
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 05 June 2023
Sec. Crop and Product Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1172354
This article is part of the Research Topic Advances in Environmental Stress Biology and Important Agronomic Traits Improvement in Non-staple Crops View all 11 articles
Legume-rhizobial symbiotic nitrogen fixation is the most efficient nitrogen assimilation system in the ecosystem. In the special interaction between organ–root nodules, legumes supply rhizobial carbohydrates for their proliferation, while rhizobials provide host plants with absorbable nitrogen. Nodule initiation and formation require a complex molecular dialogue between legumes and rhizobia, which involves the accurate regulation of a series of legume genes. The CCR4–NOT complex is a conserved multi-subunit complex with functions regulating gene expression in many cellular processes. However, the functions of the CCR4–NOT complex in rhizobia–host interactions remain unclear. In this study, we identified seven members of the NOT4 family in soybean and further classified them into three subgroups. Bioinformatic analysis showed that NOT4s shared relatively conserved motifs and gene structures in each subgroup, while there were significant differences between NOT4s in the different subgroups. Expression profile analysis indicated that NOT4s may be involved in nodulation in soybean, as most of them were induced by Rhizobium infection and highly expressed in nodules. We further selected GmNOT4-1 to clarify the biological function of these genes in soybean nodulation. Interestingly, we found that either GmNOT4-1 overexpression or down-regulation of GmNOT4-1 by RNAi or CRISPR/Cas9 gene editing would suppress the number of nodules in soybean. Intriguingly, alterations in the expression of GmNOT4-1 repressed the expression of genes in the Nod factor signaling pathway. This research provides new insight into the function of the CCR4–NOT family in legumes and reveals GmNOT4-1 to be a potent gene for regulating symbiotic nodulation.
Nitrogen is one of the essential macroelements for plant growth and development (Jia et al., 2017). Therefore, improving nitrogen utilization efficiency and appropriately applying nitrogen fertilizer are important guarantees for a high and stable yield of crops (Chun et al., 2005). In the ecosystem, given that leguminous plants have a high demand for nitrogen, they evolved an additional special root organ–root nodules—to fix atmospheric nitrogen and provide nitrogen to improve legume development (Ren et al., 2019).
Nodulation is a complex biological process involving direct interactions between rhizobial and legume signals (Howard, 1991). Nodulation is initiated by the flavonoids secreted by the legume roots, which were perceived by the surrounding compatible rhizobium strains and stimulated them to synthesize lipochitin oligosaccharides, called Nod factors (NF, Patra et al., 2017), which were sensed by Nod factor receptors (NFRs) (e.g., NF Perception [NFP] in M. truncatula, NF Receptors 1 and 5 [NFR1/5] in L. japonicus, and NFR1/5α in soybean) located on legume root hairs. This interaction then stimulated the consequent NF signaling pathway, which promoted root hair deformation, infection thread formation, outer cortical cell division, and root nodule primordia formation (Gao et al., 2002; Chou and Wei, 2010). Nodulation is a high-energy-consumption biological and host-dominant process; thus, legumes evolved an auto-regulation of nodulation (AON) mechanism; in brief, when the root nodules reach a certain number, the host will generate the CLE peptides as a signal molecular (Carroll et al., 2016), which would transmit to the legume shoot and be perceived by the NARK receptor. Furthermore, this recognition will generate shoot-derived molecular signals (e.g., cytokinin, miRNA2111), which transmit back to the roots and attenuate the nodulation process (Chou and Wei, 2010; Yuan et al., 2016).
Nodules are produced via a complex genetic program to allow rhizobial recognition and nodule formation, a series of transcription factors modulate the downstream responses to NF signaling, including NIN (L. Yuan et al., 2016; Wang et al., 2019), IPD3 (Interacting Protein of DMI3; Horváth, 2011), ERN1/ERN2/ERN3 (ERFs Required for Nodulation; Andriankaja et al., 2007; Middleton et al., 2007), NF-YA1 (Nuclear Factor-Y Subunit A1; Laloum et al., 2013), the NSP1 (Nodulation Signaling Pathway1) and NSP2 (Kaló et al., 2005; Smit et al., 2005; Oldroyd & Downie, 2008; Heckmann et al., 2011), NNC1 (Nodule number control1), and the DELLAs (Fonouni-Farde et al., 2016; Jin et al., 2016).
The ubiquitin–proteasome system is the most efficient and specific protein degradation mode for regulating plant growth and development. ASTRAY and SINAT5 encoding RING-finger domains containing E3 ubiquitin ligase have been shown to function during legume nodulation (Fujita and Kawaguchi, 2002; Nishimura et al., 2002). The no-nodule alfalfa mutants rh2 and LjnsRING, encoding RING-H2 domain-containing proteins (Shimomura et al., 2006), and LIN (in Medicago) and CERBERUS (in Lotus) shared 86% homology and both contained a U-box domain (Kiss et al., 2010). Plant U-box protein1 (PUB1) has the activity of an E3 ligase and interacts with LYK3/NFR1/DMI2/SYMRK to inhibit the infection of rhizobia and mycorrhizal fungi through its ubiquitination activity (Vernié et al., 2016).
The carbon catabolite repression 4–negative on TATA-less (CCR4–NOT) complex multi-subunit complex functions as a major regulator of gene expression homeostasis through ubiquitination (CCR4 and CAF1 [CCR4 associated factor 1]) and deadenylation (NOT4) in eukaryotes. In plants, core components of the CCR4–NOT complex were identified and revealed that Arabidopsis possesses AtCCR4–NOT complexes involved in mRNA recognition, AtCCR4-CAF1 has mRNA deadenylase activity to regulate environmental stresses (Liang et al., 2008; Walley et al., 2010; Suzuki et al., 2015), and NOT9B and CCR4–NOT can respond to far-red light and are involved in phyA-modulated gene expression. However, the identification of NOT4 in the complex and its function in legumes are yet to be unveiled.
In this study, genome-wide systematic characterization, including protein properties, chromosome distribution, phylogenetic relationship, protein motif, and gene expression pattern, was performed to study the soybean CCR4–NOT complex gene family. We selected GmNOT4-1, a member of the CCR4–NOT complex gene family that is highly expressed in nodules and significantly stimulated by Rhizobium infection, to determine its function in symbiotic nodulation using overexpression, RNAi, and CRISPR/Cas9. We found that in transgenic hairy roots harboring GmNOT4-1-overexpressing or in roots carrying GmNOT4-1-RNAi and CRISPR/Cas9-GmNOT4-1, the number of root nodules was significantly inhibited, and the marker genes for both the NF and AON signaling pathways were repressed. In conclusion, this study reports for the first time the function of the key eukaryotic gene expression regulatory complex CCR4–NOT in legume nodulation and identifies a distinguished nodulation regulator, NOT4-1, whose expression balance is relevant to soybean symbiosis.
In this study, soybean [G. max (L.) Merrill cv. Williams 82] and Agrobacterium rhizogenes strain K599 were used for the hairy root transformation. The hairy root transformation procedure was previously described (Wang et al., 2014) with some modifications. The positive transformed composite plants were cultured in a low-nitrogen nutrient solution for 5 days for recovery; after that, the plants were transferred to vermiculite for inoculation with a suspension of B. japonicum strain USDA110 (30 ml, OD600 = 0.08). Nodule numbers were evaluated at 28 DAI (days after inoculation).
Genome data, protein sequence, and genome annotation files of Glycine max were downloaded from the Phytozome database (https://phytozome-next.jgi.doe.gov/). Protein families in the PANTHER database (http://www.pantherdb.org) were applied to download the Hidden Markov model (HMM) of the NOT transcription complex related family (PTHR12603). The soybean genome was searched using hmm search in HMMER (http://hmmer.org/) to identify NOT4 candidate genes (the screening threshold was 1.0e−10). The resulting candidate sequences were submitted to Intel Pro Scan (https://www.ebi.ac.uk/interpro/search/sequence-search) to check the PTHR12603 structure domain. Soybean GmNOT4 family members were named according to their location distribution on soybean chromosomes.
NOT4 protein sequences from Medicago truncatula, Phaseolus vulgaris, Arabidopsis thaliana, soybean [G. max (L.)], and rice (Oryza sativa) NOT4 were downloaded from the Phytozome database (https://phytozome-next.jgi.doe.gov/). The phylogenetic tree of the GmNOT4 gene family members of the five species was constructed by MEGA-X software and calculated by the NJ (neighbor-joining) method. The parameters were set to self-expanding and repeated 1,000 times. Chromosome distribution was analyzed using TBtools.
The MEME Suite online software (https://meme-suite.org/meme/) was used for conserved motif analysis, and the parameter was set to 10. The gene structure information of GmNOT4s was extracted from the soybean gene information GFF file, and the gene structures were visualized by TBtools.
RNAprep Plant Plus Trizol Kit was used to extract RNA from collected samples, and the first-strand cDNA was synthesized using the Super Mix Kit (Hifair II 1 strand cDNA Synthesis SuperMix, gDNA digester plus) (Yeasen Biotech Co. Ltd., Shanghai, China). qPCR was performed using SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich). GmCYP2 was used as an internal control (Jian et al., 2008). The primers used in this study are shown in Table S1.
For the GmNOT4-1 overexpression construct, the GmNOT4-1 CDS fragment was inserted into the pCAMBIA1300-GFP vector through seamless cloning using the BamH1 restriction site; for the GmNOT4-1-RNAi construct, the GmNOT4-1 CDS fragment was ligated into the pDONOR207 entry vector, and then the target sequence was cloned into the pK7GWIW-GFP vector through the gateway LR reaction. For the GmNOT4-1 CRISPR/Cas9 knock-out construct, sgRNAs were designed using the software Crispr-P (http://cbi.hzau.edu.cn/crispr/), and the top two reliable sgRNAs, CAAGGTGCGGTGAAGAGCA and TCGTCCTCTTCGCCTCTGC, were selected. Then, vector pCBC-DT1T2 was used as a template to clone the two CRISPR fragments, and the two obtained products were inserted into vector pKSE401-GFP.
GraphPad Prism 7 (GraphPad Software, La Jolla, CA, USA) was used to analyze the data in this study. A Student’s t-test was performed to generate P-values. The statistical differences are marked as follows: *, P <0.05; **, P <0.01; ***, P <0.001.
Seven GmNOT4s were identified from the soybean genome using BLAST and PANTHER searches and named GmNOT4-1–GmNOT4-7 according to their positions on six chromosomes (Figure S1). The chromosomal distribution of NOT4s indicated that they were dispersed on chromosomes 5, 10, 12, 15, and 17. Except for GmNOT4-4 and GmNOT4-5, which are on chromosome 13, each of the other five chromosomes contains one family member. The physical and chemical properties of GmNOT4s were analyzed, and it was found that the amino acid residues encoded by seven GmNOT4s ranged from 232 (GmNOT4-1) to 1,046 (GmNOT4-6), and the corresponding molecular weight ranged from 35,386.84 (GmNOT4-1) to 115,070.36 (GmNOT4-6) Da. The theoretical isoelectric points of seven GmNOT4 family members ranged from 4.76 (GmNOT4-7) to 6.40 (GmNOT4-4), all of which belonged to weakly acidic proteins (Table S2). The total mean hydrophobic index was less than 0, indicating that they were hydrophilic proteins. To understand the functional characteristics and evolutionary relationship of the NOT4 gene family, NOT4 protein sequences in soybean, M. truncatula, common bean, A. thaliana, and rice were retried to construct the phylogenetic tree. The results showed that the members of the NOT4 gene family were divided into three subgroups (Groups I–III), in which groups I and III contained 10/11 members, and group II had three members. In the same subgroups, NOT4 proteins cluster together within species (Figure 1). Soybean NOT4 family members were evenly distributed in three groups, with three members in Group I, two members in Group II, and two members in Group III.
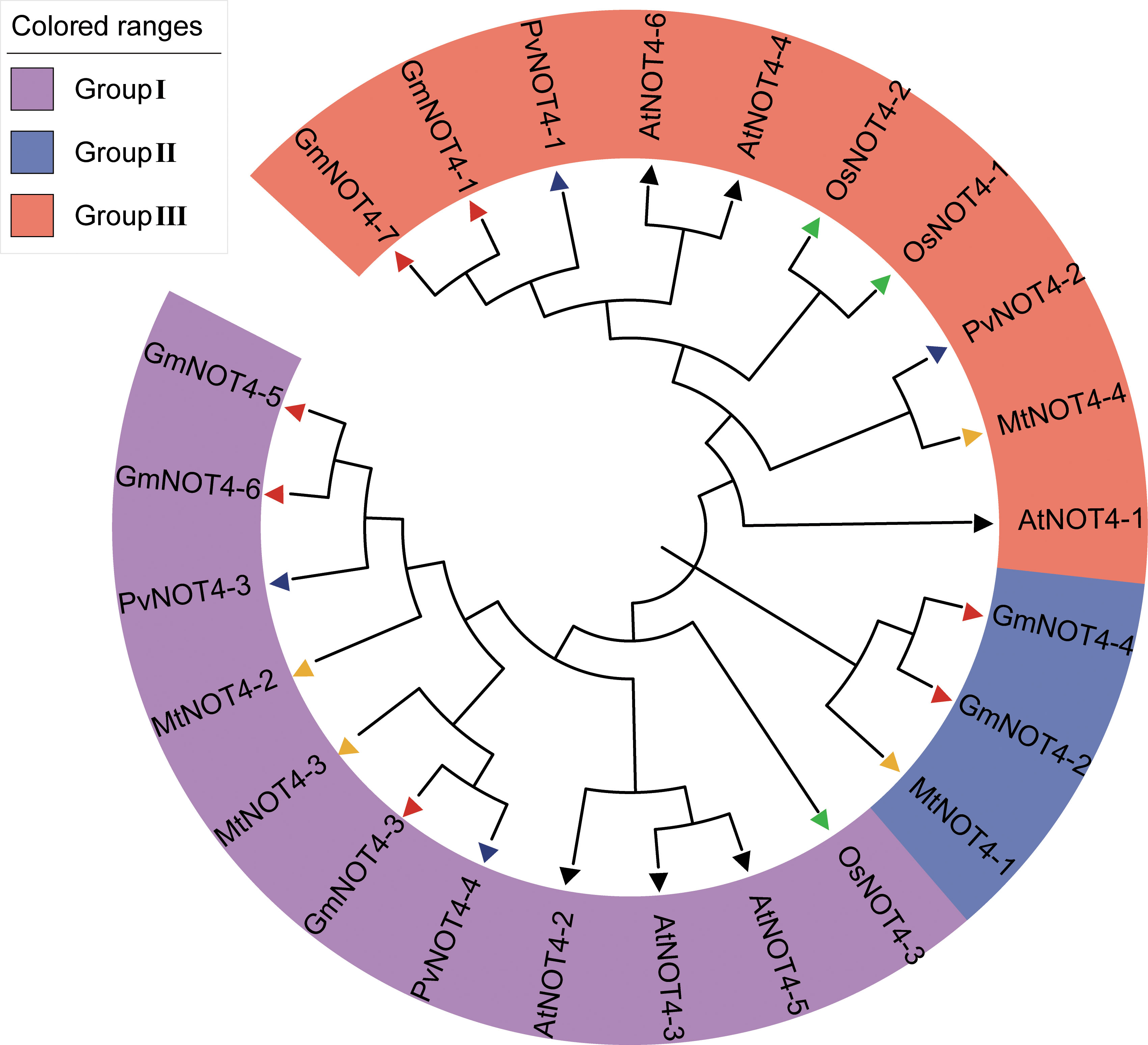
Figure 1 Phylogenetic tree of NOT4 family members. NOT4 Protein sequences of Medicago truncatula (Mt), Phaseolus vulgaris (Pv), Arabidopsis thaliana (At), Glycine max (Gm) and Oryza sativa (Os) were divided into Group I to Group III and colored in purple, blue, and orange, respectively. Protein sequences were clustered using CLASTALW in MEGA 11.0. The phylogenetic tree was constructed by MEGA 11.0 using a bootstrap neighbor join method.
To get a hint about the function of NOT4s, we first performed conserved motif analysis and gene structure analysis. There are 10 conserved motifs obtained from the GmNOT4 protein, and the number of motifs of GmNOT4s in different subgroups varied significantly, ranging from 2 to 10. Three members of Group I contain 10 motifs; there were eight motifs in GmNOT4-2 and seven for GmNOT4-4 in Group II and the members of Group III contain only two motifs. Among the 10 motifs, Motif1 was the most conserved one and was present in all GmNOT4 proteins (Figure 2). The variation in the types and amounts of conserved motifs in GmNOT4s reflects the functional diversity of these proteins. In addition, we have confirmed the conservation of motifs in NOT4 proteins in other plant species; we found these motifs are consistently present among Arabidopsis, rice, Medicago, and the common bean (Figure S2).
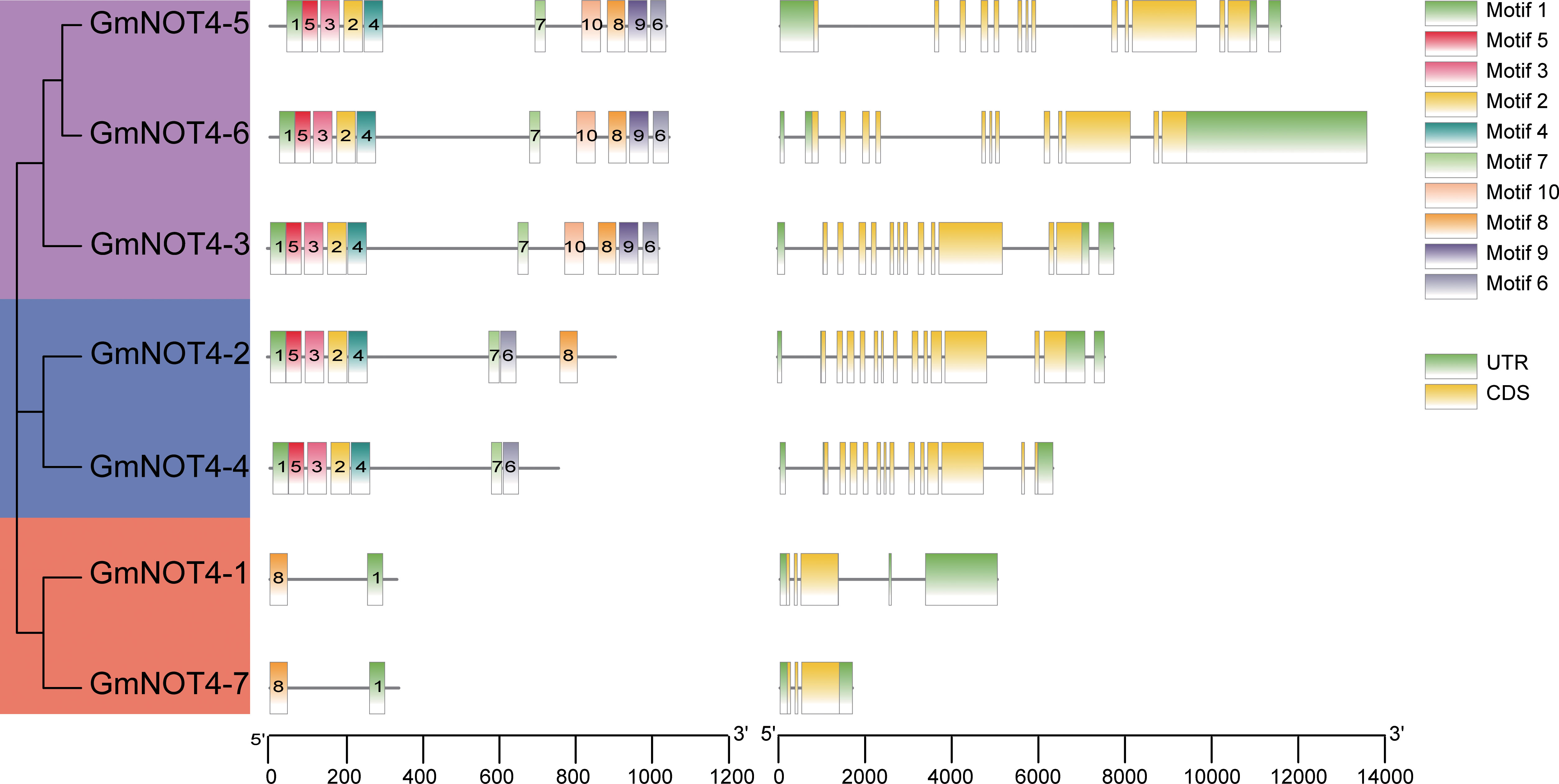
Figure 2 Gene structure and conserved motif distribution of GmNOT4s. MEME online software was used to identify conserved motifs. A total of 10 motifs were found in GmNOT4s genes and represented by different colored boxes and corresponding motif number, conserved amino acid sequence themes were shown below. (The left panel) shown the phylogenetic tree of group I–III genes in purple, blue, and orange colors; (The middle panel) shown the conserved motifs distribution in GmNOT4s; (The right panel) shown the gene structure of untranslated regions (UTRs) of the GmNOT4s gene are shown as green, yellow boxes and black lines.
Diagram of GmNOT4s gene structures shows the exon number variation of GmNOT4s ranged from 3 to 13 (Figure 2). Combined with phylogenetic tree analysis, we found that genes with close genetic relationships mostly had similar gene structures. For example, all Group I GmNOT4s contained 12 exons, while Group III GmNOT4s contained three exons.
Most transcriptional factors function in gene expression regulation by binding specific cis-acting elements in gene promoters to modulate gene expression. To predict the functions of GmNOT4 genes, 2-kilobase pairs upstream of the translational initiation site were selected as the promoter sequence and submitted to PlantCARE online software to predict the cis-acting elements (Figure S3). A total of 63 cis-acting elements were identified in seven GmNOT4 promoters, while 54 cis-acting elements were related to plant hormones, stress, growth, and development. Firstly, core cis-acting elements exist in almost all promoters and consist of AT-TATA-box, CAAT-box, TATA-box, and TATA. The second group contains 26 cis-acting elements related to plant growth and development; all GmNOT4 promoters contain photoreactive elements, which coincide with the CCR4–NOT complex response to far-red light. However, only GmNOT4-1 and GmNot4-7 contain cis-regulatory elements for flavonoid biosynthesis, which play a vital role in rhizobia attraction and symbiosis construction. Plant hormones are important for legume nodulation; 13 kinds of plant hormone related cis-acting elements were harbored in GmNOT4 promoters; all GmNOT4 promoters contained abscisic acid and ethylene-related cis-acting elements; and auxin response elements (TGA-elements), gibberellin response elements (P-box, GARE-motif) existed in most GmNOT4s promoters. Low oxygen conditions were very important for nitrogen fixation in nodules, and we found that among the stress response cis elements, GmNOT4s harbored the most anaerobic induction and stress-related cis elements. The specific cis-acting element distributions in GmNOT4 promoters suggest their different biological functions.
To investigate the potential function of NOT4s during soybean nodulation, we examined the temporal and spatial expression patterns of GmNOT4s by qRT-PCR.
We found that all but NOT4-7 were highly expressed in root nodules (Figures 3A–F). Further study has shown that the expression of GmNOT4-1, GmNOT4-2, GmNOT4-3, GmNOT4-4, and GmNOT4-7 was dramatically stimulated by rhizobium infection at 1 DAI (Figures 3H–K, N). The expression of GmNOT4-1 and GmNOT4-2 was induced by rhizobium USDA110 while GmNOT4-3 was repressed at 3 DAI (Figure 3H–J). The expression of GmNOT4-1, GmNOT4-5, and GmNOT4-6 was induced by rhizobium, while GmNOT4-4 and GmNOT4-7 were repressed at 6 DAI (Figures 3H, L, M). These results show that all GmNOT4s responded to rhizobium at least at one point. Combining the above tissue expression pattern to prove the function of GmNOT4s in legume root nodule symbiosis, we selected GmNOT4-1 for further study, whose expression was induced at all checkpoints within a short period of inoculation (1, 3, and 6 DAI).
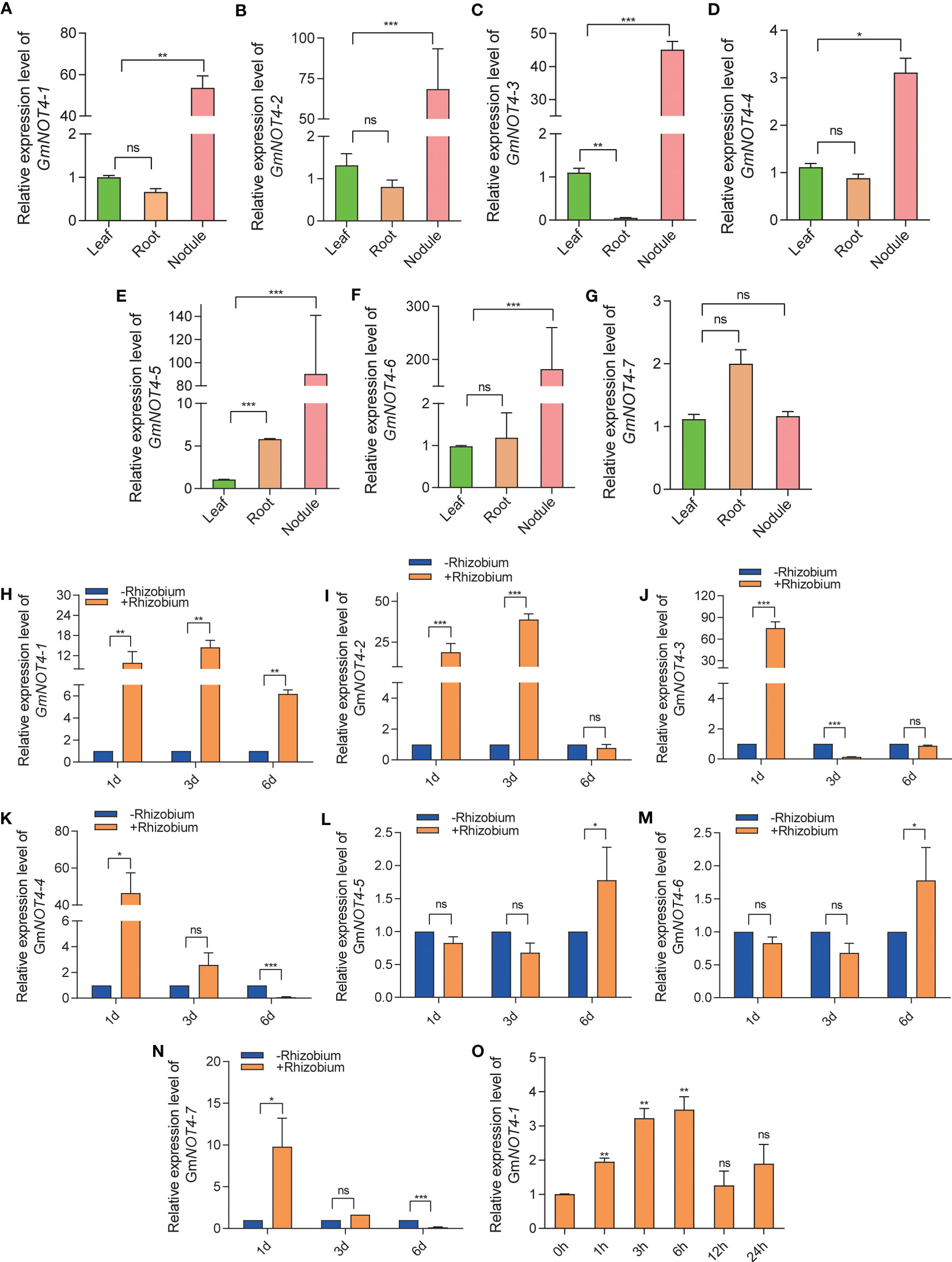
Figure 3 Expression pattern of soybean GmNOT4s. (A–G) show the relative expression level of GmNOT4-1, GmNOT4-2, GmNOT4-3, GmNOT4-4, GmNOT4-5, GmNOT4-6, and GmNOT4-7 in soybean leaf, root, and nodule at 28 DAI, respectively. (H–N) show the relative expression level of GmNOT4-1, GmNOT4-2, GmNOT4-3, GmNOT4-4, GmNOT4-5, GmNOT4-6, and GmNOT4-7 at 1, 3, and 6 DAI were validated by qPCR. (O) expression level of GmNOT4-1 in soybean roots at 0, 1, 3, 6, 12, and 24 HAI (hours after inoculation). GmCYP2 was used as an internal control. (A–O), n = 12, Student’s t-test; *P <0.05; **P <0.01; ***P <0.001, ns, no significance).
To genetically explore whether GmNOT4-1 is involved in the regulation of soybean nodulation, overexpression and knockout/knockdown analyses of GmNOT4-1 were performed using the hairy root transformation system. Firstly, we constructed 35S:GmNOT4-1 and obtained overexpressing GmNOT4-1 (GmNOT4-1-OX) roots by qPCR analysis (Figure 4A). The effects of GmNOT4-1 overexpression on the early and late stages of nodulation were evaluated at 1, 6, and 28 days after inoculation (DAI) (Figure S4). Firstly, the expression of rhizobial infection-related genes, including GmRPG, GmNPL, GmVPY, GmCYCLOPS, and GmSCARN, was validated. As shown in Figure S5, all rhizobial infection-related genes we checked in this study were significantly inhibited in GmNOT4-1 overexpressing roots. Then, we found the number of root hairs showing deformation was markedly decreased in GmNOT4-1-OX hairy roots at 6 DAI (Figure S6). Finally, the nodule numbers were quantified at 28 DAI. The average nodule number per GmNOT4-1 overexpressed root was about 3.6, while empty vector (EV) control roots produced an average of 11.8 nodules per root, with an approximately 69% reduction by GmNOT4-1 overexpression. These data suggest that GmNOT4-1 plays a negative role in regulating soybean nodulation (Figures 4B, C).
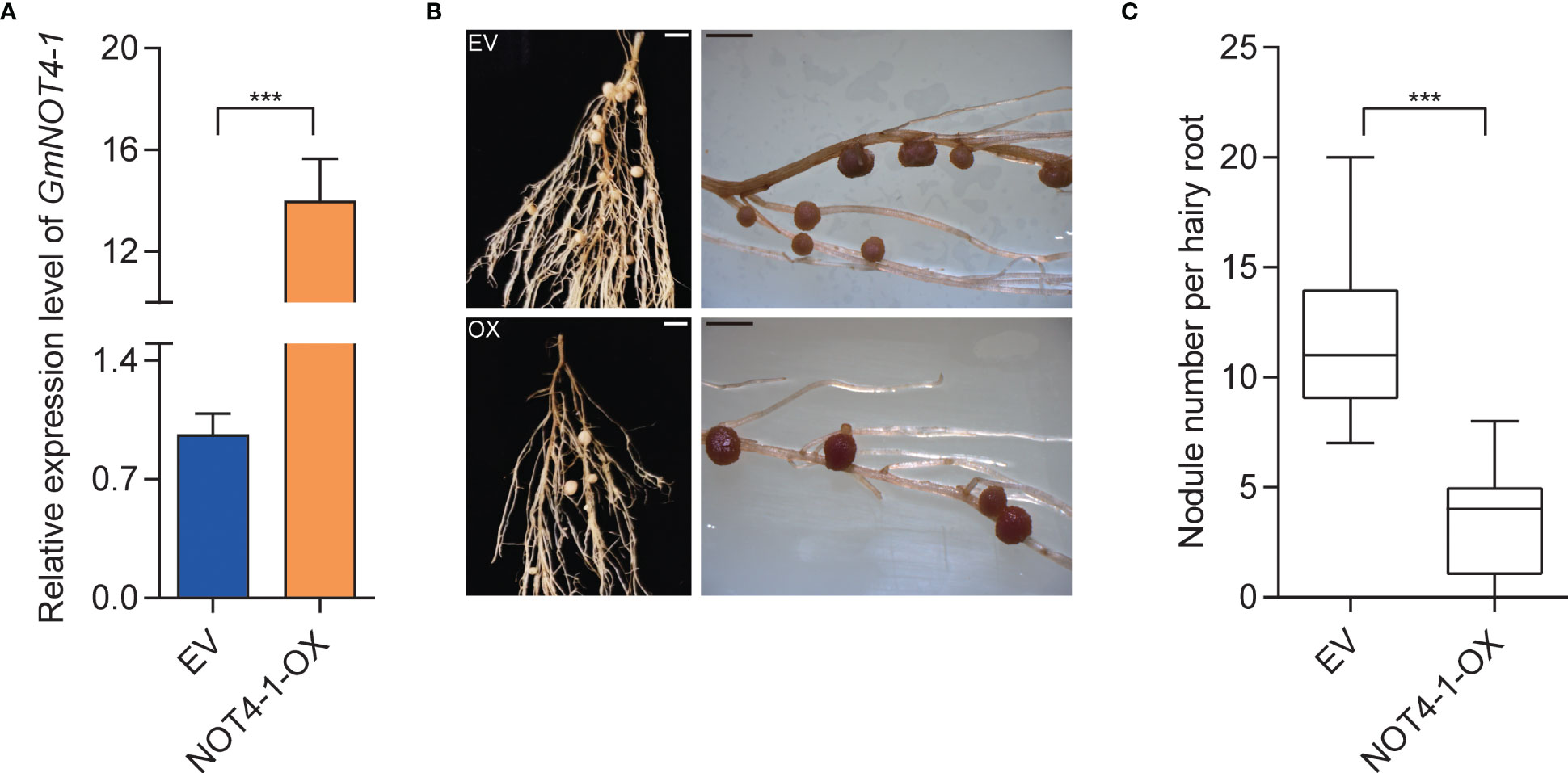
Figure 4 Phenotypic analysis of GmNOT4-1 overexpression. (A) Expression level of transgenic hairy roots harboring empty vector and 35S:GmNOT4-1. The expression levels were normalized against the housekeeping gene of soybean GmCYP2. Student’s t-test was performed (***p <0.001, n = 15). (B) Nodule status of individual transgenic roots expressing EV1 and 35S:GmNOT4-1 at 28 DAI. Bar = 2 mm. (C), Quantitative analysis of nodule number per hairy root carrying EV and 35S: GmNOT4-1 at 28 DAI. Values are the mean ± SD. A total of 20 hairy roots were collected for each biological replicate (n = 12, Student’s t-test; ***p <0.001).
To further validate the function of GmNOT4-1 during nodulation, we constructed the NOT4-1-RNAi to analyze the soybean nodulation phenotype when the expression of GmNOT4-1 was reduced (Figure S4). As shown in Figures 5B, C, transgenic roots harboringGmNOT4-1-RNAi produced fewer nodules (6.9 nodules per root) than empty vector (EV) control roots (19.5 nodules per root), so that the nodule number per GmNOT4-1-silenced root was reduced by approximately 64.6%. Further, the CRISPR/Cas9 system was also applied to knock out GmNOT4-1 in soybean hairy roots (Figures S8, S9), and the gene editing was verified by sequencing (Table S3). The expression of rhizobial infection-related genes, including GmRPG, GmNPL, GmVPY, GmCYCLOPS, and GmSCARN, was significantly inhibited in GmNOT4-1-KO roots. The number of deformed root hairs was markedly decreased in GmNOT4-1 edited knock-out hairy roots at 6 DAI compared to the vector control (Figure S6). Finally, we found that the GmNOT4-1 edited roots produce functional nodules (Figure S7) with a significantly reduced nodule number, which showed the same result as the GmNOT4-1-RNAi roots (Figures 5B, C).
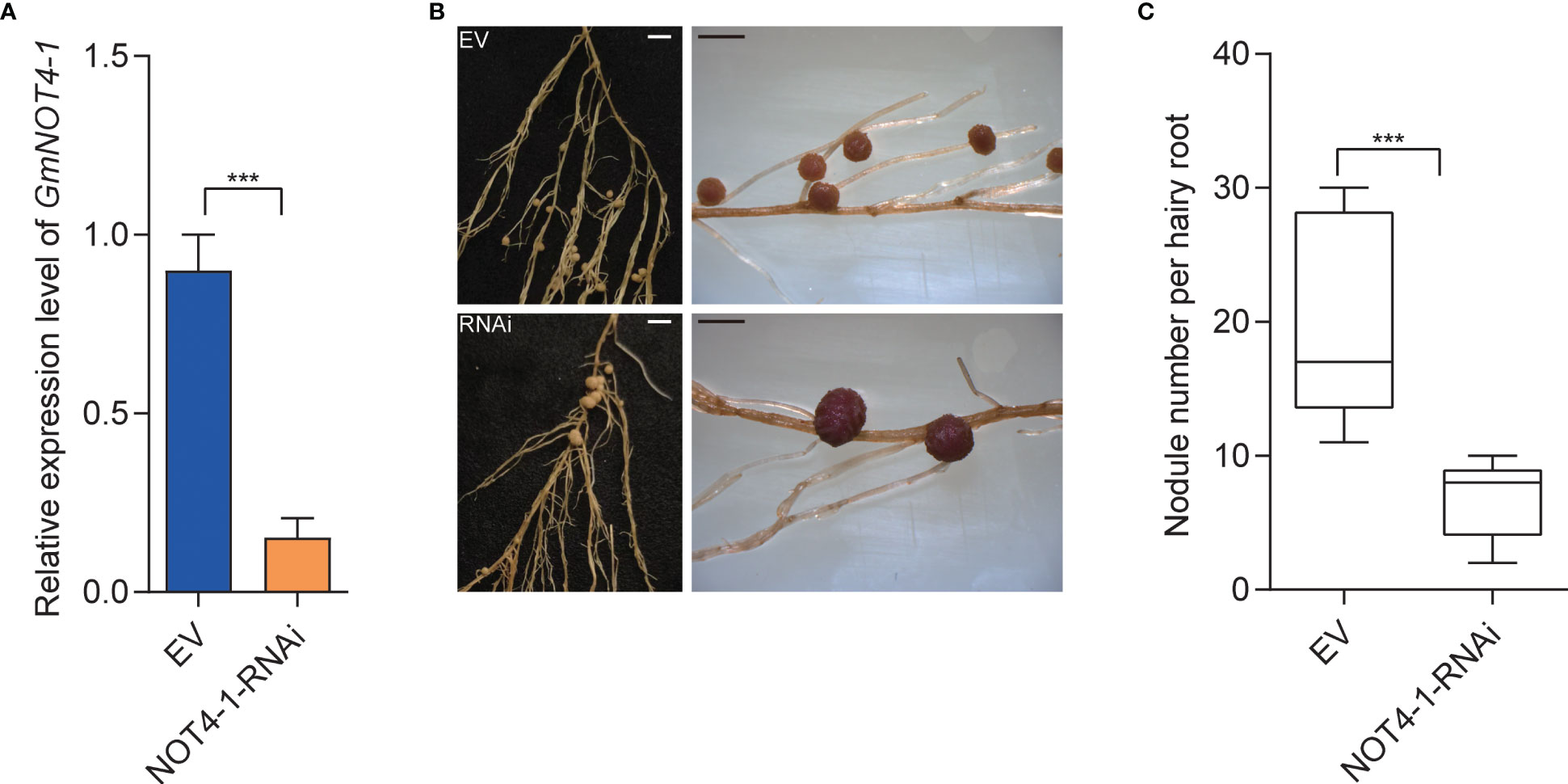
Figure 5 Knocking down GmNOT4-1 inhibits nodulation. (A) qRT-PCR analysis of transgenic hairy roots harboring empty vector and GmNOT4-1-RNAi. The expression levels were normalized against the housekeeping gene of soybean GmCYP2. Student’s t-test was performed (***p <0.001, n = 15). (B) Nodule status of individual transgenic roots expressing empty vector and GmNOT4-1-RNAi at 28 DAI. Bar = 2 mm. (C) Quantitative analysis of nodule number per hairy root carrying empty vector and GmNOT4-1-RNAi at 28 DAI. Values are the mean ± SD. A total of 20 hairy roots were collected for each biological replicate (n = 12, Student’s t-test; ***p <0.001).
Combined with the overexpression results, it is suggested that homeostasis of the expression of GmNOT4-1 is critical for the regulation of soybean nodulation.
Nodule number was mainly modulated by both the NF signaling and AON signaling pathways. Given the phenotype that either overexpression or knockdown (out) of GmNOT4-1 significantly decreased soybean nodule number, it is worth checking whether GmNOT4-1 regulates nodulation through the above pathways. Thus, we examined the expression level of several nodulation marker genes in soybean, including NF signaling pathway genes ENOD40, GmNINa, NSP1, HAP2-1, and HAP2-2, and AON signaling pathway genes GmRIC1/2. In general, we found that either overexpression or silence of GmNOT4-1 resulted in reduced expression of nodulation marker genes compared with the empty vector control roots (Figure 6).
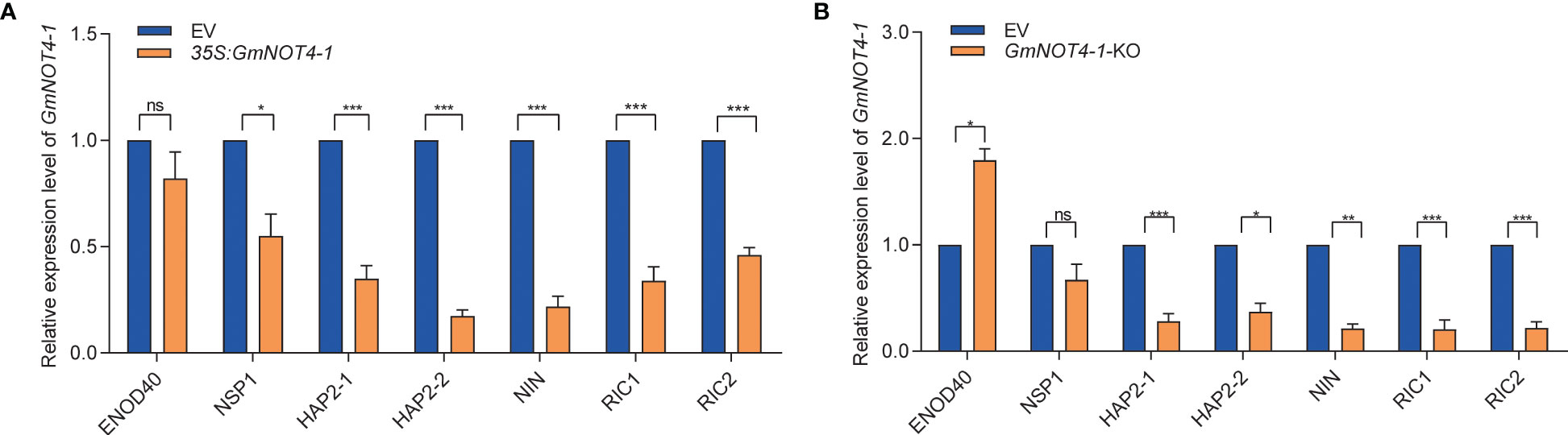
Figure 6 GmNOT4-1 expression alliterating inhibit the transcript levels of nodulation-related genes. (A) qRT-PCR analysis of ENOD40, GmNINa, NSP1, HAP2-1, HAP2-2, GmRIC1, and GmRIC2 in roots transformed with empty vector and GmNOT4-1 at 6 DAI (n = 6). (B) qRT-PCR analysis of ENOD40, GmNINa, NSP1, HAP2-1, HAP2-2, GmRIC1, and GmRIC2 in roots transformed with empty vector and GmNOT4-1 knock out at 6 DAI (n = 6). We set transcript level of the ENOD40, GmNINa, NSP1, HAP2-1, HAP2-2, GmRIC1, and GmRIC2 at 6 DAI EV hairy roots as “1.” The transcript amounts in each sample were normalized to those of GmCYP2 (n = 6, Student’s t-test; *p <0.05, **p <0.01, and ***p <0.001; ns, no significance).
Legumes plants can specifically interact with the phylogenetically diverse group of soil bacteria-rhizobia to form nodules. However, symbiotic nitrogen fixation is a highly energy-intensive biological process; thus, host legumes have evolved a root–shoot–root long-distance auto-regulation of nodulation (AON) system to refine the number of nodules (Patra et al., 2017; Ren et al., 2019). NODULE INCEPTION (NIN) induces the expression of CLE ROOT SIGNAL1 (CLE-RS1) and CLE-RS2 to activate AON to inhibit excessive nodulation (Chun et al., 2005). NIN also modulates almost all nodulation processes, including nodule initiation, nodule organogenesis, and nitrogen fixation. Thus, NIN can act as a bifunctional transcription factor, fine-tuning legume nodulation. Interestingly, in this study, we observed that GmNOT4-1 also function as a biofunctional regulator in nodulation, both overexpression and CRISPR/Cas9 knock out of GmNOT4-1 inhibited the number of nodules in soybean. Except for GmNOT4-1, we found all GmNOT4s responded to rhizobium at least at one time point (Figure 3). We do not exclude the possibility that different family members might have a role in a particular stage of root nodule symbiosis.
In previous studies, a series of transcriptional factors and microRNAs were identified (NIN, NSP1/2, miR172c, etc.) that modulate the expression of some key nodulation regulators (ERN1, ENOD40, RICs, etc.) and finally legume nodule number. In eukaryotic cells, except for transcriptional factors and miRNAs, the multiprotein complex CCR4–NOT also plays a vital role in regulating gene expression via the shortening of poly(A) tails of messenger RNA. In this study, we identified GmNOT4-1, one component of the soybean CCR4–NOT complex, whose expression homeostasis is important for soybean nodule initiation and subsequent nodule number (Figures 4, 5; S4-S6). We found that either overexpression or silencing of GmNOT4-1 resulted in reduced expression of nodulation marker genes compared with empty vector control roots (Figure 6). RIC1/2 was induced by nodule primordial formation to inhibit excessive nodulation; GmNOT4-1 may function before nodule primordial formation; thus, the resulting RIC1/2 induction and AON signaling were blocked by the overexpression or silencing of GmNOT4-1. Further study is needed to clarify the other components of the CCR4–NOT complex, including GmNOT4 family members in nodulation, and construct a direct link between the CCR4–NOT complex and nodulation signaling pathway genes. In addition, GmNOT4-1 encodes a RING domain containing an E3 ligase; it may function by adjusting substrate protein levels. Identification of the target of GmNOT4-1 can further unveil its role in the process.
In this research, we identified seven members of the NOT4 family in soybeans and found that GmNOT4-1 was mainly expressed in soybean nodules. Interestingly, we observed that both overexpression and downregulation of GmNOT4-1 inhibited the number of nodules in soybean. The CRISPR/Cas9 system was applied to validate this phenotype. Finally, we demonstrated that alterations in GmNOT4-1 expression level repressed the expression of genes in the Nod factor signaling pathway. To our best knowledge, this is the first research to study the CCR4–NOT complex in legume nodulation. Like the founder transcription factor NIN (NODULE INCEPTION), essential for nodulation, the gene expression level of NIN was rigidly regulated for different processes of nodulation, including rhizobial infection, nodule organogenesis, and AON signaling. GmNOT4-1 showed the same phenotype pattern as the NIN gene; overexpression and knockdown (out) both inhibit the proper nodulation. The further detailed phenotype and mechanism study will provide a better understanding of GmNOT4-1 and its roles in nodulation.
This research will provide novel insight into the function of the CCR4–NOT family in legumes and reveal GmNOT4-1 to be a valuable gene in regulating symbiotic nodulation.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
WLX, GSJ, and DWJ designed and conceived the study. ZJT cloned the gene and constructed the vector, grew the seedlings, and harvested them for quantitative detection. SLL completed the bioinformatics analysis. WDM and HL participated in the expression analysis. All authors contributed to the article and approved the submitted version.
This work is supported by the Natural Science Foundation of Henan Province (Grant No. 202300410056), the Science and Technology Innovation Young Talent Team of Shanxi Province (202204051001020), the Basic Research Program of Shanxi Province (20210302123365, 202103021224146), the National Natural Science Foundation of China (Grant No. 32241046), the National Key Research and Development Program (Grant No. 2021YFD1600605-10), and the National Laboratory of Minor Crops Germplasm Innovation and Molecular Breeding (In preparation) (Grant Nos. 202105D121010-23 and 202204010910001-33).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1172354/full#supplementary-material
Supplementary Figure 1 | Chromosomal distribution of NOT4 gene family members 7 GmNOT4s were identified from soybean genome and named GmNOT4-1 to GmNOT4-7 according to their distribution positions on chromosomes.
Supplementary Figure 2 | Motif distribution of GmNOT4s NOT4 Protein sequences of Medicago truncatula (Mt), Phaseolus vulgaris (Pv), Arabidopsis thaliana (At), Glycine max (Gm) and Oryza sativa (Os) were subjected into MEME online software to identify conserved motifs. 10 motifs were found in NOT4s genes and represented by different colored boxes and corresponding motif number, conserved amino acid sequence themes were shown below. (The left panel) shown the phylogenetic tree of group I~III genes in purple, blue, and orange colors; (The middle panel) shown the conserved motifs distribution in NOT4s; (The right panel) shown the gene structure of untranslated regions (UTRs) of the NOT4s gene are shown as green, yellow boxes and black lines.
Supplementary Figure 3 | Cis-acting elements of soybean GmNOT4s gene promoter Promoter sequences (2000 base pair upstream of start codon site) of GmNOT4s were submitted to PlantCARE database to identify cis-acting elements. Boxes with different colors indicate various cis-acting elements located in the promoter regions of GmNOT4s. Abbreviations: AT~TATA-box, CAAT-box, TATA-box, TATA: core cis-elements; Box 4, GT1-motif, ACE, G-box: optical response element; CAT-box, CCGTCC-box: associated with meristem expression; circadian: circadian rhythm control; GCN4-Motif: endosperm expression; HD-Zip1: cell differentiation in palisade tissue; AC-I: xylem synthesis; MBSI: flavonoid biosynthesis; TGA-element: Auxin responsive element; ABRE, AAGAA-motif, ABRE3a: abscisic acid reaction; TCA, TCA-element: salicylic acid responsive elements; P-box, GARE-motif: gibberellin response element; TGACG-Motif, CGTCA-motif: methyl jasmonate responsive elements; ERE: ethylene response element; ARE: anaerobic induction element; MYB, Myb-binding site, MYC, MBS: Drought induction elements; LTR: low temperature element; TC rich repeats, CCAAT-box, as-1, STRE: stress reaction elements; WRE3, WUN-motif, W box: traumatic stress elements; A-box, CARE, CCGTCC motif, AT-rich sequence: unknown function.
Supplementary Figure 4 | No combined and off-target effects existed in GmNOT4-1 overexpression or knoct-out roots (A), qRT-PCR analysis of GmNOT4-2, GmNOT4-3, GmNOT4-4, GmNOT4-5, GmNOT4-6 and GmNOT4-7 in roots transformed with empty vector and GmNOT4-1 overexpression at 6 DAI (n = 6). (B), qRT-PCR analysis of GmNOT4-2, GmNOT4-3, GmNOT4-4, GmNOT4-5, GmNOT4-6 and GmNOT4-7 in roots transformed with empty vector and GmNOT4-1-KO at 6 DAI (n = 6). We set transcript level of the GmNOT4-2, GmNOT4-3, GmNOT4-4, GmNOT4-5, GmNOT4-6 and GmNOT4-7 at 6 DAI EV hairy roots as “1”. The transcript amounts in each sample were normalized to those of GmCYP2 (n = 12, Student’s t-test; “ns” =No significance).
Supplementary Figure 5 | GmNOT4-1 regulate nodulation though affecting rhizobial infection (A), qRT-PCR analysis of GmNPL, GmRPG, GmVPY, GmCYCLOPS and GmSCARN in roots transformed with empty vector and GmNOT4-1 overexpression at 6 DAI (n = 6). (B), qRT-PCR analysis of GmNPL, GmRPG, GmVPY, GmCYCLOPS and GmSCARN in roots transformed with empty vector and GmNOT4-1-KO at 6 DAI (n = 6). The transcript amounts in each sample were normalized to those of GmCYP2 (n = 12, Student’s t-test; *p < 0.05, **p < 0.01, and ***p < 0.001).
Supplementary Figure 6 | GmNOT4-1 overexpression and knock out decreases the number of deformed root hairs (A), Expression level of transgenic hairy roots harboring empty vector and 35S:GmNOT4-1. The expression levels were normalized against the housekeeping gene of soybean GmCYP2. Student’s t-test was performed (***p < 0.001, n = 20). (B), At 6 DAI, 2 cm root segments of hairy roots overexpressing GmNOT4-1 or expressing EV below the root-hypocotyl junction were cut and stained with 1% (w/v) methylene blue. Deformed root hairs were counted (n=20). (B) Root hair deformation in transgenic roots harboring EV and 35S:GmNOT4-1 vector. Bar=40 μm. (C), Quantification of deformed root hairs in the transgenic lines (n=10 to 12). Values are averages ± SD from three independent experiments. Asterisks represent statistically significant differences. (n = 20, Student’s t-test; ***p < 0.001). (D), Expression level of transgenic hairy roots harboring empty vector and GmNOT4-1-KO. The expression levels were normalized against the housekeeping gene of soybean GmCYP2. Student’s t-test was performed (***p < 0.001, n = 20). (E), Root hair deformation in transgenic roots harboring EV and GmNOT4-1-KO. Bar=40 μm. (F), Quantification of deformed root hairs in the transgenic root harboring EV and GmNOT4-1-KO (n=20). Values are averages ± SD from three independent experiments. Asterisks represent statistically significant differences. (n = 20, Student’s t-test; ***p < 0.001).
Supplementary Figure 7 | Nodules of GmNOT4-1 overexpression and RNAi roots are functional Nodule performance of mature nodule in roots expressing empty vector, overexpression, and GmNOT4-1-KO at 28 DAI. Bar =2 mm.
Supplementary Figure 8 | Phenotypic analysis of GmNOT4-1 gene editing (A), Relative expression level of GmNOT4-1 in GmNOT4-1 edited roots; The expression levels were normalized against the housekeeping gene of soybean GmCYP2. Student’s t-test was performed (***p < 0.001, n = 15). (B), Nodule performance of individual transgenic roots expressing empty vector and GmNOT4-1-KO at 28 DAI. Bar =2 mm. (C), Quantitative data of nodule number per hairy root carrying empty vector and GmNOT4-1-KO at 28 DAI. Values are the mean ± SD. 20 hairy roots were collected for each biological replicate. (Student’s t-test; ***p < 0.001).
Supplementary Figure 9 | Alignment of GmNOT4-1 gRNAs with other GmNOT4 family members (A), The front gRNA sequence aligned with the corresponding region of other GmNOT4 family members. (B), The back gRNA sequence aligned with the corresponding region of other GmNOT4 family members, the software Daneman was applied for the sequence alignment.
Andriankaja, A., Boisson-Dernier, A., Frances, L., Sauviac, L., Jauneau, A., Barker, D. G., et al. (2007). AP2-ERF transcription factors mediate nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19 (9), 2866–2885. doi: 10.1105/tpc.107.052944
Carroll, R. K., Weiss, A., Broach, W. H., Wiemels, R. E., Mogen, A. B., Rice, K. C., et al. (2016). Genome-wide annotation, identification, and global transcriptomic analysis of regulatory or small RNA gene expression in staphylococcus aureus. mBio 7 (1), e01990–e01915. doi: 10.1128/mBio.01990-15
Chou, M. X., Wei, X. Y. (2010). Review of research advancements on the molecular basis and regulation of symbiotic nodulation of legumes. Chin. J. Plant Ecol. 7), 876–888. doi: 10.3773/j.issn.1005-264x.2010.07.013
Chun, L., Mi, G. H., Li, G. S., Chen, F. G., Zhang, F. S. (2005). Genetic analysis of maize root characteristics in response to low nitrogen stress. Plant Soil 276 (1-2), 369–382. doi: 10.1007/s11104-005-5876-2
Fonouni-Farde, C., Diet, A., Frugier, F. (2016). Root development and endosymbioses: DELLAs lead the orchestra. Trends Plant Sci. 21 (11), 898–900. doi: 10.1016/j.tplants.2016.08.012
Fujita, H., Kawaguchi, M. (2002). A lotus basic leucine zipper protein with a RING-finger motif negatively regulates the developmental program of nodulation. Proc. Natl. Acad. Sci. United States America 99 (No.23), 15206–15210. doi: 10.1073/pnas.222302699
Gao, L. F., Hu, Z. A., Wang, H. X., Yang, W. L. (2002). Structure and function of Rhizobium Nod factors. Chinese Bulletin of Life Sci. (01), 17–19.
Heckmann, A. B., Sandal, N., Bek, A. S., Madsen, L. H., Jurkiewicz, A., Nielsen, M. W., et al. (2011). Cytokinin induction of root nodule primordia in lotus japonicus is regulated by a mechanism operating in the root cortex. Mol. Plant-Microbe Interact. 24 (No.11), 1385–1395. doi: 10.1094/mpmi-05-11-0142
Horváth, B., Yeun, L. H., Domonkos, A., Halász, G., Gobbato, E., Ayaydin, F., et al. (2011). Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Molecular Plant-Microbe Interactions : MPMI 24 (11), 1345. doi: 10.1094/MPMI-01-11-0015
Howard, J. (2010). A phylogenetic approach to examining symbiotic specificity and evolution in the legume-rhizobia nitrogen-fixing symbiosis. Diss. Arizona State University.
Jia, Z. R., Zhang, M. J., Yang, W. D. (2017). Effects of nitrogen fertilizer on nitrogen uptake and utilization of intercropped maize. Shanxi Agric. Sci. 45 (12), 1960–1964. doi: 10.3969/j.issn.1002-2481.2017.12.16
Jian, B., Liu, B., Bi, Y., Hou, W., Wu, C., Han, T. (2008). Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol. Biol. 9 (1), 1–14. doi: 10.1186/1471-2199-9-59
Jin, Y., Liu, H., Luo, D., Yu, N., Dong, W., Wang, C., et al. (2016). DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat. Commun. 7, 12433. doi: 10.1038/ncomms12433
Kaló, P., Gleason, C., Edwards, A., Marsh, J., Mitra, R. M., Hirsch, S., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Sci. (New York N.Y.) 308 (No.5729), 1786–1789. doi: 10.1126/science.1110951
Kiss, E., Olaíh, B. R., Kaloí, P. T., Morales, M., Heckmann, A. B., Borbola, A., et al. (2010). LIN, a novel type of U-box/WD40 protein, controls early infection by rhizobia in legumes. Plant Physiol. 151 (No.3), 1239–1249. doi: 10.1104/pp.109.143933
Laloum, T., De Mita, S., Gamas, P., Baudin, M., et al. (2013). CCAAT-box binding transcription factors in plants: y so many? Trends Plant Sci. 18 (No.3), 157–166. doi: 10.1016/j.tplants.2012.07.004
Liang, W., Li, C., Liu, F., Jiang, H., Li, S., Sun, J., et al. (2008). The arabidopsis homologs of CCR4-associated factor 1 show mRNA deadenylation activity and play a role in plant defence responses. Cell Res. 19, 307–316. doi: 10.1038/cr.2008.317
Middleton, E. T., Steel, S. A., Doherty, S. M. (2007). The effect of prior bisphosphonate exposure on the treatment response to teriparatide in clinical practice. Calcif Tissue Int. 81 (5), 335–340. doi: 10.1007/s00223-007-9066-5
Nishimura, R., Ohmori, M., Kawaguchi, M. (2002). The novel symbiotic phenotype of enhanced-nodulating mutant of lotus japonicus: astray mutant is an early nodulating mutant with wider nodulation zone. Plant Cell Physiol. 43 (No.8), 853–859. doi: 10.1093/pcp/pcf098
Oldroyd, G. E. D., Downie, J. A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59 (No.1), 519–546. doi: 10.1146/annurev.arplant.59.032607.092839
Patra, R. K., Pant, L. M., Pradhan, K. (2017). Response of soybean to inoculation with rhizobial strains: effect on growth, yield, n uptake and soil n status. World J. Agric. Sci. 8 (1), 28–31. doi: 10.11942/j.issn1002-2767.2017.08.0028
Ren, B., Wang, X., Duan, J., Ma, J. (2019). Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science 365 (6456), 919–922. doi: 10.1126/science.aav8907
Shimomura, K., Nomura, M., Tajima, S., Kouchi, H. (2006). LjnsRING, a novel RING finger protein, is required for symbiotic interactions between mesorhizobium loti and lotus japonicus. Plant &Cell Physiol. 47 (No.11), 1572–1581. doi: 10.1093/pcp/pcl022
Smit, P., Raedts, J., Portyanko, V., Debellé, F., Gough, C., Bisseling, T., et al. (2005). NSP1 of the GRAS protein family is essential for rhizobial nod factor-induced transcription. Sci. (New York N.Y.) 308 (No.5729), 1789–1791. doi: 10.1126/science.1111025
Suzuki, Y., Arae, T., Green, P. J., Yamaguchi, J., Chiba, Y. (2015). AtCCR4a and AtCCR4b are involved in determining the Poly(A) length of granule-bound starch synthase 1 transcript and modulating sucrose and starch metabolism in arabidopsis thaliana. Plant Cell Physiol. 56 (No.5), 863–874. doi: 10.1093/pcp/pcv012
Vernié, T., Camut, S., Camps, C., Rembliere, C., Carvalho-Niebel, F., Mbengue, M., et al. (2016). PUB1 interacts with the receptor kinase DMI2 and negatively regulates rhizobial and arbuscular mycorrhizal symbioses through its ubiquitination activity in Medicago truncatula. Plant Physiol. 170 (No.4), 2312–2324. doi: 10.1104/pp.15.01694
Walley, J. W., Kelley, D. R., Nestorova, G. (2010). Arabidopsis deadenylases AtCAF1a and AtCAF1b play overlapping and distinct roles in mediating environmental stress responses. Plant Physiol. 152 (No.2), 866–875. doi: 10.1104/pp.109.149005
Wang, L., Sun, Z., Su, C., Wang, Y., Yan, Q., Chen, J., et al. (2019). A GmNINa-miR172c-NNC1 regulatory network coordinates the nodulation and autoregulation of nodulation pathways in soybean. Mol. Plant 12 (9), 1211–1226. doi: 10.1016/j.molp.2019.06.002
Wang, Y., Wang, L., Zou, Y., Chen, L., Cai, Z., Zhang, S., et al. (2014). Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. PLANTCELL 26 (No.12), 4782–4801. doi: 10.1105/tpc.114.131607
Keywords: legume symbiotic nitrogen fixation, Ccr4-not complex, root nodule (symbiotic), CRISPR/Cas9, nod factor signalling
Citation: Zheng J, Sun L, Wang D, He L, Du W, Guo S and Wang L (2023) Roles of a CCR4–NOT complex component GmNOT4-1 in regulating soybean nodulation. Front. Plant Sci. 14:1172354. doi: 10.3389/fpls.2023.1172354
Received: 23 February 2023; Accepted: 02 May 2023;
Published: 05 June 2023.
Edited by:
Meng Kou, Xuzhou Institute of Agricultural Sciences in Jiangsu Xuhuai District, ChinaReviewed by:
Oswaldo Valdes-Lopez, National Autonomous University of Mexico, MexicoCopyright © 2023 Zheng, Sun, Wang, He, Du, Guo and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lixiang Wang, bHh3YW5nQHN4YXUuZWR1LmNu; Shujin Guo, c3huZGdzakAxNjMuY29t
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.