- 1Shanghai Agrobiological Gene Center, Shanghai Academy of Agricultural Sciences, Shanghai, China
- 2Key Laboratory of Grain Crop Genetic Resources Evaluation and Utilization, Ministry of Agriculture and Rural Affairs, Shanghai, China
Phosphate (Pi) is indispensable for the growth and development of plant, and low-Pi stress is a major limitation for crop growth and yield worldwide. The tolerance to low-Pi stress varied among rice germplasm resources. However, the mechanisms underlying the tolerance of rice to low-Pi stress, as a complex quantitative trait, are not clear. We performed a genome-wide association study (GWAS) through a diverse worldwide collection of 191 rice accessions in the field under normal-Pi and low-Pi supply in two years. Twenty and three significant association loci were identified for biomass and grain yield per plant under low-Pi supply respectively. The expression level of OsAAD as a candidate gene from a associated locus was significantly up-regulated after low-Pi stress treatment for five days and tended to return to normal levels after Pi re-supply in shoots. Suppression of OsAAD expression could improve the physiological phosphorus use efficiency (PPUE) and grain yields through affecting the expression of several genes associated with GA biosynthesis and metabolism. OsAAD would be a promising gene for increasing PPUE and grain yield in rice under normal- and low-Pi supply via genome editing.
Introduction
Phosphorus (P) is one of the indispensable macronutrients for crops and is a major factor constraining the growth and development of crop worldwide in soil (Raghothama, 1999). Excess P fertilizers are applied to meet the high demands of rice due to the high P-fixing capacity of the soils (Vance et al., 2003). However, the non-renewable and finite reserves of phosphate resources would be exhausted not far in the future due to high inputs of P fertilizers. The low recovery rates of P fertilizers in season lead to increasing the cost of rice production and water eutrophication (Conley et al., 2009). Therefore, breeding and application of P-efficient crops would be a necessary strategy for sustainable crop production.
Phosphorus efficiency has typically been consist of P acquisition efficiency from native soil and internal P use efficiency (Veneklaas et al., 2012). The crops have developed many adaptive mechanisms to absorb P from soil, including increasing the number of root and change root architecture, secretion of organic acid to mobilization of sparingly soluble P in soil, the expression of genes related with Pi signal and uptake in roots, and symbiotic association with mycorrhizal fungi (Raghothama and Karthikeyan, 2005; Wu et al., 2013). Many genes related with rice Pi signal, uptake or translocation have been studied in detail. But overexpression or mutation of a Pi transporter gene or its regulatory gene often lead to excessive Pi accumulation with negative effects on plant growth, especially under Pi-replete conditions (Gu et al., 2015). A set of backcrossed inbred lines derived from a japonica x indica rice cross was used to identify a major quantitative trait locus (QTL) named Pup1 on chromosome 12 of rice (Wissuwa et al., 2002). A protein kinase gene (OsPSTOL1) in the candidate region was cloned that enhanced P acquisition through increasing root growth in low-P soils (Gamuyao et al., 2012). QTL for P acquisition efficiency has received significant attention (Wang et al., 2010; Wu et al., 2013). However, physiological PUE has received less attention and QTLs are not available for breeding crops with enhanced PUE as it was difficult to measure (Rose et al., 2011). Improved PPUE may be realized by producing high yields of crops with lower P tissue concentrations to maintain the growth and development or by improving the efficiency of P remobilization from old organs to young, developing organs (Dissanayaka et al., 2018). Twenty six QTLs for PUE traits were detected in a two-year field experiment under normal and low P supply through a recombinant inbred population of rice derived from Minghui 63 and Zhenshan 97 (Wang et al., 2014). PUE loci were mapped on chromosomes 1, 4, 11 and 12 by using a rice germplasm panel of 292 genotypes of diverse plant types and origins that include all five subpopulations of Oryza sativa under hydroponic system that assured equal plant P uptake (Wissuwa et al., 2015).
Advances in sequencing technologies have made it possible to characterize the genetic variation presented in an increasing number of crop gene bank accessions. Genome wide association studies (GWAS) can investigate the populations with wide range of natural variation, and discovery larger numbers of important loci, especially for complicated agronomic traits (Huang et al., 2010; Zhao et al., 2011). In present study, we conducted GWAS using a diverse worldwide collection of 191 rice accessions which were re-sequenced on the Illumina HiSeq 2000 (Wu et al., 2015). Twenty loci associated with biomass and 3 loci associated with grain yield at the ripening stage were identified in the diverse germplasm panel that was grown under P deficiency condition in the field for two years. Candidate genes underlying significant associated regions were further analyzed. Six known genes which were responsible for Pi uptake, translocation, signaling and homeostasis were identified. We also discovered a new gene, OsAAD, encoding an amino acid dehydrogenase family protein. OsAAD enhances physiological PUE and grain yield by adjusting rice tillering. Thus, OsAAD turns out to be a promising gene for increasing the yield in rice under low and normal Pi supply via genome editing or marker-assisted breeding.
Materials and methods
Plant material and field experiment
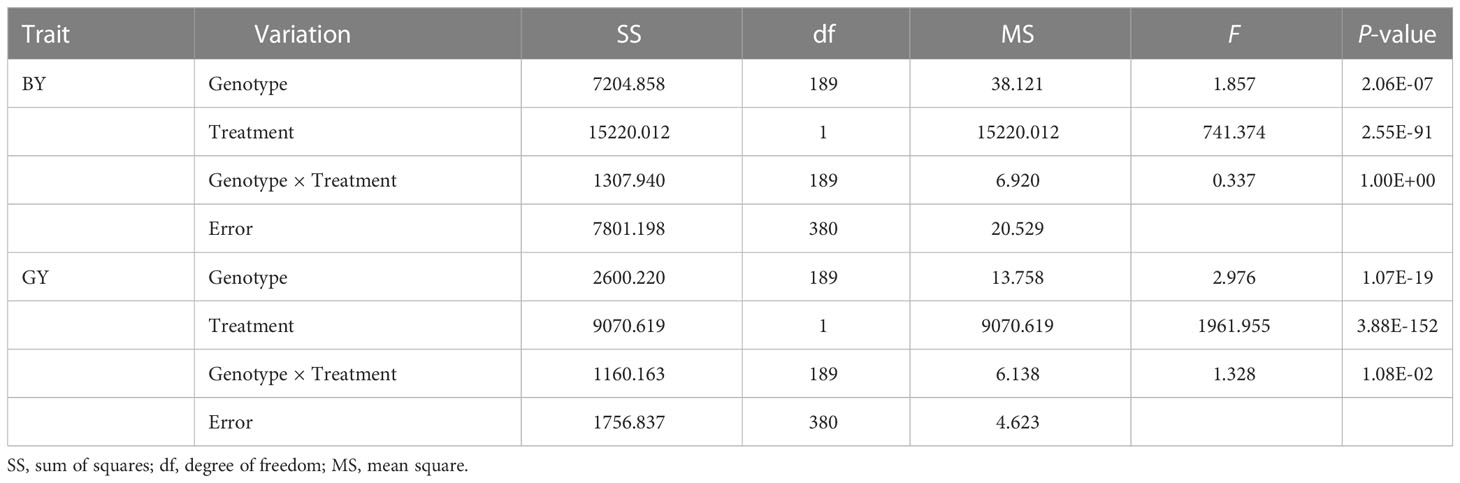
Two subsets of rice germplasm resource described in detail by Wu et al. (2015) were used in this study. After excluding varieties with too short or too long growth periods, 191 accessions were included (Supplementary Table 1). The field experiments were carried out in Hainan experimental station of Shanghai Academy of Agricultural Sciences (SAAS) in Lingshui, Hainan, China (18°33′ N, 110°04′ E) in 2014 and 2015. Each line was transplanted with a spacing of 20 cm × 20 cm to plots with an area of 0.72 m2. Each plot included 3 rows with 7 hills per row. The plots were arranged following a randomized complete block design with four replicates. The total P concentration of the soil was 0.18 mg/g. Pi-fertilizer application amount was 26 kg P2O5/ha for low P treatment and 105 kg P2O5/ha for normal P treatment, respectively. All the Pi-fertilizer was applied as basal fertilizer in the form of calcium superphosphate. To achieve high grain yield, a total of 210 kg N ha−1 in the form of urea was applied twice: 105 kg ha−1 as basal fertilizer, 105 kg ha−1 20 days after transplanting. Potassium sulfate (100 kg K ha−1) and zinc sulfate heptahydrate (5 kg Zn ha−1) were applied as basal fertilizer. A flood-irrigation system was adopted, which followed high-yielding agricultural practices. At the maturity of the plants from each plot, three uniform plants were sampled. Biomass per plant (BY) and grain yield per plant (GY) were measured.
Genome wide association studies
The GWAS was conducted by using the CMLM algorithm implemented in GAPIT R package (Zhang et al., 2010). A total of 3,038,555 SNP markers were used for GWAS. The genome-wide threshold was first set at false discovery rate (FDR) = 0.05, and then it was set at p = 3.29E-07, calculated via the formula: 1/total number of SNPs, which was widely used in plant GWAS studies (Wen et al., 2014; Wang et al., 2015; Bai et al., 2016). We furthermore evaluated the extent of local linkage disequilibrium (LD) for each significant SNP. The extended region, where LD between nearly SNPs and lead SNP (with the lowest p-value) decayed to r2 = 0.6, was defined as the local LD-based QTL interval (Yano et al., 2016).
RNA extraction, cDNA synthesis and RT-qPCR
Total RNA was extracted using RNAiso Plus (TaKaRa) according to the manufacturer’s instructions. First-strand cDNAs were synthesized from total RNA using EasyScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech). Reverse transcription quantitative PCR was performed with the PerfectStart Green qPCR SuperMix (TransGen Biotech) on the CFX96™ Real-Time System (BIO-RAD, USA). The relative level of expression was calculated by the equation 2-ΔΔCt using housekeeping gene OsActin1 (LOC_Os03g50885) as an internal reference (Feng et al., 2017). The primers used are listed in Supplementary Table 2.
Production and identification of aad mutant lines
For the generation of aad mutant, the target sequence (Supplementary Table 2) was synthesized and ligated with respective sgRNA catastases and then were sequentially cloned into the CRISPR/Cas9 binary vectors pYLCRISPR/Cas9Pubi-H as previously described (Ma et al., 2015). The DNA sequence of OsAAD was amplified by PCR using gene specific primers (Supplementary Table 2), and PCR products were sequenced and aligned with wild-type sequences. Two homozygous knockout mutants (aad-6 and aad-7) with different mutation sites were identified and used for subsequent physiological analysis (Supplementary Figure 1).
For hydroponic experiments, seeds were surface sterilized with 10% (v: v) H2O2 for 30 min, and rinsed thoroughly with deionized water. The sterilized seeds were germinated on PCR plate mounted in plastic containers for 1 week. The seedlings were transferred to a half-strength Kimura B solution with pH 5.6 (Yamaji et al., 2013) and grown in artificial climate chamber with a 14 h: 10 h as light: dark photoperiod and 30°C: 22°C as day: night temperature, and the relative humidity was controlled at c. 60%.
Pot experiments were conducted with four replications in artificial climate chamber using the soil collected from Hainan experimental station of SAAS. The acid soil contained 6.5 mg Pi kg-1 extracted by the Bray I method (Bray and Kurtz, 1945). One wild-type plant and one aad mutants were grown in each pot containing 6 kg of air-dried soil. The Pi fertilizer were 40 and 160 mg Pi kg-1 soil as low and normal supply levels, respectively.
Measurement of total P concentration
For total P measurement, the shoots and roots of rice plants were dried to a constant weight at 70℃. Dried samples (0.5 g) were pre-digested in 100 ml glass tubes with 5 ml concentrated sulfuric acid (H2SO4) for 2 h. The tubes were then heated to 280℃ for 30 min, and 50 μl H2O2 was added every 10 min until the solution became colorless. The digestion was continued for another 30 min. The dilution of the digested solution was analyzed by the molybdenum blue method according to the procedure of Ames (1966).
Definition of PPUE
The P concentration of shoot and root is the P accumulation divided by its corresponding dry weight. The physiological phosphorus use efficiency (PPUE) is tissue dry weight divided by its P concentration under low or normal-Pi supply (g2 DW g-1P).
RNA-seq analysis
Four-leaf-old seedlings of WT, aad-6 and aad-7 mutants grown under low-Pi supply for two weeks were harvested for subsequent RNA-seq analysis. Total RNA was extracted from the shoots and roots of three biological replicates (5 seedlings per replicate) using RNAiso Plus (TaKaRa). RNA-seq library was constructed with the TruSeq RNA Sample Preparation v2 Guide, and RNA sequencing was performed using Illumina HiSeq 2500 by Shanghai Personal Biotechnology Co., Ltd. After filtering adapters and low-quality reads, the paired-end reads were then aligned to the reference genome of rice using HISAT2 v2.1.0 (Kim et al., 2019). Fragments per kilobase per million mapped (FPKM) reads was then used to estimate the expression level of the genes. The genes with the parameter of false discovery rate (FDR) below 0.01 was considered as differentially expressed genes (DEGs). Function of the DEG was predicted using Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/) databases (Xie et al., 2011). The raw reads were deposited in the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) with the accession number of PRJNA306542.
Results
Statistics analysis of phenotypic traits
Two traits varied widely among genotypes under both normal and low Pi supply in the field in two years. Variation for biomass in the P-deficient plot and P-replete plot ranged from 2.30 g to 18.30 g and from 3.93 g to 28.17 g in two years, respectively. Variation for grain yield in the P-deficient plot and P-replete plot ranged from 1.30 g to 8.35 g and from 2.87 g to 17.48 g in two years, respectively (Table 1). Under P-deficiency, the phenotypic mean values of the population were seriously reduced. P deficiency reduced biomass accumulation in Pi-deficient plots by 60.49% and 60.72% in 2014 and 2015, respectively. P deficiency reduced grain yield in Pi-deficient plots by 61.63% and 66.76% in 2014 and 2015 (Table 1; Supplementary Figure 2). The ANOVA results revealed significant effects from genotypes and treatments on both traits (Table 2). The high phenotypic ranges in this natural population indicated that these germplasm accessions contained rich genetic variations related to P-efficiency.
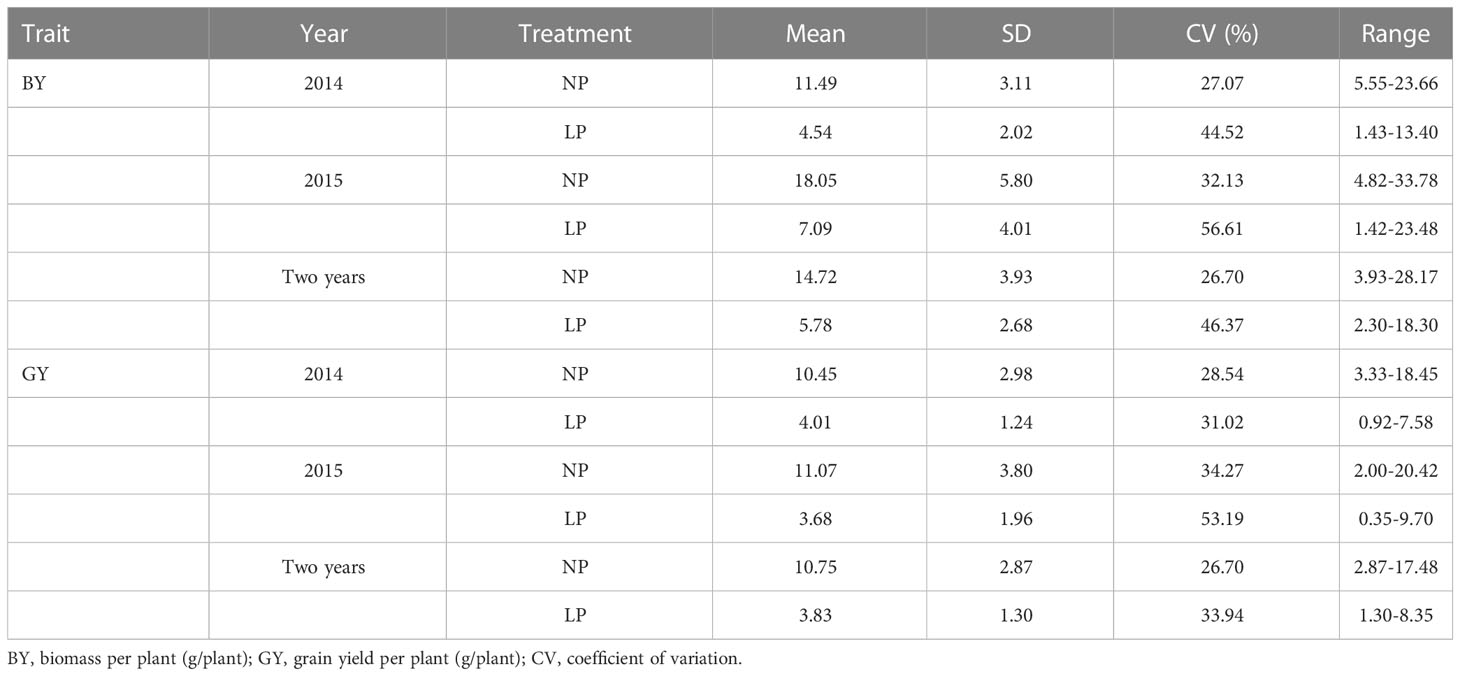
Table 1 Descriptive statistical results for traits under normal and low Pi supply in the natural population in the field experiments conducted in 2014 and 2015.
Genome-wide association study
As the population was used in GWAS of mesocotyl elongation, a two-subpopulation structure corresponding to indica and japonica subspecies has been reported in our previous publication (Wu et al., 2015). Using 3,038,555 SNPs covering the entire rice genome, a total of 20 loci associated with biomass per plant under low-Pi supply were identified under a Compressed Mixed Linear Model. Those associated loci were located on chromosome 1, 2, 3, 4, 6, 7, 9, 10, 11 and 12 (Figure 1B, Supplementary Table 3). However the significant loci associated with biomass per plant under normal-Pi supply was not detected (Figure 1A). Four Pi uptake genes (OsPHT1;1, 1;2, 1;7, 1;12), Pi signaling gene (OsPHR2) and Pi homeostasis gene (OsSPX3) were located within three QTLs intervals (Zhou et al., 2008; Ai et al., 2009; Sun et al., 2012; Dai et al., 2022; Figure 1B). Only three loci on chromosome 1, 7 and 12 associated with grain yield per plant under low-Pi supply were detected, but none of loci associated with grain yield per plant under normal-Pi supply was identified (Figures 1C, D). We found that qGY7 and qBY7.2 were in the same QTLs intervals which contained OsPHR2 regulating Pi uptake (Figures 1B, D; Zhou et al., 2008). The candidate genes from 23 GWAS loci associated with biomass and grain yield per plant (QTL interval, r2 of LD > 0.6) were further screened according to their expression levels in response to phosphate starvation and recovery (Secco et al., 2013). Among them, the expression levels of four genes were significantly up-regulated after 7-day Pi starvation and then tended to return to normal levels after Pi re-supply (Supplementary Table 4). These results indicate that these four genes might be good candidates for the GWAS loci. All four genes were knockdown by CRISPR/Cas9 in the Nipponbare, but only the phenotype of aad (LOC_Os09g15810) mutants was different from wild-type plants. The function of OsAAD in response to low-Pi stress then was investigated in detail.
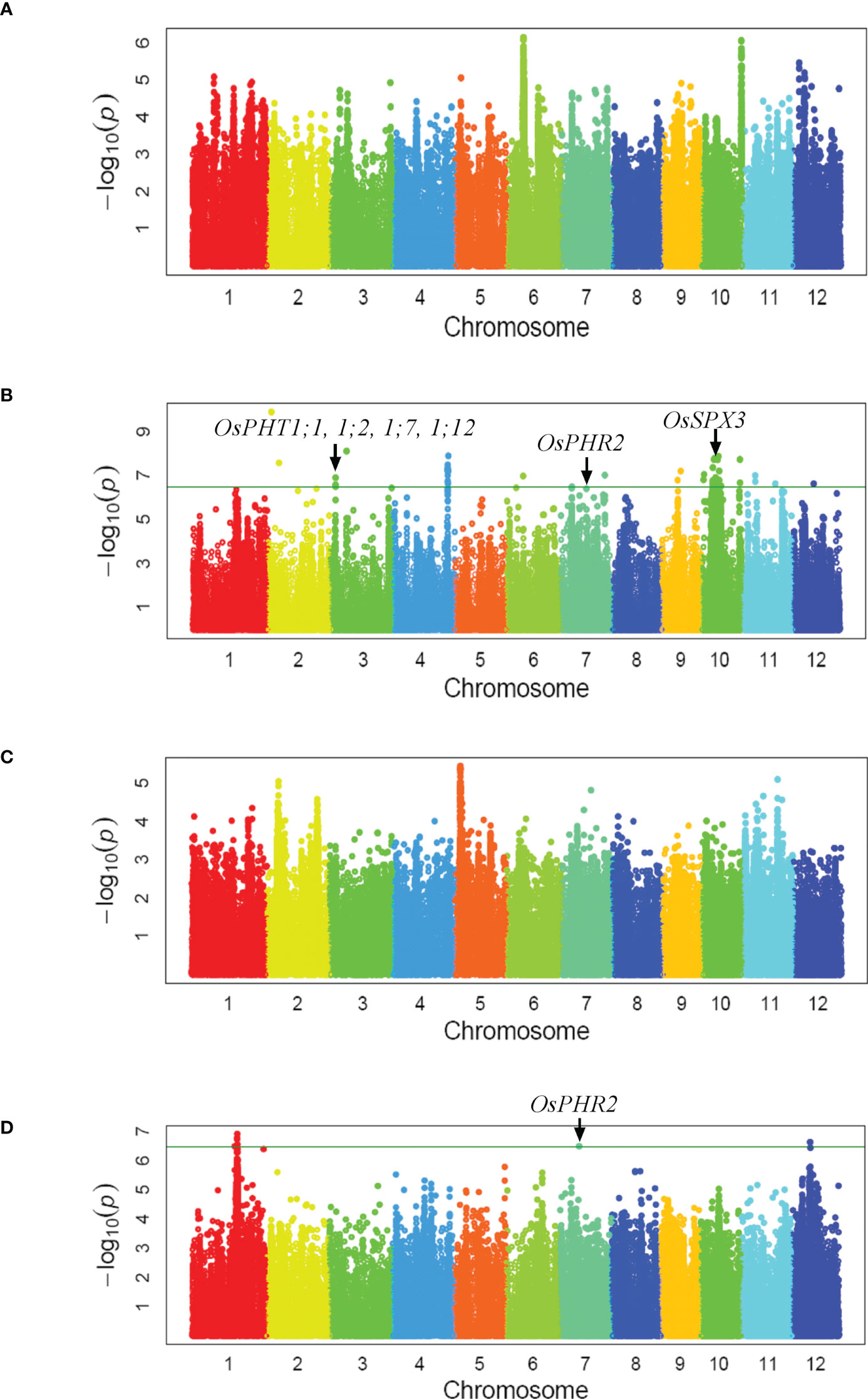
Figure 1 Genome-wide association study. Manhattan plots of GWAS mapping for two-year averages of biomass per plant (A, B) and grain yield per plant (C, D) measured under normal (A, C) and low Pi-fertilizer supply (B, D), respectively.
Expression patterns of OsAAD is responsive to low Pi
To check the spatial expression patterns of OsAAD in rice, reverse transcription quantitative (RT-qPCR) analysis was conducted for roots, shoot basal region, leaf sheath and leaf blade of seedlings supplied with normal Pi. As shown in Figure 2A, OsAAD was much more abundant in leaf blade than in other parts. A time-course treatment was carried out to check the transcriptional expression of OsAAD in response to low Pi stress. In shoots, OsAAD was significantly induced by low Pi treatment from day 5 to 14. Resupply of Pi for 1-day inhibited the expression of OsAAD to a level comparable with normal Pi supply (Figure 2B). In roots, the moderate induction of OsAAD expression was only observed after 7-day low Pi treatment that was completely abolished by 1-day normal Pi resupply (Figure 2B).
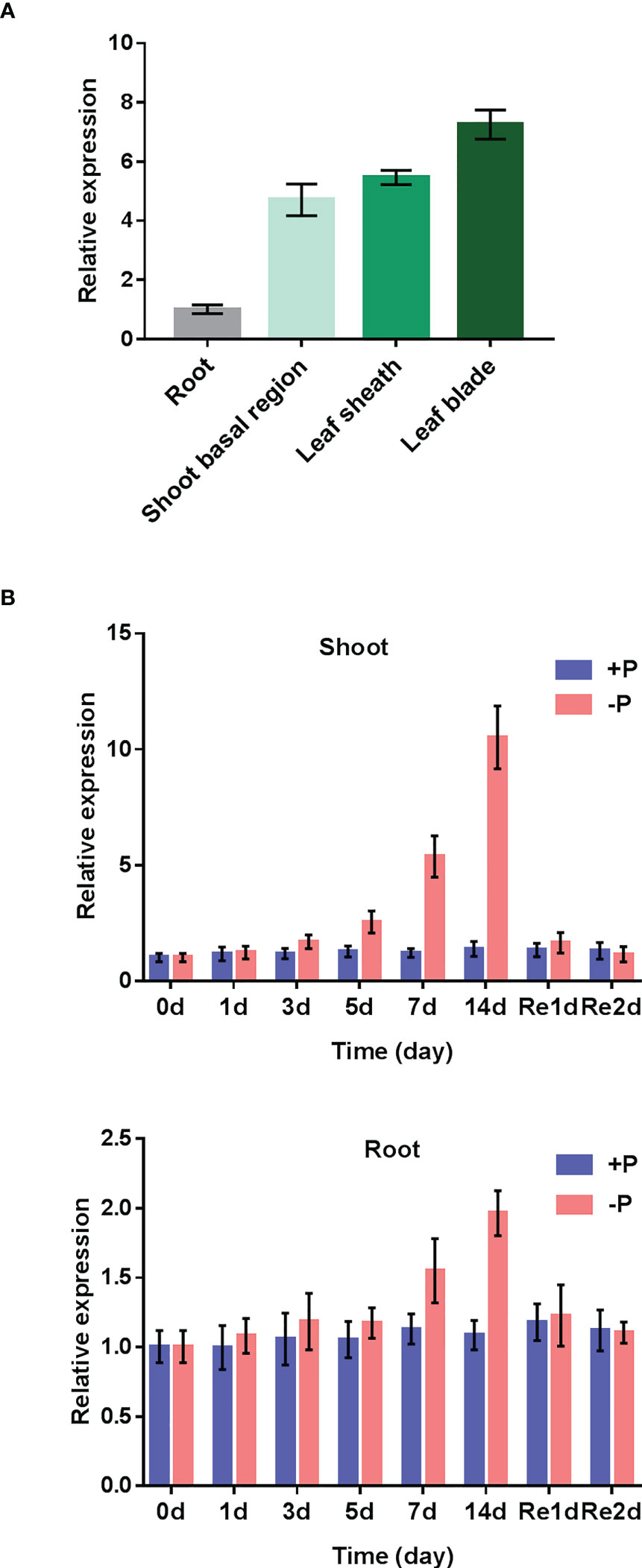
Figure 2 Expression patterns of OsAAD gene. (A) Gene expression in roots, shoot basal region, leaf sheath and leaf blade that were separately sampled at the six-leaf-old stage under normal Pi supply. (B) Four-leaf-old seedlings transferred into nutrient solution supplied with normal (200 m M Pi) or low (10 m M Pi) for 14 d, and then resupplied with normal Pi for 2 d. The shoots and roots were sampled at the beginning (0 d) and 1, 3, 5, 7 and 14 d after the treatment, and 1 and 2 d after the resupply of Pi. Values represent means ± SD of three biological replicates.
Knockdown OsAAD enhanced PPUE of shoot and root
To investigate the physiological role of OsAAD in rice plants, corresponding mutant plants were obtained using the CRISPR-Cas9 system. Two independent lines (aad-6 and aad-7) were used in further studies (Supplementary Figure 1). One-week-old aad mutants and wild-type seedling were cultivated in the hydroponic solution containing a normal and low level of Pi for 30 d. Two lines of aad mutants showed better growth than wild-type plants under both normal and low Pi supply (Figure 3A). The shoot and root dry weight of two aad mutants was significantly higher than wild-type plants under both normal and low Pi supply (Figure 3B). Total Pi concentration in shoots and roots of two aad mutants was lower than wild-type plants when grown in both normal and low Pi solution, except no difference in roots under normal Pi supply (Figure 3C). The PPUE of shoot and root in two aad mutants was higher than in the wild-type plants incubated in both normal and low Pi solution (Figure 3D). This suggested that aad mutants with lower tissue P concentrations could use less P-fertilizer to maintain the growth and development of rice.
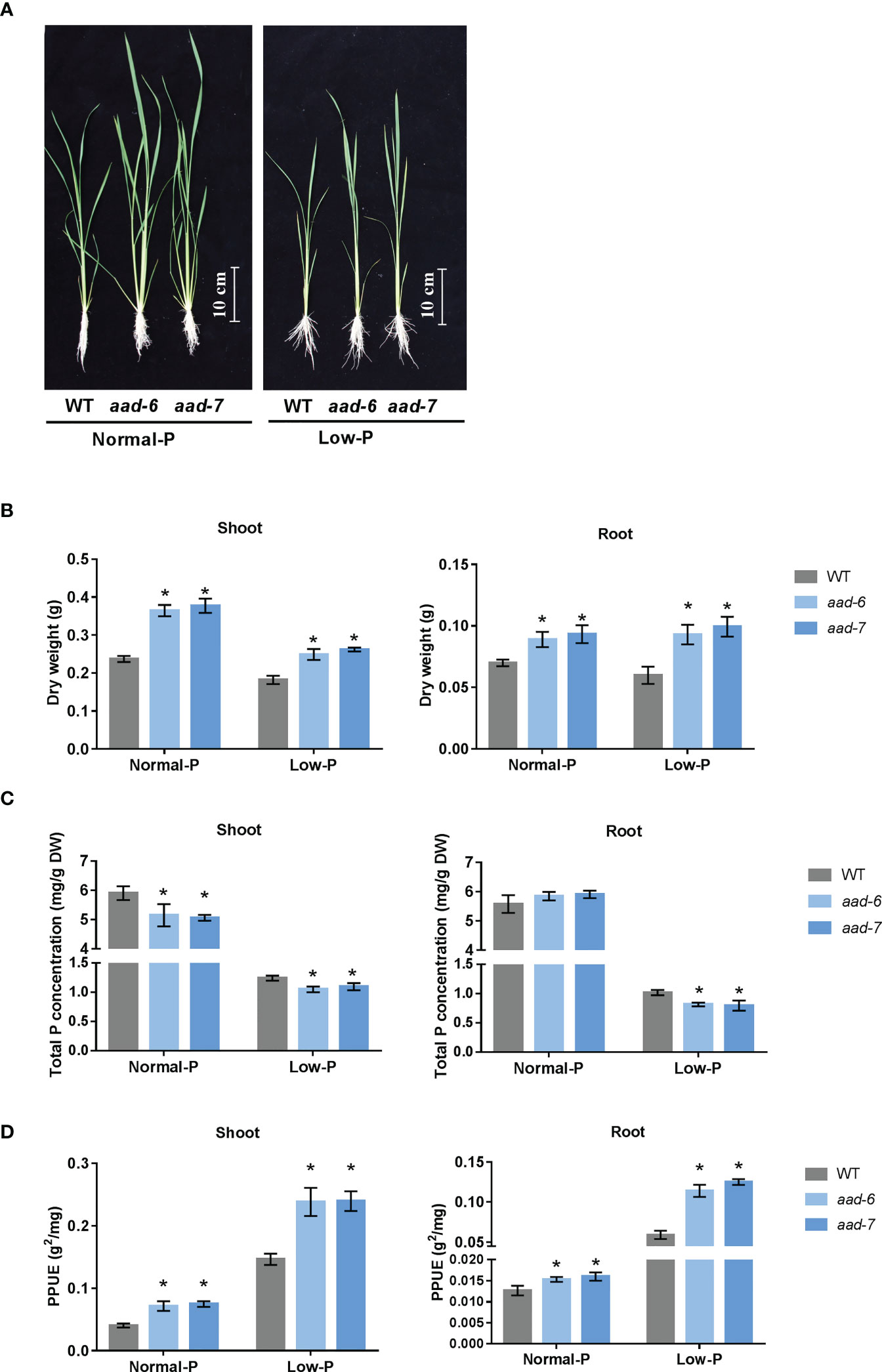
Figure 3 Phenotypic characteristics and total P concentration analysis of wild-types and aad mutants. (A) Growth performance of wild-type and aad mutants grown in a solution culture with normal Pi (200 m M) or low Pi (10 m M) supply for 30d. (B) Shoot and root dry weight of wild-type and aad mutants. (C) Total P concentration in shoot and root. (D) Physiological phosphorous use efficiency. Values represent means ± SD of three biological replicates *P < 0.01.
Mutation of OsAAD increases effective tiller number and grain yield
To determine the function of OsAAD during the entire plant growth period, soil pot experiments were conducted in artificial climate chamber. We observed that two lines of aad mutants grew better than wild-type at 40 and 160 mg fertilizer Pi kg-1 soil. The effective tiller number, shoot dry weight per plant and grain yield per plant were measured at the harvest stage. The effective tiller number, biomass and grain yield were higher in aad mutants than in the wild-type plants under both Pi-deficient and Pi-replete soil (Figure 4). Furthermore, the grain yields of aad mutants under low Pi fertilizer treatment was almost equal to that of the wild-type under Pi-replete conditions (Figure 4C). Taken together, these results indicate that OsAAD would be a promising candidate gene for breeding rice with high P-efficiency under normal and low Pi supply.
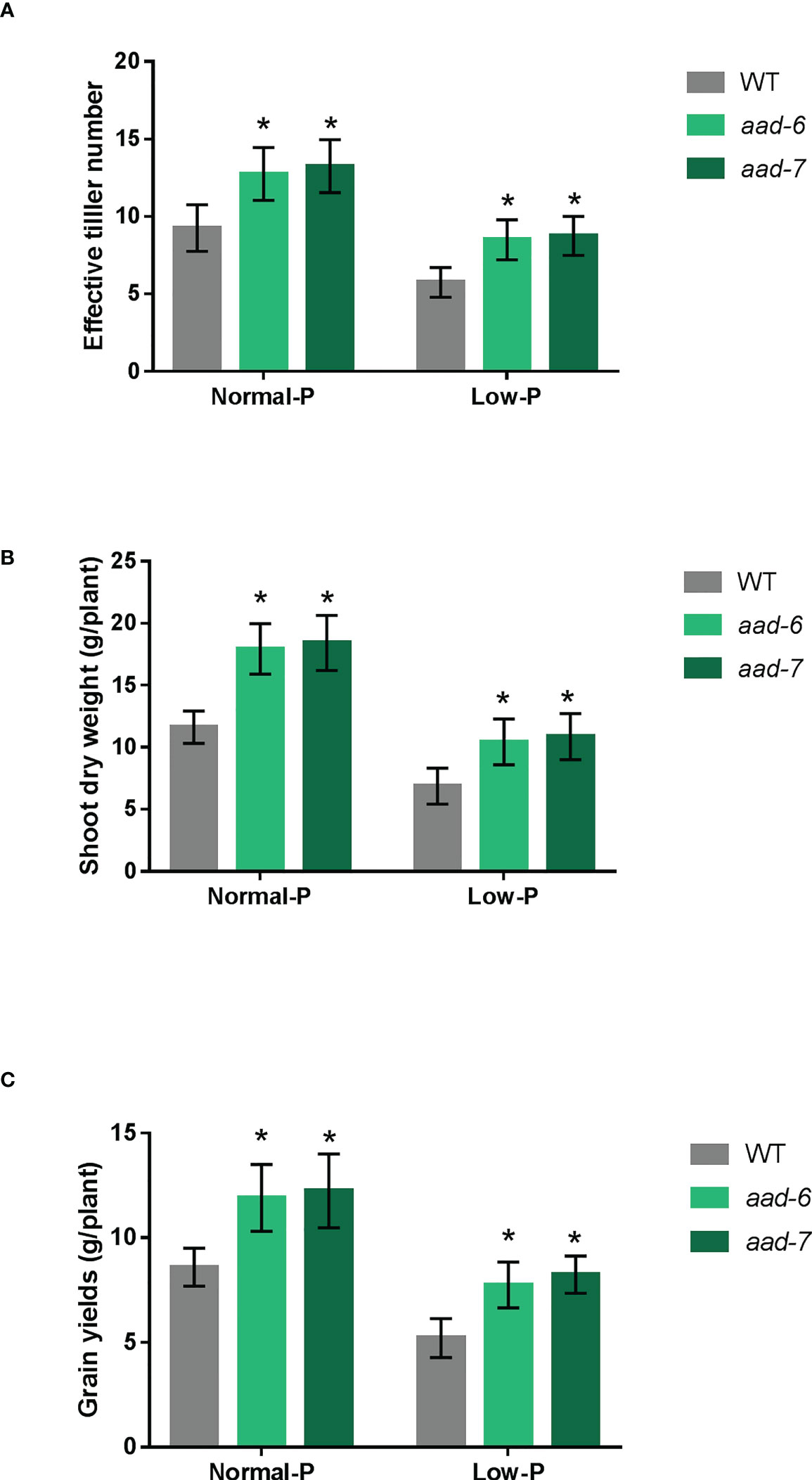
Figure 4 Growth performances of the wild-type and aad mutants under normal and low Pi supply in a pot experiment. (A) Effective tiller number per plant. (B) Shoot dry weight per plant. (C) Grain yields per plant. Values represent means ± SD of four biological replicates *P < 0.01.
The OsAAD affects several genes involved in maintaining rice tillering
To investigate the molecular mechanism underlying OsAAD-regulated rice tillering, a transcriptomic analysis was performed using the shoots and roots of two aad mutants and WT plants under low Pi condition for 2 weeks. Twenty and 18 genes were significantly upregulated in shoot and root, while 64 and 62 genes were downregulated in shoot and root, respectively, in two aad lines compared with the WT (Supplementary Table 5). Unexpectedly, 41 known genes associated with Pi uptake, translocation and signal were not differently expressed in aad mutants against the WT (Supplementary Table 6). Kyoto Encyclopedia for Genes and Genomes (KEGG) enrichment analysis revealed that diterpenoid biosynthesis, cysteine and methionine metabolism were significantly enriched in shoot, and phenylpropanoid biosynthesis was markedly enriched in root of both aad-6 and aad-7 mutants (Figure 5A). Among these differently expressed genes, the expression of OsGA2ox5 gene which negatively regulate rice tiller was not detected, while the CYP714B2 gene which is for GA metabolic enzymes was significantly downregulated in the shoots of two aad mutants (Figure 5B). The expression level of OsGA2ox5 and CYP714B2 in shoots of WT and two aad mutants was confirmed by qRT-PCR (Figure 5C). These results suggest that the OsAAD influenced tillering by the regulation of GA related gene expression.
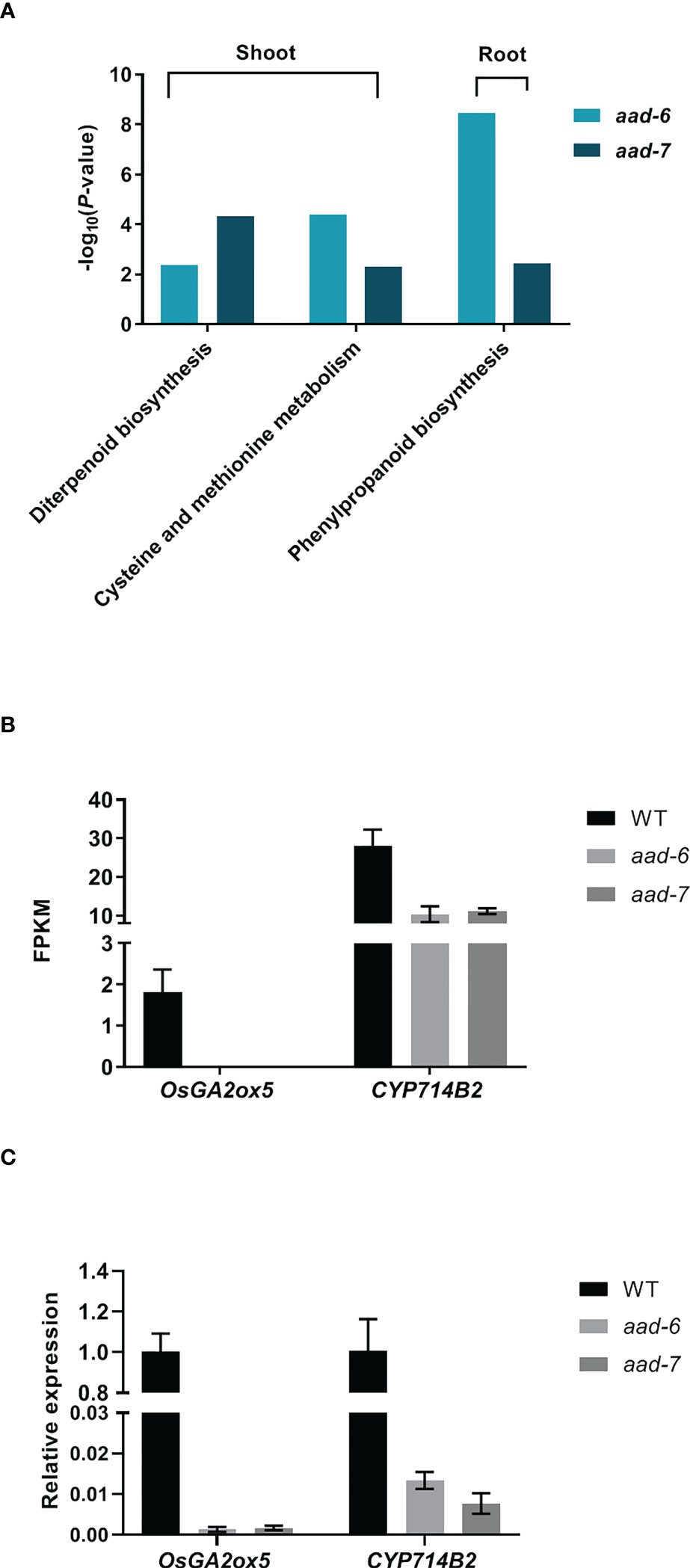
Figure 5 Transcriptome analysis of shoots and roots in two lines of aad mutants under low-Pi treatment for 2 weeks. (A) Histogram of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment results. The X-axis is the name of the pathways, and the Y-axis is -log10 (p-value) enriched by each pathway. (B) and (C) FPKM and relative expression level of two genes related with GA pathway in shoots of two aad mutants and WT, respectively. Values represent means ± SD of three biological replicates.
Natural variation of OsAAD might be tolerant to low Pi stress
To uncover the natural variation of OsAAD and identify elite alleles, 191 rice varieties were used for haplotype analysis (Supplementary Table 1). Twenty-three SNPs were detected in 2k-bp promoter, two SNPs were located in the first intron, and only one SNP occurred in the exon of OsAAD. Based on these SNPs, four main haplotypes which contained at least five accessions were identified (Figure 6A). We found that hap1 and hap3 are belong to indica subgroup, while hap4 is a japonic-specific haplotype (Figure 6A). The shoot dry weight of hap1 was significantly higher than that of the other haplotypes under both normal and low Pi supply. Compared with normal Pi supply, the shoot dry weight of hap1 and hap2 under low Pi supply was decreased by 44.8% and 61.5%, respectively (Figure 6B). These results indicated that the tolerance of hap1 to low Pi stress was stronger than that of hap2. The grain yields of hap1 was also higher than that of the other haplotypes under both normal and low Pi supply. Among these four haplotypes, the grain yields of hap4 was lowest under both normal and low Pi treatment (Figure 6C). All these results suggested that hap1 would be a potential haplotype for breeding variety tolerant to low Pi stress through marker-assisted selection.
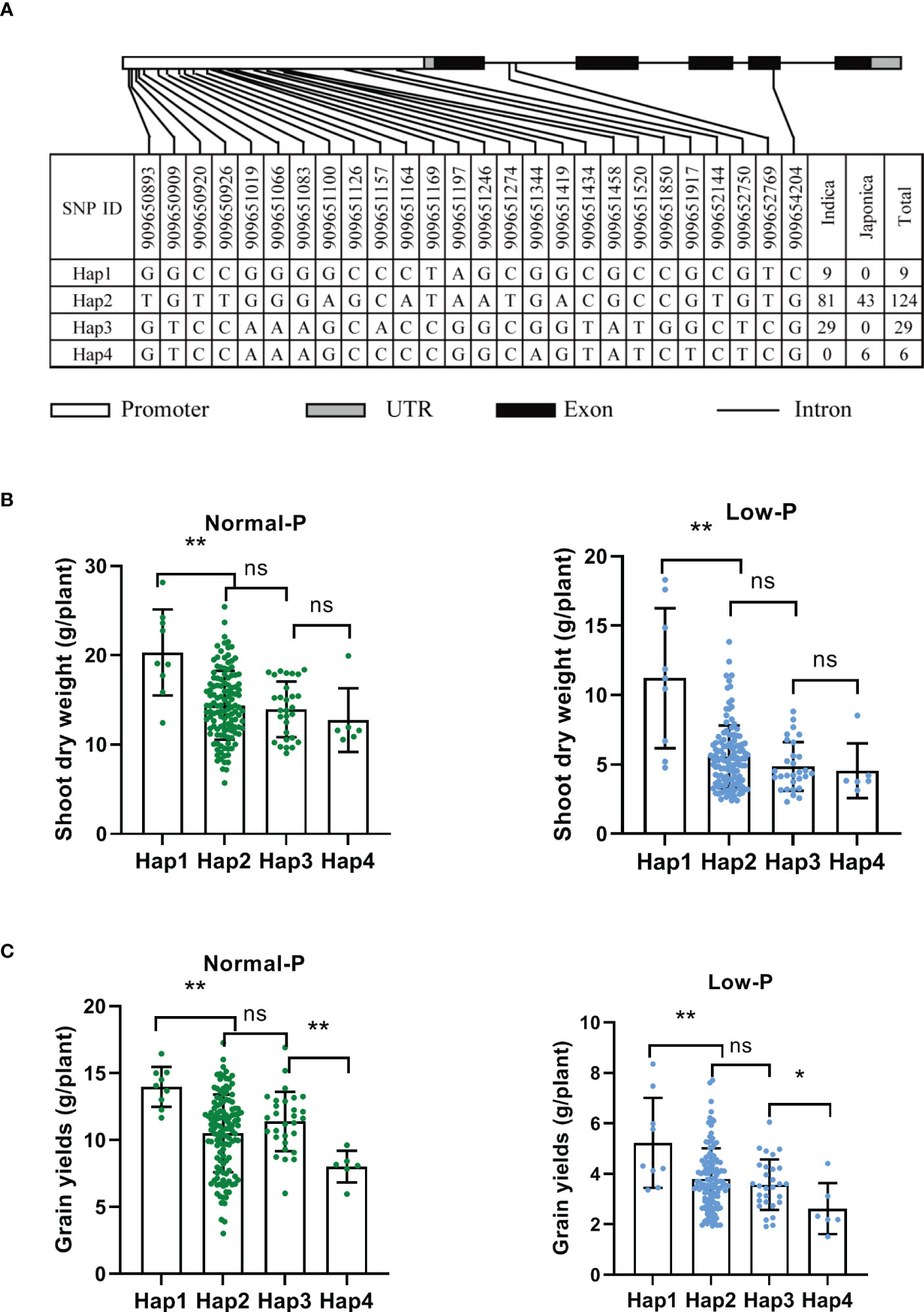
Figure 6 Haplotype analysis of OsAAD. (A) The gene structure of OsAAD. Four main haplotypes were detected by single nucleotide polymorphism (SNP) variations. The numbers of varieties in two subspecies of each haplotype are shown in the right column. (B, C) Shoot dry weight and grain yields per plant were compared among four haplotypes under normal and low Pi supply, respectively. All data are presented as the means ± SD. *P < 0.01; **P < 0.001, ns, not significant.
Discussion
Tolerance to low-Pi stress is a complicated trait with low heritability and is strongly affected by both environment and genetic materials (Veneklaas et al., 2012). Based on reverse genetics, a lot of genes responsive for Pi signal and uptake were cloned and investigated in detail, but those genes were difficult to use for breeding P efficient variety in practical application (Gu et al., 2015). In this study, the tolerance to low-Pi stress of 191 rice germplasm accessions was evaluated under normal- and low-Pi supply in the Pi-deficit field in two years (Supplementary Figure 2; Supplementary Table 1). Twenty loci associated with biomass and three loci with grain yield under low-Pi supply was identified by using the average values measured in two years (Figure 1, Supplementary Table 3). One QTL (qBY12) on rice chromosome 12 located at the same region as the Pup1 locus which was the most influential QTL mapped by using recombinant inbred line of Nipponbare and Kasalath in phosphorus-deficient soil (Wissuwa et al., 1998; Supplementary Table 3). It was subsequently shown to improve Pi uptake by promoting root growth (Gamuyao et al., 2012). Another QTL (qBY3.1) was mapped to chromosomal region containing four known P transporters (PHT1;1, PHT1;2, PHT1;7 and PHT1;12) of the PHT1 family which were in responsive for Pi uptake and translocation (Figure 1; Supplementary Table 3; Ai et al., 2009; Sun et al., 2012; Dai et al., 2022). Genes related to Pi signaling (PHR2) and Pi homeostasis (SPX3) were previously detected in QTL (qBY7.2, qGY7) and QTL (qBY10.2) interval, respectively (Zhou et al., 2008; Shi et al., 2014; Figure 1; Supplementary Table 3). All these results indicated that field experiments under Pi-deficit condition of diverse rice germplasm panel and GWAS for biomass and grain yield could serve as an efficient stretagy to identify genetic loci for tolerance to low P stress. These significant associated loci could be beneficial for understanding the molecular mechanism of the tolerance to low P and could be used for breeding new variety tolerant to low P stress through marker-assisted selection.
Via an integrative approach combining genomic mapping in this study and previous reported transcriptomic results (Secco et al., 2013), OsAAD was cloned as a potential candidate gene related to low-Pi stress tolerance. The expression level of OsAAD was induced by Pi starvation and tended to return to normal levels quickly after Pi re-supply in shoots (Figure 2B), in agreement with the reported RNA sequencing data (Secco et al., 2013). Growth performance of the wild-type, aad-6 and aad-7 mutants in nutrient hydroponic solutions and pot experiments under normal- and low-Pi supply showed that inhibition of OsAAD not only enhances shoot dry weight, effective tiller number and grain yield, but also increases physiological phosphorus use efficiency under normal- and low-Pi supply (Figures 3 and 4). The lateral root length of high PPUE cultivar was longer than that of low PPUE cultivar (Akhtar et al., 2008). The shoots and roots biomass of the OsAVP1DOX line grown in both Pi sufficient and Pi deficient conditions were higher than controls (Yang et al., 2007). We also found that the root dry weight of aad-6 and aad-7 mutants was higher than that of wild-type plants (Figure 3B). The total P concentration of shoot in two lines of aad mutants was lower than that of wild-type plants (Figure 3C). This suggested that aad muants were likely to use less Pi to maintain growth and tolerate to low Pi stress better. Compared with low-PUE genotype, high-PUE groups were able to reduce tissue P concentration to a lower level (Veneklaas et al., 2012; Adem et al., 2020). Meanwhile, significant changed expression level of other known genes related with Pi signaling, uptake, translocation and homeostasis was not detected in aad mutants (Supplementary Table 6). This indicated that low P concentration in the aad mutants was not due to the change of genes associated with Pi signaling, uptake, translocation and homeostasis.
The OsAAD encoded an amino acid dehydrogenase family protein according to genome annotation, consistent with the enriched cysteine and methionine metabolism pathway by KEGG analysis (Figure 5A). The substrate of OsAAD was still unknown and needed to be further explored in future. Diterpenoid biosynthesis was also significantly enriched in aad mutants by KEGG analysis (Figure 5A). The expression levels of OsGA2ox5 gene which negatively regulate rice tiller numbers was not detectable, while the CYP714B2 gene which is for GA metabolic enzymes was significantly downregulated in the leaf of two aad mutants (Figure 5B; Lo et al., 2008; Magome et al., 2013). This suggested that suppression of OsAAD expression increased tiller numbers by affecting the expression of genes related with GA biosynthesis and metabolism. Phosphorus played an important role in rice tillering which was inhibited by low available Pi in soil (Takehisa and Sato, 2019). The effective tiller numbers of aad mutants was much more than that of wild-type plants under normal and low Pi supply (Figuer 4A). The nucleotide diversity of OsAAD was characterized which revealed that hap1 was most tolerant to low P stress (Figure 6). The decrease of hap1 biomass caused by low P treatment was lower than that of other haplotypes. The germplasm accessions harbored elite hap1 in this study could be valuable for rice breeding. Taken together, OsAAD would be a promising gene for molecular breeding of rice cultivars with high yield and using less Pi to maintain the growth and development under low and normal Pi supply through genome editing.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioProject, PRJNA306542.
Author contributions
HM, LL and MY designed this study. MY, XX, QL, LC and AZ performed the field experiment. MY, HM and FF analyzed the data. MY and PF studied the function of candidate genes. MY and HM drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31401939), the Natural Science Foundation of Shanghai (22ZR1455300) and the Earmarked Fund for China Agriculture Research System (CARS-01).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1153967/full#supplementary-material
References
Adem, G. D., Ueda, Y., Hayes, P. E., Wissuwa, M.. (2020). Genetic and physiological traits for internal phosphorus utilization efficiency in rice. PloS One 15, e0241842. doi: 10.1371/journal.pone.0241842
Ai, P., Sun, S., Zhao, J., Fan, X., Xin, W., Guo, Q., et al. (2009). Two rice phosphate transporters, OsPht1; 2 and OsPht1; 6, have different functions and kinetic properties in uptake and translocation. Plant J. 57, 798–809. doi: 10.1111/j.1365-313X.2008.03726.x
Akhtar, M. S., Oki, Y., Adachi, T. (2008). Genetic variability in phosphorus acquisition and utilisation efficiency from sparingly soluble p-sources by brassica cultivars under p-stress environment. J. Agron. Crop Sci. 194, 380–392. doi: 10.1111/j.1439-037X.2008.00326.x
Ames, B. N. (1966). Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8, 115–118. doi: 10.1016/0076-6879(66)08014-5
Bai, X., Zhao, H., Huang, Y., Xie, W., Han, Z., Zhang, B., et al. (2016). Genome-wide association analysis reveals different genetic control in panicle architecture between indica and japonica rice. Plant Genome 9, 1–10. doi: 10.3835/plantgenome2015.11.0115
Bray, R. H., Kurtz, L. T. (1945). Determination of total, organic, and available forms of phosphorus in soils. Soil Sci. 59, 39–45. doi: 10.1097/00010694-194501000-00006
Conley, D. J., Paerl., H. W., Howarth, R. W., Boesch, D. F., Seitzinger, S. P., Havens, K. E., et al. (2009). Ecology. controlling eutrophication: nitrogen and phosphorus. Science 323, 1014–1015. doi: 10.1126/science.1167755
Dai, C., Dai, X., Qu, H., Men, Q., Liu, J., Yu, L., et al. (2022). The rice phosphate transporter OsPHT1;7 plays a dual role in phosphorus redistribution and anther development. Plant Physiol. 188, 2272–2288. doi: 10.1093/plphys/kiac030
Dissanayaka, D. M. S. B., Plaxton, W. C., Lambers, H., Siebers, M., Marambe, B., Wasaki, J. (2018). Molecular mechanisms underpinning phosphorus-use efficiency in rice. Plant Cell Environ. 41, 1483–1496. doi: 10.1111/pce.13191
Feng, F. J., Mei, H. W., Fan, P. Q., Li, Y. N., Xu, X. Y., Wei, H. B., et al. (2017). Dynamic transcriptome and phytohormone profling along the time of light exposure in the mesocotyl of rice seedling. Sci. Rep. 7, 11961. doi: 10.1038/s41598-017-12326-2
Gamuyao, R., Chin, J. H., Pariasca-Tanaka, J., Pesaresi, P., Catausan, S., Dalid, C., et al. (2012). The protein kinase Pstol1 from traditional rice confers tolerance of phosphorus deficiency. Nature 488, 535–539. doi: 10.1038/nature11346
Gu, M., Chen, A. Q., Sun, S. B., Xu, G. H. (2015). Complex regulation of plant phosphate transporters and the gap between molecular mechanisms and practical application: What is missing? Mol. Plant 9, 396–416. doi: 10.1016/j.molp.2015.12.012
Huang, X., Wei, X., Sang, T., Zhao, Q., Feng, Q., Zhao, Y., et al. (2010). Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42, 961–967. doi: 10.1038/ng.695
Kim, D., Paggi, J. M., Park, C., Bennett, C., Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Lo, S., Yang, S., Chen, K., Hsing, Y., Zeevaart, J., Chen, L., et al. (2008). A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell. 20, 2603–2618. doi: 10.1105/tpc.108.060913
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Magome, H., Nomura, T., Hanada, A., Takeda-Kamiya, N., Ohnishi, T., Shinma, Y., et al. (2013). CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. PNAS 110, 1947–1952. doi: 10.1073/pnas.1215788110
Raghothama, K. G. (1999). Phosphate acquisition. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 665–693. doi: 10.1146/annurev.arplant.50.1.665
Raghothama, K. G., Karthikeyan, A. S. (2005). Phosphate acquisition. Plant Soil 274, 37–49. doi: 10.1007/s11104-004-2005-6
Rose, T. J., Rose, M. T., Pariasca-Tanaka, J., Heuer, S., Wissuwa, M. (2011). The frustration with utilization: why have improvements in internal phosphorus utilization efficiency in crops remained so elusive? Front. Plant Sci 2, 73. doi: 10.3389/fpls.2011.00073
Secco, D., Jabnoune, M., Walker, H., Shou, H., Wu, P., Poirier, Y., et al. (2013). Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell 25, 4285–4304. doi: 10.1105/tpc.113.117325
Shi, J., Hu, H., Zhang, K. M., Zhang, W., Yu, Y., Wu, Z., et al. (2014). The paralogous SPX3 and SPX5 genes redundantly modulate pi homeostasis in rice. J. Exp. Bot. 65, 859–870. doi: 10.1093/jxb/ert424
Sun, S., Gu, M., Cao, Y., Huang, X., Zhang, X., Ai, P., et al. (2012). A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiol. 159, 1571–1581. doi: 10.1104/pp.112.196345
Takehisa, H., Sato, Y. (2019). Transcriptome monitoring visualizes growth stage-dependent nutrient status dynamics in rice under field conditions. Plant J. 97, 1048–1060. doi: 10.1111/tpj.14176
Vance, C. P., Uhde-Stone, C., Allan, D. (2003). Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 157, 423–447. doi: 10.1046/J.1469-8137.2003.00695.X
Veneklaas, E. J., Lambers, H., Bragg, J., Finnegan, P. M., Lovelock, C. E., Plaxton, W. C., et al. (2012). Opportunities for improving phosphorus-use efficiency in crop plants. New Phytol. 195, 306–320. doi: 10.1111/j.1469-8137.2012.04190.x
Wang, K., Cui, K. H., Liu, G. L., Xie, W. B., Yu, H. H., Pan, J. F., et al. (2014). Identification of quantitative trait loci for phosphorus use efficiency traits in rice using a high density SNP map. BMC Genet. 15, 155. doi: 10.1186/s12863-014-0155-y
Wang, X., Shen, J., Liao, H. (2010). Acquisition or utilisation, which is more critical for enhancing phosphorus efficiency in modern crops? Plant Sci. 179, 302–306. doi: 10.1016/j.plantsci.2010.06.007
Wang, Q., Xie, W., Xing, H., Yan, J., Meng, X., Li, X., et al. (2015). Genetic architecture of natural variation in rice chlorophyll content revealed by a genome-wide association study. Mol. Plant 8, 946–957. doi: 10.1016/j.molp.2015.02.014
Wen, W., Li, D., Li, X., Gao, Y., Li, W., Li, H., et al. (2014). Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat. Commun. 5, 3438. doi: 10.1038/ncomms4438
Wissuwa, M., Kondo, K., Fukuda, T., Mori, A., Rose, M. T., Pariasca-Tanaka, J., et al. (2015). Unmasking novel loci for internal phosphorus utilization efficiency in rice germplasm through genome-wide association analysis. PloS One 10, e0124215. doi: 10.1371/journal.pone.0124215
Wissuwa, M., Wegner, J., Ae, N., Yano, M. (2002). Substitution mapping of Pup1: a major QTL increasing phosphorus uptake of rice from a phosphorus-deficient soil. Theor. Appl. Genet. 105, 890–897. doi: 10.1007/s00122-002-1051-9
Wissuwa, M., Yano, M., Ae, N. (1998). Mapping of QTLs for phosphorus-deficiency tolerance in rice (Oryza sativa l.). Theor. Appl. Genet. 97, 777–783. doi: 10.1007/s001220050955
Wu, J. H., Feng, F. J., Lian, X. M., Teng, X. Y., Wei, H. B., Yu, H. H., et al. (2015). Genome-wide association study (GWAS) of mesocotyl elongation based on re-sequencing approach in rice. BMC Plant Biol. 15, 218. doi: 10.1186/s12870-015-0608-0
Wu, P., Shou, H. X., Xu, G. H., Lian, X. M. (2013). Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr. Opin. Plant Biol. 16, 205–212. doi: 10.1016/j.pbi.2013.03.002
Xie, C., Mao, X. Z., Huang, J. J., Ding, Y., Wu, J. M., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identifcation of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Yamaji, N., Xia, J., Mitani-Ueno, N., Yokosho, K., Ma, J. F. (2013). Preferential delivery of zinc to developing tissues in rice is mediated by p-type heavy metal ATPase OsHMA2. Plant Physiol. 162, 927–939. doi: 10.1104/pp.113.216564
Yang, H., Knapp, J., Koirala, P., Rajagopal, D., Peer, W. A., Silbart, L., et al. (2007). Enhanced phosphorus nutrition in monocots and dicots overexpressing a phosphorus-responsive type I h+-pyrophosphatase. Plant Biotechnol. J. 5, 735–745. doi: 10.1111/j.1467-7652.2007.00281.x
Yano, K., Yamamoto, E., Aya, K., Takeuchi, H., Lo, P. C., Hu, L., et al. (2016). Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat. Genet. 48, 927–934. doi: 10.1038/ng.3596
Zhang, Z. W., Ersoz, E., Lai, C. Q., Todhunter, R. J., Tiwari, H. K., Gore, M. A., et al. (2010). Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42, 355–360. doi: 10.1038/ng.546
Zhao, K., Tung, C. W., Eizenga, G. C., Wright., M. H., Ali., M. L., Price, A. H., et al. (2011). Genome-wide association mapping reveals a rich genetic architecture of complex traits in oryza sativa. Nat. Commun. 2, 467. doi: 10.1007/s00122-017-2957-6
Keywords: GWAS, OsAAD, physiological phosphorus use efficiency, biomass, grain yields
Citation: Yan M, Feng F, Xu X, Fan P, Lou Q, Chen L, Zhang A, Luo L and Mei H (2023) Genome-wide association study identifies a gene conferring high physiological phosphorus use efficiency in rice. Front. Plant Sci. 14:1153967. doi: 10.3389/fpls.2023.1153967
Received: 30 January 2023; Accepted: 01 March 2023;
Published: 14 March 2023.
Edited by:
Jianlong Xu, Institute of Crop Sciences (CAAS), ChinaReviewed by:
Wen-Xue Li, CAAS, ChinaKeke Yi, Institute of Agricultural Resources and Regional Planning (CAAS), China
Copyright © 2023 Yan, Feng, Xu, Fan, Lou, Chen, Zhang, Luo and Mei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanwei Mei, aG1laUBzYWdjLm9yZy5jbg==; Lijun Luo, bGlqdW5Ac2FnYy5vcmcuY24=
†These authors have contributed equally to this work
 Ming Yan
Ming Yan Fangjun Feng
Fangjun Feng Xiaoyan Xu1,2
Xiaoyan Xu1,2 Qiaojun Lou
Qiaojun Lou Liang Chen
Liang Chen Anning Zhang
Anning Zhang Lijun Luo
Lijun Luo Hanwei Mei
Hanwei Mei