
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 April 2023
Sec. Functional and Applied Plant Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1184276
This article is part of the Research Topic Genetics, Genomics and Breeding of Plant Architecture, Biomass, Grain Quality and Grain Yield Traits in Rice and Wheat View all 15 articles
 Xing Huo†
Xing Huo† Jian Wang†
Jian Wang† Luo Chen
Luo Chen Hua Fu
Hua Fu Tifeng Yang
Tifeng Yang Jingfang Dong
Jingfang Dong Yamei Ma
Yamei Ma Lian Zhou
Lian Zhou Jiansong Chen
Jiansong Chen Dilin Liu
Dilin Liu Bin Liu
Bin Liu Junliang Zhao
Junliang Zhao Shaohong Zhang*
Shaohong Zhang* Wu Yang*
Wu Yang*Grain chalkiness is the main factor determining the market value of rice. Reducing chalkiness is an important breeding goal for genetic improvement of high quality rice. Identification of QTLs or genes controlling chalkiness is the prerequisite for molecular breeding in rice. Here, we conducted a genome-wide association study to identify QTLs associated with grain chalkiness including percentage of grains with chalkiness (PGWC) and degree of endosperm chalkiness (DEC) in 450 rice accessions consisting of 300 indica and 150 japonica rice in two environments. A total of 34 QTLs were identified, including 14 QTLs for PGWC and 20 QTLs for DEC. Among them, seven QTLs were commonly identified in two environments, and eight QTLs were simultaneously related to two traits. Based on the haplotype analysis, LD decay analysis, RNA-sequencing, qRT-PCR confirmation and haplotype comparisons, four genes (LOC_Os10g36170, LOC_Os10g36260, LOC_Os10g36340 and LOC_Os10g36610) were considered as the candidate genes for qDEC-10c1w,2wj, which could be identified in both environments and had the most significant p-value among the newly identified QTLs. These results provided new insight into the genetic basis of grain chalkiness and gene resources for improving quality by molecular breeding in rice.
Rice is one of the most important food crops in the world. Improving rice yield and quality is the main goal of rice breeding. At present, with the improvement of people’s living standard, high quality rice is the primary concern of rice breeders and consumers. Rice quality mainly includes milling, appearance and cooking quality. Grain chalkiness, as an important character of appearance quality, is highly undesirable in rice breeding and marketing (Zhao et al., 2022). Percentage of grains with chalkiness (PGWC) and degree of endosperm chalkiness (DEC) are two indexes to evaluate grain chalkiness. Rice with high PGWC and DEC is easily broken during milling, and thus reducing the yield of head milled rice. Furthermore, the taste of chalky grain declines after cooking (Wu et al., 2022). Therefore, reducing chalkiness has been the key trait selected for released rice cultivars.
Understanding the genetic basis of grain chalkiness can greatly improve breeding efficiency. Grain PGWC and DEC are quantitatively inherited, controlled by multiple QTLs or genes, and affected by environments (Wang et al., 2017; Qiu et al., 2021). In past decade, bi-parental populations, rice mutants and reverse genetics have been widely used in QTL mapping and gene cloning for the two traits, and many QTLs have been reported and some genes have been cloned. Chalk5 is cloned from natural variants and encodes a vacuolar H+-translocating pyrophosphatase. Elevated expression of Chalk5 increases the endosperm chalkiness (Li et al., 2014). Recently, WCR1 was identified by map-based cloning and up-regulated expression of WCR1 significantly reduces white-core rate (Wu et al., 2022). Besides, some chalkiness-related genes are cloned from floury endosperm mutants, most of which are related to starch synthesis or starch quality. The starch synthase IIIa (SSIIIa) regulates short amylopectin elongation, and knockout of SSIIIa causes the formation of white core (Ryoo et al., 2007). The GIF1 encodes a cell-wall invertase required for carbon partitioning during early grain-filling, and the gif1 mutant shows loose starch grain structure and increased chalkiness (Wang et al., 2008). OsLTPL36 is specially expressed in developing rice seed, and suppressed expression of this gene increases grain chalkiness rate (Wang et al., 2015). Fan et al. found that reduced expression of an autophagy gene OsATG8b results in chalky endosperm and poor seed quality (Fan et al., 2020).
In recent years, genome-wide association study (GWAS) based on linkage disequilibrium (LD) in a large germplasm resources has been widely used to identify QTLs that control complex traits, such as appearance quality in rice, on a large scale (Qiu et al., 2015; Wang et al., 2017; Qiu et al., 2021). Using a worldwide collection of indica rice germplasm, Qiu et al. identified two QTLs for PGWC and five QTLs for DEC, and two QTLs, qPGWC5.1 and qDEC5.2 on chromosome 5 were co-located with GW5 (Qiu et al., 2015). Furthermore, they identified 12 QTLs for DEC in two years by using a larger panel mainly including indica and japonica from the 3K Rice Genomes Project, but none could be detected in both years (Qiu et al., 2021). Wang et al. detected four QTLs affecting DEC and four QTLs affecting PGWC, and haplotype analysis suggested that Os07g0604500 was the candidate gene underlying qDEC7 (Wang et al., 2017). Recently, OsbZIP60 was identified as a vital regulator of grain chalkiness by GWAS, and knockout of OsbZIP60 results in high grain chalkiness through regulating the expression of key genes related to grain chalkiness (Yang et al., 2022a). Although progress has been made in the identification and molecular mechanism of QTLs or genes associated with grain chalkiness in rice, but few of them have been applied to rice breeding. Therefore, it is necessary to further explore the stable QTLs or genes related to grain chalkiness in rice.
In the present study, PGWC and DEC in a diverse panel containing 450 accessions selected from the RDP2 (McCouch et al., 2016) were evaluated. GWAS was conducted to identify QTLs for the two traits. In total, 34 QTLs were identified including 11 QTLs co-localized with the reported QTLs or genes and 23 newly identified QTLs. Based on the linkage disequilibrium (LD) decay analysis, RNA-sequencing, qRT-PCR confirmation and haplotype comparisons, four genes were considered as candidate genes underlying qDEC-10c1w,2wj, which could be identified in both environments and had the most significant p-value among the newly identified QTLs. This study provides new insight into the genetic basis of grain chalkiness and contributes to molecular breeding for high-quality rice.
The 450 rice accessions (Table S1) were used for phenotype evaluation and GWAS in this study. These rice accessions were selected from RDP2 (McCouch et al., 2016) based on their origins and diversity, including 300 indica and 150 japonica rice.
The 450 rice accessions from RDP2 had been genotyped by 700K SNPs (McCouch et al., 2016). The phylogenetic tree was constructed by SNPhylo using SNP data. The figure of phylogenetic tree was drawn by ITOL (Interaction Tree Of Life, https://itol.embl.de/).
The 450 rice accessions were planted in the experimental fields of Guangzhou (2016GZ, 23°8’ N and 113°17’ E) and Yangjiang (2018YJ, 21°89’ N and 111°90’ E) in Guangdong Province, China, in the second cropping season in 2016 and 2018, respectively. The experiments were performed in a randomized complete block design with two replicates in both environments. The field management, including irrigation, fertilization, and disease and pest control, followed the conventional or local practice in rice production. At complete maturity, the grains were harvested and dried naturally, and then stored in storage at 15 °C. 20 g grains were de-husked by a huller (JLG-III, Chengdu, China) and milled by a polisher (JNM, Chengdu, China). PGWC and DEC of head rice were detected by the Rice Appearance Quality Determination Instrument (SC-E, Hangzhou, Chian). PGWC was the percentage of grains with chalkiness, and DEC was calculated as the product of PGWC and chalkiness size (the area of chalkiness divided by the area of whole grain). All measurements were conducted using three independent samples from both two replicates, and the average values were used for subsequent GWAS analysis.
GWAS was conducted as described in our previous study (Zhao et al., 2018; Yang et al., 2021) by using GAPIT version 2 software. Briefly, SNPs were filtered using the criteria of having less than 30% of missing data and minor allele frequency (MAF) > 0.05. The mixed linear model (MLM) with kinship matrix was used for GWAS, and the principal component was set to 3 in GAPIT. Manhattan and QQ plots were produced by R-package qqman (Turner, 2018). A region containing two or more than two significant SNPs (p < 0.0001) within 200 kb is considered as one QTL.
The SNP information of about 900 kb (18.98-19.88 Mb) for qDEC-10c1w,2wj was used for LD Decay analysis. The LD heatmap were drawn by the R package LD heatmap.
Based on haplotype analysis and consistency of flowering time, six accessions with consistent flowering time, including three accessions with High DEC (accessions 787, 806 and 854) and three accessions with low DEC (accessions 730, 937 and 1046) were selected to conduct RNA-sequencing. The spikes were sampled on the 15th day after flowering, and total RNA was extracted using Trizol reagent (Takara, Dalian, China). RNA-sequencing was conducted by the Annoroad Gene Technology (Beijing, China), and data analysis was conducted as described in our previous study (Yang et al., 2022b). Genes with read count less than 30 were regarded as no expression. The differentially expressed genes between two sets of contrasting accessions were identified according to the criteria of p-value ≤ 0.01 and fold change of pairwise comparison ≥2 or ≤0.5.
The leaves of rice seedlings were collected and DNA extraction was conducted by CTAB method. The experiment was conducted according to the standard protocol provided by Illumina. The qualified genomic DNA was fragmented by ultrasonication. These fragments were then for purification, terminal repair and addition of A at the 3’ end, ligation and sequencing, and then fragmentation by agarosegel electrophoresis. Selection of appropriate DNA fragments and PCR amplification were performed to construct a sequencing library, and the library was sequenced using the Illumina NovaSeq6000 platform (BerryGenomics, Beijing, China). The raw sequence data have been reported in our previous study (Wang et al., 2023).
The RNA samples for RNA-sequencing assays were used to confirm the expression level of candidate genes in this study. The cDNA synthesis was conducted using the PrimeScriptTM RT reagent kit (Takara, Japan). The qRT-PCR analysis was performed by qRT-PCR (Biorad CFX96, Pleasanton, CA, USA). The primers were designed by the Primer designing tool on NCBI. The gene-specific primers are listed in Supplementary Table S5. The EF1α was used as the normalized genes for mRNA. All reactions were repeated three times.
Phylogenetic analysis based on their genotypes determined by the 700 K SNPs (McCouch et al., 2016) indicated that the 450 rice accessions could be roughly clustered into two groups, consisting of 300 indica and 150 japonica rice (Table S1 and Figure 1A).
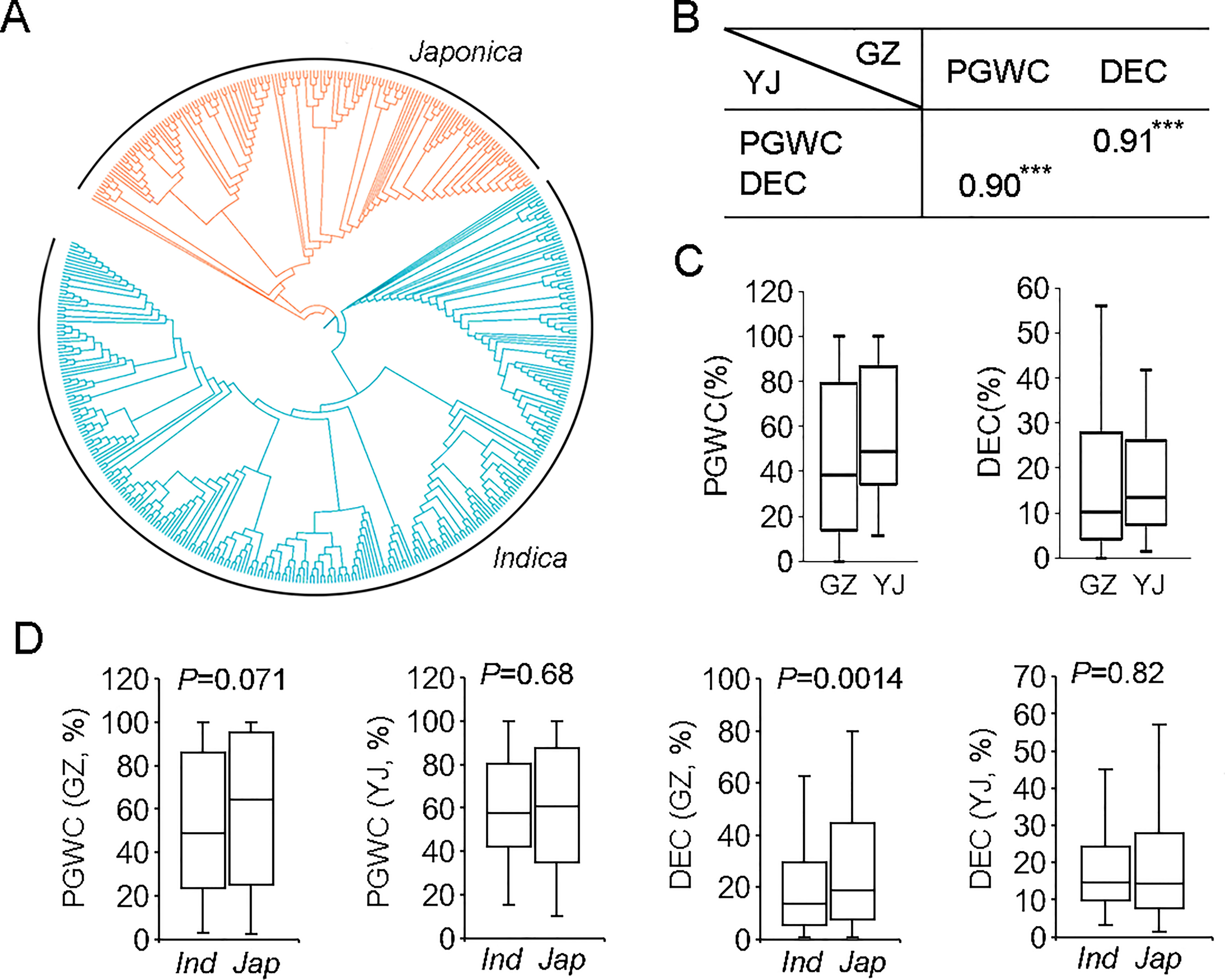
Figure 1 The phylogenetic tree and phenotypic comparison of 450 rice accessions used in this study. (A) The phylogenetic tree of 450 rice accessions. (B) Correlation analysis of PGWC and DEC in two environments. ***p<0.001. (C) Box plots of PGWC and DEC in two environments. (D) Phenotypic comparison between indica and japonica rice. Statistical comparison was performed by one-side t-test. GZ, Guangzhou; YJ, Yangjiang; PGWC, Percentage of grains with chalkiness; DEC, Degree of endosperm chalkiness.
PGWC and DEC of the 450 rice accessions were evaluated in two environments (2016GZ and 2018YJ). The phenotype pairwise correlations between two traits were positively correlated in both environments, with correlation coefficients of 0.91 and 0.90 in 2016GZ and 2018YJ, respectively (Figure 1B). The panel showed a large variations for the two traits (Figure 1C). By comparing the phenotypes of indica and japonica rice, there were no significant differences between the two subpopulations in both environments (p>0.05) except that the DEC of japonica rice was significantly higher than that of indica rice in 2016GZ (p<0.01) (Figure 1D).
A total of 34 QTLs (14 for PGWC and 20 for DEC) were identified in the two environments (Figure 2; Figure S1 and Table 1). Among them, seven QTLs could be identified in both environments, while 15 and 12 QTLs were detected only in 2016GZ and 2018YJ, respectively. These QTLs were identified in different populations, including 11 QTLs in the whole population, 7 QTLs in the indica population, 3 QTLs in the japonica population, and 13 QTLs in more than one population.
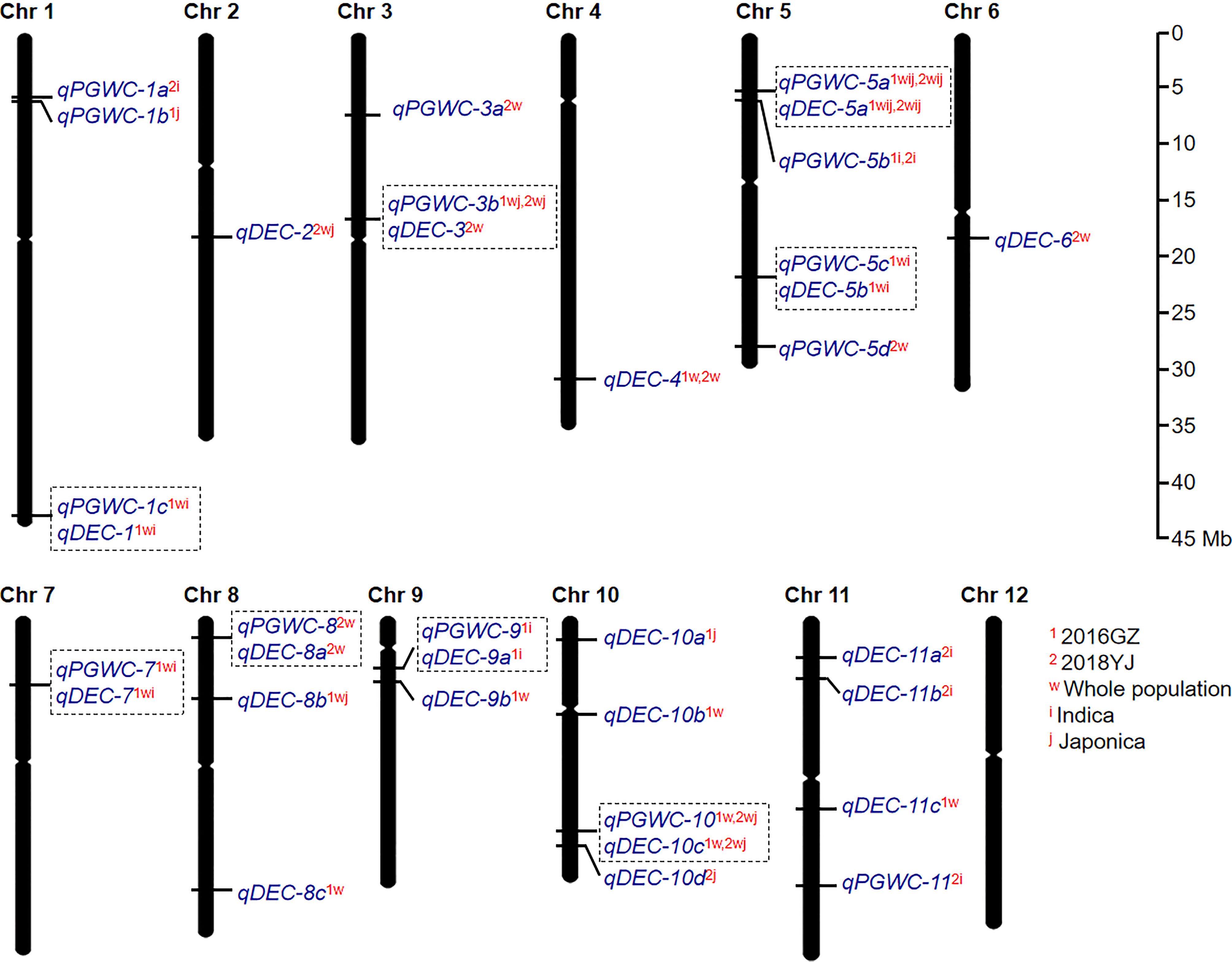
Figure 2 The global view of QTL mapping for PGWC and DEC by GWAS. The co-located QTLs were surrounded with dotted boxes. The superscript letters (1 and 2) refer to the two environments (2016GZ and 2018YJ, respectively). The superscript letters (w, i and j) refer to the three populations (whole, indica and japonica, respectively). For example, qDEC-10c1w,2wj indicates that qDEC-10c can be identified in 2016GZ by the whole population and in 2018YJ by the whole and japonica population.
Besides, it was found that some QTLs for PGWC and DEC were co-located on the same chromosomal regions. They were qPGWC-1c1wi and qDEC-11wi on chromosome 1; qPGWC-3b1wj,2wj and qDEC-32w on chromosome 3; qPGWC-5a1wij,2wij and qDEC-5a1wij,2wij on chromosome 5; qPGWC-5c1wi and qDEC-5b1wi on chromosome 5; qPGWC-71wi and qDEC-71wi on chromosome 7; qPGWC-82w and qDEC-8a2w on chromosome 8; qPGWC-91i and qDEC-9a1i on chromosome 9; qPGWC-101w,2wj and qDEC-10c1w,2wj on chromosome 10. While the other 18 QTLs were associated with one trait (Figure 2).
Compared with the previous studies, 11 QTLs identified in this study were co-localized with the reported QTLs or cloned genes (Table 1). In particular, qPGWC-3b1wj,2wj and qDEC-32w were co-located with GS3, a major gene regulating grain length (Mao et al., 2010); qPGWC-5a1wij,2wij and qDEC-5a1wij,2wij were co-located with GW5, a major gene regulating grain width (Weng et al., 2008) and several QTLs for chalky traits (Weng et al., 2008; Mao et al., 2010; Bian et al., 2014; Peng et al., 2014; Yun et al., 2016; Zhao et al., 2016; Wang et al., 2017; Misra et al., 2021; Qiu et al., 2021). The other 23 QTLs were newly identified in the present study.
For the 34 QTLs identified in the present study, qDEC-10c1w,2wj was co-located with qPGWC-101w,2wj and they could be identified in the whole populations in both environments (Figure 2). Besides, qDEC-10c1w,2wj and qPGWC-101w,2wj had the most significant p-value among the newly identified QTLs (Table 1). For qDEC-10c1w,2wj and qPGWC-101w,2wjloci, both PGWC and DEC were significantly different between accessions with different haplotypes (Figure 3A). Based on the SNP for GWAS, the LD decay analysis was used to narrow the region of qDEC-10c1w,2wj. There was a significant disconnection at 19.28 Mb and 19.68 Mb of chromosome 10 (Figure 3B). Because the rice linkage disequilibrium block is about 100-200 kb (Wang et al., 2020). So the qDEC-10c1w,2wj was delimited to an approximately 400 kb region (from 19.28 to 19.68 Mb) on chromosome 10.
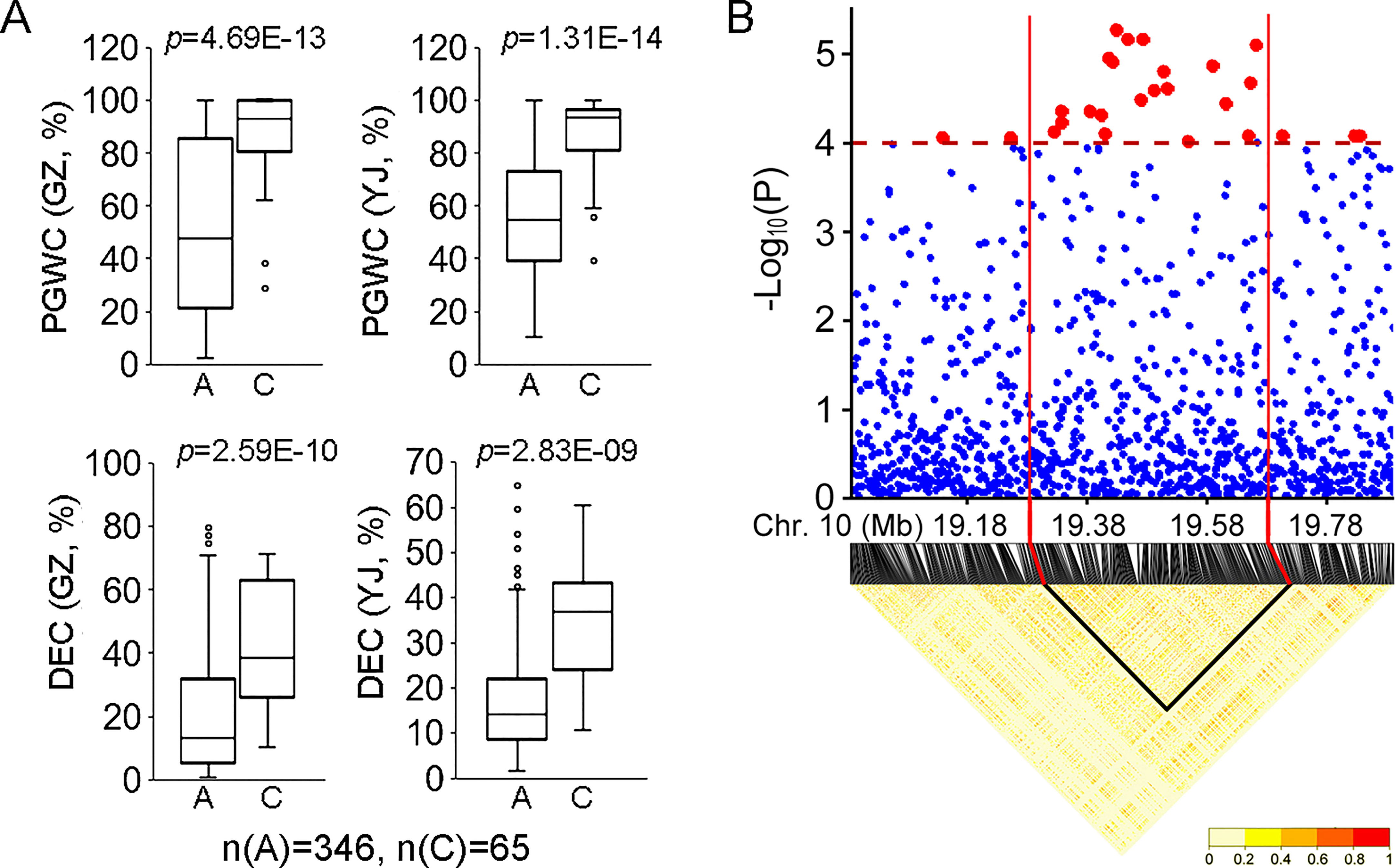
Figure 3 Phenotype and region analysis of qDEC-10c1w,2wj.(A) Phenotypic comparison of PGWC and DEC between accessions with different haplotypes for qDEC-10c1w,2wj. Statistical comparison was performed by one-side t-test. (B) Local manhattan plot of GWAS and linkage disequilibrium heatmap for qDEC-10c1w,2wj.
There are 51 annotated genes within the qDEC-10c1w,2wj region (Table S2) based on release 7 of the MSU Rice Genome Annotation Project (Kawahara et al., 2013). Since the grain filling stage is the key period for seed development and chalkiness formation, and the differentially expressed genes during this stage may result in variant quality (Yang et al., 2022c; Zhao et al., 2022). To reduce the number of candidate genes, three accessions with high DEC haplotype and three accessions with low DEC haplotype based on haplotype analysis of qDEC-10c1w,2wj were selected for gene differential expression analysis at grain filling stage (15 days after flowering). RNA-sequencing revealed that 28 genes were expressed, of which five genes (LOC_Os10g36170, LOC_Os10g36260, LOC_Os10g36270, LOC_Os10g36340 and LOC_Os10g36610) were differentially expressed between the two sets of contrasting accessions (Table S2). qRT-PCR assays confirmed that the expression patterns of LOC_Os10g36170, LOC_Os10g36260, LOC_Os10g36340 and LOC_Os10g36610 were consistent with the RNA-sequencing result (Figure 4), while LOC_Os10g36270 showed inconsistent differential expression compared with the RNA-sequencing results (Figure S2). The expression level of LOC_Os10g36170 in the accessions with high DEC was lower than that in accessions with low DEC (p < 0.01), while expression levels of LOC_Os10g36260, LOC_Os10g36340 and LOC_Os10g36610 in the accessions with high DEC was higher than that in accessions with low DEC (p < 0.01) (Figure 4).
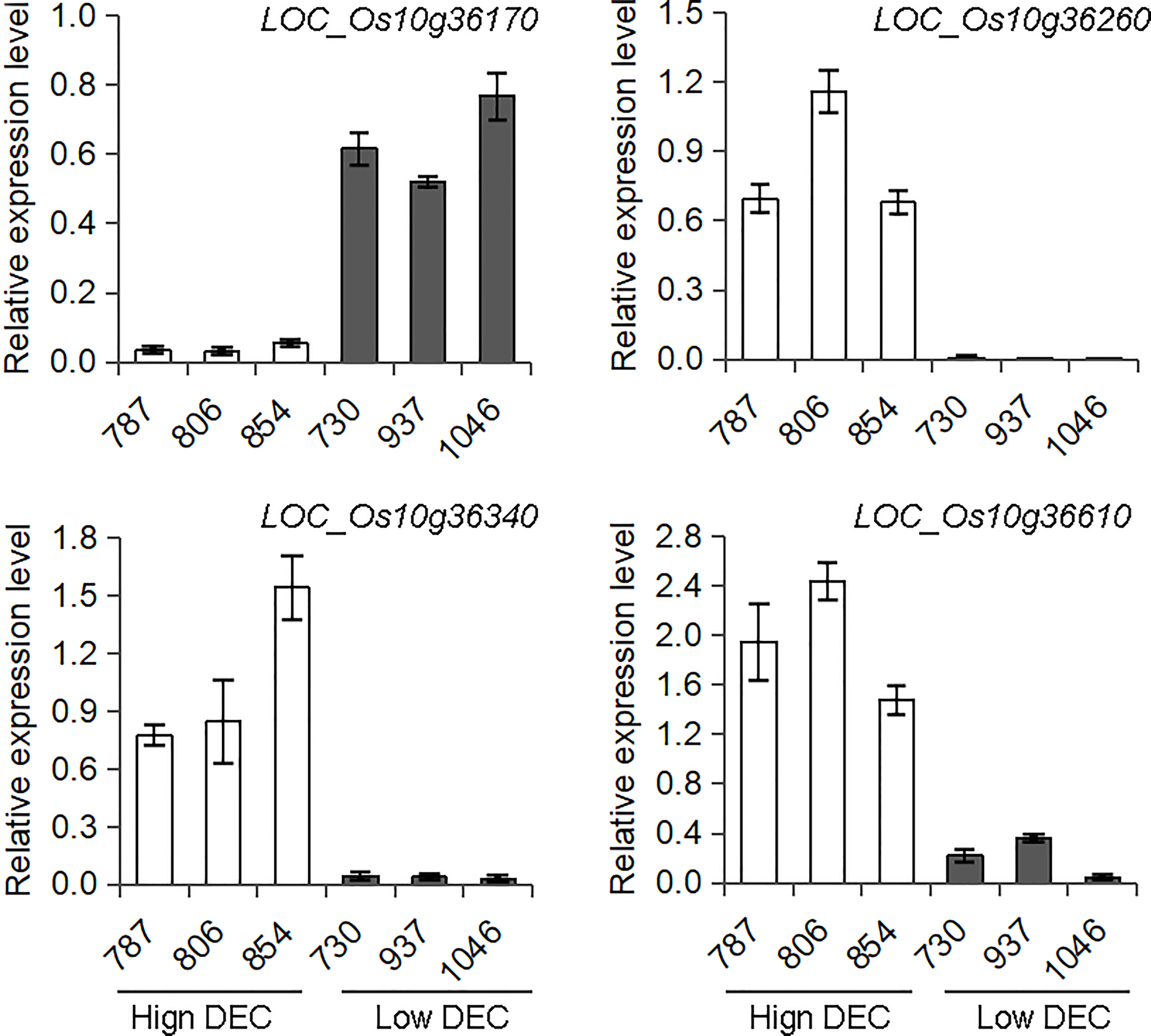
Figure 4 Expression analysis of candidate genes for qDEC-10c1w,2wj. The spikes were sampled on the15th day after flowering of three accessions with high DEC (accessions 787, 806 and 854) and three accessions with low DEC (accessions 730, 937 and 1046) were selected based on haplotype analysis to conduct RNA-sequencing and qRT-PCR. Different letters indicate significant difference at P < 0.01.
We further analyzed the promoter region (2 kb from ATG) of the four candidate genes using re-sequencing data of the 450 accessions, and conducted integration analysis between the four major haplotypes (Hap 1 to 4) of each gene and phenotypes. For LOC_Os10g36170, accessions with Hap 1 had higher PGWC and DEC than other haplotypes, and three sequence variations in the promoter region may be the difference between Hap 1 and the other haplotypes (Figure 5A; Table S3). For LOC_Os10g36260, accessions with Hap 1 had higher PGWC and DEC than other haplotypes, and 11 sequence variations in the promoter region may be the difference between Hap 1 and the other haplotypes (Figure 5B; Table S3). For LOC_Os10g36340, accessions with Hap 1 and Hap 2 had higher PGWC and DEC than Hap 3 and Hap 4, and the sequence deletion or existence in the promoter region may account for their differences (Figure 5C; Table S3). For LOC_Os10g36610, accessions with Hap 1 had higher PGWC and DEC than the other haplotypes, but no possible differences were found in the sequence comparison of their promoter regions (Figure 5D, Table S3).
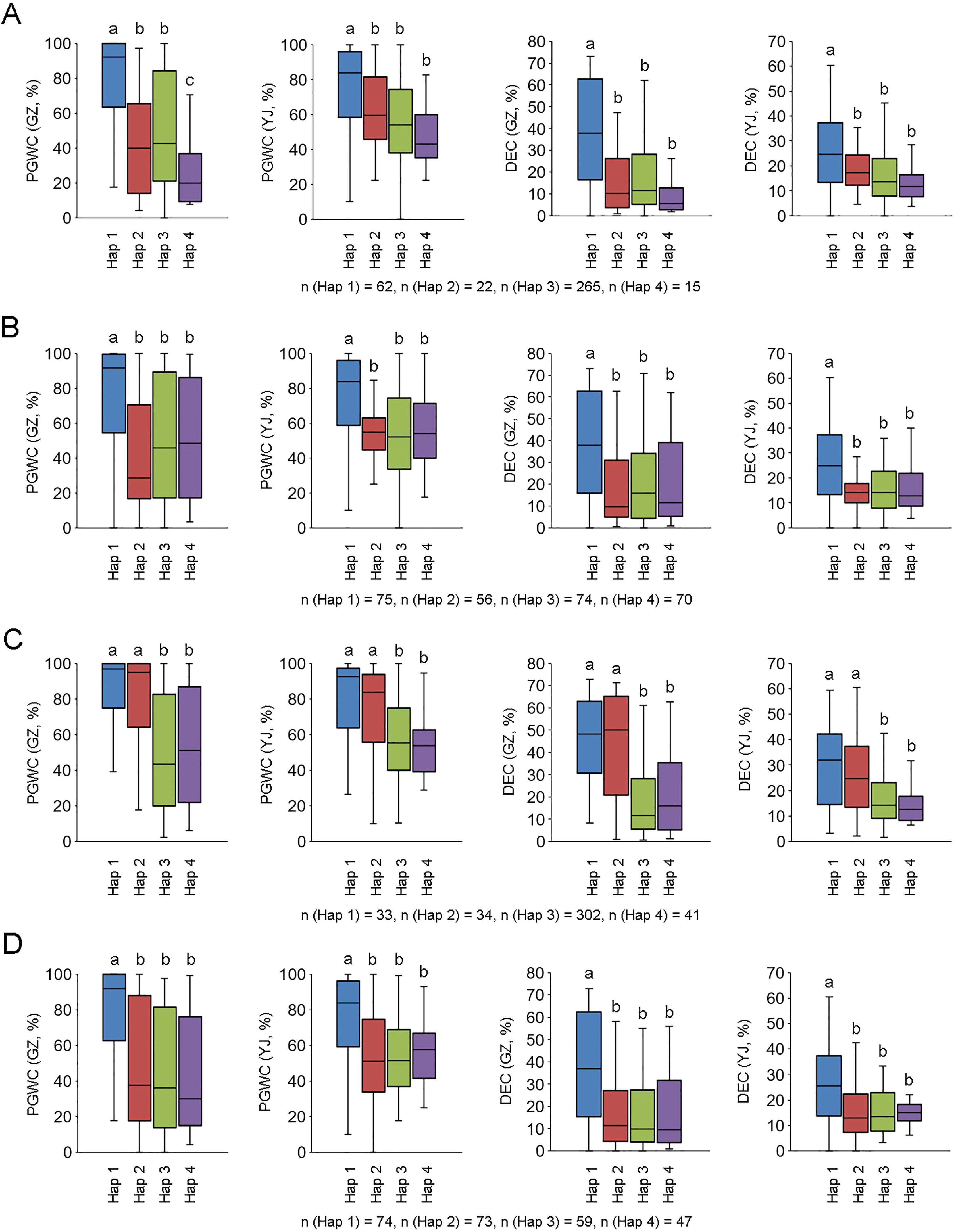
Figure 5 Phenotype comparisons among different haplotypes of four candidate genes. (A–D) are LOC_Os10g36170, LOC_Os10g36260, LOC_Os10g36340 and LOC_Os10g36610, respectively. Different letters indicate significant difference at P < 0.01.
PGWC and DEC are two important indexes to evaluate rice appearance quality, and some previous studies have reported that there is a high correlation between them (Wang et al., 2017; Qiu et al., 2021). In the present study, the correlation between PGWC and DEC was high (Figure 1B). The co-located QTLs identified for PGWC and DEC further confirmed this (Figure 2). Therefore, PGWC and DEC could be simultaneously improved by molecular breeding without fear of genetic drag. However, 18 out of 34 QTLs were only associated with one trait (Figure 2), indicating that there were different regulatory mechanisms among the two traits. Research on the specific regulation mechanism between PGWC and DEC will further enrich the genetic mechanism of grain chalkiness.
Among the 34 QTLs identified in this study, 11 QTLs were co-localized with the reported QTLs or genes (Table 1), indicating the reliability of GWAS analysis and diversity of rice accessions used in this study. Among them, qPGWC-3b1wj,2wj and qDEC-32w were co-located with GS3, and qPGWC-5a1wij,2wij and qDEC-5a1wij,2wij were co-located with GW5 (Table 1). In several studies, GS3 and GW5 loci have been reported to control grain shape and chalkiness simultaneously (Qiu et al., 2015; Wang et al., 2017), indicating that the two genes have multiple effects. However, the regulation intensity and direction of different alleles of GS3 and GW5 on multiple quality traits are worthy of careful study, which can provide support for the cooperative improvement of grain shape and chalkiness.
In addition to the four QTLs mentioned above, the other seven QTLs were co-located with QTLs that had not yet been cloned. qDCE-41w,2w could be identified in the two environments (Figure 2) and co-located with PGC4.4 (Misra et al., 2021)and qWCR4 (Yun et al., 2016). Misra et al. demonstrated that PGC4.4 is an important locus affecting chalkiness, and LOC_Os04g52230 and LOC_Os04g52240 may be the optimal candidate genes for this QTL by haplotype analysis (Misra et al., 2021). Other QTLs are co-located with some stable or unstable QTLs (Table 1). These information suggest that the stable expression of some QTLs is relative, greatly influenced by the environment, and may also be related to the background of research materials.
Because PGWC and DEC are highly positively correlated, it is possible to identify QTLs of PGWC and DEC in the similar region by using the same materials and environments. Wang et al. detected four QTLs affecting DEC and four QTLs affecting PGWC. Among them, qDEC3 is co-located with qPGWC3 and qDEC8 is co-located with qPGWC8 (Wang et al., 2017). Among the newly identified QTLs in this study, qPGWC-101w,2wj was co-located with qDEC-10c1w,2wj on chromosome 10, and they were stably detected in both environments and had the most significant p-value (Table 1). Interestingly, a QTL controlling grain width (qGW10.2) (Wang et al., 2017) was also located in the qDEC-10c1w,2wjregion. Since there is a high positive correlation between grain width and chalkiness (Qiu et al., 2015; Wang et al., 2017), whether qDEC-10c1w,2wj regulates both grain width and chalkiness needs further investigation. We further mined the candidate genes for qDEC-10c1w,2wj. By integrated analysis of interval definition, differential gene expression and haplotype comparison, four candidate genes were identified for qDEC-10c1w,2wj. Three genes (LOC_Os10g36260, LOC_Os10g36340, LOC_Os10g36610) encoding unknown functional proteins and one gene (LOC_Os10g36170) encoding a lipid transfer protein (LTP) were considered as its candidate genes. LTPs play diverse roles in various biological processes, such as pollen development, stress response, plant signaling and seed maturation (Zhang et al., 2010; Wang et al., 2012). A previous study indicated that down-regulated expression of OsLTPL36 leads to chalky endosperm and impaired seed germination in rice (Wang et al., 2015). Our results also exhibited that the expression level of LOC_Os10g36170 in the accessions with high DEC was significantly lower than that in accessions with low DEC (Figure 4), which speculates that LOC_Os10g36170 may functions on regulating the formation of grain chalkiness. In addition, we found some possible key sequence variants related to the expression and phenotype in the promoter regions of three genes by integrating phenotype, haplotype and sequence variation analysis (Table S3 and Figure 5). Further studies are needed to confirm the functions of these candidate genes in grain chalkiness through gain or loss-of function analysis. The identification of novel QTLs and candidate genes for rice appearance quality provides new sources for molecular breeding and cloning of genes associated with rice quality.
A total of 34 QTLs for grain chalkiness were identified by GWAS in 450 rice accessions. Based on the haplotype analysis, LD decay analysis, RNA-sequencing and promoter sequences analysis, four genes were considered as the candidate genes underlying the potential QTL (qDEC-10c1w,2w). These results will enrich the genetic bases of grain chalkiness and contributes to molecular breeding for high-quality rice.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA948558.
SZ, WY and JZ designed and supervised this work. XH, JW analyzed the data and wrote the article. LC, HF, TY and JD investigated the phenotype, and performed sample detection. YM and LZ performed the images. JC, DL and BL participated in the revision of the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Research and Development Plan of Key Fields in Guangdong Province(2018B020202004), the National Natural Science Foundation of China (32272085), the Guangdong Basic and Applied Basic Research Foundation (2023A1515010400), the Special Fund for Scientific Innovation Strategy-construction of High Level Academy of Agriculture Science (R2018YJ-YB1001 and R2021PY-QF002), the Project of Collaborative Innovation Center of GDAAS (XT202218), and the Guangdong Key Laboratory of New Technology in Rice Breeding (2020B1212060047).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1184276/full#supplementary-material
Supplementary Figure 1 | Manhattan plots of GWAS for PGWC and DEC using different populations in two environments.
Supplementary Figure 2 | The expression level of LOC_Os10g36270 measured by qRT-PCR.
Supplementary Table 1 | Information of the 450 rice accessions used in this study.
Supplementary Table 2 | The read count of genes within qDEC-10c1w,2wj region measured by RNA-seq.
Supplementary Table 3 | Sequence variations in promoter regions of the four candidate genes for qDEC-10c1w,2wj.
Supplementary Table 4 | Primers used in this study.
Bian, J., Li, C., He, H., Shi, H., Yan, S. (2014). Identification and analysis of QTLs for grain quality traits in rice using an introgression lines population. Euphytica 195, 83–93. doi: 10.1007/s10681-013-0976-1
Fan, T., Yang, W., Zeng, X., Xu, X., Xu, Y., Fan, X., et al. (2020). A rice autophagy gene OsATG8b is involved in nitrogen remobilization and control of grain quality. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00588
Kawahara, Y., de la Bastide, M., Hamilton, J. P., Kanamori, H., McCombie, W. R., Ouyang, S., et al. (2013). Improvement of the oryza sativa nipponbare reference genome using next generation sequence and optical map data. Rice (N Y) 6, 4. doi: 10.1186/1939-8433-6-4
Li, Y., Fan, C., Xing, Y., Yun, P., Luo, L., Yan, B., et al. (2014). Chalk5 encodes a vacuolar h(+)-translocating pyrophosphatase influencing grain chalkiness in rice. Nat. Genet. 46, 398–404. doi: 10.1038/ng.2923
Mao, H., Sun, S., Yao, J., Wang, C., Yu, S., Xu, C., et al. (2010). Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. U S 107, 19579–19584. doi: 10.1073/pnas.1014419107
McCouch, S. R., Wright, M. H., Tung, C. W., Maron, L. G., McNally, K. L., Fitzgerald, M., et al. (2016). Open access resources for genome-wide association mapping in rice. Nat. Commun. 7, 10532. doi: 10.1038/ncomms10532
Misra, G., Badoni, S., Parween, S., Singh, R. K., Leung, H., Ladejobi, O., et al. (2021). Genome-wide association coupled gene to gene interaction studies unveil novel epistatic targets among major effect loci impacting rice grain chalkiness. Plant Biotechnol. J. 19, 910–925. doi: 10.1111/pbi.13516
Peng, B., Wang, L., Fan, C., Jiang, G., Luo, L., Li, Y., et al. (2014). Comparative mapping of chalkiness components in rice using five populations across two environments. BMC Genet. 15, 49. doi: 10.1186/1471-2156-15-49
Qiu, X., Pang, Y., Yuan, Z., Xing, D., Xu, J., Dingkuhn, M., et al. (2015). Genome-wide association study of grain appearance and milling quality in a worldwide collection of indica rice germplasm. PloS One 10, e0145577. doi: 10.1371/journal.pone.0145577
Qiu, X., Yang, J., Zhang, F., Niu, Y., Zhao, X., Shen, C., et al. (2021). Genetic dissection of rice appearance quality and cooked rice elongation by genome-wide association study. Crop J. 9, 1470–1480. doi: 10.1016/j.cj.2020.12.010
Ryoo, N., Yu, C., Park, C. S., Baik, M. Y., Park, I. M., Cho, M. H., et al. (2007). Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white-core floury endosperm in rice (Oryza sativa l.). Plant Cell Rep. 26, 1083–1095. doi: 10.1007/s00299-007-0309-8
Turner, S. D. (2018). Qqman: An r package for visualizing GWAS results using q-q and manhattan plots. J. Open Source Software 3, 731. doi: 10.21105/joss.00731
Wang, H. W., Hwang, S. G., Karuppanapandian, T., Liu, A., Kim, W., Jang, C. S., et al. (2012). Insight into the molecular evolution of non-specific lipid transfer proteins via comparative analysis between rice and sorghum. DNA Res. 19, 179–194. doi: 10.1093/dnares/dss003
Wang, X., Pang, Y., Wang, C., Chen, K., Zhu, Y., Shen, C., et al. (2017). New candidate genes affecting rice grain appearance and milling quality detected by genome-wide and gene-based association analyses. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01998
Wang, Q., Tang, J. L., Han, B., Huang, X. H. (2020). Advances in genome-wide association studies of complex traits in rice. Theor. Appl. Genet. 133, 1415–1425. doi: 10.1007/s00122-019-03473-3
Wang, E., Wang, J., Zhu, X., Hao, W., Wang, L., Li, Q., et al. (2008). Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 40, 1370–1374. doi: 10.1038/ng.220
Wang, J., Yang, W., Zhang, S., Hu, H., Yuan, Y., Dong, J., et al. (2023). A pangenome analysis pipeline provides insights into functional gene identification in rice. Genome Biol. 24, 19. doi: 10.1186/s13059-023-02861-9
Wang, X., Zhou, W., Lu, Z., Ouyang, Y., O, C. S., Yao, J. (2015). A lipid transfer protein, OsLTPL36, is essential for seed development and seed quality in rice. Plant Sci. 239, 200–208. doi: 10.1016/j.plantsci.2015.07.016
Weng, J., Gu, S., Wan, X., Gao, H., Guo, T., Su, N., et al. (2008). Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 18, 1199–1209. doi: 10.1038/cr.2008.307
Wu, B., Yun, P., Zhou, H., Xia, D., Gu, Y., Li, P., et al. (2022). Natural variation in WHITE-CORE RATE 1 regulates redox homeostasis in rice endosperm to affect grain quality. Plant Cell 34, 1912–1932. doi: 10.1093/plcell/koac057
Yang, W., Chen, L., Zhao, J., Wang, J., Li, W., Yang, T., et al. (2022b). Genome-wide association study of pericarp color in rice using different germplasm and phenotyping methods reveals different genetic architectures. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.841191
Yang, W., Jiang, X. Y., Xie, Y. L., Chen, L., Zhao, J. L., Lui, B., et al. (2022c). Transcriptome and metabolome analyses reveal new insights into the regulatory mechanism of head milled rice rate. Plants-Basel 11, 2838. doi: 10.3390/plants11212838
Yang, W., Xu, P., Zhang, J., Zhang, S., Li, Z., Yang, K., et al. (2022a). OsbZIP60-mediated unfolded protein response regulates grain chalkiness in rice. J. Genet. Genomics 49, 414–426. doi: 10.1016/j.jgg.2022.02.002
Yang, W., Zhao, J., Zhang, S., Chen, L., Yang, T., Dong, J., et al. (2021). Genome-wide association mapping and gene expression analysis reveal the negative role of OsMYB21 in regulating bacterial blight resistance in rice. Rice (N Y) 14, 58. doi: 10.1186/s12284-021-00501-z
Yun, P., Zhu, Y., Wu, B., Gao, G., Sun, P., Zhang, Q., et al. (2016). Genetic mapping and confirmation of quantitative trait loci for grain chalkiness in rice. Mol. Breed. 36, 162. doi: 10.1007/s11032-016-0600-x
Zhang, D., Liang, W., Yin, C., Zong, J., Gu, F., Zhang, D. (2010). OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 154, 149–162. doi: 10.1104/pp.110.158865
Zhao, X., Daygon, V. D., McNally, K. L., Hamilton, R. S., Xie, F., Reinke, R. F., et al. (2016). Identification of stable QTLs causing chalk in rice grains in nine environments. Theor. Appl. Genet. 129, 141–153. doi: 10.1007/s00122-015-2616-8
Zhao, J., Yang, W., Zhang, S., Yang, T., Liu, Q., Dong, J., et al. (2018). Genome-wide association study and candidate gene analysis of rice cadmium accumulation in grain in a diverse rice collection. Rice (New York N.Y.) 11, 61. doi: 10.1186/s12284-018-0254-x
Keywords: rice, grain chalkiness, genome-wide association study, quantitative trait loci (QTL), candidate gene
Citation: Huo X, Wang J, Chen L, Fu H, Yang T, Dong J, Ma Y, Zhou L, Chen J, Liu D, Liu B, Zhao J, Zhang S and Yang W (2023) Genome-wide association mapping and gene expression analysis reveal candidate genes for grain chalkiness in rice. Front. Plant Sci. 14:1184276. doi: 10.3389/fpls.2023.1184276
Received: 11 March 2023; Accepted: 27 March 2023;
Published: 14 April 2023.
Edited by:
Yue Feng, China National Rice Research Institute (CAAS), ChinaReviewed by:
Jw Wu, South China Agricultural University, ChinaCopyright © 2023 Huo, Wang, Chen, Fu, Yang, Dong, Ma, Zhou, Chen, Liu, Liu, Zhao, Zhang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaohong Zhang, c3poYW5nZ3pAdG9tLmNvbQ==; Wu Yang, eWFuZ3d1QGdkYWFzLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.