
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 20 June 2023
Sec. Functional and Applied Plant Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1152468
 Niaz Ahmad1
Niaz Ahmad1 Samia Fatima1
Samia Fatima1 Muhammad Aamer Mehmood2
Muhammad Aamer Mehmood2 Qamar U. Zaman3,4
Qamar U. Zaman3,4 Rana Muhammad Atif5,6
Rana Muhammad Atif5,6 Weijun Zhou7
Weijun Zhou7 Mehboob-ur Rahman1*
Mehboob-ur Rahman1* Rafaqat Ali Gill8*
Rafaqat Ali Gill8*CRISPR-mediated genome editing has emerged as a powerful tool for creating targeted mutations in the genome for various applications, including studying gene functions, engineering resilience against biotic and abiotic stresses, and increasing yield and quality. However, its utilization is limited to model crops for which well-annotated genome sequences are available. Many crops of dietary and economic importance, such as wheat, cotton, rapeseed-mustard, and potato, are polyploids with complex genomes. Therefore, progress in these crops has been hampered due to genome complexity. Excellent work has been conducted on some species of Brassica for its improvement through genome editing. Although excellent work has been conducted on some species of Brassica for genome improvement through editing, work on polyploid crops, including U’s triangle species, holds numerous implications for improving other polyploid crops. In this review, we summarize key examples from genome editing work done on Brassica and discuss important considerations for deploying CRISPR-mediated genome editing more efficiently in other polyploid crops for improvement.
Brassica is an important genus of the Brassicaceae family, also known as Cruciferae or the mustard family. It is the 5th largest monophyletic family among the angiosperms (Mun et al., 2009). The renowned Brassica “U’s triangle” (Nagaharu and Nagaharu, 1935) comprises three diploid species, including Brassica rapa (AA), B. nigra (BB), B. oleracea (CC), and their derivatives, three allotetraploid species, namely B. napus (AACC), B. juncea (AABB), and B. carinata (BBCC) (Cheng et al., 2017). Brassica crops are an important source of vegetable oils (Lu et al., 2018) and contribute significantly to global vegetable crop production, providing protein-rich feed for animals, as well as raw materials for biofuel production (Yang et al., 2018). Typically, Brassica seed contains 40–48% oil and 38–40% protein in the oil-free seed meal (Rahman et al., 2013). Oil extracted from Brassica species is considered one of the healthiest edible oils due to its low saturated fat contents (~7%), with the remaining 93% comprised of mono and poly-unsaturated fatty acids (Orsavova et al., 2015; Sharafi et al., 2015).
Polyploidy is a heritable state where a crop acquires one or more additional sets of genomes, either similar to the earlier one (autopolyploidy) or different ones (allopolyploidy), during the course of its evolution (Mason and Wendel, 2020). Positive consequences of polyploidy include gene redundancy for masking the effect of deleterious alleles, neo-functionalization of duplicated genes, emergent self-fertilization, fixed heterosis, and the capability of asexual reproduction, all of which have contributed significantly to the success of polyploid crops (Comai, 2005). Polyploidization has played a crucial role in the domestication of crop species through the creation of novel gene functions, combinations, interactions, and epigenetic changes, resulting in the enhanced adaptability of these crop species (Udall and Wendel, 2006). The beneficial effects of polyploidy are well elucidated, such as higher photosynthetic rates in polyploid species compared to diploids (Coate et al., 2012), increased fibre quality of allotetraploid cotton (Jiang et al., 1998), and grain hardness in tetra- and hexaploid wheat (Chantret et al., 2005).
Polyploidy is a significant contributor to speciation in plants, and almost every plant species displays signs of polyploidization. While polyploidy can create new variations that enhance a plant’s genetic potential, polyploid crops experience significant genome rearrangements, including multiple gene homologues, extensive patches of repetitive DNA, and high levels of heterozygosity – resulting in highly complex genomes. As a result, genetic improvement of field crops has become increasingly challenging (Xiong et al., 2011; Chen et al., 2019). However, the recent emergence of precise genome editing tools has revolutionized plant biotechnology. Genome editing entails creating site-specific mutations in the genome, such as insertions, deletions, or base substitutions (Manghwar et al., 2019). All genome editing tools, including Mega nucleases (MNs), Zinc finger nucleases (ZFNs), Transcription activator-like effector nucleases (TALENs), and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), operate by creating double-stranded breaks (DSBs) followed by DNA repair via either through intrinsic NHEJ (non-homologous end joining) or homologous recombination (HR) with the provision of an extrinsic DNA fragment (Symington and Gautier, 2011; Ahmad et al., 2020).
The CRISPR system has completely transformed the genome editing landscape and has been extensively used in humans, animals, and plants for various applications, such as base editing, epigenetic modifications, prime editing, multiplexed genome editing, gene regulation, gene replacement, and labelling of specific chromosomal segments (Larson et al., 2013; Li et al., 2013; Nekrasov et al., 2013; Sander and Joung, 2014; Schaeffer and Nakata, 2015; Gallego-Bartolomé et al., 2018; Lowder et al., 2018; Anzalone et al., 2019; Wang et al., 2019; Zhang et al., 2019b). Compared to previous protein-DNA-based genome editing systems, CRISPR technology is cost-effective, robust, simple, and able to target multiple genes simultaneously (Cong et al., 2013; Shan et al., 2013). In plants, CRISPR technology has primarily been used to eliminate gene products that negatively affect plant traits through small indels, resulting in frame-shift mutations or premature stop codons (Ahmad and Mukhtar, 2017; Zhang et al., 2018b). CRISPR/Cas type II is the most widely used genome editing system (Hsu et al., 2014), consisting of two main components: Cas9 and guide RNA (gRNA). Cas9 is the nuclease responsible for creating DSBs at the target site, while gRNA is a chimeric RNA molecule formed by the homologous pairing of Precursor-CRISPR RNA (pre-crRNA) and Trans-activating RNA (tracrRNA) after transcription (Kumar et al., 2019; El-Mounadi et al., 2020). The gRNA contains 20 nucleotides that are complementary to a specific region in the target DNA, followed by a 3-bp consensus PAM (protospacer adjacent motif) sequence. The 3’ end of gRNA forms a loop structure, which helps fix the guide sequence to the target DNA, and interacts with Cas9 protein to form a ribonucleoprotein (RNP) complex responsible for creating DSBs at the target DNA location (Deltcheva et al., 2011).
The world’s population is expected to reach 10 billion by 2050, necessitating a significant increase in global food production (Hickey et al., 2019). To meet this challenge, various international organizations, including the UN and WHO, have called for a 60–70% increase in food production by 2050 (van Dijk et al., 2021). The improvement of Brassica crops could help meet these demands, as they are consumed in a variety of ways by both humans and animals and have many applications in industry. However, these crops face many challenges, including susceptibility to heat, cold, drought, shattering, insect/pests, deadly blackleg disease, and late maturation in the case of the widely cultivated B. napus (Augustine et al., 2014; Raman et al., 2014; Zhu et al., 2016; Wrucke et al., 2019; Kourani et al., 2022). B. juncea, another Brassica crop, is early maturing and tolerant to harsh climatic conditions but it contains high levels of anti-nutrients such as erucic acid and glucosinolates (Nour-Eldin et al., 2017). Therefore, the development of need-based superior Brassica crops that offer better yield potential, resilience to harsh climatic conditions, disease and insect pest resistance, and meet either edible or non-edible requirements has been a long-standing objective in Brassica breeding, which can be achieved more effectively with advanced biotech tools like CRISPR technology. The focus of this article is to review the latest research on the use of CRISPR technology for improving Brassica crops. This article also highlights the lessons learned from Brassica genome editing that can be applied to improve polyploid crops through CRISPR.
Earlier efforts to improve Brassica involved traditional breeding approaches. However, due to its polyploidy, achieving breeding objectives through traditional methods has been quite challenging (Mason and Snowdon, 2016). Modern genetic tools have become necessary to develop high-yielding crop varieties on a sustainable basis against changing climates, given the complex genome of polyploids like Brassica. In cases where genetic variability is not present in the crossable germplasm for a specific trait, spontaneous natural mutations or induced mutagenesis can also be an option. However, finding the desired edits can be complicated and time-consuming, and sometimes the desired mutations may negatively impact the desired characteristics such as yield. As a result, breeders have to undergo multiple laborious and time-taking crossing and selfing procedures to combine mutant copies in a single line (Schouten and Jacobsen, 2007).
An alternative approach to creating desired traits in field crops is genetic engineering. This approach has been used to engineer several desirable traits in plants, including herbicide tolerance (Blackshaw et al., 1994; Rong et al., 1997; Qing et al., 2000), oil quality improvement (Stoutjesdijk et al., 2000; Liu et al., 2001; Naeem et al., 2020), insect/pest resistance (Kanrar et al., 2002; Wang et al., 2005; Liu et al., 2011; Rani et al., 2017), salt tolerance (Prasad et al., 2000; Zhang et al., 2001), cold tolerance (Jaglo et al., 2001), shattering resistance (Østergaard et al., 2006), phytase enzyme production (Ponstein et al., 2002; Wang et al., 2013), and development of male sterile and fertility restorer transgenic lines (Jagannath et al., 2001; Jagannath et al., 2002). However, genetic transformation approaches are associated with health and environmental concerns, and genetically modified crops have low acceptance by the public (See Ahmad and Mukhtar, 2017 for review). The controversy surrounding the development of transgenic plants through conventional genetic engineering approaches has led to the ban of open-field cultivation in many countries, such as France, Germany, Greece, Italy, Turkey, Ecuador, Russia, Madagascar, Netherlands, Algeria, Saudi Arabia, Venezuela, Ukraine, Austria, Hungary and Poland (Jaganathan et al., 2018). Genome editing tools, particularly CRISPR, allow the development of transgene-free edited plants that can pass the regulatory framework easily and increase public acceptance of GM crops (Figure 1). Consequently, CRISPR has become a tool of choice for developing edited plants with desired traits (Chen and Gao, 2014). In the following sections, we will discuss different studies reporting the successful deployment of the CRISPR/Cas platform for the improvement of Brassica.
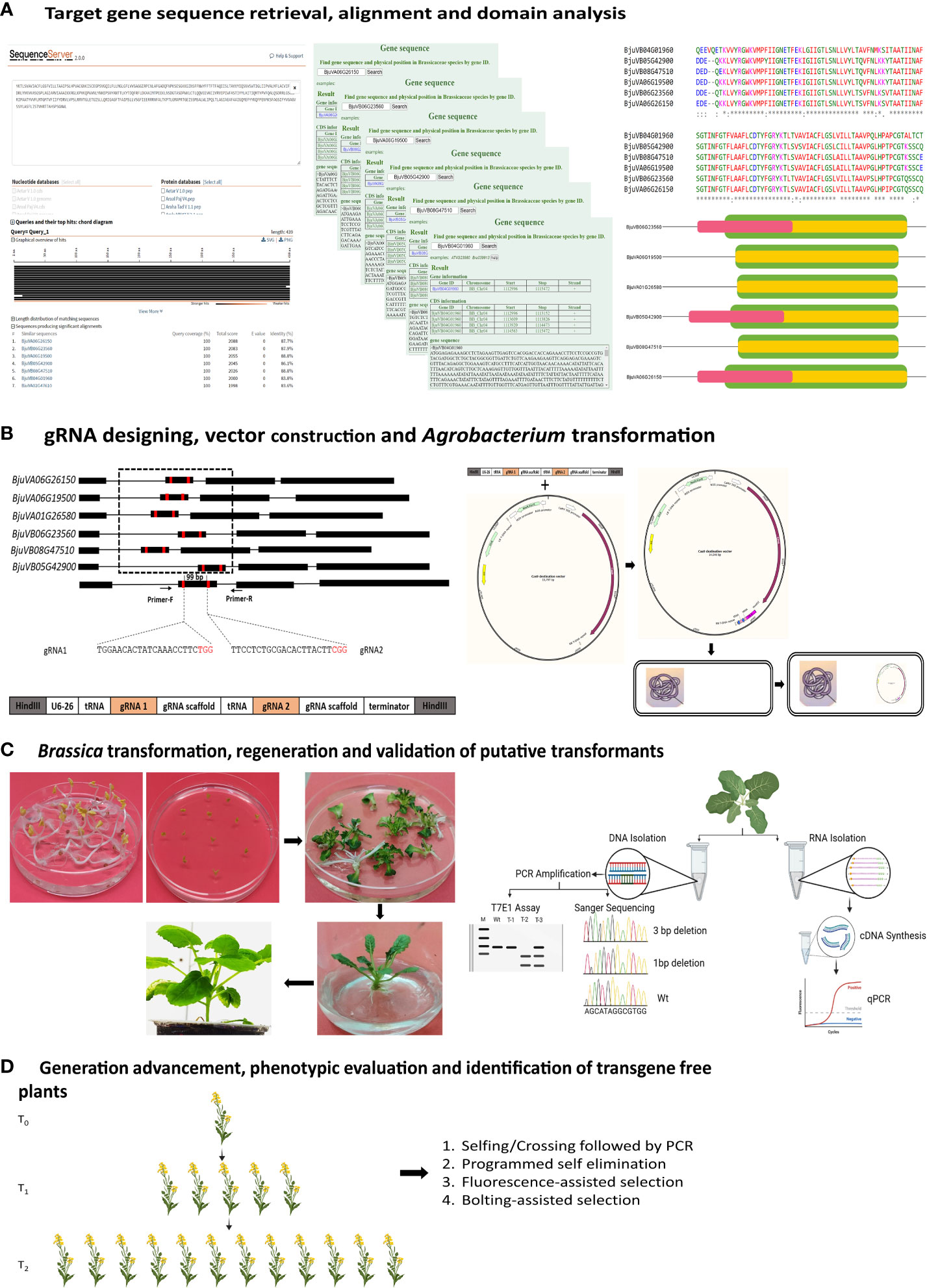
Figure 1 Schematic illustration of the steps involved in the development of transgene-free genome-edited Brassica plants. (A) shows the identification and retrieval of target gene sequences from public databases, such as GenBank or The Brassicaceae Database (BRAD V3.0; http://brassicadb.cn). The sequences are aligned to identify potential domains for designing gRNA. (B) shows the gRNA designing, construction of plant expression vectors followed by transformation of that construct into Agrobacterium strain for plant transformation. (C) represents Agrobacterium¬- mediated Brassica transformation using cotyledonary explants, shoot regeneration on the selection medium, root induction, establishment of the plants in compost, and screening of the putative transformants using techniques such as T7 endonuclease I assay, Sanger sequencing, or qPCR. (D) illustrates the process of eliminating marker genes to obtain transgene-free edited plants through Mendelian segregation. Transgene-free edited plants can also be obtained by programmed self-elimination, using suicidal genes, and can be immediately identified using fluorescent genes. Another way of identification of transgene-free edited plants is by incorporating genes that confer distinct phenotypes, such as early flowering (Bolting assisted selection) or fluorescence. Some parts of the picture were created using BioRender.com.
Yield is a complex trait that depends on several parameters including plant height, number of siliques, number of branches, multilocular siliques, and seed shattering. Researchers have targeted these primary traits to increase yield. For instance, when a gene ALCATRAZ (ALC) involved in valve margin development, was edited, not only was pod-shattering reduced, but the silique length was also significantly improved (Braatz et al., 2017). Likewise, editing the CLAVATA (CLV1, CLV2 and CLV3) genes involved in multilocular silique phenotype by regulating stem cell function, cell fate, and proliferation in Brassica increased the number of seeds as well as the seed weight (Yang et al., 2018). Repressing all five copies of JAGGED (JAG) genes in B. napus resulted in disturbed plant phenotypes, such as unorganized cell identity in floral primordia leading to pod deformation and serrated leaves. Transgenic plants with edits in BnJAG.A08 showed alterations in the dehiscence zone development only, with increased replum area, decreased pod length and improved pod shattering resistance (Zaman et al., 2019a). Sriboon et al. (2020) edited all five copies of BnaTFL1, a flowering inhibitor gene also implicated in the regulation of plant architectural traits. The authors showed that plants with edits in all gene copies displayed significant changes in the plant architecture, such as reduced plant height, branch initiation height, branch number, silique number, and the number of seeds/siliques on the main inflorescence. The most severe alterations in the plant architecture were observed in the BnaC03.tfl1 edited plant. This suggests that BnaC03.TFL1 not only plays a significant role in flowering mechanism but also plant architecture traits along with other BnaTFL1 gene homologues. In a recent study, Khan et al. (2021) edited four homologues of Brassica napus gene BnaEOD3, resulting in plants with shorter siliques, with a smaller yet increased number of seeds/siliques. The study further illustrated the differential and quantitative involvement of BnaEOD3 gene homologues in seed development. For instance, BnaC04.EOD3b and BnaA04.EOD3 were found to play the most significant and minor role in seed development, respectively.
Plant architecture traits such as plant height, branch number, and root architecture are associated with yield. Therefore, understanding the function of genes governing these traits can help improve them and ultimately increase yield. Lawrenson et al. (2015) edited the GIBBERELLIN 3-OXIDASE1 (BolC.GA4.a) gene in B. oleracea and found it to be involved in regulating the gibberellic acid pathway. Kirchner et al. (2017) investigated the role of the BcFLA gene in B. carinata and found that edited plants developed shorter root hairs. Leaf morphology is an important trait that indirectly contributes to yield increase by affecting evapotranspiration rates, sunlight penetration, and insect/pest preference. In Brassica species, diverse leaf morphology is observed, including entire, serrated and lobed leaves. The lobed leaf phenotype is preferred for high-density crop cultivation, mechanized harvesting, and reduced insect and disease incidence. Hu et al. (2018) studied the role of the BnA10.LMI1 gene in the formation of lobed leaves in B. napus and reported that the LMI1 gene encodes for an HD-Zip I transcription factor that regulates the expression of the LLA10 gene, which is responsible for the lobed leaf shape. Zheng et al. (2020) developed edited plants with mutations in BnaMAX1. The resulting plants displayed a semi-dwarf phenotype with enhanced branching and siliques, resulting in a significant yield increase. Cheng et al. (2021) used an A3A-PB3 base editing system to replace C with T in BnRGA and BnIAA7 genes separately and obtained dwarf plant types in both cases, suggesting that both of these genes are involved in regulating plant height. Stanic et al. (2021) edited the BnD14 gene and observed a significant increase in the number of branches (200%) and the number of flowers per plant (37%), suggesting it is a single major potential candidate gene for improving yield-related traits. Fan et al. (2021) developed semi-dwarf B. napus plants with compact architecture and without any undesirable traits by targeting BnaA03.BP gene, concluding that BnaA03.BP alone could be a potential candidate for a moderate phenotype.
Improvement in total oil content, along with a better fatty acid profile, is a major goal in improving oilseed Brassica crops. The manipulation of these traits has been achieved through the successful application of CRISPR/Cas9 technology. Studies have shown that yellow seed coat colour is associated with enhanced oil content and better fatty acid composition in Brassica species. Double knockout yellow seed phenotypes were produced with enhanced seed oil (51.8%) and protein content (16.97%) with a modified fatty acid profile without causing any severe defects in yield-related traits by targeting TRANSPARENT TESTA8 (BnTT8) (Zhai et al., 2020). Similarly, Xie et al. (2020) edited the TRANSPARENT TESTA2 (BnTT2) gene in B. napus, resulting in a yellow-seeded phenotype with improved oil content up to 45–47% and better fatty acid composition with enhanced linoleic and linolenic acid.
The nutritional quality of the oil is primarily determined by the proportion of different fatty acids present. CRISPR technology has also been used to develop Brassica plants with a better fatty acid profile. Oleic acid is a monounsaturated acid that not only increases the shelf life of vegetable oil but also protects it from damage due to oxidation. High oleic acid B. napus lines were produced by introducing a novel edited allele of the FATTY ACID DESATURASE 2 (FAD2) gene involved in the conversion of oleic acid into linoleic acid (Okuzaki et al., 2018; Huang et al., 2020).
In addition to being rich in oil contents, Brassica seeds are also a good source of proteins with a good amino acid profile. However, the presence of certain anti-nutrients called glucosinolates reduces the utilization of seed meal as animal feed. Neequaye et al. (2021) targeted three gene copies of the MYB28 gene involved in the regulation and biosynthesis of aliphatic glucosinolates in B. oleracea. Editing was observed in only C2 and C9 copies of the gene but not in the C7 gene copy. The knockout plants showed notable downregulation of aliphatic glucosinolates biosynthesis and accumulation in the leaves and florets of the edited plants. This implies that a significant reduction in the aliphatic glucosinolates accumulation can be achieved by editing two of the three gene copies and the C7 copy may have a subfunctional role in glucosinolates accumulation, or it may evolve into a pseudogene in the course of evolution.
Phytic acid is another important anti-nutrient present in Brassica seed that blocks the absorption of minerals and proteins in monogastric animals. Sashidhar et al. (2020) edited the members of the BnITPK gene family (four from BnITPK1 and two from the BnITPK4 sub-family) involved in the biosynthesis of phytic acid. They observed a significant reduction (~35%) in the phytic acid content in edited lines.
Brassica seed, particularly those of yellow mustard (Sinapis alba) and brown mustard (B. juncea), contains proteins that can trigger allergic reactions, such as urticaria, wheezing, abdominal pain, dyspnea, and life-threatening anaphylactic shocks (Matsuo et al., 2015). One of the proteins, BrajI, found in B. juncea seeds, cause allergic reactions. The development of allergen-free food could offer a reliable solution to this issue. Assou et al. (2022) employed CRISPR/Cas9 genome editing in B. juncea and successfully eliminated BrajI from the seeds.
Biotic and abiotic stresses are major factors that depress yield in field crops, including Brassica. The CRISPR platform is increasingly being used to develop crops that are resilient to these stresses. Sclerotinia sclerotiorum is a fungal disease that causes significant yield losses in Brassica. Sun et al. (2018) edited BnWRKY11 and BnWRKY70 genes in B. napus. The lines with edited BnWRKY70 showed enhanced resistance to S. sclerotiorum while those with BnWRKY11 showed no significant difference. Verticillium longisporum (Vl43) is another serious fungal disease of Brassica responsible for significant yield losses due to the absence of effective genetic resistance in the crop. Pröbsting et al. (2020) developed knock-out plants of B. napus for the BnCRT1 gene using CRISPR and observed a significant drop in the fungal susceptibility of the crop against Vl43. Zhang et al. (2022a) elucidated the mechanism of rapeseed resistance against S. sclerotiorum using CRISPR/Cas9 technology. They identified a positive role of MITOGEN-ACTIVATED PROTEIN KINASES, BnaA03.MKK5-BnaA06.MPK3/BnaC03.MPK3 module in resistance against S. sclerotiorum. The double knock out plants showed an enhanced susceptibility to the disease.
Wu et al. (2020) mutated BnaA6.RGA and the resulting plants showed increased drought tolerance. However, the quadruple bnarga edited plant line exhibited reduced drought tolerance.
Mineral deficiencies, such as boron, are significant factors involved in stunted growth and yield reduction in Brassica. However, the mechanism involved in the adaptation of plants in low boron environments is not clear. Feng et al. (2020) elucidated the function of the BnWRKY47 transcription factor under boron stress using CRISPR technology. They found that BnWRKY47 was responsible for the adaptation of B. napus to low boron environments by up-regulating the boric acid channel gene BnaA3.NIP5;1.
As biotic and abiotic stresses cause major yield losses in almost all major field crops including polploid crops, therefore, utilizing CRISPR could be an effective strategy for developing stress-tolerant crops.
In polyploid crop species, the most important traits are controlled by multiple genes, making it necessary to identify and select genes that significantly contribute to the trait of interest for manipulation. In polyploid crops like Brassica, the situation becomes more complicated due to the presence of many copies of a single gene in the genome and their functional redundancy. Thus, functional genomics and proper validation of target genes is crucial for improving particular traits. Genes that control a trait can be divided into two classes: structural (protein coding) and regulatory (transcription factors). Structural genes, with a direct role in crop traits, provide an attractive target for precise genome editing. Regulatory genes, mostly consisting of transcription factors and other regulatory elements, are involved in regulating structural genes. They are induced by certain biological processes such as biotic and abiotic stress signals, and hence, control the expression of many downstream structural genes. Therefore, targeting regulatory genes may help improve crops against complex stress phenomena.
CRISPR/Cas9 technology can be effectively employed to gain insights into the functional roles of specific genes by producing knockout or loss-of-function alleles. Pod shattering is a widely investigated subject in the case of the Brassica crop because of its significant role in yield reduction. Previously, ALCATRAZ (ALC) and INDEHISCENT (IND) gene homologues were found to have a significant role in pod dehiscence. Zhai et al. (2019) assessed the function of these genes in pod shattering in B. napus using CRISPR/Cas9 technology. The results from the phenotypic evaluation of homozygous knockout plants showed the conserved and essential role of the BnIND gene, whereas the BnALC gene showed limited potential for B. napus pod-shattering resistance. Additionally, BnIND gene homologues exhibited functional redundancy, with a major contribution from BnA03.IND.
The CRISPR platform has also been used in Brassica to carry out functional genomic studies on genes with multiple copies. For example, Zhang et al. (2019a) used CRISPR/Cas9 to understand the functions of LYSOPHOSPHATIDIC ACID ACYLTRANSFERASE (LPAT2) and (LPAT5) family genes, both of which have seven and five homologues, respectively. These genes encode several key enzymes in the Kennedy pathway, which catalyze the fatty acid chains into 3-phosphoglycerate and promote oil production in the form of triacylglycerols. The edited lines showed enlarged oil bodies but lesser oil content than the wild type, suggesting that the LPAT2/5 gene families play an important role in fatty acid biosynthesis.
Heat stress severely impacts seed production in Brassica by altering the normal anatomy of floral organs. Unfortunately, the molecular mechanism underlying this phenomenon is poorly understood. One mutation produces novel pistil-like flowers in B. rapa, where four of the five sepals merge to form a ring structure that encapsulates abnormal stamens and a pistil, leading to poor seed production. The mutation is called sepal carpal modification (scm) and is found in the BrAP2 gene homologues, which are orthologues of the Arabidopsis APETALA (AP2) gene. To investigate the function of four BnAP2 gene homologues in sepal and petal development, rapeseed knockout plants were generated using the CRISPR/Cas9 tool (Zhang et al., 2018a). The quadruple knockout plants showed an scm-like appearance, confirming the functional conservation of the AP2 gene in Brassica. This study also provides information on the modification of floral organs by genome manipulation for yield improvement (Zhang et al., 2018a). Jedličková et al. (2022) studied the function of the TRYPTOPHAN AMINOTRANSFERASE (BnaTAA1) gene in the auxin biosynthesis pathway.
Yang et al. (2017) simultaneously targeted 12 genes from different families, including the RGA, FUL, DA1, and DA2 gene families. Most of the plant lines exhibited a mutation frequency of 5.3% to 100%, with the development of homozygous and bi-allelic mutations stably inherited to successive generations, highlighting the fact that single gene editing in polyploid species did not yield any significant phenotypic differences. Xiong et al. (2019) functionally characterized the gene involved in pollen growth and development in B. compestris. They targeted three homologous PECTIN METHYLESTERASE (PME) genes using CRISPR/Cas9 technology and explored that Bra003491 had a significant role in pollen growth and development, while the other two homologues, BRA007665 and Bra014410, may function redundantly. Wang et al. (2022b) employed the CRISPR/Cas9 system to explore the function of STARCH BRANCHING ENZYME (BnaSBE) genes in starch structure and overall throughput of B. napus. The analysis revealed the least activity of the starch-binding enzyme, binding frequency, higher starch-bound phosphate content, and altered pattern of amylopectin chain length distribution in sextuple edited plants compared to the wild-type.
The development of early maturing B. napus has long been a breeding objective due to the crop’s ability to escape disease and aphid attack, both of which can significantly reduce crop yields. Jiang et al. (2018) investigated the role of methyl transferase in two SET DOMAIN GROUP 8 (BnSDG8.A and BnSDG8.C) genes in rapeseed using CRISPR/Cas9 technology. The SDG8 gene is pleiotropic in nature and regulates multiple biological processes, such as flowering time and plant height. The mutations significantly reduced flowering time compared to the wild type. Jeong et al. (2019) developed early-flowering B. rapa (Chinese cabbage) through targeted editing of the four homologous FLOWERING LOCUS C (BrFLC) genes, eliminating the need for vernalization for flower induction. In a similar effort to reduce flowering time, Sriboon et al. (2020) targeted five gene copies of TERMINAL FLOWER 1 (BnaTFL1), a flowering inhibitor gene and also regulate plant architectural traits, using CRISPR/Cas9 technology. They obtained knockout plants for all gene copies and a knockout plant for BnaC03.tfl1/BnaC09.tfl1. However, only the single knockout BnaC03.tfl1 and double knockout BnaC03.tfl1/BnaC09.tfl1 displayed an early flowering phenotype, demonstrating the significant role of BnaC03.TFL1 in the flowering mechanism. Vernalization treatment has been found to reduce the transcript levels of the FLC gene in Arabidopsis, thereby inducing flowering. Hong et al. (2021) targeted three gene homologues of VERNALIZATION 1 (VRN1) using CRISPR/Cas9 technology to develop late flowering phenotypes in B. rapa. VRN1 is known to be involved in the negative regulation of FLC gene expression. In an effort to attain early maturity, Ahmar et al. (2022) targeted SHORT VEGETATIVE PHASE (BnaSVP) genes using CRISPR. Homozygous transgene-free lines showed a significant decrease in flowering time, with the shortest flowering time in the quadruple edited plant as compared to that of the wild type, confirming the quantitative contribution of BnaSVP gene copies in the flowering time trait in a polyploid species.
Brassica crops are also consumed as vegetables, and cabbage (B. oleracea var. capitata) is well-known for its better nutritional profile, anti-cancer, and anti-inflammatory properties. However, early flowering affects the yield and quality of cabbage by reducing vegetative growth. To address this, Park et al. (2019) targeted two alleles of the flowering time regulator BoGIGANTEA (GI) gene using CRISPR, resulting in a significant increase in flowering time in the edited lines. Li et al. (2018) targeted five gene copies of SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE 3 (BnSPL3) gene, a key floral activator involved in the plant’s transition from vegetative to the reproductive stage, in oilseed rape. They obtained lines with all five edited gene copies that displayed significantly delayed phenotypes compared to wild-type control plants.
Brassica plants have also been cultivated as ornamental crops in some countries, such as China. Flower colour is an important aesthetic value of the crop, and Liu et al. (2020) developed phenotype with orange-coloured flowers by knocking out the ZEAXANTHIN EPOXIDASE (BnaA09.ZEP and BnaC09.ZEP) genes involved in the increased lutein content and decreased violaxanthin content in petals, specifically giving orange colour to petals.
Pod shattering is a significant factor that reduces yield in Brassica. The SHATTERPROOF (SHP1 and SHP2) genes regulate lignin content in the dehiscence zone, which contributes to seed shattering in Brassica. To address yield losses due to pod shattering, Zaman et al. (2021) targeted six BnSHP1 and two BnSHP2 homologues to develop edited lines with multiple edited gene copies. The phenotypic evaluation showed that BnSHP1A09 may have a significant role in regulating lignin content in the dehiscence zone, with BnSHP1A09/C04-B/A04 and BnSHP2A05/C04-A showing the most reduction in lignin and separation layer adjacent to valves and replum. To confirm the functional redundancy of these genes, a single knockout line was crossed with a quadruple knockout line to develop a line with mutations at all five homologues, namely, BnSHP1A09, BnSHP1C04-B, BnSHP1A04, BnSHP2A05, and BnSHP2C04-A. The resulting five-homologues knockout plant showed significantly increased resistance to pod shattering.
Brassica yields are also affected by weed infestations, resulting in significant yield losses and quality deterioration. Effective weed management involves the use of herbicides, which is labour and cost-effective. However, Brassica crops tolerant to herbicides, developed through traditional genetic engineering approaches, fall under strict GMO regulations and hence cannot be cultivated in many countries. Wang et al. (2021) recently reported the development of glyphosate-tolerant B. napus using a CRISPR/Cas9-based geminiviral donor DNA replication system. They used a bacterial Cys4-based single RNA processing system to replace the endogenous 5-ENOLPYRUVYLSHIKIMATE-3-PHOSPHATE SYNTHASE (EPSPS) gene, involved in the synthesis of branched amino acids, with its modified form. Additionally, Wang et al. (2022a) used the modified CRISPR/Cas9-based editing system to induce a point mutation (C to T) in the ACETOLACTATE SYNTHASE (ALS) gene, which encodes for a key enzyme involved in the biosynthesis of branched-chain amino acids, resulting in the creation of herbicide-tolerant cauliflower plants. The ALS gene presents a potential target site for several important herbicides. Table 1 provides a summary of different traits edited in Brassica using CRISPR.
CRISPR/Cas9 technology has immense potential and advantages, but it also poses significant challenges when it comes to genome editing of crops with complex polyploid genomes like Brassica. While discussing the successful deployment of CRISPR in various fields, it is important to consider the potential difficulties of editing the genomes of polyploid species with complex genomes.
One of the primary challenges is genome complexity. Polyploid crops, including Brassica species, have undergone complex genomic rearrangements, including at least one whole genome duplication event in their evolutionary history. The triplication events undergone by Brassica species after separation from Arabidopsis resulted in highly complex genomes. For instance, Yang et al. (2018) found three gene copies of BnCLV3 in the released B. napus genome while targeting BnCLV gene homologues, but they were able to identify only two gene copies in the pure line J9707. Similarly, in an attempt to edit BnSBE2 gene homologues, Wang et al. (2022b) retrieved a total of six genes (four BnSBE2.1 and two BnSBE2.2) from the released B. napus genome database, but they found only three gene copies of BnSBE2.1 and one gene of BnSBE2.2 in the cultivar DH12075. This high level of genome complexity and gene redundancy makes gene editing a challenging task in polyploid crops, particularly when dealing with recessive traits that require the elimination of all alleles (Khan et al., 2021; Assou et al., 2022). It is worth mentioning that for traits that require editing of all genes, as is the case with polyploid crops, the phenotype may quickly revert when grown in the field, particularly in open-pollinated species (Wang et al., 2014). The reversion frequency in the case of recessive alleles has been proposed to be 50% higher than dominant genes (Ahmad et al., 2020). Therefore, it is important to undertake preventive strategies such as introducing buffer zones and maintaining the original edited versions in isolated blocks.
The availability of complete, well-annotated, and elucidated genome sequence information, along with functional genomic studies, is a prerequisite for genome editing technology, as it facilitates the direct identification of candidate genes for editing. However, the highly repetitive and complex nature of polyploid genomes poses risks for computational biologists, such as phasing, full chromosome assembly, gene annotations, and differentiation between homo- and homeologues during the development of polyploid genome sequences (Kyriakidou et al., 2018). Additionally, chromosomal rearrangements and epigenetic modifications in polyploid plant species often result in transcriptional changes, including the activation of transposable elements, neo-functionalization of duplicated genes, and biased homeologues expression (Wendel et al., 2018), which can make it difficult to link genotype to phenotype and molecular characterization of edited genes in polyploid species. Due to a sequence resource deficit, and biased gene expression, genome sequencing is often limited to a model plant within a species. This can pose difficulties in selecting target genes, which can be addressed by using translational genomics approaches to develop genotype-phenotype connections (May et al., 2023).
The actual number of homologue and homeologue genes underlying a trait and their functionality status remains unclear in polyploid crops. However, gene dosage phenomena have been observed for several traits in polyploid crops, where each gene has a small contribution to the execution of a trait (Schaart et al., 2021).
Designing gRNA is a critical step in CRISPR/Cas9 genome editing, as it needs to be highly specific to the target gene. In a polyploid crop, where multiple genes are targeted simultaneously using a single gRNA, specific gRNA design becomes a challenging task (Zaman et al., 2019b). Although various online tools have been developed for gRNA designing and predicting gRNA efficiency in polyploid crops, they may not guarantee the efficiency of a particular gRNA in living cells (Okada et al., 2019). Therefore, gRNA in polyploid crops like Brassica are manually designed after multiple sequence alignments on the conserved regions of all targeted gene copies. If a conserved region is not present in all the gene homologues, these homologues are grouped, and gRNAs are designed on the conserved region of each group (Zaman et al., 2019b). Apart from gRNA specificity, gRNAs with a GC content of 50–60% and a relatively shorter length of 18 bp, preferably starting from A or G, exhibit high on-target efficiencies (Feng et al., 2014; Tsai et al., 2015; Pan et al., 2016).
An important challenge for gRNA design is its strict dependence on PAM, which is NGG in the case of SpCas9 (Jinek et al., 2012). This PAM dependency limits the choice of a target site in the target gene. The problem is further exacerbated when designing gRNA in polyploid species. The development of altered or near PAM-less systems may present a much-needed solution to this challenge and can also enhance the utilization of CRISPR technology in functional genomics and precision breeding. Scientists have modified existing Cas enzymes and identified new Cas enzymes that have different sets of PAM sequences (Steinert et al., 2015). These newly discovered Cas enzymes include SpCas9 NG, VQR, and VRER versions, which recognize NG, NGA, and NGCG PAMs, respectively, while xCas9 3.7 can recognize GAT, GAA, and NG PAMs with increased specificity (Kleinstiver et al., 2015; Zhong et al., 2019). Walton et al. (2020) developed the Cas9 variant SpG with PAM NGN and further optimized it to create a near PAM-less Cas9 variant named SpRY (NRN and NYN), which can recognize almost all PAMs. Ren et al. (2021) utilized the SpRY nuclease and base editor in rice and developed herbicide-tolerant lines with high editing efficiency. The application of these near PAM-less Cas enzymes has been limited to the model species to date, such as rice and Arabidopsis (Endo et al., 2019; Yamamoto et al., 2019; Qin et al., 2020), and has not been reported in polyploid crops.
Off-target effects pose a significant challenge to CRISPR-based genome editing as a whole. Although plants have comparatively fewer off-target effects compared to animals or humans, they can still lead to undesired changes in plant phenotype. Therefore, eliminating off-targets effects is crucial for precise and efficient genome editing. The gRNA should be highly specific to the target site, as non-specific gRNA can edit the non-target sites in the genome, resulting in unpredictable changes in the genome and phenotype. It has been reported that the number of off-targets increases with the number of gRNAs used, which is often the case in polyploid crops where editing of multiple copies is required. This may explain the high number of off-targets observed in Brassica. For example, in a study by Ahmar et al. (2022), using four gRNAs in a multiplexed editing approach for editing four gene homologues of the BnaSVP gene resulted in the gRNA targeting all the homologues having the most number of off-target sites. Zhai et al. (2019) found 26 off-target sites in total corresponding to three designed gRNAs targeting two genes. Assou et al. (2022) predicted 24 off-target sites in two edited lines in B. juncea, while Huang et al. (2020) found 40 off-target sites for two gRNAs for editing of BnFAD2 gene homologues in B. napus. In another study, Yang et al. (2018) reported 57 off-target sites while editing BnCLV gene homologues. The number of off-target sites increases when using a smaller seed sequence in gRNA, for example, less than 7 bp, in the genome for the off-target effects.
Designing gRNAs with the least or no off-targets in polyploid crops becomes challenging. However, off-target effects can be avoided by selecting gRNAs with optimum GC content (50-60%) and relatively shorter gRNAs (18-20bp), avoiding mismatches and bulge formation in the 7-10 bp and 12 bp adjacent to PAM, respectively (Lin et al., 2014). Another strategy to reduce the off-target effects is to use a low concentration of gRNA/Cas9 complex. However, the concentration should not be too low that the on-target efficiency is also compromised (Jia and Wang, 2014). Hence, choosing a suitable promoter for driving the expression of gRNA and Cas9 genes is of crucial importance. Direct delivery of CRISPR components, such as RNP complexes comprising gRNA and Cas9 protein, into plant cells can substantially reduce the off-target effects as they are not stably transformed into the plant genome (Subburaj et al., 2016; Ahmad et al., 2020). The plant’s endogenous system quickly degrades these RNP complexes after their function, reducing the chance of targeting and editing other unintended locations. Off-target effects can also be reduced by using high-fidelity SpCas9 variants such as eSpCas9(1.0), eSpCas9(1.1), and SpCas9-HF1 (Slaymaker et al., 2016; Wang et al., 2017). These modified versions of the Cas9 protein show fewer off-targets as they require a precise 20-nucleotide guide sequence for identifying and editing targets.
The efficiency of genome editing tools, particularly in polyploid crops, has been found to be very low (Zaman et al., 2019b). Several factors affect editing efficiency, including the number of delivered gRNAs, the activity and expression levels of the gRNA and Cas9 protein, the GC content of gRNA, the secondary structure of gRNA, and the target environment (Abe et al., 2019). While a single gRNA, can induce frameshift mutation in a single gene, multiple gRNAs may generate large deletions between targeted sites, thereby increasing the chance of creating multiallelic knockout mutations. However, the co-expression of multiple gRNAs often results in low editing efficiency (Jansing et al., 2019). The use of native promoters for gRNA and Cas9 expression is associated with higher editing efficiencies and co-editing events in polyploid crops (Johansen et al., 2019; Li et al., 2020). However, in species where native promoter sequences have not been discovered, the promoter of other relative species can be used, although editing efficiency may be compromised. For example, in Brassica genome editing experiments, the Arabidopsis U3 and U6 promoters are being used (Zheng et al., 2020). Editing efficiency may also be improved by codon optimization of Cas9-coding sequences and the use of enhancers (Kusano et al., 2018; Brauer et al., 2020; Grützner et al., 2021; Zhang et al., 2022b).
The target environment also plays an important role in efficient genome editing. Genome bias has been observed for gRNAs, which edit one genome more effectively than the other one in polyploids, as in the case of simultaneous editing of multiple BnaSVP gene copies. A comparatively larger number of edits were obtained for copies present in the subgenome A rapeseed as compared to that in the subgenome C at all target sites (Ahmar et al., 2022). Huang et al. (2020) also observed this phenomenon of marked preference for a gRNA to edit a specific gene more efficiently than others. They designed a gRNA with high sequence similarity to the four gene homologues of the BnFAD2 gene located on two different subgenomes, A and C. However, most of the editing was observed in the BnFAD2.A5 gene present in the subgenome A only.
Other factors affecting editing efficiency in Brassica, such as the incubation period of explants on regeneration medium and the choice of explants, have also been investigated. Lawrenson et al. (2015) reported high editing efficiency in B. oleracea calli kept on regeneration media for seven weeks as compared to those kept for four weeks with no off-target effects. Mikami et al. (2015) also reported higher editing efficiencies in prolonged tissue culture conditions in the case of rice. Editing efficiency may also vary with the target explant used for transformations. Jeong et al. (2019) designed eight gRNAs and tested the editing efficiency of CRISPR/Cas9 and gRNA complex by delivering them into the protoplasts of B. rapa leaves and selected four gRNAs with higher editing efficiencies to transform into hypocotyl explants. Surprisingly, only one out of the four gRNAs exhibited editing in regenerated plants, and the gRNA with the highest editing efficiency in protoplast culture gave no mutations. This could be also due to differences in the insertion position of T-DNA, which might affect their expression.
Molecular characterization of targeted mutations in polyploid crops is also challenging compared to diploids. Medium-throughput PCR-based assays, such as cleaved amplified polymorphic sequences (CAPS), T7 endonuclease 1 (T7E1), high-resolution melting analysis (HRMA), and capillary electrophoresis, which are typically used for molecular characterization of edited genes in diploid crops, may not provide a detailed characterization of the edited alleles/genes in polyploids. High sequence depth and longer read lengths are required to distinguish between the number of edited alleles/genes, leading to increased costs and labour involved in the genotyping of polyploids (May et al., 2023). Sanger and Illumina high-throughput sequencing are commonly used for the characterization of the type and extent of mutation(s) in polyploid crops. Computational tools like tracking of indels by decomposition (TIDE) (Brinkman et al., 2014) and Inference if CRISPR edits (ICE; Conant et al., 2022) are being employed for the quantitative assessment of the type and extent of mutations in case of Sanger sequencing for crops with low to moderate ploidy levels (Park et al., 2017; Liu et al., 2019). To determine the number of co-edited alleles/genes, the amplicons, amplified by allele-specific primers, are first cloned into a sequencing vector followed by monoclonal sequencing using the Sanger platform. This technique can obtain long read lengths, accurately distinguishing between alleles/genes, and assessing the editing efficiency of multiple gRNAs within a single allele/gene (Li et al., 2017; Eid et al., 2021).
Although genome editing has become a tool of choice for gene functional studies and precision breeding, its applications are still limited to diploid crops due to factors such as genome complexity, gene redundancy, the challenge of designing efficient gRNA and finding allele-specific mutations. It is expected that developments, such as finding new Cas protein variants, and advanced analytical approaches to identify edits, are likely to expand the utility of this toolbox for genome editing in polyploid crops. Once complete genome information is available, CRISPR/Cas technology can be effectively used to develop genome-wide mutant libraries of Brassica crops, as done in other crops such as rice. Besides crop improvement, the domestication process of wild Brassica species can be accelerated using CRISPR/Cas genome editing by targeting domestication-related genes, as evident in wild rice and tomato. CRISPR-based gene editing technology has emerged as a powerful tool for precision agriculture and will be effectively utilized for enhancing yield, biotic/abiotic stress tolerance, improving quality and plant architecture, haploid induction, and de novo domestication of Brassica crops. It is hoped that regulatory standards related to gene-edited crops will also be reformed to facilitate their fast commercialization and public acceptance.
Aggrevating the changing climate are the biotic and abiotic factors which result in significant yields losses in field crops. In addition to yield losses, biotic factors including fungus, bacteria, viruses, nematodes and insect attacks also deteriorate the crop produce quality (Giudice et al., 2021). The changing climatic patterns (which includes early and long high temperature regimes, unexpected heavy rainfalls, cloud bursts, severe winters) coupled with drought, salinity and heavy metal toxicity altogether pose a severe threat to global agricultural production and food security (Rahman et al., 2022). Therefore, development of stress-resilient crops is the need of the hour to ensure global food security and meet the UN’s target of a 60-70% increase in the agricultural crop production by 2050. CRISPR/Cas9 technology can be effectively employed to develop stress resilient crops by targeting genes involved in these stress related pathways. The resistance or tolerance to these stresses can be incorporated in two ways i.e., by knocking those genes in which are involved in resistance (R genes) or by knock down of genes involved in crop’s susceptibility (S genes). A more advanced CRISPRa (CRISPR activation) of R genes and CRISPRi (CRISPR interference) of S genes can also be employed to incorporate stress resilience in crop plants (Zafar et al., 2020). One of the main reasons for the crop failure due to sudden disease outbreak or changing environmental conditions is their minimized genetic diversity which have been lost in the process of intensive selection and domestication. Therefore, synthetic directed evolution (SDE) presents a promising solution to increase the localized sequence diversification of specific genes (e.g., stress related genes) and then selection of gene variants exhibiting better resistance to biotic and abiotic stresses (Zhang and Qi, 2019; Rao et al., 2021).
NA, M-UR, and RG designed the study; NA and SF wrote the first draft; MM, QZ, RA, WZ, M-UR, and RG critically revised the study. All authors contributed to the article and approved the submitted version.
This work was supported by the Science and Technology Department of Zhejiang Province (2021C02064-2), the Collaborative Innovation Center for Modern Crop Production co-sponsored by Province and Ministry (CIC-MCP), and the Agriculture and Rural Affairs Department of Zhejiang Province (2021XTTGLY0202). We also gratefully acknowledge the support of ICGEB to NA via grant No. CRP/PAK20-02.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Østergaard, L., Kempin, S. A., Bies, D., Klee, H. J., Yanofsky, M. F. (2006). Pod shatter-resistant Brassica fruit produced by ectopic expression of the FRUITFULL gene. Plant Biotechnol. J. 4 (1), 45–51. doi: 10.1111/j.1467-7652.2005.00156.x
Abe, F., Haque, E., Hisano, H., Tanaka, T., Kamiya, Y., Mikami, M., et al. (2019). Genome-edited triple-recessive mutation alters seed dormancy in wheat. Cell Rep. 28 (5), 1362–1369. doi: 10.1016/j.celrep.2019.06.090
Ahmad, N., Mukhtar, Z. (2017). Genetic manipulations in crops: challenges and opportunities. Genomics 109 (5-6), 494–505. doi: 10.1016/j.ygeno.2017.07.007
Ahmad, N., Rahman, M. U., Mukhtar, Z., Zafar, Y., Zhang, B. (2020). A critical look on CRISPR-based genome editing in plants. J. Cell. Physiol. 235 (2), 666–682. doi: 10.1002/jcp.29052
Ahmar, S., Zhai, Y., Huang, H., Yu, K., Khan, M. H. U., Shahid, M., et al. (2022). Development of mutants with varying flowering times by targeted editing of multiple SVP gene copies in Brassica napus L. Crop J. 10 (1), 67–74. doi: 10.1016/j.cj.2021.03.023
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576 (7785), 149–157. doi: 10.1038/s41586-019-1711-4
Assou, J., Zhang, D., Roth, K. D., Steinke, S., Hust, M., Reinard, T., et al. (2022). Removing the major allergen bra j I from brown mustard (Brassica juncea) by CRISPR/Cas9. Plant J. 109 (3), 649–663. doi: 10.1111/tpj.15584
Augustine, R., Arya, G. C., Nambiar, D. M., Kumar, R., Bisht, N. C. (2014). Translational genomics in Brassica crops: challenges, progress, and future prospects. Plant Biotechnol. Rep. 8, 65–81. doi: 10.1007/s11816-013-0298-8
Blackshaw, R. E., Kanashiro, D., Moloney, M. M., Crosby, W. L. (1994). Growth, yield and quality of canola expressing resistance to acetolactate synthase inhibiting herbicides. Can. J. Plant Sci. 74 (4), 745–751. doi: 10.4141/cjps94-133
Braatz, J., Harloff, H. J., Mascher, M., Stein, N., Himmelbach, A., Jung, C. (2017). CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 174 (2), 935–942. doi: 10.1104/pp.17.00426
Brauer, E. K., Balcerzak, M., Rocheleau, H., Leung, W., Schernthaner, J., Subramaniam, R., et al. (2020). Genome editing of a deoxynivalenol-induced transcription factor confers resistance to Fusarium graminearum in wheat. Mol. Plant-Microbe Interact. 33 (3), 553–560. doi: 10.1094/MPMI-11-19-0332-R
Brinkman, E. K., Chen, T., Amendola, M., Van Steensel, B. (2014). Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42 (22), e168–e168. doi: 10.1093/nar/gku936
Chantret, N., Salse, J., Sabot, F., Rahman, S., Bellec, A., Laubin, B., et al. (2005). Molecular basis of evolutionary events that shaped the hardness locus in diploid and polyploid wheat species (Triticum and aegilops). Plant Cell 17 (4), 1033–1045. doi: 10.1105/tpc.104.029181
Chen, K., Gao, C. (2014). Targeted genome modification technologies and their applications in crop improvements. Plant Cell Rep. 33, 575–583. doi: 10.1007/s00299-013-1539-6
Chen, K., Wang, Y., Zhang, R., Zhang, H., Gao, C. (2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. doi: 10.1146/annurev-arplant-050718-100049
Cheng, H., Hao, M., Ding, B., Mei, D., Wang, W., Wang, H., et al. (2021). Base editing with high efficiency in allotetraploid oilseed rape by A3A-PBE system. Plant Biotechnol. J. 19 (1), 87–97. doi: 10.1111/pbi.13444
Cheng, F., Liang, J., Cai, C., Cai, X., Wu, J., Wang, X. (2017). Genome sequencing supports a multi-vertex model for Brassiceae species. Curr. Opin. Plant Biol. 36, 79–87. doi: 10.1016/j.pbi.2017.01.006
Coate, J. E., Luciano, A. K., Seralathan, V., Minchew, K. J., Owens, T. G., Doyle, J. J. (2012). Anatomical, biochemical, and photosynthetic responses to recent allopolyploidy in Glycine dolichocarpa (Fabaceae). Am. J. Bot. 99 (1), 55–67. doi: 10.3732/ajb.1100465
Comai, L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6 (11), 836–846. doi: 10.1038/nrg1711
Conant, D., Hsiau, T., Rossi, N., Oki, J., Maures, T., Waite, K., et al. (2022). Inference of CRISPR edits from Sanger trace data. CRISPR J. 5 (1), 123–130. doi: 10.1089/crispr.2021.0113
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339 (6121), 819–823. doi: 10.1126/science.1231143
Deltcheva, E., Chylinski, K., Sharma, C. M., Gonzales, K., Chao, Y., Pirzada, Z. A., et al. (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471 (7340), 602–607. doi: 10.1038/nature09886
Eid, A., Mohan, C., Sanchez, S., Wang, D., Altpeter, F. (2021). Multiallelic, targeted mutagenesis of magnesium chelatase with CRISPR/Cas9 provides a rapidly scorable phenotype in highly polyploid sugarcane. Front. Genome Editing 3, 654996. doi: 10.3389/fgeed.2021.654996
El-Mounadi, K., Morales-Floriano, M. L., Garcia-Ruiz, H. (2020). Principles, applications, and biosafety of plant genome editing using CRISPR-Cas9. Front. Plant Sci. 11, 56. doi: 10.3389/fpls.2020.00056
Endo, M., Mikami, M., Endo, A., Kaya, H., Itoh, T., Nishimasu, H., et al. (2019). Genome editing in plants by engineered CRISPR–Cas9 recognizing NG PAM. Nat. Plants 5 (1), 14–17. doi: 10.1038/s41477-018-0321-8
Fan, S., Zhang, L., Tang, M., Cai, Y., Liu, J., Liu, H., et al. (2021). CRISPR/Cas9-targeted mutagenesis of the BnaA03. BP gene confers semi-dwarf and compact architecture to rapeseed (Brassica napus L.). Plant Biotechnol. J. 19 (12), 2383. doi: 10.1111/pbi.13703
Feng, Y., Cui, R., Wang, S., He, M., Hua, Y., Shi, L., et al. (2020). Transcription factor BnaA9. WRKY47 contributes to the adaptation of Brassica napus to low boron stress by up-regulating the boric acid channel gene BnaA3. NIP5; 1. Plant Biotechnol. J. 18 (5), 1241–1254. doi: 10.1111/pbi.13288
Feng, Z., Mao, Y., Xu, N., Zhang, B., Wei, P., Yang, D. L., et al. (2014). Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in arabidopsis. Proc. Natl. Acad. Sci. 111 (12), 4632–4637. doi: 10.1073/pnas.1400822111
Food and Agriculture Organization (2021) FAOSTAT statisticaldatabase. Available at: https://www.fao.org/statistics/en/.
Gallego-Bartolomé, J., Gardiner, J., Liu, W., Papikian, A., Ghoshal, B., Kuo, H. Y., et al. (2018). Targeted DNA demethylation of the arabidopsis genome using the human TET1 catalytic domain. Proc. Natl. Acad. Sci. 115 (9), E2125–E2134. doi: 10.1073/pnas.1716945115
Giudice, G., Moffa, L., Varotto, S., Cardone, M. F., Bergamini, C., De Lorenzis, G., et al. (2021). Novel and emerging biotechnological crop protection approaches. Plant Biotechnol. J. 19 (8), 1495–1510. doi: 10.1111/pbi.13605
Grützner, R., Martin, P., Horn, C., Mortensen, S., Cram, E. J., Lee-Parsons, C. W., et al. (2021). High-efficiency genome editing in plants mediated by a Cas9 gene containing multiple introns. Plant Commun. 2 (2), 100135. doi: 10.1016/j.xplc.2020.100135
Hickey, L. T., Hafeez, N., Robinson, H., Jackson, S. A., Leal-Bertioli, S. C., Tester, M., et al. (2019). Breeding crops to feed 10 billion. Nat. Biotechnol. 37 (7), 744–754. doi: 10.1038/s41587-019-0152-9
Hong, J. K., Suh, E. J., Park, S. R., Park, J., Lee, Y. H. (2021). Multiplex CRISPR/Cas9 mutagenesis of BrVRN1 delays flowering time in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Agriculture 11 (12), 1286. doi: 10.3390/agriculture11121286
Hsu, P. D., Lander, E. S., Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157 (6), 1262–1278. doi: 10.1016/j.cell.2014.05.010
Hu, L., Zhang, H., Yang, Q., Meng, Q., Han, S., Nwafor, C. C., et al. (2018). Promoter variations in a homeobox gene, BnA10. LMI1, determine lobed leaves in rapeseed (Brassica napus L.). Theor. Appl. Genet. 131, 2699–2708. doi: 10.1007/s00122-018-3184-5
Huang, H., Cui, T., Zhang, L., Yang, Q., Yang, Y., Xie, K., et al. (2020). Modifications of fatty acid profile through targeted mutation at BnaFAD2 gene with CRISPR/Cas9-mediated gene editing in Brassica napus. Theor. Appl. Genet. 133, 2401–2411. doi: 10.1007/s00122-020-03607-y
Jaganathan, D., Ramasamy, K., Sellamuthu, G., Jayabalan, S., Venkataraman, G. (2018). CRISPR for crop improvement: an update review. Front. Plant Sci. 9, 985. doi: 10.3389/fpls.2018.00985
Jagannath, A., Arumugam, N., Gupta, V., Pradhan, A., Burma, P. K., Pental, D. (2002). Development of transgenic barstar lines and identification of a male sterile (barnase)/restorer (barstar) combination for heterosis breeding in Indian oilseed mustard (Brassica juncea). Curr. Sci. 82 (1), 46–52.
Jagannath, A., Bandyopadhyay, P., Arumugam, N., Gupta, V., Burma, P. K., Pental, D. (2001). The use of a spacer DNA fragment insulates the tissue-specific expression of a cytotoxic gene (barnase) and allows high-frequency generation of transgenic male sterile lines in Brassica juncea L. Mol. Breed. 8, 11–23. doi: 10.1023/A:1011916216191
Jaglo, K. R., Kleff, S., Amundsen, K. L., Zhang, X., Haake, V., Zhang, J. Z., et al. (2001). Components of the arabidopsis c-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 127 (3), 910–917. doi: 10.1104/pp.010548
Jansing, J., Sack, M., Augustine, S. M., Fischer, R., Bortesi, L. (2019). CRISPR/Cas9-mediated knockout of six glycosyltransferase genes in Nicotiana benthamiana for the production of recombinant proteins lacking β-1, 2-xylose and core α-1, 3-fucose. Plant Biotechnol. J. 17 (2), 350–361. doi: 10.1111/pbi.12981
Jedličková, V., Mácová, K., Štefková, M., Butula, J., Staveníková, J., Sedláček, M., et al. (2022). Hairy root transformation system as a tool for CRISPR/Cas9-directed genome editing in oilseed rape (Brassica napus). Front. Plant Sci. 13, 2514. doi: 10.3389/fpls.2022.919290
Jeong, S. Y., Ahn, H., Ryu, J., Oh, Y., Sivanandhan, G., Won, K. H., et al. (2019). Generation of early-flowering Chinese cabbage (Brassica rapa spp. pekinensis) through CRISPR/Cas9-mediated genome editing. Plant Biotechnol. Rep. 13, 491–499. doi: 10.1007/s11816-019-00566-9
Jia, H., Wang, N. (2014). Targeted genome editing of sweet orange using Cas9/sgRNA. PloS One 9 (4), e93806. doi: 10.1371/journal.pone.0093806
Jiang, L., Li, D., Jin, L., Ruan, Y., Shen, W. H., Liu, C. (2018). Histone lysine methyltransferases bna SDG 8. a and bna SDG 8. c are involved in the floral transition in Brassica napus. Plant J. 95 (4), 672–685. doi: 10.1111/tpj.13978
Jiang, C. X., Wright, R. J., El-Zik, K. M., Paterson, A. H. (1998). Polyploid formation created unique avenues for response to selection in Gossypium (cotton). Proc. Natl. Acad. Sci. 95 (8), 4419–4424. doi: 10.1073/pnas.95.8.4419
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337 (6096), 816–821. doi: 10.1126/science.1225829
Johansen, I. E., Liu, Y., Jørgensen, B., Bennett, E. P., Andreasson, E., Nielsen, K. L., et al. (2019). High efficacy full allelic CRISPR/Cas9 gene editing in tetraploid potato. Sci. Rep. 9 (1), 1–7. doi: 10.1038/s41598-019-54126-w
Kanrar, S., Venkateswari, J., Kirti, P., Chopra, V. (2002). Transgenic Indian mustard (Brassica juncea) with resistance to the mustard aphid (Lipaphis erysimi kalt.). Plant Cell Rep. 20, 976–981. doi: 10.1007/s00299-001-0422-z
Khan, M. H., Hu, L., Zhu, M., Zhai, Y., Khan, S. U., Ahmar, S., et al. (2021). Targeted mutagenesis of EOD3 gene in Brassica napus L. regulates seed production. J. Cell. Physiol. 236 (3), 1996–2007. doi: 10.1002/jcp.29986
Kirchner, T. W., Niehaus, M., Debener, T., Schenk, M. K., Herde, M. (2017). Efficient generation of mutations mediated by CRISPR/Cas9 in the hairy root transformation system of Brassica carinata. PloS One 12 (9), e0185429. doi: 10.1371/journal.pone.0185429
Kleinstiver, B. P., Prew, M. S., Tsai, S. Q., Topkar, V. V., Nguyen, N. T., Zheng, Z., et al. (2015). Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523 (7561), 481–485. doi: 10.1038/nature14592
Kourani, M., Mohareb, F., Rezwan, F. I., Anastasiadi, M., Hammond, J. P. (2022). Genetic and physiological responses to heat stress in Brassica napus. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.832147
Kumar, R., Kaur, A., Pandey, A., Mamrutha, H. M., Singh, G. P. (2019). CRISPR-based genome editing in wheat: a comprehensive review and future prospects. Mol. Biol. Rep. 46 (3), 3557–3569. doi: 10.1007/s11033-019-04761-3
Kusano, H., Ohnuma, M., Mutsuro-Aoki, H., Asahi, T., Ichinosawa, D., Onodera, H., et al. (2018). Establishment of a modified CRISPR/Cas9 system with increased mutagenesis frequency using the translational enhancer dMac3 and multiple guide RNAs in potato. Sci. Rep. 8 (1), 13753. doi: 10.1038/s41598-018-32049-2
Kyriakidou, M., Tai, H. H., Anglin, N. L., Ellis, D., Strömvik, M. V. (2018). Current strategies of polyploid plant genome sequence assembly. Front. Plant Sci. 9, 1660. doi: 10.3389/fpls.2018.01660
Larson, M. H., Gilbert, L. A., Wang, X., Lim, W. A., Weissman, J. S., Qi, L. S. (2013). CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat. Protoc. 8 (11), 2180–2196. doi: 10.1038/nprot.2013.132
Lawrenson, T., Shorinola, O., Stacey, N., Li, C., Østergaard, L., Patron, N., et al. (2015). Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 16, 1–13. doi: 10.1186/s13059-015-0826-7
Li, C., Hao, M., Wang, W., Wang, H., Chen, F., Chu, W., et al. (2018). An efficient CRISPR/Cas9 platform for rapidly generating simultaneous mutagenesis of multiple gene homoeologs in allotetraploid oilseed rape. Front. Plant Sci. 9, 442. doi: 10.3389/fpls.2018.00442
Li, J. F., Norville, J. E., Aach, J., McCormack, M., Zhang, D., Bush, J., et al. (2013). Multiplex and homologous recombination–mediated genome editing in arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31 (8), 688–691. doi: 10.1038/nbt.2654
Li, X., Sandgrind, S., Moss, O., Guan, R., Ivarson, E., Wang, E. S., et al. (2021). Efficient protoplast regeneration protocol and CRISPR/Cas9-mediated editing of glucosinolate transporter (GTR) genes in rapeseed (Brassica napus L.). Front. Plant Sci. 12, 680859. doi: 10.3389/fpls.2021.680859
Li, C., Unver, T., Zhang, B. (2017). A high-efficiency CRISPR/Cas9 system for targeted mutagenesis in cotton (Gossypium hirsutum L.). Sci. Rep. 7 (1), 1–10. doi: 10.1038/srep43902
Li, J., Wang, Z., He, G., Ma, L., Deng, X. W. (2020). CRISPR/Cas9-mediated disruption of TaNP1 genes results in complete male sterilityin bread wheat. J. Genet. Genomics 47 (5), 263–272. doi: 10.1016/j.jgg.2020.05.004
Lin, Y., Cradick, T. J., Brown, M. T., Deshmukh, H., Ranjan, P., Sarode, N., et al. (2014). CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 42 (11), 7473–7485. doi: 10.1093/nar/gku402
Liu, J. W., DeMichele, S., Bergana, M., Bobik, E., Hastilow, C., Chuang, L. T., et al. (2001). Characterization of oil exhibiting high γ-linolenic acid from a genetically transformed canola strain. J. Am. Oil Chemists’ Soc. 78, 489–493. doi: 10.1007/s11746-001-0291-2
Liu, H., Guo, X., Naeem, M. S., Liu, D., Xu, L., Zhang, W., et al. (2011). Transgenic Brassica napus L. lines carrying a two gene construct demonstrate enhanced resistance against Plutella xylostella and Sclerotinia sclerotiorum. Plant Cell Tissue Organ Culture (PCTOC) 106, 143–151. doi: 10.1007/s11240-010-9902-6
Liu, Q., Wang, C., Jiao, X., Zhang, H., Song, L., Li, Y., et al. (2019). Hi-TOM: a platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 62, 1–7. doi: 10.1007/s11427-018-9402-9
Liu, Y., Ye, S., Yuan, G., Ma, X., Heng, S., Yi, B., et al. (2020). Gene silencing of BnaA09. ZEP and BnaC09. ZEP confers orange color in Brassica napus flowers. Plant J. 104 (4), 932–949. doi: 10.1111/tpj.14970
Lowder, L. G., Zhou, J., Zhang, Y., Malzahn, A., Zhong, Z., Hsieh, T. F., et al. (2018). Robust transcriptional activation in plants using multiplexed CRISPR-Act2. 0 and mTALE-act systems. Mol. Plant 11 (2), 245–256. doi: 10.1016/j.molp.2017.11.010
Lu, S., Sturtevant, D., Aziz, M., Jin, C., Li, Q., Chapman, K. D., et al. (2018). Spatial analysis of lipid metabolites and expressed genes reveals tissue-specific heterogeneity of lipid metabolism in high-and low-oil Brassica napus L. seeds. Plant J. 94 (6), 915–932. doi: 10.1111/tpj.13959
Ma, C., Zhu, C., Zheng, M., Liu, M., Zhang, D., Liu, B., et al. (2019). CRISPR/Cas9-mediated multiple gene editing in Brassica oleracea var. capitata using the endogenous tRNA-processing system. Horticult. Res. 6. doi: 10.1038/s41438-018-0107-1
Manghwar, H., Lindsey, K., Zhang, X., Jin, S. (2019). CRISPR/Cas system: recent advances and future prospects for genome editing. Trends Plant Sci. 24 (12), 1102–1125. doi: 10.1016/j.tplants.2019.09.006
Mason, A. S., Snowdon, R. J. (2016). Oilseed rape: learning about ancient and recent polyploid evolution from a recent crop species. Plant Biol. 18 (6), 883–892. doi: 10.1111/plb.12462
Mason, A. S., Wendel, J. F. (2020). Homoeologous exchanges, segmental allopolyploidy, and polyploid genome evolution. Front. Genet. 11, 1014. doi: 10.3389/fgene.2020.01014
Matsuo, H., Yokooji, T., Taogoshi, T. (2015). Common food allergens and their IgE-binding epitopes. Allergol. Int. 64 (4), 332–343. doi: 10.1016/j.alit.2015.06.009
May, D., Paldi, K., Altpeter, F. (2023). Targeted mutagenesis with sequence-specific nucleases for accelerated improvement of polyploid crops: progress, challenges, and prospects. Plant Genome, e20298. doi: 10.1002/tpg2.20298
Mikami, M., Toki, S., Endo, M. (2015). Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol. Biol. 88 (6), 561–572. doi: 10.1007/s11103-015-0342-x
Mun, J. H., Kwon, S. J., Yang, T. J., Seol, Y. J., Jin, M., Kim, J. A., et al. (2009). Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 10 (10), 1–18. doi: 10.1186/gb-2009-10-10-r111
Naeem, I., Munir, I., Durrett, T. P., Iqbal, A., Aulakh, K. S., Ahmad, M. A., et al. (2020). Feasible regeneration and agro bacterium-mediated transformation of Brassica juncea with euonymus alatus diacylglycerol acetyltransferase (EaDAcT) gene. Saudi J. Biol. Sci. 27 (5), 1324–1332. doi: 10.1016/j.sjbs.2019.12.036
Nagaharu, U.J.J.J.B., Nagaharu, N. (1935). Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Japanese J. Bot. 7 (7), 389–452.
Neequaye, M., Stavnstrup, S., Harwood, W., Lawrenson, T., Hundleby, P., Irwin, J., et al. (2021). CRISPR-Cas9-mediated gene editing of MYB28 genes impair glucoraphanin accumulation of Brassica oleracea in the field. CRISPR J. 4 (3), 416–426. doi: 10.1089/crispr.2021.0007
Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J. D., Kamoun, S. (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31 (8), 691–693. doi: 10.1038/nbt.2655
Nour-Eldin, H. H., Madsen, S. R., Engelen, S., Jørgensen, M. E., Olsen, C. E., Andersen, J. S., et al. (2017). Reduction of antinutritional glucosinolates in Brassica oilseeds by mutation of genes encoding transporters. Nat. Biotechnol. 35 (4), 377–382. doi: 10.1038/nbt.3823
Okada, A., Arndell, T., Borisjuk, N., Sharma, N., Watson-Haigh, N. S., Tucker, E. J., et al. (2019). CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol. J. 17 (10), 1905–1913. doi: 10.1111/pbi.13106
Okuzaki, A., Ogawa, T., Koizuka, C., Kaneko, K., Inaba, M., Imamura, J., et al. (2018). CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol. Biochem. 131, 63–69. doi: 10.1016/j.plaphy.2018.04.025
Orsavova, J., Misurcova, L., Vavra Ambrozova, J., Vicha, R., Mlcek, J. (2015). Fatty acids composition of vegetable oils and its contribution to dietary energy intake and dependence of cardiovascular mortality on dietary intake of fatty acids. Int. J. Mol. Sci. 16 (6), 12871–12890. doi: 10.3390/ijms160612871
Pan, C., Ye, L., Qin, L., Liu, X., He, Y., Wang, J., et al. (2016). CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 6 (1), 24765. doi: 10.1038/srep24765
Park, J., Lim, K., Kim, J. S., Bae, S. (2017). Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics 33 (2), 286–288. doi: 10.1093/bioinformatics/btw561
Park, S. C., Park, S., Jeong, Y. J., Lee, S. B., Pyun, J. W., Kim, S., et al. (2019). DNA-Free mutagenesis of GIGANTEA in Brassica oleracea var. capitata using CRISPR/Cas9 ribonucleoprotein complexes. Plant Biotechnol. Rep. 13, 483–489. doi: 10.1007/s11816-019-00585-6
Ponstein, A. S., Bade, J. B., Verwoerd, T. C., Molendijk, L., Storms, J., Beudeker, R. F., et al. (2002). Stable expression of phytase (phyA) in canola (Brassica napus) seeds: towards a commercial product. Mol. Breed. 10 (1-2), 31–44. doi: 10.1023/A:1020326219687
Prasad, K. V. S. K., Sharmila, P., Kumar, P. A., Saradhi, P. P. (2000). Transformation of Brassica juncea L. czern with bacterial codA gene enhances its tolerance to salt stress. Mol. Breed. 6, 489–499. doi: 10.1023/A:1026542109965
Pröbsting, M., Schenke, D., Hossain, R., Häder, C., Thurau, T., Wighardt, L., et al. (2020). Loss of function of CRT1a (calreticulin) reduces plant susceptibility to Verticillium longisporum in both Arabidopsis thaliana and oilseed rape (Brassica napus). Plant biotechnol. J. 18 (11), 2328–2344. doi: 10.1111/pbi.13394
Qin, R., Li, J., Liu, X., Xu, R., Yang, J., Wei, P. (2020). SpCas9-NG self-targets the sgRNA sequence in plant genome editing. Nat. Plants 6 (3), 197–201. doi: 10.1038/s41477-020-0603-9
Qing, C. M., Fan, L., Lei, Y., Bouchez, D., Tourneur, C., Yan, L., et al. (2000). Transformation of pakchoi (Brassica rapa L. ssp. chinensis) by Agrobacterium infiltration. Mol. Breed. 6, 67–72. doi: 10.1023/A:1009658128964
Rahman, H., Harwood, J., Weselake, R. (2013). Increasing seed oil content in Brassica species through breeding and biotechnology. Lipid Technol. 25 (8), 182–185. doi: 10.1002/lite.201300291
Rahman, M. U., Zulfiqar, S., Raza, M. A., Ahmad, N., Zhang, B. (2022). Engineering abiotic stress tolerance in crop plants through CRISPR genome editing. Cells 11 (22), 3590. doi: 10.3390/cells11223590
Raman, H., Raman, R., Kilian, A., Detering, F., Carling, J., Coombes, N., et al. (2014). Genome-wide delineation of natural variation for pod shatter resistance in Brassica napus. PloS One 9 (7), e101673. doi: 10.1371/journal.pone.0101673
Rani, S., Sharma, V., Hada, A., Koundal, K. R. (2017). Efficient genetic transformation of Brassica juncea with lectin using cotyledons explants. Int. J. Adv. Biotechnol. Res. 7, 1–12.
Rao, G. S., Jiang, W., Mahfouz, M. (2021). Synthetic directed evolution in plants: unlocking trait engineering and improvement. Synthetic Biol. 6 (1), ysab025. doi: 10.1093/synbio/ysab025
Ren, Q., Sretenovic, S., Liu, S., Tang, X., Huang, L., He, Y., et al. (2021). PAM-less plant genome editing using a CRISPR–SpRY toolbox. Nat. Plants 7 (1), 25–33. doi: 10.1038/s41477-020-00827-4
Rong, Z., Feng, Z., Yu-le, L., Sheng-guo, L., Liang-yi, K., Peng, L. (1997). Oilseed rape transformation and the establishment of a bromoxynil resistant transgenic oilseed rape. J. Integr. Plant Biol. 39 (1), 22–27.
Sander, J. D., Joung, J. K. (2014). CRISPR-cas systems for genome editing, regulation and targeting. Nat. Biotechnol. 32 (4), 347. doi: 10.1038/nbt.2842
Sashidhar, N., Harloff, H. J., Potgieter, L., Jung, C. (2020). Gene editing of three BnITPK genes in tetraploid oilseed rape leads to significant reduction of phytic acid in seeds. Plant Biotechnol. J. 18 (11), 2241–2250. doi: 10.1111/pbi.13380
Schaart, J. G., van de Wiel, C. C., Smulders, M. J. (2021). Genome editing of polyploid crops: prospects, achievements and bottlenecks. Transgenic Res. 30 (4), 337–351. doi: 10.1007/s11248-021-00251-0
Schaeffer, S. M., Nakata, P. A. (2015). CRISPR/Cas9-mediated genome editing and gene replacement in plants: transitioning from lab to field. Plant Sci. 240, 130–142. doi: 10.1016/j.plantsci.2015.09.011
Schouten, H. J., Jacobsen, E. (2007). Are mutations in genetically modified plants dangerous? J. Biomed. Biotechnol. doi: 10.1155/2007/82612
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-cas system. Nat. Biotechnol. 31 (8), 686–688. doi: 10.1038/nbt.2650
Sharafi, Y., Majidi, M. M., Goli, S. A. H., Rashidi, F. (2015). Oil content and fatty acids composition in Brassica species. Int. J. Food Properties 18 (10), 2145–2154. doi: 10.1080/10942912.2014.968284
Slaymaker, I. M., Gao, L., Zetsche, B., Scott, D. A., Yan, W. X., Zhang, F. (2016). Rationally engineered Cas9 nucleases with improved specificity. Science 351 (6268), 84–88. doi: 10.1126/science.aad5227
Sriboon, S., Li, H., Guo, C., Senkhamwong, T., Dai, C., Liu, K. (2020). Knock-out of TERMINAL FLOWER 1 genes altered flowering time and plant architecture in Brassica napus. BMC Genet. 21 (1), 1–13. doi: 10.1186/s12863-020-00857-z
Stanic, M., Hickerson, N. M., Arunraj, R., Samuel, M. A. (2021). Gene-editing of the strigolactone receptor BnD14 confers promising shoot architectural changes in Brassica napus (canola). Plant Biotechnol. J. 19 (4), 639. doi: 10.1111/pbi.13513
Steinert, J., Schiml, S., Fauser, F., Puchta, H. (2015). Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J. 84 (6), 1295–1305. doi: 10.1111/tpj.13078
Stoutjesdijk, P. A., Hurlestone, C., Singh, S. P., Green, A. G. (2000). High-oleic acid Australian Brassica napus and B. juncea varieties produced by co-suppression of endogenous Δ12-desaturases. Biochem. Soc. Trans. 28 (6), 938–940. doi: 10.1042/bst0280938
Subburaj, S., Chung, S. J., Lee, C., Ryu, S. M., Kim, D. H., Kim, J. S., et al. (2016). Site-directed mutagenesis in Petunia× hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 35, 1535–1544. doi: 10.1007/s00299-016-1937-7
Sun, Q., Lin, L., Liu, D., Wu, D., Fang, Y., Wu, J., et al. (2018). CRISPR/Cas9-mediated multiplex genome editing of the BnWRKY11 and BnWRKY70 genes in Brassica napus L. Int. J. Mol. Sci. 19 (9), 2716. doi: 10.3390/ijms19092716
Symington, L. S., Gautier, J. (2011). Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45, 247–271. doi: 10.1146/annurev-genet-110410-132435
Tsai, S. Q., Zheng, Z., Nguyen, N. T., Liebers, M., Topkar, V. V., Thapar, V., et al. (2015). GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-cas nucleases. Nat. Biotechnol. 33 (2), 187–197. doi: 10.1038/nbt.3117
Udall, J. A., Wendel, J. F. (2006). Polyploidy and crop improvement. Crop Sci. 46, S–3. doi: 10.2135/cropsci2006.07.0489tpg
van Dijk, M., Morley, T., Rau, M. L., Saghai, Y. (2021). A meta-analysis of projected global food demand and population at risk of hunger for the period 2010–2050. Nat. Food 2 (7), 494–501. doi: 10.1038/s43016-021-00322-9
Walton, R. T., Christie, K. A., Whittaker, M. N., Kleinstiver, B. P. (2020). Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368 (6488), 290–296. doi: 10.1126/science.aba8853
Wang, J., Chen, Z., Du, J., Sun, Y., Liang, A. (2005). Novel insect resistance in Brassica napus developed by transformation of chitinase and scorpion toxin genes. Plant Cell Rep. 24, 549–555. doi: 10.1007/s00299-005-0967-3
Wang, C., Liu, Q., Shen, Y., Hua, Y., Wang, J., Lin, J., et al. (2019). Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat. Biotechnol. 37 (3), 283–286. doi: 10.1038/s41587-018-0003-0
Wang, M., Lu, Y., Botella, J. R., Mao, Y., Hua, K., Zhu, J. K. (2017). Gene targeting by homology-directed repair in rice using a geminivirus-based CRISPR/Cas9 system. Mol. Plant 10 (7), 1007–1010. doi: 10.1016/j.molp.2017.03.002
Wang, Z., Wan, L., Xin, Q., Zhang, X., Song, Y., Wang, P., et al. (2021). Optimising glyphosate tolerance in rapeseed (Brassica napus L.) by CRISPR/Cas9-based geminiviral donor DNA replicon system with Csy4-based single-guide RNA processing. J. Exp. Bot. 72 (13), 4796–4808. doi: 10.1093/jxb/erab167
Wang, L., Wang, Y., Makhmoudova, A., Nitschke, F., Tetlow, I. J., Emes, M. J. (2022b). CRISPR–Cas9-mediated editing of starch branching enzymes results in altered starch structure in Brassica napus. Plant Physiol. 188 (4), 1866–1886. doi: 10.1093/plphys/kiab535
Wang, T., Wei, J. J., Sabatini, D. M., Lander, E. S. (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science 343 (6166), 80–84. doi: 10.1126/science.1246981
Wang, Y., Ye, X., Ding, G., Xu, F. (2013). Overexpression of phyA and appA genes improves soil organic phosphorus utilisation and seed phytase activity in Brassica napus. PloS One 8 (4), e60801. doi: 10.1371/journal.pone.0060801
Wang, G., Zong, M., Liu, D., Wu, Y., Tian, S., Han, S., et al. (2022a). Efficient generation of targeted point mutations in the Brassica oleracea var. botrytis genome via a modified CRISPR/Cas9 system. Hortic. Plant J. 8 (4), 527–530. doi: 10.1016/j.hpj.2022.01.005
Wendel, J. F., Lisch, D., Hu, G., Mason, A. S. (2018). The long and short of doubling down: polyploidy, epigenetics, and the temporal dynamics of genome fractionation. Curr. Opin. Genet. Dev. 49, 1–7. doi: 10.1016/j.gde.2018.01.004
Wrucke, D. F., Mamidi, S., Rahman, M. (2019). Genome-wide association study for frost tolerance in canola (Brassica napus L.) under field conditions. J. Plant Biochem. Biotechnol. 28, 211–222. doi: 10.1007/s13562-018-0472-8
Wu, J., Yan, G., Duan, Z., Wang, Z., Kang, C., Guo, L., et al. (2020). Roles of the Brassica napus DELLA protein BnaA6. RGA, in modulating drought tolerance by interacting with the ABA signaling component BnaA10. ABF2. Front. Plant Sci. 11, 577. doi: 10.3389/fpls.2020.00577
Xie, T., Chen, X., Guo, T., Rong, H., Chen, Z., Sun, Q., et al. (2020). Targeted knockout of BnTT2 homologues for yellow-seeded Brassica napus with reduced flavonoids and improved fatty acid composition. J. Agric. Food Chem. 68 (20), 5676–5690. doi: 10.1021/acs.jafc.0c01126
Xiong, Z., Gaeta, R. T., Pires, J. C. (2011). Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. 108 (19), 7908–7913. doi: 10.1073/pnas.1014138108
Xiong, X., Liu, W., Jiang, J., Xu, L., Huang, L., Cao, J. (2019). Efficient genome editing of Brassica campestris based on the CRISPR/Cas9 system. Mol. Genet. Genomics 294, 1251–1261. doi: 10.1007/s00438-019-01564-w
Yamamoto, A., Ishida, T., Yoshimura, M., Kimura, Y., Sawa, S. (2019). Developing heritable mutations in Arabidopsis thaliana using a modified CRISPR/Cas9 toolkit comprising PAM-altered Cas9 variants and gRNAs. Plant Cell Physiol. 60 (10), 2255–2262. doi: 10.1093/pcp/pcz118
Yang, H., Wu, J. J., Tang, T., Liu, K. D., Dai, C. (2017). CRISPR/Cas9-mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus. Sci. Rep. 7 (1), 7489. doi: 10.1038/s41598-017-07871-9
Yang, Y., Zhu, K., Li, H., Han, S., Meng, Q., Khan, S. U., et al. (2018). Precise editing of CLAVATA genes in Brassica napus L. regulates multilocular silique development. Plant Biotechnol. J. 16 (7), 1322–1335. doi: 10.1111/pbi.12872
Zafar, S. A., Zaidi, S. S. E. A., Gaba, Y., Singla-Pareek, S. L., Dhankher, O. P., Li, X., et al. (2020). Engineering abiotic stress tolerance via CRISPR/Cas-mediated genome editing. J. Exp. Bot. 71 (2), 470–479. doi: 10.1093/jxb/erz476
Zaman, Q. U., Chu, W., Hao, M., Shi, Y., Sun, M., Sang, S. F., et al. (2019a). CRISPR/Cas9-mediated multiplex genome editing of JAGGED gene in Brassica napus L. Biomolecules 9 (11), 725. doi: 10.3390/biom9110725
Zaman, Q. U., Li, C., Cheng, H., Hu, Q. (2019b). Genome editing opens a new era of genetic improvement in polyploid crops. Crop J. 7 (2), 141–150. doi: 10.1016/j.cj.2018.07.004
Zaman, Q. U., Wen, C., Yuqin, S., Mengyu, H., Desheng, M., Jacqueline, B., et al. (2021). Characterization of SHATTERPROOF homoeologs and CRISPR-Cas9-mediated genome editing enhances pod-shattering resistance in Brassica napus L. CRISPR J. 4 (3), 360–370. doi: 10.1089/crispr.2020.0129
Zhai, Y., Cai, S., Hu, L., Yang, Y., Amoo, O., Fan, C., et al. (2019). CRISPR/Cas9-mediated genome editing reveals differences in the contribution of INDEHISCENT homologues to pod shatter resistance in Brassica napus L. Theor. Appl. Genet. 132, 2111–2123. doi: 10.1007/s00122-019-03341-0
Zhai, Y., Yu, K., Cai, S., Hu, L., Amoo, O., Xu, L., et al. (2020). Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotechnol. J. 18 (5), 1153–1168. doi: 10.1111/pbi.13281
Zhang, H. X., Hodson, J. N., Williams, J. P., Blumwald, E. (2001). Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc. Natl. Acad. Sci. 98 (22), 12832–12836. doi: 10.1073/pnas.231476498
Zhang, Y., Huang, S., Wang, X., Liu, J., Guo, X., Mu, J., et al. (2018a). Defective APETALA2 genes lead to sepal modification in Brassica crops. Front. Plant Sci. 9, 367. doi: 10.3389/fpls.2018.00367
Zhang, R., Liu, J., Chai, Z., Chen, S., Bai, Y., Zong, Y., et al. (2019b). Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat. Plants 5 (5), 480–485. doi: 10.1038/s41477-019-0405-0
Zhang, Y., Massel, K., Godwin, I. D., Gao, C. (2018b). Applications and potential of genome editing in crop improvement. Genome Biol. 19, 1–11. doi: 10.1186/s13059-018-1586-y
Zhang, K., Nie, L., Cheng, Q., Yin, Y., Chen, K., Qi, F., et al. (2019a). Effective editing for lysophosphatidic acid acyltransferase 2/5 in allotetraploid rapeseed (Brassica napus L.) using CRISPR-Cas9 system. Biotechnol. Biofuels 12 (1), 1–18. doi: 10.1186/s13068-019-1567-8
Zhang, Y., Qi, Y. (2019). CRISPR enables directed evolution in plants. Genome Biol. 20 (1), 1–4. doi: 10.1186/s13059-019-1693-4
Zhang, S., Wu, S., Hu, C., Yang, Q., Dong, T., Sheng, O., et al. (2022b). Increased mutation efficiency of CRISPR/Cas9 genome editing in banana by optimized construct. PeerJ 10, e12664. doi: 10.7717/peerj.12664
Zhang, K., Zhuo, C., Wang, Z., Liu, F., Wen, J., Yi, B., et al. (2022a). BnaA03. MKK5-BnaA06. MPK3/BnaC03. MPK3 module positively contributes to Sclerotinia sclerotiorum resistance in Brassica napus. Plants 11 (5), 609. doi: 10.3390/plants11050609
Zheng, M., Zhang, L., Tang, M., Liu, J., Liu, H., Yang, H., et al. (2020). Knockout of two bna MAX 1 homologs by CRISPR/Cas9-targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.). Plant Biotechnol. J. 18 (3), 644–654. doi: 10.1111/pbi.13228
Zhong, Z., Sretenovic, S., Ren, Q., Yang, L., Bao, Y., Qi, C., et al. (2019). Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting Cas9-NG. Mol. Plant 12 (7), 1027–1036. doi: 10.1016/j.molp.2019.03.011
Keywords: genome editing, CRISPR, Brassica, polyploid crops, crop improvement
Citation: Ahmad N, Fatima S, Mehmood MA, Zaman QU, Atif RM, Zhou W, Rahman M-u and Gill RA (2023) Targeted genome editing in polyploids: lessons from Brassica. Front. Plant Sci. 14:1152468. doi: 10.3389/fpls.2023.1152468
Received: 27 January 2023; Accepted: 11 April 2023;
Published: 20 June 2023.
Edited by:
Muhammad Qasim Shahid, South China Agricultural University, ChinaReviewed by:
Jw Wu, South China Agricultural University, ChinaCopyright © 2023 Ahmad, Fatima, Mehmood, Zaman, Atif, Zhou, Rahman and Gill. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mehboob-ur Rahman, bWVoYm9vYl9wYmRAeWFob28uY29t; Rafaqat Ali Gill, ZHJyYWdpbGxAY2Fhcy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.