
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 27 June 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1187605
This article is part of the Research Topic Plant Signaling in Response to Environmental Stresses View all 7 articles
Introduction: With the warming global climate, drought stress has become an important abiotic stress factor limiting plant growth and crop yield. As the most rapidly drought-sensing organs of plants, roots undergo a series of changes to enhance their ability to absorb water, but the molecular mechanism is unclear.
Results and methods: In this study, we found that PLT1 and PLT2, two important transcription factors of root development in Arabidopsis thaliana, are involved in the plant response to drought and are inhibited by BR signaling. PLT1- and PLT2-overexpressing plants showed greater drought tolerance than wild-type plants. Furthermore, we found that BZR1 could bind to the promoter of PLT1 and inhibit its transcriptional activity in vitro and in vivo. PLT1 and PLT2 were regulated by BR signaling in root development and PLT2 could partially rescue the drought sensitivity of bes1-D. In addition, RNA-seq data analysis showed that BR-regulated root genes and PLT1/2 target genes were also regulated by drought; for example, CIPK3, RCI2A, PCaP1, PIP1;5, ERF61 were downregulated by drought and PLT1/2 but upregulated by BR treatment; AAP4, WRKY60, and AT5G19970 were downregulated by PLT1/2 but upregulated by drought and BR treatment; and RGL2 was upregulated by drought and PLT1/2 but downregulated by BR treatment.
Discussion: Our findings not only reveal the mechanism by which BR signaling coordinates root growth and drought tolerance by suppressing the expression of PLT1 and PLT2 but also elucidates the relationship between drought and root development. The current study thus provides an important theoretical basis for the improvement of crop yield under drought conditions.
With global warming, reduced rainfall, and other factors, drought has become one of the most serious abiotic stresses. Drought causes a series of impairments to the physiological, biochemical, and metabolic processes of plants, thereby causing plant growth retardation, cell damage, declines in crop yield and quality, and even plant death (Blum, 2009; Hu and Xiong, 2014; Lorts and Lasky, 2020). Therefore, studying the mechanism by which plants respond to drought is essential for the enhancement of plant drought tolerance and the improvement of crop yield.
Being sessile, plants have root systems that enable them to anchor themselves in the soil and obtain nutrients and water from their environments. Thus, the roots are important organs in the initial response to drought stress (Koevoets et al., 2016; Dinneny, 2019; Kim et al., 2020; Qin et al., 2022). When the roots of plants sense drought stress, their root tip stem cells, meristems, and vascular tissues coordinate with each other to affect the morphological structure of the roots and ultimately increase the area or length of the roots in order to enhance the root water absorption capacity (Gupta et al., 2020). Recent studies have found that drought, if not too severe, enhances root growth, which allows plants to absorb water from deeper soils, and roots with smaller diameters and greater lengths have larger surface areas in contact with water (Xu et al., 2013; Gupta et al., 2020; Kim et al., 2020). However, whether the components regulating root length are involved in the drought stress response is unclear. Furthermore, the molecular mechanism by which roots perceive drought and its downstream signaling pathway still needs further exploration.
Root length is determined by cell number and cell size. The root apical meristem produces a series of cells that can elongate and differentiate after multiple divisions. Therefore, the activity of the meristem is a key factor determining root length (Beemster and Baskin, 1998; Petricka et al., 2012). The PLETHORAs (PLTs), a class of transcription factors containing the APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) domain, play an important role in root development (Aida et al., 2004; Galinha et al., 2007; Mähönen et al., 2014). Among them, PLT1 and PLT2 are highly homologous. When the two genes encoding these proteins are mutated, the root length of the plant is severely shortened, and the number of apical meristem cells is reduced; thus, the activity of PLT1 and PLT2 is crucial for the maintenance of the apical meristem (Aida et al., 2004; Galinha et al., 2007). However, whether and how these two genes participate in the drought stress response remains unknown.
Brassinosteroids (BRs) play important roles in several plant life processes, including cell elongation and division, photomorphogenesis, plant senescence, and responses to various biological and abiotic stresses (Clouse, 1998; Müssig et al., 2003; Clouse, 2011; Yang et al., 2011). BR can be recognized by the membrane receptor BRASSINOSTEROID INSENSITIVE 1 (BRI1), which promotes the reciprocal phosphorylation of BRI1 with the coreceptor BRI1-ASSOCIA TED RECEPTOR KINASE 1 (BAK1/SERK3), inhibits the activity of the negative regulator GSK3/Shaggy-like kinase (BRINSENSITIVE2, BIN2) and promotes the degradation of BIN2 (Vert and Chory, 2006; Peng et al., 2008). Thus, BIN2 is unable to phosphorylate the downstream transcription factors BRASSINAZOLE RESISTANT 1 (BZR1) and BRI1-EMS-SUPPRESSOR 1 (BES1). Phosphorylated BZR1/BES1 is dephosphorylated by PROTEIN PHOSPHATASE 2A (PP2A) and binds to downstream target genes to regulate various growth and developmental processes in plants (Sun et al., 2010; Tang et al., 2011; Yu et al., 2011; Kono and Yin, 2020).
BRs are important participants in both root development and drought stress response (González-García et al., 2011; Chaiwanon and Wang, 2015; Nolan et al., 2017; Ye et al., 2017). BR can influence the size of the root meristem by regulating the self-renewal ability of the quiescent center (QC) and the expression of cell cycle-related genes (Vilarrasa-Blasi et al., 2014; Planas-Riverola et al., 2019). Interestingly, depending on the plant species and the BR signaling- or biosynthesis-related genes studied, BR has been found to play both positive and negative roles in regulating plant drought response (Sahni et al., 2016; Ye et al., 2017; Fàbregas et al., 2018; Cui et al., 2019; Nie et al., 2019). The paradoxical results observed in these studies suggest that BR signaling regulates plant drought response via multiple mechanisms. Therefore, identifying the tissue- and plant-specific drought-regulating components, such as root-specific drought-regulating proteins, that can be controlled via the BR signaling pathway is essential to understanding the role of BR signaling in regulating plant drought resistance.
In this study, to reveal the relationship between drought stress and plant root growth and development, we first explored whether PLT1 and PLT2 are involved in the drought response. We found that the drought tolerance of PLT1- and PLT2-overexpressing plants was significantly enhanced compared with that of the wild type, indicating that these genes might involved in the response to drought. We also found that BZR1 could directly bind to the promoter of PLT1 and repress its expression. BR application reduced the expression levels of PLT1 and PLT2 in roots, but the PLT2-overexpressing plants were less BR-sensitive than the wild-type plants during primary root growth. In contrast, the double mutant plt1plt2 had enhanced sensitivity to BR, suggesting that PLT1 and PLT2 are regulated by the BR signaling pathway and function redundantly. Genetic experiments revealed that PLT2 overexpression suppressed the drought-sensitive phenotype of bes1-D, suggesting that PLT2 acts downstream of the BR signaling pathway to regulate the drought response. Further RNA-seq analysis showed that some drought- and BR-related genes were coregulated by PLT1/2. Our study not only reveals that the root growth regulatory factors PLT1 and PLT2 can regulate the drought stress response but also further clarifies that BR participates in root development and the drought stress response by inhibiting the expression of PLT1 and PLT2.
The Arabidopsis mutant and transgenic plants used in this study, including bzr1-1D, bes1-D, plt1 plt2, pPLT1:PLT1-YFP/Col-0, and pPLT2:PLT2-YFP/Col-0, are in the Col-0 background and have been reported previously (An et al., 2018). To check bes1-D and PLT2-OX/bes1-D in different backgrounds, we performed MspI Digest and western blot. The plants were grown at 22°C and 55% relative humidity under long-day conditions (16 h of light at 100 μmol/m2s, followed by 8 h of darkness) either in a growth chamber or in a growth room equipped with T5 4000K fluorescent tubes (Philips). Double or triple mutants generated by genetic crossing and F3-segregated homozygous plants were used for phenotypical or biochemical analysis.
The different concentrations of eBL treatment were described previously (Zhao et al., 2021). 10 days Col-0 seedlings were immersed in 1 μM eBL for 0-9 h, and roots were placed into liquid nitrogen immediately after treatment. 14 days Col-0, bzr1-1D, and bes1-D seedlings were grown with 0.25 μM PCZ, and roots were placed into liquid nitrogen immediately after treatment. Then, RNA was extracted to detect DWF4 (BR synthesis gene), PLT1, and PLT2 expression. For the drought-stress treatments, the seedlings with uniform growth were either watered as usual (control) or stopped watering for 18 days and rewatered 12 days before taking photos for phenotype comparison. Each pot is considered as one biological replicate. In general, at least 10 biological replicates collected from three to four independent experiments were used for survival rate quantification.
First-strand cDNA was synthesized from approximately 1 µg total RNA using M-MLV Reverse Transcriptase (Takara Bio Inc., Otsu, Japan). Quantitative real-time PCR was performed according to the standard protocol using a Bio-Rad CFX Connect real-time PCR machine (Bio-Rad Laboratories, Hercules, CA, United States) using the SYBR Premix Ex TaqTM system (Takara Bio Inc.). The primers used are listed in Supplementary Table S5. UBC30 as an internal reference. Averaged value from at least three biological replicates was presented.
EMSA was performed using recombinant MBP-BZR1 proteins purified from E. coli and biotin-labeled double-stranded oligonucleotide probes containing BRRE sequences chosen from the PLT1 promoter. Probe Primers used are listed in Supplementary Table S5. EMSAs was performed as described by Zhao et al. (2021) (Zhao et al., 2021).
The wild-type and mutation forms of the 1884 bp promoter sequences of PLT1 were incorporated into pGreenII 0800-LUC dual luciferase vector (Hellens et al., 2005) using LR Clonase (Invitrogen). Arabidopsis thaliana protoplast transformations were performed according to the methods described by Yang et al. (2016) (Yang et al., 2016). Relative promoter activity was calculated by dividing the PLT1 promoter-driven firefly luciferase signal by the 35S promoter-driven renilla signal which serves as the internal normalization control. The experiment has been repeated at least three times with similar results.
A one-way analysis of variance (ANOVA) or two-way ANOVA with Tukey’s honestly significant difference test was performed to evaluate the differences across sample groups or treatments. Statistically significant differences are indicated by different lowercase letters (P < 0.05, ANOVA test), one asterisk (P < 0.05, Student’s t-test), or two asterisks (P < 0.01, Student’s t-test).
To investigate whether root-specific regulatory components play an important role in the response to drought stress, we performed drought stress experiments on mutants and on plants overexpressing genes that regulate root growth and development. We found that the plants overexpressing PLT1 and PLT2 (PLT1-OX and PLT2-OX), which are important transcription factors regulating root growth and development, showed significantly higher survival rates and better drought resistance after drought treatment than the wild-type Col-0 plants (Figure 1), suggesting that PLT1 and PLT2 positively regulate the plant response to drought.
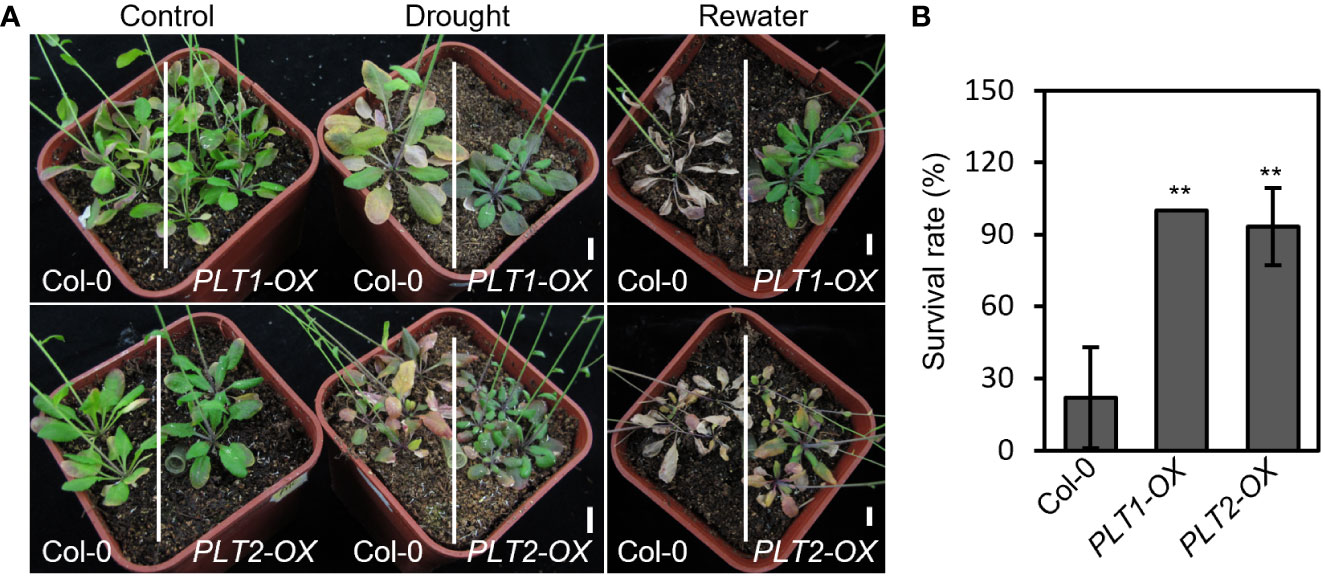
Figure 1 Overexpression of PLT1 and PLT2 enhances drought tolerance in Arabidopsis. (A) The phenotypes of wild type (Col-0), PLT1-OX, and PLT2-OX plants that were subjected to well-watered (control), drought, and rewater conditions. (B) Quantitation of the survival rates of the drought-treated plants shown in (A) after 12 days of re-watering, n≥40. Error bars indicate mean ± SD. Statistically significant differences are indicated by asterisks (Student’s t-test, *p < 0.05, **p < 0.01).
The BR signaling pathway can regulate both root development and plant drought response, but whether BR regulates the drought stress response by regulating root-specific components remains unclear. Our previous findings revealed that the BZR1 homolog in foxtail millet, SiBZR1, recognizes and binds to the BR response element (BRRE) in the promoter region of the PLT1 homolog SiPLT-L1 and downregulates its expression to affect root development (Zhao et al., 2021). However, transcriptional activity assays in vivo and genetic experiments had yet to be completed. To further characterize the mechanism, we analyzed the promoter sequence of PLT1 in Arabidopsis and found that the 2000 bp promoter and 5’-UTR of PLT1 contained a BRRE (CGTGTG) cis-element (Figure 2A).
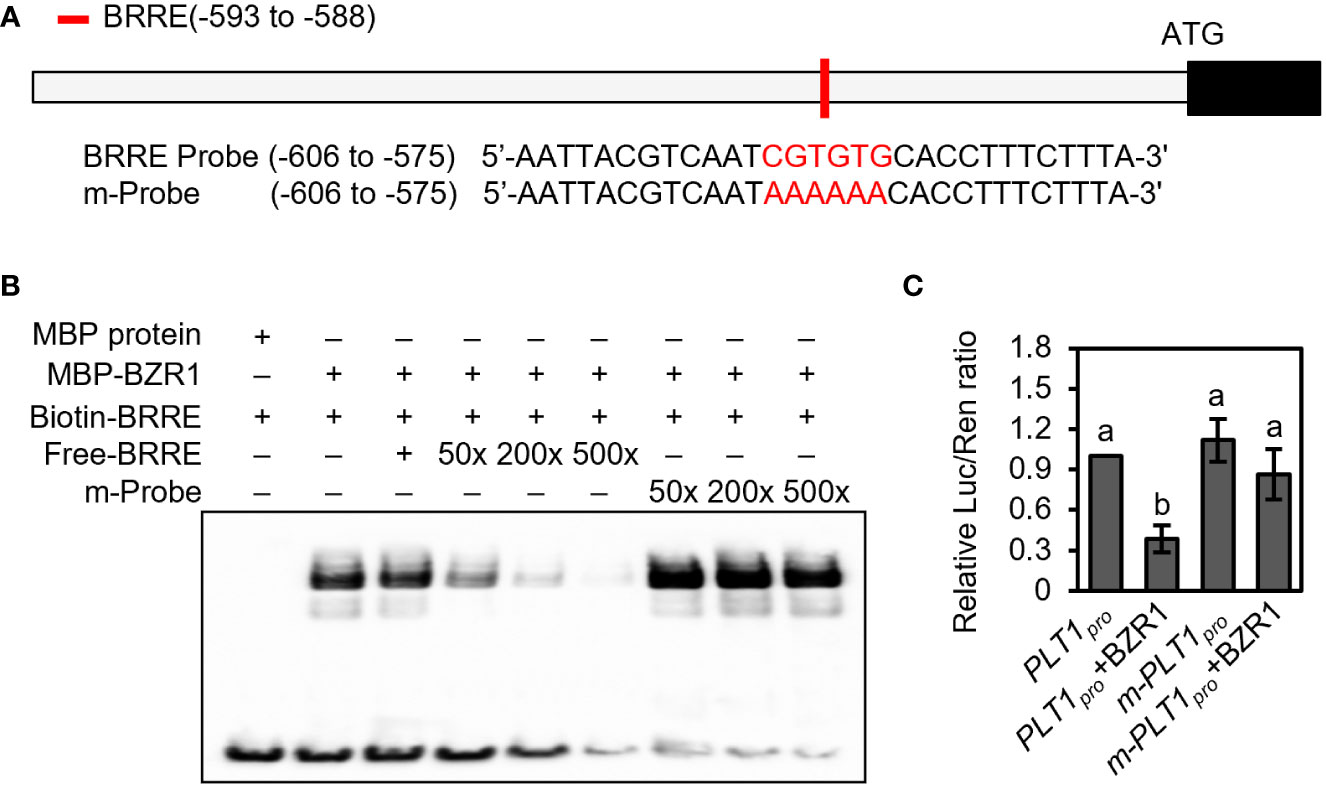
Figure 2 BZR1 binds to the promoter of PLT1 and represses its expression. (A) Schematic diagram of the PLT1 promoter and probe sequences. The red box represents the BRRE motif, and the red letters represent the BRRE sequence and the mutant sequence. (B) BZR1 bound the BRRE cis-element in the PLT1 promoter. EMSA was performed using recombinant proteins in vitro as described in the Materials and Methods. (C) BZR1 represses PLT1 expression in vivo. Arabidopsis leaf protoplasts were co-transformed with PLT1pro:luciferase (PLT1pro) or PLT1promBRRE:luciferase (m-PLT1pro) vector plasmids and 35Spro : BZR1-GFP (BZR1) or 35Spro : GFP vector plasmids, and the relative luciferase activity was measured by Luc/Ren ratio, n=3. Error bars indicate mean ± SD. Statistically significant differences are indicated by different lowercase letters (one-way ANOVA, p < 0.05).
Next, we performed an EMSA, which showed that MBP-BZR1 was able to bind to DNA probes containing the BRRE of the PLT1 promoter in vitro. When unlabeled probes were added to the reaction system, the binding of biotin-labeled probes to MBP-BZR1 was reduced, and the binding of biotin-labeled probes to MBP-BZR1 was weakened with increased concentrations of unlabeled probes (Figure 2B). When the BRRE sequence was mutated to AAAAAA, the mutant probe exhibited a significantly reduced ability to compete with the biotin-labeled probe (Figure 2B). This suggests that BZR1 can specifically recognize and bind to the BRRE cis-element of PLT1. In transient transformation experiments in Arabidopsis protoplasts, we further showed that BZR1 was able to repress the expression of pPLT1:Luciferase, and mutating the BRRE of the PLT1 promoter significantly attenuated the repressive effect (Figure 2C). These results suggest that BZR1 binds to the BRRE-acting element on the PLT1 promoter and represses PLT1 gene expression in Arabidopsis and that the regulatory mechanism is conserved in the dicotyledonous plant Arabidopsis and the monocotyledonous foxtail millet. These results also indicate that BR may regulate Arabidopsis root growth and development through PLT-family genes.
PLT1 and PLT2 are two highly homologous genes that participate in the growth and development of plant roots in a functionally redundant manner (Aida et al., 2004). To determine whether BR are involved in PLT1- and PLT2-related regulation of root growth and development in Arabidopsis, we first explored whether PLT1 and PLT2 are involved in the response to BR. We treated wild-type Col-0 plants with BR at different times and examined the expression levels of PLT1 and PLT2 in the roots. We found that BR treatment for 30 min significantly suppressed the transcript levels of PLT1 and PLT2 and that the expression levels of PLT1 and PLT2 reached the lowest value after BR treatment for 1-3 h. When the treatment time was extended to 6-9 h, the transcript levels of PLT1 and PLT2 partially recovered but were still lower than those in the control group without BR treatment (Figure 3A). Similarly, the transcript levels of PLT1 and PLT2 were significantly lower in the BR gain-of-function mutants bzr1-1D and bes1-D than in Col-0 (Figure 3B). These results suggest that PLT1 and PLT2 are indeed regulated by the BR signaling pathway.
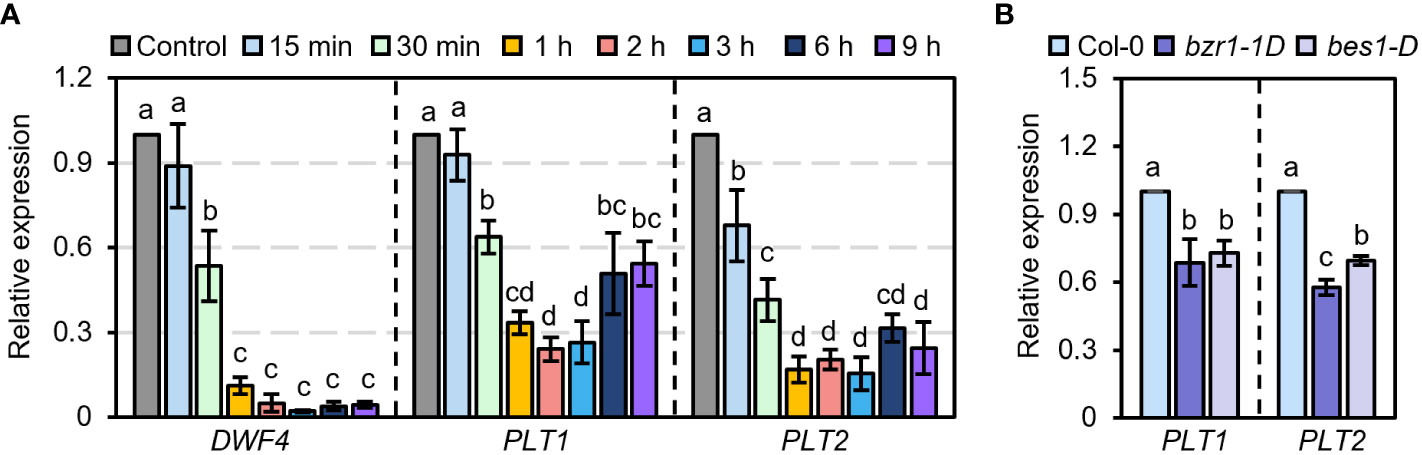
Figure 3 PLT1 and PLT2 are involved in the BR signaling pathway. (A) The relative expression levels of DWF4, PLT1, and PLT2 in the roots of light-grown 10 days Col-0 plants treated with 1 µM eBL for the indicated time. (B) The relative expression levels of PLT1 and PLT2 in the roots of Col-0, bzr1-1D, and bes1-D plants that were grown in 0.25 μM PCZ for 14 days, n=3. Error bars indicate mean ± SD. Statistically significant differences are indicated by different lowercase letters (two-way ANOVA, p < 0.05).
Since the expression of PLT2-OX was significantly higher than that of PLT1-OX, making it easier to observe (Supplementary Figure S1), we next selected PLT2-OX plants for subsequent experiments. To further determine the role of PLT1 and PLT2 in BR-mediated root growth and development, Col-0, PLT2-OX, plt1plt2, and bzr1-1D plants were grown in 1/2 MS medium containing different concentrations of epibrassinolide (eBL), and the root phenotypes of 7-day-old seedlings were observed (Figure 4A). Root elongation was promoted at low concentrations of eBL, but the root length of Col-0 plants gradually decreased as the eBL concentration increased, which was mainly due to shortening of the meristem and a decrease in the number of cortex cells in the meristem (Figures 4B, C). This was consistent with results reported previously (González-Garcia et al., 2011; Zhao et al., 2021). Compared with Col-0 roots, PLT2-OX roots were significantly less sensitive to eBL. Under the same concentration of eBL, the root length of PLT2-OX was significantly greater than that of the wild type. Under 100 nM eBL growth conditions, the relative meristem length and the number of cortex cells in the meristem were also significantly higher than those of the wild type (Figures 4B–E). In contrast, the roots of plt1plt2 and bzr1-1D had enhanced sensitivity to eBL, and they were significantly shorter than those of the wild type under the same conditions. Moreover, the plt1plt2 and bzr1-1D root apical meristem cells were disordered under 100 nM eBL growth conditions, making them difficult to observe (Figures 4B–E). Taken together, these results suggest that PLT1 and PLT2 participate in the BR signaling pathway and that BZR1/BES1 inhibits root growth by suppressing PLT1 and PLT2 expression in Arabidopsis.
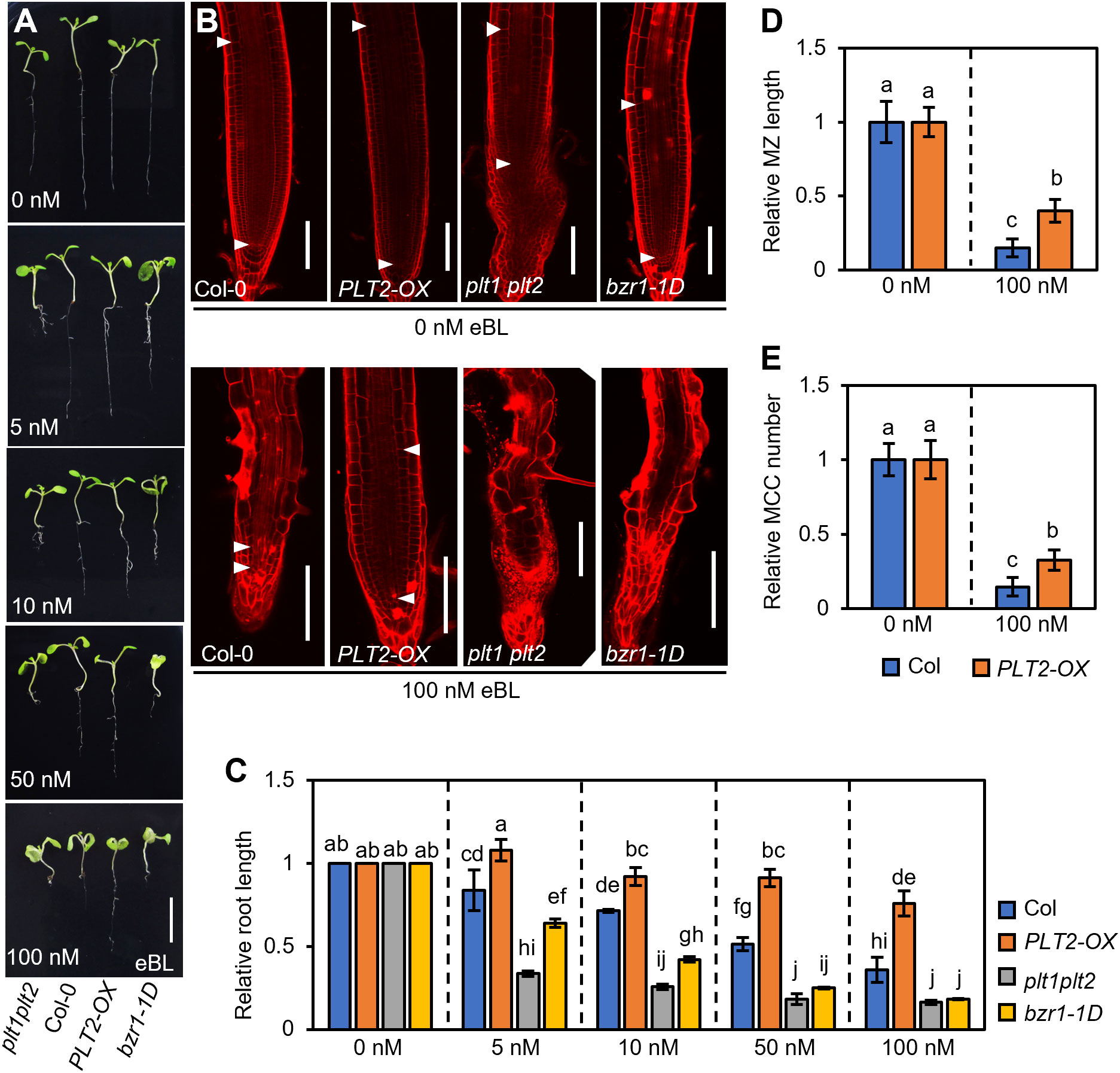
Figure 4 PLT1 mediates BR-regulated primary root elongation in Arabidopsis. (A) Root phenotypes of Col-0, PLT2-OX, plt1plt2, and bzr1-1D plants that were grown on vertical 1/2MS plates supplied with the indicated concentration of eBL for 7 days. Scale bar, 1 cm. (B) Propidium iodide (PI)-stained root meristems of the plants shown in (A) The white arrowheads mark the meristematic zone (MZ). Scale bar, 100 μm. (C) Quantitation of relative root length for the plants shown in (A). (D, E) Quantitation of the relative MZ length (D) and relative meristematic cortex cell (MCC) number (E) for the plants shown in (B), n>10. Error bars indicate mean ± SD. Statistically significant differences are indicated by different lowercase letters (two-way ANOVA, p < 0.05).
BR also plays an important role in the response to drought, and BZR1/BES1 can inhibit root growth by repressing the expression of PLT1 and PLT2, so we investigated whether BR regulates the drought response through a similar pathway. To answer this question, PLT2-OX plants were crossed with the BR gain-of-function mutant bes1-D, which is hypersensitive to drought (Ye et al., 2017), to obtain PLT2-OX/bes1-D plants (Supplementary Figure S2). The drought tolerance of related plants was then tested. Consistent with previous reports, the survival rate of bes1-D was significantly lower than that of Col-0 under drought conditions, while the survival rate of PLT2-OX was significantly higher than that of Col-0 (Figure 5). However, the survival rate of PLT2-OX/bes1-D was close to that of Col-0 under drought stress (Figure 5), this suggests that PLT2 overexpression suppresses the drought-sensitive phenotype of bes1-D and that BES1 suppresses PLTs transcription expression partially contribute to regulate drought tolerance in Arabidopsis.
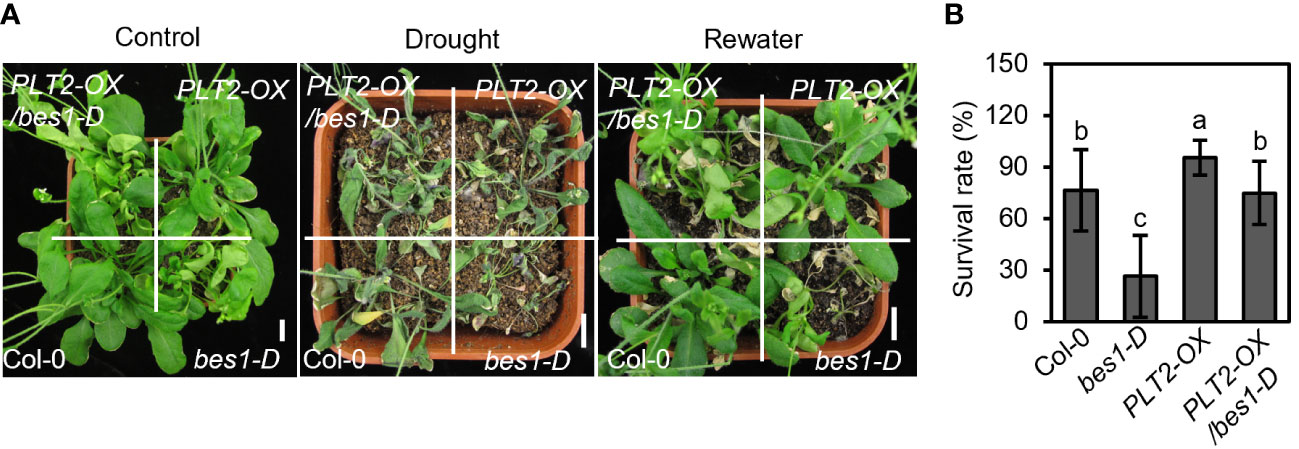
Figure 5 PLT2-OX enhances drought tolerance of bes1-D. (A) The phenotype of wild type (Col-0), PLT1-OX, bes1-D, and PLT2-OX/bes1-D plants that were subjected to well-watered (control), drought, and rewater conditions. (B) Quantitation of the survival rates of the drought-treated plants shown in (A) after 12 days of re-watering. n=11. Error bars indicate mean ± SD. Statistically significant differences are indicated by different lowercase letters (One-way ANOVA, p < 0.05).
To investigate the mechanism by which PLT1 and PLT2 regulate drought resistance in plants, we analyzed the downstream-regulated target genes of PLT1 and PLT2 by comparing the 3645 PLT1/2-targeted genes (Santuari et al., 2016) and 5310 BR-regulated Arabidopsis root genes (Chaiwanon and Wang, 2015) with 2900 drought-regulated genes (Wang et al., 2021) (Supplementary Table S1). We found 335 genes that were coregulated by BR, PLT1/2, and drought (Figure 6A and Supplementary Table S2). Further comparison of these 335 genes showed that 87 genes were downregulated by BR but were activated by PLT1/2; in contrast, 143 genes were upregulated by BR but were repressed by PLT1/2 (Figure 6B). GO analysis of these 230 genes showed enrichment in organic acid transport, chemical stimulation, abiotic stimulation, oxidative response, nitrogen transport, and hormone stimulation responses (including those for auxin, gibberellin, and abscisic acid [ABA]) (Figure 6C and Supplementary Table S3). We next searched for BR-regulated drought stress-related downstream genes via PLT1 and PLT2. We found by comparison that 19 of 87 BR-downregulated but PLT-activated genes were upregulated by drought, while 68 were downregulated by drought; in addition, 87 of 143 BR-upregulated but PLT-suppressed genes were upregulated by drought, while 56 were downregulated by drought (Figure 6D; Supplementary Figure S3, and Supplementary Table S4). Among these were several genes that are known to regulate the plant drought response, such as CIPK3 (Sanyal et al., 2017), RCI2A (Medina et al., 2005), PIP1;5 (Alexandersson et al., 2010), ERF61 (Bano et al., 2022), and WRKY60 (Chen et al., 2010; Arjmand et al., 2023). Through qRT−PCR, these genes were indeed found to be repressed in PLT1-OX and PLT2-OX (Figure 7). PCaP1 is specifically expressed in endodermal cells of the root elongation zone and plays an important role in the hydrotropic response (Tanaka-Takada et al., 2019). PCaP1 was downregulated by drought, repressed by PLT and upregulated by BR treatment and verified to be downregulated in PLT1-OX and PLT2-OX (Figure 7). AAP4, which is downregulated by dehydration, and AT5G19970, which is involved in hormone metabolic processes and root development, were repressed by PLT1/2 but upregulated by drought and BR treatment, and it was verified that these genes were indeed downregulated in PLT1-OX and PLT2-OX (Figure 7). RGL2, which encodes a DELLA protein that restrains the cell proliferation and expansion that drives plant growth, was upregulated by drought and PLT1/2 but downregulated by BR treatment, and it was verified that these genes were indeed upregulated in PLT1-OX and PLT2-OX (Figure 7). These findings suggest that in response to BR-mediated drought stress, the transcription factor PLT1/2 not only regulates the expression of genes corresponding to important drought or dehydration responses but also may regulate the expression of root growth and development genes and cell proliferation and expansion genes (Figure 8).
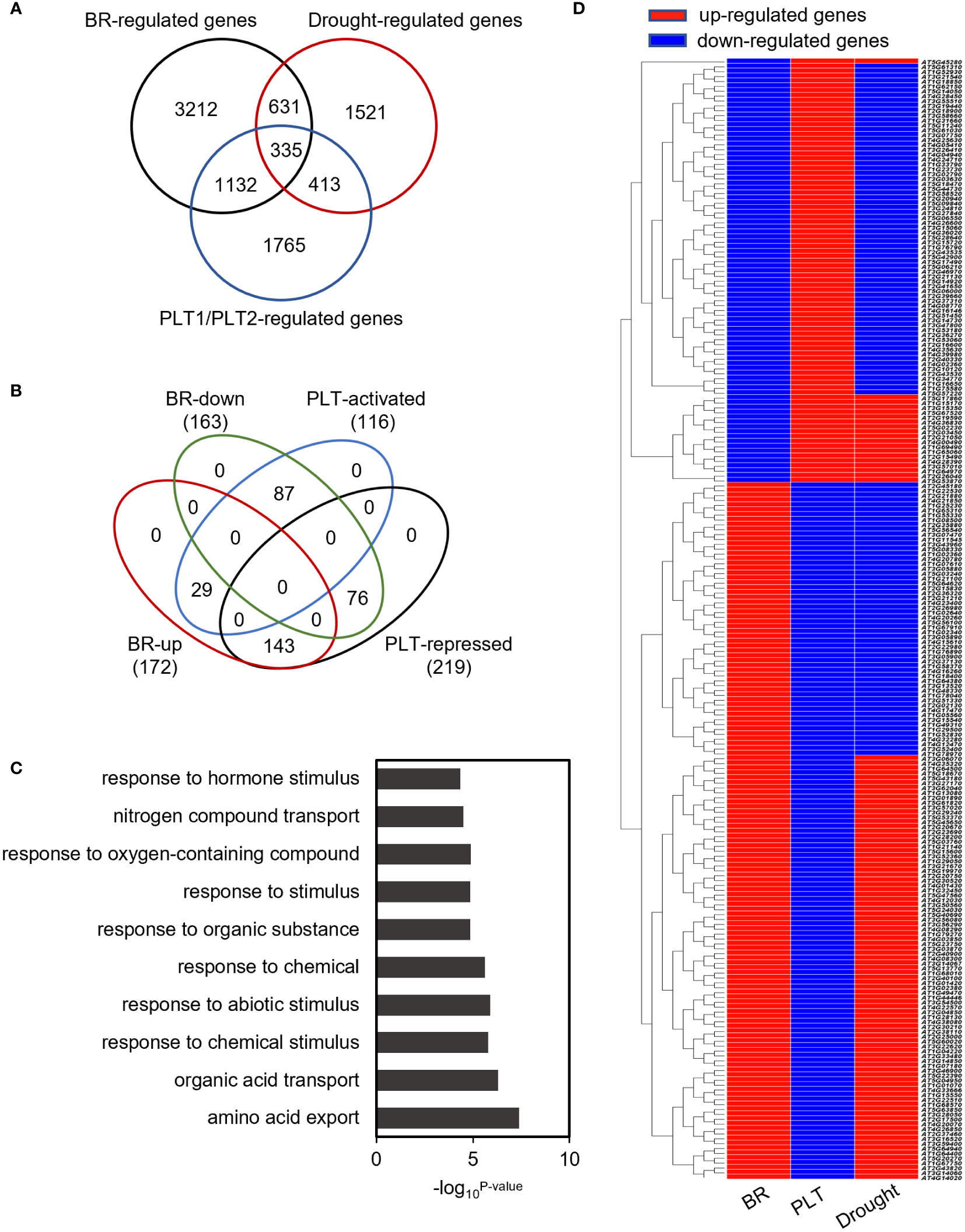
Figure 6 Co-regulated genes analysis of BR, PLT1, PLT2, and drought. (A) Venn diagrams of BR-regulated genes, drought-regulated genes, and PLT1/2 target genes. (B) Venn diagrams of BR up or down and PLT1-PLT2 activated or repressed of 335 regulated genes. (C) GO classification of the 230 regulated genes. (D) The expression heatmap showing the 230 regulated genes antagonistically regulated in BR, PLT1-PLT2, and drought. Red indicates up or activated genes, and blue indicates down or repressed genes.
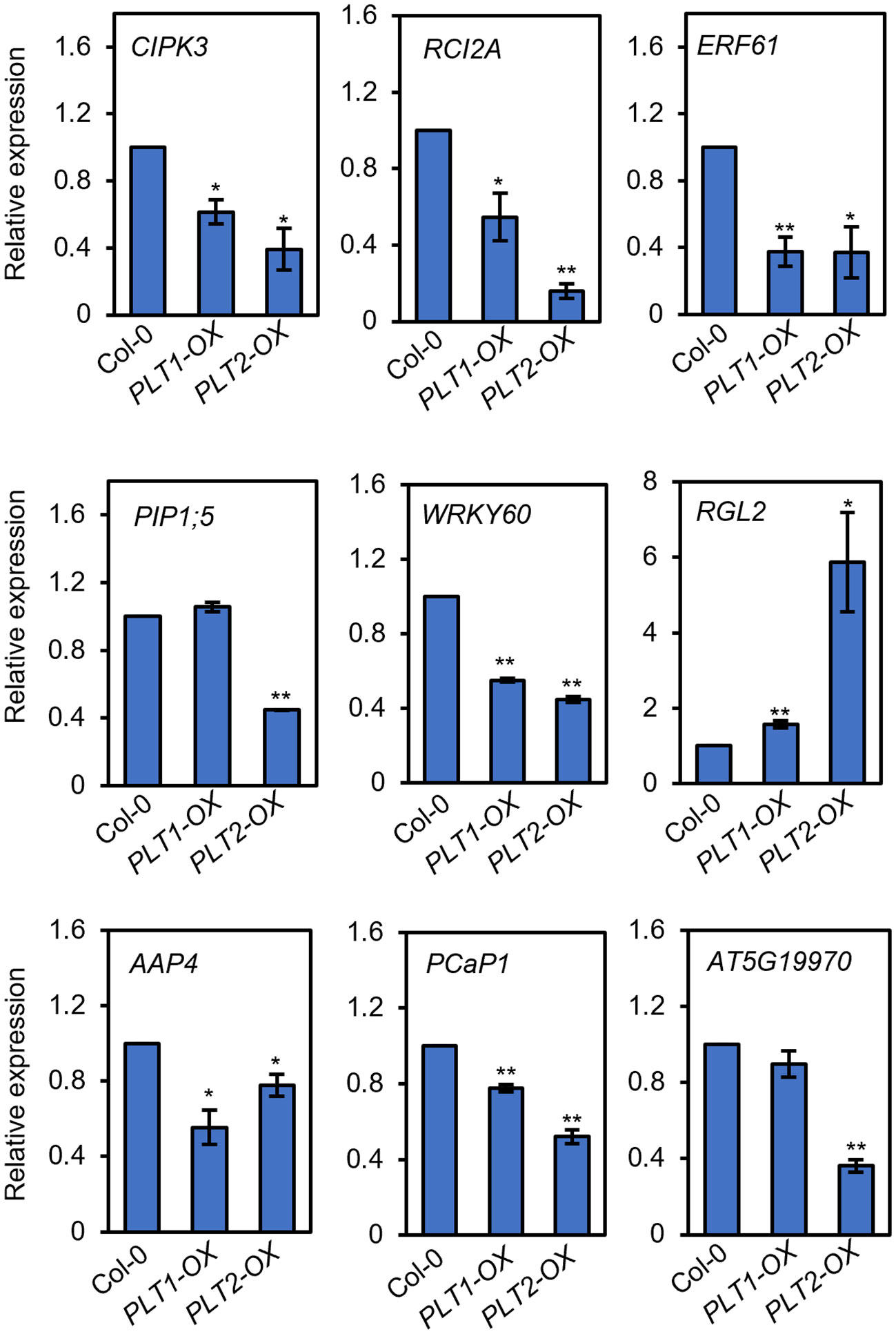
Figure 7 PLTs regulate the expression of drought-responsive genes. The relative expression levels of drought-responsive genes in roots of Col-0, PLT1-OX, and PLT2-OX plants. Error bars indicate the mean ± SD. Statistically significant differences are indicated by asterisks (Student’s t-test, *p < 0.05, **p < 0.01).
Roots are the main organs that plants use to absorb water and nutrients. When drought occurs, the morphological structure of roots changes to enhance the water uptake capacity of plants. Longer roots with fewer branches are conducive to the survival of plants in a soil environment with dry surfaces but sufficient deep water. In contrast, shorter roots with more branches are more conducive to the survival of plants in areas with less rainfall (Dinneny, 2019; Gupta et al., 2020; Karlova et al., 2021). However, at present, we know very little about the molecular mechanisms by which drought regulates plant root morphogenesis.
Root length is an important factor in plant adaptation to drought and is significantly influenced by the activity of the root meristem (Beemster and Baskin, 1998). Several transcription factors and hormonal regulatory networks have been reported to play important roles in the development of the meristematic zone. Among them, WUSCHEL-RELATED HOMEOBOX-5 (WOX5), SHORT ROOT (SHR), SCARECROW (SCR), and PLTs are four important transcription factors/families that reportedly participate in regulating the development of the root meristem (Xu et al., 2020), but their relationship with drought stress has rarely been reported thus far. In this study, we screened different plants for drought sensitivity and found that PLT1-OX and PLT2-OX overexpression plants had elevated resistance to drought stress (Figure 1). Further comparison of PLT1 and PLT2 target genes and drought-regulated genes revealed 748 coregulated genes (Figure 6A). The drought-regulated genes CIPK3, RCI2A, PIP1;5, ERF61, and WRKY60 were found to be repressed in PLT1-OX and PLT2-OX (Figure 7). These results suggest that PLT1 and PLT2, important transcription factors regulating root growth and development, are also involved in the drought stress response. This evidence expands the current understanding of the functions of PLT1/PLT2 and lays a foundation for further study of the relationship between root length and drought responses.
BR plays an important roles in Arabidopsis root growth and development; for example, the short-root phenotype is observed in the BR synthesis-deficient mutant dwf4, the signaling pathway-deficient mutant bri1-116, and the gain-of-function mutant bes1-D (Müssig et al., 2003; Kim et al., 2007; González-García et al., 2011; Chaiwanon and Wang, 2015), while bzr1-1D-CFP and bzr1-1D exhibit a partially alleviated short-root phenotype of dwf4 and bri1-116 (Chaiwanon and Wang, 2015). The transcription factors BZR1 and BES1 function mainly through transcriptional regulation. It has been reported that BZR1/BES1 can regulate meristem size by regulating the expression of ERF115 and BRAVO, but it is unclear whether BZR1 and BES1 can also regulate the development of the root meristem by regulating other genes and how they affect the overall development of Arabidopsis roots.
In this study, we found that PLT1 and PLT2 are involved in the BR signaling pathway and that BZR1/BES1 represses the expression of PLT1 and PLT2 to regulate the development of the Arabidopsis root meristem. First, we found that BZR1 can bind to the promoter of PLT1 and repress PLT1 expression in vivo, which confirms that PLT1 is a target gene of BZR1 (Figure 2). Second, we found that the expression of PLT1 and PLT2 in roots is repressed in response to BR treatment as well as in the BR gain-of-function mutants bzr1-1D and bes1-D, which indicates that the BR signaling pathway can indeed regulate the gene expression of PLT1/PLT2 through the transcription factor BZR1/BES1 (Figure 3). Our results are consistent with previous reports that PLT1 and PLT2 expression is repressed after BR treatment (Chaiwanon and Wang, 2015; Vragović et al., 2015). Third, we found that compared with that of Col-0, the sensitivity of PLT2-OX to eBL was significantly decreased, while the sensitivity of plt1 plt2 and bzr1-1D to eBL was enhanced, and the root meristem was disordered (Figure 4). This suggests that the role of PLT1/PLT2 in root growth and development is regulated by the BR signal transduction pathway and is genetically redundant. We have also previously found that SiBZR1 inhibits SiPLT-L1 expression and affects root development in foxtail millet (Zhao et al., 2021), suggesting that the mechanism by which BRs regulate root elongation through PLTs is relatively conserved in both monocotyledons and dicotyledons. The receptor BRI1 recognizes BRs and conveys signals to the transcription factor BZR1/BES1. BZR1/BES1 binds directly to the promoter of PLT1 and represses its expression, thereby regulating the development of the root apical meristem.
BR also plays a role in regulating plant responses to drought stress (Anwar et al., 2018; Nolan et al., 2019). Recent studies in Arabidopsis have shown that BRs inhibit the expression of RD26 and its homolog but promote the expression of WRKY54 through BZR1/BES1, thereby negatively regulating the plant drought response (Chen et al., 2017; Ye et al., 2017). However, it has also been found that exogenous application of 1 μM eBL enhances drought tolerance in Arabidopsis (Kagale et al., 2007) and that the BR receptor BRL3 enhances drought tolerance in plants upon excessive accumulation in root vascular tissues (Fàbregas et al., 2018; Nolan et al., 2019). These conflicting results suggest that BR may affect plant drought tolerance in a tissue-specific manner through multiple regulatory mechanisms, but the molecular mechanisms involved need to be further explored. In this study, we mainly investigated whether PLTs are involved in BR-mediated drought stress response, but since BZRs family transcription factors, which had six paralog genes in the Arabidopsis genome, and they function redundantly in regulating Arabidopsis development and BR signaling. No obvious phenotype can be observed for BZRs single mutants, double mutants and quadruple mutant (Chen et al., 2019). Unfortunately, it is hard for us to construct the bzr1 bes1 beh1 beh2 beh3 beh4 plt1 plt2. Therefore, we used the gain-of-function mutant bes1-D and PLTs overexpression plants for the study. We found that PLT1-OX and PLT2-OX plants had enhanced tolerance to drought stress (Figure 1) and that PLT2 overexpression alleviated the bes1-D drought-sensitive phenotype (Figure 5). These results suggest that BR can negatively regulate plant drought tolerance by suppressing the expression of PLTs through BZR1/BES1. Our results identify new target genes of BR, PLT1/PLT2, in the drought stress response. However, PLT2-OX only partially recovered the drought sensitive phenotype of bes1-D might be that bes1-D regulates a number of downstream target genes, including PLT1/2, and then regulates plant drought responses. Whether PLTs can interact with RD26 or WRKY54 to regulate drought tolerance in plants and the regulatory relationship between PLTs and the receptors BRI1 and BRL3 need to be further investigated.
Taken together, our findings thus far revealed that BR can regulate root growth and plant responses to drought through PLTs. However, how does BZR1/BES1-PLT affect plant drought tolerance? Does BZR1/BES1-PLT respond to drought stress by regulating root morphogenesis? To answer these questions, we performed a comprehensive analysis by comparing the downstream-regulated target genes of PLT1 and PLT2 (Santuari et al., 2016), BR-regulated Arabidopsis root genes (Chaiwanon and Wang, 2015), and drought-regulated genes (Wang et al., 2021). First, the analysis revealed that BR and PLTs coregulated the expression of some drought-related genes in opposite ways (Figure 6 and Supplementary Figure S3), and we selected some of them for validation. We found that the expression levels of CIPK3, RCI2A, PIP1;5, ERF61, AAP4, and WRKY60 were suppressed by PLT1/2, while that of RGL2 was enhanced by PLT1/2 (Figure 7). The results suggest that BZR1/BES1-PLTs may participate in drought stress control by regulating the expression of downstream drought-responsive genes. Moreover, we found that PCaP1 and AT5G19970, which are involved in root development, were downregulated by PLT1/2 (Figure 7). We also focused on 75 genes that were coregulated by drought, PLTs, and BR treatment. We found that they were significantly enriched in root morphogenesis (Supplementary Figure S3), and many of them were genes that are known to regulate root development and are related to the auxin signaling pathway. This indicates that BR and auxin may engage in crosstalk in root morphogenesis under drought conditions. In addition, PIP1;5 and AT5G19970 were downregulated in PLT2-OX, while there were no significant changes in PLT1-OX (Figure 7). This may have been due to the specificity of PLT1 and PLT2 in the regulation of homologous gene families of downstream target genes, such as the downregulation of SiPIP2;1 and SiPIP2;2 mediated by the PLT1-homologous gene SiPLT-L1 in foxtail millet (Zhao et al., 2021).
In conclusion, our results suggest that after BR treatment, the expression levels of PLTs in roots are reduced, the root system is weak, and water uptake ability is reduced, resulting in drought sensitivity. However, whether BZR1/BES1-PLTs can regulate plant drought tolerance by increasing plant root length and the root phenotypes of related plants under drought conditions, especially their phenotypes in soil, still need to be further studied. Such research will enhance understanding of how BRs mediate root elongation under drought and will be necessary to improve the drought resistance of crops in the future.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
WT and BZ designed research; ZZ performed most of the research; W and HG performed the qRT-PCR and genetic experiment; ZZ, XW and BZ wrote the manuscript. All authors have read and approved the final manuscript.
This work was supported by the National Science Foundation of China (Grant No. 31700242 to BZ), the China Postdoctoral Science Foundation (2022M713434 to ZZ), the Science and Technology Project of Hebei Education Department (Grant No. BJ2021029 to BZ), the Natural Science Foundation for Excellent Youth Scholars of Hebei Province of China (Grant No. C2021205011 to BZ).
We thank American Journal Experts (www.aje.com) for their linguistic assistance during the preparation of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1187605/full#supplementary-material
Aida, M., Beis, D., Heidstra, R., Willemsen, V., Blilou, I., Galinha, C., et al. (2004). The PLETHORA genes mediate patterning of the arabidopsis root stem cell niche. Cell 119, 109–120. doi: 10.1016/j.cell.2004.09.018
Alexandersson, E., Danielson, J. A. D., Råde, J., Moparthi, V. K., Fontes, M., Kjellbom, P., et al. (2010). Transcriptional regulation of aquaporins in accessions of arabidopsis in response to drought stress. Plant J. 61, 650–660. doi: 10.1111/j.1365-313X.2009.04087.x
An, Z., Liu, Y., Ou, Y., Li, J., Zhang, B., Sun, D., et al. (2018). Regulation of the stability of RGF1 receptor by the ubiquitin-specific proteases UBP12/UBP13 is critical for root meristem maintenance. Proc. Natl. Acad. Sci. U. S. A. 115, 1123–1128. doi: 10.1073/pnas.1714177115
Anwar, A., Liu, Y., Dong, R., Bai, L., Yu, X., Li, Y. (2018). The physiological and molecular mechanism of brassinosteroid in response to stress: a review. Biol. Res. 51, 46. doi: 10.1186/s40659-018-0195-2
Arjmand, M. P., Lahiji, H. S., Golfazani, M. M., Biglouei, M. H. (2023). New insights on the regulatory network of drought-responsive key genes in Arabidopsis thaliana. Genetica 151, 29–45. doi: 10.1007/s10709-022-00177-3
Bano, N., Fakhrah, S., Mohanty, C. S., Bag, S. K. (2022). Transcriptome meta-analysis associated targeting hub genes and pathways of drought and salt stress responses in cotton (Gossypium hirsutum): a network biology approach. Front. Plant Sci. 13, 818472. doi: 10.3389/fpls.2022.818472
Beemster, G. T., Baskin, T. I. (1998). Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol. 116, 1515–1526. doi: 10.1104/pp.116.4.1515
Blum, A. (2009). Effective use of water (EUW) and not water-use efficiency (WUE) is the target of crop yield improvement under drought stress. Field Crops Res. 112, 119–123. doi: 10.1016/j.fcr.2009.03.009
Chaiwanon, J., Wang, Z. Y. (2015). Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in arabidopsis roots. Curr. Biol. 25, 1031–1042. doi: 10.1016/j.cub.2015.02.046
Chen, L. G., Gao, Z., Zhao, Z., Liu, X., Li, Y., Zhang, Y., et al. (2019). BZR1 family transcription factors function redundantly and indispensably in BR signaling but exhibit BRI1-independent function in regulating anther development in arabidopsis. Mol. Plant 12 (10), 1408–1415. doi: 10.1016/j.molp.2019.06.006
Chen, H., Lai, Z., Shi, J., Xiao, Y., Chen, Z., Xu, X. (2010). Roles of arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 10, 281. doi: 10.1186/1471-2229-10-281
Chen, J., Nolan, T. M., Ye, H., Zhang, M., Tong, H., Xin, P., et al. (2017). Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell. 29, 1425–1439. doi: 10.1105/tpc.17.00364
Clouse, S. D. (1998). And sasse, J BRASSINOSTEROIDS, essential regulators of plant growth and development. M.Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 427–451. doi: 10.1146/annurev.arplant.49.1.427
Cui, X. Y., Gao, Y., Guo, J., Yu, T. F., Zheng, W. J., Liu, Y. W., et al. (2019). BES/BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1. Plant Physiol. 180, 605–620. doi: 10.1104/pp.19.00100
Dinneny, J. R. (2019). Developmental responses to water and salinity in root systems. Annu. Rev. Cell Dev. Biol. 35, 239–257. doi: 10.1146/annurev-cellbio-100617-062949
Fàbregas, N., Lozano-Elena, F., Blasco-Escámez, D., Tohge, T., Martínez-Andújar, C., Albacete, A., et al. (2018). Overexpression of the vascular brassinosteroid receptor BRL3 confers drought resistance without penalizing plant growth. Nat. Commun. 9, 4680. doi: 10.1038/s41467-018-06861-3
Galinha, C., Hofhuis, H., Luijten, M., Willemsen, V., Blilou, I., Heidstra, R., et al. (2007). PLETHORA proteins as dose-dependent master regulators of arabidopsis root development. Nature. 449, 1053–1057. doi: 10.1038/nature06206
González-García, M. P., Vilarrasa-Blasi, J., Zhiponova, M., Divol, F., Mora-García, S., Russinova, E., et al. (2011). Brassinosteroids control meristem size by promoting cell cycle progression in arabidopsis roots. Development 138, 849–859. doi: 10.1242/dev.057331
Gupta, A., Rico-Medina, A., Caño-Delgado, A. I. (2020). The physiology of plant responses to drought. Science. 368, 266–269. doi: 10.1126/science.aaz7614
Hellens, R. P., Allan, A. C., Friel, E. N., Bolitho, K., Grafton, K., Templeton, M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13. doi: 10.1186/1746-4811-1-13
Hu, H., Xiong, L. (2014). Genetic engineering and breeding of drought-resistant crops. Annu. Rev. Plant Biol. 65, 715–741. doi: 10.1146/annurev-arplant-050213-040000
Kagale, S., Divi, U. K., Krochko, J. E., Keller, W. A., Krishna, P. (2007). Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 225, 353–364. doi: 10.1007/s00425-006-0361-6
Karlova, R., Boer, D., Hayes, S., Testerink, C. (2021). Root plasticity under abiotic stress. Plant Physiol. 187, 1057–1070. doi: 10.1093/plphys/kiab392
Kim, Y., Chung, Y. S., Lee, E., Tripathi, P., Heo, S., Kim, K. H. (2020). Root response to drought stress in rice (Oryza sativa l.). Int. J. Mol. Sci. 21, 1513. doi: 10.3390/ijms21041513
Kim, T. W., Lee, S. M., Joo, S. H., Yun, H. S., Lee, Y., Kaufman, P. B., et al. (2007). Elongation and gravitropic responses of arabidopsis roots are regulated by brassinolide and IAA. Plant Cell Environ. 30, 679–689. doi: 10.1111/j.1365-3040.2007.01659.x
Koevoets, I. T., Venema, J. H., Elzenga, J. T., Testerink, C. (2016). Roots withstanding their environment, exploiting root system architecture responses to abiotic stress to improve crop tolerance. Front. Plant Sci. 7, 1335. doi: 10.3389/fpls.2016.01335
Kono, A., Yin, Y. (2020). Updates on BES1/BZR1 regulatory networks coordinating plant growth and stress responses. Front. Plant Sci. 11, 617162. doi: 10.3389/fpls.2020.617162
Lorts, C. M., Lasky, J. R. (2020). Competition × drought interactions change phenotypic plasticity and the direction of selection on arabidopsis traits. New Phytol. 227, 1060–1072. doi: 10.1111/nph.16593
Mähönen, A. P., Ten Tusscher, K., Siligato, R., Smetana, O., Díaz-Triviño, S., Salojärvi, J., et al. (2014). PLETHORA gradient formation mechanism separates auxin responses. Nature 515, 125–129. doi: 10.1038/nature13663
Medina, J., Rodríguez-Franco, M., Peñalosa, A., Carrascosa, M. J., Neuhaus, G., Salinas, J. (2005). Arabidopsis mutants deregulated in RCI2A expression reveal new signaling pathways in abiotic stress responses. Plant J. 42, 586–597. doi: 10.1111/j.1365-313X.2005.02400.x
Müssig, C., Shin, G. H., Altmann, T. (2003). Brassinosteroids promote root growth in arabidopsis. Plant Physiol. 133, 1261–1271. doi: 10.1104/pp.103.028662
Nie, S., Huang, S., Wang, S., Mao, Y., Liu, J., Ma, R., et al. (2019). Enhanced brassinosteroid signaling intensity via SlBRI1 overexpression negatively regulates drought resistance in a manner opposite of that via exogenous BR application in tomato. Plant Physiol. Biochem. 138, 36–47. doi: 10.1016/j.plaphy.2019.02.014
Nolan, T. M., Brennan, B., Yang, M., Chen, J., Zhang, M., Li, Z., et al. (2017). Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell. 41, 33–46. doi: 10.1016/j.devcel.2017.03.013
Nolan, T. M., Vukašinović, N., Liu, D., Russinova, E., Yin, Y. (2019). Brassinosteroids, multidimensional regulators of plant growth, development, and stress responses. Plant Cell. 32, 295–318. doi: 10.1105/tpc.19.00335
Peng, P., Yan, Z., Zhu, Y., Li, J. (2008). Regulation of the arabidopsis GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 through proteasome-mediated protein degradation. Mol. Plant 1, 338–346. doi: 10.1093/mp/ssn001
Petricka, J. J., Winter, C. M., Benfey, P. N. (2012). Control of arabidopsis root development. Annu. Rev. Plant Biol. 63, 563–590. doi: 10.1146/annurev-arplant-042811-105501
Planas-Riverola, A., Gupta, A., Betegón-Putze, I., Bosch, N., Ibañes, M. (2019). And caño-Delgado, a Brassinosteroid signaling in plant development and adaptation to stress. I.Development. 146, dev151894. doi: 10.1242/dev.151894
Qin, T., Kazim, A., Wang, Y., Richard, D., Yao, P., Bi, Z. (2022). Root-related genes in crops and their application under drought stress resistance. Int. J. Mol. Sci. 23, 11477. doi: 10.3390/ijms231911477
Sahni, S., Prasad, B. D., Liu, Q., Grbic, V., Sharpe, A., Singh, S. P., et al. (2016). Overexpression of the brassinosteroid biosynthetic gene DWF4 in brassica napus simultaneously increases seed yield and stress tolerance. Sci. Rep. 6, 28298. doi: 10.1038/srep28298
Santuari, L., Sanchez-Perez, G. F., Luijten, M., Rutjens, B., Terpstra, I., Berke, L., et al. (2016). The PLETHORA gene regulatory network guides growth and cell differentiation in arabidopsis roots. Plant Cell. 28, 2937–2951. doi: 10.1105/tpc.16.00656
Sanyal, S. K., Kanwar, P., Samtani, H., Kaur, K., Jha, S. K., Pandey, G. K. (2017). Alternative splicing of CIPK3 results in distinct target selection to propagate ABA signaling in arabidopsis. Front. Plant Sci. 8, 1924. doi: 10.3389/fpls.2017.01924
Sun, Y., Fan, X. Y., Cao, D. M., Tang, W. Q., He, K., Zhu, J. Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in arabidopsis. Dev. Cell. 19, 765–777. doi: 10.1016/j.devcel.2010.10.010
Tanaka-Takada, N., Kobayashi, A., Takahashi, H., Kamiya, T., Kinoshita, T., Maeshima, M. (2019). Plasma membrane-associated Ca2+-binding protein PCaP1 is involved in root hydrotropism of Arabidopsis thaliana. Plant Cell Physiol. 60, 1331–1341. doi: 10.1093/pcp/pcz042
Tang, W. Q., Yuan, M., Wang, R. J., Yang, Y., Wang, C., Oses-Prieto, J. A., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13, 124–131. doi: 10.1038/ncb2151
Vert, G., Chory, J. (2006). Downstream nuclear events in brassinosteroid signalling. Nature 441, 96–100. doi: 10.1038/nature04681
Vilarrasa-Blasi, J., González-García, M. P., Frigola, D., Fàbregas, N., Alexiou, K. G., López-Bigas, N., et al. (2014). Regulation of plant stem cell quiescence by a brassinosteroid signaling module. Dev. Cell. 30, 36–47. doi: 10.1016/j.devcel.2014.05.020
Vragović, K., Sela, A., Friedlander-Shani, L., Fridman, Y., Hacham, Y., Holland, N., et al. (2015). Translatome analyses capture of opposing tissue-specific brassinosteroid signals orchestrating root meristem differentiation. Proc. Natl. Acad. Sci. U. S. A. 112, 923–928. doi: 10.1073/pnas.1417947112
Wang, Q., Liu, P., Jing, H., Zhou, X. F., Zhao, B., Li, Y., et al. (2021). JMJ27-mediated histone H3K9 demethylation positively regulates drought-stress responses in arabidopsis. New Phytol. 232, 221–236. doi: 10.1111/nph.17593
Xu, M., Gu, X., Liang, N., Bian, X., Wang, H., Qin, Y., et al. (2020). Intersected functional zone of transcriptional regulators patterns stemness within stem cell niche of root apical meristem. J. Integr. Plant Biol. 62, 897–911. doi: 10.1111/jipb.12881
Xu, W., Jia, L., Shi, W., Liang, J., Zhou, F., Li, Q., et al. (2013). Abscisic acid accumulation modulates auxin transport in the root tip to enhance proton secretion for maintaining root growth under moderate water stress. New Phytol. 197, 139–150. doi: 10.1111/nph.12004
Yang, X. R., Bai, Y., Shang, J. X., Xin, R., Tang, W. Q. (2016). The antagonistic regulation of abscisic acid-inhibited root growth by brassinosteroids is partially mediated via direct suppression of ABSCISIC ACID INSENSITIVE 5 expression by BRASSINAZOLE RESISTANT 1. Plant Cell Environ. 39, 1994–2003. doi: 10.1111/pce.12763
Yang, C. J., Zhang, C., Lu, Y. N., Jin, J. Q., Wang, X. L. (2011). The mechanisms of brassinosteroids’ action, from signal transduction to plant development. Mol. Plant 4, 588–600. doi: 10.1093/mp/ssr020
Ye, H., Liu, S., Tang, B., Chen, J., Xie, Z., Nolan, T. M., et al. (2017). RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat. Commun. 8, 14573. doi: 10.1038/ncomms14573
Yu, X., Li, L., Zola, J., Aluru, M., Ye, H., Foudree, A., et al. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65, 634–646. doi: 10.1111/j.1365-313X.2010.04449.x
Keywords: PLT1, PLT2, brassinosteroids, drought, root
Citation: Zhao Z, Wu S, Gao H, Tang W, Wu X and Zhang B (2023) The BR signaling pathway regulates primary root development and drought stress response by suppressing the expression of PLT1 and PLT2 in Arabidopsis thaliana. Front. Plant Sci. 14:1187605. doi: 10.3389/fpls.2023.1187605
Received: 16 March 2023; Accepted: 02 May 2023;
Published: 27 June 2023.
Edited by:
Tae-Hwan Kim, Chonnam National University, Republic of KoreaReviewed by:
Arif Ashraf, University of Massachusetts Amherst, United StatesCopyright © 2023 Zhao, Wu, Gao, Tang, Wu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuedan Wu, d3V4dWVkYW4yMDE3QDE2My5jb20=; Baowen Zhang, Ynd6aGFuZzIwMTlAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.