
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 29 March 2023
Sec. Crop and Product Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1157836
This article is part of the Research TopicGenetics, Breeding and Engineering to Enhance Oil Quality and YieldView all 11 articles
 Na Lin1
Na Lin1 Mu Wang1
Mu Wang1 Jiayi Jiang1
Jiayi Jiang1 Qinyuan Zhou1
Qinyuan Zhou1 Jiaming Yin1
Jiaming Yin1 Jiana Li1,2
Jiana Li1,2 Jianping Lian1
Jianping Lian1 Yufei Xue1
Yufei Xue1 Yourong Chai1,2*
Yourong Chai1,2*MYB transcription factors are major actors regulating plant development and adaptability. Brassica napus is a staple oil crop and is hampered by lodging and diseases. Here, four B. napus MYB69 (BnMYB69s) genes were cloned and functionally characterized. They were dominantly expressed in stems during lignification. BnMYB69 RNA interference (BnMYB69i) plants showed considerable changes in morphology, anatomy, metabolism and gene expression. Stem diameter, leaves, roots and total biomass were distinctly larger, but plant height was significantly reduced. Contents of lignin, cellulose and protopectin in stems were significantly reduced, accompanied with decrease in bending resistance and Sclerotinia sclerotiorum resistance. Anatomical detection observed perturbation in vascular and fiber differentiation in stems, but promotion in parenchyma growth, accompanied with changes in cell size and cell number. In shoots, contents of IAA, shikimates and proanthocyanidin were reduced, while contents of ABA, BL and leaf chlorophyll were increased. qRT-PCR revealed changes in multiple pathways of primary and secondary metabolisms. IAA treatment could recover many phenotypes and metabolisms of BnMYB69i plants. However, roots showed trends opposite to shoots in most cases, and BnMYB69i phenotypes were light-sensitive. Conclusively, BnMYB69s might be light-regulated positive regulators of shikimates-related metabolisms, and exert profound influences on various internal and external plant traits.
MYB is a large family of transcription factors (TFs) (Feller et al., 2011). It has been a hotspot in the study of the plant transcription factor function because of its large number of genes and different functions. The first identified MYB TF in plants was COLORED1 (C1), which is involved in the biosynthesis of anthocyanins in Zea mays (Paz-Ares et al., 1987). MYB TFs contain a highly conserved DNA binding domain (DBD) located at the N-terminus (Baldoni et al., 2015; Wei et al., 2017; Zhuang et al., 2021). The MYB domain is composed of 1-4 imperfect repeats (R1-R4) each containing approximately 50-53 amino acids (aa). Each repeat consists of three α-helices, of which the second and third ones form a helix-turn-helix (HTH) motif (Baldoni et al., 2015). The HTH intercalates with the major groove of DNA. MYB TFs have three spaced Trp residues forming a hydrophobic core that stabilizes the structure (Ogata et al., 1995). The C-terminus is responsible for the distinct regulatory activities of MYB TFs (Jin and Martin, 1999).
MYB TFs are divided into four groups, 1R-MYB, R2R3-MYB, 3R-MYB, and 4R-MYB, based on the number of repeats (Dubos et al., 2010). R2R3-MYB transcription factors (TFs) have been shown to play important roles in plants, including cell fate and identity, developmental processes, and responses to biotic and abiotic stresses (Hajiebrahimi et al., 2017). Although members of the MYB superfamily have been annotated in Arabidopsis thaliana and many other plants, most of them have not been functionally characterized.
In the past decade, R2R3-MYB genes have been extensively studied. The R2R3-MYB proteins play important roles in diverse biological processes including growth and development, primary and secondary metabolism such as flavonoid and anthocyanin biosynthesis as well as abiotic and biotic stress responses (Yao et al., 2020). Plant development and secondary metabolisms are related to cell differentiation including the change of cell size, cell number and cell wall. It was well known that the plant cell wall is a dynamic barrier against pathogens invasion (Underwood, 2012). The cell walls of vascular bundles deposit a large amount of cellulose, lignin and pectin, which are closely related to the support and resistance of plants. Lignin plays an important role in plant growth and development, lodging resistance and structural support (Jiang et al., 2020). The deposition of unpolymerized lignin monomers is considered to present a physical barrier that prevents infection in the interaction between plant and pathogen, and its metabolism is also actively involved in plant lodging resistance and in response to various environmental stresses (Jiang et al., 2020).
Many studies have shown that MYB69, which is from subgroup S21, is a SCW (secondary cell wall) activator (Kranz et al., 1998; Zhong et al., 2007; Dubos et al., 2010; Zhu et al., 2017). It has been demonstrated by transcriptional activation analysis that MYB69 and other transcription factors are able to activate the expression of the HIS3 and b-GAL reporter genes in yeast, confirming that they are indeed transcriptional activators (Zhong et al., 2008). AtMYB69 is a positive regulator dedicated to cell wall thickening of fiber cells in A. thaliana (Dubos et al., 2010). Zhao and Bartley (2014) have shown that AtMYB69 is an activator whose dominant repression reduces SCW thickening of both interfascicular fibers and xylem fibers in stems. These studies indicate that MYB69 may be related to the growth of plant cell. Studies have also shown by cluster analysis that MYB105, MYB110, MYB117, MYB56, MYB52, MYB54 and MYB69 belong to the same subgroup, so it is probable that their functions are similar (Kranz et al., 1998; Dubos et al., 2010; Zhu et al., 2017). MYB20, MYB63, MYB69, SND2 and SND3 are also hypothesized to be part of the SND1/NST1 transcriptional network regulating secondary wall biosynthesis (McCarthy et al., 2010). Xu et al. (2021) reported that FvMYB3, FvMYB9, FvMYB11, FvMYB22, FvMYB64, and FvMYB105 mostly expressed at green stage of fruit development in woodland strawberry, aligned with proanthocyanidins accumulation. Wang et al. (2022) found that both 35S::MYB10 and MYB110 can upregulate anthocyanin biosynthesis in Actinidia chinensis fruit. Hong et al. (2021) have identified MYB117 as a negative regulator in controlling flowering time through regulating the expression of Flowering Locus T in Arabidopsis. Other researchers showed that SiMYB56 enhanced ABA synthesis under drought conditions and activated the ABA signaling pathway, contributing to the enhanced drought tolerance of transgenic rice (Xu et al., 2020). It has also been demonstrated that dominant repression of MYB69 and SND2, SND3, MYB103, MYB85, MYB52, MYB54, and KNAT7 causes a severe reduction in secondary wall thickening of both interfascicular fibers and xylary fibers in inflorescence stems (Zhong et al., 2008). It was reported that ERF4 and MYB52 transcription factors played antagonistic roles in regulating homogalacturonan de-methylesterification in Arabidopsis seed coat mucilage (Ding et al., 2021). For MYB46 downstream transcription factors, MYB43, MYB52, MYB54, MYB58, MYB63 and KNAT7 are direct targets of MYB46 (Zhong and Ye, 2012), which was a master transcriptional regulator of secondary wall biosynthesis (Kim et al., 2012). And it was reported that MYB46 is negatively regulated by MPK6 during salt stress (Im et al., 2021). These studies have revealed that the function of MYB69 was not only related to regulating secondary wall biosynthesis but also probably related with some metabolisms and resistances in plant.
B. napus is an important oil crop worldwide and is negatively impacted by a variety of biotic and abiotic stresses, particularly lodging and S. sclerotiorum stem rot, which seriously reduce the yield and affect the quality of B. napus seeds. Lodging is related to stem and root lodging (Li and Qi, 2017; Zhang et al., 2021). The flexibility of stems and roots is related to secondary wall thickening and lignin accumulation. Studying the function of MYB69 in B. napus can both elucidate whether it contributes to lodging resistance and antimicrobial resistance in B. napus and systematically analyze how MYB69 regulates plant cell growth and development.
In this work, downregulation of BnMYB69 causes an increase in the biomass and susceptibility to S. sclerotiorum, and related biochemical, physiological and gene expression characterization provides insight into BnMYB69 function in B. napus.
The material of gene cloning and expression characterization was cv. Zhongyou 821 (ZY821) of B. napus. The explant material of genetic transformation is cv. Zhongshuang 10 (ZS10) of B. napus. These plants were from growth chambers, green houses or isolated field farms at Southwest University and cultivated by standard agronomic management. Nicotiana benthamiana plants for protein localization study were cultured in room at 28°C under a 16 h/8 h light/dark cycle.
The strain of S. sclerotiorum in this experiment was provided by Dr. Jiaqin Mei at Southwest University. S. sclerotiorum was grown up on potato media (PDA) at 22°C with 85% humidity.
Total RNA was isolated from ZY821 organs using EASYspin plant RNA rapid extraction kit (Biomed, China). Total DNA was extracted from ZY821 leaves using traditional CTAB method. The quality and quantity of RNA and DNA were detected by agarose gel electrophoresis and NanoDrop 2000c (Thermo Fisher, USA).
In-silico cloning of Brassica napus MYB69s was performed on NCBI (https://blast.ncbi.nlm.nih.gov) using A. thaliana MYB69 (AtMYB69) mRNA as Query in BLASTn against B. napus (taxid:3708) nr/nt, refseq_genomes, refseq_rna, est, and TSA databases. Sequences orthologous to AtMYB69 were downloaded and multi-aligned on Vector NTI Advance 11.51, and primers for RACE (rapid-amplification of cDNA ends) cloning were designed (Table S1).
Using the SMARTer RACE Amplification Kit (Clontech, USA), 1 µg of total RNA mixture from various organs of ZY821 was used for synthesis of first-strand total cDNA. The primer pairs RBnMYB69-51 + LUPM and RBnMYB69-52 + NUP were used for the primary and the nested PCRs in 5’-RACE of BnMYB69s, while primer pairs FBnMYB69-31 + LUPM and FBnMYB69-32 + NUP were used for primary and nested PCRs in 3’-RACE of BnMYB69s. Based on sequencing results of 5’- and 3’-RACE colonies, primers FBnMYB69-1 + RBnMYB69-1, FBnMYB69-2 + RBnMYB69-2, FBnMYB69-3 + RBnMYB69-3 and FBnMYB69-4 + RBnMYB69-4 were used to amplify full-length cDNAs and gDNAs of BnMYB69-1, BnMYB69-2, BnMYB69-3 and BnMYB69-4, respectively. Electrophoresis of PCR products, gel recovery, TA cloning, Escherichia coli transformation, colony culture and other experiments abided by routine methods. Bioinformatics Analyses of BnMYB69s genes and proteins were performed on local Vector NTI Advance 11.51 and websites of NCBI, Expasy, Softberry, etc. (See Supplementary Tables and Figures for more information).
The total RNA of each organ or tissue sample from material was reverse transcribed by the PrimeScript 1st Strand cDNA Synthesis Kit (TaKaRa, Dalian, China) to obtain the corresponding first-strand total cDNA. All cDNA samples were diluted 20-fold using sterile water for qRT-PCR. The overall and member-specific expression levels of B. napus genes were detected by Gene-specific primers for qRT-PCR, respectively.
Primers are listed in Table S1, and the 25SrRNA gene was used for the internal standard. The CFX Connect Real-Time PCR Detection System (Bio-Rad, Berkeley, CA, USA) with FastStart Universal SYBR Green Master reagents (Roche, Basel, Switzerland) was performed for qRT−PCR analysis. The program was that firstly, the sample was put in 95°C 10 min, followed by 45 cycles of amplification (95°C 10 s, 58-64°C 30 s). Secondly, the temperature was raised from 65 to 95°C, and the specificity of the amplification was confirmed by the melting curve. The detection was repeated three times. Thirdly, CFX Manager 3.1 (Bio-Rad, Berkeley, CA, USA) by the 2-△△CT method analyzed all data.
The coding sequence of BnMYB69-1(BnaA01.MYB69) was amplified using the primer pair FBnMYB69SL + RBnMYB69SL (Table S1), and it was cloned and designated BnMYB69SL. Then BnMYB69SL was subcloned into pEGAD by EcoRI+BamHI double digestion, and enhanced green fluorescent protein (EGFP) was joined into the N-terminus to form the subcellular localization expression vector pEGAD-BnMYB69SL (Figure S4A). The vector was introduced in Agrobacteriu tumefaciens strain LBA4404 and transformed into tobacco leaves. Then the plant was cultured at 28°C for two days, stained its leaves with nuclear dye (DAPI) and photographed with a confocal microscope (LSM 800, ZEISS, Jena, Germany).
Using the primer pair FBnMYB69I + RBnMYB69I (Table S1), a 429 bp C-terminus coding region of cDNA for the BnMYB69 family (BnMYB69s) was amplified from the cDNA library. This cDNA fragment had >86.7% identity match with all BnMYB69 genes, and no identity match with other sequences which were found by BLASTing the entire B. napus genome (http://www.ncbi.nlm.nih.gov/genome/?term=Brassica+napus). An antisense fragment of BnMYB69I (BnMYB69IA) was subcloned into pFGC5941M using NcoI+AatII double digestion to form the intermediate vector pFGC5941M-BnMYB69IA (Figure S4B). And a sense fragment of BnMYB69I (BnMYB69IS) was subcloned into pFGC5941M-BnMYB69IA using BamHI+XbaI double digestion to form the RNAi vector pFGC5941M-BnMYB69I. CaMV35S was the promoter of the vector. pFGC5941M-BnMYB69I plasmids were introduced into LBA4404 (A. tumefaciens) by the freeze-thaw transformation method. The engineering strain harboring pFGC5941M-BnMYB69I was transformed into hypocotyl segments of ZS10 using the method described by Cardoza and Stewart (2003). The positive transgenic plants were detected by DNA electrophoresis after PCR amplification (Figure S4C).
Transgenic and nontransgenic B. napus (WT) plants were grown in a greenhouse, agronomic traits were investigated at the bolting stage, full flowering stage and pod setting stage, and dry matter weight, chlorophyll content, and 1000 seed weight per plant were measured at the harvest stage.
The method of leaf inoculation was from Godoy et al. (1990). The third or fourth leaf from the top in each plant at the 9 to 12-leaf stage was excised with at least 6 leaves per different line. The excisions of these leaves were wrapped with wet paper to keep moisture, and were placed into porcelain plate paved with wet filter paper. 7-mm-diameter PDA discs containing S. sclerotiorum hyphae were inoculated at the center of the left and right abdomens in leaf. Then, the whole plate was covered with cling film and kept at 22°C. After 48 h, the long diameter (a) and the short diameter (b) of the lesion were measured. The lesion area was gained by this calculation formula: S = (π×a×b)/4.
The method of stem inoculation was from Mei et al. (2015). The stems were inoculated at the full-bloom stage with 6 plants per different line. Stem fragments (45 cm long) were cut off at 20 cm from the ground, and both ends also were wrapped with wet paper and covered with cling film. 7-mm-diameter PDA discs containing S. sclerotiorum hyphae were inoculated at two points distanced with 10 cm on the stem segment. Lesion lengths were measured after 72 h.
During S. sclerotiorum infection, leaves were taken at different time points for the determination of relevant biochemical components and key enzymes, and RNA was extracted for qRT−PCR (as described above).
The methods of Mäule staining, phloroglucinol-HCl staining and fluorescent brightener dyeing of lignin were from the literature (Zhong et al., 2007; Yin et al., 2017). Cellulose staining with Fast Green FCF and pectin staining by the hydroxylamine method (Leagene, Beijing, China) were performed on the stems and roots of transgenic and WT plants. Stained sections were observed with a stereo light microscope (Olympus SZX2-FOA, Tokyo, Japan) and a fluorescence microscope (Nikon Eclipse E600 W, Tokyo, Japan). The tissues and organs were observed by scanning electron microscopy (Hitachi SU3500, Japan).
At the harvest stage, the middle part of mature stems was cut off and dried at 70°C to constant weight. Then, these stems were powdered with a grinder, and screened with a 100-mesh sieve. The cellulose, lignin and pectin contents from the cell walls of transgenic and WT plants were extracted and determined. The method of extraction and determination of cellulose and lignin was from Foster et al. (Foster et al., 1996). The method of extraction and quantification of pectin was from Blumenkrantz and Asboe-Hansen (1973). The absorbance of cellulose and lignin was determined by an Infinite M2000 Pro Microplate Reader (Tecan, Männedorf, Switzerland). The contents protopectin and soluble pectin were determined according to the instructions of the Protopectin and Soluble Pectin Content Determination kits from Shanghai Haling Biotechnology Co., Ltd.
At bolting stage, the middle-part samples of stems, leaves and roots were fetched and stored in a -80°C freezer. The frozen samples were ground into powder with liquid nitrogen to test the activity of CAT, MDA, SOD and POD, as well as the contents of chlorophyll, reducing sugars, and total amino acids. The extraction and determination of these biochemical components followed the kit instructions from Beijing Leagene Biotech Co., Ltd.
At the bolting stage, the mid-region fragments of stems and leaves were sampled and stored in a -80°C refrigerator. Frozen samples were sent to Qingdao Kechuang Quality Inspection Company and Nanjing Zhongding Biotechnology Co., Ltd. to detect the changes in hormone components using gas chromatography or liquid chromatography, respectively.
Cultured in 1/3 X Hoagland nutrient solution, WT and BnMYB69i lines were treated with 100 µmol/L IAA, with non-addition as the experimental control (CK). They were cultured on Petri dishes (light 16 h/22°C; dark 8 h/18°C) and then in large pots after two weeks with 40 seedlings per pot. The water was changed every 10 days. There were 2 lines, 2 treatments and 3 repetitions. The morphological, physiological and biochemical indices were measured at the cotyledon stage (20 days), 3 true leaves stage (60 days) and 5 true leaves stage (120 days). Three plants were randomly selected from each line for the measurement of plant height, stem diameter and physiological and biochemical indices, and the average values of each parameter were calculated. Plant height was defined as the length from the cotyledonary node to the highest part of the entire plant (Gai, 2005); stem diameter of the stem base at the cotyledon mark was measured with a vernier caliper.
WT and transgenic plants were cultured in high light (8000 lx) and low light (4000 lx) incubators. They grew to the three-leaf stage under light for 16 h at 22°C and under darkness for 8 h at 18°C, and the chlorophyll content of the third true leaf was measured. The methods were performed according to instructions of the Beijing Leagene Biotechnology kit and the experimental guidance of Plant Physiology handbook (College of Agriculture/Department of Plant Physiology, Shanxi Agricultural University).
The transgenic and control plants were treated in the dark three times for 20 min each at different stages. The chlorophyll fluorescence kinetic parameters were determined with a Spectral Fluorescence Imager and Fluor Scan software.
A 0.1 g sample of B. napus leaves was frozen in liquid nitrogen and 300 μL of 1% hydrochloric acid methanol solution was added after proofing, mixed evenly and shaded at 4°C overnight. The next day, 200 μL distilled water was added along with 200μL chloroform, the mixture was vortexed for 20 s at room temperature and then centrifuged for 5 min at 17000 g. The extracted supernatant was added into a 96-well plate and determined using an Enzyme Labeling Instrument at 530 nm and 657 nm. The proanthocyanidin content (mg/g) was calculated as (A530-A657)/fresh weight (g).
The IAA content was determined according to the instructions of the plant indole-3-acetic acid ELISA kit (Nanjing Camilo Bioengineering Co., Ltd.), and the method of quantifying protopectin and amino acids was as described above.
In this study, at least three biological replications (n≥3) were performed for each experiment. All values are presented as the mean ± standard erro (SE). Asterisks indicate significant or extremely significant differences from WT (*0.01≤ p < 0.05, ** for p < 0.01) using a one-way ANOVA.
Using mixture total RNA from various organs of ZY821 as start material, our RACE-PCR based cloning isolated four MYB69 genes from B. napus, i.e., BnMYB69-1 (BnaA01.MYB69), BnMYB69-2 (BnaC01.MYB69), BnMYB69-3 (BnaA08.MYB69) and BnMYB69-4 (BnaC03.MYB69), with respective gene lengths of 1110, 1418, 1296 and 1215 bp, mRNA lengths (not including polyA) of 1015, 1320, 1191 and 1110 bp, and coding region (open reading frame, ORF, including stop codon) lengths of 756, 753, 732, and 756 bp, respectively. BnMYB69-1, BnMYB69-2, BnMYB69-3 and BnMYB69-4 showed high homologies to AtMYB69, with gene level identities of 84.2%, 78.3%, 77.8% and 80.0%, mRNA level identities of 85.5%, 80.0%, 79.5% and 80.8%, and ORF level identities of 86.5%, 86.2%, 84.8 and 87.0%, respectively (Table S2). Only one intron exists in each of the four BnMYB69 genes. 5’UTR length of BnMYB69 genes is 28-132 bp, and their 3’UTR length is 127-494 bp. Every BnMYB69 gene shows alternative transcription start sites (TSS) and alternative polyadenylation sites, thus multiple versions of mRNAs could be cloned with differed length of 5’UTRs and 3’UTRs. One feature is that in most cases the TSS base is a purine (A or G), and in most cases the base before the polyA tail is a pyrimidine (T or C). Another feature is that as BnMYB69-1 could generate long 5’UTRs, its 3’UTRs are relatively shorter, while the other three BnMYB69 genes have the opposite trend (Table S2).
The predicted BnMYB69-1, BnMYB69-2, BnMYB69-3, BnMYB69-4 proteins have a length of 251, 250, 243, 251 aa, a MW of 29.01, 28.90, 28.16, 29.04 kD, and a pI of 9.58, 9.70, 9.72, 9.56 implying that they are basic proteins. All of them have no signal peptide and no transmembrane helix, and are predicted to be located in the nuclear. Their predicted phosphorylation sites are 19-22 for S (serine), 8-9 for T (threonine), and 1-2 for Y (tyrosine). The N-terminal half of each BnMYB69 protein has two predicted HTH MYB-type domain regions at 14-65/64 and 66-120, on which there are two HTH motif DNA-binding regions at 42-63 and 93-116 (Table S3). The front HTH model belongs to the R2 subdomain and has three conserved tryptophan residues (W), which are separated by 18 or 19 amino acids, and the latter HTH model belongs to the R3 subdomain (Ogata et al., 1994). In the secondary structure of BnMYB69s, α-helix, extended strand, β-turn, and random coil occupy 23.11%-31.87%, 6.77%-12.75%, 4.38%-6.40%, and 54.40%-59.76%, respectively (Figure S1). SWISS-MODEL-predicted tertiary structures of BnMYB69s conform to conserved R2R3-MYB domain (Figure S2). Thus, the four BnMYB69 genes cloned here most probably encode typical R2R3-MYB transcription factors.
In multiple alignment, BnMYB69 proteins have high similarities to AtMYB69 on the whole protein level. Though they show high similarities to MYB52, MYB54, MYB56, MYB105, MYB117, MYB110 and MYB89 of S21 in the N-terminal half, the similarities in the C-terminal half are distinctly lower with conservation only at a C-terminal motif and several dispersed residues (Figure 1A). Phylogenetic analysis showed that the distances among BnMYB69 proteins are small, and they clustered tightly with AtMYB69. BnMYB69-1 and BnMYB69-2 are close, while BnMYB69-3 and BnMYB69-4 are close, but these two sister pairs are a little far from each other. In the tree, the distances of BnMYB69s to other S21 AtMYB proteins are much larger than to AtMYB69 (Figure 1B). These results clearly show that all BnMYB69 genes isolated here belong to S21 MYBs and are orthologous genes of AtMYB69.
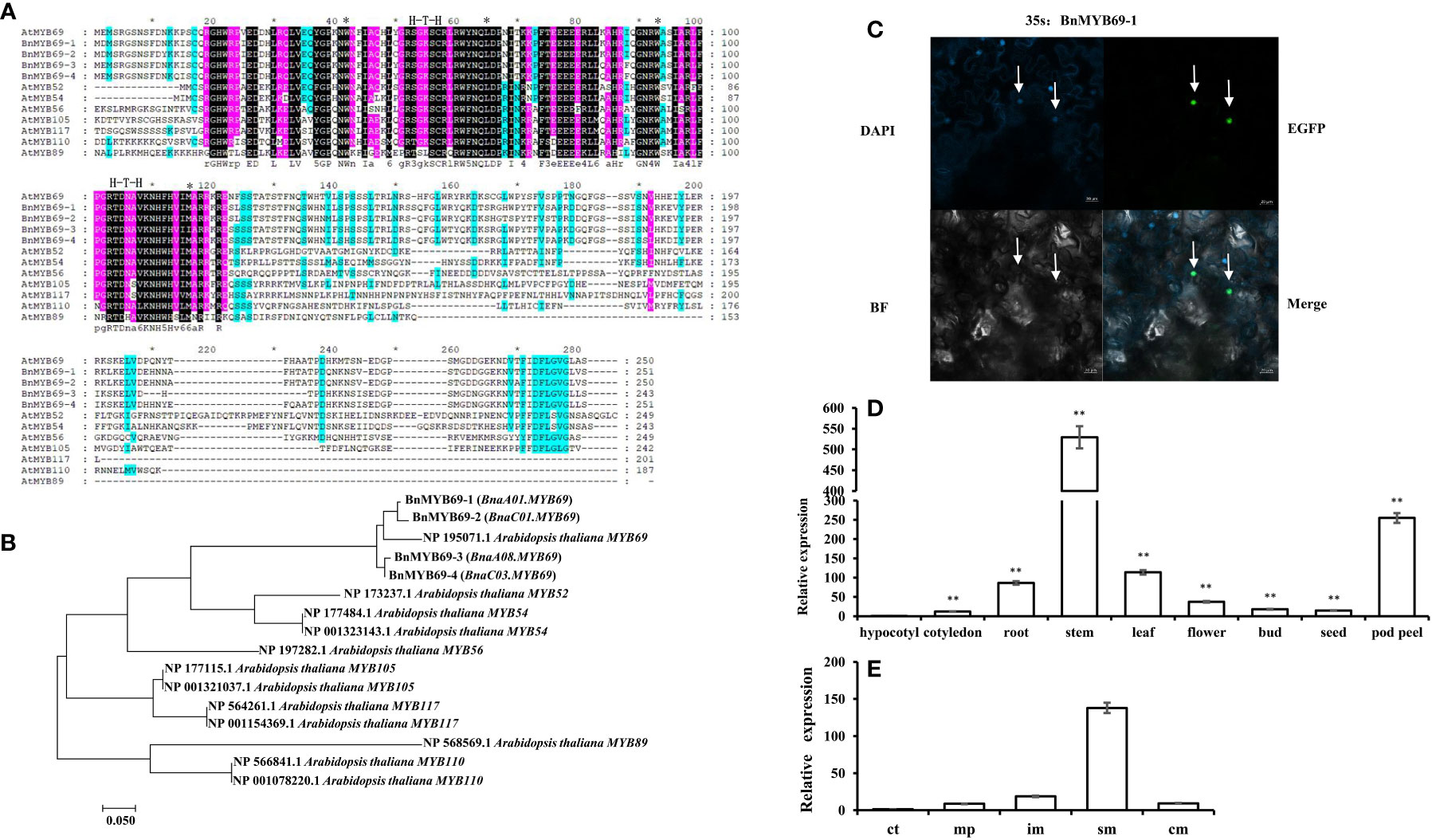
Figure 1 Cloning and analysis of BMYB69s. (A) Sequence alignment analysis of AtMYB69 and S21-AtMYB proteins. (*Between them are amino acid residues with H-T-H motif structure) (B) Molecular phylogenetic analysis by maximum likelihood method of BnMYB69s with S21 AtMYBs. (C) Subcellular localization of BnMYB69-1. (The arrow refers to the nucleus) (D) BnMYB69s overall expression in different organs of B. napus. (E) Expression of BnMYB69s in different parts of the main stem of B. napus. Note, (ct) growth cone tip. (mp) main-stem primordium. (im) initially lignified main-stem, (sm) semi-lignified main-stem. (cm) completely lignified main-stem. At least three biological replications (n≥3) were conducted for each experiment. All values are presented as the mean ± standard deviation (SD). Asterisks indicate significant or extremely significant differences from the first series (*0.01<p < 0.05; **p< 0.01) using one-way ANOVA.
To confirm the subcellular localization of BnMYB69s, experimental validation was carried out. Consistent with the bioinformatics prediction and its predicted role as a transcription factor, the expression of enhanced green fluorescent protein (EGFP)-labeled BnMYB69-1 and blue fluorescent nucleic acid dye (DAPI) coincided in tobacco leaves, showing that they were both localized in the nucleus (Figure 1C).
In qRT-PCR results, overall BnMYB69 expression could be detected in all organs of B. napus but varied greatly among them. It was highly expressed in stems, moderately expressed in pod peels, leaves and roots, and lowly expressed in seeds, flowers, buds, cotyledons, and hypocotyls (Figure 1D). At bolting stage, different stem sections were detected for BnMYB69 expression, which showed an increase trend in parallel with the stem vascularization/lignification process, with little expression in growth cone tip and highest expression in semi-lignified main-stem. However, its expression in the completely lignified main-stem dropped down (Figure 1E). We also detected the organ-specificity of the four individual member genes, and all of them showed typical stem-specific expression patterns (Figure S3). Overlapping result of the four genes’ expression patterns was not perfectly coincided to the overall expression pattern, suggesting that there might be other member genes that have not been cloned in this study. We further checked the BLAST results, and found that the B. napus genome might contain another two BnMYB69 genes beyond these four genes.
Compared with the WT plants, the BnMYB69i plants had larger leaves, a higher chlorophyll content, heavier 1000-seed weight and shorter height when grown in the field (Figures 2, 3). The leaf area of transgenic plants at the bolting stage was significantly larger than that of the wild type (Figure 2C). The dry weight of BnMYB69i plants at the early bolting stage, full bloom stage and harvest stage increased by 36.4%, 44.5% and 53.2%, respectively, compared with those of the WT (Figure 2D). The 1000 seed weight of transgenic plants increased by 7.35% to 55.57% compared with that of WT plants (Figure 2E).
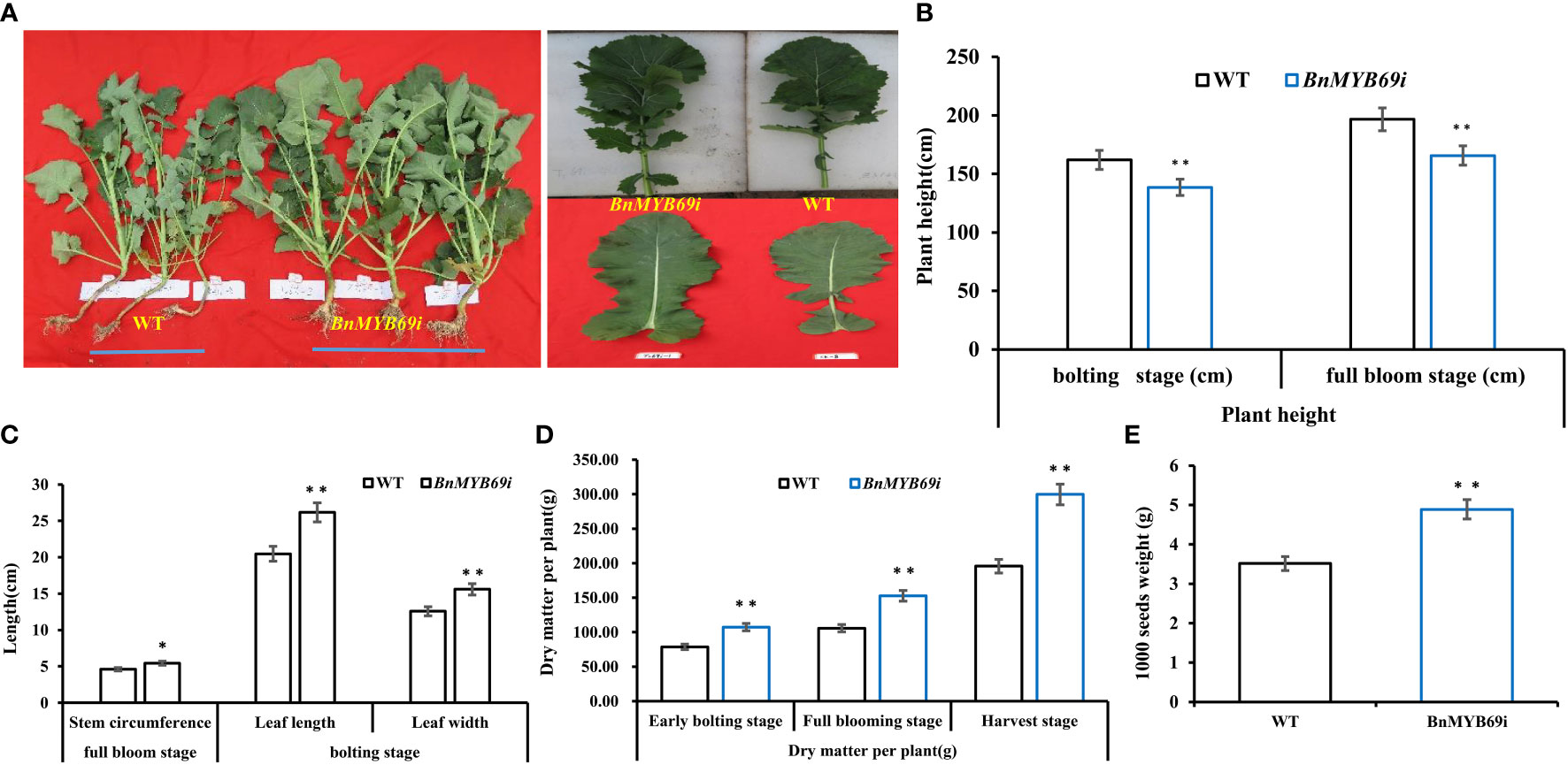
Figure 2 Morphological observation and dry matter weight of transgenic and WT plants. (A) Comparison of plant morphology between transgenic and WT plants in each generation. (B) Comparison of plant height of transgenic and WT plants at different stages. (C) Comparison of stem circumference and leaf of transgenic and WT plants. (D) Comparison of dry matter weight of transgenic and WT plants at three growth stages. (E) Comparison of 1000 seeds weight of transgenic and WT plants. Note, (WT) wild type Brassica napus cv. Zhongshuang 10 (BnMYB69i) BnMYB69 RNA interference rapeseed (the same below). Asterisks indicate significant or extremely significant differences from WT.
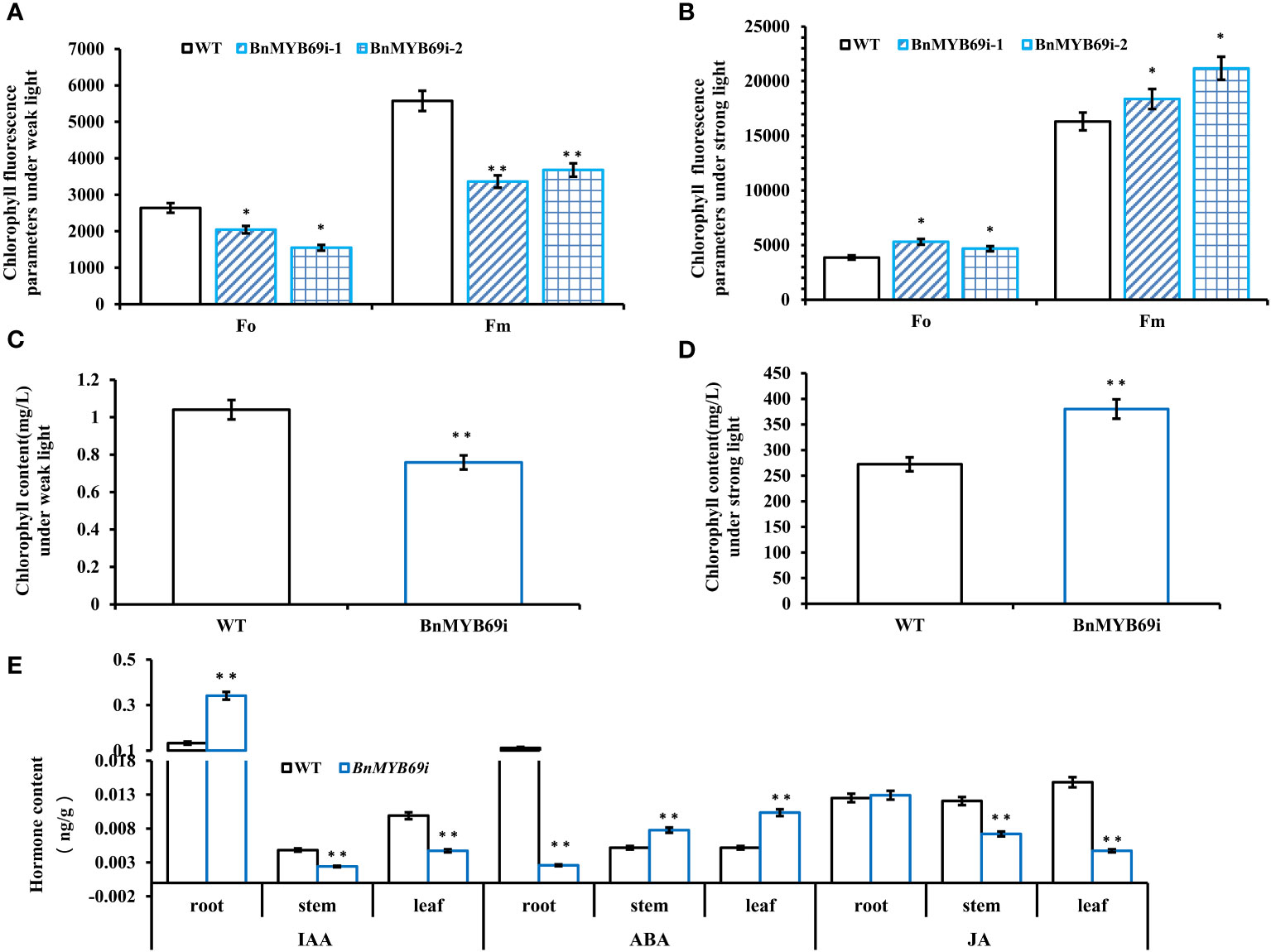
Figure 3 Analysis of chlorophyll content in different materials under different light intensities. (A) Chlorophyll fluorescence parameters under weak light. (B) Chlorophyll fluorescence parameters under strong light. (C) Chlorophyll content under weak light. (D) Chlorophyll content under strong light. (E) Comparison of three hormones in different organs between transgenic and WT plants. (root) hormone content of root. (stem) hormone content of stem. (leaf) hormone content of leaf. Asterisks indicate significant or extremely significant differences from WT (*0.01≤p < 0.05; **p < 0.01) using one-way ANOVA.
The chlorophyll fluorescence analysis showed that BnMYB69-silenced plants were very sensitive to light intensity (Figures 3A, B). Furthermore, the analysis of the chlorophyll content of BnMYB69i plants under different light conditions showed that chlorophyll synthesis in BnMYB69i plants may be related to light intensity (Figures 3C, D).
The hormone auxin (IAA) and 6-deoxocastasterone (6DS) in stems decreased significantly (Tables 1A, B); in contrast, abscisic acid (ABA) and brassinolide (BL) increased significantly (Tables 1A, B). In addition, IAA decreased and ABA increased in both stems and leaves, but the opposite was true in roots (Figure 3E). These results may explain the changes in morphological characteristics of transgenic plants.
BnMYB69 is mainly expressed in vascular organs, particularly xylem (Figure 4A). BnMYB69i plants showed a phenotypic change related to vascular organ growth retardation (Figure 4C). The stem circumference of the transgenic plant was significantly larger than that of the WT (Figure 2C), and the staining of xylem and the vascular bundle showed that the total number of xylem and vascular bundles in the cross-section of the stem increased (Figures 4Cb, c, e, f), but the number per unit area decreased, and this structural change caused the stem to become loose and soft (Figures 4B, C). The xylem in the longitudinal section of the stem decreased significantly, and the width was only 30% of that of the WT (Figures 4B, Ci, l). The number of pith cells increased, and the number per unit area increased both horizontally and vertically (Figures 4Ca, d, i, l). In addition, compared with the WT, the root xylem of transgenic plants was stained deeper, indicating that the lignin content was increased (Figure 4E).
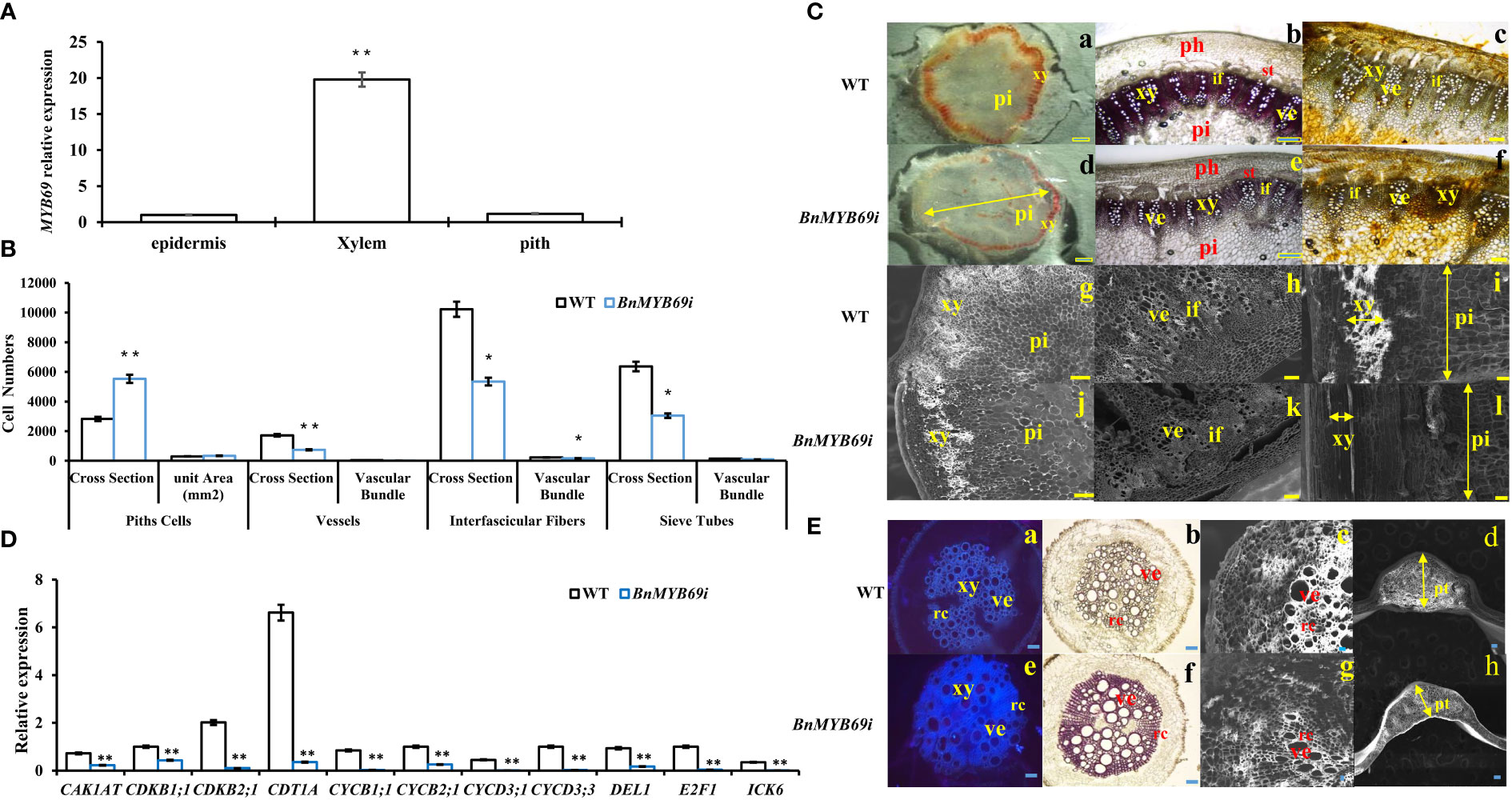
Figure 4 BnMYB69s Expression and analysis in stem tissues and microscopic observation of stem and root. (A) Expression of BnMYB69 in different parts of main stem of rapeseed. (B) Number of cells of piths, sclerenchyma, vessels, interfascicular fibers and sieve tubes of stems cross-section of BnMYB69I transgenic plants. Cross Section is the whole cross section for Pith Cells, and is a microscopic vision of the cross section for Vessels, Interfascicular Fibers and Sieve Tubes. (C) Histochemical staining and electron microscopic observation of cross-section of stem. (a-c) Histochemical staining of stems of WT. (d-f) Histochemical staining of stems of BnMYB69 silenced plants. (a, d) Mäule staining of lignin in whole cross-sections. (b, e) Phloroglucinol-HCl staining of lignin in partial cross-sections. (c, f) Hydroxylammonium staining of pectin in partial cross-sections. (g-h) Electron microscopic cross-section of stems of WT. (i) Electron microscopic longitudinal section of stem of WT. (j-k) Electron microscopic cross-section of stems of BnMYB69 silenced plants, (l) Electron microscopic longitudinal section of stem of BnMYB69 silenced plants. pi, Piths; xy, Xylem; ph, Phloem; ve, Vessel; if, Interfascicular fibers; st, Sieve tube. Bar, 2 mm (a,d), 1 mm (b, c, e, f, g-l). (D) The cell cycle related genes expression in stems of transgenic plant and WT. (E) Comparison of root, petiole and leaf morphology between transgenic and WT. (a, e) Fluorescent brightener dyeing of lignin in whole root cross-sections, (b, f) Phloroglucinol-HCl staining of lignin in whole root cross-sections, (c, g) Electron microscopic observation of root partial cross-sections, (d, h) Electron microscopic observation of petiole cross-section of transgenic and control plants. xy, Xylem; ve, Vessel; rc, Ray cell; pt, Petiole thickness. Bar, 1 mm (a, b, e, f), 200 μm (c, g),1 mm (d, h). Asterisks indicate significant or extremely significant differences from WT (*0.01≤ p < 0.05; **p< 0.01) using one-way ANOVA.
Statistical analysis showed that the number of pith cells in the unit area of transgenic plant stems increased by 14%, and the size and number of cells in cross section changed significantly (Figure 4B). In transgenic plants, the single xylem layer was loose and was approximately 43% thinner than that in the WT (Figure 4B), and the number of vessels in the vascular bundle of a microscopic vision of the cross section was also reduced by approximately 57% (Figure 4B). More significantly, the number of interfascicular fibers of transgenic plants in the cross-section decreased by 47.7%, and the number of interfascicular fibers between two vascular bundles decreased by 30% (Figure 4B). In addition, the number of pith cells in the cross-section of stem of transgenic plants increased extremely significantly by 95.6% (Figure 4B). The total number of vascular bundles in a microscopic vision of the cross section decreased significantly by 50.1% (Figure 4B). Furthermore, the sieve tube number in a microscopic vision of the cross section also decreased significantly in BnMYB69i plants (Figure 4B). Due to the changes in the number and size of xylem and pith cells in the cross-section of the stem, the cell cycle-related genes of the stem were analyzed by RT-PCR. The cell cycle-related genes CAK1AT, CDKB1;1, CDKB2;1, CDT1A, CYCB1;1, CYCB2;1, CYCD2;1, CYCD3;1, CYCD3;3, DEL1, E2F1 and ICK6 were significantly downregulated (Figure 4D).
Because of the changes in the stem xylem of transgenic plants, the resistance to S. sclerotiorum and stem strength were tested. After S. sclerotiorum inoculation, BnMYB69 expression was upregulated at 3 and 9 h, distinctly upregulated at 24 h, and returned to a slightly higher than base level at 48 h, showing a dynamic change during the interaction between B. napus and S. sclerotiorum (Figure 5). This implies that the BnMYB69 gene family may be involved in the interaction with S. sclerotiorum in oilseed rape. BnMYB69i plants were evaluated for decreasing S. sclerotiorum resistance. The stem and leaf lesion sizes were increased by 81.94% and 65.91%, respectively, compared with those of the WT, implying a substantial decrease in resistance to S. sclerotiorum stem rot disease (Figures 5B, C). It is clear that the suppression of BnMYB69 significantly but solely weakened the resistance to S. sclerotiorum in vascular tissues in which innate BnMYB69 is dominantly expressed.
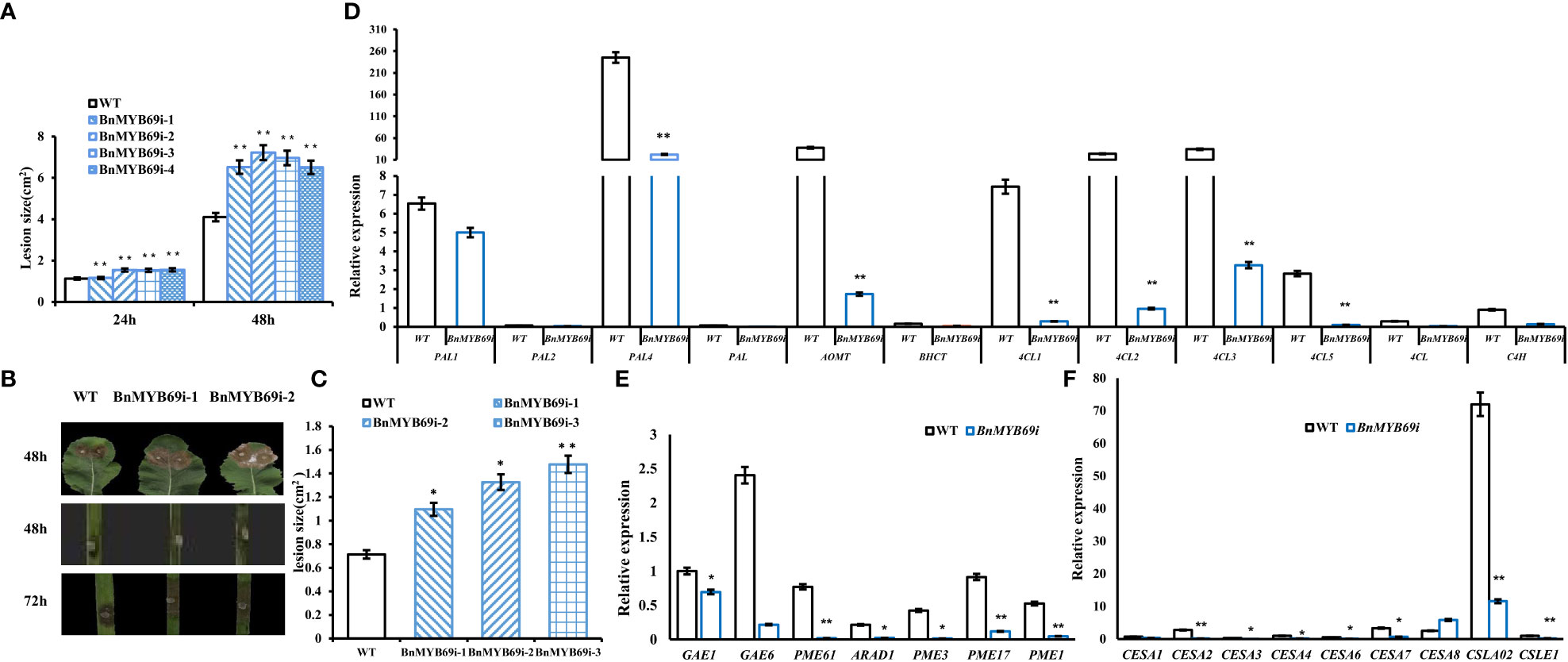
Figure 5 Expression of BnMYB69s and secondary wall biosynthesis genes in B. napus leaf and stem after S. sclerotiorum inoculation. (A) Statistics of leaf lesion area at different times after inoculation. (B) Lesion plaques on leaves and stems after S. sclerotiorum inoculation. (C) Statistics of stem lesion area at 48h after inoculation. (D) Lignin biosynthesis related genes expression in stems of transgenic plant and WT. (E) Pectin biosynthesis related genes expression in stems of transgenic plant and WT. (F) Cellulose biosynthesis related genes expression in stems of transgenic plant and WT. Asterisks indicate significant or extremely significant differences from WT (*0.01≤ p < 0.05; **p< 0.01) using one-way ANOVA.
In addition, the breaking resistance of the upper, middle and lower stems of transgenic plants was decreased by 47.31%, 11.93% and 37.65%, respectively, with an average value of 32.3% for the entire stem (Table 2), which indicates that the stem is fragile and the lodging resistance is reduced. In the stems of transgenic plants, compared with those of WT plants, the contents of neutral detergent fiber (NDF), acid detergent fiber (ADF) and lignin (ADL) decreased significantly by 17.17%, 13.48% and 30.04%, respectively (Table 3), whereas the contents of protopectin and soluble pectin decreased by 19.75% and 42.4%, respectively (Table 4). There was no significant difference in the pectin content of leaves between transgenic plants and WT plants (Table 4). In contrast, in the roots of transgenic plants, the contents of neutral detergent fiber (NDF), acid detergent fiber (ADF) and lignin (ADL) were significantly increased by 10.8%, 11.5%, and 9.3%, respectively (Table 3).
Rapeseed stems mainly contain S and G lignin types, and we found that the ratio of S/G increased significantly by 7.35% (Table 3), implying G lignin biosynthesis was more serviously impaired than S lignin biosynthesis. However, the S/G ratio of roots in transgenic plants decreased significantly, implying a trend opposite to the stem. These results showed that the silencing of BnMYB69 significantly reduced the differentiation of lignification tissues, the deposition of stem secondary cell wall ingredients (such as lignin, cellulose and protopectin), and the mechanical strength in the stem, with little influence on leaf lignification and an increase in root lignification (Table 3).
The lignin biosynthesis-related genes PAL1, PAL2, PAL4, PAL, C4H, 4CL1, 4CL2, 4CL3, 4CL5, 4CL, COMT, and HCT were downregulated in the stems of transgenic plants (Figure 5D). The pectin biosynthesis-related genes GAE1, GAE6, and ARAD1 were significantly downregulated. Additionally, the pectin methylesterase genes PME1, PME3, PME17, and PME61 were significantly downregulated (Figure 5E). In transgenic plants, the genes CESA1, CESA2, CESA3, CESA4, CESA6, CESA7, CSLE1 and CSLA02, which are related to cellulose biosynthesis, were significantly downregulated (Figure 5F), and only CESA8 was upregulated. The results showed that BnMYB69 regulated the synthesis of lignin, cellulose and pectin, resulting in changes in breaking resistance and disease resistance.
Phenotypic observation of the growth changes of BnMYB69i plants at the cotyledon stage and the three- and five-leaf stages showed that IAA treatment significantly increased the plant height of transgenic plants and inhibited the increase in stem circumference. The BnMYB69i plants recovered to the same size as the WT plants at the seedling stage in the growth chamber (Figures 6A, B).
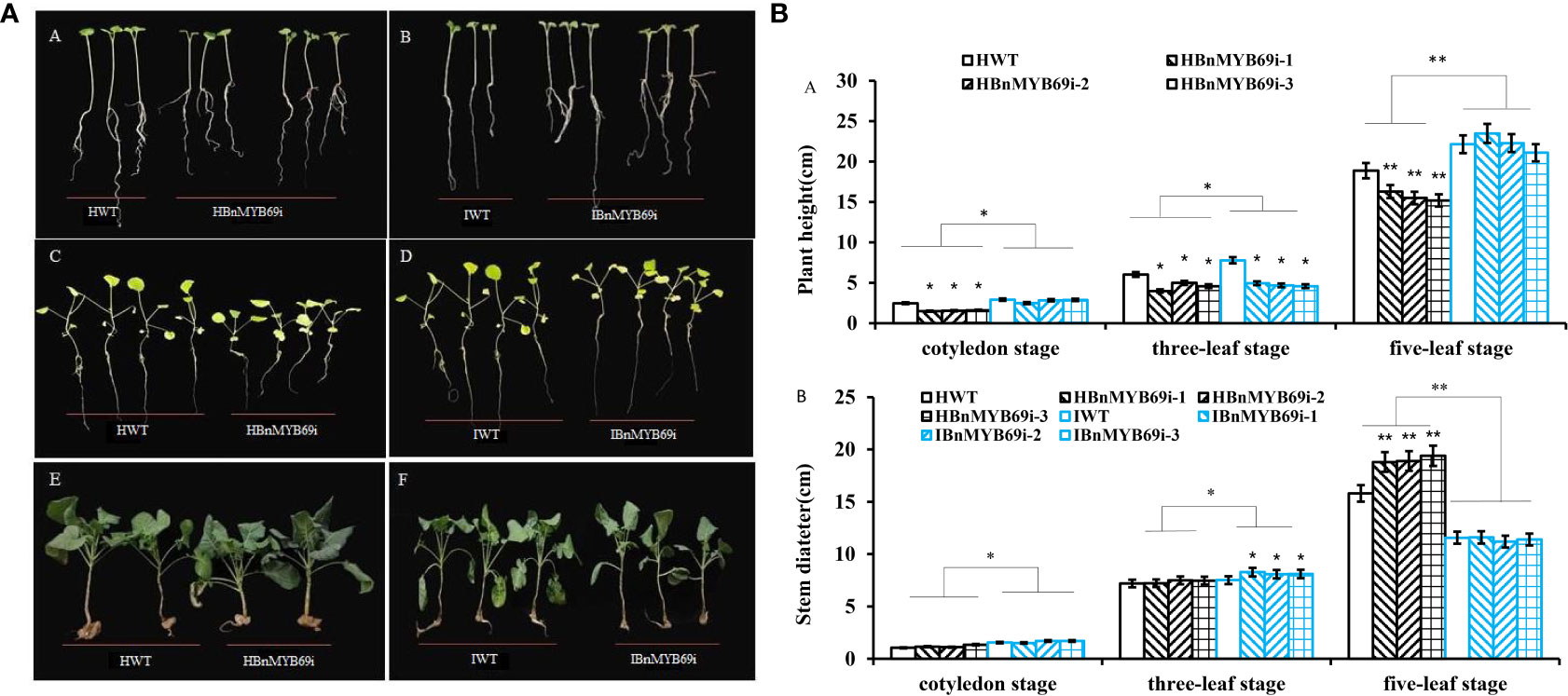
Figure 6 Phenotypic observation of transgenic plants and WT treated with H2O and IAA. (A) Growth of BnMYB69 transgenic plants and WT under different treatments at different stages. (A, B) cotyledon stage. (C, D) three-leaf stage. (E, F) five-leaf stage. HWT, hydroponic WT plant; HBnMYB69i, hydroponic BnMYB69 interfering plant; IWT, WT plant with IAA treatment; IBnMYB69i, BnMYB69 interfering plant with IAA treatment (the same below). (B) Plant height and stem diameter of RNAi plants and WT under different treatments at different periods. (A) Change of plant height. (B) Change of stem diameter. Asterisks indicate significant or extremely significant differences from WT (*0.01≤ p < 0.05; **p< 0.01) using one-way ANOVA.
qRT-PCR analysis of the expression of key genes in the IAA synthesis pathway showed that the expression of the TAA1, YUC2, TAR1, TAR2, CYP79B3, NIT2, AMI1, AAO1 and AAO2 genes decreased significantly in the untreated BnMYB69i plants (P < 0.05), while the expression of CYP79B2 increased significantly (P < 0.05). The expression levels of TAR1, TAR2, YUC2, and NIT2 in BnMYB69i plants treated with IAA were significantly higher than those in untreated plants, but there was no significant difference in TAA1, CYP79B2, or CYP79B3. AMI1, AAO1, and AAO2 were significantly lower than in untreated plants (P < 0.05) (Figure 7A).
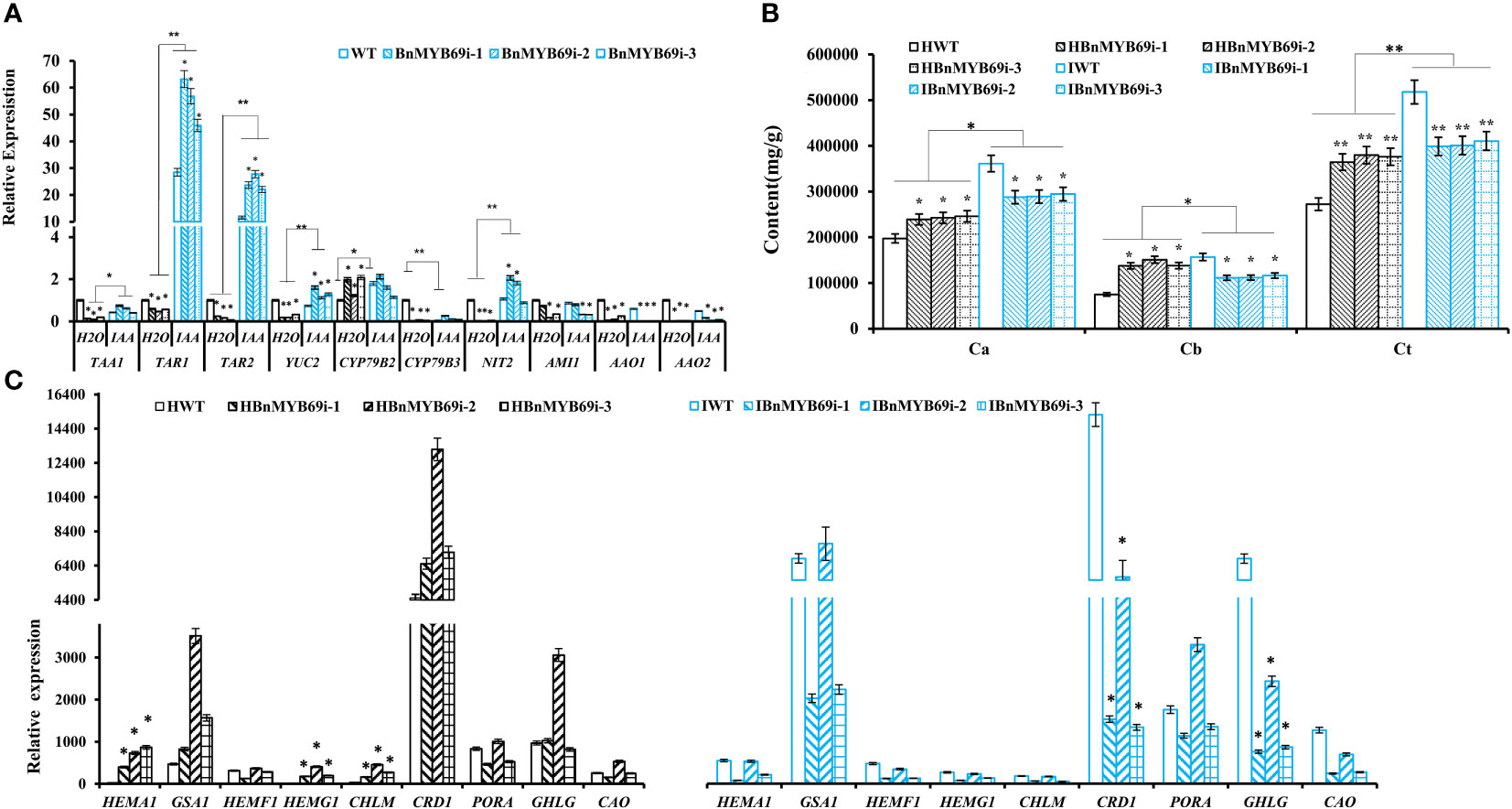
Figure 7 Changes of IAA and chlorophyll synthesis in transgenic and WT plants under different treatments. (A) Expression analysis of genes related to IAA synthesis pathway in transgenic and WT plants under different treatment. (B) Analysis of chlorophyll content difference of BnMYB69 transgenic plants under different treatments. Ca, chlorophyll a; Cb, chlorophyll b; Ct, total chlorophyll. (C) Differential expression of key enzymes in chlorophyll synthesis pathway under different treatments. (H2O) Control plant with water instead of IAA. (IAA) plant with IAA treatment (the same below). Asterisks indicate significant or extremely significant differences from WT (*0.01≤ p < 0.05; **p< 0.01) using one-way ANOVA.
The chlorophyll content of various lines under strong light was determined. The results showed that the total chlorophyll (Ct) of BnMYB69i plants increased significantly under strong light with an increase mainly in leaf chlorophyll a and a decrease in chlorophyll b (Figure 7B).
The expression of chlorophyll synthesis-related genes under IAA treatment was checked with qRT-PCR (Figure 7C). The expression levels of HEMA1, HEMG1, CHLM, CRD1, and CHLG in transgenic plants were significantly downregulated after treatment (P < 0.05), but the expression levels of GSA1, PORA and CAO were significantly upregulated in the transgenic plants (P < 0.05) (Figure 7C). There was no significant difference in HEMF1 gene expression (Figure 7C). It was also shown that BnMYB69 probably regulates the gene expression of HEMA1, HEMG1, CHLM, CRD1, and CHLG in some way.
Our above results indicated that BnMYB69 is involved in the regulation of various metabolites, such as lignin, cellulose, pectin, phytohormones, and chlorophylls. The upstream reasons should be diverse and complicated, in this study we first detected the changes of shikimate-related pathways, because shikimate is directly upstream to the lignin pathway which is the major target influenced by BnMYB69 downregulation.
qRT-PCR showed that the expression of key genes of the glycolytic pathway (HKL1 and PKT7) was significantly upregulated in transgenic plants, and IAA treatment completely mitigated this effect (P < 0.01) (Figure 8A). The expression of key genes of the pentose phosphate pathway (GP6D5 and tktA) (Figure 8B) and shikimic acid pathway (ADT2) were significantly downregulated in BnMYB69i plants (P < 0.01) (Figure 8C). IAA treatment completely recovered expression levels of ADT2 and GP6D5, over-compensated the expression of DHD/SDH and SK2, but had little recovery on tktA (Figure 8C). These results show that the pentose phosphate pathway and shikimic acid pathway, in coordination with the upstream glucose metabolic pathway, are most likely to be regulated by BnMYB69 together with IAA signaling, and different loci have differed responses.
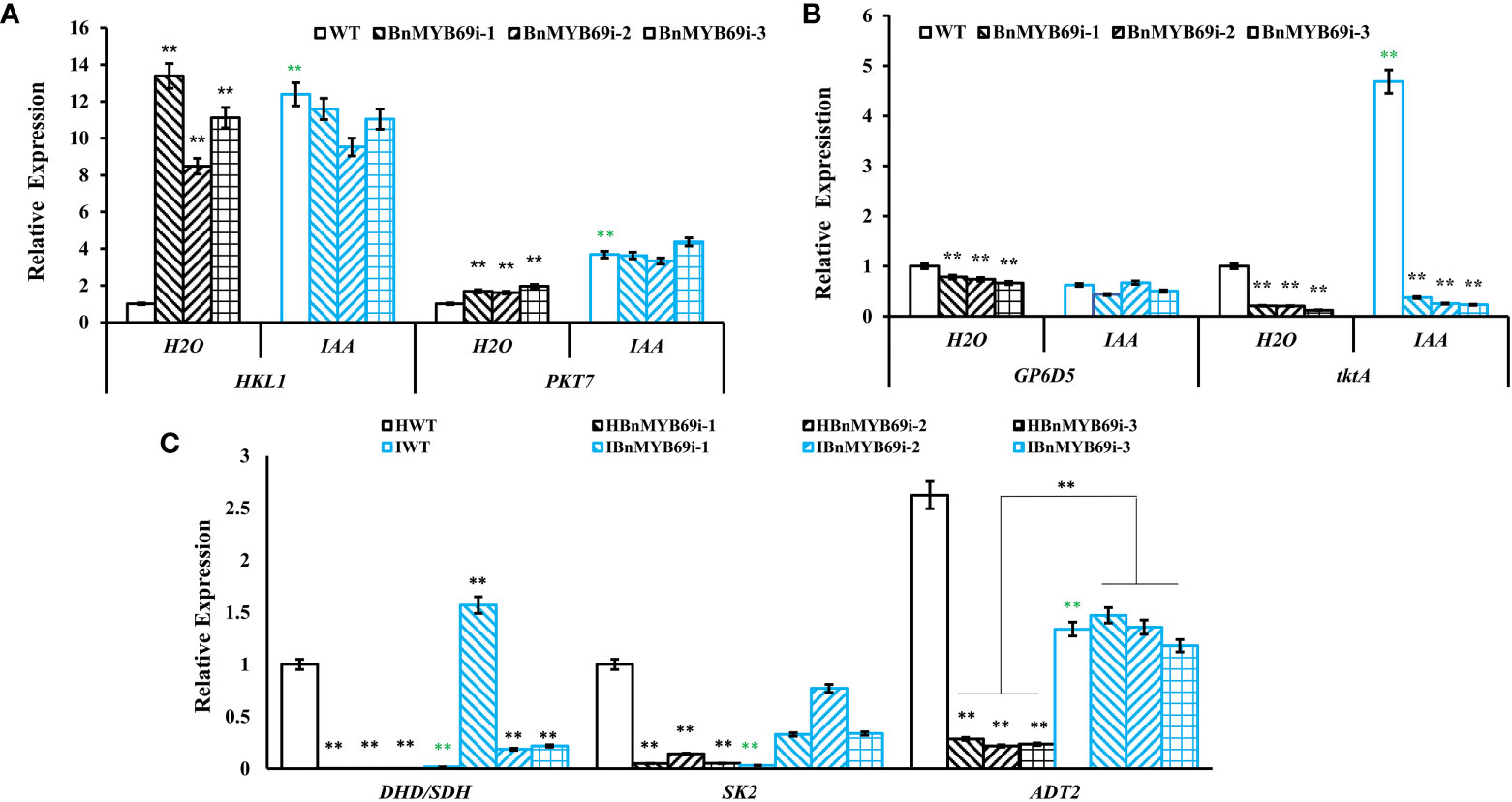
Figure 8 Expression of key genes of related metabolic pathways under different treatments. (A) Expression of key genes of glycolysis pathway in different materials. (B) Expression of key genes of pentose phosphate pathway in different materials. (C) Expression of DHD/SDH, SK2 and ADT2 in shikimate pathway in different materials. Asterisks * or ** indicate significant or extremely significant differences from HWT (*0.01<p < 0.05; **p< 0.01) using one-way ANOVA.
Downstream the shikimate pathway, flavonoid-proanthocyanidin (PA) pathway is neighboring to lignin pathway and these two pathways have competition in metabolic flux. Thus, we detected the influence of BnMYB69 downregulation on the flavonoid-PA pathway. Like lignin content, the PA content in the leaves of BnMYB69i plants was also significantly lower than WT, and IAA treatment exerted a little compensation (Figure 9A). It was confirmed that shikimates, lignins, and PAs are all decreased by BnMYB69 downregulation.
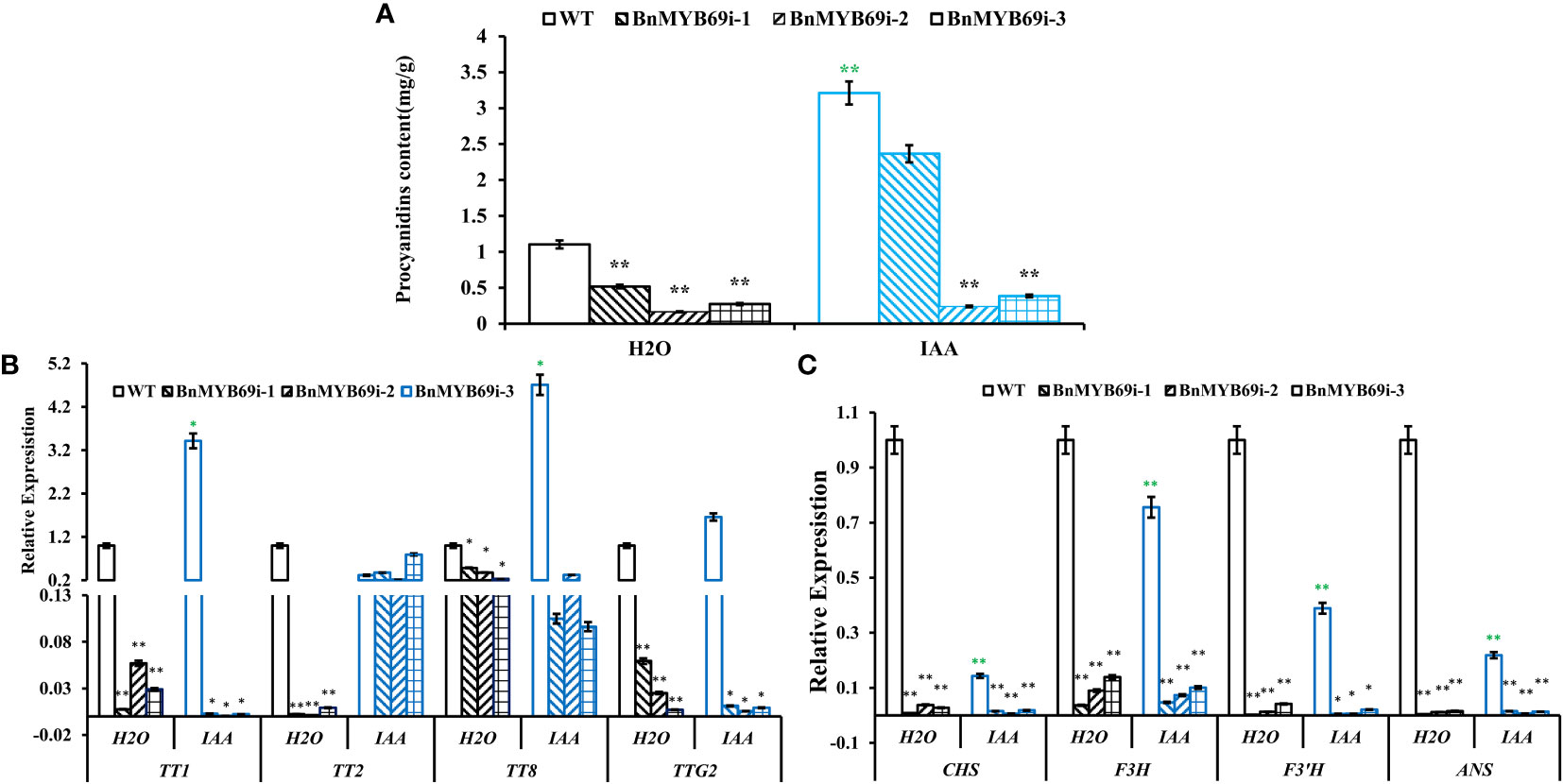
Figure 9 Changes of proanthocyanidin (PA) synthesis genes in transgenic and WT plants under different treatments. (A) Changes of PA content under different treatments. (B, C) Expression of PA synthesis related genes of different materials under different treatments. Asterisks indicate significant or extremely significant differences from WT (*0.01≤ p < 0.05; **p< 0.01) using one-way ANOVA.
The expression of key genes involved in PA biosynthesis was also analyzed using qRT-PCR (Figures 9B, C). In BnMYB69i plants, expression levels of core flavonoid biosynthesis genes CHS, F3H and F3’H, anthocyanin biosynthesis gene ANS, and PA pathway regulatory genes TT1, TT2, TT8 and TTG2 were all significantly lower than those of WT plants (P < 0.05). IAA treatment did not exert recovery effect on most of these loci, but TT2 expression was completely recovered. These results showed that BnMYB69 regulated the shikimate-flavonoid-PA compound pathway, independent of or downstream IAA signaling, but IAA is involved in BnMYB69 regulation on TT2 expression.
The R2R3-MYB family is the largest and most widely studied family of MYB transcription factors. In addition to the similar functions of 1R-MYB family members, the members of this family are widely involved in plant life processes such as primary metabolism and biological and abiotic stress responses (Ito, 2005). Researchers believe that most members of the R2R3 MYB protein family are involved in regulating plant-specific physiological and biochemical processes such as plant secondary metabolism and cell morphogenesis (Wan and Li, 2002). In addition, many members of the R2R3 MYB gene family isolated from different species regulate anthocyanin biosynthesis (Czemmel et al., 2009; Machado et al., 2009; Feng et al., 2010; Lin-Wang et al., 2011). However, most MYB transcription factors have been cloned, analyzed and studied in Arabidopsis, some researchers have found that MYB TFs play vital roles in plant development and anthocyanin metabolism, and PAP1/2 can promote the expression of anthocyanin biosynthesis genes (Muleke et al., 2021). In fact, MYB transcription factor members play a role in regulating growth and development in different parts of different time periods. For example, AtMYB115 and AtMYB188/PGA37 participate in regulating the seed germination rate at the seed germination stage (Wang et al., 2009); AtMYB30, AtMYB38, and AtMYB18/LAF1 regulate hypocotyl elongation (Yang et al., 2009; Froidure et al., 2010); and AtMYB21, AtMYB24, AtMYB26, ATMYB35, AtMYB57, GhMYB80, and AtMYB99 participate in anther development (Millar and Gubler, 2005; Xu et al., 2014). There are also some MYB transcription factors, such as BnMYB43 and GbMYBR1, involved in lateral organ development and stem morphogenesis, which regulate the number of plant branches and the development of plant higher organs (Jiang et al., 2020; Su et al., 2020).
In this study, four BnMYB69 genes were isolated, characterized, and functionally elucidated. Results of systemic analysis on the structure and homology features of their genes and proteins all indicated that they encoded a S21 R2R3-MYB transcription factor, orthologous to AtMYB69. There are few reports on the function of MYB69. Previous studies prove that AtMYB69 is a transcriptional activator dedicated to secondary cell wall thickening of fiber cells (Dubos et al., 2010). According to other reports on the function of MYB transcription factors similar to ATMYB69 protein, these transcription factors are not only related to secondary wall synthesis (Zhong et al., 2008; Kim et al., 2012), but also related to anthocyanin metabolism (Peng et al., 2019; Xu et al., 2021), plant resistance (Xu et al., 2020; Im et al., 2021), etc. Therefore, systematic study of the function of MYB69 is helpful to providing insight into the biofunction of MYB gene family.
The MYB69 transcription factor is related to xylem formation and is strongly expressed in stems. Many researchers have found that the expression of key genes in lignin synthesis is related to lodging traits, particularly COMT (Ma, 2009; Li et al., 2013), F5H (Li and Qi, 2011; Li et al., 2013), 4CL (Li and Qi, 2011; Huang et al., 2013; Li et al., 2013) and PAL (Huang et al., 2013). Our results showed that silencing BnMYB69 reduced the lodging resistance of rapeseed. Cytological observation of cross-sections and longitudinal sections of stems and the expression of key genes of lignin metabolism in transgenic plants revealed a positive correlation with lignin synthesis. This shows that BnMYB69 is an activator of lignin synthesis in B. napus.
In addition to lignin, plant cell wall components also contain cellulose and pectin. Cellulose and pectin content analysis showed that the cell wall of BnMYB69i plants developed poorly. This might be the reason that the resistance to S. sclerotiorum in BnMYB69i plants was significantly decreased compared with that in WT plants.
The effect of BnMYB69s on roots is quite different from or even opposite to its effect on stems, particularly on monolignol types. The proportion of S/G in BnMYB69i plants increases in stems and decreases in roots, which may contribute to the different physical characteristics of the roots and the stems. Furthermore, lignin, cellulose and pectin increased in roots, though decreased in the stems, indicating that there was a significant difference between the aboveground and underground parts of BnMYB69i plants. Since the basic difference between aboveground and underground is dark or in light, this phenomenon implies that BnMYB69i functioning depends on light condition.
The offspring of BnMYB69i were generally shorter than those of WT plants. Pith cells and unit volume were greater based on electron microscopy and staining, and the expression of most cell cycle-related genes in the stems of transgenic plants was significantly lower than that in the stems of WT plants. This indicates that cell division in transgenic plants was not vigorous and rapid, and the total number of cells should be reduced, which was consistent with the results of cells in the previous cross-cut stems. In addition, the IAA content in the stems of transgenic plants was lower than that of WT plants, which confirmed that plant cell division was not vigorous. Furthermore, the increase in ABA content led to plant dwarfing and root development changes. Antisense transgenic plants and overexpression plants of the rice dwarf gene OsDWARF48 were analyzed, and the IAA content of the dwarf plants was lower than that of WT plants (Li et al., 2018).
qRT-PCR results confirmed that auxin can also promote cell division by degrading the expression of CDK inhibitor protein Kip-related proteins (KRPs). The statistical results of the number of cells (Figure 4B) in the stems of transgenic plants also show that the change in cell division caused a change in the number of xylem cells and then caused a change in stem height and stem circumference. Since BnMYB69 is mainly expressed in tissues under lignification, thus it is logical that the confinement of BnMYB69 downregulation on cell cycle and cell division is mainly on the secondary growth and plant height, and the promotion effect on parenchyma cells, lateral growth and total biomass might be because of more flux branch-off from suppressed secondary metabolism.
The results of the hormone content tests on the stems and leaves of the offspring of transgenic plants showed that the content of IAA was significantly lower than that of WT, indicating that BnMYB69 affected the synthesis of IAA. The precursor of IAA is tryptophan, which regulates the growth, development and resistance of plants. Researchers have found that expression of the BnMYB193 gene in B. napus was also induced by IAA, ABA and other hormones to promote the development of lateral roots of rapeseed and improve the lodging resistance of plants (Wu et al., 2018).
In the IAA compensation experiment, BnMYB69 affected the shikimic acid pathway upstream of IAA synthesis. In addition, the pentose phosphate pathway upstream of the shikimic acid pathway was also affected by BnMYB69. It can be inferred that BnMYB69 has regulatory functions on metabolic pathways since sugar metabolism, although sugar synthesis is affected by photosynthesis. The up and down regulation of the hexokinase gene in plants leads to extensive plant phenotypic changes such as overexpression of AtHKL1 in tomato, inhibition of plant growth, reduction of chlorophyll content and photosynthetic rate in leaves, and photochemical quantum efficiency reduction (Dai et al., 1999). Inhibiting AtG6PD5 expression causes plants to have short stems and leaves, curly leaves and light leaf color (Wang, 2013). BnMYB69i transgenic plants indeed had lower plant height, which also confirmed the regulatory function of BnMYB69.
In a study of B. napus, TT1 gene silencing led to the downregulation of flavonoid pathway genes such as TT3, TT4, TT5, TT6, TT7 and TT18. The interference of BrTT1 gene expression in yellow seed B. napus causes the abnormal or absent expression of several important BrTT1 genes in the flavonoid biosynthesis pathway, which then blocks the accumulation of PAs resulting in a transparent seed coat. The BnTT1 gene in B. napus also regulates the PA and flavonoid metabolism pathways (Lian et al., 2017).
ANS is the most critical gene in the anthocyanin biosynthesis pathway. UFGT and ANS cooperate to transform unstable anthocyanins into stable anthocyanins in the cytoplasm and vacuole. Among the five genes of the anthocyanin biosynthesis pathway, only ANS and DFR are regulated by TTG2. Studies on tobacco show that TTG2 and ARF8 work together to control the degree of flower color by regulating the expression of ANS and DFR in the anthocyanin biosynthesis pathway (Li et al., 2017). ARF8 relies on TTG2 to regulate the expression of ANS and DFR, which affects anthocyanin accumulation. TTG2 inhibition prevents plant trichome formation, anthocyanin accumulation and seed color pigmentation. Similarly, AtTT8 is also involved in the regulation of anthocyanin and PA biosynthesis in Arabidopsis (Baudry et al., 2006; Xu et al., 2013). Complementation analysis in Arabidopsis TT8 mutant showed that NnTT8 in Nelumbo nucifera could function similarly to AtTT8 in regulating anthocyanin and proanthocyanin biosynthesis (Deng et al., 2021). The downregulated expression of TT1, TTG2, TT8, ANS and DFR in BnMYB69i transgenic plants also confirmed its impact on flavonoid compounds and the anthocyanin synthesis pathway, which explains the increase in surface trichomes of transgenic plants.
The entire process of chlorophyll synthesis requires 15 enzymatic reactions. Among them, HEMA is the coding gene that catalyzes ALA synthesis. The HEMA family has three members in Arabidopsis, and the expression of HEMA1 is induced by light (McCormac et al., 2001), whereas HEMA2 and HEMA3 are not induced by light, and the HEMA3 gene is not even expressed. However, the HEMA1 gene is expressed in all plant tissues, and its expression is higher in photosynthetic tissues (Tanaka et al., 1996). CHLM and CRD can catalyze the transformation of protoporphyrin IX into chlorophyll ester a, and knock out of CRD in Arabidopsis resulted in slow plant growth and development and chlorosis of leaves (Bang et al., 2008). CHLG acts to form chlorophyll a, and CAO catalyzes Chla oxidation to form Chlb. Under strong light, chlorophyll synthesis increases, and the PSII reaction intensifies, such that the expression of the two coding genes increases. The PORA gene, a member of the POR family, is a coding gene for protochlorophyll ester oxidoreductase. The PORA gene has a photoinduction effect, and it is also the main enzyme of plant leaf chlorophyll accumulation and the only confirmed gene in the phytochrome A-mediated mechanism for the greening blocks (Sperling et al., 1997). Some researchers have shown that PORA overexpression is sufficient to guide the synthesis of a large amount of chlorophyll and the development of normal plants in the absence of PORB and PORC (Paddock et al., 2010). However, the expression of PORA decreased in that experiment. It was speculated that PORA was affected by the BnMYB69 gene, and the expressions of PORB and PORC may be increased. Mechanism of BnMYB69 effect on chlorophyll synthesis and light reaction will be deeply studied in the future.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
YC: Designing the research, analyzing some data, and revising the manuscript. NL: Designing and doing some experiments, analyzing most data, and writing the original manuscript. MW and JJ: Doing some experiments. QZ and JY: Providing writing suggestions. JLi: Providing some plant material. JLian and YX: Providing some technologies of the experiment. All authors: reading this manuscript and agreeing with publication. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (NSFC, with grant numbers of 31701335, 31171177 and 32272015).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1157836/full#supplementary-material
Baldoni, E., Genga, A., Cominelli, E. (2015). Plant MYB transcription factors: Their role in drought response mechanisms. Int. J. Mol. Sci. 16 (7), 15811–15851. doi: 10.3390/ijms160715811
Bang, W. Y., Jeong, I. S., Kim, D. W., Im, C. H., Ji, C., Hwang, S. M., et al. (2008). Role of Arabidopsis CHL27 protein for photosynthesis, chloroplast development and gene expression profiling. Plant Cell Physiol. 49 (9), 1350–1363. doi: 10.1093/pcp/pcn111
Baudry, A., Caboche, M., Lepiniec, L. (2006). TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 46 (5), 768–779. doi: 10.1111/j.1365-313X.2006.02733.x
Blumenkrantz, N., Asboe-Hansen, G. (1973). New method for quantitative determination of uronic acids. Anal. Biochem. 54 (2), 484–489. doi: 10.1016/0003-2697(73)90377-1
Cardoza, V., Stewart, C. N. (2003). Increased Agrobacterium-mediated transformation and rooting efficiencies in canola (Brassica napus L.) from hypocotyl segment explants. Plant Cell Rep. 21 (6), 599–604. doi: 10.1007/s00299-002-0560-y
Czemmel, S., Stracke, R., Weisshaar, B., Cordon, N., Harris, N. N., Walker, A. R., et al. (2009). The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol. 151 (3), 1513–1530. doi: 10.1104/pp.109.142059
Dai, N., Schaffer, A., Petreikov, M., Shahak, Y., Giller, Y., Ratner, K., et al. (1999). Overexpression of Arabidopsis hexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence. Plant Cell 11 (7), 1253–1266. doi: 10.2307/3870747
Deng, J., Li, J., Su, M., Lin, Z., Chen, L., Yang, P. (2021). A bHLH gene NnTT8 of Nelumbo nucifera regulates anthocyanin biosynthesis. Plant Physiol. Biochem. 158, 518–523. doi: 10.1016/j.plaphy.2020.11.038
Ding, A., Tang, X., Yang, D., Wang, M., Ren, A., Xu, Z., et al. (2021). ERF4 and MYB52 transcription factors play antagonistic roles in regulating homogalacturonan de-methylesterification in Arabidopsis seed coat mucilage. Plant Cell 33 (2), 381–403. doi: 10.1093/plcell/koaa031
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15 (10), 573–581. doi: 10.1016/j.tplants.2010.06.005
Feller, A., Machemer, K., Braun, E. L., Grotewold, E. (2011). Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 66 (1), 94–116. doi: 10.1111/j.1365-313X.2010.04459.x
Feng, S., Wang, Y., Yang, S., Xu, Y., Chen, X. (2010). Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 232 (1), 245–255. doi: 10.1007/s00425-010-1170-5
Foster, T. J., Ablett, S., McCann, M. C., Gidley, M. J. (1996). Mobility-resolved 13C-NMR spectroscopy of primary plant cell walls. Biopolymers 39 (1), 51–66. doi: 10.1002/(sici)1097-0282(199607)39:1<51::Aid-bip6>3.0.Co;2-u
Froidure, S., Roby, D., Rivas, S. (2010). Expression of the Arabidopsis transcription factor AtMYB30 is post-transcriptionally regulated. Plant Physiol. Biochem. 48 (8), 735–739. doi: 10.1016/j.plaphy.2010.04.012
Gai, J. Y. (2005). Segregation analysis of genetic system of quantitative traits in plants. Yi Chuan (in Chinese) 27 (1), 130–136. doi: 10.1007/s11515-005-0001-8
Godoy, G., Steadman, J. R., Dickman, M. B., Dam, R. (1990). Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris. Physiol. Mol. Plant Pathol. 37 (3), 179–191. doi: 10.1016/0885-5765(90)90010-U
Hajiebrahimi, A., Owji, H., Hemmati, S. (2017). Genome-wide identification, functional prediction, and evolutionary analysis of the R2R3-MYB superfamily in Brassica napus. Genome 60 (10), 797–814. doi: 10.1139/gen-2017-0059
Hong, L., Niu, F., Lin, Y., Wang, S., Chen, L., Jiang, L. (2021). MYB117 is a negative regulator of flowering time in Arabidopsis. Plant Signaling Behav. 16 (5), e1901448. doi: 10.1080/15592324.2021.1901448
Huang, J.-H., LI, W., Qu, C.-M., Liu, L.-Z., Xu, X.-F., Wang, R., et al. (2013). Expression characteristics of key genes in lignin pathway among different lodging resistance lines of Brassica napus L. Acta Agronomica Sin. 39 (8), 1339–1344. doi: 10.3724/sp.j.1006.2013.01339
Im, J. H., Ko, J.-H., Kim, W.-C., Crain, B., Keathley, D., Han, K.-H. (2021). Mitogen-activated protein kinase 6 negatively regulates secondary wall biosynthesis by modulating MYB46 protein stability in Arabidopsis thaliana. PloS Genet. 17 (4), e1009510. doi: 10.1371/journal.pgen.1009510
Ito, M. (2005). Conservation and diversification of three-repeat myb transcription factors in plants. J. Plant Res. 118 (1), 61–69. doi: 10.1007/s10265-005-0192-8
Jiang, J., Liao, X., Jin, X., Tan, L., Lu, Q., Yuan, C., et al. (2020). MYB43 in oilseed rape (Brassica napus) positively regulates vascular lignification, plant morphology and yield potential but negatively affects resistance to Sclerotinia sclerotiorum. Genes (Basel) 11 (5), 581. doi: 10.3390/genes11050581
Jin, H., Martin, C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41 (5), 577–585. doi: 10.1023/a:1006319732410
Kim, W. C., Ko, J. H., Han, K. H. (2012). Identification of a cis-acting regulatory motif recognized by MYB46, a master transcriptional regulator of secondary wall biosynthesis. Plant Mol. Biol. 78 (4-5), 489–501. doi: 10.1007/s11103-012-9880-7
Kranz, H. D., Denekamp, M., Greco, R., Jin, H., Leyva, A., Meissner, R. C., et al. (1998). Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16 (2), 263–276. doi: 10.1046/j.1365-313x.1998.00278.x
Li, P., Chen, X., Sun, F., Dong, H. (2017). Tobacco TTG2 and ARF8 function concomitantly to control flower colouring by regulating anthocyanin synthesis genes. Plant Biol. 19 (4), 525–532. doi: 10.1111/plb.12560
Li, Y., H.i, G., Cun-kou, Q. (2013). Expression of lignin synthesis key genes of F5H,4CL and COMT on lodging resistance of Brassica napusror L. Chin. J. Oil Crop Sci. (in Chinese) 35 (1), 24–28. doi: 10.7505/j.issn.1007-9084.2013.01.004
Li, Y. C., Qi, C. K. (2011). Lignin content and key gene expression in lignin synthesis of Brassica napus L. with lodging resistance. Jiangsu J. Agric. Sci. (in Chinese) 3), 481–487. doi: 10.3969/j.issn.1000-4440.2011.03.004
Li, J. B., Qi, H. X. (2017). The advancement of genetic research for lodging resistance in rice. Hubei Agri. Sci. (in Chinese) 56 (23), 4450–4453. doi: 10.14088/j.cnki.issn.0439-8114.2017.23.005
Li, Z., Wang, Q. G., Pan, J. W., Liu, W. (2018). Functional analysis of height regulation of cyptochrome P450 gene OsDWARF48 in rice. Shandong Agric. Sci. (in Chinese) 50 (11), 1–9. doi: 10.14083/j.issn.1001-4942.2018.11.001
Lian, J., Lu, X., Yin, N., Ma, L., Lu, J., Liu, X., et al. (2017). Silencing of BnTT1 family genes affects seed flavonoid biosynthesis and alters seed fatty acid composition in Brassica napus. Plant Sci. 254, 32–47. doi: 10.1016/j.plantsci.2016.10.012
Lin-Wang, K., Micheletti, D., Palmer, J., Volz, R., Lozano, L., Espley, R., et al. (2011). High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ. 34 (7), 1176–1190. doi: 10.1111/j.1365-3040.2011.02316.x
Ma, Q. H. (2009). The expression of caffeic acid 3-o-methyltransferase in two wheat genotypes differing in lodging resistance. J. Exp. Bot. 60 (9), 2763–2771. doi: 10.1093/jxb/erp132
Machado, A., Wu, Y., Yang, Y., Llewellyn, D. J., Dennis, E. S. (2009). The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J. 59 (1), 52–62. doi: 10.1111/j.1365-313X.2009.03847.x
McCarthy, R. L., Zhong, R., Fowler, S., Lyskowski, D., Piyasena, H., Carleton, K., et al. (2010). The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol. 51 (6), 1084–1090. doi: 10.1093/pcp/pcq064
McCormac, A. C., Fischer, A., Kumar, A. M., Soll, D., Terry, M. J. (2001). Regulation of HEMA1 expression by phytochrome and a plastid signal during de-etiolation in Arabidopsis thaliana. Plant J. 25 (5), 549–561. doi: 10.1046/j.1365-313x.2001.00986.x
Mei, J., Liu, Y., Wei, D., Wittkop, B., Ding, Y., Li, Q., et al. (2015). Transfer of sclerotinia resistance from wild relative of Brassica oleracea into Brassica napus using a hexaploidy step. Theor. Appl. Genet. 128 (4), 639–644. doi: 10.1007/s00122-015-2459-3
Millar, A. A., Gubler, F. (2005). The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are MicroRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17 (3), 705–721. doi: 10.1105/tpc.104.027920
Muleke, E., Wang, Y., Zhang, W.-t., Xu, L., Ying, J.-l., Karanja, B. K., et al. (2021). Genome-wide identification and expression profiling of MYB transcription factor genes in radish (Raphanus sativus L.). J. Integ. Agr. 20 (11), V–V. doi: 10.1016/S2095-3119(20)63308-1
Ogata, K., Morikawa, S., Nakamura, H., Hojo, H., Yoshimura, S., Zhang, R., et al. (1995). Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-myb. Nat. Struct. Biol. 2 (4), 309–320. doi: 10.1038/nsb0495-309
Ogata, K., Morikawa, S., Nakamura, H., Sekikawa, A., Inoue, T., Kanai, H., et al. (1994). Solutin structure of a specific DNA complex of the MYB DNA-binding domain with cooperative recognition helices. Cell 79 (4), 639–648. doi: 10.1016/0092-8674(94)90549-5
Paddock, T. N., Mason, M. E., Lima, D. F., Armstrong, G. A. (2010). Arabidopsis protochlorophyllide oxidoreductase a (PORA) restores bulk chlorophyll synthesis and normal development to a porB porC double mutant. Plant Mol. Biol. 72 (4-5), 445–457. doi: 10.1007/s11103-009-9582-y
Paz-Ares, J., Ghosal, D., Wienand, U., Peterson, P. A., Saedler, H. (1987). The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 6 (12), 3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x
Peng, Y., Kui, L.-W., Cooney, J. M., Wang, T., Espley, R. V., Allan, A. C. (2019). Differential regulation of the anthocyanin profile in purple kiwifruit (Actinidia species). Hortic. Res. 6, 3. doi: 10.1038/s41438-018-0076-4
Sperling, U., van Cleve, B., Frick, G., Apel, K., Armstrong, G. A. (1997). Overexpression of light-dependent PORA or PORB in plants depleted of endogenous POR by far-red light enhances seedling survival in white light and protects against photooxidative damage. Plant J. 12 (3), 649–658. doi: 10.1046/j.1365-313x.1997.00649.x
Su, X., Xia, Y., Jiang, W., Shen, G., Pang, Y. (2020). GbMYBR1 from ginkgo biloba represses phenylpropanoid biosynthesis and trichome development in Arabidopsis. Planta 252 (4), 68. doi: 10.1007/s00425-020-03476-1
Tanaka, R., Yoshida, K., Nakayashiki, T., Masuda, T., Tsuji, H., Inokuchi, H., et al. (1996). Differential expression of two HEMA mRNAs encoding glutamyl-tRNA reductase proteins in greening cucumber seedlings. Plant Physiol. 110 (4), 1223–1230. doi: 10.1104/pp.110.4.1223
Underwood, W. (2012). The plant cell wall: A dynamic barrier against pathogen invasion. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00085
Wan, X. R., Li, L. (2002). MYB proteins in plants. Plant Physiol. Commun. (in Chinese) 38 (2), 165–170. doi: 10.13592/j.cnki.ppj.2002.02.031
Wang, W. Q., Moss, S. M. A., Zeng, L., Espley, R. V., Wang, T., Kui, L.-W., et al. (2022). The red flesh of kiwifruit is differentially controlled by specific activation-repression systems. New Phytol. 235 (2), 630–645. doi: 10.1111/nph.18122
Wang, X. (2013). Cloning and functional analysis of Arabidopsis glucose-6-phosphate 1-dehydrogenase gene (G6PD5). [master's thesis]. [Haerbing]: Heilongjiang University. doi: 10.7666/d.Y2322537
Wang, X., Niu, Q. W., Teng, C., Li, C., Mu, J., Chua, N. H., et al. (2009). Overexpression of PGA37/MYB118 and MYB115 promotes vegetative-to-embryonic transition in arabidopsis. Cell Res. 19 (2), 224–235. doi: 10.1038/cr.2008.276
Wei, X., Shan, T., Hong, Y., Xu, H., Liu, X., Zhang, Z. (2017). TaPIMP2, a pathogen-induced MYB protein in wheat, contributes to host resistance to common root rot caused by Bipolaris sorokinian. Sci. Rep. 7, 1754. doi: 10.1038/s41598-017-01918-7
Wu, Y. W., Ke, Y. Z., Ran, F., Wen, J., Guo, P. C., Sun, L. P., et al. (2018). Cloning and expression analysis of BnMYB193 gene in Brassica napus. J. Agric. Biotechnol. (in Chinese) 26 (7), 1155–1163. doi: 10.3969/j.issn.1674-7968.2018.07.007
Xu, W., Grain, D., Le Gourrierec, J., Harscoet, E., Berger, A., Jauvion, V., et al. (2013). Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression, in Arabidopsis. New Phytol. 198 (1), 59–70. doi: 10.1111/nph.12142
Xu, Y., Iacuone, S., Li, S. F., Parish, R. W. (2014). MYB80 homologues in Arabidopsis, cotton and Brassica: Regulation and functional conservation in tapetal and pollen development. BMC Plant Biol. 14, 278. doi: 10.1186/s12870-014-0278-3
Xu, W., Tang, W., Wang, C., Ge, L., Sun, J., Qi, X., et al. (2020). SiMYB56 confers drought stress tolerance in transgenic rice by regulating lignin biosynthesis and ABA signaling pathway. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00785
Xu, P., Wu, L., Cao, M., Ma, C., Xiao, K., Li, Y., et al. (2021). Identification of MBW complex components implicated in the biosynthesis of flavonoids in woodland strawberry. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.774943
Yang, S. W., Jang, I. C., Henriques, R., Chua, N. H. (2009). FAR-RED ELONGATED HYPOCOTYL1 and FHY1-LIKE associate with the Arabidopsis transcription factors LAF1 and HFR1 to transmit phytochrome a signals for inhibition of hypocotyl elongation. Plant Cell 21 (5), 1341–1359. doi: 10.1105/tpc.109.067215
Yao, L., Yang, B., Xian, B., Chen, B., Yan, J., Chen, Q., et al. (2020). The R2R3-MYB transcription factor BnaMYB111L from rapeseed modulates reactive oxygen species accumulation and hypersensitive-like cell death. Plant Physiol. Bioch. 147, 280–288. doi: 10.1016/j.plaphy.2019.12.027
Yin, N. W., Li, J. N., Liu, X., Lian, J. P., Fu, C., Li, W., et al. (2017). Lignification response and the difference between stem and root of Brassica napus under heat and drought compound stress. Acta Agronomica Sin. (in Chinese) 43 (11), 1689–1695. doi: 10.3724/SP.J.1006.2017.01689
Zhang, M., Mo, Z., Liao, J., Pan, S., Chen, X., Zheng, L., et al. (2021). Lodging resistance related to root traits for mechanized wet-seeding of two super rice cultivars. Rice Sci. 28 (2), 200–208. doi: 10.1016/j.rsci.2021.01.009
Zhao, K., Bartley, L. E. (2014). Comparative genomic analysis of the R2R3 MYB secondary cell wall regulators of Arabidopsis, poplar, rice, maize, and switchgrass. BMC Plant Biol. 14, 135. doi: 10.1186/1471-2229-14-135
Zhong, R., Lee, C., Zhou, J., McCarthy, R. L., Ye, Z. H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20 (10), 2763–2782. doi: 10.1105/tpc.108.061325
Zhong, R., Richardson, E. A., Ye, Z. H. (2007). The MYB46 transcription factor is a direct target of SND1 and regulates secondary wall biosynthesis in Arabidopsis. Plant Cell 19 (9), 2776–2792. doi: 10.1105/tpc.107.053678
Zhong, R., Ye, Z.-H. (2012). MYB46 and MYB83 bind to the SMRE sites and directly activate a suite of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol. 53 (2), 368–380. doi: 10.1093/pcp/pcr185
Zhu, X. B., Zhang, G. F., Chen, P. (2017). Research progress in the transcriptional regulation of secondary cell wall thickening(Review). Plant Physiol. J. (in Chinese) 53 (9), 1598–1608. doi: 10.13592/j.cnki.ppj.2017.0203
Keywords: Brassica napus, MYB69, shikimates, lignin, Sclerotinia sclerotiorum, IAA, chlorophyll, biomass
Citation: Lin N, Wang M, Jiang J, Zhou Q, Yin J, Li J, Lian J, Xue Y and Chai Y (2023) Downregulation of Brassica napus MYB69 (BnMYB69) increases biomass growth and disease susceptibility via remodeling phytohormone, chlorophyll, shikimate and lignin levels. Front. Plant Sci. 14:1157836. doi: 10.3389/fpls.2023.1157836
Received: 03 February 2023; Accepted: 07 March 2023;
Published: 29 March 2023.
Edited by:
Hongbo Chao, Zhengzhou University, ChinaReviewed by:
Zhen Huang, Northwest A&F University, ChinaCopyright © 2023 Lin, Wang, Jiang, Zhou, Yin, Li, Lian, Xue and Chai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yourong Chai, Y2hhaXlvdXJvbmdAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.