- National Citrus Engineering Research Center, Citrus Research Institute, Southwest University, Chongqing, China
Citrus chlorotic dwarf-associated virus (CCDaV) is a Citlodavirus species in the Geminiviridae family that causes tremendous economic loss to the citrus industry in China. Some proteins encoded by geminiviruses are crucial for the interaction between the virus and its host plant. However, the exact functions of CCDaV-encoded proteins such as CCDaV-RepA have not been investigated. This study presents evidence that CCDaV-RepA elicits a hypersensitive response (HR)-like cell death in Nicotiana benthamiana that was accompanied by the production of H2O2 and ion leakage, which suggested that CCDaV-RepA is a potential recognition target for inducing host defense responses. Furthermore, the rolling-circle replication motifs of CCDaV-RepA are associated with triggering HR-like cell death in N. benthamiana. Confocal microscopy and deletion mutagenesis assays showed that CCDaV-RepA was located in the nucleus, while the first eight amino acids (aa) at the N terminus and two regions located between aa residues 122-263 and 220-264 of RepA were not associated with nuclear localization. Tobacco rattle virus-induced gene silencing of the key signaling cascade components revealed that HR-like cell death induced by RepA was inhibited in WRKY1-silenced N. benthamiana. Moreover, WRKY1 expression was upregulated in RepA-GFP infiltrated Overall, the results suggest that NbWRKY1 positively regulated CCDaV-RepA -induced cell death in N. benthamiana. These findings provide novel information for further research on the interactions between CCDaV and the host plant.
1 Introduction
Plants are particularly susceptible to infection by some viral diseases that can cause enormous economic loss by reducing yield and quality (Macho & Zipfel, 2014). Plants rely on several sophisticated antiviral immune responses for their survival and reproduction. The innate plant immune response is initiated when a plant-encoded resistance (R) protein is recognized by pathogens that carry avirulence (avr) genes, such as viral movement or coat proteins (Martin et al., 2003; Park & Kim, 2012). This recognition often leads to a hypersensitive response (HR) that involves rapid programmed cell death at the site of invasion (Zhou & Zhang, 2020). The HR is characterized by the release of reactive oxygen species (ROS) and nitric oxide (NO), ion fluxes, protein phosphorylation, the accumulation of signaling molecules such as salicylic acid (SA), ethylene (ET), and jasmonic acid (JA) and the activation of multiple defense genes, including pathogenesis-related genes (Mur et al., 2008).
Geminiviruses have one or two circular single-stranded DNA genomes in twinned icosahedral particles and are transmitted by a variety of insects, which can result in devastating crop losses worldwide (Navas-Castillo et al., 2011). According to their inferred genome organization, insect vectors, and host ranges, geminiviruses are classified into 14 genera (Roumagnac et al., 2022). Replication associated protein A (RepA) is unique to some geminivirus genera, including Citlodavirus, Becurtovirus, Capulavirus, Mulcrilevirus, Grablovirus, Mastrevirus, and Topilevirus (Sun et al., 2022). RepA is a multifunctional protein responsible for the transactivation of virus-sense open reading frames (ORFs), repressing its own expression as well as that of Rep, viral replication, and leaf development and senescence (Gutierrez et al., 2004; Hefferon et al., 2006). Recently, RepA was reported to be encoded by oat dwarf virus (ODV), bean yellow dwarf virus (BYDV), and mulberry mosaic dwarf-associated virus (MMDaV) and found to have the ability to induce HR-like cell death in Nicotiana benthamiana (Qian et al., 2016; Diamos & Mason, 2019; Sun et al., 2022).
Citrus chlorotic dwarf-associated virus (CCDaV) is a member of the genus Citlodavirus and was first observed in Turkey in the 1980s (Kersting et al., 1996). The host range of CCDaV is restricted to citrus and related species (Yang et al., 2022) and has been found in China and Thailand, where it has caused significant losses to lemon and pummelo yields (Guo et al., 2015; Yang et al., 2020; Yang et al., 2022). The genome of CCDaV ranges from 3639 to 3763 nucleotides, including four ORFs that encode the predicted coat proteins V1, V2, and V3, and the putative movement protein V4 in the virion strand, as well as RepA (C1) and Rep (C2) on the complementary strand (Roumagnac et al., 2022).
Because CCDaV is a newly discovered virus and studying woody plant-virus interactions is complex, the function of CCDaV-RepA and its effect on molecular and physiological responses in the host plant have not been characterized. To identify the interaction between CCDaV-RepA and plant defense responses. In this study, RepA was transiently expressed in N. benthamiana, and induced a HR-like cell death. The ability of RepA to localize to the nucleus was required for inducing a HR-like cell death. Further study suggested that WRKY1 could positively regulate RepA-induced cell death.
2 Results
2.1 CCDaV-encoded RepA and Rep induce HR-like cell death in N. benthamiana
To evaluate whether CCDaV-encoded proteins induce HR-like cell death, all six proteins encoded by CCDaV were transiently expressed in N. benthamiana using the potato virus X (PVX) vector carrying the green fluorescent protein (PVX-GFP). At 4 days post-infiltration (dpi), RepA-GFP and Rep-GFP induced cell death in infiltrated leaves, which was similar to the HR-based cell death triggered by the pro-apoptotic mouse BCL2-associated X protein (PVX-Bax) positive control (Figure 1A). Severe necrosis and collapse symptoms were observed in N. benthamiana emerging leaves that were infiltrated with RepA-GFP and Rep-GFP at 7 dpi and 9 dpi, respectively. No necrosis was found in N. benthamiana infiltrated with the strain GV3101 pSOUP carrying V1-GFP, V2-GFP, V3-GFP, V4-GFP, or PVX-GFP after 13 dpi (Figure 1A). To exclude the possible effect of PVX on cell death activity induced by CCDaV-encoded proteins, all six CCDaV proteins were also transiently expressed using the pNmGFPer vector (kindly provided by Prof. Xiuping Zou, Southwest University, China) in N. benthamiana. At 9 dpi, CCDaV-Rep and RepA-induced HR-like cell death in N. benthamiana leaves (Figure S2), while the other four CCDaV proteins did not trigger these symptoms up to 13 dpi.
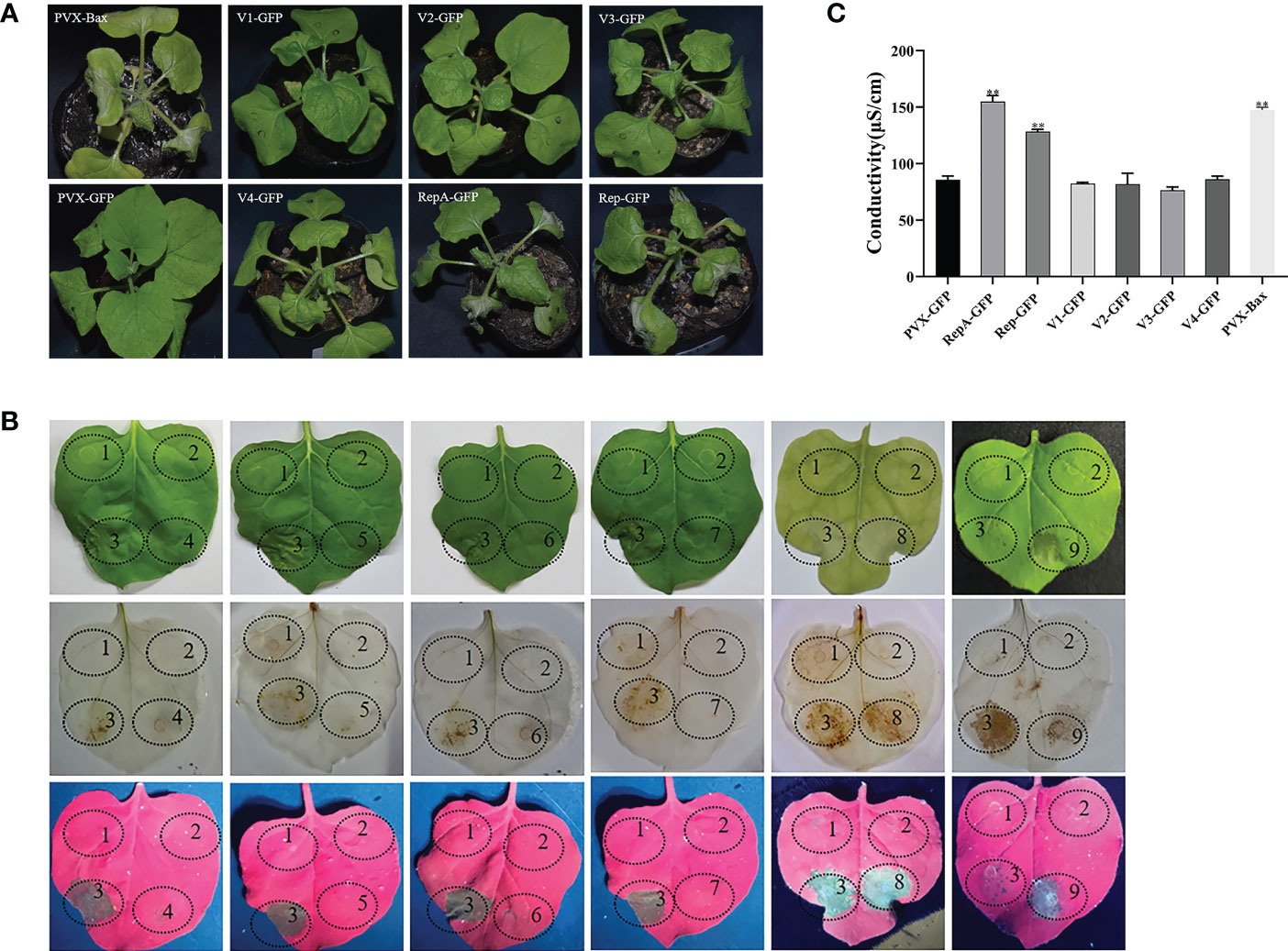
Figure 1 Identification of cell death induced by CCDaV-encoded proteins in Nicotiana benthamiana. (A) Disease symptoms observed in N. benthamiana leaves infiltrated with PVX-Bax, PVX-GFP, or recombinant PVX-GFP vectors expressing individual CCDaV proteins at 5 days post-infiltration (dpi). (B) CCDaV-encoded proteins transiently expressed in N. benthamiana. Photos were taken under white light (top panels); after 3,3′-diaminobenzidine staining (middle panels); and under ultraviolet light at 5 dpi (bottom panels). 1, PVX-GFP; 2, Buffer; 3, PVX-Bax; 4, V1-GFP; 5, V2-GFP; 6, V3-GFP; 7, V4-GFP; 8, RepA-GFP; 9, Rep-GFP. (C) Conductivity detection at 3 dpi. Significant differences (**p < 0.01) were determined by Student’s t test. Each experiment was repeated four times with at least six independent biological replicates.
To exclude the possible influence of GFP on cell death-inducing activity, the six CCDaV proteins were transiently expressed using the PVX vector into N. benthamiana. The results showed that only Rep and RepA could induce cell death in the infiltrated leaves, with no significant difference found in the titer of PVX or PVX derivatives infiltrated into N. benthamiana (Figure S1).
The accumulation of ROS (i.e., H2O2) and increased ion leakage are typical features of HR (Mur et al., 2008). At 5 dpi, RepA-GFP- and Rep-GFP-infiltrated N. benthamiana leaves typically stained brown after 3,3′-diaminobenzidine (DAB) treatment, indicating that RepA and Rep induced peroxidase activity and subsequent H2O2 production (Figure 1B). To quantify cell death, electrolyte leakage was also analyzed. The results showed that the conductivity of N. benthamiana leaves infiltrated with RepA-GFP or Rep-GFP was significantly higher (p < 0.001) than that of N. benthamiana infiltrated with other GFP-fusion structures (V1-GFP, V2-GFP, V3-GFP, V4-GFP, and PVX-GFP) at 3 dpi (Figure 1C). Taken together, CCDaV-RepA and Rep elicited HR-like cell death in N. benthamiana. Considering that RepA produced more severe symptoms than Rep, RepA was selected for subsequent analyses.
2.2 Mapping the key domains for RepA-induced HR-like cell death
The functional domains of CCDaV-RepA were predicted using SMART (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1) and Interpro (http://www.ebi.ac.uk/inter) software (Sun et al., 2022), and seven conserved domains were predicted in RepA (Figure 2A). To explore the key domain(s) associated with RepA-induced HR-like cell death, eight truncated proteins were transiently expressed in N. benthamiana. The results demonstrated that RepADM1-GFP (amino acid 1-8 deleted), RepADM7-GFP (amino acid 220-264 deleted), and RepADM8-GFP (amino acid 1-8, 122-263 deleted) could still produce HR-like cell death and H2O2 in N. benthamiana similar to that of the full-length RepA-GFP. However, the three rolling-circle replication motif-deletion mutants, RepADM4-GFP (amino acid 16-20 deleted), RepADM5-GFP (amino acid 57-63 deleted), and RepADM6-GFP (amino acid 103-108 deleted), as well as RepADM2-GFP without the catalytic domain (amino acid 8-120 deleted) or RepADM3-GFP without the central domain (amino acid 122-220 deleted), produced leaf curling in the infiltrated N. benthamiana and no H2O2 deposition was observed (Figure 2B). These results confirmed that the rolling-circle replication motifs of CCDaV-RepA were associated HR-like cell death induction in N. benthamiana.
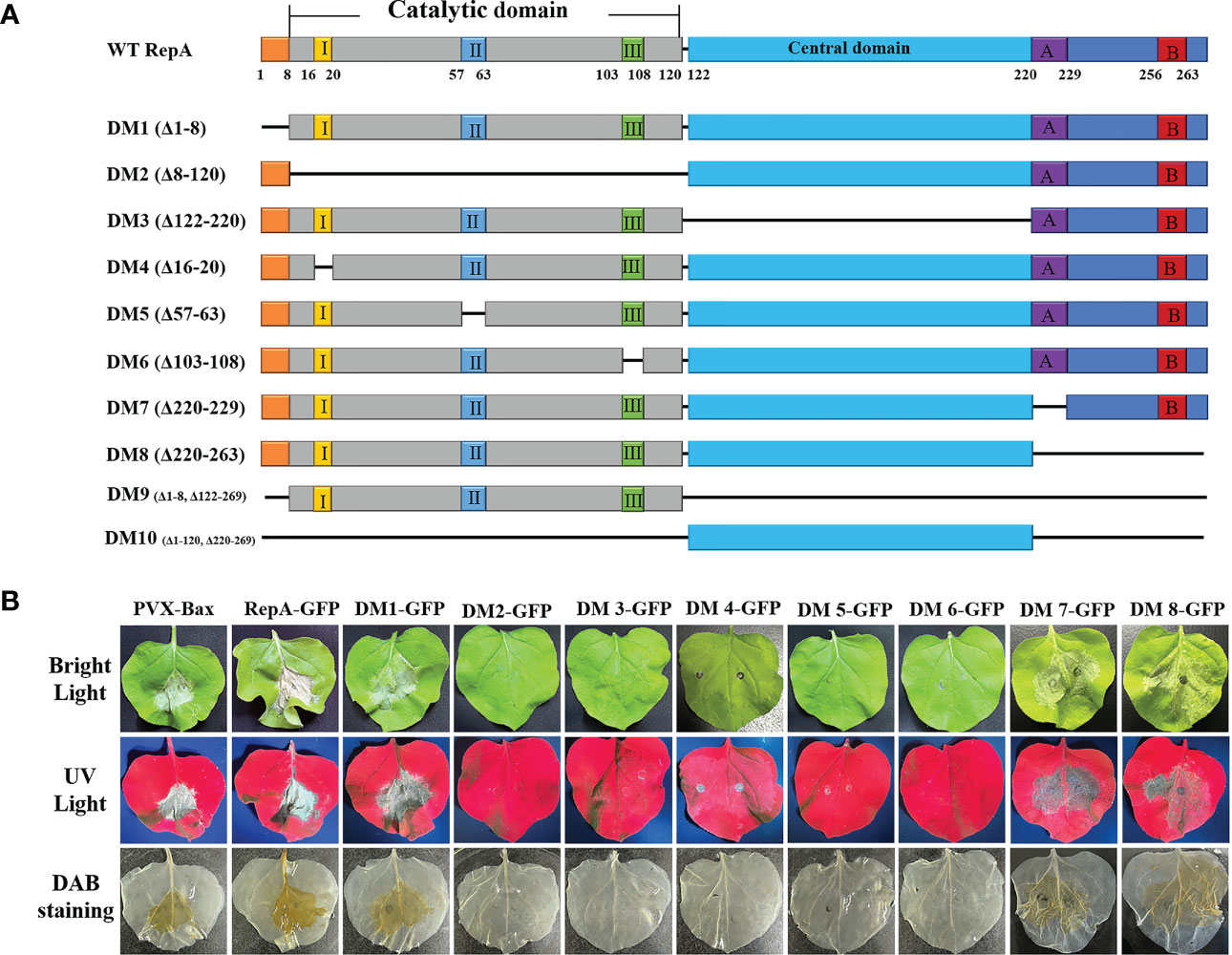
Figure 2 Mapping the key domains in CCDaV-RepA associated with HR-like cell death. (A) Schematic representation of RepA and its derivatives used in this study. I, II, and III indicate the conserved motifs in the catalytic domain contains. A and B indicate the Walker A and B motifs, respectively. (B) RepA-GFP and RepADM1-8-GFP were transiently expressed in N. benthamiana and imaged under bright light (top panels), ultraviolet light (middle panels), and after staining with 3,3′-diaminobenzidine (DAB) (bottom panels) at 5 days post-infiltration (dpi). All sample measurements were repeated four times.
2.3 Subcellular localization of RepA and its deletion mutants
Identification of the subcellular localization of a viral protein is an important step in understanding its putative functions (Zhan et al., 2018; Diamos & Mason, 2019). Confocal microscopy analysis suggested that RepA-GFP localized exclusively in the nucleus of epidermal cells at 48 h post-inoculation (hpi). As with RepA-GFP, RepADM1-GFP, RepADM7-GFP, and RepADM8-GFP localized in the nucleus at 28-36 hpi. However, RepADM2-GFP, RepADM3-GFP RepADM4-GFP, RepADM5-GFP, and RepADM6-GFP were predominantly found in the cytoplasm (Figure 3). These results showed that the first eight amino acids at the N terminus, amino acids 122-263, and amino acids 220-264 of RepA were not associated with its nuclear localization. Furthermore, no basic amino acids with a consensus nuclear localization signal sequence were predicted using the PSORT online software, indicating the presence of an atypical nuclear localization signal.
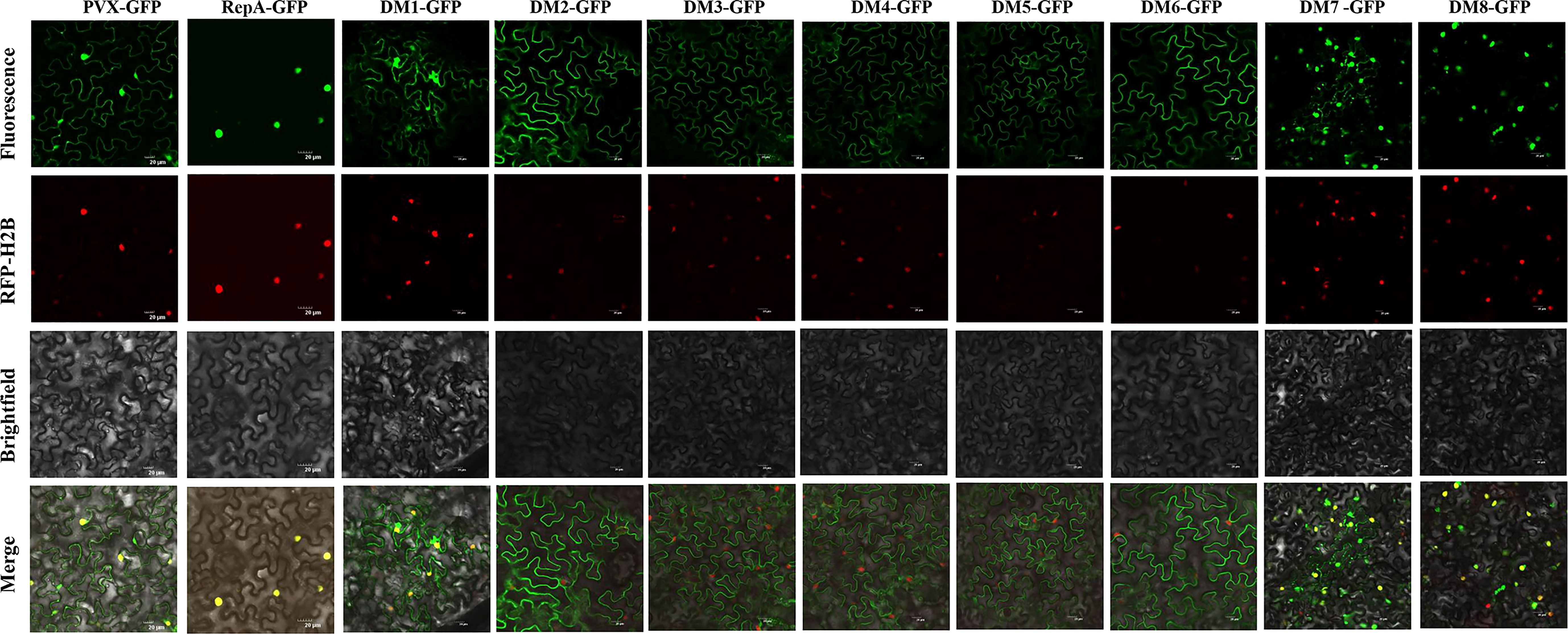
Figure 3 Subcellular localization analyses of CCDaV-RepA and its derivatives in Nicotiana benthamiana epidermal cells. Red fluorescent protein-histone 2B (RFP-H2B) was co-infiltrated with PVX-GFP or PVX-GFP-fusion constructs into N. benthamiana leaves. Each experiment was repeated four times and at least 40 cells (average 10 cells per field) were observed per plant. Scale bars = 20 µm.
2.4 RepA elicits plant immune responses in N. benthamiana
The innate immune system of plants has an interconnected two-layered system of pathogen-associated molecular patterns (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI), with the JA and SA signaling pathways also playing roles in plant defense (Jones & Dangl, 2006; Zhang et al., 2022). To further determine the association between RepA-induced cell death and plant immune responses, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was used to quantify the key defense response-related genes involved in the SA (NbPR1, NbPR2), JA (NbPR3, NbPR4 and NbLOX), and ET (NbERF1) pathways, as well as marker genes for HR (HIN1) and PAMP-triggered immunity (NbWRKY7, NbWRKY8, and NbACRE31) (Du et al., 2022; Sun et al., 2022). The results showed that the levels of NbPR1, NbPR2, NbPR3, NbPR4, NbWRKY7, NbWRKY8, and NbACRE31 were significantly upregulated in N. benthamiana infiltrated with RepA-GFP, with a 4-100-fold increase at 3 dpi when compared with the negative control. Furthermore, the expression of NbNPR1, which is the upstream regulator of the SA signaling pathway, also increased (Figure 4). These results showed that CCDaV-RepA induced HR-related plant defense responses.
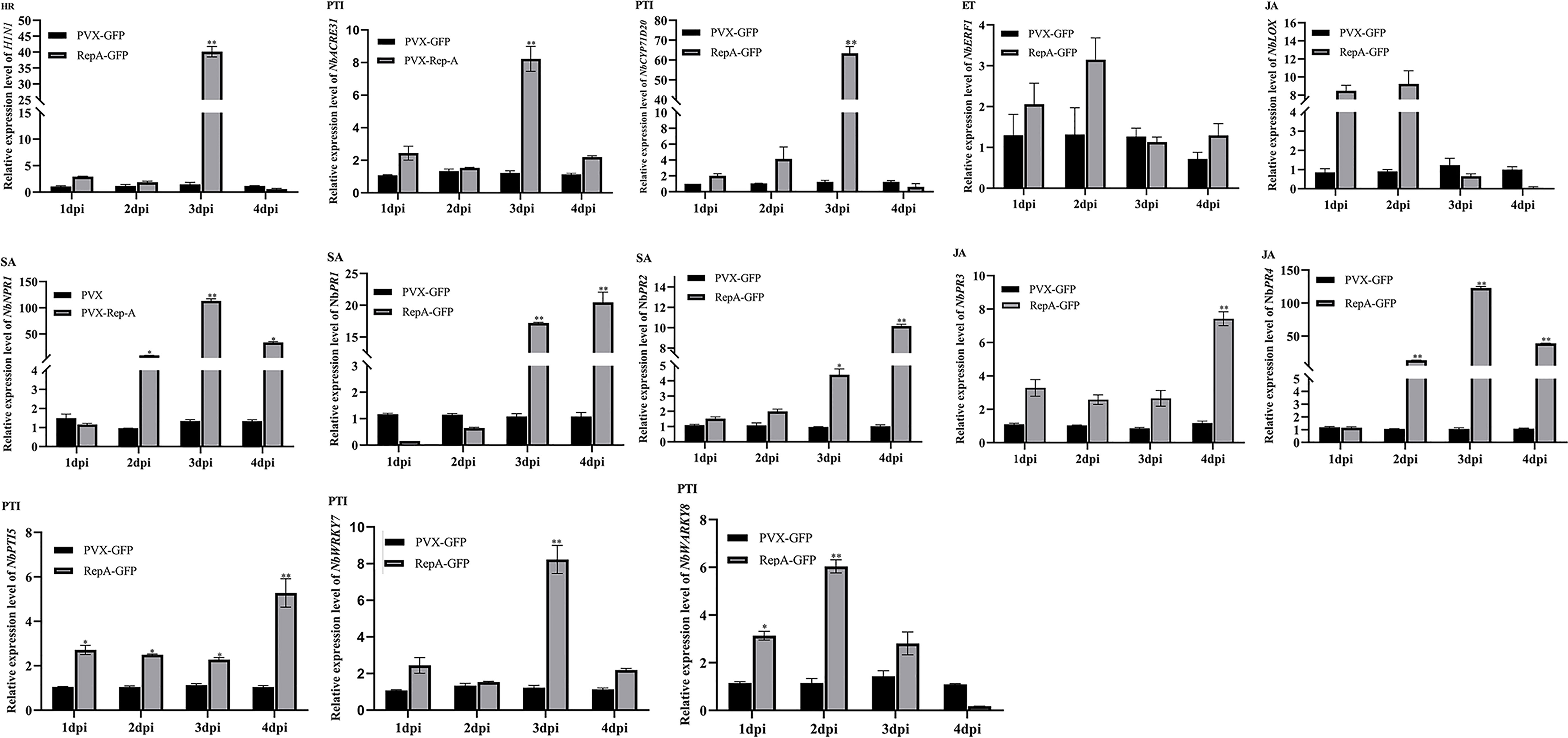
Figure 4 Changes in the expression level of defense-related genes in Nicotiana benthamiana leaves inoculated with PVX-GFP or RepA-GFP. NbACTIN was used as the housekeeping gene. The expression of each gene was quantified using the 2−ΔΔCt method. Statistical differences were calculated with the Student’s t-test, where *p < 0.05 and **p < 0.01. The experiment was repeated using four independent biological replicates and four technical replicates.
2.5 Other CCDaV-encoded proteins do not prevent CCDaV-RepA-induced HR-like cell death
Several studies showed that some plant virus-encoded proteins can inhibit HR-induced cell death to support virus survival (Mubin et al., 2010). However, the cell death symptoms in N. benthamiana produced by the co-expression of RepA with other CCDaV-encoded proteins (V1, V2, V3, V4, and Rep) were the same as those produced by RepA alone (Figure 5). Furthermore, no significant difference in the accumulation of RepA among the different inoculated leaves was observed.

Figure 5 CCDaV-encoded proteins inhibited cell death induced by RepA. Each experiment was repeated three times and at least five plants were used per biological replicate.
2.6 WRKY1 positively regulates RepA-induced cell death in N. benthamiana
Previous studies showed that signaling cascade components, such as WRKY1, NPR1, AOX, COI, CTR, NDR1, RAR1, NTF6, and MEK2 play important roles in cell death (Zhang et al., 2012; Meng & Zhang, 2013; Adachi et al., 2016). Therefore, these signaling cascade components were silenced by using the tobacco rattle virus (TRV) vector in N. benthamiana to assess whether they are involved in RepA-induced cell death. At 9 dpi, RepA-GFP was infiltrated in the emerging leaves of the silenced plants (Figure 6A). RT-qPCR analysis showed that the relative transcript level of RAR1, COI, CTR, NTF6, NPR1, MEK2, and AOX in silenced N. benthamiana plants was reduced by approximately 62.68 - 92.62% (Figure S4) in these plants, which exhibited significant cell death similar to that induced by RepA alone. The relative transcript level of WRKY1 and NDR1 in silenced N. benthamiana plants was reduced by approximately 88.35% and 88.14%, respectively (Figures 6B, C). Conversely, no apparent cell death was observed in WRKY1-silenced plants after RepA was transiently expressed, while NDR1-silenced N. benthamiana produced mild cell death symptoms.
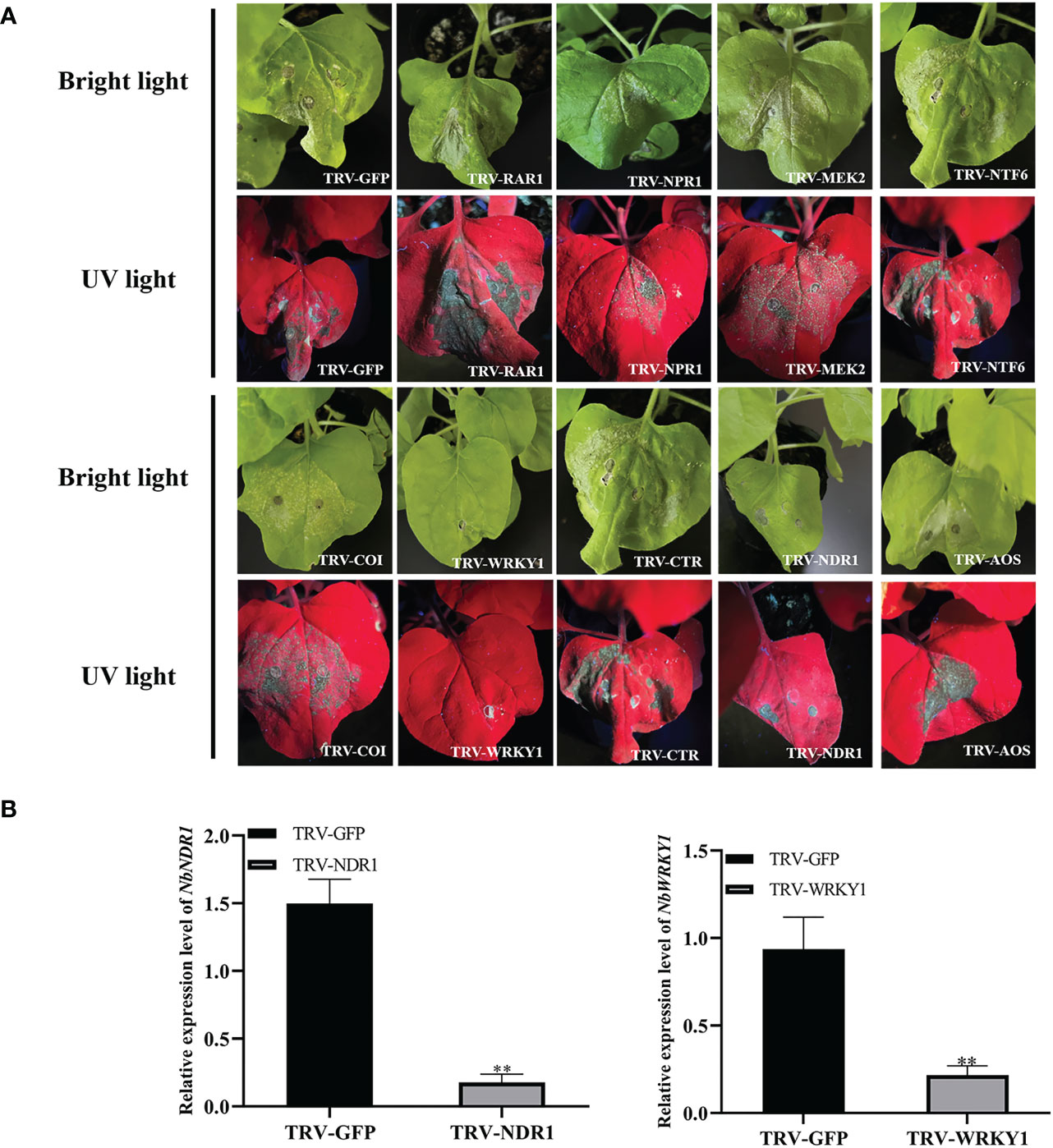
Figure 6 Effect of signaling cascade silencing on RepA-induced cell death in Nicotiana benthamiana. (A) Disease symptoms in silenced N. benthamiana plants at 3 days post-infiltration (dpi) with RepA-GFP. (B) The expression level of NbWRKY1 was calculated using the 2−ΔΔCt method with the housekeeping gene NbACTIN (Student’s t-test, **p < 0.01). Each experiment was repeated four times and at least five plants were used per biological replicate.
2.7 NbWRKY1 is upregulated by RepA
At 2 dpi, NbWRKY1 transcripts were increased approximately 2.78-fold and 2.17-fold in N. benthamiana infiltrated with PVX-RepA compared to the control plants infiltrated with RepADM2-GFP and PVX-GFP, respectively (Figure 7). These results suggested that CCDaV-RepA upregulated the expression of NbWRKY1.
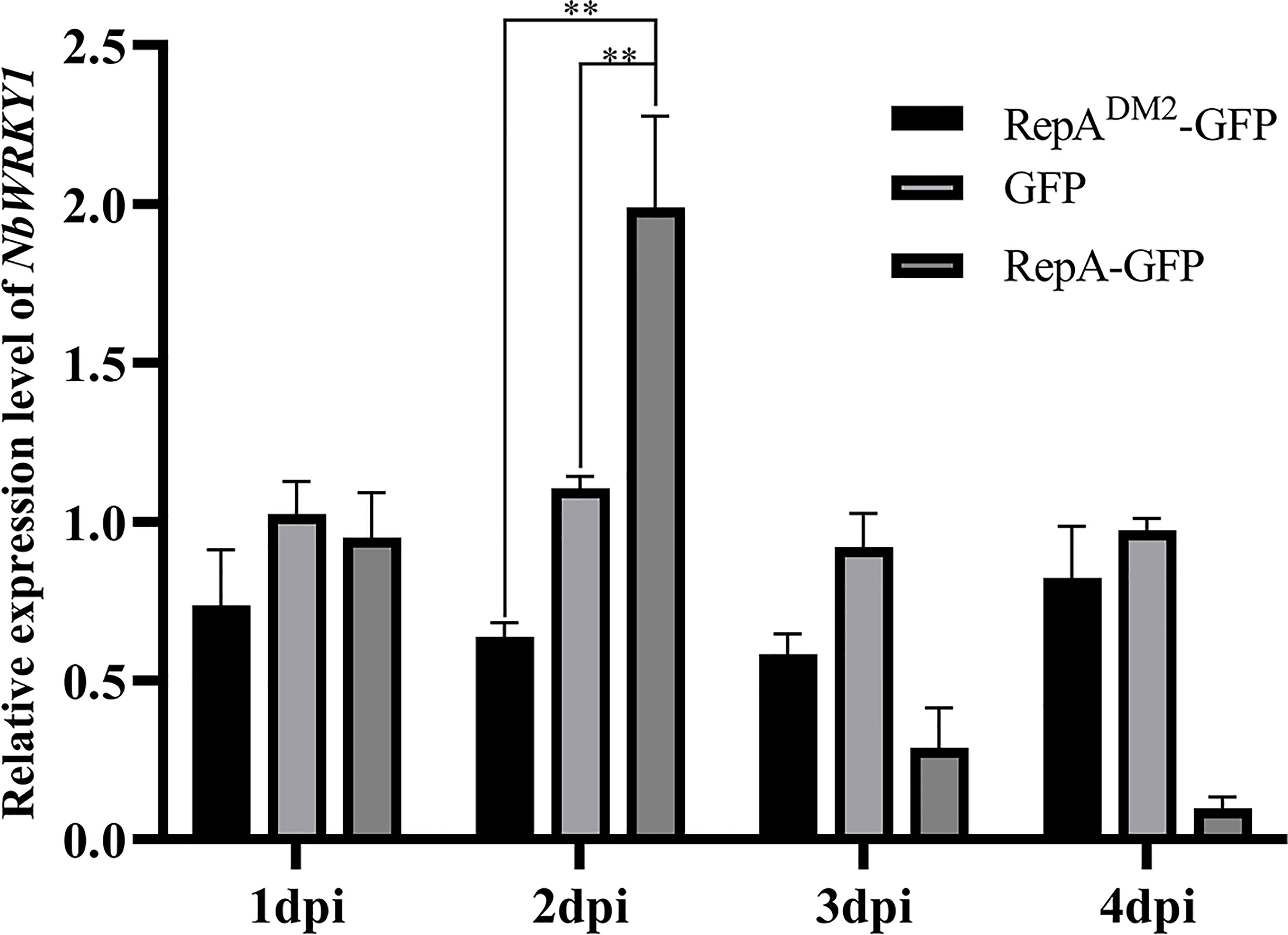
Figure 7 Relative expression of NbWRKY1 in response to transient expression of RepADM2-GFP, GFP, and RepA-GFP. Total RNA extracted at 4 days post-infiltration (dpi) was used for reverse transcription quantitative PCR (RT-qPCR) analysis. Significant differences (**p < 0.01) were determined with the Student’s t-test. The experiment was repeated four times with four independent biological replicates.
As NbWRKY1 and RepA localize to the nucleus (Sun et al., 2022), yeast two-hybrid (Y2H), firefly luciferase complementation imaging (LCI), and bimolecular fluorescence complementation (BiFC) assays were used to determine whether there was a direct interaction between NbWRKY1 and CCDaV-RepA. However, the results showed RepA could not directly interact with WRKY1.
3 Discussion
RepA is a multifunctional protein with an essential role in the viral DNA replication of geminiviruses (Varsani et al., 2017). In addition, RepA is a pathogenicity factor and potent inducer of HR in plants in some geminiviruses, including soybean geminivirus A, MMDaV, and ODV (Dodds & Rathjen, 2010; Qian et al., 2016; Yang et al., 2018; Sun et al., 2022). In this study, transient expression of CCDaV-RepA induced severe necrosis and collapse in N. benthamiana, indicating its function as a potential key virulence factor. Furthermore, CCDaV-RepA elicited HR-like cell death in N. benthamiana. HR is generally associated with the recognition of specific pathogen-encoded Avr proteins produced by the plant-encoded R gene product (Martin et al., 2003). Therefore, CCDaV-RepA may act as an Avr gene and be recognized by an unknown host R gene product that elicits a defense reaction that can potentially attenuate virus infection.
Previous reports revealed that deletion of the N-terminal amino acid compromises the ability of ODV-RepA to induce HR-like cell death in N. benthamiana (Qian et al., 2016). In contrast, in this study, only the rolling-circle replication motifs were associated with HR-like cell death, which was more consistent with findings reported for MMDaV (Sun et al., 2022). These results reconfirmed that not all positional homologs of geminivirus-encoded proteins are made equal (Luna & Lozano-Durán, 2020).
Previous studies showed that the nuclear localization of the transcription activation protein of tomato yellow leaf curl China virus (TYLCV), East African cassava mosaic virus (EACMV), and the RepA of MMDaV was essential for inducing HR-like cell death (Dong et al., 2003; Chowda-Reddy et al., 2009; Sun et al., 2022). In this study, nuclear localization of CCDaV-RepA was involved in its induction of HR-like cell death. Furthermore, sequence prediction results suggested that the CCDaV-RepA protein may contain an atypical nuclear localization signal. Considering RepA may be transported to the nucleus through protein-protein interaction, further research will be carried out in the future.
The WRKY family is one of the largest transcription factor (TF) families in plants (Yue et al., 2016). Although several studies have elucidated the role of WRKY TFs in bacterial and fungal pathogen responses (Hu et al., 2012; Kim et al., 2014; Abbas et al., 2020), very few plant virus-responsive WRKYs have been reported. Some studies showed that WRKY TFs positively regulated the defense response to tobacco mosaic virus (TMV) and cucumber mosaic virus (CMV) infection, and participated in disease resistance through the transcriptional reprogramming of pathogenesis-related gene expression (Huh et al., 2015; Zou et al., 2019). While WRKY8 and MeWRKY81 are involved in the negative regulation of crucifer-infecting TMV (TMV-cg) and South African cassava mosaic virus (SACMV), respectively (Chen et al., 2013; Freeborough et al., 2021). In this study, the expression of NbWRKY1 was upregulated by the expression of CCDaV-RepA in N. benthamiana. Moreover, silencing WRKY1 inhibited RepA-induced cell death, indicating that NbWRKY1 is important for RepA-induced cell death by regulating the downstream genes associated with antiviral defense. NbWRKY1 was reported to be located in nuclear (Sun et al., 2022). However, no direct interaction between NbWRKY1 and CCDaV-RepA was found using multiple interaction assays. These results indicated that an indirect interaction may exist between CCDaV-RepA and NbWRKY1.
In conclusion, CCDaV-RepA elicited HR-like cell death and plant immune responses in N. benthamiana. RepA upregulated the expression level of NbWRKY1 and NbWRKY1, which positively regulated CCDaV-RepA-induced cell death. These findings provide novel insight into the interactions between CCDaV and the host plant. In the future, the role of the NbWRKY1 regulatory network in the antiviral response of citrus will be elucidated.
4 Materials and methods
4.1 Virus source and plant material
The CCDaV isolate ULK-1 was maintained on Eureka lemon (C. limon) and used as the virus source for all experiments. N. benthamiana plants were grown in an incubator at 25°C under a 16:8 h light/dark cycle.
4.2 Expression constructs
The PVX-GFP-containing pGR106 vector (N-terminal GFP-fusion) was constructed as described previously (Earley et al., 2006). Full-length RepA, Rep, V1, V2, V3, and V4 genes encoded by CCDaV were amplified by using specific primer pairs (Table S1) and individually cloned into the pGEM-T Easy vector (Promega). These plasmids were digested using Cla I and Sal I, and ligated into PVX-GFP to generate RepA-GFP, Rep-GFP, V1-GFP, V2-GFP, V3-GFP, and V4-GFP. The PVX-V1, PVX-V2, PVX-V3, PVX-V4, PVX-RepA, and PVX-Rep clones were constructed as described by Yang et al. (2018).
To explore the key domain(s) of RepA, eight deletion mutations were amplified (Table S1) and cloned into PVX-GFP to yield RepADM1-GFP to RepADM8-GFP by homologous recombination (Sun et al., 2022). All constructs were verified by sequencing.
4.3 Agrobacterium-mediated infiltration
All constructs were transformed into Agrobacterium tumefaciens GV3101 as described by Du et al. (2022). The A. tumefaciens cultures harboring the constructs were adjusted to a final OD600 0.6-0.8 and incubated in the dark for 2-3 h before infiltration. The suspensions were then infiltrated into at least six fully expanded leaves of four-week-old N. benthamiana.
4.4 H2O2 detection in N. benthamiana
N. benthamiana leaves were collected at 5 dpi and soaked in 1 mg/mL DAB-HCl solution (pH 3.8). The samples were incubated in the dark for 8 h at room temperature and then bleached in boiling 96% ethanol for 10 min (Hou et al., 2014).
4.5 Measurement of electrolyte leakage
Five N. benthamiana leaf discs (9 mm in diameter) were collected from infiltrated areas at 3 dpi. The tissues were incubated in 10 mL distilled water at 28°C for 1 h (Pitino et al., 2016). The solution conductivity was measured using an FE30 conductivity meter (Mettler-Toledo Company), and the electrolyte leakage was calculated by the ratio of the conductivity to total conductivity.
4.6 Protein extraction and western blot analysis
The total protein from infiltrated N. benthamiana leaves exhibiting transient expression was extracted using a Plant Total Protein Extraction Kit (Solarbio). Western blot analysis was performed as described previously (Du et al., 2022) with an anti-GFP monoclonal primary antibody (Proteintech) and anti-mouse IgG coupled with horseradish peroxidase (HRP) (Proteintech) as primary and secondary antibodies, respectively. The products were visualized using a chemiluminescence detection reagent (Everbright).
4.7 Enzyme-linked immunosorbent assay
Systemic leaf samples of N. benthamiana infiltrated with PVX or derivatives of PVX were detected using ELISA as previously described (Wang et al., 2014; Qian et al., 2016).
4.8 Laser scanning confocal microscopy
For subcellular localization, mCherry-H2B and RFP-plasma membrane were used as nuclear and plasma membrane markers, respectively. At 48 hpi, fluorescence signals of the epidermal cells from infiltrated N. benthamiana leaves were observed using an FV3000 confocal microscope (Olympus) (Du et al., 2022).
4.9 Virus-induced gene silencing assay in N. benthamiana
The TRV silencing vector was kindly provided by Dr. Wanxia Shen from the Southwest University, China and used for the VIGS assay in N. benthamiana previously described (Senthil-Kumar & Mysore, 2014). NbWRKY1, NbNDR1, NbNPR1, NbCOI1, NbCTR1, NbNTF6, NbRAR1, and NbMEK2 were amplified (Table S1) and cloned into TRV2. After sequencing verification, TRV1 and its derivatives were co-inoculated into N. benthamiana using GV3101 infiltration (Sun et al., 2022).
4.10 Total RNA extraction and RT-qPCR analysis
To evaluate the silencing efficiency, total RNA was extracted from systemically infected leaves of N. benthamiana at 10 dpi using TRIzol (Tiangen). To quantify the expression of host defense-related genes responsive to CCDaV-RepA, total RNA was extracted from infiltrated leaves at 3 dpi. RT-qPCR was performed using The BlasTap™2×RT-qPCR Master MIX (ABM). The housekeeping gene NbActin was used as an internal control for RNA quantification (Ananthakrishnan et al., 2010). Each experiment included samples run in triplicate, including the internal control gene. The expression of target genes was calculated using the formula described by Pfaffl (2001). The primers for RT-qPCR are listed in Table S1.
4.11 Yeast-two-hybrid assay
The Matchmaker Yeast Two-Hybrid System (Weidi Biotechnology) was used for Y2H assays to verify the interaction between RepA and NbWRKY1. The Y2H screening was performed as described previously (Shi et al., 2007).
4.12 Firefly luciferase complementation imaging assay
The full-length RepA and NbWRKY1 were amplified with specific primers (Table S1) and inserted into PCCL-P9 and PHNL vectors, which were kindly provided by Professor Jianmin Zhou, University of Chinese Academy of Sciences, Beijing, to produce PCCL-P9-CCDaV-RepA and PHNL-NbWRKY1, respectively. The constructs were introduced into A. tumefaciens GV3101 pSOUP and agroinfiltrated into N. benthamiana leaves. The infiltrated leaves were detached at 48 hpi and sprayed with 1 mM luciferin (Macklin). The tissues were kept in dark for 6 min before the LUC activity was detected by using IVIS Lumina Series III system (PerkinElmer).
4.13 Bimolecular fluorescence complementation assay
RepA and NbWRKY1 were amplified with specific primers (Table S1) and cloned into pSPYCE-35S and pSPYNE-35S, respectively. After sequencing verification, the recombinant vectors were transformed into A. tumefaciens GV3101 pSOUP and infiltrated to N. benthamiana leaves at the four- to six-leaf stages. At 48 hpi, the fluorescence signal was observed by FV3000 scanning confocal microscope (Olympus).
4.14 Statistical analyses
GraphPad Prism 9 was used for statistical analyses, and the data were expressed as the means ± standard deviation (SD) of three biological replicates. Significant differences in gene expression were evaluated using Student’s test at *P < 0.05, **P < 0.01.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
YQ, CZ, and YZ designed the experiments. YQ, XY, and JZ performed the experiments. YQ and JW analyzed the data, and YQ wrote the draft manuscript. YZ revised and polished the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was partially supported by National Key Research and Development Program of China (2019YFD1001802) and China Agriculture Research System of MOF and MARA (CARS-26-05B), Overseas Expertise Introduction Project for Discipline Innovation, Ministry of Education of the People’s Republic of China (CN) (B18044).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1164416/full#supplementary-material
Supplementary Figure 1 | Detection of Potato virus X (PVX) content after RepA from the citrus chlorotic dwarf-associated virus was transiently expressed in Nicotiana benthamiana using the PVX vector. The error bars indicate the standard deviation within one representative experiment (n = 3). NG indicates uninoculated N. benthamiana plants. Each experiment was repeated three times and at least five plants were used per biological replicate.
Supplementary Figure 2 | Identification of cell death induced by CCDaV-encoded proteins in Nicotiana benthamiana by using pNmGFPer vector. (A) Disease symptoms observed in N. benthamiana leaves expressing individual CCDaV proteins at 9 days post-infiltration (dpi). (B) Images of infiltrated leaves were captured under bright light and UV light at 9 dpi. The 3,3′-diaminobenzidine (DAB) staining was performed to visualize the production of H2O2. Each experiment was repeated three times and at least five plants were used per biological replicate.
Supplementary Figure 3 | Mapping the key domains of CCDaV-RepA associated with HR-like cell death. 35S-RepA-GFP, 35S-RepADM1-GFP, 35S-RepADM2-GFP, 35S-RepADM7 -GFP, and 35S-RepADM8-GFP were transiently expressed in N. benthamiana and then imaged under bright light (top panels), ultraviolet light (middle panels), and after 3,3′-diaminobenzidine (DAB) treatment (bottom panels) at 9 days post-infiltration.
Supplementary Figure 4 | Effect of signaling cascade component silencing on RepA-induced cell death in Nicotiana benthamiana. The expression of NbWRKY1 was calculated using the formula 2 −ΔΔCt with the housekeeping gene, NbACTIN (Student’s t-test **p < 0.01).
References
Abbas, H. M. K., Ahmad, A., Dong, W. B., Xiang, J. S., Iqbal, J., Ali, S., et al. (2020). Heterologous WRKY and NAC transcription factors triggered resistance in Nicotiana benthamiana. J. King Saud Univ. Sci. 32, 3005–3013. doi: 10.1016/j.jksus.2020.08.005
Adachi, H., Ishihama, N., Nakano, T., Yoshioka, M., Yoshioka, H. (2016). Nicotiana benthamiana MAPK-WRKY pathway confers resistance to a necrotrophic pathogen Botrytis cinerea. Plant Signaling Behav. 11 (6), e1183085. doi: 10.1080/15592324.2016.1183085
Ananthakrishnan, G., Venkataprasanna, T., Roy, A., Brlansky, R. N. (2010). Characterization of the mixture of genotypes of a Citrus tristeza virus isolate by reverse transcription-quantitative real-time PCR. J. Virolog Methods 164, 75–82. doi: 10.1016/j.jviromet.2009.12.001
Chen, L. G., Zhang, L. P., Li, D. B., Wang, F., Yu, D. Q. (2013). WRKY8 transcription factor functions in the TMV-cg defense response by mediating both abscisic acid and ethylene signaling in arabidopsis. Proc. Natl. Acad. Sci. United States America 110 (21), E1963–E1971. doi: 10.1073/pnas.1221347110
Chowda-Reddy, R. V., Dong, W. B., Felton, C., Ryman, D., Ballard, K., Fondong, V. N. (2009). Characterization of the Cassava geminivirus transcription activation protein putative nuclear localization signal. Virus Res. 145, 270–278. doi: 10.1016/j.virusres.2009.07.022
Diamos, A. G., Mason, H. S. (2019). Modifying the replication of geminiviral vectors reduces cell death and enhances expression of biopharmaceutical proteins in Nicotiana benthamiana leaves. Front. Plant Sci. 9, 1974. doi: 10.3389/fpls.2018.01974
Dodds, P. N., Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Dong, X. L., van Wezel, R., Stanley, J., Hong, Y. G. (2003). Functional characterization of the nuclear localization signal for a suppressor of posttranscriptional gene silencing. J. Virol. 77, 7026–7033. doi: 10.1128/JVI.77.12.7026-7033.2003
Du, J., Wang, Q. Y., Zeng, C. H., Zhou, C. Y., Wang, X. F. (2022). A prophage-encoded nonclassical secretory protein of "Candidatus liberibacter asiaticus" induces a strong immune response in nicotiana benthamiana and citrus. Mol. Plant Pathol. 23, 1022–1034. doi: 10.1111/mpp.13206
Earley, K. W., Haag, J. R., Pontes, O., Opper, K., Juehne, T., Song, K. M., et al. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. doi: 10.1111/j.1365-313X.2005.02617.x
Freeborough, W., Gentle, N., Rey, M. E. C. (2021). Wrky transcription factors in cassava contribute to regulation of tolerance and susceptibility to cassava mosaic disease through stress responses. Viruses 13, 1820. doi: 10.3390/v13091820
Guo, J., Lai, X. P., Li, J. X., Yue, J. Q., Zhang, S. Y., Li, Y. Y., et al. (2015). First report on citrus chlorotic dwarf associated virus on lemon in dehong prefecture, yunnan, China. Plant Dis. 99, 1287–1287. doi: 10.1094/PDIS-01-15-0011-PDN
Gutierrez, C., Ramirez-Parra, E., Castellano, M. M., Sanz-Burgos, A. P., Luque, A., Missich, R. (2004). Geminivirus DNA replication and cell cycle interactions. Vet Microbiol. 98, 111–119. doi: 10.1016/j.vetmic.2003.10.012
Hefferon, K. L., Moon, Y. S., Fan, Y. (2006). Multi-tasking of nonstructural gene products is required for bean yellow dwarf geminivirus transcriptional regulation. FEBS J. 273, 4482–4494. doi: 10.1111/j.1742-4658.2006.05454.x
Hou, P. Q., Lee, Y. I., Hsu, K. T., Lin, Y. T., Wu, W. Z., Lin, J. Y., et al. (2014). Functional characterization of Nicotiana benthamiana chromomethylase 3 in developmental programs by virus-induced gene silencing. Physiologia Plantarum 150, 119–132. doi: 10.1111/ppl.12071
Hu, Y. R., Dong, Q. Y., Yu, D. Q. (2012). Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen. Pseudomonas syringae. Plant Sci. 185, 288–297. doi: 10.1016/j.plantsci.2011.12.003
Huh, S. U., Lee, G. J., Jung, J. H., Kim, Y., Kim, Y. J., Paek, K. H. (2015). Capsicum annuum Transcription factor WRKYa positively regulates defense response upon TMV infection and is a substrate of CaMK1 and CaMK2. Sci. Rep. 5, 7981. doi: 10.1038/srep07981
Jones, J. D. G., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kersting, U., Korkmaz, S., Cinar, A., Ertugrul, B., Onelge, N., Garnsey, S. M. (1996). “Citrus Chlorotic Dwarf: A new whitefly-transmitted disease in the eastern mediterranean region of Turkey,” in Proceedings of the 13th conference of international organization of citrus virologist. (California, USA: IOCV, Riverside) 220–225.
Kim, H. S., Park, Y. H., Nam, H., Lee, Y. M., Song, K., Choi, C., et al. (2014). Overexpression of the Brassica rapa transcription factor WRKY12 results in reduced soft rot symptoms caused by Pectobacterium carotovorum in Arabidopsis and Chinese cabbage. Plant Biol. 16, 973–981. doi: 10.1111/plb.12149
Luna, A. P., Lozano-Durán, R. (2020). Geminivirus-encoded proteins: not all positional homologs are made equal. Front. Microbiol. 11, 878. doi: 10.3389/fmicb.2020.00878
Macho, A. P., Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272. doi: 10.1016/j.molcel.2014.03.028
Martin, G. B., Bogdanove, A. J., Sessa, G. (2003). Understanding the functions of plant disease resistance proteins. Annu. Rev. Plant Biol. 54, 23–61. doi: 10.1146/annurev.arplant.54.031902.135035
Meng, X. Z., Zhang, S. Q. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol Vol 51 51, 245–266. doi: 10.1146/annurev-phyto-082712-102314
Mubin, M., Amin, I., Amrao, L., Briddon, R. W., Mansoor, S. (2010). The hypersensitive response induced by the V2 protein of a monopartite begomovirus is countered by the C2 protein. Mol. Plant Pathol. 11, 245–254. doi: 10.1111/j.1364-3703.2009.00601.x
Mur, L. A. J., Kenton, P., Lloyd, A. J., Ougham, H., Prats, E. (2008). The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 59, 501–520. doi: 10.1093/jxb/erm239
Navas-Castillo, J., Fiallo-Olive, E., Sanchez-Campos, S. (2011). Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 49, 219–248. doi: 10.1146/annurev-phyto-072910-095235
Park, S.-H., Kim, K.-H. (2012). Virus-induced silencing of the WRKY1 transcription factor that interacts with the SL1 structure of Potato virus X leads to higher viral RNA accumulation and severe necrotic symptoms. Plant Pathol. J. 28, 40–48. doi: 10.5423/PPJ.OA.11.2011.0226
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, 2003–2007. doi: 10.1093/nar/29.9.e45
Pitino, M., Armstrong, C. M., Cano, L. M., Duan, Y. P. (2016). Transient expression of Candidatus liberibacter asiaticus effector induces cell death in Nicotiana benthamiana. Front. Plant Sci. 7, 982. doi: 10.3389/fpls.2016.00982
Qian, Y. J., Hou, H. W., Shen, Q. T., Cai, X. Z., Sunter, G., Zhou, X. P. (2016). Repa protein encoded by Oat dwarf virus elicits a temperature-sensitive hypersensitive response-type cell death that involves jasmonic acid-dependent signaling. Mol. Plant-Microbe Interact. 29, 5–21. doi: 10.1094/MPMI-07-15-0149-R
Roumagnac, P., Lett, J. M., Fiallo-Olive, E., Navas-Castillo, J., Zerbini, F. M., Martin, D. P., et al. (2022). Establishment of five new genera in the family geminiviridae: Citlodavirus, Maldovirus, Mulcrilevirus, Opunvirus, and Topilevirus. Arch. Virol. 167, 695–710. doi: 10.1007/s00705-021-05309-2
Senthil-Kumar, M., Mysore, K. S. (2014). Tobacco rattle virus-based virus-induced gene silencing in Nicotiana benthamiana. Nat. Protoc. 9, 1549–1562. doi: 10.1038/nprot.2014.092
Shi, Y., Chen, J., Hong, X., Chen, J., Adams, M. J. (2007). A potyvirus P1 protein interacts with the rieske Fe/S protein of its host. Mol. Plant Pathol. 8, 785–790. doi: 10.1111/j.1364-3703.2007.00426.x
Sun, S. S., Ren, Y. X., Wang, D. X., Farooq, T., He, Z. F., Zhang, C., et al. (2022). A group I WRKY transcription factor regulates mulberry mosaic dwarf-associated virus-triggered cell death in Nicotiana benthamiana. Mol. Plant Pathol. 23, 237–253. doi: 10.1111/mpp.13156
Varsani, A., Roumagnac, P., Fuchs, M., Navas-Castillo, J., Moriones, E., Idris, A., et al. (2017). Capulavirus And Grablovirus: two new genera in the family geminiviridae. Arch. Virol. 162, 1819–1831. doi: 10.1007/s00705-017-3268-6
Wang, Y. Q., Dang, M. Q., Hou, H. W., Mei, Y. Z., Qian, Y. J., Zhou, X. P. (2014). Identification of an RNA silencing suppressor encoded by a mastrevirus. J. Gen. Virol. 95, 2082–2088. doi: 10.1099/vir.0.064246-0
Yang, X. L., Ren, Y. X., Sun, S. S., Wang, D. X., Zhang, F. F., Li, D. W., et al. (2018). Identification of the potential virulence factors and RNA silencing suppressors of mulberry mosaic dwarf-associated geminivirus. Viruses 10, 472. doi: 10.3390/v10090472
Yang, Z., Zhang, L., Zhao, J., Li, T., Liu, Q., Cao, M., et al. (2020). First report of citrus chlorotic dwarf-associated virus on pomelo in nakhon, Thailand. Plant Dis. 104, 1262. doi: 10.1094/PDIS-10-19-2093-PDN
Yang, Z., Zhang, L., Zhao, J. F., Zhang, X. K., Wang, Y., Li, T. S., et al. (2022). New geographic distribution and molecular diversity of Citrus chlorotic dwarf-associated virus in China. J. Integr. Agric. 21, 293–298. doi: 10.1016/S2095-3119(20)63601-2
Yue, H., Wang, M., Liu, S. Y., Du, X. H., Song, W. N., Nie, X. J. (2016). Transcriptome-wide identification and expression profiles of the WRKY transcription factor family in broomcorn millet (Panicum miliaceum l.). BMC Genomics 17, 1–11. doi: 10.1186/s12864-016-2677-3
Zhan, B. H., Zhao, W. Y., Li, S. F., Yang, X. L., Zhou, X. P. (2018). Functional scanning of apple geminivirus proteins as symptom determinants and suppressors of posttranscriptional gene silencing. Viruses 10, 488. doi: 10.3390/v10090488
Zhang, H. J., Li, D. Q., Wang, M. F., Liu, J. W., Teng, W. J., Cheng, B. P., et al. (2012). The Nicotiana benthamiana mitogen-activated protein kinase cascade and WRKY transcription factor participate in Nep1(Mo)-triggered plant responses. Mol. Plant-Microbe Interact. 25, 1639–1653. doi: 10.1094/MPMI-11-11-0293
Zhang, B. M., Liu, M. T., Wang, Y. C., Yuan, W. Y., Zhang, H. T. (2022). Plant NLRs: evolving with pathogen effectors and engineerable to improve resistance. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.1018504
Zhou, J. M., Zhang, Y. L. (2020). Plant immunity: danger perception and signaling. Cell 181, 978–989. doi: 10.1016/j.cell.2020.04.028
Keywords: citrus chlorotic dwarf-associated virus (CCDaV), RepA, cell death, geminivirus, WRKY
Citation: Qin Y, Zhao J, Wang J, Ye X, Zhou C and Zhou Y (2023) Regulation of Nicotiana benthamiana cell death induced by citrus chlorotic dwarf-associated virus-RepA protein by WRKY 1. Front. Plant Sci. 14:1164416. doi: 10.3389/fpls.2023.1164416
Received: 12 February 2023; Accepted: 10 April 2023;
Published: 25 April 2023.
Edited by:
Yasir Iftikhar, University of Sargodha, PakistanReviewed by:
Tariq Mukhtar, Pir Mehr Ali Shah Arid Agriculture University, PakistanSagheer Atta, Ghazi University, Pakistan
Copyright © 2023 Qin, Zhao, Wang, Ye, Zhou and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Zhou, emhvdXlhbkBjcmljLmNu
 Yangyang Qin
Yangyang Qin Jinfa Zhao
Jinfa Zhao Jiajun Wang
Jiajun Wang Changyong Zhou
Changyong Zhou Yan Zhou
Yan Zhou