
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 31 March 2023
Sec. Plant Bioinformatics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1174955
This article is part of the Research Topic Bioinformatics, Big Data and Agriculture: A Challenge for the Future View all 6 articles
A correction has been applied to this article in:
Corrigendum: Comprehensive evolutionary analysis of growth-regulating factor gene family revealing the potential molecular basis under multiple hormonal stress in Gramineae crops
 Wei Wang1,2†
Wei Wang1,2† Mingxing Cheng1,2†
Mingxing Cheng1,2† Xiao Wei1,2
Xiao Wei1,2 Ruihua Wang1,2
Ruihua Wang1,2 Fengfeng Fan1,2
Fengfeng Fan1,2 Zhikai Wang3
Zhikai Wang3 Zhihong Tian3
Zhihong Tian3 Shaoqing Li1,2*
Shaoqing Li1,2* Huanran Yuan1,2*
Huanran Yuan1,2*Growth-regulating factors (GRFs) are plant-specific transcription factors that contain two highly conserved QLQ and WRC domains, which control a range of biological functions, including leaf growth, floral organ development, and phytohormone signaling. However, knowledge of the evolutionary patterns and driving forces of GRFs in Gramineae crops is limited and poorly characterized. In this study, a total of 96 GRFs were identified from eight crops of Brachypodium distachyon, Hordeum vulgare, Oryza sativa L. ssp. indica, Oryza rufipogon, Oryza sativa L. ssp. japonica, Setaria italic, Sorghum bicolor and Zea mays. Based on their protein sequences, the GRFs were classified into three groups. Evolutionary analysis indicated that the whole-genome or segmental duplication plays an essential role in the GRFs expansion, and the GRFs were negatively selected during the evolution of Gramineae crops. The GRFs protein function as transcriptional activators with distinctive structural motifs in different groups. In addition, the expression of GRFs was induced under multiple hormonal stress, including IAA, BR, GA3, 6BA, ABA, and MeJ treatments. Specifically, OjGRF11 was significantly induced by IAA at 6 h after phytohormone treatment. Transgenic experiments showed that roots overexpressing OjGRF11 were more sensitive to IAA and affect root elongation. This study will broaden our insights into the origin and evolution of the GRF family in Gramineae crops and will facilitate further research on GRF function.
Growth-regulating factors (GRFs) are plant-specific transcription factors that are widely distributed in the plant kingdom. OsGRF1 was the first identified member of the GRF family in plants and is known to play a crucial role in early stem elongation (Knaap and Kende, 2000). Since its discovery, GRF proteins have been gradually identified in other plant species, such as Arabidopsis thaliana, Brassica napus, Glycine max, and Solanum tuberosum (Greco et al., 2012; Kim et al., 2012; Omidbakhshfard et al., 2015; Zhang et al., 2018). GRF proteins typically range in size from 20 to 60 kD and commonly contain two highly conserved domains: QLQ and WRC (Vercruysse et al., 2020). However, the composition and number of amino acid residues in the C-terminal region vary greatly (Kim and Tsukaya, 2015). The QLQ domain, consisting of 36 amino acid residues with highly conserved Gln-Leu-Gln (QX3LX2Q) residues, is a crucial motif for protein-protein interactions (Knaap and Kende, 2000). In contrast, the WRC domain is typically associated with DNA binding and contains a putative nuclear localization signal (Omidbakhshfard et al., 2015). This domain consists of 44 amino acid residues and is characterized by a Trp-Ar-Cys motif and a conserved distance between three cysteine residues and one histidine residue (CX9CX10CX2H; C3H motif) (Kim and Tsukaya, 2015).
Transcription factors are essential regulators of gene expression, and rice possesses approximately 2147 transcription factors (Jin et al., 2017). Among them, GRFs are major transcription factors in plants that play a crucial role in various developmental processes, including regulating the growth of plant roots, leaves, and floral organ GRF9, a negative regulator of leaf growth, reduces leaf development when overexpressed, whereas grf9 mutants produce larger leaf primordia, rosette leaves, and petals. GRF9 activates the expression of LEUCINE-ZIPPER (bZIP) type transcription factor OBP3-RESPONSIVE GENE3 (ORG3) to regulate leaf growth. In flowers, APETALA1 (AP1) and SEPALLATA3 (SEP3) are the main controlling factors. (Kaufmann et al., 2009). They interact specifically with members of the GRF family to regulate floral organ development (Pajoro et al., 2014). AtGRF2 and AtGRF5 are highly expressed in floral organ meristem and primary primordium, while AtGRF8 is expressed in floral organ differentiation (Pajoro et al., 2014). In maize, overexpression of ZmGRF10 reduced leaf size and plant height by significantly decreasing the expression of PIF4 (phytochrome interaction factor 4), TATA-binding protein-related factor 5 (TAF5) and REVISRADIX (BRX) genes (Wu et al., 2014).
In rice, the GRFs are regulated by OsmiR396, including OsGRF4, OsGRF6, and OsGRF10, which play important roles in regulating grain size, grain yield, and inflorescence development (Liu et al., 2014; Che et al., 2015; Duan et al., 2015; Gao et al., 2015; Hu et al., 2015; Li et al., 2016; Tang et al., 2018). Down-regulation of target OsGRFs by 35S: osa-miR396 leads to a dehiscent outer shell of the floret, elongated and sterile lemma, abnormal number of anthers, and stigma in progeny plants (Liu et al., 2014). In OEmiR396b plants, miR396 inhibits the expression of OsGRF6 by cleaving its transcripts, leading to shortened branches, clusters, and decreased yield (Gao et al., 2015). OsGRF4 is regulated by miR396c, and the mutation of OsGRF4 suppresses the regulation of the OsmiR396 target. Additionally, the direct interaction between OsGRF4 and OsGIF1 results in larger grains and improved yield of rice (Li et al., 2016).
One of the most important regulators of plant growth and development, GRFs have been reported in several cereal crops, including rice, wheat, and corn (Zhang et al., 2008; Greco et al., 2012; Kim et al., 2012; Fİlİz et al., 2014; Omidbakhshfard et al., 2015). However, a systematic understanding of the evolution of the GRF family in Gramineae crops is still lacking. In this study, the GRFs in the Gramineae crops of Brachypodium distachyon, Hordeum vulgare, Oryza sativa L. ssp. indica, Oryza rufipogon, Oryza sativa L. ssp. japonica, Setaria italic, Sorghum bicolor and Zea mays were identified, and the exon-intron structure, chromosomal distribution and the evolution of GRFs were systematically characterized. This study will help to deepen our understanding of the function and evolutionary relationship of GRFs in the Gramineae crops.
A total of eight genomes of Gramineae crops, including Brachypodium distachyon, Hordeum vulgare, Oryza sativa L. ssp. indica, Oryza rufipogon, Oryza sativa L. ssp. japonica, Setaria italic, Sorghum bicolor and Zea mays, were downloaded from Ensembl Plants release-41 (http://plants.ensembl.org/index.html) and Phytozome v.12 (https://phytozome-next.jgi.doe.gov/) (Goodstein et al., 2012; Bolser et al., 2016). To identify GRFs in the eight Gramineae crops, PF08879 and PF08880 were downloaded from InterPro (www.ebi.ac.uk/interpro). Then all the GRF candidate genes were separately identified by HMMER 3.2.1 and BLASTP (Camacho et al., 2009; Finn et al., 2011). Finally, the sequences were further verified by using InterPro (www.ebi.ac.uk/interpro) and SMART (http://smart.embl-heidelberg.de) (Malik et al., 2020; Cheng et al., 2021).
To identify the exon/intron of the conserved GRFs protein, we extracted eight genomes annotation GFF files from Ensembl Plants website (Bolser et al., 2016). The software MEME Suite v.5.3.3 was used to identify conserved motifs with a maximum number of 20, the phylogenetic tree was constructed by MEGA and then visualized and managed by EvolView (He et al., 2016), motif arrangement and the exon/intron structures were shown by TBtools (Chen et al., 2020a).
The full-length amino acid sequence of the GRFs from the eight plant genomes were aligned with the ClustalW program (version 2.0) and constructed the neighbor-joining (NJ) tree with 1000 bootstrap replicates using the Poisson substitution model in MEGA 7.0 (Edgar and Sjolander, 2004; Kumar et al., 2016).
Synteny relationship between duplicated gene pairs from eight Gramineae crops genomes was analyzed by using MCScanX software and diagrammatical results were visualized by using simple Circos-0.69 software (http://circos.ca/) (Krzywinski et al., 2009; Wang et al., 2012).
The orthogroup was identified using OrthoFinder v.2.5.4 with a cut-off e-value of 1×10−3 (Emms and Kelly, 2019). Then, STAG and STRID algorithms were used to rebuild the phylogenetic tree of the selected species based on the detected orthogroup (Emms and Kelly, 2019).
To investigate the collinearity and analyze the syntenic relationship of GRFs in eight Gramineae crops, the whole genome sequences and corresponding genome annotation files GRFs were analyzed by the MCScanX tool (Wang et al., 2012). For the calculation of the selection pressure of GRFs among eight species, synonymous (Ks) and non-synonymous (Ka) substitution rates and their ratios were estimated using the Ka/Ks calculator in TBtools (Cheng et al., 2021). And the divergence time (T) was estimated by T = Ks/(2ⅹ9.1ⅹ10−9)ⅹ10−6 million years ago (MYa) (Cheng et al., 2021).
According to the DNA sequence 2000 bp upstream of the GRFs in japonica rice, the cis-acting elements were predicted from the PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) (Rombauts et al., 1999) and categorized based on their annotated functions. Furthermore, the other miRNAs except for miR396 potentially target to GRFs were predicted using the website psRNATarget (https://www.zhaolab.org/psRNATarget/home).
The rice cultivar Zhong-Hua-11 (ZH11, Oryza sativa L. ssp. japonica) was used to analysis the expression of GRFs. The rice plants were soaked in 100 µM hormone (IAA, BR, GA3, 6BA, ABA, and MeJ) at the three-leaf-one-heart stage for various time points (0 h, 6 h, 9 h, 12 h, 24 h). All the seedlings were cultured at 28 °C for 12 days with 14-h light/10-h dark and 60% relative humidity (Cheng et al., 2021).
All RNAs of samples were extracted using TRIzol Reagent according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA), And the first-strand cDNA was synthesized from RNA treated with a 2 µg DNA enzyme using HiScript III First Strand cDNA Synthesis Kit (+gDNA Wiper) (Vazyme, Nanjing, China). The RT-qPCR analysis of GRFs in rice was performed by using the HieffqPCR SYBR Green Master Mix (No Rox) (Yeasen, Shanghai, China), and the relative expression level of the 12 GRFs was analyzed with the 2−ΔΔCT method. GRFs specific primers used for RT-qPCR analysis were listed in Supplementary Table 8.
The open reading frames (ORFs) of five GRFs in ZH11 (Oryza sativa L. ssp. japonica) were amplified from their cDNA and inserted into the modified HBT-eGFP vector to produce 35S: GRF: GFP fusion vector. The structure is driven by the CaMV 35S promoter. The recombinant plasmids were transformed into DH5α chemically competent cells (Weidi Biotechnology, Shanghai). The recombinant competent cells were transinfected into rice protoplasts. The empty HBT vector was used as a control, and all the protoplasts were cultured at 28 °C overnight. Then the protoplasts were observed for GFP signals by using the FV1000 confocal system. The GRFs specific primers for subcellular localization were listed in Supplementary Table 9.
To investigate the transcriptional activity of GRFs, the full-length coding sequences of GRFs were amplificated into pGBKT7, pGADT7, and GAL4DB vectors. The recombinant plasmids pGBKT7 and pGADT7 were transformed into yeast strain AH109 chemically competent cells (Weidi Biotechnology, Shanghai). The chemically competent cells were cultured on the corresponding defective medium for 3-5 days for observation. Then GRF-GAL4DB fusion proteins were transiently expressed in rice protoplasts to measure transcriptional activity by using the dual-luciferase reporter assay system (Promega, Madison, WI, USA) (Ren et al., 2016). The empty GAL4DB vector was used as a control. The specific primers used are listed in Supplementarys Table 10, 11.
The coding genes for GRF transcription factors were identified in Brachypodium distachyon, Hordeum vulgare, Oryza sativa L. ssp. indica, Oryza rufipogon, Oryza sativa L. ssp. japonica, Setaria italic, Sorghum bicolor and Zea mays by searching for the WRC domain (Pfam accession no. PF08879, InterPro accession no. IPR014977) and QLQ domain (Pfam accession no. PF008880, InterPro accession no. IPR014978) using Pfam (Mistry et al., 2021). A total of 96 GRFs were detected, with 12, 13, 13, 12, 12, 8, 10, and 16 GRFs identified in Brachypodium distachyon, Hordeum vulgare, Oryza sativa L. ssp. indica, Oryza rufipogon, Oryza sativa L. ssp. japonica, Setaria italic, Sorghum bicolor and Zea mays respectively (Supplementary Table 1). The number of GRFs is relatively stable among these eight Gramineae crops.
To understand the phylogenetic relationship of the GRFs among Gramineae crops, the phylogenetic tree was constructed and classified the identified GRF protein sequences into three subfamilies: Group 1, Group 2, and Group 3 (Figure 1). All subgroups showed a clear expansion of gene numbers, with Group2 having the highest number of gene members, followed by Group1 and Group3 (Figure 1 and Supplementary Table 2). In Setaria italic, Sorghum bicolor and Zea mays Group1 had fewer members (Figure 1, Supplementary Table 2). To better understand the evolutionary pattern of GRFs in Gramineae crops, we performed orthogroup (OG) clustering using OrthoFinder software 9, which divided the GRFs into five orthogroups: OG00, OG01, OG02, OG03, and OG04 (Figure 1). The number of genes varied greatly among the orthogroups, ranging from 3 to 42 (Figure 1, Supplementarys Table 3, 4). Notably, the OG04 contained only three GRFs from Hordeum vulgare, indicating unequal loss and expansion of GRFs among orthogroups during the evolution of Gramineae crops.
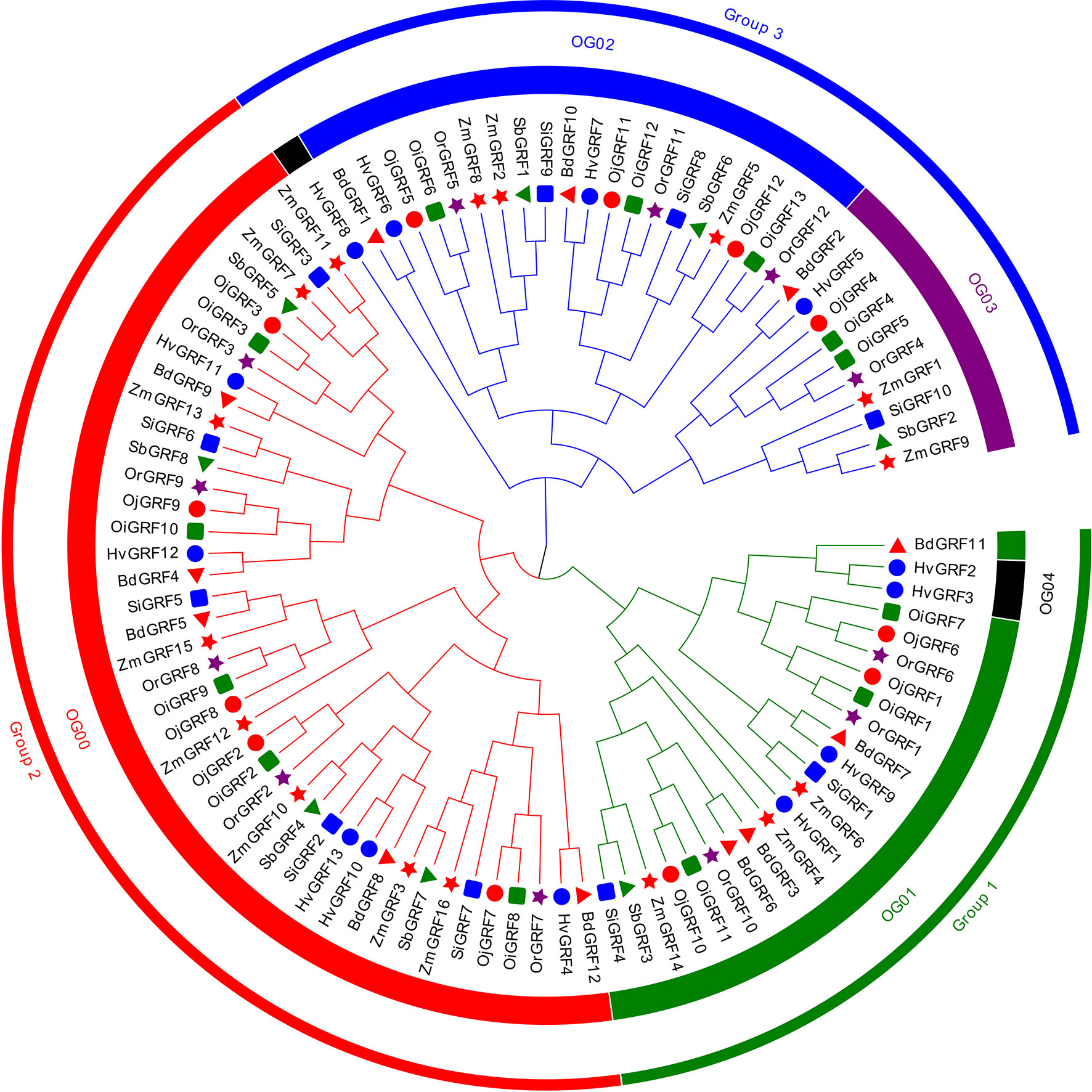
Figure 1 The neighbor-joining (NJ) phylogeny tree of the GRFs based on protein sequences from eight Gramineae crops. The colors of the outside circles represent different subfamilies, including Group1, Group2, and Group3. The colors of the inside circles represent different orthogroups (OGs). The different colors and shapes indicate the different species, the red triangle means Brachypodium distachyon, blue filled circles denote Hordeum vulgare, green squares represent Oryza sativa L. ssp. indica, red filled circles show Oryza sativa L. ssp. japonica, brown pentagon stand for Oryza rufipogon, blue squares mean Sorghum bicolor, the green triangle represents Sorghum bicolor, red pentagon denotes Zea mays.
To gain insight into the expansion mechanism of GRFs among the eight Gramineae crops, we further analyzed the chromosomal distribution and duplication modes of the GRFs. The GRFs were found to be mainly distributed on 3-4 chromosomes in each species (Figure 2), with chromosome 2 having the largest proportion, while chromosome 8 had only one GRF (Supplementary Table 1). In Brachypodium distachyon, nearly half of the GRFs were located on chromosome 1 (Figure 2), whereas in Oryza sativa L. ssp. japonica, the GRFs were almost evenly distributed on chromosomes 2, 3, 4, 6, 7, 11, and 12 (Figure 2 and Supplementary Table 1). Additionally, a total of 33 duplicated gene pairs were identified in the eight species (Table 1), which were generated from either segmental or tandem duplication. However, tandem duplications were found only in Hordeum vulgare and Oryza sativa L. ssp. indica (Table 1). Importantly, the GRF number and the duplication mode were the same between Oryza sativa L. ssp. Japonica and Oryza rufipogon, indicating a close evolutionary relationship and supporting the notion that Oryza rufipogon is the direct ancestor of Oryza sativa (Londo et al., 2006).
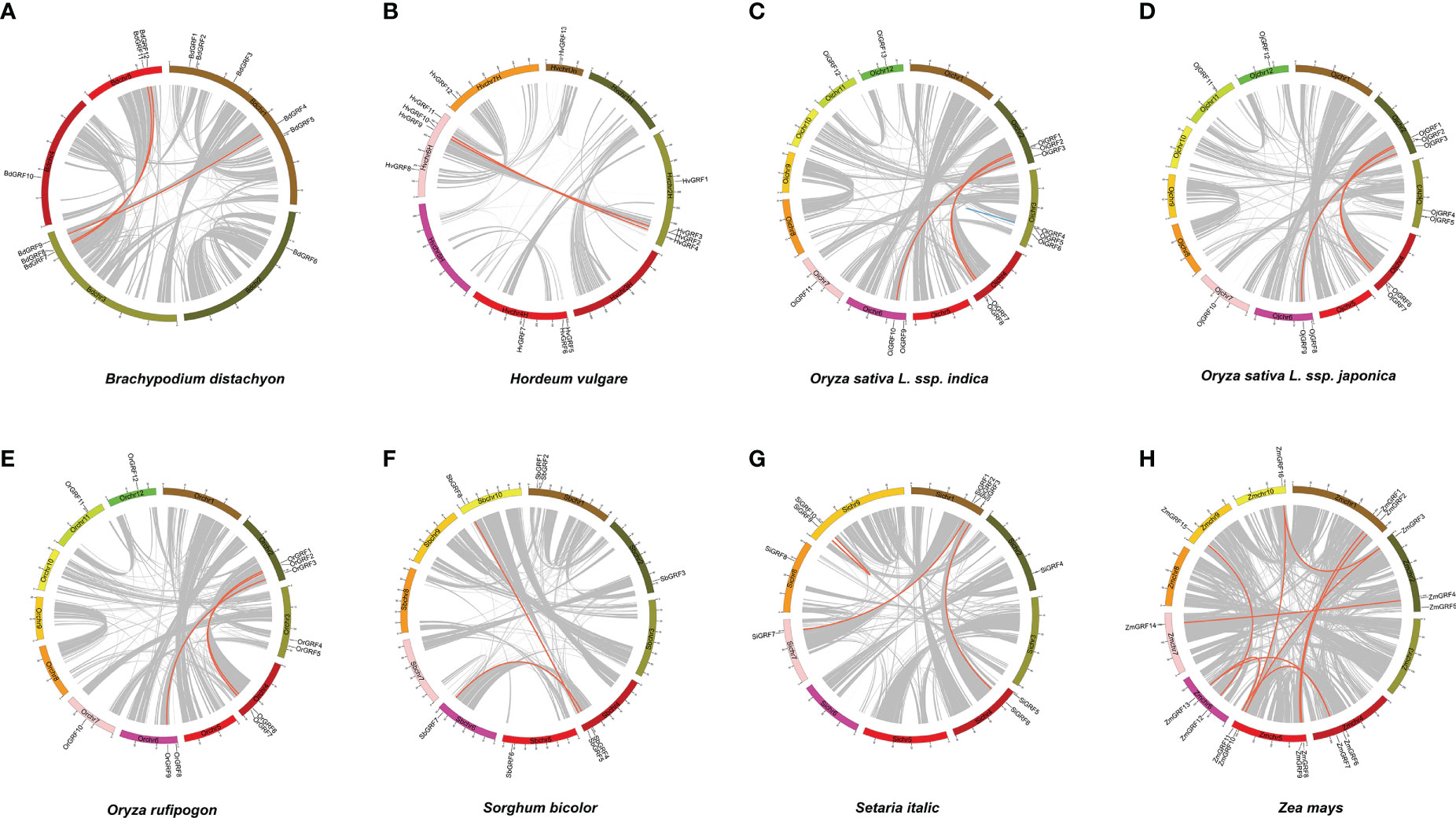
Figure 2 The chromosome location and duplication events of GRFs in eight Gramineae crops. The position of each GRF gene was marked using Circos software, and whole-genome duplication (WGD) or fragment duplication/tandem duplication gene pairs were connected by red/blue lines. (A) Brachypodium distachyon. (B) Hordeum vulgare. (C) Oryza sativa L. ssp. indica. (D) Oryza rufipogon. (E) Oryza sativa L. ssp. japonica. (F) Setaria italic. (G) Sorghum bicolor. (H) Zea mays.
The investigation of divergence revealed a wide range of divergent times for the duplicated gene pairs, ranging from 0.23 to 122.24 Mya (Table 1), indicating significant variation in the origin of GRFs among Gramineae crops. For instance, the divergence time ranged from 25.68 to 122.24 Mya in Setaria italic, and 0.23 to 31.38 Mya in Oryza sativa L. ssp. indica. Ka and Ks analysis demonstrated that the Ka/Ks values were all less than 1 in the Gramineae crops (Table 1), suggesting that GRFs have been subjected to strong negative selection during the evolution of Gramineae crops.
To further elucidate the evolutionary relationship of the GRF family within the Gramineae crops, we analyzed the synteny of GRFs using the MCScanX toolkit. Reference with the genome of japonica rice Nipponbare, there were 17, 14, 18, 18, 15, 15 and 22 collinear gene pairs being detected in the genomes of Brachypodium distachyon, Hordeum vulgare, Oryza sativa L. ssp. indica, Oryza rufipogon, Setaria italic, Sorghum bicolor, and Zea mays (Figure 3). The collinearity of GRFs between the Oryza sativa L. ssp. japonica and Oryza sativa L. ssp. indica, Oryza rufipogon and Zea mays were closer than in the other species (Figure 3), consistent with their closer evolutionary ship. These results suggest the syntenic relationship of the GRFs is highly conserved among Gramineae crops.
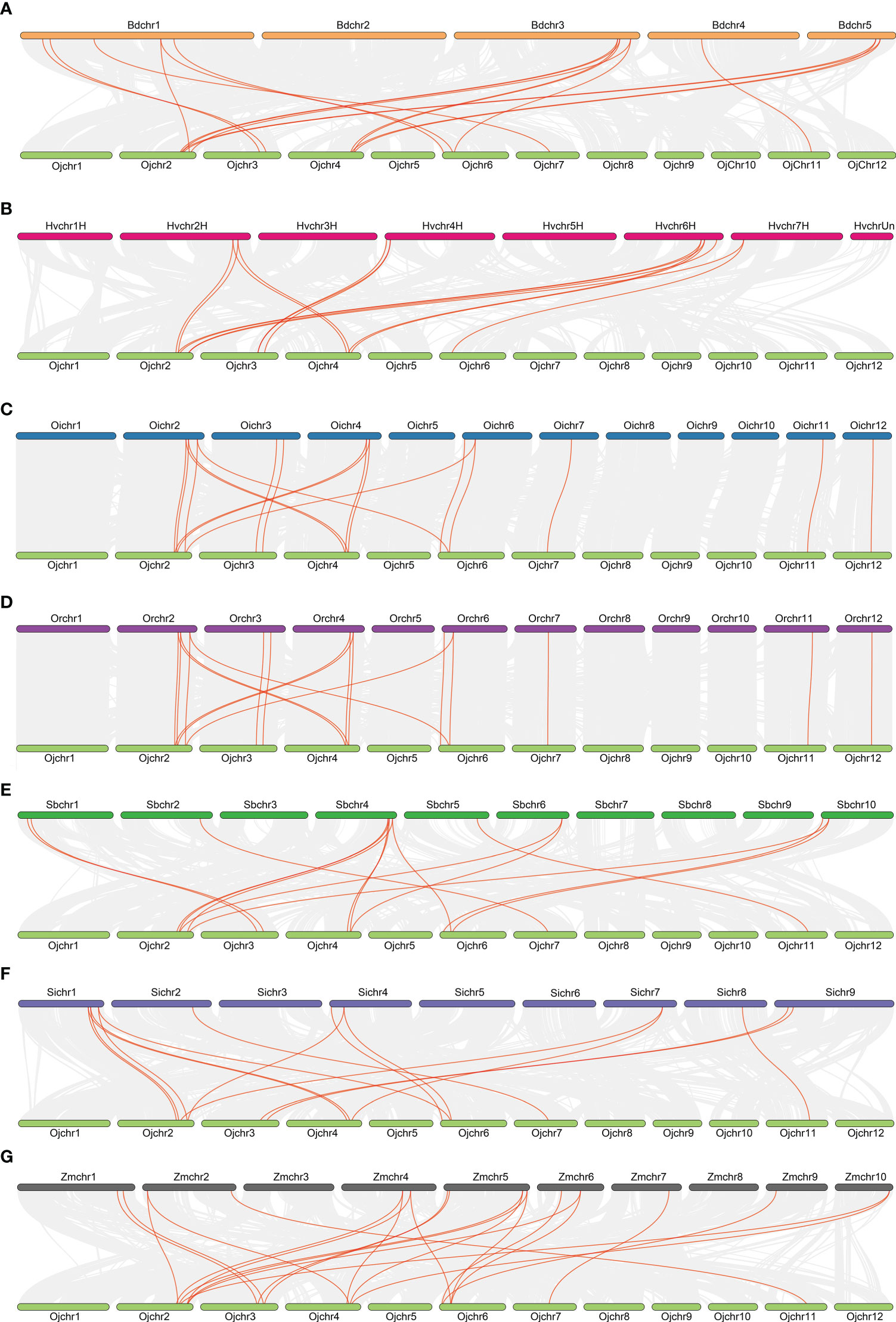
Figure 3 Collinearity relationships of GRFs in eight species or subspecies. Brachypodium distachyon (A), Hordeum vulgare (B), Oryza sativa L. ssp. indica, (C), Oryza sativa L. ssp. japonica (D), Sorghum bicolor (E), Setaria italic (F), Zea mays (G).
To elucidate the functional relationships of among members of the GRF family during evolution, the conserved protein motifs and structures of the 96 GRFs were analyzed. Results showed that the exon number of GRFs in Gramineae crops ranged from 2 to 5, except OiGRF6 had 12 exons (Figure 4C). Moreover, more than 70% of GRFs consisted of three or four exons in each species (Figures 4A, C).
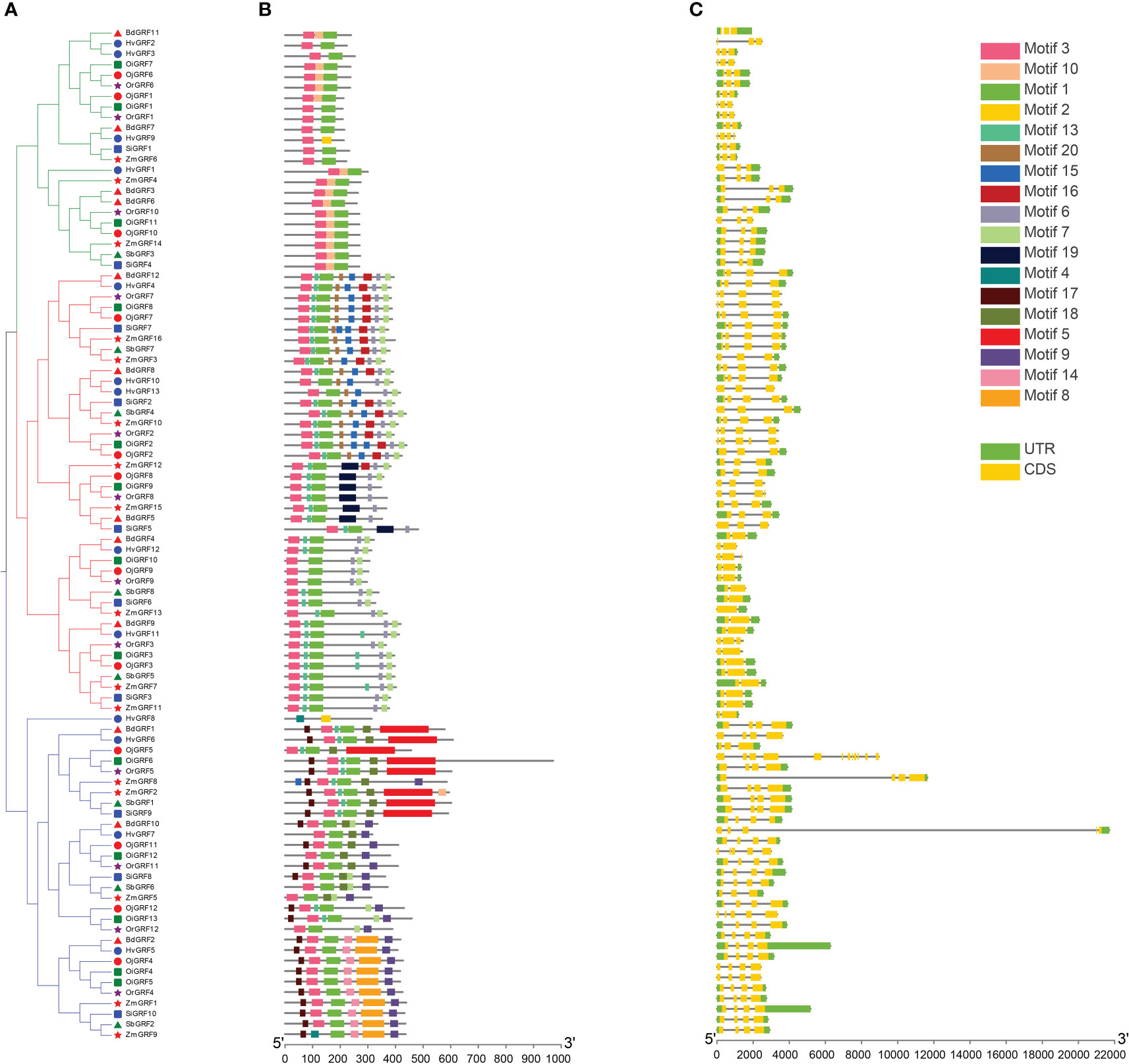
Figure 4 Phylogenetic relationships, gene structure, and the protein conserved motif of GRFs in eight Gramineae crops. (A) According to the NJ method in MEGA software, a phylogenetic tree of the GRF family from the eight Gramineae crops. (B) Distribution of the conserved motifs of the GRFs elucidated by TBtool Software. The different colored and numbered boxes represented different conserved motifs. (C) Exon-intron structures of GRF family. The green boxes represent exon and the gray line represents intron.
The secondary structure analysis showed the GRFs protein were mainly composed of Alpha helix, Extended strand, Beta turn, and Random coil (Supplementary Table 5). Specifically, Random coil accounted for 51.67~70.79%, followed by Alpha helix (13.76~33.46%), Extended strand (4.98~14.83%), and Beta turn (1.97~7.2%) (Supplementary Table 5). The three-dimensional structure analysis of the GRFs protein in rice with Alphafold2 (Jumper et al., 2021) showed that the GRFs protein were highly similar, with a simple arrangement lacking complex helical folded structures (Figure 5).
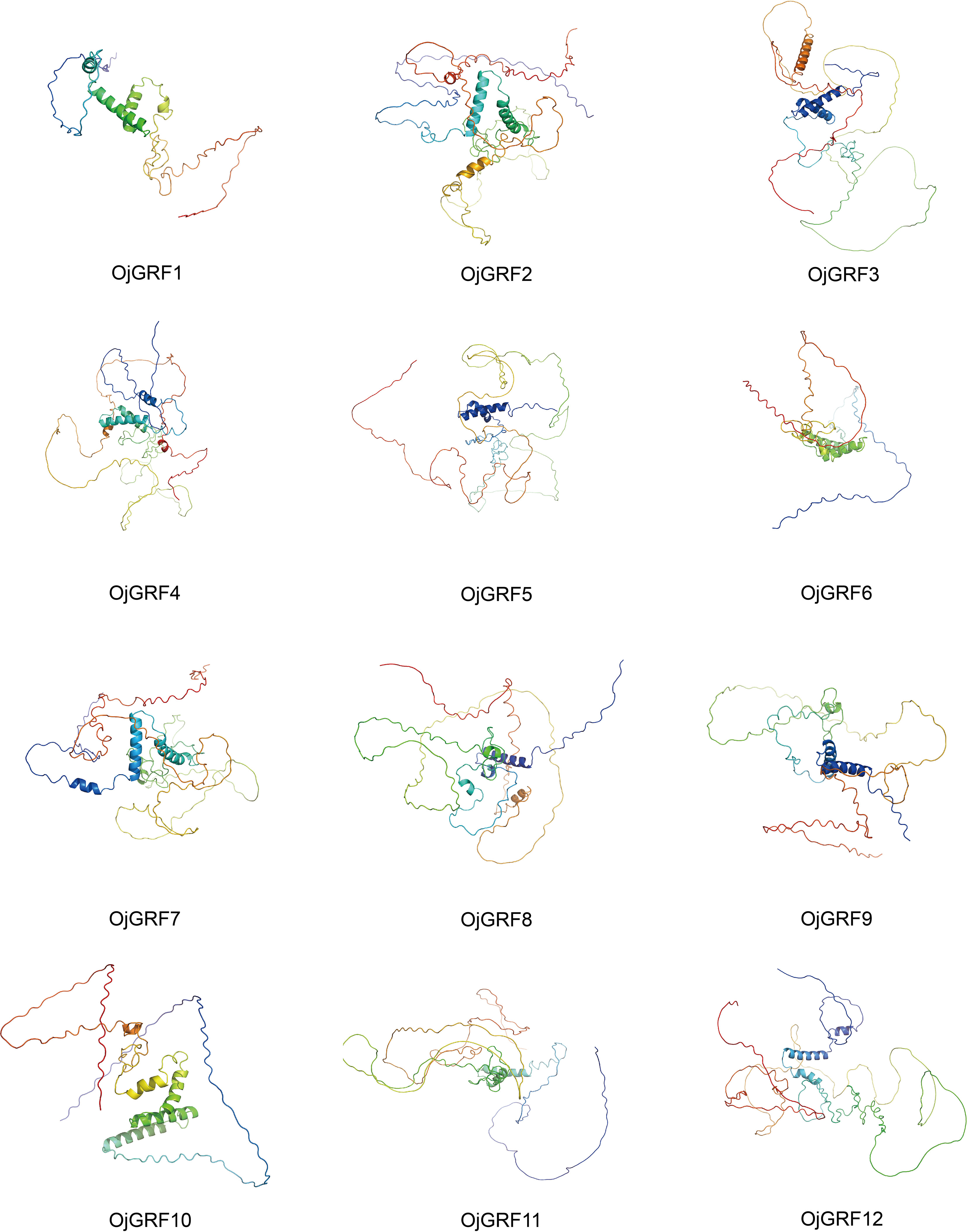
Figure 5 Prediction of the three-dimensional structure of GRFs protein in rice. The three-dimensional structure of 12 GRFs protein in rice was predicted by using the Alphafold2, and the results were output by PyMol software.
Functional motif analysis revealed that GRFs contain a total of 20 conserved motifs (Figure 4B). Most GRFs contain motif 1 and motif 3, and those with similar protein motif patterns were clustered into the same clade (Figures 4A, B). This is consistent with the phylogenetic analysis in Figure 1, indicating the sequence and functional conservation of GRFs within the same clade of Gramineae crops.
Transcription factors typically contain DNA binding domains (BD), transcriptional activation domains (AD), and nuclear location signal regions, which often form dimers to perform functions in the nucleus (Miura et al., 2010). As GRFs have been identified as important transcription factors in plants, it is of interest to determine whether all GRF proteins are localized in the nucleus. We predicted the 12 GRFs in japonica rice and found that all of them contained nuclear localization signals (Supplementary Table 6). Then, the GRFs were transiently expressed in the rice protoplast under driven by the CaMV 35s promoters, and found that the OjGRF1, OjGRF2, OjGRF3, OjGRF5 and OjGRF12 from japonica rice had green fluorescence signals in the nucleus (Figure 6), indicating that these GRFs are located in the nucleus.
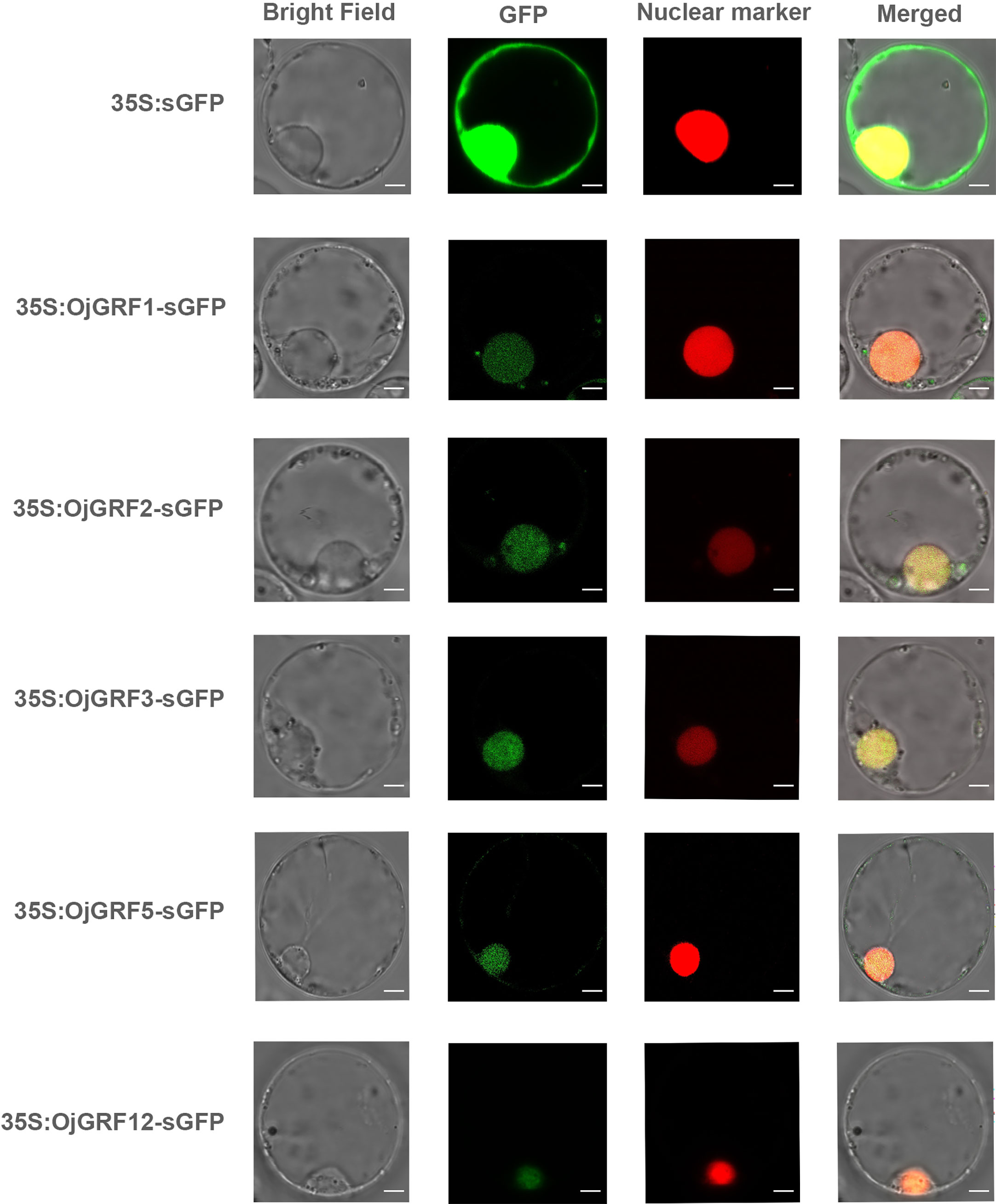
Figure 6 The subcellular localization of GRFs protein in rice protoplasts. The GRFs were fused to the GFP signal protein and the RFP signal represents the nucleus. The empty vector, HBT-sGFP, was used as a control. Scale bars correspond to 5 mm.
To investigate the transcriptional activity of GRFs, their coding regions were cloned into the pGBKT7 and pGADT7 vectors and tested by transforming yeast. Expression assay revealed that, except for OjGRF1, OjGRF4, OjGRF6 and OjGRF7 (Figure 7A), the other eight GRFs from japonica rice exhibited transcriptional self-activating activity (Figure 6A and Supplementary Figure 1). Moreover, nine GRFs protein from japonica rice were able to form dimers, except for OjGRF1, OjGRF6, and OjGRF10 (Figure 7B and Supplementary Figure 2). Transient expression assays with a dual-luciferase reporter in rice protoplasts showed that all the GRFs had transcriptional activity (Figures 7C, D). These results demonstrated that GRFs function as transcriptional activators.
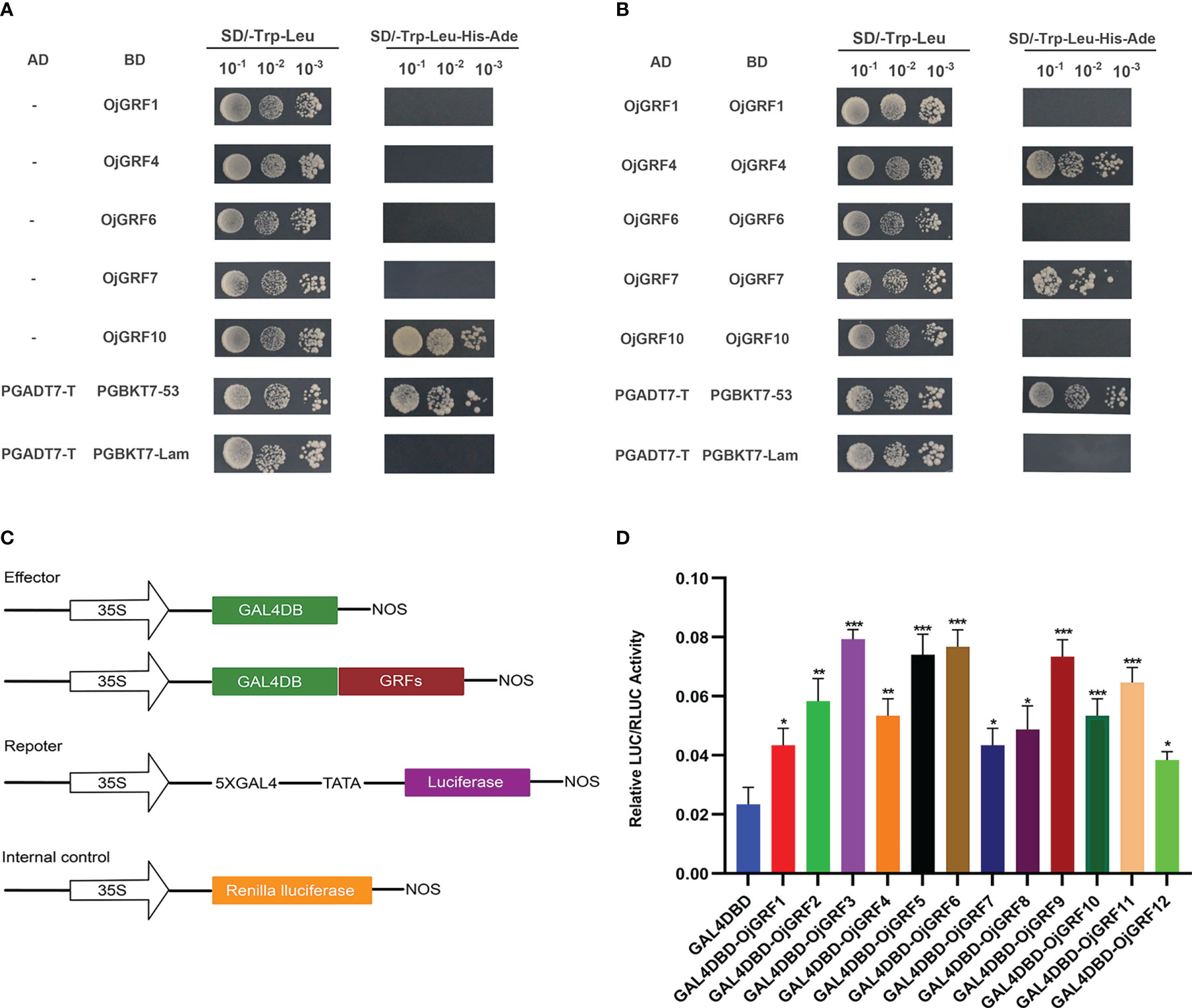
Figure 7 Transcriptional activity assay of GRFs. (A) Analysis of GRFs protein transcriptional self-activating activity in yeast; (B) Analysis of GRFs protein dimers in yeast; (C) The vector constructs are used in the dual-luciferase assay; (D) Dual-luciferase assay in rice protoplasts. The error bars show the standard deviations of the three independent biological replicates. Student’s t-test was used in this experiment; *; *p < 0.05, **p < 0.01, ***p < 0.001.
Cis-acting elements in the promoters usually play an important role in responding to diverse environments and regulating gene-specific expression (Wang et al., 2018). Thus, the potential cis-acting elements in the promoter regions 2,000 bp upstream from the RNA start site of GRFs were identified using the PlantCARE database (Lescot et al., 2002). A total of 41 types of cis-acting elements were detected in the GRF promoters of the eight Gramineae crops, which were mainly classified into three categories: growth and development, phytohormone response, and stress response (Figure 8A).
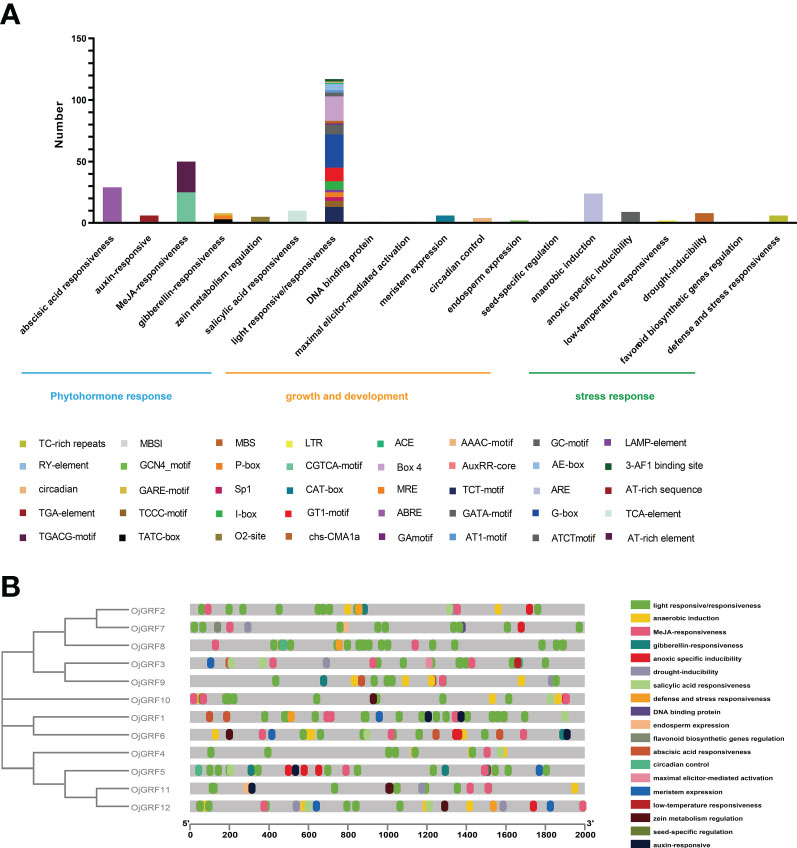
Figure 8 Identification of cis-acting elements in all GRFs of japonica rice. (A) Distribution of cis-acting elements in a different category. (B) Cis-acting elements of all GRFs in the phylogenetic tree. The differently colored boxes mean different promoter elements in each GRF gene.
The growth and development category contained 18 types of cis-acting elements including several elements involved in light responsiveness (3-AF1 binding site, AAAC-motif, ACE, AE-box, AT1-motif, ATCT-motif, Box 4, chs-CMA1a, GA-motif, GATA-motif, G-box, GT1-motif, I-box, LAMP-element, MRE, Sp1, TCCC-motif, TCT-motif). Among them, the G-box (23.1%) and Box 4 (17.1%) were the most abundant elements in this category, indicating that light responsiveness elements widespread distribution throughout the promoter region (Figure 8A). In the phytohormone response category, almost half of the predicted cis-acting elements were related to the MeJA-responsiveness, including CGTCA-motif and TGACG-motif. In addition, part of the elements related to abscisic acid responsiveness (ABRE), auxin-responsive (AuxRR-core and TGA-element), gibberellin-responsiveness (GARE-motif, P-box, and TATC-box), salicylic acid responsiveness (TCA-element), and zein metabolism were also observed (Figure 8A), implying that the GRFs play multiple roles in hormone response. Further analysis revealed that the cis-acting elements were unevenly distributed among the GRFs, and some cis-acting elements, such as GCN4_motif, MBSI, AT-rich sequence, and RY-element, were concentrated in a few GRFs (Figure 8B), reflecting the functional differentiation of the GRFs.
In plants, miRNAs play a crucial role in regulating various biological processes by binding to their target genes. The 12 GRFs in rice were predicted as the potential targets of 91 miRNAs. Among these, the miR167, miR1861, miR395, miR399 and miR395, except miR396, were all predicted targeting to GRFs (Supplementary Table 7), reflecting the regulatory complexity of GRFs at RNA level.
To better understand the potential function of GRFs in rice, we analyzed public rice transcriptomic data (http://expression.ic4r.org/search) from different tissues, including the aleurone, antler, callus, leaf, panicle, pistil, root, seed and shoot, and found that the 12 GRFs ubiquitously expressed in rice (Figure 9), with particularly high expression in young inflorescence, implying that GRFs play an important role in inflorescence development (Figure 9). Additionally, OjGRF4, OjGRF5, and OjGRF7 were highly expressed in leaf, anther, and callus, respectively, while OjGRF10 was highly expressed throughout the growth period of rice (Figure 9), indicating the functional differentiation of GRFs in rice development.
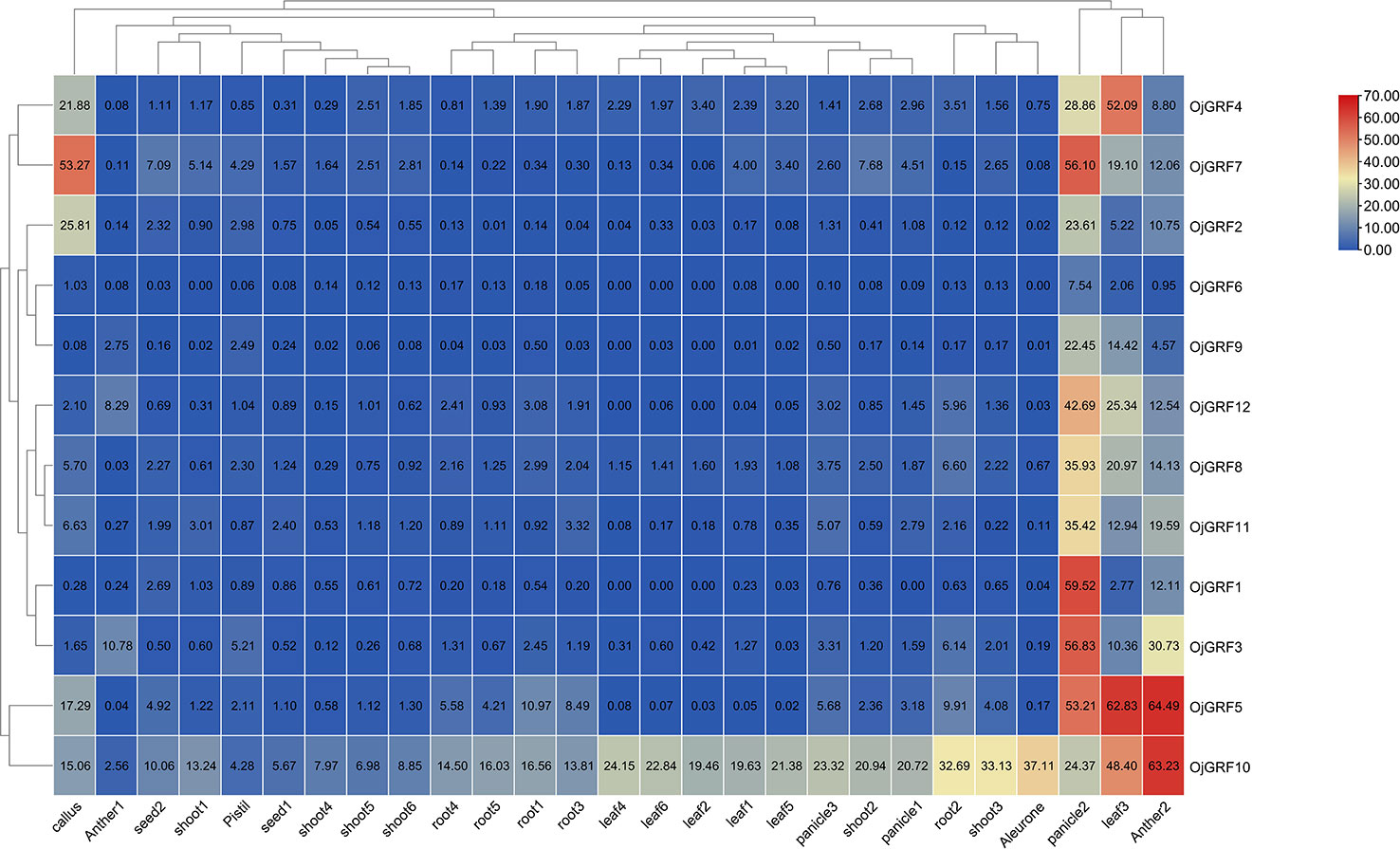
Figure 9 Expression profiles of the GRFs in the various rice tissues at different developmental stages. The heatmap based on the data of public rice transcriptomic by log2 FPKM represents the relative expression in 27 tissues. The GRFs are clustered according to hierarchical clustering. The blue and red color indicates the transcripts’ high and low expression levels, respectively.
The analysis of cis-elements revealed that the GRF promoters contain a large number of cis-acting elements associated with ABA, IAA, MeJ, SA, and GA responses (Figure 10). Then, we investigated the expression of GRFs of rice under treatment of IAA, BR, GA3, 6BA, ABA, and MeJ. Results showed that all of the GRFs were significantly induced by the six hormones (Figure 10). Relatively, OjGRF6 and OjGRF11 was most sensitive to IAA, BR, and ABA (Figure 10F), while OjGRF3, OjGRF4, and OjGRF9 were most sensitive to IAA (Figures 10C, D, I). The expression of OjGRF4 and OjGRF6 showed a steep rise from the 6 h to the 24 h after hormone treatment, while the other ten GRFs had two apparent expression peaks after treatment (Figures 10D, F). OjGRF1, OjGRF2, and OjGRF7 were induced to reach their highest after 9 h with ABA treatment, then began to decline (Figures 10A, B, G).
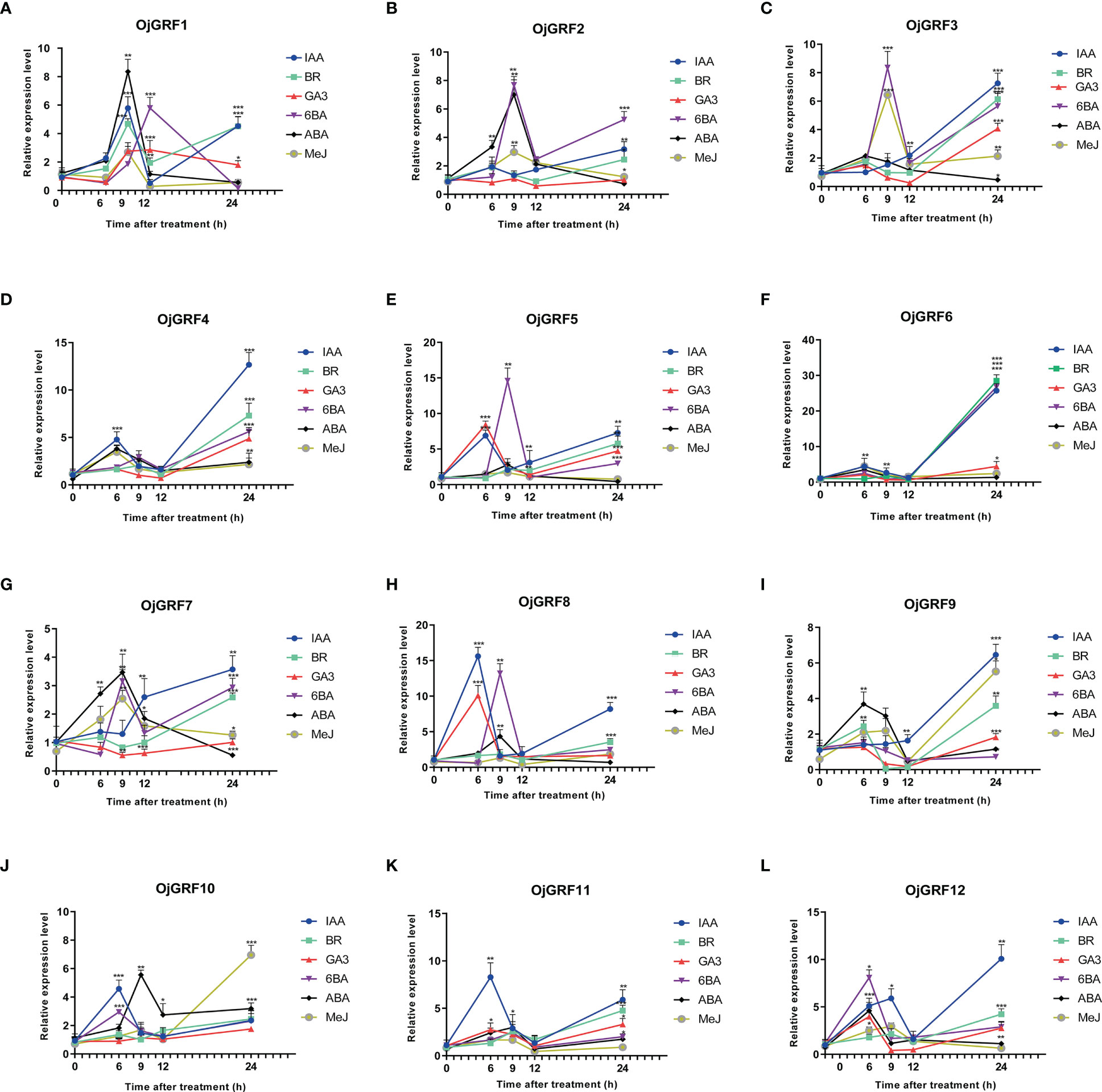
Figure 10 Analysis of the different expression patterns of GRF family members in rice under various hormonal treatments. (A–L) The expression of 12 GRFs in rice was calculated at 6, 9, 12, and 24 h of treatments compared with the expression value at 0h. The expression level of GRF was tested by qRT-PCR estimated by the 2ΔΔCT method. The error bars show the standard deviation of the three biological replicates. Student’s t-test was used in this experiment; *p < 0.05; **p < 0.01; ***p < 0.001.
The above results showed that OjGRF11 was the one of earliest gene to responded to IAA treatment. Previous studies have shown that auxin signaling genes are involved in root development (Notaguchi et al., 2012; Yu et al., 2015; Cui et al., 2017; Ye et al., 2021). To investigate whether OjGRF11 regulates root development through the IAA signaling pathway, an overexpression construct of OjGRF11 was introduced into YB (Oryza sativa L. ssp. indica), and the OjGRF11 transcript level was significantly increased, as assessed by qRT-PCR (Figure 11C). The statistical analysis of the root length of OjGRF11 overexpression lines in 10-day-old seedlings showed that it was shorter than that of the WT/YB under hydroponic conditions (Figures 11A, B) indicating that OjGRF11 negatively regulates root elongation.
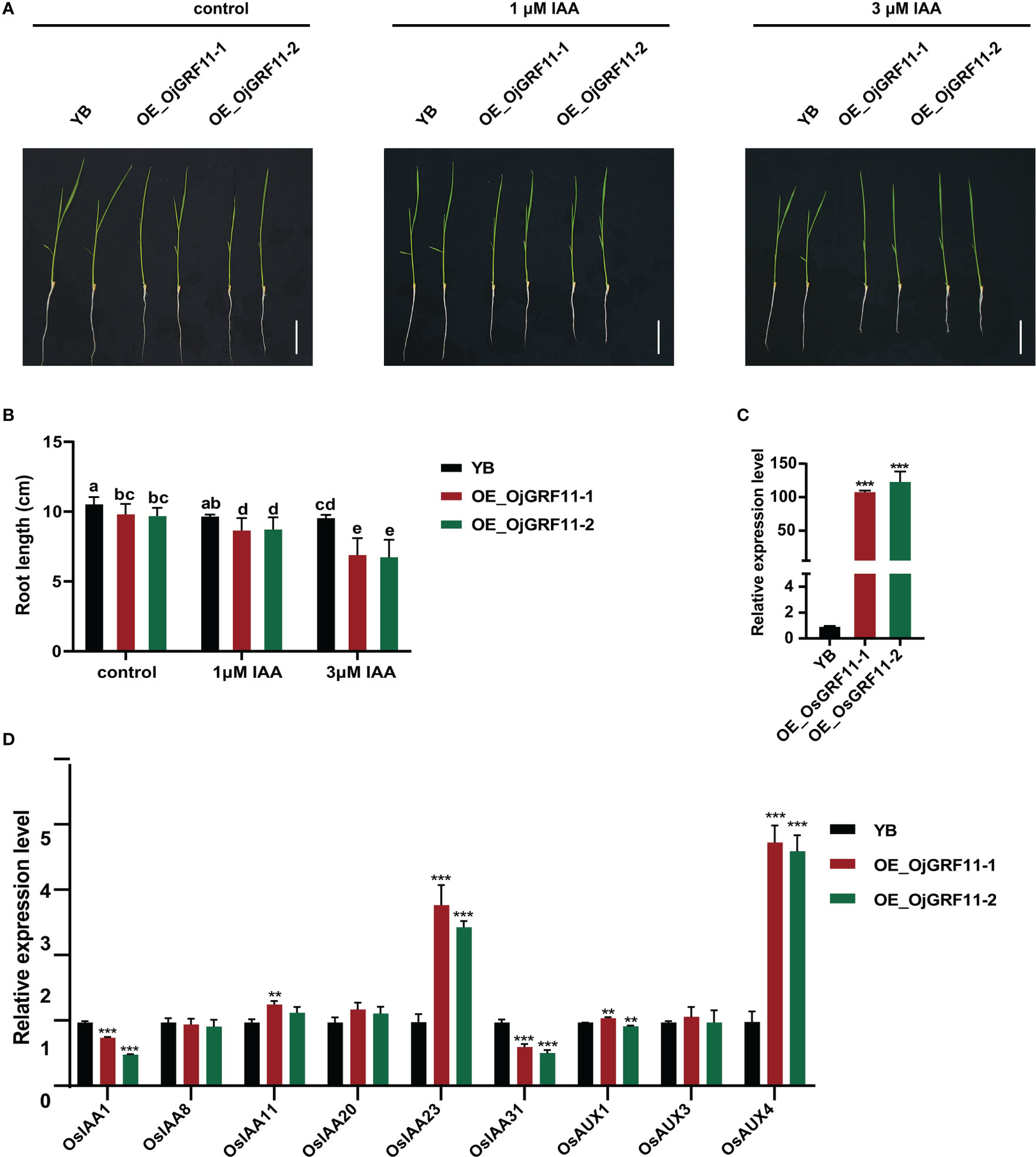
Figure 11 OjGRF11 are insensitive to auxin treatment. (A) Characterization of root length in wild‐type (WT/YB) and GRF11 over-expression lines under control, l μM IAA, and 3 μM IAA treatments for 7 d. Scale bars = 5 cm. (B) Statistical analysis of root length of in the WT/YB and GRF11 overexpression transgenic plant under control, l μM IAA, and 3 μM IAA treatments for 7 d. The error bars show the standard deviation of the ten biological replicates. Different letters indicate significant differences (P < 0.05) determined by Duncan’s multiple range test. (C) Expression levels of GRF11 in the WT/YB and OjGRF11 overexpression transgenic plants. (D) Relative expression levels of the OjGRF11target genes in the transgenic plants compared with in the YB. In (C, D) the expression level was tested by qRT-PCR estimated by the 2ΔΔCT method. The error bars show the standard deviation of the three biological replicates. ns: p > 0.05, **: p < 0.01, ***: p < 0.001.
To determine whether root length of OjGRF11 over-expression lines respond to auxin treatment, these seedlings were grown in 0 μM IAA, 1 μM IAA, and 3 μM IAA nutrient solution. After 7 d of IAA treatment, the root length of OjGRF11 over-expression lines was more suppressed than that of WT/YB (Figures 11A, B). Furthermore, an expression analysis verified that OjGRF11 could be induced by an IAA treatment (Supplementary Figure 3). Thus, OjGRF11regulates the plants’ t root elongation.
To further confirm the effect of OjGRF11 on root elongation through the IAA signal pathway, we examined several crucial genes in the IAA signaling pathway. The qRT-PCR results demonstrated that the expression levels of OsIAA23 and OsAUX4 were upregulated (Figure 11D). OsIAA23 and OsAUX4 are known to play critical roles in the development of root elongation (Jun et al., 2011; Ye et al., 2021). These findings indicate that OjGRF11 is essential for regulating rice root development through the IAA pathway.
Previous results reflected that GRFs were specific in plant and play key roles in various plants growth and development (Fİlİz et al., 2014; Cao et al., 2016). Thus, it is necessary to systematically describe the evolutionary patterns of GRFs in different species. In this study, a total of 96 GRFs were identified in eight Gramineae crops (Figure 1, Supplementary Table 2), which were derived from two main types of gene duplication including tandem duplication and WGD/segmental duplication (Table 1), and the numbers of duplication gene pairs varied greatly among Gramineae crops (Table 1). However, except for two tandem duplicated gene pairs of HvGRF2/HvGRF3 and GRF4/GRF5 in indica rice (Table 1), the other paralogous GRFs were all homologs, they were derived from WGD/segmental duplication. Correspondingly, the GRFs had similar gene structure and function within the same category (Figures 4, 5). For instance, the homologs GRF4 in indica rice and GRF5 in japonica rice were classified into Group 3 (Figure 1), which is known to be involved in auxiliary branch and spikelet development in rice (Gao et al., 2015). The 96 GRFs in Gramineae crops were categorized into three groups based on their structure, and each group showed some differences in terms of member composition, distribution of conserved motifs, and expression patterns (Figure 4), revealing the differentiation of GRFs in structure and function.
Transcription factors are considered to be key regulators of gene expression and coordinate the relationship between plants and the environment (Chen et al., 2021). Previous studies have demonstrated that GRFs, an important transcription factor in plants, is regulated by miR396 (Liu et al., 2014; Gao et al., 2015; Omidbakhshfard et al., 2015). In our study, we have newly identified that GRF5 in japonica rice is targeted by miR166, miR395 and miR6248 (Supplementary Table 7). Importantly, miR166 and miR395 have been implicated in cadmium stress response and disease resistance, respectively (Ding et al., 2018; Yang et al., 2022). Combined with the anoxic elements in the promoters of OjGRF5 in japonica rice (Figure 8), it suggests that GRF5 is possibly involved in both biological and abiotic stresses in rice.
Cis-acting elements prediction has been utilized in exploring gene function in various species (Cheng et al., 2021; Wang et al., 2021a; Wang et al., 2021b). In this study, a total of 41 cis-acting elements were identified in the GRF promoters. G-box (23.1%) and Box 4 (17.1%) occupied the largest number of light-responsive/responsiveness subcategories, which belong to the growth and development category (Figure 8). Recent reports indicate that BnGRF2 regulates cell number and plant photosynthesis to enhance seed oil production in Brassica napus (Liu et al., 2012; Vercruyssen et al., 2015), and AtGRF5 stimulates chloroplast division and photosynthesis in Arabidopsis (Liu et al., 2012; Vercruyssen et al., 2015). These findings suggest that GRFs are potentially involved in light-responsiveness and photosynthesis.
Increasing evidence shows that GRFs play critical roles in phytohormone response during plant growth and development (Knaap and Kende, 2000; Liu et al., 2014; Gao et al., 2015; Tang et al., 2018; Chen et al., 2020b). Our results showed that several motifs related to phytohormone response were identified (Figure 8), and all the GRFs in japonica rice were induced by IAA, BR, GA3, 6BA, ABA, and MeJ, although they showed various response patterns under different hormones (Figure 10). Notably, roots overexpressing OjGRF11 were more sensitive to IAA (Figure 11). Furthermore, results showed that OjGRF11 regulates root development by influencing genes in the auxin signaling pathway, particularly OsIAA23 and OsAUX4, whose expression levels were significantly altered (Figure 11). However, further validation is needed to elucidate the molecular mechanisms underlying their interactions. Considering the high expression pattern of GRFs in young inflorescence, we can deduce that the GRFs are widely involved in the plant inflorescence development and responses to diverse environmental stresses in Gramineae crops.
In this study, we found that the number and structure of GRFs in Gramineae crops were relatively stable and highly conserved. These proteins were highly expressed in the leaf and young inflorescence and contained rich cis-elements related to hormone response in their promoters. Furthermore, we observed that the expression of GRFs was induced under multiple hormonal stresses, including IAA, BR, GA3, 6BA, ABA, and MeJ treatments. Our investigation of OjGRF11 revealed that this gene is significantly induced after 6 hours of IAA treatment and overexpression of this gene increases sensitivity to IAA and affects root elongation. These findings provide a comprehensive understanding of the molecular characteristics and evolutionary patterns of the GRF family in Gramineae crops.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
WW and HY designed the research; WW, MC carried out the bioinformatic analyses. WW performed subcellular localization and partial quantitative PCR experiments. XW, RW, FF and ZW participated in other molecular experiments; MC, WW and HY wrote the manuscript. ZT, SL and HY was responsible for revising the manuscript. All authors have read and agreed to the published version of the manuscript.
This project was supported by the National Key Research and Development Program, the National Natural Science Foundation of China (31870322, 32172074), Hubei Hongshan Laboratory (2021hszd010).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1174955/full#supplementary-material
Bolser, D., Staines, D. M., Pritchard, E., Kersey, P. (2016). Ensembl plants: Integrating tools for visualizing, mining, and analyzing plant genomics data. Methods Mol. Biol. 1374, 115–140. doi: 10.1007/978-1-4939-3167-5_6
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: Architecture and applications. BMC Bioinf. 10, 421. doi: 10.1186/1471-2105-10-421
Cao, Y., Han, Y., Jin, Q., Lin, Y., Cai, Y. (2016). Comparative genomic analysis of the GRF genes in Chinese pear (Pyrus bretschneideri rehd), poplar (Populous), grape (Vitis vinifera), arabidopsis and rice (Oryza sativa). Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01750
Che, R., Tong, H., Shi, B., Liu, Y., Fang, S., Liu, D., et al. (2015). Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants 2, 15195. doi: 10.1038/nplants.2015.195
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020a). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Y., Dan, Z., Gao, F., Chen, P., Fan, F., Li, S. (2020b). Rice GROWTH-REGULATING FACTOR7 modulates plant architecture through regulating GA and indole-3-Acetic acid metabolism. Plant Physiol. 184, 393–406. doi: 10.1104/pp.20.00302
Chen, Y., Shen, J., Zhang, L., Qi, H., Yang, L., Wang, H., et al. (2021). Nuclear translocation of OsMFT1 that is impeded by OsFTIP1 promotes drought tolerance in rice. Mol. Plant 14, 1297–1311. doi: 10.1016/j.molp.2021.05.001
Cheng, M., Yuan, H., Wang, R., Zou, J., Liang, T., Yang, F., et al. (2021). Genome-wide identification and analysis of the metallothionein genes in oryza genus. Int. J. Mol. Sci. 22, 9651. doi: 10.3390/ijms22179651
Cui, P., Liu, H., Ruan, S., Ali, B., Gill, R. A., Ma, H., et al. (2017). A zinc finger protein, interacted with cyclophilin, affects root development via IAA pathway in rice. J. Integr. Plant Biol. 59, 496–505. doi: 10.1111/jipb.12531
Ding, Y., Gong, S., Wang, Y., Wang, F., Bao, H., Sun, J., et al. (2018). MicroRNA166 modulates cadmium tolerance and accumulation in rice. Plant Physiol. 177, 1691–1703. doi: 10.1104/pp.18.00485
Duan, P., Ni, S., Wang, J., Zhang, B., Xu, R., Wang, Y., et al. (2015). Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants 2, 15203. doi: 10.1038/nplants.2015.203
Edgar, R. C., Sjolander, K. (2004). A comparison of scoring functions for protein sequence profile alignment. Bioinformatics 20, 1301–1308. doi: 10.1093/bioinformatics/bth090
Emms, D. M., Kelly, S. (2019). OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238. doi: 10.1186/s13059-019-1832-y
Fİlİz, E., KoÇ, İ., Tombuloğlu, H. (2014). Genome-wide identification and analysis of growth regulating factor genes in brachypodium distachyon: in silico approaches. Turkish J. Biol. 38, 296–306. doi: 10.3906/biy-1308-57
Finn, R. D., Clements, J., Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Gao, F., Wang, K., Liu, Y., Chen, Y., Chen, P., Shi, Z., et al. (2015). Blocking miR396 increases rice yield by shaping inflorescence architecture. Nat. Plants 2, 15196. doi: 10.1038/nplants.2015.196
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Greco, M., Chiappetta, A., Bruno, L., Bitonti, M. B. (2012). In posidonia oceanica cadmium induces changes in DNA methylation and chromatin patterning. J. Exp. Bot. 63, 695–709. doi: 10.1093/jxb/err313
He, Z., Zhang, H., Gao, S., Lercher, M. J., Chen, W. H., Hu, S. (2016). Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44, W236–W241. doi: 10.1093/nar/gkw370
Hu, J., Wang, Y., Fang, Y., Zeng, L., Xu, J., Yu, H., et al. (2015). A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant 8, 1455–1465. doi: 10.1016/j.molp.2015.07.002
Jin, J., Tian, F., Yang, D. C., Meng, Y. Q., Kong, L., Luo, J., et al. (2017). PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045. doi: 10.1093/nar/gkw982
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Jun, N., Gaohang, W., Zhenxing, Z., Huanhuan, Z., Yunrong, W., Ping, W. (2011). OsIAA23-mediated auxin signaling defines postembryonic maintenance of QC in rice. Plant J. 68, 433–442. doi: 10.1111/j.1365-313X.2011.04698.x
Kaufmann, K., Muino, J. M., Jauregui, R., Airoldi, C. A., Smaczniak, C., Krajewski, P., et al. (2009). Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the arabidopsis flower. PloS Biol. 7, e1000090. doi: 10.1371/journal.pbio.1000090
Kim, J. S., Mizoi, J., Kidokoro, S., Maruyama, K., Nakajima, J., Nakashima, K., et al. (2012). Arabidopsis growth-regulating factor7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. Plant Cell 24, 3393–3405. doi: 10.1105/tpc.112.100933
Kim, J. H., Tsukaya, H. (2015). Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J. Exp. Bot. 66, 6093–6107. doi: 10.1093/jxb/erv349
Knaap, E., Kende, K. H. (2000). A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 122, 695–704. doi: 10.1104/pp.122.3.695
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, S., Gao, F., Xie, K., Zeng, X., Cao, Y., Zeng, J., et al. (2016). The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol. J. 14, 2134–2146. doi: 10.1111/pbi.12569
Liu, H., Guo, S., Xu, Y., Li, C., Zhang, Z., Zhang, D., et al. (2014). OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4. Plant Physiol. 165, 160–174. doi: 10.1104/pp.114.235564
Liu, J., Hua, W., Yang, H.-L., Zhan, G.-M., Li, R.-J., Deng, L.-B., et al. (2012). The BnGRF2 gene (GRF2-like gene from brassica napus) enhances seed oil production through regulating cell number and plant photosynthesis. J. Exp. Bot. 63, 3727–3740. doi: 10.1093/jxb/ers066
Londo, J. P., Chiang, Y.-C., Hung, K.-H., Chiang, T.-Y., Schaal, B. A. (2006). Phylogeography of Asian wild rice, oryza rufipogon, reveals multiple independent domestications of cultivated rice, oryza sativa. Proc. Natl. Acad. Sci. 103, 9578–9583. doi: 10.1073/pnas.0603152103
Malik, W. A., Wang, X., Wang, X., Shu, N., Cui, R., Chen, X., et al. (2020). Genome-wide expression analysis suggests glutaredoxin genes response to various stresses in cotton. Int. J. Biol. Macromol 153, 470–491. doi: 10.1016/j.ijbiomac.2020.03.021
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Miura, K., Ikeda, M., Matsubara, A., Song, X. J., Ito, M., Asano, K., et al. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549. doi: 10.1038/ng.592
Notaguchi, M., Wolf, S., Lucas, W. J. (2012). Phloem-mobile Aux/IAA transcripts target to the root tip and modify root architecture. J. Integr. Plant Biol. 54, 760–772. doi: 10.1111/j.1744-7909.2012.01155.x
Omidbakhshfard, M. A., Proost, S., Fujikura, U., Mueller-Roeber, B. (2015). Growth-regulating factors (GRFs): A small transcription factor family with important functions in plant biology. Mol. Plant 8, 998–1010. doi: 10.1016/j.molp.2015.01.013
Pajoro, A., Madrigal, P., Muino, J. M., Matus, J. T., Jin, J., Mecchia, M. A., et al. (2014). Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. 15, R41. doi: 10.1186/gb-2014-15-3-r41
Ren, D., Rao, Y., Leng, Y., Li, Z., Xu, Q., Wu, L., et al. (2016). Regulatory role of OsMADS34 in the determination of glumes fate, grain yield, and quality in rice. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01853
Rombauts, S., Déhais, P., Van Montagu, M., Rouzé, P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296. doi: 10.1093/nar/27.1.295
Tang, Y., Liu, H., Guo, S., Wang, B., Li, Z., Chong, K., et al. (2018). OsmiR396d affects gibberellin and brassinosteroid signaling to regulate plant architecture in rice. Plant Physiol. 176, 946–959. doi: 10.1104/pp.17.00964
Vercruysse, J., Baekelandt, A., Gonzalez, N., Inze, D. (2020). Molecular networks regulating cell division during arabidopsis leaf growth. J. Exp. Bot. 71, 2365–2378. doi: 10.1093/jxb/erz522
Vercruyssen, L., Tognetti, V. B., Gonzalez, N., Van Dingenen, J., De Milde, L., Bielach, A., et al. (2015). GROWTH REGULATING FACTOR5 stimulates arabidopsis chloroplast division, photosynthesis, and leaf longevity. Plant Physiol. 167, 817–832. doi: 10.1104/pp.114.256180
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, J., Zhang, Y., Xu, N., Zhang, H., Fan, Y., Rui, C., et al. (2021a). Genome-wide identification of CK gene family suggests functional expression pattern against Cd(2+) stress in gossypium hirsutum l. Int. J. Biol. Macromol 188, 272–282. doi: 10.1016/j.ijbiomac.2021.07.190
Wang, X., Zheng, Y., Chen, B., Zhi, C., Qiao, L., Liu, C., et al. (2021b). Genome-wide identification of small heat shock protein (HSP20) homologs in three cucurbit species and the expression profiles of CsHSP20s under several abiotic stresses. Int. J. Biol. Macromol 190, 827–836. doi: 10.1016/j.ijbiomac.2021.08.222
Wang, J., Zhou, L., Shi, H., Chern, M., Yu, H., Yi, H., et al. (2018). A single transcription factor promotes both yield and immunity in rice. Science 361, 1026–1028. doi: 10.1126/science.aat7675
Wu, L., Zhang, D., Xue, M., Qian, J., He, Y., Wang, S. (2014). Overexpression of the maize GRF10, an endogenous truncated growth-regulating factor protein, leads to reduction in leaf size and plant height. J. Integr. Plant Biol. 56, 1053–1063. doi: 10.1111/jipb.12220
Yang, Z., Hui, S., Lv, Y., Zhang, M., Chen, D., Tian, J., et al. (2022). miR395-regulated sulfate metabolism exploits pathogen sensitivity to sulfate to boost immunity in rice. Mol. Plant 15, 671–688. doi: 10.1016/j.molp.2021.12.013
Ye, R., Wu, Y., Gao, Z., Chen, H., Jia, L., Li, D., et al. (2021). Primary root and root hair development regulation by OsAUX4 and its participation in the phosphate starvation response. J. Integr. Plant Biol. 63, 1555–1567. doi: 10.1111/jipb.13142
Yu, C., Sun, C., Shen, C., Wang, S., Liu, F., Liu, Y., et al. (2015). The auxin transporter, OsAUX1, is involved in primary root and root hair elongation and in cd stress responses in rice (Oryza sativa l.). Plant J. 83, 818–830. doi: 10.1111/tpj.12929
Zhang, D.-F., Li, B., Jia, G.-Q., Zhang, T.-F., Dai, J.-R., Li, J.-S., et al. (2008). Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in maize (Zea mays l.). Plant Sci. 175, 809–817. doi: 10.1016/j.plantsci.2008.08.002
Keywords: GRFS, Gramineae crops, evolutionary analysis, transcriptional activators, hormone treatments
Citation: Wang W, Cheng M, Wei X, Wang R, Fan F, Wang Z, Tian Z, Li S and Yuan H (2023) Comprehensive evolutionary analysis of growth-regulating factor gene family revealing the potential molecular basis under multiple hormonal stress in Gramineae crops. Front. Plant Sci. 14:1174955. doi: 10.3389/fpls.2023.1174955
Received: 27 February 2023; Accepted: 20 March 2023;
Published: 31 March 2023.
Edited by:
Muhammad Waseem, Hainan University, ChinaReviewed by:
Zhi Gang Meng, Biotechnology Research Institute of CAAS, ChinaCopyright © 2023 Wang, Cheng, Wei, Wang, Fan, Wang, Tian, Li and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huanan Yuan, aHVhbnJhbnl1YW5Ad2h1LmVkdS5jbg==; Shaoqing Li, c2hhb3FpbmdsaUB3aHUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.