
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 06 October 2023
Sec. Plant Breeding
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1176914
Modern sugarcane cultivars (Saccharum spp., 2n = 100~120) are complex polyploids primarily derived from interspecific hybridization between S. officinarum and S. spontaneum. Nobilization is the theory of utilizing wild germplasm in sugarcane breeding, and is the foundation for utilizing S. spontaneum for stress resistance. However, the exact chromosomal transmission remains elusive due to a lack of chromosome-specific markers. Here, we applied chromosome-specific oligonucleotide (oligo)-based probes for identifying chromosomes 1-10 of the F1 hybrids between S. officinarum and S. spontaneum. Then, S. spontaneum-specific repetitive DNA probes were used to distinguish S. spontaneum in these hybrids. This oligo- fluorescence in situ hybridization (FISH) system proved to be an efficient tool for revealing individual chromosomal inheritance during nobilization. We discovered the complete doubling of S. officinarum-derived chromosomes in most F1 hybrids. Notably, we also found defective S. officinarum-derived chromosome doubling in the F1 hybrid Yacheng75-4191, which exhibited 1.5n transmission for all nonhomologous chromosomes. Altogether, these results highlight the presence of variable chromosome transmission in nobilization between S. officinarum and S. spontaneum, including 1.5n + n and 2n + n. These findings provide robust chromosome markers for in-depth studies into the molecular mechanism underlying chromosome doubling during the nobilization, as well as tracing chromosomal inheritance for sugarcane breeding.
Sugarcane (Saccharum spp.), a member of the family Poaceae, is a complex heterozygous polyploid plant, exhibiting up to decaploidy or even higher (Grivet and Arruda, 2002; Piperidis and D’Hont, 2020). Sugarcane has been received more attention and is used to produce sugar as well as energy. S. officinarum (2n = 80 or 81, x = 10) exhibits a high sugar content, but poor environmental resistance (Irvine, 1999; Aitken et al., 2006; Piperidis et al., 2010). In contrast, S. spontaneum (2n = 40 ~ 128, x = 8, 9, 10), which is distributed widely from the Mediterranean to the Pacific, is highly stress-resistance (Panje and Babu, 1960). S. spontaneum is typically hybridized with S. officinarum (Price, 1957; BT, 1972). Thus, sugarcane is an allopolyploid produced by repeated interspecific hybridization. Allopolyploidy, which results in genetic redundancy, is a source of new variation. In sugarcane, such allopolyploidy results in increased drought tolerance, pest resistance, and biomass production etc. (Osborn et al., 2003; Chen, 2007). Despite its economic importance, the complexity of the sugarcane genetic background has limited classical genetic studies (Barnes and Bester, 2000). The high ploidy level of interspecific Saccharum hybrids, and the variability of the hybridization environment, further complicate sugarcane genetic research.
The complex genetic background of cultivated sugarcane is the result of multiple interspecific hybridization events, particularly between S. officinarum and S. spontaneum. The first artificial interspecific hybrids between these two species were created to overcome disease outbreaks and were followed by repeated backcrossing using S. officinarum as the recurrent female parent to restore high sucrose content. This process, known as ‘nobilization’, has been central to sugarcane genetic improvement (D'Hont et al., 1996; Ming et al., 2010). Jeswiet has bred a series of excellent parental materials over the past few decades, including the ‘POJ’ series (Heinz, 1987). This strategy combines the stress resistance of S. spontaneum with the high yield and sugar content of S. officinarum (Jackson, 2005). Grivet et al. found that 15~25% lineages of the sugarcane cultivar R570 were derived from recombination between S. officinarum and S. spontaneum (Grivet et al., 1996). Recent cytogenetic studies have shed light on the basic chromosome number of S. spontaneum and chromosomal inheritance during sugarcane breeding (Cuadrado et al., 2004; Edmé et al., 2006). A variety of chromosomal inheritance patterns have been suggested, including “2n + n” and “n + n” (Lalitha and Premachandran, 2007; Paterson et al., 2013). Studies suggest that higher-yielding varieties are the result of “2n+n” heredity (D'Hont et al., 1998). However, the limited chromosomal markers, gDNA probes, and repetitive probes hinders the further exploration of the cytogenetic mechanism underlying sugarcane nobilization.
Fluorescence in situ hybridization (FISH) is being applied in many biological research and has become an indispensable cytogenetic tool (Hu et al., 2014). Through the use of fluorescence, FISH allows the targeting and visualization of specific DNA within whole chromosomes or chromosomal regions (Younis et al., 2015). However, the exact individual chromosome identification is still challenging in plants due to a lack of chromosome-specific markers. Improvements in genomic sequencing have allowed the development of a novel FISH technique called oligonucleotide (oligo)-based FISH (Beliveau et al., 2012; de Oliveira Bustamante et al., 2021). In contrast to conventional FISH, oligo-FISH utilizes bioinformatics-designed probes, which has broadened the scope of cytogenetic researches (Schmutz et al., 2014; Han et al., 2015). Oligo-FISH has been used to study chromosomes and structural genomic variants in several crop species, including potato (Braz et al., 2018), maize (Braz et al., 2021), cucumber (Zhao et al., 2022), and rapeseed (Boideau et al., 2022) etc. Here, we applied oligo-based probes to identify the individual chromosomes of S. officinarum and S. spontaneum, and study the chromosomal heredity of their F1 hybrids. The results of this study provide a cytological basis for a better understanding of the nobilization in sugarcane breeding.
Plant materials were obtained primarily from Fujian Agriculture and Forestry University and Yunnan Academy of Agricultural Sciences, China. The study materials, including S. officinarum, S. spontaneum, and their F1 hybrids, are reviewed in Table 1.
Root tips (~1 cm long) were pretreated with 1,4-dichlorobenzene-α-naphthylbromide for 2.5 h, then fixed with freshly-made Carnoy’s fixative (ethanol:glacial acetic acid, 3:1) for 24 h at 4 °C. The fixed samples were subsequently rinsed with distilled water and stored in 70% ethanol at -20°C. Root tips were digested with an enzymatic solution (1% pectolase Y-23, 2% pectolyase, 2% cellulase RS, 4% macerozyme R-10) for 3~4 h at 37°C, according to a previously-published protocol (Schwarzacher and Leitch, 1994). The digested root tip suspension was dropped on a slide and then an additional Carnot’s fixative was added to disperse the cells. Slide was used for microscopic examination and then the perfect metaphase cells were selected for further oligo-FISH.
All oligo probe sequence libraries are available in Dataset S1(nph17905-sup-0001-Dataset1.xlsx) (Yu et al., 2022). Oligo probes were synthesized and labeled with either TAMARA- or FAM-, according to the method of Yu et al. (Yu et al., 2022). S. spontaneum-specific repetitive DNA probes (SsRetro1-SsRetro4) were directly-labeled with Cy3-dUTP, according to Huang et al. (Huang et al., 2020). Oligo-FISH was performed according to Braz et al. (Braz et al., 2020), with minor modifications. The treated slides were dehydrated in 75% and 100% ethanol sequentially for each 3 min. Briefly, slides were treated for 1.5 h with pepsin then fixed with 4% formaldehyde for 5 min and with 2× Saline Sodium Citrate (SSC) buffer for 3 min each. The treated slides were sequentially dehydrated with 75% and 100% ethanol for 3 min each. Next, the slides were denatured at 60°C for 90 s with 70% formamide (FD) and rapidly cooled to -20°C. The hybridization solutions (100% FD, 20 × SSC, 50% dextran sulphate (DS) and ~400 ng oligo probes) was added on the center of each slide. Hybridization was carried out at 37°C overnight. Next, the slides were washed using 2 × SSC for 5 min and 10 min, and twice with 1 × PBS for 5 min. The washed slides were then dried and counterstained with the 5 μg/mL 4′,6-diamidino-2- phenylindole (DAPI). Chromosome images were captured by Olympus BX53 camera epifluorescence microscope. The contrast of FISH images was adjusted and merged using Photoshop CS6 software.
Oligo-based probes were designed based on the S. officinarum LA Purple genome and were anchored to chromosomes 1-10 (Yu et al., 2022). The ten chromosome-specific probes (Chr1-Chr10) were labeled using fluorescent dyes (TAMRA-, FAM-, or Cy5-) and hybridized to the somatic metaphase chromosomes prepared from three female S. officinarum parents (Badila-CN, Fiji and Yuenan Niuzhe) (Figure 1). Labeled Chr1-Chr10 probes will be expected to generate chromosome-specific fluorescence signals corresponding to chromosomes 1-10. For example, the Chr1, Chr2, and Chr8 painting probes, when hybridized to a metaphase cell, worked to identify chromosomes 1, 2, and 8, respectively (Figure 1A1). The slide was then washed prior to applying the Chr3 and Chr4 painting probes (Figure 1B1). All 10 chromosomes were successfully identified after five sequential rounds of FISH, and each probe hybridized to eight copies of a chromosome in Badila-CN (Figures 1A1–E1), Fiji (Figures 1A2–E2) and Yuenan Niuzhe (Figures 1A3–E3). These results indicate that all three S. officinarum have the same chromosomal composition and are autooctaploid with 2n = 8x = 80.
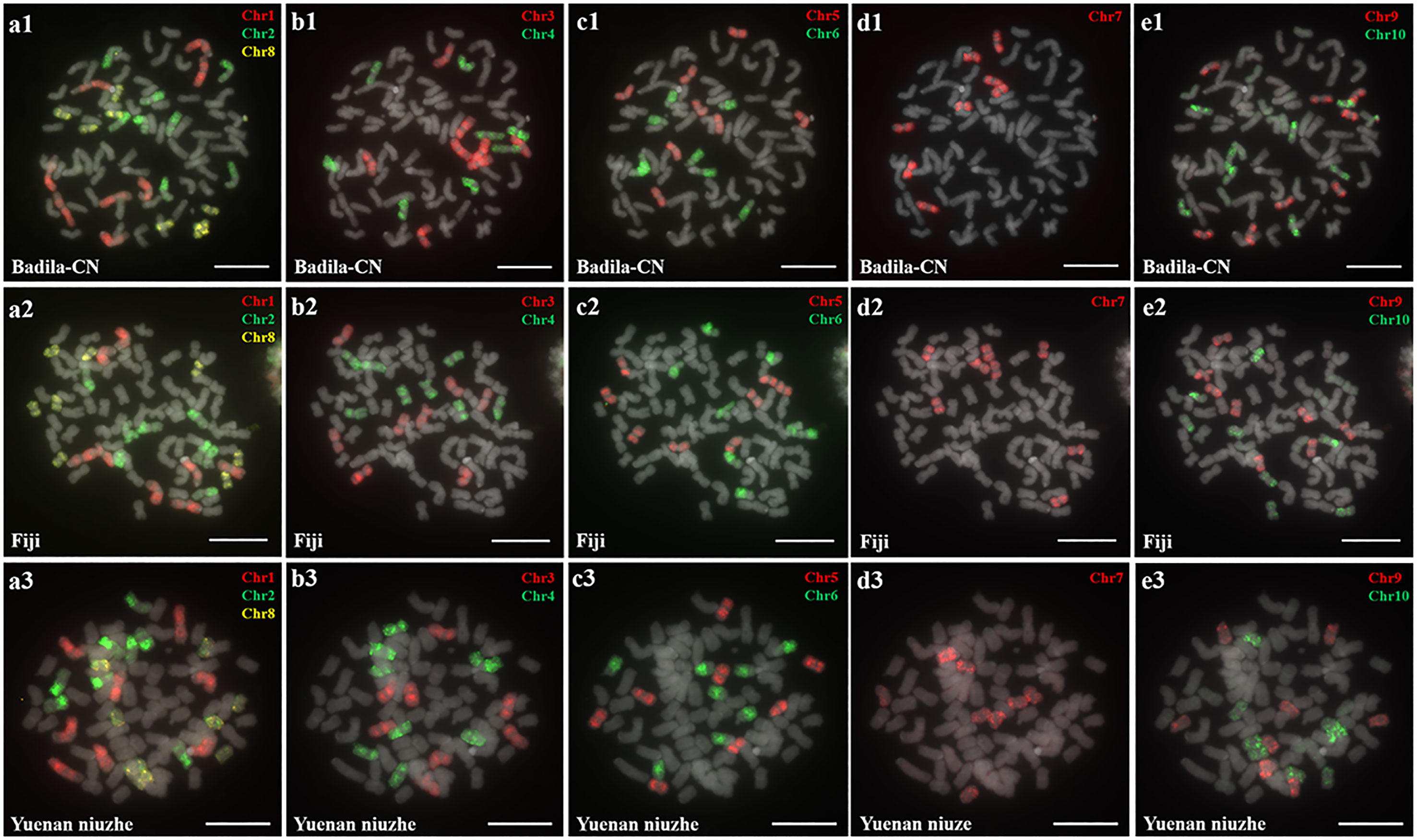
Figure 1 Metaphase chromosome identification of S. officinarum. (A) Chr1(red), Chr2 (green) and Chr8 (yellow); (B) Chr3 (red) and Chr4 (green); (C) Chr5 (red) and Chr6 (green); (D) Chr7 (red); (E) Chr9 (red) and Chr10 (green). Chromosome-specific probes were hybridized to somatic metaphase chromosomes prepared from Badila-CN (A1–E1), Fiji (A2–E2), and Yuenan niuzhe (A3–E3). The gray chromosomes are counterstained by DAPI. Bars = 10 µm.
Two S. spontaneum accessions, Yacheng-spon (2n = 80) and Yunnan75-2-11 (2n = 64), were used for individual chromosome analysis. Again, we used 10 S. officinarum-derived chromosome-specific painting probes hybridized to meiotic S. spontaneum chromosomes. Each of the 10 painting probes produced obvious FISH signals in both Yacheng-spon and Yunnan75-2-11. After six sequential rounds of chromosome painting, Yacheng-spon was confirmed to be a decaploid with a basic chromosome number x = 8 (Figure 2; Table 2). SsChr5 was hybridized with both the Chr5 and Chr6 probes, indicating that the S. officinarum 5-like chromosome was broken and fused with the S. officinarum 6-like chromosome to form SsChr5 of Yacheng-spon (Figure 2C), as well as the S. officinarum 7-like chromosome to form SsChr6 of Yacheng-spon (Figure 2D). Additionally, S. officinarum 8-like was broken and fused with S. officinarum 2-like and 9-like to form SsChr2 and SsChr7 of Yacheng-spon, respectively (Figures 2E, F). Individual chromosome of Yunnan75-2-11 were also identified, with similar fusions occurring in SsChr2, SsChr5, SsChr6 and SsChr7 (Figure 3). These results indicate that Yunnan75-2-11 is an octoploid with a basic chromosome number x = 8 (Figure 3; Table 2).
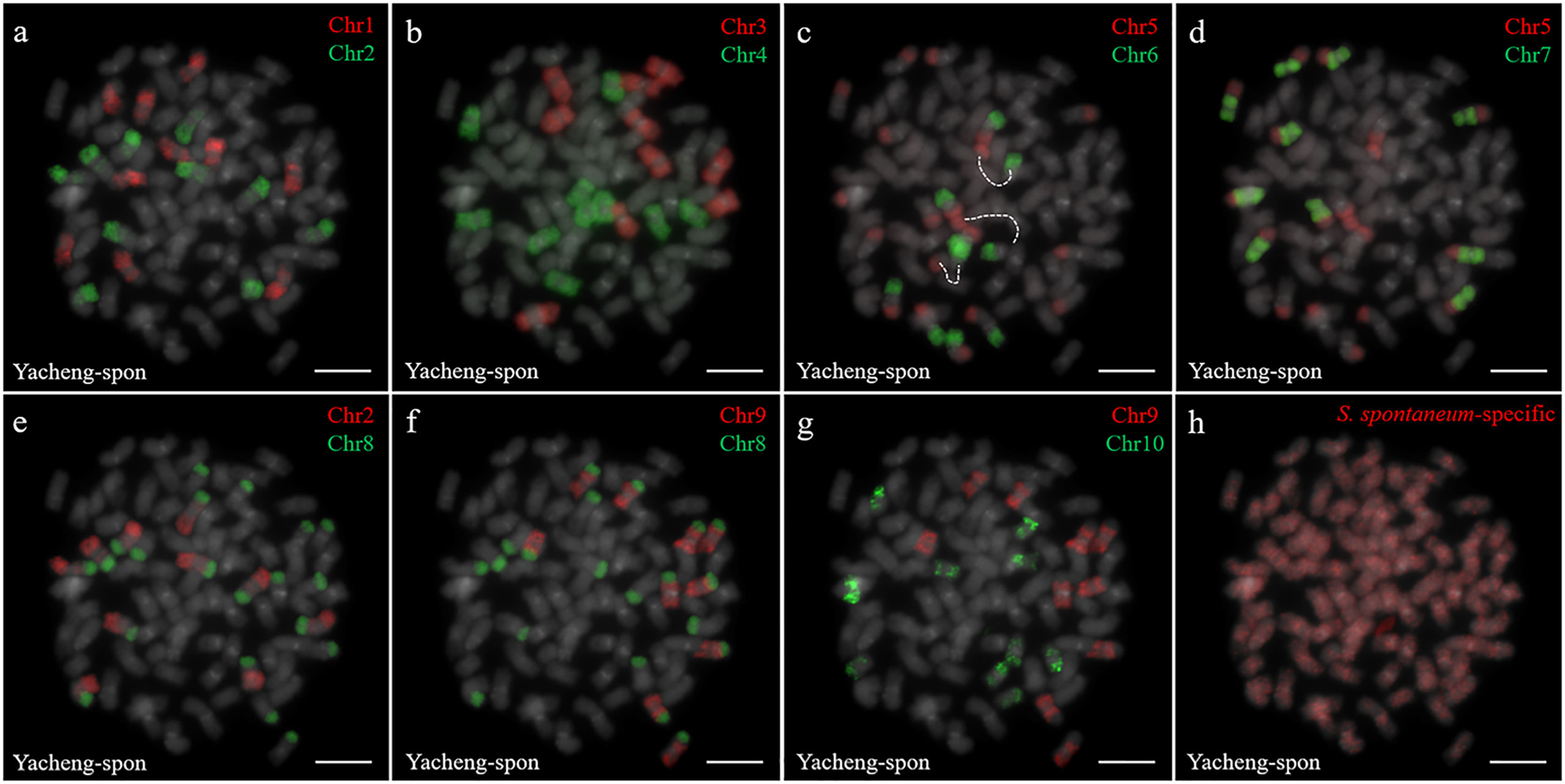
Figure 2 Metaphase chromosome identification of S. spontaneum Yacheng-spon. (A) Chr1(red) and Chr2 (green); (B) Chr3 (red) and Chr4 (green); (C) Chr5 (red) and Chr6 (green), with dotted lines linking the fusion chromosome SsChr5; (D) Chr5 (red) and Chr7 (green); (E) Chr2 (red) and Chr8 (green); (F) Chr9 (red) and Chr8 (green); (G) Chr9 (red) and Chr10 (green); (H) S. spontaneum-specific probe (red). The gray chromosomes are counterstained by DAPI. Bars = 10 µm.
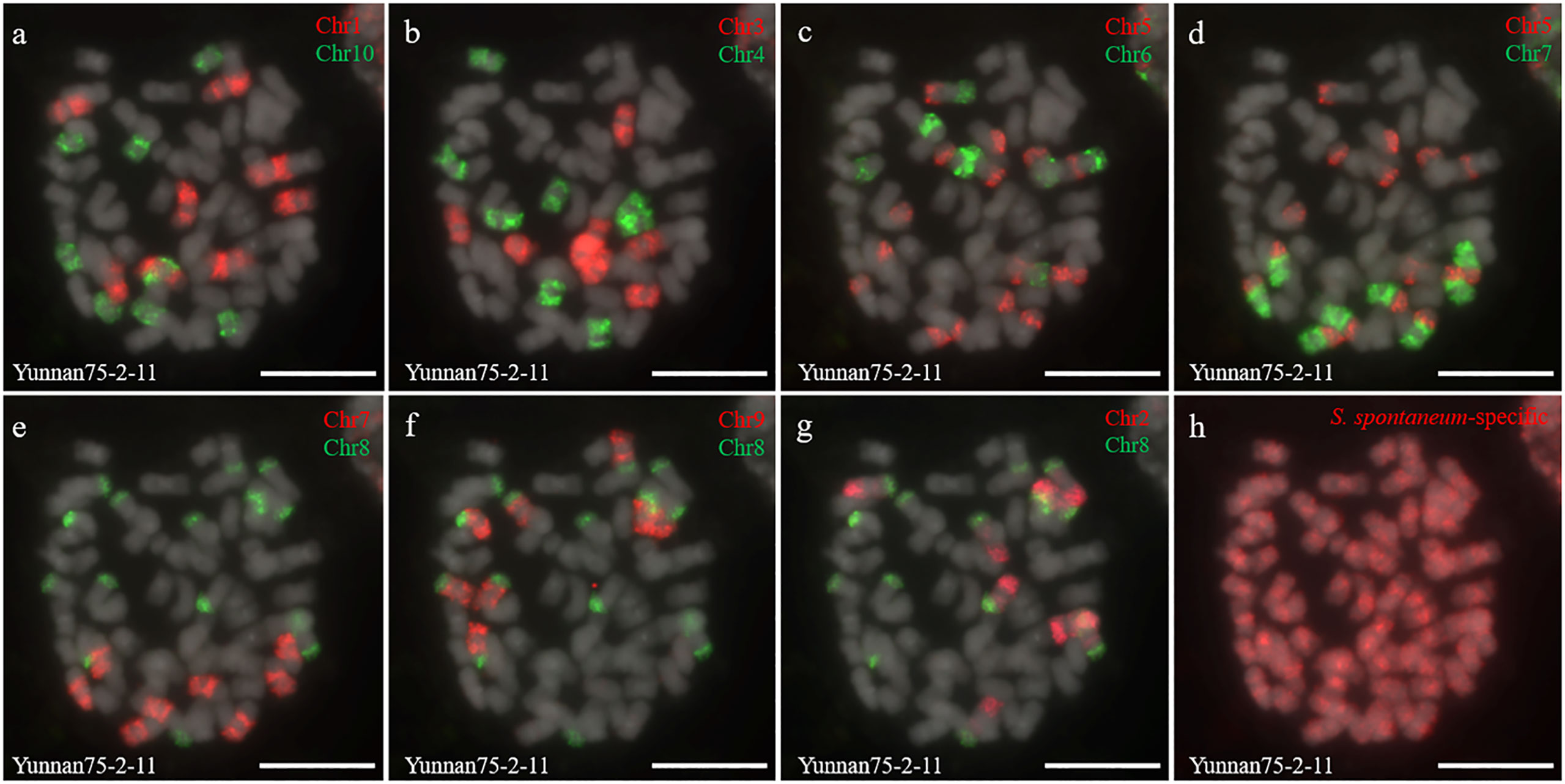
Figure 3 Metaphase chromosome identification of S. spontaneum Yunnan75-2-11. (A) Chr1(red) and Chr10 (green); (B) Chr3 (red) and Chr4 (green); (C) Chr5 (red) and Chr6 (green); (D) Chr5 (red) and Chr7 (green); (E) Chr7 (red) and Chr8 (green); (F) Chr9 (red) and Chr8 (green); (G) Chr2 (red) and Chr8 (green); (H) S. spontaneum-specific probe (red). The gray chromosomes are counterstained by DAPI. Bars = 10 µm.
The 10 chromosome-specific painting probes allow us essentially to identify each nonhomologous chromosomes in S. officinarum and S. spontaneum. Thus, we are interested in knowing how the individual chromosomes inheritance, as well as introgression of the S. spontaneum lineage into S. officinarum. Hence, four F1 hybrids between S. officinarum and S. spontaneum, Yacheng82-108, Yacheng58-43, Yacheng75-4191 and Yacheng75-409, were selected for chromosomal painting.
We first identified all chromosomes through multiple rounds of oligo-FISH, and then distinguished S. spontaneum-specific chromosomes using S. spontaneum-specific probes applied to the same cell. In Yacheng82-108 (Badila-CN × Yunnan75-2-11), each of the 10 oligo probes generated symmetrical signals through multiple rounds of FISH (Figure 4). The chromosomal colocalization results indicated that Yacheng82-108 contains a total of 112 chromosomes, including 80 derived from S. officinarum and 32 derived from S. spontaneum (Figure 4; Table 2). Each set of nonhomologous chromosomes was uniformly inherited from their parents.
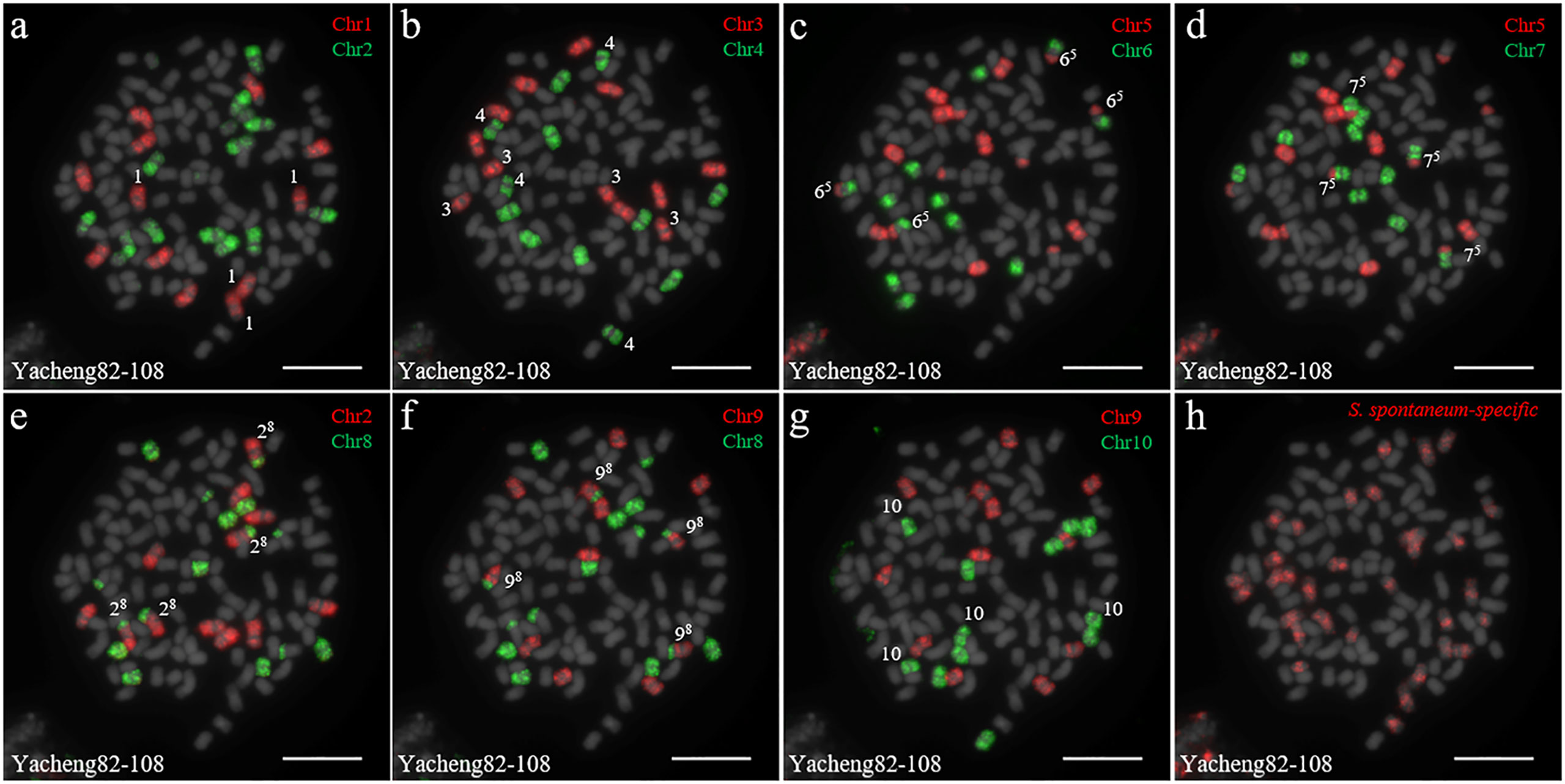
Figure 4 Chromosome identification of F1 hybrid Yacheng82-108. (A) Chr1(red) and Chr2 (green); (B) Chr3 (red) and Chr4 (green); (C) Chr5 (red) and Chr6 (green); (D) Chr5 (red) and Chr7 (green); (E) Chr2 (red) and Chr8 (green); (F) Chr9 (red) and Chr8 (green); (G) Chr9 (red) and Chr10 (green); (H) S. spontaneum-specific probe (red). The gray chromosomes are counterstained by DAPI. The Arabic numerals indicate S. spontaneum chromosomes. Xy indicates a fused chromosome in which chromosome Y was broken and fused with chromosome X. Bars = 10 µm.
In addition, we chose three other F1 hybrids from the same male parent Yacheng-spon but with different female S. officinarum parents. For Yacheng58-43, the painting probes generated distinct signals (Figure 5). FISH indicated that Yacheng58-43 (2n) contained 120 somatic chromosomes, including 40 derived from S. spontaneum chromosomes (in which half of each chromosome was derived from the male parent) and 80 derived from S. officinarum. The same chromosomal composition was also found for Yacheng75-409 (Figure 6). Yacheng75-4191 contained 40 chromosomes derived from S. spontaneum (Figure 7, labeled with Arabic numerals), but a different number of chromosomes derived from S. officinarum. Each of the 10 painting probes hybridized to six S. officinarum chromosomes in Yacheng75-4191, suggesting that 1.5n of female parent (S. officinarum) chromosomes were transmitted to Yacheng75-4191 (Figure 7; Table 2). Finally, FISH results indicated that the chromosomal transmission of S. spontaneum is “n” in all F1 hybrids.
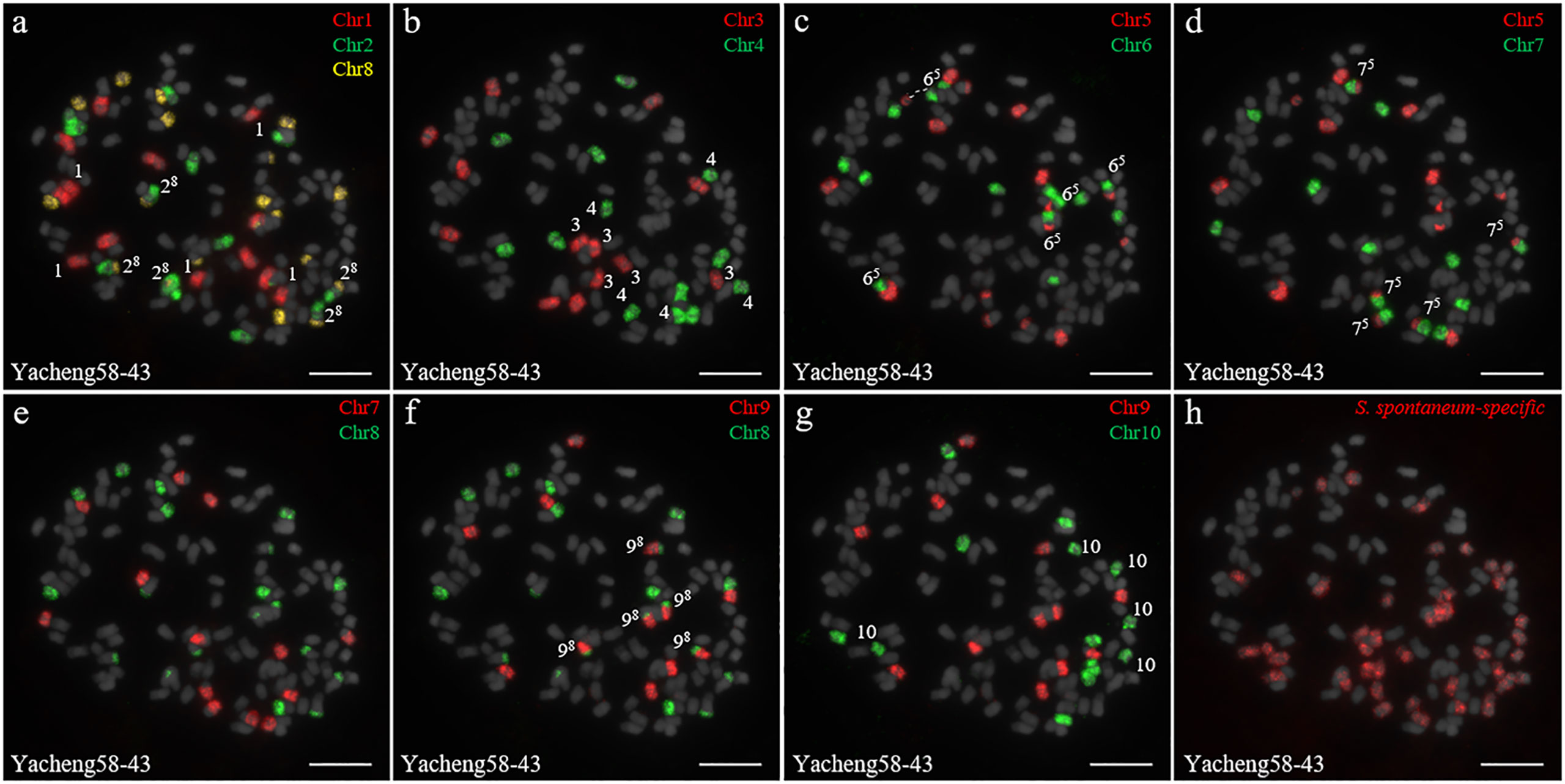
Figure 5 Chromosome identification of F1 hybrid Yacheng58-43. (A) Chr1(red) Chr2 (green) and Chr8 (yellow); (B) Chr3 (red) and Chr4 (green); (C) Chr5 (red) and Chr6 (green), with dotted lines linking the fusion chromosome SsChr5; (D) Chr5 (red) and Chr7 (green); (E) Chr7 (red) and Chr8 (green); (F) Chr9 (red) and Chr8 (green); (G) Chr9 (red) and Chr10 (green); (H) S. spontaneum-specific probe (red). The gray chromosomes are counterstained by DAPI. The Arabic numerals indicate S. spontaneum chromosomes. Xy indicates a fused chromosome in which chromosome Y was broken and fused with chromosome X. Bars = 10 µm.
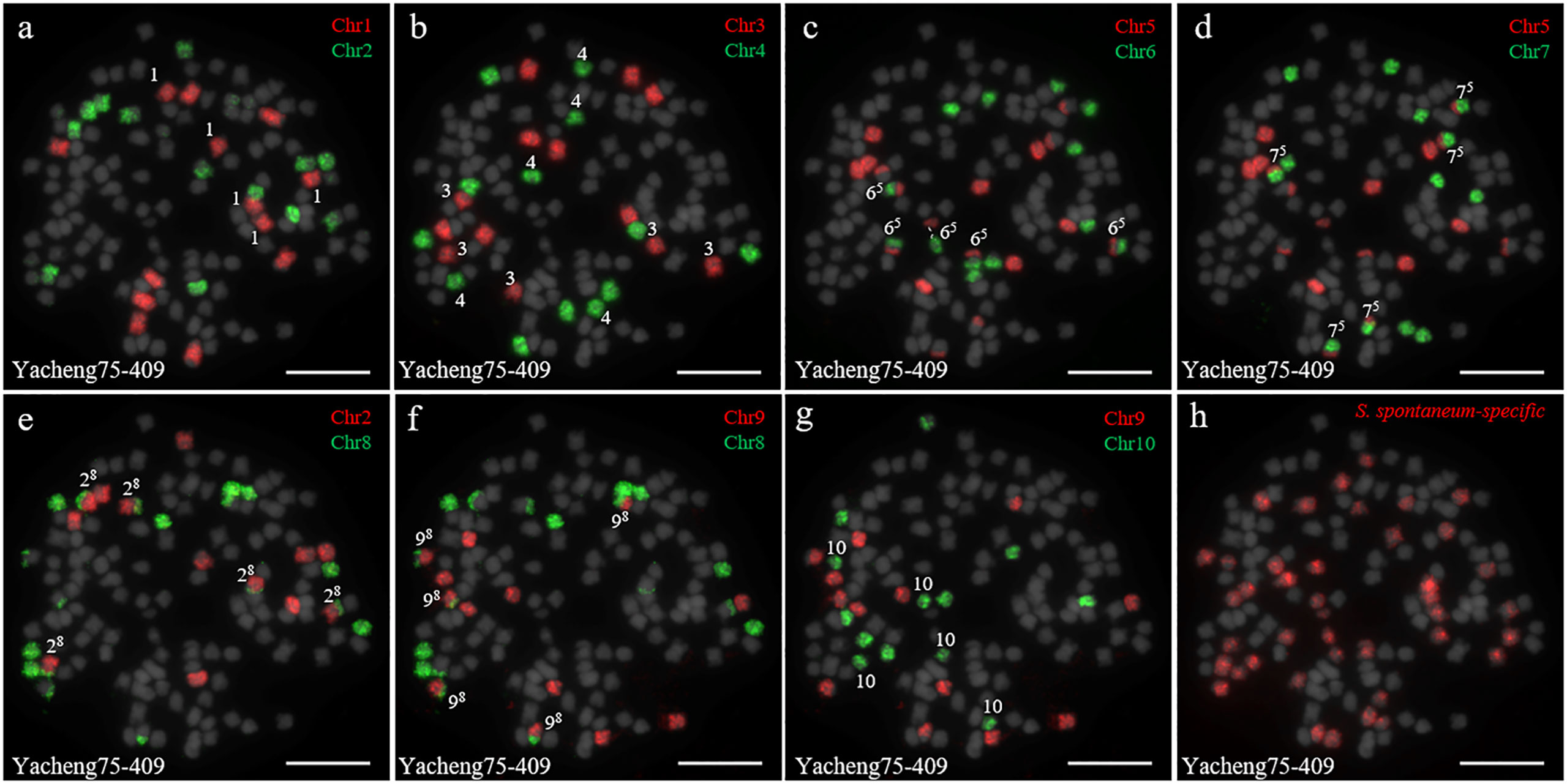
Figure 6 Chromosome identification of F1 hybrid Yacheng75-409. (A) Chr1(red) and Chr2 (green); (B) Chr3 (red) and Chr4 (green); (C) Chr5 (red) and Chr6 (green), with dotted lines linking the fusion chromosome SsChr5; (D) Chr5 (red) and Chr7 (green); (E) Chr2 (red) and Chr8 (green); (F) Chr9 (red) and Chr8 (green); (G) Chr9 (red) and Chr10 (green); (H) S. spontaneum-specific probe (red). The gray chromosomes are counterstained by DAPI. The Arabic numerals indicate S. spontaneum chromosomes. Xy indicates a fused chromosome in which chromosome Y was broken and fused with chromosome X. Bars = 10 µm.
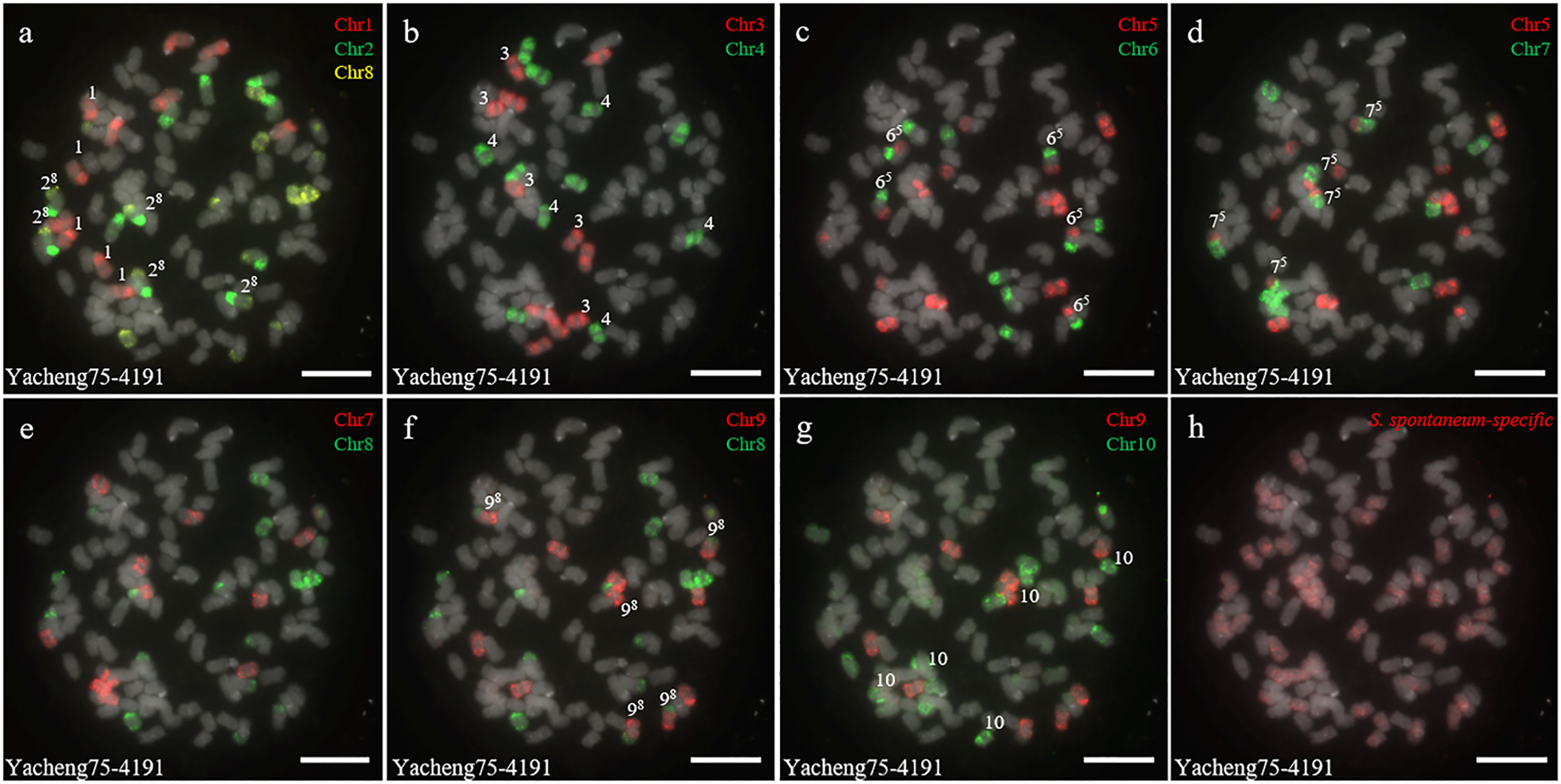
Figure 7 Chromosome identification of F1 hybrid Yacheng75-4191. (A) Chr1(red) Chr2 (green) and Chr8 (yellow); (B) Chr3 (red) and Chr4 (green); (C) Chr5 (red) and Chr6 (green); (D) Chr5 (red) and Chr7 (green); (E) Chr7 (red) and Chr8 (green); (F) Chr9 (red) and Chr8 (green); (G) Chr9 (red) and Chr10 (green); (H) S. spontaneum-specific probe (red). The gray chromosomes are counterstained by DAPI. The Arabic numerals indicate S. spontaneum chromosomes. Xy indicates a fused chromosome in which chromosome Y was broken and fused with chromosome X. Bars = 10 µm.
Identification of individual chromosome is essential to understand chromosome organization and evolution of related species (Jiang and Gill, 2006; Xiong and Pires, 2011). In plants, chromosome painting with oligo-based probes has been used for karyotyping as well as studies of chromosomal rearrangement, meiotic pairing and recombination (Jiang, 2019). Sugarcane has one of the most complex genetic backgrounds, and exhibits high degrees of polyploidy as well as aneuploidy (Heinz et al., 1969). The use of chromosome-specific barcodes and painting probes has revealed the complex chromosomal structure and evolutionary history of sugarcane and related species (Meng et al., 2020; Meng et al., 2022; Yu et al., 2022). Sugar-rich S. officinarum typically has either 2n = 80 or 81 chromosomes, with plants containing more than 81 chromosomes likely to be hybrids (Bremer, 1961; Piperidis et al., 2010; Piperidis and D’Hont, 2020). In this study, we found that S. officinarum Fiji and Yuenan niuzhe each possessed eight copies of homologous chromosomes, suggesting that they are typical octaploids (Figure 1). These results support the classical view that S. officinarum is characterized as an autooctaploid with x = 10 (Figure 1). For S. spontaneum, previous cytological evidence indicated that the x = 8 cytotype was derived from x = 10 through consecutive chromosomal breaking and fusion. In this study we used chromosome-specific paints based on the S. officinarum genome and found that both S. spontaneum Yunnan75-2-11 and Yacheng-spon were x = 8. Consistent with previous results, we also found that the S. officinarum 5-like and S. officinarum 8-like chromosomes had been broken and fused. Additionally, we found that SsChr5 is most susceptible to breakage at the S. officinarum-like chromosome 5-6 fusion point in S. spontaneum Yacheng-spon (Figure 2). Notably, this phenomenon is genetically transmitted to the offspring (Figures 5, 6), potentially leading to meiotic instability.
S. spontaneum has been widely used for improving sugarcane genetics due to its excellent agronomic characteristics and the increasing demand for stress-resistant sugarcane cultivars (De Morais et al., 2015; Racedo et al., 2016). In nobilization, the retention of S. spontaneum-derived resistance genes is accompanied by the presence of unreduced S. officinarum gametes, leading to high sugar content (Lu et al., 1994). The varied cytotypes and rich genetic diversity of S. spontaneum have hampered cytogenetic studies in sugarcane (Zhang et al., 2018). Previous studies suggest that either “2n + n” or “n + n” transmission occurs in the F1 hybrids between S. officinarum and S. spontaneum (Price, 1957; Piperidis et al., 2010). However, the available cytogenetic probes, 5S rDNA, 35S rDNA, and genomic DNA, have been unable to identify all non-homologous chromosomes in these hybrids. Chromosomal inheritance in F1 hybrids between S. officinarum and S. spontaneum was preliminarily studied based on chromosome numbers (D'Hont et al., 1998; Yu et al., 2018). However, the precise mechanism of individual chromosome transmission remained largely unknown. In this study, we combined chromosome-specific oligo-based probes and S. spontaneum-specific repeat probes to identify all ten sugarcane chromosomes and determine whether they were derived from S. officinarum or S. spontaneum (Figures 4–7). Our system proved highly efficient for tracing the precise chromosomal inheritance pattern in hybrids of S. officinarum and S. spontaneum, and will aid in efforts to further utilize S. spontaneum in sugarcane breeding.
Based on nobilization theory, sugarcane breeders have carried out many researches for improving the genetic background of sugarcane. However, chromosome inheritance mechanism of S. officinarum is still confused and dubious in the hybrids between S. officinarum and S. spontaneum. Previously, “2n + n” chromosome transmission has been found by the method of only using genomic in situ hybridization, but this technique cannot verify whether the S. officinarum chromosomes were fully doubled (Price, 1957; Piperidis et al., 2010; Yu et al., 2018). In this study, by combining S. officinarum-derived chromosome paints and S. spontaneum-specific probes, we that all three studied F1 hybrids (Yacheng82-108, Yacheng58-43, Yacheng75-409) had “2n + n” chromosome transmission (Figures 4–6). These results suggest that each non-homologous S. officinarum chromosomes was completely doubled. Interestingly, we found one F1 hybrid (Yacheng75-4191) exhibited “1.5n + n” transmission, suggesting that 1.5 × of S. officinarum chromosomes were inherited (Figure 7). Altogether, these results will broaden our understanding of the sugarcane nobilization between S. officinarum and S. spontaneum. Furthermore, these chromosome-specific identification results will be useful for detailed physical mapping for use in guiding breeders to select suitable parents for sugarcane breeding.
In the present study, the chromosome-specific painting probes derived from S. officinarum can be used for accurately identifying individual chromosome and chromosomal heredity of F1 hybrids during sugarcane nobilization. We discovered that nonhomologous S. officinarum chromosomes were completely doubling in most F1 hybrids. However, the F1 hybrid Yacheng75-4191 exhibited defective chromosome doubling with 1.5n of S. officinarum chromosomes transmission. These results support previous genetic studies of S. spontaneum and provide more useful molecular cytogenetic data for its F1 hybrids. In addition, these results provide robust chromosome markers for in-depth studies into the molecular mechanism underlying chromosome doubling during the nobilization, as well as tracing chromosomal inheritance for sugarcane breeding.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
ZD and FY designed the research. JC, LX and JL performed the experiments. WY provided the plant resources. MZ, ZD, and FY analyzed the results and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was funded by the Sugarcane Research Foundation of Guangxi University (No. 2022GZB006), an independent fund of Guangxi Key Laboratory of sugarcane biology (GXKLSCB-20190201) and supported by the China Agriculture Research System of MOF and MARA (No. CARS-17-1-04, CARS-17- 05).
This is a short text to acknowledge the contributions of specific colleagues, institutions, or agencies that aided the efforts of the authors.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aitken, K., Li, J.-C., Jackson, P., Piperidis, G., McIntyre, C. (2006). AFLP analysis of genetic diversity within Saccharum officinarum and comparison with sugarcane cultivars. Aust. J. Agric. Res. 57, 1167–1184. doi: 10.1071/AR05391
Barnes, J., Bester, A. (2000). Genetic mapping in sugarcane: prospects and progress in the South African sugar industry. Proc. Proc. S Afr Sugar Tech Assoc. 74.
Beliveau, B. J., Joyce, E. F., Apostolopoulos, N., Yilmaz, F., Fonseka, C. Y., McCole, R. B., et al. (2012). Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc. Natl. Acad. Sci. 109, 21301–21306. doi: 10.1073/pnas.1213818110
Boideau, F., Richard, G., Coriton, O., Huteau, V., Belser, C., Deniot, G., et al. (2022). Epigenomic and structural events preclude recombination in Brassica napus. New Phytologist. 234, 545–559. doi: 10.1111/nph.18004
Braz, G. T., He, L., Zhao, H., Zhang, T., Semrau, K., Rouillard, J.-M., et al. (2018). Comparative oligo-FISH mapping: an efficient and powerful methodology to reveal karyotypic and chromosomal evolution. Genetics. 208, 513–523. doi: 10.1534/genetics.117.300344
Braz, G. T., Yu, F., Vale Martins, Ld., Jiang, J. (2020). “Fluorescent in situ hybridization using oligonucleotide-based probes,” in In situ hybridization protocols (New York, NY: Springer), 71–83.
Braz, G. T., Yu, F., Zhao, H., Deng, Z., Birchler, J. A., Jiang, J. (2021). Preferential meiotic chromosome pairing among homologous chromosomes with cryptic sequence variation in tetraploid maize. New Phytologist. 229, 3294–3302. doi: 10.1111/nph.17098
Bremer, G. (1961). Problems in breeding and cytology of sugar cane. Euphytica. 10, 59–78. doi: 10.1007/BF00037206
Chen, Z. J. (2007). Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol 58, 377–406. doi: 10.1146/annurev.arplant.58.032806.103835
Cuadrado, A., Acevedo, R., Moreno Díaz de la Espina, S., Jouve, N., de la Torre, C. (2004). Genome remodelling in three modern S. officinarum× S. spontaneum sugarcane cultivars. J. Exp. Botany. 55, 847–854. doi: 10.1093/jxb/erh093
De Morais, L., Aguiar, M., Albuquerque e Silva, Pd., Câmara, T. M. M., Cursi, D. E., Júnior, A. R. F., et al. (2015). “Breeding of sugarcane,” in Industrial crops (New York: Springer), 29–42.
D'Hont, A., Grivet, L., Feldmann, P., Glaszmann, J., Rao, S., Berding, N. (1996). Characterisation of the double genome structure of modern sugarcane cultivars (Saccharum spp. ) by molecular cytogenetics. Mol. Gen. Genet. MGG. 250, 405–413. doi: 10.1007/BF02174028
D'Hont, A., Ison, D., Alix, K., Roux, C. (1998). Determination of basic chromosome numbers in the genus Saccharum by physical mapping of ribosomal RNA genes. Genome 41, 221–225. doi: 10.1139/g98-023
de Oliveira Bustamante, F., do Nascimento, T. H., Montenegro, C., Dias, S., do Vale Martins, L., Braz, G. T., et al. (2021). Oligo-FISH barcode in beans: A new chromosome identification system. Theor. Appl. Genet. 134, 3675–3686. doi: 10.1007/s00122-021-03921-z
Edmé, S., Glynn, N., Comstock, J. (2006). Genetic segregation of microsatellite markers in Saccharum officinarum and S. spontaneum. Heredity. 97, 366–375. doi: 10.1038/sj.hdy.6800888
Grivet, L., Arruda, P. (2002). Sugarcane genomics: depicting the complex genome of an important tropical crop. Curr. Opin. Plant Biol. 5, 122–127. doi: 10.1016/S1369-5266(02)00234-0
Grivet, L., D'Hont, A., Roques, D., Feldmann, P., Lanaud, C., Glaszmann, J. C. (1996). RFLP mapping in cultivated sugarcane (Saccharum spp. ): genome organization in a highly polyploid and aneuploid interspecific hybrid. Genetics. 142, 987–1000. doi: 10.1093/genetics/142.3.987
Han, Y., Zhang, T., Thammapichai, P., Weng, Y., Jiang, J. (2015). Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics. 200, 771–779. doi: 10.1534/genetics.115.177642
Heinz, D. J., Mee, G. W., Nickell, L. (1969). Chromosome numbers of some Saccharum species hybrids and their cell suspension cultures. Am. J. Botany. 56, 450–456. doi: 10.1002/j.1537-2197.1969.tb07556.x
Hu, L., Ru, K., Zhang, L., Huang, Y., Zhu, X., Liu, H., et al. (2014). Fluorescence in situ hybridization (FISH): an increasingly demanded tool for biomarker research and personalized medicine. biomark. Res. 2, 1–13. doi: 10.1186/2050-7771-2-3
Huang, Y., Chen, H., Han, J., Zhang, Y., Ma, S., Yu, G., et al. (2020). Species-specific abundant retrotransposons elucidate the genomic composition of modern sugarcane cultivars. Chromosoma. 129, 45–55. doi: 10.1007/s00412-019-00729-1
Irvine, J. E. (1999). Saccharum species as horticultural classes. Theor. Appl. Genet. 98, 186–194. doi: 10.1007/s001220051057
Jackson, P. A. (2005). Breeding for improved sugar content in sugarcane. Field Crops Res. 92, 277–290. doi: 10.1016/j.fcr.2005.01.024
Jiang, J. (2019). Fluorescence in situ hybridization in plants: recent developments and future applications. Chromosome Res. 27, 153–165. doi: 10.1007/s10577-019-09607-z
Jiang, J., Gill, B. S. (2006). Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome. 49, 1057–1068. doi: 10.1139/g06-076
Lalitha, R., Premachandran, M. N. (2007). Meiotic Abnormalities in Intergeneric Hybrids between Saccharum spontaneumand Erianthus arundinaceus (Gramineae). Cytologia. 72, 337–343. doi: 10.1508/cytologia.72.337
Lu, Y., D'Hont, A., Paulet, F., Grivet, L., Arnaud, M., Glaszmann, J. (1994). Molecular diversity and genome structure in modern sugarcane varieties. Euphytica. 78, 217–226. doi: 10.1007/BF00027520
Meng, Z., Han, J., Lin, Y., Zhao, Y., Lin, Q., Ma, X., et al. (2020). Characterization of a Saccharum spontaneum with a basic chromosome number of x= 10 provides new insights on genome evolution in genus Saccharum. Theor. Appl. Genet. 133, 187–199. doi: 10.1007/s00122-019-03450-w
Meng, Z., Wang, F., Xie, Q., Li, R., Shen, H., Li, H. (2022). Reconstruction of karyotypic evolution in Saccharum spontaneum species by comparative oligo-FISH mapping. BMC Plant Biol. 22, 599. doi: 10.1186/s12870-022-04008-7
Ming, R., Moore, P. H., Wu, K. K., D’Hont, A., Glaszmann, J. C., Tew, T. L., et al. (2010). Sugarcane improvement through breeding and biotechnology. Plant Breed. Rev. 27, 15–118. doi: 10.1002/9780470650349.ch2
Osborn, T. C., Chris Pires, J., Birchler, J. A., Auger, D. L., Jeffery Chen, Z., Lee, H.-S., et al. (2003). Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19, 141–147. doi: 10.1016/S0168-9525(03)00015-5
Panje, R. R., Babu, C. N. (1960). Studies in Saccharum spontaneum distribution and geographical association of chromosome numbers. Cytologia 25, 152–172. doi: 10.1508/cytologia.25.152
Paterson, A. H., Moore, P. H., Tew, T. L. (2013). “The gene pool of Saccharum species and their improvement,” in Genomics of the saccharinae (New York, NY: Springer), 43–71.
Piperidis, N., D’Hont, A. (2020). Sugarcane genome architecture decrypted with chromosome-specific oligo probes. Plant J. 103, 2039–2051. doi: 10.1111/tpj.14881
Piperidis, G., Piperidis, N., D’Hont, A. (2010). Molecular cytogenetic investigation of chromosome composition and transmission in sugarcane. Mol. Genet. Genomics 284, 65–73. doi: 10.1007/s00438-010-0546-3
Price, S. (1957). Cytological studies in Saccharum and allied genera. III. Chromosome numbers in interspecific hybrids. Botanical Gazette. 118, 146–159. doi: 10.1086/335939
Racedo, J., Gutiérrez, L., Perera, M. F., Ostengo, S., Pardo, E. M., Cuenya, M. I., et al. (2016). Genome-wide association mapping of quantitative traits in a breeding population of sugarcane. BMC Plant Biol. 16, 1–16. doi: 10.1186/s12870-016-0829-x
Schmutz, J., McClean, P. E., Mamidi, S., Wu, G. A., Cannon, S. B., Grimwood, J., et al. (2014). A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 46, 707–713. doi: 10.1038/ng.3008
Schwarzacher, T., Leitch, A. R. (1994). “Enzymatic treatment of plant material to spread chromosomes for in situ hybridization,” in Protocols for nucleic acid analysis by nonradioactive probes (Springer), 153–160.
Xiong, Z., Pires, J. C. (2011). Karyotype and identification of all homoeologous chromosomes of allopolyploid Brassica napus and its diploid progenitors. Genetics. 187, 37–49. doi: 10.1534/genetics.110.122473
Younis, A., Ramzan, F., Hwang, Y.-J., Lim, K.-B. (2015). FISH and GISH: molecular cytogenetic tools and their applications in ornamental plants. Plant Cell Rep. 34, 1477–1488. doi: 10.1007/s00299-015-1828-3
Yu, F., Wang, P., Li, X., Huang, Y., Wang, Q., Luo, L., et al. (2018). Characterization of chromosome composition of sugarcane in nobilization by using genomic in situ hybridization. Mol. Cytogene. 11, 1–8. doi: 10.1186/s13039-018-0387-z
Yu, F., Zhao, X., Chai, J., Ding, X., Li, X., Huang, Y., et al. (2022). Chromosome-specific painting unveils chromosomal fusions and distinct allopolyploid species in the Saccharum complex. New Phytol. 233, 1953–1965. doi: 10.1111/nph.17905
Zhang, J., Zhang, X., Tang, H., Zhang, Q., Hua, X., Ma, X., et al. (2018). Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat. Genet. 50, 1565–1573. doi: 10.1038/s41588-018-0237-2
Keywords: sugarcane, interspecific hybridization, nobilization, chromosomal inheritance, oligo-FISH
Citation: Chai J, Xue L, Lei J, Yao W, Zhang M, Deng Z and Yu F (2023) All nonhomologous chromosomes and rearrangements in Saccharum officinarum × Saccharum spontaneum allopolyploids identified by oligo-based painting. Front. Plant Sci. 14:1176914. doi: 10.3389/fpls.2023.1176914
Received: 06 April 2023; Accepted: 01 September 2023;
Published: 06 October 2023.
Edited by:
Jian Zeng, Sichuan Agricultural University, ChinaReviewed by:
Wellington Ronildo Clarindo, Universidade Federal de Viçosa, BrazilCopyright © 2023 Chai, Xue, Lei, Yao, Zhang, Deng and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zuhu Deng, ZGVuZ3p1aHVAMTYzLmNvbQ==; Fan Yu, eXVmYW5reUAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.