- 1Department of Plant Pathology, College of Agricultural, Human, and Natural Resource Sciences, Washington State University, Pullman, WA, United States
- 2Molecular Plant Sciences Program, Washington State University, Pullman, WA, United States
When cells experience acute mechanical distress, they release ATP from their cellular compartment into the surrounding microenvironment. This extracellular ATP (eATP) can then act as a danger signal—signaling cellular damage. In plants, cells adjacent to damage detect rising eATP concentrations through the cell-surface receptor kinase, P2K1. Following eATP perception, P2K1 initiates a signaling cascade mobilizing plant defense. Recent transcriptome analysis revealed a profile of eATP-induced genes sharing pathogen- and wound-response hallmarks—consistent with a working model for eATP as a defense-mobilizing danger signal. To build on the transcriptional footprint and broaden our understanding of dynamic eATP signaling responses in plants, we aimed to i) generate a visual toolkit for eATP-inducible marker genes using a β-glucuronidase (GUS) reporter system and ii) evaluate the spatiotemporal response of these genes to eATP in plant tissues. Here, we demonstrate that the promoter activities of five genes, ATPR1, ATPR2, TAT3, WRKY46, and CNGC19, were highly sensitive to eATP in the primary root meristem and elongation zones with maximal responses at 2 h after treatment. These results suggest the primary root tip as a hub to study eATP-signaling activity and provide a proof-of-concept toward using these reporters to further dissect eATP and damage signaling in plants.
Introduction
Although typically regarded as an intracellular metabolite and potential energy storage molecule, the high concentration of ATP within the cell (mM) and its relatively low extracellular concentration (~nM) make it a potent signal molecule. In both animals and plants, wounding (Song et al., 2006), physical stimulation (Sauer et al., 2000; Jeter et al., 2004), and microbe interactions (Torres et al., 2008; Nizam et al., 2019) cause ATP to be released from its cellular container into the extracellular space. Once in the extracellular space, ATP becomes a danger signal (Tanaka et al., 2014). As a signal, extracellular ATP (eATP) belongs to a group of defense molecules known as damage-associated molecular patterns or DAMPs (Tanaka and Heil, 2021).
The first plant receptor for extracellular ATP identified is a plasma membrane-localized legume-type lectin receptor kinase (LecRK), also known as P2 Kinase 1, or P2K1, which was identified from the does not respond to nucleotides1 (dorn1) mutant (Choi et al., 2014). P2K1 elicits secondary messenger reactive oxygen species (ROS) and cytosolic calcium (Ca2+) influx following eATP perception (Jeter et al., 2004; Song et al., 2006; Demidchik et al., 2009; Tanaka et al., 2010; Choi et al., 2014; Chen D. et al., 2017; Wang et al., 2018). P2K1-dependent eATP signaling impacts a diverse range of plant–pathogen interactions, including those involving viruses, bacteria, fungi, oomycetes, nematodes, and even insects (Bouwmeester et al., 2011; Bouwmeester et al., 2014; Wang et al., 2014; Balagué et al., 2017; Kumar et al., 2020; Jewell et al., 2022). Recently, it was reported that the indolic glucosinolate pathway can be induced by eATP in Arabidopsis thaliana (Arabidopsis), serving as a biochemical defense against pathogens and pests (Jewell et al., 2022). Although the impact of eATP signaling in plants is well documented, characterizing the molecular mechanisms that connect early eATP signaling activities by the receptor P2K1 (i.e., ROS and Ca2+) to plant immunity is a work in progress.
Recent headway has been made by revealing P2K1-dependent eATP-responsive transcriptional activities, which showed expression of many genes involved in wound and defense response that are linked to jasmonic acid (JA) signaling, a key plant defense hormone—suggesting that eATP could employ JA-mediated signal amplification (Tripathi et al., 2018; Jewell et al., 2019). To gain a deeper understanding of P2K1-mediated signaling activity through the eATP-induced transcriptome, we created a kit of ATP-responsive marker tools using the β-glucuronidase (GUS) reporter system in Arabidopsis. We hypothesized that evaluating the expression patterns of eATP-responsive markers would reveal previously unknown spatial and temporal aspects of eATP signaling in plants. Additionally, we expected that eATP-responsive reporter tools would allow for the visualization of eATP-responsive signaling with a high resolution of spatiotemporal precision, for example, making them useful for high-throughput forward genetic screens. Five highly responsive eATP marker genes were selected from the published transcriptome (Jewell et al., 2019) to generate promoter::reporters for TAT3, WRKY46, CNGC19, and two uncharacterized genes, which have been named ATP RESPONSIVE 1 (ATPR1; AT5G19110) and ATP RESPONSIVE 2 (ATPR2; AT3G44870), respectively. Each of the five markers displayed maximal eATP-responsive expression in the root tip of the Arabidopsis seedlings. These tools will allow us to unlock additional insights into the mechanisms underlying eATP-mediated signaling.
Methods
Plant growth
Arabidopsis plants of wild type (ecotype Columbia-0 or Col-0), dorn1-3 (Choi et al., 2014), and transgenic lines were each surface- sterilized and sown on a solid medium containing half-strength Murashige and Skoog (MS) (Murashige and Skoog, 1962) with vitamins (MSP09; Casson, Smithfield, UT, USA), 1% (w/v) sucrose, and 1% (w/v) agar Daishin (RPI, Mt. Prospect, IL, USA) and buffered in 0.05% (w/v) MES-KOH (pH 5.7) in a square petri dish. Seeds were cold- stratified in the dark for 3 days at 4°C and grown vertically in a 22°C growth chamber (CMP6010; Conviron, Winnipeg, MB, Canada) with a 12-h photoperiod (~100 µmol photons m−2 s−1).
ATP treatment
Working ATP stock solution (100 mM) was prepared by dissolving ATP (Sigma-Aldrich, St. Louis, MO, USA) in 2 mM of MES. The pH was adjusted to 5.7 with KOH, and the solution was filter-sterilized and stored in aliquots at −20°C indefinitely. For the GUS reporter and RT-qPCR experiments, Arabidopsis plants were grown for 5 or 7 days, respectively. After the vertical growth period, the seedlings were transferred to six-well plates (15–20 seedlings per well) containing half-strength MS (as above) without agar (“liquid media”). Once transferred to a liquid medium, the seedlings were returned to the growth chamber to equilibrate overnight. The following day, the medium in each well was replaced with 1 ml of liquid medium (mock) or ATP solution (in liquid medium) at a final concentration of 500 μM. For the RT-qPCR experiments, the seedlings were blotted dry and snap-frozen in liquid nitrogen 30 min after treatment. In the GUS reporter experiments, treatment durations were variable and the seedlings were harvested and fixed in ice-cold 90% (v/v) acetone.
Transgenic material
The transgenic Arabidopsis lines pP2K1::GUS (Cho et al., 2017) and proCNGC19::GUS #4 (Kugler et al., 2009) were graciously provided by Dr. Gary Stacey at the University of Missouri and Dr. Petra Dietrich at the University of Erlangen, Germany, respectively. To construct GUS reporters for the other eATP marker genes [CML39 (AT1G76640), TAT3 (AT2G24850), WRKY46 (AT2G46400), ATPR1 (AT5G19110), and ATPR2 (AT3G44870)], 800 –2,500-bp non-coding regions upstream of each gene were amplified from genomic DNA using primers designed with the 5′-tails containing restriction sites of HindIII and NcoI, respectively (Table S1). Once amplified, the promoter fragments were ligated into pCAMBIA1305.2, yielding a promoter linked to the β-glucuronidase (GUS) reporter. After the transformation and propagation of plasmids in E scherichia coli (DH5α), the promoter::GUS constructs were validated by Sanger sequencing (Elim Biopharmaceuticals, Inc., Hayward, CA, USA) and introduced to Agrobacterium (GV3101 pMP90) for plant transformation (Koncz and Schell, 1986). Arabidopsis plants (Col-0 or dorn1-3) were transformed using the floral dip method (Clough and Bent, 1998) and selected in the next generation on hygromycin-containing media to obtain independent T3 homozygous lines. All transgenic lines created are listed in Table S2.
Histochemical staining and imaging
After ATP treatment, the transgenic Arabidopsis plants expressing GUS reporter constructs were harvested in ice-cold 90% (v/v) acetone for 20 min. The seedlings were blotted dry, washed once with sterile deionized water to remove residual acetone, and returned to a clean six-well plate. In the dark, the seedlings were incubated with 3 ml of the GUS staining solution (2 mM of potassium ferricyanide, 2 mM of potassium ferrocyanide, 0.25% (v/v) Triton X-100, 50 mM of sodium phosphate pH 7.2, 1 mM of X-Gluc), placed under vacuum at room temperature for 20 min, and transferred to a 37°C incubator for an additional 60 min. The GUS staining solution was replaced with 3 ml of 70% ethanol, and the samples were heated to 70°C for 30 min. Afterward, fresh 70% (v/v) ethanol was added and left overnight at room temperature. The next day, the samples were heated again to 70°C for 30 min, and the ethanol was replaced with fresh 70% ethanol. This was repeated a third time the following day with 90% (v/v) ethanol and finally replaced with 100% (v/v) ethanol. The samples were then stored in 100% (v/v) ethanol at 4°C until imaged, no more than 48 h later. Primary root zones were defined as previously described (Beemster and Baskin, 1998; Dello Ioio et al., 2008; Perilli et al., 2012; Petricka et al., 2012). Microscopic images were obtained with an inverted phase-contrast microscope (Leica DMI6000) or stereo microscope (Leica MZ10 F).
RT-qPCR
After the samples were snap-frozen in liquid nitrogen, they were stored in a −80°C freezer. Frozen tissue was homogenized using the Mini-Beadbeater™ (BioSpec Products, Bartlesville, OK, USA), and RNA isolation was performed with the Quick-RNA isolation kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s protocols. RNA concentration was measured with an Eppendorf μCuvette® G1.0 on the BioPhotometer® D30 (Eppendorf, Hamburg, Germany), and 1 μg of RNA was used for cDNA synthesis (iScript; Bio-Rad, Hercules, CA, USA). The cDNA was 10-fold diluted with sterile deionized water, of which 2 μl was used in a 20-μl SYBR Green reaction mix (SsoAdvanced; Bio-Rad). All reactions were run in a CFX96 thermocycler (Bio-Rad). Gene expression was normalized to PP2A and SAND with similar results (Czechowski et al., 2005; Zhang et al., 2015). All primers used are listed in Table S1.
Image processing
Adobe Photoshop 2020 (v21.2.10.118) was used for processing microscope image data. To improve visual interpretation, post-capture white balancing and removal of background noise were done to provide a fair comparison between experiments performed on different dates. All image data presented in this study are representative depictions of at least two and, in most cases, three independent stable transgenic lines (not including lines previously published) and have been observed across hundreds of samples in no less than five independent experiments.
Results
Spatial expression of the eATP receptor P2K1
The expression patterns of the extracellular ATP receptor P2K1 have previously been described using the GUS reporter line pP2K1::GUS (Cho et al., 2017). We re-evaluated the expression of P2K1 in our lab using the same reporter line. In 4-day-old plants, P2K1 was previously shown to be highly expressed throughout various tissue types (Cho et al., 2017), while, here, the expression in 7-day-old plants was confined to leaf mesophyll and vasculature, root junction, root vasculature, and root tip tissues (Figure 1A). Our findings also revealed that the expression pattern of P2K1 did not show any significant difference 1 or 5 h of treatment with 500 μM of ATP, in comparison to its pattern under mock treatment, i.e., treatment with liquid media (Figure 1B). This is a novel result that has not been previously reported. This result suggested that the eATP receptor P2K1 is expressed in the whole seedling, whereas the expression level was stable even after ATP treatment. Based on our data about the stable expression of P2K1 throughout all tissues, we expected that spatial analysis of the expression of eATP-responsive genes would provide further insight into eATP signaling in plants, including the yet- described tissue-specific responses (Figure 1C).
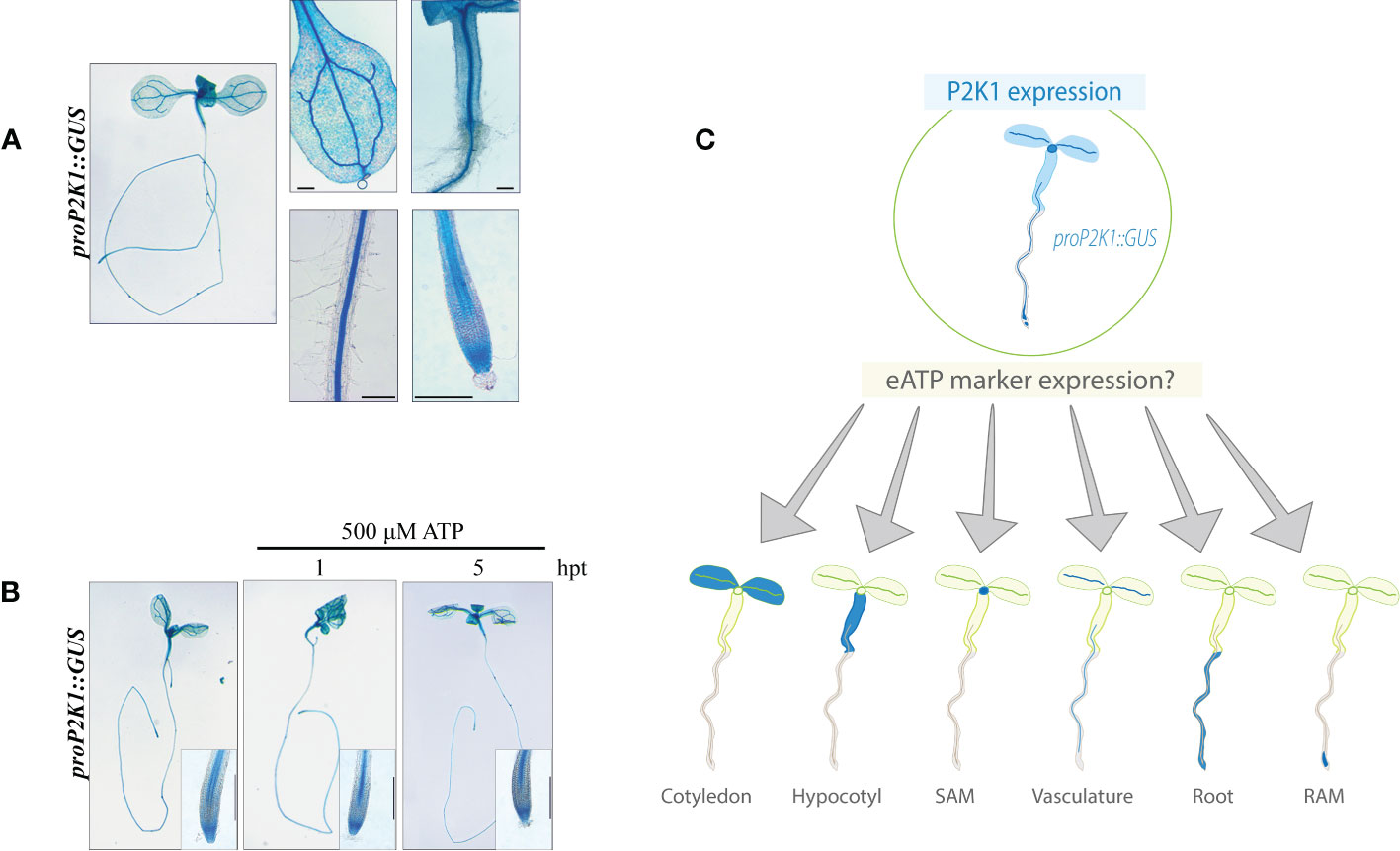
Figure 1 P2K1 promoter activity is not responsive to ATP treatment. (A) Histochemical β-glucuronidase (GUS) staining in pP2K1::GUS transgenic plants. The P2K1 promoter is active in various tissues of 7-day-old seedlings. (B) Seven-day-old pP2K1::GUS seedlings were incubated with MS or MS plus 500 μM of ATP for 1 or 5 h post-treatment (hpt). Scale bar = 300 μm. (C) Graphical research question: What is the spatiotemporal expression pattern of eATP marker genes? RAM, root apical meristem; SAM, shoot apical meristem.
Selection of eATP-responsive marker genes
Recently, we described eATP-induced transcriptional responses in Arabidopsis (Jewell et al., 2019). Putative eATP marker genes were first selected based on fold change (FC) and false discovery rate (FDR, with Bonferroni correction) in the transcriptomics data set (Figures 2A, B). Fold change and FDR cutoffs for putative marker genes were 2.5 log2 FC and 10 −log10 FDR (Figures 2A, B)—which reduced the number of candidates to 10. Finally, the genes with less than 1 CPM in mock-treated samples and greater than 10 CPM in ATP-treated samples were considered, leaving seven putative genes as the eATP markers: CML39 (AT1G76640), ATPR2 (AT3G44870), CNGC19 (AT3G17690), MAPKKK21 (AT4G36950), ATPR1 (ATG5G19110), WRKY46 (AT2G46400), and TAT3 (AT2G24850).
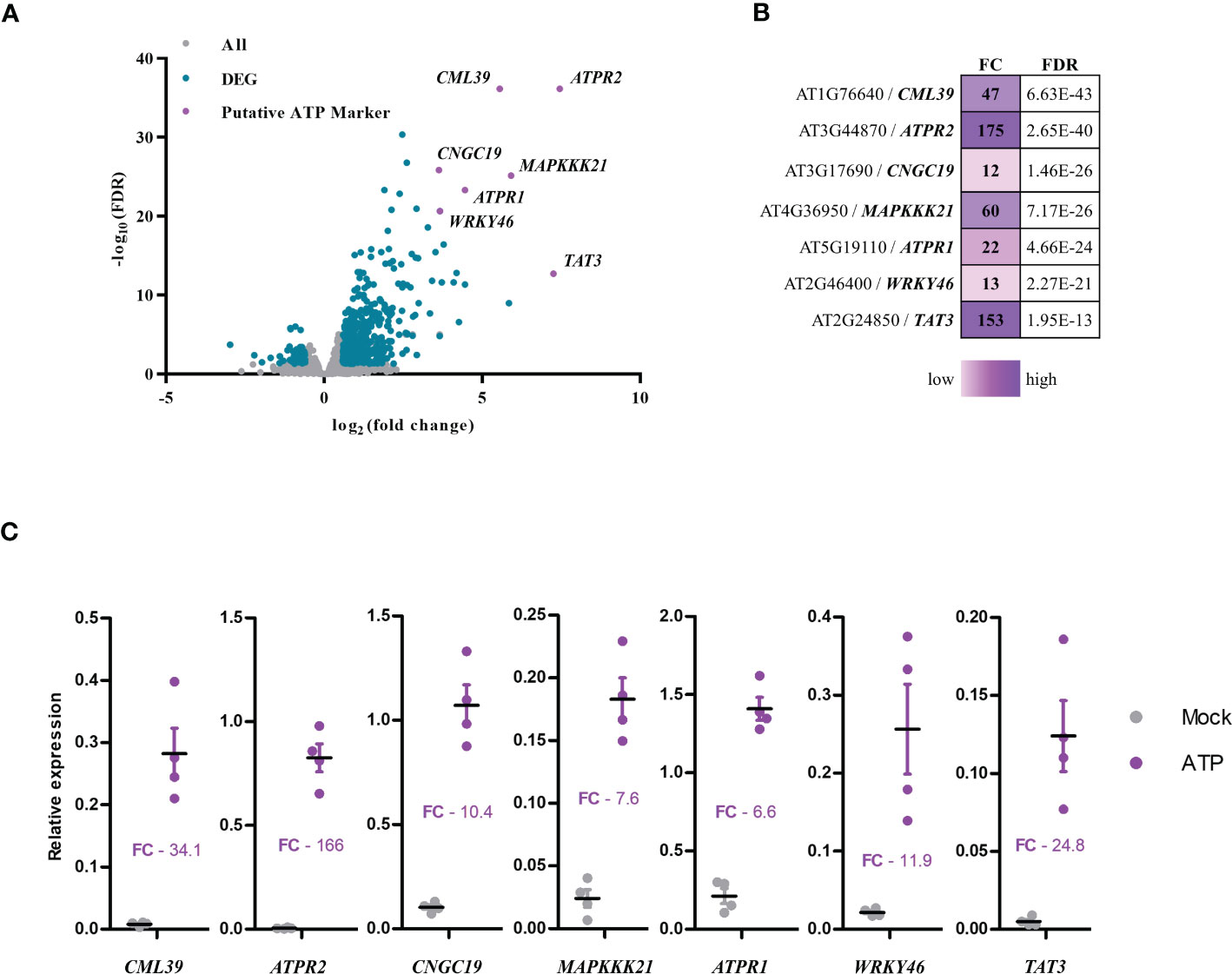
Figure 2 Selection of extracellular ATP (eATP)-sensitive markers. (A) Volcano plot of differentially expressed genes in the eATP-induced transcriptome in BioProject accession no. PRJNA494862 (Jewell et al., 2019) plotted as false discovery rate (−log10FDR) over fold change (log2). (B) Table of putative eATP-sensitive markers was shown with fold change (FC) and false discovery rate (FDR) following 500 μM of ATP treatment for 30 min in the transcriptome data. (C) Measurement of changes in gene expression of the putative eATP-responsive markers in 7-day-old Col-0 seedlings treated with or without 500 μM of ATP for 30 min. Gene expression was analyzed by RT-qPCR and normalized to PP2A (and SAND with similar results). Individual replicates are shown with mean and standard error bars (n = 4). Fisher’s LSD (P < 0.05) was used as a statistical cutoff for the comparison of ATP- and mock-treated samples. All the markers were expressed significantly higher following ATP treatment (repeated at least five times with similar results).
Next, we performed RT-qPCR to validate the seven eATP markers and tested the expression of these genes after ATP treatment by RT-qPCR. After 30 min of ATP treatment, transcripts of CML39, ATPR2, CNGC19, MAPKKK21, ATPR1, and WRKY46 were more abundant relative to the mock-treated (liquid media without ATP) samples (Figure 2C). These data were nearly comparable to those in the transcriptomics data (Figure 2B). For example, the ATP-induced fold change of RT-qPCR vs. transcriptomics data was as follows: 34.1 vs. 47 for CML39, 166 vs. 175 for ATPR2, 10.4 vs. 12 for CNGC19, 7.6 vs. 60 for MAPKKK21, 6.6 vs. 22 for ATPR1, 11.9 vs. 13 for WRKY46, and 24.8 vs. 153 for TAT3 (Figures 2B, C). These results confirmed the validity of these seven genes as the eATP markers that we selected. While distinct RNA processing and normalization techniques in RNA-seq and RT-qPCR experiments can introduce some variation when comparing their data (Everaert et al., 2017), the results generally agree that these seven genes are significantly and reliably upregulated by eATP treatment as previously described (Jewell et al., 2019).
Basal expression patterns of eATP marker genes
After generating the eATP marker GUS reporter germplasm, two to five representative lines were selected for each of the promoter/genotype combinations, except for pCNGC19::GUS #4, already described (Kugler et al., 2009). We were unsuccessful in cloning the promoter region for MAPKKK21. We could create pCML39::GUS transgenic plants. However, its GUS staining was highly saturated before ATP treatment. Therefore, these two genes were undesirable for our purpose and excluded from further experiments. First, basal expression (no treatment of any kind) patterns for the reporters in 7-day-old wild-type plants were recorded for a representative line of pATPR1::GUS (line #2.5), pATPR2::GUS (line #4.5), pWRKY46::GUS (line #1.1), and pTAT3::GUS (line #12.1) along with pCNGC19::GUS #4 (Figure 3). Supplemental lines had similar expression patterns and are shown in Figure S2. Three of the reporters for ATPR1, ATPR2, and WRKY46 had detectable expression in the cotyledon leaf, i.e., the leaf vasculature (Figure 3), while two reporters for ATPR2 and WRKY46 showed expression in the hypocotyl, near the root junction (Figure 3). All five markers were expressed in at least one region of the root tip and the distal primary root vasculature (Figure 3). In addition to distal vasculature, ATPR2 expression was identified throughout root tip tissues approximately corresponding to the primary root meristematic zone, while TAT3 was also expressed in the meristematic zone that was limited to the most distal portion near or overlapping the stem cell niche (Figure 3). No expression was detectable in columella cells.
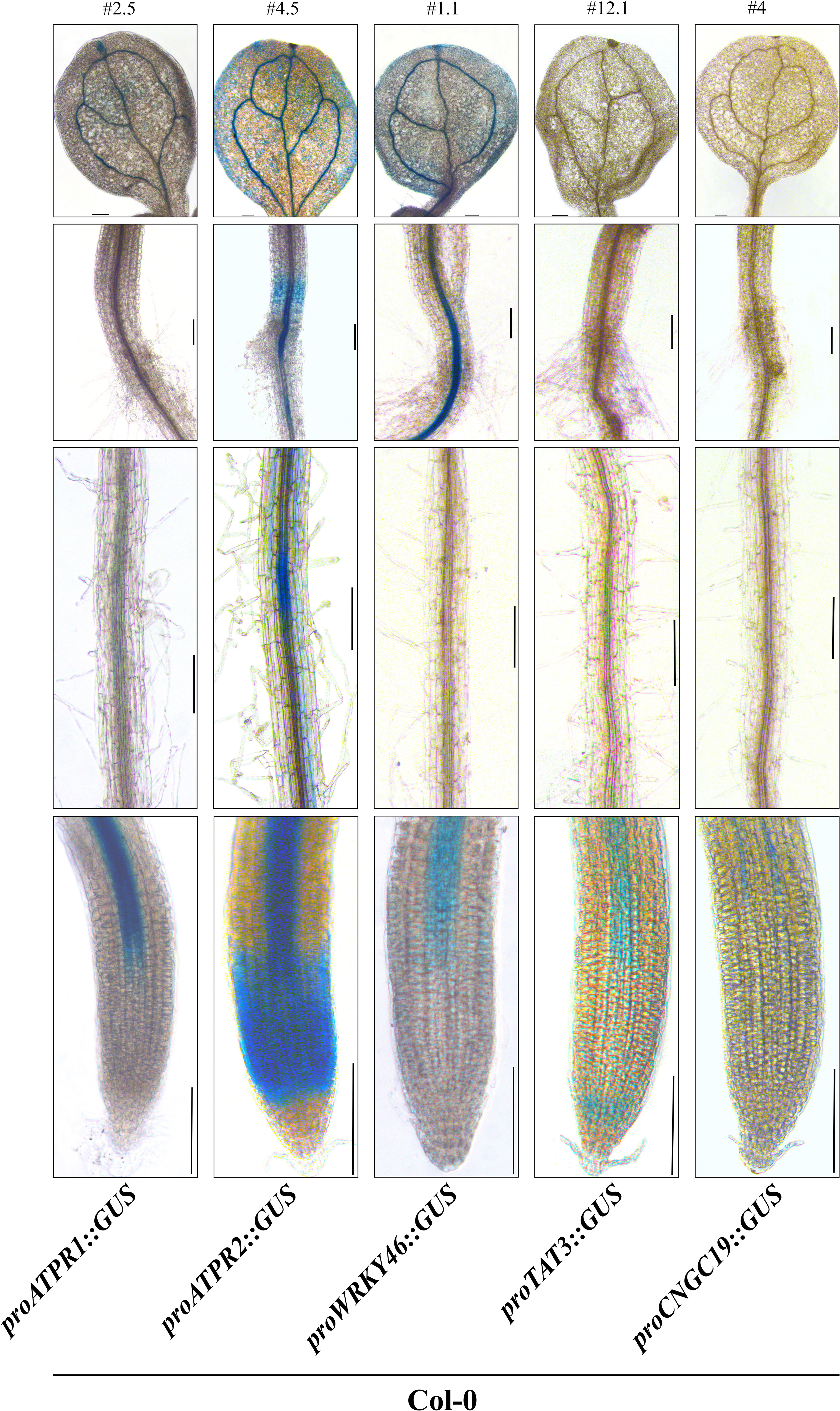
Figure 3 ATP marker genes have a shared pattern of vascular activity. Representative β-glucuronidase (GUS) staining of five eATP marker gene promoter::GUS fusions (ATPR1, ATPR2, WRKY46, TAT3, and CNGC19) in stably transformed 7-day-old seedlings in the wild-type background. Each line (# above the top panel) is representative of promoter activity observed in at least three independent transformants. Panels from top to bottom: cotyledon, hypocotyl/root junction, distal root, primary root tip. All images were obtained on an inverted phase- contrast microscope (Leica DMI6000). Scale bars = 150 μm.
eATP-induced reporter expression in a time-dependent manner
To identify an ATP treatment duration maximizing reporter intensity, the reporter line pCNGC19::GUS as a representative was treated with 500 μM of ATP for 0–90 h (Figure S1). Maximum GUS reporter responses to ATP treatment were identified between 1 and 5 h following treatment (Figure S1), and 2 h was selected as the optimal treatment duration for further evaluation. While the average maximal expression among the markers was 2 h post- eATP treatment, minor deviations included TAT3, which was highly induced as early as 1 h after treatment, and ATPR1, which peaked between 2 and 5 h (Figure S1). Next, to provide a comparison of ATP-sensitive promoter activities, the duration of GUS staining was performed identically for each promoter–reporter line. Staining for 1 h at 37°C was selected as the ideal incubation period since it allowed low promoter activity to be detected (e.g., proCNGC19), without allowing highly active reporters to become oversaturated (data not shown). It is worth noting that prolonged incubations in GUS staining buffer (e.g., overnight at room temperature) revealed additional expression patterns in some cases. For example, expression of CNGC19 was detected in the root cap of ATP-treated seedlings but only after prolonged GUS staining (Figure S1).
eATP-induced reporter expression in the root tip
Interestingly, for each of the GUS reporters, ATP-dependent expression was confined to the distal portion of the root vasculature, root tip, or both 2 h after ATP treatment (Figures 4A, B). ATPR1, ATPR2, and TAT3 shared a similarly remarkable ATP response pattern throughout the root tip, with some notable differences, while the responses for WRKY46 and CNGC19 were more subtle. ATP-induced ATPR1 expression was observed throughout the root meristem zone tissues, including the stem cell niche (SCN) and columella cells, after 60 min (Figure S2) and extended to the transition and elongation zones after 2 h (Figure 4B; S2). Similarly, the induction of TAT3 expression by ATP occurred throughout the root tip meristem after 60 min and extended into the transition and elongation after 2 h (Figure 4B; S2). Meanwhile, ATPR2 expression was already detectable throughout the meristem before ATP treatment and was noticeably induced throughout the primary root transition and elongation zones after ATP treatment, but not in the most distal root tip cell files, in contrast to ATPR1 and TAT3 (Figure 4). After ATP treatment, WRKY46 expression increased throughout the distal third of the root vasculature and in the root meristem, where it had been undetectable before ATP treatment (Figure 4B). Similarly, the CNGC19 expression was slightly upregulated in the distal root vasculature (Figure 4B) and root tip, with the latter only detectable after prolonged GUS staining (Figures S1, S2). Overall, all five reporters were responsive to ATP treatment in the root tip of the seedlings.
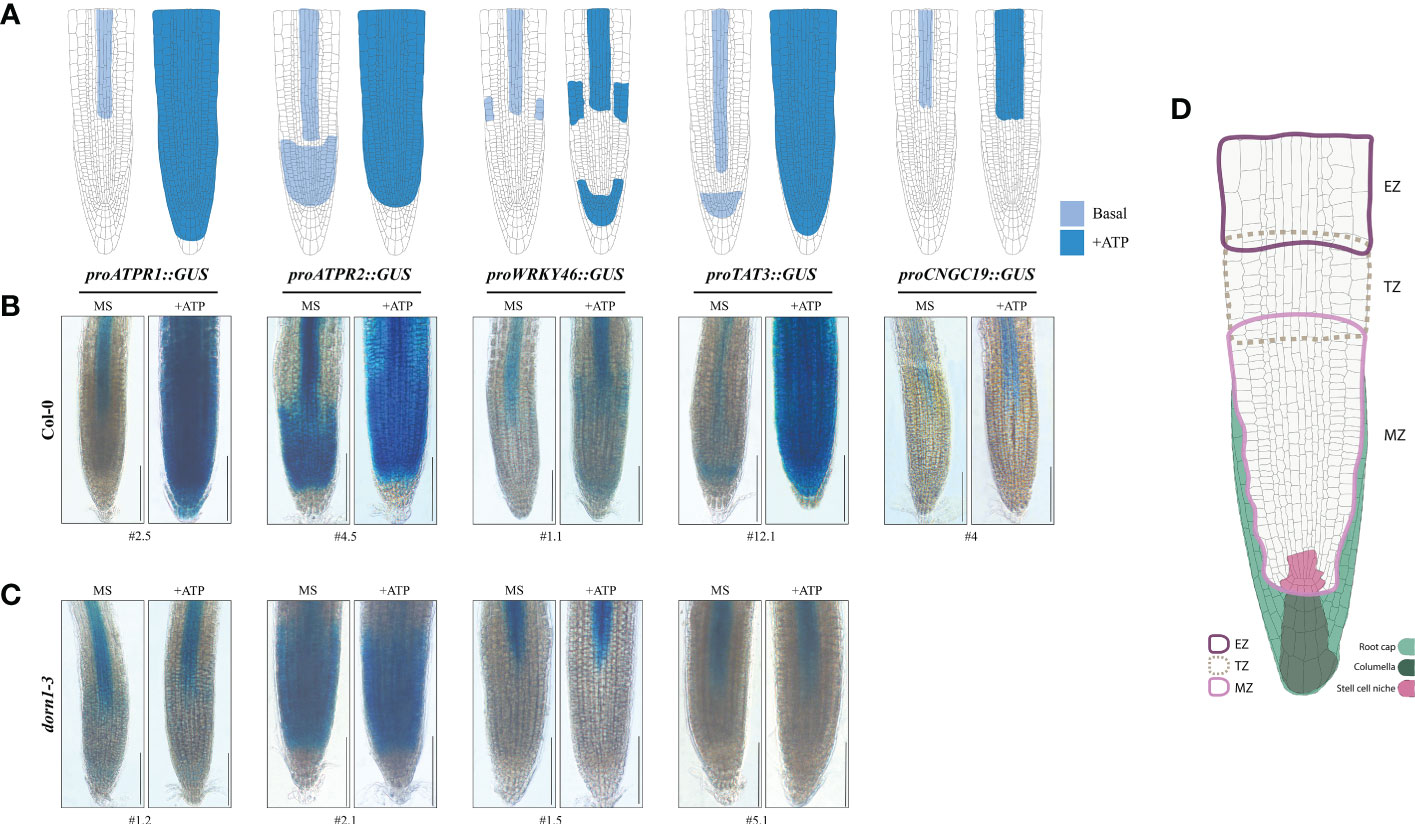
Figure 4 The eATP reporters are activated in the primary root tip and require P2K1. (A) Graphical summary of ATP-responsive eATP marker expression patterns in the primary root tip of the promoter::GUS reporter lines. (B, C) GUS staining in the 7-day-old wild-type or dorn1-3 mutant plants stably transformed with ATP-sensitive promoter::GUS reporters treated with MS (mock) or MS plus 500 μM of ATP (+ATP) for 2 h. pCNGC19::GUS was not generated in the dorn1-3 background. Scale bars = 150 μm. (D) The graphic guide for primary root morphology shows the meristematic zone (MZ; solid border in light pink), transition zone (TZ; dotted border in light brown), elongation zone (EZ; solid border in purple), root cap (filled in light green) including the columella (dark green), and the stem cell niche (filled in pink) for reference.
To determine whether the eATP receptor P2K1 is implicated in the spatial expression patterns of eATP marker genes, we examined the individual GUS reporters in a P2K1 null mutant, dorn1-3. Basal expression patterns of ATP-responsive GUS reporters were similar in the dorn1-3 plants compared with the wild-type plants (Figures 4B, C; S3). However, some subtle exceptions were observed. For example, basal expression of ATPR1 in dorn1-3 induced low levels of expression in approximate primary root SCN and transition zone cell files which were not observed in the wild-type background (Figure 4). Meanwhile, TAT3 was not expressed in the meristem region in dorn1-3, which was also contrary to that in the wild-type background (Figure 4). Notably, the expression patterns of all GUS reporters in dorn1-3 remained unaltered upon ATP addition. In conclusion, our results indicate that P2K1 is essential for normal basal and eATP-responsive expressions.
Discussion
In the present study, we selected the five genes ATPR1, ATPR2, WRKY46, TAT3, and CNGC19 from the transcriptomics data previously reported (Jewell et al., 2019) as the eATP-responsive markers that were highly upregulated upon ATP treatment. Through the use of the GUS reporter system, we discovered that the five eATP marker genes responded significantly to ATP treatment, particularly in the root tip tissue, where the functional P2K1 receptor was essential for their complete response to eATP (Figure 4).
ATPR1 is an uncharacterized gene (annotated as putative dermal glycoprotein-like or aspartyl protease family protein). In an Arabidopsis N-myristoylome, ATPR1 was identified with the myristoylation motif, MGSSLTRLLV (Boisson et al., 2003). In another report, ATPR1 was identified in cell wall protein isolates (Irshad et al., 2008). To our knowledge, ATPR1 has not been described in detail, though the Arabidopsis eFP browser (https://bar.utoronto.ca/efp_arabidopsis/cgi-bin/efpWeb.cgi) indicates that it is transcriptionally activated by Pseudomonas syringae inoculation, salt stress, and methyl jasmonate treatment (Kilian et al., 2007; Winter et al., 2007). Interestingly, our data showed that ATPR1 is induced by eATP without full dependence on the defense signaling hormones jasmonic acid (JA), salicylic acid (SA), or ethylene (ET) since it still responds to ATP in the signaling mutants of those hormones (Figure S4; Table S3). Thus, the pATPR1::GUS reporter has the potential to be a unique tool for the study of eATP signaling without the full requirement of inputs from plant stress hormones (i.e., JA, SA, and ET).
Prolonged GUS staining demonstrated WRKY46 promoter activity throughout the root vasculature (Figure S4), in agreement with past reports (Ding et al., 2014; Ding et al., 2015; Di et al., 2021). The promoter activity was further upregulated upon ATP treatment (Figure 4; S1). Previous studies have shown brassinosteroids, drought, and salt stress-induced WRKY46 expression (Ding et al., 2014; Sheikh et al., 2016; Chen J. et al., 2017), while abscisic acid repressed WRKY46 expression (Ding et al., 2015). Furthermore, suppression of two apyrase genes, APY1 and APY2, leads to the upregulation of WRKY46 (Lim et al., 2014)—presumably the result of eATP accumulation in the absence of apyrase-mediated hydrolysis. This report corroborates WRKY46 as an eATP-responsive marker. Like ATPR1, WRKY46 was induced by eATP independently of signaling pathways associated with JA, SA, and ET (Figure S4; Table S3), indicating that it can be used as a useful tool for the study of eATP signaling with any confounding influence from these hormones. While it is known that the induction of WRKY46 expression by SA requires the SA receptor NPR1 (Zhang et al., 2021), eATP was still able to induce WRKY46 in the npr1-3 null mutant (Figure S4; Table S3), suggesting that WRKY46 activation by eATP is independent of SA. It will be prudent to evaluate the downstream impact of WRKY46 activation on eATP signaling in the near term.
ATPR2 is a predicted Farnesoic Acid Methyl Transferase-Like (FAMT-L) gene that has not been the subject of dedicated research. One report found that ATPR2 expression was induced 24 h following treatment with alamethicin, a membrane-disrupting agent, and in response to herbivory by Plutella xylostella (Chen et al., 2003). Although disparate stress signaling pathways may lead to upregulation of ATPR2, the requirement of P2K1 for eATP-induced ATPR2 upregulation (Figure 4) could suggest that eATP accumulation after wounding (i.e., 24 h after alamethicin treatment or herbivory) accounts for previous observations. The transcriptional activation of ATPR2 was found to be dependent on JA signaling (Figure S4; Table S3), suggesting a possible convergence of ATP- and JA-dependent wound signals in the regulation of ATPR2. The ATPR2 promoter spans 1,180 base pairs upstream of the gene. It is noteworthy that three gene loci, namely, ATG3G44870 (ATPR2), ATG3G44880 (ACCELERATED CELL DEATH1), and ATG3G44890 (RIBOSOMAL PROTEIN BL9C), are arranged in tandem on the third chromosome of Arabidopsis. It cannot be ruled out that the intergenic sequence upstream of ATPR2 may regulate one or more of these genes.
TAT3 is a putative tyrosine aminotransferase (TAT) and is transcriptionally activated in response to JA and wounding, which depends on the intact JA signaling pathway (Yan et al., 2007; Riewe et al., 2012). Similarly, our data indicate that TAT3 expression in response to eATP treatment was dependent on JA perception (Figure S4; Table S3). Interestingly, it has been reported that the expression of TAT3 in response to JA treatment was dependent on the SA receptor NPR1, based on a reduced fold induction of TAT3 in the npr1-1 mutant after JA treatment (Brosché and Kangasjärvi, 2012). In partial agreement, our transcriptomics data (Jewell et al., 2019) showed that differential expression of TAT3 was reduced in the npr1-3 null mutant (FC = 46) compared with Col-0 (FC = 153) but remained highly upregulated (Figure S4; Table S3). Considering the antagonistic relationship between SA and JA, it could be that NPR1 has a greater contribution toward regulating TAT3 in response to JA, compared with eATP. Notably, eATP-induced TAT3 response was completely dependent on the JA signaling pathway since the response disappeared in the coi1 mutant (Figure S4; Table S3). The reciprocal modulation between signaling pathways linked to other hormones is still not well understood. The pTAT3::GUS reporter has the potential to be a valuable tool for exploring the intricate connections between eATP and other plant hormones.
The pCNGC19::GUS reporter was originally used to demonstrate transcriptional regulation of CNGC19 by NaCl (Kugler et al., 2009). Interestingly, CNGC19 was transiently upregulated by salt treatment in the roots at early time points (~30 min) but was only induced in the shoots at later time points, from 6 to 72 hpt (Kugler et al., 2009). In contrast, our data showed that eATP induced the expression of CNGC19 in the root tip after 1 h of treatment which continued for at least 90 h after treatment (Figure S1). In addition, eATP-induced CNGC19 expression does not require the hormonal signaling pathways JA, SA, or ET (Figure S4; Table S3). Meanwhile, there was no difference in CNGC19 expression in the shoots after eATP treatment (Figure S2). Similar to eATP treatment, boron starvation was reported to induce CNGC19 expression in the root tip (Quiles-Pando et al., 2013), along with CPK28, another ATP-responsive gene (Choi et al., 2014). In the leaves and roots, CNGC19 is a permeable channel for Ca2+ mobilization in response to herbivory or colonizing fungi (Meena et al., 2019; Jogawat et al., 2020). One of the initial signaling responses to eATP perception is cytosolic Ca2+ influx mediated by P2K1 (Choi et al., 2014). Thus, it is possible that the expression of CNGC19 primes local tissues by increasing sensitivity to Ca2+ flux, amplifying any response to a second wave of stimuli (e.g., more damaged cells releasing eATP). Future studies on the spatiotemporal correlation between Ca2+ levels and gene expression using pCNGC19::GUS would enhance our understanding of the role of CNGC19-dependent priming in response to wounding and eATP treatment.
In conclusion, the five eATP-responsive markers we selected—ATPR1, ATPR2, WRKY46, TAT3, and CNGC19—are highly responsive to eATP in the root tip. These results suggest that the primary root tip is a critically important hub for eATP signaling. The physiological role of P2K1 in the primary root tip has not been extensively explored and requires further investigation. Transcriptional reporters have been a powerful tool for characterizing signaling pathways in plants; auxin is a prime example (Jedličková et al., 2022). Thus, the multigene eATP reporter system we developed can serve as a valuable tool to explore and assess new aspects of eATP signaling in plants.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
KT conceptualized the project. KT and JS designed the experiments. JS performed the experiments and prepared the manuscript. KT and JS edited and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the NIH Protein Biotechnology Training Fellowship awarded to JS and by the NSF (no. IOS-1557813) and USDA NIFA (Hatch project no. 1015621) to KT.
Acknowledgments
Special thanks to Dr. Jeremy Jewell for providing technical guidance and mentoring to JS. We thank Dr. Gary Stacey at the University of Missouri for providing the proP2K1::GUS Arabidopsis plants and Dr. Petra Dietrich at the University of Erlangen in Germany for the proCNGC19::GUS seeds.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1183335/full#supplementary-material
References
Balagué, C., Gouget, A., Bouchez, O., Souriac, C., Haget, N., Boutet-Mercey, S., et al. (2017). The arabidopsis thaliana lectin receptor kinase LecRK-I.9 is required for full resistance to pseudomonas syringae and affects jasmonate signalling. Mol. Plant Pathol. 18, 937–948. doi: 10.1111/mpp.12457
Beemster, G. T. S., Baskin, T. I. (1998). Analysis of cell division and elongation underlying the developmental acceleration of root growth in arabidopsis thaliana. Plant Physiol. 116, 1515–1526. doi: 10.1104/pp.116.4.1515
Boisson, B., Giglione, C., Meinnel, T. (2003). Unexpected protein families including cell defense components feature in the n-myristoylome of a higher eukaryote*. J. Biol. Chem. 278, 43418–43429. doi: 10.1074/jbc.M307321200
Bouwmeester, K., de Sain, M., Weide, R., Gouget, A., Klamer, S., Canut, H., et al. (2011). The lectin receptor kinase LecRK-I.9 is a novel phytophthora resistance component and a potential host target for a RXLR effector. PloS Pathog. 7, e1001327. doi: 10.1371/journal.ppat.1001327
Bouwmeester, K., Han, M., Blanco-Portales, R., Song, W., Weide, R., Guo, L.-Y., et al. (2014). The arabidopsis lectin receptor kinase LecRK-I.9 enhances resistance to phytophthora infestans in solanaceous plants. Plant Biotechnol. J. 12, 10–16. doi: 10.1111/pbi.12111
Brosché, M., Kangasjärvi, J. (2012). Low antioxidant concentrations impact on multiple signalling pathways in arabidopsis thaliana partly through NPR1. J. Exp. Bot. 63, 1849–1861. doi: 10.1093/jxb/err358
Chen, D., Cao, Y., Li, H., Kim, D., Ahsan, N., Thelen, J., et al. (2017). Extracellular ATP elicits DORN1-mediated RBOHD phosphorylation to regulate stomatal aperture. Nat. Commun. 8, 2265. doi: 10.1038/s41467-017-02340-3
Chen, F., D’Auria, J. C., Tholl, D., Ross, J. R., Gershenzon, J., Noel, J. P., et al. (2003). An arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J. 36, 577–588. doi: 10.1046/j.1365-313X.2003.01902.x
Chen, J., Nolan, T. M., Ye, H., Zhang, M., Tong, H., Xin, P., et al. (2017). Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 29, 1425–1439. doi: 10.1105/tpc.17.00364
Cho, S.-H., Choi, J., Stacey, G. (2017). Molecular mechanism of plant recognition of extracellular ATP. Protein Rev. 19, 233–253. doi: 10.1007/5584_2017_110
Choi, J., Tanaka, K., Cao, Y., Qi, Y., Qiu, J., Liang, Y., et al. (2014). Identification of a plant receptor for extracellular ATP. Science 343, 290LP–29 294. doi: 10.1126/science.343.6168.290
Clough, S. J., Bent, A. F. (1998). Floral dip a simplified method for agrobacterium-mediated transformation of arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Czechowski, T., Stitt, M., Altmann, T., Udvardi, M. K., Scheible, W.-R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in arabidopsis. Plant Physiol. 139, 5–17. doi: 10.1104/pp.105.063743
Dello Ioio, R., Linhares, F. S., Sabatini, S. (2008). Emerging role of cytokinin as a regulator of cellular differentiation. Curr. Opin. Plant Biol. 11, 23–27. doi: 10.1016/j.pbi.2007.10.006
Demidchik, V., Shang, Z., Shin, R., Thompson, E., Rubio, L., Laohavisit, A., et al. (2009). Plant extracellular ATP signalling by plasma membrane NADPH oxidase and Ca 2+ channels. Plant J. 58, 903–913. doi: 10.1111/j.1365-313X.2009.03830.x
Di, D.-W., Sun, L., Wang, M., Wu, J., Kronzucker, H. J., Fang, S., et al. (2021). WRKY46 promotes ammonium tolerance in arabidopsis by repressing NUDX9 and indole-3-acetic acid-conjugating genes and by inhibiting ammonium efflux in the root elongation zone. New Phytol. 232, 190–207. doi: 10.1111/nph.17554
Ding, Z. J., Yan, J. Y., Li, C. X., Li, G. X., Wu, Y. R., Zheng, S. J. (2015). Transcription factor WRKY46 modulates the development of arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 84, 56–69. doi: 10.1111/tpj.12958
Ding, Z. J., Yan, J. Y., Xu, X. Y., Yu, D. Q., Li, G. X., Zhang, S. Q., et al. (2014). Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in arabidopsis. Plant J. 79, 13–27. doi: 10.1111/tpj.12538
Everaert, C., Luypaert, M., Maag, J. L. V., Cheng, Q. X., Dinger, M. E., Hellemans, J., et al. (2017). Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci. Rep. 7, 1559. doi: 10.1038/s41598-017-01617-3
Irshad, M., Canut, H., Borderies, G., Pont-Lezica, R., Jamet, E. (2008). A new picture of cell wall protein dynamics in elongating cells of arabidopsis thaliana confirmed actors and newcomers. BMC Plant Biol. 8, 94. doi: 10.1186/1471-2229-8-94
Jedličková, V., Ebrahimi Naghani, S., Robert, H. S. (2022). On the trail of auxin reporters and sensors. Plant Cell 34, 3200–3213. doi: 10.1093/plcell/koac179
Jeter, C. R., Tang, W., Henaff, E., Butterfield, T., Roux, S. J. (2004). Evidence of a novel cell signaling role for extracellular adenosine triphosphates and diphosphates in arabidopsis. Plant Cell 16, 2652. doi: 10.1105/tpc.104.023945
Jewell, J. B., Berim, A., Tripathi, D., Gleason, C., Olaya, C., Pappu, H. R., et al. (2022). Activation of indolic glucosinolate pathway by extracellular ATP in arabidopsis. Plant Physiol. 190, 1574–1578. doi: 10.1093/plphys/kiac393
Jewell, J. B., Sowders, J. M., He, R., Willis, M. A., Gang, D. R., Tanaka, K. (2019). Extracellular ATP shapes a defense-related transcriptome both independently and along with other defense signaling pathways. Plant Physiol. 179, pp.01301.2018. doi: 10.1104/pp.18.01301
Jogawat, A., Meena, M. K., Kundu, A., Varma, M., Vadassery, J. (2020). Calcium channel CNGC19 mediates basal defense signaling to regulate colonization by piriformospora indica in arabidopsis roots. J. Exp. Bot. 71, 2752–2768. doi: 10.1093/jxb/eraa028
Kilian, J., Whitehead, D., Horak, J., Wanke, D., Weinl, S., Batistic, O., et al. (2007). The AtGenExpress global stress expression data set protocols, evaluation and model data analysis of UV-b light, drought and cold stress responses. Plant J. 50, 347–363. doi: 10.1111/j.1365-313X.2007.03052.x
Koncz, C., Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of agrobacterium binary vector. Mol. Gen. Genet. MGG 204, 383–396. doi: 10.1007/BF00331014
Kugler, A., Köhler, B., Palme, K., Wolff, P., Dietrich, P. (2009). Salt-dependent regulation of a CNG channel subfamily in arabidopsis. BMC Plant Biol. 9, 140. doi: 10.1186/1471-2229-9-140
Kumar, S., Tripathi, D., Okubara, P. A., Tanaka, K. (2020). Purinoceptor P2K1/DORN1 enhances plant resistance against a soilborne fungal pathogen, rhizoctonia solani. Front. Plant Sci. 11, 1479. doi: 10.3389/fpls.2020.572920
Lim, M. H., Wu, J., Yao, J., Gallardo, I. F., Dugger, J. W., Webb, L. J., et al. (2014). Apyrase suppression raises extracellular ATP levels and induces gene expression and cell wall changes characteristic of stress responses. Plant Physiol. 164, 2054–2067. doi: 10.1104/pp.113.233429
Meena, M. K., Prajapati, R., Krishna, D., Divakaran, K., Pandey, Y., Reichelt, M., et al. (2019). The Ca2+ channel CNGC19 regulates arabidopsis defense against spodoptera herbivory. Plant Cell 31, 1539–1562. doi: 10.1105/tpc.19.00057
Murashige, T., Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nizam, S., Qiang, X., Wawra, S., Nostadt, R., Getzke, F., Schwanke, F., et al. (2019). Serendipita indica E5′NT modulates extracellular nucleotide levels in the plant apoplast and affects fungal colonization. EMBO Rep. 20, e47430. doi: 10.15252/embr.201847430
Perilli, S., Di Mambro, R., Sabatini, S. (2012). Growth and development of the root apical meristem. Curr. Opin. Plant Biol. 15, 17–23. doi: 10.1016/j.pbi.2011.10.006
Petricka, J. J., Winter, C. M., Benfey, P. N. (2012). Control of arabidopsis root development. Annu. Rev. Plant Biol. 63, 563–590. doi: 10.1146/annurev-arplant-042811-105501
Quiles-Pando, C., Rexach, J., Navarro-Gochicoa, M. T., Camacho-Cristóbal, J. J., Herrera-Rodríguez, M. B., González-Fontes, A. (2013). Boron deficiency increases the levels of cytosolic Ca2+ and expression of Ca2+-related genes in arabidopsis thaliana roots. Plant Physiol. Biochem. 65, 55–60. doi: 10.1016/j.plaphy.2013.01.004
Riewe, D., Koohi, M., Lisec, J., Pfeiffer, M., Lippmann, R., Schmeichel, J., et al. (2012). A tyrosine aminotransferase involved in tocopherol synthesis in arabidopsis. Plant J. 71, 850–859. doi: 10.1111/j.1365-313X.2012.05035.x
Sauer, H., Hescheler, J., Wartenberg, M. (2000). Mechanical strain-induced Ca2+waves are propagated via ATP release and purinergic receptor activation. Am. J. Physiol. Physiol. 279, C295–C307. doi: 10.1152/ajpcell.2000.279.2.C295
Sheikh, A. H., Eschen-Lippold, L., Pecher, P., Hoehenwarter, W., Sinha, A. K., Scheel, D., et al. (2016). Regulation of WRKY46 transcription factor function by mitogen-activated protein kinases in arabidopsis thaliana. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00061
Song, C. J., Steinebrunner, I., Wang, X., Stout, S. C., Roux, S. J. (2006). Extracellular ATP induces the accumulation of superoxide via NADPH oxidases in arabidopsis. Plant Physiol. 140, 1222. doi: 10.1104/pp.105.073072
Tanaka, K., Choi, J., Cao, Y., Stacey, G. (2014). Extracellular ATP acts as a damage-associated molecular pattern (DAMP) signal in plants. Front. Plant Sci. 5, 446. doi: 10.3389/fpls.2014.00446
Tanaka, K., Heil, M. (2021). Damage-associated molecular patterns (DAMPs) in plant innate immunity applying the danger model and evolutionary perspectives. Annu. Rev. Phytopathol 59, 53–75. doi: 10.1146/annurev-phyto-082718-100146
Tanaka, K., Swanson, S. J., Gilroy, S., Stacey, G. (2010). Extracellular nucleotides elicit cytosolic free calcium oscillations in arabidopsis. Plant Physiol. 154, 705. doi: 10.1104/pp.110.162503
Torres, J., Rivera, A., Clark, G., Roux, S. J. (2008). Participation of extracellular nucleotides in the wound response of dasycladus vermicularis and acetabularia acetabulum (Dasycladales, chlorophyta). J. Phycol. 44, 1504–1511. doi: 10.1111/j.1529-8817.2008.00602.x
Tripathi, D., Zhang, T., Koo, A. J., Stacey, G., Tanaka, K. (2018). Extracellular ATP acts on jasmonate signaling to reinforce plant defense. Plant Physiol. 176, 511. doi: 10.1104/pp.17.01477
Wang, Y., Bouwmeester, K., Beseh, P., Shan, W., Govers, F. (2014). Phenotypic analyses of arabidopsis T-DNA insertion lines and expression profiling reveal that multiple l-type lectin receptor kinases are involved in plant immunity. Mol. Plant-Microbe Interact. 27, 1390–1402. doi: 10.1094/MPMI-06-14-0191-R
Wang, L., Wilkins, K. A., Davies, J. M. (2018). Arabidopsis DORN1 extracellular ATP receptor; activation of plasma membrane k+-and Ca2+-permeable conductances. New Phytol. 218, 1301–1304. doi: 10.1111/nph.15111
Winter, D., Vinegar, B., Nahal, H., Ammar, R., Wilson, G. V., Provart, N. J. (2007). An “Electronic fluorescent pictograph” browser for exploring and analyzing Large-scale biological data sets. PloS One 2, e718. doi: 10.1371/journal.pone.0000718
Yan, Y., Stolz, S., Chételat, A., Reymond, P., Pagni, M., Dubugnon, L., et al. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19, 2470–2483. doi: 10.1105/tpc.107.050708
Zhang, K., Han, Y.-T., Zhao, F.-L., Hu, Y., Gao, Y.-R., Ma, Y.-F., et al. (2015). Genome-wide identification and expression analysis of the CDPK gene family in grape, vitis spp. BMC Plant Biol. 15, 1–19. doi: 10.1186/s12870-015-0552-z
Keywords: transcriptional histochemical reporters, damage-associate molecular patterns (DAMPs), purinoceptor P2K1, purinergic sigaling, extracellular ATP (eATP), signal transduction, root tip
Citation: Sowders JM and Tanaka K (2023) A histochemical reporter system to study extracellular ATP response in plants. Front. Plant Sci. 14:1183335. doi: 10.3389/fpls.2023.1183335
Received: 10 March 2023; Accepted: 09 May 2023;
Published: 02 June 2023.
Edited by:
Takuya Furuichi, Hagoromo International University, JapanReviewed by:
Kenji Hashimoto, Tokyo University of Science, JapanGreg Clark, The University of Texas at Austin, United States
Copyright © 2023 Sowders and Tanaka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kiwamu Tanaka, a2l3YW11LnRhbmFrYUB3c3UuZWR1
 Joel M. Sowders
Joel M. Sowders Kiwamu Tanaka
Kiwamu Tanaka