
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 06 July 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1183445
This article is part of the Research TopicCrop Physiological Responses to Abiotic StressView all 22 articles
 Yang Shi1
Yang Shi1 Nan Jiang1
Nan Jiang1 Mengting Wang1
Mengting Wang1 Zhiye Du1
Zhiye Du1 Ji Chen2
Ji Chen2 Yanyan Huang3
Yanyan Huang3 Mingyu Li1
Mingyu Li1 Yufan Jin1
Yufan Jin1 Jiahao Li1
Jiahao Li1 Jian Wan2
Jian Wan2 Xiaowan Jin2
Xiaowan Jin2 Lang Zhang2
Lang Zhang2 Jin Huang1*
Jin Huang1*Introduction: Heavy metal-associated isoprenylated plant proteins (HIPPs) play vital roles in metal absorption, transport and accumulation in plants. However, so far, only several plant HIPPs have been functionally analyzed. In this study, a novel HIPP member OsHIPP17, which was involved in the tolerance to copper (Cu) was functionally characterized.
Methods: In this study, qRT-PCR, Yeast transgenic technology, Plant transgenic technology, ICP-MS and so on were used for research.
Results: OsHIPP17 protein was targeted to the nucleus. The Cu concentration reached 0.45 mg/g dry weight due to the overexpression of OsHIPP17 in yeast cells. Meanwhile, the overexpression of OsHIPP17 resulted in the compromised growth of Arabidopsis thaliana (Arabidopsis) under Cu stress. The root length of Oshipp17 mutant lines was also significantly reduced by 16.74- 24.36% under 25 mM Cu stress. The roots of Oshipp17 rice mutant showed increased Cu concentration by 7.25%-23.32%. Meanwhile, knockout of OsHIPP17 decreased the expression levels of OsATX1, OsZIP1, OsCOPT5 or OsHMA5, and increased the expression levels of OsCOPT1 or OsHMA4. Antioxidant enzyme activity was also reduced in rice due to the knockout of OsHIPP17. Moreover, the expression levels of cytokinin-related genes in plants under Cu stress were also affected by overexpression or knockout of OsHIPP17.
Discussion: These results implied that OsHIPP17 might play a role in plant Cu toxic response by affecting the expression of Cu transport genes or cytokinin-related genes. Simultaneously, our work may shed light on the underlying mechanism of how heavy metals affect the plant growth and provide a novel rice genetic source for phytoremediation of heavy metal-contaminated soil.
Copper (Cu), which plays key roles in many biological activities such as energy production, response to oxidative stress and cell wall formation is an essential micronutrient for all living organisms including plants (Huang et al., 2016). Although the deficiency of Cu in plants may cause growth limitation, chlorosis or other development defects, excessive Cu is toxic to plants, in which case, inhibition of plant root growth or cellular damage has been observed (Guan et al., 2022; Li et al., 2023). Nevertheless, Cu is also required for human health, for example, Cu deficiency may cause immune defects or anemia (Torkian et al., 2019; Guan et al., 2022). It has been estimated that over two billion people worldwide are in the trouble of micronutrient deficiencies, which are mainly caused by deficient intake of Cu or iron (Fe) (Huang et al., 2016). Dietary intake is the main supplement resource of Cu for the human body. Therefore, improving Cu content in the crops, such as rice, by regulating absorption or transport related genes, may be an optional approach to improve dietary Cu for these people who lack Cu. Meanwhile, the development of industrialization and urbanization has also increased farmland pollution by releasing heavy metals including Cu. It has been reported that Cu is one of the major pollution sources of farmland in China (Zhao et al., 2015). Therefore, developing rice cultivars resistant to Cu and accumulating appropriate levels of Cu in the grain is crucial.
To maintain proper Cu content, plants have developed a fine-tuned Cu homeostasis system in their cells (Cai and Peng, 2013). In eukaryotes, when metallic ions enter the cell, they are chelated by some specific molecules such as proteins or small sized ligands (Guo et al., 2013). Nonetheless, to function as structural components or co-factors of enzymes, specific localization is usually required for these metallic ions. To this end, the transport task of these metallic ions is undertaken by a group of proteins named metallochaperones. These proteins contain conserved domains called heavy metal-associated (HMA) domains, which are able to directly bind metal ions (Zhao et al., 2020). Usually, these proteins exert their functions in metal homeostasis via their ability of metal binding (Zhang et al., 2020). According to the protein domain composition, plant metallochaperones are divided into two subfamilies, those proteins that contain only HMA domains are classified as heavy metal associated plant proteins (HPPs), and the proteins bearing both HMA domains and C-terminal isoprenylation motifs are termed as heavy metal associated isoprenylated plant proteins (HIPPs) (de Abreu-Neto et al., 2013). Studies have shown that members of the HIPPs family affect the uptake, transport and accumulation of heavy metals of plants (Tehseen et al., 2010). For example, the expression of HIPP26 (AtFP6) is induced by cadmium (Cd) and zinc (Zn) in Arabidopsis, and HIPP26 protein bind Cd, lead (Pb) or Cu (Gao et al., 2009). In rice, OsHIPP29 locus has been proven to enhance Cd-tolerance by reducing Cd accumulation in the plant (Zhao et al., 2020). Although rice HIPPs family has a large number of members, only a few of these HIPP genes have been functionally studied (Khan et al., 2019; Khan et al., 2020; Zhao et al., 2020; Chen and Xiong, 2021).
So far, most studies have focused on the function of HIPP genes in plants in response to heavy metal stress. Beyond this, HPP/HIPP genes have been reported to regulate cytokinin signaling pathways, and cytokinin homeostasis is implicated in the regulation of root development in plants (Guo et al., 2021). At the same time, endogenous cytokinin concentration in plants varies due to abiotic stress conditions, which implies that cytokinin is implicated in stress response (Brien and Benkova, 2013). But, so far, whether HIPP genes may affect abiotic stress resistance in plants by regulating the cytokinin signaling pathway is still unclear.
In our previous work, we have obtained a number of rice heavy metal-responsive genes by screening the transcriptome databases (unpublished data), as one of these genes, OsHIPP17(LOC_Os09g09930) was chosen for further study. Although OsHIPP17 is responsive to heavy metals, its biological function has not been characterized yet. In this study, we further analyzed the function and potential mechanism of OsHIPP17 mediated Cu stress response in yeast, Arabidopsis or rice. The purpose of this study is to further elucidate the diverse biological roles of the HIPP genes and to explain the mechanism of HIPP mediated heavy metals affected plant growth.
The rice genome data (accession date: 26th November 2018) and gff3 annotation files were obtained from EnsemblPlant database (http://plants.ensembl.org/index.html), and the protein database was constructed using TBtools software (version 1.098769). The sequence of the HMA domain (pfam 00403.23) from Pfam database (https://pfam.xfam.org/) was used as a reference sequence to compare with the protein database to obtain proteins that may contain the HMA domain. The retrieved proteins were further validated by using Web CD-Search Tool of NCBI website (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) and the bulk search tool of Pfam website. Finally, HIPP or HPP proteins were identified based on the presence or absence of C-terminal isoprenylation motif. Phylogenetic analysis was conducted by using MEGA7.0 software (version 7.0.26). The Neighbor-Joining (NJ) method and bootstrap replications of 1000 were employed (Zhang et al., 2018a).
For the analysis of the expression level of OsHIPP17 under Cu stress, rice seedlings were grown at 30°C with a 16-hours-light/8-hours-dark cycle. The seedlings of 5-day-old rice plants were transferred to 1/2 Murashige and Skoog (1/2 MS) liquid medium containing 25, 50 or 100 μM CuSO4 (Murashige and Skoog, 1962). According to the reported method, the shoots and roots of Cu-treated rice plants were sampled after treatment with time gradients (1, 6 or 12 hours) and kept at -80°C for further analysis (Lee et al., 2007; Chen and Xiong, 2021). The shoots and roots of Cu-treated rice plants were used for RNA extraction. Total RNAs were extracted by using EASYspin Plus reagent (Aidlab Biotechnologies Co. Ltd, Beijing, China). The cDNA was obtained by using a reverse transcription kit (Thermo fisher, USA) according to the manufacturer’s instructions. Triplicate quantitative assays were performed by using T5 Fast qPCR Mix kit (Aidlab Biotechnologies Co. Ltd, Beijing, China). The rice Ubiquitin gene (LOC_Os05g06770) was used as a housekeeping gene. The qRT-PCR primers were summarized in Table S1.
Seeds of wild type (Control) and OsHIPP17 overexpression Arabidopsis (OE-3, OE-7 or OE-8) were placed on 1/2 MS media (30 g/L sucrose + 7 g/L agar) containing different concentrations of CuSO4 (0 or 50 μM) for 15 days. Total RNA extraction from plant roots and cDNA synthesis were performed as described above. The expression levels of AtCKX1, AtIPT1 and AtARR5 were analyzed by quantitative real-time polymerase chain reaction (qRT-PCR). The thermocycler was set as follows: 95°C for 3 min, 40 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min. The Arabidopsis Ubiquitin (At5g53300) was used as a housekeeping gene to normalize the other genes.
Rice Oshipp17 knockout mutants were generated by CRISPR-Cas9 technology. To generate Oshipp17 mutant rice plants, Wimi Biotechnology company (http://www.wimibio.com/) was entrusted for the construction of knockout vector and rice transformation. Seven-days-old wild-type (Control) and Oshipp17 mutant (Oshipp17-1 and Oshipp17-2) rice seedlings were placed in 1/2 MS liquid medium containing different concentrations of CuSO4 (0 or 25 μM) for another fifteen days. Total RNA extraction from plant roots and cDNA synthesis were performed as described above. The expression levels of OsCKX1, OsIPT1, OsRR1, OsATX1, OsZIP1, OsCOPT1, OsCOPT5, OsHMA4 and OsHMA5 were analyzed by qRT-PCR. The thermocycler was set as follows: 95°C for 3 min, 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min. The rice Ubiquitin was used as a housekeeping gene to normalize the other genes.
The galactose induced expression vector pYES2 has been used to verify the function of heavy metal-responsive genes in Saccharomyces cerevisiae (yeast) (Khan et al., 2019; Chen and Xiong, 2021). Yeast strain BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0) has been widely used to explore the function of plant genes under heavy metal stress (Polina et al., 2016; Wang et al., 2019). Therefore, the pYES2 empty vector (Control) or pYES2::OsHIPP17 was transformed into the yeast strain BY4743, via a polyethylene glycol (PEG)-lithium acetate-based transformation method (Gietz and Woods, 2006). The transformed yeast cells were selected on SD-Ura medium plates. For yeast spotting assays, a single colony of the yeast transformant was cultured in liquid SD-Ura medium for 24 hours at 30°C, and the liquid medium containing the yeast cells was diluted for 10 folds with fresh SD-Ura medium. Then the diluted yeast cells were further cultivated till the values of OD600 increased to 0.5~0.8, and the yeast cells were collected and diluted with sterile water. The 5 μL drops containing diluted yeast cells (OD600 = 1.0, 0.1, 0.01 or 0.001) were spotted onto the surfaces of SD-Ura medium with 2% galactose and 0, 2 or 2.5 mM CuSO4. The spotted plates were incubated for 3 days at 30°C. For Cu toxicity growth curve assay, the yeast cells with an OD600 of 1.0 were collected and then diluted with 20 mL liquid SD medium containing 2% galactose and 0 or 2.5 mM CuSO4 to an OD600 of 0.01. The yeast cells were cultured in a shaking incubator at 200 rpm at 30°C. The OD600 values of these yeast cells were determined after 6, 12, 18, 24 or 30 hours.
To determine the effect of OsHIPP17 on the Cu absorption capability in the yeast cells, the Cu contents of the control and transgenic yeast cells were measured. The control and transgenic yeast cells were grown in SD-Ura liquid medium containing 2% glucose overnight. After centrifugation, yeast cells were washed three times with sterile water and diluted with 250 mL SD-Ura liquid medium containing 2% galactose to an OD of 0.1, and these cells were cultured in a constant temperature shaking incubator at 30°C until the OD reached 0.4-0.5. Then, CuSO4 was added into the liquid medium to a final concentration of 10 μM, and the control and transgenic yeast cells were incubated for another 12 hours. These yeast cells were collected and washed five times with deionized water. The yeast cells treated with CuSO4 in the liquid medium were collected and dried at 85°C to constant weight. The dried yeast cells were digested with HNO3 and H2O2. The Cu content of the samples was determined by using inductively coupled plasma-mass spectrometry (ICP-MS) (Zhang et al., 2018b).
To measure Cu content in rice tissues, shoots and roots of wild-type (Control) and Oshipp17 mutant (Oshipp17-1 and Oshipp17-2) rice plants treated with 25 μM CuSO4 for fifteen days were collected. These rice tissues were washed five times with deionized water in order to remove metal ions from the surface. Then, the rice tissues were dried at 85°C to constant weight. The Cu content in rice tissues was determined with reference to the above method. Moreover, the translocation factor of Cu in rice plants was calculated using the methods described previously (Meng et al., 2022).
Plasmid pHB, a plant binary expression vector, effectively increases the expression level of exogenous genes due to CaMV 35S promoter. Fragments of the coding sequences of OsHIPP17 and eGFP were ligated into the binary vector pHB by using seamless homologous recombination technique for the expression of OsHIPP17-eGFP fusion protein in plants. ClonExpress II One Step Cloning Kit from Vazyme (Vazyme, Nanjing, China) was used in the construction of all recombinant vectors in this study. The gene expression of OsHIPP17-eGFP or eGFP was driven by CaMV 35S promoter. Agrobacterium tumefaciens(Agrobacterium) containing eGFP, OsHIPP17-eGFP and Agrobacterium harboring the gene encoding the silencing inhibitor protein p19 were diluted to OD = 0.8 by the infection solution (10 mM MgCl2, 10 mM 2-Morpholinoethanesulfonic acid, 100 μM Acetosyringone, pH = 5.7). Tobacco (Nicotiana benthamiana) leaves were used for Agrobacterium agroinfiltration. The agroinfiltrated plants were moved to a plant culture room of 25°C. For GFP or GFP-fused protein imaging, the fluorescence emitted by eGFP or OsHIPP17-eGFP was observed by using a confocal laser scanning microscope (Nikon A1i90, LSCM, Japan) 24 hours after infiltration. The nucleus staining was conducted with 4’,6-diamidino-2-phenylindole (Dapi) by dipping the tobacco leaves into Dapi solution for at least 30 min before fluorescence observation (Shinohara et al., 2014).
To generate OsHIPP17 overexpression Arabidopsis plants, the binary vector pHB::OsHIPP17 was constructed. The pHB::OsHIPP17 plasmid was transferred into Agrobacterium strain GV3101, and Arabidopsis was infected with the Agrobacterium containing the binary vector plasmids. The transformation was carried out following the reported protocol with minor modifications (Clough et al., 1998). The Agrobacterium containing pHB::OsHIPP17 plasmid were diluted to OD = 0.8 by the infiltration solution (1/2 MS, 0.03% sucrose, 1 mg/ml, 6-Benzylaminopurine, 0.01% Silwet l-77, pH = 5.7). For floral dipping, the Arabidopsis was inverted into the infection solution containing Agrobacterium to submerge all flowers for 30 s and then pulled out, and the procedure was repeated after 10 min. The infected Arabidopsis were placed in moist black plastic bags for 24 hours before removing the covers and moving to normal culture condition of 16-hours-light/8-hours-dark cycle at 23°C. The selection medium (1/2 MS, 30 g/L sucrose, 7 g/L agar, 10 mg/L hygromycin B) was used to screen positive plants. The T2 generation transgenic plants were used for Cu stress analysis. The primer pairs used for transgene confirmation were summarized in Table S1.
To determine whether the sensitivity to Cu of transgenic Arabidopsis plants was affected, the surface sterilized seeds of wild type (Control) and overexpression plants (OE-3, OE-7 or OE-8) were placed on the surface of 1/2 MS agar media containing different concentration of Cu (0, 35 μM CuSO4) at 4°C. After three days, these seeds were moved to plant culture room (23°C, 16 hours of light and 8 hours of darkness). After another 15 days, the length of plant roots was measured by using ImegaJ software (version: 1.52a) in five independent plants (Xian et al., 2020). Moreover, the effect of overexpression OsHIPP17 to germination rate of Arabidopsis seeds was analyzed under 250 μM Cu treatment based on previous studies (Tejpal et al., 2012).
To explore the effect of OsHIPP17 mutation on rice plants under Cu stress, seeds of wild-type (Control) and Oshipp17 mutants (Oshipp17-1 and Oshipp17-2) were sterilized and placed in 1/2 MS liquid medium grown in plant culture room for seven days. The 7-day-old rice seedlings were grown in 1/2 MS liquid medium with different Cu concentrations (0, 25 μM CuSO4) for another fifteen days, and the medium was changed every day. Then, the phenotype of wild-type and Oshipp17 mutant rice plants was observed. The length of rice roots and shoots was measured by using ImegaJ software in three independent plants.
The Arabidopsis (whole seedling) and rice (leaves at the same position) treated with different concentrations of Cu for fifteen days were frozen with liquid nitrogen and ground. Extraction buffer (0.1 M K2HPO4-KH2PO4, 1 mM EDTA, 0.3% Triton X-100, 2% polyvinylpyrrolidone, pH = 7.6) was added to the grinded tissues to prepare enzyme solution. Superoxide dismutase (SOD), peroxidase (POD) and catalase (CAT) activities in plants were analyzed according to the published protocol with minor modification (Chen and Zhang, 2016).
All results in this study were derived from the average of at least three biological replications. All values in the chart are represented by “averages and standard deviations.” The data were statistically analyzed by t-tests method (p< 0.05). The statistical analysis was conducted by using Graphpad Prism software (version: 9.0.2). The online site Hiplot (https://bukesci.com/sites/527.html) was used to calculate the values of pearson.
The function of proteins is tightly related with their protein structure and domain composition. To better understand the function of OsHIPP17 and HPP/HIPP proteins in rice, bioinformatics analysis was carried out. The amino acid sequence of HMA domain (pfam 00403.23) was employed to retrieve HPP/HIPP proteins from the rice genome (Khan et al., 2019). There were 54 HMA containing proteins were identified (Table S2). Phylogenetic analysis showed that these rice HPP/HIPP proteins could be divided into five distinct subfamilies (Figure 1A). Among these rice HMA containing proteins, OsHPP4 showed the closest relation to OsHIPP17. Besides the study of protein phylogenetic relation, the conserved motifs of these rice HIPP proteins were also analyzed by using MEME website (http://meme-suite.org/tools/meme). The results indicated that among these HIPP proteins, there were total ten conserved motifs. It was noticed that the motif composition significantly differs between HIPP protein members. However, similar motif composition was shown among the members belonging to the same subfamily, indicating their conserved protein functions (Figure 1B). Furthermore, an uneven distribution pattern of these 54 rice HPP/HIPPs on the 12 chromosomes of rice was observed. For example, the chromosome number one (Chr. 1) contained as much as 12 HPP/HIPP genes (Figure 1C).
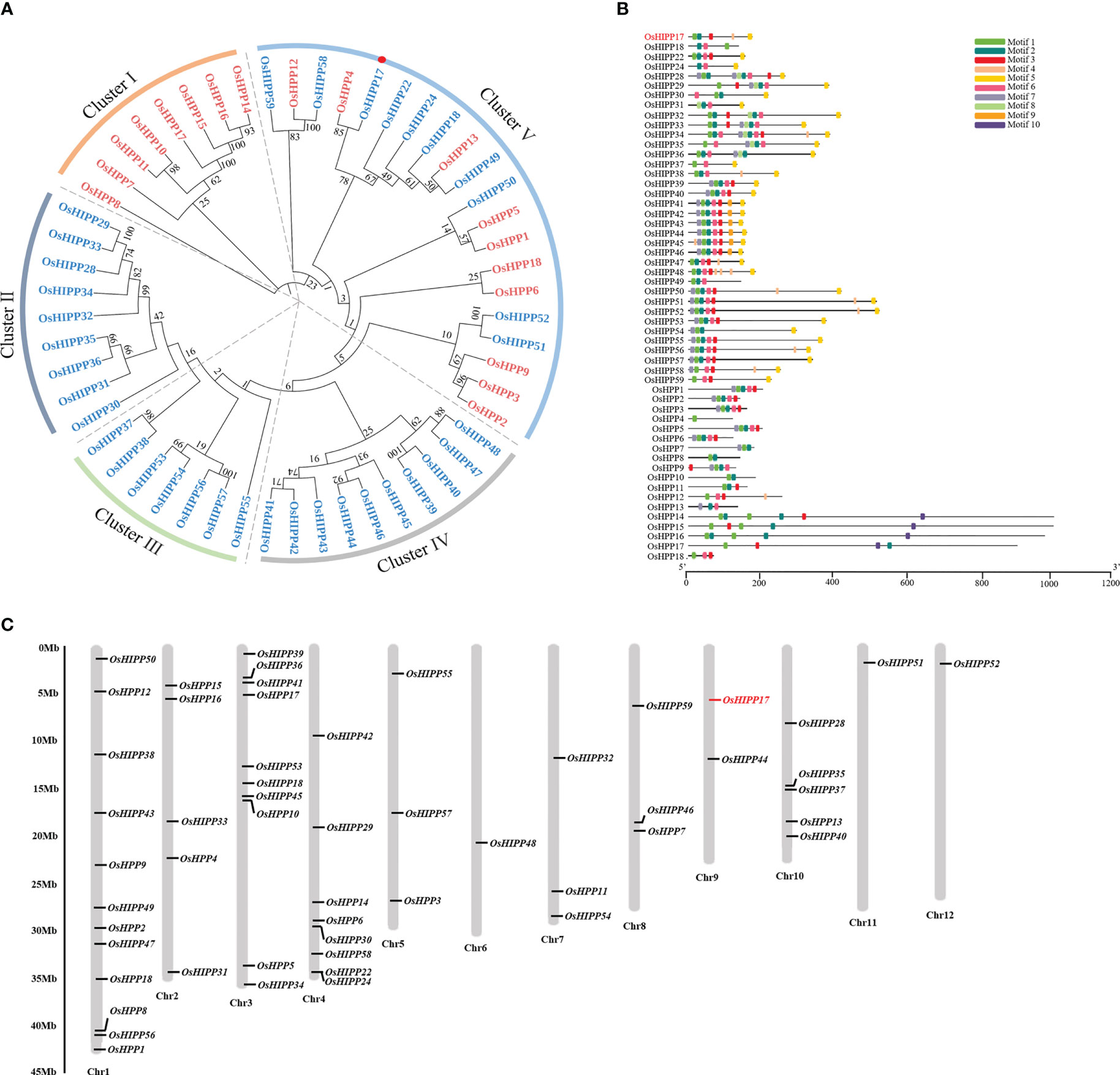
Figure 1 Bioinformatics analysis of HPP and HIPP protein family. (A) Phylogenetic analysis of HPP and HIPP proteins in rice. The HPP proteins were written in red and the HIPP proteins were written in blue. (B) Conserved motif analysis of rice HPP and HIPP proteins. The analysis was performed by using MEME program. (C) Chromosomal mapping of HPP and HIPP genes of rice. The chromosome numbers were indicated at the bottom of each chromosome.
To further confirm if OsHIPP17 is involved in the Cu stress response, the gene expression of OsHIPP17 was analyzed by using the rice plants treated with series of concentrated CuSO4. The results of qRT-PCR analysis showed that 100 μM Cu treatment significantly induced the expression of OsHIPP17 up to 199 folds in the shoots after 1 hour treatment (Figure 2D). Meanwhile, the expression levels of OsHIPP17 in roots were increased after 12 hours of treatment of 25 or 50 μM Cu (Figures 2B, C). In addition, the expression levels of OsHIPP17 in roots were increased after 1 hours of treatment of 100 μM Cu, but were decreased after 12 hours (Figure 2D). In addition, the expression levels of OsHIPP17 differ with growth stage (Figure 2A).
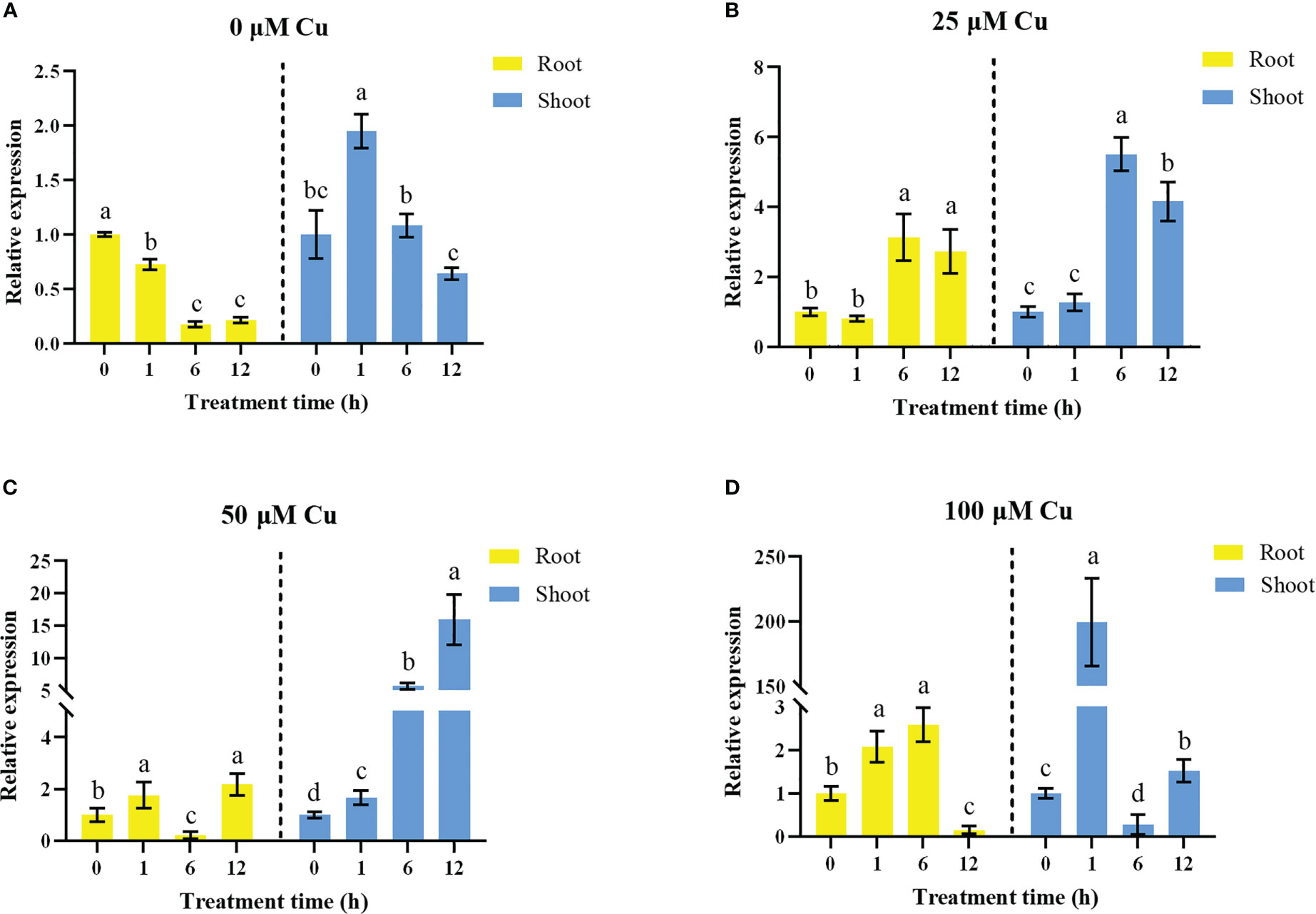
Figure 2 Expression of OsHIPP17 when treated with differently concentrated CuSO4. The 5-day-old rice plants were exposed to CuSO4 (0, 25, 50 or 100 mM) for 0, 1, 6 or 12 hours. (A) 0 μM; (B) 25 μM; (C) 50 μM; (D) 100 μM. Error bars represent standard deviation of three independent plants. Statistical comparison was performed by t-tests. Different letters (a-d) indicate significant differences (p< 0.05).
To determine the effect of OsHIPP17 on Cu tolerance of yeast cells, the growth of a yeast BY4743 strain containing OsHIPP17 gene was compared with that of the strain containing empty pYES2 vector under Cu treatment. When supplied with 2.5 mM Cu, OsHIPP17 significantly repressed the growth of yeast compared with the control, suggesting that OsHIPP17 decreased Cu tolerance of yeast (Figure 3A). To further confirm and quantify the effect of OsHIPP17 on the Cu tolerance of yeast, we measured the growth rate of yeast cells transformed with OsHIPP17 or empty vector in SD-Ura liquid medium containing 0, 2.5 mM Cu. The results showed that the growth of transgenic yeast containing OsHIPP17 was significantly repressed compared with that of the control in the medium containing 2.5 mM Cu after 36 hours (Figure 3B). Nevertheless, more Cu content was detected in the yeast cells containing OsHIPP17 than that in the control (Figure 3C).
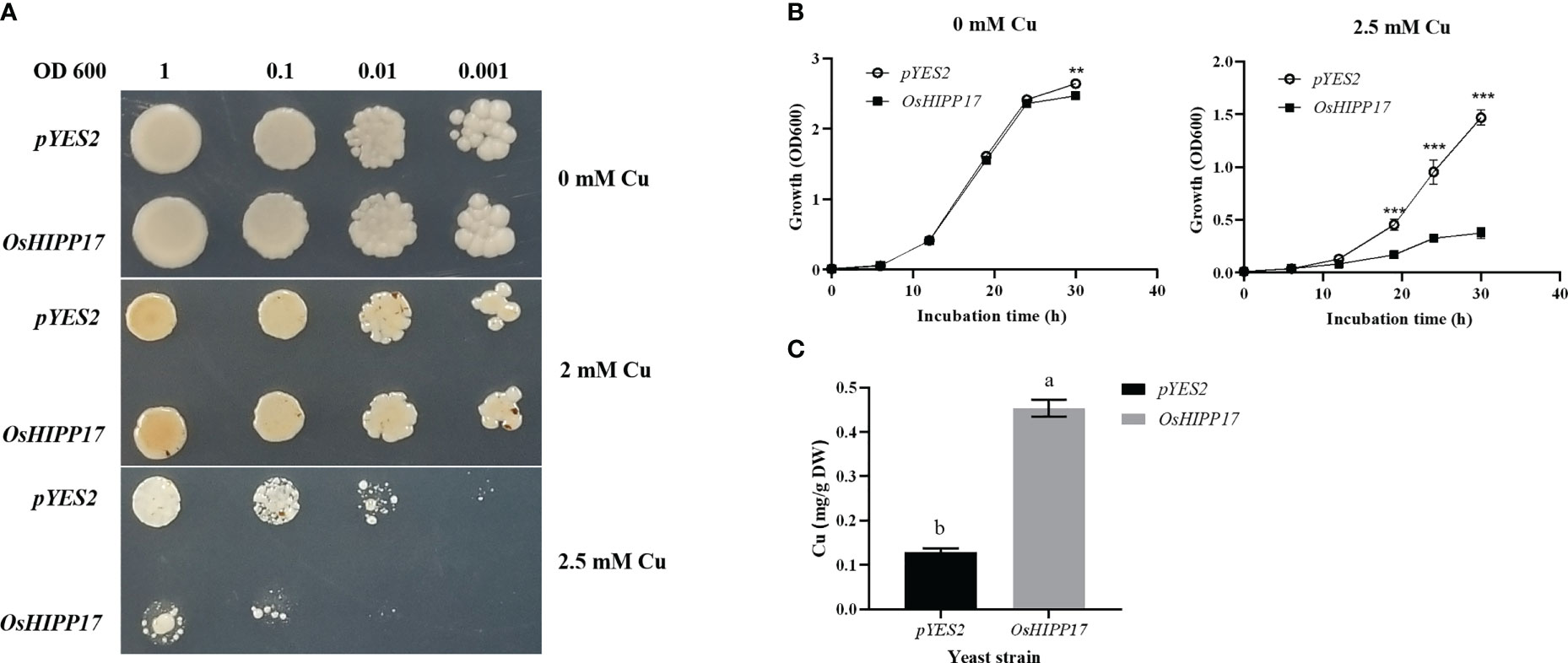
Figure 3 Functional analysis of OsHIPP17 in yeast. (A) The effects of OsHIPP17 on the sensibility of Cu in yeast. (B) The growth rate of yeast cells when treated with Cu. (C) The Cu contents in the yeast cells treated with 10 μM CuSO4 for 12 hours. Statistical comparison was performed by t-tests. Different letters (a, b). ** and *** indicate significant differences at p < 0.05 and p < 0.001, respectively.
To determine the subcellular localization of OsHIPP17 proteins in yeast and plant cells, the construct of eGFP fused to the C-terminus of OsHIPP17 which was under the control of GAL1 promoter (GAL1::OsHIPP17-eGFP) or 35S promoter (35S::OsHIPP17-eGFP) was use for transformation. The fluorescence signals in yeast cells or tobacco leaves were observed. In the yeast cells, eGFP signals were observed in the nucleus (Figure 4A). Meanwhile, OsHIPP17-eGFP proteins were located in the nucleus of tobacco leave epidermal cells compared to the localization of eGFP alone (Figure 4B).
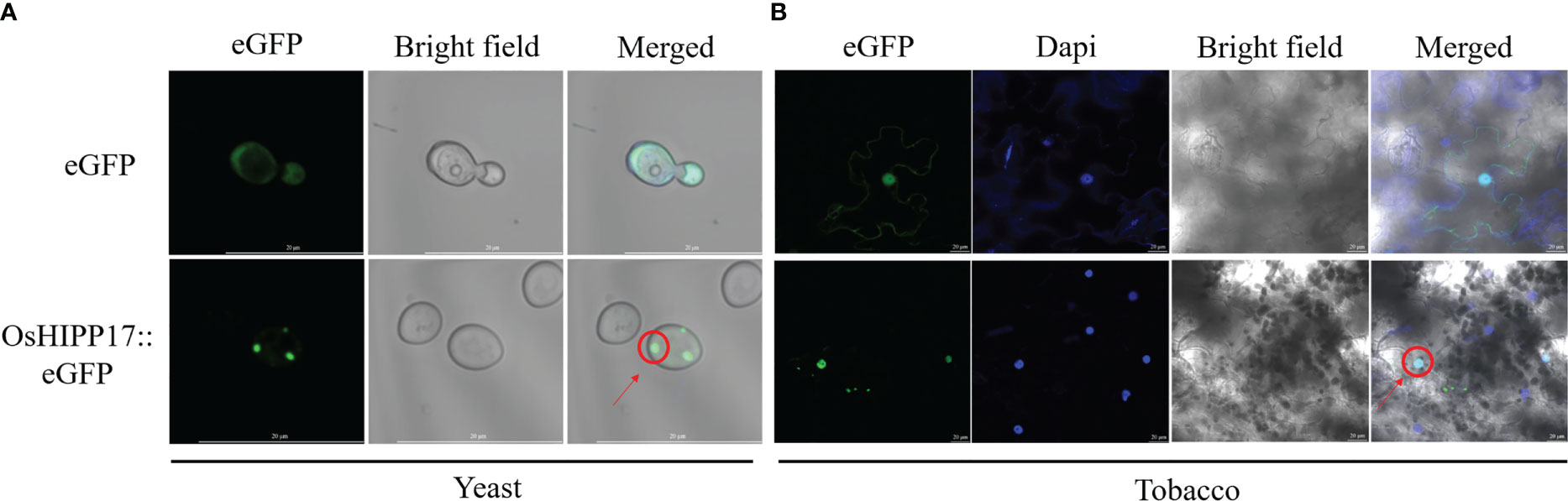
Figure 4 Subcellular localization of OsHIPP17 proteins. (A) Subcellular localization of OsHIPP17 proteins in yeast cells. Scale bars in A =20 µm. The pYES2::eGFP (for control) or pYES2::OsHIPP17-eGFP (for obtaining OsHIPP17 subcellular localization information) were transferred into yeast strain BY4743. After incubation of the transgenic yeast in SD-Ura (containing 2% galactose) liquid medium to OD = 0.7-1, images were collected by confocal laser scanning microscope with an excitation wavelength of 488 nm. (B) Subcellular localization of OsHIPP17 proteins in tobacco epidermic cells. Scale bars in A =20 µm. The pHB::eGFP (for control) or pHB::OsHIPP17-eGFP, for obtaining OsHIPP17 subcellular localization information) were transferred into Agrobacterium GV3101. Agrobacterium containing the recombinant plasmid was introduced into 5-week-old tobacco leaves by using injection method. After 24 hour of injection, tobacco leaves were immersed in 10 μg/mL Dapi solution (marker for nucleus) for 3 hours. Images were collected by confocal laser scanning microscope with excitation wavelengths of 340 nm (excitation wavelength of Dapi) or 488 nm (excitation wavelength of eGFP).
The OsHIPP17 overexpression Arabidopsis plants were constructed to further explore the effect of OsHIPP17 on plants under Cu treatment. The overexpression levels of OsHIPP17 were confirmed by RT-PCR analysis (Figure 5A). The results showed that without Cu stress, the roots of overexpression plants were significantly longer than those of wild-type plants (Figures 5B, C). Yet when treated with 35 μM Cu, the root length of OE-7, OE-8 or OE-3 was significantly reduced comparing to that of wild type (Figures 5B, C). Meanwhile, the overexpression of OsHIPP17 also reduced the germination rate of Arabidopsis seeds under Cu stress by 1.8-2.2% (Figure S1).
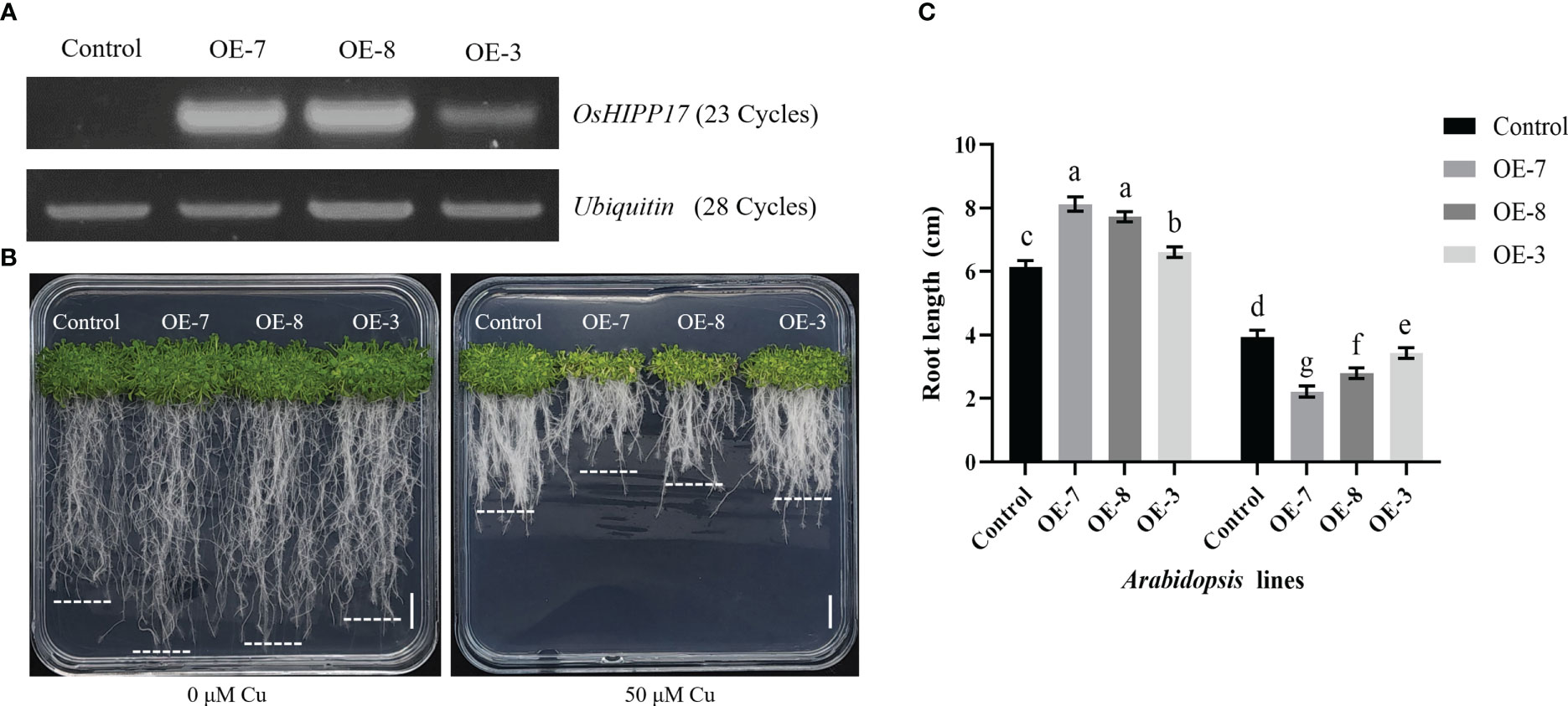
Figure 5 Phenotypic analysis of OsHIPP17 transgenic Arabidopsis lines under CuSO4 treatment. (A) The expression levels of OsHIPP17 overexpression lines were analyzed by using RT-PCR. Control represents wild type Arabidopsis plants, OE-7, OE-8 or OE-3 represent three independent OsHIPP17 transgenic Arabidopsis lines and Ubiquitin was used as the internal reference gene. Numbers in parentheses are the cycles of PCR. (B) The growth phenotypes of OsHIPP17 transgenic and control lines after 15 days under Cu treatment. Scale bars in B = 1 cm. (C) The root length OsHIPP17 transgenic and control lines under Cu treatment. Values are mean ± standard deviation (SD) from five independent plants. Statistical comparison was performed by t-tests. Different letters (a, b, c, d, e, f, g) indicate significant differences (p< 0.05).
To further elucidate the function of OsHIPP17 in rice under Cu stress, Oshipp17 knockout mutants of rice were constructed. Two knockout mutant lines generated by deleting one or two nucleotides in the OsHIPP17 coding region were used for following analysis (Figure 6A). Even without Cu treatment, the results showed that the loss-of-function of OsHIPP17 reduced the length of rice shoots by 15.60-23.98% (Figures 6B, C). The root length of Oshipp17 mutant lines was also significantly reduced by 16.74-24.36% under 25 μM Cu stress compared with the control (Figure 6D). The Cu concentrations in the shoots and roots of wild-type and mutant lines were also measured by ICP-MS. Results showed that an increased Cu content was detected in the roots of the mutant lines (Figure 6E). However, interestingly, the Cu concentration in the shoots of Oshipp17 mutant lines was less than that in the shoot of the wild type (Figure 6E).
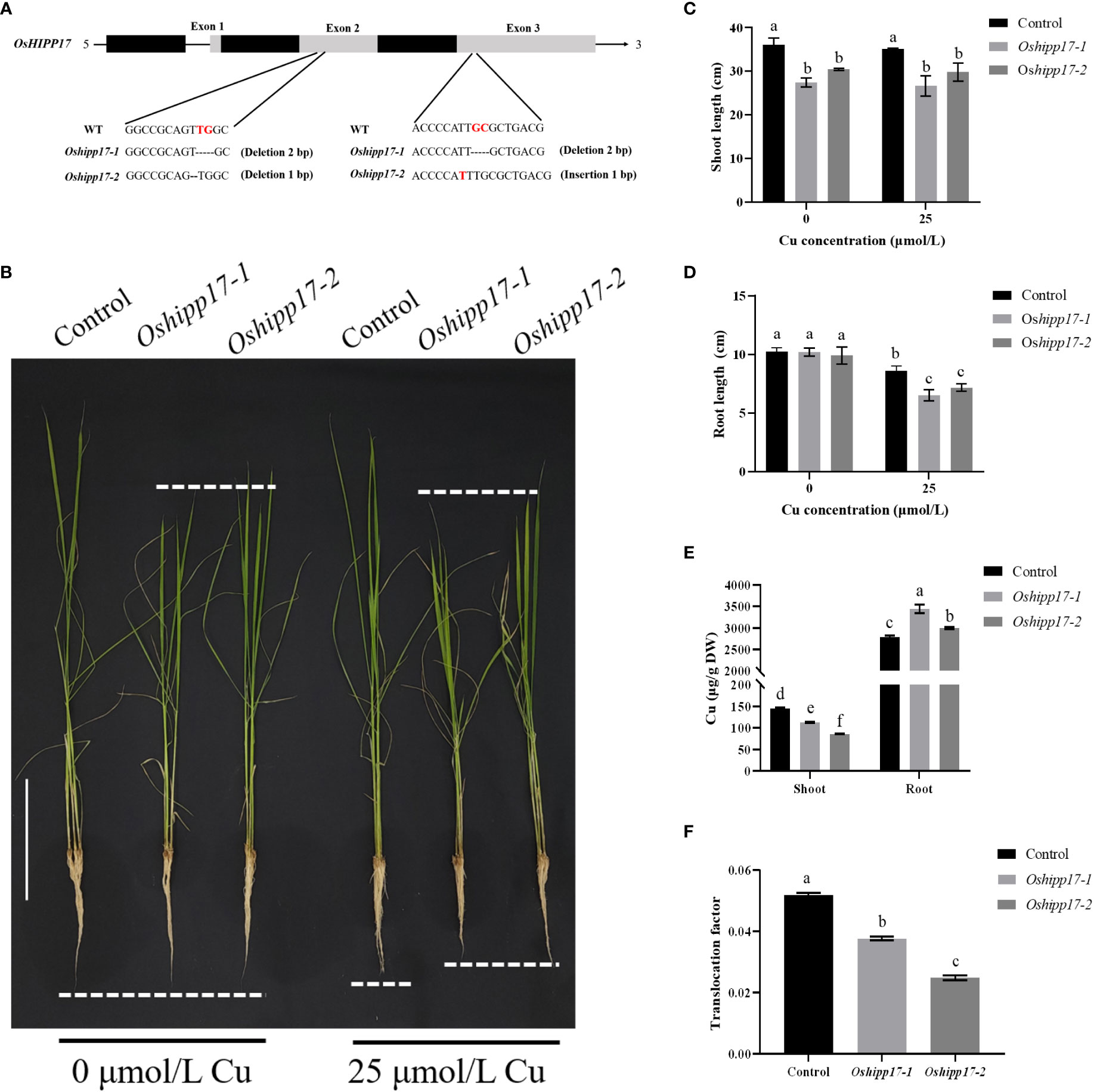
Figure 6 Growth phenotypes of rice wild type (Control) and Oshipp17 mutant lines (Oshipp17-1 and Oshipp17-2) under Cu stress. (A) Rice Oshipp17 knockout mutants created by CRISPR/Cas9 system. Two mutant lines showed frame-shift mutations with one or two nucleotide deletions in the coding region of OsHIPP17. (B) Growth phenotype of seven-day-old wild-type and Oshipp17 mutants rice treated with 0 or 25 μM CuSO4 for fifteen days. Scale bars in B = 10 cm. (C, D) Length of shoots and roots of wild-types and Oshipp17 mutants. (E) Cu content of shoots and roots of wild types and Oshipp17 mutants. (F) Translocation factor of Cu in wild-type and Oshipp17 mutants. Values are mean ± standard deviation (SD) from three independent plants. Statistical comparison was performed by t-tests. Different letters (a, b, c, d, e, f) indicate significant differences (p< 0.05).
Previous studies have showed that Cu accumulation is regulated by absorption or transport proteins in rice, such as OsATX1, OsZIP1, OsCOPT1, OsCOPT5, OsHMA4 or OsHMA5 (Yuan et al., 2010; Huang et al., 2016; Zhang et al., 2018c; Liu et al., 2019; Navarro et al., 2021). In this study, functional loss of OsHIPP17 resulted in the increased accumulation of Cu in rice (Figure 6E). Therefore, OsHIPP17 correlation with Cu absorption and transport genes was analyzed for rice in this study. The results showed that OsHIPP17 showed negative correlation with OsHMA4 or OsCOPT1, and positive correlation with OsATX1, OsZIP1, OsCOPT5 or OsHMA5 (Figure 7A). Meanwhile, the effect of knockout of OsHIPP17 on the expression levels of these genes was analyzed. Results showed that the knockout of OsHIPP17 decreased the expression levels of OsATX1, OsZIP1, OsCOPT5 or OsHMA5, and increased the expression levels of OsCOPT1 or OsHMA4 (Figures 7B-G). These results indicated that OsHIPP17 may regulate Cu content in rice by influencing the expression levels of Cu absorption and transport genes.

Figure 7 Effect of OsHIPP17 on the expression levels of Cu transporter genes in rice. (A) Correlation analysis of expression levels of OsHIPP17 and Cu transporter genes under Cu stress. The expression levels of (B) OsATX1, (C) OsZIP1, (D) OsCOPT1, (E) OsCOPT5, (F) OsHMA4 or (G) OsHMA5 in control and mutants under Cu treatment. Values are mean ± standard deviation (SD) from three independent plants. Statistical comparison was performed by t-tests. Different letters (a, b) indicate significant differences (p< 0.05).
The activity of three antioxidant enzyme were measured in this study to reveal if the antioxidant mechanism was involved in the Cu toxic response in transgenic Arabidopsis or Oshipp17 rice mutants. The results showed that the enzymatic activities of SOD, POD or CAT were significantly increased in OsHIPP17 overexpression Arabidopsis compared with the ones of control without Cu treatment (Figures 8A-C). Meanwhile, knockout of OsHIPP17 resulted in decreased POD or CAT activity in rice with or without Cu treatment (Figures 8E, F). However, SOD activity in the Oshipp17 mutant showed only a slightly decreased (Figure 8D). The enzymatic activities of SOD and POD were increased in the transgenic Arabidopsis than the ones of control under Cu stress, as well (Figure 8A, B). Moreover, exactly as in OsHIPP17 overexpression Arabidopsis, Cu treatment enhanced SOD activity in Oshipp17 mutant rice (Figures 8A, D). The results indicated that OsHIPP17 affected the activity of antioxidant enzymes in Arabidopsis or rice.
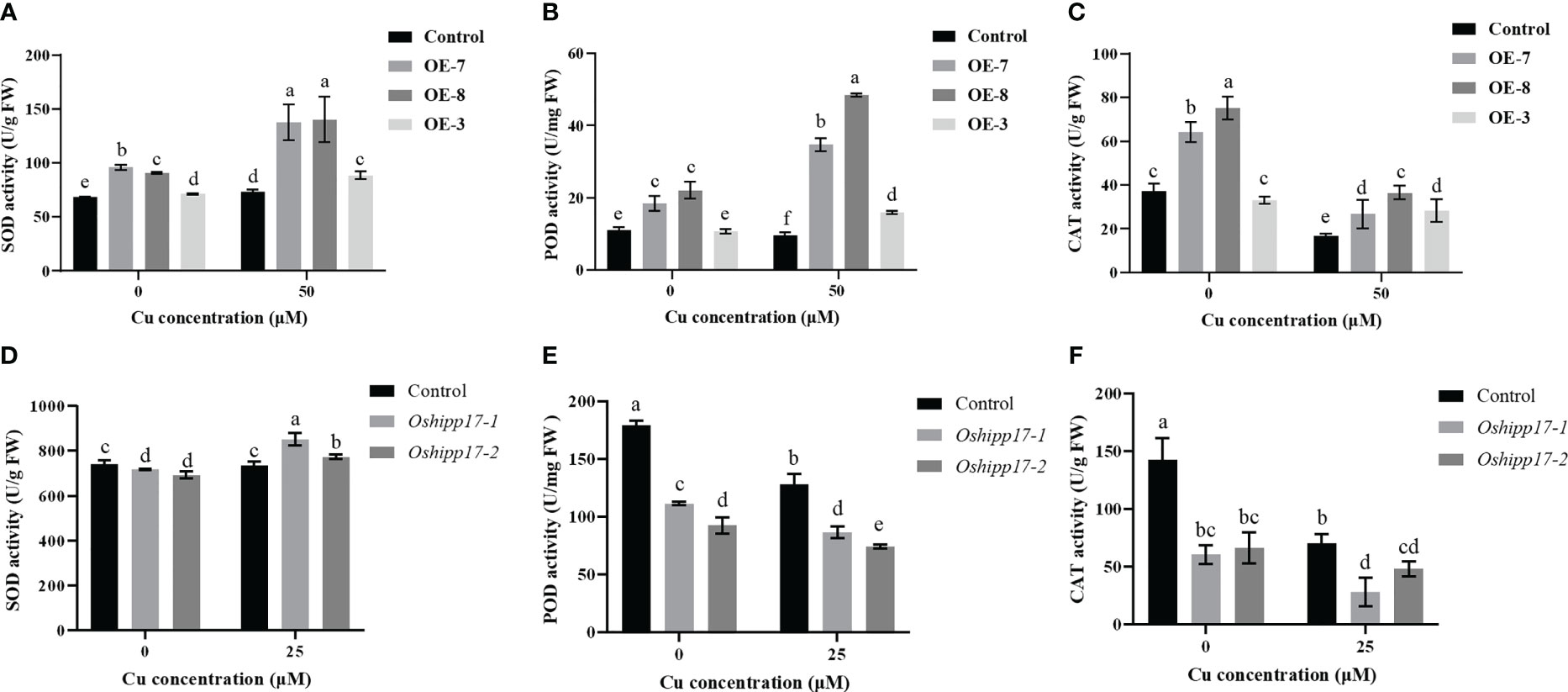
Figure 8 Activity analysis of antioxidant enzymes in OsHIPP17 overexpression Arabidopsis or Oshipp17 mutants treated with different Cu concentrations. Activities of antioxidant enzymes in wild-type and OsHIPP17 overexpression Arabidopsis was analyzed after 0 or 50 μM CuSO4 treatment for fifteen days. (A-C) SOD, POD or CAT in wild-type and OsHIPP17 overexpression Arabidopsis. Activities of antioxidant enzymes in wild-type and Oshipp17 rice mutants were analyzed after 15 days of treatment with 0 or 25 μM CuSO4. (D-F) SOD, POD or CAT in wild-type and Oshipp17 mutants. Values are mean ± standard deviation (SD) from three independent plants. Statistical comparison was performed by t-tests. Different letters (a, b, c, d, e, f) indicate significant differences (p< 0.05).
HPP/HIPP genes are reported to regulate cytokinin signaling pathways, and cytokinin homeostasis is implicated in root development control in plants (Laplaze et al., 2007; Guo et al., 2021). In order to further explore the cause of the affected root development of transgenic plants under Cu stress, the expression of three vitals genes that were implicated in the biosynthesis, degradation or signaling of cytokinin were determined in OsHIPP17 overexpression Arabidopsis or Oshipp17 rice mutants. The expression levels of cytokinin degradation gene (AtCKX1), cytokinin biosynthesis gene (AtIPT1) or cytokinin primary response gene (AtARR5) in transgenic Arabidopsis were significantly increased under the condition without Cu treatment compared to the corresponding ones of the control (Figures 9A-C). Meanwhile, loss-of-function of Oshipp17 resulted in a significant decrease in the expression levels of cytokinin-related genes in rice without Cu treatment compared to the control (Figures 9D, E). Interestingly, an opposite trend in the expression levels of cytokinin-related genes under Cu treatment conditions was observed in the OsHIPP17 overexpression Arabidopsis or Oshipp17 rice mutants (Figure 9). These results implicated that OsHIPP17 may be involved in Cu toxicity responses by affecting the expression levels of cytokinin-related genes in plants.
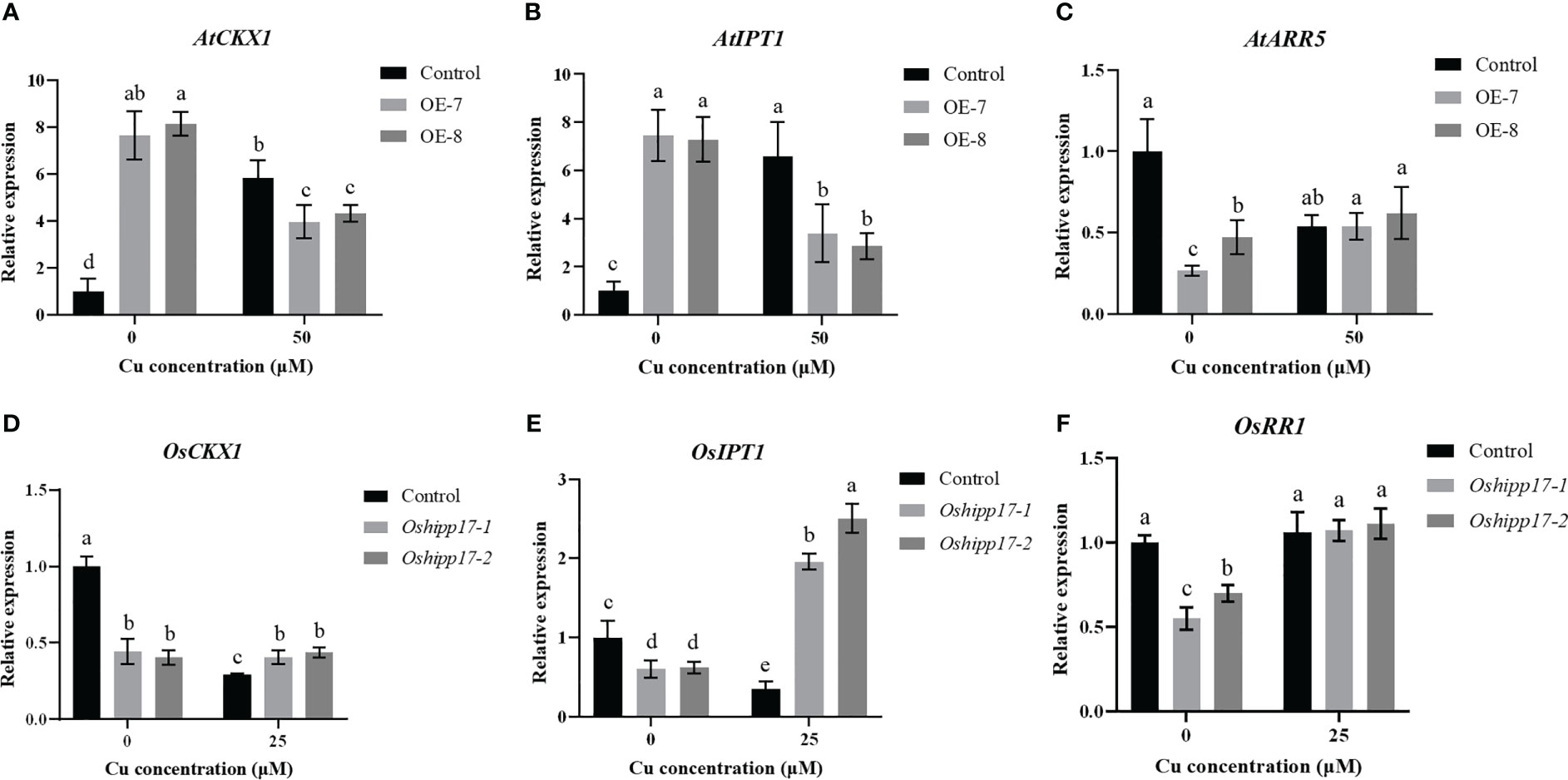
Figure 9 The expression level of cytokinin related genes in OsHIPP17 overexpression Arabidopsis or Oshipp17 mutants treated with Cu. The expression levels of (A) AtCKX1, (B) AtIPT1, (C) AtARR5, (D) OsCKX1, (E) OsIPT1 or (F) OsRR1. Values are mean ± standard deviation (SD) from three independent plants. Statistical comparison was performed by t-tests. Different letters (a, b, c, d) indicate significant differences (p< 0.05).
The growing global population and developing economy have intensified the pollution caused by heavy metals (Järup and Åkesson, 2009). The wide-spreading heavy metal contamination decreases farm land and food availability and causes increased threat to human health as well (Midhat et al., 2018). Cu is one of the most common heavy metal elements, but is a necessary trace element for plants as well. However, excessive supply of Cu leads to toxic effects on plants. Accumulated heavy metals in plant cells is mediated by some metal transporters including HMAs (heavy metal ATPases) and ZIP proteins which play key roles in heavy metal efflux or cellular sequestration (Deng et al., 2013; Liu et al., 2019). Although HIPPs are not transporters, a number lines of proof have shown that they may function in transport and detoxification of heavy metals in plants (Zhang et al., 2018a; Khan et al., 2020; Chen and Xiong, 2021). So far, only a few of plant genes belonging to HPPs and HIPPs have been functionally studied (Khan et al., 2019). Therefore, it is necessary to further study the functions of plant HPP and HIPP gene. Genes that encode HIPP proteins in rice have been identified by exhaustive genome-wide bioinformatics screening by using publicly available databases (Table S2). The HPP and HIPP proteins could be phylogenetically classified into five independent subfamilies (Figure 1A). The allele duplication and diversification events were implied by the presence of HIPPs in all subfamilies (Zhang et al., 2017). It is a quite useful strategy for an organism to deal with various of environmental stresses by duplicating its genes to expand the genome (Lynch and Conery, 2000).
In this study, a putative locus encoding OsHIPP17 was identified and functionally analyzed. There are some study cases have shown that some plant HIPP genes are able to increase the resistance of yeast or plants to heavy metals. For example, OsHIPP42 and OsHIPP16 confer Cd tolerance in yeast (Khan et al., 2019). On the contrary, our data showed that OsHIPP17 was involved in the decreased heavy metal tolerance in yeast cells. The repressed growth rate of the yeast cells transformed with OsHIPP17 gene on the solid plates or in the liquid medium containing Cu indicated that OsHIPP17 gene reduces the tolerance of Cu in the cells. Meanwhile, the Cu content in the yeast cells was significantly increased by OsHIPP17 (Figure 3C). This suggested that in the yeast cell, OsHIPP17 induced Cu tolerance decrease may be due to the increased toxicity resulted from increased cellular Cu content.
The subcellular localization of proteins is closely related to their functions (Feng et al., 2020). Studies have shown that some HIPPs are nucleus or cytoplasm localized (Chen and Xiong, 2021). In Arabidopsis, HIPP26 interacts with a zinc finger transcription factor ATHB29 in the nucleus to jointly regulate the drought stress response of Arabidopsis (Barth et al., 2009). Interestingly, the subcellular localization of OsHIPP17 was observed in nucleus in both tobacco and yeast cells (Figure 4). Therefore, we cannot exclude the possibility that OsHIPP17 may regulate the tolerance of yeast or plant cells by “sensing” and passing the Cu signal to other regulators instead of a direct effect of binding or absorbing Cu. Therefore, identification of OsHIPP17 interacting factors especially transcription factors may help to elucidate the mechanism of OsHIPP17 regulated Cu tolerance in yeast and plants.
To further reveal the function of OsHIPP17 in plants in response to Cu stress, OsHIPP17 overexpression Arabidopsis and Oshipp17 knockout rice mutants were constructed. Overexpression of OsHIPP17 increased the sensitivity of Arabidopsis to Cu (Figure 5B). This phenotype is similar to that of OsHIPP24 in response to Cu stress (Chen and Xiong, 2021). We have known that members of HIPPs gene family play diverse functions under heavy metal stress in rice. It has been proven that OsHIPP29 improves the tolerance of plants to heavy metals (Zhang et al., 2020). The different functions of HIPPs in response to heavy metal stress may be due to the presence of HMA and isoprenylation motifs. Meanwhile, the knockout of OsHIPP17 affected the Cu content in rice (Figure 6E). It was shown that Cu content in rice was strictly regulated by Cu transport proteins (Zhang et al., 2018c). In this study, different correlations patterns were shown between the expression levels of OsHIPP17 and variant Cu transporter genes (Figure 7A). Meanwhile, the knockout of OsHIPP17 increased the expression level of OsCOPT1 (Figure 7D). Studies have indicated that overexpression of OsCOPT1 leads to increased Cu content in rice roots (Yuan et al., 2010). Also, OsATX1, OsZIP1 and OsHMA5 showed decreased expression levels in the Oshipp17 mutant compared to those of the control (Figure 7B, C). Studies have showed that knockout of OsATX1, OsZIP1 or OsHMA5 increased Cu content in rice roots (Zhang et al., 2018c; Liu et al., 2019). Therefore, the Cu content change resulted from the knockout of OsHIPP17 may be associated with the expression level changes of Cu transporter genes. Yet, whether the underlying relationship between OsHIPP17 and Cu transporter genes needs to be further explored. On the other hand, the knockout of Oshipp17 influenced the growth of shoots in rice, which may be due to the reduced Cu translocation from roots to shoots. Related studies have shown that OsHIPP24 may plays a crucial role in long-distance transport of Cu ion from roots to shoots of plants (Chen and Xiong, 2021). Our yeast two-hybrid analysis result also showed that OsHIPP17 and OsHIPP24 may interact with each other (Unpublished data). Therefore, it is still necessary to further explore whether OsHIPP17 and OsHIPP24 together influence the translocation of Cu from roots to shoots. Meanwhile, OsHIPP17 may affected cytokinin signaling in rice (Figures 9D-F). Therefore, it is a worthwhile topic to study whether the impaired shoot growth of Oshipp17 rice mutants was mediate by the changes of cytokinin level. Moreover, further study on the function of OsHIPP17 may be expected to reveal the mechanisms involved in the growth differences under Cu deficiency in rice.
Antioxidant enzymes such SOD, POD or CAT are crucial for plants to deal with stress (Tatiana et al., 2017). Therefore, the activities of these antioxidant enzymes could be used to estimate the status of plants under specific stress (Tatiana et al., 2017). In this study, overexpression and knockout of OsHIPP17 even without Cu treatment conditions affected the activity of antioxidant enzymes in plants (Figure 8). Studies have shown that the activities of antioxidant enzymes are affected by endogenous cytokinin in plants as well (Lubovská et al., 2014; Wang et al., 2015). Therefore, based on the result that the expression levels of cytokinin-related genes were influenced by OsHIPP17 in plants, we cannot exclude the possibility that the activities of antioxidant enzymes were affected by cytokinin. Meanwhile, the overexpression of OsHIPP17 in Arabidopsis resulted in the increased antioxidant enzymatic activities compared with those of control under Cu stress (Figure 8). It has been shown that although plant growth was inhibited by heavy metal stress, plants still had higher antioxidant enzymatic activities compared with the control (Ali et al., 2008). Therefore, this results implied that the increased content and toxicity of Cu induced the increased antioxidant enzymatic activities in OsHIPP17 overexpression Arabidopsis plants. Moreover, knockout of OsHIPP17 slightly decreased SOD activity in rice without copper stress (Figure 8D). Although this slight decrease showed a statistically significant difference, it does not seem to be biologically significant for the plant.
Under heavy metal stress, plant growth and development are regulated by HIPP proteins. The roots of OsHIPP17 overexpression Arabidopsis showed a stunting phenotype under Cu stress (Figure 5B). Studies have shown that HIPP proteins regulate the cytokinin signaling in plants (Guo et al., 2021). In this study, the expression levels of AtCKX1 or AtIPT1 were significantly reduced in transgenic Arabidopsis treated with Cu (Figures 9A, B). It has been shown that the inhibition of Arabidopsis root growth due to Cu stress may be attributed to the affected cytokinin homeostasis (Lequeux et al., 2010). Therefore, we conclude that overexpression of OsHIPP17 may affect the expression levels of plant cytokinin-related genes to result in abnormal root development in overexpression or knockout mutant plants. However, whether cytokinin content is affected by OsHIPP17 overexpression or knockout deserves further investigation. Moreover, the expression levels of AtARR5 or OsRR1 were insensitive to Cu (Figures 9C, E). It has shown that the expression level of OsARR5 is variably regulated by different Cu treatment concentrations (Lequeux et al., 2010). Therefore, we suppose that the expression levels of AtARR5 or OsRR1 may be determined by various factors such as the stage of plant growth, Cu treatment concentration and duration and so on. Meanwhile, AtARR5 or OsRR1, as members of the Type-A RRs gene family, may have feedback effect on their own expression (Kieber and Schaller, 2018).
Cytokinin plays a crucial role in maintaining the activity of shoot and root meristem, which may be based on the uptake and translocation of essential nutrients by plants (Gao et al., 2019). It is remarkable that knockout of OsHIPP17 restricts the translocation of Cu from rice roots to shoots (Figure 6F). Studies have shown that the transporters responsible for internal Zn transport in rice are tightly controlled by cytokinin (Gao et al., 2019). Currently, there are few studies on the link between Cu transporters and cytokinin. however, it should be noted that some Zn uptake or translocation proteins are also responsible for the regulation of Cu transport in plants (Liu et al., 2019). In addition, the knockout of OsHIPP17 increased the expression levels of OsCKX1 and OsIPT1 in rice under Cu treatment (Figure 9D, E). Therefore, a hypothesis is put forward in this study that the roots of Oshipp17 rice mutants exhibiting Cu sensitivity might be due to the high level of Cu accumulated in the roots. However, in this process, whether cytokinin or Cu transporters which are responsible for Cu translocation from roots to shoots are involved still needs further investigation.
In this study, we proved that both overexpression and knockout of OsHIPP17 affected the tolerance of plants to Cu. OsHIPP17 affects the activities of SOD, POD or CAT enzymes in plants under Cu stress. Meanwhile, OsHIPP17 regulates the expression levels of Cu transport genes or cytokinin-related genes in plants. In addition, knockout of OsHIPP17 also affected the translocation of Cu from rice roots to shoots. Overall, we conclude that OsHIPP17 is involved in the regulation of tolerance to Cu stress in rice, and this regulatory process may involve antioxidant enzymatic activity, Cu translocation and expression of cytokinin-related genes.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
JH, JC and YS contributed to conception and design of the study. YS, MW and NJ: data analysis. YS wrote the first draft of the manuscript. JL and YJ performed the statistical analysis. NJ, YH and ML wrote sections of the manuscript. LZ, JW and XJ: resources. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (No. 31870383).
We appreciate Farhan Afzal for the comments and suggestion on our manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1183445/full#supplementary-material
Ali, B., Hasan, S. A., Hayat, S., Hayat, Q., Hayat, S., Yadav, Q. (2008). A role for brassinosteroids in the amelioration of aluminium stress through antioxidant system in mung bean (Vigna radiata l. wilczek). Environ. Exp. Bot. 62, 153–159. doi: 10.1016/j.envexpbot.2007.07.014
Barth, O., Vogt, S., Uhlemann, R., Zschiesche, W., Humbeck, K. (2009). Stress induced and nuclear localized HIPP26 from arabidopsis thaliana interacts via its heavy metal associated domain with the drought stress related zinc finger transcription factor ATHB29. Mol. Biol. Rep. 69, 213–226. doi: 10.1007/s11103-008-9419-0
Brien, J. A. O., Benkova, E. (2013). Cytokinin cross-talking during biotic and abiotic stress responses. Front. Plant Sci. 4, 451. doi: 10.3389/fpls.2013.00451
Cai, H., Peng, F. (2023). Knockdown of copper chaperone antioxidant-1 by RNA interference inhibits copper-stimulated proliferation of non-small cell lung carcinoma cells. Oncol. Rep. 30, 269–275. doi: 10.3892/or.2013.2436
Chen, G., Xiong, S. (2021). OsHIPP24 is a copper metallochaperone which affects rice growth. J. Plant Biol. 64, 145–153. doi: 10.1007/s12374-020-09287-x
Chen, T., Zhang, B. (2016). Measurements of proline and malondialdehyde contents and antioxidant enzyme activities in leaves of drought stressed cotton. Bio-Protocol 6, e1913. doi: 10.21769/BioProtoc.1913
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
de Abreu-Neto, J. B., Turchetto-Zolet, A. C., de Oliveira, L. F., Zanettini, M. H., Margis-Pinheiro, M. (2013). Heavy metal-associated isoprenylated plant protein (HIPP): characterization of a family of proteins exclusive to plants. FEBS. J. 280, 1604–1616. doi: 10.1111/febs.12159
Deng, F. L., Yamaji, N., Xia, J., Ma, J. F. (2013). A member of the heavy metal p-type ATPase OsHMA5 is involved in xylem loading of copper in rice. Plant Physiol. 163, 1353–1362. doi: 10.1104/pp.113.226225
Feng, S. J., Liu, X. S., Ma, L. Y., Khan, I. U., Rono, J. K., Yang, Z. M. (2020). Identification of epigenetic mechanisms in paddy crop associated with lowering environmentally related cadmium risks to food safety. Environ. pollut. 256, 113464. doi: 10.1016/j.envpol.2019.113464
Gao, W., Xiao, S., Li, H., Tsao, S. W., Chye, M. L. (2009). Arabidopsis thaliana Acyl-CoAbinding protein ACBP2 interacts with heavy-metal-binding farnesylated protein AtFP6. New Phytol. 181, 89–102. doi: 10.1111/j.1469-8137.2008.02631.x
Gao, S. P., Xiao, Y. H., Xu, F., Gao, X. K., Cao, S. Y., Zhang, F. X., et al. (2019). Cytokinin-dependent regulatory module underlies the maintenance of zinc nutrition in rice. New Phytol. 224, 202–215. doi: 10.1111/nph.15962
Gietz, R. D., Woods, R. A. (2006). Yeast transformation by the LiAc/SS carrier DNA/PEG method. Methods Mol. Biol. 313, 107–120. doi: 10.1385/1-59259-958-3:107
Guan, M. Y., Zhang, W. Y., Xu, P., Zhao, Q., Chen, M. X., Cao, Z. Z. (2022). Mapping and functional analysis of high-copper accumulation mutant oshc1 in rice. J. Hazard. Mater. 426, 128063. doi: 10.1016/j.jhazmat.2021.128063
Guo, T. Q., Weber, H., Niemann, M. C. E., Theisl, L., Leonte, G., Novák, O., et al. (2021). Arabidopsis HIPP proteins regulate endoplasmic reticulum-associated degradation of CKX proteins and cytokinin responses. Mol. Plant 14, 1918–1934. doi: 10.1016/j.molp.2021.07.015
Guo, J., Xu, L., Su, Y., Wang, H., Gao, S. W., Xu, J. S., et al. (2013). ScMT2-1-3, A metallothionein gene of sugarcane, plays an important role in the regulation of heavy metal tolerance/accumulation. Biomed. Res. Int. 23, 904769. doi: 10.1155/2013/904769
Huang, X. Y., Deng, F., Yamaji, N., Pinson, S. R.-M., Fujii-Kashin, M., Danku, J., et al. (2016). A heavy metal p-type ATPase OsHMA4 prevents copper accumulation in rice grain. Nat. Commun. 7, 12138. doi: 10.1038/ncomms12138
Järup, L., Åkesson, A. (2009). Current status of cadmium as an environmental health problem. Toxicol. Appl. Pharmacol. 238, 201–208. doi: 10.1016/j.taap.2009.04.020
Khan, I. U., Rono, J. K., Liu, X. S., Feng, S. J., Li, H., Chen, X., et al. (2020). Functional characterization of a new metallochaperone for reducing cadmium concentration in rice crop. J. Clean. Prod. 272, 123152. doi: 10.1016/j.jclepro.2020.123152
Khan, I. U., Rono, J. K., Zhang, B. Q., Liu, X. S., Wang, M. Q., Wang, L. L., et al. (2019). Identification of novel rice (Oryza sativa) HPP and HIPP genes tolerant to heavy metal toxicity. Ecotoxicol. Environ. Safety. 175, 9–18. doi: 10.1016/j.ecoenv.2019.03.040
Kieber, J. J., Schaller, G. E. (2018). Cytokinin signaling in plant development. Development 145, 149344. doi: 10.1242/dev.149344
Laplaze, L., Benkova, E., Casimiro, I., Maes, L., Vanneste, S., Swarup, R., et al. (2007). Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell Online. 19, 3889–3900. doi: 10.1105/tpc.107.055863
Lee, S., Kim, Y.-Y., Lee, Y., An, G. (2007). Rice P1B-type heavy-metal ATPase, OsHMA9, is a metal efflux protein. Plant Physiol. 145, 831–842. doi: 10.1104/pp.107.102236
Lequeux, H., Hermans, C., Lutts, S., Verbruggen, N. (2010). Response to copper excess in Arabidopsis thaliana: impact on the root system architecture, hormone distribution, lignin accumulation and mineral profile. Plant Physiol. Biochem. 48, 673–682. doi: 10.1016/j.plaphy.2010.05.005
Li, X. B., Fan, Y. J., Ma, J., Gao, X. Y., Wang, G., Wu, S. D., et al. (2023). Cerium improves the physiology and medicinal components of dendrobium nobile lindl. under copper stress. J. Plant Physiol. 280, 153896. doi: 10.1016/J.JPLPH.2022.153896
Liu, X. S., Feng, S. J., Zhang, B. Q., Wang, M. Q., Cao, H. W., Rono, J. K., et al. (2019). OsZIP1 functions as a metal efflux transporter limiting excess zinc, copper and cadmium accumulation in rice. BMC. Plant Biol. 19, 283. doi: 10.1186/s12870-019-1899-3
Lubovská, Z., Dobrá, J., Helena, S., Wilhelmová, N., Vanková, R. (2014). Cytokinin oxidase/dehydrogenase overexpression modifies antioxidant defense against heat, drought and their combination in nicotiana tabacum plants. J. Plant Physiol. 171, 1625–1633. doi: 10.1016/j.jplph.2014.06.021
Lynch, M., Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Meng, X. X., Li, W. F., Shen, R. F., Lan, P. (2022). Ectopic expression of IMA small peptide genes confers tolerance to cadmium stress in arabidopsis through activating the iron deficiency response. J. Hazard. Mater. 422, 126913. doi: 10.1016/j.jhazmat.2021.126913
Midhat, L., Ouazzani, N., Hejjaj, A., Ouhammou, A., Mandi, L. (2018). Accumulation of heavy metals in metallophytes from three mining sites (Southern centre Morocco) and evaluation of their phytoremediation potential. Ecotoxicol. Environ. Safety. 169, 150–160. doi: 10.1016/j.ecoenv.2018.11.009
Murashige, T., Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 15, 473–476. doi: 10.1111/j.1399-3054.1962.tb08052.x
Navarro, B. B., Del Frari, B. K., Dias Pedro, V., da., C., Lemainski, L. E., Mario, R. B., et al. (2021). The copper economy response is partially conserved in rice (Oryza sativa). Plant Physiol. Bioch. 158, 113–124. doi: 10.1016/j.plaphy.2020.11.051
Polina, G., Rotem, K., Nakonechny, F., Aronov, S., Marina, N. (2016). Increased copper bioremediation ability of new transgenic and adapted Saccharomyces cerevisiae strains. Environ. Sci. pollut. R. 23, 19613–19625. doi: 10.1007/s11356-016-7157-4
Shinohara, M., Kibi, M., Riley, I. R., Chiang, N., Dalli, J., Kraft, B. D., et al. (2014). Cell-cell interactions and bronchoconstrictor eicosanoid reduction with inhaled carbon monoxide and resolvin D1, am. J. Physiol. Lung. Cell Mol. Physiol. 307, 746–757. doi: 10.1152/ajplung.00166.2014
Tatiana, R., do Nogueira, A. M., Carvalho, B. L., Lopes, V. I., Bolacel, B. E. J., de Martins, M. J. A., et al. (2017). Gene expression and activity of antioxidant enzymes in rice plants, cv. BRS AG, under saline stress. Physiol. Mol. Biol. Plants. 23, 865–875. doi: 10.1007/s12298-017-0467-2
Tehseen, M., Cairns, N., Sherson, S., Cobbett, C. S. (2010). Metallochaperone-like genes in Arabidopsis thaliana. Metallomics 2, 556–564. doi: 10.1039/c003484c
Tejpal, G., Vivek, D., Sanjay, K., Singh, A. P., Yelam, S. (2012). Protein dynamics during seed germination under copper stress in Arabidopsis over-expressing Potentilla superoxide dismutase. J. Plant R. 125, 165–172. doi: 10.1007/s10265-011-0421-2
Torkian, S., Khanjani, N., Mahmoodi, M.-R., Khosravi, V. (2019). A review of copper concentrations in Iranian populations. Environ. Monit. Assess. 191, 537. doi: 10.1007/s10661-019-7633-7
Wang, C., Chen, X., Yao, Q., Long, D., Fan, X., Kang, H. Y., et al. (2019). Overexpression of TtNRAMP6 enhances the accumulation of cd in arabidopsis. Gene 696, 225–232. doi: 10.1016/j.gene.2019.02.008
Wang, Y. P., Shen, W. Z., Chan, Z. L., Wu, Y. (2015). Endogenous cytokinin overproduction modulates ROS homeostasis and decreases salt stress resistance in Arabidopsis Thaliana. Front. Plant Sci. 6, 1004. doi: 10.3389/fpls.2015.01004
Xian, P. Q., Cai, Z. D., Cheng, Y. B., Lin, R. B., Lian, T. X., Ma, Q. B., et al. (2020). Wild soybean oxalyl-CoA synthetase degrades oxalate and affects the tolerance to cadmium and aluminum stresses. Int. J. Mol. Sci. 21, 8869. doi: 10.3390/ijms21228869
Yuan, M., Chu, Z. H., Li, X. H., Xu, C. G., Wang, S. Q. (2010). The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. The. Plant Cell. 22, 3164–3176. doi: 10.1105/tpc.110.078022
Zhang, Y., Chen, K., Zhao, F. J., Sun, C., Jin, C., Shi, Y., et al. (2018c). OsATX1 interacts with heavy metal P1B-type ATPases and affects copper transport and distribution. Plant Physiol. 178, 329–344. doi: 10.1104/pp.18.00425
Zhang, B. Q., Liu, X. S., Feng, S. J., Zhao, Y. N., Wang, L. L., Rono, J. K., et al. (2020). Developing a cadmium resistant rice genotype with OsHIPP29 locus for limiting cadmium accumulation in the paddy crop. Chemosphere 247, 125958. doi: 10.1016/j.chemosphere.2020.125958
Zhang, X. D., Meng, J. G., Zhao, K. X., Chen, X., Yang, Z. M. (2017). Annotation and characterization of cd-responsive metal transporter genes in rapeseed (Brassica napus). Biometals 31, 107–121. doi: 10.1007/s10534-017-0072-4
Zhang, X. D., Sun, J. Y., You, Y. Y., Song, J. B., Yang, Z. M. (2018a). Identification of cd-responsive RNA helicase genes and expression of a putative BnRH 24 mediated by miR158 in canola (Brassica napus). Ecotoxicol Environ. Saf. 157, 159–168. doi: 10.1016/j.ecoenv.2018.03.081
Zhang, X. D., Wang, Y., Li, H. B., Yang, Z. M. (2018b). Isolation and identification of rapeseed (Brassica napus) cultivars for potential higher and lower cd accumulation. J. Plant Nutr. Soil Sci. 181, 479–487.
Zhao, F. J., Ma, Y., Zhu, Y. G., Tang, Z., McGrath, S. P. (2015). Soil contamination in China: current status and mitigation strategies. Envron. Sci. Technol. 49, 750. doi: 10.1021/es504709
Keywords: HIPP, metallochaperone, heavy metal, rice, gene
Citation: Shi Y, Jiang N, Wang M, Du Z, Chen J, Huang Y, Li M, Jin Y, Li J, Wan J, Jin X, Zhang L and Huang J (2023) OsHIPP17 is involved in regulating the tolerance of rice to copper stress. Front. Plant Sci. 14:1183445. doi: 10.3389/fpls.2023.1183445
Received: 10 March 2023; Accepted: 17 April 2023;
Published: 06 July 2023.
Edited by:
Rangjian Qiu, Wuhan University, ChinaReviewed by:
Giovanni DalCorso, University of Verona, ItalyCopyright © 2023 Shi, Jiang, Wang, Du, Chen, Huang, Li, Jin, Li, Wan, Jin, Zhang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin Huang, aHVhbmdqaW4xOEBjZHV0LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.