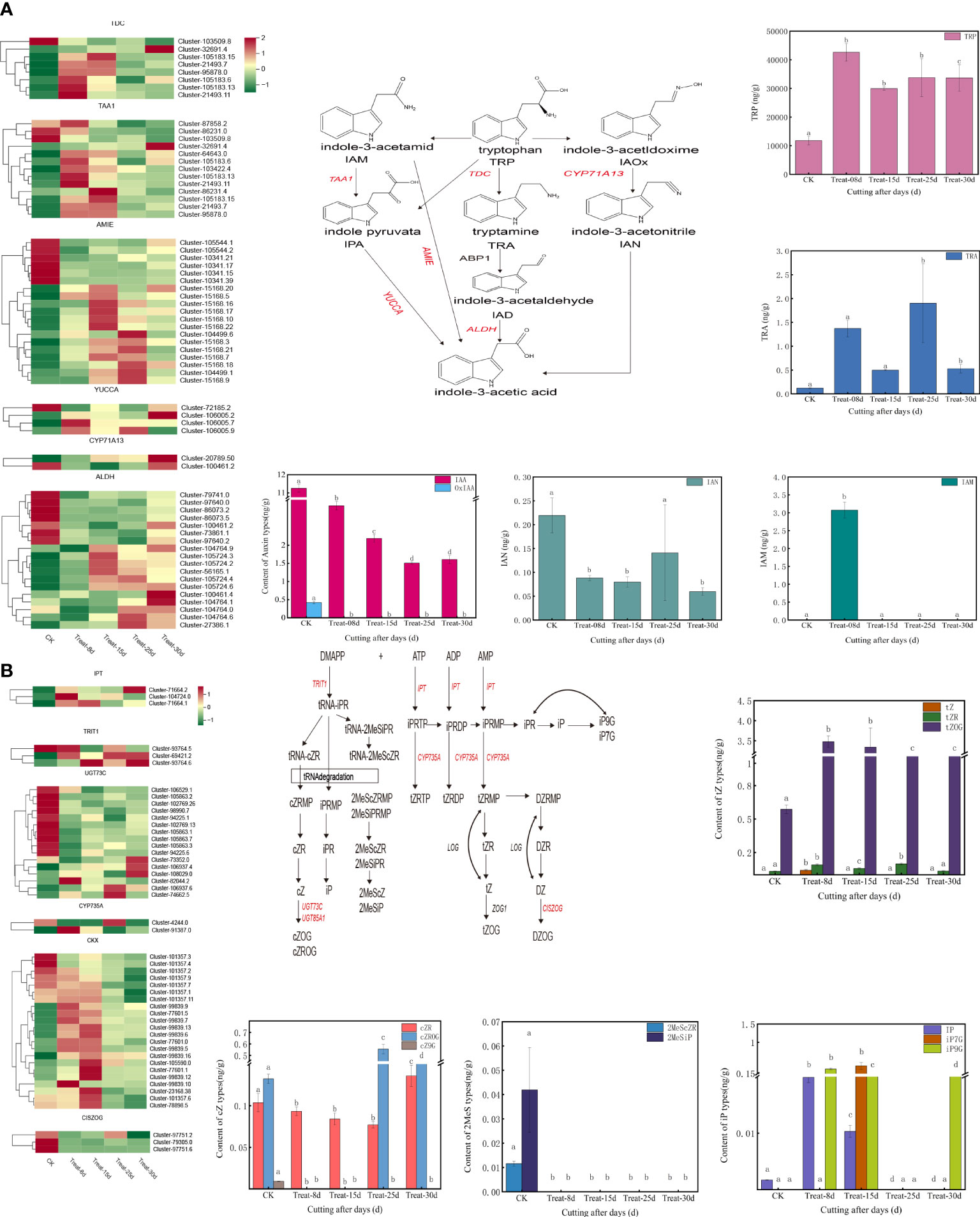
Figure 5 (A) Changes in expression of genes and metabolites of the auxin biosynthesis pathway. (B) Changes in the expression of genes and metabolites of the cytokinin biosynthetic pathway.
A total of 84 DEGs are involved in the CK biosynthetic pathway, with DEGs including IPT [adenylate dimethylallyl transferase (cytokinin synthase)], CYP735A (cytokinin trans-hydroxylase), TRIT1 (tRNA dimethylallyltransferas), UGT73C (UDP-glucosyltransferase 73C), CKX (cytokinin dehydrogenase), CISZOG (cis-zeatin O-glucosyltransferase), and others (Figure 5B). Among them, one IPT gene, one TRIT1 gene, one UGT73C gene, one CYP735A gene, and one CKX gene were significantly upregulated at 8d callus stage. One TRIT1 gene and six CKX genes were significantly activated at 15d. Two UGT73C genes and one CYP735A gene were significantly upregulated at 25d. One IPT gene, two TRIT1 gene, and three UGT73C genes were significantly induced at 30d. One IPT gene, two TRIT1 genes, and three UGT73C genes were significantly upregulated at 30d. Notably, the TRIT1 gene (Cluster-69421.2) remained highly expressed at 25d and 30d.
The jasmonic acid synthesis pathway involved in 109 DEGs. They included LOX2S (lipoxygenase), AOS (hydroperoxide dehydratase), AOC (allene oxide cyclase), OPR (12-oxophytodienoic acid reductase), OPCL1 (OPC-8:0 CoA ligase 1), ACOX1/ACOX3 (acyl-CoA oxidase), JMT (jasmonate O-methyltransferase), and others (Figure 6A). The expression of six LOX2S genes, eight AOC genes, and two OPR genes were significantly upregulated at 8d. Four AOS genes were significantly upregulated at 15d. Three LOX2S genes, one OPR gene, and four ACOX1/ACOX3 genes were significantly upregulated at 25d. One OPR gene and one OPCL1 gene were significantly increased at 30d. Three LOX2S genes, one OPR gene, and four ACOX1/ACOX3 genes were significantly upregulated at 25d.
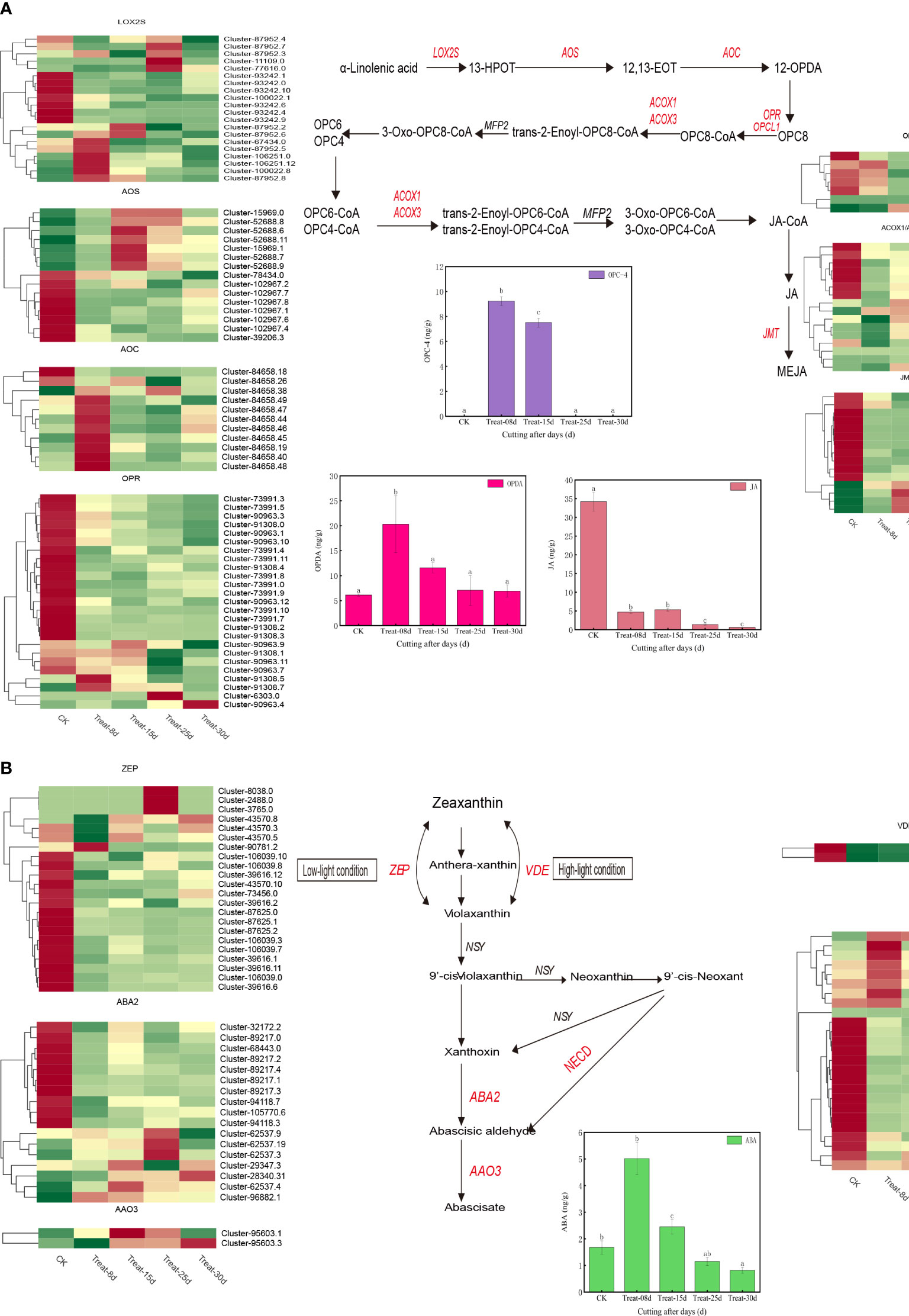
Figure 6 (A) Changes in expression of genes and metabolites of the auxin biosynthesis pathway. (B) Changes in the expression of genes and metabolites of the cytokinin biosynthetic pathway.
Finally, 68 DEGs were involved in the ABA biosynthetic pathway and included the ZEP/ABA1 (zeaxanthin epoxidase), VDE (violaxanthin de-epoxidase), NECD (9-cis-epoxycarotenoid dioxygenase) ABA2 (xanthoxin dehydrogenase), AAO3 (abscisic-aldehyde oxidase), and others (Figure 6B). Among them, one ZEP gene, three NECD genes were significantly activated at 8d. One AAO3 gene was significantly upregulated at 15d. Three ZEP genes, one NECD gene, two ABA2 genes were significantly induced at 25d and one AAO3 gene was significantly induced at 30d.
3.4.4 Analysis of transcription factors during callus formation
The TF (transcription factor) expression levels were further analyzed during the callus formation process, as TF-mediated regulation is crucial for all stages of callus development. We identified 2,659 TF genes from all DEGs. Based on the TF gene expression levels, nine TF families with high expression were further analyzed, mainly including bHLH (basic helix-loop-helix basic helix-loop-helix, 169 members), AP2/ERF-ERF (ethylene response factor, 190 members), C2H2 (cysteine-2/histidine-2, 90members), WARK (WRKY transcription factor, 141 members), CH3 (54 members), NAC (87 members), bZIP (59 members), MYB-related (myeloblastosis related, 81 members), and AUX/IAA (auxin-responsive protein IAA, 102 members).
We identified 67 bHLH transcription factors genes during the callus formation (Figure 7A), including K16189-photosensitive pigment signaling interactions factor (PIF4), K13422-transcriptional factor MYC2, K1212-transcriptional factor MYC2, K1212-transcriptional protein IAA, and K1212-transcriptional protein IAA factor MYC2, K12126-photosensitive pigment interaction factor 3 (PIF3), and others. Only one PIF4 was strongly expressed at 15d. The MYC2 TF (Cluster-79911.2) was strongly activated at 25d. Among AP2/ERF-ERF, seven genes were identified (Figure 7B), of which two TFs were strongly activated at 8d, especially the K14516-ethylene response transcription factor 1 (ERF1). In C2H2 TF family (Figure 7C), two genes were found to be associated with K13463-coronin insensitive protein 1 (COI1). Nineteen bZIP genes were found (Figure 7D), including the K14432-ABA response element binding factor (ABF) and K14431-transcription factor TGA. One hundred ten AUX/IAA genes were identified (Figure 7E), including the K14486-auxin response factor (ARF), K14484-auxin-responsive protein IAA (IAA), most of which were significantly expressed in control (CK). Additionally, four ARF TFs were significantly highly expressed at 8d, and only one was significantly upregulated at 15d. Moreover, four IAA TFs were upregulated at 8d, and two TFs were significantly upregulated at 30d.
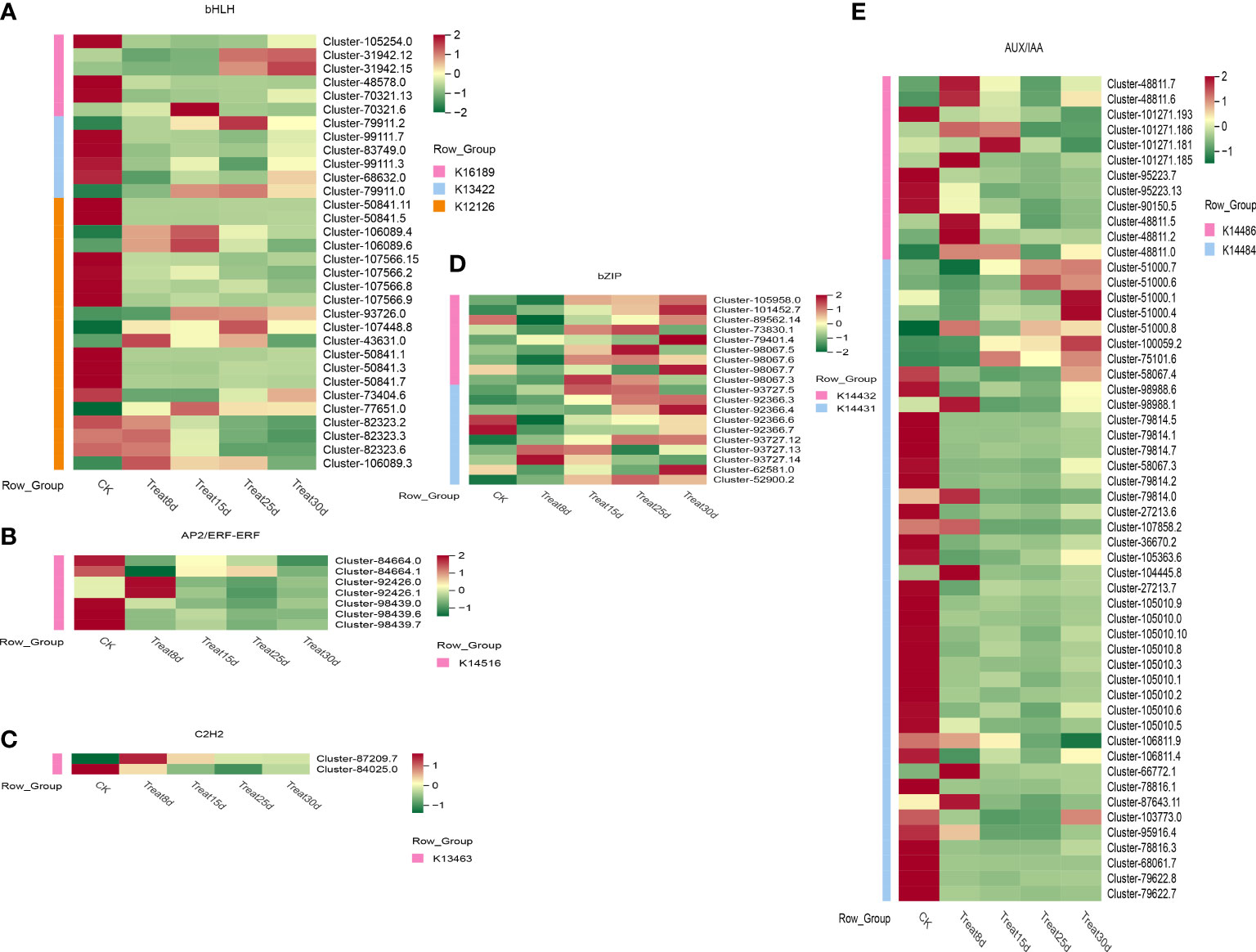
Figure 7 Changes in gene expression in phytohormone signaling in developing callus. (A) Expression levels of the bHLH TFs family differentially expressed genes (DEGs). (B) Expression levels of the AP2/ERF-ERF TFs family DEGs. (C) Expression levels of the C2H2 TFs family DEGs. (D) Expression levels of the bZIP TFs family DEGs. (E) Expression levels of the AUX/IAA TFs family DEGs.
3.5 Quantitative polymerase chain reaction expression analysis of genes involved in plant hormone biosynthesis and signaling pathways
To experimentally confirm the expression of the unigenes identified by sequencing and computational analysis, seven representative DEGs related to plant hormone synthesis and signaling pathways including AUX/IAA, ARF, A-ARR, COI1, JAZ, AOC, and OPCL1 were selected for qRT-PCR analysis across the five sequential developmental stages in P. ternata hydroponic cuttings.
Based on the analyzed qRT-PCR data (Supplementary Figure S7; Supplementary Table S9), these DEGs were expressed at varying levels in different stages. Moreover, the qRT-PCR results were generally consistent with the RNA-seq data, indicating transcriptome analysis the reliability. These results also suggest that the evaluated genes potentially play important roles in plant hormone synthesis, signal transduction and distribution, which lead to callus induction.
4 Discussion
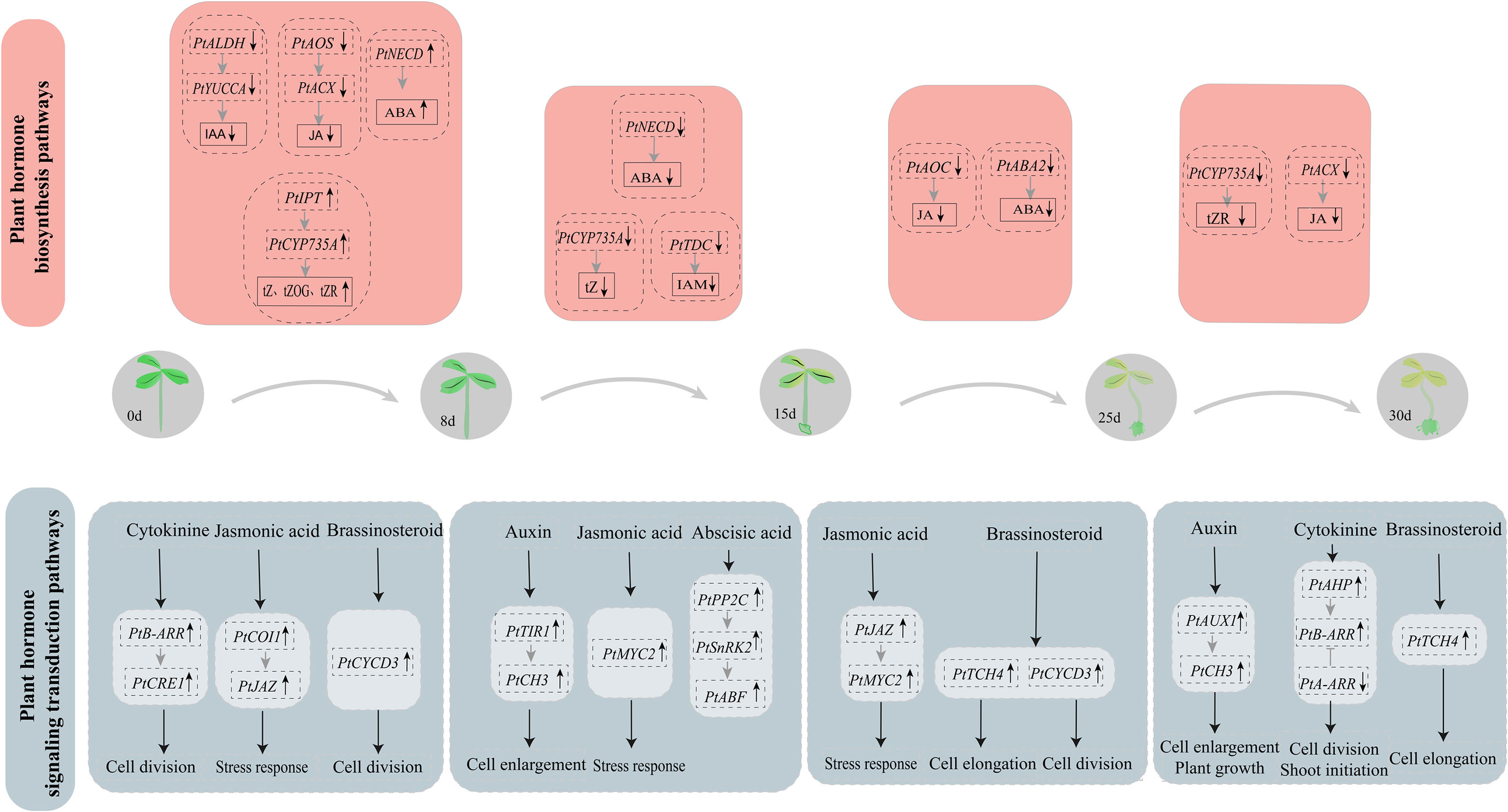
Our group has developed the first efficient cultivation technique of “hydroponic cuttings from P. ternata” using liquid as the cultivation medium. Callus induction is the key link of this technique, but the mechanism of callus formation and development has thus far remained unclear. Therefore, in order to further investigate the molecular mechanisms of callus formation and development in hydroponic cuttings, we provided P. ternata hormone metabolism and transcriptome data. These data included the hormone metabolic compounds and DEGs involved in hormone metabolism and signal transduction pathways of the callus in the hydroponic cuttings at different stages (Figure 8).
The process of callus development is closely linked to its endogenous hormone homeostasis and dynamic changes. We found that CKs, auxin, JA, and ABA-type derivatives are present mainly in biologically active form throughout callus development from hydroponic cuttings (Figures 2, 3; Supplementary Figures S3, S4). This confirms that callus development is a highly dynamic process under hormonal control (Perez-Garcia and Moreno-Risueno, 2018). All hormones reached high concentrations throughout callus development, with a major peak occurring at 15d and a lower peak in concentration at 8d (Supplementary Figure S8). This also indicates again that endogenous hormones play an indispensable role in the early stages of callus formation (Perez-Garcia and Moreno-Risueno, 2018).
In previous studies, auxin was shown to play an important role in tissue culture, promoting callus formation and adventitious root formation in cuttings (Che et al., 2002). The content of IAA increased 1–3 days after the flowering stems of Arabidopsis were cut (Cui et al., 2021). However, in this study, we showed that auxin content was regulated by ALDH, YUCCA, and TAA1, gradually decrease during callus formation and during callus proliferation, indicating that low auxin concentrations were beneficial to callus formation as well as its development. In terms of auxin signal activation, three major primary response gene families were induced: AUX/IAA genes, GH3 genes (Hayashi et al., 2021), and SAUR genes (Che et al., 2002). The AUX/IAA family members are short-lived nuclear proteins that form homo- and heterodimers with other AUX/IAA and ARF genes and repress transcription of downstream auxin response genes (Chen et al., 2019). Eight AUX/IAA genes were differentially expressed in P. ternata cuttings during callus formation, and only one was significantly expressed at 30d. The SAUR family is the largest known family of early auxin-inducible genes (Ren and Gray, 2015), and OsSAUR39 has been reported to regulate auxin biosynthesis and transport negatively. Interestingly, one SAUR gene was differentially expressed at 30d late in P. ternata callus formation, suggesting that this gene facilitates late callus differentiation to form shoots. Therefore, their role in this process needs further investigation. Several auxin biosynthetic genes regulated the decreasing IAA concentration at the gene expression level. Overall, the low level of IAA is beneficial to the formation of calli.
CKs are essential in grafted plants, especially in callus formation and vascular system development. Notably, CK responses were elevated in Arabidopsis explants (Iwase et al., 2011). In contrast, CKs regulate plant growth and development through a two-component regulatory system (Che et al., 2002). CKs were found to control the shoot meristem development by stimulating the upregulation of A-ARR genes and suppressing WUS expression. In addition, CRE1, AHP, and B-ARR act as upstream regulators of A-ARR (Buechel et al., 2010). One CRE1 gene and five A-ARR genes were significantly upregulated during P. ternata callus induction, suggesting they play an important role in CK signaling and callus formation. The hormone metabolism levels gradually increased for most of the CKs and were regulated by several biosynthetic pathway genes, such as IPT, CYP735A, and CKX. However, cZROG, which may be regulated by the UGT73C gene, decreased during the early callus formation stage and then increased at the differentiation stage of shoot formation. Moreover, cZROG may be a key factor in shoot formation. In conclusion, CK is the core hormone controlling the formation of calli in P. ternata hydroponic cuttings.
JA is well-known as a stress-induced hormone. Previous studies found that JA prevents plant growth (Yan et al., 2007) and inhibits cell proliferation and expansion to counter-regulate organ development (Noir et al., 2013). In the present study, three JAZ genes were significantly upregulated. The level of JA inhibition was low during the process of P. ternata callus formation, indicating that low concentrations of JA can promote rapid callus proliferation and development, consistent with the results of garlic callus proliferation studies (Mostafa et al., 2020). ABA negatively regulates plant growth by inhibiting cell proliferation under stress conditions (Wang et al., 1998). The ABA levels, and key genes for ABA synthesis, were regulated at 8d, suggesting ABA plays an important role in the rapid P. ternata callus proliferation, contrary to previous studies.
Plant endogenous hormones form a complex signaling network that affects cell growth and development by regulating key genes for cell proliferation (Ahuja, 1965; Hofhuis et al., 2013). Plants can respond rapidly to external stimuli, leading to changes in gene transcriptional levels. Moreover, a large number of transcription factors are responsive to external stresses. We identified a differential expression of transcription factors such as PIF, MYC, ERF, TGA, and ARF was found, suggesting they may be involved in gene network regulation during P. ternata callus formation from hydroponic cuttings. PIF4 can positively regulate elongation growth in plants by regulating the expression of auxin genes encoding, affecting downstream signal transduction pathways (Sun et al., 2013). In the present study, early callus grew faster, and the expression of PIF TFs was suppressed, resulting in decrease IAA content. ARF transcription factors are key components in the auxin signaling pathway and repress or activate the expression of downstream genes by binding to upstream response elements of auxin response genes. Eight ARF genes were significantly upregulated in the callus at the 8d stage, suggesting their potential involvement in callus induction and development. MYC transcription factors are JA-related gene expression activators or repressors that regulate plant growth and development (Aleman et al., 2016). ERF transcription factors occur in plants and are involved in plant responses to biotic and abiotic stresses (Ding et al., 2021). In Cercis canadensis, taxol biosynthesis is regulated by ERF transcription factors and is dependent on JA signaling. It is possible that the expression of MYC TFs was repressed, resulting in reduced JA content. TGA TFs are members of the bZIP family and play an important role in developing stress tolerance (Dröge-Laser et al., 2018; Wang et al., 2020; Ke et al., 2022). In Arabidopsis, TGA TFs enhance resistance by regulating the auxin signaling (Choi et al., 2010). In this study, TGA expression may be maintained at low levels to promote callus development.
5 Conclusion
In conclusion, our study evaluated the endogenous hormone content and performed transcriptome sequencing at different stages of callus of P. ternata hydroponic cuttings. Genes co-expressed in phytohormone synthesis and transduction were analyzed at each stage and were combined with hormone metabolite data and transcriptomics to reveal the molecular mechanism of callus formation from P. ternata hydroponic cuttings. CKs were the hormones with the highest concentration during the whole callus development. In the early stage of callus formation (8d), CK levels were gradually increased through the regulation of IPT and CYP735A genes. AHP positively regulated B-ARRs and CRE1, promoting CK signaling and facilitating callus cell differentiation. The brassinosteroid signaling gene Cyclin D3 (CYCD3) was upregulated, facilitating the initiation of callus cell differentiation. The B-ARR gene promoted cell differentiation and thus induced callus formation through regulation of the ALDH and YUCCA genes, which reduced the IAA levels, facilitating the formation and development of callus. AOS, ACX negatively regulated JA synthesis, reducing JA levels, and promoting callus formation in the cutting wound site. Moreover, COI1 and JAZ positively regulated JA signaling and promoted plant stress and wound responses at the wound site. During the middle and late stages of callus formation (15d, 25d, and 30d), the endogenous hormone content in the callus was decreasing. However, specific hormone signaling functions positively regulated the formation and development of the callus. At 15d, the auxin signaling genes TIR1 and CH3 were upregulated, promoting callus cell proliferation. The upregulation of MYC transcription factors involved in JA signaling also indicated that JA regulated the response to mechanical damage of P.ternata hydroponic cuttings. At 25d, the brassinosteroid signaling genes TCH4 and CYCD3 were upregulated, promoting the differentiation and proliferation of callus cells. At 30d, the auxin-signaling genes AUX1 and CH3 positively regulated callus development and promoted cell proliferation and plant growth. The CK signaling genes AHP and B-ARR were upregulated to promote continuous cell differentiation and shoot initiation. Plant endogenous hormones are particularly important for callus formation and development. The results of this study elucidate for the first time the molecular mechanism of the callus formation process in P. ternata cuttings grown in hydroponic, providing new insights and approaches for expanding P. ternata resources and propagation practices.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA942460.
Author contributions
XD, HC, FW and YH designed the experiments and contributed to writing and revising the manuscript. XD and LC performed the experiments. XD and LC analyzed the data. YL supervised this study. All authors contributed to this paper and have approved the submission.
Funding
This work was supported by Science & Technology Department of Sichuan Province in China (Nos. 2021YFN0109).
Acknowledgments
This research would like to acknowledge Wuhan Servicebio Technology Co., Ltd (Wuhan, China) for kind assistance of technical support of Morphological and histological analysis , acknowledge Wuhan Metware Biotechnology Co., Ltd. (Wuhan, China) for kind assistance of technical support of Plant hormone determination and transcriptomic sequencing and Beijing Tsingke Biotech Co., Ltd. (Beijing, China) for kind assistance of technical support of q-PCR assay.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1189499/full#supplementary-material
Footnotes
- ^ UniProtKB/Swiss-Prot - SIB Swiss Institute of Bioinformatics | Expasy, n.d
- ^ http://www.metware.cn/
- ^ https://github.com/trinityrnaseq/trinityrnaseq
- ^ https://www.kegg.jp/kegg/kegg1.html
- ^ http://www.omicshare.com/tools
- ^ https://bioconductor.org/packages/DESeq2/, version3.10
References
Ahuja, M. R. (1965). Genetic control of tumor formation in higher plants. Q. Rev. Biol. 40, 329–340. doi: 10.1086/404744
Aleman, F., Yazaki, J., Lee, M., Takahashi, Y., Kim, A. Y., Li, Z., et al. (2016). An ABA-increased interaction of the PYL6 ABA receptor with MYC2 transcription factor: a putative link of ABA and JA signaling. Sci. Rep. 6, 28941. doi: 10.1038/srep28941
Bai, J., Qi, J., Yang, L., Wang, Z., Wang, R., Shi, Y. (2022). A comprehensive review on ethnopharmacological, phytochemical, pharmacological and toxicological evaluation, and quality control of pinellia ternata (Thunb.) breit. J. Ethnopharmacol. 298, 115650. doi: 10.1016/j.jep.2022.115650
Birnbaum, K. D., Alvarado, A. S. (2008). Slicing across kingdoms: regeneration in plants and animals. Cell 132, 697–710. doi: 10.1016/j.cell.2008.01.040
Buechel, S., Leibfried, A., To, J. P. C., Zhao, Z., Andersen, S. U., Kieber, J. J., et al. (2010). Role of a-type ARABIDOPSIS RESPONSE REGULATORS in meristem maintenance and regeneration. Eur. J. Cell Biol. 89, 279–284. doi: 10.1016/j.ejcb.2009.11.016
Che, P., Gingerich, D. J., Lall, S., Howell, S. H. (2002). Global and hormone-induced gene expression changes during shoot development in arabidopsis. Plant Cell 14, 2771–2785. doi: 10.1105/tpc.006668
Chen, L., Tong, J., Xiao, L., Ruan, Y., Liu, J., Zeng, M., et al. (2016). YUCCA -mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in arabidopsis. EXBOTJ 67, 4273–4284. doi: 10.1093/jxb/erw213
Chen, L., Zheng, X., Guo, X., Cui, Y., Yang, H. (2019). The roles of Aux/IAA gene family in development of Dendrocalamus sinicus (Poaceae: Bambusoideae) inferred by comprehensive analysisand expression profiling. Molecular Biol. Reports 46, 1625–1634. doi: 10.1007/s11033-019-04611-2
Choi, J., Huh, S. U., Kojima, M., Sakakibara, H., Paek, K.-H., Hwang, I. (2010). The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in arabidopsis. Dev. Cell 19, 284–295. doi: 10.1016/j.devcel.2010.07.011
Chu, Z., Chen, J., Sun, J., Dong, Z., Yang, X., Wang, Y., et al. (2017). De novo assembly and comparative analysis of the transcriptome of embryogenic callus formation in bread wheat (Triticum aestivum l.). BMC Plant Biol. 17, 244. doi: 10.1186/s12870-017-1204-2
Cui, Q., Xie, L., Dong, C., Gao, L., Shang, Q. (2021). Stage-specific events in tomato graft formation and the regulatory effects of auxin and cytokinin. Plant Sci. 304, 110803. doi: 10.1016/j.plantsci.2020.110803
Creelman, Tierney, M. L., Mullet, J. E. (1992). Jasmonic acid/methyl jasmonate accumulate in wounded soybean hypocotyls and modulate wound gene expression. Proc. Natl. Acad. Sci. USA 89, 4938–1.
Dai, X., Liu, N., Wang, L., Li, J., Zheng, X., Xiang, F., et al. (2020). MYB94 and MYB96 additively inhibit callus formation via directly repressing LBD29 expression in arabidopsis thaliana. Plant Sci. 293, 110323. doi: 10.1016/j.plantsci.2019.110323
Dammann, C., Rojo, E., Sanchez-Serrano, J. J. (1997). Abscisic acid and jasmonic acid activate wound-inducible genes in potato through separate, organ-specific signal transduction pathways. Plant J. 11, 773–782. doi: 10.1046/j.1365-313X.1997.11040773.x
Ding, A., Tang, X., Yang, D., Wang, M., Ren, A., Xu, Z., et al. (2021). ERF4 and MYB52 transcription factors play antagonistic roles in regulating homogalacturonan de-methylesterification in arabidopsis seed coat mucilage. Plant Cell 33, 381–403. doi: 10.1093/plcell/koaa031
Dröge-Laser, W., Snoek, B. L., Snel, B., Weiste, C. (2018). The arabidopsis bZIP transcription factor family [[/amp]]mdash; an update. Curr. Opin. Plant Biol. 45, 36–49. doi: 10.1016/j.pbi.2018.05.001
Fan, M., Xu, C., Xu, K., Hu, Y. (2012). LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in arabidopsis regeneration. Cell Res. 22, 1169–1180. doi: 10.1038/cr.2012.63
Ferdausi, A., Chang, X., Jones, M. (2021). Transcriptomic analysis for differential expression of genes involved in secondary metabolite production in narcissus pseudonarcissus field derived bulb and in vitro callus. Ind. Crops Products 168, 113615. doi: 10.1016/j.indcrop.2021.113615
Floková, K., Tarkowská, D., Miersch, O., Strnad, M., Wasternack, C., Novák, O. (2014). UHPLC–MS/MS based target profiling of stress-induced phytohormones. Phytochemistry 105, 147–157. doi: 10.1016/j.phytochem.2014.05.015
Hayashi, K., Arai, K., Aoi, Y., Tanaka, Y., Hira, H., Guo, R., et al. (2021). The main oxidative inactivation pathway of the plant hormone auxin. Nat. Commun. 12, 6752. doi: 10.1038/s41467-021-27020-1
Hofhuis, H., Laskowski, M., Du, Y., Prasad, K., Grigg, S., Pinon, V., et al. (2013). Phyllotaxis and rhizotaxis in arabidopsis are modified by three PLETHORA transcription factors. Curr. Biol. 23, 956–962. doi: 10.1016/j.cub.2013.04.048
Ikeuchi, M., Iwase, A., Rymen, B., Lambolez, A., Kojima, M., Takebayashi, Y., et al. (2017). Wounding triggers callus formation via dynamic hormonal and transcriptional changes. Plant Physiol. 175, 1158–1174. doi: 10.1104/pp.17.01035
Ikeuchi, M., Ogawa, Y., Iwase, A., Sugimoto, K. (2016). Plant regeneration: cellular origins and molecular mechanisms. Development 143, 1442–1451. doi: 10.1242/dev.134668
Ikeuchi, M., Sugimoto, K., Iwase, A. (2013). Plant callus: mechanisms of induction and repression. Plant Cell 25, 3159–3173. doi: 10.1105/tpc.113.116053
Iwase, A., Harashima, H., Ikeuchi, M., Rymen, B., Ohnuma, M., Komaki, S., et al. (2017). WIND1 promotes shoot regeneration through transcriptional activation of ENHANCER OF SHOOT REGENERATION1 in arabidopsis. Plant Cell 29, 54–69. doi: 10.1105/tpc.16.00623
Iwase, A., Mitsuda, N., Koyama, T., Hiratsu, K., Kojima, M., Arai, T., et al. (2011). The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in arabidopsis. Curr. Biol. 21, 508–514. doi: 10.1016/j.cub.2011.02.020
Ke, D., He, Y., Fan, L., Niu, R., Cheng, L., Wang, L., et al. (2022). The soybean TGA transcription factor GmTGA13 plays important roles in the response to salinity stress. Plant Biol. J. 24, 313–322. doi: 10.1111/plb.13360
Kong, X., Yang, M., Khalid Abbas, H. M., Wu, J., Li, M., Dong, W. (2018). Antimicrobial genes from Allium sativum and Pinellia ternate revealed by a Bacillus subtilis expression system. Scientific Reports 8, 14514. doi: 10.1038/s41598-018-32852-x
Kumari, A., Ray, K., Sadhna, S., Pandey, A. K., Sreelakshmi, Y., Sharma, R. (2017). Metabolomic homeostasis shifts after callus formation and shoot regeneration in tomato. PloS One 12, e0176978. doi: 10.1371/journal.pone.0176978
Li, Y., Zhou, C., Yan, X., Zhang, J., Xu, J. (2016). Simultaneous analysis of ten phytohormones in sargassum horneri by high-performance liquid chromatography with electrospray ionization tandem mass spectrometry. J. Sep Sci. 39, 1804–1813. doi: 10.1002/jssc.201501239
Mao, R., He, Z. (2020). Pinellia ternata (Thunb.) breit: a review of its germplasm resources, genetic diversity and active components. J. Ethnopharmacol. 263, 113252. doi: 10.1016/j.jep.2020.113252
Mostafa, H. H. A., Wang, H., Song, J., Li, X. (2020). Effects of genotypes and explants on garlic callus production and endogenous hormones. Sci. Rep. 10, 4867. doi: 10.1038/s41598-020-61564-4
Nakayama, H., Fukushima, K., Fukuda, T., Yokoyama, J., Kimura, S. (2014). Molecular phylogeny determined using chloroplast DNA inferred a new phylogenetic relationship of <i<Rorippa aquatica</i< (Eaton) EJ palmer & steyermark (Brassicaceae)–lake cress. AJPS 05, 48–54. doi: 10.4236/ajps.2014.51008
Noir, S., Bömer, M., Takahashi, N., Ishida, T., Tsui, T.-L., Balbi, V., et al. (2013). Jasmonate controls leaf growth by repressing cell proliferation and the onset of endoreduplication while maintaining a potential stand-by mode. Plant Physiol. 161, 1930–1951. doi: 10.1104/pp.113.214908
Pēna-Cortés, H., Sánchez-Serrano, J. J., Mertens, R., Willmitzer, L., Prat, S. (1989). Abscisic acid is involved in the wound-induced expression of the proteinase inhibitor II gene in potato and tomato. Proc. Natl. Acad. Sci. U.S.A. 86, 9851–9855. doi: 10.1073/pnas.86.24.9851
Pena-Cortes, H., Willmitzer, L., Sanchez-Serrano, J. J. (1991). Abscisic acid mediates wound induction but not developmental-specific expression of the proteinase inhibitor II gene family. Plant Cell, 963–972. doi: 10.1105/tpc.3.9.963
Peng, W., Li, N., Jiang, E., Zhang, C., Huang, Y., Tan, L., et al. (2022). A review of traditional and current processing methods used to decrease the toxicity of the rhizome of pinellia ternata in traditional Chinese medicine. J. Ethnopharmacol. 299, 115696. doi: 10.1016/j.jep.2022.115696
Peng, W., Wei, D.-N., Liu, Y.-J., Zhang, M.-M., Hu, M.-B., Wu, C.-J. (2019). Comparative research of the curative effects of pinelliae rhizoma and pinelliae rhizoma praeparatum cum alumine on ovalbumin-induced allergic asthma in rats. Phcog Mag 15, 29. doi: 10.4103/pm.pm_397_18
Perez-Garcia, P., Moreno-Risueno, M. A. (2018). Stem cells and plant regeneration. Dev. Biol. 442, 3–12. doi: 10.1016/j.ydbio.2018.06.021
Ren, H., Gray, W. M. (2015). SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol. Plant 8, 1153–1164. doi: 10.1016/j.molp.2015.05.003
Sugimoto, K., Jiao, Y., Meyerowitz, E. M. (2010). Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 18, 463–471. doi: 10.1016/j.devcel.2010.02.004
Sun, J., Qi, L., Li, Y., Zhai, Q., Li, C. (2013). PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in arabidopsis. Plant Cell 25, 2102–2114. doi: 10.1105/tpc.113.112417
Wang, P., Nolan, T. M., Yin, Y., Bassham, D. C. (2020). Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis. Autophagy 16, 123–139. doi: 10.1080/15548627.2019.1598753
Wang, H., Qi, Q., Schorr, P., Cutler, A. J., Crosby, W. L., Fowke, L. C. (1998). ICK1, a cyclin-dependent protein kinase inhibitor from arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 15, 501–510. doi: 10.1046/j.1365-313x.1998.00231.x
Xu, C., Cao, H., Zhang, Q., Wang, H., Xin, W., Xu, E., et al. (2018). Control of auxin-induced callus formation by bZIP59–LBD complex in arabidopsis regeneration. Nat. Plants 4, 108–115. doi: 10.1038/s41477-017-0095-4
Yan, Y., Stolz, S., Chételat, A., Reymond, P., Pagni, M., Dubugnon, L., et al. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19, 2470–2483. doi: 10.1105/tpc.107.050708
Záveská Drábková, L., Pokorná, E., Dobrev, P. I., Kůrková, J., Steinbachová, L., Honys, D., et al. (2021). Hormonome dynamics during microgametogenesis in different nicotiana species. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.735451
Keywords: P. ternate, histomorphological, phytohormone, transcriptome, plant hormone signaling and synthesis pathways
Citation: Duan X, Chen L, Liu Y, Chen H, Wang F and Hu Y (2023) Integrated physicochemical, hormonal, and transcriptomic analysis reveals the underlying mechanism of callus formation in Pinellia ternata hydroponic cuttings. Front. Plant Sci. 14:1189499. doi: 10.3389/fpls.2023.1189499
Received: 19 March 2023; Accepted: 30 May 2023;
Published: 20 June 2023.
Edited by:
Wenying Zhang, Yangtze University, ChinaReviewed by:
Prakash Babu Adhikari, Huazhong Agricultural University, ChinaAbdullah Shalmani, Huazhong Agricultural University, China
Copyright © 2023 Duan, Chen, Liu, Chen, Wang and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongping Chen, Y2hlbi1ob25ncGluZ0AxMjYuY29t; Fu Wang, d2FuZ2Z1Ym95MjAxOUAxMjYuY29t; Yuan Hu, aHV5dWFuQGNkdXRjbS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Xiaoqing Duan
Xiaoqing Duan Lin Chen
Lin Chen Youping Liu
Youping Liu Hongping Chen
Hongping Chen Fu Wang
Fu Wang