
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 12 June 2023
Sec. Plant Development and EvoDevo
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1167458
This article is part of the Research Topic Stress-induced Flowering in Plants View all 7 articles
Litchi (Litchi chinensis) is an economically important fruit tree in southern China and is widely cultivated in subtropical regions. However, irregular flowering attributed to inadequate floral induction leads to a seriously fluctuating bearing. Litchi floral initiation is largely determined by cold temperatures, whereas the underlying molecular mechanisms have yet to be identified. In this study, we identified four CRT/DRE BINDING FACTORS (CBF) homologs in litchi, of which LcCBF1, LcCBF2 and LcCBF3 showed a decrease in response to the floral inductive cold. A similar expression pattern was observed for the MOTHER OF FT AND TFL1 homolog (LcMFT) in litchi. Furthermore, both LcCBF2 and LcCBF3 were found to bind to the promoter of LcMFT to activate its expression, as indicated by the analysis of yeast-one-hybrid (Y1H), electrophoretic mobility shift assays (EMSA), and dual luciferase complementation assays. Ectopic overexpression of LcCBF2 and LcCBF3 in Arabidopsis caused delayed flowering and increased freezing and drought tolerance, whereas overexpression of LcMFT in Arabidopsis had no significant effect on flowering time. Taken together, we identified LcCBF2 and LcCBF3 as upstream activators of LcMFT and proposed the contribution of the cold-responsive CBF to the fine-tuning of flowering time.
Litchi (Litchi chinensis Sonn.) is a subtropical evergreen fruit tree native in China (Hu et al., 2022), and is also widely cultivated in subtropical regions due to its high nutritional values, attractive appearance, and striking tastes. Litchi plants mostly flower and bear in the terminal shoots, whereas its irregular flowering attributed to the inadequate floral induction results in serious fluctuating production (Menzel, 1983). Perception of a period of low temperature is largely required for litchi floral initiation, and temperatures above 20°C are highly detrimental to the floral induction (Menzel and Simpson, 1988). Although the floral initiation of litchi can be affected by drought that appropriate drought prior to cold induction can reduce the requirement of low temperatures for litchi floral induction, but litchi plants cannot complete floral induction in the absence of prolonged exposure to low temperatures (Stern and Gazit, 2010; Shen et al., 2016).
Flowering is an important transition from the vegetative to reproductive phase during the plant’s life cycle in response to both environmental and endogenous signals, such as temperature, photoperiod, and age (Amasino, 2010; Song et al., 2013; Wang, 2014). Based on the genetic and physiological analyses in Arabidopsis thaliana, floral induction is regulated by four major genetic pathways, including day-length, vernalization, autonomous, and gibberellin-dependent pathways. These pathways fine-tune a set of genes to switch on/off the biosynthesis of florigen encoded by FLOWERING LOCUS T (FT), eventually determining the exact flowering time (Amasino, 2010; Song et al., 2013; Wang, 2014).
FT is a member of the phosphatidylethanolamine-binding protein (PEBP) family that can be clustered into three subfamilies, including FT-like, TERMINAL FLOWER 1 (TFL1)-like, and MOTHER OF FT AND TFL1 (MFT)-like clades. The MFT-like clade is proposed to be an ancestor of the family (Hedman et al., 2009). In general, PEBP members mostly interact with FLOWERIING LOCUS D (FD) protein, instead of directly bind to DNA, to manipulate the expression of downstream targets (Abe et al., 2005; Hanano and Goto, 2011; Wickland and Hanzawa, 2015). TFL1 and its homologs are best known for their opposite roles to FT in regulating flowering time, and act as key players in the formation of inflorescence architecture (Hanano and Goto, 2011; Wickland and Hanzawa, 2015). In contrast, MFT is strongly expressed in seeds and promotes seed dormancy (Xi et al., 2010; Nakamura et al., 2011), while the role of MFT in the determination of flowering time is still ambiguous. For instance, overexpression of MFT in Arabidopsis resulted in slightly early flowering, but the loss of AtMFT function barely led to an obvious phenotype in flowering, proposing a redundant role for MFT in Arabidopsis flowering (Yoo et al., 2004). In kiwifruit, two MFT-like genes show no influence on flowering time (Voogd et al., 2017). Interestingly, ectopic overexpression of AcMFT, a MFT homolog from a non-flowering plant Adiantum capillus-veneris, promotes the floral initiation and partially rescues the late-flowering defect in ft-1 mutants in Arabidopsis (Hou and Yang, 2016). However, OsMFT1 clearly acts as a suppressor of flowering time but a promoter of panicle architecture in rice (Song et al., 2018). Similar studies demonstrated that ectopic overexpression of HbMFT1 (Bi et al., 2016) and DnMFT (Li et al., 2012) in Arabidopsis causes delayed flowering. Despite the importance of PEBP members in plant development, their upstream regulators remain largely unknown.
Cold activates a range of so-called cold-regulated (COR) genes encoding proteins that enhance the freezing tolerance of plants. Such COR genes share a cis-acting DNA regulatory element in their promoters, namely, the C-repeat/dehydration responsive elements (CRT/DRE). CRT/DRE BINDING FACTORS (CBFs) serve as the key upstream regulators of the cold response pathway in plants (Stockinger et al., 1997). CBF/DREB1 is a class of AP2/ERF (APETALA2) transcription factor proteins that are extensively involved in responses of plants to external stresses such as cold and drought (Novillo et al., 2004; Thomashow, 2010; Zhou et al., 2011; Nakashima et al., 2014). There are four CBF homologs identified in Arabidopsis, of which CBF1, 2, and 3 can be significantly induced by cold, and improve freezing tolerance (Stockinger et al., 1997; Gilmour et al., 2004). In contrast to the three CBF homologs, AtCBF4 is up-regulated by drought instead of by low temperatures (Haake et al., 2002). It is noteworthy that AtCBF1 and AtCBF3 are modulated distinctly than AtCBF2, and AtCBF2 can negatively regulates the expression of AtCBF1 and AtCBF3 (Novillo et al., 2004; Novillo et al., 2007).
CBFs not only play an important role in plant response to abiotic stresses, but also participate in plant flowering regulation. In Arabidopsis, constitutively expression of AtCBF1 or AtCBF4 leads to late-flowering (Haake et al., 2002; Achard et al., 2008). The condition of short or intermittent low temperatures delays the flowering of Arabidopsis thaliana through the activation of the expression of FLC (FLOWERING LOCUS C) induced by CBFs, and overexpression of CBF1/2/3 in Arabidopsis thaliana can significantly induce the expression of FLC (Gilmour et al., 2004; Seo et al., 2009). Furthermore, AtFLC was proved to be directly activated by INDUCER OF CBF EXPRESSION 1 (ICE1), an upstream activator of CBFs, revealed by the analysis of ice1 mutants in which AtFLC was significantly down-regulated, and the time to flowering was significantly promoted (Lee et al., 2015). However, the CBF-dependent regulatory pathways in plant flowering are not fully understood.
In this research, the CBF homologs were isolated and identified in the litchi genome. The expression profiles of CBF genes in responses to low temperature were investigated. Changes in the expression of flowering genes were also examined during the floral induction. We showed the transcriptional regulation of LcMFT by LcCBFs. Taken together, we propose a new strategy for litchi floral induction in a CBF-dependent manner.
Potted air-layered litchi trees (Litchi chinenesis cv. Feizixiao) grown in the greenhouse at South China Agricultural University in Guangzhou were used in this study. The terminal shoots of these plants were pruned and turned to be fully mature prior to the cold treatment. The plants were treated at 15°C with the approximately 25% soil moisture for 40 days under nature lights. After the floral induction treatment, the temperature increased to 25°C for flower development. Leaves in the terminal shoots were collected 0, 10, 30 and 40 days during the cold treatment, and the leaf tissues were frozen in the liquid nitrogen as soon as possible, subsequently stored at -75°C for further studies.
To obtain overexpression constructs, the coding sequence of LcCBFs and LcMFT was linked to the expression vector pCAMBIA-1302 using the homologous recombination method. The recombinant plasmid was introduced into the Agrobacterium tumefaciens strain GV3100. Subsequently, the positive bacteria were transformed into Arabidopsis thaliana ecotype Columbia (Col) using the pollen tube introduction method to obtain transgenic Arabidopsis plants. Seeds of the transformed plants were germinated and selected on Murashige and Skoog (MS) agar media in the presence of Hygromycin (Hyg). One-week-old seedlings were transferred into soil (nutrient soil: vermiculite: perlite=4:1:1), and plants were grown in growth chambers at 23°C under long daylength condition (light/dark for 16h/8h). Homozygous transgenic plants were used in the following experiments.
The total RNA of the collected plant materials was extracted using the RNA Prep Pure Polysaccharide Polyphenol Plant TOTAL RNA Extraction kit (Beijing Tiangen Biochemical Technology). The first strand of cDNA was reverse-transcribed with the HiScript® II qRT SuperMix kit. cDNA samples were used as templates for the following real-time quantitative PCR (Q-PCR) assays. Triplicate quantitative assays and three biological replicates were performed on Bio-Rad iQ5 Optical System using the SYBR Green PCR Master Mix (Bio-Rad). The specificity of each pair of primers was evaluated before Q-PCR analysis and primers were listed in the Supplementary Table 1. The relative gene expression was calculated using the 2-△△CT method (Livak and Schmittgen, 2001), and values are indicated as mean ± standard error.
The freezing tolerance test of transgenic Arabidopsis plants over-expressing LcCBFs was conducted as previously documented (Li et al., 2018). 4-week-old seedlings grown in the soil were adopted to cold acclimation at 4°C for 3 d, and then were transferred into a freezing container set at 0°C with a decrease of 1°C/h to -8°C for 3 h. After the freezing treatment, the plants were put at 4°C in the dark for 12 h to recover and then were transferred to 22°C. The survival rate was scored three days later. Two transgenic lines for each gene were used in the experiment and at least three biological replicates per line were examined.
The drought tolerance test of transgenic Arabidopsis plants over-expressing LcCBFs is performed according to Siddiqua’s method with slight modifications (Siddiqua and Nassuth, 2011). 30-day-old Arabidopsis plants from wild-type (WT) and transgenic groups were used in this study. The soil substrates in the pots were saturated with water, and 12 h later plants were subjected to a 16 day-drought treatment without any watering. During the drought treatment, the environmental conditions were maintained at 23°C with the relative humidity is approximately 50%. After that, drought-treated plants were re-watered, and the survival ratio and phenotypes were analyzed after 7 days. By contrast, the control plants were watered for the whole duration of the experiment. Two transgenic lines for each gene in the experiment and at least three biological replicates per line were examined.
The CBF and PEBP protein sequences of Arabidopsis and litchi were obtained from TAIR (http://www.arabidopsis.org/) and SapBase (http://www.sapindaceae.com/) databases, respectively. The well-known CBF and PEBP sequences in Arabidopsis were used as queries to search candidate homologs by BLAST against the litchi genome with TBtools software (v1.09854; https://github.com/CJ-Chen/TBtools/releases). In addition, the CBF and PEBP Strubbelig-Receptor Family (SRF) domain (PF00847 and PF00319, respectively) were used to identify the genes using a Hidden Markov Model Search (Guan et al., 2021). All the predicted sequences were further confirmed using Conserved Domain Database (CDD) (http://www.ncbi.nlm.nih.gov/cdd/) and Simple Modular Architecture Research Tool (SMART, http://smart.embl-heidelberg.de/) to search for conserved domains. Finally, CBF and PEBP candidate genes were manually examined to remove redundant and incomplete sequences.
Fv/Fm (ratio of variable to maximum fluorescence, Fm, fluorescence maximum) value, reflecting the quantum efficiency of photosystem II, is widely used to illustrate the effects of environmental stresses on the plant. In this study, Fv/Fm values of leaves from plants subjected to cold or drought treatments were examined using a chlorophyll fluorescence imaging system (Imaging-PAM, Walz, Germany). Prior to the measurements, the whole plant was adapted to darkness for 20 min. Three individual plants of each line were used for Fv/Fm measurements.
For measuring of electrolyte leakage of leaves, two fresh leaves (approximately 0.2 g) of Arabidopsis plants from each group were cut and put into a 15 mL centrifuge tube with 5 mL double distilled H2O. The tubes were shaken at 150 rpm and 25°C for 2 h, and then the electrolyte leakage of the solution (L1) was measured using a conductivity meter (CON2700, Eutech instrument). After that, the solutions were boiled for 20 min, and the electrolyte leakage of which (L2) was measured again after they were cooled down to room temperature under running water. The relative electrolyte leakage was calculated as follows: EL (%) = (L1/L2) × 100%.
The luciferase assay was conducted in vitro according to the method as described (Shan et al., 2014). The coding sequence (CDS) of LcCBFs and the promoter of LcMFT were amplified with the primers listed in Supplementary Table 1. The CDS of LcCBFs were cloned into the pGreen II 0029 62-SK vector, while the promoter of LcMFT was recombined into the pGreen II 0800-LUC vector. The two sets of recombinant plasmids were electroporated into Agrobacterium tumefaciens (GV3101). Thereafter, positive bacterial cultures were selected and prepared with the infection buffer (200 mM acetosyringone, 10 mM MES, 10 mM MgCl2, pH 5.6) to an OD600 of 0.8 at 28°C. Mixtures of transformed A. tumefaciens expressing the promoter of LcMFT and the full CDS of LcCBFs were co-infiltrated into fully expanded leaves of approximately 3-week-old tobacco (Nicotiana benthamiana) plants with a needless syringe, while the mixtures of empty pGreen II 0029 62-SK and LcCBF-Luc vectors were co-infiltrated into the other half leaves as negative controls. The transformed tobacco plants were grown at 23°C in a growth chamber (16 h daylight). Three days later, the infected tobacco leaves were collected for measurements of renilla luciferase and firefly luciferase activities using a dual luciferase assay system (Promega, Madison, WI, USA), and the measurements were conducted on a GLO-MAX 20/20 luminometer (Promega, Madison, WI, USA). For each DNA-protein binding assay, two independent experiments and at least three biological replicates per experiment were performed.
Yeast-one-hybrid (Y1H) assays were conducted as previously described by Xiao et al. (2019) to detect the potential DNA-protein interaction, using the Matchmaker™ Gold Yeast One-Hybrid System kit (Clontech, CA, USA). The domain of the LcMFT promoter (1119 bp), including one CBF recognition core element (CCGAC), was amplified and inserted into the pAbAi plasmid. Thereafter, the recombinant plasmid was transformed into the competent Y1H Gold strain. The positive yeasts were selected and co-transformed with pGADT7-AD inserted with the coding sequence of the LcCBF genes. The binding of transcription factor and promoter was determined based on the growth ability of the yeast cells harboring these co-transformants on SD/-Leu medium in the presence of aureobasidin A (AbA).
Electrophoretic mobility shift assays were performed as described by Shan et al. (2014). The coding sequence of LcCBFs was amplified and fused into pGEX-4T-1 to generate the LcCBF-GST fusion construct. The fusion construct was transformed into E. coli strain BL21 cells. After growing to saturation, the transformed cells were collected and treated with 0.2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), and incubated at 37°C for 3 h to express the fusion protein. The LcCBF-GST fusion protein was purified and collected using a Pierce GST spin purification kit (Thermo Scientific) according to the instructions. The promoter fragments of LcMFT used as the probes were synthesized and biotin-labeled using a LightShift chemiluminescent EMSA kit (Thermo Scientific). LcCBF-GST protein and biotin-labeled probes were incubated at room temperatures, while unlabeled cold probes, mutated probes and the GST protein alone acted as competitors and negative controls, respectively. All of the biotin-labeled binding DNA was analyzed by 6% native polyacrylamide gel electrophoresis, and was detected using the chemiluminescence method on a ChemiDoc MP imaging system (Bio-Rad). The primers used in the EMSA assays are listed in Supplementary Table 1.
T-test was used for two-sample statistical comparisons in this study. For multiple comparisons, one way analysis of variance (ANOVA) was performed using SPSS software (v.22, IBM) followed by a Duncan test. The significant statistical differences are indicated in each figure.
Since CBFs play important roles both in freezing tolerance and thermos sensory-pathway regulated flowering, CBF genes were identified and investigated in this study. Four litchi CBF homologs were identified on the basis of BLAST searches against the litchi genome using Arabidopsis CBF genes. The length of proteins encoded by LcCBFs arranged from 192 to 259 amino acid residues (Figure 1A). According to the multiple alignment of protein sequences from litchi and Arabidopsis, all litchi CBF proteins contain a unique AP2 DNA-binding domain of about 60 amino acid residues, which is evolutionally conserved in plants (Figure 1A). Furthermore, as shown by the phylogenetic analysis of protein sequences, litchi CBFs are distinguished from the Arabidopsis CBF group, whereas LcCBF1 and 2 were classified into a subgroup with tomato CBF homologs that has showed a role in enhancing freezing tolerance (Figure 1B) (Zhang et al., 2004; Zhang et al., 2020). Taken together, these results show that the identified litchi CBF sequences are Arabidopsis orthologs, but have species-related features.
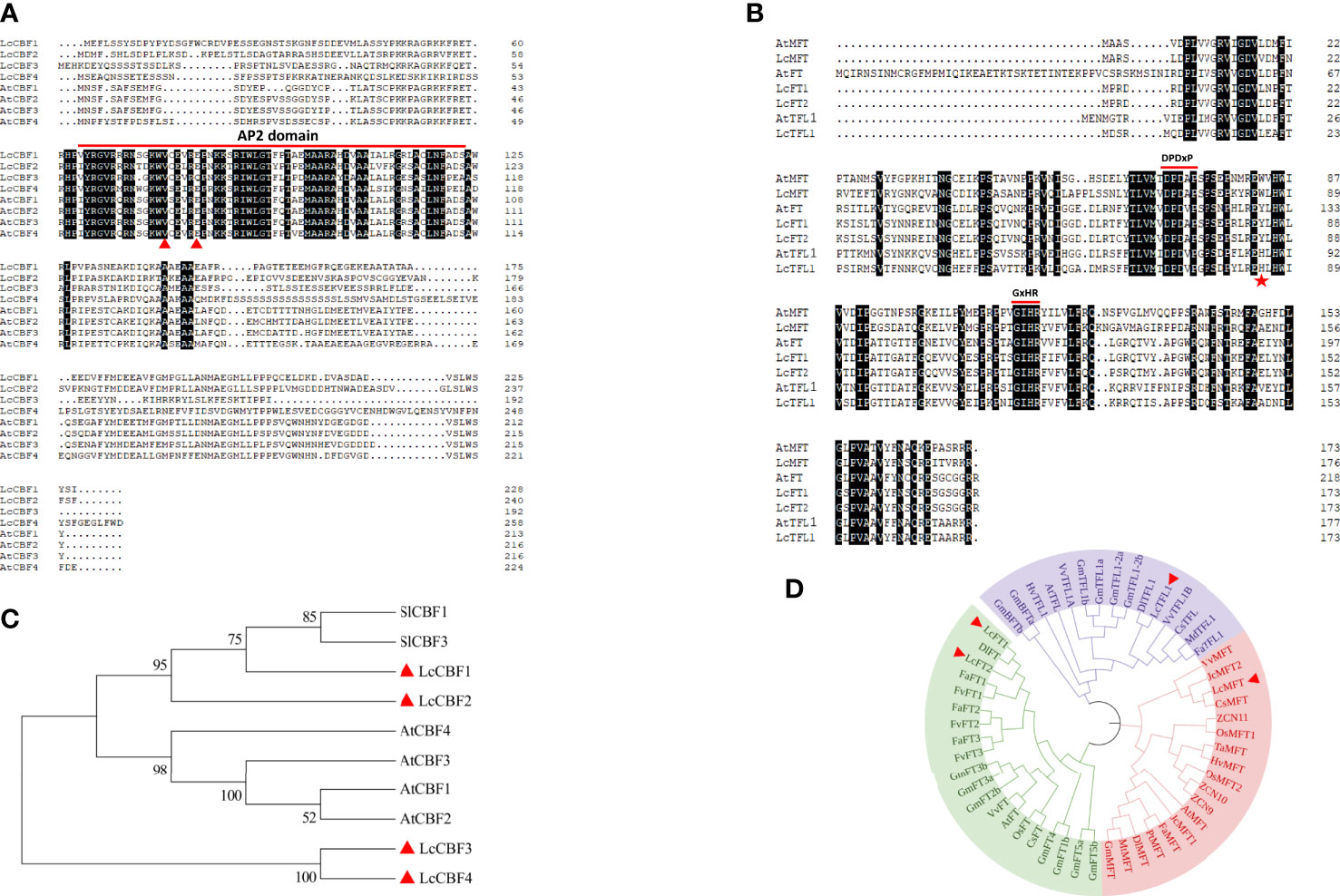
Figure 1 Alignment and sequence analysis of protein sequences encoded by LcCBFs and LcMFT. (A) Alignment of CBF protein sequences from litchi and Arabidopsis, and the conserved AP2 domain was indicated with a red line. (B) Phylogenetic analysis of CBF protein sequences from litchi (indicated with red triangles), Arabidopsis thaliana and tomato. Accession numbers of these protein sequences are shown in Supplementary Table 2. (C) Alignment of putative PEBP protein sequences from litchi and their homologs. The highly conserved motifs of the PEBP family including D-P-D-x-P and G-x-H-R were indicated with red lines. The conserved position of each subfamily was indicated with a red pentagram. (D) Phylogenetic analysis of PEBP protein sequences from litchi (indicated with red triangles) and other species. Accession number and species name of these protein sequences are shown in Supplementary Table 2.
The PEBP family can be mainly divided into three subclades, including FT, MFT, and TFL1 that primarily act as floral pathway integrators (Hanano and Goto, 2011; Wickland and Hanzawa, 2015). Previously, two members of the FT subgroup were identified and studied in litchi (Ding et al., 2015; Xiao et al., 2019). Here, homologs in the TFL1 and MFT subgroups were identified via protein homology searches of the litchi genome using the full-length protein sequences of PEBP members in Arabidopsis and the genome annotation, and found one member for each subclade. Overall comparisons of PEBP protein sequences between litchi and Arabidopsis showed that highly conserved motifs including D-P-D-x-P and G-x-H-R, were present in all litchi putative homologs (Figure 1C) (Banfield and Brady, 2000). Consistent with the previous study, histidine 88 of TFL1, tyrosine 86 of FT, and tryptophan of MFT exclusively exist at the corresponding positions (indicated with a red pentagram) in each subclade (Figure 1C) (Hanzawa et al., 2005). In addition, phylogenetic analysis showed that PEBP family could be clearly divided into three groups, in which litchi MFT and TFL1 exhibited closer genetic relationship to homologs from citrus species than others (Figure 1D).
After an exposure to 40 days of low temperature floral induction, all litchi plants were competent to flower. Therefore, the expression of critical flowering-related genes was investigated. As shown by Q-PCR results, the expression of LcFT1 gene significantly increased during the cold-dependent floral induction, and had approximately fifty times increase at the complete of induction (Figure 2A). By contrast, the abundance of LcMFT transcript decreased in response to the cold and showed a significant difference 30 days after the treatment. Interestingly, both the expression of LcFLC and LcSOC1 (SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1) showed a slight incline during the floral induction, but the changes were not statistically significant (Figure 2A).
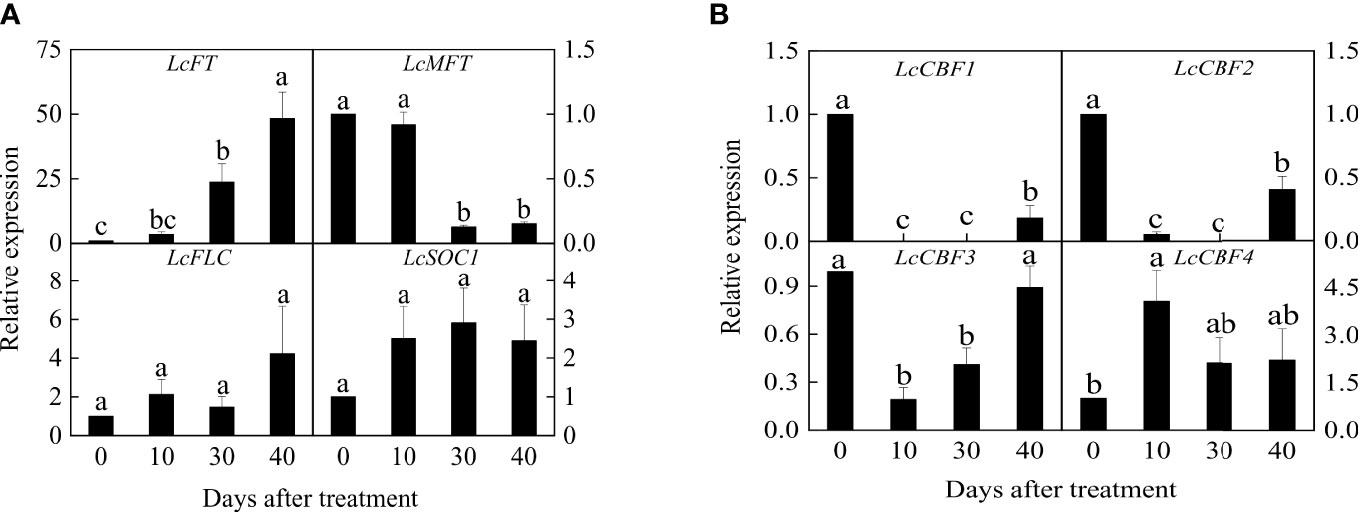
Figure 2 Expression profiles of flowering genes (A) and LcCBFs (B) during the floral induction. Expression values of each gene are calculated as a ratio relative to the beginning of floral induction (0 day), which was set at 1. Values with different letters for gene expression are significantly different at p < 0.05. Error bars indicate standard error from six replicates at each time point.
In addition, the key cold-responsive genes CBF homologs were examined in litchi leaves. LcCBF1 and LcCBF2 showed a rapid and significant decrease during the floral induction (Figure 2B). Similarly, LcCBF3 decreased in response to the early cold treatment, whereas its expression level was restored at the end of the induction. LcCBF4 had a significant increase during the ten days, and then decreased, eventually exhibiting no significant changes (Figure 2B).
Despite the few upstream regulators of PEBP family members and the similar expression patterns of LcCBFs and LcMFT, the regulation of LcMFT by LcCBFs was investigated. Yeast one-hybrid (Y1H) analysis was first performed to this aim, due to that there was one CBF recognition element (CCGAC) in the promoter of LcMFT. However, there was strong basal activity in yeast cells, when a whole region of the promoter (1119 base nucleotides prior to the transcription start site) was cloned in front of the reporter gene AUR1-C. Hence, the promoter of LcMFT was truncated into two domains covering the recognition element (Figure 3A). After the Y1H reporter strains constitutively expressing the LcCBF2 or LcCBF3 effector, the growth of yeast cells including the LcMFT promoters was observed in the presence of aureobasidin A to examine the DNA-protein binding. The yeast cells harboring the LcMFT promoter could grow well when co-transformed with LcCBF2 or LcCBF3 effectors (Figure 3B). As evidenced by the Y1H analysis, LcCBF2 or LcCBF3 can directly bind to the promoter of LcMFT.
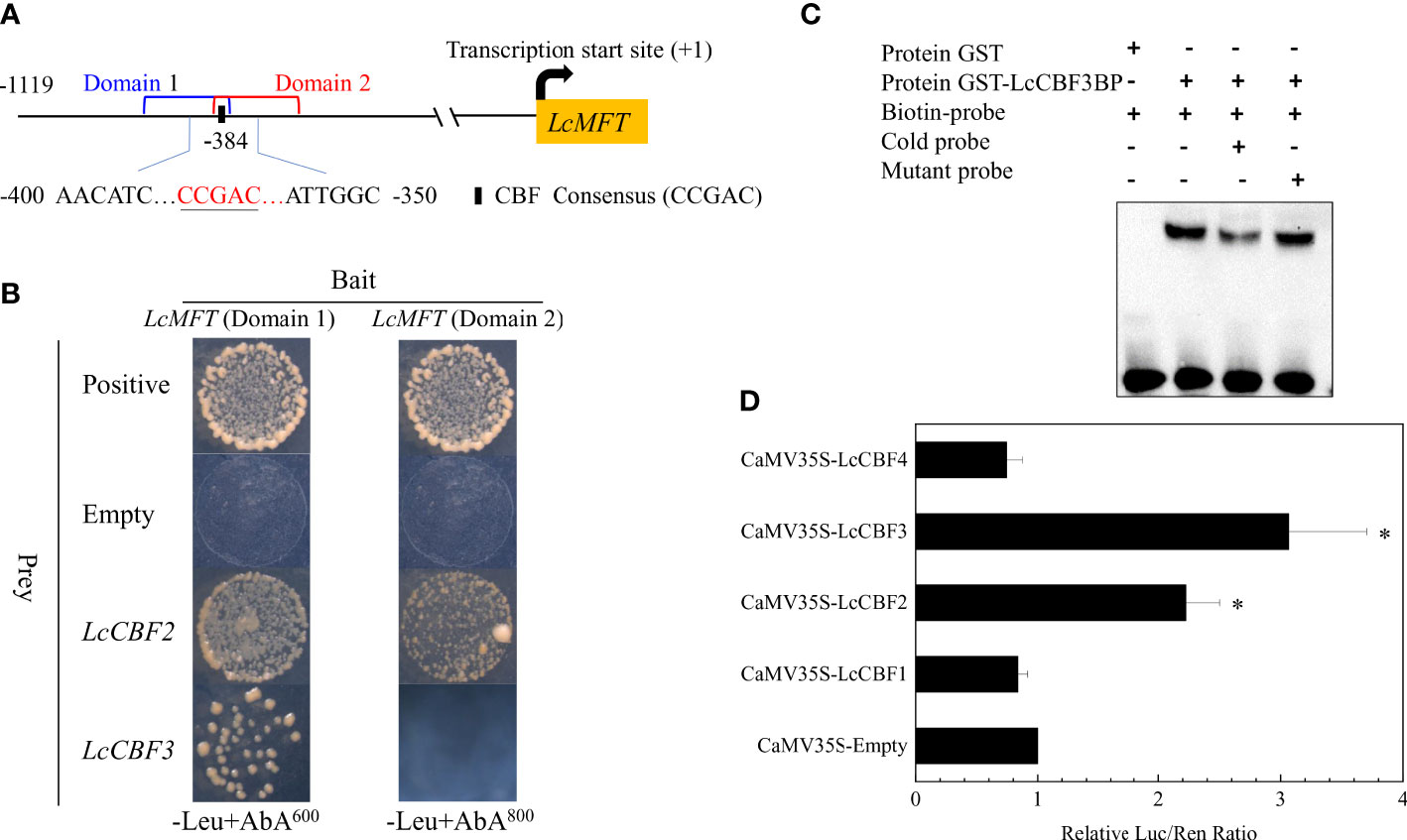
Figure 3 Analysis of interaction of LcCBF proteins and the promoter of LcMFT. (A) the left panel demonstrates a schematic representation of CBF recognition site (CBFRS) in the promoter of LcMFT. (B) yeast-one-hybrid (Y1H) assay for direct binding of LcCBF2/3 proteins to the promoter of LcMFT. The fragment of the promoter of LcMFT was integrated into the reporter gene AUR1-C (an Aureobasidin A (AbA) resistance gene in yeast). The Y1H reporter strains were transformed with fused plasmids constitutively expressing LcCBF2 and LcCBF3 effector, empty (negative control) and P53 effector (positive control). Interactions between LcCBFs and the promoter of LcMFT were determined based on the growth ability of the transformed yeasts on the selective SD medium without Leu in the presence of AbA. (C) Electrophoretic mobility shift assays (EMSA) showing the GST-LcCBF3 fusion protein binds to DNA probe from the LcMFT promoter in vitro. Biotin-labeled probes containing CBFRS and mutated CBFRS, as well as the cold probes (biotin-labeled) were incubated with GST-LcCBF3 proteins, and the DNA-protein complexes were separated on 6% polyacrylamide gels. The probe containing the mutated CBFRS was used to test binding specificity. (D) LcCBF2 and LcCBF3 activate the activity of LcMFT promoter in the dual Luciferase complementation assay. Transient co-expression of LcCBFs and the promoter of LcMFT were performed in tobacco (N. benthamiana) leaves as the reporter and effector vectors, respectively. Data show the ratio of firefly luciferase (LUC) and renilla luciferase (REN) activities, and the ratios of LUC to REN of the empty vector (SK) plus the promoter was used as the control and set as 1. The significant difference was determined by Student’s t test and indicated with asterisks at p < 0.05.
Furthermore, electrophoretic mobility shift assay (EMSA) assays were performed using purified recombinant proteins of LcCBFs to confirm the DNA-protein interaction. While no mobility shift was detected in the presence of GST alone, LcCBF3 proteins could bind to the labeled probes including CBF recognition sites derived from the promoter of LcMFT, leading to mobility shifts (Figure 3C). Additionally, the shifted bands were effectively competed by the addition of unlabeled probes (cold probes), but not by the mutated probes (Figure 3C). Unfortunately, we failed in the purification of the LcCBF2-GST protein to conduct the similar EMSA assays.
To verify the DNA-protein binding between LcCBF2/3 and the promoter of LcMFT in vivo, we performed a dual-luciferase transient co-transformational assay in tobacco leaves. The interaction between LcCBF2/3 and the promoter of LcMFT caused significant increases in Luc activities (Figure 3D), indicating that LcCBF2 and LcCBF3 could interact with the promoter of LcMFT to activate its transcription.
To confirm the putative functions of LcCBF2, LcCBF3, and LcMFT, the constructs containing LcCBF2/3 or LcMFT was driven by the constitutive cauliflower mosaic virus 35S promoter (CaMV 35S), and was then transformed into wild-type Arabidopsis (Col-0) to obtain overexpression transgenic plants. Two transgenic T3 lines per gene were selected based on the expression level for further analysis (Figure 4A). Furthermore, downstream cold and drought tolerance-related genes in the CBF-dependent regulatory pathway, including AtCOR6.6, AtCOR15a, AtCOR47 and AtCOR78 were examined in transgenic Arabidopsis plants (Kasuga et al., 1999; Siddiqua and Nassuth, 2011). As expected, over-expression of LcCBF2/3 significantly induced the expression level of all four or most of these COR genes (Figure 4B).
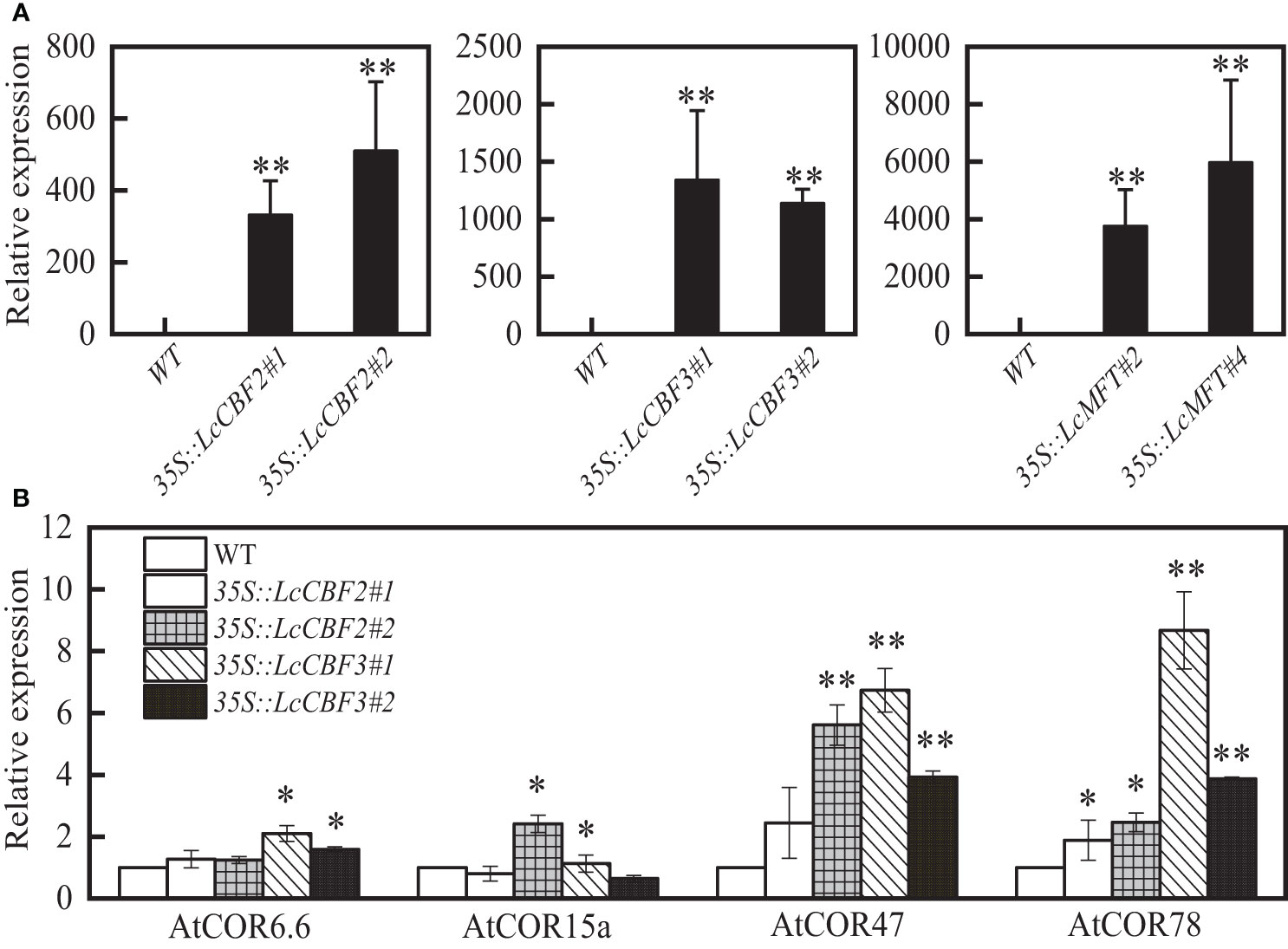
Figure 4 Ectopic expression of LcCBF2, LcCBF3 and LcMFT in the wild-type Arabidopsis thaliana (Col-0). Identification of the expression level of these litchi genes (A) and abiotic tolerance-related Arabidopsis genes (B) in transgenic plants were determined by Q-PCR. Asterisks indicate significant difference between wild-type and transgenic Arabidopsis plants at P < 0.05 (*) or P < 0.01 (**).
The 35S::LcCBF2/3 transgenic Arabidopsis plants were subjected to a freezing treatment to investigate their roles in modulating cold resistance. First, 2-week-old transgenic Arabidopsis seedlings grown on MS agar medium were subjected to the freezing treatment. After the freezing treatment, approximately half of wild-type plants died after a further 3 d under the non-stressed conditions (Figure 5A), whereas the 35S::LcCBF2 and 35S::LcCBF3 transgenic seedlings had a significantly higher survival rate at the same conditions (Figure 5B). Furthermore, a similar freezing treatment was applied to 4-week-old wild-type and transgenic plants, and chlorophyll fluorescence (indicates as Fv/Fm values to monitor PSII activity) and electrolyte leakage values of leaves were quantified to determine the freezing damage. The average Fv/Fm values detected with the chlorophyll fluorescence test were 0.75 for both the wild-type and LcCBF transgenic Arabidopsis lines at regular growing conditions (Figure 5C). After the freezing treatment, the Fv/Fm values remarkedly decreased in wild-type Arabidopsis leaves and showed the lowest value, followed by the values for 35S::LcCBF2 plants, whereas the 35S::LcCBF3 transgenic plants exhibited no significant differences in Fv/Fm values across the freezing treatment (Figure 5C). Accordingly, the average electrolyte leakage value of leaves from the 35S::LcCBF3 transgenic plants was the lowest after the treatment, followed by the values for 35S::LcCBF2 plants, whereas the wild-type plants showed a significantly higher values at the complete of freezing treatment (Figure 5D). It would be concluded that an increase of LcCBF2 and/or LcCBF3 expression could improve the freezing tolerance of plants.
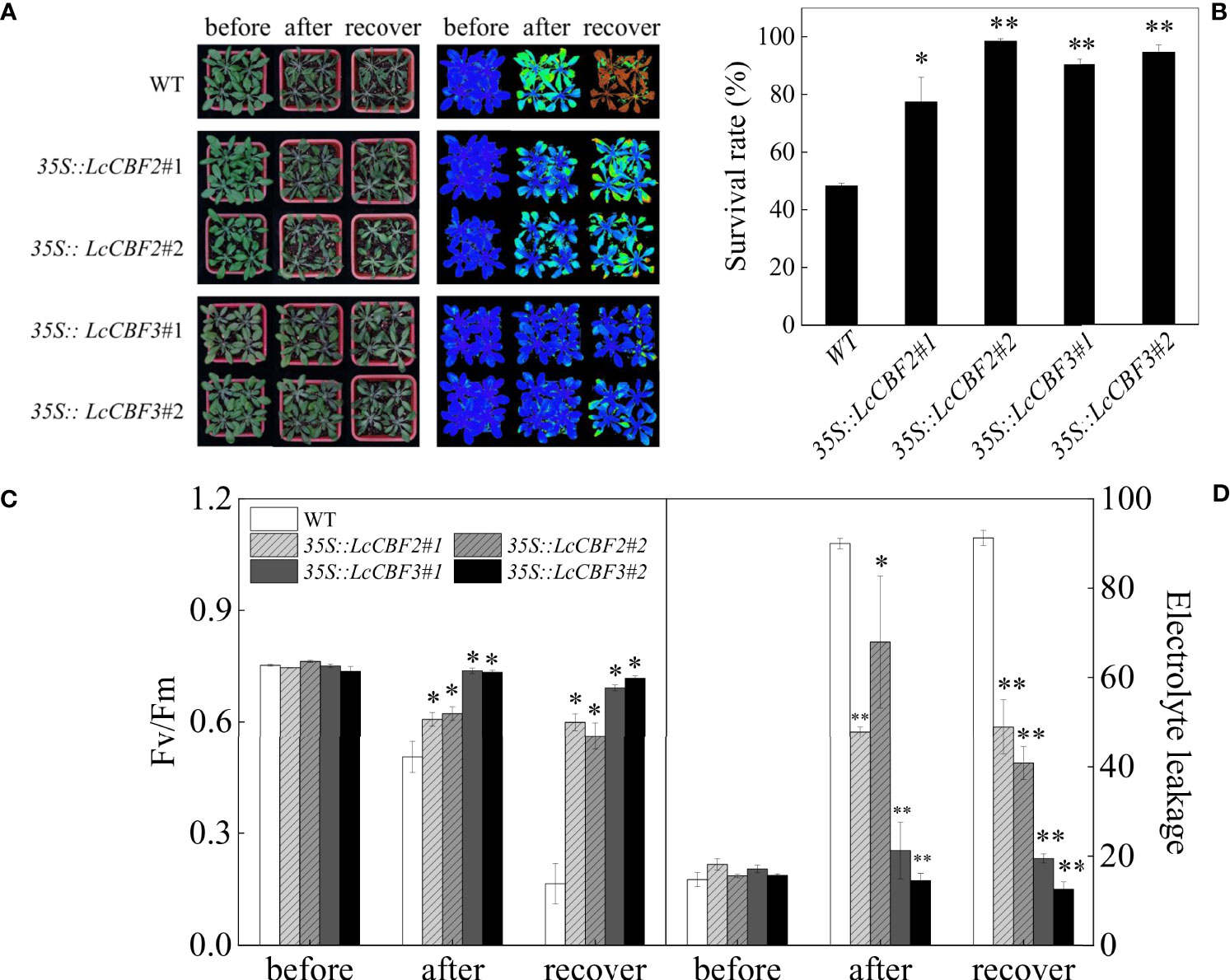
Figure 5 Overexpression of LcCBF2 and LcCBF3 improved the freezing tolerance of transgenic Arabidopsis thaliana plants. (A) response of wild-type (WT) and transgenic Arabidopsis thaliana over-expressing LcCBF2 and LcCBF3 to the freezing treatment; (B) Survival rates of plants after the freezing treatment; (C) comparisons of Fv/Fm and electrolyte leakage values (D) of freezing-treated WT and transgenic plants. The significant difference was determined by t test and indicated with * (at p < 0.05) and ** (at p < 0.01), respectively.
In addition, the effect of overexpression of LcCBF2 and 3 on drought tolerance was tested. Individually potted wild-type and the selected 35S::LcCBF2 and 35S::LcCBF3 transgenic lines were subjected to a drought treatment by removal of watering for 16 d, and all lines were analyzed after a further 7 d of rewatering. More than 60% of wild-type plants were dead (Figure 6A), whereas the overexpression transgenic lines had a significantly higher survival rate at the completion of the treatment, indicating that both LcCBF2 and LcCBF3 could increase drought tolerance (Figure 6B). The reduction in Fv/Fm values was observed in both wild-type and transgenic plants attributed to the drought treatment, whereas the average Fv/Fm values remarkedly decreased in wild-type plants and could not be restored by rewatering (Figure 6C). Drought treatment also induced serious damage in membrane stability, resulting in a significant increase in electrolyte leakage in wild-type plants (Figure 6D). Taken together, the overexpression of LcCBF2 or LcCBF3 could improve the drought tolerance of transgenic Arabidopsis thaliana.
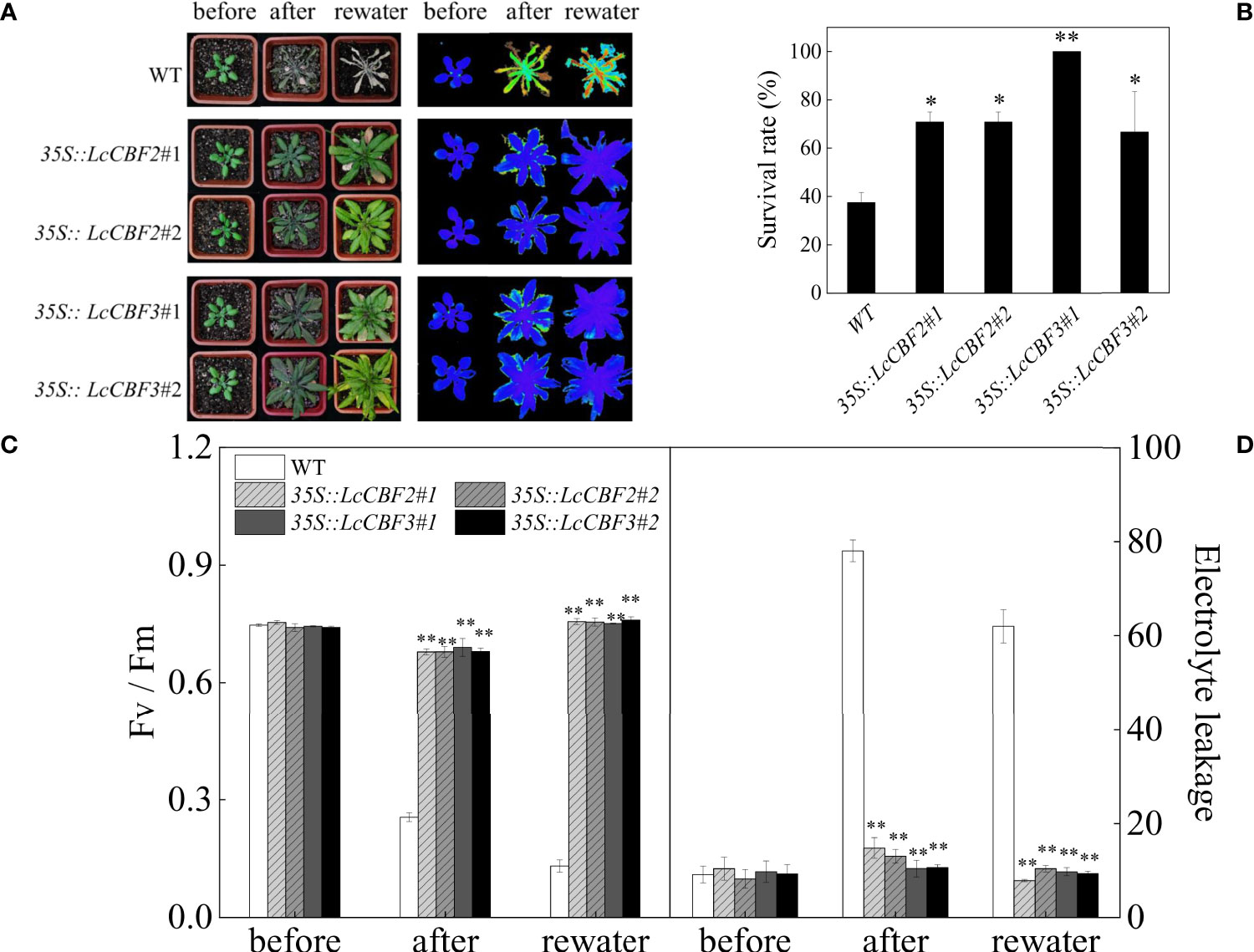
Figure 6 Overexpression of LcCBF2 and LcCBF3 improved the drought tolerance of transgenic Arabidopsis thaliana plants. (A) response of wild-type (WT) and transgenic Arabidopsis thaliana over-expressing LcCBF2 and LcCBF3 to the drought treatment; (B) Survival rates of plants after the drought treatment; (C) comparisons of Fv/Fm and electrolyte leakage values (D) of drought-treated WT and transgenic plants. The significant difference was determined by t test and indicated with * (at p < 0.05) and ** (at p < 0.01), respectively.
Moreover, the putative function of LcCBFs and LcMFT in the regulation of flowering time was analyzed. The time of bolting and the number of rosette leaves at that time were calculated as indicators of flowering time. Interestingly, overexpression of LcMFT could not influence the flowering time or alter the number of rosette leaves at flowering, although LcMFT is also a member of the PEBP family. By contrast, 35S::LcCBF2 and 35S::LcCBF3 transgenic lines took significant longer to bolt and had more cauline leaves at flowering under long-day conditions compared with the wild-type plants (Figure 7A). However, the morphological features of main inflorescence and rosette leaves were unaffected in those transgenic plants (Figure 7B). In conclusion, overexpression of LcCBF2 and LcCBF3, instead of LcMFT, delayed the flowering time of Arabidopsis thaliana.
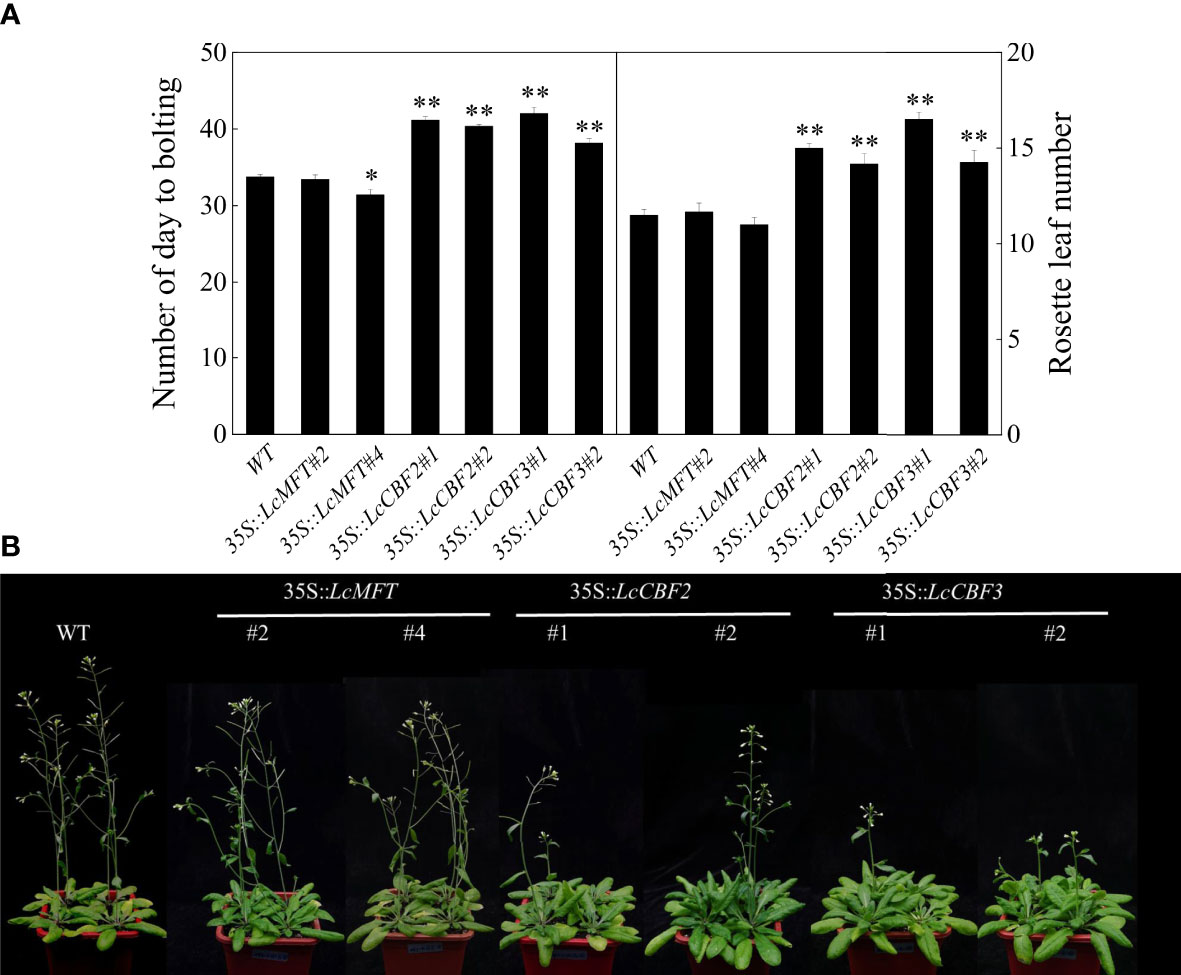
Figure 7 Effect of ectopic over-expressing LcMFT and LcCBFs on flowering. (A) Comparison of flowering time and number of rosette leaves at flowering between wild-type (WT) and 35S::LcMFT and 35S::LcCBFs transgenic lines; (B) Flowering phenotypes of WT and 35S::LcMFT and 35S::LcCBFs transgenic lines. The significant difference was determined by t test and indicated with * (at p < 0.05) and ** (at p < 0.01), respectively.
As a subtropical fruit tree, low ambient temperatures (< 18°C) or intermittent cold can activate the floral initiation in litchi (O’Hare, 2002). Consistently, in this study the potted litchi plants complete floral induction after 40-day exposure to 15°C in a green house. During low temperature-dependent litchi floral induction, the expression level of LcCBF2 and LcCBF3 decreased (Figure 2). It has been reported that the endogenous CBF protein levels are extremely low under warm growth conditions, but they dramatically accumulated in response to a 4°C treatment, reaching a peak at 6 h approximately (Lv et al., 2021). However, the accumulated effects of low ambient temperature result in a decreased expression of CBFs in Arabidopsis, due to the degradation of ICE1 protein (the upstream activator of CBFs) by the RING finger protein HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 (HOS1) (Dong et al., 2006; Lee et al., 2012). HOS1 can physically interact with ICE1 and causes the ubiquitination and subsequent degradation of ICE1 protein (Dong et al., 2006). HOS1 protein predominantly resides in the cytoplasm at normal growth temperatures, whereas HOS1 transfers into the nucleus and accumulated there in the presence of low temperature (Lee et al., 2001; Dong et al., 2006). Moreover, the recessive hos1 mutation leads to enhanced induction of the CBF genes by low temperature and their downstream genes, while overexpression of HOS1 represses the expression of CBFs as well as of their downstream cold-responsive genes, thereby supporting the reduction of CBF expression in litchi in response to floral inductive low temperature (Lee et al., 2001; Dong et al., 2006).
In addition, ectopic overexpression of LcCBF2 or LcCBF3 in Arabidopsis resulted in delayed flowering time and increased freezing and drought tolerance (Figures 5–7). In accordance with the notion, it has been demonstrated that overexpression of CBF1, CBF2, and CBF3 in Arabidopsis causes delayed flowering and dwarf phenotypes, as well as alterations in indicators involved in increased freezing tolerance, such as accumulations of proline and sugar and transcriptional activation of COR genes (Gilmour et al., 2004). The delayed flowering may be partially attributed to the activation of FLC by the increased expression of CBFs, subsequently repressing the two flowering pathway integrators FT and SOC1 (Seo et al., 2009). As the upstream regulator of CBFs, ICE1 also directly activates the gene encoding FLC, which represses SOC1 expression. By contrast, SOC1 play as a transcriptional repressor of ICE1 and CBF genes, inducing flowering with a reduction of freezing tolerance under floral promotive conditions (Seo et al., 2009; Lee et al., 2015).
In this report, we also showed the direct binding of LcCBF2/3 proteins to the LcMFT in vivo and vitro (Figure 3), as well as the involvement of the CBF-MFT module in fine-tuning of litchi flowering (Figure 7). The CBF/DREB1 transcription factors are able to specifically recognize a CRT/DRE (CCGAC) motif within the promoters of COR genes to mediate their expression. In Arabidopsis, according to the ChIP-seq analysis on a genome-wide scale, the binding region peaks of the three CBFs were all located 1,500-2,000 bp upstream of the transcription start site (TSS), with the highest frequency of peaks around the TSS itself. However, more than 50% of the binding sites for each CBF protein were located within the 1,000-bp promoter regions of the target genes (Song et al., 2021). Furthermore, RNA-seq data derived from individual cbf1, cbf2 and cbf3 mutants showed the over-lapping and distinct regulated genes by each CBF protein (Jia et al., 2016; Zhao et al., 2016; Shi et al., 2017; Song et al., 2021). In this study, one CRT/DRE cold response element was found in the proximal region of the promoter of LcMFT, and the protein-DNA interaction was exclusively observed between LcCBF2/3 and LcMFT promoter as indicated by the analysis of EMSA and Dual luciferase complementation assay (Figure 3).
MFT belongs to the PEBP family that includes FT-like subfamily, TFL1-like subfamily and FT-like subfamily. However, members within this family show distinct or opposite functions in regulating flowering time (Ahn et al., 2006). LcMFT, similar to the LcFT1 and LcTFL1, has been demonstrated to physically interact with the LcFD protein (Data submitted). The decrease of LcCBF and its target (LcMFT) during the litchi floral induction may inhibit the competitive interaction between LcMFT and LcFD, enhancing litchi flowering. Similar observations in other species also demonstrate the negative role of MFT homologs in controlling flowering time (Li et al., 2012; Bi et al., 2016). However, overexpression of LcMFT in Arabidopsis did not alter the flowering time significantly, indicating the potential weakened protein-protein interaction between LcMFT and AtFD, and/or the presence of other downstream player(s) of CBF protein involved in flowering (Yoo et al., 2004; Hou and Yang, 2016).
In conclusion, four CRT/DRE BINDING FACTORS (CBF) homologs were identified in litchi, of which LcCBF1, LcCBF2 and LcCBF3 decreased in response to floral inductive cold. A similar expression pattern was observed for the MOTHER OF FT AND TFL1 homolog (LcMFT) in litchi. Furthermore, both LcCBF2 and LcCBF3 were found to bind to the promoter of the LcMFT gene to activate its expression. Ectopic overexpression of LcCBF2 and LcCBF3 in Arabidopsis caused delayed flowering and increased freezing and drought tolerance. Therefore, prolonged low temperatures facilitate litchi floral initiation partially by the suppression of LcCBF2 and LcCBF3.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
JS, HC, and XS contributed to the design of this work. Material preparation, data measurements and analyses were performed by XS, YY, SW, CW, and WS. The first draft of the manuscript was written by XS. The latest version of the manuscript was written by XS and JS. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China under Grant (31601705), the National Key R & D Program of China under Grant (2020YFD1000103) and China Agriculture Research System under grant (CARS-32).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1167458/full#supplementary-material
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. doi: 10.1126/science.1115983
Achard, P., Gong, F., Cheminant, S., Alioua, M., Hedden, P., Genschik, P., et al. (2008). The cold-inducible CBF1 factor–dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20, 2117–2129. doi: 10.1105/tpc.108.058941
Ahn, J. H., Miller, D., Winter, V. J., Banfield, M. J., Lee, J. H., Yoo, S. Y., et al. (2006). A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 25, 605–614. doi: 10.1038/sj.emboj.7600950
Amasino, R. (2010). Seasonal and developmental timing of flowering. Plant J. 61, 1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x
Banfield, M. J., Brady, R. L. (2000). The structure of Antirrhinum centroradialis protein (CEN) suggests a role as a kinase regulator. J. Mol. Biol. 297, 1159–1170. doi: 10.1006/jmbi.2000.3619
Bi, Z. H., Li, X., Huang, H. S., Hua, Y. (2016). Identification, functional study, and promoter analysis of HbMFT1, a homolog of MFT from rubber tree (Hevea brasiliensis). Int. J. Mol. Sci. 17, 247. doi: 10.3390/ijms17030247
Ding, F., Zhang, S., Chen, H., Su, Z., Zhang, R., Xiao, Q. S., et al. (2015). Promoter difference of LcFT1 is a leading cause of natural variation of flowering timing in different litchi cultivars (Litchi chinensis sonn.). Plant Sci. 241, 128–137. doi: 10.1016/j.plantsci.2015.10.004
Dong, C. H., Agarwal, M., Zhang, Y., Xie, Q., Zhu, J. K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. PNAS 103, 8281–8286. doi: 10.1073/pnas.0602874103
Gilmour, S. J., Fowler, S. G., Thomashow, M. F. (2004). Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 54, 767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4
Guan, H., Wang, H., Huang, J., Liu, M., Chen, T., Shan, X., et al. (2021). Genome-wide identification and expression analysis of MADS-box family genes in litchi (Litchi chinensis sonn.) and their involvement in floral sex determination. Plants (Basel Switzerland) 10 (10), 1–17. doi: 10.3390/plants10102142
Haake, V., Cook, D., Riechmann, J., Pineda, O., Thomashow, M. F., Zhang, J. Z., et al. (2002). Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 130, 639–648. doi: 10.1104/pp.006478
Hanano, S., Goto, K. (2011). Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant Cell 23, 3172–3184. doi: 10.1105/tpc.111.088641
Hanzawa, Y., Money, T., Bradley, D. (2005). A single amino acid converts a repressor to an activator of flowering. PNAS 102, 7748–7753. doi: 10.1073/pnas.0500932102
Hedman, H., Källman, T., Lagercrantz, U. (2009). Early evolution of the MFT-like gene family in plants. Plant Mol. Biol. 70, 359–369. doi: 10.1007/s11103-009-9478-x
Hou, C. J., Yang, C. H. (2016). Comparative analysis of the pteridophyte Adiantum MFT ortholog reveals the specificity of combined FT/MFT c and n terminal interaction with FD for the regulation of the downstream gene AP1. Plant Mol. Biol. 91, 563–579. doi: 10.1007/s11103-016-0489-0
Hu, G. B., Feng, J. T., Xiang, X., Wang, J. B., Salojärvi, J., Liu, C. M., et al. (2022). Two divergent haplotypes from a highly heterozygous lychee genome suggest independent domestication events for early and late-maturing cultivars. Nat. Genet. 54, 73–83. doi: 10.1038/s41588-021-00971-3
Jia, Y. X., Ding, Y. L., Shi, Y. T., Zhang, X. Y., Gong, Z. Z., Yang, S. H., et al. (2016). The cbfs triple mutants reveal the essential functions of CBFs in cold acclimation and allow the definition of CBF regulons in Arabidopsis. New Phytol. 212, 345–353. doi: 10.1111/nph.14088
Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. doi: 10.1038/7036
Lee, J. H., Jung, J. H., Park, C. M. (2015). INDUCER OF CBF EXPRESSION 1 integrates cold signals into FLOWERING LOCUS c-mediated flowering pathways in Arabidopsis. Plant J. 84, 29–40. doi: 10.1111/tpj.12956
Lee, J. H., Kim, J. J., Kim, S. H., Cho, H. J., Kim, J., Ahn, J. H., et al. (2012). The E3 ubiquitin ligase HOS1 regulates low ambient temperature-responsive flowering in Arabidopsis thaliana. Plant Cell Physiol. 53, 1802–1814. doi: 10.1093/pcp/pcs123
Lee, H., Xiong, L., Gong, Z., Ishitani, M., Stevenson, B., Zhu, J. K., et al. (2001). The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 15, 912–924. doi: 10.1101/gad.866801
Li, R. H., Wang, A., Sun, S. L., Liang, S., Wang, X. J., Ye, Q. S., et al. (2012). Functional characterization of FT and MFT ortholog genes in orchid (Dendrobium nobile lindl) that regulate the vegetative to reproductive transition in Arabidopsis. Plant Cell. Tissue Organ Cult. 111, 143–151. doi: 10.1007/s11240-012-0178-x
Li, J., Wang, Y. Q., Yu, B., Song, Q. P., Liu, Y., Chen, T. H. H., et al. (2018). Ectopic expression of StCBF1 and ScCBF1 have different functions in response to freezing and drought stresses in Arabidopsis. Plant Sci. 270, 221–233. doi: 10.1016/j.plantsci.2018.01.015
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lv, J., Liu, J. Y., Ming, Y. H., Shi, Y. T., Song, C. P., Gong, Z. Z., et al. (2021). Reciprocal regulation between the negative regulator PP2CG1 phosphatase and the positive regulator OST1 kinase confers cold response in Arabidopsis. J. Integr. Plant Biol. 63, 1568–1587. doi: 10.1111/jipb.13100
Menzel, C. M. (1983). The control of floral initiation in lychee: a review. Sci. Hortic. 21, 201–215. doi: 10.1016/0304-4238(83)90093-6
Menzel, C. M., Simpson, D. R. (1988). Effect of temperature on growth and flowering of litchi (Litchi chinensis sonn.) cultivars. J. Hortic. 60, 349–360. doi: 10.1080/14620316.1988.11515869
Nakamura, S., Abe, F., Kawahigashi, H., Nakazono, K., Tagiri, A., Matsumoto, T., et al. (2011). A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell 23, 3215–3229. doi: 10.1105/tpc.111.088492
Nakashima, K., Yamaguchi-Shinozaki, K., Shinozaki, K. (2014). The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00170
Novillo, F., Alonso, J. M., Ecker, J. R., Salinas, J. (2004). CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. PNAS 101, 3985–3990. doi: 10.1073/pnas.0303029101
Novillo, F., Medina, J., Salinas, J. (2007). Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. PNAS 104, 21002–21007. doi: 10.1073/pnas.0705639105
O’Hare, T. J. (2002). Interaction of temperature and vegetative flush maturity influences shoot structure and development of lychee (Litchi chinensis sonn.). Sci. Hortic. 95, 203–211. doi: 10.1016/S0304-4238(02)00035-3
Seo, E., Lee, H., Jeon, J., Park, H., Kim, J., Noh, Y., et al. (2009). Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell 21, 3185–3197. doi: 10.1105/tpc.108.063883
Shan, W., Kuang, J. F., Lu, W. J., Chen, J. Y. (2014). Banana fruit NAC transcription factor MaNAC1 is a direct target of MaICE1 and involved in cold stress through interacting with MaCBF1. Plant Cell Environ. 37, 2116–2127. doi: 10.1111/pce.12303
Shen, J. Y., Xiao, Q. S., Qiu, H. J., Chen, C. J., Chen, H. B. (2016). Integrative effect of drought and low temperature on litchi (Litchi chinensis sonn.) floral initiation revealed by dynamic genome-wide transcriptome analysis. Sci. Rep. 6, 1–11. doi: 10.1038/srep32005
Shi, Y. H., Huang, J. Y., Sun, T. S., Wang, X. F., Zhu, C. Q., Ai, Y. X., et al. (2017). The precise regulation of different COR genes by individual CBF transcription factors in Arabidopsis thaliana. J. Integr. Plant Biol. 59, 118–133. doi: 10.1111/jipb.12515
Siddiqua, M., Nassuth, A. (2011). Vitis CBF1 and Vitis CBF4 differ in their effect on Arabidopsis abiotic stress tolerance, development and gene expression. Plant Cell Environ. 34, 1345–1359. doi: 10.1111/j.1365-3040.2011.02334.x
Song, Y. H., Ito, S., Imaizumi, T. (2013). Flowering time regulation: photoperiod-and temperature-sensing in leaves. Trends Plant Sci. 18, 575–583. doi: 10.1016/j.tplants.2013.05.003
Song, S., Wang, G. F., Hu, Y., Liu, H. Y., Bai, X. F., Qin, R., et al. (2018). OsMFT1 increases spikelets per panicle and delays heading date in rice by suppressing Ehd1, FZP and SEPALLATA-like genes. J. Exp. Bot. 69, 4283–4293. doi: 10.1093/jxb/ery232
Song, Y., Zhang, X. Y., Li, M. Z., Yang, H., Fu, D. Y., Lv, J., et al. (2021). The direct targets of CBFs: in cold stress response and beyond. J. Integr. Plant Biol. 63, 1874–1887. doi: 10.1111/jipb.13161
Stern, R. A., Gazit, S. (2010). The reproductive biology of the lychee. Hortic. Rev. 393-453. doi: 10.1002/9780470650851.ch8
Stockinger, E. J., Gilmour, S. J., Thomashow, M. F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the c repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. PNAS 94, 1035–1040. doi: 10.1073/pnas.94.3.1035
Thomashow, M. F. (2010). Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 154, 571–577. doi: 10.1104/pp.110.161794
Voogd, C., Brian, L. A., Wang, T., Allan, A. C., Varkonyi-Gasic, E. (2017). Three FT and multiple CEN and BFT genes regulate maturity, flowering, and vegetative phenology in kiwifruit. J. Exp. Bot. 68, 1539–1553. doi: 10.1093/jxb/erx044
Wang, J. W. (2014). Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 65, 4723–4730. doi: 10.1093/jxb/eru246
Wickland, D. P., Hanzawa, Y. (2015). The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: functional evolution and molecular mechanisms. Mol. Plant 8, 983–997. doi: 10.1016/j.molp.2015.01.007
Xi, W. Y., Liu, C., Hou, X. L., Yu, H. (2010). MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 22, 1733–1748. doi: 10.1105/tpc.109.073072
Xiao, Q. S., Su, Z. X., Chen, H. B., Shen, J. Y. (2019). Genome-wide identification and involvement of litchi SPL genes in flowering in response to cold and leaf maturity. J. Hortic. Sci. Biotech. 94, 428–440. doi: 10.1080/14620316.2018.1543557
Yoo, S. Y., Kardailsky, I., Lee, J. S., Weigel, D., Ahn, J. H. (2004). Acceleration of flowering by overexpression of MFT. Mol. Cells 17 (1), 95–101.
Zhang, X., Fowler, S. G., Cheng, H. M., Lou, Y. G., Rhee, S. Y., Stockinger, E. J., et al. (2004). Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J. 39, 905–919. doi: 10.1111/j.1365-313X.2004.02176.x
Zhang, L. Y., Jiang, X. C., Liu, Q. Y., Ahammed, G. J., Lin, R., Wang, L. Y., et al. (2020). The HY5 and MYB15 transcription factors positively regulate cold tolerance in tomato via the CBF pathway. Plant Cell Environ. 43, 2712–2726. doi: 10.1111/pce.13868
Zhao, C. Z., Zhang, Z. J., Xie, S. J., Si, T., Li, Y. Y., Zhu, J. K., et al. (2016). Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 171, 2744–2759. doi: 10.1104/pp.16.00533
Keywords: litchi, flower, cold, CBF, MFT
Citation: Shan X, Yang Y, Wei S, Wang C, Shen W, Chen H-b and Shen J-y (2023) Involvement of CBF in the fine-tuning of litchi flowering time and cold and drought stresses. Front. Plant Sci. 14:1167458. doi: 10.3389/fpls.2023.1167458
Received: 16 February 2023; Accepted: 25 May 2023;
Published: 12 June 2023.
Edited by:
Jian Wu, China Agricultural University, ChinaReviewed by:
Xin Zhao, China Agricultural University, ChinaCopyright © 2023 Shan, Yang, Wei, Wang, Shen, Chen and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ji-yuan Shen, anlzaGVuQHNjYXUuZWR1LmNu
†ORCID: Ji-yuan Shen, orcid.org/0000-0002-3472-2402
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.