- 1Agricultural College, Heilongjiang Bayi Agricultural University, Daqing, Heilongjiang, China
- 2National Cereals Technology Engineering Research Center, Daqing, Heilongjiang, China
- 3Heilongjiang Bayi Agricultural University, Key Laboratory of Ministry of Agriculture and Rural Affairs of Soybean Mechanized Production, Daqing, Heilongjiang, China
- 4Qiqihar Branch of Heilongjiang Academy of Agricultural Sciences, Qiqihar, Heilongjiang, China
- 5Research Center of Saline and Alkali Land Improvement Engineering Technology in Heilongjiang Province, Daqing, Heilongjiang, China
Cytokinin oxidase/dehydrogenase (CKX) irreversibly degrades cytokinin, regulates growth and development, and helps plants to respond to environmental stress. Although the CKX gene has been well characterized in various plants, its role in soybean remains elusive. Therefore, in this study, the evolutionary relationship, chromosomal location, gene structure, motifs, cis-regulatory elements, collinearity, and gene expression patterns of GmCKXs were analyzed using RNA-seq, quantitative real-time PCR (qRT-PCR), and bioinformatics. We identified 18 GmCKX genes from the soybean genome and grouped them into five clades, each comprising members with similar gene structures and motifs. Cis-acting elements involved in hormones, resistance, and physiological metabolism were detected in the promoter regions of GmCKXs. Synteny analysis indicated that segmental duplication events contributed to the expansion of the soybean CKX family. The expression profiling of the GmCKXs genes using qRT-PCR showed tissue-specific expression patterns. The RNA-seq analysis also indicated that GmCKXs play an important role in response to salt and drought stresses at the seedling stage. The responses of the genes to salt, drought, synthetic cytokinin 6-benzyl aminopurine (6-BA), and the auxin indole-3-acetic acid (IAA) at the germination stage were further evaluated by qRT-PCR. Specifically, the GmCKX14 gene was downregulated in the roots and the radicles at the germination stage. The hormones 6-BA and IAA repressed the expression levels of GmCKX1, GmCKX6, and GmCKX9 genes but upregulated the expression levels of GmCKX10 and GmCKX18 genes. The three abiotic stresses also decreased the zeatin content in soybean radicle but enhanced the activity of the CKX enzymes. Conversely, the 6-BA and IAA treatments enhanced the CKX enzymes’ activity but reduced the zeatin content in the radicles. This study, therefore, provides a reference for the functional analysis of GmCKXs in soybean in response to abiotic stresses.
1 Introduction
Soybean (Glycine max L.) is an important food and oil crop worldwide, with its seed oil accounting for approximately 30% of the global vegetable oil consumption (Zhan et al., 2020). Soybean seeds contain various substances beneficial to human health, which have been proven important in preventing and treating cancer, atherosclerosis, osteoporosis, and coronary heart disease (Malenčić et al., 2012; Zhang et al., 2014). Soybeans also have a variety of industrial uses and are considered a potential crop for biodiesel production (Woyann et al., 2019). In the US, 30% of printed matter uses soybean ink, and many city buses have switched to an environmentally friendly blend of soybean oil and diesel (Frederick et al., 2001). Brazil, the world’s top soybean producer, reportedly produced 124.5 million tons during 2019–2020 (USDA, 2020). Furthermore, soybean is one of the most common crops in arid and semi-arid areas where its growth and yield are easily affected by various abiotic stresses (Silvente et al., 2012; Gavili et al., 2019). Therefore, there is a need to explore the molecular mechanism involved in the soybean response to abiotic stress.
Cytokinin (CTK) is an important plant hormone that regulates many plant growth and development processes. CTKs naturally occurring in plants are derivatives of adenine and contain an isoprene-derived side chain or an aromatic side chain at the N6 end, called isoprenoid CTKs and aromatic CTKs (Figure S1), respectively (Sakakibara, 2006). In plants, CTKs are distributed mainly in the dividing cells of stem and root tips, immature and germinating seeds, and growing fruits, promoting cell division and regulating differentiation. In tissue culture, the high ratio of CTKs to auxin benefits shoot differentiation, while a low ratio promotes root differentiation. The phytohormone also delays protein and chlorophyll degradation and plays a role in response to biological and abiotic stresses (Hwang et al., 2012; Jameson & Song, 2016; Wybouw & De Rybel, 2019).
Cytokinin oxidation/dehydrogenase (CKX) enzymes are encoded by a family of CKX genes that can specifically degrade unsaturated side chains at the N6 position in CTKs and catalyze their irreversible degradation (Brownlee et al., 1975; McGaw & Horgan, 1983; Schmülling et al., 2003). Multiple gene families encoding CKX proteins (Werner et al., 2006) have been identified, and their evolution has been extensively studied in Arabidopsis thaliana (Werner et al., 2003), Oryza sativa (Ashikari et al., 2005; Rong et al., 2022), Nicotiana tabacum (Rong et al., 2022), Zea mays (Zalabák et al., 2014), Triticum aestivum (Mameaux et al., 2012), Brassica rapa (Mameaux et al., 2012), Brassica napus (Liu et al., 2018), Brassica oleracea (Zhu et al., 2022), and Vitis vinifera (Yu et al., 2021). The CKX genes also have several other functions in plants. For example, compared with the wild type, atckx3/ckx5 double mutant showed increased stem apex meristem and silique number of Arabidopsis (Bartrina et al., 2011), while the inhibition of the expression of OsCKX2 promotes rice growth by increasing its tiller number and yield (Gao et al., 2014; Yeh et al., 2015). The OsCKX11 gene also regulates leaf senescence and grain number and coordinates the source–sink relationship in rice (Zhang et al., 2021). The CKX genes are also involved in plant responses to various biological and abiotic stresses. For example, suppressing the expression of the CKXs gene can significantly enhance Arabidopsis resistance against Verticillium wilt and fungal infection (Reusche et al., 2013). Bol020547, Bol028392, and Bol045724 are important in determining cabbage (B. oleracea var. capitata) tolerance to Plasmodiophora brassicae (Zhu et al., 2022). In maize, most CKX genes were upregulated under salt stress (Vyroubalová et al., 2009); the overexpression of CKXs genes also enhanced Arabidopsis and tobacco tolerance to drought, salt, and abscisic acid stress (Nishiyama et al., 2011; Werner et al., 2011).
In this study, the CKX gene family in the whole soybean genome was identified and analyzed by bioinformatics techniques. The gene structure, chromosome distribution, cis-regulatory elements, gene replication, collinearity, and spatiotemporal expression patterns of the GmCKX genes were further analyzed. In addition, the key GmCKX genes that respond to salt, drought, salt combined with drought stress, 6-benzylaminopurine (6-BA), and indole-3-acetic acid (IAA) were screened. The results of this study lay the foundation for the study of GmCKXs gene function and provide important information for elucidating the evolutionary roles of CKXs.
2 Materials and methods
2.1 Identification and analysis of the GmCKX genes
The information of the reference genome and annotated proteins of soybean (Glycine max Wm82.a2.v1) was obtained from Ensembl Plants (http://plants.ensembl.org/index.html). The hidden Markov model (HMM) profile (http://hmmer.janelia.org/) and the Pfam database (http://pfam-legacy.xfam.org/) were used to screen candidate GmCKX proteins (PF01565 and PF09265). The CKXs protein sequence files were obtained from Ensembl Plants database (http://plants.ensembl.org/index). The InterPro (http://www.ebi.ac.uk/interpro/) (Finn et al., 2017) and SMART (http://smart.embl-heidelberg.de/) (Letunic et al., 2015; Han et al., 2019) software were used to further confirm the reliability of the CKX domain prediction. Then, the integrity of CKX domains was confirmed by Prosite (http://prosite.expast.org/) and WoLF PSORT (http://wolfpsort.hgc.jp/). All identified GmCKX genes were mapped according to their reference genome and named according to their locations on the chromosome using TBtools (Chen et al., 2020a). The CKX protein sequences derived from Arabidopsis, maize (Z. mays L.), and rice (O. sativa L.) were obtained from Ensembl Plants by searching CKX domains and used for phylogenetic analysis. A phylogenetic tree was constructed using the maximum likelihood method with 1,000 bootstrap replicates and the JTT+G model by MEGA X (version X-10.1.8, Mega Limited, Auckland, New Zealand).
The exon–intron structure of GmCKX genes was analyzed by the GSDS platform (http://gsds.cbi.pku.edu.cn/) (Guo, 2007). Gene-wise (Birney et al., 2004) was used to detect the correspondence between DNA and protein sequences. Then, the CKX domain coordinates in the protein sequence were converted to the coordinates in the nucleotide sequence using in-house perl script. The conserved motifs of CKX proteins were analyzed using MEME tool (http://meme.nbcr.net/meme/) (Bailey et al., 2009) with the following parameters: the motif length set at 10–50 amino acids and E value < 1e−20. The upstream regions (1,500 bp) of GmCKX genes were extracted and used as the gene promoter sequence. The cis-regulatory elements were analyzed by the PlantCare database (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The Multiple Collinearity Scan toolkit (MCScanX) was used to analyze the synteny and collinearity of GmCKX genes (Wang et al., 2012). Subsequently, the collinearity of the duplicated genes was visualized by Circos software (version 0.69) (Krzywinski et al., 2009). The expression data of GmCKXs in different tissues came from the Phytozome database (https://phytozome-next.jgi.doe.gov) and Soybean ePF Browser database (http://bar.utoronto.ca/efpsoybean/cgi-bin/efpWeb.cgi), respectively. The heatmap was generated using TBtools (Chen et al., 2020a).
2.2 Plant materials and treatments
The soybean seeds (Heike68) were obtained from the National Coarse Cereals Engineering Research Center, Daqing, Heilongjiang, China. The surface-sterilized soybean seeds were placed on a petri dish measuring 9 cm in diameter and incubated in the dark at 28°C until germination, which was indicated by the emergence of radicles. After 5 days of germination under distilled water treatment (CK), samples of soybean cotyledons, radicles, and hypocotyls were harvested, frozen in liquid nitrogen for 5 min, and then stored at −80°C for tissue-specific expression analysis of GmCKXs using quantitative real-time PCR (qRT-PCR). After 4 days of germination, we selected seedlings with consistent growth to explore the response of their GmCKXs to different abiotic stress. The experimental treatments consisted of seedlings exposed to 150 mM NaCl (SS, simulated salt stress), 20% (W/V) PEG 6000 (D, simulated drought stress), 150 mM NaCl +20% (W/V) PEG 6000 (SS+D), 10 µM IAA, and 10 µM 6-BA. The seedlings were exposed to treatments as described previously (Liu et al., 2018), with those treated with distilled water (CK) alone as controls. We then incubated the treated seedlings at 28°C for 24 h in the dark, harvested radicle samples from the treated seedlings in liquid nitrogen, and then stored them at −80°C before further use.
Average-sized soybean seedlings without disease symptoms or insect spots were selected and sown in a polypropylene pot (upper diameter = 21 cm, lower diameter = 15 cm, and height = 19 cm). The pots were filled with peat-soil mixed with vermiculite at a volume ratio of 3:1 and pH 7.0 and maintained in a controlled environmental chamber with a light regime of 16 h/8 h (light/dark) and relative humidity of 50%–55% at 28 ± 2°C until the V1 stage. The seedlings were thinned to three per pot to obtain uniform seedlings and then treated with 50 ml of each CK (control), 75 mM NaCl (SS), 20% (W/V) PEG 6000 (D), and SS+D. After 5 days of treatment, the soybean root and shoot tissues were separated and collected in liquid nitrogen for 5 min, then stored at −80°C for RNA extraction and transcriptome analysis. The soybean seedlings in the same pot/petri dish were considered one experimental unit. All experiments were repeated three times.
2.3 RNA extraction, transcriptome analysis, and gene expression by qRT-PCR
Total RNA of soybean root and leaf samples were extracted using the Trizol reagent (Invitrogen, CA, USA), and their quality and purity were checked using the NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, DE). The RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA) was used to detect RNA integrity. The sequencing libraries were constructed by Biomarker Technologies Corporation (Beijing, China) on the Illumina HiSeq2500 as recommended by the manufacturer. After deleting the low-quality bases, the clean reads were mapped to the soybean genome (Glycine max Wm82.a2.v1). The differentially expressed genes (DEGs) with an adjusted p-value < 0.01 found by DESeq2 and FDR < 0.01 were assigned as differentially expressed.
The single-stranded cDNA of soybean seedling samples was synthesized using a 5 × HiScript SuperMix II according to the manufacturer’s (Vazyme, Nanjing, China) instructions. The GmCKXs primers (Table S1) were designed with Primer 5.0 (Primer, Canada). Soybean TUBULIN A (NM_001250372) and ACTIN (NM_001289231) were used as the internal control genes. The qRT-PCR reaction was conducted using SYBR qPCR Master Mix (Vazyme, Nanjing, China) and run using the Roche Cycler 480II system (Roche, Roche Diagnostics, Switzerland). Relative expression levels for each CKX gene were calculated using the operational formula 2−ΔΔCt (Livak and Schmittgen, 2001). Three technical replicates and three biological replicates were performed for each reaction for each sample in this study.
2.4 Determination of CKX enzyme activity and zeatin content
The CKX enzyme activity of samples was detected using the ELISA kit (10894, Meibiao, Jiangsu, China) according to the instructions. The zeatin content was determined using high-performance liquid chromatography (HPLC-MS/MS) (AB SCIEX, ShimadzuLc-20AD, AB5500 Massachusetts, USA) at the Customs Quality Inspection Center (Dalian, Liaoning, China).
2.5 Statistical analysis
Results were analyzed using one-way analysis of variance (ANOVA) and the Duncan’s multiple range tests within the SPSS 19.0 (SPSS Inc., Chicago, IL, United States). Differences in values were considered statistically significant at p < 0.05.
3 Results
3.1 Identification and physicochemical property analysis of GmCKX genes in soybean
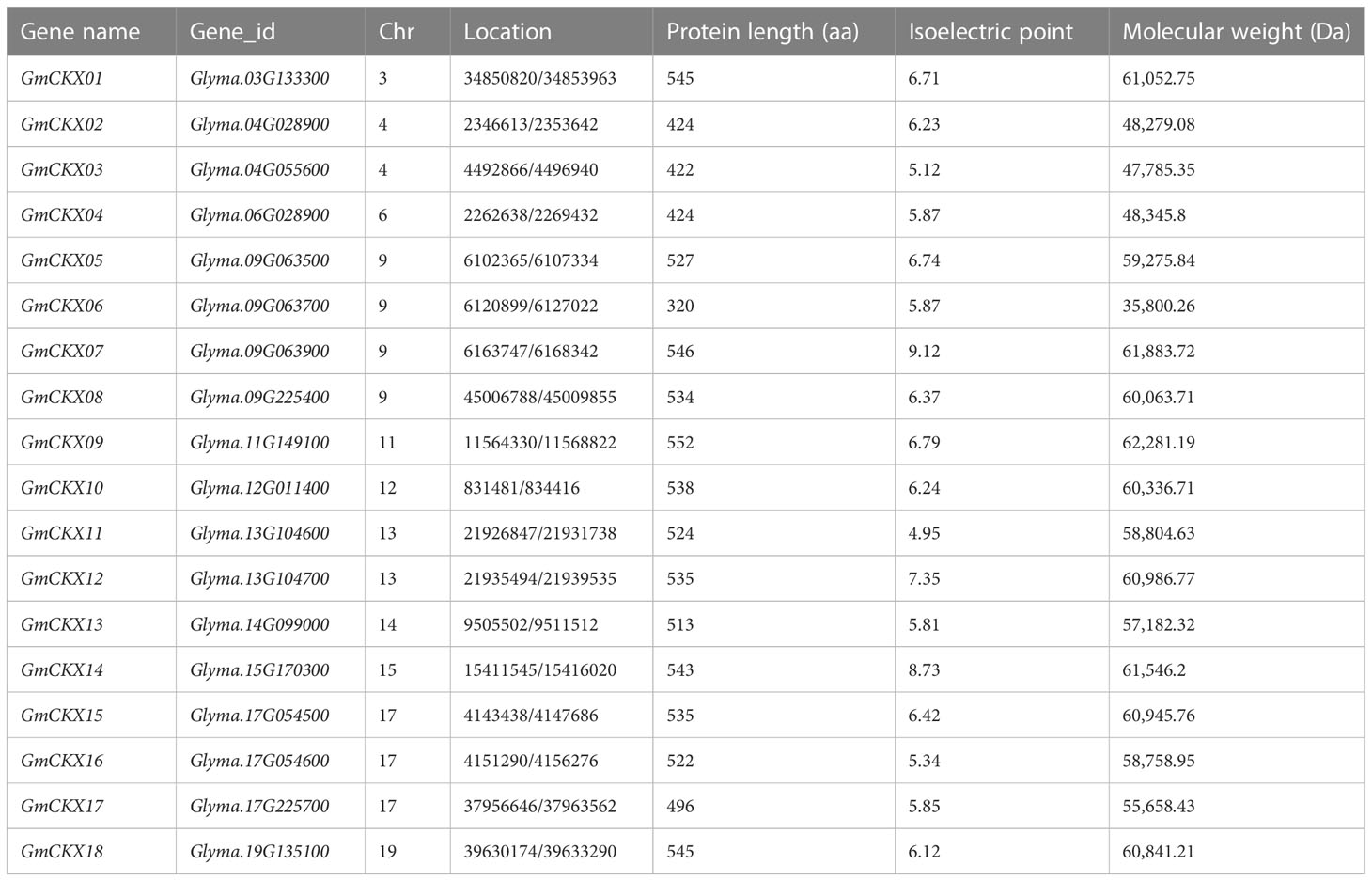
A total of 18 GmCKX genes were identified according to the result of an HMM profile, InterPro, and SMART analysis (Table 1). The results showed that the 18 GmCKX proteins contained amino acids (aa) ranging from 320 in GmCKX06 to 552 aa in GmCKX09, with the lowest isoelectric point (IP) in GmCKX11 (4.95) and the highest IP of 9.12 in GmCKX07 and a low molecular weight (MW) of 35,800.26 Da in GmCKX06 and a high MW of 62,281.19 Da in GmCKX09.
As shown in Figure 1, 18 GmCKX genes were unevenly distributed on 11 chromosomes: one gene on chromosome 3 (5.56% of the total), two genes on chromosome 4 (11.11% of the total), one gene on chromosome 6 (5.56% of the total), four genes on chromosome 9 (22.22% of the total), one gene on chromosome 11 (5.56% of the total), one gene on chromosome 12 (5.56% of the total), two genes on chromosome 13 (11.11% of the total), one gene on chromosome 14 (5.56% of the total), one gene on chromosome 15 (5.56% of the total), three genes on chromosome 17 (16.66% of the total), and one gene on chromosome 19 (5.56% of the total). In addition, GmCKXs are mostly distributed at both ends of the chromosomes.
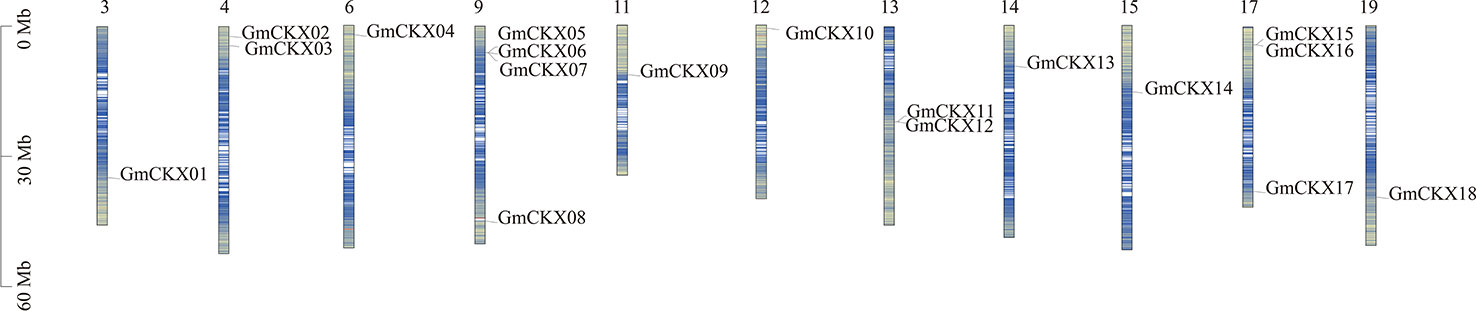
Figure 1 Localization of the GmCKXs on the soybean chromosomes. The chromosomal position of each GmCKX gene is shown on the corresponding chromosome from top to bottom according to the soybean genome. The blue line shows the gene density. The darker the color, the more dense the gene. The value on the Y-axis represents the position of the chromosome. The chromosome number is shown at the top of each bar.
3.2 Phylogenetic analysis of GmCKX proteins
To understand the evolution and development of the CKX gene family members in different species, 7 AtCKX, 11 OsCKX, 13 ZmCKX, and the 18 GmCKX proteins were assessed in a phylogenetic tree (Figure 2). The different CKXs were divided into six major clades (I–VI), with those from soybean only distributed in five subfamilies. Among the GmCKX proteins, clade I contained eight proteins, namely, GmCKX5, GmCKX6, GmCKX7, GmCKX11, GmCKX12, GmCKX14, GmCKX15, and GmCKX16; clade II had GmCKX2 and GmCKX4; clade III contained GmCKX3, GmCKX13, and GmCKX17; clade IV had GmCKX1 and GmCKX18; and clade V contained GmCKX8, GmCKX9, and GmCKX10 (Figures 2, S2).
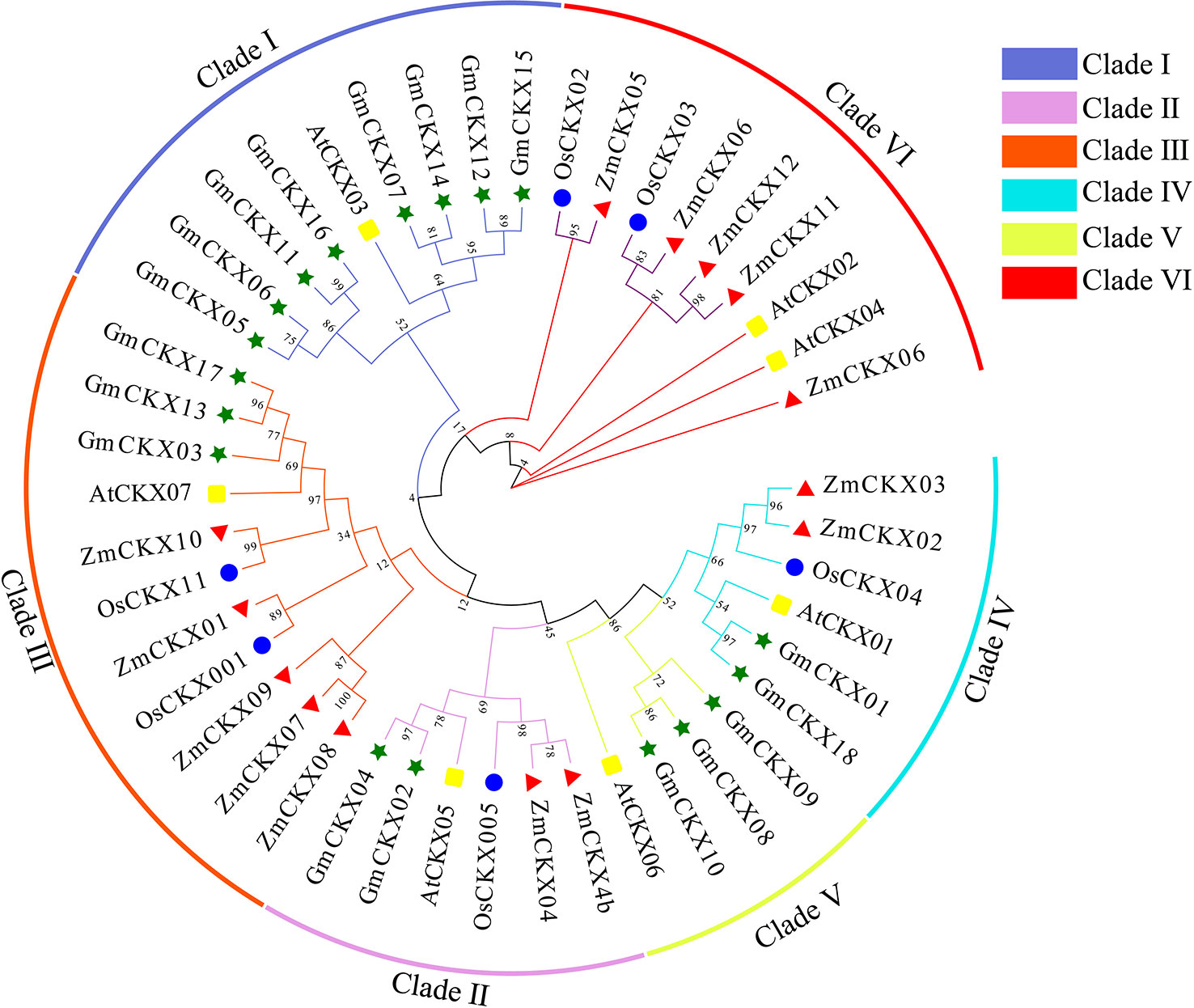
Figure 2 Phylogenetic analysis of CKX proteins in soybean, Arabidopsis, maize, and rice. The green, blue, red, and yellow circle represent soybean (G. max L.), A. thaliana, maize (Z. mays L.), and rice (O. sativa L.) respectively.
3.3 Conserved motifs and gene structure analysis
The online software MEME was used to analyze the conservative motifs of GmCKXs. A total of 10 conserved motifs were obtained from the 18 GmCKXs, designated as Motifs 1 to 10 (Figures 3A, B). The GmCKX members in the same subfamily had similar motif characteristics but differed among GmCKXs in other subfamilies. Most GmCKX members contained 10 motifs each, with GmCKX6 found in clade I containing five motifs, GmCKX2 and GmCKX4 (clade II), and GmCKX3 (clade III) containing eight motifs each. We analyzed the exon–intron structures of the GmCKX members on the GSDS website (Figure 3C) and found similar exon–intron structures for GmCKXs in the same subfamily, with differences among GmCKXs in different subfamilies. The number of exons in the 18 GmCKX genes ranged from four to seven, with the majority containing six exons each. The least number of exons was found in GmCKX3, while the highest number was found in GmCKX8.
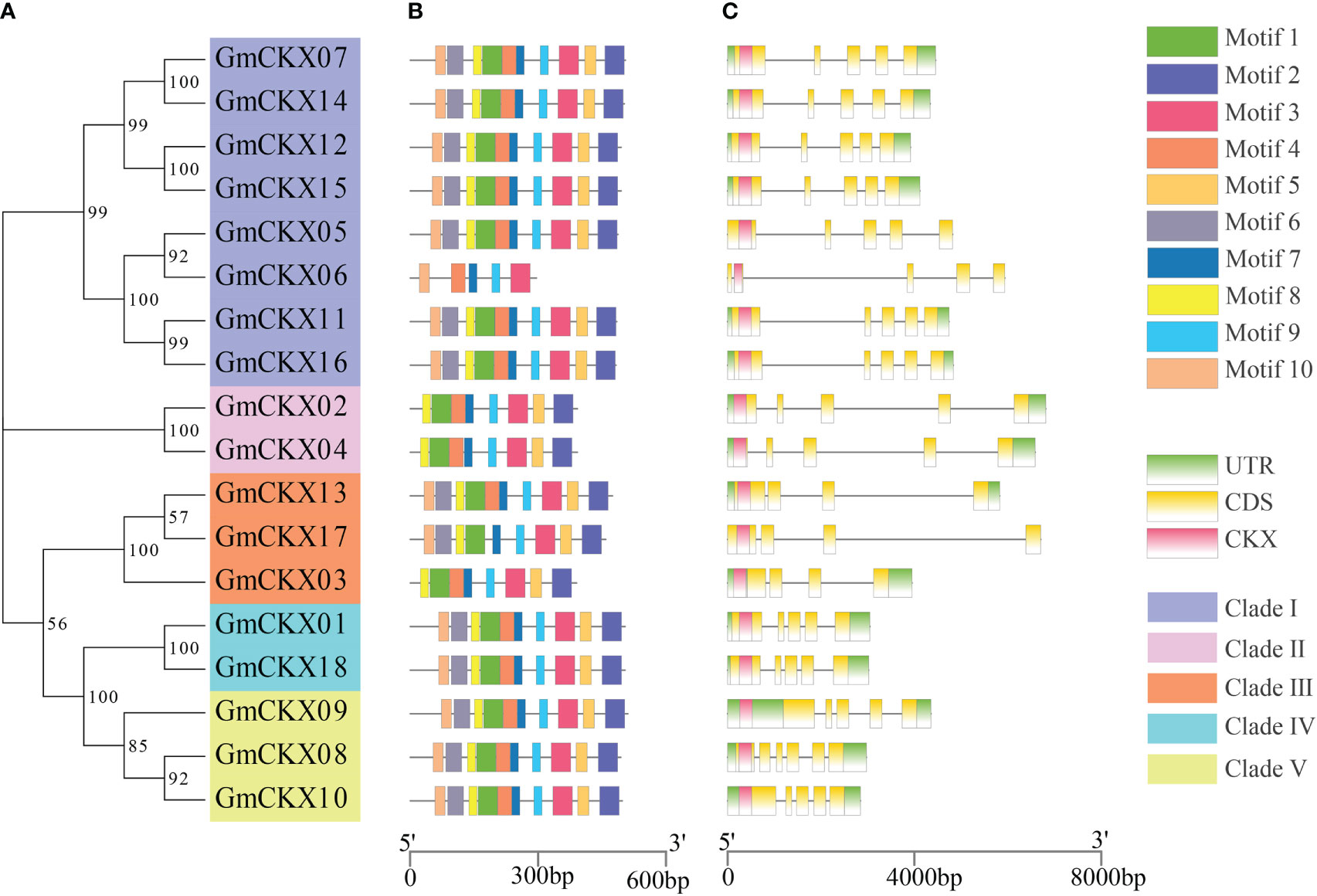
Figure 3 Gene structure and conserved motifs of the GmCKXs. (A) The phylogenetic classification of GmCKXs. (B) Conserved motif analysis of GmCKXs; different colored rectangles represented different motifs. (C) Gene structure analysis of GmCKXs. UTR regions (green rectangles), exons (yellow rectangles), CKX domains (pink rectangles), and introns (black lines).
3.4 Analysis of promoter cis-regulatory elements
To further study the regulatory mechanism of the GmCKX family in response to abiotic stress, the upstream 1.5-kb sequences of each of the 18 GmCKXs were extracted and used to analyze the cis-regulatory elements (Figure 4; Table S2). We identified 13 cis-regulatory elements and divided them into three groups: hormone-, resistance-, and physiological metabolism-related elements. The hormone-related elements consisted of the P-box, ABRE, TGA-element, TCA-element, GARE-motif, AuxRR-core, and TATC-box. The resistance-related elements included LTR, ARE, GC-motif, and MBS, while the physiological metabolism-related elements had only the MBSI and CAT-box.
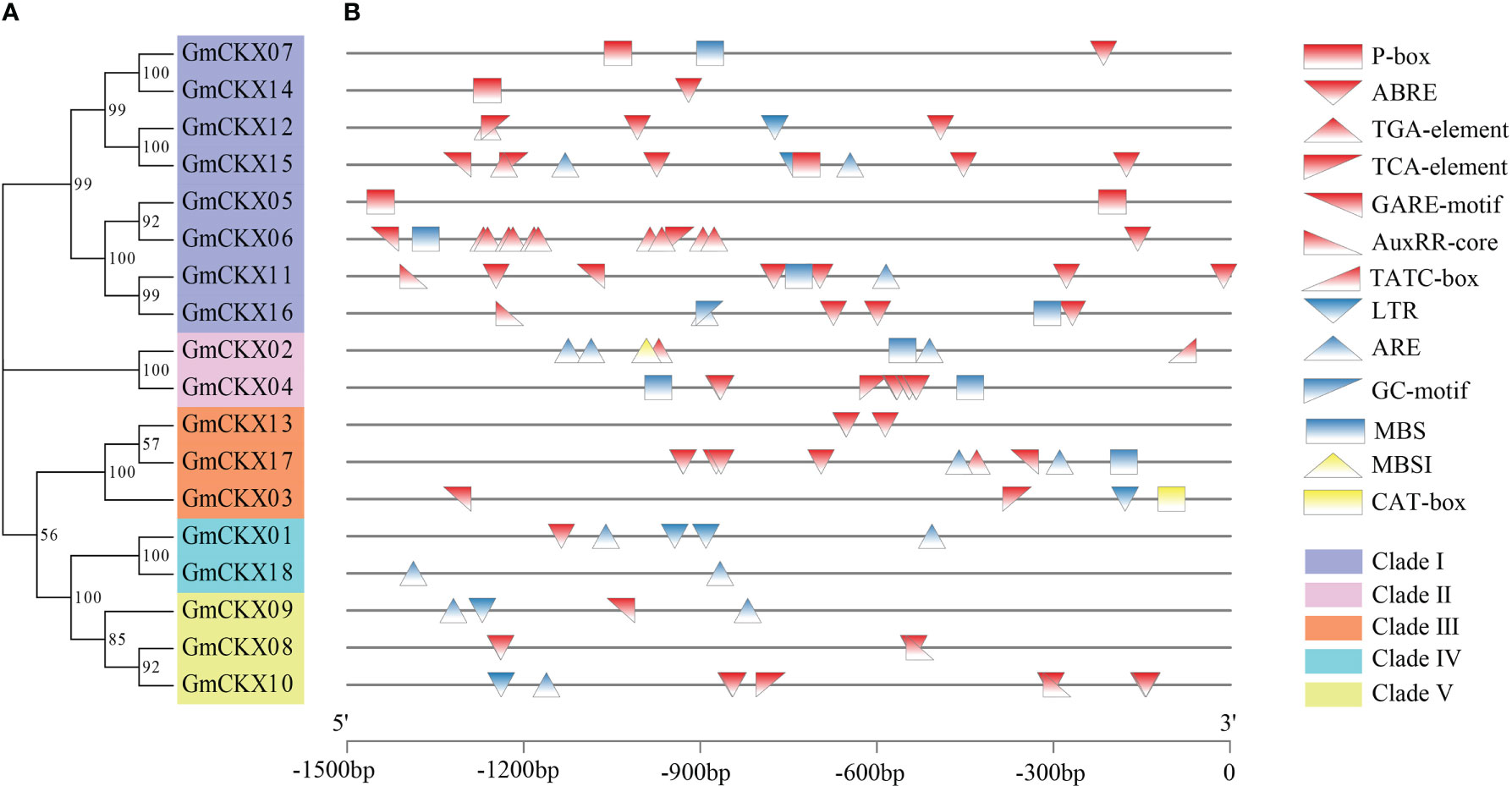
Figure 4 Promoter cis-regulatory element analysis of the GmCKXs. (A) The phylogenetic classification of GmCKXs. (B) Cis-element analysis of the promoter regions of GmCKXs genes. P-box: gibberellin-responsive element; ABRE: abscisic acid elements; TGA-element: auxin responsive element; TCA-element: salicylic acid elements; GARE-motif: gibberellin responsive element; AuxRR-core: auxin response promoter element; TATC-box: gibberellin responsive element; LTR: low-temperature responsiveness; ARE: anaerobic responsiveness; GC-motif: enhancer-like element involved in anoxic specific inducibility; MBS: drought stress inducibility element; MBSI: flavonoid biosynthesis regulation; CAT-box: meristem expression element.
3.5 Collinearity analysis
The origins of duplicates for CKX genes were detected by MCScanX and used to analyze the distribution and arrangement of its homologs within or between species. We identified six GmCKX duplicate gene pairs in the soybean genome, all characterized as segmental duplication events (Figure 5A; Table S3). Subsequently, we performed nonsynonymous and synonymous substitution ratio (Ka and Ks) analyses of duplicated genes to examine the driving forces of the soybean CKX gene family. The results showed that all six GmCKX gene pairs underwent purification selection with the Ka/Ks < 1. The collinearity analysis of GmCKXs with Arabidopsis was further used to explore the evolutionary mechanisms of the soybean CKX gene family. The results identified five orthologous gene pairs between soybean and Arabidopsis as collinear pairs, including GmCKX1/AtCKX1, GmCKX2/AtCKX5, GmCKX5/AtCKX3, GmCKX6/AtCKX3, and GmCKX6/AtCKX3 (Figure 5B).
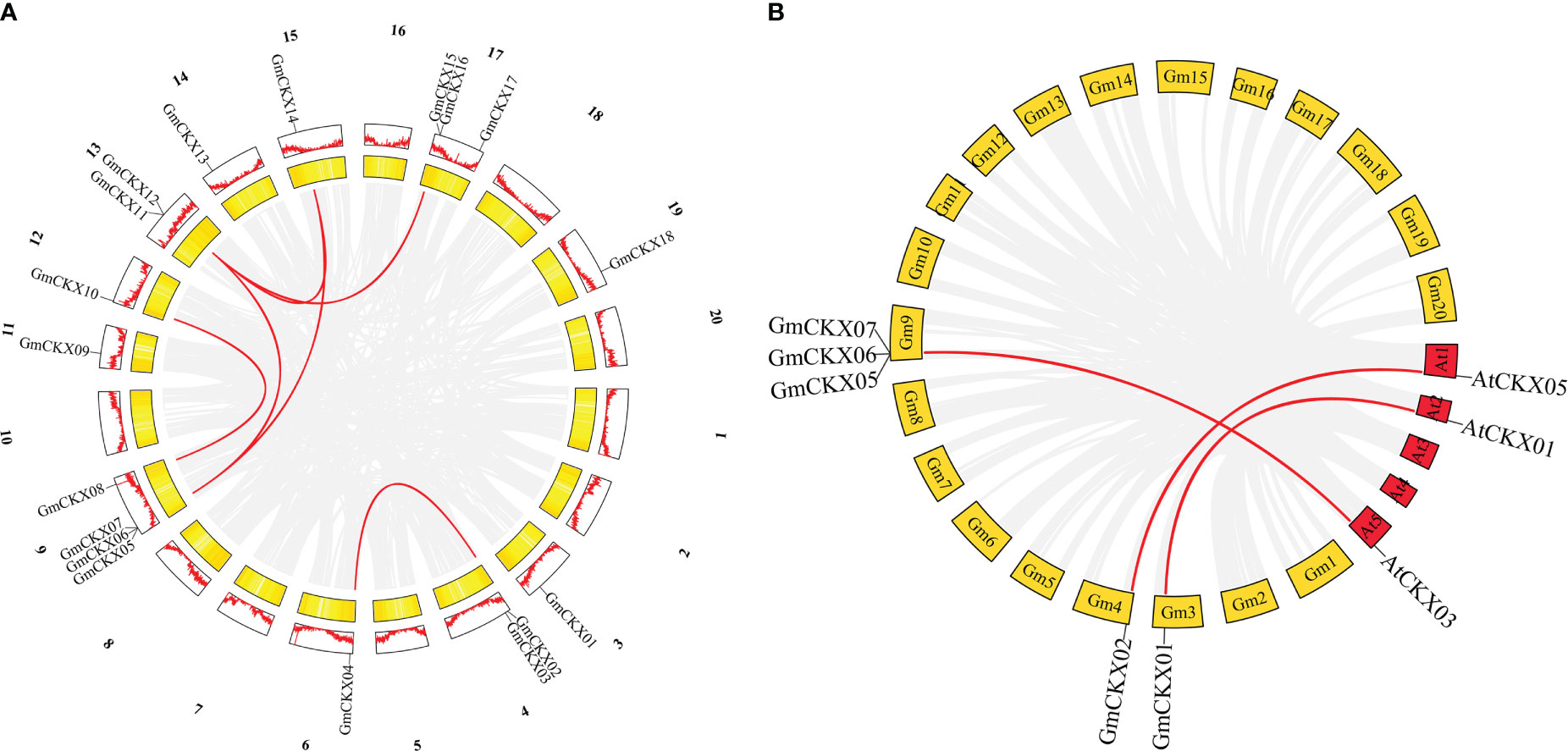
Figure 5 Collinearity analysis of the GmCKXs. (A) Duplicated gene pairs in soybean genome. Red lines indicate the duplication of GmCKXs gene pairs. (B) Collinearity analysis of GmCKX genes with A. thaliana. Red lines connect fragments of repeated gene pairs between soybean and A. thaliana.
3.6 Expression profile analysis of GmCKX genes in soybean tissues
To explore the spatiotemporal expression patterns of soybean GmCKX genes, we compared the transcript abundances of all the 18 GmCKX genes using two publicly available RNA-Seq data from the Phytozome and Soybean ePF Browser database, respectively (Figures 6, 7). The Phytozome dataset contained root, root tip, lateral root, stem, shoot tip, leaf, flower, and nodules. In contrast, the Soybean ePF Browser dataset consisted of the root hair, shoot apical meristem (SAM), flower, green pods, leaf, nodule, root and root tip, and also the treatment and control root hair tissue after Bradyrhizobium japonicum infection at three different time points (Libault et al., 2010a; Libault et al., 2010b). Most GmCKX genes were preferentially expressed in more than one tissue, with GmCKX7 and GmCKX8 highly expressed and GmCKX5 and GmCKX6 genes lowly expressed or undetected in both two datasets. The data also showed that over 40% of the highly expressed GmCKX genes occurred in the floral organs of soybean.
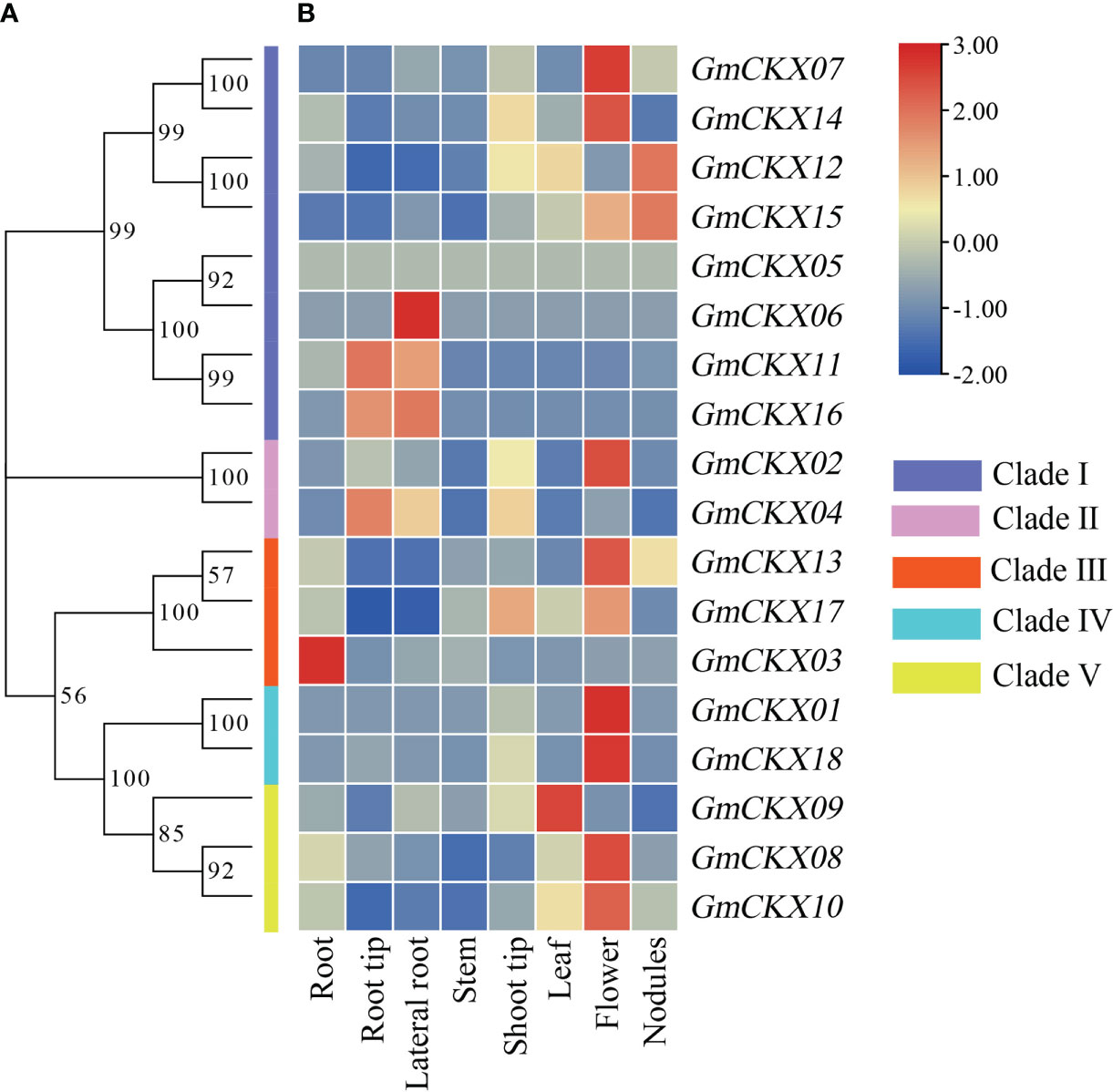
Figure 6 Heatmaps of the expression profiling of 18 GmCKX genes. (A) The phylogenetic classification of GmCKXs. (B) Expression profiling of GmCKXs in different tissues based on the Phytozome database. The color scale represents expression levels from high (red) to low (blue).
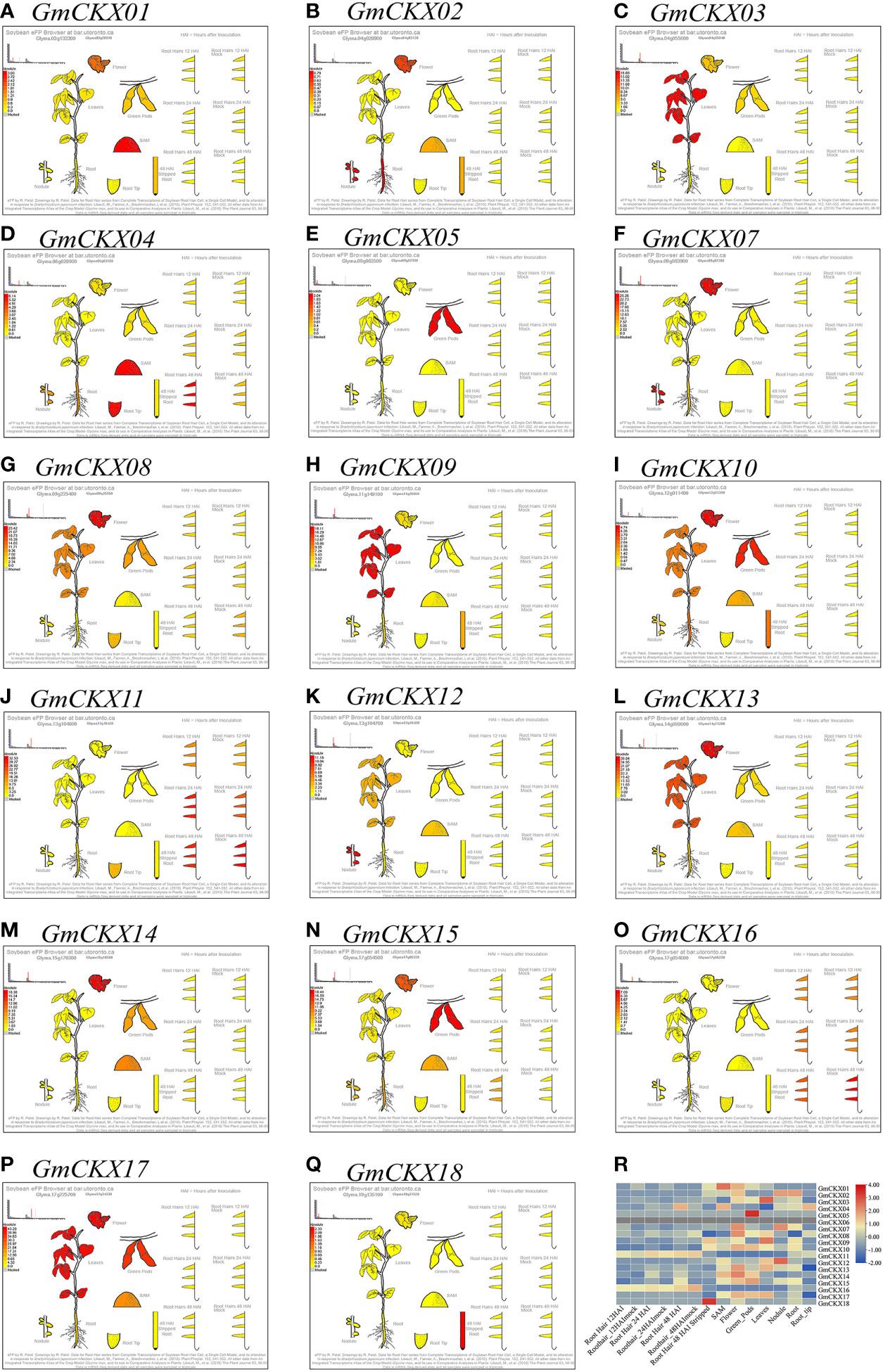
Figure 7 Expression profiling of GmCKXs in different tissues based on the Soybean eFP Browser database. (A–Q) Diagram showing the different soybean tissues. Red represents high expression and yellow represents low expression. (R) The heatmap were drawn by TBtools. Red represents high expression and blue represents low expression.
3.7 Expression analysis of GmCKX genes in soybean seeds during the germination stage
Because seed germination is an important growth stage in the plant life cycle, we used qRT-PCR to investigate the expression of GmCKXs in the soybean seed’s radicle, hypocotyl, and cotyledon during this stage (Figure 8). Some genes displayed tissue-specific expression. For example, GmCKX3, GmCKX17, and GmCKX18 were found in the radicle; GmCKX7, GmCKX9, and GmCKX10 were found in hypocotyl; and GmCKX6, GmCKX8, GmCKX11, and GmCKX12 were found in the cotyledon.

Figure 8 Expression pattern of soybean GmCKX genes in different tissues by qRT-PCR analysis during the germination stage. (A) The phylogenetic classification of GmCKXs. (B) Expression profiling of GmCKXs in radicle, hypocotyl, and cotyledon tissues. The color scale represents expression levels from high (red) to low (blue).
3.8 Expression patterns of GmCKXs under abiotic stress
To explore the roles of specific GmCKX genes in response to different abiotic stresses, transcriptome expression patterns of all soybean GmCKX genes were analyzed in the leaves and roots of soybean seedlings under salt (SS), drought (D), and salt combined with drought stress (SS+D) (Figures 9A, B). The assembled gene dataset was deposited at the National Center for Biotechnology Information with the accession number PRJNA930177. Our data showed different expression profiles of GmCKX genes in different stress treatments and tissues. For example, compared to the control, three stress treatments significantly upregulated the expression level of GmCKX13 in leaf and root but downregulated the expression level of GmCKX3 and GmCKX8. The GmCKX14 gene in soybean leaves was highly upregulated under the three stress treatments, while its expression level in the roots was significantly downregulated. The expression levels of GmCKX9 in leaf and root were downregulated considerably under D and SS+D treatment, but SS treatment had no significant effects on its expression level. In the leaf, the GmCKX15 gene was highly expressed under SS treatment but downregulated under D and SS+D treatment. At the same time, all three stress treatments significantly downregulated the expression of GmCKX15 in the roots. The accuracy of transcriptome data was verified by qRT-PCR of six randomly selected GmCKX genes (Figures 9C–H).
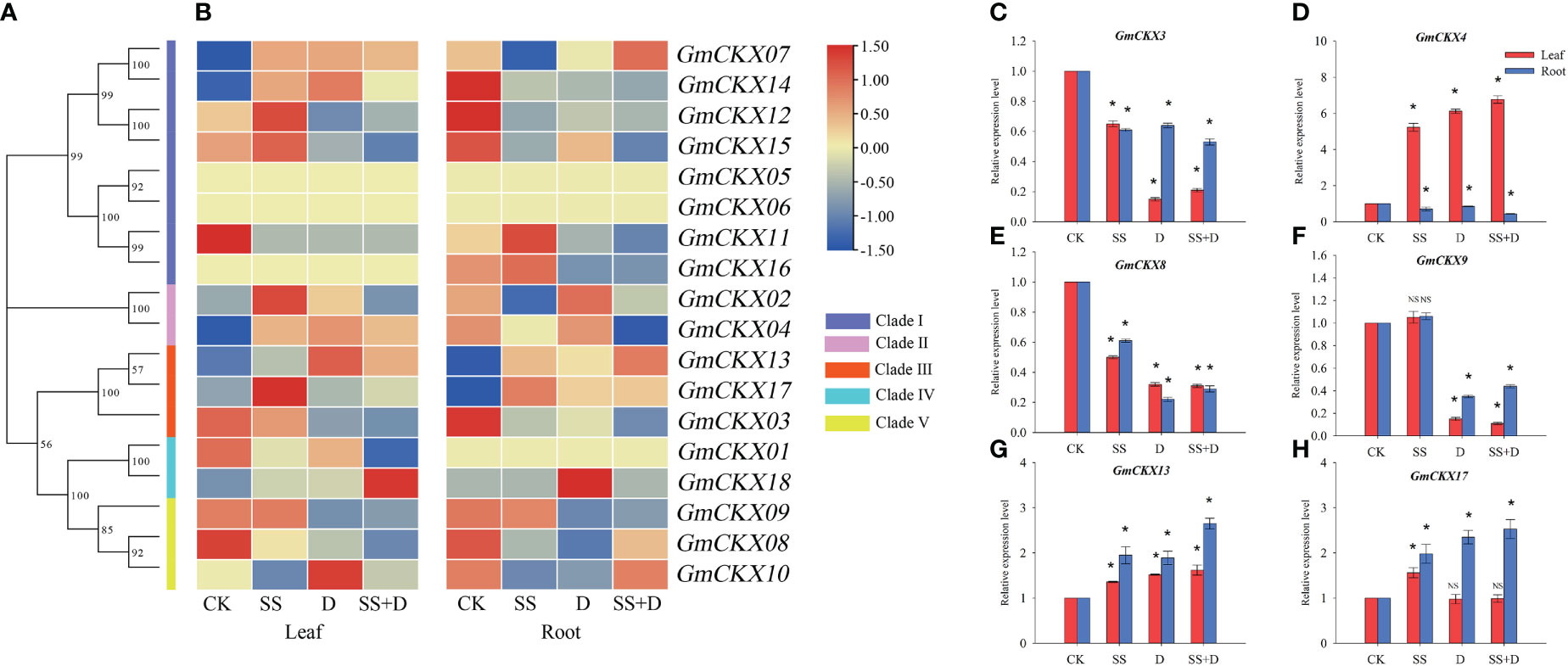
Figure 9 Expression profiles of soybean GmCKXs under different abiotic stresses at the seedling stage. (A) The phylogenetic classification of GmCKXs. (B) Expression profiling of GmCKXs in leaf and root under different abiotic stresses. CK, control condition; SS, salt stress; D, drought stress; SS+D, salt combined with drought stress. The color scale represents expression levels from high (red) to low (blue). (C–H) qRT-PCR analysis of six selected GmCKX genes in leaf and root under different abiotic stresses. Values are the means ± standard deviations of three replicates. * indicates significant difference between CK and treatment condition (p < 0.05). NS indicates no significant differences between CK and treatment condition.
To further investigate whether GmCKXs participate in response to the abiotic stresses during the germination stage, soybean seed radicles treated with SS, D, and SS+D were collected for qRT-PCR (Figure 10). We found that the expression of most GmCKX genes differed under different stress treatments. For example, compared with the control, the expression of GmCKX1 and GmCKX3 was significantly upregulated under SS treatment but significantly downregulated under D and SS+D treatments. The GmCKX2 and GmCKX8 were upregulated considerably under SS and D treatments but were significantly downregulated under SS+D treatment. All three stress treatments also significantly downregulated genes such as GmCKX4, GmCKX7, GmCKX12, GmCKX13, GmCKX14, GmCKX15, and GmCKX17, but significantly upregulated GmCKX16.
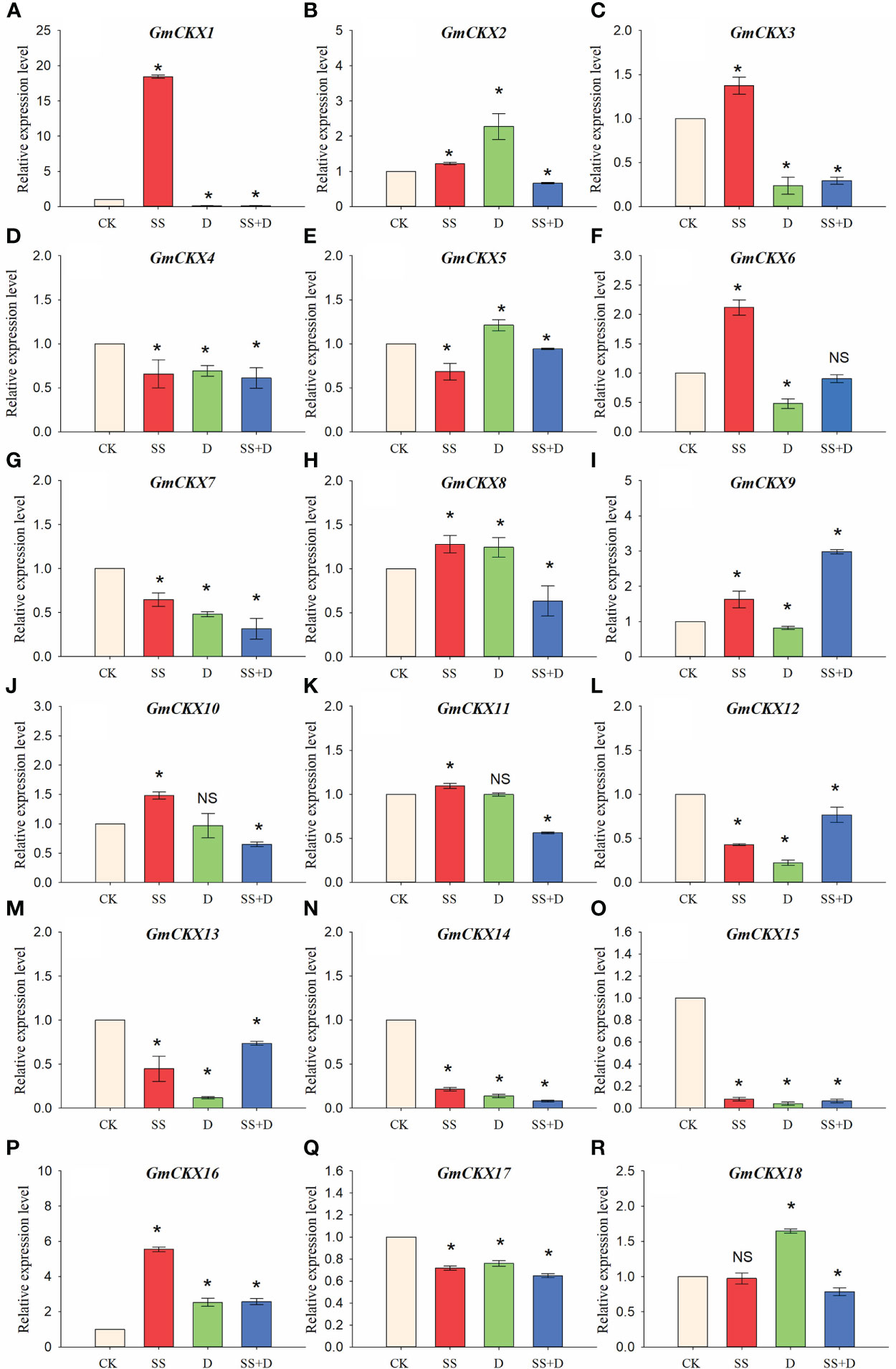
Figure 10 Relative expression levels of soybean GmCKX genes under SS, D, and SS+D treatments at the germination stage (A–R). Values are the means ± standard deviations of three replicates. * indicates significant difference between CK and treatment condition (p < 0.05). NS indicates no significant differences between CK and treatment condition.
3.9 Hormone-induced patterns of expression of the GmCKX genes
We analyzed the relative expression level of GmCKXs in radicles treated with 6-BA and IAA using qRT-PCR to explore the hormone-induced patterns of expression of the GmCKX genes (Figure 11) and found differential expression of the GmCKXs under different hormone treatments. Compared to the control, the 6-BA and IAA significantly upregulated GmCKX10 and GmCKX18 genes but highly repressed the GmCKX1, GmCKX6, and GmCKX9 genes. The GmCKX2, GmCKX3, GmCKX7, GmCKX12, GmCKX13, GmCKX14, GmCKX15, GmCKX16, and GmCKX17 genes were upregulated considerably under 6-BA treatment but downregulated under IAA treatment. The GmCKX4, GmCKX8, and GmCKX11 genes were significantly upregulated under 6-BA treatment but remained unaffected under IAA treatment. The IAA hormone also affected the expression of the GmCKX5 gene by significantly upregulating it.
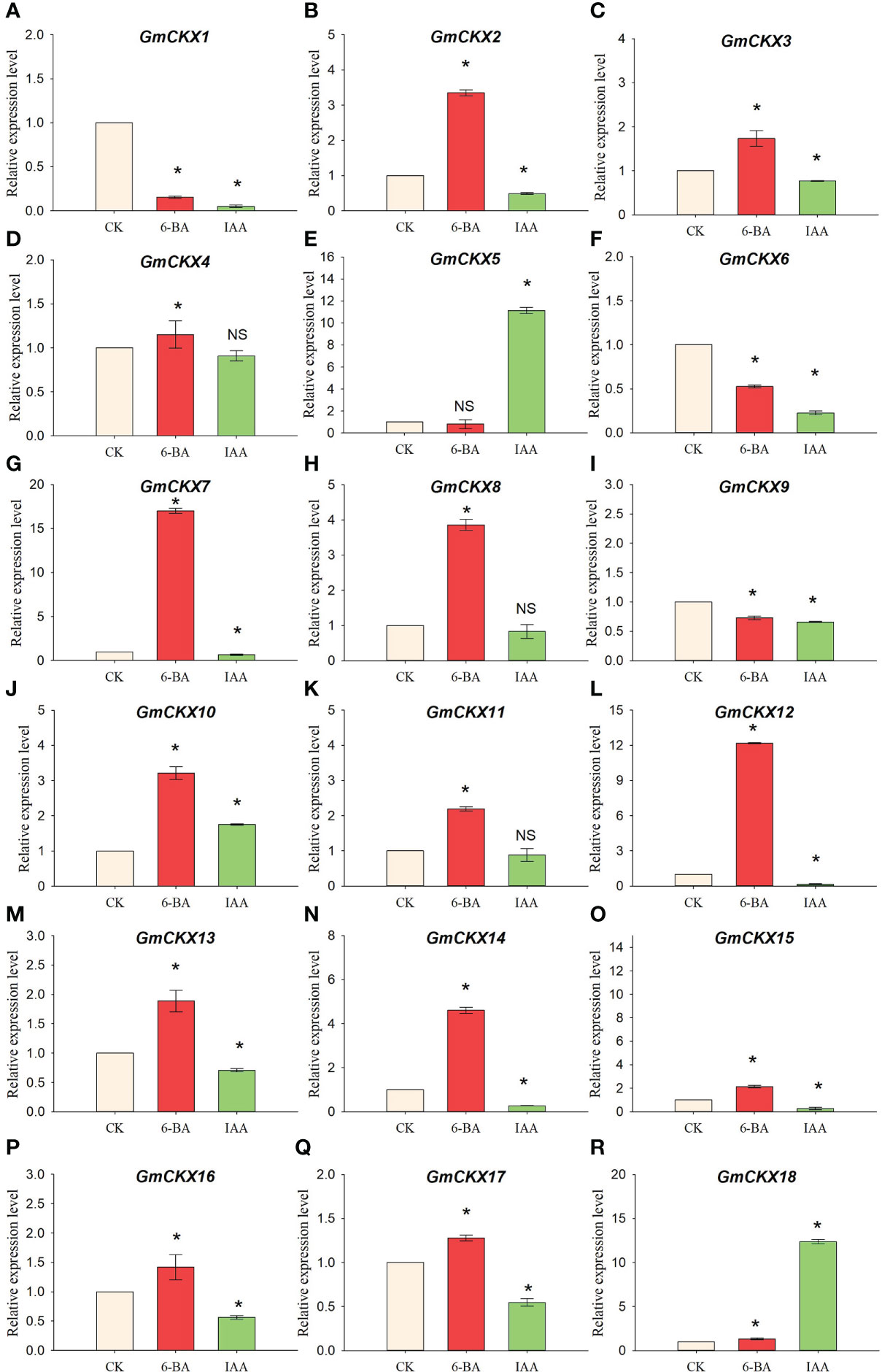
Figure 11 Relative expression levels of soybean GmCKX genes under IAA and 6-BA treatments (A–R). Values are the means ± standard deviations of three replicates. * indicates significant difference between CK and treatment condition (p < 0.05). NS indicates no significant differences between CK and treatment condition.
3.10 Abiotic stress and hormone-induced changes of CKX enzyme activity and zeatin content
We determined the zeatin content and CKX enzyme activity in soybean radicles under SS, D, SS+D, 6-BA, and IAA treatments to analyze the relationship between CKX enzyme activity and CTK content under abiotic stress (Figure 12). Compared with the control, SS, D, and SS+D stress treatments significantly decreased zeatin content in soybean radicles by 32.02%, 44.2%, and 54.31%, respectively. In comparison, 6-BA and IAA treatments significantly increased the zeatin content in soybean radicles by 274.79% and 199.81%, respectively. Compared to the control, SS, D, and SS+D significantly increased CKX enzyme activity in soybean radicles by 32.9%, 39.35%, and 73.46%, respectively, while 6-BA and IAA significantly decreased CKX enzyme activity in soybean radicles by 38.33% and 26.9%, respectively.
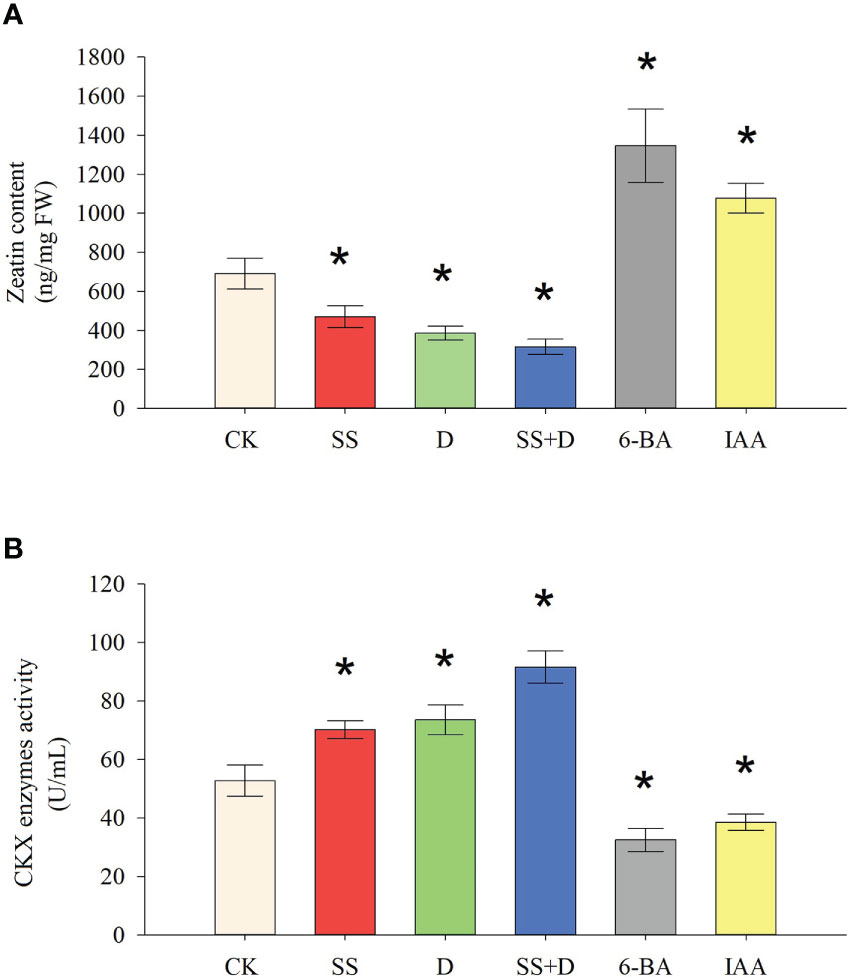
Figure 12 Zeatin content and CKX enzyme activity under different abiotic stresses and hormone treatments. (A) Zeatin content under SS, D, SS+D, 6-BA, and IAA treatments. (B) CKX enzyme activity under SS, D, SS+D, 6-BA, and IAA treatments. Values are the means ± standard deviations of three replicates. * indicates significant difference between CK and treatment condition (p < 0.05). NS indicates no significant differences between CK and treatment condition.
4 Discussion
CTKs play an important role in numerous plant physiology processes, such as fatty acid biosynthesis in seed (Thien Nguyen et al., 2016), the development of plant floral organs and pod setting (Nonokawa et al., 2012), leaf senescence (Merewitz et al., 2010), and seed yield (Jameson and Song, 2016; Chen et al., 2020b). The hormone also helps the plants to respond to a variety of abiotic stresses, including drought (Hai et al., 2020), heat (Prerostova et al., 2020), and salt (Yu et al., 2022). Several members of the CKX gene family, which regulate the endogenous CKs, have been identified in various crops, including 11 members in rice (Mameaux et al., 2012), 13 in maize (Zalabák et al., 2014), 36 in cabbage (Zhu et al., 2022), 23 in oilseed rape (Liu et al., 2018), 5 in potato (Suttle et al., 2014), and 12 members in apple (Tan et al., 2018). In this study, we identified 18 CKX family members with complete domains in the entire genome of soybean, named GmCKX1-GmCKX18, according to their position on the chromosome. The variation in the number of CKX genes in different species is possibly due to genome evolution and replication, which causes the production of homologous genes and an increase in their numbers (Kaltenegger et al., 2018; Wang et al., 2021). We further divided the soybean CKXs into five subgroups (I–V) based on a phylogenetic tree containing Arabidopsis and soybean CKX protein sequences. Detailed molecular characterization of the GmCKX genes showed that CKX protein members had different physicochemical properties, such as protein length, pI, and MW, indicating a high diversity of these gene family members. These insights will help investigate the function of GmCKXs.
The exon–intron structure provides important information for exploring the evolutionary relationship of genes (Ahmad et al., 2019). In general, the number of exons plays an important role in gene evolution (Xu et al., 2012), and the number of introns determines the rate of gene expression (Jeffares et al., 2008; Shaul, 2017). Genes with similar exon–intron structures have similar gene functions. Therefore, gene function can be predicted by analyzing its structure (Li et al., 2019b). In the current study, most GmCKX genes in the same subfamilies had similar numbers of exons and introns, suggesting that they might have similar functions. However, we also found some GmCKX genes with different exon and intron numbers within the same subfamily, a phenomenon that has also been reported in oilseed rape (Liu et al., 2018) and cabbage (Zhu et al., 2022), which might be due to the functional diversity of genes throughout evolution.
The chromosomal localization analysis showed that the GmCKX genes were distributed non-homogeneously on chromosomes, showing a cluster distribution, which may be attributed to the non-uniform replication event of soybean chromosome fragments. Gene duplication is an important mechanism that promotes the expansion and diversification of gene families. Synteny analyses revealed that segmental duplication contributed to the expansion and diversification of the soybean GmCKX gene family. Similar results have been observed in maize (Gu et al., 2010) and Chinese cabbage (Liu et al., 2013). The study also showed that all six GmCKX gene pairs had Ka/Ks < 1, indicating that GmCKXs underwent purification selection under environmental stress, which is consistent with the results of previous studies (Yu et al., 2021; Zhu et al., 2022). Genome comparison is considered a relatively fast and effective method to study the potential characteristics and functions of genes (Lyons and Freeling, 2008). Therefore, the possible role of CKX homologous genes in the soybean can be inferred by analyzing the information on CKX genes in the model plant, such as Arabidopsis. This is supported by the location of five orthologous gene pairs in syntenic genomic regions between soybean and Arabidopsis genomes. For example, the atCKX1 gene is expressed in root tissues and participates in lateral root formation (Chang et al., 2015). Ectopic expression of the AtCKX1 gene in tobacco enhanced drought and heat stress tolerance (Macková et al., 2013). In Arabidopsis, AtCKX3 and AtCKX5 genes were expressed in reproductive meristems. The ckx3 ckx5 double mutant could delay the differentiation of reproductive meristem cells and exhibit more and larger flower and silique numbers (Bartrina et al., 2011). Based on the reported function of the CKX gene (AtCKX1, AtCKX3, and AtCKX5) in Arabidopsis, we could predict the possible role of the soybean GmCKX genes. However, their functional roles need to be further confirmed in future reverse genetics studies.
Germination is the initial stage of soybean growth, which is also the most sensitive to environmental stress. The expression levels of genes in tissues and organs are closely related to their functions. We used two sets of public databases and qRT-PCR data from germination soybeans to detect the expression levels of the 18 GmCKX genes in different soybean tissues. The expression of the GmCKX genes differed among the soybean tissues, indicating that the GmCKX genes had different biological functions and were involved in soybean growth regulation and various tissue development processes. The expression patterns of individual GmCKXs in soybean were shown to be tissue and development specific. For example, GmCKX7 and GmCKX8 were highly expressed in all organs, while GmCKX5 was mainly expressed in hypocotyl and cotyledon, and GmCKX6 was highly expressed in cotyledon. These results are consistent with studies of CKX genes in other species (Song et al., 2012; Chen et al., 2020b; Yu et al., 2021).
CTK is a physiological hormone that widely exists in plants. As a key enzyme, which degrades endogenous CTK, CKX plays an important role in maintaining intracellular CTK homeostasis and for adaption to environmental stress (Vyroubalová et al., 2009; Le et al., 2012). Cis-acting elements play an important role in signal transduction and regulation of gene transcription initiation. Analysis of cis-acting elements in the promoters of GmCKXs demonstrated that the GmCKXs play a role in response to the hormone, plant growth, and biotic and abiotic stress responses. Similar findings were found in the CKX gene families in oilseed rape, maize, and Arabidopsis. For example, the overexpression of MsCKX genes in Arabidopsis exhibited stronger salt tolerance (Li et al., 2019a), while ZmCKX1 was strongly induced by CTKs, abscisic acid, and abiotic stress in maize (Brugière et al., 2003). In oilseed rape, the expression level of BnCKX7-1 was downregulated by the exogenous supply of 6-BA (Liu et al., 2018). In the current study, soybean GmCKX genes showed various roles in response to salt, drought, salt combined with drought stresses, 6-BA, and IAA. Our results demonstrate that each GmCKX gene is expressed differently in response to salt and drought stress and the exogenous supply of 6-BA and IAA hormones. The analysis of the GmCKX gene family at seedling and germination stages of soybean under salt, drought, and salt combined with drought stress showed that GmCKX14 was downregulated both in root at the seedling stage and in hypocotyl at the germination stage under the three abiotic stress treatments. The results of the evolutionary analysis showed that GmCKX14 and AtCKX3 were homologous genes. The overexpression of the AtCKX3 gene increased the growth rate of primary roots and reduced the number of flowers in transgenic Arabidopsis (Werner et al., 2003). Transgenic tomatoes with overexpressed AtCKX3 gene maintained the plants in a higher water state by reducing transpiration under drought treatment, thus enhancing their drought resistance (Farber et al., 2016). Our results suggested that soybean GmCKX14 might be an important negative regulatory gene in abiotic stress such as salt and drought. The analysis of zeatin content and CKX enzyme activity in radicle under salt, drought, and salt combined with drought stress confirmed that abiotic stress enhanced CKX enzyme activity but reduced zeatin content.
In bread wheat, exogenous hormone treatment significantly induced TuCKXs gene expression within 3 h (Shoaib et al., 2019). Our results show that an exogenous supply of 6-BA and IAA could dramatically reduce the CKX enzyme activity of soybean radicle and enhance zeatin content. However, the soybean GmCKXs showed different expression patterns in response to 6-BA and IAA. After the exogenous supply of 6-BA and IAA, the GmCKX1, GmCKX6, and GmCKX9 genes were all repressed, while the GmCKX10 and GmCKX18 genes were upregulated. Combined with physiological analysis results, the GmCKX1, GmCKX6, and GmCKX9 genes could be used as positive regulatory factors, and GmCKX10 and GmCKX18 could be used as negative regulatory factors to participate in CK metabolism in response to exogenous 6-BA and IAA. However, this regulatory effect still requires further confirmation of its functional role in future reverse genetics studies. Our results also provide a reference for studying the function of the CKX gene under abiotic stress and hormonal regulation.
5 Conclusion
In this study, the 18 GmCKX genes were identified from the soybean genome, and their evolutionary relationship, chromosomal location, gene structure, motifs, cis-regulatory elements, collinearity, and gene expression patterns were analyzed by bioinformatics tools, RNA-seq, and qRT-PCR methods. The GmCKXs members were divided into five clades according to the phylogenetic tree. Synteny analyses revealed that the expansion of the GmCKXs gene family is mainly due to fragment replication. The analysis of Phytozome and Soybean ePF Browser databases and qRT-PCR showed that GmCKX genes had tissue-specific expression patterns. In addition, GmCKXs genes were differentially regulated in response to salt, drought, salt combined with drought stress, 6-BA, and IAA treatments. Expression of GmCKX14 was downregulated both in root at the seedling stage and in radicle at the germination stage under salt, drought, and salt combined with drought stress treatments. Under 6-BA and IAA treatments, the expressions of GmCKX1, GmCKX6, and GmCKX9 decreased, while the expressions of GmCKX10 and GmCKX18 increased. Finally, physiological analysis results showed that GmCKX genes could respond to abiotic stress and regulate the activity of CKX enzymes and the zeatin content.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA930177.
Author contributions
YD, ZZ, YG, JD, and QZ participated in the experimental design. YD, ZZ, WL, WW, XY, YZ, and MY performed material sampling, gene expression experiments, and physiology experiments. YD, ZZ, YG, and JD contributed to the data collection and data analysis. YD wrote the manuscript. ZZ, JD, and QZ revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program (2021YFD1201603-3), the Heilongjiang Bayi Agricultural University Support Program for San Zong (ZRCQC202102 and ZRCQC202201), the Talent Introduction Project of Heilongjiang Bayi Agricultural University (XYB202006), the Postdoctoral General Project of Heilongjiang Province (LBH-Z21196), and the Innovative Research Project for Graduate Students of Heilongjiang Bayi Agricultural Reclamation University (YJSCX2022-Y01).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1163219/full#supplementary-material
References
Ahmad, B., Zhang, S., Yao, J., Rahman, M. U., Hanif, M., Zhu, Y., et al. (2019). Genomic organization of the B3-domain transcription factor family in grapevine (Vitis vinifera l.) and expression during deed development in deedless and deeded cultivars. Int. J. Mol. Sci. 20, 4553. doi: 10.3390/ijms20184553
Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309, 741–745. doi: 10.1126/science.1113373
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bartrina, I., Otto, E., Strnad, M., Werner, T., Schmülling, T. (2011). Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23, 69–80. doi: 10.1105/tpc.110.079079
Birney, E., Clamp, M., Durbin, R. (2004). Gene wise and genomewise. Genome Res. 14, 988–995. doi: 10.1101/gr.1865504
Brownlee, B. G., Hall, R. H., Whitty, C. D. (1975). 3-Methyl-2-butenal: an enzymatic degradation product of the cytokinin, N6 -(Δ 2 -isopentenyl) adenine. Can. J. Biochem. 53, 37–41. doi: 10.1139/o75-006
Brugière, N., Jiao, S., Hantke, S., Zinselmeier, C., Roessler, J. A., Niu, X., et al. (2003). Cytokinin oxidase gene expression in maize is localized to the vasculature, and is induced by cytokinins, abscisic acid, and abiotic stress. Plant Physiol. 132, 1228–1240. doi: 10.1104/pp.102.017707
Chang, L., Ramireddy, E., Schmülling, T. (2015). Cytokinin as a positional cue regulating lateral root spacing in Arabidopsis. J. Exp. Bot. 66, 4759–4768. doi: 10.1093/jxb/erv252
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020a). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, L., Zhao, J., Song, J., Jameson, P. E. (2020b). Cytokinin dehydrogenase: a genetic target for yield improvement in wheat. Plant Biotechnol. J. 18, 614–630. doi: 10.1111/pbi.13305
Farber, M., Attia, Z., Weiss, D. (2016). Cytokinin activity increases stomatal density and transpiration rate in tomato. J. Exp. Bot. 67, 6351–6362. doi: 10.1093/jxb/erw398
Finn, R. D., Attwood, T. K., Babbitt, P. C., Bateman, A., Bork, P., Bridge, A. J., et al. (2017). InterPro in 2017–beyond protein family and domain annotations. Nucleic Acids Res. 45, D190–D199. doi: 10.1093/nar/gkw1107
Frederick, J. R., Camp, C. R., Bauer, P. J. (2001). Drought-stress effects on branch and mainstem seed yield and yield components of determinate soybean. Crop Sci. 41, 759–763. doi: 10.2135/cropsci2001.413759x
Gao, S., Fang, J., Xu, F., Wang, W., Sun, X., Chu, J., et al. (2014). CYTOKININ OXIDASE/DEHYDROGENASE4 integrates cytokinin and auxin signaling to control rice crown root formation. Plant Physiol. 165, 1035–1046. doi: 10.1104/pp.114.238584
Gavili, E., Moosavi, A. A., Kamgar Haghighi, A. A. (2019). Does biochar mitigate the adverse effects of drought on the agronomic traits and yield components of soybean? Ind. Crops Products 128, 445–454. doi: 10.1016/j.indcrop.2018.11.047
Gu, R., Fu, J., Guo, S., Duan, F., Wang, Z., Mi, G., et al. (2010). Comparative expression and phylogenetic analysis of maize cytokinin dehydrogenase/oxidase (CKX) gene family. J. Plant Growth Regul. 29, 428–440. doi: 10.1007/s00344-010-9155-y
Guo, A.-Y. (2007). GSDS: a gene structure display server. HEREDITAS 29, 1023. doi: 10.1360/yc-007-1023
Hai, N. N., Chuong, N. N., Tu, N. H. C., Kisiala, A., Hoang, X. L. T., Thao, N. P. (2020). Role and regulation of cytokinins in plant response to drought stress. Plants 9, 422. doi: 10.3390/plants9040422
Han, Z., Liu, Y., Deng, X., Liu, D., Liu, Y., Hu, Y., et al. (2019). Genome-wide identification and expression analysis of expansin gene family in common wheat (Triticum aestivum l.). BMC Genomics 20, 101. doi: 10.1186/s12864-019-5455-1
Hwang, I., Sheen, J., Müller, B. (2012). Cytokinin signaling networks. Annu. Rev. Plant Biol. 63, 353–380. doi: 10.1146/annurev-arplant-042811-105503
Jameson, P. E., Song, J. (2016). Cytokinin: a key driver of seed yield. J. Exp. Bot. 67, 593–606. doi: 10.1093/jxb/erv461
Jeffares, D. C., Penkett, C. J., Bähler, J. (2008). Rapidly regulated genes are intron poor. Trends Genet. 24, 375–378. doi: 10.1016/j.tig.2008.05.006
Kaltenegger, E., Leng, S., Heyl, A. (2018). The effects of repeated whole genome duplication events on the evolution of cytokinin signaling pathway. BMC Evolutionary Biol. 18, 76. doi: 10.1186/s12862-018-1153-x
Krzywinski, M., Schein, J., Birol, İ., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Le, D. T., Nishiyama, R., Watanabe, Y., Vankova, R., Tanaka, M., Seki, M., et al. (2012). Identification and expression analysis of cytokinin metabolic genes in soybean under normal and drought conditions in relation to cytokinin levels. PLoS One 7, e42411. doi: 10.1371/journal.pone.0042411
Letunic, I., Doerks, T., Bork, P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. doi: 10.1093/nar/gku949
Li, S., An, Y., Hailati, S., Zhang, J., Cao, Y., Liu, Y., et al. (2019a). Overexpression of the cytokinin oxidase/dehydrogenase (CKX) from Medicago sativa enhanced salt stress tolerance of arabidopsis. J. Plant Biol. 62, 374–386. doi: 10.1007/s12374-019-0141-z
Li, Y., Chen, D., Luo, S., Zhu, Y., Jia, X., Duan, Y., et al. (2019b). Intron-mediated regulation of β-tubulin genes expression affects the sensitivity to carbendazim in Fusarium graminearum. Curr. Genet. 65, 1057–1069. doi: 10.1007/s00294-019-00960-4
Libault, M., Farmer, A., Brechenmacher, L., May, G. D., Stacey, G. (2010a). Soybean root hairs: A valuable system to investigate plant biology at the cellular level. Plant Signaling Behav. 5, 419–421. doi: 10.4161/psb.5.4.11283
Libault, M., Farmer, A., Joshi, T., Takahashi, K., Langley, R. J., Franklin, L. D., et al. (2010b). An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 63, 86–99. doi: 10.1111/j.1365-313X.2010.04222.x
Liu, Z., Lv, Y., Zhang, M., Liu, Y., Kong, L., Zou, M., et al. (2013). Identification, expression, and comparative genomic analysis of the IPT and CKX gene families in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genomics 14, 594. doi: 10.1186/1471-2164-14-594
Liu, P., Zhang, C., Ma, J.-Q., Zhang, L.-Y., Yang, B., Tang, X.-Y., et al. (2018). Genome-wide identification and expression profiling of cytokinin oxidase/dehydrogenase (CKX) genes reveal likely roles in pod development and stress responses in oilseed rape (Brassica napus l.). Genes 9, 168. doi: 10.3390/genes9030168
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lyons, E., Freeling, M. (2008). How to usefully compare homologous plant genes and chromosomes as DNA sequences: How to usefully compare plant genomes. Plant J. 53, 661–673. doi: 10.1111/j.1365-313X.2007.03326.x
Macková, H., Hronková, M., Dobrá, J., Turečková, V., Novák, O., Lubovská, Z., et al. (2013). Enhanced drought and heat stress tolerance of tobacco plants with ectopically enhanced cytokinin oxidase/dehydrogenase gene expression. J. Exp. Bot. 64, 2805–2815. doi: 10.1093/jxb/ert131
Malenčić, D., Cvejić, J., Miladinović, J. (2012). Polyphenol content and antioxidant properties of colored soybean seeds from central Europe. J. medicinal Food 15 (1), 89–95. doi: 10.1093/jxb/ert131
Mameaux, S., Cockram, J., Thiel, T., Steuernagel, B., Stein, N., Taudien, S., et al. (2012). Molecular, phylogenetic and comparative genomic analysis of the cytokinin oxidase/dehydrogenase gene family in the poaceae. Plant Biotechnol. J. 10, 67–82. doi: 10.1111/j.1467-7652.2011.00645.x
McGaw, B. A., Horgan, R. (1983). Cytokinin catabolism and cytokinin oxidase. Phytochemistry 22, 1103–1105. doi: 10.1016/0031-9422(83)80200-3
Merewitz, E. B., Gianfagna, T., Huang, B. (2010). Effects of SAG12-ipt and HSP18.2-ipt expression on cytokinin production, root growth, and leaf senescence in creeping bentgrass exposed to drought stress. J. Am. Soc. Hortic. Sci. 135, 230–239. doi: 10.21273/JASHS.135.3.230
Nishiyama, R., Watanabe, Y., Fujita, Y., Le, D. T., Kojima, M., Werner, T., et al. (2011). Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23, 2169–2183. doi: 10.1105/tpc.111.087395
Nonokawa, K., Nakajima, T., Nakamura, T., Kokubun, M. (2012). Effect of synthetic cytokinin application on pod setting of individual florets within raceme in soybean. Plant Production Sci. 15, 79–81. doi: 10.1626/pps.15.79
Prerostova, S., Dobrev, P. I., Kramna, B., Gaudinova, A., Knirsch, V., Spichal, L., et al. (2020). Heat acclimation and inhibition of cytokinin degradation positively affect heat stress tolerance of arabidopsis. Front. Plant Sci. 11, 87. doi: 10.3389/fpls.2020.00087
Reusche, M., Klásková, J., Thole, K., Truskina, J., Novák, O., Janz, D., et al. (2013). Stabilization of cytokinin levels enhances Arabidopsis resistance against Verticillium longisporum. Mol. Plant-Microbe Interactions® 26, 850–860. doi: 10.1094/MPMI-12-12-0287-R
Rong, C., Liu, Y., Chang, Z., Liu, Z., Ding, Y., Ding, C. (2022). Cytokinin oxidase/dehydrogenase family genes exhibit functional divergence and overlap in rice growth and development, especially in control of tillering. J. Exp. Bot. 73, 3552–3568. doi: 10.1093/jxb/erac088
Sakakibara, H. (2006). CYTOKININS: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57, 431–449. doi: 10.1146/annurev.arplant.57.032905.105231
Schmülling, T., Werner, T., Riefler, M., Krupková, E., Bartrina y Manns, I. (2003). Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, arabidopsis and other species. J. Plant Res. 116, 241–252. doi: 10.1007/s10265-003-0096-4
Shaul, O. (2017). How introns enhance gene expression. Int. J. Biochem. Cell Biol. 91, 145–155. doi: 10.1016/j.biocel.2017.06.016
Shoaib, M., Yang, W., Shan, Q., Sajjad, M., Zhang, A. (2019). Genome-wide identification and expression analysis of new cytokinin metabolic genes in bread wheat (Triticum aestivum l.). PeerJ 7, e6300. doi: 10.7717/peerj.6300
Silvente, S., Sobolev, A. P., Lara, M. (2012). Metabolite adjustments in drought tolerant and sensitive soybean genotypes in response to water stress. PLoS One 7, e38554. doi: 10.1371/journal.pone.0038554
Song, J., Jiang, L., Jameson, P. E. (2012). Co-Ordinate regulation of cytokinin gene family members during flag leaf and reproductive development in wheat. BMC Plant Biol. 12, 78. doi: 10.1186/1471-2229-12-78
Suttle, J. C., Huckle, L. L., Lu, S., Knauber, D. C. (2014). Potato tuber cytokinin oxidase/dehydrogenase genes: Biochemical properties, activity, and expression during tuber dormancy progression. J. Plant Physiol. 171, 448–457. doi: 10.1016/j.jplph.2013.11.007
Tan, M., Li, G., Qi, S., Liu, X., Chen, X., Ma, J., et al. (2018). Identification and expression analysis of the IPT and CKX gene families during axillary bud outgrowth in apple (Malus domestica borkh.). Gene 651, 106–117. doi: 10.1016/j.gene.2018.01.101
Thien Nguyen, Q., Kisiala, A., Andreas, P., Neil Emery, R. J., Narine, S. (2016). Soybean seed development: fatty acid and phytohormone metabolism and their interactions. Curr. Genomics 17, 241–260. doi: 10.2174/1389202917666160202220238
USDA (2020). United states department of agriculture (USDA) [WWW document]. Foreing Agric. Service World Agric. Production.
Vyroubalová, Š., Václavíková, K., Turečková, V., Novák, O., Šmehilová, M., Hluska, T., et al. (2009). Characterization of new maize genes putatively involved in cytokinin metabolism and their expression during osmotic stress in relation to cytokinin levels. Plant Physiol. 151, 433–447. doi: 10.1104/pp.109.142489
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49–e49. doi: 10.1093/nar/gkr1293
Wang, C., Wang, H., Zhu, H., Ji, W., Hou, Y., Meng, Y., et al. (2021). Genome-wide identification and characterization of cytokinin oxidase/dehydrogenase family genes in Medicago truncatula. J. Plant Physiol. 256, 153308. doi: 10.1016/j.jplph.2020.153308
Werner, T., Köllmer, I., Bartrina, I., Holst, K., Schmülling, T. (2006). New insights into the biology of cytokinin degradation. Plant Biol. 8, 371–381. doi: 10.1055/s-2006-923928
Werner, T., Motyka, V., Laucou, V., Smets, R., Van Onckelen, H., Schmülling, T. (2003). Cytokinin-deficient transgenic arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15, 2532–2550. doi: 10.1105/tpc.014928
Werner, T., Nehnevajova, E., Köllmer, I., Novák, O., Strnad, M., Krämer, U., et al. (2011). Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 22, 3905–3920. doi: 10.1105/tpc.109.072694
Woyann, L. G., Meira, D., Zdziarski, A. D., Matei, G., Milioli, A. S., Rosa, A. C., et al. (2019). Multiple-trait selection of soybean for biodiesel production in Brazil. Ind. Crops Products 140, 111721. doi: 10.1016/j.indcrop.2019.111721
Wybouw, B., De Rybel, B. (2019). Cytokinin - a developing story. Trends Plant Sci. 24, 177–185. doi: 10.1016/j.tplants.2018.10.012
Xu, G., Guo, C., Shan, H., Kong, H. (2012). Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. 109, 1187–1192. doi: 10.1073/pnas.1109047109
Yeh, S.-Y., Chen, H.-W., Ng, C.-Y., Lin, C.-Y., Tseng, T.-H., Li, W.-H., et al. (2015). Down-regulation of cytokinin oxidase 2 expression increases tiller number and improves rice yield. Rice 8, 36. doi: 10.1186/s12284-015-0070-5
Yu, Y., Li, Y., Yan, Z., Duan, X. (2022). The role of cytokinins in plant under salt stress. J. Plant Growth Regul. 41, 2279–2291. doi: 10.1007/s00344-021-10441-z
Yu, K., Yu, Y., Bian, L., Ni, P., Ji, X., Guo, D., et al. (2021). Genome-wide identification of cytokinin oxidases/dehydrogenase (CKXs) in grape and expression during berry set. Scientia Hortic. 280, 109917. doi: 10.1016/j.scienta.2021.109917
Zalabák, D., Galuszka, P., Mrízová, K., Podlešáková, K., Gu, R., Frébortová, J. (2014). Biochemical characterization of the maize cytokinin dehydrogenase family and cytokinin profiling in developing maize plantlets in relation to the expression of cytokinin dehydrogenase genes. Plant Physiol. Biochem. 74, 283–293. doi: 10.1016/j.plaphy.2013.11.020
Zhan, Y., Li, H., Sui, M., Zhao, X., Jing, Y., Luo, J., et al. (2020). Genome wide association mapping for tocopherol concentration in soybean seeds across multiple environments. Ind. Crops Products 154, 112674. doi: 10.1016/j.indcrop.2020.112674
Zhang, W., Peng, K., Cui, F., Wang, D., Zhao, J., Zhang, Y., et al. (2021). Cytokinin oxidase/dehydrogenase OsCKX11 coordinates source and sink relationship in rice by simultaneous regulation of leaf senescence and grain number. Plant Biotechnol. J. 19, 335–350. doi: 10.1111/pbi.13467
Zhang, S., Shi, Y., Zhang, S., Shang, W., Gao, X., Wang, H. (2014). Whole soybean as probiotic lactic acid bacteria carrier food in solid-state fermentation. Food Control 41, 1–6. doi: 10.1016/j.foodcont.2013.12.026
Zhu, M., Wang, Y., Lu, S., Yang, L., Zhuang, M., Zhang, Y., et al. (2022). Genome-wide identification and analysis of cytokinin dehydrogenase/oxidase (CKX) family genes in Brassica oleracea l. reveals their involvement in response to Plasmodiophora brassicae infections. Hortic. Plant J. 8, 68–80. doi: 10.1016/j.hpj.2021.05.003
Keywords: abiotic stress, CKX gene family, soybean, expression analysis, growth and development
Citation: Du Y, Zhang Z, Gu Y, Li W, Wang W, Yuan X, Zhang Y, Yuan M, Du J and Zhao Q (2023) Genome-wide identification of the soybean cytokinin oxidase/dehydrogenase gene family and its diverse roles in response to multiple abiotic stress. Front. Plant Sci. 14:1163219. doi: 10.3389/fpls.2023.1163219
Received: 10 February 2023; Accepted: 30 March 2023;
Published: 17 April 2023.
Edited by:
Zhengjun Xia, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Ya-nan Jin, Inner Mongolia Minzu University, ChinaYiqiong Huo, Shanxi Agricultural University, China
Copyright © 2023 Du, Zhang, Gu, Li, Wang, Yuan, Zhang, Yuan, Du and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jidao Du, ZGpkYnluZEAxNjMuY29t; Qiang Zhao, enFpYW5nMDQxNkBob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Yanli Du
Yanli Du Zhaoning Zhang1†
Zhaoning Zhang1† Yuxian Zhang
Yuxian Zhang Ming Yuan
Ming Yuan Jidao Du
Jidao Du