
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 15 May 2023
Sec. Plant Biotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1169984
This article is part of the Research Topic Plant Biotechnology and Genetics for Sustainable Agriculture and Global Food Security View all 12 articles
Introduction: Empirical research has refined traditional herbal medicinal systems. The traditional market is expanding globally, but inadequate regulatory guidelines, taxonomic knowledge, and resources are causing herbal product adulteration. With the widespread adoption of barcoding and next-generation sequencing, metabarcoding is emerging as a potential tool for detecting labeled and unlabeled plant species in herbal products.
Methods: This study validated newly designed rbcL and ITS2 metabarcode primers for metabarcoding using in-house mock controls of medicinal plant gDNA pools and biomass pools. The applicability of the multi-barcode sequencing approach was evaluated on 17 single drugs and 15 polyherbal formulations procured from the Indian market.
Results: The rbcL metabarcode demonstrated 86.7% and 71.7% detection efficiencies in gDNA plant pools and biomass mock controls, respectively, while the ITS2 metabarcode demonstrated 82.2% and 69.4%. In the gDNA plant pool and biomass pool mock controls, the cumulative detection efficiency increased by 100% and 90%, respectively. A 79% cumulative detection efficiency of both metabarcodes was observed in single drugs, while 76.3% was observed in polyherbal formulations. An average fidelity of 83.6% was observed for targeted plant species present within mock controls and in herbal formulations.
Discussion: In the present study, we achieved increasing cumulative detection efficiency by combining the high universality of the rbcL locus with the high-resolution power of the ITS2 locus in medicinal plants, which shows applicability of multilocus strategies in metabarcoding as a potential tool for the Pharmacovigilance of labeled and unlabeled plant species in herbal formulations.
The herbal commodity market is thriving, owing to the widespread belief that traditional medicine is natural and thus safer, thereby promoting good health and sustainable life policies. But as the market expands, the shortage of genuine resources and lack of taxonomic knowledge challenge the authenticity of herbal drugs and increase the incidence of economically motivated or unintentional adulterations and/or substitutions (Raclariu et al., 2018a). Strict pharmacovigilance is necessary to retain the trust and safety of consumers and their health. However, the regulatory guidelines for medicinal plants often blur the line between foods and therapeutics and vary from nation to nation. To close this research gap, regulatory bodies must implement more reliable, universal, and robust detection methods (Shetti et al., 2011).
Nowadays, various pharmacopeia advocate DNA-based methods such as DNA barcoding and species-specific PCR assay to authenticate herbal raw material. DNA is more stable, unaffected by external factors, and invariably present in almost all plant tissues (Wu and Shaw, 2022). In addition, the DNA-based results are independent of seasonal variations in the age of the plant, which in the case of chemical marker-based methods vary significantly (Parveen et al., 2016). Therefore, the results of DNA-based methods are free from subjectivity, accurate, and provide a universally accepted platform for the authentication of botanicals in a wide range of food and herbal products (Lo and Shaw, 2019). The advent of DNA barcoding is the first step in this direction, as barcoding gives resolution up to species level (Hebert et al., 2003). There are 17 potential barcode regions (matK, rbcL, ITS, ITS2, psbA-trnH, atpF-atpH, ycf5, psbKI, psbM-trnD, coxI, nad1, trnL-F, rpoB, rpoC1, atpF-atpH, and rps16) for plants, having different degrees of universality, specificity, and taxa resolution power that extensively used in the authentication and identification of medicinal plants (Parveen et al., 2016; Kress, 2017). However, DNA barcoding (Vassou et al., 2016) and species-specific assays (Sharma and Shrivastava, 2016; Travadi et al., 2022a; Travadi et al., 2022b) cannot resolve presence of multiple plant species in a single sample (Mishra et al., 2016), that limitation could be overcome by DNA metabarcoding.
DNA metabarcoding combined the strengths of next-generation sequencing and barcoding for detecting multiple taxa in samples (Coghlan et al., 2012). Using a single plant barcode for species-level identification has proven challenging due to the great diversity, relatively slow molecular evolution, frequent cross-pollinations, and hybridization in the plant kingdom; henceforth, different barcodes show different degrees of taxon specificity (Fazekas et al., 2009). To precisely identify the plant species in the sample, multi-barcode approaches have become more prevalent. Xin et al. (2018) and Frigerio et al. (2021) employed a multi-barcode approach of ITS2 and trnL for various Chinese medicine and herbal teas. However, these studies also highlighted the limitations of DNA metabarcoding applications for authentication of herbal products due to variability in degrees of universality and resolution power of barcodes for specific taxa, a lack of a curated database, and a robust bioinformatics pipeline. To overcome these constraints, there is a need for screening of new barcodes and new variable regions within the same barcode for authentication of the herbal products.
Therefore, the aims of the present study were: 1) to develop a new rbcL and ITS metabarcode for the detection of medicinal plant species, 2) to validate the primers specificity and efficiency using mock controls and 3) to see whether a multi-barcoding approach could be used for the pharmacovigilance of the herbal products? (17 different single plant formulation and 15 polyherbal market formulations in this study), and 3) to see whether a multi-barcoding approach could detect targeted plant species in herbal formulations?
Reference plant materials were collected with the aid of a taxonomist from the Maharaja Sayajirao University (MSU), Vadodara (Gujarat, India) and the Directorate of Medicinal and Aromatic Plants Research (DMAPR), Anand (Gujarat, India). As described earlier, reference plant materials were authenticated by Sanger sequencing of rbcL gene (Pandit et al., 2021), and sequences were submitted to the NCBI database (accession number MW628906 to MW628936). Voucher specimens were developed and deposited in our institutional herbarium.
32 herbal products were collected by blind sampling from the local market and e-commerce, with 17 single drugs and 15 polyherbal formulations. Single drugs include four Tulsi (Ocimum tenuiflorum) powders, five Gokhru (Tribulus terrestris) powders, three Shatavari (Asparagus racemosus) powders, two Vasa (Justicia adhatoda) products, and one each of Bhringraj (Eclipta alba), Ashwagandha (Withania somnifera), and Arjuna (Terminalia arjuna) powder. Polyherbal formulations include three market samples of Trikatu (has three plant species), three samples of Sitopladi [comprises five constituents, only three of which are plant species; the other two are sugar and Vanshlochan (the female bamboo exudate), hence these two constituents were not considered while analyzing the data expecting absence of DNA for these two], four samples of Rasayana (has three plant species), four samples of Hingwashtak (has seven plant species), and one sample of Talisadi [comprises eight constituents, only six of which are plant species; the other two are sugar and Vanshlochan (the female bamboo exudate), hence these two constituents were not considered while analyzing the data expecting absence of DNA for these two].
To design the metabarcodes for ITS2 gene, ITS2 sequences of Magnoliophyta from the BOLD database were downloaded and curated, particularly for the length. For the rbcL gene, we used 1,776 sequences of our in-house sequencing project submitted to the BOLD database. To design universal barcodes, rbcL gene sequences were filtered by length between 450-600 bp. At the end, 1,465 and 1,701 ITS2 and rbcL sequences were retained to design the metabarcode. These sequences were preceded for multiple sequence alignment separately (ITS2 and rbcL) using BioEdit 7.2. HYDEN (HighlY DEgeNerate primers) software (Linhart and Shamir, 2005) to design degenerate primers, where the maximum of 3 degeneracy per primer was allowed. The designed primers were checked for amplicon length using NCBI primer BLAST (Altschul et al., 1990). rbcL reverse primer sequence was designed in this study, while forward primer sequence was obtained from Maloukh et al. (2017). To synthesize fusion primers, forward primers of rbcL and ITS2 were tagged with the Ion torrent adapter and a ten bp multiplex identifier barcode. In contrast, reverse primers were tagged with the P1 adapter. Nucleotide sequence of the designed primers and their amplicon length are shown in Table 1.
The library preparation process became a single-step process with barcoded fusion primers. The PCR optimization with each barcoded fusion primer was done with 45 different plant DNA listed in Supplementary Information Table S1. Thermal cycler conditions, especially primer annealing temperature, were optimized for rbcL and ITS2 primer pairs with the following conditions. PCR mixture containing 10 µL Emerald Master mix (2X) (TaKaRa), 2 µL total genomic DNA (10-15 ng/µL), 1 µL of forward (5 pmol), 1 µL of reverse primer (5 pmol), 1 µL BSA (2 mg/mL) and 5 µL PCR grade water with the following thermal cycling conditions. Initial denaturation 95°C for 5 minutes, followed by 30 cycles of 95°C for 1 minute, for primer annealing a temperature gradient of 50°C to 60°C with an interval of 2°C for 30 seconds and 72°C for 1 minute, and final extension 72°C for 5 minutes. 2.4 Preparation of different mock controls
Three different types of controls were prepared as follows: Control 1) genomic DNA from plant leaves from different genera belonging to diverse families has been first isolated and pooled into three different groups as mentioned below, Control 2) simulated plant biomass controls (blended formulations) in which individual plant part having medicinal value has been mixed in three groups as control one and subjected to DNA isolation, and Control 3) genomic DNA (Isolated from plant leaves) pool from different species of the two genera (Figure 1). As mentioned above, three different groups were prepared for the first type of control with the plant species of a different genus. Group one comprised DNA of five species in equal proportion (5P), and further, in groups 2 and 3, DNA was added from ten (10P) and fifteen different plant species (15P) (Figure 1, 2A). High-quality DNA of all the species have been isolated individually from leaf tissue and pooled together in equal proportion to make these groups. The group’s diversity has been increased by adding species from diverse genera that belong to diverse families to evaluate the resolution power and universality of the primers for the maximum number of species. For the second type of control, the same three groups of plants were used in the first controls (labeled as 5S, 10S, 15S). Still, simulated blended plant parts containing bioactive therapeutics were mixed in equal proportion (biomass admixture controls). These controls can be used to comprehend biases introduced during the DNA extraction and PCR dynamics under the influence of secondary metabolites on PCR amplification. For the third type of control, two groups were prepared. Group one comprises six plant species of the two genera, including Asparagus and Terminalia (Figure 2B). The second group includes seven plant species of the two genera, including Piper and Phyllanthus (Figure 2B). Similar to the first control, high-quality DNA was individually isolated from each species and pooled in equal amounts. These controls were utilized to obtain insight into the resolving strength of our newly designed rbcL and ITS2 metabarcodes at lower taxa levels.
Figure 1 Schematic representation of different types of mock controls prepared in this study. First and second type of mock controls has 3 different groups comprising 5, 10, and 15 plant species. Third type of mock control has 2 different groups, one with a gDNA pool of six plant species from genera Asparagus and Terminalia and another with a gDNA pool of seven plant species from genera Piper and Phyllanthus.
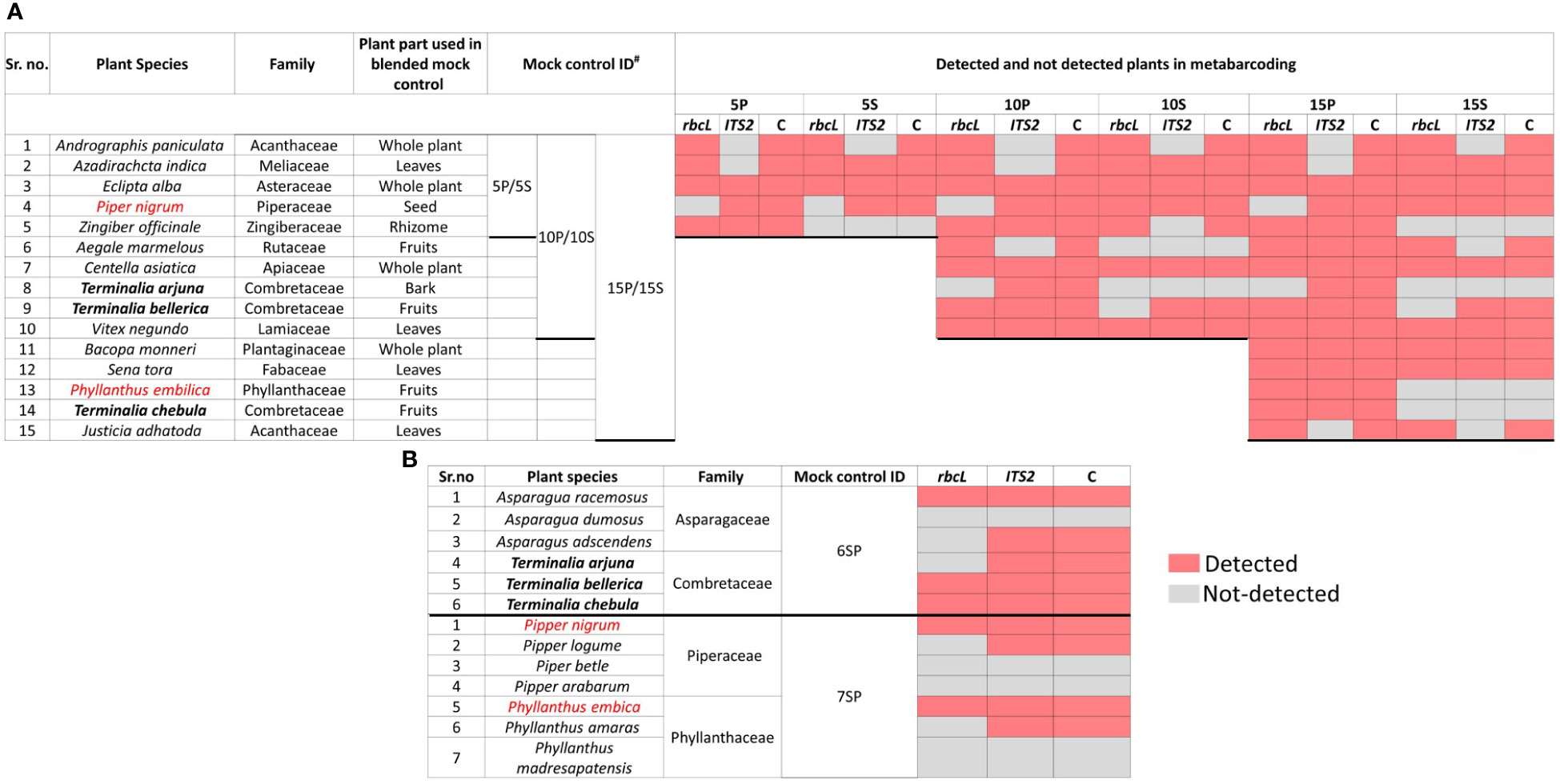
Figure 2 The distribution of predefined herbal species detected in each mock control with rbcL, ITS2, and combined metabarcoding approach. (A) Detected and undetected plant species with rbcL, ITS2, and combined approach in type one mock control, i.e., genomic DNA pool of different genera, and type two mock control, i.e., simulated plant biomass control (blended formulations) comprising three groups having five plant species (5P/5S), ten plant species (10P/10S) and 15 plant species (15P/15S). (B) Detected and undetected plant species with rbcL, ITS2, and combined approach in a third type of mock controls (i.e., genomic DNA pooled from different species of the genus) comprising two different groups, one having six plant species (6SP) from genera Asparagus and Terminalia and another having seven plant species (7SP) from genera Piper and Phyllanthus. Detected plant species are represented in pink, while undetected plant species are represented in grey. Plant species, their family, plant parts used in type two mock controls (simulated plant biomass controls or blended formulations), and mock control IDs are also indicated in the figure. P: gDNA plant pool controls, S: simulated blended plant pool (biomass control), C: a combined approach.
DNA from plant materials, blended formulations, and herbal products ‘was extracted in duplicate using DNeasy Plant Mini Kit (QIAGEN, Germany) following themanufacturer’s instructions. The library was prepared from each DNA sample using rbcL and ITS2 fusion primers with optimized PCR conditions. The libraries were purified using AMPure XP beads (Beckman Coulter, CA, USA), and the quality of some of the libraries was checked using Agilent high-sensitivity DNA kit on Agilent 2100 Bioanalyzer. For each sample, libraries from two replicates were pooled. Further, all libraries were diluted to 100 pM and pooled in equimolar concentration. Emulsion PCR was carried out using Ion 520™ & Ion 530™ Kit-OT2 with 400 bp chemistry (Thermo Fisher Scientific, MA, USA). Sequencing was performed on the Ion S5 system using a 520/530 chip (Thermo Fisher Scientific, MA, USA). The data have been submitted to the National Centre for Biotechnology Information (NCBI) BioProject database under accession number PRJNA960808 (https://www.ncbi.nlm.nih.gov/bioproject/).
Optimization was done with three parameters for establishing the metabarcoding data analysis pipeline. 1) filtering criteria includes discarding reads with length <280 and >350 bp, <300 and >350 bp, <320 and >350 bp for rbcL and for ITS2 discarding reads <280 and >300 bp; 2) OTU clustering with 97, 98 and 99% similarity in reads; 3) discarding OTU clusters having <5 and <10 reads. Obtained reads were filtered based on the quality score (Q >=25 for rbcL and Q >=20 for ITS) and read length using PRINSEQ (Schmieder and Edwards, 2011). Clustering filtered reads were performed using CD-HIT-EST (Huang et al., 2010). After that, the taxonomic assignment of OTU clusters having ≥5 and ≥10 reads was done using BLASTn (Altschul et al., 1990) (NCBI) with minimum E value 10E-5. For each sequence, ten hits were retrieved, and each hit was inspected and evaluated manually for the assigned plant genus and species. To analyze read abundance of each plant species, the number of reads was normalized by considering a total number of reads obtained after discarding clusters with <5 reads and <10 reads as 100%. The following formula was calculated for detection efficiency (%) for both metabarcodes. (Total number of detected targeted or labeled plant species/total number of plant species present in herbal formulation) × 100. Fidelity of detection (absolute) can be defined as the total number of samples or herbal formulations in which targeted plant species were detected per the total number of samples or herbal formulations in which targeted plant species were present (Seethapathy et al., 2019). The relative fidelity of detection (%) was calculated using the following formula. (Total number of samples or herbal formulations in which targeted plant species detected/total number of samples or herbal formulations in which targeted plant species are present) × 100. Fidelity of detection (absolute or relative) was calculated only where plant species that present in more than one group of the same type of control (n>1) and for market samples, sample size is more than one (n>1).
Coghlan et al. (2012) first introduced a metabarcoding approach for detecting plant and animal raw materials used in 15 traditional Chinese medicines using a P-loop region of the plastid trnL gene and 16S mtDNA marker, respectively. Later, several studies reported application of metabarcoding for authentication and detection of plant materials in herbal medicines with single and multi-barcode approaches. For instance, Yao et al. (2022) and Cheng et al. (2014) employed a multi-barcode approach of ITS2 and trnL in metabarcoding for detection of plant species in various traditional Chinese medicine (TCM). Urumarudappa et al. (2020) used ITS2 and rbcL barcode in metabarcoding for detection of plant species in herbal medicines of Thailand. Using ITS1 and ITS2 barcodes in metabarcoding, Raclariu et al. (2017a; 2017b; 2018b) reported presence of unlabeled species by 89, 68, and 15% in single drugs of Echinacea species, Hypericum perforatum, and Veronica officinalis, respectively sold in the European market.
In 2014 and 2015, the total commercial market for herbal materials in India was estimated to be more than 512,000 tonnes, with a market value of USD 1 billion (Ved and Goraya, 2007). India has over 8,000 authorized medicinal product manufacturing units, and the market growth for herbal products is outstripping supply capacity for some plant species (Ved and Goraya, 2007). However, to date, detection of raw plant materials of Indian-marketed herbal medicine using metabarcoding approach is not well established. Ichim (2019) demonstrated 31% adulteration in 752 Indian-marketed herbal products with DNA barcoding and species-specific marker-based approach but not via metabarcoding. Earlier, we reported presence of unspecified plant species in four polyherbal formulations of the Indian market using rbcL minibarcode via metabarcoding approach (Pandit et al., 2021). Here, we have used a multi-barcode strategy to identify raw plant components in single drugs and polyherbal formulations of the Indian market using newly designed rbcL and ITS2 metabarcodes.
Minimal criteria, such as universal amplifiabilities and minimum intraspecific but maximum interspecific divergence at the taxon level, must be followed in the search for the appropriate barcode region. Hence, degenerated rbcL and ITS2 metabarcode primers were designed for high amplification efficiency, universality, and resolution power. In total, 45 medicinal plants from diverse families, genera, and species were taken to confirm and optimize the newly designed rbcL and ITS2 primer sets for PCR amplification experimentally (Supplementary Information Table S1). The annealing temperature was optimized, and the results showed that the rbcL and ITS2 primer sets performed optimum at 56°C (data not shown). Among newly designed rbcL and ITS2 metabarcodes, rbcL is very robust and universal and gives a 100% amplification efficiency within selected 45 plants, but ITS2 metabarcode gives 88.9% amplification efficiency and is not able to provide amplification in 5 plant species (11.1%) include Ailanthus excelsa, Andrographis paniculata, A. vasica, O. tenuiflorum, and Ocimum canum (Supplementary Information Table S1). However, due to the greater species-level discrimination power of ITS2 in medicinal plants (Newmaster et al., 2013; Yao et al., 2022), ITS2 metabarcode was taken together with rbcL metabarcode. The amplification effectiveness of “fusion primers” (tagged with Ion torrent adapter and barcodes) of rbcL and ITS2 metabarcodes remained unchanged. However, the appearance of non-specific amplification in some barcodes suggests that 56°C annealing temperature is not optimal for the fusion primers. Therefore, further optimization of annealing temperature revealed that non-specific amplification was overcome by increasing the annealing temperature to 60°C (data not shown). At 60 °annealing temperature the amplification efficiency remained unaffected.
The first type of mock control, i.e., gDNA pooled controls of a different genus, was used to establish the metabarcoding data analysis pipeline. The first parameter is filtering criteria; for ITS2, the best filtering criteria was to remove reads with length < 300 bp, and for rbcL, discarding reads with length <300 and >350 bp (data not shown here). The second parameter is percentage similarity for the reads clustering (OTU picking), where in case of rbcL metabarcode, a greater number of plant species was detected when the reads were clustered at 99% identity. In the case of ITS2 metabarcode, clustering the reads at 97% and 98% similarity were equally capable of resolving the plant species (Supplementary Information Figure S1). Therefore, for ITS2 metabarcode, subsequently, for OTU clustering, i.e., 98% with a greater percentage was selected. The third parameter is the discarding OTU clusters having <5 or <10 reads, in which we observed that discarding OTU clusters having <5 reads was able to detect a greater number of plant species in the case of both metabarcodes (Supplementary Information Figure S2). Based on these findings, while selected read lengths were between 300 to 350 bp, 99% OTU clustering, and discarding of OTU clusters having <5 reads for the rbcL metabarcode and read lengths of >300 bp, 98% OTU clustering, and discarding of OTU clusters comprising <5 reads for the ITS2 metabarcode for analyzing the metabarcoding data of other mock controls and commercial herbal formulations.
Total reads obtained after filtering and percentage of reads analyzed (from filtered reads) after discarding OTU clusters having <5 reads for each mock control are shown in Supplementary Information Table S2 for the first type of control, which is gDNA pooled controls of different genera comprising 5 (5P), 10 (10P), and 15 (15P) plant species total of 18657 and 452380 reads were obtained for rbcL and the ITS2, respectively (Supplementary Information Table S2). Zingiber officinale had the highest percentage of reads with rbcL metabarcode in 5P (45.9%) and 10P (30.9%), whereas Senna tora had the highest percentage (21.3%) in 15P (Supplementary Information Figure S3A). Eclipta prostrata had the highest percentage of reads with ITS2 metabarcode in 5P (97.02%) and 10P (79.5%), whereas Phyllanthus emblica had the highest percentage (40.7%) in 15P (Supplementary Information Figure S3A). In 5P, out of five total four target plants were detected with rbcL metabarcode, and three were detected with ITS2. In 10P, out of ten, nine targeted plants were detected with rbcL, and seven targeted plants were detected with ITS2. In 15P, out of fifteen, thirteen targeted plants were detected with rbcL, and twelve targeted plants were detected with ITS2 (Figure 2; Supplementary Information Figure S3A). On the whole, for the gDNA pooled controls of a different genus, detection efficiency of rbcL was observed at 80% for 5P and 10P, 86.7% for 15P. While detection efficiency of ITS2 was observed at 80%, and combined detection efficiency of both metabarcodes was observed at 100% for all three gDNA pooled mock controls (Figures 2A, 3). Five plant species in all three gDNA pooled mock controls had 80% average fidelity, and the other five were present in two groups, i.e., 10P and 15P, had 90% average fidelity with rbcL metabarcode (Table 2). ITS2 metabarcode exhibited 66.7% average fidelity for five plant species present in all three gDNA pooled mock controls and 90% average fidelity for other five plant species present in 10P and 15P. Combined average fidelity with both barcodes was 100% for gDNA pooled control (Table 2).
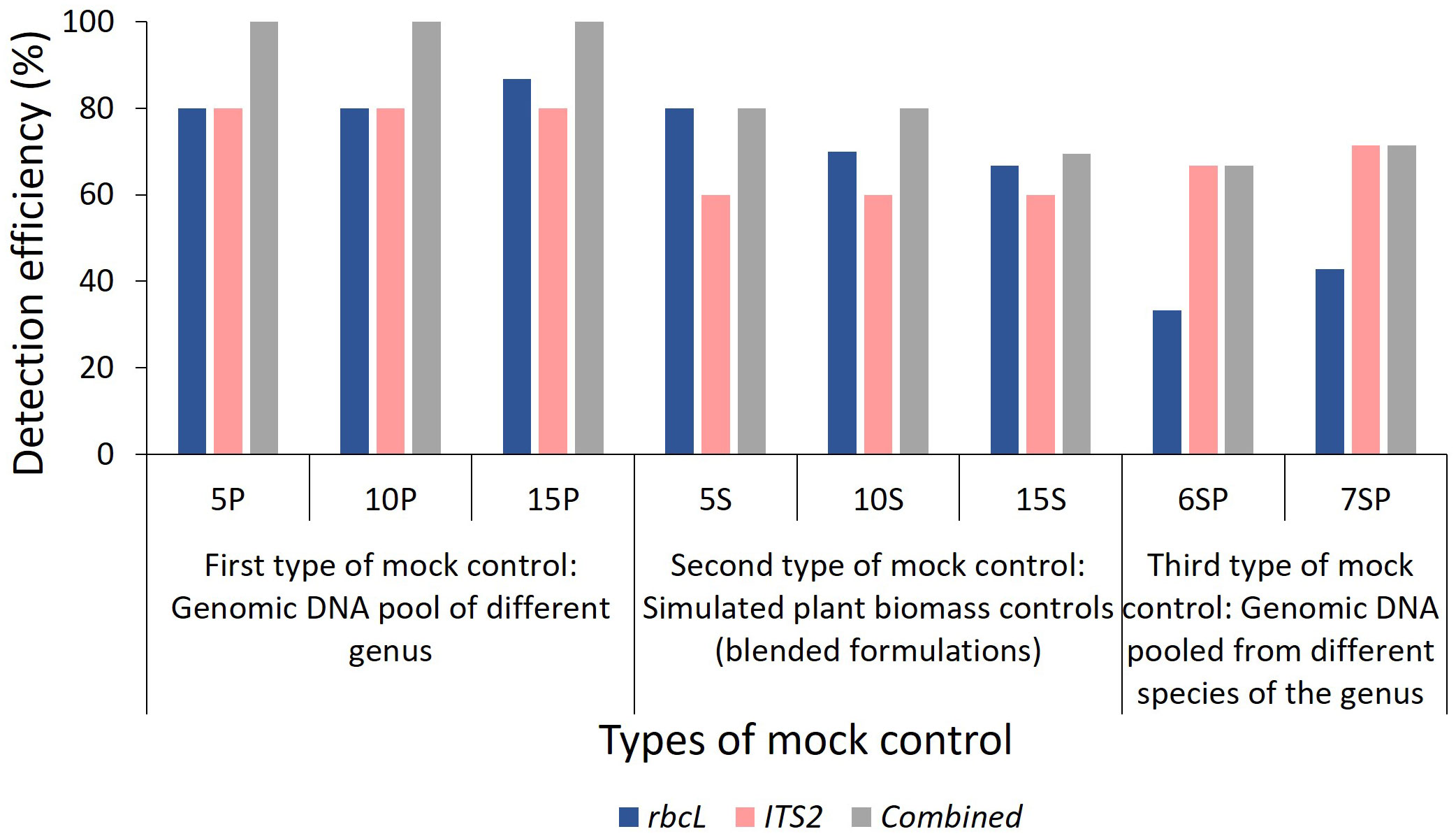
Figure 3 Detection efficiency obtained in mock controls by rbcL, ITS2, and combined approach. Detection efficiency (%) was calculated using a formula described in the Metabarcoding data analysis section of Materials and Methods. 5P: genomic DNA pool of five plant species, 10P: genomic DNA pool of ten plant species, 15P: genomic DNA pool of fifteen plant species, 5S: simulated blended plant pool of five plant species, 10S: simulated blended plant pool of ten plant species, 15S: simulated blended plant pool of fifteen plant species, 6SP: genomic DNA pool of six plant species from genera Asparagus and Terminalia, 7SP: genomic DNA pool of seven plant species from genera Piper and Phyllanthus. The detailed list of plant species used in each mock control is described in Figure 2.
For the simulated plant biomass controls (i.e., blended formulations or second type of mock controls), 42140 and 334017 reads were obtained after filtering for rbcL and the ITS2, respectively (Supplementary Information Table S2). The highest percentage of reads was observed for Azadirachta indica in 5S (62.5%) and 10S (34.7%), whereas, in 15S Justicia adhatoda (24.8%) showed highest percentage of reads using rbcL metabarcode (Supplementary Information Figure S3B). While in the case of ITS2, Eclipta prostrata had the highest percentage of reads in all simulated plant biomass controls (Supplementary Information Figure S3B). In 5S, 10S, and 15S total of 3, 7, and 10 targeted plants were detected by rbcL, and a total of 3, 6, and 8 targeted plants were detected by ITS2, respectively (Figure 2A; Supplementary Information Figure S3B). Detection efficiency for simulated plant biomass controls was observed 80% for 5S, 70% for 10S, and 66.7% for 15S with rbcL metabarcode. While detection efficiency of ITS2 was observed at 60% for all three groups of simulated plant biomass control, and combined detection efficiency of both metabarcodes was observed 80% for 5S and 10S and 69.4% for 15S (Figure 3). Five plant species that were present in all three groups of simulated plant biomass control had 80% average fidelity, and other five plant species that were present in 10S and 15S had 70% average fidelity with rbcL metabarcode (Table 2). ITS2 metabarcode exhibited 53.3% average fidelity for five plant species in all three groups and 70% average fidelity for five other plants present in two groups, i.e., 10S and 15S. Combined average fidelity with both barcodes was observed at 80% for simulated plant biomass controls (Table 2).
For the third type of control, gDNA pooled from different species of two genera, 10265 and 164358 reads were obtained for rbcL and the ITS2, respectively (Supplementary Information Table S2). In 6SP, out of six plant species total of three plant species include Asparagus racemosus (81.1%), Terminalia bellirica (11.8%), and Terminalia chebula (6.8%), were detected by rbcL while ITS2 metabarcode was able to resolve five plant species except for Asparagus dumosus (Figure 2B; Supplementary Information Figure S3C). In 7SP, out of seven plant species, two plant species include Piper nigrum (24.7%) and Phyllanthus emblica (72.3%) were detected by rbcL. In comparison, ITS2 metabarcode was able to resolve four plant species, including Piper nigrum (0.8%), Piper longum (21%), Phyllanthus emblica (62.5%), Phyllanthus amaras (13.5%) (Figure 2B; Supplementary Information Figure S3C). rbcL showed 33.3%, and ITS2 showed 66.7% detection efficiency in species-level control with a combined detection efficiency of 66.7% (Figures 2B, 2). These two species-level controls indicate a greater resolution spectrum of ITS2 metabarcode than rbcL metabarcode. This finding corroborates with earlier reports in which authors demonstrated that ITS2 metabarcode has greater species-level discrimination power than rbcL while rbcL has greater universality (Newmaster et al., 2013; Yao et al., 2022).
Here, in the first and third types of mock controls, DNA was pooled in equal proportions, and for the second type of simulated plant biomass controls, the equivalent weight of each plant species part with therapeutic importance was mixed. The primary aim of all three mock controls was to evaluate the read abundance, detection efficiency, and fidelity differences introduced under the influence of secondary metabolites, primer fit compatibilities, and PCR dynamics. The impact was observed with percentage read variation of the same plant in a different control (Arulandhu et al., 2017). Terminalia arjuna, T. chebula, and Phyllanthus emblica were not detected in simulated plant biomass control might be due to variability in quality and quantity of DNA extracted from each plant species from the mixtures as different parts of plants, i.e., rhizome, fruits, leaves, and bark have been added into plant biomass controls (Ivanova et al., 2016). In addition to that, ITS2 metabarcode is unable to resolve A. paniculata and J. adhatoda in all mock controls because the newly designed ITS2 metabarcode is impotent in amplifying target sequence from these two plant species (Figure 1; Supplementary Information Table S1). Despite the mentioned limitations, the combined rbcL and ITS2 metabarcoding approach could resolve plant species with high fidelity (Table 2) and can be implemented to detect plant species in herbal products.
Tulsi (O. tenuiflorum) (The Ayurvedic Pharmacopoeia of India, Vol. II, 1999), Gokhru (Tribulus terrestris) (The Ayurvedic Pharmacopoeia of India, Vol. I,1990) Shatavari (Asparagus racemosus) (The Ayurvedic Pharmacopoeia of India, Vol. III, 2001), Vasa (Justicia adhatoda), Ashwagandha (Withania somnifera), Bhringraj (Eclipta alba), and Arjuna (Terminalia arjuna) were among the 17 single drugs that were collected. Total reads, reads obtained after filtering, and percentage of analyzed reads (from filtered reads) after discarding OTU clusters having <5 reads for each single drug are shown in Supplementary Information Table S3. For all single drugs, 135505 total raw reads for rbcL and 1390098 total raw reads for ITS2 metabarcode were obtained (Supplementary Information Table S3). On an average, 12.6% (0-99.8%) reads have been obtained for non-targeted plant species with rbcL metabarcode and 46.5% (0-100%) reads have been obtained for non-targeted plant species with ITS2 metabarcode in single drugs (Supplementary Information Table S4). Cross-contamination with allied species, harvesting process, pollen contamination, misidentification due to cryptic taxonomy, polynomial vernacular identification, manufacturing and packing procedure may contribute to the presence of non-targeted plant species (Seethapathy et al., 2019).
In tulsi powder (labeled as O.tenuiflorum), rbcL detected O.tenuiflorum ranging between 99.8% to 64.4%. Along with O. tenuiflorum, substituted Ocimum basilicum (Travadi et al., 2022b) was also observed in three herbal products with 17.7, 14.4, and 1.4% reads with rbcL metabarcode, respectively (Figure 4A). O. tenuiflorum could not be detected by ITS2 metabarcode in any samples. However, O. basilicum was found in one sample with 11.6% reads (Figure 4B). That could be due to inability of new ITS2 metabarcode to amplify O. tenuiflorum (Supplementary Information Table S1), which is proof of PCR biases toward the unintentional and low level of contamination present in samples and leads to the high number of reads for non-targeted plant species.
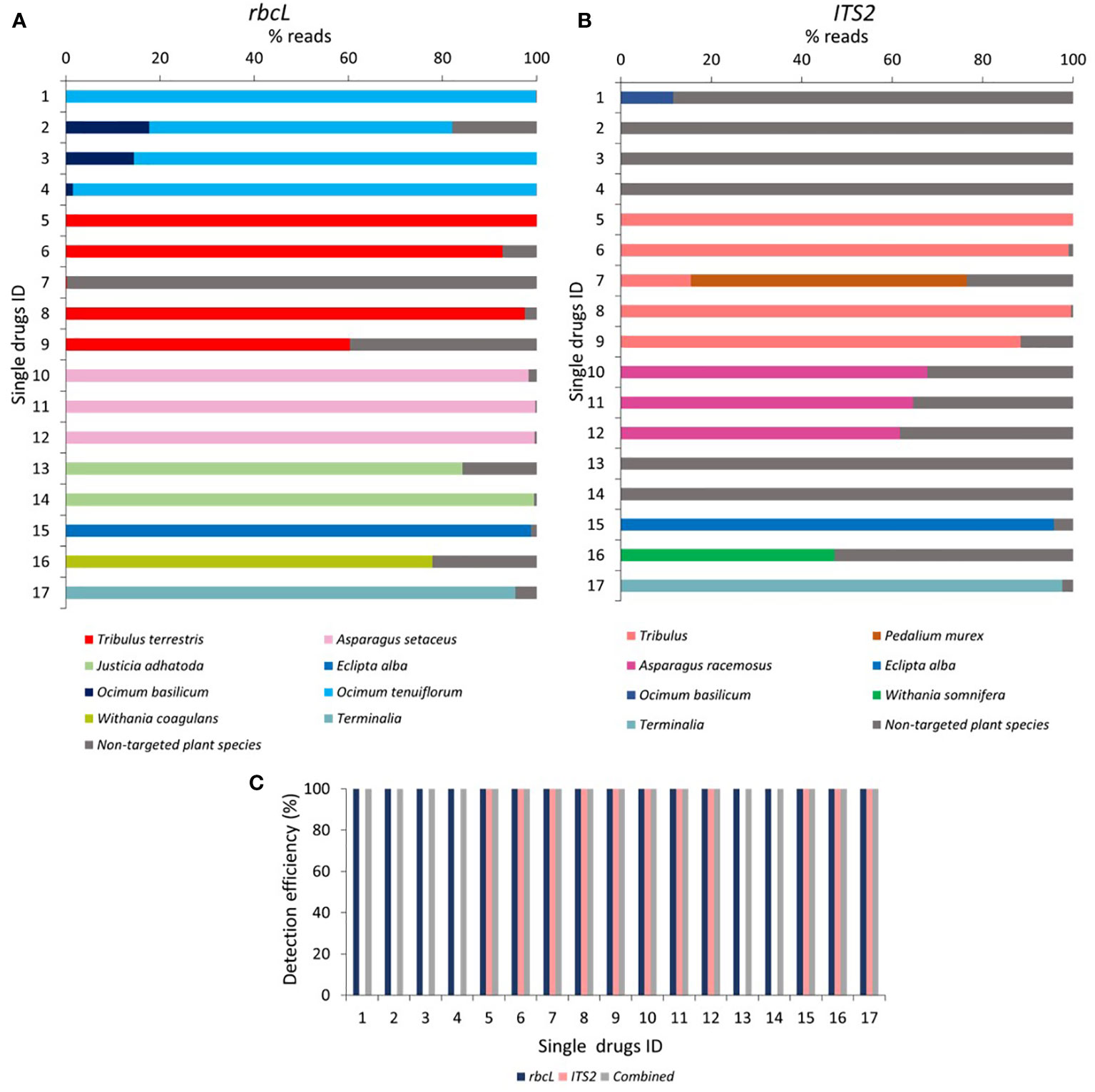
Figure 4 Relative abundance of the plant species and detection efficiency in single drugs through rbcL and ITS2 metabarcoding. (A) Relative abundance (% reads) of the plant species detected in single drugs through rbcL metabarcoding. Relative abundance (% reads) of non-targeted plant species is reported in Supplementary Information Table S4. (B) Relative abundance (% reads) of the plant species detected in single drugs through ITS2 metabarcoding. The relative abundance of non-targeted plant species is reported in Supplementary Information Table S4. (C) Detection efficiency obtained in single drugs by rbcL, ITS2, and combined metabarcoding approach. Single drugs ID 1 to 4 for Tulsi (Ocimum tenuiflorum) powder, 5 to 9 for Gokhru (Tribulus terrestris) powder, 10 to 12 for Shatavari (Asparagus racemosus) powder, 13 and 14 for Vasa (Justicia adhatoda) powder, 15 for Bhringraj (Eclipta alba) powder, 16 for Ashwagandha (Withania somnifera) powder, and 17 for Arjuna (Terminalia arjuna) powder.
In Gokhru powder, T.terrestris (Chota Gokhru) was detected up to species level by rbcL with 100% reads in sample 5, 92.7% in sample 6, 0.2% in sample 7, 97.4% in sample 8, and 60.3% in sample 9 (Figure 4A). The ITS2 metabarcode resolved T. terrestris only at the genus level with 100% in sample 5, 99% in sample 6, 15.5% in sample 7, 99.6% in sample 8, and 88.4% in sample 9. ITS2 revealed the presence of 61% of Pedalium murex (Bada Gokhru) in sample 7 (Figure 4B), which is a commonly substituted species and labeled as Gokhru in Indian marketed herbal products (Choudhary et al., 2021).
In all three Shatavari powders, rbcL metabarcode detected Asparagus setaceus instead of A. racemosus with 98.3% reads in sample 10, 99.7% in sample 11, and 99.6% reads in sample 12. By ITS2, 67.7% to 61.7% reads for A. racemosus were obtained in all three Shatavari powders. In vasa powder, J.adhatoda was detected by rbcL with 84.2% reads in sample 13 and 99.4% in sample 14. In comparison, ITS2 could not resolve J.adhatoda because of the amplification inability of our ITS2 metabarcode (Figure 4, Supplementary Information Table S1). This result was consistent with the mock control 15P and 15S. In Ashwagandha powder (sample 16), Withania coagulans (77.9% reads) instead of W. somnifera was detected by rbcL. While with ITS2, W. somnifera was detected with 47.2% reads.
In Bhringraj powder, both rbcL and ITS2 metabarcode detected E.alba with 98.8% and 95.7% reads, respectively. In Arjuna powder, rbcL metabarcode identified Terminalia arjuna only up to genus level with 95.5% reads, while ITS2 metabarcode was identified T. arjuna at species level with 97.6% reads (Figure 4).
For Tulsi, Gokhru, Shatavari, and Vasa powder, 100% fidelity was obtained with rbcL metabarcode, while ITS2 metabarcode exhibited 0% fidelity for Tulsi and Vasa powder and 100% fidelity for Gokhru and Shatavari powder (Table 3A). Suggesting that, for Tulsi and Vasa powder authentication, our rbcL metabarcode works efficiently but not ITS2. While rbcL metabarcode was not suitable for identifying P. murex in Gokhru powder, rbcL sequence for Pedalium murex was unavailable in NCBI (database was accessed on December 15, 2022). In addition, T. arjuna was resolved only up to genus level by rbcL, and T. terrestris was resolved up to only at the genus level by ITS2 metabarcode. This observation is in concordance with mock controls and could be due to low interspecific variability of barcode sequence covered by our metabarcodes for resolving species to be identified. Although, combined metabarcoding approach provides 100% detection efficiency with 100% fidelity for single drugs by overcoming the limitations of individual barcodes due to PCR biases, low interspecific variability, or the absence of the corresponding sequences in the database. Seethapathy et al. (2019) demonstrated 67% fidelity for targeted plant species present in single drugs of the European market using ITS1 and ITS2 barcodes. We obtained 100% fidelity for targeted plant species within single drugs. The overall results of the single drugs revealed that a multi-barcode metabarcoding approach could be used to assess the prevalence of widespread adulterated and substituted plant material in single drugs and implement more stringent supply chain precautionary measures at primary level.
Trikatu, Sitopaladi, Rasayana, Hingwashtak, and Talisadi (Talisadya) (Joshi et al., 2017) were among the 15 polyherbal formulations collected. Total reads obtained after filtering and percentage of analyzed read (from filtered reads) after discarding OTU clusters having <5 reads for each polyherbal formulation are shown in Supplementary Information Table S5. A total of 53087 and 1429238 reads were obtained by rbcL and ITS2 metabarcode for polyherbal formulation, respectively (Supplementary Information Table S5). On average, 1.4% (0-3.9%) reads have been obtained for non-targeted plant species with rbcL metabarcode, and 16.5% (0.1-87.4%) reads were obtained for non-targeted plant species with ITS2 metabarcode in polyherbal formulations (Supplementary Information Table S6).
In Trikatu, 28.2%, 4.6%, and 57% read for P. nigrum with rbcL, and 8.3%, 14.7 and 37.9% reads with ITS2 was observed in sample 18, 19, and 20, respectively. P.longum comprised 2%, 37.1%, and 47.8% reads with rbcL and 0%, 42.1%, and 19.7% with ITS2; Z. officinale possess 67.6%, 56.5%, and 43.6% with rbcL and 4.3%, 18.8% and 2.4% with ITS2 in sample 18, 19, and 20 respectively (Figures 5A, B). All three targeted plants were detected (i.e., 100% detection efficiency) in all three Trikatu samples (i.e., 100% fidelity) using a combined approach (Figure 5C, Table 3B). Nevertheless, ITS2 showed the higher percentage of non-targeted reads (87.4% in sample 18, 24.4% in sample 19, and 40% in sample 20) might be due to technical bias that can be introduced during DNA extraction and PCR towards the unintentional cross-contamination happens during the supply chain (Figure 5B).

Figure 5 Relative abundance of the plant species and detection efficiency in polyherbal formulations through rbcL and ITS2 metabarcoding. (A) Relative abundance (% reads) of the plant species detected in polyherbal formulations through rbcL metabarcoding. Relative abundance (% reads) of non-targeted plant species is reported in Supplementary Information Table S6. (B) Relative abundance (% reads) of the plant species detected in polyherbal formulations through ITS2 metabarcoding. Relative abundance (% reads) of non-targeted plant species is reported in Supplementary Information Table S6. (C) Detection efficiency obtained in polyherbal formulation by rbcL, ITS2, and combined metabarcoding approach. Polyherbal formulations ID 18 to 20 for Trikatu powder, 21 to 23 for Sitopaladi powder, 24 to 27 for Rasayana powder, 28 to 31 for Hingwashtak powder, and 32 for Talisadi powder.
Sitopaladi powder is primarily composed of five constituents; with the exclusion of sacrarium (sugar) and vanshlochan (the female bamboo exudate) and from the total number of designated species, the aim was to detect C. cassia, P. longum, and Elettaria cardamom. C. cassia was detected by rbcL metabarcode with 0.2% reads only in sample 22. P. longum exhibited 1.8, 66.6, and 83.3% of reads with rbcL and 0.1, 6.6, and 31.9% of reads with ITS2 in samples 21, 22, and 23, respectively. E. cardamomum showed 96.3%, 30.2%, and 15.9% reads with rbcL and 72.4%, 43.7%, and 65.1% with ITS2 in samples 21, 22, and 23, respectively (Figures 5A, B). Overall, the combined approach showed 66.7% detection efficiency in sample 21, 100% in sample 22, and 66.7% in sample 23, and average of 77.8% fidelity (Figure 5C, Table 3B).
In Rasayana, rbcL metabarcode exhibited 91% to 98% reads for T. teresteris, while ITS2 metabarcode exhibited 98 to 100% reads in all samples (samples 24 to 27). In all samples, T. cordifolia was resolved by rbcL with 1.7% to 9.1% reads, while ITS2 metabarcode could not (Figures 5A, B). P. emblica was not detected by both metbarcode except in sample 24 (ITS2 obtained 0.01% reads were), possibly due to DNA extraction biases as DNA extraction from amla fruits is difficult due to high acidic nature and high tannin content (Warude et al., 2003). The combined metabarcoding approach showed 100% detection efficiency in sample 24 and 66.7% in remaining other samples with average of 75% fidelity (Figure 5C, Table 3B).
Hingwashtak powder (samples 28 to 31) comprised seven ingredients, including Zinger officinale, P. nigrum, P. longum, C. cyminum, C. carvi [C. cyminum, and C. carvi commonly substituted with Bunium persicum (Syn. Elwendia persica) (Johri, 2011; Singh et al., 2017; Bansal et al., 2018)], Apium leptophyllum (majority of commercial products comprised/labeled Apium graveolens instead of A. leptophyllum; further these plant species commonly substituted with T. ammi (Pushpendra et al., 2016)) and Ferula foetida. From these seven ingredients, rbcL metabarcode was able to resolve Z. officinale in all samples with 0.37 to 5.5% reads, P. nigrum in sample 29 (2.7% reads) and 30 (1.0% reads), P. longum in sample 30 (8.9% reads), C. cyminum in all samples with 32.7% to 94.1% reads, A. graveolens in sample 29 (2.9% reads). C. carvi commonly substituted with B. persicum (Elwendia persica), neither C. carvi nor B. persicum was detected by rbcL in all Hingwashtak samples (Figure 5A). ITS2 metabarcode exhibited a high prevalence of C. cyminum in all samples with 51.8% to 95.4% reads, B. persicum in sample 28 with 37.9% reads, T. ammi (substitution of A. leptophyllum or A. graveolens) in sample 28 with 10% reads (Figure 5B). In addition, ITS2 metabarcode also detected Trachyspermum ammi in samples 29 and 31. rbcL metabarcode showed reads for Ligusticum jeholense (Chinese medicinal herb from the Apiaceae family) instead of Trachyspermum ammi in sample 28, 30, 31 and A. graveolens in sample 29. This could be due to our rbcL metabarcode unable to resolve T. ammi and detect L. jeholense falling under the same family. Overall, the combined metabarcoding approach showed average 72.6% detection efficiency with 63.3% fidelity for Hingwashtak powders (Figure 5C and Table 3B).
Talisadi/Talisadya powder (sample 32) comprises eight constituents including Abies webbiana, P. longum, P. nigrum, Z. officinale, E. cardamomum, Cinnamomum Zeylanicum, Vanshlochan (the female bamboo exudate), and sugar (sugar and Vanshlochan are excluded for metabarcoding analysis). From these six ingredients, only three plant species which include P. longum (0.8% reads), P. nigrum (21.3% reads), and Z. officinale (23.8% reads) were detected by rbcL metabarcode. Three plant species which include P. nigrum (3.79% reads), Z. officinale (0.03% reads), and E. cardamomum (0.6% reads) were detected by ITS2 metabarcode. A. webbiana and C. Zeylanicum were not detected by either of the barcodes. The combined metabarcode approach detected four plant species (66.7% detection efficiency) out of six (Figure 5C). In Talisadi/Talisadya powder, C. cyminum (27.9% reads with rbcL and 51.3% reads with ITS2), B. persicum (32.0% reads with ITS2), L. jeholense (26.2% reads with rbcL) and T. ammi (11.8% reads with ITS2) was detected might be due to unintentional cross-contamination happen during sample processing as a collection of Hingwashtak powder sample 28 and Talisadi powder sample 32 were done from the same company. In addition, a high percentage of reads were covered by C. cyminum, T. ammi, L. jeholens, B. persicum (all plant species belonging to Apiaceae family), then Z. officinale, P. longum, and P. nigrum. That could be because of technical bias introduced during DNA extraction and PCR.
Up to this point, the fidelity of plant species per number of mock controls or herbal formulations has been calculated and discussed. Here, we have estimated the fidelity of the targeted plant species included within different mock controls and polyherbal formulations to get a better perspective of species discrimination capabilities and reliabilities of single and multi-barcode approaches. Both the rbcL and ITS2 metabarcodes resolved 19 (46.7%) of the 39 listed plant species at the species level. However, plants detected at the species level were different, and a multi-barcode approach provided species-level resolution for 27 (69.23%) species, leading to a 20.5% increment in the whole (Table 4). This observation confirms robustness for our newly designed metabarcodes in detecting plant species at lower taxonomic levels. In addition, 100% fidelity was observed for T. bellirica, A. paniculata, A. indica, E. alba, and C. asiatica within gDNA controls, biomass controls, and cumulative analysis by the combined approach of rbcL and ITS2. However, the combined approach of rbcL and ITS2 exhibited 100% fidelity for Z. officinale, V. negundo, P. nigrum, A. marmelous, T. arjuna, and P. embilica only within gDNA controls and having lower fidelity in biomass controls and cumulative analysis (Table 4). That could be due to biases in the DNA isolation process; yielding equal proportional DNA from the poly formulation is impossible due to genome size differences and differences in plant parts and amounts of secondary metabolites.
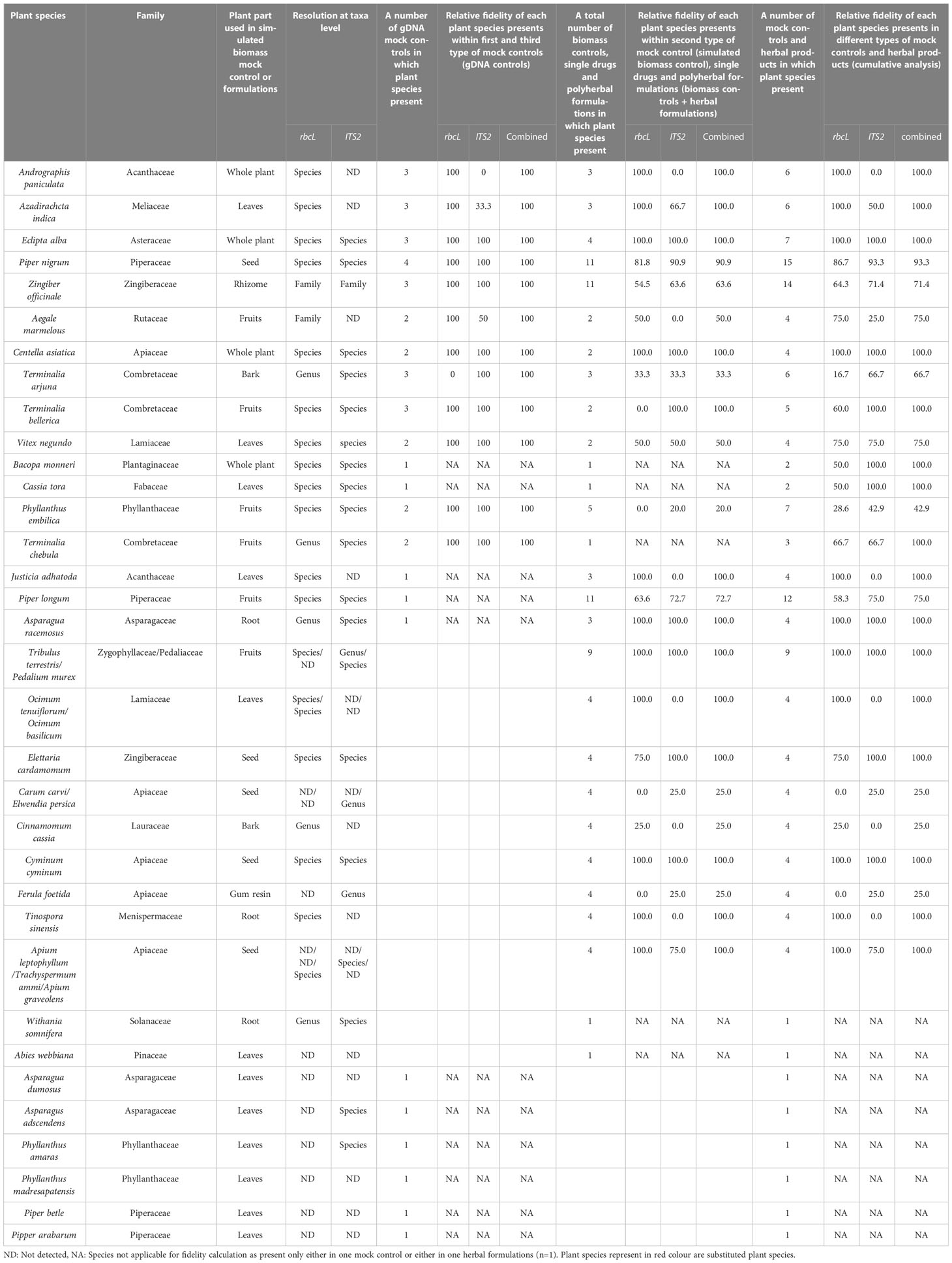
Table 4 Fidelity of targeted plant species present within mock controls as well herbal formulations.
Furthermore, the extracted DNA is degraded because herbal products are intensively processed. The PCR conditions and reactions will also have a significant impact on the primer fit and PCR bias of the mixture. That was demonstrated by comparing the combined fidelity of gDNA controls, biomass controls, and cumulative analysis (Table 4). On average, 83.6% fidelity was observed for targeted plant species in the cumulative analysis. This result confirmed the high reliability of our multi-barcode sequencing approach.
On the whole, our findings suggest that the multi-barcode DNA metabarcoding method assessed in this study can provide a composition of more diverse sets of single drugs and polyherbal formulations listed in the Ayurvedic Pharmacopoeia of India. We obtained 100% average detection efficiency and relative fidelity of targeted plants for single drugs and 79% for polyherbal formulations through the multi-barcode sequencing approach. We have primarily focused on detected plant species in herbal products rather than undetected plant species because many steps, such as DNA extraction biases, PCR biases, and manufacturing processes, that can lead to DNA degradation or loss beyond detectable limits, failing to detect plant species. The presence of non-targeted plant species in herbal products could be due to unintentional contamination of the supply chain, economically motivated adulteration, and/or admixture of other species. Our study showed that the rbcL metabarcode had better detection ability for certain plant species, e.g., O. tenuiflorum, J. adhatoda, and A. paniculata, while ITS2 had better discrimination power for certain plant species, e.g., species of the genus Terminalia, Asparagus, Piper, Phyllanthus, and Pedalium murex. Thus, the complementary approach of both metabarcodes is a promising tool for quality evaluation of herbal products and pharmacovigilance. However, the development of standardized methods for metabarcoding sequencing and bioinformatics analysis pipeline and curated database is needed for effective use as a regulatory tool to authenticate herbal products in combination with advanced chemical methods to identify bioactive therapeutics.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
TT: performed experiments, established metabarcoding data analysis pipeline, data analysis, writing and editing manuscript, and validation of final manuscript. AS: performed experiments, data analysis, writing and editing manuscript, and validation of final manuscript. RP: designed primers for metabarcoding, performed experiments, established metabarcoding data analysis pipeline, and manuscript editing. SS: performed experiments, and manuscript editing. CJ: project administration, methodology, supervision, and review & editing. MJ: principal investigator, conceptualization, methodology, supervision, and review & editing. All authors contributed to the article and approved the submitted version.
Gujarat State Biotechnology Mission (GSBTM), Gandhinagar, Gujarat, India, has provided financial support for the project under the Research Support Scheme, grant ID GSBTM/JDRD/584/2018/204.
The authors would like to thank Prof. Padamnabhi S. Nagar, The Maharaja Sayajirao University of Baroda, Gujarat, India and Director, DMAPR, Gujarat, India for helping us in plant collection and authentication. The authors would like to thank Dr. Darshan Parmar, M.D. (Rasashastra and Bhaishajya Kalpana), Government Ayurvedic College, Vadodara (Gujarat, India) for providing some of the herbal formulations and Mr. Nitin Savaliya, Technical Assistant from Thermo Fisher Scientific, for NGS instrument handling and run setup.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1169984/full#supplementary-material
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Arulandhu, A. J., Staats, M., Hagelaar, R., Voorhuijzen, M. M., Prins, T. W., Scholtens, I., et al. (2017). Development and validation of a multi-locus DNA metabarcoding method to identify endangered species in complex samples. Gigascience 6 (10), gix080. doi: 10.1093/gigascience/gix080
Bansal, S., Thakur, S., Mangal, M., Mangal, A. K., Gupta, R. K. (2018). DNA Barcoding for specific and sensitive detection of cuminum cyminum adulteration in bunium persicum. Phytomedicine 50, 178–183. doi: 10.1016/j.phymed.2018.04.023
Cheng, X., Su, X., Chen, X., Zhao, H., Bo, C., Xu, J., et al. (2014). Biological ingredient analysis of traditional Chinese medicine preparation based on high-throughput sequencing: the story for liuwei dihuang wan. Sci. Rep. 4, 1–12. doi: 10.1038/srep05147
Choudhary, S., Kaurav, H., Chaudhary, G. (2021). Gokhru (tribulus terrestris and pedalium murex): medicinal importance of chota gokhru and bada gokhru in ayurveda and modern science. Asian J. Pharm. Clin. Res. 14, 6–13. doi: 10.22159/ajpcr.2021.v14i6.41366
Coghlan, M. L., Haile, J., Houston, J., Murray, D. C., White, N. E., Moolhuijzen, P., et al. (2012). Deep sequencing of plant and animal DNA contained within traditional Chinese medicines reveals legality issues and health safety concerns. PloS Genet. 8 (4), e1002657. doi: 10.1371/journal.pgen.1002657
Fazekas, A. J., Kesanakurti, P. R., Burgess, K. S., Percy, D. M., Graham, S. W., Barrett, S. C. H., et al. (2009). Are plant species inherently harder to discriminate than animal species using DNA barcoding markers? Mol. Ecol. Resour. 9, 130–139. doi: 10.1111/j.1755-0998.2009.02652.x
Frigerio, J., Agostinetto, G., Mezzasalma, V., De Mattia, F., Labra, M., Bruno, A. (2021). Dna-based herbal 'teas' authentication: an ITS2 and psba-trnh multi-marker dna metabarcoding approach. Plants 10, 1–14. doi: 10.3390/plants10102120
Hebert, P. D. N., Cywinska, A., Ball, S. L., DeWaard, J. R. (2003). Biological identifications through DNA barcodes. Proc. R. Soc B Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Huang, Y., Niu, B., Gao, Y., Fu, L., Li, W. (2010). CD-HIT suite: a web server for clustering and comparing biological sequences. Bioinformatics 26, 680–682. doi: 10.1093/bioinformatics/btq003
Ichim, M. C. (2019). The DNA-based authentication of commercial herbal products reveals their globally widespread adulteration. Front. Pharmacol. 10. doi: 10.3389/fphar.2019.01227
Ivanova, N. V., Kuzmina, M. L., Braukmann, T. W. A., Borisenko, A. V., Zakharov, E. V. (2016). Authentication of herbal supplements using next-generation sequencing. PloS One 11, 1–24. doi: 10.1371/journal.pone.0156426
Johri, R. K. (2011). Cuminum cyminum and carum carvi: an update. Pharmacogn. Rev. 5, 63–72. doi: 10.4103/0973-7847.79101
Joshi, V. K., Joshi, A., Dhiman, K. S. (2017). The ayurvedic pharmacopoeia of India, development and perspectives. J. Ethnopharmacol. 197, 32–38. doi: 10.1016/j.jep.2016.07.030
Kress, W. J. (2017). Plant DNA barcodes: applications today and in the future. J. Syst. Evol. 55, 291–307. doi: 10.1111/jse.12254
Linhart, C., Shamir, R. (2005). The degenerate primer design problem: theory and applications. J. Comput. Biol. 12, 431–456. doi: 10.1089/cmb.2005.12.431
Lo, Y. T., Shaw, P. C. (2019). Application of next-generation sequencing for the identification of herbal products. Biotechnol. Adv. 37, 107450. doi: 10.1016/j.biotechadv.2019.107450
Maloukh, L., Kumarappan, A., Jarrar, M., Salehi, J., El-wakil, H., Rajya Lakshmi, T. V. (2017). Discriminatory power of rbcL barcode locus for authentication of some of united Arab Emirates (UAE) native plants. 3 Biotech. 7, 1–7. doi: 10.1007/s13205-017-0746-1
Mishra, P., Kumar, A., Nagireddy, A., Mani, D. N., Shukla, A. K., Tiwari, R., et al. (2016). DNA Barcoding: an efficient tool to overcome authentication challenges in the herbal market. Plant Biotechnol. J. 14, 8–21. doi: 10.1111/pbi.12419
Newmaster, S. G., Grguric, M., Shanmughanandhan, D., Ramalingam, S., Ragupathy, S. (2013). DNA Barcoding detects contamination and substitution in north American herbal products. BMC Med. 11, 222. doi: 10.1186/1741-7015-11-222
Pandit, R., Travadi, T., Sharma, S., Joshi, C., Joshi, M. (2021). DNA Meta-barcoding using rbcL based mini-barcode revealed presence of unspecified plant species in ayurvedic polyherbal formulations. Phytochem. Anal. 32, 804–810. doi: 10.1002/pca.3026
Parveen, I., Gafner, S., Techen, N., Murch, S. J., Khan, I. A. (2016). DNA Barcoding for the identification of botanicals in herbal medicine and dietary supplements: strengths and limitations. Planta Med. 82, 1225–1235. doi: 10.1055/s-0042-111208
Pushpendra, P., Sunil Kumar, K. N., Priyadarshini, P., Holla, B. S., Ravishankar, B., Yashovarma, B. (2016). Quality standards for hutabhuga¯di cu¯ra (Ayurvedic formulary of India). J. Tradit. Complement. Med. 6, 78–88. doi: 10.1016/j.jtcme.2014.11.019
Raclariu, A. C., Heinrich, M., Ichim, M. C., de Boer, H. (2018a). Benefits and limitations of DNA barcoding and metabarcoding in herbal product authentication. Phytochem. Anal. 29, 123–128. doi: 10.1002/pca.2732
Raclariu, A. C., Mocan, A., Popa, M. O., Vlase, L., Ichim, M. C., Crisan, G., et al. (2017a). Veronica officinalis product authentication using DNA metabarcoding and HPLC-MS reveals widespread adulteration with veronica chamaedrys. Front. Pharmacol. 8, 378. doi: 10.3389/fphar.2017.00378
Raclariu, A. C., Paltinean, R., Vlase, L., Labarre, A., Manzanilla, V., Ichim, M. C., et al. (2017b). Comparative authentication of hypericum perforatum herbal products using DNA metabarcoding, TLC and HPLC-MS. Sci. Rep. 7, 8–10. doi: 10.1038/s41598-017-01389-w
Raclariu, A. C., Ţebrencu, C. E., Ichim, M. C., Ciupercă, O. T., Brysting, A. K., de Boer, H. (2018b). 'What's in the box? authentication of echinacea herbal products using DNA metabarcoding and HPTLC. Phytomedicine 44, 32–38. doi: 10.1016/j.phymed.2018.03.058
Schmieder, R., Edwards, R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864. doi: 10.1093/bioinformatics/btr026
Seethapathy, G. S., Raclariu-Manolica, A.-C., Anmarkrud, J. A., Wangensteen, H., De Boer, H. J. (2019). DNA Metabarcoding authentication of ayurvedic herbal products on the European market raises concerns of quality and fidelity. Front. Plant Sci. 10, 68. doi: 10.3389/fpls.2019.00068
Sharma, S., Shrivastava, N. (2016). Internal transcribed spacer guided multiplex PCR for species identification of convolvulus prostratus and evolvulus alsinoides. Acta Pharm. Sin. B 6, 253–258. doi: 10.1016/j.apsb.2016.02.003
Shetti, S., Kumar, C. D., Sriwastava, N. K., Sharma, I. P. (2011). Pharmacovigilance of herbal medicines: current state and future directions. Pharmacogn. Mag. 7, 69–73. doi: 10.4103/0973-1296.75905
Singh, R. P., H.V., G., K, M. (2017). Cuminum cyminum {\textendash} a popular spice: an updated review. Pharmacogn. J. 9, 292–301. doi: 10.5530/pj.2017.3.51
The ayurvedic pharmacopoeia of India, 1s, Vol. I. (1990). Department of AYUSH, Ministry of Health and Family Welfare, Government of India.
The ayurvedic pharmacopoeia of India, 1st, Vol. II. (1999). Department of AYUSH, Ministry of Health and Family Welfare, Government of India.
The ayurvedic pharmacopoeia of India, 1st, Vol. III. (2001). Department of AYUSH, Ministry of Health and Family Welfare, Government of India.
Travadi, T., Shah, A. P., Pandit, R., Sharma, S., Joshi, C., Joshi, M. (2022a). Detection of carica papaya adulteration in piper nigrum using chloroplast DNA marker-based PCR assays. Food Anal. Methods. 16, 107–114. doi: 10.1007/s12161-022-02395-z
Travadi, T., Sharma, S., Pandit, R., Nakrani, M., Joshi, C., Joshi, M. (2022b). A duplex PCR assay for authentication of ocimum basilicum l. and ocimum tenuiflorum l in tulsi churna. Food Control 137, 108790. doi: 10.1016/j.foodcont.2021.108790
Urumarudappa, S. K. J., Tungphatthong, C., Prombutara, P., Sukrong, S. (2020). DNA Metabarcoding to unravel plant species composition in selected herbal medicines on the national list of essential medicines (NLEM) of Thailand. Sci. Rep. 10, 1–11. doi: 10.1038/s41598-020-75305-0
Vassou, S. L., Nithaniyal, S., Raju, B., Parani, M. (2016). Creation of reference DNA barcode library and authentication of medicinal plant raw drugs used in ayurvedic medicine. BMC Complement. Altern. Med. 16, 9–15. doi: 10.1186/s12906-016-1086-0
Ved, D. K., Goraya, G. S. (2007). Demand and supply of medicinal plants in India. NMPB, New Delhi & FRLHT, Bangalore, India. 18 (85), 210–52.
Warude, D., Chavan, P., Joshi, K., Patwardhan, B. (2003). DNA Isolation from fresh and dry plant samples with highly acidic tissue extracts. Plant Mol. Biol. Rep. 21, 467. doi: 10.1007/BF02772600
Wu, H. Y., Shaw, P. C. (2022). Strategies for molecular authentication of herbal products: from experimental design to data analysis. Chin. Med. (United Kingdom) 17, 1–15. doi: 10.1186/s13020-022-00590-y
Xin, T., Xu, Z., Jia, J., Leon, C., Hu, S., Lin, Y., et al. (2018). Biomonitoring for traditional herbal medicinal products using DNA metabarcoding and single molecule, real-time sequencing. Acta Pharm. Sin. B 8, 488–497. doi: 10.1016/j.apsb.2017.10.001
Keywords: authentication, DNA metabarcoding, herbal medicines, next generation sequencing, pharmacovigilance
Citation: Travadi T, Shah AP, Pandit R, Sharma S, Joshi C and Joshi M (2023) A combined approach of DNA metabarcoding collectively enhances the detection efficiency of medicinal plants in single and polyherbal formulations. Front. Plant Sci. 14:1169984. doi: 10.3389/fpls.2023.1169984
Received: 20 February 2023; Accepted: 17 April 2023;
Published: 15 May 2023.
Edited by:
Yinglong Chen, University of Western Australia, AustraliaReviewed by:
Shuiming Xiao, China Academy of Chinese Medical Sciences, ChinaCopyright © 2023 Travadi, Shah, Pandit, Sharma, Joshi and Joshi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Madhvi Joshi, bWFkaHZpbWljcm9iaW9AZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.