- 1State Key Laboratory of Herbage Improvement and Grassland Agro-ecosystems, College of Pastoral Agriculture Science and Technology, Lanzhou University, Lanzhou, China
- 2National Engineering Laboratory for Volatile Organic Compounds Pollution Control Material & Technology, University of Chinese Academy of Sciences, Beijing, China
As important components of the two-component regulatory system, response regulatory proteins (RRPs) play a crucial role in histidine phosphorylation-mediated signal transduction in response to environmental fluctuations. Accumulating evidence has revealed that RRPs play important roles in plant growth and stress response. However, the specific functions of RR genes (RRs) in cultivated alfalfa remain ambiguous. Therefore, in this study, we identified and characterized the RR family genes in the alfalfa genome using bioinformatics methods. Our analysis revealed 37 RRs in the alfalfa genome of Zhongmu No.1 that were unevenly distributed on the chromosomes. Cis-elements analysis revealed the involvement of RRs in responses to light, stress, and various plant hormones. Expression analysis of RRs in different tissues revealed their distinct tissue expression patterns. These findings provide preliminary insights into the roles of RRs in plant responses to abiotic stress, which can be used to improve the stress tolerance of autotetraploid-cultivated alfalfa plants via genetic engineering.
Introduction
Adverse environmental conditions, such as drought, high salinity, and low temperatures (Hasanuzzaman et al., 2020; Imran et al., 2021), can greatly affect plant growth and crop production. To adapt to these challenges, plants have developed complex molecular response pathways for their survival (Razi and Muneer, 2021; Sun and Guo, 2022). The two-component system is one such response pathway that converts external stimuli into internal molecular signals in various prokaryotes, fungi, and plants (Hwang et al., 2002; Gao et al., 2008; Huo et al., 2020). The two-component system functions as a growth regulator during responses to stress factors, such as ethylene, cytokinins, light, and osmotic stress (Jain et al., 2008; Geng et al., 2022).
As important components of the two-component system, response regulators regulate the expression of downstream genes by receiving phosphate groups from receptors on conserved Asp residues in response to environmental stimuli (Kieber and Schaller, 2018; Hu et al., 2022). Various plant genome sequencing projects have improved our understanding of the functions of response regulatory (RR) genes in plants (Zhang et al., 2012). RRPs are encoded by a large gene family in plants (Wang et al., 2007), which can be divided into three subtypes based on their domain structure and amino acid sequence: A, B, and P. Type-A RRs only possess a receiver domain, and their transcripts are induced by cytokinins and nitrates (West and Stock, 2001; Hass et al., 2004). Type-B RRs are transcription factors that can activate their target genes, including type-A RRs, and act as positive regulators of the cytokinins response (Mason et al., 2005). Type-P RRs lack the Asp phosphorylation sites required to maintain their activity and may affect the biological clock (McClung, 2006).
To date, RRs have been identified on a genome-wide scale in various plant species, such as Arabidopsis thaliana (D’Agostino et al., 2000), Oryza sativa (Tsai et al., 2012), Zea mays (Wang et al., 2022), Gossypium hirsutum (Zhao et al., 2022), and Glycine max (Le et al., 2011). Although the number of identified RRs varies among plants, their functions are conserved in these different species. Many stress tolerance-related genes have been identified in different plants. For instance, OsRR6, GmRR01, GmRR02, GmRR25, and GmRR32 have been reported as positive regulators of stress response, increasing the plant resistance to drought and salt stress (Le et al., 2011). In contrast, OsRR9 and OsRR10 negatively regulate salt tolerance (Wang et al., 2019; Bhaskar et al., 2021). Z. mays ZmRR1 is the only gene reported to be involved in cold tolerance (Zeng et al., 2021). Some RRs have been found to play vital roles in developmental and environmental signals mediated by light, cytokinins, and ethylene (Doi et al., 2004; Ito and Kurata, 2006). For instance, the expression patterns of O. sativa Ehd1 and ZmRR3 are related to cytokinins (Pareek et al., 2006), and those of GhRR41, SlPRR1, SlPRR2, SlPRR3, SlPRR4, SlPRR5, and SlPRR6 are mainly related to the circadian rhythm and regulation of flowering time in plants (He et al., 2016).
Alfalfa, the most widely cultivated legume forage in the world with a planting area of 32 million hectares (Ren et al., 2021), is known as the “queen of forage” because of its high nutritional value as a source of proteins, vitamins, and other nutrients for livestock. It also exhibits potential for ethanol production (Liu et al., 2021). Alfalfa is widely cultivated in the arid and semiarid areas of the Northeast, Northwest, and Central North regions of China, and its production is strongly constrained by various biotic and abiotic stresses (Dong et al., 2023). Although the stress-tolerance-related functions of RRs have been widely reported in other crops, only a few studies have investigated their roles in the stress response of cultivated alfalfa plants. Therefore, in this study, we systematically identified and characterized the RRs to understand their biological functions, physicochemical properties, gene structures, evolutionary relationships, and location on the chromosome. We also analyzed the expression patterns of RRs under different abiotic stresses using quantitative reverse transcription-polymerase chain reaction (qRT-PCR). Our results provide a basic understanding of the roles of RRs in cultivated alfalfa and lay the foundation for further breeding studies of this plant via genetic engineering.
Materials and methods
Plant growth, treatments, and tissue collection
Alfalfa (Zhongmu No.1) seeds from the College of Grassland Agricultural Science and Technology of Lanzhou University (humidity: 80%) were germinated in a culture dish and grown in half-concentration Murashige and Skoog nutrient solution (pH 5.8) under 16/8 h light/dark conditions at 22 °C and 80% relative humidity for 15 d. For the stress treatment, fifteen groups of alfalfa seedlings were separately treated with abscisic acid (ABA; 10 µM), NaCl (250 mM), and mannitol (400 mM) for different time points (0, 1, 3, 6, and 24 h), and each treatment group contained five plants. Whole plants from different treatments were separately stored at –80 °C in an ultra-low temperature refrigerator for subsequent analysis. Total RNA was extracted with the TRIzol Total RNA Extraction Kit (Shenggong Bioengineering Co., Ltd., Shanghai), and reverse transcribed into cDNA using the TIANScript II RT Kit (Tiangen Biochemical Technology Co., Ltd., Beijing) for gene expression verification.
Identification of RRs in alfalfa
Alfalfa genome coding sequence (CDS) and protein sequences were downloaded from the website (http://47.92.172.28:12088/). RR protein sequences of A. thaliana from the TAIR (https://www.arabidopsis.org/) were used as query sequences to identify candidate RRs in the alfalfa genome using the Protein Basic Local Align Search Tool (BLASTP) with an E-value cutoff of 10-5. A hidden Markov model (HMM) for the response regulator receiver domain, PF00072, from the PFAM protein database (http://pfam.xfam.org/) was used to further filter the previously identified RRs, and the remaining protein sequences were considered to be members of RR gene family.
Gene characteristics, structure, conserved motif, and subcellular localization analyses
Physical parameters of putative proteins, including the lengths of amino acid sequences, molecular weights (MWs), theoretical isoelectric points (pIs), and instability index, were calculated using the online ExPASy tool (http://www.expasy.org/tools/protparam.html), and the gene structure display server program (http://gsds.cbi.pku.edu.cn/) was used to illustrate the exon–intron structures by alignment of the CDSs of individual RRs. For motif structures analysis of RRPs, the online MEME program (http://meme-suite.org/tools/meme) was used with the default parameters, and the optimum motif widths were set at 6 residues. For subcellular localization analyses, the CELLO webtool (http://cello.life.nctu.edu.tw/) was used to predict the subcellular location of RRPs.
Chromosomal localization and collinearity analysis of RRs
To better recognize the genomic distribution of RRs, the R package RIdiogram was used to draft the chromosomal location map of RRs based on the genome annotation files of alfalfa. For gene duplication event analysis, we used the DupGen_finder pipeline (https://github.com/qiao-xin/DupGen_finder) to identify different modes of gene duplication (tandem duplication [TD], transposed duplication [TRD], whole genome duplication [WGD], proximal duplication [PD], and dispersed duplication [DSD]) using the default settings (Qiao et al., 2019). For Ka, Ks, and Ka/Ks calculations, the KaKs_Calculator 2.0 was used based on the Tamura–Nei model model (Wang et al., 2010).
Phylogenetic and cis-regulatory element analyses
To analyze the evolutionary relationships among RRs in alfalfa, a phylogenetic tree containing 37 alfalfa RRs and 32 Arabidopsis RRs was constructed with MEGA software (version 6.0; https://www.megasoftware.net/egasoftware.net) using the neighbor-joining method, and the bootstrap coefficient was set to 1000 times. PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to determine potential cis-regulatory elements within 2000 bp promoter sequences of RRs.
Expression analysis of RRs
Transcriptome data from the CADL-Gene Expression Atlas (https://www.alfalfatoolbox.org/) and National Center for Biotechnology Information Sequence Read Archive database (http://www.ncbi.nlm.nih.gov; accession numbers SRX4079528–SRX4079572) were used to explore the expression profiles of RRs in different tissues (leaves, flowers, pre-elongated stems, elongated stems, roots, and nodules) and with different treatments (ABA, NaCl, and mannitol). Subsequently, the expression patterns of three RRs under different treatments in alfalfa were explored using the 2 x SG Fast qPCR Master Mix (Sangon Biotech, Shanghai, China) on a CFX96 Real-Time PCR Detection System (Bio-Rad, Los Angeles, CA, USA). The reaction system consisted of 10 μL volume (0.2 μL of each primer, 2.6 μL of ddH2O, and 1 μL of DAF buffer, 1 μL of cDNA, and 5 μL of 2 x SG Fast qPCR Master Mix). PCR reaction conditions were as follows: 94 °C for 30 s, followed by 20 cycles of 94 °C for 5 s and 54 °C for 30 s. The actin gene from alfalfa was selected as an internal reference, and the relative expression level of each gene was calculated using the 2-ΔΔCT method. Three biological replicates were used, and the significant differences between the treatment and control groups were determined at the same time point using a t-test. A histogram with error lines was drawn using R package ggplot2. All qRT-PCR validation primers used in the present study are listed in Table S1.
Results
Identification and gene characteristic analyses of the RR gene family in alfalfa
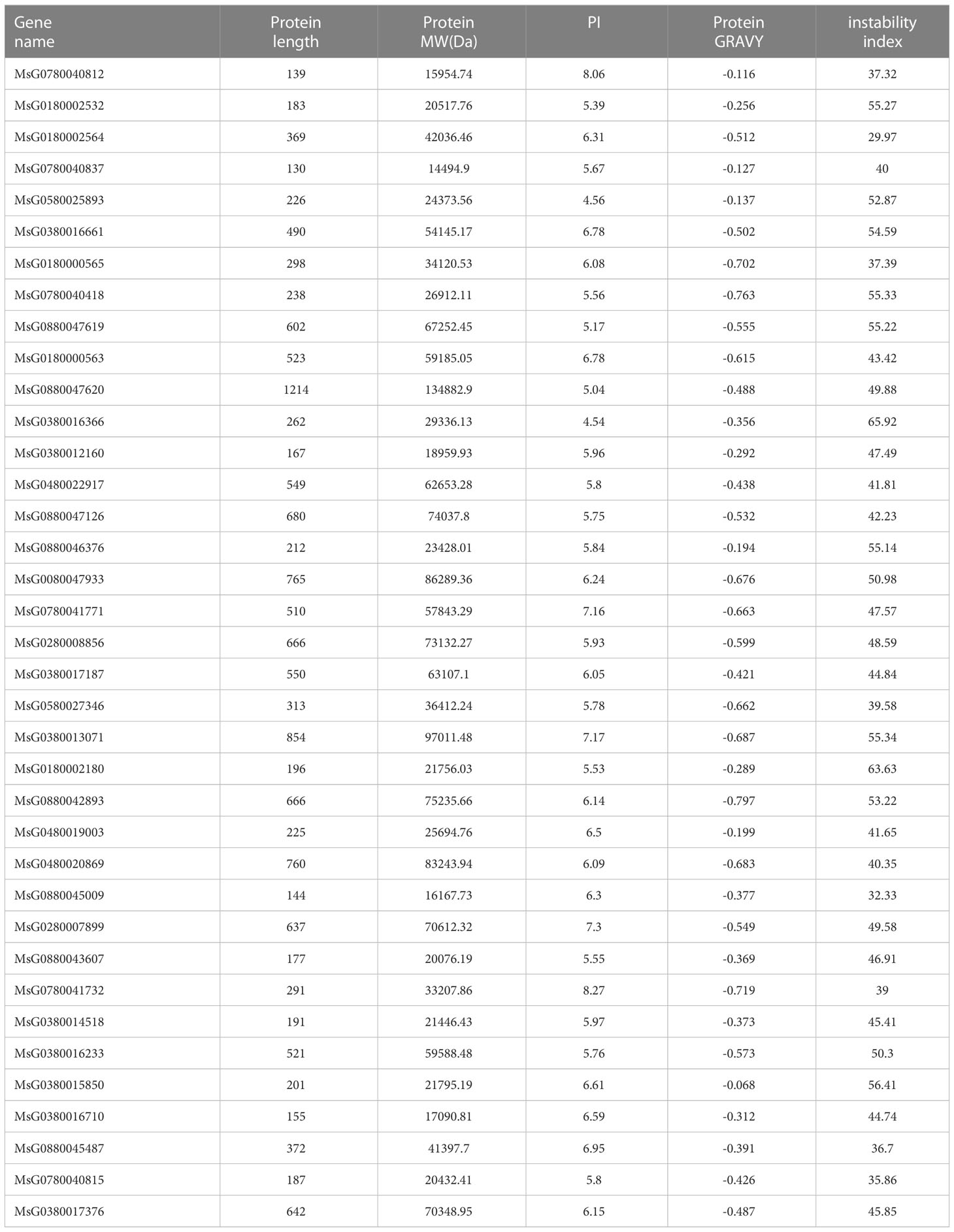
We identified 37 RRs in alfalfa based on a homology-based prediction approach using BLASTP. Further analysis of the evolutionary relationship between the structural specificity and conservation of these RRs led to the identification of 10 type-A, 16 type-B, and 11 type-P RRs (Table 1). The number of RRs was close to the RR gene family size observed in other plant species, such as A. thaliana (32 genes) and O. sativa (36 genes) (Ramírez-Carvajal et al., 2008).
In addition to the identification of RRs in alfalfa, we characterized the physicochemical properties of RRPs. Detailed information is provided in Table 1. The lengths of the identified RRPs ranged from 130 to 1214 amino acids (AAs), and the pIs varied from 4.54 to 8.27 (Table 1). These RRPs also displayed a wide range of MWs from 14.49 to 134.88 kDa (Table 1). Notably, most of these proteins were soluble hydrophilic proteins as the overall mean hydropathicity for all RRPs was negative (< 0) (Table 1).
Phylogenetic, gene structure, and protein domain analyses of RRPs
To confirm the evolutionary relationships among RRPs, we aligned the full-length RRPs of alfalfa and A. thaliana to construct a phylogenetic tree using MEGA 6.0. Based on the phylogenetic tree, 69 RRs (32 RRs in A. thaliana and 37 RRs in alfalfa) were divided into three subfamilies: 20 type-A, 32 type-B, and 17 type P-RRs (Figure 1B). The structural diversity of genes drives the evolution of multigene families. To better understand the structural diversity of RRs, we plotted exon–intron organization maps and found that all RRs contained more than one intron. The number of exons in the 37 genes varied from 2 to 12. Some genes, such as MsG0880047620, MsG0380017187, and MsG0380013071, contained more than 10 exons, and others had less than 10 introns. Notably, some type-A and type-B RRs contained untranslated regions (UTRs), but none of the type-P RRs had any UTRs (Figure 1A).
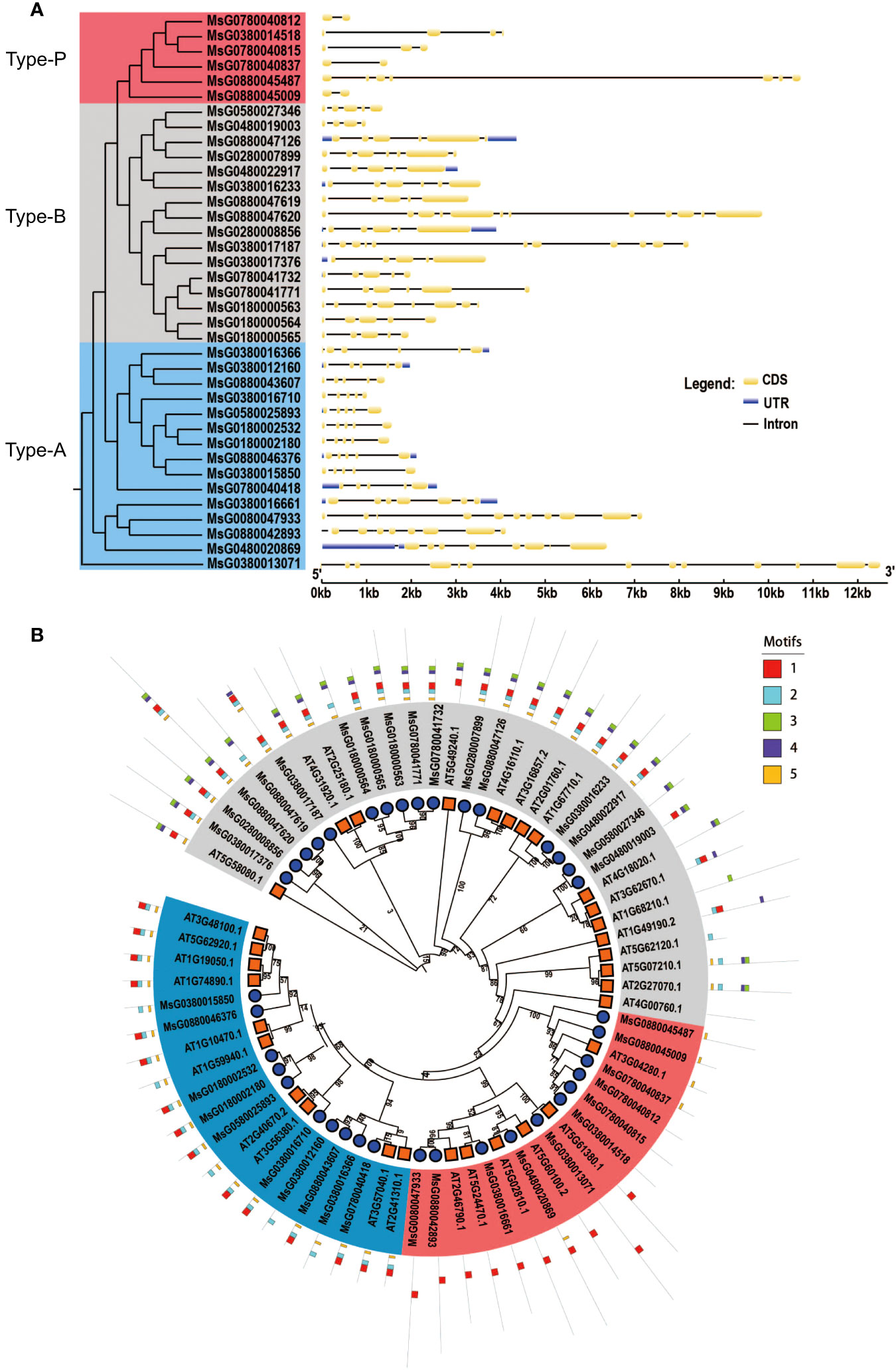
Figure 1 Gene structure, phylogenetic tree and meme structure analyses of RRs, Blue, type-A; Gray, type-B; Pink, type-P. (A) The rooted phylogenetic tree was constructed with 37 RR amino acid sequences of alfalfa. Exon-intron organization of corresponding of the RRs. The exons and introns are represented by yellow boxes and black lines, untranslated regions (UTRs) are represented by blue boxes, respectively. The length of each RR gene is shown as a proportion. (B) Phylogenetic tree and meme structure of RRs between alfalfa and A. thaliana. The phylogenetic tree was constructed using MEGA 6.0 by the neighbor-joining method with 1000 bootstrap replicates. The tree was divided into 3 subgroups. Members of different subgroups are denoted by solid circles with red, blue and gray colors, respectively. The conserved motifs in the RPPs were identified by MEME. The black lines represent no conserved sequences. Each motif is indicated by a colored box numbered at the bottom. Details are listed in Figure S1.
To further investigate the distribution and structural diversification of conserved motifs in alfalfa RRPs, we used the MEME software and found that most closely related members shared common motifs, but the conserved motifs among the subfamilies were significantly different, with some exceptions. For example, all type-A RRPs, except MsG0380016366, MsG0380012160, and MsG0380015850, contained motifs 1, 2, and 5; all type-B RRPs, except MsG0880047619, MsG0180000564, MsG0580027346, and MsG0480019003, contained motifs 1, 2, 3, 4, and 5 (Figure 1B; Figure S1).
Chromosomal distribution and subcellular localization of RRs
To study the orientation of RRs on each chromosome, we obtained accurate information about the initiation sites to construct the chromosomal location map (Figure 2A). Our results showed that the RR family members were distributed unevenly across the eight chromosomes of alfalfa, with the number of RRs varying among the chromosomes. Almost half of the RRs (18 RRs) were present on Chr3 and Chr8 of alfalfa, but there were no RR genes on Chr6, and RRs were less distributed on Chr2, Chr4, and Chr5, with only two, three, and two genes found in these chromosomes, respectively. Interestingly, most of the RRs were located in the distal centromere region, indicating that these RRs may have experienced long-term evolution in alfalfa.
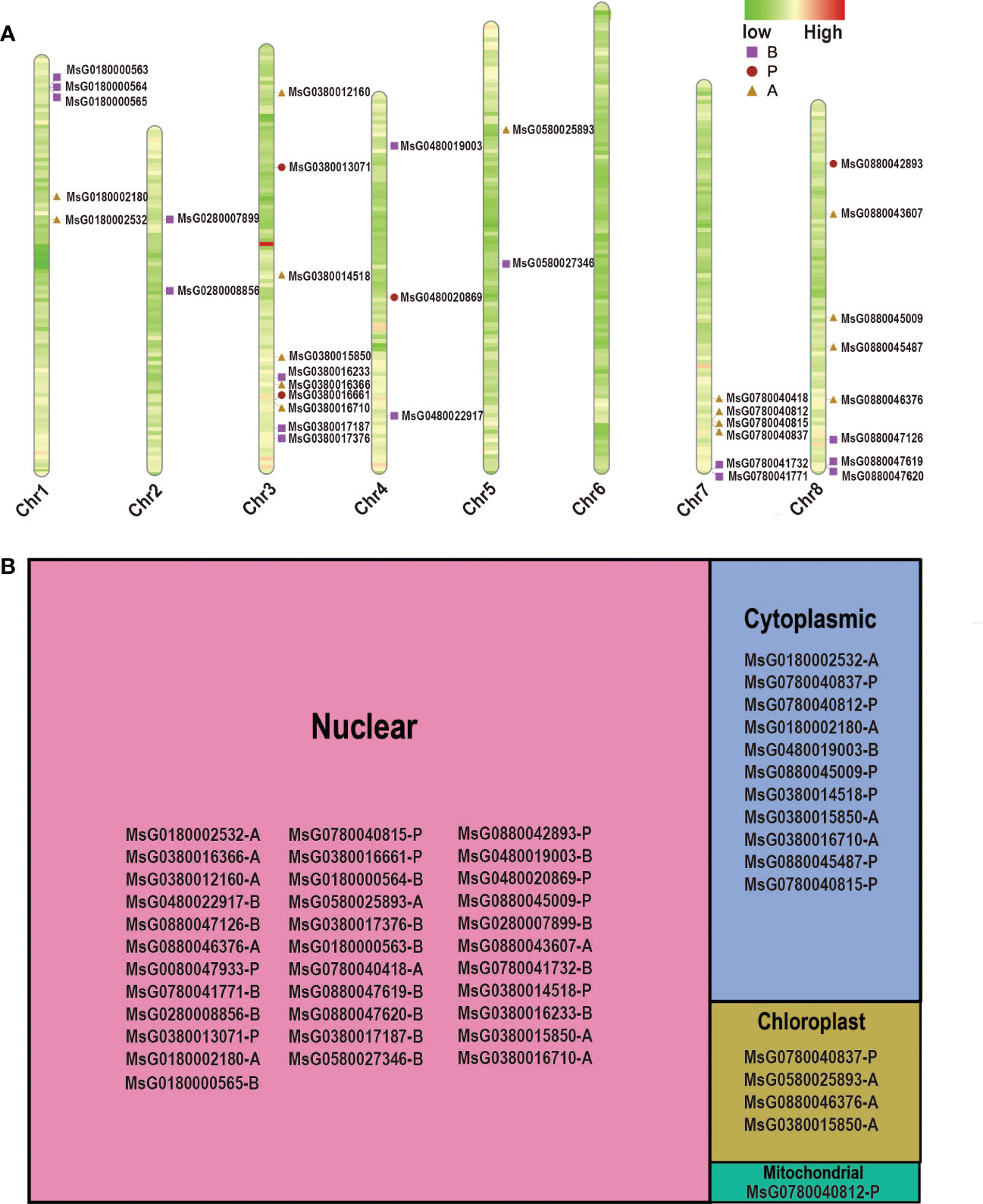
Figure 2 Chromosome distribution and subcellular localization of RRPs. (A) Chromosome distributions of different subtypes RRPs in alfalfa. The chromosome number is indicated at the bottom of each bar. (B) Tree map showing the subcellular localization of different subtypes RRPs in alfalfa.
Information on subcellular localization is crucial for understanding the functions of proteins. The CELLO web server was used to determine the subcellular localization of RRPs. We found that most RRPs were mainly located in the nucleus and mitochondria (Figure 2B). This is not surprising as some RRPs can function as transcription factors. Furthermore, only a few RR members were predicted to be located in the chloroplast and one was found to be targeted to the mitochondria. Notably, some RRPs exhibit more than one subcellular localization. For example, MsG0780040812 is located in both the mitochondria and cytoplasm, whereas MsG0380015850 is distributed in almost all subcellular compartments, except for the mitochondria (Figure 2B). Therefore, these proteins may need to be trafficked to different subcellular organelles under different conditions to perform their functions.
Cis-acting element analysis of the RR promoter
Cis-acting elements located in the promoter regions are recognized by transcription factors that regulate the spatial and temporal expression patterns of their target genes. To identify these cis-regulating elements in the promoter sequence (2000 bp) of each RR gene, we used the PlantCARE online tool and detected many cis-elements in the promoter regions of RRs (Figure 3). These cis-elements included abiotic stress-related, light, and plant growth and development response elements. As shown in Figure 3, almost all RRs had abiotic stress-related response elements, such as the ABA-responsive element (ABRE), MeJA-responsive element (TGACG-motif), and salicylic acid-responsive element (TCA-element), suggesting that all RRs have the ability to perform their functions under stress. However, not all RRs showed high levels of expression under stress conditions. Some genes, including MsG0180002180, MsG0480022917, MsG0380016710, MsG0180000565, MsG0880045009, MsG0880045487, MsG0780041771, MsG0780041732, MsG0780040837, MsG0780040812, MsG0780040815, MsG0580027346, MsG0180002532, MsG0380012160, MsG0880047619, MsG0480019003, MsG0380014518, MsG0180000563, MsG0180000564, and MsG0880047620, exhibit low expression or no expression under ABA, mannitol, and NaCl treatments (Figure 3). This may be caused by heterochromatin-mediated gene silence because most of RRs showed telomere-near-distribution. In addition, only 16 and 13 RRs had light response elements in plant growth and development in their promoter regions (Figure 3), respectively, indicating that some RRs are also associated with light response and plant growth and development.
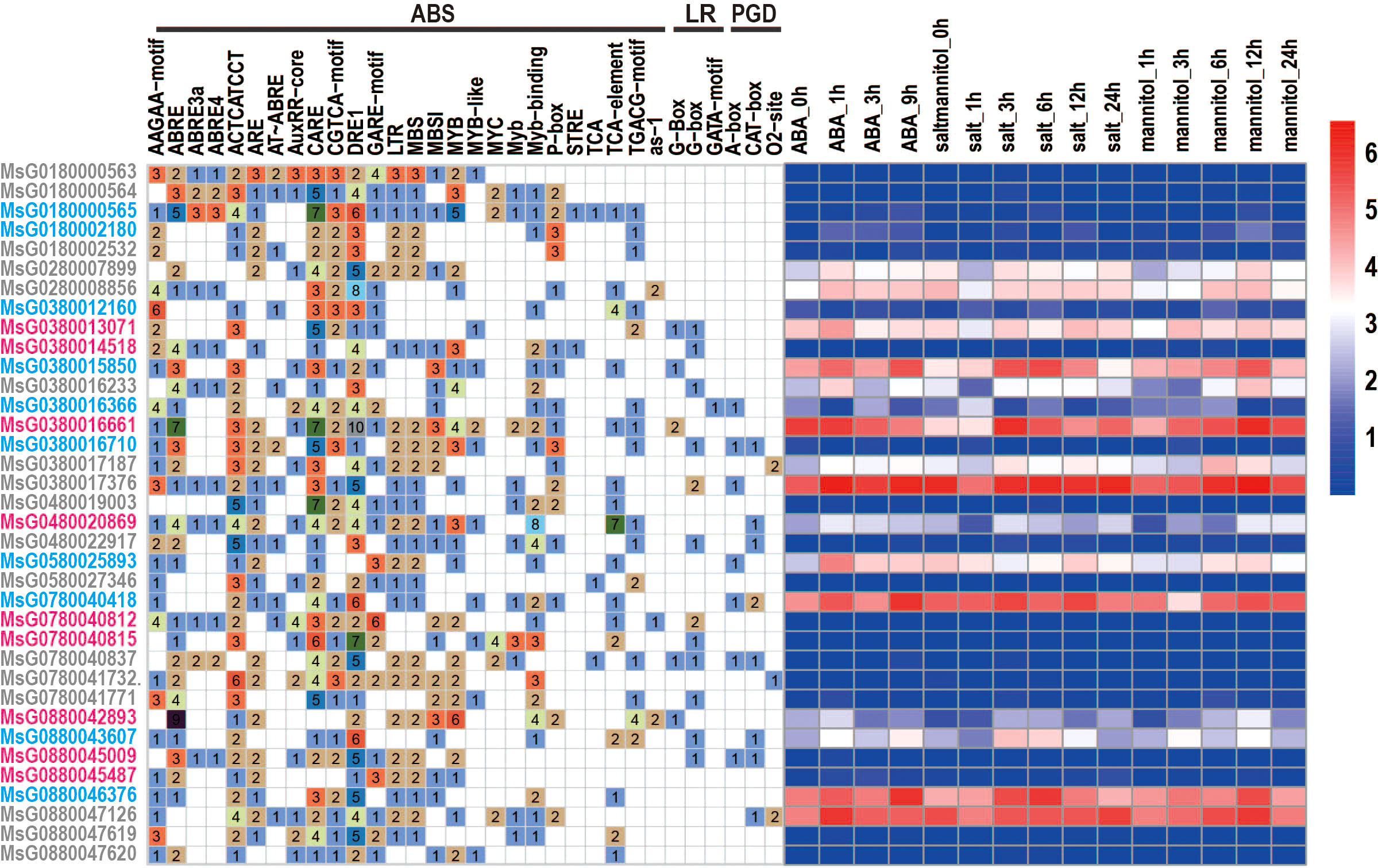
Figure 3 Cis-acting elements in the promoter region of RRs and the expression profiles of RRs under different stress treatments in alfalfa. ABS: abiotic stress related response elements; LR: light response elements; PGD: plant growth and development response elements. The expression levels of gene are showed in different colors; red and blue represent high and low expression level, respectively. Figure 3. Gene duplications and collinearity analysis.
Gene duplication and collinearity analysis of RRs
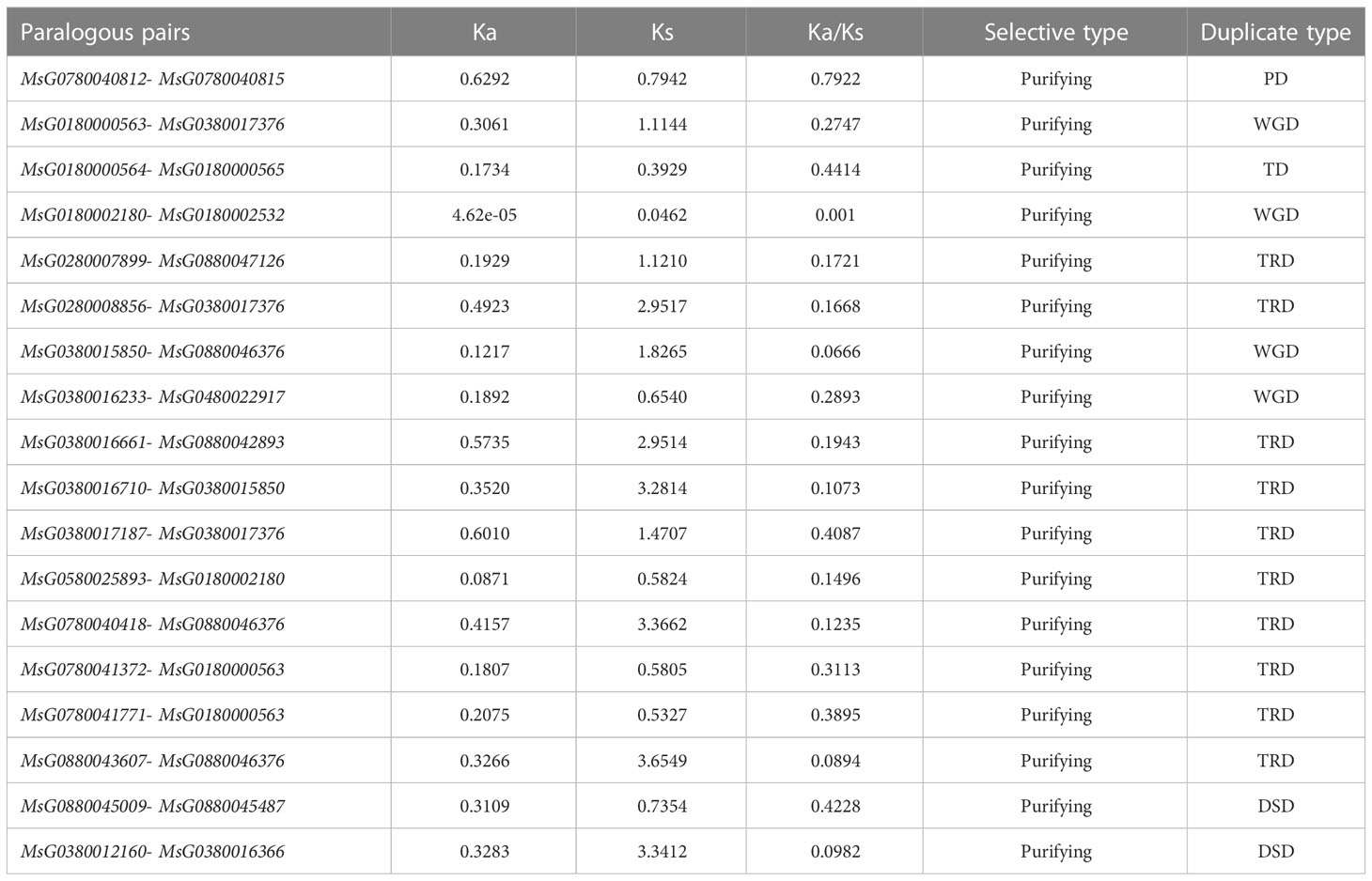
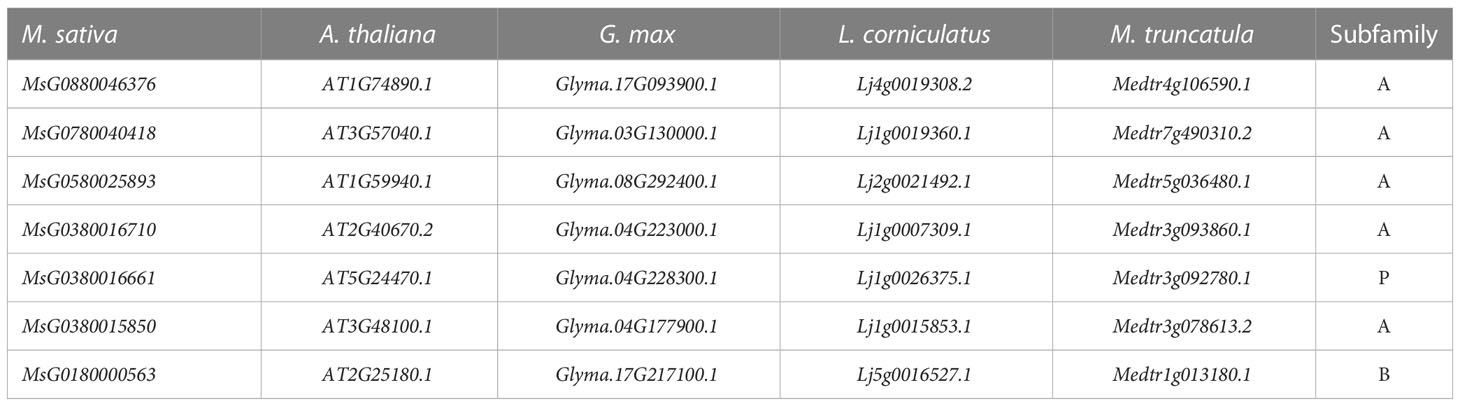
WGDs along with single gene duplications, such as DSDs, TRDs, PDs, and TDs, are the main driving forces for gene family evolution. In this study, we identified nine TRD events, four WGD events, two DSD events, one TD event, and one PD event (Figure 4; Table 2) in the RR subfamily. To further understand the evolutionary constraints acting on duplicated RRs, we calculated Ka (non-synonymous substitution rate), Ks (synonymous substitution rate), and the Ka/Ks ratio for each duplicated RR gene pair. Our results showed that all Ka/Ks ratios for the 17 segmentally duplicated gene pairs were < 1 (Table 2), indicating that the RRs primarily evolved under the influence of purifying selection. To further determine the origin and evolution of RR gene family members, four comparative synteny maps were constructed between autotetraploid alfalfa and A. thaliana, Medicago truncatula, G. max, and Lotus corniculatus. These results showed that 30 RRs of Zhongmu No. 1 were collinear with G. max, followed by M. truncatula (19), L. corniculatus (19), and A. thaliana (18). Among these orthologous pairs, seven RRs (MsG0780040418, MsG0580025893, MsG0880046376, MsG0380016710, MsG0380016661, MsG0380015850, and MsG0180000563) had their corresponding orthologs both in G. max, L. corniculatus, M. truncatula, and A. thaliana (Figure 5; Table 3), suggesting that these genes may play an important role in the evolution of RRs. The collinear relationships between the RRs of Chr3 and those of the other four plants were significantly greater than those of the other chromosomes.
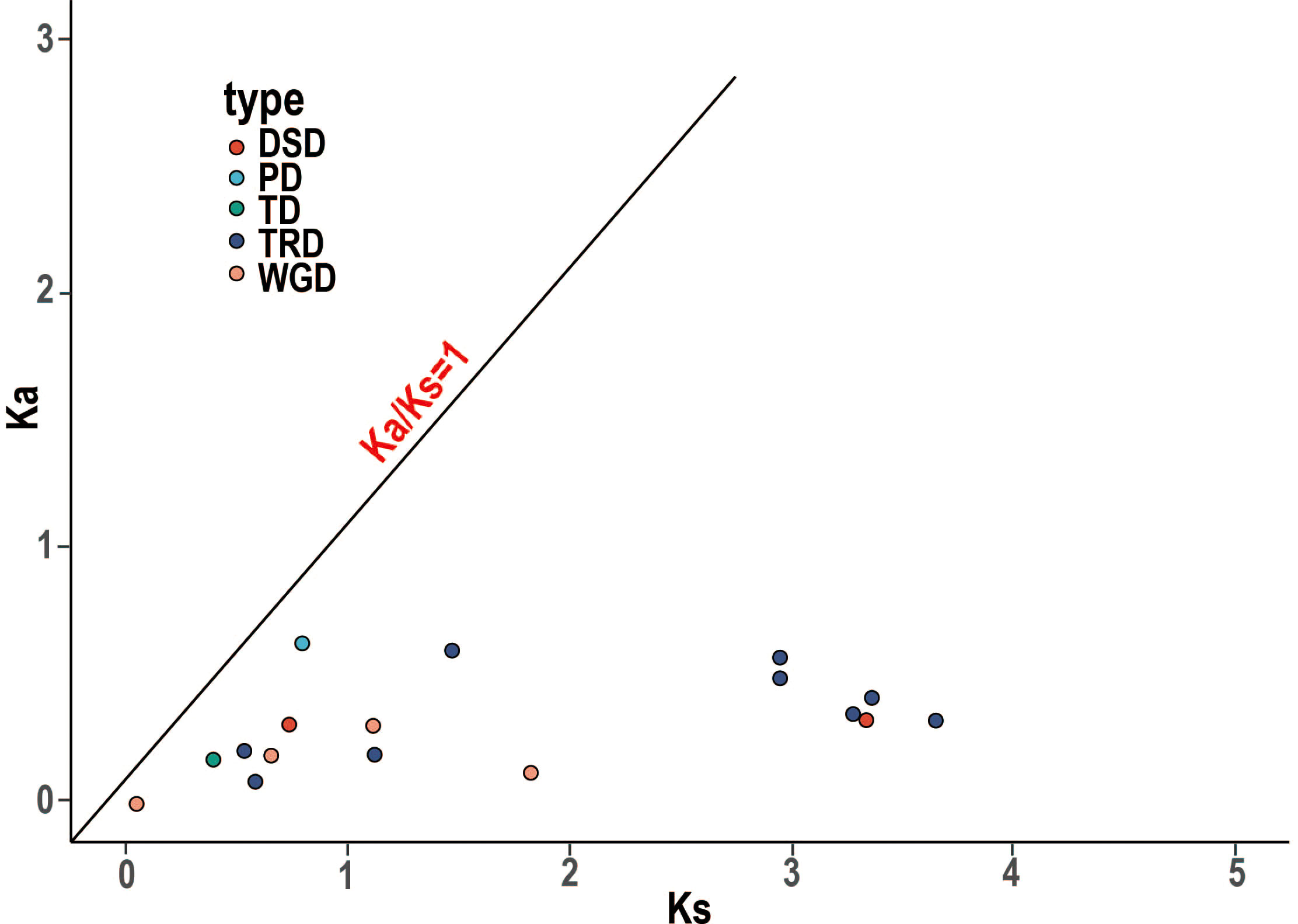
Figure 4 Ka/Ks ratios of duplicated RR gene pairs. Gene pairs from different duplication types are indicated by different scatter. The y and x axes denote the Ka and Ks values for each pair and the black line shows a Ka/Ks ratio = 1. The detail of Ka, Ks, and Ka/Ks listed in Table S2.
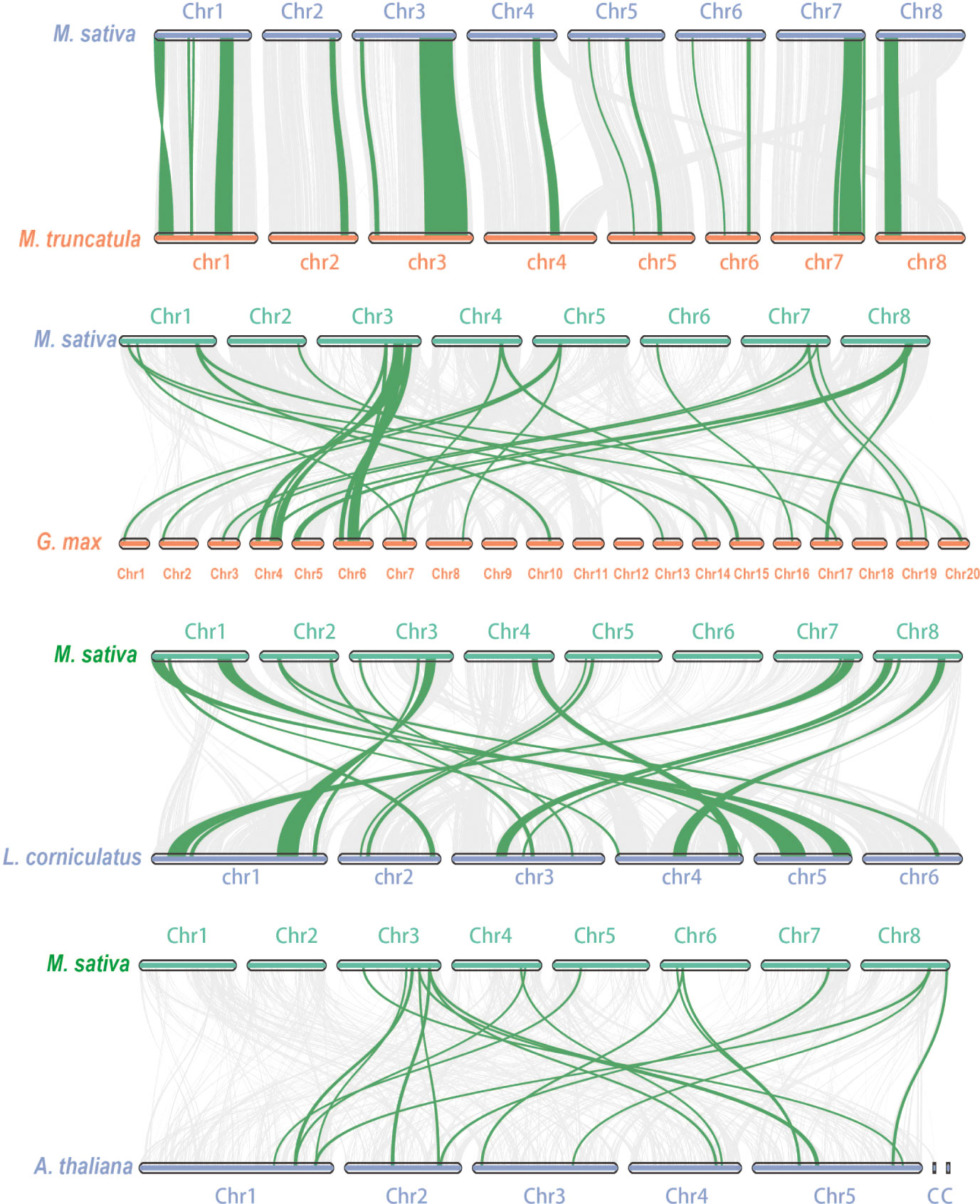
Figure 5 Collinearity analysis between alfalfa and A. thaliana/M. truncatula/G. max/L. corniculatus. The gray lines in the background indicate the collinear blocks between alfalfa and A. thaliana/M. truncatula/G. max/L. corniculatus, while the blue lines highlight the syntenic RR gene pairs.
Tissue expression patterns of RRs in alfalfa
Tissue expression profiles are useful in determining the potential roles of target genes and their specific properties in a particular tissue. To gain a deeper understanding of the potential function of RRs, we analyzed their expression profiles in six tissues (leaves, flowers, pre-elongated stems, elongated stems, roots, and nodules) using RNA-sequencing data from alfalfa and found that the expression patterns of RRs were distinct. Most RRs showed a constitutive expression pattern, but nearly half of the genes were expressed at low levels in all tissues. This may be caused by the telomere-near-distribution due to the heterochromatin-mediated epigenetic regulation of gene expression (Matzke et al., 2009; To et al., 2011). Furthermore, MsG0380013071, MsG0280008856, MsG0880047126, MsG0480020869, and MsG0380017187 were highly expressed in all six tissues. Interestingly, we also observed tissue-specific expression patterns of some RRs. For example, MsG0880043607 was expressed only in the stem, while MsG0580025893 was highly expressed in leaves, stems, and flowers (Figure 6; Figure S2).
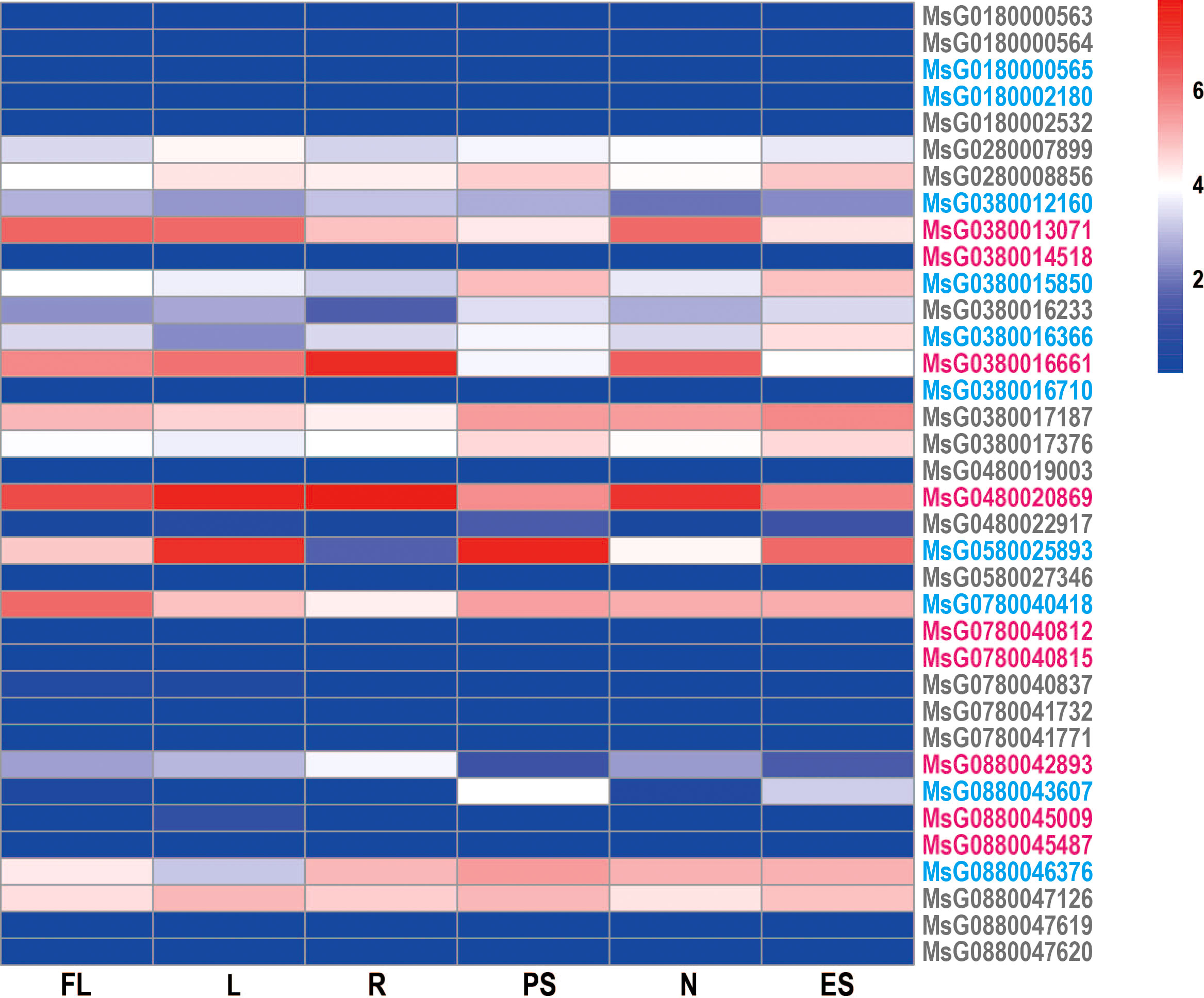
Figure 6 Heatmap displaying the RRs expression in various tissues of alfalfa. Transcriptome data from 6 tissues (FL, flower; L, leaf; R, root; PS, pre-elongated stem; N, nodule; ES, elongated stem) were used to construct the expression patterns of RRs in alfalfa. The bar to the right of the heat map represents the relative expression values.
Discussion
In response to abiotic stress, RRPs obtain phosphoric groups from histidine phosphate transfer proteins to stimulate the transcription of downstream functional genes. Previous studies have reported that RRs in various plants, such as A. thaliana, O. sativa, Pyrus bretschneideri, Prunus persica, Fragaria vesca, and Brassica rapa ssp. Pekinensis (Liu et al., 2014; Bhaskar et al., 2021), are crucial for the plant response to abiotic stress (Imamura et al., 1999), which can be applied in alfalfa molecular breeding for drought tolerance. It is also important to elucidate the RR-related genes and their functions in alfalfa and other legumes. This will expand our knowledge of RRs and provide a foundation for further investigation of their specific roles and functions. In this study, 37 RRs were identified in alfalfa, which is approximately equivalent to the RR family size in A. thaliana (32 genes) and O. sativa (36 genes) (Ramírez-Carvajal et al., 2008). The close relationship between RRs of A. thaliana and alfalfa, as observed in the phylogenetic tree, suggests that the functions of these RRs in alfalfa may be similar to those observed in A. thaliana. Based on evolutionary relationships and structural specificity, the 37 RRs can be clustered into three subfamilies: type-A, type-B, and type-P RRs (Figure 1B). Protein domain analysis revealed that the conserved motifs within each subfamily were similar, but those among different subfamilies were significantly different. Specifically, motifs 1 and 5 were present in the members of type-P RRPs, while motifs 1, 2, and 5 were present in the members of the type-A RRPs, and all motifs were present in most members of type-B RRPs (Figure 1B). These differences in characteristics among the different subfamilies may indicate the diverse functions of the RR family members in alfalfa.
Gene duplication is a fundamental source of new genes in the evolutionary process, facilitating the successful evolution of genes and contributing to the rapid expansion of gene families (Van de Peer et al., 2009; Wang et al., 2017). RR-related duplication events have been observed in many species, including A. thaliana, Malus domestica, and Gossypium species (Li et al., 2017). Our gene duplication analysis revealed that RRs in alfalfa also experienced different types and numbers of gene duplication events. In Gossypium species, no tandem duplication gene pairs were detected in the RR gene family, and WGD seemed to contribute more to alfalfa RR gene family expansion than single gene duplication events; however, our analysis documented the presence of TD events and revealed that TRD events, not WGD events, played a major role in the expansion of the RR gene family in alfalfa (Figure 4). These conflicting findings may be attributed to the independent evolution of genes.
RRs have been reported to be involved in stress resistance and response in various plants, such as A. thaliana (Nakamichi et al., 2016) and O. sativa (Karan et al., 2009; Rehman et al., 2022), and several RRs in alfalfa were also found to respond to different types of abiotic stresses (Figure 3). For example, MsG0380016233, MsG0580025893, MsG0780040418, MsG0280008856, MsG0380013071, MsG0380017187, MsG0480020869, and MsG0280007899 were significantly up-regulated by ABA treatment, some of which were confirmed by qRT-PCR analysis (Figure S3). Cis-element analysis indicated that ABREs were located in the promoter regions of these genes, and subcellular localization predicted analysis revealed that they were only localized in the nucleus (Figure 2B). Taken together, these results suggest that these genes may be directly regulated by ABA in response to drought stress (Figure 3) and should be transferred into the nucleus to regulate the expression of some genes in response to ABA stimuli. Interestingly, MsG0580025893 was predicted to be distributed in both the nucleus and chloroplast, and was highly expressed in leaves, stems, and flowers (Figure 2B; Figure 6), suggesting that it may function in abiotic stress by regulating both nuclear and chloroplast gene expression to enhance photosynthesis. In addition, we also identified several RRs, including MsG0180002180, MsG0480022917, MsG0380016710, MsG0180000565, MsG0880045009, MsG0880045487, MsG0780041771, MsG0780041732, MsG0780040837, MsG0780040812, MsG0780040815, MsG0580027346, MsG0180002532, MsG0380012160, MsG0880047619, MsG0480019003, MsG0380014518, MsG0180000563, MsG0180000564, and MsG0880047620, which were not expressed under ABA, NaCl, and mannitol treatments (Figure 3). Interestingly, ABREs were still detected in the promoter region of MsG0180000565. This could be partly explained by the presence of other regulatory mechanisms, such as heterochromatin-mediated gene silence, that compete with ABA induction during transcription (Peng et al., 2008). RRs also play important roles in plant immune response and disease resistance (Cheval et al., 2017; Bacete et al., 2020). In line with this, we found that some genes, such as MsG0780040815 and MsG0380016661, have gibberellin, salicylic acid, and methyl jasmonate response elements in their promoter regions (Figure 3).
Circadian rhythms are produced by the internal clock or oscillator of many organisms and control the timing of genetic, metabolic, and physiological processes, including photosynthesis (Dodd et al., 2005; Chow and Kay, 2013), growth rate, and flowering time (Woelfle et al., 2004). Photoperiodic regulation of the flowering period is one of the most common circadian events (Yanovsky and Kay, 2003). In A. thaliana, the APRR family has been reported to be associated with the circadian clock (Murakami et al., 2004). The expression patterns of some orthologs of AtPRRs in Populus exhibit circadian waves (Liu et al., 2013), but the circadian rhythm function of alfalfa type-P RRs has not yet been reported. Notably, photoperiod-responsive elements, including G-box, GATA-motif, and methyl jasmonate, were also detected in the promoter region of type-P RRs, suggesting that members of the RR family may also be related to the circadian rhythm (Figure 3). Subcellular localization analysis of A. thaliana and Glycyrrhiza uralensis revealed that type-P RRPs are mainly localized in the nucleus (Fujiwara et al., 2008). However, in our predicted results, type-P RRPs were found to exist not only in the nucleus but also in the cytoplasm, mitochondria, and chloroplasts (Figure 2B). This suggests that type-P RRs in alfalfa have roles other than the maintenance of circadian rhythm, which require further exploration future studies.
Conclusion
In this study, we comprehensively and systematically analyzed the response regulator gene family in the autotetraploid-cultivated alfalfa genome. We identified 37 RR family genes unevenly distributed across eight chromosomes in the alfalfa genome and further grouped them into three subfamilies, type-A, type-B, and type-P RRs, based on their evolutionary relationship, structural specificity, and degree of conservation. We also analyzed the physicochemical properties, phylogenetic relationships, exon–intron structures, conserved motifs, chromosomal location, gene duplication, cis-regulatory elements, and tissue-specific expression patterns of these genes. Our findings shed light on the significant roles of alfalfa RR gene family in hormone induction and abiotic stress tolerance and have great scientific and practical application in alfalfa molecular breeding.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
YQ, XH and LF designed the experiments and wrote the manuscript. YQ, SL, JZ, MT, XW, TL analyzed the data. LF, ZL and ZPL edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (32201463), Major Program of Agricultural Biology Breeding (2022ZD04011) and the Fundamental Research Fund for the Central Universities (lzujbky-2021-ct21).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1149880/full#supplementary-material
References
Bacete, L., Mélida, H., López, G., Dabos, P., Tremousaygue, D., Denancé, N., et al. (2020). Arabidopsis response regulator 6 (ARR6) modulates plant cell-wall composition and disease resistance. Mol. Plant Microbe In. 33, 767–780. doi: 10.1094/MPMI-12-19-0341-R
Bhaskar, A., Paul, L. K., Sharma, E., Jha, S., Jain, M., Khurana, J. P. (2021). OsRR6, a type-a response regulator in rice, mediates cytokinin, light and stress responses when over-expressed in Arabidopsis. Plant Physiol. Bioch. 161, 98–112. doi: 10.1016/j.plaphy.2021.01.047
Cheval, C., Perez, M., Leba, L. J., Ranty, B., Perochon, A., Reichelt, M., et al. (2017). PRR2, a pseudo-response regulator, promotes salicylic acid and camalexin accumulation during plant immunity. Sci. Rep. 7, 1–13. doi: 10.1038/s41598-017-07535-8
Chow, B. Y., Kay, S. A. (2013). Global approaches for telling time: omics and the Arabidopsis circadian clock. Semin. Cell. Dev. Biol. 24, 383–392. doi: 10.1016/j.semcdb.2013.02.005
D’Agostino, I. B., Deruere, J., Kieber, J. J. (2000). Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 124, 1706–1717. doi: 10.1104/pp.124.4.1706
Dodd, A. N., Salathia, N., Hall, A., Kévei, E., Tóth, R., Nagy, F., et al. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309, 630–633. doi: 10.1126/science.111558
Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z., et al. (2004). Ehd1, a b-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Gene. Dev. 18, 926–936. doi: 10.1101/gad.1189604
Dong, X. M., Han, B. C., Yin, X. Y., Mao, P., Luo, D., Zhou, Q., et al. (2023). Genome-wide identification of the GRAS transcription factor family in autotetraploid cultivated alfalfa (Medicago sativa l.) and expression analysis under drought stress. Ind. Crop Prod. 194, 116379. doi: 10.1016/j.indcrop.2023.116379
Fujiwara, S., Wang, L., Han, L. Q., Suh, S. S., Salomé, P. A., McClung, C. R., et al. (2008). Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 283, 23073–23083. doi: 10.1074/jbc.M803471200
Gao, J. P., Chao, D. Y., Lin, H. X. (2008). Toward understanding molecular mechanisms of abiotic stress responses in rice. Rice 1, 36–51. doi: 10.1007/s12284-008-9006-7
Geng, X. C., Zhang, C., Wei, L. N., Lin, K., Xu, Z. F. (2022). Genome-wide identification and expression analysis of cytokinin response regulator (RR) genes in the woody plant Jatropha curcas and functional analysis of JcRR12 in Arabidopsis. Int. J. Mol. Sci. 23, 11388. doi: 10.3390/ijms231911388
Hasanuzzaman, M., Bhuyan, M. B., Zulfiqar, F., Raza, A., Mohsin, S. M., Mahmud, J. A., et al. (2020). Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 9, 681. doi: 10.3390/antiox9080681
Hass, C., Lohrmann, J., Albrecht, V., Sweere, U., Hummel, F., Yoo, S. D., et al. (2004). The response regulator 2 mediates ethylene signalling and hormone signal integration in Arabidopsis. EMBO J. 23, 3290–3302. doi: 10.1038/sj.emboj.7600337
He, Y. J., Liu, X., Ye, L., Pan, C. T., Chen, L. T., Zou, T., et al. (2016). Genome-wide identification and expression analysis of two-component system genes in tomato. Int. J. Mol. Sci. 17, 1204. doi: 10.3390/ijms17081204
Hu, Y. F., Cui, H. M., Xia, P. L., Liu, G. S., Wu, X. Y., Li, Y. T., et al. (2022). Genome-wide mining of b-type cytokinin response regulators in wheat reveals the involvement of TaRR5. 1-6A in drought and salt tolerance. Crop Pasture Sci. 73, 997–1010. doi: 10.1071/CP21766
Huo, R. X., Liu, Z. N., Yu, X. L., Li, Z. Y. (2020). The interaction network and signaling specificity of two-component system in Arabidopsis. Int. J. Mol. Sci. 21, 4898. doi: 10.3390/ijms21144898
Hwang, I., Chen, H. C., Sheen, J. (2002). Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 129, 500–515. doi: 10.1104/pp.005504
Imamura, A., Hanaki, N., Hussain, A., Nakamura, B. G., Suzuki, T., Taniguchi, M., et al (1999). Compilation and characterization of Arabiopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol. 40, 733–742. doi: 10.1093/oxfordjournals.pcp.a029600
Imran, Q. M., Falak, N., Hussain, A., Mun, B. G., Yun, B. W. (2021). Abiotic stress in plants; stress perception to molecular response and role of biotechnological tools in stress resistance. Agronomy 11, 1579. doi: 10.3390/agronomy11081579
Ito, Y., Kurata, N. (2006). Identification and characterization of cytokinin-signalling gene families in rice. Gene 382, 57–65. doi: 10.1016/j.gene.2006.06.020
Jain, M., Tyagi, A. K., Khurana, J. P. (2008). Differential gene expression of rice two-component signaling elements during reproductive development and regulation by abiotic stress. Funct. Integr. Genomic. 8, 175–180. doi: 10.1007/s10142-007-0063-6
Karan, R., Singla-Pareek, S. L., Pareek, A. (2009). Histidine kinase and response regulator genes as they relate to salinity tolerance in rice. Funct. Integr. Genomic. 9, 411–417. doi: 10.1007/s10142-009-0119-x
Kieber, J. J., Schaller, G. E. (2018). Cytokinin signaling in plant development. Development 145, 149344. doi: 10.1242/dev.149344
Le, D. T., Nishiyama, R., Watanabe, Y., Mochida, K., Yamaguchi-Shinozaki, K., Shinozaki, K., et al. (2011). Genome-wide expression profiling of soybean two-component system genes in soybean root and shoot tissues under dehydration stress. DNA Res. 18, 17–29. doi: 10.1093/dnares/dsq032
Li, Y. M., Zhang, D., Zhang, L. Z., Zuo, X. Y., Fan, S., Zhang, X., et al. (2017). Identification and expression analysis of cytokinin response-regulator genes during floral induction in apple (Malus domestica borkh). Plant Growth Regul. 83, 455–464. doi: 10.1007/s10725-017-0311-2
Liu, S. X., Qin, B., Fang, Q. X., Zhang, W. J., Zhang, Z. Y., Liu, Y. C., et al. (2021). Genome-wide identification, phylogeny and expression analysis of the bZIP gene family in alfalfa (Medicago sativa). Biotechnol. Biotec. Eq. 35, 905–916. doi: 10.1080/13102818.2021.1938674
Liu, W., Zhang, W., Du, M., Sha, Y., Yu, X., Ohtani, M., et al. (2013). Diurnal and circadian expression of clock-associated pseudo-response regulators in Populus trichocarpa. Plant Biotechnol. 30, 517–521. doi: 10.1007/s10725-017-0311-2
Liu, Z. N., Zhang, M., Kong, L. J., Lv, Y. X., Zou, M. H., Lu, G., et al. (2014). Genome-wide identification, phylogeny, duplication, and expression analyses of two-component system genes in Chinese cabbage (Brassica rapa ssp. pekinensis). DNA Res. 21, 379–396. doi: 10.1093/dnares/dsu004
Mason, M. G., Mathews, D. E., Argyros, D. A., Maxwell, B. B., Kieber, J. J., Alonso, J. M., et al. (2005). Multiple type-b response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17, 3007–3018. doi: 10.1105/tpc.105.035451
Matzke, M., Kanno, T., Daxinger, L., Huettel, B., Matzke, A. J. (2009). RNA-Mediated chromatin-based silencing in plants. Curr. Opin. Cell Biol. 21, 367–376. doi: 10.1016/j.ceb.2009.01.025
Murakami, M., Yamashino, T., Mizuno, T. (2004). Quintet of circadian associated APRR1/TOC1 family in Arabidopsis thaliana: Characterization of a transgenic line aberrantly overexpressing APRR3. Plant Cell Physiol. 45, S69–S69. doi: 10.1093/pcp/pch065
Nakamichi, N., Takao, S., Kudo, T., Kiba, T., Wang, Y., Kinoshita, T., et al. (2016). Improvement of Arabidopsis biomass and cold, drought and salinity stress tolerance by modified circadian clock-associated PSEUDO-RESPONSE REGULATORs. Plant Cell Physiol. 57, 1085–1097. doi: 10.1093/pcp/pcw057
Pareek, A., Singh, A., Kumar, M., Kushwaha, H. R., Lynn, A. M., Singla-Pareek, S. L. (2006). Whole-genome analysis of Oryza sativa reveals similar architecture of two-component signaling machinery with Arabidopsis. Plant Physiol. 142, 380–397. doi: 10.1104/pp.106.086371
Peng, J. C., Karpen, G. H. (2008). Epigenetic regulation of heterochromatic DNA stability. Curr. Opin. Genet. Dev. 18, 204–211. doi: 10.1016/j.gde.2008.01.021
Qiao, X., Li, Q. H., Yin, H., Qi, K. J., Li, L. T., Wang, R., et al. (2019). Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 20, 1–23. doi: 10.1186/s13059-019-1650-2
Ramírez-Carvajal, G. A., Morse, A. M., Davis, J. M. (2008). Transcript profiles of the cytokinin response regulator gene family in populus imply diverse roles in plant development. New Phytol. 177, 77–89. doi: 10.1111/j.1469-8137.2007.02240.x
Razi, K., Muneer, S. (2021). Drought stress-induced physiological mechanisms, signaling pathways and molecular response of chloroplasts in common vegetable crops. Crit. Rev. Biotechnol. 41, 669–691. doi: 10.1080/07388551.2021.1874280
Rehman, O. U., Uzair, M., Chao, H., Fiaz, S., Khan, M. R., Chen, M. (2022). Role of the type-b authentic response regulator gene family in fragrant rice under alkaline-salt stress. Physiol. Plant. 174, e13696. doi: 10.1111/ppl.13696
Ren, L., Bennett, J. A., Coulman, B., Liu, J., Biligetu, B. (2021). Forage yield trend of alfalfa cultivars in the Canadian prairies and its relation to environmental factors and harvest management. Grass Forage Sci. 76, 390–399. doi: 10.1111/gfs.12513
Sun, J., Guo, C. (2022). Genome-wide identification and expression analysis of RR-type MYB-related transcription factors in tomato (Solanum lycopersicum l.). Horticulturae 8, 399. doi: 10.3390/horticulturae8050399
To, T. K., Kim, J. M., Matsui, A., Kurihara, Y., Morosawa, T., Ishida, J., et al. (2011). Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PloS Genet. 7, e1002055. doi: 10.1371/journal.pgen.1002055
Tsai, Y. C., Weir, N. R., Hill, K., Zhang, W., Kim, H. J., Shiu, S. H., et al. (2012). Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 158, 1666–1684. doi: 10.1104/pp.111.192765
Van de Peer, Y., Maere, S., Meyer, A. (2009). he evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 10, 725–32. doi: 10.1038/nrg2600
Wang, W. C., Lin, T. C., Kieber, J., Tsai, Y. C. (2019). Response regulators 9 and 10 negatively regulate salinity tolerance in rice. Plant Cell Physiol. 60, 2549–2563. doi: 10.1093/pcp/pcz149
Wang, D., Pei, K., Fu, Y., Sun, Z., Li, S., Liu, H., et al. (2007). Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394, 13–24. doi: 10.1016/j.gene.2007.01.006
Wang, R., Ming, M., Li, J., Shi, D., Qiao, X., Li, L., et al (2017). Genome-wide identification of the MADS-box transcription factor family in pear (Pyrus bretschneideri) reveals evolution and functional divergence. PeerJ 5, e3776. doi: 10.7717/peerj.3776
Wang, C., Wang, L., Liu, Q., Zhang, Y., Dong, K. (2022). Genome-wide identification and characterization of PRR gene family and their diurnal rhythmic expression profile in maize. Int. J. Genomics, 6941607. doi: 10.1155/2022/6941607
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., Yu, J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinf. 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3
West, A. H., Stock, A. M. (2001). Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26, 369–376. doi: 10.1016/S0968-0004(01)01852-7
Woelfle, M. A., Ouyang, Y., Phanvijhitsiri, K., Johnson, C. H. (2004). The adaptive value of circadian clocks: An experimental assessment in cyanobacteria. Curr. Biol. 14, 1481–1486. doi: 10.1016/j.cub.2004.08.023
Yanovsky, M. J., Kay, S. A. (2003). Living by the calendar: how plants know when to flower. nat. rev. mol. Cell Biol. 4, 265–276. doi: 10.1038/nrm1077
Zeng, R., Li, Z. Y., Shi, Y. T., Fu, D. Y., Yin, P., Cheng, J. H., et al. (2021). Natural variation in a type-a response regulator confers maize chilling tolerance. Nat. Commun. 12, 1–13. doi: 10.1038/s41467-021-25001-y
Zhang, Y. C., Gao, M., Singer, S. D., Fei, Z. J., Wang, H., Wang, X. P. (2012). Genome-wide identification and analysis of the TIFY gene family in grape. PloS One 7, e44465. doi: 10.1371/journal.pone.0044465
Keywords: alfalfa, response regulators, stress response, gene family, expression analysis
Citation: Qiang Y, He X, Li Z, Li S, Zhang J, Liu T, Tursunniyaz M, Wang X, Liu Z and Fang L (2023) Genome-wide identification and expression analysis of the response regulator gene family in alfalfa (Medicago sativa L.) reveals their multifarious roles in stress response. Front. Plant Sci. 14:1149880. doi: 10.3389/fpls.2023.1149880
Received: 23 January 2023; Accepted: 23 February 2023;
Published: 14 March 2023.
Edited by:
Jing Zhang, Nanjing Agricultural University, ChinaReviewed by:
Mingna Li, Chinese Academy of Agricultural Sciences (CAAS), Institute of Animal Sciences, ChinaRui Dong, Guizhou University, China
Copyright © 2023 Qiang, He, Li, Li, Zhang, Liu, Tursunniyaz, Wang, Liu and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Longfa Fang, RmFuZ2xmQGx6dS5lZHUuY24=; Zhipeng Liu, bHpwQGx6dS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Yuqin Qiang1†
Yuqin Qiang1† Zhipeng Liu
Zhipeng Liu Longfa Fang
Longfa Fang