
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 26 April 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1160102
This article is part of the Research Topic Identification and Functional Analysis of Differentially Expressed Genes in Plant Response to Abiotic Stresses View all 24 articles
Heat shock transcription factor (HSF) is an important TF that performs a dominant role in plant growth, development, and stress response network. In this study, we identified a total of 30 HSF members from poplar, which are unevenly distributed on 17 chromosomes. The poplar HSF family can be divided into three subfamilies, and the members of the same subfamily share relatively conserved domains and motifs. HSF family members are acidic and hydrophilic proteins that are located in the nucleus and mainly carry out gene expansion through segmental replication. In addition, they have rich collinearity across plant species. Based on RNA-Seq analysis, we explored the expression pattern of PtHSFs under salt stress. Subsequently, we cloned the significantly upregulated PtHSF21 gene and transformed it into Populus simonii × P. nigra. Under salt stress, the transgenic poplar overexpressing PtHSF21 had a better growth state and higher reactive oxygen scavenging ability. A yeast one-hybrid experiment indicated PtHSF21 could improve salt tolerance by specifically binding to the anti-stress cis-acting element HSE. This study comprehensively profiled the fundamental information of poplar HSF family members and their responses to salt stress and specifically verified the biological function of PtHSF21, which provides clues for understanding the molecular mechanism of poplar HSF members in response to salt stress.
Plants are frequently affected by adverse environments in growth and development, such as high temperature, salt injury, drought, cold, and other abiotic threats, insect pests, viruses, and other biotic stresses (Huang et al., 2021). Nowadays, saline-alkali hazards have become universal problems worldwide (Polle and Chen, 2015). Soil salinization can change soil properties, reduce soil water potential, and lead to soil consolidation, which restricts plant growth and development (Evelin et al., 2019). As sessile organisms, plants cannot actively escape adverse environments due to their own fixed nature, so they rely on physiological and biochemical mechanisms to survive under stress (Shao et al., 2007; Abdullah et al., 2018). Therefore, plants have evolved a series of complex and effective strategies to maintain normal physiological metabolism and stress-resistant growth (Zhou et al., 2009). Transcription factors (TF) are sequence-specific DNA-binding protein molecules that can regulate downstream gene expression by binding to specific cis-acting elements in promoter regions (Shao et al., 2015). Increasing reports have indicated that many kinds of TF carry out a significant role in plant stress resistance. For example, heat shock factor (HSF) can activate transcriptional responses to salinity and oxidative defense in Populus euphratica (Shen et al., 2013). MYB-related TF is widely involved in the phosphate starvation response and the tolerance to extreme cold, drought, and salt stress (Sun et al., 2018). The PsnHDZ63 gene can improve salt tolerance of poplar (Guo et al., 2021a). ABRE binding factor and MYC are directly involved in the signaling pathways of abscisic acid (ABA) and jasmonic acid (JA) (Yoon et al., 2020). WRKY proteins recognize and activate a variety of plant defense genes (Yu et al., 2001).
In particular, HSF is a type of TF that widely exists in eukaryotes and carries out an important role in signal reception and transmission, downstream gene regulation, and stress resistance in plants (Ohama et al., 2017). Highly conserved HSF protein is composed of five basic functional domains: DNA binding domain (DBD), adjacent dimer oligomeric domain (OD), nuclear localization signal (NLS), nuclear export signal (NES), and C-terminal activating peptide protein (CTD) (Li et al., 2014; Guo et al., 2016). DBD is the most conserved domain in HSF, which forms a compact sphere consisting of three α-helical bundles and four inversely parallel β-folded layers. DBD ensures that HSF members specifically bind to cis-acting elements in the target gene promoter, therefore regulating target gene transcription (Sangster and Queitsch, 2005). OD consists of two hydrophobic heptapeptide repeat regions, HR-A and HR-B. According to the differences between HR-A and HR-B, HSF is divided into three subfamilies (HSFA, HSFB, and HSFC) (Nover et al., 2001; Baniwal et al., 2004). NLS is composed of basic amino acids, and NES is rich in leucine (Lyck et al., 1997; Heerklotz et al., 2001). CTD is the least conserved region, which consists of AHA motifs and exhibits the characteristics of transcriptional activation. The first plant HSF gene was cloned from a tomato in 1990. So far, more and more HSFs have been found in many species. For instance, 21 HSF members were identified in Arabidopsis thaliana, 25 in rice, 27 in potato, 82 in wheat, 25 in corn, and 19 in grape (Wang et al., 2009; Lin et al., 2011; Guo et al., 2016; Tang et al., 2016; Yang et al., 2016; Duan et al., 2019). Previous studies on individual HSF subfamilies have shown that HSF members are widely involved in plant development and stress responses. Overexpression of HsfA1b could significantly increase plant yield, harvest index, and plant resistance to pathogens (Bechtold et al., 2013). AtHSFA7b regulates the expression of the target gene by combining with E-box-like elements to mediate a series of physiological changes, including maintaining cell ion homeostasis, lowering water loss rate, reducing reactive oxygen species accumulation, and regulating osmotic pressure, to improve plant salt tolerance (Zang et al., 2019). Overexpression of the AtHsfA1b and AtHsfA1d genes can enhance drought and heat tolerance in tomatoes (Higashi et al., 2013). In addition, heterologous overexpression of BcHsfA1 or HmHsp70 genes in tobacco can enhance heat tolerance (Zhu et al., 2018; Xu et al., 2020). Overexpression of the HsfA1 gene from Lilium brownii can upregulate the heat shock and betaine synthase genes in A. thaliana (Gong et al., 2014). HsfA1d transgenic pea decreased the content of H2O2 under heat stress and significantly increased the activity of antioxidant enzymes (Shah et al., 2020).
As perennial woody plants, poplar has experienced various environmental pressures and seasonal changes. The genome of Populus trichocarpa was first published in 2006, which makes it an ideal genetic model for forest genetics and breeding (Tuskan et al., 2006). Populus simonii × Populus nigra is a hybrid of P. simonii and P. nigra, which has the advantages of rapid growth, excellent quality, easy survival of cutting, strong adaptability and high development potential. However, salinized soil seriously restricts the growth range of P. simonii × P. nigra (Guo et al., 2021a). How to improve the salt tolerance of woody plants has become an important research topic. In this study, we identified 30 HSF members from poplar and analyzed their phylogeny, gene structure, conserved domain, promoter cis-acting elements, chromosome location, collinearity, and evolution pattern. In addition, we profiled their expression pattern under salt stress by RNA-Seq and screened out a highly salt-induced PsnHSF21 gene. We confirmed it was targeted to the nucleus and had self-activating activity, and it can specifically bind to the HSE element. Moreover, we obtained transgenic P. simonii × P. nigra over-expressing PsnHSF21 and verified the transgenic poplar displayed growth and physiological advantages under high salt stress. These results are helpful to understand the mechanism of salt resistance and the genetic improvement of HSF members in poplar.
The genome information of poplar HSF family genes and their conserved domains was obtained from Phytozome (https://phytozome-next.jgi.doe.gov/). The hidden Markov model (HMM) (Evalue <1 × 10−5) of HSF (PF00447) was downloaded from the Pfam database (https://pfam.xfam.org/) (Finn et al., 2011). Poplar HSF sequences were obtained by BLASTP-HMMER, and sequence accuracy was further verified by Pfam and NCBI-CDD (https://www.ncbi.nlm.nih.gov/cdd) (Finn et al., 2016). Multiple sequence alignment was carried out through CLUSTALX 2.0 (Zhao et al., 2020) and visualized with DNAMAN and Web Logo (Guo et al., 2021b). Subcellular localization was predicted by PSORT (http://psort1.hgc.jp/form.html) with default parameters. Physical and chemical parameters were predicted by ExPASy (https://web.expasy.org/protparam/), including molecular weight (MW), isoelectric point (PI), instability index (II), and the average hydrophilic coefficient of proteins (Huang and Xu, 2008).
The sequences of 24 HSF members of A. thaliana were downloaded from the TAIR database (https://www.arabidopsis.org/). Multiple-sequence alignment was conducted by CLUSTALX 2.0. A phylogenetic tree was constructed by the NJ method in MEGA7.0, and the reliability of branches was evaluated with 1,000 Bootstrap repeated samplings (Kumar et al., 2016). Motif identification was predicted by MEME (https://meme.suite.org/meme/info/status.Service=MEME&id=app.MEME5.5.016727959406951083280962) with the maximum motif number of 10 (Bailey et al., 2009). The conserved domain was analyzed by the NCBI-CD-Search program (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). The information about UTR, intron, and CDS was obtained from the GFF annotation of the poplar genome and visualized by TBtools (Chen et al., 2020).
Genome information for A. thaliana and Oryza sativa was obtained from the Ensembl Plants database (http://plants.ensembl.org/index.html). Tandem repeatability of the poplar HSF gene and gene collinear relationships across plant species were identified by BLASTP and the multiple collinear scanning tool (MCScanX) (Wang et al., 2012). The chromosome location of the poplar HSF gene was shown by TBtools (Tuskan et al., 2006). Ka/Ks replacement rate of homologous HSF genes and evolution time T (T = Ks/2λ, λ = 9.1 × 10−9) were also calculated by TBtools (Chen et al., 2020).
The PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to predict the cis-acting elements in the upstream 2,000 bp promoter sequences of poplar HSF genes. Furthermore, the results were visualized by TBtools (Chen et al., 2020).
P. simonii × P. nigra seedlings from the same poplar clone at Northeast Forestry University were cultured in water under the light/dark cycle of 16/8 h and 26 °C/22 °C, until new roots and new leaves sprouted (Yao et al., 2019). A total of 60 hydroponic seedlings with similar growth states were selected for salt treatment: 30 for 150 mM NaCl solution and 30 for hydroponic control. After 24 h, the roots, stems, and leaves were collected and preserved in liquid nitrogen, then transported to GENEWIZ Company (www.GENEWIZ.com) for RNA-Seq on the IIIumina HiSeq 2500 platform. The analysis of differentially expressed genes (DEGs) was carried out by the DESeq method in R software, and the standards of significant DEGs include |log2 fold change (FC)| ≥1 and a p-value ≤0.05. A Venn diagram was drawn by Venny2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny/index.html) (Anders and Huber, 2010). The relative expression levels of significant differentially expressed PtHSFs screened by RNA-Seq were verified by RT-qPCR. The internal reference was Actin, and the primers of PtHSFs for RT-qPCR are listed in Supplementary Table S1. The relative expression level in transgenic poplar was calculated by the 2−ΔΔ Ct method as previously described (Guo et al., 2021b). Furthermore, the LSD test (P <0.05) of SPSS software was performed to examine the significance of gene expression levels (Zhao et al., 2021).
Total RNA was extracted using the TaKaRa MiniBEST Plant RNA Extraction Kit (Takara, Dalian), and cDNA was synthesized using the PrimeScript™ reverse RT reagent kit (Takara, Dalian). The cDNA fragment of the PtHSF21 gene was cloned by PCR with specific primers (Supplementary Table S2) (Goodstein et al., 2012), and it was sequenced by Sangon (http://www.sangon.com/). The sequence was compared by Blastn (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The conserved domain of PtHSF21 was analyzed by the NCBI-CD-Search program (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), and the protein structure of PtHSF21 was predicted by Swiss Model (https://swissmodel.expasy.org/interactive/NNZZXxy/model/) (Zhang et al., 2022). The homologous proteins of PtHSF21 were screened by NCBI-blast, and their phylogenetic tree was represented by the NJ method in MEGA7.0, with the reliability of branches being evaluated with 1,000 Bootstrap repeated samplings (Kumar et al., 2016). The potential proteins interacting with PtHSF21 were predicted by String (https://cn.string-db.org/).
The coding region sequence of PsnHSF21 without a stopping codon was introduced into the pBI121-GFP vector by SalI and SpeI restriction sites. The PCR primers can be found in Supplementary Table S3. The fusion vectors pBI121-PsnHSF21-GFP and pBI121-GFP as controls were transferred into Agrobacterium tumefaciens GV3101, respectively. Moreover, the Agrobacterium solutions were injected into the lower epidermis of the strong leaves, which belong to one-month-old tobacco soil seedlings after transplanting. The injected leaves were cultured in the dark for 24 h and then observed for a green fluorescence signal under a Zeiss laser confocal microscope (ZEISS LSM 800).
The tissue culture poplar seedlings were used for gene transformation by the leaf disk transformation method. The leaves were immersed in solutions of GV3101 containing pBI121-PsnHSF21-GFP for 10 min and cultured in the dark for 2–3 days. Then the leaves were transferred to a differentiation medium composed of WPM, 0.1 mg/L NAA, 0.2 mg/L 6-BA, 0.5 mg/L Gas, and 50 mg/L Kan to induce adventitious buds. The 2-cm buds can be transferred to 1/2 MS rooting medium with 50 mg/L Kan. The Kan-resistant seedlings were confirmed by molecular detection with WT DNA as a negative control and plasmid as a positive control. In addition, the relative expression levels of PsnHSF21 in transgenic poplar were calculated by RT-qPCR with three biological repeats per sample.
Two-month-old transgenic poplars (OE1, OE2, OE3, and OE4) and WT plants as controls were irrigated with 200 mM NaCl solution for 7 days. The growth indexes, such as fresh weight, plant height, main root length, and lateral root number, were measured. Furthermore, leaves in three to four layers were collected for the determination of physiological indexes, including POD, proline, and MDA, using the Nanjing Jiancheng Bioengineering Research Institute kit. The relative expression levels of stress resistance genes such as SOD, POD, HRG, ABA, and GA were analyzed by RT-qPCR. The relevant primers can be found in Supplementary Table S4. Histochemical staining of DAB and Evans blue was used to predict the activities of hydrogen peroxide and superoxide in plants. Tissue culture seedlings at one month old were soaked in 200 mM NaCl for 24 h, and the leaves were immersed in DAB and Evans blue solution in the dark for 24 h and then decolorized with ethanol (Sekulska-Nalewajko et al., 2016).
The CDS sequence of the PsnHSF21 gene was fused into pGBKT7 to form pGBKT7-PsnHSF21 at two restriction sites, NdeI and SmaI. The related primers can be found in Supplementary Table S5. The fusion vectors pGBKT7-PsnHSF21, pGBKT7 (negative control), and pGBKT7-53/pGADT7-T (positive control) were respectively transferred into the Y2H Gold yeast strain according to the Yeast Maker yeast transformation system and then inoculated on SD/-Trp and SD/-Trp/-His/X-α-Gal solid medium to detect the self-activating activity of the PsnHSF21 protein.
The stress-resistant element HSE (GAATTC) was refolded in series three times and then inserted into the pAbAi vector to form the bait vector AB-HSE, which was prepared for AB-HSE-Y1H competent cells. The CDS sequence of the PsnHSF21 gene was inserted into pGADT7 to form the bait vector AD-PsnHSF21 by restriction sites NdeI and BamHI. The primers are listed in Supplementary Table S5. The AD-PsnHSF21 vector was transferred into yeast cells and cultured on SD/-Leu/AbA (200 ng/ml) for 3–5 days. The positive clones were diluted and presented in culture medium.
Allowing the hidden Markov model (HMM) of HSF (PF00447) as a clue, we searched the poplar genome and identified a total of 30 HSF proteins by Pfam and NCBI-CDD multiple verification. The repetitive regions of the 30 PtHSF proteins were extracted for multiple sequence alignment and visualization (Figure 1). The DBD domains of PtHSF proteins include three α-helical bundles and four inversely parallel β-folded layers. PtHSF proteins have 210–596 aa, with a protein molecular weight of 24,028.43–65,369.32 Da. The isoelectric points of the 30 PtHSF proteins are different. The isoelectric points of PtHSF06, PtHSF08, PtHSF11, PtHSF21, PtHSF25, and PtHSF30 are more than 7; these are basic proteins, of which PtHSF21 is the largest with 9.35. The values of the other 24 PtHSFs are all less than 7 and are acidic proteins, of which PtHSF20 is the smallest with 4.7. The instability index of PtHSF proteins varies greatly, in which the value of PtHSF15 and PtHSF16 is low and their stability is poor. PtHSFs are all hydrophilic proteins, of which PtHSF10 has the weakest hydrophilicity (−0.914) and PtHSF06 has the strongest hydrophilicity (−0.468). Based on the prediction of subcellular localization, all poplar HSF family proteins are in the nucleus, except PtHSF16, which is also found in the cytoplasm (Table 1).
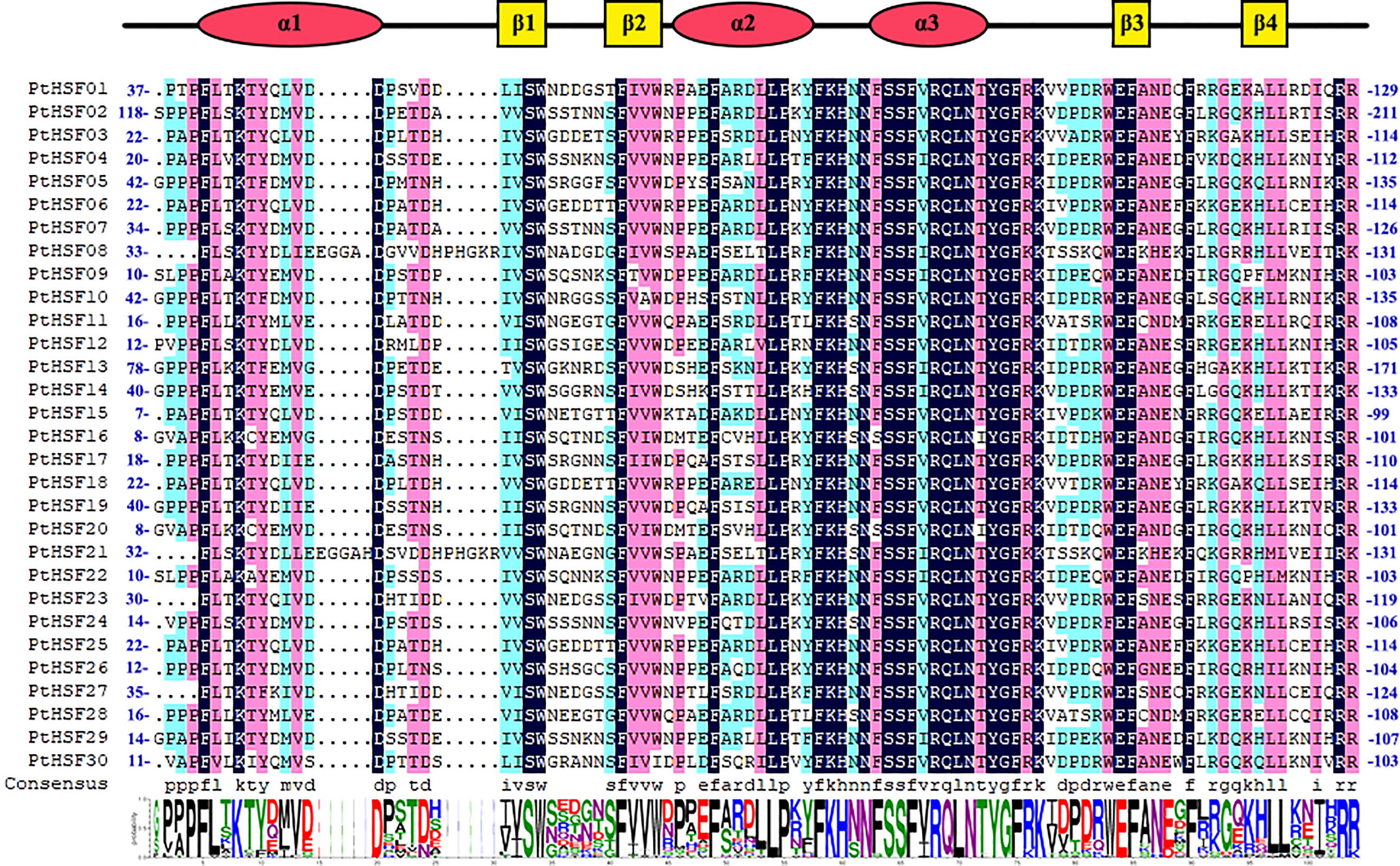
Figure 1 Comparison of the HSF domain of PtHSFs. Secondary structure elements of the DBDs (α1-β1-β2-α2-α3-β3-β4) based on JNet structure predictions are shown above the alignment; α-helices are indicated by red circular legends, and β-folds are indicated by yellow square legends. The total height of each column of letters in the Web logo section indicates the conservation of each position, and the height of each letter represents the relative rating of the corresponding amino acids.
In the study, 30 poplar HSF proteins and 24 HSF proteins from A. thaliana were used for constructing a phylogenetic tree (Figure 2). The similarity between PtHSFs and AtHSFs is 70%–100%. HSF proteins are divided into three subfamilies, including Classes A, B, and C. Among them, Class A is the largest with 17 PtHSF proteins; Class B is the smallest, which merely has PtHSF06 (Potri.T137400.1); and Class C accounts for 40% of poplar HSF members.
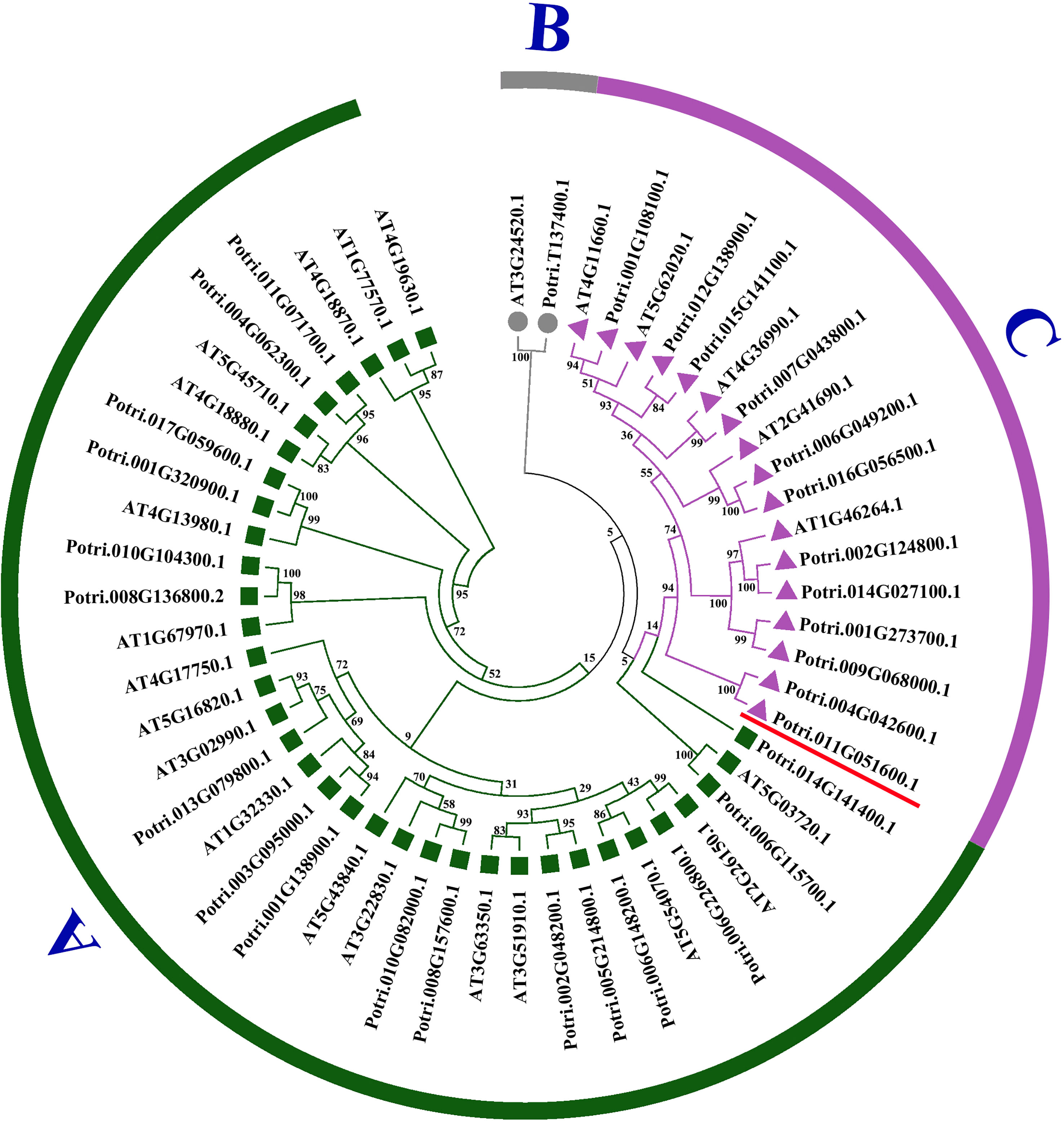
Figure 2 Phylogenetic tree of PtHSFs and AtHSFs. The rootless phylogenetic tree of the HSF protein sequence of poplar and Arabidopsis thaliana was drawn by the NJ neighbor method in MEGA7.0. The phylogenetic tree was divided into three groups: (A–C), with each color remaining in one group. Potri.011G051600.1 (PtHSF21) was marked with a red “–.”.
As shown in the gene structure map, all PtHSF members have a typical HSF conserved domain, while there are differences in the UTR, CDS, domain, and intron structures among PtHSFs (Figure 3). Broadly speaking, the PtHSF members in the same subfamily have similar gene structures except for the HSF domain, Classes A and B members also contain some additional conservative domains. For example, in Class A, PtHSF04 contains the DivlC domain, PtHSF05 contains the Macoilin domain, PtHSF10 contains the hadR domain, PtHSF09, PtHSF22, and PtHSF26 all contain the Mplase_alph_rch domain, and PtHSF02, PtHSF07, and PtHSF12 all contain the SMC_prok_B domain. PtHSF30 in Class C contains the DUF5320 domain. In addition, PtHSFs contain 1–2 introns; 93% of PtHSFs contain one intron, while PtHSF14 and PtHSF30 contain two introns. MEME prediction results indicated PtHSF proteins contain 4–9 conserved motifs, among which the members in Class A contain the most motifs (6–9) and PtHSF01, PtHSF23, and PtHSF27 in Class C have the least, with only four motifs, including motifs 1, 2, 3, and 10. All members include three conserved motifs, including motifs 1–3. Except for the above three motifs, Class A members contain motifs 4 and 5, and Class C members contain motif 10. The information for motifs 1 to 10 is given in Supplementary Figure 1.
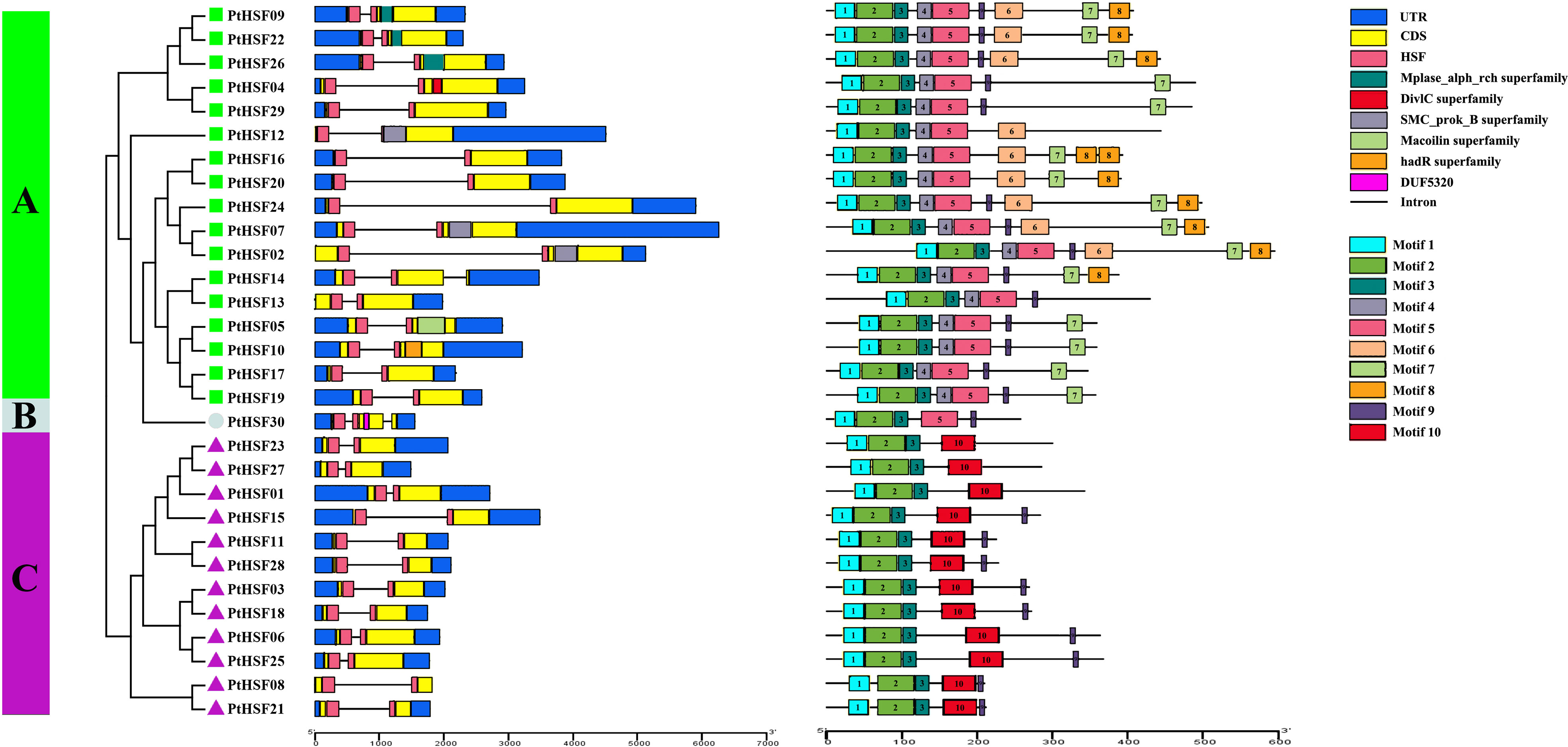
Figure 3 Analysis of HSF gene structure and conserved motif of poplar. The phylogenetic tree was constructed based on 30 HSF protein sequences from poplar, which were divided into three subfamilies. In the gene structure analysis, blue represents the UTR, yellow represents the CDS, pink represents the HSF region, and “–” represents the intron. The conservative motif of the HSF gene is predicted by the MEME website. The number in the color block (1–10) represents motifs 1–10, respectively, and the length of the color block indicates the size of the motif.
The 30 PtHSF members have chromosomes that are unevenly distributed on poplar chromosomes. It is unusual there exists a PtHSF30 distributed on the KZ623489 scaffold. Chromosomes 1 and 6 contain four HSF genes; chromosomes 2, 4, 8, 10, and 11 contain two HSF genes; chromosomes 18 and 19 have no HSF genes; and the other chromosomes contain one HSF gene (Figure 4A). Phylogenetic analysis indicated that the whole genome duplication of HSF family genes may have occurred in poplar, which were then analyzed by TBtools-McSxanX. There were 11 duplicate pairs in PtHSFs (Figure 4A), but no tandem repeats appeared, indicating that fragment repetition remains the main factor of gene expansion in the poplar HSF family. To further study the evolutionary constraints of PtHSFs genes, we analyzed the non-synonymous substitution rate Ka, synonymous substitution rate Ks, and Ka/Ks values of the 11 homologous PtHSFs gene pairs (Table 2). The non-synonymous substitution rate Ka indicates that the amino acid changes were caused by base replacement, while the synonymous substitution rate Ks indicates that the amino acid changes did not occur due to base replacement. The synonymous substitution rate Ks is used to predict the occurrence time of genome-wide repetitive events, which are between 0.2397 and 1.9035. So large-scale gene repetition events may have occurred about 104.58887 MAY (million years ago) in poplar. The most recent event happened at 13.16965 MAY (million years ago). Moreover, the Ka/Ks values of PtHSFs gene pairs are all less than 1, indicating that poplar HSF family genes have experienced strong purification selection.
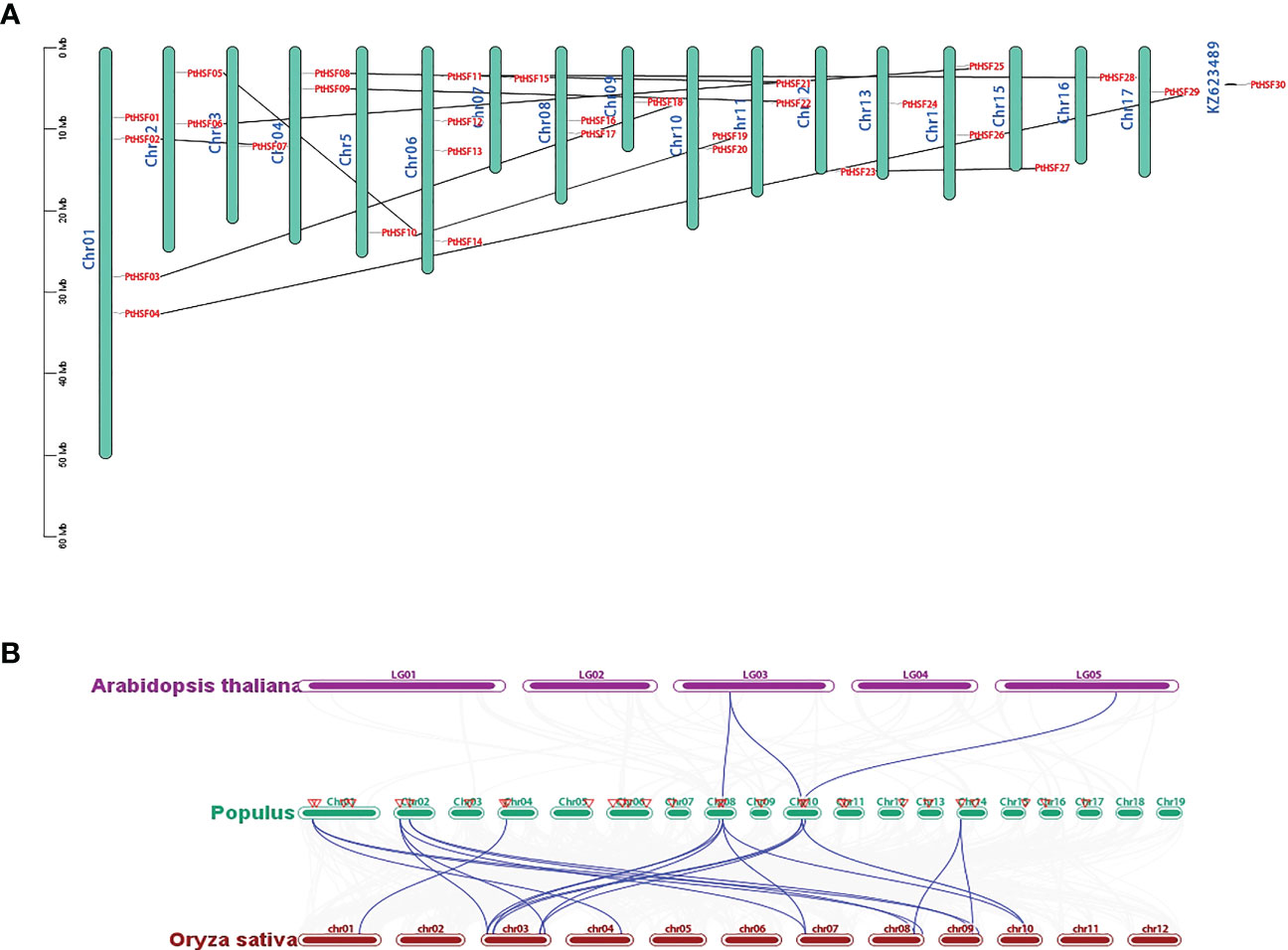
Figure 4 Repetition and collinearity analysis of PtHSFs fragments. (A) Approximately 30 HSF genes were unevenly distributed on poplar chromosomes, and the segmental repetition relationships were represented by black lines. (B) Collinearity analysis of HSF genes in poplar, Arabidopsis, and rice. The blue line represents the HSF lineal homologous gene pairs, and the red triangle indicates PtHSFs..
To further confirm the phylogenetic relationship of HSF family members, the collinear relationships of nine PtHSFs from poplar, two AtHSFs from A. thaliana, and twelve OsHSFs from rice were analyzed. The HSF family members constitute 23 lineal homologous gene pairs, including three PtHSFs-AtHSFs pairs and twenty PtHSFs-OsHSFs pairs (Figure 4B; Supplementary Table S6). Ordinarily, PtHSFs can correspond to one or two HSF genes from other plant species. Interestingly, PtHSF19 corresponds to two AtHSFs (AT3G22830.1 and AT5G43840.1) and three OsHSFs (LOC_Os03g06630.1, LOC_Os03g53340.1, and LOC_Os10g28340.1), indicating that this gene is critically valuable in the evolution of the poplar HSF family.
Cis-acting elements are the core part of gene promoters, which carry out a significant regulatory function in gene expression. We analyzed the cis-acting elements in the upstream 2,000 bp promoter sequences of PtHSFs by PlantCARE. Among these genes, the PtHSF01 promoter contains the most cis-acting elements (37), while the PtHSF23 promoter contains the least cis-acting elements (13) (Figure 5). Additionally, these promoters include many elements that regulate development, such as meristem-specific expression regulatory elements, seed-specific expression regulatory elements, endosperm-specific expression regulatory elements, palisade mesophyll cell differentiation regulatory elements, and so on. Many abiotic stress response elements, such as light response elements, low temperature response elements, salt response elements, drought response elements, and wound response elements, and hormone synthesis-related elements, such as jasmonic acid response elements, gibberellin response elements, and auxin response elements, were also present in PtHSFs promoters.
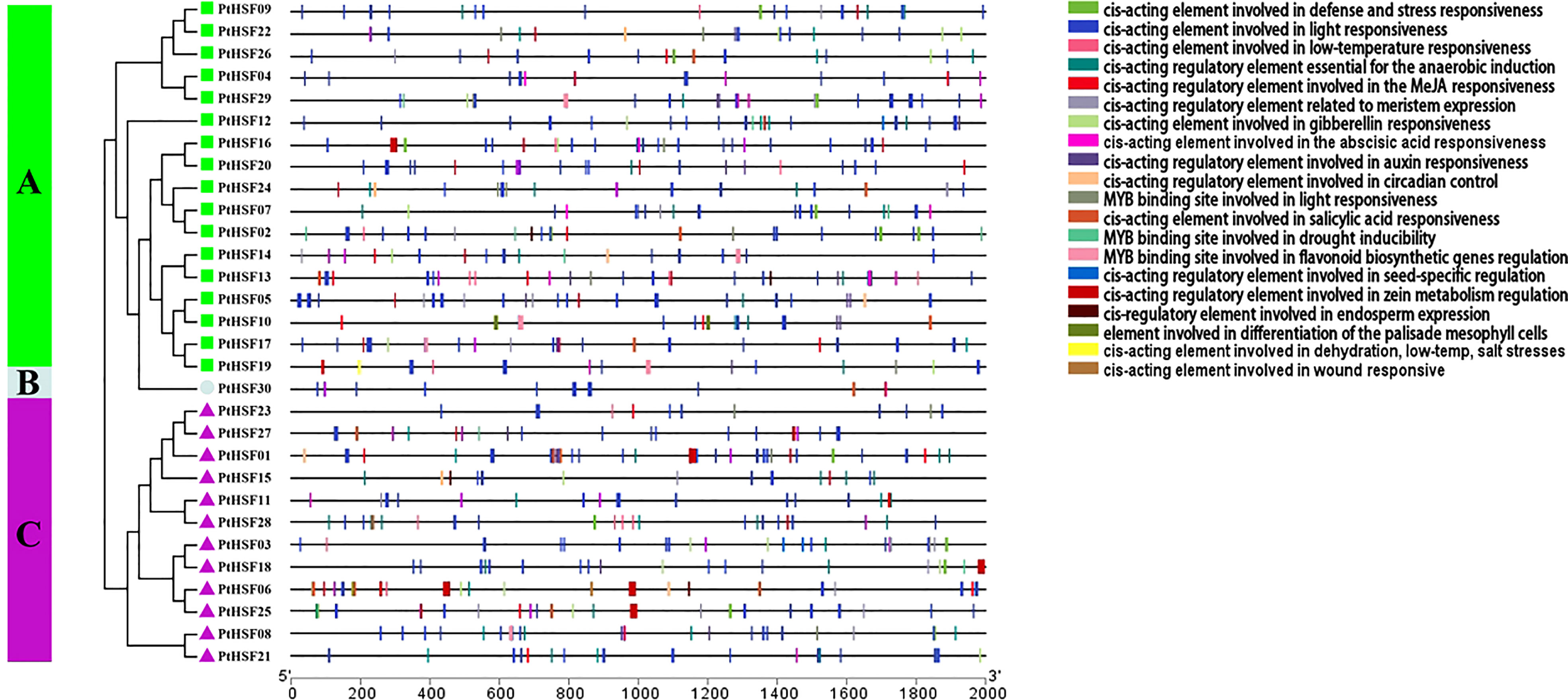
Figure 5 Analysis of Cis-acting elements in the HSF gene promoter in poplar. The upstream 2,000 bp sequences of 30 HSF genes were truncated and analyzed, and the blocks of different colors represented cis-acting elements with diverse functions.
The expression pattern of PtHSFs in the root, stem, and leaf of poplar under salt stress was analyzed by RNA-Seq (Supplementary Sheet). In the root, twenty PtHSFs responded to salt stress, including nine upregulated genes and eleven downregulated genes. There were ten salt-induced PtHSFs in the stem; two were upregulated and eight were downregulated. There were seven PtHSFs in the leaf of poplar in response to salt stress, including five upregulated genes and two downregulated genes. Simultaneously, we drew a Venn diagram of the differentially expressed genes in the three tissues. PtHSF03 and PtHSF05 were differentially expressed in the three tissues. PtHSF28 was upregulated in leaf and root, PtHSF26 was upregulated in stem and root, and PtHSF21 was upregulated in leaf and stem. PtHSF03 was downregulated in three tissues; PtHSF06, PtHSF18, PtHSF25, and PtHSF27 were downregulated, in stem and root; PtHSF05 was downregulated in the stem and leaf (Figure 6A). RT-qPCR of 6 DEGs in different tissues was carried out to check the RNA-Seq results, which proved to be consistent. PtHSF3, PtHSF6, PtHSF18, and PtHSF25 were all downregulated in all three tissues (Figure 6B). It is worth paying attention to that one-third of the genes were upregulated in all three tissues, and the expression of PtHSF28 changed less, while PtHSF21 was significantly upregulated in both stems and leaves, which was selected as the key research object in the study.
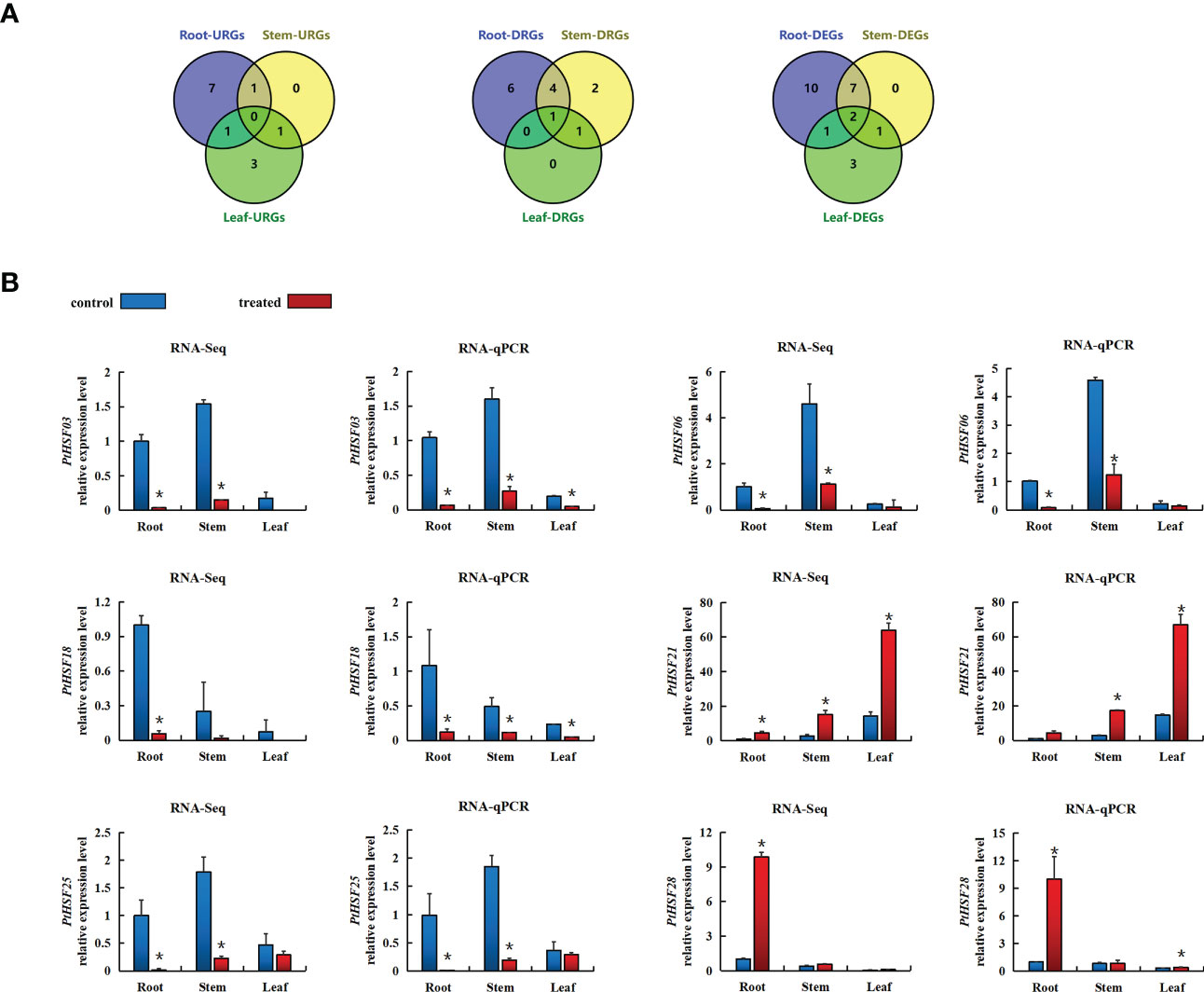
Figure 6 (A) VENN diagram of differential gene expression in the roots, stems, and leaves of poplar after salt stress. The figure indicates the number of DEGs in response to salt stress in different tissues, upregulating the number of DEGs and downregulating the number of DEGs. DEG means differentially expressed gene, URG means represent upregulating differentially expressed gene, and DRG means represent downregulating differentially expressed gene. (B) After salt stress, the expression pattern of the HSF gene in different poplar tissues was based on RNA-Seq and RT-qPCR analysis. Blue indicates the control group, and red indicates the salt treatment. Based on the expression level of corresponding genes in the roots of the control treatment, the relative expression level of each gene under salt stress was calculated. The error line represents the standard deviation (SD) of biological replication. “*” indicates there is a significant difference (P <0.05).
We cloned the PsnHSF21 CDS with a length of 636 bp from P. simonii × P. nigra, encoding 212 amino acids with a highly conserved HSF domain (Figure 7A), and its protein sequence similarity to PtHSF21 was 96.21%. The protein has three α-helical bundles and four inverted parallel β-folded layers, which is consistent with the protein characteristics of the HSF family (Figure 7B). According to the results of NCBI-blast (Figure 7C), there were nine proteins highly homologous to PsnHSF21, including HSFs from P. trichocarpa, Hevea brasiliensis, Prunus avium, Manihot esculenta, Juglans microcarpa × Juglans regia, Abrus precatorius, Quercus lobata, Pistacia vera, and Carya illinoinensis. Among them, HSF (XP_006377347) from P. trichocarpa was most closely related to PsnHSF21, while it was farthest for the HSFs from P. avium (XP_021825794.1) and P. vera (XP_031258981.1). The prediction results of the protein–protein interaction network (Figure 7D) show there were ten proteins interacting with PsnHSF21, namely Potri.001G212200.1, Potri.001G285100.1, Potri.001G286700.1, Potri.005G241100.1, Potri.006G150300.1, Potri.006G230500.1, Potri.010G025000.1, Potri.015G078000.1, Potri.014G131700.1, and Potri.018G084900.1. The average node degree of the network is 4, the local clustering coefficient is 0.791, and the enrichment P value is 0.719.
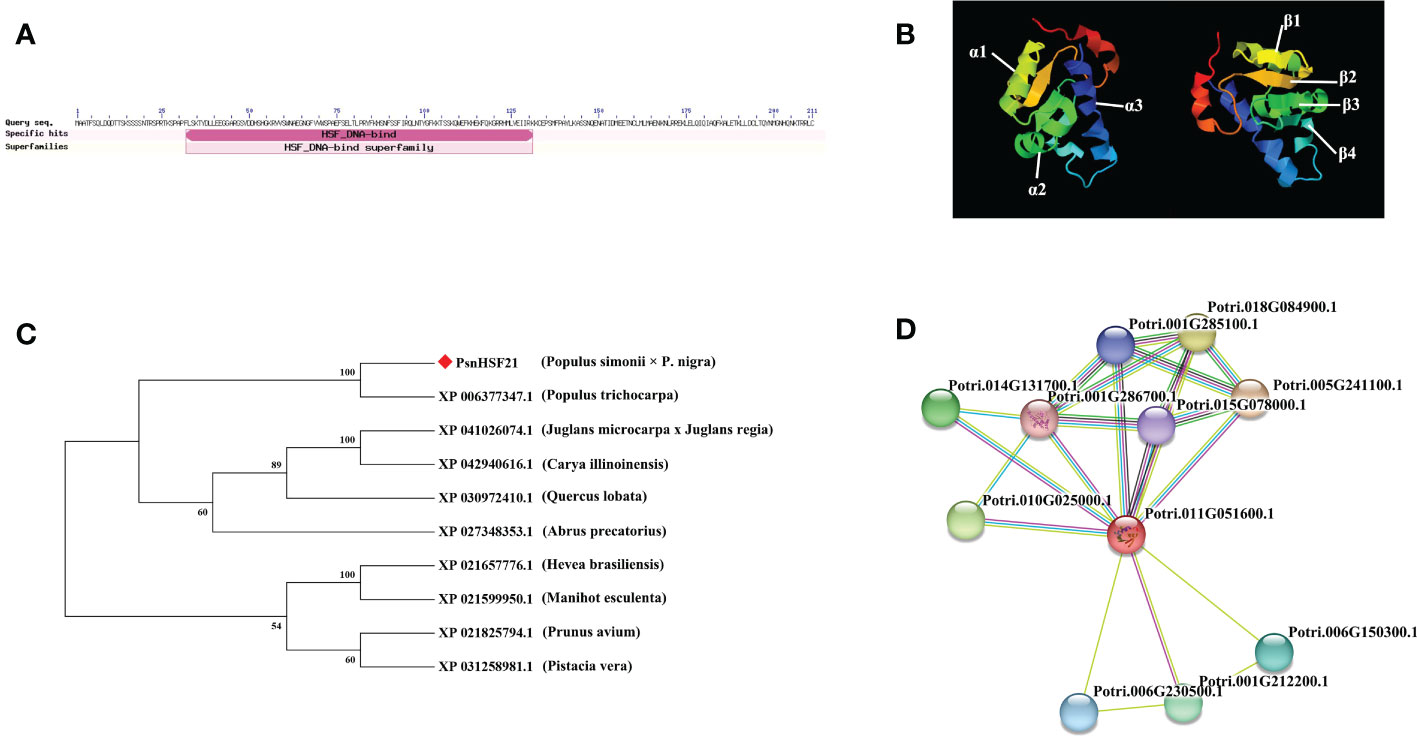
Figure 7 (A) Exhibition of the conserved domain of PsnHSF21 protein. (B) The schematic diagram of the tertiary structure of the PsnHSF21 protein. (C) Phylogenetic relationship between PsnHSF21 and HSFs in various species. (D) Prediction of PsnHSF21 interaction protein.
The subcellular localization of PsnHSF21 protein was analyzed in tobacco leaves by transient infection. As shown in Figure 8, the green fluorescence signal of pBI121-PsnHSF21-GFP was only observed in the nucleus, whereas it was expressed throughout the cells for the control vector pBI121-GFP, indicating that PsnHSF21 is a nuclear localization protein.
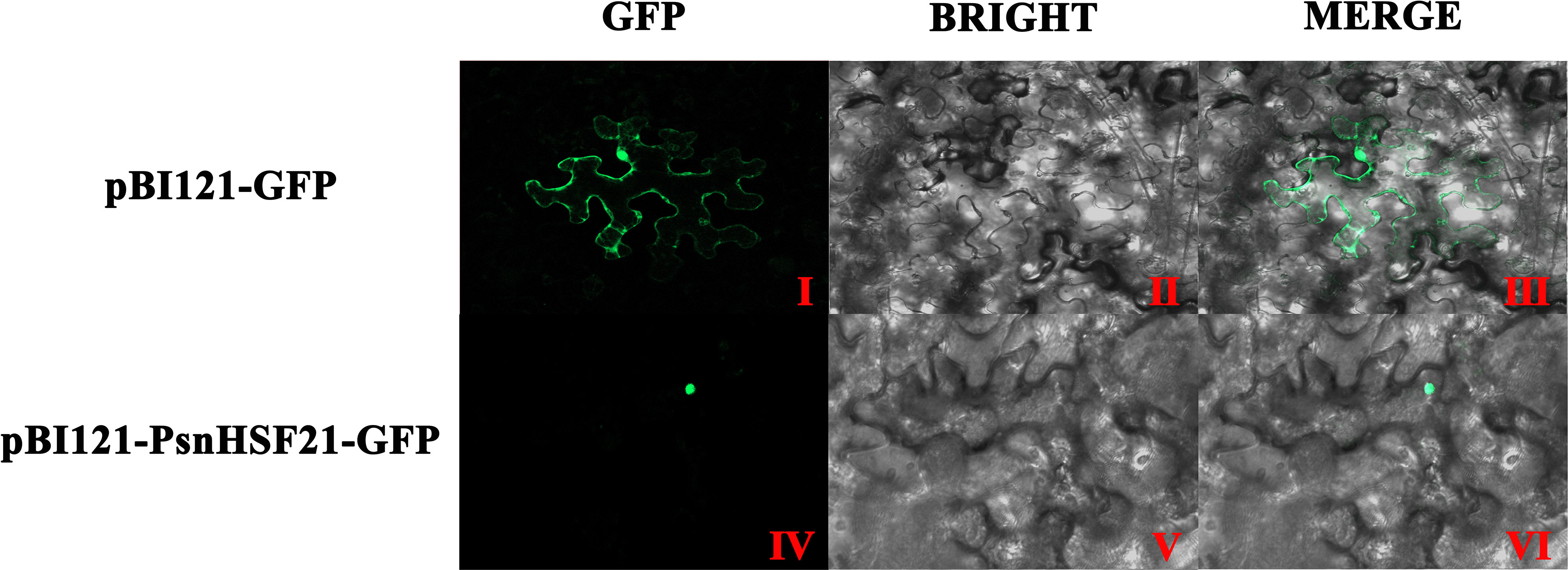
Figure 8 Subcellular localization of PsnHSF21 protein. The pBI121-GFP-PsnHSF21 fusion vector and pBI121-GFP control vector were transferred into tobacco leaf cells by instantaneous transformation. After 24–36 h of transformation, the pictures were imaged by the laser confocal microscope. (I, IV) are GFP fluorescence detection, (II, V) are bright fields, and (III, VI) are superposition fields.
Seven transgenic poplar lines overexpressing PsnHSF21 were obtained by the leaf disc transformation method, which was confirmed by RT-qPCR (Supplementary Figure 2). OE1-OE4 with high expression levels were selected for functional analysis. Under normal conditions, there was no obvious morphological difference between WT and OE. Under salt stress, severe salt spots and obvious wilting appeared on the leaves and stems of WT. Compared with WT, the injury degree of transgenic lines was lower (Figure 9A). Furthermore, plant height, fresh weight, taproot length, and number of lateral roots of transgenic lines were 17%, 6%, 14%, and 41% higher than those of WT, respectively (Figure 9B).
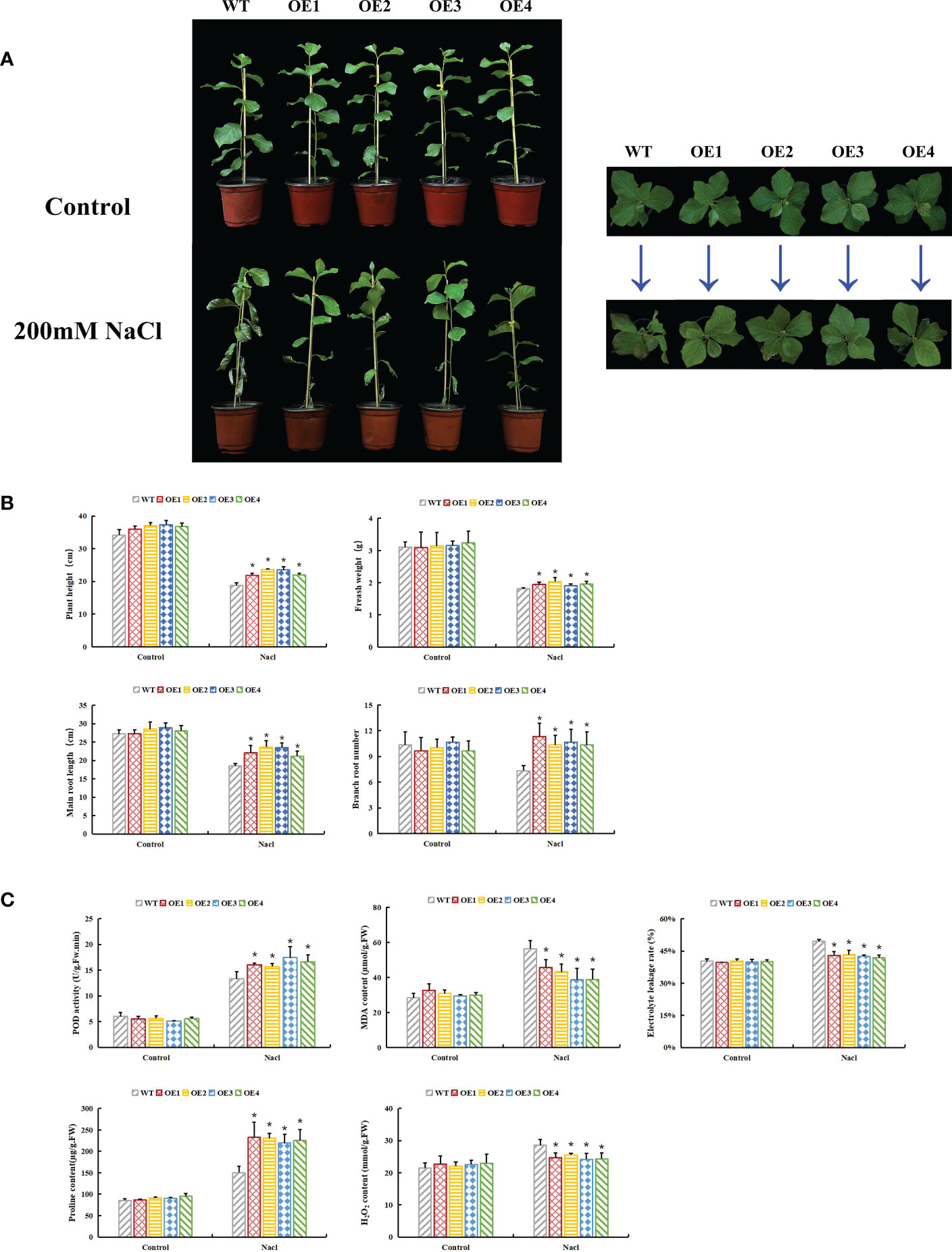
Figure 9 Functional analysis of PsnHSF21 under salt stress. The seedlings of WT and overexpressed PsnHSF21 poplar were stressed with water and 200 mM NaCl for 7 days. Control was water and treatment was 200 mM NaCl. (A) Display of the growth phenotype. (B) The growth index shows plant height, fresh weight, main root length, and lateral root number, three biological repeats. (C) Physiological index displays: POD, MDA, proline, electrical conductivity, and H2O2 content, three biological repeats. The error line represents the standard deviation (SD) of biological replication. “*” indicates there is a significant difference (P <0.05).
Under salt stress, plants produce a large amount of ROS, and the toxic effect of ROS can inhibit plant development. The activities of peroxidase (POD) and superoxide dismutase (SOD) indirectly reflect the ROS scavenging ability of plants. H2O2 remains the core component of ROS, which can also oxidize DAB into brown precipitates. Physiological index measurements indicated POD activity, H2O2 content, MDA content, electrical conductivity, Evans blue, and DAB staining degrees of leaves that displayed no difference between OE and WT under normal conditions. After salt stress, the POD activity of OE was 18%–31% higher than that of WT, and the H2O2 content of OE was 10%–16% lower than that of WT. MDA content and electrical conductivity of OE decreased by 19%–31% compared with WT. The Evans blue and DAB staining degrees of OE leaves were weaker than those of WT (Figures 9C, 10). Proline is also an important index to measure plant stress resistance. High proline content indicates improved stress tolerance. Under salt stress, the proline content of OE was 1.44–1.55 times higher than that of WT. RT-qPCR analysis showed that the relative expression levels of POD and SOD genes in OE were higher than those in WT (Supplementary Figure 3). The results indicated that PsnHSF21 transgenic poplar lines had stronger scavenging ability for ROS than WT. In addition, the relative expression levels of ABA, GA, and HRG genes associated with stress resistance were higher in OE than those in WT (Supplementary Figure 3). The above results showed that overexpression of PsnHSF21 could improve the salt resistance of transgenic poplars.

Figure 10 Histochemical staining analysis of poplar leaves. The tissue culture seedling leaves of WT and overexpressed PsnHSF21 poplar were treated with 150 mM NaCl and water for 24 h, then stained with DAB (diaminobenzidine) and Evans Blue, respectively.
To identify the transcriptional activity of PsnHSF21, we performed a Y2H assay. It was found that BD-PsnHSF21 could grow normally on both media and turn blue on SD/-Trp/-His/X-α-Gal, indicating it had self-activating activity and could be efficiently transcribed (Figure 11A). We divided PsnHSF21 into four segments, including BD-I (1–96 aa), BD-II (97–212 aa), BD-III (97–149 aa), and BD-IV (150–212 aa). The vectors contain four differential segments, which were respectively transformed into Y2H cells to determine the self-activating region of PsnHSF21. As shown in Figure 11A, all fragments can survive on SD/-Trp, but only BD-II and BD-IV can turn blue on SD/-Trp/-His/X-α-Gal. Therefore, the self-activated region of the PsnHSF21 protein is located at the C-terminal 150–212 aa, and the amino acid residues in this region are essential for PsnHSF21 activity.
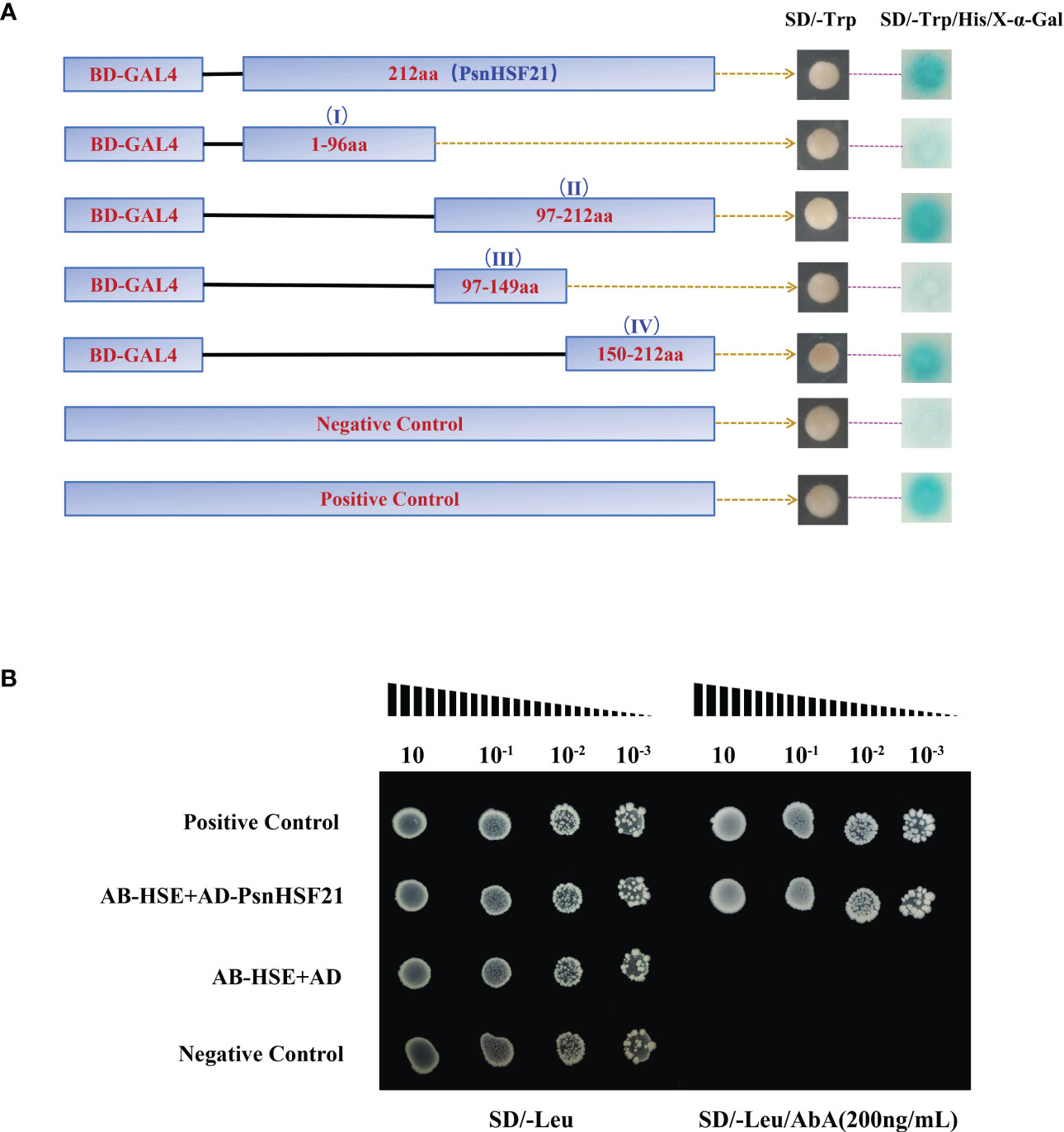
Figure 11 (A) Analysis of the PsnHSF21 protein’s self-activating activity. The pGBKT7-PsnHSF21 fusion vector was constructed and transferred into Y2H yeast cells. Transformed cell dots were plated on both SD/-Trp and SD/-Trp/-His/X-α-Gal medium. BD indicates pGBKT7. pGBKT7 was loaded empty as a negative control and pGBKT7-53/pGADT7-T as a positive control. (B) Analysis of resistant cis-acting elements identified by PsnHSF21. PsnHSF21 can be specifically bound to HSE (GAATTC) in yeast one-hybrid assays. AB-HSE indicates pAbAi-HSE, AD indicates pGADT7, and AD-PsnHSF21 indicates pGADT7-PsnHSF21 Positive conversion was determined after being diluted with positive yeast on a Leu plate (200 ng/ml) supplemented with AbA (aureobasidin A). The pAbAi-p53/pGADT7-p53 served as positive controls, and the pAbAi-53/pGADT7 served as negative controls.
Many studies have proved that the stress-resistant element HSE can bind to HSF family TFs in particular. Therefore, we carried out a yeast one-hybrid experiment with PsnHSF21. The results showed that all combinations could grow normally on SD/-Leu, but only AD-PsnHSF21/AB-HSE and the positive control could grow normally on SD/-Leu/AbA (200 ng/ml) (Figure 11B). Therefore, PsnHSF21 may regulate the expression of downstream stress-related genes by specifically binding to their HSE cis-acting element in promoter regions.
HSF is an important TF family in the plant kingdom that has been identified in A. thaliana, rice, tomato, corn, pepper, cabbage, and other plants. There are 21 HSF members in A. thaliana, which are divided into three subfamilies. AtHSFA6b in Class A not only participates in the high temperature stress response but also acts as a positive regulator of the stress response pathway mediated by the ABA signal, which significantly improves plant salt and drought tolerance (Huang et al., 2016). There are 25 HSF members in rice, and OsHsfB2b negatively regulates drought and salt tolerance in rice (Xiang et al., 2013). A total of 41 HSF members were identified from Phyllostachys pubescens, whose promoters contain many cis-acting elements related to stress. PeHSFs can be induced by gibberellin (GA) and naphthylacetic acid (NAA) and are highly expressed in panicles and young shoots, which may play a significant role in reproductive growth and organ development. As many as 49 HSF members were identified in tobacco, among which NtHSF03 and NtHSF12 were induced by high temperatures (Guo et al., 2022). Zizania aquatica has 28 HSF members, and 14 of them were highly expressed under high temperature stress (Cai et al., 2022).
In this study, we identified a total of thirty HSF members with conserved HSF domains from poplar, which all belong to acidic and hydrophilic proteins. All the PtHSFs were in nucleus, but PtHSF16 was also located in the whole cell, which was interesting. PtHSFs can be divided into three subfamilies (Classes A–C) (Huang et al., 2016). The structure of PtHSFs in the same subfamily is similar, while there are some differences among individual members. For example, PtHSF14 (Potri.006G226800.1) and PtHSF30 (Potri.T137400.1) have two intron structures, while the others contain one intron. Therefore, PtHSF14 and PtHSF30 may have experienced the cutting or insertion of gene fragments in the process of evolution (Staiger and Brown, 2013; Li et al., 2016). In addition to the common HSF conserved domain, PtHSF04 also contains a unique DivlC superfamily domain, which is a sporangium-forming domain of Bacillus subtilis. The SMC_prok_B domain represents an important domain for the maintenance of chromosome structure in B. subtilis (Soppa et al., 2002), and has an obvious inhibitory effect on plant pathogenic bacteria (Moo-Koh et al., 2022), which are present in PtHSF02, PtHSF07, and PtHSF12. In addition, all PtHSF members contain motifs 1–3, which may be the core components of the HSF domain. Class A had the largest number of PtHSF members with the largest number of motifs and the most complex conserved domain, which may have evolved from other functions.
Gene replication represents a general process of species evolution that can produce new functional genes and promote species evolution (Lynch and Conery, 2000). There are three modes of conventional gene replication: segmental replication, tandem repetition, and translocation events (Cannon et al., 2004). Rice has nine fragment repeat pairs in the HSF family (Guo et al., 2008), and P. pubescens has two tandem repeat pairs and twenty-seven fragment repeat pairs (Huang et al., 2021). In the study, we detected eleven fragment repeat pairs in PtHSFs, so segmental repetition dominates the expansion of the HSF family in poplar. Ka/Ks values of poplar HSF fragment repeats were all far less than 1, indicating these homologous genes underwent synonym mutation that was subjected to not only natural selection but also strong purification selection. In addition, every HSF fragment duplication gene belongs to the same subfamily in poplar, which may perform similar functions. There was a significant collinearity of HSF genes between poplar and Arabidopsis, like PtHSF17 (Potri.008G157600.1)/AtHSF6b (At3G22830.1) and PtHSF19 (Potri.010G08200.1)/AtHSF6b (At5G43840.1) (Figure 4B; Supplementary Table S6). Studies have shown that AtHSF6b mediates the ABA pathway to negatively regulate the drought resistance of A. thaliana (Wenjing et al., 2020), so it can be inferred that PtHSF17 and PtHSF19 may also be related to drought stress. In addition, PtHSF19 also corresponds to three rice HSF genes (LOC_Os03g06630.1, LOC_Os03G53340.1, and LOC_Os10G28340.1), which are sensitive to high temperature, cold, and oxidative stress (Mittal et al., 2009; Mittal et al., 2012). The direct homologous gene of PtHSF01 (Potri.001G1081.1) is OsHsfB2b (LOC_Os08g43334.1), which negatively regulates drought and salt tolerance in rice (Xiang et al., 2013). The direct homologous gene of PtHSF09 (Potri.004G062300.1) is OsHSF9 (LOC_Os01g54550.1), which is sensitive to temperature and improves cadmium resistance in rice (Shim et al., 2009). OsHsfA9 (LOC_Os03G12370.1) is the direct homologous gene of PtHSF16 (Potri.008G136800.2) and PtHSF20 (Potri.010G104300.1), which can not only promote reproductive development of plants but also improve stress resistance (Chauhan et al., 2011). Therefore, these homologous PtHSFs may carry out similar functions in stress responses.
The promoters of poplar HSF family members contain many elements with various functions. It can be inferred that these genes may be involved in multiple signaling pathways and carry out a critical role in regulating development, abiotic stress, and hormone synthesis in poplar. The HSF gene is well-known to respond to high temperature stress (Guo et al., 2016). In this study, we explored the biological function of poplar HSF family members under salt stress. Among the 30 HSF family members, 25 PtHSFs respond to salt stress in different tissues, with thirteen upregulated genes and fourteen downregulated genes. Among them, PtHSF21 was upregulated 4.5 times, 5.5 times, and 4.2 times in roots, stems, and leaves, respectively. This gene may perform a significant role in responding to salt stress; it was selected for further function validation.
We cloned PsnHSF21 from P. simonii × P. nigra, which belongs to the Class C subfamily in poplar and is accurately located in the nucleus. PsnHSF21 contains three α helical bundles and four inverted parallel β folds, which are important DBD structures in the HSF domain (Sangster and Queitsch, 2005). Among the 10 interacting proteins of PsnHSF21, Hsp81.4 (Potri.001G286700.1) and HSC70-5 (Potri.015G078000.1) have been proved to be significantly induced by heat stress, and HSC70-5 is involved in seed development. HSP90.7 (Potri.005G241100.1) can improve drought and salt tolerance in plants through the ABA pathway (Song et al., 2009). Therefore, PsnHSF21 may perform a similar function or cooperate with the above proteins to promote plant development and participate in the stress response.
In this study, we obtained seven transgenic poplar lines overexpressing PsnHSF21 (OE). Under normal conditions, there was no obvious difference in morphology between OE and WT. Under salt stress, the growth state of OE was significantly better than WT. After external stimulation, plants will undergo a series of physiological changes, especially the accumulation of ROS dominated by hydrogen peroxide (H2O2), the hydroxyl radical (OH−), and the superoxide anion (Erpen et al., 2018). SOD and POD are antioxidant enzymes that efficiently scavenge reactive oxygen species (ROS) (Feng et al., 2016; Survila et al., 2016). In this study, POD activity in OE was higher than that in WT, and SOD and POD gene expressions in OE were also higher than those in WT. This indicated that OE had better ROS scavenging ability. The H2O2 content of OE was lower than that of WT after salt stress, and the OE leaves with weaker DAB staining also indicated that there was less H2O2 content in OE. Free proline is an important index to detect plant growth status and evaluate plant stress resistance (Abrahám et al., 2010). Electrical conductivity, MDA content, and Evans blue staining are also indicators to evaluate the degree of cell damage. After salt stress, the proline content of OE was higher than that of WT, while the electrical conductivity and MDA content of OE were lower than those in WT. It is well recognized that abscisic acid (ABA) is widely involved in the stress response network to various environmental and abiotic stresses like drought, salinity, osmotic stress, and low temperature (Hamisch et al., 2016; Yu et al., 2017). Hydroxyproline glycoprotein (HRG) can improve plant antibacterial and disease resistance (Showalter et al., 2016). Gibberellin (GA) is a key hormone involved in plant development and stress response (Liu et al., 2018). Interestingly, the expression of the above stress-resistant genes in OE was higher than that in WT. Overall, the above results illustrated that PsnHSF21 could improve the salt resistance of OE by regulating these stress-related genes.
PsnHSF21 had self-activating activity, and the function region was at the C-terminal 150–212 aa, indicating the gene could form a stable transcription initiation complex and exert protein efficiently (Figure 11A). In animals and plants, HSE elements can be specifically recognized and bound by helix-transduction-helix motifs in the DNA binding domain (DBD) of HSF (Harrison et al., 1994; Schultheiss et al., 1996). When plants are subjected to high temperatures or other stresses, HSFs form HSF active trisomies through binding to hydrophobic OD, which accurately recognizes and binds to HSE elements in the gene promoter of the heat shock protein HSP gene, molecular chaperone, or other downstream genes to activate their transcription (Döring et al., 2000). For example, CmHSFA4 with transcriptional activation activity was identified in Chrysanthemum, which could respond to salt stress in combination with HSE elements (Li et al., 2018). In the study, the PsnHSF21 protein was proved to specifically bind to HSE elements, which indicates PsnHSF21 may regulate the expression of downstream salt-resistant genes by binding to HSE elements in their promoters.
In this study, a total of 30 HSF family genes were identified in poplar, all of which had HSF conserved domains. We systematically analyzed the gene structure, chromosome distribution, promoter cis-acting elements, gene duplication, and gene evolution of 30 PtHSFs and explored their expression pattern under salt stress by RNA-Seq and RT-qPCR. Among them, we identified a significantly salt-induced and nucleus-localized gene, PsnHSF21. We obtained seven transgenic poplar lines overexpressing PsnHSF21 (OE) by the leaf-disk method. OE displayed morphological and physiological advantages under salt stress, for example, higher plant height and root length, enhanced ROS scavenging ability, and reduced cell damage compared to WT. At the molecular level, the relative expression levels of stress-related genes such as ABA and GA were significantly higher in OE. Moreover, PsnHSF21 was proven to specifically bind to the stress-resistant element HSE, and the self-activated region was at the C-terminal 150–212 aa. In conclusion, PsnHSF21 regulates stress-related genes by specifically binding to the HSE element in their promoter regions to improve salt tolerance in poplar. This study provides a basis for understanding the biological function of the HSF gene in poplar under salt stress.
The original contributions presented in the study are included in the article/Supplementary material. Further inquiries can be directed to the corresponding authors.
TJ and GQ designed the research. QG and RW conducted the experiments, analyzed the data, and wrote the manuscript. MX, JJ, and XM performed the data analysis. WY revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Key Research and Development Program of China (No. 2021YFD2200203), the Fundamental Research Funds for the Central Universities (2572018CL03), and the Innovation Project of State Key Laboratory of Tree Genetics and Breeding (Northeast Forestry University) (No. 2015A02).
QG and RW wish to thank all their laboratory team for their support and suggestions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1160102/full#supplementary-material
Abdullah, M., Cao, Y., Cheng, X., Shakoor, A., Su, X., Gao, J., et al. (2018). Genome-wide analysis characterization and evolution of SBP genes in Fragaria vesca, Pyrus bretschneideri, Prunus persica and Prunus mume. Front. Genet. 9. doi: 10.3389/fgene.2018.00064
Abrahám, E., Hourton-Cabassa, C., Erdei, L., Szabados, L. (2010). Methods for determination of proline in plants. Methods Mol. Biol. 639, 317–331. doi: 10.1007/978-1-60761-702-0_20
Anders, S., Huber, W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. doi: 10.1186/gb-2010-11-10-r106
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Baniwal, S. K., Bharti, K., Chan, K. Y., Fauth, M., Ganguli, A., Kotak, S., et al. (2004). Heat stress response in plants: a complex game with chaperones and more than twenty heat stress transcription factors. J. Biosci. 29, 471–487. doi: 10.1007/BF02712120
Bechtold, U., Albihlal, W. S., Lawson, T., Fryer, M. J., Sparrow, P. A. C., Richard, F., et al. (2013). Arabidopsis HEAT SHOCK TRANSCRIPTION FACTORA1b overexpression enhances water productivity, resistance to drought, and infection. J. Exp. Bot. 64, 3467–3481. doi: 10.1093/jxb/ert185
Cai, L., Tang, M., Xu, J., Qi, Z., Fan, F., Zhou, Y.. (2022). Identification and analysis of heat shock transcription factor gene in Zizania latifolia. Journal of Zhejiang University (Agriculture and Life Sciences) 7 (15). doi: 10.3785/j.issn.1008-9209.2022.03.281
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10
Chauhan, H., Khurana, N., Agarwal, P., Khurana, P. (2011). Heat shock factors in rice (Oryza sativa l.): genome-wide expression analysis during reproductive development and abiotic stress. Mol. Genet. Genomics 286, 171–187. doi: 10.1007/s00438-011-0638-8
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Döring, P., Treuter, E., Kistner, C., Lyck, R., Chen, A., Nover, L. (2000). The role of AHA motifs in the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell. 12, 265–278. doi: 10.1105/tpc.12.2.265
Duan, S., Liu, B., Zhang, Y., Li, G., Guo, X. (2019). Genome-wide identification and abiotic stress-responsive pattern of heat shock transcription factor family in Triticum aestivum l. BMC Genomics 20, 257. doi: 10.1186/s12864-019-5617-1
Erpen, L., Devi, H. S., Grosser, J. W., et al. (2018). Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell Tissue Organ Cult. Int. J. Vitr. Cult. High. Plants. 132, 1–25. doi: 10.1007/s11240-017-1320-6
Evelin, H., Devi, T. S., Gupta, S., Kapoor, R. (2019). Mitigation of salinity stress in plants by arbuscular mycorrhizal symbiosis: current understanding and new challenges. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00470
Feng, K., Yu, J., Cheng, Y., Ruan, M., Wang, R., Ye, Q., et al. (2016). The SOD gene family in tomato: identification, phylogenetic relationships, and expression patterns. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01279
Finn, R. D., Clements, J., Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. doi: 10.1093/nar/gkr367
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Gong, B., Yi, J., Wu, J., Sui, J., Khan, M. A., Wu, Z., et al. (2014). LlHSFA1, a novel heat stress transcription factor in lily (Lilium longiflorum) can interact with LlHSFA2 and enhance the thermotolerance of transgenic Arabidopsis thaliana. Plant Cell Rep. 33, 1519–1533. doi: 10.1007/s00299-014-1635-2
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Guo, Q., Jiang, J., Yao, W., Li, L., Zhao, K., Cheng, Z., et al. (2021a). Genome-wide analysis of poplar HD-zip family and over-expression of PsnHDZ63 confers salt tolerance in transgenic Populus simonii × p.nigra. Plant Sci. 311. doi: 10.1016/J.PLANTSCI.2021.111021
Guo, Q., Li, L., Zhao, K., Yao, W., Cheng, Z., Zhou, B., et al. (2021b). Genome-wide analysis of poplar squamosa-promoter-binding protein (Sbp) family under salt stress. Forests 12. doi: 10.3390/f12040413
Guo, M., Liu, J. H., Ma, X., Luo, D. X., Gong, Z. H., Lu, M. H. (2016). The plant heat stress transcription factors (HSFs): structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00114
Guo, C., Wang, Q., Li, X., et al. (2022). Genome-wide identification and systemic analysis of the hsf gene family in Nicotiana tabacum l. Chin. TOBACCO SCIENCE. 43, 47–56. doi: 10.13496/j.issn.1007-5119.2022.03.008
Guo, J., Wu, J., Ji, Q., Wang, C., Luo, L., Yuan, Y., et al. (2008). Genome-wide analysis of heat shock transcription factor families in rice and Arabidopsis. J. Genet. Genomics 35, 105–118. doi: 10.1016/S1673-8527(08)60016-8
Hamisch, D., Kaufholdt, D., Kuchernig, J. C., Bittner, F., Mendel, R. R., Hnsch, R., et al. (2016). Transgenic poplar plants for the investigation of ABA-dependent salt and drought stress adaptation in trees. Am. J. Plant Sci. 07 (09), 1337–1356. doi: 10.4236/ajps.2016.79128
Harrison, C. J., Bohm, A. A., Nelson, H. C. (1994). Crystal structure of the DNA binding domain of the heat shock transcription factor. Science 263, 224–227. doi: 10.1126/science.8284672
Heerklotz, D., Döring, P., Bonzelius, F., Winkelhaus, S., Nover, L. (2001). The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Mol. Cell. Biol. 21, 1759–1768. doi: 10.1128/MCB.21.5.1759-1768.2001
Higashi, Y., Ohama, N., Ishikawa, T., Katori, T., Shimura, A., Kusakabe, K., et al. (2013). HsfA1d, a protein identified via FOX hunting using Thellungiella salsuginea cDNAs improves heat tolerance by regulating heat-stress-responsive gene expression. Mol. Plant 6, 411–422. doi: 10.1093/mp/sst024
Huang, B., Huang, Z., Ma, R., Chen, J., Zhang, Z., Yrjälä, K. (2021). Genome-wide identification and analysis of the heat shock transcription factor family in moso bamboo (Phyllostachys edulis). Sci. Rep. 11, 1–19. doi: 10.1038/s41598-021-95899-3
Huang, Y. C., Niu, C. Y., Yang, C. R., Jinn, T. L. (2016). The heat stress factor HSFA6b connects ABA signaling and ABA-mediated heat responses. Plant Physiol. 172, 1182–1199. doi: 10.1104/pp.16.00860
Huang, B., Xu, C. (2008). Identification and characterization of proteins associated with plant tolerance to heat stress. J. Integr. Plant Biol. 50, 1230–1237. doi: 10.1111/j.1744-7909.2008.00735.x
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, J., Meng, X., Zong, Y., Chen, K., Zhang, H., Liu, J., et al. (2016). Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9. Nat. Plants. 2, 16139. doi: 10.1038/nplants.2016.139
Li, P. S., Yu, T. F., He, G. H., Chen, M., Zhou, Y. B., Chai, S. C., et al. (2014). Genome-wide analysis of the hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genomics 15, 1009. doi: 10.1186/1471-2164-15-1009
Li, F., Zhang, H., Zhao, H., Gao, T., Song, A., Jiang, J., et al. (2018). Chrysanthemum CmHSFA4 gene positively regulates salt stress tolerance in transgenic chrysanthemum. Plant Biotechnol. J. 16, 1311–1321. doi: 10.1111/pbi.12871
Lin, Y. X., Jiang, H. Y., Chu, Z. X., Tang, X. L., Zhu, S. W., Cheng, B. J. (2011). Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genomics 12, 76. doi: 10.1186/1471-2164-12-76
Liu, L. L., Lu, T. T., Wang, S., Li, M. L., Wei, Z. G. (2018). ). effects of poplar PsnGA20ox1 overexpression on leaf development of tobacco. J. Beijing For. Univ. 40, 22–30. doi: 10.13332/j.1000-1522.20170245
Lyck, R., Harmening, U., Höhfeld, I., Treuter, E., Scharf, K. D., Nover, L. (1997). Intracellular distribution and identification of the nuclear localization signals of two plant heat-stress transcription factors. Planta 202, 117–125. doi: 10.1007/s004250050110
Lynch, M., Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 5494. doi: 10.1126/science.290.5494.1151
Mittal, D., Chakrabarti, S., Sarkar, A., Singh, A., Grover, A. (2009). Heat shock factor gene family in rice: genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol. Biochem. PPB. 47, 785–795. doi: 10.1016/j.plaphy.2009.05.003
Mittal, D., Madhyastha, D. A., Grover, A. (2012). Gene expression analysis in response to low and high temperature and oxidative stresses in rice: combination of stresses evokes different transcriptional changes as against stresses applied individually. Plant Sci. 197, 102–113. doi: 10.1016/j.plantsci.2012.09.008
Moo-Koh, F. A., Cristóbal-Alejo, J., Tun-Suárez, J. M., Medina-Baizabal, I. L., Arjona-Cruz, A. A., Gamboa-Angulo, M. (2022). Activity of aqueous extracts from native plants of the Yucatan peninsula against fungal pathogens of tomato in vitro and from croton chichenensis against corynespora cassiicola on tomato. Plants (Basel Switzerland). 11. doi: 10.3390/plants11212821
Nover, L., Bharti, K., Döring, P., Mishra, S. K., Ganguli, A., Scharf, K. D. (2001). Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need. Cell Stress Chaperones. 6, 177–189. doi: 10.1379/1466-1268(2001)006<0177:aathst>2.0.co;2
Ohama, N., Sato, H., Shinozaki, K., Yamaguchi-Shinozaki, K. (2017). Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 22, 53–65. doi: 10.1016/j.tplants.2016.08.015
Polle, A., Chen, S. (2015). On the salty side of life: molecular, physiological and anatomical adaptation and acclimation of trees to extreme habitats. Plant Cell Environ. 38, 1794–1816. doi: 10.1111/pce.12440
Sangster, T. A., Queitsch, C. (2005). The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Curr. Opin. Plant Biol. 8, 86–92. doi: 10.1016/j.pbi.2004.11.012
Schultheiss, J., Kunert, O., Gase, U., Scharf, K. D., Nover, L., Rüterjans, H. (1996). Solution structure of the DNA binding domain of the tomato heat-stress transcription factor HSF24. Eur. J. Biochem. 236, 911–921. doi: 10.1111/j.1432-1033.1996.00911.x
Sekulska-Nalewajko, J., Gocławski, J., Chojak-Koźniewska, J., Kuźniak, E. (2016). Automated image analysis for quantification of reactive oxygen species in plant leaves. Methods 109, 114–122. doi: 10.1016/j.ymeth.2016.05.018
Shah, Z., Iqbal, A., Khan, F. U., Khan, H., Ahmad, M. Z.. (2020). Genetic manipulation of pea (Pisum sativum L.) with Arabidopsis’s heat shock factor HsfA1d improves ROS scavenging system to confront thermal stress. Genet. Resour. Crop Evol. 67, 2119–2127. doi: 10.1007/s10722-020-00966-9
Shao, H. B., Guo, Q. J., Chu, L. Y., Zhao, X. N., Su, Z. L., Hu, Y. C., et al. (2007). Understanding molecular mechanism of higher plant plasticity under abiotic stress. Colloids Surf. B. Biointerf 54, 37–45. doi: 10.1016/j.colsurfb.2006.07.002
Shao, H., Wang, H., Tang, X. (2015). NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00902
Shen, Z., Ding, M., Sun, J., Deng, S., Zhao, R., Wang, M., et al. (2013). Overexpression of PeHSF mediates leaf ROS homeostasis in transgenic tobacco lines grown under salt stress conditions. Plant Cell. Tissue Organ Cult. 115, 299–308. doi: 10.1007/s11240-013-0362-7
Shim, D., Hwang, J. U., Lee, J., Lee, S., Choi, Y., An, G., et al. (2009). Orthologs of the class A4 heat shock transcription factor HsfA4a confer cadmium tolerance in wheat and rice. Plant Cell. 21, 4031–4043. doi: 10.1105/tpc.109.066902
Showalter, A. M., Keppler, B. D., Liu, X., Lichtenberg, J., Welch, L. R. (2016). Bioinformatic identification and analysis of hydroxyproline-rich glycoproteins in Populus trichocarpa. BMC Plant Biol. 16, 229. doi: 10.1186/s12870-016-0912-3
Song, H., Zhao, R., Fan, P., Wang, X., Chen, X., Li, Y. (2009). Overexpression of AtHsp90.2, AtHsp90.5 and AtHsp90.7 in Arabidopsis thaliana enhances plant sensitivity to salt and drought stresses. Planta 229, 955–964. doi: 10.1007/s00425-008-0886-y
Soppa, J., Kobayashi, K., Noirot Gros, M. F., Oesterhelt, D., Ehrlich, S. D., Dervyn, E., et al. (2002). Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the bacillus subtilis family members ScpA and ScpB. Mol. Microbiol. 45, 59–71. doi: 10.1046/j.1365-2958.2002.03012.x
Staiger, D., Brown, J. W. S. (2013). Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell. 25, 3640–3656. doi: 10.1105/tpc.113.113803
Sun, X., Matus, J. T., Wong, D. C. J., Wang, Z., Chai, F., Zhang, L., et al. (2018). The GARP/MYB-related grape transcription factor AQUILO improves cold tolerance and promotes the accumulation of raffinose family oligosaccharides. J. Exp. Bot. 69, 1749–1764. doi: 10.1093/jxb/ery020
Survila, M., Davidsson, P. R., Pennanen, V., Kariola, T., Broberg, M., Sipari, N., et al. (2016). Peroxidase-generated apoplastic ROS impair cuticle integrity and contribute to DAMP-elicited defenses. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01945
Tang, R., Zhu, W., Song, X., Lin, X., Cai, J., Wang, M., et al. (2016). Genome-wide identification and function analyses of heat shock transcription factors in potato. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00490
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. doi: 10.1126/science.1128691
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, C., Zhang, Q., Shou, H. (2009). Identification and expression analysis of OsHsfs in rice. J. Zhejiang Univ. Sci. B. 10, 291–300. doi: 10.1631/jzus.B0820190
Wenjing, W., Chen, Q., Singh, P. K., Huang, Y., Pei, D. (2020). CRISPR/Cas9 edited HSFA6a and HSFA6b of Arabidopsis thaliana offers ABA and osmotic stress insensitivity by modulation of ROS homeostasis. Plant Signal. Behav. 15, 1816321. doi: 10.1080/15592324.2020.1816321
Xiang, J., Ran, J., Zou, J., Zhou, X., Liu, A., Zhang, X., et al. (2013). Heat shock factor OsHsfB2b negatively regulates drought and salt tolerance in rice. Plant Cell Rep. 32, 1795–1806. doi: 10.1007/s00299-013-1492-4
Xu, L., Gao, J., Guo, L., Yu, H. (2020). Heat shock protein 70 (HmHsp70) from Hypsizygus marmoreus confers thermotolerance to tobacco. AMB Express. 10, 12. doi: 10.1186/s13568-020-0947-6
Yang, X., Zhu, W., Zhang, H., Liu, N., Tian, S. (2016). Heat shock factors in tomatoes: genome-wide identification, phylogenetic analysis and expression profiling under development and heat stress. PeerJ 4, e1961. doi: 10.7717/peerj.1961
Yao, W., Zhou, B., Zhang, X., Zhao, K., Cheng, Z., Jiang, T. (2019). Transcriptome analysis of transcription factor genes under multiple abiotic stresses in Populus simonii × p.nigra. Gene 707, 189–197. doi: 10.1016/j.gene.2019.04.071
Yoon, Y., Seo, D. H., Shin, H., Kim, H. J., Kim, C. M., Jang, G. (2020). The role of stress-responsive transcription factors in modulating abiotic stress tolerance in plants. Agronomy 10, 1–23. doi: 10.3390/agronomy10060788
Yu, D., Chen, C., Chen, Z. (2001). Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell. 13, 1527–1540. doi: 10.1105/tpc.010115
Yu, J., Ge, H., Wang, X., Tang, R., Wang, Y., Zhao, F., et al. (2017). Overexpression of pyrabactin resistance-like abscisic acid receptors enhances drought, osmotic, and cold tolerance in transgenic poplars. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01752
Zang, D., Wang, J., Zhang, X., Liu, Z., Wang, Y. (2019). Arabidopsis heat shock transcription factor HSFA7b positively mediates salt stress tolerance by binding to an e-box-like motif to regulate gene expression. J. Exp. Bot. 70, 5355–5374. doi: 10.1093/jxb/erz261
Zhang, X., Cheng, Z., Fan, G., Yao, W., Li, W., Chen, S., et al. (2022). Functional analysis of PagNAC045 transcription factor that improves salt and ABA tolerance in transgenic tobacco. BMC Plant Biol. 22, 1–14. doi: 10.1186/s12870-022-03623-8
Zhao, K., Chen, S., Yao, W., Cheng, Z., Zhou, B., Tingbo, T. (2021). Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol. 1). doi: 10.1186/S12870-021-02879-W
Zhao, K., Cheng, Z., Guo, Q., Yao, W., Liu, H., Zhou, B., et al. (2020). Characterization of the poplar R2R3-MYB gene family and over-expression of PsnMYB108 confers salt tolerance in transgenic tobacco. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.571881
Zhou, R., Li, B., Liu, H., Sun, D. (2009). Progress in the participation of Ca2+ calmodulin in heat shock signal transduction. Prog. Nat. Sci. 19(10):1201–1208. doi: 10.1016/j.pnsc.2008.12.011
Keywords: Populus simonii × P. nigra, heat shock transcription factor, salt stress, PsnHSF21, transcriptional regulation
Citation: Guo Q, Wei R, Xu M, Yao W, Jiang J, Ma X, Qu G and Jiang T (2023) Genome-wide analysis of HSF family and overexpression of PsnHSF21 confers salt tolerance in Populus simonii × P. nigra. Front. Plant Sci. 14:1160102. doi: 10.3389/fpls.2023.1160102
Received: 06 February 2023; Accepted: 28 March 2023;
Published: 26 April 2023.
Edited by:
Waqar Afzal Malik, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Longbiao Guo, China National Rice Research Institute (CAAS), ChinaCopyright © 2023 Guo, Wei, Xu, Yao, Jiang, Ma, Qu and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guanzheng Qu, Z3pxdUBuZWZ1LmVkdS5jbg==; Tingbo Jiang, dGJqaWFuZ0BuZWZ1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.